- Key Laboratory for Northern Urban Agriculture of Ministry of Agriculture and Rural Affairs, College of Bioscience and Resources Environment, Beijing University of Agriculture, Beijing, China
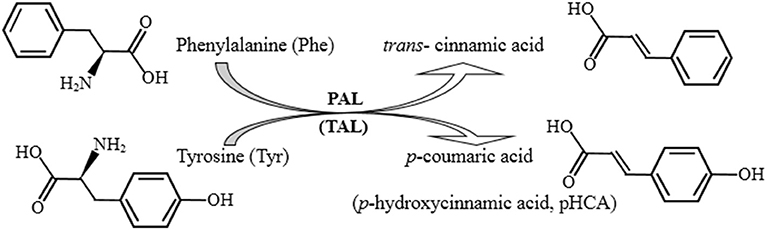
PAL (phenylalanine ammonia lyase) is important for secondary metabolite production in plants and microorganisms. There is broad interest in engineering PAL for its biocatalytic applications in industry, agriculture, and medicine. The production of quantities of high-activity enzymes has been explored by gene cloning and heterogeneous expression of the corresponding protein. Here, we cloned the cDNA of Rhodotorula glutinis PAL (RgPAL) and introduced codon optimization to improve protein expression in Escherichia coli and enzyme activities in vitro. The RgPAL gene was cloned by reverse transcription and named pal-wt. It had a full-length of 2,121 bp and encoded a 706-amino-acid protein. The pal-wt was inefficiently expressed in E. coli, even when the expression host and physical conditions were optimized. Therefore, codon optimization was used to obtain the corresponding gene sequence, named pal-opt, in order to encode the same amino acid for the RgPAL protein. The recombinant protein encoded by pal-opt, named PAL-opt, was successfully expressed in E. coli and then purified to detect its enzymatic activity in vitro. Consequently, 55.33 ± 0.88 mg/L of PAL-opt protein with a specific activity of 1,219 ± 147 U/mg and Km value of 609 μM for substrate L-phenylalanine was easily obtained. The enzyme protein also displayed tyrosine ammonia lyase (TAL)–specific activity of 80 ± 2 U/mg and Km value of 13.3 μM for substrate L-tyrosine. The bifunctional enzyme RgPAL/TAL (PAL-opt) and its easy expression advantage will provide an important basis for further applications.
Introduction
Phenylalanine ammonia lyase (PAL, EC 4.3.1.5) is the first enzyme of the general phenylpropanoid pathway catalyzing ammonia elimination from phenylalanine (Phe) to give trans-cinnamic acid, or tyrosine (Tyr) deamination to form p-coumaric acid (p-hydroxycinnamic acids), indicating its additional tyrosine ammonia lyase (TAL) activity (Figure 1) (Jun et al., 2018). It plays an important role in the synthesis of secondary metabolites with high biological value and has been of great interest in the food industry, agriculture, and medicine (Wang et al., 2016; Jun et al., 2018; Levy et al., 2018; Lin et al., 2018; Otto et al., 2019; Mays et al., 2020). Initially discovered as a plant enzyme, PAL has also been subsequently found in some microorganisms (Ogata et al., 1967; Barron et al., 2017; Levy et al., 2018). Among the PAL-producing microorganisms, yeasts, especially red yeasts, have garnered great interest regarding potential enzyme production (MacDonald and D'Cunha, 2007; Cui et al., 2014). Furthermore, the genus Rhodotorula has been the primary commercial source of enzyme (D'Cunha et al., 1996; D'Cunha, 2005; Cui et al., 2008; Barron et al., 2017). Studies on improving Rhodotorula PAL stability and activity have been focused. To increase enzymatic stability, the immobilized Rhodotorula PAL has been evaluated (Cui et al., 2015, 2017). To increase enzymatic activity, the cloning and heterogeneous expression of Rhodotorula PAL in recombinant Escherichia coli have typically been used (Cui et al., 2008; Jia et al., 2008; Babich et al., 2013; Zhu et al., 2013; Vargas-Tah et al., 2015; Rowles et al., 2016; Levy et al., 2018). Some methods, such as induction by the addition of amino acids, organic solvents, and surfactants (Cui et al., 2008), directed evolution by site-specific mutagenesis (Rowles et al., 2016; Mays et al., 2020), and coexpression of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase and transketolase (Vargas-Tah et al., 2015), have been further used to enhance recombinant PAL production. Here, we cloned the full-length R. glutinis PAL (RgPAL) gene and provided an efficient expression of the recombinant enzyme in E. coli by codon optimization. This low-cost and easy method reported here for obtaining abundant recombinant RgPAL with high activity offers an effective and sustainable PAL production source.
Materials and Methods
Microbial Strains, Plasmids, and Reagents
The strains used in this work included Rhodotorula glutinis CGMCC2258 and E. coli BL21 (DE3), Rosetta-Gami 2 (DE3), and DH5α. The plasmids PMD-18T and pET-30a were used for gene cloning and expression, respectively. TransStart FastPfu DNA polymerase DNA kit, Plasmid Extraction kit, HisPur™ Ni-NTA Resin, and BCA Protein Assay kit were purchased from Beijing Solarbio Science & Technology Co., Ltd (Beijing, China). The restriction endonucleases (HindIII and EcoRI) and the rapid amplification of cDNA end (RACE) kits (3′-Full RACE Core Set and 5′-Full RACE) were purchased from Takara (Shiga, Japan). Yeast extract and peptone were purchased from Oxoid (Basingstoke, Hampshire, UK). All solvents for high-performance liquid chromatography (HPLC) analysis were purchased from Fisher Scientific (Fair Lawn, NJ, USA). All chemicals, including standards of trans-cinnamic acid and p-coumaric acid, were purchased from Sigma–Aldrich (St. Louis, MO, USA) unless indicated otherwise.
