- 1Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Chinese Academy of Sciences, Beijing, China
The outbreak of invasive ascidian Molgula manhattensis has negatively affected marine and coastal ecosystems and caused huge economic loss in various industries such as aquaculture. In mariculture systems usually characterized by high ammonia nitrogen, the capacity of M. manhattensis to defend against drastic ammonia elevation plays a crucial role in its survival and subsequent invasions. However, ammonia coping strategies and associated genes/proteins remain largely unknown. Here we investigated rhesus glycoproteins (Rh)-mediated ammonia transport by identifying all Rh proteins and exploring their mRNA expression regulations under ammonia stress. Three types of primitive Rh proteins were identified, and all contained conserved amino acid residues and functional domains. Ammonia stress largely suppressed the expression of immune-related genes, but rapidly induced the increased expression of Rh genes. Ammonia was converted into glutamine as indicated by the increased expression of glutamine synthetase gene, rather than urea as illustrated by the stable expression of arginase gene. Collectively, M. manhattensis mitigates ammonia challenge by enhancing ammonia excretion through Rh channels and detoxifying ammonia into glutamine. Our results provide insights into the molecular mechanisms underlying high tolerance and invasion success to high ammonia environments by invasive ascidians.
Introduction
Biological invasion in marine and coastal ecosystems has severely threatened the local ecology and economy globally (Pimentel et al., 2000). Among diverse notorious species, the solitary tunicate Molgula manhattensis (DeKay, 1843), which is considered to be native to the northwest Atlantic Ocean, has successfully invaded coastal zones globally, such as the southwest coast of the Atlantic Ocean, the Mediterranean Sea, and the southwest, northwest, and northeast coasts of the Pacific Ocean in the past several decades (Lambert, 2003; Zvyagintsev et al., 2003; Hewitt et al., 2004; Haydar et al., 2011; Zhan et al., 2017; Chen et al., 2018; Fofonoff et al., 2018). Such high invasiveness is, at least partially, derived from its strong tolerance to various environmental stressors, including temperature, salinity, dissolved oxygen, and chemical pollution (Zvyagintsev et al., 2003; Pyo et al., 2012; Chen et al., 2018). In China, for example, it has successfully invaded coasts from the northern temperate to the southern subtropical, occupying diverse environmental habitats (Zheng, 1988; Chen et al., 2018). In addition to its significantly negative ecological impacts along coasts, of particular concern is the enormous economic loss caused by M. manhattensis’ rapid establishment and expansion in the aquaculture industry, leading to significant reduction in harvest in many species such as sea cucumbers, oysters, and abalones (Anderson, 1971; Osman and Whitlatch, 1995; Haydar et al., 2011; Chen et al., 2018; Fofonoff et al., 2018). Typically, large populations of M. manhattensis intensively attach to aquaculture facilities such as cages, water flow channels, oxygen pipelines, and even the farmed organisms’ body surface, to compete with cultured animals for food, oxygen, and space. Modern high-intensity aquaculture often results in the significant increase of ammonia nitrogen concentration in water, which can exert toxic effects on organismal survival (Ip et al., 2001; Randall and Tsui, 2002; Ip and Chew, 2010; Cheng et al., 2015; Guo et al., 2020). Even worse, the thriving M. manhattensis populations would excrete a large amount of nitrogenous metabolite, which could further increase the ammonia concentration, causing death of more cultured animals (Osman and Whitlatch, 1995; Haydar et al., 2011; Pyo et al., 2012; Chen et al., 2018). In contrast, M. manhattensis can survive in such harsh environments and even completely replace cultured species finally (Osman and Whitlatch, 1995; Haydar et al., 2011; Chen et al., 2018; Fofonoff et al., 2018). However, rapid response and acclimation mechanisms of M. manhattensis to the high ammonia stress have not yet been studied.
In aquaculture farms, ambient ammonia, known as a common environmental toxicant, is the prevailing environmental stressor caused by animal excretion and ammonification of unconsumed food (Randall and Tsui, 2002; Wang et al., 2014; Guo et al., 2020). Two forms of ammonia nitrogen exist in aquaculture seawater, unionized ammonia (NH3) and ammonium ion (NH4+), depending on the pH value, salinity, and temperature (Emerson et al., 1975). Unless specified, “ammonia” henceforth refers to the total ammonia as the sum of NH3 and NH4+. It has been reported that the ammonia concentration can reach as high as 46.11 mg⋅L–1 in intensive aquaculture systems (Chen et al., 1988; Cheng et al., 2015). In aquatic species, ammonia toxicity has been characterized by suppressed immunity, neurotoxicity, oxidative stress, reduced growth and survival, as well as various physiological dysfunctions (Randall and Tsui, 2002; Ip and Chew, 2010; Yue et al., 2010; Cheng et al., 2015; Li et al., 2016). Previous studies have discovered that most aquatic animals such as fishes and crustaceans adopt efficient ammonia excretory strategies or invest metabolic energy to convert ammonia to non- or less toxic nitrogenous compounds, such as glutamine and urea, to mitigate ammonia toxicity under high environmental ammonia loadings (Randall and Tsui, 2002; Ip and Chew, 2010; Wang et al., 2014; Weihrauch and Allen, 2018).
Ammonia excretion, which is considered as a primary strategy coping with ammonia stress, tightly involves rhesus glycoproteins (Rh) at the molecular level in most animals (Ip et al., 2004; Wright and Wood, 2009; Ip and Chew, 2010). Rh are a family of conserved proteins that play an essential role in transmembrane ammonia transport (Bakouh et al., 2006; Nakhoul and Hamm, 2014; Chng et al., 2017). According to the theories formulated on the high-resolution crystal structures of ammonium transport protein (AmtB, Rh homologous protein) of Escherichia coli (Khademi et al., 2004) and human Rh type C glycoprotein (RhCG) (Zheng et al., 2004; Gruswitz et al., 2010; Baday et al., 2015), Rh proteins recruit and deprotonate NH4+ to NH3 with conserved binding residues, followed by the diffusion of NH3 through the Rh protein channel (Chng et al., 2017; Yeam et al., 2017). However, to date, remarkably limited information is known about ammonia excretion strategies in tunicates. Relevant studies mostly focused on physiological evidence, suggesting that ascidians such as M. manhattensis, Ciona intestinalis, Styela plicata, S. partita, and S. clava excrete non-protein nitrogen mainly in the form of ammonia rather than urea (Goodbody, 1957; Markus and Lambert, 1983; Carver et al., 2006; Evans et al., 2017). An evolutionary study of the Rh protein family revealed that three types of Rh genes were identified in C. savignyi and C. intestinalis, and Ciona Rh genes were all grouped into a clade of primitive Rh proteins (Rhp1), which occurs in invertebrates and unicellular eukaryotes and is seen as the ancestral to modern Rh clades of mammalian vertebrates (Huang and Peng, 2005; Huang, 2008). However, neither molecular characterization nor functional roles of the tunicate Rh proteins were investigated.
In addition to ammonia transport mediated by Rh proteins, ammonia conversion is also recognized as an important strategy during organisms’ dealing with excessive ambient ammonia (Randall and Tsui, 2002; Ip and Chew, 2010; Wang et al., 2014; Weihrauch and Allen, 2018). Specifically, glutamine synthetase (GS) catalyzes the synthesis of glutamine from glutamate and NH4+ (Anderson et al., 2002; Ip et al., 2004; Sinha et al., 2013; Wang et al., 2014), and arginase (ARG) is known to participate in urea synthesis through the ornithine-urea-cycle (OUC) that converts ammonia to urea (Ip et al., 2001; Ip and Chew, 2010). The gene expression and enzyme activity changes of GS and ARG have been investigated in fish and crustacean (Anderson et al., 2002; Wright et al., 2007; Liu et al., 2014; Wang et al., 2014; Banerjee et al., 2018; You et al., 2018; Geng et al., 2020). However, there is a paucity of analyses of proteins participating in the ammonia conversion. Another challenge is the unresolved nitrogenous metabolic pathway in M. manhattensis, such as the incomplete nitrogen conversion involving purine and uric acid (Das, 1948; Nolfi, 1970; Saffo, 1988). It is also worth noting that the bacterial-Nephromyces-molgulid symbiosis may play a potential role in coping with ammonia (Saffo, 1988; Saffo et al., 2010; Paight et al., 2018).
As for the deleterious effects of ammonia stress, suppression of the immune system has been illustrated in various aquatic animals exposed to elevated ammonia stress (Barton and Iwama, 1991; Hurvitz et al., 1997; Li et al., 2016; Zhang et al., 2018). For example, in swimming crab, Portunus trituberculatus, immune-related genes such as crustin, anti-lipopolysaccharide factor, and lysozyme genes were regulated under short-term ammonia stress (Yue et al., 2010). Although previous studies have shown that ascidians’ innate immune response was induced under various environmental stressors (Shida et al., 2003; Ewan et al., 2005), their immune response to ammonia stress has not been studied.
In order to dissect the invasion dynamics of M. manhattensis in aquaculture environments with elevated levels of ammonia, we aimed to unmask the underlying molecular mechanisms of Rh-based coping strategy for ammonia. Firstly, we aimed to obtain the coding sequences of all Rh proteins in M. manhattensis and perform molecular characterization on their deduced amino acid sequences. Subsequently, the expression patterns of all Rh genes were evaluated under acute ammonia stress using real-time quantitative polymerase chain reaction (RT-qPCR). In order to explore the molecular mechanisms of ammonia conversion, we measured the expression levels of GS and ARG, where the former is activated to indicate ammonia conversion to glutamine while the latter is activated to convert ammonia to urea. The results obtained here are expected to expand the understanding of the causes and consequences of invasion success of M. manhattensis in high ammonia environments.
Materials and Methods
Animal Collection
Adult tunicates were collected from aquaculture ponds of sea cucumbers in Dalian, Liaoning, China, during September 2017. Collected individuals were first acclimated for a week in the filtered and aerated seawater, with an average field ammonia concentration of 0.022 mg⋅L–1 NH4+. The Total Ammonia Calculator1 was used to convert the measurement of NH4+ to 0.0225 mg⋅L–1 total ammonia (NH3 and NH4+) and 0.0005 mg⋅L–1 NH3. During the acclimation, individuals were fed with dried powder of Chlorella sp. and Spirulina sp. One third to one half of the rearing water was changed daily. After the acclimation, healthy individuals were kept for further toxicity tests and challenge experiments.
Acute Toxicity Test
To obtain the 72 h-LC50 (Lethal Concentration 50) of ammonia in M. manhattensis, we randomly assigned 150 individuals to five groups (30 individuals each). Four treatment groups were set at total ammonia concentrations of 0.67, 6.60, 16.50, and 33 mg⋅L–1 (as N) by mixing 0.50 mol⋅L–1 NH4Cl solutions with seawater, while the control group was kept at normal ambient ammonia concentration. Individuals were determined as dead when the siphon tissues bleached and inflated disproportionately, and the visceral organs leaked out of the ruptured tunics. Survival and condition of individuals were recorded at 48 and 72 h after stress exposure. Subsequently, 72 h-LC50 (total ammonia, as N) and its 95% fiducial confidence limits were calculated via the Probit method using R 4.0.2 package “ecotox” (Finney, 1971; Wheeler et al., 2006; Robertson et al., 2007). Concentrations of corresponding NH3 were calculated using the Free Ammonia Calculator2 based on salinity 28.22 ppt, pH 7.73, and temperature 23.2°C.
Ammonia Challenge Experiment
According to the acute toxicity test result, 10 mg⋅L–1 total ammonia (as N) was selected as the ammonia challenge concentration. To ensure there will be enough surviving ascidians at the end of challenge experiment, a total of 200 randomly selected individuals were transferred to the 10 mg⋅L–1 total ammonia (as N) seawater tank from the normal control seawater. For the gene expression study, siphon tissues were dissected from six replicate ascidians after 0, 2, 24, 48, and 72 h of stress treatment, respectively, immediately preserved in liquid nitrogen, and then stored at −80°C until use.
