- 1National Engineering Laboratory of Marine Germplasm Resources Exploration and Utilization, Zhejiang Ocean University, Zhoushan, China
- 2Key Laboratory of Tropical Marine Bio-resources and Ecology, Chinese Academy of Sciences, Guangzhou, China
- 3Lasbella University of Agriculture, Water and Marine Sciences, Uthal, Pakistan
- 4Center of Excellence in Marine Biology, University of Karachi, Karachi, Pakistan
Konosirus punctatus inhabits areas of significant commercial importance in Chinese coastal waters. Twelve populations distributed all along the Chinese coast were analyzed using two mtDNA genes. The cytochrome oxidase subunit I (COI) and cytochrome b (Cytb) genes were amplified from 242 and 259 individuals, respectively, and fragments of 999 and 415 bp sequence in length were used for analysis. COI and Cytb denoted a high haplotype diversity and low nucleotide diversity. The COI mtDNA gene revealed a high molecular variance within populations and low variance among populations, whereas Cytb showed a high variance among populations and low within populations. FST values obtained for COI and Cytb showed low genetic differentiation among the populations, except for the Zhongshan population where the Cytb gene showed high FST values. The constructed phylogenetic tree and haplotype network tree showed a single lineage, however, Cytb gene data showed that the Zhongshan population has a greater difference level within a population. The present investigation contributed to existing knowledge of the genetic structure of K. punctatus and can be useful for its future conservation.
Introduction
Konosirus punctatus (Temminck and Schlegel, 1846) is also known as the dotted gizzard shad or spotted sardine. It belongs to the Clupeidae family and is widely distributed in areas throughout the Indo-west Pacific Ocean, such as in Japanese coastal waters, South Korean waters, and Chinese marine waters (Zhang, 2001). The dotted gizzard shad is a pelagic fish and can remain alive even in freshwater (Whitehead, 1985; Myoung and Kim, 2014; Song et al., 2017). It breeds in shallow brackish waters (Estuaries) from April to August, takes two years to reach adulthood, and can survive for up to five years (Myoung and Kim, 2014). Generally its larval dispersal capacity is strong. The pelagic duration of its larva is 24–28 days (Kong et al., 2004; Yamada et al., 2007; Song et al., 2017). The annual global capture and aquaculture production of this species is showing a decline. The global capture production was 23,707 tonnes during 1995; subsequently, it decreased to 4300 tonnes in 2016. The global aquaculture production of this species also showed a decline. It was 2519 tonnes in 2006, which decreased to 390 tonnes in 20161.
It could potentially be seen as a commercial species and also be taken as by-catch, therefore, it has several threats to its existence, especially because of the degradation of estuarine ecosystems. Nevertheless, its larval and adult dispersal range is strong enough to enhance gene flow among different populations. In general, the decline in aquatic resources needs management and conservation plans, therefore, it is essential to have genetic investigations. So far, meager experiments have been carried out to understand the population genetic details of K. punctatus from Chinese coastal waters. For example, Song et al. (2017) used control region (CR) and cytochrome b (Cytb) to understand the genetic differences of K. punctatus distributed all along the coasts of China and Japan; they included seven populations from Chinese coastal waters and three species from Japanese coast waters, while the present investigation included 10 populations for extensive investigation. Li et al. (2012) contributed to the complete mitochondrial genome of the dotted gizzard shad (K. punctatus). Similar studies had been conducted in Korean and Japanese coast waters (Myoung and Kim, 2014; Gwak et al., 2015). To the best of our literature survey, only the above-mentioned investigations have been taken, where extended dispersal activities were shown and no phylogeographic divergence was found among populations (Song et al., 2017). Nevertheless, the capture production of this important fish is decreasing, therefore, it is deemed necessary to carry out an extensive study of the population genetics of K. punctatus.
The genetic structure is generally influenced by multiple factors, like genetic divergence and adaptation (De Luca et al., 2014). Furthermore, the longtime continuation of species isolation would result in genetic divergences and create a population unable to intermingle with the original population in terms of reproduction. This would lead to speciation (Mayr, 1942).
Marine organisms have been impacted by several factors because of the complex marine environment. The currents and certain other suitable factors are considered to be gene flow enhancers over large geographical distances (Hellberg et al., 2002; Maltagliati et al., 2002; Ferna’ndez et al., 2011). Often, the dispersal capacity of marine organisms covers an extensive range, because of their high fecundity, migration, and planktonic larval stages. These factors are gene flow enhancers (Waples, 1998; Bohonak, 1999; Fadhlaoui-Zid et al., 2012). K. punctatus is among the fish species that showed a high dispersal ability both at larval and adult stages. Active larval dispersal enhances the dispersal capability, resulting in homogenization between populations. Heterogenization or homogenization details can be obtained by investigation throughout the distribution range with extensive sampling and using conserved mtDNA molecular markers.
Mitochondrial DNA molecular markers are useful tools for population genetic analysis and are particularly accurate for tracking the population history of species with a small effective population size (Avise et al., 1987; Mortiz, 1994; Vaseeharan et al., 2013). Certain regions of mtDNA diverge rapidly while some are conserved (Eizirik et al., 1998). Therefore, it is intuitive to have appropriate molecular markers for population studies. The molecular markers Cytb and Cytochrome oxidase subunit I (COI) represent useful genetic markers for population structure and phylogenetic analyses.
In this study, we examined data of two mitochondrial genes (COI and Cytb) acquired from 242 and 259 individuals, respectively, collected from 12 locations throughout the K. punctatus dispersal area along the Chinese coast. The sequences were assessed to understand the genetic diversity of K. punctatus. The results of this investigation will contribute details to researchers and fishery managers.
Materials and Methods
Ethics Statement
All methods were carried out in line with the relevant principles and directions in China.
