- 1School of BioSciences, University of Melbourne, Parkville, VIC, Australia
- 2Department of Freshwater and Marine Ecology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, Netherlands
- 3School of Biological Sciences, Victoria University of Wellington, Wellington, New Zealand
- 4Australian Institute of Marine Science, Townsville, QLD, Australia
The mutualistic symbiosis between cnidarians and photosynthetic dinoflagellates supports one of the most diverse ecosystems on the planet, coral reefs. Cnidarian-Symbiodiniaceae symbioses are broadly species-specific, but little is known about the mechanisms underpinning this specificity. Here, we explored the ability of three genotypes of the sea anemone Exaiptasia diaphana (Aiptasia) – a model organism for the cnidarian-dinoflagellate symbiosis – from the Great Barrier Reef (GBR), to take up and maintain seven different Symbiodiniaceae strains. A method to track the number of symbiont cells by quantitative microscopy of algal chlorophyll auto-fluorescence in the anemone tentacles was developed. Breviolum minutum, the homologous (i.e., native) symbiont in these anemones, was the most successful of the seven algal types tested at colonizing aposymbiotic anemones of all three genotypes. The heterologous (i.e., non-native) but compatible species Cladocopium goreaui was also able to colonize GBR anemones, albeit at lower cell densities. Durusdinium trenchii, Fugacium kawagutii, “Symbiodinium F5.1,” and “Symbiodinium G3” showed little or no ability to colonize any E. diaphana genotype, and Symbiodinium tridacnidorum, isolated from clams, apparently killed the anemones. Histology localized the homologous and compatible heterologous symbionts within the endodermis of the host, but appreciable numbers of C. goreaui cells were not fully internalized by anemone cells. Colonization dynamics were influenced by symbiont type and host genotype, suggesting that a mechanism of recognition and incorporation has components in both symbiont and host. The matrix of different host–symbiont compatibilities described here can be used to explore the molecular mechanisms of recognition and establishment of cnidarian-Symbiodiniaceae symbiosis.
Introduction
Symbiotic associations that are of mutual advantage to both partners are common and widespread in nature – so common in fact that probably all macroorganisms rely on microbial symbionts for optimal health and function (McFall-Ngai et al., 2013). Mutualistic interactions underpin a significant amount of biodiversity and can become the basis of entire ecosystems, as is the case for coral reefs. In coral reef symbioses, the autotrophic capacity of the photosynthetic symbiont (Symbiodiniaceae) combines with the heterotrophic capacity of the cnidarian host to create a mixotrophic holobiont in which nutrient acquisition, conservation, and cycling enable very high productivity in oligotrophic environments (Margulis, 1971; Lipschultz and Cook, 2002; Yellowlees et al., 2008).
The family Symbiodinaceae comprises at least seven different genera showing various morphologies, physiologies, ecological niches and lifestyles (LaJeunesse et al., 2018). These protists have been intensively studied in the past 40 years, revealing an enormous inter-species diversity and a phylogenetic placement close to apicomplexan parasites within the sub-Kingdom Alveolata (Lenaers et al., 1991; Coats, 2002; Adl et al., 2012).
The majority of newborn coral offspring are aposymbiotic and must acquire their Symbiodinaceae algae from the environment. Although there is some flexibility in the choice of a mutualistic partner, various cnidarian-algal pairings show a high level of specificity, particularly in the adult stages (Baker, 2003; Thornhill et al., 2013). This specificity demonstrates the existence of a recognition mechanism between symbiotic partners. In other symbioses there typically exists an intricate series of inter-partner molecular signals that allow suitable partners to be identified at the onset of symbiosis (Nyholm and McFall-Ngai, 2004), but how this works for cnidarians and Symbiodinaceae is largely unknown (Davy et al., 2012; Parkinson et al., 2018).
Much progress in science has come from research on model organisms (Davis, 2004), and the sea anemone Exaiptasia diaphana (formerly Exaiptasia pallida and commonly referred to as “Aiptasia”) is increasingly used by biologists investigating the cnidarian-dinoflagellate symbiosis and its role in coral reef function. Symbiosis studies of E. diaphana usually employ one or more of five clonal lines: two from the Atlantic Ocean (CC7 and F003), one from the Pacific Ocean (H2 from Hawaii), one from an unknown location in the Indo-Pacific (NZ1), and one from the Okinawa region. E. diaphana has been divided into a lineage from the Atlantic Ocean and a network of genotypes from other localities (Thornhill et al., 2013).
These genetically distinct populations of E. diaphana are further defined by their homologous Symbiodiniaceae symbiont. CC7 anemones naturally harbor Symbiodinium linucheae (ITS2 type A4), whereas anemones from the Indo-Pacific region contain Breviolum minutum (ITS2 type B1) as their native (homologous) symbiont species (Thornhill et al., 2013). It is common for E. diaphana to have affinities for heterologous symbionts (Table 1). For instance, anemone genotype CC7 exhibits high compatibility for the homologous algae of E. diaphana from other regions (Hambleton et al., 2014; Wolfowicz et al., 2016), and the genetic lines of anemones from both Atlantic and Indo-Pacific regions are able to establish symbiosis with members of the Durusdinium genus (Wolfowicz et al., 2016; Gabay et al., 2018; Medranos et al., 2019; Sproles et al., 2019). This matrix of compatibility/incompatibility between hosts and symbionts probably represents an underlying system of genetic match-ups that regulates which partnerships occur in any given region, but the underlying cellular mechanisms are completely unknown.
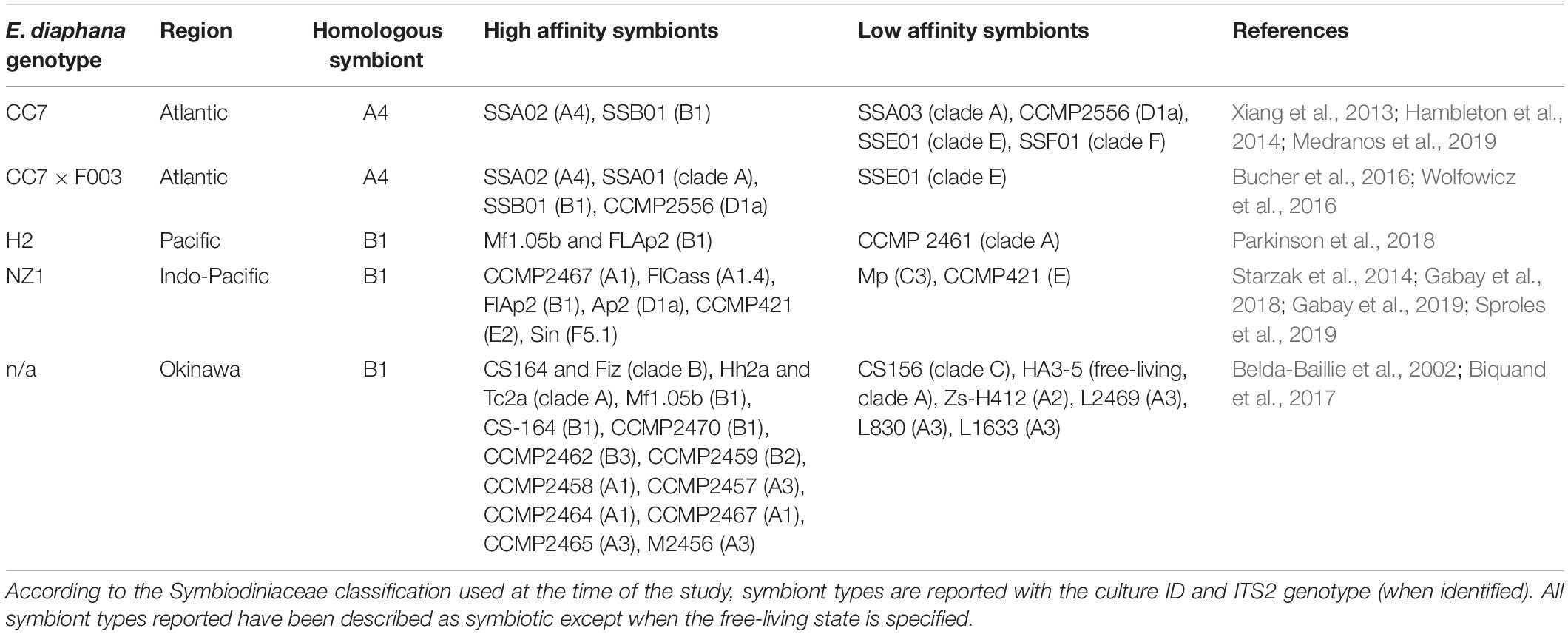
Table 1. Literature describing affinity of E. diaphana genetic lines from different localities for homologous and heterologous symbionts.
