Abstract
Cells interact with their microenvironment by constantly sensing mechanical and chemical cues converting them into biochemical signals. These processes allow cells to respond and adapt to changes in their environment, and are crucial for most cellular functions. Understanding the mechanism underlying this complex interplay at the cell-matrix interface is of fundamental value to decipher key biochemical and mechanical factors regulating cell fate. The combination of material science and surface chemistry aided in the creation of controllable environments to study cell mechanosensing and mechanotransduction. Biologically inspired materials tailored with specific bioactive molecules, desired physical properties and tunable topography have emerged as suitable tools to study cell behavior. Among these materials, synthetic cell interfaces with built-in sensing capabilities are highly advantageous to measure biophysical and biochemical interaction between cells and their environment. In this review, we discuss the design of micro and nanostructured biomaterials engineered not only to mimic the structure, properties, and function of the cellular microenvironment, but also to obtain quantitative information on how cells sense and probe specific adhesive cues from the extracellular domain. This type of responsive biointerfaces provides a readout of mechanics, biochemistry, and electrical activity in real time allowing observation of cellular processes with molecular specificity. Specifically designed sensors based on advanced optical and electrochemical readout are discussed. We further provide an insight into the emerging role of multifunctional micro and nanosensors to control and monitor cell functions by means of material design.
Introduction
Cell adhesion is a critical aspect of the constitution of tissues and organs. The complex organization of tissues relies on a precise control over the formation of adhesive contacts between cells and the extracellular matrix (ECM) (Gumbiner, 1996). From regenerative medicine to development biology there is a great interest in comprehending the mechanisms that control the assembly of cells into tissues and organs (Keung et al., 2010; Gaharwar et al., 2020). Increasing evidence has shown that these processes are regulated not only by biochemical signals but also by biophysical cues from the environment. For example, it has been found that cells are able to sense and respond to the topography (Curtis and Riehle, 2001; Spatz and Geiger, 2007), rigidity (Discher et al., 2005; Engler et al., 2006), and anisotropy (Théry et al., 2006; Xia et al., 2008) of their environment.
To reveal the mechanosensory elements involved in cell-cell and cell-ECM interactions, ECM-inspired materials have been developed (Mager et al., 2011; Xi et al., 2019). Advancements in material science and surface chemistry made it possible to create materials that mimic both physical (e.g., stiffness and topography) and chemical cues (adhesive and soluble) of the extracellular environment. These materials are highly desired because cell development and behavior can be studied under conditions similar to those found in the cell microenvironment in vivo. As materials incorporate biochemical and biophysical cues from the natural ECM, the information they provide allows a closer estimation of the in vivo situation. Among biomaterials, those with built-in sensing properties are particularly attractive to obtain quantitative information on how cells probe and respond to relevant physicochemical cues of the ECM. The pioneering work of Dembo and Wang (1999) on deformable elastic materials with embedded fluorescent tracers was one the first material of this class capable of deciphering cell contractile forces. Similarly, in the field of electrochemical sensors, the work of Giaever and Keese (1984, 1986, 1991) set the foundations for the development of materials capable of probing the cell adhesion interface in real time. Both approaches profited from their label-free capabilities. Since then, a myriad of synthetic responsive biointerfaces with unprecedented temporal and spatial resolution was engineered to study cell signaling and behavior in real time and with high sensitivity.
In this review, we will focus on the design of micro and nanostructured biomaterials that resemble the complex properties of the ECM, and that play an active role in measuring cell-adhesion related processes. Materials developed to study the impact of different properties of the ECM on cell behavior, but only provide adhesion support for cells are covered elsewhere (Mager et al., 2011; Rosales and Anseth, 2016). Emphasis will be placed on label-free sensing schemes. These approaches have advantages over those that require labeling. Label-free sensors offer real-time measurements, less or no sample preparation, and low non-specific response, reducing the risk of generating artifacts and false positives in the measurements. Commonly used labels like fluorescent or colorimetric dyes are often cytotoxic and hamper further use of the cultured cells, which is particularly desired for regenerative tissue applications. Thus, biomaterials with sensing capabilities hold great potential to bridge the gap between traditional cell binding assays and in vivo studies.
We aim to offer readers an overview of the latest sensing biomaterials and their main advantages and applications in order to guide the selection of the most appropriate platforms for specific purposes. We will describe the specific features that have been provided to materials and how these characteristics have contributed to reveal key aspects of the cellular adhesion mechanism. Although most of the systems described in this review are research-oriented, commercial applications are possible especially in biomedical diagnosis (Suhito et al., 2018; Tutar et al., 2019) and tissue engineering (Mitrousis et al., 2018; Gaharwar et al., 2020).
Cells Adhesion to the Microenvironment
The ECM is a complex and dynamic mixture of proteins and polysaccharides that provides not only support for cells but also is involved in regulating many important cellular processes including proliferation, survival, differentiation, and apoptosis (Frantz et al., 2010). Cells sense and respond to changes in topological, physical, and chemical properties of the ECM through a sophisticated system that allows them to adapt their behavior by converting these cues into biochemical signals. The interaction of cells with the ECM is mostly mediated by a family of cell transmembrane receptors called integrins that are responsible for cell attachment, and connect the cell-matrix adhesions with the cell cytoskeleton (Geiger and Yamada, 2011) (Figure 1A). Integrins undergo conformational changes upon biochemical and mechanical interactions leading to outside-in and inside-out mechanotransduction. Although it is well known that integrins have a crucial role in regulating diverse adhesion-related functions, the mechanism by which cells translate extracellular stimuli into biological responses remains unclear.
FIGURE 1

Molecular complexes involved in cell adhesion processes. Cells form bonds with the ECM and with other cells by protein receptors in the plasma membrane (A). These receptors are linked to the cytoskeleton by adaptor proteins. In (B), a scheme of a FA where integrins (formed by α and β subunits) recognize specific ligands in the ECM. In the cytoplasm, talin, kindling, and vinculin among other proteins (“adaptor proteins” in the image) associate with the integrins forming a complex that acts as a molecular clutch. The adaptor proteins connect the integrins with the cytoskeleton and participate actively in mechanosensing. In (C), a scheme of an adherent junction where cadherins in the extracellular medium establish bonds with cadherins in the other cell. The link between cadherins and the actin cytoskeleton is mediated by adaptor proteins α and β catenins.
Integrins are heterodimers constituted by two transmembrane protein subunits α and β which bind to specific ligands located in the ECM proteins or in membrane of other cells (I-CAM and V-CAM receptors) (Sun et al., 2019). Integrins adopt closed or open conformations characterized by low or high-affinity states, respectively. The transition from the closed to the open conformation is crucial for integrin activation and can be induced from the extracellular medium or from the cytoplasm (Luo et al., 2007). Upon activation, integrins form clusters and associate with adaptor proteins like talin, kindling, and vinculin among others. These proteins connect the integrins to F-actin fibers forming a molecular “clutch” that mediates mechanical forces between the membrane and the cytoskeleton (Figure 1B) (Sun et al., 2019). The macromolecular complex of integrins and adaptor proteins constitute the focal adhesions (FA). It has been shown that FA act as mechanosensory machines, translating multiple environmental cues to cellular responses (Geiger et al., 2009). The integrin-mediated binding to the ECM, even though being one of the most important adhesion mechanisms is not the only one. Syndecans and lectins also participate in cell adhesion, although their role in mechanosensing is not totally clear (Gumbiner, 1996; Mager et al., 2011; Guilluy and Dolega, 2020).
Cell–cell contacts are mediated by different types of junctions: adherent junctions, tight junctions, and desmosomes (Gumbiner, 1996). Adherent junctions are one of the most important sites of intercellular mechanical coupling (Ladoux and Mège, 2017). Cadherins are integral membrane proteins that participate in the formation of adherent junctions. Their extracellular domains mediate the adhesion to neighbor cells, whereas their intracellular regions are connected to the actin cytoskeleton by adaptor proteins α and β-catenin (Figure 1C). Cadherins associate between them and with adaptor proteins to form clusters constituting the mature adherent junction (Mège and Ishiyama, 2017). Cadherin complexes respond to tension load by the actomyosin. When pulling forces are applied to the adherent junction through the actin cytoskeleton, α catenin unfolds and associates with vinculin, which strengthens the adherent junction (Le Duc et al., 2010; Yonemura et al., 2010; Buckley et al., 2014). Therefore, mechanosensing in cell-cell bonds is mediated by the same structures that mediate cell–cell adhesions. There is also evidence that tight junctions mediate mechanosensing in epithelia by association with the actin cytoskeleton (Tornavaca et al., 2015). Desmosomes are tightly associated with intermediary filaments and participate in mechanosensing (Weber et al., 2012), although their role in mechanotransduction is complex as intermediary filament proteins are diverse and their expression is tissue specific (Ladoux and Mège, 2017). An excellent review on cell-to-cell association and the dynamics of collective cell behaviors has been published by Ladoux and Mège (2017).
Cell-to-cell adhesion is also important in cell communication processes. During antigen presentation, antigen presenting cells (APC) associate with T or B lymphocytes forming the immunological synapse. Evidence has shown that this process is mechanosensitive, although cell-to-cell bonds and mechanosensing is mediated by the interaction of the peptide major histocompatibility complex (pMHC) in the APC with the specific T or B cell receptor in the lymphocyte (Bashoura et al., 2014; Liu B. et al., 2014; Liu et al., 2016).
Relevance of Biosensors in Cell Adhesion Studies
Much of the information we have so far on how cells adhere to the ECM and other cells have come from a wide set of bioanalytical tools. Genetic modification like gene knockout (Elosegui-Artola et al., 2016; Strohmeyer et al., 2017), protein expression modulation by siRNA (Plotnikov et al., 2012; Bazellières et al., 2015) and the use of externally added inhibitors (Bashoura et al., 2014; Collins et al., 2017) are important methods to investigate the role of different proteins in the cell sensing mechanism. Cell behavior as a consequence of these modifications is often monitored by optical microscopy-based techniques like immunofluorescence (Engler et al., 2006), in vivo observation of recombinant fluorescence proteins (Reffay et al., 2014; Oria et al., 2017), genetically incorporated molecular probes (Grashoff et al., 2010), and more recently optochemical probes (Endo et al., 2019; Ollech et al., 2020). Even though these methods have revealed important aspects of the molecular mechanisms of cellular mechanotransduction, they could not provide a complete description of the interaction between cells and their environment. In this sense, biomaterials with built-in sensing capabilities have contributed to our understanding of cell mechanosensing by providing information not previously available by other bioanalytical methods. For example, it was not until the work of Harris et al. (1980) with soft silicon substrates that traction forces generated by cells were quantified during cell spreading and migration. Decades later, advances in material science and polymer chemistry have enabled remarkable improvements of this groundbreaking approach leading to a better understanding of how cells mechanically interact with the ECM (Roca-Cusachs et al., 2017).
Noteworthy, although optical microscopy-based techniques have been fundamental for our understanding of the cell adhesion process, they often involve endpoint measurements and time-consuming sample preparation (e.g., immunostaining). This greatly restricts the temporal resolution achievable with these techniques. Genetically encoded fluorophores can overcome this drawback but require modification of cell genome, and the time span of the measurement is limited by photobleaching. Transillumination microscopy imaging like phase contrast or differential interference contrast does not have this limitation, but provide only qualitative information. In this context, non-invasive and non-destructive methods stand out as sensitive and quantitative approaches to study cell-adhesion related processes in real-time. These include electrochemistry, quartz crystal microbalance (QCM), surface enhanced Raman spectroscopy (SERS), and surface plasmon resonance (SPR) (Janshoff et al., 2010; Méjard et al., 2014; Suhito et al., 2018) (More details of these approaches can be found in sections “Cell-substrate adhesions” and “Cell adhesion biomarkers”). Analytical approaches based on these techniques provide unique information with high time resolution about the cell-matrix and cell-cell interface. For instance, electric cell-impedance sensing (ECIS) methods can easily reveal the formation of cell–cell junctions and cell-substrate contacts, and are extensively used to follow epithelial maturation (Ngok et al., 2013; Gamal et al., 2017; Van Der Stoel et al., 2020). Surface enhanced Raman spectroscopy (SERS) is ideal to obtain information about biochemical changes inside cells in the proximity of a nanostructured surface (El-Said et al., 2015; Haldavnekar et al., 2018; Rusciano et al., 2019).
In cell culture, substrate materials provide the physical scaffold that supports cells allowing their adhesion and proliferation. Therefore, these materials have to be biocompatible, meaning that they have to comply with determined characteristics in terms of topology, stiffness, and chemical composition. However, substrates are not necessarily relegated to being a physical support, on the contrary, by incorporating a transducer element these materials can reveal crucial aspects that regulate cell behavior. This type of responsive biomaterials can be classified as biosensors. Conventionally, biosensors are devices able to provide selective quantitative analytical information using a biological recognition element and a transducer component. In the context of this review, the recognition element is often a specific cell-binding molecule (e.g., the tripeptide motif Arg-Gly-Asp (RGD) to bind αvβ3 integrin), while the transducer can be optical (SERS, traction force microscopy), electrochemical (ECIS), or piezoelectric (QCM). Because of its pivoting role, the transducer element will define important aspects of the analytical response as spatial and time resolution, signal to noise ratio, and selectivity.
Despite all the progress made in the study on how cells sense and react to physicochemical properties of their environment, understanding the mechanisms by which cells can transduce mechanical signals into biochemical events is still a challenge. Moreover, the relationship between these events and cell differentiation, physiological function, and pathology have not been elucidated (Mohammed et al., 2019). In the field of regenerative medicine, a scenario like this raises important questions on how to ensure the efficiency of materials to promote cell differentiation. In this regard, biomaterials with cell-instructive characteristics and built-in sensing capabilities could provide valuable information about cell-material interaction. Devices capable of providing new sources of information will be key to elucidate how different properties of the cell matrix affect cell behavior. In order to discuss the relevance of the biosensors in cell behavior studies, each section of the review nucleates biosensors in terms of the biological variable about which they provide information.
Biological Applications of Biosensors
Cell-Substrate Adhesions
After a cell in suspension makes the first contact with a substrate, it carries out successive steps of attachment, adhesion, spreading, and in some cases followed by migration and proliferation. To allow cell adhesion, substrates require the presence of adhesion-promoting proteins or ligands immobilized on the substrate (Janshoff et al., 2010). There are two options to achieve substrate modification to elicit cell adhesion: the adhesive molecules are incorporated during the substrate synthesis or they are secreted by the cells in the adhesion process. In the second case, the material has to allow protein absorption. Surface wettability and topography have a major influence on this process (Prime and Whitesides, 1991; Janshoff et al., 2010).
Most cells have excellent insulating properties. When cells adhere on an electrode surface, they modify the environment at the solution-electrode interface affecting the charge transfer events at the surface (Giaever and Keese, 1993; Ding et al., 2008). This phenomenon was exploited by Giaever and Keese (1984, 1991) when they created the first electrodes to study cell adhesion using electric impedance. In their design, the substrate incorporated working electrodes and a counter electrode. The working and counter electrodes were connected to a lock-in amplifier, and the culture medium completed the circuit. The authors monitored cell adhesion events by applying an alternating sinusoidal voltage and monitoring current. When cells adhered and spread on the working electrode they generated an impedance increase as a consequence of the formation of an insulating layer (Giaever and Keese, 1993). This technique was called electric cell impedance sensing (ECIS) and due to its high sensitivity, it has been used for monitoring cells attachment (Han et al., 2011; Xue et al., 2011), spreading (Wegener et al., 2000; Arias et al., 2010; Pradhan et al., 2014), locomotion (Giaever and Keese, 1991; Wang et al., 2008), and apoptosis (Arndt et al., 2004; Liu et al., 2009). Impedance measurements depend on the number of cells seeded on the electrode, their morphology, motility, and on the formation of cell–cell interactions (for further details see section “Cell-Cell adhesion”). Analysis of data is aided by mathematical models that allow calculating cell morphological parameters (Giaever and Keese, 1991; Lo et al., 1995). For more detailed reviews on ECIS please see Janshoff et al. (2010) and Hong et al. (2011). The use of ECIS substrates for monitoring cell behavior has advantages compared to traditional optical microscopy methods. ECIS is a non-invasive and non-destructive technique capable of providing information without the needing for cell staining (Suhito et al., 2018). In microscopy, quantification of the cell adhesion and spreading requires tedious data processing compared to the straightforward information provided by impedance measurements. Besides, impedance can be registered on cells over days with a temporal resolution of seconds (Hong et al., 2011). Moreover, transparent ECIS substrates can be excellent complements to optical microscopy as ECIS can provide information not easily accessible to visualization like the formation of cell-cell junctions, or cell micromotion (Giaever and Keese, 1991; Lo et al., 1995) (Table 1).
