- 1Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences, Guangzhou, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China
- 4Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 5Sanya National Marine Ecosystem Research Station, Tropical Marine Biological Research Station in Hainan, Chinese Academy of Sciences, Sanya, China
- 6Key Laboratory of Tropical Marine Biotechnology of Hainan Province, Sanya Institute of Oceanology, South China Sea Institute of Oceanology (SCSIO), Sanya, China
Coral reefs are undergoing global phase shifts from coral-dominated to algae-dominated stages. The negative effects of this substratum shift on the diversity and abundance of fish have been well documented, but the influence on fish feeding is less studied, which may limit a deeper understanding of trophic pathways in such a disturbed system. In this study, we investigated the feeding response of a numerically dominant fish species Ctenochaetus striatus to different substrate types, including hard coral, short algal turfs (SATs, <5 mm), and long algal turfs (LATs, >5 mm), on reefs in the South China Sea. The biomass of C. striatus showed an inverted U-shaped relationship with coral coverage and a significant positive correlation with SAT coverage (p < 0.05), indicating that rising SAT coverage associated with moderate coral loss provoked a feeding response in C. striatus. Stomach contents of C. striatus, analyzed using high-throughput sequencing (HTS), were dominated by algal sequences (relative read abundance, RRA > 80.0%), including macroalgae, filamentous algae, and microalgae (e.g., Symbiodinium and Prorocentrum). The sequence number and diversity of microalgae (mainly dinoflagellates) tended to be abundant (RRA 13.5–36.5%) with increased SAT cover, but brown algae sequences (RRA 17.2–57.8%) or green algae sequences (RRA > 50.7% except one site) dominated the stomach content DNA in reefs with high coral cover and high LAT or macroalgal cover, respectively. Considering the limited ability of C. striatus to remove mature algae, macroalgal DNA might be from algal debris. Our results indicate that C. striatus populations respond positively to conditions of moderate coral loss through increases in body condition identified as increased biomass. These responses are correlated to the expansion of SAT’s as coral cover declined, however, this relationship reverses if coral loss is high due to the succession of LAT’s over SAT’s and a corresponding decrease in the quality of food available. Our use of HTS has nevertheless identified the importance of detritivory in the flow of energy through reefs in the Anthropocene which are increasingly becoming depauperate in hard coral.
Introduction
Coral reefs are degenerating worldwide under anthropogenic and natural stresses, causing phase shifts from coral-dominated to algae-dominated reefs (Bellwood et al., 2006; Cheal et al., 2010). In several cases, this has led to a rise in coverage of highly productive and stress-tolerant algal turfs (Connell et al., 2014). Algal turfs contain filamentous algae, sediments, microalgae, and detrital material and can occupy 30–80% of the reef substratum after coral degradation (Wilson and Bellwood, 1997; Goatley and Bellwood, 2011; Kramer et al., 2014; Tebbett et al., 2020). These substratum changes will not only influence coral recruitment (Bellwood and Fulton, 2008; Spear et al., 2019; Tebbett and Bellwood, 2020), but also influence consumers relying on benthic primary productivity, such as detritivorous/herbivorous fishes (Bellwood and Fulton, 2008; Marshell and Mumby, 2015; Bellwood et al., 2019). Given shifting benthic configurations on coral reefs toward algal turfs (Goatley et al., 2016), detritivorous/herbivorous fishes feeding on algal turf are becoming critical in Anthropocene coral reefs (Max et al., 2013; Bellwood et al., 2019).
Although extensive coral degradation will exert negative effects on the diversity and abundance of many reef fishes, species with resource utilization patterns less dependent on live coral may differ in their responses to disturbances (Pratchett et al., 2011). Compared with coral-dependent species, detritivorous/herbivorous fishes may benefit from moderate coral reduction (Wilson et al., 2006). Russ et al. (2018) found that detritivorous surgeonfishes typically feed on dead substrata and thus increase in density with the reduction in live hard coral cover after large environmental disturbances. Several recent studies have also shown that the distribution of detritivorous/herbivorous fishes is often correlated with their food resources, such as algal turfs and fleshy macroalgae (Russ et al., 2015; Tootell and Steele, 2016). Indeed, coral loss will usually be followed by the expansion of benthic algae and other non-coral organisms, and food resources for detritivorous/herbivorous fishes may become more available than on coral-dominated reefs. However, previous studies have paid more attention to the distribution or variation in abundance of fish, with the effect of these substratum changes on the feeding of fish less studied, limiting insights into the adaptability, trophic interactions, food webs and functional delivery of reef fish (Wilson et al., 2006; Brandl and Bellwood, 2014; Leal and Ferrier-Pages, 2016; Tebbett et al., 2020).
Extensive coral reef degeneration will also exert negative effects on detritivorous/herbivorous fishes, not only due to the decline in habitat and topographical complexity (Pratchett et al., 2011), but also due to the change in food quality and palatability (Tebbett et al., 2020). Algal turfs have a spectrum of forms from short productive algal turfs (SATs) to long sediment-laden algal turfs (LATs), with the latter more abundant on disturbed reefs (Goatley et al., 2016; Tebbett and Bellwood, 2020). Compared with LATs, SATs have a higher detrital proportion and fewer sediments, which make them more palatable for detritivorous fish (Tebbett and Bellwood, 2020). A recent study found that high sediment loads on algal turfs would limit the abundance and feeding activity of C. striatus (Tebbett et al., 2020), a major detritivore on tropical reefs (Trip et al., 2008; Cheal et al., 2012). Ctenochaetus striatus is involved in a series of ecosystem functions, such as detritivory and sediment removal (Goatley and Bellwood, 2010; Marshell and Mumby, 2015), but it is sensitive to sediments remaining in algal turfs (Tebbett et al., 2017a,b), and LATs are also not suitable for its distinctive “brushing” feeding habit (Tebbett et al., 2017c). It is interesting that while C. striatus is sensitive to substratum traits it is widely distributed on various disturbed reefs, and more details about the dietary changes behind substratum shifting may provide important information for revealing the responses and adaptability of this species to disturbances (Tebbett et al., 2017b, 2020; Bellwood et al., 2019). However, few studies have investigated the correlation between the specific species within the algal turfs that C. striatus feeds on, which may limit an extensive analysis of the function and trophic roles of this important detritivore on Anthropocene reefs (Bellwood et al., 2019; Tebbett et al., 2020).
Obstacles to documenting the diversity of the C. striatus diet include the complex dietary sources and the high level of mechanical processing during the digestive process of this fish (Choat et al., 2004). It is difficult and time-consuming to identify the stomach contents of C. striatus. Most previous studies have classified food items into different morphological types, such as turf algae, macroalgae, sediments, and other unidentified materials (mostly categorized as detritus; Kelly et al., 2016; Tebbett et al., 2017c), without identifying the specific species or components in the diets of C. striatus. For example, microbes and microalgae that are often classified as detritus are also a potential nutrition source for C. striatus and other detritivores (Wilson et al., 2003; Tebbett and Bellwood, 2020). However, the species information of these cryptic organisms has often been deficient, which may ignore some subtle but important variations in the resource utilization of detritivores.
In recent years, high-throughput sequencing (HTS) has been widely used for trophic analyses of animals, including fish and other marine animals, because of its accuracy, comprehensiveness, and ability to process large databases (O’Rorke et al., 2012). HTS is highly suitable for screening certain cryptic species, which are difficult to precisely identify using traditional gut morphological identification methods (Leal and Ferrier-Pages, 2016), such as microalgae in the diets of gelatinous zooplankter, ciliates ingested by juvenile fish and dietary partitioning among cryptobenthic species (Lin et al., 2018a; Walters et al., 2019; Brandl et al., 2020). Meanwhile, the sources of detritus in the algal turfs are from a variety of organisms (Wilson et al., 2003), which implies that the DNA of these organisms remaining in the detritus could provide some useful information about the possible sources of detritus. Hence, we applied HTS to investigate the specific components of the diets of C. striatus and how this varies across coral reef habitats in the South China Sea with differing levels of degradation. The study objectives were to (1) identify the exact species removed or ingested by C. striatus and (2) determine whether and how the abundance, biomass, and specific food species of C. striatus are influenced by the substratum. These results will offer an improved and more detailed perspective on the diets of detritivorous fish and take a key step forward in understanding the influence of coral degeneration on detritivory and trophic interactions on coral reefs.
Materials and Methods
Study Design and Location
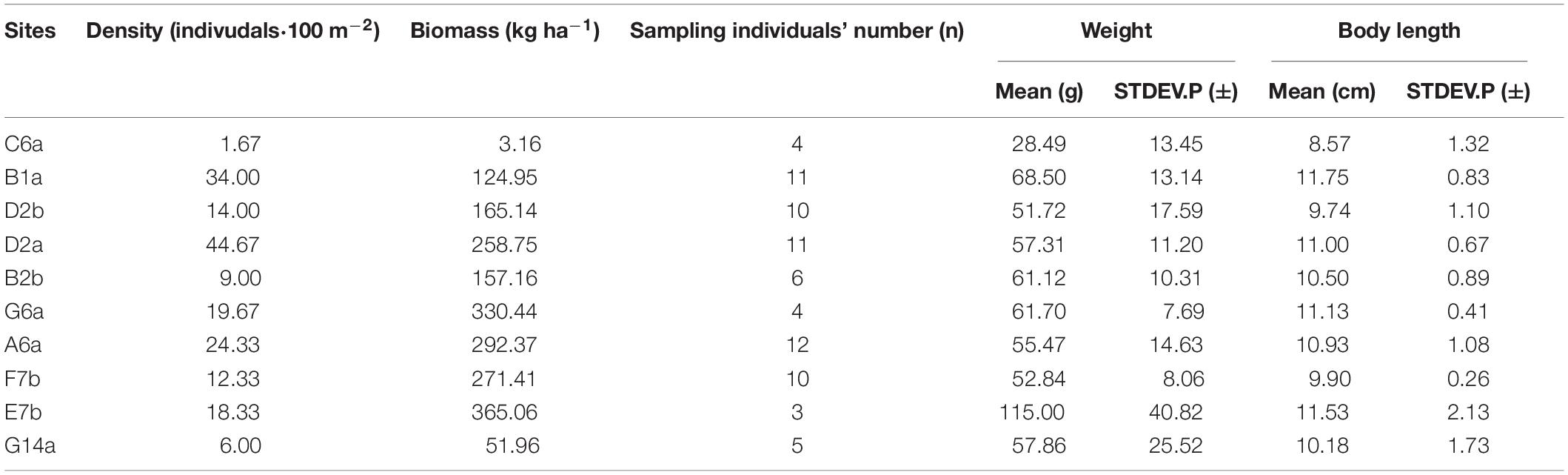
Seven reefs with different levels of hard coral coverage located at the same latitude and relatively far away from the mainland (N: 8°51′–10°57′ and E: 112°50′–115°35′, Figures 1A,B), were surveyed in the southern part of the South China Sea in May 2016 and 2017. Considering that detritivorous/herbivorous fishes may benefit from moderate coral loss, three reef categories characterized by different coral dominance levels were classified: HC reefs (the highest coverage of hard living coral with more than 35%), IC reefs (intermediate coral coverage of 15–30%), and LC reefs (the lowest coral coverage ≤ 15%). The classification standards were mainly based on the historical data of surveyed reefs and relatively healthy reefs in other sea areas (Kingman Reef and Palmyra Atoll reef; Huang et al., 2012; Williams et al., 2013; Zhao et al., 2013). For site selection, at least three sites from each reef category were included. Surveys were conducted on the seaward reef crest (between 2 and 4 m depth; Figures 1C,D) to ensure that all sites had similar geographical structures and environmental conditions. Finally, a total of ten sites met all of the criteria and were selected for field surveying and fish sampling: three sites for HC reefs, four sites for IC reefs, and three sites for LC reefs.
