- 1Department of Marine Sciences and Convergent Technology, Hanyang University ERICA Campus, Ansan, South Korea
- 2Korea Polar Research Institute, Incheon, South Korea
- 3Petroleum and Marine Resources Research Division, Korea Institute of Geoscience and Mineral Resources, Daejeon, South Korea
- 4Monterey Bay Aquarium Research Institute, Moss Landing, CA, United States
- 5Department of Earth and Environmental Sciences, Pukyong National University, Busan, South Korea
During the ARA08C expedition in 2017, sediment push cores were collected at an active mud volcano (420 m water depth) in the Canadian Beaufort Sea from two visually discriminative siboglinid tubeworm (ST) habitats that were colonized densely and less densely (ST1 and ST2, respectively). In this study, we investigated the biogeochemical and microbial community characteristics at ST1 by analyzing the geochemical properties, microbial lipids, and nucleic acid signatures, and comparing them with the data previously reported from ST2. The two ST sites showed distinct differences in vertical geochemical gradients [methane, sulfate, dissolved inorganic carbon (DIC), total organic carbon, and total sulfur], with a higher methane flux recorded at ST1 (0.05 mmol cm–2 y–1) than at ST2 (0.01 mmol cm–2 y–1). Notably, the δ13C values of DIC were more depleted at ST1 than at ST2, resulting in a higher proportion of DIC derived from the anaerobic oxidation of methane (AOM) at ST1 than at ST2. Moreover, both the ST1 and ST2 sites revealed the dominance of AOM-related lipid biomarkers (especially sn-2-hydroxyarchaeol), showing highly 13C-depleted values. The 16S rRNA analyses showed the presence of AOM-related archaea, predominantly anaerobic methanotrophic archaea (ANME)-3 at ST1 and ST2. Our results suggest that AOM-related byproducts (sulfide and DIC) potentially derived from ANME-3 were more abundant at ST1 than at ST2. This variation was attributed to the intensity and persistence of ascending methane. Therefore, our study suggests that AOM-derived byproducts are possibly an essential energy source for tubeworms during chemosynthetic metabolism, shaping different colony types on the seafloor.
Introduction
Cold seeps can host diverse faunal and microbial communities (e.g., siboglinid tubeworms (STs), mytilid and vesicomyid bivalves, and giant sulfide-oxidizing bacteria; Levin, 2005; Jørgensen and Boetius, 2007; Vanreusel et al., 2009; Niemann and Boetius, 2010). STs (referred to as typical chemosynthetic organisms) lack a digestive organ and thus rely entirely on their endosymbionts for nutrient and energy supply (Cavanaugh et al., 1981; Felbeck et al., 1981; Southward et al., 1986 and references therein). In most cases, bacterial symbionts are embedded within host cells in the interior of the trophosomes (highly vascularized tissue) (Bright and Giere, 2005). For instance, the bottom sections (i.e., roots) of tubeworms have a distinct function for taking up the reduced sulfur species (Julian et al., 1999; Freytag et al., 2001; Cordes et al., 2005), and transport various substrates to the endosymbionts mainly inhabiting trophosomes (Hilário et al., 2011; Duperron et al., 2014). Furthermore, the seawater-exposed tube of tubeworms may act as a direct conduit of sulfate from seawater to sediment layers (Duperron et al., 2014). Therefore, the growth of these tubeworms inhabiting cold seeps may provide the basis for chemosynthetic ecosystems that are decoupled from photosynthesis (Sibuet and Olu, 1998).
In methane-rich seeping systems, one of the most dominant microbial processes in sediments is the anaerobic oxidation of methane (AOM) associated with sulfate reduction (SR), which produces byproducts, such as sulfide and dissolved inorganic carbon (DIC) (Reeburgh, 1996; Hinrichs and Boetius, 2002). Tubeworms take up sulfide compounds, oxidize these compounds into sulfate, and then release sulfate by the posterior end to sediments, which may enhance the AOM (Felden et al., 2010; Hilário et al., 2011). Therefore, the long-term development of tubeworm colonization may play an important ecological role in the reduction of methane venting (Sibuet and Olu, 1998; Cordes et al., 2005; de Beer et al., 2006; Boetius and Wenzhöfer, 2013). The aerobic oxidation of methane (MOx) is another biological sink for ascending methane. The oxidized surface sediments may support specific MOx-related bacterial groups, which depend on the availability of oxygen (Omoregie et al., 2009; Boetius and Wenzhöfer, 2013). Such communities not only form dense mats at the sediment-water interface, but also coexist as endosymbiotic associations with benthic fauna such as bivalves and tubeworms (Fisher, 1990; Duperron et al., 2007; Petersen and Dubilier, 2009; Rodrigues et al., 2011). Therefore, the chemosynthetic interaction between benthic organisms and methanotrophs may play an essential role in efficiently reducing methane emissions from the seafloor.
Since methane assimilation performed by AOM- and MOx-related microbial communities is influenced by varying gas-rich fluid fluxes (Niemann et al., 2006), the ecological function of chemosynthetic ecosystems appears to be driven by local environmental factors, such as morphologic setting and physico-chemical conditions (Rossel et al., 2011). In this context, active mud volcanoes (MVs) discovered in the Canadian Beaufort Sea can be considered as a natural laboratory for studying the biogeochemical dynamics of methane in sediments displaying various chemosynthetic habitats (Paull et al., 2015). In such settings, the activities of benthic organisms potentially control methane emissions from the seafloor (Boetius and Wenzhöfer, 2013). In this regard, Lee et al. (2019b) recently investigated vertical variations in AOM- and MOx-related methanotrophic communities in the mud volcano MV420 (420 m water depth) in the Canadian Beaufort Sea using sediment push cores collected from visually discriminative habitats devoid of megafauna and microbial mats to the naked eye, covered with bacterial mats, or colonized by STs. They showed that a niche diversification in MV420 shaped distinct methanotrophic communities due to the availability of electron acceptors in association with varying degrees of methane flux. However, our knowledge of the primary processes controlling the biological succession of STs in this region remains limited.
In this study, we investigated the biogeochemical and microbial community characteristics in a densely colonized ST field (ST1) in the MV420 of the Canadian Beaufort Sea by analyzing geochemical properties, microbial lipids, and nucleic acid signatures. The data from ST1 were then compared with those previously obtained from a less densely colonized ST field (ST2, Lee et al., 2019b) to constrain the primary biological process shaping the visually discriminative ST habitats in an active MV in the Canadian Beaufort Sea.
Materials and Methods
Sample Collection
In this study, one MV located at a water depth of 420 m (MV420) was mapped in detail (Figure 1) using an autonomous underwater vehicle during the ARA08C expedition of the Korean ice-breaker RV Araon in September 2017. Based on the observation of tubeworm patches dominated by Oligobrachia sp. at this MV (Paull et al., 2011, 2015; Lee et al., 2019a), we collected two push cores (ARA08C-DIVE104-1 and -10) using the remotely operated vehicle (ROV) “Mini” from a densely colonized ST site for investigation (ST1; 70.792°N, 135.556°W) (Figure 1). Similar to a previous study conducted at a less densely colonized ST site (ST2; 70.790°N, 135.565°W; Lee et al., 2019b), the first sediment core was subsampled at 2–4 cm intervals and the subsamples were transferred into serum vials containing saturated NaCl solution for gas analysis. After subsampling, these vials were immediately sealed with butyl rubber stoppers to prevent gas exchange. The second sediment core was used to extract pore waters at 4-cm intervals using the Rhizon soil moisture sampler. The extracted pore water was filtered through a 0.20 μm disposable polytetrafluorethylene filter before collection for onboard and post-cruise analyses. For the sulfate, DIC, and nutrient (NH4+ and PO43–) analyses, an aliquot of pore water was stored at −20 °C until ion chromatography analysis. Another aliquot was collected in a 2-mL septum screw-lid glass vial and preserved with 20 μL of saturated mercuric chloride (HgCl2) solution for DIC carbon isotope analysis (δ13CDIC). Afterward, the same core was sliced at 1-cm intervals for the analyses of bulk elements, lipid biomarkers, and 16S rRNA gene sequences. Sediment samples were transferred into aluminum sheets and stored at −80°C until analysis.
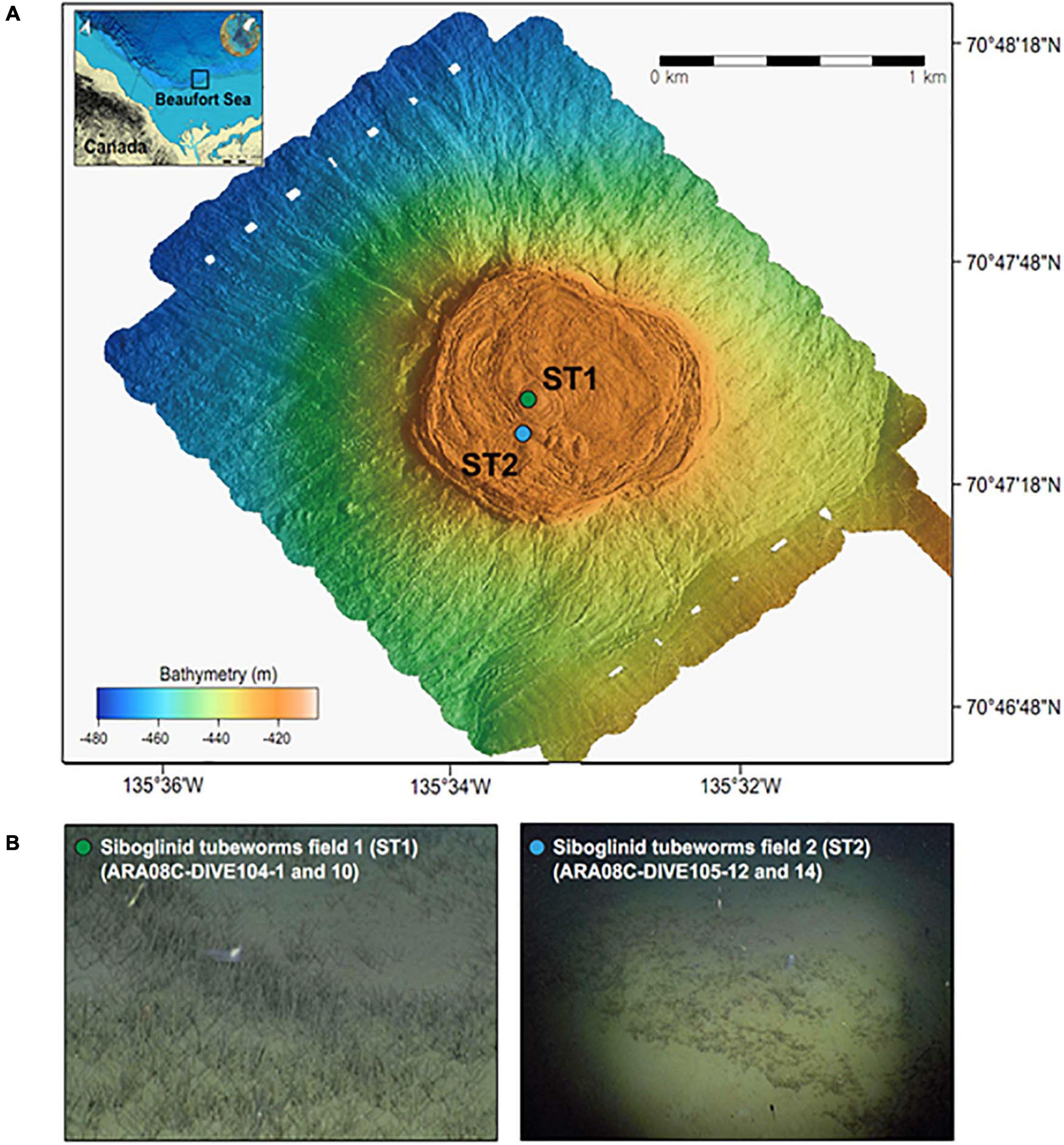
Figure 1. (A) Multibeam maps indicating a mud volcano at a water depth of 420 m in the Canadian Beaufort Sea and (B) ROV photos showing the sampling sites with different densities of siboglinid tubeworms (identified as Oligobrachia haakonmosbiensis, Paull et al., 2015).
