- 1Department of Life Sciences, Ben-Gurion University of the Negev, Beersheba, Israel
- 2Department of Natural Sciences, University of Alaska Southeast, Juneau, AK, United States
- 3Department of Life Sciences, Achva Academic College, Arugot, Israel
- 4The National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beersheba, Israel
The insulin-like androgenic gland hormone (IAG) secreted from the androgenic gland (AG) is a unique endocrine controller (the IAG-switch) of crustacean sexual differentiation. However, while previous studies of the IAG-switch focused mainly on sexual differentiation at early developmental stages of gonochoristic species, this mechanism is yet to be deciphered during naturally occurring sexual shifts in hermaphrodite species. The Northern spot shrimp, Pandalus platyceros, is a protandric hermaphrodite species, native to the North Pacific Ocean. We collected four stages of spot shrimp in Southeast Alaska including: juveniles, adult males, transitionals, and adult females. The AG was dissected from each stage and characterized histologically. Additionally, the IAG mRNA was sequenced. The function of the IAG-switch during the life history of this protandric species was demonstrated through monitoring IAG gene expression. Transcript levels were highest at the juvenile stage, then decreased significantly in mature males and became negligible in the transitional and female stages. Moreover, manipulating the IAG-switch via IAG loss of function in males through RNAi, induced the expected masculine to feminine sexual transformation that naturally occurs in this species. This included reduction in IAG transcript levels in males, elevation of vitellogenin gene expression in hepatopancreas and transformation of the gonad from an ovotestis containing both ovarian and testicular tissue to a true ovary with vitellogenic oocytes. Furthermore, a transcriptomic library from tissues associated with the endocrine axis upstream and downstream the IAG-switch yielded 1,801, 1,707, 1,946, and 182 differentially expressed genes between males, transitionals, and females in the AG, eyestalk, gonad, and hepatopancreas, respectively. Among these genes, the transcriptional pattern of six of them (all of them in the AG), between males, transitionals and females, had similar or inverted transcriptional pattern to that of IAG in P. platyceros. Five of these putatively IAG-switch associated genes are downregulated and one is upregulated throughout the P. platyceros shift from maleness to femaleness. Homolog protein sequences for the above novel genes were found in 17 other decapod species suggesting that they might represent conserved factors associated with the IAG-switch and involved in universal crustacean sexual differentiation mechanisms.
Introduction
A key player governing the crustacean sexual differentiation process is the androgenic gland (AG), a unique crustacean endocrine gland that controls masculine differentiation and development (Cronin, 1947; Charniaux-Cotton, 1954). The AG is known to secrete the insulin-like androgenic gland (IAG) hormone (Manor et al., 2007), which is suggested as a universal sex-differentiating mechanism, termed the ’IAG-switch’ (Levy et al., 2018; Levy and Sagi, 2020), in which the synthesis and secretion of IAG from the AG induces masculinization. The absence of IAG secretion results in feminization (Ventura et al., 2011). Until now, the IAG-switch and its role in sexual differentiation was mostly studied in gonochoristic species but never in the light of sex-transformation in hermaphrodite species (Levy and Sagi, 2020). The IAG-switch has been thoroughly described in many species and its manipulation in many crustacean species resulted in partial or even full sexual shifts (Charniaux-Cotton, 1958; Katakura, 1960; Payen, 1969; Nagamine et al., 1980a,b; Nagamine and Knight, 1987; Malecha et al., 1992; Aflalo et al., 2006; Ventura et al., 2012; Levy et al., 2016). Even in isopods, in which an elevated bacterial dosage of Wolbachia resulted in feminization, it was postulated that Wolbachia feminized males through intervention in the IAG-switch by instigating dysfunctional insulin receptors (Herran et al., 2020). However, while few other genes were found to be associated with the expression of IAG [MIH, GIH, CHH, CFSH, DMRT, and GEM (Li et al., 2015; Jin et al., 2019; Zhong et al., 2019; Jiang et al., 2020)], genes (other than the IAG itself) that act directly within the core of the IAG-switch cascade, have yet to be found.
Next generation sequencing techniques provide powerful tools to study the upstream controlling factors for the mechanism of sexual differentiation. RNA libraries generated from tissues involved with reproduction and sexual differentiation can, after assembly and annotation, provide general information about potential upstream controlling factors for sexual differentiation, and more specifically for the IAG-switch. Applying the above next generation transcriptomic approach in gonochoristic crustacean species is complicated since sexual differentiation occurs at an early developmental stage (Levy et al., 2018; Levy and Sagi, 2020), when the size of the animal is usually too small to perform high-resolution dissections of target tissues. Therefore, using a hermaphrodite organism as a model, in which sexual shifts occur naturally in mature stages when the animal is relatively large (Ghiselin, 1969; Benvenuto and Weeks, 2020), may overcome this obstacle.
The Northern spot shrimp, Pandalus platyceros, a member of the Pandalidae family, is a protandric hermaphrodite in which each individual first matures as a functional male, and then going through a transitional phase followed by maturation as a functional female (Berkeley, 1930; Hoffman, 1969; Levy et al., 2020). Distinguishing between the different developmental stages could be done through observation of the secondary sexual characteristics like appendix masculine (AM) which is developed during the maturation of a juvenile into a male stage, then gradually disappears when the shrimp transforms to a female (Levy et al., 2020). This species is widely distributed in the North Pacific Ocean (Butler, 1964), constitutes the main target species for shrimp fishery in Southeast Alaska (Smith and Gray, 2017) and is considered a highly important species for the ecosystem’s health while serving as a putative indicator for climate change (Anderson, 2000).
Due to its pivotal role in crustacean sexual differentiation (Levy et al., 2018; Levy and Sagi, 2020), understanding the IAG-switch in P. platyceros will shed new light on its role in reproductive physiology and potentially will benefit better conservation efforts, support fishery management practices and maybe even enable sustainable P. platyceros mariculture. Moreover, since the sexual differentiation mechanism in P. platyceros is not limited to early developmental stages, but is an integral to the life history as shrimp transform between maleness and femaleness (Bergstrom, 2000), the results of this study could be relevant also to other crustacean species.
In the present study we characterized the AG histologically, sequenced the IAG and followed its activity during the life cycle of P. platyceros, and tested, in vivo, its function in the course of sexual transformation from maleness to femaleness. Moreover, we constructed RNA libraries from tissues related to reproduction and its control (AG, eyestalk, gonad, and hepatopancreas) in order to discover genes that are associated with the IAG-switch and are correlated with the IAG transcriptional pattern during the P. platyceros life-cycle.
Materials and Methods
Animals and Life Stage Definition
Pandalus platyceros were collected using baited pots during the annual shrimp survey conducted by the Alaska Department of Fish and Game. Animals were transferred to the University of Alaska Southeast marine laboratory in Juneau and maintained in 750-L tanks in flow-through seawater with ambient temperature (4–11°C), salinity (20–33 ppt) and dark conditions (tanks were covered) throughout this study. The animals were fed ad libitum (herring chunks, squid or clams) throughout the entire study. In P. platyceros, the AM located on the 2nd pleopod develops as the juvenile shrimp matures as a male and gradually decreases in length as the shrimp transforms into a female (Hoffman, 1972; Levy et al., 2020). Therefore, for the entire study, the following stages were determined using morphological parameters such as body size and appendage lengths [i.e., appendix masculina (AM) and appendix interna (AI)]. Juveniles (J) were small animals (total body length of 4 to 8 cm) with AM/AI ≤ 1. Males (M) were medium sized animals (total body length of 12 to 16 cm) AM/AI ≈ 2. The transitional phase during which the male transforms into the female was separated into two stages based on the AM/AI ratio: early transitional (T1), represented the first stage that occurs one molt after the M phase (the first molt in which the AM becomes shorter), with AM/AI ≈ 1 and late transitional (T2), is the phase that occurs one molt after the T1 stage and the AM/AI is < 1. Mature females (F) were large animals (over 16 cm total body length) with visible developed ovary and absence, or nearly absence of AM (AM/AI ≈ 0).
Characterizing the AG and Sequencing the IAG in P. platyceros
Fifth pereiopods, the prominent location of the AG (Charniaux-Cotton, 1962), were dissected from P. platyceros males. One pereiopod was fixed for histology in 4% buffered formalin for 48 h in room temperature. Samples were gradually dehydrated through a series of increasing alcohol concentrations (70, 80, 90, and 100%), incubated with xylene, and embedded in Paraplast (Kendall, Mansfield, MA, United States). Consecutive sections of 5 μm were placed on silane-coated slides (Menzel-Gläser, Brunswick, Germany) and stained with hematoxylin and eosin (H&E) for morphological observations as follows: slides were dipped in Xylene for 5 min × 2, then 100, 90, 80, and 70% of EtOH for 1 min each. Later, in tap water for 1 min, Hematoxylin for 5 min, washing in tap water for 4 min and removal of background staining in Acidic 70% EtOH for 10 s. The final stage included dipping the slides in Eosin, 95 and 100% EtOH for 5 min × 2 each, Xylene for 5 min × 2 and then cover the section with cover slip.
In order to sequence the IAG mRNA in P. platyceros (Pnp-IAG), total RNA was extracted from the other male 5th pereiopod using EZ-RNA Isolation kit (BI, Cromwell, CT, United States). A forward degenerative primer (5′-GATCAGRTHGACTTYGACTGYGG-3′) was designed on the basis of the conserved amino acids sequence DFDCG in the IAG B-chains as previously described (Levy et al., 2017). The Pnp-IAG mRNA sequence was obtained by the rapid amplification of cDNA ends (RACE) method using a SMARTer RACE cDNA Amplification kit (Clontech, Mountain View, CA, United States), using the total RNA extraction from the fifth pereiopod as a template and the above-mentioned forward degenerative primer and the Universal Primers Mix (UPM) from the RACE kit (including the long universal primer: 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACG CAGAGT-3′, and the short universal primer: 5′-CTAATACGACTCACTATAGGGC-3′). The latter reaction amplified the 3′ end of the mRNA sequence (including the poly-A tail). Additional PCR amplification of the 5′ region was performed using specific primer (5 RACE_Rev: 5′-AGGGTTCGACTGGAGCATCA-3′) and the UPM with the above mentioned RACE kit. After sequencing the full Pnp-IAG mRNA, the predicted structure of the protein was inferred from its deduced amino acids sequence.