Here, we introduce for the first time E. diaphana from the Great Barrier Reef (GBR) with its homologous symbiont B. minutum. Four distinct anemone genotypes (AIMS1, AIMS2, AIMS3, AIMS4) are maintained in our laboratory at the University of Melbourne, offering an ideal opportunity to perform true biological replication of host/symbiont compatibility experiments. To explore the specificity of Australian E. diaphana strains during onset of symbiosis, three anemone genotypes (AIMS2, AIMS3, AIMS4) were chemically bleached to achieve the aposymbiotic state and challenged with seven different Symbiodinaceae strains representing five genera (Symbiodinium, Breviolum, Cladocopium, Durusdinium, and Fugacium) and two “Symbiodinium” clades (F and G) as distinguished by the former ITS2 nomenclature. We quantified the ability of hosts to establish symbiosis with homologous and heterologous symbionts over 30 days using a method to measure symbiont colonization rate that does not require the sacrifice of the experimental unit. A grid of compatibilities across this collection of seven potential symbionts and three potential hosts was defined, thereby creating an excellent system with which to begin dissecting some of the underlying cellular mechanisms of host/symbiont compatibility.
Materials and Methods
Experimental Cnidarian Host
E. diaphana polyps were of central GBR origin and were sourced from the Australian Institute of Marine Science (AIMS), Townsville, Australia. The anemones used in this study represent three different genotypes (AIMS2, AIMS3, AIMS4), distinguished based on genome-wide SNP analysis (Dungan et al., in preparation). Anemones were maintained in 0.2 μm-filtered, reconstituted seawater (FRSW) prepared with Red Sea Salt (Red Sea) dissolved in deionized water (34 ppt) and were fed ad libitum twice per week with freshly hatched Artemia sp. nauplii. Aposymbiotic anemones were created by 6 weeks of chemical-induced bleaching (Matthews et al., 2016) in a growth chamber (LE-509, Thermoline Scientific) under constant temperature (26°C), 12 h:12 h light:dark photoperiod cycle, and 15 μmol photons m–2s–1 irradiance (white + red LED lights; EDOLED).
Experimental Algal Symbionts
B. minutum was isolated from GBR E. diaphana tissue to establish a laboratory culture line (MMSF 01), which we refer to as the homologous algal symbiont. An anemone was anesthetized by immersion in a 1:1 mixture of 0.37 M MgCl2 and FRSW, and a tentacle removed and homogenized with a micro-pestle. Cells were pelleted and washed four times in FRSW and resuspended in 1× IMK+ culture medium (Daigo’s IMK, 1% w/v) prepared in FRSW (34 ppt), and supplemented with antibiotics (penicillin, streptomycin, nystatin 100 μg/ml each, amphotericin 2.5 μg/ml) to minimize bacterial growth, and germanium dioxide (GeO2 50 μM) to minimize diatom growth (Beltran et al., 2012). Algae were transferred to 24-well plates and incubated in growth chambers (740FHC LED, HiPoint) under constant temperature (26°C), 12 h:12 h light:dark photoperiod cycle, and 60 μmol photons m–2s–1 of light. Cultures were checked for cell growth and, when confirmed, the culture medium was refreshed every day (to minimize proliferation of ciliate protozoans) and algae sub-cultured in a new well. The 1× IMK+ was used for 10 additional subcultures, and then replaced by 1× IMK– (antibiotics and GeO2 free). After 1 month of incubation under the conditions described above, algae were transferred to culture flasks with 0.2 μm membrane vented caps, and maintained under constant temperature, photoperiod and irradiance.
Cultures of Symbiodinium tridacnidorum (ITS2 type A3c), Cladocopium goreaui (ITS2 type C1), Durusdinium trenchii (ITS2 type D1), Fugacium kawagutii (ITS2 type F1), “Symbiodinium F5.1” (ITS2 type F5.1), and “Symbiodinium G3” (ITS2 type G3) were obtained from AIMS. Algal cultures were maintained under the conditions reported above. The identity and source of the cultures used in this experiment are provided in Supplementary Table 1.
Experimental Design
Three anemone genotypes (AIMS2, AIMS3, AIMS4) were used in this study. Bleaching was performed on 288 anemones with an oral disk diameter of ∼3 mm, each of which was isolated in a single well of a 12-well plate. Each 12-well plate contained n = 4 anemone genotype replicates, organized randomly to minimize well effect. Each 12-well plate had n = 3 technical replicates, all inoculated with the same symbiont type. Seven Symbiodiniaceae cultures were sub-cultured in 25 cm2 flasks, 6 weeks prior to inoculation of anemones. Every 2 weeks, 1× IMK– medium was refreshed (to keep the cells in the exponential growth phase), and cultures were sub-cultured to bigger flasks (75, 150, and 225 cm2). A total of seven different algal inoculations were performed (one per each Symbiodiniaceae strain). Anemones in negative control plates did not receive algal cells. Three anemone replicates per each genotype and symbiont treatment were selected randomly at every time point post-inoculation for tentacle sampling, and three tentacles were sampled from each anemone. At 30 days post-inoculation (dpi), three anemone replicates per genotype and symbiont treatment were selected randomly for symbiont DNA extraction or histology. The experimental design is illustrated in Supplementary Figure 1.
Inoculation of Aposymbiotic Anemones With Algal Symbionts
Bleaching of anemones was assessed microscopically and, at the end of the menthol-Diuron treatment, experimental anemones were kept in FRSW and – 1 week prior to inoculation with symbionts – feeding of anemones was discontinued. For each algal culture, a sample was fixed, and cell density was determined with an automated cell counter (CountessTM II FL). New aliquots were then prepared by concentration of the algal cells and 1 ml of 1 × 106 algal cells/ml was added to each anemone well with a sterilized glass pipette, followed by 10 μl of freshly hatched brine shrimp to stimulate a feeding response (Davy et al., 1997). Four hours post-inoculation (4 hpi), a second dose of algae was provided following the same procedure. At 24 hpi, 500 μl of FRSW were added to each well to improve oxygenation and re-suspend the symbiont cells. At 48 hpi, a total medium exchange was performed, and standard feeding of anemones resumed.
