- 1New Industry Creation Hatchery Center, Tohoku University, Sendai, Japan
- 2Laboratory of Environmental Interface Technology of Filamentous Fungi, Kyoto University, Kyoto, Japan
- 3Medical Mycology Research Center, Chiba University, Chiba, Japan
- 4Laboratory of Applied Microbiology, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
- 5Laboratory of Enzymology, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
- 6Department of Biotechnology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 7Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, Tokyo, Japan
- 8Terrestrial Microbial Ecology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
Many eukaryotic histidine-to-aspartate (His-Asp) phosphorelay systems consist of three types of signal transducers: a His-kinase (HK), a response regulator (RR), and a histidine-containing phosphotransfer intermediate (HPt). In general, the HPt acts as an intermediate between the HK and the RR and is indispensable for inducing appropriate responses to environmental stresses. In a previous study, we attempted but were unable to obtain deletion mutants of the ypdA gene in order to characterize its function in the filamentous fungus Aspergillus nidulans. In the present study, we constructed the CypdA strain in which ypdA expression is conditionally regulated by the A. nidulans alcA promoter. We constructed CypdA strains with RR gene disruptions (CypdA-sskAΔ, CypdA-srrAΔ, and CypdA-sskAΔsrrAΔ). Suppression of YpdA induced by ypdA downregulation activated the downstream HogA mitogen-activated protein kinase cascade. YpdA suppression caused severe growth defects and abnormal hyphae, with features such as enhanced septation, a decrease in number of nuclei, nuclear fragmentation, and hypertrophy of vacuoles, both regulated in an SskA–dependent manner. Fludioxonil treatment caused the same cellular responses as ypdA suppression. The growth-inhibitory effects of fludioxonil and the lethality caused by ypdA downregulation may be caused by the same or similar mechanisms and to be dependent on both the SskA and SrrA pathways.
Introduction
Histidine-to-aspartate (His-Asp) phosphorelay (or two-component) systems are common in bacteria, slime molds, plants, and fungi and are involved in a wide variety of cellular responses to environmental stimuli (Hoch and Silhavy, 1995). These systems commonly consist of (i) a His-kinase (HK) that serves as a sensor for certain stimuli, (ii) a response regulator (RR) containing a phospho-accepting receiver domain, and (iii) a histidine-containing phosphotransfer intermediate (HPt). Eukaryotic HKs are generally hybrid-type kinases that contain a kinase and a receiver domain. A phosphate group is transmitted as follows: hybrid HK (His) → hybrid HK (Asp) → HPt (His) → RR (Asp). This type of sequential phosphorelay is generally referred to as a multistep phosphorelay (Appleby et al., 1996). Numerous instances of such His-Asp phosphorelay systems have been identified and characterized in many fungi (Maeda et al., 1994; Alex et al., 1996; Yamada-Okabe et al., 1999; Buck et al., 2001). In a previous study, we focused on the His-Asp phosphorelay system of a model filamentous fungus, Aspergillus nidulans, which is a representative of related species of interest, such as A. fumigatus (a serious pathogen) and A. oryzae (a fungus used for industrial fermentation). Based on genome sequence data, A. nidulans has 15 HKs, four RRs, and only one HPt (Hagiwara et al., 2007a). The physiological functions of four HKs (TcsA, TcsB, FphA, and NikA) and four RRs (SskA, SrrA, SrrB, and SrrC) have been investigated so far (Appleyard et al., 2000; Furukawa et al., 2002; Blumenstein et al., 2005; Hagiwara et al., 2007a,b). The two-component signaling system in A. nidulans is summarized in Supplementary Figure 1.
In Saccharomyces cerevisiae, the His-Asp phosphorelay system has been intensively studied by many researchers. Saccharomyces cerevisiae has a single transmembrane HK, Sln1p; one HPt, Ypd1p; and two RRs, Ssk1p and Skn7p. A typical multistep phosphorelay pathway consisting of Sln1p (hybrid HK)–Ypd1p (HPt)–Ssk1p (RR) is crucial to the osmotic stress response; this pathway modulates the downstream high osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) cascade consisting of Ssk2p/Ssk22p (MAPKKK), Pbs2p (MAPKK), and Hog1p (MAPK) (Maeda et al., 1994; Posas et al., 1996; Posas and Saito, 1998). Thus, when cells are exposed to high osmotic stress, the HOG MAPK pathway is activated via the His-Asp phosphorelay system. Activation of the HOG pathway results in the induction of genes required for osmotic adaptation, including glycerol biosynthesis genes such as glycerol-3-phosphate dehydrogenase (GPD1) and glycerol-3-phosphatase (GPP2) (Akhtar et al., 1997; Gustin et al., 1998; Hohmann, 2002). In S. cerevisiae, the RR Skn7p is also regulated via Sln1p-Ypd1p (Li et al., 1998). Skn7p is a transcription factor involved in multiple physiological processes, such as maintenance of cell wall integrity and oxidative stress response (Brown et al., 1993, 1994).
Based on these findings for S. cerevisiae, the signaling network of the His-Asp phosphorelay system and the HOG MAPK cascade were investigated in the filamentous fungus A. nidulans. When A. nidulans cells are exposed to high osmotic stress, HogA (Hog1 ortholog), also called SakA, is phosphorylated in a manner dependent on PbsB, SskB, and SskA (orthologs of Pbs2p, Ssk2p/Ssk22p, and Ssk1p, respectively), but not on TcsB (an ortholog of Sln1p) (Furukawa et al., 2005). Deletion mutants of HogA and SskA are similarly sensitive to osmotic stress (Furukawa et al., 2005), and their conidia are sensitive to oxidative and heat stress (Kawasaki et al., 2002; Hagiwara et al., 2007a). The HogA MAPK cascade is involved in light sensing via the interaction between the phytochrome FphA and the HPt protein YpdA in A. nidulans (Yu et al., 2016). In response to white or red light, SakA (HogA) translocates from the cytoplasm to the nucleus in an FphA-dependent manner (Yu et al., 2016). FphA and TcsB are also involved in temperature sensing (Yu et al., 2019). FphA could act as a thermosensor and is involved in SakA translocation, although the temperature response of SakA is not completely FphA dependent (Yu et al., 2019). Thus, the His-Asp phosphorelay system appears to regulate the HogA MAPK cascade and to play a specific role in A. nidulans response to environmental changes (Supplementary Figure 1). Through this signaling network, genes encoding stress-related factors, such as glycerol-3-phosphate dehydrogenase (GfdB) (Han and Prade, 2002), trehalose-6-phosphate synthase (TpsA), and conidia-specific catalase (CatA) (Navarro and Aguirre, 1998), are regulated in response to osmotic stress (Hagiwara et al., 2007a, 2009). The light- and temperature-regulated genes, conJ, ccgA, and ccgB, are also regulated in a HOG pathway–dependent manner (Yu et al., 2016, 2019). Previously, we performed transcriptome analyses using DNA microarrays, which revealed that the His-Asp phosphorelay system and the HogA MAPK cascade are responsible for the transcriptional responses to both fungicide and osmotic stress in A. nidulans (Hagiwara et al., 2009).
Phenylpyrrole- or dicarboximide-type fungicides such as fludioxonil or iprodione, respectively, are agronomically useful. These fungicides appear to act via the signaling network of the His-Asp phosphorelay system and the HOG MAPK cascade (Ochiai et al., 2001; Yoshimi et al., 2004, 2005; Motoyama et al., 2005; Viaud et al., 2006; Hagiwara et al., 2007b). Fludioxonil converts Drk1 (an HK from Blastomyces dermatitidis, expressed in S. cerevisiae) into a phosphatase, resulting in Ypd1 dephosphorylation and constitutive activation of the HOG pathway (Lawry et al., 2017). Brandhorst et al. (2019) have suggested the molecular mechanism of this conversion. Fludioxonil could modify the action of triosephosphate isomerase, inducing elevation of methylglyoxal in the Drk1-expressing yeast. Methylglyoxal stress could act on Drk1 and convert it to a phosphatase, inducing constitutive activation of the HOG pathway and cell death (Brandhorst et al., 2019).
To better understand the molecular mechanisms of the His-Asp phosphorelay system, we attempted to characterize the A. nidulans HPt, YpdA. Despite many attempts to obtain a ypdA disruptant, we could not isolate it (Furukawa et al., 2005). Because deletion of Ypd1p is lethal in S. cerevisiae, deletion of A. nidulans YpdA was also presumed to be lethal (Posas et al., 1996). Vargas-Perez et al. (2007) also tried to obtain an A. nidulans ypdA gene deletion strain and obtained ypdA−/ypdA+ heterokaryons, but could not segregate the desired ypdA− haploid strain, demonstrating that the ypdA− allele could only be maintained in heterokaryons. Vargas-Perez et al. (2007) concluded that ypdA deletion is probably lethal in A. nidulans. In Neurospora crassa, the hpt-1 gene encoding a single ortholog of Ypd1p could be disrupted in the os-2 (hog1 ortholog gene) mutant but not in the wild-type strain (Banno et al., 2007), indicating that the HPt gene is essential in wild-type N. crassa, but not in the os-2 mutant background. Collectively, these data indicate that HPt may be an essential component and may play a crucial role in His-Asp phosphorelay signaling systems in filamentous fungi. Although many studies have suggested that ypdA and its homologs are essential in many species of fungi, as described above, the involvement of elements downstream of YpdA remains unclear.
In filamentous fungi such as A. nidulans (Hagiwara et al., 2007b) and Cochliobolus heterostrophus (Yoshimi et al., 2004), resistance to the fungicide fludioxonil is known to depend on two-component signaling. Fludioxonil strongly inhibits growth of wild-type strains of these two fungi, and mutants with the two RR genes disrupted (sskAΔsrrAΔ) are fully resistant to fludioxonil (Hagiwara et al., 2007b; Izumitsu et al., 2007). Because fludioxonil treatment increases the levels of phosphorylated Hog1-type MAP kinases (Kojima et al., 2004), the toxicity of fludioxonil and the lethality of ypdA disruption, which is thought to cause hyper-activation of HogA, may share some components of their mechanisms (Furukawa et al., 2005; Vargas-Perez et al., 2007). Although the essentiality of the ypdA gene and the toxicity of fludioxonil are known, as described above, the detailed intracellular events caused by ypdA downregulation or fludioxonil treatment remain unclear.
In the present study, we constructed a strain of A. nidulans in which ypdA was conditionally regulatable, with or without deletion of sskA and/or srrA, which encode RRs in the two-component signaling system. We downregulated expression of ypdA in each strain and demonstrated that ypdA downregulation causes severe growth defects and abnormal hyphae, with features such as enhanced septation, a decrease in number of nuclei, and hypertrophy of vacuoles. The growth defects caused by ypdA downregulation were restored by sskAΔsrrAΔ double disruption. Fludioxonil treatment of wild-type cells produced phenotypes similar to those caused by ypdA downregulation. The intracellular phenomena caused by ypdA downregulation and fludioxonil treatment are discussed from the viewpoint of the events downstream of YpdA signaling.
Results
Construction of the alcA Promoter-Driven ypdA Strain (CypdA) and Its Response Regulator Gene Disruptants (CypdA-sskAΔ, CypdA-srrAΔ, and CypdA-sskAΔsrrAΔ Strains)
To assess the essentiality of the ypdA gene and investigate the physiological role of YpdA in A. nidulans, we generated a strain, in which the transcription of ypdA was under the control of the A. nidulans alcA promoter. The native ypdA promoter (−1,000 bp from the start codon) was replaced by the alcA promoter and the pyrG selectable marker in the ABPU1 (ΔligD) strain (Supplementary Figure 2, upper panel). The genomic structure of the resulting conditional-ypdA (CypdA) strain was confirmed by PCR (Supplementary Figure 2, lower panel). From the CypdA strain, we constructed sskA single disruptants (CypdA-sskAΔ), srrA single disruptants (CypdA-srrAΔ), and sskA-srrA double disruptants (CypdA-sskAΔsrrAΔ). Two other RR genes of A. nidulans, srrB and srrC, were excluded from the present study because deletion of either of these genes had no effect on phenotype (Hagiwara et al., 2007a). Disruption of the sskA and srrA genes in the CypdA-sskAΔ, CypdA-srrAΔ, and CypdA-sskAΔsrrAΔ strains was confirmed by PCR (Supplementary Figure 3).
Regulation of the Expression Level of the ypdA Gene
To confirm the regulation of ypdA expression in the CypdA strain and its derivatives CypdA-sskAΔ, CypdA-srrAΔ, and CypdA-sskAΔsrrAΔ, we performed transcriptional analysis. The alcA promoter activates gene expression in the presence of carbon sources such as ethanol, glycerol, and threonine, and represses it in the presence of glucose (Gwynne et al., 1987). In addition, the presence of yeast extracts often affects the expression of genes controlled by the alcA promoter (Nikolaev et al., 2002). First, the composition of CD medium was optimized to produce CDTFY medium (50 mM threonine, 0.2% fructose, 0.2% yeast extract) to decrease the transcription levels of ypdA in the CypdA strain to less than three times that in the wild-type strain ABPU1 (Supplementary Figure 4A). Next, using this medium, we evaluated the relative expression levels of ypdA in CypdA and its derivative strains, and found that they did not differ significantly among strains under these growth conditions (Supplementary Figure 4B).
To repress the transcription of ypdA, CDY medium (2% glucose, 0.1% yeast extract) was used for cultures of CypdA and its derivatives. Transcription levels of ypdA in the wild-type strain remained stable for 36 h after transfer from CDTFY (0 h) to CDY (Figure 1A). In contrast, after 12 h on CDY medium, transcription levels of ypdA in the CypdA and its derivative strains were ≤10% of those in the wild-type strain (Figure 1A). Two-way ANOVA revealed a significant interaction between strain and time; thus, multiple-comparison tests of the ypdA expression level were conducted. At each time point from 12 h after transfer to ypdA-repressing medium (CDY), the expression levels of ypdA in all the mutant strains were significantly lower than those in the wild-type strain (Supplementary Table 1). To confirm that ypdA expression levels affected YpdA protein levels, we performed western blot analysis using an anti-YpdA antibody (Figure 1B). After transfer from ypdA-inducing to ypdA-repressing medium, YpdA protein was hardly detectable in CypdA and its derivative strains.
