- 1Marine Biology Research Division, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA, United States
- 2Laboratoire d’Océanographie Microbienne, Observatoire Océanologique de Banyuls s/mer, Sorbonne Universités, UPMC, Banyuls-sur-Mer, France
- 3Department of Biological and Environmental Sciences, Nanoscience Center, University of Jyväskylä, Jyväskylä, Finland
Understanding the temporal variations and succession of bacterial communities involved in the turnover of one-carbon and methylated compounds is necessary to better predict bacterial impacts on the marine carbon cycle and air-sea carbon fluxes. The ability of the local bacterioplankton community to exploit one-carbon and methylated compounds as main source of bioavailable carbon during a productive and less productive period was assessed through enrichment experiments. Surface seawater was amended with methanol and trimethylamine-N-oxide (TMAO), and bacterial abundance, production, oxygen consumption, as well as methanol turnover and growth rates of putative methylotrophs were followed. Bacterial community structure and functional diversity was examined through amplicon sequencing of 16S rRNA and methanol dehydrogenase (mxaF) marker genes. 2-fold increase in oxygen consumption and bacterial growth rates, and up to 4-fold higher methanol assimilation were observed in the amended seawater samples. Capacity to drawdown the substrates was similar between both experiments. In less productive conditions, methanol enriched obligate methylotrophs, especially Methylophaga spp., accounted for ∼70% of bacterial cells analyzed by fluorescence in situ hybridization and 16S rRNA gene sequencing, while TMAO enriched taxa belonged to Oceanospirillales and putative β- and γ-Proteobacterial methylotrophs. In the experiment performed during the more productive period, bacterial communities were structurally resistant, suggesting that facultative organisms may have dominated the observed methylotrophic activity. Moreover, enrichment of distinct methylotrophic taxa but similar activity rates observed in response to different substrate additions suggests a functional redundancy of substrate specific marine methylotrophic populations. Marine bacterioplankton cycling of one-carbon and methylated compounds appears to depend on the system productivity, and hence may have predictable temporal impacts on air-sea fluxes of volatile organic compounds.
Introduction
Marine bacteria play key roles in food webs and biogeochemical cycles of coastal surface waters (e.g., Ducklow et al., 1986). Bacterial community abundance, composition and function are tightly coupled with spatiotemporal gradients of dissolved organic carbon composition and concentration (Azam and Malfatti, 2007) and depend on the metabolic capacities of bacterial taxa (Mou et al., 2008; Poretsky et al., 2010). For instance, methylotrophic bacteria that use one carbon and methylated (hereafter C1) compounds as their sole source of carbon and energy are widespread in the ocean (Chistoserdova et al., 2009; Chistoserdova, 2015). C1 compounds are ubiquitous in the ocean (Beale et al., 2011; Carpenter et al., 2012; Beale and Airs, 2016) and therefore a potential carbon source for marine bacteria. Most of the C1 compounds are volatile and have the potential to influence atmospheric chemistry and climate (Williams et al., 2004). Hence, it is important to understand how marine bacteria interact with these compounds to better assess air-sea fluxes for climatic predictions.
Indeed, the directions of net air-sea fluxes of C1 compounds may vary depending on utilization by marine methylotrophs. Methanol is among the most abundant oxygenated volatile organic compounds in the atmosphere, where it can influence atmospheric chemistry through oxidation and ozone formation (Singh et al., 2000). Methanol is a source of carbon and energy for diverse marine bacteria (Murrell et al., 1993), and surface methylotrophs may act as oceanic sink of methanol (Dixon et al., 2011a,b, 2012). However, it is unclear if the ocean acts as a net source or sink of atmospheric methanol (Singh et al., 2000, 2003; Dixon et al., 2013). Measurements in some oceanic regions found them to be net sink of methanol (Yang et al., 2013, 2014a,b). On the other hand, methylamine (MA), can form aerosols and act as cloud condensation nuclei, are constantly emitted from the ocean (Facchini et al., 2008; Müller et al., 2009; Sorooshian et al., 2009). MA are products of the methylotrophic degradation of precursors trimethylamine-N-oxide (TMAO) and glycine betaine. Methylotrophs may thus act as a source of atmospheric aerosols derived from MA production through, for instance, TMAO demethylation (Lidbury et al., 2017).
Methanol and TMAO are ubiquitous in the ocean (Beale et al., 2011), as are bacteria able to use them. Bacteria use methanol and TMAO either only as energy source, such as the abundant surface α-Proteobacteria of the SAR11clade (Sun et al., 2011), or as both carbon and energy source (Chen et al., 2011; Lidbury et al., 2014a,b). In the ocean, these two C1 compounds are produced by phytoplankton (Nieder et al., 2014; Mincer and Aicher, 2016) and other organisms. Their concentrations in the ocean vary temporally (Beale et al., 2015) depending on sources, e.g., phytoplankton bloom, atmospheric wet and dry deposition, and photooxidation. Methylotrophic metabolism in the coastal ocean is also expected to follow a similar seasonal dynamic (Sargeant et al., 2016), with different feedbacks on the biogeochemical cycles and air-sea fluxes.
Here, we investigated the capacity of bacterioplankton community experimentally enriched with methanol and TMAO to assimilate and grow on C1 compounds during less productive winter and the following more productive summer. We measured carbon drawdown through bacterial production and respiration, as well as followed changes in the microbial community structure using 16S rRNA gene sequencing. The methylotrophic capacities of the bacterioplankton was assessed in terms of methanol assimilation and respiration, FISH enumeration of potential methylotrophs and composition of the methylotrophic population through sequencing of methanol dehydrogenase gene mxaF.
Materials and Methods
Experimental Setup
Enrichment experiments were performed to examine the activity and structure of enriched coastal bacterial communities able to utilize C1 compounds during low productivity winter (February 2015) and higher productivity summer (July 2015). Aged (3 months) GF/F filtered seawater, kept in the dark at near in situ temperature (16 ± 0.5°C) was gently filtered through rinsed 0.2 μm filters (Supor 200; Pall Laboratory) and distributed randomly into 1 L bottles. This 0.2 μm filtered water was then inoculated (20% v/v) with freshly collected seawater gently filtered through 0.65 μm Isopore (Millipore) filters. The seawater was sampled on the day of the experiment from coastal surface (∼1 m below the surface) off the Pier of Scripps Institution of Oceanography, La Jolla, CA, United States (32°53′N, 117°15′W). Experimental setup consisted of triplicate 1 L bottles divided into five treatments: (1) control (no addition; hereafter called Control); (2) Control plus inorganic nutrients (10 μM NaNO3; 10 μM NH4Cl; 3 μM NaH2PO4; 20 pM vitamin B12), hereafter called Control + Nutrients; (3) methanol plus inorganic nutrients (20 μM methanol), MetOH; (4) Trimethylamine N-oxide (20 μM TMAO plus inorganic nutrients), TMAO; and (5) mixture of methanol and TMAO (10 μM each plus inorganic nutrients), hereafter called MetOH + TMAO. The bottles were incubated for 7 days in the dark at near in situ temperature (ca. 17°C in winter and ca. 21°C in summer). All materials in contact with the samples were acid washed in 10% HCl and repeatedly rinsed with Q-grade water (Millipore) prior to use.
Samples for bacterial abundance, fluorescence in situ hybridization (winter samples), bacterial production, methanol assimilation and respiration (summer samples) were taken at T0, 24, 72, 120 h and at the end of the incubation (168 h). Dissolved oxygen concentration was measured daily and samples for bacterial community composition were filtered at the start and at the end of the experiment.
Bacterial Abundance
Samples (1.5 ml) for bacterial enumeration were fixed with EM grade glutaraldehyde (EMS; 1% v/v), flash frozen and stored at -80°C. The samples were stored for less than 2 months before further analyses. Thawed samples were filtered onto 0.2 μm white polycarbonate filters (Isopore, Millipore), stained with DAPI (Porter and Feig, 1980) and enumerated by epifluorescence microscopy on an Olympus BX51 microscope at 1000x magnification. At least 20 fields were counted. Cell biovolumes were measured with Fiji (Schindelin et al., 2012) and averaged over several time points and treatments. Bacterial carbon content was calculated according to Bratbak (1985) and Simon and Azam (1989).
Fluorescence in situ Hybridization (FISH)
Duplicate samples (12 ml) were fixed with formaldehyde (3% final) overnight at 4°C and filtered onto 0.2 μm white polycarbonate filters (Isopore, Millipore). Preparations were stored at -20°C until further processing. All hybridization were carried out under the conditions previously described by Glöckner et al. (1996). The samples were hybridized for 2 h at 46°C and washed at 48°C. No formamide was added to the hybridization buffer (Tsien et al., 1990; Janvier et al., 2003). Pieces of filters (1/4) were double hybridized with fluorescent 16S rRNA targeting probes, FAM-MPH-730 (5′-CAGTAATGGCCCAGTGAGTCGCC-3′) complementary to Methylophaga spp. specific 16S rRNA region (Janvier et al., 2003) and cy3-10-γ (5′-GGTCCGAAGATCCCCCGCTT-3′) complementary to the 16S rRNA region of the methylotrophic species of β- and γ-Proteobacteria with the RuMP pathway (Tsien et al., 1990). Another piece of each filter was hybridized with 16S rRNA targeting probes, FAM-9-α (5′-CCCTGAGTTATTCCGAAC-3′) complementary to the 16S rRNA region of methylotrophic α- Proteobacteria with the serine pathway (Tsien et al., 1990). Images were analyzed with Fiji (Schindelin et al., 2012). The effect of time was tested in different treatments using a two-way ANOVA.
