- Department of Oral Health Sciences, School of Dentistry, University of Washington, Seattle, WA, United States
Orthodontic treatment is an appliance-intensive endeavor, where an array of mechanical devices is used to bring about tooth movement. By virtue of remaining in close proximity to the enamel, gingiva and periodontal ligament intra-orally over a prolonged period of time, orthodontic appliances have a significant impact on the paradental tissues, oral environment and oral microbiome. Orthodontic appliances, by acting as anchors for biofilm and plaque formation, accumulate bacteria and other microorganisms in amounts unfavorable for maintenance of healthy equilibrium. The resulting loss of balance in the oral microbiome causes dysbiosis, which manifests clinically as increased enamel demineralization, dental caries and periodontal disease. Mechanical removal of the accumulated plaque by maintaining rigorous oral hygiene has been proven to be the single most important factor to mitigate the harmful effects of dysbiosis. This review investigates how each of the various components of orthodontic appliances, different types of appliances and unique surface properties of biomaterials have contributory effects at the interface of orthodontic biomaterials and oral biology. The information thus obtained will be critical in instituting the best diagnostic and therapeutic measures at the clinical level. It will also be instrumental in devising improvements and providing new directions for future research in general and precision orthodontics in particular.
Introduction
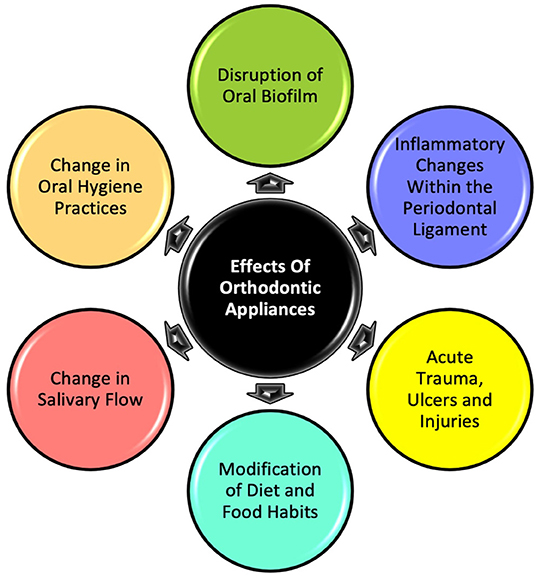
Orthodontic treatment with oral appliances—either removable or fixed—is a significant disruptor of the oral environment. Through their continual presence to exert the force needed to bring about orthodontic tooth movement (OTM), the appliances make a significant biological impact (Figure 1), with effects lasting at least until treatment completion, which could be anywhere between 1 and 3 years on average (1). The mechanical, chemical and biological upheaval that occurs at the interface between orthodontic biomaterials and the oral ecosystem is responsible for heightened risks associated with orthodontic treatment, such as enamel decalcification, dental caries and periodontal disease (2). There are enormous humanitarian and economic costs associated with this because, according to the American Association of Orthodontists, at least 3.2 million Americans undergo orthodontic treatment, on which they collectively spend around 17 billion dollars in just one year (3). Hence, investigating what happens at the interface between orthodontic biomaterials and oral biology is necessary not only from a clinical and research standpoint but also from a public health perspective.
Pathogen-mediated oral diseases occur mostly when the health-promoting regulatory mechanisms are overrun by strong or persistent environmental assaults. Otherwise, under normal circumstances, a human body—termed as a “holobiont”—is a super-organism where almost equal parts (~3 × 1013) of eukaryotic cells and prokaryotic microorganisms co-exist symbiotically in health (4, 5). The oral environment is second only to the gut in the complexity and diversity of the micro-organisms it hosts, referred to variously as oral microflora, oral microbiota and most recently—“oral microbiome” (6). This is reflected in the expanded Human Oral Microbiome Database (eHOMD), which encompasses a total of 775 microbial species and 2074 oral/nasal genomes, representing 529 taxa (6) including bacteria, viruses, fungi, protozoa and archaea. The term microbiome, which encompasses the totality of the microorganisms, their genomes and ecosystems (7), was originally coined by Nobel Laureate, Joshua Lederberg “to signify the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space and have been all but ignored as determinants of health and disease” (8). The inseparability of the human and the microbiome components in determining oral health and disease, is what necessitates investigating the changes precipitated by orthodontic appliances and biomaterials on the oral microbiome in the pursuit of minimizing or eliminating adverse effects on dental tissues during orthodontic treatment.
During orthodontic treatment, maintaining a healthy, symbiotic relationship between the host and oral microbiome is critical to minimizing adverse effects such as enamel demineralization, dental caries and periodontal disease. However, OTM is a biological process which is inherently dependent on inflammatory mediators within the periodontal ligament (9). In combination with additional factors such as increased biofilm formation, change in composition of plaque, difficulty in oral hygiene maintenance and modification of dietary habits, this can cause an imbalance in the native state of the oral microbiome, referred to as dysbiosis (Figure 2) (4). A dysbiotic microbiome is defined as “one in which the diversity and relative proportions of species or taxa within the microbiota are disturbed” (10). Dysbiosis of oral microbiome is also primarily responsible for enamel demineralization or white spot lesions and periodontal disease—significantly problematic sequelae of orthodontic treatment (2). Additionally, change in oral bacterial composition accompanying the dysbiosis has been associated with serious systemic diseases such as Alzheimer's disease, rheumatoid arthritis, meningitis, cardiovascular diseases, brain, lung, liver or splenic abscesses, appendicitis, pneumonia diabetes, and colorectal disease (4, 6, 11). The link between oral microbiome and its systemic effects was demonstrated when oral administration of Porphyromonas gingivalis in animals directly affected the gut microbiome composition and caused inflammatory changes in tissues and organs (12). Considering this intimate association of oral microbiome with several systemic health conditions, its impact on quality of life can be significant. Importantly, the oral microbiome is also associated with mental health issues that adversely affect quality of life. For instance, in adolescents with depression and anxiety an abundance of certain microbial species like Spirochaetaceae, Actinomyces, Treponema, Fusobacterium and Leptotrichia spp. (13) regulated by high levels of cortisol and C-reactive protein was found in the oral microbiome. Similarly, a difference in oral microbiota between healthy controls and adults with panic disorders was found, mediated by metabolic and inflammatory pathways (14). Thus, we can infer that, when orthodontic treatment-induced oral dysbiosis intersects with the various biological pathways specific to systemic health conditions which alter the nature, diversity and species abundance of the oral microbiome, the potential fallout on patients' quality of life can be substantial. Hence, exploring the biogenesis and mechanisms of orthodontic appliance-induced modifications of the oral environment and the oral microbiome is the main objective of this review. Through our review, we aim to provide a brief overview of the oral microbiome, outline techniques for microbiome analysis, and discuss orthodontic appliance-mediated dysbiosis, its effects, and mitigation measures. This will be pivotal in mitigating the potentially harmful local effects of orthodontic treatment-induced dysbiosis and lifesaving if the knowledge helps in resolving, minimizing or preventing systemic effects of the dysbiosis. Identifying the exact nature and mechanistic details of microbiome changes is also a critical preliminary step toward future implementation of precision dentistry in general and personalized orthodontics in particular.
