- College of Resources and Environmental Science, Nanjing Agricultural University, Nanjing, China
Diatoms play a key role in water quality assessments and algae bloom. However, taxonomic confusion often exists for diatoms, and morphological characters are extremely diverse for species identification. DNA barcoding with multiple genetic markers can contribute much to diatom diversity investigation. In this study, we employed sequences of four genetic markers (COI, rbcL, SSU, and LSU) to discriminate diatom strains from both marine and freshwater environments of China, by tree, distance, and character-based barcoding methods. The available published diatom sequences were also incorporated into our new sequences. A total of 93 rbcL, 81 COI, 83 SSU, and 75 LSU sequences of diatom samples were obtained in this study. The multiple genetic markers discriminated most species clearly. The identification of species by micrographic observation was generally consistent with the DNA barcoding analysis except that some potential cryptic species were revealed by DNA barcoding. The COI, rbcL, and LSU sequences all showed high taxonomic resolution at the species level by phylogenetic and character-based analysis. Some potential identification errors in public diatom sequences were also found. The phylogenetic and character-based analysis revealed consistent species identification and showed clearer species discrimination than the distance-based method. In conclusion, our study evaluated the efficiency of four genetic markers in barcoding 11 genera within Bacillariophyta isolated from China and complemented many diatom reference sequences to public databases.
Introduction
Diatoms are photosynthetic secondary endosymbionts found throughout marine and freshwater environments and are believed to be responsible for around one-fifth of primary productivity on earth and the occurrence of blooms (Bowler et al., 2008; Casteleyn et al., 2010). Diatoms are also frequently used for water quality assessments for marine as well as freshwater environments (Kawecka and Olech, 1993; Spaulding and McKnight, 1999). While some diatom species have broad ecological plasticity, others, including closely related species, are adapted to specific environmental conditions (Vanelslander et al., 2009). There are, estimated 200,000 diatom species, living in terrestrial, freshwater, and marine systems as benthos or phytoplankton (Dam et al., 1994; Potapova and Charles, 2007; Zalack et al., 2010; Hamsher et al., 2011). Diatom-based indices require unambiguous identification at the species level. However, the species identification of diatoms is time-consuming and needs in-depth knowledge of organisms under investigation, such as bacteria (Zhang et al., 2018). Thus, taxonomic confusion often exists for diatoms, while a large number of morphological characters are extremely diverse (Evans et al., 2007).
The identification of diatoms has been somewhat improved by molecular tools, e.g., the discovery of cryptic diversity (Medlin et al., 1991; Behnke et al., 2004; Beszteri et al., 2005; Sarno et al., 2005; Amato et al., 2007; Evans et al., 2007; Poulíčková et al., 2010). For many years DNA barcoding has been proved as a promising approach for species identification and detection of cryptic species, particularly for microbial communities (Hebert et al., 2003a,b; Zou et al., 2016a,b, 2018). Our previous studies have shown that it is important to combine different analytical tools for the DNA barcoding of microalgae (Zou et al., 2016a,b, 2018). While the tree-based approach uses neighbor-joining (NJ), Bayesian, or maximum-likelihood trees for species identification, the distance-based approach calculates a genetic distance between species and assigns a cutoff value (the “barcode gap”) to discriminate species. The character-based approach discriminates species by the fundamental concept that members of a given taxonomic group share diagnostic characters (more than three bases) that are absent from comparable groups (Rach et al., 2008; Sarkar et al., 2008). A program based on the Characteristic Attributes Organization System (CAOS) algorithm (Sarkar et al., 2002a,b) was developed to implement a character-based approach for DNA barcoding (Sarkar et al., 2008). CAOS is an automated systematic method for discovering conserved character states from cladograms (i.e., trees) or groups of categorical information, and defines attribute tests at each node in a phylogenetic tree, similar to decision tree algorithms. Character states, called “attribute tests” in decision trees, are termed “Characteristic Attributes” (CAs) in CAOS (Sarkar et al., 2008). Although it remains argued which analytical method of DNA barcoding is more precise, it is unquestionable that comparison of multiple analytical methods would be important for taxonomic assignments.
While there is no single conserved gene that could be used for barcoding all phytoplankton taxa, multiple genetic markers (like rbcL and SSU) have been proposed as potential markers for barcoding diatoms (Mónica and Kaczmarska, 2009; Hamsher et al., 2011; Tamura et al., 2011; Guo et al., 2015; Li et al., 2015). Within Bacillariophyta, it was indicated that ITS was a potential marker for the DNA barcoding of Thalassiosirales and that COI could just barcode some genera (Guo et al., 2015). Trobajoa et al. (2011) showed that although COI was more variable than LSU and rbcL for barcoding Nitzschiapalea, it was difficult to recover cox1 sequences. Hamsher et al. (2011) suggested that rbcL-3P should be used as the primary marker for barcoding Sellaphora. Within Chlorophyta, recommended that tufA be adopted as the standard marker for the routine barcoding of green marine macroalgae (excluding the Cladophoraceae). Thus, genetic markers that have universal primers for PCR easy amplification and are variable enough for species discrimination should be further selected. Another issue is that the current reference database is incomplete so some molecular sequences cannot be matched to species level or even higher level. In this case, new DNA marker sequences of various taxa need to be added to the public reference library. In recent years, metabarcoding has developed as a new identification tool for environmental samples (Zimmermann et al., 2015; David and Jed, 2016). For example, Liu et al. (2020a) employed metabarcoding to identify forensic discrimination of drowning incidents. However, one substantial limitation of metabarcoding is exactly the limited reference sequences in public libraries that are used for read assignments (Liu et al., 2020b).
China has large sea areas and many freshwater lakes. Algae bloom in China is becoming a serious environmental problem (Qin et al., 2011; Duan et al., 2015). The cyanobacteria, Chlorophyta and Bacillariophyta, are the main microalgae for bloom. While most researchers focused on the cyanobacteria diversity study in China, the taxonomy of Chlorophyta and Bacillariophyta is lagged by molecular tools. Our previous studies have just identified some genera of Chlorophyta by DNA barcoding (Zou et al., 2016a). The identification of comprehensive species of diatoms from China is important for aquatic ecology.
In this study, we employed sequences of four genetic markers (COI, rbcL, SSU, and LSU) to barcode diatoms from a wide distribution of marine and freshwater environments from China by tree-, distance-, and character-based analytical methods. The available published diatom sequences were also incorporated into our new sequences for better analysis. We aim to (1) evaluate the efficiency of the four genetic markers in barcoding some genera within Bacillariophyta collected by us in this study; (2) contribute new reference sequences of multiple genetic markers of various diatoms species to the public database.
Materials and Methods
Sample Collection and Culture
We collected diatoms from both marine and lake environments in Qingdao, Nantong, Wuhan, and Zhoushan, China, where the locations in Qingdao, Nantong, Zhoushan, Lianyungang, and Ningbo were marine regions and the location in Wuhan was a lake region (Supplementary Table 1; Supplementary Figure 1). Following Andersen (2005), the diatom strains collected were isolated first. After isolation, the strains were cultured in a 250-ml flask containing a medium. Then, the cultured strains were identified using an electron microscope (40 × zoom), where we assigned the strains to species first by their general shape characteristics and then compared the micrographic observations with the barcoding identification. The detailed sampling information, including GenBank numbers, is shown in Supplementary Table 1 for all the diatom strains. The detailed sampling locations included in Supplementary Table 1 are shown in Supplementary Figure 1.
