- 1The Agricultural Center, Louisiana State University, Baton Rouge, LA, United States
- 2School of Renewable Natural Resources, Louisiana State University, Baton Rouge, LA, United States
- 3College of the Coast and Environment, Louisiana State University, Baton Rouge, LA, United States
- 4Louisiana Sea Grant Oyster Research Laboratory, Grand Isle, LA, United States
Biomineralization is a complexed process by organisms producing protective and supportive structures. Employed by mollusks, biomineralization enables creation of external shells for protection against environmental stressors. The shell deposition mechanism is initiated in the early stages of development and is dependent upon the concentration and availability of calcium carbonate ions. Changes in concentrations of the critical ions required for shell formation can result in malformation of shells. As pCO2 concentrations in the atmosphere continue to increase, the oceans are becoming more acidified. This process, known as ocean acidification (OA), has demonstrated adverse effects on shell formation in calcifying organisms across taxa. Although OA is known to inhibit the shell deposition in mollusks, the impact of OA on the gene regulation of calcium deposition remains unknown. Here we show the responses of four calcium-binding protein genes, caltractin (cetn), calmodulin (calm), calreticulin (calr), and calnexin (canx), to CO2-derived OA using a Crassostrea virginica mantle cell (CvMC) culture model and a larval C. virginica model. These four genes were cloned from C. virginica and the three-dimensional structures of the proteins encoded by these four genes were fully characterized using homolog modeling methods. Although an acidified environment by increased atmospheric pCO2 (1,000 ppm) did not result in significant effects on CvMC proliferation and apoptosis, lower environmental pH induced upregulations of all four calcium-binding protein genes in CvMCs. Similarly, increased pCO2 did not affect the growth of larval C. virginica in the early stages of development. However, elevated pCO2 concentrations enhanced the expression of these calcium-binding protein genes at the protein level. The four calcium-binding protein genes demonstrated responsive expression profiles to an acidified environment at both cellular and individual levels. Further investigation of these genes may provide insight into the molecular regulation of mollusk biomineralization under OA stress.
Introduction
Mollusca have developed hard external structures (shells) to protect their soft tissues against pathogens, predation, and other environmental stressors (Marin et al., 2012; Arivalagan et al., 2017). These protective structures are commonly formed by a biologically controlled process called biomineralization. Mollusk shells contain approximately 95% calcium carbonate (CaCO3) and 5% organic matter (Lowenstam and Weiner, 1989). The major forms of environmental CaCO3 are aragonites, calcites, or vaterites. Although CaCO3 is also the main component, the shells formed by molluscan species are thousands fold tougher than any of these environmental CaCO3 polymorphs (Currey, 1977), which suggests a unique biomineralized structure within shells (Harper, 2000). The hypothesis of the synthesis of this unique structure suggests that shell fabrication is regulated by specialized cells in the host where activities are controlled by a cascade of genes (Mann, 1983).
Biomineralization occurs primarily in the extrapallial cavity, which contains extrapallial fluid with a substantially higher concentration of calcium and carbonate ions compared to the surrounding environment. Numerous calcium-binding proteins, passive calcium transporters, and active calcium pumps are involved in calcium transport and ion regulation to maintain a high CaCO3 saturation state within the extrapallial fluid (Ip et al., 2006). Therefore, the concentrations and availability of environmental calcium and carbonate ions are critical to the shell formation mechanism.
The availability of calcium and carbonate ions in the ocean can be manipulated by a number of environmental factors. Atmospheric CO2 levels are continuing to increase due to anthropogenic activities. The oceans are currently the sink for ~30% of anthropogenic carbon dioxide (Sabine et al., 2004). The relationship between atmospheric CO2 concentration and carbonate ion concentration in water is represented with the following chemical equation (Orr et al., 2005; Zeebe, 2012):
As a result of increased atmospheric CO2, the balance of inorganic carbon is shifting toward generating more bicarbonate, carbonate, and H+, a process known as ocean acidification (OA). Meanwhile, increased H2CO3 compete with calcifiers on carbonate to generate more bicarbonate, which leads insufficient CaCO3 deposition during biomineralization. As a result, the homeostasis of carbonate and calcium ions in the extrapallial fluid of mollusks is disrupted, which ultimately inhibits the biomineralization process.
It is well known that the early-life history stages of bivalves, including gametes, embryos and larvae are vulnerable to environmental stressors. Although studies have revealed the potential impact of ocean acidification on larval stages of oysters (Parker et al., 2010; Gazeau et al., 2011; Waldbusser et al., 2013), efforts in understanding the mechanism of this impact are limited. Several critical developmental phases, along with significant morphological changes, are involved in the oyster life cycle following gamete fertilization. The first vital development event is initiated 24 h post fertilization in larva when the shell begins to form. In this stage, bivalve larvae are particularly sensitive to decreases in environmental pH (Waldbusser et al., 2013). Within the ocean, pH, dissolved inorganic carbon (DIC), and mineral solubility are regulated by the balance of pCO2 and carbonate ions (Waldbusser et al., 2014). Bivalve larvae utilize dissolved calcium and carbonate ions to deposit their shells through calcification processes. Fluctuation of dissolved CO2 results in equilibrium disruption of pH, mineral solubility and DIC, which disrupts the mechanisms of larval bivalve shell calcification.
Numerous genes that are potentially involved in biomineralization have been identified from various mollusk species with the completion of genome sequencing and the development of high-throughput sequencing. Zhang et al. predicted over 250 shell formation related genes from the genome of Crassostrea gigas by looking for the classic calcium-binding domain, E-F hand structures (Friedberg, 1988) in translated amino acid sequences (Zhang et al., 2012). Another study using a Pinctada fucata microarray containing over 58,000 transcript represented probes suggested that 31 gene regulation pathways were likely to be involved in shell formation during larval development. One identified pathway was directly related to calcium signaling in the database of Kyoto Encyclopedia of Genes and Genomes (KEGG, ko04020) (Liu et al., 2015). The most recent genomic study obtained a group of shell formation related genes from 14 released molluscan genomes (Takeuchi, 2017). In addition to genes identified by high-throughput techniques, many biomineralization-related individual genes were also recognized as calcium-binding protein encoding genes (Weiss et al., 2000; Hattan et al., 2001; Huang et al., 2007; Yan et al., 2007; Takagi and Miyashita, 2010; Su et al., 2013). Unfortunately, most of the calcium-binding protein encoding genes have not been sequenced or identified in the model species of the current study, C. virginica. Therefore, the first goal of this study is to clone and fully characterize the structures of the full length of some calcium-binding protein genes from C. virginica.
Four calcium binding proteins were investigated in the current study. Calmodulin (CALM) is a well-studied calcium-binding protein in mollusk biomineralization. Several calmodulins and the homologs have been identified and cloned from various mollusk species, including Pinctada fucata (Li et al., 2004; Yan et al., 2007), Hyriopsis schlegelii (Peng et al., 2017), and C. gigas (Li et al., 2016). Through studying bivalve organs and tissues in mollusks, it was determined that mantle tissue produced the highest number of calmodulins in P. fucata (Li et al., 2005). The recombinant calmodulin in vitro was established as a protein that modifies calcite formation and nucleate aragonite during biomineralization (Yan et al., 2007). Less frequently studied in mollusk biomineralization, caltractin (CETN), also known as centrin, was recognized as a calmodulin-related protein (Nishi et al., 2013). Identified from various species ranging from yeast to human, caltractins were believed to participate in numerous calcium-related cell activities (Nishi et al., 2013). Calreticulin (CALR) and calnexin (CANX) were additionally studied in mollusk species as biomineralization-related genes. Originally discovered in the cell endoplasmic reticulum, CALR has also been located in the cell surface (Arosa et al., 1999), bloodstream (Sueyoshi et al., 1991), and extracellular matrix (Somogyi et al., 2003) in many cell types and species (Coppolino and Dedhar, 1998). These are consistent to another finding which suggests that the biomineralization proteins can be provided through other resources, such as hemolymph (Mount et al., 2004). Similarly, CANX, a close relative to calreticulin, demonstrated strong calcium binding abilities in various signaling pathways in cells (Corbett and Michalak, 2000; Michalak et al., 2002).