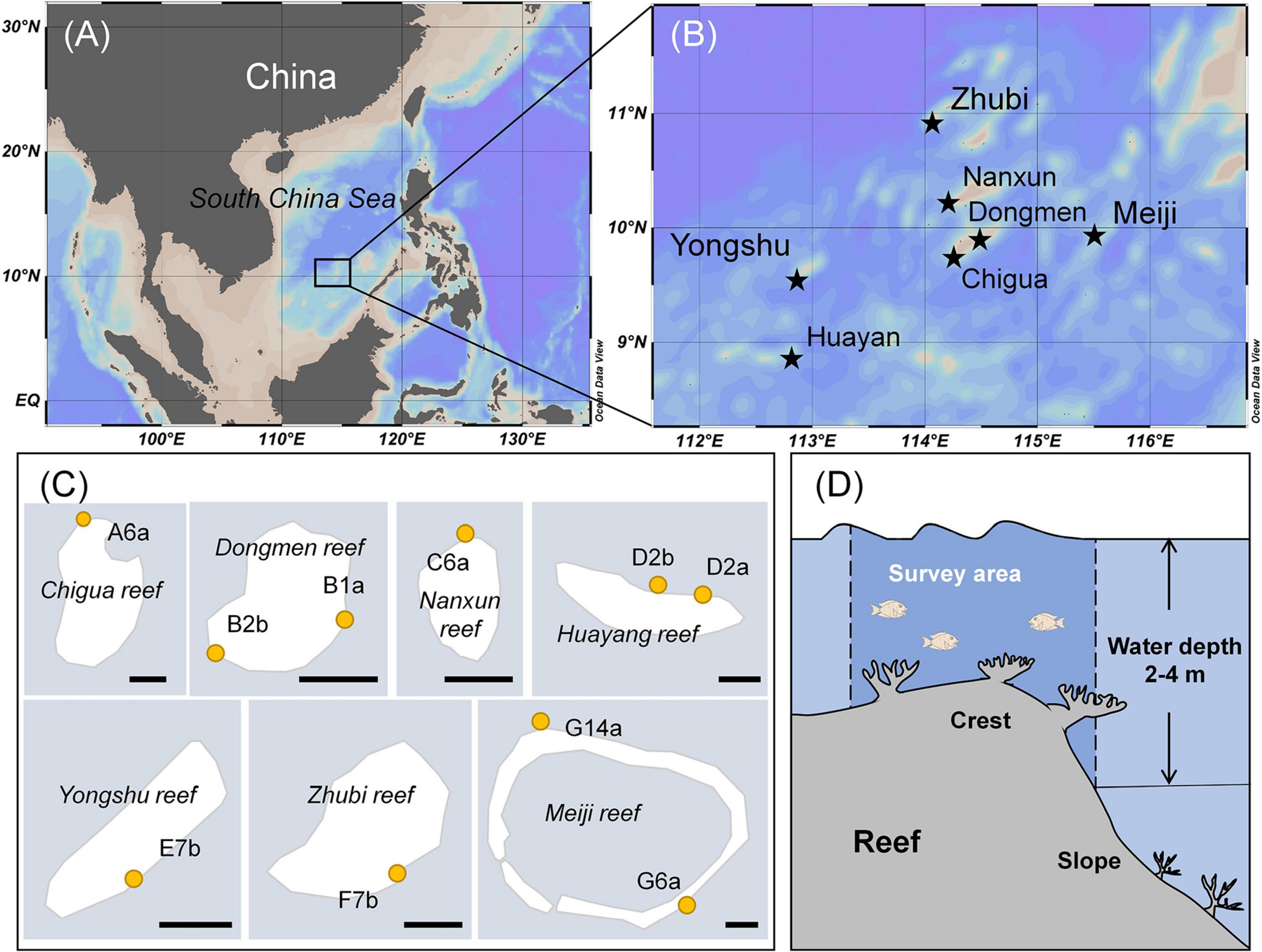
Figure 1. Location of surve yed sites and area on the reefs in the southern part of the South China Sea. Seven reefs were surveyed (A,B) and all the sites were located on the windward side of reefs. The solid line on the scale at the bottom right of the figure represents 1 km (C), and surveyed areas were the seaward reef crests at a depth of 2–4 m (D).
Fish and Benthic Surveys
At each site, a 60 m long transect line was laid parallel to the crest between depths of 2–4 m. The abundance and biomass of C. striatus was quantified first at each site by an experienced surveyor using underwater visual census along the 2.5 × 60 m belt transects. All individuals of C. striatus were counted and placed into 5 cm size categories. Biomass of C. striatus was estimated from the individual fish-length data through length-weight relationships extracted from FISHBASE (Froese and Pauly, 2020). The calculated results were compared with those calculated from fish sampled at the same reefs to ensure the accuracy of fish weight data.
Substrate surveys were carried out by another diver after the visual fish counts at the same transects. Thirty quadrats (0.5 m × 0.5 m) were placed randomly along each transect to maximize spatial coverage and minimize frame overlaps. The substrate and associated benthic community were recorded in each quadrat using an underwater video camera (Olympus TG-4). The quadrats were divided into a 4 × 4 grid using strings placed at 12.5 cm intervals along the vertical and horizontal axes. Substrate identifications were summarized into the following general categories, hard living coral, fleshy macroalgae, crustose coralline algae (CCA), sands, short algal turf (SATs, algal turf length ≤ 5 mm), and long algal turf (LATs, algal turf length > 5 mm). To distinguish LATs from SATs, algal turf length was measured using a depth probe of vernier calipers in situ (Tebbett and Bellwood, 2019) and recorded using a digital camera. The substrate identifications were compared with the algal turf length records to classify the forms of algal turf. Partial substrate composition was done in situ and was used to regulate the results identified from the photos. The percentage cover per category and transect variance were automatically generated using Excel Extensions v.3.0 (Kohler and Gill, 2006).
Fish Sampling
Fish sampling was conducted at the same area where transect surveys were conducted. Adult individuals of C. striatus were caught using a trammel net (10 m length × 1.5 m height, the maximum and minimum mesh sizes were 9.0 and 3.0 cm, respectively). Fish were captured between 0900 and 1100 h, which is when feeding intensities of C. striatus are higher (Zemke-White et al., 2002), and stored at −20°C within 1 h. After completing the weight and body length measurements, stomach contents were carefully collected and preserved in 95% ethanol at 4°C before dietary analysis. The stomach contents of fish samples were identified at two magnifications: 4 × 10 magnification using a dissecting microscope and 10 × 40 magnification under higher power before DNA extraction.
Identification of Stomach Contents Using High-Throughput Sequences
At least three fish were randomly selected from the samples at each site. Their stomach contents were ground and homogenized with silicate sand in a FastPrep® instrument (MP Biomedicals, Santa Ana, United States) for 40 s at a speed setting of 6.0 m/s to ensure that all material was completely broken and homogenized. The DNA of the stomach contents was extracted using the FastDNA® Spin Kit for Feces (MP Biomedicals, Santa Ana, United States) following the manufacturer’s instructions and eluted in 30 μL of 10 mM Tris–HCl (pH 8.0). DNA quality was assessed by spectrophotometry at a 260/280 nm ratio, and DNA concentrations were quantified using a Nanodrop® spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). DNA samples were stored at −20°C until further use.
Because the exact dietary components of C. striatus were not clear, whole DNA extracts were amplified using the universal TAReuk454FWD1-TAReukREV3 primer pair (TAReuk454FWD1: 5-CCAGCASCYGCGGTAATTCC-3; TAReukREV3: 5-ACTTTCGTTCTTGATYRA-3), which targets the V4 region (∼380 bp) of eukaryotic 18S rDNA (Stoeck et al., 2010). 18S rDNA was selected because it has been widely used for the identification of eukaryotic organisms and has a comprehensive reference library (Corse et al., 2010). In addition, the accuracy of its semiquantitative data on the dietary analysis of fish feeding on benthic organisms was shown in our previous study (Lin et al., 2018a,b). However, considering the low species resolution of 18S rDNA V4 metabarcoding (Lin et al., 2018b), the highest identification resolution and associated analyses were limited to the genus level in this study. PCR was conducted in a 20 μL reaction volume composed of 4 μL 5 × FastPfu Buffer, 2 μL 2.5 mM dNTPs, 0.8 μL each of 5 μM universal forward and reverse primers, 0.4 μL FastPfu Polymerase, and 10 ng genomic DNA. The PCR conditions were as follows: an initial denaturation step at 95°C for 5 min; 27 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s; and a final elongation step at 72°C for 10 min. The PCR products were prepared for sequencing at 10°C, examined by electrophoresis with a 2% agarose gel, and then sequenced using the Illumina HiSeq platform.
The raw data obtained from the Illumina HiSeq platform were subjected to quality control in accordance with the Illumina HiSeq platform workflow (Pompanon et al., 2012). After removing the primer sequences, data splitting, and splicing of paired-end reads, the sequences were filtered and intercepted. Only high-quality, long (>300 bp) sequences were retained for further analysis. To evaluate the composition and diversity of feeding items, the effective sequences were clustered into operational taxonomic units (OTUs) using Uparse v7.0.1001 software with a 97% threshold.