PCR Amplification, Sequencing, and Sequence Alignment
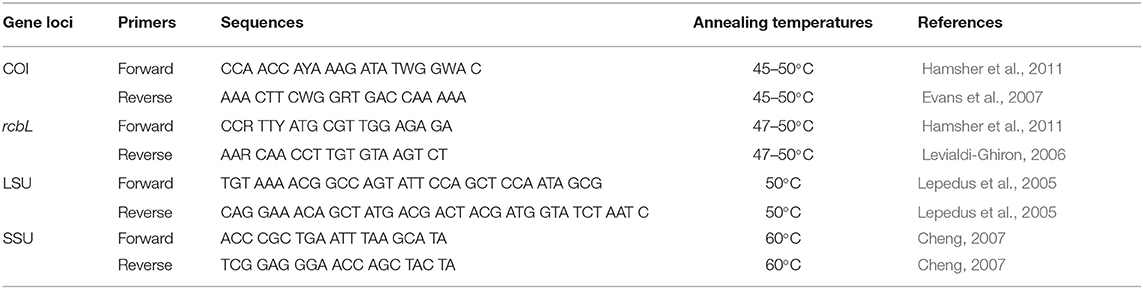
After DNA extraction with the Qiagen DNEasy Plant Extraction kit (Qiagen Inc., Valencia, CA, United States), each marker of COI, rbcL, SSU, and LSU was amplified with multiple primers (Table 1). PCR reactions and conditions also followed Zou et al. (2016a,b), with different annealing temperatures (Table 1). A 1.5% agarose gel was used to confirm PCR products producing a single band, and the products were sent to the Beijing Genomics Institute (BGI) for bidirectional sequencing. A set of publicly available sequences of diatom for each gene marker downloaded from GenBank was added to the new sequences produced in this study to be analyzed together. MAFFT (Katoh et al., 2009) was employed for alignment and trimming. The sequences of the four genetic markers were also joined together as an integrated target (COI + rbcL + SSU + LSU) for barcoding analysis.
Barcoding Assignments
The phylogenetic- distance- and character-based barcoding analyses were conducted for each of the four genetic markers and the combined fragment (COI + rbcL + SSU + LSU). Neighbor-joining (NJ), Bayesian, and maximum-likelihood (ML) were employed for phylogenetic barcoding analysis, where NJ trees were constructed based on the Kimura two-parameter (K2P) distance model (Hebert et al., 2003a) with MEGA (Tamura et al., 2011); Bayesian analyses were performed with MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003); and ML searches were performed with PhyML 3.0 (Guindon et al., 2010). jModeltest v.0.1.1 (Posada, 2008) was used to estimate the most appropriate models for both Bayesian and ML tree construction. The most appropriate models for rbcL, COI, LSU, and SSU were GTR + G, TVMef + I + G, GTR + G, and GTR + G, respectively. The distance-based barcoding analysis was performed for each of the four markers in MEGA (Tamura et al., 2011), where the intraspecific and interspecific distances were analyzed. The character-based analysis was performed for each of the four genetic markers and the combined target in Characteristic Attribute Organization System (CAOS) and CAOS-Analyzer (Sarkar et al., 2008). The datasets in NEXUS files and their DNA data matrices were produced in MacClade v4.0659 (Mindell, 1994), which were carried out in the CAOS system to get the characteristic attributes at the nucleotide positions (Bergmann et al., 2009).
Results
A total of 93 rbcL, 81 COI, 83 SSU, and 75 LSU sequences of diatom samples were obtained in this study (Supplementary Table 1). The new sequences from this study were submitted to GenBank with accession numbers MT684603-MT684690 (COI), MT644354-MT644461(LSU), MT680465-MT680611 (rbcL), and MT634264-MT634387 (SSU). Additional published sequences of rbcL, COI, SSU, and LSU from NCBI were downloaded and added to each new set of sequences (Supplementary Table 1).
Generally, the identification of species for each strain by micrographic observations was consistent with the identification by DNA barcoding of all the four gene loci, except that some potential cryptic species were found within some species. We also found some misidentifications of diatom sequences from public databases. The detailed barcoding results for each gene locus are shown below individually, where the names of species for our newly-obtained sequences in the phylogenetic trees were based on micrographics observations. Some potential cryptic species revealed in the phylogenetic trees are indicated as species names (I, II, III…). For the sequences downloaded from NCBI, their GenBank numbers are shown beside the name of a strain. The species for character-based analysis were from the phylogenetic trees for each gene locus, where the cryptic species were included.
A total of 11 species, 10 genera, 10 families, seven orders, and three classes were recovered from all the samples collected from each location (Supplementary Table 2). It was indicated that the diversity of species was high in Lianyungang, Jiangsu (Supplementary Figure 2).
rbcL Barcoding Assignments
The phylogenetic analysis of rbcL recovered a generally clear assignment resolution within Bacillariophyta (Figure 1). At the species level, most species analyzed were distinguished as separate clades. The rbcL sequences of the 11 species were newly obtained in this study, including Asteroplanus karianus, Cerataulina pelagica, Chaetoceros muellerii, Cyclotella sp., Entomoneis sp., Licmophora paradoxa, Melosira varians, Navicula bottnica, Phaeodactylum tricornutum, Skeletonema costatum, and Thalassiosira gravida. The strains within C. muellerii from different sea areas clustered together as one clade (Figure 1). For species whose data were downloaded from GenBank, most of them could be assigned as monophyletic clades, but some of them clustered together as one group (e.g., A. karianus, Asterionellopsis glacialis, and Asterionellopsis socialis). Sequences of L. paradoxa from this study clustered together with that from published papers. Sequences of Navicula ramosissima from this study were also separated clearly from a published sequence. Additionally, T. rotula, T. gravida, and Thalassiosira delicata, including samples from this study and GenBank, were closely related in the phylogenetic trees. At the genus level, all the genera that were analyzed clustered as monophyletic clades, except for Licmophora, Thalassiosira, and Asteroplanus, which gathered as paraphyletic clades, (Figure 1).
