- 1School of Interdisciplinary Bioscience and Bioengineering, Pohang University of Science and Technology, Pohang, South Korea
- 2Department of Convergence IT Engineering, Pohang University of Science and Technology, Pohang, South Korea
- 3Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, South Korea
- 4Institute of Convergence Science, Yonsei University, Seoul, South Korea
A wide variety of experimental models including 2D cell cultures, model organisms, and 3D in vitro models have been developed to understand pathophysiological phenomena and assess the safety and efficacy of potential therapeutics. In this sense, 3D in vitro models are an intermediate between 2D cell cultures and animal models, as they adequately reproduce 3D microenvironments and human physiology while also being controllable and reproducible. Particularly, recent advances in 3D in vitro biomimicry models, which can produce complex cell structures, shapes, and arrangements, can more similarly reflect in vivo conditions than 2D cell culture. Based on this, 3D bioprinting technology, which enables to place the desired materials in the desired locations, has been introduced to fabricate tissue models with high structural similarity to the native tissues. Therefore, this review discusses the recent developments in this field and the key features of various types of 3D-bioprinted tissues, particularly those associated with blood vessels or highly vascularized organs, such as the heart, liver, and kidney. Moreover, this review also summarizes the current state of the three categories: (1) chemical substance treatment, (2) 3D bioprinting of lesions, and (3) recapitulation of tumor microenvironments (TME) of 3D bioprinting-based disease models according to their disease modeling approach. Finally, we propose the future directions of 3D bioprinting approaches for the creation of more advanced in vitro biomimetic 3D tissues, as well as the translation of 3D bioprinted tissue models to clinical applications.
Introduction
The establishment of effective methods for disease treatment and prevention requires a clear understanding of pathophysiological phenomena. Various experimental models that mimic human physiology have been developed to test the safety and efficacy of potential therapeutics (Benam et al., 2015; Weinhart et al., 2019). Conventional 2D cell culture models are the most widely used tool in laboratories and have thus provided critical insights into various research fields by reproducing fundamental biological functions or features in vitro. This approach is not only easy to control but also renders robust results in a quick and cost-efficient manners (Langhans, 2018). Moreover, the advent of stem cell engineering has enabled the generation of various lineages of human cells with specific genetic characteristics (Hoes et al., 2019). Nonetheless, cells on a 2D surface do not behave in the same way as they naturally do in a 3D microenvironment (Amelian et al., 2017; Langhans, 2018). On the other hand, animal models can be used to characterize complex pathophysiological mechanisms in vivo. However, although these models have greatly contributed to our current understanding of various diseases and potential treatments, in vivo experiments are less reproducible due to inter-individual variations and are more cost- and time-consuming compared to cell culture experiments (Healy, 2018). Efforts have been made to bridge the gap between humans and animals using humanized mouse models, which are implanted with functional human cells and tissues; however, this approach still entails important limitations due to species-specific differences (e.g., residual innate immune system, cytokines, and humoral responses) (Walsh et al., 2017). Moreover, ethical concerns exist regarding the use of animal models (Walker and Eggel, 2020). Therefore, the pressing need for alternative platforms to investigate human pathophysiology in vitro has led to the development of 3D in vitro tissue models.
Because 3D models are based on cell culture, those are not only easy to control and reproduce experimental conditions but also provide a 3D microenvironment that mimics the physiological microenvironment, thereby allowing cell-cell and cell-matrix interactions akin to the physiological ones. More importantly, this approach enables the modeling of human physiology when human-derived cell sources are used. Therefore, 3D in vitro tissues exhibit more natural cellular behaviors, morphology, and functions compared to conventional 2D cell culture models by mimicking the native microenvironment and cell scaffolding structures (Kim J. et al., 2020). Multicellular spheroid structures (hereinafter referred to simply as “spheroids”) allow the modeling of heterogeneous cell-cell interactions by aggregating multiple types of cells (Langhans, 2018; Ide et al., 2020; Kim M. et al., 2020). Although spheroids can resemble the characteristics of native tissue in terms of cellular behaviors including proliferation, differentiation, maturation, and migration, simulating complex structures using these models can be quite challenging (Chatzinikolaidou, 2016; Hoes et al., 2019; Salaris and Rosa, 2019). 3D in vitro models constructed using microengineering such as micropattern and microfluidic channels enable the spatial organization of cells with micron-scale precision (Bhatia and Ingber, 2014; Laurent et al., 2017). This approach allows cells to self-organize by providing the geometric cues of the native tissue. However, more advanced fabrication methods are required to guide geometric cell morphology such as convoluted tubules, chamber-like structures, and lobule-like structures, which would further enhance the structural maturity and function of 3D bioprinted tissues by reproducing the characteristics of tissue-specific analogs (Jang et al., 2018; Kim M. et al., 2020; Yong et al., 2020). Given these requirements, 3D bioprinting has been considered a promising fabrication method capable of placing biomaterials and cell-laden biomaterials, bioink, in the desired locations. Many studies have reported that 3D bioprinted tissue improved tissue/organ function by mimicking complex native tissue architectures (Choi et al., 2019; Kim M. et al., 2020; Kim J. et al., 2020; Yong et al., 2020).
Vascularization plays a pivotal role in achieving sizeable and complex in vitro models, because the tissue models with a thickness larger than 400 μm require vasculature to ensure cell viability. In addition, recapitulation of blood vessel is important to improve the similarity of tissue models by enriching the microenvironment and is important for observing crosstalk between blood vessels and organs (Auger et al., 2013; Pellegata et al., 2018). In this respect, 3D bioprinting technique facilitates the generation of vascularized in vitro tissue models because endothelial cells (ECs) can be positioned to form lumen structure effortlessly.
This review covers the latest trends in the fabrication of in vitro 3D bioprinted tissue models, as well as strategies to reproduce natural tissues/organs. Particularly, this review focused on blood vessels and highly vascularized organs such as the heart, liver, and kidney, as these organs are inherently related to the vascular system due to their specific metabolic activities and functions. Moreover, we provide a brief introduction to the representative functions and features of each organ and describe their relationship with blood vessels. We will then discuss 3D bioprinting approaches for the fabrication of 3D in vitro tissue models and the advantages of 3D bioprinting techniques for the recreation of physiological features of native organs. Furthermore, this review will also summarize the current state of disease modeling methods based on 3D bioprinted in vitro tissue models. Specifically, modeling approaches will be divided into three categories to facilitate their discussion: (1) chemical substance treatment, (2) 3D bioprinting of lesions, and (3) recapitulation of tumor microenvironments (TME). Finally, we discuss the remaining challenges of 3D bioprinting and propose potential strategies to achieve more realistic human pathophysiological features in vitro and translate 3D bioprinted tissue models to clinical applications.
Mimicking Physiological Characteristics Using 3D Bioprinted in vitro Models
Perfusable and Multi-Layered Blood Vessel Models in Various Sizes and Shapes
Blood vessels are responsible for the transport of substances such as nutrients, oxygen, hormones, and drugs to the cells, as well as the elimination of the metabolized substances from the cells. Moreover, blood vessels connect the organs of our body and allow them to interact. Blood vessels are typically classified as arteries, veins, and capillaries, and each vessel has its own unique characteristics in wall thickness, elasticity, lumen diameter, etc. (Dewhirst and Secomb, 2017; Jarvis, 2018). Existing in vitro blood vessel models were developed focusing on the transport of substances, and their functions are typically evaluated in terms of the barrier function and perfusion of vascular ECs. Although microfluidic techniques have been utilized to fabricate blood vessels by perfusing ECs through hollow channels, rectangular channels are not well-suited to mimic natural vascular structures as they lack structural complexity (Wong et al., 2012; Abudupataer et al., 2020; Cao et al., 2020).
3D bioprinting technology has enabled the fabrication of more realistic blood vessel models. Using sacrificial biomaterial ink, cylindrical hollow tubules were generated with perfusable vascular channels and multi-scale vascular networks (Bertassoni et al., 2014; Kolesky et al., 2014, 2016; Lee et al., 2014a,b). Moreover, ECs have been directly printed onto multi-layered blood vessels of different diameters using a co-axial printing approach (Jia et al., 2016; Gao et al., 2017; Pi et al., 2018).
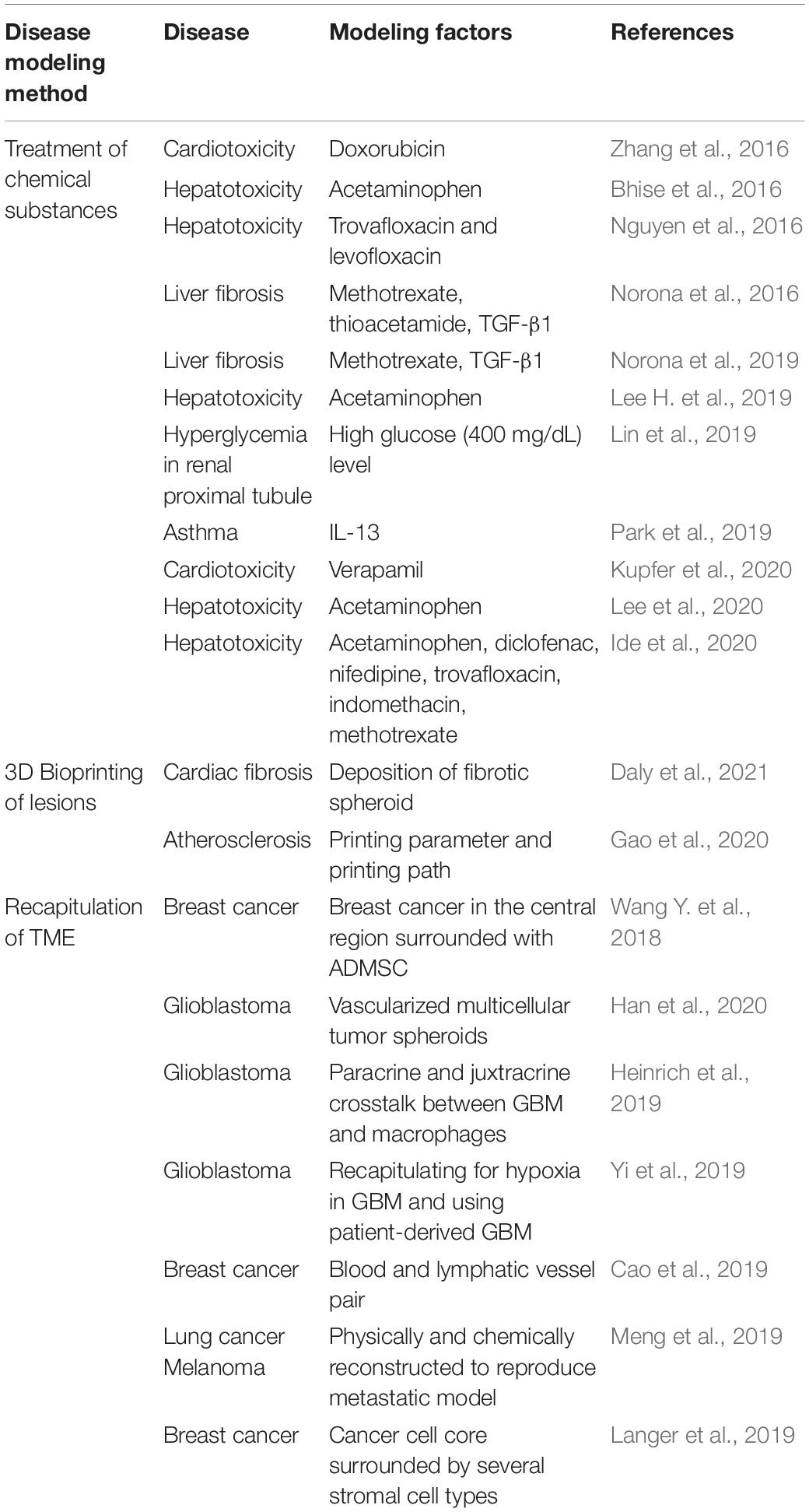
Gao et al. (2018) established a freestanding, perfusable, and functional in vitro vascular model using co-axial printing and blood vessel-derived ECM bioink. This approach allowed not only for the creation of straight structures but also complex endothelium patterns according to predesigned geometry (Figure 1A). Shear stress can also be induced by providing perfusion through the hollow channel, resulting in improved selective permeability, thrombogenic quiescence, and self-remodeling. The authors further induced angiogenic sprouting and endothelium dysfunction by treating the 3D printed tissues with proangiogenic growth factors and inflammatory cytokines, respectively, thus mimicking native blood vessel pathophysiology (Gao et al., 2018). This vascular model was further developed to create separate endothelial and muscular layers using tri-axial printing, which more closely mimics the native vessel layers (Gao et al., 2019). More recently, the authors employed 3D in-bath coaxial cell printing to construct a triple-layered vascular model composed of connective tissue, smooth muscle, and endothelium, thereby enhancing barrier function (Figure 1B; Gao et al., 2020).
