- School of Dental Medicine, University of Pittsburgh, Pittsburgh, PA, United States
There is great interest in identifying the subset of individuals in the population that are more susceptible to dental caries. We proposed that a portion of these particular individuals are more susceptible to dental caries due to changes in dental enamel that are related to amelogenin genomic variation. However, apparently amelogenin function can be impacted by inflammation, and this can lead to small changes in the structure of the dental enamel that later in life increases the risk of dental caries.
Introduction
Dental caries is highly preventable but continues to affect a large proportion of populations. Identification of risk factors for the disease that will allow for preventive strategies are of great interest. Aspects related to the individual have been less studied and are likely to provide the new insight needed. We believe that factors that cause deviations of normal dental enamel development should be the focus of future research that aims to unveil new targets for dental caries prevention.
Dental Caries Epidemiology
The epidemiology of dental caries has changed, and it is common to be able to identify clusters of individuals that are caries-free. At the same time, certain individuals appear to have most of the disease of the population (1). It has become a need to be able to identify early these individuals who are at higher risk for dental caries. We have suggested that individual genetic variation may be a factor playing a role in these specific individuals that appear to have most of the burden of disease. One plausible gene that may influence individual risk to dental caries is amelogenin, which is mutated in X-linked forms of amelogenesis imperfecta and accounts for 5% of all cases of the condition (2). We and others have hypothesized that variation in enamel formation genes could explain in part individual susceptibility to dental caries (1, 3). Further, genetic variation in amelogenin was associated with caries experience (4–11), erosive tooth wear (12), variation in microscopic enamel structure (13), salivary calcium levels (14), and molar–incisor hypomineralization (MIH) (8) in a number of cohorts with distinct geographic origins, and with incisor enamel microhardness in amelogenin-impaired mouse models (15).
Amelogenesis Imperfecta
Amelogenesis imperfecta is a collective designation for the variety of inherited conditions displaying isolated enamel malformations. However, the designation of amelogenesis imperfecta is also used to indicate the presence of an enamel phenotype in syndromes (16). There is great variability in the clinical presentation of amelogenesis imperfecta. It has been suggested for specific subpopulations that the most prevalent in the amelogenesis imperfecta group are autosomal recessive hypomature amelogenesis imperfecta and autosomal dominant hypoplastic amelogenesis imperfecta (17). Amelogenesis imperfecta is typically classified into four phenotypes: hypoplastic type I, hypomatured type II, hypocalcified type III, and hypoplastic-hypomaturation type IV (18). This broad spectrum of conditions associates to different mutations and it is possible that some of them may increase individual susceptibility to dental caries whereas others may have a protective effect against the disease.
Patients with hypoplastic amelogenesis imperfecta appear to have lower levels of caries experience because of an alkaline pH of saliva enriched with Bacillus spp., Enterococcus faecalis, and Enterococcus faecium (19). An analysis of the enamel of individuals born with amelogenesis imperfecta submitted in laboratory to high cariogenic conditions may provide insight on the higher susceptibility of dental caries that some in the population have.
The dental enamel of a tooth affected by amelogenesis imperfecta shows under the scanning electron microscope extensive irregular, disorganized rough superficial enamel layer. The enamel appears parallel or irregularly decussate with filamentous prisms accompanied by small, irregularly rounded formations (20, 21). The challenge of these findings continues to be that the histopathology does not appear to clearly correlate to a specific type of amelogenesis imperfecta (hypoplastic, hypocalcified, or hypomature) or a particular mutation. However, calcium levels in enamel of amelogenesis imperfecta, as well as in unaffected teeth, differed significantly between anterior and posterior teeth, indicating that the factors that influence normal mineralization in different regions of the arch are not altered due to amelogenesis imperfecta (20). The susceptibility to dental caries due to amelogenin genetic variation may therefore come from small differences in chemical content and/or physical structure of the enamel. Differences in physical structure of the enamel have been hypothesized as a mechanism underlying MIH (22).
Discussion
Variation in amelogenin that impacts enamel development that ultimately increases an individual susceptibility to dental caries may be through inflammation. Amelogenin contributes to early resolution of inflammation. It suppresses major histocompatibility class II (MHC II) gene expression. Apparently, it downmodulates the interferon gamma–induced cell surface expression of MHC II molecules in macrophages (23). Although these experiments were done using murine macrophages, this mechanism is apparently conserved between species (24, 25). This ability to enhance initial macrophage response and to reduce the duration of inflammation allows for providing the hydrophobic environment necessary for the initiation and growth of calcium hydroxyapatite crystals during enamel development (23). This hydrophobic environment allows for amelogenin to self-assemble into nanospheres and nanosphere ribbons to form the scaffold that will guide the growth of the crystals in the maturation stage of enamel development. These apparent different processes may be executed by the amelogenin alternative splice variants such as leucine-rich and tyrosine-rich amelogenin peptides, which are suggested to induce distinct cellular functions. We believe the consequences of amelogenin genetic variation in enamel development fall in a spectrum that ranges from amelogenesis imperfecta to dental caries [(26); Figure 1] and may explain a subset of cases of dental caries higher susceptibility. More localized enamel defects in comparison with amelogenesis imperfecta are seen in MIH. Proteins exclusively found in saliva of individuals with MIH include pathways for neutrophil-mediated adaptive immunity, the activation of the classical pathway of complement activation, extracellular matrix degradation, heme scavenging as well as glutathione and drug metabolism. In comparison, individuals without MIH had proteins that belong to adaptive immunity related to platelet degranulation and the lysosome (27).
