- 1School of Computer Science and Technology, Anhui University, Hefei, China
- 2Islet Pathophysiology, Department of Clinical Science, Lund University Diabetes Centre, Malmö, Sweden
Robust skin lesion segmentation of dermoscopic images is still very difficult. Recent methods often take the combinations of CNN and Transformer for feature abstraction and multi-scale features for further classification. Both types of combination in general rely on some forms of feature fusion. This paper considers these fusions from two novel points of view. For abstraction, Transformer is viewed as the affinity exploration of different patch tokens and can be applied to attend CNN features in multiple scales. Consequently, a new fusion module, the Attention-based Transformer-And-CNN fusion module (ATAC), is proposed. ATAC augments the CNN features with more global contexts. For further classification, adaptively combining the information from multiple scales according to their contributions to object recognition is expected. Accordingly, a new fusion module, the GAting-based Multi-Scale fusion module (GAMS), is also introduced, which adaptively weights the information from multiple scales by the light-weighted gating mechanism. Combining ATAC and GAMS leads to a new encoder-decoder-based framework. In this method, ATAC acts as an encoder block to progressively abstract strong CNN features with rich global contexts attended by long-range relations, while GAMS works as an enhancement of the decoder to generate the discriminative features through adaptive fusion of multi-scale ones. This framework is especially good at lesions of varying sizes and shapes and of low contrasts and its performances are demonstrated with extensive experiments on public skin lesion segmentation datasets.
1 Introduction
Skin cancer is listed as one of the fastest-growing cancers in the world (Jemal, 2017) and dermatologists usually identify lesions visually from dermoscopy images captured by dermoscopy. However, manual identification is usually tedious and time-consuming. Therefore, automatic skin lesion segmentation is badly needed in clinical practice, which can assist dermatologists in further analysis.
Skin lesions often have a vast variety of lesion shapes and sizes and are often with low contrasts (Figure 1). It means both global and local contexts are important for an effective feature abstraction, which is also why some methods (Wu et al., 2022; Zhang et al., 2021; Xu et al., 2021; Chen et al., 2021) combine both convolution neural network (CNN) and Transformer (Vaswani et al., 2017): CNN gets features with rich local information while Transformer captures the long-range relationships. They often fuse the two types of feature serially (Chen et al., 2021), or after the last stage of the Transformer branch (Zhang et al., 2021; Xu et al., 2021; Wu et al., 2022).
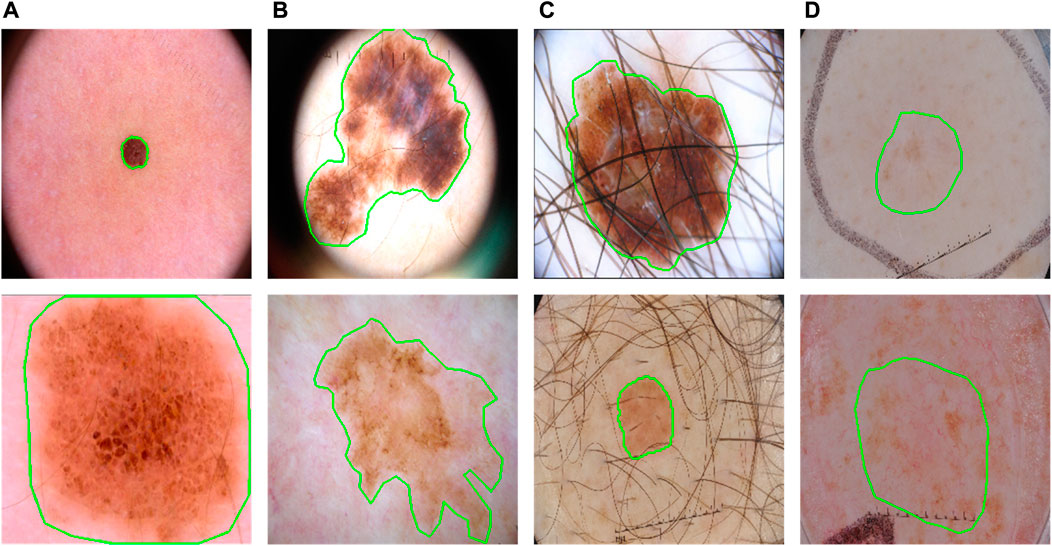
FIGURE 1. Some typical cases in dermoscopic images for skin lesion segmentation. (A) Large variety in sizes; (B) large variety in shapes; (C) hair occlusion; (D) low contrast between lesions and backgrounds.
However, such fusions may not utilize the Transformer effectively. Transformer in principle computes the affinities as attention for long-range relationships. The size and shape variations are significant symbols of lesions (Figure 1), which means a more effective fusion of them can be obtained if applying Transformer as an augmentation to different scales at different encoding stages during the progress of CNN. This progressive boost is very important, especially when facing the low-contrast appearances of lesions.
Therefore, we argue that the better way is to take the Transformer as a progressive attention tool to enhance the long-range information gradually and consequently the feature responses of lesions will be significantly enhanced. Accordingly, a new feature fusion module, the Attention-based Transformer-And-CNN fusion module (ATAC), is proposed. It can fulfill the attention-based fusion progressively in multiple scales which is different from the traditional fusion applied in tandem or after the last stage.
Effectively decoding from the strong features is also important for a successful segmentation, where the fusion of features from different scales is often considered an effective idea. Recent studies show that different scales may have different weights in fusion and features at suboptimal scales may reduce segmentation accuracy (Chen et al., 2016; Shi et al., 2018), e.g., large scales are more important for bigger lesions. Recent methods (Xu et al., 2021; Dai et al., 2022) fuse the multi-scale features with weights computed from several additional convolutions and thus increase the computation complexity.
We prefer a light-weighted scheme to fuse the multi-scale features. Considering that a gating mechanism is effective in filtering the features with fewer parameters, this paper proposes a new multi-scale fusion module, the GAting-based Multi-Scale fusion module (GAMS), to aggregate the multi-scale features adaptively by the weights from gating.
The two fusion modules ATAC and GAMS lead to a new skin lesion segmentation method. Built on the popular U-Net (Ronneberger et al., 2015) structure, it takes ATAC as an encoder block for the effective abstraction of features from both global and local contexts while adopting GAMS as an enhancement to the decoder for robust exploration of the multi-scale features. Experiments show that this method can accurately locate the lesions of different lesion shapes and sizes and low contrasts.
The main contributions can be summarized as follows.
• A novel CNN and Transformer fusion module, ATAC, which takes Transformer features as affinity estimation to attend CNN features for progressively boosting the global contexts.
• A novel multi-scale fusion module, GAMS, which takes weighted contributions from multi-scale features by gating to fuse information from different contexts.
• A new encoder-decoder-based skin lesion segmentation network for single images, which integrates both ATAC and GAMS as the encoder block and decoder enhancement separately and thus can reach robust segmentation of skin lesions without the affection of size and shape variations and low contrasts.
2 Related work
2.1 Skin lesion segmentation
Traditional skin lesion segmentation methods are mainly based on manually defined traditional features, such as color (Azad et al., 2015; Ashour et al., 2018), shape (Riaz et al., 2018; Silveira et al., 2009), and threshold (Garcia-Arroyo and Garcia-Zapirain, 2019; Pereira et al., 2019), which are not robust and stable. Nowadays many CNN-based methods have been explored for skin lesion segmentation (Yuan and Lo, 2017; Goyal et al., 2020; Yuan et al., 2017; Bi et al., 2017). The popular way is the U-Net (Ronneberger et al., 2015) based idea (Taghanaki et al., 2019; Zhang et al., 2019; Azad et al., 2019; Jha et al., 2020). For example, DoubleU-Net (Jha et al., 2020) uses two U-Net architectures in sequence. Azad et al. (2019) encoded densely connected convolutions into the bottleneck of the encoder-decoder.
