- Division of Applied Microbiology, Department of Chemistry, Faculty of Engineering, Lund University, Lund, Sweden
We report on the development of a method based on recombinant yeast Saccharomyces cerevisiae to produce nonivamide, a capsaicinoid and potent agonist of the pain receptor TRPV1. Nonivamide was produced in a two-step batch process where yeast was i) grown aerobically on glucose and ii) used to produce nonivamide from vanillin and non-anoic acid by bioconversion. The yeast was engineered to express multiple copies of an amine transaminase from Chromobacterium violaceum (CvTA), along with an NADH-dependent alanine dehydrogenase from Bacillus subtilis (BsAlaDH) to enable efficient reductive amination of vanillin. Oxygen-limited conditions and the use of ethanol as a co-substrate to regenerate NADH were identified to favour amination over the formation of the by-products vanillic alcohol and vanillic acid. The native alcohol dehydrogenase ADH6 was deleted to further reduce the formation of vanillic alcohol. A two-enzyme system consisting of an N-acyltransferase from Capsicum annuum (CaAT), and a CoA ligase from Sphingomonas sp. Ibu-2 (IpfF) was co-expressed to produce the amide. This study provides proof of concept for yeast-based production of non-ivamide by combined transamination and amidation of vanillin.
1 Introduction
Capsaicinoids are plant alkaloids with strong agonist activity on TRPV1 (Transient Receptor Potential Cation Channel Subfamily V Member 1). They have several applications in the food industry, e.g. for colouring, seasoning and flavour enhancement, and in the pharmaceutical industry, especially for their anti-inflammatory and analgesic properties (Baenas et al., 2019; Koivisto et al., 2021). Production in chili pepper is impacted by pre- and post-harvest factors, such as environmental, genotypical and storage conditions, which affect the amount and composition of the capsaicinoids (Uarrota et al., 2021). The development of a fermentation process with metabolically engineered yeast may enable more efficient production compared to the plant (Muratovska et al., 2022b). Baker’s yeast, Saccharomyces cerevisiae, is a well-known production host within industrial microbiology and several methods have been developed recently for its use to biosynthesize natural products such as terpenoids (cannabinoids), flavonoids and betalains (Romero-Suarez et al., 2022).
In the plant, capsaicinoid biosynthesis involves the phenylpropanoid pathway to vanillylamine (derived from vanillin), and the fatty acid synthesis pathway to CoA-activated fatty acid, which are condensed by an amide-forming N-acyltransferase (capsaicin synthase) (Ogawa et al., 2015). De novo biosynthesis of capsaicinoids in yeast has to date not been developed and would require functional expression of several heterologous genes and re-routing of endogenous biochemical pathways. Previously, yeast has been engineered to produce vanillin and vanillin glycoside from sugar via the shikimic acid pathway (Hansen et al., 2009; Strucko et al., 2015). There are also examples of engineered yeast for production of medium-chain fatty acids (Gajewski et al., 2017; Zhu et al., 2020). Yeast expression of N-acyltransferase and CoA-ligase from Capsicum annuum (CaAT) (Kang et al., 2005) and Sphingomonas sp. Ibu-2 (IpfF) (Murdoch and Hay, 2013), respectively, was recently shown to enable production of nonivamide (a model capsaicinoid) from supplemented vanillylamine and non-anoic acid (Muratovska et al., 2022a), as the last step in capsaicinoid synthesis.
A key step in the pathway is the reductive amination of vanillin to form vanillylamine, which has not been shown previously in yeast. The reaction is hampered by an unfavourable thermodynamic equilibrium that lies in the direction of the aldehyde when using endogenous amine donors (such as L-alanine); however, this can be influenced by several strategies. Vanillin aminotransaminase from Capsicum sp. (Weber et al., 2014b), as well as Chromobacterium violaceum amine transaminase (CvTA), have been shown to be functional in yeast. However, these have only been applied for whole-cell production of chiral amines by kinetic resolution of racemic amines (amine to ketone), since the reverse direction (ketone to amine) has been proven to be exceptionally difficult to achieve in vivo (Weber et al., 2014a; Weber et al., 2014b; Knudsen et al., 2016; Weber et al., 2017). This was though recently achieved for vanillylamine production in engineered Escherichia coli (Fu et al., 2021) and Pseudomonas putida (Manfrao-Netto et al., 2021), both expressing CvTA and alanine dehydrogenase from Bacillus subtilis (BsAlaDH) (Koszelewski et al., 2008). Key to success was the co-expression of CvTA and BsAlaDH in combination with appropriate reaction conditions in favour of amine formation. The transaminase reaction competes with endogenous activity of alcohol dehydrogenases and reductases resulting in conversion of vanillin to the by-products vanillic acid, and vanillic alcohol (Wang et al., 2018). Therefore, a strategy to suppress formation of these is necessary. In yeast, the deletion of alcohol dehydrogenase 6 (ADH6) was previously shown to decrease (but not totally remove) formation of vanillic alcohol in strains engineered to produce vanillin-glucoside (Hansen et al., 2009). Another strategy to suppress the activity of NADPH-dependent vanillin reductases and not affect reductive amination, which are typically dependent on regeneration of NADH, may be to selectively reduce NADPH regeneration. This strategy has previously been applied in yeast to selectively favour NADH-dependent reductases over competing NADPH-dependent enzymes, thereby improving stereoselectivity of whole-cell bioreduction of ketones to chiral alcohols (Kratzer et al., 2008).
Here, yeast strains co-expressing CvTA, BsAlaDH, CaAT and IpF, and with deleted ADH6 were constructed and evaluated for reductive amination and amidation of vanillin (Figure 1). The use of glucose and ethanol as co-substrate were compared under both aerobic and anaerobic conditions to determine environmental conditions favouring the formation of vanillylamine over the alcohol and acid by-products. Using a combination of genetic and process engineering we demonstrate the use of yeast for production of nonivamide from supplemented vanillin and non-anoic acid in well controlled bioreactors.