Gas and Porewater Analyses
The hydrocarbon composition in the headspace gas was measured using a gas chromatograph (GC) 7890A (Agilent Technologies, CA, United States) with a flame ionization detector (FID) at the Korea Institute of Geoscience and Mineral Resources (KIGAM). Precision from the repeated standard analysis was better than 5%. The δ13C values of methane (δ13CCH4) were obtained using an isotope ratio mass spectrometer (IRMS; Finnigan MAT 252, Thermo Fisher Scientific, Waltham, MA, United States) with a Combustion III interface (Thermo Fisher Scientific) at Nagoya University. The δ13C values were calibrated using methane standards with certified isotopic values (−35.2‰) obtained from the National Institute of Standards and Technology. The analytical errors were ± 0.2‰ for δ13CCH4. Sulfate concentrations were measured using ion chromatography (761 compact ion chromatograph; Metrohm, Ionenstrasse, Switzerland). DIC concentrations were analyzed by titration using HCl at KIGAM. Reproducibility was monitored by repeated titrations of < 2% standard seawater supplied by the International Association of Physical Sciences of the Oceans for DIC. The δ13CDIC values were obtained using an IRMS (Finnigan DELTAplusXL, Thermo Fisher Scientific, Waltham, MA, United States) with a Finnigan GasBench-II headspace autosampler at Oregon State University. The δ13C values were calibrated using analyses of the Wiley CaCO3 DIC standard with certified isotopic values (−0.4‰). The analytical error was ± 0.02‰ for δ13CDIC. Phosphate and ammonium concentrations were measured at KIGAM using colorimetric methods with a spectrophotometer (UV-2450; Shimadzu, Kyoto, Japan) at 885 and 640 nm (Gieskes et al., 1991). Data on methane and sulfate fluxes were calculated as reported in a previous study (Lee et al., 2019b), which includes a detailed description of the corresponding calculation.
Bulk Element Analysis
Sediment samples were freeze-dried and homogenized using an agate mortar prior to the bulk element analyses. For total organic carbon (TOC), sediment samples (∼1 g) were treated with 8 mL of 1 N HCl to remove carbonates. The TOC contents and their isotopic compositions were measured using an elemental analyzer (vario PYRO cube, Elementar, Germany) connected to an IRMS (Isoprime visION, Isoprime, Manchester, United Kingdom). The stable carbon isotopic values of TOC (δ13CTOC) were reported in standard delta (δ) notation relative to the Vienna Pee-Dee Belemnite (VPDB) scale per mil. The δ13C values were calibrated using the International Atomic Energy Agency (IAEA) CH3 standard with a certified isotopic value of −24.7‰. The analytical precisions were ± 0.2 wt.% for TOC and ± 0.1‰ for δ13CTOC. The total sulfur (TS) content and its isotopic composition were measured using an elemental analyzer (EA1110, Thermo) connected to an IRMS (Dual-pumped 20-20-S, Secon) in organic elemental analysis labs in the United Kingdom. The stable sulfur isotopic values of TS (δ34STS) were reported in standard delta (δ) notation relative to the Vienna Canyon Diablo Troilite (VCDT) scale per mil. The δ34CTS values were calibrated using the IAEA S-2 and S-3 standards, with certified isotopic values of 22.7 and −32.3‰, respectively. The analytical precisions were ± 0.3 wt.% for TS and ± 0.4‰ for δ34CTS.
Lipid Biomarker Analysis
Lipid analytical procedures were performed as described by Lee et al. (2018). Briefly, total lipids were extracted using an accelerated solvent extractor (Dionex ASE 200, Dionex Corporation, Sunnyvale, CA, United States) and separated into apolar and polar fractions over an Al2O3 column (activated for 2 h at 150°C). After column separation, 40 μL of 5α-androstane (10 μg mL–1) was added to the apolar fraction as an internal standard. The polar fraction was divided into two aliquots to which either C22 7, 16-diol (10 μg mL–1), or C46 glycerol dialkyl glycerol tetraethers (GDGTs; 10 μg mL–1) were added as internal standards. Half of the polar fraction containing C22 7,16-diol was dried and silylated with 25 μL of N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) and 25 μL of pyridine before heating to 60°C for 20 min to form trimethylsilyl derivatives. The second half of the polar faction containing C46 GDGT was re-dissolved by sonication (5 min) in hexane:isopropanol (99:1, v:v) and filtered with a 0.45 μm polytetrafluoroethylene filter. Subsequently, an aliquot of the filtered fraction was treated with iodic acid following the procedure described by Kaneko et al. (2011) to cleave ether bonds from the GDGTs. All the apolar and polar fractions were analyzed using a GC equipped with an FID, GC connected to a mass spectrometer (MS), and high-performance liquid chromatography-atmospheric pressure positive-ion chemical ionization-MS (HPLC-APCI-MS). The δ13C values of selected compounds were determined by an IRMS connected to a GC via a combustion interface [glass tube packed with copper oxide (CuO), operated at 850°C]. Isotopic values were expressed as δ13C values per mil relative to the VPDB. The δ13C values were corrected for the introduction of additional carbon atoms during salivation. The analytical errors were less than ± 0.4‰.
Nucleic Acid Analysis
Genomic DNA was extracted using an Exgene Soil SV mini kit (Cambio, United Kingdom) at GeneAll (Seoul, South Korea). The V4-V5 regions (coverage of 85–95%) of bacterial 16S rRNA genes and the V6-V8 regions (coverage of 66–88%) of archaeal 16S rRNA genes were amplified by polymerase chain reaction using primer pairs 515F/926R and A956F/A1401R, respectively, at the Integrated Microbiome Resource (IMR) at Dalhousie University, Canada1 (Walters et al., 2016). Sequencing of the amplicons was conducted at IMR using the paired-end (2 × 300 bp) Illumina MiSeq system (Illumina, United States). Paired-end sequences were trimmed based on quality scores using Sickle (Schirmer et al., 2015) followed by error corrections with BayesHammer (Nikolenko et al., 2013). The resultant quality-trimmed and error-corrected paired-end sequences were assembled using PANDAseq (Masella et al., 2012). Sequence-processing steps were performed by following the Miseq SOP in mothur. The assembled sequences were aligned against a SILVA alignment (version 1322; Pruesse et al., 2007) and subsequently denoised using the “pre.cluster” command. Chimeric sequences were removed using the “chimera.uchime” command in de novo mode (Edgar et al., 2011). The sequences were further clustered to operational taxonomic units (OTUs) at a 97% sequence similarity level using the OptiClust clustering algorithm. Sequences were normalized by rarefying sequences per sample to the smallest library size (17,667 for bacteria and 6,768 for archaea). Taxonomic assignments of each OTU (2,286 bacterial OTUs and 2,758 archaeal OTUs) were determined by sequence similarity searches against the EzBioCloud database (Yoon et al., 2017). All sequence data used in this study were deposited in the Sequence Read Archive at the National Center for Biotechnology Information under the accession numbers PRJNA559003 and PRJNA643655. Phylogenetic trees of major archaeal OTUs of Methanomicrobia and bacterial OTUs of Gammaproteobacteria and Deltaproteobacteria with greater than 3% relative abundance were constructed using the maximum-likelihood algorithm (Felsenstein, 1981) and the General Time Reversible evolutionary model with MEGA X (Kumar et al., 2018). The robustness of the tree topologies was assessed by bootstrap analyses based on 1,000 replications of the sequences. To infer the putative sulfate-reducing partners of ANMEs, co-occurrence between two dominant archaeal OTUs of ANMEs and 8 bacterial OTUs of Deltaproteobacteria with greater than 3% relative abundances was investigated by calculating Pearson correlation coefficients using the “cor” function of the base R package.
Results
Geochemical Profiles
The gas and porewater properties varied along the sediment depths of ST1 (Figure 2 and Supplementary Table 1). The methane concentrations were below 5.4 mM, and the δ13CCH4 values ranged from −42.8 to −15.0‰. The sulfate concentrations varied between 2.9 and 27.8 mM. The measured DIC concentrations and δ13CDIC values ranged from 2.6 to16.1 mM and from −25.9 to −7.8‰, respectively. The phosphate concentrations were in the range of 0.3–0.8 mM, while the ammonium concentrations were below 0.1 mM. For the bulk elements, the TOC contents and δ13CTOC values ranged from 1.2 to.4 wt.% and from −27.0 to −26.4‰, respectively (Figure 2). The TS contents and δ34STS values varied between 0.3 and 0.5 wt.% and ranged from −18.3 to −2.4‰, respectively (Figure 2).
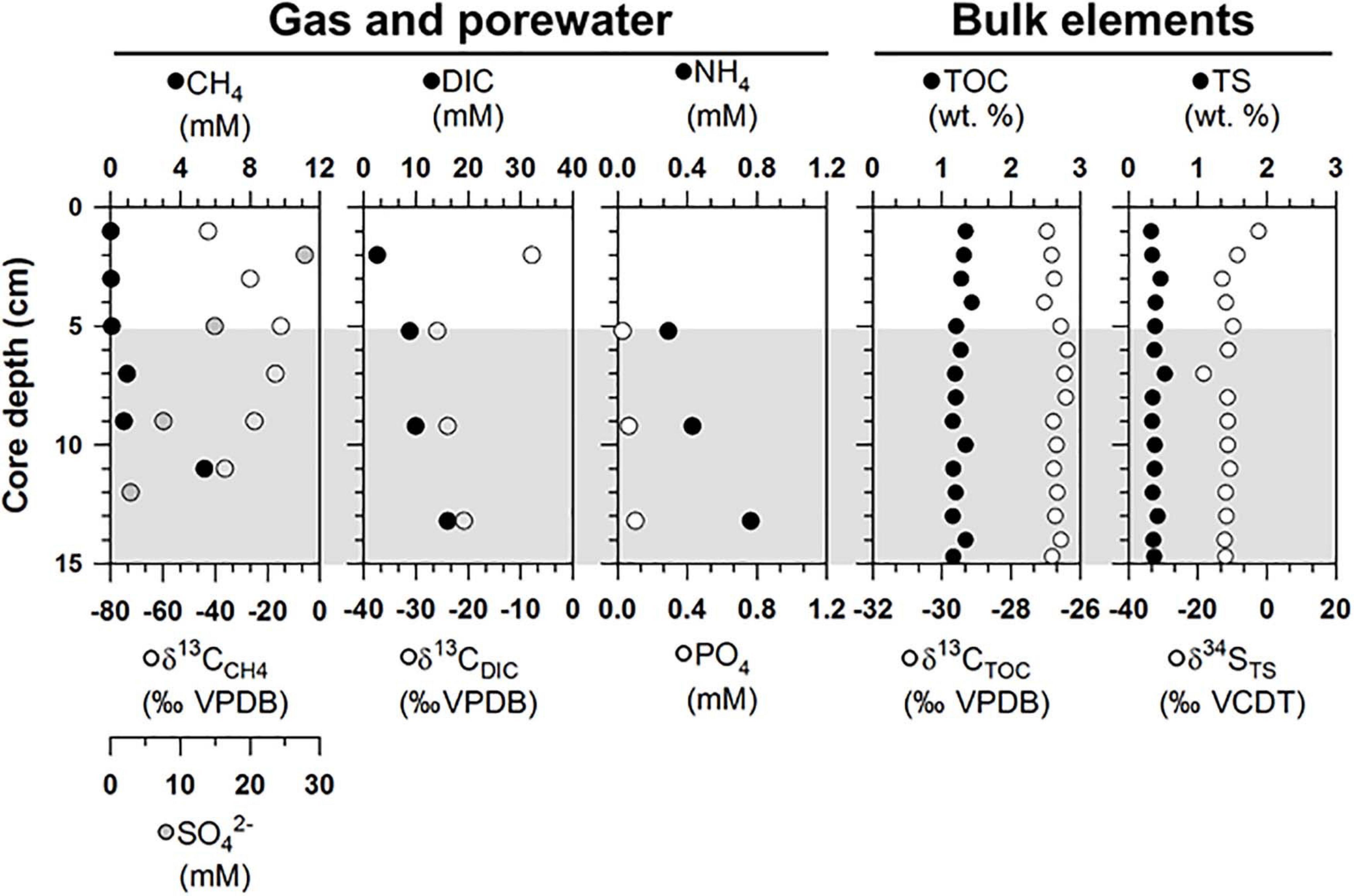
Figure 2. Depth profiles of gas (methane), porewater [sulfate, dissolved inorganic carbon (DIC), nutrients], and bulk elements [total organic carbon (TOC) and total sulfur (TS) contents] and isotopic compositions of methane, DIC, TOC, and TS (δ13CCH4, δ13CDIC, δ13CTOC, and δ34STS) obtained from the ST1 sites. Note that the gray shading indicates potential SMTZ layers.