More recently, Transformers (Vaswani et al., 2017; Dosovitskiy et al., 2020) have been demonstrated extraordinary capabilities for skin lesion segmentation (Wu et al., 2022; Zhang et al., 2021; Xu et al., 2021; Wang J. et al., 2022, 2021; Reza et al., 2022; Cao et al., 2022). For example, Wang J. et al. (2022), Wang et al. (2021) used boundary information to address ambiguous boundary problems of skin lesion segmentation. Chen et al. (2021) combined CNN and Transformer serially, which may miss some important information required by the successive modules.
Parallel adaption of both CNN and Transformer is also proposed (Wu et al., 2022; Zhang et al., 2021; Xu et al., 2021). Possible fusion methods include concatenation (Wu et al., 2022) and some attention-inspired mechanisms, such as convolution-based attention (Zhang et al., 2021) or direct attention-based supervision (Xu et al., 2021). However, all these fusions happen after the last stages of the Transformer branch and thus may not fully explore the rich contexts from the multi-scale features robustly.
2.2 Multi-scale feature aggregation
Some nature image oriented methods (Zhao et al., 2017; Chen et al., 2018; Lin et al., 2017) first extract multi-scale features by pyramid pooling module (PPM), pyramid atrous convolutions, or feature pyramid network (FPN) and then combine these features to predict segmentation results. For skin lesion segmentation, researchers usually first extract multi-scale features by atrous convolution or standard convolution and then fuse them using concatenation or element-wise addition (Zhang et al., 2019; Liu et al., 2019; Cui et al., 2019). Recently, Xu et al. (2021) and Dai et al. (2022) fused multi-scale features by learned weights which are computed by additional convolutions. However, their methods increase the training parameters and consequently the computational complexity.
3 Methods
Overall, our proposed framework (Figure 2) takes the U-shaped encoder-decoder structure. The encoder adopts ATAC as a building block, which gradually fuses the Transformer and CNN features for feature abstraction. The decoder consists of the normal decoder and its enhancement GAMS. The normal decoder is skipped and connected from the encoder as the typical U-Net, while GAMS takes the features from the decoder for adaptive fusion.

FIGURE 2. The pipeline of the proposed method. Building on the U-Net and incorporating both CNN and Transformer, it includes two new fusion modules, ATAC and GAMS, into the encoder and decoder respectively for progressively boosting the feature during abstraction and adaptively combining the features for classification respectively. Note: PVT stands for PVT v2, which supplies the Transformer features to ATAC.
The CNN features of images input to the encoder are attended by the Transformer features stage by stage. Gradually, globally augmented CNN features can be obtained. Then the normal decoder is applied to fulfill the final classification (Prediction 1), while the multi-scale decoder features are also input to GAMS so that effective features aggregated by adaptive fusion are generalized (Prediction 2). The final prediction is based on the results from both predictions.
The four-stage PVT v2 (Wang W. et al., 2022) supplies the Transformer features to ATAC. The normal decoder is made up of up-sampling and two 3 × 3 convolution, as the decoder of UNet (Ronneberger et al., 2015).
Now let’s discuss the details of ATAC and GAMS.
3.1 The attention-based transformer-and-CNN fusion module
3.1.1 Why progressive attention?
Generally, CNN is good at capturing features with rich local details, while Transformer can capture long-range dependence vital to distinguish the target from the background. Therefore, to aggregate information on features from both CNN and Transformer, there are two typical ways of fusion (Figures 3A,B). One can be called serial fusion which treats either CNN or Transformer as neighboring branches and then serially fuses their features. The other can be called last-stage fusion which treats the CNN and Transformer as two parallel branches and fuses their features finally after the last stage of the Transformer branch.
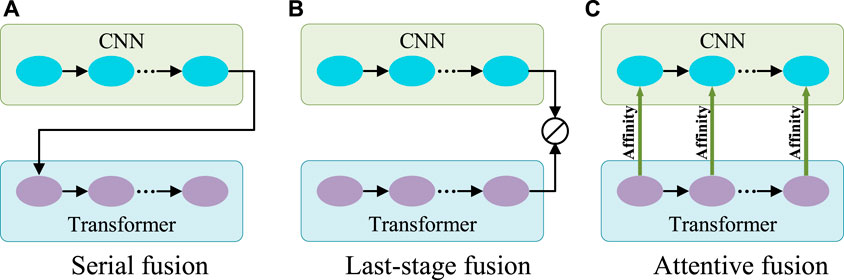
FIGURE 3. Three different fusion schemes. (A) Serial fusion; (B) Last-stage Fusion; (C) Attentive fusion. Note: 1) CNN and Transformer can change their orders in the serial fusion; 2) ⊘ in (B) represents the fusion operation.
However, serial fusion may not obtain robust features after the second branch because of the possible information loss brought by the filtering effect of the first branch. The last-stage fusion tries to keep all information from the two branches. But the combination after the last stages of the Transformer branch may mess up the information from different scales of the Transformer and cannot effectively utilize it. A more efficient utilization of Transformer features is expected.
Transformer is different from CNN in computing the features. Transformer is configured with the multi-head self-attention for learning the long-range dependencies of image patches. This attention mechanism means that Transformer actually captures the patch affinities globally. The multi-scale Transformer branch supplies rich affinity information from different scales and thus can be used to boost the CNN features progressively as the general attention mechanism for more robust exploration and fusion. Therefore, a novel fusion method called attentive fusion can be obtained (Figure 3C).
Let’s revisit the principle of the multi-head self-attention mechanism in Transformer.
Given a set of N tokens
Then it computes the attention matrix Ai ∈ RN×N, representing affinities between tokens,
where
The final feature map L is obtained by a linear layer after concatenating
This principle shows that Transformer essentially models the patch affinity. Therefore, its features can be treated as affinities whose global information can be utilized to boost CNN features.
Consequently, to better benefit from Transformer, the proposed attentive fusion is installed as the Transformer attended module ATAC to boost the long-range relations inside the CNN features from different stages and thus progressively dig up significant large-scale contexts. This design fits well with lesions: Their varying sizes and shapes need global contexts to capture, especially considering their possibly low contrasts.
3.2 The structure of ATAC
ATAC is designed as follows (Figure 4). First, the feature maps F is 3 × 3 convoluted twice for extracting CNN feature Fc, and then regular max-pooling (pooling size is 2) is applied to integrate features as
where
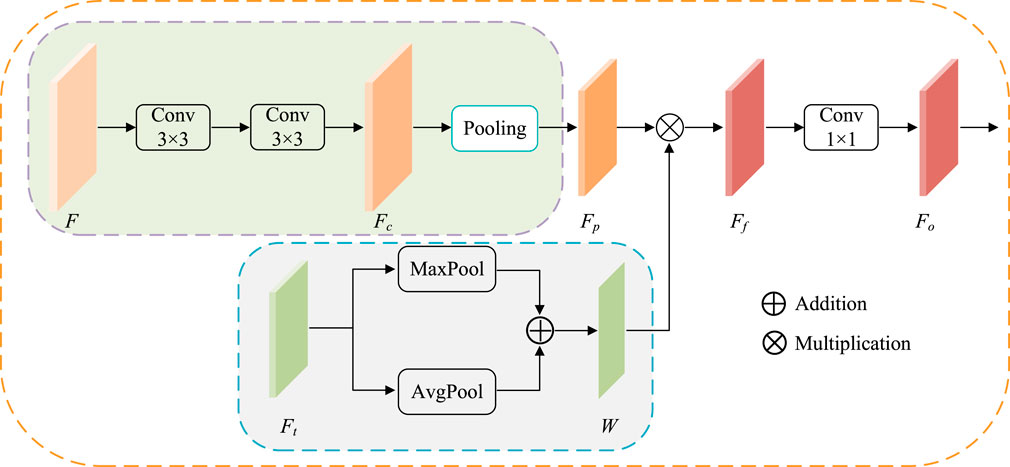
FIGURE 4. The structure of ATAC. Pooling represents the regular max-pooling operation with the pooling size 2. Maxpool and AvgPool represent max-pooling operation and average-pooling operation along the channel axis respectively.
