- 1Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
- 2Department of Biology, Temple University, Philadelphia, PA, United States
- 3Department of Biological Sciences, Lehigh University, Bethlehem, PA, United States
Resource extraction from the ocean comes with ecosystem-wide risks, including threats to its biota such as the habitat forming corals that support elevated biomass and biodiversity. Despite catastrophic incidents like the Deepwater Horizon oil spill (DWHOS) disaster that occurred in 2010, offshore oil and gas drilling continues to occur around the world. Previous work investigating the toxicity of oil and the chemical dispersant used in an attempt to mitigate the effects of the DWHOS revealed that the dispersant elicits a stronger, negative physiological response than oil alone. However, little is known regarding the specific ways in which these anthropogenic pollutants impact organisms at the cellular level. To investigate the impacts of each pollutant and their synergistic effects on corals, we analyzed the transcriptional responses of the deep-sea octocorals Callogorgia delta and Paramuricea type B3 following 12 h of exposure to oil, dispersant, and mixtures of oil and dispersant. Analyses revealed that the highest levels of significant differential gene expression were found among the treatments containing dispersant, which corresponds to the significant effects observed in physiological experiments. Functional analyses of annotated transcripts further suggest both species- and colony-specific responses to the exposures, likely due to underlying cellular and physiological differences. However, some commonalities were observed among the responses to chemical stress across treatments and species, including immune and cellular stress responses, altered energy metabolism, and oxidative stress, elucidating how corals respond to chemical pollutants. As resource extraction is an ongoing threat, this study demonstrates the importance of considering the varied and diverse responses of biota to anthropogenic disturbances and the implications of introducing chemicals into vulnerable ecosystems like those associated with deep-sea corals.
Introduction
Marine ecosystems are increasingly threatened by resource extraction as oil and gas exploration expands offshore. Despite catastrophic incidents like the Deepwater Horizon oil spill (DWHOS) disaster which occurred in the Gulf of Mexico (GoM) in 2010, drilling continues in deep waters [>200 meters (m)] worldwide, with active ultra-deep (>1000 m) drilling advancing in the GoM (Fisher et al., 2014b; Cordes et al., 2016) and beyond. Drilling activities can be detrimental to deep-sea communities in various ways including physical disturbances by mooring anchors and pipelines (Ulfsnes et al., 2013), exposure to toxic drill cuttings, muds, and fluids (Gray et al., 1990; Larsson et al., 2013), and accidental oil spills. In the United States alone there was an oil spill over 160,000 L on average every 1.75 years between 1971 and 2010 (Anderson et al., 2012). Oil and gas extraction in deeper waters also increases the likelihood of accidental spills (Muehlenbachs et al., 2013) like the DWHOS blowout of the Macondo well that occurred at approximately 1500 m depth. This was one of the worst environmental disasters in U.S. history, and the largest accidental oil spill world-wide, releasing approximately 5 million barrels of crude oil into the deep waters of the GoM (McNutt et al., 2012). During active well discharge, approximately 7 million liters of chemical dispersants (Corexit 9500A and 9527; Barron, 2012) were also applied in an attempt to mitigate the impacts of the oil release, both at the sea surface and at depth, without a comprehensive understanding of how these chemicals would affect deep-sea fauna.
Spill impacted deep-sea coral communities were initially found at a depth of 1370 m, approximately 11 km from the Macondo well at a site in the Mississippi Canyon (MC) 294 lease block (White et al., 2012). When first discovered, coral colonies were covered in a brown flocculent material (floc) containing Macondo well oil as well as dispersant constituents (White et al., 2014). Impacted coral species at this site, primarily the deep-sea octocoral Paramuricea biscaya, exhibited various phenotypic indicators of impact including abnormal skeletal (sclerite) development, excess mucous production, and tissue death (necrosis), as well as complete colony mortality (White et al., 2012). Corals showing signs of sublethal impacts exhibited subsequent declines in health (i.e., tissue and branch loss) and limited recovery potential (Hsing et al., 2013; Girard and Fisher, 2018). Additional impacted deep-sea coral communities were later identified based on their similarity in appearance to the impacted communities at the original site (Fisher et al., 2014a, b).
A recent study investigating the in situ transcriptional response of P. biscaya colonies collected in December 2010 at MC294, revealed further impacts at the gene expression level (DeLeo et al., 2018). These included elevated expression of genes associated with oxidative, immune, and metabolic stress responses, as well as wound repair mechanisms (i.e., hyper-melanization). Contrastingly, expression of the polycyclic aromatic hydrocarbon biomarker gene, cytochrome P450 (CYP) was depressed, which may indicate an inhibition of the corals’ ability to process xenobiotics at the time of sampling. Further, the study revealed variability among the global expression patterns of the individual spill-impacted colonies (DeLeo et al., 2018).
Because P. biscaya occurs in the GoM between 1200 and 2600 m and rarely survives the trip to the surface, it is not amenable to controlled laboratory experiments aimed at testing the toxicity of crude oil and dispersants. A close relative of P. biscaya, Paramuricea type B3 (Doughty et al., 2014), is also found in the GoM at shallower depths between 800 and 1100 m. This species can survive careful collection and thus serves as the best available model to understand toxicity effects underlying the impacts observed in P. biscaya.
Laboratory toxicity tests with P. type B3, and with the more distantly related octocoral Callogorgia delta, revealed that exposure to dispersant or dispersant/crude oil mixtures is more toxic to these corals than is exposure to crude oil alone (DeLeo et al., 2016). This was particularly evident following exposure to the dissolved, water-accommodated fractions (WAFs) of the dispersants. Phenotypic responses observed in P. type B3 were similar to those observed in P. biscaya at MC294, including excess mucous release, tissue damage, necrosis and morality (DeLeo et al., 2016). The octocoral C. delta however exhibited less severe health declines than P. type B3, suggestive of taxon-specific responses.
The objective of this study was to investigate the transcriptional responses of the corals subjected to the laboratory toxicity tests performed by DeLeo et al. (2016) and compare them to the in situ transcriptional responses of P. biscaya (DeLeo et al., 2018). It is important to investigate the early transcriptional responses to oil, dispersant, and oil/dispersant mixtures in these corals, prior to the onset of visible damage and mortality in order to understand how deep-sea corals respond to, and potentially endure, acute pollution events from marine oil spills. The gene expression patterns of P. type B3 and C. delta were examined after 12 h of exposure to (1) bulk oil and dispersant treatments (heterogeneous solutions, with dissolved and undissolved chemical components), and (2) treatments solely containing the dissolved, or WAF of oil and dispersant. As higher mortality was observed in the WAF exposures during the 96 h toxicity tests, we anticipated a greater degree of differential gene expression in response to the short-term WAF exposures, corresponding to more severely impacted biological processes. Expression profiles were analyzed to investigate the effect of treatment type – oil-only, dispersant-only, and oil/dispersant mixtures – on both coral species. We hypothesized that the corals would have a more pronounced genome-wide response following exposure to dispersant, relative to the crude oil, given the strong physiological response observed in the toxicity tests and the natural occurrence of crude oil in their habitat (Quattrini et al., 2013). Gene expression data were further explored to identify early onset genome-wide impacts underlying the health declines and mortality observed during the full 96 h toxicological assays described by DeLeo et al. (2016). These findings improve our understanding of how resource extraction impacts deep-sea biota, and will aid in the development of diagnostic biomarkers for future spill monitoring efforts.
Materials and Methods
Sample Collection and Acclimation
All samples were collected from two sites in the GoM. P. type B3 colonies were collected from a large population of corals at approximately 1050 m depth at Atwater Valley (AT) 357 (27°58.6′N, 89°70.4′W; Doughty et al., 2014). C. delta was collected from the Viosca Knoll (VK) 826 site at a depth of approximately 500 m (29°09.5′N, 88°01.0′W; Cordes et al., 2008; Davies et al., 2010). At each site, corals were haphazardly collected with the remotely operated vehicles (ROV) Global Explorer MK3 or Hercules for experimentation. For each species, five coral colonies were collected alive for the experimental exposures described in DeLeo et al. (2016). Three of the largest colonies were fragmented for use in this study. Samples were collected several meters apart from conspecific colonies to reduce the likelihood of sampling genetic clones. Corals were visually identified using live video from cameras attached to each ROV, before being collected with a manipulator arm and secured in an insulated biobox and/or sealable collection quivers. When possible, branches of large colonies were sampled to avoid whole-colony mortality. Collection of individual branches from large coral colonies produces little or no phenotypic response from the coral colony (EEC unpublished data). Additional coral specimens from the same sites, previously preserved in situ on the seafloor using collection quivers filled with RNALater, were used for RNA sequencing. This included fragments from two P. type B3 (2011, ROV Schilling UHD) and four C. delta colonies (2010, ROV Jason II).
At the surface, live corals were immediately transferred to containers with filtered seawater of the species-appropriate temperature and salinity (35 psu). P. type B3 was maintained at approximately 5°C and C. delta at 8°C (the average in situ temperatures at depth) for the duration of the experiment. Temperature in the holding vessels was continuously monitored using temperature probes (Hobo® Data Loggers). Corals were allowed to acclimate for approximately 12 h prior to experimentation.
Sublethal Oil and Dispersant Exposures
Partial coral colonies (n = 3 genets per species, per exposure series; six genets total) were fragmented into nubbins and exposed to oil and dispersant solutions (Figure 1) prepared according to the protocols described in DeLeo et al. (2016). In brief, corals were exposed to treatments made using either (1) heterogenous bulk oil, dispersant and oil-dispersant mixtures containing both dissolved and undissolved chemical components, hereafter referred to as the Bulk-exposure series, or (2) using only the dissolved or water-accommodated oil (and dispersant) fractions (WAF), referred to as the WAF-exposure series. In each set of exposures, fragments were placed in the following treatments: artificial seawater (ASW; control), oil-only, dispersant-only or an oil/dispersant mixture (to represent dispersed oil). ASW was chosen as the control for comparison to the oil and dispersant treatments as all mixtures (Bulk and WAF) were made using sterile ASW (made using Instant OceanTM at the in situ salinity of 35 psu). Coral nubbins were exposed to sublethal concentrations of oil and dispersant for a duration of 12 h and preserved prior to the onset of visible tissue damage. For the Bulk-exposure series this corresponds to the “High” concentration of oil and dispersant (∼25 ppm) described by DeLeo et al. (2016), and for the WAF-exposure series this corresponds to the “Low” concentration of oil (∼50 μM) and dispersant (∼35.3 mg/L). Nubbins in these concentrations were chosen for sequencing for several reasons: (1) there were clear phenotypic differences observed across treatments during the full 96 h exposures implying the mixtures elicited a physiological response, (2) the concentrations were deemed sublethal as severe stress responses and mortality were not induced across all treatment-types within those 96 h, and (3) to minimize the differences between the target concentrations of the Bulk and WAF exposure series to facilitate comparisons. After the 12 h exposures, fragments were fixed in RNAlater to preserve changes in gene expression. Samples were then frozen and returned to the lab at Temple University for further processing.
