- 1State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
- 2College of Resources and Environment, Huazhong Agricultural University, Wuhan, China
Anthropogenic activities have released various contaminants into soil that pose a serious threat to the ecosystem and human well-being. Compared to conventional analytical methodologies, microbial cell-based bioreporters are offering a flexible, rapid, and cost-effective strategy to assess the environmental risks. This review aims to summarize the recent progress in the application of bioreporters in soil contamination detection and provide insight into the challenges and current strategies. The biosensing principles and genetic circuit engineering are introduced. Developments of bioreporters to detect and quantify heavy metal and organic contaminants in soil are reviewed. Moreover, future opportunities of whole-cell bioreporters for soil contamination monitoring are discussed.
Introduction
Soil is a central ecosystem that sustains humans, plants, and animals. However, the increasing industrial and agricultural activities have released various contaminants such as aromatic compounds and heavy metals into soil, which severely deteriorated soil health and sustainability (Bispo et al., 1999; Gu and Chang, 2001; Wu et al., 2018; Bae et al., 2020). The detection and monitoring of soil contaminants are vital for the security of humans and other biota on earth (Lu et al., 2015).
The conventional techniques for soil contaminant analysis include gas chromatography–mass spectrometry (GC-MS), high-performance liquid chromatography (HPLC), atomic absorption spectroscopy (AAS), atomic fluorescence spectroscopy (AFS), inductively coupled plasma mass spectrometry (ICP-MS), and X-ray absorption spectroscopy (Branyikova et al., 2020; Wang et al., 2020). These instrumental-based analyses enable soil contamination measurements with high sensitivity and accuracy. However, the conventional chemical approaches measure chemical availability of soil contaminants rather than the bioavailability (Hou et al., 2014). Based on the definition proposed by the National Research Council’s (NRC) committee and the ISO/TC 190 working group, bioavailability is a dynamic process that comprises multiple steps (NRC, 2003; IOS, 2008). It includes the environmental availability of contaminants in soil, the contaminant uptake by organisms, and accumulation and toxic effects of contaminants within organisms (Harmsen, 2007; Kim et al., 2015). Therefore, a biological approach is required to complement chemical analyses to assess the actual ecotoxicological and health risks.
Whole-cell bioreporter, a promising technique to address this issue, allows a rapid detection of bioavailable contaminants in soil (Liu et al., 2011; Song Y. et al., 2014; Francisco et al., 2019). The bioreporter technology uses microbial cells as molecular recognition elements to sense analytes in the environment and exhibits a measurable signal (Su et al., 2011; Turdean, 2011; Bilal and Iqbal, 2019). Due to the simplicity, portability, better detection limit, rapid response, and cost-effectiveness, bioreporters are ideal approaches to assess the bioavailable fraction of contaminants (Wang X. et al., 2014; Bilal and Iqbal, 2019). This review summarized the current state of whole-cell bioreporter in soil contaminant monitoring. The principles and configurations of whole-cell bioreporters are briefly introduced. The applicability of bioreporters in soil contaminant detection is discussed. The recent advances in the application of bioreporters to assess the contamination status of heavy metals and organic pollutants were surveyed. Future prospects of whole-cell bioreporters are further discussed.
Whole-Cell Microbial Bioreporter Designs for Contaminant Measurement
A whole-cell bioreporter comprises three parts: the sensor, the genetic logic circuit, and the actuator (Figure 1; Kaur et al., 2015; Gupta et al., 2019). The biological sensor element responds to target compounds and triggers a cascade of biological reactions (Bilal and Iqbal, 2019). Genetic logic circuits link the sensors and actuators and convert the bioreaction into a detectable signal proportional to the analyte concentration (Su et al., 2011; Jia et al., 2019).
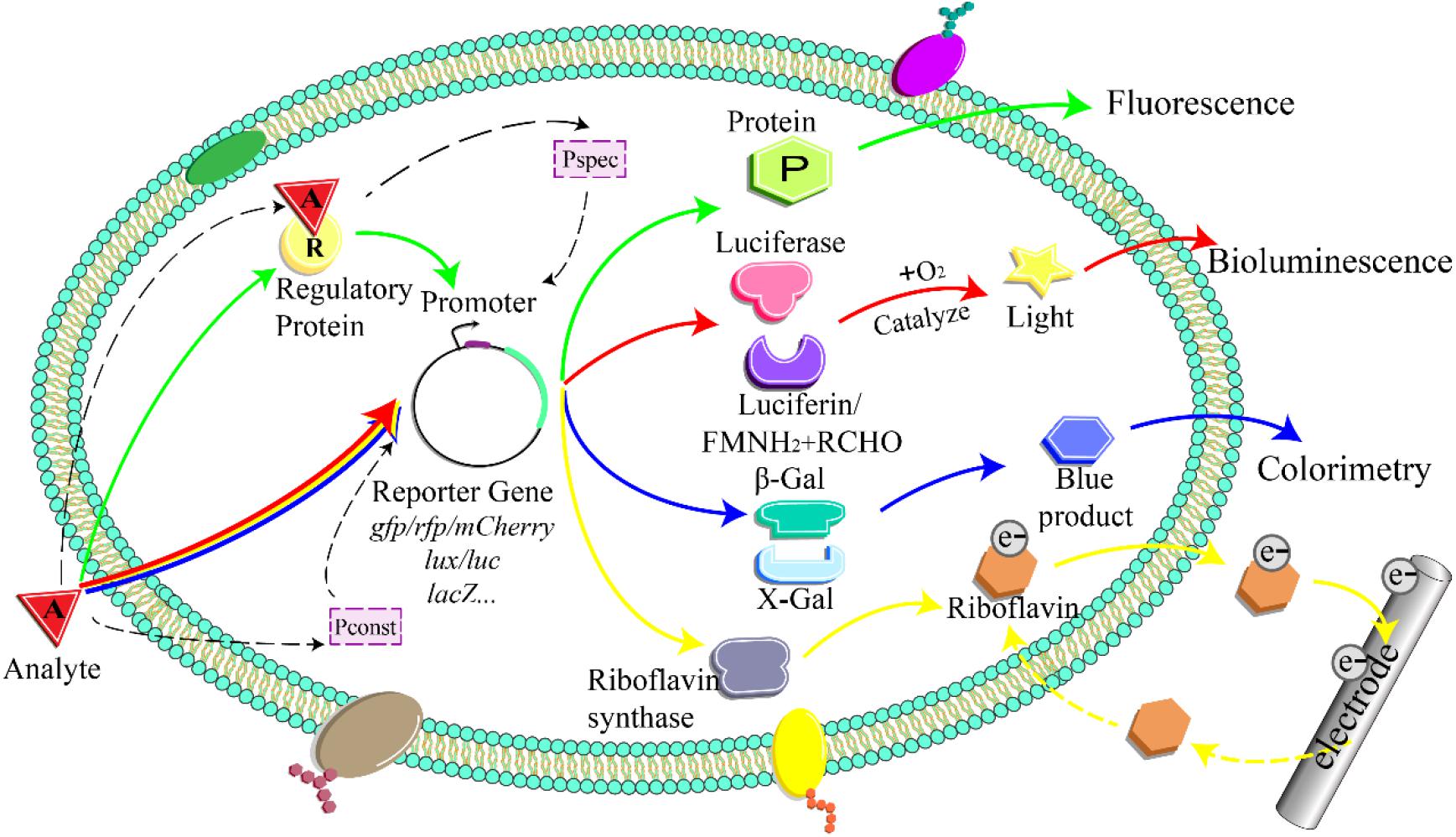
Figure 1. Detection principle of whole-cell microbial bioreporter. The analyte activates or inhibits the expression of the specific promoter and downstream reporter gene. Pspec, specific promoter; Pconst, constitutive promoter; FMNH2, reduced flavin mononucleotide; RCHO, aldehyde; β-Gal, β-galactosidase; X-Gal, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside; e–, electron.
Sensor Elements
The specificity and sensitivity of bioreporters are mainly determined by sensor elements (Shin, 2011). Based on the specificity, bioreporters can be categorized into non-specific and specific bioreporters (Jarque et al., 2016). The non-specific bioreporter responds to toxic substances that cause stress (Sorensen et al., 2006). The presence of toxic contaminants suppresses the transcriptional activity of a constitutive promoter to decrease the output signal or triggers the transcription of the reporter genes through stress-responsive promoters. For example, a non-specific bioreporter was deigned via fusing the reporter gene to a constitutive promoter PpsbA (Elväng et al., 2001; Gu and Chang, 2001). When exposed to toxic chemicals, the expression of the reporter gene is inhibited, which induces a decrease in signal intensity. Another example is a non-specific stress-responsive bioreporter constructed via fusing the bioluminescent reporter luxCDABE from Photorhabdus luminescens with SOS-controlled recA promoter in Acinetobacter baylyi ADP1 (Sorensen et al., 2006; Song et al., 2009). When exposed to genotoxic contaminants, DNA damage triggered an SOS response to activate recA promoter for the expression of lux genes (Song Y. et al., 2014).