Representative OTU sequences were aligned with the GenBank database using the basic local alignment search tool (BLAST). A species name was assigned only if a single best hit achieved 100% similarity. A genus name was accepted only if the similarities of the five best hits were ≥98%. A family name was retained only if the similarities of all the best hits were ≥95%. A phylum or subphylum name was retained only if the similarities of all the best hits were ≥90%. Sequences with maximum similarities <90% were labeled ‘‘No Account (NA).’’ These were most likely due to the GenBank deficiency. The number of effective sequences was then calculated for each phylum. Sequences that were identified as of fish and human origin, and low-frequency OTU sequences (sequence abundance in each sample was less than 10) were removed from each data set. To reduce the risk of misidentification at the lower taxonomic levels, a proportion of the different OTUs was returned for each level and those labeled ‘‘NA’’ were removed prior to dietary composition analysis. To calculate the alpha diversity, two metrics were calculated (Chao1 and OTU numbers) and rarefaction curves were generated based on the filtered OTU table using the QIIME software1.
Data Analysis
The relationship between substrate categories and C. striatus distribution was analyzed by using Pearson’s product–moment and investigating the correlation of the percentage of substratum coverage with the density and biomass of C. striatus. The correlation matrix and corresponding significance level were calculated using the “Hmisc” package in R statistical software (version 4.0.3; Harrell, 2019; R Core Team, 2019).
To measure the dissimilarity of the diets of C. striatus among reefs and the relationships between relative read abundance (RRA) and substrate categories, redundancy analysis (RDA) was performed using the “vegan” R package software (Oksanen et al., 2009). However, in some cases, the RRA-based data from fecal or stomach content samples were affected by recovery biases, and occurrence data were included to present results that were more reliable (Deagle et al., 2019). A minimum sequence percentage threshold (1%) was defined to determine occurrences, which balanced the data, thereby maximizing the inclusion of real diet sequences and excluded low-level background noise (secondary predation, contamination, and sequencing errors). Following this, a relative abundance read table was converted to a presence/absence matrix. Non-metric multidimensional scaling (NMDS) ordination was calculated using the Jaccard metric within the vegan package to provide a graphical representation of different species relations (and thus differences in diet) based on the presence/absence matrix.
To compare the feeding diversity and specific food taxa of C. striatus among reefs, the food items in the presence/absence matrix were selected, heatmaps were created based on the RRA of these food items, and the maximum-likelihood tree of 18S rDNA sequences was established using MEGA 4.0 software. All graphic visualizations were prepared using the “ggplot2” package in R (Wickham, 2016). Differences were considered significant at p < 0.05, and highly significant at p < 0.001.
This study was reviewed and approved by the Laboratory Animal Management and Ethics Committee of the South China Sea Institute of Oceanology, Chinese Academy of Sciences.
Results
Substrate Composition and Correlation With the Abundance/Biomass of C. striatus
Apart from being higher in coral cover, the reefs categorized as HC had low algal turf coverage and a considerable amount of CCA compared with other reefs (Figure 2A and Table 1). Substrates in the IC reefs were mostly covered by SATs (Figure 2D) and had a low coverage of macroalgae (Figure 2B). In the LC reefs, LATs were the dominant group with a coverage of up to 55% (Figure 2C and Table 1) and containing a large amount of sediment (Figure 2E).
Figure 2. Benthic coral reef conditions of the different study reefs. The 0.25 m2 quadrats were divided into a 4 × 4 grid using strings at 12.5 cm intervals along the vertical and horizontal axes. (A) Coral-dominated reefs with a complex benthic structure and high coral coverage; (B) Reefs with low macroalgae cover had a large area and flat, hard substratum. (C) Reefs with the lowest coral cover and highest coverage of dense algal turfs and macroalgae; (D) The “dead hard substratum” was primarily covered by short (length was approximately 2 mm) and sparse algal turfs (SATs). (E) Long (length was more than 5 mm) and dense algal turfs (LATs) laden with large amounts of sediments dominated in low coral cover reefs.
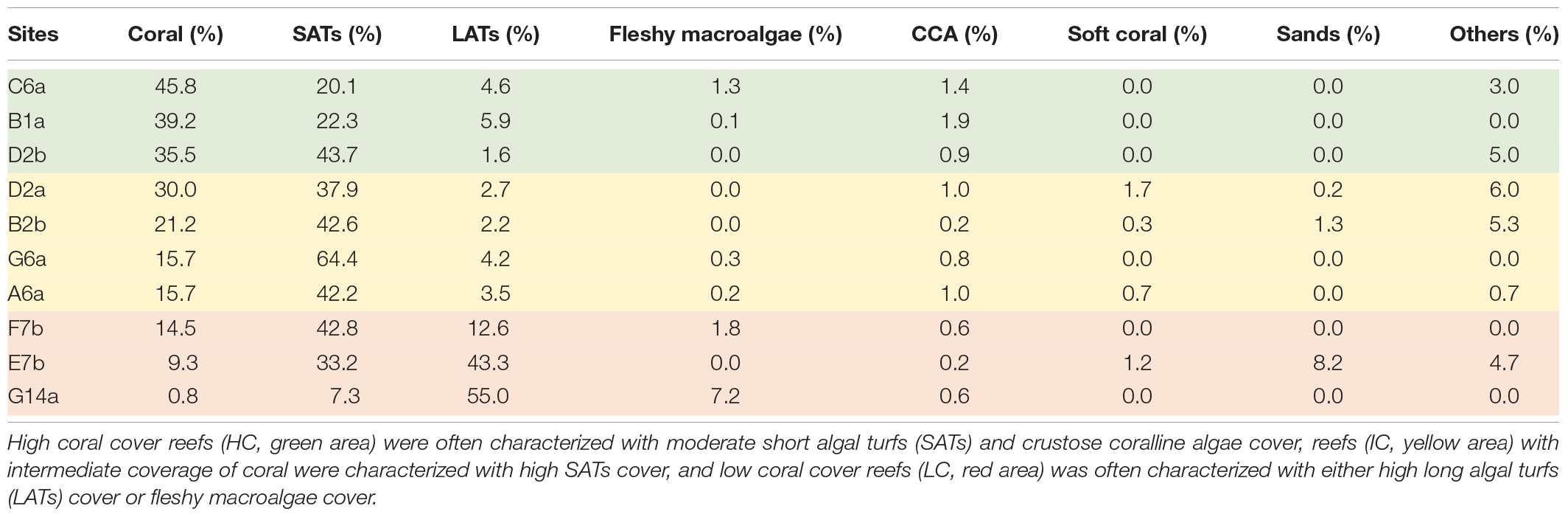
Table 1. Benthic community composition of the study sites revealed different habitat characteristics among the reefs.
The highest individual density of C. striatus (44.7 individuals 100 m–2) was found at site D2a, while the highest biomass of 365.1 kg ha–1 was found at site E7b (Table 2). Both individual density and biomass of C. striatus tended to increase with moderate coral reduction (Figures 3A,B). There was no significant correlation (p = 0.56) between individual density of C. striatus and the percentage ratio of SAT cover (Figure 3C), however, there was a significant positive correlation between the biomass of C. striatus and SAT coverage (p < 0.05, Figure 3D). There was no significant correlation between the abundance of C. striatus and LAT cover (Figures 3E,F), as with the other substrate categories (Supplementary Figure 1).
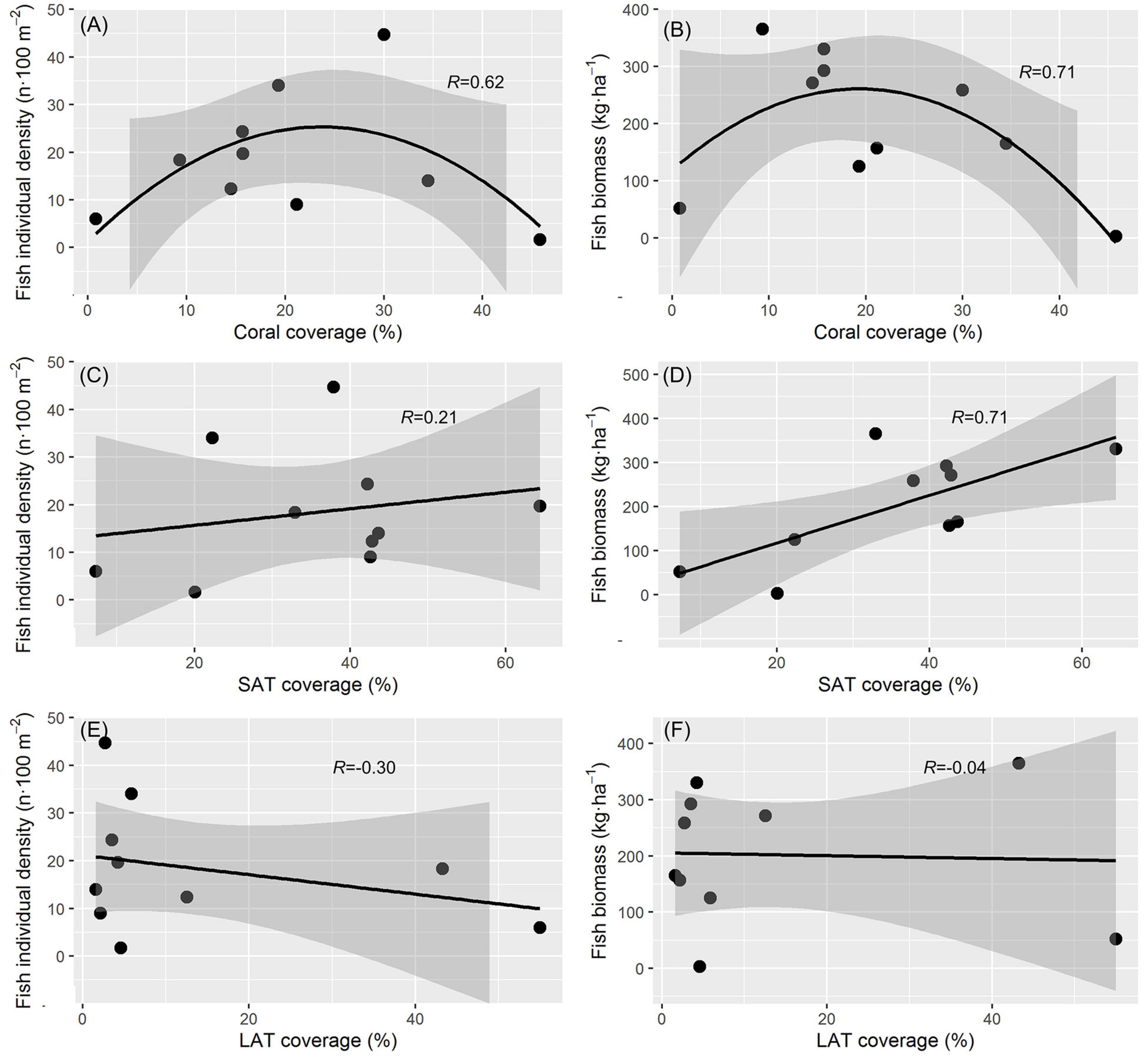
Figure 3. Relationship of Ctenochaetus striatus with benthic conditions. Hard living coral coverage and (A) individual abundance, (p = 0.18) (B) biomass (p = 0.08) of C. striatus. The linear relationship between short algal turfs (SATs) coverage and (C) individual abundance (p = 0.56) and (D) biomass of C. striatus (p = 0.02). The linear relationship between SATs coverage and (E) the individual abundance (p = 0.40) and (F) biomass of C. striatus (p = 0.91).
