- 1Muping Coastal Environment Research Station, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 2Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Shandong Marine Resource and Environment Research Institute, Yantai, China
- 5Marine Science Research Institute of Shandong Province, Qingdao, China
Although defensins have been isolated from a variety of metazoan, their role in cellular immunity has not been answered. In the study, we found that the hemocytes of the Manila clams Ruditapes philippinarum release defensin (designated as Rpdef3) in response to Vibrio parahaemolyticus challenge. The antimicrobial Rpdef3 was proved to be involved in the extracellular traps (ETs) that hemocytes released in response to Vibrio challenge. Scanning electron microscopy observation proved the patterns how ETs eliminate invading bacteria. Furthermore, Rpdef3 involved in ETs had broad-spectrum antimicrobial effect on both Gram-negative bacteria and Gram-positive bacteria. ELISA assay revealed that Rpdef3 could bind lipopolysaccharides and peptidoglycan in a dose-dependent manner. As concerned to the antibacterial mechanisms, Rpdef3 can cause bacterial membrane permeabilization, leading to cell death. As a result, Rpdef3 might contribute to the trap and the elimination of invading Vibrio in clam ETs. Taken together, our study suggest that the formation of ETs is a defense mechanism triggered by bacterial stimulation, coupled with antibacterial defensin.
Introduction
Antimicrobial peptides (AMPs) are essential part of the innate immune system in most organisms (Bulet et al., 2004). AMPs could be roughly divided into several groups based on their molecular structures and origins. The best-known ones included defensins, cecropins, crustins, and anti-lipopolysaccharide factors (Tharntada et al., 2008). Defensins are the first line of host innate immunity and eliminate invasive pathogens (Ayabe et al., 2004; Tincu and Taylor, 2004). Usually, they can be activated by interacting with the negatively charged membranes of bacteria, and leaded to the disruption of bacterial membrane and metabolic process (Boman et al., 1993; Park et al., 1998). Moreover, other immune functions of defensin have been also demonstrated in invertebrates, such as inducing inflammation, suppressing inflammatory responses, and modulating immune responses by forming complexes with cellular molecules including proteins, nucleic acids and carbohydrates (Schneider et al., 2005).
Extracellular traps (ETs) are web-like DNA structures released by activated immune cells (Nathan, 2006). This new type of cell death process with antibacterial effect is called ETosis (Guimaraes-Costa et al., 2012). The structures are decorated with antimicrobial proteins, such as histones, antimicrobial proteolytic enzymes, which contributes to trap and kill bacteria extracellularly (Papayannopoulos et al., 2010). In addition, defensins have also been proven to be one of the antibacterial protein components produced by ETs in human (Saitoh et al., 2012; Porto and Stein, 2016). Usually, ETs are released in response to invading pathogens, such as bacteria, viruses, fungi, and parasites (Von Kockritz-Blickwede and Nizet, 2009; Schonrich and Raftery, 2016; Silva et al., 2016). In the process, ETs might first entrap microbes and prevent their spread, and then concentrate antimicrobials to kill microbes (Papayannopoulos and Zychlinsky, 2009; McDonald et al., 2012; Yipp et al., 2012).
Ruditapes philippinarum is a widely distributed species in many countries. The pathogen infection and environmental pollution can cause mass death of Manila clams (Allam et al., 2002). Therefore, the study of the immune response characteristics of clams provides an important basis for disease control in aquaculture. In recent years, ETs have been found effective in combating against a variety of pathogenic bacteria and even the intracellular parasites (Neeli et al., 2009). Moreover, formation of ETs usually accompanies the production of higher levels of antimicrobial molecules that kill invaded pathogens (Hazeldine et al., 2014). Since the first discovery in mammalian neutrophils, ETosis has been found in immune cells of a variety of animals like mice, cats, chickens and fish. However, the presence of ETosis and its roles in the cell-mediated immune responses has not been explored in detail in invertebrates. As far as we know, clear evidence of the antibacterial peptides involved in ETosis have not been reported in Manila clams so far. In this study, a preliminary study was conducted to investigate the roles of ETs, associated with defensin production in response to Vibrio challenge in R. philippinarum. These findings will perhaps provide evidence for a better understanding innate immunity to pathogen invasion in clam.
Materials and Methods
Hemocyte ET Formation
Ruditapes philippinarum with a shell length of 3.0–4.0 cm were collected from an aquaculture farm of Yantai, China and acclimated in the laboratory for a week. The clams were cultivated in tanks of aerated seawater at 20–22°C and fed with a mixture of Phaeodactylum tricornutum and Isochrysis galbana. After acclimation, hemolymph was withdrawn from the pericardial cavity into pre-cooled anticoagulant solution, which was prepared according to Han et al. (2021). The collected hemocytes (1 × 106 cells/mL) were mixed with cytochalasin D (10 μg/mL, Aladdin, China) to inhibit phagocytosis and cultured with Vibrio parahaemolyticus (1 × 103 and 1 × 106 CFU/mL) for 1 h. Then hemocytes were incubated with or without DNase I (1 U/mL, Promega, United States) for 15 min after bacterial challenge. Hemocytes only added with cytochalasin D served as the control group. 1 μM SytoX Green (Invitrogen, United States) was used to detect the DNA structure of formed ETs. After washing with PBS, the images were captured using Leica-DMi8 microscope. Fluorescence intensity was quantified as relative fluorescence units (RFU) by using a fluorescence plate reader (Thermo Fisher, Fluoroskan Ascent, United States) at an excitation wavelength of 485 nm and an emission wavelength of 525 nm.
Co-occurrence of DNA Traps and Rpdef3
Solid-Phase Peptide Synthesis of Rpdef3 and Antibody Preparation
The Rpdef3 (accession no. AFP49946) peptide was synthesized by using an automated solid peptide synthesizer as described previously (Yang et al., 2018). For antibody preparation, 6-weeks-old mice were intraperitoneally injected with Rpdef3 in complete Freund’s adjuvant (Sigma, United States). The method of the immunization of mice and the acquisition of antibody was performed as described previously (Yang et al., 2019b). Western blotting analysis was performed as described previously for the detection of antibody specificity (Yang et al., 2019a).
Detection of DNA Traps and Rpdef3 by Confocal Microscopy
The immunofluorescence staining assay was measured as proposed by Ng et al. (2013) with some modification. In brief, hemocytes (1 × 106 cells/mL) were co-cultured with V. parahaemolyticus (0, 1 × 103 and 1 × 106 CFU/mL) on glass slides at 4°C for 1 h. Then the adherent hemocytes on the slides were gently rinsed and fixed with 4% formaldehyde. Hemocytes were sequentially blocked with bovine serum albumin (5%, BSA) and incubated with anti-Rpdef3 antibody (1:1,000) and Alexa Fluor 488-labeled goat anti-mouse IgG (1:1,000, Beyotime, China). Cells were stained with DAPI to detect DNA. After washing with PBS, the fluorescence images of samples were observed using a confocal microscope (Carl Zeiss, Oberko-chen, Germany).
Antibacterial Activities of ETs and Rpdef3
Antibacterial Activity of ETs
The antibacterial activities of ETs were examined as reported previously with some modification (Zhao et al., 2017). To eliminate the interference of phagocytosis, cytochalasin D (10 μg/mL) was added into hemocytes before the experiment. The hemocytes treated with 1 U/mL DNase I were served as a control. V. parahaemolyticus (1 × 103 CFU/mL) were incubated with hemocytes for 0, 0.5, 1, and 2 h and transferred to 2216E agar plates. The colony counting method was used to count the number of viable bacteria.