TABLE 1
| Method | Principle for monitoring cell adhesion | Advantages | Disadvantages | Resolution | References |
| ECIS | Changes in the impedance at the electrode-medium interface | • Cell-surface or cell-cell adhesion monitoring depending on current frequency • Sensing surface can be controlled with electrode design • Surface can be modified with micro and nanostructures | • Obtaining cell morphological parameters requires mathematical modeling | • Classical ECIS senses bulk cell population (no spatial resolution) • Single cell resolution using MEA • Spatial sub μmeter resolution in combination with SPR or optical microscopy. | Giaever and Keese, 1993; Lo et al., 1995; Ding et al., 2008; Wang et al., 2011; Susloparova et al., 2015 |
| QCM | Changes in resonance frequency of the piezoelectric substrate | • Changes in ΔD/Δf ratio as a fingerprint for cell-surface adhesion process • Versatile chemical functionalization of the substrate | • Non-rigid coatings complicates interpretation of data • Lack of direct correlation between signal and cell morphology | • Bulk cell population (no spatial resolution) | Fredriksson et al., 1998; Janshoff et al., 2010; Xi and Chen, 2013; Zhang et al., 2018 |
| SPR | Changes in the refractive index of the substrate surface | • High sensitivity to events within 200 nm of the surface • Sensitive to cell morphology | • Low sensitivity to events 200 nm above the surface • Limited to thin coatings • Relative more expensive | • Classical SPR senses bulk cell population (no spatial resolution) • SPR microscopy allows μmeter resolution | Rothenhäusler and Knoll, 1988; Willets and Van Duyne, 2007; Chabot et al., 2009 |
Comparison between label-free methods to measure cell-substrate adhesion.
Coatings on substrate materials for ECIS can offer better control of adhesive cell behavior without hampering the sensing capabilities of the electrodes (Giaever and Keese, 1986). Different strategies, including adherent polymer coatings (Choi et al., 2015), self-assembled monolayers (SAM) (Parviz et al., 2017), metallic nanoparticles (Kim et al., 2013; Pallarola et al., 2017a), carbon nanotubes (Srinivasaraghavan et al., 2014), and silicon nanowires (Abiri et al., 2015) have been reported. Susloparova et al. (2015) created new substrates for ECIS using open gate field-effect transistors instead of gold electrodes, which allowed to obtain single-cell resolution of the impedance measurements. Decker et al. (2019) employed 3D nanostructured multielectrode arrays to study cell adhesion. Using nanoimprint lithography, the authors created an electrode with incorporated nanostructures in different forms, dimensions, or pitch lengths in a reproducible way. By changing the synthesis parameters, especially the time during electroplating, the height and shape of the nanostructures could be modulated (Figures 2A,B). The authors created a multi-electrode array with half of the electrodes with nanostructured patterns and half without them. The authors tested different type of nanostructures with a pillar shape modifying the distances between nanostructures and their shape. Cells could attach to both the nanopatterned and unpatterned electrodes, although the nanostructured ones displayed a lower impedance (Figure 2C). However, upon cell adhesion some nanostructures showed increased cell-nanostructure coupling and increased impedance change as consequence of cell adhesion (Figure 2D). This work showed how nanostructured topologies on electrodes could improve ECIS biosensing capabilities. Moreover, ECIS sensitivity could be enhanced using redox probes. In this approach, the probe (for example [Fe(CN)6]3–) is incorporated in the culture medium (Ding et al., 2008). Cell adhesion and spreading on the electrode forms a barrier that hinders the access of the probe to the electrode, decreasing electron transfer. Thus, high sensitivity to the area covered by the cell can be achieved (Ding et al., 2007). A modification of this strategy was used in a recent work from Du et al. (2020) where they created a biosensor using nanocomposite materials to follow the epithelial-mesenchymal transition of A459 lung cancer cells.
FIGURE 2
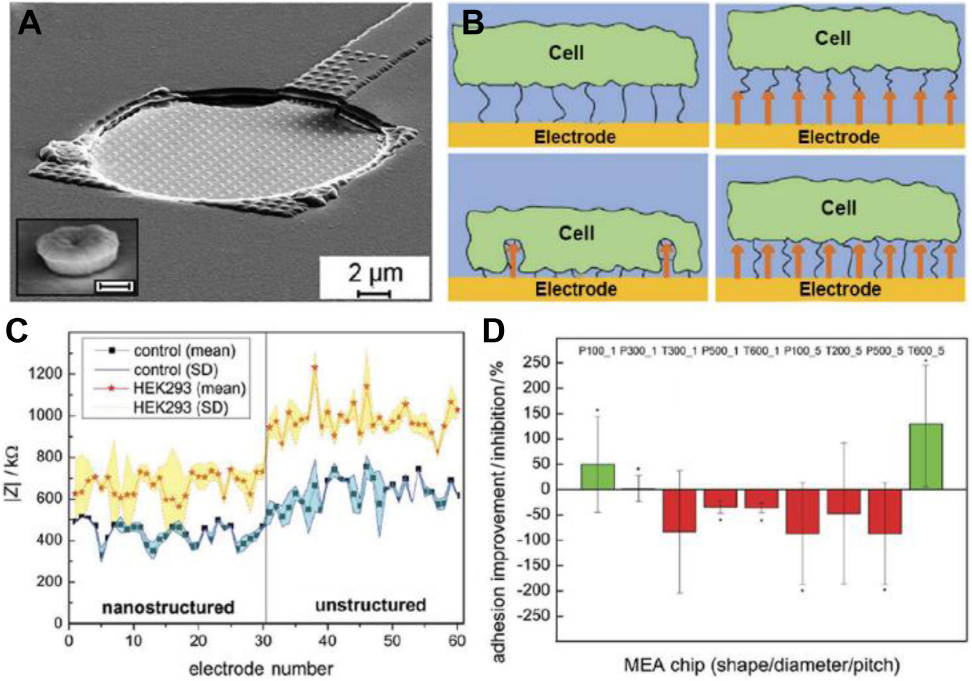
A nanostructured biosensor for the detection of cell-adhesion. In (A), a SEM image of a nanostructured electrode with incorporated nanostructures separated by 1 μm (inset: magnification of a tube-like nanostructure, scale bar: 200 nm). In (B), the image shows, the possible cell adhesion mechanisms of the cell to the surfaces. Compared to an unstructured electrode (image at top left) the cell can bind to the top of the nanostructures increasing cell–electrode distance and lowering impedance (top right). Alternatively, they can promote cell adhesion resulting in a closer cell–electrode distance and higher impedance signals after cell adhesion (bottom left). Finally, it is possible that nanostructures have no effect on cell adhesion (bottom right). In (C), a representative example of recordings showing the magnitude of impedance (| Z|), before (control) and after cell attachment on the nanostructured (no. 1–30), or unstructured electrodes (no. 31–60). In (D), the normalized ratio of the increase/decrease of the impedance ΔZ for nanostructured electrodes compared to the unstructured ones. P-value < 0.05 compared to unstructured electrodes. Figures were adapted from Decker et al. (2019) with permission. Copyright 2019 John Wiley and Sons.
Piezoelectric materials can also be employed for monitoring cell adhesion. Quartz microbalance experiments are performed on sensor materials made of an α-quartz disk sandwiched between two metal electrodes. Due to the piezoelectric nature of α-quartz, any mechanical deformation of the crystal creates an electrical potential at the quartz surface, and vice versa (Janshoff et al., 2010; Chen et al., 2018). In most common approaches, an alternating current is applied between the electrodes allowing measuring the resonance frequency of the crystal over time. Cells can adhere and grow on the resonator surface, which produces changes in its resonance frequency (Δf). Moreover, other materials can be deposited on the resonator surface to assess the cell adhesion to them. It has been shown that Δf changes correlate with cell coverage on the sensor surface (Redepenning et al., 1993; Wegener et al., 1998; Tagaya et al., 2011). Hence, monitoring Δf as a function of time was employed to follow cell adhesion, spreading, and proliferation (Ishay et al., 2015). However, due to the viscous nature of cells and culture mediums, the changes in vibrational energy dissipation (ΔD) of the sensor can also provide relevant information on cell behavior. Nonetheless, the link between ΔD and the physical characteristics of cells that elicit these changes has not been understood (Xi and Chen, 2013). The ratio ΔD/Δf has been regarded as a fingerprint of the cell adhesion process, as different cell lines display different ΔD and Δf behaviors during adhesion and proliferation (Fredriksson et al., 1998). QCM sensors can be modified with coatings to provide enhanced cell adhesion, although the nature of the coating could influence the responses of the sensor (Lord et al., 2006). Despite the needing for non-common materials (α-quartz sensors), QCM is an inexpensive and valuable technique to monitor cell adhesion dynamics. Table 1 summarizes the advantages and disadvantages of QCM in cell adhesion study.
Materials with surface plasmonic properties allow the implementation of an evanescent wave-based optical technique, surface plasmon resonance (SPR). SPR can be employed for monitoring cell adhesion processes in the proximity of the substrate surface (Chabot et al., 2009; Peterson et al., 2009; Wang et al., 2012). SPR sensors consist of a glass substrate (LaSFN9 or BK7) coated with a thin gold layer (∼50-nm). These sensors allow monitoring cell-substrate interactions occurring in the proximity of the first few hundreds nanometers over the gold layer due to the evanescent decay of the plasmon perpendicular to the surface interface (Willets and Van Duyne, 2007). The molecules in the proximity of the interface interact with the confined electromagnetic wave, resulting in changes in the refractive index at the metal surface (Homola, 2003; Willets and Van Duyne, 2007). Therefore, events occurring on the substrate surface such as cell adhesion and spreading modify the local refractive index of the surface, which can be followed in real time (Yashunsky et al., 2010; Borile et al., 2019). The changes in the refractive index are measured by irradiating the gold surface through a high refractive index prism in an angle that yields total internal reflection. The reflected light is interrogated by varying the angle of incidence at a fixed wavelength or by changing the wavelength at a fixed angle (Willets and Van Duyne, 2007). Also, spatial resolution of the cell-substrate contacts can be obtained using SPR microscopy (Rothenhäusler and Knoll, 1988). SPR has the advantage of detecting changes in cell morphology, outperforming ECIS and QCM in this aspect (Table 1). Noteworthy, SPR can be combined with electrochemical measurements as the local effective refractive index on a determined spot depends on the local charge density of the surface. Changes in the electrode interface by cell adhesion processes modify the local impedance of the surface, which is translated into changes in the local SPR signal. Using this phenomenon, Wang et al. developed electrochemical impedance microscopy (EIM) using a transparent conductive substrate with plasmonic properties (Wang et al., 2011). Mapping the local impedance on the surface allowed obtaining high-resolution images of the cell-substrate contacts. This technology is an example of how recent advances in the use of biosensor substrates allow characterizing the cell-substrate interaction with increasing time and spatial resolution.
Cell–Cell Adhesion
Direct interactions between cells are often mediated by a set of ligands and receptors expressed by both cells. When cells have to build long-term bonds between them, they assemble different types of junctions: adherent junctions, tight junctions, and desmosomes (Ladoux and Mège, 2017). In other cases, cell–cell adhesions have to be transient like those formed by the natural killers lymphocytes and their target cells (Orange, 2008).
Recently, Pallarola et al. (2017a) developed a nanostructured electrochemical sensor exhibiting high sensitivity to the constitution of cell–cell adhesion interactions. The sensing platform consisted of a 100-μm-diameter ITO microelectrode patterned with an ordered array of AuNPs, and surrounded by a SiO2-insulating layer (Figure 3A). The sensor was built by a combination of diblock copolymer micelle nanolithography (Pallarola et al., 2014) and photolithography. The use of a gold nanopatterned surface allowed a precise control over the distribution of cell adhesion ligands on a non-adhesive PEG-passivated background (Pallarola et al., 2017b). Cell behavior was monitored over several hours by simultaneous electrochemical impedance spectroscopy and optical microscopy (Figure 3B). The surface of the electrode exhibited high sensitivity toward early events of cell interaction. Resistance and capacitance recordings were used to study the behavior of different cell types. It was observed that cell lines expressing lower levels of E-cadherins registered lower resistance values at low frequency (429 Hz) (Figures 3C,D). This was in agreement with the fact that increased cell-to-cell adhesion results in a lower paracellular current (Hong et al., 2011). This feature allowed for distinguishing between different cell types based on the density of adherent junctions, as observed for MCF-7 cells, in comparison with MCF-10A cells. This approach is a powerful tool to study the dynamics of cell–cell contact formation and remodeling of junctions under specifically engineered environments in a highly sensitive, instantaneous, and non-destructive manner.
FIGURE 3
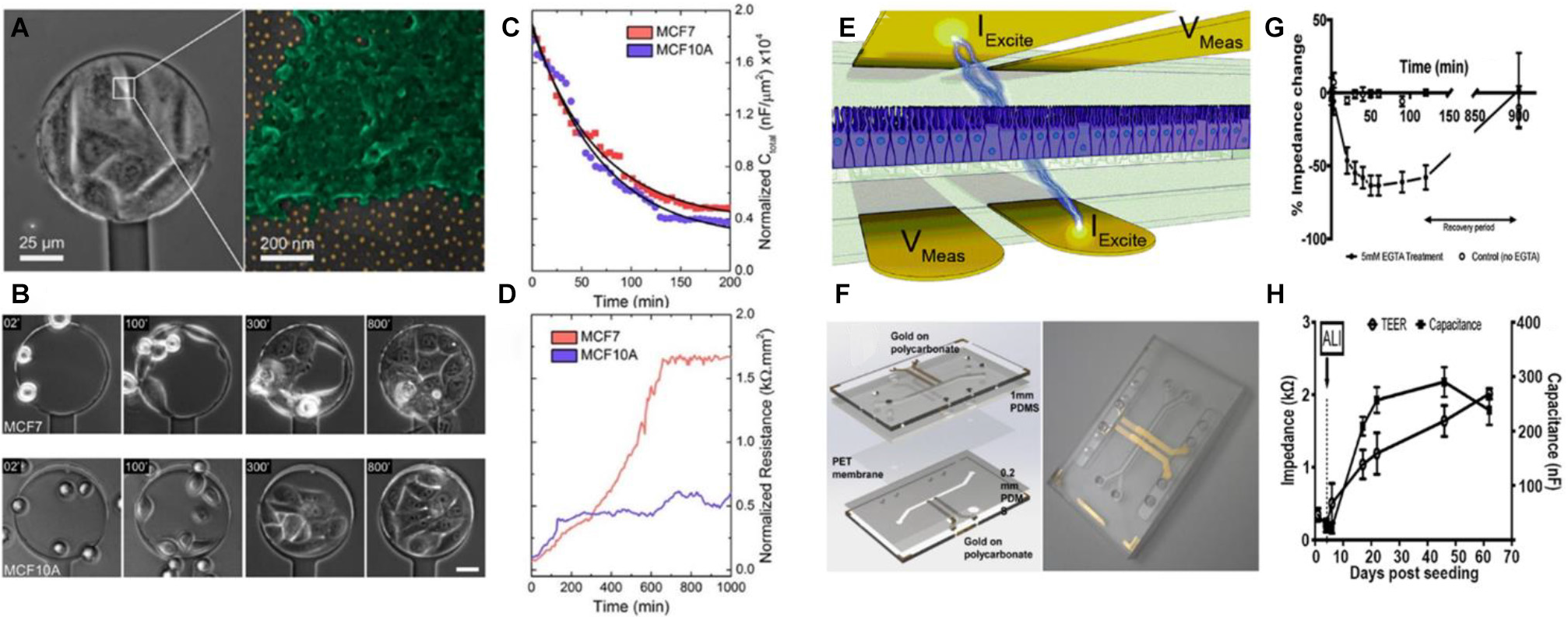
Biosensors based on electrochemical readout for monitoring cell-cell junctions. In (A), the image on the left shows an optical microscopy image of a cell-coated sensor. The magnification shows a cell on a nanopatterned surface, where the golden dots correspond to the AuNP in a quasi-hexagonal array. In (B), phase-contrast microscopy images show cell adhesion dynamics on a nanopatterned electrode (53 nm spacing between AuNP). AuNP were coated with specific αvβ3-ligands. Scale bar: 30 μm. In (C,D), the graphs show resistance and capacitance respectively, as a function of time for MCF7 and MCF10A cell lines. The results indicate that MCF7 cells induce a higher change in the electrode resistance in comparison with MCF10A cells. This can be attributed to the differential expression of E-cadherin. Figures (A–D) were adapted from Pallarola et al. (2017a) with permission. Copyright 2017 American Chemical Society. In (E), the image shows a schematic representation of the device for measuring transepithelial electrical resistance. The IExcite electrodes apply a small current of 10 μA and the VMeas electrodes measure the respective potential across the cell epithelium. In (F), the left panel shows a CAD model of the chip employed, while on the right panel a photograph of the assembled chip is shown. Gold electrodes were patterned on polycarbonate substrates. The polyethylene membrane where the cells adhered was placed between two PDMS layers that formed the microfluidic channels. The assembly was placed between two polycarbonate layers to assemble the chip. The graph in (G) shows the relative impedance change as a function of time of intestinal epithelial cells in the absence or presence of 5 mM EGTA. The decrease in impedance in the presence of EGTA was attributed to the disassembly of the cell to cell junctions. The graph in (H) shows the TEER and capacitance of a culture of airway epithelial cells as a function of time. In this case, the cell line was cultivated submerged in liquid medium on both sides of the epithelium. Then, on day 6, the liquid of the upper chamber was removed. Thus, an air-liquid interface was generated (ALI) emulating the natural environment of the cells. Figures (E–H) were adapted from Henry et al. (2017) with permission. Copyright 2017 Royal Society of Chemistry.
