- School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan, South Korea
The present study elaborates on the propionic acid (PA) production by the well-known microbial cell factory Pseudomonas putida EM42 and its capacity to utilize biomass-derived levulinic acid (LA). Primarily, the P. putida EM42 strain was engineered to produce PA by deleting the methylcitrate synthase (PrpC) and propionyl-CoA synthase (PrpE) genes. Subsequently, a LA-inducible expression system was employed to express yciA (encoding thioesterase) from Haemophilus influenzae and ygfH (encoding propionyl-CoA: succinate CoA transferase) from Escherichia coli to improve the PA production by up to 10-fold under flask scale cultivation. The engineered P. putida EM42:ΔCE:yciA:ygfH was used to optimize the bioprocess to further improve the PA production titer. Moreover, the fed-batch fermentation performed under optimized conditions in a 5 L bioreactor resulted in the titer, productivity, and molar yield for PA production of 26.8 g/L, 0.3 g/L/h, and 83%, respectively. This study, thus, successfully explored the LA catabolic pathway of P. putida as an alternative route for the sustainable and industrial production of PA from LA.
Highlights
• P. putida was engineered to produce propionic acid from levulinic acid.
• A substrate-based inducible system was used to express heterologous genes to avoid using chemical inducers.
• Carbon flux to propionic acid was promoted by expressing ygfH.
• Engineered P. putida produced 26.8 g/L propionic acid with 83% molar yield in a 5 L fed-batch bioreactor.
Introduction
Propionic acid (PA) is a C3 carboxylic acid and a promising second-tier group of building block candidates recommended by the U.S. Department of Energy (Werpy & Petersen, 2004). However, the petrochemical-based process for PA production requires the application of high temperatures and pressures along with toxic chemicals such as ethylene, carbon monoxide, and metal catalysts. This makes the whole process non-renewable and unsustainable, generating many environmental pollutants (Liu et al., 2020). Hence, the conversion of biomass-based substrates into PA by microbial cell factories has emerged as an alternative eco-friendly strategy (Eş et al., 2017). However, biorefinery-based bioprocessing has technical limitations related to the bioconversion process and the efficiency of potential pathways in the selected microbial candidates for large-scale biomanufacturing of PA (Ammar & Philippidis, 2021).
To date, high-level PA production has been achieved through the Wood–Werkman cycle of several candidates belonging to the genus Propionibacteria and the reductive acrylate pathway of Clostridium propionicum under anaerobic conditions using glucose or glycerol as the carbon source (Gonzalez-Garcia et al., 2017a; Collograi et al., 2022). However, Propionibacteria or Clostridium-based PA production has several limitations, including slow growth rates and costly downstream processing due to the high amount of by-products generated during fermentation (Ranaei et al., 2020). A co-culture approach was also performed for PA production. In this approach, Lactobacillus zeae convert glucose to lactate which is the substrate of Veillonella criceti, producing PA with a high productivity rate (Dietz et al., 2013). In addition to these conventional microbial candidates, few attempts have been made to engineer microbial cell factories such as Escherichia coli to produce PA (Akawi et al., 2015; Gonzalez-Garcia et al., 2017a; Gonzalez-Garcia et al., 2017b). Mostly in E. coli, engineering the native sleeping beauty mutase (Sbm) operon or expression of a heterogeneous Wood–Werkman pathway from Propionibacterium has resulted in higher PA production under anaerobic conditions (Srirangan et al., 2013; Gonzalez-Garcia et al., 2020). However, this manufacturing process is adversely affected by the low titer and high by-product generation (Ammar & Philippidis, 2021). Moreover, expensive nitrogen flushing is required to maintain the anaerobic conditions throughout the fermentation process, making the complete process economically unviable (Liu et al., 2016; Li et al., 2017). Recently, L-threonine catabolism was engineered in Pseudomonas putida to produce PA under aerobic conditions (Ma et al., 2020; Mu et al., 2021). This pathway could thus be explored for the production of PA under aerobic conditions. Moreover, a synthetic propionate pathway has been constructed in Saccharomyces cerevisiae by engineering the L-threonine catabolism, thereby resulting in a strain capable of producing the PA from glucose. However, the resultant PA titer (1.05 g/L) was determined to be quite low compared to those reported in other studies (Kidd et al., 2021). L-threonine is the third-most sold amino acid used in livestock feed, pharmaceuticals, and cosmetics (Liu et al., 2019). As L-threonine is mainly produced via fermentative bioprocesses, it cannot be considered the best substrate to produce PA. Although, this cost issue was tried to address by adopting a sequential fermentation−biotransformation process to produce PA directly from the fermentation broth containing unpurified L-threonine (Mu et al., 2021). In a PA biorefinery, the cost of raw materials is approximately 50% of the total cost (Dishisha et al., 2013). Hence, there is a genuine need to select a low-cost and sustainable substrate for PA production. Levulinic acid (LA) is a γ-keto acid (C5) platform chemical that can be obtained by the acid-catalyzed dehydration and hydrolysis of sugars obtained from lignocellulosic biomass substrates (Isoni et al., 2018). As LA can be produced without the use of expensive hydrolytic enzymes, it decreases the total cost of bioprocesses based on lignocellulosic biomass (Habe et al., 2020). The market price of biologically produced PA is 2.00–3.00 $/kg (Ammar & Philippidis, 2021). On the other hand, using biomass is considered a potential way to decrease the LA price by less than 1 $/kg, through Biofine (Hayes et al., 2006) and other processes (Kang et al., 2018; Meramo Hurtado et al., 2021). Thus, LA can serve as a sustainable product that can revitalize the cellulosic biomass-based biorefinery industry. Furthermore, the production cost of LA from biomass has been reduced by the development of LA production technology (Isoni et al., 2018). Complex sugars obtained from the lignocellulosic biomass are used for microbial fermentation, and their production efficiency decreases through long-term process operations owing to sequential sugar application. However, the use of the LA metabolic pathway offers several advantages, such as the lack of complex metabolic regulations and the production of central metabolic intermediates (Kim et al., 2019). Notably, the metabolic pathway of LA utilization was discovered in P. putida KT2440 and further employed in the LA-based biorefineries to produce biologically diverse chemicals (Rand et al., 2017; Sathesh-Prabu & Lee, 2019; Cha et al., 2020).
