- 1Department of Biotechnology, Deen Dayal Upadhyaya Gorakhpur University, Gorakhpur, Uttar Pradesh, India
- 2Department of Biotechnology, School of Engineering and Technology, Sandip University, Nashik, Maharashtra, India
- 3Division of Plant Molecular Biology and Biotechnology, Department of Biosciences, Rajagiri College of Social Sciences, Kochi, Kerala, India
- 4ICAR-Vivekananda Institute of Hill Agriculture, Almora, Uttarakhand, India
- 5Department of Botany, Deen Dayal Upadhyaya Gorakhpur University, Gorakhpur, Uttar Pradesh, India
Introduction: The Pyrabactin Resistance 1-like (PYL) receptors, a family of proteins in plants, play a vital role in abscisic acid (ABA) signalling, assisting plants in managing abiotic stresses. Finger millet (Eleusine coracana (L.) Gaertn) is a naturally drought tolerant crop, yet the receptor proteins involved in its stress signalling pathways remain poorly understood.
Method: This study employed bioinformatics, machine learning, and molecular approaches to identify, characterize, and profile the expression of PYL receptors in response to drought and salinity stress.
Results: The study identified 14 PYL genes in the finger millet genome, irregularly distributed across four of the nine mapped chromosomes. Phylogenetic analysis grouped these genes into three subfamilies. Machine learning analysis highlighted five putative PYL genes—EcPYL4-2A, EcPYL7-2B, EcPYL11-5A, EcPYL12-5A, and EcPYL14-5B with expression levels exceeding 70% under drought and salinity stress. These genes were further validated through qRT-PCR, confirming their expression under stress conditions, though expression levels varied across tissues and genes.
Discussion: The identification of PYL genes responsive to drought and salt stress provides valuable insights into the stress-signalling mechanisms of finger millet. Among the identified genes, EcPYL7-2B and EcPYL12-5A emerged as promising candidates for further characterization through genome editing and molecular approaches. This study highlights the potential of these genes in enhancing the stress resilience of finger millet, contributing to its role in improving food and nutritional security under challenging environmental conditions.
Introduction
Finger millet (Eleusine coracana L. Gaertn.), an ancient grain, has considerable historical, cultural, and nutritional significance, especially in Asia and Africa (Sood et al., 2019). Farmers widely cultivate it in arid and semi-arid regions for their daily diet and to fulfill its nutritional properties for their daily life (Goron and Raizada, 2015). Finger millet is widely recognized for its capacity to thrive in resource-scarce environments and its exceptional nutritional value. Amidst climate change and population expansion, the stability of finger millet production is essential for ensuring global food security and socio-economic development (Rani et al., 2023; Sanjay Kumar et al., 2024).
The growth and yield of finger millet are primarily influenced by biotic and abiotic stresses (Rani et al., 2023). As finger millet has evolved over time, it has developed complex signal networks which regulate a number of biochemical and physiological processes imparting tolerance to biotic and abiotic stresses (Xiong et al., 2002; Hittalmani et al., 2017). Studying the expression pattern and role of key candidate genes associated with stress tolerance will be helpful in developing new stress responsive finger millet genotypes.
Abscisic acid (ABA) is a phytohormone that has a huge impact on plant development and growth. It is an important hormone that controls organ senescence and morphogenesis, stomatal movement, and transpiration (Cutler et al., 2010; Zhu, 2016). In addition to acting as a stress indicator for plants, ABA may also be used to identify drought, salinity, or pathogen infection (Zhu, 2002; Cutler et al., 2010). In response to abiotic stress, the accumulation of ABA increases in plants. Generally, plants perceive ABA through the Pyrabactin Resistance 1 (PYR1)/PYR1-like (PYL)/Regulatory component of the ABA receptor (RCAR) protein family. After binding to PYL, ABA changes the conformation of PYL, which facilitates its binding to type 2C protein phosphatases (PP2Cs). This allows SnRK2s to be released from PP2C inhibition. Yadav et al. (2020) reported that ABA stimulates downstream targets like transcription factors (TF) and other regulatory proteins because it turns on SnRK2. The stimulated TF then enters the nucleus, up-regulating downstream genes associated with ABA-induced stress. Hence, land plants require PYLs, PP2Cs, and SnRK2s for ABA signalling to survive under abiotic stresses like drought, salinity, and others (Cutler et al., 2010).
PYLs have been identified and characterized in Arabidopsis (Gonzalez-Guzman et al., 2012), rice (Kim et al., 2012; Yadav et al., 2020), sorghum (Dalal and Inupakutika, 2014), maize (Fan et al., 2018; Wang et al., 2018) and wheat (Lei et al., 2021). Some plants, like Arabidopsis, have a gene called AtPYR1 that codes for a cyclase subfamily member and is part of the star related lipid transfer (START) domain superfamily (Park et al., 2009). AtPYL (PYR1-like) genes AtPYL1 to AtPYL13 are genes that share the same function as AtPYR1. In Arabidopsis PYL family, there are 14 members, each having a START domain. These 14 PYLs are divided into three subfamilies (Park et al., 2009; Park et al., 2010). It has been observed that some AtPYLs form monomers which interact with phosphatase 2C (Bai et al., 2013) protein without the involvement of ABA. In addition, some AtPYLs can form homodimers that only interact with PP2C after binding to ABA (Hao et al., 2011). It has been reported that overexpression of AtPYL4 enhanced drought tolerance in Arabidopsis. More interestingly, overexpression of OsPYL5 in rice improved drought and salinity tolerance (Pizzio et al., 2013; Kim et al., 2014). These results suggest that the PYL family plays an important role in plant development and abiotic stress response. However, the role of PYL receptors in finger millet under abiotic stresses is still unknown, and needs to be deciphered. Identification and characterization of PYL genes across the finger millet genome can provide an insight into the response of finger millet towards stresses and role of ABA signaling pathways.
Several studies have reported that finger millet is drought-tolerant, yet further research is required to identify how genes and regulatory proteins contribute to stress impartation in finger millet (Mukami et al., 2019; Talwar et al., 2020; Rani et al., 2024a; Rani et al., 2024c; Rani et al., 2024b). The draft and annotated genome sequence of finger millet has been made public and openly accessible (Hittalmani et al., 2017; Hatakeyama et al., 2018); however, many genes are still unannotated and need to be studied using in silico and wet lab methods. To the best of our knowledge, the role of PYL receptors in finger millet, has not been reported. The ultimate aim of the study is to reveal the role of PYL receptors in finger millet through in silico, machine learning and molecular approaches.
The advancement of bioinformatics and computational tools has enabled the integration of machine learning (ML) into stress biology, offering new opportunities for predicting gene expression patterns under adverse environmental conditions. This study provides an insight into the role of Pyrabactin-Like Receptors (PYL) in finger millet under drought and salinity stress with involvement of ABA signaling pathways using a comprehensive approach comprising of in silico, machine learning, and molecular tools. The finger millet genome was mined for several bioinformatics attributes like phylogenetic analysis, chromosomal localization, gene structure mapping, motif analysis, cis-regulatory element identification, protein-protein interaction analysis, miRNA target predictions and digital expression profiling using machine learning algorithm of ASRpro web server (Meher et al., 2024). Further, the most promising PYL candidate genes, likely to be expressed under drought and salinity stress, has been identified revealing the possible plant stress adaptation mechanisms. Furthermore, qRT-PCR validation was conducted for the identified candidate genes in shoots and roots of finger millet exposed to drought and salinity stress. The identification of key PYL receptors in finger millet could reveal the possible molecular mechanisms underlying its resilience to abiotic stresses.
Materials and methods
Collection and characterization of PYL genes of finger millet
Firstly, we obtained the PYL full-length protein sequence of rice from the NCBI (http://www.ncbi.nlm.nih.gov/nucleotide/) (Yadav et al., 2020). The rice PYLs gene sequences were used as query sequences in the Phytozome database (https://phytozome-next.jgi.doe.gov/) to find PYLs in the finger millet genome. This was done by using BLASTp searches (http://blast.ncbi.nlm.nih.gov/) against the finger millet genome E. coracana v1.1 (Goodstein et al., 2012; Hittalmani et al., 2017; Hatakeyama et al., 2018). The appeared duplicate and redundant sequences were eliminated by venny2.1 (https://bioinfogp.cnb.csic.es/tools/venny/) (Oliveros, 2016). Then, the NCBI open reading frames (ORF) Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to determine ORFs. On the other hand, we used Pfam (https://www.ebi.ac.uk/interpro/entry/pfam/) to search for the polyketide cyclase 2/START domain (PF10604), which is a characteristic of the PYR/PYL protein sequence. Further, multiple sequence alignment by MEGA11 was used to identify conserved amino acid residues among all putative PYL sequences of finger millet (Tamura et al., 2021). Genes that lack specific conserved domains were sorted and removed. Further the domain position, physicochemical attributes and subcellular localization were predicted by HMMSCAN (http://www.ebi.ac.uk/Tools/hmmer/search/hmmscan, ExPaSy Protparam (https://web.expasy.org/protparam/) and Wolf-Psort online tool (https://www.genscript.com/tools/wolf-psort) respectively (Gasteiger et al., 2005; Horton et al., 2007).
Evolutionary analysis, gene structure display and motif prediction of PYL genes
The PYL gene family in finger millet has been designated as EcPYL1-1A, which indicates for the first PYL genes of E. coracana and is found on chromosome 1A. Similarly, other finding was also annotated and used for evolutionary study. An evolutionary analysis was made between putative PYLs genes of finger millet and the PYLs genes of rice (Yadav et al., 2020), sorghum (Dalal and Inupakutika, 2014), and foxtail millet (Duarte et al., 2019), which were retrieved from the Phyozome and NCBI database. All of the retrieved sequences of the PYL genes family were aligned through ClustalW, and the phylogenetic tree was constructed through MEGA11 using the Maximum Likelihood method at 1,000 bootstrap levels (Larkin et al., 2007; Tamura et al., 2021). MapGene2 Chromosome v2.1 (http://mg2c.iask.in/mg2c_v2.1/) was used for physical mapping of PYLs gene of finger millet (Devos et al., 2023), and TB tool was used to display the exon-intron structure (Chen et al., 2020; Chao et al., 2021). The MEME Suite Vs. 5.5.3 (https://meme-suite.org/meme/tools/meme) predicted the conserved motif within each finger millet protein sequence. The maximum number of motifs was set to 6, keeping others at default parameters (Bailey et al., 2015).
Ka/Ks and gene duplication analysis of PYLs gene family of finger millet
A ratio of non-synonymous (Ka) versus synonymous (Ks) substitution rates of nucleotides was calculated among finger millet PYLs gene family by using Ka/Ks calculation tool (http://services.cbu.uib.no/tools/kaks) and the gene duplication event was calculated by using T = Ks/2λ), where (λ)= (6.5 × 10−9 (Gaut et al., 1996; Zhang et al., 2006). It measures the selective pressures placed on genes and can be used to identify pairs of genes where encoded proteins may have changed their function. When the ratio of Ka/Ks > 1, it indicates that there is a positive selective pressure or Darwinian selection. If the ratio is 1, it indicates neutral evolution and if the ratio is less than 1, then it indicates stabilizing selection, i.e., the protein sequence is pressured to be conserved.
Prediction of cis-regulatory elements in the promoters of finger millet
The 2,000 bp upstream sequences from the translations tart site of all of the EcPYLs genes were obtained from finger millet genome at Phytozome database (Goodstein et al., 2012) and the putative cis regulatory elements were predicted by using the PlantCARE web server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to know the putative function associated with the genes (Lescot et al., 2002). According to the cis-regulatory elements, PYL genes were categorized into three groups based on their response to phytohormones, stress, and growth and development.
miRNA-PYLs target prediction and protein-protein interaction and gene ontology analysis
For the prediction of miRNA target sites in the EcPYL genes, the miRNA dataset of finger millet was retrieved from PmiREN server (https://www.pmiren.com/) (Guo et al., 2020) and cDNA sequence of EcPYLs genes were examined against miRNA dataset using the pSRNATarget server (https://www.zhaolab.org/psRNATarget/) (Dai and Zhao, 2011). To determine the interrelationships among the 14 putative EcPYLs genes, the STRING database (https://string-db.org/) (Szklarczyk et al., 2021) was used at confidence level (0.07) against rice (Yu et al., 2002), Arabidopsis (Cheng et al., 2017), sorghum (McCormick et al., 2018) and foxtail millet (Zhang et al., 2012) genome one by one. Using the combination of Cytoscape v. 3.10.3 with MCODE (Molecular Complex Detection), the most accurate prediction of the interaction network was displayed. Additionally, the integration of Cytoscape v. 3.10.3 with BinGO program were used for Gene Ontology (GO), functional annotation and gene enrichment profiling of EcPYL genes of finger millet (Paul et al., 1971; Bader and Hogue, 2003).
Machine learning approach for preselection of candidate genes
After characterization of EcPYL genes, machine learning algorithm was used to predict the expression pattern of these genes. For the prediction of probability of expression of EcPYL genes under drought and salinity stresses we used ASRPro webserver (http://iasri-sg.icar.gov.in/asrpro/result.php) (Meher et al., 2024). This web server utilized machine learning algorithms to assess the likelihood of gene expression under the given stress conditions, and generated a probability score for each gene. Genes with a probability score of ≥0.70 were considered highly likely to be expressed in response to drought and salinity stress. On the basis of the probability of gene expression, we constructed a heatmap and scatter plot to predict the candidate genes for qRT-PCR expression profiling.