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from a mixture of two siphons using Trizol reagent (Ambion, United States) according to the manufacturer’s instruction. The integrity of extracted RNA was tested by visual inspection of the 18S and 28S ribosomal bands using 1.5% agarose gel electrophoresis. The quantity of extracted RNA was assessed using NanoDrop 2000 spectrophotometer (Nanodrop Technologies, United States). The first strands of cDNA were synthesized from 3 μg of total RNA using M-MLV reverse transcriptase (Takara, Japan) with the Oligo-dT primer according to standard procedures. The cDNA was stored at −20°C until use.
cDNA Sequence Cloning of Rh Genes
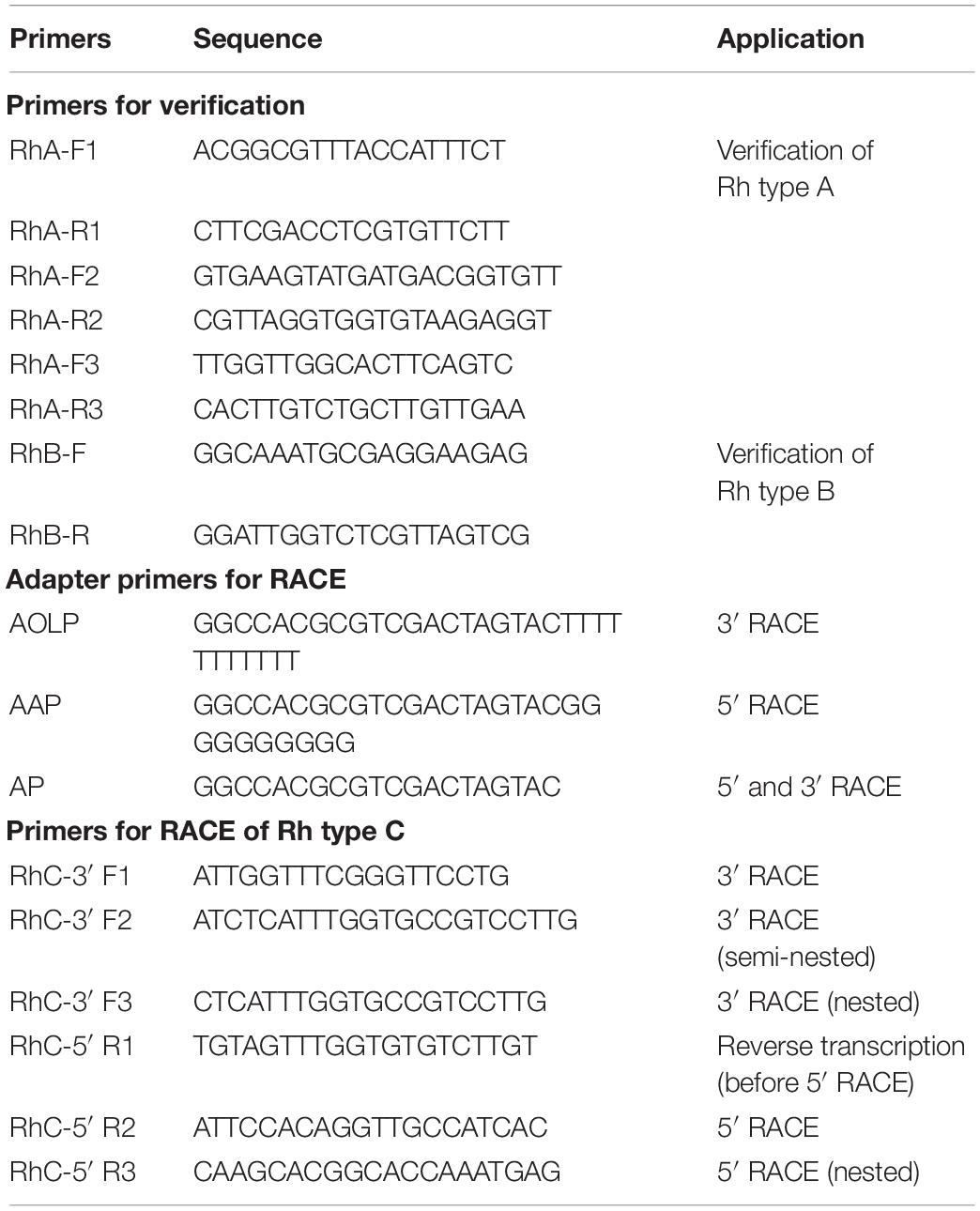
With primers designed based on the sequences from transcriptome datasets (Chen et al., 2021), the coding sequences (CDSs) of Rh type A and B genes were obtained, assembled, and then verified by NCBI BLAST with reference to complete CDSs in other ascidian species (C. intestinalis and C. savignyi). Based on Rh type C gene’s partial sequence from transcriptome datasets, RACE (rapid amplification of cDNA ends) approach was adopted to obtain the 3′ and 5′ ends of cDNA using adapter primers and gene-specific primers (Table 1). Three rounds of 3′ RACE reactions were performed using the following pairs of primers: RhC-3′ F1 and AOLP, RhC-3′ F2 and AP, RhC-3′ F3 and AP, with the last two rounds each using product (10 × dilution) of its previous round of RACE reaction. For 5′ RACE, total RNA was reverse transcribed by M-MLV reverse transcriptase (Takara, Japan) with gene-specific primer RhC-5′ R1. The obtained cDNA was purified with SanPrep DNA purification kit (Sangon Biotech, China) and tailed with poly (C) at the 3′ end by terminal deoxynucleotidyl transferase (TdT). Next, 5′ RACE reaction was performed with primer RhC-5′ R2 and AAP using the tailed cDNA as template, followed by nested PCR with RhC-5′ R3 and AP using product (10 × dilution) of the first round of 5′ RACE reaction. Rhesus glycoprotein type C gene’s complete CDS was assembled from Sanger sequenced 3′ and 5′ RACE PCR products and then verified by NCBI BLAST as described previously. Primers used were designed using Primer Premier 5.0.
Sequence Analysis and Phylogenetic Analysis
Gene/Protein Properties and Sequence Analysis
Nucleic sequence identities were investigated using MUSCLE3 server. Open reading frames (ORFs) of Rh type A-C genes were determined using NCBI ORF Finder4. The corresponding amino acid sequences were deduced using the ExPASy Translate5 tool. The protein physicochemical properties were determined using the ProtParam5 tool. Transmembrane helices (TMHs) were predicted and confirmed using TMHMM Server v.2.06, Phobius7 Normal Prediction, and PolyPhobius. Potential phosphorylation sites were identified using NetPhos 3.18. Potential N-glycosylation sites were identified using NetNGlyc 1.09. Ligand binding sites were predicted by GalaxySite using the GalaxyWEB10 server. The deduced amino acid sequences were aligned and compared with selected Rh proteins from various animal species using Clustal Omega11. Aligned sequences were annotated using ESPript 3.0 (Robert and Gouet, 2014). Protter12 (Omasits et al., 2014) was used to visualize these predicted features.
Motif and Phylogenetic Analyses
The amino acid sequences of Rh proteins were retrieved with corresponding sequence accession numbers (Supplementary Table 1) from the protein database using the NCBI protein search, including tunicates (C. intestinalis, C. savignyi, Phallusia mammillata), lancelets (Branchiostoma belcheri, B. floridae), crustaceans (Carcinus maenas, Metacarcinus magister, Daphnia magna), anurans (Nanorana parkeri, Xenopus tropicalis), primates (H. sapiens), and bony fishes (Oncorhynchus mykiss, Takifugu rubripes, Kryptolebias marmoratus). Conserved domains of deduced amino acid sequences were identified by NCBI CD-search13. The protein superfamily was annotated by EMBL-EBI Pfam 33.1 database14. Conserved motif analysis was performed using MEME Suite 5.1.115 with default parameters. Gene ontology (GO) analysis was performed using PANNZER 216.
Aiming to trace the evolutionary relationship of Rh proteins, a phylogenetic tree was reconstructed using the deduced Rh amino acid sequences. Multiple sequence alignments of the amino acid sequences were performed using ClustalW (Thompson et al., 1994). The pairwise deletion was used to eliminate poorly aligned or missing regions from the multiple alignments. Phylogenetic analysis was performed with the Neighbor-Joining (NJ) method, Poisson model, and 1000 bootstrap replications using MEGA 7 (Kumar et al., 2016). The phylogenetic tree was displayed and annotated using the online tool Interactive Tree Of Life (iTOL v5).
Real-Time Quantitative PCR (RT-qPCR)
Using Primer Premier 5.0, RT-qPCR primers (Table 2) were designed based on the partial sequences (β-ACTIN, ITGAL, C3, ARG, and Rh type A genes) from available transcriptomic datasets (Chen et al., 2021) and the coding sequences (Rh type B and C genes) from later cloning experiments. The housekeeping gene β-ACTIN was selected as the reference gene. Especially, RT-qPCR primers for Rh type A-C genes were designed based on their non-homologous sequence segments identified by alignment analysis.
RT-qPCR was conducted to quantify the target genes’ expression levels under ammonia stress using Light Cycler 96 System (Roche, Germany) with a total volume of 10 μL, which contained 5 μL SYBR Green Master Mix (Roche, Germany), 1 μL cDNA (10 × dilution), and 0.5 μM each primer. The specificity and efficiency of primers were verified before the RT-qPCR experiment. Amplification products were tested using regular PCR, 1.5% agarose gel electrophoresis, and Sanger sequencing. The efficiency of each primer pair was calculated by the slope of its standard curve, which was generated using five 10-fold dilutions of the same cDNA samples. Each amplification was performed in triplicate. The PCR program was 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s. To confirm that only a single specific PCR product was amplified and detected, the melt-curve analysis was performed at the end of each program.
The relative expression levels of target genes were normalized with the reference gene β-ACTIN and calculated with the 2−ΔΔCq (Cq, quantification cycle) method (Livak and Schmittgen, 2001). RefFinder17 was used to assess the gene expression stability. Data transformation and calculation were carried out using Microsoft Excel 16.39.
Statistical Analysis
Gene expression results were reported as the mean of three replicates ± standard error of the mean (S.E.M.). Significant differences between all groups were analyzed with the one-way ANOVA test (two-tailed), followed by Dunnett’s multiple comparison test to compare each of the challenged groups (2, 24, 48, 72 h) to control (0 h) (R 4.0.2). Normality assumption was checked by analyzing the ANOVA model residuals with the Shapiro-Wilk test. Homogeneity of variance assumption was checked using Levene’s test. P < 0.05 was considered statistically significant.
Results
Ammonia Acute Toxicity to M. manhattensis
In the acute toxicity study, M. manhattensis individuals were exposed to four ammonia concentration gradients, with each treatment group containing 30 individuals. After 72 h of treatment, no individuals died in the control solution and 0.67 mg⋅L–1 group, whereas 36.7, 66.7, and 100% mortality occurred in the 6.6, 16.5, and 33 mg⋅L–1 group, separately (Figure 1A). Using the Probit method, 72 h-LC50 of ammonia in M. manhattensis was determined as 9.42 mg⋅L–1 total ammonia (as N, equals to 12.12 mg⋅L–1), with a 95% lower confidence limit of 6.84 mg⋅L–1 (as N, equals to 8.64 mg⋅L–1) and a higher confidence limit of 11.89 mg⋅L–1 (as N, equals to 15.01 mg⋅L–1) (Figure 1B). Equally, 72 h-LC50 was calculated to be 0.257 mg⋅L–1 NH3 (confidence limits of 0.183–0.319 mg⋅L–1).