The integrity of epithelia relies on the ability to form strong junctions between cells. Another suitable approach to monitor the formation of cell-cell contacts is to measure the electrical impedance across an epithelium placed between two electrodes. Measurements of trans-epithelial electrical resistance (TEER) is a valuable method for evaluating in vitro barrier tissue integrity. TEER measurements of cell cultures were widely utilized in research (Ferruzza et al., 1999; Huh et al., 2010; Lippmann et al., 2012) and for a more detailed review on this topic see the article of Srinivasan et al. (2015). In particular, the integration of immobilized TEER electrodes with microfluidics holds great potential to study cell barrier functions and cell behaviors in cell-mimetic environments. For example, Henry et al. (2017) developed a robust approach to fabricate organ-on-chip based on microfluidics with fully integrated electrodes (Figures 3E,F). The assembly of tight junctions was monitored by measuring the TEER and capacitance at low frequency. The authors demonstrated that when cells established tight junctions between them, capacitance reaches its maximum. A classical experiment to prove the ability of electrochemical devices to measure the formation of tight junctions is to add EGTA to the culture medium (Lo et al., 1995). The authors also showed that impedance decreased over time when Ca2+ is sequestered due to of disassembly of the tight junction (D’Angelo Siliciano and Goodenough, 1988). After EGTA is removed, tight junctions are assembled again and impedance is recovered (Figure 3G). Also, the authors could employ the chip to follow the behavior of airway epithelial cells cultured in an air-liquid interface (Figure 3H). The work established a standard and reproducible protocol for the fabrication of organ-on-chip systems with TEER-based sensing capabilities. This kind of platforms displays promising applications as TEER measurements are frequently used to follow epithelial integrity and differentiation in organs-on-chips (van der Helm et al., 2019; De Gregorio et al., 2020).
Cell and Tissue Architecture
When cells constitute tissues, cell–ECM and cell–cell adhesions are orchestrated to achieve proper collective structure and organization. During tissue formation, cells selectively form bonds between them, change shape, migrate, and synthesize ECM (Gumbiner, 1996). In this context, materials with the ability to control topological and geometrical properties of cells are desired in order to induce the same cell architecture found in normal tissues. Due to the frequent complex 3D topological organization of cells in tissues, analyzing the response of single cells in vivo is challenging (Ladoux and Mège, 2017). Therefore, smart in vitro strategies are necessary to mimic as close as possible the conditions of cells in multicellular organizations. The incorporation of cell geometry aspects adds a layer of complexity to the design of biosensors, particularly if the nanometric topologies are also incorporated into the substrate material.
The ability to produce precisely engineered scaffolds can provide a way to control cell architecture during culture. Conventional cell cultures lack the ability to control cell spatial organization, therefore micropatterning techniques have been developed to create 2D patterns to control cell-substrate interactions and cell behavior. Micropatterning can be done by microcontact patterning (μCP), which consist in creating micro-stamps to deposit an “ink” material on it and print the material onto a substrate, resulting in a 2D pattern of the “ink” material on the surface (Alom Ruiz and Chen, 2007). The first micropatterning methods were developed in the late 60s (Carter, 1967; Harris, 1973). However, they became more available with the increased accessibility to photolithography techniques. The use of polydimethylsiloxane elastomer (PDMS) to prepare molds made the process easier, allowing to print patterns of proteins on substrates (López et al., 1993b), and to employ them to study cell adhesion in different patterns (Whitesides et al., 2001). There are other methods to create micropatterns on surfaces like photolithography (Bélisle et al., 2009; Kim et al., 2010), microwritting (López et al., 1993a), and microfiltration (Kailas et al., 2009), although μCP remains as one of the most common methods (Théry, 2010). Micropatterning was also successfully combined with nanopatterning to create substrate materials controlling topography aspects in the micro and nanometer range (Ren et al., 2017). New strategies were developed for orthogonally functionalized surfaces with two different adhesion ligands, controlling each nanodistribution independently (Polleux et al., 2011; Guasch et al., 2016; Yüz et al., 2018). Surfaces could be tailored with Au or TiO2 nanoparticles that act as anchor points of cell adhesive ligands. A similar approach was used to functionalize orthogonally micropatterns on a surface with two ligands with specificity for either α5β1 or αVβ3 (Guasch et al., 2015). Using photolithography and metal sputtering, a micropatterned surface consisting of stripes of Au or TiO2 was synthesized. The different Au or TiO2 areas could be functionalized selectively by using integrin ligands with a thiol or phosphonic acid group respectively. These examples show how adhesive ligand distribution of a substrate can be precisely controlled to build micro and nanopatterned substrates.
The creation of micropatterns on different materials opens the possibility to introduce topographical features on surfaces with built-in sensing capabilities. Micropatterning can be applied to sensing materials in order to study the role of cell architecture on mechanobiology. Ribeiro et al. (2015) created Matrigel microtopographies on polyacrylamide substrates with embedded fluorescent beads to study the differentiation of cardiomyocytes from human pluripotent stem cells (hPSC-CM). The material design allowed to: (i) control the 2D cellular aspect ratio on the substrate; (ii) modulate the stiffness of the substrate; (iii) measure the contractile force of cardiomyocytes by traction force microscopy (TFM). hPCS-CM were cultivated on isolated rectangular micropatterns with different aspect ratios. The authors showed that myofibril alignment and contractile forces along the major axes of the cell were greatest in high aspect ratios (7:1) and physiological stiffness (10 kPa). These conditions indicated a more differentiated phenotype compared to cells growing on micropatterns with different aspect ratios and/or substrate stiffness. The results were also supported by the characterization of Ca2+ signaling, mitochondrial organization, and protein expression in the cells. The ability of this functional material to modulate cell aspect ratio, substrate stiffness, and force transduction constituted a powerful tool to control hPCS-CM differentiation.
However, despite the versatility of micropatterning techniques on 2D substrates, they fail to mimic in vivo-like 3D microenvironment and organization. Several strategies have been developed to create biomimetic 3D scaffolds like solid porous substrates (Lai et al., 2012), hydrogels matrixes (McKinnon et al., 2013), microconduits (Anderson et al., 2016), and microtracks (Kraning-Rush et al., 2013). The use of two photon polymerization allowed to create very complex 3D scaffolds for cell culture (Turunen et al., 2017). Nevertheless, the incorporation of controlled nanopatterning and built-in sensing capabilities is still a challenge in 3D culture matrixes. To create a 3D biosensor that could transduce cell behavior in 3D environments, Pitsalidis et al. (2018) designed an organic biotransistor, in which the conductive polymer poly(3,4-ethylenedioxythiophene) was doped with poly(styrene sulfonate) (PEDOT:PSS) to create a suitable scaffold for cell growth. The addition of collagen and single-walled carbon nanotubes (SWCNTs) for improving biocompatibility and electrical performance, respectively, was also studied. Then, the polymer scaffold was fixed between two electrodes in a chamber filled with aqueous solution containing a gate electrode. In these conditions, the organic polymer scaffold was in direct contact with two electrodes and indirectly with the gate through the aqueous medium. The system behaved as a transistor, the two electrodes induced a current that went through the scaffold, which depended on the gate voltage. Due to the porous nature of the scaffold, cells could attach and grow in it. Madin-Darby canine kidney cells II (MDCKII) proliferation inside the porous scaffold induced a decrease in transconductance, which allowed monitoring cell growth in real-time. Also, after 3 days of culture MDCKII cells exhibited less transconductance compared with telomerase immortalized fibroblast (TIF). The authors explained these results by the presence of a higher density of cell-cell junctions in MDCKII compared to TIF. Although this device is not yet fully biomimetic, it represents a step forward in creating electrochemical sensing platforms for 3D cell culture studies in real-time.
Cell Adhesion Biomarkers
Biochemical signals have a critical role in tissue formation. They can be found free in the extracellular medium, embedded in the ECM or at the surface of other cells like the Notch signaling pathway (Chacón-Martínez et al., 2018). At the same time, cell differentiation is followed by the release of biochemical mediators like neurotransmitters in neurons (Kim et al., 2015; Kruss et al., 2017), hormones in endocrine cells (Lund et al., 2016; Hunckler and García, 2020), and ECM components (Mehlhorn et al., 2006; Zhang et al., 2020). Moreover, differentiation induces changes in cell metabolism that modifies the concentrations of intermediates in metabolic pathways (Quinn et al., 2013; Carey et al., 2015). The creation of active materials capable of detecting molecules released by cells, although is highly valuable, yet remains a challenge. This can be attributed to the fact that, in many cases, the sensor material is passivated by the same molecules released by cells (Spégel et al., 2007), and because of the complex mixtures of biochemicals that can interfere in the sensing process (Huang et al., 2011). Furthermore, high sensitivity should be provided, since often the concentration of the target molecule reaches low concentrations and for short periods of time (Amatore et al., 2008).
Electrochemical biosensors provide a versatile and sensitive means to probe the content of biological environments (Ding et al., 2008). Wang et al. (2018) created stretchable photocatalytically renewable electrodes for nitric oxide (NO) sensing by functionalizing PDMS films with a nanonetwork of Au nanotubes (NTs) and TiO2 nanowires (NWs). The Au NTs rendered electrochemical sensing performance, while the TiO2 NWs provided photocatalytic activity to recover the performance of the sensor after UV irradiation. In addition, electrochemical biosensors can measure neurotransmitters released by neurons, measuring the redox reaction of a neurotransmitter with the electrode. Kim et al. (2015) created a nanopatterned electrochemical sensor to monitor the dopaminergic differentiation of human neural stem cells (hNSCs). The nanopatterned surface showed increased cell adhesion and spreading of a dopaminergic cell line (PC12) and enhanced sensitivity toward dopamine compared to a planar gold or ITO electrode. Spatial resolution of neurotransmitter release could be obtained by using microelectrode arrays (MEAs). Wang et al. (2013) created subcellular MEAs ranging from 4 to 16 μm2 to record the release of dopamine across single cells and PC12 cell clusters.
In addition to sensors based on electrochemical transduction, sensors based on optical readout have been proposed to monitoring molecule release by cells (Kim et al., 2018; Dinarvand et al., 2019; Liu et al., 2020). Kruss et al. (2017) developed single-walled carbon nanotubes wrapped with short DNA sequences (DNA-wrapped SWCNT) to measure dopamine release from PC12 cells. These DNA-wrapped SWCNT displayed near-infrared fluorescence and changed their fluorescence emission spectrum in presence of specific organic molecules (Figure 4A). The authors optimized the DNA sequences to improve the selectivity and sensitivity toward dopamine. Then, sensors were immobilized on a glass surface and PC12 cells were seeded on it. PC12 cells dopamine release was measured by fluorescence microscopy. Surface immobilized DNA-wrapped SWCNT fluorescence emission depended on the local dopamine concentration on the surface. The authors recorded fluorescence images of the cells and divided the image into discrete pixels. A fitting algorithm was developed for the normalized fluorescence intensity traces of the pixel groups. Using this method the authors could localize transient peaks in fluorescence recordings due to dopamine exocytic events (Figure 4B). Results showed that PC12 cells secreted dopamine at determined exocytosis sites or “hot spots,” instead of a release of neurotransmitters at random locations on the membrane (Figures 4C,D). Notably, the authors demonstrated that the substrate functionalized with DNA-wrapped SWCNTs could render higher spatial resolution compared with MEAs and similar time resolution to that of cyclic voltammetry-based sensors.
FIGURE 4
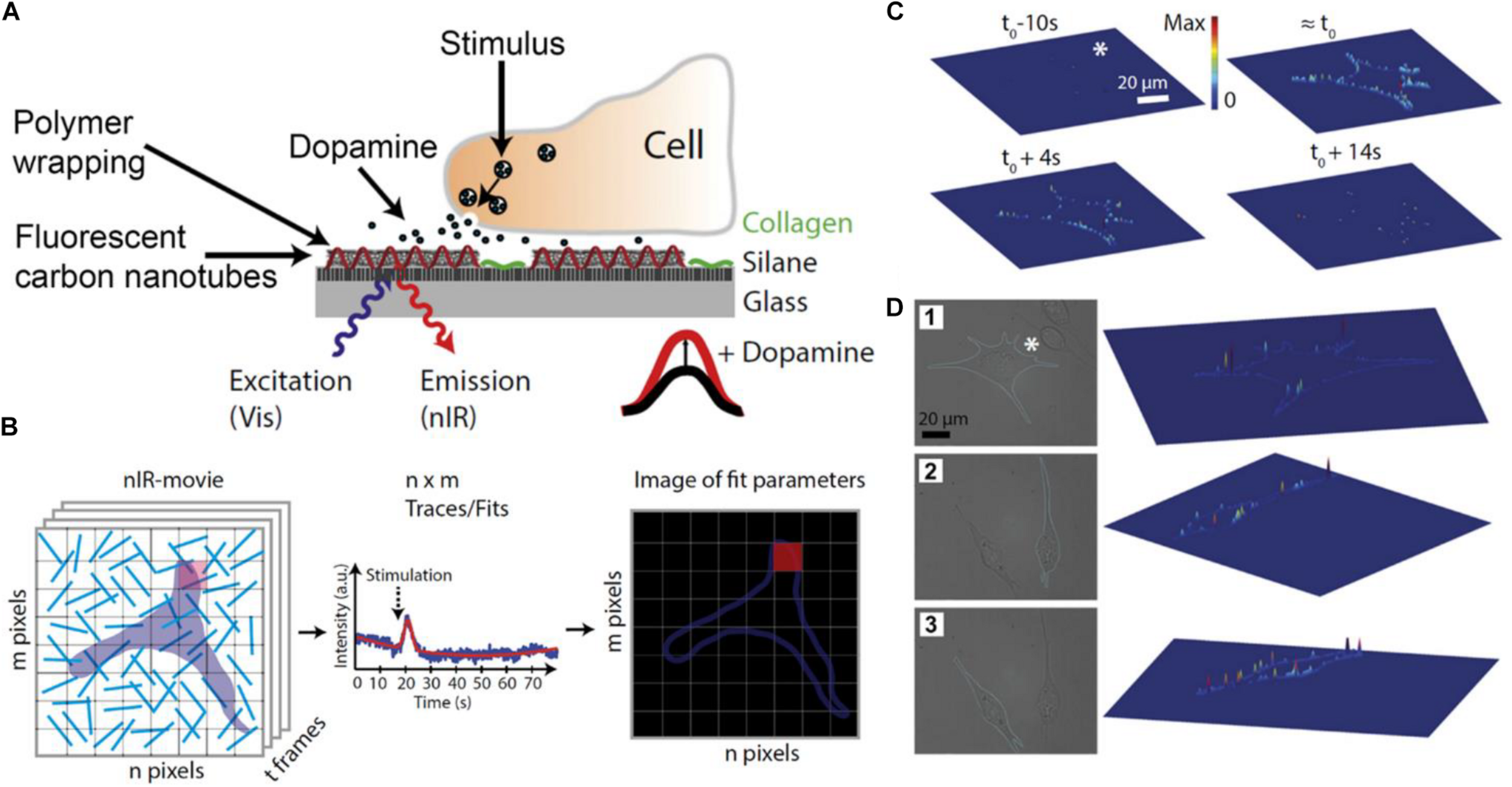
Detection of dopamine released by PC12 with nanosensor arrays. In (A), a scheme of the biosensor surface where DNA-wrapped SWCNTs were immobilized on a glass surface. Surfaces were coated with collagen to facilitate cell adhesion. The DNA-wrapped SWCNTs modified their fluorescence spectrum as a consequence of non-covalent dopamine binding. In (B), the scheme shows the analysis of experimental images. Each pixel of the images corresponded to a region containing one or more DNA-wrapped SWCNT nanosensors. Each pixel of the fluorescence movies produces a trace that contains information about the local dopamine concentration. A function was fitted to the data of each pixel to obtain the amplitude, width, and time of the signal. The fitted parameters can be represented in false-color images. In (C), the image shows released dopamine profiles across the border of different cells. For this analysis, only pixels on the cell border were considered. On the top of (C), 3D plots of fitted sensor signal responses at different times before or after stimulation (t0) are shown. Height and color of the 3D plot surfaces indicate the relative fluorescence change in pixels in the cell border normalized to the maximum fluorescent change in the same cell (values between 0 and 1). Results show that the maximum response is acquired at t0, then signal decreases. In (D), images 1, 2, and 3 indicate different cells and their respective 3D plots. In this case, the height of 3D surface plots shows the maximum dopamine response obtained in each pixel, showing that dopamine is released at particular locations or “hot spots” on the cell membrane. Figures were adapted from Kruss et al. (2017) with permission. Copyright 2017 National Academy of Sciences.
Among existing materials for detecting the release of molecules from cells, metal nanopatterned substrates have a remarkable advantage. Metal nanostructured surfaces with plasmonic properties can enhance the Raman scattering of molecules, a phenomenon known as surface enhanced Raman scattering (SERS). Raman scattering occurs because of the inelastic scattering of photons by molecules. Due to their different vibrational modes, molecules produce a spectrum of Raman scattered light, which contains information about the chemical identity of the molecule (Kneipp et al., 2010). The Raman scattering is often weak, but when the molecules are very close to the metal nanostructured surface, their Raman scattering can be enhanced several orders of magnitude. The detailed mechanisms by which molecules enhance their Raman scattering near metal nanostructures are beyond the scope of this review, but the reader can refer to these excellent articles for more details (Haynes et al., 2005; Kneipp et al., 2010; Schlücker, 2014).