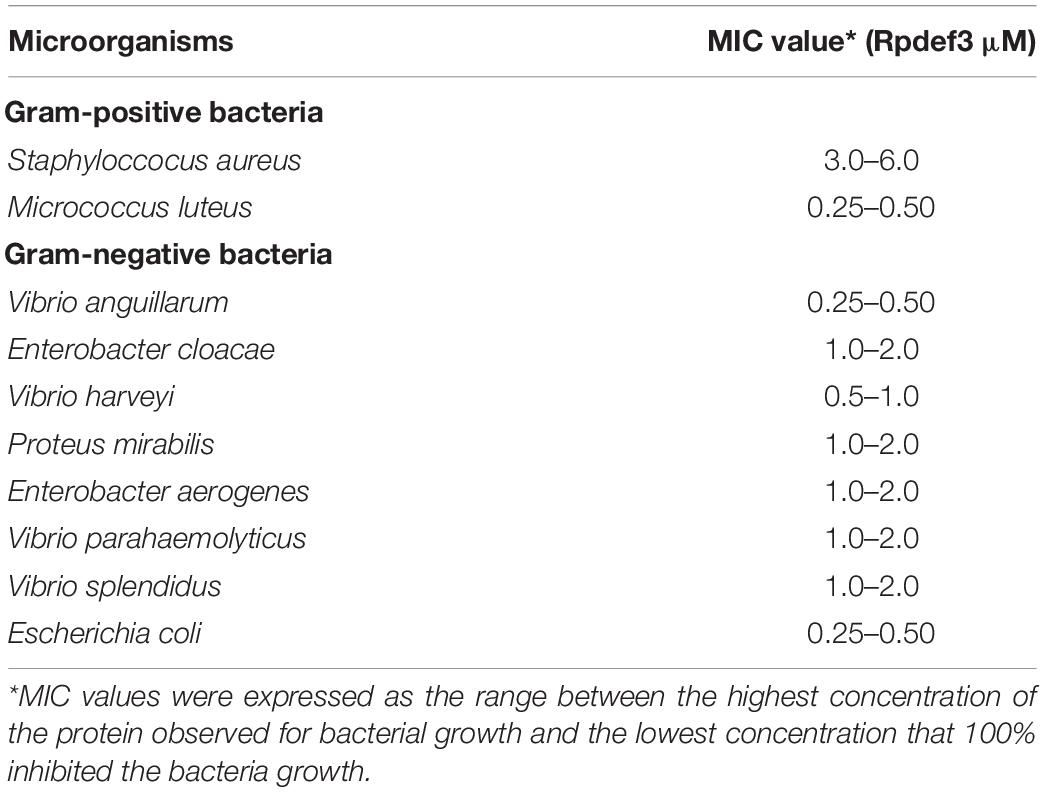
Minimum Inhibitory Concentration
The antibacterial activity of Rpdef3 was determined by the detecting MIC of Rpdef3 against Gram-positive (Staphylococcus aureus and Micrococcus luteus) and Gram-negative bacteria (Vibrio anguillarum, Enterobacter cloacae, Vibrio harveyi, Proteus mirabilis, Enterobacter aerogenes, V. parahaemolyticus, Vibrio splendidus, and Escherichia coli). Briefly, twofold serially diluted Rpdef3 was added to sterile 96-well microtiter plates. The bacteria mentioned above were cultivated to an OD600 of 1.0 and diluted by 1,000 times. The bacterial suspension (10 μL) and broth medium (0.1 mL) were added to the plates. Those without Rpdef3 were used as blank control. The MIC was detected using a microplate reader (Tecan, Infinite M2000 pro, Switzerland) according to the method of Hancock1. The MIC was defined as the range between the highest concentration of the purified Rpdef3 observed for bacterial growth and the lowest concentration resulting in complete inhibition of bacteria growth.
Bactericidal Kinetics of Rpdef3
Escherichia coli was used as the test stain to determine the bacterial kinetics of Rpdef3 according to the previous method (Lv et al., 2020). Briefly, the cultured E. coli suspension was mixed with Rpdef3 (0.5 and 5 μM) and the samples were taken at 0, 10, 30, 60, 160, 400, and 1,000 min, respectively. Bacteria co-incubated with PBS were used as control. The mixtures were serially diluted and transferred to LB agar plates. The viable bacteria were counted by colony counting method.
Biofilm-Forming Ability
The formation of attached biofilms was measured using crystal violet staining method (Pratt and Kolter, 1998). E. coli MG1655 was co-cultured with Rpdef3 (0.005 and 0.05 μM) for 8 h and detected the absorbance at 620 nm. The biofilms were stained by crystal violet and the absorbance was immediately detected at 540 nm. Biofilm formation was detected by dividing the stained biofilm (OD540) by the growth of bacteria (OD620).
Antibacterial Mechanism of Rpdef3
Scanning Electron Microscopy Analysis
Hemocytes (1 × 106 cells/mL) were treated with LPS (500 ng/mL) for 30 min to form ETs. ETs isolated from hemocytes were dispersed into PBS by sonication, referring to the method of Song et al. (2019). The ETs suspension was diluted to 50 μg/mL and incubated with V. parahaemolyticus (1 × 106 CFU/mL) for 1 h or not. The suspension was dropped onto the slides and fixed with 2.5% glutaraldehyde. The samples were processed by ethanol gradient dehydration, critical point drying, gold spraying and observed in a scanning electron microscope (HITACHI, S-4800, Japan). In addition, to determine the effect of Rpdef3 on V. parahaemolyticus, V. parahaemolyticus were cultured and incubated with Rpdef3 (0.5 μM). SEM analysis of V. parahaemolyticus morphology was also performed as described above.
PAMP Binding Assay of Rpdef3
The binding activity of Rpdef3 to pathogen-associated molecular pattern (PAMP) was measured by ELISA (Yu et al., 2007). Briefly, peptidoglycan (PGN) or lipopolysaccharides (LPS) were added to the microtiter plate for 6 h, followed by a BSA (3%, 200 μL/well) blocked treatment. Then the plate was successively incubated with anti-Rpdef3 antibody (100 μL, 1:1,000) and goat-anti-mouse Ig-alkaline phosphatase conjugate (100 μL, 1:5,000, Southern Biotech, United States). PBS was added in blank control group, and pre-immune serum was added in negative control group. The samples were incubated with 0.1% p-nitrophenyl phosphate (100 μL, Sigma, United States) in dark for 30 min. The absorbance of the samples was detected at 405 nm using a microplate reader (Tecan, Infinite M2000 pro, Switzerland). Samples with P (sample)-B (blank)/N (negative)-B (blank) > 2.1 were considered positive (Zhang et al., 2020).
Membrane Permeability Assay of Rpdef3
The permeability of the outer membrane was measured by 1-N-phenylnaphtylamine (NPN) uptake method (Nievagomez et al., 1976). Shortly, E. coli ATCC 25922 were washed, resuspended and added with NPN (10 μM). The fluorescence value of the samples was recorded at an excitation wavelength of 350 nm and an emission wavelength of 420 nm using a fluorescence microplate reader (Thermo Fisher Scientific, United States). Then the samples were treated with Rpdef3 (0.5, 1.25, 2.5, and 5.0 μM) and recorded the fluorescence value. NPN uptake factor was calculated as the fluorescence value of the NPN uptake in the presence of Rpdef3 minus the NPN uptake in the absence of Rpdef3.
Electrochemical Assay of Rpdef3
Supported bilayer lipid membrane (s-BLMs) was constructed by coating freezing method (White, 1974; Ding et al., 1996). The self-assembled monolayer (SAM) was assembled by immersing the gold electrode into 2 mmol/L 1, 2-dipalmitoyl-sn-glycero-3-phosphothioethanol solution. The gold electrode was washed, dried and dripped with decane containing 5 μL of 1, 2-dipalmitoyl-sn-glycero-3-phosphocholine solution. After frozen at –20°C, the electrode was immersed in potassium chloride solution and washed with water to prepare s-BLMs. The mixture of Rpdef3 and electrolyte (PBS) interacted with s-BLMs, and electrochemical measurement was performed using a three-electrode system by 660C Electrochemical Analyzer (CHI Instruments).