Atmospheric CO2 is expected to reach 1,000 ppm by the year 2100, which is more than double the amount of current CO2 in the air (400 ppm), according to the Intergovernmental Panel on Climate Change (IPCC, 2007). Therefore, it is critical to identify the potential impacts of increased atmospheric CO2 on the biomineralization process of calcifying mollusks, many of which are important both economically and ecologically in coastal areas. The second goal of this study is to investigate the effect of elevated pCO2 on the regulation of these proteins in biomineralization. Expression levels of four calcium-binding protein encoding genes, caltractin (calt), calmodulin (calm), calreticulin (calr), and calnexin (canx) were investigated during the development of larval C. virginica at two atmospheric pCO2 concentrations. Cells cultured from the mantle of C. virginica were established and utilized to study the regulation of these four calcium-binding proteins in response to elevated pCO2 since over 80% biomineralization-related genes in C. gigas are mantle tissue specific (Zhang et al., 2012). Our results indicate that CO2-derived OA alters the larval mollusk shell formation through the regulation of calcium-binding proteins. These calcium-binding proteins can potentially be used as indicators of successful development and stress levels in larval mollusks, and as selective breeding markers for the improvement of OA resistant lines in mollusk aquaculture.
Materials and Methods
Cloning and Bioinformatics Analyses of Calcium-Binding Protein Genes From C. virginica
All the oysters used in this study were maintained at the Michael C. Voisen Louisiana Sea Grant Oyster Research Laboratory. The original oyster individuals were collected from the Barataria Bay near Grand Isle, Louisiana. The current individuals were generated from those original individuals through several passages. The water condition in the Oyster Research Laboratory was created to mimic the natural environment near the coast of Grand Isle (pH8.5; salinity 15 ppt).
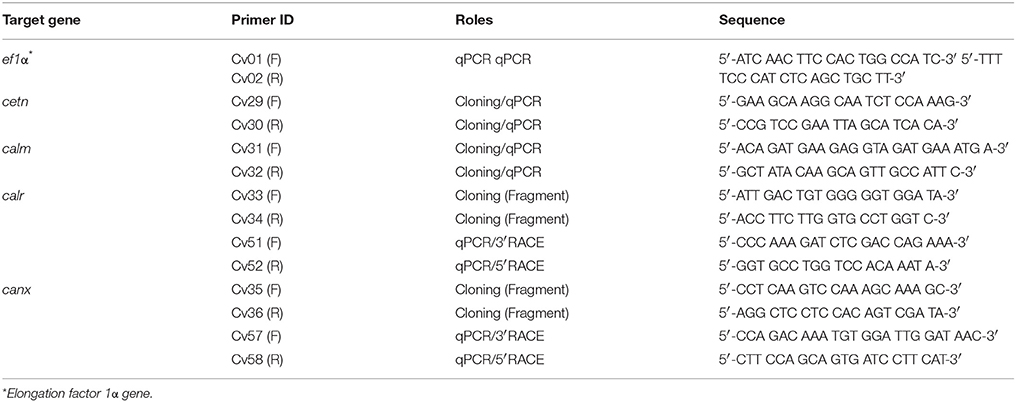
Four calcium-binding protein encoding genes, caltractin (cetn), calmodulin (calm), calreticulin (calr), and calnexin (canx), were cloned. Briefly, a pair of cloning primers were designed based on expressed sequence tag (EST) in the GenBank database from Crassostrea spp. (Table 1). The C. virginica individuals used for gene cloning were obtained from Grand Isle, Louisiana. The total RNA was isolated from homogenized C. virginica mantle tissues using TRIzol® Reagent (Life Technologies, Carlsbad, CA) followed by DNA removal with TURBO DNA-free™ Kit (Thermo-Fisher Scientific, Plaquemine, LA). The total RNA was then used for cDNA synthesis with SuperScript IV Reverse Transcriptase (Thermo-Fisher Scientific) following the manufacturer's instructions. Fragments of the four calcium-binding protein genes were amplified using the gene cloning primers and GoTaq® DNA Polymerase (Promega Biotechnology Co., Madison, WI) using regular PCR cycles (94°C 5 min followed by 32 cycles of 94°C 30 s-55°C 30 s-72°C 30 s, and a final extension step at 72°C for 7 min). The PCR product was then purified from a 7% polyacrylamide-TBE gel using a DNA dialysis collection method following the standard protocol (Sambrook and Russell, 2006b). The purified PCR products were cloned to the pGEM®-T Easy Vector Systems (Promega Biotechnology Co.) and transformed into DH5α competent Escherichia coli following a standard protocol (Sambrook and Russell, 2006a). The bacterial suspension was spread on LB plate containing 100 ng/ml ampicillin overnight at 37°C. Colonies were randomly picked, verified, and submitted for sequencing at the Gene Lab of the Louisiana State University, School of Veterinary Medicine (Baton Rouge, LA). For plasmids isolated from each bacterial inoculate, the inserts of the plasmids were sequenced from two directions with T7 and SP6 primers, respectively. The two reads of each insert were aligned using BLASTn. The aligned sequences were trimmed with VecScreen to remove the sequence of pGEM-T Easy vector.
With the obtained fragment of calr and canx, new primers were designed using the primer design software provided by the IDT DNA website. These specific primers were used for both full-length gene cloning and qPCR. Cloning of full length sequences of the four genes was performed using the Rapid Amplification of cDNA Ends (RACE) system from Invitrogen (Life Technologies). PCR products for 3′ or 5′ end gene fragments were loaded and run on a 0.8% agarose TAE gel and purified using Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA) following the manufacturer's directions. The purified PCR products were submitted for sequencing at the Gene Lab using the gene specific primers for each gene.
The obtained 3′ and 5′ end sequences of each gene along with the originally cloned fragment in step 2 were assembled using BLASTn for two sequences. The protein sequence of each gene was predicted by the mRNA translation tool in Expert Protein Analysis System (ExPaSy) (Artimo et al., 2012). The open reading frame (ORF) of each gene was identified based on the length of predicted ORF and the length of the ORFs in homologous sequences in GenBank database. The translated protein sequence for each gene was first submitted to GenBank for homologous sequence searching using BLASTp. Then motifs and signature sequences of each protein were predicted using the simple modular architecture research tool (SMART) (Schultz et al., 1998; Letunic et al., 2015). Finally, the three-dimensional structure of each protein was predicted based on protein structure homology-modeling system (SWISS-MODEL, Figure 1) (Arnold et al., 2006; Kiefer et al., 2009; Biasini et al., 2014). To select the best structure from multiple predictions for each protein, we used GMQE (Global Model Quality Estimation) and the QMEAN as parameters. The 3D structure with the highest GMQE and QMEAN scores was chosen for each protein.