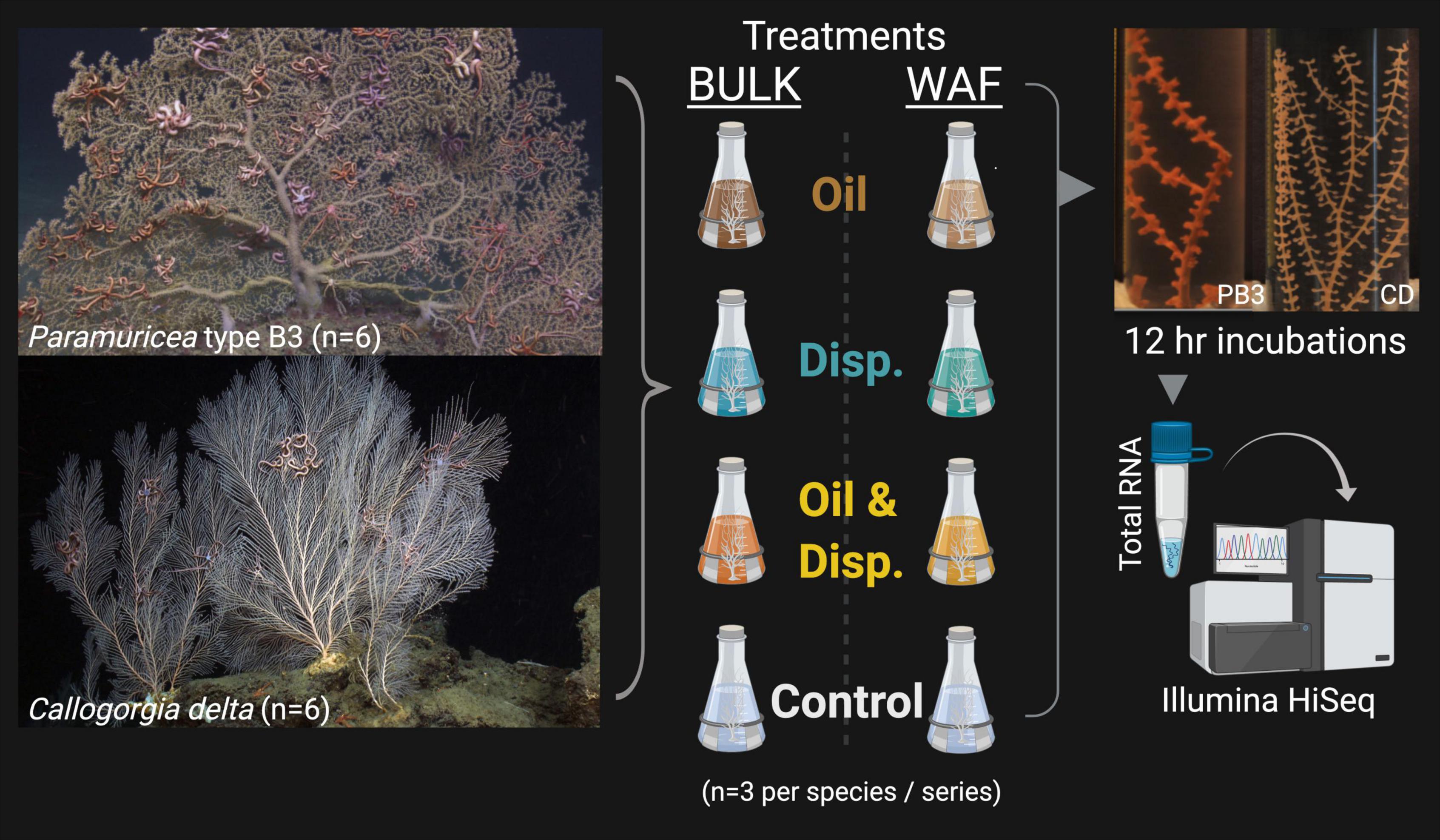
Figure 1. Experimental setup for the deep-sea octocorals Paramuricea type B3 (PB3) and Callogorgia delta (CD) (n = 6 genets total per species, 3 genets per exposure series). Corals were exposed to both Bulk (heterogenous) and Water-Accommodated Fractions (WAF) of oil and/or dispersant (center). Coral fragments were maintained at sub-lethal concentrations for 12 h, as determined in DeLeo et al. (2016) (right). For the Bulk exposure series, oil and dispersant concentrations each correspond to ∼25 ppm. For the WAF exposure series, oil concentrations correspond to ∼50 μM and dispersant ∼35.3 mg/L. Control coral fragments were also incubated in the artificial seawater used to make the aforementioned treatments of each exposure series. Fragments were preserved (RNAlater) after 12 h incubations in the treatments, for total RNA extraction and sequencing (1 × 100 bp). Created with BioRender.com.
Sample Sequencing and Processing
Total RNA was isolated using a Qiagen RNeasy Kit or a modified Trizol/Qiagen RNeasy protocol (described in Polato et al., 2010; Burge et al., 2013), from both the in situ preserved samples and the experimental nubbins sampled after 12 h of exposure. RNA concentrations were determined using a NanoDrop® ND-1000 and RNA integrity was evaluated with gel electrophoresis and an Agilent Bioanalyzer. Library preparations, including a poly-A selection step to target primarily eukaryotic (coral host) RNAs, were performed by the sequencing facilities on high quality (RIN values > 7) RNA samples (Illumina Truseq RNA library kit). In situ preserved specimens were sequenced on an Illumina HiSeq2000 at the University of Wisconsin-Madison Biotechnology Center (UWBC, Madison WI, United States) to acquire 150 base-pair (bp) paired-end reads for use in assembling de novo reference transcriptomes for downstream analyses. The experimental samples were further multiplexed and sequenced across Illumina HiSeq2500 rapid flow cells at the Fox Chase Cancer Center (FCCC, Philadelphia, PA, United States) sequencing facility to acquire 100 bp single-end reads for use in gene expression analyses, at approximately 20–30 million reads per sample.
Raw reads were quality checked using FastQC v0.11.5 (Andrews, 2010). This output was further used to find overrepresented sequences, which were queried against GenBank using Blastx (e-value: e–15) to determine whether they represented bacterial contamination. Trimmomatic v0.36 (Bolger et al., 2014) was then used to filter for quality, with a Phred score threshold of 30, leading and trailing bases dropped if the score was below 3 (LEADING: 3 TRAILING: 3), and excluding reads shorter than 50 bp (MINLEN:50). Cutadapt v1.15 (Martin, 2011) was used to trim adaptors and cut overrepresented sequences that appeared to be bacterial contamination with an overlap of 10 and a minimum-length of 60.
Transcriptome Assemblies
The filtered and cut paired-end sequences were further processed with Rcorrector (Song and Florea, 2015) using the k-mer counter Jellyfish v2.2.6 (Marcais and Kingsford, 2012), to remove erroneous k-mers. Read pairs deemed unfixable were discarded. Microbial contaminants were further identified and removed using the standard database in Kraken v1.0 (Wood and Salzberg, 2014). Sequences not classified as bacterial, microbial, archaeal, or viral in Kraken were used to generate de novo reference assemblies with Trinity v2.4.0 (Grabherr et al., 2011). Transcriptome quality was assessed with Transrate v1.0.3 (Smith-Unna et al., 2016) and the Benchmarking Universal Single-Copy Orthologs (BUSCO v3.0.2) program (Simão et al., 2015; Zdobnov et al., 2017) using the metazoa dataset (n = 978 orthologs). Assemblies were further annotated via dammit v1.01 using the corresponding P-FamA, RFam, OrthoDB, BUSCO, and Uniref90 databases (Scott, 2016). Gene ontology (GO) annotations were also generated using standard parameters (seed ortholog e-value: 0.01; seed ortholog score: 60; min% of query cov.: 20; min% of subject cov.: 0) in eggNOG (Huerta-Cepas et al., 2016) using Github support scripts2.
Gene Expression Analyses
Processed reads from P. type B3 and C. delta experimental samples were mapped to their respective reference transcriptomes using Salmon (Patro et al., 2017). The resulting count files were used as input for DESeq2 (Love et al., 2014), using R v3.6.2, to analyze the gene expression patterns of the corals in each exposure series (Bulk or WAF) and treatment. Expression was compared to the corresponding ASW control (of each species) using the tximport package (Soneson et al., 2015), which obtains gene-level expression by summing all transcript-level count estimates corresponding to a given gene. Transcripts were collapsed into genes for analysis at the gene level with a DESeq dataset design of colony + treatment, which was applied discretely to each exposure series. Transcripts were considered over-expressed if they had an adjusted p-value < 0.05 and a log-2 fold change (lfc) > 1, and under-expressed if they had an adjusted p-value of <0.05 and a lfc < –1. An adonis (permutational multivariate analysis of variance using distance matrices) test, implemented using the Vegan package in R, was used to analyze the influence of each factor (colony, treatment, and the interaction of colony and treatment) on the variation in gene expression (Oksanen et al., 2013). A Principal Coordinate Analysis (PCoA) was also carried out to visualize how the samples grouped in multivariate space, and whether treatment-type and/or colony genet could explain their distribution.
KEGG Functional Analyses
In order to further understand the influence of each treatment on higher-level functions and biological processes, differentially expressed genes from each treatment were assessed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Mapper v3.1 tool (Kanehisa et al., 2016). This was done to decipher the molecular interaction networks associated with the genes (KEGG pathway mapping; reconstruct), based on all available KEGG Orthology (KO) assignments designated by eggNOG (Huerta-Cepas et al., 2016). These data were compared to investigate cellular processes and pathways that were impacted in both species in the Bulk and WAF exposures and to elucidate species-specific differences in the early (12 h) responses to pollutant introduction.
In order to compare the initial responses of the experimentally exposed corals to corals impacted by the DWHOS in situ, direct comparisons were made between the closely related octocorals P. type B3 and P. biscaya. This was done by comparing the KEGG pathway annotations for the genes differentially expressed in this study to the KO assignments for genes differentially expressed among P. biscaya exposed to DWHOS oil and dispersant laden floc in situ (DeLeo et al., 2018) via KEGG mapper reconstruct. While KEGG pathways are comprised of various components, each with unique KO identifiers, only the overlapping KO identifiers were reported as shared impacts observed for Paramuricea spp.
Gene Ontology Analysis
To examine significantly enriched GO categories among the over- and under-expressed genes that correspond to either the biological process, molecular function, or cellular component sub-ontologies, a rank-based gene ontology (GO) analysis with adaptive clustering was performed using the Gene Ontology Mann–Whitney U (GO_MWU) program (Wright et al., 2015). This program utilizes Mann–Whitney U (MWU) tests and a global-ranked list of genes and their associated GO terms to identify enriched GO categories using a continuous measure of significance, –log(p-value), calculated with DESeq2. Hierarchical clustering of GO categories was based on the number of shared genes (clusterCutHeight = 0.25). Discrete species-specific tests were carried out for the Bulk and WAF exposure series to analyze expression patterns related to all treatments (the factor “treatment” in the DESeq design matrix) as well as each treatment individually. GO terms significantly enriched among the over- (p.adj < 0.05, delta rank > 0) or under-expressed (p.adj < 0.05, delta rank < 0) transcripts were reported.
Results
De novo Transcriptome Assemblies
The final de novo transcriptome assembly for C. delta contained 39,475 transcripts with a contig N50 of 1,467 bases (Table 1). Of the assembled contigs, 94.9% of metazoan orthologs (BUSCOs) were present, indicating a relatively complete reference assembly. The final reference transcriptome for P. type B3 contained 60,989 transcripts with a N50 of 1,451 bases (Table 1). Among these contigs, 94.5% of metazoan BUSCOs were present. The TransRate scores were comparable to the optimal assembly scores, which considers only the “good” transcripts based on a learned cutoff value.
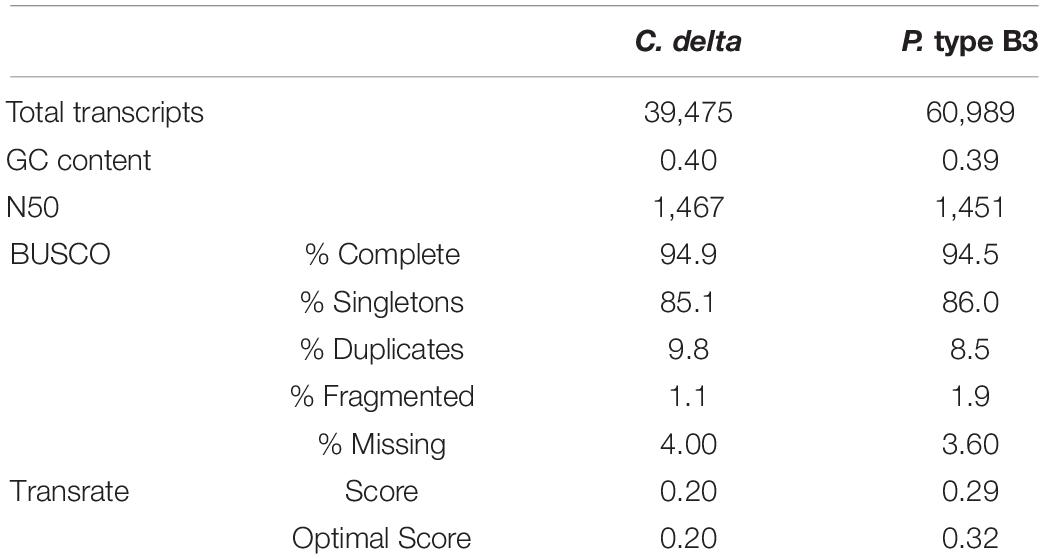
Table 1. Summary statistics for the de novo transcriptome assemblies of Callogorgia delta and Paramuricea type B3.