Lipid Biomarker Profiles
Archaeal lipids (archaeol and sn-2-hydroxyarchaeol) were found at ST1 (Figure 3 and Supplementary Table 2). Both lipids were below 0.09 μg g–1 in dry weight (dw). Moreover, site ST1 showed the presence of GDGTs containing zero to three cyclopentane moieties (GDGT-0 to GDGT-3) and crenarchaeol, which contains a cyclohexane moiety (Supplementary Table 2). Overall, the isoprenoidal GDGTs were dominated by GDGT-0 and crenarchaeol, with concentrations of 0.05–0.19 μg g–1 dw and 0.03–0.24 μg g–1 dw, respectively, whereas GDGT-1, −2, and −3 showed substantially lower concentrations (≤ 0.01 μg g–1 dw) (Figure 3). In the apolar fractions, we did not identify any isoprenoid hydrocarbons typically associated with AOM-related methanotrophs, that is, the C20 compound 2,6,11,15-tetramethylhexadecane (crocetane) or the C25 compound 2,6,10,15,19-pentamethylicosane (PMI). In contrast to specific archaeal lipid profiles, MOx-related bacterial lipids (i.e., 4α-methyl sterols) were predominantly detected in the upper sediment layers at ST1 (Figure 3). Their concentrations ranged from 0.08 to 0.18 μg g–1 dw. Bacterial fatty acids (i.e., C16:1ω8 and C16:1ω5) were also more abundant in the upper sediment layers, ranging from 0.01 to 0.14 μg g–1 dw and from 0.01 to 0.02 μg g–1 dw, respectively (Figure 3). The δ13C values of archaeol and sn-2-hydroxyarchaeol varied between −56.5 and −51.7‰ and between −102.3 and −78.6‰, respectively (Figure 3). The δ13C values of biphytanes (BPs) derived from the isoprenoid GDGTs were in the range of −54.7 to −19.2‰ (Figure 3 and see Supplementary Table 2). Among them, the δ13C values of BP-1 (on average −46.6 ± 6.5‰, n = 5) were slightly lower than those of BP-3 (on average −28.1 ± 6.7‰, n = 6). The δ13C values of 4α-methyl sterols varied between −49.5 and −36.5‰, whereas those of the fatty acids ranged from −44.5 to −40.0‰ (Figure 3).
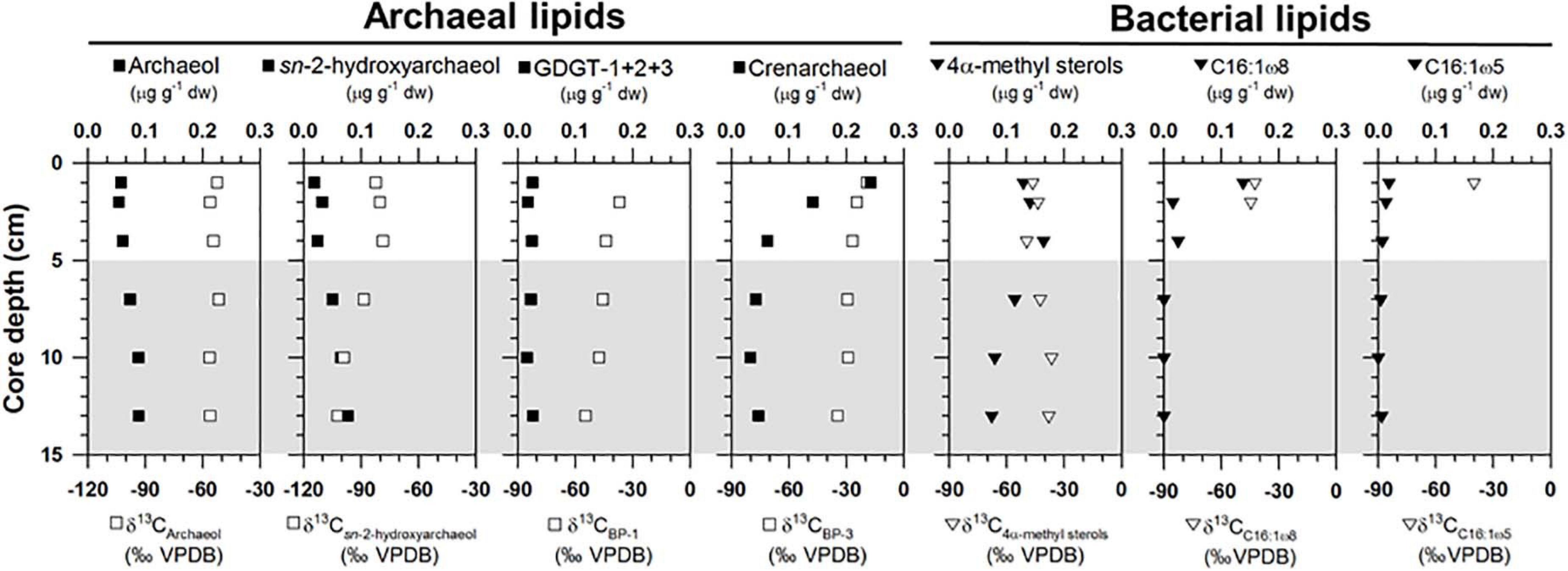
Figure 3. Depth profiles of concentrations and isotopic compositions of selected lipid biomarkers (archaeol, sn-2-hydroxyarchaeol, GDGTs, 4α-methyl sterols, and bacterial fatty acids). Note that the gray shading indicates potential SMTZ layers.
Microbial Composition and Occurrence of Methanotrophs
The archaeal phyla detected at ST1 were Euryarchaeota, Bathyarchaeota, Thaumarchaeota, and Lokiarchaeota (Figure 4). Except for relatively higher proportions of Thaumarchaeota near the surface, Euryarchaeota was predominant below the 2-cm sediment layers, accounting for 18.0–99.0% of the total archaeal sequences. Among them, ANME-3 (A_OTU001) was dominant (up to 96.4%) along the sediment depths (Supplementary Table 3). Another archaeal OTU (A_OTU008) belonging to ANME-2 was less than 0.4% in the sediment layers of ST1 (Supplementary Table 3). For the bacterial communities, Proteobacteria was most predominant in all sediment samples of ST1. Other bacterial communities (i.e., Bacteroidetes, Ca. Atribacteria, Planctomycetes, Firmicutes, and Ca. WM88) showed minor abundances along the sediment core of ST1 (Figure 4). The proportion of Proteobacteria was higher in the deeper sediment layers (6–10 cm). In contrast, the proportions of Bacteroidetes and Planctomycetes were relatively higher at the surface, while those of Ca. Atribacteria, Firmicutes, and Ca. WM88 increased along the sediment core depth. The order Methylococcales of Gammaproteobacteria (B_OTU004) showed relatively higher proportions (0.9–4.1%) near the surface (∼5 cm) (Figure 5 and Supplementary Table 3). The Deltaproteobacteria, belonging to the family Desulfobacteraceae or Desulfobulbaceae (nine major OTUs), showed low similarity with the cultured representatives (Supplementary Table 3). Overall, Desulfobulbaceae (B_OTU010) showed relatively higher proportions (1.4–7.2%) at 1–8 cm sediment depths (Figure 5 and Supplementary Table 3). According to the Pearson correlations (Supplementary Table 4), at the ST1 site, both archaeal OTUs showed significant positive correlations (r = 0.71–0.82, p < 0.01) with four deltaproteobacterial OTUs (B_OTU002, B_OTU011, B_OTU021, and B_OTU029) (Supplementary Tables 3,4). At the ST2 site, A_OTU001 showed significant positive correlations (r = 0.68–0.92, p < 0.01) with B_OTU002, B_OTU011, and B_OTU021 while A_OTU008 showed significant positive correlations (r = 0.71, p < 0.01) with B_OTU018 and B_OTU029 (Supplementary Tables 3,4).
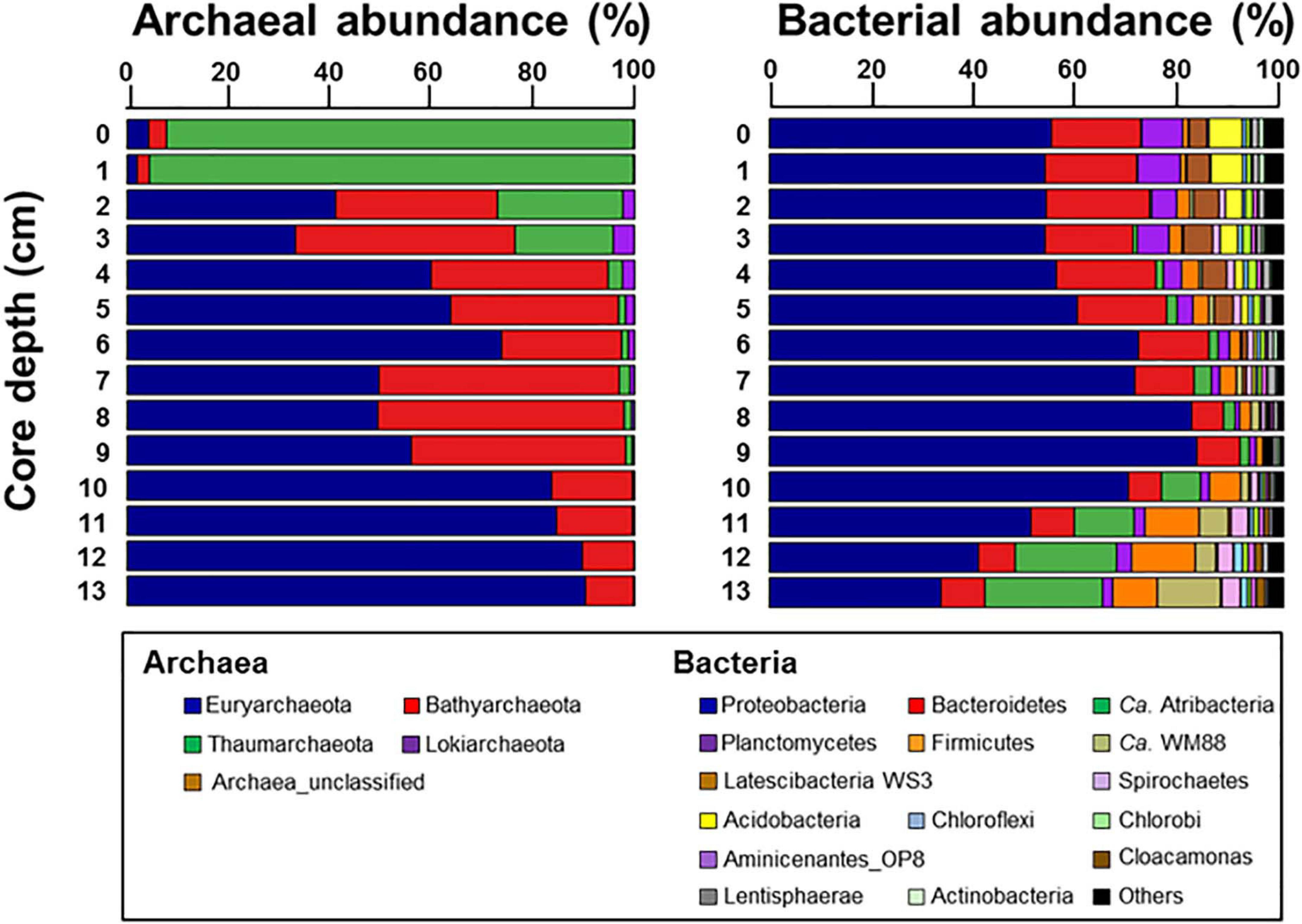
Figure 4. Relative abundances of archaea taxa at the class level and bacteria taxa at the phylum level with the exception of Proteobacteria which was represented at the class level along the sediment core depths.
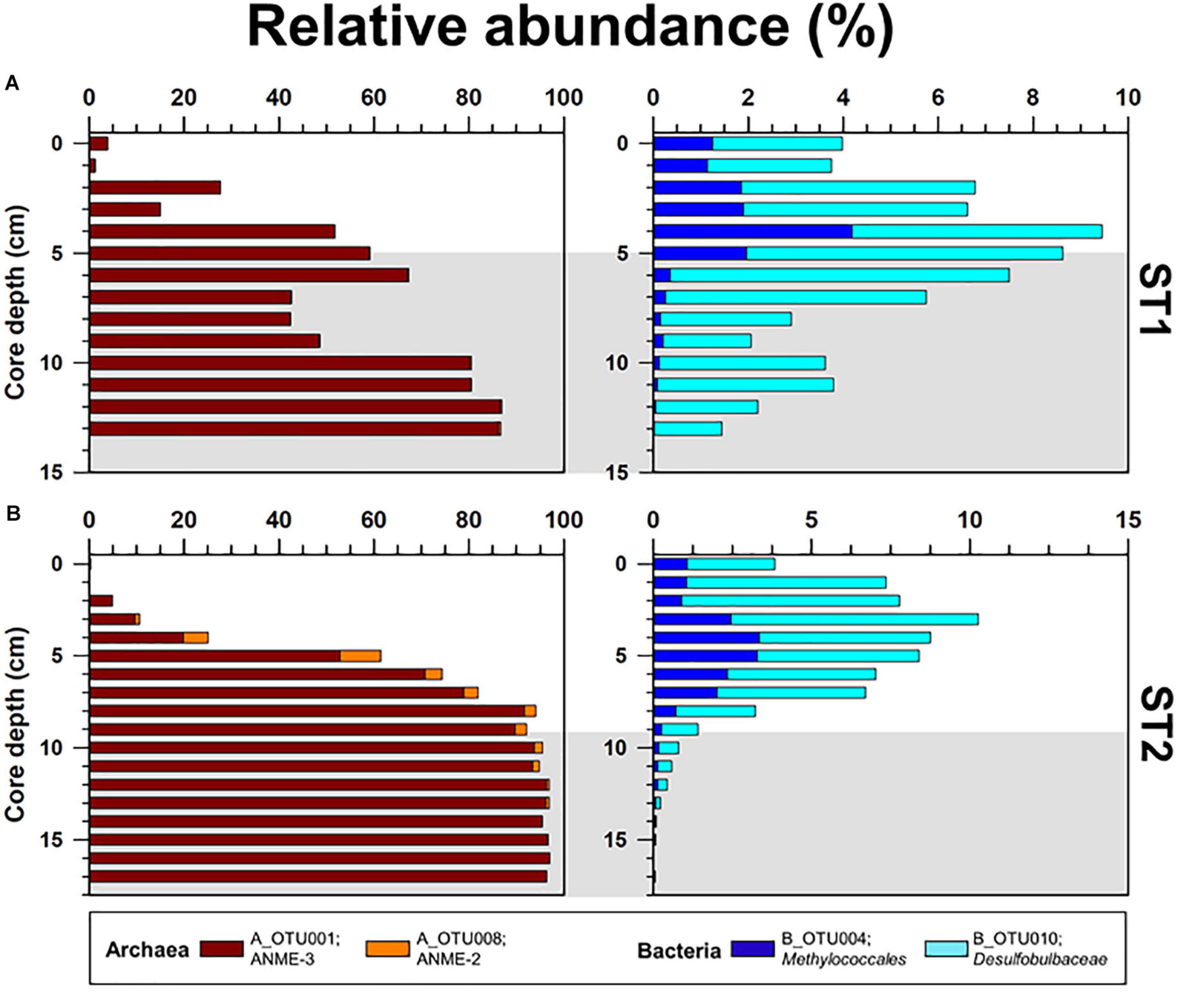
Figure 5. Depth profiles of the relative abundances of major OTUs (Methanomicrobia; A_OTU001 and A_OTU008, and Methylococcales; B_OTU004 and B_OTU010) obtained from the (A) ST1 and (B) ST2 sites. The ST2 data were obtained from Lee et al. (2019b). Gray shading indicates potential SMTZ layers.