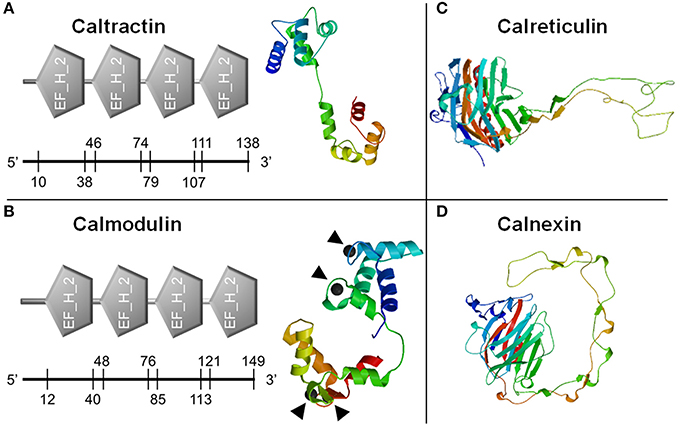
Figure 1. Prediction of calcium-binding domains and three dimensional structures of calcium binding proteins. Classic calcium-binding domains, E-F hands (EF_H_2), were identified from caltractin (A, left) and calmodulin (B, left). Four calcium-binding locations in calmodulin were also predicted (▴). The three dimensional structures of caltractin (A, right), calmodulin (B, right), calreticulin (C), and calnexin (D) were simulated. The numbers on the axis under each domain were starting and ending amino acid sites in each protein.
C. virginica Mantle Cell (Cv-MC) Culture
Adult eastern oysters were obtained from the Michael C. Voisen Louisiana Sea Grant Oyster Research Laboratory, where they were maintained in off bottom culture cages. Oysters were then transferred to the indoor facility with recirculating systems containing artificial seawater and acclimated to 25ppt salinity at 22°C. Prior to cell culture, selected oysters were kept in sterilized Ca2+ and Mg2+ free artificial seawater solution (CMFSS, including 436 mM NaCl, 10.7 mM KCl, 21.2 mM Na2HPO4, 16.7 mM Glucose, and 12.0 mM HEPES) supplemented with 100 units/mL of penicillin and 100 μg/mL of streptomycin (CMFF-Pen/Strep) overnight at 4°C. The sterilized oysters were dissected and the connective tissues along the edge of the mantle were collected (Figure 2a). The collected C. virginica mantle tissues were thoroughly washed with CMFF-Pen/Strep and digested with 0.2% collagenase I (Thermo-Fisher Scientific) in CMFF-Pen/Strep solution overnight at room temperature. The digested tissue suspension was mixed with oyster cell culture medium (OCCM, Leibovitz-L15 basal medium with additional 345.6 mM NaCl, 7.2 mM KCl, 5.4 mM CaCl2, 4.0 mM MgSO4•7H2O, 19.2 mM MgCl2•6H2O, and 4.0 mM L-Glutamine) supplemented with 10% (v/v) chicken serum (Life Technologies, Carlsbad, CA), 100 units/mL of penicillin and 100 μg/mL of streptomycin (OCCM-Pen/Strep). The mixture was then filtered with a 100 μm cell strainer (VWR International, Radnor, PA) to remove the non-digested tissue. The flow-through solution was centrifuged at 500 × g for 5 min to collect cells and the supernatant was discarded. Fresh OCCM-Pen/Strep medium was added to the cell pellet to re-suspend the cells. The fully suspended C.virginica mantle cells (CvMCs) in medium were then seeded in a 24-well cell culture plate pre-coated with 0.01% Poly-L-lysine in CMFSS solution. All seeded plates were incubated at 27°C for 24–48 h to allow the adhesion of cells to the bottom of the plate. Cells were observed under a microscope to ensure attachment and to confirm the cell confluence at ~80% (Figure 2b).
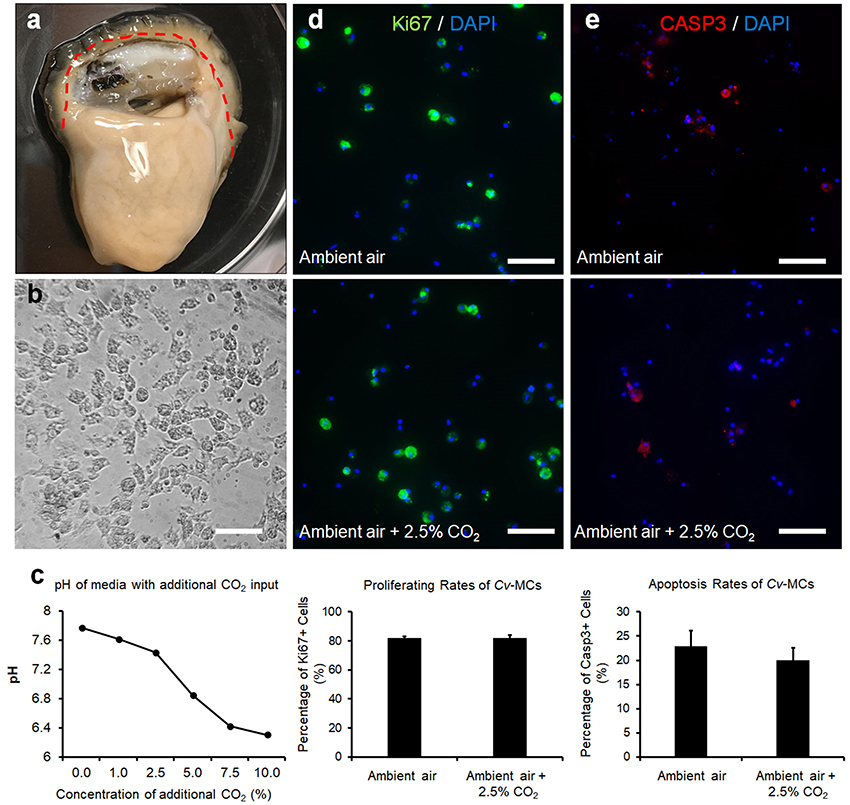
Figure 2. CvMC proliferation and apoptosis assays. The location of mantle tissue used for cell culture was indicated by dashed line (a). CvMCs after 48-h culture was shown (b). Changes of medium pH in response to increased atmospheric CO2 (n = 4), (c). Results of IFs with Ki-67 (d) and caspase 3 (e) were also quantified with the cells treated by ambient air and increased CO2 concentration (2.5%). n = 8. Scale bar: 50 μm. Error bar: standard error of mean.
Stimulation of CvMCs With Increased pCO2
CvMCs from each C. virginica individual were cultured in two 24-well plates until reaching 80% confluences. Medium in each well was replaced with fresh medium prior to the treatment. All the cell cultures were kept in a CO2 cell culture incubator. The concentration of CO2 in the input air can be regulated by the incubator. The increased CO2 concentration in the input air mimics the increased atmospheric CO2 level in nature. One plate of CvMCs were treated with ambient air (Control) and the cells in the other plate were treated with additional 1, 1.5, or 2.5% CO2 input. All cells were kept at 27°C for 24 h prior to harvesting. Half of the wells were harvested using TRIzol® Reagent for RNA isolation while the other half were fixed in 4% paraformaldehyde (PFA) for immunofluorescent staining. The cell culture medium was collected from each cell culture and the pH of each medium sample was measured immediately using a bench top pH meter (VWR International). Cells from eight C. virginica individuals were used as eight biological replicates for this study.
Stimulation of Larval C. virginica With Increased pCO2
The production of C. virginica larvae was performed at the Michael C. Voisen Louisiana Sea Grant Oyster Research Laboratory following a previously established protocol. Briefly, individual sexually matured C. virginica were maintained at 22°C, until spawning was thermally induced through exposure of estuarine water from Grand Isle, Louisiana raised to 34°C. Once spawning concluded, isolated eggs and sperm were pooled and thoroughly mixed to initiate fertilization. Successfully fertilized eggs, determined by light microscopy, were used for the experiment 2 h post fertilization. Following fertilization, triplicate larval cultures were established for each treatment at a density of 100 eggs/ml. After 48 h, the larvae were restocked at 50 larvae/ml to meet the culture standards and were established in new pre-conditioned estuarine water.