Gene Expression Analyses
The adonis analysis, conducted to investigate the influence of each factor – colony (or genet), treatment, and colony + treatment – on the variation in gene expression, was not significant. However, in both exposure series, “colony” explained the largest proportion of the variation in global gene expression patterns. Further, when analyzing global expression patterns using the PCoA, the samples grouped largely by colony (Figure 2).
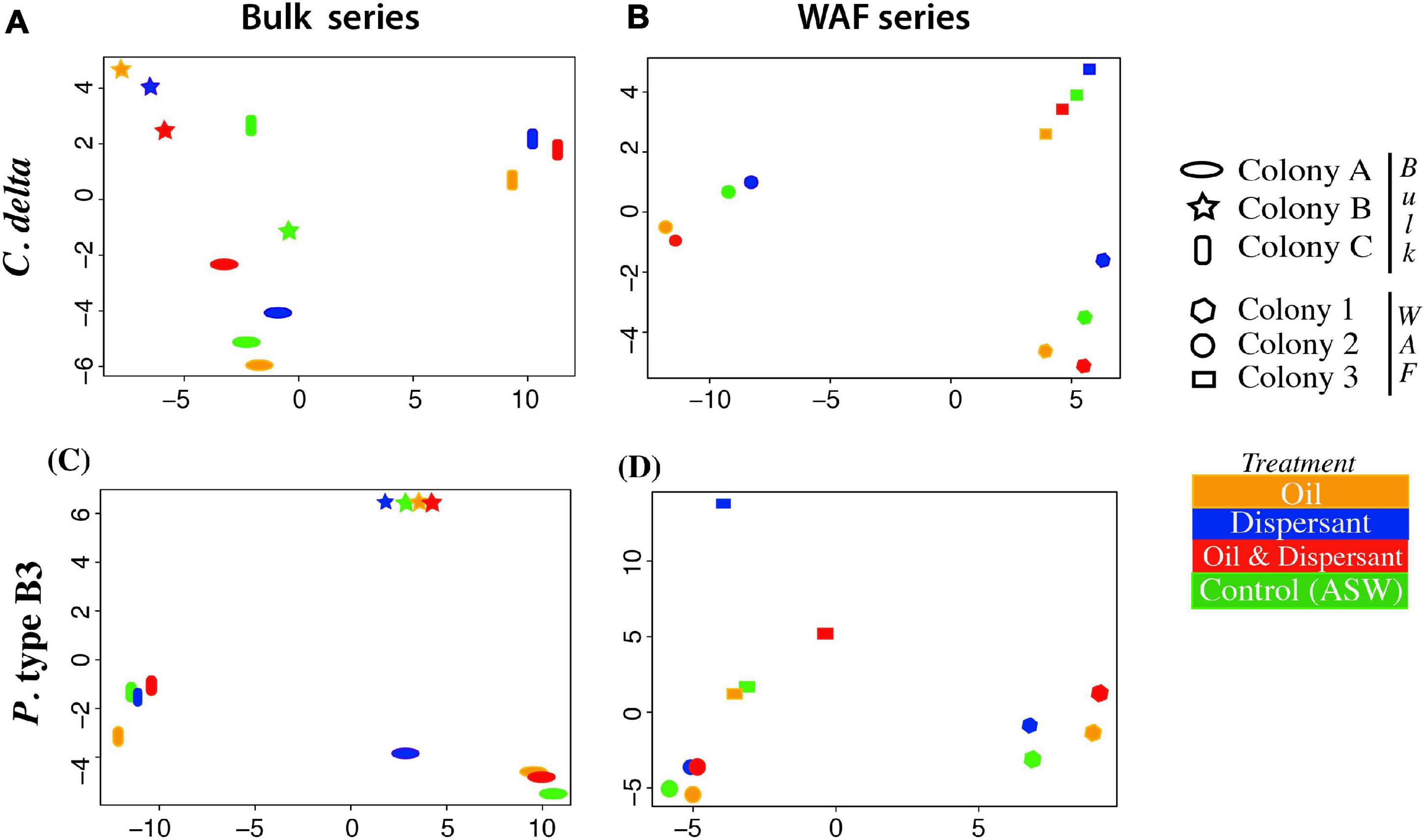
Figure 2. Principal coordinate analysis estimated from genome-wide transcript expression patterns (DESeq2) from C. delta in (A) bulk oil treatments and (B) WAF treatments, and P. type B3 in (C) bulk oil treatments and (D) WAF treatments. Colors represent treatment and shape represent colony/genet.
Bulk Exposure Series
After 12 h in the Bulk exposure series, the greatest number of significant differentially expressed genes (adj. p-value < 0.05, absolute lfc > 1) was observed among P. type B3, with a total of 140 genes differentially expressed relative to the 66 genes differentially expressed in C. delta (Table 2 and Supplementary Table 1). For both species, the highest differential expression response was observed in the Bulk dispersant-only treatment. P. type B3 had 2.9–34 times as many differentially expressed transcripts in the dispersant-only treatment, relative to the oil/dispersant mixture and oil-only treatments, respectively; C. delta had 4.8–6.0 times as many differentially expressed transcripts in the respective treatments. See Supplemental Information for more detailed results on gene expression and functional analyses.
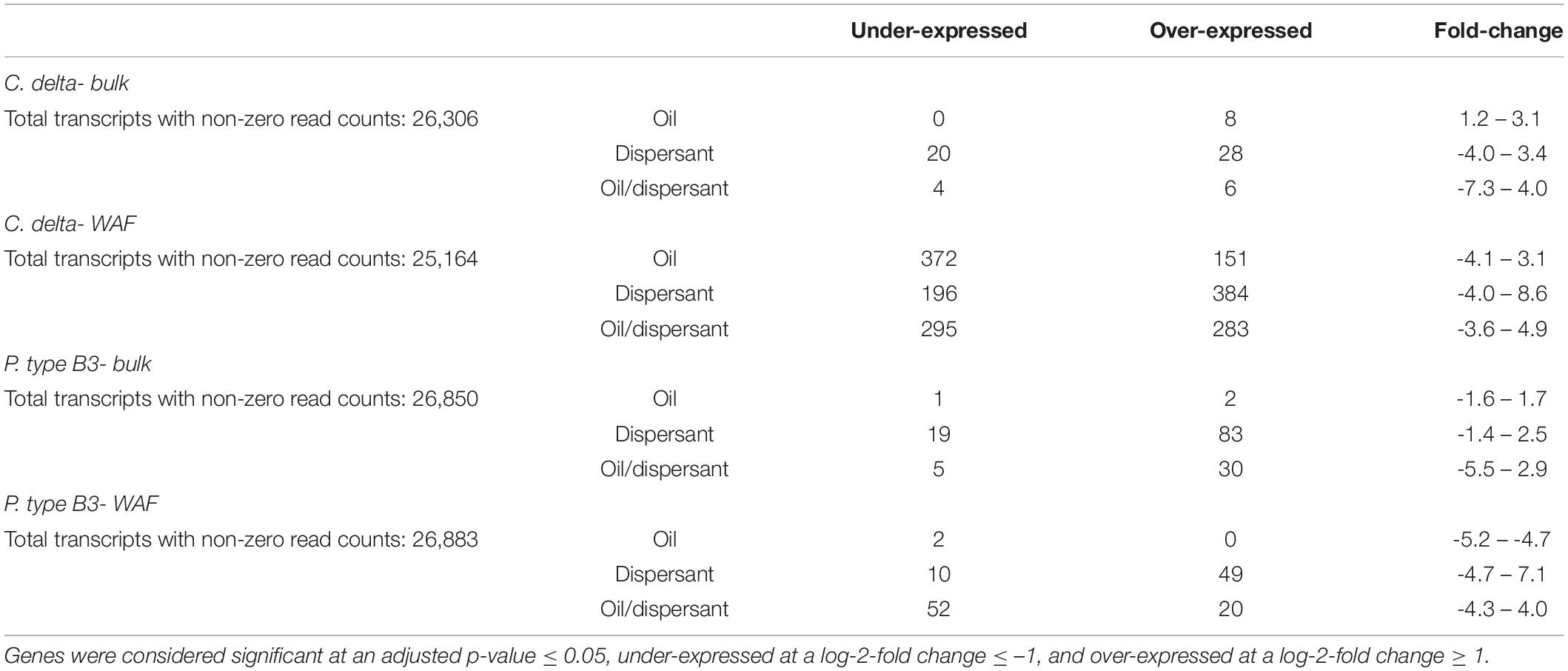
Table 2. Summary of the differentially expressed genes in C. delta and P. type B3 by treatment – oil, dispersant, and oil and dispersant mixtures- relative to control expression (seawater) in both the WAF (dissolved) and Bulk (heterogenous dissolved/undissolved) exposure series.
Among the differentially expressed genes in both octocorals, 2 putative genes (annotated as cartilage matrix protein-like and uncharacterized protein) were shared between C. delta and P. type B3 exposed to the Bulk-dispersant treatment (Supplementary Table 1). No putative genes with annotations were shared among the two species in the Bulk-oil or oil/dispersant mixture treatments.
WAF Exposure Series
After 12 h in the WAF exposure series, the greatest number of significant differentially expressed genes (adj. p-value < 0.05, absolute lfc > 1) was observed among C. delta with a total of 1,681 genes differentially expressed relative to the 133 genes differentially expressed in P. type B3 (Table 2 and Supplementary Table 2). For C. delta, the highest differential expression response was observed in the dispersant-only treatment, though this number was comparable across treatments with at most 1.1 times as many differentially expressed transcripts relative to the oil/dispersant mixture and oil-only treatments. For P. type B3, the highest differential expression response was observed in the WAF oil/dispersant mixture treatment, with 1.2–36 times as many differentially expressed transcripts relative to the dispersant-only and oil-only treatments.
Among the differentially expressed genes, one putative gene (annotated as lactoperoxidase-like), was shared between C. delta and P. type B3 exposed to the WAF-oil treatment, with an additional 14 putative genes shared among the two species in the WAF oil/dispersant mixture treatment, including but not limited to: a Slit homolog 1 protein involved in negative chemotaxis, a metalloendopeptidase involved in wound repair pathways, the growth factor bone morphogenetic protein 6 (BMP6) involved in skeletogenesis and oogenesis, the interferon-induced protein (IFI35) involved in innate immune responses, and the tumor suppressor RNA-binding protein (RBM47) that responds to DNA damage (Supplementary Table 2). No putative genes were shared among the WAF dispersant-only treatment after 12 h of exposure.
KEGG and GO Functional Analyses
Bulk Exposure Series
KEGG pathway maps revealed that the genes differentially expressed among both coral species in the Bulk dispersant-only treatment were associated with: the immune (complement and coagulation cascades) and digestive (protein digestion and absorption) systems, environmental information processing (intracellular signaling and ECM-receptor interactions), and cellular processes (focal adhesion) (Table 3 and Supplementary Table 3). A response was also elicited in these pathways among P. type B3 fragments in the bulk oil/dispersant treatment, in addition to several metabolic pathways (e.g., xenobiotic biodegradation and metabolism). There were no KEGG assignments associated with the genes differentially expressed in the Bulk oil-only treatment or for C. delta exposed to oil/dispersant.