Statistical Analysis
All the data were presented as the means ± standard deviation of at least three replicates. The normality of data was warranted by Shapiro-Wilk test, and the homogeneity of variance was analyzed by Levene’s test. Means were analyzed with One-way analysis of variance (one-way ANOVA) followed by Duncan test using SPSS 17.0 software. The differences were considered as significant if P < 0.05.
Results
Peptide Synthesis and Western Blotting Assay of Rpdef3
The Rpdef3 peptide was synthesized by solid-phase chemical synthesis technology and the intact lyophilized peptide was purified by reversed phase high performance liquid chromatography. The purity of the synthetic Rpdef3 peptide was approximately 95% (Supplementary Figure 1A). In addition, the products also matched the amino acid sequence of Rpdef3 (Supplementary Figure 1B). A clear band was detected in western blotting assay, suggesting that the antibody could work well with Rpdef3 protein (Supplementary Figure 1C).
The Formation of ETs Induced by V. parahaemolyticus
Clam hemocytes were treated with V. parahaemolyticus, and the formation of ETs was determined by SytoX Green fluorescent staining. It was observed that V. parahaemolyticus obviously stimulated the formation of ETs-like structures in hemocytes of Manila clams. When V. parahaemolyticus-induced ETs was incubated with DNase I, the ET structure was totally disappeared (Figure 1A). These results suggested that DNA was an essential component of ETs. As revealed in Figure 1B, the value of released DNA from hemocytes increased along with bacterial concentrations, indicating that V. parahaemolyticus induced the formation of ETs in a dose-dependent manner (P < 0.01). In addition, DNase I significantly inhibited the increase of DNA value induced by V. parahaemolyticus at the concentration of 1 × 106 CFU/mL (Figure 1B).
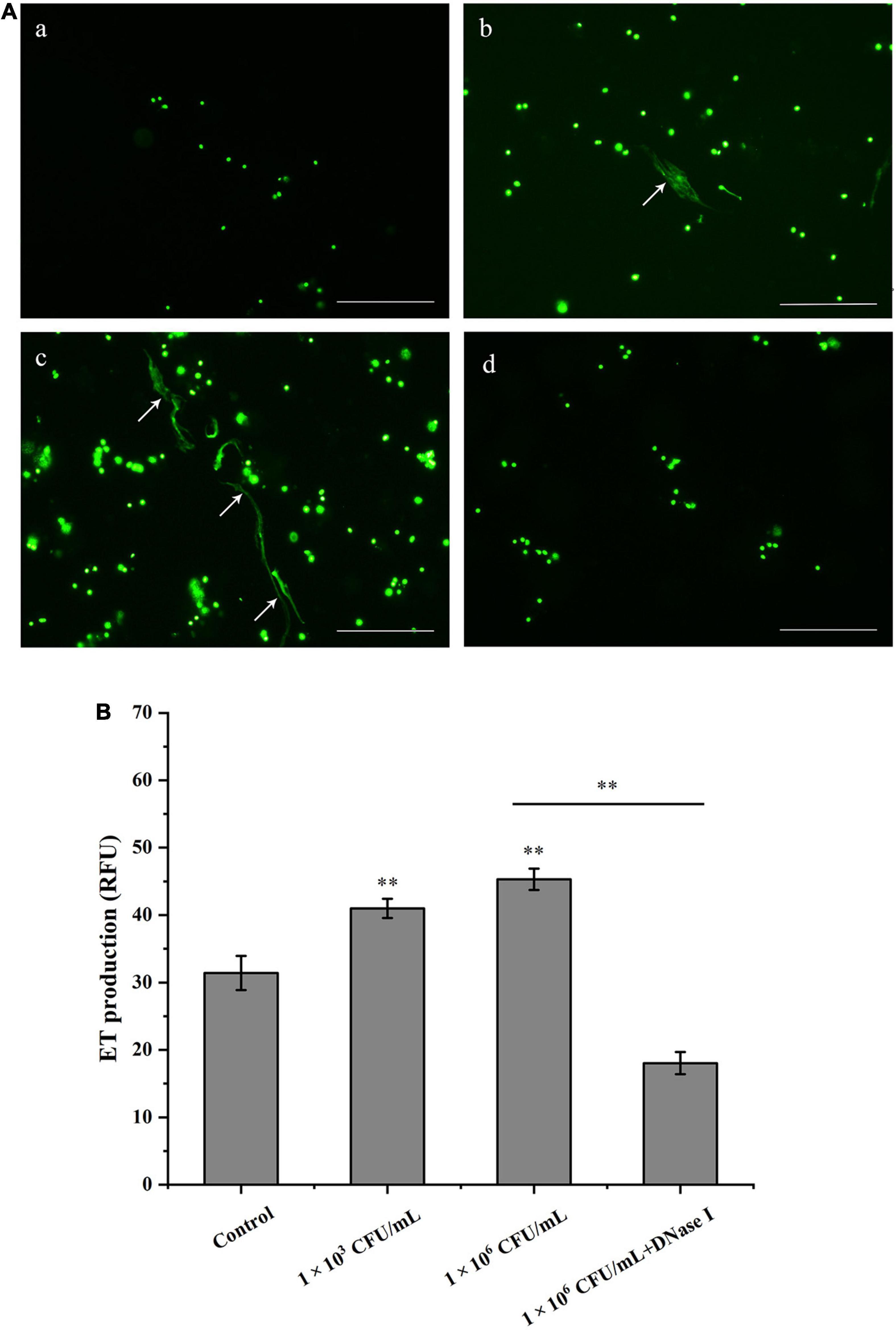
Figure 1. Hemocytes release DNA in response to Vibrio parahaemolyticus stimulation. (A) V. parahaemolyticus induced ETs formation. Hemocytes treated with V. parahaemolyticus (1 × 103 and 1 × 106 CFU/mL) for 1 h. After SytoX staining for 5 min, the DNA released from hemocytes was observed using fluorescence microscope (b,c). Hemocytes were treated with DNase I after stimulated with V. parahaemolyticus (1 × 106 CFU/mL), and DNA released from hemocytes was stained by SytoX fluorescent dye and observed using fluorescence microscope (d). Unstimulated hemocytes were used as a blank control group (a). Arrows indicate ETs. Bar, 100 μm. (B) Quantitation of V. parahaemolyticus-inducted ETs. Hemocytes were stimulated with V. parahaemolyticus (1 × 103 and 1 × 106 CFU/mL) for 1 h, then the formation of ETs was examined with a fluorescence plate reader using an excitation wavelength of 485 nm and an emission wavelength at 525 nm. Date was presented as mean ± SD (N = 6). ∗ ∗P < 0.01.
Detection of DNA Traps and Rpdef3
As shown in Figure 2, DNA traps were released by hemocytes after the stimulation of V. parahaemolyticus (1 × 103 and 1 × 106 CFU/mL). More importantly, Rpdef3 was obviously observed around the formed DNA traps (Figure 2). The results proved that the ETs induced by V. parahaemolyticus were accompanied with the production of Rpdef3.