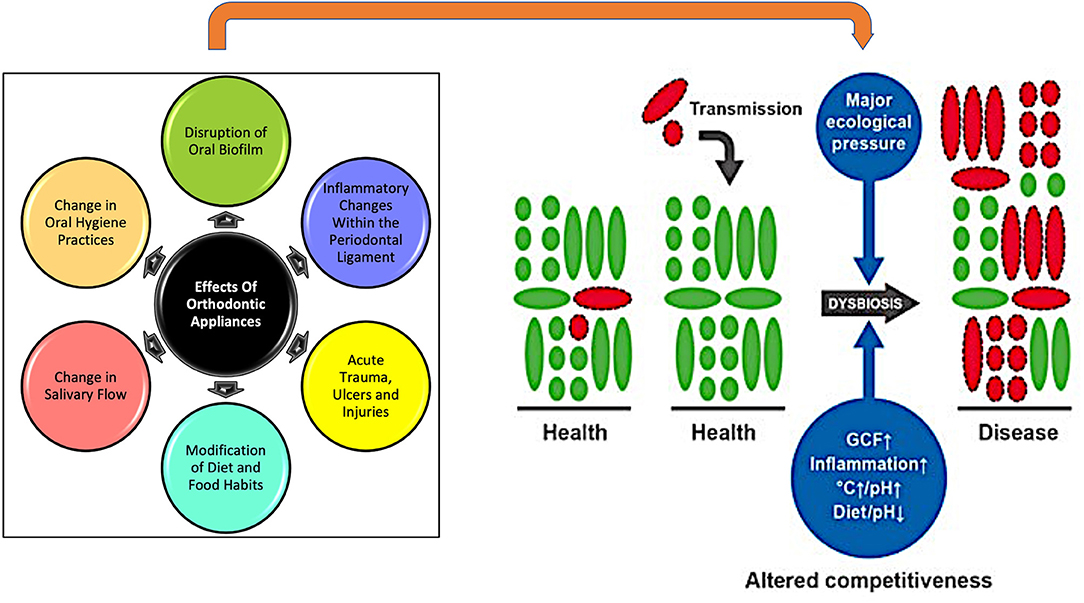
Figure 2. The intra-oral effects of having an orthodontic appliance in place act as significant disruptors. They exert major ecological pressure, causing dysbiosis and a shift in oral microbiome from predominantly healthy microbiota (green) to predominantly pathogenic bacteria (red). [Adapted from Kilian et al. (4)].
Techniques for Oral Microbiome Analysis
Conventional techniques for assessing microbial composition have involved either culturing microorganisms, microscopically observing them or detecting their functional activities using enzymatic or immunoassays (Table 1). These techniques, though reliable, were limited in scope for assessing the vast array of microorganisms constituting the oral microbiome. Especially, since many species of the abundant oral microbes are uncultivable, the culture-dependent techniques become ineffective. Molecular techniques to identify and classify uncultivable oral microbial species at a smaller scale have been in use for many years, like restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA fingerprinting (RPAD), denaturing gradient gel electrophoresis (DGGE), quantitative real-time polymerase chain reaction (qPCR), microarrays, checkerboard hybridization etc. (16). The field of culture-independent microbial sequencing was revolutionized with the introduction of high-throughput, next-generation sequencing (NGS), which made it possible to directly sequence the total DNA of microorganisms obtained from an environmental sample on a massive scale, referred to as metagenomics. Metagenomic techniques like 16S ribosomal RNA (rRNA) sequencing, pyrosequencing and shotgun sequencing have been crucial in identifying hundreds of oral microbiome species in all their diversity and abundance levels (Table 1). Using NGS techniques, the eHOMD was established which provides a comprehensive curated information on bacterial species inhabiting the human mouth and aerodigestive tract, including the pharynx, nasal passages, sinuses and esophagus (17). Of the 774 identified bacterial species, 58% are officially named, 16% are unnamed but cultivated and 26% are unnamed and known only as uncultivated phylotypes, as naming requires growth in culture and full phenotypic characterization (6) (http://www.homd.org/). In a healthy human mouth, the predominant bacterial phyla are Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes and Fusobacteria which account for 80–95% of species detected (6, 18). To assess the whole range of diverse microbial populations, including fungi and viruses, a more comprehensive technique called whole-genome shotgun sequencing (WGSS) has been used. Using WGSS, Caselli et al. (18) reported bacteriophages and viruses of the Herpesviridae family to constitute around 0.03% of the oral microbiome and fungi predominantly made up of Candida and other Saccharomicetales species, around 0.004% of total microbiome. Importantly, the microbiome composition was found to be unique for various oral microenvironments in an individual like tongue dorsum, hard palate, buccal mucosa, keratinized gingiva, and supragingival and subgingival plaque (18).
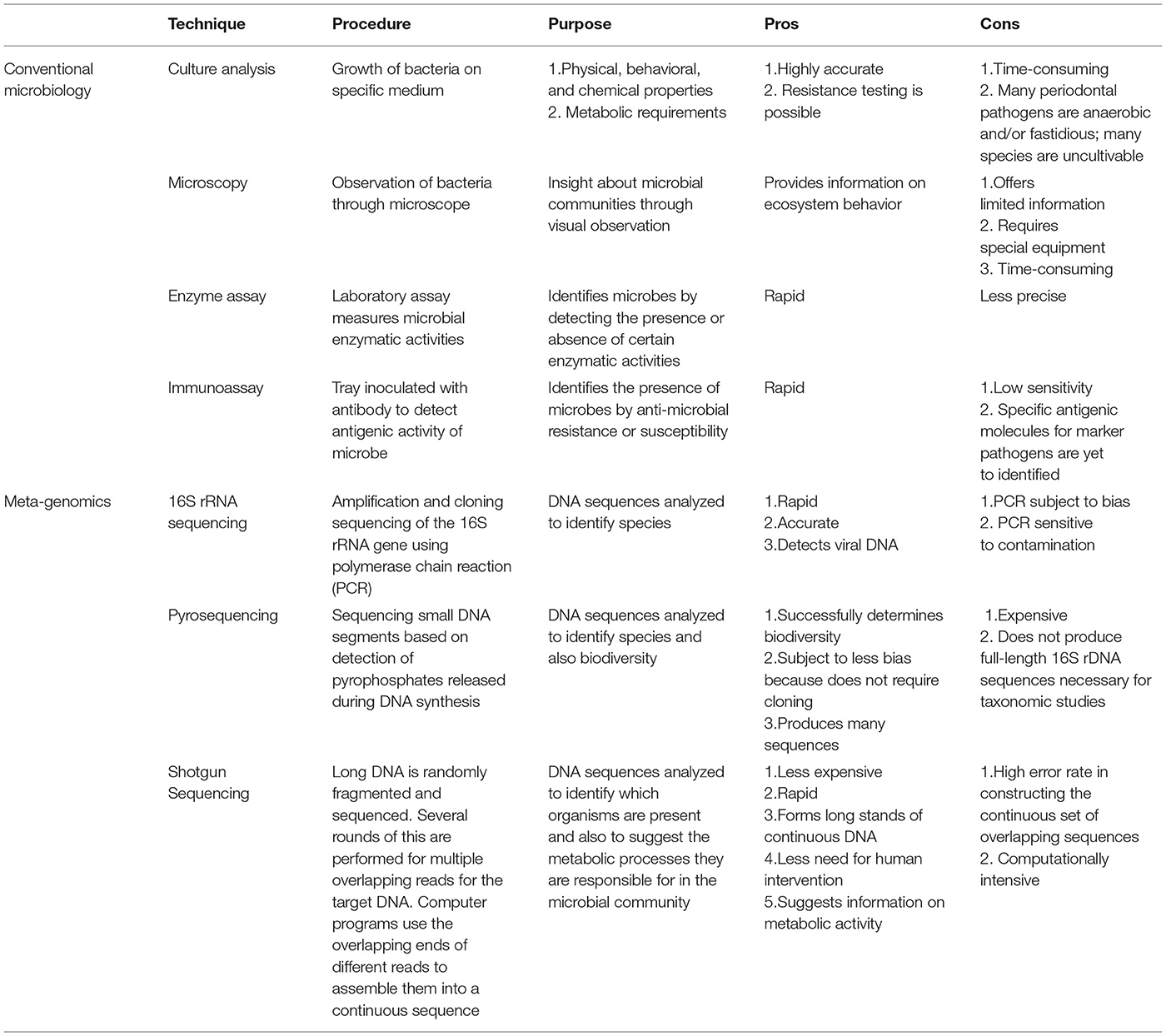
Table 1. Methodologies for microbiome analysis [Reused with permission from Zarco et al. (15)].