Bacterial Production, Respiration and Growth Efficiency
Bacterial production was measured by [H3]-leucine incorporation (Kirchman et al., 1985) modified for microcentrifugation (Smith and Azam, 1992). Triplicate 1.7 ml aliquots were incubated with [H3]-leucine (20 nM final conc.) in sterile 2.0 ml polypropylene tubes for 1 h at in situ temperature. Samples with 5% trichloroacetic acid added prior to [H3]-leucine addition served as blanks. Leucine incorporation was converted to carbon production assuming 3.1 kg C (mol leucine)-1 (Simon and Azam, 1989).
Dissolved oxygen was measured according to Kragh et al. (2008) using a fiber-optic oxygen mini-sensor (Fibox 4, PreSens Precision Sensing GmbH), which utilizes fluorescence quenching of the sensor by oxygen. A sensor spot (Pst3; 5 mm) inside each bottles allowed for readings with a fiber-optic probe from outside the bottle. A “control” bottle with Milli-Q water was used to ensure no inherent O2 consumption. No leaching or O2 consumption was observed in the control incubator.
O2 consumption (ΔO2) and cumulative time-integrated bacterial production (BPcum) were used to calculate bacterial carbon demand (BCD) and bacterial growth efficiency (BGE), with BCD = -ΔO2 + BPcum and BGE = BPcum/BCD. ΔO2 and BPcum were calculated over 0–120 h for the winter and 0–72 h for the summer experiment following the ΔO2 leveling off for all treatments. The percentage of bioavailable carbon from the added substrate (bDOC) was calculated as the average of (BCDtreatment-BCDcontrol)/DOC added.
Statistical differences between treatments were calculated with a Student’s t-test (two samples, assuming unequal variances).
Methanol Assimilation and Respiration
Bacterial methanol assimilation was measured by [C14]-methanol incorporation. Triplicate 1.7 ml aliquots were incubated with [C14]-methanol (100 nM final conc.) in sterile 2.0 ml polypropylene tubes for 24 h, at in situ temperature. Samples with 5% trichloroacetic acid added prior to methanol addition served as blanks. Following the incubation samples were centrifuged for 10 min at 16000 × g. The pellets were dried, scintillation cocktail (Ultima Gold, Perkin Elmer) was added and the samples were radioassayed in a liquid scintillation counter (Beckman LS6000).
Bacterial methanol respiration was measured by [C14]-methanol respiration. Triplicate 1.7 ml aliquots were incubated with [C14]-methanol (100 nM final) in sterile closed culture tubes for 24 h at in situ temperature. Samples with 4% formaldehyde added prior to the radioisotope served as blanks. After 24 h the samples were acidified with concentrated H2SO4 and a center well with a 2 × 2 cm piece of fluted filter soaked with β-phenylethylamine was added to the tube (Karl et al., 1998). After 72 h the fluted filters were dried in a dessicator and trapped14CO2 was measured.
Bacterial Community Composition
For each incubation and T0 samples, cells from 800 mL of water were collected by gentle filtration onto 0.2 μm, 47 mm Supor membrane filters (Pall). Filters were subsequently frozen at -80°C. DNA was extracted using ZR Fungal/Bacterial DNA MiniPrep kit (ZYMO research) following manufacturer guidelines after a proteinase K digestion (20 mg ml-1 final) at 55°C for 2 h. PCR amplification with bacterial 16S rRNA gene (V1-V3 region) specific primers 27F and 338R was followed by a nested PCR with barcoded adapter primers to identify different samples. The products were then purified with Agencourt AmPure XP magnetic beads (Beckman Coulter). Ion Torrent PGM sequencing was performed as described by Stark et al. (2015). Methanol dehydrogenase genes (mxaF) were amplified using primers 1003F (McDonald and Murrell, 1997) and 1555R (Neufeld et al., 2007). PCR products were gel purified using GenElute DNA Gel extraction kit (Sigman-Aldrich) and barcodes were added to the PCR products through the M13-linker using the protocol described by Mäki et al. (2016). 16S rRNA and mxaF raw datasets were deposited to NCBI Sequence Read Archive under Bioproject PRJNA447999.
16S rRNA and mxaF Gene Sequence Analysis
All reads were processed using the Quantitative Insight Into Microbial Ecology pipeline (QIIME v1.9; Caporaso et al., 2010a). For 16S rRNA gene sequences, all reads with mismatches in the primer or lengths below 200 bp and quality scores < 25 were discarded. Reads were quality controlled and chimeras were removed using UPARSE (Edgar, 2013). Reads were clustered into operational taxonomic units (OTUs) at 99% pairwise identity using the UPARSE and representative sequences from each bacterial OTU aligned to SILVA 128 reference alignment (Quast et al., 2013) using PyNAST (Caporaso et al., 2010b). Taxonomy assignments were made using UCLUST algorithm (Edgar, 2010) against the database SILVA 128.
For mxaF sequences, all reads with mismatches in the primer or lengths below 250 bp and quality score < 25 were discarded. Reads were quality controlled and chimeras were removed using UPARSE (Edgar, 2013). Reads were clustered into OTUs at 77% pairwise identity using UPARSE. This cutoff for mxaF corresponds to a 97% pairwise identity for 16S rRNA genes, based on pairwise comparison of both genes from different isolates described by Stacheter et al. (2012).
Alpha and beta-diversity indices were estimated for all samples after randomized subsampling to 1047 reads for 16S rRNA gene and to 1212 reads for mxaF sequences. Alpha diversity analyses (Richness: observed OTUs and diversity: Shannon index) were run in QIIME. Beta-diversity was investigated with Primer V.6 software package (Clarke and Warwick, 2001) on Bray Curtis dissimilarity. Differences between communities were tested with PERMANOVA (Permutational Multivariate Analysis of Variance) with pairwise test and 9999 permutations in Primer V.6 (Clarke et al., 2014). The mxaF OTUs contributing most to the dissimilarity between samples were identified using SIMPER (similarity percentage) on normalized samples, with Primer V.6. The sequences of OTUs contributing to 90% of the dissimilarity between samples were aligned with references sequences on their amino acid sequences, and maximum likelihood tree with bootstrap test was constructed using MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 (Kumar et al., 2016).
Results
In situ Parameters and System Productivity
The productivity of the system between winter and summer sampling was determined as the biological productivity differences between the two sampling points. The colder winter water was characterized by lower productivity compared to the more productive summer waters with higher Chl a, POC and bacterial production (Table 1). Moreover, the summer sample followed the spring phytoplankton bloom observed in May – June1. Overall, bacterial production and methanol assimilation were higher during the whole summer period compared to winter (J. Dinasquet, unpublished time series). At the time points sampled for this experiment bacterial production and methanol assimilation were 2.3-fold and 5.6-fold higher in summer compared to winter while bacterial abundances were similar for the two seasons (Table 1).

TABLE 1. In situ conditions off Scripps Pier at the time of sampling (mean ± standard deviation for n = 3).
Bacterial Utilization of Added Substrates
Enhanced bacterial growth and oxygen consumption showed that coastal bacteria responded to substrates additions both in the winter and summer experiment (Figure 1). Increase in the bacterial production was higher in the winter than in the summer, but started later. The active parts of the experiment were determined as 0–120 h in the winter and 0–72 h in the summer incubations before oxygen consumption leveled off and bacterial abundance decreased (Figures 1A–D). Although samples were originally filtered, grazing affected the bacterial abundance during the later time points. At the time of sampling, the concentration of heterotrophic nanoflagellates in situ was also higher in summer (5300 cells ml-1) compared to winter (2500 cells ml-1), which may explain the highest mortality in the summer experiment. Substrate additions doubled the O2 consumption in both the winter and summer experiments (Figures 1A,B and Table 2). The bacterial growth efficiency with the substrate additions was ∼1.6-fold higher in the winter experiment (Table 2). Of the studied C1 compounds, carbon drawdown was greater in winter in the methanol treatment, while in summer it was greater in the TMAO treatment. The drawdown of carbon in the substrate mixture fitted the combined carbon drawdown in the separate carbon source treatments.
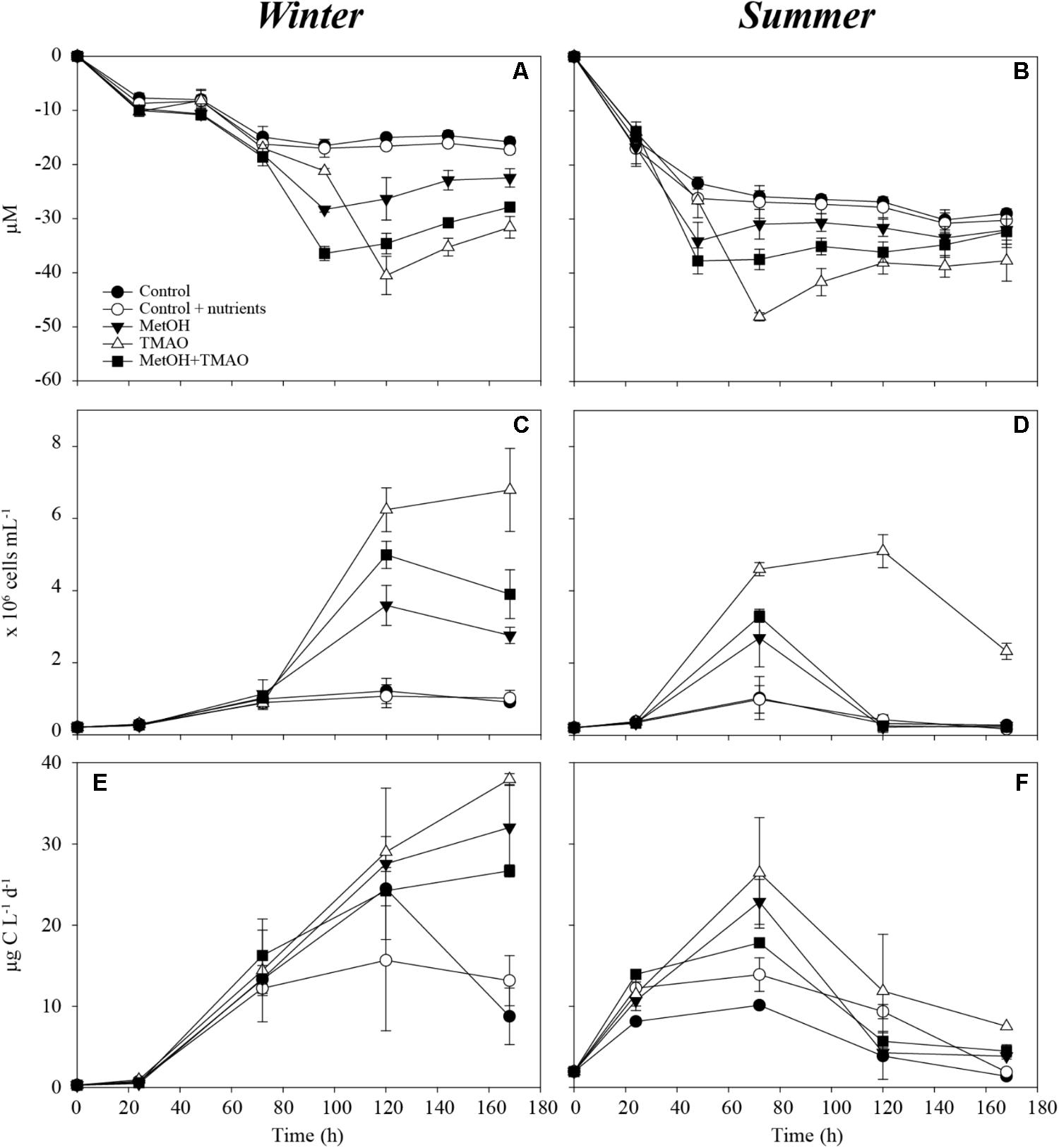
FIGURE 1. Oxygen consumption over the course of the experiment in winter (A) and summer (B), bacterial abundance over the course of the experiment in winter (C) and summer (D) and bacterial production over the course of the experiment in winter (E) and summer (F). Mean ± standard deviation, n = 3.