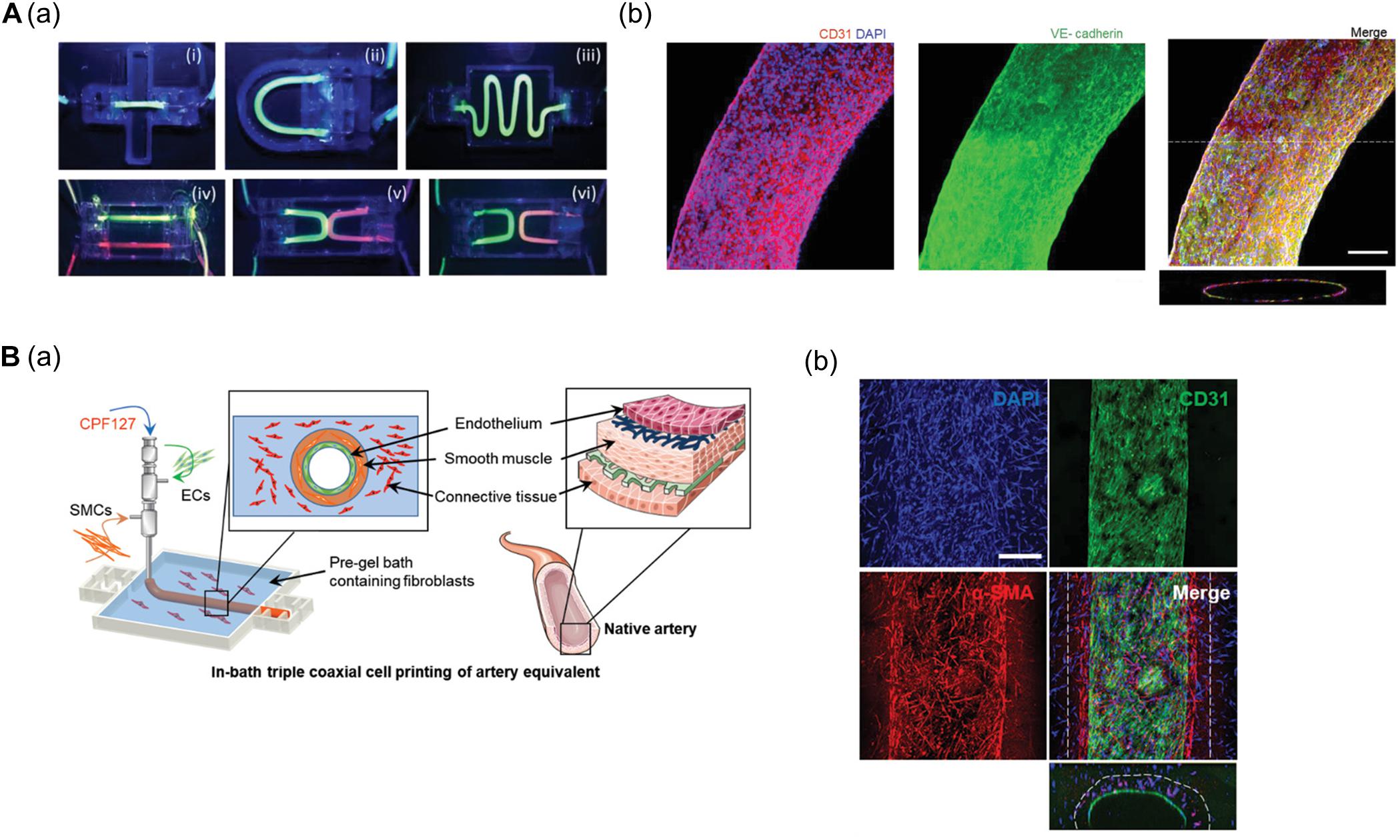
Figure 1. (A) Construction of perfusable and functional in vitro vascular models using co-axial cell printing. (a) Fabrication of various patterns of in vitro vascular models is demonstrated by perfusion of fluorescent dyes. (b) Expression of the cell-cell junction marker VE-cadherin is confirmed on day 7 in the printed vessels, indicating maturation of the vessels. Reprinted with permission from Gao et al. (2018). (B) Formation of triple-layered vascular model via in-bath triple co-axial cell printing. (a) Schematic describes printing procedure to produce triple-layered vascular model. (b) Immunofluorescence images show multi-layered blood vessels with endothelial cells (CD31) on the inside and surrounded by smooth muscle cells (α-SMA). Reprinted with permission from Gao et al. (2020).
Extrusion-based bioprinting produces anatomically relevant vascular structures; however laser-based printing technologies [e.g., selective laser sintering (SLS), stereolithography (SLA), digital light processing (DLP), and 2-photon polymerization (2PP)] are more suitable for the production of capillary-sized microvessels that require higher printing resolution (Moroni et al., 2018a,b; Miri et al., 2019; Mota et al., 2020). Therefore, several studies have employed LAB (laser-associated bioprinting) to fabricate capillary-sized microvascular networks (Arakawa et al., 2017; Miri et al., 2019). Furthermore, dynamic conditions such as the cyclic movements of alveoli and coronary arteries are also important features of the physiological environment of blood vessels, and efforts have been made to mimic these movements in vitro (Huh et al., 2010; Stucki et al., 2018; Nam et al., 2020). For instance, Grigoryan et al. (2019) created a vascularized alveolar model that mimics complex microvessel networks using LAB and reproduces blood flow and oxygen delivery according to cyclic alveolar movements. The authors perfused deoxygenated red blood cells (RBCs) through the vessel inlet and allowed them to flow to an adjacent channel supplemented with oxygen via cyclic alveolar movements. The color of the RBCs shifted from dark to bright red, which indicated that the cells successfully performed gas exchange (Grigoryan et al., 2019).
As described above, 3D bioprinting techniques have proven to be a crucial strategy to mimic multi-layered blood vessel models. Particularly, these techniques allow for the production of blood vessel models with great structural complexity, as well as the generation of capillary-sized microvessel networks under tissue-specific microenvironments. Vascular morphology may vary due to tissue-specific differences; therefore, the capacity of ECs to generate a barrier varies dramatically among organs involved in absorption and filtration. Moreover, organ-specific vascular systems differ not only in permeability but also in their ability to deliver nutrients to tissues. For example, in energy-intensive organs such as the heart, ECs control nutrient delivery by adjusting capillary density. For example, EC permeability in the blood-brain barrier is highly selective, whereas endothelial permeability is very high in liver sinusoidal ECs (Potente and Mäkinen, 2017). Therefore, the production of organ-specific blood vessels and structurally robust and physiologically enhanced blood vessels can be a promising tool to connect 3D bioprinted organs. This approach would allow for a deeper understanding of organ-organ crosstalk and other physiological conditions such as absorption, distribution, metabolism, excretion, and cancer metastasis.
Chamber-Like Cardiac Tissue Models Exhibiting Volume-Pressure Relationships
The heart is inherently related to the body’s blood vessel network. This organ is responsible for blood circulation throughout the body, supplying nutrients and oxygen and removing waste products through arteries and veins. To circulate blood, the myocardial muscle bundles are uniquely oriented. The myocardium consists of the endocardium, mid-wall, and epicardium, and these three parts are aligned in different directions to maximize the contraction. Moreover, given that the heart walls are composed of thick muscle bundles, they need to be supplied with sufficient oxygen and nutrients through the coronary arteries (Loukas et al., 2009; Lee A. et al., 2019; Noor et al., 2019). Many in vitro engineered heart tissue (EHT) models that recapitulate the function and physiology of cardiac tissues have been developed. These models generally exhibit the contractility and electrophysiological properties of native cardiac tissues (Leonard et al., 2018; Zhao et al., 2019; Goldfracht et al., 2020). However, the ECs used for the vascularization of cardiac tissues should be configured to create more biomimetic vasculatures. Furthermore, the volumetric features that recapitulate cardiac chambers must also be reproduced to understand the heart function and diseases associated with blood pumping (e.g., ejection fraction) (Li et al., 2018; Macqueen et al., 2018; Lee A. et al., 2019; Kupfer et al., 2020).
3D printing contributed to the fabrication of EHTs with precisely located cardiomyocytes (CMs) and stromal cells [e.g., ECs (Zhang et al., 2016; Arai et al., 2018; Maiullari et al., 2018) and fibroblasts (Arai et al., 2018; Anil Kumar et al., 2019; Daly et al., 2021)], thus mimicking the structural features of cardiac muscles and vasculatures (Wang Z. et al., 2018; Das et al., 2019; Yu et al., 2019). Furthermore, many studies have successfully developed volumetric cardiac chambers via support bath printing (Lee A. et al., 2019; Noor et al., 2019; Kupfer et al., 2020).
Noor et al. (2019) generated a 3D cardiac chamber using a support bath strategy coupled with CT images to aid in the design of the printed structures (Figure 2A). The resulting cardiac chamber featured two separate chambers and a major blood vessel structure composed of induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) and human umbilical vein endothelial cells (HUVECs), respectively. The separation of the chambers was demonstrated by filling them with different colored dyes. The authors also fabricated complex coronary artery networks using CAD data obtained from CT images. These experiments demonstrated that the resulting anatomically similar structure could supply oxygen to every area, taking into account the diffusion limits (Figure 2B; Noor et al., 2019). More recently, Kupfer et al. (2020) reported more advanced cardiac chamber constructs with two chambers and a vessel inlet and outlet with a high cell density. They supplemented photo-crosslinkable bioink (gelatin methacrylate and collagen methacrylate) with ECM components such as laminin-511/111 and fibronectin to regulate induced pluripotent stem cell (iPSC) behaviors (e.g., proliferation and differentiation). iPSCs were then embedded in modified bioink and directly printed to construct the cardiac chamber. The iPSCs then proliferated to a sufficient cell density and differentiated into CMs in situ. Remarkably, the cardiac chamber exhibited 3D features of native cardiac tissue including perfusion between chambers, volume-pressure relationships, and electromechanical functions, all of which are essential to the study of cardiac pathophysiology (Kupfer et al., 2020).
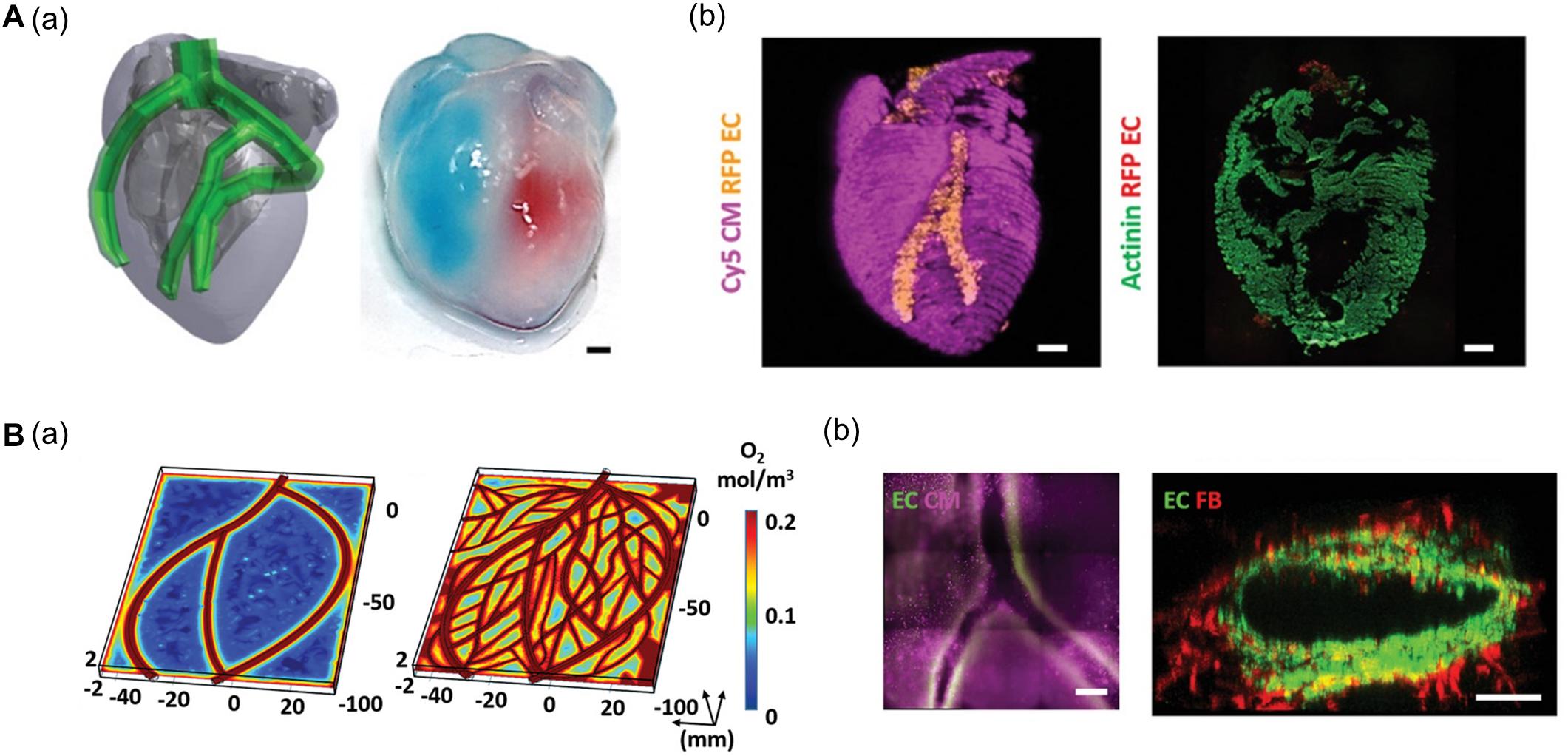
Figure 2. (A) Representative images of cardiac models composed of chamber and major vessel. (a) CAD design and printed structure of the cardiac chamber show the compartment of the left and right ventricle. (b) Confocal image of printed cardiac chamber indicates the spatial organization of CM (pink) and EC (orange) and cross-sectioned immunostaining showed internal compartmented CM (green) and EC (red) structure. (B) Characteristic of coronary artery network. (a) Supplemented blood vessels of coronary artery network exhibit improved oxygen diffusion via oxygen concentration profiles. (b) Immunofluorescence images show the branching and lumen structure of the coronary artery structure. Reprinted with permission from Noor et al. (2019).
