- LOHABE, Department of Oceanography, Chonnam National University, Gwangju, South Korea
Phagotrophy of the harmful benthic dinoflagellates of the genus Ostreopsis has long been inferred based on observations of food particles present inside cells, but the prey has not yet been identified. This study aimed to investigate the seasonal dynamics of benthic dinoflagellates Ostreopsis spp. in temperate Korean coastal sites, with special emphasis on their phagotrophy. Further, prey species were identified by extracting the ingested food particles from single Ostreopsis cells and determining their rbcL and psaA gene sequences. High concentration of Ostreopsis cells was observed between June and October at all sites, when the water temperatures were higher than 19°C, exhibiting a marked temporal seasonality. The percentage of Ostreopsis cells containing ingested food particles exhibited large spatial and temporal variations among sampling sites, ranging from undetectable level to 29.5%, and was not always associated with Ostreopsis cell abundance. Phylogenetic analyses performed using both plastid-encoded rbcL and psaA genes revealed that all sequences obtained from the ingested food particles of Ostreopsis cells grouped within the class Florideophyceae, Rhodophyta. Our result clearly demonstrates that Ostreopsis species consume various macroalgae from Rhodophyta, but not protists, which have long been thought to be the potential prey. The results of this study provide a basis for better understanding the mixotrophic behavior and nutritional ecology of the harmful benthic dinoflagellate Ostreopsis species.
Introduction
The genus Ostreopsis belonging to the family Ostreopsidaceae (Dinophyceae) has a worldwide distribution from tropical to temperate marine waters (Rhodes, 2011; Parsons et al., 2012; Accoroni and Totti, 2016). At present, the genus Ostreopsis includes 11 species with type species of O. siamensis (Schmidt, 1901; Parsons et al., 2012; Accoroni et al., 2016; Verma et al., 2016), of which several species are known to produce toxins of the palytoxin group that could cause serious human and environmental health problems (e.g., Yasumoto et al., 1987; Ukena et al., 2001; Tichadou et al., 2010). Ostreopsis species have been found in various of habitats, including on macroalgae, hard substrates (e.g., rocks, sand, and mollusk shells), and in the water column (Schmidt, 1901; Faust et al., 1996; Vila et al., 2001; Laza-Martinez et al., 2011). They often occur in association with other toxic or potentially toxic benthic dinoflagellates Gambierdiscus spp., Amphidinium spp., Prorocentrum spp., and Coolia monotis (Penna et al., 2005; Aligizaki and Nikolaidis, 2006; Parsons et al., 2012).
Although great progress has been made in the study of Ostreopsis biology, ecology, and toxicology over the past several years, some aspects about the nutritional ecology associated with this genus still remain unclear. For example, Faust and her colleagues (Faust and Morton, 1995; Faust et al., 1996; Faust, 1998) reported reddish round inclusions inside Ostreopsis cells collected in Belize through light microscopic observations and speculated that they were the ingested prey that might originate from cyanobacteria, centric diatoms, ciliates, and small microalgae, although the feeding behavior was not reported. Since then, some studies (e.g., Aligizaki and Nikolaidis, 2006; Selina and Orlova, 2010; Accoroni et al., 2014) also observed the presence of red bodies in Ostreopsis cells collected in field samples, and Fraga et al. (2012) suggested that the presence of these red bodies might be related to the mixotrophic behavior of Ostreopsis species. Very recently, Almada et al. (2017) also reported that only a small percentage (0.4%) of O. cf. ovata cells in Brazil showed evidence of phagotrophy by using epifluorescence microscopy with DAPI and LysoSensor staining, although the prey inside the cells were not discernible.
The present study aimed to investigate the seasonal dynamics of the benthic dinoflagellates Ostreopsis spp.in temperate Korean coastal waters, with special emphasis on their phagotrophy. To further identify the prey, the ingested food particles were extracted from single Ostreopsis cells and their rbcL and psaA genes sequences were determined. These chloroplast-encoded genes have often been used to identify food particles inside protists such as dinoflagellates and ciliates (e.g., Hackett et al., 2003; McManus et al., 2004; Nishitani et al., 2010; Kim et al., 2012; Fawcett and Parrow, 2014). For example, McManus et al. (2004) found through rbcL gene sequencing from chloroplasts found in the marine ciliates Strombidium oculatum and S. stylifer that the ciliates ingest ulvaceous green macroalgal swarmers and retain their chloroplasts. The psbA gene has also been applied to infer the origin and diversity of chloroplasts found in the mixotrophic dinoflagellate Dinophysis (e.g., Hackett et al., 2003; Nishitani et al., 2010; Kim et al., 2012). Thus, the application of these chloroplast-encoded gene sequences on red bodies found in Ostreopsis would be expected to provide a clue about the identity of prey.
Materials and Methods
Sampling and Sample Treatment
Macroalgal samples were collected monthly from August 2016 to December 2017 during low tide from 4 sites on the Korean southeastern coast (Figure 1). The study area is among the regions within the Korean peninsula, where the benthic and epiphytic dinoflagellate Ostreopsis has been previously reported to occur (Baek, 2012), and is known to be seasonally affected by the Tsushima Warm Current (Park et al., 2013), which could be responsible for introduction of the species. Therefore, the area was considered as appropriate sampling sites for monitoring the seasonal dynamics of the species and its phagotrophy. Additional planktonic Ostreopsis cells were collected using a plankton net (20 μm) from Chuja Island on September 14, 2017 and Jeju Island on November 21, 2017. During the sampling, surface water temperature and salinity were measured using a Yellow Spring Instrument (YSI, Ohio, USA). Two macroalgae of the most common macroalgal species at each sampling time were chosen; each was collected in duplicate (Table S1) and transferred to a zip lock bag along with some adjacent seawater. After the samples were transported to the laboratory, they were vigorously shaken to detach the epiphytic Ostreopsis cells, filtered through a 200 μm mesh to remove large particles, and then concentrated using a 20 μm mesh. The concentrates on the second mesh were resuspended in filtered seawater (50 ml volume). The aliquots (5 ml) of the 50 ml volume were fixed with glutaraldehyde (final 1%) to determine the abundance of Ostreopsis cells. Macroalgal thalli were weighed to determine fresh weight (g fw).