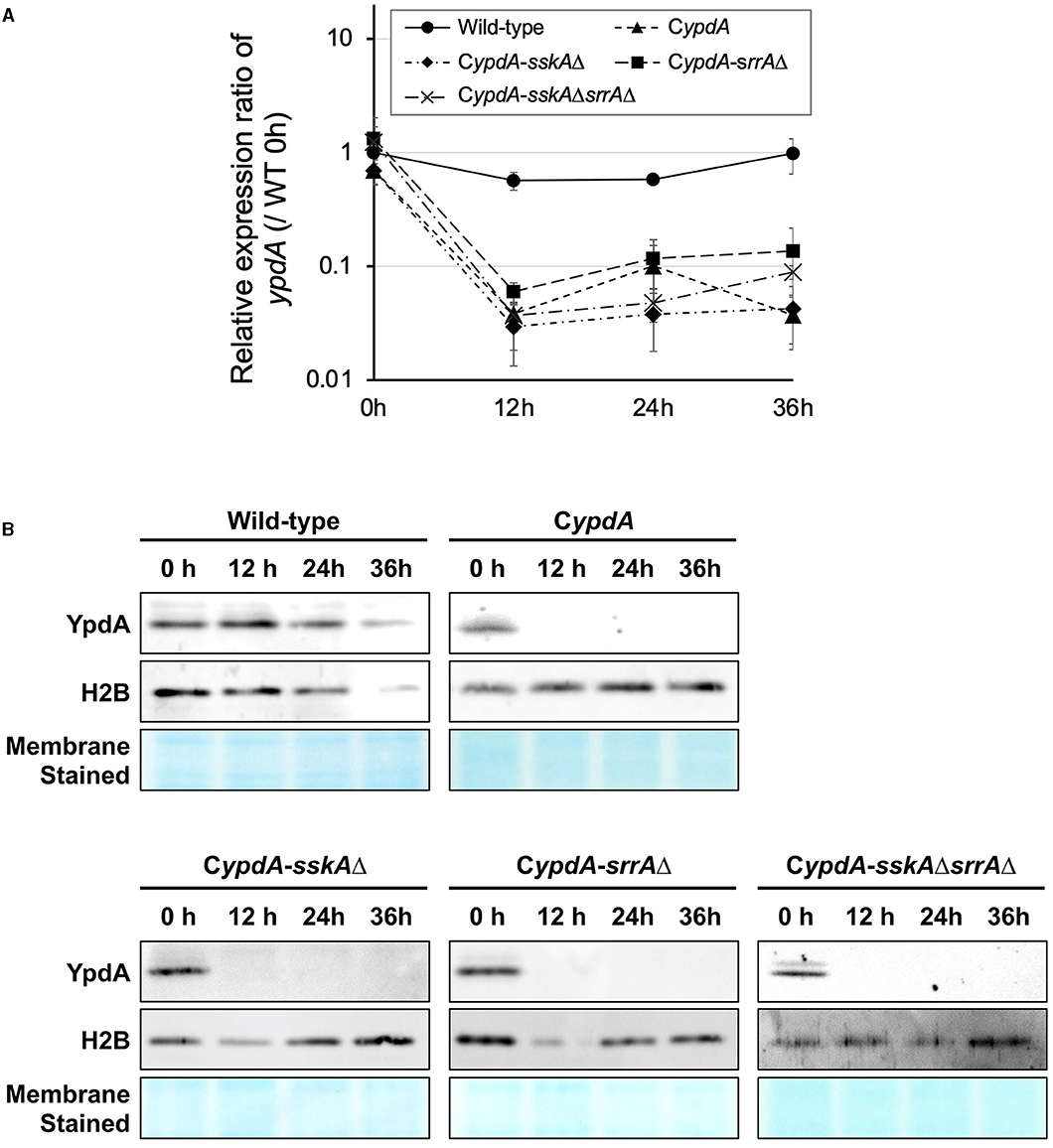
Figure 1. Downregulation of the ypdA gene (A) and YpdA protein levels (B) in Aspergillus nidulans wild-type, CypdA, and CypdA-derivative strains. (A) Time course of ypdA expression after transfer from ypdA-inducing medium (CDTFY; 0 h) to ypdA-repressing medium (CDY). Conidia (final concentration, 5 × 105 conidia/ml) were inoculated into ypdA-inducing medium and cultured for 20 h before transfer to ypdA-repressing medium, at 0 h. Samples were prepared from mycelia at 12, 24, and 36 h, and ypdA expression was quantified by RT-PCR. The expression level of the ypdA gene in each sample was normalized to that of the histone H2B gene (internal control) in the same sample. Each value represents the ypdA expression ratio relative to expression in the wild-type strain cultured in CDTFY medium for 20 h (WT 0 h). Error bars represent standard deviations from three independent replicates. (B) Western blot analysis of YpdA protein with histone H2B protein as an internal standard and stained membrane for confirmation of protein transfer. Samples were prepared from mycelia at 12, 24, and 36 h after transfer from ypdA-inducing medium (CDTFY; 0 h) to ypdA-repressing medium (CDY). Each lane was loaded with 15 μg protein.
Thus, in ypdA-repressing medium, both ypdA transcription and YpdA protein levels were stable in the wild type but were downregulated in CypdA and its derivative strains.
Downregulation of ypdA Induces HogA Pathway–Regulated Genes
In His-Asp phosphorelay systems, HPt factors mediate phosphotransfer between HKs and RRs, so we investigated whether a decreased level of YpdA affects downstream signaling pathways. Because the HogA MAPK cascade is regulated downstream of SskA, one of the RRs of interest, we investigated the expression of the HOG pathway–regulated gene catA (Hagiwara et al., 2007a). On ypdA-inducing (CDTFY) medium, no significant differences were detected in the relative expression ratio of catA among strains (Figure 2A). After transfer to ypdA-repressing medium, transcription levels of catA tended to be upregulated in the CypdA strain (Figure 2B, Supplementary Table 2) and were significantly upregulated in the CypdA-srrAΔ strain but not in the wild-type, CypdA-sskAΔ, or CypdA-sskAΔsrrAΔ (Figure 2B, Supplementary Table 2), suggesting that downregulation of ypdA activated the HogA pathway.
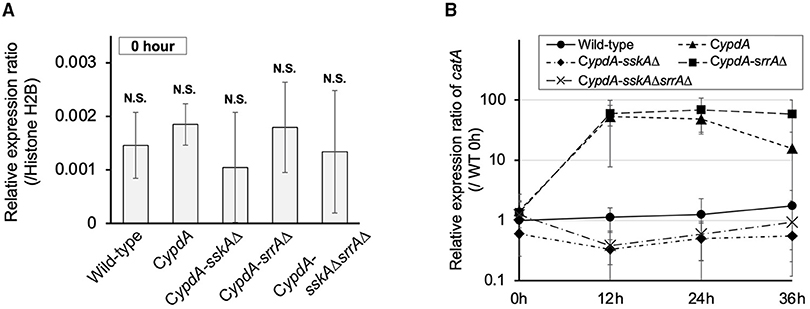
Figure 2. Expression levels of catA in Aspergillus nidulans wild-type, CypdA, and CypdA-derivative strains. (A) Quantitative real-time PCR analysis. Conidia (final concentration, 5 × 105 conidia/ml) were inoculated into ypdA-inducing CDTFY medium and cultured for 20 h before transfer to ypdA-repressing CDY medium, at 0 h. Transcript levels of catA were quantified by RT-PCR. Each value represents the expression ratio relative to the gene encoding H2B in each strain. Error bars represent standard deviations from three independent replicates. N.S., not significant. (B) Time course of catA expression after transfer from ypdA-inducing medium (CDTFY; 0 h) to ypdA-repressing medium (CDY). Samples were prepared from mycelia at 12, 24, and 36 h, and expression of catA was quantified by RT-PCR. The expression level of the catA gene in each sample was normalized to that of the histone H2B gene (internal control) in the same sample. Each value represents the catA expression ratio relative to expression in the wild-type strain cultured in CDTFY medium for 20 h (WT 0 h). Error bars represent standard deviations from three independent replicates.
Downregulation of ypdA Causes an Increase in Total and Phosphorylated HogA and Overexpression of hogA
To determine whether HogA was activated (i.e., phosphorylated) under ypdA-repressing conditions, we performed western blot analysis to detect HogA phosphorylation. HogA was weakly phosphorylated in wild-type cells after transfer to ypdA-repressing (CDY) medium (Figure 3A). In CypdA and CypdA-srrAΔ cells, HogA was strongly expressed and phosphorylated after transfer to ypdA-repressing medium (Figure 3A). In contrast, in CypdA-sskAΔ and CypdA-sskAΔsrrAΔ cells, HogA was scarcely expressed or phosphorylated after transfer to ypdA-repressing medium. These results suggested that ypdA downregulation caused hyper-expression and phosphorylation of HogA in an SskA–dependent manner. The dependence of HogA expression on SskA signaling was also supported by transcription levels of hogA (Figures 3B,C). During vegetative growth, no significant differences were detected in the relative expression ratio of hogA among the wild-type, CypdA, and CypdA-srrAΔ strains, whereas the levels were significantly lower in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains (Figure 3B). After transfer from ypdA-inducing (CDTFY) to ypdA-repressing medium, transcription levels of hogA were nearly constant in the wild-type strain but were increased significantly in the CypdA and CypdA-srrAΔ strains (Figure 3C, Supplementary Table 3). On both media types, transcription levels of hogA were consistently lower in CypdA-sskAΔ and CypdA-sskAΔsrrAΔ cells than in the wild-type strain (Figure 3C, Supplementary Table 3).
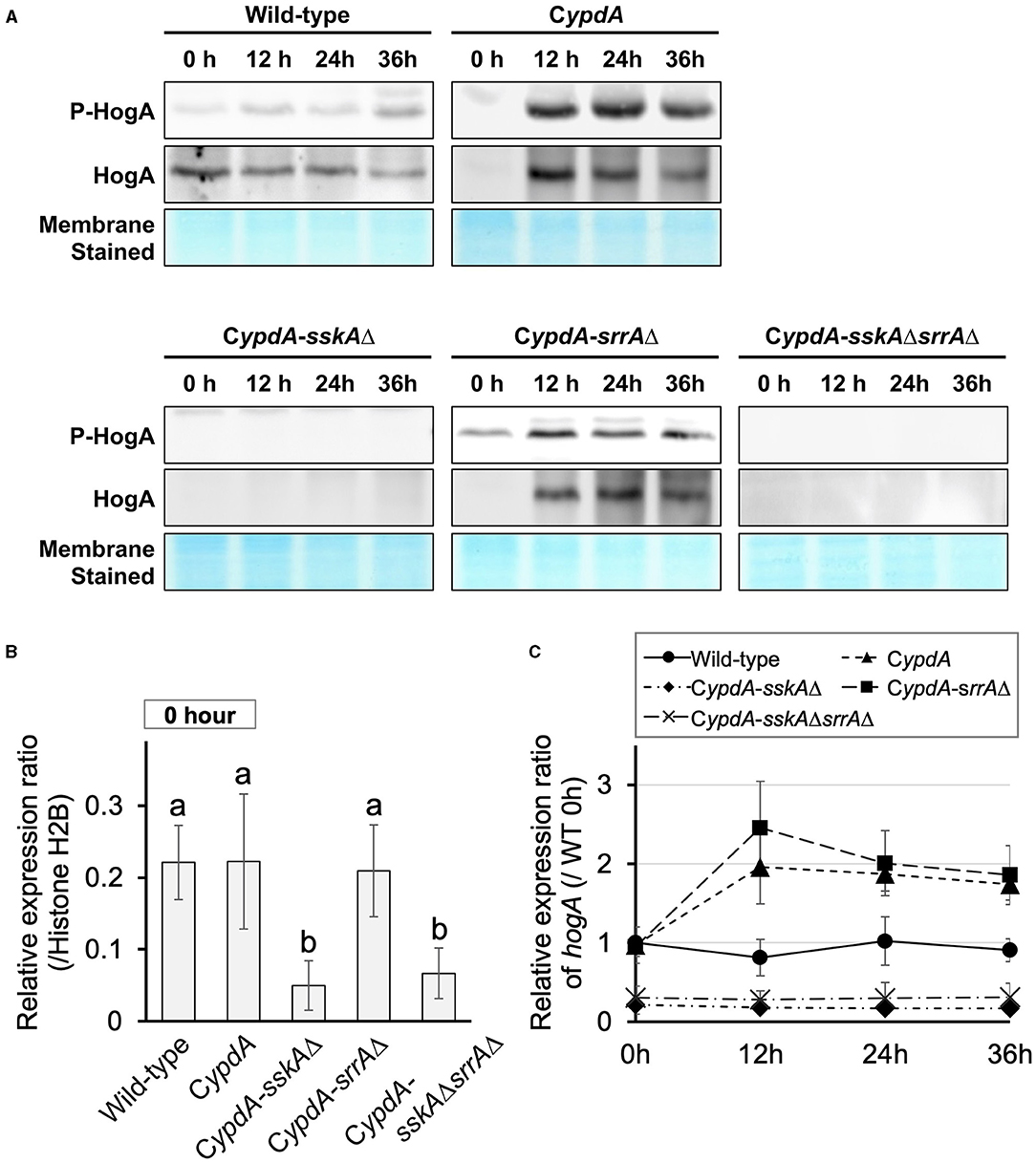
Figure 3. Phosphorylation of HogA protein (A) and gene expression of hogA (B,C) in Aspergillus nidulans wild-type, CypdA, and CypdA-derivative strains. (A) Western blot analysis of phospho-HogA. HogA protein and stained membrane for confirmation of protein transfer are shown. Samples were prepared from mycelium at 12, 24, and 36 h after transfer from ypdA-inducing medium (CDTFY; 0 h) to ypdA-repressing medium (CDY). Each lane was loaded with 15 μg protein. (B) Quantitative real-time PCR analysis. Conidia (final concentration, 5 × 105 conidia/ml) were cultured in ypdA-inducing CDTFY for 20 h before transfer to ypdA-repressing CDY medium, at 0 h. Transcript levels of hogA were quantified by RT-PCR. Each value represents the expression ratio relative to that of the gene encoding histone H2B in each strain. Error bars represent standard deviations from three independent replicates. Different letters indicate significant differences by Tukey–Kramer's test (P < 0.05). (C) Time course of hogA expression after transfer from ypdA-inducing medium (CDTFY; 0 h) to ypdA-repressing medium (CDY). Samples were prepared from mycelia at 12, 24, and 36 h, and expression of hogA was quantified by RT-PCR. The expression level of the hogA gene in each sample was normalized to that of the histone H2B gene (internal control) in the same sample. Each value represents the hogA expression ratio relative to expression in the wild-type strain cultured in CDTFY medium for 20 h (WT 0 h). Error bars represent standard deviations from three independent replicates.
