- 1Department of Plant and Soil Sciences, University of Pretoria, Pretoria, South Africa
- 2Centre for Microbial Ecology and Genomics, Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria, South Africa
- 3Department of Science and Innovation – National Research Foundation Centre of Excellence in Food Security, Pretoria, South Africa
Introduction: Several postharvest strategies have been explored to prevent postharvest losses of fruit and vegetables in small-scale production environments that are caused by fungal spoilage during storage; however, the losses remain persistent. In the bell pepper industry for instance, approximately 40% of the total global production is lost annually, highlighting a need to understand pathogen dynamics at the preharvest stage that could contribute to tissue breakdown at storage. This study therefore aimed to explore fungal community shifts during bell pepper fruit development from flowering to storage.
Methods: The samples of flowers, small fruits, mature fruits at harvest and storage were processed to identify the fungal composition using next-generation sequencing of the internal transcribed spacer region.
Results: The results showed that bell pepper harbored 346 fungal genera across all fruit stages mainly from the phyla Ascomycota (83.9%) and Basidiomycota (15.3%). The fungal community comprised both pathogenic and beneficial taxa: Cladosporium, Alternaria, and Fusarium were among the most abundant probable pathogenic taxa, while Aureobasidium, Filobasidium, and Sampaiozyma represented potential biocontrol agents (BCAs). Trend and correlation analysis showed an antagonistic relationship between the BCAs and pathogenic fungi, possibly explaining their dynamic composition across the fruit stages.
Discussion: The analysis showed interaction likelihood between pathogenic taxa, giving insights into co-infection, as well as among beneficial taxa with biocontrol potential, highlighting their synergistic effect against pathogens. Based on redundancy analysis, fruit physiological changes across the developmental stages may have accounted for approximately 8.53% of the total microbial variation observed and could favor growth of spoilage pathogens at storage. The overall analysis confirmed that primary infection at the early fruit developmental stage was the source of the bell pepper decay at postharvest. This highlights the critical need to refocus postharvest spoilage management on reducing preharvest infection, particularly those relating to quiescent infections. The antagonistic characteristics in the bell pepper mycobiome can be harnessed for the development of biocontrol consortia targeting dual/multiple infections. These findings offer a new approach to the management of postharvest losses while aligning with sustainable agricultural production and food security by promoting the use of naturally occurring beneficial microbes for crop protection.
1 Introduction
Bell pepper (Capsicum annuum L.), a widely cultivated and economically significant crop in the Solanaceae family, was characterized by a rise in global production, from 29.63 million tonnes in 2010 to 36.09 million tonnes in 2020 (Anaya-Esparza et al., 2021; Krasnow and Ziv, 2022; Tridge, 2023). Its economic contribution is estimated to be approximately USD 4.2 billion which is projected to increase to USD 6.6 billion by 2028 (Business-Research, 2023). Despite this economic importance, approximately 40% of the global produce is lost at postharvest (Anaya-Esparza et al., 2021; Tiamiyu et al., 2023) due to fungal spoilage which may be introduced during flowering and early fruit developmental stages (Singh et al., 2021).
Common bell pepper fungal diseases reported globally include black rot disease caused by Alternaria species, gray mold caused by Botrytis cinerea Pers (Tiamiyu et al., 2023), anthracnose disease caused by Colletotrichum spp. and Fusarium rots caused by Fusarium spp (Frans et al., 2015). Although these pathogens typically affect fruit at postharvest, latent infections occurs during earlier stages of fruit development (Goudarzi et al., 2021; Morales-Cedeño et al., 2021). Several control strategies have therefore been developed to minimize postharvest bell pepper losses caused by fungal pathogen infection (Tiamiyu et al., 2023). These strategies involve both preharvest chemical treatments, e.g., boscalid, azoxystrobin, cyprodinil, fludioxonil (Shim et al., 2023), or postharvest non-chemical approaches, such as the use of edible coatings, essential oils, hot water treatment, modified atmosphere/active packaging, ultraviolet (UV-C) irradiations, and ozone gas treatment (Glowacz and Rees, 2016; Rybak et al., 2021; Tiamiyu et al., 2023). Despite these efforts, postharvest losses due to rot-causing pathogens remain high (Anaya-Esparza et al., 2021). This can largely be attributed to insufficient understanding of the dynamic microbial interactions between microbes and the host plant (Santos and Olivares, 2021). Consequently, there is a limited grasp of the optimal intervention point to target these pathogens (Liu et al., 2022).
Microbiome studies have provided some insight into the interactions between microbiota and their plant host, revealing the potential for microbiome-driven disease management (Enespa and Chandra, 2022). With the possibility of quiescent preharvest infections being the major cause of bell pepper spoilage at storage, this study aimed to provide an overview of the bell pepper mycobiome dynamics across the different fruit developmental stages, including storage, how these trends could be influencing the development of decay at postharvest and to elucidate the possible pathogen entry points for timely intervention.
2 Materials and methods
2.1 Study location and sample collection
The study was conducted at two representative smallholder bell pepper farms located in the Tshwane (farm E), and Germiston (farm G) regions of Gauteng Province, South Africa. The province falls in the summer rainfall area and receives 600–700 mm of rain per annum. It has average annual maximum temperature ranges of 22 and 25°C with an altitude between 1350 and 1800 m above sea level (Dyson, 2015). Production in the selected farms were under shade nets. Asymptomatic bell pepper samples were collected during the September 2022 to April 2023 growing season for four different stages of fruit development. The samples included flowers (FL; n=3 composite samples, each comprising of 15 individual flowers), small fruits (SF; n=3 composite samples of five fruits each), mature fruits at harvest (MF1; n=3 composite samples of three fruits each), and fruits that completed 10 days of storage (ST; n=3 composite samples of three fruits each), resulting in a total of n=24 samples. All samples were randomly collected within the central rows under the shade nets (Figure 1). The samples were transported in cooler boxes containing ice packs to the Plant Pathology laboratory at the University of Pretoria for processing. A selection of fruit was stored in the collection boxes at room temperature for 10 days to simulate postharvest storage. All samples were processed within 24 hours after sampling.
2.2 Assessment of fruit physiological attributes and analysis
Physiological characteristics were measured at SF, MF1, and ST fruit stages, and included total soluble solids (TSS), pH, and defense-related phytochemicals; total phenolic content (TPC), total flavonoid content (TFC), and ascorbic acid (AA). The TSS content (% Brix) of the fruit was measured using a hand digital refractometer (PR-32, Atago, TSS 0–32%, Palette, Japan) following the method described by Nisansala et al. (2015). The fruit pH was measured as described by Abudi et al. (2020), using a Jenway 3510 pH meter (Bibby Scientific Ltd., Stone, UK) with a flat surface electrode combination (Extech Instruments, Waltham, MA, USA).
Plant material (fresh weight) from each fruit stage was prepared for the assessment of defense-related phytochemicals by cutting the samples into small pieces and snap-freezing them in liquid nitrogen. The frozen samples were then aseptically ground to powder form using a sterile electric coffee blender (Kambrook) packaged in sterile resealable bags and stored at −80°C until use. To test for TPC, the extraction was done as described by Boeing et al. (2014) with some minor modifications: sample powder (1 g) was suspended in 5 mL acetone water (Minema Chemicals, Johannesburg, South Africa) (1:1 v/v). This was followed by vigorous mixing using a vortex before incubation of the mixture for 6 hours at room temperature. The mixture was then centrifuged at 3000 × g for 10 mins, after which the supernatant/plant extract was transferred into clean tubes and used for fruit total phenolic estimation following the Folin-Ciocalteu method (Sellamuthu et al., 2013). In triplicates, the supernatant (9 µL) was mixed with 109 µL of 10% Folin-Ciocalteu reagent (Sigma-Aldrich, Darmstadt, Germany) and incubated at room temperature for 3 mins. This was followed by adding 180 µL of 7.5% sodium bicarbonate solution (NaCO3) (Minema Chemicals), and the reaction mixture was incubated at 50 °C for 5 mins. The optical density was measured at 760 nm using an ultraviolet (UV) spectrophotometer (ThermoScientific, Multiskan Go, Waltham, MA USA). The absorbance values of the samples were then corrected using the corresponding blank readings prior to concentration calculations. Gallic acid (Sigma-Aldrich) was used as the standard, and results were presented as mg of gallic acid equivalent (GAE)/100 g of bell pepper fruit powder.
The total flavonoid content was measured by a colorimetric assay, as described by Zhishen et al. (1999). Plant extraction was done according to Alipieva et al. (2010) with a few modifications, where 1 g of the sample was suspended in 5 mL of 60% methanol (Sigma-Aldrich). The mixture was then incubated at 50°C in a water bath for 30 mins before centrifuging at 3000 × g for 10 mins. In triplicates, the resultant supernatant (0.15 mL) was added to 1 mL of distilled water, followed by the addition of 0.15 mL of 5% sodium nitrite (NaNO2) (Minema Chemicals). After 5 min, 0.15 mL of 10% aluminium chloride (AlCl3) (Sigma-Aldrich) was added to the solution and left to stand for 6 min at room temperature, before adding 0.5 mL of 1 M sodium hydroxide (NaOH) (Minema Chemicals) and vortexing. The absorbance was determined at 510 nm and the sample reading corrected using the respective blanks before calculating the concentrations. Catechin (Sigma-Aldrich) was used as the standard for the calibration curve and TFC was expressed as mg of catechin equivalent (CE)/100 g of bell pepper fruit powder.
Ascorbic acid was determined using the indophenol method described by Nielsen (2017) with minor modifications. Fruit powder (4 g) was mixed with 8 mL of metaphosphoric acid (HPO3) (Sigma-Aldrich), vortexed and incubated at room temperature for 10 mins. The mixture was then centrifuged at 6000 × g for 10 mins, and the resultant supernatant was used for the ascorbic acid assay. In triplicates, 2 mL of the supernatant was mixed with 5 mL HPO3 in volumetric flasks and titrated against the prepared 2,6-dichloroindophenol dye (Sigma-Aldrich) until a light, but distinct rose-pink color persisted for more than five seconds. The initial and final burette readings were recorded before and after each titration to calculate the amount of the dye used. The same procedure was repeated for standard ascorbic acid and the blank solutions. Ascorbic acid was calculated as described by Nielsen (2017) (Supplementary Material 1).
The data collected was analyzed using R statistical software version 4.3.1 in RStudio (R Core Team, 2023). Means and standard errors for TPC, TFC, AA, TSS, and pH were calculated, and analysis of variance (ANOVA) at a 95% confidence interval was used to test for significant differences of the means. Post hoc analysis was performed using the least significant difference (LSD) test for pairwise comparison.
2.3 Bell pepper sample preparation, DNA extraction and Illumina sequencing analysis
The collected flower and fruit samples were suspended in sterile double-distilled water containing 0.1% Tween 80 (Sigma-Aldrich) to remove microfloral epiphytes. This was followed by sonication in an ultrasonic water bath (Labotech, Johannesburg) for five minutes, as described by Gomba et al. (2016). These micro-floral washes were filtered through a cellulose nitrate filter paper (0.45 µm pore size; Sartorius, Gottingen, Germany), and the filters containing wash residue (microflora epiphytes) aseptically stored at −20 °C before DNA extraction for the total epiphyte metagenomic DNA.
Total metagenomic DNA extractions of samples at the different pepper developmental stages were done using the Quick-DNA™ Fungal/Bacterial Miniprep kit (Zymo-Research, California, USA), following the manufacturer’s instructions. The DNA concentrations were quantified using a Nanodrop ND2000 Spectrophotometer (Thermo Fischer Scientific, Dreieich, Germany). DNA were stored as aliquots at −20°C, with a separate aliquot prepared for submission to Molecular Research LP (MR DNA; Texas, USA) for paired-end (2×300 bp) high-throughput Illumina MiSeq (Illumina, Inc. USA) sequencing, using the bTEFAP® method (Dowd et al., 2008). The internal transcribed spacer (ITS) region was targeted, using primers ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and ITS2R (5’-GCTGCGTTCTTCATCGATGC-3’), as described by Tedersoo et al. (2015).
