- 1Key Laboratory of Marine Hazards Forecasting, Ministry of Natural Resources, Hohai University, Nanjing, China
- 2Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 3Key Laboratory of Sustainable Utilization of Technology Research for Fisheries Resources of Zhejiang Province, Marine Fisheries Research Institute of Zhejiang, Zhoushan, China
Climate-driven shifts of coastal species’ ranges constitute a key factor shaping both the vegetation composition and biodiversity of coastal ecosystems. Although phylogeographic studies relying on genetic data have shed light on the evolutionary history of macroalgae along the Northwest Pacific (NW-Pacific) coast, their distribution dynamics are less understood and require interdisciplinary examination. Here, we used a combination of species distribution models (SDMs) and genetic data to explain the causes of population persistence and genetic differentiation and their consequences for the brown seaweed Sargassum horneri in the NW-Pacific. In this region, the phylogeographic structure of S. horneri was analyzed by screening 72 populations spawning across the entire coastal distribution and obtaining their mtDNA cox3 data. Population genetic structure and SDMs based on paleoclimatic data consistently revealed that southern coasts of the Sea of Japan, North-Pacific-Japan, and the northern part of Okinawa Trough might have served as potential refugia for S. horneri during the Last Glacial Maximum (LGM). Furthermore, we projected the distribution dynamics of S. horneri under future climate scenarios. The range of S. horneri was predicted to move northward, with a significant loss of suitable habitat, under the high emissions scenario (RCP 8.5). By contrast, projected range shifts were minimal under the low emissions scenario (RCP 2.6). Furthermore, North-Pacific-Japan was projected to be long-term persistence habitat for S. horneri under future climatic conditions, thus including this area in conservation planning could help mitigate for climate change implications. Our results enable a better understanding of the impacts of climate change on the spatio-temporal distribution of macroalgae and how this can inform coastal management and marine conservation planning.
Introduction
Understanding the impacts of global warming on seaweed communities is now a central challenge for marine species conservation. Due to anthropogenic emissions of greenhouse gases, the major consequence of climate change is an increase in both atmospheric and sea surface temperatures (SST) (Foo and Byrne, 2016), with SSTs expected to increase by 1.2–3.2°C by the year 2100 (IPCC, 2013). The Northwest Pacific (hereon NW-Pacific) is among the most important and vulnerable marine regions for biodiversity, having lost most of the biomass because of global warming (Zhang et al., 2019; Sudo et al., 2020). Over the last three decades, from the Bering Strait to Southwestern Japan, the coastal SST has risen at a rate of 0.24 ± 0.11°C per decade, while along the coasts of the Chinese marginal seas, the temperature has increased much faster (by 0.5°C per decade) (Lima and Wethey, 2012), suggesting that global warming is far more complex than a simple monotonic increase of average temperatures. Identifying climate refugia and areas that harbor high biodiversity is essential to operationalize and enforce marine conservation under ongoing climate change (Rilov et al., 2019).
Canopy-forming seaweed has experienced noticeable regression because of climatic changes and anthropogenic activities (Casado-Amezúa et al., 2019). Canopy-forming species, such as large brown seaweeds support elevated levels of biodiversity through their complex structure (holdfast and surrounding substratum), thus the regression has negatively affected these associated marine species (Verdura et al., 2018; Sudo et al., 2020). Climate change has also altered the composition of canopy-forming seaweeds, with implications for ecosystem functions and services (Leclerc et al., 2015; Casado-Amezúa et al., 2019). For example, Pessarrodona et al. (2019) investigated how climatic-driven substitutions of two foundation species, Laminaria hyperborea (cold-affinity) and L. ochroleuca (warm-affinity), influenced community, trophic structure and functioning through processes associated with the cycling of organic matter (including biomass production, detritus flow, herbivory, and decomposition).
Distribution patterns of genetic diversity are often related to past demographic processes and range shifts (Dolby et al., 2016; Assis et al., 2018b). In Europe, the predicted south-to-north recolonization-expansions in response to paleoclimatic oscillations were verified in several seaweeds, e.g., Fucus serratus (Hoarau et al., 2007), Palmaria palmata (Li et al., 2015), and Ascophyllum nodosum (Olsen et al., 2010). By contrast, the postglacial expansion of warm-temperate seaweed in the NW-Pacific is affected by both complex coastal topography and ocean currents (Li et al., 2017b). Genetic diversification of marine species in this area is often related to the marginal sea basins (Hu et al., 2015, 2017), in that a changed sea level due to paleoclimatic oscillations has caused marginal seas to split apart (Wang, 1999). When the marginal seas reunited due to postglacial sea-level rise, populations that survived glaciation started to expand outward, with coastal currents (e.g., China Coastal Current and Kuroshio Current) possibly helping to accelerate gene flow between regions. So populations in this region have undergone multiple range contractions and expansions (Boo et al., 2019; Ng et al., 2019). Ancient isolation in separated glacial refugia is a key factor shaping the phylogeographic pattern of marine species in the NW-Pacific (Hu et al., 2017). Such ancient refugia were often rich in unique phylogeographic lineages that are potentially essential for their adaptation to environmental change (Gurgel et al., 2020). Most of these refugial populations, especially those at low latitude ranges, will likely become extinct under varying scenarios of climate change, resulting in the loss of this ancient genetic diversity (Assis et al., 2018a).
Climate refugia could provide comparatively stable environments wherein macroalgae could persist (Ban et al., 2016; Rilov et al., 2019), and these may be identified by species distribution models (SDMs) run under climate extremes, such as the Last Glacial Maximum (LGM, 20 Kya) and the warmer Mid-Holocene (MH, 6 Kya). SDMs are powerful tools for predicting the suitable distribution area of species and to understand how they may respond to climate change (see Renaud et al., 2019). Studies applying marine-based SDMs were predominantly carried out in the temperate North Atlantic (45%) followed by studies done at the global scale (11%) and those focused on temperate Australasia (10%) (reviewed in Robinson et al., 2017). In stark contrast, species’ distribution dynamics and corresponding environmental drivers are far less understood for the NW-Pacific.
Sargassum horneri is a warm-temperate and habitat-forming species found in the coastal ecosystem of the NW-Pacific, where it provides substrate for epiphytic communities and habitat for fish and invertebrates (Komatsu et al., 2014). However, the natural resource of S. horneri is heavily threatened by human activity (e.g., over-harvesting) and climatic changes (e.g., ocean warming). As a prominent species forming the “golden tide” in the NW-Pacific, S. horneri can stay on the sea surface for ca. 1–5 months (Yatsuya, 2008). Ocean currents such as the Kuroshio Current could facilitate the long-distance dispersal of multiple marine species from south to north (Yamasaki et al., 2014; Li et al., 2017a). In particular, non-buoyant seaweed and/or invertebrates could hitchhike on floating rafts formed by positive buoyant seaweeds (e.g., Sargassum, Durvillaea, Macrocystis), to colonize to newly available suitable habitats (Fraser et al., 2013; Boo et al., 2014; Guillemin et al., 2014; van Hees et al., 2019). Thus the abundance and distribution of positive buoyant seaweeds can strongly influence the expansion of coastal species under climate change (Macreadie et al., 2011; van Hees et al., 2019).
Any range shifts of habitat-forming species found would have important consequences for community constitution and ecosystem functioning, with implications for marine management and conservation. In this study, we assessed how past climate changes produced significant biogeographical shifts and predicted the distribution of potential climatic refugia for S. horneri. We further estimated the threat posed on ancient refugia and rear edge populations by future climatic change under two contrasting RCPs (Representative Concentration Pathway scenarios).
Materials and Methods
Phylogeographic Structure of Sargassum horneri
Previous studies have amplified the cox3 sequences for S. horneri populations ranging from Hokkaido, Japan (43°12′N) to Zhanjiang, China (20°53′N), including 54 sites in Japan (Uwai et al., 2009; Watanabe et al., 2019), 14 sites in Korea (Byeon et al., 2019), and 8 sites in China (Hu et al., 2011). In total, the mtDNA cox3 of 996 individuals yielded 62 haplotypes, which were downloaded from NCBI1 (Supplementary Table S1). These sequences were aligned and trimmed using the MEGA v6.06 tool (Tamura et al., 2013). To evaluate the relationships among haplotypes, a median-joining network was built using NETWORK v4.5.1 software (Bandelt et al., 1999). Then we used the jModeltest (Posada and Crandall, 1998) to determine the most appropriate model for sequence evolution; according to the Bayesian Information Criterion (BIC), this was revealed to be Hasegawa-Kishino-Yano with a gamma distribution (HKY + G). We conducted a bootstrapped (1000 replicates) maximum-likelihood (ML) analysis using the PhyML v3.0 program (Guindon et al., 2010) and used the neighbor-joining (NJ) method in MEGA v6.06 (Tamura et al., 2013) to construct the phylogenetic trees. For these, the congeneric species Sargassum muticum (GenBank accession no. KJ938301.1), S. natans (no. KY084907.1), and S. fluitans (no. KY084909.1) were chosen to serve as the out-groups.