Species Detected in the Stomach Contents of C. striatus
Most visual matter consisted of sand, pieces of algae, and unidentified organic matter, which provided limited information about the feeding preferences of C. striatus (Supplementary Figure 2). In total, 511,684 raw sequences were generated from the stomach contents of C. striatus (Supplementary Table 1). After removing sequences identified as fish and low-frequency OTU sequences, a total of 316,687 effective sequences remained. These sequences belonged to 764 OTUs and 29 biological taxa. Most of the sequences and OTUs were identified at the genus or family level, and 14,708 remained unassigned (“NA”). To reduce the risk of misidentification at the lower taxonomic levels, a proportion of different OTUs was returned for each level, and those labeled “NA” were removed prior to dietary composition analysis. The alpha diversity (Chao1 and OTU numbers) and rarefaction curves generated based on the OTU table showed that the sequencing number covered the range of species diversity (Supplementary Figure 3).
Most of the identified taxa were algae (RRA > 80%, proportion of total number of OTUs > 45.0%), with microalgae (Dinoflagellata) being the most abundant taxa, followed by Rhodophyta, Chlorophyta, and Ochrophyta (Supplementary Figure 4). There were also a few taxa of micro-invertebrates, such as Arthropoda, Platyhelminthes, and Nematoda. Rhodophyta showed the highest diversity, including genus Hypnea and Champia, followed by Dinoflagellata, mainly from orders Suessiales and Prorocentrales, with genera such as Symbiodinium. There was also a high diversity of arthropod species, with no obvious dominant species. Chlorophyta and Ochrophyta had relatively low diversities, but were often abundant. For example, the RRA of brown algae Sphacelaria and Lobophora reached 55.5 and 51.2% at some sites, respectively. Similarly, green algae Neomeris and Parvocaulis also reached 36.3 and 73.4% at some sites, respectively (Figure 4).
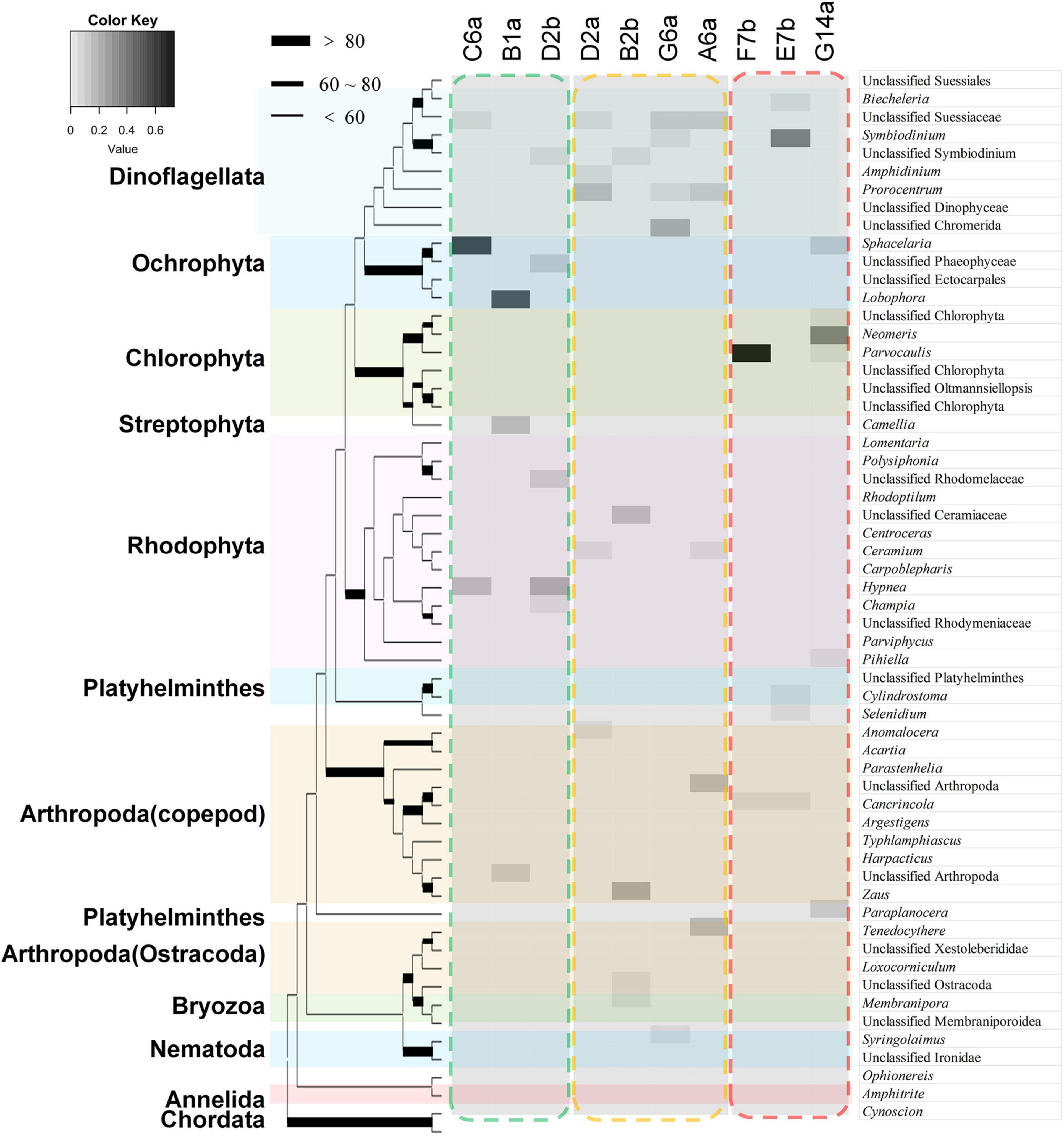
Figure 4. The main species in the stomach contents of Ctenochaetus striatus (as revealed by DNA analysis) at the genus level showed distinct divergences among reefs. The characters above the picture represent the IDs of the sampled sites. The taxonomic species in the green dashed frame was the main species detected in the stomach contents of C. striatus sampled from high coral cover (HC) reefs, the orange dashed frame surrounds the main feeding items on intermediate coral cover (IC) reefs, and the red dashed frame represents the detected species on the samples of low coral cover reefs. Brown and red algal species were more frequently detected in the stomach contents within HC reefs. On IC reefs, the stomach content DNA tended to be more diverse and was mainly comprised of dinoflagellates. Green algae were the most abundant taxa on the reefs with high macroalgae or long algal turfs coverage, except site E7b.
Correlation Between Stomach Contents and Different Substrate Categories
Based on the RRA data of stomach content DNA, the RDA showed that the composition of stomach contents differed significantly according to the different benthic conditions (Figure 5A). The divergence among sites was reflected in a few groups, including dinoflagellates and red, brown, and green algae. Sites with the highest coral cover showed high similarity, with stomach content DNA in these sites dominated by brown and red algae. Stomach content DNA sampled from G14a and F7b sites, which had high macroalgal cover and low coral cover, was dominated by green algae. Dinoflagellates tended to be diverse and dominated the stomach content DNA from sites with higher SAT coverage and intermediate levels of coral (Table 3 and Figure 5A). The Jaccard metric, which is based on data presence/absence, also showed that samples of different reefs separated well in NMDS plots, with IC reefs showing less variance between sites than the other reef groups (Figure 5B). The main species in the stomach content DNA showed distinct divergence among the reefs (Figure 4). Ochrophyta species belonging to genus Sphacelaria and Lobophora, were abundant in the HC reef sites, with RRAs of up to 55.0 and 51.2%, respectively. The dominant species on the reefs with high SATs mostly belonged to Dinoflagellata. The order Suessiales belonging to Dinoflagellata was abundant at all sites; however, some Suessiales were only identified to the family level (RRA ranged from 9.4 to 14.6%), followed by the genus Symbiodinium (RRA 5.1–6.6%). Genus Prorocentrum, belonging to Dinoflagellata, was also abundant (RRA up to 17.6%). Food items belonged to Arthropoda also had high diversity, especially genus Anomalocera. There were different species along different sites on the reefs with high macroalgae or LAT coverage. Chlorophyta species, such as genus Neomeris and Parvocaulis, were dominant in the G14a and F7b sites. However, the dominant taxa at the E7b site were mainly dinoflagellates of the genus Symbiodinium (38.0%).