Raw sequence data received from MR DNA were processed using the Quantitative Insights Into Microbial Ecology 2 (QIIME 2) platform (Bolyen et al., 2019) on VirtualBox version 6.1.32 (Oracle Corporation, 2022). This involved demultiplexing, quality filtering by adapter trimming, denoising, paired read merging and chimera removal using the DADA2 analysis pipeline (Callahan et al., 2016). Feature tables were then generated from the cleaned amplicon sequence variants (ASVs) and taxonomic classification was done using a self-trained classifier database developed from the UNITE QIIME release version 9.0 (Abarenkov et al., 2023) at a 99% sequence similarity threshold.
Comprehensive fungal community and statistical analysis, including visualization of the generated datasets, were performed in R statistical software version 4.3.1 in RStudio (R Core Team, 2023). Fungal community structure and alpha diversity were calculated from non-rarefied sequences, while sequences rarefied to a minimum library size of 38–231 reads were used to determine beta diversity as described by Cameron et al. (2021). The alpha diversity indices measured at genus level included the Chao1 richness index, Shannon, and Gini-Simpson diversity indices. These were visualized using boxplots generated using the ggplot2 function in R.
The Shapiro–Wilk normality test was used to determine the distribution of the dataset for downstream statistical testing based on fruit developmental stage and sample origin (González-Estrada et al., 2022). Where normal distribution was observed, an analysis of variance (ANOVA) was carried out, followed by a post hoc Tukey’s Honestly Significant Difference (HSD) test for pairwise comparison (Sawyer, 2009). Alternatively, the Kruskal–Wallis test was used for samples not normally distributed, followed by a post hoc Dunn’s test (MacFarland and Yates, 2016). Beta diversity was determined using the Bray-Curtis dissimilarity matrix and visualized through principal coordinates analysis (PCoA) ordination (Xia and Sun, 2023). Statistical differences in beta diversity between fruit developmental stages and origin were inferred with permutational multivariate analysis of variance (PERMANOVA, 999 permutations) using the adonis function (Xia and Sun, 2023).
To assess ecological importance, common fungal taxa were determined at a minimum detection threshold of 1% relative abundance at a minimum sample prevalence of 0.9 (de Souza et al., 2016). Interaction of common mycobial organisms occurring in at least 90% of the total samples was determined using Spearman rank correlation by carrying out a sparse correlation for compositional data (SPARCC) using the phyloseq package in R (Kurtz et al., 2015; Neu et al., 2021). The fungal co-occurrence network generated was visualized using the igraph package (Csardi and Nepusz, 2006). To assess the postharvest pathogen profile, ITS amplicon sequence data of taxa with putative pathogenic nature based on literature were extracted from the processed files and their identity confirmed using NCBI BLASTn (Altschul et al., 1990). The abundance trends of potential pathogen and biological control taxa across the fruit developmental stages were evaluated using ggplot package in R. A redundancy analysis (RDA) was carried out on Hellinger transformed data to determine the influence of fruit physiological parameters on common fungal composition across the fruit developmental stages (Legendre and Gallagher, 2001). The RDA plot was generated using the ggplot.
3 Results
3.1 Assessment of bell pepper quality from small fruit to storage
Fruit physiological parameters (TPC, TFC, AA and TSS) increased with fruit maturity (SF-ST) in farm E (Table 1), while fruit pH, decreased with maturity. The changes in physiological parameters were significantly different between the small fruits and mature fruits at harvest and storage (p < 0.05, Table 1). Farm G’s fruits also depicted a general increase in TPC and TFC concentrations between the small fruits and mature fruits at storage, despite a notable drop at MF1 stage (Table 1). Ascorbic acid concentration and % brix (TSS) significantly increased with fruit maturity, while fruit pH declined (p < 0.05, Table 1).
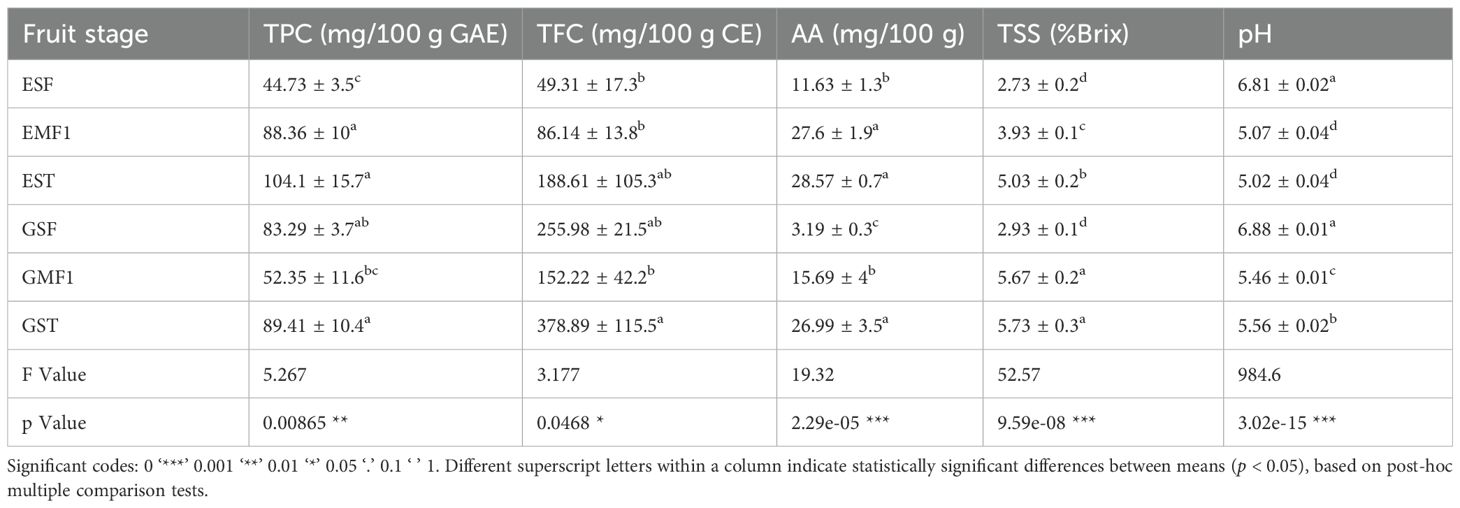
Table 1. Changes in the polyphenol (TPC), flavonoid (TFC), ascorbic acid (AA), total soluble solids (TSS), and pH content across the bell pepper fruit developmental stages: SF, small fruit; MF1, mature fruit at harvest; and ST, mature fruit after storage for 10 days; letters E and G in the fruit stage represent farms.
3.2 Fungal community composition, diversity and interactions
A total of 1,552,622 fungal reads (max. number of reads =101,632, min. number of reads = 38,231, and avg. number of reads = 64,692.58) were retrieved from the 24 bell pepper samples. Data processing yielded a total of 1,778 ASVs comprising of five phyla, 24 classes, 64 orders, 158 families and 346 genera. Alpha diversity measures at genus level ASVs across all the developmental stages (Figure 2a; Table 2) for farm E showed that small fruits had a significantly lower mean Chao1 ASV richness compared to the flowering stage (ANOVA, p = 0.00153). The ASV richness, however, increased with fruit maturity (SF-MF1). Similarly, for farm G, the ASV richness dropped from the FL to SF stage and again peaked with fruit maturation. Despite the variance in ASV richness for farm G, the values were not significantly different across the fruit developmental stages. In both farms, there was a drop in ASV richness at storage (Figure 2a; Table 2).
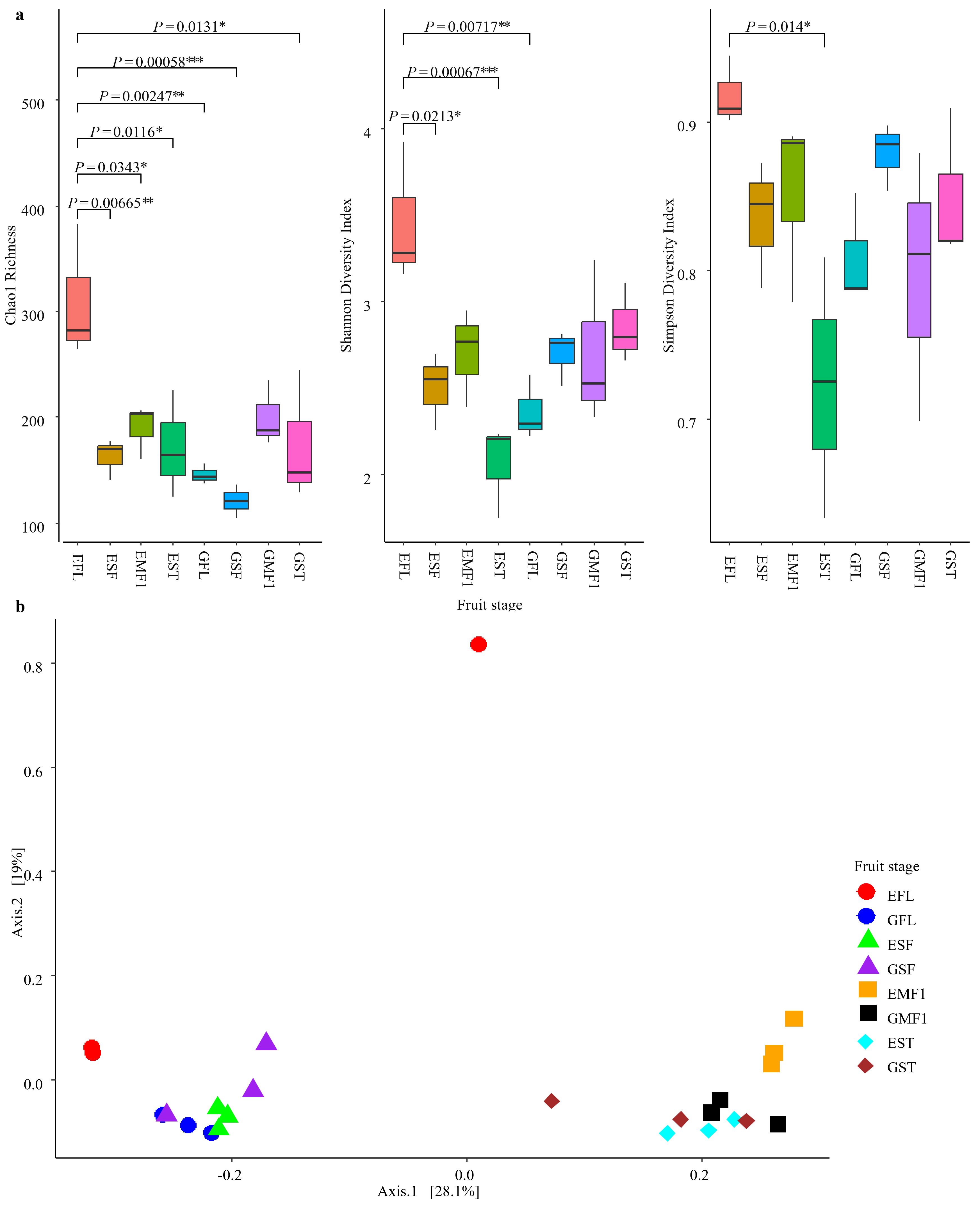
Figure 2. Diversity of fungal communities from the different bell pepper fruit developmental stages: FL, flowering; SF, small fruit; MF1, mature fruit at harvest; and ST, fruit stored for 10 days. (a) Alpha diversity showing the Chao1 richness index, Shannon, and Gini-Simpson diversity indices. Tukey HSD analyses with significant p values indicate the 95 % confidence level. (b) The principal coordinates analysis using the Bray-Curtis dissimilarity of the bell pepper epiphytic fungal community are shown, categorized by developmental stage. PC1 and PC2 accounted for 28.1% and 19% of the variance respectively. Letters E and G in the fruit stage represent farms.