Measurement of Symbiont Cell Densities in hospite
To monitor symbiont uptake and colonization rate in the anemones, we extended methods from earlier works (Berner et al., 1993; Neubauer et al., 2016; Chakravarti et al., 2017; Parkinson et al., 2018) by including a calibration curve that allows symbiont coverage to be expressed as in hospite cell density. Our approach takes advantage of fluorescent microscopy and symbiont red chlorophyll auto-fluorescence. Anemone replicates were randomly selected, anesthetized for 30 min with MgCl2 solution and tentacles excised. Because tentacle excision is non-lethal, this assay allows multiple time-points to be sampled from one individual. The interval in between sampling from the same anemone was 20 days. Excised tentacles were fixed in 4% paraformaldehyde (PFA)/phosphate buffered saline 1× (PBS) for 1 h, then stored overnight at 4°C in PBS 1×. The day after, fixed tentacles were mounted onto microscope slides in mounting medium (90% glycerol/PBS 1×) and imaged with a Leica M205 FA dissecting microscope equipped with a Leica DFC450 C camera, using the software LAS X Life Science Leica. Microscope and software settings were constant over the course of the experiment. Bright field (BF) images were captured to visualize tentacles, and algal chlorophyll auto-fluorescence within the tentacles was observed with Green Fluorescence Protein Longpass emission (GFP-LP). To monitor the uptake during the early stages of symbiosis establishment, tentacles were sampled and analyzed every 5 dpi for a total of 30 days (5, 10, 15, 20, 25, and 30 dpi). Image analysis was performed with FiJi software (Schindelin et al., 2012). Regions of interest (ROI) were determined on BF images by delineating the outline of each tentacle and calculating the area within the polygon selection (Figure 1a). The ROI was then overlaid on the corresponding GFP-LP image (Figure 1b), and a threshold was applied to the red channel of the fluorescent picture (Figure 1c) to quantify the tentacle surface area covered by algal autofluorescence. To calibrate this visual method, single tentacles of aposymbiotic anemones inoculated with the homologous alga were homogenized and symbiont cells counted in each replicate by quadruplicate hemocytometer counts. In this case, to record all the stages of symbiosis establishment, triplicate measurements were taken at 2, 5, 7, 9, 12, 15, 19, 22, 25, 30 dpi and on a permanently symbiotic anemone. The number of cells in each tentacle was plotted against the red fluorescent surface area within the same tentacle to establish a calibration curve allowing transformation of red fluorescent surface area into algal cell number (Figure 1d). Linear regression analysis was performed in R Core Team (2017) and plots were obtained using the package ggplot2 (Wickham, 2016). The detailed protocol can be accessed on at dx.doi.org/10.17504/protocols.io.2i6gch..
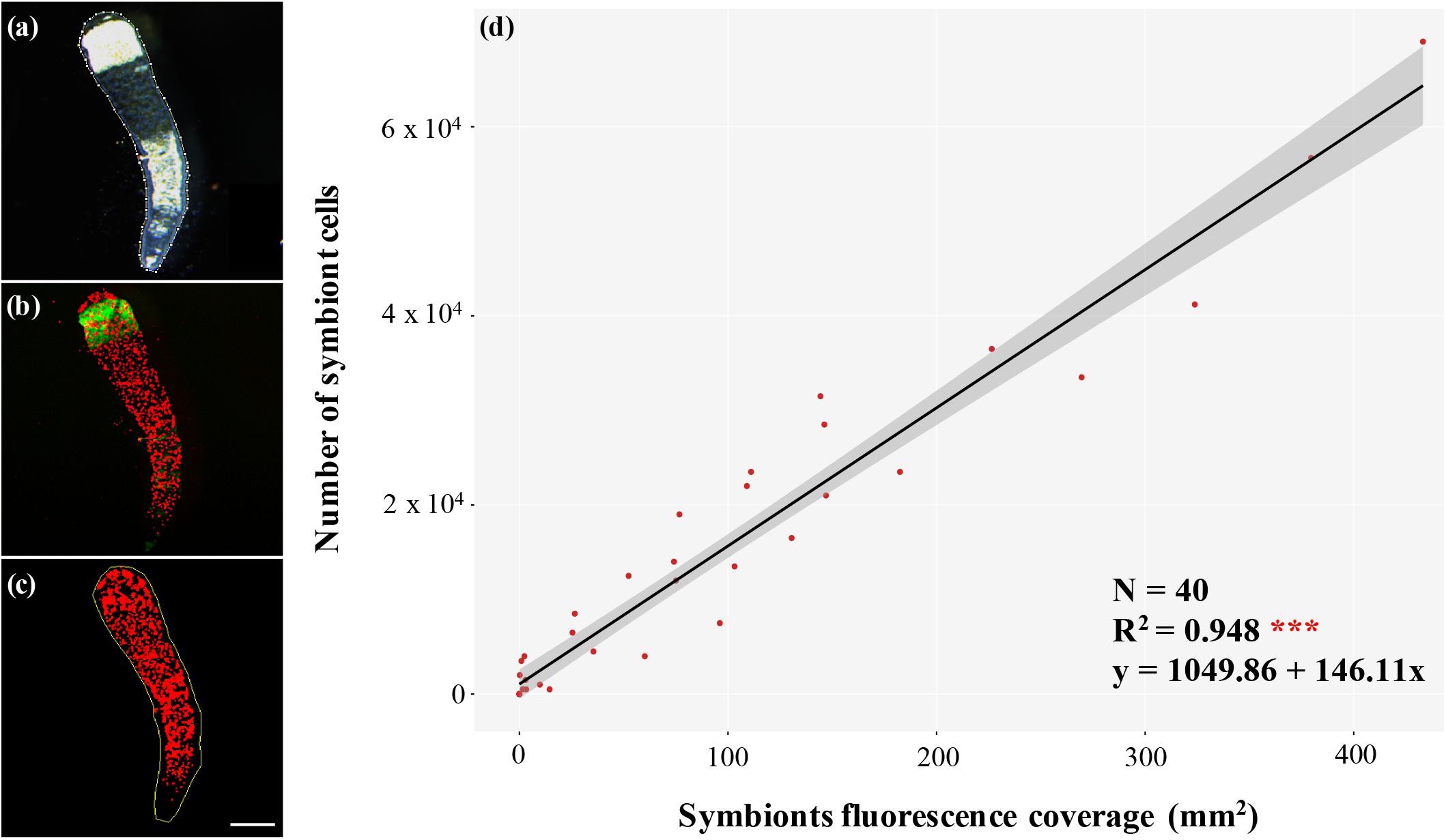
Figure 1. Accurate, non-lethal symbiont quantitation method. (a) Bright field image of a tentacle with region of interest (ROI) selected in FiJi software (dotted white line); (b) fluorescent image of the same tentacle showing algal chlorophyll autofluorescence in red and host autofluorescence in green; (c) superimposition of ROI on the red channel to which a threshold has been applied to score the pixels as either red (symbiont present) or black (symbiont absent), which allows a simple estimate of percentage area colonized, which we show is proportional to the number of symbiont cells per tentacle in panel (d); scale bar 250 μm; (d) calibration curve of the number of symbiont cells and the corresponding fluorescent area occupied within single tentacles from N = 40 anemones at 2, 5, 7, 9, 12, 15, 19, 22, 25, 30 dpi and a permanently symbiotic anemone with the homologous alga B. minutum. The two values correlate significantly (R2 = 0.948), and the resulting equation has been used to obtain symbiont density from the measured autofluorescence coverage.
