- Department of Life Sciences and Chemistry, Jacobs University Bremen, Bremen, Germany
Diverse microalgae-bacteria interactions play important roles for nutrient exchange processes and marine aggregate formation leading to the cycling, mineralization, or sedimentation of organic carbon in the Oceans. The main goal of this study is to report an alternative way to assess a distinct diatom-bacteria interaction. To study this at the cellular scale, an in vitro interaction model system consisting of the diatom, Thalassiosira weissflogii, and the gamma-proteobacterium, Marinobacter adhaerens HP15, had previously been established. HP15 is able to attach to T. weissflogii cells, to induce transparent exopolymeric particle formation, and to increase marine aggregation formation. Several bacterial genes important during this interaction have been studied thus far, but genes specifically expressed in co-culture remained unknown. To identify such bacterial genes, an in vivo Expression Technology (IVET) screen was employed. For this, a promoter-trapping vector containing a fusion between a promoter-less selection marker gene and a promoterless reporter gene was constructed. Construction of a library of plasmids carrying genomic fragments upstream of the fusion and its subsequent transformation into a selection marker mutant allowed detection of 30 bacterial promoters specifically expressed during the interactions with T. weissflogii. Their sequence analyses revealed that the corresponding genes could be involved in many processes such as biochemical detection of diatom cells, bacterial attachment, metabolic exchange of nitrogen compounds, and resistance toward heavy metals. Identification of genes potentially involved in branched-chain amino acid uptake and utilization confirmed our previous results of a proteomics analysis. Since our current approach identified several additional genes to be induced in co-culture, use of IVET might be a valuable complementing strategy to proteomic or transcriptomic analysis of the diatom-bacteria crossplay. The current IVET approach demonstrated that the interaction between M. adhaerens HP15 and T. weissflogii is multifactorial and is likely to involve a complex network of physiological processes.
Introduction
In the Ocean, aggregate formation from organic matter, fecal pellets, detritus, and living cells is an important ecological mechanism that mediates settling of organic carbon to the depth (Alldredge and Silver, 1988). Aggregation in form of marine snow is fundamental for marine biogeochemical cycles since it greatly affects the efficiency of the biological pump (Fowler and Knauer, 1986; Jahnke, 1996). Initially mediated by either simple collision events or diverse chemical reactions, diatom-bacteria interactions initialize, or reinforce these processes by formation of extra-cellular polysaccharides affecting the stickiness and hence size of aggregates (Alldredge et al., 1993; Logan et al., 1995; Passow, 2002). In the last decade, considerable progress has been made in understanding algae-bacteria interactions at both, the ecological as well as the cellular and molecular levels (Wang et al., 2014; Amin et al., 2015; Durham et al., 2015, 2017; Needham et al., 2017; van Tol et al., 2017). In this way, different types of interactions were discovered, which were either microbial population driven or dictated by individual bacterial organisms. Recently, Behringer et al. (2018) demonstrated that bacterial communities on diatom surfaces displayed strong conservation across diatom strains. Seymour et al. (2017) elegantly summarized the main corresponding mechanisms and processes, which show an impressive array of diversity and complexity.
Irrespective of their origin, marine particles and aggregates represent surfaces for bacterial colonization (Dang and Lovell, 2016). These colonization processes have been studied intensively (Grossart et al., 2006b; Thiele et al., 2015; Busch et al., 2017). Interactions of bacteria with microalgae or detritus will result in a series of biochemical or physical events, which change the characteristics of the colonized particle, let it grow or shrink in size, and alter its density, porosity, stickiness, and sinking velocity (Alldredge and Youngbluth, 1985; Alldredge et al., 1986; Biddanda and Pomeroy, 1988; Kiørboe and Jackson, 2001; Kiørboe et al., 2002). The actual outcome of each individual aggregation event will depend on both, the biochemical properties of the particle(s) and the composition of the colonizing microbial populations (DeLong et al., 1993; Kiørboe et al., 2003; Bach et al., 2016), which in turn depend on each other making marine aggregate formation a highly dynamic event.
To contribute to a better understanding of the molecular basis of diatom-bacteria interactions and in particular to gain knowledge about the bacteria-mediated initial processes of diatom aggregation, we established an in vitro interaction model system consisting of the axenic diatom, Thalassiosira weissflogii, and the marine gamma-proteobacterium, Marinobacter adhaerens HP15 (Kaeppel et al., 2012), which belongs to a ubiquitously occurring marine bacterial genus. The two organisms are separately maintained and can be combined in co-culture. Strain HP15 was shown to preferentially attach to T. weissflogii cells as opposed to other bacteria, to induce transparent exopolymeric particle (TEPs) production in the diatom, and to increase aggregation during co-culture with T. weissflogii (Gärdes et al., 2011). This bacterial strain had initially been isolated from marine particles in the German Wadden Sea (Grossart et al., 2004), and its particular ability to interact with diatom cells had been screened from a pool of almost 90 different marine bacterial isolates (Gärdes et al., 2011). HP15's genome was sequenced and fully annotated (Gärdes et al., 2010). Furthermore, HP15 is genetically accessible and several genes potentially important for diatom-bacteria interactions have been investigated by mutagenesis (Sonnenschein et al., 2011). For instance, a variety of chemotaxis-deficient mutants were generated to study the chemotactic behavior of HP15 toward T. weissflogii in detail (Sonnenschein et al., 2012). The corresponding mutants showed a reduced attachment to live diatoms suggesting that chemotaxis is a substantial initial step for the interaction. The co-culture model system was further used to assess the impact of temperature and acidification on aggregate formation demonstrating that aggregation rates and sinking velocities decrease in warmer and more acidified seawater (Seebah et al., 2014). In order to get first hints on gene products potentially important for the interaction, a proteomics analysis of HP15 in response to the diatom was conducted (Stahl and Ullrich, 2016) suggesting a distinct nutrient supply for M. adhaerens during co-cultivation. The bacterium seemed to prefer amino acids as main carbon and nitrogen source but not polysaccharides or sugars as one would suspect from a TEP-consuming bacterial organism. Aside of this, other bacterial genes that are specifically and directly induced during the interaction remained unknown. The identification and characterization of such novel genes will in turn shed light on the nature of diatom-bacteria interaction and its underlying cellular and biogeochemical processes.
The in vivo expression technology (IVET) screen is a promoter-trapping technique that allows the selection of genes specifically expressed in vivo (Mahan et al., 1993). This technique has been applied to study gene expression in a wide range of microorganisms, providing new insights into the role of bacteria during various types of symbioses (Rediers et al., 2005; Dudley, 2008; Roberfroid et al., 2016). Although a comprehensive experimental comparison of IVET with other screening techniques such as transcriptomics or proteomics has not been done yet, IVET is unique in focusing on one of the two interacting partners thereby excluding signals from the other interaction partner. The approach is based on the principle that an auxotrophic bacterial mutant will only multiply and thus survive inside or in close proximity of a host organism if a host-inducible bacterial promoter drives a complementing essential growth factor gene provided on a separate plasmid (in trans). Thus, the method is qualitative and not quantitative as are screenings done with transcriptomics or proteomics. Herein, for the first time, an IVET screening was applied to identify bacterial genes relevant during diatom-bacteria interactions using the bacterial strain HP15 and T. weissflogii as an interaction model system.
In the present study and in order to complement and extend on our previous proteomics work with the diatom-bacteria co-culture system, an IVET screen was applied for co-cultured M. adhaerens and T. weissflogii. To this end, an IVET plasmid containing a promoterless essential growth factor gene and an in vitro reporter gene was constructed and loaded with a genomic library of short DNA fragments, some of which contained promoters. The IVET library was inserted into an essential growth factor mutant, which was unable to survive in co-culture with the diatom, and transformants were screened for survival during co-culture. Surviving mutant transformants, which were not expressing the in vitro reporter gene after re-isolation from the co-culture, indicated presence of co-culture specific promoter sequences. The corresponding genes were analyzed with respect to homologies with data base entries in order to learn about their potential functions and to propose their putative roles during the diatom-bacteria interaction.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
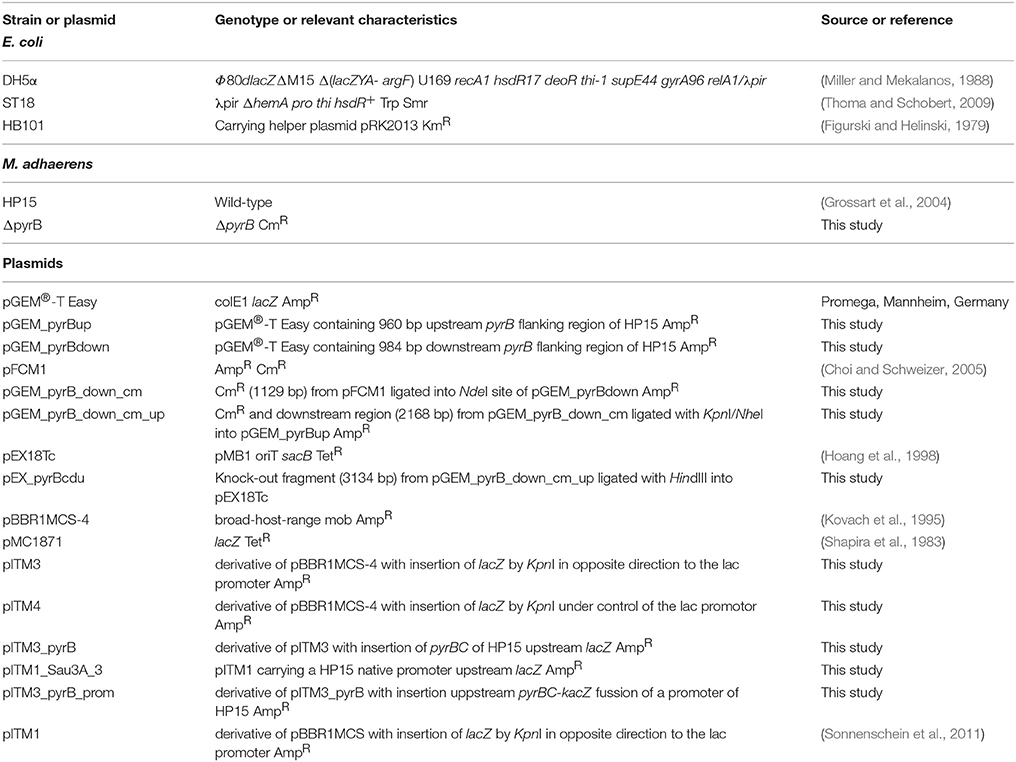
Bacterial strains and plasmids used are listed in Table 1. HP15 was isolated from particles collected from surface waters of the German Bight (Grossart et al., 2004). The bacterium was grown in marine broth (MB) medium (ZoBell, 1941) using sterile North-Sea water at 28°C. MB medium was supplemented with uracil (5 μg ml−1) as a pyrimidine source for growth of HP15 pyrimidine-deficient auxotrophs. During the in vitro screening, MB medium was supplemented with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) (50 μg ml−1) allowing for blue/white colony selection. f/2 GLUT medium was made with f/2 medium (Guillard and Ryther, 1962) supplemented with 5 g l−1 glutamate and was used as minimal medium to confirm auxotrophy of the HP15 mutant ΔpyrB. f/2 medium was prepared with pre-filtered (0.2 μm pore size) and autoclaved North-Sea water. All herein reported CFU/ml values for bacteria were derived by determining the optical density at 600 nm (OD600) and deducing CFU/ml from a previously established calibration curve of OD600 vs. CFU/ml derived from serial dilution platings.