Specific bioreporters contain a regulatory protein responsible for activating or repressing the promoter activities as the sensing domain (Burmølle et al., 2006; Sorensen et al., 2006). When the target contaminants are present, the regulatory protein binds with the contaminants and undergoes a conformational change, which activates or inhibits its DNA binding and modulates the expression of reporter genes. The proteins from MerR family recognize specific target metal ions, such as the mercury-sensing regulatory protein MerR and lead-sensing regulatory protein PbrR (Brown et al., 2003). For example, mercury-specific bioreporters are constructed via fusing reporter genes such as luxAB and gfp to the downstream of the merR promoter (Branyikova et al., 2020).
Genetic Logic Circuits
Genetic circuits connect sensor apparatuses and actuators that program cells to respond to signal inputs and generate precise behavioral actions (Wang and Buck, 2012; Saltepe et al., 2018). Genetic circuitry is capable of integrating multiple input signals and resulting in specific biochemical behaviors that can enhance the specificity and sensitivity of bioreporters (Lim, 2010). Based on the basal transcriptional activity of promoters, there are two types of non-specific bioreporters (Jiang et al., 2020). The first one comprises a constitutively expressed promoter to maintain a high basal transcriptional activity. When exposed to toxic contaminants, the activity of the promoter is suppressed to decrease the output signal. In the second non-specific bioreporter, the reporter gene is fused with a stress-responsive promoter, whose activation induces the expression of the reporter gene under stress conditions (Burmølle et al., 2006). Specific bioreporters are composed of specific regulatory proteins responsible for promoter activities. The binding of target contaminants with the regulatory protein results in the induction or repression of reporter gene expression (Reyes-Caballero et al., 2011). The regulator proteins are generally continuously expressed to function as repressors that inhibit the expression of reporter genes. When contaminants are present, the repressor protein interacts with the contaminants and activates the reporter gene expression. For example, the regulator alkR in the bioreporter sensing alkanes is constitutively transcribed to inhibit the signal output. Its interaction with alkanes activates the promoter PalkM and expression of downstream reporter genes (Zhang et al., 2012).
Most bioreporters were designed to target a single contaminant. To sense multiple contaminants concurrently, logic gates were incorporated in bioreporters. The typical logic gates include amplifier, NOT, AND, and OR (Silva-Rocha and de Lorenzo, 2008; Katz, 2019). When the input signal is present, the amplifier generates one output and the NOT gate suppresses the output. The AND and OR gates are capable of computing multiple inputs and generating one output. For example, a dual-input “AND gate” can respond to two inputs and only be expressed when both inputs are detected, while the OR gate generates output when either one of the inputs is present (Song H. et al., 2014). A triple-input AND logic gated bioreporter system was engineered to detect three different heavy metals by coupling two dual-input AND gates (Wang et al., 2013). The first dual-input AND gate can detect arsenic and mercury via two specific inducible promoters (ParsR and PmerT). The output of the first bioreporter is the quorum-sensing molecule 3OC6HSL. The second AND gate comprises quorum-sensing promoter PluxI and copper-responsive promoter PcusC, which can sense quorum-sensing molecule and copper ion with rfp as output readout. Consequently, the AND gated triple-input bioreporter system based on two AND gates can detect and integrate arsenic, mercury, and copper concentrations in parallel.
Genetic logic gates can also function as a biological filter and an amplifier (Kim et al., 2016; Jia et al., 2018, 2019; Saltepe et al., 2018). A positive feedback amplifier using LuxR auto tuning elements improved the sensitivity and specificity of an arsenic whole-cell bioreporter. The arsenic-inducible promoter Pars and its regulatory gene arsR were coupled to the reporter transcriptional activator luxR in the first plasmid. The mCherry gene and luxR gene were placed downstream of the promoter PluxI in the second plasmid to form a positive feedback loop. When arsenic ion binds to ArsR to alleviate its inhibition of Pars promoter, the expression of LuxR in the first plasmid is activated, which initiates the expression of mCherry and luxR in the second plasmid. Its expression product LuxR in the second plasmid activates the expression of its own mCherry (Jia et al., 2019). The minimum detection limit was reduced by one order of magnitude. Similarly, a lead whole-cell bioreporter containing positive feedback amplifiers was 15–20 times more sensitive than the one without amplifiers (Jia et al., 2018). Compared with the constitutively expressed regulator, the positive feedback regulator enhances the specificity and sensitivity of bioreporters (Jia et al., 2019). However, the main concern of the positive feedback regulator is the high noise level and the false-positive signal from leaky promoters (Bansal et al., 2010).
Actuator Elements
Genetic logic circuits respond to the input signal and convert it into a detectable signal output (Figure 1) (e.g., fluorescence, bioluminescence, colorimetry). Reporter genes play a crucial role in the signal transduction process. Optical-based reporters are mostly commonly used in soil contamination detection (Burmølle et al., 2006; Xu et al., 2013). The target analyte induces or represses the expression of the fluorescent and luminescent reporter genes, such as lux, lacZ, and gfp (Shin, 2011). Although the actuators based on visual detection are characterized by simple operation, good stability, high sensitivity, and fast response speed, they come with limitations (Burmølle et al., 2006; Jarque et al., 2016).
The lacZ gene of Escherichia coli encoding β-galactosidase was one widely used reporter element. The color change during enzyme assay allows the colorimetric detection of β-galactosidase activity. Compared with bacterial luciferase and green fluorescent protein, β-galactosidase can be expressed with low metabolic activities and the colorimetric reaction of β-galactosidase does not require oxygen. Another key strength of the β-galactosidase-based actuator is the high sensitivity. The detection limit of a β-galactosidase-based cadmium bioreporter is 10 pM, which is three orders of magnitude lower than that of a gfp-based construct in the same host strain (Shetty et al., 2003). However, the colorimetric assay of β-galactosidase requires disruption of host cells and addition of substrates. Therefore, chromogenic reporter genes like lacZ were not frequently employed in soil contamination detection.
Bioluminescent and fluorescent actuators emit light or fluorescence upon soil contaminants without cell disruption and substrate addition. Due the ease of visualization, luminescent and fluorescent reporter genes were preferable than chromogenic reporter genes. The bacterial luciferase encoded by luxCDABE operon from Aliivibrio fischeri or luxCDABE operon from Photobacterium leiognathi converts fatty aldehyde to fatty acid coupled with the oxidation of FMNH2 and emits a blue light as readout (Sorensen et al., 2006). The luciferase substrate, aldehyde, is subsequently regenerated via the multienzyme reductase complex encoded by luxCDE. However, as the bacterial luciferase reaction requires oxygen, lux reporter genes are not applicable for anaerobic environment. Additionally, it is noteworthy that light output is an energy-demanding process that is dependent on the metabolic state of host cells. Fluorescent proteins, especially green fluorescent protein (GFP) from jellyfish Aequorea victoria, are also widely applied in bioreporter design (Daunert et al., 2000). The measurement of GFP fluorescence only requires light excitation. The green fluorescent signal allows single-cell detection via microscopy or flow cytometry (Gutierrez et al., 2015; Kaur et al., 2015). The GFP protein also requires the presence of oxygen to fold and fluoresce properly. However, natural GFP takes a long time to fold and is difficult to denature, which may cause false-positive results. Therefore, the lux-based actuator is preferred in constructing bioreporters with a fast response time (Valkova et al., 1999; Hernández-Sánchez et al., 2016).
Due to the opacity of the soil matrix, the optical-based bioreporter requires preparation of soil solution or soil extract. The electrochemical bioreporter system featured by minimal sample pretreatment and rapid detection is a promising candidate to improve the measurement. The principles of electrochemical measurement are based on the electrochemical properties of electroactive products catalyzed by reporter enzymes or biological interaction processes, which generate a measurable electrochemical signal. For instance, ampere-based whole-cell electrochemical bioreporters can be constructed via lacZ or phoA reporter gene (Paitan et al., 2004; Cortes-Salazar et al., 2013). Expression of β-galactosidase or alkaline phosphatase converts p-aminophenyl-β-D-galactopyranoside (PAPG) or p-aminophenol phosphate (PAPP) into p-aminophenol (PAP). PAP is oxidized at the electrode and can be converted to a current signal using the chronoamperometric technique (Paitan et al., 2004). One recent study fuses PzntA promoter with zntR, rib, and oprF, which respond to Zn and trigger riboflavin and porin production in E. coli. The production of outer membrane porin and electron shuttles enhanced extracellular electron transfer, which exhibited a linear relationship between the maximum voltage and Zn concentration (Khan et al., 2020). Microbial cells can also function as an adsorbent to immobilize heavy metal ions on bioelectrode. The contaminants can be then measured via differential pulse anodic stripping voltammetry analysis (Dali et al., 2018). Moreover, as an electrochemical bioreporter can complete the detection process on-site, it greatly avoids the influence of subjective factors caused by sampling and has high sensitivity and strong specificity.