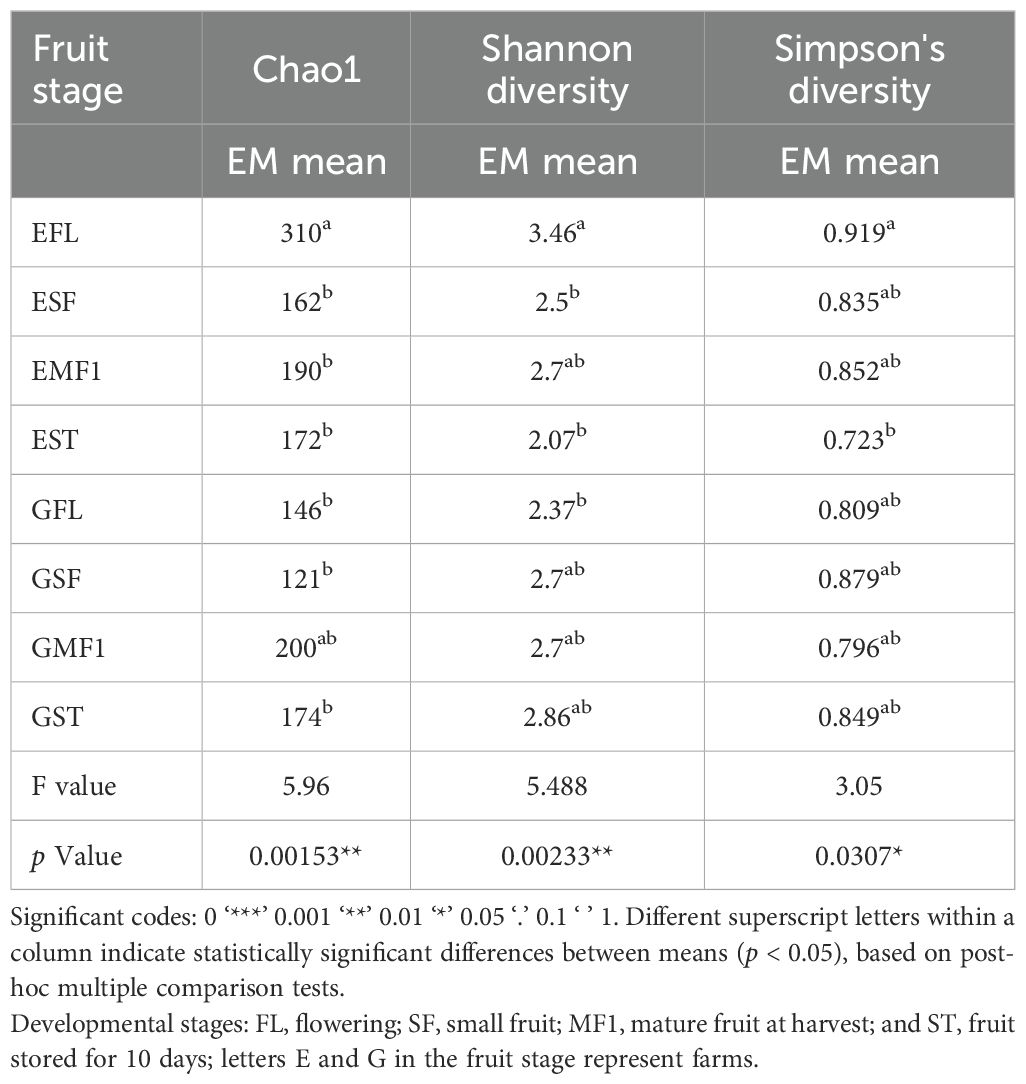
Table 2. Chao1, Shannon and Simpson fungal ASV community richness and diversity using estimated marginal mean (EM mean).
Farm E had a similar trend in Shannon and Simpson diversity as the ASV richness, where the floral stage had a significantly higher diversity compared to the other fruit developmental stages (ANOVA, p = 0.0023, and p = 0.0307 respectively). According to the two indices, the diversity decreased at the small fruit stage, increased with fruit maturation, and decreased at fruit storage (Figure 2a; Table 2). In contrast, farm G had a linear increase in Shannon diversity from floral stage to fruits at storage. Diversity according to the Simpson index increased from FL to SF with a slight decrease towards maturity and subsequent increase in storage. Despite the observed variations, there was no statistical difference in diversity across the developmental stages in farm G (ANOVA, p > 0.05, and p >0.05) for Shannon and Simpson diversity indices respectively (Figure 2a; Table 2).
Beta diversity analysis using PCoA ordination showed clear clustering of the fungal community based on fruit development stage (Figure 2b). Floral and small fruit fungal communities from both farms clustered together except for EFL, which was separate and sparsely distributed from the rest. Fungal communities from mature fruit stages also clustered together, except for those from EMF1, which formed a slightly separate group (Figure 2b). A total of 47% of the variation in the fungal community was accounted for within the first two PC’s in the dataset, with a strong and significant developmental stage influence (PERMANOVA: R2 = 0.3760, F=4.0164, P=0.001) on the fungal microbiota between early and late fruit maturity (28%).
Further analysis at genus level showed that 57 (19.1%) of the total taxa in farm E were shared across the four fruit stages. A total of 91 (30.5%), 9 (3.0%), 28 (9.4%), and 23 (7.7%) of the taxa identified were exclusive to EFL, ESF, EMF, and EST stages, respectively. Farm G samples indicated that 54 (25.7%) of the total taxa were shared across the four fruit stages. The GFL, GSF, GMF1 and GST samples exclusively accounted for 16 (7.6%), 7 (3.3%), 51 (24.3%), and 19 (9.0%) of the taxa respectively.
Ascomycota and Basidiomycota were the most abundant phyla across all samples, accounting for 83.9% and 15.3% of the total fungal community, respectively. The remaining 0.8% of the total fungal community population was accounted for by Chytridiomycota (0.08%), Mortierellomycota (0.02%), Incertae sedis/uncultured fungi (0.4%), and unclassified fungi (0.3%). Cladosporium (37%) and Alternaria (16%) were ranked as the most abundant genera across all four different fruit developmental stages. These were distantly followed by Fusarium, accounting for 5% of the total relative abundance (Figure 3). Other common and abundant genera included Sampaiozyma, Psedopithomyces, Naganishia, Filobasidium, Plectospharella, Aureobasidium, Hohenbuehella, Sporobolomyces, Exserohilum, Acremonium, Coprinellius, Rhodotorula and Xenodidymella.
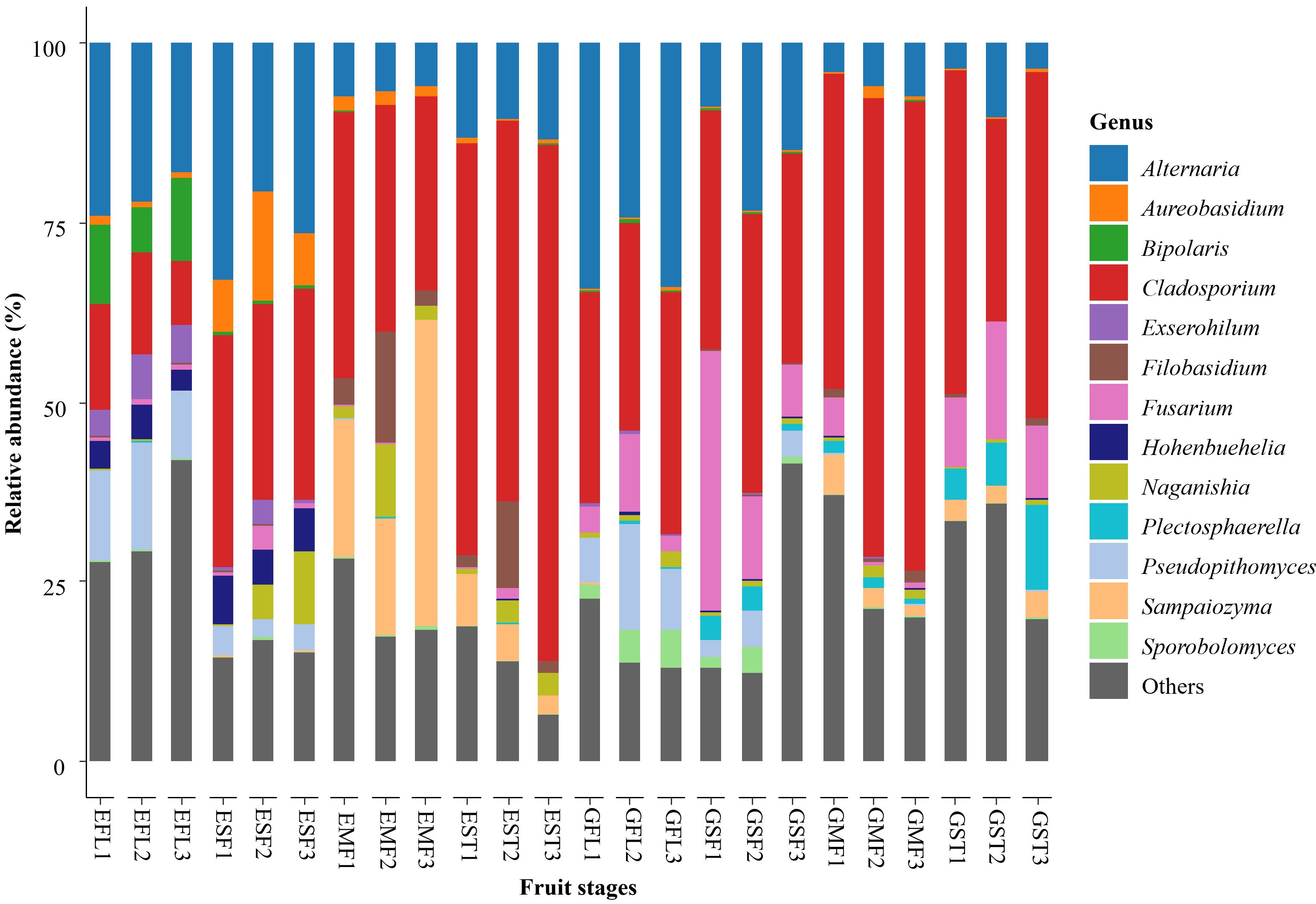
Figure 3. Taxonomic relative abundance of fungal ASVs at genus level identified on bell pepper flower and fruit surfaces at four different developmental stages. The listed genera met the threshold of at least 1% of the total abundance in the specific sample. Others- aggregated ASVs of all taxa accounting for less than 1 % of the total community abundance per sample. Developmental stages: FL, flowering; SF, small fruit; MF, mature fruit; and ST, fruit stored for 10 days; letters E and G in the fruit stage represent farms.
Among the common genera across all the fruit developmental stages were Alternaria, Cladosporium and Fusarium, previously reported as pathogens of bell pepper (Supplementary Material 2). The abundance and distribution of Alternaria were almost similar in both farms while Fusarium was more abundant in farm G (Figure 3). Colletotrichum was also present at all fruit developmental stages in farm G and only present on mature fruits (MF1 and ST) in farm E. According to the NCBI BLASTn taxonomy database, the species-level identity for Alternaria ASVs were mainly A. alternata, A. arborescens, A. cinerarie, A. japonica, A. solani, and A. sonchi. Fusariam comprised of F. oxysporum, F. graminearum, F. equiseti, F. domesticum, F. dimerum, F. delphinoides, F. citri, F. solani, F. tricinctum, F. proliferatum, F. penzigii, and F. chlamydosporum. Colletotrichum ASVs included C. gloeosporioides, C. truncatum and C. cirnanams.