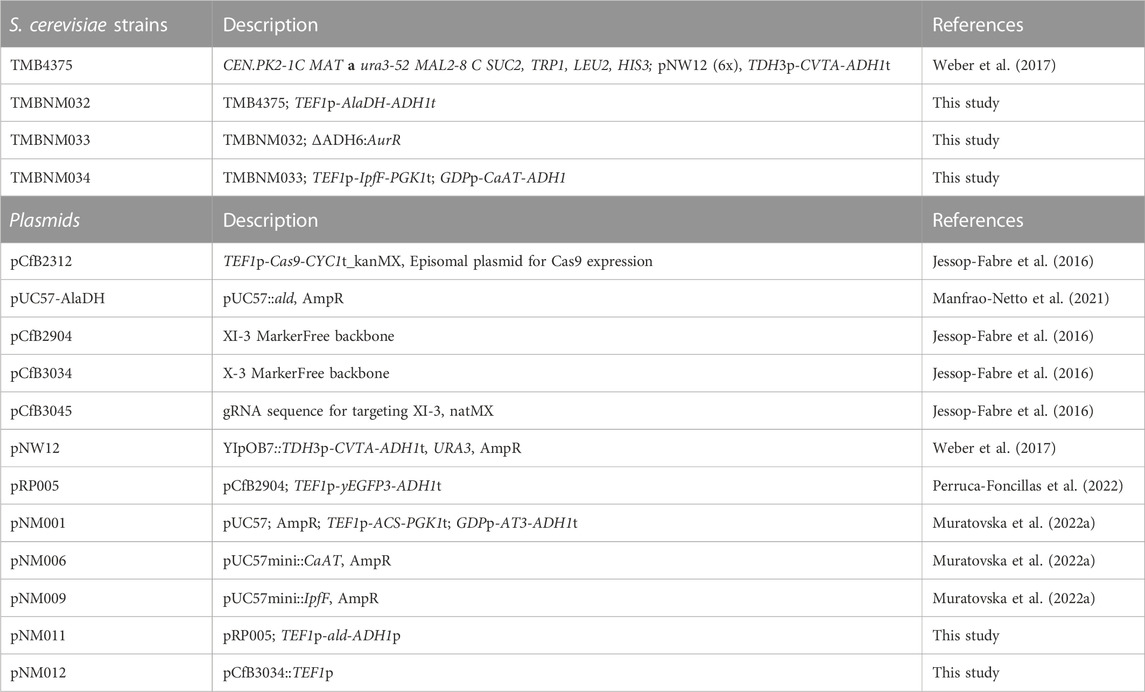
FIGURE 1. Production of nonivamide from non-anoic acid and vanillin in recombinant yeast overexpressing IpfF (Spingomonas sp. Ibu-2 CoA-ligase), CaAT (Capsicum annuum N-acyltransferase), CvTA (Chromobacterium violaceum amine transaminase) and BsAlaDH (Bacillus subtilis alanine dehydrogenase), and with deleted ADH6 (alcohol dehydrogenase 6). The by-products vanillyl acid and vanillic alcohol formed by endogenous enzymes VDH (vanillin dehydrogenase), and ADH6 (and other oxidoreductases), respectively, are suppressed under oxygen-limited conditions and with ethanol as co-substrate to regenerate NADH.
2 Material and methods
2.1 Strains and media
Saccharomyces cerevisiae strains were maintained on Yeast Peptone Dextrose (YPD) medium (20 g L−1 peptone, 10 g L−1 yeast extract, 20 g L−1 glucose; 20 g L−1 agar for plates). Geneticin (200 mg L−1) and nourseothricin (100 mg L−1) were supplemented in the media when needed for selection of Cas9 (KanMX) and gRNA (natMX) plasmids, respectively. Sub-cloning was done in electro-competent Escherichia coli DH5α. Lysogeny Broth (LB) medium (10 g L−1 tryptone, 5 g L−1 yeast extract, 10 g L−1 NaCl; 15 g L−1 agar for plates) with 100 mg L−1 ampicillin was used for E. coli liquid cultivation and selection on solid plates. All strains were stored in 25% (v/v) glycerol at −80°C. Defined mineral medium (Verduyn et al., 1992) was used as basis for the bioconversions, buffered at pH 6.5.
2.2 Plasmid and strain constructions
Strains and plasmids used in this work are listed in Table 1. Standard molecular biology methods were used (Sambrook and Russell, 2001). Preparative and diagnostic PCR for genome engineering was performed using Physion and DreamTaq polymerase (Thermo Scientific, US), respectively. All modifications were verified by colony PCR using primers listed in Supplementary Table S1. Primers were designed using SnapGene® (GSL Biotech LLC. Version 3.03, US) and purchased from Eurofins (Eurofins Genomics, Germany). Agarose (0.8%) gel electrophoresis was used for estimating size of the DNA fragments. All plasmids (Table 1) were purified using GeneJet plasmid MiniPrep Kit (Thermo Scientific, US) after propagation in E. coli. Alanine dehydrogenase gene (Genbank ID 936557) from Bacillus subtilis (BsAlaDH) was codon optimized for expression in S. cerevisiae and obtained in a pUC57 vector (Genescript, US). BsAlaDH gene was amplified with primers flanked with restriction sites SfaAI/XhoI and subsequently cloned into pRP005 between TEF1p promotor and ADH1t terminator flanked by homologous regions for chromosomal integration, generating pNM011. pNM012 was constructed by cloning TEF1p into the backbone plasmid pCfB3034. Correct DNA sequence of constructed plasmids were verified by Sanger sequencing (Eurofins Genomics, Germany). S. cerevisiae strains were engineered by the previously described CRISPR-Cas9 toolkit (Jessop-Fabre et al., 2016). The lithium acetate-based protocol (Gietz and Schiestl, 2007) with the addition of DMSO for improving cell permeability, was used for generating competent yeast cells. TMB4375 strain carrying six copies of Chromobacterium violaceum amine transaminase gene (CvTA) (Weber et al., 2017) was transformed with the pCfB2312 plasmid, carrying Streptococcus pyogenes gene, SpCas9, and used as background strain for the strain engineering. Transformation with a linearized pNM011 (using NotI restriction enzyme) and pCfB3045 (gRNA) plasmid generated strain TMBNM032. Deletion of ADH6 was achieved by transforming TMBNM032 with a PCR product carrying AUR gene cassette for aureobasidin A (AurA) resistance and ∼50bp homologous flanks to the target locus, generating strain TMBNM033. Transformants resistant to AurA were selected in solid medium and correct gene knockout was analysed with diagnostic PCR. Codon optimized genes coding for CaAT (Tyramine N-(hydroxycinnamoyl) transferase from Capsicum annum, and IpfF (Ibuprofen CoA ligase from Sphingomonas sp. Ibu-2)) were integrated using a method developed previously (Muratovska et al., 2022a), generating strain TMBNM034 (Table 1).