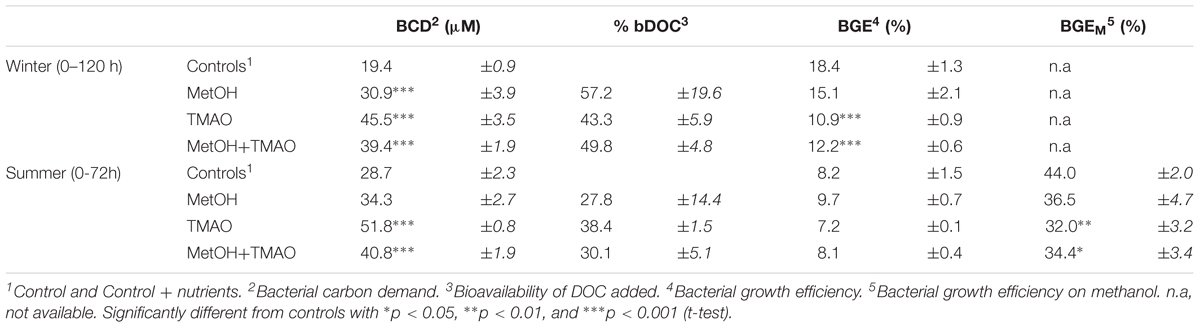
TABLE 2. Summary of data during the active phases of the enrichment experiments, 0–120 h and 0–72 h of incubation in winter and summer, respectively (mean ± standard deviation for n = 3 for enrichments and n = 6 for controls).
In the winter experiment, bacterial growth efficiency (BGE) was significantly lower in the TMAO treatments (11–12%) than in the controls or methanol treatment alone (15–18%). There was no differences in the BGE in the summer experiment (Table 2).
At the end of the experiments, C1 treatments had increased methanol assimilation 2- to 4-fold, when compared to winter and summer controls, respectively (Figures 2A,B). Methanol respiration was also about 4-fold higher in all C1 treatments at the end of the summer experiment (Figure 2C). The BGE based on added [C14]-methanol (BGEM) was higher in the controls compared to the C1 treatments in summer. Growth efficiency on methanol (34.4 ± 2.3%) was not significantly different between different substrate addition treatments (Table 2).
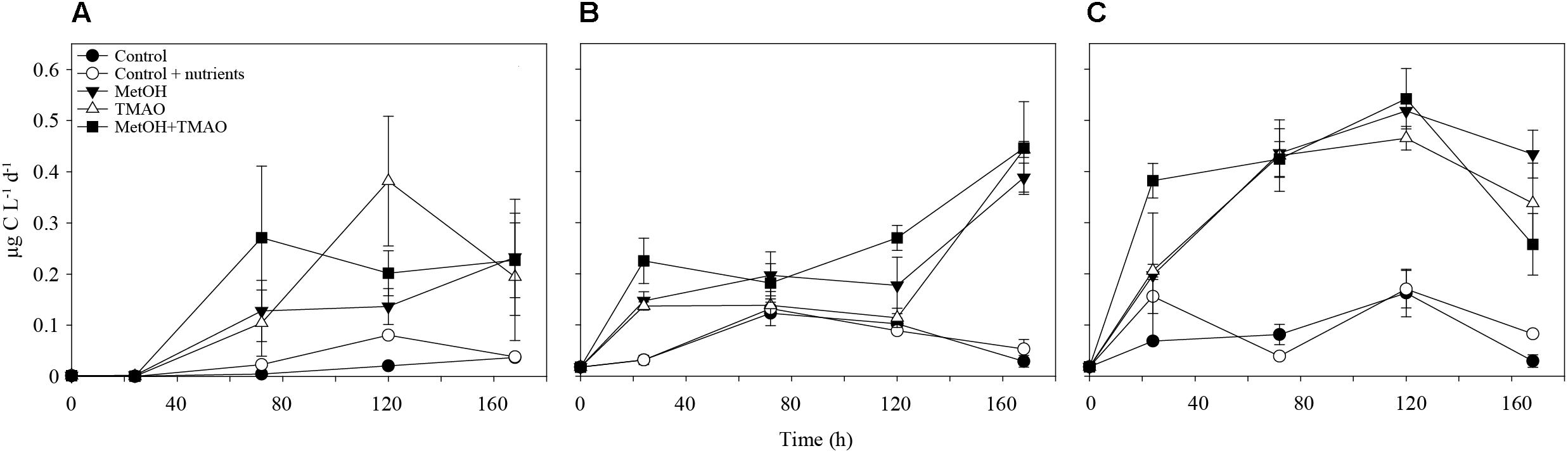
FIGURE 2. Methanol assimilation over the course of the experiment in winter (A) and summer (B) and methanol respiration in summer (C). Mean ± standard deviation, n = 3.
Bacterial Community Composition in Response to Substrates Addition
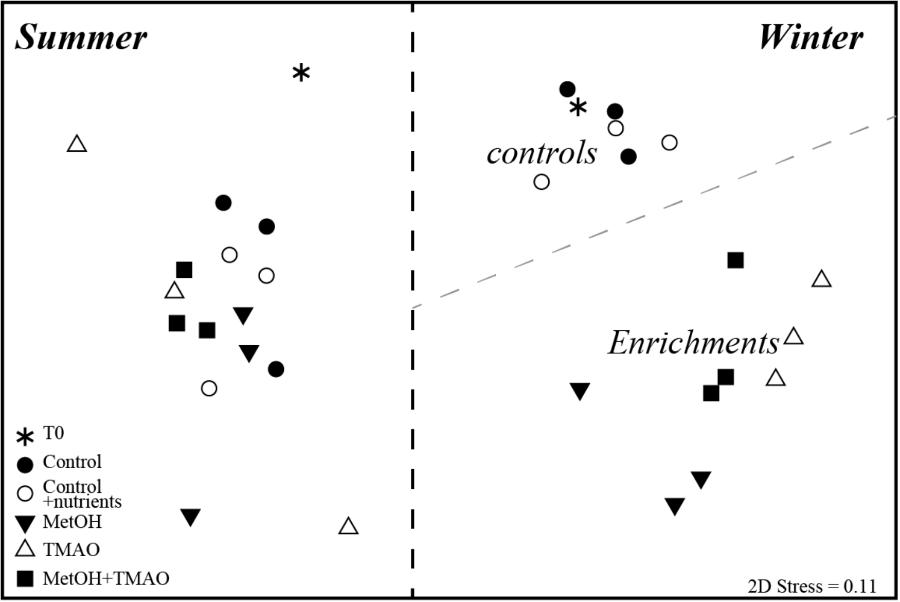
A total of 300,618 16S rRNA gene sequences remained after quality control, and were clustered to 5995 OTUs (99% identity). Original bacterial community composition displayed higher richness and diversity in summer than in winter (Supplementary Table S1). In winter, richness (as measured by observed OTUs and Chao1 index) and diversity (Shannon Index) decreased further in all C1 treatments when compared to the initial water, while diversity remained constant in the control samples. The richness and diversity indexes were significantly lower (∼0.5-fold) in the methanol treatments compared to the controls, and slightly lower in the TMAO treatments (Supplementary Table S1). In summer, richness and diversity did not significantly change between experiment start and end communities and between treatments, with the exception of TMAO treatments where it was slightly lower (Supplementary Table S1). The bacterial community composition in the 32 samples was compared with non-metric multidimensional scaling (nMDS) based on Bray-Curtis dissimilarity of rarefied data (Figure 3). The nMDS plot showed that winter and summer communities independent of time and treatment were significantly different. Incubation without carbon addition did not change the winter communities (PERMANOVA, p = 0.53 T0 to C and p = 0.37 T0 to CN), while control and C1 treatments communities were significantly different at the end of the experiment (PERMANOVA, control to methanol, p = 0.012, to TMAO p = 0.01 and to MetOH + TMAO p = 0.013). Furthermore, methanol communities were distinct from the two other C1 treatments (PERMANOVA, methanol to TMAO p = 0.004 and to MetOH + TMAO p = 0.03), while TMAO and MetOH + TMAO communities were similar (p = 0.18). On the other hand, neither time nor carbon addition affected the overall bacterial community structure in summer (Figure 3).