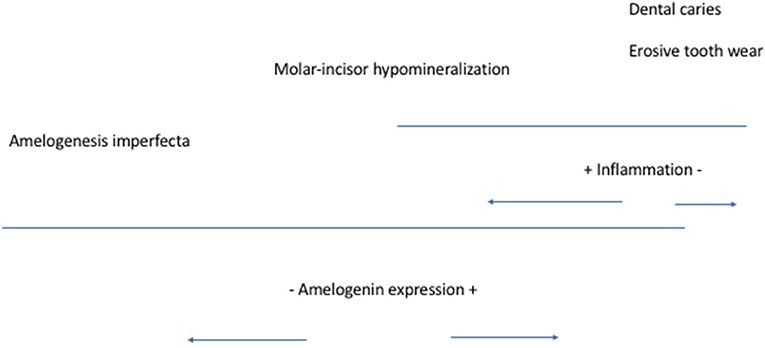
Figure 1. Amelogenin expression spectrum of influences [modified from (26)].
We believe enamel development happens under a catabolic environment that can be changed by inflammation and that leads to increased individual susceptibility later in life to dental caries. To test this hypothesis, longitudinal studies enrolling subjects at birth and recording all infections and allergic reactions are necessary to demonstrate that these events in the presence of certain molecular markers associate with higher caries experience for individuals who are exposed to cariogenic challenges.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
AV and AM conceived the idea, wrote first draft, and critically revised the final submitted version. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Vieira AR, Modesto A, and Marazita ML. Caries: review of human genetics research. Caries Res. (2014) 48:491–506. doi: 10.1159/000358333
2. Backman B, and Holmgren G. Amelogenesis imperfecta: a genetic study. Hum Hered. (1988) 38:189–206. doi: 10.1159/000153785
3. Nibali L, Di Iorio A, Tu YK, and Vieira AR. Host genetics role in the pathogenesis of periodontal disease and caries. J Clin Periodontol. (2017) 44:S52–S78. doi: 10.1111/jcpe.12639
4. Deeley K, Letra A Rose EK, Brandon CA, Resick JM, Marazita ML, and Vieira AR. Possible association of amelogenin to high caries experience in a Guatemalan-Mayan population. Caries Res. (2008) 42:8–13. doi: 10.1159/000111744
5. Patir A, Seymen F, Yildirim M, Deeley K, Cooper ME, Marazita ML, et al. Enamel formation genes are associated with high caries experience in Turkish children. Caries Res. (2008) 42:394–400. doi: 10.1159/000154785
6. Kang SW, Yoon I, Lee HW, and Cho J. Association between AMELX polymorphisms and dental caries in Koreans. Oral Dis. (2011) 17:399–406. doi: 10.1111/j.1601-0825.2010.01766.x
7. Shimizu T, Ho B, Deeley K, Briseño-Ruiz J, Faraco IM Jr, Schupack BI, et al. Enamel formation genes influence enamel microhardness before and after cariogenic challenge. PLoS ONE. (2012) 7:e45022. doi: 10.1371/journal.pone.0045022
8. Jeremias F, Koruyucu M, Küchler EC, Bayram M, Tuna EB, Deeley K, et al. Genes expressed in dental enamel development are associated with molar-incisor hypomineralization. Arch Oral Biol. (2013) 58:1434–42. doi: 10.1016/j.archoralbio.2013.05.005
9. Bayram M, Deeley K, Reis MF, Trombetta VM, Ruff TD, Sencak RC, et al. Genetic influences on dental enamel that impact caries differ between the primary and permanent dentitions. Eur J Oral Sci. (2015) 123:327–34. doi: 10.1111/eos.12204
10. Saha R, Sood PB, Sandhu M, Diwaker A, and Upadhyaye S. Association of amelogenin with high caries experience in Indian children. J Clin Pediatr Dent. (2015) 39:458–61. doi: 10.17796/1053-4628-39.5.458
11. Gerreth K, Zaorska K, Zabel M, Borysewicz-Lewicka M, and Nowicki M. Chosen single nucleotide polymorphisms (SNPs) of enamel formation genes and dental caries in a population of Polish children. Adv Clin Exp Med. (2017) 26:899–905. doi: 10.17219/acem/63024
12. Søvik JB, Vieira AR, Tveit AB, and Mulic A. Enamel formation genes associated with dental erosive wear. Caries Res. (2015) 49:236–42. doi: 10.1159/000369565
13. Kelly AM, Kallistova A, Küchler EC, Romanos HF, Lips A, Costa MC, et al. Measuring the microscopic structures of human dental enamel can predict caries experience. J Pers Med. (2020) 10:5. doi: 10.3390/jpm10010005