Although 3D bioprinted cardiac models have been proven too closely reproduce the volumetric features of natural tissues, their capacity to produce a volume differential is insufficient to mimic the pumping motion of the heart. Therefore, strategies to reproduce blood ejection are required. In this context, increasing the contractile force of cardiac tissue can enhance the movement of the cardiac chamber. Moreover, improving the maturity of CMs and increasing the thickness of the cardiac tissue can enhance the contractility of the cardiac chamber models at the cellular and tissue levels, respectively. Moreover, mimicking the anisotropic orientation of the myocardium can elevate the contractile capacity of cardiac chambers. Therefore, reproducing these features would facilitate the study of the pathophysiology related to blood pump function and hemodynamics.
Hepatic Lobule-Like Models as Functional Units
The liver is an important organ that is largely responsible for the metabolic functions of the body (e.g., molecular anabolism, catabolism, and conversion and regulation of the energy balance), as well as detoxification and bile production. Two major blood vessels supply blood to the liver: 1) the hepatic artery and 2) the hepatic portal vein (Corsini and Bortolini, 2013; Kim J. et al., 2020; Ma et al., 2020). The liver is comprised of hexagonal structural and functional units called hepatic lobules which consist of a portal triad, hepatocytes arranged along a network of capillaries, and hepatic veins. Hepatic functions are highly specialized in each of the three zones of the hepatic lobules (zone 1: periportal; zone 2: midzonal area; zone 3: perivenous) (Ahn et al., 2019). Moreover, each zone has a unique microenvironment due to an oxygenation differential, which is determined by the distance from the hepatic arteries.
Current 3D in vitro liver tissue models have been developed to reproduce the 3D cell-cell/cell-matrix interaction, blood flow, and basic anatomical features of the liver, including hepatic zonation (Ahn et al., 2019; Collins et al., 2019). However, these features have been recreated as simple features or linear structures rather than complex and hexagonal structures. Therefore, additional studies are required to more properly reproduce the structural features of hepatic lobules including their complex vascular networks, zonal regions, and hexagonal functional units (Sacchi et al., 2020).
Using 3D bioprinting techniques, several studies have successfully fabricated hexagonal and compartmentalized structures to mimic the anatomy of hepatic lobules (Ma et al., 2016; Grix et al., 2018; Yu et al., 2019; Kang et al., 2020; Mao et al., 2020). Kang et al. (2020) generated a vascularized hepatic lobule structure using preset extrusion bioprinting (Figure 3A). The resulting hepatic tissue contained hepatocytes surrounded and compartmentalized by ECs with a lumen structure mimicking the vascular network of a native structure. Unlike simple 3D mixtures of hepatic and ECs, the printed lobule exhibited higher liver function (e.g., albumin secretion and urea production), albumin, MRP2, and CD31 expression, and CYP3A4 and CYP1A1 enzyme activities. Furthermore, they used this printed lobule as a structural unit to produce larger hepatic lobule arrays using layer-by-layer printing (Figure 3B).
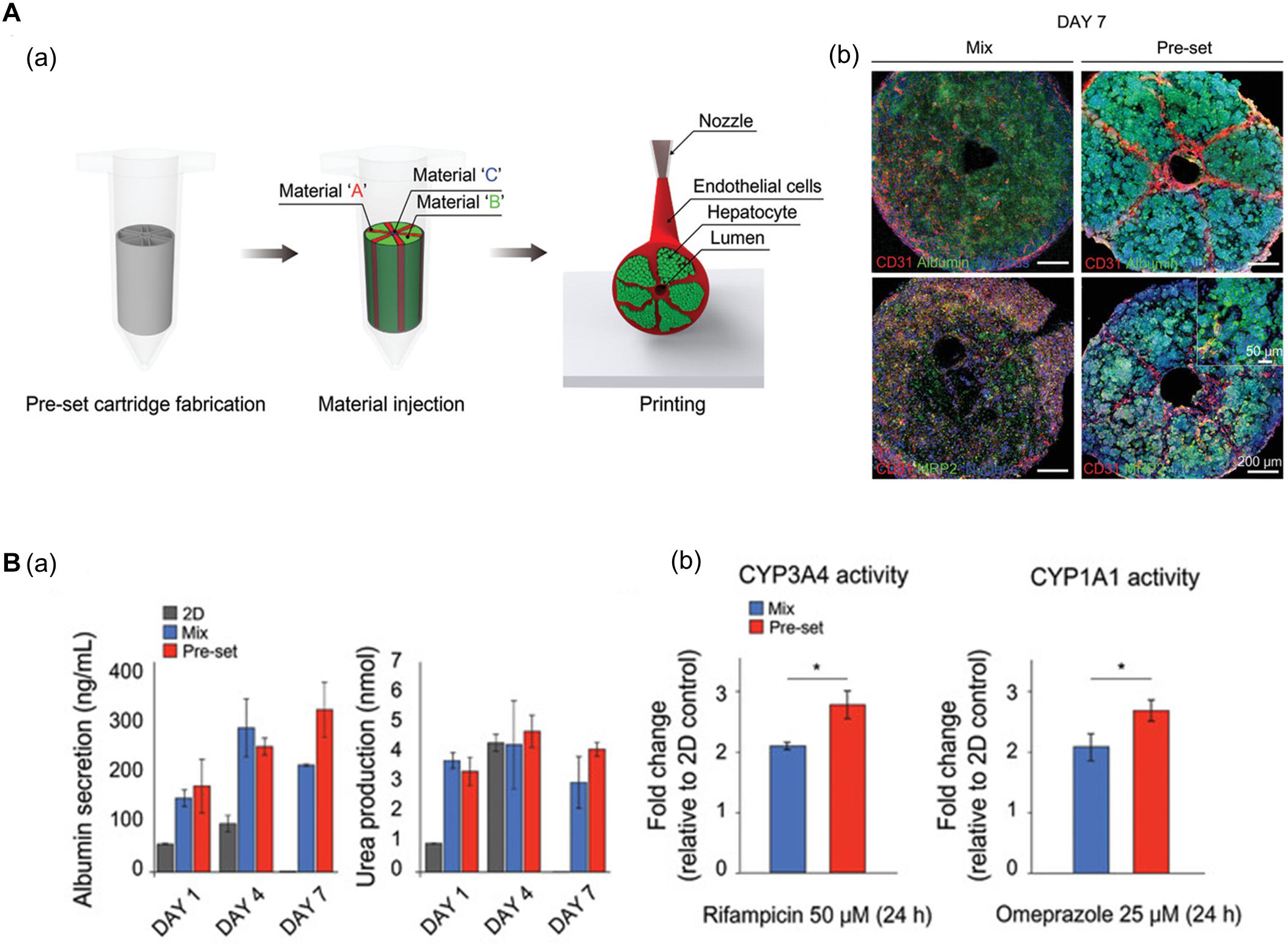
Figure 3. (A) Representative images of vascularized hepatic lobule structure. (a) The schematic of the preset extrusion bioprinting describes the printing strategy for hepatic lobules. (b) Immunofluorescence images of hepatic lobule construct using preset bioprinting exhibit well-preserved structural integrity not seen in the Mix group. (B) Evaluation of hepatic functions. (a) Albumin secretion and urea production measured by ELISA are enhanced in the printed hepatic lobule. (b) CYP3A4 and CYP1A1 enzyme activities are upregulated in printed lobule construct (*p < 0.05). Reprinted with permission from Kang et al. (2020).
Furthermore, Mao et al. (2020) employed a digital light processing (DLP)-based 3D bioprinting approach to generate liver microtissues using tissue-specific bioink. The hydrogel of decellularized ECM (dECM) from porcine liver was mixed with gelatin methacrylate to obtain a tissue-specific microenvironment. Moreover, the microtissues were designed to recapitulate the microenvironment for the cellular function of the internal hepatocytes, which are derived from human fibroblasts, by increasing the contact area to mimic the environment of a capillary network. The authors concluded that the use of tissue-specific bioink and DLP-based 3D bioprinting resulted in a corresponding reproduction of the microenvironment and structural features of native liver tissue, thus enhancing cell viability and liver functions such as albumin and urea secretion (Mao et al., 2020).
The structural and functional features of liver lobules and vascular networks have been well developed using 3D bioprinting technology. However, mimicking the heterogeneous functions of hepatocytes according to zonal location and the intricate network of blood vessels that irrigate the liver (e.g., hepatic arteries and hepatic portal veins) are still challenging. Differentiation of hepatocytes with functional heterogeneity could be achieved via an oxygen gradient (van Wenum et al., 2018; Tonon et al., 2019). Therefore, 3D bioprinting strategies to create hepatic lobule models should incorporate oxygen gradients. Specifically, the generation of perfusable vascular channels, which deliver oxygenated blood to hepatocytes, in anatomically accordant positions would enable the differentiation of hepatocytes into specialized cells. Furthermore, simulating other liver functions such as bile production would result in more realistic in vitro liver models.
Convoluted Renal Proximal Tubule Models With Vascular Interfaces
The kidney plays a major role in the filtration of various molecules and homeostasis regulation (Gupta et al., 2020). Nephrons, the functional units of the kidney, consist of Bowman’s capsule glomerulus, and tubules, such as the proximal and distal convoluted tubules and loop of Henle. Nephrons closely interact with blood vessels, such as the renal artery and vein (Wragg et al., 2019; Gupta et al., 2020). Given that kidney models are typically based on fluidic systems, several techniques are employed to manufacture 3D in vitro kidney models, including soft lithography, molding, and hollow fibers (Wilmer et al., 2016; Wragg et al., 2019; Zanetti, 2019; Singh et al., 2020). Although these models exhibited microfluidic conditions, they cannot fully mimic the complex tubule structure that may affect renal tubule cell performance in vitro (Wilmer et al., 2016; Wragg et al., 2019; Singh et al., 2020). 3D bioprinting research has largely focused on the fabrication of perfusable systems with convoluted structures to recapitulate the architecture and functions of native proximal tubules (Homan et al., 2016; Lin et al., 2019; Singh et al., 2020). Moreover, multiple renal cells could be elaborately positioned to mimic the complex cellular composition and functions of the native kidney tissues.
Homan et al. (2016) developed a convoluted proximal tubule using sacrificial Pluronic F-127 (PF-127) bioink (Figure 4A). A suspension of human proximal tubule epithelial cells (PTECs) was perfused through a channel after the removal of PF-127 to form the lumen structure. The PTECs formed a polarized epithelium and biologically relevant morphology (e.g., enhanced brush borders and cell height) under continuous flow which mimicked physiological shear stresses. Unlike PTEC monolayer models, the developed proximal tubule exhibited an improved albumin uptake, which is important for body fluid homeostasis (Homan et al., 2016).