Symbiont DNA Extraction and ITS2 Amplicon Sequencing
To confirm that inoculated anemones did indeed become colonized by the strain present in the inoculum, re-colonized anemones were snap-frozen at 30 dpi and DNA extracted according to Wilson et al. (2002) but modified with the inclusion of 15 min incubation with 20 μl of 10 mg/ml lysozyme along with 750 μl of lysis buffer, and 60 s bead beating at 30 Hz (Qiagen Tissue-Lyser II) with 100 mg of sterile glass beads (Sigma G8772). Polymerase chain reaction (PCR) was performed using the Symbiodiniaceae-specific ITS2 forward primer (ITS2-F) 5′-GTGAATTGCAGAACTCCGTC-3′ and reverse primer (ITS2-R) 5′-CCTCCGCTTACTTATATATGCTT-3′ (Pochon et al., 2012). Reactions were performed in a total volume of 25 μl using MangoMix (Bioline 25034) with amplification profile: 1 cycle of 5 min at 94°C; 35 cycles of 45 s at 94°C, 45 s at 55°C, and 45 s at 72°C; 1 cycle of 5 min at 72°C; and a final hold temperature of 15°C. Both strands of PCR products were Sanger sequenced with the forward and reverse primers mentioned above at the Australian Genome Research Facility (Melbourne, VIC, Australia). Taxonomic identification of sequences (CodonCode Aligner V.8.0.1) was followed by a BLASTn search performed at the National Centre for Biotechnology Information (NCBI) against the nr/nt database to taxonomically assign the sequences.
Localization of Symbionts Within Host Tissues
To verify that the algal cells were incorporated in the gastrodermal (=endodermal) tissue of the host, 30 dpi anemone tentacles were fixed in 4% PFA/PBS overnight at 4°C, and then processed at the Melbourne Histology Platform (Melbourne, VIC, Australia).
Statistical Analyses
To test the effect of host genotype (AIMS2, AIMS3, AIMS4), symbiont type, and time (as days post-inoculation, i.e., dpi) on symbiont uptake by the host (as number of symbiont cells/mm2 of anemone tentacle), a linear mixed effects model (LMEM) was used for those symbiont species for which an increase of in hospite cell densities over time was observed. Anemone genotype, time and their interaction were treated as fixed effects, plate and well placement of the organism were considered as nested random effects, and symbiont uptake was the response variable of the model. Analyses were conducted in R v. 3.4.3 (R Core Team, 2017) with the packages lme4 (Bates et al., 2012) and lmerTest (Kuznetsova et al., 2017). The lmer specification for the model was:
Inspection of residual plots did not reveal any obvious deviation from homoscedasticity or normality for the data where an increase in number of algal cells in host could be observed. The model was checked with a correlation test between observed and predicted data by using Pearson’s product moment correlation coefficient (confidence level = 0.95). Best model selection was performed by comparing the full model with all the effects against the model without each of the effects in question, and confirmed using the Akaike Information Criterion (AIC). Analysis of variance was used for the significance of the overall fixed effects fitted in the model. Post hoc tests for multiple comparisons were carried out using Tukey’s test (p > 0.05).
Results and Discussion
To explore the mechanisms involved in recognition and establishment of symbiosis, a set of host–symbiont pairs with different levels of colonization and compatibility would provide an invaluable experimental system with which to dissect the mechanism(s) of specificity. Here we describe a tractable approach to assay the number of algal cells in a host over 30 days after inoculation with prospective symbionts. We used our method to quantify the ability of three genotypes of E. diaphana from the GBR to establish symbioses with seven different strains of Symbiodiniaceae to develop a grid of compatible and incompatible matchings for further exploration.
An Accurate, Non-lethal Symbionts Quantitation Method
The number of algae internalized and retained by the host cells, especially when measured at early stages of colonization, is an index of the progress of symbiosis establishment. Although several techniques have been developed to quantify the density of in hospite symbionts, they often require the sacrifice of the organism, which prevents continuous monitoring and limits the scale of experiments.
To develop an accurate symbiont quantitation method that, while preserving host viability also provides a reasonable throughput, we took advantage of the ability of anemones to survive tentacle excision. Since tentacles are among the first areas to be colonized by symbionts in E. diaphana (Gabay et al., 2018), our approach seems especially well suited to documenting the initial stages of algal colonization. We validated our method by comparing the number of microalgae within host tentacles with the relative fluorescence area measured and showed that these measures are proportional (R2 = 0.948), which allowed us to generate a calibration curve (Figure 1d), and thus extrapolate the number of symbiont cells represented by the measured fluorescence coverage with a simple equation:
B. minutum Is the Homologous Symbiont of GBR Anemones
We confirmed findings from a previous report (Thornhill et al., 2013) that E. diaphana from the GBR harbors B. minutum. We established a unialgal culture (MMSF 01) with algae extracted from GBR anemones, and ITS2 sequencing identified it as B. minutum. We refer to this as the homologous type, whereas the six other strains used here are referred to as heterologous.
Confirmation That the Symbionts Reside in the Anemone Gastrodermis and Are Descendants of the Inoculated Strain
Histology confirmed that symbionts quantified using the fluorescence area assay were localized predominantly within gastrodermal cells of the anemones, thereby confirming symbiotic uptake and establishment of bona fide endosymbiosis (Supplementary Figure 2). Sequencing of the diagnostic ITS2 region of the symbionts at the end of the colonization experiment confirmed that in most cases the established symbiotic population was derived from the inoculum provided. In select cases, anemones inoculated with heterologous types turned out to be colonized by the homologous B. minutum; namely, two out of 12 replicates inoculated with D. trenchii, nine out of 12 replicates inoculated with F. kawagutii and four out of 12 replicates inoculated with “Symbiodinium G3”. For this reason, these samples where not considered in the analyses. Whether colonization by B. minutum resulted from incomplete bleaching or from spurious introduction is not known, but it is consistent with relatively poor colonization rates by these heterologous symbionts (see below). Anemones of the negative control showed no algae at the end of the experiment (Supplementary Figure 3). From all these observations, we conclude that the inoculation, and quantitation protocols used provide a valid method to assess symbiotic compatibility between these algae and animal hosts.
Homologous Symbionts Are the Most Effective Colonizers
The densities of six viable Symbiodiniaceae strains at the end of the 30-days experiment exhibited considerable variation among the three E. diaphana genotypes (Figures 2, 3). The homologous B. minutum was the most effective colonizer (Figure 3). At five dpi, the density of B. minutum was greater than for any other symbiont (70 ± 10 cells/mm2). B. minutum density increased further over time, reaching a maximum of 127 ± 2 cells/mm2 at 30 dpi. These results are congruent with our identification of B. minutum as the homologous symbiont in GBR anemones and are consistent with previous works showing that the homologous symbiont is the most efficient colonizer of other E. diaphana genotypes (Belda-Baillie et al., 2002; Xiang et al., 2013; Gabay et al., 2018, 2019; Parkinson et al., 2018; Medranos et al., 2019; Sproles et al., 2019). Such an affinity profile is not exclusive to anemones, as it has also been reported in several corals (Weis et al., 2001; Rodriguez-Lanetty et al., 2003, 2004; Wolfowicz et al., 2016; Lin et al., 2019). Importantly, this consistency in preference for homologous symbionts lends further weight to the putative existence of a recognition mechanism that shapes cnidarian specificity for particular endosymbionts (Weis et al., 2008). The system described here is in line with prior findings and confirms the possibility of using homologous symbionts as positive controls in future investigations on recognition in symbiosis.