Acidification treatments included a current pCO2 of 400 ppm and a future by year 2100 pCO2 of 1,000 ppm based on IPCC (IPCC, 2007). Compressed zero air tanks with added CO2 where prepared gravimetrically and certified to the specified CO2 concentrations within a 2% analytical tolerance (AirGas, Baton Rouge, LA). Air was bubbled into the estuarine water through an air stone. The estuarine water was bubbled with the proper pCO2 for at least a 3-day period to ensure proper dissolution of CO2 (pre-conditioned water). Bubbling of the treatment CO2 was continued in the pre-conditioned water and culturing vessel after C. virginica were introduced. Bubbling rates were adjusted to ensure consistent turbulence between biological replication.
Larval samples from each treatment group were collected at 0, 24, and 48 h post fertilization. The larvae provided with a daily algal mixture of Tisochrysis lutea, Cisochrysis galbana, Chaetoceros calcitrans, Chaetoceros muelleri, and Pavlova luthen, at concentrations of 40,000 cells mL−1. Temperature, pH, and salinity were measured daily (Orion 370 PerpHecT benchtop pH/ORP/ISE/T meter, Thermo Fisher Scientific, Waltham, MA). Total alkalinity was measured from water that had been treated with supersaturated 0.02% mercuric chloride and frozen prior to analysis. Briefly, total alkalinity samples were titrated with 0.097N hydrochloric acid (HCl) to achieve a pH of 3.5 using a Schott Titroline easy titrator (General Laboratory Supply, Pasadena, TX) (Dickson et al., 2007). Samples were then left to de-gas for 3 min and titrated step-wise at 20 s intervals in 0.05 ml increments until pH 3.0 to create a Gran Line, using the SeaCarb (http://CRAN.Rproject.org/package=seacarb) program in R Studio (Boston, MA). Larval samples were harvested and maintained in 4% PFA for microscopy and immunofluorescence (IF). The shell length of each larva was measured as the maximum distance between two points on the edge of the shell.
Quantitative PCR (qPCR)
Total RNAs from CvMC were isolated and used for cDNA synthesis as described above. qPCRs with synthesized cDNAs were performed using Sybr Green method in a 7900HT Real-Time PCR System (Applied Biosystems, Grand Island, NY). The qPCR reaction was run at 50°C for 2 min, 94°C for 2 min, followed by 40 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 30 s. The cycle threshold (Ct) values generated from the system for all samples were used for relative expression analysis using 2−ΔΔCt method (Rao et al., 2013).
Immunofluorescence With CvMCs and Larval C. virginica
Immunofluorescence (IF) was used to visualize and quantify the proliferation and apoptosis of mantle cells in both the control and treatment. Cells or larval C. virginica fixed in 4% PFA were washed with phosphate-buffered saline (PBS). Samples were then blocked for 2 h at room temperature with 10% lamb serum in PBS in each well. After 3 washes with PBS, a primary antibody diluted in PBS was applied to cover the samples in each well and incubated at 4°C overnight. Following removal of primary antibody and three washes with PBS, the secondary antibody conjugated with either Alexa flour 594 (red) or Alexa flour 488 (green) dye, was diluted and allocated into the wells. The plates were incubated in a dark area for 2 h. After secondary antibody incubation and three washes in PBS, a 10-min counterstaining with a nuclear dye, 4′,6-diamidino-2-phenylindole (DAPI, blue, Biotium Inc., Fremont, CA) was performed on each sample with 0.1 μg/ml DAPI in PBS. Primary antibodies used in this study include mouse-anti-human Ki-67 (Novus Biologicals, Littleton, CO), rabbit-anti-human caspase 3 (Casp3, Boster Biological Technology, Pleasanton, CA), and three mouse originated calcium-binding protein antibodies (all from the Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA). The use of Ki-67 and Casp3 antibodies was to test cell proliferation and apoptosis, respectively. The ideal pCO2 concentrations created in our study were expected to induce the differential gene expression without significantly changing cell proliferation and apoptosis. Therefore, the IF with these two antibodies was to avoid overdosing the cells with excessive CO2 input.
The secondary antibodies were all from Life Technologies. Dilutions for primary antibodies varied based on the optimized results: 1:500 for Ki-67 antibody, 1:200 for caspase 3 antibody, and 1:100 for all three calcium-binding protein antibodies. All dilutions for secondary antibodies were 1:500. To test autofluorescence of the background, we tested the secondary antibodies on each sample with the absence of primary antibodies (Supplementary Figure 5). Four calcium-binding primary antibodies (CALR, CALM, CETN, and CANX) were tested before the application in this study. Only the antibodies for CALR, CALM, and CANX were confirmed to be reliable in C. virginica tissues and cells. The samples were observed under the Nikon Eclipse Ti fluorescent microscope (Nikon USA) immediately after IF staining. Pictures were taken for each observation under various colored filters. The green and blue channels or red and blue channels of each observation field were merged using ImageJ (Schneider et al., 2012). To eliminate the effects of autofluorescence during quantification, we adjusted the exposure time of the image to minimize the signal from the negative control of each antibody. Same exposure time was used on all photos of each antibody.
Statistical Analyses
For IF of Ki-67 and caspase 3, the total number of cells (only blue signals) and Ki-67 positive cells (cells with both blue and green signals or with both blue and red signals) were counted using ImageJ and the proliferating/apoptosis rate of each cell culture was calculated as:
For the IF of calcium-binding proteins in CvMCs, the total calcium-binding protein signal intensity of each photo was measured using ImageJ and the total number of cells was obtained by counting the number of nuclei as described above (DAPI signals). The mean calcium-binding protein level of the cells in each photo was calculated as:
The average calcium-binding production in cells with each treatment was normalized using average calcium-binding protein production in control cells. Similarly, IF signal intensities of calcium binding proteins in larval C.virginica were also digitalized using ImageJ. The intensity values were all normalized against control samples to generate relative calcium-binding protein productions. Comparisons of cell proliferation, cell apoptosis, and calcium-binding protein productions in cells and larvae were made between control and treated groups using t-tests.
Results
Calcium Binding Protein Sequence Analyses
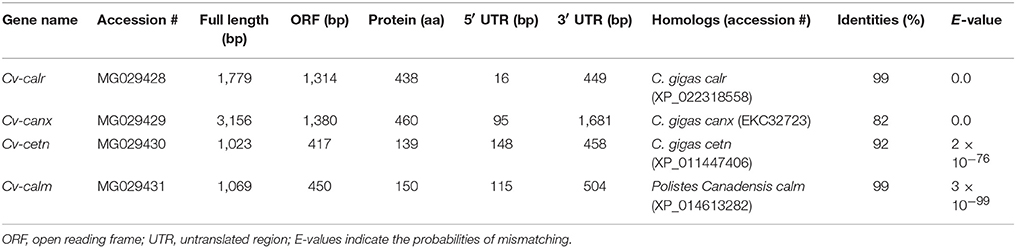
Full sequences of the four calcium-binding protein genes were obtained through cloning and fragment assembly. All four gene sequences have been submitted to NCBI GenBank database with accession numbers (MG029428-MG029431) with detailed information of the sequences in Table 2. The homologous sequences of the four calcium-binding protein genes were identified by GenBank database searching using BLASTx. Their homologs are listed in Table 2. The translated protein sequences of calcium-binding proteins were screened in ScanProsite for signature domain analyses (de Castro et al., 2006). The proteins CETN (Figure 1A) and CALM (Figure 1B) both include four E-F hand motifs, which primarily consist of α-helixes. Four calcium binding sites from CALM were located by SWISS MODEL (Figure 1B) while no obvious calcium-binding sites were predicted in CETN (Figure 1A). Protein CALR and CANX also demonstrated similarities in 3D structures (Figures 1C,D). Each one of them had a long low complexity fragment in protein sequence. In high complexity areas, β-sheets are the major secondary structures in both CALR and CANX (Figures 1C,D). No clear calcium-binding sites were identified from CALR or CANX. The detailed amino acid sequence in each α-helix or β-sheet from each sequence was highlighted in the sequence (Supplementary Figures 1–4).