At the same time, the corresponding scale Transformer features
Then, the attention is applied. Here, fused Transformer features W are embedded into CNN features Fp by element-wise multiplication to get enhanced CNN features Ff. Here, Fp and W have the same width and height, so the element-wise multiplication is broadcasted along each channel.
where ⊙ indicates Hadamard product. The output features Fo is finally obtained by 1 × 1 convolution of Ff.
3.3 The GAting-based multi-scale fusion module
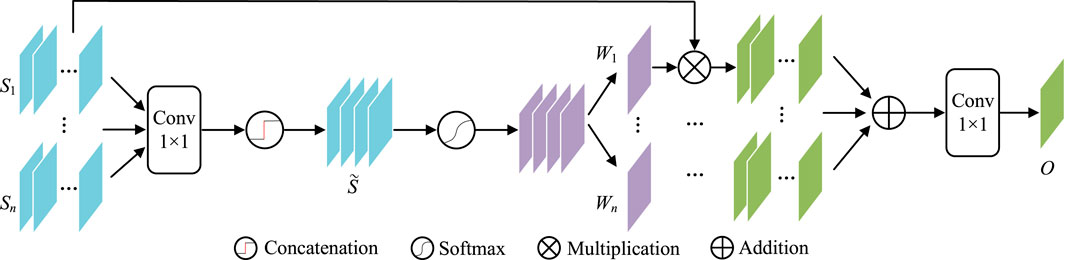
Contexts from different scales may have different influences on object perception. For example, their large scales are more important for bigger lesions and vice versa. It is better to have a weighting scheme to automatically utilize such differences. Considering gating is a very multi-scale filter for such a purpose, this paper introduces GAMS (Figure 5) to improve the feature discrimination.
In GAMS, the input feature maps are first rescaled to the same scale as Si(i ∈ {1, … , n}) by bilinear upsampling (In our experiment, n is set to four according to the four stages of the normal decoder). Then 1 × 1 convolution
Then, the gating map W can be obtained by activation function
W in Eq. 9 is further divided into W1, W2, … , Wn as the corresponding weights for the n scales. These weights are used to weighted all input features, which are further convoluted by 1 × 1 as the aggregated output features O,
3.4 Loss function
The overall loss is set to be the weighted average of the losses from both predictions as shown in Figure 2,
where: 1) λ denotes the weight (λ = 0.2 in the experiment); and 2) LGAMS and LNormal are the losses from GAMS and the normal decoder respectively. Each loss Li (i ∈ {GAMS, Normal}) is estimated by the combination of both weighted binary cross-entropy (WBCE) and weighted Intersection over Union (WIOU),
where: 1) p and
4 Experiments
4.1 Setup
The system is built by PyTorch with a single NVIDIA GeForce GTX 2080Ti GPU. The epoch is 100 and Adam is the optimizer with an initial learning rate of 10–4. For PH2, the batch size is set to 8. And for the other three datasets, the batch size is set to 16. All images are re-sized to 256 × 256 as input with various data augmentations, including vertical and horizontal flip, and random rotation.
The proposed method is evaluated on four public skin lesion segmentation datasets, ISIC 2018 (Codella et al., 2019), ISIC 2017 (Codella et al., 2018), ISIC 2016 (Gutman et al., 2016) and PH2 (Mendonça et al., 2013), where the dataset division for ISIC 2017 is the same as the previous study (Reza et al., 2022) with those of the other three following the setting in FAT-Net (Wu et al., 2022). Details of four datasets used in our experiments are described below.
• ISIC 2016 is provided by the international skin imaging collaboration (ISIC). There are a total of 1279 RGB skin lesions images, of which 900 are used for training and 379 are used for testing.
• ISIC 2017 is also provided by ISIC, which includes 2000 RGB skin lesion images as the training set with masks for segmentation. We randomly divide the original dataset into a training set, validation set, and testing set in a ratio of 7:1:2.
• ISIC 2018 is also collected by ISIC, which contains 2594 RGB skin lesions images. Like the ISIC 2017 data set division, we use 1815 samples for the training set, 259 samples for the validation set, and 520 samples for the testing set.
• PH2 is provided by the dermatology service of hospital Pedro Hispano, Matosinhos, Portugal, which includes 200 RGB skin lesions images. Like ISIC 2017 and ISIC 2018 data set division, we randomly divide them into 140 images as the training set, 20 images as the validation set, and 40 images as the testing set.
The proposed method are compared with some state-of-the-arts methods, including eight CNN-based models (U-Net (Ronneberger et al., 2015), AttU-Net (Schlemper et al., 2019), CPFNet (Feng et al., 2020), DAGAN (Lei et al., 2020), MCGU-Net (Asadi-Aghbolaghi et al., 2020), (Asadi-Aghbolaghi et al., 2020), SBPS (Lee et al., 2020), iFCN (Öztürk and Özkaya, 2020) and CKDNet (Jin et al., 2021)) and three Transformer-based models (TransUNet (Chen et al., 2021), FAT-Net (Wu et al., 2022) and TMUNet Reza et al. (2022)). Among CNN-based models, U-Net and AttU-Net are basic medical image segmentation frameworks. DAGAN, iFCN, and CKDNet are specially designed for skin lesion segmentation. CPFNet, MCGU-Net, and SBPS are excellent segmentation networks in recent years, solving the problems of large size and structure variation and boundary ambiguities, which can be applied to various types of medical images. Among Transformer-based models, TMUNet and FAT-Net fuse CNN and Transformer features at the last stage, while TransUNet fuses CNN and Transformer features serially.
4.2 Evaluation metrics
Five widely used metrics are employed to quantitatively evaluate the segmentation performances, including the Sensitivity (SE) (Yerushalmy, 1947), (Yerushalmy, 1947), Specificity (SP) (Yerushalmy, 1947), Intersection over Union (IoU) (Everingham et al., 2015), Dice Similarity Coefficient (DSC) (Dice, 1945), (Dice, 1945)and Accuracy (ACC) (per a la Normalització, 1994). They are defined as:
where: 1) TP (True-Positive) represents the number of pixels that are correctly classified as lesions; 2) TN (True Negative) represents the number of pixels that are correctly classified as backgrounds; 3) FP (False Positive) represents the number of pixels which are falsely classified as lesions; and 4)FN (False Negative) represents the number of pixels which are falsely classified as backgrounds.
4.3 Ablation studies
4.3.1 General ablation study
First, the general ablation study for the proposed modules and method for skin lesion segmentation is conducted. U-Net is taken as the baseline. ATAC and GAMS are added to the baseline as different configurations which run on the same environment with the same data augmentations for a fair comparison.