SERS is a promising strategy to analyze the chemical composition near a nanostructured surface in a non-invasive and sensitive way. Cells can be seeded on SERS substrates, and once they adhered and spread, their chemical composition near the surface can be studied by irradiating the substrate and measuring the Raman spectra. Over the years, a wide diversity of metal nanostructured surfaces have been created for SERS, such as nanoparticles attached to a surface (Freeman et al., 1995; Zhai et al., 2009; Lussier et al., 2016), nanopillars (Kang et al., 2015; Li et al., 2016), nanoholes (Abdelsalam et al., 2005; Luo et al., 2019), and others (Yüksel et al., 2017; Yao et al., 2020). A detailed review on the fabrication of SERS substrates can be found in Fan et al. (2011). El-Said et al. (2015) built a nanopatterned surface for in vitro monitoring of neural stem cell (NSC) differentiation. The authors electrochemically deposited Au on an ITO surface to obtain an array of Au nanostars. PC12 cells adhered and spread on this substrate, and could be electrically stimulated due to the conductive characteristics of the material. The Raman spectrum was recorded at different stages of the cells differentiation process and exhibited different peaks that could be attributed to the presence of specific functional groups in biomolecules. Although it is difficult to link the changes in Raman spectra with specific changes in cell biochemistry composition, the Raman spectra can be used as a fingerprint to follow the cell differentiation process in a non-invasive manner.
Excitatory Cell Adhesion
Changes in membrane potential are a critical aspect of neurons and myocytes function. Action potentials travel through the membrane of these cells allowing communication of signals between different parts of the cell, eliciting the release of signaling molecules like neurotransmitters, and triggering contractile activity of muscle cells. Thus, proper cell electrical activity is an important characteristic of neural and cardiac differentiated tissues (Gunhanlar et al., 2017; Karbassi et al., 2020). Nowadays, neural and myocyte single-cell electrical activity can be measured by patch clamp techniques, optical imaging using genetically encoded or extrinsic fluorophores and substrate-integrated MEAs (Spira and Hai, 2013).
In particular, MEAs provide non-invasive monitoring of electrical activity and stimulation of multiple neurons in vitro and in vivo (Hutzler et al., 2006; Berdondini et al., 2009). At the beginning, electrodes were only capable of registering extracellular potential of cells (Thomas et al., 1972; Gross et al., 1977; Pine, 1980; Csicsvari et al., 2003), however, modifications of the material topology allowed to record the intracellular action potential. This could be achieved by the generation of protrusions of different shapes like mushrooms (Hai et al., 2010) and pillars (Robinson et al., 2012) where electrodes are inserted into the cell. Another configuration was created by Desbiolles et al. (2019). These authors fabricated a surface containing nanovolcanoes with an electrode in their interior. Cells fused spontaneously with the nanovolcano allowing the electrodes to be in contact with the intracellular medium (Desbiolles et al., 2019). Other approaches include kinked nanowires (Tian et al., 2010) or nanotubes (Duan et al., 2012). In these cases, the nanostructures allow a field-effect transistor to gain access to the intracellular medium. Protrusive nanostructures disrupt the membrane and insert into the cytoplasm spontaneously (Desbiolles et al., 2019), by electroporation (Xie et al., 2012), or by chemical functionalization (Duan et al., 2012).
Planar patch clamp chips are other type of substrates used to measure intracellular potential, constituting a protrusion-free approach. Using this strategy, Martina et al. developed a planar substrate with holes from 2 to 4 μm in diameter that connected to a microfluidic channel under the surface (Martina et al., 2011). Neurons adhered to the substrate and spread, covering the holes. Then, negative pressure was applied in the channel, which breaks the cell membrane and connects the cytoplasm with the microfluidic channel, similarly to a whole-cell patch-clamp configuration. Thus, the intracellular potential could be registered through the microfluidic conduits.
Usually, electrodes in MEAs are made of metallic conductors like gold, titanium nitride, platinum, and alloys like iridium oxide (IrOx). Electrode surface can be modified with porous materials from platinum, gold nanostructures, CNTs, and conductive polymers like poly(3,4-ethylenedioxythiophene) (PEDOT) (Obien et al., 2015) to increase the effective surface area of electrodes. In the last years, the use of complementary metal-oxide-semiconductor (CMOS) technology provided an increase of electrode density in MEAs (Obien et al., 2015). Despite the promising sensitive capabilities of protruding conductive electrodes to measure intracellular action potential, their effects on cells are not fully characterized. For example, it has been reported that protrusions that insert into cells could alter intracellular trafficking (Zhao et al., 2017). In the case of non-protrusive strategies like planar patch clamp chips, the measurement time could be limited by the perfusion of microfluidic liquid into the cell. Because of this, a different approach was employed by Dipalo et al. (2018) consisting of MEAs made of platinum or gold porous electrodes (Figures 5A,B). The electrodes acted as plasmonic antennas, which under infrared light illumination generated acoustic waves that transiently porate the cell membrane at the illuminated spot, a process named “optoacoustic poration.” A platinum porous electrode was placed on top of the aluminum surface of the CMOS-MEA electrodes. Cardiomyocytes derived from human-induced pluripotent stem cells (hiPSCs-derived cardiomyocytes) were cultured on the MEA. After optoacoustic poration of cardiomyocytes the intracellular action potentials could be recorded (Figure 5C). Interestingly, membrane potential recordings were not affected by repeated optoacoustic porations on the same spot or different spots on the same cell (Figures 5D,E). The material took advantage of the nanostructured properties of the porous electrode to facilitate the access to the intracellular medium and provided a robust and reliable way to measure intracellular action potentials.
FIGURE 5
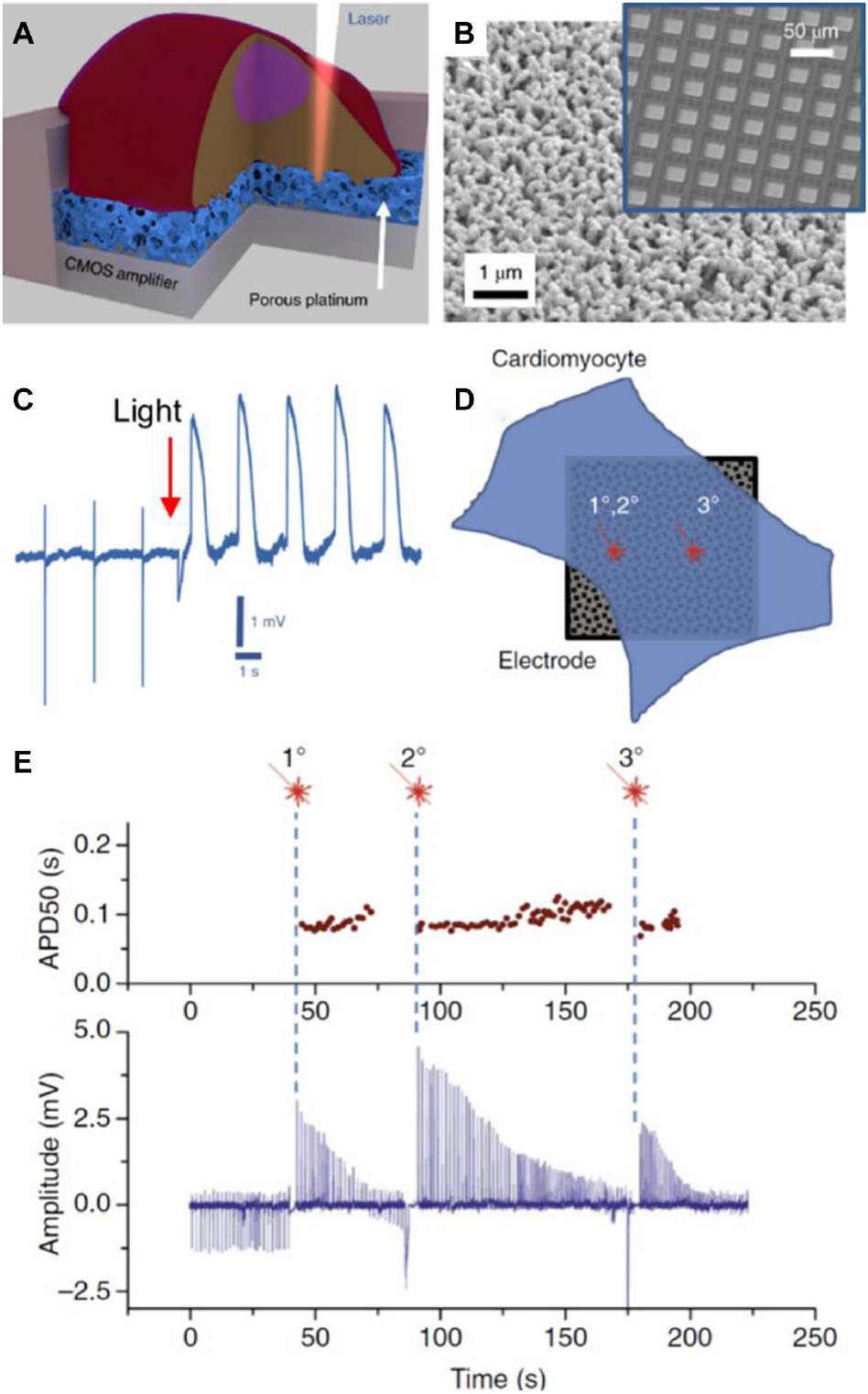
In (A), a scheme of the porous platinum electrode on the CMOS amplifier. Due to the structure of the platinum electrode, cell poration can be done with light (opto-poration). In (B), the image shows a surface electron microscopy image of the porous platinum surface of an electrode. In the inset, an image of the CMOS-MEA employed by the authors is depicted. In (C), a recording of the electrode signal where the instantaneous transition from the extracellular to intracellular recording takes place. In (D), the image represents the opto-poration of a cardiomyocyte in the same spot twice (1° and 2°) or in a different spot (3°). Results of electrical activity are shown in (E). At the top, action potential duration at 50% amplitude (APD50) as a function of time after opto-poration. At the bottom, intracellular signals as a function of time. Signal amplitude decreased as a function of time due to membrane re-sealing. However, action potential duration (APD50) remained at the same value, no matter if opto-poration was performed twice on the same spot or other places of the cell. Figures were adapted from Dipalo et al. (2018) with permission Copyright 2018 Springer Nature.
Measurements of Cell-Generated Forces
Upon adhesion, cells generate contractile forces to their supporting material. On materials of soft composition, the forces transmitted from the cells cause substrate deformation. This phenomenon can be exploited to assess the link between the mechanical properties of cells and the stiffness of the extracellular environment. In the beginning, forces were estimated by counting wrinkles as a consequence of material deformation by the cell (Harris et al., 1980). Forces could be quantified by a calibration curve created by correlation of forces of known magnitude with the length of the wrinkles they produce (Burton and Taylor, 1997). However, this methodology was limited in terms of spatial and temporal resolution as wrinkles are usually larger than cells, they develop slowly, and are intrinsically non-linear (Dembo and Wang, 1999). A major improvement was introduced in the work of Dembo and Wang (1999), where fluorescent beads were embedded in the soft polymer layer on which cells grow, establishing the basis of traction force microscopy (TFM). In classic TFM, forces exerted by the cells on the soft material are calculated by tracking the displacement of the tracer particles by fluorescent microscopy. An image of the substrate in a stress-free reference state is compared with the image of the substrate in the presence of cells.
Over the last few years, TFM has been implemented on diverse elastic materials like silicone, polyacrylamide, and polyethylene, being polyacrylamide one of the most used due to its transparency and elastic properties (Mohammed et al., 2019). Regarding elasticity, substrate materials in TFM are generally linearly elastic, i.e., stress is directly proportional to strain, which facilitates force calculation. However, the ECM is a fibrous network, and therefore elasticity is not linear. For a more detailed description of the mechanical properties of the ECM see the recent review from Polacheck and Chen (2016). The first attempts to measure forces in non-linear elastic biopolymers were implemented by Steinwachs et al. (2016). Often, substrate materials are coated with ECM components like collagen, fibronectin, laminin, or peptide mimetic ligands of these proteins to provide cell-adhesion sites on the substrate. A detailed protocol and guideline to perform TFM can be found in a manuscript from Plotnikov et al. (2014).
In a recent work, Oria et al. (2017) fabricated polyacrylamide hydrogels of different stiffness embedded with fluorescent nanobeads and decorated with a quasi-hexagonal array of gold nanoparticles on their surface (Figure 6A). Nanometer scale distribution of integrin ligands (cyclic arginine-glycine-aspartate, cRGD) was achieved by functionalization of the gold nanoarray, while the tracking of the fluorescent beads allowed the measurement of cell forces. The created substrates offered a versatile platform for studying how cells sense spatial and physical information at the nanoscale. Interestingly, the authors showed that ligand spacing and substrate stiffness had opposite effects on cell behavior. Increased adhesion was produced at long ligand spacing on less rigid surfaces (200 nm, 1.5 kPa) or at short ligand spacing on more rigid surfaces (50 nm, 30 kPa). The authors measured the length of FAs on cells that expressed a paxillin labeled with a green fluorescent protein (GFP-paxillin), which was an indicator of the maturation of the FA. Results showed how adhesion depended on the ligand spacing and surface stiffness (Figure 6B). At higher substrate stiffness, the cell exerted higher tensile forces on the surface (Figure 6C). Moreover, cell adhesion was dependent on whether ligand distribution was ordered or not (homogenous ligand spacing or random distribution). These results ruled out a molecular-rule hypothesis in which there is an optimal ligand spacing for FA assembly and growth. On the contrary, the authors explained their results by a model that takes into account pulling forces exerted by myosin on actin filaments, a force threshold that triggers FA growing, and a maximum integrin recruitment in the FA. On soft surfaces, ligand spacing has to be long, so that actin-pulling forces are distributed on fewer integrins. Thus, the force on each integrin is high enough to trigger FA growing. As more rigid surfaces were employed, FA can grow until the maximum number of integrins in the cluster is reached. If stiffness is too high, the pulling force exerted by the cell overpass the FA binding force to the substrate and adhesion collapse. This work is an exquisite example of how materials can be engineered to mimic different properties of the cellular environment, and at the same time provide sensitive means for probing cell mechanics.
FIGURE 6
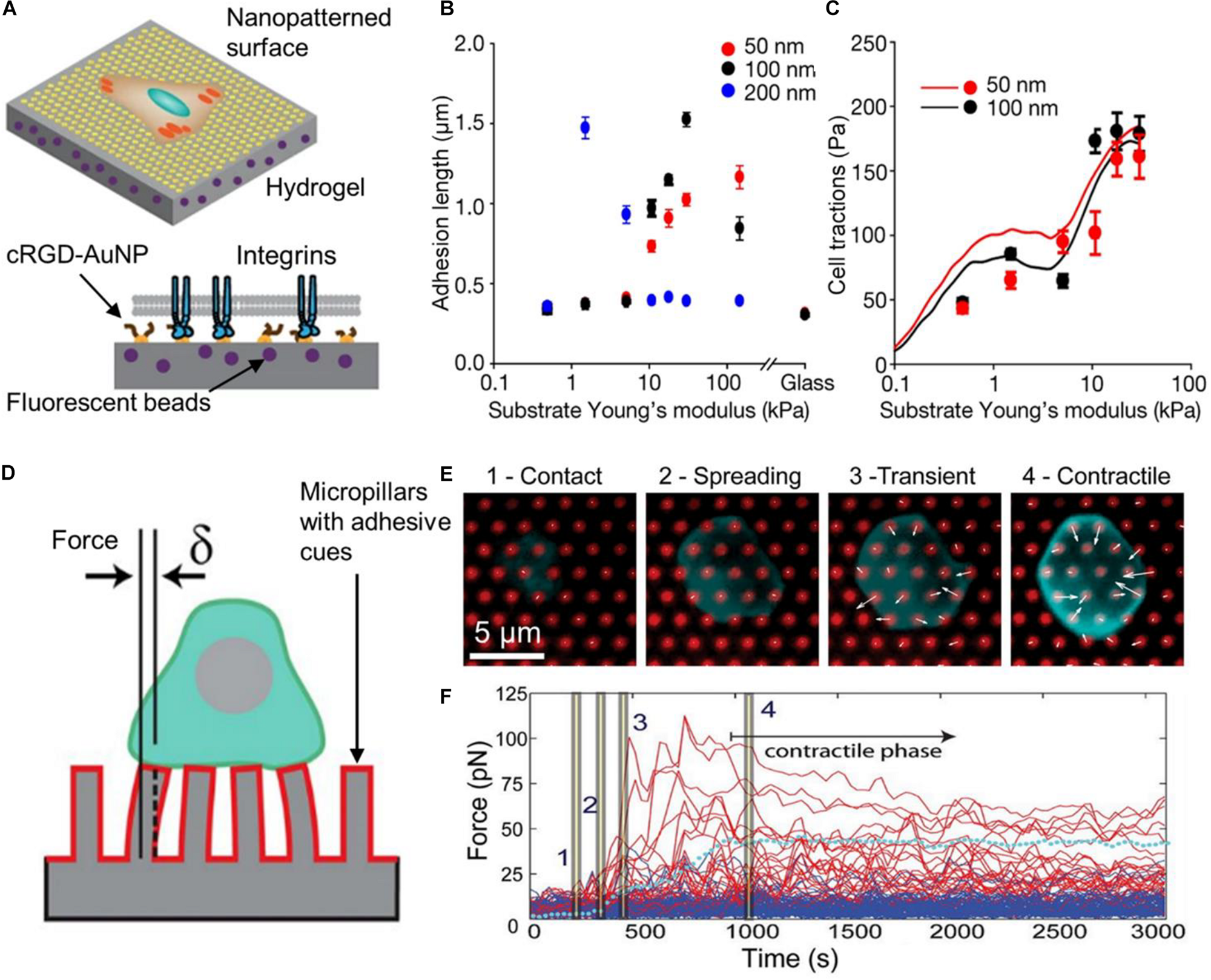
Substrate materials for measuring cell-exerted forces. In (A), the scheme represents the nanopatterned substrate employed by Oria et al. (2017). Hydrogels with different stiffness and coated with AuNP at different distances were employed. AuNP were functionalized a cRGD-based ligand (integrin ligand). Due to steric hindrance only one integrin could bind to each AuNP. The length of FA (adhesion length) was measured using cells that expressed GFP-paxillin. In (B), results show that greater FA were assembled on soft substrates with high spacing between ligands (200 nm, blue dots). On stiffer substrates, longer FA were detected at shorter ligand spacing (50 nm, red dots). In (C), the graph shows averaged cell tractions as a function of substrate stiffness. The continuous lines represent the predictions of the model created by the authors to explain the results. Figures in (A–C) were adapted from Oria et al. (2017) with permission. Copyright 2017 Springer Nature. In (D), a scheme of a T cell on an array of elastomer pillars is presented. These pillars were coated with antibodies that activated the TCR and the CD28 (red coating on the pillars). Forces exerted by cells were calculated by measuring the pillar displacement (δ). In this type of material, pillar displacement is proportional to the force applied to them (for small displacements). In (E), the images show different phases of the T cell interaction with the substrate. White arrows indicate the direction of forces and their magnitude. An arrow of the same length of the 5 μm scale bar represents a force of 250 pN. In (F), the graph shows the force applied to individual pillars as a function of time. Red lines represent the forces exerted on pillars in contact with the cells whereas blue lines indicate forces on pillars not in contact with the cell. The dotted cyan line represents the average pillar force within the same cell. Time points indicated with bars in the trajectory correspond to the images in (E). Figures in (D–F) were adapted from Bashoura et al. (2014) with permission. Copyright 2014 National Academy of Sciences.