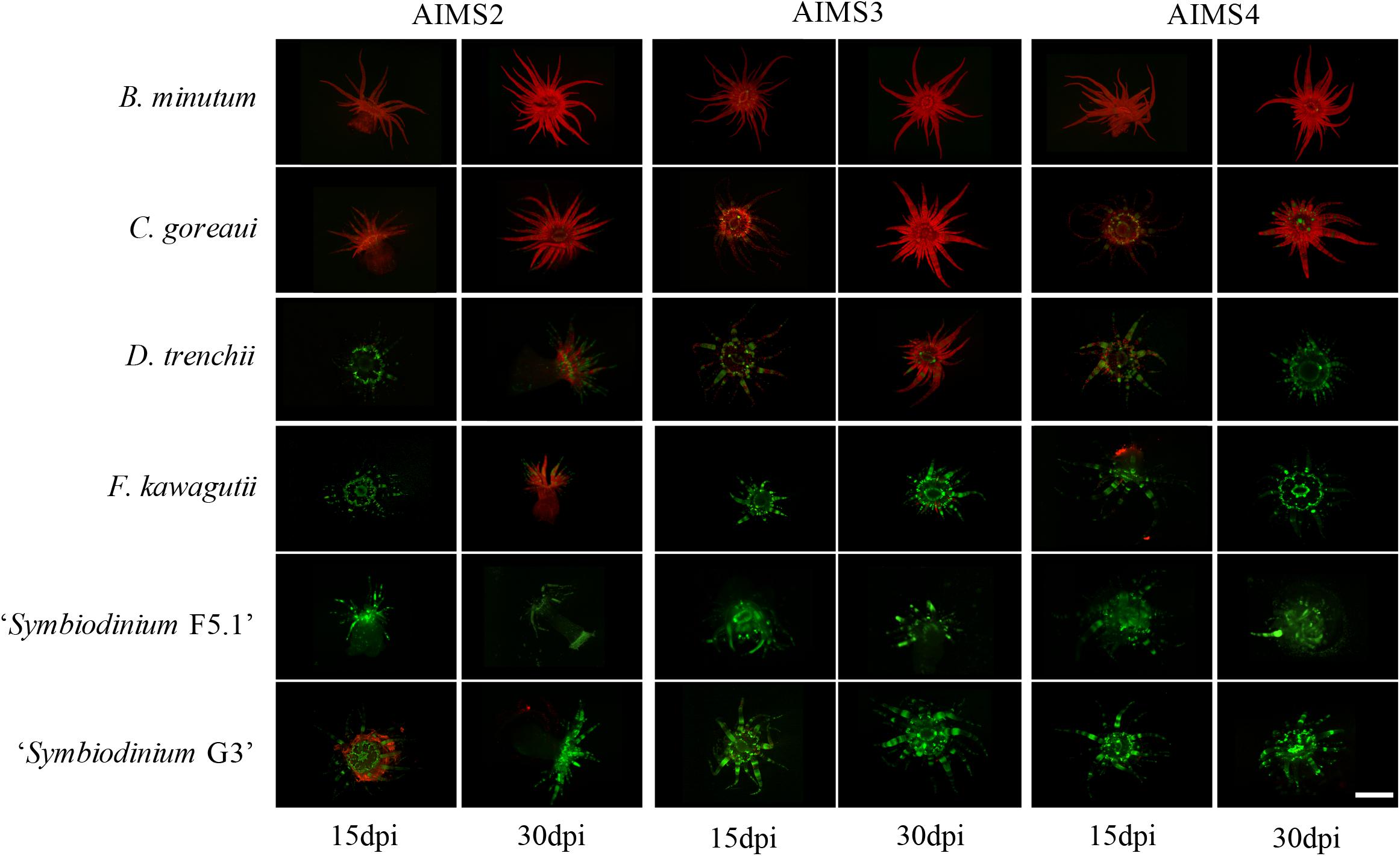
Figure 2. Representative fluorescence microscopy of the three E. diaphana genotypes (AIMS2, AIMS3, AIMS4) colonized by six Symbiodiniaceae strains (B. minutum, C. goreaui, D. trenchii, F. kawagutii, “Symbiodinium F5.1,” and “Symbiodinium G3”) at 15 and 30 dpi. Symbiodiniaceae chlorophyll autofluorescence is in red, and E. diaphana autofluorescence is in green. Pictures of anemones inoculated with S. tridacnidorum are not shown as exposure to this strain resulted in the death of all host organisms. Scale bar 3 mm.
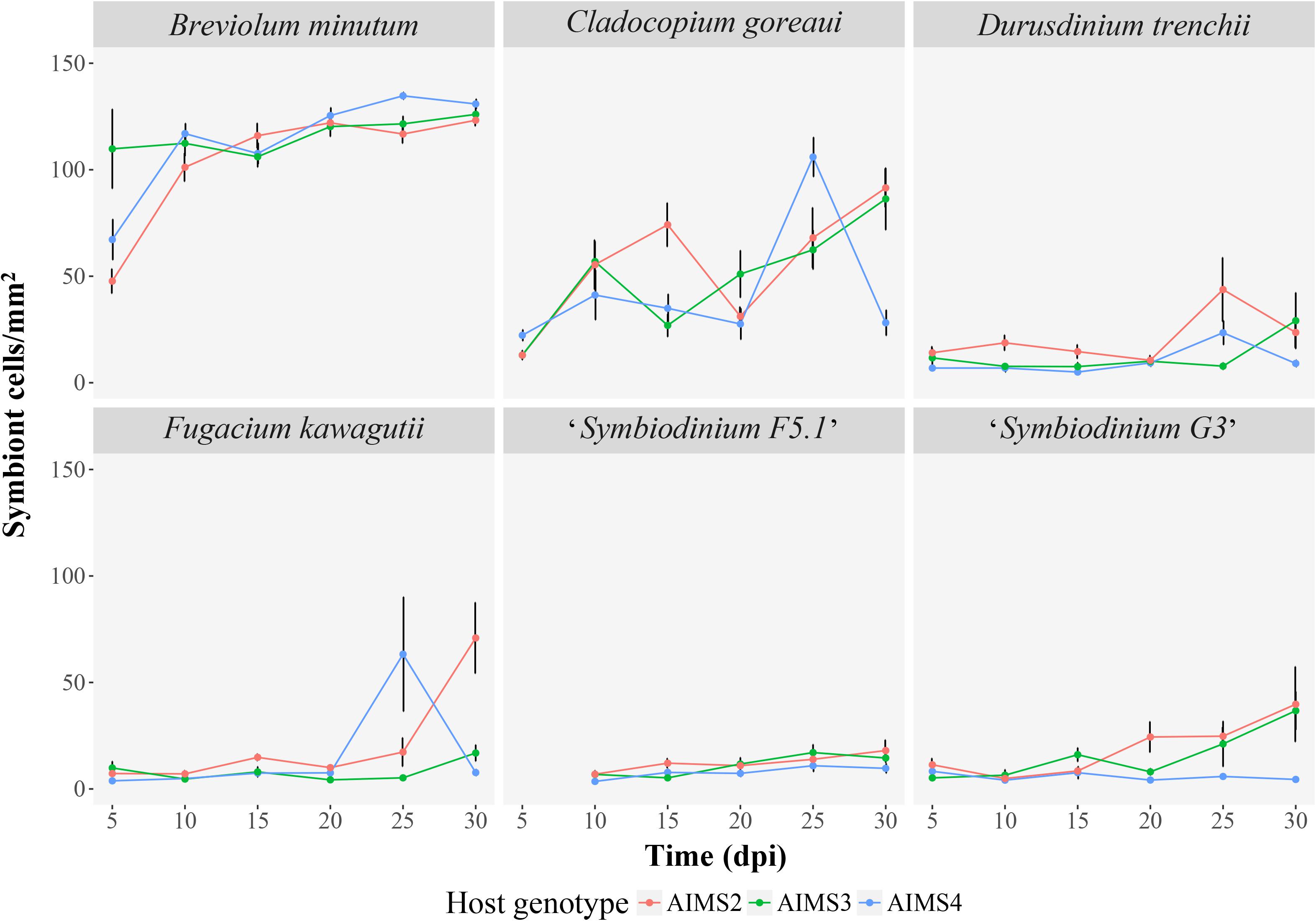
Figure 3. Colonization of three E. diaphana genotypes (AIMS2, AIMS3, AIMS4) with six Symbiodiniaceae (B. minutum, N = 146; C. goreaui, N = 156; D. trenchii, N = 157; F. kawagutii, N = 138; “Symbiodinium F5.1”, N = 135; “Symbiodinium G3”, N = 144) during the 30-days experiment. Each panel of the graph represents the mean (±SEM) density of one algal symbiont type in tentacles of three anemones per genotype at each time point post-inoculation (5, 10, 15, 20, 25, 30 dpi). Where error bars are not visible, they are small and hidden by the symbols. Exposure of anemones to Symbiodinium tridacnidorum resulted in the decease of all organism hence it was not possible to sample the tentacles for analysis. For “Symbiodinium F5.1” it was not possible to perform the sampling at 5 dpi.