Gene Cloning and Plasmid Construction
Total RNA of R. glutinis was extracted with TRIzol (Zhu et al., 2013). The first-strand cDNA was synthesized using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Based on the nucleotide sequences of Rhodotorula PAL published in NCBI (GenBank accession no. KF770992.1, DQ013364.1, and X13094.1), the specific primers F1 and R1 were designed to match the conserved region. F2 (gene-specific primer based on the amplified sequence above) and R2 (3′ RACE out primer based on the 3′ RACE adaptor) were applied for 3′ RACE to obtain the 3′ fragment. Degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR) further proceeded using the F3 and R3 primers. The primers F4, R4, F5, and R5 were used in a nested PCR strategy for the 5′ RACE to obtain the 5′ fragment. Based on the sequence information obtained above, the specific primers F6 and R6 were further designed to amplify the full-length cDNA, which was named pal-wt. All of the amplified fragments were subcloned into the plasmid PMD-18T for identification by sequencing. The cloning process is shown in Supplementary Figure 1.
To construct the recombinant plasmid, the F7 and R7 primers containing the restriction endonuclease sites of HindIII and EcoRI, respectively, were used to amplify pal-wt. All primers used are shown in Table 1. The amplified products were purified and ligated into the PMD-18T vector to get PMD-18T-pal-wt for identification by restriction enzyme digestion and sequencing. The expected pal-wt fragment was extracted and ligated into the pET-30a vector with the His6-tag that had been previously digested with HindIII and EcoRI to yield the recombinant plasmid pET-30a-pal-wt. The empty vector and ligated products were then transformed into E. coli BL21 (DE3) and E. coli Rosetta-Gami 2 (DE3) for expression identification.
Codon Optimization
Without changing the corresponding amino acid sequence, the pal-wt gene was optimized by replacing the codons to balance its G + C content and avoid restriction endonuclease sites, the ribosome binding site, and the rare codons of E. coli. Named pal-opt, the optimized gene introduced with the restriction sites (HindIII and EcoRI) was synthesized by Synbio Technologies (Suzhou, China). The synthetic pal-opt gene was digested and ligated into the pET-30a vector to generate the recombinant plasmid pET-30a-pal-opt. The empty vector and resulting plasmid were transformed into E. coli BL21 (DE3) for expression identification.
Protein Expression and Purification
The recombinant E. coli with pET-30a, pET-30a-pal-wt, or pET-30a-pal-opt was cultured in lysogeny broth (LB) medium at 37°C until the expected OD600 was reached. Expression of the recombinant gene was induced by the addition of 0.5 mM Isopropyl β-D-Thiogalactoside (IPTG), followed by an incubation process at the designed temperature and time. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using 12% SDS–polyacrylamide gels was performed to determine the approximate molecular mass and expression quantity of the recombinant PAL. The optimal induction conditions achieved above were further used for protein expression in large quantities. Centrifugation (10,000 × g, 10 min, 4°C) proceeded after the cells were disrupted by sonication (40 × 5 s) in ice bath. The supernatant was passed through ProteiIso Ni-NTA resin. The target PAL-opt tagged with 6 × His was purified by eluting with different imidazole concentrations (20, 200, and 500 mM). The collected fractions were analyzed by SDS-PAGE. Protein samples were then concentrated in ultracentrifuge tubes (MM Amicon Uitracel-30K) and purified with a PD-10 desalting column (Amersham Bioscience, Little Chalfont, UK). The purified PAL-opt was stored at 4°C until further analysis and application.
Enzyme Yield and Activity Assay
The enzyme yield was assessed by calculating the purified protein content per liter of culture broth. The protein concentration was measured with a BCA Protein Assay kit after purification.
PAL activity was assayed by monitoring the formation of trans-cinnamic acid catalyzed by the purified protein. The reaction mixture included 4 μg purified protein and 2.5 mL × 50 mM L-Phe, with 25 mM Tris-HCl buffer (pH 8.8) to obtain a total volume of 5 mL. The reaction was performed at 40°C for 30 min, with 5 mL methanol added to be terminated. The reaction product was evaporated at 60°C, following which it was redissolved in 5 mL methanol. After centrifugation (12,000 × g, 10 min), the supernatant was filtered through a nylon membrane (0.22 μm) to obtain the sample for HPLC analysis. Five microliters of sample was analyzed by HPLC at 290 nm with a C18 column (4.6 × 250 mm, 5 μm, Agela Innoval, CA, USA) and a gradient elution using a mobile phase comprising (A) methanol and (B) 1.5% acetic acid in water, starting at 30% A and increasing linearly to 50% until 20 min. One unit of enzyme catalyzed 1.0 μmol of trans-cinnamic acid synthesis per minute under the assay conditions. The assay of Michaelis–Menten kinetic parameter (Km) was performed with different concentrations of L-Phe from 1 to 40 mM. To obtain the optimal reaction pH and temperature, the PAL activity was measured at different temperatures (from 30 to 60°C) and pH values (from 6 to 10).