• Baseline The backbone network using U-Net;
• Baseline + ATAC Baseline but replacing its encoder block with ATAC;
• Baseline + GAMS Baseline with the additional GAMS in the decoder;
• Baseline + ATAC + GAMS Our full method.
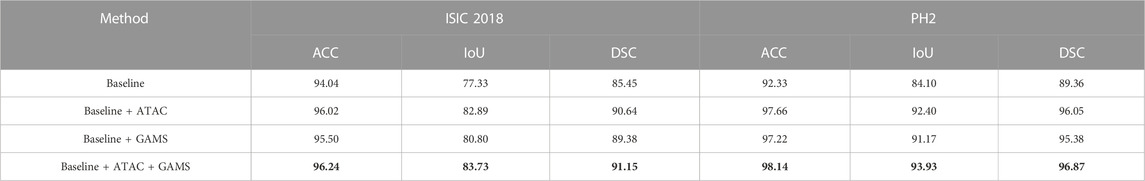
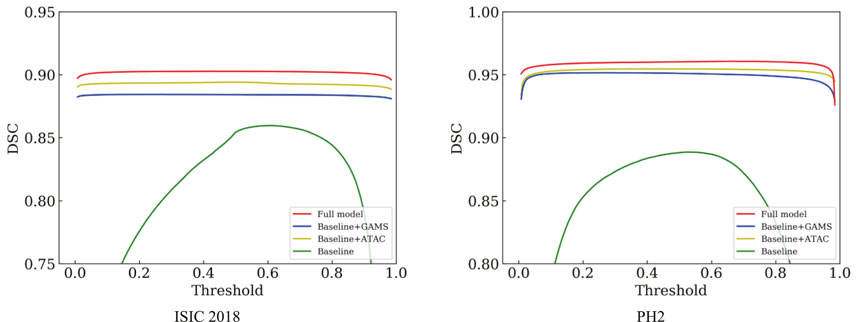
Table 1 shows that either ATAC or GAMS improves the performance of the Baseline, demonstrating the effectiveness of each individual component. Our full model further obtains about 0.84% or 2.93% improvements than the model with ATAC or GAMS alone respectively in IoU on ISIC 2018. Similar observations can also be found on PH2. The DSC values under various thresholds are also accumulated (Figure 6), which demonstrates the performance gains by ATAC, GAMS, and the full model over Baseline with the full model being the best among all methods.
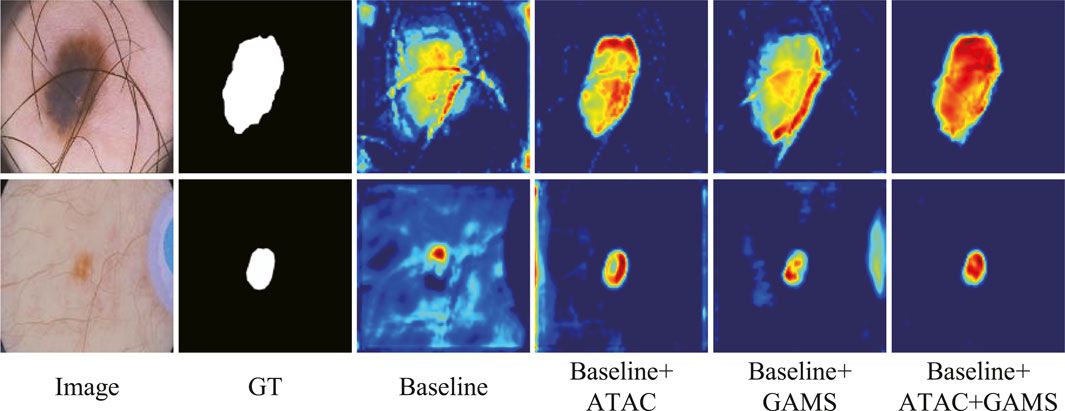
The feature maps output by the third stage of the normal decoder in different configurations are also visualized (Figure 7). We randomly selected one-channel feature maps for different configurations, which are uniformly resized to 128 × 128 for better display. The lesions are of different sizes and shapes with the smaller ones in low contrast. ATAC can significantly remove the background distractions because of the global enhancement from Transformer, while GAMS further improves the object responses, especially for the smaller lesion, thanks to its varying weight scheme. Their combination, i.e., the full model, obtains the best result with the strongest maps.
4.3.2 Ablation study on the fusion method in ATAC
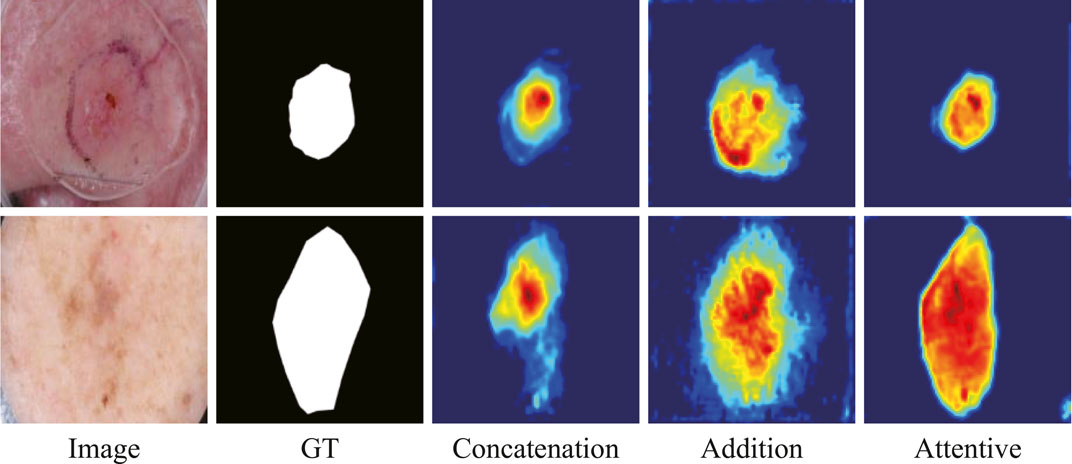
The ablation study on the fusion method in ATAC is also undertaken (Table 2). Two widely used fusion methods, concatenation and addition, are compared with our proposed attentive fusion, where multiplication operations of ATAC are substituted with concatenation or addition separately. The attentive method achieves the best performances on both ISIC 2018 and PH2 among all methods.
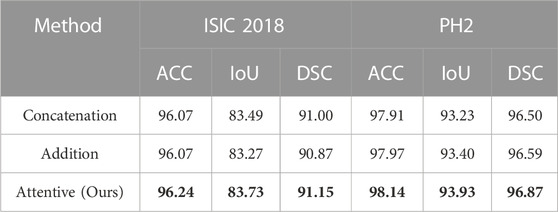
TABLE 2. Quantitative results for the ablation study on the fusion method in ATAC. The best results are shown in bold.
The features abstracted with different fusion operations are also extracted (Figure 8). The method of feature visualization is the same as in Figure 7. The responses from attentive fusion are stronger and more focused than the other two operations, which also demonstrates the importance of attentive fusion for robust lesion segmentation.