2.3 Whole-cell bioconversion in shake flasks and serum vials
Pre-cultures were prepared from a single colony in 5 ml YPD grown overnight. Shake flask with 25 mL defined mineral medium (Verduyn et al., 1992) and glucose (20 g L−1) was inoculated to initial optical density (OD620nm) ∼0.5 and incubated at 30°C in 180 rpm shaking incubator for 24–26 h to generate biomass. Subsequently, yeast cells were harvested by centrifugation at 3220 g for 10 min (Eppendorf Centrifuge 5810 R, Germany) and re-suspended at OD620nm ∼5 in 50 mL of reaction medium. The reaction medium was based on defined mineral medium buffered at 6.5 pH, and supplemented with vanillin (5 mM), glucose (10 g L−1) or ethanol (0.5 or 10 g L−1) as co-substrate, and with or without L-alanine (50 mM) and ammonium chloride (200 mM) as nitrogen source. Whole-cell bioconversions were performed under aerobic or anaerobic conditions. For the aerobic conditions, bioconversions were carried out in 500 mL shake flasks containing 50 mL medium and incubated at 30°C in 180 rpm shaking incubator as described above. Anaerobic bioconversions were performed in sealed serum vials containing 50 mL medium and sparged with nitrogen gas for 1 h before the start of the reaction and incubated at 30°C in 180 rpm shaking incubator.
2.4 Batch bioreactor cultivations
Pre-cultures were prepared from a single colony inoculated in a shake flask with 50 mL defined mineral medium incubated under shaking (180 rpm) at 30°C for around 24 h. The pre-culture was used to inoculate 3 L bench-top bioreactor (Minifors 2. Infors HT, Bottmingen, Switzerland) at OD620nm = 0.1 in 1 L of defined mineral medium with 50 g L−1 glucose. The pH was kept at 6.5 by automated addition of 3 M KOH, the stirring was constant at 300 rpm, and the temperature was set at 30°C. Yeast cells were first grown under aerobic conditions with air sparging set to 500 ml/min (0.5 vvm) for approximately 20 h, and the whole-cell bioconversions were subsequently started by adding the substrates (vanillin and non-anoic acid) to the media. For the non-sparged condition, aeration was stopped to achieve oxygen-limited conditions after the initial growth phase, and directly prior to the supplementation of substrates.
2.5 Analytical methods
Samples were collected with a syringe regularly from cell cultures for analysis of yeast cell concentration, yeast cell viability, and extracellular metabolite analysis. Yeast cell concentration was estimated by measuring optical density at 620 nm (OD620nm) with a spectrophotometer (Ultrospec 2,100 pro UV/Visible spectrophotometer, Amersham Biosciences, Buckinghamshire, United Kingdom). Yeast cell viability was monitored by flow cytometry (Accuri C+, BD, US) using the fluorescent dyes SYBR Green I and propidium iodide, as described previously (Rao et al., 2021). Instrument settings were adjusted to the following: medium flow rate, FSC-H threshold at 80,000, and acquisition of 10,000 events. FCM data analysis was made with FlowJo® (United States). Gating of yeast cells was based on FSC-H and SSC-H signals to distinguish cells from background noise, and gating of damaged and intact cells were based on SYBR Green I detected with FL1-H (530/30 nm), and PI detected with FL3-H (≥670 nm) signals. Yeast cells treated with 70% ethanol for 30 min were used as control to identify damaged cells and to adjust the gates. For the analysis of glucose, ethanol, glycerol and acetate, a Waters HPLC system equipped with an Aminex HPX-87 H ion-exchange column (7.8 × 300 mm, Bio-Rad, Hercules, United States), and a refractive index detector (Waters 2,414, Milford, United States) were used, as described previously (Weber et al., 2014b). The mobile phase was 5 mM H2SO4 with a flow rate of 0.6 mL/min, and the column was kept at 60°C. For reverse-phase HPLC analysis of vanillin, vanillylamine, vanillic alcohol and vanillic acid, a Select C18 column (4.6 × 150 mm) with mobile phase consisting of ultrapure water and 0.1% trifluoroacetic acid (TFA) (A) and acetonitrile (B). The method was isocratic with 65% A and 35% B for 10 min with a flow of 1 ml min−1, at room temperature and the monitored wavelength was at 281 nm, as described previously (Manfrao-Netto et al., 2021). Prior to HPLC analysis of nonivamide, product extraction was performed from 20 ml samples from the bioreactors. Supernatant was collected after centrifugation at 3220 g for 10 min, transferred to an extraction funnel and extracted with one volume of ethyl acetate. The organic phase was collected and evaporated using a rotary vacuum evaporator. The resulting viscous oil was dissolved in 1 mL of methanol and HPLC analysis were performed using the same Select C18 column (4.6 × 150 mm) as previously described (Muratovska et al., 2022a).
2.6 Calculations
The rate ratio is defined as the vanillylamine formation rate (mmol h-1) divided by the by-product (vanillic alcohol and vanillic acid) formation rate (mmol h-1) for the initial 6 h. Molar yield is defined as moles of product (vanillylamine) or by-product (vanillic alcohol or vanillic acid) per moles of consumed substrate (vanillin) after 48 h. The specific vanillylamine productivity (µmol OD−1 h−1) is calculated for the initial 6 h.
3 Results and discussion
3.1 Comparison of reaction conditions for reductive amination with yeast over-expressing CvTA
Yeast carrying six copies of the CvTA gene expressed under control of the strong constitutive TDH3 promoter was evaluated under different reaction conditions for whole-cell conversion of vanillin to vanillylamine (Supplementary Figure S1). Most vanillin reductases are NADPH-dependent, and vanillin dehydrogenase is NAD + -dependent; therefore, the redox balance significantly influence by-product formation. In Crabtree positive yeast, such as Saccharomyces cerevisiae, glucose is readily fermented to ethanol in both aerobic and anaerobic conditions, while ethanol is slowly respired in aerobic condition and stays practically inert in anaerobic condition (De Deken, 1966). The availability of NADPH is limited with the use of ethanol as co-substrate, especially under anaerobic condition resulting in suppression of the activity of NADPH-dependent reductases (but not NADH-dependent reductases) (Kratzer et al., 2008). In contrast, NADPH is efficiently regenerated in the pentose phosphate pathway during glucose assimilation (Parachin et al., 2009). Another important factor is the abundance of pyruvate formed both as central carbon metabolite and from the amine donor L-alanine in yeast (Weber et al., 2014b; Weber et al., 2017). Pyruvate is a strong inhibitor of CvTA and its removal shifts the equilibrium to the amine (Sánchez-Moreno et al., 2012; Weber et al., 2017). Here, four conditions were evaluated: (i) aerobic with glucose as carbon source, (ii) aerobic with ethanol as carbon source, (iii) anaerobic with glucose as carbon source, and (iv) anaerobic with ethanol as carbon source. Pre-cultures were made in the same way and whole-cell bioconversions were performed at relatively high cell density (OD620nm = 5); therefore, the expression level of endogenous genes was the same at start of the reaction, and any differences in product spectra would be due to the specific applied reaction environment. Significant differences were observed for the different conditions, with the highest vanillin consumption rate (0.11 ± 0.00 mmol OD−1 h−1) under aerobic conditions and with ethanol used as a co-substrate (Supplementary Figure S1). This condition resulted in more production of vanillylamine (3.21 ± 0.56 mM) compared to the rest, but also of the by-products vanillic alcohol (4.11 ± 1.48 mM) and vanillic acid (1.72 ± 0.40 mM) (Supplementary Figure S1). To compare the activity of CvTA and endogenous enzymes acting on vanillin, the rate ratio, defined as vanillylamine formation rate per by-product formation rate, was calculated (Table 2). A higher rate ratio (5-7- fold) was observed with ethanol as co-substrate, both under aerobic and anaerobic condition. The difference in rate ratio under aerobic conditions was mostly due to an increased formation of vanillylamine with the use of ethanol as the co-substrate. A significant amount of vanillic alcohol and vanillic acid was formed for both glucose and ethanol as co-substrate under aerobic conditions, which was expected due to balanced levels of NAD(P)H/NAD(P)+ ratios under respiration (Murray et al., 2011). Under anaerobic conditions, the transamination was less efficient, however, alcohol and acid formation were also significantly lower. Worth noting is the almost complete suppression of vanillic acid formation under anaerobic conditions, possibly due to low availability of NAD+, or NADP+ (Supplementary Figure S1C, D). In aerobic conditions, the vanillin consumption rate was generally faster, regardless on the choice of co-substrate; however, which end products were formed from vanillin depend on the choice of co-substrate and oxygen availability.
