- 1Laboratory of Environmental Interface Technology of Filamentous Fungi, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 2Laboratory of Terrestrial Microbial Ecology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 3Terrestrial Microbiology and Systematics, Graduate School of Global Environmental Studies, Kyoto University, Kyoto, Japan
- 4Graduate School of Sciences and Technology for Innovation, Yamaguchi University, Yamaguchi, Japan
- 5Laboratory of Forest Biochemistry, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
Hydrophobins are small amphiphilic proteins secreted by filamentous fungi. These proteins confer hydrophobic properties to the hyphae and conidia. Bipolaris maydis is the causal agent of southern corn leaf blight; the biological function of its hydrophobins is not clear. In the present study, we focused on the broad function of hydrophobins in the life cycle of this fungus. We found that the B. maydis genome encodes four hydrophobins—Hyp1 of class I, and Hyp2, Hyp3 and Hyp4 of class II—and all of them are expressed. We generated single disruptants of each gene, as well as triple and quadruple disruptants. No differences were detected between the wild type and any of disruptants in mycelial growth, conidiation, stress tolerance, virulence, or sexual reproduction. The colony hydrophobicity of all disruptant strains was similar to that of the wild-type strain. Complementation of a null Aspergillus nidulans mutant of dewA, which showed a significantly reduced colony hydrophobicity, with each of the four B. maydis hydrophobin genes restored the hydrophobic phenotype, although the degree of hydrophobicity varied among them. Despite the absence of any significant phenotypic changes in the B. maydis mutants generated, results strongly suggest that all four hydrophobins have retained their function in hydrophobicity. Furthermore, the results of this study suggest that the role of hydrophobins might change depending on the fungal species.
1 Introduction
Hydrophobins are low-molecular-weight, amphipathic proteins secreted by filamentous fungi (Wösten, 2001). Their amino acid sequences have low similarity, but their hydrophobic patterns are similar. These proteins have a highly conserved pattern of eight cysteine residues (-C-CC-C-C-CC-C-) capable of forming four disulfide bonds (Sehuren and Wessels, 1990; Wösten, 2001). A characteristic feature of hydrophobins is their ability to self-assemble at air–water interfaces (Wösten, 2001). On the basis of their hydrophobic patterns and the solubility of their aggregates in solvents, hydrophobins are primarily classified into classes I and II (Wösten, 2001). Class I hydrophobins are insoluble in sodium dodecyl sulphate (SDS) even at high temperatures but soluble in trifluoroacetic acid, whereas class II hydrophobins are soluble in SDS (Wessels et al., 1991).
Hydrophobins are secreted to the surface of the cell wall of aerial hyphae and conidia and play crucial roles in fungal biological processes (Wösten, 2001). Hyphae of many filamentous fungi are hydrophobic: they repel water and create a hydrophobic barrier. Hydrophobins are key contributors to the hydrophobic properties of a wide range of filamentous fungi including ascomycetes and basidiomycetes, suggesting that this is a general function of these proteins (Wösten, 2001). In Aspergillus oryzae, hydrophobins interact with cutinases to facilitate efficient degradation of solid polymers (Takahashi et al., 2005). Similar findings in the plant pathogen Pyricularia oryzae (syn. P. grisea, Magnaporthe oryzae and M. grisea) suggest that hydrophobins contribute to the degradation of plant wax layers (Pham et al., 2016). Class I hydrophobin Mpg1 in P. oryzae is important for appressorium formation and pathogenicity (Beckerman and Ebbole, 1996; Talbot et al., 1996). The class II hydrophobin Mhp1 of this fungus is also required for virulence (Kim et al., 2005). In Verticillium dahliae, class II hydrophobins VDH1 and VdHP1 are required for microsclerotia formation (Klimes et al., 2008; Zhang et al., 2022). Deletion of VDH1 does not significantly affect virulence, but deletion of VdHP1 increases crude toxin content and enhances virulence (Zhang et al., 2022). In contrast, hydrophobins are not required for pathogenicity in Cladosporium fulvum or Botrytis cinerea (Whiteford and Spanu, 2001; Mosbach et al., 2011). These data suggest that the functions of hydrophobins vary among fungal species.
Most filamentous fungi have multiple hydrophobins. The genome of A. nidulans is currently known to encodes six, that of Fusarium graminearum encodes five, that of Botrytis cinerea encodes three, and that of P. oryzae encodes two (Talbot et al., 1993; Kim et al., 2005; Mosbach et al., 2011; Grünbacher et al., 2014; Quarantin et al., 2019). In B. cinerea, the deletion of at least two hydrophobins leads to aberrant development of apothecia, but the loss of a single one does not affect sexual reproduction (Terhem and van Kan, 2014). These data point to functional redundancy.
The necrotrophic pathogen Bipolaris maydis (syn. Cochliobolus heterostrophus) causes southern corn leaf blight. Turgeon et al. (2008) reported that the lack of a gene encoding the non-ribosomal peptide synthetase 4 (Nps4) leads to a wettable phenotype (i.e. low hydrophobicity) in this fungus. Colony hydrophobicity is significantly higher in the wild type than in a strain lacking the CHK1 gene for mitogen-activated protein kinase (MAPK) (Degani et al., 2013). The genome of B. maydis is predicted to encode at least four hydrophobins (Degani et al., 2013). These authors used BLAST searches to identify hydrophobins in this fungus but might not have identified all of them due to the low sequence homology among hydrophobins. Furthermore, the effect of the deletion of these genes on morphogenesis and pathogenicity have not been analyzed, and it remains unclear whether hydrophobins contribute to the hydrophobic properties of the cell surface in this fungus.
In this study, to find previously uncharacterized hydrophobins of B. maydis, we conducted a comprehensive in silico analysis using publicly available amino acid sequence data. To investigate the biological functions of the predicted hydrophobins, we generated single-null mutants as well as mutants combining deletions of the identified genes. We also examined the interspecies compatibility of hydrophobins by heterologous expression of a B. maydis hydrophobin in A. nidulans.
2 Materials and methods
2.1 Strains and culture conditions
Bipolaris maydis HITO7711 (mating type MAT1-2) was used as the wild-type strain throughout this study. Albino strain alb3 (mating type MAT1-1) was used in a crossing assay (Tanaka et al., 1991). The strain lacking the CHK1 gene (Δchk1) was used as a negative control in a hydrophobicity assay (Izumitsu et al., 2009; Degani et al., 2013). All gene disruptants of B. maydis were derived from HITO7711. MASHIKI2-2 (MAT1-1) was used as an alternative wild-type strain in a following crossing. For the crossing assay, the quadruple disruptant of hydrophobin genes (MAT1-1) was obtained by crossing the quadruple disruptant (MAT1-2) and MASHIKI2-2.
Aspergillus nidulans ABPU1 strain (ΔligD), which is highly efficient for gene disruption via homologous recombination, was used as a parental strain to obtain a dewA gene-disruptant (Hagiwara et al., 2007; Yoshimi et al., 2013). The BPU1 strain carrying the argB gene was used as a control (CNT) strain for phenotypic comparisons (Hagiwara et al., 2007; Miyazawa et al., 2021).
All strains were maintained on complete medium agar (CMA; Tanaka et al., 1991) or vegetable juice agar (VJA) medium at 25°C. VJA contained vegetable juice (Kagome, Tokyo, Japan) instead of V8 juice but otherwise had the same composition as V8 agar medium (Ribeiro, 1978); no difference in the growth of B. maydis was detected between V8 agar and VJA (data not shown). In the crossing assay, Sachs’s agar medium was used. In a stress tolerance assay, CMA was used. Czapek–Dox (CD) medium was used for culturing A. nidulans (Fujioka et al., 2007).
2.2 Identification of hydrophobins in B. maydis
Hydrophobins are small secreted proteins with a signal peptide and a conserved characteristic C-terminal sequence. To identify the hydrophobins of B. maydis, we downloaded the translated sequence from gene catalog of Cochliobolus heterostrophus C5 v2.0 from the website of the DOE Joint Genome Institute (JGI, https://mycocosm.jgi.doe.gov/CocheC5_3/CocheC5_3.home.html, Ohm et al., 2012; Condon et al., 2013). In this dataset, we first searched for proteins that were cysteine rich (≥4) and small (≤250 amino acids). Next, we checked whether those proteins possess signal peptides using SignalP 5.0 (https://services.healthtech.dtu.dk/services/SignalP-5.0/). From the refined data, we visually selected proteins with a cysteine pattern specific to hydrophobins (-C-CC-C-C-CC-C-). Additionally, we performed the same search using the updated v3.0 dataset (https://mycocosm.jgi.doe.gov/CocheC5_4m/CocheC5_4m.home.html, Haridas et al., 2023). Hydropathy plots were investigated using ProtScale (https://web.expasy.org/protscale/) based on the Kyte and Doolittle (1982) method. The amino acid sequences of B. maydis and P. oryzae hydrophobins were aligned with MUSCLE using MEGA X (Edgar, 2004; Kumar et al., 2018). In the alignment and the analysis of the hydropathy plots, the amino acid sequences of P. oryzae Mpg1 (class I) and Mhp1 (class II) were used as references. For in silico characterization, InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/) was used to analyze each B. maydis hydrophobin.