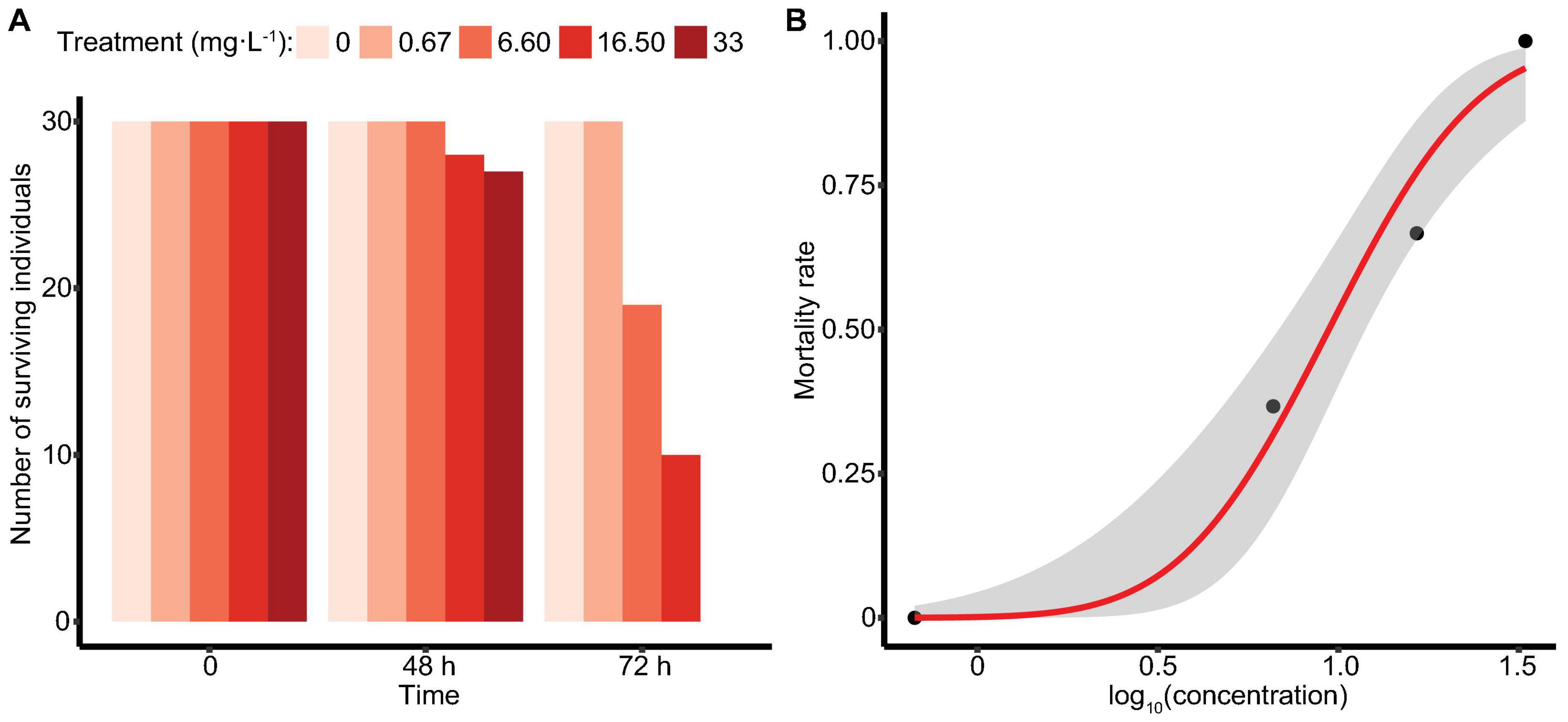
Figure 1. Survival of M. manhattensis (A) at 48 and 72 h (mg⋅L–1 as N); (B) concentration-response curve at 72 h in the ammonia acute toxicity test. The concentration-response was determined by plotting the Probit transformed percent mortality against log concentration (mg⋅L–1 as N total ammonia). The gray area indicates the 95% confidence intervals.
Analysis of Rh Coding Regions/Protein Sequences of M. manhattensis
Details of Coding Regions/Proteins
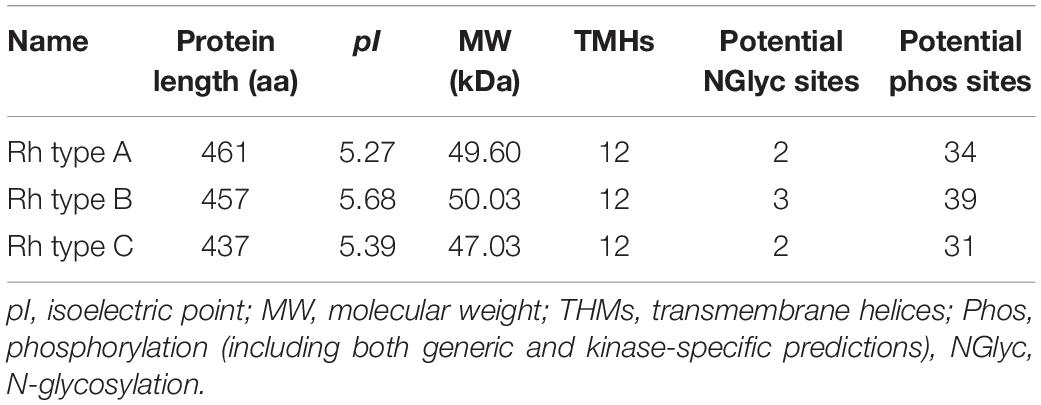
Rhesus glycoproteins type A-C genes of M. manhattensis contained an open reading frame (ORF) of 1,386, 1,374, and 1,314 nt, respectively, encoding 461, 457, and 437 amino acid residues. The complete CDSs of Rh proteins of M. manhattensis have been deposited to GenBank with the following accession numbers MW464206 (Rh type A), MW464207 (Rh type B), and MW464208 (Rh type C). Rhesus glycoproteins type B and C shared 62.84% identity in their nucleotide sequences and 58.85% identity in their amino acid sequences. Rhesus glycoprotein type A shared low similarity with Rh type B and C, with 57.48 and 58.14% identity, respectively, for nucleotide sequences and 46.33 and 49.54% identity at the amino acid level. Low similarity among these three genes indicates their potential functional differences.
Molecular weight and isoelectric point (pI) value of Rh proteins ranged from 47.03 to 50.03 kDa and from 5.27 to 5.68, respectively (Table 3). Transmembrane topology prediction showed that deduced Rh type A-C proteins of M. manhattensis comprised 12 putative transmembrane regions, respectively. Generic and kinase-specific predictions indicated 31–39 phosphorylation sites in the deduced amino acid sequences of M. manhattensis Rh proteins, which were predicted to be potentially phosphorylated by kinases, including PKA, PKC, CKII, DNAPK, CDK5, PKG, INSR, CDC2, P38MAPK, and CKI. Visualization of the predicted transmembrane regions was annotated with molecular features (Figure 2).
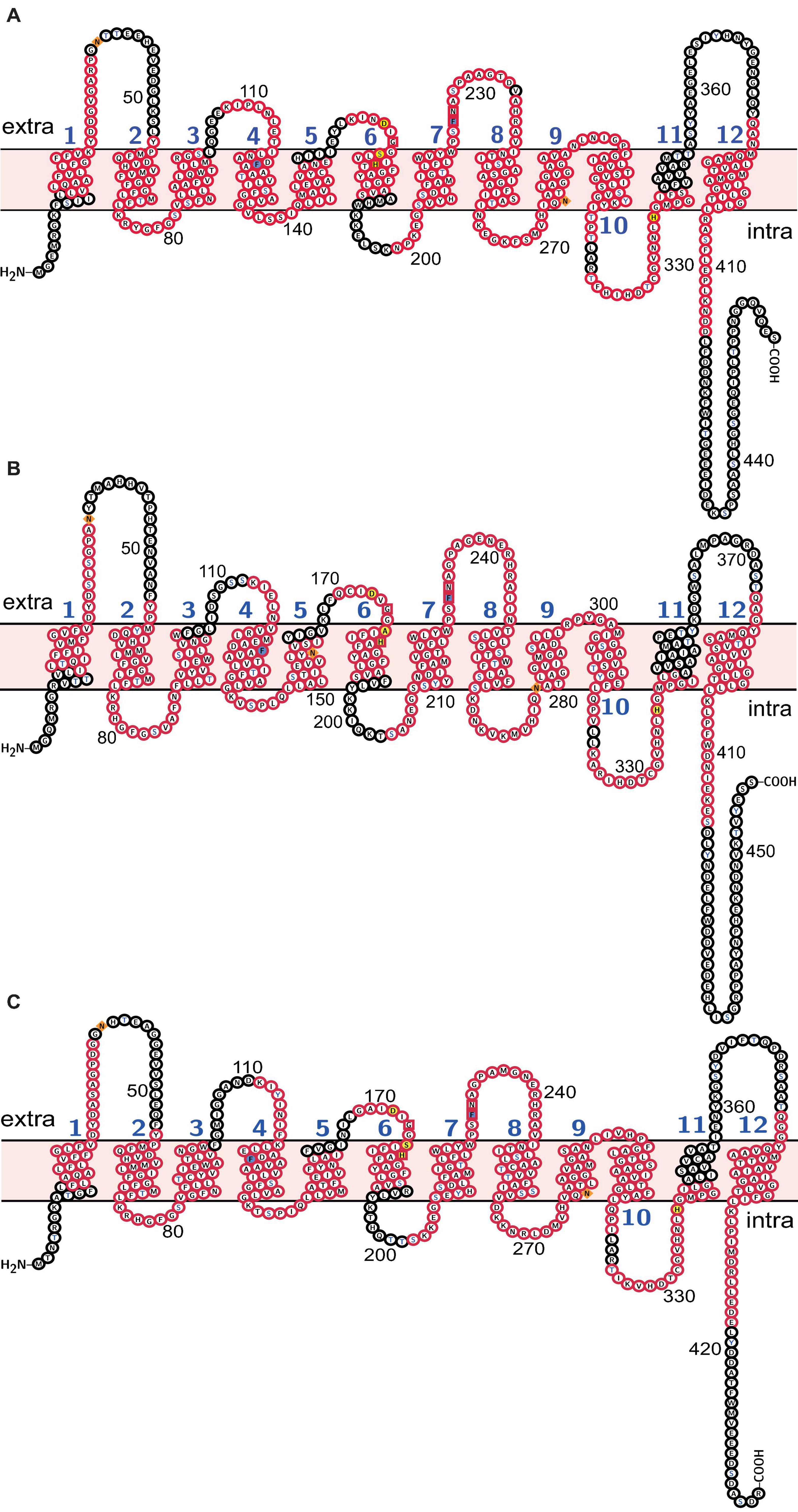
Figure 2. Molecular characterization of (A) Rh type A; (B) Rh type B; (C) Rh type C of M. manhattensis. Transmembrane domains are labeled in blue numbers (1–12). Membrane topology is in light pink. Potential N-glycosylation sites are diamond-shaped and filled with orange. Potential phosphorylation sites are of blue characters. Phenylalanine gates of the ammonia channel are filled with light blue. Residues involved in NH4+ recruitment are box-shaped. Residues involved in deprotonation are filled with yellow. Conserved motifs predicted by MEME are in red frames.