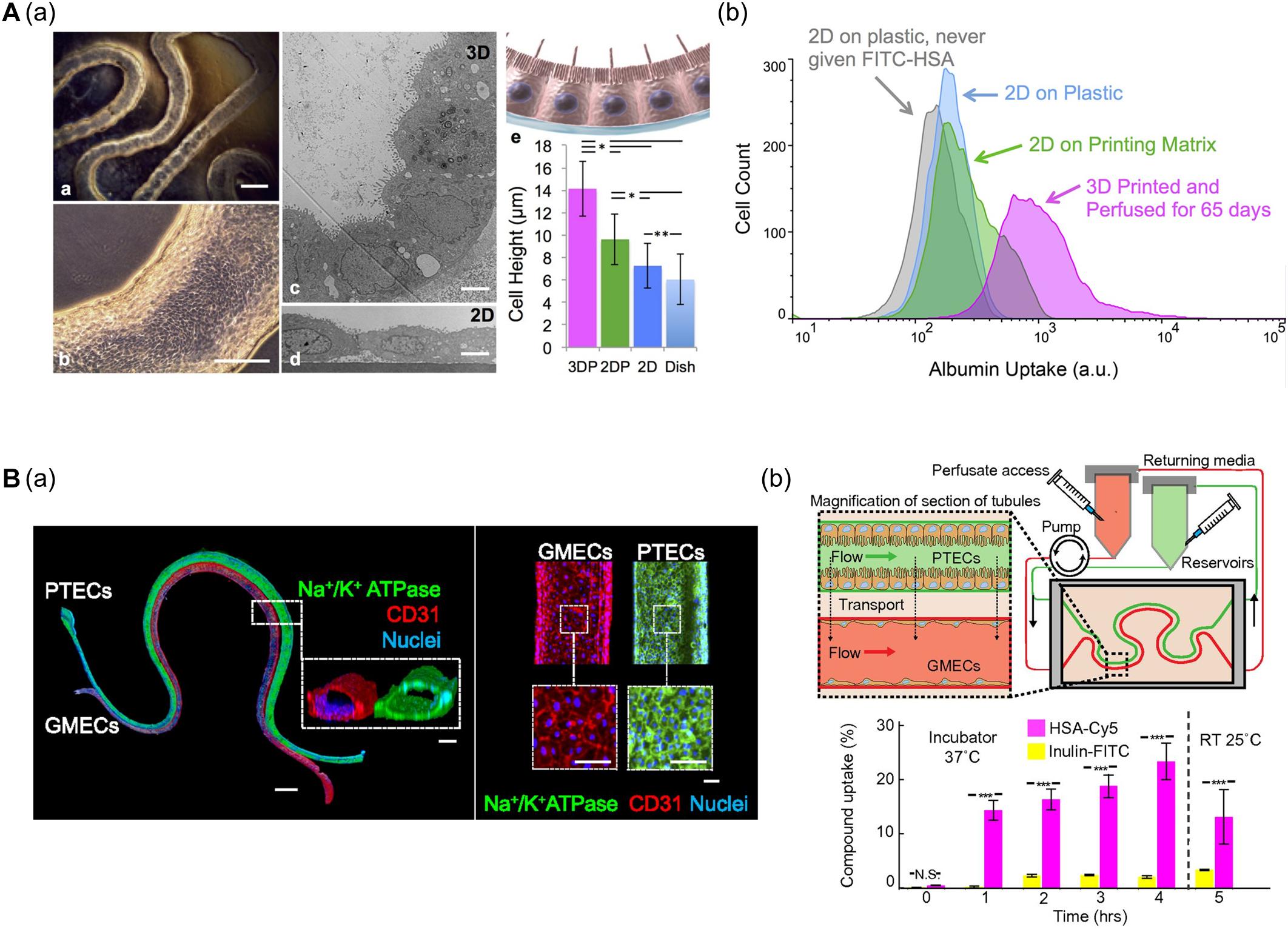
Figure 4. (A) Characteristics of 3D bioprinted convoluted renal proximal tubule structure. (a) Phase contrast and TEM images of printed proximal tubule models and enhanced cell height in 3D proximal tubule model. (b) Flow cytometry data demonstrated increased albumin uptake compared to 2D conditions (*p < 0.001; **p < 0.02). Reprinted with permission from Homan et al. (2016). (B) 3D renal proximal tubule with glomerular microvascular channels. (a) Whole-mount immunofluorescence images show the co-localized epithelium and endothelium with a distance of approximately 70 μm. (b) Schematic depicts a closed-loop perfusion system for assessment of renal resorption, and time-dependent measurements of albumin and insulin uptake performed by 3D printed proximal tubules indicate selective transport of albumin from PTEC to GMEC channels (N.S.: not significant; ***p < 0.0001). Reprinted with permission from Lin et al. (2019).
Lin et al. (2019) (i.e., the same research group) fabricated vascularized proximal tubule models that exhibited renal reabsorption of albumin and glucose via proximal tubule-glomerular microvascular exchange (Figure 4B). Using a similar strategy, proximal tubules and glomerular microvascular channels were created using channel-specific cells. The epithelium and endothelium were co-localized with a separation distance of approximately 70 μm. Active reabsorption was demonstrated by circulating specific solutes and drugs through each channel. Importantly, the proximal tubule selectively reabsorbed albumin when simultaneously perfused with albumin and inulin (Lin et al., 2019).
It has also been reported that the kidney cells are embedded into kidney tissue-derived dECM bioink to recapitulate a tissue-specific microenvironment, thereby improving the sensitivity of the 3D kidney models to drug-induced toxicity (Wilmer et al., 2016). Recently, Singh et al. (2020) introduced a co-axial nozzle-based direct cell printing strategy for proximal tubule fabrication using kidney dECM bioink. Based on this approach, renal PTEC- and HUVEC-laden bioinks were printed into hollow tubes. Moreover, this approach could potentially be used to fabricate a complex hollow tubule with both mono- and bi-layers in a single step. This strategy could be further utilized for the construction of complex renal tissues including both monolayered proximal tubules and bilayer glomerulus structures. Consistent with other studies, each hollow channel exhibited upregulation of tissue-specific gene expression compared to their 2D culture counterparts. Furthermore, both tubes displayed an adequate barrier function and albumin transport from the proximal tubules to the blood vessels (Singh et al., 2020).
The in vitro models of renal proximal tubules with perfusable and complex tubular constructs have been developed using 3D bioprinting technologies. These studies have mainly focused on reabsorption and transportation functions, both of which occur in proximal tubules and glomeruli. 3D bioprinting provides a promising means to construct a complete in vitro nephron model consisting not only of proximal tubules but also other structures such as Bowman’s capsule, the loop of Henle, and distal tubules. 3D bioprinting technology has been developed to utilize multiple bioinks, thus enabling the creation of models using bioinks corresponding to each part of the nephron. In addition, printing parameters can be adjusted to simulate various diameters and shapes (e.g., straight or convoluted) for each tubule. Furthermore, a 3D bioprinting approach employing a co-axial nozzle can generate double- or triple-layer tubules to create multilayer tubule structures. Taken together, generation of complete in vitro nephron models can provide important insights into the pathophysiological characteristics of native kidney tissues.
Disease Models
As described above, 3D bioprinting enables the creation of structurally and physiologically comparable 3D in vitro tissue models. Based on the advantages of 3D bioprinted tissue models, many researchers have exposed chemicals (e.g., drugs and cytokines), construct lesions, and use patient-derived cells to investigate and test the pathophysiological properties of the disease (Table 1).
Chemically Induced Disease Modeling
Exposure to bioactive compounds such as drugs and cytokines to induce tissue injuries (e.g., cardiotoxicity, hepatotoxicity, and renal toxicity) or other diseases (e.g., fibrosis and inflammation) is the easiest and most widely used method for disease modeling. Disease modeling methods based on drug-induced toxicity can be used to ensure that the 3D in vitro models respond appropriately to drugs or other bioactive compounds, thus highlighting the potential of 3D bioprinted tissue as a promising drug testing platform. Therefore, compounds that are already known to induce organ dysfunction or anticancer drugs with severe side effects are often used (Bhise et al., 2016; Nguyen et al., 2016; Norona et al., 2016, 2019; Zhang et al., 2016; Lee H. et al., 2019; Lee A. et al., 2019; Lee et al., 2020; Ide et al., 2020; Kupfer et al., 2020). Similarly, inflammatory factors such as inflammatory cytokines and other chemical substances are used to stimulate tissue models and induce pathological features such as inflammation and fibrosis (Norona et al., 2016, 2019; Lin et al., 2019; Park et al., 2019).
Several recent studies have reported that 3D tissue models can be used to realistically mimic disease states. Norona et al. (2016) generated a 3D bioprinted liver tissue model composed of compartmentalized hepatocytes and stromal cells including stellate cells and ECs. This model was then treated with several fibrogenic compounds such as TGF-β1, methotrexate, and thioacetamide to induce liver fibrosis (Norona et al., 2016). The authors further enhanced the biomimetic properties of the liver tissue model by adding Kupffer cells, a type of inflammatory cell that mainly populates intact liver tissues and is responsible for homeostasis (Figure 5A). Subsequently, the author observed the response of the liver tissue model to the integrated Kupffer cells after inducing fibrogenesis as described in previous studies. The Kupffer cells in the liver-tissue model were observed to behave almost similarly as the Kupffer cells in vivo, behaving as immune cells under pathophysiological conditions. Concretely, the presence of Kupffer cells shortened the release of lactate dehydrogenase, increased collagen deposition, and altered cytokine responses (Norona et al., 2019).
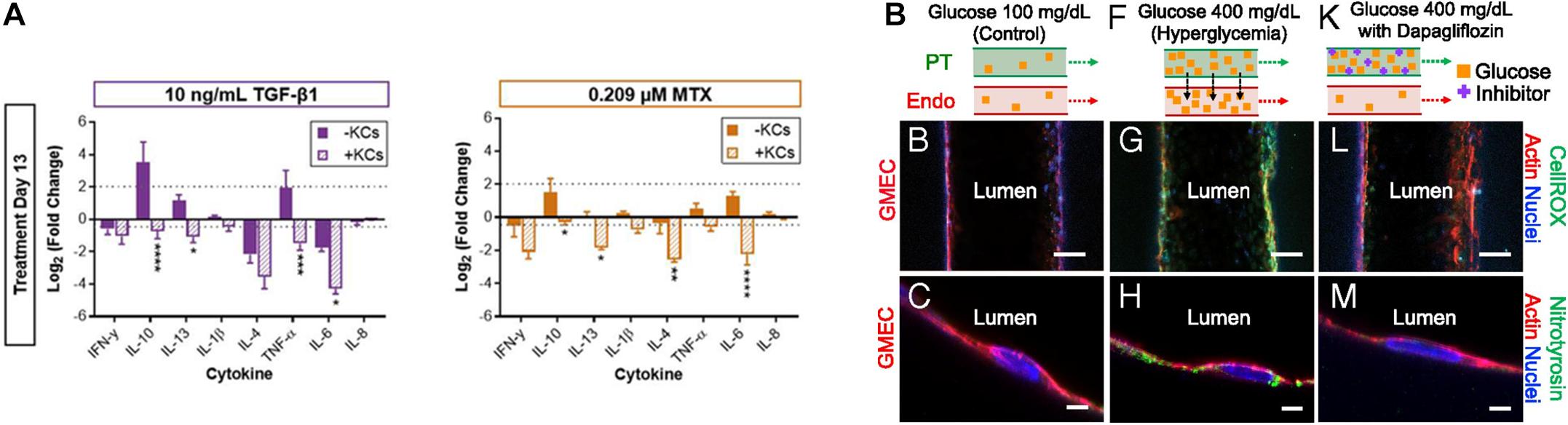
Figure 5. (A) Impact of Kupffer cells on the chemically induced inflammatory response of 3D bioprinted liver tissue models (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Reprinted with permission from Norona et al. (2019). (B) Pathophysiological features of GMEC lumen under normo- and hyperglycemic conditions indicated by immunofluorescen staining. Reprinted with permission from Lin et al. (2019).
As mentioned in the previous kidney tissue modeling section, Lin et al. (2019) established an in vitro proximal tubule model with a glomerular microvascular channel interface (Figure 5B). Furthermore, a reduction in glucose reabsorption was observed when Dapagliflozin [i.e., a type 2 diabetes medication known to inhibit glucose transporter (SGLT2) receptors] was administered, thus confirming that their model responded properly to the drug stimulus. They also induced their models to hyperglycemia by circulating a high glucose medium into the proximal tubule. Damage in both tubular channels was then observed, including a disruption of the cell-cell junction and lowered PTEC height. Interestingly, the vascular channel was restored when the tissue was treated with Dapagliflozin, which blocks glucose transport from the proximal tubule to the vasculature. These results indicated that the developed 3D printed tissues could serve as a platform for diabetes research.
3D Bioprinting of Disease Factors to Mimic Pathological Structures
In vitro disease models can be constructed by recreating the defining features of a specific disease using 3D bioprinting technology. Using this approach, spheroids can be deposited at precise locations as building blocks to create larger and more complex structures (Kizawa et al., 2017; Arai et al., 2018; Kim et al., 2018). Moreover, spheroids have also been utilized to model a variety of diseases such as fibrosis and tumor models with high cell densities. This approach has been used to generate oxygen gradients to recapitulate the disease microenvironment (Kim et al., 2018; Sacchi et al., 2020; Daly et al., 2021).
In this context, Daly et al. (2021) fabricated a ring-shaped EHT by arranging cardiac spheroids in a self-healing hydrogel bath and inducing the fusion of spheroids (Figure 6A). To mimic healthy and fibrotic heart tissue, they generated spheroids with varying ratios of iPSC-CMs and cardiac fibroblasts (CFs) (4:1 iPSC-CMs to CFs ratio for healthy tissues and 1:4 for scarred tissue). The authors then deposited spheroids (8 healthy spheroids for normal tissue; 1 scarred spheroid among 7 healthy spheroids for the disease model) to fabricate a ring-shaped EHT. The cardiac fibrosis model showed reduced contraction amplitudes and disrupted electrophysiological synchronization of EHTs. Additionally, the developed EHTs were used to study microRNA (miRNA) therapeutics aimed at cardiac regeneration after myocardial infarction. The cardiac fibrosis models to screen a range of treatment durations and assess the effects of miRNA treatment. Interestingly, the miRNA treatment enhanced iPSC-CM proliferation, leading to improvement of contractility and electrophysiological integration of scarred heart tissue (Daly et al., 2021).