Upon analysis of the trends at genus level across the fruit developmental stages, Cladosporium, Filobasidium, Acremonium, and Plectosphaerella had increased abundance from flowering to storage. Alternaria, Sampaiozyma, Fusarium, Naganishia, Aerobasidium, Hohenbuehelia, Rhodotorula, and Xenodidymella had fluctuating abundance over the fruit developmental stages, which differed by farm. Alternaria abundance in farm E, for instance, was highest at the small fruit stage, reduced with fruit maturity, and increased at storage. Alternaria abundance in farm G was highest at the floral stage and reduced as the fruit matured. Fusarium in both farms had the highest abundance at the small fruit stage, reduced towards harvest and increased at storage. In both farms, the abundance of Sampaiozyma increased as Alternaria decreased (Table 3) with a Pearson correlation analysis showing significant negative correlation between the two taxa across the fruit developmental stages (r = −0.63, p< 0.05, Supplementary Material 3). The abundance of Hohenbuehelia and Exserohilum were inversely proportional to that of Sampaiozyma across the fruit developmental stages (r = −0.6, p <0.05). Sporobolomyces and Exserohilum decreased in abundance from flowering to storage (Table 3). Sporobolomyces in farm E peaked at the small fruit stage, while flowers recorded the highest abundance in farm G. Exserohilum in both farms had reduced abundance from flowering to stored stages.
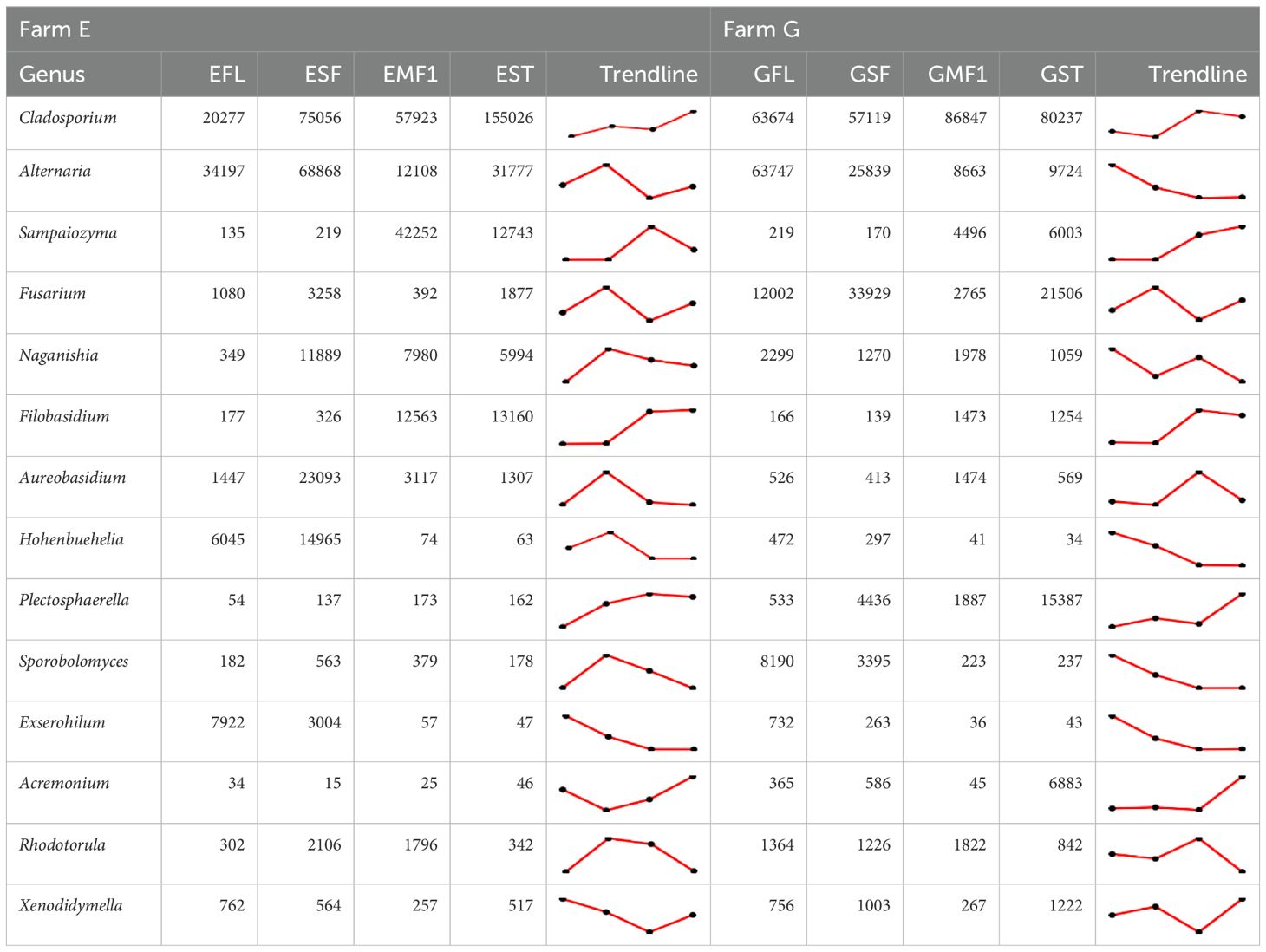
Table 3. Changes in absolute abundance trends of the top common fungal genera across the fruit developmental stages: SF, small fruit; MF1, mature fruit at harvest; and ST, mature fruit after storage for 10 days; letters E and G in the fruit stage represent farms.
Co-occurrence network analysis based on statistical correlation showed the positive (green arrows) and negative (red arrows) correlations between the fungal ASVs in the community (Figure 4). Strong positive correlations were observed between Alternaria and the genera; Exserohilum, Hohenbuehelia, Pseudopithomyces, Sporobolomyces, Xenodidymella and Aureobasidium. On the other hand, Alternaria had a strong negative correlation with Sampaiozyma and moderate negative correlation with Coprinellus and Filobasidium (Figure 4). Fusarium was depicted to have a strong positive correlation with Acremonium, Plectosphaerella and a moderate positive correlation with Sporobolomyces. It had a strong negative correlation with Aureobasidium, Coprinellus and Filobasidium (Figure 4).
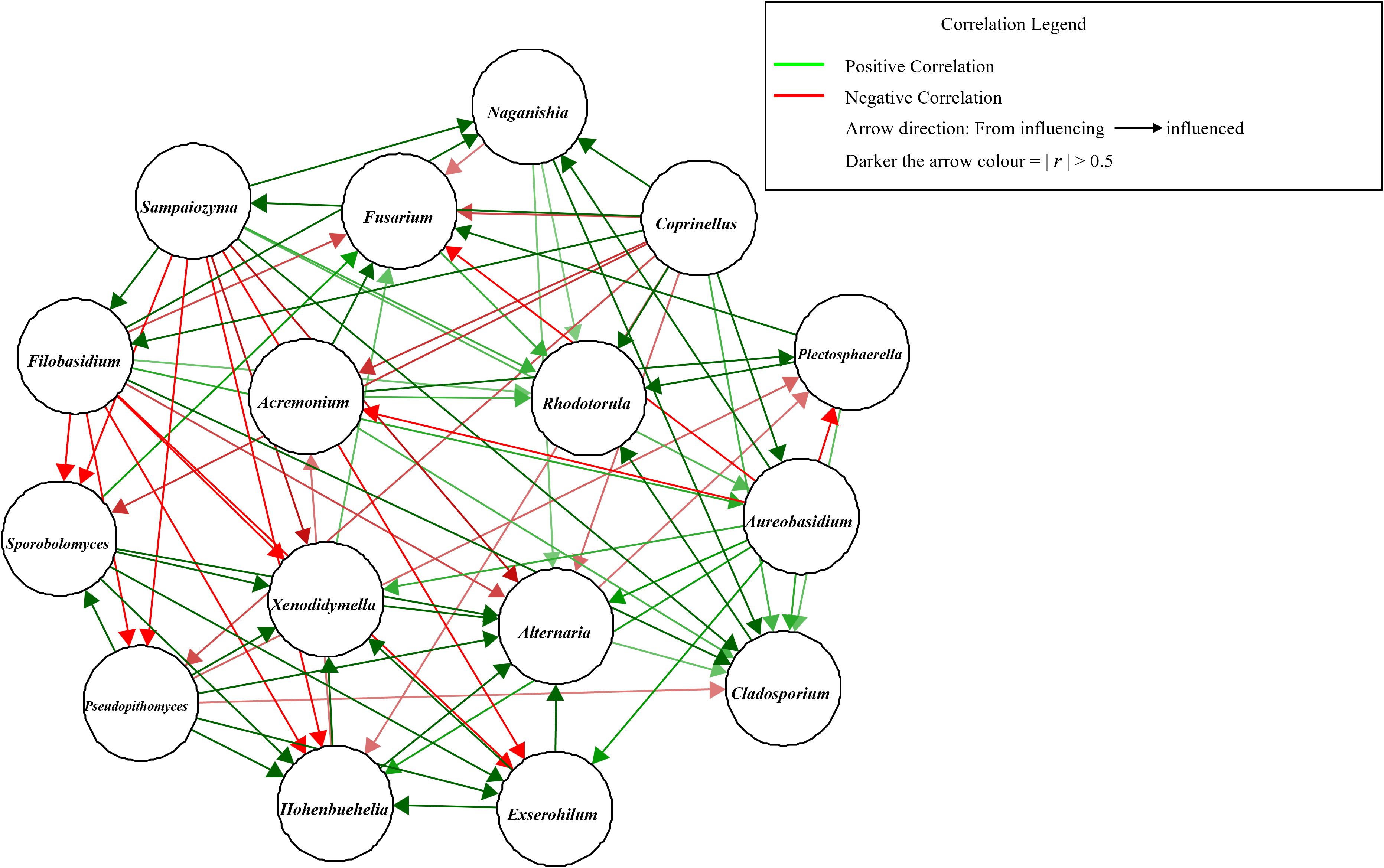
Figure 4. Co-occurrence network of significantly abundant bell pepper fungal microbiota. Nodes represent genera, while edges denote correlations with a threshold of >0.3. Green = positive correlation, red = negative correlation. The color intensity of the arrow indicates a stronger correlation. Generated using SPARCC (5–000 permutations).
Acremonium had a strong negative correlation with Aureobasidium, Corprinellus. Plectosphaerella had a strong positive correlation with Acremonium and a strong negative correlation with Aureobasidium. The network also depicted that Cladosporium positively correlated with almost all taxa around it with Pseudopithomyces interaction being the only negative, albeit weak, correlations. Naganishia also had a strong positive correlation with all the surrounding taxa; Aureobasidium, Coprinellus, Filobasidium and Sampaiozyma.
Aureobasidium, Coprinellus, Filobasidium, and Sampaiozyma were also included in this positive correlation hub. However, they additionally showed negative correlations with selected ASVs. For instance, Coprinellus negatively correlated towards Pseudopithomyces and Sporobolomyces. It also had a positive correlation towards Aureobasidium, Cladosporium, Filobasidium, Naganishia and Sampaiozyma. Sampaiozyma on the other hand strongly correlated negatively towards Alternaria, Exserohilum, Hohenbuehelia, Pseudopithomyces and Sporobolomyces but had strong positive correlations with Cladosporium, Filobasidium and Naganishia (Figure 4). Filobasidium was strongly negatively correlated towards Alternaria, Exserohilum, Fusarium, Hohenbuehelia, Pseudopithomyces, Sporobolomyces and Xenodidymella, and positively correlated with Cladosporium and Naganishia. Despite Auerobasidium having a strong negative correlation towards Acremonium, Fusarium and Plectosphaerella, it interestingly had a strong positive correlation with Alternaria, Cladosporium, Exserohilum, Hohenbuehelia and Xenodidymella.