Applicability in Soil Contaminant Detection
Soil is a complex matrix that is composed of minerals, organic matter, and organisms. Although the previous study demonstrated that the single bioassay cannot interpret the overall quality of soil, microbial cells exhibited good sensitivity to assess heavy metals and organic pollutants in soil extract (Bierkens et al., 1998). The main obstacle to the application of bioreporter in soil is the opacity of the soil matrix and background luminescence of minerals, which interfere with the visualization of the bioreporter (Rasmussen et al., 2000; Kim et al., 2020). Although bioreporter cells showed good performance in laboratory conditions, the functionality and reliability of bioreporters are difficult to retain under edaphic environments. The insufficient nutrient supply and toxicity of contaminants suppress microbial activities and cause interference (Haft et al., 2009; Wu et al., 2016, 2020b). The direct contact between bioreporter cells with soil particles can enhance the accuracy of bioreporter detection (Magrisso et al., 2009; Jiang et al., 2017), but minerals were found to impact the microbial cell robustness (Cai et al., 2018). For example, iron oxides and manganese oxides have been found to trigger an SOS response and deactivate the bioreporter cells (Cai et al., 2019; Liu et al., 2020).
There are several current strategies to improve the applicability of bioreporters in soil. Soil was mixed with minimal salt solutions to enable visualization of fluorescent and luminescent signals (Song Y. et al., 2014; Yoon et al., 2016d). Soil samples are generally diluted about 5–20 times and vigorously shaken to break down soil aggregates. The resultant mixture and the supernatant can be subject to fluorescence or luminescence measurement. To enhance the adaptability of bioreporter cells in soil, indigenous soil microorganisms, such as Pseudomonas putida and Acinetobacter baylyi, are preferred to the model strain E. coli in bioreporter designs (Zhang et al., 2012).
However, the pretreatment process alters the bioavailability of soil contaminants. The addition of minimal salt solutions solubilizes metals and organics, which generates an overestimated concentration for the bioavailable contaminants (Ivask et al., 2002). The pretreatment of soil samples by sonication or shaking homogenizes and emulsifies hydrophobic organic pollutants (Jiang et al., 2017). The physiochemical properties of the soil environment like pH and moisture vary widely within micrometer distance, which cause the heterogeneous distribution and speciation of contaminants in soil. Previous studies using synchrotron-based X-ray fluorescence spectroscopy found hundred- and thousand-fold differences in concentration of heavy metals like Cu and Ni occurring over micrometer distances (Jacobson et al., 2007; Schmid et al., 2016; Qu et al., 2019). As contaminants heterogeneously distributed at spatial scales relevant to microbes, they trigger a heterogeneous response of bioreporter cells. Single-cell detection is required to assess the spatial heterogeneity of soil contaminants.
To address these issues, attempts have been made to recover the bioreporter cells from the soil matrix with density gradient centrifugation (Hurdebise et al., 2015). Previous studies also functionalized bioreporter cells with magnetic nanoparticles like Fe3O4, by which bioreporter cells can be isolated under a magnetic field (Jia et al., 2016). Moreover, the immobilization of bioreporter cells on or in solid media enhances their stability in the soil environment, which enables in situ measurement. For example, bioreporter cells can be encapsulated into hydrogel beads (Bae et al., 2020). The alginate beads were deployed on the soil surface to detect toluene, and bioluminescence was measured via a fiber-optic probe. Moreover, bioreporters can be immobilized on optical fibers, which further simplifies the bioreporter system (Polyak et al., 2001; Kumar et al., 2006). The portability of bioreporter systems requires further research to allow on-site measurements.
Recent Advances in Soil Contaminant Detection
Heavy Metals
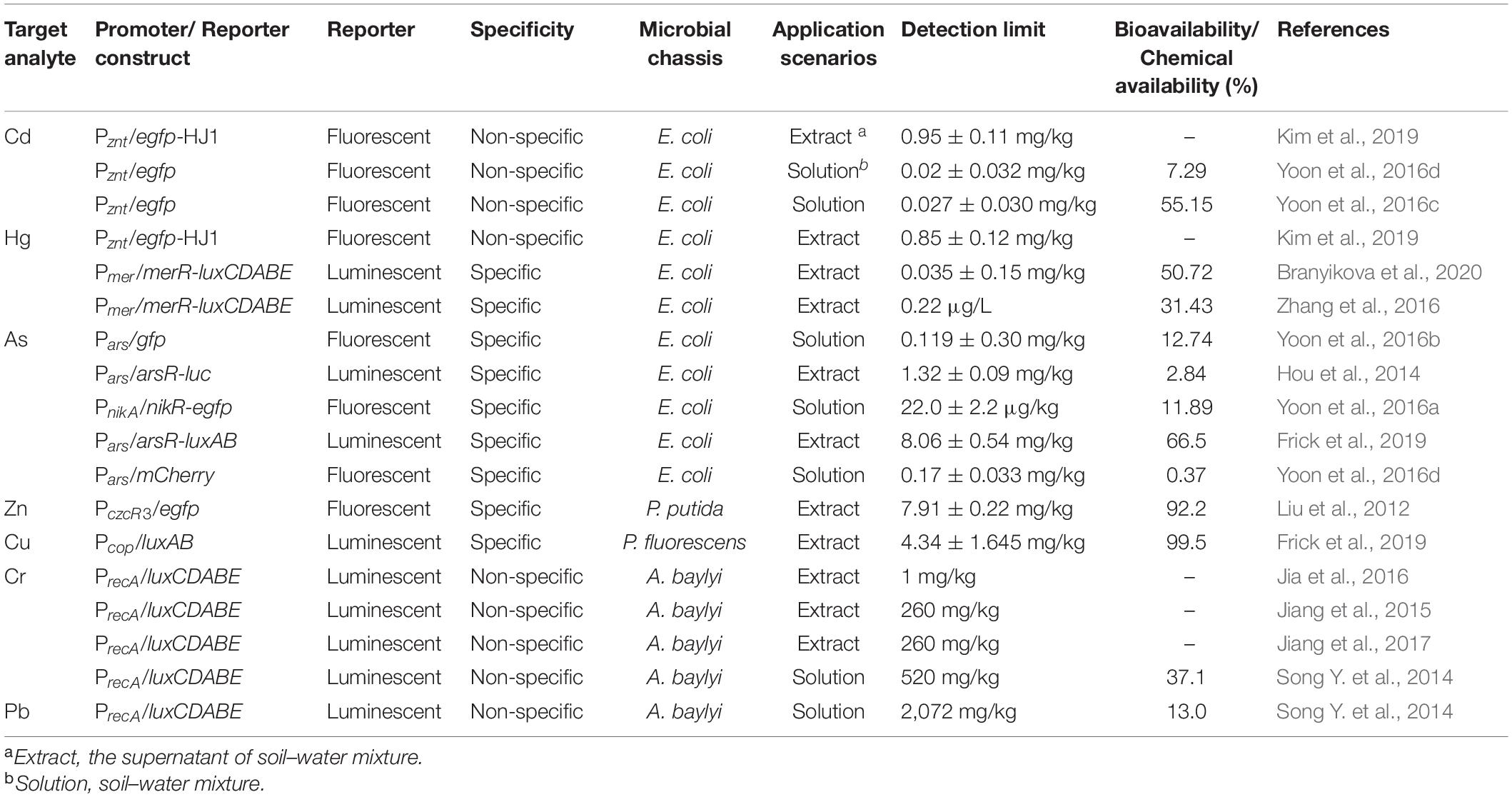
Early studies have demonstrated metal-sensing bioreporters with a nanomolar detection limit in aqueous samples (Selifonova et al., 1993; Corbisier, 1997). Recent works employed bacteria such as E. coli, P. putida, and A. baylyi to detect heavy metals in the soil environment (Table 1; Bilal and Iqbal, 2019). Both specific and non-specific metal-sensing bioreporters were constructed to measure Cd, Hg, As, Zn, Cu, Cr, and Pb in soil–water mixture (Table 1). Non-specific bioreporters using the stress-responsive promoter recA can respond to a range of heavy metal contaminants based on genotoxicity (Song Y. et al., 2014). The non-specific bioreporter with recA as sensing element has a detection limit of 1 mg/kg chromium (Jia et al., 2016). The sensitivity of non-specific bioreporters can be enhanced by using metalloproteins as sensor apparatuses. ZntR is one of the most commonly used non-specific transcriptional regulators, which responds to Zn2+, Cd2+, and Pb2+ (Table 1; Brown et al., 2003). E. coli bioreporter with PzntA-egfp has a detection limit as low as 0.02 mg/kg for Cd2+ in soil samples (Yoon et al., 2016c,d). With the aid of synthetic biology, the selectivity of non-specific regulators can be further enhanced via replacing the metal-binding loop. The bioreporters can recognize Cd2+ or Hg2+ specifically when the metal-binding loop of ZntR was replaced by cadmium- or mercury-binding sequences (Kang et al., 2018). In one recent work, the metal-binding loop (CTTCGCG) was inserted into a loop region of egfp to construct a split-protein system (Kim et al., 2019). Upon exposure to Cd and Hg, the binding of metal ions to the metal-binding loop induces a conformational change and restores the fluorescence.