Gene expression analysis
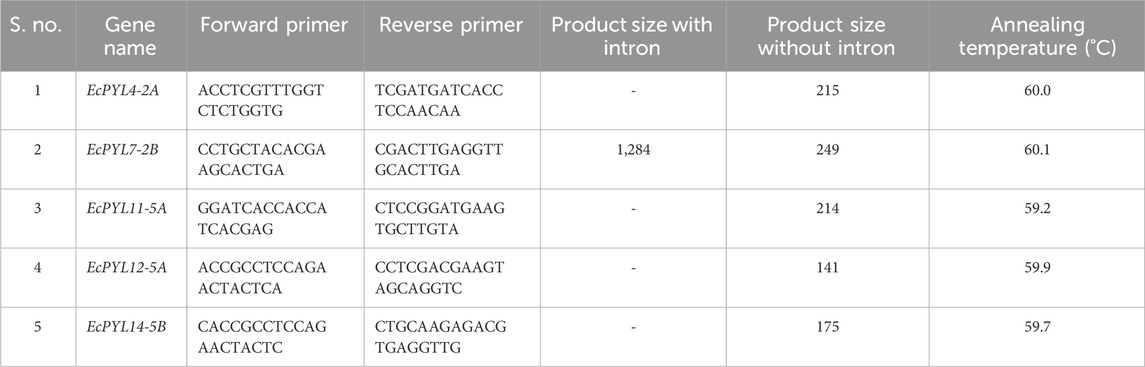
Five PYL genes (EcPYL4-2A, EcPYL7-2B, EcYL11-5A, EcPYL12-5A, and EcPYL14-5B) with probability score of 0.70 were selected for qRT-PCR analysis. Primers for each gene were designed using Primer3 input software (https://bioinfo.ut.ee/primer3-0.4.0/), and the primers specificity was done with genome sequences of finger millet to ensure that primers only bind to the intended target sequence. The primer sequences, annealing temperature, and product size of each primer were listed in Table 1. Healthy and viable seeds of finger millet genotype (IC87469) was selected for this experiment, and grown under drought (15% PEG-induced drought stress) and salinity (200 mM NaCl) stress conditions according to previous protocols (Rakkammal et al., 2023b; Rakkammal et al., 2023a). Both shoot and root tissues were harvested after 28 days of growth, and they were used for gene expression analysis. Total RNA was isolated from shoot and root tissues using the Trizol method (Rakkammal et al., 2023a; Rakkammal et al., 2023b), and cDNA was synthesized using the QuantiTect Reverse transcription kit (Qiagen, Germany). EF-1α was used as a reference gene to normalize the expression pattern of each PYL gene. Cycle threshold values of each PYL genes were analyzed using the formula 2–∆∆Ct. A total of three biological and three technical replicates were used for each treatment. Whereas, statistical analysis were performed through origin 2021 software (https://www.originlab.com/origin) using ANOVA, followed by Tukey test for multiple comparisons for significantly difference genes (p ≤ 0.05).
Homology modeling of finger millet PYLs genes
The SWISS MODEL predicted the in silico structure of EcPYLs proteins (Waterhouse et al., 2018). PDBsum (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html) and the SAVES v6.0 server (https://saves.mbi.ucla.edu/) were used to validate, analyze, and estimate the quality of the build model in two dimensions (Laskowski et al., 2018). Whereas, ResProx (http://www.resprox.ca/) (Berjanskii et al., 2012) was used to estimate the resolution of the model and SuperPose v. 1 (http://superpose.wishartlab.com/) and Biovia Discovery Studio 2019 was used for superposition and visualization of model, respectively.
Results
Sequence analysis of PYL genes in finger millet
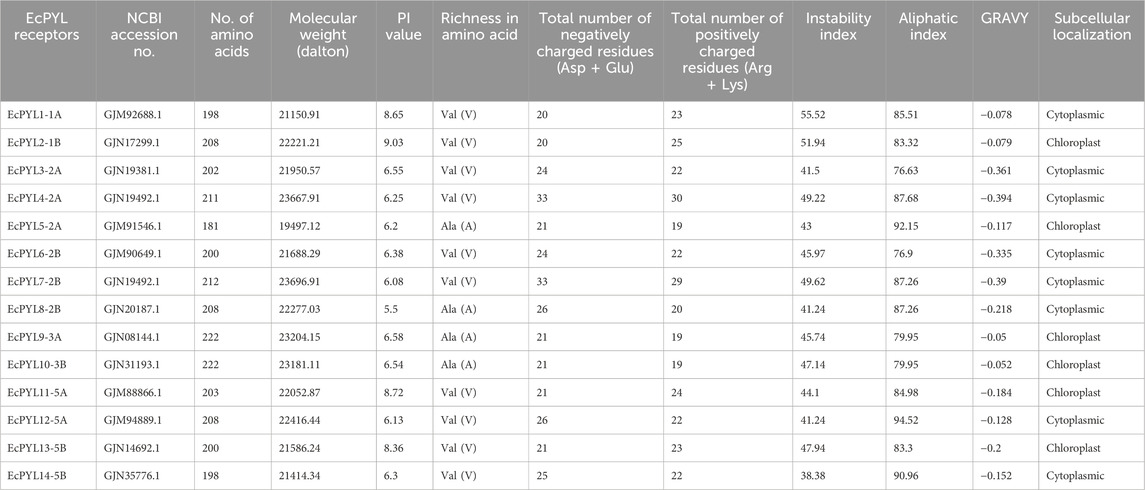
A total of 27 PYL genes were retrieved from the finger millet genome (Supplementary Table S1). Moreover, based on conserved domain analysis, the redundant sequences were removed, and 14 PYL genes were considered for further in silico studies. Table 2 shows the gene position, ORF length, domain position, and chromosome location of these 14 PYL genes. These genes have the polyketide cyclase 2/START domain and have the Pfam accession number PF10604. These 14 putative EcPYLs genes were unevenly distributed among four out of nine sets of chromosomes (Figure 1). The chromosomes 2A and 2B possess three EcPYL genes, chromosomes 5A and 5B has two EcPYL genes while chromosomes 1A and 1B and 3A and 3B possess only one EcPYL gene. EcPYL genes were not observed on chromosomes 4, 6, 7, 8, and 9. In general, both A and B sub-genomes have seven EcPYL genes, showing that there is not much difference in the number of PYL genes at the sub-genome level. For a better understanding, we named the possible genes EcPYL (E. coracana PYL) followed by the number of chromosomes located, for example, EcPYL_1A. We identified 14 EcPYL genes in finger millet and sequentially named them as EcPYL1 to EcPYL14, followed by their respective chromosomal numbers. The physicochemical attributes of putative PYLs of finger millet (Table 3) revealed that EcPYLs ranged from 181 to 222 aa. Accordingly, the molecular weight ranged from 19.5 to 23.7 kDa, and PI ranged from 5.5 to 9.3. The ORF lengths predicted by the ORF finder for EcPYLs ranged from 546 to 669 bp (Table 2). On the other hand, WolfsPort’s predicted subcellular localization of EcPYLs genes revealed that the majority of the putative PYLs genes of finger millet were located in the cytoplasm and chloroplast (Table 3). Further, all the putative PYLs of finger millet were hydrophilic in nature as they possess negative value of Grand Average of Hydropathicity.
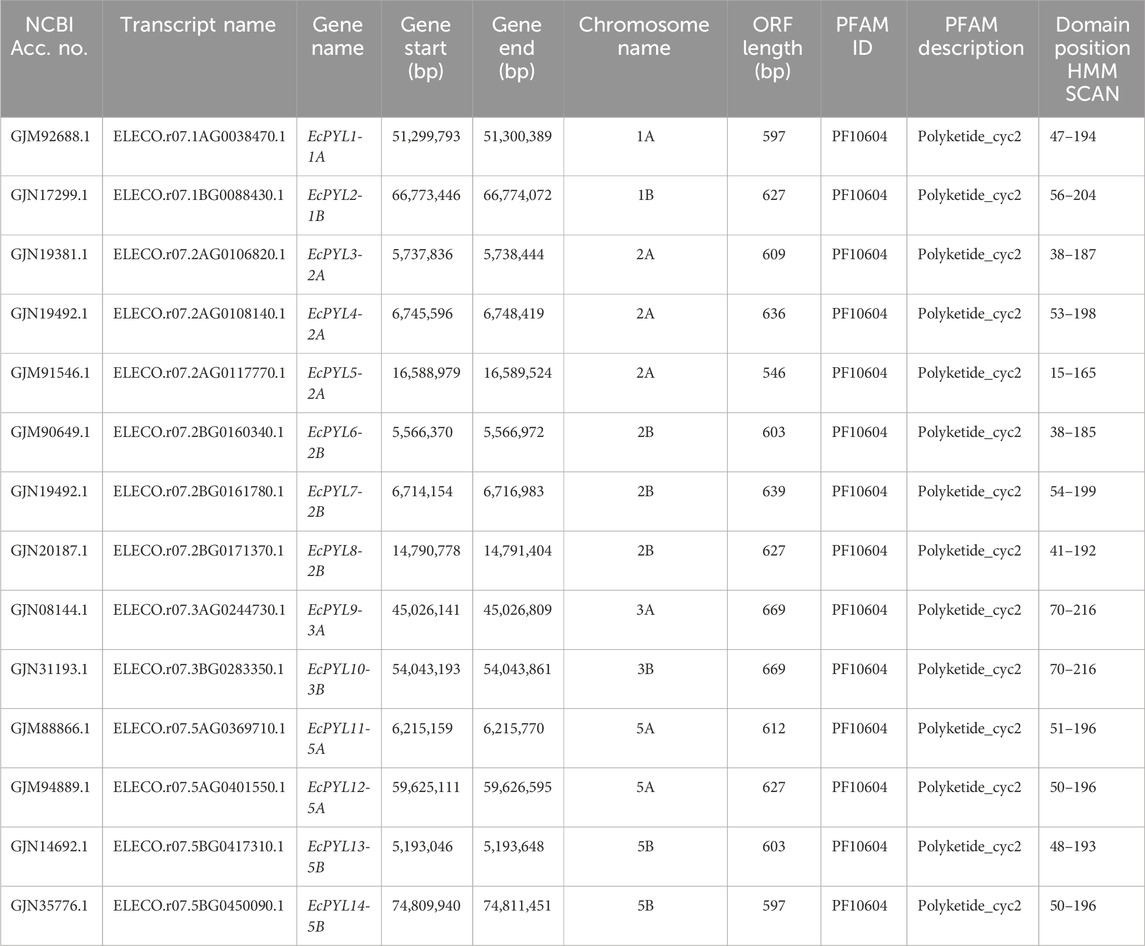
Table 2. Gene position, ORF length, domain position and chromosome location of putative PYLs genes of finger millet.
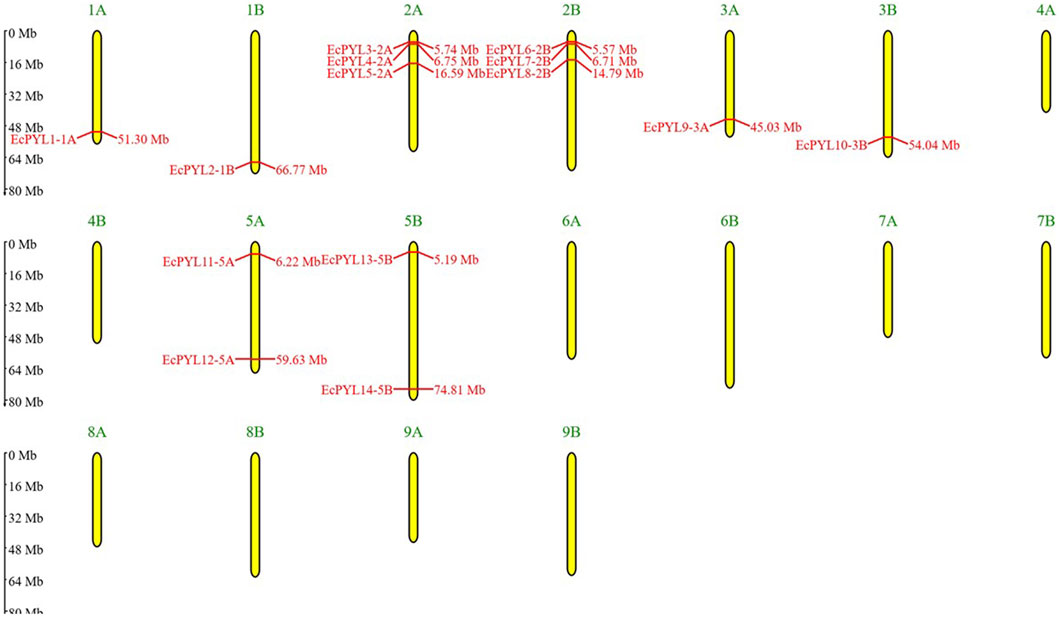
Figure 1. Distributions of 14 EcPYL genes on finger millet chromosomes. Physical map of putative EcPYL genes across nine chromosome groups found on the finger millet genome, each with a homologous chromosome (A,B). All finger millet chromosomes are shown to scale based on their actual length.