Discussion
Geochemical Signatures of Methane Oxidation
Overall, the methane concentrations were lower in the upper 5 cm of the sediment core at ST1 (Figure 2), which was comparable to those previously reported at ST2 (Lee et al., 2019b, see Supplementary Figure 1). This depleted profile of methane concentration at ST1 was also similar to those at other cold seepages (e.g., Haakon Mosby MV and Japan Trench) which were characterized by tubeworm patches and clam colonies (de Beer et al., 2006; Felden et al., 2014). Moreover, the δ13CCH4 profile investigated at ST1 was enriched only to a sediment depth of 5 cm, in contrast to the relatively consistent δ13CCH4 values observed at ST2 (Figure 2 and see also Supplementary Figure 1). The enriched degree of δ13CCH4 values appears to be related to methane oxidation in connection with the penetrated sulfate depths, due to selective isotopic fractionation (Whiticar et al., 1986; Boetius et al., 2000; Knittel and Boetius, 2009). In such cases, most of the methane is reduced via AOM, which occurs at sulfate-penetrated depths within the sulfate-methane transition zone (SMTZ) (Reeburgh, 1980), indicating that the remaining methane can approach relatively enriched isotope values (Pohlman et al., 2009). Notably, the signature of 13C-enriched methane could be in part attributed to the contribution of methylotrophic methanogenesis, as shown by the metabolism of Methanococcoides that was cultivated from seeping sediments (Wegener et al., 2016). It is also worth noting that tubeworms live in symbiosis with thiotrophic bacteria that utilize sulfide as energy source and release sulfate into sediments (Lösekann et al., 2007; Felden et al., 2010). Hence, the 13C-depleted value of DIC at 5 cm sediment depth might be associated with the enhanced AOM. The depth and thickness of SMTZ within tubeworm habitats can be thus controlled by various factors, such as the depth of the methane production zone, bioirrigation, the flux of methane and sulfate, and their consumption rates (Jørgensen and Kasten, 2006; Knittel and Boetius, 2009; Felden et al., 2010; Meister et al., 2013). Considering the different methane fluxes calculated at both ST sites (Table 1), the SMTZ may have formed below a sediment depth of approximately 5 cm at ST1, whereas it may occur at deeper sediment depths (i.e., below ca. 9 cm) at ST2. With respect to the incomplete depletion of methane and sulfate in the deeper sediment layers, we could not determine the exact SMTZ layers at both sites. Nevertheless, together with 13C-enriched methane, the most 13C-depleted DIC (∼-25.9‰) occurred at a 5-cm sediment depth at ST1 (Figure 2), reflecting the potential increase of AOM-derived byproducts within the sediments (de Beer et al., 2006; Felden et al., 2010). The relative contribution of the AOM-derived DIC along the sediment depths was approximately estimated based on the isotopic mass balance (cf. Coffin et al., 2013), assuming that the δ13C signatures of the parent methane and seawater DIC in the Canadian Beaufort Sea were −64 and −0.4‰, respectively (Paull et al., 2015). Our results revealed that 11.7–40.2% of the total DIC was contributed by the AOM-derived DIC source at both ST sites (see also Supplementary Figure 2 and Table 1). In particular, the highest contribution of AOM-derived DIC in the depth profiles at ST1 was consistent with the most depleted δ13CDIC value at a 5-cm sediment depth, compared to the relatively lower contributions (below 30%) at ST2. Furthermore, the increasing nutrient profiles in the deeper sedimentary layers at both ST sites were typically indicative of the presence of byproducts derived from the degradation of organic matter (Figure 2).
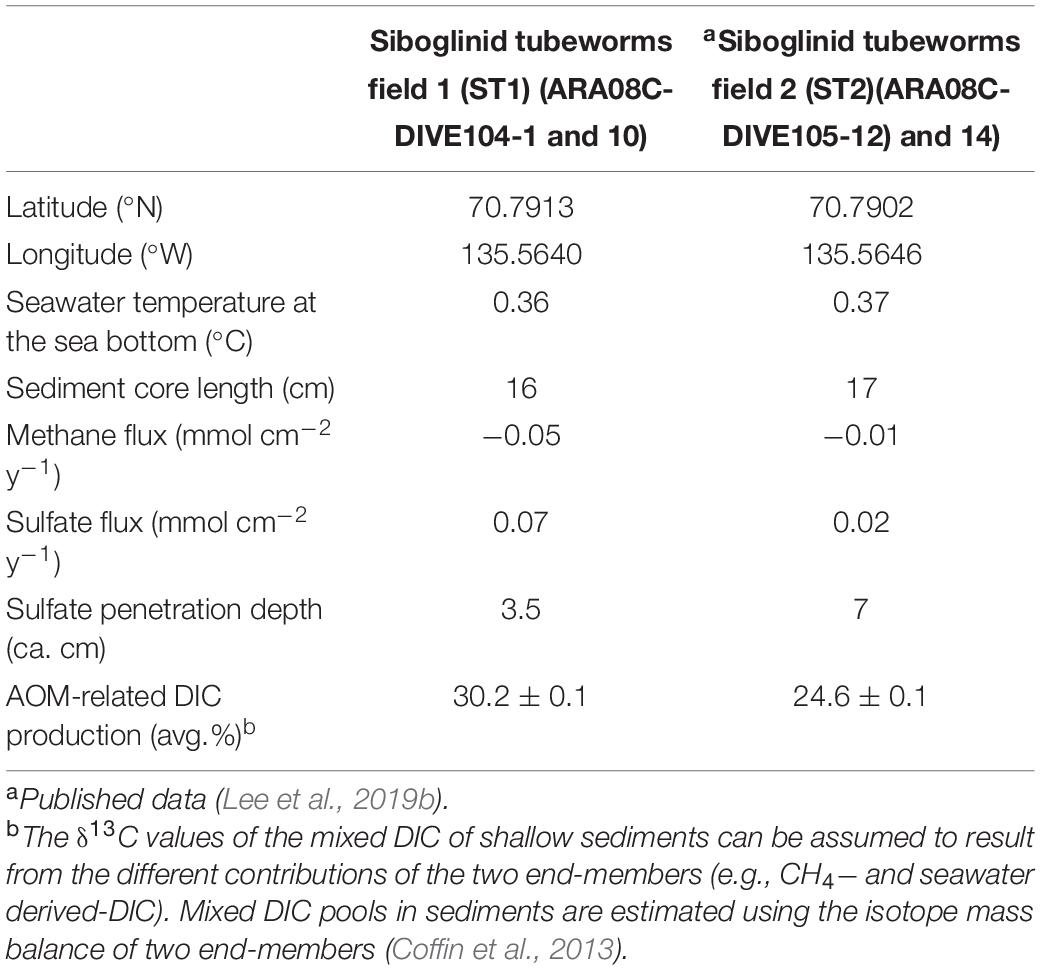
Table 1. Geographic coordinates (latitude and longitude), water temperature at the sea bottom (°C), sediment core length, and methane and sulfate fluxes (mmol cm–2 y–1) calculated from concentration profiles and penetration depths (cm) of sulfate and AOM-related DIC production (%).
The bulk element (C and S) contents and their isotopic values showed different vertical variations at both ST sites (Figure 2 and see also Supplementary Figure 1). The origin of 13C-depleted isotopic signatures is commonly regarded as terrestrial plants (Peters et al., 1978; Fry, 1999; Perdue and Koprivnjak, 2007). However, 13C-depleted TOC has also been found in seeping sediments, likely resulting from the assimilation of methane into bacterial biomasses (Haese et al., 2003; Elvert et al., 2005). In this regard, the bacterial biomass can be incorporated and preserved in solid-phase sediment, and through time can constitute a significant portion of the organic carbon pool (Canuel and Martens, 1993; Gong and Hollander, 1997). As AOM occurs at deeper sediment layers under low methane fluxes (Niemann et al., 2006; Lee et al., 2019b), bacterial biomass involved in MOx are incorporated to some degree into the sedimentary carbon pool at the surface of ST2. Under this setting, the isotopic shift in the TS profiles may also provide indirect evidence for the supply of electron acceptors (sulfate and oxygen) derived from seawater (Canfield and Thamdrup, 1994). Assuming that seawater sulfate with a δ34S value of + 21‰ was supplied to the sediments (Rees et al., 1978), the significant isotopic shift of δ34STS at ST2 can potentially be regarded as the active penetration of downward sulfate with oxygen from seawater. Conversely, with respect to the significant AOM signals at ST1, the depleted δ34S signatures (−12.0 ± 2.0‰) along depth profiles may indicate that sulfide or sulfur species are likely produced via microbial SR coupled with methane oxidation (Boetius et al., 2000; Jørgensen et al., 2004; Reeburgh, 2007). Indeed, the δ34S values of sulfide may be depleted by as much as 46‰ relative to those of seawater sulfate during SR (Hoefs, 2007). With respect to the input of seawater-derived sulfate and the oxidation of sulfide to intermediates (e.g., elemental sulfur and thiosulfate), the sulfide pool at ST1 appears to be, at least in part, associated with AOM-derived SR. Although more complicated sulfur cycles involved in alternative pathways may occur in sediments, the range of δ34STS values at both sites suggests that SR coupled with AOM was more active under higher methane fluxes. Therefore, we argue that the different methane fluxes may be potentially associated with the discriminative biogeochemical signals observed at both ST sites.
Molecular Biogeochemical Signatures of Methane Oxidation
For AOM-related biomarkers, we identified sn-2-hydroxyarchaeol at ST1 and ST2 (Figure 3 and see also Supplementary Figure 3). The sn-2-hydroxyarchaeol concentrations were generally higher in the SMTZ than in the surface layers at both sites (Figures 6A,C), with 13C-depleted values compared to the δ13CCH4 values (δ13CCH4; −27.1 ± 10.8‰ (n = 6) at ST1 and −59.2 ± 4.5‰ (n = 4) at ST2). The difference in the δ13C values between sn-2-hydroxyarchaeol and methane (i.e., Δδ13C) in the SMTZ was, on average, −69.5 ± 7.2‰ (n = 3) at ST1 and −48.7 ± 2.4‰ (n = 2) at ST2 (Figure 6B). The methane-derived microbial biomass was generally depleted in 13C compared to the source methane as a result of isotopic fractionation during methane assimilation (Whiticar, 1999). The negative Δδ13C values of sn-2-hydroxyarchaeol in the SMTZ at both sites were thus indicative of the AOM occurrence in sediments where sulfate was present (Pancost et al., 2000; Elvert et al., 2005; Niemann et al., 2005; Bradley et al., 2009). Furthermore, the Δδ13C values of predominant archaeol at ST1 (−27.7 ± 2.7‰, n = 3) were also negative (Figures 6A,B), providing additional evidence for an active AOM in the SMTZ at ST1. It is worth noting that the Δδ13C values of both sn-2-hydroxyarchaeol and archaeol were more depleted at ST1 than at ST2, suggesting that the AOM activities were stronger at ST1 than at ST2. Interestingly, the SMTZ Δδ13C values of archaeol were more enriched in the Håkon Mosby Mud Volcano (HMMV) setting, which was also covered by beggiatoa organisms, than at the Beaufort sites. Enriched 13C-archaeol has typically been associated with the lipid production of methanogens that use CO2 as their carbon source (Belyaev et al., 1983). Accordingly, the isotopic compositions of methanogenic biomarkers are approximately equal to those of methane. Thus, considering the isotopic fractionation involved in methanogenesis, the 13C-enriched archaeol at the HMMV was attributed to the contribution of methane-producing, rather than methane-consuming organisms (Whiticar, 1999; Niemann et al., 2006).