2.3 RT-PCR analysis of hydrophobin genes in B. maydis
The wild-type strain was incubated under three different conditions: liquid culture, on solid medium, or on a host plant (maize leaves). In liquid culture, the conidia (final concentration, 105 conidia/mL) were added to 100 mL CM liquid medium and incubated with shaking. On solid medium, 100 conidia were inoculated onto cellophane membrane placed on the top of Petri plates filled with CMA. On the host plant, conidial suspension (20 μL of 5.0 × 105 conidia/mL) was inoculated at five points on a single detached 1-month-old leaf. Subsequently, five lesion areas were cut off from leaves and collected. All incubations were carried out at 25°C for 3 days in the dark and were repeated 3 times. Total RNA was extracted from mycelia or leaf lesion areas using Trizol, according to Yoshida and Tanaka (2019). cDNA was synthesized from the total RNA with SuperScript IV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) using the (dT)21VN primer.
2.4 Generation of disruptants of B. maydis
The procedure for generating disruptants and locations of primers to confirm gene disruptions are shown in Supplementary Figure S1A. All primers are listed in Supplementary Table S1. A disruption cassette was constructed for each gene encoding a putative hydrophobin. PCR to construct these cassettes was performed with Takara Ex Taq (Takara Bio, Kusatsu, Japan). All PCR confirmations of gene disruption were performed with SapphireAmp Fast PCR Master Mix (Takara Bio).
HYP1 disruptant (Δhyp1): The genomic DNA of B. maydis HITO7711 was obtained as described by Izumitsu et al. (2012). The upstream (primer set Hyp1-f1/Hyp-r1) and downstream regions (Hyp-f2/Hyp-r2) of the HYP1 gene ORF were amplified from genomic DNA. The neomycin phosphotransferase II gene (NPTII) cassette was amplified from the pZGenI plasmid (primer set NPTandHPH-f1/NPTII-r1; Sumita et al., 2017). The three amplified products were fused by fusion-PCR (Hyp1-f3/Hyp1-r3 primer set). The resulting cassette was purified by ethanol precipitation and used for HYP1 disruption. The transformation was based on the PEG-mediated protoplast transformation method described in Izumitsu et al. (2009). The replacement of HYP1 by the disruption cassette was confirmed by PCR with three primer sets (Hyp1-f1/NPTII-chk1, Hyp1-f4/Hyp1-r4 and NPTandHPH-chk2/Hyp1-r1). A similar approach was used to disrupt HYP2–HYP4 individually and to generate Δhyp2–Δhyp4 strains.
Double, triple, and quadruple disruptants: To generate a double disruptant, Δhyp2Δhyp3, the HYP2 gene was replaced in the parental Δhyp3 strain using a disruption cassette with a nourseothricin-resistance gene (NAT). The NAT gene was amplified from the pZNAT1 plasmid with the primer set NATandBAR-f1/NAT-r1 (Izumitsu et al., 2009). To generate the triple disruptant, Δhyp2Δhyp3Δhyp4, the HYP4 gene of the double disruptant Δhyp2Δhyp3 was replaced with the bialaphos-resistance gene (BAR) cassette. The BAR gene cassette was amplified from the pCB1546 plasmid with the primer set NATandBAR-f1/BAR-r1 (Sweigard et al., 1997). To generate the quadruple disruptant, Δhyp1Δhyp2Δhyp3Δhyp4, the HYP1 gene of the triple disruptant was replaced with a hygromycin B phosphotransferase gene (HPH) cassette. The HPH cassette was amplified from the plasmid pCB1004 (Carroll et al., 1994).
2.5 Stress tolerance assay
CMA was supplemented with 0.05% SDS, 0.015% H2O2, or 25 μg/mL Calcofluor White (CFW; Sigma-Aldrich, Saint Louis, MO, USA). Each strain was incubated for 7 days at 25°C, and their colony diameters were measured. To quantify the reduction in radial-growth of colonies, we calculated the percentage decrease relative to the untreated control.
2.6 Pathogenicity assay
Maize plants (Zea mays cv. Takanestar) were grown at 25°C for 1 month. Freshly detached leaves were placed in plastic cases lined with wet paper towels. Conidia of each strain were harvested from colonies grown on VJA for 7 days. Twenty μL of 5.0 × 105 conidia/mL suspensions (in 0.01% Tween-20) were dropped onto the leaves and incubated in the dark at 25°C for 3 days. We measured and photographed the lesions on the leaves.
2.7 Crossing assay
Bipolaris maydis forms a characteristic sexual organ called a pseudothecium; usually, it is black in the wild-type strain and light tawny in the albino strain. The ability of any strain to form pseudothecia can be evaluated by using the albino strain in a crossing assay. The crossing assay was performed according to Tanaka et al. (1991). Dried maize leaves were placed on Sachs’s agar medium, then each disruptant (MAT1-2) and the albino strain (MAT1-1) were inoculated diagonally across the leaves and incubated at 25°C in the dark for 30 days. The pseudothecia were photographed. Both black and light tawny pseudothecia were harvested and dissected in a drop of 10% glycerol on a glass slide to observe the asci and ascospores within the pseudothecia, respectively, under a microscope (Leica DML, Leica Microsystems, Wetzlar, Germany).
2.8 Generation of complementation strains of A. nidulans
The gene disruption strategies and locations of primers to confirm the insertion are shown in Supplementary Figure S2. PCR for plasmid construction was performed with PrimeSTAR GXL DNA polymerase (Takara Bio). All PCR confirmations were performed with Takara Ex Premier DNA Polymerase (Takara Bio). NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA, USA) was used for fusions. Fungal transformations were performed using the PEG-mediated protoplast transformation method as described in Yoshimi et al. (2013).
To construct a disruption cassette of the A. nidulans dewA gene (for class I hydrophobin), the upstream and downstream regions of this gene were amplified from genomic DNA with the primer sets dewA-f1/dewA-r1 and dewA-f2/dewA-r2. The argB gene of A. oryzae (used in this work as selection marker) was amplified from the pAORB plasmid with a primer set AoargB-f1/AoargB-r1 (Furukawa et al., 2002). The three amplicons were fused into a BamHI-linearized pUC19 plasmid. The dewA gene disruption cassette was amplified from the resulting plasmid with the primer set pUC-chk1/pUC-chk2, purified by ethanol precipitation and integrated into the ABPU1 (ligD) strain. The candidate disruptants were inoculated and incubated on selective CD medium without arginine. The replacement of the dewA gene with the argB gene was confirmed with the dewA-f3/AoargB-chk1, dewA-f4/dewA-r4, and AoargB-chk2/dewA-r3 primer sets.
For complementation assays with the dewA and B. maydis hydrophobin genes, we constructed a plasmid to reintroduce the dewA gene and four plasmids to introduce B. maydis hydrophobin genes into the ΔdewA strain. To generate the pUC-pyrG plasmid, the entire A. nidulans pyrG gene with the putative promoter was amplified with the primer set pyrG-f1/pyrG-r1, and the PCR fragment was fused into a BamHI-linearized pUC19 vector. To generate the dewA complementation plasmid, the entire dewA gene with the promoter and ORF was amplified with the primer set dewA-pyrG-f1/dewA-pyrG-r1 and fused into a SmaI-linearized pUC-pyrG plasmid.
To generate the HYP1 complementation plasmid, the HYP1 gene with the 3′ untranslated region from the start codon was amplified from B. maydis genomic DNA with the primer set HYP1-start-f1/HYP1-pyrG-r1. The putative promoter region (ca. 1000 bp upstream of dewA ORF) was amplified with the primer set dewA-pyrG-f1/dewA-HYP1-r1. These two PCR fragments were fused into the SmaI-linearized pUC19-pyrG plasmid. Complementation plasmids for the remaining three B. maydis hydrophobins were constructed in a similar fashion.
To generate dewAcomp and B. maydis hydrophobin-complemented strains (HYP1–4comp), the corresponding complementation plasmids were introduced into the ΔdewA strain. Resulting candidate strains were inoculated and incubated on selective CD medium without arginine, uridine, and uracil. Insertion of each gene was confirmed with the corresponding primer set f4/r4.
2.9 RT-PCR analysis of hydrophobin genes in A. nidulans
The CNT, ΔdewA, dewAcomp, and HYP1–4comp strains were incubated on CD medium for 1 week. Conidia (final concentration, 105 conidia/mL) from each colony were added to 20 mL of CD liquid medium and incubated with shaking for 24 hours. The extraction of total RNA from the mycelia and the synthesis of cDNA were performed as described above. The histone H2B gene was used as internal control (Yoshimi et al., 2013).
2.10 Hydrophobicity assays in B. maydis and A. nidulans
Colonies of the wild-type strain and all disruptants of B. maydis were grown on CMA at 25°C for 7 days. The colonies were punched 1 cm from their edge with a cork borer (diameter: 1 cm), and 10 µL of a solution containing 0.2% SDS and 50 mM ethylenediaminetetraacetic acid (EDTA) was dropped onto the mycelial disks. We took photos of these droplets every 10 min for 30 min to record their changes.
