- 1College of Life Science, Shaoxing University, Shaoxing, China
- 2College of Agriculture and Bioengineering, Jinhua Polytechnic, Jinhua, China
Immune response plays an important role in the pearl sac formation and pearl quality. However, there is little knowledge about the mode and mechanism of the immune responses in the pearl sac formation of the pearl mussel Hyriopsis cumingii. In this study, we monitored the process of pearl sac formation by histology examination for 21 days after grafting. The results showed that a large number of hemocytes aggregated in the pearl sac cavity on day 8 after grafting and provided organic substrate for the deposition of extracellular matrix and CaCO3 crystal and mediated the initiation of pearl forming on day 12 after grafting. These results revealed the important role of hemocytes on the early pearl sac and pearl formations. In the transcriptome analysis of pearl sac at the key time-points of day 0 and days 6, 8, 12 after grafting, the expression profiles of the immune-related genes revealed the immune response pattern of H. cumingii after grafting. The significant up-regulation in the hydrolytic enzymes, acute phase proteins, and antimicrobial peptides of humoral defense factors mRNA expression on day 6 after grafting suggested that these immune-related gene products were the main immune defense during the early wound healing; Meanwhile, the continuous mRNA up-regulation of antioxidant enzymes such as Cu-Zn SOD, Se-GPX, and tyrosinase in prophenoloxidase (proPO) system on days 8 and 12 suggested the important role of these immune effectors in oxidation-reduction on the later stage of pearl sac and pearl formations in H. cumingii. Moreover, the results of KEGG analysis of differentially expressed genes suggested the potential regulatory function of the signaling pathways of proPO system and complement system during pearl sac formation. These results provided valuable new insights into the roles and functions of the immune system in pearl sac and pearl formation.
Introduction
Pearl is not only one of the expensive organic jewelry but also is used as a precious traditional Chinese medicine. Compared with the nucleated pearls produced by seawater pearl oyster species of Pinctada sp. and Pteria penguin, the freshwater non-nucleated pearls mainly produced by the pearl mussel Hyriopsis cumingii in China is the largest production of cultured pearl in the world (Zhu et al., 2019). However, although non-nucleated pearl achieved more than 1500 tons of productivity, accounting for over 98% of the world’s pearl production in the past 10 years, it corresponds to only 5% of the output value due to the extremely high proportion of low-quality pearls (FAO, 2018). Most of the low-quality freshwater pearls are used as one of the active ingredients in the cosmetics industry. Thus, improvement of the pearl quality has been one of the most important research contents in the freshwater pearl industry.
The cultured pearl from a seawater or freshwater pearl mussel is formed with the nacre secreted by the pearl sac of host mussel which is developed after a grafting operation to insert a mantle piece (also known as saibo) from a donor mussel into the gland or the connective tissue of the pallial mantle of a host mussel. After the grafting operation, a strong immune response is immediately occurred at the incision site to heal the wound. The immune response remains active during the process of pearl sac formation to tolerate the nucleus and allograft implantation (Li et al., 2010; Kishore and Southgate, 2015a; Wang W. et al., 2017). However, excessive inflammatory reaction will increase the nucleus rejection and mortality in the marine pearl oysters Pinctada margaritifera and P. maxima (Cochennec-Laureau et al., 2010) and the freshwater pearl mussel H. cumingii (Li et al., 2010). Histological analysis showed that at the initiation of pearl sac formation, a large number of hemocytes accumulated at the incision site to seal the damaged area and triggered the subsequent immune process of wound healing (Li et al., 2010; Kishore and Southgate, 2015b). Moreover, the hemocytes, initially surrounding in the pearl sac, would infiltrate into the nucleus cavity (Acosta-Salmon and Southgate, 2006; Eddy et al., 2015; Atsumi et al., 2018), but the excessive accumulation of hemocytes could cause bulges in the pearl sac, resulting in an irregularly shaped pearl (Kishore and Southgate, 2015a).
Comparing to the production of seawater pearls which both mantle pieces and nucleus are inserted into the sex gland of seawater pearl oysters, the production of the freshwater non-nucleated pearl only needs to graft the mantle pieces into the pallial mantle of H. cumingii. Thus, the incision site of non-nucleated pearl grafting is usually smaller than that with nucleus implantation. Although the pearl sac development of nucleus pearl had widely studied in the pearl mussels such as black-lip pearl oyster, Pinctada margaritifera (Cochennec-Laureau et al., 2010; Kishore and Southgate, 2016), P. maxima (Eddy et al., 2015), P. martensii (Jiao et al., 2010), and H. cumingii (Shi et al., 1985), that of non-nucleated pearl is still unclear, and the roles of hemocytes in pearl sac formation are also unknown and needed to be further studied.
Recently, using the methods of transcriptome and proteomics, some differentially expressed genes of immune effectors and signaling pathways were identified to be related to the immunological molecular mechanism of nucleus implantation in Pinctada sp. species (Wei et al., 2017a, b; Wang et al., 2019). In H. cumingii, many immune-related molecules were detected in H. cumingii and proven to involve in the immune response in wound healing and pearl formation (Bai et al., 2009; Li et al., 2010; Huang et al., 2019). These molecules include the pathogen pattern recognition receptors (PRRs) such as Toll-like receptors, galectins and peptidoglycan recognition proteins (PGRPs) (Zhang et al., 2017; Zhao et al., 2018; Huang and Ren, 2019), antimicrobial peptides (AMPs) such as defensin and theromacin (Xu et al., 2010; Ren et al., 2011), lysozyme (Ren et al., 2012), etc. Nevertheless, despite the importance of the immune response to the pearl sac formation and the pearl quality in H. cumingii, there is little knowledge on expression pattern of immune-related genes after grafting and their regulated signaling pathways in the initial stage of pearl formation in this species. Thus, the effective immunological methods to improve the pearl quality are limited in pearl production.
In this study, we used histological analysis to monitor the changes of hemocytes in the process of pearl sac and pearl formation until day 21 after non-nucleus grafting. Our results showed a characteristic immune response to the allografts in the freshwater pearl mussel H. cumingii. Meanwhile, comparative transcriptome analysis of the pearl sacs on key time-point of days 0, 6, 8, and 12 after grafting was performed to identify important genes and pathways involved in the immune responses to the allografts. Our results revealed the response pattern of H. cumingii immune system in non-nucleus pearl operation and the potential regulation signaling pathways in pearl sac formation. These results will be benefit for the mechanism of pearl sac formation of H. cumingii and enlighten a potential pathway of improving the pearl quality in the near future.
Materials and Methods
Grafting Operation and Mussel Culture
A total of 280 1-year-old mussels with shell length about 8–10 cm was selected from a seed multiplication farm (Jinhua Jewel Pearl Institute, Jinhua, China), and divided into two equal parts as host and donor mussels respectively. The grafting operation was performed according to the following procedure with minor modifications. The outer epithelium of the posterior mantle pallial from a donor mussel was cut into 3 cm × 2.5 cm rectangular pieces (saibo). Nine grafting saibos were inserted into the posterior mantle on each side of the host mussel. The size of surgical incision in pallial mantle was about 3 mm in width and 3.5 mm in depth. The operation was performed by an experienced technician. After a temporary culture of 6 h in clean flowing water, the grafted mussels were released into a greenhouse pond whose temperature was fluctuated between 24 and 28°C.
Pearl Sac Collection
Fourteen mussels were harvested on days 2, 4, 6, 8, 10, 12, 15, 18, and 21 after grafting. The pearl sacs and adjacent mantle tissue of five mussels were isolated and fixed in 4% paraformaldehyde solution at 4°C for histological examination, and the other 9 mussels were frozen in liquid nitrogen and then stored at −80°C for transcriptome analysis. The saibo alone was used as the control group (day 0).
Histological Analysis
After 2 months of preservation in 4% paraformaldehyde solution, a total of 15 the pearl sacs at each time-point were selected and embedded in paraffin blocks, further sectioned to a thickness of 3 μm and stained with conventional hematoxylin and eosin (H&E) kit (Beyotime Institute of Biotechnology, Haimen, China) following the manufacturer’s instructions. To obtain the distribution of calcium carbonate crystal across the tissues, sections from the same peal sac were also stained with Alizarin Red S (Sangon Biotechnology Co., Ltd., Shanghai, China). Sections were examined under a Nikon 80i microscope (Nikon, Tokyo, Japan) and images were captured by Nikon DS-Fi3 camera. The length of epithelial cell of saibo or pearl sac was measured using the NIS-Elements software (Nikon), and about 80 epithelial cells around each saibo or pearl sac were measured randomly.
Comparative Transcriptome Analysis
According to the results of histological analysis (see details in the result of section “Histomorphological Characteristics of the Pearl Sac Development”), days 6, 8, and 12 after grafting were selected as key time-points of pearl sac formation. A total of three samples of each time-point were used for comparative transcriptome analysis, and each sample was mixed with pearl sac tissues isolated from three host mussels. The isolated saibo was used as control (day 0). Total RNA was extracted using RNAiso Plus Reagent (Takara, Kusatsu, Japan) according to the manufacturer’s instructions, and quantified with Agilent 2100 Bioanalyzer (Agilent, Santa Clara, United States). The complementary DNA (cDNA) libraries were synthesized with TruSeq RNA sample preparation kit V2 (Illumina Corp., San Diego, United States) and then sequenced using Illumina HiSeq 2500 at BGI-Shenzhen, China. The de novo assembly of the cleaned reads was performed with Trinity v2.0.6 software. The functional annotation and classification of unigenes were performed by BLASTx (National Center for Biotechnology Information, Bethesda, United States) against the public database of non-redundant (Nr), Nt, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and EuKaryotic Orthologous Groups (KOG), Interpro, and Gene ontology (GO) (E-value < 10–5). Differentially expressed genes (DEGs) in the tissues of four time-points were identified with DEGseq software (Wang et al., 2010) with a setting of fold change ≥ 2 and false discovery rate (FDR) ≤ 0.001. GO term enrichment and KEGG pathway analysis of DEGs were performed using Phyper function in R packages.