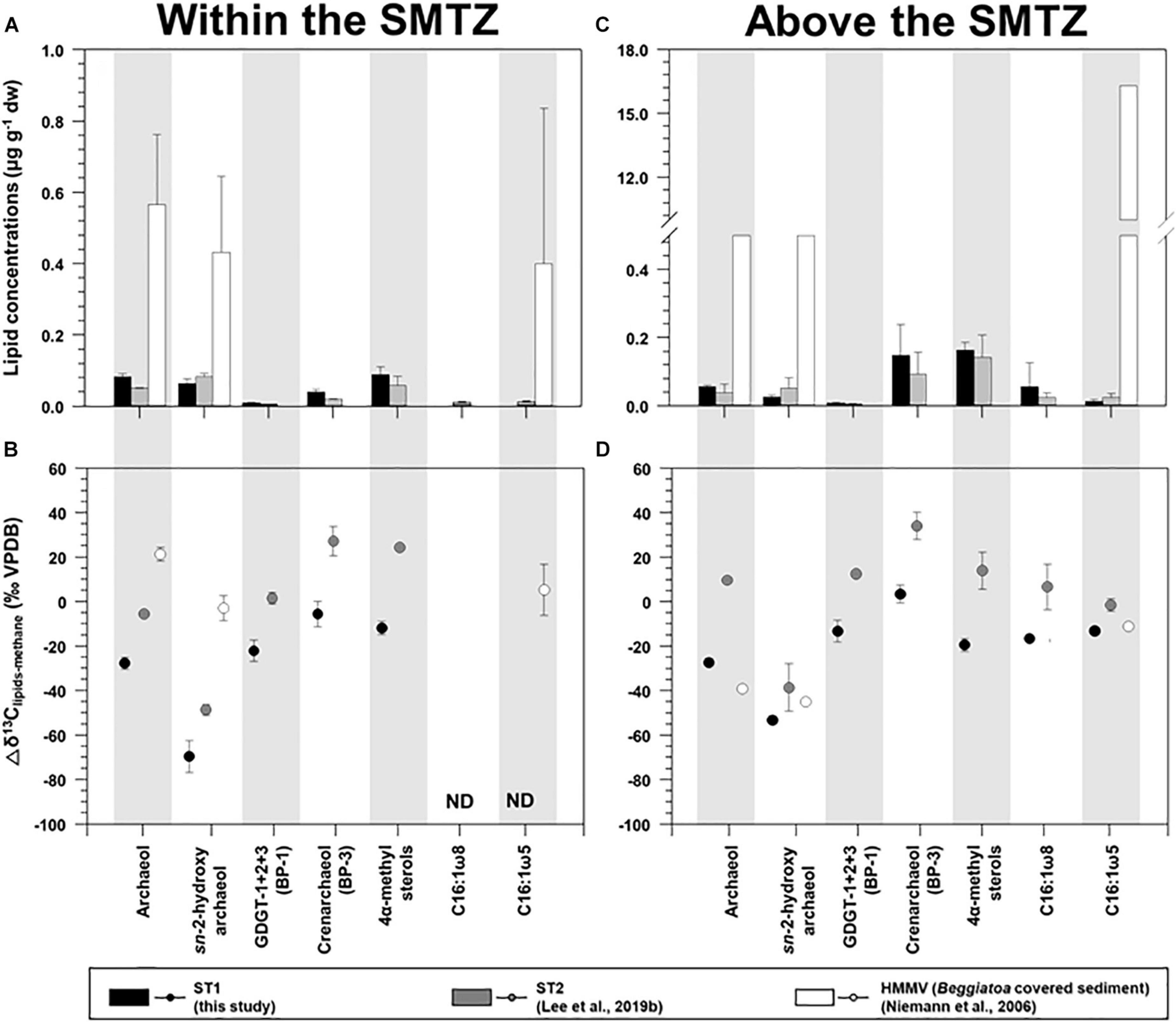
Figure 6. Comparison of the concentrations and isotopic compositions (Δδ13C) of selected lipid biomarkers (archaeol, sn-2-hydroxyarchaeol, biphytanes derived from GDGTs, 4α-methyl sterols, and bacterial fatty acids) obtained from ST1, ST2, and the Haakon Mosby mud volcano (HMMV): sites (A,B) for the sediment layers within the SMTZ, and (C,D) for the sediment layers above the SMTZ. Note that the ST2 and HMMV data were obtained from Niemann et al. (2006) and Lee et al. (2019b), respectively. Δδ13C values were calculated by subtracting the δ13C values of methane from those of selected lipid biomarkers.
Isoprenoidal GDGTs were found in all the sediments investigated (see Supplementary Figure 3 and Supplementary Table 2). The concentrations of GDGT-1, -2, and -3 remained constant along the core depth; however, both GDGT-0 and crenarchaeol were more abundant near the surface layers at both sites. Moreover, the isoprenoidal GDGT distributions did not show a clear dominance of GDGT-1, −2, and −3 over GDGT-0, which is an indicator of the contribution of methanotrophic Euryarchaeota (Schouten et al., 2013). The values of other methanotroph-related indices such as GDGT-0 to crenarchaeol (Liu et al., 2011) and GDGT-2 to crenarchaeol (Weijers et al., 2011), and the methane index values (Zhang et al., 2011) were also low, with ranges of 0.4–1.6, 0.0–0.2, and 0.1–0.4, respectively. Accordingly, it appears that marine pelagic Thaumarchaeota (e.g., Pearson et al., 2001; Wuchter et al., 2005; Schouten et al., 2013) were the major contributors to the total GDGT pool in the Canadian Beaufort Sea MV, in contrast to other cold seeps such as the Hydrate Ridge (Elvert et al., 2005) and the Marmara Sea (Chevalier et al., 2013). With respect to carbon metabolism at the molecular level, BP-1 has one cyclopentane ring that may originate from GDGT-1, -2, or -3 (Schouten et al., 2013). If methanotrophs are responsible for synthesizing these three GDGT compounds, then the isotopic signatures derived from BP-1 are subject to the presence of AOM (Niemann et al., 2005). The Δδ13C values of BP-1 in the SMTZ at ST1 were similar to those of archaeol (Figure 6B), suggesting the influence of AOM-related methanotrophs. However, the contribution of the AOM-related methanotrophs should be minor, considering the low methanotroph-related indices mentioned above. In contrast, BP-3 is produced only by crenarchaeol, and thus contains the pelagic Thaumarchaeota signature (Schouten et al., 2013). The 13C-enriched isotopic signatures of BP-3 relative to other compounds in the SMTZ at both ST sites (Figure 6B) were comparable to those previously reported in non-seeping marine environments (Hoefs et al., 1997). This indicates that marine pelagic Thaumarchaeota using seawater derived-DIC as a carbon source were the primary contributors to the GDGT pool at both sites (e.g., Hoefs et al., 1997; Könneke et al., 2005; Wuchter et al., 2005).
For MOx-related biomarkers, we identified 4α-methyl sterols and C16:1ω8 at ST1 and ST2 (Figure 3 and see also Supplementary Figure 3). Overall, the concentrations of 4α-methyl sterols increased toward the upper sediment layers at both sites (Figure 3 and see also Supplementary Figure 3). Their occurrences have been associated with aerobic methanotrophs (e.g., Methylococcales belonging to alphaproteobacteria) near the surface where oxygen as an electron acceptor is likely available (Elvert and Niemann, 2008). The δ13C values of the 4α-methyl sterols were more depleted near the surface at both sites (Figure 3 and see also Supplementary Figure 3). Thus, it appears that the MOx process predominantly occurred in the upper sediment layers at both sites. The biosynthesis of 4α-methyl sterols might occur during the oxidative removal of methyl carbons at C-4 and C-14 of lanosterol or of another, yet undescribed, protosterol (Pearson et al., 2003). Furthermore, a previous study suggested that Methylococcus capsulatus uses an alternative enzymatic pathway for the C-4 demethylase reaction (Summons et al., 2006). Therefore, the predominance of these MOx-related methanotrophs near the surface at both STs seems to be influenced by more effective use of nutrients and substrates (e.g., oxygen/nitrate and methane), in contrast to those of AOM-related archaeal and sulfur-oxidizing bacterial communities. Notably, the average Δδ13C value of 4α-methyl sterols investigated at ST1 (−19.4 ± 3.0‰, n = 3, Figure 6D) was more depleted than that of ST2 (14.1 ± 8.3‰, n = 4, Figure 6D), and most likely resulted from the stronger MOx activities at ST1 than at ST2. As supporting evidence, we also found higher abundances and depleted δ13C values of C16:1ω8 in the surface layers at ST1 and ST2 (Figure 3). However, the Δδ13C values of C16:1ω8 were lower at ST1 (−16.4 ± 1.5‰, n = 2) than at ST2 (−19.4 ± 3.0‰, n = 3), similar to those of the 4α-methyl sterols (Figure 6D). With respect to the synthesis of C16:1 fatty acids with ω8 double bond positions (Bowman et al., 1991, 1993; Fang et al., 2000), the 13C-depleted C16:1ω8 values in the near-surface layers at ST1 suggest the presence of MOx-related bacteria derived from type I methanotrophs (Makula, 1978; Nichols et al., 1985). In particular, the negative Δδ13C values of C16:1ω8 are indicative of the involvement of type I methanotrophs in the RuMP pathway for methane oxidation (Jahnke et al., 1999).
As evidence for other bacterial communities, we found C16:1ω5 at both sites, which were also more abundant in the upper sediment layers where the 4α-methyl sterols were predominant (Figure 3 and see also Supplementary Figure 3). The Δδ13C values of C16:1ω5, associated with sulfate-reducing bacteria (Elvert et al., 2003; Blumenberg et al., 2004; Niemann et al., 2006), were lower in the upper sediment layers at ST1 (−12.9‰, n = 1, Figure 6D) than at ST2 (-1.4 ± 2.8‰, n = 3, Figure 6D). The Δδ13C values of C16:1ω5 at both sites were more depleted than those (Δδ13C values: 30–40‰) in non-seeping Arctic environments (Birgel et al., 2004). Given that the MOx-related methanotrophs relative to the AOM-related methanotrophs were more abundant near the surface layers, as discussed above, the isotopic signature of C16:1ω5 at ST1 might be related to the assimilation of MOx-derived DIC. Hence, the Δδ13C pattern of bacterial fatty acids (i.e., C16:1ω8 and C16:1ω5) at ST1 and ST2 was in line with that of the MOx-related biomarker, that is, the 4α-methyl sterols, which suggests that MOx-related processes were stronger at ST1 than at ST2.
Possible Mechanism Shaping Siboglinid Tubeworm Fields
The composition and δ13C values of microbial lipids provide insight into the chemotaxonomic composition of microbes involved in sulfate-dependent AOM (Niemann and Elvert, 2008). PMIs that are produced by anaerobic methanotrophic archaea (ANME-1, ANME-2, and ANME-3) (Niemann and Elvert, 2008) were not present at either ST site. Crocetane, which is structurally similar to PMIs but diagnostic for ANME-2 (Elvert et al., 1999), was also not detected in any of the sediments investigated. However, archaeol, which serves as an indicator of methanogenic archaea in a wide range of environments, including MVs (e.g., de Rosa and Gambacorta, 1988; Koga et al., 1993, 1998; Pancost et al., 2011), and sn-2-hydroxyarchaeol, which has only been found in certain orders of methanogens (e.g., Kushwaha and Kates, 1978; Koga et al., 1993, 1998; Koga and Morii, 2005), were detected in all sediments, as mentioned in section of molecular signatures. However, the increase of 13C-depleted sn-2-hydroxyarchaeol in the deeper sediment layers at both ST1 and ST2 provided evidence for the occurrence of methanotrophic activities under anaerobic conditions (e.g., Hinrichs et al., 2000; Stadnitskaia et al., 2008). AOM-related microbial communities dominated by ANME-2 and ANME-3 contain higher concentrations of sn-2-hydroxyarchaeol relative to archaeol (Blumenberg et al., 2004; Niemann and Elvert, 2008). Therefore, the ratio of isotopically depleted sn-2-hydroxyarchaeol relative to archaeol suggests that ST2, with a value range of 0.6–1.8, is associated with ANME-2 and ANME-3, and not ANME-1, which has lower ratios (0:0.8) (Niemann et al., 2006; Niemann and Elvert, 2008). In contrast, this ratio was relatively low at ST1 (0.3–0.9), indicating relatively small contributions from ANME-2 (e.g., Blumenberg et al., 2004; Niemann and Elvert, 2008). In general, GDGT-1, -2, and -3 derived from ANME-2 (except for ANME-2c) and ANME-3 are minor components, whereas ANME-1 produces more abundant GDGT-1, -2, and -3 (Blumenberg et al., 2004; Elvert et al., 2005; Stadnitskaia et al., 2005, 2008; Chevalier et al., 2013; Kurth et al., 2019). As a result, the ratio of GDGT-0 over GDGT-1, -2, and -3 is lower for ANME-1 (< 0.5, Stadnitskaia et al., 2008; Gontharet et al., 2009) than for ANME-2 and ANME-3 (> 2, Elvert et al., 2005). Thus, although the dominance of GDGT-0 and crenarchaeol at ST1 and ST2 was most likely introduced by Thaumarchaeota (Schouten et al., 2013), together with the ratio of sn-2-hydroxyarhcaeol and archaeol, the GDGT distribution with high ratios of GDGT-0 over GDGT-1, -2 and -3 (2.0-14.4) at our ST sites provides further supporting evidence that ANME-2 and ANME-3 were more actively involved in AOM than ANME-1 at both ST sites, which is in line with a previous study in the Beaufort Sea (Lee et al., 2018).