Analysis of molecular variance (AMOVA) conducted in Arlequin 3.5 (Excoffier and Lischer, 2010) was used to assess the spatial partitioning of genetic variance among lineages. The software IMa2 (Hey, 2010) was used to estimate divergence time (t) of lineages. The final runs included a total of 1 × 105 genealogies and a burin-in period of 1 × 106 steps. The generation time, g (1 yr) and the mutation rate of cox3 (2.6–4.1%/Myr) (Uwai et al., 2009) were used to convert the output units into years. The departure from population demographic equilibrium was tested using Tajima’s D (Tajima, 1989) and Fu’s FS (Fu, 1997) in Arlequin 3.5 (Excoffier and Lischer, 2010) with 1,000 coalescent simulations. Under the assumption of neutrality, negative values represent population expansions, while positive values are considered a signature of recent bottlenecks (Excoffier and Lischer, 2010). Mismatch distributions implemented in Arlequin 3.5 were also used to test demographic history (Rogers and Harpending, 1992). Recent demographic expansion is characterized by smooth unimodal distribution (Rogers and Harpending, 1992).
Study Area and Occurrence Records
Our study area ranged from the Leizhou Peninsula in Guangzhou, China, to Sakhalin Bay in Russia (20–55°N, 109–150°E), thus covering the entire distribution of S. horneri in the NW-Pacific. We collected presence-only occurrence records of S. horneri from the Global Biodiversity Information Facility (GBIF)2, the AlgaeBase3, the journal articles, and regional experts (Supplementary Table S2). In this way, a total of 171 occurrence records of S. horneri in the NW-Pacific were finally compiled (Figure 1A, Supplementary Table S2). Presence records were geo-referenced onto a map using ArcGIS v10.2, at a resolution of 0.1°; any overlapping records within a cell were only considered once, so as to reduce spatial autocorrelation in the niche modeling.

Figure 1. (A) Occurrence points of brown seaweed Sargassum horneri in the Northwest Pacific. Dash line represent northward-flowing Kuroshio Current. Biogeographic regions around Japanese Seas were defined based on Hu et al. (2011). (B) Median-joining network inferred from mtDNA cox3. The size of each circle is proportional to the frequency of haplotypes, and each line between the main haplotypes represents one mutation step (equal to 1 where unnoted). Species distribution models for Sargassum horneri for (C) the Last Glacial Maximum (LGM) and (D) Mid-Holocene (MH).
Environmental Data Layers
Environment variables were downloaded as raster layers from Bio-ORACLE4 at a 5-arcmin resolution. We selected 13 relevant variables that could affect the distribution of macroalgae in their natural environment (Supplementary Table S3). In particular, extreme environment conditions, such as the long-term average (LTA) of the hottest summer and the coldest winter month for sea surface temperature (SST) and the respective LTA of the maximum (summer) and minimum (winter) month for surface salinity (SSS), were also included. To avoid overfitting the models to the occurrence records, we used the R package ‘MaxentVariableSelection’ (Jueterbock, 2015) to excluding a set of environmental variables with a relative contribution score < 5% or a correlation of > 0.7 with other variables (Supplementary Figures S1, S2, Table S4). Collinearity of the retained environmental variables were assessed with the variance inflation factor (VIF < 10). VIF was assessed using the R package usdm (Naimi et al., 2014).
Environmental variables (temperature and salinity) for the LGM were derived from PaleoMARSPEC data layers (Sbrocco and Barber, 2013). To project these into future climate conditions, we chose two widely recognized representative concentration pathways (RCPs): RCP 2.6 and RCP 8.5, the former being the most optimistic climate change scenario with a predicted CO2 concentration of 421 ppm by 2100, whereas the latter is a high emission “business-as-usual” scenario, with CO2 reaching a concentration of 936 ppm by 2100 (Moss et al., 2010). For present-day time, the greenhouse gas CO2 concentration used was 410 ppm (d.d. May 2018; www.esrl.noaa.gov) (Westmeijer et al., 2019). Apart from temperature, the monthly maximum and minimum average sea surface RCP scenarios also included simulations of salinity changes arising from alterations to the hydrological cycle. Both SST and SSS were each derived for the mid-century (2040–2050) and end-century (2090–2100) in the two RCP scenarios (from Bio-ORACLE).
All the environmental layers were gridded to a 1-arcmin resolution (approximately 0.016° at the equator), by using bilinear interpolation. In all cases, the projected distribution area was restricted in space by a 17.5-km buffer zone from the coastline.
Species Distribution Models (SDMs)
To generate SDMs for S. horneri, we applied Maximum Entropy Modeling in MAXENT v3.3.3 (Phillips et al., 2006) and Genetic Algorithm for Rule-set Prediction (GARP) model in openModeller v1.2 (Muñoz et al., 2011). These algorithms are compatible with the low numbers of available information on the natural occurrence of our study species. We randomly selected 20% of the distribution records for training, reserving the remaining 80% for testing. MAXENT models the environmental conditions in the distributional range of S. horneri according to 10 000 background sites positioned randomly along the coastline. Then MAXENT was run 15 times, and each run was convergent within 1000 iterations. According to the output results, the variables with a relative contribution score less than 5% in the model were excluded. For the GARP model, we ran it with 10 000 replicate enforcing a low omission (E = 5%), and selected 10 best models out 100 runs. Default settings were otherwise used for all other parameters in each model. A jackknife test was used to rank the importance of each environmental variable in both models; the variable with the smallest contribution was dropped, and this process repeated until only one variable remained which we inferred was most important variable for predicting suitable habitat. The final model was run using all occurrence data and selected variables to project the suitable habitat within the study area.
The models’ algorithm performance was evaluated using the predicted area under the curve (AUC), provided by the ROC curve (Receiver Operator Characteristic) (Phillips et al., 2006), ROC-derived sensitivity (presences correctly predicted) and specificity (absences correctly predicted) (Fielding and Bell, 1997), and the true skill statistic (TSS) (Allouche et al., 2006). For AUC, models with value > 0.90 were considered “excellent” and in the range 0.7–0.9 “reasonable predictions”. For TSS, models with value > 0.8 were considered “excellent” and in the range 0.4–0.8 “good” (de la Hoz et al., 2019). MAXENT showed better performance (AUC = 0.97; TSS = 0.60) in the validation than did GARP (AUC = 0.87; TSS = 0.70), so we used MAXENT to generate the distribution models using all presence points of the seaweed species. Then we projected these models onto ancient and future climatic layers. Additionally, model overfitting was calculated as the difference between training and testing AUC, and higher values (AUC Diff.) indicate over-parameterizing (Jiménez-Alfaro et al., 2018).
Distribution maps with threshold values represent habitat suitability (range: 0–1) were built up by MAXENT. To illustrate the changes to suitable habitat in response to climate changes, we compared the future suitable area and southern ranges with its present distribution (Figure 2). The probability grid data were projected in UTM (Universal Transverse Mercator Grid System) to calculate the area of suitable habitat (species’ presence probability > 0.5) location arising under different periods and RCP scenarios.