Conserved Domain Analysis and Identification of Key Residues
Search against the conserved domain and Pfam database confirmed that all the deduced M. manhattensis Rh proteins contained the ammonium transporter domain structure (Superfamily: Ammonium_transp, PF00909). MEME server detected nine conserved motifs (Figure 2 and Supplementary Figure 1). By comparing Rh proteins of M. manhattensis with E. coli AmtB to identify essential residues, all three Rh proteins of M. manhattensis lacked the π caption binding tryptophan W148 identified in E. coli AmtB (Khademi et al., 2004; Zheng et al., 2004). Alignment of Rh type A-C of M. manhattensis with E. coli AmtB and the corresponding Rh sequences from tunicate (C. intestinalis), crustacean (D. magna), frog (X. tropicalis), fish (T. rubripes), and human (accession numbers listed in Supplementary Table 1) revealed highly conserved residues (Figures 3–5). These residues are involved in the phenylalanine gating of the ammonia channel (F122 and F226 for Rh type A; F127 and F231 for Rh type B; F126 and F230 for Rh type C), NH4+ binding (G171, H177, F226, and N227 for Rh type A; G176, H182, F231, and N232 for Rh type B; G175, H181, F230, and N231 for Rh type C), and deprotonation of entering NH4+ ions (D169, S173, H177, and H335 for Rh type A; D173, S177, H181, and H338 for Rh type C). In Rh type B of M. manhattensis, residues involved in the deprotonation of entering NH4+ ions (D174, H182, and H338) were highly conserved, except for a replacement of A178 in Rh type B of M. manhattensis with S171 in human RhBG (Figure 4). In the meantime, using the GalaxyWEB server, NH3 ligand-binding sites were predicted to be F68, W223, and H335 for Rh type A; M64, F69, A130, H182, and W228 for Rh type B; F68, H181, W227, and F230 for Rh type C (Supplementary Table 2). Conservation of the key residues across different species indicates that Rh proteins should be functionally conserved.
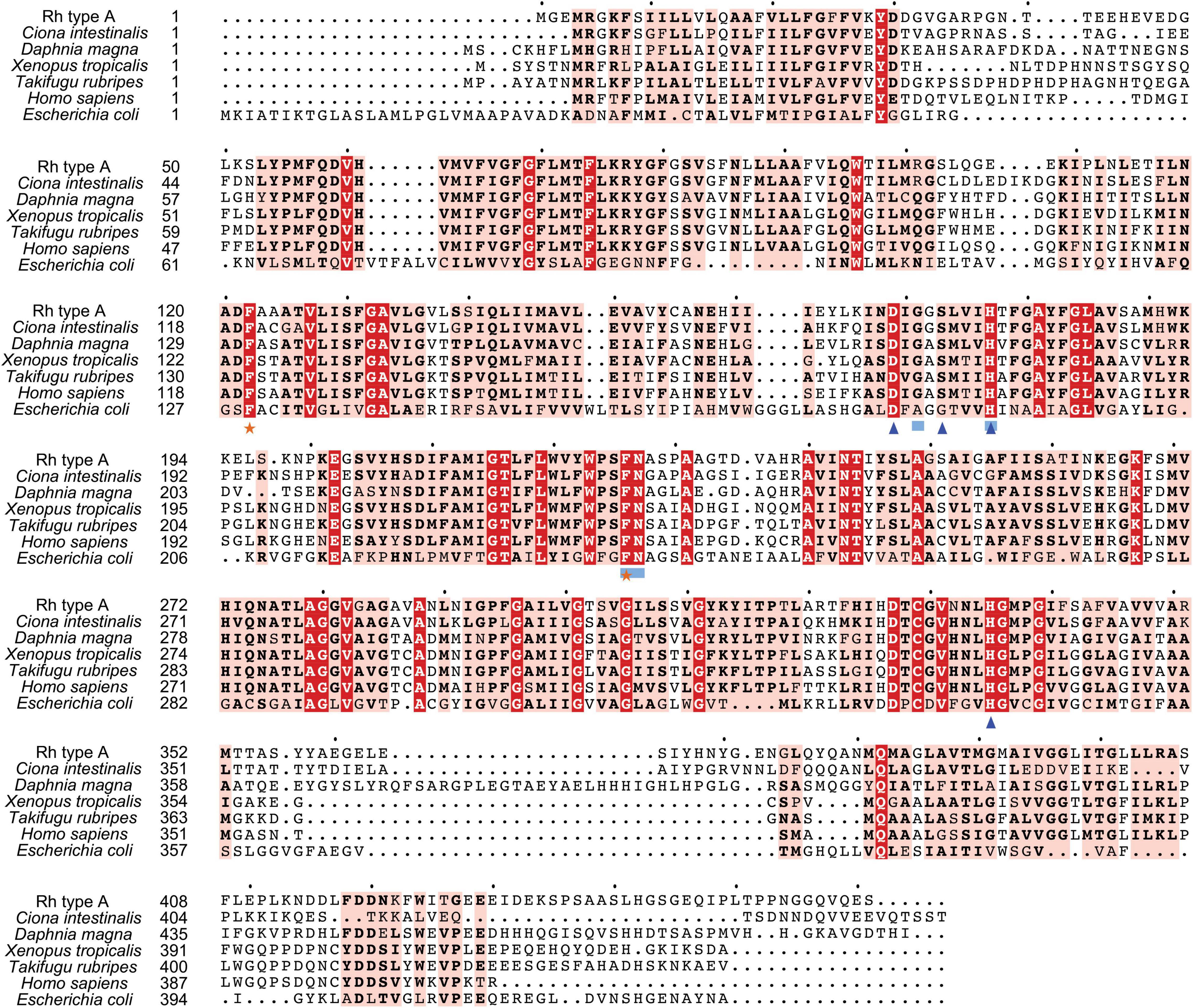
Figure 3. Multiple amino acid alignment of deduced Rh type A of M. manhattensis with E. coli AmtB and corresponding sequences of selected species. Identical amino acid residues are in bold white and filled with red. Strongly similar amino acid residues are in bold black and filled with light red. Phenylalanine gates of the ammonia channel are indicated by black stars. Residues involved in NH4+ recruitment are indicated by light blue strokes. Residues involved in deprotonation are indicated by blue triangles.
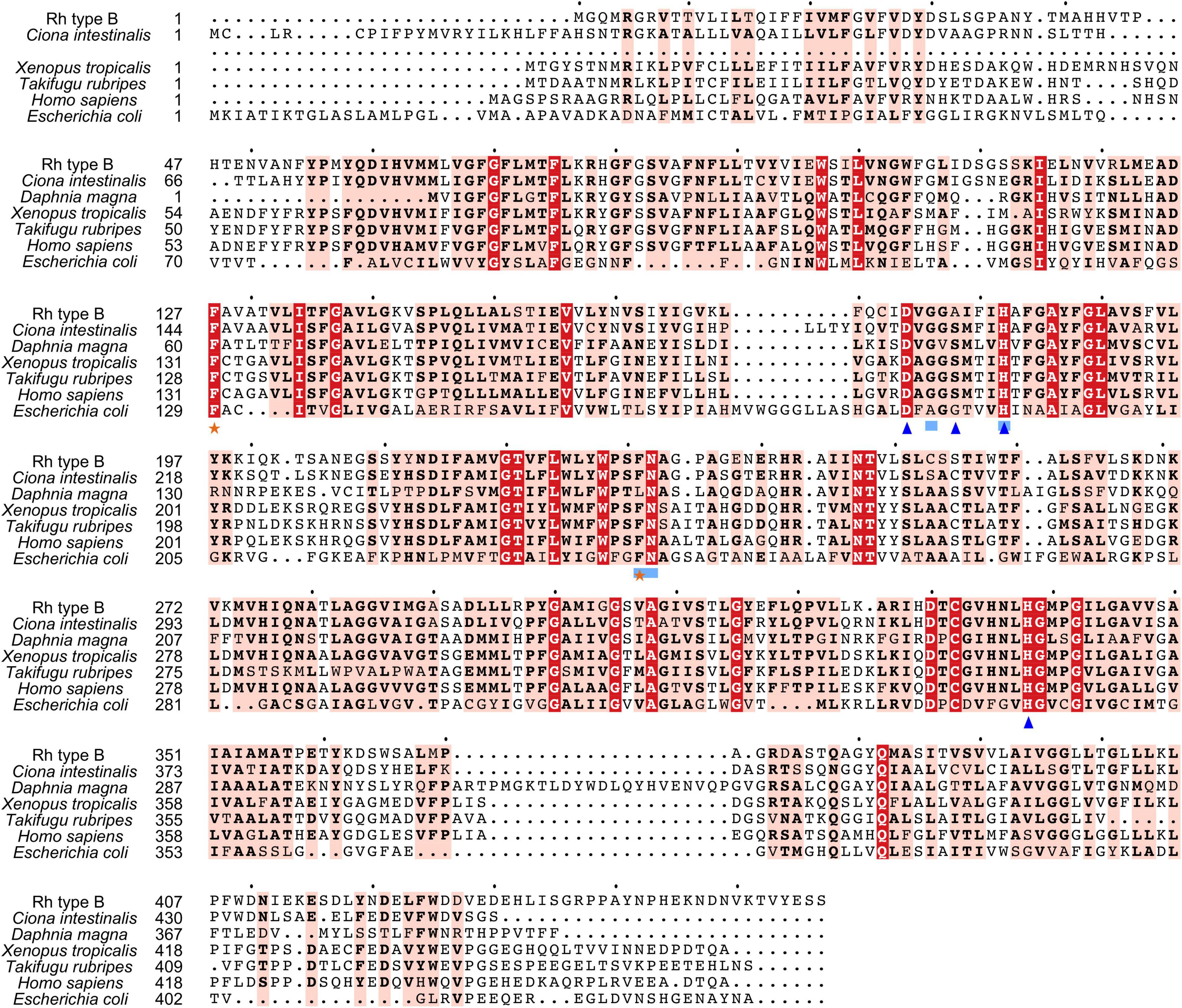
Figure 4. Multiple amino acid alignment of deduced Rh type B of M. manhattensis with E. coli AmtB and corresponding sequences of selected species. Identical amino acid residues are in bold white and filled with red. Strongly similar amino acid residues are in bold black and filled with light red. Phenylalanine gates of the ammonia channel are indicated by black stars. Residues involved in NH4+ recruitment are indicated by light blue strokes. Residues involved in deprotonation are indicated by blue triangles.