Cell Defects Caused by ypdA Downregulation Depend on Both SskA and SrrA Pathways
On ypdA-inducing (CDTFY) agar plates, no significant differences in colony growth were detected among the wild-type, CypdA, CypdA-sskAΔ, and CypdA-srrAΔ strains, but CypdA-sskAΔsrrAΔ strain showed severe growth retardation (Figure 4A, Supplementary Table 4). On ypdA-repressing (CDY) agar plates, colonies of the CypdA, CypdA-sskAΔ, and CypdA-srrAΔ strains were significantly smaller than that of the wild-type strain, but growth of the CypdA-sskAΔsrrAΔ strain was almost completely restored (Figure 4B, Supplementary Table 4), suggesting that the growth defects caused by ypdA downregulation depend on both SskA and SrrA. Although conidial formation is affected in the sskAΔ and srrAΔ strains (Hagiwara et al., 2007a), no difference in conidial formation was observed between the wild-type and CypdA strains, regardless of whether they were grown on ypdA-inducing or -repressing medium (Supplementary Figure 5A). No effect of ypdA repression was observed on conidial germination in the wild-type or CypdA strain (Supplementary Figure 5B). We also tested the growth responses to osmotic stress in the wild-type, CypdA, and its derivative strains together with the sskAΔsrrAΔ strain (Supplementary Figure 6). Consistent with a previous report (Vargas-Perez et al., 2007), both RRs were required for proper responses to osmotic stress, whether ypdA is conditional or not.
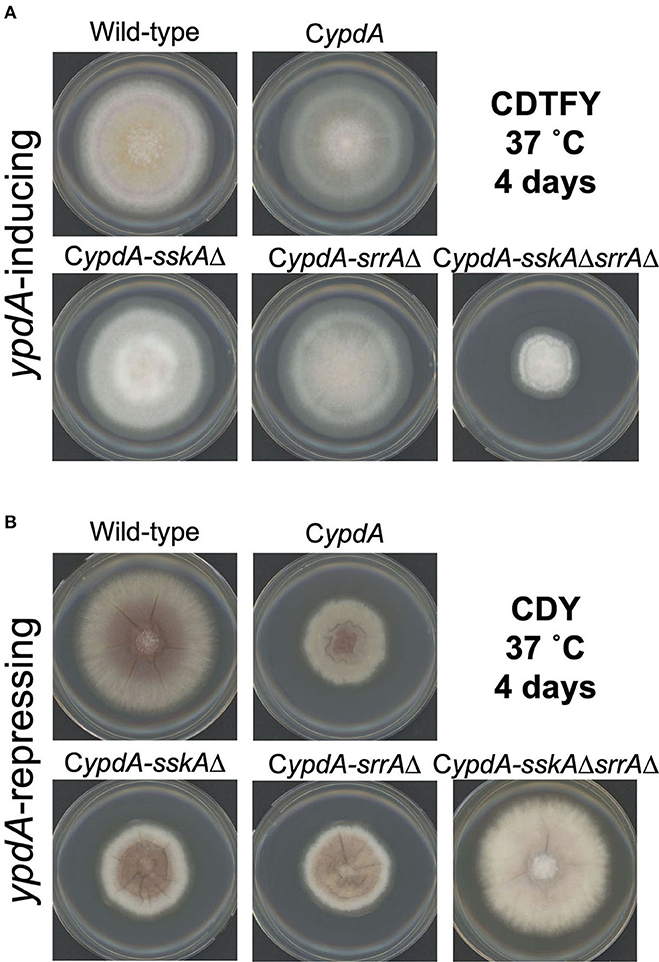
Figure 4. Mycelial growth of Aspergillus nidulans wild-type, CypdA, and its derivative strains on ypdA-inducing and -repressing plates. Conidia (1 × 104) of each strain were inoculated at the center of agar plates with ypdA-inducing CDTFY medium (A) or ypdA-repressing CDY medium (B) and incubated at 37°C for 4 days.
Next, we performed detailed microscopic analysis of the growth defects of CypdA, CypdA-sskAΔ, CypdA-srrAΔ, and CypdA-sskAΔsrrAΔ (Figures 5A,B). We compared the average lengths of hyphal cells and distance between septa among the wild-type, CypdA, and CypdA-derivative strains (n = 100) (Figure 5C, Supplementary Table 5). Average lengths of hyphal cells were significantly shorter after transfer to ypdA-repressing medium than those before the transfer in CypdA and CypdA-srrAΔ, but was not significantly altered in the wild-type and CypdA-sskAΔ strains (Figure 5C). Hyphal cell length was the same or slightly higher in CypdA-sskAΔsrrAΔ (Figure 5C, Supplementary Table 5). In addition to being shorter, cells of the CypdA and CypdA-srrAΔ strains were balloon-shaped under ypdA-repressing conditions, bulging outward between septa (Figure 5B).
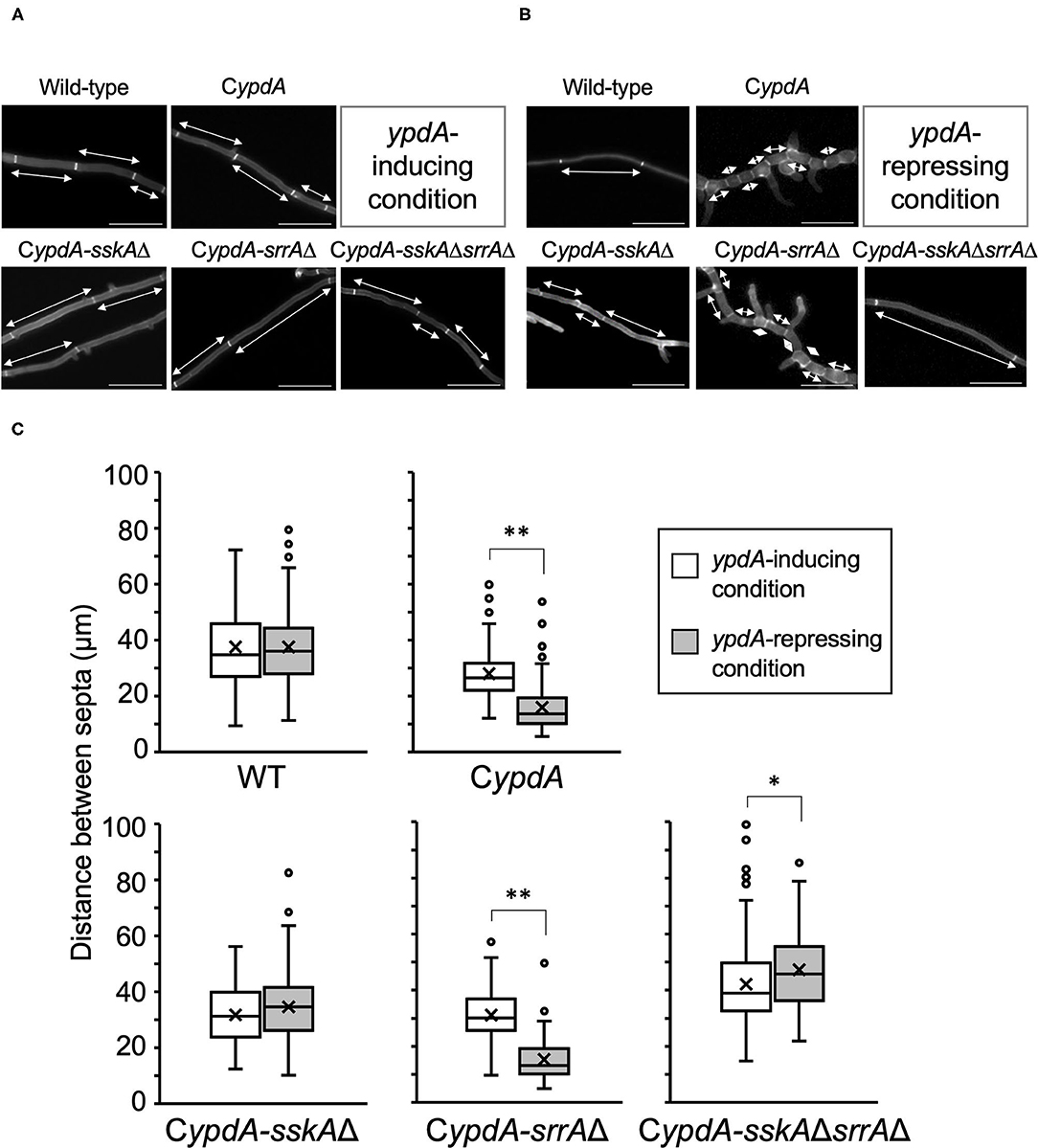
Figure 5. Hyphal morphology (A,B) and distance between septa (C) of the wild-type, CypdA, and its derivative strains under ypdA-inducing and -repressing conditions. Each strain was cultured in liquid ypdA-inducing CDTFY (A) or ypdA-repressing CDY (B) medium and stained with Calcofluor White. Double-headed arrows indicate distance between septa. Scale bars, 35 μm. (C) White and gray boxes indicate the distribution of distances between septa under the ypdA-inducing and ypdA-repressing conditions, respectively. Lines in boxes indicate medians, and crosses indicate averages. White circles indicate outliers. Error bars represent the range of maximum and minimum values excluding the outliers. *P < 0.05, **P < 0.01; Welch's t-test.
Disorders of Nuclear Distribution Caused by ypdA Downregulation
Because the average length of hyphal cells of CypdA and CypdA-srrAΔ was significantly shorter under ypdA downregulation, we examined the distribution of nuclei between septa in the wild-type, CypdA, and CypdA-derivatives under the ypdA-inducing and -repressing conditions by using an enhanced green fluorescent protein fused histone H2B (H2B-EGFP) expressing strains (Supplementary Figure 7, Figure 6). The number of nuclei between septa was similar in the wild-type and CypdA-sskAΔ strains regardless of culture conditions (Figure 6). In contrast, the number of nuclei between septa of hyphae grown after transfer to the repressing medium was significantly lower than before the transfer in the CypdA and CypdA-srrAΔ strains (Figure 6, Supplementary Table 6). In these two strains, the median number of nuclei between septa decreased (from 4 to 2 in both strains) (Figures 6C,D, Supplementary Table 6). In the CypdA-sskAΔsrrAΔ strain, the nuclear number of hyphae grown after transfer to the repressing medium was slightly greater than before the transfer (Figures 6C,D, Supplementary Table 6). These results suggest that the decrease in nuclear number caused by ypdA downregulation is SskA-dependent.
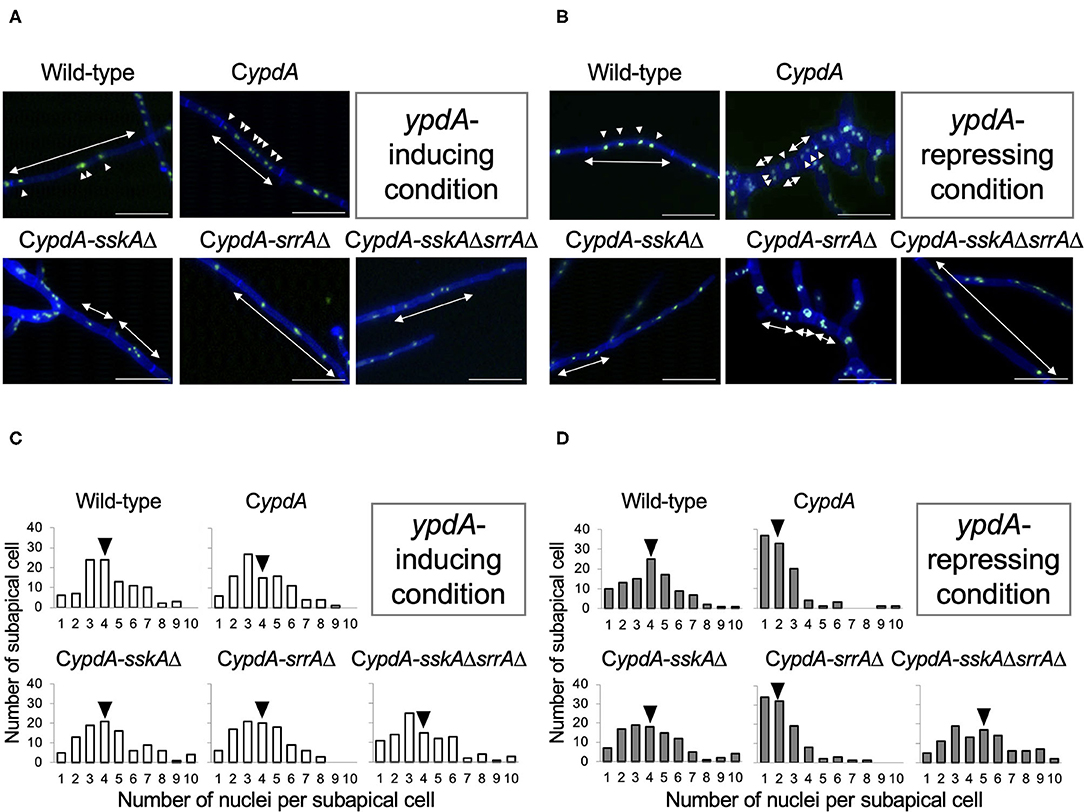
Figure 6. Fluorescent observation of nuclei (A,B) and distribution of nuclei between septa (C,D) in the wild-type, CypdA, and its derivative strains under ypdA-inducing and -repressing conditions. (A,B) Nuclei were visualized by using GFP-fused histone H2B under ypdA-inducing (A) and -repressing (B) conditions. Double-headed arrows indicate distance between septa. Arrowheads indicate nuclei. Scale bars, 35 μm. (C,D) Nuclei between septa were counted in 100 cells in each strain under ypdA-inducing (C) and -repressing (D) conditions. Arrowheads indicate the median.