Collection
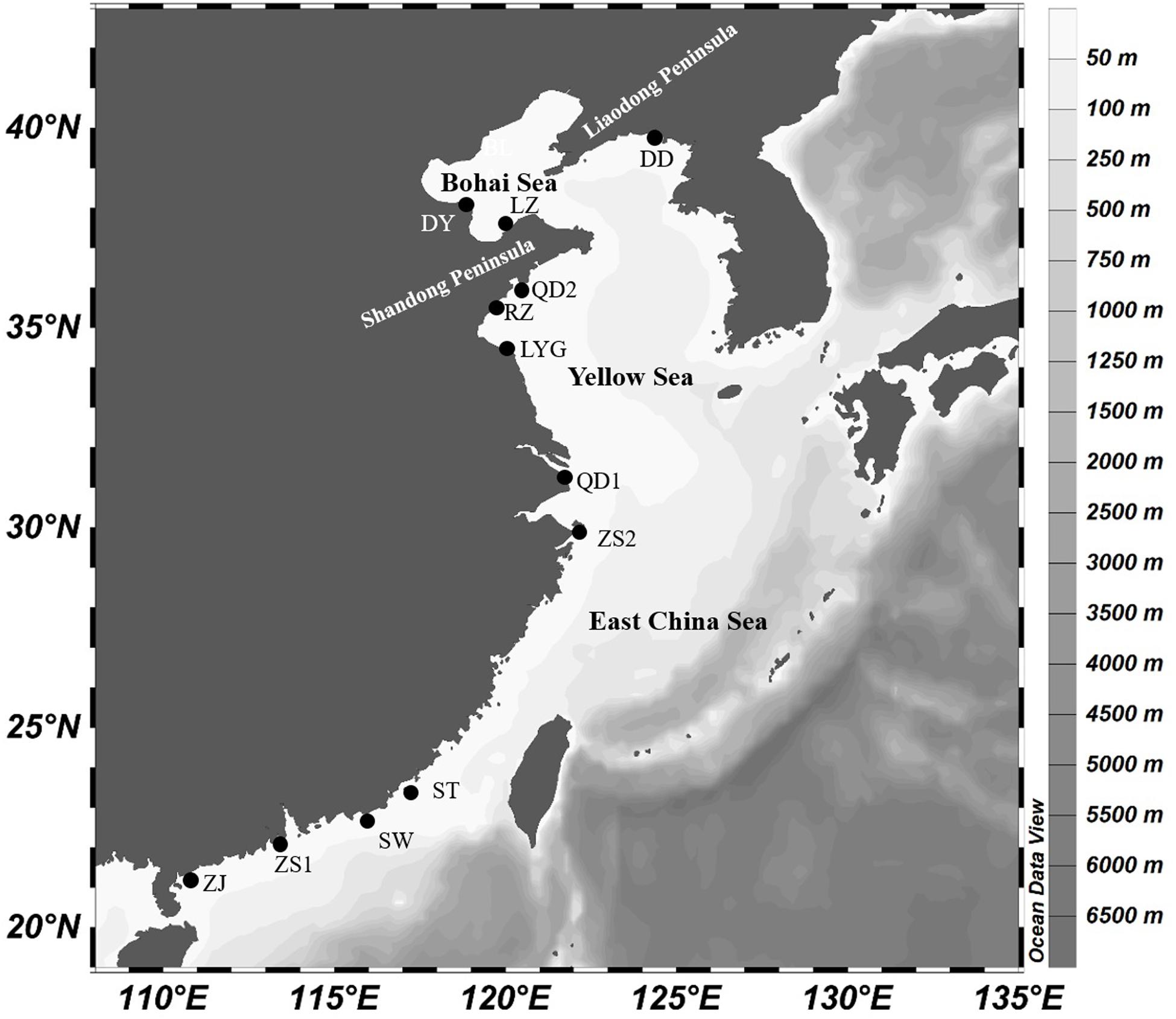
In total, 242 and 259 specimens of K. punctatus were sequenced from 12 geographic locations across Chinese coastal waters (Figure 1 and Table 1) for COI and Cytb genes respectively. Samples were collected by hiring local fishing boats. Only adult individuals were chosen for experiments. Muscle tissues were preserved in 90% ethanol and transported to the laboratory.
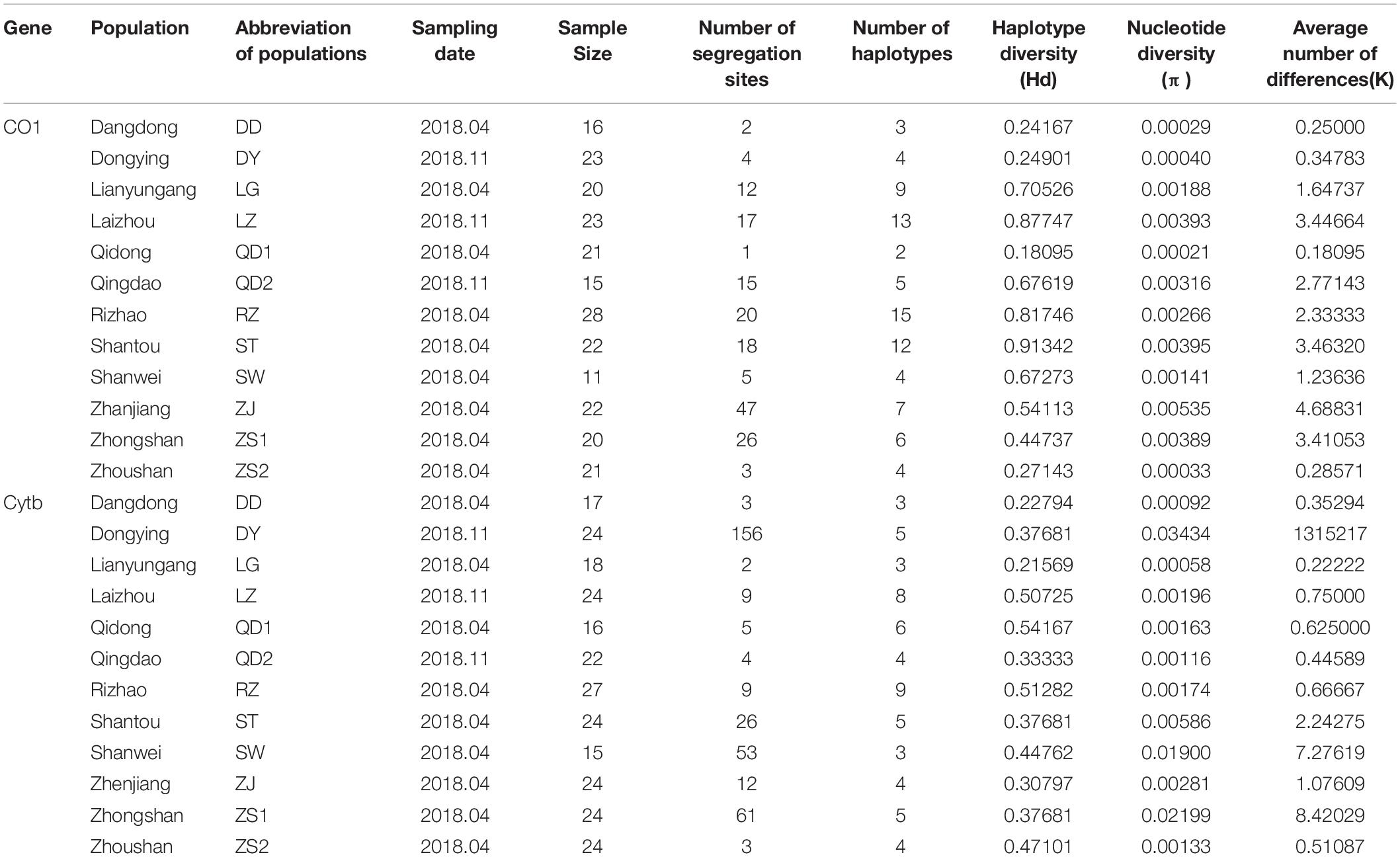
Table 1. Location, sample size, and summary statistics of the genetic variability of Konosirus punctatus as revealed by two mtDNA molecular markers.