TAL activity was assayed by monitoring the formation of p-coumaric acid catalyzed by the purified protein. The reaction mixture included 10 μg purified protein and 1 mL × 50 mM Tyr, with 25 mM Tris-HCl buffer (pH 7.5) to obtain a total volume of 5 mL. The reaction was performed at 40°C for 30 min, with 5 mL methanol added to terminate the reaction. The reaction product was freeze-dried and extracted with 5 mL methanol. The extract solution was centrifuged (10,000 × g, 3 min), and the supernatant was filtered through a nylon membrane (0.22 μm) to prepare the sample for HPLC analysis. Ten microliters of sample was analyzed by HPLC at 310 nm with an isocratic elution using a mobile phase comprising 20% A (acetonitrile) and 80% B (3.5% acetic acid in water) over 10 min. One unit of enzyme catalyzed 1.0 μmol of p-coumaric acid synthesis per minute under the assay conditions. The assay of Km was performed with different concentrations of L-Tyr from 1 to 25 mM. To obtain the optimal reaction pH and temperature, the TAL activity was measured at different temperatures (from 30 to 60°C) and pH values (from 6 to 10).
Results and Discussion
Results of Gene Cloning and Recombinant Strain Construction
Some Rhodotorula PAL gene sequences have been reported in the NCBI database. However, it is still not easy to obtain the full-length cDNA because of the significant differences in sequences, especially at the two ends 3′ and 5′ (Supplementary Figure 2).
Therefore, the full-length RgPAL cDNA was obtained using reverse transcription–PCR, DOP-PCR, and RACE technology. The total RNA extracted from R. glutinis had three obvious bands (28S, 18S, and 5S), as shown in Figure 2A. The quality and quantity assessment of the extracted RNA indicated an OD260/OD280 of 1.9 and a concentration of 2.5 μg/μL. As shown in Figure 2B, a 450-bp fragment was obtained based on the conserved sequence of known PALs in Rhodotorula (GenBank accession no. KF770992.1, DQ013364.1, and X13094.1). Subsequently, fragments of 1,252, 1,922, and 357 bp were amplified using 3′ RACE, DOP-PCR, and 5′ RACE (Figures 2C–E). The full-length RgPAL cDNA sequence, named pal-wt, was eventually isolated (Figure 2F). Its recombinant plasmid pMD-18T-pal-wt was identified, as shown in Figure 2G, after digestion by HindIII and EcoRI. The recombinant strains E. coli BL21 (DE3) and E. coli Rosetta-Gami 2 (DE3) with plasmid pET-30a-pal-wt were constructed and identified by colony PCR, as indicated in Figures 2H,I.
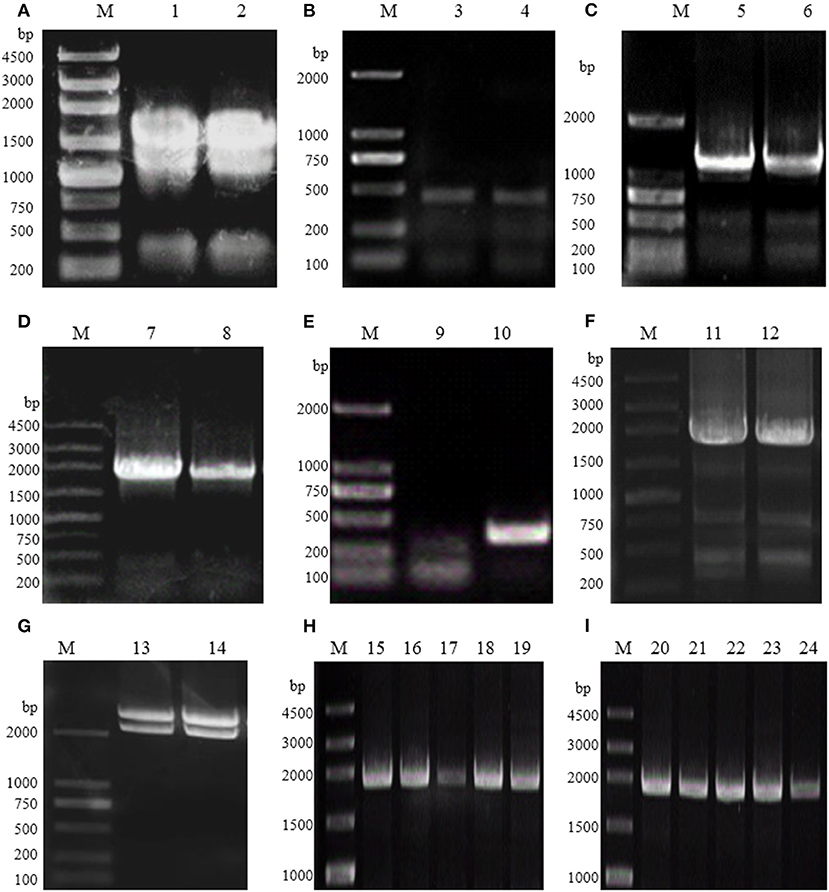
Figure 2. Electrophoresis analysis of R. glutinis PAL gene cloning and recombinant strain construction. (A) Total RNA from R. glutinis. (B) The conserved region of PAL. (C) Product of 3′ RACE. (D) Product of DOP-PCR. (E) Product of 5′ RACE. (F) Full-length of pal-wt. (G) Double-enzyme digestion of PMD-18T-pal-wt. (H) Products of colony PCR of recombinant E. coli BL21 (DE3). (I) Products of colony PCR of recombinant strains E. coli Rosetta-Gami 2 (DE3). M, DNA Marker; 1–2, total RNA; 3–12, PCR products; 13–14, double-enzyme digestion products; 15–24, colony PCR products.