Escherichia coli strains were maintained in Luria-Bertani (LB) agar medium supplemented with appropriate antibiotics. E. coli DH5α was used for maintaining the HP15 genomic library generated in the IVET vector. E. coli ST18 (Thoma and Schobert, 2009) was used as a donor strain during bi-parental conjugation and grown in LB medium containing 50 μg/ml 5-aminolevulinic acid (ALA). E. coli HB101 (pRK2013) was used as a helper strain during tri-parental conjugation (Figurski and Helinski, 1979). The following antibiotics were added to media when needed (in μg ml−1): chloramphenicol, 25; kanamycin, 50; and ampicillin, 50.
Diatom Culture Conditions
Axenic cultures of T. weissflogii (CCMP 1336) were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (Maine, USA). Diatom cultures were grown at 16°C in f/2 medium, with a 12-h photoperiod at 115 μmol photons m−2 s−1 and were checked regularly for bacterial contaminations by plating on LB agar medium plates and by frequent microscopic observations. Diatom cell numbers were determined by cell counts in a Sedgewick Rafter Counting Chamber S50 (SPI Supplies, West Chester, PA, USA).
DNA Techniques
Plasmid preparation, DNA extraction, PCR, and other standard DNA techniques were performed as previously described (Sambrook et al., 1989). Restriction enzymes and DNA-modifying enzymes were used as recommended by the manufacturer (Fermentas, St. Leon-Rot, Germany). Nucleotide sequencing was carried out at Eurofins MWG (Ebersberg, Germany). DNA sequences analysis and oligonucleotide primer design were done using Vector NTI®; Software 10.3.0 (Invitrogen Corporation, Carlsbad, CA, USA). Nucleotide and amino acid sequences were compared using the Basic Local Alignment Search Tool BLAST provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al., 1990) and InterProScan Sequence Search provided by the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/pfa/iprscan/). The oligonucleotides used in this study are listed in Table 2.
IVET Vector Construction
The first step to implement the IVET strategy is the construction of an IVET vector (Supplementary Figure 1); the remaining steps in the strategy are summarized in Figure 1. The broad-host-range plasmid pBBR1MCS-4 (Kovach et al., 1995) was used as a backbone to construct the IVET vector (Supplementary Figure 1). The full-sized E. coli promoter-less lacZ gene was amplified from plasmid pMC1871 with the primers LacZF/LacZR (Smirnova and Ullrich, 2004). The resulting 3.1-kb PCR fragment was treated with KpnI and ligated into KpnI-treated pBBR1MCS-4 in opposite direction of the vector-borne lac promoter resulting in plasmid pITM3. A second plasmid, pITM4, was generated with the 3.1-kb PCR fragment containing lacZ cloned in the same orientation as that of the lac promoter of pBBR1MCS-4. Plasmids pITM3 and pITM4 were transformed into E. coli ST18 and subsequently transferred to HP15 by conjugation. For this, cells of donor and recipient (~109 cell ml−1 each) were mixed in a ratio of 1:2, spotted on LB agar plates, and incubated for 24 h at 28°C. The cell mass was then scraped off and re-suspended in MB medium for subsequent dilution plating. Transconjugants were selected on MB agar supplemented with ampicillin and X-Gal after incubation at 28°C for 2–3 days. Presence of IVET plasmids in transconjugants was confirmed by plasmid DNA isolation using the Midi Prep Kit according to the manufacturer's recommendations (Macherey-Nagel, Düren, Germany) and agarose gel electrophoresis.
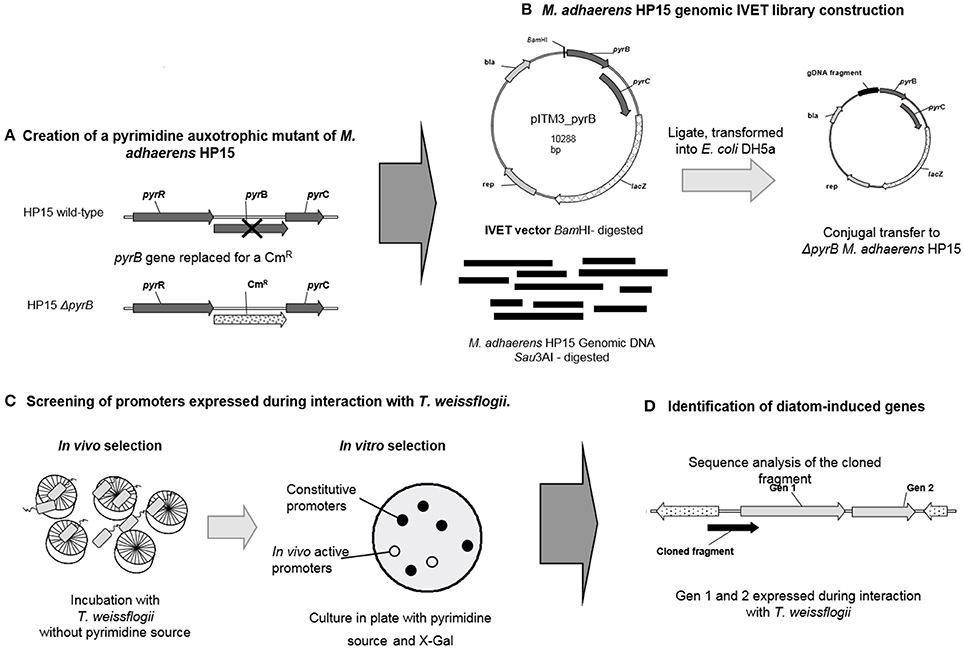
Figure 1. Schematic representation of IVET strategy. (A) An auxotroph mutant in M. adhaerens HP15 was created by replacing the gene pyrB with a chloramphenicol resistant cassette (CmR). This mutant is unable to grow in the absence of a pyrimidine source; (B) A genomic library was generated in the pITM3-pyrB plasmid by digesting the genomic DNA of HP15 and cloning the resulting fragments into the BamHI site of the plasmid. The resulting plasmids were transformed into HP15 by triparental conjugation; (C) For the selection of promoters active during the interaction the trasnconjugants of HP15 ΔpyrB carrying the genomic library in pITM3-pyrB were incubated with T. weissflogii, without the addition of a pyrimidine source. Then the survival trasnconjugants were plated in agar media supplemented with pyrimidine and X-Gal, the brownish (non-blue) colonies, which contain active in co-culture promoter, were selected; (D) The plasmid DNA was extracted from the positive colonies, the fragment upstream the fusion pyrB-lacZ was PCR amplified and sequenced. The genes under the control of the promoter found in the DNA fragment were identified.
Primers pyrBF and pyrBR were designed to amplify the promoter-less pyrBC locus from HP15. This 2,285-bp PCR product was restricted with HindIII and XhoI and cloned into HindIII/XhoI-treated pITM3 to generate pITM3_pyrB. Plasmid pITM3_pyrB was transformed into HP15 wild-type and ΔpyrB mutant by bi-parental conjugation as described above. The transconjugants were selected in MB media containing X-Gal and ampicillin.
Generation of a Pyrimidine Auxotrophic Mutant of M. adhaerens HP15
A gene-specific mutagenesis based on homologous recombination was conducted to generate a ΔpyrB mutant in HP15 according to Hoang et al. (1998) (Figure 1A). A mutagenic plasmid was constructed in which a chloramphenicol resistance (CmR) cassette was flanked by DNA fragments obtained from upstream and downstream regions of the pyrB gene as follows: 960 bp upstream and 984 bp downstream of the pyrB gene were PCR-amplified using the primer pairs pyrB_upF/pyrB_upR and pyrB_down2F/pyrB_down2R, respectively. Both flanking fragments were sub-cloned into pGEM®-T Easy vector (Promega, Mannheim, Germany) resulting in plasmids, pGEM_pyrBup and pGEM_pyrBdown, respectively. A DNA fragment of 1,129 bp carrying a CmR cassette was amplified with the primers cm2R/cm2F from pFCM1. The fragment was treated with NdeI and sub-cloned into NdeI-treated pGEM_pyrBdown resulting in plasmid pGEM_pyrB_down_cm. From this plasmid, the 2,168-bp fragment containing the downstream region and the CmR cassette was restricted with KpnI/NheI and ligated into KpnI/NheI-treated pGEM_pyrB_up, resulting in plasmid pGEM_pyrB_down_cm_up. From this plasmid, a 3,134-bp fragment containing both flanking regions and the CmR cassette was excised with HindIII and ligated into the HindIII-treated vector pEX18Tc. The generated conjugable mutagenic construct pEX_pyrBcdu was then transformed into HP15 by bi-parental conjugation. Resulting mutants were selected on chloramphenicol-containing MB agar plates. A successful double cross-over event for the ΔpyrB mutant was confirmed by PCR using the primers mut_pyrBF/mut_pyrBR resulting in the expected 1,205 and 1,410-bp amplification fragments for the wild-type and the mutated pyrB gene, respectively.