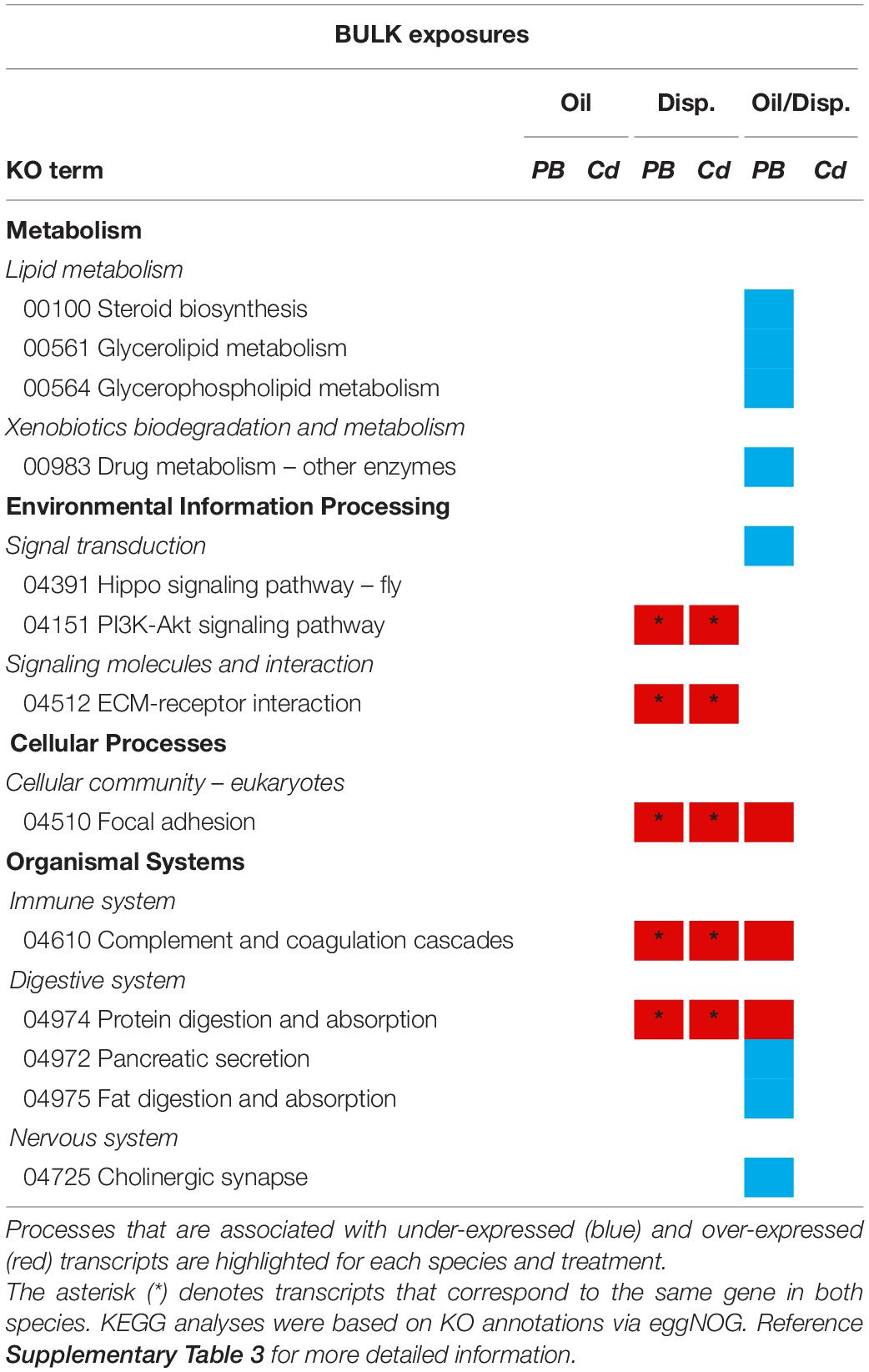
Table 3. Summary of the KEGG pathway analyses for Paramuricea type B3 (PB) and Callogorgia delta (Cd) in the Bulk oil and dispersant exposure series.
There were 104 enriched GO terms shared among both octocorals exposed to the Bulk dispersant treatment (Figure 3 and Supplementary Table 4). Among the shared GO terms were “innate immune responses,” “response to tumor necrosis factor (TNF),” and “regulation of immune effector process,” further revealing elevated immune responses among C. delta and P. type B3 fragments exposed to dispersant. C. delta exposed to the oil and oil/dispersant treatments had additional enrichment of unique immunity-related GO terms including “positive regulation of immune system process” and “negative regulation of wound healing” (Supplementary Table 4). Shared GO enrichment analyses revealed additional impacts to cellular processes including, but not limited to, elevated “response to oxidative stress” and “regulation of apoptotic signaling pathway” and diminished “DNA repair.” There were no GO terms shared among C. delta and P. type B3 in the Bulk oil-only and oil/dispersant treatments. See Supplementary Table 4 for additional details on GO enrichment and for details on the differences between species.
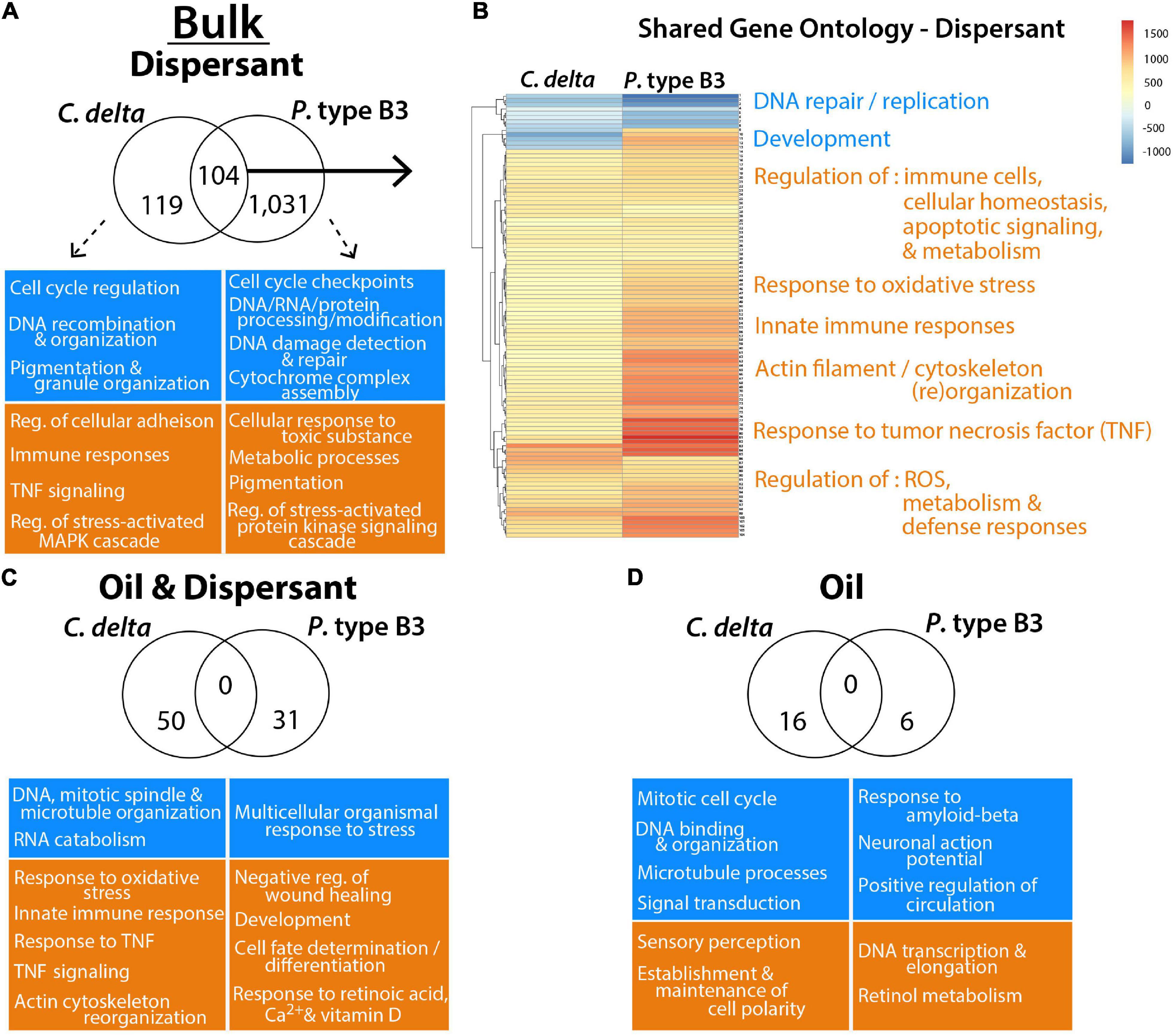
Figure 3. Gene Ontology (GO) terms significantly enriched for over- (orange) or under- (blue) expressed transcripts from C. delta and P. type B3 experimentally exposed to bulk oil and dispersant. Highlighted GO terms represent a snapshot summary of some of the impacted processes/functions either unique to (A,C,D) or shared (B) among species in relation to the dispersant (A,B), oil and dispersant mixture (C) or oil-only (D) treatments. The Venn diagrams represent the total number of shared and unique GO terms for each treatment, with representative GO terms of interest, unique to each species, highlighted in the boxes underneath. (B) The heatmap of the shared GO terms from the dispersant exposure illustrates the corresponding level of expression for each term (row) and compares expression between each coral species (column). Processes (orange/blue) are summarized to highlight impacted biological pathways. More details on GO enrichment can be found in Supplementary Table 4, including row-specific heatmap terms (B).
WAF Exposure Series
KEGG pathway maps indicated that the gene differentially expressed among both coral species in the WAF oil-only exposures was associated with metabolism (peroxidase) (Table 4 and Supplementary Table 5). Common impacts were also observed in the WAF oil/dispersant treatment, which were further associated with genetic information processing (ribosome), environmental information processing (signal transduction), cellular processes (cell growth and death), and organismal systems (development and regeneration).
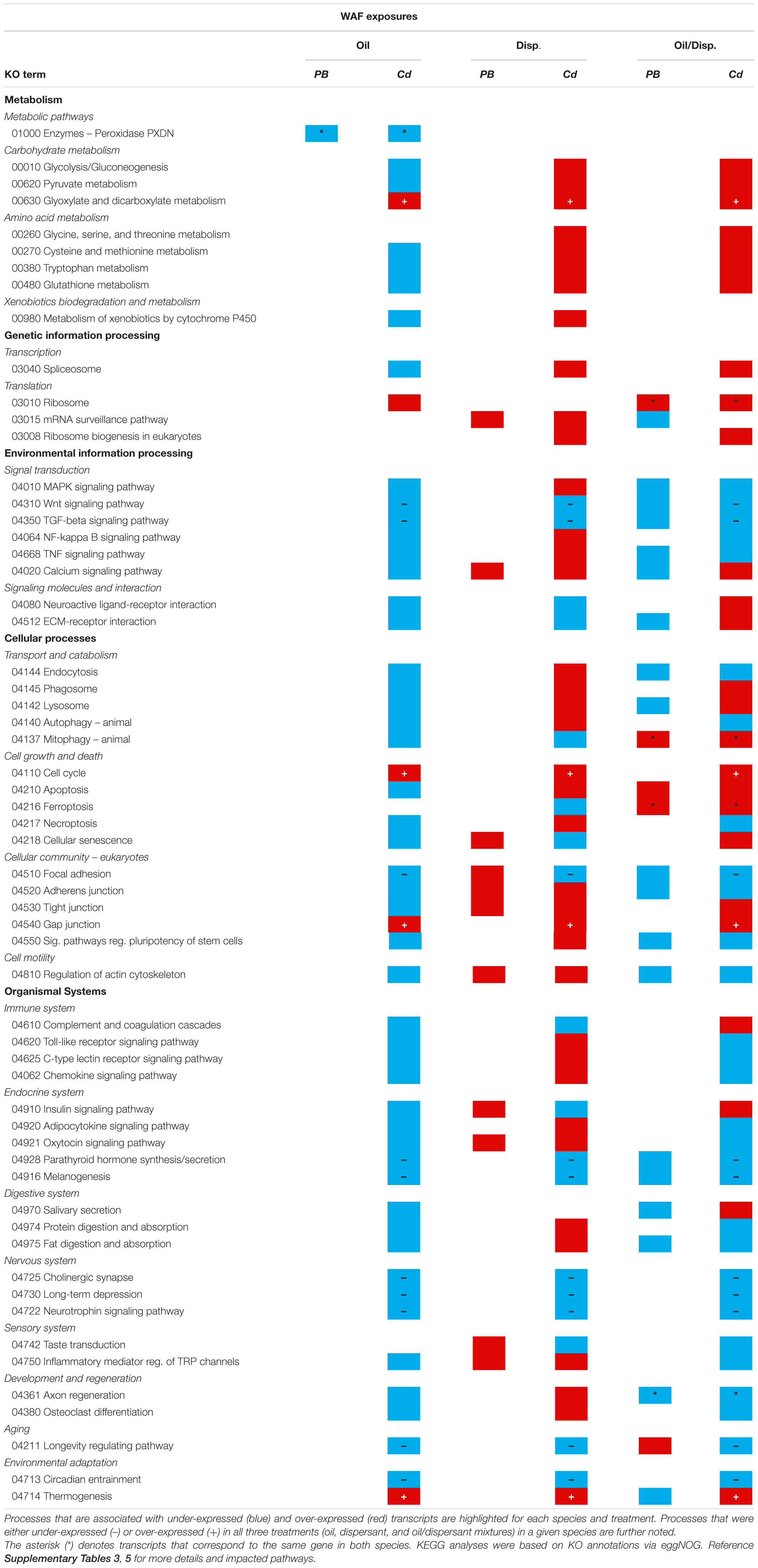
Table 4. Snapshot summary of the KEGG pathway analyses for Paramuricea type B3 (PB) and Callogorgia delta (Cd) in the WAF oil and dispersant exposure series.
