- 1School of Diagnostic and Applied Health Sciences, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
- 2Strathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, Glasgow, United Kingdom
Endophytic actinobacteria offer great potential as a source of novel bioactive compounds. In order to investigate the potential for the production of secondary metabolites by endophytes, we recovered a filamentous microorgansism from the tree Antidesma neurocarpum Miq. After phenotypic analysis and whole genome sequencing we demonstrated that this organism, SUK42 was a member of the actinobacterial genus Kitasatospora. This strain has a small genome in comparison with other type strains of this genus and has lost metabolic pathways associated with Stress Response, Nitrogen Metabolism and Secondary Metabolism. Despite this SUK42 can grow well in a laboratory environment and encodes a core genome that is consistent with other members of the genus. Finally, in contrast to other members of Kitasatospora, SUK42 encodes saccharide secondary metabolite biosynthetic gene clusters, one of which with similarity to the acarviostatin cluster, the product of which displays α-amylase inhibitory activity. As extracts of the host plant demonstrate this inhibitory activity, it suggests that the potential medicinal properties of A. neurocarpum Miq might be provided by the endophytic partner and illustrate the potential for exploitation of endophytes for clinical or industrial uses.
Introduction
Natural products hold great potential for novel drug discovery and members of the actinobacteria are amongst the most prolific producers of bioactive natural compounds (Barka et al., 2016). Despite their abundance in soil, more recently alternative environments such as plant tissues have been investigated for their discovery (Barka et al., 2016). Such organisms include symbiotic, endophytic actinobacteria that occupy an ecological niche within plant tissues and have recently begun to generate great interest as a potential source of novel natural products with industrial, environmental or clinical applications (Zin et al., 2007; Singh and Dubey, 2018). Endophytes enjoy mutualistic or antagonistic, but not parasitic, relationships with their host and can have important effects on plant growth through the production of hormones or other growth-influencing factors. They are also able to enhance increase resistance to abiotic and biotic stresses, as well as providing resistance to pests and pathogens. In exchange, the host plant offers food and shelter for these symbiotic organisms (Singh and Dubey, 2018). Endophytic actinobacteria, especially from the genus Streptomyces, are known to synthesize active compounds which make these bacteria a rich source of natural products particularly for clinical applications (Strobel, 2003), such as antimalarial (Ahmad et al., 2021) or antibacterial agents (Alshaibani et al., 2017). For example, the endophytic strain Streptomyces SUK25 can produce various diketopiperazine derivatives, with bioactivity against pathogenic bacteria such as MRSA and Enterococcus raffinosus whilst possessing low toxicity against HepaRG cells (Alshaibani et al., 2017) and illustrates the rich prospects offered by drug discovery from endophytes.
The family Streptomycetaceae includes the genera Streptomyces and Streptacidiphilus (Nouioui et al., 2018), as well as Kitasatospora (Omura et al., 1982; Takahashi, 2017). Like Streptomyces, members of the genus Kitasatospora are known to produce active compounds such as setamycin (bafilomycin B1) and bafilomycin A1, a proteasome inhibitor and an anti-fungal agent (Momose et al., 2001; Yoon et al., 2006; Ichikawa et al., 2010). Members of the Kitasatospora are non-fastidious in their nutritional requirements and form leathery colonies with both vegetative and aerial hyphae. The latter develop further to form 20 or more spores per chain. As such, their development closely resembles that of members of Streptomyces (Takahashi, 2017). Although once a subject of debate (Wellington et al., 1992), the classification of the Kitasatospora has been resolved by the application of 16S rRNA sequencing (Labeda et al., 2012) and whole genome sequencing (Nouioui et al., 2018; Li et al., 2021). The genus Kitasatospora is also distinguishable from Streptomyces on the basis of cell wall composition (Ichikawa et al., 2010), although more recently, another criterion that can be used to distinguish between these two genera is through the use of the SsgB amino acid sequence (Keijser et al., 2003; Willemse et al., 2011). SsgB is a member of the SALPs (SsgA-like proteins), which are present in all members of the Streptomyceteaceae. SALPs play an important role in the control of cell division and morphogenesis in actinobacteria with complex life cycles (Traag and van Wezel, 2008). SsgB recruits the cytokinetic protein FtsZ, which is responsible in initiating sporulation-specific cell division in an SsgA-dependent manner (Willemse et al., 2011). Importantly, SsgB displays conservation within a single genus while between genera variation is as low as 40–50% (Girard et al., 2013). This makes SsgB ideal as a tool for molecular systematic differentiation of Streptomyces and Kitasatospora.
Earlier work reported the isolation and classification of Streptomyces kebangsaanensis sp. from a Malaysian ethnomedicinal plant (Sarmin et al., 2013). In order to investigate genome mining for natural product discovery from endophytic bacteria from indigenous Malaysian plants, we isolated one organism, SUK42, from the plant Antidesma neurocarpum Miq with reported α-glucosidase inhibitory activity (Elya et al., 2012). This strain was initially believed to be a member of the Streptomyces, however further analysis revealed that this strain was a member of the rare genus Kitasatopsora. As such, we decided to confirm this initial identification, carry out comparative genomics with other members of this genus and explore the strains potential for production of secondary metabolites through genome mining.
Methods
Isolation and Growth Conditions
SUK42 was isolated from the internal tissue of stem from the plant, Antidesma neurocarpum Miq, collected at the UKM forest reserve Bangi, Malaysia. The plant sample was first subjected to surface sterilisation (Coombs and Franco, 2003) in order to remove epiphytic organisms. The outer layer of plant stem was removed and the inner tissue was excised and plated on water agar (Coombs and Franco, 2003), pH 7.2 and supplemented with cycloheximide (50 μg ml−1) and nystatin (50 μg ml−1) before incubation at 27°C. SUK42 was purified and maintained on ISP2- agar and stored using 20% (v/v) glycerol at −80°C.
Phenotypic Characterisation
Production of melanin was examined on ISP-7 agar after 14 days incubation at 30°C. Agar without inoculation was used as a control and production of a dark brown to black pigmentation produce recorded as positive. Tolerance to NaCl (1–15%), pH (3–12) and temperature (10, 30, 37, and 50°C) were analyzed according to (Gordon et al., 1974) on modified ISP-2 agar incubated for 14 days at 30°C. Degradation of adenine, hypoxanthine, L-tyrosine, and xanthin was analysed according to Gordon et al. (1974). Carbon source utilization growth on the isomeric form of diaminopimelic acid was determined by growth on ISP-9 agar supplemented with 1% (w/v) carbon sources (Shirling and Gottlieb, 1966) after incubation at 30°C. Hydrolysis of aesculin and starch and decomposition of casein and urea was carried out using established procedures (Gordon et al., 1974) and nitrate reduction determined by inoculation into nitrate broth supplemented with sulphanilic acid (Gottlieb, 1961).
Cell wall analysis were done following growth on ISP-2 agar at 28°C for 14 days. Cells were harvested by centrifugation at 12,000 × g, washed twice with sterile dH2O and dried before dissolving in 1 ml 6N HCL as described previously (Staneck and Roberts, 1974). Standard solutions of DAP (DL-diamonipimelic acid) and LL-DAP were and thin layer chromatography plates developed with ninhydrin solution before drying at 80°C) (Staneck and Roberts, 1974).
Genomic DNA Extraction, Sequencing and Assembly
Genomic DNA from SUK42 was obtained from cultivation of SUK42 in ISP-2, extracted according to established procedures and 16S rRNA sequence amplified using universal bacterial 16S rRNA gene primers (Coombs and Franco, 2003). PCR products were sequenced and compared to existing sequences using EzBioCloud (Yoon et al., 2017). Whole genome sequencing of strain SUK42 were done using an Illumina MiSeq system (first base Company) with 301 bp paired-end reads. The sequence was assembled using Unicycler version 0.4.8 (Wick et al., 2017) and assembly quality assessed using Quast (Mikheenko et al., 2018) and viewed in CG viewer (Grant and Stothard, 2008).