To further clarify the phylogenetic position within the class Methanomicrobia, phylogenies of the two dominant Methanomicrobia OTUs (A_OTU001 and A_OTU008) were inferred from 16S rRNA gene sequences (Supplementary Figure 4 and Supplementary Table 3). ANME-3 (A_OTU001) accounted for more than 50% of the archaeal sequences at 5–17 cm sediment depths at both sites (Figure 5). ANME-2 (A_OTU008) was mostly detected at 4–9 cm sediment depths at ST2, accounting for 2.4–8.7% of total OTUs (Figure 5). Together with our lipid data, ANME-2 and ANME-3 groups were identified at 4–11 cm sediment depths at ST2, in contrast to the predominant occurrence of ANME-3 at ST1. To investigate the relationship between ANMEs and SRB, the presence of the sulfate-reducing Deltaproteobacteria was determined by analyzing deltaproteobacteria OTUs (Desulfobacteraceae and Desulfobulbaceae) at both ST sites. Several OTUs of Desulfobacteraceae showed positive Pearson correlations with dominant ANME-2 and ANME-3. However, deltaproteobacterial OTUs obtained from the Beaufort Sea which have positive correlations with ANME-2 and ANME-3 are phylogenetically distant from previously known SRB of Desulfosarcina-Desulfococcus or Desulfobulbus (Knittel et al., 2005; Niemann et al., 2006). Hence, to better assess ANME-SRB consortia, further studies in the Beaufort Sea are required by applying fluorescence in situ hybridization. Notably, both ST sites contained the 4α-methyl sterols, which are putatively specific for bacterial groups involved in MOx. The 13C-depleted 4α-methyl sterols were more abundant in the upper sediment layers at both ST sites in conjunction with high proportions of OTUs belonging to the order Methylococcales. The vertical distributions of Methylococcales varied from those of ANME-2 and ANME-3, showing higher proportions in the shallower sediment layers at both ST sites. Thus, the vertically discriminative trends between the AOM- and MOx-related methanotrophs may be related to the supply of available electron acceptors (i.e., sulfate and oxygen) under the bioirrigation activities of STs.
At MV420, the distinct development of ST patches investigated at ST1 appeared to correlate with the active AOM process associated with mainly ANME-3, while the co-occurrence of ANME-2 and ANME-3 was observed at ST2 which was characterized by lower methane flux (Figure 5 and see also Supplementary Figure 4). In this regard, ANME-3 has been found only in cold seepages (including MVs) with high methane partial pressures and temperatures of < 20 °C (Niemann et al., 2006; Lösekann et al., 2007; Ruff et al., 2015; Gründger et al., 2019). For instance, ANME-3 was reported at the HMMV in the Barents Sea (at a water depth of 1,250 m), Kazan MV in the Eastern Mediterranean (at a water depth of 1,700 m), and surface sediments from the Guaymas Basin in the Gulf of California (at a water depth of 1,500 m) (Niemann et al., 2006; Heijs et al., 2007; Pachiadaki et al., 2010; Vigneron et al., 2013). The bioirrigation of STs sustains the supply of available electron acceptors (especially sulfate) in sediment layers, resulting in increasing AOM rates within the SMTZ (Treude et al., 2003; Niemann et al., 2006; Ruff et al., 2013, 2018). Therefore, ANME-3 might have been more strongly forced to tolerate high sulfide concentrations produced during the AOM at ST1 than at ST2, thereby supporting the metabolism of sulfur-oxidizing endosymbionts inhabiting STs (Hilário et al., 2011; Duperron et al., 2014; Lee et al., 2019a). Notably, ANME-2 (mainly ANME-2c) often occurs in sediments bioirrigated by benthic organisms, such as at the Hydrate Ridge in the Northeast Pacific and the REGAB pockmark in the Congo Basin (Elvert et al., 2005; Knittel et al., 2005; Pop Ristova et al., 2012), and in low fluid flux regimes (Elvert et al., 2005; Wegener et al., 2008). Therefore, ANME-2 detected at ST2 may have a preferential niche in relatively low–methane fluxes. With respect to the relatively small contribution of AOM-derived DIC at ST2, AOM activities by ANME-2 and -3 may be weakened for the production of sulfide and DIC, causing the low density of tubeworm patches. Furthermore, the commonly increased abundance of Methylococcales near the surface of both sites may additionally contribute to the DIC supply related to tubeworm metabolism.
A previous MV420 study in the Canadian Beaufort Sea suggested that visually discriminative habitats that were devoid of megafauna and microbial mats (DM) to the naked eye, covered with bacterial mats (BM), or colonized by STs were shaped in association with varying degrees of methane flux (Lee et al., 2019b). Notably, the BM (0.04 mmol cm–2 y–1) and ST1 (0.05 mmol cm–2 y–1) sites showed similar methane fluxes. Therefore, it appears that the methane flux is not a unique environmental factor for niche diversification of chemosynthetic organisms within an MV system. In this regard, the discriminative colonization of chemosynthetic organisms might be related to substrate availability, according to the mass transfer limitation of porewater fluids in stagnant sediments (Boetius and Suess, 2004; de Beer et al., 2006). Under slower up-flow fluids that permit the diffusional penetration of sulfate, AOM communities can be developed, producing sulfide, which is then aerobically oxidized by chemosynthetic bacteria (e.g., Beggiatoa and tubeworms). In contrast to DM site which was characterized by the recent, active methane outgassing, the sediments at the BM and ST1 sites were influenced by up-flowing methane for a longer period, providing sufficient time for a microbial and faunal colonization associated with the AOM. However, ST2 may have had insufficient time for its formation to be colonized by slower growing STs. Therefore, we infer that the differences in colonization between ST1 and ST2 may be related to the varying persistence of ascending methane, together with varied methane fluxes. Our data thus provide a new clue about a succession of potentially competing tubeworm communities over methane exposure time on the same MV.
Conclusion
Integrated biogeochemical and microbial investigations were performed on the sediment push cores retrieved from densely (ST1) and less densely (ST2) colonized tubeworm patches at MV420 in the Canadian Beaufort Sea. Compared to ST2, which recorded a low methane flux (0.01 mmol cm–2 y–1), the significantly depleted δ13CDIC values measured in the depth profiles of ST1 can be regarded as active AOM evidence, indicating the relatively highest contribution (40.2%) of AOM-derived DIC within the SMTZ of this site. Similarly, the mostly depleted δ34STS values recorded at ST1 also appear to reflect active SR involved in AOM. More specifically, we found isotopically 13C-depleted lipid biomarkers (sn-2-hydroxyarchaeol and 4α-methyl sterols) and related OTUs, belonging to ANME-2/-3 and Methylococcales, mediating AOM and MOx in the sediments of both STs. Notably, the prevalence of ST patches observed at ST1 may be associated with the supply of AOM-related byproducts (sulfide and DIC) performed mostly by ANME-3 as a result of active AOM. Conversely, at ST2, sulfide and DIC production via AOM may not be well facilitated by ANME-2 and -3, resulting in the low density of ST patches. Taking into consideration the common biogeochemical signatures investigated at another habitat under a similar methane flux as the Canadian Beaufort MV, a possible scenario explaining the biological succession of benthic communities is likely the change in the up-flow persistence of methane-rich and sulfate-depleted pore water. Therefore, we infer that the various byproducts produced by methanotrophic activities play an important role in the life strategy of STs involved in chemosynthetic metabolism. Consequently, our study reveals that the interaction between methanotrophs and benthic organisms is critical for understanding the ecology, diversity, and biogeochemistry of MVs and other types of cold seep ecosystems.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DHL, YML, and JHK designed and coordinated the study and led the writing and interpreted the data. YKJ, CP, and KHS contributed to sample acquisition. DHL conducted experiments for gas, porewater, and lipid biomarkers, and YML analyzed microbial diversity. J-HK provided the methane and DIC concentration data. All authors commented on the manuscript and contributed to the writing.
Funding
This study was supported by the KOPRI projects (KIMST Grant 202106322) funded by the Ministry of Oceans and Fisheries. This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (MSIT) of Korea (NRF-N2021M1A5A1075512) and by a KIGAM project (KIGAM-NP2018-022).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely appreciated the two reviewers for their valuable comments. We would like to thank the captain and crews of the R/V Araon for their help at sea. We also thank S. Kang, D. Kim, and S. Kim for their analytical assistance in the laboratory at Hanyang University and D. Graves, F. Flores, and R. Gwiazda at the Monterey Bay Aquarium Research Institute for remotely operated vehicle operation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.656171/full#supplementary-material
Footnotes
References
Belyaev, S. S., Wolkin, R., Kenealy, W. R., Deniro, M. J., Epstein, S., and Zeikus, J. G. (1983). Methanogenic bacteria from the bondyuzhskoe oil field: general characterization and analysis of stable-carbon isotopic fractionation. Appl. Environ. Microbiol. 45, 691–697. doi: 10.1128/AEM.45.2.691-697.1983
Birgel, D., Stein, R., and Hefter, J. (2004). Aliphatic lipids in recent sediments of the Fram Strait/Yermak Plateau (Arctic Ocean): composition, sources and transport processes. Mar. Chem. 88, 127–160. doi: 10.1016/j.marchem.2004.03.006
Blumenberg, M., Seifert, R., Reitner, J., Pape, T., and Michaelis, W. (2004). Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. U.S.A. 101, 11111–11116. doi: 10.1073/pnas.0401188101
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Boetius, A., and Suess, E. (2004). Hydrate ridge: a natural laboratory for the study of microbial life fueled by methane from near-surface gas hydrates. Chem. Geol. 205, 291–310. doi: 10.1016/j.chemgeo.2003.12.034
Boetius, A., and Wenzhöfer, F. (2013). Seafloor oxygen consumption fuelled by methane from cold seeps. Nat. Geosci. 6, 725–734. doi: 10.1038/ngeo1926
Bowman, J. P., Sly, L. I., Nichols, P. D., and Hayward, A. C. (1993). Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., Emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 43, 735–753. doi: 10.1099/00207713-43-4-735
Bowman, J. P., Skerratt, J. H., Nichols, P. D., and Sly, L. I. (1991). Phospholipid fatty acid and lipopolysaccharide fatty acid signature lipids in methane-utilizing bacteria. FEMS Microbiol. Lett. 85, 15–22. doi: 10.1111/j.1574-6968.1991.tb04693.x
Bradley, A. S., Fredricks, H., Hinrichs, K., and Summons, R. E. (2009). Structural diversity of diether lipids in carbonate chimneys at the Lost City Hydrothermal Field. Org. Geochem. 40, 1169–1178. doi: 10.1016/j.orggeochem.2009.09.004
Bright, M., and Giere, O. (2005). Microbial symbiosis in Annelida. Symbiosis 38, 1–45. doi: 10.1016/bs.aiip.2020.04.003
Canfield, D. E., and Thamdrup, B. (1994). The production of 34S-depleted sulfide during bacterial disproportionation of elemental sulfur. Science 266, 1973–1975. doi: 10.1126/science.11540246
Canuel, E. A., and Martens, C. S. (1993). Seasonal variations in the sources and alteration of organic matter associated with recently-deposited sediments. Org. Geochem. 20, 563–577. doi: 10.1016/0146-6380(93)90024-6
Cavanaugh, C. M., Gardiner, S. L., Jones, M. L., Jannasch, H. W., and Waterbury, J. B. (1981). Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213, 340–342. doi: 10.1126/science.213.4505.340
Chevalier, N., Bouloubassi, I., Birgel, D., Taphanel, M. H., and López-García, P. (2013). Microbial methane turnover at Marmara Sea cold seeps: a combined 16S rRNA and lipid biomarker investigation. Geobiology 11, 55–71. doi: 10.1111/gbi.12014
Coffin, R. B., Smith, J. P., Plummer, R. E., Yoza, B., Larsen, R. K., Millholland, L. C., et al. (2013). Spatial variation in shallow sediment methane sources and cycling on the Alaskan Beaufort Sea Shelf/Slope. Mar. Petrol. Geol. 45, 186–197. doi: 10.1016/j.marpetgeo.2013.05.002
Cordes, E. E., Arthur, M. A., Shea, K., Arvidson, R. S., and Fisher, C. R. (2005). Modeling the mutualistic interactions between tubeworms and microbial consortia. PLoS Biol. 3:e77. doi: 10.1371/journal.pbio.0030077
de Beer, D., Sauter, E., Niemann, H., Kaul, N., Foucher, J. P., Witte, U., et al. (2006). In situ fluxes and zonation of microbial activity in surface sediments of the Håkon Mosby Mud Volcano. Limnol. Oceanogr. 51, 1315–1331. doi: 10.4319/lo.2006.51.3.1315
de Rosa, M., and Gambacorta, A. (1988). The lipids of archaebacteria. Prog. Lipid Res. 27, 153–175. doi: 10.1016/0163-7827(88)90011-2
Duperron, S., Gaudron, S. M., Lemaitre, N., and Bayon, G. (2014). A microbiological and biogeochemical investigation of the cold seep tubeworm escarpia southwardae (Annelida: Siboglinidae): symbiosis and trace element composition of the tube. Deep Sea Res. I 90, 105–114. doi: 10.1016/j.dsr.2014.05.006
Duperron, S., Sibuet, M., MacGregor, B. J., Kuypers, M. M., Fisher, C. R., and Dubilier, N. (2007). Diversity, relative abundance and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussel species from cold seeps in the Gulf of Mexico. Environ. Microbiol. 9, 1423–1438. doi: 10.1111/j.1462-2920.2007.01259.x
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Elvert, M., Boetius, A., Knittel, K., and Jørgensen, B. B. (2003). Characterization of specific membrane fatty acids as chemotaxonomic markers for sulfate-reducing bacteria involved in anaerobic oxidation of methane. Geomicrobiol. J. 20, 403–419. doi: 10.1080/01490450303894
Elvert, M., Hopmans, E. C., Treude, T., Boetius, A., and Suess, E. (2005). Spatial variations of methanotrophic consortia at cold methane seeps: implications from a high-resolution molecular and isotopic approach. Geobiology 3, 195–209. doi: 10.1111/j.1472-4669.2005.00051.x
Elvert, M., and Niemann, H. (2008). Occurrence of unusual steroids and hopanoids derived from aerobic methanotrophs at an active marine mud volcano. Org. Geochem. 39, 167–177. doi: 10.1016/j.orggeochem.2007.11.006
Elvert, M., Suess, E., and Whiticar, M. J. (1999). Anaerobic methane oxidation associated with marine gas hydrates: superlight C-isotopes from saturated and unsaturated C20 and C25 irregular isoprenoids. Naturwissenschaften 86, 295–300. doi: 10.1007/s001140050619
Fang, J., Barcelona, M. J., and Semrau, J. D. (2000). Characterization of methanotrophic bacteria on the basis of intact phospholipid profiles. FEMS Microbiol. Lett. 189, 67–72. doi: 10.1111/j.1574-6968.2000.tb09207.x
Felbeck, H., Childress, J. J., and Somero, G. N. (1981). Calvin-Benson cycle and sulphide oxidation enzymes in animals from sulphide-rich habitats. Nature 293, 291–293. doi: 10.1038/293291a0
Felden, J., Ruff, S. E., Ertefai, T., Inagaki, F., and Hinrichs, K. U. (2014). Anaerobic methanotrophic community of a 5346-m-deep vesicomyid clam colony in the Japan Trench. Geobiology 12, 183–199. doi: 10.1111/gbi.12078
Felden, J., Wenzhöfer, F., Feseker, T., and Boetius, A. (2010). Transport and consumption of oxygen and methane in different habitats of the Håkon Mosby Mud Volcano (HMMV). Limnol. Oceanogr. 55, 2366–2380. doi: 10.4319/lo.2010.55.6.2366
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Fisher, C. R. (1990). Chemoautotrophic and methanotrophic symbioses in marine invertebrates. Rev. Aquat. Sci. 2, 399–436.