Recently, TFM substrates were improved by the generation of precise arrays of fluorescent quantum dots inside a silicone elastomer by electrohydrodynamic nanodrip-printing. The creation of controlled arrays of fluorescent particles removes the necessity of an image of the field after cell detachment (Bergert et al., 2016). Moreover, the use of super resolution fluorescence microscopy increased the resolution of the force map on the substrate (Colin-York et al., 2016; Stubb et al., 2020). However, calculation of the force maps from the microscopy images is not a trivial process and requires complex algorithms. For a more detailed review on this matter see Style et al. (2014), Schermelleh et al. (2019).
TFM-optimized elastic materials have been created to study mechanobiology in 3D. The deconvolution in 3D of forces exerted by the cell on a surface (Maskarinec et al., 2009; Legant et al., 2013) or inside 3D matrixes (Legant et al., 2010) was developed. Although the deconvolution of the force map in 3D environments is still challenging (Polacheck and Chen, 2016), these approaches hold great promise for measuring cell forces and mimic the ECM in geometries that more closely resemble those found in normal tissues. Vorselen et al. (2020) created a sophisticated version of 3D TFM by using uniform hydrogel particles with tunable size and stiffness. These particles were incubated in the cell medium and suffer deformation as a consequence of the forces applied to them. A computational method was developed by the authors to infer the mechanical forces from the deformation of the hydrogel particles. Thus, the pressure exerted by the cell to the particle could be measured with high resolution. This approach moves away from the classical TFM concept as hydrogel particles are not fixed around the cells but free to move and interact with them. For example, the authors could measure the spatial distribution of forces applied by a macrophage that phagocyted a hydrogel particle. Besides, particles could be tailored with pMHC ligands for the T cell receptor (TCR) of lymphocytes and I-CAM. These ligands induced the adhesion of cytotoxic T lymphocytes to the particle, simulating the interaction of T lymphocytes with their target cells, thus allowing to monitor forces induced by the T cell. Remarkably, the authors introduced hydrogel particles as micrometer tension probes breaking the general concept of TFM limited to 2D surfaces.
Other class of elastic materials have been developed to study cell mechanical properties and behavior. The use of micropillars to measure cell pulling forces was first introduced by Tan and coworkers (Tan et al., 2003). In this strategy, an array of cylindrical micrometer-scale cantilevers called micropillars is fabricated on polyacrylamide or polydimethylsiloxane (PDMS) substrates. The micropillars top is functionalized with ECM molecules, allowing cells to adhere on them. Therefore, when cells spread and maturate, they exert forces on the micropillars supporting them. The magnitude of these forces can be calculated from the micropillar displacement from their undeflected positions. This has the advantage that there is no need for cell detachment processes as in classical TFM (Roca-Cusachs et al., 2017; Banda et al., 2019). Besides, force calculation is simpler than with TFM, because small deformations follow a linear regime: micropillar deflection is directly proportional to the force (Du Roure et al., 2005). A third advantage is that micropillar stiffness can be modulated to some extent by modifying pillar geometry (Saez et al., 2007). This feature was exploited to create a wide range of stepped gradients by combining different pillar geometries (Lee et al., 2015). However, a major limitation of pillar arrays is that the substrate is not continuous. Cells can bind only to discrete spots (the top of the pillars) which can affect the morphology of cell-ECM adhesion.
Researchers have been exploring the use of micropillars with magnetic properties to exert controlled forces on cells (Sniadecki et al., 2007; Le Digabel et al., 2011) or the generation of nanopatterns on top of the micropillar surface to control ligand spacing and the selective binding of integrin types (Rahmouni et al., 2013). Recently, Bashoura et al. (2014) assessed the role of receptors involved in the interactions of T cell lymphocytes with APC. A micropillar pattern was coated with antibodies that bind to the TCR or the CD28 proteins to mimic the activation exerted by the ligands of these proteins (Figure 6D). Thus, the micropillar surface simulates the membrane activation by the APC. Pillars were also coated with fluorescent molecules to facilitate the force map generation. Using antibodies or ligands for the TCR or CD28, the authors could demonstrate that T lymphocytes exert pulling forces through the TCR (Figures 6E,F), whereas CD28 on its own does not mediate forces. Instead, CD28 is important for signal transduction.
Other approaches have been developed where forces can be measured without the need for soft deformable substrates. In 2012, Salaita’s group developed a sensitive approach to spatially and temporally map forces exerted by cells. Similar to protein tension probes expressed by cells (Grashoff et al., 2010), they implemented molecular probes as force transducer sensors (Stabley et al., 2012). The sensor consisted of a flexible polyethylene glycol (PEG) linker covalently bound to a ligand at one terminus and anchored onto a surface. The ligand and the surface were functionalized with a fluorophore and a quencher molecule, respectively. In a resting state, the linker is in a collapsed conformation, allowing the fluorophore to be quenched due to proximity with the quencher. When a force is applied to the probe, the linker stretches, separating the fluorophore from the quencher and increasing the fluorescence yield of the probe. Thus, knowing the quenching efficiency at a particular spot on the surface, it is possible to determine the distribution of collapsed/extended probes. As a proof of concept, the authors used an epidermal growth factor (EGF) ligand to map forces associated with initial uptake and trafficking of the EGF receptor (EGFR) upon binding to its cognate ligand. This work was motivated by the need for functional materials that can surpass the limitations of TFM in terms of sensitivity, spatial, and temporal resolution. Later on, the same group applied a similar concept to quantify the innate forces involved in the binding of TCR to the pMHC (Liu et al., 2013, 2016). The force sensor consisted of a DNA hairpin labeled with a fluorophore-quencher pair immobilized onto a gold nanoparticle (AuNP). Both the molecular quencher and AuNP supported on glass quench the fluorophore by FRET and plasmon effect, respectively. The dual quenching mechanism provided increased sensitivity and lower background signal.
The use of molecular tension probes was also implemented on nanopatterned surfaces to study the molecular biophysics of integrin ligand clustering (Liu Y. et al., 2014) (Figure 7A). Nanopatterned surfaces were prepared to create an array of AuNP, in which the distance between AuNP is precisely controlled (between 30 and 300 nm). Molecular tension probes were bound to AuNP on one extreme and had an integrin ligand [cyclic Arg-Gly-Asp-dPhe-Lys, c-(RGDfK)] on the other. A Cy3B fluorophore was located next to the integrin ligand (Figure 7B), so in the resting state, the fluorescence was quenched by the plasmon of the AuNP via nanometal surface energy transfer (NSET) (Figure 7C). The authors studied the adhesion of NIH/3T3 fibroblasts that expressed a recombinant GFP-paxillin (a protein expressed at the FA). A relationship between integrin ligand density and the magnitude of forces exerted by NIH/3T3 fibroblast was found. At low ligand density (100 nm of ligand spacing) cells showed less adhesion, smaller FA after 30 minutes, and lower pulling forces compared to cells cultured at higher ligand density (50 nm of ligand spacing) (Figure 7D). In the latter case, forces exerted by cells increased until a maximum value (at 1 h) after which forces remained constant. This could indicate that integrin ligand density has to be high enough to harness actin and myosin driven tension, which is necessary for FA maturation. Using cells expressing GFP-paxillin, the authors could measure the size of FA (Figure 7E). Results showed a similar behavior of the forces and FA size as a function of time. At higher ligand densities, cells exerted higher forces and assembled bigger FAs.
FIGURE 7

Molecular fluorescence tension microscopy substrates. In (A), the image shows a drawing of a cell adhered on the nanostructured biosensor surface. In (B), the chemical structure of the tension probe synthesized by the authors is shown. The probe is composed of three main components, a cyclic RGDfK which constitutes the integrin ligand (represented by a blue triangle), a Cy3B dye (represented by a red dot), and a PEG chain with a thiol group at the end (represented by a gray line). The probe binds to the AuNP through the thiol group and acquires a collapsed conformation in the absence of pulling forces on the probe. Each AuNP has on average 2.5 probes. The image in (C) shows the scheme of the molecular probe. In a rest position the probe acquires a collapsed conformation where the Cy3B fluorescence is quenched by the AuNP. Upon exertion of pulling forces by the cell, the linker stretches and the fluorescent probe is no longer quenched. In (D), the graph shows the mean tension per ligand across one entire cell on substrates coated with AuNP at different interparticle distances. After 1 h only cells on surfaces with high ligand density (50 nm spacing) could exert forces higher than 5 pN. The graph in (E) shows the GFP-paxillin cluster size (which indicates FA size) as a function of time. The increase in FA size was correlated with high tension as observed in (D). Figures were adapted from Liu Y. et al. (2014) with permission. Copyright 2014 American Chemical Society.
Summary and Future Perspectives
In the last years, advances in material synthesis have allowed the incorporation of sensing capabilities into cell substrate materials, which expanded the experimental information obtained from cell adhesion studies. This trend continued as novel synthetic strategies to tailor specific properties to materials were in constant development, leading to improvements in terms of sensitivity, spatial, and time resolution. In addition, modulation of topographical and chemical features allowed more control over cell functions. Surface nanopatterning was crucial to reproduce the physicochemical characteristics of the cellular microenvironment, creating accurate synthetic ECM analogs. Novel synthetic strategies together with sophisticated analysis techniques contributed to address important aspects of the mechanism by which cells interact with their microenvironment. For instance, accurate localization of fluorescent nanotracer inside soft elastic materials was crucial to spatially and dynamically map cell forces by stimulated emission depletion (STED) microscopy (Bergert et al., 2016; Stubb et al., 2020). Adding multiple features to the functional material provides more realistic physiological conditions and higher information output as demonstrated using precisely distributed nanometer-scale arrays of ECM ligands on TFM substrates of variable rigidity (Oria et al., 2017). 3D soft architectures with programmed physicochemical properties incorporating a transducer element were also demonstrated (Pitsalidis et al., 2018; Vorselen et al., 2020), which constitute a necessary step towards a new paradigm for in vitro studies of cell processes.
3D synthetic microenvironments provide a platform for cell culture and cell analysis ex vivo where cells behave more natively (Weigelt et al., 2014). Advances in 3D culture platforms that merge biosensing capabilities with sophisticated biochemical and biophysical properties as those found in the native ECM allow the real-time study of mammalian tissues (Shamir and Ewald, 2014). This type of stimuli-responsive functional materials would be ideal toward understanding how cells change their phenotype and acquire specific functions as a consequence of the cues from the environment. This is an important aspect of tissue physiology, and a critical step to understanding alterations that lead to tissue pathophysiology (Bhatia et al., 2014; Pickup et al., 2014; Gaetani et al., 2020). In this context, biomimetic 3D biosensors for cell culture will increasingly contribute to physiology, histology, and physiopathology studies.
3D matrices that combine essential biophysical and biochemical aspects of the native cellular microenvironment with biosensing features will also bring benefits to the field of tissue regeneration and healing. A major goal in this field is to improve the biointegration of orthopedic and dental implants with the surrounding tissues. During decades, numerous surface modification strategies based on chemical coatings and nanotopographical features were developed (Mas-Moruno et al., 2019). However, despite the research efforts, implants still nowadays fail at an unacceptable rate (Raphel et al., 2016). This, in part, is due to the lack of critical information about the mechanisms governing the material biointegration process. In this perspective, ECM mimetics able to monitor cell adhesion dynamics quantitatively in real time hold great promise toward a rational design of ‘smart’ implants that possess cell-instructive characteristics.
A further challenge should be devoted to the development of novel approaches that could provide information in real time of the cell behavior in vivo. Mammalian tissues and organs are particularly difficult to study by direct optical observation (Shamir and Ewald, 2014). This is the case for a number of diverse 3D culture formats including organoids (Simian and Bissell, 2017). Electrochemistry could provide the means to accomplish this goal. Conducting polymers, for instance, are excellent building blocks for the creation of electrically responsive hydrogels (Zhang et al., 2019; Yang et al., 2020), although improvements in conductivity are desired to achieve highly sensitive cell sensing. Alternatively, the integration of hydrogels with conductive nanomaterials could overcome those limitations (Li et al., 2018). A fundamental requirement is to maintain not only the morphology of the hydrogel, but also the electrostatic and biocompatible properties, which will be essential for observing cells on a controllable nature-loyal microenvironment.
The electronic industry had advanced in the creation of complex circuits in very small dimensions. Adapting these advances to the field of cell biology emerges as a promising way to improve biosensor sensing capabilities. For example, the incorporation of CMOS-based MEAs in biosensors increased the density of electrodes on a surface (Obien et al., 2015), and with it the spatial resolution. Moreover, electrochemical biosensors can enhance biocompatibility by the creation of flexible electrodes (Song et al., 2020).
Finally, substrates with built-in sensing capabilities are suited for multiparametric cell monitoring. For example, transparent electrodes allow simultaneous microscopy observation and ECIS recordings (Pallarola et al., 2017a; Parviz et al., 2017), whereas nanostructured conductive substrates allow the implementation of ECIS and SERS (Zong et al., 2015). Often the result is more than the sum of its parts; SPR conductive substrates allowed the creation of electric impedance microscopy which added spatial resolution to ECIS measurements (Wang et al., 2011). Multiparametric approaches increase the information output from cell culture experiments. A further challenge will be the creation of computational models and simulations that could help to interpret and understand the multi-scale information obtained by multiparametric biosensors.
Statements
Author contributions
DP and NS designed the content of the manuscript. All the authors performed the literature survey, wrote the manuscript, and edited and reviewed the manuscript before submission. NS and DP prepared the figures.
Funding
This work was financially supported by the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement no. 872869. DP and NS acknowledge financial support from Max-Planck-Gesellschaft (Max Planck Partner Group Nanoelectronics for Cellular Interfaces INS/MPI-MR).