The Heterologous Symbiont C. goreaui Can Also Colonize GBR Anemones
Although C. goreaui was never recovered from our source stocks of GBR E. diaphana animals, it was able to colonize and proliferate in these hosts (Figures 2, 3). C. goreaui achieved densities of 91 ± 9 cells/mm2 in AIMS2, and 86 ± 15 cells/mm2 in AIMS3 anemones at 30 dpi, which approaches the densities achieved by the homologous symbiont (see above). AIMS4 anemones inoculated with C. goreaui showed a peak of 106 ± 9 cells/mm2 at 25 dpi, which later dropped to 28 ± 6 cells/mm2 at 30 dpi. Localization of the symbionts in these colonizations revealed that not all the C. goreaui cells were in the endodermis, with appreciable numbers observed in the gastrovascular cavity of the anemones at 30 dpi (Supplementary Figure 2b). Other studies also observed good affinity of E. diaphana for select heterologous symbionts (Belda-Baillie et al., 2002; Xiang et al., 2013; Hambleton et al., 2014; Starzak et al., 2014; Biquand et al., 2017; Gabay et al., 2018). Cell surface characteristics may perhaps be shared between two or more algal types, thus tricking the lock-and-key mechanism that allows uptake of the symbiont by the host. In our system, the comparison between B. minutum and C. goreaui cell densities at each time-point highlights somewhat reduced colonization by the heterologous species over time (Figure 3). For instance, B. minutum had colonized more than the 70% of host tissue after just 10 days, and the number of cells gradually increased over time. By contrast, the in hospite density of C. goreaui fluctuated over the 30-days period, achieving a maximum of just 60% colonization at the end of the experiment. Colonization patterns in E. diaphana were, indeed, significantly influenced by symbiont taxonomic identity (LMEM, F = 248.12, df = 1, p < 0.001), time post-inoculation (LMEM, F = 35.89, df = 5, p < 0.001) and the interaction of the two effects (LMEM, F = 3.19, df = 5, p < 0.05). The different colonization kinetics may be attributed to differences in host efficiency to internalize homologous versus heterologous symbionts. Perhaps the presence of C. goreaui cells expanded to the gastrovascular cavity of the host (Supplementary Figure 2b) might indicate subtle differences in the uptake and symbiosis establishment of homologous versus heterologous algae. We encourage further research on the spatial and temporal dynamics of Symbiodiniaceae uptake by E. diaphana.
Several Heterologous Symbionts Are Poor Colonizers of GBR Anemones
Inoculations of E. diaphana with D. trenchii, F. kawagutii, “Symbiodinium F5.1,” and “Symbiodinium G3” resulted in these algal types achieving only relatively low host densities compared to B. minutum and C. goreaui inoculations (Figures 2, 3). Hence, it was not possible to meet the model assumptions, so a LMEM was not fitted to these data. These heterologous symbionts are apparently unable to strongly colonize and grow within E. diaphana from the GBR within the 30 days of the experiment and might need longer to reach in hospite densities similar to the homologous and heterologous-compatible types (Starzak et al., 2014; Gabay et al., 2018).
Giant Clam Symbionts Are Apparently Lethal to GBR Anemones
S. tridacnidorum was apparently lethal to the anemones just 24 h after the inoculation. All anemone genotypes inoculated with S. tridacnidorum became dark-colored, shriveled and appeared dead (G. Tortorelli, personal observation); thus, it was not possible to consider this algal species in the analysis, but it deserves further investigation. Certain non-native Symbiodiniaceae have been shown to be symbiotically disadvantageous and harmful to the host (Starzak et al., 2014; Matthews et al., 2017). Indeed, the high energetic cost of some symbionts’ rapid proliferation may impact symbiosis functionality and eventually lead to the death of anemones colonized by these algae.
Host Genotype Influences Symbiont Compatibility
While symbiont taxonomy was a determinant in the success or failure of new associations with E. diaphana from the GBR over the 30-days experiment, host genotype also played a significant role in defining patterns of symbiosis establishment. The in hospite density of the homologous B. minutum and the heterologous but compatible C. goreaui over 30 days was affected by host genotype (LMEM, F = 4.55, df = 10, p < 0.001), and by its interaction with symbiont type and time post-inoculation (LMEM, F = 2.54, df = 10, p < 0.01). The leading role of host genotype in determining both abundance and diversity of its symbionts, or even its entire microbiome, has been reported in a variety of organisms, ranging from cnidarians (Grajales et al., 2015; Grajales and Rodríguez, 2016; Quigley et al., 2016; Goldsmith et al., 2018) to mammals (Zoetendal et al., 2001; Campbell et al., 2012; Liu et al., 2015). In previous studies, host genetics have been shown to be responsible for modeling the physiological flexibility and hence the tolerance of host and symbiont to stress events, especially temperature-driven bleaching through symbiont loss (Manzello et al., 2018). The involvement of host genotype in both establishment and disruption of symbiotic associations is thus further evident. Our findings showed that anemone genotype influences the onset of symbiosis by dictating, in part, the pace at which new associations are established. The expression of genes responsible for the synthesis of molecules (e.g., lectins; Davy et al., 2012) of the lock-and-key symbiont recognition mechanism may perhaps differ among anemone genotypes, creating variations in symbiosis establishment. Our system of colonization measurement should now enable experimental testing of such a hypothesis.