The limitations of metagenomic techniques like their inability to identify species due to short segments of DNA sequences, contamination with small amounts of human DNA with the microbial genome, lack of differentiation between dead and living microbes and the inability to determine activity and physiological state of microorganisms inherent in the DNA sequencing technique, necessitate development of newer methods. Additionally, the critical need to grow micro-organisms and characterize their phenotype and functional states has led to the rebirth of the culture technique in the current era through “culturomics”. Culturomics is a hybrid technique that combines high-throughput culture method with matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (19). The process consists of growing specimen aliquots under various culture conditions that encourage growth of rarer, fastidious microbes while suppressing the majority populations and then subjecting cultured colonies to a rapid and cost-effective MALDI-TOF mass spectroscopic analysis to identify microbes at a genus and species level. Although culturomics has been instrumental in vastly increasing the repertoire of known gut microbiome species, we found only one study where this technique was used on an oral microbiome. Using culturomics in four patients, Martellacci et al. (20) analyzed the peri-implant, subgingival plaque microbiome and detected 12 novel bacterial species which had previously never been identified in the oral cavity. As evidenced in the gut microbiome studies and the peri-implant pilot study, culturomics seems to be quite promising in providing greater clarity into the oral microbiome composition and functions with further research studies. Furthermore, the evolution of diagnostics and therapeutics with newer technological tools being made available has given rise to the emerging field of research that investigates therapeutic interventions by targeting the microbiome, referred to as microbiomics (15). Combining oral microbiome characterization and application of microbiomics can be critical to not only treating oral diseases but also preventing oral dysbiosis before it can cause downstream deleterious effects on dental tissues. We identify this as a major area of future research which can greatly transform oral health and mitigate the global health burden imposed by oral diseases.
Influence of Orthodontic Appliances on Oral Biofilm Formation and Bacterial Adherence
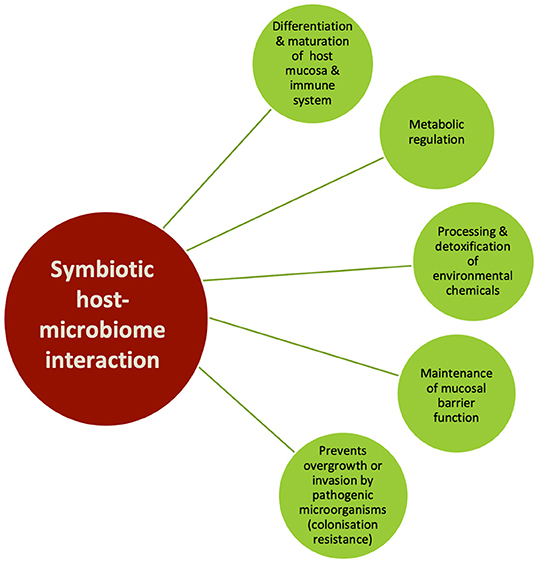
The oral environment is inherently highly dynamic by virtue of being the gateway for food and fluid ingestion for the body across its lifespan. As such, the orodental tissues have to be in a state of dynamic equilibrium to constantly readjust to the changing oral environment. A positive host–oral microbiome relationship in such an ecosystem provides critical metabolic, physiological and immunological support (Figure 3) (21). In its natural habitat, the oral microbiome is found as a multicellular aggregate embedded in an extracellular polysaccharide and protein matrix, adherent to hard and soft tissue surfaces within the oral cavity (including oral appliances and dental materials), called a biofilm. Biofilms have been defined as matrix-embedded microbial populations adherent to each other and/or to surfaces or interfaces (22). Natural biofilms have a highly diverse microflora where the component species are not randomly distributed but are spatially and functionally organized (21). The binding of microbes to each other and to host receptors via a biofilm triggers gene expression changes in both microbes and host cells. This is a critical determinant of important clinical outcomes like disease progression, host response, and anti-microbial sensitivity (21, 23). Dental plaque is a specific type of oral biofilm, in which a diverse community of microorganisms found on the tooth surface as a biofilm is embedded in an extracellular matrix of polymers of both host and microbial origin (24). The mere physical presence of orthodontic appliances, accompanied by the chemical, biological and lifestyle changes triggered by them, significantly changes the nature, quantity and areas of oral biofilm and plaque formation. This in turn shifts the balance of the oral microbiome away from health, necessitating implementation of preventative and therapeutic measures during orthodontic treatment.
Bracket Material
Orthodontic appliances lend themselves readily as convenient, new retentive surfaces for plaque and microbiota. The intricate topology of brackets superimposed with multitudes of nooks and crannies created by wires and the elastomeric or ligature ties used to secure them, together serve as definitive plaque magnets. However, the role played by physico-chemical properties of bracket materials in biofilm accumulation is not as clear. Eliades et al. (25) hypothesized that since adhesion of microorganisms to surfaces is a result of specific lectinlike reactions, secondary electrostatic interactions, and van der Waals forces, bracket material properties like interfacial surface free energy, hydrophobicity, and total work of adhesion would be critical. They found stainless steel had the highest critical surface tension and total work of adhesion, whereas polycarbonate and ceramic alumina materials had the lowest. Accordingly, stainless steel appliances would have greater plaque-retaining capacity compared to the other two materials (25), but future studies would show much different results. One study found that stainless steel surface accumulated less initial biofilm as compared to gold or ceramic materials (26). Lipopolysaccharides from gram-negative bacteria found stainless steel to be the most adherent (27), but Candida albicans and Streptococcus mutans were most adherent to composite, ceramic and metallic brackets in decreasing order (28). Au contraire, Papaioannou et al. consistently found no differences in the adherence of S. mutans to stainless steel, ceramic, or plastic brackets (29). This despite the observation that polymer materials (acrylics, composites and glass ionomers) provide excellent incubation niches for micro-organisms in the pores and defects in their surfaces as compared to smoother surfaces metals, ceramics and enamel (30). Such contradictory findings have led investigators to hypothesize that other factors such as diet, salivary flow, oral hygiene practices, and the presence of metal bands, archwires, elastomeric modules and resins may affect bacterial adhesion more than just the nature of bracket material itself.