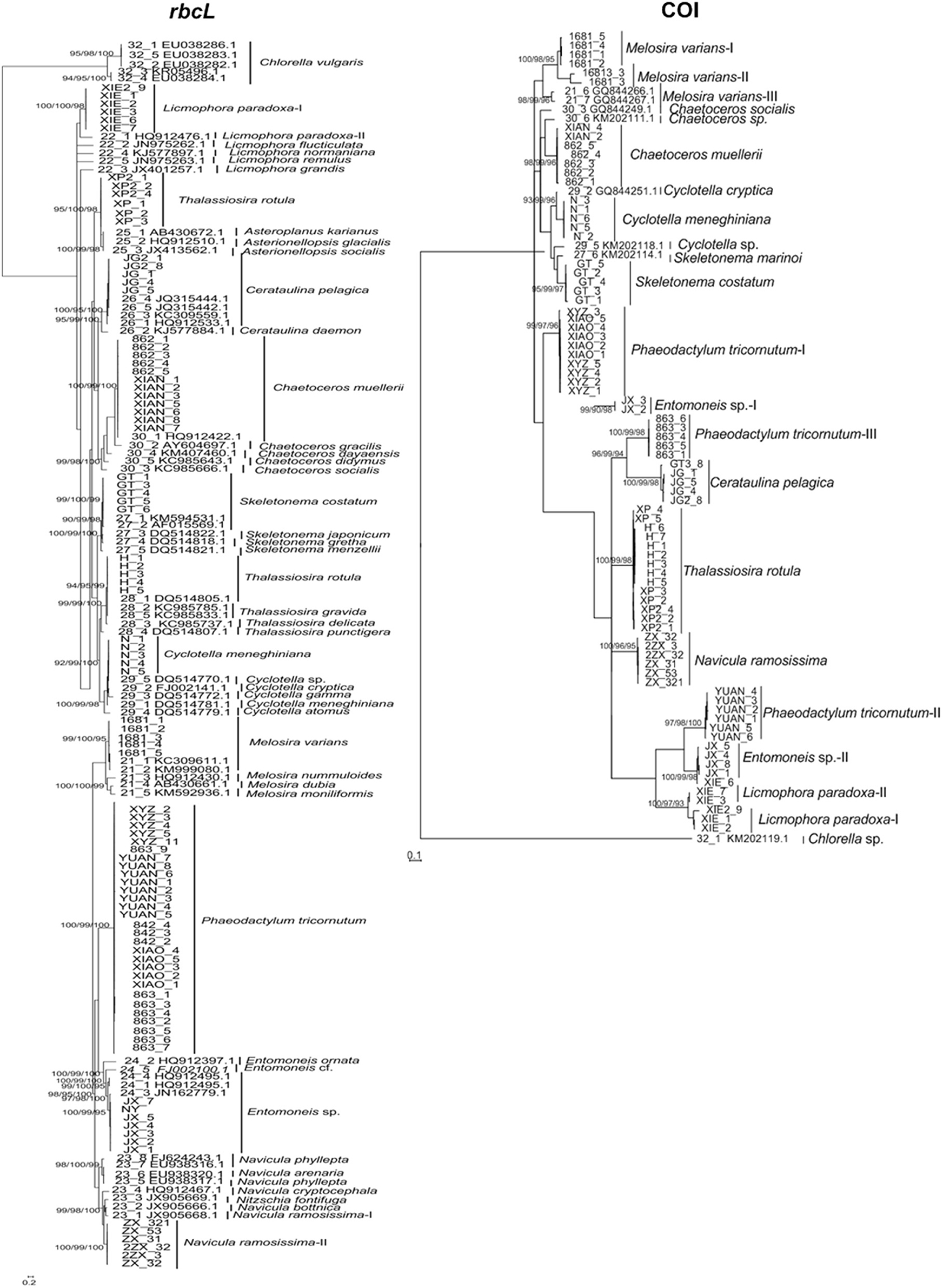
Figure 1. Bayesian phylogenetic trees for the rbcL and COI genes. The NJ, Bayesian, and Maximum Likelihood bootstraps are indicated for species clades recovered, where the order is NJ/Bayesian/ML. Some potential cryptic species revealed are indicated by species name (I, II, III…). For sequences downloaded from NCBI, their GenBank numbers are shown besides the name of strain.
The intraspecific and interspecific distances were calculated separately (Figure 2). Most interspecific distances were higher than 0.02. However, no apparent barcoding gap existed between the intraspecific and interspecific distances, and several species within certain genera were separated by interspecific distances lower than 0.02, such as Skeletonema and Thalassiosira. On the other hand, most of the species had intraspecific distances lower than 0.02, as expected.
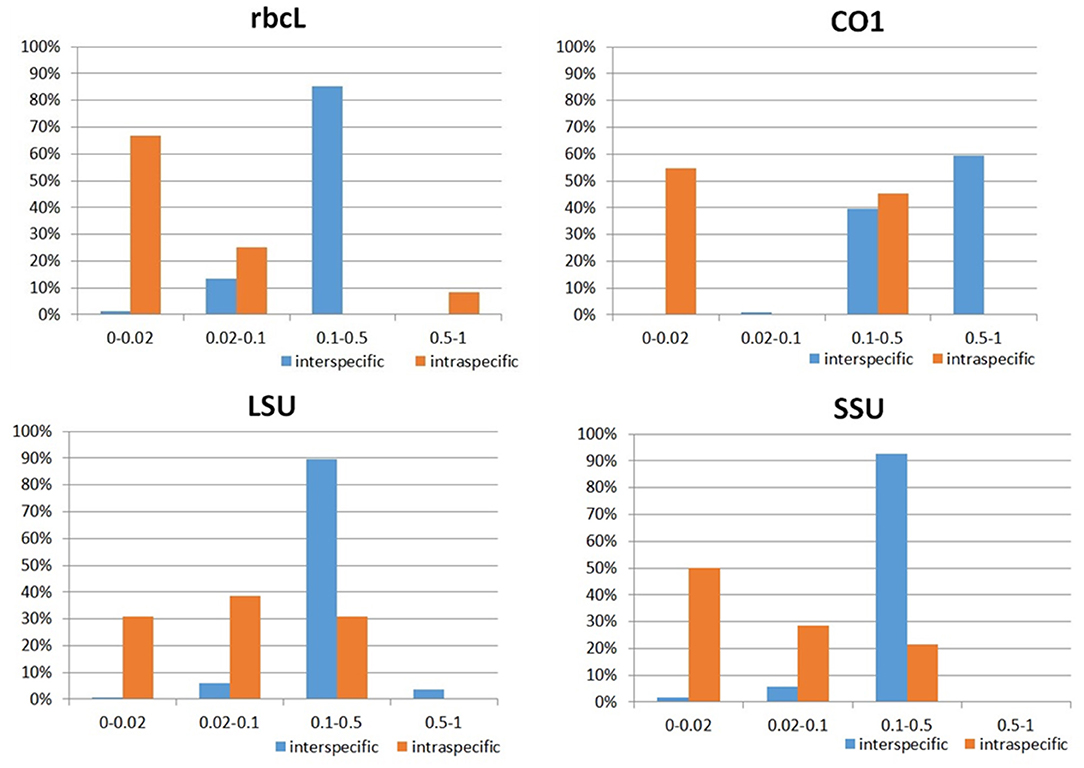
Figure 2. Proportion of intraspecific and interspecific distances for rbcL, COI, LSU, and SSU, respectively.
The character analysis showed general consistent taxonomic assignments with the phylogenetic-based identification (Table 2). Species that were clearly assigned as monophyletic clades in the phylogenetic trees were also separated with more than three characters attributes (CAs), such as M. varians, Melosira nummuloides, Licmophora normanina, and Entomonesis ornata. L. paradoxa and Navicula ramosissma, which were divided into two clades in the phylogenetic trees, were also separated as two clades, which showed more than three CAs. For species that could not be distinguished by tree-based barcoding, the character analysis also shows the same CAs for them, e.g., A. karianus, A. glacialis, and A. socialis.