In vitro Expression of Pnp-IAG in Different Stages
Total RNA was extracted as described above and cDNA was synthetized using qScript cDNA Synthesis kit (Quanta, Beverly, MA, United States) from the AG of P. platyceros animals at different stages: J (n = 10), M (n = 8), T1 (n = 7), T2 (n = 6), and F (n = 8). Additionally, RNA was extracted from a male hepatopancreas (which does not express IAG) as a control to normalize the transcript levels of the qPCR assay. Relative quantification of transcript levels was performed using Roche Diagnostics FastStart Universal Probe Master Mix (Basel, Switzerland) and Roche Universal Probe Library probes. The following primers and probe were used: qPnp-IAG F (5′-CTTGACACGACACGCAGAAG-3′) and qPnp-IAG R (5′-GCGTCTTTGAAGAGACACTGGT-3′), and Probe #98. P. platyceros 18S, which served for normalization, was also quantified by means of qPCR using the primers, qPnp-18S F (5′-CCCTAAACGATGCTGACTAGC-3′) and qPnp-18S R (5′-TACCCCCGGAACTCAAAGA-3′), and Probe #152. The qPCR reactions were performed in the QuantStudio 1 Real-Time PCR System, Applied Biosystems (Foster City, CA, United States).
dsRNA Preparation for Pnp-IAG Loss of Function Experiments
Using plasmid containing the Pnp-IAG open-reading frame (ORF) coding sequence, two PCR products of Pnp-IAG were generated using a T7 promoter anchor (T7P: 5′-TAATACGACTCACTATAGGG-3′) attached to one of the two primers used to amplify each product. For Pnp-IAG sense RNA synthesis: primer T7P forward (5′-T7PAACTTCCACGATGGGACGAG-3′) and primer reverse (5′-GCCTGAATTAGTGCAGTTGTTG-3′). For Pnp-IAG antisense RNA synthesis: primer forward (5′-AACTTCCACGATGGGACGAG-3′) and primer T7P reverse (5′-T7PGCCTGAATTAGTGCAGTTGTTG-3′). The two PCR products were electrophoresed on a 1.5% agarose gel, visualized with SYBR Safe DNA Gel Stain, Thermo Fisher Scientific (Waltham, MA, United States) and blue light, excised from the gel, and purified with a NucleoSpin Gel and PCR Clean-up Kit, Machery-Nagel (Düren, Germany). Single-stranded RNA of each PCR product was synthesized with TranscriptAid T7 High Yield Transcription Kit, Thermo Fisher Scientific (Waltham, MA, United States) according to the manufacturer’s instructions. The two strands were hybridized at 70°C for 15 min, 65°C for 15 min followed by incubation in room temperature for 30 min.
Short Term Reproductive Effects Following Pnp-IAG Knockdown
To test the effects of Pnp-IAG during sexual transformation and masculine maintenance in P. platyceros, Pnp-IAG loss of function experiment, using RNAi, was performed. All animals received 5 μg/g body weight of dsPnp-IAG prepared from a 1 μg/μL solution of dsPnp-IAG. Ten males with average weight ± SD of 22.7 ± 3.8 g were injected in the abdominal muscle with dsPnp-IAG and 10 males (23.9 ± 3.9 g) served as controls and were injected with crustacean saline solution (Pantin, 1934). Previous RNAi experiments performed in our laboratory used injection of dsRNA of exogenous gene (Ventura et al., 2012; Lezer et al., 2015; Sharabi et al., 2015; Abehsera et al., 2017, 2018) or water (Rosen et al., 2010) as a control. Since none of them resulted in off-target effects nor were different from the solvent itself, in recent years we have changed our protocols and commonly use crustacean saline as a control. Two days post-injection, total RNA was extracted from the AG and the hepatopancreas of each animal and cDNA was synthetized as described above. Relative quantifications of Pnp-IAG in the AG and P. platyceros vitellogenin (Pnp-Vg) in the hepatopancreas were performed. Pnp-IAG transcript levels were quantified to test the dsPnp-IAG silencing efficiency and Pnp-Vg levels were quantified to examine initiation of vitellogenesis that occurs when the animal transforms from maleness to femaleness (Levy et al., 2020). The qPCR assay for Pnp-IAG was performed as described above while for Pnp-Vg the same assay was performed using the following primers: qPnp-Vg F (5′-TGTGCAACTAAGGGAGTTATGGA-3′) and qPnp-Vg R (5′-GGTGAGTGCCAAAGAAGAGTG-3′), and Probe #89.
Long Term Reproductive Effects Following Pnp-IAG Knockdown
To test whether Pnp-IAG knockdown will induce the sex-transformation that naturally occurs in this protandric species, an additional Pnp-IAG loss of function experiment, using RNAi, was performed. Twenty males (22.5 ± 4.4 g) were injected with dsPnp-IAG and 14 males (21.6 ± 5.1 g) were injected with crustacean saline solution for a control, as described above. Injections of dsPnp-IAG or crustacean saline were repeated a week and 2 weeks after the first injection. All animals were tagged and maintained in separate tanks for 7 months. Molting events and mortality were monitored daily. After the 7-month long experiment, 11 and 8 animals remained alive in the dsPnp-IAG injected and the control group, respectively. For the remaining animals: (i) each animal was weighed, (ii) gonad was dissected, weighed and fixed for histology as described above, (iii) gonadosomatic index (GSI; i.e., the proportion of the gonad weight out of the total body weight) was calculated as previously described (Levy et al., 2016) and (iv) RNA was extracted from the AG and the hepatopancreas of each animal and cDNA was synthetized for qPCR of Pnp-IAG and Pnp-Vg as described above. Low quality RNA samples were discarded from the qPCR analysis. Later, histological sections of the gonads were stained with H&E and oocyte diameters were measured as previously described (Levy et al., 2020).
RNA Library Preparation of P. platyceros Different Reproductive Stages
In order to characterize transcriptional patterns of genes in tissues related to reproduction in different stages during the P. platyceros life-cycle, RNA was extracted, as described above, from AG, gonad, hepatopancreas and eyestalks of males (n = 10), late transitionals (T2; n = 7), and females (n = 10). While the choice of the gonad to study reproduction is obvious, the AG was chosen since it is a prominent crustacean masculine endocrine organ (Charniaux-Cotton, 1962), the hepatopancreas was chosen due to its involvement in vitellogenin synthesis (Levy et al., 2020) and the eyestalk was chosen since it functions as a controlling endocrine factor over the reproductive system in crustaceans (Khalaila et al., 2002). Due to the location of the AG (Hoffman, 1969), in order to avoid the loss of AG cells and collect the entire AG, as commonly practiced we dissected the entire area of the terminal ampule, which, in males, contains the AG (Nagamine et al., 1980a). For males and females, 2 pools of RNA were made for each tissue, each contained RNA from five animals, while for the transitionals, the first and second pools of each tissue were made from 3 and 4 animals, respectively. All of the 24 pooled RNA samples (4 tissues × 2 replicates × 3 stages) were sent for sequencing (Novogene, Hong Kong) using Illumina NovaSeq 6000 platform.
Assembly and Annotation of P. platyceros RNA Library
The bioinformatics analysis was carried out at the Bioinformatics Core Facility of Ben-Gurion University using NeatSeq-Flow version 1.6.0 (Sklarz et al., 2018). The sequenced RNA samples generated 1,024 million high-quality paired-end 150 bp-long reads. Reads were quality-trimmed with TrimGalore! version 0.4.21, which removed less than 1% of the reads. Trinity version 2.5.1 (Grabherr et al., 2011) was used to de novo assemble all clean reads. The quality of the assembly was determined with quast version 4.5 (Gurevich et al., 2013). BUSCO analysis (Simão et al., 2015) was performed on the transcriptome against the Metazoa database (metazoa_odb9). Representative transcripts per gene were selected with the filter_low_expr_transcripts.pl script from the Trinity software suite. The representative transcripts were annotated using Trinotate pipeline version 3.1.12.
Differential Expression Analysis of Genes From the RNA Library
Mapping of samples to the reference transcriptome was conducted with bowtie2 version 2.3.5 (Langmead and Salzberg, 2012) and transcript abundance was estimated with RSEM version 1.3.0 (Li and Dewey, 2011). DESeq2 (Love et al., 2014) was used to identify the differentially expressed (DE) genes between the stages (namely males, transitionals, and females) in each of the tissues (namely, AG, eyestalks, gonad, and hepatopancreas) separately. Genes were considered as DE if they had FDR adjusted P-value < 0.05 and absolute linear fold change > 1.3. Hierarchical clustering of the DE genes, after z-scoring of their variance stabilized expression values, was carried out using R’s pheatmap function to generate a heatmap representing the transcriptional pattern of the DE genes in each tissue. Venn diagrams were generated to compare the lists of the DE genes between each of the stages in each tissue.
Mining for Genes Associated With Pnp-IAG Transcriptional Pattern
According to the transcriptional pattern of Pnp-IAG during the life-history of P. platyceros, DE genes that are correlated or negatively correlated with Pnp-IAG transcription levels between the different reproductive stages (males, transitionals, and females) were searched in the above RNA library. SMART (Schultz et al., 2000) was used to predict domains of the deduced amino acid sequences inferred from the genes correlated with Pnp-IAG transcription levels. To examine whether those genes are conserved among decapods, blastx alignment (i.e., translated nucleotide to protein sequences) to decapod protein databases [i.e., Non-redundant protein sequences (nr), Transcriptome Shotgun Assembly proteins (tsa_nr), and Protein Data Bank proteins (pdb); Organism: Decapoda (taxid: 6683)] in the NCBI server was performed.
Statistical Analyses
All data used for statistical analysis in this study was first tested for residuals’ normality using the Shapiro–Wilk test and for variance’s homogeneity using the Levene’s test and, if needed, was transformed as to fit proper statistical analysis. For the qPCR relative transcript levels of Pnp-IAG and Pnp-Vg, as well as for the oocyte diameters, the data was first logarithmically transformed. For the GSI measurements, reciprocal transformation (1/x) of the data was performed. The transformed data was then analyzed using one-way ANOVA followed by a post hoc Tukey’s HSD test, for Pnp-IAG quantification in different stages, or using t-test, to compare all the tested parameters between the two groups (silenced and control) in the short and long term silencing experiments. All statistical analyses were performed using Statistica v13.3 software (StatSoft, Ltd., Tulsa, OK, United States).