The winter inoculation consisted of significantly (t-test p < 0.05) higher abundance of α- and γ-Proteobacteria and lower abundance of Bacteroidetes than summer inoculation, as well as different OTUs of the clade SAR11. In winter the difference between controls and C1 treatments was mainly driven by a significant decrease (t-test p < 0.05) in relative abundance of α-Proteobacteria (especially SAR11 related OTUs) and a significant increase (t-test p < 0.05) of γ-Proteobacteria (especially Methylophaga sp. related OTUs in the methanol treatment, and Oceanospirillales, Pseudomonadales and Methylophaga sp. in the TMAO and MetOH + TMAO treatments). In summer, minor differences were observed such as an increase in δ- and γ-Proteobacteria and decrease in α-Proteobacteria at the end of the incubations compared to the start water. There was a small enrichment in Methylophaga sp. in the enrichment treatments compared to the control (Figures 4A–C).
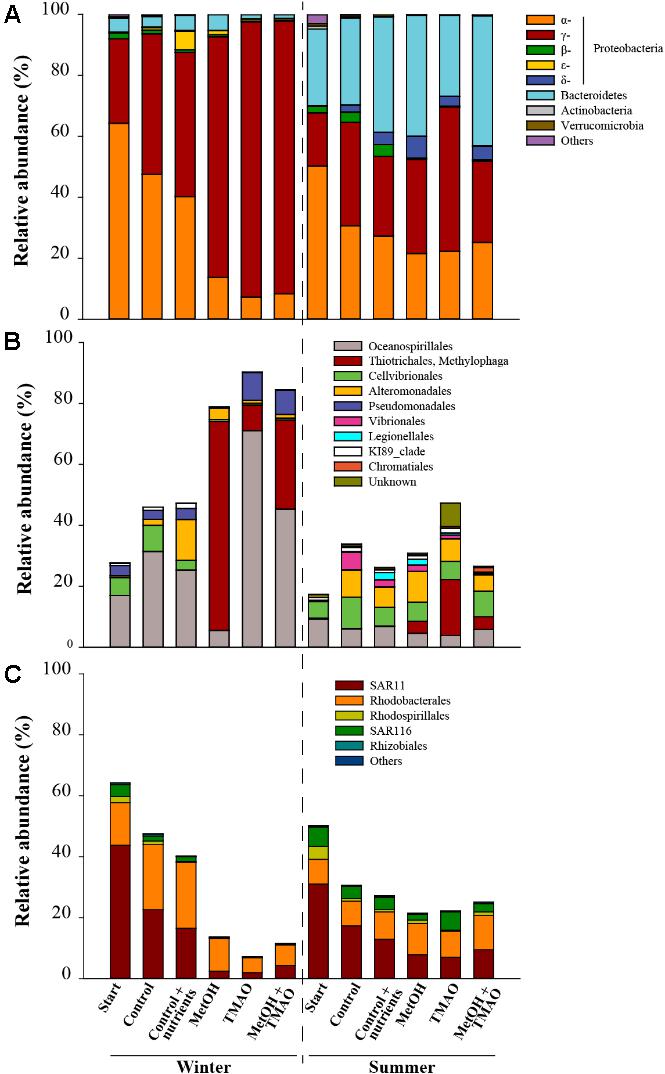
FIGURE 4. Relative abundance of the main bacterial phyla and subclasses (A), γ-Proteobacteria (B) and α-Proteobacteria (C) in winter and summer experiments. Relative abundances are expressed as % of total 16S rRNA gene sequences in the original assemblage or as a mean of the treatment triplicates at the end of the experiment.
Functional Community of Methanol Users in Response to Substrate Addition
A total of 40,000 sequences shared between 41 OTUs (at 77% identity) were recovered from sequencing of the methanol dehydrogenase gene mxaF. Richness and diversity of the methanol users (as defined by mxaF- methanol dehydrogenase sequences) were slightly lower in summer when compared to winter, and in the C1 treatments compared to the controls (Supplementary Table S2). Based on the nMDS plot (Supplementary Figure S1), the functional diversity was slightly different between winter and summer and between winter C1 treatments and controls, and MetOH+TMAO treatment and controls in summer. Nevertheless, these differences were not significant (PERMANOVA, p > 0.05, community was 78.4% similar in winter between all treatments and 83.2% similar in summer). Few main mxaF-OTUs were driving over 70% of the dissimilarity of the functional community between treatment and controls in winter and summer (Figure 5). Some of the mxaF-OTUs were related to known mxaF sequences from β-, α- and γ-Proteobacteria (closely related to Methylophilales, Methylobacterium and Methylophaga respectively) but also from unknown putatively new methanol dehydrogenases from Rhodobacterales (Confluentimicrobium sp. and Ruegiria sp.) and Oceanospirillales (Halomonas sp., Figure 6).
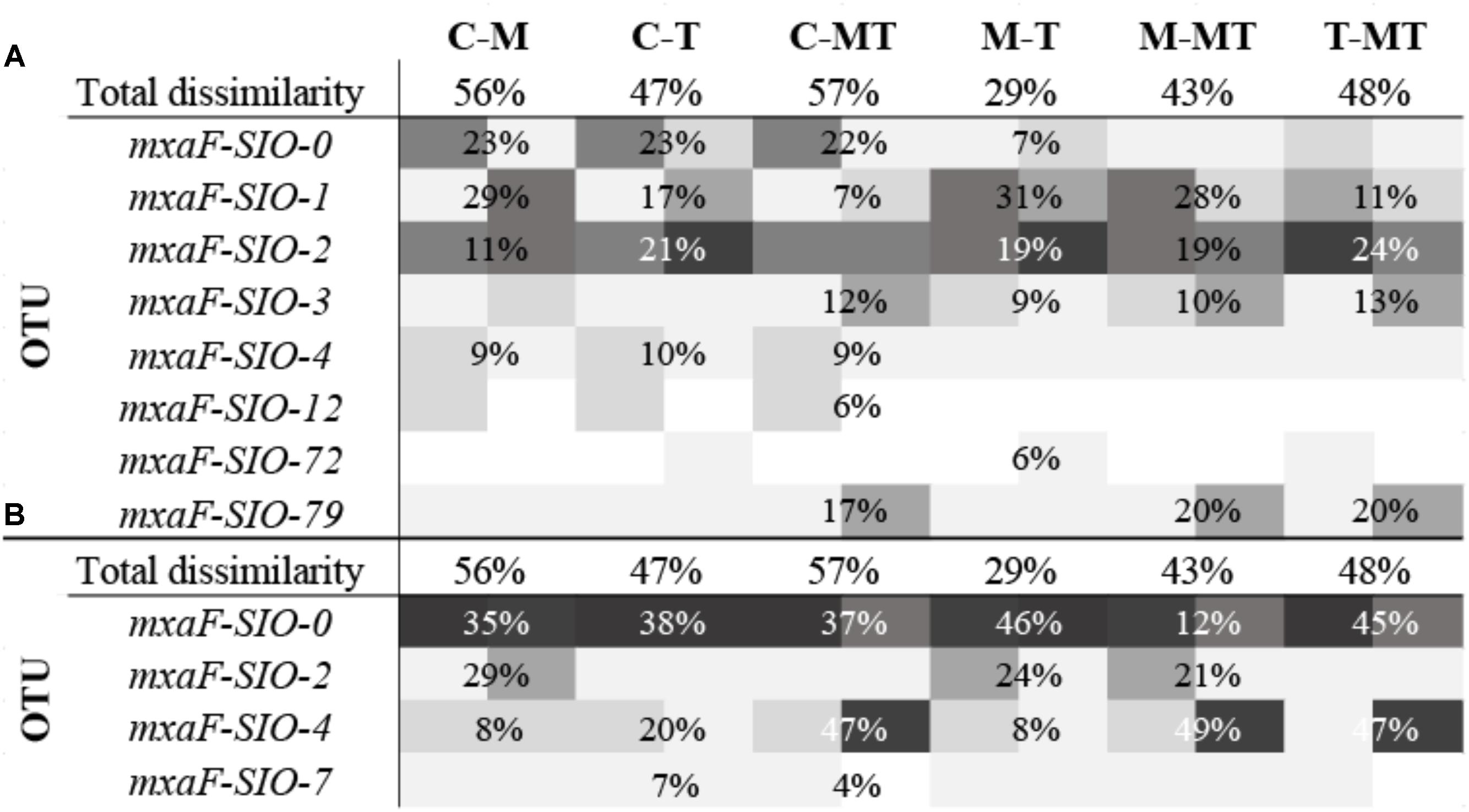
FIGURE 5. Heatmap of percent relative abundance of bacterial OTUs responsible for more than 70% of the dissimilarity between two samples (calculated by Bray Curtis, SIMPER analysis), in winter (A) and in summer (B). The percentages represent the average contribution of an OTU to the dissimilarity between the samples. Total dissimilarity includes the whole communities of the sample duplicates. C-M, controls to MetOH; C-T, controls to TMAO; C-MT, controls to metOH + TMAO; M-T, MetOH to TMAO; M-MT, MetOH to MetOH + TMAO; T-MT, TMAO to MetOH + TMAO.
