- 1Center for Craniofacial Molecular Biology, University of Southern California, Los Angeles, CA, United States
- 2State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Chengdu, China
The homeostasis of adult tissues, such as skin, hair, blood, and bone, requires continuous generation of differentiated progeny of stem cells. The rodent incisor undergoes constant renewal and can provide an extraordinary model for studying stem cells and their progeny in adult tissue homeostasis, cell differentiation and injury-induced regeneration. Meanwhile, cellular heterogeneity in the mouse incisor also provides an opportunity to study cell-cell communication between different cell types, including interactions between stem cells and their niche environment. More importantly, the molecular and cellular regulatory mechanisms revealed by the mouse incisor have broad implications for other organs. Here we review recent findings and advances using the mouse incisor as a model, including perspectives on the heterogeneity of cells in the mesenchyme, the niche environment, and signaling networks that regulate stem cell behavior. The progress from this field will not only expand the knowledge of stem cells and organogenesis, but also bridge a gap between animal models and tissue regeneration.
Introduction
Stem cells have an unlimited ability for self-renewal and the capability to differentiate into specialized cell types (1). Therefore, stem cells are the engine for supporting tissue homeostasis and regeneration because they are responsible for tissue renewal under normal conditions and in injury repairs (2, 3). Extensive progress has been made in recent decades regarding the mechanisms that regulate stem cell behavior (4, 5). Accumulating evidence suggests that stem cells are tightly regulated by both extrinsic signals from the surrounding niche cells, other tissue structures and intrinsic signals from the stem cells themselves (6, 7). Numerous organs have been utilized as models to investigate the function of stem cells in various scenarios (8).
At the cellular level, stem cells are highly heterogeneous and their interactions with the neighboring cells and structures will determine whether these stem cells will proliferate or differentiate into different cell types. Multiple signaling pathways have been shown to control the fate of stem cells during development, tissue homeostasis and repair. Specifically, FGF, BMP, Notch, WNT, and Hh signaling networks play crucial role in regulating the fate of stem cells and providing important feedback to them (9–11). Transcription factors strategically teaming up with growth factor signaling mediators can activate or repress downstream signaling pathways to control the fate of stem cells in a cell type dependent manner (12). Furthermore, growing evidence has shown that epigenetic modifications play crucial roles in regulating the fate of stem cells in development, tissue homeostasis, injury repair and diseases (13, 14). Different tissues and organs have been used to investigate the regulatory mechanism of stem cell fate determination. Unlike most organs in adult animals, rodent incisors can continuously grow throughout the lifespan of the animal, making this organ an attractive model to study the role of stem cells in tissue homeostasis and repair.
Significant advances have been made using the incisor as a model to identify dental stem cells, understand their functions and characterize the molecular and cellular mechanisms that regulate their dynamic fate decisions (15–17). In the incisor, stem cells are compartmentalized into epithelial stem cells (ESCs), which reside in the cervical loop to support epithelial tissue renewal; and mesenchymal stem cells (MSCs), which encompass the territory between the cervical loops to fuel the homeostasis of the mesenchymal tissue in the incisor. Like in many tissues and organs, there is an undifferentiated population of cells that undergo mitosis, termed transit amplifying cells (TACs), positioned between the stem cells and terminally differentiated cells (18, 19). As the stem cells exit their quiescent state they immediately give rise to TACs, which then will differentiate into ameloblasts in the dental epithelium and odontoblasts and dental pulp cells in the dental mesenchyme of adult mouse incisor. The differentiation of these stem cells is highly organized along the proximal to distal axis, making incisor an ideal model to track stem cell fate determination and differentiation process, and to investigate the mechanisms through which stem cells support tissue homeostasis and repair. Lessons learned from stem cell studies in adult mouse incisor are highly informative for investigation of stem cells in other organs in maintaining tissue homeostasis and repair (20, 21). Furthermore, recent studies have clearly identified genes that are specifically expressed in stem cells, or TACs within the adult mouse incisor, making it possible to utilize these genes as markers to identify stem cells or TACs in vivo and perform cell lineage analysis in order to test the fate determination process of stem cells in maintaining tissue homeostasis or repair (19, 22, 23). Here we review the progress and current understanding of the rodent incisor as a model to investigate the regulatory mechanism and feedback on stem cells. We highlight some outstanding questions for future investigation. Specifically, we will address epithelial and mesenchymal stem cells in the adult mouse incisor, focusing more on the MSCs and their niche environment, signaling pathways and epigenetic regulators that control the fate of MSCs, and offer perspectives on how the adult mouse incisor will continue to serve as a highly informative model to advance our understanding of the regulatory mechanisms of stem cells in supporting tissue homeostasis, repair and regeneration.
Epithelial and mesenchymal stem cells in the mouse incisor
Epithelial stem cells in the mouse incisor follow a classical stem cell paradigm, in which a few slow-cycling quiescent stem cells reside in the proximal region of the labial cervical loop (laCL), in particular the structures known as the outer enamel epithelium (OEE) and stellate reticulum (SR). Several signaling pathways have been shown to regulate the fate of these epithelial stem cells, including Hh, FGF, BMP and Activin, Hippo and others (24). This epithelial stem cell population gives rise to TACs in the inner enamel epithelium (IEE), which subsequently differentiate into ameloblasts to form enamel (25). Cell lineage and transplantation studies have definitively validated the stem cell status of these epithelial cells (16). The transition from epithelial stem cells to TACs, then to pre-ameloblasts and finally to fully differentiated ameloblasts provides a well-organized cell lineage differentiation process that can serve as a model to investigate the regulation of epithelial stem cells in maintaining tissue homeostasis and injury repair.
Recently, the identity of mouse incisor epithelial stem cells was characterized from different perspectives using a combination of single-cell RNA sequencing (scRNA-seq) and computational approaches. These studies revealed that dividing cells in the incisor IEE appear to undergo self-renewal during incisor epithelial tissue homeostasis and both ameloblasts and adjacent non-ameloblast cells are differentiated from these actively cycling epithelial progenitors, suggesting that these dividing cells orchestrate the homeostasis and repair of the incisor epithelium (26). Though well established, our understanding of the identity of epithelial stem cells in the incisor is still evolving, and future technologies may be utilized to study this question from different perspectives.
