- 1Department of Environmental Health Science, University of Georgia, Athens, GA, United States
- 2Odum School of Ecology, University of Georgia, Athens, GA, United States
- 3US Geological Survey, St. Petersburg, FL, United States
- 4Marine Meteorology Division, Naval Research Laboratory, Monterey, CA, United States
Vibrio is a cosmopolitan genus of marine bacteria, highly investigated in coastal and estuarine environments. Vibrio have also been isolated from pelagic waters, yet very little is known about the ecology of these oligotrophic species. In this study we examined the relative change in bacterial abundance and more specifically the dynamics of Vibrio in the tropical North Atlantic in response to the arrival of pulses of Saharan dust aerosols, a major source of biologically important nutrients for downwind marine surface waters. Aerosol and surface water samples were collected over 1 month coinciding with at least two distinct dust events. Total bacterial counts increased by 1.6-fold correlating with the arrival of Saharan dust (r = 0.76; p = 0.001). Virus-like particles (VLP) also followed this trend and were correlated with bacterial counts (r = 0.67; p = 0.01). Vibrio specific qPCR targeting the 16S rRNA gene ranged from below detection limits to a high of 9,145 gene copies ml−1 with the arrival of dust. This increase equated to 6.5 × 102−1.5 × 103 individual genome equivalents ml−1 based on the known range of 16S rRNA copies among this genus. Vibrio exhibited bloom-bust cycles potentially attributed to selective viral lysis or bloom depletion of organic carbon. This work is one of the few studies to examine the open ocean ecology of Vibrio, a conditionally rare taxon, whose bloom-bust lifestyle likely is a contributing factor in the flow of nutrients and energy in pelagic ecosystems.
Introduction
The pelagic ocean is one of the largest habitats on earth. Surface water microbes in the ocean drive the majority of biogeochemical cycling and are a critical linkage in the ocean-atmosphere exchange of gases that affect global climate (Falkowski et al., 2008). An understanding of microbial community composition and function—who is doing what—is a key aspect in marine microbial ecology research (Lauro et al., 2009; Brown et al., 2014; Shilova et al., 2014). Through metagenomic analysis of surface ocean prokaryotes, two different adaptive strategies have been identified (Polz et al., 2006; Yooseph et al., 2010). Globally, the most abundant group, adapted to nutrient poor conditions of the open ocean, is characterized by small size and streamlined genomes, resulting in slow but sustained growth with little metabolic plasticity and low capability to respond to nutrient influx (Yooseph et al., 2010). An opposing adaptive strategy is characterized by the rare opportunistic group, with greater genomic and metabolic diversity, able to survive in low numbers, but capable of rapidly exploiting spatially and temporally variable nutrient resources, thus occasionally becoming highly abundant in the community (Yooseph et al., 2010). This latter group of conditionally rare taxa can have a disproportionate effect on ecosystem functioning during periodic bloom conditions by providing important ecosystem services that are disproportionate to their initial low abundance (Shade et al., 2014). Their susceptibility to predation and viral lysis also make them important players in food web dynamics and biogeochemical cycling (Yooseph et al., 2010), effectively influencing the trophic transfer of key elements and trace metals. Because of their rarity in ocean surveys, little is known about conditionally rare taxa and what environmental drivers induce an increase in their abundance, especially in remote pelagic systems.
The genus Vibrio includes a group of heterotrophic marine bacteria with a worldwide distribution, but characterized as conditionally rare in the open ocean because of their infrequent detection and low numerical abundance (Yooseph et al., 2010). Although perhaps best known as pathogens, of the ~150 currently described species, only 12 are typically associated with human disease; however, all play potentially important roles in nutrient and chemical cycling in the marine environment (Grimes et al., 2009). Vibrio have a broad metabolic and genomic potential, are able to take advantage of ephemeral pulses of nutrients through chemotaxis and motility, and show rapid growth in response to nutrient addition (Worden et al., 2006; Takemura et al., 2014; Westrich et al., 2016). Vibrio are often found in close association with plankton and marine particulates where they degrade a broad range of substrates with extracellular enzymatic proteins such as chitinases, proteases, and lipases (Thompson and Polz, 2006), but can also survive in a free-living state (Worden et al., 2006; Hunt et al., 2008). Vibrio tend to be biogeographically associated with nutrient-rich, copiotrophic environments of near-shore systems (Takemura et al., 2014). Vibrio have been well-studied in coastal habitats in part because of their potential to cause disease in humans and marine organisms. Although they can be isolated from oligotrophic open ocean environments, little is known about the ecology of these pelagic species and their population dynamics (Thompson and Polz, 2006).
In a recent study, Vibrio were shown to be highly responsive to nutrient and trace metal inputs through atmospheric deposition of Saharan dust delivered to downwind coastal surface waters in the Caribbean (Westrich et al., 2016). Saharan dust is entrained in the atmosphere from source regions in northern Africa and is blown across the Atlantic Ocean and deposited, providing essential nutrients and trace metals, like iron, to marine ecosystems (Mahowald et al., 2009; Boyd and Ellwood, 2010). Several studies suggest that fast responding heterotrophs in oligotrophic waters can out compete autotrophic organisms for Saharan dust associated nutrients and the degree of response increases with the degree of oligotrophy (Pulido-Villena et al., 2008; Marañón et al., 2010); however, there has been little work on how specific rare taxa respond in highly oligotrophic systems, such as open ocean gyres. We hypothesized, that Vibrio, as a representative conditionally rare taxon, would increase in abundance with the arrival of pulses of Saharan dust in the open ocean of the tropical North Atlantic.