Acknowledgments
NS acknowledges CONICET for a postdoctoral fellowship. DP is staff researcher of CONICET.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AbdelsalamM. E.BartlettP. N.BaumbergJ. J.CintraS.KelfT. A.RussellA. E. (2005). Electrochemical SERS at a structured gold surface.Electrochem. Commun.7740–744. 10.1016/j.elecom.2005.04.028
2
AbiriH.AbdolahadM.GharooniM.Ali HosseiniS.JanmalekiM.AzimiS.et al (2015). Monitoring the spreading stage of lung cells by silicon nanowire electrical cell impedance sensor for cancer detection purposes.Biosens. Bioelectron.68577–585. 10.1016/j.bios.2015.01.057
3
Alom RuizS.ChenC. S. (2007). Microcontact printing: a tool to pattern.Soft Matter.3168–177. 10.1039/b613349e
4
AmatoreC.ArbaultS.GuilleM.LemaîtreF. (2008). Electrochemical monitoring of single cell secretion: vesicular exocytosis and oxidative stress.Chem. Rev.1082585–2621. 10.1021/cr068062g
5
AndersonM. A.BurdaJ. E.RenY.AoY.O’SheaT. M.KawaguchiR.et al (2016). Astrocyte scar formation AIDS central nervous system axon regeneration.Nature532195–200. 10.1038/nature17623
6
AriasL. R.PerryC. A.YangL. (2010). Real-time electrical impedance detection of cellular activities of oral cancer cells.Biosens. Bioelectron.252225–2231. 10.1016/j.bios.2010.02.029
7
ArndtS.SeebachJ.PsathakiK.GallaH. J.WegenerJ. (2004). Bioelectrical impedance assay to monitor changes in cell shape during apoptosis.Biosens. Bioelectron.19583–594. 10.1016/S0956-5663(03)00269-0
8
BandaO. A.SabanayagamC. R.SlaterJ. H. (2019). Reference-free traction force microscopy platform fabricated via two-photon laser scanning lithography enables facile measurement of cell-generated forces.ACS Appl. Mater. Interfaces1118233–18241. 10.1021/acsami.9b04362
9
BashouraK. T.GondarenkoA.ChenH.ShenK.LiuX.HuseM.et al (2014). CD28 and CD3 have complementary roles in T-cell traction forces.Proc. Natl. Acad. Sci. U.S.A.1112241–2246. 10.1073/pnas.1315606111
10
BazellièresE.ConteV.Elosegui-ArtolaA.Serra-PicamalX.Bintanel-MorcilloM.Roca-CusachsP.et al (2015). Control of cell-cell forces and collective cell dynamics by the intercellular adhesome.Nat. Cell Biol.17409–420. 10.1038/ncb3135
11
BélisleJ. M.KunikD.CostantinoS. (2009). Rapid multicomponent optical protein patterning.Lab. Chip93580–3585. 10.1039/b911967a
12
BerdondiniL.ImfeldK.MacCioneA.TedescoM.NeukomS.Koudelka-HepM.et al (2009). Active pixel sensor array for high spatio-temporal resolution electrophysiological recordings from single cell to large scale neuronal networks.Lab. Chip92644–2651. 10.1039/b907394a
13
BergertM.LendenmannT.ZündelM.EhretA. E.PanozzoD.RichnerP.et al (2016). Confocal reference free traction force microscopy.Nat. Commun.7:12814. 10.1038/ncomms12814
14
BhatiaS. N.UnderhillG. H.ZaretK. S.FoxI. J. (2014). Cell and tissue engineering for liver disease.Sci. Transl. Med.6:245sr2. 10.1126/scitranslmed.3005975
15
BorileG.RossiS.FilippiA.GazzolaE.CapaldoP.TregnagoC.et al (2019). Label-free, real-time on-chip sensing of living cells via grating-coupled surface plasmon resonance.Biophys. Chem.254:106262. 10.1016/j.bpc.2019.106262
16
BuckleyC. D.TanJ.AndersonK. L.HaneinD.VolkmannN.WeisW. I.et al (2014). The minimal cadherin-catenin complex binds to actin filaments under force.Science346:1254211. 10.1126/science.1254211
17
BurtonK.TaylorD. L. (1997). Traction forces of cytokinesis measured with optically modified elastic substrata.Nature385450–454. 10.1038/385450a0
18
CareyB. W.FinleyL. W. S.CrossJ. R.AllisC. D.ThompsonC. B. (2015). Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells.Nature518413–416. 10.1038/nature13981
19
CarterS. B. (1967). Haptotactic islands.Exp. Cell Res.48189–193. 10.1016/0014-4827(67)90298-4
20
ChabotV.CuerrierC. M.EscherE.AimezV.GrandboisM.CharetteP. G. (2009). Biosensing based on surface plasmon resonance and living cells.Biosens. Bioelectron.241667–1673. 10.1016/j.bios.2008.08.025
21
Chacón-MartínezC. A.KoesterJ.WickströmS. A. (2018). Signaling in the stem cell niche: regulating cell fate, function and plasticity.Development145:dev165399. 10.1242/dev.165399
22
ChenJ. Y.PennL. S.XiJ. (2018). Quartz crystal microbalance: sensing cell-substrate adhesion and beyond.Biosens. Bioelectron.99593–602. 10.1016/j.bios.2017.08.032
23
ChoiY.YagatiA. K.ChoS. (2015). Electrochemical characterization of poly-L-lysine coating on indium tin oxide electrode for enhancing cell adhesion.J. Nanosci. Nanotechnol.157881–7885. 10.1166/jnn.2015.11229
24
Colin-YorkH.ShresthaD.FelceJ. H.WaitheD.MoeendarbaryE.DavisS. J.et al (2016). Super-resolved traction force microscopy (STFM).Nano Lett.162633–2638. 10.1021/acs.nanolett.6b00273
25
CollinsC.DenisinA. K.PruittB. L.NelsonW. J. (2017). Changes in E-cadherin rigidity sensing regulate cell adhesion.Proc. Natl. Acad. Sci. U.S.A.114E5835–E5844. 10.1073/pnas.1618676114
26
CsicsvariJ.HenzeD. A.JamiesonB.HarrisK. D.SirotaA.BarthóP.et al (2003). Massively parallel recording of unit and local field potentials with silicon-based electrodes.J. Neurophysiol.901314–1323. 10.1152/jn.00116.2003
27
CurtisA.RiehleM. (2001). Tissue engineering: the biophysical background.Phys. Med. Biol.4646–65. 10.1088/0031-9155/46/4/201
28
D’Angelo SilicianoJ.GoodenoughD. A. (1988). Localization of the tight junction protein, ZO-1, is modulated by extracellular calcium and cell-cell contact in Madin-Darby canine kidney epithelial cells.J. Cell Biol.1072389–2399. 10.1083/jcb.107.6.2389
29
De GregorioV.CorradoB.SbresciaS.SibilioS.UrciuoloF.NettiP. A.et al (2020). Intestine-on-chip device increases ECM remodeling inducing faster epithelial cell differentiation.Biotechnol. Bioeng.117556–566. 10.1002/bit.27186
30
DeckerD.HempelmannR.NatterH.PirrungM.RabeH.SchäferK. H.et al (2019). 3D nanostructured multielectrode arrays: fabrication, electrochemical characterization, and evaluation of cell–electrode adhesion.Adv. Mater. Technol.4:1800436. 10.1002/admt.201800436
31
DemboM.WangY. L. (1999). Stresses at the cell-to-substrate interface during locomotion of fibroblasts.Biophys. J.762307–2316. 10.1016/S0006-3495(99)77386-8
32
DesbiollesB. X. E.De CoulonE.BertschA.RohrS.RenaudP. (2019). Intracellular recording of cardiomyocyte action potentials with nanopatterned volcano-shaped microelectrode arrays.Nano Lett.196173–6181. 10.1021/acs.nanolett.9b02209
33
DinarvandM.NeubertE.MeyerD.SelvaggioG.MannF. A.ErpenbeckL.et al (2019). Near-infrared imaging of serotonin release from cells with fluorescent nanosensors.Nano Lett.196604–6611. 10.1021/acs.nanolett.9b02865
34
DingL.DuD.WuJ.JuH. (2007). A disposable impedance sensor for electrochemical study and monitoring of adhesion and proliferation of K562 leukaemia cells.Electrochem. Commun.9953–958. 10.1016/j.elecom.2006.11.040
35
DingL.DuD.ZhangX.JuH. (2008). Trends in cell-based electrochemical biosensors.Curr. Med. Chem.153160–3170. 10.2174/092986708786848514
36
DipaloM.MelleG.LovatoL.JacassiA.SantoroF.CaprettiniV.et al (2018). Plasmonic meta-electrodes allow intracellular recordings at network level on high-density CMOS-multi-electrode arrays.Nat. Nanotechnol.13965–971. 10.1038/s41565-018-0222-z
37
DischerD. E.JanmeyP.WangY. L. (2005). Tissue cells feel and respond to the stiffness of their substrate.Science3101139–1143. 10.1126/science.1116995
38
DuX.ZhangZ.ZhengX.ZhangH.DongD.ZhangZ.et al (2020). An electrochemical biosensor for the detection of epithelial-mesenchymal transition.Nat. Commun.11:192. 10.1038/s41467-019-14037-w
39
Du RoureO.SaezA.BuguinA.AustinR. H.ChavrierP.SiberzanP.et al (2005). Force mapping in epithelial cell migration.Proc. Natl. Acad. Sci. U.S.A.1022390–2395. 10.1073/pnas.0408482102
40
DuanX.GaoR.XieP.Cohen-KarniT.QingQ.ChoeH. S.et al (2012). Intracellular recordings of action potentials by an extracellular nanoscale field-effect transistor.Nat. Nanotechnol.7174–179. 10.1038/nnano.2011.223
41
Elosegui-ArtolaA.OriaR.ChenY.KosmalskaA.Pérez-GonzálezC.CastroN.et al (2016). Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity.Nat. Cell Biol.18540–548. 10.1038/ncb3336
42
El-SaidW. A.KimS. U.ChoiJ. W. (2015). Monitoring in vitro neural stem cell differentiation based on surface-enhanced Raman spectroscopy using a gold nanostar array.J. Mater. Chem. C33848–3859. 10.1039/c5tc00304k
43
EndoM.IwawakiT.YoshimuraH.OzawaT. (2019). Photocleavable cadherin inhibits cell-to-cell mechanotransduction by light.ACS Chem. Biol.142206–2214. 10.1021/acschembio.9b00460
44
EnglerA. J.SenS.SweeneyH. L.DischerD. E. (2006). Matrix elasticity directs stem cell lineage specification.Cell126677–689. 10.1016/j.cell.2006.06.044
45
FanM.AndradeG. F. S.BroloA. G. (2011). A review on the fabrication of substrates for surface enhanced Raman spectroscopy and their applications in analytical chemistry.Anal. Chim. Acta6937–25. 10.1016/j.aca.2011.03.002
46
FerruzzaS.ScarinoM. L.RotilioG.CirioloM. R.SantaroniP.MudaA. O.et al (1999). Copper treatment alters the permeability of tight junctions in cultured human intestinal Caco-2 cells.Am. J. Physiol. Gastrointest. Liver Physiol.277G1138–G1148. 10.1152/ajpgi.1999.277.6.g1138
47
FrantzC.StewartK. M.WeaverV. M. (2010). The extracellular matrix at a glance.J. Cell Sci.123(Pt 24), 4195–4200. 10.1242/jcs.023820
48
FredrikssonC.KihlmanS.RodahlM.KasemoB. (1998). The piezoelectric quartz crystal mass and dissipation sensor: a means of studying cell adhesion.Langmuir14248–251. 10.1021/la971005l
49
FreemanR. G.GrabarK. C.AllisonK. J.BrightR. M.DavisJ. A.GuthrieA. P.et al (1995). Self-assembled metal colloid monolayers: an approach to SERS substrates.Science2671629–1632. 10.1126/science.267.5204.1629
50
GaetaniR.ZizziE. A.DeriuM. A.MorbiducciU.PesceM.MessinaE. (2020). When stiffness matters: mechanosensing in heart development and disease.Front. Cell Dev. Biol.8:334. 10.3389/fcell.2020.00334
51
GaharwarA. K.SinghI.KhademhosseiniA. (2020). Engineered biomaterials for in situ tissue regeneration.Nat. Rev. Mater.5686–705. 10.1038/s41578-020-0209-x
52
GamalW.TreskesP.SamuelK.SullivanG. J.SillerR.SrsenV.et al (2017). Low-dose acetaminophen induces early disruption of cell-cell tight junctions in human hepatic cells and mouse liver.Sci. Rep.7:37541. 10.1038/srep37541
53
GeigerB.SpatzJ. P.BershadskyA. D. (2009). Environmental sensing through focal adhesions.Nat. Rev. Mol. Cell Biol.1021–33. 10.1038/nrm2593
54
GeigerB.YamadaK. M. (2011). Molecular architecture and function of matrix adhesions.Cold Spring Harb. Perspect. Biol.3:a005033. 10.1101/cshperspect.a005033
55
GiaeverI.KeeseC. R. (1984). Monitoring fibroblast behavior in tissue culture with an applied electric field.Proc. Natl. Acad. Sci. U.S.A.813761–3764. 10.1073/pnas.81.12.3761
56
GiaeverI.KeeseC. R. (1986). Use of electric fields to monitor the dynamical aspect of cell behavior in tissue culture.IEEE Trans. Biomed. Eng.33242–247. 10.1109/TBME.1986.325896
57
GiaeverI.KeeseC. R. (1991). Micromotion of mammalian cells measured electrically.Proc. Natl. Acad. Sci. U.S.A.887896–7900. 10.1073/pnas.88.17.7896
58
GiaeverI.KeeseC. R. (1993). A morphological biosensor for mammalian cells.Nature366591–592. 10.1038/366591a0
59
GrashoffC.HoffmanB. D.BrennerM. D.ZhouR.ParsonsM.YangM. T.et al (2010). Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics.Nature466263–266. 10.1038/nature09198
60
GrossG. W.RieskeE.KreutzbergG. W.MeyerA. (1977). A new fixed-array multi-microelectrode system designed for long-term monitoring of extracellular single unit neuronal activity in vitro.Neurosci. Lett.6101–105. 10.1016/0304-3940(77)90003-9
61
GuaschJ.ConingsB.NeubauerS.RechenmacherF.EndeK.RolliC. G.et al (2015). Segregation versus colocalization: orthogonally functionalized binary micropatterned substrates regulate the molecular distribution in focal adhesions.Adv. Mater.273737–3747. 10.1002/adma.201500900
62
GuaschJ.DiemerJ.RiahinezhadH.NeubauerS.KesslerH.SpatzJ. P. (2016). Synthesis of binary nanopatterns on hydrogels for initiating cellular responses.Chem. Mater.281806–1815. 10.1021/acs.chemmater.5b04910
63
GuilluyC.DolegaM. E. (2020). Syndecan-4 forces integrins to cooperate.Nat. Mater.19587–588. 10.1038/s41563-020-0689-5
64
GumbinerB. M. (1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis.Cell84345–357. 10.1016/S0092-8674(00)81279-9
65
GunhanlarN.ShpakG.van der KroegM.Gouty-ColomerL. A.MunshiS. T.LendemeijerB.et al (2017). A simplified protocol for differentiation of electrophysiologically mature neuronal networks from human induced pluripotent stem cells.Mol. Psychiatry231336–1344. 10.1038/mp.2017.56
66
HaiA.ShappirJ.SpiraM. E. (2010). Long-term, multisite, parallel, in-cell recording and stimulation by an array of extracellular microelectrodes.J. Neurophysiol.104559–568. 10.1152/jn.00265.2010
67
HaldavnekarR.VenkatakrishnanK.TanB. (2018). Non plasmonic semiconductor quantum SERS probe as a pathway for in vitro cancer detection.Nat. Commun.9:3065. 10.1038/s41467-018-05237-x
68
HanE.DingL.JinS.JuH. (2011). Electrochemiluminescent biosensing of carbohydrate-functionalized CdS nanocomposites for in situ label-free analysis of cell surface carbohydrate.Biosens. Bioelectron.262500–2505. 10.1016/j.bios.2010.10.044
69
HarrisA. (1973). Behavior of cultured cells on substrata of variable adhesiveness.Exp. Cell Res.77285–297. 10.1016/0014-4827(73)90579-X
70
HarrisA. K.WildP.StopakD. (1980). Silicone rubber substrata: a new wrinkle in the study of cell locomotion.Science208177–179. 10.1126/science.6987736
71
HaynesC. L.McFarlandA. D.Van DuyneR. P. (2005). Surface-enhanced: Raman spectroscopy.Anal. Chem.77338A–346A. 10.1021/ac053456d
72
HenryO. Y. F.VillenaveR.CronceM. J.LeineweberW. D.BenzM. A.IngberD. E. (2017). Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function.Lab. Chip172264–2271. 10.1039/c7lc00155j
73
HomolaJ. (2003). Present and future of surface plasmon resonance biosensors.Anal. Bioanal. Chem.377528–539. 10.1007/s00216-003-2101-0
74
HongJ.KandasamyK.MarimuthuM.ChoiC. S.KimS. (2011). Electrical cell-substrate impedance sensing as a non-invasive tool for cancer cell study.Analyst136237–245. 10.1039/c0an00560f
75
HuangY.CaiD.ChenP. (2011). Micro- and nanotechnologies for study of cell secretion.Anal. Chem.834393–4406. 10.1021/ac200358b
76
HuhD.MatthewsB. D.MammotoA.Montoya-ZavalaM.Yuan HsinH.IngberD. E. (2010). Reconstituting organ-level lung functions on a chip.Science3281662–1668. 10.1126/science.1188302
77
HuncklerM. D.GarcíaA. J. (2020). Engineered biomaterials for enhanced function of insulin-secreting β-Cell organoids.Adv. Funct. Mater.3:2000134. 10.1002/adfm.202000134
78
HutzlerM.LambacherA.EversmannB.JenknerM.ThewesR.FromherzP. (2006). High-resolution multitransistor array recording of electrical field potentials in cultured brain slices.J. Neurophysiol.961638–1645. 10.1152/jn.00347.2006
79
IshayR.Ben, Kapp-BarneaY.GrigoriantzI.TeblumE.LelloucheJ. P. (2015). Real time acoustic profiling of a live cancerous cell monolayer using QCM.Sens. Actuat. B Chem.215373–381. 10.1016/j.snb.2015.03.091
80
JanshoffA.KunzeA.MichaelisS.HeitmannV.ReissB.WegenerJ. (2010). Cell adhesion monitoring using substrate-integrated sensors.J. Adhes. Sci. Technol.242079–2104. 10.1163/016942410X507939
81
KailasL.RatcliffeE. C.HayhurstE. J.WalkerM. G.FosterS. J.HobbsJ. K. (2009). Immobilizing live bacteria for AFM imaging of cellular processes.Ultramicroscopy109775–780. 10.1016/j.ultramic.2009.01.012
82
KangG.MatikainenA.StenbergP.FärmE.LiP.RitalaM.et al (2015). High aspect-ratio iridium-coated nanopillars for highly reproducible surface-enhanced raman scattering (SERS).ACS Appl. Mater. Interf.711452–11459. 10.1021/acsami.5b02206
83
KarbassiE.FenixA.MarchianoS.MuraokaN.NakamuraK.YangX.et al (2020). Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine.Nat. Rev. Cardiol.17341–359. 10.1038/s41569-019-0331-x
84
KeungA. J.KumarS.SchafferD. V. (2010). Presentation counts: microenvironmental regulation of stem cells by biophysical and material cues.Annu. Rev. Cell Dev. Biol.26533–556. 10.1146/annurev-cellbio-100109-104042
85
KimE. H.ChinG.RongG.PoskanzerK. E.ClarkH. A. (2018). Optical probes for neurobiological sensing and imaging.Acc. Chem. Res.511023–1032. 10.1021/acs.accounts.7b00564
86
KimM.ChoiJ. C.JungH. R.KatzJ. S.KimM. G.DohJ. (2010). Addressable micropatterning of multiple proteins and cells by microscope projection photolithography based on a protein friendly photoresist.Langmuir2612112–12118. 10.1021/la1014253
87
KimT. H.El-SaidW. A.AnJ. H.ChoiJ. W. (2013). ITO/gold nanoparticle/RGD peptide composites to enhance electrochemical signals and proliferation of human neural stem cells.Nanomed. Nanotechnol. Biol. Med.9336–344. 10.1016/j.nano.2012.08.006
88
KimT.-H.YeaC.-H.ChuengS.-T. D.YinP. T.-T.ConleyB.DardirK.et al (2015). Nanoelectrodes: large-scale nanoelectrode arrays to monitor the dopaminergic differentiation of human neural stem cells (Adv. Mater. 41/2015).Adv. Mater.276306–6306. 10.1002/adma.201570275
89
KneippJ.KneippH.WittigB.KneippK. (2010). Novel optical nanosensors for probing and imaging live cells.Nanomed. Nanotechnol. Biol. Med.6214–226. 10.1016/j.nano.2009.07.009
90
Kraning-RushC. M.CareyS. P.LampiM. C.Reinhart-KingC. A. (2013). Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D.Integr. Biol.5606–616. 10.1039/c3ib20196a
91
KrussS.SalemD. P.VukovićL.LimaB.EndeE.Vanderet al (2017). High-resolution imaging of cellular dopamine efflux using a fluorescent nanosensor array.Proc. Natl. Acad. Sci. U.S.A.1141789–1794. 10.1073/pnas.1613541114
92
LadouxB.MègeR. M. (2017). Mechanobiology of collective cell behaviours.Nat. Rev. Mol. Cell Biol.18743–757. 10.1038/nrm.2017.98
93
LaiY.ChengK.KisaalitaW. (2012). Three dimensional neuronal cell cultures more accurately model voltage gated calcium channel functionality in freshly dissected nerve tissue.PLoS One7:e45074. 10.1371/journal.pone.0045074
94
Le DigabelJ.BiaisN.FresnaisJ.BerretJ. F.HersenP.LadouxB. (2011). Magnetic micropillars as a tool to govern substrate deformations.Lab. Chip112630–2636. 10.1039/c1lc20263d
95
Le DucQ.ShiQ.BlonkI.SonnenbergA.WangN.LeckbandD.et al (2010). Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner.J. Cell Biol.1891107–1115. 10.1083/jcb.201001149
96
LeeS.SahaB.LeeJ. (2015). “Cell motility regulation on stepped micro pillar array device (SMPAD) with discrete stiffness gradient,” in Proceedings of the 28th IEEE International Conference on Micro Electro Mechanical Systems (MEMS), Estoril, 628–631.