Validation of DEGs by Real-Time Quantitative PCR
To validate the expression patterns in the pearl sac development, 10 immune-related gene transcripts, namely ferritin, heat shock protein 90 (HSP90), complement component C3 (C3), cathepsin-L, galectin, defensin, matrix metalloproteinase-19 (MMP-19), C1q domain containing protein (C1qDC), C-type lectin, allograft inflammatory factor 1 (AIF1), were selected for qPCR analysis. The RNA samples used to construct the RNA-Seq library were used for qRT-PCR analysis. cDNA synthesis and amplification procedure were performed as the published reference previously (Ren et al., 2014). The primers used for amplification were listed in Supplementary Table S1 and β-actin was used as an endogenous reference gene. A melting curve analysis (80 cycles of 10 s, increasing the temperature by 0.5°C step from 55 to 95°C) was performed to confirm the presence of single amplification products of each primer pair. The relative expression levels of the target genes were normalized to β-actin transcripts using the following formula N = 2(Ct β–actin–Ct target gene). The results were expressed as fold changes of mRNA expression level between adjacent time point after grafting.
Statistical Analysis
The differences in the length of epithelial cells of saibo and pearl sac were analyzed by one-way ANOVA with further comparisons using Tukey’s HSD test. Differences were considered statistically very significant at P < 0.01. All the statistics were performed with SPSS 25.0 software (SPSS Inc., Chicago, IL, United States).
Results
Histomorphological Characteristics of the Pearl Sac Development
During the process of pearl sac formation, the saibo was clearly free in the incision site of mantle connective tissue of host mussel on days 2 to 6 after grafting (Figures 1A–C); then the connective tissue of saibo gradually integrated with the connective tissue of the host mussel on days 8 to 10 after grafting (Figures 2A,B), and the integration was full completed on day 12 after grafting (Figure 2C). Meanwhile, only a small number of hemocytes were present and surrounding the saibo, and the number of hemocytes invaded into the interspace of saibo was increased gradually on days 2 to 6 after grafting (Figures 1D–F); Then, a large number of hemocytes filled in the pearl sac cavity and formed hemocytes clot on days 8 to 10 after grafting (Figures 2D,E); whereas, hemocytes secrete organic matrices and formed a thin layer of organic substrate to cover on the clot of hemocytes on day 12 after grafting (Figures 2C,F); moreover, the CaCO3 crystals stained blue by HE staining and red by Alizarin Red S staining, were found deposition in the hemocytes (Figures 3B,E) and the clots of hemocytes debris (Figures 3C,F) in the pearl sacs; In addition, the clots comprised with hemocytes debris, CaCO3 vesicles, outer organic layer formed the core of pearl. On days 15 to 21 after grafting, CaCO3 crystals continually deposited and grew on the core of pearl (Figure 4). Finally, with the development of pearl sac, morphology of epithelial cell was changed from short columnar shape in saibo on day 2 after grafting to long columnar shape in pearl sac on day 12 after grafting, but changed to flat shape on day 21 after grafting. The average length of epithelial cells of saibo or pearl sac increased gradually from days 2 to 8 after grafting, and reached its maximum on day 12 after grafting, then decreased until day 21 after grafting (Figure 5).
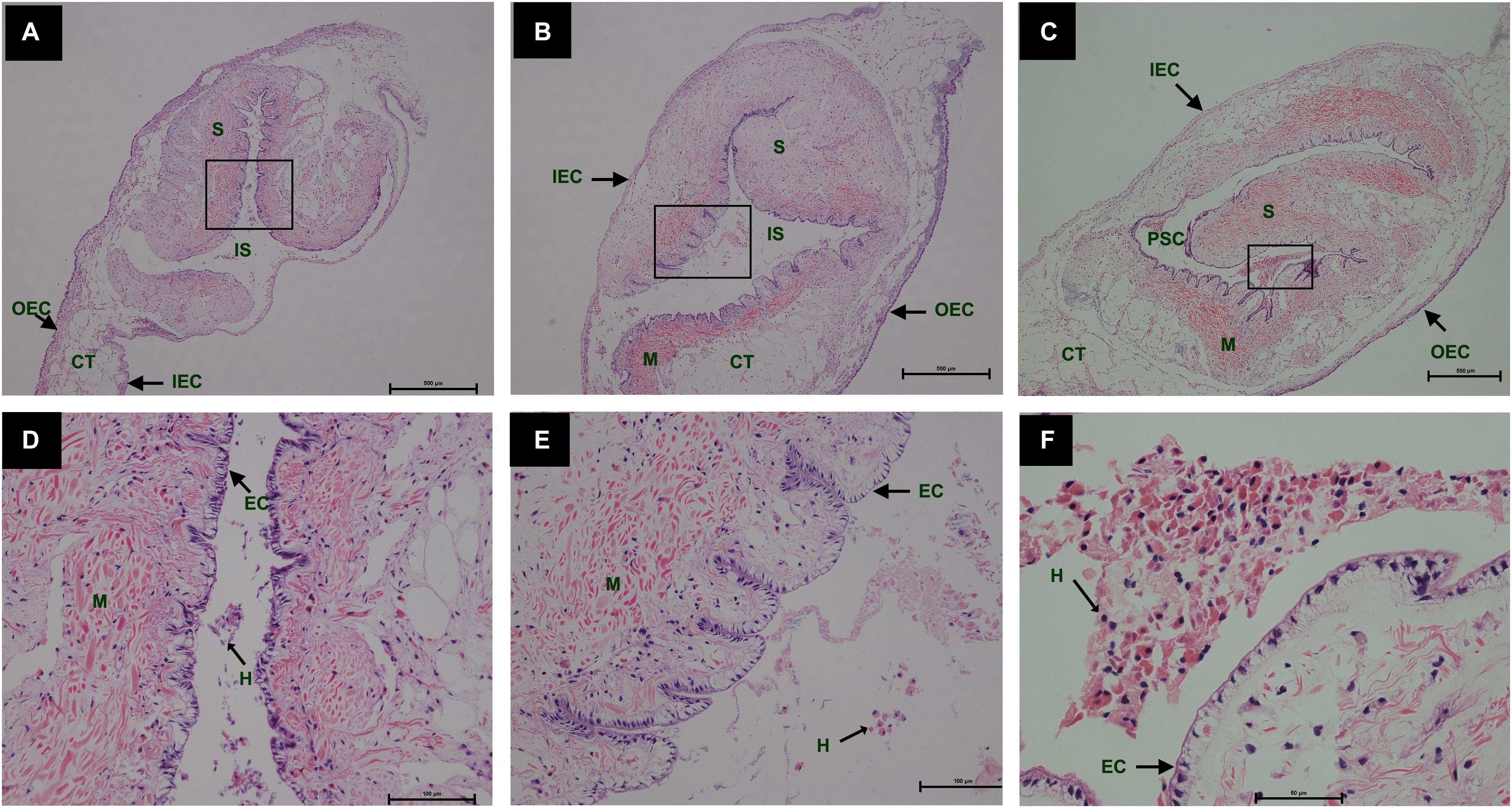
Figure 1. Histology of pearl sac development in Hyriopsis cumingii on days 2, 4 and 6 after grafting by Haematoxylin-Eosin staining. (A–C) are overview of free saibo implanted in the connective tissue of pallial mantle of host mussel on days 2, 4, and 6, respectively. The black boxes indicate the areas shown at higher magnification. (D–F) are the high magnification views of partial area of saibo in panels (A–C), respectively. The abbreviations indicate S, saibo, H, hemocyte, CT, connective tissue, EC, epithelial cell of saibo, IS, interspace in saibo, OEC, outer epithelial cell of pallial mantle, IEC, inner epithelial cell of pallial mantle, M, muscle. Scale bars are 500 μm in panels (A–C), 100 μm in panels (E–G).