To clarify how the timing of septal formation is related to the reduction in nuclear number, we analyzed the number of septa per germling newly formed in 4 h on either ypdA-inducing or -repressing medium after transfer from ypdA-inducing medium (n = 100) (Figure 7, Supplementary Table 7). In the wild-type strain, the number of septa formed after transfer to either medium was not significantly different. In contrast, in CypdA and CypdA-srrAΔ the number of septa formed after transfer to ypdA-repressing medium was significantly higher than that after transfer to ypdA-inducing medium, and in CypdA-sskAΔ and CypdA-sskAΔsrrAΔ the number formed after transfer to ypdA-repressing medium was significantly lower than that after transfer to ypdA-inducing medium (Figure 7). These observations suggest that the decreases in hyphal length and number of nuclei between septa are caused by accelerated septum formation in response to ypdA downregulation.
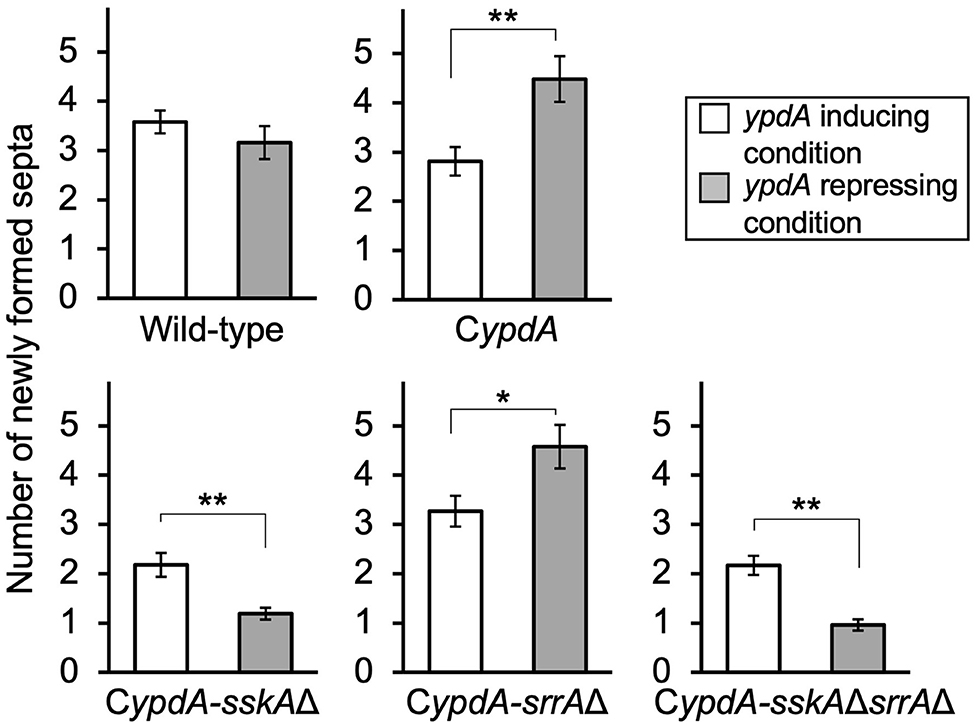
Figure 7. Number of newly formed septa after medium transfer in the wild-type and CypdA strains. The number of newly formed septa in mycelia germinated from a single conidium was calculated by subtracting the average number of septa produced during preculture (CDTFY at 37°C for 10 h, ypdA-inducing) from that of septa counted after incubation for 4 h after transfer to ypdA-inducing or -repressing medium (n = 100 mycelia for each strain in each medium). Error bars represent the standard errors of the mean. *P < 0.05, **P < 0.01; Welch's t-test.
Fludioxonil Has the Same Effects as ypdA Downregulation
Fludioxonil is an inhibitor of group III HKs (Catlett et al., 2003) in fungi; the HAMP domain is thought to be the target of fludioxonil (Ochiai et al., 2001, Yoshimi et al., 2004). Fludioxonil treatment of A. nidulans wild-type strains leads to activation of the HOG MAP kinase cascade via inhibition of the HK NikA. Therefore, fludioxonil treatment leads to constitutive activation of HogA as well as ypdA downregulation in A. nidulans (Lara-Rojas et al., 2011; Figure 3). When wild-type cells were exposed to fludioxonil, they showed changes similar to those caused by ypdA repression, depending on the fludioxonil concentration (Figures 6B, 8). No obvious changes in cell length and number of nuclei between septa were observed after treatment with 0.01 μg/ml fludioxonil for 16 h, but cells became shorter, with a balloon-like shape and fewer nuclei between septa, after treatment with 0.04 μg/ml fludioxonil for 16 h (Figure 8, Supplementary Table 8). Balloon-shaped cells and fragmented nuclei were observed in some cells after treatment with 0.1 μg/ml fludioxonil for 6 or 16 h (Figure 8, Supplementary Table 8). Treatment with 0.1 μg/ml fludioxonil increased the percentage of cells without nuclei (Supplementary Table 8). In the CypdA and CypdA-srrAΔ strains, treatment with 0.1 μg/ml fludioxonil for 16 h reduced the number of nuclei between septa and fragmented nuclei, but the percentage of cells with fragmented nuclei was reduced in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains (Figure 9, Supplementary Table 9). At 16 h after transfer to ypdA-repressing medium, the CypdA strain showed reduced numbers of nuclei and fragmented nuclei (Supplementary Figure 8, Supplementary Table 10). This phenomenon was also observed in the CypdA-srrA strain under ypdA repression, but no morphological changes in nuclei were observed in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains (Supplementary Figure 8, Supplementary Table 10). The CypdA-sskAΔ strain showed reduced septum formation compared to the CypdA-srrAΔ strain in the presence of fludioxonil (Supplementary Figure 9) or ypdA repression (Figure 7). These observations suggest that fludioxonil treatment produced the same effects as ypdA downregulation, in an SskA–dependent manner. As in the case of the sskAΔsrrAΔ strain, the CypdA-sskAΔsrrAΔ strain grew vigorously even in the presence of fludioxonil (Supplementary Figure 10), which is consistent with the restoration of its growth under ypdA-repressing conditions (Figure 4).
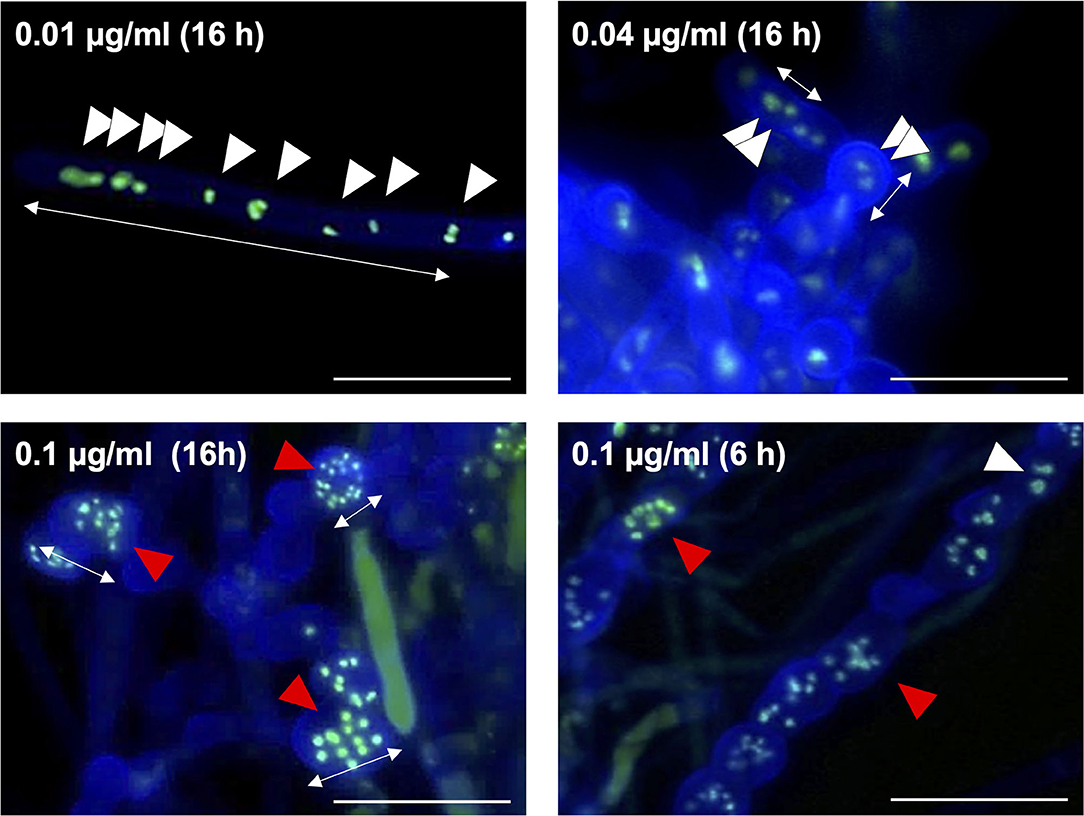
Figure 8. Morphology of hyphae and nuclei after treatment with fludioxonil in the wild-type strain. Conidia (1 × 107 cells) were incubated in liquid CDTFY medium (67 ml) for 20 h at 37°C. Mycelia were transferred to CDTFY medium with the indicated concentrations of fludioxonil and incubated for 6 or 16 h at 37°C. Lines with arrows indicate distance between septa. White arrowheads indicate nuclei. Red arrowheads indicate fragmented nuclei. Scale bars, 35 μm.
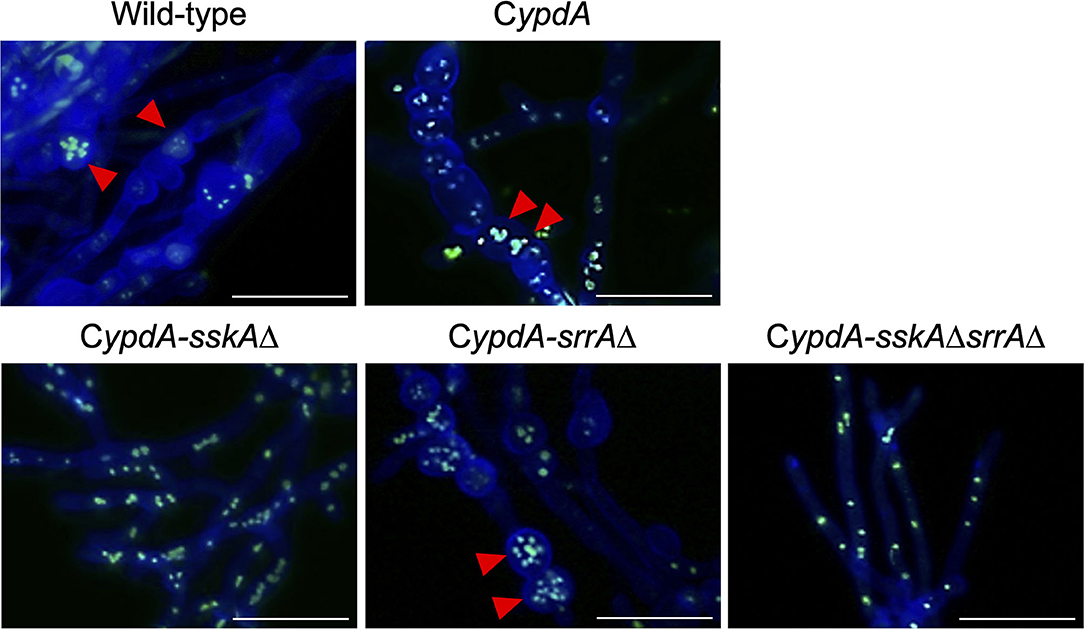
Figure 9. Morphology of hyphae and nuclei under ypdA-inducing + 0.1 μg/ml fludioxonil conditions in the wild-type and CypdA strains. Strains were cultured for 20 h in liquid CDTFY (ypdA-inducing) medium at 37°C, then transferred to CDTFY medium with fludioxonil (0.1 μg/ml) and incubated for 16 h at 37°C. Red arrowheads indicate fragmented nuclei. Bars, 35 μm.
Vacuole Development Caused by ypdA Downregulation and Fludioxonil Treatment
Since ypdA downregulation increased the ratio of cells with reduced numbers of nuclei in the CypdA and CypdA-srrAΔ strains, we microscopically observed the intracellular membranes by FM4-64 staining. All CypdA derivatives and the wild-type strain showed normal size of vacuoles under ypdA-inducing conditions (Figure 10A). Downregulation of ypdA transcription resulted in formation of many large vacuoles in the CypdA and CypdA-srrAΔ strains, but not in the CypdA-sskAΔ or CypdA-sskAΔsrrAΔ strains (Figure 10B). We also examined whether fludioxonil treatment of A. nidulans induces formation of large vacuoles (Figure 11). Under ypdA induction, fludioxonil treatment led to formation of many large vacuoles in the wild-type, CypdA, and CypdA-srrAΔ strains but not in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains (Figure 11). These results suggest that fludioxonil treatment and ypdA downregulation increased the formation of large vacuoles in a HOG pathway–dependent manner.