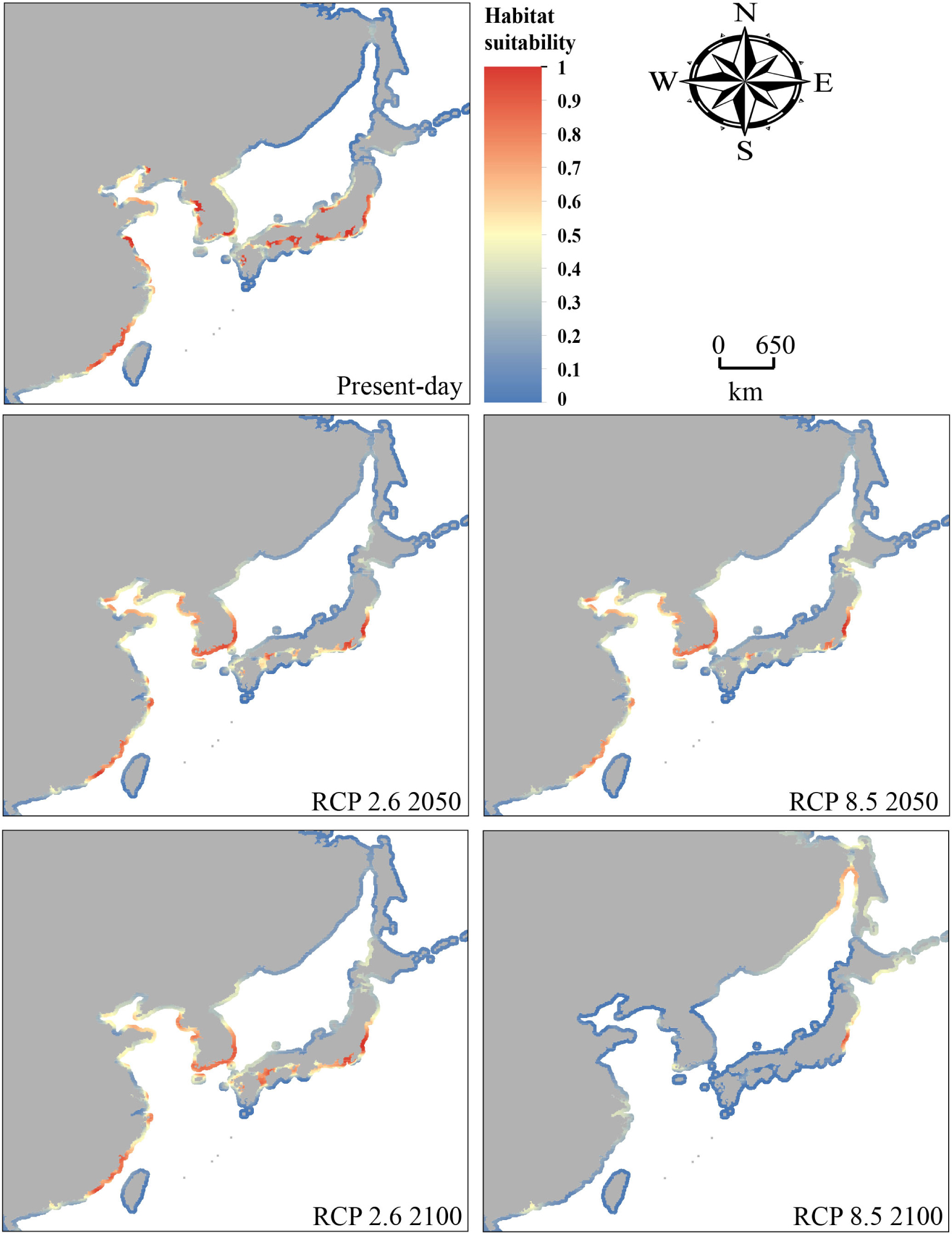
Figure 2. Range projections for Sargassum horneri for present-day and the future (years 2050 and 2100) under different climate scenarios (RCP 2.6 and RCP 8.5).
Results
Genetic Structure
The haplotype network revealed deep genetic splits among populations along the NW-Pacific (Figures 1A,B). The ML and NJ phylogeographic trees separated the 62 haplotypes into three genetic lineages (labeled here I, II, and III), with moderate-to-high support for each node as indicated by their bootstrap values (Supplementary Figure S3). Lineage I dominated the populations from the North-Pacific-Japan; Lineage II dominated coasts of Pacific-Japan and Korea; Lineage III occurred in the populations from South-Pacific-Japan, China, Korea, and Sea of Japan (Supplementary Table S1, Figures 1A,B). In particular, 98.5% samples from South-Pacific-Japan grouped in lineage III, and the rest samples were found in lineage II. Notably, all the haplotypes found in China and the Sea of Japan belonged to lineage III (Figure 1B). For AMOVA analysis, 84.10% of genetic variance occurred between lineages (FST = 0.84170, P < 0.0001; Supplementary Table S5).
Most haplotypes were represented by individuals located in one biogeographic region, apart from the three haplotypes H5, H29, and H50 (Figure 1B, Supplementary Table S1). Of the latter, H5 (JF461023) in lineage III was found in samples from China (=haplotype 3, in Hu et al., 2011) and South-Pacific-Japan (=haplotype D, in Watanabe et al., 2019; Figure 1B, Supplementary Table S1). A haplotype in the center of a star-like topology is often regarded as being an ancient haplotype, and H29 (AB430550) in lineage III fit this description and predominated among the samples from South-Pacific-Japan (=haplotype A, in Watanabe et al., 2019), Korea (=haplotype 1, in Byeon et al., 2019), China and Sea of Japan (=haplotype 7, in Hu et al., 2011; Figure 1B). We found the H50 (AB430580) in lineage II among samples from North-Pacific-Japan (=haplotype 39, in Hu et al., 2011), South-Pacific-Japan (=haplotype 2, in Byeon et al., 2019), and Korea (=haplotype O, in Watanabe et al., 2019; Figure 1B, Supplementary Table S1).
The divergence time between lineage I and II was c. 0.175 Mya (95% HPD 0.067–0.301 Mya) (Supplementary Figure S5), corresponding to the Saale glacial period (0.130–0.191 Mya). The estimated split time between lineage I and III was c. 0.205 Mya (95% HPD 0.035–0.414 Mya), and the divergence time between lineage II and III was c. 0.266 Mya (95% HPD 0.094–0.432 Mya) (Supplementary Figure S5). For lineage I and II, Tajima’s D and Fu’s Fs values were statistically non-significant, and the mismatch distribution of the two lineages showed no signs of recent demographic or range expansion (Supplementary Figure S6). The Tajima’s D and Fu’s Fs values estimated for lineage III were significantly negative (p < 0.05) (Supplementary Figure S6). In addition, unimodal distribution was observed in the mismatch distribution for lineages III (Supplementary Figure S6), indicating a demographic expansion scenario.
Environment Variables and Model Performance
The model with the lowest AIC was built based on the following three uncorrelated environmental variables (Supplementary Figure S1, S2, Table S4): mean primary productivity, LTA of maximum sea surface temperature (MaxSST) and LTA of minimum surface sea salinity (MinSSS) (Table 1). For GARP, variables with an omission error above the 95% basic omission error (=6.33) could change the potential distribution area significantly (Table 1). The ranking of relative contribution of the three variables was the same in both MAXENT and GARP models (Table 1). Mean primary productivity was the most important variable (44.7% model contribution, Table 1) in discriminating suitable from non-suitable habitats. MaxSST had a contribution of 28.2%, and MinSSS had a contribution of 27.1% in Maxent (Table 1). Response curves showed that mean primary productivity was positively correlated with the habitat suitability of S. horneri whereas MinSSS was negatively correlated with it (Supplementary Figure S4). The MaxSST in the range of 24.5°C–28°C was generally predicted as optimal for the species (Supplementary Figure S4).
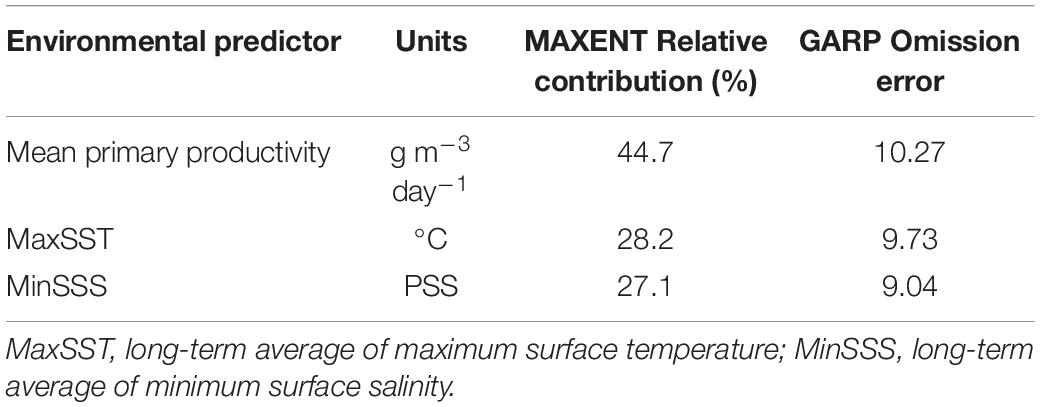
Table 1. Environmental predictors used in species distribution models (MAXENT and GARP) of the seaweed Sargassum horneri.
The final predictive MAXENT model had high performance (AUC = 0.97; TSS = 0.6; Sensitivity = 0.62; Specificity = 0.98). The difference between training and testing AUC (AUC Diff.) was 0.0128 (Supplementary Table S4), without overfitting risk.
Ancient, Present, and Future Projections of Habitat Suitability
When projected back in time, to the conditions of the LGM (20 Kya), both models indicated the southern Sea of Japan were potential refugia for S. horneri (Figure 1C). In addition, the northern Okinawa Trough and North-Pacific-Japan were suboptimal (with probability of 0.5). During the MH (6 Kya) period, S. horneri began to expand outside refugia as sea levels rose, moving westward to the Bohai Bay and northward to the coasts of Hokkaido, Japan, the Russian coast of the Japan Sea, the Sakhalin Bay and also around the Kuril Islands (Figure 1D).