To confirm the auxothrophy of mutant ΔpyrB, it was grown in f/2 GLUT medium with and without uracil. Subsequently, to determine whether the pyrB gene is a suitable selection marker for promoters expressed during the interaction with T. weissflogii, mutant ΔpyrB and the wild-type were separately co-cultivated with the diatom in the absence of uracil. Bacteria were grown overnight in MB liquid medium at 18°C and the cells were harvested by centrifugation at 4,000 rpm for 15 min. To avoid carrying over of nutrients from MB medium the cells were washed twice with f/2 medium. Cell numbers were adjusted to ~105 CFU ml−1 and mixed with ~104 cells ml−1 of T. weissflogii in exponential growth stage. The cultures were incubated for 6 days at 16°C, with a 2-h photoperiod at 115 μmol photons m−2 s−1, and shaking at 50 rpm. Dilutions series were done daily in appropriate media, and CFU numbers were determined.
To complement the pyrB mutant, a functional native promoter from the HP15 genome was cloned upstream the pyrBC-lacZ fusion in pITM3_pyrB. Such a native promoter was selected by partial restriction of the genomic DNA of HP15 with Sau3AI and cloning of a pool of restriction fragments into BamHI-treated plasmid pITM1 (Sonnenschein et al., 2011). The resulting genomic library was transformed into HP15 wild-type by bi-parental conjugation. Five blue colonies were selected and the region upstream the pyrBC-lacZ fusion from one of those clones (pITM1_Sau3A_3) was sequenced thereby identifying the native promoter. This DNA region was excised from pITM1_Sau3A_3 with XbaI and HindIII and cloned into XbaI/HindIII-treated pITM3_pyrB, resulting in the plasmid pITM3_pyrB_prom, which was transformed into ΔpyrB HP15 by bi-parental conjugation. Transconjugants were selected after incubation on f/2 GLUT agar supplemented with ampicillin and X-Gal without uracil at 28°C for 2–3 days.
M. adhaerens HP15 Genomic IVET Library Construction
A genomic IVET library of HP15 was constructed in pITM3_pyrB as follows (Figure 1B): Total genomic DNA of HP15 was partially digested with Sau3AI; DNA fragments with sizes ranging from 0.5 to 1.5 kb were selected, and ligated into the BamHI site of pITM3_pyrB. The IVET library was then transformed into E. coli DH5α. To verify the randomness of inserts, 24 clones of E. coli DH5α were randomly picked and the region upstream the pyrBC-lacZ fusion was amplified with the primers T7 and PyrB_out by colony PCR. The IVET library was then transferred into the mutant ΔpyrB by tri-parental conjugation. Briefly, E. coli DH5α carrying the IVET library was used as a donor and E. coli HB101 (pRK2013) served as a helper strain. Cells of donor, helper, and recipient (~1010 cells ml−1) were mixed in a ratio of 1:1:2, spotted on LB ALA agar plates, and incubated for 24 h at 28°C. Cell mass was scraped off and re-suspended in MB medium for subsequent dilution plating. The resulting transformants were selected on MB agar plates containing chloramphenicol, ampicillin, uracil, and X-Gal. To verify the randomness of plasmids transformed into mutant ΔpyrB, 24 clones were randomly picked and subjected to colony PCR as described above. Although, cloning by partial digestion using an enzyme with frequent recognition sites does not comprehensively cover the entire genome of M. adhaerens, the randomness of insert sizes allows to conclude that a genome-wide array of promoter sequences had been obtained.
Screening of Promoters Expressed During Interaction With T. weissflogii
For the in co-culture selection of promoters, co-cultivation of mutant ΔpyrB transconjugants carrying the IVET library with diatom cells was conducted (Figure 1C). Briefly, 10,000 cells ml−1 of T. weissflogii in exponential growth stage were mixed with the bacterial transconjugants (1 × 105 CFU ml−1) in cell culture flasks with a final volume of 35 ml of f/2 media and incubated at room temperature and a 2-h photoperiod for 24 h. Next, 5 ml of the co-cultures were transferred to 30 ml fresh f/2 medium containing T. weissflogii in exponential growth stage (10,000 cells ml−1) and incubated under the same conditions as above for additional 24 h. This procedure was repeated five times in order to enrich for positive clones. After 6 days of incubation, transconjugants were recovered and subjected to in vitro selection. For this, cultures initiated on day 4 and 5 were individually centrifuged at 4,000 rpm for 20 min and dilution series were performed with recovered cells in MB agar plates containing chloramphenicol, ampicillin, uracil and X-Gal at 28°C. After 3–4 days of incubation brownish colonies (non-blue) were selected and re-streaked.
A control experiment was performed to observe the growth pattern of the mutant ΔpyrB carrying plasmid pITM3_pyrB without any insertion upstream the pyrBC-lacZ fusion (empty IVET vector). 10,000 cells ml−1 of T. weissflogii in exponential growth stage were mixed with ΔpyrB transconjugants carrying pITM3_pyrB (1 × 105 CFU ml−1) in cell culture flasks in f/2 medium and incubated at room temperature and a 2-h photoperiod for 5 days, which was used in all previous studies on this model interaction system (Gärdes et al., 2011) because the diatom aggregate formation was optimal under this light regime. Dilution series were carried out daily from the culture and the number of CFU was calculated.
Identification of Diatom-Induced Genes
Brownish colonies of ΔpyrB transconjugants were selected after co-inoculation with T. weissflogii, and subjected to colony PCR with the primers T7/pyrB_out to check for the presence of an insert upstream of the pyrBC-lacZ fusion in pITM3_pyrB. Transconjugants giving individually different PCR products of sizes larger than 570 bp were selected and their PCR product sequences determined. The obtained sequences were analyzed by BLAST and potential promoters regions as well as downstream located genes identified. The positive transconjugants were individually co-inoculated with T. weissflogii to confirm their survival in presence of diatoms. For this, bacteria were grown overnight in MB liquid cultures at 18°C, the cells were harvested by centrifugation at 4,000 rpm for 15 min, washed twice with f/2 medium, their concentration adjusted to 1 × 105 CFU ml−1, and mixed with 10,000 cells ml−1 of T. weissflogii in exponential growth stage. The co-cultures were incubated for 8 days at 18°C, with a 2 h photoperiod. Dilutions series were done in appropriate media and CFU numbers were determined.
Results
Pyrimidine Auxotrophic Mutant of M. adhaerens HP15
For the identification of bacterial promoters, the gene pyrB was used as selection marker. pyrB is essential for survival encoding for aspartate transcarbamoylase (ATCase; EC 2.1.3.2), an enzyme that is necessary for biosynthesis of pyrimidines (Schurr et al., 1995). The availability of pyrimidines in many natural environments is limited, which renders pyrB a successful selection marker (Lee and Cooksey, 2000). To verify its potential as a selection marker in our system, a pyrB mutant in HP15 was created. This mutant should be unable to grow in the absence of a pyrimidine source. A mutagenic construct was transformed into HP15 wild-type and the pyrB gene was replaced by a CmR cassette via homologous recombination. A clone that had undergone a double crossover of the CmR cassette as demonstrated by PCR was selected. Subsequently, the auxotrophy of the pyrB deletion mutant was confirmed by testing its growth on agar minimal medium with or without uracil as the sole pyrimidine source. Mutant ΔpyrB was not able to grow without uracil thus proving its expected phenotype (data not shown). In addition, growth of mutant ΔpyrB and HP15 wild-type in liquid f/2 GLUT medium were indistinguishable from each other when uracil was supplemented indicating that the mutation did not affect bacterial growth in general (Supplementary Figure 2). However and as expected, mutant ΔpyrB did not grow without uracil.
To determine whether lack of pyrB expression is a suitable selection criterion during the interaction with T. weissflogii and to test whether the diatom provides any pyrimidines to functionally complement the mutant's genotype, mutant ΔpyrB was cultivated with diatoms in f/2 medium with and without any uracil. Mutant ΔpyrB cell numbers dramatically decrease during the incubation time when compared to the growth of the wild-type when no uracil was added to the medium (Figure 2). In accordance with the in vitro growth, mutant ΔpyrB grew similar to the wild type in presence of diatoms and uracil. These result clearly demonstrated that the diatom is not providing any pyrimidines proving pyrB's suitability as a selection marker gene for this study.
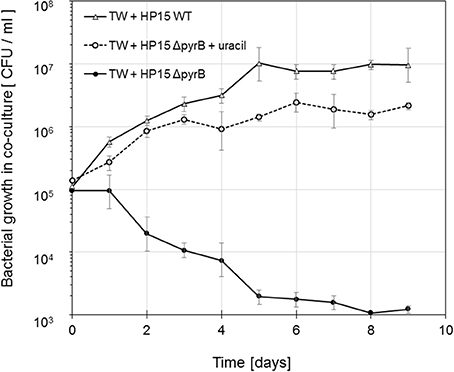
Figure 2. Growth of M. adhaerens HP15 ΔpyrB in co-culture with T. weissflogii in F/2 media with and without uracil (5 μg ml−1). M. adhaerens HP15 wild-type was used as a positive control. Experiment was conducted with three biological replicates, error bars represent standard deviation.