3.3 Influence of fruit physiological attributes on microbial composition
The redundancy analysis showed that fruit physiological attributes (AA, TPC, TFC and TSS) had a significant influence on fungal composition, contributing to 8.53% of the total variance (ANOVA, Variance = 0.085335, F = 4.2068, p = 0.001, Table 4). Total flavonoid content (TFC) had the highest influence on fungal composition (ANOVA, Variance = 0.021337, F= 5.2594, p = 0.0002) while TPC had the lowest effect (ANOVA, Variance = 0.009587, F= 2.3631, p = 0.066, Table 4).
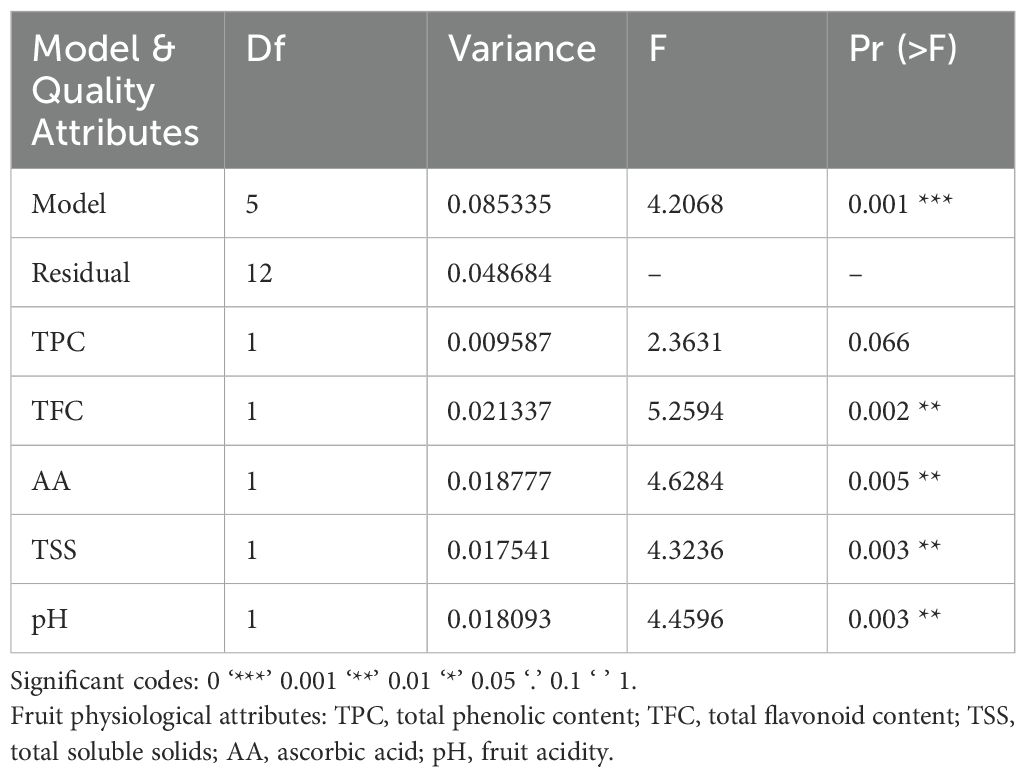
Table 4. Redundancy analysis results showing overall and individual model significance of fruit physiological attributes on fungal composition.
The ordination (Figure 5) indicated that 58.61% of the observed variance was captured in RDA1 and RDA2. The taxa Acremonium and Plestospharella clustered closer to TFC, TPC and TSS. Fusarium was shown to strongly correlate with the changes in fruit pH. Cladosporium and Filobasidium depicted a strong positive correlation to the increase in ascorbic acid and was also negatively correlated to TFC, and TSS increase. Alternaria, Aureobasidium, Exserohilum, Naganishia, Sporobolomyces, Rhodoturula and Xenodidymella had moderate correlation with all the tested physiological attributes (AA, TFC, TPC, TSS and pH), as shown by their central positioning (Figure 5).
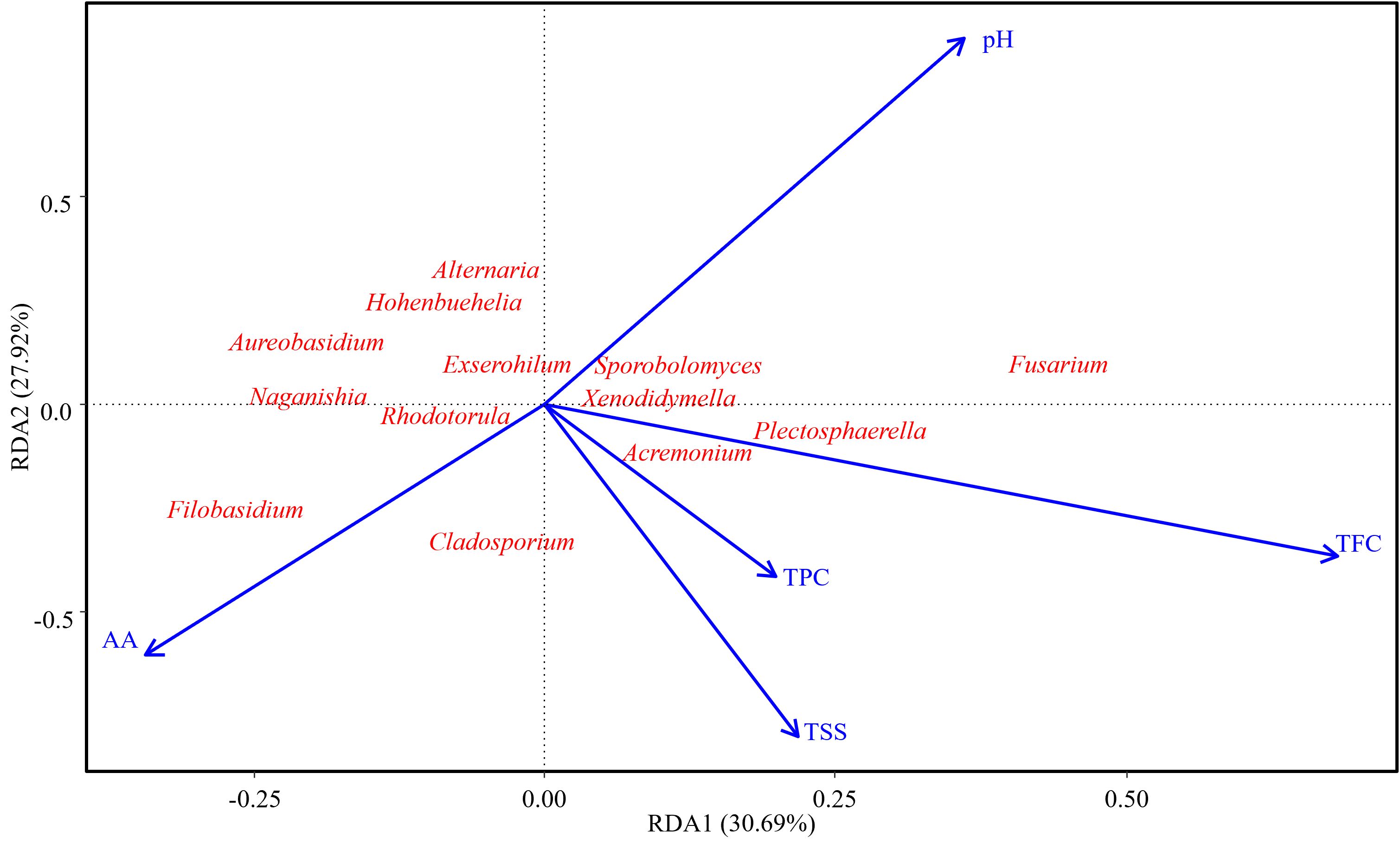
Figure 5. A redundancy analysis (RDA) plot showing the influence of fruit physiological attributes (vectors) on common bell pepper fungi. Fruit physiological attributes: TPC, total phenolic content; TFC, total flavonoid content; TSS, total soluble solids; AA, ascorbic acid; pH, fruit acidity. RDA1 and RDA2 explained 30.69 % and 27.92 % of the observed variance.
4 Discussion
4.1 Bell pepper microbial composition and diversity
The main objective of the present study was to characterize the bell pepper mycobiome across different fruit developmental and storage stages. This understanding can help identify possible pathogen entry points guiding timely intervention to reduce postharvest decay and spoilage. The study showed Ascomycota and Basidiomycota as the most abundant phyla across all bell pepper fruit developmental stages, accounting for a cumulative 99% of the total surface fungi. This agrees with the study findings by Khwantongyim et al. (2021), which showed that these two phyla were the most abundant on apple fruits, accounting for 77.6% and 22.1% of the total counts, respectively. The dynamics of Ascomycota are specifically important as this dominant phylum includes the pre- and postharvest pathogenic genera, namely, Alternaria, Fusarium and Colletotrichum, as identified in the current study. The dominance of Ascomycota could also be associated with their intrinsically higher genomic potential for resource utilization, competition, and tolerance to stress when compared to saprobic Basidiomycota (Egidi et al., 2019). The consistent presence of Basidiomycota such as Sampaiozyma, Filobasidium, Naganishia, Hohenbuehelia suggests a key, yet lesser-understood role of this phylum in fungal dynamics, potentially influencing rot development during storage.
Genus-level analysis revealed further insights into the dynamic shifts in fungal communities across developmental stages. For instance, less than 26% of the observed genera on each farm were shared across all fruit developmental stages, indicating that most of the observed fungal genera were unique to specific stages of development. During the early stages, the bell pepper community was dominated by fungal genera such as Cladosporium, Alternaria, and Fusarium, which are typically associated with leaf blight diseases on several hosts (Ghoneem et al., 2025; Manathunga et al., 2024). At maturity and during storage, Cladosporium became more prominent. The increase in Cladosporium counts on mature fruits supports the finding by Swett et al. (2019) that fruit susceptibility increases with maturity. In their study, Cladosporium infection of raspberry increased from 2.5% at preharvest to over 50% after harvest, compared to the present study where the relative abundance increased from 26.7% at preharvest to 47.7% after harvest. At postharvest, Alternaria and Fusarium also slightly increased, while populations of some taxa, for instance Aureobasidium, Naganishia and Rhodoturula, declined. This reinforces a potential link between microbial competition, the role of these fungi in temporal community dynamics and their contribution to the onset of decay during storage (Verma et al., 2022; Fink and Manhart, 2023). This can also be associated with the observed decline in community diversity at storage as the pathogenic taxa possibly outcompeted other fungal genera in the community (Zhang et al., 2021). Zhang et al. (2021) reported that fungal communities are more sensitive to rot, resulting in a significant decline in abundance during fruit decay. Fungal community structure in terms of genus types was similar in the two farms but only varied in abundance across the developmental stages. For instance, Fusarium were slightly more in farm G compared to farm E and this could be attributed to farmer agronomic practices ranging from seed sources, planting substrate and routine management practices.
PCoA ordinations support the observed alpha diversity trends by clearly distinguishing fungal communities between early and later bell pepper development stages. The distinction could be a result of the physiological changes in fruit over time, as indicated by the tested physiological parameters and the RDA analysis in the current study. Previous studies have shown that physiological changes during fruit development result in biochemical shifts, which in turn influence microbial community assembly or colonization patterns and may further affect the entire plant holobiont (Lone et al., 2024; Zhimo et al., 2022). According to Baglioni et al. (2024) and Pang et al. (2021), plant secondary metabolites shape the endophyte and epiphyte microbial composition by acting as defense compounds against some microbes and as signaling molecules or source of energy for other microbes.