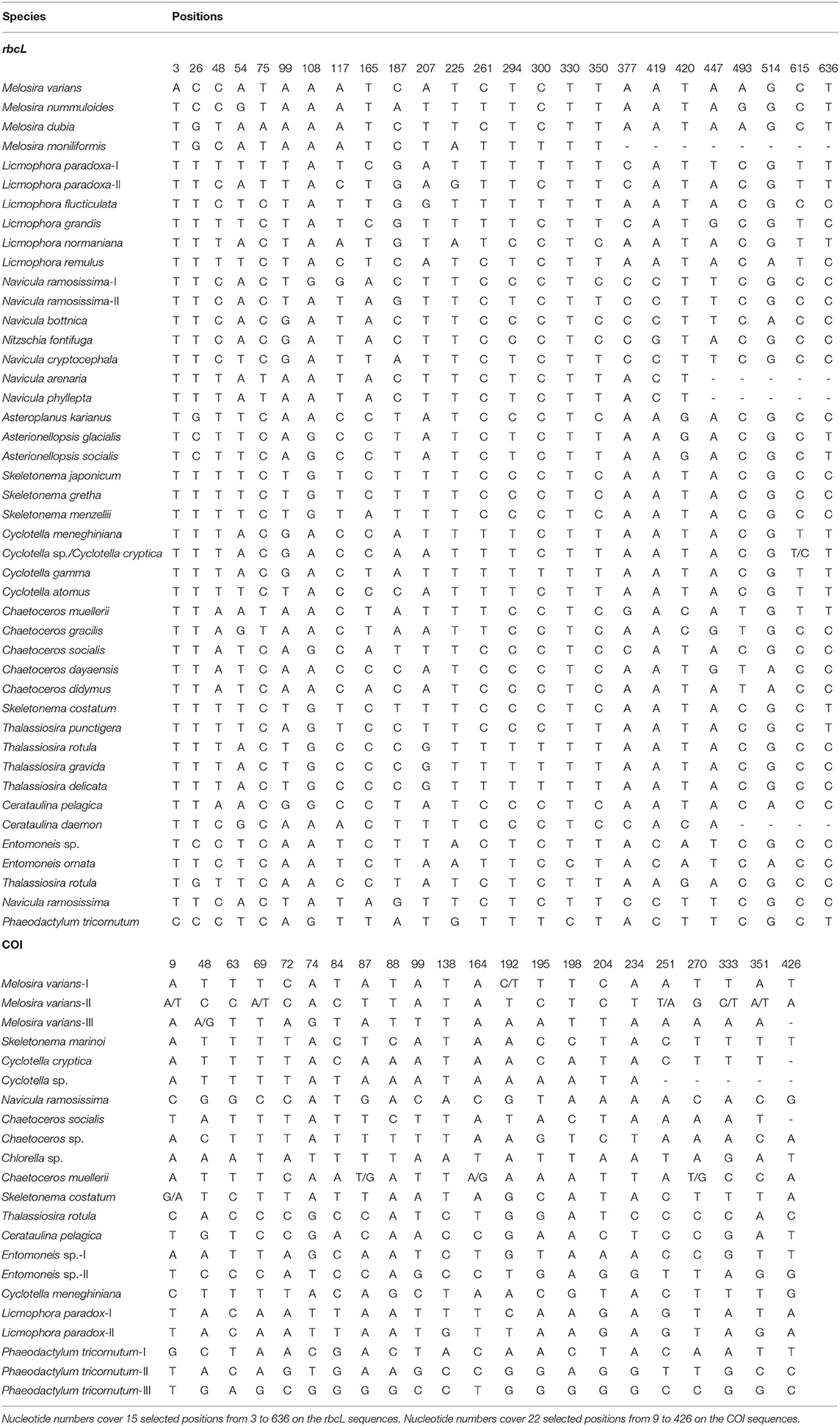
Table 2. Combinations of diagnostic nucleotides for species assignments in Figure 3 (LSU and SSU) by Characteristic Attributes Organization System (CAOS) analysis.
COI Barcoding Assignments
Most species included in COI were separated clearly in the NJ, ML, and Bayesian trees, and all formed monophyletic clades with high support, including species assigned from this study, such as C. muellerii, Cyclotella meneghiniana, and S. costatum (Figure 1). These species were also discriminated by more than three CAs from positions 9 to 426 of the COI fragments (Table 2). However, several species were divided into separate clades that could be cryptic species, e.g., M. varians and P. tricornutum. These potential cryptic species were also shown as separate clades in the character analysis where they were distinguished by more than three characters (Table 2). For example, we identified all strains of P. tricornutum I, II, III in Figure 1 as P. tricornutum by micrographic observation. However, all their sequences were assigned to separate clades, which did not cluster with any other species. Thus, we consider the separated clades as cryptic P. tricornutum species that need to be noticed and confirmed in future studies related to species identification. It was also shown that the separated clades of P. tricornutum were from different sea areas, e.g., the strains of P. tricornutum II were from Zhoushan, Zhejiang, and the strains of P. tricornutum III were from Lianyungang, Jiangsu. At the genus level, for all the genera analyzed, Chaetoceros, Cyclotella, and Skeletonema were assigned as paraphyletic clades (Figure 1).
Compared with rbcL, the COI marker also produced higher distances for both intraspecific and interspecific comparisons, and no gap appeared between the intraspecific and interspecific distances (Figure 2). Almost all the interspecific distances were higher than the threshold of 0.02, except for T. rotula and A. karianus, which had an interspecific distance of 0.0189. For the intraspecific distance, three species (M. varians, L. paradoxa, and Entomoneis sp) had values higher than 0.02, and all the rest had values lower than 0.02.
LSU Barcoding Assignments
At the species level, while most species clustered as monophyletic clades, several species were divided into separate groups (e.g., Thalassiosia rotula) and available GenBank sequences clustered as one group (e.g., T. gravida, T. delicate) (Figure 3). These phylogenetic assignments were consistent with the character analysis, where C. pelagica and Entomoneis. sp. and T. rotula were also separated as different clades with more than three CAs, and T. gravida, T. delicate, and T. punctigera showed the same CAs from positions 35 to 532 of the fragment (Table 3). At the genus level, almost all the genera clustered as monophyletic clades except for the cryptic species in Thalassiosira (Figure 3).