Gene structure and conserved motifs analysis
It was demonstrated that EcPYL can be classified into two distinct clades based on the intron/exon structure analyzed using TB tool’s (Figure 2). In subfamilies I and II, no introns were detected, whereas all members of subfamily III except EcPYL14-5B contain two introns (Figure 2). As shown in Figure 3a, motifs 1, 2 and 3 were conserved across all EcPYL genes. Among the 14 EcPYLs studied, motif 1 contains the conserved characteristic Gate–Latch domain (Figure 4). However, motif 4 was found to be conserved across all receptors except EcPYL3/5/8. Motif 5 was found to be present only in the EcPYL3/6/9/10 genes, whereas EcPYL11 and EcPYL13 of the 5A and 5B chromosomes contained Motif 6. The members of the subfamily share conserved motifs and possess unique motifs (Figure 3a). The unique motifs could indicate that each member performs a unique or specialized biological function, while similar motifs reveal functional similarities. Figure 3b displays the logos of these identified motif groups, indicating the frequency of amino acid occurrence at that site.
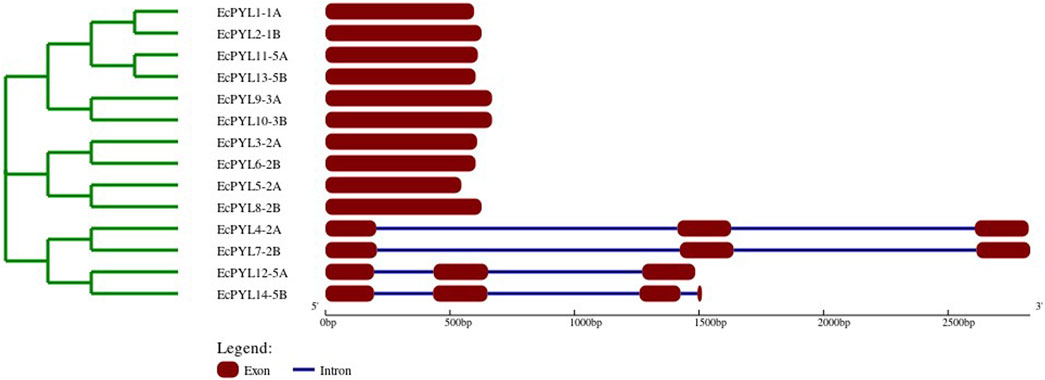
Figure 2. Evolutionary analysis and gene structure organizations of EcPYL genes, Exons and introns are represented by maroon boxes and blue lines, respectively. Scale represents the sizes of exons and introns.

Figure 3. Evolutionary relation with reference to conserved motifs of PYL genes in finger millet. (a) evolutionary analysis classified EcPYL genes into three subfamilies, Subfamily-I (Red), subfamily-II (Blue) and Subfamily-III (Green), with their conserved motif. The different colours represent the 6 identified motifs. Motif 1-3 (red, cyan and green) are conserved across all 14EcPYLs, whereas motif 4 (blue) is conserved among subfamily I and III. Motif 5 (orange) is conserved for EcPYL-9-3A/10-3B/3-2A/6-2B of subfamily I and II), and Motif 6 (brown) is only present in EcPYL11-5A and EcPYL13-5B. (b) The six highly conserved sequence logos in EcPYLs. The sequence logos are the motifs which found to be conserved in 14 EcPYL proteins based on full-length alignments. The height of each letter in logo represents frequency of its occurrence at that site.
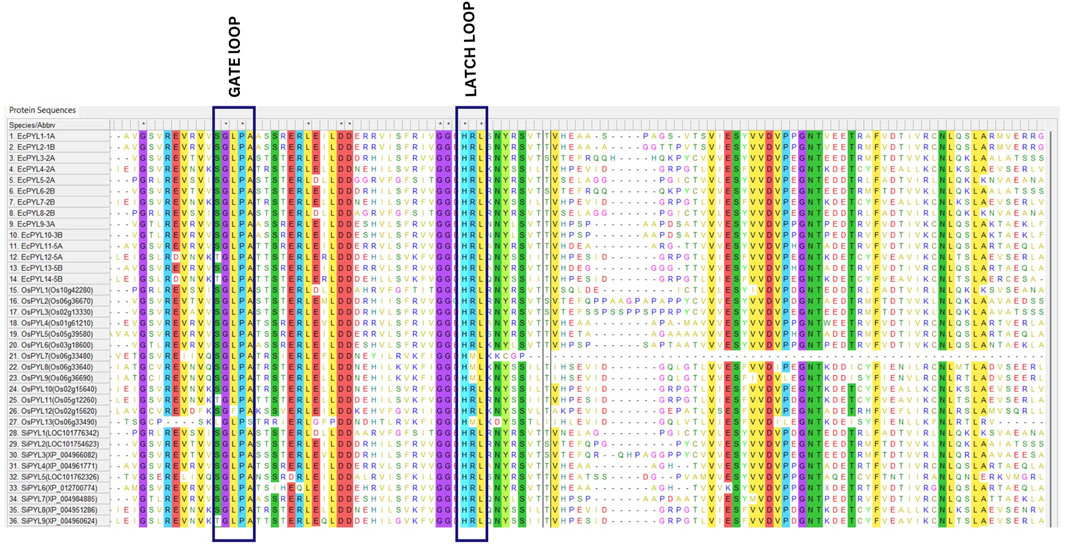
Figure 4. Multiple sequence alignment of PYL genes of finger millet, rice, and foxtail millet. MSA of putative PYLs genes of finger millet, rice and foxtail millets illustrate conserved GATE (CL2 loop) and LATCH (CL3 loop), essential for abscisic acid signaling.
Evolutionary analysis
In terms of sequence similarity, EcPYL gene can be grouped in three major subfamilies, namely, subfamily-I, subfamily-II, and subfamily-III (Figure 5). Among the 14 EcPYL genes, EcPYL1/2/11/13/9/10 (six PYLs distributed among chromosomes 1A,1B, 5A, 5B and 3A, 3B) belong to subfamily I; EcPYL3/6/5/8 belong to subfamily II (four PYLs distributed among chromosomes 2A and 2B), while EcPYL4/7/12/14 belong to subfamily III (four PYLs distributed among chromosomes 2A, 2B and 5A,5B). A comparative evolutionary tree (Figure 5) shows that the PYL genes of rice (13 OsPYL), sorghum (8 SbPYL), finger millet (9 SiPYL) and foxtail millet (14 EcPYL) are grouped together into subfamilies I, II, and III. The genes EcPYL11/13 in subfamily I are closely related to one another and distantly related to OsPYL5, suggesting that these genes might be involved in signaling pathways that help plants resist the rice blast fungus (Yadav et al., 2020). Along with this, EcPYL10 may also play a similar role because it is closely related to OsPYL6 and has the same inhibitory phosphatase activity (Kim et al., 2014). EcPYL9 is closely related to SiPYL7/4 and also has some evolutionary similarities with EcPYL10-B from finger millet and OsPYL6 from rice. These evolutionary similarities are made up of conserved amino acid residues. EcPYL3/6 belong to subfamily II of the evolutionary tree and are closely related to foxtail millet SiPYL2 as well as to sorghum SbPYL2. EcPYL4/7 are distantly related to rice OsPYL10, suggesting that EcPYL4/7 could inhibit phosphatase activity without ABA (He et al., 2014). In addition, the OsPYL7/8/9/13 appear in different subclades (Figure 5) as a result of mutations in conserved amino acid residues of the latch loop (HRL), which are replaced by HML amino acid residues (Figure 4) in the same evolutionary tree.
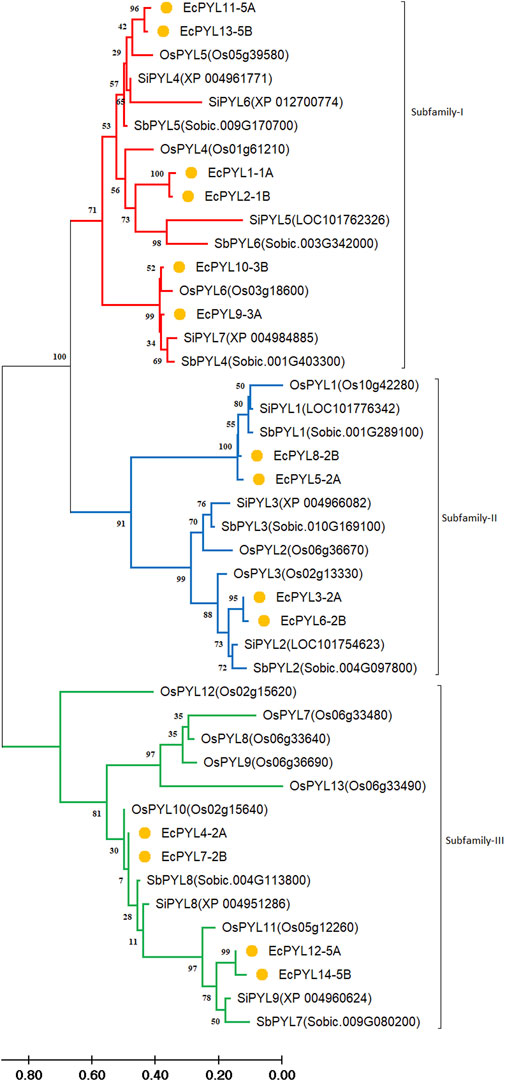
Figure 5. Comparative Phylogenetic analysis between finger millet, rice, foxtail millet and Sorghum. Tree was constructed by MEGA11 using Maximum Likelihood method with 1000 bootstraps Three subfamilies are represented in different colours: Subfamily-I (Red), subfamily-II (blue), and subfamily-III (Green). At the same time, the genes of finger millet throughout the phylogenetic are marked by orange nodes. The genes appear in the same clade of one subfamily having higher similarity, whereas all are evolutionarily connected. The branch length represents the magnitude of genetic change.
Non-synonymous (Ka)/synonymous (Ks) analysis
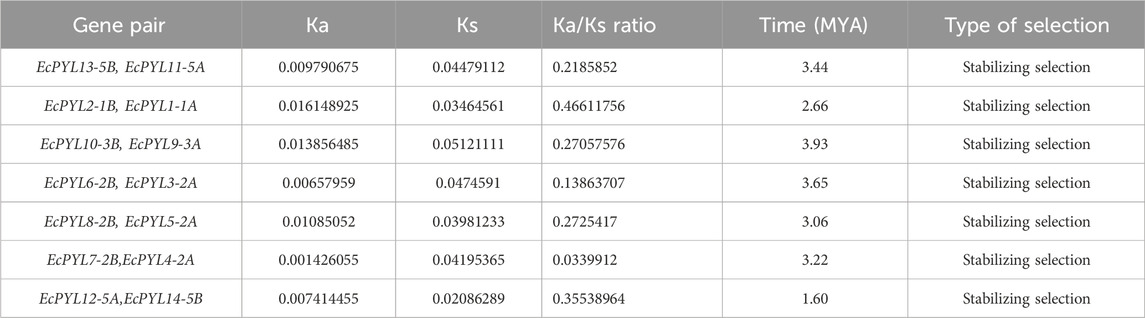
To determine the degree of selection pressure during the divergence of PYL genes, we evaluated the (Ka) versus (Ks) analysis for the paralogous gene pairs of finger miller PYL receptor protein. Based on the Ka versus Ks calculation tool, seven pairs of paralogous genes with their position on the chromosome were identified for finger millet PYLs proteins, as shown in Table 4 and Figure 6. The selective pressure on genes due to which encoded protein shows functional alteration can be predicted by nucleotide substitution rate, i.e., the ratio of Ka (non-synonymous) versus Ks (synonymous) (Table 4). All gene pairs with Ka/Ks < 1 indicate the possibility of synonymous substitution and stabilizing selection during evolution. Moreover, the evolutionary time study of these paralogous genes revealed that they evolved approximately 3.1 million years ago.
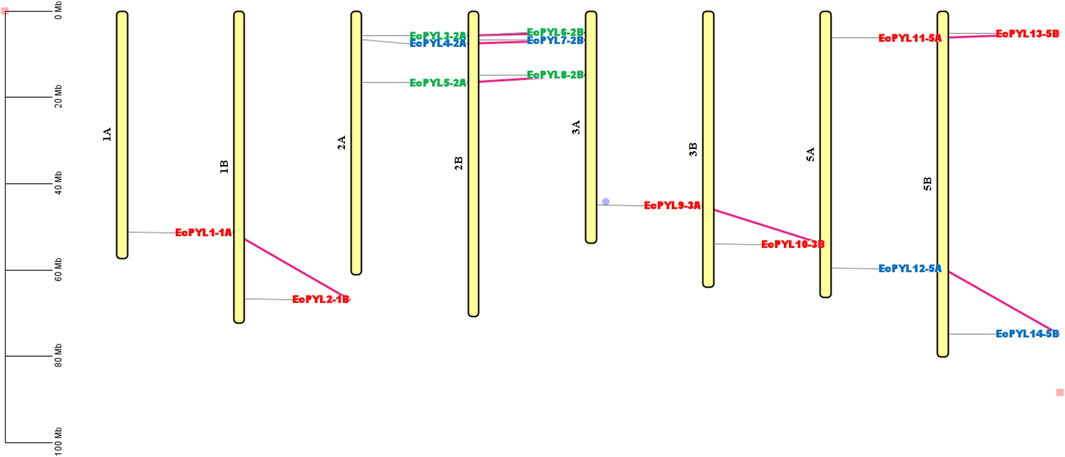
Figure 6. Physical mapping of homologous genes in finger millet genome. The labelled EcPYL genes in different colours represent different subfamilies; genes labeled with red belong to Subfamily-I, blue belong to subfamily-II, and green belong to subfamily-III. The violet connecting line represents a homologous pair. At the same time, the scale represents the actual length of the chromosome.