Concluding Remarks
The exploration of host–symbiont specificities is fundamental to understanding mechanisms of symbiosis establishment. The present study used algal coverage in anemone tentacles as a tool to measure the capacity of seven Symbiodiniaceae strains to colonize E. diaphana from the GBR. The onset of this mutualistic relationship appears to be a synergistic process governed by complex specificities (Hambleton et al., 2014) and our results confirm that the association is shaped by both symbiont and host features. The differences in host-algal affinity allowed us to create a matrix of symbiotic compatibilities that we can strategically use to investigate mechanisms of recognition and establishment in the cnidarian-Symbiodiniaceae symbiosis system. Importantly, E. diaphana genetics was shown to have an influence on the onset of the relationship. Different host–symbiont genotypic combinations better mirror the performance of the mutualistic units during early stages of the association and, therefore, we encourage the use of true biological replication in future investigations. Further research should explore variability in the expression of symbiosis genes among the different E. diaphana genotypes and consider the effect of host genetics in symbiosis establishment and dysfunction studies.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
GT, MO, GM, and SD designed the experiments. MO and GM developed the concept of this study. GT and RB conducted the experiments. GT performed the analyses, wrote the first draft of the manuscript with input from MO and GM. All authors contributed to the final version of the manuscript.
Funding
This research was funded by the Australian Government through the Australian Research Council Discovery Project DP160101539. GT thanks the Botany Foundation at the University of Melbourne for awarding the Protist Systematics research fund that supported Symbiodiniaceae ITS2 sequencing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Carlos Alvarez-Roa for providing Symbiodiniaceae cultures and support with algal culturing. We thank the Biological Optical Microscopy Platform (BOMP) at the University of Melbourne for assistance with confocal microscopy. GT acknowledges receipt of the University of Melbourne International Research Scholarship and Fee Remission Scholarship. MO acknowledges the Australian Research Council Laureate Fellowship FL180100036.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00833/full#supplementary-material
References
Adl, S. M., Simpson, A. G., Lane, C. E., Lukeš, J., Bass, D., Bowser, S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–514.
Baker, A. C. (2003). Flexibility and Specificity in Coral-Algal Symbiosis: diversity, Ecology, and Biogeography of Symbiodinium. Ann. Rev. Ecol. Evol. Systemat. 34, 661–689. doi: 10.1111/mec.12801
Bates, D., Maechler, M., and Bolker, B. (2012). lme4: linear mixed-effects models using S4 classes. R Package Vers. 1, 1–23.
Belda-Baillie, C. A., Baillie, B. K., and Maruyama, T. (2002). Specificity of a model cnidarian-dinoflagellate symbiosis. Biol. Bull. 202, 74–85. doi: 10.2307/1543224
Beltran, V. H., Dunlap, W. C., and Long, P. F. (2012). “Comparison of the photosynthetic bleaching response of four coral species common to the central GBR,” in Procedings of the 12th International Coral Reef Symposium, (Cairns, QLD), 1–6.
Berner, T., Baghdasarian, G., and Muscatine, L. (1993). Repopulation of a sea anemone with symbiotic dinoflagellates: analysis by in vivo fluorescence. J. Exp. Mar. Biol. Ecol. 170, 145–158. doi: 10.1016/0022-0981(93)90149-i
Biquand, E., Okubo, N., Aihara, Y., Rolland, V., Hayward, D. C., Hatta, M., et al. (2017). Acceptable symbiont cell size differs among cnidarian species and may limit symbiont diversity. ISME J. 11, 1702–1712. doi: 10.1038/ismej.2017.17
Bucher, M., Wolfowicz, I., Voss, P. A., Hambleton, E. A., and Guse, A. (2016). Development and Symbiosis Establishment in the Cnidarian Endosymbiosis Model Aiptasia sp. Sci. Rep. 6:19867.
Campbell, J. H., Foster, C. M., Vishnivetskaya, T., Campbell, A. G., Yang, Z. K., Wymore, A., et al. (2012). Host genetic and environmental effects on mouse intestinal microbiota. ISME. J. 6, 2033–2044. doi: 10.1038/ismej.2012.54
Chakravarti, L. J., Beltran, V. H., and Van Oppen, M. J. H. (2017). Rapid thermal adaptation in photosymbionts of reef-building corals. Glob. Change Biol. 23, 4675–4688. doi: 10.1111/gcb.13702
Coats, D. (2002). Dinoflagellate life-cycle complexities. J. Phycol. 38, 417–419. doi: 10.1046/j.1529-8817.2002.03832.x
Core Team, R. (2017). R: A Language And Environment For Statistical Computing. Vienna: R Foundation for Statistical Computing.
Davy, S., Turner, J., and Lucas, I. (1997). “The nature of temperate anthozoan-dinoflagellate symbioses,” in Procedings of the 8th International Coral Reef Symposium, (Panama), 1307–1312.
Davy, S. K., Allemand, D., and Weis, V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mole. Biol. Rev. 76, 229–261. doi: 10.1128/mmbr.05014-11
Gabay, Y., Parkinson, J. E., Wilkinson, S. P., Weis, V. M., and Davy, S. K. (2019). Inter-partner specificity limits the acquisition of thermotolerant symbionts in a model cnidarian-dinoflagellate symbiosis. ISME J. 13, 2489–2499. doi: 10.1038/s41396-019-0429-5
Gabay, Y., Weis, V. M., and Davy, S. K. (2018). Symbiont Identity Influences Patterns of Symbiosis Establishment, Host Growth, and Asexual Reproduction in a Model Cnidarian-Dinoflagellate Symbiosis. Biol. Bull. 234, 1–10. doi: 10.1086/696365
Goldsmith, D. B., Kellogg, C. A., Morrison, C. L., Gray, M. A., Stone, R. P., Waller, R. G., et al. (2018). Comparison of microbiomes of cold-water corals Primnoa pacifica and Primnoa resedaeformis, with possible link between microbiome composition and host genotype. Sci. Rep. 8:12383. doi: 10.1038/s41598-018-30901-z
Grajales, A., and Rodríguez, E. (2016). Elucidating the evolutionary relationships of the Aiptasiidae, a widespread cnidarian–dinoflagellate model system (Cnidaria: anthozoa: Actiniaria: Metridioidea). Mole. Phylogen. Evol. 94, 252–263. doi: 10.1016/j.ympev.2015.09.004
Grajales, A., Rodríguez, E., and Thornhill, D. J. (2015). Patterns of Symbiodinium spp. associations within the family Aiptasiidae, a monophyletic lineage of symbiotic of sea anemones (Cnidaria, Actiniaria). Coral Reefs 35, 345–355. doi: 10.1007/s00338-015-1352-5
Hambleton, E. A., Guse, A., and Pringle, J. R. (2014). Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. J. Exp. Biol. 217, 1613–1619. doi: 10.1242/jeb.095679
Kuznetsova, A., Brockhoff, P. B., and Christensen, R. H. B. (2017). lmerTest package: tests in linear mixed effects models. J. Statist. Softw. 82, 1–26.