Generation of the IVET Library
The suitability of lacZ as an in vitro reporter gene in HP15 had previously been described (Sonnenschein et al., 2011), thus this gene was used for the construction of the IVET vector. As an in co-culture selection marker the promoter-less pyrBC locus of HP15 was chosen since it had been previously shown that the gene pyrB alone was not able to complement pyrB deletion mutants and that the pyrC gene located downstream of pyrB was required for this purpose (Schurr et al., 1995; Lee and Cooksey, 2000). In order to be suitable tools, the ΔpyrB mutant must not grow unless provided by uracil while the IVET plasmid must not complement the ΔpyrB mutant as long as it does not carry a functional promoter sequence upstream of the in co-culture selection marker gene. The ΔpyrB mutant carrying the IVET plasmid pITM3_pyrB did not grow in f/2 GLUT medium without uracil. In contrast, this transconjugant grew well in form of brownish colonies in f/2 GLUT medium supplemented with uracil and X-Gal (data not shown) confirming that the pyrBC-lacZ fusion present in pITM3_pyrB is not expressed. Furthermore, the introduction of a functional promoter derived from the genome of HP15 into the IVET plasmid and its subsequent transformation into ΔpyrB mutant allowed its full complementation. Corresponding transformants grew on agar medium without uracil and formed LacZ-expressing (blue) colonies when X-Gal was provided (data not shown). In summary, these tested parameters demonstrated that the IVET plasmid and the ΔpyrB mutant were suitable tools for the identification of diatom contact-induced genes of HP15.
A genomic library of HP15 was constructed in pITM3_pyrB and transformed into E. coli DH5α plated on 50 agar plates each containing 200–300 colonies resulting in a total of roughly 10,000 to 15,000 clones. The randomness of inserts was confirmed by PCR with 190 randomyl chosen clones showing that 85% of the analyzed E. coli DH5α transformants had an insert in the IVET plasmid. Importantly, all of the insert sizes were different from each other (data not shown) demonstrating that the IVET library contained a random number of different DNA fragments. After conjugation of the IVET library into mutant ΔpyrB, a total of 4.5 × 106 CFU ml−1 were obtained, with roughly 40% of the transconjugants forming LacZ-positive blue colonies on X-Gal agar medium. To demonstrate the randomness of IVET plasmids in the transconjugant pool, a second round of PCR tests was done. In this case 82% of the analyzed 200 clones had an insert, and all inserts had different sizes (data not shown) suggesting that roughly one fifth of all transferred IVET plasmids did not contain an insert. In conclusion, there results hint at a randomness of DNA fragments inserted in the IVET plasmid but do not allow speculations about the completeness of the screen since the very frequently occuring recognition sites of the used endonuclease Sau3A might have been used unevenly during the applied partial digest.
In Co-culture Selection of Promoters During Interaction With T. weissflogii
In order to test whether or not mutant transconjugants with empty IVET plasmids could survive the in co-culture selection, first a control experiment was conducted, in which a ΔpyrB transconjugant carrying an empty IVET plasmid was grown together with the diatom. From an initial inoculum of 1.2 × 105 CFU ml−1 only 395 CFU ml−1 were recovered after 2 days and 22 CFU ml−1 after 3 days. However, after day 4 no colonies were observed. These results showed that transconjugant ΔpyrB (pITM3_pyrB) is not able to survive for more than 4 days in the presence of the diatom. Consequently, the following diatom-bacteria co-cultures were performed in such a way that positive clones were enriched while negative clones or clones with empty IVET plasmids were eliminated. To frequently refresh potential diatom-borne selecting factors or signals, co-cultures of ΔpyrB mutants carrying the IVET library with diatom cells were diluted every 24 h by transferring 5 ml of the co-culture into f/2 medium containing fresh diatom cells. Co-cultures were repeated several times to increase the number of positive clones. Determination of CFUs was carried out daily (data not shown), and randomly chosen brownish colonies (non-blue) were selected while blue colonies, indicating a constitutive expression of lacZ, were discarded. Brownish colonies represented transconjugants with IVET plasmids carrying a promoter sequence inactive under in vitro conditions. However, since these transconjugants had survived several diatom co-cultures, their promoter sequences must have been active in co-culture. All obtained brownish colonies were checked for inserts by PCR. All analyzed colonies from 4- or 5-day-old cultures had an insert demonstrating the suitability of the selection regime.
Potential in Co-culture Induced Promoters
In total, 97 brownish colonies were obtained which carried a DNA insert (confirmed by PCR) upstream of the pyrBC-lacZ fusion in pITM3_pyrB. These transconjugants were subsequently cultivated in f/2 GLUT solid medium with or without uracil to confirm their auxotrophy in culture media. From these 97 transconjugants, 23 cultures did not show the expected lack of growth suggesting a false-positive result. The PCR products for the remaining 74 colonies were sequenced. Nucleotide sequence analysis indicated that almost one third of the sequences occurred in duplicates so that a total of 41 individual and unique insert DNAs could be obtained. The corresponding IVET plasmids were isolated from the transconjugants and used for further characterization.
Interestingly, from the 41 insert DNAs, 30 inserts were in the same 5′-3′ orientation in terms of their downstream reading frames and the pyrBC-lacZ fusion of the IVET plasmid, while 11 were found in the opposite orientation on the antisense strand of an annotated coding region. In addition to the in co-culture induced clones, a randomly chosen blue colony representing a constitutively expressed pyrBC-lacZ phenotype was sequenced and used as a control in the following experiments. The insert of this transconjugant showed sequence identities to the upstream sequence of a gene encoding aldehyde dehydrogenase (Locus tag: HP15_943).
To confirm that the 30 cloned insert sequences were specifically allowing expression of pyrBC-lacZ during the interaction with T. weissflogii, the growth patterns of 13 randomly chosen in vitro uracil-deficient brown transconjugants (colonies 1, 10, 15, 22, 26, 28, 30, 68, 90, 100, 161, 168, and 194), and the blue transconjugant were analyzed in individual co-cultures with the diatom in f/2 medium without uracil (Figure 3). A transconjugant carrying the empty IVET plasmid was used as a negative control. All 13 positive transconjugants were able to survive the diatom co-incubation reaching cell numbers comparable to that of the wild-type or the blue transconjugant, which harbors a constitutively expressed promoter sequence. After 4 days of incubation, there was a more than three orders of magnitude difference in viable cell numbers of the positive clones and the negative control demonstrating that the identified promoter sequences were induced and enabled the ΔpyrB mutant to survive during the interaction with T. weissflogii.
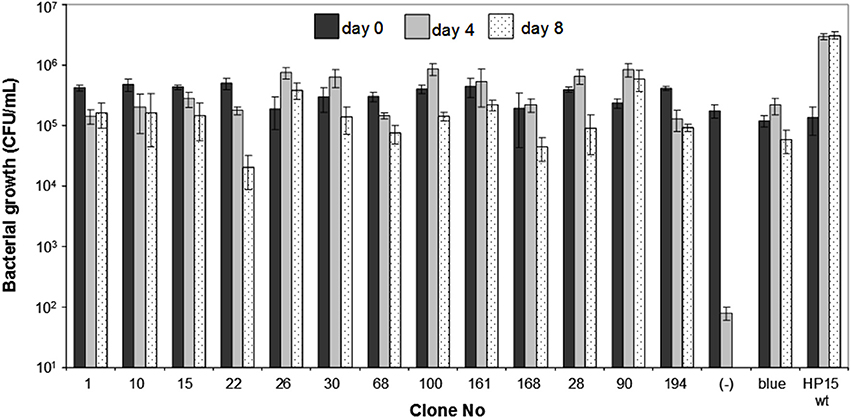
Figure 3. Growth of individual clones of M. adhaerens HP15 ΔpyrB carrying potential in co-culture induced promoters and T. weissflogii. Wild-type and a blue colored mutant were used as a positive control. ΔpyrB HP15 (pITM3_pyrB) was used as a negative control.
In order to rule out any possible genomic alterations in the 41 mutant transconjugants, re-isolated IVET plasmids were individually re-introduced to the ΔpyrB mutant for a second time. Novel transconjugants were again tested for growth in f/2 GLUT solid medium with or without uracil but with X-Gal: These novel transconjugants did not grow when uracil was missing but grew well on uracil-containing medium indicating that the corresponding promoter sequences were not active and that the mutant transconjugants remained auxotrophic in vitro (Data not shown). Colonies on agar plates with uracil were brownish in color just like those of the wild type of HP15. While this result was expected for the 30 IVET plasmids, which contained promoter sequences in the same orientation as the annotated downstream gene, the corrrspondingly positive results for the remaining 11 IVET plasmid with DNA sequences in opposite direction to annotated genes remained obscure. One can only speculate that the automatic annotation of the HP15 genome (accession no. CP001978.1) might still be partially incomplete or incorrect (Gärdes et al., 2010).
DNA Sequence Analysis
M. adhaerens HP15 genes under the control of the promoters identified as specifically induced during interaction with T. weissflogii are listed in Table 3. All accession numbers are from Genbank entries CP001978.1 (M. adhaerens HP15 genome) or CP001978.1 (M. adhaerens HP15 187-kb indigenous plasmid). The possible functions of the correspondingly down-stream located genes were deduced by analyzing its amino acid sequences and determining the presence of conserved domains or similarities with reported proteins in the GenBank. Genes identified in the 11 transcriptional fusions orientated in the opposite direction to the in vitro reporter gene were not considered any further.