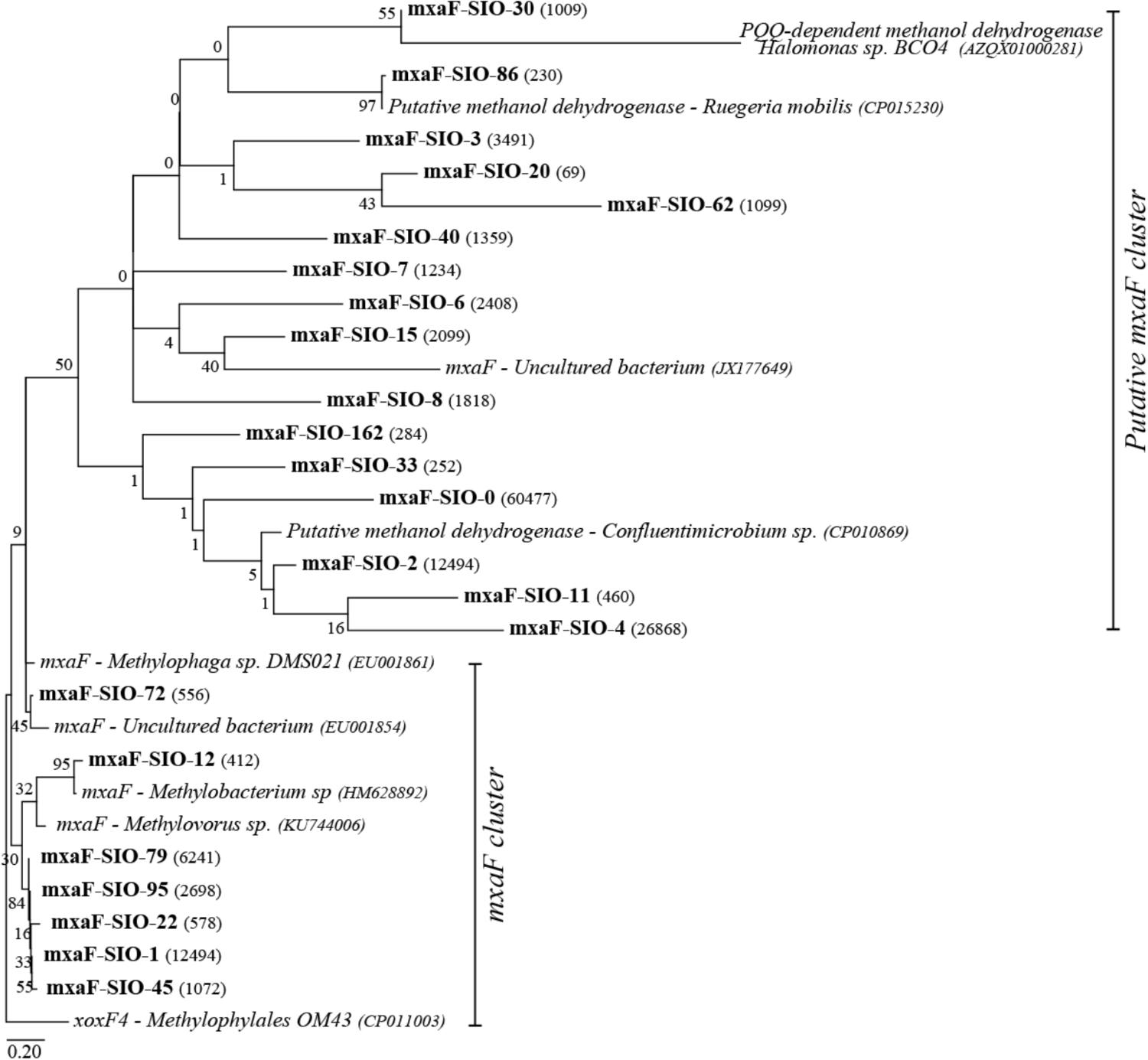
FIGURE 6. Maximum likelihood tree of the main mxaF sequences (OTU0.77), number of reads per OTUs are represented in parenthesis. Reference sequences are indicated in italics. Bootstrap values (n = 1000) are indicated at nodes; the scale bar represents changes per positions.
Putative Methylotroph Abundance and Growth Rate in Winter
The abundance of the obligate methylotroph Methylophaga spp. increased significantly over the course of the experiment in all treatments (ANOVA p < 0.001), especially in the MetOH and MetOH + TMAO treatments, which had 28-fold higher growth rates than the controls and 1.8-fold higher growth rates than TMAO treatment (Table 3 and Figure 7). Abundance of methylotrophic β- and γ-Proteobacteria with RumP pathway (except Methylophaga spp., Janvier et al., 2003) increased significantly in all treatments (ANOVA, p < 0.001). While growth rates were similar in the control and MetOH treatments, rates were 2-fold higher in the TMAO and MetOH + TMAO treatment compared to controls (Table 3 and Figure 7). The total growth rates of methylotrophic bacteria in winter was similar in all enrichments despite being driven by different methylotrophs. No methylotrophic α-Proteobacteria with serine pathways were observed in any of the samples.
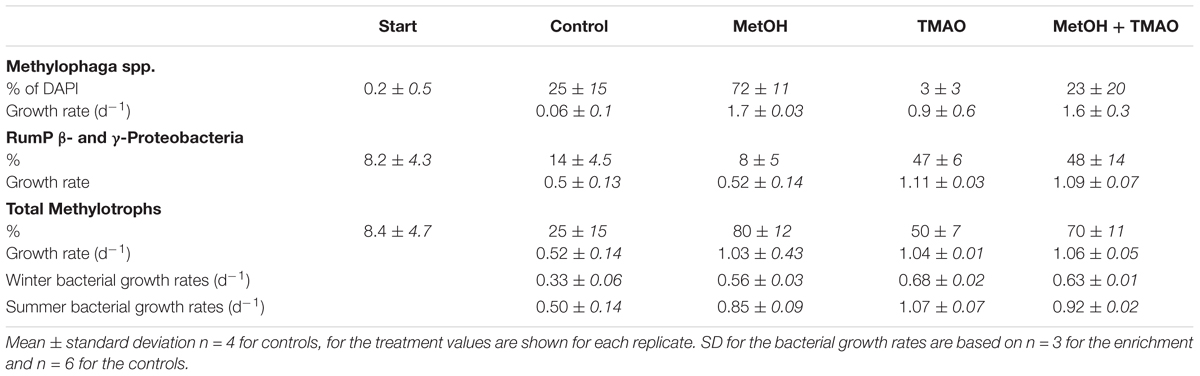
TABLE 3. Winter contribution of methylotrophs (as a % of hybridized cells to DAPI cells at 120 h, standard deviation for n = 10 fields of view) to total bacterial abundance, and total bacterial growth rates during the active phase of the winter and summer experiments.
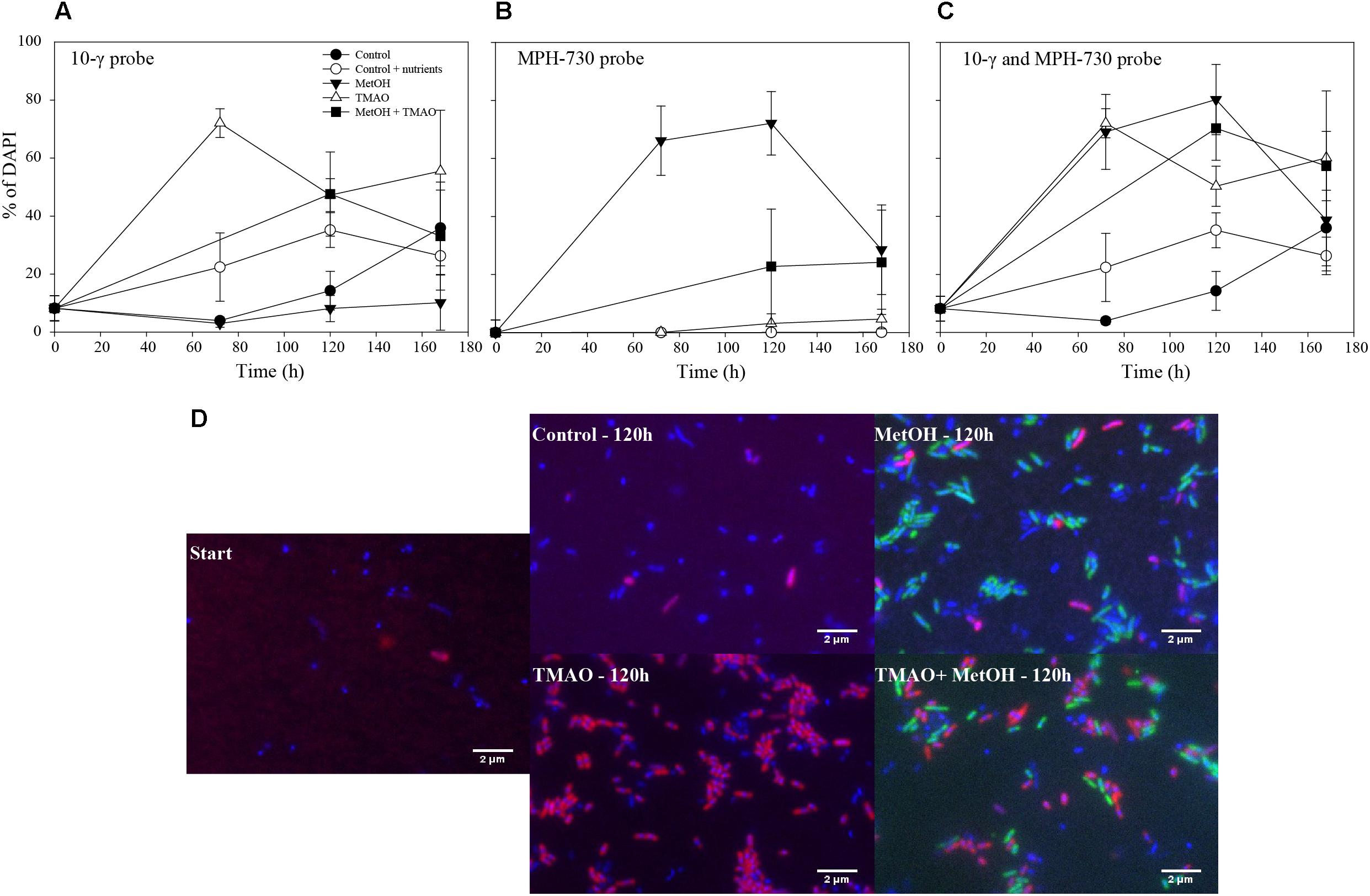
FIGURE 7. Percentage of hybridization over time for the 10-γ probe (A), MPH-730 probe (B) and the sum of both probes (C). Merged epifluorescence images of fluorescence in situ hybridization for the 10-γ probe (red) accounting for γ- and β-Proteobacterial methylotrophs with the RumP pathway, MPH-730 probes (green) accounting for Methylophaga spp. bacteria and DAPI-stained organisms (blue) (D).