Freytag, J. K., Girguis, P. R., Bergquist, D. C., Andras, J. P., Childress, J. J., and Fisher, C. R. (2001). A paradox resolved: sulfide acquisition by roots of seep tubeworms sustains net chemoautotrophy. Proc. Natl. Acad. Sci. U.S.A. 98, 13408–13413. doi: 10.1073/pnas.231589498
Fry, B. (1999). Using stable isotopes to monitor watershed influences on aquatic trophodynamics. Can. J. Fish. Aquat. Sci. 56, 2167–2171. doi: 10.1139/f99-152
Gieskes, J., Gamo, T., and Brumsack, H. (1991). Chemical Methods for Interstitial Water Analysis Aboard JOIDES Resolution. Chem. Methods Interstitial Water Anal. Aboard JOIDES Resolut. College Station, TX: Ocean Drilling Program, 1–60. doi: 10.2973/odp.tn.15.1991
Gong, C., and Hollander, D. J. (1997). Differential contribution of bacteria to sedimentary organic matter in oxic and anoxic environments, Santa Monica Basin, California. Org. Geochem. 26, 545–563. doi: 10.1016/S0146-6380(97)00018-1
Gontharet, S., Stadnitskaia, A., Bouloubassi, I., Pierre, C., and Damsté, J. S. S. (2009). Palaeo methane-seepage history traced by biomarker patterns in a carbonate crust, Nile deep-sea fan (Eastern Mediterranean Sea). Mar. Geol. 261, 105–113. doi: 10.1016/j.margeo.2008.11.006
Gründger, F., Carrier, V., Svenning, M. M., Panieri, G., Vonnahme, T. R., Klasek, S., et al. (2019). Methane-fuelled biofilms predominantly composed of methanotrophic ANME-1 in Arctic gas hydrate-related sediments. Sci. Rep. 9:9725. doi: 10.1038/s41598-019-46209-5
Haese, R. R., Meile, C., Van Cappellen, P., and De Lange, G. J. (2003). Carbon geochemistry of cold seeps: methane fluxes and transformation in sediments from Kazan mud volcano, eastern Mediterranean Sea. Earth Planet. Sci. Lett. 212, 361–375. doi: 10.1016/S0012-821X(03)00226-7
Heijs, S. K., Haese, R. R., van der Wielen, P. W. J. J., Forney, L. J., and Van Elsas, J. D. (2007). Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a Mediterranean cold seep. Microb. Ecol. 53, 384–398. doi: 10.1007/s00248-006-9172-3
Hilário, A., Capa, M., Dahlgren, T. G., Halanych, K. M., Little, C. T., Thornhill, D. J., et al. (2011). New perspectives on the ecology and evolution of siboglinid tubeworms. PLoS One 6:e16309. doi: 10.1371/journal.pone.0016309
Hinrichs, K. U., and Boetius, A. (2002). “The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry,” in Ocean Margin Systems, eds G. Wefer, D. Billett, D. Hebbeln, B. B. Jørgensen, M. Schlüter, and T. C. E. van Weering (Berlin: Springer-Verlag), 457–477. doi: 10.1007/978-3-662-05127-6_28
Hinrichs, K. U., Summons, R. E., Orphan, V., Sylva, S. P., and Hayes, J. M. (2000). Molecular and isotopic analysis of anaerobic methane- oxidizing communities in marine sediments. Org. Geochem. 31, 1685–1701. doi: 10.1016/S0146-6380(00)00106-6
Hoefs, J. (2007). Stable Isotope Geochemistry. Translatio. Berlin: Springer-Verlag, 35–92. doi: 10.1007/978-3-540-70708-0
Hoefs, M., Schouten, S., De Leeuw, J. W., King, L. L., Wakeham, S. G., Damste, J., et al. (1997). Ether lipids of planktonic archaea in the marine water column. Appl. Environ. Microbiol. 63, 3090–3095. doi: 10.1128/AEM.63.8.3090-3095.1997
Jahnke, L. L., Summons, R. E., Hope, J. M., and Des Marais, D. J. (1999). Carbon isotopic fractionation in lipids from methanotrophic bacteria II: the effects of physiology and environmental parameters on the biosynthesis and isotopic signatures of biomarkers. Geochim. Cosmochim. Acta 63, 79–93. doi: 10.1016/S0016-7037(98)00270-1
Jørgensen, B. B., and Boetius, A. (2007). Feast and famine – microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781. doi: 10.1038/nrmicro1745
Jørgensen, B. B., Böttcher, M. E., Lüschen, H., Neretin, L. N., and Volkov, I. I. (2004). Anaerobic methane oxidation and a deep H2S sink generate isotopically heavy sulfides in Black Sea sediments. Geochim. Cosmochim. Acta 68, 2095–2118. doi: 10.1016/j.gca.2003.07.017
Jørgensen, B. B., and Kasten, S. (2006). “Sulfur cycling and methane oxidation,” in Marine Geochemistry, eds H. D. Schulz and M. Zabel (Berlin: Springer), 149–154. doi: 10.1007/3-540-32144-6_8
Julian, D., Gaill, F., Wood, E., Arp, A. J., and Fisher, C. R. (1999). Roots as a site of hydrogen sulfide uptake in the hydrocarbon seep vestimentiferan Lamellibrachia sp. J. Exp. Biol. 202, 2245–2257. doi: 10.1242/jeb.202.17.2245
Kaneko, M., Kitajima, F., and Naraoka, H. (2011). Stable hydrogen isotope measurement of archaeal ether-bound hydrocarbons. Org. Geochem. 42, 166–172. doi: 10.1016/j.orggeochem.2010.11.002
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Knittel, K., Lösekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/AEM.71.1.467-479.2005
Koga, Y., and Morii, H. (2005). Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci. Biotechnol. Biochem. 69, 2019–2034. doi: 10.1271/bbb.69.2019
Koga, Y., Morii, H., Akagawa-matsushita, M., and Ohga, M. (1998). Correlation of polar lipid composition with 16S rRNA phylogeny in methanogens. Further analysis of lipid component parts. Biosci. Biotechnol. Biochem. 62, 230–236. doi: 10.1271/bbb.62.230
Koga, Y., Nishihara, M., Morii, H., and Akagawa-Matsushita, M. (1993). Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. Microbiol. Rev. 57, 164–182. doi: 10.1128/MMBR.57.1.164-182.1993
Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurth, J. M., Smit, N. T., Berger, S., Schouten, S., Jetten, M. S. M., and Welte, C. U. (2019). Anaerobic methanotrophic archaea of the ANME-2d clade feature lipid composition that differs from other ANME archaea. FEMS Microbiol. Ecol. 95:fiz082. doi: 10.1093/femsec/fiz082
Kushwaha, S. C., and Kates, M. (1978). 2,3-Di-O-phytanyl-sn-glycerol and prenols from extremely halophilic bacteria. Phytochemistry 17, 2029–2030. doi: 10.1016/S0031-9422(00)88759-2
Lee, D. H., Kim, J.-H., Lee, Y. M., Jin, Y. K., Paull, C. K., Kim, D., et al. (2019a). Chemosynthetic bacterial signatures in Frenulata tubeworm Oligobrachia sp. in an active mud volcano of the Canadian Beaufort Sea. Mar. Ecol. Prog. Ser. 628, 95–104. doi: 10.3354/meps13084
Lee, D. H., Kim, J.-H., Lee, Y. M., Stadnitskaia, A., Jin, Y. K., Niemann, H., et al. (2018). Biogeochemical evidence of anaerobic methane oxidation on active submarine mud volcanoes on the continental slope of the Canadian Beaufort Sea. Biogeosciences 15, 7419–7433. doi: 10.5194/bg-15-7419-2018
Lee, D. H., Lee, Y. M., Kim, J. H., Jin, Y. K., Paull, C., Niemann, H., et al. (2019b). Discriminative biogeochemical signatures of methanotrophs in different chemosynthetic habitats at an active mud volcano in the Canadian Beaufort Sea. Sci. Rep. 9:17592. doi: 10.1038/s41598-019-53950-4
Levin, L. A. (2005). Ecology of cold seep sediments: interactions of fauna with flow, chemistry and microbes. Oceanogr. Mar. Biol. Annu. Rev. 43, 1–46. doi: 10.1201/9781420037449.ch1
Liu, X., Lipp, J. S., and Hinrichs, K. U. (2011). Distribution of intact and core GDGTs in marine sediments. Org. Geochem. 42, 368–375. doi: 10.1016/j.orggeochem.2011.02.003
Lösekann, T., Knittel, K., Nadalig, T., Fuchs, B., Niemann, H., Boetius, A., et al. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73, 3348–3362. doi: 10.1128/AEM.00016-07
Makula, R. A. (1978). Phospholipid composition of methane-utilizing bacteria. J. Bacteriol. 134, 771–777. doi: 10.1128/JB.134.3.771-777.1978
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: paired−end assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31
Meister, P., Liu, B., Ferdelman, T. G., Jørgensen, B. B., and Khalili, A. (2013). Control of sulphate and methane distributions in marine sediments by organic matter reactivity. Geochim. Cosmochim. Acta 104, 183–193. doi: 10.1016/j.gca.2012.11.011
Nichols, P. D., Glen, A. S., Antworth, C. P., Hanson, R. S., and White, D. C. (1985). Phospholipid and lipopolysaccharide normal and hydroxy fatty acids as potential signatures for methane-oxidizing bacteria. FEMS Microbiol. Lett. 31, 327–335. doi: 10.1111/j.1574-6968.1985.tb01168.x
Niemann, H., and Boetius, A. (2010). “Mud volcanoes,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. N. Timmis (Berlin: Springer-Verlag), 206–214. doi: 10.1007/978-3-540-77587-4_13
Niemann, H., and Elvert, M. (2008). Diagnostic lipid biomarker and stable carbon isotope signatures of microbial communities mediating the anaerobic oxidation of methane with sulphate. Org. Geochem. 39, 1668–1677. doi: 10.1016/j.orggeochem.2007.11.003
Niemann, H., Lösekann, T., de Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby Mud Volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Niemann, H., Elvert, M., Hovland, M., Orcutt, B., Judd, A., Suck, I., et al. (2005). Methane emission and consumption at a North Sea gas seep (Tommeliten area). Biogeosci. Discuss. 2, 1197–1241. doi: 10.5194/bgd-2-1197-2005
Nikolenko, S. I., Korobeynikov, A. I., and Alekseyev, M. A. (2013). BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14(Suppl. 1):S7. doi: 10.1186/1471-2164-14-S1-S7
Omoregie, E. O., Niemann, H., Mastalerz, V., de Lange, G. J., Stadnitskaia, A., Mascle, J., et al. (2009). Microbial methane oxidation and sulfate reduction at cold seeps of the deep eastern Mediterranean Sea. Mar. Geol. 261, 114–127. doi: 10.1016/j.margeo.2009.02.001
Pachiadaki, M. G., Lykousis, V., Stefanou, E. G., and Kormas, K. A. (2010). Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan Mud Volcano, east Mediterranean Sea). FEMS Microbiol. Ecol. 72, 429–444. doi: 10.1111/j.1574-6941.2010.00857.x
Pancost, R. D., McClymont, E. L., Bingham, E. M., Roberts, Z., Charman, D. J., Hornibrook, E. R. C., et al. (2011). Archaeol as a methanogen biomarker in ombrotrophic bogs. Org. Geochem. 42, 1279–1287. doi: 10.1016/j.orggeochem.2011.07.003
Pancost, R. D., Sinninghe Damsté, J. S., de Lint, S., Van der Maarel, M. J. E. C., and Gottschal, J. C. (2000). Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. The Medinaut Shipboard Scientific Party. Appl. Environ. Microbiol. 66, 1126–1132. doi: 10.1128/aem.66.3.1126-1132.2000
Paull, C. K., Dallimore, S. R., Caress, D. W., Gwiazda, R., Melling, H., Riedel, M., et al. (2015). Active mud volcanoes on the continental slope of the Canadian Beaufort Sea. Geochem. Geophys. Geosyst. 16, 3160–3181. doi: 10.1002/2015GC005928
Paull, C. K., Dallimore, S., Hughes, J., Blasco, S., Lundsten, E., and Ussler, W. III, et al. (2011). “Tracking the decomposition of submarine permafrost and gas hydrate under the shelf and slope of the Beaufort Sea,” in Proceedings of the 7th International Conference on Gas Hydrates, Edinburgh, 1689–1699.