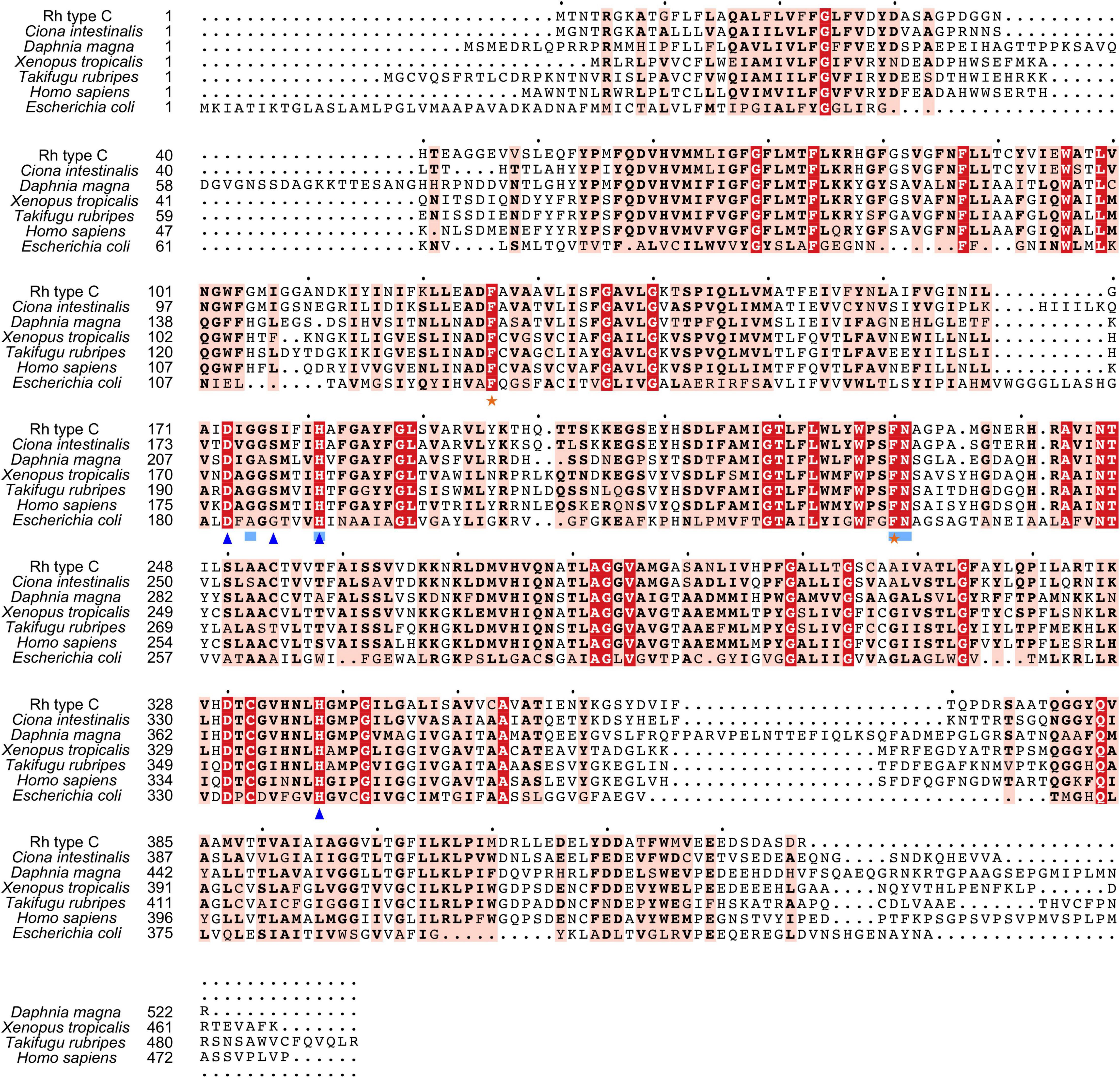
Figure 5. Multiple amino acid alignment of deduced Rh type C of M. manhattensis with E. coli AmtB and corresponding sequences of selected species. Identical amino acid residues are in bold white and filled with red. Strongly similar amino acid residues are in bold black and filled with light red. Phenylalanine gates of the ammonia channel are indicated by black stars. Residues involved in NH4+ recruitment are indicated by light blue strokes. Residues involved in deprotonation are indicated by blue triangles.
Sequence Similarity Analysis
A comparison of Rh type A from M. manhattensis with Rh type A/RhAG/Rh-like proteins from other animal species indicated that it had the highest sequence identity with Rh type A of tunicates (59.60–61.19%), followed by those of lancelets (49.56–49.88%), bony fishes (48.82–49.18%), anurans (47.74–48.57%), crustaceans (43.76–48.12%), primates (46.19%), and flies (44.03%) (Supplementary Table 3A). A comparison of Rh type B from M. manhattensis with Rh type B/RhBG/Rh-like proteins from other animal species indicated that it had the highest sequence identity with Rh type B of tunicates (59.14–61.79%), followed by those of anurans (42.66–44.44%), primates (42.89%), crustaceans (42.44–42.89%), flies (41.72%), and bony fishes (37.90–40.90%) (Supplementary Table 3B). A comparison of Rh type C from M. manhattensis with Rh type C/RhCG/Rh-like proteins from other animal species indicated that it had the highest sequence identity with Rh type C of tunicates (68.52–69.75%), followed by those of lancelets (57.89%), crustaceans (47.56–49.88%), anurans (46.28–47.66%), primates (46.28%), flies (45.09%), and bony fishes (42.26–44.52%) (Supplementary Table 3C).
Altogether, the order of the respective sequence similarities with other species groups is not consistent among Rh type A-C genes, suggesting different evolutionary histories of these three genes.
Phylogenetic Analysis of Rh Proteins
Aiming to trace Rh proteins’ evolutionary relationships, a phylogenetic tree was reconstructed using the deduced amino acid sequences of selected species, including tunicates, lancelets, crustaceans, bony fishes, and primates (Figure 6). Overall, invertebrates were well separated from vertebrates, with Rh type A-C genes exhibiting different evolutionary patterns in each group. For vertebrates, including primates and bony fishes, three sub-groups were identified as RhAG, RhBG, and RhCG, respectively. RhBG and RhCG formed a cluster first, which then clustered with RhAG. In contrast, invertebrates, including lancelets, crustaceans, and tunicates, were clustered together, separated from vertebrates’ RhAG, RhBG, and RhCG clades. This group corresponded to the Rh primitive clade (Rhp1) previously identified by Huang and Peng (2005). Species of the Rhp1 each exhibited one to three types of Rh proteins. Here, three types of Rh proteins were obtained in M. manhattensis, likely as a result of two steps of gene duplication events as previously proposed in Ciona (Huang and Peng, 2005).
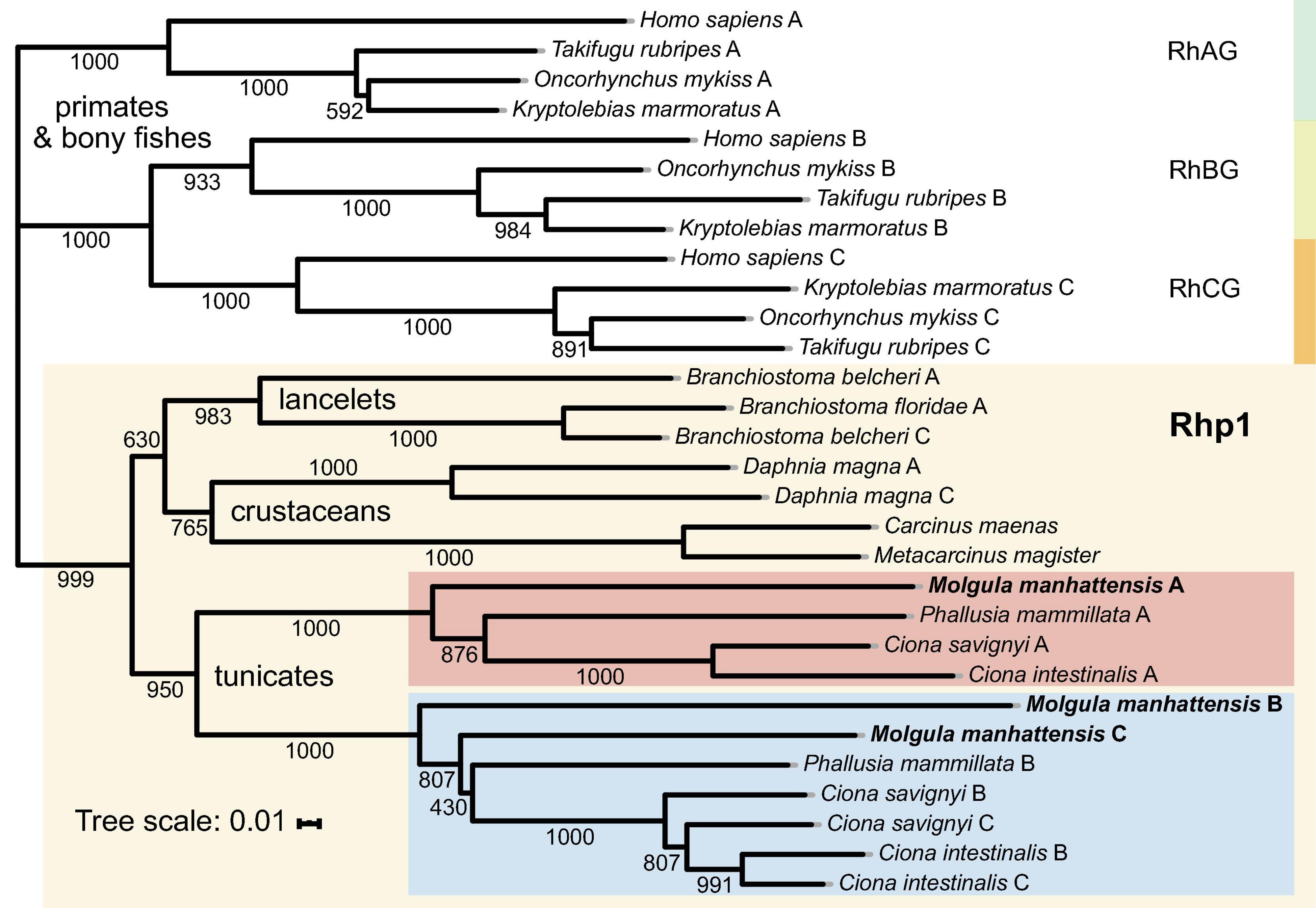
Figure 6. Phylogenetic tree of amino acid sequences of the Rh gene family of selected species. Each amino sequence is presented by the name of the species from which the amino acid sequence originated. Numbers presented at each branch point represent bootstrap values from 1,000 replicates. The scale bar represents a genetic distance of 0.01 amino acid substitutions per site.
The three types of Rh proteins of M. manhattensis were clustered together within the larger Rhp1 group and found to be present in one clade together with other tunicate Rh proteins. The Rh type B and C of M. manhattensis and P. mammillata Rh type B shared the same sub-cluster with Rh type B and C of C. intestinalis and C. savignyi. In contrast, M. manhattensis Rh type A and P. mammillata Rh type A were clustered together in another sub-clade with Rh type A of C. intestinalis and C. savignyi. This phylogeny shows the divergence of Rh type A from Rh type B and C in tunicates, whereas Rh type B and C were not yet separated.
Target Gene Expression in Response to Ammonia Stress
The mean Cq ± standard deviation (S.D.) of the reference gene β-ACTIN was 23.76 ± 0.47 across all samples. Comprehensive rankings by RefFinder identified β-ACTIN as the most stable gene, with the lowest geomean of ranking values of 1.19, followed by ARG, ITGAL, Rh type A, GS, C3, Rh type B, and C, with geomeans of ranking values of 2.11, 3.34, 3.98, 4.12, 5.05, 6.51, and 7.48, respectively.
The mRNA expression levels of Rh type A-C were calculated compared with the control (Figure 7A). The results reveal different regulation patterns among three Rh proteins. Rhesus glycoprotein type A gene expression significantly increased at 24 and 48 h, then returned to the control level at 72 h. For Rh type B, mRNA expression levels significantly increased at first, but drastically decreased at 48 h, followed by another dramatic increase to threefold. Unlike Rh type A and B, Rh type C gene expression was downregulated at 2 h, yet drastically upregulated to 2.8-fold at 48 h, then again downregulated by 45% at 72 h.