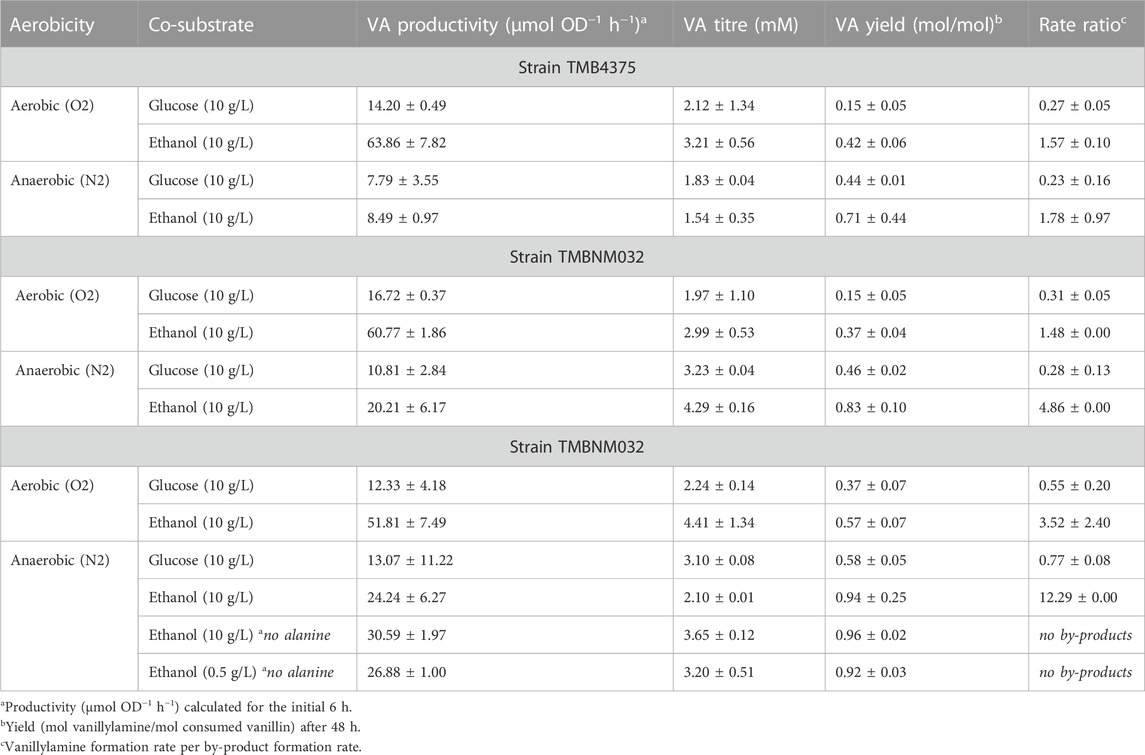
TABLE 2. Process parameters calculated for the different reaction conditions and strains during whole-cell conversion of vanillin to vanillylamine (VA).
The highest molar yield of vanillylamine was reached under the anaerobic condition with ethanol as co-substrate (0.71 ± 0.44 mol/mol), while the lowest molar yield of vanillylamine per consumed vanillin after 48 h was on glucose in the aerobic condition (0.15 ± 0.05 mol/mol) (Table 2). However, a problem during the anaerobic condition was incomplete conversion of vanillin. A reason for this could be inhibition of the transaminase by pyruvate accumulation, or low limitation in amine donor availability. Alanine was supplied in excess and gets converted to pyruvate by both CvTA, as well as by endogenous transaminases, for example by alanine transaminase (encoded by ALT1) (García-Campusano et al., 2009). Pyruvate could be removed from the reaction system through conversion to ethanol by pyruvate decarboxylase and alcohol dehydrogenase. During respiration, pyruvate is converted to acetyl-CoA by pyruvate dehydrogenase, and further oxidised in the tricarboxylic acid (TCA) cycle to CO2 and generating NADH (Flikweert et al., 1996). When glucose is present during the bioconversion, there would be higher intracellular pyruvate levels (Weber et al., 2014b; Weber et al., 2017), which could explain why conditions with ethanol gave better transamination yields.
3.2 Co-expression of BsAlaDH with the CvTA improves vanillylamine production
To relieve potential pyruvate inhibition and improve the amine donor supply, an NADH-dependant alanine dehydrogenase from Bacillus subtilis, BsAlaDH, was expressed in strain TMBNM032 under control of the TEF1 promoter. Co-expression of CvTA and BsAlaDH resulted in almost complete conversion of vanillin within 48 h (Supplementary Figure S2), which could indicate that pyruvate accumulation and/or alanine limitation was the issue in the parent strain TMB4375. Vanillin consumption rate for this strain as well as the vanillylamine titres after 48 h was almost double when compared to strain TMB4375 in the anaerobic conditions (Table 2; Supplementary Figures S1C, D vs. Supplementary Figures S2A, B). The rate ratios (vanillylamine formed per by-products formed) were not noticeably different compared to TMB4375 (Table 2), except under anaerobic conditions with ethanol as co-substrate (4.86 rate ratio) Here, only low amounts of vanillic alcohol were detected after 48 h (0.9 mM; Supplementary Figure S2B). Compared to TMB4375 in the same condition, the formation of vanillic alcohol decreased with the expression of AlaDH by around a third, and the vanillylamine yields were increased from 0.71 to 0.83 mol/mol (Table 2).