Cis-regulatory elements (CRE) analysis
CREs in the promoter region are known to influence transcriptional regulation greatly. Across fourteen EcPYL genes, 37 putative CREs were identified and mapped on the promoter region of EcPYL genes (Figure 7a; Supplementary Table S2). Several cis-elements were found to have different functions in finger millet. These included role in hormonal signaling (ABRE, TGACG/CGTCA-motif, P-box, GARE-motif, AuxRR-core TGA-element, and TCA-element), developmental processes (RY-element and circadian), light-responsiveness (G-box, C-box, Box 4, ACE, GATA motif MRE and Sp1) and various stresses (TC-rich repeats, LTR, MBS) in finger millet (Figure 7b). In addition to transcription-related CRE, a lot of the ABREs (abscisic acid-responsive element) and MeJA (methyl jasmonate/TGACG-motif/CGTCA-motif), which are linked to plant defense systems, were found. Furthermore, the fact that several EcPYL genes have GARE (gibberellic acid-responsive elements) and ARE (auxin-responsive elements) in their promoter region suggests that hormones may interact at the level of ABA receptor expression. Among stress-responsive factors, MBS (MYB binding site involved in drought-inducibility) and LTR (low-temperature responsive elements) are commonly observed in finger millet. Further, one additional CRE has been observed to be ubiquitously present in all the PYL genes of finger millet, called CCAAT. This five nucleotide base motif is associated with controlling flowering in plants (Wenkel et al., 2006) and contains a binding region for Nuclear Factor-Y transcription factor (NF-Y), which has been reported to regulate several biotic and abiotic stress factors (Rani et al., 2023; Rani et al., 2024a; Rani et al., 2024c). Hence, the presence of different CREs (8% stress responsive, 25% light responsive, 29% growth and development related and 38% hormone responsive) in the promoter region of finger millet provides an insight into its putative role in plant development and stress tolerance (Figure 7c).
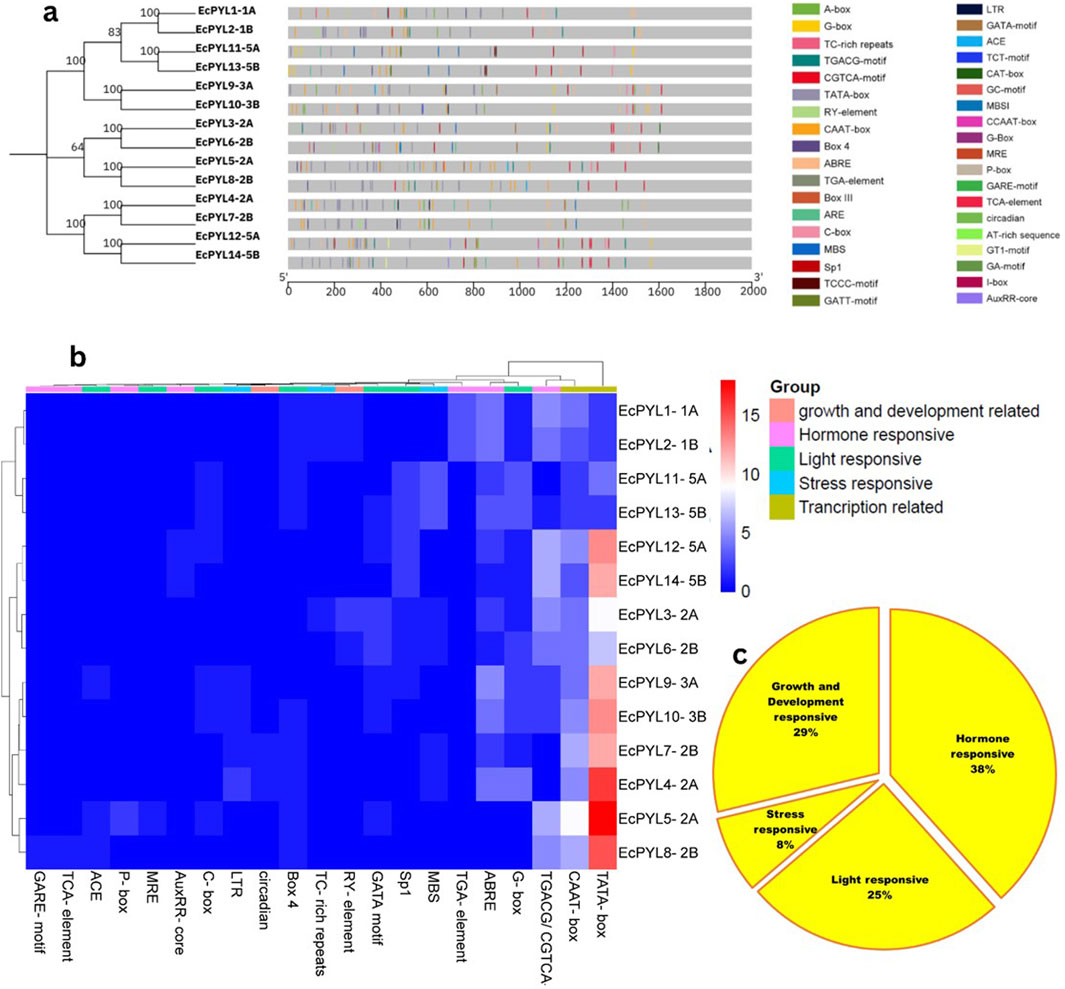
Figure 7. The Cis-regulatory element (CRE) analysis of EcPYL gene promoters. (a) The locations of CREs in the 14 EcPYL gene promoters. The grey rectangular horizontal bar represents the 2 kb promoter regions, and the different color boxes correspond with the different kinds of CREs. The horizontal values represent the position of CRE in 2KB promoter length. (b) A heatmap of CREs in the 14EcPYL gene promoters. The different colour boxes indicate the number of CREs in different EcPYL gene promoters. The dark blue colour boxes represented no corresponding CRE, and the red boxes represent eighteen corresponding CREs (highest number). The phylogenetic tree present at upper side of heat map showing 5 colour box represent different groups of the CRE responsible for specific functions. Whereas left side of tree represent subfamily of EcPYL genes. (c) Pie chart represent abundance of CRE in promoter of PYL genes of finger millet playing role in hormone response, stress response, light response and plant growth and development in EcPYL genes
Protein-protein interaction and EcPYL-miRNA interaction analysis
The PPI results reveal a total of 44 nodes and 289 edges, and the MCODE of Cytoscope classifies them into three clusters. There are four EcPYL genes in cluster 3, and they interact with nine Arabidopsis PYL genes and 35 other genes performing the similar biological functions (Figure 8). In most cases, EcPYLs interact with PP2C, SnRK2 (the core ABA signaling complex), and the MYB transcription factor, which are vital components of the ABA-auxin signaling complex (Figure 8). Based on functional annotation, a group of EcPYLs proteins was discovered to play a role in different biological functions (BF), molecular functions (MF), cellular component (CC) (Figure 9).
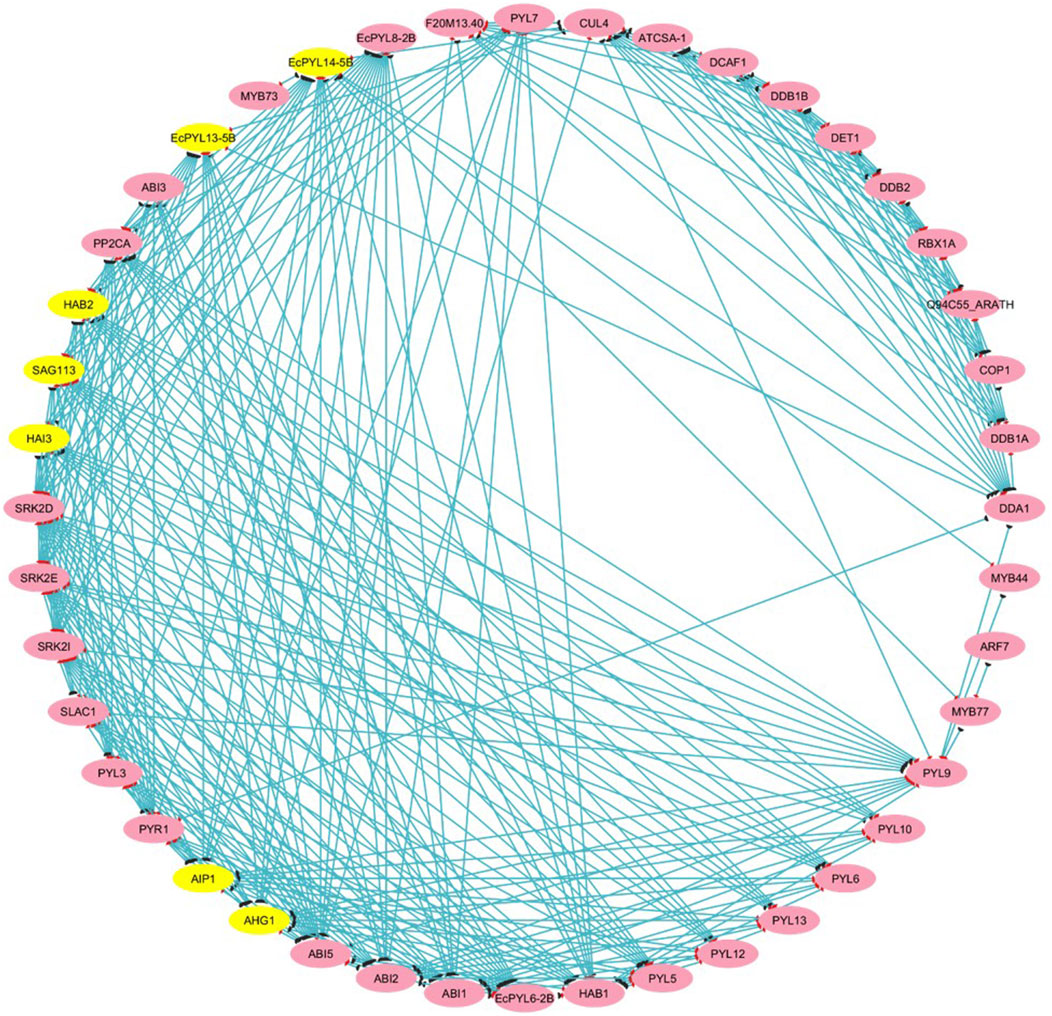
Figure 8. Protein-Protein (P-P) interaction analysis between putative PYLs of finger millet. The P-P interaction network is drawn using STRING tool and visualized using Cytoscape v. 3.10.3. The yellow highlights represent major genes that participated in interaction with other proteins or transcription factors that appeared in clusters analyzed through MCODE. At the same time, pink highlights represent (targets). The blue line represents the interacting string.
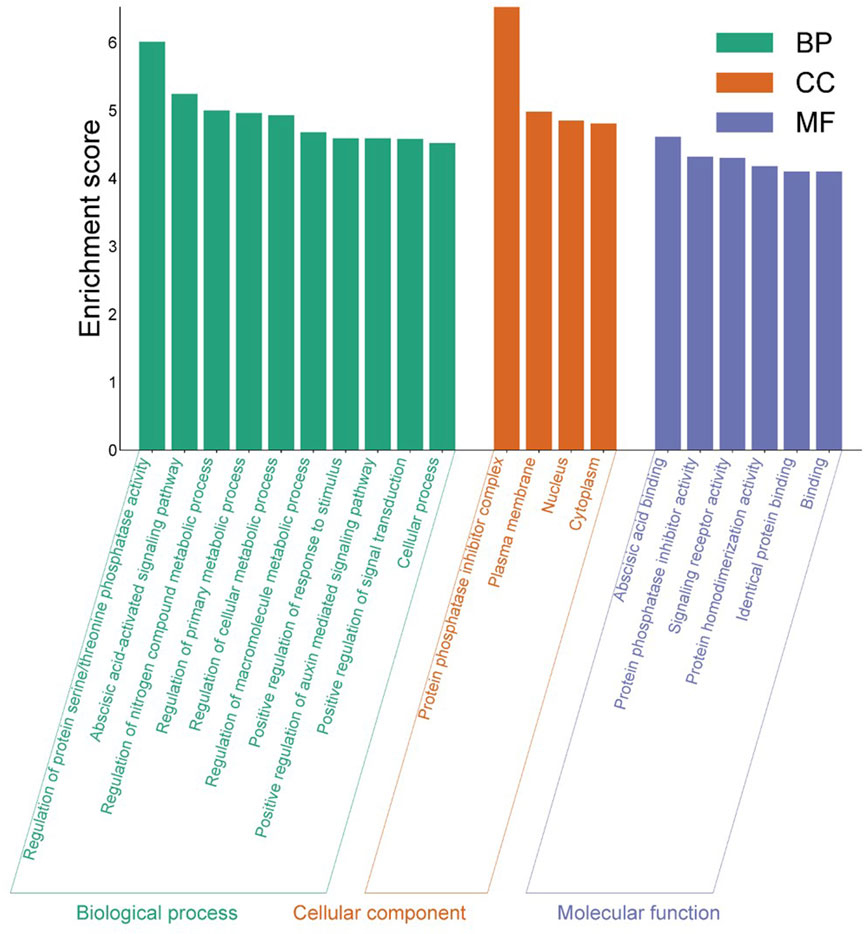
Figure 9. Gene Ontology analysis of identified EcPYL gene. The enrichment analysis shows the involvement of EcPYL in biological processes, molecular functions, and cellular components. In all, 20 GO terms were over-represented by > 4-fold enrichment value, with p-values <0.05.