The specific metal-binding regulators can increase both the selectivity and sensitivity of the bioreporter significantly. The metal-sensing repressors like ArsR and MerR function as sensing apparatuses to control the transcription of downstream operons (Kaur et al., 2015; Guo et al., 2019). Selective mercury-sensing system was incorporated in E. coli via using regulator protein MerR and promoter mer (Branyikova et al., 2020). The detection limit of the mercury-selective bioreporter is 0.035 mg/kg, which is not interfered by other toxic heavy metal ions (Branyikova et al., 2020). The arsenic-specific bioreporter with regulators NikR and ArsR exhibits good sensitivity with a detection limit of 22 μg/kg (Yoon et al., 2016a; Frick et al., 2019).
Due to the toxicity of heavy metal, detoxification genes are fused into regulatory regions to enhance the chassis heavy metal tolerance. For example, after incorporation of the metal transport protein-coding genes, such as zntA or cadA, microbes can adapt to severely contaminated environments (Biran et al., 2000). Fusion of Pb-binding protein PbrR with the outer membrane protein can allocate heavy metal-binding proteins on the cell surface, which alleviated the toxicity of accumulated Pb in the cell (Wei et al., 2014). On the contrary, deletion of metal exporter genes like copA can accumulate heavy metal intracellularly, which enhances the sensitivity of bioreporters with low metal exposure (Kang et al., 2018).
The signal transduction of microbes can be accomplished via electrochemical system (Dali et al., 2018). The electrochemical arsenic reporter was constructed in E. coli DH5α, which harbors a plasmid carrying ars operon promoter-controlled lacZ genes. With the presence of arsenate, β-galactosidase transforms p-aminophenyl-β-D-galactopyranoside into p-aminophenol, which can be detected electrochemically outside the cell (Cortes-Salazar et al., 2013). Whole-cell bioreporters can also be incorporated in microbial fuel cells to enable metal measurement. Zn-responsive bioreporter was constructed via fusing PzntA promoter with zntR, rib, and oprF. The presence of Zn triggered the riboflavin and porin production, which demonstrated a linear relationship between the maximum voltage with Zn concentration (Khan et al., 2020).
The bioreporters with metalloregulators like MerR and ZntR can detect μg/L of heavy metal ions in the soil–water mixture (Yoon et al., 2016d; Zhang et al., 2016). The corresponding detection limits of bioreporters for heavy metals like As, Hg, and Cd in soil are below mg/kg, which is comparable to that of inductively coupled plasma-atomic emission spectroscopy (ICP-AES) (Martin et al., 1992). The detection limits of whole-cell bioreporters are 10 to 100 times lower than the Chinese regulatory standards and in the same order of magnitude as the US EPA standards (EPA, 2002, 2012; MEE, 2018). Moreover, although the current soil quality standards are established based on chemical analysis, the bioavailable fraction of contaminants determines ecological and toxicological relevance. Previous studies have demonstrated that the bioavailability of heavy metals in soil was only about 10∼50% of chemical-extractable fractions (Table 1). Therefore, the whole-cell bioreporter is sufficiently sensitive and reliable to assess soil heavy metal contamination.
Organic Pollutants
Different from heavy metals, types of organic contaminants in soil are highly diverse. The first whole-cell bioreporter was constructed by inserting the lux operon into a naphthalene catabolic plasmid in P. fluorescens (King et al., 1990). The luminescence was induced when exposed to naphthalene or the regulatory inducer metabolite salicylate. Similarly, an alkane bioreporter was constructed using alkane-degrading A. baylyi ADP1 carrying transcriptional regulator alkR, promoter alkM, and reporter luxCDABE (Zhang et al., 2012). AlkR recognizes alkanes and activates luminescent gene expression. The bioreporter can stick to the interface of oil and water and emulsify crude oil into micron droplets, which facilitates the detection of the oil spills (Zhang et al., 2012).
Due to the diverse organic contaminants, the contaminant concentration can also be evaluated via the central metabolite. For example, most polycyclic aromatic hydrocarbon (PAH) metabolisms occur via the salicylate pathway. A whole-cell microbial bioreporter containing nahAD and luxCDABE gene was constructed to degrade and detect naphthalene (Table 2; Sun et al., 2017). The constitutive promoter Ptet is fused to catabolic gene nahAD to enhance the degradation of naphthalene to salicylate. Salicylate binds to the SalR regulator and activates the Psal promoter, which triggers the expression of salAR operon and luxCDABE gene (Sun et al., 2017). The bioreporter targeting salicylate exhibits good sensitivity, with a detection limit of 0.018 mg/kg. As bioreporter construction for different PAH molecules is laborious, quantification of the parent molecules via detection of metabolic intermediates is a feasible approach.
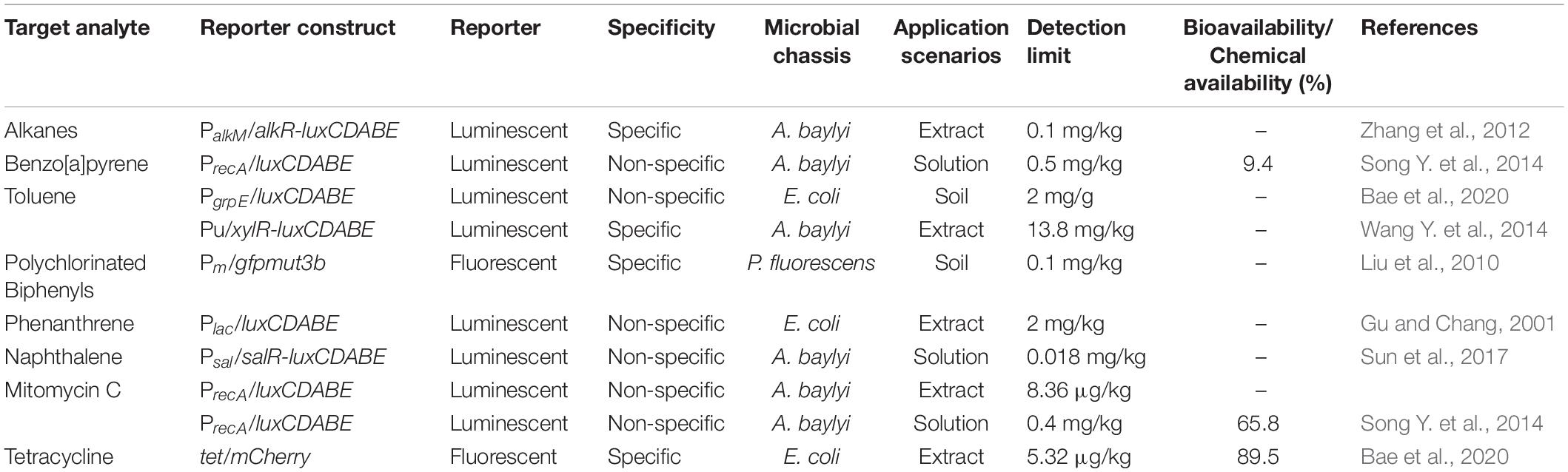
Table 2. Current microbial bioreporters designated for the detection of organic pollutants (PAHs, PCBs, pesticides, antibiotics).
Advances in cell immobilization are beneficial for in situ measurement. In one recent study, bioreporter cells are encapsulated into alginate beads to enhance the stability of bioreporters in the soil environment (Bae et al., 2020). The bioreporter can sense toluene via stress-responsive promoter PgrpE. The alginate beads were deployed on the surface of the soil to detect volatile contaminants like toluene, and the bioluminescence was measured via a fiber-optic probe. Bioelectrochemical systems can also be employed in organic contaminant detection, in which bioreporters were immobilized on electrodes (Boron et al., 2020). Recently, Boron et al. (2020) have developed a herbicide bioreporter based on the suppression of photosynthesis by atrazine. In this system, microalgae Monoraphidium contortum are fixed on the screen-printed graphite electrode. The decreased oxygen production can be measured by electrochemical methods (Boron et al., 2020).