Among C. delta nubbins in the WAF exposure series, several KEGG pathways were similarly impacted by all three treatments, potentially indicative of a generalized, early (∼12 h) response to anthropogenic pollutants. These pathways fell into four main categories: metabolism (carbohydrate metabolism), environmental information processing (signal transduction), cellular processes (cell growth/death and focal adhesion), and organismal systems (including the immune, endocrine and nervous systems, and environmental adaptation) (Table 4 and Supplementary Table 5). KEGG analyses further revealed common impacts to crucial cellular stress response pathways (e.g., NF-kappa B, TNF, and chemokine signaling pathways) among the octocorals exposed to oil (alone or dispersed). Additional sub-lethal impacts were observed among corals exposed to chemical dispersant components (alone or with oil) including elevated calcium signaling and apoptotic processes. See Supplementary Table 3 for full details on the KEGG analyses and Supplementary Information for detailed comparisons between treatments.
There were 10 GO terms shared among C. delta and P. type B3 in the WAF oil treatment, 64 terms shared in the dispersant-only treatment and 435 terms shared among the oil/dispersant mixtures (Figure 4). GO terms shared among the corals in treatments containing dispersant (alone or with oil) include “pigment cell differentiation,” “regulation of cytokine production,” “mucosal immune response,” and “regulation of inflammatory response,” which further revealed impairments to immune responses after the 12 h exposures. GO enrichment analyses uncovered additional impacts to cellular processes following dispersant exposure including, but not limited to, elevated “ribosomal biogenesis” and “DNA repair/modifications” (Figure 4 and Supplementary Table 4).
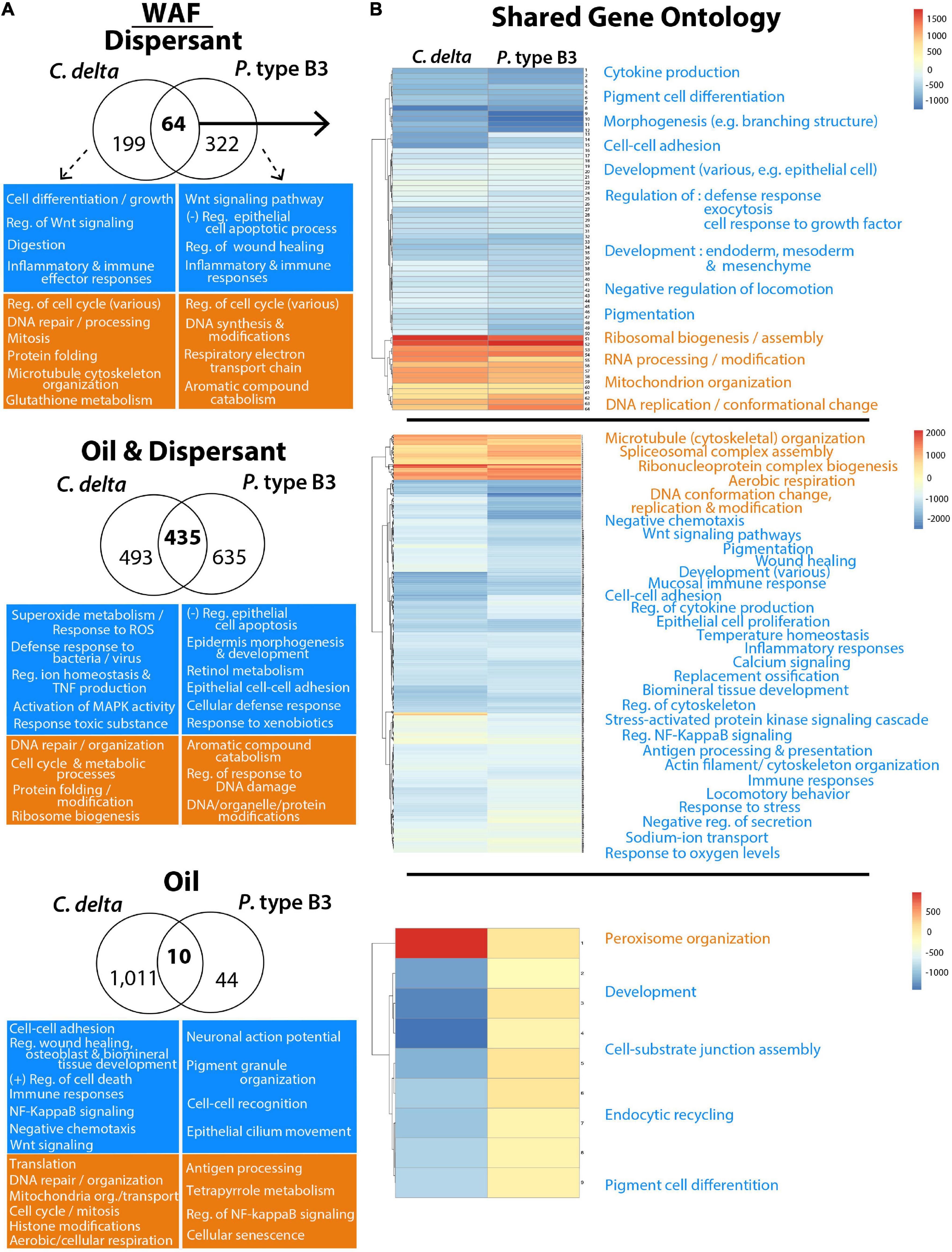
Figure 4. Gene Ontology (GO) terms significantly enriched for over- (orange) or under- (blue) expressed transcripts from C. delta and P. type B3 experimentally exposed to WAF oil and dispersant. Highlighted GO terms represent a snapshot of some of the impacted processes/functions either unique to (A) or shared (B) among species in relation to the dispersant (top), oil and dispersant mixture (middle), or oil-only (bottom) treatments. The Venn diagrams (A) represent the total number of shared and unique GO terms for each species in the different treatments, with representative GO terms of interest, unique to each species, highlighted in the boxes underneath. (B) The heatmaps of the shared GO terms from each treatment illustrates the corresponding level of expression for each term (rows) and compares expression between each coral species (column). Processes (orange/blue) are summarized to highlight various impacted biological pathways. More details on GO enrichment can be found in Supplementary Table 4.
Shared Impacts: DWHOS and Experimental
KEGG pathway analyses revealed that both in situ impacted P. biscaya and P. type B3, exposed to the Bulk treatments containing dispersant (alone or dispersed), exhibited similar impacts to environmental information processing pathways, specifically genes associated with signal transduction (i.e., Hippo and PI3K signaling pathways) and ECM-receptor interactions. Common impacts were also observed among pathways involved in cellular processes including focal adhesion and protein digestion and absorption.
Similar impacts were likewise observed among DWHOS impacted P. biscaya and P. type B3 experimentally exposed to WAFs containing dispersants. This includes impacts to metabolism, translation, protein folding and degradation, environmental information processing pathways (i.e., ABC membrane transporters) and various signal transduction pathways. Common impacts were also observed among pathways involved in crucial cellular processes including endocytosis and actin cytoskeleton regulation, as well as the digestive system (digestive secretions). See Supplementary Table 6 for more detailed information.
Discussion
This study provides novel transcriptomic data for two species of deep-sea octocorals, including a close relative of P. biscaya – the most severely impacted deep-sea species in the aftermath of the DWHOS (Deepwater Horizon Natural Resource Damage Assessment, 2016). Similar to in situ impacted P. biscaya (DeLeo et al., 2018), these results highlight the variability among coral responses to anthropogenic pollutant exposure, both inter- and intra-specific, and the utility of expression-level investigations of deep-sea fauna to elucidate sub-lethal impacts. These data add to the sparse amount of high-throughput sequencing data available for octocorals and deep-sea species, and provide added resources for studying and monitoring coral and deep-sea ecosystems in the face of future anthropogenic impacts and global ocean change.
Past transcriptomic investigations of P. biscaya revealed genome-wide impacts following in situ exposure to oil and dispersant laden floc (DeLeo et al., 2018). However, the full extent and duration of the exposure to oil and dispersant during the spill is unknown. There were multiple conceivable routes of exposure as oil and chemical dispersants were released at depth (∼1500 m; Hazen et al., 2010) and ultimately persisted as a deep-water oil plume around ∼1,100 m (Camilli et al., 2010). These alternative exposure routes may have elicited early onset stress responses following initial pollutant contact, which may not have been detectable among floc-exposed P. biscaya at the time of sampling. The transcriptional investigations of spill-impacted P. biscaya colonies by DeLeo et al. (2018) were also limited in terms of biological replication and temporal sampling due to restrictions on site access and sampling in the aftermath of the DWHOS. The experimental transcriptomic profiles presented here elucidate these early onset responses of deep-sea octocorals to oil and dispersants with increased replication, biological controls (genets), and interspecific comparisons. However, the authors note that limitations in obtaining and maintaining live deep-sea corals for laboratory experimentation at the time of sampling, as well as the partial colony sampling done to prevent adverse impacts in situ following the DWHOS, also limited the sample size, technical replication, and statistical robustness of this study. Regardless, these data show that the experimental oil and dispersant treatments that ultimately elicited mortality in these coral species during the full 96 h of exposure (DeLeo et al., 2016), particularly in the WAF treatments, also elicited large genome-wide expression changes of crucial stress response genes after just 12 h. The largest number of differentially expressed genes and the highest magnitude of gene expression change was found in treatments containing dispersants, relative to the treatments only containing naturally occurring crude oil. This study improves our understanding of how different species of corals respond to, and cope with, environmental contaminant exposure in the short-term, as well as the variability in these responses.
Common Responses to Pollutants
While gene expression profiles revealed the unique influences of different chemical fractions (Bulk/heterogenous vs. WAF/dissolved) and treatments (oil-only, dispersant-only, and oil/dispersant mixtures) on the corals, they also revealed mutual responses elicited across various treatments in each exposure series.