Similar to epithelial stem cells, MSCs residing in the proximal region are quiescent stem cells that can be activated into TACs, which differentiate into odontoblasts and dental pulp cells in the mouse incisor. MSCs are located close to the neurovascular bundle from which they receive signals to regulate their fate decision process (19). Using scRNA-seq analysis, studies have shown that MSCs are highly heterogeneous and may play multiple roles in maintaning incisor tissue homeostasis and repair (23). The anatomical locations of MSCs and TACs in the incisor, which are in close proximity to each other, are well defined, making the incisor an excellent model for studying the functions of these cell populations. Our study has also shown that MSCs in the proximal region of the adult incisor take about four weeks to populate the entire dental mesenchyme and reach the distal end of the incisor (19). This knowledge has provided the opportunity to measure the rate by which MSCs give rise to cells of the incisor mesenchyme, such as odontoblasts and dental pulp cells, which has facilitated the analysis of how genetic mutations, for example, may affect the migration of MSC progeny and ultimately affect the fate of these cells in supporting tissue homeostasis and repair (27, 28). Similarly, the rate of odontoblast differentiation from the MSCs/TACs can also be measured. Because TACs serve as important intermediates between MSCs and pre-odontoblasts, we have a unique system to investigate MSC-TAC interaction and feedback in maintaining tissue homeostasis (29). Equally important, mouse incisor MSCs share some common characteristics with MSCs in the long bone. However, unlike in the long bone, the distribution of incisor MSCs, TACs, pre-odontoblasts, odontoblasts and dental pulp cells is well-organized along the proximal to distal axis of the incisor, providing an ideal environment to perform stem cell lineage tracing and differentiation analysis. Finally, because of the easy access to and well defined molecular markers in adult mouse incisors, we can measure the growth rate of the incisor and its ability to repair following injury. These analyses can be linked with the dynamic changes in MSCs or TACs in the proximal region of the incisor, making it possible to comprehensively evaluate the molecular and cellular processes involved in tissue homeostasis and repair (27, 28).
Stem cell heterogeneity in the mouse incisor
Recent studies have shown that stem cells in the mouse incisor are very heterogeneous. This is especially the case in the dental mesenchyme. Different stem cell populations support the homeostasis and repair of the mouse incisor. The in vivo identities of these stem cells have been validated by lineage tracing experiments, which are the gold standard for identifying stem cells. For example, Ng2+ cells arising from pericytes can become odontoblasts after incisor damage (30). Gli1+ perivascular cells in the dental mesenchyme are Shh-responsive and can also contribute to incisor homeostasis and injury repair (19). To date, Gli1+ cells in adult mouse incisor represent the MSC population that can give rise to the entire dental mesenchyme while maintaining self-renewal in supporting incisor tissue homeostasis, highlighting the importance of the perivascular population in the incisor dental mesenchyme. Nerve associated Plp1+ glial cells can also give rise to odontoblasts and dental pulp cells during incisor growth and repair (31), suggesting the direct contribution of the nerve cells to the dental stem cells. Moreover, Thy1+ cells can contribute to incisor growth but not tissue homeostasis. Interestingly, a group of quiescent Celsr1+ cells in the dental mesenchyme can be activated and replenish Thy1+ cells upon injury (22). Recently, single-cell analysis of the mouse incisor has revealed that Foxd1 + cells near the labial cervical loop appear to possess self-renewal ability and thus can be considered another stem cell subpopulation in the incisor mesenchyme (23), suggesting that dental stem cell populations in the mouse incisor are quite heterogeneous.
Multiple epithelial stem cell populations in the proximal region of the OEE of the mouse incisor have been identified. For example, Sox2+cells can contribute to all the epithelial lineages in the mouse incisor (32). Bmi1+stem cells are regulated by Bmi1-Ink4a/Arf axis (25). Shh-responsive Gli1+cells in the incisor epithelium are also stem cells that contribute to incisor growth (33). Interestingly, the Acta2+cell population can also contribute into the homeostasis of the incisor epithelium (23). In addition, Igfbp5+and Lrig1+cells in the mouse incisor reportedly give rise to the epithelial lineages in this organ (34).
Incisor MSC niche
Stem cells are tightly controlled by their niche, which regulates how they participate in tissue homeostasis and repair. The neurovascular bundle serves as an important niche environment that integrates signals to mediate the balanced response of MSCs to the needs of homeostasis and repair. Recently, nerves have been found to regulate the MSC niches in other organs, such as skull and long bone. For example, Ngf is abundant in mesenchymal cells of cranial sutures, and directs sensory nerve transit and promotes nerve survival at suture sites (35). Moreover, sensory nerves secrete Fstl1 to regulate cell fate in the cranial suture and maintain MSCs in an undifferentiated state (36).
The interaction between MSCs and the neurovascular bundle niche component also creates the dynamic system necessary for sustaining tissues in the incisor. For example, Shh secreted by the inferior alveolar nerve activates Gli1+ cells in the mouse incisor to support its homeostasis. Interestingly, we found that a Runx2+ subpopulation of Gli1+ cells constitutes an important portion of the niche that regulates incisor homeostasis (27). These Runx2+ cells are not stem cells. Moreover, Axin2+ TACs function as a niche component, interacting with Gli1+ MSCs to regulate incisor homeostasis (29) (Figure 1).
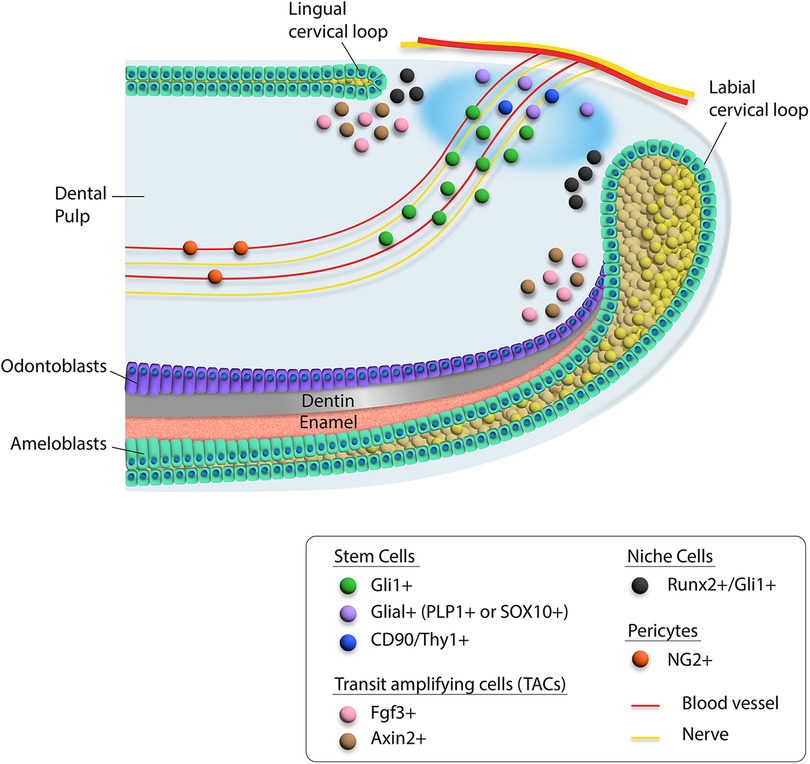
Figure 1. Schematic diagram of heterogeneity of cells in the proximal region of the mouse incisor. Different cell types are indicated by colored dots. Mesenchymal stem cells including Gli1+, Glial+, and CD90/Thy1+ cells are located around the neurovascular bundle, while Runx2+/Gli1+ niche cells are adjacent to the cervical loop. The transit amplifying cells are indicated by Fgf3+ and Axin2+ cells.
Stem cell and niche cell populations were identified in the mouse incisor.