Archwires and Their Securing Mechanisms
Orthodontic archwires were found to harbor significant amounts of microorganisms after clinical use (31). Orthodontic archwires are made of stainless steel, nickel titanium (NiTi) and cobalt-chromium. These metallic archwires may be coated with esthetic materials such as teflon, epoxy resin, and rhodium to improve their appearance. Surface roughness and surface free energy of these archwire materials are thought to be critical determinants of bacterial adhesion (32). In accordance with the thermodynamic principle that materials with higher surface free energy will attract greater bacterial load, Kim et al. found that the NiTi wires had the highest free surface energy, hence the greatest amount of bacterial adhesion, followed by stainless steel, and lastly, the aesthetic wires. Both in vivo and in vitro studies have shown that surface roughness of the archwires increases with time (31, 33). Intra-oral aging, change in mechano-chemical properties of wire due to exposure to saliva, abrasion due to toothbrushing and eating, and interactions between the archwires, brackets, and ligatures are some of the factors increasing surface roughness. Surprisingly, studies have leaned more toward showing a lack of correlation between surface roughness of orthodontic archwires and bacterial adhesion, and there appears to be a greater correlation between surface free energy and bacterial adhesion (31–33). However, the landmark review by Bollenl et al. (34) concluded that an increase in both supra- and subgingival surface roughness resulted in a higher rate of bacterial surface colonization and plaque maturation, thus increasing the risk for caries and periodontal inflammation. They recommended an Ra value, a measure of surface roughness, of 0.2 microns or lower for all intra-oral hard surfaces, as below this there was no significant reduction of bacterial load and above this value plaque accumulation increased proportionately (34).
In fixed orthodontic appliances, archwires are secured within the brackets using one of three mechanisms—ligation with steel wires, elastomeric ligatures, or self-ligating brackets (Figure 4). All three methods provide a favorable substratum for biofilm formation, but ligation with steel wires was found to retain less biofilm and bacteria compared to elastomeric ties (35, 36). However, a study by Turkkahraman et al. (37) showed no difference between these two methods. Conventional brackets with elastomeric ties were also found to have greater biofilm formation as compared to self-ligating appliances. Self-ligating brackets were found to have statistically significant lower loads of total bacteria and oral streptococci compared to the elastomerically tied conventional brackets, although results ought to be interpreted with caution due to the low sample size of 14 in this study (38). A systematic review and meta-analysis found non-significant difference in the plaque index of patients undergoing orthodontic treatment with either conventional (involving ligation with steel wires and/or elastomeric ligatures) or self-ligating brackets (39).
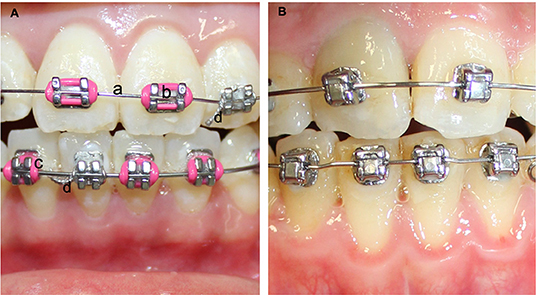
Figure 4. (A) Methods to secure archwire to conventional brackets. (a) archwire, (b) bracket, (c) elastomeric tie to secure archwire in place, (d) ligature tie to secure archwire; (B) self-ligating bracket.
Salivary and Dietary Changes Due to Orthodontic Appliances
The mere physical presence of the orthodontic appliance has shown to increase stimulated salivary flow, buffering capacity and pH at 1 and 3 months after appliance insertion, which has been implied to indicate an adaptive ability of the oral environment to adverse situations (40, 41). This is disputed by studies which found that salivary flow rate, pH and buffer capacity did not differ significantly from the baseline in patients with fixed appliances even over a 1-year period (42, 43). The different observations were explained on the basis that since introduction of a foreign body causes a physiological increase in salivation, salivary parameters are found to rise when observed immediately after appliance placement. In the long term, adaptation to the foreign body serves to bring back the levels to normal (42). A study by Arab et al. (44), however, found that even though the salivary flows did not change over an 18-week period, salivary pH did drop significantly. In the Zogakis et al. (45) study, there was a significant decrease in the pH immediately after bonding fixed orthodontic appliances, which returned to normal levels after 4–6 weeks. This was attributed to the usage of 37% phosphoric acid for enamel etching prior to bonding of the orthodontic appliances.
Salivary pH and buffering capacity are inexorably linked to diet. The dietary change of switching to softer and non-sticky foods, in addition to being the orthodontist's recommendations for appliance homecare, was also found to be the patients' preferred choice owing to pain and discomfort associated with fixed appliances (46). In a qualitative study of adolescents undergoing fixed orthodontic treatment, patients reported difficulty in biting and chewing hard foods, causing them to either eat less, change what they ate or change the way they ate. In a study of 67 adolescents undergoing treatment with fixed appliances and 70 controls with no treatment, 90% of the patients in both groups consumed chocolate daily, more than 97% of patients in both groups said they consume sweets at frequent intervals on a regular basis and more than half in both groups reported consuming sweets and various soft drinks every single day (47). Such regular intake of sweets and sugary drinks has the potential to reduce salivary pH, which, accompanied by increased plaque retention by fixed appliances, creates extremely favorable conditions for caries development, failing the institution of remedial measures. Regular reinforcement of the importance of good homecare to maintain excellent oral hygiene in patients, interspersed with professional oral prophylaxis at regular intervals, is critical in avoiding the very real pitfalls of enamel demineralization and caries formation during orthodontic treatment.
Influence of Orthodontic Appliances on Composition of the Oral Microbiome
Orthodontic appliances significantly altered the oral microbiome within one month after the start of orthodontic treatment, independent of the type of appliance used, according to a systematic review that included 52 studies with moderate-to-high quality of evidence. (48). Studies variously tried to ascertain levels of cariogenic bacteria such as Streptococcus mutans and Lactobacillus species, periodontopathogenic bacteria such as Tannerella forsythia, Treponema denticola, Fusobacterium nucleatum, Aggregatibacter actinomycetemcomitans, Porphyromans gingivalis, Prevotella intermedia, and Prevotella nigrescens, and Candida albicans. Overall, studies on conventional brackets detected high levels of both gram-positive bacteria such as Streptococcus mutans and Lactobacillus spp. and gram-negative bacteria such as A. actinomycetemcomitans. Several studies also showed an increase in Candida during treatment (48). Another systematic review and meta-analysis of four periodontopathogens in subgingival plaque revealed a pattern of temporary increase in pathogens for a short time (0 to 3 months) after orthodontic appliance placement, which returned to pretreatment levels six months later (49).