Materials and Methods
Sample Collection
All samples were collected during the Integrated Ocean Drilling Program Expedition 336 (16 September−16 November 2011) aboard the R/V JOIDES Resolution. Surface water samples used in this study were collected when on station at the North Pond area on the western flank of the Mid-Atlantic Ridge at 22°45′ N and 46°05′ W (Figure 1) during the period of 5 October−5 November 2011. Surface water grabs were collected daily between 1,045 and 1,200 h (local time) from a port/bow location (under the bridge) at a maximum depth of ~0.25 m. For each sample, two 1-ml subsamples were centrifuged at ~13,000 × g for 20 min; after centrifugation, the supernatant fluid was removed and discarded using a micropipette. Boström et al. (2004) reported ~99% recovery using this approach to concentrate bacteria from seawater. Resulting pellets were stored at −80°C for Vibrio qPCR analyses, which we used to estimate population changes at the genus level. Two 10 ml subsamples were fixed in formalin and used for bacteria and virus like particle direct counts (Noble and Fuhrman, 1998).
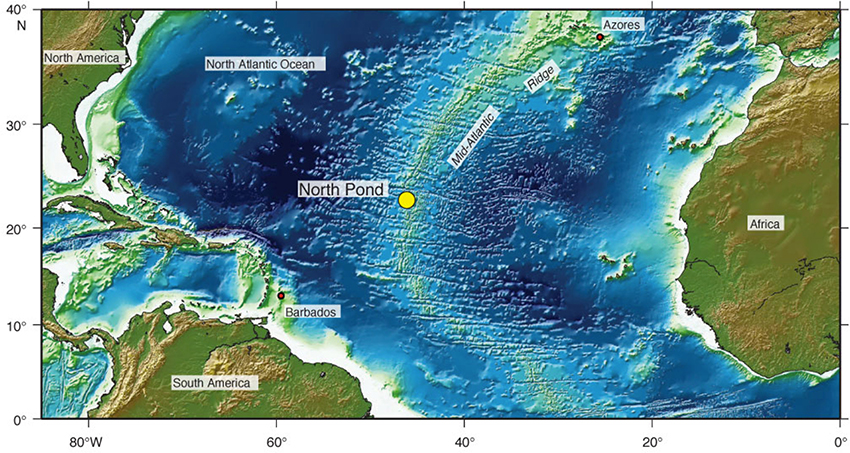
Figure 1. Sample collection location at North Pond on the western flank of the Mid-Atlantic Ridge at 22°45′ N and 46°05′ W (used with permission, Expedition 336 Scientists, 2012).
DNA Extraction
DNA was extracted from the pelleted seawater samples using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol for Gram-positive bacteria using AE buffer as the elution medium. Two duplicate elutions of 25 μl each were combined for a final eluent volume of 50 μl. Extracts were stored at −80°C until use.
Construction of Quantitative PCR (qPCR) Standard
PCR standards were prepared from genomic DNA extracted from V. alginolyticus (American Type Culture Collection strain 33839). Briefly, a culture was grown overnight in LBS (Miller Broth [Fisher, BP1426] supplemented with 2% NaCl and 0.05 M Tris Buffer) then pelleted by centrifugation for 10 min at 8,000 × g. Supernantant fluid was discarded and the genomic DNA (gDNA) in the final pellet was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol for Gram-negative bacteria. A PCR product was generated from the gDNA using Vibrio group specific primers targeting a variable region of the 16S rRNA gene, 567F, 5′GGCGTAAAGCGCATGCAGGT-3′ and 680R, 5′-GAAATTCTACCCCCCTCTACAG-3, (the PCR cycling conditions were described by Thompson et al. and outlined below; Thompson et al., 2004). The PCR amplicon was cleaned (QIAquick PCR purification kit; Qiagen) and checked by agarose gel electrophoresis to verify it was a single amplicon of the correct size (113 bp). The amplicon was inserted into a PCR-4 vector and cloned into E. coli using a TA-TOPO kit (Life Technologies Grand Isle, NY). The plasmid was extracted (QIAquick Spin Miniprep kit; Qiagen) and the cloned region sequenced to verify the correct insert. The plasmid was linearized with NotI (Roche, Indianapolis, IN) after cleanup (QIAquick PCR purification kit; Qiagen) and was quantified using a Qubit Fluorometer (Thermo Fisher Scientific, Grand Island, NY). The linearized standard was serially diluted in AE and run with each qPCR assay.
Quantitative PCR
Total Vibrio concentrations were quantified using a SYBR green qPCR method utilizing the Vibrio specific primers 567F and 680R (described above) (Thompson et al., 2004); this methods has previously been found to track very closely to culturable Vibrio counts in tropical marine water (Westrich et al., 2016). Briefly, DNA template (5 μl) was added to 10 μl SYBR SELECT PCR Master Mix (Applied Biosystems, Foster City, CA), 4.76 μl of PCR-grade water (Thermo Fisher Scientific), and primers were added to a final concentration of 0.16 μM in a total reaction volume of 20 μl. All reactions were run on a StepOne real-time PCR system (Life Technologies, Grand Isle, NY) with the following cycling conditions: 2 min at 50°C for UDG activation and 95°C for 2 min to activate AmpliTaq polymerase and UP, followed by 40 cycles of 95°C for 3 s for denaturation and 60°C for 30 s for annealing and extension. Each run was followed by a dissociation step (95°C for 15 s and 60°C for 1 min and 95°C for 15 s) to determine a melt curve for analysis of specificity. Each sample was analyzed in triplicate (technical replication) and at least two replicate positive controls and two replicate negative (no template) controls were included with every run. PCR inhibition tests were conducted by the addition of 5 × 103 gene copies of linearized standard to each sample. If inhibition was detected, the sample was diluted 1:10 and amplification was repeated. Samples that continued to show inhibition were excluded from the analysis. Cycle threshold (CT) values for all qPCR assays were compared to a standard curve representing 101–106 gene copies per reaction volume. The standard curve linear regression (R2 = 0.994) resulted in an equation of y = −3.3x + 40.4 with an efficiency of 100.4%. Linearized standards could be detected to a level of 2 copies μl−1, corresponding to a CT value of 37. Samples were considered not detectable if the mean CT of technical replicates for each sample was above this value. Vibrio concentrations in seawater were expressed as gene copies ml−1. To give biological context and because copy numbers of 16S rRNA are highly variable across the genus Vibrio, the range of Vibrio genome equivalents (GE) was also determined, by dividing the sample copy number by the currently reported Vibrio 16S rRNA gene copy numbers found in the Ribosomal RNA Database (rrnDB). The rrnDB is an online, curated resource for current ribosomal operon (rrn) copy number information for bacteria (Stoddard et al., 2015). The database currently contains 122 genomes from the genus Vibrio, with a range of 6–14 16S rRNA gene copy numbers per species and a mean of 10 copies per genome. GE ml−1 are reported for the low and high ends of this range, in other words, providing a range of possible cell numbers (genomes) based on 14 copies per genome (to give lower end) and 6 copies per genome (to give higher end).