C. virginica Mantle Cells (CvMCs) in Response to Increased pCO2 Stimulation
A decline in the pHs of cell culture medium was observed with increasing input of CO2 in the cell culture incubator (Figure 2c). The pH levels of media decreased from 7.78 to 7.44 when the atmospheric CO2 level was increased from ambient concentration to 2.5%. A more dramatic decrease of pH values was observed when atmospheric CO2 was over 2.5%. The pH levels of media at 5 and 7.5% CO2 input dropped to 6.85 and 6.43, respectively, and was dropped further to 6.31 when the concentration of CO2 reached 10% (Figure 2c). The CvMCs cultured in 5% CO2 and above demonstrated minimum numbers of cells attached on the bottom of the plate during culture. The proliferation of CvMC cultures was estimated by IF with Ki-67 antibody. Under control conditions, the average proliferating rate of CvMCs was 82 ± 1.6%. With a 2.5% CO2 input, the CvMCs showed an 82 ± 2.0% proliferating rate, which was not significantly different than cells in control conditions (p = 0.92, Figure 2d). Cell apoptosis was also estimated using caspase 3 (Casp3) as a marker. There were 23 ± 3.3% Casp3 positive CvMCs in control conditions while 20 ± 2.6% CvMCs were Casp3 positive in 2.5% CO2 conditions (p = 0.50, Figure 2e). Although there was nearly 3% difference in cell apoptosis between control and 2.5% CO2 treated cells, the difference was not statistically different (p = 0.50). This suggested that stresses from CO2 under 2.5% in the atmosphere did not cause lethal effect on CvMCs.
Expression of Calcium-Binding Proteins in CvMCs in Elevated Atmospheric CO2
Expressions of the four calcium-binding protein encoding genes at the mRNA level were all significantly upregulated based on qPCR analysis (Figure 3a). Expressions of calm in CvMCs at mRNA level were upregulated by 1.9 (p = 0.031) and 1.8 (p = 0.049) fold under 1 and 2.5% CO2 conditions, respectively, compared to control condition with an ambient CO2 level. Upregulation of calt expression was also observed at the mRNA level. The amount of mRNAs in cells cultivated at 1 and 2.5% CO2 treatment was upregulated by 4.7 (p = 0.042) and 3.4 (p = 0.041) fold, respectively compared to control cells. (Figure 3a). The calr expression displayed upregulation at both the mRNA and protein levels in cells stimulated with elevated pCO2. Although mRNAs of calr were only upregulated by 2.1 fold (p = 0.031) with 1% CO2 stimulation, a 9.7 (p = 0.006) fold increase of mRNA level was seen in CvMCs at 2.5% CO2 conditions (Figure 3a). The elevation of canx was found at the mRNA level in CO2 treated CvMCs as well. However, only the 2.6-fold increase in 1% CO2 induced cells was statistically significant (p = 0.018, Figure 3a), while the expression levels of canx in cells treated with 2.5% CO2 was not statistically different than in control cells (p = 0.283).
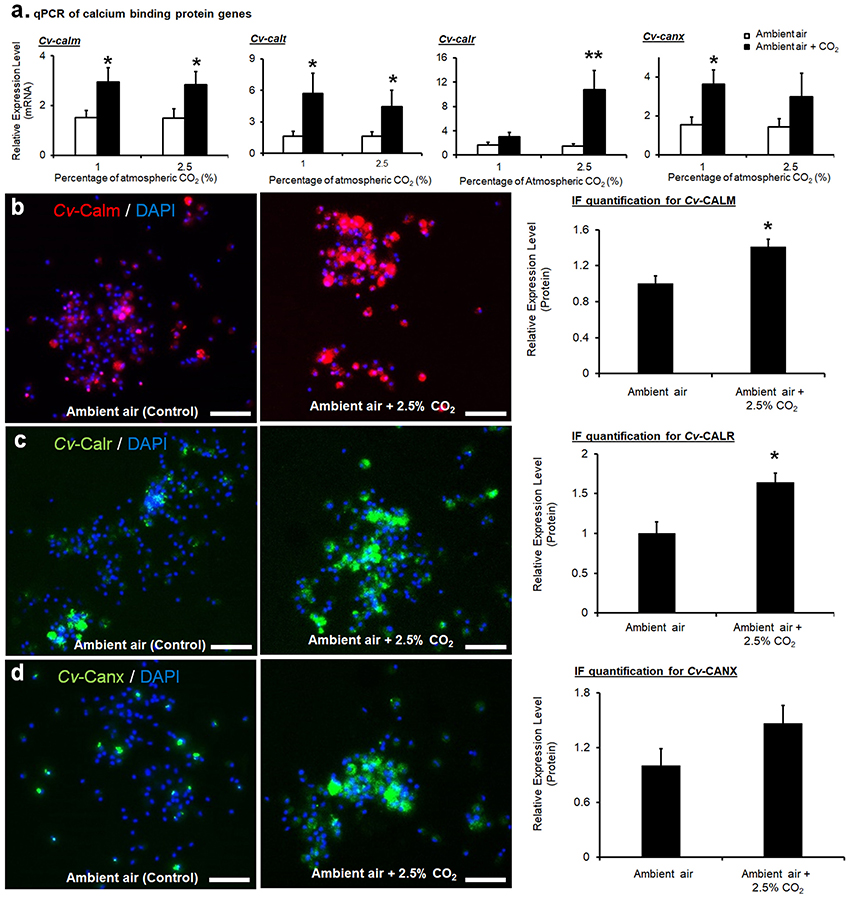
Figure 3. Expressions of calcium-binding proteins in CvMCs. The relative amount of mRNA of cetn, calm, calr, and canx, was quantified and compared between ambient air and increased CO2 treatments (n = 16–24), (a). Protein levels of CALM (b), CALR (c), and CANX (d) with control and 2.5% increase of CO2 treatments were estimated using IF, and the results were quantified using ImageJ (n = 8). All treatments were applied on cells for 24 h. *0.01 < p < 0.05; **p < 0.01. Scale bar: 50 μm. Error bar: standard error of mean.
Similarly, higher calm expression at the protein level was also detected in CvMCs cultivated in 2.5% atmospheric CO2 compared to cells cultured in ambient CO2 conditions (41% more in 2.5% CO2 treated cells than in control cells, p = 0.040, Figure 3b). Accordingly, a 64% (p = 0.034) elevation in CALR protein was induced by 2.5% CO2 (Figure 3c). Statistical analyses also did not show any significant difference of CANX protein production in CvMCs responding to 2.5% CO2 stimulation compared to control condition (Figure 3d, p = 0.273).
Production of Calcium-Binding Proteins in Larval C. virginica Under Increased pCO2
In the larval C. virginica development assay, elevated pCO2 treatment (1,000 ppm) in water decreased the water pH by 0.15 [F(1, 28) = 23.49, p < 0.0001] compared to the current pCO2 conditions. No significant changes in total alkalinity, temperature, and salinity were observed in elevated pCO2. Additionally, there was also no significant difference in the mortality and shell length of larval C. virginica between the two different pCO2 treatments. Maximum shell length was 69.9 ± 0.75 μm at 400 ppm pCO2 vs. 69.8 ± 0.60 μm at 1,000 ppm pCO2, F(1, 28) = 0.02, p = 0.89. However, differential expression of calm, calr, and canx at the protein level was identified between larvae in different pCO2 treatments. Productions of CALM, CALR, and CANX in larvae cultivated at 1,000 pCO2 concentrations were elevated by 38% (p = 0.024, Figure 4a), 52% (p = 0.033, Figure 4b), and 26% (p = 0.025, Figure 4c), respectively. According to the statistical analysis (Figure 4d).