Lingual orthodontic appliances were associated with greater plaque retention, more gingival inflammation and higher S. mutans and A. actinomycetemcomitans counts as compared to labial brackets (43, 50). However, the two did not differ in Lactobacillus counts, the salivary flow rate, and saliva buffer capacity (43). Sfondrini et al. (51) also did not find any significant difference between the lingual and labial brackets in a 30-day randomized clinical trial with respect to periodontal pocket depth, bleeding on probing and number of CFUs of either streptococci or anaerobic bacteria.
Orthodontic appliances, such as clear aligners and thermoplastic retainers, have the advantage of being removable. This promotes ease of eating, without food getting stuck in the appliance as would happen in a fixed appliance, and also makes maintenance of oral hygiene much easier for patients. This is reflected in clinical studies which have shown that patients using Invisalign had much better clinical parameters for gingival and periodontal health compared to those with fixed appliances (52–55). However, a high-throughput analysis of the 16S rRNA gene of the oral microbiome showed that both Invisalign and fixed orthodontic appliances caused microbial dysbiosis, with no statistically significant difference between the two (56). However, there was a statistically significant difference in microorganism abundance between the two groups. Invisalign had significantly higher relative abundance of TM7 and Neisseria at the genus level. Literature shows that TM7 is associated with gingivitis and periodontitis, whereas Neisseria is associated with better oral health and reduced gingivitis. The authors concluded that effect of these changes with microbiome on oral health was inconsistent and that the inferred microbial function of the Invisalign group suggested this group was more predisposed to periodontal diseases. Thus, the improved clinical outcome for periodontal health in Invisalign is not because of favorable oral microbiome changes, but instead due to ease of oral hygiene maintenance. Newer, more sophisticated microbiome-detecting technologies (Table 1) (15) have the potential to shed better light on the exact nature of dysbiosis caused by orthodontic appliances. Additionally, orthodontists can also keep an eye on the effects of dysbiosis during treatment, especially on the more troublesome and frequently occurring white spot lesions. Techniques like digital subtraction radiography, digital image enhancement, fiber optic transillumination (FOTI), quantitative light-induced fluorescence (QLF), digital image fiber optic transillumination (DiFOTI), laser fluorescence measurement (DiagnoDent), electrical conductance/impedance measurement, ultrasonic caries detector etc., can help in early detection and remediation of white spot lesions secondary to appliance-induced dysbiosis (57).
Anti-Microbial Effect of Orthodontic Appliances on Oral Microbiome
The predilection of orthodontic appliances to accumulate plaque and cause microbial dysbiosis has led to exploration of anti-microbial bonding agents, brackets, ligatures and archwires to mitigate the said effects. A 2018 systematic review and meta-analysis assessed 32 studies on anti-microbial orthodontic bonding systems (58). The range of anti-microbial agents found in this review consisted of silver nanoparticles, benzalkonium chloride, chlorhexidine, triclosan, cetylpyridinium chloride, Galla chinensis extract, acid ursolic, dimethylaminododecyl methacrylate, dimethylaminohexadecyl methacrylate, 2-methacryloyloxyethyl phosphorylcholine, 1,3,5-triacryloylhexahydro-1,3,5-triazine, zinc oxide and titanium oxide. Some bonding agents incorporate fluoride; however, its effect is considered to be complementary as it serves to reduce enamel demineralization but does not have direct anti-microbial action per se. The bonding systems that incorporated the anti-microbials showed significantly enhanced anti-microbial activity compared to the control group (agar diffusion test: overall standardized mean difference: 3.71; 95% CI 2.98–4.43; optical density tests: 0.41; 95% CI −0.05–0.86). These anti-microbial agents demonstrated inhibition of S. mutans, S. sobrinus and Lactobacillus spp. growth without compromising their adhesive properties. However, the studies failed to show antibacterial effect in in vitro biofilm assays with single species. The authors recommend using microcosm methodology which reflects the oral microbiome more closely in its complexity and precision instead of the vitro biofilm assays with single species. Additionally, long-term, in vivo studies with focus on both anti-microbial properties and bond strength are recommended to draw stronger and more accurate conclusions (58).
Brackets with an anti-microbial, nanofilm coating of silver and titanium oxide have shown promising preliminary results, wherein bacterial colonies accumulated were greatly reduced compared to non-coated brackets which had an exponential increase in S. mutans colonies (59). Other studies on brackets, archwires and retainers coated with nano-copper oxide, nano-zinc oxide, zinc oxide, silver-platinum, silver ions, nitrogen-doped titanium oxide and polytetrafluoroethylene have similarly exhibited a beneficial anti-microbial activity (60–67). As coating the bracket slots with titanium or silver also has the undesirable effect of increased friction with the archwire, leaving the slots uncoated while deriving the anti-microbial benefits by coating the rest of the bracket would be a better design option (59). Similarly, for archwires, adjusting their composition and selecting an appropriate coating technology can help obtain optimal porosity, surface roughness and low-frictional characteristics while maintaining their antibacterial effects (66). Most of these studies have limitations in that they are predominantly in vitro and have been tested over a very short period of time ranging from 24 h to a week, which can be remedied by designing longer term, in vivo studies in the future (66).
Discussion
Orthodontic treatment is highly sought after by millions of children and adults across the world. When a mostly elective treatment modality is so popular and widespread among the general population, it becomes all the more important to eliminate or minimize iatrogenic or treatment-induced damage to the oral and dental tissues. A thorough research into the causation and mechanism of such adverse effects is the key. At the time when normal oral homeostasis, function and chemistry are disturbed by the insertion of relatively large and complex orthodontic appliances, it is the ongoing host-microbiome interaction which will decide if balance is restored, or a breakdown occurs leading to disease. Importantly, since orthodontic forces generate a sterile inflammatory response, their intersection with the inflammatory response generated by potential pathogens in the microbiome can have a compounding effect. For instance, proinflammatory cytokines such as interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF- α) are produced by periodontal ligament cells on orthodontic mechanical force application and also when stimulated by the lipopolysaccharide of P. gingivalis and A. actinomycetemcomitans (68–70). Additionally, activation of key orthodontic bone remodeling pathway of receptor activator of nuclear factor -κB (RANK) and its ligand (RANKL) is modulated both by mechanical and bacterial stressors (71, 72). Thus, expression of many of the biological mediators of orthodontic tooth movement can be potentiated by interactions between the orthodontically applied mechanical force and microbiome physiology. Although orthodontic interventions currently do take into account the periodontal status at a tissue level to implement appropriately accommodative treatment strategies, not much attention is paid to the intersection of mechanical force and microbiome stressors at a molecular level. Focused research in this area has the potential to impact clinical orthodontics in a major way, particularly considering the rising numbers of adult patients who may also have periodontal issues, seeking treatment.