Epigenetic regulation of stem cells in the incisor
Polycomb group (PcG) proteins are important epigenetic regulators involved in various biological processes. PcG proteins participate in two major multicomponent complexes, Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2) (38). Ring1a/b comprise the catalytic component of PRC1 complex, which mainly serves as a transcriptional repressor that deposits monoubiquitylation of histone H2A at lysine 119 (H2AK119ub1) (39). Ring1a/b are highly expressed in the proximal region of the dental mesenchyme in the incisor. Loss of Ring1a/b postnatally causes defects in the cervical loop and disturbs enamel and dentin formation, and it also causes a dramatic reduction of cell proliferation in the apical mesenchyme and cervical loop epithelium (40). Interestingly, downregulation of FGF signaling and its downstream targets is also observed in Ring1a/b mutant incisors. These results show that the PRC1 complex regulates the TACs and cell differentiation in developing mouse incisors (41).
The protein Bmi1, another member of the PRC1 family, is a well-recognized transcriptional suppressor and is capable of preventing premature senescence and maintaining the self-renewal of tissue-specific stem cells (42). Brian et al. found that Bmi1 is expressed by incisor epithelial stem cells and that deletion of Bmi1 results in diminished stem cells and defective enamel production. Mechanistically, they demonstrated that Bmi1-mediated repression of Hox genes preserves the undifferentiated state of incisor epithelial stem cells (25).
Ezh2 is one of the core enzymatic components of PRC2 and is important for the regulation of positional information of cranial neural crest cells (43). Interestingly, Ezh2 has been demonstrated to play an important function in determining the tooth root patterning of the mouse molar (44). Recently, Yu et al. found that loss of Ezh2 in Shh + epithelial progenitor cells of the mouse incisor leads to impaired epithelial regeneration upon injury, suggesting Ezh2 is indispensable for the lineage fidelity of epithelial stem cells in the mouse incisor (45).
BRG1/BRM-associated factor (BAF) is one of the most important chromatin remodelers belonging to mammalian SWI/SNF family, and plays an essential regulatory function in stem cell homeostasis (46). A recent study found that Arid1a, the largest subunit in the BAF complex, regulates mouse incisor tissue homeostasis through controlling proliferation of TACs and promoting cell cycle exit by inhibiting the Aurka-Cdk1 axis. After loss of Arid1a, the mitotic TAC population was expanded along and TAC differentiation was compromised (28).
MicroRNAs (miRNAs) are short strands of non-coding RNA that regulate protein function via post-transcriptional modifications. Selective silencing is achieved by complementary base-pairing between the miRNA and the mRNA's 3′-untranslated region (3′-UTR). Imperfect base-pairing allows miRNA to target a group of mRNA transcripts simultaneously, making miRNA a perfect tool for group-silencing related mRNA activities and maintaining stem cell homeostasis (47, 48).
Multiple studies have identified various miRNAs with roles in maintaining the tissue homeostasis of mouse incisors. The miR-200 family is one of the most studied miRNAs in the mouse incisor stem cell niche. It is highly expressed in the differentiating dental epithelial stem cells. The inhibition of the miR-200 family using the Plasmid-based miRNA Inhibitor System (PMIS) results in an expanded stem cell niche, reduced progenitor cell differentiation, and smaller incisor size (49). The miR-200 cluster maintains the stem cell homeostasis in the cervical loop through the proper compartmentalization of Sox2+ cells and epithelial differentiating cells, as well as by regulating the WNT and BMP signaling pathways (49, 50).
Signaling pathways in regulating stem cells during incisor tissue homeostasis and repair
FGF signaling
Numerous signaling pathways are involved in the tissue renewal and injury repair of the mouse incisor (Figure 2). Fibroblast growth factor (FGF) signaling induces the proliferation and differentiation of multiple cell types during embryonic development (51). Different ligands and receptors of FGF signaling participate in regulating stem cells in the incisor. For example, Fgf3 and Fgf10 are expressed in the incisor mesenchyme adjacent to the LaCL. Deletion of Fgf3 and Fgf10 leads to decreased proliferation of epithelial progenitor cells and a severely hypoplastic LaCL, suggesting functional redundancy of Fgf3 and Fgf10 in maintaining the epithelial stem cell pool in the incisor (52).
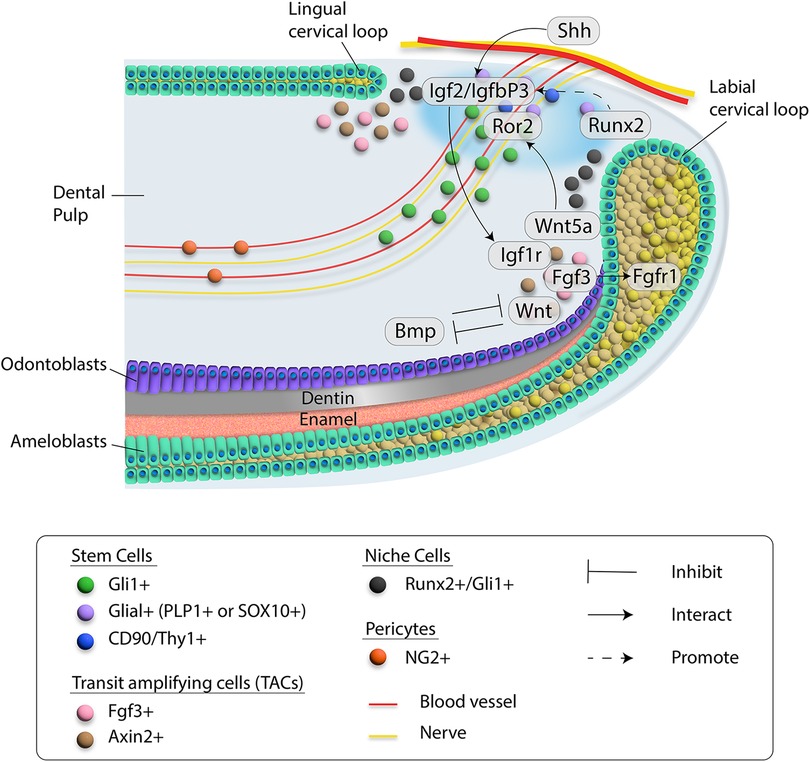
Figure 2. Schematic diagram of cell-cell communication in the proximal region of the mouse incisor. Different cell types are indicated by colored dots. Blood vessel and nerve are drawn in red and yellow, respectively. Critical molecules involved in signaling networks are listed and interactions among them are indicated by different types of lines. Details are included in the inset.
Furthermore, maintenance of epithelial stem cells in the developing incisor cervical loops is also regulated by the interaction between epithelial Fgf9 and mesenchymal FGF signaling. Loss of Fgf9 results in a lack of Fgf3 and Fgf10 expression in the dental mesenchyme (53). For this reason, it appears that Fgf9 may keep progenitor cells from differentiating in the cervical loop by protecting them from being exposed to Shh signaling. The function of FGF signaling in regulating mesenchymal stem cells and pulp cells requires future studies.