The results of sequencing analysis indicated that the full-length of pal-wt contains an open reading frame (ORF) of 2,121 bp (GenBank accession no. MG712805). It encodes 706 amino acid residues with a predicted protein molecular mass of 75.5 kDa and an isoelectric point of 6.8. It shares the highest DNA (88.75%) and amino acid (96.35%) identity with sequence no. KF770992.1, according to multiple sequence alignment performed by DNAMAN software. Its protein sequence also contains the reported Rhodotorula PAL signature motif of “GTISASGDLSPLSYIAA” (Hyun et al., 2011; Yun et al., 2015) positioned between amino acids 209 and 225 (Supplementary Figure 3). The conserved active site of cyclized tripeptide Ala213-Ser214-Gly215, constructing a prosthetic 4-methylidene imidazole-5-one (MIO) group, identified in the ORF sequence also demonstrated that pal-wt is a RgPAL gene, the protein of that could be subsequently expressed (Lin et al., 2018).
Results of Protein Expression and Codon Optimization
The recombinant strain E. coli BL21 (DE3) harboring the plasmid pET-30a (as control) or pET-30a-pal-wt was induced by the addition of IPTG to identify RgPAL expression. The SDS-PAGE analysis results (Figure 3A) indicated that the target protein did not appear at all, even after optimizing the cell concentration before induction, the cultivation time after induction, and the cultivation temperature during induction. The E. coli BL21 (DE3) was temporarily suspended and considered to be unable to express RgPAL due to codon bias problems. Rosetta™ host strains, as BL21 derivatives, are designed to enhance the expression of eukaryotic proteins that contain codons rarely used in E. coli (Yin et al., 2007; Rai et al., 2020). Subsequently, we changed the host to E. coli Rosetta-Gami 2 (DE3) with an abundance of tRNAs for rare codons and expressed the protein under different induction conditions with respect to RgPAL. However, none of these experiments resulted in the target protein band (Figure 3B).
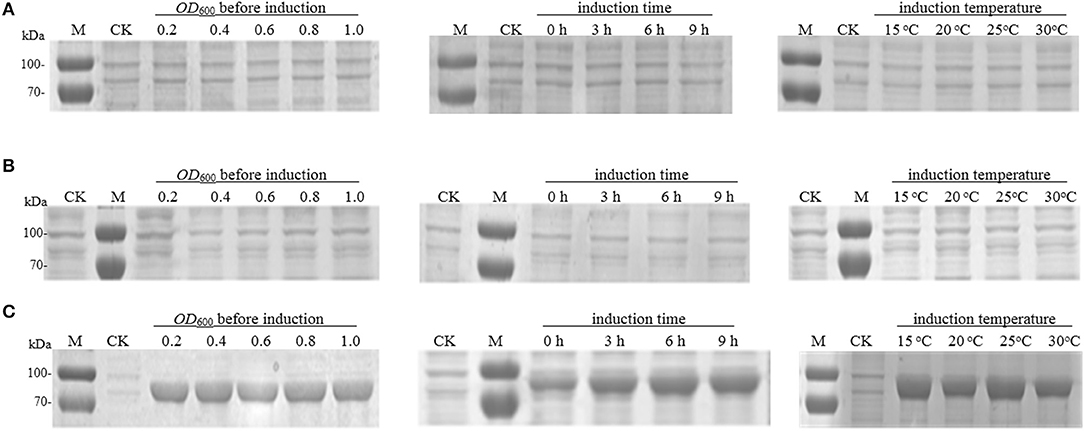
Figure 3. SDS-PAGE analysis of R. glutinis PAL expression. (A) pal-wt expression in E. coli BL21 (DE3). (B) pal-wt expression in E. coli Rosetta-Gami 2 (DE3). (C) pal-opt expression in E. coli BL21 (DE3). M, protein marker; CK, control (cell with empty vector induced at 30°C and OD600 = 0.6 for 6 h).
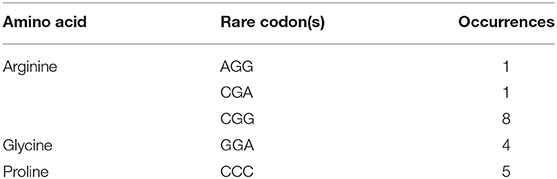
Being confronted with these results, the pal-wt sequence was further analyzed in detail based on our previous experience of recombinant protein expression (Xue et al., 2016), reported codon usage tables (Athey et al., 2017), and considerations in the use of codon optimization for recombinant protein expression (Mauro and Chappell, 2018). According to the sequence analysis, the G + C content and rare codons were considered as potential problems. The total G + C content in the pal-wt gene was as high as 65.1%, with some higher partial G + C contents. Many rarely used codons in E. coli, such as AGG, CGA, CGG, GGA, and CCC, existed in the pal-wt sequence (Table 2). Among them, 10 of the 36 arginine codons are rare in E. coli. Mistranslation errors associated with the rare arginine codon CGG in E. coli have already been observed (McNulty et al., 2003). Consequently, codon optimization was applied without changing the amino acid sequence. The G + C content was reduced to 49.6%, and the rare codons were avoided in the optimized sequence, which was named pal-opt (GenBank accession no. MK748987). The pal-opt gene was ligated into vector pET-30a to generate the plasmid pET-30a-pal-opt and then transformed into E. coli BL21 (DE3) competent cells. The resulting strain E. coli BL21 (DE3) harboring the plasmid pET-30a (as control) or pET-30a-pal-opt was induced by addition of IPTG to identify PAL-opt expression. The presence of a protein band of ~75.5 kDa in the IPTG-induced culture was in accordance with the expected size of the RgPAL protein (Figure 3C). Considerable amounts of recombinant protein were easily produced with little influence of cell concentration (OD600: 0.2–1.0) before induction, at a cultivation temperature of 15–30°C during induction and after 3 h of cultivation time after induction. A certain amount of the PAL-opt protein was produced under control of a T7 promoter even in the absence of IPTG, which is consistent with reported observations (Hartinger et al., 2010).