Bioinformatic Analysis
To place SUK42 in its phylogenetic context, related strains were identified using AutoMLST with SUK42 as the query sequence (Alanjary et al., 2019) after a concatenated alignment. Reference sequences of type strains (Supplementary Table S1) were identified using AutoMLST and used to scaffold the assembly of SUK42 with the other reference sequences with Medusa (Bosi et al., 2015). Average nucleotide identity values were determined using OrthoANI (Lee et al., 2016) and alignments of SsgB amino acid sequences were carried using Mega X (Kumar et al., 2018). Pan-genome analysis was carried using Roary version 3.13.0 following annotation by Prokka, version 1.14.6 (Seemann, 2014), available on Galaxy (Afgan et al., 2016). Outputs from Roary were displayed in Phandango (Hadfield et al., 2018). Collections of functional roles that make up metabolic pathways were identified as subsytems established by SEED-viewer analysis of genome sequences by RASTtk (Brettin et al., 2015). Secondary metabolite biosynthetic gene clusters were first identified by subjecting genome sequences to antiSMASH, Galaxy ver. 5.1.2, (Blin et al., 2019), outputs from antiSMASH were then further investigated using BiG-SCAPE (Navarro-Munoz et al., 2020) to sort clusters into higher order gene cluster families. Chord diagrams were plotted using the circlize package in R (Gu et al., 2014).
Results
Phenotypic Characterisation of SUK42
In order to identify novel endophytic actinobacteria, SUK 42 was isolated from the internal tissue of stem from the plant, Antidesma neurocarpum Miq. 16S rRNA sequencing indicated that SUK42 was most closely related to Streptomyces xanthocidicus (data not shown). This strain has now been renamed Kitasaotospora xanthocidicus (Nouioui et al., 2018). Following cultivation, macro-morphological characterization was carried out following standard protocols (Shirling and Gottlieb, 1966), aerial mycelia were observed on ISP-3 and ISP-4 whilst pigmentation was observed on all agars (Figure 1B); melanin was not produced on ISP-7 (data not shown). Microscopic observation of mycelial growth on ISP-2 was made by light microscopy and scanning electron microscopy. (Figure 1A). SUK42 could grow at temperatures up to 50°C, a pH of 12 and 10% w/v NaCl and could decompose both starch and casein. Dextrose, galactose, sucrose, maltose, glucose, rhamnose, and sorbitol were used as sole carbon, but inositol and D-mannitol were not. Hydrolysis of l-tyrosine and starch were observed, but reduction of nitrate and decomposition of urea were not. Both L,L and meso-diaminopimelic acid were detected in SUK42 when grown on ISP-2 media (data not shown).
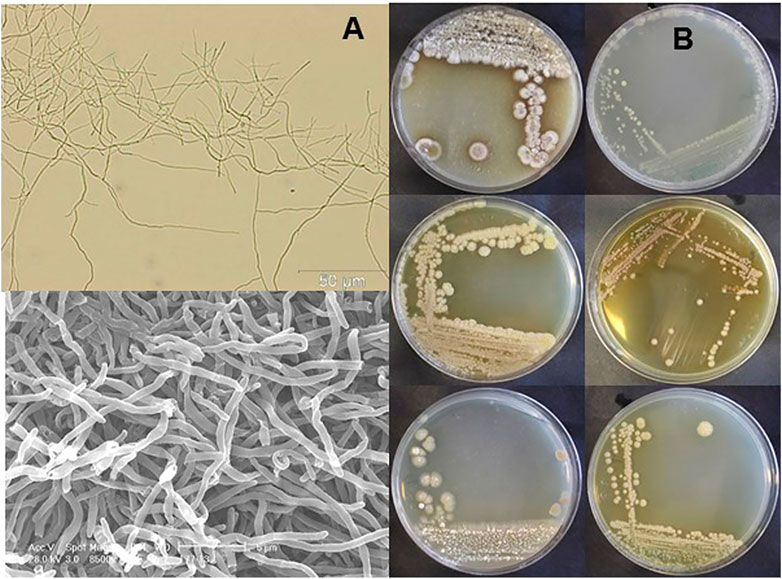
FIGURE 1. Mycelial morphology (A) and macromorphology (B) of Kitasatospora sp. SUK42. (A) Scanning electron (top) and bright field microscope images (bottom) of ISP2-grown Kitasatospora sp. SUK42 substrate mycelium. (B) Plates of Kitasatospora sp. SUK42 gown on ISP-2 (top), ISP-3, ISP-4 (left hand column) and ISP-4 (top), ISP-5, ISP6 (right hand column).
Genomic Characterisation of SUK42
Genome sequencing of SUK42 was carried out using whole-genome sequencing by 1st Base Malaysia performed on an Illumina Miseq apparatus. We obtained 51.6 million reads that, following assembly, were assembled into 35 contigs of >200 bp. The total size of the assembly was 6,568,264 with a G + C content of 72.9%. The sequence is deposited with NCBI under accession number LFMD00000000 and assembly GCA_015776785.2. Automatic functional annotation results were obtained using the NCBI Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html) and a genomic map is displayed in Supplementary Figure S1. SUK42 has a relatively small genome of 6.5 Mb compared to other strains of Kitasatopora [mean size 8.7 Mb (Li et al., 2021)].
SUK42 is a Member of the Genus Kitasatospora
In order to identify SUK42 more accurately, we analysed the assembled sequence using AutoMLST (Alanjary et al., 2019). This analysis identified eight type strains available as reference sequences that had an estimated ANI of >84.7% with SUK42 (listed in Supplementary Table S1). The next most similar type strains were members of the genus Streptacidiphilus (Streptacidiphilulus rugosus AM-16 had an ANI <81.6% compared to SUK42). The eight type strains were named as both members of the genera Streptomyces and Kitasatospora, so in order to better delineate these organisms, we subjected their genome sequences to further analysis of these strains using OrthANI (Figure 2) along with type stains of neighbouring genera (Actinospica robiniae DSM44927, Streptacidiphilus albus JL83, Streptomyces albus NBRC 13014). This demonstrated that SUK42 was most closely related to Streptomyces avellaneus of the eight type strains that were cleary distinct form the out-groups from neighbouring genera. However the presence of members of the Kitasatospora in this clade suggested that Streptomyces avellaneus, Streptomyces novoceasareae, and Streptomyces rubellomurinus might have been misclassified. Subsequently the mis-classification of these strains was confirmed in the literature (Li et al., 2021). It was noticeable that the type strains of the genera Streptomyces (Streptomyces albus NBRC 13014) and Streptacidiphilus (Streptacidiphilus albus JL83) were phylogenetically separate from SUK42 and the other eight strains (Figure 2) and confirms the grouping of SUK42 with members of Kitasatspora and distinct from the other two genera that form members of the family Streptomycetaceae. Interestingly, there were two sub-clades contained with the collection of the Kitasatospora type strains (Group 1: Kitasatospora Sp. SUK42, S. avellaneus NRRL B-3447, S. novaecaesareae NRRL B-1267, S. rubellomurinus ATCC 31215, K. purpeofusca NRRL B-1817 and Group 2: K. setae NRRL B-16185, K. setae KM-6054, K. phosalacinea NRRL B-16230, K. cheerisanensis KCTC 2395).