Results
Pnp-IAG and AG in P. platyceros
The Pnp-IAG mRNA sequence (accession number KX619617.1; Figure 1A) included fragments of 246, 504, and 241 bp for the 3′ UTR, ORF and 5′ UTR, respectively. The deduced structure of the Pnp-IAG hormone, according to the predicted ORF, contains signal peptide (27 aa), B chain (42 aa), A chain (43 aa), and C peptide (55 aa). Detailed cellular morphological changes during AG development were previously described by Hoffman (1969). As in other crustaceans, including P. platyceros (Hoffman, 1969), the histological section of the 5th pereiopod from a male (Figure 1B) revealed the typical form of the AG which consisted of cords of dense cells with oval nuclei adjacent to the terminal segment of the sperm duct.
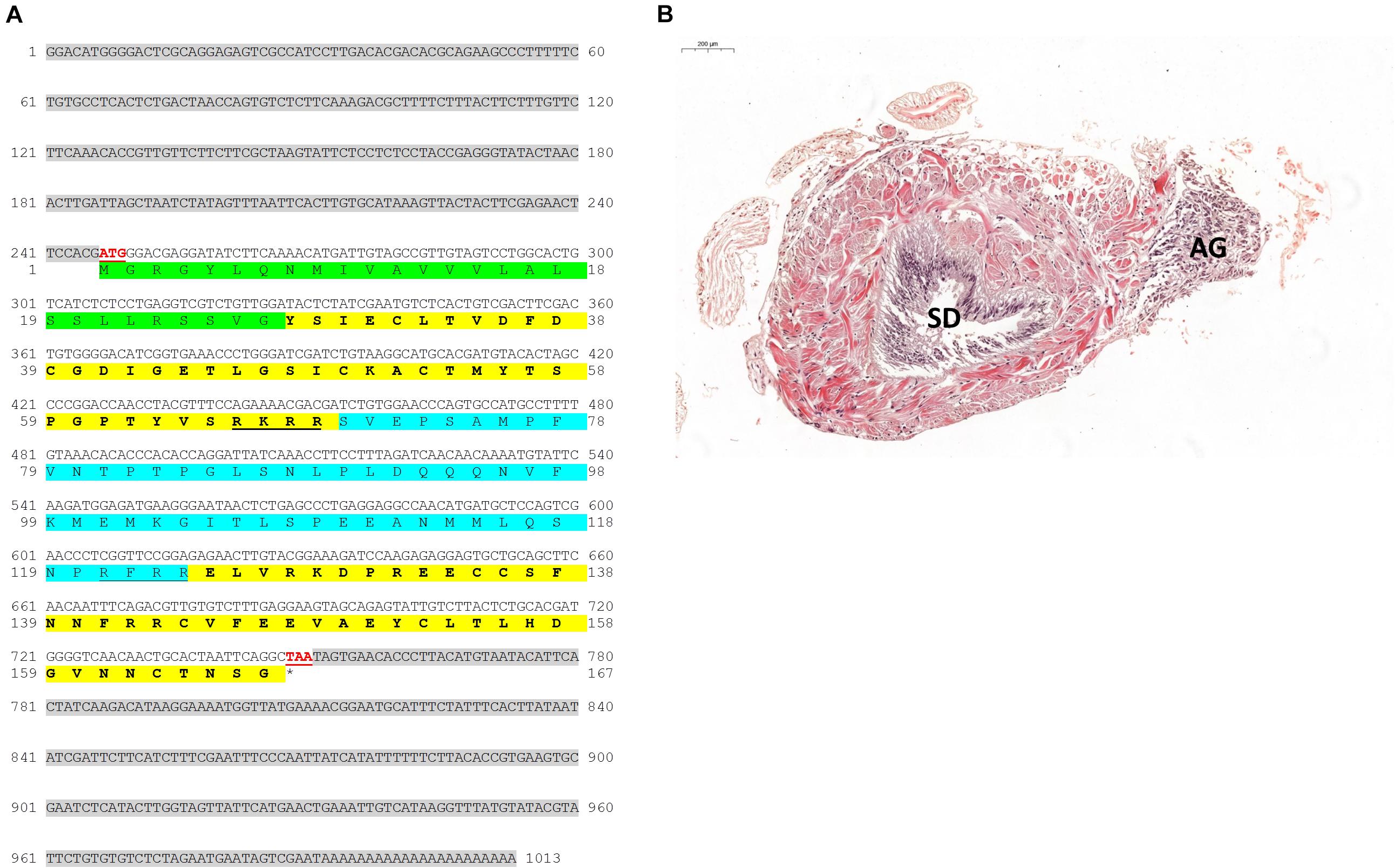
Figure 1. IAG and AG in P. platyceros. (A) Predicted structure of Pnp-IAG. The full sequence of Pnp-IAG mRNA and its open reading frame (ORF)-deduced amino acids. 5′ (first) and 3′ (second) UTR’s are highlighted with gray background. The signal peptide is highlighted with green background. B (first) and A (second) chains are highlighted with yellow background, with C peptide (blue background), including its cleavage sites (underlined), flanked between them. The start (ATG) and stop (TAA) codons are shown in red and underlined. The stop codon is also indicated with an asterisk. (B) The androgenic gland in P. platyceros (magnified × 6.1). Histological section of the fifth pereiopod base in a P. platyceros male stained with hematoxylin and eosin (H&E). The sperm duct (SD) and androgenic gland (AG) are indicated. Bar scale (200 μm) is given.
Pnp-IAG Expression Throughout the P. platyceros Life-Cycle
Pnp-IAG relative transcript levels (Figure 2A) were found to be significantly different between the stages [defined by AM/AI ratio; Figure 2B (Levy et al., 2020)] sampled in this study (ANOVA, F(4,34) = 75.72, P < 0.05). More specifically, according to the Tukey’s HSD test, IAG transcription was highest in juveniles and was significantly reduced after the animals matured as males, followed by an additional reduction in early and late transitional stages (P < 0.05). The low Pnp-IAG transcription level in females did not significantly differ from the early and late transitional stages (P > 0.05).
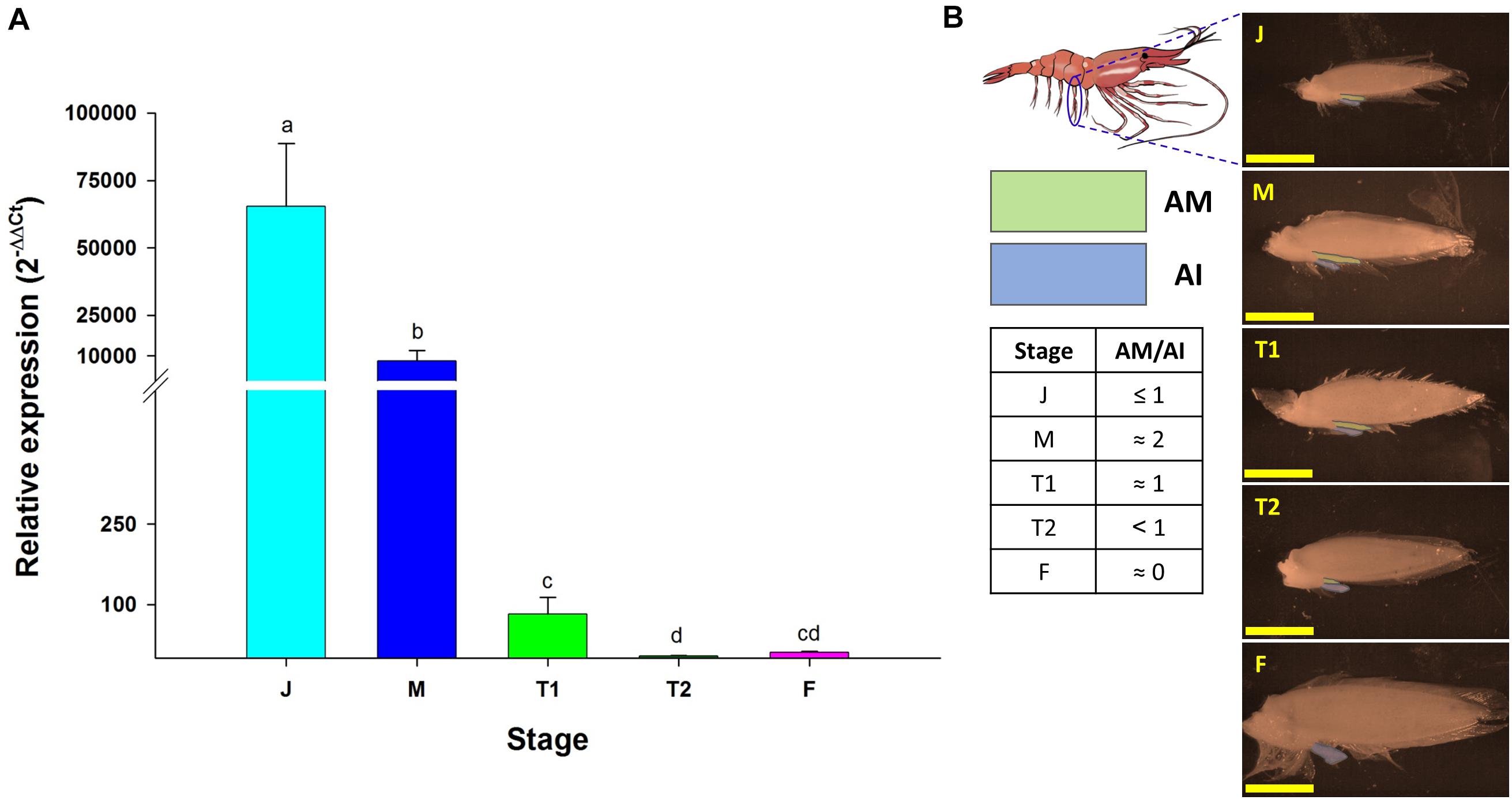
Figure 2. Pnp-IAG expression in different reproductive stages of P. platyceros. (A) Relative expression of Pnp-IAG from androgenic glands of the different reproductive stages: (J) Juvenile (n = 10), (M) Male (n = 8), (T1) Early transitional (n = 7), (T2) Late transitional (n = 6), (F) Female (n = 8). Error bars represent standard error of the means and statistical difference between the stages is distinguished with letters. (B) Second pleopods at different reproductive stages. The location of the second pleopod is shown in the blue circle (top left). Appendix masculina (AM; green) and appendix interna (AI; blue) are highlighted. Bar scales (= 5 mm) are consistent in all photos. The AM/AI ratio in the different reproductive stages are given in the table.