14. Küchler EC, Pecharki GD, Castro ML, Ramos J, Barbosa F Jr, Brancher JA, et al. Genes involved in the enamel development are associated with calcium and phosphorus level in saliva. Caries Res. (2017) 51:225–30. doi: 10.1159/000450764
15. Vieira AR, Gibson CW, Deeley K, Xue H, and Li Y. Weaker dental enamel explains dental decay. PLoS ONE. (2015) 10:e0124236. doi: 10.1371/journal.pone.0124236
16. Hu JC-C, Chun Y-H P, Al Hazzazzi T, and Simmer JP. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. (2007) 186:78–85. doi: 10.1159/000102683
17. Adorno-Farias D, Ortega-Pinto A, Gajardo P, Salazar A, Morales-Bozo I, Werlinger F, et al. Diversity of clinical, radiographic and genealogical findings in 41 families with amelogenesis imperfecta. J Appl Oral Sci. (2019) 27:e20180359. doi: 10.1590/1678-7757-2018-0359
18. Smith CEL, Poulter JA, Antanaviciute A, Kirkham J, Brookes SJ, Inglehearn CF, et al. Amelogenesis imperfecta; genes, proteins, and pathways. Front Physiol. (2017) 8:435. doi: 10.3389/fphys.2017.00435
19. Kammoun R, Zmantar T, Labidi A, Abbes I, Mansour L, and Ghoul-Mazgar S. Dental caries and hypoplastic amelogenesis imperfecta: clinical, structural, biochemical and molecular approaches. Microb Pathog. (2019) 135:103615. doi: 10.1016/j.micpath.2019.103615
20. Quevedo S, Ceballos G, Garcia JM, Rodriguez IA, Gómez de Ferraris ME, and Campos A. Scanning electron microscopy and calcification in amelogenesis imperfecta in anterior and posterior human teeth. Histol Histopathol. (2001) 16:827–32. doi: 10.14670/HH-16.827
21. Gopinath VK, Al-Salihi KAM, Yean CY, Ann MCL, and Ravichandran M. Amelogenesis imperfecta: enamel ultra-structure and molecular studies. J Clin Pediatr Dent. (2004) 28:319–22. doi: 10.17796/jcpd.28.4.27733r6m51851652
22. Vieira AR, and Manton DJ. On the variable clinical presentation of molar-incisor hypomineralization. Caries Res. (2019) 53:482–8. doi: 10.1159/000496542
23. Yotsumoto K, Sanui T, Tanaka U, Yamato H, Alshargabi R, Shinjo T, et al. Amelogenin downregulates interferon gamma-induced major histocompatibility complex class II expression through suppression of euchromatin formation in the class II transactivator promoter IV region in macrophages. Front Immunol. (2020) 11:709. doi: 10.3389/fimmu.2020.00709
24. Moradian-Oldak J, and Lakshminarayanan R. Intrinsic disorder in amelogenin. In: Goldberg M, editor. Amelogenins; Multifaceted Proteins for Dental and Bone Formation and Repair. Dubai: Bentham Books Ltd. (2010) p. 106–32.
25. Lakshminarayanan R, Yoon I, Hegde BG, Fan D, Du C, and Moradian-Oldak J. Analysis of secondary structure and self-assembly of amelogenin by variable temperature circular dichroism and isothermal titration calorimetry. Proteins. (2009) 76:560–9. doi: 10.1002/prot.22369
26. Vieira AR. Genetic basis of enamel and dentin defects. In: Vieira AR, editor. Genetic Basis of Oral Health Conditions. Cham: Springer (2020). p. 9–21.
Keywords: dental caries, amelogenesis, inflammation, AMELX, amelogenin
Citation: Vieira AR and Modesto A (2020) Amelogenesis Imperfecta Enamel Changes, Amelogenin, and Dental Caries Susceptibility. Front. Dent. Med. 1:613851. doi: 10.3389/fdmed.2020.613851
Received: 04 October 2020; Accepted: 02 November 2020;
Published: 30 November 2020.
Edited by:
Mine Koruyucu, Istanbul University, TurkeyCopyright © 2020 Vieira and Modesto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandre R. Vieira, YXJ2MTFAcGl0dC5lZHU=
 Alexandre R. Vieira
Alexandre R. Vieira Adriana Modesto
Adriana Modesto