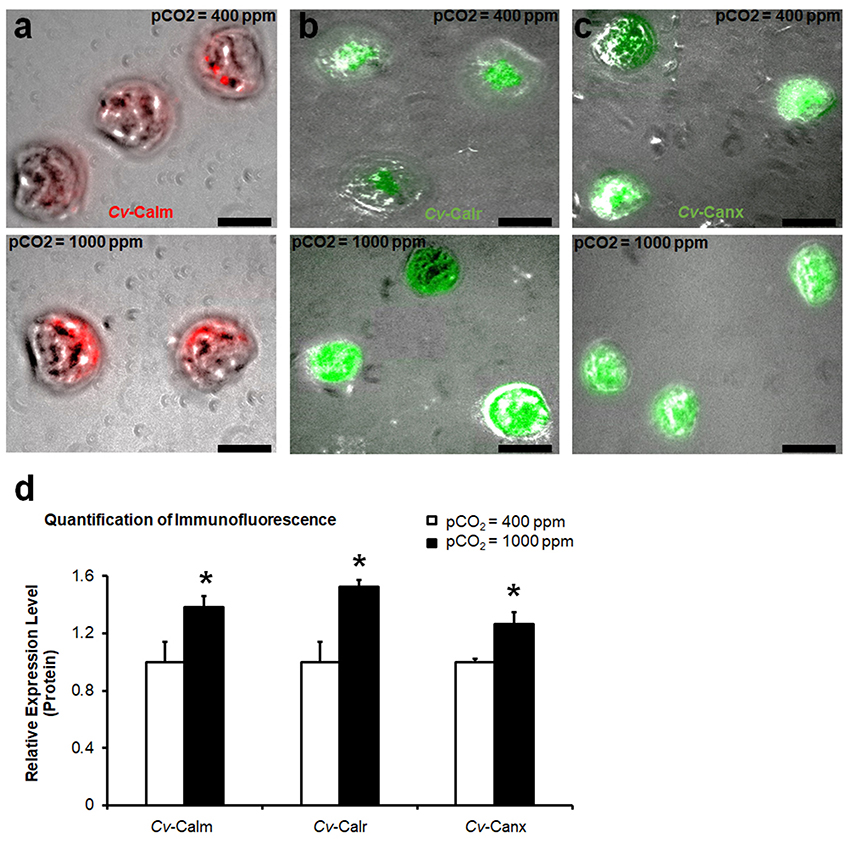
Figure 4. Production of calcium-binding proteins in larval C. virginica. The relative amount of protein levels of CALM (a), CALR (b), and CANX (c) in larvae with control (400 ppm pCO2) and increased pCO2 (1,000 ppm pCO2) treatments were estimated using IF and the results were quantified using ImageJ (n = 8), (d). Samples were collected at 48-h post treatment. *0.01 < p < 0.05. Scale bar: 50 μm.
Discussion
Populations of C. virginica endemic to Louisiana's Gulf coast flourish in unique estuarine habitats, differing from ecosystems along the Atlantic coast, that receive extremely high volumes of freshwater discharge, variable by season, from the Mississippi and Atchafalaya River systems (Guo et al., 2012). In addition to high water discharge, massive river basins input large nutrient loads and sediment into Louisiana estuaries exported from land-based runoff, contributing to alterations in carbonate chemistry and pH levels within the ecosystem (Doney et al., 2009). Oysters living in this environment may have adapted to the stressful conditions through alteration in genomics and epigenetic regulation. Responses of oysters to experimental conditions will be also determined by a variety of factors, including the evolutionary history of the source population and epigenetic changes due to previous environmental conditions. The oysters used in this study were maintained long term at the LA State Oyster Hatchery in controlled conditions similar to the average observed at the site for the source population. This minimized the alteration in genomic responses and epigenetic regulation to the long-term stresses.
The goal of our study was to understand the responses of the calcium-binding protein genes to OA during shell development in bivalves. Two different models were implemented to test our hypothesis stating that the regulation of calcium-binding protein genes involved in biomineralization are influenced by OA. The results obtained from the larval development assay clearly demonstrated the different protein productions CALM, CALR, and CANX. However, due to the difference between individuals of larval oysters, no strong conclusions were drawn based on the profile changes of mRNA levels of the calcium-binding protein genes exposed to OA stress. However, the results obtained from larval experiment were validated by the mantle cell culture assay, which suggested that the in vitro cell culture assay can be a sufficient tool for in vivo oyster studies.
Efforts were previously made to isolate and culture cells from various tissues of mollusk species, however, there was limited success in the past two decades. Only one cell line has been established from Biomphalaria glabrata, a freshwater snail (Hansen, 1976). Mantle tissue in mollusks is the key tissue involved in shell formation and was primarily used for studies of the biominearlization process. Previously, establishment of mantle cell primary cultures has been accomplished in Mytilus galloprovincialis (Cornet, 2006), P. fucata (Awaji and Suzuki, 1998), Haliotis tuberculate (Poncet et al., 2000; Sud et al., 2001), and Dreissena polymorpha (Quinn et al., 2009). Despite difficulties in maintaining the cells in sterile conditions, mantle cells with high proliferative capacity and viability can be used as a reliable tool for biomineralization studies in bivalves. The CvMCs cultured in the current study showed high proliferating rates in both ambient and increased environmental CO2 conditions. Cell apoptosis was not induced by increased atmospheric CO2 environments. Compared to the high mortality rate of larval C. virgnica cultured during the experiment, the cell culture assay provided a more efficient and convenient method to study the impact of environmental stressors on mollusk biomineralization.
In our larval developmental study, no significant difference was identified in mortality, growth, or morphology of the larvae grown in ambient and elevated pCO2 conditions. However, the substantial alteration of calcium-binding protein expression suggested that the process of biomineralization was affected by increased acidification. Increased production of calcium-binding proteins and transportation of calcium ions to overcome the adverse effects of elevated pCO2 results in higher energy demands in calcifying mollusks (Melzner et al., 2011; Thomsen et al., 2013). In addition, the shell composition as well as metabolic rates of larvae may be dramatically modified in stressful conditions. Further identifying these alterations to biomineralization in response to environmental stressors will generate a clearer understanding of the impact of OA on mollusk larval development.
The responsive expression profiles of the selected calcium-binding proteins supported our hypothesis that OA manipulates the regulation of biomineralization related genes, which are involved in calcium deposition during mollusk larval development. The four targeted calcium-binding proteins in the current study were selected according to previous reports, all of which demonstrated critical involvement of these proteins in shell formation. Based on the findings from the current study, CETN and CALM both have multiple calcium-binding domains, E-F hands, in their 3-D structures, suggesting strong association with calcium transportation. Identification of the E-F hands and 3-D structures of C. virginica CETN in this study indicated that this protein may play a similar role in biomineralization as by calmodulin. Unlike CALM and CETN, no E-F hand signature domains for calcium-binding in the 3-D structures were located in CALR and CANX. However, previous studies detected a domain near the C-terminal of CALR and CANX, which demonstrated high affinity to calcium ions (Corbett and Michalak, 2000). These two calcium-binding proteins were able to maintain a high calcium concentration in the endoplasmic reticulum through modulating Ca2+ ATPase activity (Camacho et al., 2013). Maintenance of high calcium concentration is essential to calcium capture and deposition, and necessary for shell calcification in mollusks. Therefore, CALR and CANX, in addition to CETN and CALM, are hypothesized to be heavily involved in mollusk biomineralization.