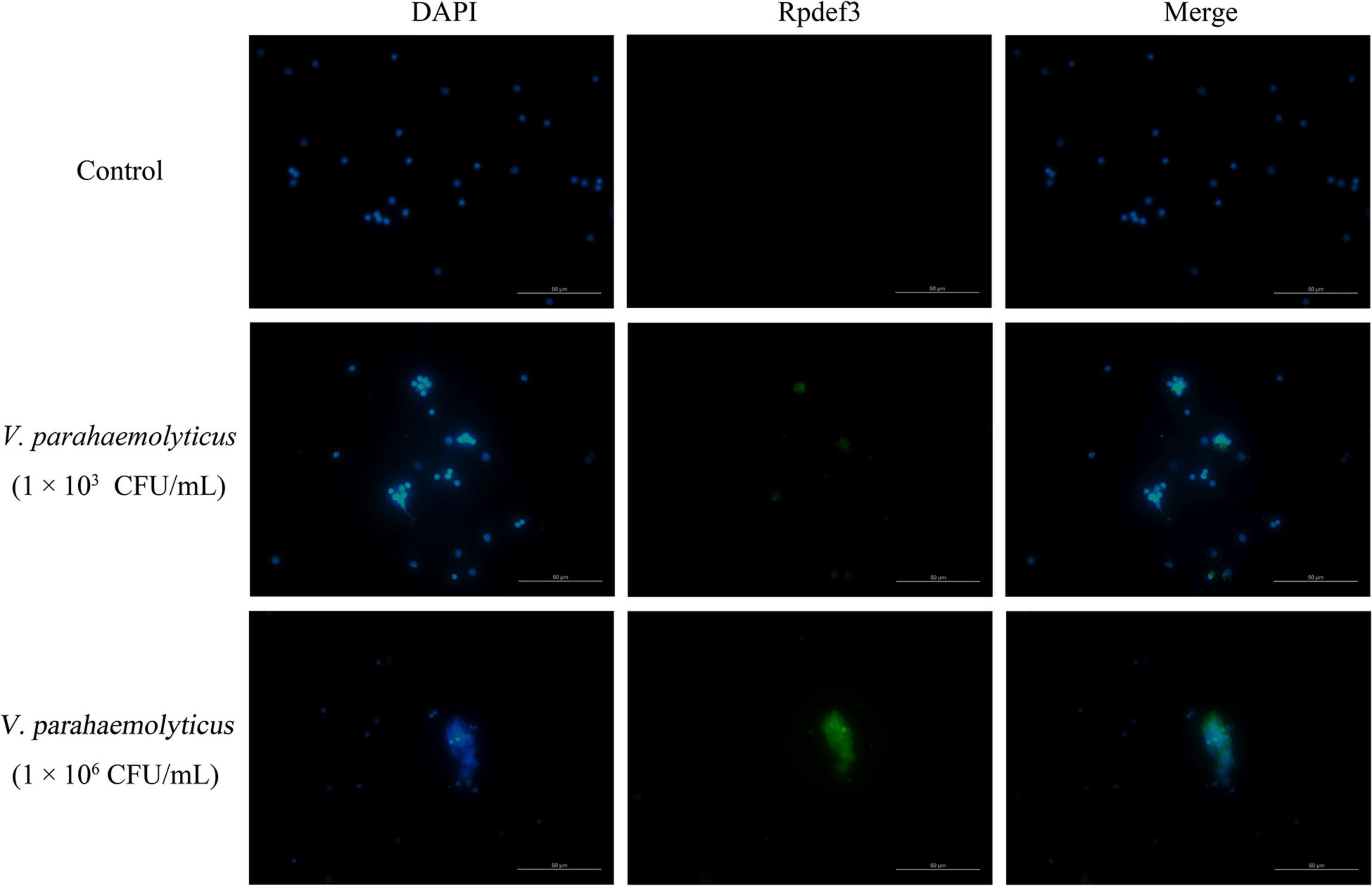
Figure 2. Immunostaining of DNA traps and Rpdef3. Clam hemocytes were treated with V. parahaemolyticus (1 × 103 and 1 × 106 CFU/mL) for 1 h and stained for DNA (blue) and defensin (green). Bar, 100 μm.
Antibacterial Activities of ETs and Rpdef3
Hemocytes were incubated with V. parahaemolyticus (ETs-positive) for various times to examine the antibacterial abilities of ETs. As shown in Figure 3A, sharp decrease of bacterial number was detected at 0.5 h (P < 0.01), 1 h (P < 0.01), and 2 h (P < 0.01), indicating that ETs exhibited significant bactericidal activities against V. parahaemolyticus. Our SEM results showed visually that the ETs network structure of hemocytes was successfully separated, and this ETs structure could capture V. parahaemolyticus (Figure 3B).Rpdef3 exhibited a broad antibacterial spectrum and showed strong microbicidal activities against M. luteus, V. anguillarum and E. coli (Table 1). The bactericidal effect of Rpdef3 was tested by the time-killing experiment of E. coli. As shown in Figure 4A, the number of bacteria in the high-dose group decreased sharply, and the bacteria were almost killed within 400 min. The results indicated that Rpdef3 could exhibit obvious bactericidal activity on the examined bacteria. In addition, Rpdef3 reduced the biofilm diffusion of E. coli, indicating that Rpdef3 significantly reduced the formation of biofilm (Figure 4B).
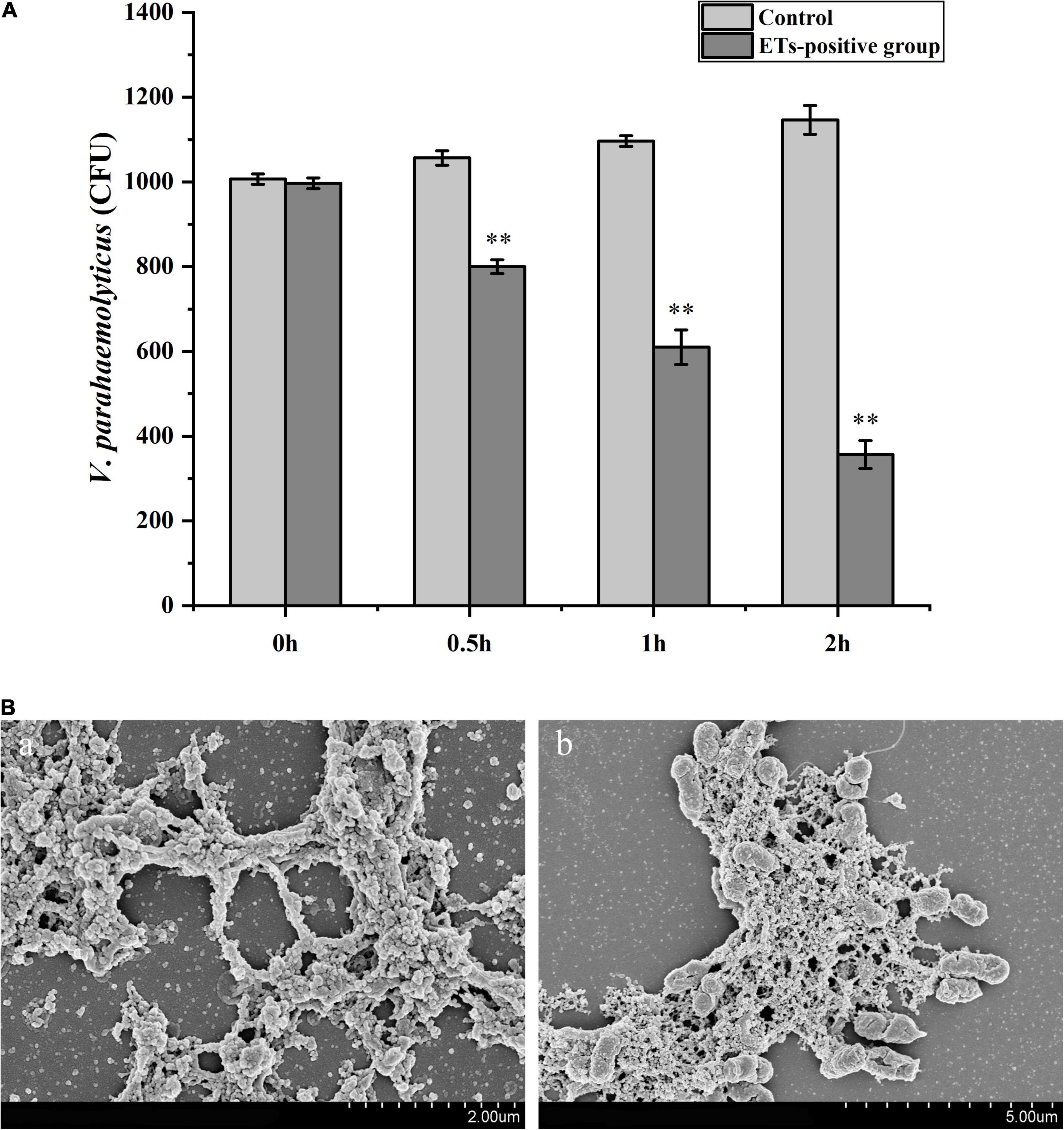
Figure 3. Antibacterial activities of ETs. (A) Hemocytes were stimulated with V. parahaemolyticus (1 × 103 CFU/mL) for ETs production (ETs-positive cells) for 0, 0.5, 1 and 2 h. Then the bacterial numbers were determined by plate count. Date was presented as mean ± S.D. (N = 3). ∗ ∗P < 0.01. (B) Scanning electron microscope (SEM) observation of ETs. The isolated ETs were incubated with V. parahaemolyticus (1 × 106 CFU/mL) (b) or not (a) for 1 h. Arrow: networks formed by ETs. Arrowheads: ET-trapped bacteria.