DNA Extraction
Total genomic DNA was extracted using the phenol–chloroform method (Sambrook et al., 1989). For polymerase chain reaction (PCR), the concentrated DNA was diluted to reach a working concentration of (20 ng/μl).
Polymerase Chain Reaction Amplification
A 999and 415 bp fragment of COI and Cytb gene sequences were amplified, respectively. The pair of primers was designed using an mtDNA gene sequence. COI was amplified with the primers C-F (5′-TGGTGCCTGAGCAGGAATAGT-3′) and C-R (5′-CAGTGCACGAATGCAGCCATA-3′); the Cytb was amplified with the primers B-F (5′-CAACGACGCAGTAGTCGACCT-3′) and B-R (5′-CGGAACGTAAGTCCTCGTTGCT-3′). A total of 20 μl PCR mixture included a master mix (10 μl), water (7 μl), forward and reverse primer (1 μl each), and 1 μl of DNA template. PCR conditions were as follows: denaturation at 94°C for 5 min, 35 cycles at 94°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 30 s, and the final extension at 72°C for 7 min. The PCR products were run on a 1.5% agarose gel. Sequencing was executed on ABI 3730 automated sequencer.
Data Analysis
The DNA sequences were aligned using ClustalW. The genetic parameters including polymorphic sites, haplotype diversity (Hd), nucleotide diversity (π), and average pairwise differences (K) was estimated using DnaSP (Librado and Rozas, 2009). Analysis of molecular variance (AMOVA), Tajim’s D, Fu’s Fs, and their corresponding P-values were obtained with software Arlequin (Excoffier and Lischer, 2010). The neighbor-joining tree was constructed using MEGA 6 (Tamura et al., 2013). The median-joining network was created using NETWORK software version 5.0.0.1 (Bandelt et al., 1999).
Results
Genetic Variation
Nine hundred and ninety-nine base pair fragments of the COI gene were examined using 242 sequences obtained from 12 populations. The analysis of sequences resulted in 108 polymorphic sites and 60 haplotypes. The Hd ranges from 0.18095 to 0.91342, nucleotide diversity (π) from 0.00021 to 0.00535, and the average number of nucleotide differences (k) from 0.18095 to 4.68831. The Shantou population showed a high Hd, while the Qidong populations revealed the lowest Hd, as shown in Table 1. Two hundred and fifty-nine sequences of Cytb, with each fragment comprised of 415bp, were analyzed to show 243 polymorphic sites and 39 haplotypes. The Hd ranges from 0.21569 to 0.54167, nucleotide diversity (π) from 0.00058 to 0.037681, and the average number of nucleotide differences (k) from 0.22222 to 13.15217. The Dongying denoted a high Hd and the Lianyungang population showed the lowest Hd (Table 1). All haplotypes were deposited in GenBank, with the Accession numbers MT381749 to MT381808 (COI sequences) and MT381809 to MT381847 (Cytb sequences).
Population Genetic Structure
The AMOVA illustrated the population structure based on both COI and Cytb. The results showed that 82.56 and 25.07% of the variation occurred within populations, while 17.44 and 74.93% of the variation occurred among populations, for COI and Cytb genes, respectively (Table 2), suggesting that COI gene supports less differentiation among populations while Cytb supports a moderate population structure. AMOVA analysis dividing by groups was also conducted; five populations (Shantou, Zhanjiang, Zhoushan, Zhongshan, and Shan Wei) were grouped for southern groups. The rest of the populations were grouped as northern populations. The results showed that 18.96 and 65.16% of the variation occurred among populations within groups, while 83.55 and 27.63% of the variation occurred within populations, for COI and Cytb genes, respectively (Supplementary Table S1). The pairwise FST values of two genes (COI and Cytb) were also suggestive of less differentiation among populations, as shown in Table 3. The developed neighbor-joining tree obtained from the data of two genes (COI and Cytb) described uniclade topology (Figures 2, 3).
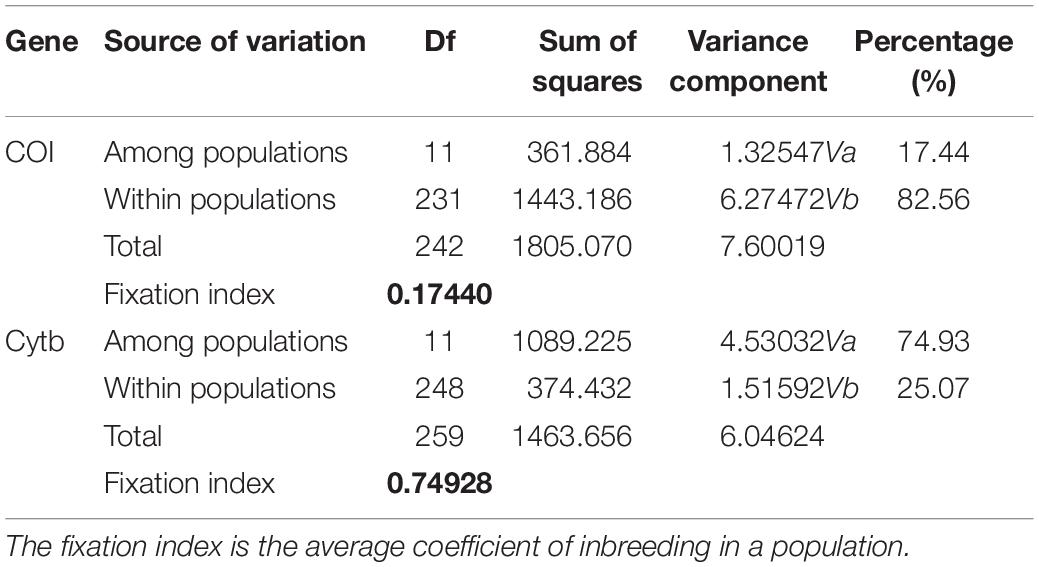
Table 2. Analysis of molecular variance among Konosirus punctatus populations using COI and Cytb mtDNA gene data.