Hedgehog signaling
In the rodent incisor, Gli1+ cells are found in peri-NVB and epithelium of the cervical loop (33). Both of these two populations contain stem cells that can contribute to the dental mesenchyme and dental epithelium. HH signaling is among the most important signaling pathways that regulate embryonic development (54). Ihh and Shh are the two most studied HH ligands in the context of tooth development. Loss of Ihh in cranial neural crest cells can lead to reverse incisor occlusion, suggesting the vital role of Ihh in regulating incisor development (55). Shh is expressed in the dental epithelium and in the inferior alveolar nerve (IAN) innervating the tooth. Numerous studies have shown that Shh is involved in tooth morphogenesis (56, 57). Importantly, Shh is the major HH ligand that can activate the Gli1+ stem cell population in the mouse incisor. Previous studies have shown that Gli1+ cells are the HH-responsive epithelial stem cells that give rise to the epithelial lineages in the mouse incisor (33). They showed that HH signaling is not required for the survival of Gli1+ stem cells but is essential for their differentiation. Moreover, Li et al. showed that the fate of dental epithelial stem cells is controlled by a BMP-HH signaling network, which partially determines the postnatal growth potential of molars and incisors (58). As mentioned earlier in this review, Shh from the IAN can also activate the Gli1+ MSCs in the incisor, highlighting the importance of HH signaling in the regulation of stem cells in the mouse incisor (19, 59). Interestingly, loss of Ptch2, which is a receptor for HH signaling, can lead to increased Gli1+ MSCs and vascularization in the mouse incisor, adding more mechanistic insight into how HH signaling regulates stem cells in the incisor (60).
BMP and TGFβ signaling
The transforming growth factor β (TGFβ) signaling family plays diverse roles in embryonic development and adult tissue homeostasis by regulating various cellular behaviors (61). The TGFβ superfamily can be subdivided into four groups: the TGFβ subfamily (TGFβ1, TGFβ2, and TGFβ3), the activin subfamily (inhibin/activin-βA, inhibin/activin-βB, and inhibin/activin-βC), the BMP family (Bmp2, Bmp4, Bmp5, Bmp6, Bmp7, Bmp8, etc.), and divergent genes (Müllerian inhibiting substance, GDF-9, inhibin-α, GDNF, screw and lefty) (62–65). TGFβ signaling also has an important role in the development of the mouse incisor. For instance, TGFβ1, which is first observed in the dental epithelium and then extends to dental mesenchyme, is important for epithelial-mesenchymal interaction (66). Tgfβ2 is reported to regulate tooth size and stage without affecting cartilage, while Tgfβ3 regulates Meckel's cartilage size without affecting tooth size and shape (67). In addition, mice lacking activin-βA are missing lower incisors (68). Meanwhile, the loss of TGF-β type I receptor (Alk5) in the dental mesenchyme leads to impaired proliferation of TA cells and the maintenance of dental epithelial stem cells (69). Guan et al. found that the mouse incisor displays “wavy” mineralized tissue caused by the premature differentiation of epithelial stem cells after the deletion of Tgfbr2 in the dental mesenchyme, highlighting the role of TGFβ signaling in mediating the cell-cell interaction between the mesenchyme and epithelium of the incisor (70).
Bone morphogenetic proteins (BMPs) are one of the largest subgroups of the TGFβ family and play an essential role in many aspects of tissue homeostasis (71). BMP signaling is indispensable for tooth development. For example, loss of Bmpr1a signaling in the mouse molar can lead to compromised odontogenic differentiation (72–74). More interestingly, the inhibition of BMP signaling early in mandible development results in a transformation of tooth identity from incisor to molar (75). Shi et al. demonstrated that antagonistic interaction between BMP signaling and WNT and FGF signaling serves as a key regulator of MSC lineage commitment in the mouse incisor. Their study showed that maintenance of quiescent MSCs requires BMP signaling in the Gli1+ cell lineage, suggesting that BMP signaling has a dual role in incisor tissue homeostasis: it regulates odontoblast differentiation as well as provides feedback to the MSC population (76).
WNT signaling
The WNT family comprises 19 WNT ligands that play essential roles during both embryonic development and tissue homeostasis by regulating stem cell behaviors including self-renewal, cell proliferation and differentiation. Members of the WNT family bind to transmembrane frizzled (FZD) receptors and various co-receptors to activate canonical (β-catenin dependent) and noncanonical (β-catenin-independent) signaling pathways (77–79). WNT signaling plays important functions in regulating various aspects of tooth development (80). Disrupting canonical WNT signaling in the incisor mesenchyme results in more apoptotic cells in the LaCL through inhibiting Fgf10 (81). Moreover, Wnt/β-catenin signaling in the dental mesenchyme can determine the number of mouse incisors (82), suggesting the importance of canonical WNT signaling at the early stage of incisor development. Noncanonical WNT signals are less well studied than canonical signals. Wnt5a is one of the typical noncanonical WNT ligands and can bind to its receptor Ror2, which belongs to the family of tyrosine-protein kinase transmembrane receptors (83, 84). Interestingly, deletion of Ror2 in the root progenitor cells of the mouse molar leads to a reduction in root length, suggesting noncanonical WNT signaling plays a vital role in tooth development (85). Wnt5a-Ror2 signaling has been reported to be involved with cell-cell interaction in the musculoskeletal system (84, 86). Our study has shown that loss of Wnt5a in Axin2+ TACs of the mouse incisor leads to diminished MSCs, suggesting Wnt5a secreted by TACs provides feedback to MSCs to regulate their maintenance. Indeed, deletion of Ror2 in MSCs of the mouse incisor recapitulates this phenotype, implying the Wnt5a-Ror2 mediated cell-cell interaction between TACs and MSCs in the incisor is important for its homeostasis (29).
Hippo signaling
The Hippo signaling pathway also regulates diverse developmental processes, although it was first appreciated for its critical role in organ size control in Drosophila (87). The Hippo pathway plays a crucial role in regulating an array of different types of stem cells during embryonic development, tissue homeostasis, repair and regeneration (88). Yap (Yes-associated protein) and Taz (transcriptional co-activator with a PDZ-binding domain) are important functional mediators for the Hippo signaling pathway. Their intracellular location and interaction with other signaling pathway molecules control cell proliferation, cell lineage determination, apoptosis, and tissue homeostasis. Yap/Taz are also sensitive to mechanical stress, which is an important part of the Hippo signaling function in regulating organogenesis and tissue homeostasis (89). In the craniofacial complex, Yap/Taz are involved in regulating different stem cells, such as DPSC (dental pulp stem cells), PDLSC (periodontal ligament stem cells) and others (90). Hu and colleagues recently found that Yap/Taz are expressed in the TACs of the incisor epithelium and loss of Yap/Taz could lead to loss of TACs in the incisor, suggesting Yap/Taz are important for the maintenance of TACs (91). Specifically, Yap/Taz act through FAK/CDC42/PP1A1 to regulate mTOR signaling to promote TACs proliferation and prevent their differentiation. Through this mechanism, Yap/Taz signaling coordinates stem cell expansion and differentiation to maintain epithelial cell homeostasis in adult mouse incisor. Further studies will explore how Yap/Taz may be involved in mediating the mechanical stimulation in adult mouse incisor.