Gene ontology and enrichment analysis reveals that EcPYL genes are enriched in plasma membrane (GO: 0005886), nucleus (GO: 0005634), protein phosphatase inhibitor complex (GO: 0062049) in terms of cellular components. Molecular function is enriched in abscisic acid binding (GO: 0010427), protein phosphatase inhibitor activity (GO: 0004864) and signaling receptor activity (GO: 0004864). Biological processes revealed that EcPYL genes are involved in regulating protein serine/threonine phosphatase activity (GO: 0080163) and the abscisic acid-activated signaling pathway (GO: 0009738). Besides these functions EcPYL genes also involved in two KEEG pathways viz. mitogen-activated protein kinase (MAPK) signaling pathway (ath04016) and plant hormone signal transduction pathway (ath04075).
In order to determine the miRNA targets and their regulation post-transcriptionally, EcPYL-miRNA interaction analysis was conducted, which revealed only three unique miRNAs targeting specific members of a particular subfamily of PYL genes in finger millet (Figure 10). Eco-miRN5585 targets EcPYL9-3A and EcPYL10-3B genes of subfamily I, while Eco-miRN34a targets EcPYL3-2A and EcPYL6-2B of subfamily-II and Eco-miRN529a targets only one gene EcPYL4-2A of subfamily III (Supplementary Table S3) through cleavage activity. Inhibition through cleavage activity appears to be the predominant mechanism by which most of the miRNAs predicted to target EcPYL members and regulating their expression post-transcriptionally either by positive or negative mechanism. However, miR529a were found to be involved in modulating panicle architecture in plants; however, miR5585 and miR34a were reported to play a crucial role as tumour suppressor cells (Farooqi et al., 2017). Thus, controlling EcPYL genes through sequence-specific interaction mediated by miRNAs could be very important for plants’ ability to respond to growth and pathogen infection.
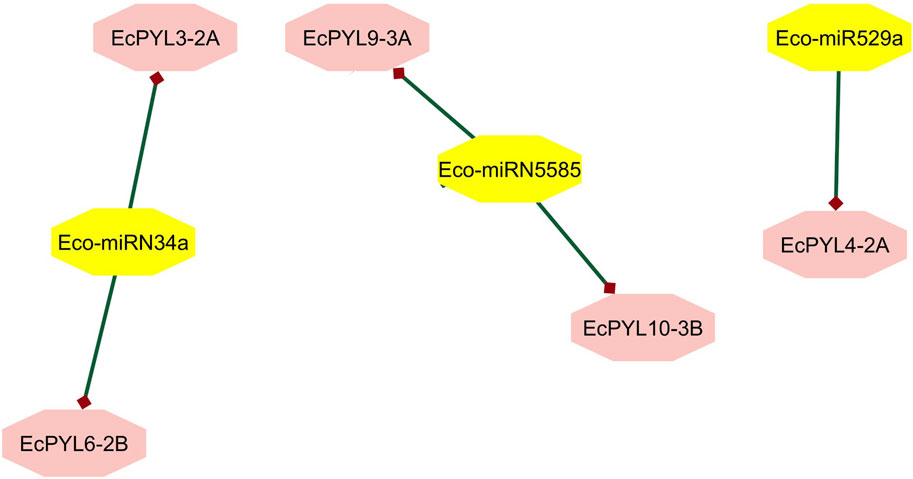
Figure 10. Finger millet, EcPYL genes, and miRNA interaction. EcPYL-miRNA interaction analysis carried out by using psRNA target and interacting network were visualized in cystoscape v. 10.3, reveals Eco-miRN5585 is targeting EcPYL9-3A and EcPYL10-3B through cleavage activity. Whereas, Eco-miRN34a is targeting two of EcPYL receptors protein EcPYL3-2A and EcPYL6-2B. Eco-miR529a is targeting EcPYL4-2A gene of finger millet.
Pre-selection of candidate genes by ML algorithm
By combining scatter plot visualization (Figure 11b), hierarchical clustering (Figure 11a), and numerical probability assessments (Table 5), the expression probabilities of these genes were analyzed under both the stress conditions. Scatter plot analysis (Figure 11b) identified distinct clusters of EcPYL genes under salt and drought conditions, each exhibiting varying degrees of expression. EcPYL14-5B, EcPYL12-5A, EcPYL11-5A and EcPYL7-2B were most likely to be expressed during drought stress, which suggests they may be involved in mechanisms that help plants deals with drought. Conversely, EcPYL4-2A and EcPYL7-2B displayed the highest expression probabilities under salt stress (≥0.72), indicating their potential role in salinity adaptation. The hierarchical clustering heatmap (Figure 11b), which grouped genes with similar expression patterns, further supported these observations. Drought stress strongly linked the groups of EcPYL14-5B, EcPYL12-5A, and EcPYL11-5A. On the other hand, EcPYL4-2A and EcPYL7-2B grouped separately, indicating their role in salinity stress. Further, EcPYL9-3A and EcPYL10-3B showed a strong preference for drought stress, while EcPYL6-2B exhibited a lower expression probability under both conditions. Table 5 shows that these trends were supported by the probability analysis. EcPYL14-5B had the highest expression probability (0.839) during drought stress, followed by EcPYL12-5A (0.818) and EcPYL11-5A (0.811). The EcPYL4-2A (0.749) and EcPYL7-2B (0.721) were also among the most highly expressed genes under salt stress. Although some genes displayed moderate expression probabilities under both stress conditions, they did not meet the threshold for high likelihood (≥0.70). For example, EcPYL5-2A showed a fairly balanced expression pattern (0.604 when exposed to salt and 0.618 when exposed to drought), which suggests it might play a small but important role in adapting to abiotic stress in general. However, genes such as EcPYL9-3A (0.13 under salt, 0.781 under drought) and EcPYL10-3B (0.164 under salt, 0.819 under drought) showed a distinct preference for drought-related expressions. Overall, our results show that EcPYL14-5B, EcPYL12-5A, EcPYL11-5A and EcPYL7-2B are important candidate genes and these genes are likely to be involved in regulating drought stress. While EcPYL4-2A and EcPYL7-2B are likely to regulate salinity stress.
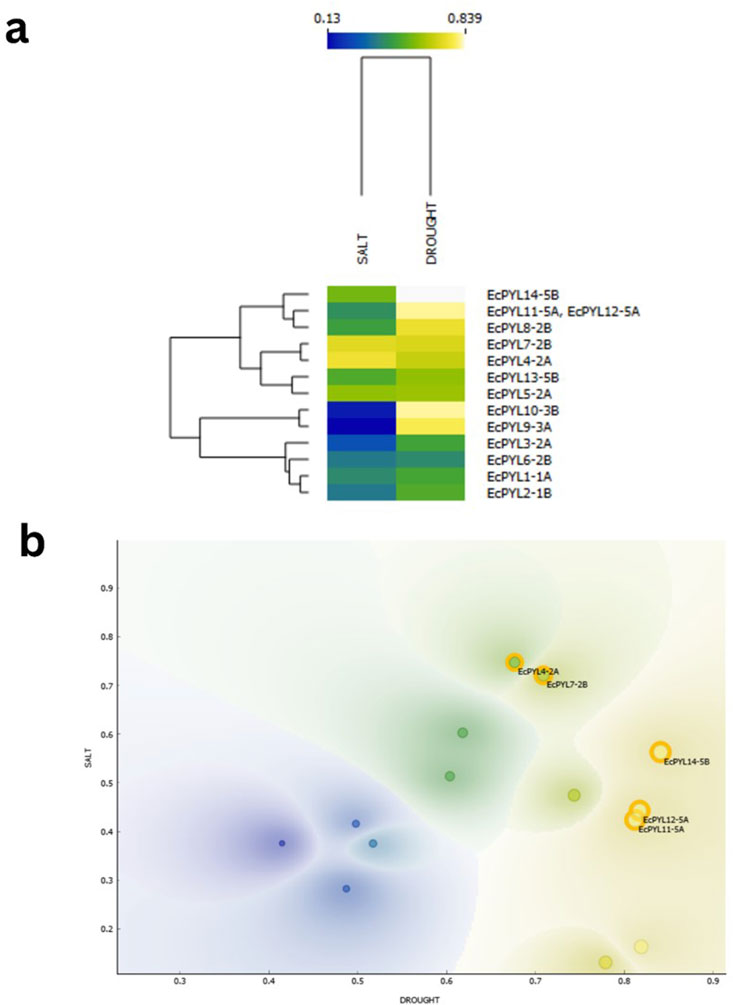
Figure 11. ML based prediction for expression of candidate genes under drought and salt stress represented by Heatmap and scatter plot. (a) Heatmap with hierarchical clustering illustrating the probability-based expression patterns of various genes under salt and drought stress conditions. The color scale represents expression probability values, ranging from 0.13 (blue) to 0.839 (yellow). Genes are clustered based on similarity in expression profiles, and their respective stress conditions (salt and drought) are indicated at the top of the dendrogram. (b) Scatter plot generated using Orange software, displaying the distribution of genes based on their expression probabilities under salt and drought conditions. Contour density shading represents regions of varying gene expression probability distributions. The best candidate genes, identified based on their significant expression in both conditions, are highlighted with orange circles. The x-axis represents drought stress probability, while the y-axis represents salt stress probability.
Expression profile of PYL genes in finger millet under drought and salinity stress conditions
Through ML based algorithm, a total of five PYL genes (EcPYL4-2A, EcPYL7-2B, EcPYL11-5A, EcPYL12-5A, and EcPYL14-5B) that respond to drought and salinity stresses were selected (Figures 11a,b), and their expression pattern was analyzed in shoot and root tissues of finger millet under drought and salinity stress (Figures 12a,b). Notably, all genes exhibited expression in both tissues under both the stress conditions (Figures 12a,b,d,f); however, their expression patterns varied between tissues and conditions (Figures 12c,e). For instance, compared to EcPYL4-2A, only EcPYL12-5A showed a 9.69-fold increase in shoot tissue under drought stress conditions (Figure 12c). When we compared the expression pattern of EcPYL12-5A in shoot tissue between salinity and drought, it was observed that drought stress increased its expression level up to 2.1-fold compared to salinity stress (Fig. d–f). The expression level of EcPYL11-5A was higher in the shoot after EcPYL12-5A under drought. Under drought stress, EcPYL7-2B increased more than 2-fold in shoot tissue compared to EcPYL4-2A and EcPYL14-5B. Three genes (EcPYL4-2A, EcPYL11-5A, and EcPYL14-5B) were comparatively less active in the shoot tissue of finger millet under salt stress (Figure 12e). However, of these, EcPYL4-2A increased its expression level by 0.76-fold higher in shoot tissue under salinity stress than drought (Figure 12f). Two genes (EcPYL7-2B and EcPYL11-5A) showed their expression level higher (>3.5-fold) in shoot tissues under salinity stress than the other three genes. In conclusion, three genes (EcPYL7-2B, EcPYL11-5A, and EcPYL12-5A) and two genes (EcPYL7-2B and EcPYL12-5A) were found to be highly expressed in shoot tissue of finger millet under drought and salinity stresses, respectively.