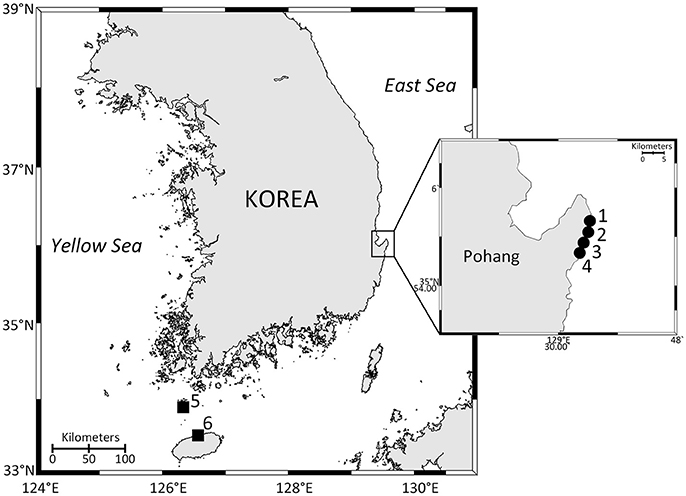
Figure 1. Map of the study area showing the sampling sites. 1, Gangsa; 2, Samjeong; 3, Guryongpo; 4, Hajeong; 5, Chuja Island; 6, Jeju harbor. Closed circles are monitoring sites. Closed squares are sites where additional Ostreopsis cells were isolated from the concentrated planktonic samples.
Determination of Ostreopsis Abundance
The cell abundance was determined by scanning all cells in a Sedgwick-Rafter counting chamber in duplicate at 100x magnification using a light microscope (Axiostar plus, Carl Zeiss Inc., Hallbergmoos, Germany). For determination of Ostreopsis cells containing the ingested food particles, all Ostreopsis cells encountered in the chamber were scored as cells containing food particles or those without food particles. Cell abundance was expressed as the number of cells per gram of fresh weight of macrophyte (cells g−1 fw).
Light Microscopy
Live Ostreopsis cells with ingested prey were individually isolated using a drawn Pasteur glass pipette and loaded on a slide glass. Light micrographs were taken at 1000 × magnification by using an AxioCam HRc (Carl Zeiss Inc., Hallbergmoos, Germany) photomicrographic system coupled to the Axio Imager A2 equipped with differential interference contrast.
DNA Extraction and PCR Amplification
A total of 60 Ostreopsis cells containing the ingested prey were isolated from the remaining 45 ml concentrated samples by using drawn Pasteur glass pipettes under an inverted microscope (AX10, Carl Zeiss Inc., Hallbergmoos, Germany) or a stereoscope (Discovery. V8, Carl Zeiss Inc., Hallbergmoos, Germany) (Table 1). First, the isolated single cells were cleaned using filtered seawater several times (at least five times) and then soaked in distilled water in order to break the cells. Shortly after cell breakage, only food particles were picked and placed into a PCR tube and stored at a −80°C for 1 day. The samples were thawed at room temperature for 10 min, and DNA was extracted using Chelex 100 resin (100–200 mesh, sodium form; Bio-Rad Laboratories, Hercules, CA, USA). Next, 20 μl of 10% Chelex solution was added to the samples and incubated at 95°C for 1 h. Subsequently, the PCR tubes were centrifuged at 8,000 rpm at 4°C for 1 min, and then the supernatant in the PCR tube was transferred to a new tube and was used as a template. Polymerase chain reaction (PCR) amplifications were performed using some primers for psaA and rbcL genes. Primer pairs for amplification and sequencing of each gene were as follows: for psaA, psaA130F-psaA1600R (Yoon et al., 2002); and for rbcL, two primer sets F57-R753 and F321-RrbcS start (Freshwater and Rueness, 1994). Each gene was amplified using DiastarTM Taq DNA polymerase (SolGent Co., Daejeon, Korea) in the following reaction mixture: 2.5 μl of 10X Taq reaction buffer (containing 25 mM MgCl2), 0.5 μl of dNTP mix (10 mM), 1 μl of each primer (10 pmole μl−1), 0.125 μl of Taq DNA polymerase (5 U μl−1), and 3 μl genomic DNA for a 25 μl reaction. The PCR for psaA was run as follows: 2 min at 95°C, followed by 39 cycles of 20 s at 95°C, 40 s at 50°C, 1 min 30 s at 72°C, and a final incubation for 5 min at 72°C. The PCR for rbcL was run as above, except for 1 min (for primer set F57-R753) and 1 min 40 s (for primer set F321-RrbcS start set) instead of 1 min 30 s at 72°C. The PCR products were analyzed using electrophoresis on 1% agarose gels and visualized under UV illumination. Subsequently, amplified PCR products were purified using a PCR purification kit (Bioneer, Daejeon, Korea) and sequenced with primers (psaA130F and psaA1600R for psaA gene, F57, F321, R753, and RrbcS start for rbcL gene) by using a Big-Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3730xl Analyzer (Applied Biosystems, Foster City, CA, USA), according to manufacturer's protocols at Cosmogenetech Corp. in Seoul, Korea. The amplicons were sequenced until at least double stranded coverage was reached. ContigExpress (Vector NTI version 10.1, Invitrogen, NY, USA) was used to assemble the individual sequence reads and edit out low quality regions. The assembled sequences were verified by comparison by using BLASTN search in the NCBI database and deposited in GenBank under the accession numbers MH200821-MH200875 for rbcL and MH200876-MH200911 for psaA.

Table 1. Summary of sampling sites, dates, and plastid genes sequenced for the ingested food particles within Ostreopsis cells isolated from field samples during this study.