Notch signaling
Canonical and non-canonical Notch signaling pathways are evolutionarily conserved and provide a mechanism that can regulate cell fate through ligand-receptor interaction (92). Previous studies of tooth development found that expression of canonical Notch receptors (Notch1, Notch2, Notch3, and Notch4) and their ligands (Jag1, Jag2, Dll1, Dll3, and Dll4) is spatiotemporally regulated during tooth formation, and is indispensable for regulating the reciprocal interactions between dental epithelium and CNC-derived mesenchymal cells. During early development of the mouse incisor (E11-E13), expression of Notch1 is located in the dental epithelium, incisor furrow and condensed dental mesenchyme. Meanwhile, expression of Notch1 is absence in the cells of epithelium adjacent to the incisor dental mesenchyme. On the other hand, expression of Notch2 is found in the labial portion of the dental epithelium, the lingual side of the dental furrow, and the condensed dental mesenchyme (93). During the bell stage (E18.5), expression of Notch1, Notch2, and Notch3 is asymmetric in the enamel (94). Ligands of Notch signaling also have unique expression patterns in the mouse incisor. Jag2 expression is limited to the inner enamel epithelial cells from E17-E18.5 (95). At E18.5, expression of Dll1 can be noticed in both the posterior outer dental mesenchyme and the lingual side of the inner dental epithelium. On the labial side, expression of Dll1 can be detected in the both ameloblasts and odontoblasts, adjacent to cells expressing Notch1, Notch2 and Notch3 (94). These expression patterns of Notch ligands and receptors indicate that Notch signaling may have a very important role in regulating tooth development. More importantly, functional studies revealed that loss of genes involved in Notch signaling results in severe defects in the mouse incisor. Lfng, a Notch signaling regulator, is expressed in the dental epithelium and defines the lingual comportment of the developing mouse incisor, while the labial comportment is defined by epithelial Notch2 (93, 96). Epithelial stem cells of the developing incisor cannot maintain their viability without Notch signaling (97). Furthermore, inhibition of Notch signaling in adult mice results in impaired interaction between ameloblasts and the underlying stratum intermedium, which causes an enamel formation defect (98). Compared to its role in the dental epithelium, the function of Notch signaling in the dental mesenchyme is understudied. However, Walker et al. showed that loss of Notch signaling in Collagen1α2-Cre;RBP-Jkappafl/fl mice results in premature differentiation of mesenchymal transit amplifying cells, highlighting the functional significance of Notch signaling in regulating dental mesenchyme (37).
Conclusion and future perspective
Although stem cell studies using the rodent incisor as a model have made substantial progress, fundamental questions regarding how stem cells are regulated within the niche environment under homeostasis and injury repair remain to be elucidated. As the niche components of stem cells evolve, the studies highlighting the function of different niche cells regulating stem cell behavior receive more attention. For example, the sensory nerve serves as an important niche player that can regulate bone homeostasis and repair through secreting neural transmitters (99, 100). Moreover, various cell types comprising blood vessels play important functions in regulating stem cells (101). Recently, the lymphatic system, including the endothelial cells in lymphatic vessels, has been found important in regulating stem cells in the intestine, shedding light on new niche cells implicated in stem cell regulation (102, 103). Therefore, functional analysis of the niche environment from these perspectives will expand our understanding of stem cell regulation.
The stem cell regions within the rodent incisor are well established, enhancing the utility of the rodent incisor as a model to study the stem cell niche environment. Tissue clearing is a powerful technique that can make the target tissue transparent while retaining fluorescent signals, making the visualization of the niche environment of stem cells possible (104). Thus, combining tissue clearing with 3D reconstruction will enable us to perform more detailed analysis of stem cell regulation. Single-cell analysis has greatly advanced our understanding of the heterogeneity and hierarchy among different stem cell populations; therefore, spatiotemporal single-cell multiomic analysis of the rodent incisor will empower the mechanistic study of stem cell regulation in various respects (23, 105, 106). Together, these newly adopted strategies will provide us with a more comprehensive understanding of how stem cells are regulated in homeostasis and injury repair.
Transit amplifying cells are important intermediates between stem cells and their progeny (18). Although several studies using the mouse incisor have investigated the contribution of TACs to homeostasis, their function as a niche component is yet to be determined (28, 29, 37, 91). Interestingly, Amnon et al. found that a group of cycling epithelial progenitors can give rise to ameloblasts and adjacent layers of non-ameloblast cells, highlighting the importance of actively proliferating cells contributing to homeostasis and injury repair (26). This finding calls into question the normal model of quiescent stem cells governing the tissue homeostasis in the mouse incisor. Therefore, the heterogeneity and function of stem cells in the mouse incisor will require more investigation.
Author contributions
J.J. and Y.C. co-wrote the paper. M.Z., T.G. and F.P. provided critical comments for this manuscript. Y.Y. participated editing for the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by funding from the National Institute of Dental and Craniofacial Research, National Institutes of Health (R01 DE025221 and R01 DE012711 to Yang Chai).