The present-day model’s results were consistent with the species’ current distribution (Figure 2). These projections showed that habitat suitability was high along the coast of East China Sea and Bohai Bay, the western and southern coasts of the Korean Peninsula, the western part of Kyushu Island and the coasts of Honshu Island (Figure 2). Compared with the Pacific coasts of Japan, a lower coverage of suitable area was detected along the Sea of Japan. Since we did not include seashore substrata in the seaweed’s niche modeling, some of the predicted suitable habitat was inconsistent with occurrence records reported for some areas. For example, in our modeling, high habitat suitability was detected along the coasts of Jiangsu Province of China (30°–35° N), but this area lacks the rocky substrata that are essential for macroalgae establishment and growth. At the present-day time, S. horneri was projected to have 16.75% suitable habitat (Figure 3). During the LGM, the suitable habitat was contracted to 15.17% of studied area (Figure 3), ranging from c. 32°–41° N (Figure 4).
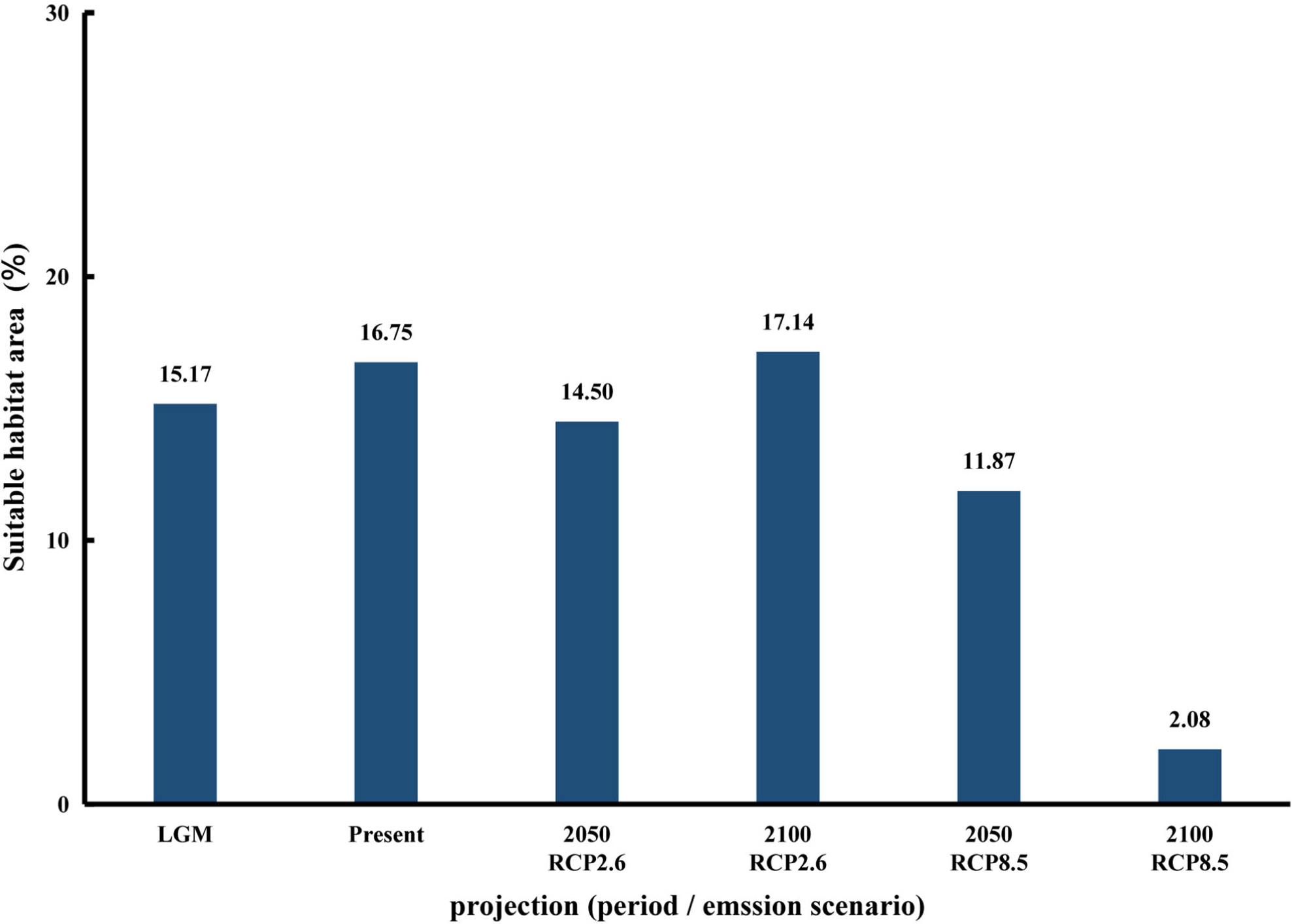
Figure 3. Changes in the suitable distribution area (%) of Sargassum horneri under the Last Glacial Maximum (LGM), present-day and future climate scenarios (RCP 2.6 and RCP 8.5).
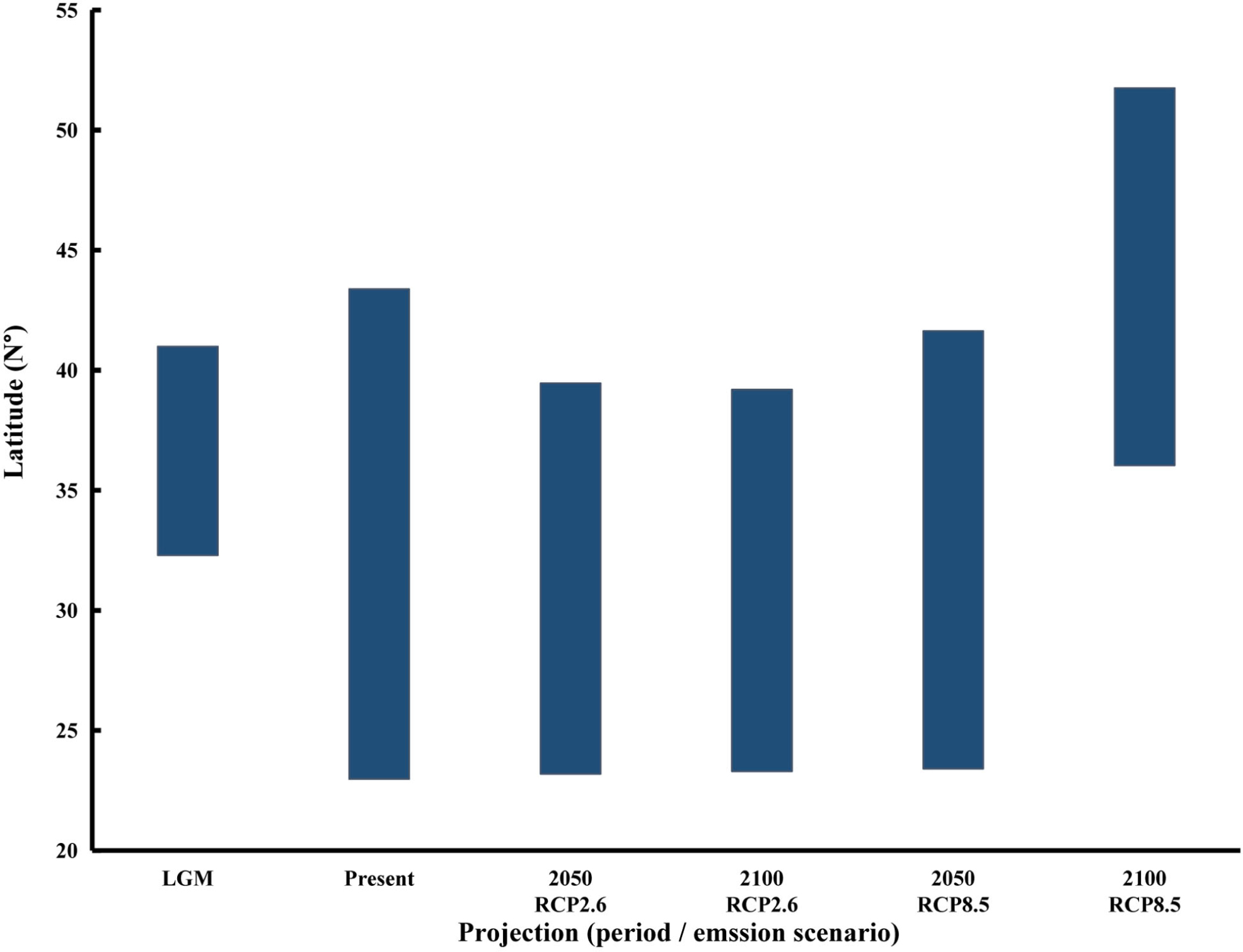
Figure 4. Changes in the distribution boundaries of Sargassum horneri under the Last Glacial Maximum (LGM), present-day and future climate scenarios (RCP 2.6 and RCP 8.5).