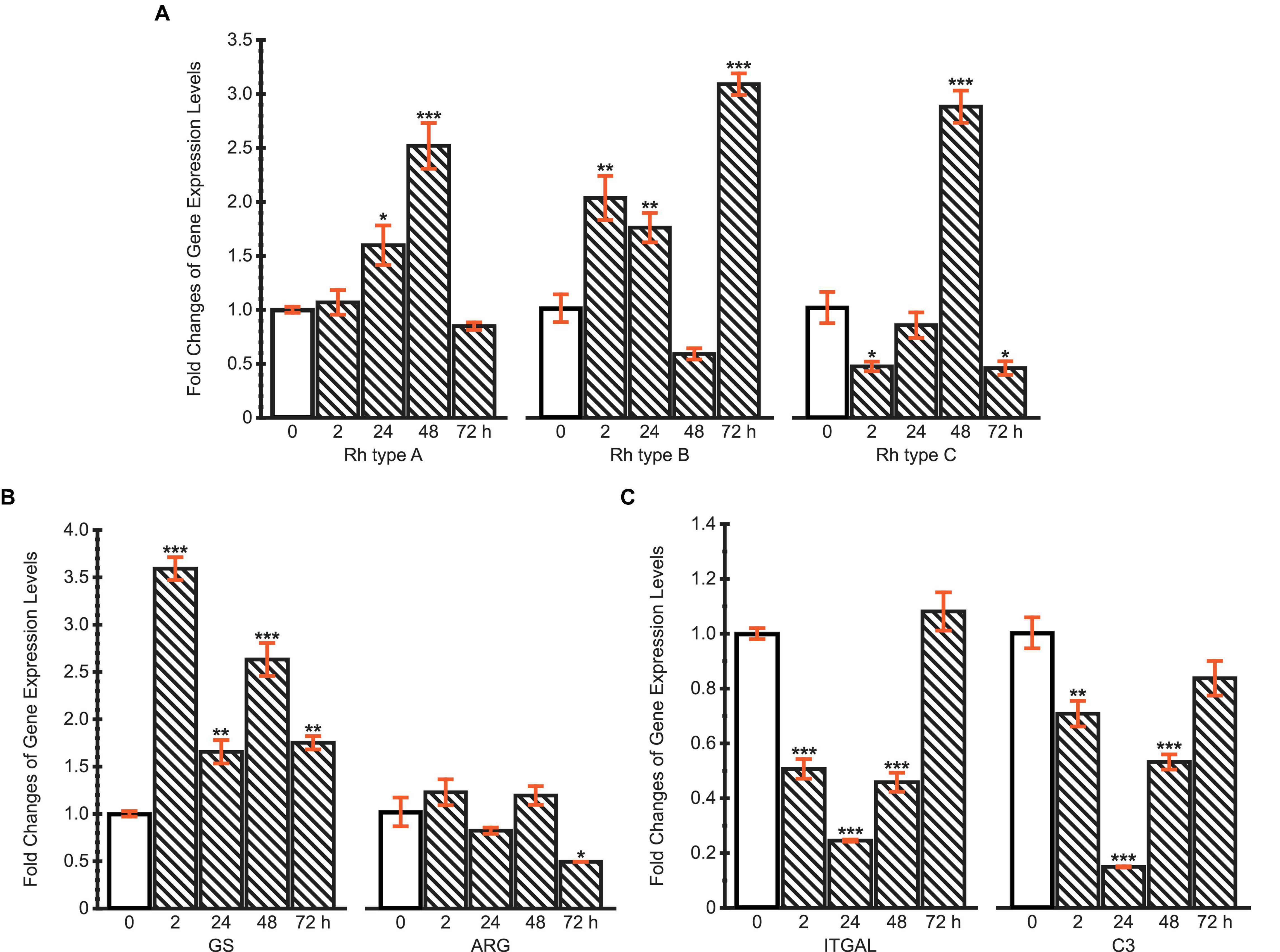
Figure 7. Fold changes of expression levels of (A) Rh type A-C genes; (B) GS and ARG genes; (C) ITGAL and C3 genes under ammonia challenge (10 mg.L−1). Values are shown as mean ± standard error of the mean (S.E.M). Asterisks (*) above the bars denote significant difference between the control (0 h) (blank pattern) and ammonia challenged groups (line pattern) (*P < 0.05, **P < 0.01, ***P < 0.001).
Glutamine synthetase and ARG genes were selected to initially examine two possible pathways of typical ammonia coping strategies, corresponding to the glutamine synthesis and urea synthesis, respectively (Figure 7B). The expression of GS gene was upregulated throughout the 72 h period, during which a maximum of 3.6-fold upregulation was achieved at 2 h. However, ARG’s mRNA expression levels remained stable during the first 48 h, then decreased by 48% at 72 h. These results indicate that M. manhattensis cope with ammonia stress primarily by excreting ammonia via Rh proteins and converting ammonia into glutamine rather than urea.
The expression changes of ITGAL and C3 genes exhibited similar dynamic patterns during 72 h of high ammonia challenge (Figure 7C). Specifically, the expression levels decreased significantly at 2, 24, and 48 h under ammonia challenge, but returned to the control level at 72 h. The recovery of the immune gene expression levels from the initial suppression further implies the roles of regulations of the Rh and GS genes’ expression shown previously.
Discussion
Molecular Characterization of Rh Proteins
The structure of a protein is critical to its function because the structure determines its interactions with other molecules. The vital significance of the molecular characteristics of a protein sequence is that the key amino acid residues/motifs dictate the protein’s unique structure, which plays a crucial role in its function (Lee et al., 2007; Sadowski and Jones, 2009). Therefore, this study first analyzed the molecular characteristics of three Rh proteins of M. manhattensis. We identified the conserved residues/motifs of Rh proteins essential for their ammonia transport function (Figures 2–5). These key residues were involved in the processes including ligand binding, ligand transport, and signal transduction.
Rhesus glycoproteins type A-C of M. manhattensis were predicted to comprise 12 transmembrane regions, with intracellular N-terminus and C-terminus, consistent with other known proteins from the Rh family (Huang and Ye, 2010; Suzuki et al., 2017). A total number of 31–39 phosphorylation sites were predicted in the Rh protein sequences of M. manhattensis. These proteins have the potential of getting phosphorylated by a variety of kinases, which may support a series of enzymatic reactions, thus assisting in regulating these proteins and their roles in various physiological processes. All structures of Rh proteins elucidated so far revealed a hydrophobic pore, the extracellular side of which is gated by two essential phenylalanine residues (Zheng et al., 2004; Gruswitz et al., 2010; Baday et al., 2015). Both phenylalanine residues (F122 and F226 for Rh type A; F127 and F231 for Rh type B; F126 and F230 for Rh type C) were conserved in the three Rh proteins of M. manhattensis (Figures 2–5).
Ammonia nitrogen exists in two forms, NH3 and NH4+. As the hydrophobic pore of Rh proteins prevents the translocation of NH4+, after being recruited at key sites near the pore, NH4+ will be deprotonated to NH3 to be transported through the pore (Baday et al., 2015). For the NH4+ recruitment, molecular dynamic simulations have illustrated that G179, H185, F235, and N236 are essential residues involved in binding NH4+ at a site near H185 in the pore of human RhCG proteins (Gruswitz et al., 2010; Baday et al., 2015). These residues were all conserved in the three Rh proteins of M. manhattensis. Subsequently, deprotonation of the recruited NH4+ facilitates the NH3 conduction. In human RhCG, proton transfer reactions involving D177, S181, H185, and H344 aid in diffusing NH3 down the pore and circulating excess protons out of the cell, thus optimizing the net transport of NH3 (Gruswitz et al., 2010; Baday et al., 2015). These residues were conserved in Rh type A and C of M. manhattensis, but for Rh type B of M. manhattensis, S181 in human RhCG is replaced with alanine (Figure 4). This replacement of S181 was also found in the RhCG of African lungfish, Protopterus annectens (Chng et al., 2017). Calculations conducted by Baday et al. (2015) suggested that the mutation of S181 could impair ammonium transport in human RhCG. However, they also pointed out that S181 should indirectly participate in the deprotonation by maintaining D177 in position to form a stable hydrogen bond from H185 to D177. Therefore, it is likely that the substitution of S181 will not altogether disable the deprotonation in Rh type B of M. manhattensis.
In the meantime, the GalaxyWEB server predicted that the ligand-binding residues should depend on the residue-ligand contact (Supplementary Table 2), which means that the distance between the predicted amino acid residues and the target ligand atom should be sufficiently close for binding. As expected, of the three Rh proteins of M. manhattensis, several predicted NH3 ligand-binding residues were consistent with the structural sites previously identified in human RhCG and E. coli AmtB. Predicted histidine residues (H182 in Rh type B, H181 in Rh type C) were characterized as related to NH4+ binding (H185 in human RhCG). F230 predicted in Rh type C was characterized as related to the gating of the channel (F235 in human RhCG). H335 predicted in Rh type A was characterized as related to the deprotonation (H344 in human RhCG). Other residues, including methionine, histidine, and tryptophan, predicted in at least one of the Rh proteins were also highly conserved across the three Rh proteins of M. manhattensis. Therefore, though they have not been characterized on a protein structural basis, these residues may also support a crucial function in ammonia transport processes via Rh proteins.
In addition to the primary structure (amino acid sequence), the secondary or higher structures and functional domains are crucial for the proper functioning of proteins. Conserved motifs related to the ammonium transport superfamily (PF00909) were identified in the Rh proteins of M. manhattensis (Supplementary Figure 1), confirming their crucial functions in ammonia transport. Comparing with Rh proteins from other species, less variability was detected in the transmembrane regions TM2–TM12 where most MEME motifs were located (∼ amino acid residue 60–400). As shown in other studies (Huang and Liu, 2001; Okuda and Kajii, 2002; Huang and Ye, 2010), C-terminus showed the highest divergence in sequence and size (Figure 2).
Overall, Rh proteins were highly conserved in essential residues/motifs that are involved in ammonia transport, but unique deviations were also detected among the three Rh proteins, which might be responsible for their functional differences (Huang, 2008). The results indicate that the three Rh proteins of M. manhattensis likely recruit and deprotonate the NH4+ in the hydrophobic pore and facilitate the diffusion of NH3 through the channel. Altogether, the predicted molecular structure implies that Rh proteins played an important role in M. manhattensis’ response to ammonia stress. However, since molecular characterization alone is not sufficient for a definite conclusion (Lee et al., 2007), experimental efforts should be made to determine whether Rh proteins of M. manhattensis functionally transport ammonia.
Transcriptional Responses Under Ammonia Challenge and Implications of Ammonia Stress Coping Strategies
For accurate normalization purpose, the expression of reference genes should have minimal variation independent of tissue types, developmental stages, and experimental treatments (Wong and Medrano, 2005; Huang et al., 2016, 2019). Indeed, such ideal universal reference genes do not exist at all (Cubero-Leon et al., 2012; Pu et al., 2020). Therefore, it is crucial to select and validate reliable reference genes for target gene expressions under specific experimental conditions in the species studied. Additionally, although multiple reference genes might perform better than a single gene, the latter is actually more preferred under the premise of its high stability for reducing the complexity, economic cost, and workload of the experiment. In this study, the comprehensive rankings of gene expression stability using RefFinder showed that β-ACTIN was the most stable gene across ammonia treatments in the siphon tissues among all the genes involved, validating it as a proper choice for subsequent analysis.
Rhesus glycoproteins are known to be involved in ammonia excretion, which has been considered an effective primary strategy to cope with high ammonia loadings (Ip et al., 2004; Wright and Wood, 2009; Ip and Chew, 2010). Our study represents the first report of Rh gene/proteins’ potential involvement in ammonia excretion in tunicates during adverse conditions with excessive external ammonia. Our data demonstrated that the expression of three types of Rh mRNA transcripts in M. manhattensis showed significant upregulations, albeit with variations in their expression patterns. All these results suggest ammonia excretion via Rh proteins should be activated when M. manhattensis is acclimating to high ammonia pressure (Figure 7A).
In addition, it has been proposed that the glutamine formation pathway is initiated as one of the classic strategies to deal with environmental ammonia increase in most aquatic animals (Anderson et al., 2002; Ip et al., 2004; Sinha et al., 2013; Wang et al., 2014). Generally, glutamine is synthesized from glutamate and NH4+, catalyzed by the enzyme GS protein. In this study, mRNA expression of GS gene showed a remarkable increase within 72 h after 10 mg⋅L–1 ammonia exposure (Figure 7B), suggesting that this protein and the glutamine formation may have a dominant role in M. manhattensis ammonia detoxification.
Arginase participates in urea synthesis through the ornithine-urea-cycle (OUC) that converts ammonia to urea (Ip and Chew, 2010). In this study, the expression of ARG remained unchanged throughout the entire process of heavy ammonia challenge (Figure 7B), indicating that the urea formation pathway may not play an active role in ammonia detoxification. Indeed, previous studies have confirmed that OUC is not functional in many aquatic animals (Sinha et al., 2013), because converting ammonia to urea via the OUC is energetically much more expensive than ammonia to glutamine, with 1 mol of ATP (adenosine triphosphate) required for producing every amide group of glutamines via GS kinase contrasting 4 mol of ATP hydrolyzed for every mol of synthesized urea through OUC (Ip et al., 2001; Ip and Chew, 2010). Moreover, a study conducted by Goodbody (1957) showed that M. manhattensis excreted non-protein nitrogen chiefly as ammonia. Other tunicate species, such as C. intestinalis, S. plicata, S. partita, and S. clava, also excrete ammonia rather than urea (Markus and Lambert, 1983; Carver et al., 2006; Evans et al., 2017).