Acknowledgments
We thank Dr. Bridget Samuels for critical editing of the manuscript. This study was supported by funding from the National Institute of Dental and Craniofacial Research, National Institutes of Health (R01 DE025221 and R01 DE012711 to Yang Chai).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Krieger T, Simons BD. Dynamic stem cell heterogeneity. Development (2015) 142:1396–406. doi: 10.1242/dev.101063
2. Nie Q, Plikus MV. Equal opportunities in stemness. Nat Cell Biol (2019) 21:921–3. doi: 10.1038/s41556-019-0366-6
3. Sagaradze GD, Basalova N. A, Efimenko AY, Tkachuk VA. Mesenchymal stromal cells as critical contributors to tissue regeneration. Front Cell Dev Biol (2020) 8:576176. doi: 10.3389/fcell.2020.576176
4. Meacham CE, DeVilbiss AW, Morrison SJ. Metabolic regulation of somatic stem cells in vivo. Nat Rev Mol Cell Bio (2022) 23:428–43. doi: 10.1038/s41580-022-00462-1
5. Sousa-Victor P, Garcia-Prat L, Munoz-Canoves P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat Rev Mol Cell Bio (2022) 23:204–26,. doi: 10.1038/s41580-021-00421-2
6. Brunet A, Goodell MA, Rando TA. Ageing and rejuvenation of tissue stem cells and their niches. Nat Rev Mol Cell Bio, (2022) doi: 10.1038/s41580-022-00510-w
7. Fuchs E, Blau HM. Tissue stem cells: architects of their niches. Cell Stem Cell (2020) 27:532–56. doi: 10.1016/j.stem.2020.09.011
8. Ge YJ, Fuchs E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet (2018) 19:311–25. doi: 10.1038/nrg.2018.9
9. Plikus MV, Wang X, Sinha S, Forte E, Thompson SM, Herzog EL, et al. Fibroblasts: origins, definitions, and functions in health and disease. Cell (2021) 184:3852–72. doi: 10.1016/j.cell.2021.06.024
10. Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol (2019) 20:303–20. doi: 10.1038/s41580-019-0103-9
11. Kann AP, Hung M, Krauss RS. Cell-cell contact and signaling in the muscle stem cell niche. Curr Opin Cell Biol (2021) 73:78–83. doi: 10.1016/j.ceb.2021.06.003
12. Mannino G, Russo C, Maugeri G, Musumeci G, Vicario N, Tibullo D, et al. Adult stem cell niches for tissue homeostasis. J Cell Physiol (2022) 237:239–57. doi: 10.1002/jcp.30562
13. Bianconi V, Mozzetta C. Epigenetic control of muscle stem cells: time for a new dimension. Trends Genet (2022) 38:501–13. doi: 10.1016/j.tig.2022.01.001
14. Cakouros D, Gronthos S. The changing epigenetic landscape of mesenchymal stem/stromal cells during aging. Bone (2020) 137:115440. doi: 10.1016/j.bone.2020.115440
15. Nagata M, Ono N, Ono W. Unveiling diversity of stem cells in dental pulp and apical papilla using mouse genetic models: a literature review. Cell Tissue Res (2021) 383:603–16. doi: 10.1007/s00441-020-03271-0
16. Yu T, Volponi AA, Babb R, An Z, Sharpe PT. Stem cells in tooth development, growth, repair, and regeneration. Curr Top Dev Biol (2015) 115:187–212. doi: 10.1016/bs.ctdb.2015.07.010
17. Sharpe PT. Dental mesenchymal stem cells. Development (2016) 143:2273–80. doi: 10.1242/dev.134189
18. Zhang B, Hsu YC. Emerging roles of transit-amplifying cells in tissue regeneration and cancer. Wiley Interdiscip Rev Dev Biol (2017) 6:5. doi: 10.1002/wdev.282
19. Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, et al. Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell (2014) 14:160–73. doi: 10.1016/j.stem.2013.12.013
20. Shi Y, He G, Lee WC, McKenzie JA, Silva MJ, Long F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat Commun (2017) 8:2043. doi: 10.1038/s41467-017-02171-2
21. Jing D, Li C, Yao K, Xie X, Wang P, Zhao H, et al. The vital role of Gli1(+) mesenchymal stem cells in tissue development and homeostasis. J Cell Physiol (2021) 236:6077–89. doi: 10.1002/jcp.30310
22. An Z, Sabalic M, Bloomquist RF, Fowler TE, Streelman T, Sharpe PT. A quiescent cell population replenishes mesenchymal stem cells to drive accelerated growth in mouse incisors. Nat Commun (2018) 9:378. doi: 10.1038/s41467-017-02785-6
23. Krivanek J, Soldatov RA, Kastriti ME, Chontorotzea T, Herdina AN, Petersen J, et al. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat Commun (2020) 11:4816. doi: 10.1038/s41467-020-18512-7
24. Yu T, Klein OD. Molecular and cellular mechanisms of tooth development, homeostasis and repair. Development (2020) 147:2. doi: 10.1242/dev.184754
25. Biehs B, Hu JK, Strauli NB, Sangiorgi E, Jung H, Heber RP, et al. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat Cell Biol (2013) 15:846–52. doi: 10.1038/ncb2766
26. Sharir A, Marangoni P, Zilionis R, Wan M, Wald T, Hu JK, et al. A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nat Cell Biol (2019) 21:1102–12. doi: 10.1038/s41556-019-0378-2
27. Chen S, Jing J, Yuan Y, Feng J, Han X, Wen Q, et al. Runx2+ niche cells maintain incisor mesenchymal tissue homeostasis through IGF signaling. Cell Rep (2020) 32:108007. doi: 10.1016/j.celrep.2020.108007
28. Du J, Jing J, Chen S, Yuan Y, Feng J, Ho TV, et al. Arid1a regulates cell cycle exit of transit-amplifying cells by inhibiting the Aurka-Cdk1 axis in mouse incisor. Development (2021) 148:8. doi: 10.1242/dev.198838
29. Jing J, Feng J, Li J, Zhao H, Ho TV, He J, et al. Reciprocal interaction between mesenchymal stem cells and transit amplifying cells regulates tissue homeostasis. Elife (2021) 10, doi: 10.7554/eLife.59459
30. Feng J, Mantesso A, De Bari C, Nishiyama A, Sharpe PT. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc Natl Acad Sci U S A (2011) 108:6503–8. doi: 10.1073/pnas.1015449108
31. Kaukua N, Shahidi MK, Konstantinidou C, Dyachuk V, Kaucka M, Furlan A, et al. Glial origin of mesenchymal stem cells in a tooth model system. Nature (2014) 513:551–4,. doi: 10.1038/nature13536
32. Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell (2012) 23:317–28. doi: 10.1016/j.devcel.2012.05.012
33. Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development (2010) 137:3753–61. doi: 10.1242/dev.056358
34. Seidel K, Marangoni P, Tang C, Houshmand B, Du W, Maas RL, et al. Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. Elife (2017) 6, doi: 10.7554/eLife.24712
35. Meyers CA, Lee S, Sono T, Xu J, Negri S, Tian Y, et al. A neurotrophic mechanism directs sensory nerve transit in cranial bone. Cell Rep (2020) 31:107696. doi: 10.1016/j.celrep.2020.107696
36. Tower RJ, Li Z, Cheng YH, Wang XW, Rajbhandari L, Zhang Q, et al. Spatial transcriptomics reveals a role for sensory nerves in preserving cranial suture patency through modulation of BMP/TGF-beta signaling. Proc Natl Acad Sci U S A (2021) 118:42. doi: 10.1073/pnas.2103087118
37. Walker JV, Zhuang H, Singer D, Illsley CS, Kok WL, Sivaraj KK, et al. Transit amplifying cells coordinate mouse incisor mesenchymal stem cell activation. Nat Commun (2019) 10:3596. doi: 10.1038/s41467-019-11611-0