3.2.1 Deleted ADH6 reduce formation of vanillic alcohol
Adh6 is an alcohol dehydrogenase with activity to both NADH and NADPH, shown previously to be largely responsible for the reduction of vanillin in yeast (Hansen et al., 2009; Wang et al., 2016). To further decrease the levels of vanillic alcohol, deletion of ADH6 was made in strain TMBNM033. In our hands, and in agreement with previous studies, the ADH6 deletion strain produced less vanillic alcohol with both glucose (3-fold less) or ethanol as co-substrate (4-fold less) than the control strain (Table 2; Figures 2A,B). The yield of vanillylamine per consumed vanillin was higher in all cases, with the highest molar yield (0.94 ± 0.25) reached in anaerobic condition with ethanol as co-substrate. The largest difference in rate ratio was observed under anaerobic condition with ethanol as co-substrate (12.3 compared to 4.9). However, under these conditions the amination of vanillin arrested around 20 h, resulting in incomplete conversion. This indicates that NADH regeneration is limiting under these conditions, which on one hand may reduce NADH-dependent reduction of vanillin from endogenous reductases, and on the other also reduce the desired conversion of pyruvate to alanine by BsAlaDH.
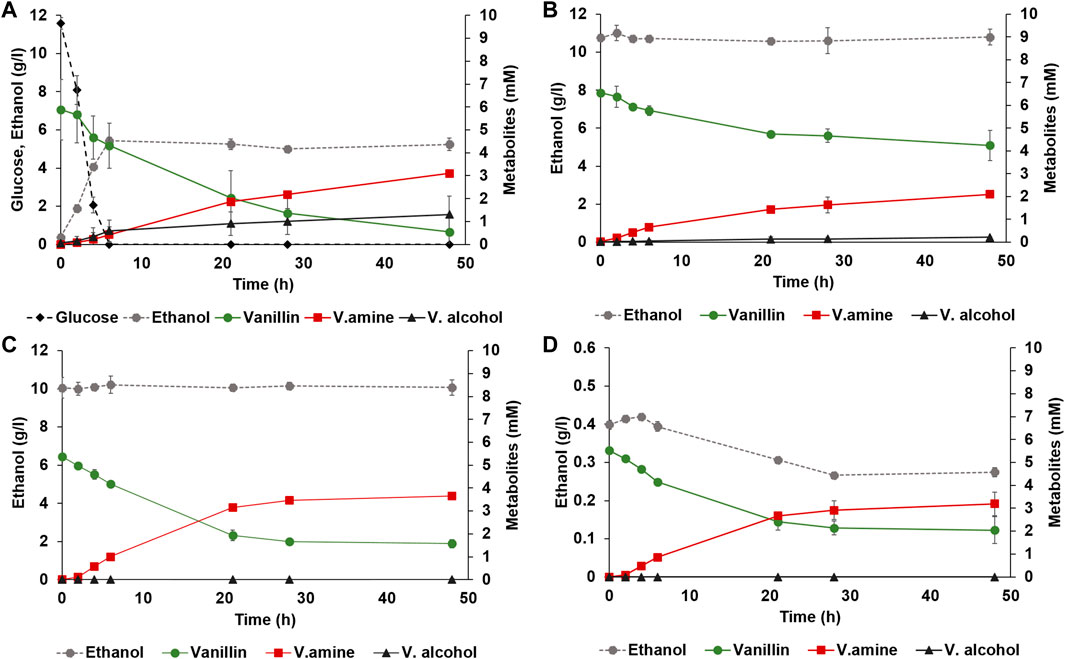
FIGURE 2. Anaerobic whole-cell bioconversions of vanillin to vanillylamine with yeast strain TMBNM033, co-expressing CvTA and BsAlaDH, and with deleted ADH6. Reactions were made in serum vials sparged with nitrogen using defined mineral medium supplemented with (A) glucose and alanine; (B) 10 g L−1 ethanol and alanine, (C) 10 g L−1 ethanol, and without alanine, or (D) 0.5 g L−1 ethanol and without alanine. Co-substrates glucose (black diamond) or ethanol (dotted line with grey circle) are shown on the left axis, and the metabolites vanillin (green circle), vanillylamine (red square) and vanillic alcohol (black triangle) are shown on the right axis.
3.2.2 Omitting alanine in the reaction broth improves vanillylamine production
Alanine is typically added in excess in biocatalytic transamination but may instead be produced de novo. In recombinant P. putida expressing CvTA and BsAlaDH, the omission of alanine resulted in improved reductive amination of vanillin, possibly due to reduced amount of intracellular pyruvate originating from the activity of endogenous alanine transaminases (Manfrao-Netto et al., 2021). To see if the same hold for recombinant yeast expressing the same CvTA and BsAlaDH, whole-cell bioconversions were performed with and without alanine with strain TMBNM033 (Figures 2C, D). The reaction was performed under anaerobic condition with ethanol as co-substrate since these conditions were found to suppress formation of vanillic alcohol and vanillic acid. The omission of alanine indeed had a noticeable positive effect on the titre and yield of vanillylamine (Figures 2C, D), in agreement with the results for P. putida (Manfrao-Netto et al., 2021). In fact, under these conditions the highest vanillylamine titre (3.65 mM) and yield from consumed vanillin (0.96 mol/mol) were obtained (Table 2). This further demonstrates that the alanine dehydrogenase efficiently converts pyruvate to alanine and drives the transamination reaction without external supplementation of amine donor.
3.2.3 Assimilation of ethanol as co-substrate correlates to the amination reaction
As mentioned before, amination of vanillin was favoured over alcohol formation the most with ethanol supplemented as a co-substrate and having anaerobic condition during the bioconversion, although the reaction arrested completely halfway through. S. cerevisiae generally cannot metabolise ethanol anaerobically because it requires the TCA and glyoxylate cycles and the electron transport chain. However, some ethanol could be converted to acetate via acetaldehyde by alcohol dehydrogenase EC 1.1.1.1 (ex. Adh1) and acetaldehyde dehydrogenase EC 1.2.1.3 (ex Ald6) (Meaden et al., 1997; de Smidt et al., 2008). This route enabled regeneration of NADH for AlaDH for ca 20 h, after which the formation of vanillylamine stopped (Figure 2C). To better see to which degree ethanol was converted during the progression of the reaction, 0.5 g L−1 ethanol instead of 10 g L−1 was used, thereby enabling the detection of an even minute decrease through HPLC analysis (Figure 2D). The ethanol titre indeed decreased under the first 25 h from 0.40 to 0.27 g L−1, or with 32%, and then it remained constant. Ethanol consumption thus coincided with the conversion of vanillin to vanillylamine, proving its importance as co-substrate for supplying reducing equivalents to the reduction reaction. Vanillic alcohol was however not detected in this condition and the vanillylamine yield was high (0.92 mol/mol), demonstrating the transamination being the dominant activity (Table 2). The arrest in ethanol assimilation, and thereby of the amination reaction, may be due to accumulation of intracellular acetaldehyde, which is inhibitory to yeast cells. Further studies could be directed to fine-tune the co-substrate for improved NADH availability for the BsAlaDH to produce higher amount of alanine (and less of pyruvate), and still avoid vanillic alcohol formation.