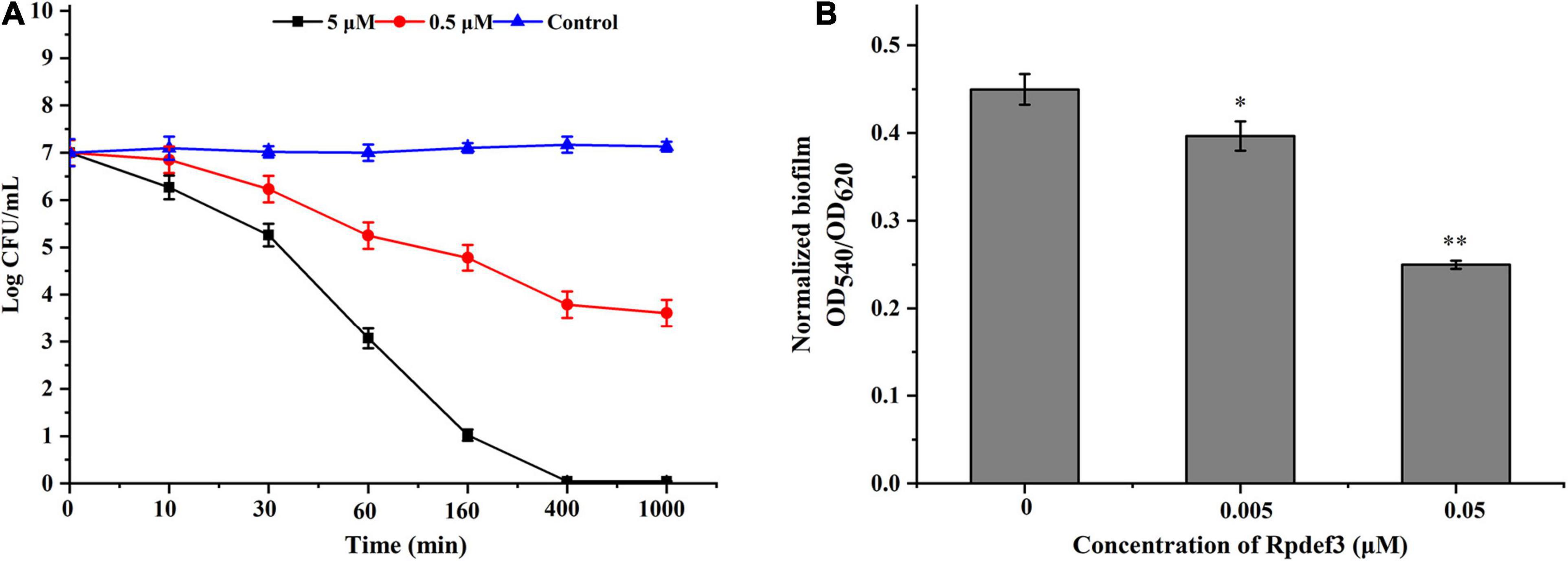
Figure 4. Antibacterial activities of Rpdef3. (A) Bacterial killing kinetics of Rpdef3 at 0.5 and 5 μM against E. coli. Each group had three replicates. (B) Biofilm formation of E. coli MG1655 treated with Rpdef3. Normalized biofilm formation (total biofilm/growth) was tested in 96-well polystyrene plates after treated with Rpdef3. Data were the average of 10 replicate wells from two independent cultures. The values were shown as mean ± S.D. (N = 10) (∗P < 0.05; ∗ ∗P < 0.01).
PAMP Binding Assay and Disruption of the Membrane Integrity
As shown in Figure 5A, Rpdef3 performed binding activity toward LPS and PGN. The permeability and integrity of the outer membrane of bacteria were detected by NPN uptake method. With the increase of Rpdef3 concentration, the NPN fluorescence gradually increased, indicating that Rpdef3 penetrated the outer membrane of E. coli (Figure 5B). In addition, SEM results showed that Rpdef3 had a destructive effect on the surface of E. coli (Figure 5C). The electrochemical assay was performed to simulate the bilayer system to explore the destructive effect of Rpdef3 on membranes. As revealed in Figure 5D, Rpdef3 could increase the charge transfer resistance (Rct) (Figure 5D).