By 2050, under the stabilizing scenario RCP 2.6, S. horneri will lose 13.43% of its current suitable habitat, but by 2100, suitable area will arise to 17.14% (Figures 2, 3). Notably, the suitable habitat of S. horneri increased along the coasts of Korea under RCP 2.6 (Figure 2), whereas that sustained at its southern boundaries changed little (Figures 3, 4). Under RCP 8.5, the more severe climate change scenario, by 2050 the highest habitat suitability was found along the coasts of Central-Pacific-Japan, and the southern and eastern coasts of Korea (Figure 2). Projections also indicated that parts of the coasts of China had suitable habitat for S. horneri. By 2100, however, S. horneri populations should be able to colonize new suitable habitat available in the north yet they will lose all their suitable habitat along the Chinese and Korean coasts, with only the coasts of Central-Pacific-Japan and northern Sea of Japan having the highest habitat suitability (Figure 2). The suitable habitat of S. horneri was contracted to11.87% and 2.08% of studied area by 2050 and 2100, respectively (Figure 3). The northward shift of southern boundaries was predicted to be ca. 1581.89 km (ca. 14.26°) by 2100 (Figure 4).
Discussion
Sargassum horneri, a warm-temperate brown algal species, forms ecosystem-structuring underwater forests. Genetic divergence was detected in S. horneri populations along the coasts of NW-Pacific, suggesting multiple ancient refugia (e.g., North-Pacific-Japan, South Sea of Japan, and northern Okinawa Trough). These refugia were supported by ancient distribution of S. horneri during the LGM. Furthermore, the loss of habitat at the southern boundaries was predicted by 2100 under high CO2 emission scenario (RCP 8.5).
Ancient Distribution and Lineage Divergence of the Seaweed
Within a species, lineage divergence reflects the occurrence of an allopatric distribution (Ni et al., 2014), and paleoclimatic oscillations are regarded as being one of the most crucial factors shaping the phylogeograhic structure of coastal species (Zhang et al., 2019). Highly structured phylogeographic patterns were detected in most marine species that occur along the NW-Pacific coasts, such as the seaweed Saccharina japonica (Zhang et al., 2019), Chondrus ocellatus (Hu et al., 2015), the yellowfin goby Acanthogobius flavimanus (Hirase et al., 2020), and the barnacle Chthamalus moro (Wu et al., 2015). Sargassum horneri populations in the NW-Pacific were also characterized by strong genetic variation among regions (Hu et al., 2011). In our study, the divergence time between lineages ranged from 0.175 Mya to 0.266 Mya, much earlier than the LGM (0.018–0.020 Mya).
Populations from North- and Central-Pacific-Japan grouped in lineage I and II, leading to a deep genetic split from other populations (Figure 1). Hu et al. (2011) presumed the existence of the northern cryptic refugia and our projections for the LGM support this view, in that North-Pacific-Japan (ca. 35–40° N) may have served as a glacial refugium for S. horneri. Similarly, an ecological niche model also predicted that another seaweed, S. japonica, survived the LGM in this same area (Zhang et al., 2019). Lineage II also occurred along the coasts of Korea. The Kuroshio Current, bifurcating at the southwestern Japan, acts as a hydrological barrier between populations in eastern Korea and Pacific coastline of Japan; thus, the occurrence of lineage II in both regions likely resulted from a much earlier mixture (e.g., pre-LGM) rather than from recent genetic exchange.
Based on our projections, the southern Sea of Japan represented a suitable distribution area during the LGM (20 Kya). In addition, S. horneri in South-Pacific-Japan and Korea was highly structured based on cox3 sequences, implying long-term persistence in this region. During the LGM, the SST was about 3°C–5°C lower than it currently is in Japanese waters (Oba et al., 1991). Since S. horneri can grow rapidly under low temperatures (10°C –15°C) (Yoshida et al., 1998, 2004), the lower SST in the past might have only shortened its growth period or slowed its growth rate instead of causing its large-scale extinction. Although the northern Okinawa Trough was projected to be low-to-moderate suitable area for S. horneri (Figure 1C), we assumed a high probability of ancient populations once existing in this area. During the LGM, the Yellow-Bohai Sea and East China Sea were reduced to Okinawa Trough, and Seto Inland Sea was terrestrial (Wang, 1999). The private haplotypes along the Chinese coasts are strongly indicative of refugia in the Okinawa Trough, as the elapsed years since the LGM (ca. 20 Kya) are too short for the formation of such a large amount of genetic variation. Furthermore, it is evident that the populations in lineage III have experienced a population expansion (Supplementary Figure S6). In line with our interpretation, Cheang et al. (2008) inferred that the “Paleo-coast” of the East China Sea could have served as glacial refugia for a congener species Sargassum hemiphyllum, leading to the vicariance of its ancestral populations.
Range Shifts and Environmental Drivers
Small environmental changes can be magnified at the rear edge of species’ range shift, which could adversely affect their distribution and diversity (Assis et al., 2016). In our study, the projected losses of suitable habitats in southern boundaries (namely South-Pacific-Japan and the East China Sea) for S. horneri were detected under RCP8.5. Similarly, southern boundaries of S. muticum were predicted to shift northward, from the coast of China and Korea to the Russian coast of the Sea of Japan, the Sakhalin Bay, and around the Kuril Islands (Chefaoui et al., 2019). Further, Westmeijer et al. (2019) estimated that some macroalgae species (Fucus serratus, Laminaria digitata, Gracilaria gracilis, Furcellaria lumbricalis, etc.) would shift 110 to 635 km from the coast of the Iberian Peninsula toward northern Brittany by 2100. The dispersal and distribution patterns of marine species are also driven by ocean currents (Li et al., 2017a). Future climate-driven oceanographic changes are likely to strengthen the Kuroshio Current, which may increase the rates of northward expansion of marine species in the NW-Pacific (Wilson et al., 2016). Non-etheless, by 2100, the forecasted suitable areas of S. horneri differed significantly between the two carbon emission scenarios. Hence, plans for mitigating CO2 emissions could be adopted to prevent the local extirpations and range shifts of this and similar species due to ocean warming.
Our findings showed that primary productivity was the most important driver shaping the distribution of S. horneri. Primary productivity, itself closely associated with nutrient concentrations, available light, and water transparency, could reliably discriminate suitable from non-suitable habitat for S. horneri populations. In particular, the concentration of nutrients seems to have been a relevant factor driving the population dynamics of S. horneri (Yoshida et al., 2001; Qi et al., 2017). Response curves showed that mean primary productivity was positively correlated with the habitat suitability of S. horneri. This is consistent with primary productivity projected as being a major variable shaping the current distribution of seaweeds Undaria pinnatifida (Báez et al., 2010), Macrocystis and Durvillaea (Bernardes Batista et al., 2018).
Extreme temperatures are likely to impact the survival of a critical phase in the life-cycle of marine species, which is more important for determining their distribution range (Martínez et al., 2018; Figueroa et al., 2019). Summer and winter SST variables are often related to shifts of southern and northern range boundaries, respectively (Neiva et al., 2015). In our study, summer temperature (MaxSST) contributed more than did winter temperature (MinSST) when projecting the distribution pattern of S. horneri. Most temperate seaweeds at the rear edge often suffer from summer isotherms above their maximum thermal thresholds (Eggert, 2012), causing their local extirpations. Summer temperature was deemed the dominant factor affecting the projected distribution boundaries of most canopy-forming seaweed in the Northwest Atlantic (Wilson et al., 2019), northern Japan (Sudo et al., 2020), and Australia (Martínez et al., 2018). The thermal thresholds of species are related to their life stages, thus integrating phenological trends should improve our ability to predict future distributional shifts at both regional and global scales. For example, Chefaoui and Serrao (2017) demonstrated that the total range area of S. muticum was expected to increase by just 1.63% when the reproductive temperature window was explicitly considered in model projections.
Seasonal salinity variation was also detected along the coasts of China due to the changes in oceanic water budget and circulation caused by river discharge (Yellow River and Yangtze River). For S. horneri, its sporophytes recruit in winter and grow rapidly from November to March (Xu et al., 2016; Zhang et al., 2018). MinSSS was projected to be a critical driver determining the distribution range of this species, perhaps by influencing its embryos development and germlings’ growth. In two earlier studies, the adults of S. muticum tolerated a wider range of salinity than did its germlings (Steen, 2004), which might also apply to S. horneri. This low salinity also affects the survival and growth of S. hemiphyllum, and it neither found in the Bohai Sea nor the coast north of the Yangtze River (Cheang et al., 2008).