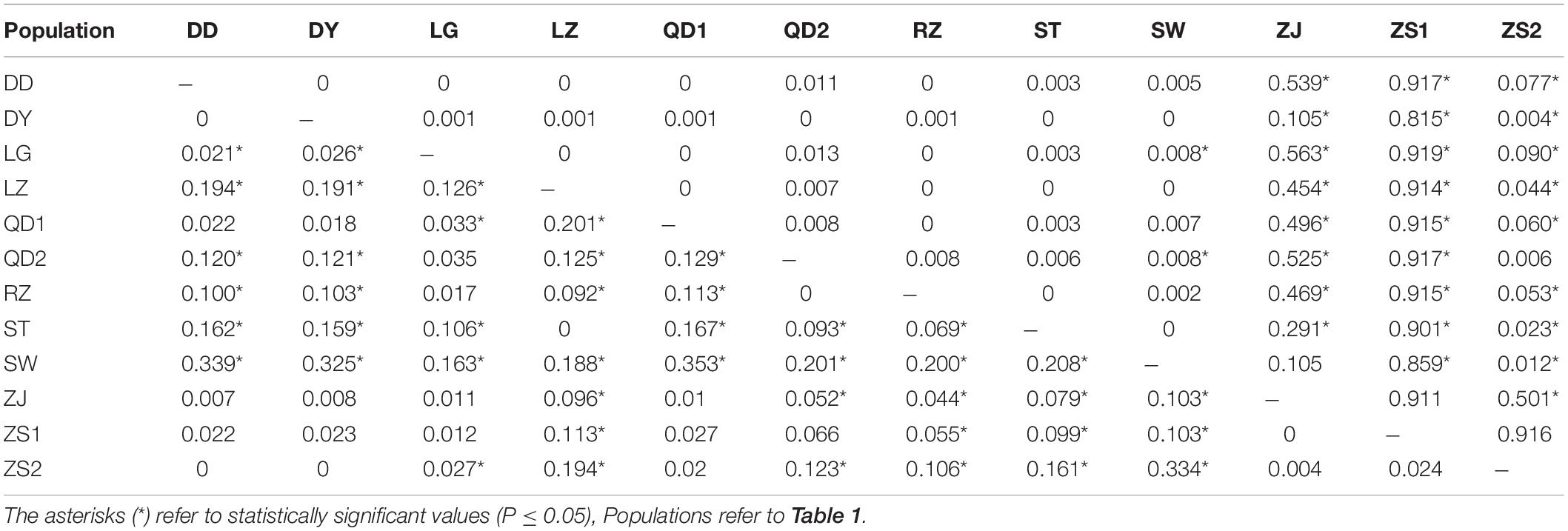
Table 3. Pairwise FST values of 12 populations of Konosirus punctatus for COI (below) and Cytb (above).
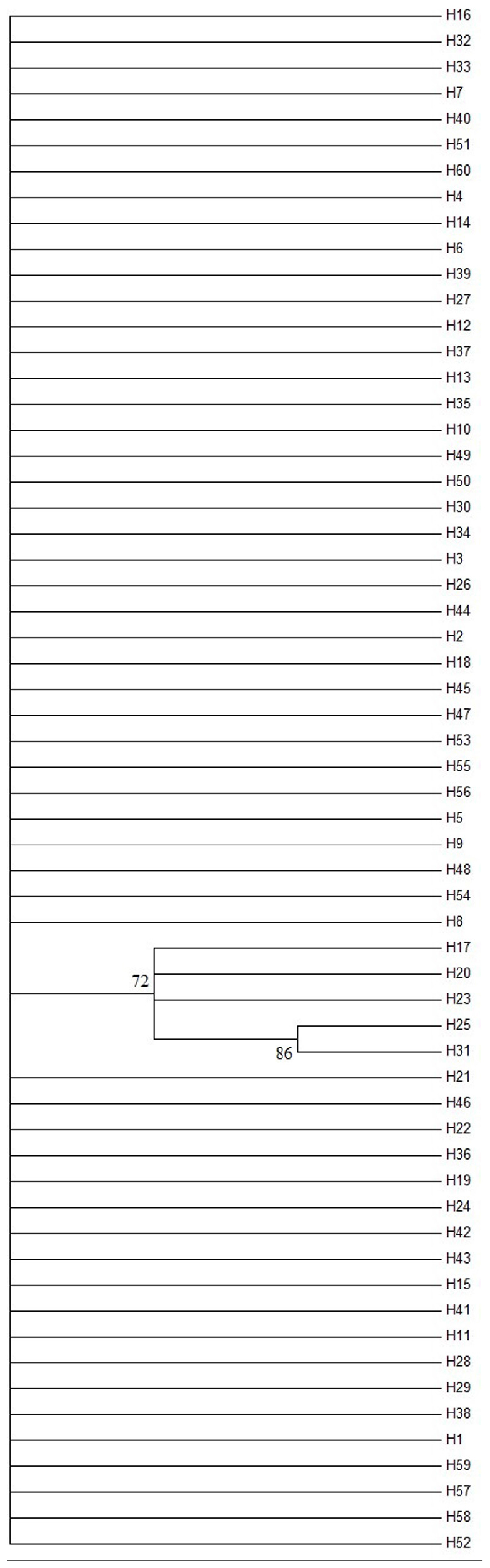
Figure 2. Neighbor-joining phylogenetic tree of Konosirus punctatus developed using COI data. The detailed haplotype representation is similar as shown in Figure 4. Branch support values (higher than 70%) in bootstrap.

Figure 3. Molecular neighbor-joining phylogenetic tree of Konosirus punctatus developed using Cytb data. The detailed haplotype representation is similar as shown in Figure 5. Branch support values (higher than 70%) in bootstrap.