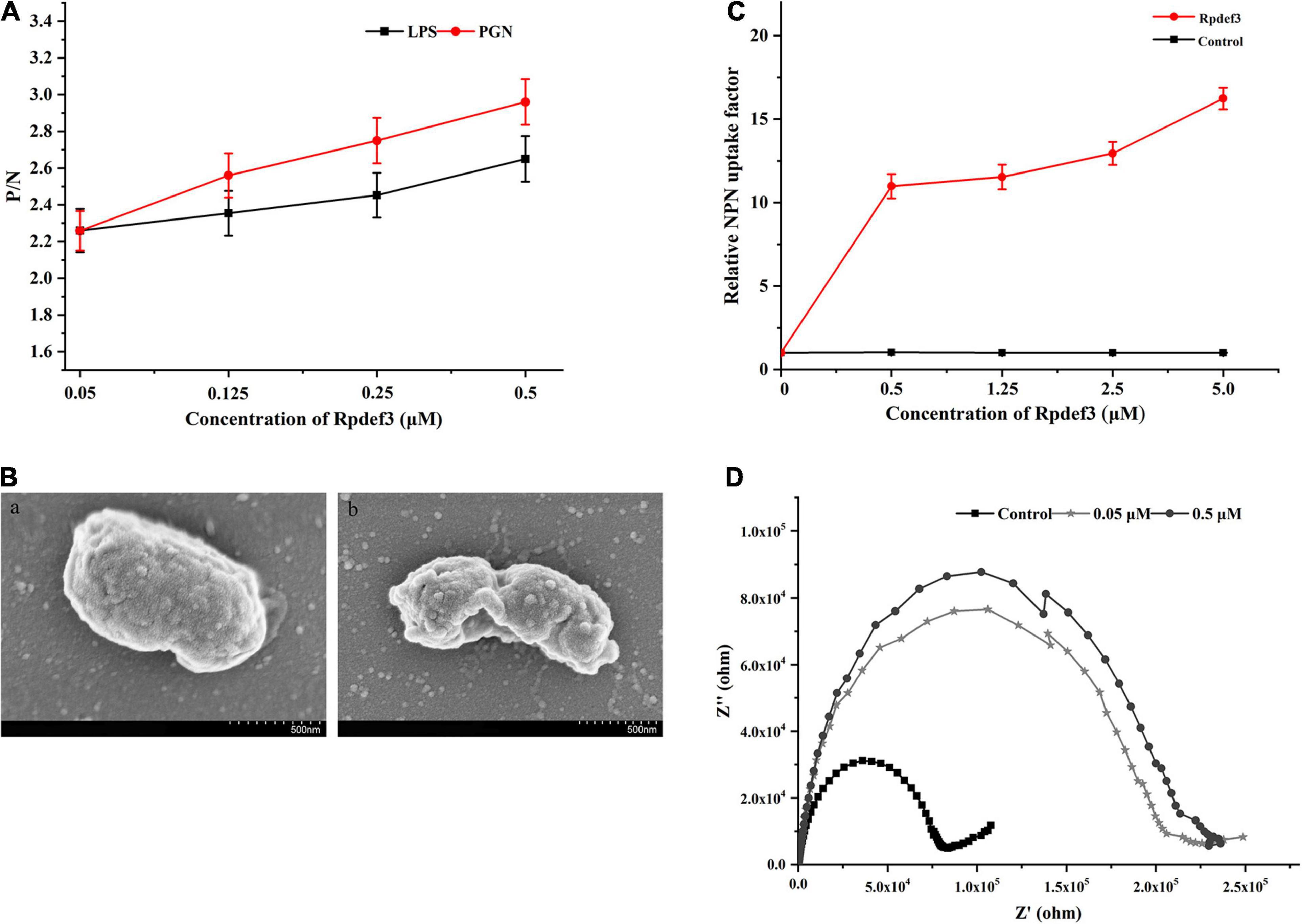
Figure 5. Antibacterial mechanism of Rpdef3. (A) ELISA assay of the interaction between Rpdef3 and PAMPs. Plates were coated with four kinds of PAMPs including LPS and PGN, and then incubated with different amounts of Rpdef3. After incubated with Rpdef3 antibody and goat-anti-mouse Ig-alkaline phosphatase conjugate, the absorbance was recorded at 405 nm. Samples with P/N > 2.1 were considered positive. Results were representative of the mean of three replicates ± S.D. (B) Membrane permeabilization of E. coli caused by Rpdef3. Each group had three replicates. (C) SEM images of E. coli in the absence (a) or presence (b) of Rpdef3 (0.5 μM) after exposure for 1 h. (D) Impedance complex plane plots of s-BLMs coated gold electrode in the presence of Rpdef3. EIS was carried out over a frequency range of 0.01 Hz–100 kHz at a signal amplitude of 10 mV.
Discussion
The composition of ETs has been well studied in vertebrates, namely, antibacterial proteins and DNA skeleton structure. It has been reported that 80 proteins have been identified in NETs induced by Pseudomonas aeruginosa (Dwyer et al., 2014). However, there are few studies on the formation of ETs in mollusks and their associated antibacterial proteins. In this study, we detect the formation of ETs in hemocytes of Manila clams induced by V. parahaemolyticus, and the potential functions of defensin involved in Vibrio-induced ETs were also investigated.
Extracellular traps formation has been found to be triggered in response to multiple stimuli, such as live bacteria, parasites and bacteria protein (Fuchs et al., 2007; Kenny et al., 2017). For example, shrimp hemocytes could form ET structure in response to E. coli stimulation (Ng et al., 2015). When exposed to Vibrio tasmaniensis and Brevibacterium stationis, ET structure was also rapidly released from oyster hemocytes (Poirier et al., 2014). In the present study, V. parahaemolyticus stimulation could induce the release of abundant DNA-containing ET structures from clam hemocytes. Similarly, the formation of ETs was also observed in hemocytes of Manila clam post V. anguillarum challenge (Han et al., 2021), indicating that different Vibrio species perhaps have similar effects on the induction of bivalve ET formation. However, it has also been reported that ET formation was not induced by bacteria such as V. splendidus and V. anguillarum in Mytilus galloprovincialis, when phagocytosis inhibitors was absent (Romero et al., 2020). This discrepancy might be related with the complex process that hemocytes eliminate invading bacteria. For example, neutrophils kill bacteria through a process of phagocytosis, degranulation, and finally ET formation (Papayannopoulos and Zychlinsky, 2009). In this sense, hemocytes may also be able to kill bacteria by phagocytosis without ET formation.
It was documented that ETs in hemocytes of Crassostrea gigas and Litopenaeus vannamei could kill microorganisms by releasing antibacterial histones (Ng et al., 2013; Poirier et al., 2014). In this study, we found that ETs triggered by V. parahaemolyticus were accompanied by the production of antibacterial protein Rpdef3. In addition, hemocyte ETs have a strong bactericidal effect on V. parahaemolyticus, and the ET structure isolated in vitro can also capture V. parahaemolyticus and destroy bacteria membrane structure. These results highlight the important role of ETs and the released defensin in the antibacterial function of Manila clam hemocytes. Consistent with the phenomenon, immune cells (e.g., neutrophils and eosinophils) have been found to maintain their natural defense capacities by producing ETs coupled with antibacterial factors (Brinkmann et al., 2004; Yousefi et al., 2012; Koiwai et al., 2016). As a releasing component companied with hemocyte ET formation, Rpdef3 has similar efficiency in antibacterial/anti-biofilm activities compared to other immune peptides, to against invading bacteria. In the process, incubation of Rpdef3 resulted in less biofilm dispersal of E. coli on polystyrene surfaces. The anti-biofilm mechanism of Rpdef3 might be performed by reducing the initial adhesion of bacteria, stimulating the movement of bacteria on the surface, as well as down-regulating of essential genes for the development and maintenance of biofilms (Singh et al., 2002; Pamp and Tolker-Nielsen, 2007; Picioreanu et al., 2007). The results supported that Rpdef3 contributed to the trap and the elimination of invading Vibrio in ETosis.
The antibacterial mechanism of ETs might be associated with their ETs-related peptides, such as defensin, histone or BPIP (Brinkmann et al., 2004; Hu et al., 2016). Previous studies have shown that some defensins could recognize PAMPs and destroy bacterial membranes or cell walls, thus performing antibacterial activity (Yang et al., 2018). In the study, the binding of Rpdef3 to PAMPs supported that defensin might take part in immune responses against invading bacteria. Notably, Rpdef3 had a disruptive effect on bacterial membrane. Similar results were also reported in the defensins identified from vertebrates (Sudheendra et al., 2015) and mollusks (Yang et al., 2018). For example, human defensin-3 adopts an α-helical structure, contributing to its activities against negative bacterial membranes. In turn, the negatively charged membranes could also increase the amphipathicity of defensin-3, leading to insert itself into phospholipid membranes effectively (Sudheendra et al., 2015). Totally, the above results supported that Rpdef3 might exert antibacterial activities by destroying the membrane permeabilization of bacteria (Shai, 2002; Nakajima et al., 2003) and then caused the leakage of cellular cytoplasmic contents (Song et al., 2010).