CNT, ΔdewA, dewAcomp, and HYP1–4comp strains of A. nidulans were incubated on CD medium at 37°C for 4 days. The colonies were punched at the edge using a cork borer, aligning the borer’s perimeter with the colony’s perimeter. The hydrophobicity of each strain was assessed as above. Photographs were taken every minute for 5 min.
3 Results
3.1 Four hydrophobins in B. maydis
By searching the gene catalog (protein data) of C. heterostrophus C5 from the JGI website, we identified four candidate proteins with a signal peptide and a cysteine pattern characteristic of hydrophobins. The total length was 117 amino acids for Hyp1, 100 amino acids for Hyp2, 157 amino acids for Hyp3, and 182 amino acids for Hyp4. Each of the hydrophobin genes had two exons. The hydropathy profile of Hyp1 was similar to that of Mpg1 of P. oryzae while that of Hyp2 was similar to that of Mhp1 (Figure 1; Supplementary Figure S3). Those of Hyp3 and Hyp4 were also similar to that of Mhp1, except some regions (Figure 1; Supplementary Figure S3B). Hyp3 had a unique long sequence (71 amino acids) after the 8th cysteine, which contained a 9th cysteine (Supplementary Figure S3B). Hyp4 had 90 amino acids, approximately half of them glycines, between the signal peptide and the first cysteine (Supplementary Figure S3B). InterProScan searches showed that Hyp1 belongs to the hydrophobin family (InterPro IRR001338; class I), whereas Hyp2–Hyp4 belong to the cerato-ulmin hydrophobin family (InterPro IRR010636; class II). Cerato-ulmin is a representative class II hydrophobin produced by Ophiostoma ulmi (Stringer and Timberlake, 1993; Sbrana et al., 2010).
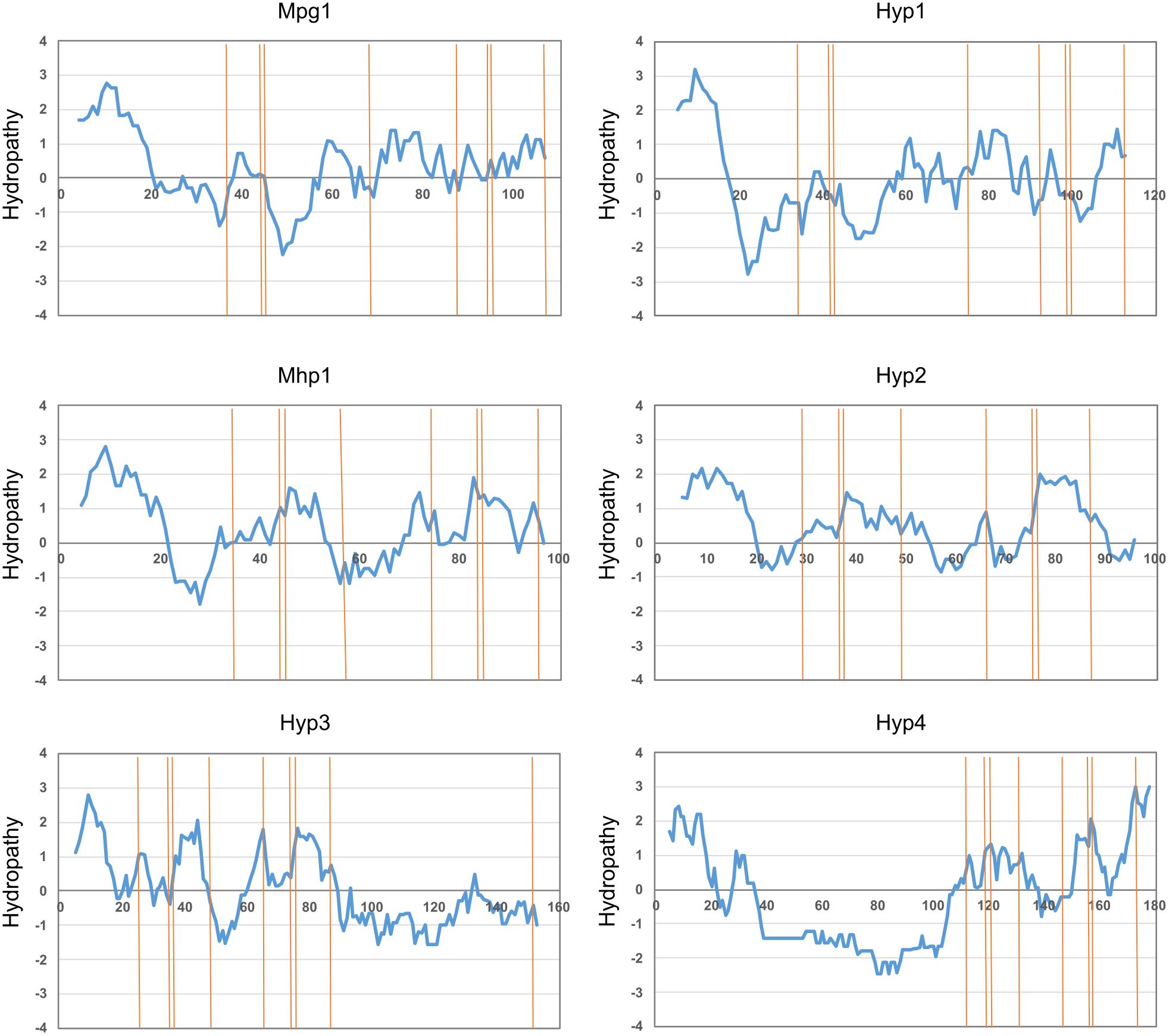
Figure 1. Hydropathy plots of B. maydis hydrophobins. Hydropathy was calculated using ProtScale with the method of Kyte and Doolittle (1982). Hydrophobins of P. oryzae Mpg1 (class I) and Mhp1 (class II) were used as controls.
3.2 Expression of the four hydrophobin genes
In mycelia of B. maydis incubated in liquid culture, on solid medium, or on maize leaves, all hydrophobin genes were expressed, but at different levels: the transcript levels were higher for HYP1 and HYP3 than for HYP2 and HYP4 (Figure 2), suggesting that this fungus uses mainly Hyp1 and Hyp3. The transcript levels of HYP1 in maize leaves and HYP3 gene in liquid culture were similar to that of β tubulin-encoding gene as an internal control. The transcript level of HYP4 was highest in maize leaves, but that of HYP2 did not differ significantly among the three conditions. The transcript levels of all hydrophobin genes, except for HYP2, differed significantly among the culture conditions, suggesting that each hydrophobin may be used differently under different conditions.
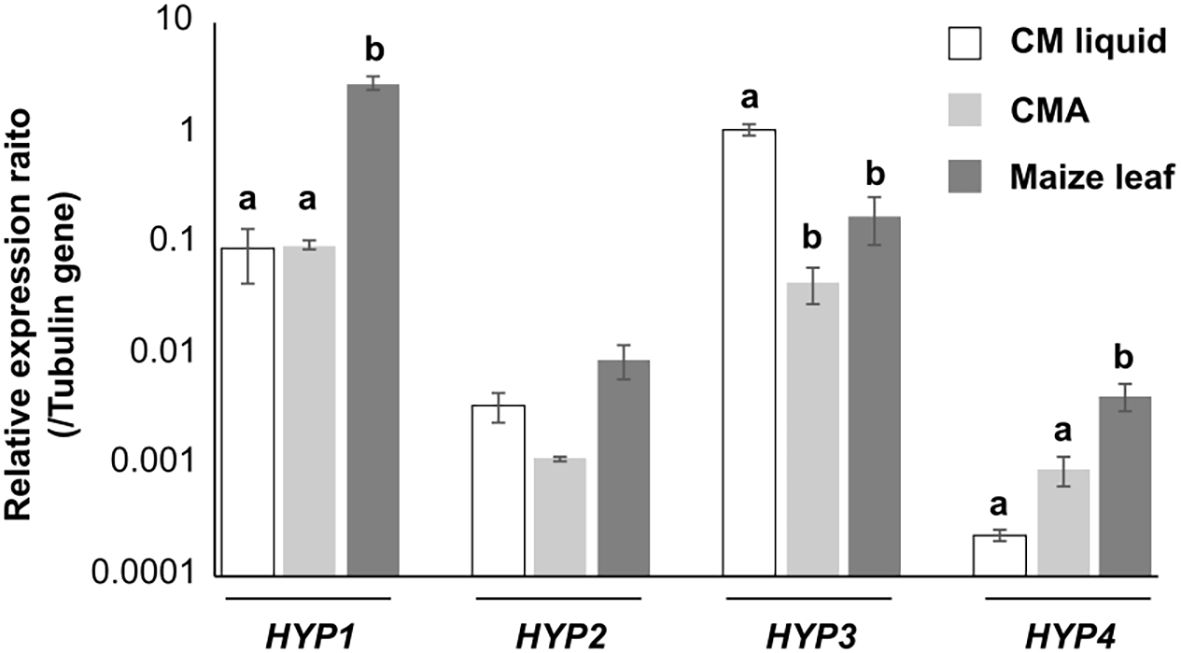
Figure 2. Transcript levels of B. maydis hydrophobin genes. Hyphae were cultured for 3 days in CM liquid, on cellophane membranes on CMA, or on the leaves of host plants (maize). RNA was extracted and used for qRT-PCR. Error bars, standard error (n = 3). Different letters within each gene indicate significant differences (Tukey’s test, P < 0.05).