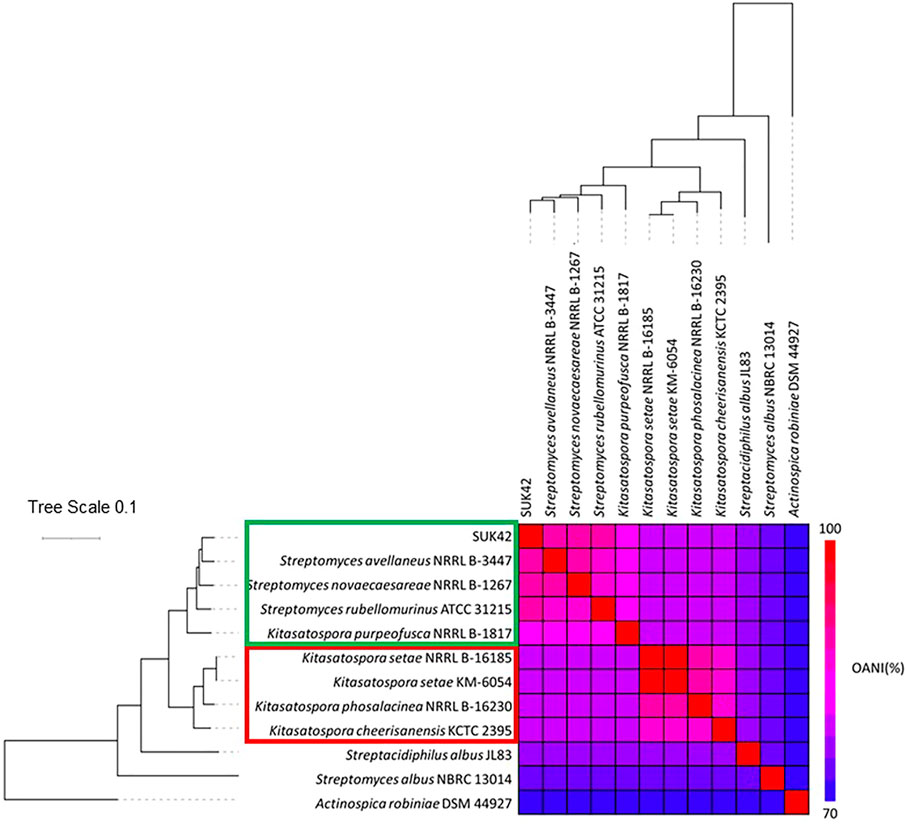
FIGURE 2. Phylogenetic relationship of putative members of the genus Kitasatospora. Eight strains with >84.7% ANI with Kitasatospora sp. SUK42 and appropriate type were used to carry out Multi-Locus Sequence Analysis (MLSA) to produce a high-resolution species tree using AutoMLST after a concatenated alignment (Alanjary et al., 2019). All branches were supported with boot strap values of 100 except the Kitasatospora sp. SUK42/S. avellaneus node shared with S. novoceasaereae (82). The average nucleotide identities (ANI) was calculated using the OrthoANI tool (Lee et al., 2016) and plotted with the heat map function in R (Ver. 4.0). Actinospica robiniae, type strain of the family Actinospicaceae and the type strains of the genera Streptomyces and Streptacidophilus, S. albus NBRC 13014 and S. albus JL83, respectively, were used as outgroups.
To confirm the phylogenetic position of SUK42 amongst the genus Kitasatospora, we extracted and analysed the amino acid sequences of the cell division protein SsgB (Girard et al., 2013) from our collection of Kitasatospora sequences in addition to the model members of the genus Streptomyces (S. albus, S. coelicolor, S. clavuligerus, and S. venezuelae). SsgB is responsible for FtsZ ring placement in streptomycetes (Keijser et al., 2003; Willemse et al., 2011). A comparison of SsgB sequences from the two genera defined three amino substitutions (V14-I14; E46-D46 and T/Q/K128–R46l Streptomyces-Kitasatospora, respectively). As shown in Figure 3, SUK42 and type other type strains show the substitutions diagnostic of Kitasatospora and demonstrate that both these organisms are indeed members of this genus. Consequently, our studies of the SsgB amino acid sequence indicate that, like SUK42, S. avellaneus, S. novoceasareae, and S. rubellomurinus should be classified as members of the genus Kitasatospora.

FIGURE 3. Alignment of SsgB sequences from Kitasatospora sp. SUK42, other putative Kitasatospora reference sequences and model strains from the genus Streptomyces. SsgB sequences were aligned using Clustal Omega in Mega (Kumar et al., 2018). Amino acid substitutions in model strains of Streptomyces are highlighted in green.
Pan-Genome of Kitasatospora
Due to the relatively small genome size of SUK42, we wanted to investigate if any specialisation to an endophytic lifestyle existed. To understand which genes were unique (accessory genome) or conserved (core) genome (Page et al., 2015) among the Kitasatospora reference sequences a comparative pan-genome analysis was performed using whole-genome sequences. The pan-genome of the eight reference sequences (Supplementary Table S1)and Kitasatopsora sp. SUK42 was evaluated using Roary set at minimum percentage identity for blastp of 80% (Corretto et al., 2017) (Table 1) to cluster the genes encoding complete protein sequences into core (hard core and soft core) and accessory (shell and cloud) genomes. It was possible to identify 29,756 gene families comprising the pan-genome and of those, 1,498 (∼5%) were present in the core genome core (>95% of strains), and the remaining 28,528(∼95%) as accessory (present <95% of strains) (Table 1). In a recent study of all available Kitasatospora sequences, but not SUK42 (Li et al., 2021), the core genome of this genus was estimated to be 1,476 gene families and the similarity of these two values suggests that SUK42 retains the essential components of the Kitasatospora core genome. When examined in a phylogenetic context, the pan-genome obtained with Roary allowed us to determine the presence or absence of genes in each genome and also to compare and identify core regions from the two Kitasatopsora groups (Figure 4). Once more, phylogenetic analysis showed that isolates within each group affiliated closely with each other but not with strains from the other group. In this way, strains from Group 1 (green) clustered closely with Kitasatospora sp. SUK42 and were distinct from Group 2. However, a core genome was observed containing shared genes between both groups. Furthermore, genes only present within one group were identified creating two distinct regions that are part of a group-specific core genome.

TABLE 1. Core and pan genome sizes of Kitasatospora sp. SUK42 and other putative Kitasatospora reference sequences Summary of the number of genes comprising the core, shell, cloud and pan-genomes of Kitasatospora sp. SUK42 and other eight putative Kitasatospora reference sequences (Supplementary Table S1) were determined using Roary (Page et al., 2015).
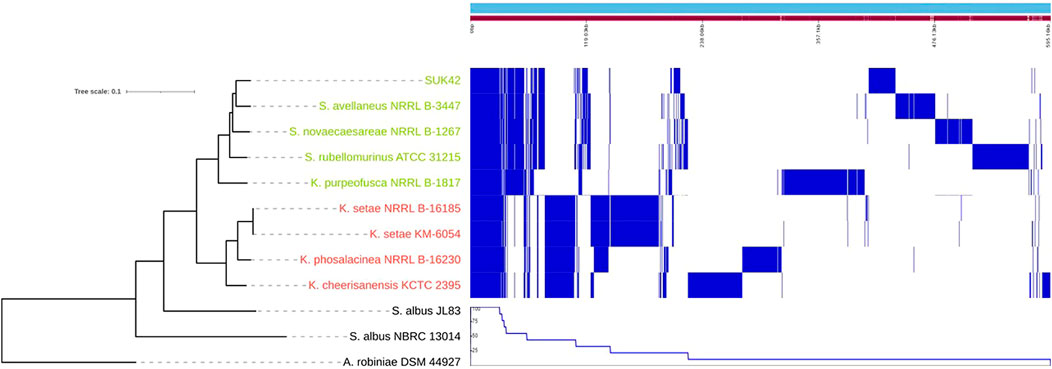
FIGURE 4. Core and pan genome of Kitasatospora sp. SUK42 and other putative Kitasatospora reference sequences. Phylogenetic tree, core, shell, cloud and pan-genomes of Kitasatospora sp. SUK42 and the other eight putative Kitasatospora reference sequences (Supplementary Table S1) were determined using Roary (Page et al., 2015) and displayed using Phandango (Hadfield et al., 2018). Group 1 Kitasatospora strains are shown in green and Group 2 in red.