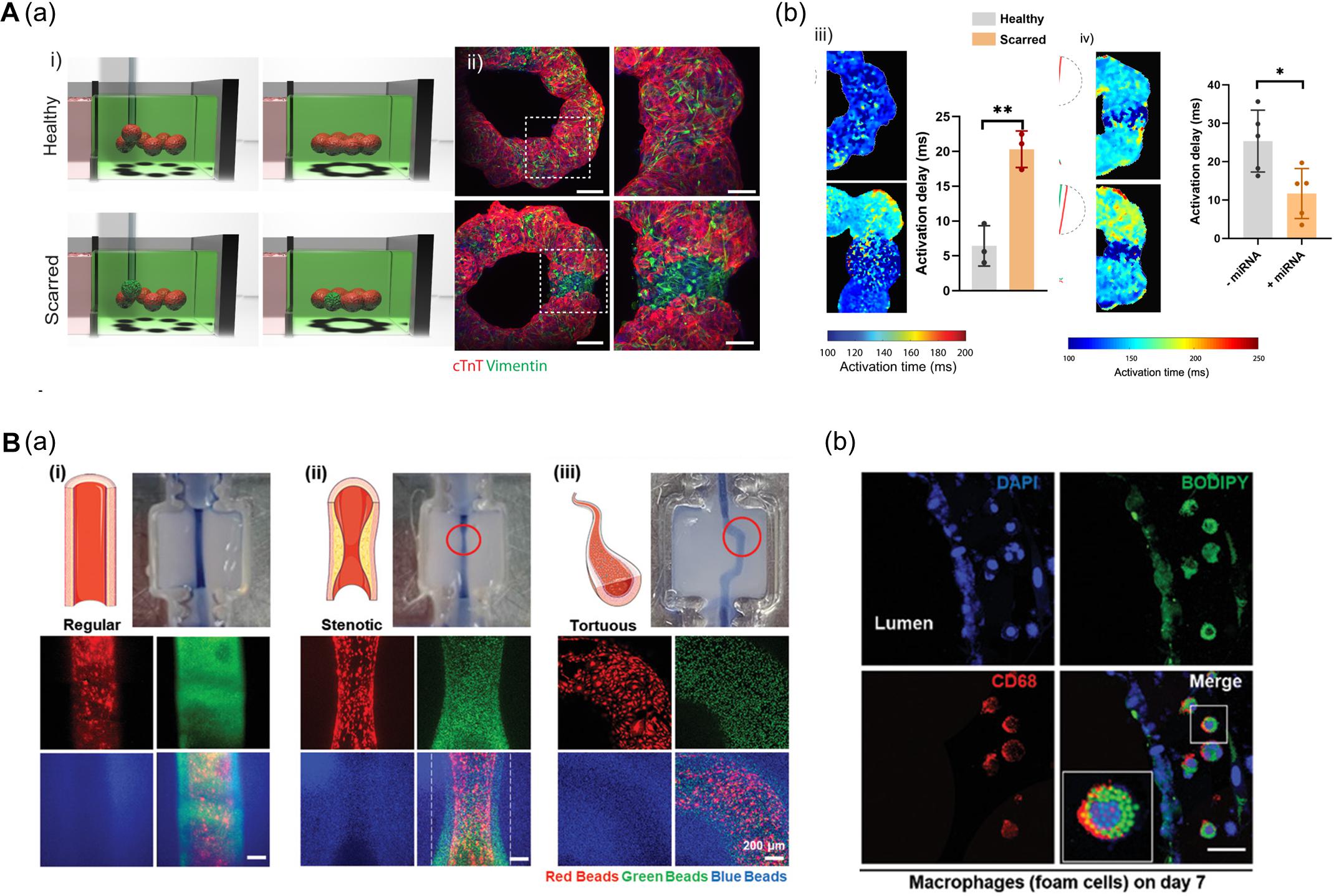
Figure 6. (A) 3D bioprinting of cardiac fibrosis models. (a) Schematic diagram of the procedure for creating a cardiac fibrosis model, and immunofluorescence image of cardiac fibrosis model. (b) Disrupted electrophysiological synchronization of cardiac models, and the therapeutic effect of miRNA treatment (*p < 0.05; **p < 0.01). Reprinted with permission from Daly et al. (2021). (B) Pathophysiological features of in vitro atherosclerosis model. (a) Structural features of the atherosclerosis model generated by adjusting the printing parameters and printing path are indicated by fluorescent beads. (b) Recapitulation of the early stages of atherosclerosis is verified by LDL accumulation and foam cell formation. Reprinted with permission from Gao et al. (2020).
Another approach to recapitulate the pathophysiological features of 3D bioprinted tissues is to adjust the printing parameters including pneumatic pressure and the nozzle moving speed. Even if a structure was printed on the same path, the details of the structure may vary due to changes in the printing parameters. Gao et al. (2020) applied a triple-layered vascular model to fabricate an atherosclerosis disease model (Figure 6B). Based on a normal vascular model, the authors generated a stenotic structure by adjusting printing parameters, and a tortuous structure was created by adjusting the printing path. Turbulent flows were generated in the stenotic and tortuous models and it was confirmed that the turbulent flows induced endothelial dysfunction. Moreover, the authors recapitulated early atherosclerosis conditions such as LDL accumulation and foam cell formation by circulating LDL and monocytes through the vessel, indicating the potential of triple-layered vascular models as promising tools for drug testing (Gao et al., 2020).
3D Bioprinting Approaches for the Recapitulation of Tumor Microenvironments
Cancer is a very complex malignant tissue that is composed of heterogeneous cell populations and a unique genetic specificity in each patient. Moreover, mutations may occur during cancer progression, thus potentially modifying the tumor tissue structure or that of surrounding tissues (Hass et al., 2020). The tumor microenvironment (TME) is a representative feature of cancer that affects several cancer properties such as proliferation, invasion, and metastasis (Spill et al., 2016). The TME is composed of many factors, including ECM, blood vessels, and cancer-associated cells (e.g., fibroblasts, macrophages), which may vary depending on the tissue and anatomical location. Therefore, recapitulating tissue-specific TMEs plays an important role in developing in vitro cancer models. Specifically, peripheral blood vessels are a crucial part of the TME, which sprout from the surrounding vessels and supply nutrients to the cancer. Additionally, metastasis occurs through the blood vessels. Therefore, reconstruction of the blood vessels is critical when developing in vitro cancer models. In vitro cancer modeling has largely focused on investigating cancer cells that interact with the TME using transwell co-culture systems, 3D hydrogel cultures, or microfluidic devices (Chung et al., 2017; Nashimoto et al., 2017; Lee et al., 2018; Nguyen et al., 2019; Haase et al., 2020). However, although these methods have successfully recapitulated both metastasis and the juxtracrine and paracrine effect induced by surrounding cells, the structural and cellular complexity of the TME remains largely unexplored.
On the other hand, 3D bioprinting techniques have been demonstrated to improve the complexity of TME models by simultaneously printing different cell types, including cancer cells, fibroblasts, immune cells, ECs, and ECM biomaterials using multiple highly precise heads (Zhao et al., 2014; Wang Y. et al., 2018; Langer et al., 2019). Moreover, mimicking tissue-specific TME heterogeneity and tumorigenesis is an important consideration for in vitro cancer modeling. For example, KRAS-mutant pancreatic ductal adenocarcinoma (PDAC) activates the epidermal growth factor receptor (EGFR) by inducing an autocrine feed-loop and amplifies the signals required for PDAC development. However, EGFR deletion in colorectal cancer cells and non-small lung cancer cells does not prevent tumor formation. Therefore, signal transduction pathway regulation may vary depending on cancer types and tissue origins (Schneider et al., 2017). Recently, bioprinted in vitro cancer models have been demonstrated to closely simulate the properties of actual cancers, including their defining phenotypes and pathological characteristics and distinct tissue-specific TME heterogeneity (Yi et al., 2019; Kim E. et al., 2020; Yoon et al., 2020).
Heinrich et al. (2019) developed a glioblastoma (GBM) model based on a mini-brain construct containing macrophages (Figure 7A). Specifically, a GBM area was printed on a section of the mini-brain construct. In this model, the authors observed the recruitment of macrophages in the GBM region and conversion to glioblastoma-associated macrophages, which was confirmed by the upregulation of typical phenotypes and related genes. Upon comparing the tissues from more than 150 patients with the 3D bioprinted tissues, it was confirmed that the behavior of macrophages in the proposed model was consistent with that of the patients, indicating its potential applicability in personalized medicine (Heinrich et al., 2019).
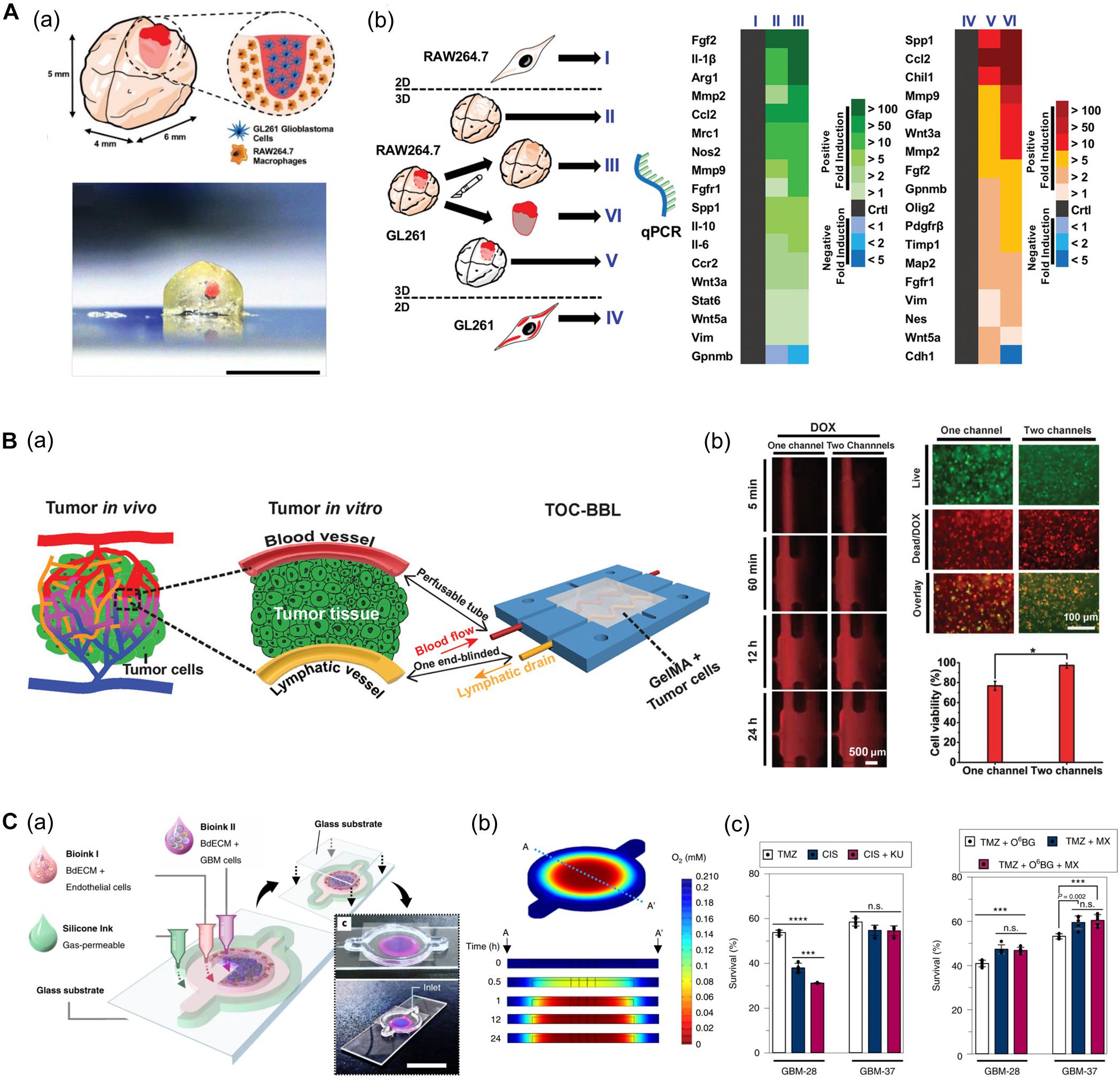
Figure 7. (A) GBM model based on a 3D bioprinted mini-brain construct containing macrophages. (a) Schematic and picture of GBM model exhibit GBM area in red on the bioprinted mini-brain. (b) Schematic illustrates the experimental conditions for the bioprinted RAW264.7/GL261 co-culture model and heatmap shows distinguished gene expression as a 2D monolayer (I and IV), 3D single bioprinted (II and V), and 3D bioprinted RAW264.7/GL261 co-culture model (III and VI) in RAW264.7 and GL261 glioblastoma. Reprinted with permission from Heinrich et al. (2019). (B) In vitro cancer model consisting of a pair of blood vessels and lymphatic vessels using 3D bioprinting. (a) Schematic shows the design of a bioprinted cancer model containing a pair of blood/lymphatic vessels that mimic actual TME. (b) Images on the left show the diffusion of DOX in a 1-channel/2-channel configuration, and the fluorescence images on the right exhibit cell viability for 24 h after DOX delivery (*p < 0.01). Reprinted with permission from Cao et al. (2019). (C) Application of patient-derived cell for in vitro GBM model mimicking hypoxic TME. (a) Schematic describes the design of GBM-on-a-chip with a compartmentalized cancer-vascular structure. (b) Colormap image of the oxygen gradient demonstrates the formation of central hypoxia in the GBM model. (c) Survival rate of patient-derived GBM treated with the drug combinations indicates patient-specific drug responses reflecting individual characteristics (n.s.: not significant; ***p < 0.001; ****p < 0.0001). Reprinted with permission from Yi et al. (2019).