In this study, the relative expression of four newly sequenced shell formation-related genes and the three linked calcium-binding proteins were evaluated after 24 h of incubation in elevated CO2 concentrations. Similarly, a previous study found gene expression upregulated in cetn, calm and various other proteins associated with calcification in C. hongkongensis larvae exposed to stressful CO2 conditions (Dineshram et al., 2015). Upregulation in expression of biomineralization-related genes and calcium-binding proteins suggested that the biomineralization mechanisms were still active in mantle cells exposed to elevated CO2 conditions. However, in comparison to normal expression of the target genes in controlled conditions, upregulation in cells cultivated in increased CO2 indicated that there were still impacts to the shell deposition mechanisms. Cells within the mantle tissue of adult C. virginica may have induced expression to combat or compensate for environmental stress. It appears that the results from primary cell culture are complimentary to the larval experiments, both of which were utilized to study the impacts of acidification on the molecular regulation of the biomineralization process. In addition, the cell culture study provides an adequate model for observing the genomic, and possible physiological responses of C. virginica.
Author Contributions
MR, WX, AM, and RE designed the experiment; MR, WX, AM, and JS performed the experiment; MR and WX performed the data analyses; MR, WX, and RE prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Drs. Jerome La Peyre and Sandra Casas Liste from LSU School of Animal Sciences for providing healthy oysters for cell culture in this study. This study was supported by the internal grant of LSU Agricultural Center.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00203/full#supplementary-material
References
Arivalagan, J., Yarra, T., Marie, B., Sleight, V. A., Duvernois-Berthet, E., Clark, M. S., et al. (2017). Insights from the shell proteome: biomineralization to adaptation. Mol. Biol. Evol. 34, 66–77. doi: 10.1093/molbev/msw219
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Arosa, F. A., de Jesus, O., Porto, G., Carmo, A. M., and de Sousa, M. (1999). Calreticulin is expressed on the cell surface of activated human peripheral blood T lymphocytes in association with major histocompatibility complex class I molecules. J. Biol. Chem. 274, 16917–16922. doi: 10.1074/jbc.274.24.16917
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Awaji, M., and Suzuki, T. (1998). Monolayer formation and DNA synthesis of the outer epithelial cells from pearl oyster mantle in coculture with amebocytes. In Vitro Cell. Dev. Biol. Anim. 34, 486–491. doi: 10.1007/s11626-998-0083-0
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Camacho, P., John, L., Li, Y., Paredes, R. M., and Roderick, H. L. (2013). Calnexin and Calreticulin, ER Associated Modulators of Calcium Transport in the ER, in: 2000-2013. Austin, TX: Landes Bioscience.
Coppolino, M. G., and Dedhar, S. (1998). Calreticulin. Int. J. Biochem. Cell Biol. 30, 553–558. doi: 10.1016/S1357-2725(97)00153-2
Corbett, E. F., and Michalak, M. (2000). Calcium, a signaling molecule in the endoplasmic reticulum? Trends Biochem. Sci. 25, 307–311. doi: 10.1016/S0968-0004(00)01588-7
Cornet, M. (2006). Primary mantle tissue culture from the bivalve mollusc Mytilus galloprovincialis: investigations on the growth promoting activity of the serum used for medium supplementation. J. Biotechnol. 123, 78–84. doi: 10.1016/j.jbiotec.2005.10.016
Currey, J. D. (1977). Mechanical properties of mother of pearl in tension. Proc. R. Soc. Lond. B 196, 443–463. doi: 10.1098/rspb.1977.0050
de Castro, E., Sigrist, C. J., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365. doi: 10.1093/nar/gkl124
Dickson, A. G., Sabine, C. L., and Christian, J. R. (2007). “Guide to best practices for ocean CO2 measurements,” in PICES Special Publication 3 (Sidney, BC: North Pacific Marine Science Organization), 73–87.
Dineshram, R., Quan, Q., Sharma, R., Chandramouli, K., Yalamanchili, H. K., Chu, I., et al. (2015). Comparative and quantitative proteomics reveal the adaptive strategies of oyster larvae to ocean acidification. Proteomics 15, 4120–4134. doi: 10.1002/pmic.201500198
Doney, S. C., Fabry, V. J., Feely, R. A., and Kleypas, J. A. (2009). Ocean acidification: the other CO2 problem. Ann. Rev. Mar. Sci. 1, 169–192. doi: 10.1146/annurev.marine.010908.163834
Friedberg, F. (1988). Calcium binding protein families: the ‘E-F Hand’ family. Biochem. Edu. 16, 35–36. doi: 10.1016/0307-4412(88)90017-9
Gazeau, F., Gattuso, J. P., Greaves, M., Elderfield, H., Peene, J., Heip, C. H., et al. (2011). Effect of carbonate chemistry alteration on the early embryonic development of the Pacific oyster (Crassostrea gigas). PLoS ONE 6:e23010. doi: 10.1371/journal.pone.0023010
Guo, X., Cai, W., Huang, W., Wang, Y., Chen, F., Murrell, M. C., et al. (2012). Carbon dynamics and community production in the Mississippi River plume. Limnol. Oceanogr. 57, 1–17. doi: 10.4319/lo.2012.57.1.0001
Hansen, E. L. (1976). “A cell line from embryos of Biomphalaria glabrata (Pulmonata): Establishment and characteristics,” in Invertebrate Tissue Culture: Research Applications, ed K. Maramorosch (New York, NY: Academic Press), 75–97.
Harper, E. M. (2000). Are calcitic layers an effective adaptation against shell dissolution in the Bivalvia? J. Zool. 251, 179–186. doi: 10.1111/j.1469-7998.2000.tb00602.x
Hattan, S. J., Laue, T. M., and Chasteen, N. D. (2001). Purification and characterization of a novel calcium-binding protein from the extrapallial fluid of the mollusc, Mytilus edulis. J. Biol. Chem. 276, 4461–4468. doi: 10.1074/jbc.M006803200
Huang, J., Zhang, C., Ma, Z., Xie, L., and Zhang, R. (2007). A novel extracellular EF-hand protein involved in the shell formation of pearl oyster. Biochim. Biophys. Acta 1770, 1037–1044. doi: 10.1016/j.bbagen.2007.03.006
Ip, Y. K., Loong, A. M., Hiong, K. C., Wong, W. P., Chew, S. F., Reddy, K., et al. (2006). Light induces an increase in the pH of and a decrease in the ammonia concentration in the extrapallial fluid of the giant clam Tridacna squamosa. Physiol. Biochem. Zool. 79, 656–664. doi: 10.1086/501061
IPCC (2007). Climate Change 2007: Synthesis Report. Contribution of Working Groups, I., II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. eds R. K. Pachauri and A. Reisinger (Geneva: Intergovernmental Panel on Climate Change).