3.3 Generation of hydrophobin gene disruptants in B. maydis
The single disruption of each hydrophobin gene in B. maydis was confirmed by PCR (Supplementary Figures S1C–F). A class II hydrophobin-deficient strain (hyp2Δhyp3Δhyp4) and a strain deficient in all four hydrophobins (Δhyp1Δhyp2Δhyp3Δhyp4) were also generated (Supplementary Figure S1A, B, G–I). All strains were inoculated on VJA and incubated at 25°C for 7 days. Radial growth was similar in all the disruptants and the wild-type strain (Figure 3; Table 1). Conidia of all strains were harvested from these colonies and analyzed. The conidia of the wild-type strain were spindle-shaped and composed of cells divided by multiple septa. No considerable differences in conidial morphology or the number of conidia were observed between the disruptants and the wild-type strain (Figure 3, Table 1). These results indicate that the loss of hydrophobins in this fungus does not affect normal radial growth and conidiation.
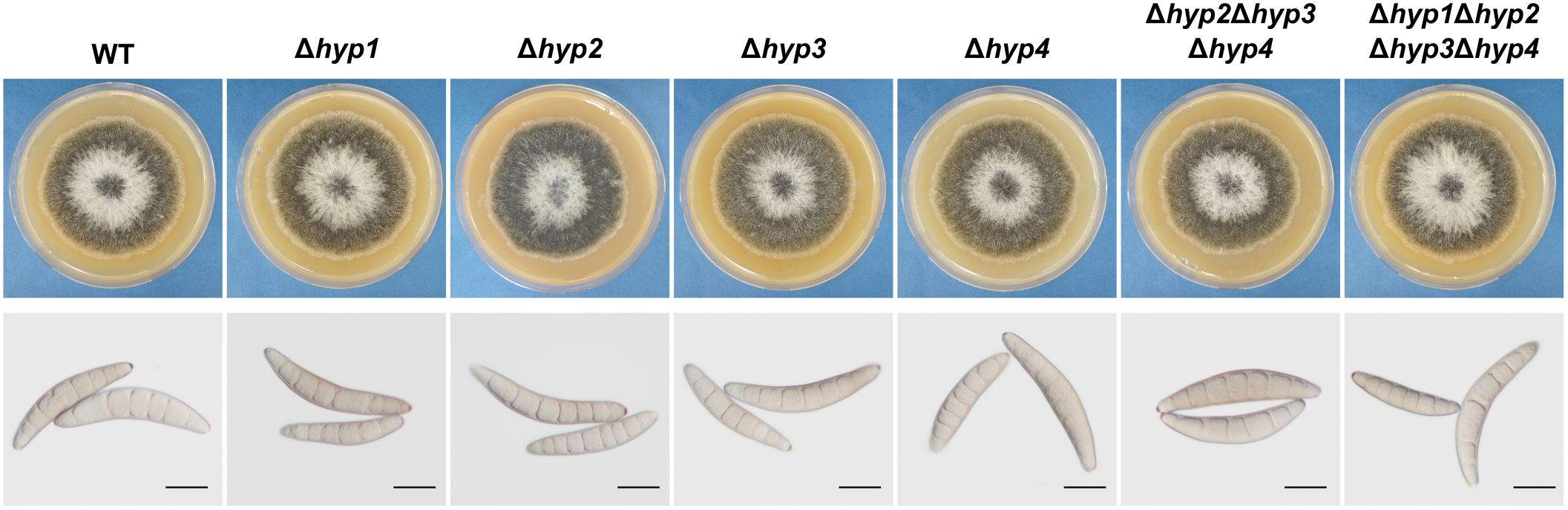
Figure 3. Colony growth and conidial morphology of the wild-type strain and disruptants of hydrophobin genes. Each strain was cultured on VJA for 7 days. Conidia were harvested from three colonies of each strain. Bars: 20 µm.
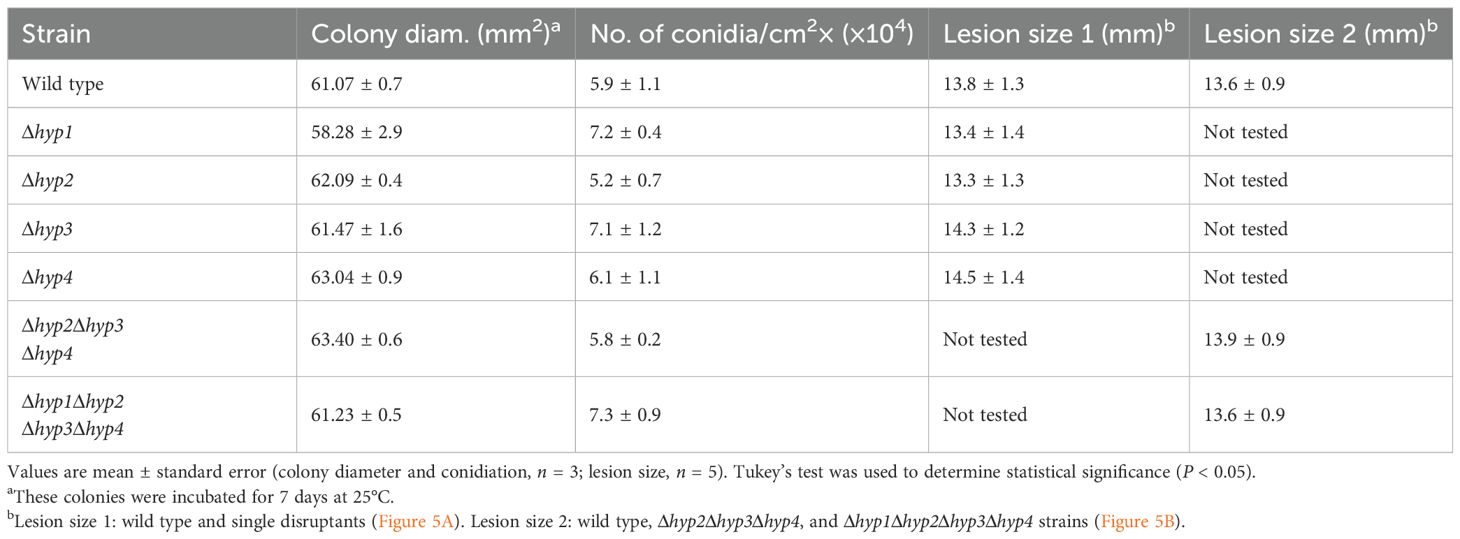
Table 1. Phenotypes of disruptants of hydrophobin genes of B. maydis: growth, conidiation, and pathogenicity.
3.4 Stress tolerance of hydrophobin gene disruptants
All strains were cultured on CMA in the absence or presence of stress-inducing compounds (CFW, NaOH, or SDS). The decrease in radial growth of all disruptants on each medium was not significantly different from those of the wild-type strain (Figure 4; Supplementary Figure S4). These results showed that hydrophobins in B. maydis do not contribute to these types of stress tolerance.