Furthermore, Meng et al. (2019) established an in vitro metastatic model via 3D bioprinting techniques, which were used to reproduce the structure of relevant peripheral blood vessels and growth factors (e.g., epidermal growth factor and vascular endothelial growth factor) released from stimuli−responsive capsules. A molecular gradient of growth factors was modulated using 3D bioprinting to mimic the biochemical characteristics of the TME. Lung cancer cells migrated to the blood vessels under the guidance of the growth factor gradient, and infiltrated the vascular system. As a result, the key features of cancer metastasis including invasion, intravasation, and angiogenesis were faithfully simulated. In addition, the usefulness of this cancer metastasis model for drug screening application was proved by assessing the anticancer efficacy of immunotoxins (Meng et al., 2019).
Among the factors that characterize the TME, lymphatic vessels are among the most closely related to cancer metastasis along with blood vessels. Moreover, microcirculation systems from blood vessels to lymphatic vessels provide a privileged recycling route to most anticancer drugs in vivo. In this regard, Cao et al. (2019) generated an in vitro cancer model consisting of a pair of blood vessels and lymphatic vessels using 3D bioprinting (Figure 7B). The pair of blood vessels and lymphatic vessels featured varying levels of diffusion properties of biomolecules and anticancer drugs in breast cancer cells. Given that anticancer drugs are delivered to the cancer cells through blood vessels and removed through the lymphatic vessels, their anticancer properties were attenuated with the incorporation of lymphatic vessels compared to when only blood vessels were included (Cao et al., 2019).
In particular, patient-derived cells retain physiological properties of in vivo lesions and represent the unique characteristics of patients. Therefore, these models have been utilized to investigate the patient-specific efficacy of and resistance to drugs and treatments. In this regard, recent studies using patient-derived cells are increasing. Yi et al. (2019) developed a bioprinted GBM model featuring a hypoxic environment, a representative feature of GBM. To achieve this, the authors printed patient-derived GBM cells that were encapsulated in brain dECM bioink concentrically on the cores, and then printed ECs around a core structure (Figure 7C). This GBM model showed a compartmentalized cancer-vascular structure that reproduced hypoxic conditions with a radial oxygen gradient. Remarkably, the results obtained with this bioprinted GBM model were consistent with the actual patient-specific resistance to chemoradiation and temozolomide (TMZ) combination therapy. Furthermore, the authors confirmed the potential utility of this model to screen for drug combinations associated with more efficient tumor resection (Yi et al., 2019).
Given that the development of treatment resistance in cancer is a process that takes several months, long-term culture of in vitro cancer models should be considered. Ozturk et al. (2019) developed a 3D vascular GBM model using sacrificial extrusion bioprinting. The researchers used 10% gelatin to generate two fluid vascular channels, after which HUVECs were seeded onto the channels. Subsequently, patient-derived GBM spheroids transduced with mCherry-expressing lentivirus were embedded between the channels. Thereafter, 100 μM of TMZ was perfused through the channels after 26 days of culture, and the authors observed tumor cell degeneration in the infiltrating area after 14 days. Vascularization enabled the tumor spheroids to grow for 26 days. Then, the spheroids had been treated with drugs for 37 days, allowing investigation of tumor behavior for up to 70 days. Interestingly, some GBM cells survived treatment and showed resistance to treatment, thus resuming proliferation and matrix invasion despite continued drug treatment. Therefore, the study demonstrated the feasibility of culturing the GBM model under vascularized TME conditions for up to 2 months, which offers important advantages for the study of long-term cancer cell behavior in vitro. Moreover, the authors demonstrated that cancer cell overgrowth is a mechanism of long-term TMZ treatment resistance (Ozturk et al., 2019).
3D bioprinting techniques have enhanced the complexity of the TME of in vitro cancer models. These 3D bioprinted in vitro cancer models have provided insights into the importance of recapitulating complex TME features in cancer research, as they allow for a more corresponding representation of patient-specific drug responsiveness and reduced drug efficacy according to TME conditions. Moreover, the combination of in vitro cancer models with multi-organ platforms can be applied to study cancer metastasis including extravasation and mesenchymal to endothelial transition, as well as endothelial to mesenchymal transition, invasion, intravasation, and angiogenesis.
Conclusion and Future Outlook
3D bioprinting techniques have recently emerged as a promising tool to fabricate 3D in vitro tissue models through the precise arrangement of biomaterials and relevant cells based on their native tissue architecture (Choi et al., 2017; Kim J. et al., 2020). This review summarized the findings of recent studies related to 3D bioprinted tissue models that mimic blood vessel and various highly vascularized tissues, such as the heart, liver, and kidney. Although uncovered by this review, there are ongoing studies aiming to generate a 3D bioprinted intestine, lung, or skin model (Supplementary Table 1). Several studies have demonstrated the capacity of 3D bioprinting to recapitulate the geometric cues and complex cellular configurations of native tissues. Additionally, 3D bioprinting facilitates the research of tissue models, as it enables rapid prototyping, reproducibility, and repeatability. With this approach, many in vitro tissue models that recapitulate pathophysiological properties have been developed focused on basic functional or structural units of target tissues/organs.
Moving forward, several factors must be addressed to generate in vitro models with more biomimetic characteristics and to transform the in vitro system into clinical application. Therefore, to improve the quality of the printed constructs, future research must focus on the development of biomaterials with improved biofunctionality and printing fidelity, as well as fully functionalized cell sources that reflect personalized genotypes.
Scaling up the 3D bioprinted tissue models is a feasible goal in the near future. This could be achieved via modular tissue engineering, where small tissue modules (i.e., well-established 3D bioprinted functional units) are assembled to build large-scale engineered tissue models. Additionally, future models should incorporate more realistic vascularization in order to supply oxygen and nutrients to deeper cell layers beyond the diffusion limits.
Along with scale-up, the complete organ should in accomplished regarding cell composition as well as the anatomical structure. For now, among the various cell types, the application of organoids with cellular diversity could potentially improve cell composition restrictions. For instance, Kupfer et al. (2020) bioprinted iPSCs directly into a cardiac chamber structure and differentiated them into CMs. They observed that the developed system contained various cell types such as smooth muscle cells and ECs, as well as CMs, but not CFs. Although complete cell composition was not achieved in this study, the authors demonstrated that the combination of 3D bioprinting and organoid technology has the potential to advance the level of complexity of in vitro tissue models.
The organs in our body do not independent but interact continuously constantly interact with other organs and the environment through a variety of signals such as electricity, biomolecules, and endocrine signals. In this regard, many efforts have been made to develop multi-organ systems (organs-on-a-chip), and future breakthroughs in 3D bioprinting technology would contribute to the advancement of these initiatives. To achieve this, bioprinted tissue research should contemplate the inclusion of various channels (e.g., nerves, blood vessels, lymph vessels) to enable the interconnection of tissue models. This approach may facilitate the study of multi-organ crosstalk including absorption, distribution, metabolism, and elimination of drugs or biomolecules, as well as cancer metastasis.
Assessment techniques for in vitro tissue models play a key role in translating developed tissue models into clinical applications. Therefore, the assessment techniques must also be advanced to validate volumetric and large-scale tissue models according to the development of 3D tissue models (Yong et al., 2020). For example, echocardiography (a clinical procedure to verify cardiac function) has been applied to measure volume changes in an in vitro cardiac chamber model (Li et al., 2018; Macqueen et al., 2018; Kupfer et al., 2020). Other studies have validated bioprinted tissues using light-sheet microscopy and the tissue-clearing method. Importantly, these methods enable the non-destructive visualization and monitoring of volumetric samples and bioprinted constructs (Ding et al., 2018; Yong et al., 2020).
In terms of regulation, it is inadequate to determine the predictive capacity by comparing the gold standards used for in vivo testing with the results from in vitro tissue models on a one-to-one basis (Pridgeon et al., 2018). Thus, it may be reasonable to establish a new gold standard by integrating various predictive approaches including in silico models. In fact, the ongoing comprehensive in vitro proarrhythmia assay (CiPA) project aims to develop a new in vitro paradigm that can more accurately predict the cardiotoxicity of new drugs (Strauss et al., 2021). Therefore, these advanced in vitro tissue models must be extensively validated and documented to enable their widespread adoption in clinical applications.
Author Contributions
DGH and YC wrote the original draft and prepared the figures. JJ supervised the work and revised the draft. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of South Korea (NRF) funded by the Ministry of Education (No. 2020R1A6A1A03047902). This work also supported by the National Research Foundation of South Korea (NRF) grant funded by the Ministry of Science and ICT (No. 2021R1A2C2004981) and the Ministry of Education (NRF-2019-Global Ph.D. Fellowship Program).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.685507/full#supplementary-material
References
Abudupataer, M., Chen, N., Yan, S., Alam, F., Shi, Y., Wang, L., et al. (2020). Bioprinting a 3D vascular construct for engineering a vessel-on-a-chip. Biomed. Microdevices 22:10. doi: 10.1007/s10544-019-0460-3
Ahn, J., Ahn, J. H., Yoon, S., Nam, Y. S., Son, M. Y., and Oh, J. H. (2019). Human three-dimensional in vitro model of hepatic zonation to predict zonal hepatotoxicity. J. Biol. Eng. 13, 1–15. doi: 10.1186/s13036-019-0148-5
Amelian, A., Wasilewska, K., Megias, D., and Winnicka, K. (2017). Application of standard cell cultures and 3D in vitro tissue models as an effective tool in drug design and development. Pharmacol. Rep. 69, 861–870. doi: 10.1016/j.pharep.2017.03.014
Anil Kumar, S., Alonzo, M., Allen, S. C., Abelseth, L., Thakur, V., Akimoto, J., et al. (2019). A visible light-cross-linkable, fibrin-gelatin-based bioprinted construct with human cardiomyocytes and fibroblasts. ACS Biomater. Sci. Eng. 5, 4551–4563. doi: 10.1021/acsbiomaterials.9b00505
Arai, K., Murata, D., Verissimo, A. R., Mukae, Y., Itoh, M., Nakamura, A., et al. (2018). Fabrication of scaffold-free tubular cardiac constructs using a Bio-3D printer. PLoS One 13:e0209162. doi: 10.1371/journal.pone.0209162