3.3 Combined reductive amination and amidation for nonivamide production
Strain TMBNM033 was further engineered by expressing CaAT and IpfF, previously shown to be effective in the biosynthesis of nonivamide from non-anoic acid and vanillylamine (Muratovska et al., 2022a). The constructed yeast strain TMBNM034, containing CvTA, BsAlaDH, ΔADH6 and CaAT/IpfF, was characterised in bench scale bioreactors for production of nonivamide from vanillin and non-anoic acid (Figure 3). The yeast cells were first grown under aerobic conditions with glucose as carbon and energy source; subsequently, the substrates vanillin and non-anoic acid were added. At this point, the cell density was high (OD620nm = 15–17) enabling higher conversion rates. Aeration was found to affect the whole-cell transamination reaction negatively, both in terms of vanillylamine formation, and by-product formation (Table 2). On the other hand, lack of oxygen may result in limited supply of ATP and CoA-SH required for the coenzyme A ligase to generate the acyl-coa for the amide-forming step. To shed light into this possible dilemma, two reaction configurations were compared: (1) air-sparged conditions (Figures 3A, B) and (2) non-sparged oxygen-limited conditions (Figures 3C, D). The supplied vanillin and non-anoic acid were converted to nonivamide in both cases, but slightly more in the presence of oxygen (0.19 vs. 0.12 μmol L−1 OD−1) (Figure 3; Table 3). The results demonstrate that amidation indeed can be combined with reductive amination in one-pot in yeast. However, nonivamide titers were low and a significant accumulation of vanillylamine (3–5 mM) was observed, showing that the amide-forming step is the bottleneck for production from vanillin. It may be speculated that increasing the amount of air in the bioreactor may improve the situation by increasing the supply of ATP and CoA-SH; however, other problems may also be at hand, e.g. CaAT, and IpfF expression levels or the specific activity of the enzymes may be low. Also, the availability of non-anoic acid may be limiting since it is supplemented in low concentration due to toxicity and low solubility (Muratovska et al., 2022a). The experiments indicate that the in vivo transamination reaction is hampered by increased aeration of the reaction broth. The rate ratio of vanillylamine formation per by-product formation was higher under non-sparged conditions (3.5 vs. 2.4). Formation of vanillic alcohol and even more so, of vanillic acid, were lower, resulting in molar yields from consumed vanillin of 0.28 ± 0.03 and 0.03 ± 0.01 at 48 h respectively. This shows that the ADH6 deletion is not sufficient to completely prevent vanillic alcohol formation under the applied conditions. Nevertheless, while ethanol assimilation was not detected under anaerobic condition, it was consumed throughout the bioconversion sparged with air, indicating functional regeneration of cofactors. Cell viability, measured by flow cytometry, remained high throughout the bioconversions with >98% intact cells for both conditions (Supplementary Figure S3 in supplementary), and could not explain the observed differences in activity.
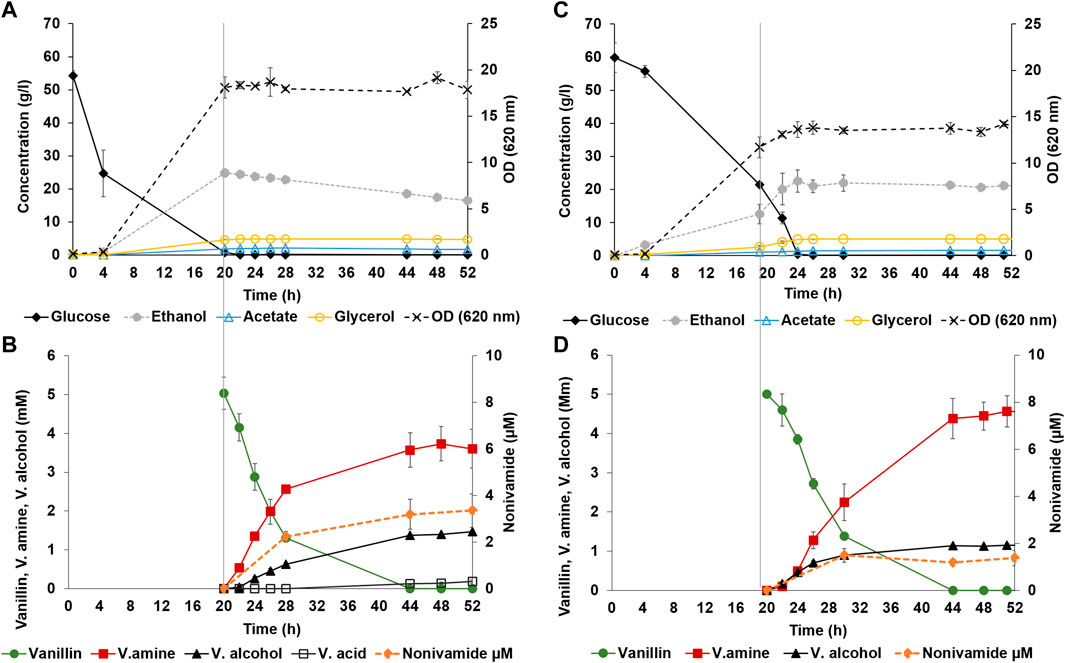
FIGURE 3. Bioreactor controlled whole-cell bioconversion of vanillin and non-anoic acid to nonivamide by yeast strain TMBNM034, co-expressing CvTA, BsAlaDH, CaAT/IpfF and with deleted ADH6. During the first 20 h both bioreactor set-ups were treated in the same way, after which (marked with vertical line) vanillin and non-anoic acid were added and aeration was kept at 0.5 vvm (A, B) or stopped (C, D). Left axis presents measured fermentation metabolites glucose (black diamond), ethanol (dotted line with grey circle), acetate (empty blue triangle), glycerol (empty yellow circle) shown in g L−1 (A, C); and bioconversion metabolites vanillin (green circle), vanillylamine (red square), vanillic alcohol (black triangle), vanillic acid (black empty square) shown in mM (B, D). Right axes present OD620nm [dotted line with black cross; (A, C)] or nonivamide in µM [orange dotted line with circle; (B, D)].