Bacterial and Viral Direct Counts
Total bacteria and virus-like particles (VLP) were enumerated following the protocol first described by Noble and Fuhrman (1997) and modified by Noble and Fuhrman (1998) and Griffin et al. (2001). In short, 10 ml aliquots of 1% formalin fixed surface water (collected daily at ~1,200 h local time) were filtered through Whatman Anodisc 0.02 μm pore-sized, 25 mm diameter membranes, in duplicate. Material was stained by placing the membrane sample-side up on top of a drop of diluted SYBR Gold nucleic-acid stain (Molecular Probes, Eugene, OR) (97.5 μl of 0.02 μm filtered water +2.5 μl of a 1:10 dilution of SYBR Gold) and incubated at room temperature in the dark for 10–15 min. Twenty-seven microliters of Prolong Antifade (Molecular Probes, Eugene, OR) was placed on a coverslip, and the coverslip placed over the filter on a glass slide. Fifteen fields per slide were counted at 1,000 × magnification under oil immersion using a Carl Zeiss, Inc. (Jena, Germany) Axioskop 40 epifluorescent microscope. For negative controls, 10 mL of autoclaved 0.02 μm filter-sterilized deionized water was filtered and stained as described above.
Regional Aerosol Modeling Analysis
The Navy Aerosol Analysis and Prediction System (NAAPS) surface-level dust and aerosol optical depth (AOD3), a vertical integral of total dust (unitless), was downloaded for the IODP sampling location (22°N and 46°W) over the full sampling time period (http://www.nrlmry.navy.mil/aerosol/; Lynch et al., 2016). The daily analyses (based on 4 6-h intervals) were averaged for each day providing one AOD value per day. This predictive model was used to estimate dust arrival into the larger geographic area at a 1° × 1° spatial resolution for comparative analysis purposes.
Local Aerosol Analyses
To monitor dust arrival and load specifically at the sampling station, a ParticleScan Pro (IQAir, La Mirada, CA) was used shipboard to determine aerosol particle counts daily at 10 min intervals. Measurements were taken on the roof of the bridge of the ship, ~10 meters above sea level. A daily average was calculated.
High-volume membrane filtration samples were collected at 24 h intervals for daily composite aerosol analyses (Haley et al., 2014). The unit flow rate was 0.57 m3 min−1. Collection began at ~0700 h each day using a Staplex TF1A filtration unit (Staplex, Brooklyn, NY) and Whatman TFAGF41 glass fiber filters. Each filter was weighed daily and stored in a labeled zip-top bag at −80° C. Filters were visually inspected for color and other features. In particular, daily filters with an orange tint, indicative of a heavy Saharan dust event (Figure 2) or those with a black to gray tint, indicative of diesel or other smoke contaminant from the ship exhaust, were noted. Local dust arrival was confirmed for specific dates using a conservative approach, which required the following observations to coincide: (1) increased daily aerosol particle counts, (2) orange-tinted filters, and (3) regional NAAPS modeling analysis indicating dust in the region. Any dates with black tinted filters were excluded from analysis because the soot obscured the ability to observe any orange coloration attributed to dust.

Figure 2. High-volume sample filters (Staplex Type TFAGF41 glass fiber filters) collected over ~24 h intervals. Analyses focused on the interval between 12 and 25 October, which included sufficient consecutive days of sampling with both dust and non-dust days and limited appearance of exhaust soot on the filters (12 and 13 October were included as pre-dust days, confirmed by NAAPS models). Filters tinted orange indicated observable Saharan dust loading (14, 15, 17, 21, 22, and 23 October). Clear days (low wind, no observable soot or dust on filters, and no dust based on NAAPS models) were noted on 19, 20, 25 October.
Statistical Analysis
Aerosol particulates, bacterial and viral direct counts, and Vibrio gene copy counts were compared for the time series with and without lag factors, using the Pearson correlation analysis, calculating a two-tailed P-value for each analysis. Samples with non-detectable Vibrio levels were assigned a value of one-half of the detection limit for statistical analyses (detection limit was 100 gene copies ml−1). Aerosol particle counts were compared between confirmed dust days (identified both by the presence of orange or red tint on high volume filters and corroborated with NAAPS modeled data) and clear days (no dust and no soot; identified by clear high volume filters and corroborated with NAAPS modeled data) using an unpaired t-test. For all tests, significance was declared when P ≤ 0.05. All tests were conducted in GraphPad Prism 5.0.