The RDA analysis showed that plant microbes associated differently with the changes in fruit physiological attributes. These differences in fungal association with specific fruit physiological stages and metabolic compounds suggest that fungal taxa exhibit selective physiological preferences, which can in turn strongly impact their colonization and fruit quality during bell pepper development and postharvest (Lievens et al., 2015; Palmieri et al., 2023). However, RDA explains only 8.53% of fungal variation, with the remaining 91.47% possibly being accounted for by other variables not tested in this study. This includes abiotic factors, such as environmental conditions (Verma et al., 2022), and handling practices (Abdelfattah et al., 2016; Bill et al., 2021).
4.2 Potential pathogenic and beneficial fungal genera in bell pepper microbiome
Among the taxa consistently present across all bell pepper developmental stages were Alternaria, and Fusarium. These fungal phylotypes have a wide distribution in agricultural environments and exhibit variable abundance (Egidi et al., 2019; Yadav et al., 2022), providing a potential explanation for their high prevalence across the fruit developmental stages. The phylotypes have also been previously reported as pre- and postharvest pathogens of bell peppers (Tiamiyu et al., 2023). For instance, Alternaria species, particularly A. alternata, A. solani and A. tenuissima, are considered among the most aggressive fungal species affecting several horticultural crops including bell peppers globally (Balamurugan and Kumar, 2023). The significant crop losses associated with Alternaria is largely due to its high prevalence, which can be attributed to its ubiquitous nature and diverse composition, as well as its broad host range (Dang et al., 2015).
Fusarium species have also been linked with bell pepper internal fruit decay (Tiamiyu et al., 2023). The presence of Fusarium across all fruit developmental stages and at storage is consistent with previous studies on bell pepper crops at pre- and postharvest stages (Engalycheva et al., 2024; Liu et al., 2016). This can be attributed to its versatile nature with the ability to survive in almost every environment, including air, water, soil, plants, and other organic substrates (Ekwomadu and Mwanza, 2023). It also has efficient dispersal mechanisms hence its high abundance and distribution in nature (Tupaki-Sreepurna and Kindo, 2018). Some of the Fusarium species that have been reported on bell peppers include F. oxysporum, F. solani, F. verticillioides, F. commune, F. torulosum, and F. sporotrichioides (Engalycheva et al., 2024; Gilardi et al., 2019). Colletotrichum was also among the taxa identified from the retrieved ASVs across all fruit developmental stages in one of the farms, highlighting its potential to infect bell peppers both at pre and postharvest stages (Ali et al., 2016). Pathogenic Colletotrichum spp. that have been reported on Capsicum annuum globally include, C. gloeosporoides, C. truncatum, C. fruticola, C. scovillei, C. endophyticum, C. karsti, C. plurivorum, C. siamense and C. tropicale (de Silva et al., 2019; Liu et al., 2016).
The identification of pathogenic taxa Alternaria, and Fusarium from floral stage through to fruit at storage provides insights potentially linking deterioration during storage with quiescent infections at the early fruit developmental stage. This highlights the need to strengthen preharvest management, for instance timely spray cycles ideally before or at anthesis to reduce postharvest losses and maintain fruit quality. Delayed spray cycles beyond anthesis stage may result in reduced fungicide efficacy particularly against pathogens causing internal fruit rot (Paul et al., 2018). It is also essential to use synthetic fungicides judiciously, as their improper application can suppress potential biocontrol agents (BCAs), possibly enabling pathogenic taxa to proliferate (Szymanski et al., 2023), ultimately impacting fruit quality and postharvest storage outcomes. Also, among prevalent microbial taxa were Pseudopithomyces, Plectosphaerella, Bipolaris, Exserohilum, Lectera, Xenodidymella, Curvularia, and Nigrospora. Despite their existence across the fruit developmental stages, their role in the bell pepper fungal community is yet to be fully understood. These taxa, however, have been reported as pathogenic in crops such as barley, maize, rice and wheat but not in bell peppers (McDonald et al., 2018; Perelló et al., 2017; Wang et al., 2022).
Bell pepper fungal community across the different fruit developmental stages also comprised of potential BCAs. Among them were Acremonium, Aureobasidium, Filobasidium, Hohenbuehelia, Rhodotorula, Sampaiozyma, Sporobolomyces, and Wickerhamomyces. Some of these taxa exhibited an inverse relationship with pathogenic taxa. Specifically, Sampaiozyma and Filobasidium were more abundant in samples where Alternaria and Fusarium were less prevalent, whereas their abundance decreased in samples where Alternaria and Fusarium were more dominant. A similar trend was seen in the cases of Rhodoturula and Xenodidymella, highlighting their potential inhibitory effect against pathogenic taxa. Some of these taxa, for instance Aureobasidium pullulans, have been explored for biocontrol activity against B. cinerea, R. solani, Penicillium expansum and P. digitatum (Agirman and Erten, 2020; Di Francesco et al., 2021; Galli et al., 2021). These studies associated inhibition by A. pullulans to several action mechanisms including competition for space and nutrients, production of volatile organic compounds (VOCs) and secretion of extracellular lytic enzymes. Filobasidium oeirense and F. wieringae have also been reported to inhibit growth of B. cinerea and A. porri mainly by production of antifungal VOCs (Abo-Elyousr et al., 2024; Ruiz-Moyano et al., 2020). Production of VOCs by Rhodoturula species and Wickerhamomyces anomalus have also been linked to inhibition of Aspergillus flavus spore germination, mycelial growth, and toxin production (Hua et al., 2014). Apart from antibiosis, resource competition and mycoparasitism, some BCAs induce plant systemic resistance against pathogenic species (Chaudhary et al., 2024; Prasad et al., 2023). The observed inverse relationship between the potential BCAs and pathogenic taxa in this study could be due to one of these mechanisms, particularly antibiosis or resource competition. Future studies should focus on their functional level of interaction.
4.3 Fungal interactions and ecological dynamics in bell pepper development
Dynamic changes in fungal interaction on bell peppers during development and storage were noted in the current study through co-occurrence network analysis. Potential synergistic and antagonistic interactions within the bell pepper fungal community were revealed, further explaining the temporal changes within the mycobiome genera. This supports the observations of Wright et al. (2019), that microbial communities are characterized by rapid, dynamic succession that maintains ecological stability. The negative correlation of the genera Aureobasidium, Sampaiozyma, Filobasidium and Coprinellus to Alternaria and Fusarium may depict their inhibitory effect as majority of them have been reported as potential BCAs (Abo-Elyousr et al., 2024; Di Francesco et al., 2021; Elkhairy et al., 2023). Cases of positive correlation between potential pathogenic taxa e.g., Alternaria, Pseudopithomyces, Exserohilum, Xenodidymella and Hohenbuehelia, as well as that of Fusarium, Acremoniaum, Plectosphaerella and Sporobolomyces was also noted. This kind of interaction provides a possible glimpse into dual or multi-pathogen infection. This type of host interaction is less studied despite its potential to alter the course of disease and severity expression (Abdullah et al., 2017). For instance, co-infection by F. oxysporum f. sp. medicaginis and R. solani exacerbated disease severity in commercial alfalfa fields in China (Fang et al., 2021). A study by Lerch-Olson and Robertson (2020) also showed that co-inoculation with Pythium and Fusarium species greatly enhanced disease severity in soybeans compared to single pathogen inoculation.
Positive correlation was also depicted among the potential BCAs, Aureobasidium, Coprinellus, Sampaiozyma and Filobasidium, which may be interpreted as mutualistic or complementary interaction within their microbial community and can impact their inhibitory effect against pathogenic taxa. This supports the findings by Ramudingana et al. (2024), which shows that some endophytic fungi, such as Preussia africana and Coprinellus micaceus, have mutualistic associations and can be explored for combined control against fungal pathogens. Other genera such as Cladosporium positively correlated with all other fungi in the bell pepper mycobiome, affirming its ecological adaptability contributing to high abundance throughout the fruit developmental stages (Salvatore et al., 2021). Although considered a weak pathogen, its high adaptability and prevalence enable it to influence the surface microbial communities of fresh produce, potentially causing spoilage under favorable conditions. Therefore, effective postharvest management practices are essential to mitigate its impact (Bento de Carvalho et al., 2024). Also worth noting in the current study, was the negative correlation of Aureobasidium to Fusarium and a positive correlation to Alternaria. This highlights the intricate nature of microbial interactions in fruit maturation, where a single organism inhibits one pathogen while simultaneously promoting the growth of another (Knudsen and Dandurand, 2014). This complexity highlights the absence of a “silver bullet” biocontrol agent, as the efficacy of biocontrol agents can vary based on environmental conditions, target pathogens and the overall dynamic interactions on the host plant (Knudsen and Spurr, 2023).
5 Conclusion
The study findings showed that bell pepper fruit stages harbor numerous fungal genera, from the phyla Ascomycota and Basidiomycota. These genera comprise potential phytopathogens and known biocontrol taxa, all providing a dynamic microbial environment on bell pepper during the various fruit development stages. The results highlighted a distinct differentiation in fungal diversity between the early and the mature fruit development stages. Despite these differences, some taxa, including potential plant pathogens like Alternaria, Fusarium, and Cladosporium, persists from flowering to storage. This persistence suggests that these pathogens establish at early development stages i.e. flowering, and remain quiescent until storage, when rot symptoms and spoilage losses emerge. This underscores the need to adapt current postharvest disease management practices to an early field spray during flowering to mitigate postharvest losses. The study also showed that bell peppers host numerous potential BCAs such as Aureobasidium, Filobasidium, Sampaiozyma, and Coprinellus with possible inhibitory effect towards the pathogenic taxa. Mutual interaction among the BCAs is a phenomenon that should be explored for future development of biocontrol consortia with the ability to control a wider range of pathogens, especially in cases of multiple infections, since the current disease control strategies are still fixed on targeting specific pathogens only. Future research should focus on further exploring this biocontrol approach with emphasis on the functional interactions between the potential BCAs and the pathogenic species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, BioProject ID PRJNA1284057.
Author contributions
EK: Conceptualization, Writing – original draft, Methodology, Visualization, Software, Writing – review & editing, Investigation, Data curation, Project administration, Validation, Formal analysis. JG: Software, Supervision, Writing – review & editing, Conceptualization, Methodology, Formal analysis, Visualization, Data curation, Validation. NS: Project administration, Visualization, Validation, Data curation, Investigation, Methodology, Conceptualization, Supervision, Writing – review & editing, Formal analysis. TM: Investigation, Conceptualization, Data curation, Visualization, Validation, Project administration, Writing – review & editing. LK: Visualization, Resources, Project administration, Validation, Methodology, Supervision, Writing – review & editing, Conceptualization, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the Gauteng Department of Agriculture and Rural Development (GDARD) project “Assessment and mitigation of fresh produce postharvest losses among small-scale farmers in the Gauteng Province” (FY 2020/21).