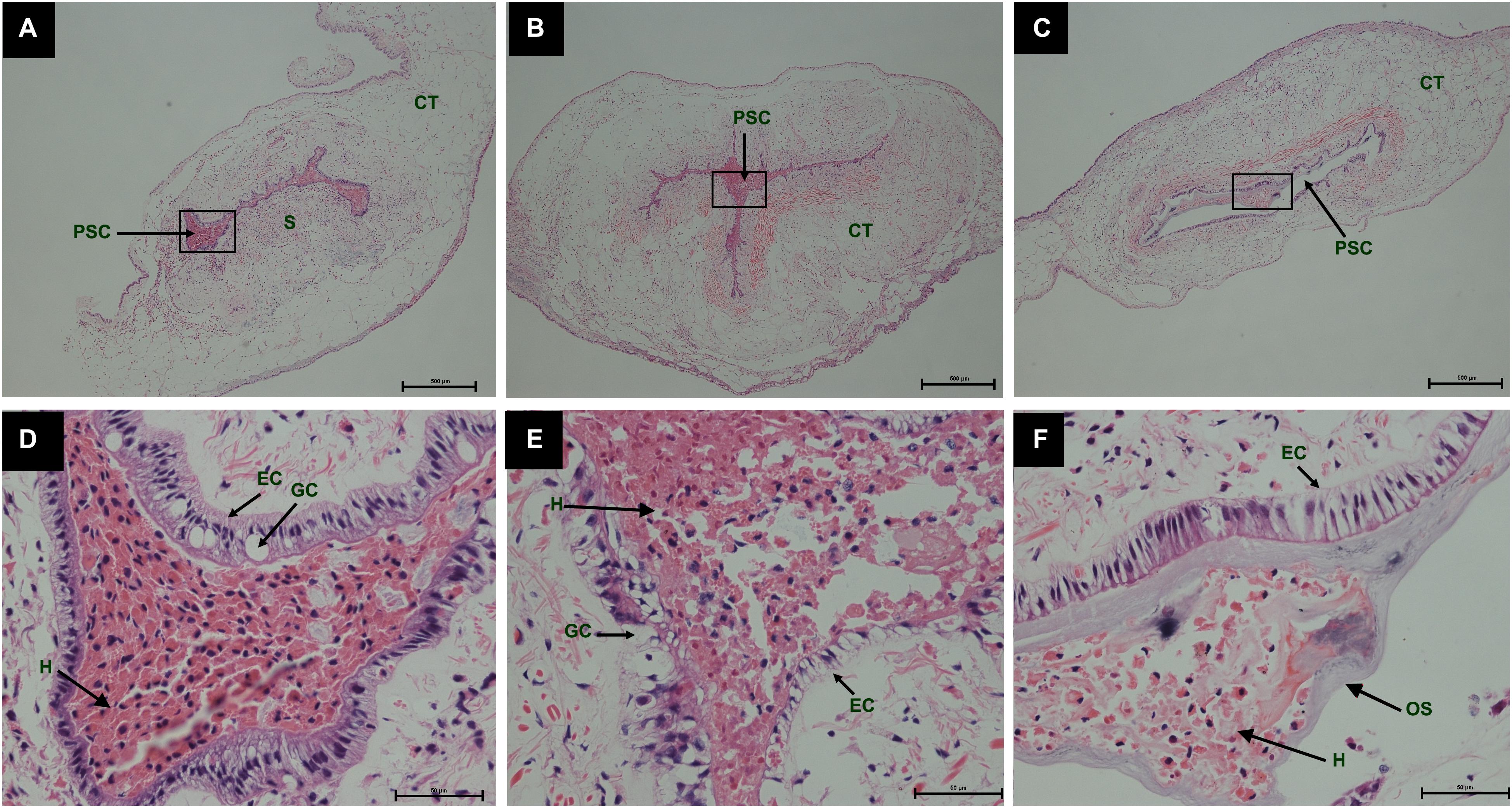
Figure 2. Histology of pearl sac development in Hyriopsis cumingii on days 8, 10 and 12 after grafting by Haematoxylin-Eosin staining. (A–C) are overview of developmental pear sac in the pallial mantle of host mussel on days 8, 10, and 12, respectively. The black boxes indicate the areas shown at higher magnification. (D–F) are the high magnification views of partial area of pearl sac in panels (A–C), respectively. The abbreviations indicate S, saibo, PSC, pearl sac cavity, H, hemocyte, CT, connective tissue, EC, epithelial cell on pearl sac, GC, gland cell on pearl sac. Scale bars are 500 μm in panels (A–C), 100 μm in panels (E–G).
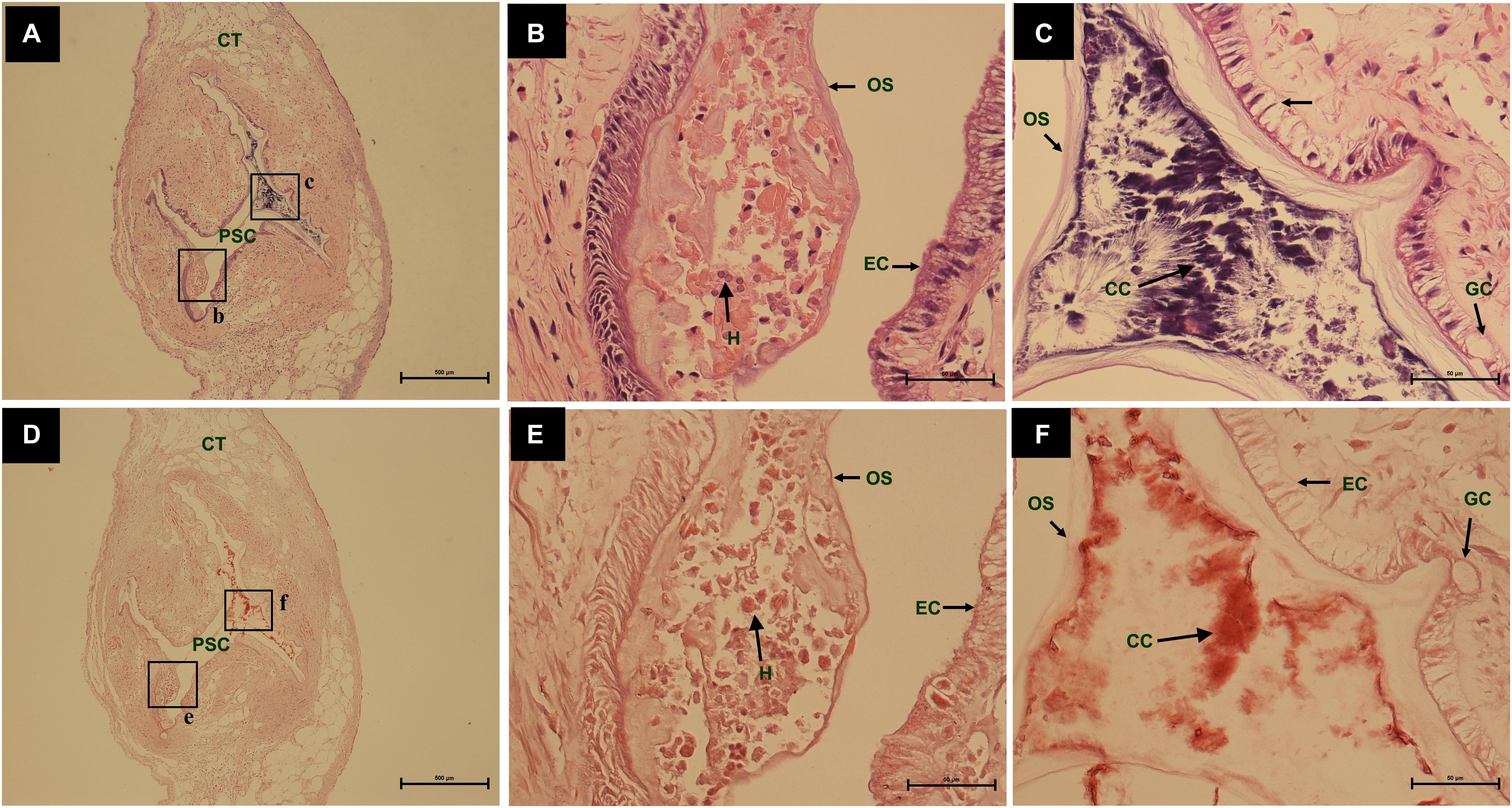
Figure 3. The comparing of histology from same pearl sac on day 12 after grafting by different stain methods. (A,D) are a low power views stained by Haematoxylin-Eosin and Alizarin Red S, respectively. The black boxes indicate the areas shown at higher magnification. (B,C) are the high magnification views of partial area of pearl sac in panels (A). (E,F) are the high magnification views of partial area of pearl sac in panel (C). The abbreviations indicate S, saibo, PSC, pearl sac cavity, H, hemocyte, CT, connective tissue, EC, epithelial cell on pearl sac, GC, gland cell on pearl sac, OS, organic substrate, CC, CaCO3 crystals. Scale bars are 500 μm in panels (A,D), 100 μm in panels (B,C,E,F).
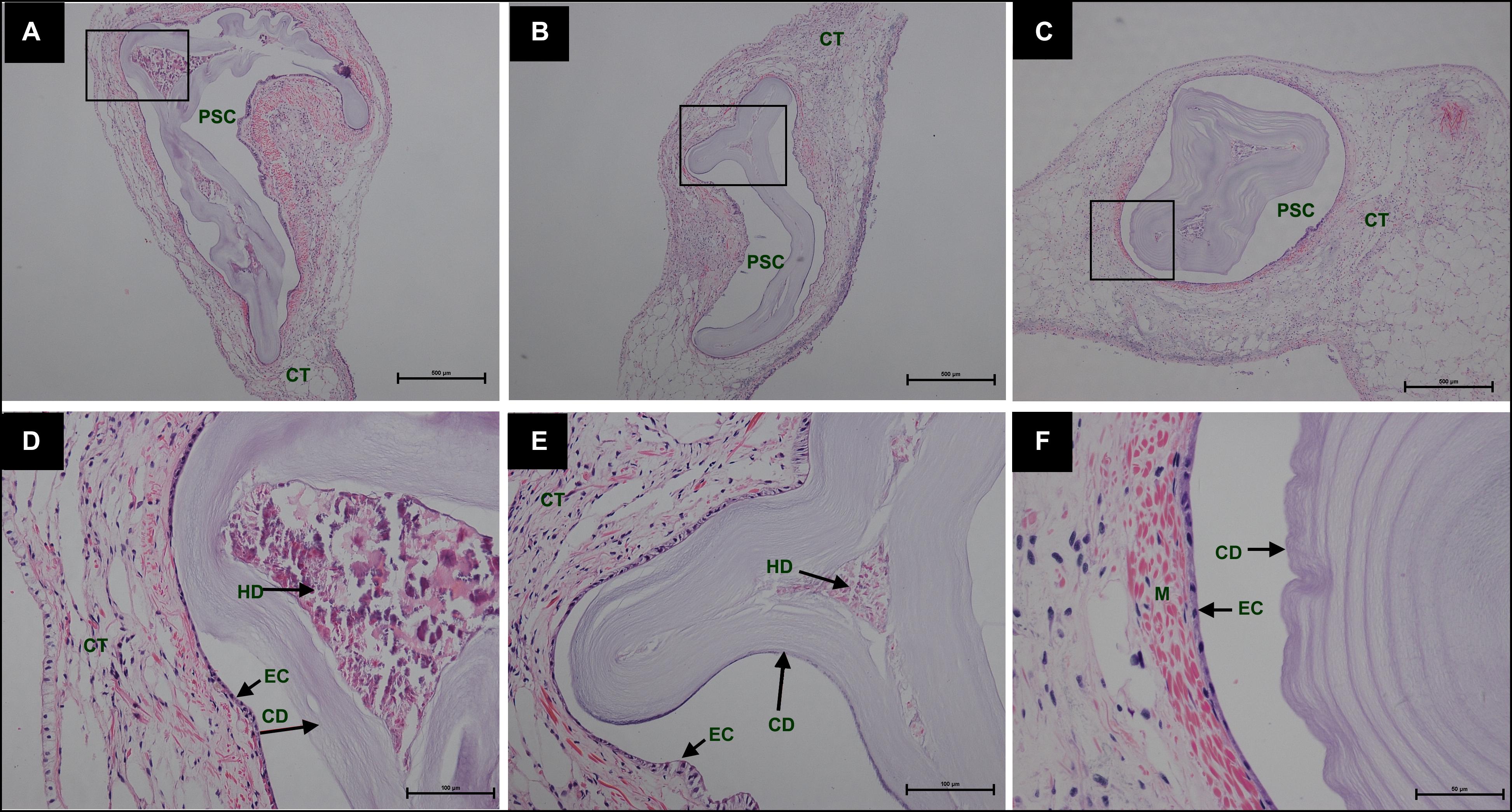
Figure 4. Histology of pearl sac development in Hyriopsis cumingii on days 15, 18, and 21 after grafting by Haematoxylin-Eosin stain. (A–C) are overview of developmental pear sac in the pallial mantle of host mussel on days 15, 18, and 21, respectively. The black boxes indicate the areas shown at higher magnification. (D–F) are the high magnification views of partial area of pearl sac in panels (A–C), respectively. The abbreviations indicate PSC, pearl sac cavity, CT, connective tissue, EC, epithelial cell on pearl sac, CD, CaCO3 crystals deposition, HD, hemocyte debris. Scale bars are 500 μm in panels (A,B), 100 μm in panels (C,D).