Results
Aerosol Particle Counts and Optical Density
Aerosol particle counts were used as a proxy for the presence of Saharan dust particulate matter in the immediate vicinity of sampling when NAAPS modeled data indicated Saharan aerosols in the region. Daily counts during the entire period of sampling averaged 1.89 × 107 particles l−1, with a range of 1.82 × 106 particles l−1 on 5 October to 4.88 × 107 particles l−1 on 22 October. The potential increase of particle counts could also be elevated on slack wind days when ship exhaust was not blown away from the ship and therefore could confound determination of dust arrival. A visual inspection of concurrently collected HV filters was made to determine those days when particle counts were compromised with possible exhaust contamination. Days with notable black particulates and little to no wind (indicating a high likelihood of originating from diesel exhaust from the ship) were noted to occur from 5 to 12 October. Conversely, a notable orange coloration, indicative of local dust intrusion, was observed on HV filters from 14, 15, 17, 21, 22, and 23 October (Figure 2). The high dust load during these dates was also confirmed by NAAPS surface level modeling with a peak dust level in the region indicated on 14–15 October and 23 October (~80 μg m−3) (Figure 3). Aerosol particle counts were significantly higher on dust days (3.487 × 107 particles l−1 ± 3.761 × 107 particles l−1 [SEM; n = 6]) than on clear days (1.163 × 107 particles l−1 ± 3.568 × 105 [SEM; n = 4]) (P = 0.0003).
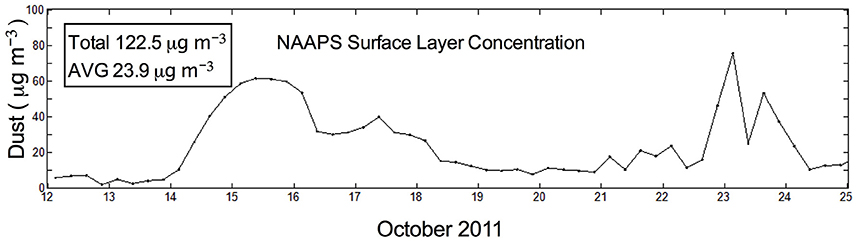
Figure 3. Naval Aerosol Analysis and Prediction System (NAAPS) model of surface level dust at the Mid-Atlantic Ridge (22°45′ N and 46°05′ W) 12–25 October 2011 (Lynch et al., 2016).
Vibrio Specific qPCR
Because of the likely influx of diesel exhaust into the HV filter between 5 and 11 October, the relationship between Vibrio and aerosol particulate counts was examined excluding this period. Two dates, 12 and 13 October, which also showed evidence of exhaust in the filters, but preceded dust arrival into the area (confirmed by NAAPS and low particle counts) were retained as low dust days in this time series (Figures 2, 3). After elimination of samples that exhibited PCR inhibition, or were chronologically isolated (making determination of Vibrio population dynamics difficult to interpret), a total of 14 samples were used for comparative analysis. This provided a 2-week long daily data set of samples collected in the mid-Atlantic 12–25 October 2011, including as noted above, two distinct increases in dust level in the region indicated on 14–15 and 23 October.
A daily average of 3,782 Vibrio 16S rRNA gene copies ml−1 were measured during this time period. Vibrio showed a high correlation with aerosol particle counts with a 24 h lag, to account for a one-day delay in Vibrio growth (r = 0.56; p = 0.04) (Figure 4). There was no significant correlation with particle counts when incorporating 48 and 72 h lag times. Additionally, Vibrio gene copy ml−1 increased with NAAPS dust aerosol optical depth (AOD) (r = 0.58; p = 0.03) (Figure 4) as did genome equivalents. We also examined possible associations between Vibrio and aerosol counts on days with no dust but soot present on filters (e.g., 5–11 October) and found no significant relationship. The highest Vibrio concentration of 9,145 gene copies ml−1 (15 October) was measured 24 h after a spike in aerosol particle counts (3.68 × 107 particles l−1; 14 October) (Figure 4).
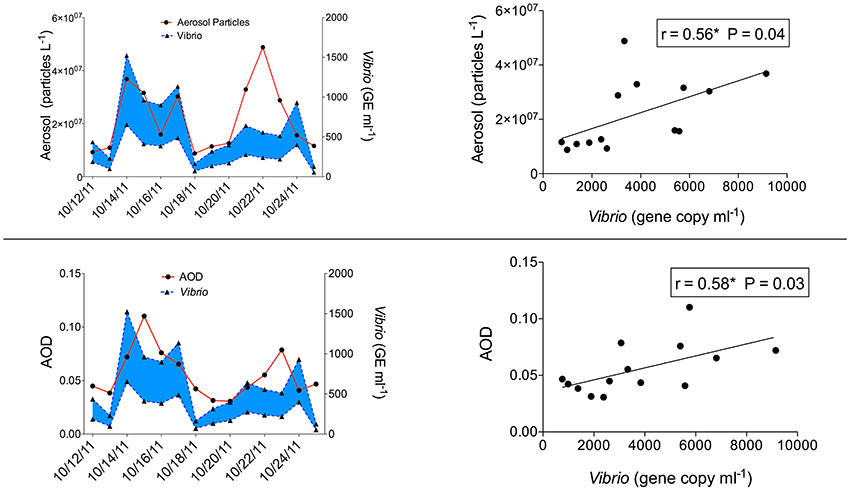
Figure 4. Time series plots (left side of figure) of Vibrio genome equivalents (GE) ml−1. (Genome equivalents are represented as a range (shown in blue shading) based on the range of 16S rRNA copy numbers documented among Vibrio species (https://rrndb.umms.med.umich.edu; Stoddard et al., 2015; see Table S1). Vibrio GE are shown shifted by 24 h to incorporate a growth response lag-time) compared to aerosol particle counts (top) and modeled Aerosol Optical Density (AOD) (bottom) for 12–25 October 2011 (including dust events). Corresponding scatter plots (and Pearson correlation) of Vibrio 16S rRNA gene copy ml−1 with aerosol particle counts and AOD are shown on the right (n = 14).