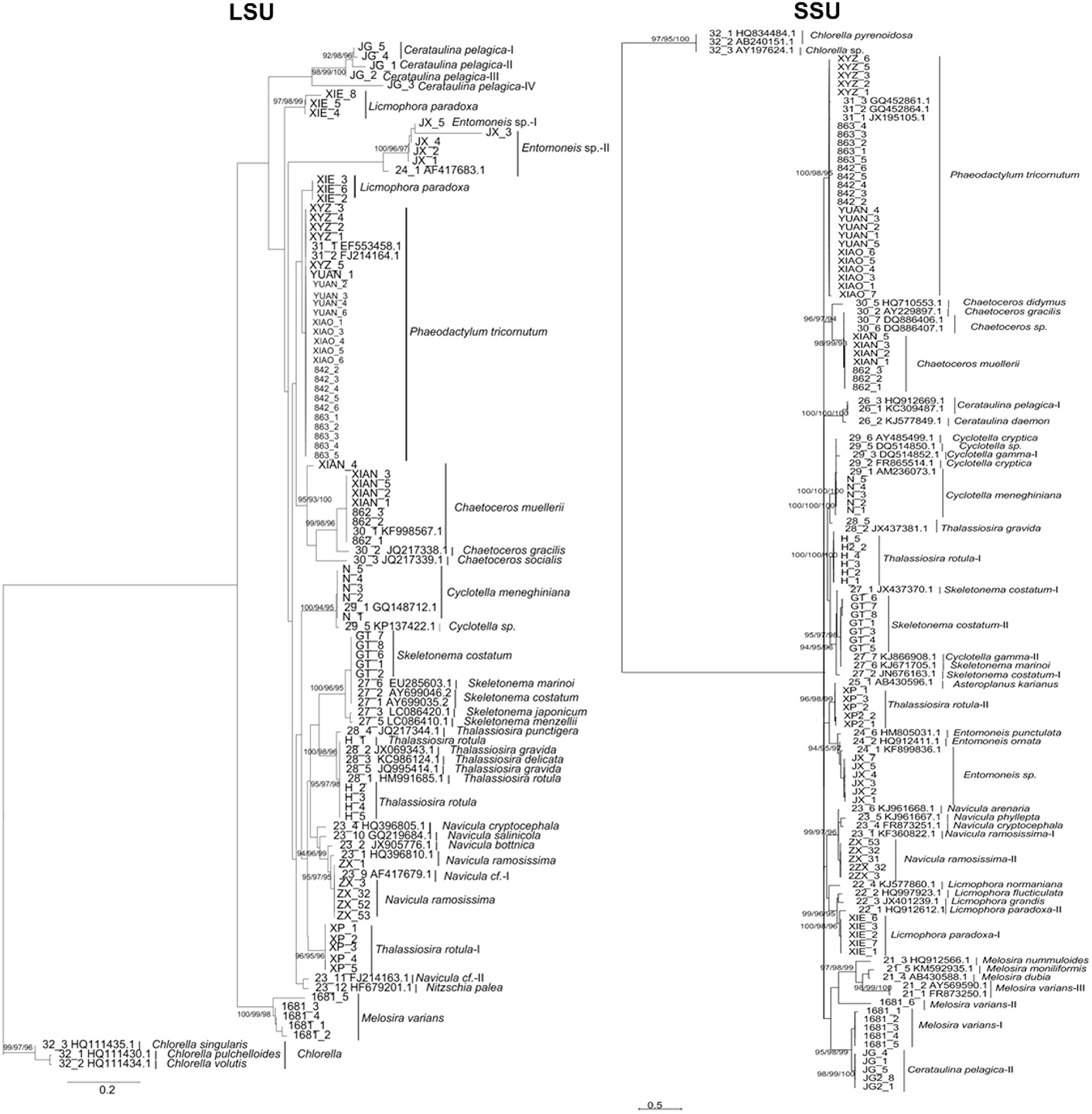
Figure 3. Bayesian phylogenetic trees for the LSU and SSU genes. The NJ, Bayesian and Maximum Likelihood bootstraps are indicated for species clades recovered, where the order is NJ/Bayesian/ML. Some potential cryptic species revealed are indicated by species name (I, II, III…). For sequences downloaded from NCBI, their GenBank numbers are shown besides the name of strain.
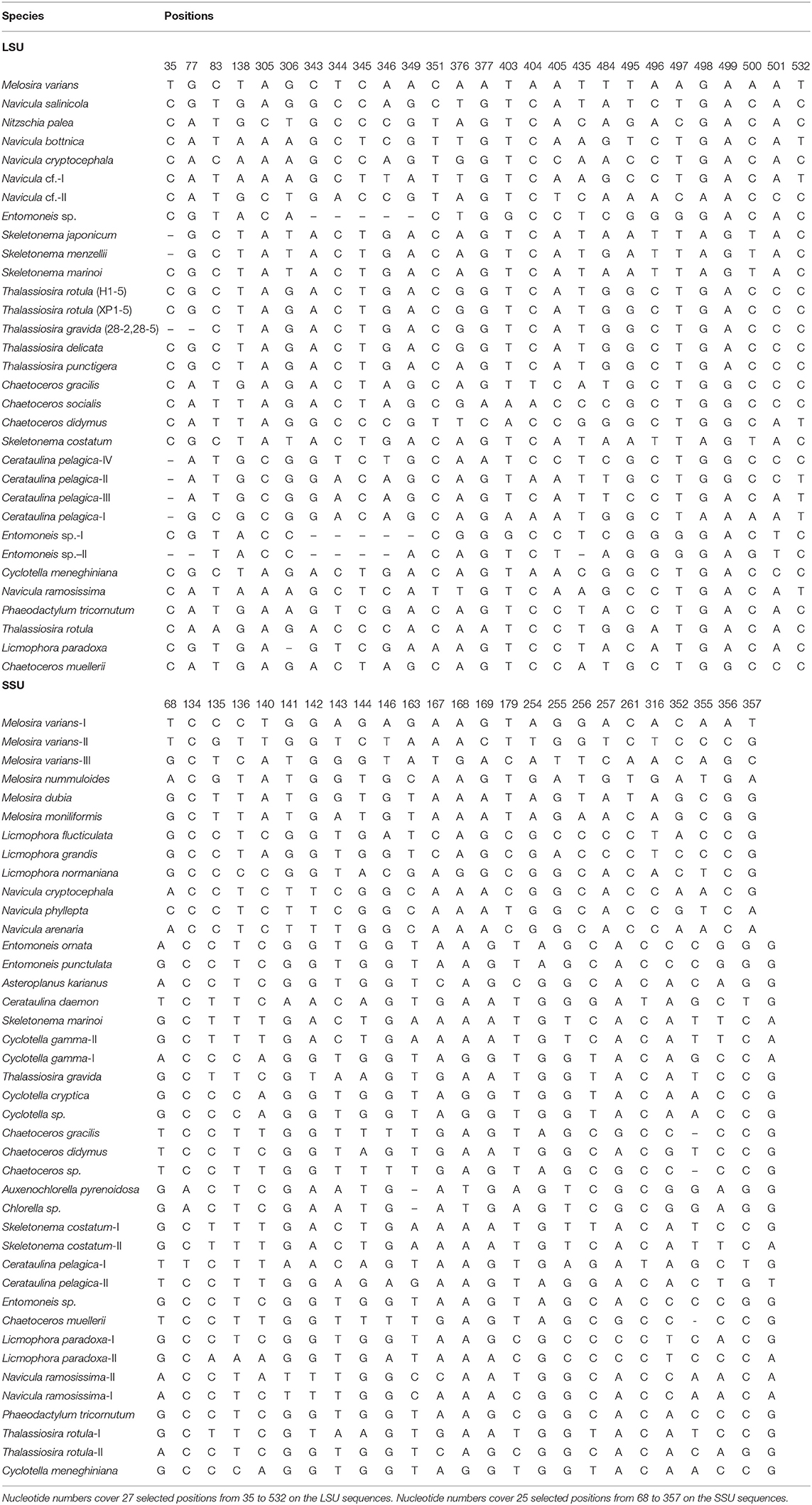
Table 3. Combinations of diagnostic nucleotides for species assignments in Figure 3 (LSU and SSU) by CAOS analysis.
For LSU, most species (96%) had interspecific distances above 0.02 (Figure 2). Of the 15 species, 6 had intraspecific distances higher than 0.02, and 9 had intraspecific distances lower than 0.02. Thus, there was an overlap between the intraspecific and interspecific distances.
SSU Barcoding Assignments
In comparison with rbcL, COI, and LSU, SSU produced less resolved tree topologies (Figure 3), where some species could not be separated clearly as phylogenetic clades (e.g., Chaetoceros gracilis and C. muellerii). However, C. gracilis and C. muellerii, and C. cryptica and C. cryptica were clearly discriminated by more than three CAs (Table 3). Some species that were divided into several separate clades in the phylogenetic trees also differed from each other by more than three CAs, such as Melosira vaians, Cyclotella gamma, and L. paradoxa (Table 3). At the genus level, many of the genera analyzed were assigned as paraphyletic clades.