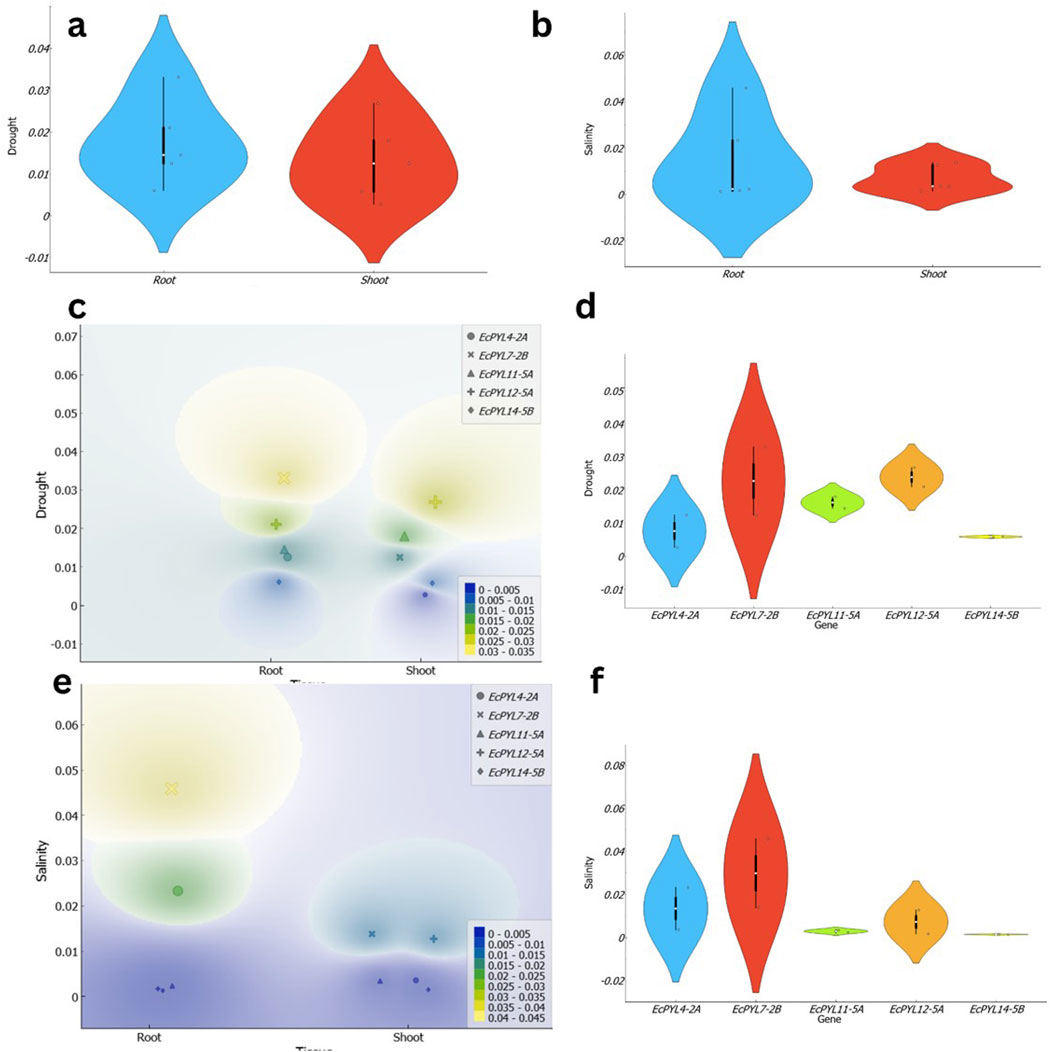
Figure 12. Illustration of the expression patterns of EcPYL genes in finger millet under drought and salinity stress across shoot and root tissues. (a,b) present violin plots depicting the distribution of EcPYL gene expression under drought and salinity stress, respectively. In (a), the expression levels under drought conditions exhibit a broader distribution in root tissues (blue) compared to shoot tissues (red), suggesting higher variability in root responses. Similarly, in (b), under salinity stress, root tissues display a wider expression range than shoot tissues, as indicated by the distribution density and black boxplots representing the median, interquartile range, and data spread. (c,e) depict density-based expression heatmaps of EcPYL genes under drought and salinity stress, respectively. In (c), the density plot illustrates the spatial expression distribution of EcPYL4-2A, EcPYL7-2B, EcPYL11-5A, EcPYL12-5A, and EcPYL14-5B across root and shoot tissues under drought stress. The color gradient represents expression density, where yellow and green indicate high expression levels, while blue represents low expression levels. A similar pattern is observed in (e) under salinity stress, where root tissues exhibit a more pronounced variation in expression, while shoot tissues display a more localized expression response. (d,f) show violin plots representing the expression profiles of individual EcPYL genes under drought and salinity stress, respectively. Under drought stress (d), EcPYL7-2B exhibits the highest variation, while EcPYL14-5B remains relatively stable across both tissues. Similarly, under salinity stress (f), EcPYL7-2B again shows the highest expression variability, whereas EcPYL11-5A and EcPYL14-5B demonstrate limited fluctuations in expression levels.
In the root, EcPYL2-7B exhibited highest expression among the five PYLs under both stresses. Notably, expression level of EcPYL12-7B increased by more than 15-fold compared to EcPYL11-5A, EcPYL12-5A, and EcPYL14-5B under salinity stress. Similarly, the same gene demonstrated 1.96-fold higher expression in the root under salinity stress compared to EcPYL4-2A. However, EcPYL4-2A expression was up to 9.3 times higher than EcPYL11-5A, 13.5 times higher than EcPYL12-5A, and 17.6 times higher than EcPYL14-5B in the root under salinity stress. Interestingly, the expression levels of the same three genes—EcPYL11-5A, EcPYL12-5A, and EcPYL14-5B was enhanced by 6.15, 12.26, and 4.6 times in root tissues under drought stress. Moreover, EcPYL7-2B alone had highest expression in the root under drought stress among the five genes. For instance, the expression level of this gene was almost 1.5 times higher than that of the other genes. Overall, EcPYL4-2A and EcPYL7-2B showed higher expression in the root tissue, under both the stress conditions suggesting that EcPYL7-2B is the most responsive gene under drought and salinity stress, particularly in root tissues, while EcPYL12-5A and EcPYL11-5A play significant roles in shoot tissues under drought stress. This differential expression highlights the potential role of these genes in stress adaptation and provides insights for future functional studies on drought and salinity tolerance mechanisms in finger millet.
Homology modeling of finger millet PYLs genes
Protein sequences, EcPYL13-5B, EcPYL6-2B, EcPYL8-2B, and EcPYL14-5B representing each clade of the phylogenetic tree, were used for homology modeling for structural prediction (Figure 5). The widely preferred method for modeling protein structures is to compare a sequence of our interest, i.e., the target sequence (unknown structure), with a template (known structure) and look for structural similarities. The SWISS-MODEL server (https://swissmodel.expasy.org/) was used in the present study for homology modeling of EcPYL6-2B, EcPYL8-2B, EcPYL13-5B, and EcPYL14-5B (Figure 13) against best-matched template 4oic, 5ujv and 5gwo, respectively, based on the high percent of identity, as shown in Table 5. The model generated by these templates was further validated by the SAVES server by analyzing the number of amino acid residues in allowed or disallowed areas through PROCHECK, accuracy of non-bonded contact by ERRAT (Table 6). All the models had ERRAT scores greater than the acceptable value of 80%. After the accuracy measurement, the modeled structure was subjected to the degree of similarity search with the crystal structure of (PDB ID 4oic, 5ujv and 5gwo) by using SuperPose V.1. The RMSD (Root Mean Square Deviation) and NRES (Number of Overlapped Residue) were analyzed for the superposed structure as shown in Table 4. By evaluating the result of the superposition, it can be concluded that the quality of the model is good, with the least deviation of residues from the template. Further, validation of the predicted model was performed with ResProx resolution criteria (Good: 0–1.5, Middle: 1.5–2.5, Bad: >2.5), indicating the models to be of good resolution. Table 6 summarizes the detailed analysis of homology modeling using SWISS-MODEL, structural assessment, SAVES server validation, and quality and resolution estimation using SuperPOSE v.1 and ResProx. The 2-D structural analysis performed by the PDBSum server revealed that finger millet EcPYLs genes (EcPYL6-2B, EcPYL8-2B, EcPYL13-5B, and EcPYL14-5B) contains same number of strand but having different number of helices as 5:4:5:4 respectively (Table 6). Figure 13 depicts each model’s conserved residues present in CL2 (GATE loop) and CL3 (LATCH loop), specific for ABA binding.
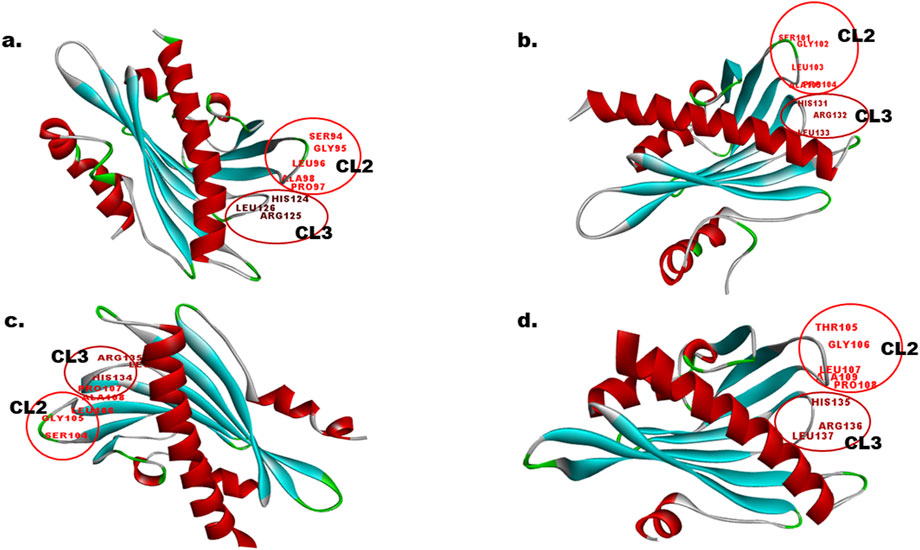
Figure 13. In-silico structural analysis of EcPYL proteins. The 3D structure modeling of EcPYL proteins was generated using the SWISS-MODEL and visualized by Biovia Discovery Studio. The alpha helix is represented by red colour, beta-sheets by cyan colour, coil by green and loops by gray colour. The conserved amino acids residue represented on loop (CL2 and CL3 loop) with their position, participating in ABA signaling (a) EcPYL6-2B, (b) EcPYL8-2B, (c) EcPYL13-5B, and (d) EcPYL14-5B.
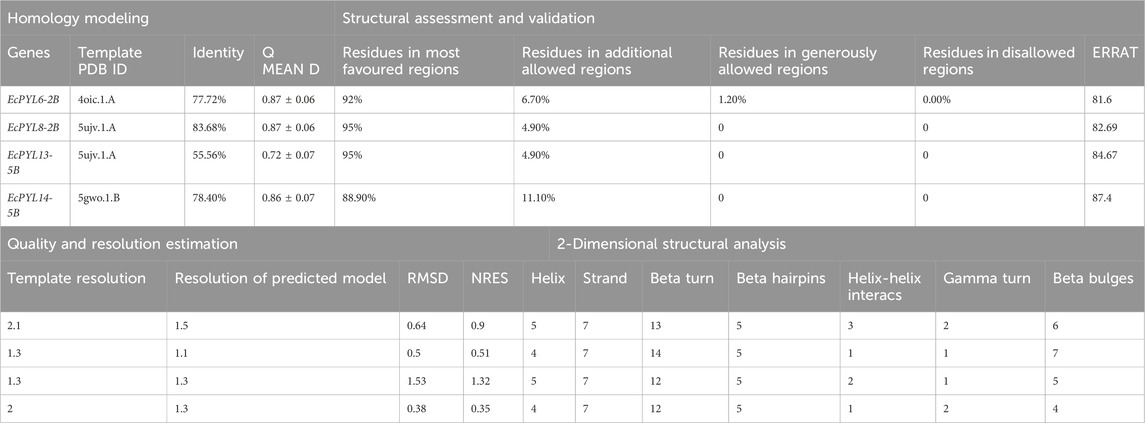
Table 6. Homology modelling, assessment, and validation of putative EcPYLs gene of finger millet, participated in Protein-Protein interaction.
Discussion
The PYL gene family is found in a variety of plant species, including Arabidopsis (Gonzalez-Guzman et al., 2012), rice (Kim et al., 2012; Yadav et al., 2020), sorghum (Dalal and Inupakutika, 2014), maize (Fan et al., 2018; Wang et al., 2018) and wheat (Lei et al., 2021). Rani et al. (2024c) reported three EcPYL genes (GJN19492.1, GJN35776.1, GJN19381.1) in finger millet based on rice homologs. However, the in-depth characterization and molecular validation of PYL receptors is yet to be explored in finger millet. A total of 14 EcPYL genes were identified in finger millet in the present study, which are distributed into three subfamilies designated as subfamily I, II and III respectively. Evolutionary analysis coupled with MEME search revealed a diversity of conserved motifs in finger millet and other cereals PYL genes. Whereas, the gene structure analysis showed presence of a common exon or intron structure in most of the genes within a subfamily. The members of EcPYLs in Subfamilies I and II are intronless and have conserved motifs 1, 2, and 3. However, only Subfamily III members contain introns and are conserved for motifs 1, 2, 3 and 4. Whereas motif 5 was observed among members of subfamily 2, and motif 6 was found only among two members of subfamily I. EcPYL genes in the same subfamily have gradually developed different protein motif structures due to adaptation to complex environmental conditions during the evolution of the finger millet genome. As compared to 14 PYLs in finger millet, there exist 13 PYLs in rice (Yadav et al., 2020), 8 PYLs in sorghum (Dalal and Inupakutika, 2014) and 9 PYLs in foxtail millet (Wu et al., 2023). The EcPYLs revealed close genetic proximity with rice and divergence with sorghum and foxtail millet PYLs. The variability could be attributed to gene duplication events resulting from polyploidization, which contribute to gene family expansion and genome evolution (Moore and Purugganan, 2005).