In the present study, the LA metabolic capacity of P. putida EM42 to produce PA was explored. Moreover, an engineered strain of the same was developed by co-expressing heterologous thioesterase (YciA) and propionyl-CoA: succinate-CoA transferase (YgfH) under a substrate (LA)-based inducible promoter system to avoid the requirement of other chemical inducers throughout the bioprocess. Furthermore, the assessment of titer and PA productivity along with a large-scale fermentation process was performed to confirm the efficiency of the engineered strain for LA-based PA production.
Materials and methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH10B was used as the cloning host for all the experiments. P. putida EM42 strain obtained from Centro Nacional de Biotecnología (CNB-CSIC, Spain) was used as the parental strain for the gene deletion and heterologous gene expression required for PA production. The genomic DNA of Haemophilus influenzae (DSM 11121) was purchased from the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ, Germany).
Restriction enzymes (Thermo Scientific, United States), Q5 High-Fidelity DNA Polymerase (New England Biolabs, United States), and e-Taq Polymerase (SolGent, South Korea) were used for the cloning and plasmid construction. Electro-competent cells were prepared as described previously (Luo et al., 2016). The suicide plasmid pQSAK was used to construct pQSAK-prpC and pQSAK-prpE, which were used for the chromosomal in-frame gene deletion based on the sacB-negative counter-selection system (Zhou et al., 2014). To delete the prpC and prpE, the pQSAK plasmid was constructed using the Gibson assembly cloning method, containing approximately 500 bp upstream and downstream homologous regions of each gene. The plasmid construct was then transformed into P. putida EM42. After double homologous recombination, the colonies were picked from sucrose-kanamycin plates for negative selection using SacB. The deletion was confirmed by PCR followed by DNA sequencing.
In a previous study, an LA-inducible expression system was developed using the transcriptional activator LvaR and its cognate lvaA promoter from the lva operon of P. putida KT2440. Subsequently, the plasmid pPROBE_LvaR/PlvaA_egfp+ was constructed, and LA-based induction was analyzed by expressing the green fluorescent protein as a reporter protein (Sathesh-Prabu et al., 2021). To construct the pPROBE_LvaR/PlvaA_yciA and pPROBE_LvaR/PlvaA_ygfH plasmids, the yciA and ygfH genes were amplified from H. influenzae and E. coli, respectively, using the primers described in Supplementary Table S1. The amplified gene products were then cloned by digesting the pPROBE_LvaR/PlvaA_egfp+ with NdeI/HindIII and replacing the egfp+ with each gene using the Gibson assembly cloning method. The pPROBE_LvaR/PlvaA_yciA_ygfH plasmid was constructed by employing a transcriptional fusion of the two genes separated by the RBS sequence. The constructed plasmids were then transformed into the EM42:ΔCE strain to yield the EM42:ΔCE:yciA, EM42:ΔCE:ygfH, and EM42:ΔCE:yciA:ygfH strains. All the constructed plasmids were confirmed by sequencing (Macrogen, South Korea).
Media and cultivation conditions
The LA (Sigma-Aldrich) was neutralized to pH 7.0 by 10 N NaOH and sterilized by autoclaving before use. The Luria-Bertani (LB) medium (5 g/L yeast extract, 10 g/L peptone, and 10 g/L NaCl) and Terrific Broth (TB) medium (12 g/L tryptone, 24 g/L yeast extract; 9.4 g/L dibasic potassium phosphate, and 2.2 g/L monobasic potassium phosphate) were used to cultivate the E. coli and P. putida strains at 37 and 30°C, respectively, under shaking conditions at 200 rpm. To maintain the plasmid construct, the medium was supplemented with 50 μg/ml kanamycin (Km).
For the cultivation, the cells were streaked on LB agar plates (with or without Km, as required) and incubated overnight under the prescribed growth conditions. Subsequently, the colonies were inoculated into a 5 ml LB medium and incubated for 16 h under shaking conditions. The starter culture (0.5 ml) was then inoculated in a 250 ml flask containing 20 ml of TB medium containing either LA, glycerol, or both, as per the experimental conditions. Glycerol was selected as the co-substrate with LA for the production of PA. Samples were collected periodically for further analysis. All the experiments were performed in triplicate. The conversion of LA to PA was calculated on a molar basis and represented as molar yield (%), where only consumed LA was used to calculate the molar yield to avoid other unknown metabolites involved in the PA production.
For the fed-batch fermentation, experiments were performed in a 5 L bioreactor (MARADO-PDA; CNS, Daejeon, South Korea) with an initial working volume of 1800 ml. The previously described TB medium supplemented with 50 μg/ml Km, was used. The cells were initially grown rapidly on glycerol with supplementation of LA (3 g/L) for induction, and then LA was added after 12 h to initiate PA production. The temperature and agitation speed were set to 30°C and 700 rpm, respectively. In addition, the dissolved oxygen (DO) was maintained by flowing 2 vvm of air. The pH was maintained at 7.0 with 2 M NH4OH and 4 M H3PO4. The LA level was maintained between 5 g/L and 15 g/L by intermittent feeding with 50% LA. Samples were withdrawn periodically to determine the cell growth and concentrations of glycerol, LA, and PA.
Analytical methods
The growth (OD600) of the bacterial strains was observed using a spectrophotometer (Libra S22; Biochrom, UK). For the analysis of glycerol, LA, and PA, the collected samples were diluted, and 20 µl aliquots were injected into an HPX-87H column (Bio-Rad) at 0.5 ml/min and a column temperature of 35°C. The analysis was performed on a Shimadzu HPLC station equipped with a refractive index detector (Shimadzu) and SIL-20A auto-sampler (Shimadzu).