Discussion
Marine bacterial communities exhibit strong temporal variations (Fuhrman et al., 2015; Bunse and Pinhassi, 2017) that are partly driven by the productivity of the system and variability in bioavailable carbon sources (Alonso-Saez and Gasol, 2007; Lønborg et al., 2009; Osterholz et al., 2016). Here, we show that the coastal bacterioplankton is rapidly responding to an addition of methylated compounds and that enrichment of coastal bacterioplankton with methylotrophic capacity varies with the productivity of the system. This also implies that methylotrophy may have seasonally varying effects on carbon cycling and air-sea fluxes in coastal waters.
Bacterial Carbon Drawdown in Response to Methanol and Trimethylamine-N-oxide Additions
Marine bacteria can use methanol and TMAO as a source of carbon for biomass and/or energy (e.g., Sun et al., 2011; Dixon et al., 2012; Lidbury et al., 2014b). In marine waters, methanol and TMAO may be produced by phytoplankton (Nieder et al., 2014; Mincer and Aicher, 2016) among others. Thus, their concentration may change depending on the productivity of the system. We hypothesized that marine bacteria would respond differently to C1 compounds during periods of different biological productivity. The colder less productive (lower Chl a, POC and bacterial production) winter water was characterized by lower bacterial activities compared to the warmer water that followed the spring bloom. Bacteria responded to the input of carbon in both experiments, but the lag phase was slightly longer in winter, which is typical for slower metabolism in colder waters (Degerman et al., 2012). The added compounds were more labile to the winter bacteria, probably due to the lower biological productivity during the winter season and subsequent lower concentration of fresh bioavailable carbon (Lønborg et al., 2009).
In summer, the minor differences observed between the controls and treatments, as well as lower bacterial growth efficiencies when compared to winter, suggest that added substrates were mainly shunted toward respiration and used as a source of energy. This may be explained by the presence of more bioavailable ambient substrates subsequent to the higher spring and summer biological productivity of the coastal system. As suggested in other studies, one-carbon and methylated compounds may sustain energy demand, while other compounds are used for biomass production (Sun et al., 2011; Halsey et al., 2012), nevertheless, the addition of C1 compounds did not elicit enhanced utilization of ambient DOC compared to the carbon used in the control treatments, suggesting that energy sources were not limiting at this time of the year.
Trimethylamine-N-oxide may be used as an energy source when methanol is available for growth (Halsey et al., 2012), but the mix of both substrates did not trigger enhanced utilization of carbon compared to the methanol-alone treatment. TMAO may also be used as a source of nitrogen by nutrient limited bacteria (Lidbury et al., 2014a; Taubert et al., 2017), but here nutrients were added to all treatments, which enabled unbiased testing of utilization of carbon. No major differences were observed between the controls with and without nutrients either in winter or summer, suggesting that in situ communities were not nutrient limited and that TMAO was probably mainly used, as intended, as a source of carbon.
Bacterial Taxa Enriched During Methanol and TMAO Incubations
In winter, the initial bacterial community composition was typical of assemblages usually observed in coastal waters at this time of the year (e.g., Fuhrman et al., 2006; Gilbert et al., 2010). The main differences between the controls and the initial water was the slight increase in γ-Proteobacteria, mainly related to Oceanospirillales and Alteromonadales, which are often observed in incubation experiments responding to bottle effect (Dinasquet et al., 2013). No major community changes were observed with the addition of nutrients. The main responses observed with the C1 additions was an increase in γ- and a decrease in α-Proteobacteria. The major group enriched in the methanol treatment was related to Methylophaga spp. which is an obligate marine methylotroph (Janvier et al., 1985; Neufeld et al., 2007, 2008). Oceanospirillales also increased in the treatments with TMAO and they are known to be capable of using this substrate (Kube et al., 2013). Therefore, it appears that in winter the lag phase observed in bacterial activity in response to the addition of C1 carbon sources may be due to an adaptation of the community to the input of C1.
The lack of obvious bottle effect in summer as well as very similar bacterial community between start and end incubations in all treatments, points toward the in situ presence of many generalists taxa capable of using different carbon substrates. The main difference was a decrease in β-Proteobacteria in the C1 treatments compared to the controls. This is unexpected considering that one of the most abundant coastal methylotrophs, the β-Proteobacteria OM43 (Giovannoni et al., 2008; Halsey et al., 2012; Gifford et al., 2016) is usually observed in summer and during incubation experiments (Sosa et al., 2015). Nevertheless, it appears that this oligotrophic methylotroph might have been outcompeted here. The observed low abundance of OM43 may also be the results of primer bias (Parada et al., 2016). Nevertheless, a moderate response to the added carbon compounds was observed at lower taxonomic resolutions as previously observed during incubation experiments (Dinasquet et al., 2013). Methylophaga spp. related OTUs increased in all C1 treatments, especially in the TMAO-only treatment, which was different from the pattern observed in winter. This might be due to the simultaneous utilization of TMAO between N users and C1 users (McCarren et al., 2010; Lidbury et al., 2014a) in the TMAO-only treatment where Methylophaga did not have to compete with other C1 users, which may have been the case in the mixed treatment. The relative abundance of SAR11, the most abundant bacterial clade observed in marine ecosystems (Morris et al., 2002), decreased in all treatments and seasons. Despite its capacity to use C1 compounds as a source of energy (Sun et al., 2011) SAR11 is an oligotrophic clade, which is easily outcompeted in incubation experiments (Giovannoni, 2017).
Overall, it seems that while the winter bacterial community became enriched in specialist methylotrophs in response to the addition of methanol and TMAO, the already diverse and potentially generalist community was resistant to these perturbations during the productive period (summer).
Methylotrophic Tracers During Methanol and TMAO Incubations
Methanol incorporation measurements have been used as indicators of methylotrophic activities (Dixon et al., 2011b) and the marker gene for the methanol dehydrogenase large subunit mxaF has been used as a tracer for enrichment of methylotrophic taxa (McDonald and Murrell, 1997; Neufeld et al., 2007). However, mxaF may not be the most common gene marker for methylotrophy in marine environments, where xoxF is sometimes the most abundant methanol dehydrogenase (Ramachandran and Walsh, 2015; Taubert et al., 2015). Moreover, TMAO methylotrophic users do not all possess the mxaF gene and the capacity to assimilate methanol. Therefore, the tracers used in this study are probably underestimating the methylotrophic activity in the TMAO and mixed treatments (Chen, 2012). Sequencing the genes related to TMAO demethylation (Neufeld et al., 2007; Chen, 2012) and using radiolabeled TMAO marker could help better resolve the diversity and activities of TMAO users.
Methanol assimilation increased in both experiments, suggesting that both winter and summer enriched bacterial assemblages had the capacity to use methanol for biomass production. The measured assimilation rates were lower than in some coastal areas (Dixon et al., 2012; Halsey et al., 2012) but higher than in the English Channel (Sargeant et al., 2016) and overall in the range of methanol utilization (BGEM) observed in the coastal areas (Dixon et al., 2012). Increase in methanol utilization was more remarkable in winter when compared to summer. In winter, methanol represented up to 1.2% of the carbon used for biomass production by bacteria, while it was only up to 0.01% of bacterial production in summer (average ratio of the methanol assimilation:bacterial production in the treatments with carbon addition integrated over the active phase of the experiment). In summer, the high methanol respiration observed in the treatments with carbon enrichment concords with the utilization of C1 compounds as energy source, as was suggested above.
The observed change in bacterial community composition in winter in response to the addition of carbon source was reflected in the change in functional community. While Methylophilaceae are known methylotrophs found in surface waters (Giovannoni et al., 2008; Halsey et al., 2012), 16S rRNA gene sequence related to β-Proteobacteria were low. However, the contribution of mxaF-OTUs related to Methylophilaceae increased especially in the treatments with methanol, suggesting that despite low overall abundance based on 16S rRNA genes, they may have an important functional role in the system. In summer, the functional community appeared to be especially related to unknown/uncultured putative methylotrophs related to α- and γ-Proteobacteria. For instance, no Oceanospirillales representative have been previously shown to grow on methanol as their sole source of carbon, but they can use TMAO (Kube et al., 2013); the presence of mxaF-OTU related to Oceanospirillales in our treatment suggest that some may also be able to use methanol as a carbon source. This new capacities could have been acquired through horizontal gene transfer of mxaF between bacterial groups (Vuilleumier et al., 2009).
The growth of the specific winter methylotrophic population was further assessed through FISH over the course of the experiment. The enrichment of Methylophaga spp. observed at the end of the experiment through 16S rRNA gene sequencing was confirmed with FISH, showing a higher growth rate of Methylophaga spp. in the treatments with methanol. Other γ- and β-Proteobacteria methylotrophs grew faster in the treatments with TMAO. This is also consistent with the sequencing results. Overall, it appears that the change in the functional community observed in winter, but not in summer, was related to the enrichment of substrate specific methylotrophs. Nevertheless, the final growth and production rates of these different TMAO or methanol enriched populations were similar, suggesting a functional redundancy of the substrate specific methylotrophic populations.