38. Blackledge NP, Klose R. J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol (2021) 22:815–33. doi: 10.1038/s41580-021-00398-y
39. Barbour H, Daou S, Hendzel M, Affar EB. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat Commun (2020) 11:5947. doi: 10.1038/s41467-020-19722-9
40. Lapthanasupkul P, Feng J, Mantesso A, Takada-Horisawa Y, Vidal M, Koseki H, et al. Ring1a/b polycomb proteins regulate the mesenchymal stem cell niche in continuously growing incisors. Dev Biol (2012) 367:140–53. doi: 10.1016/j.ydbio.2012.04.029
41. An Z, Akily B, Sabalic M, Zong G, Chai Y, Sharpe PT. Regulation of mesenchymal stem to transit-amplifying cell transition in the continuously growing mouse incisor. Cell Rep (2018) 23:3102–11. doi: 10.1016/j.celrep.2018.05.001
42. Yang D, Liu HQ, Yang Z, Fan D, Tang QZ. BMI1 In the heart: novel functions beyond tumorigenesis. EBioMedicine (2021) 63:103193. doi: 10.1016/j.ebiom.2020.103193
43. Minoux M, Holwerda S, Vitobello A, Kitazawa T, Kohler H, Stadler MB, et al. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science (2017) 355:6332. doi: 10.1126/science.aal2913
44. Jing J, Feng J, Li J, Han X, He J, Ho TV, et al. Antagonistic interaction between Ezh2 and Arid1a coordinates root patterning and development via Cdkn2a in mouse molars. Elife (2019) 8, doi: 10.7554/eLife.46426
45. Yu F, Li F, Zheng L, Ye L. Epigenetic controls of Sonic hedgehog guarantee fidelity of epithelial adult stem cells trajectory in regeneration. Sci Adv (2022) 8:eabn4977. doi: 10.1126/sciadv.abn4977
46. Ho PJ, Lloyd S. M, Bao X. Unwinding chromatin at the right places: how BAF is targeted to specific genomic locations during development. Development (2019) 146:19. doi: 10.1242/dev.178780
47. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet (2004) 5:522–31. doi: 10.1038/nrg1379
48. Foshay KM, Gallicano GI. Small RNAs, big potential: the role of MicroRNAs in stem cell function. Curr Stem Cell Res Ther (2007) 2:264–71. doi: 10.2174/157488807782793781
49. Sweat M, Sweat Y, Yu W, Su D, Leonard RJ, Eliason SL, et al. The miR-200 family is required for ectodermal organ development through the regulation of the epithelial stem cell niche. Stem Cells (2021) 39:761–75. doi: 10.1002/stem.3342
50. Cao H, Jheon A, Li X, Sun Z, Wang J, Florez S, et al. The Pitx2:miR-20°c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development (2013) 140:3348–59. doi: 10.1242/dev.089193
51. Xie Y, Su N, Yang J, Tan Q, Huang S, Jin M, et al. FGF/FGFR signaling in health and disease. Signal Transduct Target Ther (2020) 5:181. doi: 10.1038/s41392-020-00222-7
52. Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, et al. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol (2007) 5:e159. doi: 10.1371/journal.pbio.0050159
53. Kurosaka H, Islam MN, Kuremoto K, Hayano S, Nakamura M, Kawanabe N, et al. Core binding factor beta functions in the maintenance of stem cells and orchestrates continuous proliferation and differentiation in mouse incisors. Stem Cells (2011) 29:1792–803. doi: 10.1002/stem.722
54. Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, Christensen ST. Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol (2019) 15:199–219. doi: 10.1038/s41581-019-0116-9
55. Amano K, Okuzaki D, Aikawa T, Kogo M. Indian Hedgehog in craniofacial neural crest cells links to skeletal malocclusion by regulating associated cartilage formation and gene expression. FASEB J (2020) 34:6791–807. doi: 10.1096/fj.201903269R
56. Seppala M, Fraser GJ, Birjandi AA, Xavier GM, Cobourne MT. Sonic hedgehog signaling and development of the dentition. J Dev Biol (2017) 5:2. doi: 10.3390/jdb5020006
57. Hosoya A, Shalehin N, Takebe H, Shimo T, Irie K. Sonic hedgehog signaling and tooth development. Int J Mol Sci (2020) 21:5. doi: 10.3390/ijms21051587
58. Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, et al. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev Cell (2015) 33:125–35. doi: 10.1016/j.devcel.2015.02.021
59. Ishikawa Y, Nakatomi M, Ida-Yonemochi H, Ohshima H. Quiescent adult stem cells in murine teeth are regulated by shh signaling. Cell Tissue Res (2017) 369:497–512. doi: 10.1007/s00441-017-2632-x
60. Juuri E, Tikka P, Domanskyi A, Corfe I, Morita W, McKinnon PJ, et al. Ptch2 is a potential regulator of mesenchymal stem cells. Front Physiol (2022) 13:877565. doi: 10.3389/fphys.2022.877565
61. Jia S, Meng A. TGFbeta family signaling and development. Development (2021) 148:5. doi: 10.1242/dev.188490
62. Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev (1994) 8:133–46. doi: 10.1101/gad.8.2.133
63. Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, et al. Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature (1996) 381:151–5. doi: 10.1038/381151a0
64. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature (1997) 387:83–90. doi: 10.1038/387083a0
65. Nakashima M, Toyono T, Murakami T, Akamine A. Transforming growth factor-beta superfamily members expressed in rat incisor pulp. Arch Oral Biol (1998) 43:745–51. doi: 10.1016/s0003-9969(98)00046-6
66. Vaahtokari A, Vainio S, Thesleff IR. Associations between transforming growth factor-beta-1 Rna expression and epithelial mesenchymal interactions during tooth morphogenesis. Development (1991) 113:985–94. doi: 10.1242/dev.113.3.985
67. Chai Y, Mah A, Crohin C, Groff S, Bringas P Jr, Le T, et al. Specific transforming growth factor-beta subtypes regulate embryonic mouse Meckel's Cartilage and tooth development. Dev Biol (1994) 162:85–103. doi: 10.1006/dbio.1994.1069
68. Matzuk MM, Kumar TR, Vassalli A, Bickenbach JR, Roop DR, Jaenisch R, et al. Functional analysis of activins during mammalian development. Nature (1995) 374:354–6. doi: 10.1038/374354a0
69. Zhao H, Li S, Han D, Kaartinen V, Chai Y. Alk5-mediated transforming growth factor beta signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol Cell Biol (2011) 31:2079–89. doi: 10.1128/MCB.01439-10
70. Yang G, Zhou J, Teng Y, Xie J, Lin J, Guo X, et al. Mesenchymal TGF-beta signaling orchestrates dental epithelial stem cell homeostasis through wnt signaling. Stem Cells (2014) 32:2939–48. doi: 10.1002/stem.1772
71. Zinski J, Tajer B, Mullins MC. TGF-beta family signaling in early vertebrate development. Cold Spring Harb Perspect Biol (2018) 10:6. doi: 10.1101/cshperspect.a033274
72. Feng J, Jing J, Li J, Zhao H, Punj V, Zhang T, et al. BMP Signaling orchestrates a transcriptional network to control the fate of mesenchymal stem cells in mice. Development (2017) 144:2560–9. doi: 10.1242/dev.150136