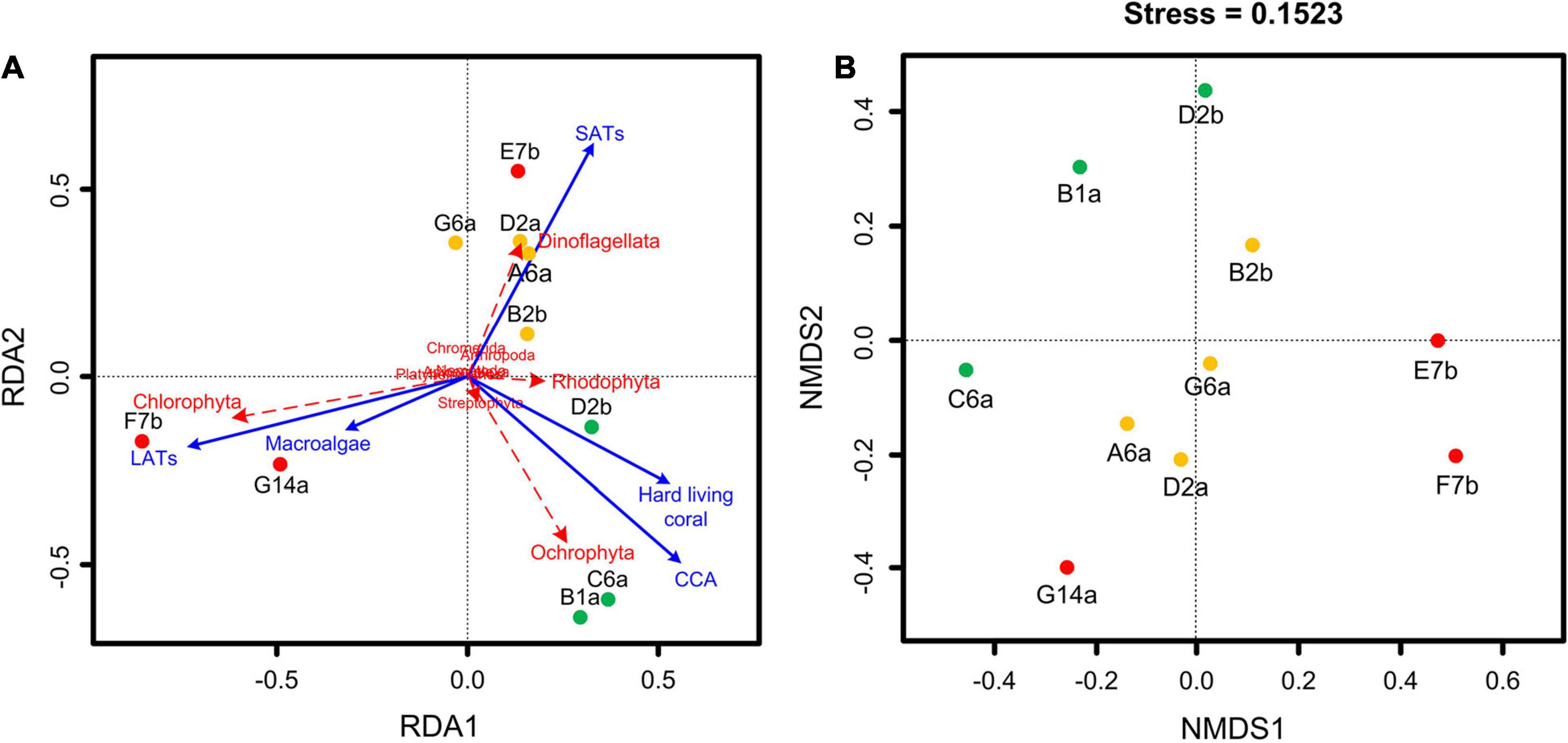
Figure 5. Ordination results of (A) redundancy analysis (RDA) and (B) non-metric multidimensional scaling (NMDS). RDA was based on the relative read abundance of food taxa DNA and different substrate-type coverage. NMDS was based on a presence/absence matrix of stomach content genera. Sites with the same colors represent similar benthic conditions. The green points indicate high coral cover reefs, the red points indicate low coral cover reefs and the orange points represent sites from reefs with intermediate coral cover.
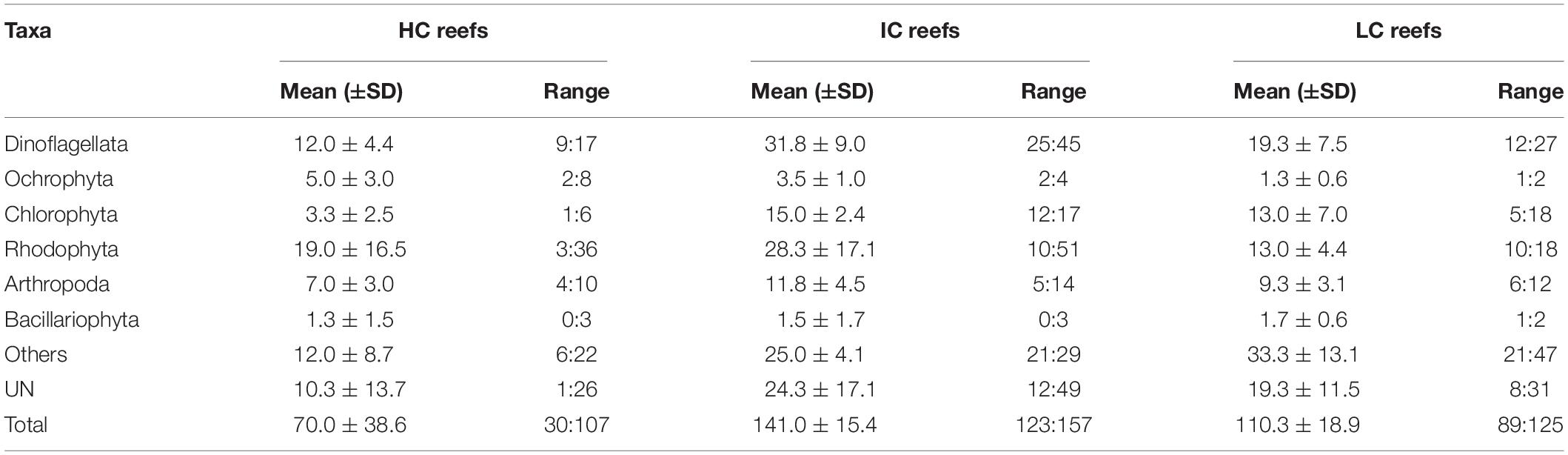
Table 3. Number of operational taxonomic units (OTUs) detected in the stomach contents of Ctenochaetus striatus from different reefs.
Discussion
The High Biomass of C. striatus Related to High SAT Cover
In our study, both the abundance and biomass of C. striatus showed an inverted U-shaped relationship with coral coverage and were highest with a moderate reduction in live hard coral cover (Figures 3A,B). Similarly, Russ et al. (2018) found that the density of C. striatus had a negative relationship with hard coral cover. Where coral cover was very low, LATs tended to be abundant on reefs (Table 1). Although both the abundance and biomass of C. striatus were not directly correlated with LATs, sediments trapped by LATs constrain the area suitable for C. striatus grazing (Tebbett and Bellwood, 2020), with LATs also less nutritious and palatable to C. striatus than SATs (Tebbett et al., 2017c). Indeed, site G14a which had the lowest hard coral coverage, highest LATs coverage and lowest coverage of SATS, had a low density and biomass of C. striatus (Table 1). Russ et al. (2018) also found that the density of C. striatus had a strong positive relationship with the amount of dead substratum. The “dead substratum” on coral reefs was often covered by SATs (Figures 2D,E), and the biomass of C. striatus had a significant correlation with the coverage of SATs (Figure 3D). However, the correlation between the density of C. striatus and SATs coverage was slightly but not significantly positive (Figure 3C), which may be due to the limited number of survey sites. Hence, the increase in SAT cover following coral cover reduction may be the actual factor for attracting C. striatus. Although the intensive presence of C. striatus with sparse algal turfs may have been found on other trophic reefs of the world (Russ et al., 2018), these results are mainly based on the abundance of C. striatus, and biomass data are often deficient. Our results indicated that the biomass of C. striatus may be a more sensitive indicator of changes than abundance, when changes in resources are involved.
Possible Dietary Resources for C. striatus
A previous study revealed that the diets of C. striatus consisted of plant cellular fragments, meiofaunal elements, very fine sediment grains, and relatively more unidentified material (Choat et al., 2002). Instead of merely classifying these unidentified contents as organic matter or algal debris, by using HTS multiple eukaryotic organisms were identified at the family or genus level in the stomach contents of C. striatus. Ctenochaetus striatus was confirmed to have a complex dietary intake comprised of arthropods, dinoflagellates, and red, brown, and green algae. Although DNA from these organisms was detected in stomach contents, this does not imply that they were consumed directly, at least not in a living state. Marshell and Mumby (2012) found that C. striatus consistently fed more intensively on SATs and that their removal from artificial substrates was more effective than removal by another surgeonfish. However, this removal effect was limited to natural reefs, and C. striatus was observed to consume only detritus and sediments, leaving mature algal turfs relatively intact (Purcell and Bellwood, 1993; Tebbett et al., 2017c). Although the RRA of macroalgae/filamentous algae in the diets of C. striatus was high, with an average ratio of 46.71% per site (Supplementary Figure 4 and Supplementary Table 2; Guiry and Guiry, 2020), the special bristle-like teeth of C. striatus would be more suitable for “brushing” organic matter and debris from algal turfs instead of directly feeding on mature algae (Purcell and Bellwood, 1993; Tebbett et al., 2017c). This “brushing” feeding process over algal turfs has the possibility for C. striatus to ingest debris or early settlers of algae nevertheless. McMahon et al. (2016) found that macroalgae provide an average of 14 ± 7% (up to 30%) of the C consumed and assimilated by C. striatus. Therefore, the DNA of macroalgae/filamentous algae detected in the present study is also likely to be from debris or early successional communities of these algae and consumed by C. striatus.
A considerable number of microalgae sequences were found in the stomach contents of C. striatus, and some were identified for the first time in this study. These consisted of mainly dinoflagellates (RRA 2.3–53.8%), for example, Symbiodinium and Prorocentrum. Most published studies generally agree that C. striatus targets detritus in algal turfs and possibly includes microalgae (Polunin and Klumpp, 1989; Choat et al., 2002; Wilson et al., 2003; Clements et al., 2017; Tebbett et al., 2017c). However, there is less information about microalgae species that can be drawn from previous studies. Microscopic observations have not revealed significant amounts of microalgae previously (Choat et al., 2002), even though diatoms and dinoflagellates are generally common in the marine environment and are easily recognizable (Hatcher, 1988; Luo et al., 2017). It is possible that the digestion process may have destroyed the cell structures of microalgae (Choat et al., 2004). Additionally, C. striatus is considered to be a high risk species for contracting ciguatera poisoning (Mak et al., 2013; Rongo and van Woesik, 2013) and a primary vector of water-soluble toxins produced by benthic dinoflagellates (Yasumoto et al., 1976; Lewis, 2006), which indicates that C. striatus can intake and ingest microalgae.
The dominant microalgae genera detected in this study were mainly from the order Suessiales and the genus Prorocentrum (Figure 4). The representative genera were Symbiodinium, a common symbiotic dinoflagellate in coral reef ecosystems (Hatcher, 1988), and Prorocentrum, which are also common dinoflagellates in the sand, corals, and macroalgae in the South China Sea (Luo et al., 2017). Prorocentrum concavum and P. lima often coexists with filamentous algae and may occasionally be consumed by surgeonfish (Kohler and Kohler, 1992), and coral mucus is an important source of detritus on coral reefs (Wilson et al., 2003), which might explain the prevalence of Symbiodinium in the diets of C. striatus. However, whether Symbiodinium is derived from coral mucus or is naturally associated with algal turfs needs to be confirmed. Diatoms account for 1–14% of the organic matter in detrital aggregates in algal turfs (Wilson et al., 2003), but diatom sequences detected in this study were low (even with RRA < 1% in some sites). Considerable diatom species and sequences were detected in our previous study, in which DNA extraction methods targeting phytoplankton and samples were preserved in alcohol without freezing (Lin et al., 2018a). Therefore, the underrepresentation of RRA counts on diatoms was likely a result of DNA extraction bias or the differences in the recovery of DNA from different organisms from frozen specimens (Maki et al., 2017).