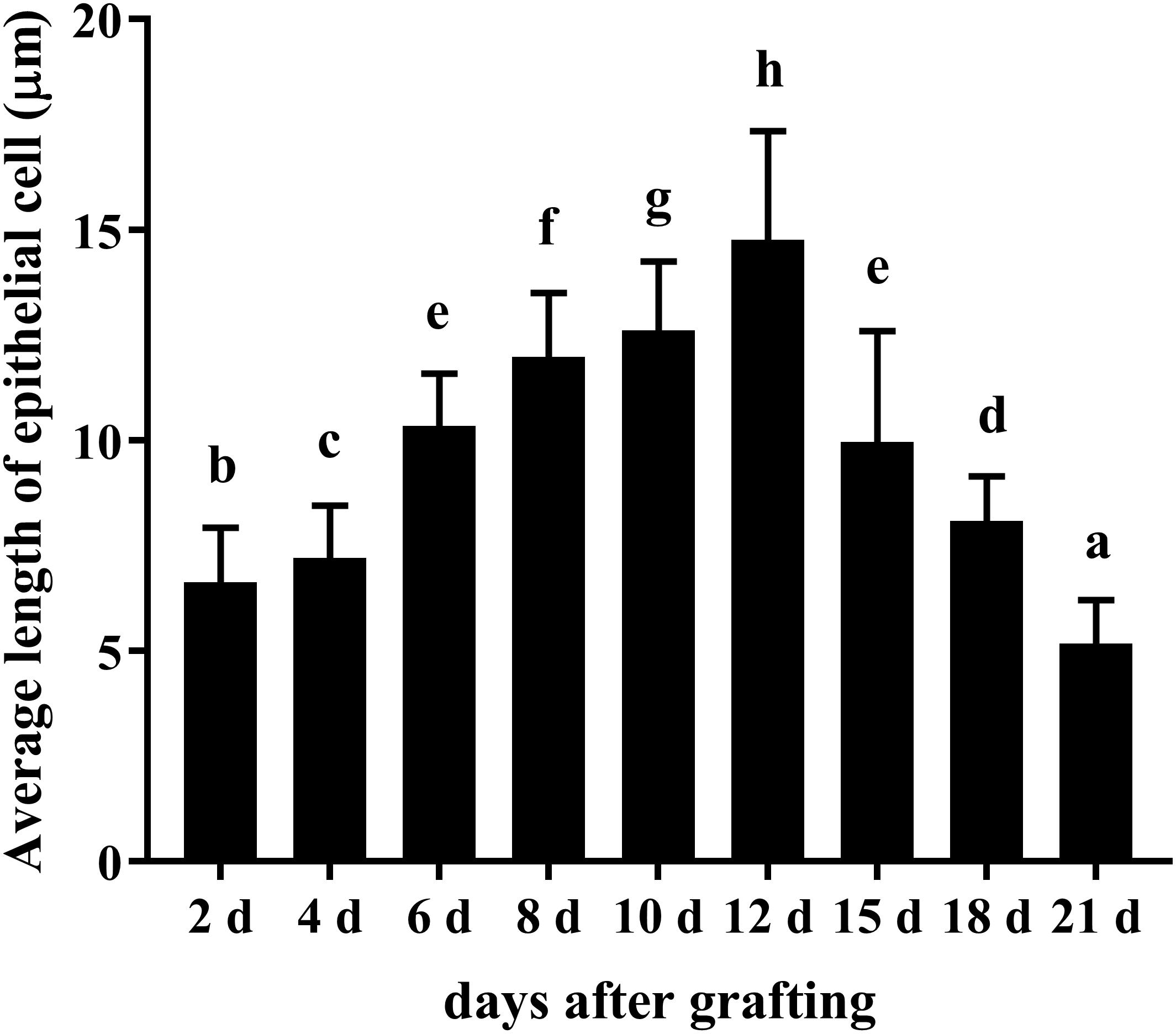
Figure 5. Changes in average length of epithelial cell of saibo and pearl sac during the pearl sac formation. Each bar represents mean ± SD. Means with different superscript letters are significantly different at P < 0.01 by the Tukey’s HSD test.
Transcriptome Assembly
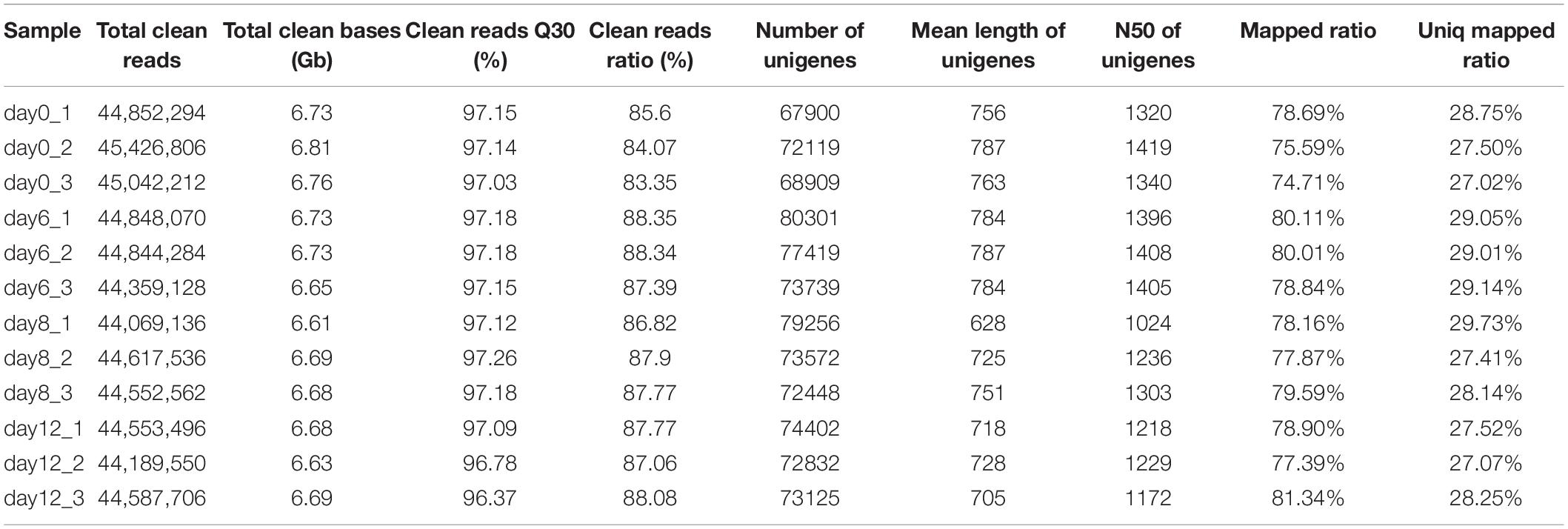
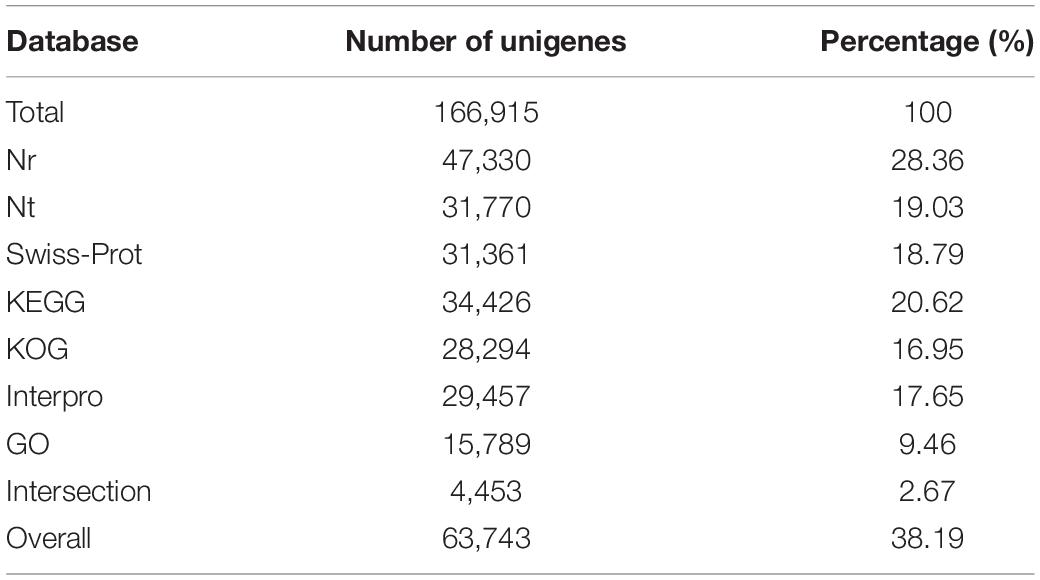
Pearl sac samples in four different developmental stages (days 0, 6, 8, 12 after grafting) were sequenced and their raw reads were deposited in the NCBI SRA database under BioProject accession PRJNA556395. The transcriptome information of data assembly and quality for each sample were summarized in Table 1. All samples had more than 44 million clean reads and the Q30 value for each sample was above 96.37%. The mean length of assembled unigenes for each sample was range from 628 to 787 bp, and the N50 values were all over 1000 bp (Table 1). A total of 63,743 unigenes were annotated functionally with seven functional databases (Nr, Nt, GO, KOG, KEGG, Swiss-Prot, and InterPro) (Table 2). Given that these transcriptome sequence generated a great deal of high-quality data which were better than transcriptome data published previously (Bai et al., 2013; Zhang et al., 2016; Huang et al., 2019), and were benefit for the subsequent transcriptome analysis.