Consequences of Range Shifts and Implications
Previous studies have noticed that canopy-forming marine species are suffering particularly strong declines worldwide (Assis et al., 2017; Wilson et al., 2019). According to work by Krumhansl et al. (2014), 38% of the world’s kelp forests have vanished in the past five decades. Sargassum horneri is one of the most important habitat-forming species in the NW-Pacific, where it annually forms large-scale floating mats (Li et al., 2020). These floating islands of biomass accumulate at the continental shelf and the Kuroshio Front, providing both shelter and food for juveniles of many fish and invertebrates (Komatsu et al., 2014; Yamasaki et al., 2014). Based on satellite data, large patches of floating S. horneri were first found near Zhejiang Province waters, and then transported northeast (Komatsu et al., 2007; Qi et al., 2017). Due to their role in supporting biodiversity and food webs, the changes in the distribution range of S. horneri may substantially impact the species diversity of coastal and pelagic ecosystems (Yamasaki et al., 2014).
Our study combined species distribution model and genetic analyses to give insight into impacts of climate change on the spatio-temporal distribution of S. horneri in the NW-Pacific. Based on our projections, the North-Pacific-Japan region may be a suitable habitat for S. horneri under ongoing global warming, which is also supported by projections done for S. japonica (Zhang et al., 2019). The cold Oyashio current in this region may mitigate the rate of ocean warming and facilitate the growth of cold-water adapted species kelps (Saccharina, Alaria, Costaria) (Sudo et al., 2020) and warm-temperate species, such as Undaria pinnatifida (Sato et al., 2016) and Sargassum thunbergii (Li et al., 2017a). Under the RCP 8.5 scenario, our projections demonstrated a significant range contraction of S. horneri with the loss of its southern limits. Rear-edge populations may embody unique adaptive strategies, which could maintain population stability so these populations should be of special interest to conservation planners (Rilov et al., 2019).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
J-JL conceived the study and coordinated all steps in finalizing the manuscript. J-JL and S-HH carried out the data analysis and wrote the manuscript. Z-YL provided guidance on the modeling. Z-YL and Y-XB interpreted the results and contributed to the writing. All the authors contributed to both the data collection and the manuscript’s revision and also contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (grant number 31700327), the Natural Science Foundation of Jiangsu Province (grant number BK20170863), and the Fundamental Research Funds for the Central Universities (grant number B200202140).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Zimin Hu, Xiangzhou Song, and Ke Ma for providing the background information supporting this study. The English language of this manuscript was edited by “The Charlesworth Group.”
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.570881/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://data.gbif.org
- ^ https://www.algaebase.org/
- ^ http://www.bio-oracle.org/
References
Allouche, O., Tsoar, A., and Kadmon, R. (2006). Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232. doi: 10.1111/j.1365-2664.2006.01214.x
Assis, J., Araujo, M. B., and Serrao, E. A. (2018a). Projected climate changes threaten ancient refugia of kelp forests in the North Atlantic. Glob. Change Biol. 24, E55–E66. doi: 10.1111/gcb.13818
Assis, J., Berecibar, E., Claro, B., Alberto, F., Reed, D., Raimondi, P., et al. (2017). Major shifts at the range edge of marine forests: the combined effects of climate changes and limited dispersal. Sci. Rep. 7:44348. doi: 10.1038/srep44348
Assis, J., Lucas, A. V., Barbara, I., and Serrao, E. A. (2016). Future climate change is predicted to shift long-term persistence zones in the cold-temperate kelp Laminaria hyperborea. Mar. Environ. Res. 113, 174–182. doi: 10.1016/j.marenvres.2015.11.005
Assis, J., Serrao, E. A., Coelho, N. C., Tempera, F., Valero, M., and Alberto, F. (2018b). Past climate changes and strong oceanographic barriers structured low-latitude genetic relics for the golden kelp Laminaria ochroleuca. J. Biogeogr. 45, 2326–2336. doi: 10.1111/jbi.13425
Báez, J. C., Olivero, J., Peteiro, C., Ferri-Yáñez, F., Garcia-Soto, C., and Real, R. (2010). Macro-environmental modelling of the current distribution of Undaria pinnatifida (Laminariales, Ochrophyta) in northern Iberia. Biol. Invasions 12, 2131–2139. doi: 10.1007/s10530-009-9614-1
Ban, S. S., Alidina, H. M., Okey, T. A., Gregg, R. M., and Ban, N. C. (2016). Identifying potential marine climate change refugia: a case study in Canada’s Pacific marine ecosystems. Glob. Ecol. Conserv. 8, 41–54. doi: 10.1016/j.gecco.2016.07.004
Bandelt, H.-J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bernardes Batista, M., Batista Anderson, A., Franzan Sanches, P., Simionatto Polito, P., Lima Silveira, T. C., Velez-Rubio, G. M., et al. (2018). Kelps’ long-distance dispersal: role of ecological/oceanographic processes and implications to marine forest conservation. Diversity 10:11. doi: 10.3390/d10010011
Boo, G., Mansilla, A., Nelson, W., Bellgrove, A., and Boo, S. (2014). Genetic connectivity between trans-oceanic populations of Capreolia implexa (Gelidiales, Rhodophyta) in cool temperate waters of Australasia and Chile. Aquat. Biol. 119, 73–79. doi: 10.1016/j.aquabot.2014.08.004
Boo, G. H., Qiu, Y. X., Kim, J. Y., Ang, P. O., Bosch, S., De Clerck, O., et al. (2019). Contrasting patterns of genetic structure and phylogeography in the marine agarophytes Gelidiophycus divaricatus and G. freshwateri (Gelidiales, Rhodophyta) from East Asia. J. Phycol. 55, 1319–1334. doi: 10.1111/jpy.12910
Byeon, S. Y., Oh, H.-J., Kim, S., Yun, S. H., Kang, J. H., Park, S. R., et al. (2019). The origin and population genetic structure of the ‘golden tide’ seaweeds, Sargassum horneri, in Korean waters. Sci. Rep. 9, 7757–7757. doi: 10.1038/s41598-019-44170-x
Casado-Amezúa, P., Araújo, R., Bárbara, I., Bermejo, R., Borja, Á, Díez, I., et al. (2019). Distributional shifts of canopy-forming seaweeds from the Atlantic coast of Southern Europe. Biodivers. Conserv. 28, 1151–1172. doi: 10.1007/s10531-019-01716-9
Cheang, C. C., Chu, K. H., and Ang, P. O. (2008). Morphological and genetic variation in the populations of Sargassum hemiphyllum (Phaeophyceae) in the northwestern Pacific. J. Phycol. 44, 855–865. doi: 10.1111/j.1529-8817.2008.00532.x
Chefaoui, R. M., Serebryakova, A., Engelen, A. H., Viard, F., and Serrão, E. A. (2019). Integrating reproductive phenology in ecological niche models changed the predicted future ranges of a marine invader. Divers. Distrib. 25, 688–700. doi: 10.1111/ddi.12910
Chefaoui, R. M., and Serrao, E. A. (2017). Accounting for uncertainty in predictions of a marine species: integrating population genetics to verify past distributions. Ecol. Model. 359, 229–239. doi: 10.1016/j.ecolmodel.2017.06.006
de la Hoz, C. F., Ramos, E., Puente, A., and Juanes, J. A. (2019). Climate change induced range shifts in seaweeds distributions in Europe. Mar. Environ. Res. 148, 1–11. doi: 10.1016/j.marenvres.2019.04.012
Dolby, G. A., Hechinger, R., Ellingson, R. A., Findley, L. T., Lorda, J., and Jacobs, D. K. (2016). Sea-level driven glacial-age refugia and post-glacial mixing on subtropical coasts, a palaeohabitat and genetic study. P. Roy. Soc. B Biol. Sci. 283, 1–8. doi: 10.1098/rspb.2016.1571
Eggert, A. (2012). “Seaweed responses to temperature,” in Seaweed Biology, eds C. Wiencke and K. Bischof (Berlin: Springer), 47–66. doi: 10.1007/978-3-642-28451-9_3
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fielding, A. H., and Bell, J. F. (1997). A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49. doi: 10.1017/S0376892997000088
Figueroa, F. L., Celis-Plá, P. S. M., Martínez, B., Korbee, N., Trilla, A., and Arenas, F. (2019). Yield losses and electron transport rate as indicators of thermal stress in Fucus serratus (Ochrophyta). Algal Res. 41:101560. doi: 10.1016/j.algal.2019.101560
Foo, S. A., and Byrne, M. (2016). Acclimatization and adaptive capacity of marine species in a Changing Ocean. Adv. Mar. Biol. 74, 69–116. doi: 10.1016/bs.amb.2016.06.001
Fraser, C. I., Zuccarello, G. C., Spencer, H. G., Salvatore, L. C., Garcia, G. R., and Waters, J. M. (2013). Genetic affinities between trans-oceanic populations of non-buoyant macroalgae in the high latitudes of the Southern Hemisphere. PLoS One 8:e0069138. doi: 10.1371/journal.pone.0069138
Fu, Y.-X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925.