Antibiotic residues in soil are emerging contaminants that impact soil microbial community and cause the accumulation of antibiotic resistance genes. Whole-cell microbial bioreporters can be constructed by fusing an antibiotic-sensitive promoter or an inducible gene promoter with a reporter gene (Parthasarathy et al., 2018; Ma et al., 2020). The previous whole-cell bioreporters were mainly used to detect tetracyclines in aqueous samples (Korpela et al., 1998; Kurittu et al., 2000). A recent tetracycline fluorescent bioreporter was constructed to analyze soil samples using tetRO tetracycline-responsive control region and gfp or mCherry reporter gene, which had a detection limit of 5.32 μg/kg soil (Bae et al., 2020). Using 96-well microplates, this fluorescent bioreporter method can measure 96 or more samples simultaneously within 6 h.
Previous studies have demonstrated that the sensitivity of bioreporters toward petroleum contaminants in aqueous samples has been improved down to the ppm and ppb range, approaching the sensitivities of chemical analysis like GC-MS (Jiang et al., 2020). The high sensitivity was retained in the soil–water mixture (Table 2; Zhang et al., 2012; Sun et al., 2017). Therefore, the bioreporter is sensitive enough to evaluate whether the concentration of contaminants like naphthalene exceeds the ppm-level regulatory standards (EPA, 2006; Kuppusamy et al., 2019). Meanwhile, the sensitivity still needs to be improved to evaluate organic pollutants (e.g., benzo[a]pyrene) with a sub-ppm regulatory limit.
Future Prospects
Whole-cell microbial bioreporters have exhibited promising performance in soil spiked with a single contaminant. But it is still challenging to differentiate multiple contaminants with a wide concentration range. The emergence of novel contaminants in soil also triggers requirements to construct new bioreporter modules. With the increasing capacity of high-throughput genomic sequencing, metagenome mining and synthetic biology provide new opportunities to explore new transcription factors or operators to improve the sensing modules (Jiang et al., 2016; Ho et al., 2018; Hicks et al., 2019). The development of novel genetic logic gates has the potential to efficiently modulate and transfer cellular signals to enhance the sensitivity and specificity while avoiding false-positive signals (Bansal et al., 2010). Applying alternative microbial chassis like basophils and halophiles enables the analysis in an extremely unfriendly environment.
In addition, significant efforts need to be undertaken to improve the measurement processes. Most previous contamination measurements were involved in soil solution or extract preparation. However, the pretreatment process alters the bioavailability of soil contaminants. For example, sonication of soil samples homogenizes and emulsifies hydrophobic organic pollutants (Jiang et al., 2017). Removal of soil particles from extracts compromised the accuracy of bioreporter detection (Magrisso et al., 2009; Jiang et al., 2017). One way to resolve this problem is to avoid the dilution/extraction step. Therefore, in situ detection based on bioelectrochemical systems and optic techniques will be necessary to advance the assessment of the actual contamination status, which soil bacteria are exposed to.
In situ measurement or monitoring in the field is difficult, which bears a risk to release genetically modified microorganisms. Different strategies have been attempted to envelop bacteria or fix microbial cells on the bottom of a microtiter plate using agar or alginate (Bjerketorp et al., 2006; Odaci et al., 2009; Ferro et al., 2012; Bae et al., 2020). Nanomaterials can be a promising alternative to further improve cell immobilization and signal transduction. For example, graphene-based materials have a large specific surface area with excellent thermal and electrical conductivity, which can adsorb organic contaminants for signal amplification (Zhang et al., 2010; Cho et al., 2020; Wu et al., 2020a). These unique properties make graphene-based electrodes a good candidate to immobilize bioreporters and transduce electrical signals (Li et al., 2017). The electrochemistry of target contaminants and electrochemically active microorganisms can further improve the selectivity and sensitivity.
The portability of bioreporter systems requires to be increased to complete all the analyses and data interpretation in the field. The flexibility of the bioreporter permits its easy adaptation to miniaturized and autonomous devices. For example, the visual signals from bioreporters in microfluidic devices can be captured by the miniaturized photomultiplier detectors (Roggo and van der Meer, 2017). The electrochemical bioreporter can be easily integrated into field-applicable electric devices (Ben-Yoav et al., 2009; Boron et al., 2020). The integration of emerging disciplines such as artificial intelligence, big data, smart mobile devices, and drones will be necessary to close the gap between research and field application and realize long-term online monitoring of soil contamination.
Author Contributions
NZ and YW reviewed the literature, designed the concept, wrote the manuscript, and prepared the figures. WC, QH, and PC revised the manuscript. All authors approved the manuscript for publication.
Funding
This work was supported by the National Key Research Program of China (2020YFC1806802), the National Natural Science Foundation of China (41807024), and the Fundamental Research Funds for the Central Universities (Program No. 52902-0900201674).
Conflict of Interest
The handling editor declared a past co-authorship with one of the authors YW.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bae, J. W., Seo, H. B., Belkin, S., and Gu, M. B. (2020). An optical detection module-based biosensor using fortified bacterial beads for soil toxicity assessment. Anal. Bioanal. Chem. 412, 3373–3381. doi: 10.1007/s00216-020-02469-z
Bansal, K., Yang, K., Nistala, G. J., Gennis, R. B., and Bhalerao, K. D. (2010). A positive feedback-based gene circuit to increase the production of a membrane protein. J. Biol. Eng. 4, 1–7.
Ben-Yoav, H., Biran, A., Pedahzur, R., Belkin, S., Buchinger, S., Reifferscheid, G., et al. (2009). A whole cell electrochemical biosensor for water genotoxicity bio-detection. Electrochim. Acta 54, 6113–6118. doi: 10.1016/j.electacta.2009.01.061
Bierkens, J., Klein, G., Corbisier, P., Van Den Heuvel, R., Verschaeve, L., Weltens, R., et al. (1998). Comparative sensitivity of 20 bioassays for soil quality. Chemosphere 37, 2935–2947. doi: 10.1016/s0045-6535(98)00334-8
Bilal, M., and Iqbal, H. M. N. (2019). Microbial-derived biosensors for monitoring environmental contaminants: recent advances and future outlook. Process Saf. Environ. Prot. 124, 8–17. doi: 10.1016/j.psep.2019.01.032
Biran, I., Babai, R., Levcov, K., Rishpon, J., and Ron, E. Z. (2000). Online and in situ monitoring of environmental pollutants: electrochemical biosensing of cadmium. Environ. Microbiol. 2, 285–290. doi: 10.1046/j.1462-2920.2000.00103.x
Bispo, A., Jourdain, M. J., and Jauzein, M. (1999). Toxicity and genotoxicity of industrial soils polluted by polycyclic aromatic hydrocarbons (PAHs). Org. Geochem. 30, 947–952. doi: 10.1016/s0146-6380(99)00078-9
Bjerketorp, J., Hakansson, S., Belkin, S., and Jansson, J. K. (2006). Advances in preservation methods: keeping biosensor microorganisms alive and active. Curr. Opin. Biotechnol. 17, 43–49. doi: 10.1016/j.copbio.2005.12.005
Boron, I., Juárez, A., and Battaglini, F. (2020). Portable microalgal biosensor for herbicide monitoring. ChemElectroChem 7, 1623–1630. doi: 10.1002/celc.202000210
Branyikova, I., Lucakova, S., Kuncova, G., Trogl, J., Synek, V., Rohovec, J., et al. (2020). Estimation of Hg(II) in soil samples by bioluminescent bacterial bioreporter E. coli ARL1, and the effect of humic acids and metal ions on the biosensor performance. Sensors 20:3138. doi: 10.3390/s20113138
Brown, N. L., Stoyanov, J. V., Kidd, S. P., and Hobman, J. L. (2003). The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163. doi: 10.1016/s0168-6445(03)00051-2
Burmølle, M., Hansen, L. H., and Sørensen, S. J. (2006). “Reporter gene technology in soil ecology; detection of bioavailability and microbial interactions,” in Nucleic Acids and Proteins in Soil, eds P. Nannipieri and K. Smalla (Berlin: Springer), 397–419. doi: 10.1007/3-540-29449-x_17
Cai, P., Liu, X., Ji, D., Yang, S., Walker, S. L., Wu, Y., et al. (2018). Impact of soil clay minerals on growth, biofilm formation, and virulence gene expression of Escherichia coli O157:H7. Environ. Pollut. 243, 953–960. doi: 10.1016/j.envpol.2018.09.032
Cai, P., Sun, X., Wu, Y., Gao, C., Mortimer, M., Holden, P. A., et al. (2019). Soil biofilms: microbial interactions, challenges, and advanced techniques for ex-situ characterization. Soil Ecol. Lett. 1, 85–93. doi: 10.1007/s42832-019-0017-7
Cho, I. H., Kim, D. H., and Park, S. (2020). Electrochemical biosensors: perspective on functional nanomaterials for on-site analysis. Biomater. Res. 24, 1–12.