Results of Protein Purification and Enzyme Characterization
After expressing the E. coli BL21 (DE3) protein harboring the plasmid pET-30a-pal-opt, purification was conducted using nickel column (ProteiIso Ni-NTA Resin) affinity chromatography. The SDS-PAGE analysis showed that the target protein eventually appeared in a single band of ~75.5 kDa (Figure 4A). The purified protein was measured by a BCA kit, resulting in a yield of 55.33 mg/L (Table 3). The enzyme activity of the purified protein was measured using L-Phe or L-Tyr as the substrate. The products of the enzyme catalysis reaction were detected by HPLC using trans-cinnamic acid or p-coumaric acid as the standards (Figure 4B). Therefore, PAL-opt is actually RgPAL/TAL as it displayed both PAL and TAL activities, leading to the formation of trans-cinnamic acid and p-coumaric acid.
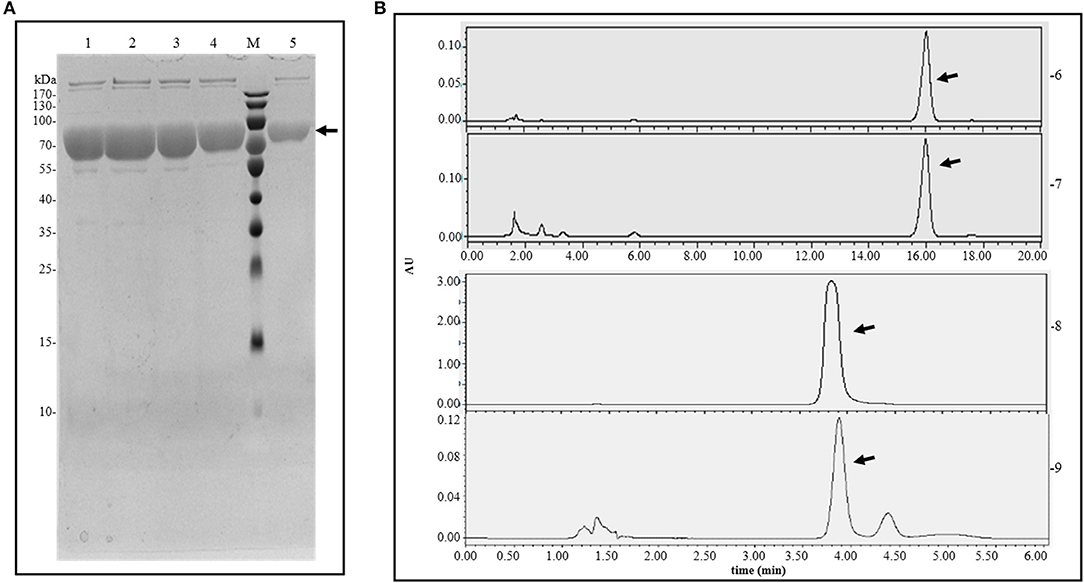
Figure 4. Protein purification and enzyme catalysis results. (A) Protein purification analyzed by SDS-PAGE. (B) Enzyme catalysis analyzed by HPLC. M, protein marker; 1–5, purified protein; 6, trans-cinnamic acid standard; 7, products of enzyme as PAL catalysis; 8, p-coumaric acid standard; 9, products of enzyme as TAL catalysis. The arrow points to the target band or peak.
As shown in Figure 5, PAL-opt exhibited a considerable high activity and relative stability. To display the activity of PAL and TAL, the optimum pH values were 8.5–9.0 and 6.5–7.5, respectively (Figure 5A), and the optimum temperature was 40°C (Figure 5B). The activities of PAL and TAL were as high as 1,219 ± 147 and 80 ± 2 U/mg, respectively.
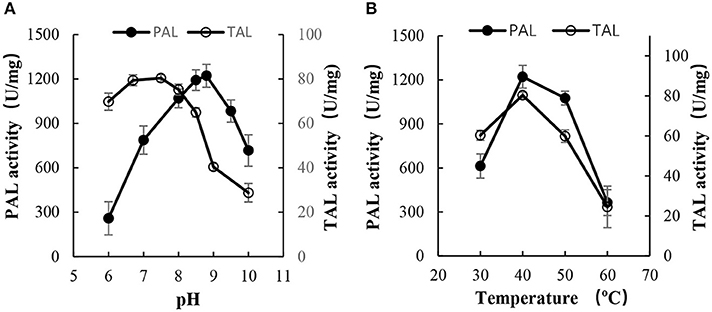
Figure 5. Effect of pH and temperature on the activity of Rg PAL/TAL. (A) Effect of pH (pH 6–7, 25 mM sodium acetate buffer; pH 7–9, 25 mM Tris-HCl buffer; pH 9–10, 25 mM sodium carbonate buffer). The reactions were performed at 40°C for 30 min to determine enzyme activity. (B) Effect of temperature. The reactions were performed for 30 min at pH 8.8 to determine PAL activity while at pH 7.5 to determine TAL activity. Each value is the mean ± SE for n = 3.