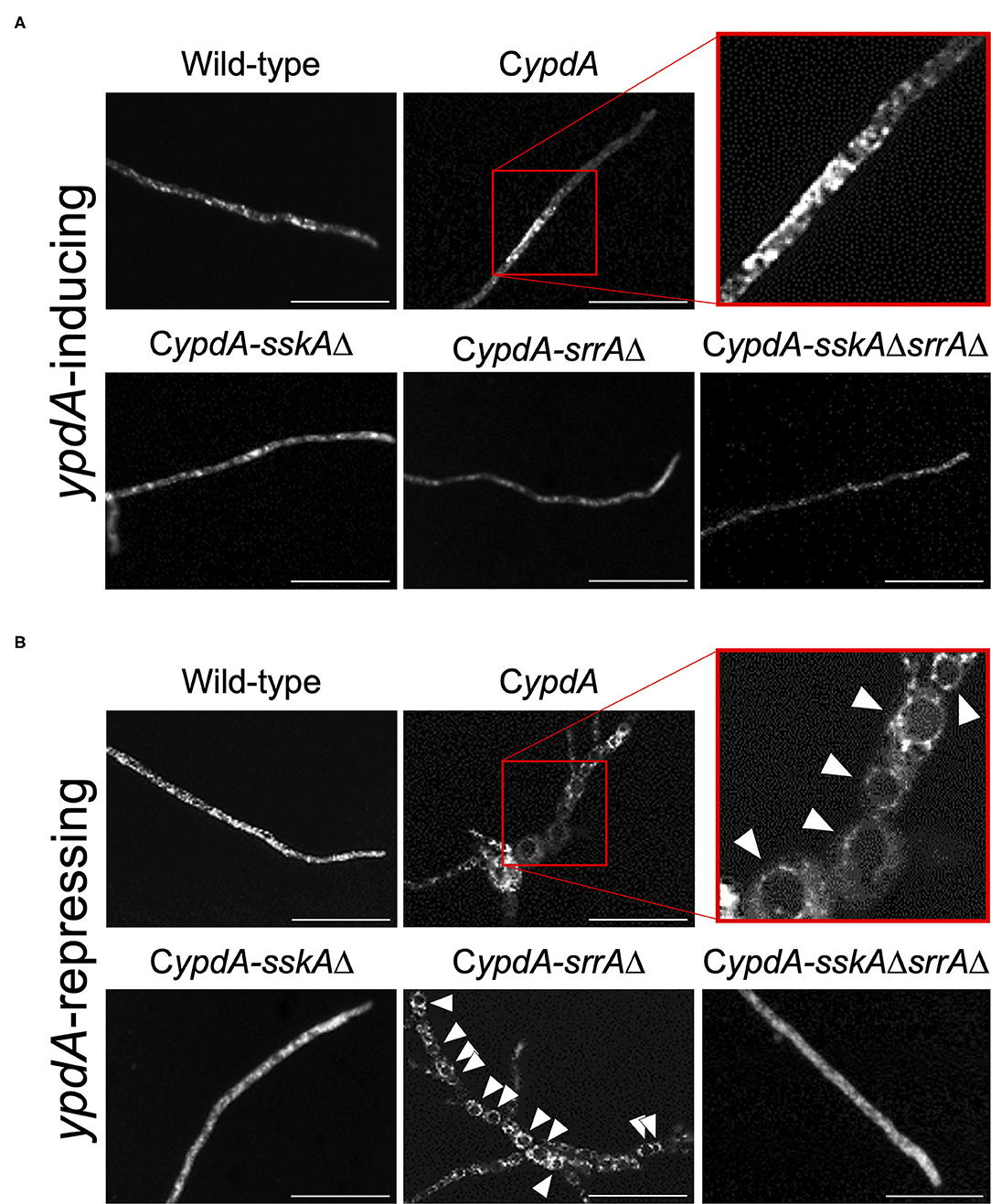
Figure 10. Vacuole morphology of the wild-type and CypdA strains under ypdA-inducing and -repressing conditions. Conidia of each strain were cultured in CDTFY (ypdA-inducing) medium for 8 h at 37°C, then transferred to CDTFY (A, ypdA-inducing) or CDY (B, ypdA-repressing) conditions and incubated for 4 h at 37°C. After incubation, mycelia were stained with FM4-64 and vacuoles were observed by fluorescence microscopy. Arrowheads indicate vacuoles. Bars, 35 μm.
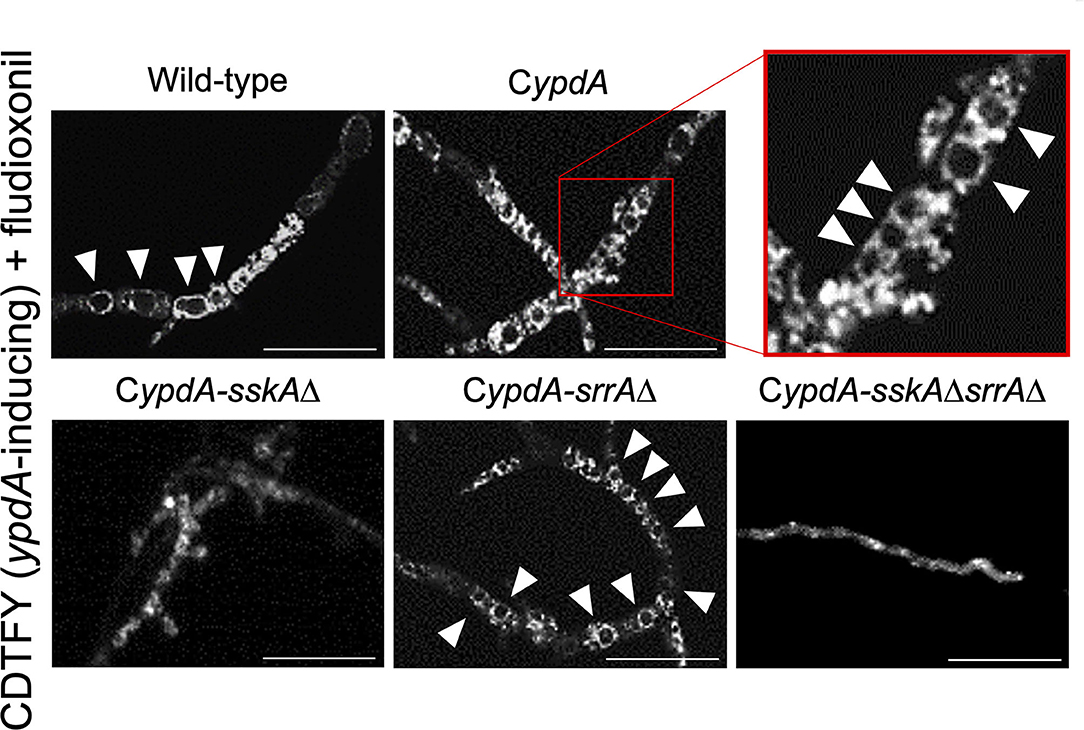
Figure 11. Vacuole morphology of the wild-type and CypdA strains treated with fludioxonil under ypdA-inducing conditions. Conidia of each strain were cultured in CDTFY (ypdA-inducing) medium for 8 h at 37°C, then transferred to CDTFY with 0.1 μg/ml fludioxonil and incubated for 4 h at 37°C. After incubation, mycelia were stained with FM4-64 and vacuoles were observed by fluorescence microscopy. Arrowheads indicate vacuoles. Bars, 35 μm.
To investigate whether vacuole development and reduction in the number of nuclei were related to autophagy, we constructed strains with the deletion of atgH, which encodes an autophagy-related ubiquitin-like protein (Shpilka et al., 2011), using the wild-type and CypdA strains (atgHΔ and CypdA-atgHΔ, respectively; Supplementary Figure 11). Under ypdA repression, the CypdA-atgHΔ strain showed fewer, smaller vacuoles than the CypdA strain, although cells of the CypdA-atgHΔ strain were balloon-like (Figure 12A, Supplementary Table 11). Cell shape remained abnormal, but atgH disruption also abrogated formation of the large vacuoles induced by fludioxonil treatment (Figure 12B, Supplementary Table 11). In addition, no significant differences were observed in fludioxonil sensitivity in the wild-type and atgHΔ strains (Supplementary Figure 12). These results suggest that the abnormal development of vacuoles is not directly related to the cell defects induced by fludioxonil treatment and ypdA-downregulation.
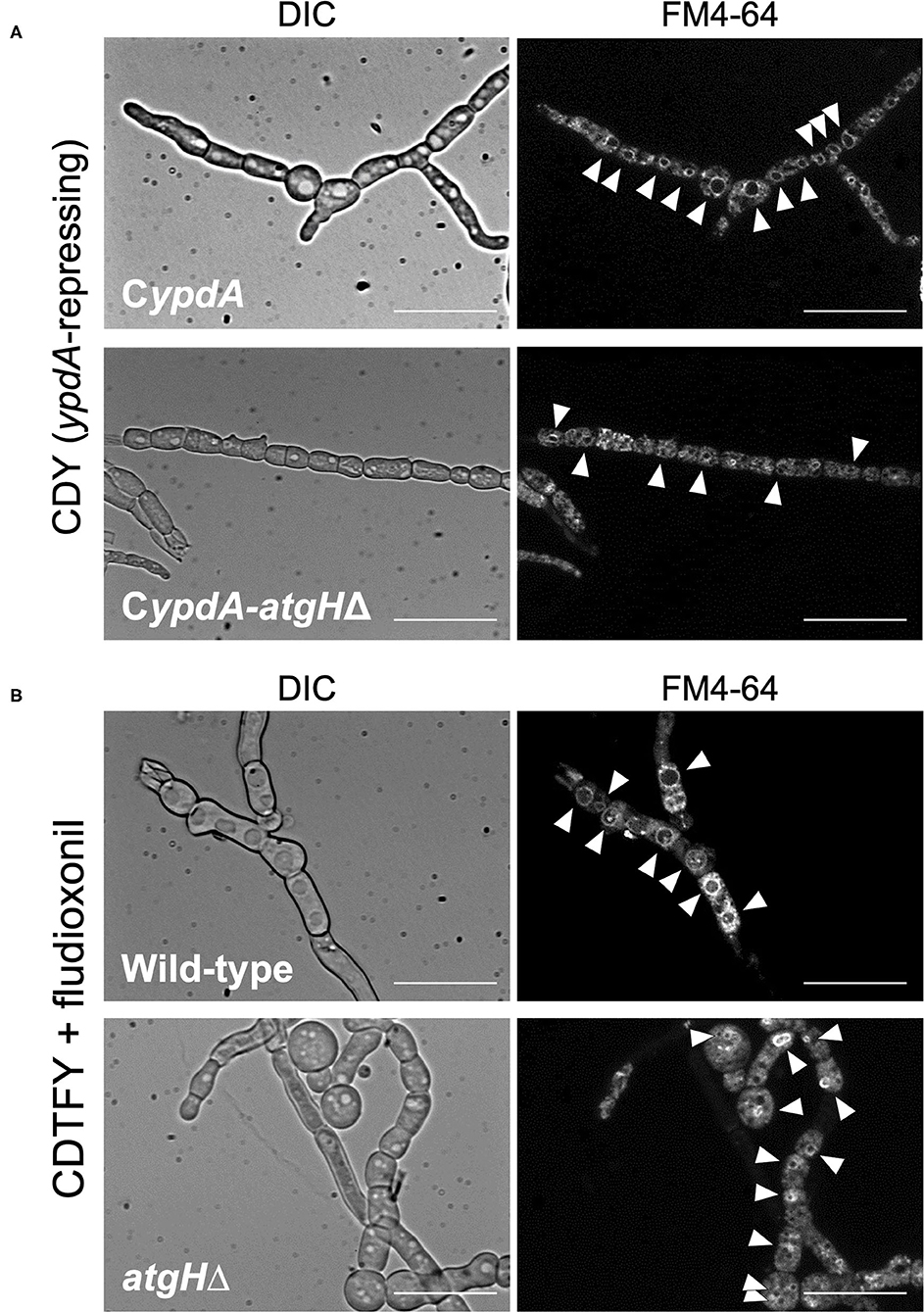
Figure 12. Vacuole morphology of atgH-disrupted strains derived from wild-type and CypdA strains under the ypdA-repressing condition and fludioxonil treatment. (A) Conidia of the CypdA and CypdA-atgHΔ strains were cultured in CDTFY (ypdA-inducing) medium for 8 h at 37°C. Mycelia were transferred to CDY medium (ypdA-repressing) and incubated for 4 h at 37°C. Mycelia were stained with FM4-64 and vacuoles were observed by fluorescence microscopy. (B) Conidia of the wild-type and atgHΔ strains were cultured in CDTFY medium for 8 h at 37°C. Mycelia were transferred to CDTFY with 0.1 μg/ml fludioxonil and incubated for 4 h at 37°C. Mycelia were stained with FM4-64 and vacuoles were observed by fluorescence microscopy. Arrowheads indicate vacuoles. Bars, 35 μm.
Discussion
In this study, we demonstrated that downregulation of ypdA transcription causes severe growth defects and abnormal hyphal morphology in A. nidulans. These results support the notion that ypdA is an essential gene, although the molecular mechanism(s) underlying the lethality caused by downregulation of ypdA remain(s) unclear. In the present study, we significantly downregulated intracellular levels of YpdA protein by downregulating ypdA transcription in CypdA and its derivatives CypdA-sskAΔ, CypdA-srrAΔ, and CypdA-sskAΔsrrAΔ (Figure 1). Loss of YpdA increased the levels of total and phosphorylated HogA in the CypdA and CypdA-srrAΔ strains (Figure 3A), which reflected hyper-activation of HOG MAPK signaling. The activation of HOG MAPK signaling was maintained for at least 36 h, and the transcription of the HOG pathway–regulated gene catA (Figure 2) and hogA itself (Figures 3B,C) were persistently upregulated. The increased level of hogA transcript reflected the high levels of HogA protein (Figure 3A). Taking into consideration that the hogA expression level was lower in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains in ypdA-repressing medium (CDY), these results suggested that the transcription of hogA was regulated by the HOG pathway via autoregulation. Although autoregulation of MAP kinase gene expression has been reported in the cell wall integrity signaling pathway in A. nidulans (Fujioka et al., 2007), this is the first clear demonstration of the autoregulation of the HOG pathway in filamentous fungi.
The growth defects caused by ypdA downregulation were restored by double disruption of the SskA and SrrA pathways, but not by disruption of either pathway alone (Figure 4B). This result strongly suggests that the lethality caused by downregulation of ypdA is not dependent on either the SskA or SrrA pathway alone, but on both pathways. In this respect, our findings differ from those in S. cerevisiae (Posas et al., 1996). Downregulation of S. cerevisiae YPD1 results in lethal, constitutive activation of the Hog1 MAPK cascade, which is almost completely suppressed by deletion of SSK1, HOG1, or PBS2 (Posas et al., 1996). Consequently, the lethality caused by downregulation of YPD1 expression fully depends on the Hog1 MAPK cascade in S. cerevisiae. Interestingly, even under ypdA-inducing conditions, severe growth retardation was observed only in the CypdA-sskAΔsrrAΔ strain (Figure 4A). Thus, the activation of one response regulator appears to be sufficient for the phosphate group on YpdA to be processed at normal levels. However, deletion of both response regulators may hamper phosphate group transfer, leading to excessive accumulation of phosphorylated YpdA and to severe growth retardation in the CypdA-sskAΔsrrAΔ strain. Further study is needed to elucidate the relationship between over-accumulation of phosphorylated YpdA and cell toxicity.