Subsystems Involved in Carbohydrate and Amino Acid Metabolism Are Enriched in Kitasatospora SUK42
In order to further investigate the functional categories of the genes encoded by Kitasatospora SUK42, we subjected this strain and the eight Kitasatospora type strains to analysis by RASTtk (Brettin et al., 2015) that provided high-quality assessments of gene functions and an initial metabolic reconstruction. Annotation data in the SEED (Overbeek et al., 2005), where genes are organized into sets of logically related functional roles (subsystems) allowed us to compare subsystems between the nine Kitasatospora genome sequences (Figure 5). Although the relatively small genome size of Kitasatospora SUK42 led us to predict that this strain would show reduced numbers (Table 2) of subsystems than the average for Kitasatospora reference sequences (Table 2) some subsystems were more or less represented than others. SUK42 encodes 94.25% of the average number of subsystems per Kitasatospora genome, however subsystems involved in Cell Wall and Capsule formation; Dormancy and Sporulation; Phosphorus Metabolism; Metabolism of Aromatic Compounds; Regulation and Cell signalling; Potassium metabolism; Fatty Acids; Lipids; and Isoprenoids; DNA Metabolism were over-represented in respect to the average (Table 2). In contrast subsystems involved with Carbohydrates; Amino Acids and Derivatives; Protein Metabolism; Respiration; Nucleosides and Nucleotides; Miscellaneous; Membrane Transport; Sulphur Metabolism; RNA Metabolism; Cofactors, Vitamins, Prosthetic Groups, Pigments; Iron acquisition and metabolism; Virulence, Disease and Defense; Phages, Prophages, Transposable elements, Plasmids; Stress Response; Nitrogen Metabolism; Secondary Metabolism were under-represented with respect to the average (Table 2). Consequently, if the endophytic life-style of SUK42, has led to its small genome size, it seems that the disproportionate loss of the latter subsystems has contributed to the reduction in genome size.
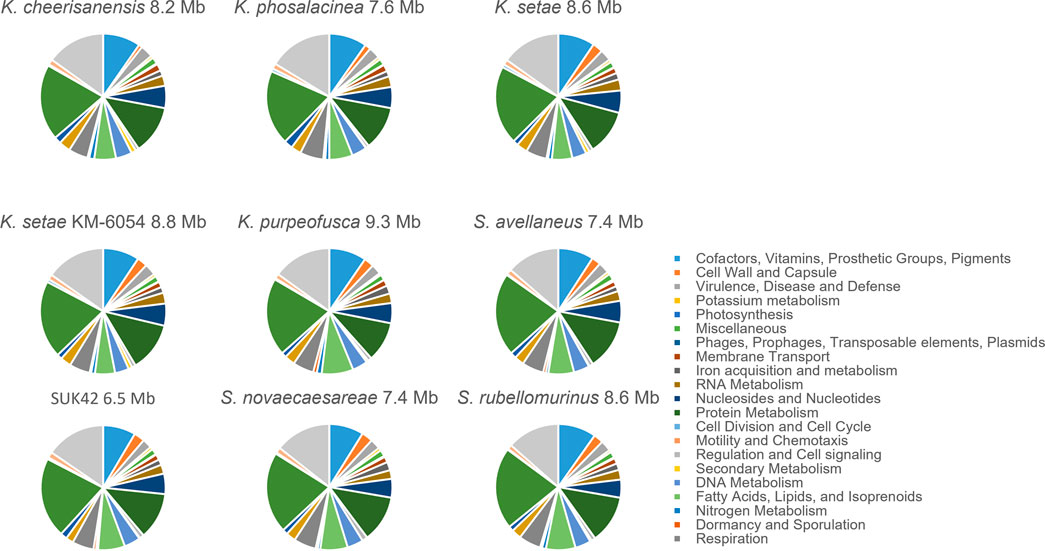
FIGURE 5. Sub-system comparison of Kitasatospora sp. SUK42 and other putative Kitasatospora reference sequences. Pie charts showing SEED sub-systems determined for Kitasatospora sp. SUK42 and eight other Kitasatospora type strains using RASTtk (Brettin et al., 2015).
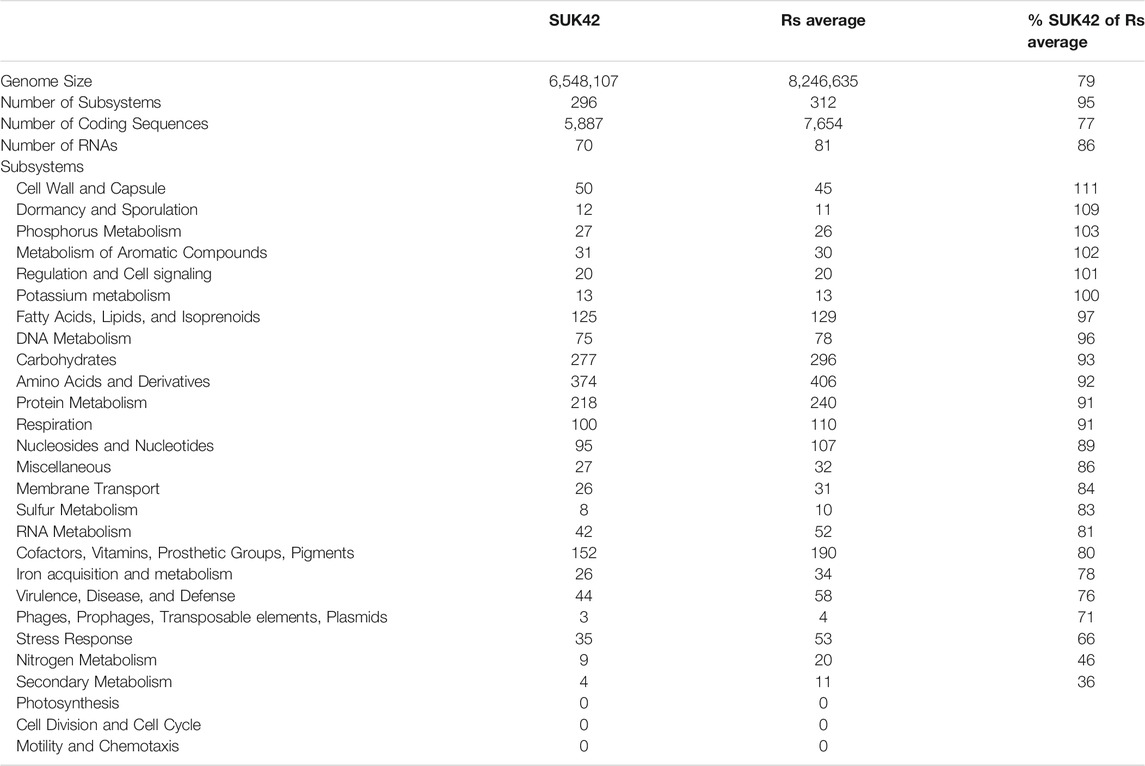
TABLE 2. Subsystems from SUK42 and Kitasatospora reference sequences. SUK42 and other Kitasatospora reference sequences (RS average) were analysed by RASTtk to establish collections of functional roles and were identified as subsytems by SEED-viewer analysis. The percentage of each SUK42 subsystem was calculated with respect to the RS average.