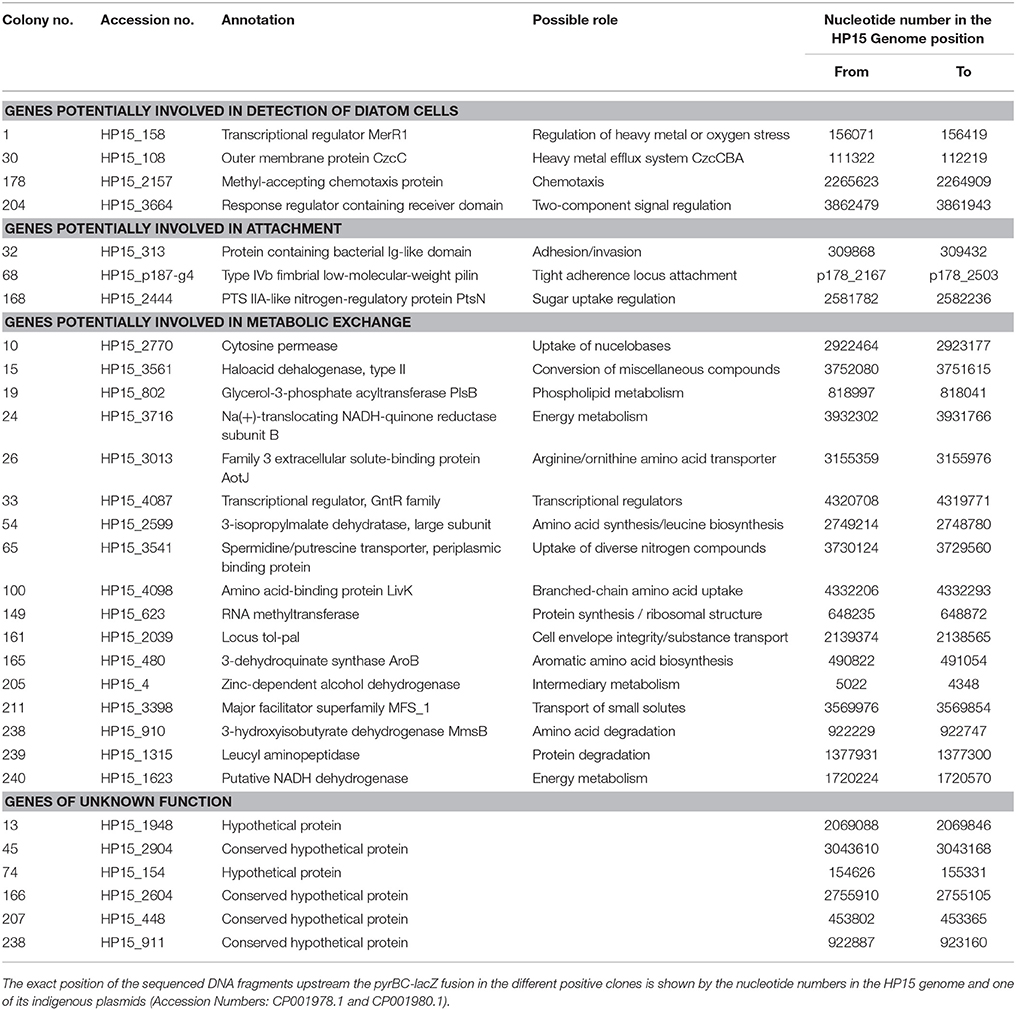
Table 3. List of bacterial genes identified as being under control of the identified promoters during interaction with T. weissflogii.
Detected genes were grouped depending on their potential ecological role during the interaction as follows: Genes that encode enzymes required in (i) detection of diatom cells; (ii) attachment to the diatom cells; (iii) putative metabolic exchange with diatom cells; and (iv) unknown functions (Table 3). Their potential role(s) will be discussed below.
Discussion
Identifying bacterial genes expressed during diatom-bacteria interactions is a fundamental pre-requisite for understanding the functional mechanisms governing bacteria-induced algal aggregation and bacterial organic matter transformation in the oceans. Therefore, an IVET strategy was employed to identify M. adhaerens HP15 genes specifically induced during its interaction with T. weissflogii. IVET is an economically feasible, cultivation-based approach, which allows the use of DNA and standard molecular techniques. DNA is generally more stable than RNA and thus less vulnerable to biasing differential signal degradation in course of multi-step procedures. In addition, IVET avoids some of the potential next-generation sequencing biases, such as problems associated with high-throughput cDNA generation and high-throughput nucleotide sequencing. Although IVET itself has biases such as being rather qualitative than quantitative and that the required absolute length of insert DNA in the IVET library or low level of expression from some promoters might exclude certain genes or groups of genes, the herein applied approach complements transcriptomics work of others (Durham et al., 2015, 2017) and our own proteomics work (Stahl and Ullrich, 2016). Others had shown previously that IVET can be used to identify novel genes important for the respective analyzed interactions (Rediers et al., 2005; Dudley, 2008). Naturally, those will differ from interaction to interaction. With our approach, for the first time we were able to evaluate the potential importance of key genes for diatom-bacteria interactions using a model system and promoter trapping. This strategy allowed the identification of HP15 genes coding for functions such as detection of diatom cells, attachment, and metabolic exchange. Additionally, six gene loci encoding conserved hypothetical proteins without known functions were identified, which were expressed during this interaction. Two of these were located in close proximity to other identified genes hits. In the current study, co-culture-inducible promoter activities were detected. Although it is widely accepted that promoters drive the expression of directly downstream located genes, our study did not detect the expression of those genes. However, finding active promoters for some genes, which code for proteins previously detected by a proteomics approach (Stahl and Ullrich, 2016) as well as finding gene homologs found by transcriptomics studies of others (Durham et al., 2017) are strong hints for a successful screening approach. In the next paragraphs, the possible ecological roles and importance of the identified genes are discussed.
Genes Involved in Detection of Diatom Cells
Chemotaxis is a mechanism that bacteria use to chemically sense their environment and move toward attractants or away from repellents (Baker et al., 2005). In this process, chemoreceptors interact with specific chemicals—often amino acids—and interact with key chemotaxis proteins to control flagellar or pilus-mediated motility (Eisenbach, 1996). Chemoreceptors have a methyl-accepting chemotaxis domain, a homolog of which was found in the putative HP15 protein encoded by the gene downstream of the promoter sequence found in col. 178 (Table 3). Therefore, our current results support the previous finding that chemotaxis of HP15 is important for the diatom-bacteria interaction (Sonnenschein et al., 2012). Another example supporting this finding is col. 204 representing a genetic locus encoding a response regulator containing a receiver domain (HP15_3664) and a histidine kinase (HP15_3665). These proteins belong to the two-component signal transduction systems used by bacteria to respond to different stimuli and to regulate diverse cellular processes (Capra and Laub, 2012). They could be involved in chemotaxis since HP15_3664 shares similarities to chemotaxis-associated CheY-like response regulators (Stock et al., 1990). With new elegant technical approaches now available (Lambert et al., 2017) and our mutant arsenal in the model organism M. adhaerens HP15 (Sonnenschein et al., 2012), it is planned to investigate in depth the chemotactic behavior of HP15 in future studies.
An example of a regulatory gene potentially involved in sensing the diatom was found in col. 1 encoding the MerR1 transcriptional regulator (HP15_158) which has been described to mediate responses to environmental stimuli such as heavy metals, drugs, antibiotics, or oxidative stress (Brown et al., 2003). During the diatom's photosynthesis or photolysis of dissolved organic matter many reactive oxygen species are released (Asada, 2006). The ability to detoxify reactive oxygen species represents a selective advantage in the aquatic environment (Glaeser et al., 2010). Hünken et al. (2008) had shown that certain diatom-interacting bacteria protect the diatom by detoxifying oxygen species produced during the diatom's growth. However, it remains to be tested in future studies whether MerR1 is important for sensing reactive oxygen species or other potential threads such a heavy metals. The later possibility is supported by the IVET clone no. 30 containing the sequence of a genetic locus encoding the outer membrane component CzcC (HP15_108) of the heavy metal efflux pump CzcCBA (Nies, 2003). Interestingly, M. adhaerens HP15 possesses two highly homologous copies of fully functional czcCBA clusters prompting speculations about its pronounced role in detoxification processes (Stahl et al., 2015) and suggesting future studies on these clusters.
Genes Involved in Attachment
To initiate a close interaction, bacteria have to attach to diatom cells. The expression of genes, which change the adhesiveness of bacterial surface and promote adhesion has been previously observed in bacterial communities associated to algal blooms by meta-transcriptomics (Rinta-Kanto et al., 2012). In addition, proteins playing a role in biofilm formation have been identified in bacterial cultures growing with diatoms (Bruckner et al., 2011). Herein, we identified two genes and one operon that might be involved in the attachment of HP15 to T. weissflogii. Firstly, the gene associated with col. 32 was annotated to encode a protein with a putative conserved bacterial Ig-like domain. Proteins carrying this domain were found to be important in host-pathogen interactions (Kelly et al., 1999; Raman et al., 2010). For example, this domain is present in intimin, an outer membrane adhesion protein present in enteropathogenic or enterohaemorrhagic E. coli (Frankel et al., 2001). Secondly, the genetic locus identified in col. 168 encodes the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) IIA-like nitrogen-regulatory protein (Cases et al., 2001). The PTS system consists of a group of phosphotransfer proteins involved in the transport of carbohydrates, in chemotaxis toward carbon sources, and in the regulation of other metabolic pathways (Postma et al., 1993). A similar PTS system, was previously identified by IVET in Erwinia chrysanthemi interacting with tomato plants (Yang et al., 2004). Mutation of this gene affected the systematic invasion capability of the plant pathogen suggesting that certain PTS systems might be pivotal for the attachment of bacteria with photosynthetic eukaryotic cells.
The third genetic locus associated with attachment is the most exciting finding: In col. 68, a plasmid-borne sequence located upstream of the gene encoding Type IV fimbrial low-molecular weight Flp pilin (Kachlany et al., 2000) was identified suggesting that expression of a corresponding pilus might be diatom-inducible. This gene is the first of the so-called tight adherence (tad) locus. The tad locus was described in a wide range of pathogenic and non-pathogenic bacteria as well as in archaea, where it plays an important role in the colonization of respective, surprisingly diverse environmental niches (Kachlany et al., 2001). In Pseudomonas aeruginosa, Flp pili contribute to adherence to abiotic surfaces and surfaces of eukaryotic cells (de Bentzmann et al., 2006). A homologous locus was also identified in Caulobacter crescentus and designated cpa (Caulobacter pilus assembly) (Bodenmiller et al., 2004). Due to its wide-spread distribution and presence on a mobile genomic island the tad locus was termed Widespread Colonization Island (WCI) (Planet et al., 2003). In the human peridontal pathogen, Aggregatibacter actinomycetemcomitans this locus is composed of the genes flp1–flp2–tadV–rcpCAB–tadZABCDEFG encoding proteins that constitute a fibril secretion system (Kachlany et al., 2000). This operon is responsible for tight adherence, autoaggregation, biofilm formation, and the production of bundled fimbria-like fibers of individual pili (Kachlany et al., 2001; Planet et al., 2003; Perez et al., 2006; Tomich et al., 2007). Interestingly, our model organism, HP15 possess the entire WCI allowing the cautious suggestion that this organism might employ similar mechanisms of attachment to diatom surfaces as pathogens use for their host cell surfaces. Future studies should focus, among others, on determining the specificity of the corresponding pili for a given attachment surface thus shedding light on role of the diatom cell wall for this interaction.