Analysis of DEGs
To reveal the candidate genes that might participate in the process of pearl sac formation in H. cumingii, a comparative transcriptome analysis was performed on the pearl sacs on days 0, 6, 8, and 12 after grafting. Hierarchical clustering was performed to examine the differential expression profiling of DEGs during the pearl sac development (Figure 6), in which day 0 was distinguishable from that in other three samples (days 6, 8, and 12), and day 6 clustered more closely with day 8 rather than with day 12. Both the number of upregulated and downregulated genes were highest in the day 0 vs. day 6 group, suggesting the strong response of gene expression at the early stage of pearl sac formation (Supplementary Figure S1).
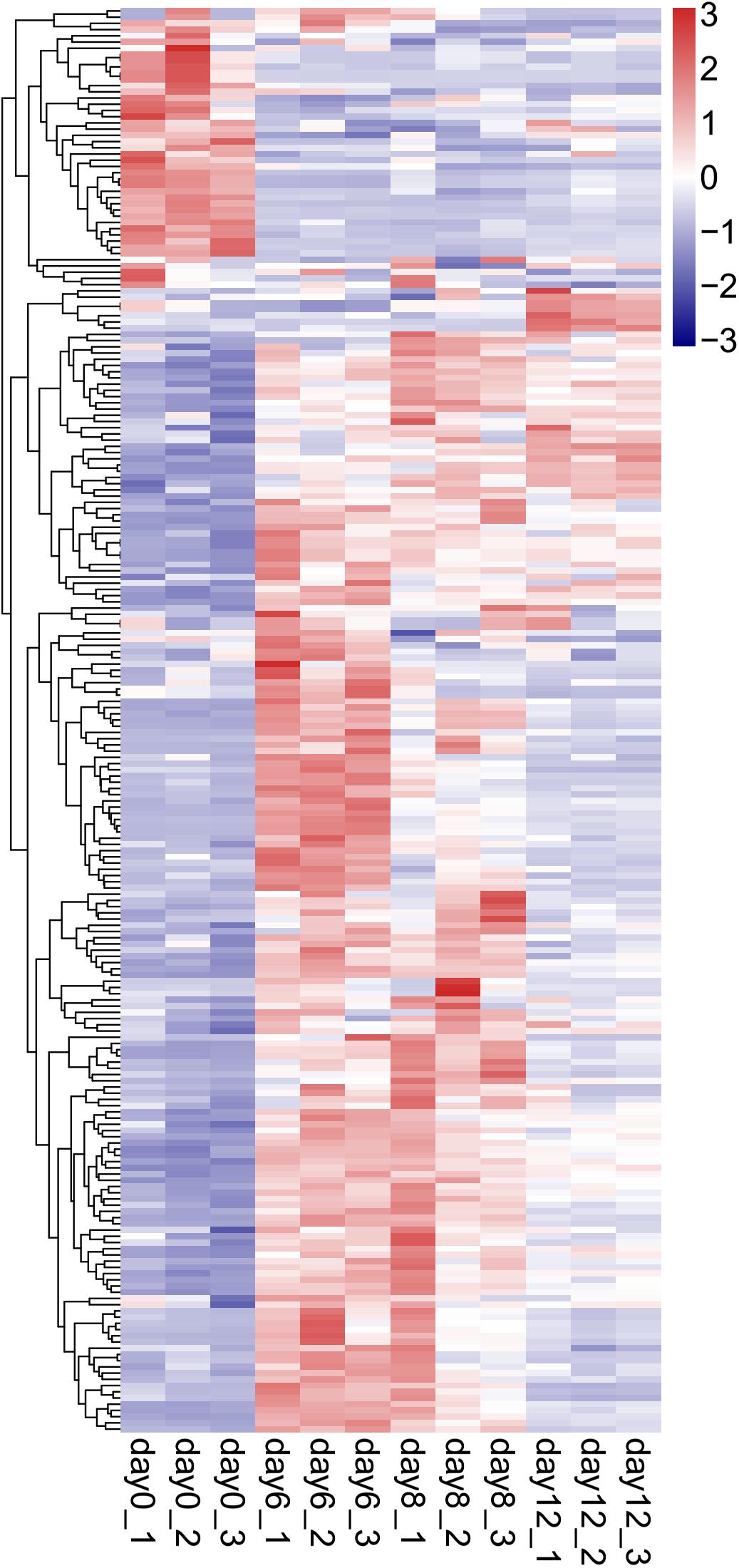
Figure 6. Hierarchical clustering analysis of DEGs differential expression genes (DEGs) in the 4 stages of developmental pearl sac of H. cumingii. The gene-normalized signal intensities are shown in the heat maps using a log10 scale.
Differentially Expressed Immune-Related Genes
Based on Nr, Nt, and Swiss-Prot annotation, various types of DEGs involved in the innate immunity of molluscs were identified. These unigenes included pattern recognition receptors, molecules in the signaling pathway of immune response, and immune effectors (Supplementary Table S2). Comparing to the control of day 0, a large number of the immune related unigenes which were annotated as the hydrolytic enzymes (such as cathepsin L and matrix metallopeptidase 19), acute phase proteins (such as ferritin and heat shock protein 70), antimicrobial peptides (such as defensin and theromacin), and pattern recognition proteins (such as galectins, C1q domain containing protein or C1qDC and scavenger receptor) were significantly up-regulated on day 6 after grafting, but on days 8 and 12 after grafting, most of them were down-regulated gradually (Figure 7). However, an opposite expression pattern was detected on the important antioxidant enzymes, including copper/zinc superoxide dismutase (Cu-Zn SOD), selenium-dependent glutathione peroxidase (Se-GPX), and the crucial enzyme in prophenoloxidase (proPO) system, tyrosinase. They were significantly down-regulated at day 6 after grafting, but up-regulated gradually on days 8 and 12 after grafting (Figure 7).

Figure 7. The changes of immune-related DEGs expression during the process of pearl sac formation. Results are shown as the log2 fold change in expression versus the mean FPKM values of immune-related DEGs (FPKM ≥ 3, log2 fold change ≥ 2) at two adjacent time points after grafting. Hierarchical clustering analysis of DEGs’ expression changes was performed by MeV 4.8.1 to identify similarly temporal expression of genes. Up-regulated DEGs are in red, while down-regulated DEGs are in green.
Gene Ontology and KEGG Analysis of DEGs
The DEGs in day 0 vs. day 6 group, day 6 vs. day 8 group, and day 8 vs. day 12 group were successfully mapped to 55, 45, and 53 GO terms, respectively. For the biological processes, top GO terms were chitin metabolic process, viral transcription, viral gene expression, and the process of biosynthesis, metabolism, translation, location of protein. For cellular components, top GO terms were enriched in membrane related terms and ribosome related terms. For molecular functions, top GO terms were mainly enriched in oxidoreductase activity, chitin binding, copper ion binding, structural constituent of ribosome, and scavenger receptor activity (Supplementary Table S3).