The human microbiome is said to be as unique to every individual as their fingerprint (4). Even though inter-individual variations in the microbiome exist, the overall general functions performed by the microbiota across individuals are consistent. Twin studies have shown that genetic variation within the individual is the dominant source of variability in composition of the microbiome (4). Current culture-independent, metagenomic methods of identifying microorganisms, such as high-throughput sequencing of the 16S rRNA gene, have made it possible to study the microbiome at a level of detail not possible before (15). The traditional microbiological assays such as the time-consuming and laborious lab culturing of microorganisms have made way for rapid, voluminous and computationally intensive methods such as next-generation sequencing (NGS) (Table 1). Such technological advances usher a paradigm shift in the research field, making it possible to identify the signature microbiome for each individual, similar to disease-specific microbiome signatures such as those for periodontitis or diabetes mellitus (73). These microbiome signatures are liable to change spatially and temporally within an individual during the course of orthodontic treatment. If microbiome signatures specific to health, dysbiosis and disease in an individual are identified based on metagenomics and NGS, chairside, rapid diagnostic tools based on microarray analysis and microfluidics can be developed for individualized, microbial monitoring of the same during orthodontic treatment. Such point-of-care (POC) diagnostic modalities would also enable the clinician to institute interventions at the first sign of shift in the microbiome toward dysbiosis and thus pre-empt the disease even before it has a chance to fully establish itself. Changes in the microbiome could also be used to facilitate patient compliance with oral hygiene practices and good dietary habits. Range of interventions could be used to treat the microbial shifts, such as dietary counseling, reinforcing good oral hygiene practices, professional oral prophylaxis to remove plaque and calculus deposits or even localized fluoride or anti-microbial therapy. Significant amount of basic science research and longitudinal clinical studies are required for the actualization and implementation of these personalized orthodontic management techniques. Recent technological advances in scientific research reassure us that we are headed in the right direction.
Summary and Conclusions
Orthodontics, as a specialty, uses an array of mechanical appliances to bring about tooth movement. Orthodontic appliances are known to attract increased amounts of plaque, biofilm and microbiota as soon as within 1 h after appliance placement, with effects being transient or lasting throughout treatment duration. They may also change salivary flow rate, pH and buffering capacity. The interfacing of these biomaterials with oral environment also causes the oral microbiome to shift into dysbiosis, wherein an increase in acidogenic, cariogenic and periodontopathogenic bacteria occurs. Whenever host responses are overrun by persistent and significant environmental assaults, the imbalance in microbiome leads to a disease process, specifically dental caries, enamel demineralization and periodontal disease during orthodontic treatment. The disease-causing shift in the microbiome can be averted by instituting rigorous oral hygiene practices at a personal and professional level while patients are undergoing orthodontic treatment. Advances in molecular biology techniques have enhanced our understanding of the exact nature of the microbiome changes. The possibility of being able to identify the exact composition and change of microbiome in each individual in the future takes us one step closer to personalized orthodontics. By designing interventions to specifically target pathogenic microorganisms, one can dramatically reduce the adverse effects of orthodontic treatment such as enamel decalcification, dental caries and periodontal disease. However, a significant amount of good, long-term studies need to be carried out before we are able to make the ideal of personalized orthodontics and targeted microbiome therapy a reality.
Author Contributions
PM and TP contributed to the conception and design of the project. PM wrote the first draft of the manuscript. TP contributed to revisions. Both authors contributed to the article and approved the submitted version.
Funding
PM was supported by the NIH/NIDCR R90 Grant #R90DE02305.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Pinto AS, Alves LS, Maltz M, Susin C, Zenkner JE. Does the duration of fixed orthodontic treatment affect caries activity among adolescents and young adults? Caries Res. (2018) 52:463–7. doi: 10.1159/000488209
2. Wishney M. Potential risks of orthodontic therapy: a critical review and conceptual framework. Aust Dent J. (2017) 62:86–96. doi: 10.1111/adj.12486
3. American Association of Orthodontists. St. Louis, (MO). 2017 Orthodontic Workforce Report (2019). Available at https://webcache.googleusercontent.com/search?q=cache:1QXPMvDJU-8J:https://www.aaoinfo.org/sites/default/files/Orthodontic%2520Workforce%2520Report_April%25202018.pdf+andcd=1andhl=enandct=clnkandgl=us (accessed February 1, 2020).
4. Kilian M, Chapple IL, Hannig M, Marsh PD, Meuric V, Pedersen AM, et al. The oral microbiome–an update for oral healthcare professionals. Br Dent J. (2016) 221:657–66. doi: 10.1038/sj.bdj.2016.865
5. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLOS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
6. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. (2010) 192:5002–17. doi: 10.1128/JB.00542-10
7. Parahitiyawa NB, Scully C, Leung WK, Jin LJ, Samaranayake LP. Exploring the oral bacterial flora: current status and future directions. Oral Dis. (2010) 16:10. doi: 10.1111/j.1601-0825.2009.01607.x
9. Li Y. Jacox LA, Little SH, K, CC. Orthodontic tooth movement: The biology and clinical implications. Kaohsiung J Med Sci. (2018) 34:207–14. doi: 10.1016/j.kjms.2018.01.007
10. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. (2012) 13:260–70. doi: 10.1038/nrg3182
11. Pedersen AM (ed). Oral Infections and General Health: From Molecule to Chairside. Vol.1. Berlin: Springer International Publishing.
12. Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep. (2014) 4:1–9. doi: 10.1038/srep04828
13. Simpson CA, Adler C, Du Plessis MR, Landau ER, Dashper SG, Reynolds EC, et al. Oral microbiome composition, but not diversity, is associated with adolescent anxiety and depression symptoms. Physiol Behav. (2020) 226:113–26. doi: 10.1016/j.physbeh.2020.113126
14. Xie Z, Jiang W, Deng M, Wang W, Xie X, Feng X, et al. Alterations of oral microbiota in patients with panic disorder. Bioengineered. (2021) 12:1, 9103–12. doi: 10.1080/21655979.2021.1994738
15. Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. (2012) 18:109–20. doi: 10.1111/j.1601-0825.2011.01851.x
16. Xu P, Gunsolley J. Application of metagenomics in understanding oral health and disease. Virulence. (2014) 5:424–32. doi: 10.4161/viru.28532
17. The Expanded Human Oral Microbiome Database. (2021). Available at http://www.homd.org/ (accessed 30th January, 2021).