Our results suggest that bacterial response to a pulse of C1 may be more important in less productive coastal waters. For instance a pulse of methanol could occur during methanol rich rainfalls (Felix et al., 2014; Mullaugh et al., 2015) more frequently in winter. And similar response could be observed at the onset of the spring bloom when waters are still carbon limited. But despite the subsequent enrichment of substrate specific methylotrophic populations their feedback on the marine carbon budget may be similar due to their functional redundancy. Nevertheless, it will be important to further study the effect of bacterial C1 drawdown and transformation, on air-sea fluxes as these will be substrate-dependent and unrelated to functional redundancy.
Author Contributions
JD designed and carried out the study and data analysis. JD and MT prepared the samples for sequencing. JD wrote the manuscript. All authors contributed to the revisions, edited the manuscript, and approved the submitted version.
Funding
This work was supported by a Marie Curie Actions-International Outgoing Fellowship (PIOF-GA-2013-629378 to JD), the European Research Council (Consolidator Grant 615146 to MT) and the Gordon and Betty Moore Foundation (Grant GBMF4827 to FA).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank M. Sc. Anita Mäki and Lionel Le Toux for helping with the sequencing and bioinformatics. We would also like to acknowledge the SCOOS program for sharing their monitoring data for Chl a off SIO Pier, as well as B. Stephens and L. Aluwihare for sharing the in situ POC and DOC values.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00307/full#supplementary-material
Footnotes
References
Alonso-Saez, L., and Gasol, J. M. (2007). Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 73, 3528–3535. doi: 10.1128/AEM.02627-06
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Beale, R., and Airs, R. (2016). Quantification of glycine betaine, choline and trimethylamine N-oxide in seawater particulates: minimisation of seawater associated ion suppression. Anal. Chim. Acta 938, 114–122. doi: 10.1016/j.aca.2016.07.016
Beale, R., Dixon, J. L., Smyth, T. J., and Nightingale, P. D. (2015). Annual study of oxygenated volatile organic compounds in UK shelf waters. Mar. Chem. 171, 96–106. doi: 10.1016/j.marchem.2015.02.013
Beale, R., Liss, P. S., Dixon, J. L., and Nightingale, P. D. (2011). Quantification of oxygenated volatile organic compounds in seawater by membrane inlet-proton transfer reaction/mass spectrometry. Anal. Chim. Acta 706, 128–134. doi: 10.1016/j.aca.2011.08.023
Bratbak, G. (1985). Bacterial biovolume and biomass estimations. Appl. Environ. Microbiol. 49, 1488–1493.
Bunse, C., and Pinhassi, J. (2017). Marine bacterioplankton seasonal succession dynamics. Trends Microbiol. 6, 494–505. doi: 10.1016/j.tim.2016.12.013
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010a). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carpenter, L. J., Archer, S. D., and Beale, R. (2012). Ocean-atmosphere trace gas exchange. Chem. Soc. Rev. 41, 6473–6506. doi: 10.1039/c2cs35121h
Chen, Y. (2012). Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ. Microbiol. 14, 2308–2322. doi: 10.1111/j.1462-2920.2012.02765.x
Chen, Y., Patel, N. A., Crombie, A., Scrivens, J. H., and Murrell, J. C. (2011). Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 108, 17791–17796. doi: 10.1073/pnas.1112928108
Chistoserdova, L. (2015). Methylotrophs in natural habitats: current insights through metagenomics. Appl. Microbiol. Biotechnol. 99, 5763–5779. doi: 10.1007/s00253-015-6713-z
Chistoserdova, L., Kalyuzhnaya, M. G., and Lidstrom, M. E. (2009). The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63, 477–499. doi: 10.1146/annurev.micro.091208.073600
Clarke, K. R., Gorley, R. N., Somerfield, P. J., and Warwick, R. M. (2014) Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 3rd Edn. Plymouth: PRIMER-E, 260.
Clarke, K. R., and Warwick, R. M. (2001). Change in Marine Communities: an Approach to Statistical Analysis and Interpretation, 2nd Edn. Plymouth: PRIMER-E, 172.
Degerman, R., Dinasquet, J., Riemann, L., De Luna, S. S., and Andersson, A. (2012). Effect of resource availability on bacterial community responses to increased temperature. Aquat. Microb. Ecol. 68, 131–142. doi: 10.3354/ame01609
Dinasquet, J., Kragh, T., Schroter, M. L., Sondergaard, M., and Riemann, L. (2013). Functional and compositional succession of bacterioplankton in response to a gradient in bioavailable dissolved organic carbon. Environ. Microbiol. 15, 2616–2628. doi: 10.1111/1462-2920.12178
Dixon, J. L., Beale, R., and Nightingale, P. D. (2011a). Microbial methanol uptake in northeast Atlantic waters. ISME J. 5, 704–716. doi: 10.1038/ismej.2010.169
Dixon, J. L., Beale, R., and Nightingale, P. D. (2011b). Rapid biological oxidation of methanol in the tropical Atlantic: significance as a microbial carbon source. Biogeosciences 8, 2707–2716. doi: 10.5194/bg-8-2707-2011
Dixon, J. L., Beale, R., and Nightingale, P. D. (2013). Production of methanol, acetaldehyde, and acetone in the Atlantic Ocean. Geophys. Res. Lett. 40, 4700–4705. doi: 10.1002/grl.50922
Dixon, J. L., Sargeant, S., Nightingale, P. D., and Murrell, J. C. (2012). Gradients in microbial methanol uptake: productive coastal upwelling waters to oligotrophic gyres in the Atlantic Ocean. ISME J. 7, 568–580. doi: 10.1038/ismej.2012.130
Ducklow, H. W., Purdie, D. A., Williams, P. J. L., and Davies, J. M. (1986). Bacterioplankton: a sink for carbon in a coastal marine plankton community. Science 232, 865–867. doi: 10.1126/science.232.4752.865
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Facchini, M. C., Decesari, S., Rinaldi, M., Carbone, C., Finessi, E., Mircea, M., et al. (2008). Important source of marine secondary organic aerosol from biogenic amines. Environ. Sci. Technol. 42, 9116–9121. doi: 10.1021/es8018385
Felix, J., Jones, S., Avery, G., Willey, J., Mead, R., and Kieber, R. (2014). Temporal and spatial variations in rainwater methanol. Atmos. Chem. Phys. Discuss. 14, 1375–1398. doi: 10.5194/acpd-14-1375-2014
Fuhrman, J. A., Cram, J. A., and Needham, D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Fuhrman, J. A., Hewson, I., Schwalbach, M. S., Steele, J. A., Brown, M. V., and Naeem, S. (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. USA 103, 13104–13109. doi: 10.1073/pnas.0602399103
Gifford, S. M., Becker, J. W., Sosa, O. A., Repeta, D. J., and Delong, E. F. (2016). Quantitative transcriptomics reveals the growth- and nutrient-dependent response of a streamlined marine methylotroph to methanol and naturally occurring dissolved organic matter. mBio 7:e01279-16. doi: 10.1128/mBio.01279-16
Gilbert, J. A., Field, D., Swift, P., Thomas, S., Cummings, D., Temperton, B., et al. (2010). The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-Omic’ study of seasonal and diel temporal variation. PLoS One 5:e15545. doi: 10.1371/journal.pone.0015545
Giovannoni, S. J. (2017). SAR11 bacteria: the most abundant plankton in the oceans. Ann. Rev. Mar. Sci. 9, 231–255. doi: 10.1146/annurev-marine-010814-015934
Giovannoni, S. J., Hayakawa, D. H., Tripp, H. J., Stingl, U., Givan, S. A., Cho, J.-C., et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ. Microbiol. 10, 1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x
Glöckner, F. O., Amann, R., Alfreider, A., Pernthaler, J., Psenner, R., Trebesius, K., et al. (1996). An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19, 403–406. doi: 10.1016/S0723-2020(96)80069-5
Halsey, K. H., Carter, A. E., and Giovannoni, S. J. (2012). Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ. Microbiol. 14, 630–640. doi: 10.1111/j.1462-2920.2011.02605.x
Janvier, M., Frehel, C., Grimont, F., and Gasser, F. (1985). Methylophaga marina gen. nov., sp. nov. and Methylophaga thalassica sp. nov., marine methylotrophs. Int. J. Syst. Evol. Microbiol. 35, 131–139. doi: 10.1099/00207713-35-2-131
Janvier, M., Regnault, B., and Grimont, P. (2003). Development and use of fluorescent 16S rRNA-targeted probes for the specific detection of Methylophaga species by in situ hybridization in marine sediments. Res. Microbiol. 154, 483–490. doi: 10.1016/S0923-2508(03)00146-3
Karl, D. M., Hebel, D. V., Bjorkman, K., and Letelier, R. M. (1998). The role of dissolved organic matter release in the productivity of the oligotrophic North Pacific Ocean. Limnol. Oceanogr. 43, 1270–1286. doi: 10.4319/lo.1998.43.6.1270
Kirchman, D. L., Knees, E., and Hodson, R. (1985). Leucine incorporation and its potential as a measure of protein-synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49, 599–607.