Acknowledgments
The authors acknowledge Dr. Mallick Bill for his valuable contributions during project conceptualization, data collection, and preprocessing, and Ms. Dina Boikanyo (GDARD project manager) for facilitating connections between the researchers and farmers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fhort.2025.1656887/full#supplementary-material
References
Abarenkov K., Zirk A., Piirmann T., Pöhönen R., Ivanov F., Henrik R., et al. (2023). UNITE QIIME release for fungi 2. doi: 10.15156/BIO/2938080
Abdelfattah A., Wisniewski M., Li Destri Nicosia M. G., Cacciola S. O., and Schena L. (2016). Metagenomic analysis of fungal diversity on strawberry plants and the effect of management practices on the fungal community structure of aerial organs. PloS One 11, e0160470. doi: 10.1371/journal.pone.0160470
Abdullah A. S., Moffat C. S., Lopez-Ruiz F. J., Gibberd M. R., Hamblin J., and Zerihun A. (2017). Host–multi-pathogen warfare: pathogen interactions in co-infected plants. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01806
Abo-Elyousr K. A., Imran M., Sallam N. M., Abdel-Aal A. M., Assiri M. E., and Abdel-Rahim I. R. (2024). Sustainable biocontrol of purple blotch disease in Allium cepa L. by biocontrol yeasts, Pichia kluyveri and Filobasidium wieringae. Egypt. J. Biol. Pest Control. 34, 11. doi: 10.1186/s41938-024-00776-6
Abudi Z. N., Hu Z., Abood A. R., Liu D., and Gao A. (2020). Effects of alkali pre-treatment, total solid content, substrate to inoculum ratio, and pH on biogas production from anaerobic digestion of mango leaves. Waste Biomass Valorization 11, 887–897. doi: 10.1007/s12649-018-0437-0
Agirman B. and Erten H. (2020). Biocontrol ability and action mechanisms of Aureobasidium pullulans GE17 and Meyerozyma guilliermondii KL3 against Penicillium digitatum DSM2750 Penicillium expansum DSM62841 causing postharvest diseases. Yeast 37, 437–448. doi: 10.1002/yea.3501
Ali A., Bordoh P. K., Singh A., Siddiqui Y., and Droby S. (2016). Post-harvest development of anthracnose in pepper (Capsicum spp): Etiology and management strategies. Crop Prot. 90, 132–141. doi: 10.1016/j.cropro.2016.07.026
Alipieva K., Petreska J., Gil-Izquierdo Á., Stefova M., Evstatieva L., and Bankova V. (2010). Influence of the extraction method on the yield of flavonoids and phenolics from Sideritis spp. (Pirin Mountain tea). Nat. Prod. Commun. 5, 1934578X1000500113. doi: 10.1177/1934578X1000500113
Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anaya-Esparza L. M., Mora Z.V.-d. l., Vázquez-Paulino O., Ascencio F., and Villarruel-López A. (2021). Bell peppers (Capsicum annum L.) losses and wastes: Source for food and pharmaceutical applications. Molecules 26, 5341. doi: 10.3390/molecules26175341
Baglioni M., Fries A., Müller J.-M., Omarini A., Müller M., Breccia J. D., et al. (2024). Acremonium sp. diglycosidase-aid chemical diversification: valorization of industry by-products. Appl. Microbiol. Biotechnol. 108, 250. doi: 10.1007/s00253-023-12957-8
Balamurugan A. and Kumar A. (2023). Postharvest fruit rot of bell pepper (Capsicum annuum L.): pathogenicity and host range of Alternaria alternata. Sci. Hortic. 319, 112156. doi: 10.1016/j.scienta.2023.112156
Bento de Carvalho T., Silva B. N., Tomé E., and Teixeira P. (2024). Preventing fungal spoilage from raw materials to final product: Innovative preservation techniques for fruit fillings. Foods 13, 2669. doi: 10.3390/foods13172669
Bill M., Chidamba L., Gokul J. K., and Korsten L. (2021). Mango endophyte and epiphyte microbiome composition during fruit development and post-harvest stages. Horticulturae 7, 495. doi: 10.3390/horticulturae7110495
Boeing J. S., Barizão É.O., e Silva B. C., Montanher P. F., de Cinque Almeida V., and Visentainer J. V. (2014). Evaluation of solvent effect on the extraction of phenolic compounds and antioxidant capacities from the berries: application of principal component analysis. Chem. Cent. J. 8, 1–9. doi: 10.1186/s13065-014-0048-1
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Business-Research (2023). Capsicum market report overview. Available online at: https://www.businessresearchinsights.com/market-reports/capsicum-market-100180 (Accessed August 10th, 2023).
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cameron E. S., Schmidt P. J., Tremblay B. J. M., Emelko M. B., and Müller K. M. (2021). Enhancing diversity analysis by repeatedly rarefying next generation sequencing data describing microbial communities. Sci. Rep. 11, 22302. doi: 10.1038/s41598-021-01636-1
Chaudhary R., Nawaz A., Khattak Z., Butt M. A., Fouillaud M., Dufossé L., et al. (2024). Microbial bio-control agents: A comprehensive analysis on sustainable pest management in agriculture. J. Agric. Food Res. 18, 101421. doi: 10.1016/j.jafr.2024.101421
Csardi G. and Nepusz T. (2006). The igraph software package for complex network research. Inter. J. Complex Syst., 1695. Available online at: https://cran.r-project.org/web/packages/igraph/index.html (Accessed November 29, 2024).
Dang H. X., Pryor B., Peever T., and Lawrence C. B. (2015). The Alternaria genomes database: a comprehensive resource for a fungal genus comprised of saprophytes, plant pathogens, and allergenic species. BMC Genomics 16, 1–9. doi: 10.1186/s12864-015-1430-7
de Silva D. D., Groenewald J. Z., Crous P. W., Ades P. K., Nasruddin A., Mongkolporn O., et al. (2019). Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus 10, 1–32. doi: 10.1186/s43008-019-0001-y
de Souza R. S. C., Okura V. K., Armanhi J. S. L., Jorrín B., Lozano N., Da Silva M. J., et al. (2016). Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 6, 28774. doi: 10.1038/srep28774
Di Francesco A., Di Foggia M., Corbetta M., Baldo D., Ratti C., and Baraldi E. (2021). Biocontrol activity and plant growth promotion exerted by Aureobasidium pullulans strains. J. Plant Growth Regul. 40, 1233–1244. doi: 10.1007/s00344-020-10184-3
Dowd S. E., Wolcott R. D., Sun Y., McKeehan T., Smith E., and Rhoads D. (2008). Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PloS One 3, e3326. doi: 10.1371/journal.pone.0003326
Dyson L. L. (2015). A heavy rainfall sounding climatology over Gauteng, South Africa, using self-organising maps. Clim. Dyn. 45, 3051–3065. doi: 10.1007/s00382-015-2523-3
Egidi E., Delgado-Baquerizo M., Plett J. M., Wang J., Eldridge D. J., Bardgett R. D., et al. (2019). A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 10, 2369. doi: 10.1038/s41467-019-10373-z
Ekwomadu T. I. and Mwanza M. (2023). Fusarium fungi pathogens, identification, adverse effects, disease management, and global food security: A review of the latest research. Agriculture 13, 1810. doi: 10.3390/agriculture13091810
Elkhairy B. M., Salama N. M., Desouki A. M., Abdelrazek A. B., Soliman K. A., Ibrahim S. A., et al. (2023). Towards unlocking the biocontrol potential of Pichia kudriavzevii for plant fungal diseases: in vitro and in vivo assessments with candidate secreted protein prediction. BMC Microbiol. 23, 356. doi: 10.1186/s12866-023-03047-w
Enespa and Chandra P. (2022). Tool and techniques study to plant microbiome current understanding and future needs: an overview. Commun. Integr. Biol. 15, 209–225. doi: 10.1080/19420889.2022.2082736
Engalycheva I., Kozar E., Frolova S., Vetrova S., Tikhonova T., Dzhos E., et al. (2024). Fusarium species causing pepper wilt in Russia: molecular identification and pathogenicity. Microorganisms 12, 343. doi: 10.3390/microorganisms12020343
Fang X., Zhang C., Wang Z., Duan T., Yu B., Jia X., et al. (2021). Co-infection by soil-borne fungal pathogens alters disease responses among diverse alfalfa varieties. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.664385
Fink J. W. and Manhart M. (2023). How do microbes grow in nature? The role of population dynamics in microbial ecology and evolution. Curr. Opin. Syst. Biol. 36, 100470. doi: 10.1016/j.coisb.2023.100470
Frans M., Aerts R., Van Calenberge B., Van Herck L., and Ceusters J. (2015). Influence of floral morphology and fruit development on internal fruit rot in bell pepper. III International Symposium on Postharvest Pathology: Using Science to Increase Food Availability. Acta Hortic. 1144, 199–206. doi: 10.17660/ActaHortic.2016.1144.29
Galli V., Romboli Y., Barbato D., Mari E., Venturi M., Guerrini S., et al. (2021). Indigenous Aureobasidium pullulans strains as biocontrol agents of Botrytis cinerea on grape berries. Sustainability 13, 9389. doi: 10.3390/su13169389
Ghoneem K. M., Rashad E. M., Al-Askar A. A., Helmy Y. A., El-Gamal S. M., Ibrahim S. D., et al. (2025). Alternaria radicina; unveiling the cause, spread, and molecular basis of a novel coriander leaf blight disease in Egypt. Heliyon 11 (1), e41081. doi: 10.1016/j.heliyon.2024.e41081
Gilardi G., Matic S., Gullino M., and Garibaldi A. (2019). First report of crown and root rot caused by Fusarium oxysporum on sweet pepper (Capsicum annuum) in Italy. Plant Dis. 103, 2946. doi: 10.1094/PDIS-04-19-0863-PDN
Glowacz M. and Rees D. (2016). Exposure to ozone reduces postharvest quality loss in red and green chilli peppers. Food Chem. 210, 305–310. doi: 10.1016/j.foodchem.2016.04.119
Gomba A., Chidamba L., and Korsten L. (2016). Prevalence and serovar diversity of Salmonella spp. in primary horticultural fruit production environments. Food Control 69, 13–19. doi: 10.1016/j.foodcont.2016.04.026
González-Estrada E., Villaseñor J. A., and Acosta-Pech R. (2022). Shapiro-Wilk test for multivariate skew-normality. Comput. Stat. 37, 1985–2001. doi: 10.1007/s00180-021-01188-y
Goudarzi A., Samavi S., Amiri Mazraie M., and Majidi Z. (2021). Fungal pathogens associated with pre-and postharvest fruit rots of mango in southern Iran. J. Phytopathol. 169, 545–555. doi: 10.1111/jph.13027
Hua S. S. T., Beck J. J., Sarreal S. B. L., and Gee W. (2014). The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 30, 71–78. doi: 10.1007/s12550-014-0189-z
Khwantongyim P., Wansee S., Lu X., Zhang W., and Sun G. (2021). Variations in the community structure of fungal microbiota associated with apple fruit shaped by fruit bagging-based practice. J. Fungi 7, 764. doi: 10.3390/jof7090764
Knudsen G. R. and Dandurand L.-M. C. (2014). Ecological complexity and the success of fungal biological control agents. Adv. Agric. 2014, 542703. doi: 10.1155/2014/542703
Knudsen G. R. and Spurr H. W. (2023). “Management of bacterial populations for foliar disease biocontrol,” in Biocontrol of Plant Diseases (Boca Raton: CRC Press), 83–92. doi: 10.1201/9780429292330-6
Krasnow C. and Ziv C. (2022). Non-chemical approaches to control postharvest gray mold disease in bell peppers. Agronomy 12, 216. doi: 10.3390/agronomy12010216
Kurtz Z. D., Müller C. L., Miraldi E. R., Littman D. R., Blaser M. J., and Bonneau R. A. (2015). Sparse and compositionally robust inference of microbial ecological networks. PloS Comput. Biol. 11, e1004226. doi: 10.1371/journal.pcbi.1004226
Legendre P. and Gallagher E. D. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280. doi: 10.1007/s004420100716
Lerch-Olson E. R. and Robertson A. E. (2020). Effect of co-inoculations with Pythium and Fusarium species on seedling disease development of soybean. Can. J. Plant Pathol. 42, 408–418. doi: 10.1080/07060661.2019.1668858
Lievens B., Hallsworth J. E., Pozo M. I., Belgacem Z. B., Stevenson A., Willems K. A., et al. (2015). Microbiology of sugar-rich environments: diversity, ecology and system constraints. Environ. Microbiol. 17, 278–298. doi: 10.1111/1462-2920.12570
Liu L., Ma L., Feng J., and Lu X. (2022). Dynamic fluctuation and niche differentiation of fungal pathogens infecting bell pepper pants. Appl. Environ. Microbiol. 88, e01003–e01022. doi: 10.1128/aem.01003-22
Liu F., Tang G., Zheng X., Li Y., Sun X., Qi X., et al. (2016). Molecular and phenotypic characterization of Colletotrichum species associated with anthracnose disease in peppers from Sichuan Province, China. Sci. Rep. 6, 32761. doi: 10.1038/srep32761
Lone R., Bhat A., Nazim N., Malla N. A., Rohella G. K., and Mohamed H. I. (2024). “Role of Phenolics in Plant–Microbe Interaction: A Review,” in Plant Phenolics in Biotic Stress Management. Eds. Lone R., Khan S., and Mohammed Al-Sadi A. (Springer, Singapore). doi: 10.1007/978-981-99-3334-1_1
MacFarland T. W. and Yates J. M. (2016). “Kruskal–Wallis H-Test for Oneway Analysis of Variance (ANOVA) by Ranks,” in Introduction to Nonparametric Statistics for the Biological Sciences Using R (Springer, Cham). doi: 10.1007/978-3-319-30634-6_6
Manathunga K. K., Gunasekara N. W., Meegahakumbura M. K., Ratnaweera P. B., Faraj T. K., and Wanasinghe D. N. (2024). Exploring endophytic fungi as natural antagonists against fungal pathogens of food crops. J. Fungi. 10, 606. doi: 10.3390/jof10090606
McDonald M. C., Ahren D., Simpfendorfer S., Milgate A., and Solomon P. S. (2018). The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol. Plant Pathol. 19, 432–439. doi: 10.1111/mpp.12535
Morales-Cedeño L. R., del Carmen Orozco-Mosqueda M., Loeza-Lara P. D., Parra-Cota F. I., de Los Santos-Villalobos S., and Santoyo G. (2021). Plant growth-promoting bacterial endophytes as biocontrol agents of pre-and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiol. Res. 242, 126612. doi: 10.1016/j.micres.2020.126612
Neu A. T., Allen E. E., and Roy K. (2021). Defining and quantifying the core microbiome: challenges and prospects. Proc. Natl. Acad. Sci. 118, e2104429118. doi: 10.1073/pnas.2104429118
Nielsen S. S. (2017). “Vitamin C Determination by Indophenol Method,” in Food Analysis Laboratory Manual. Food Science Text Series (Springer, Cham). doi: 10.1007/978-3-319-44127-6_15
Nisansala Y., Jayakod L., Sarananda H., and Somaratne S. (2015). Effect of pre-harvest potassium treatment on stem-end rot disease development of mango (Mangifera indica L.) cv. TomEJC during fruit ripening. J. Agric. Sci. (Belihuloya) 14, 119–132. doi: 10.4038/suslj.v14i2.7700
Oracle_Corporation (2022). Oracle VM VirtualBox, version 6.1.32. Available online at: https://www.virtualbox.org/ (Accessed January 22nd, 2024).