Results and discussion
Construction of engineered strain for the production of propionic acid from levulinic acid
The P. putida EM42 was derived from the platform strain P. putida KT2440 by deleting 300 genes (approximately 4.3% of the genome), resulting in increased ATP levels, oxidative stress, growth rates, and enhanced expression of heterologous genes (Martínez-García et al., 2014). As a result, the EM42 strain is more suitable for expressing heterologous genes and producing biochemicals than the KT2440 (Dvořák & de Lorenzo, 2018). P. putida strain, as it can utilize LA as the sole carbon source with the aid of proteins encoded by polycistronic genes, designated as lvaABCDEFG (Rand et al., 2017). The lva operon is upregulated by the transcriptional activator LvaR, which is induced by LA. In this assimilation pathway, LA is first activated by LvaE and a coenzyme A (CoA) thioester, levulinyl-CoA. Subsequently, the LvaD catalyzes the reduction of LA-CoA with either NADH or NADPH to yield 4-hydroxyvaleryl-CoA. The 4-hydroxyvaleryl-CoA is then phosphorylated to yield 4-phosphovaleryl-CoA by the combined action of LvaA, LvaB, and ATP. This is followed by converting 4-phosphovaleryl-CoA into 3-hydroxyvaleryl-CoA by LvaC, which is further oxidized through β-oxidation to yield acetyl-CoA and propionyl-CoA and is completely oxidized through the TCA cycle. Propionyl-CoA can be used as a precursor for PA production, and acetyl-CoA can be directed to the TCA cycle to promote cell growth. Propionyl-CoA can be consumed through the methylcitrate cycle by methylcitrate synthase (PP2335, prpC). Moreover, PA can be degraded to propionyl-CoA by propionyl-CoA synthase (PP2351, prpE). High production of PA requires an accumulation of its precursor, propionyl-CoA. As a result, prpC and prpE were deleted from the EM42 strain individually and combined to obtain the EM42:ΔC, EM42:ΔE, and EM42:ΔCE strains. Our growth curve studies demonstrated that these deletions did not affect the cell viability in the TB medium with LA (10 g/L) as the carbon source (Figure 1A). Furthermore, the assessment of PA production after 48 h revealed that while the EM42 strain cannot produce PA, the EM42:ΔCE strain exhibited maximum PA production of 0.31 g/L, accounting for a 9.6% molar yield. Our results also showed that the deletion of both genes allows the accumulation of propionyl-CoA and restricts the degradation of PA to propionyl-CoA, thereby achieving the highest PA among all strains (Figure 1B). However, the LA consumption was affected by these deletions as 48 h post-incubation, the EM42 consumed 9.6 g/L of LA, whereas the EM42:ΔC and EM42:ΔCE strains showed the consumption of only 4.6 and 4.5 g/L of LA, respectively (Figure 1C). This result, thus, showed that the deletion of the PrpC gene instigated the accumulation of propionyl-CoA by blocking the methylcitrate cycle, resulting in lower LA consumption.
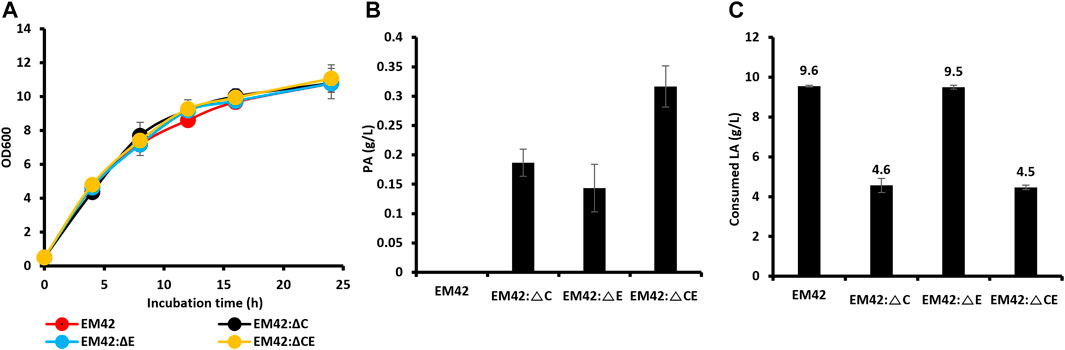
FIGURE 1. The growth pattern (A), PA production (B), and concentration of residual LA (C) by wild type (EM42) and other strains with deleted prpC and prpE. The presented value is the mean value of three experiments. The standard deviation is represented by error bars.
Heterologous expression for improved propionic acid production
Assessment of PA levels revealed that the knockout strain EM42:ΔCE showed significant production of PA as compared to that of the wild-type EM42; however, the production level was quite low. Hence, the conversion of propionyl-CoA to PA can be a limiting step catalyzed by thioesterases. To address this limitation, the well-known thioesterase (YciA) gene from H. influenzae DSM 11121 (Zhuang et al., 2008) was expressed in an LA-based inducible system (LvaR/PlvaA) from P. putida. This substrate-based induction can avoid the use of costly chemical inducers and the extra metabolic burden on the cell (Ko et al., 2020). Moreover, this promoter system has already been used for the enhanced production of 4-hydroxyvalerate from LA in P. putida KT2440 (Sathesh-Prabu & Lee, 2019). The resultant EM42:ΔCE:yciA strain utilized LA almost completely (9.8 g/L) after 48 h, which was similar to wild type EM42 and 46% higher than that noted with EM42:ΔCE. Additionally, the expression of exogenous thioesterase also improved PA production from LA by 10-fold compared to that demonstrated by the EM42:ΔCE strain, with a molar yield of 52% (Figure 2A). This result thus, confirmed that the low expression level of indigenous thioesterases was responsible for the lower PA production. Recently, a thioesterase from P. putida KT2440, encoded by PP4975, was involved in PA production (Ma et al., 2021). However, the activity of thioesterases from P. putida KT2440 was quite low in EM42:ΔCE:yciA strain.