Phylogenetic Analysis
The sequences of psaA and rbcL genes of food particles were aligned with those of Rhodophyta, Cryptophyta, Stramenopiles, Haptophyta, Chlorophyta, and Glaucophyta obtained from GenBank database by using ClustalW 1.6 (Thompson et al., 1994) and were manually refined using MacGDE 2.4 (Linton, 2005). The final alignments of 2,200 positions for the psaA region and 1,390 positions for the rbcL region were selected. Phylogenies of the psaA and rbcL regions were inferred using the maximum likelihood (ML) and Bayesian inference methods. Modeltest v.3.7 (Posada and Crandall, 1998) was used to select the most appropriate model of substitution for the ML method in PAUP. The GTRGAMMA evolution model was selected from MODELTEST by using RAxML (Stamatakis, 2006), with the rapid bootstrapping option and 2,000 replicates. Bayesian analyses were run using MrBayes v.3.1.1 (Ronquist et al., 2012). GTR + I + G (-lnL = 37064.0586) and GTR + I + G (-lnL = 29063.3262) models were selected for psaA and rbcL datasets, respectively (Posada and Crandall, 1998). Bayesian analysis was performed using MrBayes 3.1.1 (Ronquist et al., 2012) running four simultaneous Monte Carlo Markov Chains for 2,000,000 generations and sampling every 100 generations, following a burn in of 2,000 generations.
Results
Temporal Variations in Total Ostreopsis Abundance and Cells With Ingested Food Particles
During the sampling period, sea surface temperature showed a large temporal variation at 4 sampling sites, ranging from 11.3°C in February 2017 to 29.5°C in August 2016 (Figure 2). Salinity remained relatively constant (34.1 ± 0.2) throughout the study period, except for the first 3 months, when salinity ranged from 29.1 to 32.1.
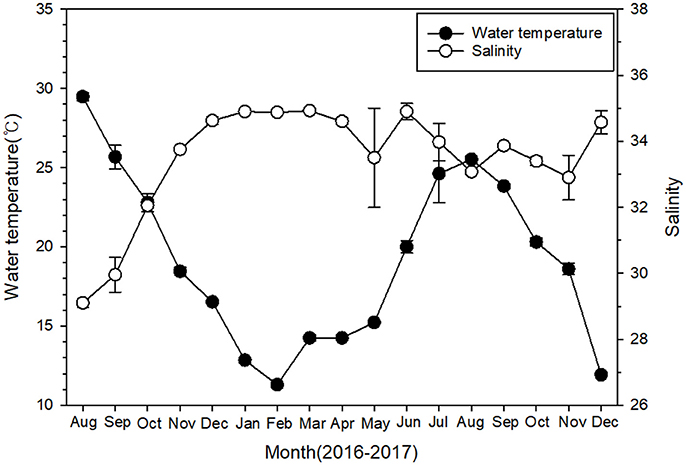
Figure 2. Temporal variations of surface water temperature and salinity at four monitoring sites near Pohang during 2016–2017. Data are given as mean ± SE.
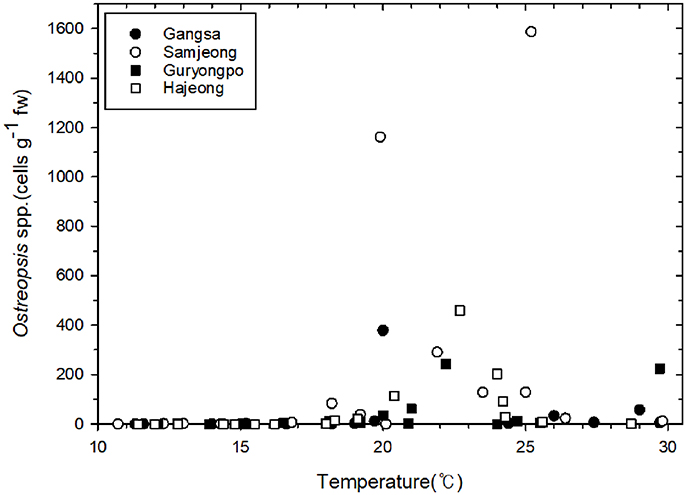
The abundance of the epiphytic dinoflagellate Ostreopsis showed spatial and temporal variations during the study period among sampling sites (Figure 3). Nonetheless, a repeated marked temporal pattern of Ostreopsis abundance was observed at each sampling site, coinciding with warm temperatures. High concentration of Ostreopsis cells was observed between June and October at all sites, when water temperatures were higher than 19°C. The highest cell abundance (1,588 cells g−1 fw) during this study was recorded in July 2017 at Samjeong site, where water temperature was 25°C (Figure 4). Between November and May, when the water temperatures ranged from 11.3° to 18.6°C, Ostreopsis cells were not detected or were present at low concentration less than 80 cells g−1 fw at all sites.
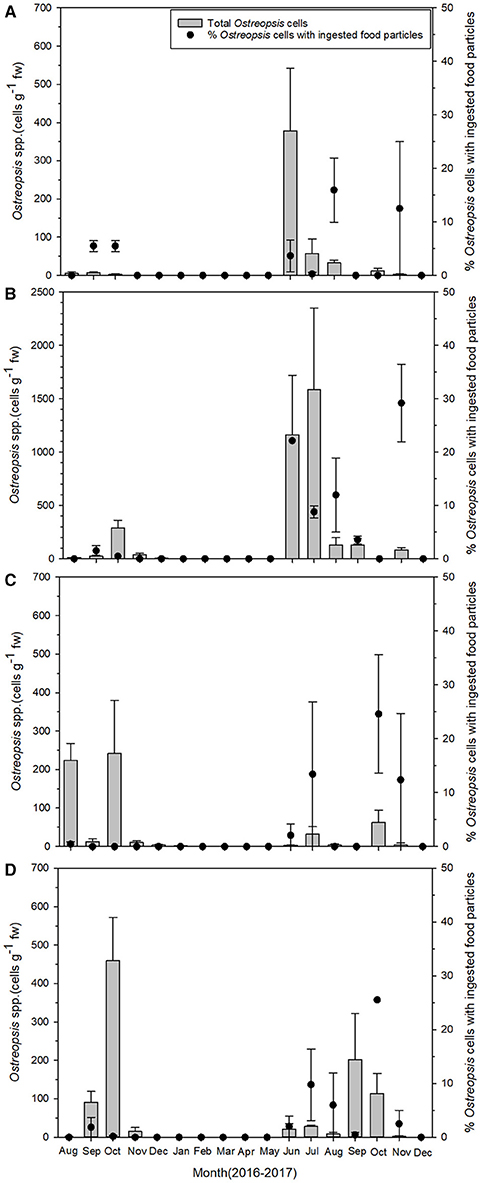
Figure 3. Temporal pattern of total Ostreopsis spp. abundance (gray bars) on macrophytes and the percentage of Ostreopsis cells with ingested food particles (closed circles) during the study period at (A) Gangsa, (B) Samjeong, (C) Guryongpo, and (D) Hajeong. Data are given as mean ± SE.