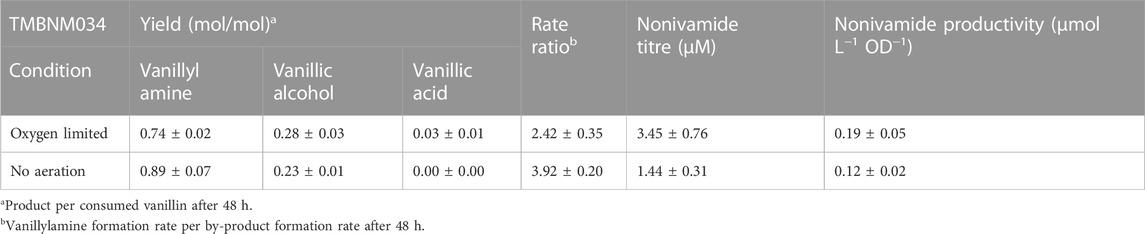
TABLE 3. Yield, rates and titers calculated for the combined amination and amidation for nonivamide production in bioreactor set-up.
4 Conclusion
S. cerevisiae was engineered for production of nonivamide from vanillin and non-anoic acid. The constructed yeast strain contains a multienzyme cascade for carrying out coupled amine and amide forming steps in vivo. Over-expression of CvTA and the NADH-dependent BsAlaDH together with ADH6 knockout, and the use of ethanol as co-substrate and oxygen-limitation to regenerate mainly NADH favoured formal reductive amination and led to an almost complete suppression of endogenous enzymes converting vanillin to vanillic acid and vanillic alcohol. The amide forming step using the CaAT/IpfF-cascade was two to three orders of magnitude less efficient than the amination, still, functionality was clear. The developed method opens the possibility for yeast-based production of capsaicinoids from vanillin, although more efforts are needed to improve efficiency.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
NM performed the experiments, analysis and drafted the manuscript. NM and MC conceived the study, designed the experiments, interpreted the data, and wrote the manuscript.
Funding
The study was financed by the Swedish Research Council under Grant 201704174.
Acknowledgments
We acknowledge the members of the Microbial Engineering Group at the department of chemistry, Lund university, for fruitful discussions during preparation of this manuscript. We would like to thank Raquel Perruca-Foncillas for providing the pRP005 plasmid.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2022.1097215/full#supplementary-material
References
Baenas, N., Belović, M., Ilic, N., Moreno, D. A., and García-Viguera, C. (2019). Industrial use of pepper (Capsicum annum L.) derived products: Technological benefits and biological advantages. Food Chem. 274, 872–885. doi:10.1016/j.foodchem.2018.09.047
De Deken, R. H. (1966). The crabtree effect: A regulatory system in yeast. J. Gen. Microbiol. 44 (2), 149–156. doi:10.1099/00221287-44-2-149
de Smidt, O., du Preez, J. C., and Albertyn, J. (2008). The alcohol dehydrogenases of Saccharomyces cerevisiae: A comprehensive review. FEMS Yeast Res. 8 (7), 967–978. doi:10.1111/j.1567-1364.2008.00387.x
Flikweert, M. T., Van Der Zanden, L., Janssen, W. M., Steensma, H. Y., Van Dijken, J. P., and Pronk, J. T. (1996). Pyruvate decarboxylase: An indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12 (3), 247–257. doi:10.1002/(SICI)1097-0061(19960315)12:3%3C247:AID-YEA911%3E3.0.CO;2-I
Fu, B., Xiao, G., Zhu, Y., Chen, Y., Lai, Y., and Yuan, J. (2021). Renewable vanillylamine synthesis from lignin-derived feedstocks. ACS Agric. Sci. Technol. 1 (5), 566–571. doi:10.1021/acsagscitech.1c00174
Gajewski, J., Pavlovic, R., Fischer, M., Boles, E., and Grininger, M. (2017). Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 8, 14650. doi:10.1038/ncomms14650
García-Campusano, F., Anaya, V.-H., Robledo-Arratia, L., Quezada, H., Hernández, H., Riego, L., et al. (2009). ALT1-encoded alanine aminotransferase plays a central role in the metabolism of alanine in Saccharomyces cerevisiae. Can. J. Microbiol. 55 (4), 368–374. doi:10.1139/w08-150
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2 (1), 31–34. doi:10.1038/nprot.2007.13
Hansen, E. H., Moller, B. L., Kock, G. R., Bunner, C. M., Kristensen, C., Jensen, O. R., et al. (2009). De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker's yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 75 (9), 2765–2774. AEM.02681-08 [pii]. doi:10.1128/AEM.02681-08
Jessop-Fabre, M. M., Jakociunas, T., Stovicek, V., Dai, Z., Jensen, M. K., Keasling, J. D., et al. (2016). EasyClone-MarkerFree: A vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-cas9. Biotechnol. J. 11 (8), 1110–1117. doi:10.1002/biot.201600147
Kang, S., Kang, K., Chung, G. C., Choi, D., Ishihara, A., Lee, D.-S., et al. (2005). Functional analysis of the amine substrate specificity domain of pepper tyramine and serotonin N-hydroxycinnamoyltransferases. Plant Physiol. 140 (2), 704–715. doi:10.1104/pp.105.071514
Knudsen, J. D., Hägglöf, C., Weber, N., and Gorwa-Grauslund, M. (2016). Increased availability of NADH in metabolically engineered baker’s yeast improves transaminase-oxidoreductase coupled asymmetric whole-cell bioconversion. Microb. Cell. Fact. 5, 37. doi:10.1186/s12934-016-0430-x
Koivisto, A.-P., Belvisi, M. G., Gaudet, R., and Szallasi, A. (2021). Advances in TRP channel drug discovery: From target validation to clinical studies. Nat. Rev. Drug Discov. 21, 41–59. doi:10.1038/s41573-021-00268-4
Koszelewski, D., Lavandera, I., Clay, D., Guebitz, G. M., Rozzell, D., and Kroutil, W. (2008). Formal asymmetric biocatalytic reductive amination. Angew. Chem. Int. Ed. Engl. 47 (48), 9337–9340. doi:10.1002/anie.200803763
Kratzer, R., Egger, S., and Nidetzky, B. (2008). Integration of enzyme, strain and reaction engineering to overcome limitations of baker's yeast in the asymmetric reduction of alpha-keto esters. Biotechnol. Bioeng. 101 (5), 1094–1101. doi:10.1002/bit.21980
Manfrao-Netto, J. H. C., Lund, F., Muratovska, N., Larsson, E. M., Parachin, N. S., and Carlquist, M. (2021). Metabolic engineering of Pseudomonas putida for production of vanillylamine from lignin-derived substrates. Microb. Biotechnol. 14, 2448–2462. doi:10.1111/1751-7915.13764