4.3.3 Ablation study on the encoder
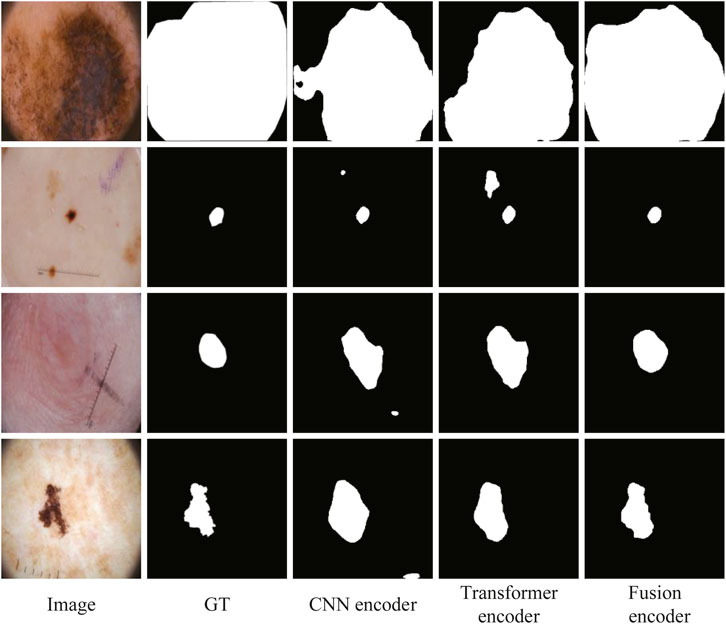
To further verify the effectiveness of fusion between CNN and Transformer features, an ablation study to compare with only CNN features or only Transformer features in the encoder is also conducted. We replace ATAC with the encoder block of U-Net for only using CNN features. And we replace ATAC with the block of PVT v2 for only using Transformer features. As can be seen in Table 3, our fused encoder achieves the best performance compared with CNN or Transformer encoder alone.
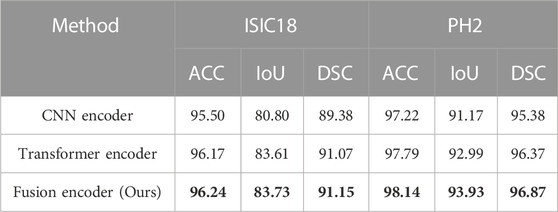
TABLE 3. Quantitative results for the ablation study on the encoder. The best results are shown in bold.
In addition, the segmentation results of some representative images are visualized in Figure 9, including the lesions with various sizes, irregular shapes, and low contrast. The first and second rows show that our fusion encoder yields the best prediction for the smallest or largest lesions. The third row shows the segmentation results for lesions with low contrast. It can be seen that both the CNN encoder and Transformer encoder exhibit over-segmentation, while our fusion encoder achieves the best performance. The last row proves that our fusion encoder segments lesions more accurately for irregularly shaped lesions.
Now, we will discuss the comparisons with the four datasets.
4.4 Evaluation on ISIC 2018
4.4.1 Quantitative results
The quantitative results of existing methods are reported by Lin et al. (2022); Wu et al. (2022); Reza et al. (2022) (Table 4). Our method achieves the highest scores in all metrics except SE with a slight decrease.
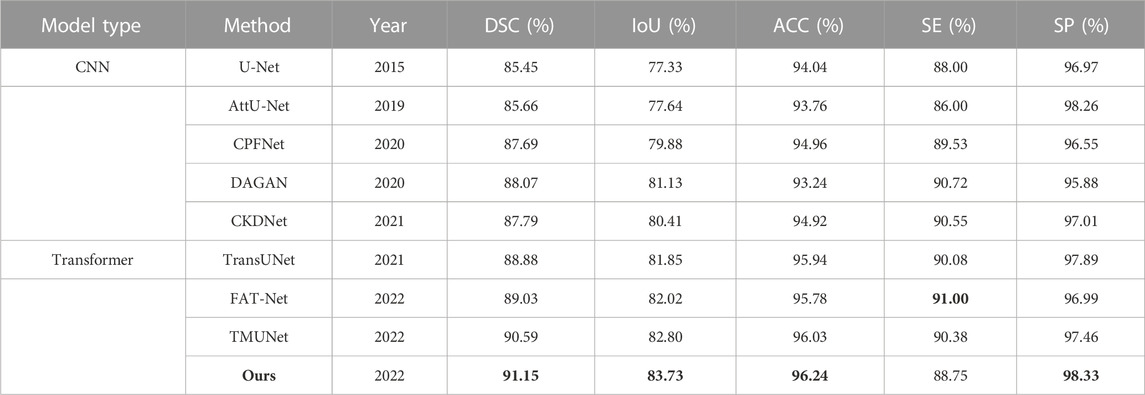
TABLE 4. Statistical comparison of the segmentation results on ISIC 2018. The best results are shown in bold.
4.4.2 Qualitative results
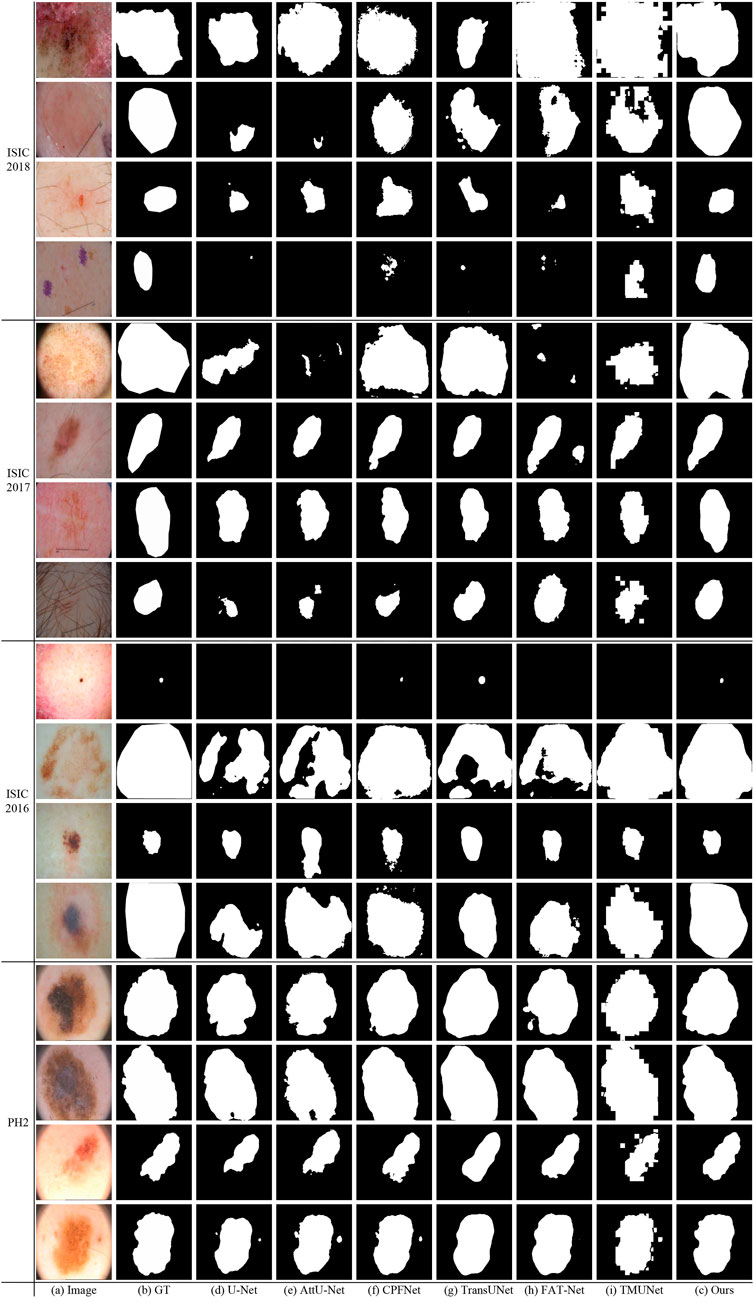
Figure 10 shows some visualization results of different methods. As can be seen, the lesion has low contrast and ambiguous boundary in the last row of ISIC 2018. The compared methods exhibit under-segmentation. In addition, FAT-Net and TransUNet can struggle to localize a complete lesion because of possible information loss brought by serial fusion and last-stage fusion. Our method benefits from information fusion at different encoder and decoder stages, which can boost feature representation, and thus our method achieves more accurate segmentation results than the compared methods.