Direct Counts of Total Bacteria and Virus-Like Particles
Bacteria and VLP direct counts were also analyzed between 12 and 25 October (Figure 5). Total bacterial counts averaged 3.59 × 105 ml−1 and ranged from 2.85 × 105 ml−1 on 14 October to a high on 22 October of 4.60 × 105 ml−1, associated with an influx of dust (NAAPS dust model; Figure 5). VLP were almost an order of magnitude higher than bacterial counts, averaging 2.66 × 106 ml−1, and ranged from 1.66 × 106 ml−1 on 12 October to a high of 3.37 × 106 ml−1 on 23 October, 24 h after the peak bacterial count. Bacterial concentrations increased with aerosol particle counts, with a 24 h lag response (r = 0.76; p = 0.001). VLP concentrations were also significantly correlated with aerosol particle counts, with a 24 h lag (r = 0.37; p = 0.02). VLP also increased with bacterial concentration (r = 0.67; p = 0.01) (Figure 5). Vibrio gene copy numbers were not significantly correlated with either the total bacterial nor VLP direct counts, with or without a time lag.
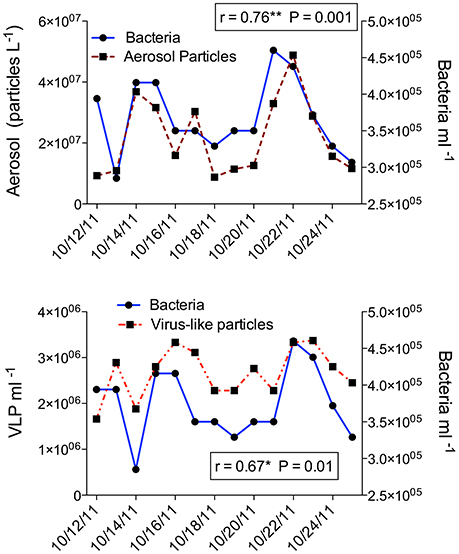
Figure 5. Direct counts of total bacteria and virus-like particles (VLP) ml−1 surface water (12–25 October 2011; including dust events). Top: Aerosol particle count l−1 air and total bacteria ml−1 surface water (shown shifted by 24 h to incorporate a growth response lag-time) with Pearson correlation (n = 14). Bottom: Virus-like particles and total bacteria ml−1 (no time shift) with Pearson correlation (n = 14).
Discussion
The genus Vibrio, most widely known for the ability of some species to cause disease, is globally distributed and important in nutrient cycling in the marine environment. Studies of Vibrio biogeography and environmental population dynamics have focused mostly on nutrient-replete coastal systems. In this study, results demonstrated that although conditionally rare in the tropical open-ocean surface water of the mid-Atlantic (Yooseph et al., 2010), an increase in Vibrio 16S rRNA gene copies was closely associated with the influx of Saharan dust, a known source of biologically important nutrients (Jickells et al., 2005; Mahowald et al., 2009) similar to previous studies conducted in coastal systems (Westrich et al., 2016). Mid-Atlantic surface water Vibrio gene copies ml−1 increased ~6-fold within 1 day of the arrival of Saharan dust (confirmed by agreement of all measures of dust presence in the vicinity). This increase in copy number equates to an increase between 653 and 1,524 individual genome equivalents ml−1 based on the known range of 16S rRNA copy numbers found in this genus (Table S1). A 24 h lag in growth response is consistent with previous findings in coastal waters, where Vibrio response to Saharan dust iron was seen 14–24 h post-dust arrival (Westrich et al., 2016). Additionally, the magnitude of response was similar to that noted previously, and brought the standing population of Vibrio in this oligotrophic open gyre system to concentrations more typical of coastal waters (Westrich et al., 2016). This is likely an endogenous response to dust nutrients or substrate given that Vibrio, which are marine in origin, have not been detected in dust aerosols (Griffin, 2007). Furthermore, the response was specific to dust aerosol influence, as soot exhaust did not support additional growth.
The dominant adaptive strategy of bacteria in pelagic systems is typically characterized by a streamlined existence of slow growing, small cells containing minimal genetic plasticity to respond to newly available nutrient sources (Yooseph et al., 2010). Here, we have shown that in contrast to this minimalistic strategy, Vibrio with larger genomes and biomass, have the metabolic capacity to survive in low numbers in the open ocean, and can mount a rapid growth response to an ephemeral nutrient pulse. Vibrio have evolved adaptations to starvation pressures and have been shown to persist for long periods under conditions of nutrient limitation (Eilers et al., 2000). Vibrio have been shown to reduce cell volume in response to starvation (Denner et al., 2002; Thompson and Polz, 2006), yet maintain a high number of rRNA copies to rapidly ramp up protein synthesis when conditions become more favorable (Eilers et al., 2000; Polz et al., 2006). This metabolic flexibility could be ecologically advantageous in the open ocean environment with varying nutrient regimes. Though not directly measured in this study, we suggest that Saharan dust associated nutrients, especially iron, drive the observed Vibrio growth response, as has been observed previously in more coastal systems (Westrich et al., 2016).