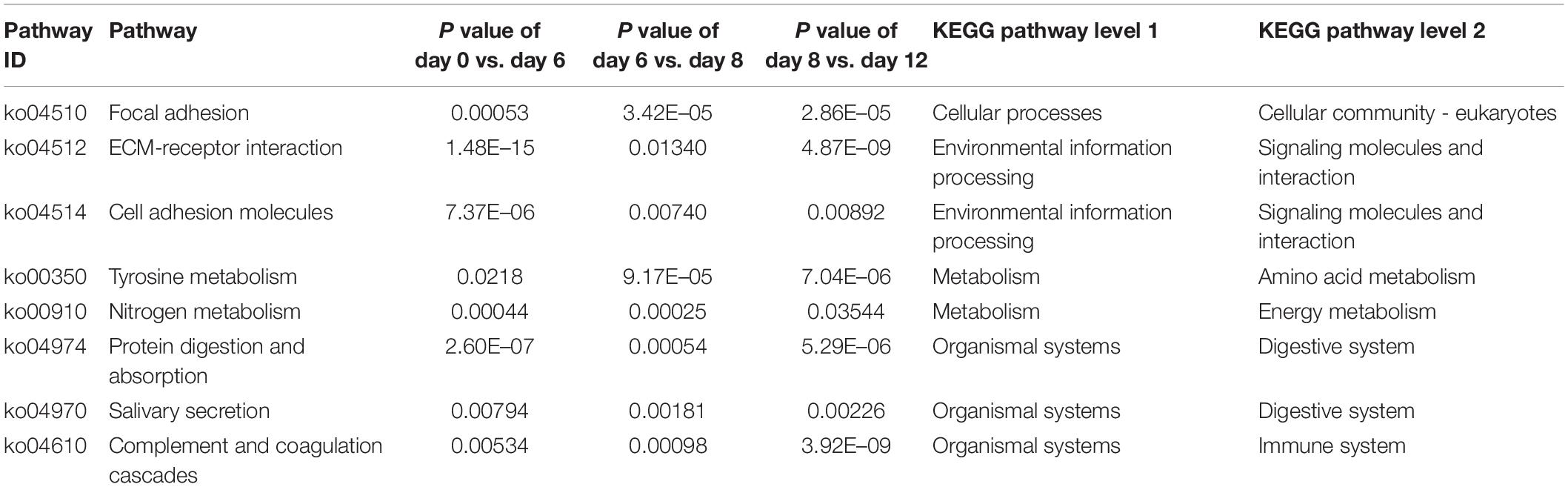
Meanwhile, all DEGs in day 0 vs. day 6 group, day 6 vs. day 8 group, and day 8 vs. day 12 group were enriched in 333, 321, and 327 KEGG pathways, respectively. Among them, eight pathways were highly enriched in all comparing groups (P < 0.05), including focal adhesion in the branch of cellular community-eukaryotes, ECM-receptor interaction and cell adhesion molecules (CAMs) in the branch of signaling molecules and interaction, tyrosine metabolism in the branch of amino acid metabolism, nitrogen metabolism in the branch of energy metabolism, protein digestion and absorption and salivary secretion in the branch of digestive system, and complement and coagulation cascades in the branch of immune system (Table 3 and Supplementary Table S4).
qRT-PCR Validation of DEGs
QRT-PCR was performed to confirm the gene expression profiles of 10 immune-related DEGs in comparative transcriptome analysis (Figure 8). The fold changes of mRNA expression level between adjacent time point after grafting detected by qRT-PCR were largely identical to those determined by transcriptome analysis in the unigenes of ferritin, complement C3, cathepsin-L, MMP-19, C-type lectin, but mostly identical in the unigenes of HSP90, galectin, defensin and C1qDC, but only but partially identical in the unigene of allograft inflammatory factor 1.
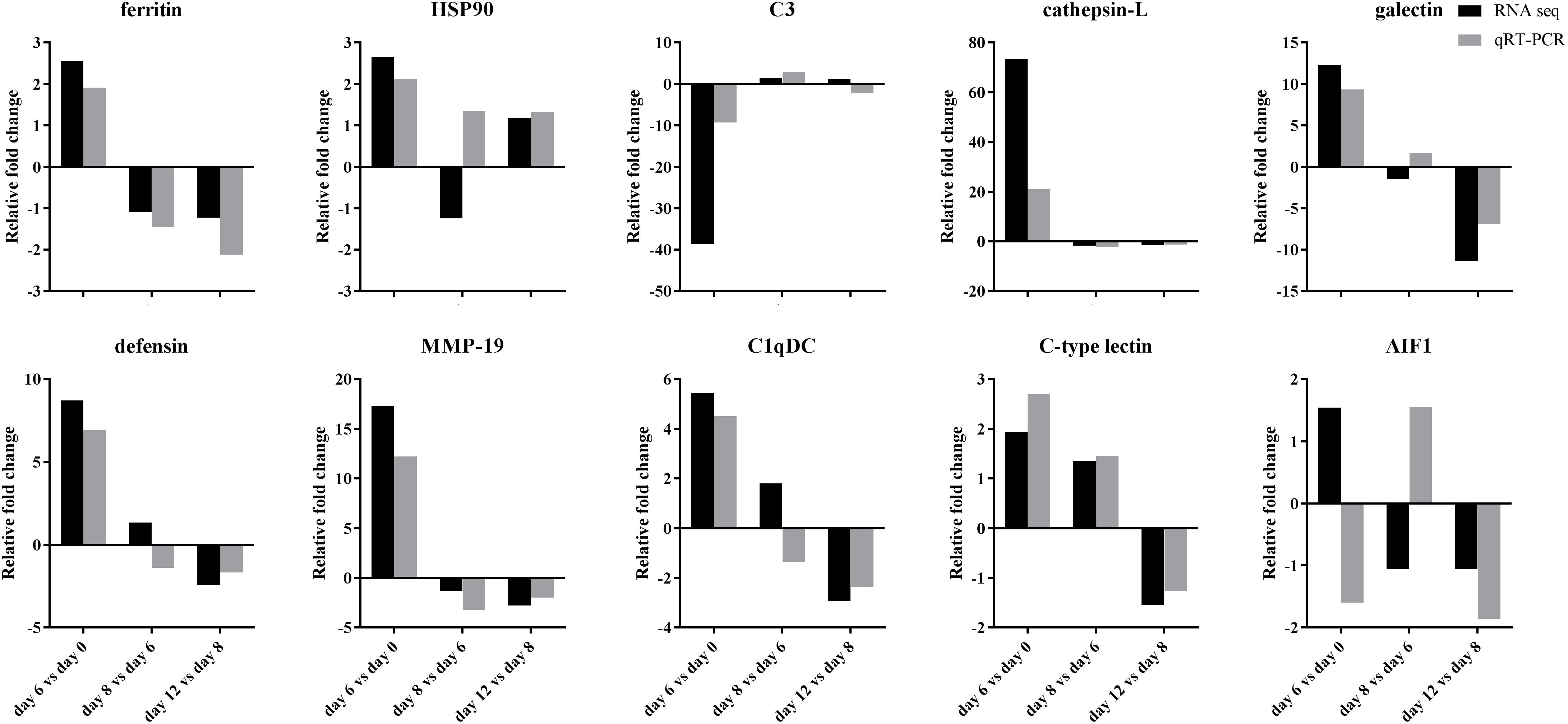
Figure 8. Comparison of fold changes of mRNA expression levels in 10 immune-related unigenes detected by transcriptome sequencing and qRT-PCR after grafting. HSP90: heat shock protein 90, C3: complement complement C3, MMP-19: matrix metalloproteinase-19, C1qDC: C1q domain containing protein, AIF1: allograft inflammatory factor 1.
Discussion
The innate immune system of Mollusca is constituted of cellular and humoral immune reactions. The cellular immune reactions performed by the hemocytes include encapsulation and phagocytosis of the pathogen, while the humoral immune responses consist of immune recognition, signal transduction, and effector synthesis (Allam and Raftos, 2015; Song et al., 2015). The immune reactions in pearl mussels during the process of wound healing are extremely crucial for post-grafting mortality, nucleus rejection, pearl sac morphology, and pearl quality (Cochennec-Laureau et al., 2010; Kishore and Southgate, 2015a, 2016; Mariom et al., 2019). In this study, histological analysis showed that there were three important time points in the process of wound healing and pearl sac formation in H. cumingii, which included (1) days 6 to 8 after grafting when the saibo integrated into the mantle connective tissue of the host mussel (Figure 1C) (2) days 8 to 10 after grafting when a large number of hemocytes aggregated in the pearl sac cavity (Figures 2A,B), and (3) day 12 after grafting when CaCO3 crystals were formed in the hemocytes clots and the epithelial cells of the pearl sac began to secrete extracellular matrix to mediate further CaCO3 crystal deposition (Figures 2F, 3). The results of our comparative transcriptome analysis were basically consistent with those of the histological analysis. For example, the highly enriched GO terms chitin metabolic process, membrane, and chitin binding, in the compared group of day 6 vs. day 8 after grafting implied a strong reaction of saibo integration at day 6, while the highly enriched GO terms peptide metabolic process, ribosomal subunit, and structural constituent of ribosome implied that the active protein metabolic activities such as the extracellular matrix (ECM) secretion happened on day 12 after grafting.
Similar to the shell, pearl secreted by the epithelial cells of pearl sac is also a kind of biomineralization product of Mollusca. Recent studies showed that hemocytes also participated in the biomineralization process of shell formation (Mount et al., 2004; Johnstone et al., 2008; Huang et al., 2018). Some special types of granulocytes contained calcium-rich vesicles were proposed to participate in the formation and transportation of CaCO3 crystal to the biomineralization sites in oysters (Johnstone et al., 2015; Li et al., 2016; Huang et al., 2018). Meanwhile, hemocytes were proven to express ECM which was associated with biomineralization in oysters (Johnstone et al., 2008; Li et al., 2016; Ivanina et al., 2017). In this study, the clots composed of apoptotic hemocytes and CaCO3 vesicles provided a surface for depositing the extracellular matrix secreted by the epithelial cells of pearl sac and formed the initial core of non-nucleus pearl for the further deposition of CaCO3 crystal (Figure 3). Thus, we proposed that the formation of the pearl core by hemocytes clot is the first step of pearl forming by providing organic substrate for deposition of ECM and CaCO3 crystal, and hemocyte aggregation in the pearl sac cavity is essential for the formation of freshwater non-nucleated pearl. Our results elucidated the complex and necessary roles of hemocytes in the pearl sac and pearl formation.