A portion of (97%) the species had interspecific distances above 0.02 (Figure 2). However, some species that could not be separated by the phylogenetic trees also had interspecific distances lower than 0.02. Thus, it is clear that there is much overlap between the intraspecific and interspecific distances.
Combined Barcoding Assignments
The phylogenetic and distance-based barcoding of the combination of rbcL, COI, LSU, and SSU was also conducted (Figure 4) for further verification. The samples that had all sequences from the four genes were collected for the combined analysis. The distance-based method was not used, because the number of samples analyzed is limited. It was indicated that the phylogenetic tree of the combined sequences showed a clear topological structure. The species analyzed were separated as monophyletic clades with higher support.
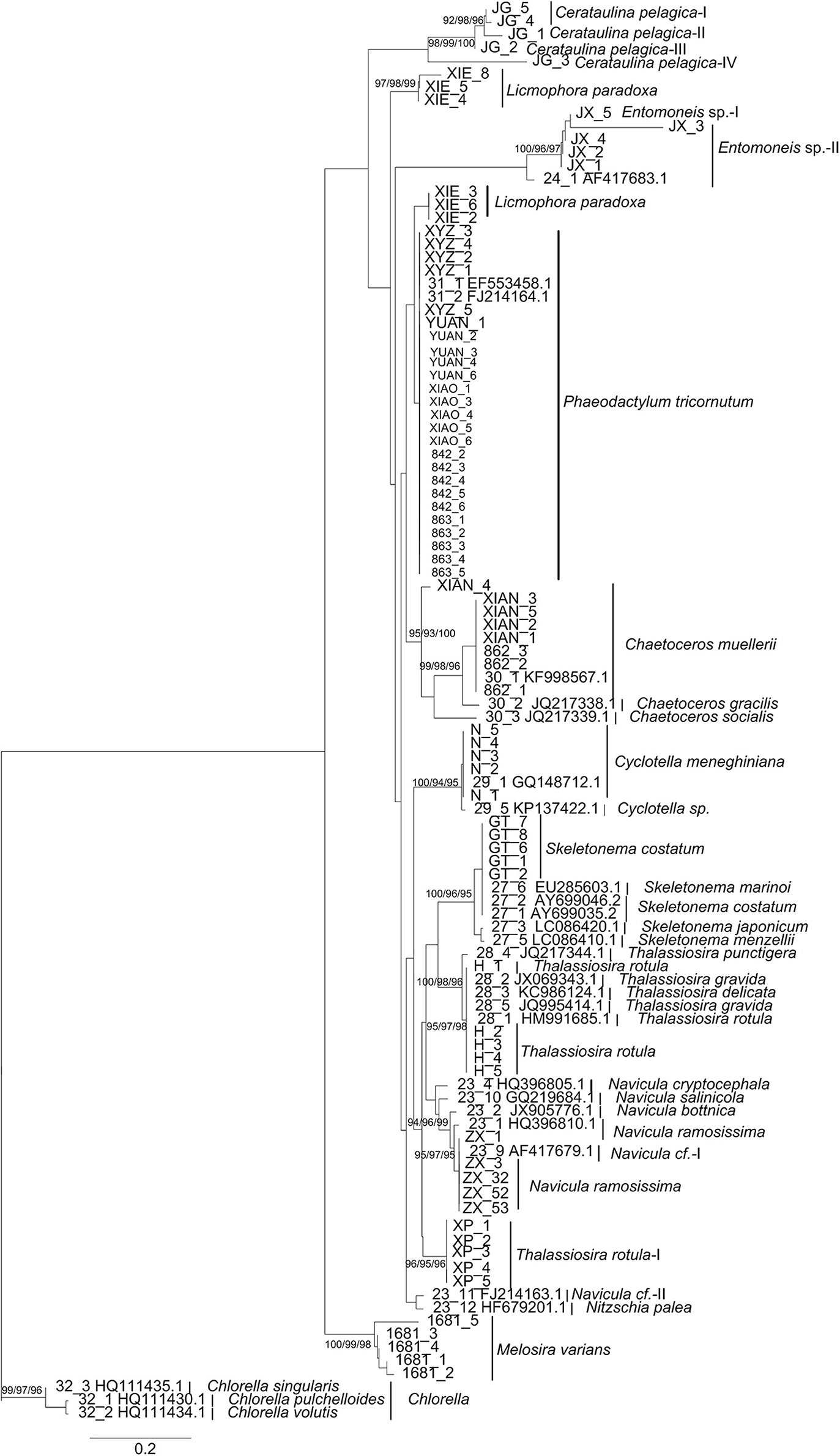
Figure 4. Bayesian phylogenetic tree for combined gene sequences of the four gene loci. The NJ, Bayesian, and Maximum Likelihood bootstraps are indicated for species.
Discussion
Although diatom species are distributed globally and play an important role in aquatic ecology (Zalack et al., 2010), many remain undiscovered or unassigned yet (Smetacek, 1999). The diatom diversity needs to be investigated globally, especially for courtiers that have large areas of water. DNA barcoding has provided a convenient tool for species identification (Hebert et al., 2003a,b; Zou et al., 2016a,b). Here, we employed four genetic markers for assigning diatoms from China with phylogenetic, distance, and character-based methods.
The identification of species for each strain by micrographic observations was generally consistent with the identification through phylogenetic-based trees by DNA barcoding. For phylogenetic-based barcoding, rbcL, COI, and LSU were able to discriminate most of the species clearly within Bacillariophyta. At the species level, both rbcL and COI phylogenetic barcoding analyses showed better resolution in discriminating all the species. Nevertheless, some available sequences from NCBI could not be separated in the rbcL and COI phylogenetic trees, which suggests that some of the sequences submitted to NCBI are possibly misidentified. Additionally, all the four genetic markers assigned some species as cryptic, which were divided into several monophyletic clades in the phylogenetic trees. The character barcoding analysis and phylogenetic barcoding analysis obtained consistent species identification accordingly. All the species identified as clearly monophyletic clades in the phylogenetic trees were also assigned as separate clades by character analysis with more than three CAs. The potential cryptic species revealed by the phylogenetic analysis were also divided into separate clades in the character analysis with more than three CAs. All the cryptic species need to be noted in future studies. While barcoding analytical methods are argued, our study suggests that the combination of phylogenetic and character analyses gives more accurate species identification results.