In filamentous fungi including aspergilli, N. crassa, and most plant pathogens, fungicides such as fludioxonil and iprodione are thought to target the His-Asp phosphorelay system (Ochiai et al., 2001; Yoshimi et al., 2004; Motoyama et al., 2005; Viaud et al., 2006; Hagiwara et al., 2007b). The growth-inhibitory effects of the fungicides have been shown to depend on group III HKs, deletion of which confers resistance. In A. nidulans, nikA encodes a group III HK, and we previously showed that a nikA deletion mutant is resistant to fludioxonil (Hagiwara et al., 2007b). The sskAΔsrrAΔ and hogAΔsrrAΔ double mutants are also resistant to fludioxonil (Hagiwara et al., 2007b). The growth defects caused by fludioxonil treatment or by ypdA downregulation are both restored by disruption of both the SskA and SrrA pathways, in a similar manner. These observations indicate that the growth inhibitory-effects of the fungicide and ypdA downregulation may be caused by disruption of His-Asp phosphorelay signaling, in which signals are transmitted from NikA to RRs (SskA and SrrA). In fact, the HogA MAPK cascade is abnormally activated by fludioxonil treatment (Furukawa et al., 2007) and ypdA downregulation (Figure 3A), both of which depends on SskA pathways (Hagiwara et al., 2007a, 2009; Figure 3A).
The abnormal cell morphology, with shorter, balloon-shaped cells caused by YpdA repression, was observed in the CypdA and CypdA-srrAΔ strains (Figure 5B). However, in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains, in which sskA was disrupted, cell length was restored to that of the wild-type strain (Figure 5), indicating that the abnormalities in cell morphology are dependent on the SskA pathway. This suggested that septum formation was regulated by a pathway downstream of SskA, for example, the HogA pathway. Phenotypes similar to the septum defects observed in the CypdAΔ and CypdA-srrAΔ strains have been reported in a study of the two-component system in the fission yeast Schizosaccharomyces pombe (Shieh et al., 1997; Shiozaki et al., 1997). Mcs4, an ortholog of SskA in A. nidulans, regulates mitotic initiation in a manner dependent on Sty1, an ortholog of HogA in A. nidulans. Cells of the mcs4Δ mutant are longer than those of the wild-type strain, a result of the delay in septum formation caused by G2 arrest, which prevents transition to the M phase. Conversely, deletion of spy1, which encodes an ortholog of YpdA, can grow but leads to reduced cell length, caused by enhanced transition to M phase. However, in the mcs4Δspy1Δ strain, cell length is restored to that of the mcs4Δ strain (Aoyama et al., 2000). Given the findings in S. pombe and the results of the present study, HogA may also regulate the transcription of cell cycle–related genes via its target transcription factor in A. nidulans.
The number of nuclei per cell was significantly decreased in the CypdA and CypdA-srrAΔ strains under the ypdA-repressing condition (Figures 6C,D). In the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains, the number of nuclei per cell was restored to that of the wild-type strain (Figures 6C,D), indicating that nuclear distribution in cells may be dependent on the SskA pathway. This suggested that nuclear distribution, as well as septum formation, is regulated by a pathway downstream of SskA. When hyphal cells of the wild-type strain were treated with 0.1 μg/ml fludioxonil for 16 h, nuclear fragmentation was observed (Figure 8). When the fludioxonil concentration or treatment time was reduced, nuclear fragmentation was suppressed and the number of nuclei per cell tended to decrease in the wild-type strain (Figure 8). In addition, when hyphal cells of the CypdA and CypdA-srrAΔ strains were treated with 0.1 μg/ml fludioxonil for 16 h under the ypdA-inducing condition, cells with reduced numbers of nuclei and fragmented nuclei were observed (Figure 9). Abnormal nuclear morphology was not observed in cells of the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains under the ypdA-inducing condition, indicating that the abnormal nuclear morphology is dependent on the SskA pathway.
Comparison of the cellular alterations caused by fludioxonil and ypdA repression showed that abnormal cells with reduced and fragmented nuclei were present in the CypdA and CypdA-srrAΔ strains under both conditions (Figure 8, Supplementary Figure 8). However, no morphological changes in nuclei were observed in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains under the ypdA-repressing condition (Supplementary Figure 8). Thus, fragmentation and decreased numbers of nuclei are common to the ypdA-repressing and fludioxonil treatments, in an SskA pathway–dependent manner.
The number of newly formed septa was not significantly different in the wild-type strain at 4 h after transfer to the ypdA-repressing condition, compared to the inducing condition (Figure 7). However, the CypdA and CypdA-srrAΔ strains showed an increased number of septa after transfer to the ypdA-repressing condition (Figure 7). This result suggested that the decreases in cell length between septa and in nuclear number, observed in the ypdA-repressing condition, were caused by enhanced septum formation. The CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains showed reduced septum formation in a certain period (4 h) compared to the CypdA-srrAΔ strain in response to ypdA repression (Figure 7), which suggests delayed formation of each septum in the strains. The delayed septum formation in the CypdA-sskAΔ and CypdA-sskAΔsrrAΔ strains suggests that septum formation is regulated in an SskA pathway–dependent manner. In studies on regulators of septum formation in A. nidulans, responses such as enhanced and delayed septum formation have been reported. ParA, a regulatory subunit of protein phosphatase 2A in A. nidulans, negatively regulates septation by dephosphorylation of septation initiation network proteins. Disruption of parA in A. nidulans enhances septum formation, and overexpression of parA leads to cessation of septum formation (Zhong et al., 2014). MztA was identified by RNA-seq and genetic analysis as a positive regulator of septum formation that acted antagonistically to ParA in A. nidulans (Jiang et al., 2018). Septum formation is delayed in the A. nidulans mztA disruptant, and the phenotypes of the parA and mztA disruptants are similar to the phenotypes observed in this study, with either delayed or accelerated septum formation. Since expression of genes involved in the regulation of septum formation, such as parA and mztA, may be altered in strains with abnormal septum formation under ypdA-repressing conditions, the relationship between changes in their expression and YpdA suppression should be clarified in future studies.
In response to fludioxonil, the CypdA-sskAΔ strain showed delayed septum formation compared to CypdA-srrAΔ (Supplementary Figure 9), as it did to ypdA repression (Figure 7). On medium containing 0.1 μg/ml fludioxonil, the CypdA-sskAΔ strain was strongly inhibited, but grew at a slightly faster than the CypdA and CypdA-srrAΔ strains, whereas the CypdA-sskAΔsrrAΔ grew well (Supplementary Figure 10). Fludioxonil treatment caused growth defects in the CypdA-sskAΔ strain without reducing hyphal length between septa. Thus, nuclear fragmentation may be one of the lethal factors, but there may also be SrrA pathway–dependent lethal factor(s). Although signaling downstream of SrrA is still obscure, YpdA was shown in our study to play a significant role in A. nidulans His-Asp phosphorelay signaling with respect to growth and morphology, besides regulation of the HogA pathway.
In the wild-type, CypdA, and CypdA-derivative strains, large vacuoles were not observed under the ypdA-inducing condition (Figure 10A). However, large vacuoles developed in the CypdA and CypdA-srrAΔ strains under the ypdA-repressing condition (Figure 10B). In response to fludioxonil, large vacuoles also developed in the wild-type, CypdA, and CypdA-srrAΔ strains (Figure 11). Vacuole development was not observed in CypdA-sskAΔ and CypdA-sskAΔsrrAΔ, even under the ypdA-repressing condition or fludioxonil treatment (Figures 10B, 11). Therefore, vacuole formation, in addition to septum formation and nuclear distribution, also appears to be regulated in an SskA pathway–dependent manner in response to either ypdA repression or fludioxonil treatment. In the CypdA and CypdA-srrAΔ strains, in which large vacuoles were observed under the ypdA-repressing condition, HogA MAP kinase was abnormally activated (Figure 3A). The HOG-MAP kinase cascade regulates glycerol biosynthetic genes in A. nidulans (Fillinger et al., 2001; Han and Prade, 2002). Therefore, we hypothesized that in the CypdA and CypdA-srrAΔ strains glycerol abnormally accumulated in the cytoplasm due to activation of HogA, and that vacuoles developed abnormally in response. This will need to be investigated in future studies.
The development of abnormally large vacuoles observed in the CypdA strain under the ypdA-repressing condition was suppressed in the CypdA-atgHΔ strain (Figure 12A). In response to fludioxonil, the development of abnormally large vacuoles was observed in the wild-type strain, whereas no vacuole development was observed in the atgHΔ strain. This suggests that AtgH may be involved in formation of abnormal vacuoles in response to fludioxonil in the wild-type and under ypdA repression in the CypdA strain. Although abnormal vacuole development was suppressed by atgH disruption, development of balloon-like cells was not (Figure 12). This indicates that abnormal development of vacuoles is not a direct cause of the balloon-like cells. We hypothesized that abnormal development of vacuoles would enhance autophagy, resulting in autophagic cell death. If this hypothesis is correct, the atgHΔ strain, in which vacuoles do not develop abnormally, should be resistant to fludioxonil. However, disruption of atgH did not alter fludioxonil sensitivity (Supplementary Figure 12). Thus, abnormal development of vacuoles does not appear to be directly related to the lethality caused by ypdA-repressing and fludioxonil treatment.
Fludioxonil may convert group III HKs into phosphatases (Lawry et al., 2017). In S. cerevisiae expressing Drk1, a group III HK from B. dermatitidis, Drk1 behaved as a phosphatase rather than as a kinase in response to fludioxonil; it dephosphorylated its downstream target, Ypd1, resulting in constitutive activation of the HOG pathway (Lawry et al., 2017). Our observations of constitutive activation of HOG pathway in response to ypdA downregulation or fludioxonil treatment, due to reduced YpdA activity and consequent perturbation of the downstream signaling pathway, are consistent with this report. The conversion of group III HKs into phosphatases by fludioxonil might also help to explain mycelial growth in the CypdA-sskAΔsrrAΔ strain on the media containing fludioxonil: in the absence of both response regulators, the phosphate group might be removed from excessively accumulated phosphorylated YpdA by fludioxonil-converted HKs, resulting in vigorous growth of this strain (Supplementary Figure 10). Investigating this hypothesis might be the object of a future study. In addition, the levels of Ypd1 protein are reportedly not affected by fludioxonil treatment (Lawry et al., 2017). Hence, fludioxonil may inhibit growth by causing HK to function as a phosphatase rather than by inhibiting YpdA expression. This would lead to abnormal activation of the HOG pathway, similar to the effects of ypdA downregulation. Thus, the YpdA function presented here, the growth-inhibitory effects of fludioxonil, and the lethality of downregulation of ypdA may act via similar or identical mechanisms, as summarized graphically in Figure 13. Additional studies, including analysis of pathways downstream of SrrA, are needed to elucidate the mechanism underlying the mode of action of phenylpyrrole- or dicarboximide-type fungicides.
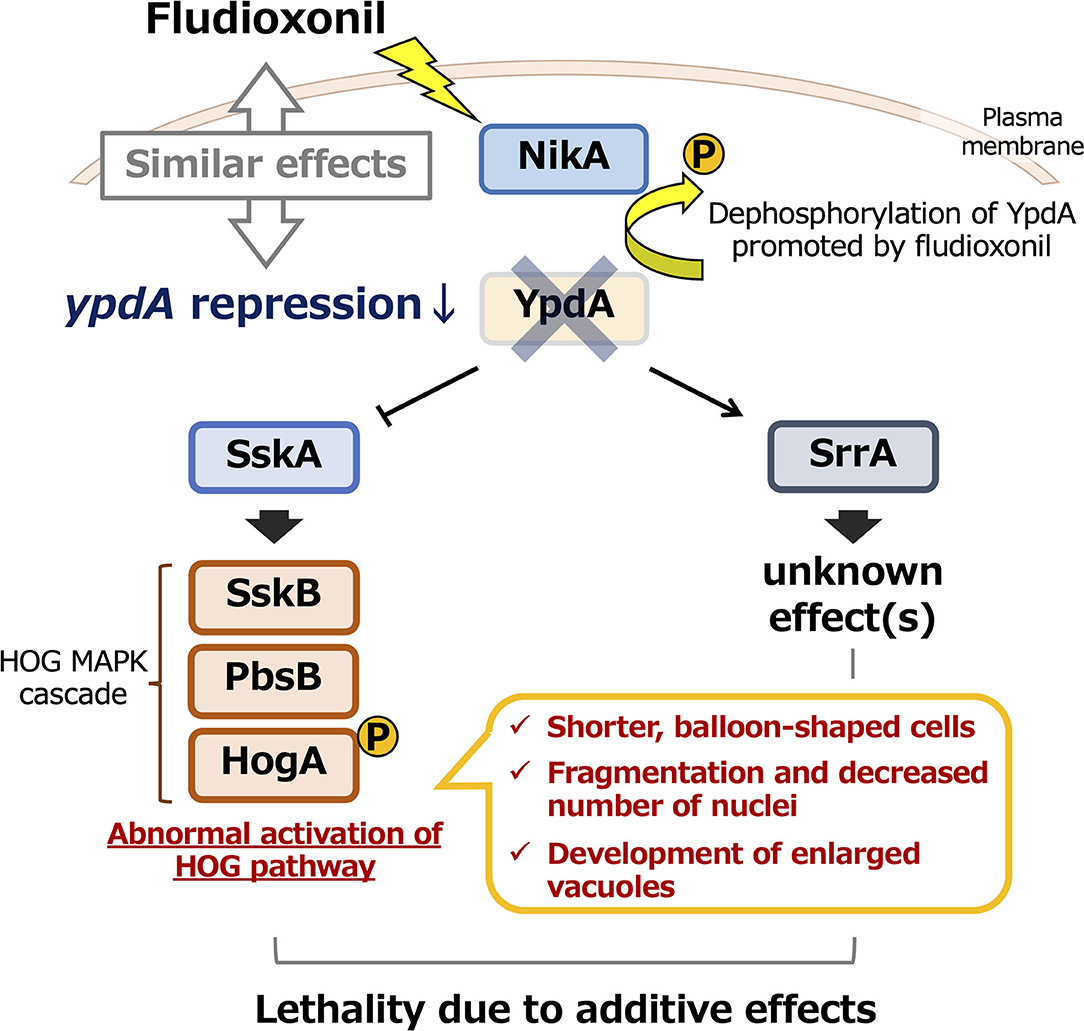
Figure 13. Model of the effects of ypdA downregulation and fludioxonil treatment on two-component signaling in Aspergillus nidulans. Downregulation of ypdA and fludioxonil treatment have similar effects. Although morphological abnormalities depend only on the SskA pathway, the lethality caused by YpdA suppression or fludioxonil treatment depends on both the SskA and SrrA pathways.