18. Caselli E, Fabbri C, D'Accolti M, Soffritti I, Bassi C, Mazzacane S, et al. Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. (2020) 20:1–9. doi: 10.1186/s12866-020-01801-y
19. Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. (2018) 16:540–50. doi: 10.1038/s41579-018-0041-0
20. Martellacci L, Quaranta G, Fancello G, D'addona A, Sanguinetti M, Patini R, et al. Characterizing peri-implant and sub-gingival microbiota through culturomics. First isolation of some species in the oral cavity. A pilot study. Pathogens. (2020) 9:365. doi: 10.3390/pathogens9050365
21. Marsh PD. Dental plaque: biological significance of a biofilm and community life-style. J Clin Periodontol. (2005) 32:7–15. doi: 10.1111/j.1600-051X.2005.00790.x
22. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. (1995) 49:711–45. doi: 10.1146/annurev.mi.49.100195.003431
23. Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. (2001) 358:135–8. doi: 10.1016/S0140-6736(01)05321-1
24. Marsh PD. Dental plaque as a microbial biofilm. Caries Res. (2004) 38:204–11. doi: 10.1159/000077756
25. Eliades T, Eliades G, Brantley WA. Microbial attachment on orthodontic appliances: I. Wettability and early pellicle formation on bracket materials. Am J Orthod Dentofacial Orthop. (1995) 108:351–60. doi: 10.1016/S0889-5406(95)70032-3
26. Dittmer MP, Hellemann CF, Grade S, Heuer W, Stiesch M, Schwestka-Polly R, et al. Comparative three-dimensional analysis of initial biofilm formation on three orthodontic bracket materials. Head Face Med. (2015) 11:1–6. doi: 10.1186/s13005-015-0062-0
27. Knoernschil KL, Rogers HM, Lefebvre CA, Fortson WM, Schuster GS. Endotoxin affinity for orthodontic brackets. Am J Orthod Dentofacial Orthop. (1999) 115:634–9. doi: 10.1016/S0889-5406(99)70288-X
28. Brusca MI, Chara O, Sterin-Borda L, Rosa AC. Influence of different orthodontic brackets on adherence of microorganisms in vitro. Angle Orthod. (2007) 77:331–6. doi: 10.2319/0003-3219(2007)0770331:IODOBO2.0.CO;2
29. Papaioannou W, Gizani S, Nassika M, Kontou E, Nakou M. Adhesion of Streptococcus mutans to different types of brackets. Angle Orthod. (2007) 77:1090–5. doi: 10.2319/091706-375.1
30. Øilo M, Bakken V. Biofilm and dental biomaterials. Materials. (2015) 8:2887–900. doi: 10.3390/ma8062887
31. Lima KC, Paschoal MA, de Araújo Gurgel J, Freitas KM, Pinzan-Vercelino CR. Comparative analysis of microorganism adhesion on coated, partially coated, and uncoated orthodontic archwires: a prospective clinical study. Am J Orthod Dentofacial Orthop. (2019) 156:611–6. doi: 10.1016/j.ajodo.2018.11.014
32. Kim IH, Park HS, Kim YK, Kim KH, Kwon TY. Comparative short-term in vitro analysis of mutans streptococci adhesion on esthetic, nickel-titanium, and stainless-steel arch wires. Angle Orthod. (2014) 84:680–6. doi: 10.2319/061713-456.1
33. Taha M, El-Fallal A, Degla H. In vitro and in vivo biofilm adhesion to esthetic coated arch wires and its correlation with surface roughness. Angle Orthod. (2016) 86:285–91. doi: 10.2319/122814-947.1
34. Bollenl CM, Lambrechts P, Quirynen M. Comparison of surface roughness of oral hard materials to the threshold surface roughness for bacterial plaque retention: a review of the literature. Dent Mater. (1997) 13:258–69. doi: 10.1016/S0109-5641(97)80038-3
35. Forsberg CM, Brattstrom VF, Malmberg E, Nord CE. Ligature wires and elastomeric rings: two methods of ligation, and their association with microbial colonization of Streptococcus mutans and lactobacilli. Eur J Orthod. (1991) 13:416–20. doi: 10.1093/ejo/13.5.416
36. Garcez AS, Suzuki SS, Ribeiro MS, Mada EY, Freitas AZ, Suzuki H. Biofilm retention by 3 methods of ligation on orthodontic brackets: a microbiologic and optical coherence tomography analysis. Am J Orthod Dentofacial Orthop. (2011) 140:e193–198. doi: 10.1016/j.ajodo.2011.04.019
37. Turkkahraman H, Sayin MO, Bozkurt FY, Yetkin Z, Kaya S, Onal S. Archwire ligation techniques, microbial colonization, and periodontal status in orthodontically treated patients. Angle Orthod. (2005) 75:231–6.
38. Pellegrini P, Sauerwein R, Finlayson T, McLead J, Covell DA, Maier T, et al. Plaque retention by self-ligation vs elastomeric orthodontic brackets: quantitative comparison or oral bacteria detection with adenosine triphosphate-driven bioluminescence. Am J Orthod Dentofacial Orthop. (2009) 135, 426.e1–9. discussion 426-7. doi: 10.1016/j.ajodo.2008.08.018
39. Arnold S, Koletsi D, Patcas R, Eliades T. The effect of bracket ligation on the periodontal status of adolescents undergoing orthodontic treatment. A systematic review and meta-analysis. J Dent. (2016) 54:13–24. doi: 10.1016/j.jdent.2016.08.006
40. Chang HS, Walsh LJ, Freer TJ. The effect of orthodontic treatment on salivary flow, pH, buffer capacity, and levels of mutans streptococci and lacto bacilli. Aust Orthod J. (1999) 15:229.
41. Lara-Carrillo E, Montiel-Bastida N-M, Sánchez-Pérez L, Alanís-Tavira J. Effect of orthodontic treatment on saliva, plaque and the levels of Streptococcus mutans and Lactobacillu. Med Oral Patol Oral Cir Bucal. (2010) 15:924–9. doi: 10.4317/medoral.15.e924
42. Bonetti GA, Parenti SI, Garulli G, Gatto MR, Checchi L. Effect of fixed orthodontic appliances on salivary properties. Prog Orthod14. (2013) 1–4. doi: 10.1186/2196-1042-14-13
43. Lombardo L, Ortan YÖ, Gorgun Ö, Panza C, Scuzzo G, Siciliani G. Changes in the oral environment after placement of lingual and labial orthodontic appliances. Prog Orthod. (2013) 14:1–8. doi: 10.1186/2196-1042-14-28
44. Arab S, Malekshah SN, Mehrizi EA, Khanghah AE, Naseh R, Imani MM. Effect of fixed orthodontic treatment on salivary flow, pH and microbial count. J Dent. (2016) 13:18.