Genes Involved in Metabolic Exchange During the Interaction
Diatoms secrete organic compounds in the form of DOC or extracellular polymeric substances (Passow, 2002), which are taken up by heterotrophic bacteria (Grossart et al., 2006a). Gene expression analysis has identified an increase of transcripts involved in DOC uptake when bacteria interact with algae, e.g., transport systems for amino acid, carboxylic acids, carbohydrates (Poretsky et al., 2005, 2010; Rinta-Kanto et al., 2012; Teeling et al., 2012; Durham et al., 2017). In the current study, several promoters driving the expression of genes involved in the uptake of organic—mainly nitrogen-containing—compounds were identified. In col. 10 a genetic locus was identified which showed high sequence similarities to the codBA operon of E. coli previously described to have a putative role in a salvage pathway for nucleobases and being inducible by nucleobase starvation (Danielsen et al., 1992). Since the HP15 ΔpyrB mutant is deficient in pyrimidines biosynthesis and there is no pyrimidine available during its growth with T. weissflogii, the first explanation for the expression of this operon might be to allow the bacteria to survive under such conditions. However, DNA uptake by heterotrophic marine bacteria has been previously reported playing an important role as a supplement for nutrients or for new DNA synthesis (Jørgensen and Jacobsena, 1996).
The gene trapped in col. 211 encodes a protein containing a major facilitator superfamily (MFS) domain. Proteins containing this domain are secondary carriers that function as transporters for small solutes, such as monosaccharides, disaccharides, peptides, amino acids, and other molecules (Pao et al., 1998). Our result thus suggested that uptake of such compounds might be induced during the interaction. Similarly, gene ybgC (found in col. 161) encodes a homolog of the Tol-Pal system-associated acyl-CoA thioesterase, the first enzyme in the Tol-Pal system of E. coli (Godlewska et al., 2009). This system consists of five proteins (TolQ, TolR, TolA, TolB, and Pal) forming a membrane associated complex, which maintains outer membrane integrity. In addition, the operon shows similarity to the TonB system, which is important for active transport of diverse substrates such as siderophores or vitamin B12 (Moeck and Coulton, 1998) indicating that HP15 might possibly use TonB-dependent outer membrane receptors for uptake of yet-to-be determined substances. In their seminal paper, Teeling et al. (2012) had pointed out that TonB-dependent transport mechanisms were instrumental for marine bacterial population during algal bloom in the German bight.
Once the yet-unknown organic compounds released by the diatom are transported into the bacterial cells, they may be (partially or fully) degraded. In this context, we identified six genetic loci, which unfortunately do not allow concluding anything directly in terms of increased catabolism or accelerated energy conservation during the co-culture (col. 15, 19, 24, 149, 205, and 240). However, these results confirmed previous metagenomic and transcriptomic studies in which genes for degradation of organic compounds and an increase in bacterial activity and cell division rates were identified during bacteria-algae interactions (Poretsky et al., 2010; Smith et al., 2013; Durham et al., 2017).
In seven of the identified transconjugants, the IVET plasmids contained upstream sequences of genes involved in amino acid uptake (col. 26, 65, and 100), amino acid or protein degradation (col. 238 and 239), or even amino acid biosynthesis (col. 54 and 165). Inserts of two additional clones physically clustered with those of col. 54 and 238 but indicated sequence similarities to conserved hypothetical proteins (col. 166 and 241). This over-proportional appearance of amino acid-associated gene products among in co-culture induced proteins prompts the assumption that HP15 might preferentially obtain and use amino acids or amino group containing substances derived from the diatom cells. A somewhat similar observation was made by Durham et al. (2017), who found that presence of T. pseudonana induced expression of uptake systems for organic nitrogen compounds in the marine bacterium Ruegeria pomeroyi DSS-3. Our previous proteomics approach combined with a functional metabolite analysis (Stahl and Ullrich, 2016) had also revealed that M. adhaerens HP15 utilizes amino acids but not sugars as preferred carbon sources.
While the co-occurrence of genetic loci encoding amino acid-degrading and amino acid-synthesizing enzymes is a bit puzzling to interpret, the simultaneous finding of three amino acid or amino compound uptake systems being required during the interaction seems to be promising: The genetic locus HP15_3541 (col. 65) encodes for the periplasmic portion of a spermidine/putrescine transporter. Diatom cell walls contain high amounts of long-chain polyamines, which are organic constituents of diatom bio-silica (Kröger et al., 2000; Kröger and Poulsen, 2008). Previously, a positive correlation was found between the concentrations of polyamines in seawater and the amount of transcripts from different groups of marine bacteria involved in the degradation of polyamines (Mou et al., 2011). Transcripts and genetic evidence involved in polyamines uptake were also found in bacteria associated to marine phytoplankton blooms (Moran et al., 2004; Poretsky et al., 2005, 2010) as well as in free-living freshwater bacteria (Garcia et al., 2013). Our data thus allowed us to speculate on a potential bacterial use of polyamines shed during diatom cell wall synthesis and prompts future experiments in this direction.
The IVET screen revealed two further genetic loci (col. 100 and 26), whose expression was associated with presence of T. weissflogii, and which encode for LivK (HP15_4098) and AotJ (HP15_3031), respectively. Interestingly, the gene product of livK was already found upregulated in our previous proteomics study (Stahl and Ullrich, 2016). LivK is part of a branched amino acid uptake system transporting leucine, isoleucine, and valine (Adams et al., 1990; Ribardo and Hendrixson, 2011). LivK's functional involvement in branched amino acid transport in M. adhaerens HP15 is supported by the close proximity of genes encoding the inner membrane translocators LivH and LivG as well as the ATP binding components LivM and LivF (Adams et al., 1990). Interestingly and in agreement with our finding, the RNAseq-based transcriptome analysis of R. pomeroyi DSS-3 revealed other branched amino acid uptake systems being upregulated in presence of diatom cells (Durham et al., 2017). Since the later study found several additional bacterial transport systems to be diatom-induced, the observed functional analogy of branched amino acid uptake systems in both organisms might reflect both, the multifactorial nature of the respective interactions and possible common metabolic interaction themes.
AotJ is an amino acid-binding protein, which belongs to the arginine/ornithine uptake system (Wissenbach et al., 1995; Nishijyo et al., 1998). AotJ's potential function is substantiated by presence of surrounding genes encoding the subunit of a histidine-lysine-arginine-ornithine transporter and two inner membrane permeases of arginine and histidine associated ABC transporters in the genome of HP15. Just like LivK, the AotJ gene product was found upregulated in M. adhaerens HP15 during exposure to T. weissflogii cells in our previous proteomics approach (Stahl and Ullrich, 2016). Both proteomics hits now substantiated by the IVET screen may hint at an important role of bacterial amino acid uptake during the bacteria-diatom interactions with a distinct focus on branched chain amino acids and arginine/ornithine, respectively.
Conclusions
This study revealed an exciting array of genes potentially important during diatom-bacteria interaction. The approach is innovative in the field and demonstrated that assessment of bacterial gene expression via determination of promoter activities without interference by mRNA degradation or protein instability is possible. From the identified genes, we can infer that chemotaxis might be important to detect diatom cells. Once the bacteria are in a close proximity, genes involved in pilus-associated attachment are expressed in order to start a cell-to-cell interaction. A benefit for the bacteria is observed by the expression of genes involved in uptake and utilization of nitrogen-containing compounds. These results are in agreement with results from the past 50 years of research and with those from recent reports (Poretsky et al., 2005, 2010; Rinta-Kanto et al., 2012; Amin et al., 2015; Durham et al., 2017) that bacteria use the DOC produced by diatoms as nutrient source. A highly interesting but puzzling result remains the identification of a heavy metal efflux pump and the MerR1 transcriptional regulator as being encoded by in co-culture induced genes allowing multiple alternative speculations. Consequently, further experiments have to be carried out to confirm whether HP15 is actively provoking the diatom to produce or release metabolites thus participating in an active exchange of signals and compounds or is saprophytically living on non-specifically released compounds because of diatom cell lysis. Our findings indicate that HP15 might participate in the cycling of nitrogen-containing DOC in the ocean and could therefore be involved in the termination of phytoplankton blooms.
Author Contributions
IT-M conducted all experiments, analyzed all data, and prepared the manuscript, all figures and tables. MU initiated this study, supervised and directed all experiments, wrote the manuscript, and submitted the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (UL169/6-1). Preliminary data and part of this manuscript have been included in the archived dissertation of the first author at Jacobs University Bremen (Torres-Monroy, 2013). The technical support by Nikki Atanasova was highly appreciated.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00200/full#supplementary-material
Supplementary Figure 1. Construction of IVET vector pITM3_pyrB. A promoter-less copy of lacZ from E. coli was ligated into KpnI-treated pBBR1MCS-4 resulting in plasmid pITM3. Then a promoter-less copy of the genes pyrBC from HP15 was cloned into HindIII/XhoI-treated pITM3 to generate pITM3_pyrB.
Supplementary Figure 2. Bacterial in vitro growth curves of M. adhaerens HP15 wild type, HP15 ΔpyrB, and HP15 ΔpyrB complemented with 5 μg ml−1 of uracil in f/2 GLUT liquid medium and constant shaking at 250 rpm.
References
Adams, M. D., Wagner, L., Graddis, T., Landick, R., Antonucci, T., Gibson, A., et al. (1990). Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J. Biol. Chem. 265, 11436–11443.