In summary, the antimicrobial activity and mode of action of the ETs were investigated in R. philippinarum. Our results revealed that R. philippinarum hemocytes could form ETs associated with antimicrobial defensin in vitro, in response to bacterial infection. We also proved that these ETs could entrap and kill invading bacteria. These findings provide descriptions of a new mode of cellular immunity, which allow us to better understand the innate immunity of R. philippinarum.
Conclusion
In this study, V. parahaemolyticus induced the formation of ETs in Manila clam hemocytes. In the process, V. parahaemolyticus could be captured and destroyed by ETs in vitro. Notably, the antibacterial agent Rpdef3 involved in ETs had been shown to be produced in ETosis. Rpdef3 has broad-spectrum antibacterial activity against both Gram-positive and Gram-negative bacteria. Especially, it cannot only bind to bacterial cell wall, but also increase bacterial membrane permeability, thus eventually causing cell death. Taken together, our data revealed a novel defense mode of ETs accompanied by defensin production in response to bacterial stimulation, leading to a better understand the innate immune system of R. philippinarum.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, AFP49946.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Chinese Academy of Sciences.
Author Contributions
DYa designed the experiments. YH and GH carried out this experiment. YC, LC, DYu, and QZ contributed to data analysis and statistics. YH and DYa wrote this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Natural Science Foundation of China (No. 41806196), the Natural Science Foundation of Shandong Province (ZR2019BD022), the Yantai Science and Technology Development Project (2020MSGY066), and the Youth Innovation Promotion Association of CAS (2019216).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.690879/full#supplementary-material
Supplementary Figure 1 | RP-HPLC profiles (A) and MALDI-MS spectra (B) of synthetic peptide Rpdef3.
Footnotes
References
Allam, B., Paillard, C., and Ford, S. E. (2002). Pathogenicity of Vibrio tapetis, the etiological agent of brown ring disease in clams. Dis. Aquat. Organ. 48, 221–231. doi: 10.3354/dao048221
Ayabe, T., Ashida, T., Kohgo, Y., and Kono, T. (2004). The role of Paneth cells and their antimicrobial peptides in innate host defense. Trends Microbiol. 12, 394–398. doi: 10.1016/j.tim.2004.06.007
Boman, H. G., Agerberth, B., and Boman, A. (1993). Mechanisms of action on Escherichia coli of cecropin-P1 and Pr-39, two antibacterial peptides from pig intestine. Infect. Immun. 61, 2978–2984. doi: 10.1128/IAI.61.7.2978-2984.1993
Brinkmann, V., Reichard, U., Goosmann, C., Fauler, B., Uhlemann, Y., Weiss, D. S., et al. (2004). Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535. doi: 10.1126/science.1092385
Bulet, P., Stocklin, R., and Menin, L. (2004). Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198, 169–184. doi: 10.1111/j.0105-2896.2004.0124.x
Ding, L., Li, J. H., Dong, S. J., and Wang, E. K. (1996). Supported phospholipid membranes: comparison among different deposition methods for a phospholipid monolayer. J. Electroanal. Chem. 416, 105–112. doi: 10.1016/S0022-0728(96)04737-7
Dwyer, M., Shan, Q., D’ortona, S., Maurer, R., Mitchell, R., Olesen, H., et al. (2014). Cystic fibrosis sputum DNA has NETosis characteristics and neutrophil extracellular trap release is regulated by macrophage migration-inhibitory factor. J. Innate Immun. 6, 765–779. doi: 10.1159/000363242
Fuchs, T. A., Abed, U., Goosmann, C., Hurwitz, R., Schulze, I., Wahn, V., et al. (2007). Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176, 231–241. doi: 10.1083/jcb.200606027
Guimaraes-Costa, A. B., Nascimento, M. T., Wardini, A. B., Pinto-Da-Silva, L. H., and Saraiva, E. M. (2012). ETosis: a microbicidal mechanism beyond cell death. J. Parasitol. Res. 2012:929743. doi: 10.1155/2012/929743
Han, Y., Chen, L., Zhang, Q., Yu, D., Yang, D., and Zhao, J. (2021). Hemocyte extracellular traps of Manila clam Ruditapes philippinarum: production characteristics and antibacterial effects. Dev. Comp. Immunol. 116:103953. doi: 10.1016/j.dci.2020.103953
Hazeldine, J., Harris, P., Chapple, I. L., Grant, M., Greenwood, H., Livesey, A., et al. (2014). Impaired neutrophil extracellular trap formation: a novel defect in the innate immune system of aged individuals. Aging Cell 13, 690–698. doi: 10.1111/acel.12222
Hu, S. C., Yu, H. S., Yen, F. L., Lin, C. L., Chen, G. S., and Lan, C. C. (2016). Neutrophil extracellular trap formation is increased in psoriasis and induces human beta-defensin-2 production in epidermal keratinocytes. Sci. Rep. 6:31119. doi: 10.1038/srep31119
Kenny, E. F., Herzig, A., Krüger, R., Muth, A., Mondal, S., Thompson, P. R., et al. (2017). Diverse stimuli engage different neutrophil extracellular trap pathways. Elife. 6:e24437. doi: 10.7554/eLife.24437
Koiwai, K., Alenton, R. R., Kondo, H., and Hirono, I. (2016). Extracellular trap formation in kuruma shrimp (Marsupenaeus japonicus) hemocytes is coupled with c-type lysozyme. Fish Shellfish Immunol. 52, 206–209. doi: 10.1016/j.fsi.2016.03.039
Lv, C., Han, Y., Yang, D., Zhao, J., Wang, C., and Mu, C. (2020). Antibacterial activities and mechanisms of action of a defensin from manila clam Ruditapes philippinarum. Fish Shellfish Immunol. 103, 266–276. doi: 10.1016/j.fsi.2020.05.025
McDonald, B., Urrutia, R., Yipp, B. G., Jenne, C. N., and Kubes, P. (2012). Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell Host Microbe 12, 324–333. doi: 10.1016/j.chom.2012.06.011
Nakajima, Y., Ishibashi, J., Yukuhiro, F., Asaoka, A., Taylor, D., and Yamakawa, M. (2003). Antibacterial activity and mechanism of action of tick defensin against Gram-positive bacteria. Biochim. Biophys. Acta. 1624, 125–130. doi: 10.1016/j.bbagen.2003.10.004
Nathan, C. (2006). Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 6, 173–182. doi: 10.1038/nri1785
Neeli, I., Dwivedi, N., Khan, S., and Radic, M. (2009). Regulation of extracellular chromatin release from neutrophils. J. Innate Immun. 1, 194–201. doi: 10.1159/000206974
Ng, T. H., Chang, S. H., Wu, M. H., and Wang, H. C. (2013). Shrimp hemocytes release extracellular traps that kill bacteria. Dev. Comp. Immunol. 41, 644–651. doi: 10.1016/j.dci.2013.06.014
Ng, T. H., Wu, M. H., Chang, S. H., Aoki, T., and Wang, H. C. (2015). The DNA fibers of shrimp hemocyte extracellular traps are essential for the clearance of Escherichia coli. Dev. Comp. Immunol. 48, 229–233. doi: 10.1016/j.dci.2014.10.011
Nievagomez, D., Konisky, J., and Gennis, R. B. (1976). Membrane changes in Escherichia coli Induced by colicin Ia and agents known to disrupt energy transduction. Biochemistry 15, 2747–2753. doi: 10.1021/bi00658a006