45. Zogakis IP, Koren E, Gorelik S, Ginsburg I, Shalish M. Effect of fixed orthodontic appliances on nonmicrobial salivary parameters. Angle Orthod. (2018) 88:806–11. doi: 10.2319/111317-773.1
46. Abed Al Jawad F, Cunningham SJ, Croft N, Johal A. A qualitative study of the early effects of fixed orthodontic treatment on dietary intake and behaviour in adolescent patients. Eur J Orthod. (2012) 34:432–6. doi: 10.1093/ejo/cjr032
47. Azaripour A, Willershausen I, Hassan M, Ebenezer S, Willershausen B. Oral hygiene and dietary habits in adolescents with fixed orthodontic appliances: A cross-sectional study. J Contemp Dent Pract. (2016) 17:179–83. doi: 10.5005/jp-journals-10024-1824
48. Lucchese A, Bondemark L, Marcolina M, Manuelli M. Changes in oral microbiota due to orthodontic appliances: a systematic review. J Oral Microbiol. (2018) 10:1476645. doi: 10.1080/20002297.2018.1476645
49. Guo R, Lin Y, Zheng Y, Li W. The microbial changes in subgingival plaques of orthodontic patients: a systematic review and meta-analysis of clinical trials. BMC Oral Health. (2017) 17:1–0. doi: 10.1186/s12903-017-0378-1
50. Demling A, Ling C, Schwestka-Polly R, Stiesch M, Heuer W. Short-term influence of lingual orthodontic therapy on microbial parameters and periodontal status: a preliminary study. Angle Orthod. (2010) 80, 480–4. doi: 10.2319/061109-330.1
51. Sfondrini MF, Debiaggi M, Zara F, Brerra R, Comelli M, Bianchi M, et al. Influence of lingual bracket position on microbial and periodontal parameters in vivo. J Appl Oral Sci. (2012) 20:357–61. doi: 10.1590/S1678-77572012000300011
52. Abbate GM, Caria MP, Montanari P, Mannu C, Orrù G, Caprioglio A, et al. Periodontal health in teenagers treated with removable aligners and fixed orthodontic appliances. J Orofac Orthop. (2015) 76:240–50. doi: 10.1007/s00056-015-0285-5
53. Azaripour A, Weusmann J, Mahmoodi B, Peppas D, Gerhold-Ay A, Van Noorden CJ, et al. Braces versus Invisalign®: gingival parameters and patients' satisfaction during treatment: a cross-sectional study. BMC Oral Health. (2015) 15:1–5. doi: 10.1186/s12903-015-0060-4
54. Levrini L, Mangano A, Montanari P, Margherini S, Caprioglio A, Abbate GM. Periodontal health status in patients treated with the Invisalign® system and fixed orthodontic appliances: a 3 months clinical and microbiological evaluation. Eur J Dent. (2015) 9:404. doi: 10.4103/1305-7456.163218
55. Lu H, Tang H, Zhou T, Kang N. Assessment of the periodontal health status in patients undergoing orthodontic treatment with fixed appliances and Invisalign system: a meta-analysis. Medicine. (2018) 97. doi: 10.1097/MD.0000000000010248
56. Wang Q, Ma JB, Wang B, Zhang X, Yin YL, Bai H. Alterations of the oral microbiome in patients treated with the Invisalign system or with fixed appliances. Am J Orthod Dentofacial Orthop. (2019) 156:633–40. doi: 10.1016/j.ajodo.2018.11.017
57. Pretty IA. Caries detection and diagnosis: novel technologies. J Dent. (2006) 34:727–39. doi: 10.1016/j.jdent.2006.06.001
58. De Almeida CM, Da Rosa WL, Meereis CT, de Almeida SM, Ribeiro JS, da Silva AF, et al. Efficacy of antimicrobial agents incorporated in orthodontic bonding systems: a systematic review and meta-analysis. J Orthod. (2018) 45:79–93. doi: 10.1080/14653125.2018.1443872
59. Ghasemi T, Arash V, Rabiee SM, Rajabnia R, Pourzare A, Rakhshan V. Antimicrobial effect, frictional resistance, and surface roughness of stainless steel orthodontic brackets coated with nanofilms of silver and titanium oxide: a preliminary study. Microsc Res Tech. (2017) 80:599–607. doi: 10.1002/jemt.22835
60. Demling A, Elter C, Heidenblut T, Bach FW, Hahn A, Schwestka-Polly R, et al. Reduction of biofilm on orthodontic brackets with the use of a polytetrafluoroethylene coating. Eur J Orthod. (2010) 32:414–8. doi: 10.1093/ejo/cjp142
61. Özyildiz F, Güden M, Uzel A, Karaboz I, Akil O, Bulut H. Antimicrobial activity of TiO2-coated orthodontic ceramic brackets against Streptococcus mutans and Candida albicans. Biotechnol Bioprocess Eng. (2010) 15:680–5. doi: 10.1007/s12257-009-3005-4
62. Ryu HS, Bae IH, Lee KG, Hwang HS, Lee KH, Koh JT, et al. Antibacterial effect of silver-platinum coating for orthodontic appliances. Angle Orthod. (2012) 82:151–7. doi: 10.2319/021411-111.1
63. Cao B, Wang Y, Li N, Liu B, Zhang Y. Preparation of an orthodontic bracket coated with an nitrogen-doped TiO(2-x)N(y) thin film and examination of its antimicrobial performance. Dent Mater J. (2013) 32:311–6. doi: 10.4012/dmj.2012-155
64. Morita Y, Imai S, Hanyuda A, Matin K, Hanada N, Nakamura Y. Effect of silver ion coating of fixed orthodontic retainers on the growth of oral pathogenic bacteria. Dent Mater J. (2014) 33:268–74. doi: 10.4012/dmj.2013-216
65. Ramazanzadeh B, Jahanbin A, Yaghoubi M, Shahtahmassbi N, Ghazvini K, Shakeri M, et al. Comparison of antibacterial effects of ZnO and CuO nanoparticles coated brackets against Streptococcus mutans. J Dent. (2015) 16:200.
66. Bacela J, Łabowska MB, Detyna J, Ziety A, Michalak I. Functional coatings for orthodontic archwires—A review. Materials. (2020) 13:3257. doi: 10.3390/ma13153257
67. Hammad SM, El-Wassefy NA, Shamaa MS, Fathy A. Evaluation of zinc-oxide nanocoating on the characteristics and antibacterial behavior of nickel-titanium alloy. Dental Press J Orthod. (2020) 25:51–8. doi: 10.1590/2177-6709.25.4.051-058.oar
68. Meikle MC. The tissue, cellular, and molecular regulation of orthodontic tooth movement: 100 years after Carl Sandstedt. Eur J Orthod. (2006) 28:221–40. doi: 10.1093/ejo/cjl001
69. Proff P, Reicheneder C, Faltermeier A, Kubein-Meesenburg D, Römer P. Effects of mechanical and bacterial stressors on cytokine and growth-factor expression in periodontal ligament cells. J Orofac Orthop. (2014) 75:191–202. doi: 10.1007/s00056-014-0212-1
70. Sun Y, Shu R, Li C-L, Zhang M-Z. (2010). Gram-negative periodontal bacteria induce the activation of Toll-like receptors 2 and 4, and cytokine production in human periodontal ligament cells. J Periodontol. 81:1488–96. doi: 10.1902/jop.2010.100004
71. Römer P, Köstler J, Koretsi V, Proff P. Endotoxins potentiate COX-2 and RANKL expression in compressed PDL cells. Clin Oral Investig. (2013) 17:2041–8. doi: 10.1007/s00784-013-0928-0
72. Schröder A, Stumpf J, Paddenberg E, Neubert P, Schatz V, Köstler J, et al. Effects of mechanical strain on periodontal ligament fibroblasts in presence of Aggregatibacter actinomycetemcomitans lysate. BMC Oral Health. (2021) 21:1–2. doi: 10.1186/s12903-021-01761-3
Keywords: orthodontic appliance, oral microbiome, oral bacteria, biomaterial, dysbiosis, demineralization/white spot lesions, periodontal disease
Citation: Mulimani P and Popowics T (2022) Effect of Orthodontic Appliances on the Oral Environment and Microbiome. Front. Dent. Med. 3:924835. doi: 10.3389/fdmed.2022.924835
Received: 20 April 2022; Accepted: 16 May 2022;
Published: 21 June 2022.
Edited by:
Akira Nifuji, Tsurumi Univesity, JapanReviewed by:
Anand Marya, University of Puthisastra, CambodiaRomeo Patini, Agostino Gemelli University Polyclinic (IRCCS), Italy
Copyright © 2022 Mulimani and Popowics. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tracy Popowics, cG9wb3dpY3NAdXcuZWR1
 Priti Mulimani
Priti Mulimani Tracy Popowics*
Tracy Popowics*