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
Lenaers, G., Scholin, C., Bhaud, Y., Saint-Hilaire, D., and Herzog, M. (1991). A molecular phylogeny of dinoflagellate protists (Pyrrhophyta) inferred from the sequence of 24S rRNA divergent domains D1 and D8. J. Mol. Evol. 32, 53–63. doi: 10.1007/bf02099929
Lin, M.-F., Takahashi, S., Forêt, S., Davy, S. K., and Miller, D. J. (2019). Transcriptomic analyses highlight the likely metabolic consequences of colonization of a cnidarian host by native or non-native Symbiodinium species. Biol. Open Bio. 8:bio038281. doi: 10.1242/bio.038281
Lipschultz, F., and Cook, C. (2002). Uptake and assimilation of 15 N-ammonium by the symbiotic sea anemones Bartholomea annulata and Aiptasia pallida : conservation versus recycling of nitrogen. Mar. Biol. 140, 489–502. doi: 10.1007/s00227-001-0717-1
Liu, C. M., Price, L. B., Hungate, B. A., Abraham, A. G., Larsen, L. A., Christensen, K., et al. (2015). Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 1:e1400216. doi: 10.1126/sciadv.1400216
Manzello, D. P., Matz, M. V., Enochs, I. C., Valentino, L., Carlton, R. D., Kolodziej, G., et al. (2018). Role of host genetics and heat tolerant algal symbionts in sustaining populations of the endangered coral Orbicella faveolata in the Florida Keys with ocean warming. Glob. Change Biol. 25, 1016–1031. doi: 10.1111/gcb.14545
Matthews, J. L., Crowder, C. M., Oakley, C. A., Lutz, A., Roessner, U., Meyer, E., et al. (2017). Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc. Nat. Acad. Sci. 114, 13194–13199. doi: 10.1073/pnas.1710733114
Matthews, J. L., Sproles, A. E., Oakley, C. A., Grossman, A. R., Weis, V. M., and Davy, S. K. (2016). Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J. Exp. Biol. 219, 306–310. doi: 10.1242/jeb.128934
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Nat. Acad. Sci. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Medranos, E., Merselis, D. G., Bellantuono, A. J., and Rodriguez-Lanetty, M. (2019). Proteomic Basis of Symbiosis: a Heterologous Partner Fails to Duplicate Homologous Success in a Novel Cnidarian-Symbiodiniaceae Mutualism. Front. Microbiol. 10:1153.
Neubauer, E. F., Poole, A. Z., Weis, V. M., and Davy, S. K. (2016). The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ. 4:e2692. doi: 10.7717/peerj.2692
Nyholm, S. V., and McFall-Ngai, M. (2004). The winnowing: establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. doi: 10.1038/nrmicro957
Parkinson, J. E., Tivey, T. R., Mandelare, P. E., Adpressa, D. A., Loesgen, S., and Weis, V. M. (2018). Subtle differences in symbiont cell surface glycan profiles do not explain species-specific colonization rates in a model cnidarian-algal symbiosis. Front. Microbiol 9:842. doi: 10.3389/fmicb.2018.00842
Pochon, X., Putnam, H. M., Burki, F., and Gates, R. D. (2012). Identifying and characterizing alternative molecular markers for the symbiotic and free-living dinoflagellate genus Symbiodinium. PLoS One 7:e29816. doi: 10.1371/journal.pone.0029816
Quigley, K. M., Willis, B. L., and Bay, L. K. (2016). Maternal effects and Symbiodinium community composition drive differential patterns in juvenile survival in the coral Acropora tenuis. R. Soc. Open Sci. 3:160471. doi: 10.1098/rsos.160471
Rodriguez-Lanetty, M., Chang, S.-J., and Song, J.-I. (2003). Specificity of two temperate dinoflagellate–anthozoan associations from the north-western Pacific Ocean. Mar. Biol. 143, 1193–1199. doi: 10.1007/s00227-003-1165-x
Rodriguez-Lanetty, M., Krupp, D. A., and Weis, V. M. (2004). Distinct ITS types of Symbiodinium in Clade C correlate with cnidarian/dinoflagellate specificity during onset of symbiosis. Mar. Ecol. Prog. Ser. 275, 97–102. doi: 10.3354/meps275097
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Sproles, A. E., Oakley, C. A., Matthews, J. L., Peng, L., Owen, J. G., Grossman, A. R., et al. (2019). Proteomics quantifies protein expression changes in a model cnidarian colonised by a thermally tolerant but suboptimal symbiont. ISME J. 13, 2334–2345. doi: 10.1038/s41396-019-0437-5
Starzak, D. E., Quinnell, R. G., Nitschke, M. R., and Davy, S. K. (2014). The influence of symbiont type on photosynthetic carbon flux in a model cnidarian–dinoflagellate symbiosis. Mar. Biol. 161, 711–724. doi: 10.1007/s00227-013-2372-8
Thornhill, D. J., Xiang, Y., Pettay, D. T., Zhong, M., and Santos, S. R. (2013). Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol. Ecol. 22, 4499–4515. doi: 10.1111/mec.12416
Weis, V. M., Davy, S. K., Hoegh-Guldberg, O., Rodriguez-Lanetty, M., and Pringle, J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23, 369–376. doi: 10.1016/j.tree.2008.03.004
Weis, V. M., Reynolds, W. S., and Krupp, D. A. (2001). Host-symbiont specificity during onset of symbiosis between the dinoflagellates Symbiodinium spp. and planula larvae of the scleractinian coral Fungia scutaria. Coral Reefs 20, 301–308. doi: 10.1007/s003380100179
Wilson, K., Li, Y., Whan, V., Lehnert, S., Byrne, K., Moore, S., et al. (2002). Genetic mapping of the black tiger shrimp Penaeus monodon with amplified fragment length polymorphism. Aquaculture 204, 297–309. doi: 10.1016/s0044-8486(01)00842-0
Wolfowicz, I., Baumgarten, S., Voss, P. A., Hambleton, E. A., Voolstra, C. R., Hatta, M., et al. (2016). Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci. Rep. 6:32366. doi: 10.1038/srep32366
Xiang, T., Hambleton, E. A., Denofrio, J. C., Pringle, J. R., and Grossman, A. R. (2013). Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity(1). J. Phycol. 49, 447–458. doi: 10.1111/jpy.12055
Yellowlees, D., Rees, T. A., and Leggat, W. (2008). Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Env. 31, 679–694. doi: 10.1111/j.1365-3040.2008.01802.x
Keywords: Exaiptasia diaphana, model system, Symbiodiniaceae, specificity, genotype, symbiosis, microscopy, fluorescence
Citation: Tortorelli G, Belderok R, Davy SK, McFadden GI and van Oppen MJH (2020) Host Genotypic Effect on Algal Symbiosis Establishment in the Coral Model, the Anemone Exaiptasia diaphana, From the Great Barrier Reef. Front. Mar. Sci. 6:833. doi: 10.3389/fmars.2019.00833
Received: 20 May 2019; Accepted: 27 December 2019;
Published: 22 January 2020.
Edited by:
Oren Levy, Bar-Ilan University, IsraelReviewed by:
Mauricio Rodriguez-Lanetty, Florida International University, United StatesLaura Fernandes De Barros Marangoni, Scientific Centre of Monaco, Monaco
Copyright © 2020 Tortorelli, Belderok, Davy, McFadden and van Oppen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giada Tortorelli, Z2lhZGF0b3J0b3JlbGxpQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Giada Tortorelli
Giada Tortorelli Roy Belderok
Roy Belderok Simon K. Davy
Simon K. Davy Geoffrey I. McFadden1†
Geoffrey I. McFadden1† Madeleine J. H. van Oppen
Madeleine J. H. van Oppen