Pnp-IAG Knockdown Effects
While the transcription level of Pnp-IAG in intact males was significantly higher than in transitionals (Figure 2A), and the level of Pnp-Vg in intact males was negligible and significantly lower than that of transitionals (Levy et al., 2020), in the short term experiment, although this result was not statistically significant (t16 = 0.95, P > 0.05), Pnp-IAG relative transcript levels in the group injected with dsPnp-IAG (i.e., the ’silenced’ group), was reduced by 55% compared to the control group (Figure 3A). However, the relative transcript levels of Pnp-Vg in the hepatopancreas (Figure 3B) was 8.5-fold higher in the silenced group compared to the control (statistically significant; t16 = −2.34, P < 0.05).
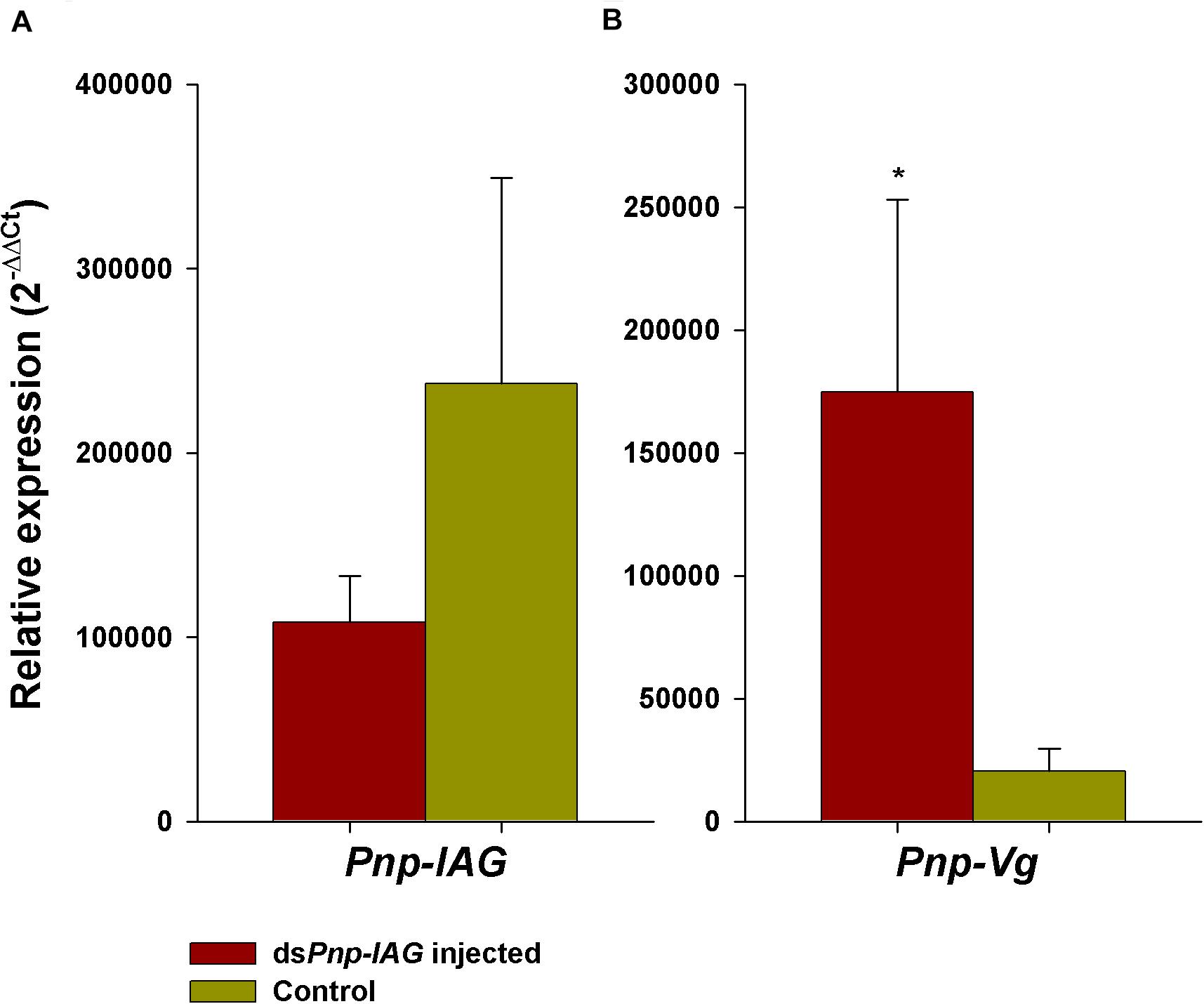
Figure 3. Short term Pnp-IAG loss of function through RNAi. (A) Relative expression of Pnp-IAG and (B) Pnp-Vg in P. platyceros males injected with dsPnp-IAG (red, n = 10) or with crustacean saline (control; green, n = 8). Error bars represent standard error of the means. *P < 0.05.
In the long term experiment, during the 7 months period, all of the 11 animals that remained alive in the silenced group injected with dsPnp-IAG, molted twice. On the other hand, in the control group that was injected with saline, among the eight animals that remained alive, six molted once, and only two molted twice. Since molting is required to facilitate transformation from a male to a female including the morphological changes in the AM/AI ratio along the transformation process (Levy et al., 2020), we followed the changes in AM/AI ratio in an amputated 2nd pleopod. It appeared that the one or two molts which occurred were not sufficient to fully regenerate the amputated pleopod and instead, the animals developed a ’regenerative-looking pleopod’ in which the AM could not be determined (Supplementary Figure S1).
Histological sections of the gonads (Figure 4A) depicted ovotestis containing both testicular and ovarian tissues in the control group where most oocytes were pre-vitellogenic. Contrary to the above, in the silenced group most animals had an ovary with vitellogenic oocytes and absence of testicular tissue. The oocytes in the gonads (Figure 4B) were significantly larger in size (t17 = −3.10, P < 0.01) in animals from the silenced group compared to the control. The average ± SD oocyte diameter measured were 298.55 ± 88.90 and 203.56 ± 32.86 μm for the silenced and control animals, respectively. However, the differences in GSI between the silenced and the control group, 0.23 ± 0.10 and 0.17 ± 0.08%, respectively, were not statistically significant (t17 = 1.09, P > 0.05).
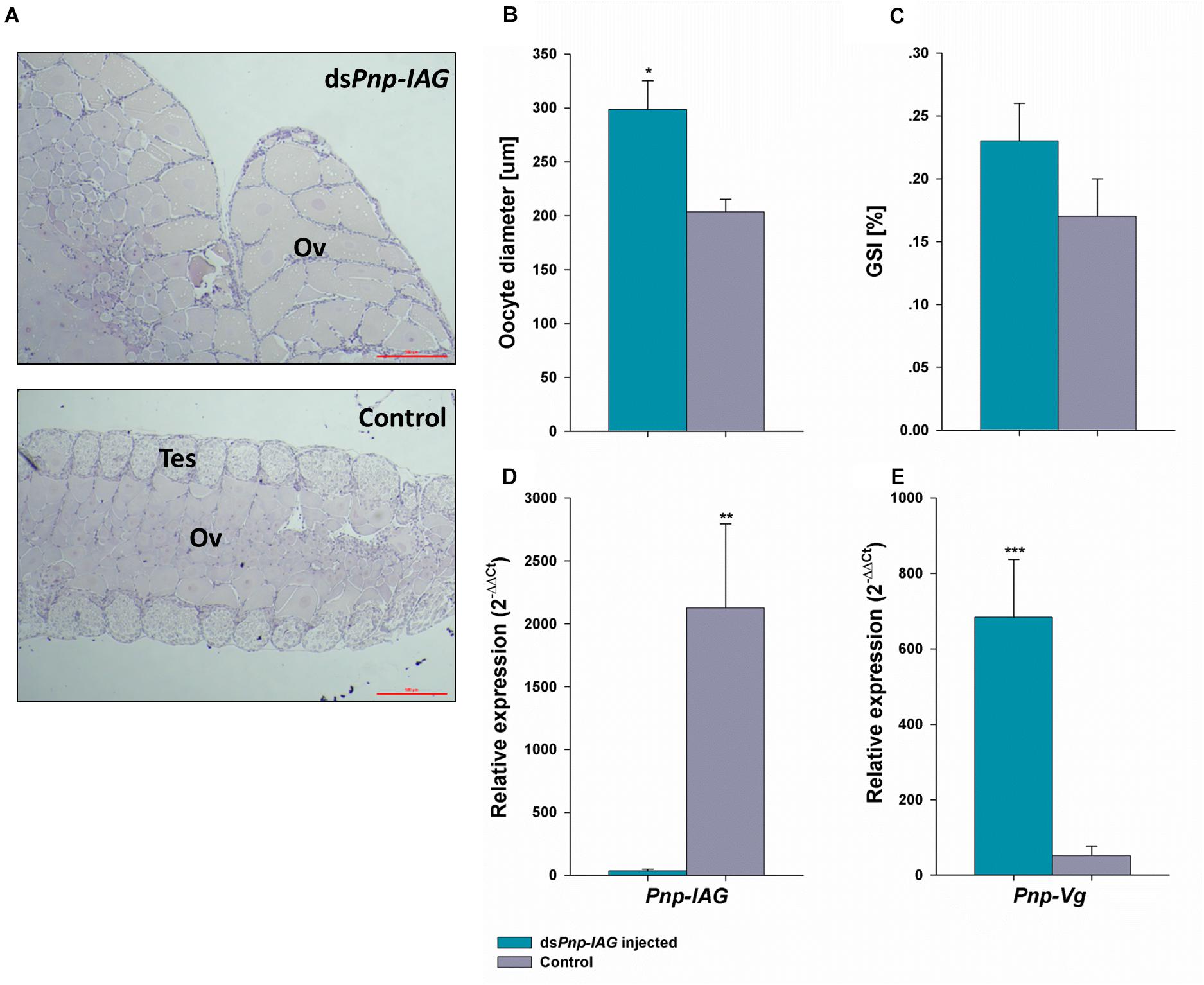
Figure 4. Long term Pnp-IAG loss of function through RNAi in terms of Pnp-IAG and Pnp-Vg transcript levels and gonad anatomy. P. platyceros males were injected with dsPnp-IAG (blue) or with crustacean saline (control; gray). (A) Representative gonad photos of dsPnp-IAG injected (top) and control (bottom) animals. Ovarian (Ov) and testicular (Tes) tissues are indicated. Bar scales (= 5 μm) are consistent in both photos. (B) Oocyte diameter and (C) Gonadosomatic index (n treatment = 11, n control = 8). (D) Pnp-IAG relative expression levels (n treatment = 9, ncontrol = 5). (E) Pnp-Vg relative expression levels (n treatment = 9, n control = 8). Error bars represent standard error of the means. P-values are denoted: *P < 0.01, **P < 0.001, ***P < 0.0001.