Saharan dust is the dominant source of iron in the North Atlantic, providing up to 87% of the dissolved iron (consider to be its most bioavailable form) to marine surface water communities (Mahowald et al., 2009; Conway and John, 2014). Vibrio spp., like other heterotrophs, have a high intracellular demand for iron in enzymatic and metabolic processes, especially in their respiratory electron transport chain (Fourquez et al., 2014). Some Vibrio spp. are capable of fixing nitrogen, which requires extremely high amounts of iron as a co-factor (Criminger et al., 2007; Chimetto et al., 2008). Iron availability has a direct effect on Vibrio growth and ability to metabolize carbon and other biologically important macronutrients like N and P (Kirchman et al., 2003; Sunda, 2012; Fourquez et al., 2014). Furthermore, Vibrio can utilize dust leachate filtered through a 0.2 μm pore size membrane (Westrich et al., 2016), suggesting that these often particle-associated microbes are not necessarily reliant on dust particulates for growth, but can utilize dust nutrients in a free-living state. Furthermore, Moreira et al. (2015) found that Vibrio operational taxonomic units (OTUs) increased significantly in the western tropical Atlantic (near the Mid-Atlantic Ridge) with nutrient enrichment experienced during turbulent mixing and iron acquisition genes dominated the functional response to these changing conditions, further supporting the importance of iron in heterotrophic fitness in Mid-Atlantic pelagic communities.
Recent investigations using experimental microcosms, have demonstrated equivocal biological response among the autotrophic and heterotrophic community to the addition of dust associated nutrients and iron (Boyd et al., 2010; Langlois et al., 2012; Pulido-Villena et al., 2014); however, few studies have been able to capture in situ response to natural dust events, which may elicit imporant differences in the microbial community response. In this study, we were able evaluate the in situ response (Figures 4, 5) to the arrival of multiple distinct dust events in the open mid-Atlantic (Figures 2, 3), providing a level of replication for such events that has not previously been reported. These results suggest that conditional rare heterotrophs, using Vibrio as a model, are responsive to the direct deposition of dust aerosols.
Heterotrophic bacteria may play a much larger role in the connections between dust and the ocean carbon cycle than previously recognized (Pulido-Villena et al., 2008). The bloom-bust cycles of Vibrio seen in this study could be attributed to both bottom-up and top-down controls. The temporary release from nutrient-limitation with dust arrival could directly drive the Vibrio increase observed with a resulting crash after exhaustion of organic carbon sources. Alternatively, the increase in virus like particles, closely associated with the increase in total bacterial counts, suggest a source of carbon and nutrients from viral-lysed cells, indirectly supporting increased Vibrio abundance (Pulido-Villena et al., 2014). Top-down controls are also known to tightly regulate Vibrio populations and could account for the 24–48 h bloom to bust dynamics we observed in this study and similarly found in other systems (Worden et al., 2006; Suttle, 2007; Westrich et al., 2016). The attenuated level of Vibrio response measured after the second dust event 21–23 October would suggest that the system may be primed with these top-down control agents following the Vibrio bloom-bust response of the first dust event (14–16 October) (Figures 3, 4). Vibrio and other fast responding Gamma-proteobacteria have been estimated to have 100 times more biomass per cell than the equivalent, globally abundant SAR 11 Alpha-proteobacteria (Yooseph et al., 2010; Cermak et al., 2016). This larger biomass is believed to be a liability, especially during bloom conditions, when they are preferentially preyed upon by eukaryotic protists and subject to viral lysis (Polz et al., 2006; Yooseph et al., 2010). The interplay between addition of iron from dust deposition, bacterial iron uptake and viral infection is interesting in light of the recent proposed Ferrojan Hypothesis (Bonnain et al., 2016). This hypothesis suggests that viruses can adsorb free iron and then utilize bacterial iron-uptake receptors for infection in the manner of a Trojan Horse.
The turnover of Vibrio and larger-biomass members of the community by top down controls, even though numerically minor, could have significant impacts on marine carbon and nutrient cycling in the pelagic ocean (Suttle, 2007). For example, primary production could benefit from lytic turnover of lysed cellular debris that is rich in P and N (Suttle, 2007). Viral lysate of V. alginolyticus has been shown to be a highly available source of iron supporting photosynthesis (Poorvin et al., 2011).
In conclusion, this study demonstrates that Vibrio populations, though typically rare in pelagic waters significantly increase in abundance with the arrival of Saharan dust. We propose that dust associated nutrients like iron, essential for Vibrio survival and fitness, drive this response in these oligotrophic systems, extending a previous finding of iron in Saharan dust influencing coastal Vibrio population blooms (Westrich et al., 2016). Ecological roles of rare microorganism are largely unknown, especially in the open ocean, but it is thought that they contribute to stability and function of an ecosystem (Shade et al., 2014). Though many uncertainties remain, identifying this response is crucial first step for more targeted studies that could have larger implications in the carbon and nutrient cycling in the oligotrophic open ocean.