Invertebrates have only innate immunity to defend themselves against biotic and abiotic stressors (Shida et al., 2003). Previous studies have indicated that ascidians’ innate immune response was induced under various environmental stressors (Shida et al., 2003; Ewan et al., 2005). Meanwhile, ITGAL and C3 genes were reported to relate to ascidians’ innate immune mechanisms (Miyazawa et al., 2001; Shida et al., 2003; Ewan et al., 2005). As expected, we reported that mRNA expression levels of the ITGAL and C3 were downregulated in M. manhattensis (Figure 7C), implying possible immune suppression caused by ammonia toxicity. However, as ammonia coping strategies such as ammonia excretion and glutamine formation took effect, the immune-related genes eventually returned to normal levels.
Collectively, these findings contributed to our understanding of ammonia stress coping strategies in M. manhattensis. However, the study did not evaluate all potential pathways. To develop a full picture, further research should be undertaken to investigate the incomplete nitrogen conversion involving purine and uric acid, the bacterial-Nephromyces-molgulid symbiosis, and their potential roles in the nitrogenous metabolism under ammonia stress (Das, 1948; Nolfi, 1970; Saffo, 1988; Saffo et al., 2010; Paight et al., 2018).
Conclusion
Here, conserved amino acid residues and domains were identified in the three types of primitive Rh proteins of M. manhattensis, supplying molecular evidence to support their conserved functionality of ammonia transport. The study also revealed for the first time that expression of these Rh genes of M. manhattensis was upregulated under ammonia stress. Meanwhile, expression levels of other genes related to ammonia conversion pathways were analyzed in response to high ammonia exposure. We found that GS gene expression was upregulated, while ARG gene expression remained stable. Taken together, the results suggest that M. manhattensis mitigate high ammonia stress by enhancing ammonia excretion via Rh proteins and detoxifying ammonia into glutamine rather than urea. The results obtained in this study will offer insights into the molecular mechanisms underlying high tolerance of invasive tunicate species M. manhattensis to adverse environments in aquaculture.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical review and approval were not required for invasive ascidians according to the regulations in China. Written informed consent for participation from the owners was not needed for the collection of wild invasive ascidian species.
Author Contributions
AZ, YuC, and XH conceived the study and designed the experiments. YiC conducted the acute toxicity test and collected the samples. YuC conducted the rest of the experiments, performed the data analysis, visualized the results, and wrote the manuscript. All authors interpreted the results. All authors contributed to manuscript revision and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32061143012 and 31772449).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Zunchun Zhou and Bei Jiang for assistance with ascidian sampling at the aquaculture farm and providing experiment sites for acclimation and treatments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.661002/full#supplementary-material
Footnotes
- ^ https://www.hamzasreef.com/Contents/Calculators/TotalAmmonia.php
- ^ https://www.hamzasreef.com/Contents/Calculators/FreeAmmonia.php
- ^ https://www.ebi.ac.uk/Tools/msa/muscle/
- ^ https://www.ncbi.nlm.nih.gov/orffinder
- ^ https://web.expasy.org/protparam/
- ^ http://www.cbs.dtu.dk/services/TMHMM-2.0/
- ^ http://phobius.sbc.su.se/index.html
- ^ http://www.cbs.dtu.dk/services/NetPhos/
- ^ http://www.cbs.dtu.dk/services/NetNGlyc/
- ^ http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=SITE
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ http://wlab.ethz.ch/protter
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ https://pfam.xfam.org/
- ^ http://meme-suite.org//tools/meme
- ^ http://ekhidna2.biocenter.helsinki.fi/sanspanz/
- ^ https://www.heartcure.com.au/reffinder/
References
Anderson, P. M., Broderius, M. A., Fong, K. C., Tsui, K. N. T., Chew, S. F., and Ip, Y. K. (2002). Glutamine synthetase expression in liver, muscle, stomach and intestine of Bostrichthys sinensis in response to exposure to a high exogenous ammonia concentration. J. Exp. Biol. 205, 2053–2065.
Anderson, R. S. (1971). Cellular responses to foreign bodies in the tunicate Molgula Manhattensis (DeKay). Biol. Bull. 141, 91–98. doi: 10.2307/1539993
Baday, S., Orabi, E. A., Wang, S., Lamoureux, G., and Bernèche, S. (2015). Mechanism of NH4+ recruitment and NH3 transport in Rh proteins. Structure 23, 1550–1557. doi: 10.1016/j.str.2015.06.010
Bakouh, N., Benjelloun, F., Cherif-Zahar, B., and Planelles, G. (2006). The challenge of understanding ammonium homeostasis and the role of the Rh glycoproteins. Transf. Clin. Biol. 13, 139–146. doi: 10.1016/j.tracli.2006.02.008
Banerjee, B., Koner, D., Bhuyan, G., and Saha, N. (2018). Differential expression of multiple glutamine synthetase genes in air-breathing magur catfish, Clarias magur and their induction under hyper-ammonia stress. Gene 671, 85–95. doi: 10.1016/j.gene.2018.05.111
Barton, B. A., and Iwama, G. K. (1991). Physiological changes in fish from stress in aquaculture with emphasis on the response and effects of corticosteroids. Annu. Rev. Fish Dis. 1, 3–26. doi: 10.1016/0959-8030(91)90019-G
Carver, C. E., Mallet, A. L., and Vercaemer, B. (2006). Biological Synopsis of the Solitary Tunicate Ciona intestinalis. Canadian Manuscript Report of Fisheries and Aquatic Sciences. Dartmouth: Fisheries and Oceans Canada.
Chen, J. C., Liu, P. C., Lin, Y. T., and Lee, C. K. (1988). Super intensive culture of red-tailed shrimp Penaeus penicillatus. J. World Aquacult. Soc. 19, 127–131. doi: 10.1111/j.1749-7345.1988.tb00940.x
Chen, Y., Gao, Y., Huang, X., Li, S., and Zhan, A. (2021). Local environment-driven adaptive evolution in a marine invasive ascidian (Molgula manhattensis). Ecol. Evol.. doi: 10.1002/ece3.7322
Chen, Y., Li, S., Lin, Y., Li, H., and Zhan, A. (2018). Population genetic patterns of the solitary tunicate, Molgula manhattensis, in invaded Chinese coasts: large-scale homogeneity but fine-scale heterogeneity. Mar. Biodiv. 48, 2137–2149. doi: 10.1007/s12526-017-0743-y
Cheng, C. H., Yang, F. F., Ling, R. Z., Liao, S. A., Miao, Y. T., Ye, C. X., et al. (2015). Effects of ammonia exposure on apoptosis, oxidative stress and immune response in pufferfish (Takifugu obscurus). Aquat. Toxicol. 164, 61–71. doi: 10.1016/j.aquatox.2015.04.004
Chng, Y. R., Ong, J. L. Y., Ching, B., Chen, X. L., Hiong, K. C., Wong, W. P., et al. (2017). Molecular characterization of three Rhesus glycoproteins from the gills of the African lungfish, Protopterus annectens, and effects of aestivation on their mRNA expression levels and protein abundance. PLoS One 12:e0185814. doi: 10.1371/journal.pone.0185814
Cubero-Leon, E., Ciocan, C. M., Minier, C., and Rotchell, J. M. (2012). Reference gene selection for qPCR in mussel, Mytilus edulis, during gametogenesis and exogenous estrogen exposure. Environ. Sci. Pollut. Res. 19, 2728–2733. doi: 10.1007/s11356-012-0772-9
Das, S. M. (1948). The physiology of excretion in Molgula (Tunicata, Ascidiacea). Biol. Bull. 95, 307–319. doi: 10.2307/1538186
Emerson, K., Russo, R. C., Lund, R. E., and Thurston, R. V. (1975). Aqueous ammonia equilibrium calculations: effect of pH and temperature. J. Fish. Res. Board Can. 32, 2379–2383. doi: 10.1139/f75-274
Evans, J. S., Erwin, P. M., Shenkar, N., and López-Legentil, S. (2017). Introduced ascidians harbor highly diverse and host-specific symbiotic microbial assemblages. Sci. Rep. 7:11033. doi: 10.1038/s41598-017-11441-4
Ewan, R., Huxley-Jones, J., Mould, A. P., Humphries, M. J., Robertson, D. L., and Boot-Handford, R. P. (2005). The integrins of the urochordate Ciona intestinalis provide novel insights into the molecular evolution of the vertebrate integrin family. BMC Evol. Biol. 5:31. doi: 10.1186/1471-2148-5-31
Fofonoff, P. W., Ruiz, G. M., Steves, B., Simkanin, C., and Carlton, J. T. (2018). National Exotic Marine and Estuarine Species Information System. Available online at: http://invasions.si.edu/nemesis/ (accessed January 13, 2021).
Geng, Z., Liu, Q., Wang, T., Ma, S., and Shan, H. (2020). Changes in physiological parameters involved in glutamine and urea synthesis in Pacific white shrimp, Litopenaeus vannamei, fed Ampithoe sp. meal and exposed to ammonia stress. Aquacult. Res. 51, 2725–2734. doi: 10.1111/are.14611
Gruswitz, F., Chaudhary, S., Ho, J. D., Schlessinger, A., Pezeshki, B., Ho, C. M., et al. (2010). Function of human Rh based on structure of RhCG at 2.1 Å. Proc. Natl. Acad. Sci. U.S.A. 107, 9638–9643. doi: 10.1073/pnas.1003587107
Guo, H., Lin, W., Wu, X., Wang, L., Zhang, D., Li, L., et al. (2020). Survival strategies of Wuchang bream (Megalobrama amblycephala) juveniles for chronic ammonia exposure: antioxidant defense and the synthesis of urea and glutamine. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 230:108707. doi: 10.1016/j.cbpc.2020.108707
Haydar, D., Hoarau, G., Olsen, J. L., Stam, W. T., and Wolff, W. J. (2011). Introduced or glacial relict? Phylogeography of the cryptogenic tunicate Molgula manhattensis (Ascidiacea, Pleurogona). Divers. Distrib. 17, 68–80. doi: 10.1111/j.1472-4642.2010.00718.x
Hewitt, C. L., Campbell, M. L., Thresher, R. E., Martin, R. B., Boyd, S., Cohen, B. F., et al. (2004). Introduced and cryptogenic species in Port Phillip Bay, Victoria, Australia. Mar. Biol. 144, 183–202. doi: 10.1007/s00227-003-1173-x
Huang, C. H. (2008). Molecular origin and variability of the Rh gene family: an overview of evolution, genetics and function. Haematologica 2, 149–157.