Corbisier, P. (1997). Bacterial metal-lux biosensors for a rapid determination of the heavy metal bioavailability and toxicity in solid samples. Res. Microbiol. 148, 534–536. doi: 10.1016/s0923-2508(97)88360-x
Cortes-Salazar, F., Beggah, S., Van Der Meer, J. R., and Girault, H. H. (2013). Electrochemical As(III) whole-cell based biochip sensor. Biosens. Bioelectron. 47, 237–242. doi: 10.1016/j.bios.2013.03.011
Dali, M., Zinoubi, K., Chrouda, A., Abderrahmane, S., Cherrad, S., and Jaffrezic-Renault, N. (2018). A biosensor based on fungal soil biomass for electrochemical detection of lead (II) and cadmium (II) by differential pulse anodic stripping voltammetry. J. Electroanal. Chem. 813, 9–19. doi: 10.1016/j.jelechem.2018.02.009
Daunert, S., Barrett, G., Feliciano, J. S., Shetty, R. S., Shrestha, S., and Smith-Spencer, W. (2000). Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev. 100, 2705–2738. doi: 10.1021/cr990115p
Elväng, A. M., Westerberg, K., Jernberg, C., and Jansson, J. K. (2001). Use of green fluorescent protein and luciferase biomarkers to monitor survival and activity of Arthrobacter chlorophenolicus A6 cells during degradation of 4–chlorophenol in soil. Environ. Microbiol. 3, 32–42. doi: 10.1046/j.1462-2920.2001.00156.x
EPA (2002). Supplemental Guidance for Developing Soil Screening Levels for Superfund Sites. Washington, DC: OSWER.
EPA (2006). Application of Equilibrium Partitioning Theory to Soil PAH concentrationS. Washington, DC: EPA.
Ferro, Y., Perullini, M., Jobbagy, M., Bilmes, S. A., and Durrieu, C. (2012). Development of a biosensor for environmental monitoring based on microalgae immobilized in silica hydrogels. Sensors 12, 16879–16891. doi: 10.3390/s121216879
Francisco, R., Branco, R., Schwab, S., Baldani, I., and Morais, P. V. (2019). Two plant-hosted whole-cell bacterial biosensors for detection of bioavailable Cr(VI). World J. Microbiol. Biotechnol. 35:129.
Frick, H., Tardif, S., Kandeler, E., Holm, P. E., and Brandt, K. K. (2019). Assessment of biochar and zero-valent iron for in-situ remediation of chromated copper arsenate contaminated soil. Sci. Total Environ. 655, 414–422. doi: 10.1016/j.scitotenv.2018.11.193
Gu, M. B., and Chang, S. T. (2001). Soil biosensor for the detection of PAH toxicity using an immobilized recombinant bacterium and a biosurfactant. Biosens. Bioelectron. 16, 667–674. doi: 10.1016/s0956-5663(01)00230-5
Guo, M., Du, R., Xie, Z., He, X., Huang, K., Luo, Y., et al. (2019). Using the promoters of MerR family proteins as “rheostats” to engineer whole-cell heavy metal biosensors with adjustable sensitivity. J. Biol. Eng. 13, 1–9.
Gupta, N., Renugopalakrishnan, V., Liepmann, D., Paulmurugan, R., and Malhotra, B. D. (2019). Cell-based biosensors: recent trends, challenges and future perspectives. Biosens. Bioelectron. 141:111435. doi: 10.1016/j.bios.2019.111435
Gutierrez, J. C., Amaro, F., and Martin-Gonzalez, A. (2015). Heavy metal whole-cell biosensors using eukaryotic microorganisms: an updated critical review. Front. Microbiol. 6:48. doi: 10.3389/fmicb.2015.00048
Haft, R. J., Mittler, J. E., and Traxler, B. (2009). Competition favours reduced cost of plasmids to host bacteria. ISME J. 3, 761–769. doi: 10.1038/ismej.2009.22
Harmsen, J. (2007). Measuring bioavailability: from a scientific approach to standard methods. J. Environ. Qual. 36, 1420–1428. doi: 10.2134/jeq2006.0492
Hernández-Sánchez, V., Molina, L., Ramos, J. L., and Segura, A. (2016). New family of biosensors for monitoring BTX in aquatic and edaphic environments. Microb. Biotechnol. 9, 858–867. doi: 10.1111/1751-7915.12394
Hicks, M., Bachmann, T. T., and Wang, B. (2019). Synthetic biology enables programmable cell-based biosensors. ChemPhysChem 21, 132–144. doi: 10.1002/cphc.201900739
Ho, J. C. H., Pawar, S. V., Hallam, S. J., and Yadav, V. G. (2018). An improved whole-cell biosensor for the discovery of lignin-transforming enzymes in functional metagenomic screens. ACS Synth. Biol. 7, 392–398. doi: 10.1021/acssynbio.7b00412
Hou, Q. H., Ma, A. Z., Lv, D., Bai, Z. H., Zhuang, X. L., and Zhuang, G. Q. (2014). The impacts of different long-term fertilization regimes on the bioavailability of arsenic in soil: integrating chemical approach with Escherichia coli arsRp::luc-based biosensor. Appl. Microbiol. Biotechnol. 98, 6137–6146. doi: 10.1007/s00253-014-5656-0
Hurdebise, Q., Tarayre, C., Fischer, C., Colinet, G., Hiligsmann, S., and Delvigne, F. (2015). Determination of zinc, cadmium and lead bioavailability in contaminated soils at the single-cell level by a combination of whole-cell biosensors and flow cytometry. Sensors 15, 8981–8999. doi: 10.3390/s150408981
IOS (2008). Soil Quality. Requirements and Guidance for the Selection and Application of Methods for the Assessment of Bioavailability of Contaminants in Soil and Soil Materials. Geneva: IOS.
Ivask, A., Virta, M., and Kahru, A. (2002). Construction and use of specific luminescent recombinant bacterial sensors for the assessment of bioavailable fraction of cadmium, zinc, mercury and chromium in the soil. Soil Biol. Biochem. 34, 1439–1447. doi: 10.1016/s0038-0717(02)00088-3
Jacobson, A. R., Dousset, S., Andreux, F., and Baveye, P. C. (2007). Electron microprobe and synchrotron x-ray fluorescence mapping of the heterogeneous distribution of copper in high-copper vineyard soils. Environ. Sci. Technol. 41, 6343–6349. doi: 10.1021/es070707m
Jarque, S., Bittner, M., Blaha, L., and Hilscherova, K. (2016). Yeast biosensors for detection of environmental pollutants: current state and limitations. Trends Biotechnol. 34, 408–419. doi: 10.1016/j.tibtech.2016.01.007
Jia, J., Li, H., Zong, S., Jiang, B., Li, G., Ejenavi, O., et al. (2016). Magnet bioreporter device for ecological toxicity assessment on heavy metal contamination of coal cinder sites. Sens. Actuators B Chem. 222, 290–299. doi: 10.1016/j.snb.2015.08.110
Jia, X., Bu, R., Zhao, T., and Wu, K. (2019). Sensitive and specific whole-cell biosensor for arsenic detection. Appl. Environ. Microbiol. 85:e00694-19.
Jia, X., Zhao, T., Liu, Y., Bu, R., and Wu, K. (2018). Gene circuit engineering to improve the performance of a whole-cell lead biosensor. FEMS Microbiol. Lett. 365:fny157.
Jiang, B., Li, G., Xing, Y., Zhang, D., Jia, J., Cui, Z., et al. (2017). A whole-cell bioreporter assay for quantitative genotoxicity evaluation of environmental samples. Chemosphere 184, 384–392. doi: 10.1016/j.chemosphere.2017.05.159
Jiang, B., Song, Y., Liu, Z., Huang, W. E., Li, G., Deng, S., et al. (2020). Whole-cell bioreporters for evaluating petroleum hydrocarbon contamination. Crit. Rev. Environ. Sci. Technol. 51, 273–322.
Jiang, B., Zhu, D., Song, Y., Zhang, D., Liu, Z., Zhang, X., et al. (2015). Use of a whole-cell bioreporter, Acinetobacter baylyi, to estimate the genotoxicity and bioavailability of chromium(VI)-contaminated soils. Biotechnol. Lett. 37, 343–348. doi: 10.1007/s10529-014-1674-3
Jiang, H., Jiang, D., Shao, J., Sun, X., and Wang, J. (2016). High-throughput living cell-based optical biosensor for detection of bacterial lipopolysaccharide (LPS) using a red fluorescent protein reporter system. Sci. Rep. 6:36987.