Micro-invertebrates, such as crustaceans and nematodes, found in the stomach contents of C. striatus (Figure 4), may be incidentally consumed during feeding. Previous studies have found that when fish remove benthic algae from reef bases, they may also consume a significant number of benthic micro-invertebrates (Polunin et al., 1995; Trip et al., 2008; Kramer et al., 2013). Many broadly categorized herbivorous fishes (including detritivorous fish) on coral reefs are functionally distinct from traditional herbivores in nutritional terms, and it is possible for these fishes to ingest mixtures of different food items to achieve adequate nutrient intake (Clements et al., 2009). Considering the wide distribution of micro-invertebrates and nutritional support for reef fishes, the contribution of micro-invertebrates to higher trophic levels may be an important topic for future research (Kramer et al., 2012).
The Positive Feeding Response of C. striatus to Disturbed Habitat
In our results, the stomach content of C. striatus was often associated with benthic conditions (Figures 4, 5). Previous studies have shown that feeding of C. striatus was less influenced by substratum organic loads, and more dependent on the availability of organic sources on the substratum (Tebbett et al., 2017a). This implied that the DNA of detected food species can possibly reflect the availability of algal resources on coral reefs. Indeed, the high RRA of macroalgae and filamentous algae often correlated with the high coverage of macroalgae or LATs on LC reefs (Figure 5). Meanwhile, with the protection of scleractinian corals, the biomass of brown algae Lobophora was often 20 times greater in branching than in planar habitats (Brandl and Bellwood, 2016), and, thus, they increase the chances that C. striatus may consume algal debris derived from “shelters” in branching habitat (HC reefs). This could explain the high RRA of Lobophora and other brown algae in the diets of C. striatus on HC reefs. Microalgae and other microbes add essential nutrients and improve the nutritional value of the detritus, and increase the palatability and digestibility of detritus for fish (Wilson et al., 2003; Tebbett et al., 2017c). Dinoflagellates in the stomach contents DNA were abundant and diverse with high SATs cover, which had a higher detrital proportion. However, RRA cannot yield results in terms of biomass without adequate preliminary work (Deagle et al., 2019). For this reason, semi-quantitative data based on 18S rDNA metabarcoding were demonstrated in our previous study (Lin et al., 2018a,b). Meanwhile, the occurrence data of food items also proved that the ingested dinoflagellate species correlated with high SAT cover reefs (Figures 4, 5B). Even with some problems in converting biomass from RRA data directly, the increasing or decreasing tendency in the RRA of specific taxa could still provide useful information on the influence of the substratum on the feeding of C. striatus, especially for the positive feeding response on microalgae on the high SAT cover reefs.
Benthic algae did not appear on the reef as mature plants, and the forces acting on the early life stages of algae may determine the ultimate development and dominance patterns of algal blooms (Lotze et al., 2000). Thus, the removal of early algal settlers and microalgae from substrata by C. striatus would also exert a top-down effect on benthic algae and potentially influence the primary succession of algal communities (Figure 6), especially considering the high biomass and intensive feeding on microalgae by C. striatus on the high SAT cover reefs after moderate coral loss. Indeed, previous studies have found that dinoflagellates, for example, Prorocentrum, often coexist with filamentous algae growing on dead coral at the early stage of coral degradation and are ingested by surgeonfish (Kohler and Kohler, 1992). In addition, this positive feeding response on microalgae may result in secondary disasters after coral degeneration, such as ciguatera poisoning (Kohler and Kohler, 1992). Rongo and van Woesik (2013) confirmed that the density increase of C. striatus was a consequence of increased algal cover after coral loss and directly coincided with increased hospital cases of ciguatera poisoning. These studies were not only consistent with the correlation between the presentation of C. striatus and SAT cover (Figure 3D), but the increasing ciguatera poisoning could also be explained by the increasing chances of dinoflagellates ingested by C. striatus on reefs with moderate coral cover (Figures 4, 5A). Even though microalgae are important food resources for consumers in other marine ecosystems (Ding et al., 2018), they are of less concern in coral reef ecosystems (Clements et al., 2017). Technically, the exact biomass or abundance of dinoflagellates ingested by C. striatus could not be converted from RRA data in this study, for example, due to the differences between dinoflagellates and other algae in copy number. The abundant sequences of dinoflagellates derived from HTS could draw more attention to this cryptic species and shed some light on the early algal succession and algal toxin transfer on coral reefs. The expansion of algal turf after coral degradation would actually promote the ecological status of microalgae in detritivory by affecting the feeding habit of C. striatus (Figure 6). This information would be helpful for risk assessment and prediction in the coral-algae phase shift.
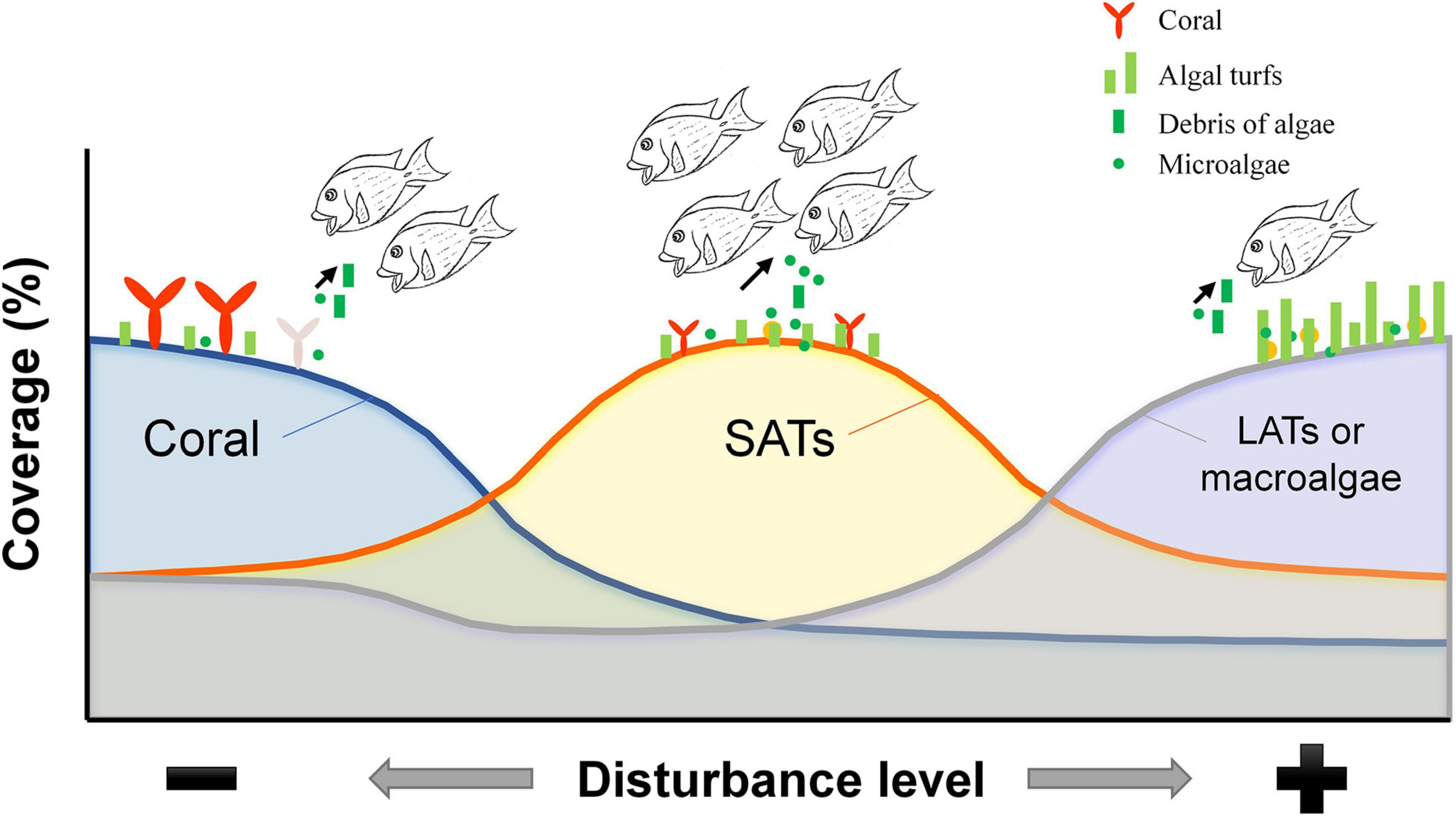
Figure 6. Schematic diagram showing the influence of the substratum on the feeding of C. striatus during algal succession. The DNA sequences of macroalgae and filamentous algae were more frequently detected in the diets of C. striatus sampled from either coral-dominated reefs or macroalgae-dominated reefs, where the biomass of C. striatus was relatively lower. Reefs with intermediate levels of coral cover have more areas of substrate exposed which are covered by short algal turfs, which C. striatus preferentially feeds on. This can be seen from the high biomass of C. striatus and the high diversity and relative read abundance of dinoflagellates detected in the stomach contents of C. striatus on these reefs.
Instead of simply classifying the contents into organic matter or algal debris, multiple eukaryotic organisms were firstly identified to the family or genus level in the diets of C. striatus using HTS. Regrettably, a weak quantitative relationship may exist between the biomass and the sequences produced (Lamb et al., 2019), and the exact proportion of different dietary taxa could not be precisely determined based on current data. A series of factors, such as DNA extraction, primer bias, and differences in dietary taxa in copy number, could influence the accuracy of RRA data. Despite some flaws and methodological problems, current data could still resolve the previous assumptions: (a) C. striatus could ingest early successional algae, such as microalgae, which may be an important process on coral reefs, and (b) C. striatus could also be influenced by benthic conditions, as its biomass was significantly positively correlated with SAT cover and its stomach contents were often associated with benthic cover (Figures 5, 6). Although massive coral loss may lead to a decline in both the diversity and biomass of reef fishes, moderate coral loss often has limited or even positive effects on generalist-feeding species (Pratchett et al., 2011), such as detritivorous reef fish. Considering the expansion of algal turfs coverage and the wide distribution and high abundance of detritivorous fish on trophic reefs, the increase in both algal turfs and detritivores will potentially change the basic energy flow after coral degradation, and HTS may provide new insight into the trophic roles of this important functional group. Ultimately, no method can possibly resolve all the problems in dietary analysis. Nevertheless, HTS can provide a comprehensive eukaryotic species list of potential dietary resources, which provides useful quantified dietary analysis of this major detritivorous reef fish for future research, especially in the collection of stable isotope characteristics of fish diets.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MN188827–MN189943.