Kiefer, F., Arnold, K., Künzli, M., Bordoli, L., and Schwede, T. (2009). The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392. doi: 10.1093/nar/gkn750
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, S., Xie, L., Ma, Z., and Zhang, R. (2005). cDNA cloning and characterization of a novel calmodulin-like protein from pearl oyster Pinctada fucata. FEBS J. 272, 4899–4910. doi: 10.1111/j.1742-4658.2005.04899.x
Li, S., Xie, L., Zhang, C., Zhang, Y., Gu, M., and Zhang, R. (2004). Cloning and expression of a pivotal calcium metabolism regulator: calmodulin involved in shell formation from pearl oyster (Pinctada fucata). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 138, 235–243. doi: 10.1016/j.cbpc.2004.03.012
Li, X. X., Yu, W. C., Cai, Z. Q., He, C., Wei, N., Wang, X. T., et al. (2016). Molecular cloning and characterization of full-length cDNA of calmodulin gene from Pacific Oyster Crassostrea gigas. Biomed Res. Int. 2016:5986519. doi: 10.1155/2016/5986519
Liu, J., Yang, D., Liu, S., Li, S., Xu, G., Zheng, G., et al. (2015). Microarray: a global analysis of biomineralization-related gene expression profiles during larval development in the pearl oyster, Pinctada fucata. BMC Genomics 16:325. doi: 10.1186/s12864-015-1524-2
Mann, S. (1983). Mineralization in biological systems. Struct. Bond. 54, 125–174. doi: 10.1007/BFb0111320
Marin, F., Le Roy, N., and Marie, B. (2012). The formation and mineralization of mollusk shell. Front. Biosci. (Schol. Ed). 4, 1099–1125. doi: 10.2741/s321
Melzner, F., Stange, P., Trübenbach, K., Thomsen, J., Casties, I., Panknin, U., et al. (2011). Food supply and seawater pCO2 impact calcification and internal shell dissolution in the blue mussel Mytilus edulis. PLoS ONE 6:e24223. doi: 10.1371/journal.pone.0024223
Michalak, M., Robert Parker, J. M., and Opas, M. (2002). Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium 32, 269–278. doi: 10.1016/S0143416002001884
Mount, A. S., Wheeler, A. P., Paradkar, R. P., and Snider, D. (2004). Hemocyte-mediated shell mineralization in the eastern oyster. Science 304, 297–300. doi: 10.1126/science.1090506
Nishi, R., Sakai, W., Tone, D., Hanaoka, F., and Sugasawa, K. (2013). Structure-function analysis of the EF-hand protein centrin-2 for its intracellular localization and nucleotide excision repair. Nucleic Acids Res. 41, 6917–6929. doi: 10.1093/nar/gkt434
Orr, J. C., Fabry, V. J., Aumont, O., Bopp, L., Doney, S. C., Feely, R. A., et al. (2005). Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437, 681–686. doi: 10.1038/nature04095
Parker, L. M., Ross, P. M., and O'connor, W. A. (2010). Comparing the effect of elevated pCO2 and temperature on the fertilization and early development of two species of oysters. Mar. Biol. 157, 2435–2452.
Peng, K., Liu, F., Wang, J., and Hong, Y. (2017). Calmodulin highly expressed during the formation of pearl sac in freshwater pearl mussel (Hyriopsis schlegelii). Thalassas. doi: 10.1007/s41208-017-0054-x
Poncet, J., Serpentini, A., Thiébot, B., Villers, C., Bocquet, J., Boucaud-Camou, E., et al. (2000). In vitro synthesis of proteoglycans and collagen in primary cultures of mantle cells from the Nacreous Mollusk, Haliotis tuberculata: a new model for study of molluscan extracellular matrix. Mar. Biotechnol. 2, 387–398. doi: 10.1007/PL00021685
Quinn, B., Costello, M. J., Dorange, G., Wilson, J. G., and Mothersill, C. (2009). Development of an in vitro culture method for cells and tissues from the zebra mussel (Dreissena polymorpha). Cytotechnology 59, 121–134. doi: 10.1007/s10616-009-9202-3
Rao, X., Huang, X., Zhou, Z., and Lin, X. (2013). An improvement of the 2∧(−deltadeltaCT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 3, 71–85.
Sabine, C. L., Feely, R. A., Gruber, N., Key, R. M., Lee, K., Bullister, J. L., et al. (2004). The oceanic sink for anthropogenic CO2. Science 305, 367–371. doi: 10.1126/science.1097403
Sambrook, J., and Russell, D. W. (2006a). Preparation and transformation of competent, E. coli using Calcium Chloride. CSH Protoc. 2006:320. doi: 10.1101/pdb.prot3932
Sambrook, J., and Russell, D. W. (2006b). Recovery of DNA from agarose and polyacrylamide gels: electroelution into dialysis bags. CSH Protoc. 2006:207. doi: 10.1101/pdb.prot4023
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Somogyi, E., Petersson, U., Hultenby, K., and Wendel, M. (2003). Calreticulin–an endoplasmic reticulum protein with calcium-binding activity is also found in the extracellular matrix. Matrix Biol. 22, 179–191. doi: 10.1016/S0945-053X(02)00117-8
Su, J., Liang, X., Zhou, Q., Zhang, G., Wang, H., Xie, L., et al. (2013). Structural characterization of amorphous calcium carbonate-binding protein: an insight into the mechanism of amorphous calcium carbonate formation. Biochem. J. 453, 179–186. doi: 10.1042/BJ20130285
Sud, D., Doumenc, D., Lopez, E., and Milet, C. (2001). Role of water-soluble matrix fraction, extracted from the nacre of Pinctada maxima, in the regulation of cell activity in abalone mantle cell culture (Haliotis tuberculata). Tissue Cell 33, 154–160. doi: 10.1054/tice.2000.0166
Sueyoshi, T., McMullen, B. A., Marnell, L. L., Du Clos, T. W., and Kisiel, W. (1991). A new procedure for the separation of protein Z, prothrombin fragment 1.2 and calreticulin from human plasma. Thromb. Res. 63, 569–575. doi: 10.1016/0049-3848(91)90184-X
Takagi, R., and Miyashita, T. (2010). Prismin: a new matrix protein family in the Japanese pearl oyster (Pinctada fucata) involved in prismatic layer formation. Zool. Sci. 27, 416–426. doi: 10.2108/zsj.27.416
Takeuchi, T. (2017). Molluscan genomics: implications for biology and aquaculture. Curr. Mol. Biol. Rep. 3, 297–305. doi: 10.1007/s40610-017-0077-3
Thomsen, J., Casties, I., Pansch, C., Körtzinger, A., and Melzner, F. (2013). Food availability outweighs ocean acidification effects in juvenile Mytilus edulis: laboratory and field experiments. Glob. Chang. Biol. 19, 1017–1027. doi: 10.1111/gcb.12109
Waldbusser, G. G., Brunner, E. L., Haley, B. A., Hales, B., Langdon, C. J., and Prahl, F. G. (2013). A developmental and energetic basis linking larval oyster shell formation to acidification sensitivity. Geophys. Res. Lett. 40, 2171–2176. doi: 10.1002/grl.50449
Waldbusser, G. G., Hales, B., Langdon, C. J., Haley, B. A., Schrader, P., Brunner, E. L., et al. (2014). Saturation-state sensitivity of marine bivalve larvae to ocean acidification. Nat. Clim. Change 5, 273–280. doi: 10.1038/nclimate2479
Weiss, I. M., Kaufmann, S., Mann, K., and Fritz, M. (2000). Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata. Biochem. Biophys. Res. Commun. 267, 17–21. doi: 10.1006/bbrc.1999.1907
Yan, Z., Fang, Z., Ma, Z., Deng, J., Li, S., Xie, L., et al. (2007). Biomineralization: functions of calmodulin-like protein in the shell formation of pearl oyster. Biochim. Biophys. Acta 1770, 1338–1344. doi: 10.1016/j.bbagen.2007.06.018
Zeebe, R. E. (2012). History of seawater carbonate chemistry, atmospheric CO2, and ocean acidification. Annu. Rev. Earth Planet. Sci. 40, 141–165. doi: 10.1146/annurev-earth-042711-105521
Keywords: biomineralization, Crassostrea virginica, ocean acidification, calcium-binding proteins, mantle cell culture
Citation: Richards M, Xu W, Mallozzi A, Errera RM and Supan J (2018) Production of Calcium-Binding Proteins in Crassostrea virginica in Response to Increased Environmental CO2 Concentration. Front. Mar. Sci. 5:203. doi: 10.3389/fmars.2018.00203
Received: 23 December 2017; Accepted: 25 May 2018;
Published: 12 June 2018.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Gary H. Dickinson, The College of New Jersey, United StatesMarta Gomez-Chiarri, University of Rhode Island, United States
Copyright © 2018 Richards, Xu, Mallozzi, Errera and Supan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Xu, d3h1QGFnY2VudGVyLmxzdS5lZHU=
 Mackenzie Richards1,2
Mackenzie Richards1,2 Wei Xu
Wei Xu