Kang, Y., Lee, W., Jang, G., Kim, B.-G., and Yoon, Y. (2018). Modulating the sensing properties of Escherichia coli-based bioreporters for cadmium and mercury. Appl. Microbiol. Biotechnol. 102, 4863–4872. doi: 10.1007/s00253-018-8960-2
Katz, E. (2019). Boolean logic gates realized with enzyme-catalyzed reactions–unusual look at usual chemical reactions. ChemPhysChem 20, 9–22. doi: 10.1002/cphc.201800900
Kaur, H., Kumar, R., Babu, J. N., and Mittal, S. (2015). Advances in arsenic biosensor development–a comprehensive review. Biosens. Bioelectron. 63, 533–545. doi: 10.1016/j.bios.2014.08.003
Khan, A., Salama, E.-S., Chen, Z., Ni, H., Zhao, S., Zhou, T., et al. (2020). A novel biosensor for zinc detection based on microbial fuel cell system. Biosens. Bioelectron. 147:111763. doi: 10.1016/j.bios.2019.111763
Kim, H., Jang, G., and Yoon, Y. (2020). Specific heavy metal/metalloid sensors: current state and perspectives. Appl. Microbiol. Biotechnol. 104, 907–914. doi: 10.1007/s00253-019-10261-y
Kim, H., Lee, W., and Yoon, Y. (2019). Heavy metal(loid) biosensor based on split-enhanced green fluorescent protein: development and characterization. Appl. Microbiol. Biotechnol. 103, 6345–6352. doi: 10.1007/s00253-019-09908-7
Kim, H. J., Lim, J. W., Jeong, H., Lee, S. J., Lee, D. W., Kim, T., et al. (2016). Development of a highly specific and sensitive cadmium and lead microbial biosensor using synthetic CadC-T7 genetic circuitry. Biosens. Bioelectron. 79, 701–708. doi: 10.1016/j.bios.2015.12.101
Kim, R. Y., Yoon, J. K., Kim, T. S., Yang, J. E., Owens, G., and Kim, K. R. (2015). Bioavailability of heavy metals in soils: definitions and practical implementation–a critical review. Environ. Geochem. Health 37, 1041–1061. doi: 10.1007/s10653-015-9695-y
King, J., Digrazia, P., Applegate, B., Burlage, R., Sanseverino, J., Dunbar, P., et al. (1990). Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science 249, 778–781. doi: 10.1126/science.249.4970.778
Korpela, M. T., Kurittu, J. S., Karvinen, J. T., and Karp, M. T. (1998). A recombinant Escherichia coli sensor strain for the detection of tetracyclines. Anal. Chem. 70, 4457–4462.
Kumar, J., Jha, S. K., and D’souza, S. (2006). Optical microbial biosensor for detection of methyl parathion pesticide using Flavobacterium sp. whole cells adsorbed on glass fiber filters as disposable biocomponent. Biosens. Bioelectron. 21, 2100–2105. doi: 10.1016/j.bios.2005.10.012
Kuppusamy, S., Maddela, N. R., Megharaj, M., and Venkateswarlu, K. (2019). Total Petroleum Hydrocarbons: Environmental Fate, Toxicity, and Remediation. Cham: Springer.
Kurittu, J., Lönnberg, S., Virta, M., and Karp, M. (2000). A group-specific microbiological test for the detection of tetracycline residues in raw milk. J. Agric. Food Chem. 48, 3372–3377. doi: 10.1021/jf9911794
Li, S., Cheng, C., and Thomas, A. (2017). Carbon-based microbial-fuel-cell electrodes: from conductive supports to active catalysts. Adv. Mater. 29:1602547. doi: 10.1002/adma.201602547
Lim, W. A. (2010). Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol. 11, 393–403. doi: 10.1038/nrm2904
Liu, C., Yong, D., Yu, D., and Dong, S. (2011). Cell-based biosensor for measurement of phenol and nitrophenols toxicity. Talanta 84, 766–770. doi: 10.1016/j.talanta.2011.02.006
Liu, P., Huang, Q., and Chen, W. (2012). Construction and application of a zinc-specific biosensor for assessing the immobilization and bioavailability of zinc in different soils. Environ. Pollut. 164, 66–72. doi: 10.1016/j.envpol.2012.01.023
Liu, X., Germaine, K. J., Ryan, D., and Dowling, D. N. (2010). Whole-cell fluorescent biosensors for bioavailability and biodegradation of polychlorinated biphenyls. Sensors 10, 1377–1398. doi: 10.3390/s100201377
Liu, Z., Mukherjee, M., Wu, Y., Huang, Q., and Cai, P. (2020). Increased particle size of goethite enhances the antibacterial effect on human pathogen Escherichia coli O157: H7: a raman spectroscopic study. J. Hazard. Mater. 2020:124174. doi: 10.1016/j.jhazmat.2020.124174
Lu, Y., Song, S., Wang, R., Liu, Z., Meng, J., Sweetman, A. J., et al. (2015). Impacts of soil and water pollution on food safety and health risks in China. Environ. Int. 77, 5–15. doi: 10.1016/j.envint.2014.12.010
Ma, Z., Liu, J., Li, H., Zhang, W., Williams, M. A., Gao, Y., et al. (2020). A fast and easily parallelizable biosensor method for measuring extractable tetracyclines in soils. Environ. Sci. Technol. 54, 758–767. doi: 10.1021/acs.est.9b04051
Magrisso, S., Belkin, S., and Erel, Y. (2009). Lead bioavailability in soil and soil components. Water Air Soil Pollut. 202, 315–323. doi: 10.1007/s11270-009-9978-y
Martin, T., Brockhoff, C., Creed, J., and Long, S. (1992). Determination of metals and trace elements in water and wastes by inductively coupled plasma-atomic emission spectrometry. Methods Determin. Metals Environ. Samples 7, 33–91.
MEE (2018). Soil Environmental Quality–Risk Control Standard for Soil Contamination of Agricultural Land (GB15618-2018). Beijing: MEE.
NRC (2003). Bioavailability of Contaminants in Soils and Sediments: Processes, Tools, and Applications. Washington, DC: National Academies Press.
Odaci, D., Timur, S., and Telefoncu, A. (2009). A microbial biosensor based on bacterial cells immobilized on chitosan matrix. Bioelectrochemistry 75, 77–82. doi: 10.1016/j.bioelechem.2009.01.002
Paitan, Y., Biran, I., Shechter, N., Biran, D., Rishpon, J., and Ron, E. Z. (2004). Monitoring aromatic hydrocarbons by whole cell electrochemical biosensors. Anal. Biochem. 335, 175–183. doi: 10.1016/j.ab.2004.08.032
Parthasarathy, R., Monette, C. E., Bracero, S., and Saha, M. (2018). Methods for field measurement of antibiotic concentrations: limitations and outlook. FEMS Microbiol. Ecol. 94:fiy105.