All the results provide us with the understanding that different barcoding genetic markers give different identification resolutions for diatoms at both high and low taxonomic levels. By comparison, rbcL, LSU, and COI proved more effective in barcoding diatoms, which is partly consistent with the previous results that rbcL should be used as the primary marker for diatom barcoding (Hamsher et al., 2011; MacGillivary and Kaczmarska, 2011). For example, MacGillivary and Kaczmarska (2011) suggested that a small rbcL fragment could be used for a dual-locus barcode with the more variable 5.8S + ITS-2 to discriminate diatom species, and Guo et al. (2015) showed that rbcL performed well in clustering some lower taxa. In Guo et al. (2015), it was also demonstrated that genetic loci had different assignment efficiency for different genera. For example, the COI region could just discriminate some genera within Bacillariophyceae, and ITS was a potential marker for barcoding some genera of Thalassiosirales (Cyclotella, Skeletonema, and Stephanodiscus). In our study, it was also indicated that different genetic loci had different identification efficiency at the genus level. Generally, LSU performed well in barcoding most of the genera within Bacillariophyta, but rbcL, COI, and SSU could not assign some of the genera as monophyletic clades, e.g., the Licmophora in rbcL phylogenetic analysis and Cyclotella in COI phylogenetic analysis. On the other hand, rbcL, COI, and SSU performed well in barcoding diatoms at the species level. Thus, we suggest the combination of rbcL, COI, LSU, and SSU for DNA barcoding the 11 genera of diatoms, since they are easily amplified by PCR and have enough variation for identifying different genera. The efficiency of barcoding entire Bacillariophyta should be tested by employing more species belonging to more different genera. We also merged the four genetic markers to conduct the phylogenetic and character analysis to verify the identification of species. The NJ, ML, and Bayesian trees of the merged sequence assigned all the species as clear monophyletic clades. The clear topology from the combined data was possible because the samples analyzed were limited. But the analysis from the combined data was generally consistent with that from the single gen. For the distance-based approach, the genetic distance of 0.02 between interspecific and intraspecific comparisons is proposed as a criterion for barcoding (Hebert et al., 2003a,b), which means that the intraspecific distance should be lower than 0.02 and the interspecific distance should be higher than 0.02. However, for all the genetic markers, some interspecific distances were lower than 0.02 and some intraspecific distances were higher than 0.02, without an obvious distance gap between the interspecific and intraspecific distances. This suggests that the distance criterion of 0.02 cannot always discriminate the species of diatoms. Thus, our study provides information that the phylogenetic and character-based methods are more effective for barcoding diatoms. In future studies, we can try to use other distance-based tools for barcoding diatoms, such as ABGD or Spider (Boyer et al., 2012; Puillander et al., 2012). However, in our previous studies, it was also indicated that the phylogenetic and character-based barcoding methods showed more advantages than the ABGD method for barcoding Chlorophyta (Zou et al., 2016a). Thus, in our opinion, we recommend the phylogenetic and character-based barcoding approaches for barcoding microalgae.
Here, we perform a comprehensive diversity investigation of diatoms from China, which will greatly contribute to the classification of diatoms. Most of the samples were collected from sea areas of the Yellow Sea and East China sea where algae bloom often occurs. The rest of the samples were collected from typical freshwater lakes in China. Therefore, the samples studied could represent the diverse diatom in China. Compared with previous studies that just used limit genetic markers or analytical methods (Mónica and Kaczmarska, 2009, 2010; Hamsher et al., 2011), we discriminated most diatom species clearly and revealed some cryptic species. For some strains from different habits (e.g., different marine sea areas and lakes) within one species, there was not much difference in their identification, such as C. muellerii and T. rotula, but for P. tricornutum, the strains from different sea areas were revealed as cryptic species. These suggest that the external habits possibly also contribute to the species diversity of diatoms. However, our study focused on accurate species identification and the complementary of diatom sequences to reference databases. The amount of the samples studied was not substantial to conduct a comprehensive diatom diversity investigation in China. In future studies, we will employ metabarcoding to monitor diatom diversity by Next Generation Sequence with a large amount of sequences. The available diatom sequences in public databases were also incorporated into our newly obtained sequences, the comprehensive analysis of which showed some possible identification errors of public diatom sequences. In conclusion, our study reports the accurate identification of diatoms from China comprehensively by DNA barcoding, which is important for well-understanding algae blooms and aquatic ecology.
Finally, with the development of Next Generation Sequencing, metabarcoding is becoming more efficient for species assignment with markers such as 16S, COI, and 18S, etc (Gogarten et al., 2020). However, metabarcoding often has a bias in accurate species identification for a large amount of reads because of incomprehensible reference sequences (Rachel et al., 2019; Gogarten et al., 2020). It is important to complement the reference sequences in public databases with more gene sequences of more species. In our study, the new sequences of multiple markers from a large number of samples provide much assistance for metabarcoding diatoms.
Data Availability Statement
The new sequences from this study were submitted to the GenBank Barcode database with accession numbers MT684603-MT684690 (COI), MT644354-MT644461 (LSU), MT680465-MT680611 (rbcL) and MT634264-MT634387 (SSU).
Author Contributions
SZ designed the experiment, analyzed the data, and wrote the manuscript. YB and XW conducted the experiment. CW helped to revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The financial support from the China Postdoctoral Science Foundation (2014M561661 and 2015T80558) and the Fundamental Research Funds for the Central Universities (KJQN201742 and Y0201600141) was gratefully acknowledged. This project was supported by the Bioinformatics Center of Nanjing Agricultural University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.698331/full#supplementary-material
Supplementary Figure 1. Sampling location from the China coast.
Supplementary Figure 2. Species diversity for each location at the species, genus, and family levels.
Supplementary Table 1. Genbank numbers of samples used in this study. Genbank numbers in bold are from published papers.
Supplementary Table 2. Statistics of taxa identified in various taxonomic level for every location of sample collection.
References
Amato, A., Kooistra, W. H. C. F., Levialdi, G. J. H., Mann, D. G., Pröschold, T., and Montresor, M. (2007). Reproductive isolation among sympatric cryptic species in marine diatoms. Protist 158, 193–207. doi: 10.1016/j.protis.2006.10.001
Behnke, A., Friedl, T., Chepurnov, V. A., and Mann, D. G. (2004). Reproductive compatibility and rDNA sequence analyses in the Sellaphora pupula species complex (Bacillariophyta). J. Phycol. 40, 193–208. doi: 10.1046/j.1529-8817.2004.03037.x
Bergmann, T., Hadrys, H., Breves, G., and Schierwater, B. (2009). Character-based DNA barcoding: a superior tool for species classification. Berl. Munch. Tierarztl. 122, 446–450.
Beszteri, E., Ács, E., and Medlin, L. K. (2005). Ribosomal DNA sequence variation among sympatric strains of Cyclotella meneghiniana complex (Bacillariophyceae) reveals cryptic diversity. Protist 156, 317–333. doi: 10.1016/j.protis.2005.07.002
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., and Jabbari, K. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Boyer, S., Brown, S. D. J., Malumbres-Olarte, J., Vink, C. J., and Cruickshank, R. H. (2012). Spider: an R package for the analysis of species identity andevolution, with particular reference to DNA barcoding. Mol. Ecol. Resour. 12, 562–565. doi: 10.1111/j.1755-0998.2011.03108.x
Casteleyn, G., Leliaert, F., Backeljau, T., Debeer, A. E., Kotaki, Y., Rhodes, L., et al. (2010). Limits to gene flow in a cosmopolitan marine planktonic diatom. P Natl. Acad. Sci. USA 107, 12952–12957. doi: 10.1073/pnas.1001380107
Cheng, J. F. (2007). The Morphylogy, Genetic Difference and Phylogenetic Analysis of Several Typical Nanoplanktonic Diatom Species in China Sea [D]. Xiamen University.