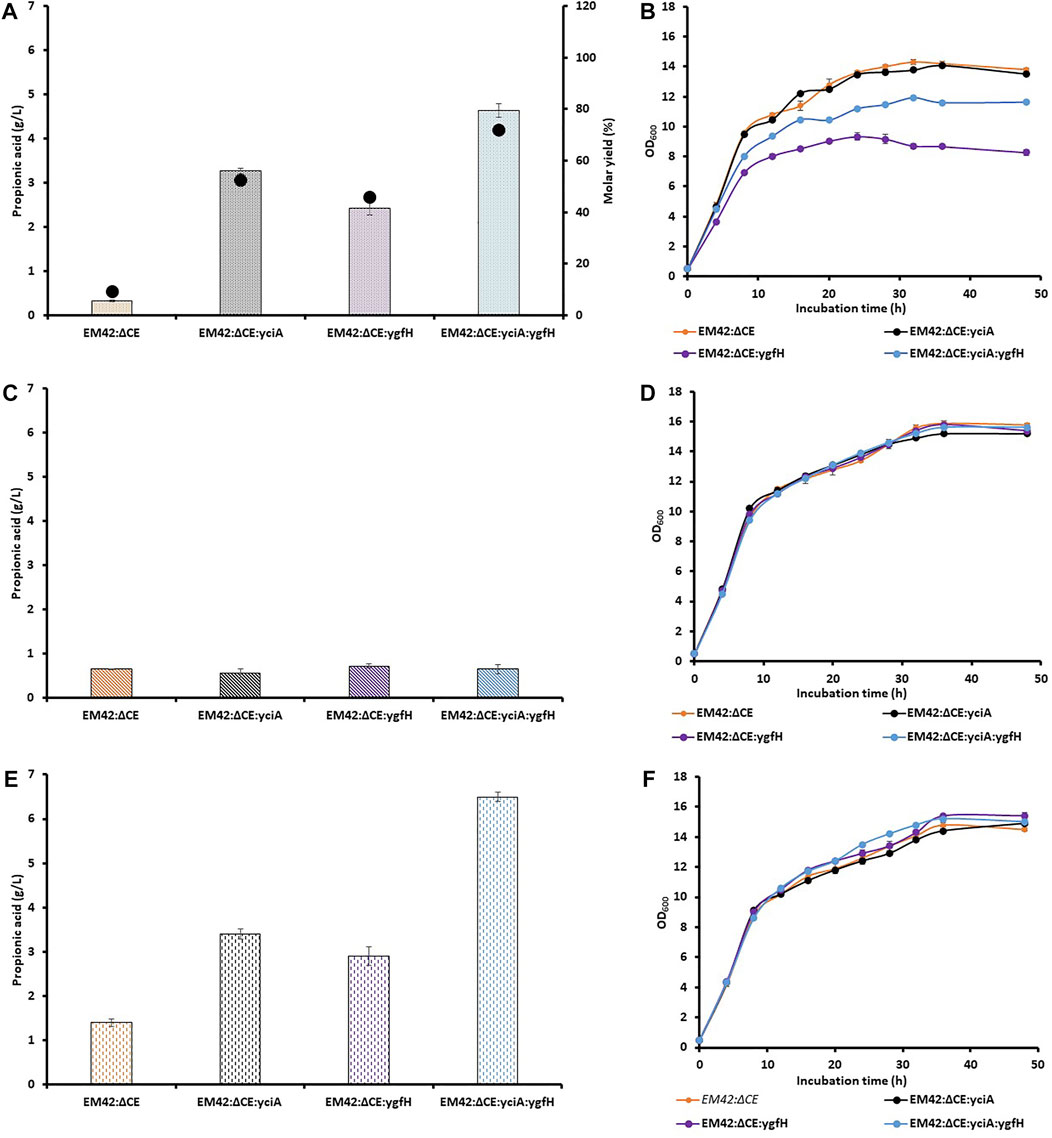
FIGURE 2. Production of PA and molar yield (%) (A,C,E), and growth pattern (B,D,F) of selected strains, including EM42:ΔCE, EM42:ΔCE:yciA, EM42:ΔCE:ygfH, and EM42:ΔCE:yciA:ygfH growing on LA (A,B), Glycerol (C,D) and LA + Glycerol (E,F). The presented value is the mean from three experiments. The standard deviation is represented by error bars.