Arakawa, C. K., Badeau, B. A., Zheng, Y., and DeForest, C. A. (2017). Multicellular vascularized engineered tissues through user-programmable biomaterial photodegradation. Adv. Mater. 29, 1–9. doi: 10.1002/adma.201703156
Auger, F. A., Gibot, L., and Lacroix, D. (2013). The pivotal role of vascularization in tissue engineering. Annu. Rev. Biomed. Eng. 15, 177–200. doi: 10.1146/annurev-bioeng-071812-152428
Benam, K. H., Dauth, S., Hassell, B., Herland, A., Jain, A., Jang, K. J., et al. (2015). Engineered in vitro disease models. Annu. Rev. Pathol. Mech. Dis. 10, 195–262. doi: 10.1146/annurev-pathol-012414-040418
Bertassoni, L. E., Cecconi, M., Manoharan, V., Nikkhah, M., Hjortnaes, J., Cristino, A. L., et al. (2014). Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab Chip 14, 2202–2211. doi: 10.1039/c4lc00030g
Bhatia, S. N., and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. doi: 10.1038/nbt.2989
Bhise, N. S., Manoharan, V., Massa, S., Tamayol, A., Ghaderi, M., Miscuglio, M., et al. (2016). A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 8:014101. doi: 10.1088/1758-5090/8/1/014101
Cao, X., Ashfaq, R., Cheng, F., Maharjan, S., Li, J., Ying, G., et al. (2019). A Tumor-on-a-Chip system with bioprinted blood and lymphatic vessel pair. Adv. Funct. Mater. 29, 1–13. doi: 10.1002/adfm.201807173
Cao, X., Maharjan, S., Ashfaq, R., Shin, J., and Shrike Zhang, Y. (2020). Bioprinting of small-diameter blood vessels. Engineering [Preprint]. doi: 10.1016/j.eng.2020.03.019
Chatzinikolaidou, M. (2016). Cell spheroids: the new frontiers in in vitro models for cancer drug validation. Drug Discov. Today 21, 1553–1560. doi: 10.1016/j.drudis.2016.06.024
Choi, Y. J., Jun, Y. J., Kim, D. Y., Yi, H. G., Chae, S. H., Kang, J., et al. (2019). A 3D cell printed muscle construct with tissue-derived bioink for the treatment of volumetric muscle loss. Biomaterials 206, 160–169. doi: 10.1016/j.biomaterials.2019.03.036
Choi, Y.-J., Yi, H.-G., Kim, S.-W., and Cho, D.-W. (2017). 3D cell printed tissue analogues: a new platform for theranostics. Theranostics 7, 3118–3137. doi: 10.7150/thno.19396
Chung, M., Ahn, J., Son, K., Kim, S., and Jeon, N. L. (2017). Biomimetic model of tumor microenvironment on microfluidic platform. Adv. Healthc. Mater. 6, 1–7. doi: 10.1002/adhm.201700196
Collins, S., Yuen, G., Tu, T., Budzinska, M., Spring, K., Bryant, K., et al. (2019). “In vitro models of the liver: disease modeling, drug discovery and clinical applications,” in Hepatocellular Carcinoma [Internet]. (Brisbane, AU: Codon Publications), 47–67. doi: 10.15586/hepatocellularcarcinoma.2019.ch3
Corsini, A., and Bortolini, M. (2013). Drug-induced liver injury: the role of drug metabolism and transport. J. Clin. Pharmacol. 53, 463–474. doi: 10.1002/jcph.23
Daly, A. C., Davidson, M. D., and Burdick, J. A. (2021). 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 12:753. doi: 10.1038/s41467-021-21029-2
Das, S., Kim, S. W., Choi, Y. J., Lee, S., Lee, S. H., Kong, J. S., et al. (2019). Decellularized extracellular matrix bioinks and the external stimuli to enhance cardiac tissue development in vitro. Acta Biomater. 95, 188–200. doi: 10.1016/j.actbio.2019.04.026
Dewhirst, M. W., and Secomb, T. W. (2017). Transport of drugs from blood vessels to tumour tissue. Nat. Rev. Cancer 17, 738–750. doi: 10.1038/nrc.2017.93
Ding, Y., Lee, J., Hsu, J. J., Chang, C.-C., Baek, K. I., Ranjbarvaziri, S., et al. (2018). Light-sheet imaging to elucidate cardiovascular injury and repair. Curr. Cardiol. Rep. 20:35. doi: 10.1007/s11886-018-0979-6
Gao, G., Kim, H., Kim, B. S., Kong, J. S., Lee, J. Y., Park, B. W., et al. (2019). Tissue-engineering of vascular grafts containing endothelium and smooth-muscle using triple-coaxial cell printing. Appl. Phys. Rev. 6:041402. doi: 10.1063/1.5099306
Gao, G., Lee, J. H., Jang, J., Lee, D. H., Kong, J. S., Kim, B. S., et al. (2017). Tissue engineered bio-blood-vessels constructed using a tissue-specific bioink and 3D coaxial cell printing technique: a novel therapy for ischemic disease. Adv. Funct. Mater. 27, 1–12. doi: 10.1002/adfm.201700798
Gao, G., Park, J. Y., Kim, B. S., Jang, J., and Cho, D. W. (2018). Coaxial cell printing of freestanding, perfusable, and functional in vitro vascular models for recapitulation of native vascular endothelium pathophysiology. Adv. Healthc. Mater. 7, 1–12. doi: 10.1002/adhm.201801102
Gao, G., Park, W., Kim, B. S., Ahn, M., Chae, S., Cho, W. W., et al. (2020). Construction of a novel in vitro atherosclerotic model from geometry-tunable artery equivalents engineered via in-bath coaxial cell printing. Adv. Funct. Mater. 31:2008878. doi: 10.1002/adfm.202008878
Goldfracht, I., Protze, S., Shiti, A., Setter, N., Gruber, A., Shaheen, N., et al. (2020). Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 11:75. doi: 10.1038/s41467-019-13868-x
Grigoryan, B., Paulsen, S. J., Corbett, D. C., Sazer, D. W., Fortin, C. L., Zaita, A. J., et al. (2019). Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 364, 458–464. doi: 10.1126/science.aav9750
Grix, T., Ruppelt, A., Thomas, A., Amler, A., Noichl, B. P., Lauster, R., et al. (2018). Advanced organ-on-a-chip applications. Genes 9:176. doi: 10.3390/genes9040176
Gupta, N., Dilmen, E., Morizane, R., Gupta, N., Dilmen, E., Morizane, R., et al. (2020). 3D kidney organoids for bench-to-bedside translation. J. Mol. Med. 99, 477–487. doi: 10.1007/s00109-020-01983-y
Haase, K., Offeddu, G. S., Gillrie, M. R., and Kamm, R. D. (2020). Endothelial regulation of drug transport in a 3D vascularized tumor model. Adv. Funct. Mater. 30:2002444. doi: 10.1002/adfm.202002444
Han, S., Kim, S., Chen, Z., Shin, H. K., Lee, S. Y., Moon, H. E., et al. (2020). 3D bioprinted vascularized tumour for drug testing. Int. J. Mol. Sci. 21, 1–14. doi: 10.3390/ijms21082993
Hass, R., von der Ohe, J., and Ungefroren, H. (2020). Impact of the tumor microenvironment on tumor heterogeneity and consequences for cancer cell plasticity and stemness. Cancers (Basel). 12, 1–20. doi: 10.3390/cancers12123716
Healy, K. (2018). Tissue-engineered disease models. Nat. Biomed. Eng. 2, 879–880. doi: 10.1038/s41551-018-0339-2
Heinrich, M. A., Bansal, R., Lammers, T., Zhang, Y. S., Michel Schiffelers, R., and Prakash, J. (2019). 3D-Bioprinted mini-brain: a glioblastoma model to study cellular interactions and therapeutics. Adv. Mater. 31, 1–9. doi: 10.1002/adma.201806590
Hoes, M. F., Bomer, N., and van der Meer, P. (2019). Concise review: the current state of human in vitro cardiac disease modeling: a focus on gene editing and tissue engineering. Stem Cells Transl. Med. 8, 66–74. doi: 10.1002/sctm.18-0052
Homan, K. A., Kolesky, D. B., Skylar-Scott, M. A., Herrmann, J., Obuobi, H., Moisan, A., et al. (2016). Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep. 6, 1–13. doi: 10.1038/srep34845
Huh, D., Matthews, B. D., Mammoto, A., Montoya-Zavala, M., Hsin, H. Y., and Ingber, D. E. (2010). Reconstituting organ-level lung functions on a chip. Science 328, 1662LP–1668LP. doi: 10.1126/science.1188302
Ide, I., Nagao, E., Kajiyama, S., and Mizoguchi, N. (2020). A novel evaluation method for determining drug-induced hepatotoxicity using 3D bio-printed human liver tissue. Toxicol. Mech. Methods 30, 189–196. doi: 10.1080/15376516.2019.1686795
Jang, J., Park, J. Y., Gao, G., and Cho, D. W. (2018). Biomaterials-based 3D cell printing for next-generation therapeutics and diagnostics. Biomaterials 156, 88–106. doi: 10.1016/j.biomaterials.2017.11.030
Jia, W., Gungor-Ozkerim, P. S., Zhang, Y. S., Yue, K., Zhu, K., Liu, W., et al. (2016). Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials 106, 58–68. doi: 10.1016/j.biomaterials.2016.07.038
Kang, D., Hong, G., An, S., Jang, I., Yun, W. S., Shim, J. H., et al. (2020). Bioprinting of multiscaled hepatic lobules within a highly vascularized construct. Small 16, 1–9. doi: 10.1002/smll.201905505
Kim, E., Choi, S., Kang, B., Kong, J. H., Kim, Y., Yoon, W. H., et al. (2020). Creation of bladder assembloids mimicking tissue regeneration and cancer. Nature 588, 664–669. doi: 10.1038/s41586-020-3034-x
Kim, J., Kong, J. S., Han, W., Kim, B. S., and Cho, D. W. (2020). 3D cell printing of tissue/organ-mimicking constructs for therapeutic and drug testing applications. Int. J. Mol. Sci. 21, 1–27. doi: 10.3390/ijms21207757
Kim, M., Hwang, D. G., and Jang, J. (2020). 3D pancreatic tissue modeling in vitro: advances and prospects. BioChip J. 14, 84–99. doi: 10.1007/s13206-020-4108-4
Kim, T. Y., Kofron, C. M., King, M. E., Markes, A. R., Okundaye, A. O., Qu, Z., et al. (2018). Directed fusion of cardiac spheroids into larger heterocellular microtissues enables investigation of cardiac action potential propagation via cardiac fibroblasts. PLoS One 13:e0196714. doi: 10.1371/journal.pone.0196714.g007
Kizawa, H., Nagao, E., Shimamura, M., Zhang, G., and Torii, H. (2017). Scaffold-free 3D bio-printed human liver tissue stably maintains metabolic functions useful for drug discovery. Biochem. Biophys. Rep. 10, 186–191. doi: 10.1016/j.bbrep.2017.04.004
Kolesky, D. B., Homan, K. A., Skylar-Scott, M. A., and Lewis, J. A. (2016). Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. U.S.A. 113, 3179–3184. doi: 10.1073/pnas.1521342113
Kolesky, D. B., Truby, R. L., Gladman, A. S., Busbee, T. A., Homan, K. A., and Lewis, J. A. (2014). 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv. Mater. 26, 3124–3130. doi: 10.1002/adma.201305506
Kupfer, M. E., Lin, W. H., Ravikumar, V., Qiu, K., Wang, L., Gao, L., et al. (2020). In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3D bioprinted, chambered organoid. Circ. Res. 127, 207–224. doi: 10.1161/CIRCRESAHA.119.316155
Langer, E. M., Allen-Petersen, B. L., King, S. M., Kendsersky, N. D., Turnidge, M. A., Kuziel, G. M., et al. (2019). Modeling tumor phenotypes in vitro with three-dimensional bioprinting. Cell Rep. 26, 608.e6–623.e6. doi: 10.1016/j.celrep.2018.12.090
Langhans, S. A. (2018). Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 9:6. doi: 10.3389/fphar.2018.00006
Laurent, J., Blin, G., Chatelain, F., Vanneaux, V., Fuchs, A., Larghero, J., et al. (2017). Convergence of microengineering and cellular self-organization towards functional tissue manufacturing. Nat. Biomed. Eng. 1, 939–956. doi: 10.1038/s41551-017-0166-x
Lee, A., Hudson, A. R., Shiwarski, D. J., Tashman, J. W., Hinton, T. J., Yerneni, S., et al. (2019). 3D bioprinting of collagen to rebuild components of the human heart. Science 365, 482–487. doi: 10.1126/science.aav9051
Lee, H., Chae, S., Kim, J. J. Y., Han, W., Kim, J. J. Y., Choi, Y., et al. (2019). Cell-printed 3D liver-on-a-chip possessing a liver microenvironment and biliary system. Biofabrication 11:025001. doi: 10.1088/1758-5090/aaf9fa
Lee, H., Kim, J., Choi, Y., and Cho, D. W. (2020). Application of gelatin bioinks and cell-printing technology to enhance cell delivery capability for 3D liver fibrosis-on-a-chip development. ACS Biomater. Sci. Eng. 6, 2469–2477. doi: 10.1021/acsbiomaterials.9b01735
Lee, J. H., Kim, S. K., Khawar, I. A., Jeong, S. Y., Chung, S., and Kuh, H. J. (2018). Microfluidic co-culture of pancreatic tumor spheroids with stellate cells as a novel 3D model for investigation of stroma-mediated cell motility and drug resistance. J. Exp. Clin. Cancer Res. 37, 1–12. doi: 10.1186/s13046-017-0654-6
Lee, V. K., Kim, D. Y., Ngo, H., Lee, Y., Seo, L., Yoo, S. S., et al. (2014a). Creating perfused functional vascular channels using 3D bio-printing technology. Biomaterials 35, 8092–8102. doi: 10.1016/j.biomaterials.2014.05.083
Lee, V. K., Lanzi, A. M., Ngo, H., Yoo, S. S., Vincent, P. A., and Dai, G. (2014b). Generation of multi-scale vascular network system within 3D hydrogel using 3D bio-printing technology. Cell. Mol. Bioeng. 7, 460–472. doi: 10.1007/s12195-014-0340-0