After 7 months, in the silenced group injected with dsPnp-IAG, Pnp-IAG relative transcript levels were significantly reduced by 99% (t12 = -5.01, P < 0.001) (Figure 4C) whereas the relative transcript levels of Pnp-Vg in the hepatopancreas (Figure 4D) was significantly higher by 13-fold in the silenced group compared to the control (t15 = 5.83, P < 0.0001).
P. platyceros RNA Library and Sex-Specific Differentially Expressed Genes
De novo assembly of all clean reads of the library resulted in 751,594 contigs representing transcripts from 416,769 putative genes. The total length of all contigs was 343,592,935 bp with N50 = 2,140 bp. BUSCO analysis performed on the gene representatives identified 92.7% of the BUSCO sequences, 78.0% in single copy and 14.7% duplicated.
Following the DESeq2 analysis, a total of 1,801, 1,707, 1,946, and 182 genes were found to be differentially expressed (FDR adjusted P-value < 0.05 and absolute linear fold change > 1.3) between the reproductive stages (male, transitional, and female) in the AG, eyestalks, gonad, and hepatopancreas, respectively. Focusing on significant differential genes between males and females, 898, 539, 1,164, and 74 genes were found in the AG, eyestalk, gonad, and hepatopancreas, respectively (2,675 in total). Three transcripts (see ’AG F vs. M’ sheet in Supplementary Table S1) were found to contain fragments that assemble the Pnp-IAG mRNA (accession number KX619617.1) and were annotated as such. Summary of all DE genes between males, transitionals and females for each tissue separately is presented in Figure 5 and Supplementary Figures S2–S5 while a detailed list of all the 2,675 DE genes between males and females in all the sampled tissues is presented in Supplementary Table S1.
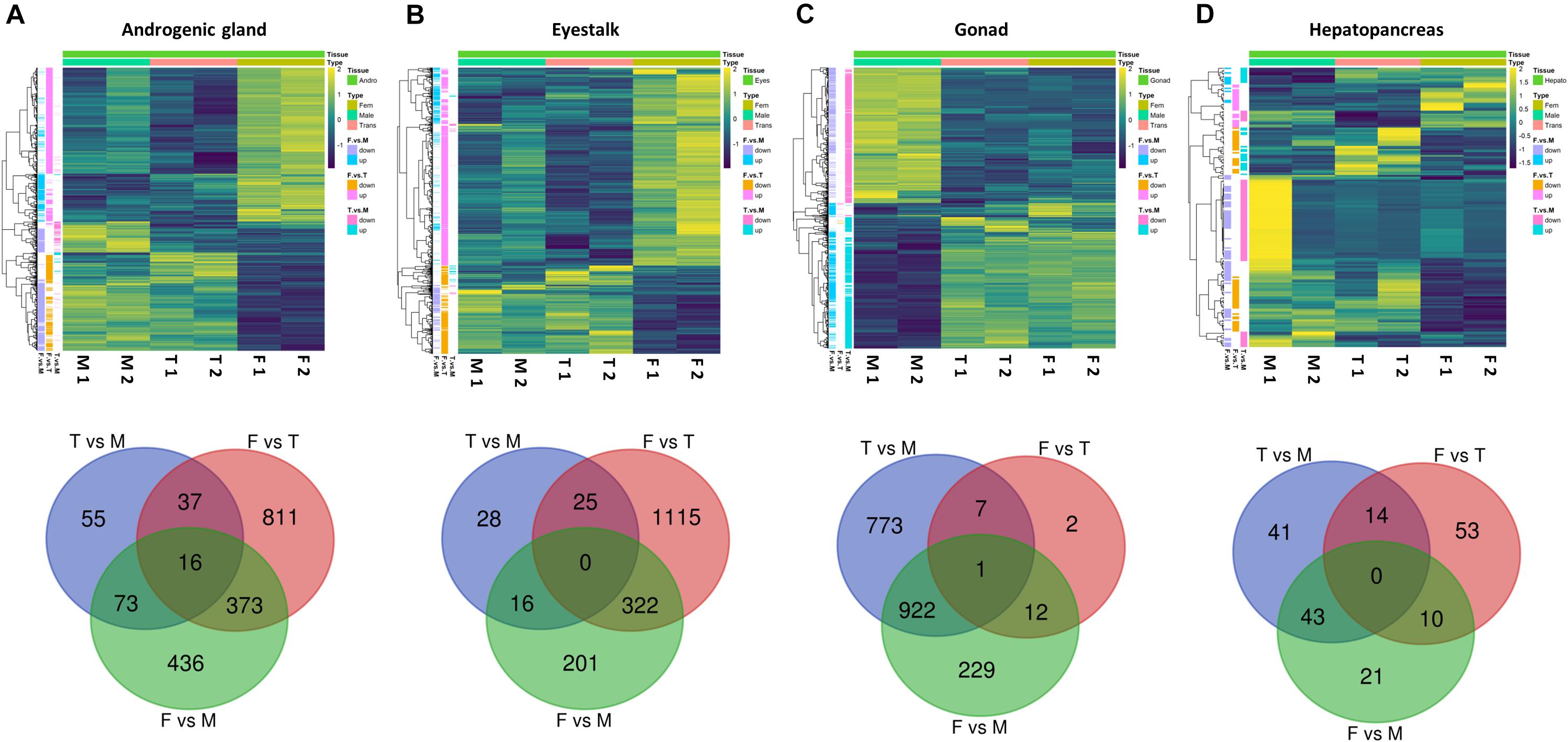
Figure 5. Differentially expressed genes at different reproductive stages of P. platyceros. Significant differential genes (FDR adjusted P-value < 0.05 and absolute linear fold change > 1.3) between males (M), transitionals (T), and females (F) in the following tissues (A) androgenic gland, (B) eyestalk, (C) gonad and (D) hepatopancreas are given. (Top) Heatmaps represent the intensity of variation in gene expression between different reproductive stages (two pooled samples in each stage). (Bottom) Venn diagrams represent the number of genes that were found to be significantly different when comparing expression patterns from the different reproductive stages in each tissue. A list of all significant differential genes between males and females in the different tissues are given in Excel table in the Supplementary Material.
Differentially Expressed Genes Correlating With the Pnp-IAG Expression Pattern
The transcriptional pattern of Pnp-IAG showed high expression at the juvenile stage and a decrease in males, with negligible levels in transitionals and female stages (Figure 2). Among the tissues sampled for transcriptomic library preparation in the present study, only the AG contained genes with similar transcriptional pattern to that of Pnp-IAG. Five of these genes were found to be positively correlated with the Pnp-IAG transcriptional pattern, i.e., downregulated in transitionals vs. males and in females vs. transitionals, and were assumed to be associated with the IAG-switch and thus termed ’Pnp-IAG-switch 1-5’ (Figure 6 and Table 1). Pnp-IAG-switch 1, 3, 4, and 5 had an ORF but none of these genes contained a signal peptide nor a typical conserved protein domain. Only a single gene, ’Pnp-IAG-switch 6,’ was found to be negatively correlated with Pnp-IAG transcriptional pattern, i.e., upregulated in transitionals vs. males and in females vs. transitionals and it was the only gene to be annotated. Pnp-IAG-switch 6 was annotated as a proclotting enzyme which contains a Trypsin-like serine protease domain (Tryp_SPc). Protein homolog sequences in 17 other decapod species, including shrimps, crabs, hermit crabs, lobsters and crayfish, were found for all the above genes but one (Pnp-IAG-switch 5), as thoroughly described in Table 2. The accession numbers of the mRNA sequences of the above six genes are given in Table 1.