Even after the successful expression of YciA and enhanced PA production, there is still scope to improve the titer and molar yield. In the engineered EM42:ΔCE:yciA strain, the propionyl-CoA was not linked to the central metabolic pathway but only led to its conversion into PA. In contrast, acetyl-CoA is available for the production of cell biomass and other metabolites. In a previous report, 55.5% of LA was presumably metabolized to CO2, cell mass, and other products in P. putida (Gorenflo et al., 2001). It is not adequate to achieve a high LA-to-PA conversion rate. Overexpression of propionyl-CoA:succinate CoA transferase in Propionibacterium freudenreichii subsp. shermanii resulted in diverted carbon fluxes toward PA and higher PA content (Wang et al., 2015). The enzyme transfers the CoA group of the propionyl-CoA product to succinate and primes the succinate to facilitate propionate decarboxylation (Haller et al., 2000). Consequently, we attempted to link the central metabolic pathway with propionyl-CoA-based PA production by overexpressing the propionyl-CoA:succinate CoA transferase (YgfH) gene from E. coli in EM42:ΔCE. The resulting EM42:ΔCE:ygfH strain produced 2.4 g/L of PA under the same culture conditions, which was 7-fold higher than that produced by EM42:ΔCE (Figure 2A). It has been reported that YgfH not only cycles the CoA pool between propionyl-CoA and succinate-CoA but also influences the carbon flow toward PA production (Wang et al., 2015). The EM42:ΔCE:ygfH strain did not consume the LA completely, and 1.8 g/L of residual LA was still available after 48 h of incubation. However, this carbon flow was not equivalent to EM42:ΔCE:yciA and hence resulted in a lower PA production (25%) and molar yield (11%) (Figure 2A). These findings thus revealed that the expression of YgfH could improve the PA production in the EM42:ΔCE strain. Furthermore, a plasmid was constructed with the transcriptional fusion of yciA and ygfH under the same LA-inducible (LvaR/PlvaA) promoter system and transformed into EM42:ΔCE to obtain the EM42:ΔCE:yciA:ygfH strain. Our results showed that the co-expression of exogenous YciA and YgfH remarkably improved the PA production by up to 4.6 g/L (Figure 2A). Moreover, as compared to EM42:ΔCE:yciA and EM42:ΔCE:ygfH, the co-expression of both genes in the EM42:ΔCE increased the PA production by 30 and 48%, respectively. The EM42:ΔCE:yciA:ygfH also improved the molar yield to 72%, which indicates the maximum carbon flow toward PA production. However, the fact that the remaining 28% of LA was not converted into PA needs to be further investigated. As discussed in the previous findings that the EM42:ΔCE also consumed 4.5 g/L of LA; however, only 0.3 g/L of PA was produced. These findings, therefore, confirmed that the consumed LA is metabolized by unknown enzymes or pathways in P. putida other than those involved in the PA production. These unknown enzymes must be revealed in future investigations to further facilitate the improved PA production in P. putida by accumulating more propionyl-CoA.
Selection of glycerol as co-substrate
The results of the previous experiments indicate a low carbon flow availability for the bacterial biomass in the case of EM42:ΔCE:yciA:ygfH-mediated production of PA. This finding was further confirmed by the reduced growth of EM42:ΔCE:yciA:ygfH as compared to that of the EM42:ΔCE:yciA, EM42:ΔCE:ygfH, and EM42:ΔCE strains (Figure 2B). Although the EM42:ΔCE:yciA:ygfH strain produced a higher PA titer and molar yield, this growth defect must be addressed before large-scale fermentation. To resolve this issue, glycerol was used as a co-substrate because it is known to provide high reducing power and has no catabolic repression with LA (Nikel et al., 2014). Accordingly, 5 g/L glycerol was initially added separately and then in combination with LA (10 g/L), and the growth, substrate(s) consumption, and PA production were estimated in all the four strains, including EM42:ΔCE, EM42:ΔCE:yciA:ygfH, EM42:ΔCE:ygfH, and EM42:ΔCE:yciA:ygfH (Figures 2C,F). Subsequent analysis revealed that the media containing glycerol alone initially showed no growth differences among the four strains (Figure 2D). However, the expression of YciA and/or YgfH showed no significant difference in PA production, possibly because glycerol could not provide sufficient propionyl-CoA as a precursor for PA production (Figure 2C). In contrast, propionyl-CoA can be generated from the degradation of the amino acids L-methionine, L-isoleucine, and L-valine and as an end-product of the β-oxidation of uneven fatty acids (Thompson et al., 2020). In this study, the nutrient-rich TB medium was used, which provided these amino acids as a source of propionyl-CoA. Hence, while using glycerol as a substrate, 0.6–0.7 g/L PA was produced from all four strains despite the expression of both enzymes. Subsequently, both substrate (LA) and co-substrate (glycerol) were added to the medium and cultivated for 48 h. Our results revealed that the addition of glycerol showed no growth defects that in this case, and all the four strains displayed a remarkably similar growth profile (Figure 2F). Furthermore, the PA production was enhanced in all four strains, but the maximum PA production was achieved by EM42:ΔCE:yciA:ygfH strain at 6.5 g/L (Figure 2E). These results validate the selection of glycerol as a co-substrate, as it is required to eliminate growth defects and improve PA production. Moreover, our results also revealed that the EM42:ΔCE:yciA:ygfH strain completely utilized glycerol and LA after 48 h of incubation. These findings again confirmed that the propionyl-CoA generated from the media source also contributed to the PA, as in the previous experiments, PA was produced although at a lower capacity when glycerol alone was used as a carbon source (Figures 2C,E).
Production of propionic acid in fed-batch bioreactor cultivation
Initially, the accumulation of propionyl-CoA in the EM42 strain was enhanced by deleting the PrpC gene responsible for its assimilation through the methylcitrate cycle. Likewise, the interconversion of PA to propionyl-CoA was blocked by the deletion of the prpE. Subsequently, the final strain, EM42:ΔCE:yciA:ygfH, was obtained after co-expressing YciA and YgfH, which are responsible for the PA production and diverting more carbon flow toward PA (Figure 3). The fed-batch fermentation was then carried out in a 5-L bioreactor using the EM42:ΔCE:yciA:ygfH strain to validate the PA production on a large scale. The biomass cultivation was initiated by adding 5 g/L glycerol and 3 g/L LA as inducers. Our growth curve assessment revealed that the OD600 reached approximately 20 after cultivation for 12 h, followed by the addition of 10 g/L of LA to initiate the PA production (Figure 4). Post-continuous LA supply as a carbon source, the maximum OD600 of the culture reached up to 35.4 after 48 h of incubation. Subsequently, the growth stopped, but the PA production continued. Further analysis revealed that the maximum PA titer of 26.8 g/L with a molar yield of 83% and productivity of 0.3 g/L/h was obtained after consuming 49.56 g of LA as a carbon source (Figure 4). By-products, such as succinate and acetate, could not be detected during the entire bioprocess. These findings are thus indicative of high molar yield at the flask scale as well as large-scale fermentation and, therefore, significant from an economic point of view. Hence, the selection of low-cost substrates such as LA and a high molar yield can enhance the profit margin for the industrial production of PA (Ahmadi et al., 2017). However, further optimization in large-scale fermentation needs to be conducted to enhance the PA titer to be comparable with Propionibacterium candidates. In this study, an enriched TB medium was used to produce PA. In future, optimization of growth components is required to render the medium cost to expand not only the sustainability of complete bioprocess but also further improve the PA productivity and titer. Moreover, compared to previous report where L-threonine was used as a substrate, the higher titer of 62 g/L and productivity of 1.07 g/L/h was achieved by the fed-batch biotransformation coupled process (Mu et al., 2021). Thus, while high-density culture can increase productivity and titer, biotransformation on an industrial scale is a difficult process.