Ostreopsis cells that contained the ingested food particles were frequently encountered at all sampling sites during this study. The ingested food particles were almost reddish brown (Figures 5A,B). In planktonic Ostreopsis cells isolated from Chuja Island in September 2017, however, food particles having either blue-green or purple color were also observed (Figures 5C,D). The size of the ingested food particles was highly variable; in some cells, the food particles occupied half or two-thirds of Ostreopsis cytoplasm (Figures 5A,B). Occasionally, an Ostreopsis cell with more than two food particles was observed (Figure 5B).
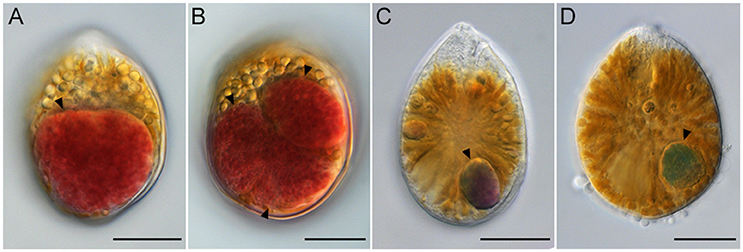
Figure 5. Light micrographs of Ostreopsis cells with the ingested food particles (arrowheads) from field samples. In (B) three food particles are visible inside an Ostreopsis cell. The ingested food particles within Ostreopsis cells were reddish brown (A,B), purple (C), and blue-green (D). Scale bars = 20 μm.
The percentage of Ostreopsis cells containing ingested food particles also exhibited large spatial and temporal variations among sampling sites (Figure 3). The percentage of Ostreopsis with ingested food particles was highly variable, ranging from undetectable level to 29.5% (in November 2017 at Samjeong site), and was not always associated with Ostreopsis cell abundance. For example, in July 2017 at Samjeong site, where the highest cell abundance (1,588 cells g−1 fw) was observed, the percentage of Ostreopsis cells with ingested food particles was 8.8%. In contrast, the highest percentage of Ostreopsis cells with ingested food particles was recorded in November 2017 at Samjeong site where relatively low cell density (83 cells g−1 fw) was observed.
Phylogenetic Analyses of the Food Particles Within Ostreopsis Cells
The sequences of the D8-D10 region of LSU rDNA suggested that all Ostreopsis species in our study area were Ostreopsis sp.1 (data not shown). A total of 36 plastid psaA and 55 rbcL sequences were determined from the food particles within 60 Ostreopsis cells (Table 1). Phylogenetic analyses inferred from psaA alignment of 2,200 sites for 80 taxa and rbcL alignment of 1,390 sites of 115 taxa showed overall similar topologies (Figures 6, 7). Phylogenetic analyses revealed that all sequences obtained from the food particles grouped within Rhodophyta, more specifically class Florideophyceae. The phylogenetic tree based on rbcL amino acid sequences also showed the same result, although the resolution was not better than the tree based on nucleotide sequences (Figure S1). Among 55 rbcL sequences, a total of 20 sequences were closely related to Melanothamnus harveyi (99% similarity), which was the most commonly detected food algal species in July 2017. Seventeen rbcL sequences were related to species belonging to the genera Lithophyllum and Pneophyllum although the sequence similarities were relatively low (86 and 88%, respectively). In addition, several sequences obtained from the food particles were related to various genera and/or species within Florideophyceae, including Acrosorium sp., Campylaephora sp., Peyssonnelia japonica, Spongites sp., Grateloupia sp., and Asparagopsis taxiformis. The rbcL sequence (cell ID OScj0914-16) obtained from blue-green food particle inside an Ostreopsis cell isolated from Chuja Island in September 2017 was closely related to Porolithon onkodes with 99% similarity. Two rbcL sequences (cell ID OScj0914-12 and OScj0914-23) obtained from purple colored food particles inside two Ostreopsis cells isolated from Chuja Island in September 2017 were related to Lithophyllum impressum, but their sequence similarities were relatively low (88%).