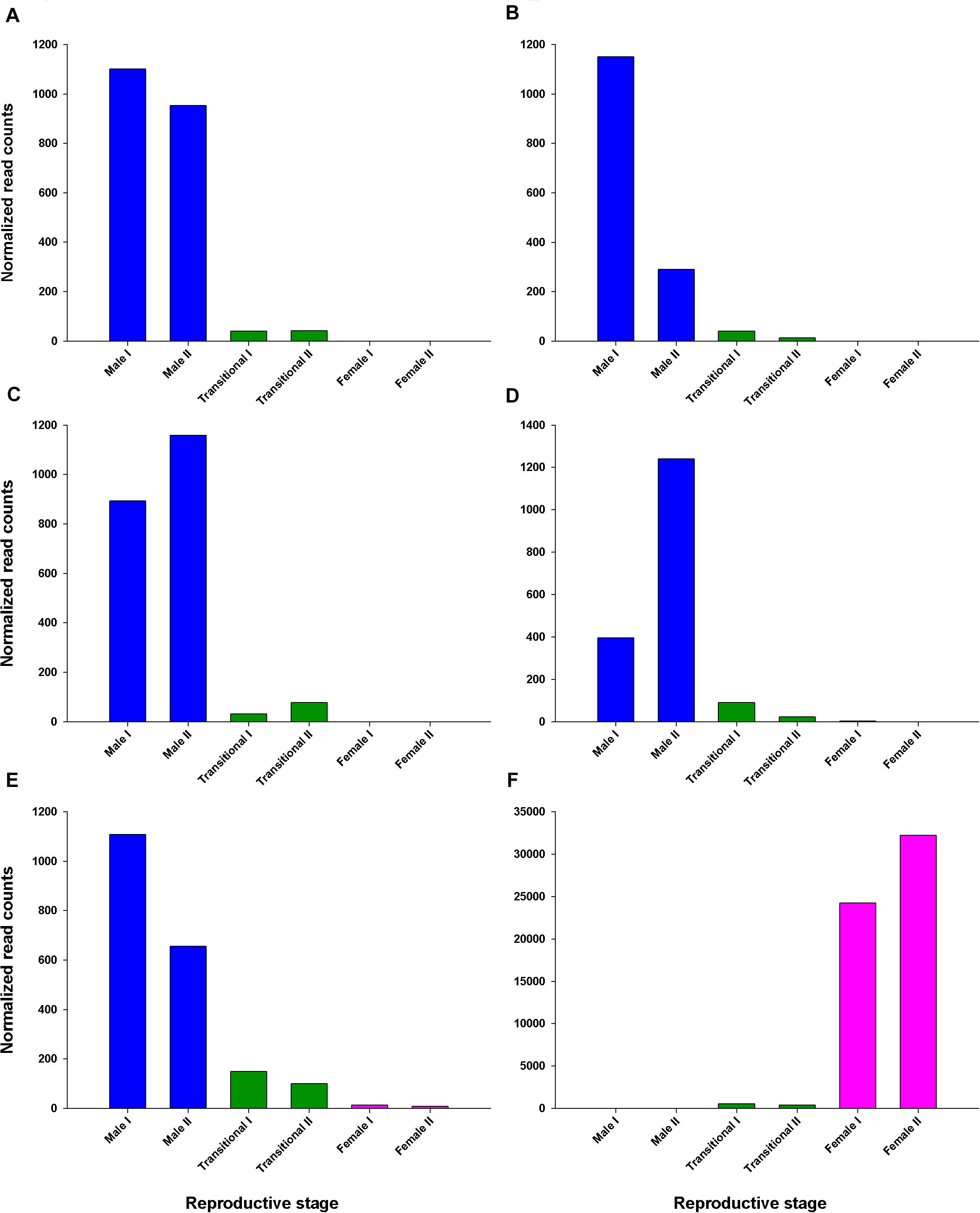
Figure 6. Read counts of AG differentially expressed genes correlated with Pnp-IAG transcriptional pattern throughout the P. platyceros life-history. Normalized read counts of each of the two replicates per reproductive stage that were sequenced to assemble the P. platyceros RNA library described in this study. (A–E) Genes with decreasing transcriptional pattern (i.e., Pnp-IAG-switch 1-5). (F) A gene with increasing transcriptional pattern (i.e., Pnp-IAG-switch 6). Genes considered significant according to the following parameters: FDR adjusted P-value < 0.05 and absolute linear fold change > 1.3. A detailed list of the above genes is given in Table 1.
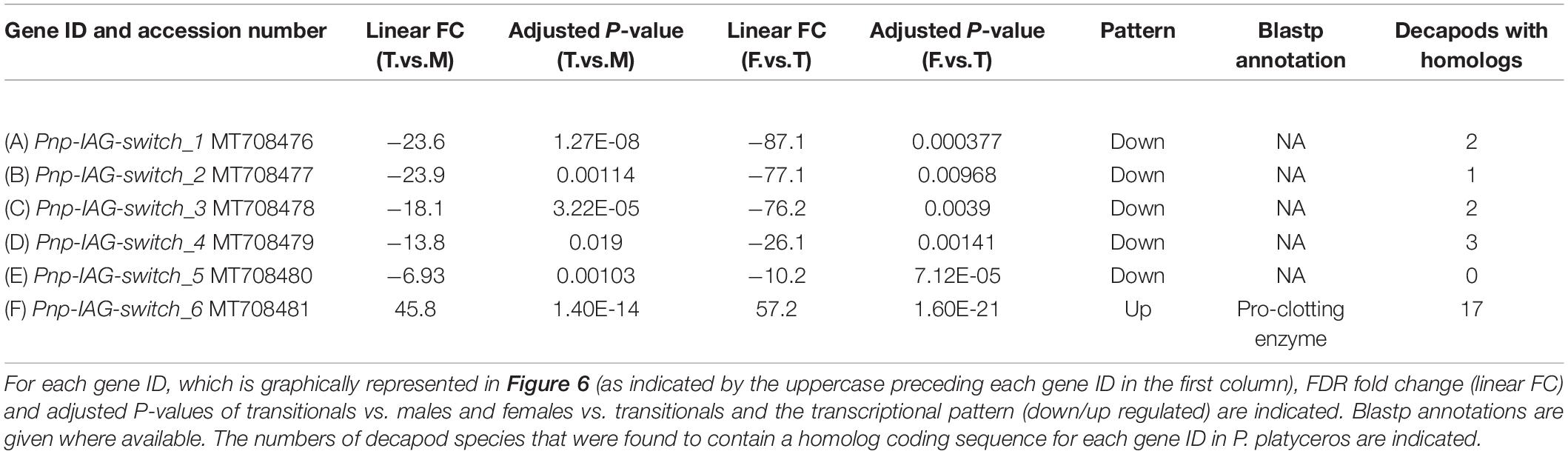
Table 1. AG differentially expressed genes correlated with Pnp-IAG transcriptional pattern throughout the P. platyceros life-history.

Table 2. Homolog sequences of the genes from Table 1 in other decapod species.
Discussion
Studying the IAG-switch and its control in gonochoristic species is complex since the sexual differentiation period is limited to early developmental stages (i.e., larval and post-larval stages) (Levy et al., 2018; Levy and Sagi, 2020) and isolation of tissues in which IAG-switch associated genes are expressed is difficult due to the small body size of the undifferentiated animal. The protandric P. platyceros used in this study is an ideal crustacean model to investigate IAG-switch-related mechanisms since the sexual differentiation process is not limited to a particular early period, but occurs when an individual matures as a male and later during its transformation from a mature male to a mature female.
To lay the foundations for the present study, we confirmed the presence of the AG at the typical location for decapods (Charniaux-Cotton, 1962), the base of the 5th pereiopod, as previously described also in P. platyceros during an histological study that followed the morphology of the AG in shrimp of different sizes and developmental stages including juveniles, reproductively active and non-active males and early transitionals (Hoffman, 1969). Moreover, while the IAG mRNA was previously obtained in a simultaneous protandric shrimp, Lysmata wurdemanni (Zhang et al., 2017), to the best of our knowledge, the Pnp-IAG mRNA obtained in this study is the first IAG to be sequenced in a sequential protandric shrimp.
The transcriptional pattern of Pnp-IAG, which was highest in juveniles, decreased significantly in males and became almost negligible in transitionals and females, could be preeminently discussed according to a previous study conducted more than four decades ago, which described the development of the AG in different stages of P. platyceros, from immature males to early transitionals (Hoffman, 1969). This histological study (Hoffman, 1969) describes juvenile males (6–8 months old) exhibiting an AG with dense proliferating cells that were later vacuolated in mature males and started degradation when the animal initiated transformation toward femaleness. Our finding of Pnp-IAG transcript levels, that are sixfold higher in juveniles than in males, implies that albeit smaller AG size in the juvenile stage (Hoffman, 1969), the proliferating cells in the AG of juveniles express more IAG than the vacuolated cells comprising the larger AG in active mature males. It has been postulated that AG and IAG activity are essential not only for crustacean male sexual differentiation (Manor et al., 2007), but also for masculine maintenance and male morphotypic differentiation in adults (Sagi et al., 1990). However, the above results of higher Pnp-IAG transcript levels in juveniles may indicate that higher IAG levels are required during the early male differentiation and maturation process rather than for masculine maintenance in adult males. To the best of our knowledge, this is the first report to compare IAG transcription between early male differentiation stage (juveniles) and mature males during the life history of the same species.
The pivotal function of IAG in this protandric species is best exemplified by the loss of function experiments. According to the short term Pnp-IAG loss of function experiment, the efficiency of the dsIAG injection on reducing the IAG transcript level was lower (55% reduction in IAG transcription level) compared to similar experiments in other gonochoristic decapods, including the crayfishes Cherax quadricarinatus and Procambarus clarkii, which showed a reduction of 97 and 98% of IAG transcription levels, respectively (Rosen et al., 2010; Savaya et al., 2020). On the other hand, while short term loss of function experiments are usually conducted as a preliminary technical step to test only the efficiency of the introduced double stranded RNA before performing a long term experiment, the results of the short term experiment in this study were surprising as even partial IAG silencing was sufficient to cause significant elevation in hepatopancreas Pnp-Vg transcript levels after 48 h. Indeed, it has been suggested that crustacean AG and IAG are considered, not only as key factors for masculine differentiation and maintenance, but also as repressors of the vitellogenesis process and transcription of the vitellogenin gene in the hepatopancreas (Sagi et al., 2002). However, as far as we know, this is the first species in which this concept is proven in such a short time frame (2 days) post dsIAG administration, thus exemplifying the high degree of sexual plasticity in protandric species.
The long term silencing experiment of Pnp-IAG revealed that the dsPnp-IAG silencing clearly induced the typical sexual transformation from maleness to femaleness that naturally occurs in protandric species. While the GSI values were larger, but not statistically significant, in the silenced group vs. the control, a comparison of the oocyte diameters in the present study to our previous study that characterized reproductive parameters in all P. platyceros stages (Levy et al., 2020), shows that the oocytes of the silenced group are quite similar to those of early transitionals (T1) sampled in the previous study (299 vs. 287 μm, respectively), whereas the control group is quite similar to mature males (204 vs. 181 μm, respectively). The latter determination of the reproductive stage (M and T1 for the control and silenced groups) is also supported by the relative quantification of Pnp-IAG transcript levels in the present study, that was 100-fold higher in the silenced compared to the control group (Figure 4D), similarly to the fold change quantified between T1 and M (Figure 2). All anatomical, morphological and molecular differences between the silenced and the control groups described in this study are consistent with the previous report regarding natural changes in the same parameters between M and T1 during a protandric sexual shift (Levy et al., 2020). While previous manipulations of the IAG-switch that were performed at early developmental stages of gonochoristic species aimed to understand the AG and IAG function in light of early sexual differentiation mechanisms (Levy and Sagi, 2020), the present study, to the best of our knowledge, is the first to describe how IAG functions as a major regulator during the sexual transformation between adult stages (i.e., mature male to mature female) in hermaphrodite species. With continuously declining P. platyceros populations in nature (Smith and Gray, 2017; Levy et al., 2020), such induction of sexual transformation, might significantly shorten the long P. platyceros life-cycle (Barr, 1973; Iversen et al., 1993; Kimker et al., 1996; Levy et al., 2020) contributing to potential establishment of sustainable mariculture for this important species and consequently, reduce the need for fishing thus allowing recovery and conservation of P. platyceros populations in nature.