Haplotype Network Analysis
The median-joining network based on the two genes illustrated one main clade, similar to the phylogenetic tree. The median-joining network of the COI gene denoted that Hap 1 was contributed to by all 12 populations, while Hap 8 was contributed to by five populations, including Shenwei, Zhongshan, Lianyungang, Qingdao, and Rizhao. Similarly, the median-joining network of the Cytb gene showed that Hap 1 was contributed to by the 12 populations, while Hap 13 was shared by six populations (Figures 4, 5). The network results suggest a boundless gene flow among populations, however, certain haplotypes of the Cytb gene, like 34, 35, 36, and 37, belonging to the Zhongshan population, cluster separately, showing differentiation within populations which may lead to a restricted gene flow of this population with their counterparts. The results of Tajima’s D showed negative values gained from both of the genes whereas, Fu’s Fs statistics showed some negative values in Cytb data, while all showed positive values in the COI data (Table 4).
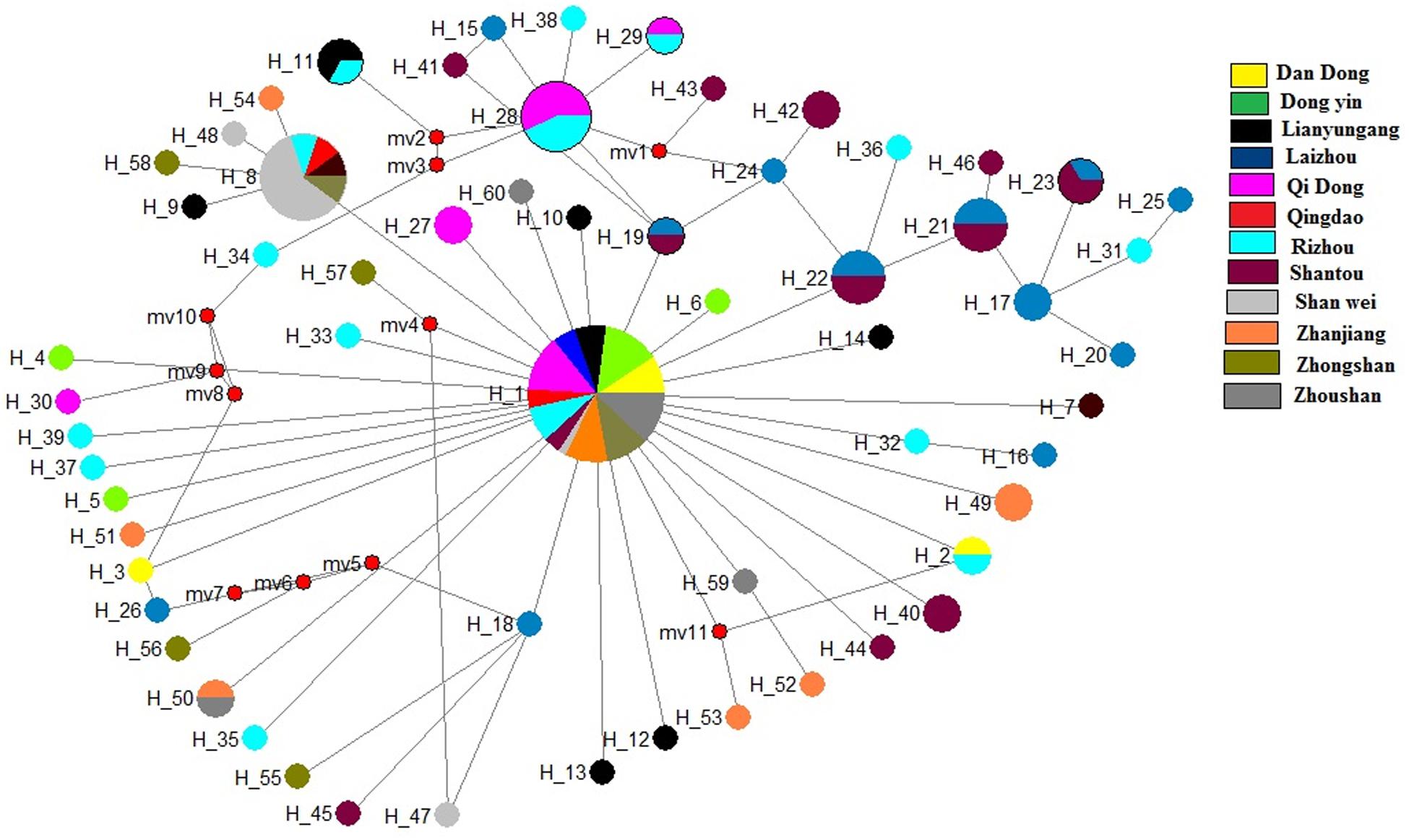
Figure 4. Median-joining networks of Konosirus punctatus developed using COI data. Color representation is showing the population frequency. Shown numbers are representing the haplotype numbers.
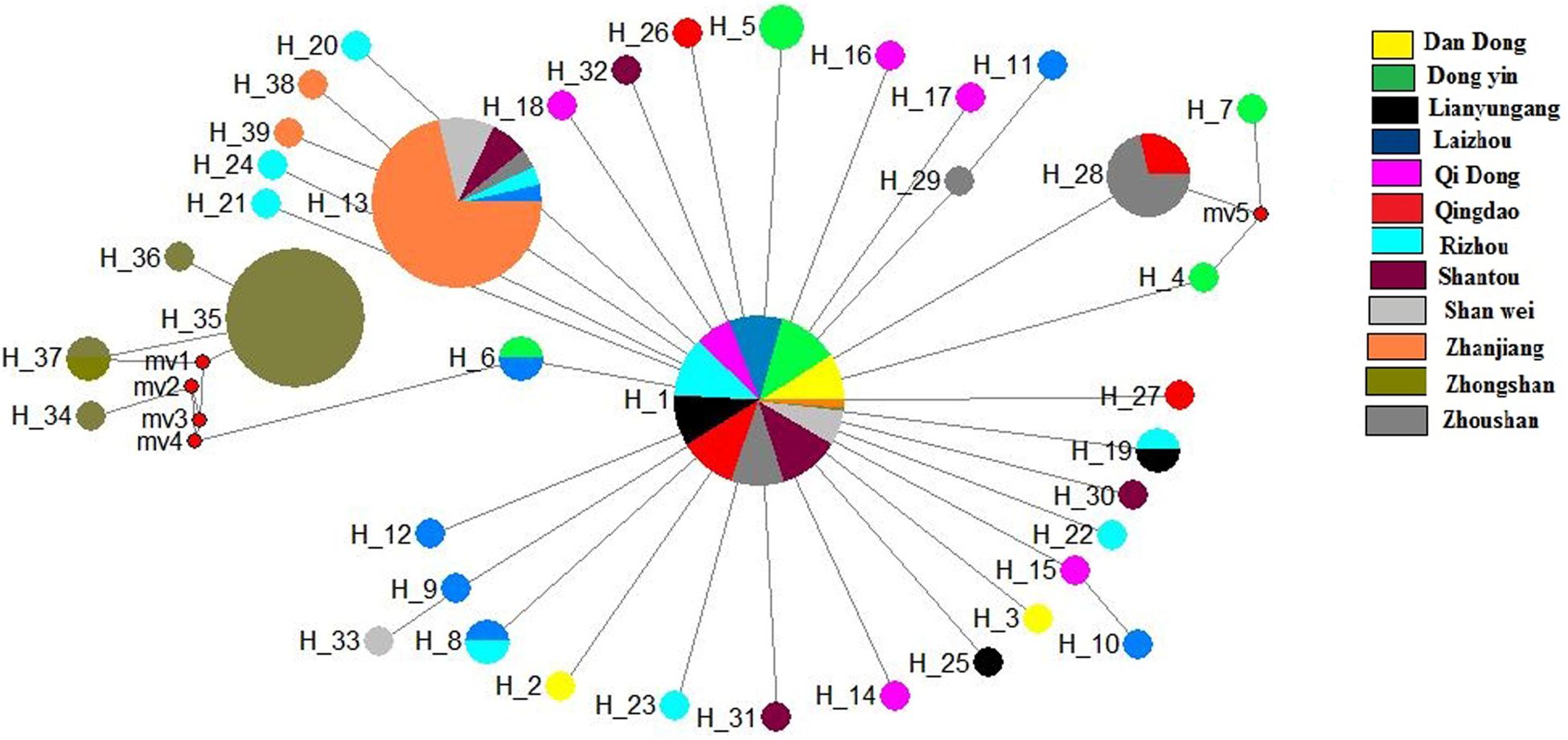