Polyak, B., Bassis, E., Novodvorets, A., Belkin, S., and Marks, R. S. (2001). Bioluminescent whole cell optical fiber sensor to genotoxicants: system optimization. Sens. Actuators B Chem. 74, 18–26. doi: 10.1016/s0925-4005(00)00707-3
Qu, C., Chen, W., Hu, X., Cai, P., Chen, C., Yu, X. Y., et al. (2019). Heavy metal behaviour at mineral-organo interfaces: mechanisms, modelling and influence factors. Environ. Int. 131:104995. doi: 10.1016/j.envint.2019.104995
Rasmussen, L. D., Sørensen, S. J., Turner, R. R., and Barkay, T. (2000). Application of a mer-lux biosensor for estimating bioavailable mercury in soil. Soil Biol. Biochem. 32, 639–646. doi: 10.1016/s0038-0717(99)00190-x
Reyes-Caballero, H., Campanello, G. C., and Giedroc, D. P. (2011). Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys. Chem. 156, 103–114. doi: 10.1016/j.bpc.2011.03.010
Roggo, C., and van der Meer, J. R. (2017). Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 45, 24–33. doi: 10.1016/j.copbio.2016.11.023
Saltepe, B., Kehribar, E. S., Su Yirmibesoglu, S. S., and Safak Seker, U. O. (2018). Cellular biosensors with engineered genetic circuits. ACS Sens. 3, 13–26. doi: 10.1021/acssensors.7b00728
Schmid, G., Zeitvogel, F., Hao, L., Ingino, P., Adaktylou, I., Eickhoff, M., et al. (2016). Submicron-scale heterogeneities in nickel sorption of various cell-mineral aggregates formed by Fe(II)-oxidizing bacteria. Environ. Sci. Technol. 50, 114–125. doi: 10.1021/acs.est.5b02955
Selifonova, O., Burlage, R., and Barkay, T. (1993). Bioluminescent sensors for detection of bioavailable Hg (II) in the environment. Appl. Environ. Microb. 59, 3083–3090. doi: 10.1128/aem.59.9.3083-3090.1993
Shetty, R. S., Deo, S. K., Shah, P., Sun, Y., Rosen, B. P., and Daunert, S. (2003). Luminescence-based whole-cell-sensing systems for cadmium and lead using genetically engineered bacteria. Anal. Bioanal. Chem. 376, 11–17. doi: 10.1007/s00216-003-1862-9
Shin, H. J. (2011). Genetically engineered microbial biosensors for in situ monitoring of environmental pollution. Appl. Microbiol. Biotechnol. 89, 867–877. doi: 10.1007/s00253-010-2990-8
Silva-Rocha, R., and de Lorenzo, V. (2008). Mining logic gates in prokaryotic transcriptional regulation networks. FEBS Lett. 582, 1237–1244. doi: 10.1016/j.febslet.2008.01.060
Song, H., Ding, M.-Z., Jia, X.-Q., Ma, Q., and Yuan, Y.-J. (2014). Synthetic microbial consortia: from systematic analysis to construction and applications. Chem. Soc. Rev. 43, 6954–6981. doi: 10.1039/c4cs00114a
Song, Y., Jiang, B., Tian, S., Tang, H., Liu, Z., Li, C., et al. (2014). A whole-cell bioreporter approach for the genotoxicity assessment of bioavailability of toxic compounds in contaminated soil in China. Environ. Pollut. 195, 178–184. doi: 10.1016/j.envpol.2014.08.024
Song, Y., Li, G., Thornton, S. F., Thompson, I. P., Banwart, S. A., Lerner, D. N., et al. (2009). Optimization of bacterial whole cell bioreporters for toxicity assay of environmental samples. Environ. Sci. Technol. 43, 7931–7938. doi: 10.1021/es901349r
Sorensen, S. J., Burmolle, M., and Hansen, L. H. (2006). Making bio-sense of toxicity: new developments in whole-cell biosensors. Curr. Opin. Biotechnol. 17, 11–16. doi: 10.1016/j.copbio.2005.12.007
Su, L., Jia, W., Hou, C., and Lei, Y. (2011). Microbial biosensors: a review. Biosens. Bioelectron. 26, 1788–1799. doi: 10.1016/j.bios.2010.09.005
Sun, Y., Zhao, X., Zhang, D., Ding, A., Chen, C., Huang, W. E., et al. (2017). New naphthalene whole-cell bioreporter for measuring and assessing naphthalene in polycyclic aromatic hydrocarbons contaminated site. Chemosphere 186, 510–518. doi: 10.1016/j.chemosphere.2017.08.027
Turdean, G. L. (2011). Design and development of biosensors for the detection of heavy metal toxicity. Int. J. Electrochem. 2011, 1–15. doi: 10.4061/2011/343125
Valkova, N., Szittner, R., and Meighen, E. A. (1999). Control of luminescence decay and flavin binding by the LuxA carboxyl-terminal regions in chimeric bacterial luciferases. Biochemistry 38, 13820–13828. doi: 10.1021/bi991407n
Wang, B., Barahona, M., and Buck, M. (2013). A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens. Bioelectron. 40, 368–376. doi: 10.1016/j.bios.2012.08.011
Wang, B., and Buck, M. (2012). Customizing cell signaling using engineered genetic logic circuits. Trends Microbiol. 20, 376–384. doi: 10.1016/j.tim.2012.05.001
Wang, D., Zheng, Y., Fan, X., Xu, L., Pang, T., Liu, T., et al. (2020). Visual detection of Hg(2+) by manipulation of pyocyanin biosynthesis through the Hg(2+)-dependent transcriptional activator MerR in microbial cells. J. Biosci. Bioeng. 129, 223–228. doi: 10.1016/j.jbiosc.2019.08.005
Wang, X., Lu, X., and Chen, J. (2014). Development of biosensor technologies for analysis of environmental contaminants. Trends Environ. Anal. Chem. 2, 25–32. doi: 10.1016/j.teac.2014.04.001
Wang, Y., Zhang, D., Davison, P. A., and Huang, W. E. (2014). Bacterial whole-cell biosensors for the detection of contaminants in water and soils. Environ. Microbiol. 5, 155–168. doi: 10.1007/978-1-62703-712-9_13
Wei, W., Liu, X., Sun, P., Wang, X., Zhu, H., Hong, M., et al. (2014). Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ. Sci. Technol. 48, 3363–3371. doi: 10.1021/es4046567
Wu, Y., Jing, X., Gao, C., Huang, Q., and Cai, P. (2018). Recent advances in microbial electrochemical system for soil bioremediation. Chemosphere 211, 156–163. doi: 10.1016/j.chemosphere.2018.07.089
Wu, Y., Mohanty, A., Chia, W. S., and Cao, B. (2016). Influence of 3-chloroaniline on the biofilm lifestyle of Comamonas testosteroni and its implications on bioaugmentation. Appl. Environ. Microbiol. 82, 4401–4409. doi: 10.1128/aem.00874-16
Wu, Y., Xia, Y., Jing, X., Cai, P., Igalavithana, A. D., Tang, C., et al. (2020a). Recent advances in mitigating membrane biofouling using carbon-based materials. J. Hazard. Mater. 382:120976. doi: 10.1016/j.jhazmat.2019.120976
Wu, Y., Zaiden, N., Liu, X., Mukherjee, M., and Cao, B. (2020b). Responses of exogenous bacteria to soluble extracellular polymeric substances in wastewater: a mechanistic study and implications on bioaugmentation. Environ. Sci. Technol. 54, 6919–6928. doi: 10.1021/acs.est.0c00015
Xu, T., Close, D. M., Sayler, G. S., and Ripp, S. (2013). Genetically modified whole-cell bioreporters for environmental assessment. Ecol. Indic. 28, 125–141. doi: 10.1016/j.ecolind.2012.01.020
Yoon, Y., Kang, Y., Chae, Y., Kim, S., Lee, Y., Jeong, S. W., et al. (2016a). Arsenic bioavailability in soils before and after soil washing: the use of Escherichia coli whole-cell bioreporters. Environ. Sci. Pollut. Res. 23, 2353–2561. doi: 10.1007/s11356-015-5457-8
Yoon, Y., Kim, S., Chae, Y., Jeong, S. W., and An, Y. J. (2016b). Evaluation of bioavailable arsenic and remediation performance using a whole-cell bioreporter. Sci. Total Environ. 547, 125–131.
Yoon, Y., Kim, S., Chae, Y., Kang, Y., Lee, Y., Jeong, S. W., et al. (2016c). Use of tunable whole-cell bioreporters to assess bioavailable cadmium and remediation performance in soils. PLoS One 11:e0154506. doi: 10.1371/journal.pone.0154506
Yoon, Y., Kim, S., Chae, Y., Kim, S. W., Kang, Y., An, G., et al. (2016d). Simultaneous detection of bioavailable arsenic and cadmium in contaminated soils using dual-sensing bioreporters. Appl. Microbiol. Biotechnol. 100, 3713–3722.
Zhang, B., Sun, G., Zhu, Y., and Paton, G. I. (2016). Quantification of the bioreactive Hg fraction in Chinese soils using luminescence-based biosensors. Environ. Technol. Innov. 5, 267–276.
Zhang, D., He, Y., Wang, Y., Wang, H., Wu, L., Aries, E., et al. (2012). Whole-cell bacterial bioreporter for actively searching and sensing of alkanes and oil spills. Microb. Biotechnol. 5, 87–97.
Keywords: microbial bioreporter, environmental monitoring, soil contamination, heavy metal, organic contaminants, biosensor
Citation: Zeng N, Wu Y, Chen W, Huang Q and Cai P (2021) Whole-Cell Microbial Bioreporter for Soil Contaminants Detection. Front. Bioeng. Biotechnol. 9:622994. doi: 10.3389/fbioe.2021.622994
Received: 29 October 2020; Accepted: 22 January 2021;
Published: 23 February 2021.
Edited by:
Bin Cao, Nanyang Technological University, SingaporeReviewed by:
Dayi Zhang, Tsinghua University, ChinaShimshon Belkin, Hebrew University of Jerusalem, Israel
Copyright © 2021 Zeng, Wu, Chen, Huang and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yichao Wu, d3V5aWNoYW9AbWFpbC5oemF1LmVkdS5jbg==
 Ni Zeng
Ni Zeng Yichao Wu
Yichao Wu Wenli Chen1
Wenli Chen1 Qiaoyun Huang
Qiaoyun Huang Peng Cai
Peng Cai