As summarized in Table 3, the specific enzyme activities of PAL and TAL of the purified PAL-opt protein were as high as 1,219 ± 147 U/mg (the equivalent of 67,045 U/L of culture broth) and 80 ± 2 U/mg, respectively. However, cell-free extracts of R. glutinis only possessed 0.0241 and 0.0143 U/mg of PAL and TAL activities (Vannellia et al., 2007). Different technologies application resulted in the improvement of the enzyme activity. Zhu et al. obtained the highest RgPAL activity of 4.2 U/mg of purified protein in recombinant E. coli BL21 (DE3) at an optimal pH and temperature (Zhu et al., 2013). Rowles et al. engineered Rhodotorula graminis PAL in recombinant E. coli BL21 (DE3) to obtain an activity of 307.28 nmol/min/mg (the equivalent of 6.15–7.68 U/L of culture broth) by site-directed mutagenesis (Rowles et al., 2016). The specific activity of RgTAL was improved by up to 7.53 U/mg via heterologous expression in E. coli W3110 (Vannellia et al., 2007). Here, the improvement of enzyme activities was contributed by purified protein in recombinant E. coli BL21 (DE3) using codon optimization and the optimum reaction system.
A wide range of specific activity and Km values for substrates L-Phe and L-Tyr has been observed in the studies of several enzymes displaying both PAL and TAL activities (Vargas-Tah and Gosset, 2015). A comparison of Rhodotorula PAL/TAL (Table 4) also revealed the difference in Km values for substrates L-Phe and L-Tyr. The difference was more significant for the enzyme of recombinant protein than the original enzyme from cell-free extract. Even if all belong to the recombinant protein of RgPAL/TAL, the Km values for substrates L-Phe and L-Tyr were quite different. Obviously, the Km for TAL observed in this study indicated its high affinity to L-Tyr substrate, which would be valuable to produce a wide range of aromatic compounds that using L-Tyr or its derivative as a precursor (Shen et al., 2020).
Why are there such significant differences in enzyme activities (Table 3) and Km values (Table 4) of PALs originated from the same genus of red yeast and expressed in E. coli? A high production and purity of the recombinant protein contributed by codon optimization (Figure 3) to remove the obstacle of nucleotide sequence (Figure 6A) as observed in this study should be one reason. An optimum reaction system mentioned above (Figure 5 and Table 3) should be another. Moreover, the differences in amino acid sequences will lead to the different enzyme activity and specificity (Figure 6B). Just as Zhu et al. (2013) reported that although R. glutinis is an anamorph of Rhodosporidium toruloides, the amino acid sequences of PALs are not the same (about 74% identity).
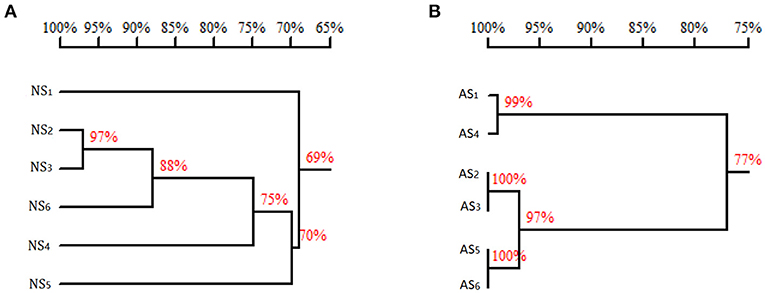
Figure 6. Homology tree of Rhodotorula PAL (TAL). (A) The homology of the nucleotide sequences. (B) The homology of the amino acid sequences. NS, nucleotide sequence; AS, amino acid sequence; 1, sequence of GenBank KF765779.1 (Rhodotorula glutinis); 2, sequence of GenBank KF770992.1 (R. glutinis); 3, sequence of GenBank X13094.1 (Rhodotorula mucilaginosa); 4, sequence of GenBank X12702.1 (Rhodotorula toruloides); 5, sequence of GenBank MK748987.1 (R. glutinis, pal-opt in this study); 6, sequence of GenBank MG712805.1 (R. glutinis, pal-wt in this study). Homology tree was constructed by DNAMAN.