Secondary Metabolism in Kitasatospora sp. SUK42
Subsystems analysis demonstrated that SUK42 contained fewer metabolic pathways associated with secondary metabolism than the average of the eight reference sequences. So, to determine the genomic potential Kitasatospora sp. SUK42, the eight Kitasatospora type strains and representatives of neighbouring genera (Actinospica robiniae DSM44927, Streptacidiphilus albus JL83, Streptomyces albus NBRC 13014), for secondary metabolite production in more detail, we examined their genomes using antiSMASH to predict biosynthetic gene clusters (BGCs) (Blin et al., 2019). When genomes sequences are comprised of many contigs, this can lead to misidentification of BGCs due to contig edge effects. With the exception of K. setae KM-6054 our collection of genome sequences consisted of more than one contig so we constructed similarity networks using BiG-SCAPE (Navarro-Munoz et al., 2020) and identified secondary metabolite biosynthetic gene cluster families (GCFs) based on Pfam similarities (Figure 6). Using antiSMASH we predict that SUK42 encodes 28 regions containing BGCs, five of which exist at contig edges (Supplementary Table S2). Of these 28 clusters, only five generated ClusterBLAST hits with similarity >75% to known BGCs. 35 GCFs were predicted using BiG-SCAPE, whilst the eight type strains encoded average of 54.125 GCFs per genome (total = 433). Of these, SUK42 encodes a smaller percentage of GCFs than the eight type strains in the following major biosynthetic classes, respectively: NRPS (20 vs. 24%), RIPPs (11 vs. 16%), Terpenes (11 vs. 12%) and PKS-NRP_Hybrids (3 vs. 5%). Conversely, a larger percentage in the GCFs in SUK42 are found in the biosynthetic categories Others (23 vs. 21%), PKSI (17 vs. 13%) and Saccharides (6 vs. 0%). It is interesting in the latter category that the only occasion when GCFs of this biosynthetic class were identified was in SUK42. We also examined BIG-SCAPE networks found in different strains (Supplementary Figure S2) and revealed that GCF FAM_00285 (Region 8.8, Supplementary Table S2) was found in all strains, including S. albus NBRC 13014 (Supplementary Figure S2B). This GCF, most accurately described in the complete genome of K. setae KM-6054, encodes the production of a putative NRP siderophore with 5% similarity to BGC0001593 (Kautsar et al., 2020), the ficellomycin biosynthetic gene cluster from Streptomyces ficellus (Liu et al., 2017). A second GCF (FAM_00311, Supplementary Figure S2B) encoding a siderophore with low similarity to the ficellomycin biosynthetic cluster was also found in Group 1 Kitasatospora strains except for SUK42. Members of FAM_00223 (Supplementary Figure S2B; Region 8.4, Supplementary Table S2), which encode γ-butryolactone biosynthesis (Kato et al., 2007), are also only found in Group 1 Kitasatospora strains, has 4% similarity to BGC0000262, the prejadomycin biosynthetic gene cluster from Streptomyces sp. PGA64 (Kallio et al., 2008). Two networks from the RIPP family (Supplementary Figure S2F) were shared among all Group 2 Kitasatospora strains, although none were found in SUK42: FAM_00111, which shares the RamAB transporters (Kodani et al., 2004) with BGC0000519, the labyrinthopeptin A2 biosynthetic gene cluster from Actinomadura namibiensis (Meindl et al., 2010). Despite FAM_00286 encoding the peptidase and lanthionine synthase enzymes, no propeptide was identified in this cluster by antiSMASH. Two Saccharide GCFs were found in SUK42 (FAM_00,205, Supplementary Figure S2G; Region 10.3, Supplementary Table S2 and FAM_00206, Figure 2G; Regions 11.1, Supplementary Table S2); the only member of this GCF found in the analysed strains. A member of the terpene class (FAM_00187, Supplementary Figure S2H; Region 4.4, Supplementary Table S2) was also found across the Kitasatospora strains; GCF, BGC0000663 (Kautsar et al., 2020) and encodes the hopene biosynthetic gene cluster first identified in S. coelicolor A3(2) (Bentley et al., 2002). Finally FAM_00218, (Supplementary Figure S2H; Supplementary Table S2, Region 4.5), encodes BGC0000804 (Kautsar et al., 2020), an acarviostatin I03 biosynthetic gene cluster from Streptomyces coelicoflavus ZG0656 in addition to a gene encoding avermitilol synthase (Guo et al., 2012).
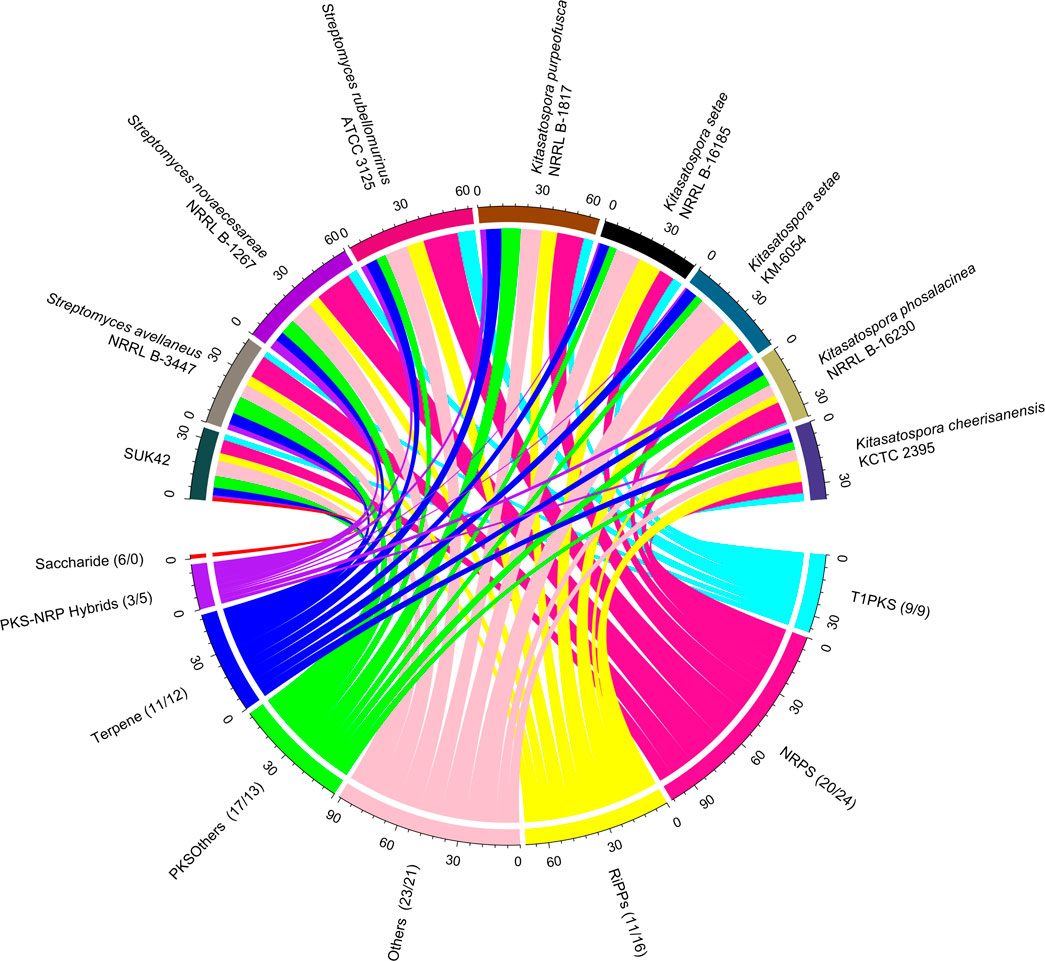
FIGURE 6. SUK42 is the only member of Kitasatospora reference sequences to encode Saccharide-type BGCs. Chord diagram illustrating the distribution of Gene Cluster Families amongst SUK42 and related strains. Of the eight, Kitasatospora sequences was the only one to encode Saccharide type clusters. Values in brackets are the percentage of clusters in a GCF that belong to SUK42 or the eight reference sequences, respectively. Gene clusters were first identified using AntiSMASH (Blin et al., 2019) before assigned to individual families using BIG-SCAPE (Navarro-Munoz et al., 2020). Plotted using the circlize package in R.