The chromosomal location of these EcPYL genes were determined and it was found that it exists as seven pairs of homologous genes present on homologous chromosomes 1, 2, 3, and 5. Most of these genes are located on chromosomes 2A and 2B. As a result of genome polyploidization, some homologous genes might have lost (Lynch and Force, 2000). Gene duplication plays an important role in expanding the PYL gene family with possibility for new biological functions (Conant and Wolfe, 2008). The analysis of cis-regulatory elements indicated that EcPYLs is associated with diverse roles such as hormone-responsive, light-responsive, stress-responsive, growth, development-responsive, and transcription-responsive genes. It has been reported that over-expression of PYL genes in Arabidopsis enhances the plant’s sensitivity to ABA during seed germination, growth, stomata regulation, and drought tolerance (Ma et al., 2009). Moreover, Rice ABA receptor OsPYL5 regulates ABA signaling, thereby improving drought and salinity tolerance and ABA sensitivity during germination and seedling development (Kim et al., 2012; Kim et al., 2014). Based on this analysis, it is evident that PYL genes may regulate finger millet growth and development and are actively involved in stress responses. Additionally, the presence of CCAAT cis-acting element in its promoter region indicates its involvement in regulation of flowering and providing binding site for nuclear factor-Y (NF-Y) transcription factor (Wenkel et al., 2006; Muthamilarasan et al., 2014; Rani et al., 2023). Further, this NF-Y-PYL/PYR module could regulate the expression of the ABA receptor and enhance ABA signal transduction via transcriptional regulation of PYR1 (Yu et al., 2021). This, in turn, promotes the expression of stress-responsive genes against drought and salt stress (Yu et al., 2021). Finger millet, extremely tolerant to drought and salt (Gupta et al., 2017; Antony Ceasar et al., 2018; Sood et al., 2019; Ceasar, 2022; Rani et al., 2023), possesses fourteen PYLs genes and shows an interaction network with several TFs (Figure 8). ABA has been found to play a crucial role in responding to abiotic stresses such as drought and salinity and ABA signaling is initiated by PYLs (Rani et al., 2024c). Many plants have been reported to have differential expression patterns of PYL genes within their tissues. Soybean seeds express many PYL genes at a relatively high level compared to other soybean tissues (Bai et al., 2013). The rubber tree, the transcript abundance of PYL genes is high in latex (Guo et al., 2017). Similarly, in rice PYLs genes are expressed at higher levels in root tissue under drought stress than in other tissues (Yadav et al., 2020). Under drought and salinity stress, finger millet exhibits a significant ABA response, but specific PYL responsible for this is still not deciphered. In the present study, EcPYL7-2B demonstrated their role in regulating both drought and salinity stress. Moreover, some of the genes like EcPYL14-5B of finger millet were found to be comparatively less expressed as compared to other genes under drought and salinity stress, which is in accordance with previous studies reported in sorghum, rice, grapes and tomato (Boneh et al., 2012; González-Guzmán et al., 2014; Yadav et al., 2020). This behavior may be explained by a negative feedback regulatory mechanism which indicates that when excessive amounts of ABA accumulate in leaves under stress, PYL expression may be inhibited (Merlot et al., 2001; Santiago et al., 2009) or the function of these genes may not be required for drought and salinity stress (Di et al., 2018). Based on the expression profiling results coupled with stress-responsive cis-elements in EcPYL promoters, it appears that some EcPYLs are responsive to drought and salinity. Hence, these PYLs may be potential candidate to enhance finger millet tolerance to abiotic stress.
An extensive network of interactions between PYL proteins of finger millet and Arabidopsis genes has been predicted. As a major component of the ABA signaling pathway, PYL interacts with SnRK and PP2C in finger millet (Figure 8). PP2CA inhibits ABA metabolism, essential to seed germination and cold tolerance (Romero et al., 2012). In guard cells that recognize ABA, ABI1 modulates both calcium channel permeability and actin reorganization within stomatal closure (Tähtiharju et al., 2002). It has been found that several finger millet PYL genes interact with PP2CA and ABI1, suggesting its possible role in seed germination and vernalization. In response to auxin stimulation, PYL8-MYB77 interaction enhances transcription activation of IAA19 in Arabidopsis (Shin et al., 2007). Similarly, it was observed that EcPYL14-5B interacts with MYB77 (Shin et al., 2007) and MYB44 (Qin et al., 2022), indicating that both proteins participate in ABA-auxin signaling and show multiple stress tolerance. Further, to decipher the relation between ABA signaling and miRNA regulation, a PYLs-miRNA interaction study was performed. The interaction between PYLs and miRNA reveals that miRNA5585 targets EcPYL10-3B and EcPYL9-3A, while miRNA34a targets EcPYL3-2A and EcPYL6-2B, as shown in Supplementary Table S3. In contrast, EcPYL4-2A was targeted by miRNA529a, suggesting that EcPYL4-2A are involved in a complex regulatory network and might be associated with the panicle architecture (Yue et al., 2017). The protein interaction networks and miRNA-mediated pathways of PYLs genes elucidated in the present study might reveal a better understanding of the ABA signal transduction pathway in finger millet.
The structural studies attempted revealed that PYR/PYL belong to the Bet v superfamily, which contains the Bet v fold or START/polypeptide cyclase2 domain (Radauer et al., 2008). A beta-sheet with seven strands flanked by two alpha helices is referred to as a helix-grip fold. As an additional characteristic of this family of ABA receptors, the PYR/PYL family contains an alpha helical segment at their N-terminus, which is not found in the Bet v fold. Therefore, this segment can be considered as a typical characteristic of this family of ABA receptors (Santiago et al., 2012). Based on the crystallographic structure analysis of Arabidopsis PYL genes, it is evident that this gene contains a seven-stranded beta-sheet flanked by two alpha-helices (α1 and α2) and an additional alpha-helical segment (α0) at its N terminus. These alpha helices and beta sheets are connected with four conserved loops: CL1 connecting (α1 and β2), CL 2 connecting (β3 and β4), CL3 connecting (β5 and β6), and CL4 connecting (β7 and α2) (Yin et al., 2009). The in silico homology modeling attempted for the four PYLs of finger millet EcPYL6-2B, EcPYL8-2B, EcPYL13-5B, and EcPYL14-5B revealed seven strands flanking with two alpha helices with the presence of additional alpha helices at N-terminal. This indicates ABA receptor family having START domain. As shown in Figure 9, the helices and strands possess conserved CL1, CL2, CL3 and CL4. As reported for Arabidopsis crystal structure (PDB ID 4oic, 5ujv and 5gwo), the CL2 loop are conserved for Ser(S), Gly(G), Leu (L), Pro(P), Ala(A) amino acid residues whereas, CL3 loops are conserved for His (H), Arg (R) and Leu (L) amino acid residue (Yin et al., 2009). The constructed models of EcPYL showed conserved amino acids in the CL2 and CL3 loops (Figure 9), except EcPYL14-5B, where Ser (S) gets replaced by Thr (T) at the CL2 loop. Similar substitution has also been observed in maize PYL genes, which contain a conserved GATE loop or CL2 loop (SGLPA replaced with TGLPA amino acid residue) (Wang et al., 2018; Yadav et al., 2020). Hence, PYL genes in finger millet possess a conserved CL2 loop, which plays a vital role in ABA binding (Yin et al., 2009) and provides valuable information regarding their biological function. In order to achieve a high standard of quality, the model should contain over 90% of the residues in the most allowed regions (Laskowski et al., 2018). The model structure of this study indicates that 92%–95% of residues are mostly in allowed regions (Supplementary Figures S1–S4; Table 5), indicating that the predicted models can be accepted. The predicted structural insights provide an opportunity for deciphering the putative functions of EcPYL receptors in finger millets, which could be explored for crop improvement after validation through extensive wet-lab based experimentations.
Conclusion
Finger millet genome mining for deciphering the putative PYL genes has been attempted in the present study. A total of 14 identified EcPYL genes have been extensively characterized in silico for multiple structural and functional properties. Chromosomal location, exon-intron structure, motif evaluation, and cis-regulatory element analysis and evolutionary analysis have been performed, revealing that the EcPYLs gene of finger millet is structurally and evolutionarily conserved among the three subfamilies. The presence of various cis-regulatory elements in the EcPYLs of finger millet may be involved in the enhancement of various stresses and developmental processes. Five genes (EcPYL4-2A, EcPYL7-2B, EcPYL11-5A, EcPYL12-5A and EcPYL145B) were found to be respond to drought and salinity stresses through machine learning approaches. Gene expression analysis revealed that under salinity and drought stress conditions, two key genes EcPYL7-2B and EcPYL12-5A were highly induced in shoot and root tissues. Further targeting of these two genes by genome editing approaches will help to identify the exact role of these genes in finger millet, which will aid in further breeding program. In addition, several CREs were found in this study from the promoter regions of EcPYLs of finger millet. Targeting drought and salinity stress responsive CREs using CRISPR activators has the potential to influence the expression of the downregulated EcPYLs genes. These significant findings could be applied for crop improvement programs by providing an insight into ABA signaling pathways associated with stress tolerance mechanisms in finger millet.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
VR: Visualization, Writing – original draft, Formal Analysis, Methodology, Investigation, Software, Data curation, Validation, Writing – review and editing, Conceptualization. TM: Validation, Methodology, Writing – review and editing, Formal Analysis, Software. SS: Data curation, Methodology, Writing – review and editing. DJ: Resources, Writing – review and editing. RG: Writing – review and editing. DY: Data curation, Writing – review and editing, Investigation, Supervision, Funding acquisition, Formal Analysis.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The financial support by the U.P. Council of Agriculture Research (UPCAR), Lucknow India (1241/06/RG/Millet/2023–24, dated 19/12/2023) to DY and RG is sincerely acknowledged).
Acknowledgments
The authors also wish to acknowledge the Head, Department of Biotechnology, D.D.U Gorakhpur University, Gorakhpur for providing the infrastructural facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1598523/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | PROCHECK analysis of EcPYL6-2B.
SUPPLEMENTARY FIGURE S2 | PROCHECK analysis of EcPYL8-2B.
SUPPLEMENTARY FIGURE S3 | PROCHECK analysis of EcPYL13-5B.
SUPPLEMENTARY FIGURE S4 | PROCHECK analysis of EcPYL14-5B.
SUPPLEMENTARY Table S1 | Retrieved PYL protein sequence (27) after BLASTP search against phytozome database in Finger millet genome.
SUPPLEMENTARY Table S2 | Cis regulatory elements in promoter region of EcPYLs genes.
SUPPLEMENTARY Table S3 | Putative miRNA and their targeted PYLs genes in finger millet.
Abbreviations
ABA, Abscisic acid; PYR1, Pyrabactin Resistance 1; PYL, Pyrabactin Resistance 1-like; RCAR, Regulatory component of the ABA receptor; PP2Cs, Type 2C protein phosphatases; SnRK2s, Sucrose non-fermenting-1-related protein kinases 2; ORFs, Open reading frames; EcPYL, Eleusine coracana Pyrabactin Resistance 1-like; CRE, Cis-regulatory elements; ABRE, Abscisic acid responsive element; MeJA, Methyl jasmonate; GARE, Gibberellic acid-responsive elements; ARE, Auxin-responsive elements; MYB, Myeloblastosis viral oncogene homolog; MBS, MYB binding site; NF-Y, Nuclear Factor-Y; TF, Transcription Factor; MCODE, Molecular Complex Detection; MAPK, Mitogen-activated protein kinase; psRNATarget, Plant Small RNA Target; RMSD, Root Mean Square Deviation; NRES, Number of Overlapped Residue; PDB, Protein Data Bank.