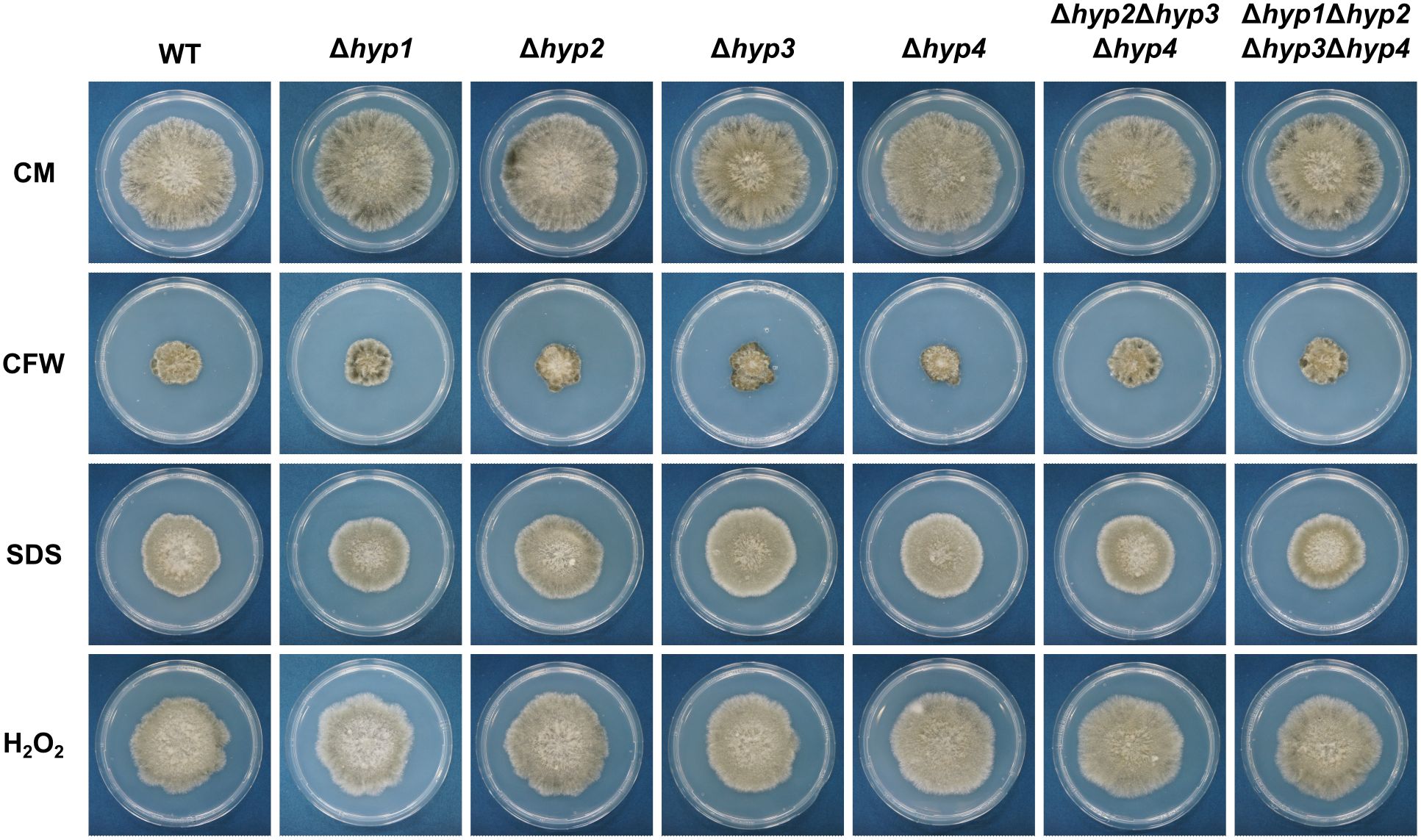
Figure 4. Sensitivity of the wild-type and hydrophobin gene disruptants to stress agents. All strains were incubated on CMA supplemented with 25 μg/mL calcofluor white (CFW), 0.05% SDS, or 0.015% H2O2 for 7 days.
3.5 Pathogenicity of hydrophobin gene disruptants
To investigate whether B. maydis hydrophobins are involved in pathogenicity, we performed a pathogenicity assay on maize leaves. Lesions formed by each single disruptant were similar to those formed by the wild-type strain (Figure 5A; Table 1). Similarly, no significant difference was observed in the size of lesions among the triple disruptant, quadruple disruptant, and the wild-type strain (Figure 5B; Table 1). These results show that the lack of hydrophobins does not affect the pathogenicity of B. maydis.

Figure 5. Pathogenicity assay on maize leaves. (A) The wild-type strain and single disruptants of each hydrophobin gene. (B) The wild-type strain, triple and quadruple disruptants of hydrophobin genes. Photos were taken 3 days after inoculation on maize leaves.
3.6 Sexual reproduction of hydrophobin gene disruptants
In the cross between the wild-type and albino strains, black pseudothecia and light tawny pseudothecia were formed along their contact line (Figure 6). When all disruptants were crossed with the albino strain, pseudothecia of black and light tawny colors were formed; therefore, the loss of hydrophobins did not affect the formation of female organs (Figure 6). To investigate whether hydrophobins are involved in ascus and ascospore development, pseudothecia of both colors were harvested and inspected. In all crosses with the albino strain, asci containing ascospores were formed in pseudothecia of both colors (Figure 6). When the quadruple disruptant was crossed with itself, asci containing ascospores were formed normally in pseudothecia (Supplementary Figure S5). These results suggested that hydrophobins in B. maydis are dispensable for sexual reproduction.
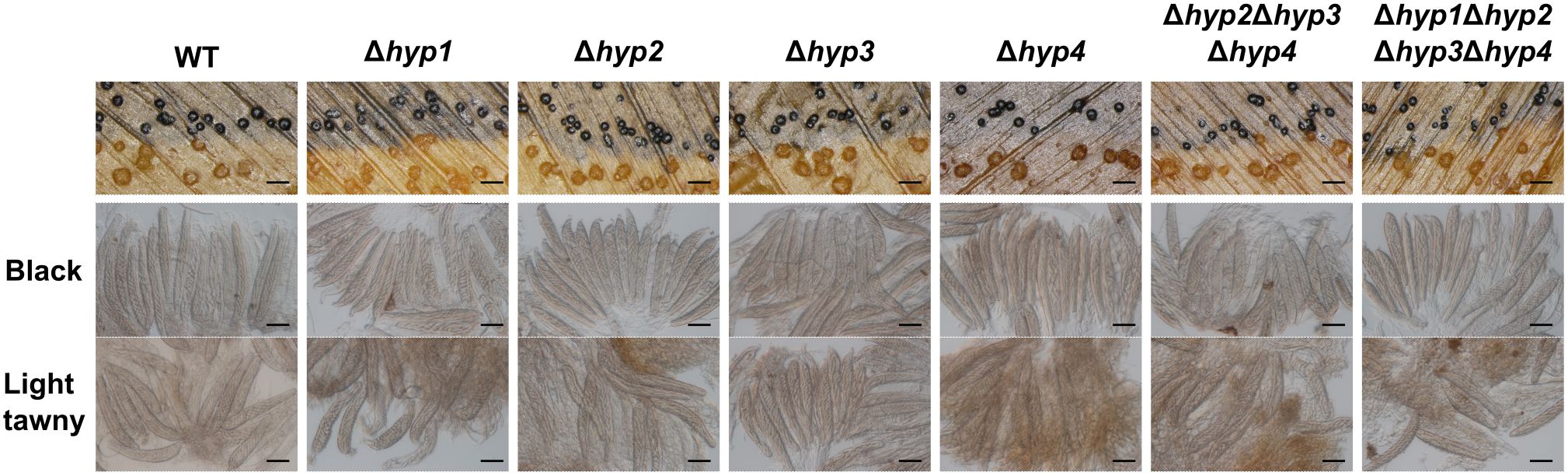
Figure 6. Sexual reproduction of disruptants of hydrophobin genes. The wild-type and each mutant were crossed with an albino strain. Black and light tawny pseudothecia were harvested 30 days after the cross and dissected to observe asci and ascospores. Bars: 1 mm (top), 50 µm (middle and bottom).
3.7 Colony hydrophobicity of hydrophobin gene disruptants
To investigate whether hydrophobins play a role in B. maydis colony hydrophobicity, solution containing 0.2% SDS and 50 mM EDTA was dropped on mycelial disks (Figure 7). The Δchk1 strain, disruptant of the CHK1 gene encoding MAPK, was used as the control because its colonies are significantly less hydrophobic than those of the wild type (Degani et al., 2013). The droplet on the mycelial disk of Δchk1 strain changed its shape after 10 min and disappeared completely after 20 min. The droplet on the wild-type strain remained unchanged after 30 min. In all disruptants, including the quadruple one, droplets also remained unchanged after 30 min (Figure 7). These results showed that B. maydis hydrophobins are not required for colony hydrophobicity.