Palmieri D., Segorbe D., López-Berges M. S., De Curtis F., Lima G., Di Pietro A., et al. (2023). Alkaline pH, low iron availability, poor nitrogen sources and CWI MAPK signaling are associated with increased fusaric acid production in Fusarium oxysporum. Toxins 15, 50. doi: 10.3390/toxins15010050
Pang Z., Chen J., Wang T., Gao C., Li Z., Guo L., et al. (2021). Linking plant secondary metabolites and plant microbiomes: a review. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.621276
Paul P. A., Bradley C. A., Madden L. V., Dalla Lana F., Bergstrom G. C., Dill-Macky R., et al. (2018). Effects of pre-and postanthesis applications of demethylation inhibitor fungicides on Fusarium head blight and deoxynivalenol in spring and winter wheat. Plant Dis. 102, 2500–2510. doi: 10.1094/PDIS-03-18-0466-RE
Perelló A., Aulicino M., Stenglein S. A., Labuda R., and Moreno M. V. (2017). Pseudopithomyces chartarum associated with wheat seeds in Argentina, pathogenicity and evaluation of toxigenic ability. Eur. J.Plant Pathol. 148, 491–496. doi: 10.1007/s10658-016-1093-5
Prasad B., Sharma D., Kumar P., and Dubey R. C. (2023). Biocontrol potential of Bacillus spp. for resilient and sustainable agricultural systems. Physiol. Mol. Plant Pathol. 128, 102173. doi: 10.1016/j.pmpp.2023.102173
Ramudingana P., Mamphogoro T. P., Kamutando C. N., Maboko M. M., Modika K. Y., Moloto K. W., et al. (2024). Antagonistic potential of endophytic fungal isolates of tomato (Solanum lycopersicum L.) fruits against post-harvest disease-causing pathogens of tomatoes: An in vitro investigation. Fungal Biol. 128, 1847–1858. doi: 10.1016/j.funbio.2024.05.006
R Core Team (2023). R (4.3.1):A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available online at: https://www.r-project.org/.
Ruiz-Moyano S., Hernández A., Galvan A. I., Córdoba M. G., Casquete R., Serradilla M. J., et al. (2020). Selection and application of antifungal VOCs-producing yeasts as biocontrol agents of grey mould in fruits. Food Microbiol. 92, 103556. doi: 10.1016/j.fm.2020.103556
Rybak K., Wiktor A., Pobiega K., Witrowa-Rajchert D., and Nowacka M. (2021). Impact of pulsed light treatment on the quality properties and microbiological aspects of red bell pepper fresh-cuts. Lwt 149, 111906. doi: 10.1016/j.lwt.2021.111906
Salvatore M. M., Andolfi A., and Nicoletti R. (2021). The genus Cladosporium: A rich source of diverse and bioactive natural compounds. Molecules 26, 3959. doi: 10.3390/molecules26133959
Santos L. F. and Olivares F. L. (2021). Plant microbiome structure and benefits for sustainable agriculture. Curr. Plant Biol. 26, 100198. doi: 10.1016/j.cpb.2021.100198
Sawyer S. F. (2009). Analysis of variance: the fundamental concepts. J. Man. Manip. Ther. 17, 27E–38E. doi: 10.1179/jmt.2009.17.2.27E
Sellamuthu P. S., Mafune M., Sivakumar D., and Soundy P. (2013). Thyme oil vapour and modified atmosphere packaging reduce anthracnose incidence and maintain fruit quality in avocado. J. Sci. Food Agric. 93, 3024–3031. doi: 10.1002/jsfa.6135
Shim J.-H., Eun J.-B., Zaky A. A., Hussein A. S., Hacimüftüoğlu A., and Abd El-Aty A. (2023). A comprehensive review of pesticide residues in peppers. Foods 12, 970. doi: 10.3390/foods12050970
Singh D., Sharma R. R., and Kesharwani A. K. (2021). “Postharvest Losses of Horticultural Produce,” in Postharvest Handling and Diseases of Horticultural Produce (CRC Press 6000 Broken Sound Parkway NW, Suite 300, Boca Raton, FL), 1–24. doi: 10.1201/9781003045502-1
Swett C. L., Hamby K. A., Hellman E. M., Carignan C., Bourret T. B., and Koivunen E. E. (2019). Characterizing members of the Cladosporium cladosporioides species complex as fruit rot pathogens of red raspberries in the mid-Atlantic and co-occurrence with Drosophila suzukii (spotted wing drosophila). Phytoparasitica 47, 415–428. doi: 10.1007/s12600-019-00734-1
Szymanski S., Longley R., Hatlen R. J., Heger L., Sharma N., Bonito G., et al. (2023). The blueberry fruit mycobiome varies by tissue type and fungicide treatment. Phytobiomes J. 7, 208–219. doi: 10.1094/PBIOMES-04-22-0028-FI
Tedersoo L., Anslan S., Bahram M., Põlme S., Riit T., Liiv I., et al. (2015). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 1–43. doi: 10.3897/mycokeys.10.4852
Tiamiyu Q. O., Adebayo S. E., and Ibrahim N. (2023). Recent advances on postharvest technologies of bell pepper: A review. Heliyon. 9 (4), e15302. doi: 10.1016/j.heliyon.2023.e15302
Tridge (2023). Fresh bell pepper production. Available online at: https://www.tridge.com/intelligences/bell-pepper/production (Accessed August 10th, 2023).
Tupaki-Sreepurna A. and Kindo A. J. (2018). Fusarium: The versatile pathogen. Indian J. Med. Microbiol. 36, 8–17. doi: 10.4103/ijmm.IJMM_16_24
Verma S., Azevedo L. C. B., Pandey J., Khusharia S., Kumari M., Kumar D., et al. (2022). Microbial intervention: an approach to combat the postharvest pathogens of fruits. Plants 11, 3452. doi: 10.3390/plants11243452
Wang S., Lu Z., Lang B., Wang X., Li Y., and Chen J. (2022). Curvularia lunata and Curvularia leaf spot of maize in China. ACS Omega. 7, 47462–47470. doi: 10.1021/acsomega.2c03013
Wright R. J., Gibson M. I., and Christie-Oleza J. A. (2019). Understanding microbial community dynamics to improve optimal microbiome selection. Microbiome 7, 1–14. doi: 10.1186/s40168-019-0702-x
Xia Y. and Sun J. (2023). “Beta Diversity Metrics and Ordination,” in Bioinformatic and Statistical Analysis of Microbiome Data: From Raw Sequences to Advanced Modeling with QIIME 2 and R (Springer Nature Switzerland AG: Springer Cham), 335–395. doi: 10.1007/978-3-031-21391-5_10
Yadav A. N., Kour D., Kaur T., Devi R., and Yadav A. (2022). Endophytic fungal communities and their biotechnological implications for agro-environmental sustainability. Folia Microbiol. 67, 203–232. doi: 10.1007/s12223-021-00939-0
Zhang Q., Shi W., Zhou B., Du H., Xi L., Zou M., et al. (2021). Variable characteristics of microbial communities on the surface of sweet cherries under different storage conditions. Postharvest Biol.Technol. 173, 111408. doi: 10.1016/j.postharvbio.2020.111408
Zhimo V. Y., Kumar A., Biasi A., Abdelfattah A., Sharma V. K., Salim S., et al. (2022). Assembly and dynamics of the apple carposphere microbiome during fruit development and storage. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.928888
Keywords: fungal microbiome, postharvest decay, biological control consortia, latent infection, food security
Citation: Karoney EM, Gokul JK, Siyoum N, Molelekoa T and Korsten L (2025) Persistence of pathogens and biocontrol potential in the bell pepper fruit mycobiome from flowering to postharvest. Front. Hortic. 4:1656887. doi: 10.3389/fhort.2025.1656887
Received: 30 June 2025; Accepted: 28 July 2025;
Published: 19 August 2025.
Edited by:
Gianfranco Romanazzi, Marche Polytechnic University, ItalyReviewed by:
Feng Zhou, Henan Institute of Science and Technology, ChinaMaria Cecilia Rasuk, CONICET Planta Piloto de Procesos Industriales Microbiológicos (PROIMI), Argentina
Copyright © 2025 Karoney, Gokul, Siyoum, Molelekoa and Korsten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lise Korsten, bGlzZS5rb3JzdGVuQHVwLmFjLnph
 Edwin M. Karoney
Edwin M. Karoney Jarishma K. Gokul
Jarishma K. Gokul Nazareth Siyoum
Nazareth Siyoum Tintswalo Molelekoa
Tintswalo Molelekoa Lise Korsten
Lise Korsten