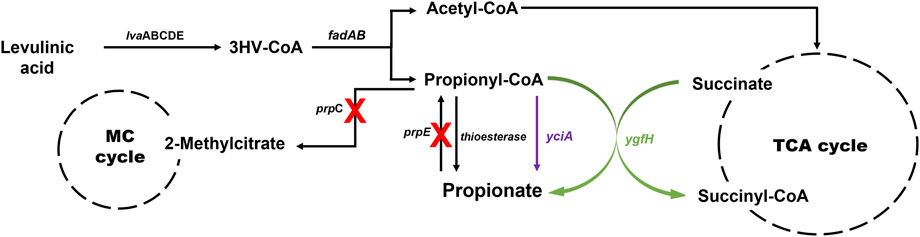
FIGURE 3. Metabolic pathway involved in PA production from LA in P. putida. The yciA (encoding thioesterase from H. influenzae) and ygfH (encoding propionyl-CoA: succinate CoA transferase from E. coli) were overexpressed, and the prpC and prpE genes encoding methylcitrate synthase and propionyl-CoA synthase, respectively, were deleted. MC cycle, methylcitrate cycle; TCA cycle, tricarboxylic acid cycle; lvaABCDE, lva operon of P. putida involved in LA degradation.
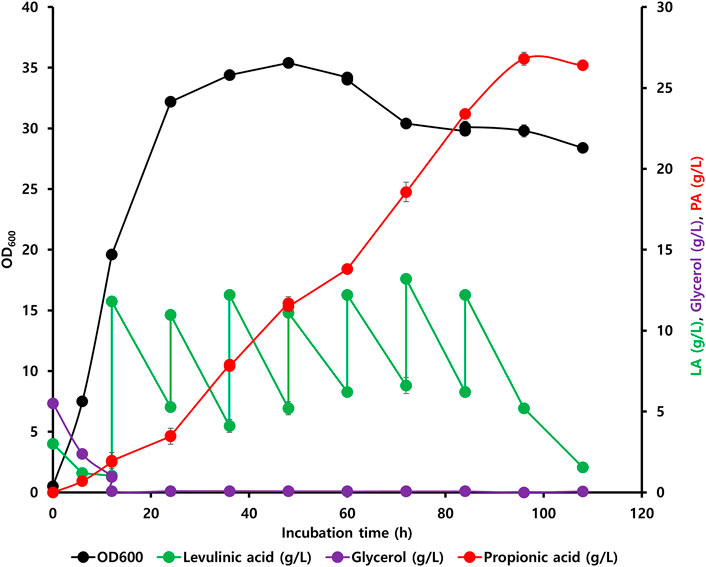
FIGURE 4. Fed-batch fermentation led by the engineered EM42:ΔCE:yciA:ygfH strain exhibiting an LA-based PA synthesis pathway.
Conclusion
In conclusion, to the best of our knowledge, the present study is the first to demonstrate the application of LA, a new renewable low-cost substrate, in the production of PA using metabolically engineered P. putida. In the reported bioproduction system, the accumulation of propionyl-CoA as a precursor for PA was confirmed by deleting alternative pathways. Moreover, the recombinant strains were developed using an LA-inducible promoter system, which eliminated the use of costly inducer chemicals and the extra metabolic burden of the engineered strain. After the selection and expression of the required thioesterase gene in the engineered strains, it was observed that the PA production level was significantly increased as compared to that achieved by the parental strains. Likewise, the molar yield and carbon flow distribution were enhanced by the co-expression of YgfH. Additionally, glycerol as a co-substrate was selected to abolish the growth defect and enhance PA titer and productivity. Furthermore, the efficiency of the engineered strain for PA production was validated by large-scale fed-batch fermentation. Our findings thus suggest that the engineered P. putida strain combined with a developed process can be used as a cost-effective and sustainable bioprocess for PA production.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
RT carried out the experimental work and prepared the draft manuscript. CS-P processed data and reviewed. SL supervised the study, reviewed, and edited the manuscript.