Pamp, S. J., and Tolker-Nielsen, T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189, 2531–2539. doi: 10.1128/JB.01515-06
Papayannopoulos, V., Metzler, K. D., Hakkim, A., and Zychlinsky, A. (2010). Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 191, 677–691. doi: 10.1083/jcb.201006052
Papayannopoulos, V., and Zychlinsky, A. (2009). NETs: a new strategy for using old weapons. Trends Immunol. 30, 513–521. doi: 10.1016/j.it.2009.07.011
Park, C. B., Kim, H. S., and Kim, S. C. (1998). Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 244, 253–257. doi: 10.1006/bbrc.1998.8159
Picioreanu, C., Kreft, J. U., Haagensen, J. A. J., Tolker-Nielsen, T., and Molin, S. (2007). Microbial motility involvement in biofilm structure formation - a 3D modelling study. Water Sci. Technol. 55, 337–343. doi: 10.2166/wst.2007.275
Poirier, A. C., Schmitt, P., Rosa, R. D., Vanhove, A. S., Kieffer-Jaquinod, S., Rubio, T. P., et al. (2014). Antimicrobial histones and DNA traps in invertebrate immunity: evidences in Crassostrea gigas. J. Biol. Chem. 289, 24821–24831. doi: 10.1074/jbc.M114.576546
Porto, B. N., and Stein, R. T. (2016). Neutrophil extracellular traps in pulmonary diseases: too much of a good thing? Front. Immunol. 7:311. doi: 10.3389/fimmu.2016.00311
Pratt, L. A., and Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Romero, A., Novoa, B., and Figueras, A. (2020). Extracellular traps (ETosis) can be activated through NADPH-dependent and -independent mechanisms in bivalve mollusks. Dev. Comp. Immunol. 106:103585. doi: 10.1016/j.dci.2019.103585
Saitoh, T., Komano, J., Saitoh, Y., Misawa, T., Takahama, M., Kozaki, T., et al. (2012). Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe. 12, 109–116. doi: 10.1016/j.chom.2012.05.015
Schneider, J. J., Unholzer, A., Schaller, M., Schafer-Korting, M., and Korting, H. C. (2005). Human defensins. J. Mol. Med. 83, 587–595. doi: 10.1007/s00109-005-0657-1
Schonrich, G., and Raftery, M. J. (2016). Neutrophil extracellular traps go viral. Front. Immunol. 7:366. doi: 10.3389/fimmu.2016.00366
Shai, Y. (2002). Mode of action of membrane active antimicrobial peptides. Biopolymers 66, 236–248. doi: 10.1002/bip.10260
Silva, L. M., Munoz-Caro, T., Burgos, R. A., Hidalgo, M. A., Taubert, A., and Hermosilla, C. (2016). Far beyond phagocytosis: phagocyte-derived extracellular traps act efficiently against protozoan parasites in vitro and in vivo. Mediators Inflamm. 2016:5898074. doi: 10.1155/2016/5898074
Singh, P. K., Parsek, M. R., Greenberg, E. P., and Welsh, M. J. (2002). A component of innate immunity prevents bacterial biofilm development. Nature 417, 552–555. doi: 10.1038/417552a
Song, L. S., Wang, L. L., Qiu, L. M., and Zhang, H. A. (2010). Bivalve Immunity. Invertebrate Immun. 708, 44–65. doi: 10.1007/978-1-4419-8059-5_3
Song, Y., Kadiyala, U., Weerappuli, P., Valdez, J. J., Yalavarthi, S., Louttit, C., et al. (2019). Antimicrobial microwebs of DNA-histone inspired from neutrophil extracellular traps. Adv. Mater. 31:e1807436. doi: 10.1002/adma.201807436
Sudheendra, U. S., Dhople, V., Datta, A., Kar, R. K., Shelburne, C. E., Bhunia, A., et al. (2015). Membrane disruptive antimicrobial activities of human beta-defensin-3 analogs. Eur. J. Med. Chem. 91, 91–99. doi: 10.1016/j.ejmech.2014.08.021
Tharntada, S., Somboonwiwat, K., Rimphanitchayakit, V., and Tassanakajon, A. (2008). Anti-lipopolysaccharide factors from the black tiger shrimp, Penaeus monodon, are encoded by two genomic loci. Fish Shellfish Immunol. 24, 46–54. doi: 10.1016/j.fsi.2007.07.010
Tincu, J. A., and Taylor, S. W. (2004). Antimicrobial peptides from marine invertebrates. Antimicrob. Agents Chemother. 48, 3645–3654. doi: 10.1128/AAC.48.10.3645-3654.2004
Von Kockritz-Blickwede, M., and Nizet, V. (2009). Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J. Mol. Med. 87, 775–783. doi: 10.1007/s00109-009-0481-0
White, S. H. (1974). Temperature-dependent structural-changes in planar bilayer membranes - solvent freeze-out. Biochim. Biophys. Acta 356, 8–16. doi: 10.1016/0005-2736(74)90289-2
Yang, D., Han, Y., Chen, L., Cao, R., Wang, Q., Dong, Z., et al. (2019a). A macin identified from Venerupis philippinarum: investigation on antibacterial activities and action mode. Fish Shellfish Immunol. 92, 897–904. doi: 10.1016/j.fsi.2019.07.031
Yang, D., Han, Y., Chen, L., Liu, Y., Cao, R., Wang, Q., et al. (2019b). Scavenger receptor class B type I (SR-BI) in Ruditapes philippinarum: a versatile receptor with multiple functions. Fish Shellfish Immunol. 88, 328–334. doi: 10.1016/j.fsi.2019.03.009
Yang, D., Zhang, Q., Wang, Q., Chen, L., Liu, Y., Cong, M., et al. (2018). A defensin-like antimicrobial peptide from the manila clam Ruditapes philippinarum: investigation of the antibacterial activities and mode of action. Fish Shellfish Immunol. 80, 274–280. doi: 10.1016/j.fsi.2018.06.019
Yipp, B. G., Petri, B., Salina, D., Jenne, C. N., Scott, B. N., Zbytnuik, L. D., et al. (2012). Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 18, 1386–1393. doi: 10.1038/nm.2847
Yousefi, S., Simon, D., and Simon, H. U. (2012). Eosinophil extracellular DNA traps: molecular mechanisms and potential roles in disease. Curr. Opin. Immunol. 24, 736–739. doi: 10.1016/j.coi.2012.08.010
Yu, Y. H., Yu, Y. C., Huang, H. Q., Feng, K. X., Pan, M. M., Yuan, S. C., et al. (2007). A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J. Immunol. 179, 8425–8434. doi: 10.4049/jimmunol.179.12.8425
Zhang, C., Xue, Z., Yu, Z., Wang, H., Liu, Y., Li, H., et al. (2020). A tandem-repeat galectin-1 from Apostichopus japonicus with broad PAMP recognition pattern and antibacterial activity. Fish Shellfish Immunol. 99, 167–175. doi: 10.1016/j.fsi.2020.02.011
Keywords: Ruditapes philippinarum, defensin, extracellular traps, antimicrobial effect, membrane permeabilization
Citation: Han Y, Hu G, Chen Y, Chen L, Yu D, Zhang Q and Yang D (2021) Antimicrobial Defensin and DNA Traps in Manila Clam Ruditapes philippinarum: Implications for Their Roles in Immune Responses. Front. Mar. Sci. 8:690879. doi: 10.3389/fmars.2021.690879
Received: 04 April 2021; Accepted: 23 June 2021;
Published: 16 July 2021.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Chuanyan Yang, Dalian Ocean University, ChinaMengqiang Wang, Ocean University of China, China
Linbao Zhang, South China Sea Fisheries Research Institute (CAFS), China
Copyright © 2021 Han, Hu, Chen, Chen, Yu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dinglong Yang, ZGx5YW5nQHlpYy5hYy5jbg==; Daode Yu, eXVkYW9kZUBzaGFuZG9uZy5jbg==
 Yijing Han1,2,3
Yijing Han1,2,3 Daode Yu
Daode Yu Qianqian Zhang
Qianqian Zhang Dinglong Yang
Dinglong Yang