Guillemin, M. L., Valero, M., Faugeron, S., Nelson, W., and Destombe, C. (2014). Tracing the trans-Pacific evolutionary history of a domesticated seaweed (Gracilaria chilensis) with archaeological and genetic data. PLoS One 9:e0114039. doi: 10.1371/journal.pone.0114039
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gurgel, C. F. D., Camacho, O., Minne, A. J. P., Wernberg, T., and Coleman, M. A. (2020). Marine heatwave drives cryptic loss of genetic diversity in underwater forests 2. Curr. Biol. 30, 1199.e2–1206.e2. doi: 10.1016/j.cub.2020.01.051
Hey, J. (2010). The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Mol. Biol. Evol. 27, 921–933. doi: 10.1093/molbev/msp298
Hirase, S., Tezuka, A., Nagano, A. J., Kikuchi, K., and Iwasaki, W. (2020). Genetic isolation by distance in the yellowfin goby populations revealed by RAD sequencing. Ichthyol. Res. 67, 98–104. doi: 10.1007/s10228-019-00709-6
Hoarau, G., Coyer, J. A., Veldsink, J. H., Stam, W. T., and Olsen, J. L. (2007). Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol. Ecol. 16, 3606–3616. doi: 10.1111/j.1365-294X.2007.03408.x
Hu, Z. M., Li, J. J., Sun, Z. M., Gao, X., Yao, J. T., Choi, H. G., et al. (2017). Hidden diversity and phylogeographic history provide conservation insights for the edible seaweed Sargassum fusiforme in the Northwest Pacific. Evol. Appl. 10, 366–378. doi: 10.1111/eva.12455
Hu, Z. M., Li, J. J., Sun, Z. M., Oak, J. H., Zhang, J., Fresia, P., et al. (2015). Phylogeographic structure and deep lineage diversification of the red alga Chondrus ocellatus Holmes in the Northwest Pacific. Mol. Ecol. 24, 5020–5033. doi: 10.1111/mec.13367
Hu, Z. M., Uwai, S., Yu, S. H., Komatsu, T., Ajisaka, T., and Duan, D. L. (2011). Phylogeographic heterogeneity of the brown macroalga Sargassum horneri (Fucaceae) in the northwestern Pacific in relation to late Pleistocene glaciation and tectonic configurations. Mol. Ecol. 20, 3894–3909. doi: 10.1111/j.1365-294X.2011.05220.x
IPCC (2013). Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. New York, NY: Cambridge University Press.
Jiménez-Alfaro, B., Suarez-Seoane, S., Chytry, M., Hennekens, S. M., Willner, W., Hajek, M., et al. (2018). Modelling the distribution and compositional variation of plant communities at the continental scale. Divers. Distrib. 24, 978–990. doi: 10.1111/ddi.12736
Jueterbock, A. (2015). R Package MaxentVariableSelection: Selecting the Best Set of Relevant Environmental Variables along with the Optimal Regularization Multiplier for Maxent Niche Modeling. Available online at: https://cran.r-project.org/web/packages/MaxentVariableSelection/index.html (accessed February 3, 2020).
Komatsu, T., Fukuda, M., Mikami, A., Mizuno, S., Kantachumpoo, A., Tanoue, H., et al. (2014). Possible change in distribution of seaweed, Sargassum horneri, in northeast Asia under A2 scenario of global warming and consequent effect on some fish. Mar. Pollut. Bull. 85, 317–324. doi: 10.1016/j.marpolbul.2014.04.032
Komatsu, T., Tatsukawa, K., Filippi, J. B., Sagawa, T., Matsunaga, D., Mikami, A., et al. (2007). Distribution of drifting seaweeds in eastern East China Sea. J. Mar. Syst. 67, 245–252. doi: 10.1016/j.jmarsys.2006.05.018
Krumhansl, K. A., Lauzon-Guay, J.-S., and Scheibling, R. E. (2014). Modeling effects of climate change and phase shifts on detrital production of a kelp bed. Ecology 95, 763–774. doi: 10.1890/13-0228.1
Leclerc, J.-C., Riera, P., Laurans, M., Leroux, C., Leveque, L., and Davoult, D. (2015). Community, trophic structure and functioning in two contrasting Laminaria hyperborea forests. Estuar. Coast. Shelf Sci. 152, 11–22. doi: 10.1016/j.ecss.2014.11.005
Li, J. J., Hu, Z. M., and Duan, D. L. (2015). Genetic data from the red alga Palmaria palmata reveal a mid-Pleistocene deep genetic split in the North Atlantic. J. Biogeogr. 42, 902–913. doi: 10.1111/jbi.12464
Li, J. J., Hu, Z. M., Gao, X., Sun, Z. M., Choi, H. G., Duan, D. L., et al. (2017a). Oceanic currents drove population genetic connectivity of the brown alga Sargassum thunbergii in the north-west Pacific. J. Biogeogr. 44, 230–242. doi: 10.1111/jbi.12856
Li, J. J., Hu, Z. M., Sun, Z. M., Yao, J. T., Liu, F. L., Fresia, P., et al. (2017b). Historical isolation and contemporary gene flow drive population diversity of the brown alga Sargassum thunbergii along the coast of China. BMC Evol. Biol. 17:10. doi: 10.1186/s12862-017-1089-6
Li, J.-J., Liu, Z.-Y., Zhong, Z.-H., Zhuang, L.-C., Bi, Y.-X., and Qin, S. (2020). Limited genetic connectivity among Sargassum horneri (Phaeophyta) populations in the Chinese marginal seas despite their high dispersal capacity. J. Phycol. 56, 994–1005. doi: 10.1111/JPY.12990
Lima, F. P., and Wethey, D. S. (2012). Three decades of high-resolution coastal sea surface temperatures reveal more than warming. Nat. Commun. 3:704. doi: 10.1038/ncomms1713
Macreadie, P. I., Bishop, M. J., and Booth, D. J. (2011). Implications of climate change for macrophytic rafts and their hitchhikers. Mar. Ecol. Prog. Ser. 443, 285–292. doi: 10.3354/meps09529
Martínez, B., Radford, B., Thomsen, M. S., Connell, S. D., Carreno, F., Bradshaw, C. J. A., et al. (2018). Distribution models predict large contractions of habitat-forming seaweeds in response to ocean warming. Divers. Distrib. 24, 1350–1366. doi: 10.1111/ddi.12767
Moss, R. H., Edmonds, J. A., Hibbard, K. A., Manning, M. R., Rose, S. K., van Vuuren, D. P., et al. (2010). The next generation of scenarios for climate change research and assessment. Nature 463, 747–756. doi: 10.1038/nature08823
Muñoz, M. E., de Giovanni, R., de Siqueira, M. F., Sutton, T., Brewer, P., Pereira, R. S., et al. (2011). openModeller: a generic approach to species’ potential distribution modelling. Geoinformatica 15, 111–135. doi: 10.1007/s10707-009-0090-7
Naimi, B., Hamm, N. A. S., Groen, T. A., Skidmore, A. K., and Toxopeus, A. G. (2014). Where is positional uncertainty a problem for species distribution modelling? Ecography 37, 191–203. doi: 10.1111/j.1600-0587.2013.00205.x
Neiva, J., Assis, J., Coelho, N. C., Fernandes, F., Pearson, G. A., and Serrao, E. A. (2015). Genes left behind: climate change threatens cryptic genetic diversity in the canopy-forming seaweed Bifurcaria bifurcata. PLoS One 10:e0131530. doi: 10.1371/journal.pone.0131530
Ng, P.-K., Chiou, Y.-S., Liu, L.-C., Sun, Z., Shimabukuro, H., and Lin, S.-M. (2019). Phylogeography and genetic connectivity of the marine macro-alga Sargassum ilicifolium (Phaeophyceae, Ochrophyta) in the northwestern Pacific. J. Phycol. 55, 7–24. doi: 10.1111/jpy.12806
Ni, G., Li, Q., Kong, L. F., and Yu, H. (2014). Comparative phylogeography in marginal seas of the northwestern Pacific. Mol. Ecol. 23, 534–548. doi: 10.1111/mec.12620
Oba, T., Kato, M., Kitazato, H., Koizumi, I., Omura, A., Sakai, T., et al. (1991). Paleoenvironmental changes in the Japan Sea during the last 85,000 years. Paleoceanogr. Paleocl. 6, 499–518. doi: 10.1029/91PA00560
Olsen, J. L., Zechman, F. W., Hoarau, G., Coyer, J. A., Stam, W. T., Valero, M., et al. (2010). The phylogeographic architecture of the fucoid seaweed Ascophyllum nodosum: an intertidal ‘marine tree’ and survivor of more than one glacial-interglacial cycle. J. Biogeogr. 37, 842–856. doi: 10.1111/j.1365-2699.2009.02262.x
Pessarrodona, A., Foggo, A., and Smale, D. A. (2019). Can ecosystem functioning be maintained despite climate-driven shifts in species composition? Insights from novel marine forests. J. Ecol. 107, 91–104. doi: 10.1111/1365-2745.13053
Phillips, S. J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Posada, D., and Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Qi, L., Hu, C., Wang, M., Shang, S., and Wilson, C. (2017). Floating algae blooms in the East China Sea. Geophys. Res. Lett. 44, 11501–11509. doi: 10.1002/2017gl075525
Renaud, P. E., Wallhead, P., Kotta, J., Wtodarska-Kowalczuk, M., Bellerby, R. G. J., Ratsep, M., et al. (2019). Arctic sensitivity? Suitable habitat for benthic taxa is surprisingly robust to climate change. Front. Mar. Sci. 6:538. doi: 10.3389/fmars.2019.00538
Rilov, G., Mazaris, A. D., Stelzenmueller, V., Helmuth, B., Wahl, M., Guy-Haim, T., et al. (2019). Adaptive marine conservation planning in the face of climate change: what can we learn from physiological, ecological and genetic studies? Glob. Ecol. Conserv. 17:e00566. doi: 10.1016/j.gecco.2019.e00566
Robinson, N. M., Nelson, W. A., Costello, M. J., Sutherland, J. E., and Lundquist, C. J. (2017). A systematic review of marine-based species distribution models (SDMs) with recommendations for best practice. Front. Mar. Sci. 4:421. doi: 10.3389/fmars.2017.00421
Rogers, A. R., and Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569.