Funding
This research was supported by Korea Research Fellowship (KRF) program funded by the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea (2018H1D3A1A01074745). This work was also supported by Basic Science Research Program through the NRF funded by the MSIT (NRF 2020R1A4A1018332).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.939248/full#supplementary-material
References
Ahmadi, N., Khosravi-Darani, K., and Mortazavian, A. M. (2017). An overview of biotechnological production of propionic acid: From upstream to downstream processes. Electron. J. Biotechnol. 28, 67–75. doi:10.1016/j.ejbt.2017.04.004
Akawi, L., Srirangan, K., Liu, X., Moo-Young, M., and Perry Chou, C. (2015). Engineering Escherichia coli for high-level production of propionate. J. Ind. Microbiol. Biotechnol. 42 (7), 1057–1072. doi:10.1007/s10295-015-1627-4
Ammar, E. M., and Philippidis, G. P. (2021). Fermentative production of propionic acid: prospects and limitations of microorganisms and substrates. Appl. Microbiol. Biotechnol. 105 (16), 6199–6213. doi:10.1007/s00253-021-11499-1
Cha, D., Ha, H. S., and Lee, S. K. (2020). Metabolic engineering of Pseudomonas putida for the production of various types of short-chain-length polyhydroxyalkanoates from levulinic acid. Bioresour. Technol. 309, 123332. doi:10.1016/j.biortech.2020.123332
Collograi, K. C., da Costa, A. C., and Ienczak, J. L. (2022). Fermentation strategies to improve propionic acid production with propionibacterium ssp.: a review. Crit. Rev. Biotechnol., 1–23. doi:10.1080/07388551.2021.1995695
Dietz, D., Sabra, W., and Zeng, A. P. (2013). Co-cultivation of Lactobacillus zeae and Veillonella criceti for the production of propionic acid. Amb. express 3 (1), 29. doi:10.1186/2191-0855-3-29
Dishisha, T., Ståhl, Å., Lundmark, S., and Hatti-Kaul, R. (2013). An economical biorefinery process for propionic acid production from glycerol and potato juice using high cell density fermentation. Bioresour. Technol. 135, 504–512. doi:10.1016/j.biortech.2012.08.098
Dvořák, P., and de Lorenzo, V. (2018). Refactoring the upper sugar metabolism of Pseudomonas putida for co-utilization of cellobiose, xylose, and glucose. Metab. Eng. 48, 94–108. doi:10.1016/j.ymben.2018.05.019
Eş, I., Khaneghah, A. M., Hashemi, S. M. B., and Koubaa, M. (2017). Current advances in biological production of propionic acid. Biotechnol. Lett. 39 (5), 635–645. doi:10.1007/s10529-017-2293-6
Gonzalez-Garcia, R. A., McCubbin, T., Navone, L., Stowers, C., Nielsen, L. K., Marcellin, E., et al. (2017a). Microbial propionic acid production. Fermentation 3 (2), 21. doi:10.3390/fermentation3020021
Gonzalez-Garcia, R. A., McCubbin, T., Wille, A., Plan, M., Nielsen, L. K., Marcellin, E., et al. (2017b). Awakening sleeping beauty: production of propionic acid in Escherichia coli through the sbm operon requires the activity of a methylmalonyl-CoA epimerase. Microb. Cell Fact. 16 (1), 121. doi:10.1186/s12934-017-0735-4
Gonzalez‐Garcia, R. A., McCubbin, T., Turner, M. S., Nielsen, L. K., and Marcellin, E. (2020). Engineering Escherichia coli for propionic acid production through the Wood–Werkman cycle. Biotechnol. Bioeng. 117 (1), 167–183. doi:10.1002/bit.27182
Gorenflo, V., Schmack, G., Vogel, R., and Steinbüchel, A. (2001). Development of a process for the biotechnological large-scale production of 4-hydroxyvalerate-containing polyesters and characterization of their physical and mechanical properties. Biomacromolecules 2 (1), 45–57. doi:10.1021/bm0000992
Habe, H., Sato, Y., and Kirimura, K. (2020). Microbial and enzymatic conversion of levulinic acid, an alternative building block to fermentable sugars from cellulosic biomass. Appl. Microbiol. Biotechnol. 104, 7767–7775. doi:10.1007/s00253-020-10813-7
Haller, T., Buckel, T., Rétey, J., and Gerlt, J. A. (2000). Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry 39 (16), 4622–4629. doi:10.1021/bi992888d
Hayes, D. J., Fitzpatrick, S., Hayes, M. H., and Ross, J. R. (2006). The biofine process–production of levulinic acid, furfural, and formic acid from lignocellulosic feedstocks. Biorefineries–Industrial Process. Prod. 1, 139–164. doi:10.1002/9783527619849.ch7
Isoni, V., Kumbang, D., Sharratt, P., and Khoo, H. (2018). Biomass to levulinic acid: A techno-economic analysis and sustainability of biorefinery processes in southeast asia. J. Environ. Manag. 214, 267–275. doi:10.1016/j.jenvman.2018.03.012
Kang, S., Fu, J., and Zhang, G. (2018). From lignocellulosic biomass to levulinic acid: A review on acid-catalyzed hydrolysis. Renew. Sustain. Energy Rev. 94, 340–362. doi:10.1016/j.rser.2018.06.016
Kidd, M., Maynard, C., and Mullenix, G. (2021). Progress of amino acid nutrition for diet protein reduction in poultry. J. Anim. Sci. Biotechnol. 12 (1), 45. doi:10.1186/s40104-021-00568-0
Kim, D., Sathesh-Prabu, C., JooYeon, Y., and Lee, S. K. (2019). High-level production of 4-hydroxyvalerate from levulinic acid via whole-cell biotransformation decoupled from cell metabolism. J. Agric. Food Chem. 67 (38), 10678–10684. doi:10.1021/acs.jafc.9b04304
Ko, Y.-S., Kim, J. W., Lee, J. A., Han, T., Kim, G. B., Park, J. E., et al. (2020). Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 49 (14), 4615–4636. doi:10.1039/D0CS00155D
Li, J., Zhu, X., Chen, J., Zhao, D., Zhang, X., Bi, C., et al. (2017). Construction of a novel anaerobic pathway in Escherichia coli for propionate production. BMC Biotechnol. 17 (1), 38. doi:10.1186/s12896-017-0354-5