Author Contributions
JW analyzed all samples and data, contributed to the experimental design, and wrote the manuscript. DG collected all samples, analyzed bacterial and viral direct counts, contributed to experimental design, and contributed to the manuscript. DW provided modeled aerosol data for the time series and reviewed the manuscript EL worked with JW on analyses, contributed to experimental design, and helped to write the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was funded through grants from the National Science Foundation (OCE-1357423 [to EL], the National Oceanic and Atmospheric Administration (Oceans and Human Health Initiative # S0867882 [to EL]), and the U.S. Geological Survey (Toxic Substances Hydrology Program [to DG]). Additional personnel, technical, and scientific support were provided to DG through the IODP (Integrated Ocean Drilling Program) Expedition 336. The development of NAAPS and the NAAPS reanalysis was supported by the Office of Naval Research Code 322 and the NASA Interdisciplinary Science Program. Blair Sterba-Boatwright (Texas A&M University—Corpus Christi) provided assistance with statistical analyses. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States government. The views and opinions expressed herein do not necessarily state or reflect those of the United States government and shall not be used for advertising or product endorsement purposes.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00012/full#supplementary-material
References
Bonnain, C., Breitbart, M., and Buck, K. (2016). The ferrojan horse hypothesis: iron-virus interactions in the ocean. Front. Mar. Sci. 3:82 doi: 10.3389/fmars.2016.00082
Boström, K. H., Simu, K., Hagström, Å., and Riemann, L. (2004). Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol. Oceanogr. Methods 2, 365–373. doi: 10.4319/lom.2004.2.365
Boyd, P. W., Mackie, D. S., and Hunter, K. A. (2010). Aerosol iron deposition to the surface ocean -Modes of iron supply and biological responses. Mar. Chem. 120, 128–143. doi: 10.1016/j.marchem.2009.01.008
Boyd, P. W., and Ellwood, M. J. (2010). The biogeochemical cycle of iron in the ocean. Nat. Geosci. 3, 675–682. doi: 10.1038/ngeo964
Brown, M. V., Ostrowski, M., Grzymski, J. J., and Lauro, F. M. (2014). A trait based perspective on the biogeography of common and abundant marine bacterioplankton clades. Mar. Genomics 15, 17–28. doi: 10.1016/j.margen.2014.03.002
Cermak, N., Becker, J. W., Knudsen, S. M., Chisholm, S. W., Manalis, S. R., and Polz, M. F. (2016). Direct single-cell biomass estimates for marine bacteria via Archimedes' principle. ISME J 11, 825–828. doi: 10.1038/ismej.2016.161
Chimetto, L. A., Brocchi, M., Thompson, C. C., Martins, R. C., Ramos, H. R., and Thompson, F. L. (2008). Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst. Appl. Microbiol. 31, 312–319. doi: 10.1016/j.syapm.2008.06.001
Conway, T. M., and John, S. G. (2014). Quantification of dissolved iron sources to the North Atlantic Ocean. Nature 511, 212–215. doi: 10.1038/nature13482
Criminger, J. D., Hazen, T. H., Sobecky, P. A., and Lovell, C. R. (2007). Nitrogen fixation by Vibrio parahaemolyticus and its implications for a new ecological niche. J. Appl. Environ. Microbiol. 73, 5959–5961. doi: 10.1128/AEM.00981-07
Denner, E. B., Vybiral, D., Fischer, U. R., Velimirov, B., and Busse, H. J. (2002). Vibrio calviensis sp. nov., a halophilic, facultatively oligotrophic 0.2 microm-fiIterabIe marine bacterium. Int. J. Syst. Evol. Microbiol. 52, 549–553. doi: 10.1099/00207713-52-2-549
Eilers, H., Pernthaler, J., and Amann, R. (2000). Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. J. Appl. Environ. Microbiol. 66, 4634–4640. doi: 10.1128/AEM.66.11.4634-4640.2000
Expedition 336 Scientists (2012). Mid-Atlantic Ridge microbiology: initiation of long-term coupled microbiological, geochemical, and hydrological experimentation within the seafloor at North Pond, western flank of the Mid-Atlantic Ridge. IODP Prel. Rept. 336. doi: 10.2204/iodp.pr.336.2012
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Fourquez, M., Devez, A., Schaumann, A., Gueneugues, A., Jouenne, T., Obernosterer, I., et al. (2014). Effects of iron limitation on growth and carbon metabolism in oceanic and coastal heterotrophic bacteria. Limnol. Oceanogr. 59, 349–360. doi: 10.4319/lo.2014.59.2.0349
Griffin, D. W. (2007). Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 20, 459–477. doi: 10.1128/CMR.00039-06
Griffin, D. W., Garrison, V. H., Herman, J. R., and Shinn, E. A. (2001). African desert dust in the Caribbean atmosphere: microbiology and public health. Aerobiologia (Bologna). 17, 203–213. doi: 10.1023/A:1011868218901
Grimes, D. J., Johnson, C. N., Dillon, K. S., Flowers, A. R., Noriea, N. F. III., and Berutti, T. (2009). What genomic sequence information has revealed about Vibrio ecology in the ocean - a review. Microb. Ecol. 58, 447–460. doi: 10.1007/s00248-009-9578-9
Haley, B. J., Kokashvili, T., Tskshvediani, A., Janelidze, N., Mitaishvili, N., Grim, C. J., et al. (2014). Molecular diversity and predictability of Vibrio parahaemolyticus along the Georgian coastal zone of the Black Sea. Front. Microbiol. 5:45. doi: 10.3389/fmicb.2014.00045
Hunt, D. E., David, L. A., Gevers, D., Preheim, S. P., Alm, E. J., and Polz, M. F. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320, 1081–1085. doi: 10.1126/science.1157890
Jickells, T. D., An, Z. S., Andersen, K. K., Baker, A. R., Bergametti, G., Brooks, N., et al. (2005). Global iron connections between desert dust, ocean biogeochemistry, and climate. Science 308, 67–71. doi: 10.1126/science.1105959
Kirchman, D. L., Hoffman, K. A., and Weaver RHutchins, D. A. (2003). Regulation of growth and energetics of a marine bacterium by nitrogen source and iron availability. Mar. Ecol. Prog. Ser. 250, 291–296. doi: 10.3354/meps250291
Langlois, R., Mills, M. M., Ridame, C., Croot, P., and LaRoche, J. (2012). Diazotrophic bacteria respond to Saharan dust additions. Mar. Ecol. Prog. Ser. 470, 1–14. doi: 10.3354/meps10109
Lauro, F. M., McDougald, D., Thomas, T., Williams, T. J., Egan, S., Rice, S., et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533. doi: 10.1073/pnas.0903507106
Lynch, P., Reid, J. S., Westphal, D. L., Zhang, J., Hogan, T. F., Hyer, E. J., et al. (2016). An 11-year global gridded aerosol optical thickness reanalysis (v1.0) for atmospheric and climate sciences. Geosci. Model. Dev. 9, 1489–1522. doi: 10.5194/gmd-9-1489-2016
Mahowald, N. M., Engelstaedter, S., Luo, C., Sealy, A., Artaxo, P., Benitez-Nelson, C., et al. (2009). Atmospheric iron deposition: global distribution, variability, and human perturbations. Ann. Rev. Mar. Sci. 1, 245–278. doi: 10.1146/annurev.marine.010908.163727
Marañón, E., Fernandez, A., Mourino-Carballido, B., Martinez-Garcia, S., Teira, E., Cermeno, P., et al. (2010). Degree of oligotrophy controls the response of microbial plankton to Saharan dust. Limnol. Oceanogr. 55, 2339–2352. doi: 10.4319/lo.2010.55.6.2339
Moreira, A. P. B., Meirelles, P. M., Santos, E. D. O., Amado-Filho, G. M., Francini-Filho, R. B., Thompson, C. C., et al. (2015). Turbulence-driven shifts in holobionts and planktonic microbial assemblages in St. Peter and St. Paul Archipelago, Mid-Atlantic Ridge, Brazil. Front. Microbiol. 6:1038. doi: 10.3389/fmicb.2015.01038
Noble, R. T., and Fuhrman, J. (1997). Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 63, 77–83.