Sato, Y., Hirano, T., Niwa, K., Suzuki, T., Fukunishi, N., Abe, T., et al. (2016). Phenotypic differentiation in the morphology and nutrient uptake kinetics among Undaria pinnatifida cultivated at six sites in Japan. J. Appl. Phycol. 28, 3447–3458. doi: 10.1007/s10811-016-0857-99
Sbrocco, E. J., and Barber, P. H. (2013). Marspec: ocean climate layers for marine spatial ecology. Ecology 94, 979–979. doi: 10.1890/12-1358.1
Steen, H. (2004). Effects of reduced salinity on reproduction and germling development in Sargassum muticum (Phaeophyceae, Fucales). Eur. J. Phycol. 39, 293–299. doi: 10.1080/09670260410001712581
Sudo, K., Watanabe, K., Yotsukura, N., and Nakaoka, M. (2020). Predictions of kelp distribution shifts along the northern coast of Japan. Ecol. Res. 35, 47–60. doi: 10.1111/1440-1703.12053
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Uwai, S., Kogame, K., Yoshida, G., Kawai, H., and Ajisaka, T. (2009). Geographical genetic structure and phylogeography of the Sargassum horneri/filicinum complex in Japan, based on the mitochondrial cox3 haplotype. Mar. Biol. 156, 901–911. doi: 10.1007/s00227-009-1136-y
van Hees, D. H., Olsen, Y. S., Mattio, L., Ruiz-Montoya, L., Wernberg, T., and Kendrick, G. A. (2019). Cast adrift: physiology and dispersal of benthic Sargassum spinuligerum in surface rafts. Limnol. Oceanogr. 64, 526–540. doi: 10.1002/lno.11057
Verdura, J., Sales, M., Ballesteros, E., Cefali, M. E., and Cebrian, E. (2018). Restoration of a canopy-forming alga based on recruitment enhancement: methods and long-term success assessment. Front. Plant Sci. 9:12. doi: 10.3389/fpls.2018.01832
Wang, P. X. (1999). Response of western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar. Geol. 156, 5–39. doi: 10.1016/s0025-3227(98)00172-8
Watanabe, K., Homma, Y., Karakisawa, H., Ishikawa, R., and Uwai, S. (2019). Haplotypic differentiation between seasonal populations of Sargassum horneri (Fucales, Phaeophyceae) in Japan. Phycol. Res. 67, 59–64. doi: 10.1111/pre.12355
Westmeijer, G., Everaert, G., Pirlet, H., De Clerck, O., and Vandegehuchte, M. B. (2019). Mechanistic niche modelling to identify favorable growth sites of temperate macroalgae. Algal Res. 41:101529. doi: 10.1016/j.algal.2019.101529
Wilson, L. J., Fulton, C. J., Hogg, A. M., Joyce, K. E., Radford, B. T. M., and Fraser, C. I. (2016). Climate-driven changes to ocean circulation and their inferred impacts on marine dispersal patterns. Global Ecol. Biogeogr. 25, 923–939. doi: 10.1111/geb.12456
Wilson, K. L., Skinner, M. A., and Lotze, H. K. (2019). Projected 21st-century distribution of canopy-forming seaweeds in the Northwest Atlantic with climate change. Divers. Distrib. 25, 582–602. doi: 10.1111/ddi.12897
Wu, T. H., Tsang, L. M., Chan, B. K. K., and Chu, K. H. (2015). Cryptic diversity and phylogeography of the island-associated barnacle Chthamalus moro in Asia. Mar. Ecol. 36, 368–378. doi: 10.1111/maec.12146
Xu, M., Sakamoto, S., and Komatsu, T. (2016). Attachment strength of the subtidal seaweed Sargassum horneri (Turner) C. agardh varies among development stages and depths. J. Appl. Phycol. 28, 3679–3687. doi: 10.1007/s10811-016-0869-5
Yamasaki, M., Aono, M., Ogawa, N., Tanaka, K., Imoto, Z., and Nakamura, Y. (2014). Drifting algae and fish: implications of tropical Sargassum invasion due to ocean warming in western Japan. Estuar. Coastl. Shelf Sci. 147, 32–41. doi: 10.1016/j.ecss.2014.05.018
Yatsuya, K. (2008). Floating period of Sargassacean thalli estimated by the change in density. J. Appl. Phycol. 20, 797–800. doi: 10.1007/s10811-007-9293-1
Yoshida, G., Arima, S., and Terawaki, T. (1998). Growth and maturation of the ‘autumn-fruiting type’ of Sargassum horneri (Fucales, Phaeophyta) and comparisons with the ‘spring-fruiting type’. Phycol. Res. 46, 183–189.
Yoshida, G., Murase, N., Arat, S., and Terawaki, T. (2004). Ecotypic differentiation in maturation seasonality among Sargassum horneri (Fucales, Phaeophyta) populations in Hiroshima Bay, Seto Inland Sea, Japan. Phycologia 43, 703–710. doi: 10.2216/i0031-8884-43-6-703.1
Yoshida, G., Yoshikawa, K., and Terawaki, T. (2001). Growth and maturation of two populations of Sargassum horneri (Fucales, Phaeophyta) in Hiroshima Bay, the Seto Inland Sea. Fish. Sci. 67, 1023–1029. doi: 10.1111/j.1440-1835.1998.tb00112.x
Zhang, J., Yao, J., Hu, Z. M., Jueterbock, A., Yotsukura, N., Krupnova, T. N., et al. (2019). Phylogeographic diversification and postglacial range dynamics shed light on the conservation of the kelp Saccharina japonica. Evol. Appl. 12, 791–803. doi: 10.1111/eva.12756
Keywords: biodiversity, climate change, environmental variables, range shift, Sargassum horneri, suitable habitat
Citation: Li J-J, Huang S-H, Liu Z-Y and Bi Y-X (2020) Climate-Driven Range Shifts of Brown Seaweed Sargassum horneri in the Northwest Pacific. Front. Mar. Sci. 7:570881. doi: 10.3389/fmars.2020.570881
Received: 09 June 2020; Accepted: 12 August 2020;
Published: 02 September 2020.
Edited by:
Christian Marcelo Ibáñez, Andres Bello University, ChileReviewed by:
Giuseppe C. Zuccarello, Victoria University of Wellington, New ZealandSylvain Faugeron, Pontificia Universidad Católica de Chile, Chile
Copyright © 2020 Li, Huang, Liu and Bi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing-Jing Li, bGlqaW5namluZ0BoaHUuZWR1LmNu
†These authors have contributed equally to this work
 Jing-Jing Li
Jing-Jing Li Sheng-Hui Huang
Sheng-Hui Huang Zheng-Yi Liu
Zheng-Yi Liu Yuan-Xin Bi
Yuan-Xin Bi