73. Omi M, Kulkarni AK, Raichur A, Fox M, Uptergrove A, Zhang H, et al. BMP-Smad Signaling regulates postnatal crown dentinogenesis in mouse molar. JBMR Plus (2020) 4:e10249. doi: 10.1002/jbm4.10249
74. Liu M, Goldman G, MacDougall M, Chen S. BMP signaling pathway in dentin development and diseases. Cells (2022) 11, doi: 10.3390/cells11142216
75. Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science (1998) 282:1136–8. doi: 10.1126/science.282.5391.1136
76. Shi C, Yuan Y, Guo Y, Jing J, Ho TV, Han X, et al. BMP signaling in regulating mesenchymal stem cells in incisor homeostasis. J Dent Res (2019) 98:904–11. doi: 10.1177/0022034519850812
77. Murillo-Garzon V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol (2017) 14:683–96. doi: 10.1038/nrurol.2017.144
78. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell (2006) 127:469–80. doi: 10.1016/j.cell.2006.10.018
79. Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol (2012) 13:767–79. doi: 10.1038/nrm3470
80. Tokavanich N, Wein MN, English JD, Ono N, Ono W. The role of wnt signaling in postnatal tooth root development. Front Dent Med (2021) 2, doi: 10.3389/fdmed.2021.769134
81. Yang Z, Balic A, Michon F, Juuri E, Thesleff I. Mesenchymal Wnt/beta-catenin signaling controls epithelial stem cell homeostasis in teeth by inhibiting the antiapoptotic effect of Fgf10. Stem Cells (2015) 33:1670–81. doi: 10.1002/stem.1972
82. Fujimori S, Novak H, Weissenbock M, Jussila M, Goncalves A, Zeller R, et al. Wnt/beta-catenin signaling in the dental mesenchyme regulates incisor development by regulating Bmp4. Dev Biol (2010) 348:97–106. doi: 10.1016/j.ydbio.2010.09.009
83. Endo M, Kamizaki K, Minami Y. The ror-family receptors in development, tissue regeneration and age-related disease. Front Cell Dev Biol (2022) 10:891763. doi: 10.3389/fcell.2022.891763
84. Kamizaki K, Endo M, Minami Y, Kobayashi Y. Role of noncanonical Wnt ligands and Ror-family receptor tyrosine kinases in the development, regeneration, and diseases of the musculoskeletal system. Dev Dyn (2021) 250:27–38. doi: 10.1002/dvdy.151
85. Ma Y, Jing J, Feng J, Yuan Y, Wen Q, Han X, et al. Ror2-mediated non-canonical Wnt signaling regulates Cdc42 and cell proliferation during tooth root development. Development (2021) 148, doi: 10.1242/dev.196360
86. Maeda K, Kobayashi Y, Udagawa N, Uehara S, Ishihara A, Mizoguchi T, et al. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat Med (2012) 18:405–12. doi: 10.1038/nm.2653
87. Zheng Y, Pan D. The hippo signaling pathway in development and disease. Dev Cell (2019) 50:264–82. doi: 10.1016/j.devcel.2019.06.003
88. Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development (2014) 141:1614–26,. doi: 10.1242/dev.102376
89. Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev (2016) 30:1–17. doi: 10.1101/gad.274027.115
90. Wang J, Martin JF. Hippo pathway: an emerging regulator of craniofacial and dental development. J Dent Res (2017) 96:1229–37. doi: 10.1177/0022034517719886
91. Hu JK, Du W, Shelton SJ, Oldham MC, DiPersio CM, Klein OD. An FAK-YAP-mTOR signaling axis regulates stem cell-based tissue renewal in mice. Cell Stem Cell (2017) 21:91–106 e106, doi: 10.1016/j.stem.2017.03.023
92. Cai X, Gong P, Huang Y, Lin Y. Notch signalling pathway in tooth development and adult dental cells. Cell Prolif (2011) 44:495–507. doi: 10.1111/j.1365-2184.2011.00780.x
93. Mucchielli ML, Mitsiadis TA. Correlation of asymmetric Notch2 expression and mouse incisor rotation. Mech Dev (2000) 91:379–82. doi: 10.1016/s0925-4773(99)00293-2
94. Mitsiadis TA, Hirsinger E, Lendahl U, Goridis C. Delta-notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev Biol (1998) 204:420–31. doi: 10.1006/dbio.1998.9092
95. Mitsiadis TA, Regaudiat L, Gridley T. Role of the Notch signalling pathway in tooth morphogenesis. Arch Oral Biol (2005) 50:137–40. doi: 10.1016/j.archoralbio.2004.10.006
96. Pouyet L, Mitsiadis TA. Dynamic lunatic fringe expression is correlated with boundaries formation in developing mouse teeth. Mech Develop (2000) 91:399–402. doi: 10.1016/S0925-4773(99)00299-3
97. Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation (2010) 80:241–8. doi: 10.1016/j.diff.2010.06.004
98. Jheon AH, Prochazkova M, Meng B, Wen T, Lim YJ, Naveau A, et al. Inhibition of notch signaling during mouse incisor renewal leads to enamel defects. J Bone Miner Res (2016) 31:152–62. doi: 10.1002/jbmr.2591
99. Hu B, Lv X, Chen H, Xue P, Gao B, Wang X, et al. Sensory nerves regulate mesenchymal stromal cell lineage commitment by tuning sympathetic tones. J Clin Invest (2020) 130:3483–98. doi: 10.1172/JCI131554
100. Chen H, Hu B, Lv X, Zhu S, Zhen G, Wan M, et al. Prostaglandin E2 mediates sensory nerve regulation of bone homeostasis. Nat Commun (2019) 10:181. doi: 10.1038/s41467-018-08097-7
101. Matsushita Y, Nagata M, Kozloff KM, Welch JD, Mizuhashi K, Tokavanich N, et al. A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat Commun (2020) 11:332. doi: 10.1038/s41467-019-14029-w
102. Palikuqi B, Rispal J, Reyes EA, Vaka D, Boffelli D, Klein O. Lymphangiocrine signals are required for proper intestinal repair after cytotoxic injury. Cell Stem Cell (2022) 29:1262–72 e1265, doi: 10.1016/j.stem.2022.07.007
103. Goto N, Goto S, Imada S, Hosseini S, Deshpande V, Yilmaz OH. Lymphatics and fibroblasts support intestinal stem cells in homeostasis and injury. Cell Stem Cell (2022) 29:1246–61 e1246, doi: 10.1016/j.stem.2022.06.013
104. Jing D, Zhang S, Luo W, Gao X, Men Y, Ma C, et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res (2018) 28:803–18. doi: 10.1038/s41422-018-0049-z
105. Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Dominguez A, et al. The bone marrow microenvironment at single-cell resolution. Nature (2019) 569:222–8. doi: 10.1038/s41586-019-1104-8
Keywords: rodent incisor, stem cell heterogeneity, stem cell niches, signaling networks, tissue homeostasis
Citation: Jing J, Zhang M, Guo T, Pei F, Yang Y and Chai Y (2022) Rodent incisor as a model to study mesenchymal stem cells in tissue homeostasis and repair. Front. Dent. Med 3:1068494. doi: 10.3389/fdmed.2022.1068494
Received: 13 October 2022; Accepted: 15 November 2022;
Published: 30 November 2022.
Edited by:
Paul Sharpe, King's College London, United KingdomReviewed by:
Brad A Amendt, The University of Iowa, United StatesAnamaria Balic, University of Helsinki, Finland
© 2022 Jing, Zhang, Guo, Pei, Yang and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Chai, eWNoYWlAdXNjLmVkdQ==
Specialty Section: This article was submitted to Regenerative Dentistry, a section of the journal Frontiers in Dental Medicine
 Junjun Jing1,2
Junjun Jing1,2 Tingwei Guo
Tingwei Guo Yang Chai
Yang Chai