Alldredge, A. L., Cole, J. J., and Caron, D. A. (1986). Production of heterotrophic bacteria inhabitating macroscopic organic aggregates (marine snow) from surface waters. Limnol. Oceanogr. 31, 68–78. doi: 10.4319/lo.1986.31.1.0068
Alldredge, A. L., Passow, U., and Logan, B. E. (1993). The abundance and significance of a class of large, transparent organic particles in the ocean. Deep Sea Res. I 40, 1131–1140. doi: 10.1016/0967-0637(93)90129-Q
Alldredge, A. L., and Silver, M. L. (1988). Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20, 41–82. doi: 10.1016/0079-6611(88)90053-5
Alldredge, A. L., and Youngbluth, M. J. (1985). The significance of macroscopic aggregates (marine snow) as sites for heterotrophic bacterial production in the mesopelagic zone of the subtropical Atlantic. Deep Sea Res. 32, 1445–1456. doi: 10.1016/0198-0149(85)90096-2
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Amin, S. A., Hmelo, L. R., van Tol, H. M., Durham, B. P., Carlson, L. T., Heal, K. R., et al. (2015). Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Asada, K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396. doi: 10.1104/pp.106.082040
Bach, L. T., Boxhammer, T., Larsen, A., Hildebrandt, N., Schulz, K. G., and Riebesell, U. (2016). Influence of plankton community structure on the sinking velocity of marine aggregates. Global Biogeochem. Cycles 30, 1145–1165. doi: 10.1002/2016GB005372
Baker, M. D., Wolanin, P. M., and Stock, J. B. (2005). Signal transduction in bacterial chemotaxis. Bioessays 28, 9–22. doi: 10.1002/bies.20343
Behringer, G., Ochsenkühn, M. A., Fei, C., Fanning, J., Koester, J. A., and Amin, S. A. (2018). Bacterial communities of diatoms display strong conservation across strains and time. Front. Microbiol. 9:659. doi: 10.3389/fmicb.2018.00659
Biddanda, B. A., and Pomeroy, L. R. (1988). Microbial aggregation and degradation of phytoplankton-derived detritus in seawater. I. Microbial. Succession. Mar. Ecol. Prog. Ser. 42, 79–88. doi: 10.3354/meps042079
Bodenmiller, D., Toh, E., and Brun, Y. V. (2004). Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 186, 1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004
Brown, N. L., Stoyanov, J. V., Kidd, S. P., and Hobman, J. L. (2003). The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163. doi: 10.1016/S0168-6445(03)00051-2
Bruckner, C. G., Rehm, C., Grossart, H. P., and Kroth, P. G. (2011). Growth and release of extracellular organic compounds by benthic diatoms depend on interactions with bacteria. Environ. Microbiol. 13, 1052–1063. doi: 10.1111/j.1462-2920.2010.02411.x
Busch, K., Endres, S., Iversen, M. H., Michels, J., Nöthig, E. M., and Engel, A. (2017). Bacterial colonization and vertical distribution of marine gel particles (TEP and CSP) in the Arctic Fram Street. Front. Mar. Sci. 4:166. doi: 10.3389/fmars.2017.00166
Capra, E. J., and Laub, M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. doi: 10.1146/annurev-micro-092611-150039
Cases, I., Lopez, J. A., Albar, J. P., and De Lorenzo, V. (2001). Evidence of multiple regulatory functions for the PtsN (IIANtr) protein of Pseudomonas putida. J. Bacteriol. 183, 1032–1037. doi: 10.1128/JB.183.3.1032-1037.2001
Choi, K. H., and Schweizer, H. P. (2005). An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30 doi: 10.1186/1471-2180-5-30
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Danielsen, S., Kilstrup, M., Barilla, K., Jochimsen, B., and Neuhard, J. (1992). Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 6, 1335–1344. doi: 10.1111/j.1365-2958.1992.tb00854.x
de Bentzmann, S., Aurouze, M., Ball, G., and Filloux, A. (2006). FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 188, 4851–4860. doi: 10.1128/JB.00345-06
DeLong, E. F., Franks, D. G., and Alldredge, A. L. (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934. doi: 10.4319/lo.1993.38.5.0924
Dudley, E. G. (2008). In vivo expression technology and signature-tagged mutagenesis screens for identifying mechanisms of survival of zoonotic foodborne pathogens. Foodborne Pathog. Dis. 5, 473–485. doi: 10.1089/fpd.2008.0104
Durham, B. P., Dearth, S. P., Sharma, S., Amin, S. A., Smith, C. B., Campagna, S. R., et al. (2017). Recognition cascade and metabolite transferrin a marine bacteria-phytoplankton model system. Environ. Microbiol. 19, 3500–3513. doi: 10.1111/1462-2920.13834
Durham, B. P., Sharma, S., Luo, H., Smith, C. B., Amin, S. A., Bender, S. J., et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 112, 453–457. doi: 10.1073/pnas.1413137112
Eisenbach, M. (1996). Control of bacterial chemotaxis. Mol. Microbiol. 20, 903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Fowler, S. W., and Knauer, G. A. (1986). Role of large particles in the transport of elements and organic compounds through the oceanic water column. Prog. Oceanogr. 16, 147–194. doi: 10.1016/0079-6611(86)90032-7
Frankel, G., Phillips, A. D., Trabulsi, L. R., Knutton, S., Dougan, G., and Matthews, S. (2001). Intimin and the host cell – is it bound to end in Tir(s)? Trends Microbiol. 9, 214–218. doi: 10.1016/S0966-842X(01)02016-9
Garcia, S. L., McMahon, K. D., Martinez-Garcia, M., Srivastava, A., Sczyrba, A., Stepanauskas, R., et al. (2013). Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J. 7, 137–147. doi: 10.1038/ismej.2012.86
Gärdes, A., Iversen, M. H., Grossart, H.-P., Passow, U., and Ullrich, M. S. (2011). Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 5, 436–445. doi: 10.1038/ismej.2010.145
Gärdes, A., Kaeppel, E., Shehzad, A., Seebah, S., Teeling, H., Yarza, P., et al. (2010). Complete genome sequence of Marinobacter adhaerens type strain (HP15), a diatom-interacting marine microorganism. Stand. Genomic Sci. 3, 97–107. doi: 10.4056/sigs.922139
Glaeser, S. P., Grossart, H. P., and Glaeser, J. (2010). Singlet oxygen, a neglected but important environmental factor, short-term and long-term effects on bacterioplankton composition in a humic lake. Environ. Microbiol. 12, 3124–3136. doi: 10.1111/j.1462-2920.2010.02285.x
Godlewska, R., Wiśniewska, K., Pietras, Z., and Jagusztyn-Krynicka, E. K. (2009). Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria, function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 298, 1–11. doi: 10.1111/j.1574-6968.2009.01659.x
Grossart, H. P., Czub, G., and Simon, M. (2006a). Algae-bacteria interactions and their effects on aggregation and organic matter flux in the sea. Environ. Microbiol. 8, 1074–1084. doi: 10.1111/j.1462-2920.2006.00999.x
Grossart, H. P., Kiørboe, T., Tang, K. W., Allgaier, M., Yam, E. M., and Ploug, H. (2006b). Interactions between marine snow and heterotrophic bacteria: aggregate formation and microbial dynamics. Aquat. Microb. Ecol. 42, 19–26. doi: 10.3354/ame042019
Grossart, H. P., Schlingloff, A., Bernhard, M., Simon, M., and Brinkhoff, T. (2004). Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol. Ecol. 47, 387–396 doi: 10.1016/S0168-6496(03)00305-2
Guillard, R. R., and Ryther, J. H. (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Hoang, T. T., Karkhoff-Schweizer, R. A. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Hünken, M., Harder, J., and Kirst, G. O. (2008). Epiphytic bacteria on the Antarctic ice diatom Amphiprora kufferathii manguin cleave hydrogen peroxide produced during algal photosynthesis. Plant Biol. 10, 519–526. doi: 10.1111/j.1438-8677.2008.00040.x
Jahnke, R. A. (1996). The global ocean flux of particulate organic carbon: areal distribution and magnitude. Global Biogeochem. Cycles 10, 71–88. doi: 10.1029/95GB03525
Jørgensen, N. G., and Jacobsena, C. S. (1996). Bacterial uptake and utilization of dissolved DNA. Aquat. Microb. Ecol. 11, 263–270. doi: 10.3354/ame011263
Kachlany, S. C., Planet, P. J., Bhattacharjee, M. K., Kollia, E., DeSalle, R., Fine, D. H., et al. (2000). Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182, 6169–6176. doi: 10.1128/JB.182.21.6169-6176.2000
Kachlany, S. C., Planet, P., and DeSalle, R. (2001). Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. l9, 429–437. doi: 10.1016/S0966-842X(01)02161-8
Kaeppel, E. C., Gärdes, A., Seebah, S., Grossart, H. P., and Ullrich, M. S. (2012). Marinobacter adhaerens sp. nov., isolated from marine aggregates formed with the diatom Thalassiosira weissflogii. Int. J. Syst. Evol. Microbiol. 62, 124–128. doi: 10.1099/ijs.0.030189-0
Kelly, G., Prasannan, S., Daniell, S., Fleming, S., Frankel, G., Dougan, G., et al. (1999). Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat. Struct. Mol. Biol. 6, 313–318.
Kiørboe, T., Grossart, H. P., Ploug, H., and Tang, K. (2002). Mechanisms and rates of bacterial colonization of sinking aggregates. Appl. Environ. Microbiol. 68, 3996–4006. doi: 10.1128/AEM.68.8.3996-4006.2002
Kiørboe, T., and Jackson, G. A. (2001). Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol. Oceanogr. 46, 1309–1318. doi: 10.4319/lo.2001.46.6.1309
Kiørboe, T., Tang, K., Grossart, H. P., and Ploug, H. (2003). Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl. Environ. Microbiol. 69, 3036–3047. doi: 10.1128/AEM.69.6.3036-3047.2003
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). 4 new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kröger, N., Deutzmann, R., Bergsdorf, C., and Sumper, M. (2000). Species-specific polyamines from diatoms control silica morphology. Proc. Natl. Acad. Sci. U.S.A. 97, 14133–14138. doi: 10.1073/pnas.260496497
Kröger, N., and Poulsen, N. (2008). Diatoms-from cell wall biogenesis to nanotechnology. Annu. Rev. Genet. 42, 83–107. doi: 10.1146/annurev.genet.41.110306.130109
Lambert, B. S., Raina, J. B., Fernandez, V. I., Rinke, C., Siboni, N., Rubino, F., et al. (2017). A microfluidics-based in situ chemotaxis assay to study the behavior of aquatic microbial communities. Nat. Microbiol. 2, 1344–1349. doi: 10.1038/s41564-017-0010-9
Lee, S. W., and Cooksey, D. A. (2000). Genes Expressed in Pseudomonas putida during colonization of a plant-pathogenic fungus. Appl. Environ. Microbiol. 66, 2764–2772. doi: 10.1128/AEM.66.7.2764-2772.2000
Logan, B. E., Passow, U., Alldredge, A. L., Grossart, H. P., and Simon, M. (1995). Rapid formation and sedimentation of large aggregates is predictable from coagulation rates (half-lives) of transparent exopolymer particles (TEP). Deep Sea Res. II 42, 203–214. doi: 10.1016/0967-0645(95)00012-F
Mahan, M. J., Slauch, J. M., and Mekalanos, J. J. (1993). Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259, 686–688. doi: 10.1126/science.8430319
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583.