Leonard, A., Bertero, A., Powers, J. D., Beussman, K. M., Bhandari, S., Regnier, M., et al. (2018). Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J. Mol. Cell. Cardiol. 118, 147–158. doi: 10.1016/j.yjmcc.2018.03.016
Li, R. A., Keung, W., Cashman, T. J., Backeris, P. C., Johnson, B. V., Bardot, E. S., et al. (2018). Bioengineering an electro-mechanically functional miniature ventricular heart chamber from human pluripotent stem cells. Biomaterials 163, 116–127. doi: 10.1016/j.biomaterials.2018.02.024
Lin, N. Y. C., Homan, K. A., Robinson, S. S., Kolesky, D. B., Duarte, N., Moisan, A., et al. (2019). Renal reabsorption in 3D vascularized proximal tubule models. Proc. Natl. Acad. Sci. U.S.A. 116, 5399–5404. doi: 10.1073/pnas.1815208116
Loukas, M., Groat, C., Khangura, R., Owens, D. G., and Anderson, R. H. (2009). The normal and abnormal anatomy of the coronary arteries. Clin. Anat. 22, 114–128. doi: 10.1002/ca.20761
Ma, L., Wu, Y., Li, Y., Aazmi, A., Zhou, H., Zhang, B., et al. (2020). Current advances on 3D-bioprinted liver tissue models. Adv. Healthc. Mater. 9:2001517. doi: 10.1002/adhm.202001517
Ma, X., Qu, X., Zhu, W., Li, Y. S., Yuan, S., Zhang, H., et al. (2016). Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U.S.A. 113, 2206–2211. doi: 10.1073/pnas.1524510113
Macqueen, L. A., Sheehy, S. P., Chantre, C. O., Zimmerman, J. F., Pasqualini, F. S., Liu, X., et al. (2018). A tissue-engineered scale model of the heart ventricle. Nat. Biomed. Eng. 2, 930–941. doi: 10.1038/s41551-018-0271-5
Maiullari, F., Costantini, M., Milan, M., Pace, V., Chirivì, M., Maiullari, S., et al. (2018). A multi-cellular 3D bioprinting approach for vascularized heart tissue engineering based on HUVECs and iPSC-derived cardiomyocytes. Sci. Rep. 8, 1–15. doi: 10.1038/s41598-018-31848-x
Mao, Q., Wang, Y., Li, Y., Juengpanich, S., and Li, W. (2020). Materials Science & Engineering C Fabrication of liver microtissue with liver decellularized extracellular matrix (dECM) bioink by digital light processing (DLP) bioprinting. Mater. Sci. Eng. C 109:110625. doi: 10.1016/j.msec.2020.110625
Meng, F., Meyer, C. M., Joung, D., Vallera, D. A., McAlpine, M. C., and Panoskaltsis-Mortari, A. (2019). 3D bioprinted in vitro metastatic models via reconstruction of tumor microenvironments. Adv. Mater. 31, 1–10. doi: 10.1002/adma.201806899
Miri, A. K., Khalilpour, A., Cecen, B., Ryon, S., Khademhosseini, A., and Maharjan, S. (2019). Biomaterials Multiscale bioprinting of vascularized models. Biomaterials 198, 204–216. doi: 10.1016/j.biomaterials.2018.08.006
Moroni, L., Boland, T., Burdick, J. A., De Maria, C., Derby, B., Forgacs, G., et al. (2018a). Biofabrication: a guide to technology and terminology. Trends Biotechnol. 36, 384–402. doi: 10.1016/j.tibtech.2017.10.015
Moroni, L., Burdick, J. A., Highley, C., Lee, S. J., Morimoto, Y., Takeuchi, S., et al. (2018b). Biofabrication strategies for 3D in vitro models and regenerative medicine. Nat. Rev. Mater. 3, 21–37. doi: 10.1038/s41578-018-0006-y
Mota, C., Camarero-Espinosa, S., Baker, M. B., Wieringa, P., and Moroni, L. (2020). Bioprinting: from tissue and organ development to in vitro models. Chem. Rev. 120, 10547–10607. doi: 10.1021/acs.chemrev.9b00789
Nam, H., Choi, Y.-M., and Jang, J. (2020). Vascularized lower respiratory-physiology-on-a-chip. Appl. Sci. 10:900. doi: 10.3390/app10030900
Nashimoto, Y., Hayashi, T., Kunita, I., and Nakamasu, A. (2017). Integrative Biology Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device †. Integr. Biol. 9, 506–518. doi: 10.1039/C7IB00024C
Nguyen, D. G., Funk, J., Robbins, J. B., Crogan-Grundy, C., Presnell, S. C., Singer, T., et al. (2016). Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS One 11:e0158674. doi: 10.1371/journal.pone.0158674
Nguyen, D. H. T., Lee, E., Alimperti, S., Norgard, R. J., Wong, A., Lee, J. J. K., et al. (2019). A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci. Adv. 5, 1–10. doi: 10.1126/sciadv.aav6789
Noor, N., Shapira, A., Edri, R., Gal, I., Wertheim, L., and Dvir, T. (2019). 3D printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. 6:1900344. doi: 10.1002/advs.201900344
Norona, L. M., Nguyen, D. G., Gerber, D. A., Presnell, S. C., and LeCluyse, E. L. (2016). Modeling compound-induced fibrogenesis in vitro using three-dimensional bioprinted human liver tissues. Toxicol. Sci. 154, 354–367. doi: 10.1093/TOXSCI/KFW169
Norona, L. M., Nguyen, D. G., Gerber, D. A., Presnell, S. C., Mosedale, M., and Watkins, P. B. (2019). Bioprinted liver provides early insight into the role of Kupffer cells in TGF-β1 and methotrexate-induced fibrogenesis. PLoS One 14:e0208958. doi: 10.1371/journal.pone.0208958
Ozturk, M. S., Lee, V. K., Zou, H., Friedel, R. H., Dai, G., and Intes, X. (2019). High resolution tomographic analysis of in vitro 3D glioblastoma tumor model under long-term drug treatment. bioRxiv[Preprint] doi: 10.1101/684019
Park, J. Y., Ryu, H., Lee, B., Ha, D. H., Ahn, M., Kim, S., et al. (2019). Development of a functional airway-on-a-chip by 3D cell printing. Biofabrication 11:015002. doi: 10.1088/1758-5090/aae545
Pellegata, A. F., Tedeschi, A. M., and De Coppi, P. (2018). Whole organ tissue vascularization: engineering the tree to develop the fruits. Front. Bioeng. Biotechnol. 6:56. doi: 10.3389/fbioe.2018.00056
Pi, Q., Maharjan, S., Yan, X., Liu, X., Singh, B., van Genderen, A. M., et al. (2018). Digitally tunable microfluidic bioprinting of multilayered cannular tissues. Adv. Mater. 30, 1–10. doi: 10.1002/adma.201706913
Potente, M., and Mäkinen, T. (2017). Vascular heterogeneity and specialization in development and disease. Nat. Publ. Gr. 18, 477–494. doi: 10.1038/nrm.2017.36
Pridgeon, C. S., Schlott, C., Wong, M. W., Heringa, M. B., Heckel, T., Leedale, J., et al. (2018). Innovative organotypic in vitro models for safety assessment: aligning with regulatory requirements and understanding models of the heart, skin, and liver as paradigms. Arch. Toxicol. 92, 557–569. doi: 10.1007/s00204-018-2152-9
Sacchi, M., Bansal, R., and Rouwkema, J. (2020). Bioengineered 3D models to recapitulate tissue fibrosis. Trends Biotechnol. 38, 623–636. doi: 10.1016/j.tibtech.2019.12.010
Salaris, F., and Rosa, A. (2019). Construction of 3D in vitro models by bioprinting human pluripotent stem cells: challenges and opportunities. Brain Res. 1723:146393. doi: 10.1016/j.brainres.2019.146393
Schneider, G., Schmidt-Supprian, M., and Saur, R. R. (2017). Tissue-specific tumorigenesis: context matters. Nat. Rev. Cancer 17, 239–253. doi: 10.1038/nrc.2017.5
Singh, N. K., Han, W., Nam, S. A., Kim, J. W., Kim, J. Y., Kim, Y. K., et al. (2020). Three-dimensional cell-printing of advanced renal tubular tissue analogue. Biomaterials 232:119734. doi: 10.1016/j.biomaterials.2019.119734
Spill, F., Reynolds, D. S., Kamm, R. D., and Zaman, M. H. (2016). Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol. 40, 41–48. doi: 10.1016/j.copbio.2016.02.007
Strauss, D. G., Wu, W. W., Li, Z., Koerner, J., and Garnett, C. (2021). Translational models and tools to reduce clinical trials and improve regulatory decision making for QTc and proarrhythmia risk (ICH E14/S7B Updates). Clin. Pharmacol. Ther. 109, 319–333. doi: 10.1002/cpt.2137
Stucki, J. D., Hobi, N., Galimov, A., Stucki, A. O., Schneider-Daum, N., Lehr, C. M., et al. (2018). Medium throughput breathing human primary cell alveolus-on-chip model. Sci. Rep. 8, 1–13. doi: 10.1038/s41598-018-32523-x
Tonon, F., Giobbe, G. G., Zambon, A., Luni, C., Gagliano, O., Floreani, A., et al. (2019). In vitro metabolic zonation through oxygen gradient on a chip. Sci. Rep. 9:13557. doi: 10.1038/s41598-019-49412-6
van Wenum, M., Adam, A. A. A., van der Mark, V. A., Chang, J. C., Wildenberg, M. E., Hendriks, E. J., et al. (2018). Oxygen drives hepatocyte differentiation and phenotype stability in liver cell lines. J. Cell Commun. Signal. 12, 575–588. doi: 10.1007/s12079-018-0456-4
Walker, R. L., and Eggel, M. (2020). From mice to monkeys? Beyond orthodox approaches to the ethics of animal model choice. Animals 10, 1–16. doi: 10.3390/ani10010077
Walsh, N. C., Kenney, L. L., Jangalwe, S., Aryee, K. E., Greiner, D. L., Brehm, M. A., et al. (2017). Humanized mouse models of clinical disease. Annu. Rev. Pathol. Mech. Dis. 12, 187–215. doi: 10.1146/annurev-pathol-052016-100332
Wang, Y., Shi, W., Kuss, M., Mirza, S., Qi, D., Krasnoslobodtsev, A., et al. (2018). 3D bioprinting of breast cancer models for drug resistance study. ACS Biomater. Sci. Eng. 4, 4401–4411. doi: 10.1021/acsbiomaterials.8b01277
Wang, Z., Lee, S. J., Cheng, H. J., Yoo, J. J., and Atala, A. (2018). 3D bioprinted functional and contractile cardiac tissue constructs. Acta Biomater. 70, 48–56. doi: 10.1016/j.actbio.2018.02.007
Weinhart, M., Hocke, A., Hippenstiel, S., Kurreck, J., and Hedtrich, S. (2019). 3D organ models—Revolution in pharmacological research? Pharmacol. Res. 139, 446–451. doi: 10.1016/j.phrs.2018.11.002
Wilmer, M. J., Ng, C. P., Lanz, H. L., Vulto, P., Suter-Dick, L., and Masereeuw, R. (2016). Kidney-on-a-Chip technology for drug-induced nephrotoxicity screening. Trends Biotechnol. 34, 156–170. doi: 10.1016/j.tibtech.2015.11.001
Wong, K. H. K., Chan, J. M., Kamm, R. D., and Tien, J. (2012). Microfluidic models of vascular functions. Annu. Rev. Biomed. Eng. 14, 205–230. doi: 10.1146/annurev-bioeng-071811-150052
Wragg, N. M., Burke, L., and Wilson, S. L. (2019). A critical review of current progress in 3D kidney biomanufacturing: advances, challenges, and recommendations. Ren. Replace. Ther. 5, 1–16. doi: 10.1186/s41100-019-0218-7
Yi, H. G., Jeong, Y. H., Kim, Y., Choi, Y. J., Moon, H. E., Park, S. H., et al. (2019). A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 3, 509–519. doi: 10.1038/s41551-019-0363-x
Yong, U., Lee, S., Jung, S., and Jang, J. (2020). Interdisciplinary approaches to advanced cardiovascular tissue engineering: ECM-based biomaterials, 3D bioprinting, and its assessment. Prog. Biomed. Eng. 2:042003. doi: 10.1088/2516-1091/abb211
Yoon, W. H., Lee, H. R., Kim, S., Kim, E., Ku, J. H., Shin, K., et al. (2020). Use of inkjet-printed single cells to quantify intratumoral heterogeneity. Biofabrication 12:035030. doi: 10.1088/1758-5090/ab9491
Yu, C., Ma, X., Zhu, W., Wang, P., Miller, K. L., Stupin, J., et al. (2019). Scanningless and continuous 3D bioprinting of human tissues with decellularized extracellular matrix. Biomaterials 194, 1–13. doi: 10.1016/j.biomaterials.2018.12.009
Zhang, Y. S., Arneri, A., Bersini, S., Shin, S. R., Zhu, K., Goli-Malekabadi, Z., et al. (2016). Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 110, 45–59. doi: 10.1016/j.biomaterials.2016.09.003
Zhao, Y., Rafatian, N., Feric, N. T., Cox, B. J., Aschar-Sobbi, R., Wang, E. Y., et al. (2019). A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176, 913.e18–927.e18. doi: 10.1016/j.cell.2018.11.042
Keywords: 3D bioprinting, 3D in vitro tissue model, vascularization, disease modeling, organ-organ crosstalk
Citation: Hwang DG, Choi Y and Jang J (2021) 3D Bioprinting-Based Vascularized Tissue Models Mimicking Tissue-Specific Architecture and Pathophysiology for in vitro Studies. Front. Bioeng. Biotechnol. 9:685507. doi: 10.3389/fbioe.2021.685507
Received: 25 March 2021; Accepted: 30 April 2021;
Published: 31 May 2021.
Edited by:
Carmine Gentile, University of Technology Sydney, AustraliaReviewed by:
Stephanie Michelle Willerth, University of Victoria, CanadaLiliang Ouyang, Tsinghua University, China
Copyright © 2021 Hwang, Choi and Jang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinah Jang, amluYWhqYW5nQHBvc3RlY2guYWMua3I=
†These authors have contributed equally to this work
 Dong Gyu Hwang
Dong Gyu Hwang Yoo-mi Choi
Yoo-mi Choi Jinah Jang
Jinah Jang