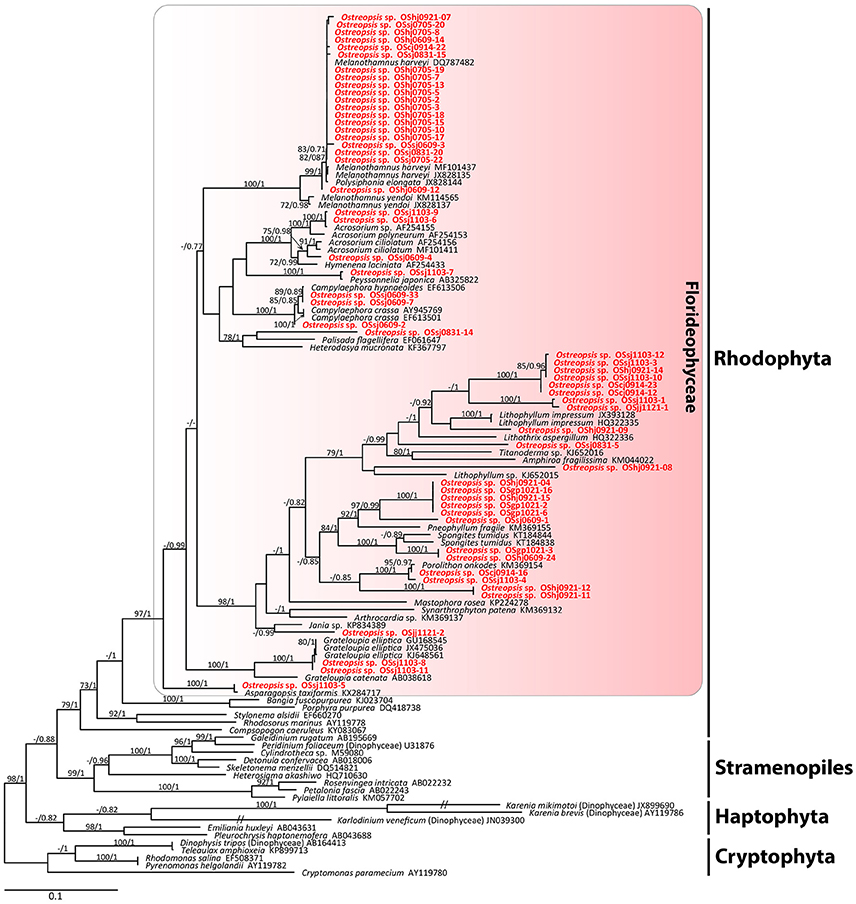
Figure 6. RAxML tree inferred from rbcL sequences of 1,390 sites and representing 115 taxa including Rhodophyta, Cryptophyta, Stramenopiles, and Haptophyta. Sequences from the ingested food particles of Ostreopsis cells isolated from this study are indicated in bold face. Bootstrap value (>70%) from maximum likelihood and a Bayesian posterior probability of 0.7 or greater are indicated at nodes. Double slash marks indicate shrink nodes length (70%).
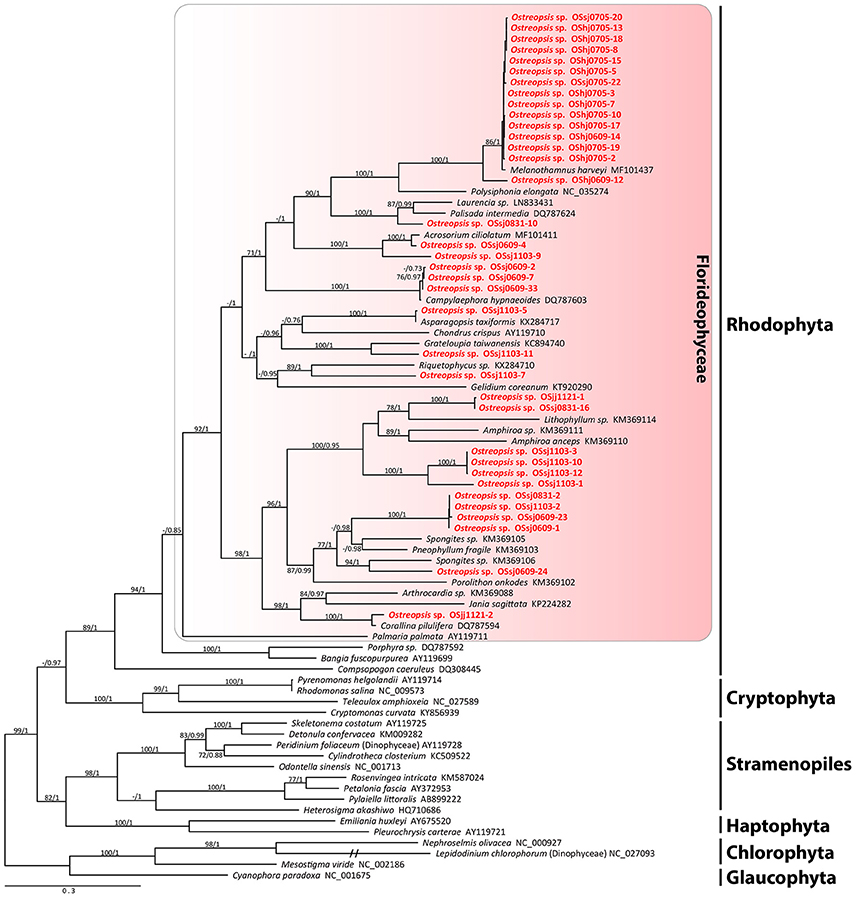
Figure 7. RAxML tree inferred from psaA sequences of 2,200 sites and representing 80 taxa including Rhodophyta, Cryptophyta, Stramenopiles, Haptophyta, Chlorophyta, and Glaucophyta. Sequences from the ingested food particles of Ostreopsis cells isolated from this study are indicated in bold face. Bootstrap value (>70%) from maximum likelihood and a Bayesian posterior probability of 0.7 or greater are indicated at nodes. Double slash marks indicate shrink nodes length (70%).
Discussion
The major findings from this study were that (1) while the total Ostreopsis cell abundance exhibited a marked temporal seasonality, mostly coinciding with warm temperatures, phagotrophy of the dinoflagellate (i.e., the percentages of cells containing ingested food particles) did not, and (2) all the ingested food particles originated from species belonging to class Florideophyceae.
While the seasonality of the epiphytic dinoflagellates Ostreopsis species has been reported in temperate waters, including the Mediterranean areas and Peter the Great Bay (NW Sea of Japan) (e.g., Aligizaki and Nikolaidis, 2006; Selina et al., 2014; Carnicer et al., 2015), their seasonal pattern seems to be highly study site-specific as well as year-specific as the peak abundances can occur from spring to autumn (Accoroni and Totti, 2016). In the present study, the Ostreopsis bloom onset was always observed at all sites in June when the water temperature reached more than 19°C, and the bloom persisted until October. Nonetheless, the bloom peak occurred at different times between June and October depending on the study sites. As in other temperate waters, the present study sites also followed a general temporal trend that Ostreopsis blooms are summer events in temperate areas although the peak can occur at different months of the year (Carnicer et al., 2015; Accoroni and Totti, 2016).