Other than IAG, genes that are correlated with the activity of the IAG-switch have yet to be found and this study is presenting a great opportunity to target such genes in the protandric P. platyceros, in which sexual differentiation processes are continuous through adulthood; while transforming from maleness to femaleness (Levy et al., 2020). According to the DESeq analysis performed on the RNA library data in the present study, 1000s of genes that are DE between the different developmental stages in the different tissues related to reproduction and its control were found (Figure 5). Interestingly, the number of DE genes in the hepatopancreas was ∼10-fold lower than in other tissues. This could be due to the fact that although the Vg gene, which is related to female reproductive activity (Okuno et al., 2002), is being transcribed in the hepatopancreas of P. platyceros (Levy et al., 2020), most of the hepatopancreas functions in crustaceans are related to metabolism as digestion, absorption and storage of nutrients, mechanisms which are not sex-specific (Vogt, 2019). Moreover, although the most dramatic change in IAG transcription levels was between male and transitional stages, most cases of DE genes in the AG and eyestalk were found between the transitional and female stages. It is noteworthy that not every sexually biased gene must be directly related to sexual development. For example, in the Pacific White shrimp Litopenaeus vannamei, genes that were annotated as 40S and 60S ribosomal proteins were expressed 10-fold higher in female larvae when compared to males, although their relation to sexual development is unclear (Wang et al., 2020). Also, in this study, most gene annotations of the DE genes between the different developmental stages in all the tissues, if they exist (Supplementary Table S1), do not necessarily reflect a function related to reproduction or sexual development. Therefore, with majority of potential genes whose annotation is not clear, the strategy was to focus on genes with similar or opposite transcription pattern to the known pivotal IAG gene. Interestingly, all of the six genes (Pnp-IAG-switch 1-6) that exhibited a very similar transcriptional pattern (or mirror image) with Pnp-IAG were found in the AG library. Thus, it may be hypothesized that those genes are also related, directly or indirectly, with the IAG-switch. Therefore, with no clear annotations we decided to term them IAG-switch related (IAG-switch 1-6). Moreover, their high conservation rate among other decapods raises assumptions that they might serve as universal crustacean IAG-switch associated factors. Those six genes may set the basis for future studies aiming to uncover controlling elements over the crustacean IAG-switch. For example, knockdown/out based functional experiments of the above six genes in P. platyceros and their homologs in other crustacean species, might yield new insights regarding the universal IAG-switch mechanism and open a new window into the heart of sexual differentiation in crustaceans.
Conclusion
Crustacean sexual differentiation and reproductive physiology have been studied for many years due to the global importance of crustaceans to the marine and freshwater ecosystems, as well as their potential of serving as a sustainable food source. With these respects, the IAG-switch is the mostly studied mechanism due to its pivotal role in crustacean sexual differentiation. However, until now, the IAG gene is the only proven key factor discovered within the IAG-switch. While previous studies focused on gonochoristic species, the novel approach of using a protandric shrimp as a model for a molecular sexual differentiation research, as described in the present study, yielded six novel significant candidate genes whose transcriptional pattern is strongly similar to that of IAG and conserved among other decapod species. We believe that further study of these genes will reveal additional pieces of the crustacean sexual differentiation puzzle and lead to better understanding of the IAG-switch mechanism.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The P. platyceros transcriptomic data is available from NCBI database under BioProject ID: PRJNA667922.
Author Contributions
TL, SLT, and AS conceived and designed the study. TL, SLT, RM, and EDA performed the research. TL and SLT collected and reared the animals for the study. MYS and VC-C performed all bioinformatics analyses. The manuscript was written by TL and reviewed and approved by all co-authors. All authors analyzed and interpreted the data.
Funding
This research was supported by a grant from the United States-Israel Binational Science Foundation (BSF) to ST and AS. Traveling to Alaska to collect the shrimp and perform the experiments described in this study was partially supported by the Graduate Student Fellowship Award No. GS-42-2018 (to TL) of the United States – Israel Binational Agricultural Research and Development (BARD) Fund, the Aharon and Ephraim Katzir study grant from the Batsheva de Rothschild Fund, the Israel Academy of Sciences and Humanities to TL, and the Prof. Rahamimoff Travel Grant for Young Scientists from the BSF to TL.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Alaska Department of Fish and Game (especially Quinn Smith and Karla Bush) for assisting us in the collection of the animals used in this study. Also, to Raz Kedem for tagging the shrimp and Jamie Musbach for helping with shrimp injections.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.587454/full#supplementary-material
Supplementary Figure 1 | ‘Regenerative-looking pleopod.’ (A) Post-molt second pleopod of a male whose original second pleopod was removed in the beginning of the long term Pnp-IAG silencing experiment. (B) Post-molt second pleopod of a male whose original second pleopod was not removed in the beginning of the same experiment. Appendix interna and appendix masculina are indicated by yellow and black arrows, respectively. Bar scales (= 1500 μm) are consistent in both photos.
Supplementary Figure 2 | Heatmaps of the DE genes in the androgenic gland. The figure represents a larger version of the heatmap given in Figure 5.
Supplementary Figure 3 | Heatmaps of the DE genes in the eyestalk. The figure represents a larger version of the heatmap given in Figure 5.
Supplementary Figure 4 | Heatmaps of the DE genes in the gonad. The figure represents a larger version of the heatmap given in Figure 5.
Supplementary Figure 5 | Heatmaps of the DE genes in the hepatopancreas. The figure represents a larger version of the heatmap given in Figure 5.
Supplementary Table 1 | Differentially expressed (DE) transcripts between males and females in P. platyceros. DE transcripts (P ≤ 0.05) from the transcriptomic library of P. platyceros (column A) and their read counts in each stage (male, transitional and female; columns B–G) are given in the excel file along with their linear fold change (column H), adjusted p-value (column I) and transcriptional pattern (down/up regulated; column J) between females and males. Blastp annotations are given in column K. Each sheet represents DE genes for a different tissue (Androgenic gland (AG), eye stalk (ES), gonad and hepatopancreas). Transcripts that were annotated as Pnp-IAG or Pnp-IAG-switch 1–6, in the AG data sheet, are marked in yellow.
Footnotes
References
Abehsera, S., Peles, S., Tynyakov, J., Bentov, S., Aflalo, E. D., Li, S., et al. (2017). MARS: a protein family involved in the formation of vertical skeletal elements. J. Struct. Biol. 198, 92–102. doi: 10.1016/j.jsb.2017.04.003
Abehsera, S., Zaccai, S., Mittelman, B., Glazer, L., Weil, S., Khalaila, I., et al. (2018). CPAP3 proteins in the mineralized cuticle of a decapod crustacean. Sci. Rep. 8:2430. doi: 10.1038/s41598-018-20835-x
Aflalo, E. D., Hoang, T. T. T., Nguyen, V. H., Lam, Q., Nguyen, D. M., Trinh, Q. S., et al. (2006). A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 256, 468–478. doi: 10.1016/j.aquaculture.2006.01.035
Anderson, P. J. (2000). Pandalid shrimp as indicators of ecosystem regime shift. J. Northw. Atlant. Fish. Sci. 27, 1–10. doi: 10.2960/j.v27.a1
Barr, L. (1973). Studies of spot shrimp, Pandalus platyceros, at Little Port Walter, Alaska. Mar. Fish. Rev. 35, 65–66.
Benvenuto, C., and Weeks, S. C. (2020). Hermaphroditism and gonochorism. Nat. Hist. Crustacea Reprod. Biol. 6:197.
Bergstrom, B. I. (2000). The biology of Pandalus. Adv. Mar. Biol. 38, 155–245. doi: 10.1016/S0065-2881(00)38003-8
Berkeley, A. A. (1930). The post-embryonic development of the common pandalids of British Columbia. Contrib. Can. Biol. Fish. 6, 79–163. doi: 10.1139/f31-006
Butler, T. H. (1964). Growth, reproduction, and distribution of pandalid shrimps in British Columbia. J. Fish. Board Can. 21, 1403–1452. doi: 10.1139/f64-121
Charniaux-Cotton, H. (1954). Discovery in, an amphipod crustacean (Orchestia gammarella) of an endocrine gland responsible for the differentiation of primary and secondary male sex characteristics. Comptes Rendus Hebdomadaires des Seances de l’Academie des Sciences 239, 780–782.
Charniaux-Cotton, H. (1958). Contrôle Hormonal De la Différenciation du Sexe et de la Reproduction Chez les Crustacés Supérieurs. Quebec: University of Quebec.
Charniaux-Cotton, H. (1962). Androgenic gland of crustaceans. Gen. Comp. Endocrinol. Suppl. 1, 241–247. doi: 10.1016/0016-6480(62)90095-3
Cronin, L. E. (1947). Anatomy and histology of the male reproductive system of Callinectes sapidus Rathbun. J. Morphol. 81, 209–239. doi: 10.1002/jmor.1050810205
Ghiselin, M. T. (1969). Evolution of hermaphroditism among animals. Q. Rev. Biol. 44, 189–208. doi: 10.1086/406066
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Herran, B., Geniez, S., Delaunay, C., Raimond, M., Lesobre, J., Bertaux, J., et al. (2020). The shutting down of the insulin pathway: a developmental window for Wolbachia load and feminization. Sci. Rep. 10:10551. doi: 10.1038/s41598-020-67428-1
Hoffman, D. L. (1969). Development of androgenic glands of a protandric shrimp. Biol. Bull. 137, 286–296. doi: 10.2307/1540101
Hoffman, D. L. (1972). Development of ovotestis and copulatory organs in a population of protandric shrimp, Pandalus platyceros Brandt from Lopez Sound, Washington. Biol. Bull. 142, 251–265. doi: 10.2307/1540229
Iversen, E. S., Allen, D. M., and Higman, J. B. (1993). Shrimp Capture and Culture Fisheries of the United States. New York, NY: Halsted Press.