Discussion
Exploitation of the biodiversity of the planet will prove key to the identification of novel natural products and generate solutions to key medical and industrial challenges that are faced in the 21st century. Many areas of the world are inhabited by understudied plants; this is especially true of tropical regions such as South East Asia with the high-rainfall necessary to generate great biodiversity. This has allowed the discovery of microbial endophytes that produce a variety of new bioactive compounds (Strobel, 2003). For example, in a study of α-glucosidase inhibitory activity of extracts of plants with reported anti-diabetic activity, crude extracts of the folium of A. neurocarpum Miq. displayed an IC50 value 4.22 μg/ml for inhibition of α-glucosidase (Elya et al., 2012). In order to determine if this plant harboured any endophytes that might possess novel bioactivity, we set out to isolate a microorganism from this plant and rather, than physically screening for bioactivity, we assessed the potential for production of natural products using a genome mining approach (Blin et al., 2019). In so doing, we isolated a microorganism that we visually identified as a member of the genus Streptomyces, well known (Barka et al., 2016) for production of secondary metabolites. However following preliminary analysis of this isolate, we identified it as a member of Kitasatospora (Li et al., 2021).
This genus was first proposed in 1982, and although they are similar to members of the genus Streptomyces, members of Kitasatospora display clear differences in cell wall composition form a well-defined cluster on the basis of phylogenetic of 16S rRNA gene sequences (Takahashi, 2017). Genome sequences of 43 strains have been published so far and range in size from 6.3 to 12.36 Mb in size; these organisms were mostly isolated from soil, but others were isolated from water or insects (Li et al., 2021). The smallest genome of this genus so far sequenced, at 6.31 Mb, belongs to Streptomyces sp. DSM 40024 isolated under orchid grass from Japan. (Li et al., 2021). As such, the genome size of SUK42, at 6.55 Mb in size, represents the second smallest of the Kitasatospora genomes sequences sequenced so far and the smallest of the reference sequences (Supplementary Table S1). As this strain is the first endophytic strain of Kitasatospora so far sequenced, it is only through further studies of other plants and strains that will be possible to determine if a small genome size is a characteristic of other endophytic members of Kitasatospora.
Phylogenetic analysis of the SUK42 genome sequence in comparison with the genome sequences of other Kitasatospora type strains demonstrated that SUK42 likely represents a novel species and form part of clade containing S. avellaneus, S. novaecaesareae, S. rubellomurinus, and K. purpeofusca, with the former being the closest relative of SUK42 (ANI 91.80%. This clade is distinct from the other Kitasatospora clade generated using reference sequences alone. These two clades likely correspond the lineage II and III, respectively, recently reported for Kitasatospora based on all available genome sequences (Li et al., 2021). Despite its small genome, there is significant core genome shared between SUK42 and the other members of this clade. For example, SUK42 has a genome size around 2/3 that of K. purpeofusca and perhaps represent specialization through genomic reduction that has allowed SUK42 to adapt to its endophytic life-style. SsgB is a unique cell division protein required for the proper placement of the cytokinetic protein FtsZ in vegetative and aerial hyphae of S. coelicolor (Grantcharova et al., 2005). Z ring placement is a major determinant of the mycelial lifestyle of streptomycetes (Jyothikumar et al., 2008) and its conserved structure illustrates its likely similar role in Kitasatospora. The shared, distinctive amino acid sequence of SsgB of SUK42 and other Kitasatospora strains confirms SUK42 as a member of this genus (Girard et al., 2013).
The pan-genome of SUK42 and the eight Kitasatospora reference sequences showed that the core genome was consistent with a recent study (1,458 vs. 1,498, respectively) (Li et al., 2021). This study also demonstrated that, like Streptomyces, Kitasatospora has an open genome. On the basis of available reference sequences, it was possible to affiliate SUK42 with a clade containing the four other reference sequences Group 1 that displayed similar core genomes that differed from that of Group 2. The recent study of Kitasatospora genomes (Li et al., 2021) also identified four major clades within the genus and, based on the related reference sequences to SUK42, this strains lies in lineage II. It will be interesting to discover if future endophytic Kitasatospora genomes are found in this lineage.
Investigation of the subsystems encoded by SUK42 in comparison to the other Kitasatospora reference sequences revealed differences in the classes of metabolic pathways encoded by different strains. Despite its relatively small genome (∼6.55 Mb compared to an average of 8.25 Mb), SUK42 has an increased proportion of subsystems such as Cell Wall and Dormancy and a reduced proportion of subsystems like Cofactors, Iron acquisition, Virulence, Stress response and especially Nitrogen and Secondary Metabolism. It is tempting to suggest that these functions might be provided by the host plant and permitted the loss of the genetic material encoding this functionality during genome reduction of this strain. If true, this suggest that SUK42 may be undergoing streamlining selection to minimize the burden of encoding unnecessary metabolic pathways. Clearly however, the fact that we could cultivate SUK42 shows that this organism is not dependent on its host, at least in the laboratory environment. The concept that endophytes have reduced genomes as compared to their relatives and might be related to the endophytic lifestyle is not new, since genome reduction is reported in endophytic bacteria from the genus Enterobacter (Lopez-Fernandez et al., 2015).
The subsystem showing the largest reduction in SUK42 was the Secondary Metabolism class, so we analysed the SUK42 and Kitasatospora reference sequences in more details using antiSMASH and BiG-SCAPE. This analysis showed that SUK42 encoded 35 BGCs and the average for the reference sequences was 54 and illustrates that SUK42 encodes fewer BGCs than the average and is perhaps consistent with the idea that an endophyte does not need the same repertoire of bioactive weapons in its arsenal as inhabitants of the heterogeneous, complex and more competitive environment of soil. Consequently, those BCGs retained by SUK42 and found in all Kitastaospora reference sequences, such as FAM_00187, encoding hopene biosynthesis (Bentley et al., 2002) likely play an important role in the life-style of this organism. Intriguingly, the only members of the saccharide class of BGCs found in the Kitasatopora sequences were found in SUK42. A member of the Terpene GCF displayed similarity to the cluster encoding the BCG for acarviostatin I03 from S. coelicoflavus ZG0656 (Guo et al., 2012). This compound displays α-amylase inhibition and suggests that the inhibition first reported for the host-plant of SUK42, A. neurocarpum Miq (Elya et al., 2012), might be as a result of this metabolite produced by the endophyte. Glucosidase inhibitors, such as Acarviostatin, are used as treatments and prophylactics for diabetes, hyperlipoproteinemia, hyperlipidemia, obesity, or other secondary symptoms caused by these diseases (Qin et al., 2011) and highlights that clinical properties of medicinal plants may be due to compounds produced by their endophytic microbial partners. In scientific terms, intriguing questions remains about whether this terpene or the saccharides are produced in planta and, if so, what role they might perform. If indeed a symbiotic relationship exists between SUK42 and A. neurocarpum, what benefits does each partner gain from the relationship?
Data Availability Statement
The datasets presented in this study can be found in online repositories and the supplementary data. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, LFMD00000000.
Author Contributions
AI, GW, and JKS isolated and characterised SUK42. DRM and PRH carried out data visualisation and data curation. PRH wrote the article whilst all authors were involved with review and editing of the article. NMZ provided the resources and, along with PRH, provided the conceptualization for the study.