Huang, C. H., and Liu, P. Z. (2001). New insights into the Rh superfamily of genes and proteins in erythroid cells and nonerythroid tissues. Blood Cells, Mol. Dis. 27, 90–101. doi: 10.1006/bcmd.2000.0355
Huang, C. H., and Peng, J. (2005). Evolutionary conservation and diversification of Rh family genes and proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 15512–15517. doi: 10.1073/pnas.0507886102
Huang, C. H., and Ye, M. (2010). The Rh protein family: gene evolution, membrane biology, and disease association. Cell Mol. Life Sci. 67, 1203–1218. doi: 10.1007/s00018-009-0217-x
Huang, X., Gao, Y., Jiang, B., Zhou, Z., and Zhan, A. (2016). Reference gene selection for quantitative gene expression studies during biological invasions: a test on multiple genes and tissues in a model ascidian Ciona savignyi. Gene 576, 79–87. doi: 10.1016/j.gene.2015.09.066
Huang, X., Li, S., and Zhan, A. (2019). Genome-wide identification and evaluation of new reference genes for gene expression analysis under temperature and salinity stresses in Ciona savignyi. Front. Genet. 10:71. doi: 10.3389/fgene.2019.00071
Hurvitz, A., Bercovier, H., and Van Rijn, J. (1997). Effect of ammonia on the survival and the immune response of rainbow trout (Oncorhynchus mykiss, Walbaum) vaccinated against Streptococcus iniae. Fish Shellf. Immunol. 7, 45–53. doi: 10.1006/fsim.1996.0062
Ip, Y. K., and Chew, S. F. (2010). Ammonia production, excretion, toxicity, and defense in fish: a review. Front. Physiol. 1:134. doi: 10.3389/fphys.2010.00134
Ip, Y. K., Chew, S. F., and Randall, D. J. (2001). “Ammonia toxicity, tolerance, and excretion,” in Fish Physiology, eds P. A. Wright and P. M. Anderson (New York, NY: Academic Press), 109–148. doi: 10.1016/s1546-5098(01)20005-3
Ip, Y. K., Chew, S. F., Wilson, J. M., and Randall, D. J. (2004). Defences against ammonia toxicity in tropical air-breathing fishes exposed to high concentrations of environmental ammonia: a review. J. Comp. Physiol. B 174, 565–575. doi: 10.1007/s00360-004-0445-1
Khademi, S., O’Connell, J. III, Remis, J., Robles-Colmenares, Y., Miercke, L. J. W., and Stroud, R. M. (2004). Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305, 1587–1594. doi: 10.1126/science.1101952
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lambert, G. (2003). New records of ascidians from the NE Pacific: a new species of Trididemnum, range extension and redescription of Aplidiopsis pannosum (Ritter, 1899) including its larva, and several non-indigenous species. Zoosystema 25, 665–679.
Lee, D., Redfern, O., and Orengo, C. (2007). Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 8, 995–1005. doi: 10.1038/nrm2281
Li, M., Gong, S., Li, Q., Yuan, L., Meng, F., and Wang, R. (2016). Ammonia toxicity induces glutamine accumulation, oxidative stress and immunosuppression in juvenile yellow catfish Pelteobagrus fulvidraco. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 183–184, 1–6. doi: 10.1016/j.cbpc.2016.01.005
Liu, S., Pan, L., Liu, M., and Yang, L. (2014). Effects of ammonia exposure on nitrogen metabolism in gills and hemolymph of the swimming crab Portunus trituberculatus. Aquaculture 432, 351–359. doi: 10.1016/j.aquaculture.2014.05.029
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Markus, J. A., and Lambert, C. C. (1983). Urea and ammonia excretion by solitary ascidians. J. Exp. Mar. Biol. Ecol. 66, 1–10. doi: 10.1016/0022-0981(83)90023-0
Miyazawa, S., Azumi, K., and Nonaka, M. (2001). Cloning and characterization of integrin α subunits from the solitary Ascidian, Halocynthia roretzi. J. Immunol. 166, 1710–1715. doi: 10.4049/jimmunol.166.3.1710
Nakhoul, N. L., and Hamm, L. L. (2014). The challenge of determining the role of Rh glycoproteins in transport of NH3 and NH4+. WIREs Membr. Transp. Signal. 3, 53–61. doi: 10.1002/wmts.105
Nolfi, J. R. (1970). Biosynthesis of uric acid in the tunicate, Molgula Manhattensis, with a general scheme for the function of stored purines in animals. Comp. Biochem. Physiol. 35, 827–842. doi: 10.1016/0010-406x(70)90078-2
Okuda, H., and Kajii, E. (2002). The evolution and formation of RH genes. Leg. Med. 4, 139–155. doi: 10.1016/S1344-6223(02)00031-7
Omasits, U., Ahrens, C. H., Müller, S., and Wollscheid, B. (2014). Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. doi: 10.1093/bioinformatics/btt607
Osman, R. W., and Whitlatch, R. B. (1995). The influence of resident adults on larval settlement: experiments with four species of ascidians. J. Exp. Mar. Biol. Ecol. 190, 199–220. doi: 10.1016/0022-0981(95)00036-Q
Paight, C., Slamovits, C. H., Saffo, M. B., and Lane, C. E. (2018). Nephromyces encodes a urate metabolism pathway and predicted peroxisomes, demonstrating that these are not ancient losses of apicomplexans. Genome Biol. Evol. 11, 41–53. doi: 10.1093/gbe/evy251
Pimentel, D., Lach, L., Zuniga, R., and Morrison, D. (2000). Environmental and economic Costs of Nonindigenous species in the United States. Bioscience 50, 53–65. doi: 10.1641/0006-35682000050[0053:EAECON]2.3.CO;2
Pu, Q., Li, Z., Nie, G., Zhou, J., Liu, L., and Peng, Y. (2020). Selection and validation of reference genes for quantitative real-time PCR in white clover (Trifolium repens L.) involved in five abiotic stresses. Plants 9:996. doi: 10.3390/plants9080996
Pyo, J., Lee, T., and Shin, S. (2012). Two newly recorded invasive alien ascidians (Chordata, Tunicata, Ascidiacea) based on morphological and molecular phylogenetic analysis in Korea. Zootaxa 3368, 211–228. doi: 10.11646/zootaxa.3368.1.10
Randall, D. J., and Tsui, T. K. N. (2002). Ammonia toxicity in fish. Mar. Pollut. Bull. 45, 17–23. doi: 10.1016/S0025-326X(02)00227-8
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Robertson, J. L., Russell, R. M., Preisler, H. K., and Savin, N. E. (2007). Bioassays with Arthropods, 2nd Edn, Boca Raton, FL: CRC Press.
Sadowski, M. I., and Jones, D. T. (2009). The sequence-structure relationship and protein function prediction. Curr. Opin. Struct. Biol. 19, 357–362. doi: 10.1016/j.sbi.2009.03.008
Saffo, M. B. (1988). Nitrogen waste or nitrogen source? Urate degradation in the renal sac of Molgulid Tunicates. Biol. Bull. 175, 403–409. doi: 10.2307/1541732
Saffo, M. B., McCoy, A. M., Rieken, C., and Slamovits, C. H. (2010). Nephromyces, a beneficial apicomplexan symbiont in marine animals. Proc. Natl. Acad. Sci. U.S.A. 107, 16190–16195. doi: 10.1073/pnas.1002335107
Shida, K., Terajima, D., Uchino, R., Ikawa, S., Ikeda, M., Asano, K., et al. (2003). Hemocytes of Ciona intestinalis express multiple genes involved in innate immune host defense. Biochem. Biophys. Res. Commun. 302, 207–218. doi: 10.1016/S0006-291X(03)00113-X
Sinha, A. K., Giblen, T., AbdElgawad, H., De Rop, M., Asard, H., Blust, R., et al. (2013). Regulation of amino acid metabolism as a defensive strategy in the brain of three freshwater teleosts in response to high environmental ammonia exposure. Aquat. Toxicol. 130–131, 86–96. doi: 10.1016/j.aquatox.2013.01.003
Suzuki, A., Komata, H., Iwashita, S., Seto, S., Ikeya, H., Tabata, M., et al. (2017). Evolution of the RH gene family in vertebrates revealed by brown hagfish (Eptatretus atami) genome sequences. Mol. Phylogenet. Evol. 107, 1–9. doi: 10.1016/j.ympev.2016.10.004
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Wang, G., Pan, L., and Ding, Y. (2014). Defensive strategies in response to environmental ammonia exposure of the sea cucumber Apostichopus japonicus: Glutamine and urea formation. Aquaculture 432, 278–285. doi: 10.1016/j.aquaculture.2014.05.006
Weihrauch, D., and Allen, G. J. P. (2018). Ammonia excretion in aquatic invertebrates: new insights and questions. J. Exp. Biol. 221:jeb169219. doi: 10.1242/jeb.169219
Wheeler, M. W., Park, R. M., and Bailer, A. J. (2006). Comparing median lethal concentration values using confidence interval overlap or ratio tests. Environ. Toxicol. Chem. 25, 1441–1444. doi: 10.1897/05-320R.1
Wong, M. L., and Medrano, J. F. (2005). Real-time PCR for mRNA quantitation. Biotechniques 39, 75–85. doi: 10.2144/05391RV01
Wright, P. A., Steele, S. L., Huitema, A., and Bernier, N. J. (2007). Induction of four glutamine synthetase genes in brain of rainbow trout in response to elevated environmental ammonia. J. Exp. Biol. 210, 2905–2911. doi: 10.1242/jeb.003905
Wright, P. A., and Wood, C. M. (2009). A new paradigm for ammonia excretion in aquatic animals: role of Rhesus (Rh) glycoproteins. J. Exp. Biol. 212, 2303–2312. doi: 10.1242/jeb.023085
Yeam, C. T., Chng, Y. R., Ong, J. L. Y., Wong, W. P., Chew, S. F., and Ip, Y. K. (2017). Molecular characterization of two Rhesus glycoproteins from the euryhaline freshwater white-rimmed stingray, Himantura signifer, and changes in their transcript levels and protein abundance in the gills, kidney, and liver during brackish water acclimation. J. Comp. Physiol. B 187, 911–929. doi: 10.1007/s00360-017-1067-8
You, X., Chen, J., Bian, C., Yi, Y., Ruan, Z., Li, J., et al. (2018). Transcriptomic evidence of adaptive tolerance to high environmental ammonia in mudskippers. Genomics 110, 404–413. doi: 10.1016/j.ygeno.2018.09.001
Yue, F., Pan, L., Xie, P., Zheng, D., and Li, J. (2010). Immune responses and expression of immune-related genes in swimming crab Portunus trituberculatus exposed to elevated ambient ammonia-N stress. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 157, 246–251. doi: 10.1016/j.cbpa.2010.07.013
Zhan, A., Ni, P., Xiong, W., Chen, Y., Lin, Y., Huang, X., et al. (2017). “Biological invasions in aquatic ecosystems in China,” in Biological Invasions and Its Management in China, eds F. Wan, M. Jiang, and A. Zhan (Dordrecht: Springer), 67–96. doi: 10.1007/978-94-024-0948-2_4
Zhang, M., Li, M., Wang, R., and Qian, Y. (2018). Effects of acute ammonia toxicity on oxidative stress, immune response and apoptosis of juvenile yellow catfish Pelteobagrus fulvidraco and the mitigation of exogenous taurine. Fish Shellf. Immunol. 79, 313–320. doi: 10.1016/j.fsi.2018.05.036
Zheng, C. (1988). The ascidians among the fouling organisms in the coast of the Yellow Sea and Bohai Sea. Acta Zool. Sin. 34, 180–188.
Zheng, L., Kostrewa, D., Berneche, S., Winkler, F. K., and Li, X. D. (2004). The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 17090–17095. doi: 10.1073/pnas.0406475101
Keywords: biological invasion, ammonia acclimation, gene expression, primitive Rh, stress response
Citation: Chen Y, Huang X, Chen Y and Zhan A (2021) Ammonia Stress Coping Strategy in a Highly Invasive Ascidian. Front. Mar. Sci. 8:661002. doi: 10.3389/fmars.2021.661002
Received: 30 January 2021; Accepted: 16 March 2021;
Published: 07 April 2021.
Edited by:
Periklis Kleitou, Marine and Environmental Research Lab (MER), CyprusReviewed by:
Xavier Turon, Consejo Superior de Investigaciones Científicas (CSIC), SpainCorina Ciocan, University of Brighton, United Kingdom
Copyright © 2021 Chen, Huang, Chen and Zhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aibin Zhan, emhhbmFpYmluQGhvdG1haWwuY29t; YXpoYW5AcmNlZXMuYWMuY24=
 Yuqing Chen
Yuqing Chen Xuena Huang
Xuena Huang Yiyong Chen1
Yiyong Chen1 Aibin Zhan
Aibin Zhan