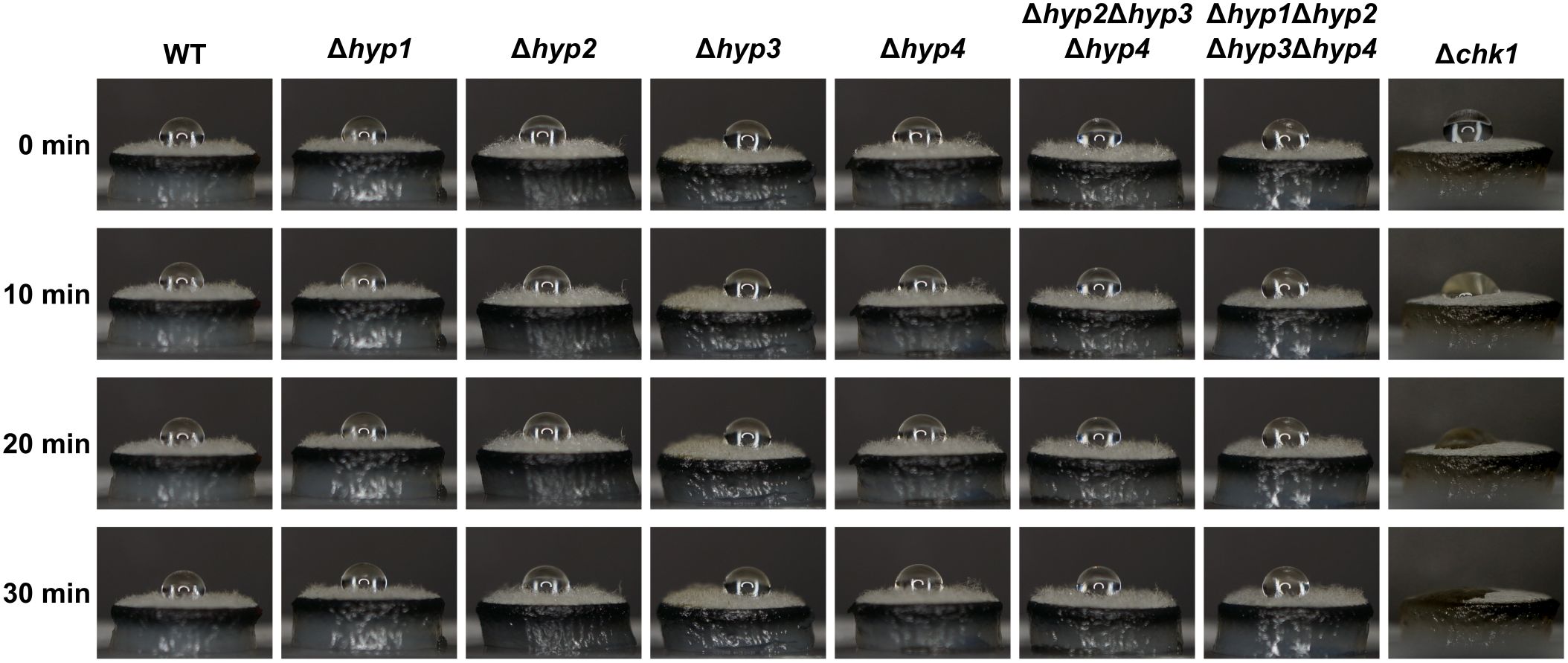
Figure 7. Hydrophobicity of the wild type and disruptants of hydrophobin genes. A 10-μL droplet of solution containing 0.2% sodium dodecyl sulfate and 50 mM ethylenediaminetetraacetic acid was placed on the mycelial disk of each strain and observed at the indicated time points.
3.8 Colony hydrophobicity of dewA gene disruptant complemented with B. maydis hydrophobin
From the above results, we hypothesized that the hydrophobins of B. maydis lost their ability to provide hydrophobicity. To test this hypothesis, we investigated whether B. maydis hydrophobin could restore the hydrophobicity of a hydrophobin-deficient mutant of A. nidulans. Aspergillus nidulans with a disruption of the dewA gene encoding a class I hydrophobin shows significant mycelial wettability (Grünbacher et al., 2014). First, we generated a dewA gene disruptant strain (Supplementary Figure S2A, B). The droplet on the CNT strain remained unchanged after 5 min, but the droplet on the ΔdewA strain disappeared completely after 2 min (Figure 8A). This result is consistent with previous studies (Stringer and Timberlake, 1995; Grünbacher et al., 2014). Next, we generated the dewAcomp strain by reintroducing the wild-type dewA gene into the ΔdewA strain and confirmed the reintroduction by PCR (Supplementary Figure S2C, D). The hydrophobicity of the dewAcomp strain was similar to that of the CNT strain (Figure 8A).
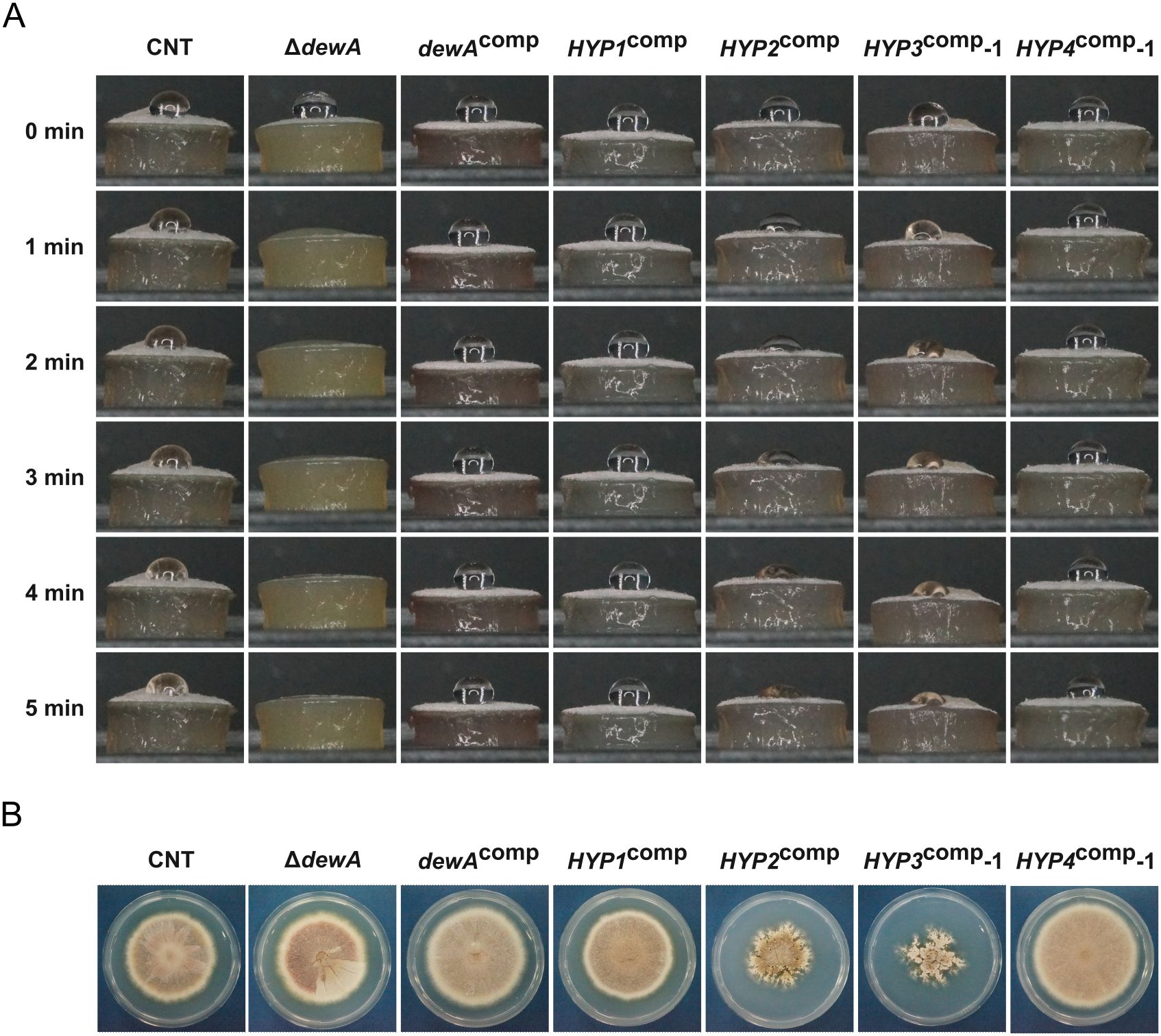
Figure 8. Colony phenotype of A. nidulans mutants: CNT, ΔdewA, dewAcomp, and ΔdewA with B. maydis hydrophobins. (A) Colony growth. Each strain was incubated on CD medium for 1 week. (B) Hydrophobicity of each strain. A 10-μL droplet of solution containing 0.2% sodium dodecyl sulfate and 50 mM ethylenediaminetetraacetic acid was placed on the mycelial disk of each strain and observed at the indicated time points.
We also constructed complemented strains, in which the B. maydis hydrophobin genes under the control of the dewA promoter were integrated into the ΔdewA strain, and obtained one strain of HYP1comp and HYP2comp each, three HYP3comp strains and two HYP4comp strains (Supplementary Figure S2C, D). While the colony growth of the HYP1comp and HYP4comp strains was similar to that of the CNT strain, HYP2comp and HYP3comp strains showed slower colony growth and an abnormal colony morphology compared to the CNT strain (Figure 8B; Supplementary Figure S6A). The expression analysis revealed that B. maydis hydrophobin expression in each complemented strain was equivalent to or even exceeded the expression level of dewA gene in the CNT strain (Supplementary Figure S6B). Similarly to the dewAcomp strain, the HYP1comp strain, complemented with class I hydrophobin HYP1 gene, regained its colony hydrophobicity (Figure 8A). Interestingly, although the degree of hydrophobicity varied, the class II hydrophobin-complemented strains (HYP2comp, HYP3comp, and HYP4comp) also showed increased hydrophobicity compared to the ΔdewA strain (Figure 8A; Supplementary Figure S6C). This result showed that the hydrophobins of B. maydis have not lost their ability to confer colony hydrophobicity.