Faust (1998) reported that the abundance of Ostreopsis species with prey ranged from 7 to 55% of all specimens examined from Belize. Very recently, Almada et al. (2017) reported only a small percentage (0.4%) of O. cf. ovata containing prey from Brazil. In the present study conducted in temperate waters, the percentages (0~30%) of Ostreopsis cells with ingested food particles were in the range reported by previous studies. Further, despite the marked seasonality of Ostreopsis abundance, no pronounced seasonality of their phagotrophy was observed in the present study. For example, in July 2017 at Samjeong site, where the highest cell abundance (1,588 cells g−1 fw) was observed, the percentage of Ostreopsis cells with ingested food particles was at most 8.8%. In contrast, the highest percentage (about 30%) of Ostreopsis cells with ingested food particles was recorded in November 2017 at Samjeong site where relatively low cell density (83 cells g−1 fw) was observed. In dinoflagellates, several potential factors, including light and nutrient limitations, organic growth factors, organic carbon, and even plastid retention, have been reported to be linked with their mixotrophic strategy (Stoecker, 1998; Mitra et al., 2016). Unfortunately, data presented from this study could not clearly explain the occurrence of phagotrophy and its role in nutritional ecology of the benthic dinoflagellate Ostreopsis. Further studies, including well-designed laboratory experiments, are necessary to assess the role of mixotrophy in benthic dinoflagellates, including Ostreopsis.
Faust and Morton (1995) reported for the first time the presence of ingested food particles with an intense red color within the cytoplasm of Ostreopsis labens cells, indicating its phagotrophic behavior, and speculated that the dinoflagellate might prey on ciliates. Faust et al. (1996) and Faust (1998) subsequently found several Ostreopsis species with ingested food particles, which were hypothesized to be centric diatoms, ciliates, and small microalgae. In the present study, we frequently observed Ostreopsis cells containing reddish brown, blue-green, or purple colored food particles, all of which corresponded to species belonging to the class Florideophyceae, Rhodophyta.
Notably, all the ingested food particles originated from species belonging to the class Florideophyceae, although red algae did not always dominate macroalgal community of the study area. For example, in July 2017 at Samjeong site, where brown (Sargassum sp.) and green (Ulva sp.) algae were dominant and collected, all Ostreopsis cells with ingested prey had reddish brown colored food particles of the Florideophyceae origin, more specifically from Melanothammus harveyi. Thus, molecular data from this study raise questions as to why and/or how the dinoflagellate Ostreopsis prefers or feeds on only species belonging to the Florideophyceae as prey among various macroalgal species. There might be several possibilities to answer these questions. First, the preferred nutritional values of red algae as prey for Ostreopsis could be attributed to the difference in cell covering between red algae and other macroalgae. While green and brown macroalgae have hard cell wall composed of cellulose, red algal cell wall is relatively flexible and is composed of extracellular matrix, which typically consists of microfibrillar and sulfated galactans (Craigie, 1990; Delattre et al., 2011). The sulfated galactans include agars and carrageenans (Domozych, 2011). Because red algae have more gelatinous material than other algae, they might be more preferable as food for the epiphytic Ostreopsis than green/brown macroalgae. This difference in cell covering might allow Ostreopsis to easily penetrate and suck out the cell material of red algae. Nonetheless, this possibility seems not always to be the case in our study because many sequences of food particles were also related to species from the order Corallinales within the class Florideophyceae, such as Lithophyllum impressum, Pneophyllum fragile, Spongites tumidus, and Porolithon onkodes. Alternatively, Ostreopsis might directly ingest carpospores, which are produced after fertilization by red algae and germinated from their carposporophyte, provided red algae are abundant or the dinoflagellate is epiphytic on red algae (West and Mcbride, 1999). Diploid carpospores are mostly larger than haploid tetraspores (Ramus, 1968; Ngan and Price, 1979) so that they might have high nutritional value as prey for Ostreopsis species on the red algae. Third, benthic dinoflagellates, including Ostreopsis, are known to be capable of producing a mucopolysaccharide matrix to attach to substrates (Honsell et al., 2013; Escalera et al., 2014). In addition to the role for attachment, the mucopolysaccharide matrix might also act as a mucus trap for Ostreopsis to attach to and feed on red algal spores, which are released into the marine environment. If this is true, this mechanism would be effective when the red algae do not dominate the macroalgal community or rarely occur even if Ostreopsis is epiphytic on green or brown algae. These possibilities need to be addressed in the future to better understand the mixotrophic behavior and nutritional ecology of harmful benthic dinoflagellate Ostreopsis species.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by a research grant funded by the National Research Foundation of Korea (NRF-2016R1A6A1A03012647) and a program entitled Management of marine organisms causing ecological disturbance and harmful effects funded by KIMST/MOF.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge BS Jeon, A Kim, and JH Park for their assistance during sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00217/full#supplementary-material
Figure S1. Phylogenetic tree based on rbcL amino acid sequences from Rhodophyta, Haptophyta, Stramenopiles, and Cryptophyta. Sequences from the ingested food particles of Ostreopsis cells isolated from this study are indicated in bold face. Bootstrap values (>70%) from maximum likelihood are indicated at nodes. Double slash marks indicate shrink nodes length (70%).
Table S1. Details of dominant macroalgal species collected at each sampling time and site in this study. R, red algae; B, brown algae; G, green algae.