Pearson, A., Budin, M., and Brocks, J. J. (2003). Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U. S. A. 100, 15352–15357. doi: 10.1073/pnas.2536559100
Pearson, A., McNIchol, A. P., Benitez-Nelson, B. C., Hayes, J. M., and Eglinton, T. I. (2001). Origins of lipid biomarkers in Santa Monica Basin surface sediment: a case study using compound-specific Δ14 C analysis. Geochim. Cosmochim. Acta 65, 3123–3137. doi: 10.1016/S0016-7037(01)00657-3
Perdue, E. M., and Koprivnjak, J. F. (2007). Using the C/N ratio to estimate terrigenous inputs of organic matter to aquatic environments. Estuar. Coast. Shelf Sci. 73, 65–72. doi: 10.1016/j.ecss.2006.12.021
Peters, K. E., Sweeney, R. E., and Kaplan, I. R. (1978). Correlation of carbon and nitrogen stable isotope ratios in sedimentary organic matter. Limnol. Oceanogr. 23, 598–604. doi: 10.4319/lo.1978.23.4.0598
Petersen, J. M., and Dubilier, N. (2009). Methanotrophic symbioses in marine invertebrates. Environ. Microbiol. Rep. 1, 319–335. doi: 10.1111/j.1758-2229.2009.00081.x
Pohlman, J. W., Kaneko, M., Heuer, V. B., Coffin, R. B., and Whiticar, M. (2009). Methane sources and production in the northern Cascadia margin gas hydrate system. Earth Planet. Sci. Lett. 287, 504–512. doi: 10.1016/j.epsl.2009.08.037
Pop Ristova, P., Wenzhöfer, F., Ramette, A., Zabel, M., Fischer, D., Kasten, S., et al. (2012). Bacterial diversity and biogeochemistry of different chemosynthetic habitats of the REGAB cold seep (West African margin, 3160 m water depth). Biogeosciences 9, 5031–5048. doi: 10.5194/bg-9-5031-2012
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Reeburgh, W. S. (1980). Anaerobic methane oxidation: rate depth distributions in Skan Bay sediments. Earth Planet. Sci. Lett. 47, 345–352. doi: 10.1016/0012-821X(80)90021-7
Reeburgh, W. S. (1996). “Soft spots’ in the global methane budget,” in Microbial Growth on C1 Compounds, eds M. E. Lidstrom and F. R. Tabita (Dordrecht: Springer), 334–342. doi: 10.1007/978-94-009-0213-8_44
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Am. Chem. Soc. Chem. Rev. 107, 486–513. doi: 10.1021/cr050362v
Rees, C. E., Jenkins, W. J., and Monster, J. (1978). The sulphur isotopic composition of ocean water sulphate. Geochim. Cosmochim. Acta 42, 377–381. doi: 10.1016/0016-7037(78)90268-5
Rodrigues, C. F., Hilário, A., Cunha, M. R., Weightman, A. J., and Webster, G. (2011). Microbial diversity in frenulata (Siboglinidae, Polychaeta) species from mud volcanoes in the Gulf of Cadiz (NE Atlantic). Antonie Van Leeuwenhoek 100, 83–98. doi: 10.1007/s10482-011-9567-0
Rossel, P. E., Elvert, M., Ramette, A., Boetius, A., and Hinrichs, K. U. (2011). Factors controlling the distribution of anaerobic methanotrophic communities in marine environments: evidence from intact polar membrane lipids. Geochim. Cosmochim. Acta 75, 164–184. doi: 10.1016/j.gca.2010.09.031
Ruff, S. E., Arnds, J., Knittel, K., Amann, R., Wegener, G., Ramette, A., et al. (2013). Microbial communities of deep-sea methane seeps at Hikurangi continental margin (New Zealand). PLoS One 8:e72627. doi: 10.1371/journal.pone.0072627
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A. (2015). Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. U.S.A. 112, 4015–4020. doi: 10.1073/pnas.1421865112
Ruff, S. E., Felden, J., Gruber-Vodicka, H. R., Marcon, Y., Knittel, K., Ramette, A., et al. (2018). In situ development of a methanotrophic microbiome in deep-sea sediments. ISME J. 13, 197–213. doi: 10.1038/s41396-018-0263-1
Schirmer, M., Ijaz, U. Z., D’Amore, R., Hall, N., Sloan, W. T., and Quince, C. (2015). Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43:e37. doi: 10.1093/nar/gku1341
Schouten, S., Hopmans, E. C., and Sinninghe Damsté, J. S. (2013). The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: a review. Org. Geochem. 54, 19–61. doi: 10.1016/j.orggeochem.2012.09.006
Sibuet, M., and Olu, K. (1998). Biogeography, biodiversity and fluid dependence of deep-sea cold-seep communities at active and passive margins. Deep Sea Res. II Top. Stud. Oceanogr. 45, 517–567. doi: 10.1016/S0967-0645(97)00074-X
Southward, A. J., Southward, E. C., Dando, P. R., Barrett, R. L., and Ling, R. (1986). Chemoautotrophic function of bacterial symbionts in small Pogonophora. J. Mar. Biol. Assoc. 66, 415–437. doi: 10.1017/S0025315400043046
Stadnitskaia, A., Ivanov, M. K., and Sinninghe Damsté, J. S. (2008). Application of lipid biomarkers to detect sources of organic matter in mud volcano deposits and post-eruptional methanotrophic processes in the Gulf of Cadiz, NE Atlantic. Mar. Geol. 255, 1–14. doi: 10.1016/j.margeo.2007.11.006
Stadnitskaia, A., Muyzer, G., Abbas, B., Coolen, M. J. L., Hopmans, E. C., Baas, M., et al. (2005). Biomarker and 16S rDNA evidence for anaerobic oxidation of methane and related carbonate precipitation in deep-sea mud volcanoes of the Sorokin Trough, Black Sea. Mar. Geol. 217, 67–96. doi: 10.1016/j.margeo.2005.02.023
Summons, R., Bradley, A., Jahnke, L., and Waldbauer, J. (2006). Steroids, triterpenoids and molecular oxygen. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 951–968. doi: 10.1098/rstb.2006.1837
Treude, T., Boetius, A., Knittel, K., Wallmann, K., and Barker Jørgensen, B. B. (2003). Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 264, 1–14. doi: 10.3354/meps264001
Vanreusel, A., Andersen, A. C., Boetius, A., Connelly, D., Cunha, M. R., Decker, C., et al. (2009). Biodiversity of cold seep ecosystems along the European margins. Oceanography 22, 110–127. doi: 10.5670/oceanog.2009.12
Vigneron, A., Cruaud, P., Pignet, P., Caprais, J. C., Cambon-Bonavita, M. A., Godfroy, A., et al. (2013). Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California). ISME J. 7, 1595–1608. doi: 10.1038/ismej.2013.18
Walters, W., Hyde, E. R., Berg-lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15
Wegener, G., Krukenberg, V., Ruff, S. E., Kellermann, M. Y., and Knittel, K. (2016). Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front. Microbiol. 7:46. doi: 10.3389/fmicb.2016.00046
Wegener, G., Niemann, H., Elvert, M., Hinrichs, K. U., and Boetius, A. (2008). Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ. Microbiol. 10, 2287–2298. doi: 10.1111/j.1462-2920.2008.01653.x
Weijers, J. W. H., Lim, K. L. H., Aquilina, A., Sinninghe Damsté, J. S., and Pancost, R. D. (2011). Biogeochemical controls on glycerol dialkyl glycerol tetraether lipid distributions in sediments characterized by diffusive methane flux. Geochem. Geophys. Geosyst. 12:Q10010. doi: 10.1029/2011GC003724
Whiticar, M. J. (1999). Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 161, 291–314. doi: 10.1016/S0009-2541(99)00092-3
Whiticar, M. J., Faber, E., and Schoell, M. (1986). Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-isotope evidence. Geochim. Cosmochim. Acta 50, 693–709. doi: 10.1016/0016-7037(86)90346-7
Wuchter, C., Schouten, S., Wakeham, S. G., and Sinninghe Damsté, J. S. (2005). Temporal and spatial variation in tetraether membrane lipids of marine Crenarchaeota in particulate organic matter: implications for TEX86 paleothermometry. Paleoceanography 20:A3013. doi: 10.1029/2004PA001110
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Keywords: mud volcano, methane oxidation, Oligobrachia haakonmosbiensis, lipid biomarkers, 16S rRNA
Citation: Lee D-H, Kim J-H, Lee YM, Kim J-H, Jin YK, Paull C, Ryu J-S and Shin K-H (2021) Geochemical and Microbial Signatures of Siboglinid Tubeworm Habitats at an Active Mud Volcano in the Canadian Beaufort Sea. Front. Mar. Sci. 8:656171. doi: 10.3389/fmars.2021.656171
Received: 20 January 2021; Accepted: 19 May 2021;
Published: 18 June 2021.
Edited by:
Il-Nam Kim, Incheon National University, South KoreaReviewed by:
Hongxiang Guan, Guangzhou Institute of Energy Conversion (CAS), ChinaCassandre Sara Lazar, Université du Québec à Montréal, Canada
Copyright © 2021 Lee, Kim, Lee, Kim, Jin, Paull, Ryu and Shin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung-Hyun Kim, amhraW0xMjNAa29wcmkucmUua3I=; Kyung-Hoon Shin, c2hpbmtoQGhhbnlhbmcuYWMua3I=
†Present address: Dong-Hun Lee, Marine Environment Research Division, National Institute of Fisheries Science, Busan, South Korea
 Dong-Hun Lee
Dong-Hun Lee Jung-Hyun Kim
Jung-Hyun Kim Yung Mi Lee
Yung Mi Lee Ji-Hoon Kim
Ji-Hoon Kim Young Keun Jin2
Young Keun Jin2 Charles Paull
Charles Paull Kyung-Hoon Shin
Kyung-Hoon Shin