4.5 Evaluation on ISIC 2017
4.5.1 Quantitative results
The experimental results of DAGAN and MCGU-Net are reported by TMUNet (Reza et al., 2022) with the rest results computed by us according to their released codes (Table 5). Our method also achieves the highest scores in most metrics. In addition, compared with the latest method TMUNet, ours is 0.83%, 1.45%, and 0.44% higher in DSC, IoU, and ACC, respectively.
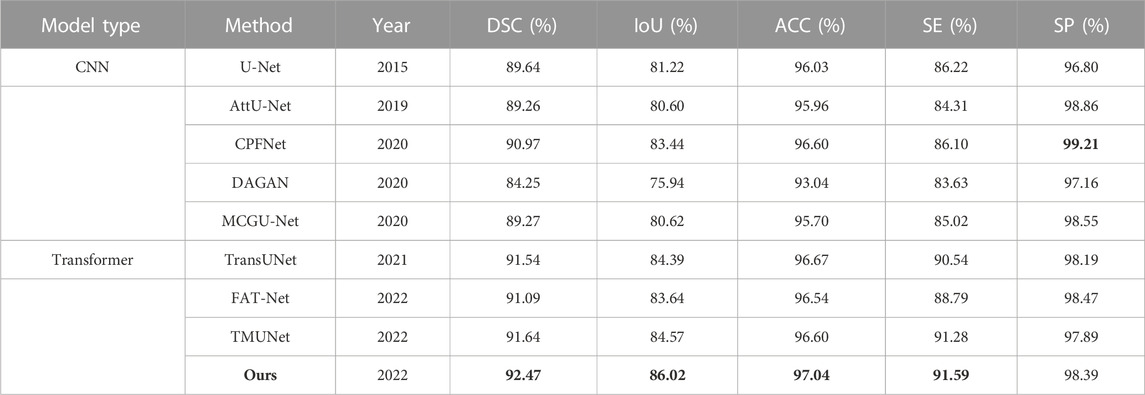
TABLE 5. Statistical comparison of the segmentation results on ISIC 2017. The best results are shown in bold.
4.5.2 Qualitative results
Figure 10 shows that our method obtains more accurate results than other methods on ISIC 2017. In the last row of ISIC 2017, the lesion has hair interference. But, apparently, our method is better than other compared methods. It is due to ATAC can effectively utilize the long-range contexts from Transformer, which helps to distinguish different classes.
4.6 Evaluation on ISIC 2016
4.6.1 Quantitative results
The quantitative results of existing methods are reported by FAT-Net (Wu et al., 2022) except that those of TransUNet and TMUNet are computed by us according to their released codes (Table 6). Ours again achieves the highest scores in most metrics.
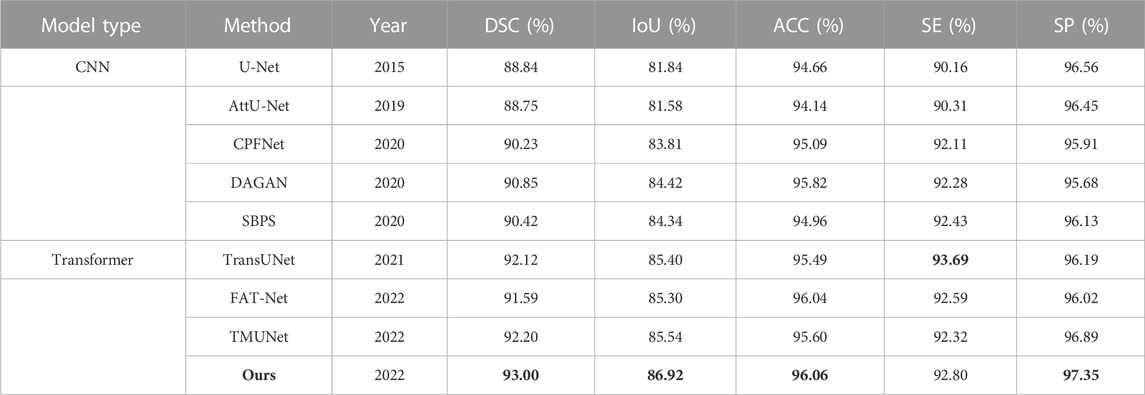
TABLE 6. Statistical comparison of the segmentation results on ISIC 2016. The best results are shown in bold.
4.6.2 Qualitative results
Figure 10 gives some visual results. As shown in the first and second rows of ISIC 2016, the lesions exhibit a large variation in sizes. But while credit should be given to the fusion of GAMS in different scales of the decoding stage, our method can detect lesions more accurately than other methods, even if they are very small or large.
4.7 Evaluation on PH2
4.7.1 Quantitative results
The quantitative results of existing methods are from FAT-Net (Wu et al., 2022), except for CPFNet, TransUNet, CPFNet, and TMUNet, which are computed by us according to their codes (Table 7). Our method again achieves the highest scores for all metrics.
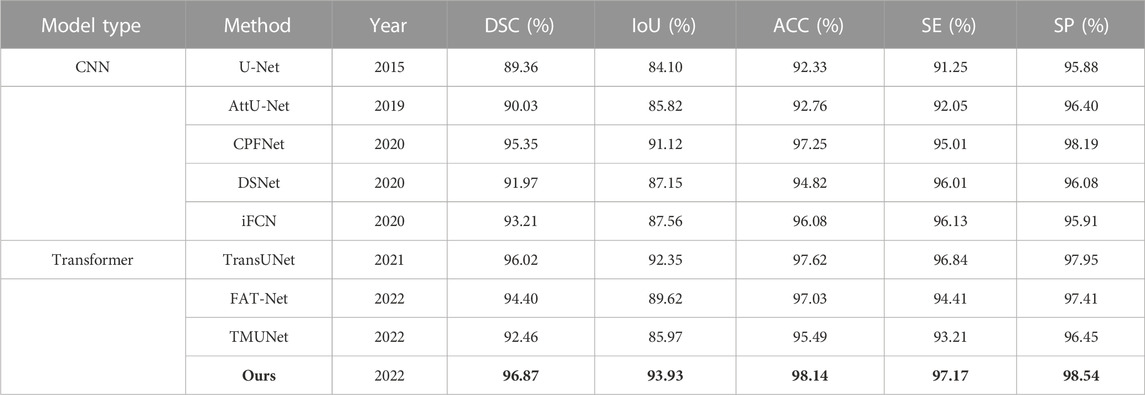
TABLE 7. Statistical comparison of the segmentation results on PH2. The best results are shown in bold.
4.7.2 Qualitative results
As can been seen in PH2 of Figure 10 there are many details around boundary of these lesions. But the boundary obtained by our method is more accurate and closer to the ground truth than other methods. This advantage depends on the strong feature representation capabilities of ATAC.