References
Accoroni, S., Romagnoli, T., Penna, A., Capellacci, S., Ciminiello, P., Dell'aversano, C., et al. (2016). Ostreopsis fattorussoi sp. nov.(Dinophyceae), a new benthic toxic Ostreopsis species from the eastern Mediterranean Sea. J. Phycol. 52, 1064–1084. doi: 10.1111/jpy.12464
Accoroni, S., Romagnoli, T., Pichierri, S., and Totti, C. (2014). New insights on the life cycle stages of the toxic benthic dinoflagellate Ostreopsis cf. ovata. Harmful Algae 34, 7–16. doi: 10.1016/j.hal.2014.02.003
Accoroni, S., and Totti, C. (2016). The toxic benthic dinoflagellates of the genus Ostreopsis in temperate areas: a review. Adv. Oceanogr. Limnol. 7, 1–15. doi: 10.4081/aiol.2016.5591
Aligizaki, K., and Nikolaidis, G. (2006). The presence of the potentially toxic genera Ostreopsis and Coolia (Dinophyceae) in the North Aegean Sea, Greece. Harmful Algae 5, 717–730. doi: 10.1016/j.hal.2006.02.005
Almada, E. V. C., Carvalho, W. F. D., and Nascimento, S. M. (2017). Investigation of phagotrophy in natural assemblages of the benthic dinoflagellates Ostreopsis, Prorocentrum and Coolia. Braz. J. Oceanogr. 65, 392–399. doi: 10.1590/s1679-87592017140706503
Baek, S. H. (2012). First report for appearance and distribution patterns of the epiphytic dinoflagellates in the korean peninsula. Korean J. Environ. Biol. 30, 355–361. doi: 10.11626/KJEB.2012.30.4.355
Carnicer, O., Guallar, C., Andree, K. B., Diogène, J., and Fernández-Tejedor, M. (2015). Ostreopsis cf. ovata dynamics in the NW Mediterranean Sea in relation to biotic and abiotic factors. Environ. Res. 143, 89–99. doi: 10.1016/j.envres.2015.08.023
Craigie, J. (1990). “Cell walls,” in Biology of the Red Algae, eds K. M Cole and R. G. Sheath (Cambridge: Cambridge University Press), 221–257.
Delattre, C., Fenoradosoa, T. A., and Michaud, P. (2011). Galactans: an overview of their most important sourcing and applications as natural polysaccharides. Braz. Arch. Biol. Technol. 54, 1075–1092. doi: 10.1590/S1516-89132011000600002
Domozych, D. S. (2011). “Algal cell walls,” in eLS (Chichester: John Wiley & Sons, Ltd). doi: 10.1002/9780470015902.a0000315.pub3
Escalera, L., Benvenuto, G., Scalco, E., Zingone, A., and Montresor, M. (2014). Ultrastructural features of the benthic dinoflagellate Ostreopsis cf. ovata (Dinophyceae). Protist 165, 260–274. doi: 10.1016/j.protis.2014.03.001
Faust, M. A. (1998). “Mixotrophy in tropical benthic dinoflagellates,” in VIII International Conference on Harmful Algae, eds B. Reguera, J. Blanco, M. L. Fernández, and T. Wyatt (Vigo: International Oceanographic Comission of UNESCO), 390–393.
Faust, M. A., and Morton, S. L. (1995). Morphology and ecology of the marine dinoflagellate Ostreopsis labens sp. nov.(Dinophyceae). J. phycol. 31, 456–463. doi: 10.1111/j.0022-3646.1995.00456.x
Faust, M. A., Morton, S. L., and Quod, J. P. (1996). Further SEM study of marine dinoflagellates: the genus Ostreopsis (Dinophyceae). J. Phycol. 32, 1053–1065. doi: 10.1111/j.0022-3646.1996.01053.x
Fawcett, R. C., and Parrow, M. W. (2014). Mixotrophy and loss of phototrophy among geographic isolates of freshwater Esoptrodinium/Bernardinium sp.(Dinophyceae). J. Phycol. 50, 55–70. doi: 10.1111/jpy.12144
Fraga, S., Rodríguez, F., Bravo, I., Zapata, M., and Marañón, E. (2012). Review of the main ecological features affecting benthic dinoflagellate blooms. Cryptogam. Algol. 33, 171–179. doi: 10.7872/crya.v33.iss2.2011.171
Freshwater, D. W., and Rueness, J. (1994). Phylogenetic relationships of some European Gelidium (Gelidiales, Rhodophyta) species, based on rbcL nucleotide sequence analysis. Phycologia 33, 187–194. doi: 10.2216/i0031-8884-33-3-187.1
Hackett, J. D., Maranda, L., Yoon, H. S., and Bhattacharya, D. (2003). Phylogenetic evidence for the cryptophyte origin of the plastid of Dinophysis (Dinophysiales, Dinophyceae). J. Phycol. 39, 440–448. doi: 10.1046/j.1529-8817.2003.02100.x
Honsell, G., Bonifacio, A., De Bortoli, M., Penna, A., Battocchi, C., Ciminiello, P., et al. (2013). New insights on cytological and metabolic features of Ostreopsis cf. ovata fukuyo (Dinophyceae): a multidisciplinary approach. PLoS ONE 8:e57291. doi: 10.1371/journal.pone.0057291
Kim, M., Kim, S., Yih, W., and Park, M. G. (2012). The marine dinoflagellate genus Dinophysis can retain plastids of multiple algal origins at the same time. Harmful Algae 13, 105–111. doi: 10.1016/j.hal.2011.10.010
Laza-Martinez, A., Orive, E., and Miguel, I. (2011). Morphological and genetic characterization of benthic dinoflagellates of the genera Coolia, Ostreopsis and Prorocentrum from the south-eastern Bay of Biscay. Eur. J. Phycol. 46, 45–65. doi: 10.1080/09670262.2010.550387
McManus, G. B., Zhang, H., and Lin, S. (2004). Marine planktonic ciliates that prey on macroalgae and enslave their chloroplasts. Limnol. Oceanogr. 49, 308–313. doi: 10.4319/lo.2004.49.1.0308
Mitra, A., Flynn, K. J., Tillmann, U., Raven, J. A., Caron, D., Stoecker, D. K., et al. (2016). Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist 167, 106–120. doi: 10.1016/j.protis.2016.01.003