Kragh, T., Søndergaard, M., and Tranvik, L. (2008). Effect of exposure to sunlight and phosphorus-limitation on bacterial degradation of coloured dissolved organic matter (CDOM) in freshwater. FEMS Microbiol. Ecol. 64, 230–239. doi: 10.1111/j.1574-6941.2008.00449.x
Kube, M., Chernikova, T. N., Al-Ramahi, Y., Beloqui, A., Lopez-Cortez, N., Guazzaroni, M.-E., et al. (2013). Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica. Nat. Commun. 4:2156. doi: 10.1038/ncomms3156
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lidbury, I., Mausz, M. A., Scanlan, D. J., and Chen, Y. (2017). Identification of dimethylamine monooxygenase in marine bacteria reveals a metabolic bottleneck in the methylated amine degradation pathway. ISME J. 11, 1592–1601. doi: 10.1038/ismej.2017.31
Lidbury, I., Murrell, J. C., and Chen, Y. (2014b). Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 2710–2715. doi: 10.1073/pnas.1317834111
Lidbury, I. D., Murrell, J. C., and Chen, Y. (2014a). Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J. 9, 760–769. doi: 10.1038/ismej.2014.149
Lønborg, C., Davidson, K., Álvarez–Salgado, X. A., and Miller, A. E. J. (2009). Bioavailability and bacterial degradation rates of dissolved organic matter in a temperate coastal area during an annual cycle. Mar. Chem. 113, 219–226. doi: 10.1016/j.marchem.2009.02.003
Mäki, A., Rissanen, A., and Tiirola, M. (2016). A practical method for barcoding and size-trimming PCR templates for amplicon sequencing. Biotechniques 60, 88–90. doi: 10.2144/000114380
McCarren, J., Becker, J. W., Repeta, D. J., Shi, Y. M., Young, C. R., Malmstrom, R. R., et al. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U.S.A. 107, 16420–16427. doi: 10.1073/pnas.1010732107
McDonald, I. R., and Murrell, J. C. (1997). The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl. Environ. Microbiol. 63, 3218–3224.
Mincer, T. J., and Aicher, A. C. (2016). Methanol production by a broad phylogenetic array of marine phytoplankton. PLoS One 11:e0150820. doi: 10.1371/journal.pone.0150820
Morris, R. M., Rappe, M. S., Connon, S. A., Vergin, K. L., Siebold, W. A., Carlson, C. A., et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810. doi: 10.1038/nature01240
Mou, X. Z., Sun, S. L., Edwards, R. A., Hodson, R. E., and Moran, M. A. (2008). Bacterial carbon processing by generalist species in the coastal ocean. Nature 451, 708–711. doi: 10.1038/nature06513
Mullaugh, K. M., Hamilton, J. M., Avery, G. B., Felix, J. D., Mead, R. N., Willey, J. D., et al. (2015). Temporal and spatial variability of trace volatile organic compounds in rainwater. Chemosphere 134, 203–209. doi: 10.1016/j.chemosphere.2015.04.027
Müller, C., Iinuma, Y., Karstensen, J., Pinxteren, D. V., Lehmann, S., Gnauk, T., et al. (2009). Seasonal variation of aliphatic amines in marine sub-micrometer particles at the Cape Verde islands. Atmos. Chem. Phys. 9, 9587–9597. doi: 10.5194/acp-9-9587-2009
Murrell, J. C., Mcgowan, V., and Cardy, D. L. N. (1993). Detection of methylotrophic bacteria in natural samples by molecular probing techniques. Chemosphere 26, 1–11. doi: 10.1016/0045-6535(93)90408-W
Neufeld, J. D., Boden, R., Moussard, H., Schafer, H., and Murrell, J. C. (2008). Substrate-specific clades of active marine methylotrophs associated with a phytoplankton bloom in a temperate coastal environment. Appl. Environ. Microbiol. 74, 7321–7328. doi: 10.1128/AEM.01266-08
Neufeld, J. D., Schafer, H., Cox, M. J., Boden, R., Mcdonald, I. R., and Murrell, J. C. (2007). Stable-isotope probing implicates Methylophaga spp and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J. 1, 480–491. doi: 10.1038/ismej.2007.65
Nieder, J. B., Hussels, M., Bittl, R., and Brecht, M. (2014). Effect of TMAO and betaine on the energy landscape of photosystem I. Biochim. Biophys. Acta Bioenerg. 1837, 849–856. doi: 10.1016/j.bbabio.2014.01.005
Osterholz, H., Singer, G., Wemheuer, B., Daniel, R., Simon, M., Niggemann, J., et al. (2016). Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 10, 1717–1730. doi: 10.1038/ismej.2015.231
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Poretsky, R. S., Sun, S. L., Mou, X. Z., and Moran, M. A. (2010). Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12, 616–627. doi: 10.1111/j.1462-2920.2009.02102.x
Porter, K. G., and Feig, Y. S. (1980). The use of dapi for identifying and counting aquatic microflora. Limnol. Oceanogr. 25, 943–948. doi: 10.4319/lo.1980.25.5.0943
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Ramachandran, A., and Walsh, D. A. (2015). Investigation of XoxF methanol dehydrogenases reveals new methylotrophic bacteria in pelagic marine and freshwater ecosystems. FEMS Microbiol. Ecol. 91:fiv105. doi: 10.1093/femsec/fiv105
Sargeant, S. L., Murrell, J. C., Nightingale, P. D., and Dixon, J. L. (2016). Seasonal variability in microbial methanol utilisation in coastal waters of the western English Channel. Mar. Ecol. Prog. Ser. 550, 53–64. doi: 10.3354/meps11705
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Simon, M., and Azam, F. (1989). Protein-content and protein-synthesis rates of planktonic marine-bacteria. Mar. Ecol. Prog. Ser. 51, 201–213. doi: 10.3354/meps051201
Singh, H., Chen, Y., Tabazadeh, A., Fukui, Y., Bey, I., Yantosca, R., et al. (2000). Distribution and fate of selected oxygenated organic species in the troposphere and lower stratosphere over the Atlantic. J. Geophys. Res. Atmos. 105, 3795–3805. doi: 10.1029/1999JD900779
Singh, H. B., Tabazadeh, A., Evans, M. J., Field, B. D., Jacob, D. J., Sachse, G., et al. (2003). Oxygenated volatile organic chemicals in the oceans: inferences and implications based on atmospheric observations and air-sea exchange models. Geophys. Res. Lett. 30, 1862–1866. doi: 10.1029/2003GL017933
Smith, D. C., and Azam, F. (1992). A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6, 107–114.
Sorooshian, A., Padró, L. T., Nenes, A., Feingold, G., Mccomiskey, A., Hersey, S. P., et al. (2009). On the link between ocean biota emissions, aerosol, and maritime clouds: airborne, ground, and satellite measurements off the coast of California. Global Biogeochem. Cycles 23:GB4007. doi: 10.1029/2009GB003464
Sosa, O. A., Gifford, S. M., Repeta, D. J., and Delong, E. F. (2015). High molecular weight dissolved organic matter enrichment selects for methylotrophs in dilution to extinction cultures. ISME J. 9, 2725–2739. doi: 10.1038/ismej.2015.68
Stacheter, A., Noll, M., Lee, C. K., Selzer, M., Glowik, B., Ebertsch, L., et al. (2012). Methanol oxidation by temperate soils and environmental determinants of associated methylotrophs. ISME J. 7, 1051–1064. doi: 10.1038/ismej.2012.167
Stark, S., Mannisto, M. K., Ganzert, L., Tiirola, M., and Haggblom, M. M. (2015). Grazing intensity in subarctic tundra affects the temperature adaptation of soil microbial communities. Soil Biol. Biochem. 84, 147–157. doi: 10.1016/j.soilbio.2015.02.023
Sun, J., Steindler, L., Thrash, J. C., Halsey, K. H., Smith, D. P., Carter, A. E., et al. (2011). One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6:e23973. doi: 10.1371/journal.pone.0023973
Taubert, M., Grob, C., Howat, A., Burns, O. J., Dixon, J. L., Chen, Y., et al. (2015). xoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ. Microbiol. 17, 3937–3948. doi: 10.1111/1462-2920.12896
Taubert, M., Grob, C., Howat, A. M., Burns, O. J., Pratscher, J., Jehmlich, N., et al. (2017). Methylamine as a nitrogen source for microorganisms from a coastal marine environment. Environ. Microbiol. 19, 2246–2257. doi: 10.1111/1462-2920.13709
Tsien, H. C., Bratina, B. J., Tsuji, K., and Hanson, R. S. (1990). Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl. Environ. Microbiol. 56, 2858–2865.
Vuilleumier, S., Chistoserdova, L., Lee, M.-C., Bringel, F., Lajus, A., Zhou, Y., et al. (2009). Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4:e5584. doi: 10.1371/journal.pone.0005584
Williams, J., Holzinger, R., Gros, V., Xu, X., Atlas, E., and Wallace, D. W. (2004). Measurements of organic species in air and seawater from the tropical Atlantic. Geophys. Res. Lett. 31:L23S06. doi: 10.1029/2004GL020012
Yang, M., Beale, R., Liss, P., Johnson, M., Blomquist, B., and Nightingale, P. (2014a). Air–sea fluxes of oxygenated volatile organic compounds across the Atlantic Ocean. Atmos. Chem. Phys. 14, 7499–7517. doi: 10.5194/acp-14-7499-2014
Yang, M., Blomquist, B. W., and Nightingale, P. D. (2014b). Air-sea exchange of methanol and acetone during HiWinGS: estimation of air phase, water phase gas transfer velocities. J. Geophys. Res. Oceans 119, 7308–7323. doi: 10.1002/2014JC010227
Keywords: C1, methanol, TMAO, bacterial community composition, mxaF, coastal ecosystem
Citation: Dinasquet J, Tiirola M and Azam F (2018) Enrichment of Bacterioplankton Able to Utilize One-Carbon and Methylated Compounds in the Coastal Pacific Ocean. Front. Mar. Sci. 5:307. doi: 10.3389/fmars.2018.00307
Received: 12 April 2018; Accepted: 10 August 2018;
Published: 06 September 2018.
Edited by:
Veronica Molina, Universidad de Playa Ancha, ChileReviewed by:
Héctor A. Levipan, Centro de Investigación Marina Quintay (CIMARQ), ChileCamila Fernandez, UMR7621 Laboratoire d’Océanographie Microbienne (LOMIC), France
Copyright © 2018 Dinasquet, Tiirola and Azam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie Dinasquet, amRpbmFzcXVldEB1Y3NkLmVkdQ==
 Julie Dinasquet
Julie Dinasquet Marja Tiirola
Marja Tiirola Farooq Azam
Farooq Azam