Figure 5. Median-joining networks of Konosirus punctatus developed using Cytb data. Color representation is showing the population frequency. Shown numbers are representing the haplotype numbers.
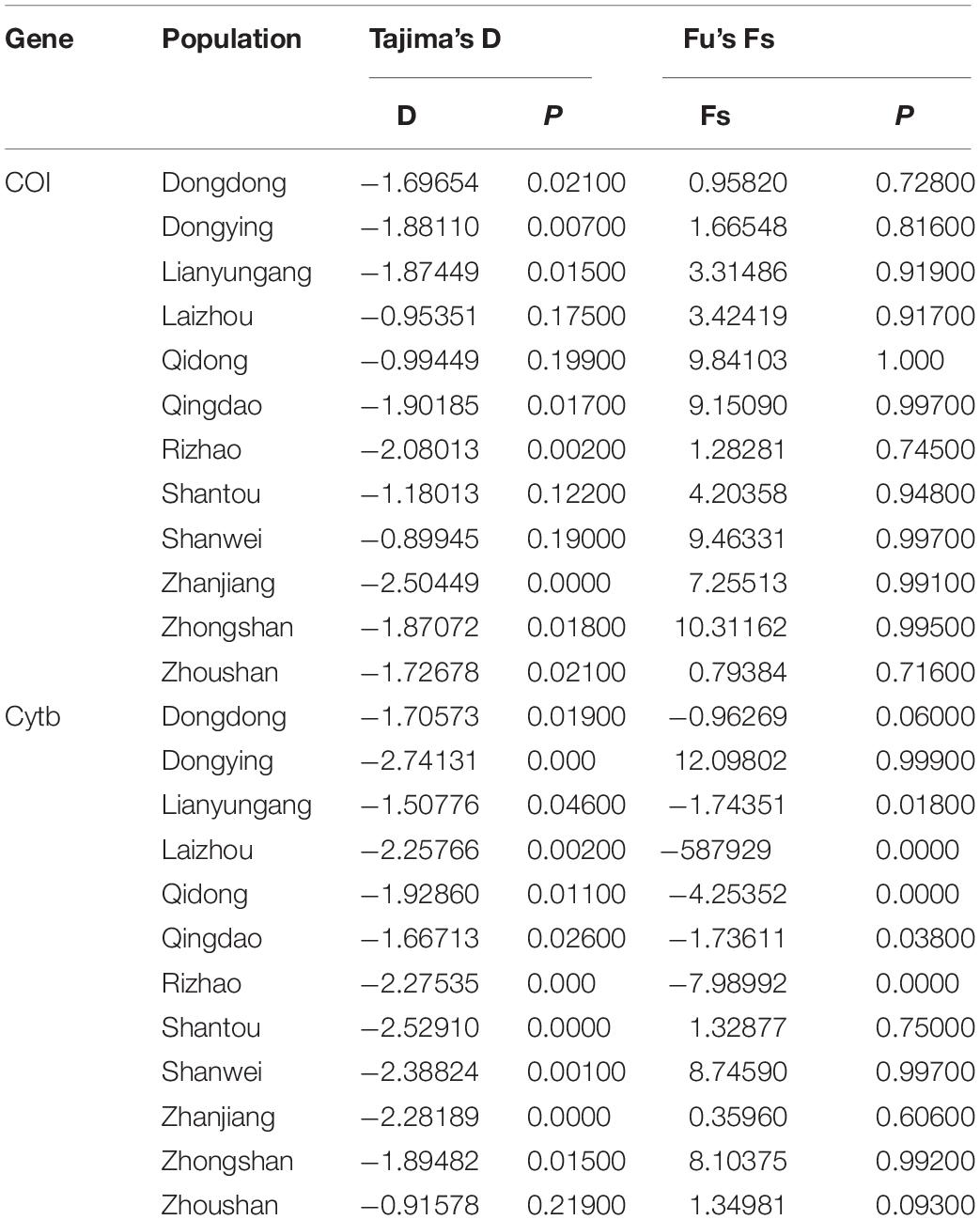
Table 4. Neutrality test results for COI and Cytb data obtained from 12 populations of Konosirus punctatus.
Discussion
Konosirus punctatus, which is commonly known as the dotted gizzard shad, is enlisted in the International Union for Conservation of Nature (Hata, 2017) red list category. This species has immense commercial importance and dwells in brackish and littoral waters of Korea (Choi et al., 2002) and many other places of the world’s marine waters. Preliminary facts are very important to knowing the genetic differentiation and population structure for fishery management and conservation (Casey et al., 2016; Melis et al., 2018). Compared to other fish species, meager information is available for the population genetics of this species (Myoung and Kim, 2014; Gwak et al., 2015; Song et al., 2017). These studies are limited to the populations of Korea, Japan, and a single study illustrating the population genetics of K. punctatus in Chinese and Japanese waters, using CR and Cytb, respectively. The mitochondrial COI is more conserved, therefore we take investigations along with the Cytb gene to understand the complete profiles of its population genetics inhabiting in a range of Chinese waters.