Bulk Exposure Series
A common response observed among the bulk treatments, which contain both dissolved and undissolved oil and dispersant chemical constituents, involved altered energy metabolism and oxidative stress responses, both of which were observed among DWHOS impacted P. biscaya (DeLeo et al., 2018). Cytochrome c oxidase (CCO) activity, a key enzyme involved in aerobic metabolism (Simon and Robin., 1971) was diminished (lfc -1.6, adj. p-value 0.02) in C. delta exposed to treatments containing dispersant, alone or mixed with (dispersed) oil. As CCO catalyzes the terminal step in aerobic oxidative metabolism, CCO inhibition is suggestive of a hypoxic response among C. delta exposed to bulk dispersants. Likewise, CCO is a crucial enzyme regulating cellular energy production (Poyton et al., 1988), further suggesting impacts to tissue energy metabolism and cytotoxicity (Khan et al., 1990) following just 12 hrs of exposure. Conversely, CCO activity was significantly elevated among floc exposed P. biscaya (lfc 1.9, adj. p-value 0.005; DeLeo et al., 2018), and among the liver/gills of fish following longer exposures (24 and 96 h) to WAF-oil, alone or dispersed (Cohen et al., 2001; Mattos et al., 2010). Elevated CCO activity among these animals may be associated with ongoing oxidative stress responses during short-term exposures to oil WAFs, that could ultimately lead to the hypoxic signature observed in our study. It is possible that exposure to bulk oil and dispersant contaminants may cause a relatively rapid hypoxic response in C. delta as compared to exposure to dissolved WAF contaminants. Likewise, succinate-cytochrome c oxidoreductase activity (part of the succinate-oxidase complex of the respiratory chain system (Khan et al., 1990), was significantly elevated (lfc 1.1, adj. p-value 0.02) in P. type B3, in addition to genes associated with oxidative stress responses (GO terms). This suggests that the bulk dispersant exposures also induced oxidative stress among P. type B3 after only 12 h, though to a different degree and possibly in a species-dependent manner.
WAF Exposure Series
The strongest genome-wide response was observed among both species exposed to WAFs, particularly WAF treatments containing dispersants. This suggests the coral species tested here are particularly sensitive to the WAFs of anthropogenic pollutants. The magnitude and severity of this response to WAF treatments containing dispersants was mirrored by the higher rates of health decline observed during the full 96 h exposure series (see DeLeo et al., 2016 for more details). These results support the growing body of evidence that chemical dispersants elicit strong, negative responses from marine invertebrates and can be more harmful than oil alone, as dispersants increase the surface area of oil-water interactions- potentially increasing toxicological impacts and bioavailability (e.g., Chandrasekar et al., 2006; Goodbody-Gringley et al., 2013; DeLeo et al., 2016). However, within the scope of this study, it is also probable that this response was inflated by concentration differences between Bulk and WAF solutions.
Dispersants are also known to impair cell membrane function (Abel, 1974; National Research Council, 1989). Therefore, strong early onset responses to dispersants at the genomic-level were not surprising, as exposure was shown to result in the increased permeability of biological membranes and loss of total membrane function and/or osmoregulating abilities (Benoit et al., 1987; Partearroyo et al., 1990). As genes associated with DNA repair, protein modification and apoptosis (programmed cell death) (Kultz, 2003; Kültz, 2005), were significantly elevated in WAF treatments containing dispersants, this further suggests that even short-term exposures can impact cellular integrity, leading to lasting impacts.
Common genome-wide responses were also observed among WAF treatments containing oil (alone or dispersed). These included an elevated expression of ribosomal proteins which was likewise observed among in situ impacted P. biscaya (DeLeo et al., 2018), and among shallow-water octocorals following periods of short-term pathogen stress (Burge et al., 2013). It has been suggested that the elevated expression of ribosomal gene products is a generalized response to environmental stressors (e.g., Burge et al., 2013; DeLeo et al., 2018) and that ribosomal proteins play an important role in regulating metabolism in invertebrates during periods of stress (Travers et al., 2010). It is possible that the elevated expression of these genes following the sub-lethal exposures to oil and dispersant was an attempt to regain cellular homeostasis following stress induced DNA and/or protein damage (Kultz, 2003).
Various structural molecules and ECM components were also inhibited among the corals experimentally exposed to WAF oil treatments. These include fibrillins that confer structural integrity to tissues (Piha-Gossack et al., 2012) and are thought to play a role in stress tolerance, albeit in plants (Singh and McNellis, 2011). Therefore, it is possible that the dissolved, water-accommodated oil and dispersant constituents may both compromise coral tissue integrity to a certain degree.
Additional putative genes involved in wound repair and inflammatory responses (i.e., metallopeptidases and peroxidasin; Massova et al., 1998), were inhibited among WAF treatments containing oil (alone or dispersed oil) as well, further suggestive of acute cytotoxicity and cellular damage (i.e., Sterchi et al., 2008). This may be linked to the tissue degradation and health decline observed during the 96 h experimental exposures (DeLeo et al., 2016). Conversely, genes coding for structural/ECM proteins and inflammatory/wound repair pathways (i.e., peroxidasin) were significantly elevated in P. biscaya exposed to DWHOS floc in situ (DeLeo et al., 2018). It is probable that natural hydrodynamic flow in situ alters and dampens exposure impacts and enables sessile invertebrates, like corals, to withstand longer exposures and/or combat these stressors at the cellular level. It is also possible that the expression signatures observed in this study reflect the prioritization (elevated expression) of other homeostatic processes as an initial defense response against chemical stressors, which in turn reduced the energetic resources allocated to maintain processes such as tissue integrity and wound-repair in the short-term. The latter scenario is likely the case, as some of the genes under-expressed here (e.g., fibrillins and peroxidasin) were over-expressed in the black coral Leiopathes glaberrima exposed to identical experimental treatments for 24 h (Ruiz-Ramos et al., 2017). While the functional role of peroxidasin is not fully understood among invertebrates, it is believed to contribute to innate immune defenses (Gotenstein et al., 2010), responses to oxidative stress, and programmed cell death (Horikoshi et al., 1999). Based on our results, it is possible that some, if not all, of these processes are initially suppressed during early responses to anthropogenic pollutants to allocate resources to other homeostatic processes.
Many GO terms related to cellular adhesion were under-expressed in all treatments (i.e., cell-cell, cell-matrix, and cell-substrate adhesion). Cell adhesion molecules are functionally diverse and are integral in a wide array of cellular processes, such as cell–cell signaling, multicellular tissue development (Gumbiner, 1996), and immune responses (Johansson, 1999; Harjunpää et al., 2019). Reduced expression of cellular adhesion molecules could therefore have wide-reaching implications, and possible associations with impairments to immunity and delayed wound healing (Harjunpää et al., 2019), though more rigorous investigation is needed to confidently draw these conclusions. Differential expression of genes associated with cellular adhesion were likewise detected among corals following temperature stress (Gates et al., 1992; Traylor-Knowles, 2019) and disease (Daniels et al., 2015; Young et al., 2020). Therefore, cellular adhesion processes appear to be commonly impacted by environmental stressors and could be used in future assessments to gain a snapshot of overall coral health/condition.
Species-Specific Differences
Our data suggest that the octocorals had distinct responses to the Bulk and WAF oil exposures after 12 h, as there was no major overlap among the differentially expressed genes that were annotated- though this may be linked to the limitations of our experimental design. It is possible that there was a delayed or altered reaction to the pollutants in one of the species. From a physiological standpoint, both C. delta and P. type B3 did surprisingly well in both oil-only treatments during the full 96 h exposure series as compared to the treatments containing dispersants (DeLeo et al., 2016). Metabolic depression was primarily observed among the corals exposed to oil in this present study, including genes associated with xenobiotic biodegradation and metabolic pathways (i.e., ABC transporters and GST). Therefore it seems unlikely that these corals were metabolizing oil in the short term as was suggested for L. glaberrima (Ruiz-Ramos et al., 2017).
Cytochrome p450 (CYP), which functions in the biotransformation and detoxification of most xenobiotics (Goldstone et al., 2006), was also not significantly differentially expressed among the corals after 12 h. CYPs are required for the efficient elimination of foreign chemicals from the body (Goldstone et al., 2006) and have become a widely used biomarker for pollutant (Devaux et al., 1998; Porte et al., 2001) and oil exposure (Garcia et al., 2012; Zhang et al., 2012; Han et al., 2014). CYPs were significantly elevated in L. glaberrima experimentally exposed to oil for 24 h (Ruiz-Ramos et al., 2017) but CYPs, along with ABC transporters and GST, were depressed among P. biscaya exposed to oil and dispersant in situ (DeLeo et al., 2018), albeit for an unknown duration. Although CYPs were not significantly expressed here, components of this same functional pathway – xenobiotic metabolism by cytochrome P450 – were significantly suppressed in C. delta exposed to WAF oil and elevated in C. delta exposed to WAF dispersant. This suggests that this pathway is impacted after just 12 h of pollutant contact, but that these impacts are dynamic and may differ depending on the species and/or the duration of exposure.
C. delta and P. type B3 experimentally exposed to WAF treatments, particularly those containing dispersants, each had a distinct overlap in gene expression with in situ impacted P. biscaya. Interestingly, this overlap appears to correlate with the treatments that elicited the strongest phenotypic response in the 96 h exposure series (see DeLeo et al., 2016). This is particularly apparent for C. delta, which had somewhat less severe rates of health decline in response to longer exposures to these treatments (DeLeo et al., 2016) and the strongest genome-wide response to the WAFs (after 12 h). It is possible that this is linked to “frontloading” or constitutive expression of genes that respond to oil exposure and/or their increased capacity for gene expression plasticity of environmental stress response (ESR) genes. For shallow-water corals, it has been hypothesized that one or both approaches may impart a certain degree of resilience to environmental stressors (i.e., Barshis et al., 2013; Kenkel and Matz, 2016). As C. delta preferentially occupies habitats near natural hydrocarbon seeps (Quattrini et al., 2013), they have likely adapted to low levels of hydrocarbon exposure potentially by employing one of these approaches.
In the case of C. delta, plastic expression may be linked to “chemical defensome” genes, evolutionarily conserved genes and proteins that are involved in numerous processes including the metabolism and biotransformation of xenobiotic toxins [i.e., polycyclic aromatic hydrocarbons (PAHs), components of oil], antioxidant systems that respond to oxidative stress, as well as protein homeostasis (Reitzel et al., 2008; Lushchak, 2011; Tarrant et al., 2014). Many of the GO terms that were unique to C. delta compared to P. type B3 exposed to oil were associated with cellular stress responses, including the regulation of stress-activated MAPK and protein kinase signaling cascades, and multicellular organismal response to stress, potentially signifying an increased capacity to respond to the stresses of oil exposure. As C. delta exhibited somewhat less severe rates of health decline in response to longer exposures to oil (DeLeo et al., 2016), it is possible this species is more resilient to short-term pollutant exposure.
The heightened cellular level response observed for C. delta, in terms of the number of genes that were differentially expressed, suggests that short-term dispersant exposure can induce cell damage in as little as 12 h. Expression differences were observed for genes involved in necroptosis (i.e., toll-like receptors), a form of necrosis or inflammatory cell death (reviewed in Dhuriya and Sharma, 2018), and C-type lectin signaling cascades that induce the production of inflammatory cytokines and chemokines (reviewed in Tang et al., 2018). Tumor necrosis factor receptor-associated factors (TRAFs), immune system receptors that elicit immune and inflammatory (e.g., necrosis) responses (Palmer and Traylor-Knowles, 2012), were also significantly differentially expressed. TRAFs were over-expressed in the WAF dispersant-only treatment after 12 h but under-expressed among C. delta in the WAF oil/dispersant mixtures, as well as P. type B3 in WAF-oil. TRAFs were similarly over-expressed in P. biscaya exposed to oil and dispersant laden floc (DeLeo et al., 2018), indicating that over-expression may be linked to the dispersant constituents. These results suggest that even short periods of dispersant exposure may influence coral immune pathways and that immune/inflammatory pathway components make good candidates for future spill monitoring efforts.