4 Discussion
Previous studies in ascomycetes and basidiomycetes show that many filamentous fungi have varying numbers of genes encoding hydrophobins. In these and other fungi, hydrophobins are involved in a variety of functions. In P. oryzae, the hydrophobins Mpg1 and Mhp1 are involved in appressorium formation and pathogenicity (Talbot et al., 1996; Kim et al., 2005). RodA on the conidia of A. fumigatus masks the cell wall components from recognition by the host immune system (Aimanianda et al., 2009). In A. oryzae, RolA interacts with the cutinase CutL1 and is involved in efficient polybutylene succinate-co-adipate degradation (Takahashi et al., 2005). In the basidiomycete Pleurotus ostreatus, hydrophobin is important for efficient lignin degradation and resistance to environmental stress (Han et al., 2023a, b). However, information about the hydrophobins of the pathogen B. maydis has been limited. In this study, we found one class I (Hyp1) and three class II hydrophobins (Hyp2–Hyp4). These number and types of hydrophobins identified in our study are consistent with the findings of Degani et al. (2013), who searched for hydrophobin genes in this fungus using a different method.
We began functional analyses by generating single-gene disruptants for each of the four hydrophobin genes. Each disruptant showed no significant differences compared to the wild-type strain in terms of hyphal growth, conidiation, environmental stress tolerance, virulence, or sexual reproduction (Figures 3-6). Disruption of a single gene out of several does not always alter the phenotype, so phenotypic analyses using a disruptant of multiple genes are also important. In B. cinerea, some double and triple disruptants, but not single disruptants, have abnormal apothecia (Terhem and van Kan, 2014). In F. graminearum, the disruption of one to five hydrophobin genes affects pathogenicity, conidiation, and secondary metabolism (Shin et al., 2022).
Here, we generated triple and quadruple disruptants. However, contrary to the reports for other fungi, the phenotype of these strains was similar to those of the wild-type strain and single disruptants. Thus, unlike in some other filamentous fungi, including ascomycetes and basidiomycetes, the hydrophobins of B. maydis appear not to play an important role in biological processes.
In general, hydrophobins are thought to be secreted into the extracellular matrix and to confer hydrophobic properties to mycelia and conidia (Wösten, 2001). The lack of class I hydrophobins DewA and RodA reduces colony hydrophobicity in A. nidulans (Grünbacher et al., 2014). Hydrophobins are also involved in colony hydrophobicity in several phytopathogenic filamentous fungi including P. oryzae and B. cinerea (Talbot et al., 1993; Kim et al., 2005; Mosbach et al., 2011). In our study, however, colony hydrophobicity did not differ significantly between the wild-type strain, single disruptants, or the quadruple disruptant (Figure 7). These results show that hydrophobins of this fungus are not essential for colony hydrophobicity. In B. maydis, hydrophobicity depends on other factors such as the Nps4, as the loss of the NPS4 gene reduces colony hydrophobicity (Turgeon et al., 2008). These authors attributed colony hydrophobicity to either Nps4 or to the regulation of hydrophobins by it. In conjunction with the present study, it is more likely that the former possibility is correct. Since the disruptant of the CHK1 gene encoding MAPK had hydrophilic colonies (Figure 7; Degani et al., 2013), Nps4 may function downstream of the MAPK pathway. To investigate the function of B. maydis hydrophobins in more detail, we introduced the HYP1, HYP2, HYP3, and HYP4 genes into the ΔdewA strain of A. nidulans.
Interestingly, all resulting complemented strains showed increased colony hydrophobicity compared to the ΔdewA strain (Figure 8A; Supplementary Figure S6C). Generally, class II hydrophobins possess lower hydrophobicity compared to class I hydrophobins (Wessels et al., 1991). Reflecting this fact, the hydrophobicity of the HYP1comp strain was significantly higher compared to that of the HYP2comp and HYP3comp strains (Figure 8A; Supplementary Figure S6C). However, the HYP4comp strain, despite being a class II hydrophobin, showed similar hydrophobicity to the HYP1comp and dewAcomp strains (Figure 8A; Supplementary Figure S6C). Furthermore, the expression level of the HYP4 gene in this strain was significantly higher than that of the dewA gene in the CNT and dewAcomp strains (Supplementary Figure S6B). This result suggests that, when their expression level is sufficiently high, class II hydrophobins can confer colonies hydrophobic properties comparable to class I hydrophobins. Although A. nidulans does not possess any endogenous class II hydrophobins (Tanaka et al., 2022), these results showed that exogenous class II hydrophobins are functionally viable in this fungus. Considering that HYP2comp and HYP3comp strains formed abnormal colony morphologies (Figure 8B; Supplementary Figure S6A), it is possible that the secretion process of the exogenous class II hydrophobins, or their behavior within the extracellular matrix causes some disadvantages to the growth of this fungus. Furthermore, unlike Hyp2 and Hyp3, the presence of the characteristic Gly-rich sequence at the N-terminus of Hyp4 may play a role in mitigating these adverse effects, explaining the normal colony morphology of the HYP4comp strain, despite it being a class II hydrophobin (Figure 1; Supplementary Figure S3). These findings strongly suggest that hydrophobins possess different functional roles depending on the fungal species.
We were unable to detect any phenotypic changes in the quadruple disruptant of hydrophobin genes in B. maydis. However, we found that the four genes encoding hydrophobins in this fungus are expressed, and that all four hydrophobins have retained their function as a hydrophobin when expressed in A. nidulans. Hence, these hydrophobins may have some functionally redundant roles in B. maydis or their role may not be the conferral of colony hydrophobicity. To clarify what their role is in B. maydis, it is necessary to analyze the hydrophobin proteins, including their localization in this fungus. Analyses of the regulation of hydrophobins in these two fungal species with different mechanisms of colony hydrophobicity may shed light on the diversification of hydrophobin function.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
KT: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. HY: Formal Analysis, Investigation, Methodology, Writing – review & editing. MS: Data curation, Formal Analysis, Methodology, Writing – review & editing. YT: Data curation, Methodology, Writing – review & editing. MK: Data curation, Funding acquisition, Resources, Writing – review & editing. YH: Data curation, Project administration, Resources, Writing – review & editing, Funding acquisition. CT: Data curation, Funding acquisition, Project administration, Resources, Conceptualization, Writing – review & editing. AY: Conceptualization, Data curation, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported partly by the Institute for Fermentation, Osaka, Japan (K-2019-002) and JSPS KAKENHI (23K23505).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2025.1604903/full#supplementary-material
References
Aimanianda V., Bayry J., Bozza S., Kniemeyer O., Perruccio K., Elluru S. R., et al. (2009). Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460, 1117–1121. doi: 10.1038/nature08264
Beckerman J. L. and Ebbole D. J. (1996). MPG1, a gene encoding a fungal hydrophobin of Magnaporthe grisea, is involved in surface recognition. Mol. Plant-Microbe Interact. 9, 450–456. doi: 10.1094/mpmi-9-0450
Carroll A. M., Sweigard J. A., and Valent B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal Genet. Rep. 41, 1–2. doi: 10.4148/1941-4765.1367
Condon B. J., Leng Y., Wu D., Bushley K. E., Ohm R. A., Otillar R., et al. (2013). Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. PloS Genet. 9, e1003233. doi: 10.1371/journal.pgen.1003233
Degani O., Lev S., and Ronen M. (2013). Hydrophobin gene expression in the maize pathogen Cochliobolus heterostrophus. Physiol. Mol. Plant Pathol. 83, 25–34. doi: 10.1016/j.pmpp.2013.03.003
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fujioka T., Mizutani O., Furukawa K., Sato N., Yoshimi A., Yamagata Y., et al. (2007). MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6, 1497–1510. doi: 10.1128/EC.00281-06
Furukawa K., Katsuno Y., Urao T., Yabe T., Yamada-Okabe T., Yamada-Okabe H., et al. (2002). Isolation and functional analysis of a gene, tcsB, encoding a transmembrane hybrid-type histidine kinase from Aspergillus nidulans. Appl. Environ. Microbiol. 68, 5304–5310. doi: 10.1128/AEM.68.11.5304-5310.2002