In this investigation, we described the genetic differentiation in 12 populations of K. punctatus collected all along the Chinese coastal waters (Yellow Sea, East China Sea, and the South China Sea) (Figure 1), taking two mtDNA molecular markers. The results showed a high haplotype and low nucleotide diversity among the species with few exceptions (Table 1), which is consistent with earlier studies (Myoung and Kim, 2014; Song et al., 2017). Results of a high haplotype and low nucleotide diversity may indicate that populations have undergone a genetic bottleneck followed by a sudden expansion (Grant and Bowen, 1998).
Analysis of molecular variance by the COI gene showed higher values within populations and lower values among populations, while the Cytb gene revealed high values among populations and low values within populations. The results of the COI gene showed agreement with the results of Song et al. (2017), while the Cytb results differ with the foresaid results (Table 2). The lower Fst values are similar to the results of Song et al. (2017), however, in the case of the Cytb gene, where the Zhongshan population showed higher values, this is in disagreement with the earlier results (Table 3). An FST value of 0.05 indicates negligible genetic variation andvalues higher than 0.25 indicate a high genetic variation within the examined population (Weir and Cockerham, 1996). Based on these criteria, the results gained in this investigation revealed less variation among the K. punctatus populations. Only the Zhongshan population revealed higher differentiation in the case of the Cytb gene, whereas, studies by Song et al. (2017) showed comparatively higher FST values. The FST values in this study illustrate that the diversity of the K. punctatus population along the Chinese coast is boundless, except for the Zhongshan population as revealed by the Cytb gene.
The neighbor-joining tree of these molecular markers (COI and Cytb) showed a single lineage for all populations. Although the Zhongshan population showed a greater differentiation within the same clade, this has only been shown by Cytb results. Excluding the Cytb gene data from the Zhongshan population, the rest of the results of this investigation are in agreement with previous studies of K. punctatus. The results of the median-joining network described no genetic variation between the populations concerning both of the investigated genes, which is an agreement with the results of Song et al. (2017) sampled from Chinese waters, however, Cytb gene data illustrated a little greater differentiation for the Zhongshan population.
The ocean current and life history characteristics plays vital roles in genetic variations. It has been reported that the Kuroshio Current does not limit the gene flow of this threatened species in this region (Song et al., 2017).
Marine fish often have a high dispersal capacity of life-history stages as compared to freshwater species, nevertheless, K. punctatus populations from Japanese waters showed restricted gene flow as reported by Song et al. (2017), although our present investigation supported earlier studies (Song et al., 2017) concerning Chinese populations. It had been suggested that some species’ dispersal strategy is to minimize the offshore transfer of larvae (Song et al., 2010). Nevertheless, K. punctatus has chosen the dispersal strategy to enhance survival and self-recruitment. The larval history has an immense impact on the survival of animals, including marine fish (Hamilton et al., 2008). Ecological strategies of early life history have an impact on phylogeographic patterns. K. punctatus has a long range of distribution areas along the Chinese coast, therefore, it has a high range of gene flow among all populations found along the Chinese coast.
This study enhances the current knowledge of K.punctatus genetic diversity and has shown the rich genetic diversity found in these populations of K. punctatus. It will furnish complementary knowledge for the species’ conservation and management. However, less genetic variation is present among populations. Continued observations would be needed to monitor the Zhongshan population for its diversity and life history characteristics.
Data Availability Statement
All haplotypes were deposited in GenBank, with the accession numbers MT381749 to MT381808 (COI sequences) and MT381809 to MT381847 (Cytb sequences: shorturl.at/GIJS1) (https://www.ncbi.nlm.nih.gov/nuccore/MT381847.1).
Ethics Statement
The animal study was reviewed and approved by Institutional Ethics Committee of Zhejiang Ocean University.
Author Contributions
BL, FM, and ZL conceived and designed the research. All authors conducted the experiments, analyzed data, wrote the manuscript, and critically reviewed and approved the manuscript.
Funding
The study was supported by the National Natural Science Foundation of China (NSFC) (Nos. 41806156 and 31702321), Open Foundation from Key Laboratory of Tropical Marine Bio-resources and Ecology, Chinese Academy of Sciences (LMB20201005), and Science and Technology Project of Zhoushan (2020C21016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00534/full#supplementary-material
Footnotes
References
Avise, J. C., Arnold, J., Ball, R. M., Bermingham, E., Lamb, T., Neigel, J. E., et al. (1987). Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Ann. Rev. Ecol. Syst. 18, 489–522. doi: 10.1146/annurev.es.18.110187.002421
Bandelt, H. J., Forster, P., and Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bohonak, A. (1999). Dispersal, gene flow and population structure. Q. Rev. Biol. 74, 21–45. doi: 10.1086/392950
Casey, J., Jardim, E., and Martinsohn, J. T. (2016). The role of genetics in fisheries management under the E.E. common fisheries policy. J. Fish. Biol. 89, 2755–2767. doi: 10.1111/jfb.13151
De Luca, D., Catanese, G., Procaccini, G., and Fiorito, G. (2014). An integration of historical records and genetic data to the assessment of global distribution and population structure in Octopus vulgaris. Front. Ecol. Evol. 2:55. doi: 10.3389/fevo.2014.00055
Eizirik, E., Bonatto, S. L., Johnson, W. E., Crawshaw, P. G. Jr., Vie, J. C., Brousset, D. M., et al. (1998). Phylogeographic Patterns and Evolution of the Mitochondrial DNA Control. J. Mol. Evol. 47, 613–624. doi: 10.1007/pl00006418
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and Windows. Mol. Ecol. Res. 10, 564–556.