4.8 Failure cases of the proposed method
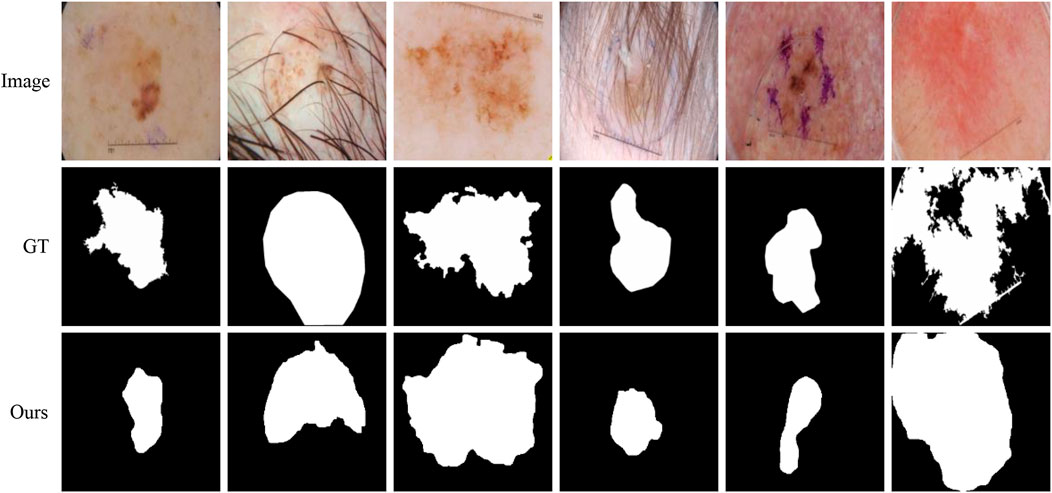
Although our method is better than the current mainstream segmentation methods, some challenges are still not solved. Figure 11 shows some failure examples. It can be observed that these lesions have very complex boundary regions (see the first, third, and sixth columns) and serious noise interference (see the second, fourth, and fifth column). Our method can basically detect the lesion locations. But in these complex scenes, our method gets poor segmentation results because it is difficult to obtain robust feature representation to distinguish different classes.
5 Conclusion
This paper aims at effective fusion policies for robust skin lesion segmentation from dermoscopic images and proposes a new method. Two new fusion modules, ATAC and GAMS, are incorporated in its encoder and decoder for robust feature abstraction and further classification separately. ATAC acts as the encoder block, which takes the Transformer to attend CNN for augmentation of global contexts in different stages. This design makes the abstracted features better fitted for the size and shape of varying lesions, especially when they are in low contrast. GAMS works as an enhancement to the normal decoder, which adaptively weights the features of multiple scales by gating. This module can help obtain features characterized for different objects in low complexity and highly discriminative for robust final inference. Quantitative and qualitative experiments demonstrate the efficacy of the proposed method.
However, ambiguous boundaries of lesions are still challenging. In addition, hair covering the lesions may also distract the model and thus affect the segmentation performances. In the future, we will study those problems and propose more robust methods accordingly.
DAS
Publicly available datasets were analyzed in this study. For PH2, please find at: https://www.dropbox.com/s/k88qukc20ljnbuo/PH2Dataset.rar; For ISIC 2016, ISIC 2017 and ISIC 2018, please find at: https://challenge.isic-archive.com/data/.
Author contributions
QG: Writing—Original Draft, Methodology, Formal analysis. XF: Writing—review and editing, Conceptualization, Supervision. LW: Writing—review and editing. EZ: Writing—review and editing. ZL: Writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by the Natural Science Foundation of Anhui Province (2108085MF210).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Asadi-Aghbolaghi, M., Azad, R., Fathy, M., and Escalera, S. (2020). Multi-level context gating of embedded collective knowledge for medical image segmentation. arXiv preprint arXiv:2003.05056.
Ashour, A. S., Hawas, A. R., Guo, Y., and Wahba, M. A. (2018). A novel optimized neutrosophic k-means using genetic algorithm for skin lesion detection in dermoscopy images. Signal, Image Video Process. 12, 1311–1318. doi:10.1007/s11760-018-1284-y
Azad, R., Ahmadzadeh, E., and Azad, B. (2015). “Real-time human face detection in noisy images based on skin color fusion model and eye detection,” in Intelligent computing, communication and devices, 435–447.
Azad, R., Asadi-Aghbolaghi, M., Fathy, M., and Escalera, S. (2019). “Bi-directional ConvLSTM U-Net with densely connected convolutions,” in ICCV workshops, 0.
Bi, L., Kim, J., Ahn, E., Kumar, A., Fulham, M., and Feng, D. (2017). Dermoscopic image segmentation via multistage fully convolutional networks. IEEE Trans. Biomed. Eng. 64, 2065–2074. doi:10.1109/tbme.2017.2712771
Cao, W., Yuan, G., Liu, Q., Peng, C., Xie, J., Yang, X., et al. (2022). ICL-Net: Global and local inter-pixel correlations learning network for skin lesion segmentation. IEEE J. Biomed. Health Inf. 27, 145–156. doi:10.1109/jbhi.2022.3162342
Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., et al. (2021). TransUNet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306.
Chen, L.-C., Yang, Y., Wang, J., Xu, W., and Yuille, A. L. (2016). “Attention to scale: Scale-aware semantic image segmentation,” in Cvpr, 3640–3649.
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., and Adam, H. (2018). “Encoder-decoder with atrous separable convolution for semantic image segmentation,” in Eccv, 801–818.
Codella, N. C., Gutman, D., Celebi, M. E., Helba, B., Marchetti, M. A., Dusza, S. W., et al. (2018). “Skin lesion analysis toward melanoma detection: A challenge,” in IEEE international symposium on biomedical imaging, 168–172.
Codella, N., Rotemberg, V., Tschandl, P., Celebi, M. E., Dusza, S., Gutman, D., et al. (2019). Skin lesion analysis toward melanoma detection 2018: A challenge. arXiv preprint arXiv:1902.03368.
Cui, Z., Wu, L., Wang, R., and Zheng, W.-S. (2019). “Ensemble transductive learning for skin lesion segmentation,” in Chinese conference on pattern recognition and computer vision, 572–581.
Dai, D., Dong, C., Xu, S., Yan, Q., Li, Z., Zhang, C., et al. (2022). Ms red: A novel multi-scale residual encoding and decoding network for skin lesion segmentation. Med. Image Anal. 75, 102293. doi:10.1016/j.media.2021.102293
Dice, L. R. (1945). Measures of the amount of ecologic association between species. Ecology 26, 297–302. doi:10.2307/1932409
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., et al. (2020). An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
Everingham, M., Eslami, S., Van Gool, L., Williams, C. K., Winn, J., and Zisserman, A. (2015). The pascal visual object classes challenge: A retrospective. Int. J. Comput. Vis. 111, 98–136. doi:10.1007/s11263-014-0733-5
Feng, S., Zhao, H., Shi, F., Cheng, X., Wang, M., Ma, Y., et al. (2020). CPFNet: Context pyramid fusion network for medical image segmentation. IEEE Trans. Med. Imaging 39, 3008–3018. doi:10.1109/tmi.2020.2983721
Garcia-Arroyo, J. L., and Garcia-Zapirain, B. (2019). Segmentation of skin lesions in dermoscopy images using fuzzy classification of pixels and histogram thresholding. Comput. Methods Programs Biomed. 168, 11–19. doi:10.1016/j.cmpb.2018.11.001
Goyal, M., Yap, M. H., and Hassanpour, S. (2020). “Multi-class semantic segmentation of skin lesions via fully convolutional networks,” in International joint conference on biomedical engineering systems and technologies, 290–295.
Gutman, D., Codella, N. C., Celebi, E., Helba, B., Marchetti, M., Mishra, N., et al. (2016). Skin lesion analysis toward melanoma detection: A challenge. arXiv preprint arXiv:1605.01397.