Liu, J., Li, H., Xiong, H., Xie, X., Chen, N., Zhao, G., et al. (2019). Two‐stage carbon distribution and cofactor generation for improving L‐threonine production of Escherichia coli. Biotechnol. Bioeng. 116 (1), 110–120. doi:10.1002/bit.26844
Liu, S., Feng, H., Li, T., Wang, Y., Rong, N., Yang, W., et al. (2020). Highly selective production of propionic acid from lactic acid catalyzed by NaI. Green Chem. 22 (21), 7468–7475. doi:10.1039/D0GC02676J
Liu, Z., Ge, Y., Xu, J., Gao, C., Ma, C., Xu, P., et al. (2016). Efficient production of propionic acid through high density culture with recycling cells of Propionibacterium acidipropionici. Bioresour. Technol. 216, 856–861. doi:10.1016/j.biortech.2016.06.023
Luo, X., Yang, Y., Ling, W., Zhuang, H., Li, Q., Shang, G., et al. (2016). Pseudomonas putida KT2440 markerless gene deletion using a combination of λ Red recombineering and Cre/loxP site-specific recombination. FEMS Microbiol. Lett. 363 (4), fnw014. doi:10.1093/femsle/fnw014
Ma, C., Mu, Q., Wang, L., Shi, Y., Zhu, L., Zhang, S., et al. (2020). Bio-production of high-purity propionate by engineering L-threonine degradation pathway in Pseudomonas putida. Appl. Microbiol. Biotechnol. 104 (12), 5303–5313. doi:10.1007/s00253-020-10619-7
Ma, C., Shi, Y. n., Mu, Q., Li, R., Xue, Y., Yu, B., et al. (2021). Unravelling the thioesterases responsible for propionate formation in engineered Pseudomonas putida KT2440. Microb. Biotechnol. 14 (3), 1237–1242. doi:10.1111/1751-7915.13804
Martínez-García, E., Nikel, P. I., Aparicio, T., and de Lorenzo, V. (2014). Pseudomonas 2.0: genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Fact. 13 (1), 159. doi:10.1186/s12934-014-0159-3
Meramo Hurtado, S. I., Puello, P., and Cabarcas, A. (2021). Technical evaluation of a levulinic acid plant based on biomass transformation under techno-economic and exergy analyses. ACS omega 6 (8), 5627–5641. doi:10.1021/acsomega.0c06088
Mu, Q., Shi, Y. N., Li, R., Ma, C., Tao, Y., Yu, B., et al. (2021). Production of propionate by a sequential fermentation–biotransformation process via L-threonine. J. Agric. Food Chem. 69 (46), 13895–13903. doi:10.1021/acs.jafc.1c05248
Nikel, P. I., Kim, J., and de Lorenzo, V. (2014). Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT 2440. Environ. Microbiol. 16 (1), 239–254. doi:10.1111/1462-2920.12224
Ranaei, V., Pilevar, Z., Mousavi Khaneghah, A., and Hosseini, H. (2020). Propionic acid: method of production, current state and perspectives. Food Technol. Biotechnol. 58 (2), 115–127. doi:10.17113/ftb.58.02.20.6356
Rand, J. M., Pisithkul, T., Clark, R. L., Thiede, J. M., Mehrer, C. R., Agnew, D. E., et al. (2017). A metabolic pathway for catabolizing levulinic acid in bacteria. Nat. Microbiol. 2 (12), 1624–1634. doi:10.1038/s41564-017-0028-z
Sathesh-Prabu, C., and Lee, S. K. (2019). Engineering the lva operon and optimization of Culture conditions for enhanced production of 4-hydroxyvalerate from levulinic acid in Pseudomonas putida KT2440. J. Agric. Food Chem. 67 (9), 2540–2546. doi:10.1021/acs.jafc.8b06884
Sathesh-Prabu, C., Tiwari, R., Kim, D., and Lee, S. K. (2021). Inducible and tunable gene expression systems for Pseudomonas putida KT2440. Sci. Rep. 11 (1), 18079. doi:10.1038/s41598-021-97550-7
Srirangan, K., Akawi, L., Liu, X., Westbrook, A., Blondeel, E. J., Aucoin, M. G., et al. (2013). Manipulating the sleeping beauty mutase operon for the production of 1-propanol in engineered Escherichia coli. Biotechnol. Biofuels 6 (1), 139. doi:10.1186/1754-6834-6-139
Thompson, M. G., Incha, M. R., Pearson, A. N., Schmidt, M., Sharpless, W. A., Eiben, C. B., et al. (2020). Fatty acid and alcohol metabolism in Pseudomonas putida: functional analysis using random barcode transposon sequencing. Appl. Environ. Microbiol. 86 (21), e01665-20. doi:10.1128/AEM.01665-20
Wang, Z., Ammar, E. M., Zhang, A., Wang, L., Lin, M., Yang, S.-T., et al. (2015). Engineering Propionibacterium freudenreichii subsp. shermanii for enhanced propionic acid fermentation: Effects of overexpressing propionyl-CoA: Succinate CoA transferase. Metab. Eng. 27, 46–56. doi:10.1016/j.ymben.2014.10.005
Werpy, T., and Petersen, G. (2004). Top value added chemicals from biomass: volume I--results of screening for potential candidates from sugars and synthesis gas. Golden, CO (US): National Renewable Energy Lab. No. DOE/GO-102004-1992.
Zhou, S., Ashok, S., Ko, Y., Kim, D.-M., and Park, S. (2014). Development of a deletion mutant of Pseudomonas denitrificans that does not degrade 3-hydroxypropionic acid. Appl. Microbiol. Biotechnol. 98 (10), 4389–4398. doi:10.1007/s00253-014-5562-5
Keywords: levulinic acid, propionic acid, Pseudomonas putida, glycerol, metabolic engineering
Citation: Tiwari R, Sathesh-Prabu C and Lee SK (2022) Bioproduction of propionic acid using levulinic acid by engineered Pseudomonas putida. Front. Bioeng. Biotechnol. 10:939248. doi: 10.3389/fbioe.2022.939248
Received: 08 May 2022; Accepted: 07 July 2022;
Published: 10 August 2022.
Edited by:
Lukasz Drewniak, University of Warsaw, PolandReviewed by:
Bo Yu, Institute of Microbiology (CAS), ChinaWael Sabra, Rhine-Waal University of Applied Sciences, Germany
Copyright © 2022 Tiwari, Sathesh-Prabu and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung Kuk Lee, c2tsZWVAdW5pc3QuYWMua3I=
 Rameshwar Tiwari
Rameshwar Tiwari Chandran Sathesh-Prabu
Chandran Sathesh-Prabu Sung Kuk Lee
Sung Kuk Lee