It is known that Toll-like receptors signaling pathway, complement system, proPO system, NF-κB signaling pathway, TNF signaling pathway, MAPK signaling pathway, and apoptosis participate in the immune response in Mollusca (Cerenius et al., 2008; Allam and Raftos, 2015; Wang et al., 2018). ProPO system is an important component of innate immune response in invertebrate involving in melanization, cytotoxic reactions, cell adhesion encapsulation, phagocytosis and self/non-self recognition (Cerenius et al., 2008; Wang et al., 2018; Bouallegui, 2019). Tyrosinase as one of type of phenoloxidases, a key member of proPO cascades of bivalves was one of the important pathways to product reactive oxygen intermediates (ROIs) to against pathogen invasion, regulation wound healing as signal molecular (Bogdan, 2007; Allam and Raftos, 2015; Wang et al., 2018). Meanwhile, researches also showed that excessive respiratory burst caused by ROIs after nuclear transplantation operation affected the pearl sac development of on pearl mussel of P. fucata (Kishore and Southgate, 2015a), suggesting the importance of oxidation-reduction process in the stress response of pearl oysters to nucleus implantation (Wei et al., 2017a; Wang et al., 2019). Recent research showed that supplying appropriate doses of dietary VD3 could enhance survival rates of host oysters P. fucata after transplantation by strengthen immunity and antioxidant capacity (Yang et al., 2019). In this study, tyrosine metabolism was highly enriched in the KEGG pathway. in which a total of 17 DEGs of tyrosinase transcripts were annotated. The various types of expression profile of tyrosine transcripts (Supplementary Table S5) suggested that the proPO system played an important role as one of the non-self-recognition systems on the innate immune responses of invertebrates (Cerenius et al., 2008; Gonzalez-Santoyo and Cordoba-Aguilar, 2012; Wang et al., 2018). The existence of a large number of tyrosinase transcripts and their differentially expression patterns implied the complexity of proPO system in participation of the immune reactions after grafting, and may play crucial roles in regulating the wound healing and the pearl sac formation in H. cumingii (Hong et al., 2006; Chen et al., 2017). Recent studies had shown that another important innate immune pathway, the complement system, played essential roles of distinguishing and eliminating non-self particles in the immune response after grafting operation in pearl mussel (Wang L. et al., 2017; Wang et al., 2018; Huang et al., 2019; Mariom et al., 2019). In this study, some core components of complement system including C1q contained domain proteins (C1qDCs) in classical pathway, galectins and C-type lectins in lectin pathway and a central element, component C3 were differential expression during the process of pearl sac formation (Figure 7). The highly enrichment of complement system in all comparing group implied that this system might be activated and it played potential important role in the regulation of the wound healing and the pearl sac formation in H. cumingii. Future studies will provide more detailed mechanism on the regulation of this process.
Previous studies reported that the immune system was reprogrammed in the process of wound healing and pearl sac formation (Wang W. et al., 2017; Wei et al., 2017b). However, the expression pattern of the immune-related genes in the immune response of mantle and nucleus grafting was still unclear. The significant up-regulation of a large number of pattern recognition proteins and immune effectors on day 6 after grafting (Figure 7) indicated that antioxidant enzymes, hydrolytic enzymes, and antimicrobial peptides of humoral defense factors were the main immune defense against potential bacterial invasion in the early period of wound healing (Schmitt et al., 2010; Allam and Raftos, 2015). Meanwhile, the significant upregulation of Cu-Zn SOD and Se-GPX of the antioxidant enzymes and tyrosinase in proPO system on days 8 and 12 after grafting (Figure 7) suggested an important role of these immune effectors in the oxidation-reduction reactions during the later period of immune resistance against allograft in H. cumingii. This result was also supported by the study in the pearl oyster Pinctada fucata against xenograft (Wei et al., 2017a). Thus, reduction the oxidative stress of host mussels after transplantation or grafting is a potential pathway to affect pearl sac formation and improve the pearl quality (Adzigbli et al., 2019; Yang et al., 2019).
Conclusion
In conclusion, we examined the changes of saibo and hemocytes in pearl sac formation of H. cumingii by histological analyses, and expression pattern of immune-related genes on days 6, 8, 12 after grafting by transcriptome analyses. The results of histological analysis suggested that the important roles of hemocytes in the wound healing and the pearl formation by transporting CaCO3 crystals and secreting extracellular matrix to mediate CaCO3 crystals deposition. The results of transcriptome analysis revealed the expression pattern of immune-related genes, the potential regulatory function of the signaling pathway proPO system, and the complement system on the immune response in pearl sac formation of H. cumingii. These findings will deepen our understanding of the immune process and its roles in pearl sac and pearl formation, and shed new lights on the potential methods of regulating the immune response to affect pearl sac formation and then improve the quality of the non-nucleus pearls.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA database under BioProject accession PRJNA556395.
Author Contributions
GR and GZ conceived and designed the experiments. WS, ZH, SX, and XX performed the experiments. WS and YH analyzed the data. GR, WS, and YH wrote and revised the manuscript.
Funding
This work was funded by Natural Science Foundation of Zhejiang Province (LY18C190009), the Public Project of Zhejiang Province, China (No. LGN19C190011), and Science and Technology Project of Shaoxing (2018C20010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00256/full#supplementary-material
References
Acosta-Salmon, H., and Southgate, P. C. (2006). Wound healing after excision of mantle tissue from the Akoya pearl oyster, Pinctada fucata. Compar. Biochem. Phys. A 143, 264–268. doi: 10.1016/j.cbpa.2005.12.006
Adzigbli, L., Wang, Z., Li, J., and Deng, Y. (2019). Survival, retention rate and immunity of the black shell colored stocks of pearl oyster Pinctada fucata martensii after grafting operation. Fish Shellfish Immunol. 98, 691–698. doi: 10.1016/j.fsi.2019.11.003
Allam, B., and Raftos, D. (2015). Immune responses to infectious diseases in bivalves. J. Invertebr. Pathol. 131, 121–136. doi: 10.1016/j.jip.2015.05.005
Atsumi, T., Aoki, H., Tanaka, S., and Komaru, A. (2018). Treatment effects during the post-operative care on the rate of pearl-sac formation in the pearl oyster Pinctada fucata. Aquaculture 483, 154–162. doi: 10.1016/j.aquaculture.2017.10.018
Bai, Z., Yin, Y., Hu, S., Wang, G., Zhang, X., and Li, J. (2009). Identification of genes involved in immune response, microsatellite, and SNP markers from expressed sequence tags generated from hemocytes of freshwater pearl mussel (Hyriopsis cumingii). Mar. Biotechnol. 11, 520–530. doi: 10.1007/s10126-008-9163-0
Bai, Z., Zheng, H., Lin, J., Wang, G., and Li, J. (2013). Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PLoS ONE 8:e53617. doi: 10.1371/journal.pone.0053617
Bogdan, C. (2007). Oxidative burst without phagocytes: the role of respiratory proteins. Nat. Immunol. 8, 1029–1031. doi: 10.1038/ni1007-1029
Bouallegui, Y. (2019). Immunity in mussels: an overview of molecular components and mechanisms with a focus on the functional defenses. Fish Shellfish Immunol. 89, 158–169. doi: 10.1016/j.fsi.2019.03.057
Cerenius, L., Lee, B. L., and Soderhall, K. (2008). The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271. doi: 10.1016/j.it.2008.02.009
Chen, X., Liu, X., Bai, Z., Zhao, L., and Li, J. (2017). HcTyr and HcTyp-1 of Hyriopsis cumingii, novel tyrosinase and tyrosinase-related protein genes involved in nacre color formation. Compar. Biochem. Phys. B 204, 1–8. doi: 10.1016/j.cbpb.2016.11.005
Cochennec-Laureau, N., Montagnani, C., Saulnier, D., Fougerouse, A., Levy, P., and Lo, C. (2010). A histological examination of grafting success in pearl oyster Pinctada margaritifera in French Polynesia. Aquat. Living Resour. 23, 131–140. doi: 10.1051/alr/2010006
Eddy, L., Affandi, R., Kusumorini, N., Sani, Y., and Manalu, W. (2015). The pearl sac formation in male and female Pinctada maxima host oysters implanted with allograft saibo. HAYATI J. Biosci. 22, 122–129. doi: 10.1016/j.hjb.2015.10.002
FAO (2018). Global Aquaculture Production. Available online at: http://www.fao.org/fishery/statistics/global-aquaculture-production/en.
Gonzalez-Santoyo, I., and Cordoba-Aguilar, A. (2012). Phenoloxidase: a key component of the insect immune system. Entomol. Exp. Appl. 142, 1–16. doi: 10.1111/j.1570-7458.2011.01187.x
Hong, X., Xiang, L., and Shao, J. (2006). The immunostimulating effect of bacterial genomic DNA on the innate immune responses of bivalve mussel, Hyriopsis cumingii Lea. Fish Shellfish Immunol. 21, 357–364. doi: 10.1016/j.fsi.2005.12.013
Huang, D., Shen, J., Li, J., and Bai, Z. (2019). Integrated transcriptome analysis of immunological responses in the pearl sac of the triangle sail mussel (Hyriopsis cumingii) after mantle implantation. Fish Shellfish Immunol. 90, 385–394. doi: 10.1016/j.fsi.2019.05.012
Huang, J., Li, S., Liu, Y., Liu, C., Xie, L., and Zhang, R. (2018). Hemocytes in the extrapallial space of Pinctada fucata are involved in immunity and biomineralization. Sci. Rep. 8:4657. doi: 10.1038/s41598-018-22961-y
Huang, Y., and Ren, Q. (2019). HcCUB-Lec, a newly identified C-type lectin that contains a distinct CUB domain and participates in the immune defense of the triangle sail mussel Hyriopsis cumingii. Dev. Comp. Immunol. 93, 66–77. doi: 10.1016/j.dci.2018.12.012
Ivanina, A. V., Falfushynska, H. I., Beniash, E., Piontkivska, H., and Sokolova, I. M. (2017). Biomineralization-related specialization of hemocytes and mantle tissues of the Pacific oyster Crassostrea gigas. J. Exp. Biol. 220, 3209–3221. doi: 10.1242/jeb.160861
Jiao, Y., Shi, S. L., Du, X. D., Lu, Y. Z., and Wang, Q. H. (2010). Histology and histochemistry on the pearl sac formation of pearl oyster Pinctada martensii. J. Guangdong Ocean Univ. 30, 7–10.