Funding
This work received financial support from the Ministry of Higher Education (Grant number ERGS/1/2013/SKK04/UKM/02/2) and the Centre for Research and Innovation Management (CRIM), Universiti Kebangsaan Malaysia (NMZ and AI). DRM was supported by a University of Strathclyde and a Tools and Resources Development Fund award from the Biotechnology and Biological Sciences Research Council (grant number BB/M018792/1) (JKS) was awarded to PRH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.740722/full#supplementary-material
References
Afgan, E., Baker, D., VAN Den Beek, M., Blankenberg, D., Bouvier, D., Čech, M., et al. (2016). The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2016 Update. Nucleic Acids Res. 44–W3-W10. doi:10.1093/nar/gkw343
Ahmad, S. J., Mohamad Zin, N., Mazlan, N. W., Baharum, S. N., Baba, M. S., and Lau, Y. L. (2021). Metabolite Profiling of Endophytic Streptomyces Spp. And its Antiplasmodial Potential. PeerJ 9, e10816. doi:10.7717/peerj.10816
Alanjary, M., Steinke, K., and Ziemert, N. (2019). AutoMLST: an Automated Web Server for Generating Multi-Locus Species Trees Highlighting Natural Product Potential. Nucleic Acids Res. 47, W276–W282. doi:10.1093/nar/gkz282
Alshaibani, M. M., Zin, N. M., Jalil, J., Sidik, N. M., Ahmad, S. J., Kamal, N., et al. (2017). Isolation, Purification, and Characterization of Five Active Diketopiperazine Derivatives from Endophytic Streptomyces SUK 25 with Antimicrobial and Cytotoxic Activities. J. Microbiol. Biotechnol. 27, 1249–1256. doi:10.4014/jmb.1608.08032
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H.-P., et al. (2016). Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi:10.1128/mmbr.00019-15
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A.-M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete Genome Sequence of the Model Actinomycete Streptomyces Coelicolor A3(2). Nature 417, 141–147. doi:10.1038/417141a
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 47, W81–W87. doi:10.1093/nar/gkz310
Bosi, E., Donati, B., Galardini, M., Brunetti, S., Sagot, M.-F., Lió, P., et al. (2015). MeDuSa: a Multi-Draft Based Scaffolder. Bioinformatics 31, 2443–2451. doi:10.1093/bioinformatics/btv171
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 5, 8365. doi:10.1038/srep08365
Coombs, J. T., and Franco, C. M. M. (2003). Isolation and Identification of Actinobacteria from Surface-Sterilized Wheat Roots. Appl. Environ. Microbiol. 69, 5603–5608. doi:10.1128/aem.69.9.5603-5608.2003
Corretto, E., Antonielli, L., Sessitsch, A., Compant, S., Höfer, C., Puschenreiter, M., et al. (2017). Complete Genome Sequence of the Heavy Metal Resistant Bacterium Agromyces Aureus AR33T and Comparison with Related Actinobacteria. Stand. Genomic Sci. 12, 2. doi:10.1186/s40793-016-0217-z
Elya, B., Basah, K., Mun'Im, A., Yuliastuti, W., Bangun, A., and Septiana, E. K. (2012). Screening of α-glucosidase Inhibitory Activity from Some Plants of Apocynaceae, Clusiaceae, Euphorbiaceae, and Rubiaceae. J. Biomed. Biotechnol. 2012, 281078. doi:10.1155/2012/281078
Girard, G., Traag, B. A., Sangal, V., Mascini, N., Hoskisson, P. A., Goodfellow, M., et al. (2013). A Novel Taxonomic Marker that Discriminates between Morphologically Complex Actinomycetes. Open Biol. 3, 130073. doi:10.1098/rsob.130073
Gordon, R. E., Barnett, D. A., Handerhan, J. E., and Pang, C. H.-N. (1974). Nocardia Coeliaca, Nocardia Autotrophica, and the Nocardin Strain. Int. J. Syst. Bacteriol. 24, 54–63. doi:10.1099/00207713-24-1-54
Gottlieb, D. (1961). An Evaluation of Criteria and Procedures Used in the Description and Characterization of the Streptomycetes. Appl. Microbiol. 9, 55–65. doi:10.1128/am.9.1.55-65.1961
Grant, J. R., and Stothard, P. (2008). The CGView Server: a Comparative Genomics Tool for Circular Genomes. Nucleic Acids Res. 36, W181–W184. doi:10.1093/nar/gkn179
Grantcharova, N., Lustig, U., and Flärdh, K. (2005). Dynamics of FtsZ Assembly during Sporulation in Streptomyces Coelicolor A3(2). J. Bacteriol. 187, 3227–3237. doi:10.1128/jb.187.9.3227-3237.2005
Gu, Z., Gu, L., Eils, R., Schlesner, M., and Brors, B. (2014). Circlize Implements and Enhances Circular Visualization in R. Bioinformatics 30, 2811–2812. doi:10.1093/bioinformatics/btu393
Guo, X., Geng, P., Bai, F., Bai, G., Sun, T., Li, X., et al. (2012). Draft Genome Sequence of Streptomyces Coelicoflavus ZG0656 Reveals the Putative Biosynthetic Gene Cluster of Acarviostatin Family α-amylase Inhibitors. Lett. Appl. Microbiol. 55, 162–169. doi:10.1111/j.1472-765x.2012.03274.x
Hadfield, J., Croucher, N. J., Goater, R. J., Abudahab, K., Aanensen, D. M., and Harris, S. R. (2018). Phandango: an Interactive Viewer for Bacterial Population Genomics. Bioinformatics 34, 292–293. doi:10.1093/bioinformatics/btx610
Ichikawa, N., Oguchi, A., Ikeda, H., Ishikawa, J., Kitani, S., Watanabe, Y., et al. (2010). Genome Sequence of Kitasatospora Setae NBRC 14216T: an Evolutionary Snapshot of the Family Streptomycetaceae. DNA Res. 17, 393–406. doi:10.1093/dnares/dsq026
Jyothikumar, V., Tilley, E. J., Wali, R., and Herron, P. R. (2008). Time-lapse Microscopy of Streptomyces Coelicolor Growth and Sporulation. Appl. Environ. Microbiol. 74, 6774–6781. doi:10.1128/aem.01233-08
Kallio, P., Liu, Z., Mäntsälä, P., Niemi, J., and Metsä-Ketelä, M. (2008). Sequential Action of Two Flavoenzymes, PgaE and PgaM, in Angucycline Biosynthesis: Chemoenzymatic Synthesis of Gaudimycin C. Chem. Biol. 15, 157–166. doi:10.1016/j.chembiol.2007.12.011
Kato, J.-Y., Funa, N., Watanabe, H., Ohnishi, Y., and Horinouchi, S. (2007). Biosynthesis of -butyrolactone Autoregulators that Switch on Secondary Metabolism and Morphological Development in Streptomyces. Proc. Natl. Acad. Sci. 104, 2378–2383. doi:10.1073/pnas.0607472104
Kautsar, S. A., Blin, K., Shaw, S., Navarro-Muñoz, J. C., Terlouw, B. R., VAN DER Hooft, J. J. J., et al. (2020). MIBiG 2.0: a Repository for Biosynthetic Gene Clusters of Known Function. Nucleic Acids Res. 48, D454–D458. doi:10.1093/nar/gkz882
Keijser, B. J. F., Noens, E. E. E., Kraal, B., Koerten, H. K., and Wezel, G. P. (2003). TheStreptomyces Coelicolor ssgBgene Is Required for Early Stages of Sporulation. FEMS Microbiol. Lett. 225, 59–67. doi:10.1016/s0378-1097(03)00481-6
Kodani, S., Hudson, M. E., Durrant, M. C., Buttner, M. J., Nodwell, J. R., and Willey, J. M. (2004). From the Cover: The SapB Morphogen Is a Lantibiotic-like Peptide Derived from the Product of the Developmental Gene ramS in Streptomyces Coelicolor. Proc. Natl. Acad. Sci. 101, 11448–11453. doi:10.1073/pnas.0404220101