Materials and Methods
Strains, Media, and Transformation
The fungal strains used in this study are listed in Table 1. The A. nidulans ABPU1 (ΔligD) strain was used as the control strain and for all genetic manipulations in the present study. For the A. nidulans conditional ypdA studies, ABPU1 cells were transformed with the gene-replacement constructs described in the “Construction of gene-replacement cassettes” subsection. Czapek–Dox (CD) medium was used for standard culture (Fujioka et al., 2007). A modified medium, designated CDTFY, CD medium containing 50 mM threonine, 0.2% (w/v) fructose, and 0.2% (w/v) yeast extract instead of 1% (w/v) glucose as a carbon source was used for up-regulation of alcA promoter–driven ypdA (Ichinomiya et al., 2002). For down-regulation of alcA promoter–driven ypdA, CD medium containing 2% (w/v) glucose and 0.2% (w/v) yeast extract (designated CDY) was used. Conidia were harvested in PBS + Tween 20 (0.1%, v/v) and counted in a hemacytometer. For mycelial growth tests, conidial suspensions of each strain (1 × 104 conidia) were spotted on the centers of CDTFY or CDY plates and incubated at 37°C for 4 days. For tests of sensitivity to fludioxonil, the same conidial suspensions were spotted on the centers of CDTFY plates containing fludioxonil (0.01, 0.05, 0.1, 0.5, 1.0 μg/ml) and incubated at 37°C for 4 days. The dose response was determined by plotting the mean diameters of the colonies on media with fludioxonil as a percentage of those on control medium (containing 0.1% DMSO as a solvent for fludioxonil). To investigate gene and protein expression, conidia (5 × 105 conidia/ml) were inoculated into liquid CDTFY medium and cultured at 37°C for 20 h. Mycelia were harvested, washed with sterile distilled water, transferred into liquid CDTFY or CDY medium, and cultured for 36 h. Mycelia were harvested at 12, 24, and 36 h. DNA extraction, RNA extraction, reverse transcriptase reactions, and fungal transformations were performed as described previously (Yoshimi et al., 2013).
Construction of Gene-Replacement Cassettes
The gene-replacement cassettes for the alcA promoter–driven ypdA strain (CypdA) and the sskA and srrA deletion mutants were constructed using a previously reported PCR-based fusion strategy (Yoshimi et al., 2013). The constructs for the H2B-EGFP expressing strains and the atgH deletion strains were constructed based on the in-fusion cloning technology as described previously (Miyazawa et al., 2018). The sequences of all primers used in this study are listed in Table 2. To construct the gene-replacement cassette for the CypdA strain, a fragment containing the 5′ non-coding region of ypdA (amplicon 1) and one including the open reading frame of ypdA (amplicon 2) were PCR amplified using A. nidulans ABPU1 genomic DNA as a template and primers listed in Table 2 (ypdA5′-F and ypdA5′-R for amplicon 1, and ypdA-F and ypdA-R for amplicon 2). In addition, the pyrG-alcA(p) cassette (Yoshimi et al., 2013), which contains the A. nidulans pyrG marker and the alcA promoter (originally derived from A. nidulans FGSC A89), was amplified by PCR using the primers pyrG-PalcA-F and pyrG-PalcA-R (amplicon 3; Table 2). These three fragments were used as the substrates for second-round PCR fusion. The resulting major PCR product was gel-purified and used to transform the ABPU1 strain.
To disrupt the genes in A. nidulans, we replaced them with either A. oryzae argB or pyroA, as a selectable marker. To disrupt sskA, a plasmid containing the 1.5- and 2-kb fragments upstream and downstream of the sskA ORF on both sides of the argB gene (Hagiwara et al., 2007a) was used as a PCR template, and the primers sskAU-U and sskAD-D (Table 2) were used for PCR amplification. The resulting sskA disruption cassette was used to transform the CypdA strain (CypdA-sskAΔ). To construct the disruption cassette for srrA in the CypdA strain, the same strategy was used. A plasmid containing the 1.5- and 2-kb fragments upstream and downstream of the srrA ORF on both sides of the argB gene (Hagiwara et al., 2007a) was used as a PCR template, with the primers srrAU-U and srrAD-D (Table 2). The resulting srrA disruption cassette was used to transform the CypdA strain (CypdA-srrAΔ). The sskAΔsrrAΔ double mutant was constructed by disruption of sskA in the CypdA-srrAΔ strain, using the sskA disruption cassette described above (CypdA-sskAΔsrrAΔ). Although sskA and srrA were disrupted by the same argB selectable marker, the CypdA-sskAΔsrrAΔ strain is highly resistant to the phenylpyrrole-type fungicide, fludioxonil, and this fungicide could be used for selection.
To construct the h2b-egfp-expressing strains, a fragment containing the promoter region of h2b and the coding region of h2b with egfp (amplicon 1) including the terminator Tnos3′ was PCR amplified using the plasmid pAH2BG (Maruyama et al., 2001) as a template and the primers, IF_H2B_Fw and IF_nos3′_Rv (Table 2). The gene fragment of pyroA (amplicon 2) and the 3′-flanking region of pyroA (amplicon 3) were PCR amplified using A. nidulans ABPU1 genomic DNA as a template and primers listed in Table 2 (IF-AnpyroA- Fw and IF-AnpyroA-Rv for amplicon 2, and IF_AN7726_Fw3 and IF_AN7726_Rv for amplicon 3). These three fragments were cloned into pUC19 using in-fusion cloning technology. The H2B-GFP expression cassette was obtained from the resulting plasmid pAH2BP by ApaI digestion (Supplementary Figure 7).
To disrupt atgH, A. oryzae argB (amplicon 1) was PCR-amplified using the plasmid for sskA disruption as a template and the primers AoargB-F and AoargB-R (Table 2). In addition, a fragment containing the 5′ non-coding region of atgH (amplicon 2) and the 3′ non-coding region of atgH (amplicon 3) were PCR-amplified using A. nidulans ABPU1 genomic DNA as a template and primers listed in Table 2 (ANatgH-LU and ANatgH-LL+Arg for amplicon 2, and ANatgH-RU+Arg and ANatgH-RL for amplicon 3). These three fragments were cloned into pUC19 using in-fusion cloning technology. The atgH replacement cassette was obtained by NotI digestion of the resulting plasmid pAAR-DatgH (Supplementary Figure 11).
Quantitative Real-Time PCR
Real-time PCR was performed using the Mini Opticon real-time PCR system (Bio-Rad Laboratories, Hercules, CA, USA) with SYBR Green detection, according to the manufacturer's instructions. The DyNAmo SYBR Green qPCR kit (Finnzymes Oy, Espoo, Finland) was used for reaction mixture preparation. Primer sets for quantifying the expression of the ypdA, catA, and hogA genes are listed in Table 2. Primer specificity was confirmed by melting curve analysis. A cDNA sample obtained from reverse transcription of an equivalent amount of total RNA was added to each reaction mixture. The gene encoding histone H2B was used as an internal control to calculate target gene expression ratios. Relative expression ratios were calculated essentially as in Hagiwara et al. (2008). A sample from ABPU1 cells before transfer to YPD medium (0 h) was used for calibration. Relative expression ratios were calculated by first calculating the threshold cycle changes in sample and calibrator as Δ = Ct(target) – Ct(histoneH2B) and Δ = Ct(target) – Ct(histoneH2B), followed by calculating the expression ratios in both sample and calibrator as Expression ratiossample = 2ΔCtsample and Expression ratioscalibrator = 2ΔCtcalibrator, and finally, Relative expression ratios = Expression ratiosample / Expression ratiocalibrator.
YpdA Antibody
For antibody production, we synthesized peptides corresponding to an internal region of YpdA (positions 61–74 and 159–172, AESTFDKMEKALED and TPSSPEESKKDAKA, respectively). These peptides were linked to a carrier protein and injected into a rabbit. The peptides and polyclonal antibody were prepared by Operon Biotechnology (Tokyo, Japan).
Western Blot Analysis
Conidia (final concentration, 1 × 105 conidia/ml) from the ABPU1 and CypdA strains were inoculated into the indicated liquid medium and cultured for 24 h. Mycelia were frozen in liquid nitrogen, ground to powder, and immediately resuspended in protein extraction buffer containing protease inhibitors (50 mM Tris-HCl [pH 7.5], 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.1 mM sodium vanadate, and a protease inhibitor cocktail [Roche Applied Science, Mannheim, Germany, 1 tablet per 50 ml]). The suspension was immediately boiled for 10 min, and cell debris were removed by centrifugation for 10 min at 16,873 × g. The protein concentration of the supernatant was determined using a reducing agent–compatible Pierce BCA protein assay kit (Pierce, Rockford, IL, USA). Total protein (15 μg) was loaded onto a 12.5% polyacrylamide SDS gel, blotted onto an Immobilon-P membrane (Millipore, Bedford, MA, USA), and processed for immunodetection of YpdA using the polyclonal antibody for YpdA, HogA using the anti-Hog1 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), phospho-HogA using the anti-phospho-p38 MAP kinase antibody (Cell Signaling Technology, Inc., Beverly, MA, USA), and histone-H2B using the anti-histone H2B antibody (Sigma-Aldrich Japan, Tokyo, Japan), and alkaline phosphatase–coupled anti-rabbit secondary antibody (GE Healthcare, Buckinghamshire, UK).
Microscopy
To analyze hyphal morphology and measure hyphal length between septa, conidia from the ABPU1 and CypdA strains were inoculated at a final concentration of 1.5 × 105 conidia/ml into liquid CDTFY and incubated statically at 37°C for 20 h (ypdA-inducing sample). To suppress the expression of ypdA, mycelia from the ypdA-inducing sample were transferred to a cell strainer (BD Biosciences, San Jose, CA, USA) and washed with CDY medium. The washed mycelia were transferred into CDY medium and incubated statically at 37°C for 16 h (ypdA-repressing sample). These ypdA-inducing or -repressing samples were washed with PBS (150 mM NaCl, 144 mM K2HPO4, 56 mM KH2PO4) and stained with Calcofluor white (0.1 μg/ml; MP Biomedicals, Inc., OH, USA) at room temperature for 10 min in the dark. After being washed with the same buffer, the samples were imaged under a FV1000-D IX81 confocal laser-scanning microscope (Olympus, Tokyo, Japan). Hyphal length between septa was determined using the Image J software (http://rsb.info.nih.gov/nih-image/about.html).
To analyze the number of nuclei per cell, samples of the ABPU1 and CypdA strains expressing h2b-egfp were prepared under ypdA-inducing or -repressing conditions, using the method described above. For fludioxonil treatment, mycelia of the ypdA-inducing samples cultured in CDTFY medium for 20 h were washed with CDTFY medium and transferred into CDTFY medium containing 0.01, 0.04, or 0.1 μg/ml fludioxonil at 37°C for appropriate time (6 or 16 h). If at least five nuclei were observed in a balloon-shaped cell, it was considered to contain fragmented nuclei.
To analyze the nuclei in the freshly grown mycelia under the ypdA-repressing condition, germinating conidia were cultured for 8 h, then collected and transferred to CDY medium. Mycelia were sampled after 3, 6, and 9 h, and the nuclei were observed under the microscope.
Statistical Analysis
Welch's t-test was used for the comparison of paired samples. The effects of two variables (strain and time) on the expression of the ypdA, catA, and hogA genes were analyzed by two-way ANOVA. Tukey–Kramer's test was used to compare multiple samples. If the treatment groups were compared with only the control group, Dunnett's test was used.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AY, DH, KF, and KA conceived and designed the experiments. YM constructed the mutant strains. YF performed transcriptional and western-blot analyses. MO performed phenotypic analysis of the mutant strains. JMaruy provided the plasmid pAH2BG and supported the nuclear distribution experiment. AY, TF, OM, NS, JMarui, YY, TN, and CT supported the experiments. AY analyzed the data. AY, KM, and KA wrote the paper. KA supervised this research and acquired funding. All authors discussed the results and commented on the manuscript.