Jemal, A., and Miller, K. D. (2017). Cancer statistics, 2017. CA: A Cancer J. Clin. 67, 7–30. doi:10.3322/caac.21387
Jha, D., Riegler, M. A., Johansen, D., Halvorsen, P., and Johansen, H. D. (2020). “DoubleU-net: A deep convolutional neural network for medical image segmentation,” in IEEE international symposium on computer-based medical systems, 558–564.
Jin, Q., Cui, H., Sun, C., Meng, Z., and Su, R. (2021). Cascade knowledge diffusion network for skin lesion diagnosis and segmentation. Appl. Soft Comput. 99, 106881. doi:10.1016/j.asoc.2020.106881
Lee, H. J., Kim, J. U., Lee, S., Kim, H. G., and Ro, Y. M. (2020). “Structure boundary preserving segmentation for medical image with ambiguous boundary,” in Cvpr, 4817–4826.
Lei, B., Xia, Z., Jiang, F., Jiang, X., Ge, Z., Xu, Y., et al. (2020). Skin lesion segmentation via generative adversarial networks with dual discriminators. Med. Image Anal. 64, 101716. doi:10.1016/j.media.2020.101716
Lin, T.-Y., Dollár, P., Girshick, R., He, K., Hariharan, B., and Belongie, S. (2017). “Feature pyramid networks for object detection,” in Cvpr, 2117–2125.
Lin, X., Yu, L., Cheng, K.-T., and Yan, Z. (2022). The lighter the better: Rethinking Transformers in medical image segmentation through adaptive pruning. arXiv preprint arXiv:2206.14413.
Liu, L., Mou, L., Zhu, X. X., and Mandal, M. (2019). “Skin lesion segmentation based on improved U-Net,” in IEEE Canadian conference of electrical and computer engineering, 1–4.
Mendonça, T., Ferreira, P. M., Marques, J. S., Marcal, A. R., and Rozeira, J. (2013). “PH 2-A dermoscopic image database for research and benchmarking,” in Annual international conference of the IEEE engineering in medicine and biology society, 5437–5440.
Öztürk, Ş., and Özkaya, U. (2020). Skin lesion segmentation with improved convolutional neural network. J. Digital Imaging 33, 958–970. doi:10.1007/s10278-020-00343-z
per a la Normalització, O. I. (1994). Accuracy (trueness and precision) of measurement methods and results. International Organization for Standardization.
Pereira, P. M., Tavora, L. M., Fonseca-Pinto, R., Paiva, R. P., Assunção, P. A. A., and de Faria, S. M. (2019). “Image segmentation using gradient-based histogram thresholding for skin lesion delineation,” in Bioimaging, 84–91.
Reza, A., Moein, H., Yuli, W., and Dorit, M. (2022). Contextual attention network: Transformer meets U-Net. arXiv preprint arXiv:2203.01932.
Riaz, F., Naeem, S., Nawaz, R., and Coimbra, M. (2018). Active contours based segmentation and lesion periphery analysis for characterization of skin lesions in dermoscopy images. IEEE J. Biomed. Health Inf. 23, 489–500. doi:10.1109/jbhi.2018.2832455
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-Net: Convolutional networks for biomedical image segmentation,” in Miccai, 234–241.
Schlemper, J., Oktay, O., Schaap, M., Heinrich, M., Kainz, B., Glocker, B., et al. (2019). Attention gated networks: Learning to leverage salient regions in medical images. Med. image Anal. 53, 197–207. doi:10.1016/j.media.2019.01.012
Shi, H., Li, H., Wu, Q., Meng, F., and Ngan, K. N. (2018). “Boosting scene parsing performance via reliable scale prediction,” in ACM multimedia, 492–500.
Silveira, M., Nascimento, J. C., Marques, J. S., Marçal, A. R., Mendonça, T., Yamauchi, S., et al. (2009). Comparison of segmentation methods for melanoma diagnosis in dermoscopy images. IEEE J. Sel. Top. Signal Process. 3, 35–45. doi:10.1109/jstsp.2008.2011119
Taghanaki, S. A., Bentaieb, A., Sharma, A., Zhou, S. K., Zheng, Y., Georgescu, B., et al. (2019). “Select, attend, and transfer: Light, learnable skip connections,” in International workshop on machine learning in medical imaging, 417–425.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., et al. (2017). Attention is all you need. NIPS 30, 6000–6010.
Wang, J., Chen, F., Ma, Y., Wang, L., Fei, Z., Shuai, J., et al. (2022). XBound-Former: Toward cross-scale boundary modeling in Transformers. arXiv preprint arXiv:2206.00806.
Wang, J., Wei, L., Wang, L., Zhou, Q., Zhu, L., and Qin, J. (2021). “Boundary-aware Transformers for skin lesion segmentation,” in Miccai, 206–216.
Wang, W., Xie, E., Li, X., Fan, D.-P., Song, K., Liang, D., et al. (2022). PVT v2: Improved baselines with pyramid vision transformer. Comput. Vis. Media 8, 415–424. doi:10.1007/s41095-022-0274-8
Wu, H., Chen, S., Chen, G., Wang, W., Lei, B., and Wen, Z. (2022). FAT-Net: Feature adaptive Transformers for automated skin lesion segmentation. Med. Image Anal. 76, 102327. doi:10.1016/j.media.2021.102327
Xu, R., Wang, C., Xu, S., Meng, W., and Zhang, X. (2021). “DC-Net: Dual context network for 2d medical image segmentation,” in Miccai, 503–513.
Yerushalmy, J. (1947). Statistical problems in assessing methods of medical diagnosis, with special reference to X-ray techniques. Public Health Rep. 62, 1432–1449. doi:10.2307/4586294
Yuan, Y., Chao, M., and Lo, Y.-C. (2017). Automatic skin lesion segmentation using deep fully convolutional networks with Jaccard distance. IEEE Trans. Med. Imaging 36, 1876–1886. doi:10.1109/tmi.2017.2695227
Yuan, Y., and Lo, Y.-C. (2017). Improving dermoscopic image segmentation with enhanced convolutional-deconvolutional networks. IEEE J. Biomed. Health Inf. 23, 519–526. doi:10.1109/jbhi.2017.2787487
Zhang, G., Shen, X., Chen, S., Liang, L., Luo, Y., Yu, J., et al. (2019). Dsm: A deep supervised multi-scale network learning for skin cancer segmentation. IEEE Access 7, 140936–140945. doi:10.1109/access.2019.2943628
Zhang, Y., Liu, H., and Hu, Q. (2021). “TransFuse: Fusing Transformers and CNNs for medical image segmentation,” in Miccai, 14–24.
Keywords: skin lesion segmentation, transformer, deep learning, robust fusion, gating mechanism
Citation: Guo Q, Fang X, Wang L, Zhang E and Liu Z (2023) Robust fusion for skin lesion segmentation of dermoscopic images. Front. Bioeng. Biotechnol. 11:1057866. doi: 10.3389/fbioe.2023.1057866
Received: 30 September 2022; Accepted: 21 February 2023;
Published: 20 March 2023.
Edited by:
Tajalli Keshavarz, University of Westminster, United KingdomReviewed by:
Zhen Ma, University of Porto, PortugalTakaaki Sugino, Tokyo Medical and Dental University, Japan
Copyright © 2023 Guo, Fang, Wang, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianyong Fang, ZmFuZ3hpYW55b25nQGFodS5lZHUuY24=
 Qingqing Guo
Qingqing Guo Xianyong Fang
Xianyong Fang Linbo Wang1
Linbo Wang1