Fadhlaoui-Zid, K., Knottweis, L., Aurelle, D., Nafkha, C., Ezzedine, S., Fiorentino, F., et al. (2012). Genetic structure of Octopus vulgaris (Cephalopoda Octopodidae) in the central Mediterranean Sea inferred from the mitochondrial COIII gene. CR. Biol. 335, 625–636. doi: 10.1016/j.crvi.2012.10.004
Ferna’ndez, M. V., Heras, S., Maltagliati, F., Turco, A., and Rolda’n, M. I. (2011). Genetic structure in the blue and red shrimp, Aristeus antennatus and the role played by present hydrographical and oceanographical barriers. Mar. Ecol.Prog. Ser. 421, 163–171. doi: 10.3354/meps08881
Grant, W. A. S., and Bowen, B. W. (1998). Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J. Hered. 89, 415–426. doi: 10.1093/jhered/89.5.415
Gwak, W. S., Lee, Y. D., and Nakayama, K. (2015). Population structure and sequence divergence in the mitochondrial DNA control region of gizzard shad Konosirus punctatus in Korea and Japan. Ichthyol. Res. 62, 379–385. doi: 10.1007/s10228-014-0450-7
Hamilton, S. L., James, R., and Warner, R. R. (2008). Postsettlement survival linked to larval life in a marine fish. Proc. Natl. Acad. Sci. U.S.A. 105, 1561–1566. doi: 10.1073/PNAS.0707676105
Hata, H. (2017). Konosirus punctatus. The IUCN Red List of Threatened Species. Gland: IUCN, doi: 10.2305/IUCN.UK.2017
Hellberg, M. E., Burton, R. S., Neigel, J. E., and Palumbi, S. R. (2002). Genetic assessment of connectivity among marine populations. Bull. Mar. Sci. 70, 273–290.
Kong, L., Kawasaki, M., Kuroda, K., Kohno, K., and Fujita, K. (2004). Spawning characteristics of the konoshiro gizzard shad in Tokyo and Sagami Bays, central Japan. Fish. Sci. 70, 116–122. doi: 10.1111/j.1444-2906.2003.00779.x
Li, M., Shi, S., Wang, M., Lun, Z., and Yang, T. (2012). Complete mitochondrial genome of the dotted gizzard shad Konosirus punctatus (Teleostei Clupeidae). Mitochondrial DNA 23, 295–297. doi: 10.3109/19401736.2012.674126
Librado, P., and Rozas, J. (2009). DnaSP v5 A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Maltagliati, F., Belcari, P., Casu, D., Casu, M., Sartor, P., Vargiu, G., et al. (2002). Allozyme genetic variability and gene flow in Octopus vulgaris (Cephalopoda, Octopodidae) from the Mediterranean Sea. Bull. Mar. Sci. 71, 473–486.
Melis, R., Vacca, L., Cuccu, D., Mereu, M., Cau, A., Follesa, M. C., et al. (2018). Genetic population structure and phylogeny of the common octopus Octopus vulgaris Cuvier, 1797 in the western Mediterranean Sea through nuclear and mitochondrial markers. Hydrobiologia 807, 277–296. doi: 10.1007/s10750-017-3399-5
Mortiz, C. (1994). Applications of mitochondrial DNA analysis in conservation: a critical review. Mol. Ecol. 3, 401–411. doi: 10.1111/j.1365-294x.1994.tb00080.x
Myoung, S. H., and Kim, J. K. (2014). Genetic diversity and population structure of the gizzard shad, Konosirus punctatus (Clupeidae, Pisces), in Korean waters based on mitochondrial DNA control region sequences. Genes. Genom. 36, 591–598. doi: 10.1007/s13258-014-0197-6
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring harbour Laboratory Press.
Song, N., Gao, T., Ying, Y., Yanagimoto, T., and Han, Z. (2017). Is the Kuroshio Current a strong barrier for the dispersal of the gizzard shad (Konosirus punctatus) in the East China Sea. Mar. Fresh. Res. 68, 810–820.
Song, N., Zhang, X. M., Sun, X. F., Yanagimoto, T., and Gao, T. X. (2010). Population genetic structure and larval dispersal potential of spottedtail goby Synechogobius ommaturus in the north-west Pacific. J. Fish Biol. 77, 388–402. doi: 10.1111/J.1095-8649.2010.02694.X
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Temminck, C. J., and Schlegel, H. (1846). “Pisces,” in Fauna Japonica, sive descriptio animalium, quae in itinere per Japoniam… suscepto annis 1823-1830 collegit, notis, observationibus et adumbrationibus illustravit Ph. Fr. de Siebold. Siebold Fauna Japonica, ed de Siebold, P. F. (Batavia: Lugduni Batavorum), 345.
Vaseeharan, B., Rajakamaran, P., Jayaseelan, D., and Vincent, A. Y. (2013). Molecular markers and their application in genetic diversity of penaeid shrimp. Aquacul. Int. 21, 219–241. doi: 10.1007/s10499-012-9582-9
Waples, R. S. (1998). Separating the wheat from the chaff: patterns of. (genetic)differentiation in high gene flow species. J. Hered 89, 438–450. doi: 10.1093/jhered/89.5.438
Weir, B. S., and Cockerham, C. C. (1996). Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer Assoc. Inc.
Whitehead, P. J. P. (1985). FAO species catalogue. Vo1.7. Clupeoid fishes of the world. An annotated and illustrated catalogue of the herrings, sardines, pilchards, sprats, anchovies and wolfherrings. Part 1 - Chirocentridae,Clupeidae and Pristigasteridae. FAO Fish. Synop. 7:303.
Yamada, U., Tokimura, M., Horikawa, H., and Nakabo, T. (2007). Fishes of the East China Sea and yellow Sea. Hadano: Tokai University Press.
Keywords: Konosirus punctatus, COI, Cytb, population genetic, population diversity
Citation: Liu B, Zhang K, Zhu K, Shafi M, Gong L, Jiang L, Liu L, Muhammad F and Lü Z (2020) Population Genetics of Konosirus punctatus in Chinese Coastal Waters Inferred From Two mtDNA Genes (COI and Cytb). Front. Mar. Sci. 7:534. doi: 10.3389/fmars.2020.00534
Received: 26 February 2020; Accepted: 11 June 2020;
Published: 30 June 2020.
Edited by:
Zhijun Dong, Yantai Institute of Coastal Zone Research (CAS), ChinaReviewed by:
Yongshuang Xiao, Institute of Oceanology (CAS), ChinaTakashi Yanagimoto, National Research Institute of Fisheries Science, Japan
Copyright © 2020 Liu, Zhang, Zhu, Shafi, Gong, Jiang, Liu, Muhammad and Lü. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingjian Liu, YmpldGJqQDE2My5jb20=; Faiz Muhammad, YmFsb3VjaF8yM0B5YWhvby5jb20=; Zhenming Lü, bmJsem1uYkAxNjMuY29t
 Bingjian Liu
Bingjian Liu Kun Zhang1
Kun Zhang1 Kehua Zhu
Kehua Zhu Faiz Muhammad
Faiz Muhammad