Funding
This study was supported by a grant from the Bio-oriented Technology Research Advancement Institution (BRAIN). The work was also supported in part by a grant-in-aid for scientific research on a priority area (applied genome; no. 17019001) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. It was also supported by a Grant-in-Aid for Scientific Research (C) (18K05384) from the Japan Society for the Promotion of Science (JSPS), and by the Institute for Fermentation, Osaka, Japan (K-2019-002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2021.675459/full#supplementary-material
References
Akhtar, N., Blomberg, A., and Adler, L. (1997). Osmoregulation and protein expression in a pbs2Δ mutant of Saccharomyces cerevisiae during adaptation to hypersaline stress. FEBS Lett. 403, 173–180. doi: 10.1016/S0014-5793(97)00048-3
Alex, L. A., Borkovich, K. A., and Simon, M. I. (1996). Hyphal development in Neurospora crassa: involvement of a two-component histidine kinase. Proc. Natl. Acad. Sci. U.S.A. 93, 3416–3421. doi: 10.1073/pnas.93.8.3416
Aoyama, K., Mitsubayashi, Y., Aiba, H., and Mizuno, T. (2000). Spy1, a histidine-containing phosphotransfer signaling protein regulates the fission yeast cell cycle through the Mcs4 response regulator. J. Bacteriol. 182, 4868–4874. doi: 10.1128/JB.182.17.4868-4874.2000
Appleby, J. L., Parkinson, J. S., and Bourret, R. B. (1996). Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86, 845–848. doi: 10.1016/S0092-8674(00)80158-0
Appleyard, M. V. C. L., McPheat, W. L., and Stark, M. J. (2000). A novel ‘two-component' protein containing histidine kinase and response regulator domains required for sporulation in Aspergillus nidulans. Curr. Genet. 37, 364–372. doi: 10.1007/s002940000123
Banno, S., Noguchi, R., Yamashita, K., Fukumori, F., Kimura, M., Yamaguchi, I., et al. (2007). Roles of putative His-to-Asp signaling modules HPT-1 and RRG-2, on viability and sensitivity to osmotic and oxidative stresses in Neurospora crassa. Curr. Genet. 51, 197–208. doi: 10.1007/s00294-006-0116-8
Blumenstein, A., Vienken, K., Tasler, R., Purschwitz, J., Veith, D., Frankenberg-Dinkel, N., et al. (2005). The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15, 1833–1838. doi: 10.1016/j.cub.2005.08.061
Brandhorst, T. T., Kean, I. R. L., Lawry, S. M., Wiesner, D. L., and Klein, B. S. (2019). Phenylpyrrole fungicides act on triosephosphate isomerase to induce methylglyoxal stress and alter hybrid histidine kinase activity. Sci. Rep. 9:5047. doi: 10.1038/s41598-019-41564-9
Brown, J., Bussey, H., and Stewart, R. (1994). Yeast Skn7p function s in a eukaryotic two-component regulatory pathway. EMBO J. 13, 5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x
Brown, J., North, S., and Bussey, H. (1993). SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175, 6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993
Buck, V., Quinn, J., Pino, T. S., Martin, H., Saldanha, J., Makino, K., et al. (2001). Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell 12, 407–419. doi: 10.1091/mbc.12.2.407
Catlett, N. L., Yoder, O. C., and Turgeon, B. G. (2003). Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2, 1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003
Fillinger, S., Ruijter, G., Tamas, M. J., Visser, J., Thevelein, J. M., et al. (2001). Molecular and physiological characterization of the NAD-dependent glycerol 3-phosphate dehydrogenase in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 39, 145–157. doi: 10.1046/j.1365-2958.2001.02223.x
Fujioka, T., Mizutani, O., Furukawa, K., Sato, N., Yoshimi, A., Yamagata, Y., et al. (2007). MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6, 1497–1510. doi: 10.1128/EC.00281-06
Furukawa, K., Hoshi, Y., Maeda, T., Nakajima, T., and Abe, K. (2005). Aspergillus nidulans HOG pathway is activated only by two-component signaling pathway in response to osmotic stress. Mol. Microbiol. 56, 1246–1261. doi: 10.1111/j.1365-2958.2005.04605.x
Furukawa, K., Katsuno, Y., Urao, T., Yabe, T., Yamada-Okabe, T., Yamada-Okabe, H., et al. (2002). Isolation and functional analysis of a gene, tcsB, encoding a transmembrane hybrid-type histidine kinase from Aspergillus nidulans. Appl. Environ. Microbiol. 68, 5304–5310. doi: 10.1128/AEM.68.11.5304-5310.2002
Furukawa, K., Yoshimi, A., Furukawa, T., Hoshi, Y., Hagiwara, D., Sato, N., et al. (2007). Novel reporter gene expression systems for monitoring activation of the Aspergillus nidulans HOG pathway. Biosci. Biotechnol. Biochem. 71, 1724–1730. doi: 10.1271/bbb.70131
Gustin, M. C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300. doi: 10.1128/MMBR.62.4.1264-1300.1998
Gwynne, D. I., Buxton, F. P., Sibley, S., Davies, R. W., Lockington, R. A., Scazzocchio, C., et al. (1987). Comparison of the cis-acting control regions of two coordinately controlled genes involved in ethanol utilization in Aspergillus nidulans. Gene 51, 205–216. doi: 10.1016/0378-1119(87)90309-X
Hagiwara, D., Asano, Y., Marui, J., Furukawa, K., Kanamaru, K., Kato, M., et al. (2007a). The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphal and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71, 1003–1014. doi: 10.1271/bbb.60665
Hagiwara, D., Asano, Y., Marui, J., Yoshimi, A., Mizuno, T., and Abe, K. (2009). Transcriptional profiling for Aspergillus nidulans HogA MAPK signaling pathway in response to fludioxonil and osmotic stress. Fungal Gent. Biol. 46, 868–878. doi: 10.1016/j.fgb.2009.07.003
Hagiwara, D., Kondo, A., Fujioka, T., and Abe, K. (2008). Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr. Genet. 54, 325–338. doi: 10.1007/s00294-008-0220-z
Hagiwara, D., Matsubayashi, Y., Marui, J., Furukawa, K., Yamashino, T., Kanamaru, K., et al. (2007b). Characterization of the NikA histidine kinase implicated in the phosphorelay signal transduction of Aspergillus nidulans, with special reference to fungicide responses. Biosci. Biotechnol. Biochem. 71, 844–847. doi: 10.1271/bbb.70051
Han, K. H., and Prade, R. A. (2002). Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 43, 1065–1078. doi: 10.1046/j.1365-2958.2002.02774.x
Hoch, J. A., and Silhavy, T. J. (1995). Two-Component Signal Transduction. Washington, DC: ASM Press, 1–473. doi: 10.1128/9781555818319
Hohmann, S. (2002). Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372. doi: 10.1128/MMBR.66.2.300-372.2002
Ichinomiya, M., Motoyama, T., Fujimura, M., Takagi, M., Horiuchi, H., and Ohta, A. (2002). Repression of chsB expression reveals the functional importance of class IV chitin synthase gene chsD in hyphal growth and conidiation of Aspergillus nidulans. Microbiology 148, 1335–1347. doi: 10.1099/00221287-148-5-1335
Izumitsu, K., Yoshimi, A., and Tanaka, C. (2007). Two-component response regulators Ssk1p and Skn7p additively regulate high-osmolarity adaptation and fungicide sensitivity in Cochliobolus heterostrophus. Eukaryot. Cell 6, 171–181. doi: 10.1128/EC.00326-06
Jiang, P., Zheng, S., and Lu, L. (2018). Mitotic-spindle organizing protein MztA mediates septation signaling by suppressing the regulatory subunit of protein phosphatase 2A-ParA in Aspergillus nidulans. Front. Microbiol. 9:988. doi: 10.3389/fmicb.2018.00988
Kawasaki, L., Sanchez, O., Shiozaki, K., and Aguirre, J. (2002). SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45, 1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x
Kojima, K., Takano, Y., Yoshimi, A., Tanaka, C., Kikuchi, T., and Okuno, T. (2004). Fungicide activity through activation of a fungal signaling pathway. Mol. Microbiol. 53, 1785–1796. doi: 10.1111/j.1365-2958.2004.04244.x
Lara-Rojas, F., Sánchez, O., Kawasaki, L., and Aguirre, J. (2011). Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 80, 436–454. doi: 10.1111/j.1365-2958.2011.07581.x
Lawry, S. M., Tebbets, B., Kean, I., Stewart, D., Hetelle, J., and Klein, B. S. (2017). Fludioxonil induces Drk1, a fungal group III hybrid histidine kinase, to dephosphorylate its downstream target, Ypd1. Antimicrob. Agents Chemother. 61, e01414–e01416. doi: 10.1128/AAC.01414-16
Li, S., Ault, A., Malone, C. L., Raitt, D., Dean, S., Johnston, L. H., et al. (1998). The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17, 6952–6962. doi: 10.1093/emboj/17.23.6952
Maeda, T., Wurgler-Murphy, S. M., and Saito, H. (1994). A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369, 242–245. doi: 10.1038/369242a0
Maruyama, J.-I., Nakajima, H., and Kitamoto, K. (2001). Visualization of nuclei in Aspergillus oryzae with EGFP and analysis of the number of nuclei in each conidium by FACS. Biosci. Biotechnol. Biochem. 65, 1504–1510. doi: 10.1271/bbb.65.1504
Miyazawa, K., Yoshimi, A., Kasahara, S., Sugahara, A., Koizumi, A., Yano, S., et al. (2018). Molecular mass and localization of α-1,3-glucan in cell wall control the degree of hyphal aggregation in liquid culture of Aspergillus nidulans. Front. Microbiol. 9:2623. doi: 10.3389/fmicb.2018.02623
Motoyama, T., Kadokura, K., Ohira, T., Ichiishi, A., Fujimura, M., Yamaguchi, I., et al. (2005). A two-component histidine kinase of the rice blast fungus is involved in osmotic stress response and fungicide action. Fung. Genet. Biol. 42, 200–212. doi: 10.1016/j.fgb.2004.11.002
Navarro, R. E., and Aguirre, J. (1998). Posttranscriptional control mediates cell type-specific localization of catalase A during Aspergillus nidulans development. J. Bacteriol. 180, 5733–5738. doi: 10.1128/JB.180.21.5733-5738.1998
Nikolaev, I., Mathieu, M., van de Vondervoort, P. J. I., Visser, J., and Felenbok, B. (2002). Heterologous expression of the Aspergillus nidulans alcR–alcA system in Aspergillus niger. Fung. Genet. Biol. 37, 89–97. doi: 10.1016/S1087-1845(02)00037-3
Ochiai, N., Fujimura, M., Motoyama, T., Ichiishi, A., Usami, R., Horikoshi, K., et al. (2001). Characterization of mutants in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag. Sci. 57, 437–442. doi: 10.1002/ps.302
Posas, F., and Saito, H. (1998). Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17, 1385–1394. doi: 10.1093/emboj/17.5.1385
Posas, F., Wurgler-Murphy, S. M., Maeda, T., Witten, E. A., Thai, T. C., and Saito, H. (1996). Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86, 865–875. doi: 10.1016/S0092-8674(00)80162-2
Shieh, J. C., Wilkinson, M. G., Buck, V., Morgan, B. A., Makino, K., and Miller, J. B. (1997). The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-Sty1 MAP kinase pathway and fission yeast cell cycle. Genes Dev. 11, 1008–1022. doi: 10.1101/gad.11.8.1008
Shiozaki, K., Shiozaki, M., and Russell, P. (1997). Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol. Biol. Cell 8, 409–419. doi: 10.1091/mbc.8.3.409
Shpilka, T., Weidberg, H., Pietrokovski, S., and Elazar, Z. (2011). Atg8: an autophagy-related ubiquitin-like protein family. Gen. Biol. 12:226. doi: 10.1186/gb-2011-12-7-226
Vargas-Perez, I., Sanchez, O., Kawasaki, L., Georgellis, D., and Aguirre, J. (2007). Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot. Cell 6, 1570–1583. doi: 10.1128/EC.00085-07
Viaud, M., Fillinger, S., Liu, W., Polepalli, J. S., Pecheur, P. L., Kunduru, A. R., et al. (2006). A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol. Plant Microbe Interact. 19, 1042–1050. doi: 10.1094/MPMI-19-1042
Yamada-Okabe, T., Mio, T., Ono, N., Kashima, Y., Matsui, M., Arisawa, M., et al. (1999). Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus Candida albicans. J. Bacteriol. 181, 7243–7247. doi: 10.1128/JB.181.23.7243-7247.1999
Yoshimi, A., Kojima, K., Takano, Y., and Tanaka, C. (2005). Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot. Cell 4, 1820–1828. doi: 10.1128/EC.4.11.1820-1828.2005
Yoshimi, A., Sano, M., Inaba, A., Kokubun, Y., Fujioka, T., Mizutani, O., et al. (2013). Functional analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major α-1,3-glucan synthase in this fungus. PLoS ONE 8:e54893. doi: 10.1371/journal.pone.0054893
Yoshimi, A., Tsuda, M., and Tanaka, C. (2004). Cloning and characterization of the histidine kinase gene Dic1 from Cochliobolus heterostrophus that confers dicarboximide resistance and osmotic adaptation. Mol. Genet. Genomics 271, 228–236. doi: 10.1007/s00438-003-0974-4
Yu, Z., Ali, A., Igbalajobi, O. A., Streng, C., Leister, K., Krau,ß, N., et al. (2019). Two hybrid histidine kinases, TcsB and the phytochrome FphA, are involved in temperature sensing in Aspergillus nidulans. Mol. Microbiol. 112, 1814–1830. doi: 10.1111/mmi.14395
Yu, Z., Armant, O., and Fischer, R. (2016). Fungi use the SakA (HogA) pathway for phytochrome-dependent light signalling. Nat. Microbiol. 1:16019. doi: 10.1038/nmicrobiol.2016.19
Zhong, G. W., Jiang, P., Qiao, W. R., Zhang, Y. W., Weo, W. F., and Lu, L. (2014). Protein phosphatase 2A (PP2A) regulatory subunits ParA and PabA orchestrate septation and conidiation and are essential for PP2A activity in Aspergillus nidulans. Eukaryot. Cell 13, 1494–1506. doi: 10.1128/EC.00201-14
Keywords: Aspergillus nidulans, His-Asp phosphorelay signaling, YpdA, response regulators, mitogen-activated protein kinases, fludioxonil, growth disorders
Citation: Yoshimi A, Hagiwara D, Ono M, Fukuma Y, Midorikawa Y, Furukawa K, Fujioka T, Mizutani O, Sato N, Miyazawa K, Maruyama J-i, Marui J, Yamagata Y, Nakajima T, Tanaka C and Abe K (2021) Downregulation of the ypdA Gene Encoding an Intermediate of His-Asp Phosphorelay Signaling in Aspergillus nidulans Induces the Same Cellular Effects as the Phenylpyrrole Fungicide Fludioxonil. Front. Fungal Biol. 2:675459. doi: 10.3389/ffunb.2021.675459
Received: 03 March 2021; Accepted: 26 July 2021;
Published: 01 September 2021.
Edited by:
Katherine A. Borkovich, University of California, Riverside, United StatesReviewed by:
Zhenzhong Yu, Nanjing Agricultural University, ChinaLori Huberman, Cornell University, United States
Copyright © 2021 Yoshimi, Hagiwara, Ono, Fukuma, Midorikawa, Furukawa, Fujioka, Mizutani, Sato, Miyazawa, Maruyama, Marui, Yamagata, Nakajima, Tanaka and Abe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keietsu Abe, a2VpZXRzdS5hYmUuYjVAdG9ob2t1LmFjLmpw
†These authors have contributed equally to this work
‡Present address: Daisuke Hagiwara, Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
Kentaro Furukawa, Department of Cellular Physiology, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
Osamu Mizutani, Department of Bioscience and Biotechnology, University of the Ryukyus, Nishihara, Japan
Junichiro Marui, Biological Resources and Post-harvest Division, Japan International Research Center for Agricultural Sciences, Tsukuba, Japan
Youhei Yamagata, Department of Applied Molecular Biology and Biochemistry, Tokyo University of Agriculture and Technology, Tokyo, Japan
 Akira Yoshimi
Akira Yoshimi Daisuke Hagiwara
Daisuke Hagiwara Miyako Ono4†
Miyako Ono4† Yura Midorikawa
Yura Midorikawa Kentaro Furukawa
Kentaro Furukawa Osamu Mizutani
Osamu Mizutani Ken Miyazawa
Ken Miyazawa Jun-ichi Maruyama
Jun-ichi Maruyama Youhei Yamagata
Youhei Yamagata Keietsu Abe
Keietsu Abe