97
LegantW. R.ChoiC. K.MillerJ. S.ShaoL.GaoL.BetzigE.et al (2013). Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions.Proc. Natl. Acad. Sci. U.S.A.110881–886. 10.1073/pnas.1207997110
98
LegantW. R.MillerJ. S.BlakelyB. L.CohenD. M.GeninG. M.ChenC. S. (2010). Measurement of mechanical tractions exerted by cells in three-dimensional matrices.Nat. Methods7969–971. 10.1038/nmeth.1531
99
LiP.JinZ.PengL.ZhaoF.XiaoD.JinY.et al (2018). Stretchable all-gel-state fiber-shaped supercapacitors enabled by macromolecularly interconnected 3D graphene/nanostructured conductive polymer hydrogels.Adv. Mater.30:1800124. 10.1002/adma.201800124
100
LiT.WuK.RindzeviciusT.WangZ.SchulteL.SchmidtM. S.et al (2016). Wafer-scale nanopillars derived from block copolymer lithography for surface-enhanced Raman Spectroscopy.ACS Appl. Mater. Interf.815668–15675. 10.1021/acsami.6b05431
101
LippmannE. S.AzarinS. M.KayJ. E.NesslerR. A.WilsonH. K.Al-AhmadA.et al (2012). Human blood-brain barrier endothelial cells derived from pluripotent stem cells.Nat. Biotechnol.30:2014. 10.1038/nbt.2247.Human
102
LiuB.ChenW.EvavoldB. D.ZhuC. (2014). Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling.Cell157357–368. 10.1016/j.cell.2014.02.053
103
LiuY.MeddaR.LiuZ.GaliorK.YehlK.SpatzJ. P.et al (2014). Nanoparticle tension probes patterned at the nanoscale: impact of integrin clustering on force transmission.Nano Lett.145539–5546. 10.1021/nl501912g
104
LiuJ.PanL.ShangC.LuB.WuR.FengY.et al (2020). A highly sensitive and selective nanosensor for near-infrared potassium imaging.Sci. Adv.6:eaax9757. 10.1126/sciadv.aax9757
105
LiuQ.YuJ.XiaoL.TangJ. C. O.ZhangY.WangP.et al (2009). Impedance studies of bio-behavior and chemosensitivity of cancer cells by micro-electrode arrays.Biosens. Bioelectron.241305–1310. 10.1016/j.bios.2008.07.044
106
LiuY.BlanchfieldL.Pui-Yan, MaV.AndargachewR.GaliorK.et al (2016). DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity.Proc. Natl. Acad. Sci. U.S.A.1135610–5615. 10.1073/pnas.1600163113
107
LiuY.YehlK.NaruiY.SalaitaK. (2013). Tension sensing nanoparticles for mechano-imaging at the living/nonliving interface.J. Am. Chem. Soc.1355320–5323. 10.1021/ja401494e
108
LoC. M.KeeseC. R.GiaeverI. (1995). Impedance analysis of MDCK cells measured by electric cell-substrate impedance sensing.Biophys. J.692800–2807. 10.1016/S0006-3495(95)80153-0
109
LópezG. P.BiebuyckH. A.FrisbieC. D.WhitesidesG. M. (1993a). Imaging of features on surfaces by condensation figures.Science260647–649. 10.1126/science.8480175
110
LópezG. P.BiebuyckH. A.HärterR.KumarA.WhitesidesG. M. (1993b). Fabrication and imaging of two-dimensional patterns of proteins adsorbed on self-assembled monolayers by scanning electron microscopy.J. Am. Chem. Soc.11510774–10781. 10.1021/ja00076a038
111
LordM. S.ModinC.FossM.DuchM.SimmonsA.PedersenF. S.et al (2006). Monitoring cell adhesion on tantalum and oxidised polystyrene using a quartz crystal microbalance with dissipation.Biomaterials274529–4537. 10.1016/j.biomaterials.2006.04.006
112
LundC.PulliK.YellapragadaV.GiacobiniP.LundinK.VuoristoS.et al (2016). Development of gonadotropin-releasing hormone-secreting neurons from human pluripotent stem cells.Stem Cell Rep.7149–157. 10.1016/j.stemcr.2016.06.007
113
LuoB.-H.CarmanC. V.SpringerT. A. (2007). Structural basis of integrin regulation and signaling.Annu. Rev. Immunol.25619–647. 10.1146/annurev.immunol.25.022106.141618
114
LuoX.XingY.GalvanD. D.ZhengE.WuP.CaiC.et al (2019). Plasmonic gold nanohole array for surface-enhanced raman scattering detection of DNA methylation.ACS Sens.41534–1542. 10.1021/acssensors.9b00008
115
LussierF.BruléT.VishwakarmaM.DasT.SpatzJ. P.MassonJ. F. (2016). Dynamic-SERS optophysiology: a nanosensor for monitoring cell secretion events.Nano Lett.163866–3871. 10.1021/acs.nanolett.6b01371
116
MagerM. D.LapointeV.StevensM. M. (2011). Exploring and exploiting chemistry at the cell surface.Nat. Chem.3582–589. 10.1038/nchem.1090
117
MartinaM.LukC.PyC.MartinezD.ComasT.MonetteR.et al (2011). Recordings of cultured neurons and synaptic activity using patch-clamp chips.J. Neural Eng.8:034002. 10.1088/1741-2560/8/3/034002
118
MaskarinecS. A.FranckC.TirrellD. A.RavichandranG. (2009). Quantifying cellular traction forces in three dimensions.Proc. Natl. Acad. Sci. U.S.A.10622108–22113. 10.1073/pnas.0904565106
119
Mas-MorunoC.SuB.DalbyM. J. (2019). Multifunctional coatings and nanotopographies: toward cell instructive and antibacterial implants.Adv. Healthc. Mater.8:e1801103. 10.1002/adhm.201801103
120
McKinnonD. D.KloxinA. M.AnsethK. S. (2013). Synthetic hydrogel platform for three-dimensional culture of embryonic stem cell-derived motor neurons.Biomater. Sci.1460–469. 10.1039/c3bm00166k
121
MègeR. M.IshiyamaN. (2017). Integration of cadherin adhesion and cytoskeleton at adherens junctions.Cold Spring Harb. Perspect. Biol.9:a028738. 10.1101/cshperspect.a028738
122
MehlhornA. T.NiemeyerP.KaiserS.FinkenzellerG.StarkG. B.SüdkampN. P.et al (2006). Differential expression pattern of extracellular matrix molecules during chondrogenesis of mesenchymal stem cells from bone marrow and adipose tissue.Tissue Eng.122853–2862. 10.1089/ten.2006.12.2853
123
MéjardR.GriesserH. J.ThierryB. (2014). Optical biosensing for label-free cellular studies.Trends Anal. Chem.53178–186. 10.1016/j.trac.2013.08.012
124
MitrousisN.FokinaA.ShoichetM. S. (2018). Biomaterials for cell transplantation.Nat. Rev. Mater.3441–456. 10.1038/s41578-018-0057-0
125
MohammedD.VersaevelM.BruyèreC.AlaimoL.LucianoM.VercruysseE.et al (2019). Innovative tools for mechanobiology: unraveling outside-in and inside-out mechanotransduction.Front. Bioeng. Biotechnol.7:162. 10.3389/fbioe.2019.00162
126
NgokS. P.GeyerR.KourtidisA.MitinN.FeathersR.DerC.et al (2013). TEM4 is a junctional Rho GEF required for cell-cell adhesion, monolayer integrity and barrier function.J. Cell Sci.1263271–3277. 10.1242/jcs.123869
127
ObienM. E. J.DeligkarisK.BullmannT.BakkumD. J.FreyU. (2015). Revealing neuronal function through microelectrode array recordings.Front. Neurosci.9:423. 10.3389/fnins.2014.00423
128
OllechD.PflästererT.ShellardA.ZambardaC.SpatzJ. P.MarcqP.et al (2020). An optochemical tool for light-induced dissociation of adherens junctions to control mechanical coupling between cells.Nat. Commun.11:1681. 10.1038/s41467-020-14390-1
129
OrangeJ. S. (2008). Formation and function of the lytic NK-cell immunological synapse.Nat. Rev. Immunol.8713–725. 10.1038/nri2381
130
OriaR.WiegandT.EscribanoJ.Elosegui-ArtolaA.UriarteJ. J.Moreno-PulidoC.et al (2017). Force loading explains spatial sensing of ligands by cells.Nature552219–224. 10.1038/nature24662
131
PallarolaD.BochenA.BoehmH.RechenmacherF.SobahiT. R.SpatzJ. P.et al (2014). Interface immobilization chemistry of cRGD-based peptides regulates integrin mediated cell adhesion.Adv. Funct. Mater.24943–956. 10.1002/adfm.201302411
132
PallarolaD.BochenA.GuglielmottiV.OswaldT. A.KesslerH.SpatzJ. P. (2017a). Highly ordered gold nanopatterned indium tin oxide electrodes for simultaneous optical and electrochemical probing cell interactions.Anal. Chem.8910054–10062. 10.1021/acs.analchem.7b02743
133
PallarolaD.PlatzmanI.BochenA.Cavalcanti-AdamE. A.AxmannM.KesslerH.et al (2017b). Focal adhesion stabilization by enhanced integrin-cRGD binding affinity.BioNanoMaterials181–2. 10.1515/bnm-2016-0014
134
ParvizM.GausK.GoodingJ. J. (2017). Simultaneous impedance spectroscopy and fluorescence microscopy for the real-time monitoring of the response of cells to drugs.Chem. Sci.81831–1840. 10.1039/c6sc05159f
135
PetersonA. W.HalterM.TonaA.BhadrirajuK.PlantA. L. (2009). Surface plasmon resonance imaging of cells and surface-associated fibronectin.BMC Cell Biol.10:16. 10.1186/1471-2121-10-16
136
PickupM. W.MouwJ. K.WeaverV. M. (2014). The extracellular matrix modulates the hallmarks of cancer.Embo Rep.151243–1253. 10.15252/embr.201439246
137
PineJ. (1980). Recording action potentials from cultured neurons with extracellular microcircuit electrodes.J. Neurosci. Methods219–31. 10.1016/0165-0270(80)90042-4
138
PitsalidisC.FerroM. P.IandoloD.TzounisL.InalS.OwensR. M. (2018). Transistor in a tube: a route to three-dimensional bioelectronics.Sci. Adv.4eaat4253. 10.1126/sciadv.aat4253
139
PlotnikovS. V.PasaperaA. M.SabassB.WatermanC. M. (2012). Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration.Cell1511513–1527. 10.1016/j.cell.2012.11.034
140
PlotnikovS. V.SabassB.SchwarzU. S.WatermanC. M. (2014). High-resolution traction force microscopy.Methods Cell Biol.123367–394. 10.1016/B978-0-12-420138-5.00020-3
141
PolacheckW. J.ChenC. S. (2016). Measuring cell-generated forces: a guide to the available tools.Nat. Methods13415–423. 10.1038/nmeth.3834
142
PolleuxJ.RaspM.LoubanI.PlathN.FeldhoffA.SpatzJ. P. (2011). Benzyl alcohol and block copolymer micellar lithography: a versatile route to assembling gold and in situ generated titania nanoparticles into uniform binary nanoarrays.ACS Nano56355–6364. 10.1021/nn201470f
143
PradhanR.MandalM.MitraA.DasS. (2014). Monitoring cellular activities of cancer cells using impedance sensing devices.Sens. Actuat. B Chem.193478–483. 10.1016/j.snb.2013.12.003
144
PrimeK. L.WhitesidesG. M. (1991). Self-assembled organic monolayers: model systems for studying adsorption of proteins at surfaces.Science2521164–1167. 10.1126/science.252.5009.1164
145
QuinnK. P.SridharanG. V.HaydenR. S.KaplanD. L.LeeK.GeorgakoudiI. (2013). Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation.Sci. Rep.3:3432. 10.1038/srep03432
146
RahmouniS.LindnerA.RechenmacherF.NeubauerS.SobahiT. R. A.KesslerH.et al (2013). Hydrogel micropillars with integrin selective peptidomimetic functionalized nanopatterned tops: a new tool for the measurement of cell traction forces transmitted through αvβ3- or α5β1-integrins.Adv. Mater.255869–5874. 10.1002/adma.201301338
147
RaphelJ.HolodniyM.GoodmanS. B.HeilshornS. C. (2016). Multifunctional coatings to simultaneously promote osseointegration and prevent infection of orthopaedic implants.Biomaterials84301–314. 10.1016/j.biomaterials.2016.01.016
148
RedepenningJ.SchlesingerT. K.MechalkeE. J.PuleoD. A.BiziosR. (1993). Osteoblast attachment monitored with a quartz crystal microbalance.Anal. Chem.653378–3381. 10.1021/ac00071a008
149
ReffayM.ParriniM. C.Cochet-EscartinO.LadouxB.BuguinA.CoscoyS.et al (2014). Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells.Nat. Cell Biol.16217–223. 10.1038/ncb2917
150
RenF.YesildagC.ZhangZ. (2017). Surface patterning of gold nanoparticles on PEG-based hydrogels to control cell adhesion.Polymers9:154. 10.3390/polym9050154
151
RibeiroA. J. S.AngY. S.FuJ. D.RivasR. N.MohamedT. M. A.HiggsG. C.et al (2015). Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness.Proc. Natl. Acad. Sci. U.S.A.11212705–12710. 10.1073/pnas.1508073112
152
RobinsonJ. T.JorgolliM.ShalekA. K.YoonM. H.GertnerR. S.ParkH. (2012). Vertical nanowire electrode arrays as a scalable platform for intracellular interfacing to neuronal circuits.Nat. Nanotechnol.7180–184. 10.1038/nnano.2011.249
153
Roca-CusachsP.ConteV.TrepatX. (2017). Quantifying forces in cell biology.Nat. Cell Biol.19742–751. 10.1038/ncb3564
154
RosalesA. M.AnsethK. S. (2016). The design of reversible hydrogels to capture extracellular matrix dynamics.Nat. Rev. Mater.1:15012. 10.1038/natrevmats.2015.12
155
RothenhäuslerB.KnollW. (1988). Surface-plasmon microscopy.Nature332615–617. 10.1038/332615a0
156
RuscianoG.SassoE.CapaccioA.ZambranoN.SassoA. (2019). Revealing membrane alteration in cellsoverexpressing CA IX and EGFR by surface-enhanced raman scattering.Sci. Rep.9:1832. 10.1038/s41598-018-37997-3
157
SaezA.GhibaudoM.BuguinA.SilberzanP.LadouxB. (2007). Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates.Proc. Natl. Acad. Sci. U.S.A.1048281–8286. 10.1073/pnas.0702259104
158
SchermellehL.FerrandA.HuserT.EggelingC.SauerM.BiehlmaierO.et al (2019). Super-resolution microscopy demystified.Nat. Cell Biol.2172–84. 10.1038/s41556-018-0251-8
159
SchlückerS. (2014). Surface-enhanced raman spectroscopy: concepts and chemical applications.Angew. Chemie Int. Ed.534756–4795. 10.1002/anie.201205748
160
ShamirE. R.EwaldA. J. (2014). Three-dimensional organotypic culture: experimental models of mammalian biology and disease.Nat. Rev. Mol. Cell Biol.15647–664. 10.1038/nrm3873