Ethics Statement
The animal study was reviewed and approved by Laboratory Animal Management and Ethics Committee, South China Sea Institute of Oceanology, Chinese Academy of Sciences.
Author Contributions
SL conceived the study. XL and SH designed the study and experimental protocols. XL performed the experiments, data analyses, and wrote the manuscript. HH contributed the habitat characterization and fish abundance data. YL, SL, SH, and LZ contributed significantly to the improvement of the manuscript and reviewed the final draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant Number XDA13020102), National Natural Science Foundation of China (Grant Number 41806188), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0404), Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences (Grant Number ISEE2018PY03), Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, Ministry of Agriculture and Rural Affairs, China (Grant Number FREU2019-03), Science and Technology Planning Project of Guangdong Province, China (Grant Number 2020B1212060058), and K. C. Wong Education Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Huang Yunlin and Zheng Dacai for their assistance with fish sampling, and Zhou Tiancheng and Zhang Chen, students from the University of Chinese Academy of Sciences, Beijing, China, for their dedication and assistance with the fish dissections. We also thank Liu Guang and Tian Yuan for their assistance with the data analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.682697/full#supplementary-material
Footnotes
References
Bellwood, D. R., and Fulton, C. J. (2008). Sediment-mediated suppression of herbivory on coral reefs: Decreasing resilience to rising sea levels and climate change? Limnol. Oceanography 53, 2695–2701. doi: 10.4319/lo.2008.53.6.2695
Bellwood, D. R., Hoey, A. S., Ackerman, J. L., and Depczynski, M. (2006). Coral bleaching, reef fish community phase shifts and the resilience of coral reefs. Glob. Change Biol. 12, 1587–1594. doi: 10.1111/j.1365-2486.2006.01204.x
Bellwood, D. R., Streit, R. P., Brandl, S. J., and Tebbett, S. B. (2019). The meaning of the term ‘function’ in ecology: A coral reef perspective. Funct. Ecol. 33, 948–961. doi: 10.1111/1365-2435.13265
Brandl, S. J., and Bellwood, D. R. (2014). Individual-based analyses reveal limited functional overlap in a coral reef fish community. J. Anim. Ecol. 83, 661–670. doi: 10.1111/1365-2656.12171
Brandl, S. J., and Bellwood, D. R. (2016). Microtopographic refuges shape consumer-producer dynamics by mediating consumer functional diversity. Oecologia 182, 203–217. doi: 10.1007/s00442-016-3643-0
Brandl, S. J., Casey, J. M., and Meyer, C. P. (2020). Dietary and habitat niche partitioning in congeneric cryptobenthic reef fish species. Coral Reefs. 39, 305–317. doi: 10.1007/s00338-020-01892-z
Cheal, A. J., MacNeil, M. A., Cripps, E., Emslie, M. J., Jonker, M., Schaffelke, B., et al. (2010). Coral-macroalgal phase shifts or reef resilience: links with diversity and functional roles of herbivorous fishes on the Great Barrier Reef. Coral Reefs 29, 1005–1015. doi: 10.1007/s00338-010-0661-y
Cheal, A., Emslie, M., Miller, I., and Sweatman, H. (2012). The distribution of herbivorous fishes on the Great Barrier Reef. Mar. Biol. 159, 1143–1154. doi: 10.1007/s00227-012-1893-x
Choat, J. H., Clements, K. D., and Robbins, W. D. (2002). The trophic status of herbivorous fishes on coral reefs - I: Dietary analyses. Mar. Biol. 140, 613–623. doi: 10.1007/s00227-001-0715-3
Choat, J. H., Robbins, W. D., and Clements, K. D. (2004). The trophic status of herbivorous fishes on coral reefs - II. Food processing modes and trophodynamics. Mar. Biol. 145, 445–454. doi: 10.1007/s00227-004-1341-7
Clements, K. D., German, D. P., Piche, J., Tribollet, A., and Choat, J. H. (2017). Integrating ecological roles and trophic diversification on coral reefs: multiple lines of evidence identify parrotfishes as microphages. Biolog. J. Linnean Soc. 120, 729–751. doi: 10.1111/bij.12914
Clements, K. D., Raubenheimer, D., and Choat, J. H. (2009). Nutritional ecology of marine herbivorous fishes: ten years on. Funct. Ecol. 23, 79–92. doi: 10.1111/j.1365-2435.2008.01524.x
Connell, S. D., Foster, M. S., and Airoldi, L. (2014). What are algal turfs? Towards a better description of turfs. Mar. Ecol. Prog. Series 495, 299–307. doi: 10.3354/meps10513
Corse, E., Costedoat, C., Chappaz, R., Pech, N., Martin, J. F., and Gilles, A. (2010). A PCR-based method for diet analysis in freshwater organisms using 18S rDNA barcoding on faeces. Mole. Ecol. Res 10, 96–108. doi: 10.1111/j.1755-0998.2009.02795.x
Deagle, B. E., Thomas, A. C., McInnes, J. C., Clarke, L. J., Vesterinen, E. J., and Clare, E. L. (2019). Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Mole. Ecol. 28, 391–406. doi: 10.1111/mec.14734
Ding, M. W., Wang, Z. K., and Dong, Y. W. (2018). Food availability on the shore: Linking epilithic and planktonic microalgae to the food ingested by two intertidal gastropods. Mar. Env. Res. 136, 71–77. doi: 10.1016/j.marenvres.2018.02.005
Froese, R., and Pauly, D. (2020). FishBase. World Wide Web electronic publication. Available online at: www.fishbase.org, version (accessed date december 2020).
Goatley, C. H. R., and Bellwood, D. R. (2010). Biologically mediated sediment fluxes on coral reefs: sediment removal and off-reef transportation by the surgeonfish Ctenochaetus striatus. Mar. Ecol. Prog. Series 415, 237–245. doi: 10.3354/meps08761
Goatley, C. H. R., and Bellwood, D. R. (2011). The Roles of Dimensionality, Canopies and Complexity in Ecosystem Monitoring. PLoS One 6:e27307. doi: 10.1371/journal.pone.0027307
Goatley, C. H. R., Bonaldo, R. M., Fox, R. J., and Bellwood, D. R. (2016). Sediments and herbivory as sensitive indicators of coral reef degradation. Ecol. Soc. 21:29. doi: 10.5751/ES-08334-210129
Guiry, M. D., and Guiry, G. M. (2020). AlgaeBase. Available online at: http://www.algaebase.org, version (12/2020).
Harrell, F. E. (2019). Hmisc: Harrell Miscellaneous. R package version 4.2-0. Available online at: https://CRAN.R-project.org/package=Hmisc (accessed date 2-February-2021).
Hatcher, B. G. (1988). Coral-reef primary productivity: a beggars banquet. Trends Ecol. Evol. 3, 106–111. doi: 10.1016/0169-5347(88)90117-6
Huang, H., Zhang, C. L., Yang, J. H., You, F., Lian, J. S., and Tan, Y. H. (2012). Scleractinian coral community characteristics in Zhubi reef sea area of Nansha Islands. J. Oceanogr. Taiwan Strait 31, 79–84.
Kelly, E. L. A., Eynaud, Y., Clements, S. M., Gleason, M., Sparks, R. T., Williams, I. D., et al. (2016). Investigating functional redundancy versus complementarity in Hawaiian herbivorous coral reef fishes. Oecologia 182, 1151–1163. doi: 10.1007/s00442-016-3724-0
Kohler, K. E., and Gill, S. M. (2006). Coral Point Count with Excel extensions (CPCe): A Visual Basic program for the determination of coral and substrate coverage using random point count methodology. Computers Geosci. 32, 1259–1269. doi: 10.1016/j.cageo.2005.11.009
Kohler, S. T., and Kohler, C. C. (1992). Dead bleached coral provides new surfaces for dinoflagellates implicated in ciguatera fish poisonings. Env. Biol. Fishes 35, 413–416. doi: 10.1007/Bf00004993
Kramer, M. J., Bellwood, D. R., and Bellwood, O. (2012). Cryptofauna of the epilithic algal matrix on an inshore coral reef, Great Barrier Reef. Coral Reefs 31, 1007–1015. doi: 10.1007/s00338-012-0924-x
Kramer, M. J., Bellwood, D. R., and Bellwood, O. (2014). Large-scale spatial variation in epilithic algal matrix cryptofaunal assemblages on the Great Barrier Reef. Mar. Biol. 161, 2183–2190. doi: 10.1007/s00227-014-2495-6
Kramer, M. J., Bellwood, O., and Bellwood, D. R. (2013). The trophic importance of algal turfs for coral reef fishes: the crustacean link. Coral Reefs 32, 575–583. doi: 10.1007/s00338-013-1009-1
Lamb, P. D., Hunter, E., Pinnegar, J. K., Creer, S., Davies, R. G., and Taylor, M. I. (2019). How quantitative is metabarcoding: A meta-analytical approach. Mole. Ecol. 28, 420–430. doi: 10.1111/mec.14920
Leal, M. C., and Ferrier-Pages, C. (2016). Molecular trophic markers in marine food webs and their potential use for coral ecology. Mar. Genom. 29, 1–7. doi: 10.1016/j.margen.2016.02.003
Lewis, R. J. (2006). Ciguatera: Australian perspectives on a global problem. Toxicon 48, 799–809. doi: 10.1016/j.toxicon.2006.07.019
Lin, X. Z., Hu, S. M., Liu, S., and Huang, H. (2018a). Unexpected prey of juvenile spotted scat (Scatophagus argus) near a wharf: The prevalence of fouling organisms in stomach contents. Ecol. Evol. 8, 8547–8554. doi: 10.1002/ece3.4380
Lin, X. Z., Hu, S. M., Liu, S., and Huang, H. (2018b). Comparison between traditional sequencing and high-throughput sequencing on the dietary analysis of juvenile fish. Chinese J. Appl. Ecol. 29, 3093–3101. doi: 10.13287/j.1001-9332.201809.005
Lotze, H. K., Worm, B., and Sommer, U. (2000). Propagule banks, herbivory and nutrient supply control population development and dominance patterns in macroalgal blooms. Oikos 89, 46–58. doi: 10.1034/j.1600-0706.2000.890106.x
Luo, Z. H., Hua, Z., Krock, B., Lu, S. H., Yang, W. D., and Gu, H. F. (2017). Morphology, molecular phylogeny and okadaic acid production of epibenthic Prorocentrum (Dinophyceae) species from the northern South China Sea. Algal Res. Biomass Biofuels Bioprod. 22, 14–30. doi: 10.1016/j.algal.2016.11.020
Mak, Y. L., Wai, T. C., Murphy, M. B., Chan, W. H., Wu, J. J., Lam, J. C. W., et al. (2013). Pacific Ciguatoxins in Food Web Components of Coral Reef Systems in the Republic of Kiribati. Environ. Sci. Technol. 47, 14070–14079. doi: 10.1021/es403175d
Maki, A., Salmi, P., Mikkonen, A., Kremp, A., and Tiirola, M. (2017). Sample preservation, DNA or RNA extraction and data analysis for high-throughput phytoplankton community sequencing. Front. Microb. 8:1848. doi: 10.3389/Fmicb.2017.01848
Marshell, A., and Mumby, P. J. (2012). Revisiting the functional roles of the surgeonfish Acanthurus nigrofuscus and Ctenochaetus striatus. Coral Reefs 31, 1093–1101. doi: 10.1007/s00338-012-0931-y
Marshell, A., and Mumby, P. J. (2015). The role of surgeonfish (Acanthuridae) in maintaining algal turf biomass on coral reefs. J. Exp. Mar. Biol. Ecol. 473, 152–160. doi: 10.1016/j.jembe.2015.09.002
Max, L. M., Hamilton, S. L., Gaines, S. D., and Warner, R. R. (2013). Benthic processes and overlying fish assemblages drive the composition of benthic detritus on a central Pacific coral reef. Mar. Ecol. Prog. Series. 482, 181–195. doi: 10.3354/meps10259
McMahon, K. W., Thorrold, S. R., Houghton, L. A., and Berumen, M. L. (2016). Tracing carbon flow through coral reef food webs using a compound-specific stable isotope approach. Oecologia 180, 809–821. doi: 10.1007/s00442-015-3475-3
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B., Simpson, G. L., Solymos, P., et al. (2009). Vegan: community ecology package. R package version 1.15−3.