Unlike C. delta, which exhibited a greater number of differentially expressed genes (and a larger average magnitude of gene expression change) when exposed to WAF treatments relative to Bulk treatments, P. type B3 had a comparable number of differentially expressed genes, albeit a higher magnitude of expression change in the WAF exposures. It is possible that this is linked to a reduced capacity for gene expression plasticity of important ESR genes among P. type B3 relative to C. delta. Studies of shallow-water corals exposed to thermal stress revealed a higher degree of thermal tolerance and reduced bleaching impacts among populations of corals that exhibited a greater capacity for gene expression plasticity related to changes in environment/condition (Kenkel and Matz, 2016). It is possible that similar interspecific differences in plasticity also exist among deep-sea octocoral species. This would suggest that C. delta may be better able to mitigate intracellular damage resulting from anthropogenic stressors, and that P. type B3 is equally vulnerable to both undissolved (Bulk) and water-accommodated (WAF) components of oil and dispersant, though further investigation is needed to elucidate these differences.
Functional enrichment analyses for P. type B3 exposed to WAFs revealed that GO terms associated with Wnt signaling were significantly enriched among the under-expressed transcripts. Wnt signaling pathways are integral cascades associated with a myriad of cellular functions including development and cell-cell interactions. Down-regulation of Wnt pathways has also been detected in shallow-water corals in response to high temperature (Polato et al., 2013; Maor-Landaw et al., 2014) and low pH/high temperature conditions (Kaniewska et al., 2015). These signaling pathways could therefore be highly susceptible to environmental stress in corals and warrant further investigation.
Conclusion
As offshore drilling continues to increase worldwide, the need to understand species and ecosystem-level impacts has become paramount. Prior to the DWHOS there was no research into how chemical dispersants would react at depth and how prolonged exposure would impact deep-sea biota. As corals are important ecosystem engineers, creating complex structures that provide habitat to diverse suites of organisms, impacts to these species will have significant consequences. This and other studies provide evidence to suggest prolonged dispersant exposure is more harmful than oil exposure alone. Our findings indicate coral fauna have variable responses to anthropogenic environmental stressors, and some species and/or genotypes may be more resilient to environmental stress. These results further highlight the possible impacts of both dissolved and undissolved anthropogenic contaminants to corals, including metabolic and hypoxic consequences and impacts to immune and wound healing processes. Exposure to bulk/heterogenous contaminants appears to cause heightened oxidative stress and metabolic responses, while exposures to dissolved/WAF contaminants appears to elicit stronger impacts to tissue/skeletal development, immune responses and cellular damage/DNA repair. Similar processes were impacted among corals damaged in situ by the DWHOS following prolonged floc exposure among other probable routes of contaminant exposure (i.e., deep-water oil plume), highlighting both the early probable inception of this damage and the complex responses of corals to oil and dispersant constituents. This improved understanding of the ways in which oil and dispersant exposure affects corals is integral to improving our ability to properly manage these ecosystems and respond to future exposure from standard operations and accidental releases.
Data Availability Statement
The RNAseq data presented in this study can be found on NCBI’s SRA database [BioProject ID: PRJNA708307].
Author Contributions
DD and EC conceived and designed the experiments. DD performed the experiments and lab work with assistance from AB. DD and AG analyzed the data. SH consulted on bioinformatic analyses. DD wrote the manuscript with input from all co-authors. All the authors contributed to the article and approved the submitted version.
Funding
This work was funded by a grant awarded to EC from The Gulf of Mexico Research Initiative to support the “Ecosystem Impacts of Oil and Gas in the Gulf” (ECOGIG) research consortium. Additional funding for coral collections was provided by (1) the Bureau of Ocean Energy Management (BOEM) and the National Oceanic and Atmospheric Administration (NOAA) contract to EC (M08PC20038) through the “Lophelia II” project administered by TDI Brooks, Inc. and (2) a NSF RAPID grant award to EC (OCE-1045079).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to the crews aboard the R/V Falkor and E/V Nautilus and both the ROV Global Explorer and ROV Hercules operators for their assistance with sample collections. Particular thanks to A. Quattrini for assistance with sample collections, J. Lunden for guidance with RNA work, and C. McNicholl, D. Ruiz-Ramos, I. Baums, A. Demopoulos, C. Fisher, and the Cordes lab for support at sea.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.649909/full#supplementary-material
Footnotes
References
Abel, P. (1974). Toxicity of synthetic detergents to fish and aquatic invertebrates. J. Fish Biol. 6, 279–298. doi: 10.1111/j.1095-8649.1974.tb04545.x
Anderson, C. M., Mayes, M., and LaBelle, R. P. (2012). Oil Spill Occurrence Rates for Offshore Spills. Herndon, DC: Bureau of Ocean Energy Management.
Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. Cambridge: Babraham Institute.
Barshis, D. J., Ladner, J. T., Oliver, T. A., Seneca, F. O., Traylor-Knowles, N., and Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U S A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Barron, M. G. (2012). Ecological impacts of the Deepwater Horizon oil spill: implications for immunotoxicity. Toxicol. Pathol. 40, 315–320. doi: 10.1177/0192623311428474
Benoit, J., Cormier, M., and Wepierre, J. (1987). Effect of proteins on the assessment of surfactant cytotoxicity by an in vitro test: possible correlations with in vivo data. Toxicol. In Vitro 1, 91–96. doi: 10.1016/0887-2333(87)90006-3
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burge, C. A., Mouchka, M. E., Harvell, C. D., and Roberts, S. (2013). Immune response of the caribbean sea fan, gorgonia ventalina, exposed to an aplanochytrium parasite as revealed by transcriptome sequencing. Front. Physiol. 4:180. doi: 10.3389/fphys.2013.00180
Camilli, R., Reddy, C. M., Yoerger, D. R., Van Mooy, BaS, Jakuba, M. V., et al. (2010). Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science 330, 201–204. doi: 10.1126/science.1195223
Chandrasekar, S., Sorial, G. A., and Weaver, J. W. (2006). Dispersant effectiveness on oil spills–impact of salinity. ICES J. Mar. Sci. 63, 1418–1430. doi: 10.1016/j.icesjms.2006.04.019
Cohen, A., Nugegoda, D., and Gagnon, M. M. (2001). Metabolic responses of fish following exposure to two different oil spill remediation techniques. Ecotoxicol. Environ. Safety 48, 306–310. doi: 10.1006/eesa.2000.2020
Cordes, E. E., Jones, D. O. B., Schlacher, T. A., Amon, D. J., Bernardino, A. F., Brooke, S., et al. (2016). Environmental Impacts of the Deep-Water Oil and Gas Industry: A Review to Guide Management Strategies. Front. Environ. Sci. 4:58. doi: 10.3389/fenvs.2016.00058
Cordes, E. E., McGinley, M. P., Podowski, E. L., Becker, E. L., Lessard-Pilon, S., Viada, S. T., et al. (2008). Coral communities of the deep Gulf of Mexico. Deep Sea Res. Part I Oceanogr. Res. Papers 55, 777–787. doi: 10.1016/j.dsr.2008.03.005
Daniels, C., Baumgarten, S., Yum, L. K., Michell, C. T., Bayer, T., Arif, C., et al. (2015). Metatranscriptome analysis of the reef-building coral Orbicella faveolata indicates holobiont response to coral disease. Front. Mar. Sci. 2:62. doi: 10.3389/fmars.2015.00062
Davies, A. J., Duineveld, G. C., van Weering, T. C., Mienis, F., Quattrini, A. M., Seim, H. E., et al. (2010). Short-term environmental variability in cold-water coral habitat at Viosca Knoll, Gulf of Mexico. Deep Sea Res. Part I Oceanogr. Res. Papers 57, 199–212. doi: 10.1016/j.dsr.2009.10.012
Deepwater Horizon Natural Resource Damage Assessment (2016). Deepwater Horizon Oil Spill: Final Programmatic Damage Assessment and Restoration Plan and Final Programmatic Environmental Impact Statement. Gulf of Mexico: Deepwater Horizon Natural Resource Damage Assessment.
DeLeo, D. M., Herrera, S., Lengyel, S. D., Quattrini, A. M., Kulathinal, R. J., and Cordes, E. E. (2018). Gene expression profiling reveals deep-sea coral response to the Deepwater Horizon oil spill. Mol. Ecol. 27, 4066–4077. doi: 10.1111/mec.14847
DeLeo, D. M., Ruiz-Ramos, D. V., Baums, I. B., and Cordes, E. E. (2016). Response of deep-water corals to oil and chemical dispersant exposure. Deep Sea Res. Part II Topical Stud. Oceanogr. 129, 137–147. doi: 10.1016/j.dsr2.2015.02.028
Devaux, A., Flammarion, P., Bernardon, V., Garric, J., and Monod, G. (1998). Monitoring of the chemical pollution of the river Rhône through measurement of DNA damage and cytochrome P4501a induction in chub (Leuciscus cephalus). Mar. Environ. Res. 46, 257–262. doi: 10.1016/s0141-1136(97)00105-0
Dhuriya, Y. K., and Sharma, D. (2018). Necroptosis: a regulated inflammatory mode of cell death. J. Neuroinflamm. 15:199.
Doughty, C. L., Quattrini, A. M., and Cordes, E. E. (2014). Insights into the population dynamics of the deep-sea coral genus Paramuricea in the Gulf of Mexico. Deep Sea Res. Part II Topical Stud. Oceanogr. 99, 71–82. doi: 10.1016/j.dsr2.2013.05.023
Fisher, C. R., Demopoulos, A. W. J., Cordes, E. E., Baums, I. B., White, H. K., and Bourque, J. R. (2014a). Coral communities as indicators of ecosystem-level impacts of the deepwater horizon spill. BioScience 64, 796–807. doi: 10.1093/biosci/biu129
Fisher, C. R., Hsing, P.-Y., Kaiser, C. L., Yoerger, D. R., Roberts, H. H., Shedd, W. W., et al. (2014b). Footprint of Deepwater Horizon blowout impact to deep-water coral communities. Proc. Natl. Acad. Sci. U S A. 111, 11744–11749. doi: 10.1073/pnas.1403492111
Garcia, T. I., Shen, Y., Crawford, D., Oleksiak, M. F., Whitehead, A., and Walter, R. B. (2012). RNA-Seq reveals complex genetic response to Deepwater Horizon oil release in Fundulus grandis. BMC Genomics 13:474. doi: 10.1186/1471-2164-13-474
Gates, R. D., Baghdasarian, G., and Muscatine, L. (1992). Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol. Bull. 182, 324–332. doi: 10.2307/1542252
Girard, F., and Fisher, C. R. (2018). Long-term impact of the Deepwater Horizon oil spill on deep-sea corals detected after seven years of monitoring. Biol. Conserv. 225, 117–127. doi: 10.1016/j.biocon.2018.06.028
Goldstone, J. V., Hamdoun, A., Cole, B. J., Howard-Ashby, M., Nebert, D. W., Scally, M., et al. (2006). The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev. Biol. 300, 366–384. doi: 10.1016/j.ydbio.2006.08.066
Goodbody-Gringley, G., Wetzel, D. L., Gillon, D., Pulster, E., Miller, A., and Ritchie, K. B. (2013). Toxicity of Deepwater Horizon source oil and the chemical dispersant, Corexit§9500, to coral larvae. PLoS One 8:e45574. doi: 10.1371/journal.pone.0045574
Gotenstein, J. R., Swale, R. E., Fukuda, T., Wu, Z., Giurumescu, C. A., Goncharov, A., et al. (2010). The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development 137, 3603–3613. doi: 10.1242/dev.049189
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29:644.