Moeck, G. S., and Coulton, J. W. (1998). TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28, 675–681. doi: 10.1046/j.1365-2958.1998.00817.x
Moran, M. A., Buchan, A., González, J. M., Heidelberg, J. F., Whitman, W. B., Kiene, R. P., et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432, 910–913. doi: 10.1038/nature03170
Mou, X., Vila-Costa, M., Sun, S., Zhao, W., Sharma, S., and Moran, M. A. (2011). Metatranscriptomic signature of exogenous polyamine utilization by coastal bacterioplankton. Environ. Microbiol. Rep. 3, 798–806. doi: 10.1111/j.1758-2229.2011.00289.x
Needham, D. M., Sachdeva, R., and Fuhrman, J. A. (2017). Ecological dynamicsand co-occurrence among phytoplankton, bacteria and myoviruses shows microdiversity matters. ISME J. 11, 1614–1629. doi: 10.1038/ismej.2017.29
Nies, D. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/S0168-6445(03)00048-2
Nishijyo, T., Park, S., Lu, C., Itoh, Y., and Abdelal, A. (1998). Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J. Bacteriol. 180, 5559–5566.
Pao, S. S., Paulsen, I. T., and Saier, M. H. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34.
Passow, U. (2002). Production of transparent exopolymer particles (TEP) by phyto- and bacterioplankton. Mar. Ecol. Prog. Ser. 236, 1–12. doi: 10.3354/meps236001
Perez, B. A., Planet, P., and Kachlany, S. (2006). Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation. J. Bacteriol. 188, 6361–6375. doi: 10.1128/JB.00496-06
Planet, P. J., Kachlany, S. C., Fine, D. H., DeSalle, R., and Figurski, D. H. (2003). The widespread colonization Island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34, 193–198. doi: 10.1038/ng1154
Poretsky, R. S., Bano, N., Buchan, A., Lecleir, G., Kleikemper, J., Pickering, M., et al. (2005). Analysis of microbial gene transcripts in environmental samples. Appl. Environ. Microbiol. 71, 4121–4126. doi: 10.1128/AEM.71.7.4121-4126.2005
Poretsky, R. S., Sun, S., Mou, X., and Moran, M. A. (2010). Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12, 616–627. doi: 10.1111/j.1462-2920.2009.02102.x
Postma, P. W., Lengeler, J. W., and Jacobson, G. R. (1993). Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594.
Raman, R., Rajanikanth, V., Palaniappan, R. U. M., Lin, Y. P., and He, H. (2010). Big domains are novel Ca2+-binding modules: evidences from big domains of Leptospira immunoglobulin-like (Lig) proteins. PLoS ONE 5:e14377. doi: 10.1371/journal.pone.0014377
Rediers, H., Rainey, P. B., Vanderleyden, J., and De Mot, R. (2005). Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69, 217–261. doi: 10.1128/MMBR.69.2.217-261.2005
Ribardo, D. A., and Hendrixson, D. R. (2011). Analysis of the LIV system of Campylobacter jejuni reveals alternative roles for LivJ and LivK in commensalism beyond branched-chain amino acid transport. J. Bacteriol. 193, 6233–6243. doi: 10.1128/JB.05473-11
Rinta-Kanto, J. M., Sun, S., Sharma, S., Kiene, R. P., and Moran, M. A. (2012). Bacterial community transcription patterns during a marine phytoplankton bloom. Environ. Microbiol. 14, 228–239. doi: 10.1111/j.1462-2920.2011.02602.x
Roberfroid, S., Vanderleyden, J., and Steenackers, H. (2016). Gene expression variability in clonal populations: causes and consequences. Crit. Rev. Microbiol. 42, 969–984. doi: 10.3109/1040841X.2015.1122571
Sambrook, J., Fritsch, E., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd Edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schurr, M. J., Vickrey, J. F., Kumar, A. P., Campbell, A. L., Cunin, R., Benjamin, R. C., et al. (1995). Aspartate transcarbamoylase genes of Pseudomonas putida: requirement for an inactive dihydroorotase for assembly into the dodecameric holoenzyme. J. Bacteriol. 177, 1751–1759.
Seebah, S., Fairfield, C., Ullrich, M. S., and Passow, U. (2014). Aggregation and sedimentation of Thalassiosira weissflogii (diatom) in a warmer and more acidified future ocean. PLoS ONE 9:e112379. doi: 10.1371/journal.pone.0112379
Seymour, J. R., Amin, S. A., Raina, J. B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:17065. doi: 10.1038/nmicrobiol.2017.65
Shapira, S. K., Chou, J., Richaud, F. V., and Casadaban, M. J. (1983). New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacA gene sequences encoding an enzymatically active carboxy-terminal portion of β-galactosidase. Gene 25, 71–82. doi: 10.1016/0378-1119(83)90169-5
Smirnova, A. V., and Ullrich, M. S. (2004). Topological and deletion analysis of CorS, a Pseudomonas syringae sensor kinase. Microbiology 150, 2715–2726. doi: 10.1099/mic.0.27028-0
Smith, M. W., Zeigler Allen, L., Allen, A. E., Herfort, L., and Simon, H. M. (2013). Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front. Microbiol. 4:120. doi: 10.3389/fmicb.2013.00120
Sonnenschein, E. C., Abebew Syit, D., Grossart, H. P., and Ullrich, M. S. (2012). Chemotaxis of Marinobacter adhaerens and its impact on attachment to the diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 78, 6900–6907. doi: 10.1128/AEM.01790-12
Sonnenschein, E. C., Gärdes, A., Seebah, S., Torres-Monroy, I., Grossart, H. P., and Ullrich, M. S. (2011). Development of a genetic system for Marinobacter adhaerens HP15 involved in marine aggregate formation by interacting with diatom cells. J. Microbiol. Methods 87, 176–183. doi: 10.1016/j.mimet.2011.08.008
Stahl, A., Pletzer, D., Mehmood, A., and Ullrich, M. S. (2015). Marinobacter adhaerens HP15 harbors two CzcCBA efflux pumps involved in zinc detoxification. Antonie Van Leeuwenhoek 108, 649–658. doi: 10.1007/s10482-015-0520-5
Stahl, A., and Ullrich, M. S. (2016). Proteomics analysis of the response of the marine bacterium Marinobacter adhaerens HP15 to the diatom Thalassiosira weissflogii. Aquat. Microbial. Ecol. 78, 65–79. doi: 10.3354/ame01804
Stock, J. B., Stock, A. M., and Mottonen, J. M. (1990). Signal transduction in bacteria. Nature 344, 395–400. doi: 10.1038/344395a0
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Thiele, S., Fuchs, B. M., Amann, R., and Iversen, M. H. (2015). Colonization in the photic zone and subsequent changes during sinking determine bacterial community composition in marine snow. Appl. Environ. Microbiol. 81, 1463–1471. doi: 10.1128/AEM.02570-14
Thoma, S., and Schobert, M. (2009). An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol. Lett. 294, 127–132. doi: 10.1111/j.1574-6968.2009.01556.x
Tomich, M., Planet, P. J., and Figurski, D. H. (2007). The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375. doi: 10.1038/nrmicro1636
Torres-Monroy, I. (2013). Identification of Bacterial Genes Required for Diatom-Bacteria Interactions. Dissertation, Jacobs University Bremen, Bremen.
van Tol, H. M., Amin, S. A., and Armbrust, E. V. (2017). Ubiquitous marine bacterium inhibits diatom cell division. ISME J. 11, 31–42. doi: 10.1038/ismej.2016.112
Wang, H., Tomasch, J., Jarek, M., and Wagner-Döbler, I. (2014). A dual-species co-cultivation system to study the interactions between Roseobacters and dinoflagellates. Front. Microbiol. 5:311. doi: 10.3389/fmicb.2014.00311
Wissenbach, U., Six, S., Bongaerts, J., Ternes, D., Steinwachs, S., and Unden, G. (1995). A 3rd periplasmic transport-system for L-arginine in Escherichia coli- molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol. Microbiol. 17, 675–686. doi: 10.1111/j.1365-2958.1995.mmi_17040675.x
Yang, S., Perna, N. T., Cooksey, D. A., Okinaka, Y., Lindow, S. E., Mark Ibekwe, A., et al. (2004). Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET Leaf Array. Mol. Plant Microbe Interact. 17, 999–1008. doi: 10.1094/MPMI.2004.17.9.999
Keywords: in vivo expression technology, diatom-bacteria interactions, Marinobacter adhaerens, marine aggregate, carbon flux, Thalassiosira weissflogii
Citation: Torres-Monroy I and Ullrich MS (2018) Identification of Bacterial Genes Expressed During Diatom-Bacteria Interactions Using an in Vivo Expression Technology Approach. Front. Mar. Sci. 5:200. doi: 10.3389/fmars.2018.00200
Received: 22 January 2018; Accepted: 18 May 2018;
Published: 05 June 2018.
Edited by:
Matthias Wietz, University of Oldenburg, GermanyReviewed by:
Irene Wagner-Doebler, Helmholtz-Zentrum für Infektionsforschung (HZI), GermanyShady A. Amin, New York University Abu Dhabi, United Arab Emirates
Mary Ann Moran, University of Georgia, United States
Copyright © 2018 Torres-Monroy and Ullrich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthias S. Ullrich, bS51bGxyaWNoQGphY29icy11bml2ZXJzaXR5LmRl
 Ingrid Torres-Monroy
Ingrid Torres-Monroy Matthias S. Ullrich
Matthias S. Ullrich