Johnstone, M. B., Ellis, S., and Mount, A. S. (2008). Visualization of shell matrix proteins in hemocytes and tissues of the Eastern oyster, Crassostrea virginica. J. Exp. Zool. Part B 310B, 227–239. doi: 10.1002/jez.b.21206
Johnstone, M. B., Gohad, N. V., Falwell, E. P., Hansen, D. C., Hansen, K. M., and Mount, A. S. (2015). Cellular orchestrated biomineralization of crystalline composites on implant surfaces by the eastern oyster, Crassostrea virginica (Gmelin, 1791). J. Exp. Mar. Biol. Ecol. 463, 8–16. doi: 10.1016/j.jembe.2014.10.014
Kishore, P., and Southgate, P. C. (2015a). Development and function of pearl-sacs grown from regenerated mantle graft tissue in the black-lip pearl oyster, Pinctada margaritifera (Linnaeus, 1758). Fish Shellfish Immunol. 45, 567–573. doi: 10.1016/j.fsi.2015.05.008
Kishore, P., and Southgate, P. C. (2015b). Haemocyte persistence after grafting for pearl production in Pinctada margaritifera (Linnaeus, 1758). Fish Shellfish Immunol. 42, 530–532. doi: 10.1016/j.fsi.2014.11.035
Kishore, P., and Southgate, P. C. (2016). A detailed description of pearl-sac development in the black-lip pearl oyster, Pinctada margaritifera (Linnaeus 1758). Aquacult. Res. 47, 2215–2226. doi: 10.1111/are.12674
Li, S., Liu, Y., Liu, C., Huang, J., Zheng, G., Xie, L., et al. (2016). Hemocytes participate in calcium carbonate crystal formation, transportation and shell regeneration in the pearl oyster Pinctada fucata. Fish Shellfish Immunol. 51, 263–270. doi: 10.1016/j.fsi.2016.02.027
Li, W., Shi, Z., and He, X. (2010). Study on immune regulation in Hyriopsis cumingii Lea: effect of pearl-nucleus insertion in the visceral mass on immune factors present in the hemolymph. Fish Shellfish Immunol. 28, 789–794. doi: 10.1016/j.fsi.2010.01.019
Mariom, Take, S., Igarashi, Y., Yoshitake, K., Asakawa, S., Maeyama, K., et al. (2019). Gene expression profiles at different stages for formation of pearl sac and pearl in the pearl oyster Pinctada fucata. BMC Genomics 20:240. doi: 10.1186/s12864-019-5579-3
Mount, A. S., Wheeler, A. P., Paradkar, R. P., and Snider, D. (2004). Hemocyte-mediated shell mineralization in the eastern oyster. Science 304, 297–300. doi: 10.1126/science.1090506
Ren, G., Wang, Y., Qing, J., Tang, J., Zheng, X., and Li, Y. (2014). Characterization of a novel carbonic anhydrase from freshwater pearl mussel Hyriopsis cumingii and its expression profiles in response to environmental conditions. Gene 546, 56–62. doi: 10.1016/j.gene.2014.05.039
Ren, Q., Li, M., Zhang, C., and Chen, K. (2011). Six defensins from the triangle-shell pearl mussel Hyriopsis cumingii. Fish Shellfish Immunol. 31, 1232–1238. doi: 10.1016/j.fsi.2011.07.020
Ren, Q., Qi, Y., Hui, K., Zhang, Z., Zhang, C., and Wang, W. (2012). Four invertebrate-type lysozyme genes from triangle-shell pearl mussel (Hyriopsis cumingii). Fish Shellfish Immunol. 33, 909–915. doi: 10.1016/j.fsi.2012.07.019
Schmitt, P., Gueguen, Y., Desmarais, E., Bachere, E., and de Lorgeril, J. (2010). Molecular diversity of antimicrobial effectors in the oyster Crassostrea gigas. BMC Evol. Biol. 10:23. doi: 10.1186/1471-2148-10-23
Shi, A., Zhang, M., Wu, Z. W., and Peng, X. F. (1985). On the formation of pearl sac in freshwater mussel. J. Fish. China 9, 247–253.
Song, L., Wang, L., Zhang, H., and Wang, M. (2015). The immune system and its modulation mechanism in scallop. Fish Shellfish Immunol. 46, 65–78. doi: 10.1016/j.fsi.2015.03.013
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, L., Song, X., and Song, L. (2018). The oyster immunity. Dev. Comp. Immunol. 80, 99–118. doi: 10.1016/j.dci.2017.05.025
Wang, L., Zhang, H., Wang, L., Zhang, D., Lv, Z., Liu, Z., et al. (2017). The RNA-seq analysis suggests a potential multi-component complement system in oyster Crassostrea gigas. Dev. Comp. Immunol. 76, 209–219. doi: 10.1016/j.dci.2017.06.009
Wang, W., Wu, Y., Lei, Q., Liang, H., and Deng, Y. (2017). Deep transcriptome profiling sheds light on key players in nucleus implantation induced immune response in the pearl oyster Pinctada martensii. Fish Shellfish Immunol. 69, 67–77. doi: 10.1016/j.fsi.2017.08.011
Wang, W., Lei, Q., Liang, H., and He, J. (2019). Towards a better understanding of allograft-induced stress response in the pearl oyster Pinctada fucata martensii: insights from iTRAQ-based comparative proteomic analysis. Fish Shellfish Immunol. 86, 186–195. doi: 10.1016/j.fsi.2018.11.044
Wei, J., Fan, S., Liu, B., Zhang, B., Su, J., and Yu, D. (2017a). Transcriptome analysis of the immune reaction of the pearl oyster Pinctada fucata to xenograft from Pinctada maxima. Fish Shellfish Immunol. 67, 331–345. doi: 10.1016/j.fsi.2017.06.030
Wei, J., Liu, B., Fan, S., Li, H., Chen, M., Zhang, B., et al. (2017b). Differentially expressed immune-related genes in hemocytes of the pearl oyster Pinctada fucata against allograft identified by transcriptome analysis. Fish Shellfish Immunol. 62, 247–256. doi: 10.1016/j.fsi.2017.01.025
Xu, Q., Wang, G., Yuan, H., Chai, Y., and Xiao, Z. (2010). cDNA sequence and expression analysis of an antimicrobial peptide, theromacin, in the triangle-shell pearl mussel Hyriopsis cumingii. Comp. Biochem. Phys. B 157, 119–126. doi: 10.1016/j.cbpb.2010.05.010
Yang, C., Du, X., Hao, R., Wang, Q., Deng, Y., and Sun, R. (2019). Effect of vitamin D3 on immunity and antioxidant capacity of pearl oyster Pinctada fucata martensii after transplantation: insights from LC–MS-based metabolomics analysis. Fish Shellfish Immunol. 94, 271–279. doi: 10.1016/j.fsi.2019.09.017
Zhang, A. J., Liu, S. L., Zhu, J. Y., Gu, Z. M., Zhou, Z. M., Zhang, G. F., et al. (2016). Transcriptome analysis of the freshwater pearl mussel, Hyriopsis cumingii (Lea) using Illumina paired-end sequencing to identify genes and markers. Iran. J. Fish. Sci. 15, 1425–1440.
Zhang, H., Huang, Y., Man, X., Wang, Y., Hui, K., Yin, S., et al. (2017). HcToll3 was involved in anti-Vibrio defense in freshwater pearl mussel, Hyriopsis cumingii. Fish Shellfish Immunol. 63, 189–195. doi: 10.1016/j.fsi.2017.02.015
Zhao, L., Hui, K., Wang, Y., Wang, Y., Ren, Q., and Li, X. (2018). Three newly identified galectin homologues from triangle sail mussel (Hyriopsis cumingii) function as potential pattern-recognition receptors. Fish Shellfish Immunol. 76, 380–390. doi: 10.1016/j.fsi.2018.02.032
Keywords: immune response, pearl sac formation, histological analysis, transcriptome analysis, Hyriopsis cumingii
Citation: Shen W, Hu Y, He Z, Xu S, Xu X, Zhang G and Ren G (2020) Histological and Comparative Transcriptome Analyses Provide Insights Into the Immune Response in Pearl Sac Formation of Hyriopsis cumingii. Front. Mar. Sci. 7:256. doi: 10.3389/fmars.2020.00256
Received: 21 July 2019; Accepted: 31 March 2020;
Published: 30 April 2020.
Edited by:
Maria Angeles Esteban, University of Murcia, SpainReviewed by:
Aldo Nicosia, Italian National Research Council, ItalyPaulina Schmitt, Pontificia Universidad Católica de Valparaíso, Chile
Shiguo Li, Research Center for Eco-environmental Sciences (CAS), China
Dahui Yu, Beibu Gulf University, China
Copyright © 2020 Shen, Hu, He, Xu, Xu, Zhang and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Ren, cmVuZ0B1c3guZWR1LmNu
†These authors have contributed equally to this work
 Wenying Shen
Wenying Shen Yiwei Hu
Yiwei Hu Zhengkai He
Zhengkai He Shuting Xu
Shuting Xu Xinyue Xu
Xinyue Xu Genfang Zhang2
Genfang Zhang2 Gang Ren
Gang Ren