Gray, J. S., Clarke, A. J., Warwick, R. M., and Hobbs, G. (1990). Detection of initial effects of pollution on marine benthos: an example from the Ekofisk and Eldfisk oilfields. North Sea Mar. Ecol. Prog. Ser. 66, 285–299. doi: 10.3354/meps066285
Gumbiner, B. M. (1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84, 345–357. doi: 10.1016/s0092-8674(00)81279-9
Han, J., Won, E. J., Hwang, D. S., Shin, K. H., Lee, Y. S., Leung, K. M. Y., et al. (2014). Crude oil exposure results in oxidative stress-mediated dysfunctional development and reproduction in the copepod Tigriopus japonicus and modulates expression of cytochrome P450 (CYP) genes. Aquat. Toxicol. 152, 308–317. doi: 10.1016/j.aquatox.2014.04.027
Harjunpää, H., Llort Asens, M., Guenther, C., and Fagerholm, S. C. (2019). Cell adhesion molecules and their roles and regulation in the immune and tumor microenvironment. Front. Immunol. 10:1078. doi: 10.3389/fimmu.2019.01078
Hazen, T. C., Dubinsky, E. A., DeSantis, T. Z., Andersen, G. L., Piceno, Y. M., Singh, N., et al. (2010). Deep-Sea Oil Plume Enriches Indigenous Oil-Degrading Bacteria. Science 330, 204–208. doi: 10.1126/science.1195979
Horikoshi, N., Cong, J., Kley, N., and Shenk, T. (1999). Isolation of differentially expressed cDNAs from p53-dependent apoptotic cells: activation of the human homologue of the Drosophila peroxidasin gene. Biochem. Biophys. Res. Commun. 261, 864–869. doi: 10.1006/bbrc.1999.1123
Hsing, P.-Y., Fu, B., Larcom, E. A., Berlet, S. P., Shank, T. M., Govindarajan, A. F., et al. (2013). Evidence of lasting impact of the Deepwater Horizon oil spill on a deep Gulf of Mexico coral community. Elementa Sci. Anthropoc. 1:000012.
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293.
Johansson, M. W. (1999). Cell adhesion molecules in invertebrate immunity. Dev. Comparat. Immunol. 23, 303–315. doi: 10.1016/s0145-305x(99)00013-0
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462.
Kaniewska, P., Chan, C.-K. K., Kline, D., Ling, E. Y. S., Rosic, N., Edwards, D., et al. (2015). Transcriptomic changes in coral holobionts provide insights into physiological challenges of future climate and ocean change. PLoS One 10:e0139223. doi: 10.1371/journal.pone.0139223
Kenkel, C. D., and Matz, M. V. (2016). Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evolut. 1, 1–6.
Khan, A. A., Schuler, M. M., Prior, M. G., Yong, S., Coppock, R., Florence, L., et al. (1990). Effects of hydrogen sulfide exposure on lung mitochondrial respiratory chain enzymes in rats. Toxicol. Appl. Pharmacol. 103, 482–490. doi: 10.1016/0041-008x(90)90321-k
Kultz, D. (2003). Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J. Exp. Biol. 206, 3119–3124. doi: 10.1242/jeb.00549
Kültz, D. (2005). Molecular and Evolutionary Basis of the Cellular Stress Response. Annu. Rev. Physiol. 67, 225–257. doi: 10.1146/annurev.physiol.67.040403.103635
Larsson, A. I., van Oevelen, D., Purser, A., and Thomsen, L. (2013). Tolerance to long-term exposure of suspended benthic sediments and drill cuttings in the cold-water coral Lophelia pertusa. Mar. Pollut. Bull. 70, 176–188. doi: 10.1016/j.marpolbul.2013.02.033
Love, M., Anders, S., and Huber, W. (2014). Differential analysis of count data–the DESeq2 package. Genome Biol. 15:10.1186.
Lushchak, V. I. (2011). Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 101, 13–30. doi: 10.1016/j.aquatox.2010.10.006
Maor-Landaw, K., Karako-Lampert, S., Ben-Asher, H. W., Goffredo, S., Falini, G., Dubinsky, Z., et al. (2014). Gene expression profiles during short-term heat stress in the red sea coral Stylophora pistillata. Glob. Change Biol. 20, 3026–3035. doi: 10.1111/gcb.12592
Marcais, G., and Kingsford, C. (2012). Jellyfish : A fast k-mer counter, 1 Edn. Maryland: University of Maryland, 1–8.
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Massova, I., Kotra, L. P., Fridman, R., and Mobashery, S. (1998). Matrix metalloproteinases: structures, evolution, and diversification. FASEB J. 12, 1075–1095. doi: 10.1096/fasebj.12.12.1075
Mattos, J. J., Siebert, M. N., Luchmann, K. H., Granucci, N., Dorrington, T., Stoco, P. H., et al. (2010). Differential gene expression in Poecilia vivipara exposed to diesel oil water accommodated fraction. Mar. Environ. Res. 69, S31–S33.
McNutt, M. K., Camilli, R., Crone, T. J., Guthrie, G. D., Hsieh, P. A., Ryerson, T. B., et al. (2012). Review of flow rate estimates of the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. 109, 20260–20267.
Muehlenbachs, L., Cohen, M. A., and Gerarden, T. (2013). The impact of water depth on safety and environmental performance in offshore oil and gas production. Energy Policy 55, 699–705. doi: 10.1016/j.enpol.2012.12.074
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R., et al. (2013). Vegan: Community Ecology Package. R package version 2.0-2. Vienna: R Core Team.
Palmer, C. V., and Traylor-Knowles, N. (2012). Towards an integrated network of coral immune mechanisms. Proc. Biol. Sci. R. Soc. 279, 4106–4114. doi: 10.1098/rspb.2012.1477
Partearroyo, M. A., Ostolaza, H., Goñi, F. M., and Barberá-Guillem, E. (1990). Surfactant-induced cell toxicity and cell lysis: A study using B16 melanoma cells. Biochem. Pharmacol. 40, 1323–1328. doi: 10.1016/0006-2952(90)90399-6
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14:417. doi: 10.1038/nmeth.4197
Piha-Gossack, A., Sossin, W., and Reinhardt, D. P. (2012). The evolution of extracellular fibrillins and their functional domains. PLoS One 7:e33560. doi: 10.1371/journal.pone.0033560
Polato, N. R., Altman, N. S., and Baums, I. B. (2013). Variation in the transcriptional response of threatened coral larvae to elevated temperatures. Mol. Ecol. 22, 1366–1382. doi: 10.1111/mec.12163
Polato, N. R., Voolstra, C. R., Schnetzer, J., DeSalvo, M. K., Randall, C. J., Szmant, A. M., et al. (2010). Location-specific responses to thermal stress in larvae of the reef-building coral Montastraea faveolata. PLoS One 5:e11221. doi: 10.1371/journal.pone.0011221
Porte, C., Biosca, X., Solé, M., and Albaigés, J. (2001). The integrated use of chemical analysis, cytochrome P450 and stress proteins in mussels to assess pollution along the Galician coast (NW Spain). Environ. Pollut. 112, 261–268. doi: 10.1016/s0269-7491(00)00104-4
Poyton, R. O., Trueblood, C. E., Wright, R. M., and Farrell, L. E. (1988). Expression and Function of Cytochrome c Oxidase Subunit Isologues: Modulators of Cellular Energy Production? a. Ann. N Y. Acad. Sci. 550, 289–307. doi: 10.1111/j.1749-6632.1988.tb35344.x
Quattrini, A. M., Georgian, S. E., Byrnes, L., Stevens, A., Falco, R., and Cordes, E. E. (2013). Niche divergence by deep-sea octocorals in the genus Callogorgia across the continental slope of the Gulf of Mexico. Mol. Ecol. 22, 4123–4140. doi: 10.1111/mec.12370
Reitzel, A. M., Sullivan, J. C., and Traylor-knowles, N. (2008). Genomic Survey of Candidate Stress-Response Genes in the Estuarine Anemone. Biol. Bull. 214, 233–254. doi: 10.2307/25470666
Ruiz-Ramos, D. V., Fisher, C. R., Baums, I. B., and Thomsen, L. (2017). Stress response of the black coral Leiopathes glaberrima when exposed to sub-lethal amounts of crude oil and dispersant. Elementa Sci. Anthropoc. 5:77.
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Simon, L. M., and Robin. (1971). Relationship of cytochrome oxidase activity to vertebrate total and organ oxygen consumption. Int. J. Biochem. 2, 569–573. doi: 10.1016/0020-711x(71)90026-7
Singh, D. K., and McNellis, T. W. (2011). Fibrillin protein function: the tip of the iceberg? Trends Plant Sci. 16, 432–441. doi: 10.1016/j.tplants.2011.03.014
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., and Kelly, S. (2016). TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144. doi: 10.1101/gr.196469.115
Soneson, C., Love, M. I., and Robinson, M. D. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4:1521. doi: 10.12688/f1000research.7563.2
Song, L., and Florea, L. (2015). Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 4:48.
Sterchi, E. E., Stöcker, W., and Bond, J. S. (2008). Meprins, membrane-bound and secreted astacin metalloproteinases. Mol. Aspects Med. 29, 309–328. doi: 10.1016/j.mam.2008.08.002
Tang, J., Lin, G., Langdon, W. Y., Tao, L., and Zhang, J. (2018). Regulation of C-type lectin receptor-mediated antifungal immunity. Front. Immunol. 9:123. doi: 10.3389/fimmu.2018.00123
Tarrant, A. M., Reitzel, A. M., Kwok, C. K., and Jenny, M. J. (2014). Activation of the cnidarian oxidative stress response by ultraviolet light, polycyclic aromatic hydrocarbons and crude oil. J. Exp. Biol. 217(Pt 9), 1444–1453. doi: 10.1242/jeb.093690
Travers, M. A., Meistertzheim, A. L., Cardinaud, M., Friedman, C. S., Huchette, S., et al. (2010). Gene expression patterns of abalone, Haliotis tuberculata, during successive infections by the pathogen Vibrio harveyi. J. Invertebr. Pathol. 105, 289–297. doi: 10.1016/j.jip.2010.08.001
Traylor-Knowles, N. (2019). Heat stress compromises epithelial integrity in the coral, Acropora hyacinthus. PeerJ 7:e6510. doi: 10.7717/peerj.6510
Ulfsnes, A., Haugland, J. K., and Weltzien, R. (2013). Monitoring of Drill Activities in Areas with Presence of Cold Water Corals. Stavanger: Det Norsk Veritas.
White, H. K., Hsing, P. Y., Cho, W., Shank, T. M., Cordes, E. E., Quattrini, A. M., et al. (2012). Impact of the Deepwater Horizon oil spill on a deep-water coral community in the Gulf of Mexico. Proc. Natl. Acad. Sci. 109, 20303–20308.
White, H. K., Lyons, S. L., Harrison, S. J., Findley, D. M., Liu, Y., and Kujawinski, E. B. (2014). Long-Term Persistence of Dispersants following the Deepwater Horizon Oil Spill. Environ. Sci. Technol. Lett. 1, 295–299. doi: 10.1021/ez500168r
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, 1–12.
Wright, R. M., Aglyamova, G. V., Meyer, E., and Matz, M. V. (2015). Gene expression associated with white syndromes in a reef building coral, Acropora hyacinthus. BMC Genomics 16:371. doi: 10.1186/s12864-015-1540-2
Young, B., Serrano, X. M., Rosales, S., Miller, M. W., Williams, D., and Traylor-Knowles, N. (2020). Innate immune gene expression in Acropora palmata is consistent despite variance in yearly disease events. PLoS One 15:e0228514. doi: 10.1371/journal.pone.0228514
Zdobnov, E. M., Tegenfeldt, F., Kuznetsov, D., Waterhouse, R. M., Simao, F. A., Ioannidis, P., et al. (2017). OrthoDB v9. 1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45, D744–D749.
Keywords: gene expression, oil spill, Deepwater Horizon, octocorals, Callogorgia, Paramuricea, RNAseq, anthropogenic
Citation: DeLeo DM, Glazier A, Herrera S, Barkman A and Cordes EE (2021) Transcriptomic Responses of Deep-Sea Corals Experimentally Exposed to Crude Oil and Dispersant. Front. Mar. Sci. 8:649909. doi: 10.3389/fmars.2021.649909
Received: 05 January 2021; Accepted: 09 March 2021;
Published: 08 April 2021.
Edited by:
Chiara Romano, Center for Advanced Studies of Blanes (CSIC), SpainReviewed by:
Rachel Michelle Wright, Harvard Medical School, United StatesMichael S. Studivan, University of Miami, United States
Copyright © 2021 DeLeo, Glazier, Herrera, Barkman and Cordes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danielle M. DeLeo, ZG1kZWxlbzE0QGdtYWlsLmNvbQ==
 Danielle M. DeLeo
Danielle M. DeLeo Amanda Glazier
Amanda Glazier Santiago Herrera
Santiago Herrera Alexandria Barkman2
Alexandria Barkman2 Erik E. Cordes
Erik E. Cordes