References
Antony Ceasar, S., Maharajan, T., Ajeesh Krishna, T. P., Ramakrishnan, M., Victor Roch, G., Satish, L., et al. (2018). Finger millet [Eleusine coracana (L.) Gaertn.] improvement: current status and future interventions of whole genome sequence. Front. Plant Sci. 9, 1054. doi:10.3389/fpls.2018.01054
Bader, G. D., and Hogue, C. W. V. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 4, 2. doi:10.1186/1471-2105-4-2
Bai, G., Yang, D.-H., Zhao, Y., Ha, S., Yang, F., Ma, J., et al. (2013). Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol. Biol. 83, 651–664. doi:10.1007/s11103-013-0114-4
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi:10.1093/nar/gkv416
Berjanskii, M., Zhou, J., Liang, Y., Lin, G., and Wishart, D. S. (2012). Resolution-by-proxy: a simple measure for assessing and comparing the overall quality of NMR protein structures. J. Biomol. NMR 53, 167–180. doi:10.1007/s10858-012-9637-2
Boneh, U., Biton, I., Zheng, C., Schwartz, A., and Ben-Ari, G. (2012). Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep. 31, 311–321. doi:10.1007/s00299-011-1166-z
Ceasar, S. A. (2022). The role of millets in attaining United Nation’s sustainable developmental goals. 345–349. doi:10.1002/ppp3.10254
Chao, J., Li, Z., Sun, Y., Aluko, O. O., Wu, X., Wang, Q., et al. (2021). MG2C: a user-friendly online tool for drawing genetic maps. Mol. Hortic. 1–4, 16. doi:10.1186/s43897-021-00020-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Cheng, C.-Y., Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S., and Town, C. D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804. doi:10.1111/tpj.13415
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi:10.1038/nrg2482
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi:10.1146/annurev-arplant-042809-112122
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi:10.1093/nar/gkr319
Dalal, M., and Inupakutika, M. (2014). Transcriptional regulation of ABA core signaling component genes in sorghum (Sorghum bicolor L. Moench). Mol. Breed. 34, 1517–1525. doi:10.1007/s11032-014-0114-3
Devos, K. M., Qi, P., Bahri, B. A., Gimode, D. M., Jenike, K., Manthi, S. J., et al. (2023). Genome analyses reveal population structure and a purple stigma color gene candidate in finger millet. Nat. Commun. 14, 3694. doi:10.1038/s41467-023-38915-6
Di, F., Jian, H., Wang, T., Chen, X., Ding, Y., Du, H., et al. (2018). Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes (Basel) 9, 156. doi:10.3390/genes9030156
Duarte, K. E., de Souza, W. R., Santiago, T. R., Sampaio, B. L., Ribeiro, A. P., Cotta, M. G., et al. (2019). Identification and characterization of core abscisic acid (ABA) signaling components and their gene expression profile in response to abiotic stresses in Setaria viridis. Sci. Rep. 9, 4028–4116. doi:10.1038/s41598-019-40623-5
Fan, W., Zhao, M., Li, S., Bai, X., Li, J., Meng, H., et al. (2018). Correction to: contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 18, 73. doi:10.1186/s12870-018-1283-8
Farooqi, A. A., Tabassum, S., and Ahmad, A. (2017). MicroRNA-34a: a versatile regulator of myriads of targets in different cancers. Int. J. Mol. Sci. 18, 2089. doi:10.3390/ijms18102089
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). Protein identification and analysis tools on the ExPASy server BT - the proteomics protocols handbook. Editor J. M. Walker (Totowa, NJ: Humana Press), 571–607. doi:10.1385/1-59259-890-0:571
Gaut, B. S., Morton, B. R., McCaig, B. C., and Clegg, M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. U. S. A. 93, 10274–10279. doi:10.1073/pnas.93.19.10274
Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24, 2483–2496. doi:10.1105/tpc.112.098574
González-Guzmán, M., Rodríguez, L., Lorenzo-Orts, L., Pons, C., Sarrión-Perdigones, A., Fernández, M. A., et al. (2014). Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 65, 4451–4464. doi:10.1093/jxb/eru219
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi:10.1093/nar/gkr944
Goron, T. L., and Raizada, M. N. (2015). Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front. Plant Sci. 6, 157. doi:10.3389/fpls.2015.00157
Guo, D., Zhou, Y., Li, H. L., Zhu, J. H., Wang, Y., Chen, X. T., et al. (2017). Identification and characterization of the abscisic acid (ABA) receptor gene family and its expression in response to hormones in the rubber tree. Sci. Rep. 7, 45157–45210. doi:10.1038/srep45157
Guo, Z., Kuang, Z., Wang, Y., Zhao, Y., Tao, Y., Cheng, C., et al. (2020). PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 48, D1114–D1121. doi:10.1093/nar/gkz894
Gupta, S. M., Arora, S., Mirza, N., Pande, A., Lata, C., Vasconcelos, M. W., et al. (2017). Finger millet: a “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Front. Plant Sci. 8, 643. doi:10.3389/fpls.2017.00643
Hao, Q., Yin, P., Li, W., Wang, L., Yan, C., Lin, Z., et al. (2011). The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell 42, 662–672. doi:10.1016/j.molcel.2011.05.011
Hatakeyama, M., Aluri, S., Balachadran, M. T., Sivarajan, S. R., Patrignani, A., Grüter, S., et al. (2018). Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. Int. J. rapid Publ. Rep. Genes Genomes 25, 39–47. doi:10.1093/dnares/dsx036
He, Y., Hao, Q., Li, W., Yan, C., Yan, N., and Yin, P. (2014). Identification and characterization of ABA receptors in oryza sativa. PLoS One 9, e95246. doi:10.1371/journal.pone.0095246
Hittalmani, S., Mahesh, H. B., Shirke, M. D., Biradar, H., Uday, G., Aruna, Y. R., et al. (2017). Genome and Transcriptome sequence of Finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 18, 465–516. doi:10.1186/s12864-017-3850-z
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi:10.1093/nar/gkm259
Kim, H., Hwang, H., Hong, J.-W., Lee, Y.-N., Ahn, I. P., Yoon, I. S., et al. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024. doi:10.1093/jxb/err338
Kim, H., Lee, K., Hwang, H., Bhatnagar, N., Kim, D.-Y., Yoon, I. S., et al. (2014). Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 65, 453–464. doi:10.1093/jxb/ert397
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi:10.1093/bioinformatics/btm404
Laskowski, R. A., Jabłońska, J., Pravda, L., Vařeková, R. S., and Thornton, J. M. (2018). PDBsum: structural summaries of PDB entries. Protein Sci. 27, 129–134. doi:10.1002/pro.3289
Lei, P., Wei, X., Gao, R., Huo, F., Nie, X., Tong, W., et al. (2021). Genome-wide identification of PYL gene family in wheat: evolution, expression and 3D structure analysis. Genomics 113, 854–866. doi:10.1016/j.ygeno.2020.12.017
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Lynch, M., and Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473. doi:10.1093/genetics/154.1.459
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi:10.1126/science.1172408
McCormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., et al. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354. doi:10.1111/tpj.13781
Meher, P. K., Sahu, T. K., Gupta, A., Kumar, A., and Rustgi, S. (2024). ASRpro: a machine-learning computational model for identifying proteins associated with multiple abiotic stress in plants. Plant Genome 17, 202599–e20313. doi:10.1002/tpg2.20259
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi:10.1046/j.1365-313X.2001.00965.x
Moore, R. C., and Purugganan, M. D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. doi:10.1016/j.pbi.2004.12.001
Mukami, A., Ngetich, A., Mweu, C., Oduor, R. O., Muthangya, M., and Mbinda, W. M. (2019). Differential characterization of physiological and biochemical responses during drought stress in finger millet varieties. Physiol. Mol. Biol. Plants 25, 837–846. doi:10.1007/s12298-019-00679-z
Muthamilarasan, M., Khandelwal, R., Yadav, C. B., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS One 9, e109920. doi:10.1371/journal.pone.0109920
Oliveros, J. C. (2016). Venny 2.1. 0. Venny. An interact tool comp list with venn’s diagrams. Available online at: https//bioinfogp.cnb.csic.es/tools/venny/ (Accessed February 15, 2016).
Park, S., Fung, P., Nishimura, N., Jensen, D. R., Zhao, Y., Lumba, S., et al. (2010). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi:10.1126/science.1173041.Abscisic
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi:10.1126/science.1173041
Paul, S., Markiel, A., Owen, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (1971). Cytoscape: a software environment for integrated models. Genome Res. 13, 426. doi:10.1101/gr.1239303.metabolite
Pizzio, G. A., Rodriguez, L., Antoni, R., Gonzalez-Guzman, M., Yunta, C., Merilo, E., et al. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163, 441–455. doi:10.1104/pp.113.224162
Qin, B., Fan, S., Yu, H. Y., Lu, Y. X., and Wang, L. F. (2022). HbMYB44, a rubber tree MYB transcription factor with versatile functions in modulating multiple phytohormone signaling and abiotic stress responses. Front. Plant Sci. 13, 893896–893913. doi:10.3389/fpls.2022.893896
Radauer, C., Lackner, P., and Breiteneder, H. (2008). The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8, 286. doi:10.1186/1471-2148-8-286
Rakkammal, K., Maharajan, T., Shriram, R. N., Ram, P. S. J., Ceasar, S. A., and Ramesh, M. (2023a). Physiological, biochemical and molecular responses of finger millet (Eleusine coracana) genotypes exposed to short-term drought stress induced by PEG-6000. South Afr. J. Bot. 155, 45–59. doi:10.1016/j.sajb.2023.01.053
Rakkammal, K., Pandian, S., Maharajan, T., Antony, S., Sohn, S. I., and Ramesh, M. (2023b). Humic acid regulates gene expression and activity of antioxidant enzymes to inhibit the salt - induced oxidative stress in finger millet. Cereal Res. Commun. 52, 397–411. doi:10.1007/s42976-023-00429-8
Rani, V., Joshi, D. C., Joshi, P., Singh, R., and Yadav, D. (2023). “Millet Models” for harnessing nuclear factor-Y transcription factors to engineer stress tolerance in plants: current knowledge and emerging paradigms. Planta 258, 29. doi:10.1007/s00425-023-04186-0
Rani, V., Rana, S., Muthamilarasan, M., Joshi, D. C., and Yadav, D. (2024a). Expression profiling of Nuclear Factor-Y (NF-Y) transcription factors during dehydration and salt stress in finger millet reveals potential candidate genes for multiple stress tolerance. Planta 259, 136. doi:10.1007/s00425-024-04417-y
Rani, V., Singh, V. K., Joshi, D. C., Singh, R., and Yadav, D. (2024b). Genome-wide identification of nuclear factor -Y (NF-Y) transcription factor family in finger millet reveals structural and functional diversity. Heliyon 10, e36370. doi:10.1016/j.heliyon.2024.e36370
Rani, V., Singh, V. K., Joshi, D. C., Singh, R., and Yadav, D. (2024c). Molecular docking insights into nuclear factor Y (NF-Y) transcription factor and pyrabactin resistance 1 (PYL) receptor proteins reveal abiotic stress regulation in finger millet. Crop Des. 3, 100051. doi:10.1016/j.cropd.2023.100051
Romero, P., Lafuente, M. T., and Rodrigo, M. J. (2012). The Citrus ABA signalosome: identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J. Exp. Bot. 63, 4931–4945. doi:10.1093/jxb/ers168
Sanjay Kumar, T., Nageswari, R., Somasundaram, S., Anantharaju, P., Baskar, M., Ramesh, T., et al. (2024). Significance of millets for food and nutritional security—an overview. Discov. Food 4, 73. doi:10.1007/s44187-024-00149-w
Santiago, J., Dupeux, F., Betz, K., Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., et al. (2012). Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant Sci. 182, 3–11. doi:10.1016/j.plantsci.2010.11.014
Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588. doi:10.1111/j.1365-313X.2009.03981.x
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., et al. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19, 2440–2453. doi:10.1105/tpc.107.050963
Sood, S., Joshi, D. C., Chandra, A. K., and Kumar, A. (2019). Phenomics and genomics of finger millet: current status and future prospects. Planta 250, 731–751. doi:10.1007/s00425-019-03159-6
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. doi:10.1093/nar/gkaa1074
Tähtiharju, S., Heino, P., and Palva, E. T. (2002). ATPP2CA negatively regulates ABA responses during cold acclimation and interacts with the potassium channel AKT3 BT - plant cold hardiness: gene regulation and genetic engineering. Editors P. H. Li, and E. T. Palva (Boston, MA: Springer US), 55–62. doi:10.1007/978-1-4615-0711-6_5
Talwar, H. S., Kumar, S., Madhusudhana, R., Nanaiah, G. K., Ronanki, S., and Tonapi, V. A. (2020). Variations in drought tolerance components and their association with yield components in finger millet (Eleusine coracana). Funct. Plant Biol. 47, 659–674. doi:10.1071/FP19274
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi:10.1093/molbev/msab120
Wang, Y.-G., Fu, F.-L., Yu, H.-Q., Hu, T., Zhang, Y.-Y., Tao, Y., et al. (2018). Interaction network of core ABA signaling components in maize. Plant Mol. Biol. 96, 245–263. doi:10.1007/s11103-017-0692-7
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi:10.1093/nar/gky427
Wenkel, S., Turck, F., Singer, K., Gissot, L., Le Gourrierec, J., Samach, A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. doi:10.1105/tpc.106.043299
Wu, C., Zhang, M., Liang, Y., Zhang, L., and Diao, X. (2023). Salt stress responses in foxtail millet: physiological and molecular regulation. Crop J. 11, 1011–1021. doi:10.1016/j.cj.2023.06.001
Xiong, L., Schumaker, K. S., and Zhu, J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. doi:10.1105/tpc.000596
Yadav, S. K., Santosh Kumar, V. V., Verma, R. K., Yadav, P., Saroha, A., Wankhede, D. P., et al. (2020). Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genomics 21, 676–727. doi:10.1186/s12864-020-07083-y
Yin, P., Fan, H., Hao, Q., Yuan, X., Wu, D., Pang, Y., et al. (2009). Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16, 1230–1236. doi:10.1038/nsmb.1730
Yu, J., Hu, S., Wang, J., Wong, G. K. S., Li, S., Liu, B., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. doi:10.1126/science.1068037
Yu, T.-F., Liu, Y., Fu, J.-D., Ma, J., Fang, Z.-W., Chen, J., et al. (2021). The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 19, 2589–2605. doi:10.1111/pbi.13684
Yue, E., Li, C., Li, Y., Liu, Z., and Xu, J. H. (2017). MiR529a modulates panicle architecture through regulating SQUAMOSA PROMOTER BINDING-LIKE genes in rice (Oryza sativa). Plant Mol. Biol. 94, 469–480. doi:10.1007/s11103-017-0618-4
Zhang, G., Liu, X., Quan, Z., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. doi:10.1038/nbt.2195
Zhang, Z., Li, J., Zhao, X.-Q., Wang, J., Wong, G. K.-S., and Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics. Proteomics Bioinforma. 4, 259–263. doi:10.1016/S1672-0229(07)60007-2
Zhu, J. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi:10.1016/j.cell.2016.08.029
Keywords: ABA receptor, ABA signaling pathways, abiotic stress, finger millet, PYL gene family
Citation: Rani V, Maharajan T, Singh S, Joshi DC, Gupta R and Yadav D (2025) Elucidating the role of pyrabactin-like receptors of finger millet under drought and salinity stress: an insight into in silico, machine learning and molecular approaches. Front. Genet. 16:1598523. doi: 10.3389/fgene.2025.1598523
Received: 23 March 2025; Accepted: 14 May 2025;
Published: 29 May 2025.
Edited by:
Rajib Roychowdhury, Institute of Plant Protection, IsraelReviewed by:
Chandra Mohan Singh, Banda University of Agriculture and Technology, IndiaMárcia Carvalho, University of Trás-os-Montes and Alto Douro, Portugal
Copyright © 2025 Rani, Maharajan, Singh, Joshi, Gupta and Yadav. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dinesh Yadav, ZGluZXNoLmJpb3RlY2hAZGR1Z3UuYWMuaW4=; D. C. Joshi, RGluZXNoLkpvc2hpQGljYXIuZ292Lmlu
 Varsha Rani
Varsha Rani Theivanayagam Maharajan
Theivanayagam Maharajan Shefali Singh
Shefali Singh D. C. Joshi
D. C. Joshi Ramwant Gupta
Ramwant Gupta Dinesh Yadav
Dinesh Yadav