Conclusion
Given increased focus on health, industry demands, agriculture development, and biotechnology, high-activity PAL is becoming increasingly desirable. Recombinant PAL is believed to be a potential source because of its optional gene origin and protein expression host, rational design, and directed evolution. PAL genes originating in the genus Rhodotorula have been shown to possess significant enzyme activity. Some PAL also displays TAL activity, in the term of PAL/TAL. In this study, when a full length of gene was cloned, an effective RgPAL expression method that does not require special physiological conditions was herein developed by codon optimization, providing RgPAL/TAL with high activity and high affinity to L-Tyr for further applications.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FX: conceptualization, methodology, investigation, resources, validation, supervision, funding acquisition, and writing–original draft preparation. ZL: codon optimization of RgPAL and protein expression. YY: gene cloning of RgPAL and recombinant plasmids construction. YW: enzyme assessment of RgPAL. YJ: kinetic characterization of RgPAL. MY: software, validation, and writing–reviewing and editing. LM: supervision, resources, and writing–reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science Foundation of China (No. 21606020) and Natural Science Foundation of Beijing Municipality (No. 2164059).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Mingcheng Zhang for his technical support and LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.610506/full#supplementary-material
References
Athey, J., Alexaki, A., Osipova, E., Rostovtsev, A., Santana-Quintero, L. V., Katneni, U., et al. (2017). A new and updated resource for codon usage tables. BMC Bioinformatics 18:391. doi: 10.1186/s12859-017-1793-7
Babich, O. O., Pokrovsky, V. S., Anisimova, N. Y., Sokolov, N. N., and Prosekov, A. Y. (2013). Recombinant L-phenylalanine ammonia lyase from Rhodosporidium Toruloides as a potential anticancer agent. Biotechnol. Appl. Biochem. 60, 316–322. doi: 10.1002/bab.1089
Barron, C. C., Sponagle, B. J., Arivalagan, P., and D'Cunha, G. B. (2017). Optimization of oligomeric enzyme activity in ionic liquids using Rhodotorula glutinis yeast phenylalanine ammonia lyase. Enzyme Microb. Technol. 96, 151–156. doi: 10.1016/j.enzmictec.2016.10.010
Cui, J. D., Jia, S. R., and Sun, A. Y. (2008). Influence of amino acids, organic solvents and surfactants for phenylalanine ammonia lyase activity in recombinant Escherichia coli. Lett. Appl. Microbiol. 46, 631–635. doi: 10.1111/j.1472-765X.2008.02364.x
Cui, J. D., Liang, L. H., Han, C., and Liu, R. L. (2015). Stabilization of phenylalanine ammonia lyase from Rhodotorula glutinis by encapsulation in polyethyleneimine-mediated biomimetic silica. Appl. Biochem. Biotechnol. 176, 999–1011. doi: 10.1007/s12010-015-1624-0
Cui, J. D., Qiu, J. Q., Fan, X. W., Jia, S. R., and Tan, Z. L. (2014). Biotechnological production and applications of microbial phenylalanine ammonia lyase: a recent review. Crit. Rev. Biotechnol. 34, 258–268. doi: 10.3109/07388551.2013.791660
Cui, J. D., Zhao, Y. M., Tan, Z. L., Zhong, C., Han, P. P., and Jia, S. R. (2017). Mesoporous phenylalanine ammonia lyase microspheres with improved stability through calcium carbonate templating. Int. J. Biol. Macromol. 98, 887–896. doi: 10.1016/j.ijbiomac.2017.02.059
D'Cunha, G. B. (2005). Enrichment of phenylalanine ammonia lyase activity of Rhodotorula yeast. Enzyme Microb. Technol. 36, 498–502. doi: 10.1016/j.enzmictec.2004.11.006
D'Cunha, G. B., Satyanarayan, V., and Nair, P. M. (1996). Stabilization of phenylalanine ammonia lyase containing Rhodotorula glutinis cells for the continuous synthesis of L-phenylalanine methyl ester/96/. Enzyme Microb. Technol. 19, 421–427. doi: 10.1016/S0141-0229(96)00013-0
Hartinger, D., Heinl, S., Schwartz, H. E., Grabherr, R., Schatzmayr, G., Haltrich, D., et al. (2010). Enhancement of solubility in Escherichia coli and purification of an aminotransferase from Sphingopyxis sp. MTA144 for deamination of hydrolyzed fumonisin B1. Microb. Cell Fact. 9:62. doi: 10.1186/1475-2859-9-62
Hyun, M. W., Yun, Y. H., Kim, J. Y., and Kim, S. H. (2011). Fungal and plant phenylalanine ammonia-lyase. Mycobiology 39, 257–265. doi: 10.5941/MYCO.2011.39.4.257
Jia, S. R., Cui, J. D., Li, Y., and Sun, A. Y. (2008). Production of L-phenylalanine from trans-cinnamic acids by high-level expression of phenylalanine ammonia lyase gene from Rhodosporidium toruloides in Escherichia coli. Biochem. Eng. J. 42, 193–197. doi: 10.1016/j.bej.2008.06.010
Jun, S. Y., Sattler, S. A., Cortez, G. S., Vermerris, W., Sattler, S. E., and Kang, C. (2018). Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase. Plant Physiol. 176, 1452–1468. doi: 10.1104/pp.17.01608
Levy, H. L., Sarkissian, C. N., and Scriver, C. R. (2018). Phenylalanine ammonia lyase (PAL): from discovery to enzyme substitution therapy for phenylketonuria. Mol. Genet. Metab. 124, 223–229. doi: 10.1016/j.ymgme.2018.06.002
Liang, J., Guo, L., Sun, P., Jiang, B., Lin, J., Guo, W., et al. (2016). A novel process for obtaining phenylpropanoic acid precursor using Escherichia coli with a constitutive expression system. Food Sci. Biotechnol. 25, 795–801. doi: 10.1007/s10068-016-0134-3