O’Rorke, R., Lavery, S., and Jeffs, A. (2012). PCR enrichment techniques to identify the diet of predators. Mole. Ecol. Resour. 12, 5–17. doi: 10.1111/j.1755-0998.2011.03091.x
Polunin, N. V. C., and Klumpp, D. W. (1989). Ecological correlates of foraging periodicity in herbivorous reef fishes of the Coral Sea. J. Exp.Mar.Biol. Ecol. 126, 1–20. doi: 10.1016/0022-0981(89)90121-4
Polunin, N. V. C., Harmelinvivien, M., and Galzin, R. (1995). Contrasts in algal food-processing among 5 herbivorous coral-reef fishes. J. Fish Biol. 47, 455–465. doi: 10.1006/jfbi.1995.0151
Pompanon, F., Deagle, B. E., Symondson, W. O. C., Brown, D. S., Jarman, S. N., and Taberlet, P. (2012). Who is eating what: diet assessment using next generation sequencing. Mole. Ecol. 21, 1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x
Pratchett, M. S., Hoey, A. S., Wilson, S. K., Messmer, V., and Graham, N. A. J. (2011). Changes in biodiversity and functioning of reef fish assemblages following coral bleaching and coral loss. Diversity 3:424. doi: 10.3390/d3030424
Purcell, S. W., and Bellwood, D. R. (1993). A functional-analysis of food procurement in 2 surgeonfish species, Acanthurus nigrofuscus and Ctenochaetus striatus (Acanthuridae). Env. Biol. Fish. 37, 139–159. doi: 10.1007/Bf00000589
R Core Team. (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna: R Core Team. doi: 10.1007/bf00000589
Rongo, T., and van Woesik, R. (2013). The effects of natural disturbances, reef state, and herbivorous fish densities on ciguatera poisoning in Rarotonga, southern Cook Islands. Toxicon 64, 87–95. doi: 10.1016/j.toxicon.2012.12.018
Russ, G. R., Payne, C. S., Bergseth, B. J., Rizzari, J. R., Abesamis, R. A., and Alcala, A. C. (2018). Decadal-scale response of detritivorous surgeonfishes (family Acanthuridae) to no-take marine reserve protection and changes in benthic habitat. J. Fish Biol. 93, 887–900. doi: 10.1111/jfb.13809
Russ, G. R., Questel, S. L. A., Rizzari, J. R., and Alcala, A. C. (2015). The parrotfish-coral relationship: refuting the ubiquity of a prevailing paradigm. Mar. Biol. 162, 2029–2045. doi: 10.1007/s00227-015-2728-3
Spear, K. E., Duran, A., Miller, M. W., and Burkepile, D. E. (2019). Sediment associated with algal turfs inhibits the settlement of two endangered coral species. Mar. Pollut. Bull. 144, 189–195. doi: 10.1016/j.marpolbul.2019.04.066
Stoeck, T., Bass, D., Nebel, M., Christen, R., Jones, M. D. M., Breiner, H. W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mole. Ecol. 19, (Suppl. 1), 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Tebbett, S. B., and Bellwood, D. R. (2019). Algal turf sediments on coral reefs: what’s known and what’s next. Mar. Pollut. Bull. 149:110542. doi: 10.1016/j.marpolbul.2019.110542
Tebbett, S. B., and Bellwood, D. R. (2020). Sediments ratchet-down coral reef algal turf productivity. Sci. Total Env. 713:136709. doi: 10.1016/j.scitotenv.2020.136709
Tebbett, S. B., Goatley, C. H. R., and Bellwood, D. R. (2017a). The effects of algal turf sediments and organic loads on feeding by coral reef surgeonfishes. PLoS One 12:169479. doi: 10.1371/journal.pone.0169479
Tebbett, S. B., Goatley, C. H. R., and Bellwood, D. R. (2017b). Fine sediments suppress detritivory on coral reefs. Mar. Pollut. Bull. 114, 934–940. doi: 10.1016/j.marpolbul.2016.11.016
Tebbett, S. B., Goatley, C. H. R., and Bellwood, D. R. (2017c). Clarifying functional roles: algal removal by the surgeonfishes Ctenochaetus striatus and Acanthurus nigrofuscus. Coral Reefs 36, 803–813. doi: 10.1007/s00338-017-1571-z
Tebbett, S. B., Goatley, C. H. R., Streit, R. P., and Bellwood, D. R. (2020). Algal turf sediments limit the spatial extent of function delivery on coral reefs. Sci. Total Env. 734:139422. doi: 10.1016/j.scitotenv.2020.139422
Tootell, J. S., and Steele, M. A. (2016). Distribution, behavior, and condition of herbivorous fishes on coral reefs track algal resources. Oecologia 181, 13–24. doi: 10.1007/s00442-015-3418-z
Trip, E. L., Choat, J. H., Wilson, D. T., and Robertson, D. R. (2008). Inter-oceanic analysis of demographic variation in a widely distributed Indo-Pacific coral reef fish. Mar. Ecol. Prog. Series 373, 97–109. doi: 10.3354/meps07755
Walters, T. L., Lamboley, L. M., Lopez-Figueroa, N. B., Rodriguez-Santiago, A. E., Gibson, D. M., and Frischer, M. E. (2019). Diet and trophic interactions of a circumglobally significant gelatinous marine zooplankter, Dolioletta gegenbauri (Uljanin, 1884). Mole. Ecol. 28, 176–189. doi: 10.1111/mec.14926
Williams, G. J., Smith, J. E., Conklin, E. J., Gove, J. M., Sala, E., and Sandin, S. A. (2013). Benthic communities at two remote Pacific coral reefs: effects of reef habitat, depth, and wave energy gradients on spatial patterns. PeerJ 1:e81. doi: 10.7717/peerj.81
Wilson, S. K., and Bellwood, D. R. (1997). Cryptic dietary components of territorial damselfishes (Pomacentridae, Labroidei). Mar. Ecol. Prog. Series 153, 299–310. doi: 10.3354/meps153299
Wilson, S. K., Bellwood, D. R., Choat, J. H., and Furnas, M. J. (2003). Detritus in the epilithic algal matrix and its use by coral reef fishes. Oceanogr. Mar. Biol. 41, 279–309.
Wilson, S. K., Graham, N. A. J., Pratchett, M. S., Jones, G. P., and Polunin, N. V. C. (2006). Multiple disturbances and the global degradation of coral reefs: are reef fishes at risk or resilient? Glob. Chang. Biol. 12, 2220–2234. doi: 10.1111/j.1365-2486.2006.01252.x
Yasumoto, T., Bagnis, R., and Jean, P. V. (1976). Toxicity of the surgeonfishes II. Properties of the principal water soluble toxin. Bull. Japan. Soc. Sci. Fish. 42, 359–365. doi: 10.2331/suisan.42.359
Zemke-White, W. L., Choat, J. H., and Clements, K. D. (2002). A re-evaluation of the diel feeding hypothesis for marine herbivorous fishes. Mar. Biol. 141, 571–579. doi: 10.1007/s00227-002-0849-y
Keywords: epilithic algal matrixes, phase shift, habitat degradation, Ctenochaetus striatus, microalgae, high-throughput sequencing
Citation: Lin X, Hu S, Liu Y, Zhang L, Huang H and Liu S (2021) Disturbance-Mediated Changes in Coral Reef Habitat Provoke a Positive Feeding Response in a Major Coral Reef Detritivore, Ctenochaetus striatus. Front. Mar. Sci. 8:682697. doi: 10.3389/fmars.2021.682697
Received: 19 March 2021; Accepted: 28 June 2021;
Published: 20 July 2021.
Edited by:
John Everett Parkinson, University of South Florida, United StatesReviewed by:
Andrew Robert Halford, Pacific Community (SPC), New CaledoniaAlma Paola Rodríguez-Troncoso, University of Guadalajara, Mexico
Copyright © 2021 Lin, Hu, Liu, Zhang, Huang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Liu, c2hsaXVAc2NzaW8uYWMuY24=; Simin Hu, aHVzaW1pbkBzY3Npby5hYy5jbg==
 Xianzhi Lin
Xianzhi Lin Simin Hu
Simin Hu Yong Liu
Yong Liu Li Zhang
Li Zhang Hui Huang
Hui Huang Sheng Liu1,2,3*
Sheng Liu1,2,3*