161
SimianM.BissellM. J. (2017). Organoids: a historical perspective of thinking in three dimensions.J. Cell Biol.21631–40. 10.1083/jcb.201610056
162
SniadeckiN. J.AnguelouchA.YangM. T.LambC. M.LiuZ.KirschnerS. B.et al (2007). Magnetic microposts as an approach to apply forces to living cells.Proc. Natl. Acad. Sci. U.S.A.10414553–14558. 10.1073/pnas.0611613104
163
SongE.LiJ.WonS. M.BaiW.RogersJ. A. (2020). Materials for flexible bioelectronic systems as chronic neural interfaces.Nat. Mater.19590–603. 10.1038/s41563-020-0679-7
164
SpatzJ. P.GeigerB. (2007). Molecular engineering of cellular environments: cell adhesion to nano-digital surfaces.Methods Cell Biol.8389–111. 10.1016/S0091-679X(07)83005-6
165
SpégelC.HeiskanenA.AcklidJ.WolffA.TaboryskiR.EmnéusJ.et al (2007). On-chip determination of dopamine exocytosis using mercaptopropionic acid modified microelectrodes.Electroanalysis19263–271. 10.1002/elan.200603720
166
SpiraM. E.HaiA. (2013). Multi-electrode array technologies for neuroscience and cardiology.Nat. Nanotechnol.883–94. 10.1038/nnano.2012.265
167
SrinivasanB.KolliA. R.EschM. B.AbaciH. E.ShulerM. L.HickmanJ. J. (2015). TEER measurement techniques for in vitro barrier model systems.J. Lab. Autom.20107–126. 10.1177/2211068214561025
168
SrinivasaraghavanV.StroblJ.WangD.HeflinJ. R.AgahM. (2014). A comparative study of nano-scale coatings on gold electrodes for bioimpedance studies of breast cancer cells.Biomed. Microdev.16689–696. 10.1007/s10544-014-9873-1
169
StableyD. R.JurchenkoC.MarshallS. S.SalaitaK. S. (2012). Visualizing mechanical tension across membrane receptors with a fluorescent sensor.Nat. Methods964–67. 10.1038/nmeth.1747
170
SteinwachsJ.MetznerC.SkodzekK.LangN.ThievessenI.MarkC.et al (2016). Three-dimensional force microscopy of cells in biopolymer networks.Nat. Methods13171–176. 10.1038/nmeth.3685
171
StrohmeyerN.BharadwajM.CostellM.FässlerR.MüllerD. J. (2017). Fibronectin-bound α5β1 integrins sense load and signal to reinforce adhesion in less than a second.Nat. Mater.161262–1270. 10.1038/nmat5023
172
StubbA.LaineR. F.MiihkinenM.HamidiH.GuzmánC.HenriquesR.et al (2020). Fluctuation-based super-resolution traction force microscopy.Nano Lett.202230–2245. 10.1021/acs.nanolett.9b04083
173
StyleR. W.BoltyanskiyR.GermanG. K.HylandC.MacminnC. W.MertzA. F.et al (2014). Traction force microscopy in physics and biology.Soft. Matter.104047–4055. 10.1039/c4sm00264d
174
SuhitoI. R.AngelineN.ChooS. S.WooH. Y.PaikT.LeeT.et al (2018). Nanobiosensing platforms for real-time and non-invasive monitoring of stem cell pluripotency and differentiation.Sensors18:2755. 10.3390/s18092755
175
SunZ.CostellM.FässlerR. (2019). Integrin activation by talin, kindlin and mechanical forces.Nat. Cell Biol.2125–31. 10.1038/s41556-018-0234-9
176
SusloparovaA.KoppenhöferD.LawJ. K. Y.VuX. T.IngebrandtS. (2015). Electrical cell-substrate impedance sensing with field-effect transistors is able to unravel cellular adhesion and detachment processes on a single cell level.Lab. Chip15668–679. 10.1039/c4lc00593g
177
TagayaM.IkomaT.TakemuraT.HanagataN.OkudaM.YoshiokaT.et al (2011). Detection of interfacial phenomena with osteoblast-like cell adhesion on hydroxyapatite and oxidized polystyrene by the quartz crystal microbalance with dissipation.Langmuir277635–7644. 10.1021/la200008z
178
TanJ. L.TienJ.PironeD. M.GrayD. S.BhadrirajuK.ChenC. S. (2003). Cells lying on a bed of microneedles: an approach to isolate mechanical force.Proc. Natl. Acad. Sci. U.S.A.1001484–1489. 10.1073/pnas.0235407100
179
ThéryM. (2010). Micropatterning as a tool to decipher cell morphogenesis and functions.J. Cell Sci.1234201–4213. 10.1242/jcs.075150
180
ThéryM.RacineV.PielM.PépinA.DimitrovA.ChenY.et al (2006). Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity.Proc. Natl. Acad. Sci. U.S.A.10319771–19776. 10.1073/pnas.0609267103
181
ThomasC. A.SpringerP. A.LoebG. E.Berwald-NetterY.OkunL. M. (1972). A miniature microelectrode array to monitor the bioelectric activity of cultured cells.Exp. Cell Res.7461–66. 10.1016/0014-4827(72)90481-8
182
TianB.Cohen-KarniT.QingQ.DuanX.XieP.LieberC. M. (2010). Three-dimensional, flexible nanoscale field-effect transistors as localized bioprobes.Science329830–834. 10.1126/science.1192033
183
TornavacaO.ChiaM.DuftonN.AlmagroL. O.ConwayD. E.RandiA. M.et al (2015). ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation.J. Cell Biol.208821–838. 10.1083/jcb.201404140
184
TurunenS.JokiT.HiltunenM. L.IhalainenT. O.NarkilahtiS.KellomäkiM. (2017). Direct laser writing of tubular microtowers for 3d culture of human pluripotent stem cell-derived neuronal cells.ACS Appl. Mater. Interf.925717–25730. 10.1021/acsami.7b05536
185
TutarR.MoteallehA.KhademhosseiniA.KehrN. S. (2019). Functional nanomaterials on 2D surfaces and in 3D nanocomposite hydrogels for biomedical applications.Adv. Funct. Mater.291–29. 10.1002/adfm.201904344
186
van der HelmM. W.HenryO. Y. F.BeinA.Hamkins-IndikT.CronceM. J.LeineweberW. D.et al (2019). Non-invasive sensing of transepithelial barrier function and tissue differentiation in organs-on-chips using impedance spectroscopy.Lab. Chip19452–463. 10.1039/c8lc00129d
187
Van Der StoelM.SchimmelL.NawazK.Van StalborchA. M.De HaanA.Klaus-BergmannA.et al (2020). DLC1 is a direct target of activated YAP/TAZ that drives collective migration and sprouting angiogenesis.J. Cell Sci.133jcs239947. 10.1242/jcs.239947
188
VorselenD.WangY.de JesusM. M.ShahP. K.FooterM. J.HuseM.et al (2020). Microparticle traction force microscopy reveals subcellular force exertion patterns in immune cell–target interactions.Nat. Commun.11:20. 10.1038/s41467-019-13804-z
189
WangJ.TrouillonR.LinY.SvenssonM. I.EwingA. G. (2013). Individually addressable thin-film ultramicroelectrode array for spatial measurements of single vesicle release.Anal. Chem.855600–5608. 10.1021/ac4009385
190
WangL.ZhuJ.DengC.XingW. L.ChengJ. (2008). An automatic and quantitative on-chip cell migration assay using self-assembled monolayers combined with real-time cellular impedance sensing.Lab. Chip8872–878. 10.1039/b804130j
191
WangW.FoleyK.ShanX.WangS.EatonS.NagarajV. J.et al (2011). Single cells and intracellular processes studied by a plasmonic-based electrochemical impedance microscopy.Nat. Chem.3249–255. 10.1038/nchem.961
192
WangW.WangS.LiuQ.WuJ.TaoN. (2012). Mapping single-cell-substrate interactions by surface plasmon resonance microscopy.Langmuir2813373–13379. 10.1021/la301712h
193
WangY. W.LiuY. L.XuJ. Q.QinY.HuangW. H. (2018). Stretchable and photocatalytically renewable electrochemical sensor based on sandwich nanonetworks for real-time monitoring of cells.Anal. Chem.905977–5981. 10.1021/acs.analchem.8b01396
194
WeberG. F.BjerkeM. A.DeSimoneD. W. (2012). A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration.Dev. Cell22104–115. 10.1016/j.devcel.2011.10.013
195
WegenerJ.JanshoffA.GallaH. J. (1998). Cell adhesion monitoring using a quartz crystal microbalance comparative analysis of different mammalian cell lines.Eur. Biophys. J.2826–37. 10.1007/s002490050180
196
WegenerJ.KeeseC. R.GiaeverI. (2000). Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces.Exp. Cell Res.259158–166. 10.1006/excr.2000.4919
197
WeigeltB.GhajarC. M.BissellM. J. (2014). The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer.Adv. Drug Deliv. Rev.69–7042–51. 10.1016/j.addr.2014.01.001
198
WhitesidesG. M.OstuniE.TakayamaS.JiangX.IngberD. E. (2001). Soft lithography in biology and biochemistry.Annu. Rev. Biomed. Eng.3335–373. 10.1146/annurev.bioeng.3.1.335
199
WilletsK. A.Van DuyneR. P. (2007). Localized surface plasmon resonance spectroscopy and sensing.Annu. Rev. Phys. Chem.58267–297. 10.1146/annurev.physchem.58.032806.104607
200
XiJ.ChenY. J. (2013). Quartz crystal microbalance in cell biology studies.J. Biochips Tissue Chipss5:1. 10.4172/2153-0777.s5-001
201
XiW.SawT. B.DelacourD.LimC. T.LadouxB. (2019). Material approaches to active tissue mechanics.Nat. Rev. Mater.423–44. 10.1038/s41578-018-0066-z
202
XiaN.ThodetiC. K.HuntT. P.XuQ.HoM.WhitesidesG. M.et al (2008). Directional control of cell motility through focal adhesion positioning and spatial control of Rac activation.FASEB J.221649–1659. 10.1096/fj.07-090571
203
XieC.LinZ.HansonL.CuiY.CuiB. (2012). Intracellular recording of action potentials by nanopillar electroporation.Nat. Nanotechnol.7185–190. 10.1038/nnano.2012.8
204
XueY.BaoL.XiaoX.DingL.LeiJ.JuH. (2011). Noncovalent functionalization of carbon nanotubes with lectin for label-free dynamic monitoring of cell-surface glycan expression.Anal. Biochem.41092–97. 10.1016/j.ab.2010.11.019
205
YangB.YaoF.YeL.HaoT.ZhangY.ZhangL.et al (2020). A conductive PEDOT/alginate porous scaffold as a platform to modulate the biological behaviors of brown adipose-derived stem cells.Biomater. Sci.83173–3185. 10.1039/c9bm02012h
206
YaoX.JiangS.LuoS.LiuB.-W.HuangT.-X.HuS.et al (2020). Uniform periodic bowtie SERS substrate with narrow nanogaps obtained by monitored pulsed electrodeposition.ACS Appl. Mater. Interf.1236505–36512. 10.1021/acsami.0c09357
207
YashunskyV.LirtsmanV.GolosovskyM.DavidovD.AroetiB. (2010). Real-time monitoring of epithelial cell-cell and cell-substrate interactions by infrared surface plasmon spectroscopy.Biophys. J.994028–4036. 10.1016/j.bpj.2010.10.017
208
YonemuraS.WadaY.WatanabeT.NagafuchiA.ShibataM. (2010). α-Catenin as a tension transducer that induces adherens junction development.Nat. Cell Biol.12533–542. 10.1038/ncb2055
209
YükselS.ZieglerM.GoerkeS.HuebnerU.WeberK.SchaafP.et al (2017). Hierarchically-designed 3D flower-like composite nanostructures as an ultrastable, reproducible, and sensitive SERS substrate.ACS Appl. Mater. Interf.938854–38862. 10.1021/acsami.7b14833
210
YüzS. G.RickenJ.WegnerS. V. (2018). Independent control over multiple cell types in space and time using orthogonal blue and red light switchable cell interactions.Adv. Sci.5:1800446. 10.1002/advs.201800446
211
ZhaiJ.WangY.ZhaiY.DongS. (2009). Rapid fabrication of Au nanoparticle films with the aid of centrifugal force.Nanotechnology20:055609. 10.1088/0957-4484/20/5/055609
212
ZhangH.ZhengX.AhmedW.YaoY.BaiJ.ChenY.et al (2018). Design and applications of cell-selective surfaces and interfaces.Biomacromolecules191746–1763. 10.1021/acs.biomac.8b00264
213
ZhangH. W.HuX. B.QinY.JinZ. H.ZhangX. W.LiuY. L.et al (2019). Conductive polymer coated scaffold to integrate 3D cell culture with electrochemical sensing.Anal. Chem.914838–4844. 10.1021/acs.analchem.9b00478
214
ZhangX.SimmonsC. A.Paul SanterreJ. (2020). Paracrine signalling from monocytes enables desirable extracellular matrix accumulation and temporally appropriate phenotype of vascular smooth muscle cell-like cells derived from adipose stromal cells.Acta Biomater.103129–141. 10.1016/j.actbio.2019.12.006
215
ZhaoW.HansonL.LouH. Y.AkamatsuM.ChowdaryP. D.SantoroF.et al (2017). Nanoscale manipulation of membrane curvature for probing endo- cytosis in live cells.Nat. Nanotechnol.12750–756. 10.1038/nnano.2017.98
216
ZongX.ZhuR.GuoX. (2015). Nanostructured gold microelectrodes for SERS and EIS measurements by incorporating ZnO nanorod growth with electroplating.Sci. Rep.5:16454. 10.1038/srep16454
Summary
Keywords
cell adhesion, electrochemistry, optical microscopy, mechanobiology, integrins, biomaterials, cell–cell adhesion, extracellular matrix
Citation
Saffioti NA, Cavalcanti-Adam EA and Pallarola D (2020) Biosensors for Studies on Adhesion-Mediated Cellular Responses to Their Microenvironment. Front. Bioeng. Biotechnol. 8:597950. doi: 10.3389/fbioe.2020.597950
Received
22 August 2020
Accepted
12 October 2020
Published
11 November 2020
Volume
8 - 2020
Edited by
Meng Deng, Purdue University, United States
Reviewed by
Yong Hou, Free University of Berlin, Germany; Zhiqin Chu, The University of Hong Kong, Hong Kong
Updates
Copyright
© 2020 Saffioti, Cavalcanti-Adam and Pallarola.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolás Andrés Saffioti, nsaffioti@unsam.edu.arDiego Pallarola, dpallarola@unsam.edu.ar
This article was submitted to Biomaterials, a section of the journal Frontiers in Bioengineering and Biotechnology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.