Meaden, P. G., Dickinson, F. M., Mifsud, A., Tessier, W., Westwater, J., Bussey, H., et al. (1997). The ALD6 gene of Saccharomyces cerevisiae encodes a cytosolic, Mg(2+)-activated acetaldehyde dehydrogenase. Yeast 13 (14), 1319–1327. doi:10.1002/(SICI)1097-0061(199711)13:14<1319:AID-YEA183>3.0.CO;2-T
Muratovska, N., Grey, C., and Carlquist, M. (2022a). Engineering Saccharomyces cerevisiae for production of the capsaicinoid nonivamide. Microb. Cell. Factories 21, 106. doi:10.1186/s12934-022-01831-3
Muratovska, N., Silva, P., Pozdniakova, T., Pereira, H., Grey, C., Johansson, B., et al. (2022b). Towards engineered yeast as production platform for capsaicinoids. Biotechnol. Adv. 59, 107989. doi:10.1016/j.biotechadv.2022.107989
Murdoch, R. W., and Hay, A. G. (2013). Genetic and chemical characterization of ibuprofen degradation by Sphingomonas Ibu-2. Microbiol. Read. 159 (3), 621–632. doi:10.1099/mic.0.062273-0
Murray, D. B., Haynes, K., and Tomita, M. (2011). Redox regulation in respiring Saccharomyces cerevisiae. Biochimica Biophysica Acta (BBA) - General Subj. 1810 (10), 945–958. doi:10.1016/j.bbagen.2011.04.005
Ogawa, K., Murota, K., Shimura, H., Furuya, M., Togawa, Y., Matsumura, T., et al. (2015). Evidence of capsaicin synthase activity of the Pun1-encoded protein and its role as a determinant of capsaicinoid accumulation in pepper. BMC Plant Biol. 15, 93. doi:10.1186/s12870-015-0476-7
Parachin, N. S., Carlquist, M., and Gorwa-Grauslund, M. F. (2009). Comparison of engineered Saccharomyces cerevisiae and engineered Escherichia coli for the production of an optically pure keto alcohol. Appl. Microbiol. Biotechnol. 84 (3), 487–497. doi:10.1007/s00253-009-1964-1
Perruca-Foncillas, R., Davidsson, J., Carlquist, M., and Gorwa-Grauslund, M. F. (2022). Assessment of fluorescent protein candidates for multi-color flow cytometry analysis of Saccharomyces cerevisiae. Biotechnol. Rep. 34, e00735. doi:10.1016/j.btre.2022.e00735
Rao, N. S., Lundberg, L., Palmkron, S., Hakansson, S., Bergenstahl, B., and Carlquist, M. (2021). Flow cytometric analysis reveals culture condition dependent variations in phenotypic heterogeneity of Limosilactobacillus reuteri. Sci. Rep. 11 (1), 23567. doi:10.1038/s41598-021-02919-3
Romero-Suarez, D., Keasling, J. D., and Jensen, M. K. (2022). Supplying plant natural products by yeast cell factories. Curr. Opin. Green Sustain. Chem. 33, 100567. doi:10.1016/j.cogsc.2021.100567
Sambrook, J., and Russell, D. W. (2001). Molecular cloning: A laboratory manual. Cold Spring Harbour: Cold Spring Harbour Laboratory Press.
Sánchez-Moreno, I., Oroz-Guinea, I., Iturrate, L., and García-Junceda, E. (2012). “7.20 multi-enzyme reactions,” in Comprehensive chirality. Editors E. M. Carreira, and H. Yamamoto (Amsterdam: Elsevier), 430–453.
Strucko, T., Magdenoska, O., and Mortensen, U. H. (2015). Benchmarking two commonly used Saccharomyces cerevisiae strains for heterologous vanillin-β-glucoside production. Metab. Eng. Commun. 2, 99–108. doi:10.1016/j.meteno.2015.09.001
Uarrota, V. G., Maraschin, M., de Bairros, Â. d. F. M., and Pedreschi, R. (2021). Factors affecting the capsaicinoid profile of hot peppers and biological activity of their non-pungent analogs (Capsinoids) present in sweet peppers. Crit. Rev. Food Sci. Nutr. 61 (4), 649–665. doi:10.1080/10408398.2020.1743642
Verduyn, C., Postma, E., Scheffers, W. A., and Van Dijken, J. P. (1992). Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8 (7), 501–517. doi:10.1002/yea.320080703
Wang, H., Li, Q., Kuang, X., Xiao, D., Han, X., Hu, X., et al. (2018). Functions of aldehyde reductases from Saccharomyces cerevisiae in detoxification of aldehyde inhibitors and their biotechnological applications. Appl. Microbiol. Biotechnol. 102 (24), 10439–10456. doi:10.1007/s00253-018-9425-3
Wang, X., Liang, Z., Hou, J., Bao, X., and Shen, Y. (2016). Identification and functional evaluation of the reductases and dehydrogenases from Saccharomyces cerevisiae involved in vanillin resistance. BMC Biotechnol. 16 (1), 31. doi:10.1186/s12896-016-0264-y
Weber, N., Gorwa-Grauslund, M., and Carlquist, M. (2014a). Engineered baker's yeast as whole-cell biocatalyst for one-pot stereo-selective conversion of amines to alcohols. Microb. Cell. Fact. 13 (1), 118. doi:10.1186/s12934-014-0118-z
Weber, N., Gorwa-Grauslund, M., and Carlquist, M. (2014b). Exploiting cell metabolism for biocatalytic whole-cell transamination by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 98 (10), 4615–4624. doi:10.1007/s00253-014-5576-z
Weber, N., Gorwa-Grauslund, M., and Carlquist, M. (2017). Improvement of whole-cell transamination with Saccharomyces cerevisiae using metabolic engineering and cell pre-adaptation. Microb. Cell. Fact. 16 (1), 3. doi:10.1186/s12934-016-0615-3
Keywords: recombinant yeast, reductive amination, vanillin, nonivamide, capsaicin, pain modulator, TRPV1, amide formation
Citation: Muratovska N and Carlquist M (2023) Recombinant yeast for production of the pain receptor modulator nonivamide from vanillin. Front. Chem. Eng. 4:1097215. doi: 10.3389/fceng.2022.1097215
Received: 13 November 2022; Accepted: 30 December 2022;
Published: 12 January 2023.
Edited by:
Vasudeo Zambare, University of Technology Malaysia, MalaysiaReviewed by:
Jingwen Zhou, Jiangnan University, ChinaXue Cai, Zhejiang University of Technology, China
Copyright © 2023 Muratovska and Carlquist. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magnus Carlquist, bWFnbnVzLmNhcmxxdWlzdEB0bWIubHRoLnNl
 Nina Muratovska
Nina Muratovska Magnus Carlquist
Magnus Carlquist