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Labeda, D. P., Goodfellow, M., Brown, R., Ward, A. C., Lanoot, B., Vanncanneyt, M., et al. (2012). Phylogenetic Study of the Species within the Family Streptomycetaceae. Antonie Van Leeuwenhoek 101, 73–104. doi:10.1007/s10482-011-9656-0
Lee, I., Ouk Kim, Y., Park, S.-C., and Chun, J. (2016). OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi:10.1099/ijsem.0.000760
Li, Y., Wang, M., Sun, Z.-Z., and Xie, B.-B. (2021). Comparative Genomic Insights into the Taxonomic Classification, Diversity, and Secondary Metabolic Potentials of Kitasatospora, a Genus Closely Related to Streptomyces. Front. Microbiol. 12. 683814, doi:10.3389/fmicb.2021.683814
Liu, Y., Li, M., Mu, H., Song, S., Zhang, Y., Chen, K., et al. (2017). Identification and Characterization of the Ficellomycin Biosynthesis Gene Cluster from Streptomyces Ficellus. Appl. Microbiol. Biotechnol. 101, 7589–7602. doi:10.1007/s00253-017-8465-4
Lòpez-Fernàndez, S., Sonego, P., Moretto, M., Pancher, M., Engelen, K., Pertot, I., et al. (2015). Whole-genome Comparative Analysis of Virulence Genes Unveils Similarities and Differences between Endophytes and Other Symbiotic Bacteria. Front. Microbiol. 6, 419. doi:10.3389/fmicb.2015.00419
Meindl, K., Schmiederer, T., Schneider, K., Reicke, A., Butz, D., Keller, S., et al. (2010). Labyrinthopeptins: a New Class of Carbacyclic Lantibiotics. Angew. Chem. Int. Ed. 49, 1151–1154. doi:10.1002/anie.200905773
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D., and Gurevich, A. (2018). Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 34, i142–i150. doi:10.1093/bioinformatics/bty266
Momose, I., Sekizawa, R., Hirosawa, S., Ikeda, D., Naganawa, H., Iinuma, H., et al. (2001). Tyropeptins A and B, New Proteasome Inhibitors Produced by Kitasatospora Sp. MK993-dF2. II. Structure Determination and Synthesis. J. Antibiot. 54, 1004–1012. doi:10.7164/antibiotics.54.1004
Navarro-Muñoz, J. C., Selem-Mojica, N., Mullowney, M. W., Kautsar, S. A., Tryon, J. H., Parkinson, E. I., et al. (2020). A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol. 16, 60–68. doi:10.1038/s41589-019-0400-9
Nouioui, I., Carro, L., García-López, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-Based Taxonomic Classification of the Phylum Actinobacteria. Front. Microbiol. 9, 2007. doi:10.3389/fmicb.2018.02007
Omura, S., Takahashi, Y., Iwai, Y., and Tanaka, H. (1982). Kitasatosporia, a New Genus of the Order Actinomycetales. J. Antibiot. 35, 1013–1019. doi:10.7164/antibiotics.35.1013
Overbeek, R., Begley, T., Butler, R. M., Choudhuri, J. V., Chuang, H. Y., Cohoon, M., et al. (2005). The Subsystems Approach to Genome Annotation and its Use in the Project to Annotate 1000 Genomes. Nucleic Acids Res. 33, 5691–5702. doi:10.1093/nar/gki866
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: Rapid Large-Scale Prokaryote pan Genome Analysis. Bioinformatics 31, 3691–3693. doi:10.1093/bioinformatics/btv421
Qin, X., Ren, L., Yang, X., Bai, F., Wang, L., Geng, P., et al. (2011). Structures of Human Pancreatic α-amylase in Complex with Acarviostatins: Implications for Drug Design against Type II Diabetes. J. Struct. Biol. 174, 196–202. doi:10.1016/j.jsb.2010.11.020
Sarmin, N. I. M., Tan, G. Y. A., Franco, C. M. M., Edrada-Ebel, R., Latip, J., and Zin, N. M. (2013). Streptomyces Kebangsaanensis Sp. nov., an Endophytic Actinomycete Isolated from an Ethnomedicinal Plant, Which Produces Phenazine-1-Carboxylic Acid. Int. J. Syst. Evol. Microbiol. 63, 3733–3738. doi:10.1099/ijs.0.047878-0
Seemann, T. (2014). Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 30, 2068–2069. doi:10.1093/bioinformatics/btu153
Shirling, E. B., and Gottlieb, D. (1966). Methods for Characterization of Streptomyces Species. Int. J. Syst. Bacteriol. 16, 313–340. doi:10.1099/00207713-16-3-313
Singh, R., and Dubey, A. K. (2018). Diversity and Applications of Endophytic Actinobacteria of Plants in Special and Other Ecological Niches. Front. Microbiol. 9, 1767. doi:10.3389/fmicb.2018.01767
Staneck, J. L., and Roberts, G. D. (1974). Simplified Approach to Identification of Aerobic Actinomycetes by Thin-Layer Chromatography. Appl. Microbiol. 28, 226–231. doi:10.1128/am.28.2.226-231.1974
Strobel, G. A. (2003). Endophytes as Sources of Bioactive Products. Microbes Infect. 5, 535–544. doi:10.1016/s1286-4579(03)00073-x
Takahashi, Y. (2017). Genus Kitasatospora, Taxonomic Features and Diversity of Secondary Metabolites. J. Antibiot. 70, 506–513. doi:10.1038/ja.2017.8
Traag, B. A., and Van Wezel, G. P. (2008). The SsgA-like Proteins in Actinomycetes: Small Proteins up to a Big Task. Antonie Van Leeuwenhoek 94, 85–97. doi:10.1007/s10482-008-9225-3
Wellington, E. M. H., Stackebrandt, E., Sanders, D., Wolstrup, J., and Jorgensen, N. O. G. (1992). Taxonomic Status of Kitasatosporia, and Proposed Unification with Streptomyces on the Basis of Phenotypic and 16S rRNA Analysis and Emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int. J. Syst. Bacteriol. 42, 156–160. doi:10.1099/00207713-42-1-156
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. Plos Comput. Biol. 13, e1005595. doi:10.1371/journal.pcbi.1005595
Willemse, J., Borst, J. W., DE Waal, E., Bisseling, T., and VAN Wezel, G. P. (2011). Positive Control of Cell Division: FtsZ Is Recruited by SsgB during Sporulation of Streptomyces. Genes Development 25, 89–99. doi:10.1101/gad.600211
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a Taxonomically United Database of 16S rRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi:10.1099/ijsem.0.001755
Yoon, T. M., Kim, J. W., Kim, J. G., Kim, W. G., and Suh, J. W. (2006). Talosins A and B: New Isoflavonol Glycosides with Potent Antifungal Activity from Kitasatospora Kifunensis MJM341. J. Antibiot. 59, 633–639. doi:10.1038/ja.2006.84
Keywords: endophyte, Kitasatospora, genome, pan-genome, Antidesma neurocarpum miq
Citation: Zin NM, Ismail A, Mark DR, Westrop G, Schniete JK and Herron PR (2021) Adaptation to Endophytic Lifestyle Through Genome Reduction by Kitasatospora sp. SUK42. Front. Bioeng. Biotechnol. 9:740722. doi: 10.3389/fbioe.2021.740722
Received: 13 July 2021; Accepted: 20 September 2021;
Published: 12 October 2021.
Edited by:
Adeline Su Yien Ting, Monash University Malaysia, MalaysiaReviewed by:
Christopher Milton Mathew Franco, Flinders University, AustraliaAshok K Dubey, Netaji Subhas University of Technology, India
Copyright © 2021 Zin, Ismail, Mark, Westrop, Schniete and Herron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul R. Herron, cGF1bC5oZXJyb25Ac3RyYXRoLmFjLnVr
 Noraziah M. Zin
Noraziah M. Zin Aishah Ismail
Aishah Ismail David R. Mark
David R. Mark Gareth Westrop
Gareth Westrop Jana K. Schniete
Jana K. Schniete Paul R. Herron
Paul R. Herron