Grünbacher A., Throm T., Seidel C., Gutt B., Röhrig J., Strunk T., et al. (2014). Six hydrophobins are involved in hydrophobin rodlet formation in Aspergillus nidulans and contribute to hydrophobicity of the spore surface. PloS One 9, e94546. doi: 10.1371/journal.pone.0094546
Hagiwara D., Asano Y., Marui J., Furukawa K., Kanamaru K., Kato M., et al. (2007). The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71, 1003–1014. doi: 10.1271/bbb.60665
Han J., Kawauchi M., Schiphof K., Terauchi Y., Yoshimi A., Tanaka C., et al. (2023a). Features of disruption mutants of genes encoding for hydrophobin Vmh2 and Vmh3 in mycelial formation and resistance to environmental stress in Pleurotus ostreatus. FEMS Microbiol. Lett. 370, 1–8. doi: 10.1093/femsle/fnad036
Han J., Kawauchi M., Terauchi Y., Yoshimi A., Tanaka C., Nakazawa T., et al. (2023b). Physiological function of hydrophobin Vmh3 in lignin degradation by white-rot fungus Pleurotus ostreatus. Lett. Appl. Microbiol. 76. doi: 10.1093/lambio/ovad048
Haridas S., González J. ,. B., Riley R., Koriabine M., Yan M., Ng V., et al. (2023). T-toxin virulence genes: unconnected dots in a sea of repeats. mBio 14, 1–14. doi: 10.1128/mbio.00261-23
Izumitsu K., Hatoh K., Sumita T., Kitade Y., Morita A., Tanaka C., et al. (2012). Rapid and simple preparation of mushroom DNA directly from colonies and fruiting bodies for PCR. Mycoscience 53, 396–401. doi: 10.1007/s10267-012-0182-3
Izumitsu K., Yoshimi A., Kubo D., Morita A., Saitoh Y., and Tanaka C. (2009). The MAPKK kinase ChSte11 regulates sexual/asexual development, melanization, pathogenicity, and adaptation to oxidative stress in Cochliobolus heterostrophus. Curr. Genet. 55, 439–448. doi: 10.1007/s00294-009-0257-7
Kim S., Ahn I. P., Rho H. S., and Lee Y. H. (2005). MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 57, 1224–1237. doi: 10.1111/j.1365-2958.2005.04750.x
Klimes A., Amyotte S. G., Grant S., Kang S., and Dobinson K. F. (2008). Microsclerotia development in Verticillium dahliae: regulation and differential expression of the hydrophobin gene VDH1. Fungal Genet. Biol. 45, 1525–1532. doi: 10.1016/j.fgb.2008.09.014
Kumar S., Stecher G., Li M., Knyaz C., and Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kyte J. and Doolittle R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi: 10.1016/0022-2836(82)90515-0
Miyazawa K., Yamashita T., Takeuchi A., Kamachi Y., Yoshimi A., Tashiro Y., et al. (2021). A glycosylphosphatidylinositol-anchored alpha-amylase encoded by amyD contributes to a decrease in the molecular mass of cell wall alpha-1,3-Glucan in Aspergillus nidulans. Front. Fungal Biol. 2. doi: 10.3389/ffunb.2021.821946
Mosbach A., Leroch M., Mendgen K. W., and Hahn M. (2011). Lack of evidence for a role of hydrophobins in conferring surface hydrophobicity to conidia and hypahe of botrytis cinerea. BMC Microbiol. 11, 1–14. doi: 10.1186/1471-2180-11-10
Ohm R. A., Feau N., Henrissat B., Schoch C. L., Horwitz B. A., Barry K. W., et al. (2012). Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PloS Pathog. 8, e1003037. doi: 10.1371/journal.ppat.1003037
Pham C. L., Rey A., Lo V., Soulés M., Ren Q., Meisl G., et al. (2016). Self-assembly of MPG1, a hydrophobin protein from the rice blast fungus that forms functional amyloid coatings, occurs by a surface-driven mechanism. Sci. Rep. 6, 25288. doi: 10.1038/srep25288
Quarantin A., Hadeler B., Kroger C., Schäfer W., Favaron F., Sella L., et al. (2019). Different hydrophobins of Fusarium graminearum are involved in hyphal growth, attachment, water-air interface penetration and plant infection. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00751
Sbrana F., Fanelli D., Vassalli M., Carresi L., Scala A., Pazzagli L., et al. (2010). Progressive pearl necklace collapse mechanism for cerato-ulmin aggregation film. Eur. Biophys. J. 39, 971–977. doi: 10.1007/s00249-009-0465-6
Sehuren F. H. J. and Wessels J. G. H. (1990). Two genes specifically expressed in fruiting dikaryons of Schizophyllum commune: homologies with a gene not regulated by mating-type genes. Gene 90, 199–205. doi: 10.1016/0378-1119(90)90180-y
Shin Y. K., Kim D. W., Lee S. W., Lee M. J., Gi Baek S., Lee T., et al. (2022). Functional roles of all five putative hydrophobin genes in growth, development, and secondary metabolism in Fusarium graminearum. Fungal Genet. Biol. 160, 103683. doi: 10.1016/j.fgb.2022.103683
Stringer M. and Timberlake W. (1993). Cerato-ulmin, a toxin involved in Dutch elm disease, is a fungal hydrophobin. Plant Cell 5, 145–146. doi: 10.1105/tpc.5.2.145
Stringer M. A. and Timberlake W. E. (1995). dewA encodes a fungal hydrophobin component of the Aspergillus spore wall. Mol. Microbiol. 16, 33–44. doi: 10.1111/j.1365-2958.1995.tb02389.x
Sumita T., Izumitsu K., and Tanaka C. (2017). Characterization of the autophagy-related gene BmATG8 in Bipolaris maydis. Fungal Biol. 121, 785–797. doi: 10.1016/j.funbio.2017.05.008
Sweigard J. A., Chumley F., Carroll A., Farrall L., and Valent B. (1997). A series of vectors for fungal transformation. Fungal Genet. Rep. 44, 52–53. doi: 10.4148/1941-4765.1287
Takahashi T., Maeda H., Yoneda S., Ohtaki S., Yamagata Y., Hasegawa F., et al. (2005). The fungal hydrophobin RolA recruits polyesterase and laterally moves on hydrophobic surfaces. Mol. Microbiol. 57, 1780–1796. doi: 10.1111/j.1365-2958.2005.04803.x
Talbot N. J., Ebbole D. J., and Hamer J. E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575–1590. doi: 10.1105/tpc.5.11.1575
Talbot N. J., Kershaw M. J., Wakley G. E., De Vries O., Wessels J., and Hamer J. E. (1996). MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8, 985–999. doi: 10.1105/tpc.8.6.985
Tanaka C., Kubo Y., and Tsuda M. (1991). Genetic analysis and characterization of Cochliobolus heterostrophus. Mycol. Res. 95, 49–56. doi: 10.1016/S0953-7562(09)81360-9
Tanaka T., Terauchi Y., Yoshimi A., and Abe K. (2022). Aspergillus hydrophobins: physicochemical properties, biochemical properties, and functions in solid polymer degradation. Microorganisms 10, 1498. doi: 10.3390/microorganisms10081498
Terhem R. B. and van Kan J. A. (2014). Functional analysis of hydrophobin genes in sexual development of Botrytis cinerea. Fungal Genet. Biol. 71, 42–51. doi: 10.1016/j.fgb.2014.08.002
Turgeon B. G., Oide S., and Bushley K. (2008). Creating and screening Cochliobolus heterostrophus non-ribosomal peptide synthetase mutants. Mycol. Res. 112, 200–206. doi: 10.1016/j.mycres.2007.10.012
Wessels J. G. H., de Vries O. M. H., Ásgeirsdóttir S. A., and Schuren F. H. J. (1991). Hydrophobin genes involved in formation of aerial hyphae and fruit bodies in schizophyllum. Plant Cell 3, 793–799. doi: 10.1105/tpc.3.8.793
Whiteford J. R. and Spanu P. D. (2001). The hydrophobin HCf-1 of Cladosporium fulvum is required for efficient water-mediated dispersal of conidia. Fungal Genet. Biol. 32, 159–168. doi: 10.1006/fgbi.2001.1263
Wösten H. A. B. (2001). Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55, 625–646. doi: 10.1146/annurev.micro.55.1.625
Yoshida H. and Tanaka C. (2019). Monitoring of in planta gene expression for xylan degradation and assimilation in the maize pathogen Bipolaris maydis. Mycoscience 60, 116–124. doi: 10.1016/j.myc.2018.10.002
Yoshimi A., Sano M., Inaba A., Kokubun Y., Fujioka T., Mizutani O., et al. (2013). Functional analysis of the alpha-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major alpha-1,3-glucan synthase in this fungus. PloS One 8, e54893. doi: 10.1371/journal.pone.0054893
Keywords: hydrophobin, hydrophobicity, Bipolaris maydis, Cochliobolus heterostrophus, Aspergillus nidulans
Citation: Tsuji K, Yoshida H, Saba M, Terauchi Y, Kawauchi M, Honda Y, Tanaka C and Yoshimi A (2025) Hydrophobins in Bipolaris maydis do not contribute to colony hydrophobicity, but their heterologous expressions alter colony hydrophobicity in Aspergillus nidulans. Front. Fungal Biol. 6:1604903. doi: 10.3389/ffunb.2025.1604903
Received: 02 April 2025; Accepted: 03 November 2025;
Published: 20 November 2025.
Edited by:
Eduardo Antonio Espeso, Spanish National Research Council (CSIC), SpainReviewed by:
Bishnu Maya Bashyal, Indian Agricultural Research Institute (ICAR), IndiaOier Etxebeste, University of the Basque Country, Spain
Copyright © 2025 Tsuji, Yoshida, Saba, Terauchi, Kawauchi, Honda, Tanaka and Yoshimi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akira Yoshimi, eW9zaGltaS5ha2lyYS44Y0BreW90by11LmFjLmpw
 Kenya Tsuji
Kenya Tsuji Hiroshi Yoshida
Hiroshi Yoshida Masafumi Saba2
Masafumi Saba2 Yuki Terauchi
Yuki Terauchi Moriyuki Kawauchi
Moriyuki Kawauchi Yoichi Honda
Yoichi Honda Akira Yoshimi
Akira Yoshimi