Jiang, Q., Lu, B., Wang, G., and Ye, H. (2020). Transcriptional inhibition of Sp-IAG by crustacean female sex hormone in the mud crab, Scylla paramamosain. Int. J. Mol. Sci. 21:5300. doi: 10.3390/ijms21155300
Jin, S., Hu, Y., Fu, H., Jiang, S., Xiong, Y., Qiao, H., et al. (2019). Potential functions of Gem-associated protein 2-like isoform x1 in the oriental river prawn Macrobrachium nipponense: cloning, qPCR, in situ hybridization, and RNAi analysis. Int. J. Mol. Sci. 20:3995. doi: 10.3390/ijms20163995
Katakura, Y. (1960). Transformation of ovary into testis following implantation of androgenous glands in Armadillidium vulgare, an isopod crustacean. Annotationes Zool. Jpn. 33, 241–244.
Khalaila, I., Manor, R., Weil, S., Granot, Y., Keller, R., and Sagi, A. (2002). The eyestalk-androgenic gland-testis endocrine axis in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 127, 147–156. doi: 10.1016/s0016-6480(02)00031-x
Kimker, A., Donaldson, W., and Bechtol, W. R. (1996). Spot shrimp growth in Unakwik Inlet, Prince William Sound, Alaska. Alaska Fish. Res. Bull. 3, 1–8.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Levy, T., Aflalo, E. D., and Sagi, A. (2018). “Sex control in cultured decapod crustaceans,” in Sex Control in Aquaculture, eds H. Wang, S. Chen, Z. G. Shen, and F. Piferrer (Hoboken, NJ: John Wiley & Sons), 689–704. doi: 10.1002/9781119127291.ch35
Levy, T., Rosen, O., Eilam, B., Azulay, D., Aflalo, E. D., Manor, R., et al. (2016). A single injection of hypertrophied androgenic gland cells produces all-female aquaculture. Mar. Biotechnol. 18, 554–563. doi: 10.1007/s10126-016-9717-5
Levy, T., Rosen, O., Simons, O., Alkalay, A. S., and Sagi, A. (2017). The gene encoding the insulin-like androgenic gland hormone in an all-female parthenogenetic crayfish. PLoS One 12:e0189982. doi: 10.1371/journal.pone.0189982
Levy, T., and Sagi, A. (2020). The ’IAG-switch’ - a key controlling element in decapod crustacean sex differentiation. Front. Endocrinol. 11:651. doi: 10.3389/fendo.2020.00651
Levy, T., Tamone, S. L., Manor, R., Bower, E. D., and Sagi, A. (2020). The protandric life history of the Northern spot shrimp Pandalus platyceros: molecular insights and implications for fishery management. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-58262-6
Lezer, Y., Aflalo, E. D., Manor, R., Sharabi, O., Abilevich, L. K., and Sagi, A. (2015). On the safety of RNAi usage in aquaculture: the case of all-male prawn stocks generated through manipulation of the insulin-like androgenic gland hormone. Aquaculture 435, 157–166. doi: 10.1016/j.aquaculture.2014.09.040
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Li, F., Bai, H., Zhang, W., Fu, H., Jiang, F., Liang, G., et al. (2015). Cloning of genomic sequences of three crustacean hyperglycemic hormone superfamily genes and elucidation of their roles of regulating insulin-like androgenic gland hormone gene. Gene 561, 68–75. doi: 10.1016/j.gene.2015.02.012
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Malecha, S. R., Nevin, P. A., Ha, P., Barck, L. E., Lamadridrose, Y., Masuno, S., et al. (1992). Sex-ratios and sex-determination in progeny from crosses of surgically sex-reversed freshwater prawns, Macrobrachium rosenbergii. Aquaculture 105, 201–218. doi: 10.1016/0044-8486(92)90087-2
Manor, R., Weil, S., Oren, S., Glazer, L., Aflalo, E. D., Ventura, T., et al. (2007). Insulin and gender: an insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. Gen. Comp. Endocrinol. 150, 326–336. doi: 10.1016/j.ygcen.2006.09.006
Nagamine, C., and Knight, A. W. (1987). Masculinization of female crayfish, Procambarus clarki (Girard). Int. J. Invertebr. Reprod. Dev. 11, 77–87. doi: 10.1080/01688170.1987.10510268
Nagamine, C., Knight, A. W., Maggenti, A., and Paxman, G. (1980a). Effects of androgenic gland ablation on male primary and secondary sexual characteristics in the Malaysian prawn, Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae), with first evidence of induced feminization in a nonhermaphroditic decapod. Gen. Comp. Endocrinol. 41, 423–441. doi: 10.1016/0016-6480(80)90048-9
Nagamine, C., Knight, A. W., Maggenti, A., and Paxman, G. (1980b). Masculinization of female Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae) by androgenic gland implantation. Gen. Comp. Endocrinol. 41, 442–457. doi: 10.1016/0016-6480(80)90049-0
Okuno, A., Yang, W. J., Jayasankar, V., Saido-Sakanaka, H., Huong, D. T. T., Jasmani, S., et al. (2002). Deduced primary structure of vitellogenin in the giant freshwater prawn, Macrobrachium rosenbergii, and yolk processing during ovarian maturation. J. Exp. Zool. 292, 417–429. doi: 10.1002/jez.10083
Payen, G. (1969). Experiences de graffes de glandes androgenes sur la femelle pubere du Crabe Rhithropanopeus harrisii (Gould)(Crustace, Decapode). Comptes Rendus Hebdomadaires des Seances de l’Academie des Sciences 268, 393–396.
Rosen, O., Manor, R., Weil, S., Gafni, O., Linial, A., Aflalo, E. D., et al. (2010). A sexual shift induced by silencing of a single insulin-like gene in crayfish: ovarian upregulation and testicular degeneration. PLoS One 5:e15281. doi: 10.1371/journal.pone.0015281
Sagi, A., Cohen, D., and Milner, Y. (1990). Effect of androgenic gland ablation on morphotypic differentiation and sexual characteristics of male fresh-water prawns, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 77, 15–22. doi: 10.1016/0016-6480(90)90201-V
Sagi, A., Manor, R., Segall, C., Davis, C., and Khalaila, I. (2002). On intersexuality in the crayfish Cherax quadricarinatus: an inducible sexual plasticity model. Invertebr. Reprod. Dev. 41, 27–33. doi: 10.1080/07924259.2002.9652732
Savaya, A., De Leo, G., Aalto, E., Levy, T., Rosen, O., Manor, R., et al. (2020). The IAG gene in the invasive crayfish Procambarus clarkii – towards sex manipulations for biocontrol and aquaculture. Manag. Biol. Invasions 11, 237–258. doi: 10.3391/mbi.2020.11.2.05
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P., and Bork, P. (2000). SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. doi: 10.1093/nar/28.1.231
Sharabi, O., Manor, R., Weil, S., Aflalo, E. D., Lezer, Y., Levy, T., et al. (2015). Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 157, 928–941. doi: 10.1210/en.2015-1391
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Sklarz, M. Y., Levin, L., Gordon, M., and Chalifa-Caspi, V. (2018). Neatseq-flow: a lightweight high throughput sequencing workflow platform for non-programmers and programmers alike. BioRxiv [Preprint]. doi: 10.1101/173005
Smith, Q. T., and Gray, D. (2017). 2018 Report to the Board of Fisheries on Region 1 Shrimp Fisheries. Alaska: Alaska Department of Fish and Game, Division of Sport Fish, Research and Technical Services.
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Rosen, O., and Sagi, A. (2012). Timing sexual differentiation: full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 86, 1–6. doi: 10.1095/biolreprod.111.097261
Ventura, T., Rosen, O., and Sagi, A. (2011). From the discovery of the crustacean androgenic gland to the insulin-like hormone in six decades. Gen. Comp. Endocrinol. 173, 381–388. doi: 10.1016/j.ygcen.2011.05.018
Vogt, G. (2019). Functional cytology of the hepatopancreas of decapod crustaceans. J. Morphol. 280, 1405–1444. doi: 10.1002/jmor.21040
Wang, Y., Yu, Y., Li, S., Zhang, X., Xiang, J., and Li, F. (2020). Sex-specific transcriptome sequencing of zoea I larvae and identification of sex-linked genes using bulked segregant analysis in Pacific White shrimp Litopenaeus vannamei. Mar. Biotechnol. 22, 423–432. doi: 10.1007/s10126-020-09962-7
Zhang, D., Sun, M., and Liu, X. (2017). Phase-specific expression of an insulin-like androgenic gland factor in a marine shrimp Lysmata wurdemanni: implication for maintaining protandric simultaneous hermaphroditism. PLoS One 12:e0172782. doi: 10.1371/journal.pone.0172782
Keywords: androgenic gland, IAG-switch, insulin-like androgenic gland hormone, Pandalus platyceros, protandric hermaphrodite, reproductive physiology, sexual differentiation, spot shrimp
Citation: Levy T, Tamone SL, Manor R, Aflalo ED, Sklarz MY, Chalifa-Caspi V and Sagi A (2020) The IAG-Switch and Further Transcriptomic Insights Into Sexual Differentiation of a Protandric Shrimp. Front. Mar. Sci. 7:587454. doi: 10.3389/fmars.2020.587454
Received: 26 July 2020; Accepted: 12 October 2020;
Published: 28 October 2020.
Edited by:
Valerio Matozzo, University of Padua, ItalyReviewed by:
Haihui Ye, Xiamen University, ChinaHidekazu Katayama, Tokai University, Japan
Naoaki Tsutsui, Mie University, Japan
Copyright © 2020 Levy, Tamone, Manor, Aflalo, Sklarz, Chalifa-Caspi and Sagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sherry L. Tamone, c2x0YW1vbmVAYWxhc2thLmVkdQ==; Amir Sagi, c2FnaWFAYmd1LmFjLmls
†ORCID: Tom Levy, orcid.org/0000-0003-1484-0310; Sherry L. Tamone, orcid.org/0000-0002-7839-3567; Amir Sagi, orcid.org/0000-0002-4229-1059
 Tom Levy
Tom Levy Sherry L. Tamone2*†
Sherry L. Tamone2*† Vered Chalifa-Caspi
Vered Chalifa-Caspi Amir Sagi
Amir Sagi