Lin, W., Liu, A., Weng, C., Li, H., Sun, S., Song, A., et al. (2018). Cloning and characterization of a novel phenylalanine ammonia-lyase gene from Inonotus baumii. Enzyme Microb. Technol. 112, 52–58. doi: 10.1016/j.enzmictec.2017.10.010
MacDonald, M. J., and D'Cunha, G. B. (2007). A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 85, 273–282. doi: 10.1139/O07-018
Mauro, V. P., and Chappell, S. A. (2018). “Considerations in the use of codon optimization for recombinant protein expression,” in Recombinant Protein Expression in Mammalian Cells, ed D. L. Hacker (New York, NY: Humana Press), 275–288. doi: 10.1007/978-1-4939-8730-6_18
Mays, Z. J., Mohan, K., Trivedi, V. D., Chappell, T. C., and Nair, N. U. (2020). Directed evolution of Anabaena variabilis phenylalanine ammonia-lyase (PAL) identifies mutants with enhanced activities. Chem. Commun. 56, 5255–5258 doi: 10.1039/D0CC00783H
McNulty, D. E., Claffee, B. A., Huddleston, M. J., and Kane, J. F. (2003). Mistranslational errors associated with the rare arginine codon CGG in Escherichia coli. Protein Express. Purif. 27, 365–374. doi: 10.1016/S1046-5928(02)00610-1
Ogata, K., Uchiyam, K., and Yamada, H. (1967). Metabolism of aromatic amino acid in microorganisms. Agric. Biol. Chem. 31, 200–206. doi: 10.1080/00021369.1967.10858795
Otto, M., Wynands, B., Lenzen, C., Filbig, M., Blank, L. M., and Wierckx, N. (2019). Rational engineering of phenylalanine accumulation in Pseudomonas taiwanensis to enable high-yield production of trans-cinnamate. Front. Bioeng. Biotechnol. 7:312. doi: 10.3389/fbioe.2019.00312
Rai, V., Upmanyu, V., Mohd, G., Kumar, R., Koppad, S., Sachdeva, H. S., et al. (2020). Comparing the efficiency of different Escherichia coli strains in producing recombinant capsid protein of porcine circovirus type 2. Mol. Cell. Probes 52:101556. doi: 10.1016/j.mcp.2020.101556
Rowles, I., Groenendaal, B., Binay, B., Malone, K. J., Willies, S. C., and Turner, N. J. (2016). Engineering of phenylalanine ammonia lyase from Rhodotorula graminis for the enhanced synthesis of unnatural L-amino acids Tetrahedron 72, 7343–7347. doi: 10.1016/j.tet.2016.06.026
Shen, Y. P., Niu, F. X., Yan, Z. B., Fong, L. S., Huang, Y. B., and Liu, J. Z. (2020). Recent advances in metabolically engineered microorganisms for the production of aromatic chemicals derived from aromatic amino acids. Front. Bioeng. Biotechnol. 8:407. doi: 10.3389/fbioe.2020.00407
Vannelli, T., Xue, Z., Breinig, S., Qi, W. W., and Sariaslani, F. S. (2007). Functional expression in Escherichia coli of the tyrosine-inducible tyrosine ammonia-lyase enzyme from yeast Trichosporon cutaneum for production of p-hydroxycinnamic acid. Enzyme Microb. Technol. 41, 413–422. doi: 10.1016/j.enzmictec.2007.03.013
Vannellia, T., Qib, W. W., Sweigardc, J., Gatenbyd, A. A., and Sariaslanid, F. S. (2007). Production of p-hydroxycinnamic acid from glucose in Saccharomyces cerevisiae and Escherichia coli by expression of heterologous genes from plants and fungi. Metab. Eng. 9, 142–151. doi: 10.1016/j.ymben.2006.11.001
Vargas-Tah, A., and Gosset, G. (2015). Production of cinnamic and p-hydroxycinnamic acids in engineered microbes. Front. Bioeng. Biotechnol. 3:116. doi: 10.3389/fbioe.2015.00116
Vargas-Tah, A., Martínez, L., Hernández-Chávez, G., Rocha, M., Martínez, A., Bolívar, F., et al. (2015). Production of cinnamic and p-hydroxycinnamic acid from sugar mixtures with engineered Escherichia coli. Microb. Cell Fact. 14, 1–12. doi: 10.1186/s12934-014-0185-1
Wang, Z., Li, J. Y., Jia, C. H., Li, J. P., Xu, B. Y., and Jin, Z. Q. (2016). Molecular cloning and expression of four phenylalanine ammonia lyase genes from banana interacting with Fusarium oxysporum. Biol. Plant. 60, 459–468. doi: 10.1007/s10535-016-0619-1
Xue, F., Guo, H., Hu, Y., Liu, R., Huang, L., Lv, H., et al. (2016). Expression of codon-optimized plant glycosyltransferase UGT72B14 in Escherichia coli enhances salidroside production. BioMed Res. Int. 2016:9845927. doi: 10.1155/2016/9845927
Yin, J., Li, G., Ren, X., and Herrler, G. (2007). Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 127, 335–347. doi: 10.1016/j.jbiotec.2006.07.012
Yun, Y. H., Koo, J. S., Kim, S. H., and Kong, W. S. (2015). Cloning and expression analysis of phenylalanine ammonia-lyase gene in the mycelium and fruit body of the edible mushroom Flammulina velutipes. Mycobiology 43, 327–332. doi: 10.5941/MYCO.2015.43.3.327
Keywords: phenylalanine ammonia lyase, codon optimization, Rhodotorula glutinis, Escherichia coli, tyrosine ammonia lyase
Citation: Xue F, Liu Z, Yu Y, Wu Y, Jin Y, Yang M and Ma L (2021) Codon-Optimized Rhodotorula glutinis PAL Expressed in Escherichia coli With Enhanced Activities. Front. Bioeng. Biotechnol. 8:610506. doi: 10.3389/fbioe.2020.610506
Received: 26 September 2020; Accepted: 23 December 2020;
Published: 03 February 2021.
Edited by:
Wen-Yong Lou, South China University of Technology, ChinaReviewed by:
Anwar Sunna, Macquarie University, AustraliaJian Dong Cui, Tianjin University of Science and Technology, China
Copyright © 2021 Xue, Liu, Yu, Wu, Jin, Yang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feiyan Xue, ZmVpeWFueHVlQGJ1YS5lZHUuY24=; Lanqing Ma, bHFtYUBidWEuZWR1LmNu
 Feiyan Xue
Feiyan Xue Zihui Liu
Zihui Liu Yuxin Jin
Yuxin Jin