Noble, R. T., and Fuhrman, J. A. (1998). Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14, 113–118. doi: 10.3354/ame014113
Polz, M. F., Hunt, D. E., Preheim, S. P., and Weinreich, D. M. (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 361, 2009–2021. doi: 10.1098/rstb.2006.1928
Poorvin, L., Sander, S. G., Velasquez, I., Ibisanmi, E., Lecleir, G. R., and Wilhelm, S. W. (2011). A comparison of Fe bioavailability and binding of a catecholate siderophore with virus-mediated lysates from the marine bacterium Vibrio alginolyticus PWH3a. J. Exp. Mar. Biol. Ecol. 399, 43–47. doi: 10.1016/j.jembe.2011.01.016
Pulido-Villena, E., Baudoux, A.-C., Obernosterer, I., Landa, M., Caparros, J., Catala, P., et al. (2014). Microbial food web dynamics in response to a Saharan dust event: results from a mesocosm study in the oligotrophic Mediterranean Sea. Biogeosciences 11, 5607–5619. doi: 10.5194/bg-11-5607-2014
Pulido-Villena, E., Wagener, T., and Guieu, C. (2008). Bacterial response to dust pulses in the western Mediterranean: implications for carbon cycling in the oligotrophic ocean. Glob. Biogeochem. Cycles 22:BG1020. doi: 10.1029/2007GB003091
Shade, A., Jones, S. E., Caporaso, J. G., Handelsman, J., Knight, R., Fierer, N., et al. (2014). Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. MBio 5:e01371–14. doi: 10.1128/mBio.01371-14
Shilova, I. N., Thompson, A. W., Hewson, I., and Zehr, J. P. (2014). “Ocean Gyres, Metagenomics of,” in Encyclopedia of Metagenomics, ed K. E. Nelson (New York, NY: Springer), 468–484.
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K., and Schmidt, T. M. (2015). rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598. doi: 10.1093/nar/gku1201
Sunda, W. G. (2012). Feedback interactions between trace metal nutrients and phytoplankton in the ocean. Front. Microbiol. 3:204. doi: 10.3389/fmicb.2012.00204
Suttle, C. A. (2007). Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812. doi: 10.1038/nrmicro1750
Takemura, A. F., Chien, D. M., and Polz, M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5:38. doi: 10.3389/fmicb.2014.00038
Thompson, J. R., and Polz, M. F. (2006). “Dynamics of Vibrio populations and their role in environmental nutrient cycling,” in The Biology of Vibrios, eds F. L. Thompson, B. Austin, and J. Swings (Washington, DC: ASM Press), 190–203.
Thompson, J. R., Randa, M. A., Marcelino, L. A., Tomita-Mitchell, A., Lim, E., and Polz, M. F. (2004). Diversity and dynamics of a North Atlantic coastal Vibrio community. J. Appl. Environ. Microbiol 70, 4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004
Westrich, J. R., Ebling, A. M., Landing, W. M., Joyner, J. L., Kemp, K. M., Griffin, D. W., et al. (2016). Saharan dust nutrients promote Vibrio bloom formation in marine surface waters. Proc. Natl. Acad. Sci. U.S.A. 113, 5964–5969. doi: 10.1073/pnas.1518080113
Worden, A. Z., Seidel, M., Smriga, S., Wick, A., Malfatti, F., Bartlett, D., et al. (2006). Trophic regulation of Vibrio cholerae in coastal marine waters. Environ. Microbiol. 8, 21–29. doi: 10.1111/j.1462-2920.2005.00863.x
Keywords: conditionally rare taxa, oligotrophic conditions, aerosols, desert dust, Vibrio, marine ecology
Citation: Westrich JR, Griffin DW, Westphal DL and Lipp EK (2018) Vibrio Population Dynamics in Mid-Atlantic Surface Waters during Saharan Dust Events. Front. Mar. Sci. 5:12. doi: 10.3389/fmars.2018.00012
Received: 19 May 2017; Accepted: 16 January 2018;
Published: 02 February 2018.
Edited by:
Télesphore Sime-Ngando, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Daniela Ceccarelli, Wageningen Bioveterinary Research (WBVR), NetherlandsIsabelle C. Biegala, Institut de Recherche pour le Développement (IRD), France
Copyright © 2018 Westrich, Griffin, Westphal and Lipp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin K. Lipp, ZWxpcHBAdWdhLmVkdQ==
 Jason R. Westrich1,2
Jason R. Westrich1,2 Dale W. Griffin
Dale W. Griffin Douglas L. Westphal
Douglas L. Westphal Erin K. Lipp
Erin K. Lipp