Ngan, Y., and Price, I. R. (1979). Systematic significance of spore size in the Florideophyceae (Rhodophyta). Br. phycol. J. 14, 285–303. doi: 10.1080/00071617900650311
Nishitani, G., Nagai, S., Baba, K., Kiyokawa, S., Kosaka, Y., Miyamura, K., et al. (2010). High-level congruence of Myrionecta rubra prey and Dinophysis species plastid identities as revealed by genetic analyses of isolates from Japanese coastal waters. Appl. Environ. Microbiol. 76, 2791–2798. doi: 10.1128/AEM.02566-09
Park, K. A., Park, J. E., Choi, B. J., Byun, D. S., and Lee, E. I. (2013). An oceanic current map of the east sea for science textbooks based on scientific knowledge acquired from oceanic measurements. Sea 18, 234–265. doi: 10.7850/jkso.2013.18.4.234
Parsons, M. L., Aligizaki, K., Bottein, M.-Y. D., Fraga, S., Morton, S. L., Penna, A., et al. (2012). Gambierdiscus and Ostreopsis: reassessment of the state of knowledge of their taxonomy, geography, ecophysiology, and toxicology. Harmful Algae 14, 107–129. doi: 10.1016/j.hal.2011.10.017
Penna, A., Vila, M., Fraga, S., Giacobbe, M. G., Andreoni, F., Riobó, P., et al. (2005). Characterization of Ostreopsis and Coolia (Dinophyceae) isolates in the Western Mediterranean Sea based on morphology, toxicity and internal transcribed spacer 5.8 S rDNA sequences. J. Phycol. 41, 212–225. doi: 10.1111/j.1529-8817.2005.04011.x
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Ramus, J. S. (1968). The Developmental Sequence of the Marine Red Alga Pseudogloiphloea Confusa in Culture. Berkeley, CA: University of California.
Rhodes, L. (2011). World-wide occurrence of the toxic dinoflagellate genus Ostreopsis Schmidt. Toxicon 57, 400–407. doi: 10.1016/j.toxicon.2010.05.010
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schmidt, J. (1901). Preliminary report of the biological result of the Danish Expedition to Siam (1899-1900). Botanisk Tidsskrift. 24, 212–221.
Selina, M. S., Morozova, T. V., Vyshkvartsev, D. I., and Orlova, T. Y. (2014). Seasonal dynamics and spatial distribution of epiphytic dinoflagellates in Peter the Great Bay (Sea of Japan) with special emphasis on Ostreopsis species. Harmful Algae 32, 1–10. doi: 10.1016/j.hal.2013.11.005
Selina, M. S., and Orlova, T. Y. (2010). First occurrence of the genus Ostreopsis (Dinophyceae) in the Sea of Japan. Botanica Marina 53, 243–249. doi: 10.1515/BOT.2010.033
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stoecker, D. K. (1998). Conceptual models of mixotrophy in planktonic protists and some ecological and evolutionary implications. Eur. J. Protistol. 34, 281–290. doi: 10.1016/S0932-4739(98)80055-2
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tichadou, L., Glaizal, M., Armengaud, A., Grossel, H., Lemée, R., Kantin, R., et al. (2010). Health impact of unicellular algae of the Ostreopsis genus blooms in the Mediterranean Sea: experience of the French Mediterranean coast surveillance network from 2006 to 2009. Clin. Toxicol. 48, 839–844. doi: 10.3109/15563650.2010.513687
Ukena, T., Satake, M., Usami, M., Oshima, Y., Naoki, H., Fujita, T., et al. (2001). Structure elucidation of ostreocin D, a palytoxin analog isolated from the dinoflagellate Ostreopsis siamensis. Biosci. Biotechnol. Biochem. 65, 2585–2588. doi: 10.1271/bbb.65.2585
Verma, A., Hoppenrath, M., Dorantes-Aranda, J. J., Harwood, D. T., and Murray, S. A. (2016). Molecular and phylogenetic characterization of Ostreopsis (Dinophyceae) and the description of a new species, Ostreopsis rhodesae sp. nov., from a subtropical Australian lagoon. Harmful algae 60, 116–130. doi: 10.1016/j.hal.2016.11.004
Vila, M., Garcés, E., and Masó, M. (2001). Potentially toxic epiphytic dinoflagellate assemblages on macroalgae in the NW Mediterranean. Aquat. Microb. Ecol. 26, 51–60. doi: 10.3354/ame026051
West, J., and Mcbride, D. (1999). Long-term and diurnal carpospore discharge patterns in the Ceramiaceae, Rhodomelaceae and Delesseriaceae (Rhodophyta). Hydrobiologia 398, 101–114. doi: 10.1023/A:1017025815001
Yasumoto, T., Seino, N., Murakami, Y., and Murata, M. (1987). Toxins produced by benthic dinoflagellates. Biol. Bull. 172, 128–131. doi: 10.2307/1541612
Keywords: mixotroph, phagotrophy, red algae, phylogeny, seasonality
Citation: Lee B and Park MG (2018) Genetic Analyses of the rbcL and psaA Genes From Single Cells Demonstrate a Rhodophyte Origin of the Prey in the Toxic Benthic Dinoflagellate Ostreopsis. Front. Mar. Sci. 5:217. doi: 10.3389/fmars.2018.00217
Received: 13 February 2018; Accepted: 31 May 2018;
Published: 19 June 2018.
Edited by:
Matthew D. Johnson, Woods Hole Oceanographic Institution, United StatesReviewed by:
Jelena Godrijan, Bigelow Laboratory for Ocean Sciences, United StatesPunyasloke Bhadury, Indian Institute of Science Education and Research Kolkata, India
Copyright © 2018 Lee and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Myung G. Park, bXBhcmtAY2hvbm5hbS5hYy5rcg==
 Bora Lee
Bora Lee Myung G. Park
Myung G. Park