Dam, H. V., Mertens, A., and Sinkeldam, J. (1994). A coded checklist and ecological indicator values of freshwater diatoms from the Netherlands. Neth. J. Aquat. Ecol. 28, 117–133. doi: 10.1007/BF02334251
David, M. N., and Jed, A. F. (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1:16005. doi: 10.1038/nmicrobiol.2016.5
Duan, H., Loiselle, S. A., and Li, Z. (2015). Distribution and incidence of algae blooms in Lake Tai. Aquat Sci. 77, 9–16. doi: 10.1007/s00027-014-0367-2
Evans, K. M., Wortley, A. H., and Mann, D. G. (2007). An Assessment of Potential Diatom “Barcode” Genes (cox1, rbcL, 18S and ITS rDNA) and their Effectiveness in Determining Relationships in Sellaphora (Bacillariophyta). Protist 158, 349–364. doi: 10.1016/j.protis.2007.04.001
Gogarten, J. F., Calvignac-Spencer, S., Nunn, C. L., and Saiepour, N. (2020). Metabarcoding of eukaryotic parasite communities describes diverse parasite assemblages spanning the primate phylogeny. Mol. Ecol. Resour. 20, 204–215. doi: 10.1111/1755-0998.13101
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, L., Sui, Z., Zhang, S., Ren, Y., and Liu, Y. (2015). Comparison of potential diatom ‘barcode’ genes (the 18S rRNA gene and ITS, COI, rbcL) and their effectiveness in discriminating and determiningspecies taxonomy in the Bacillariophyta. Int. J. Syst. Evol. Micr. 65, 1369–1380. doi: 10.1099/ijs.0.000076
Hamsher, S. E., Evans, K. M., Mann, D. G., and Aloisie, P. (2011). Barcoding diatoms: exploring alternatives to coi-5p. Protist 162, 405–422. doi: 10.1016/j.protis.2010.09.005
Hebert, P. D. N., Cywinska, A., Ball, S.L., and deWaard, J. R. (2003a). Biological identifications through DNA barcodes. Proc. RSoc. Lond B 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert, P. D. N., Ratnasingham, S., and deWaard, J. R. (2003b). Barcoding animal life: cytochrome c oxidase subunit 1divergences among closely related species. P Roy Soc. Lond. BBio. 270:S96. doi: 10.1098/rsbl.2003.0025
Katoh, K., Asimenos, G., and Toh, H. (2009). In bioinformatics for DNA sequence analysis. Methods Mol Biol 537:39–64. doi: 10.1007/978-1-59745-251-9_3
Kawecka, B., and Olech, M. (1993). Diatom communities in the Vanishing and Ornithologist Creek, King George Island, South Shetlands, Antarctica. Hydrobiologia 269, 327–333. doi: 10.1007/BF00028031
Lepedus, H., Schlensog, M., and Muller, L. (2005). Function and molecular organisation of photosystem II in vegetative buds ad mature needles of Norway spruce during the dormancy. Biologia 60, 89–92.
Levialdi-Ghiron, J. H. (2006). Plastid phylogeny and chloroplast inheritance in the planktonic pennate dia-tom Pseudo-nitzschia (Bacillariophyceae). Doctoral thesis, Universita Degli Studi Di Messina.
Li, X., Yang, Y., Robert, J., Henry, R. M., Wang, Y., and Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Liu, M., Zhao, Y., Sun, Y., Li, Y., Wu, P., Zhou, S., et al. (2020b). Comparative study on diatom morphology and molecular identification in drowning cases. Forensic. Sci. Int. 317:110552. doi: 10.1016/j.forsciint.2020.110552
Liu, M., Zhao, Y., Sun, Y., Wu, P., Zhou, S., and Ren, L. (2020a). Diatom DNA barcodes for forensic discrimination of drowning incidents. FEMS Microbiol. Lett. 367:145. doi: 10.1093/femsle/fnaa145
MacGillivary, M. L., and Kaczmarska, I. (2011). Survey of the efficacy of a short fragment of the rbcL gene as a supplemental DNA barcode for diatoms. J. Eukaryot. Microbiol. 58, 529–536. doi: 10.1111/j.1550-7408.2011.00585.x
Medlin, L. K., Elwood, H. J., Stickel, S., and Sogin, M. L. (1991). Morphological and genetic variation within the diatom Skeletonema costatum (Bacillariophyta): evidence for a new species, Skeletonema pseudocostatum. J. Phycol. 27, 514–524. doi: 10.1111/j.0022-3646.1991.00514.x
Mindell, D. P. (1994). MacClade: analysis of phylogeny and character evolution. Auk 111, 1035–1036. doi: 10.2307/4088848
Mónica, B. J., and Kaczmarska, I. (2010). Barcoding of diatoms: nuclear encoded ITS Revisited. Protist 161, 7–34. doi: 10.1016/j.protis.2009.07.001
Mónica, B. J. M., and Kaczmarska, I. (2009). Barcoding diatoms: is there a good marker?. Mol. Ecol. Resour. 9, 65–74. doi: 10.1111/j.1755-0998.2009.02633.x
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Potapova, M., and Charles, D. F. (2007). Diatom metrics for monitoring eutrophication in rivers of the United States. Ecol. Indicat. 7, 48–70. doi: 10.1016/j.ecolind.2005.10.001
Poulíčková, A., Veselá, J., Neustupa, J., and Škaloud, P. (2010). Pseudocryptic diversity versus cosmopolitanism in diatoms: a case study on Navicula cryptocephala Kütz. (Bacillariophyceae) and morphologically similar taxa. Protist 161, 353–369. doi: 10.1016/j.protis.2009.12.003
Puillander, N., Lambert, A., Brouillet, S, and Achaz, G. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Qin, B. Q., Zhu, G. W., Gao, G., et al. (2011). A drinking water crisis in Lake Taihu, China: linkage to climatic variability and lake management. Environ. Manage. 451, 105–112. doi: 10.1007/s00267-009-9393-6
Rach, J., DeSalle, R., Sarkar, I. N., Schierwater, B., and Hadrys, H. (2008). Character-based DNA barcoding allows discrimination of genera, species and populations in Odonata. PRoy Soc B-Biol. Sci. 275, 237–247. doi: 10.1098/rspb.2007.1290
Rachel, S. M., Emily, E. C., Teia, S., Zack, G., Dannise, R. R., Sabrina, S., et al. (2019). The California EnvironMantel DNA “CALeDNA” Program. bioRxiv. Cold Spring Harbor Laboratory.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sarkar, I. N., Planet, P. J., and Bael, T. E. (2002a). Characteristic attributesin cancer microarrays. J. Biomed. Inform. 35, 111–122. doi: 10.1016/S1532-0464(02)00504-X
Sarkar, I. N., Planet, P. J., and Desalle, R. (2008). CAOS software for use in character-based DNA barcoding. Mol. Ecol. Resour. 8, 1256–1259. doi: 10.1111/j.1755-0998.2008.02235.x
Sarkar, I. N., Thornton, J., Planet, P. J., Schierwater, B., and DeSalle, R. (2002b). A systematic method for classification of novel homeoboxes. Mol. Phylogenet. Evol. 24, 388–399. doi: 10.1016/S1055-7903(02)00259-2
Sarno, D., Kooistra, W. H. C. F., Medlin, L. K., Percopo, I., and Zingone, A. (2005). Diversityin the genus Skeletonema (Bacillariophyceae). II. an assessment of the taxonomy of s. costatum-like species with the description of four new species. J. Phycol. 41, 151–176. doi: 10.1111/j.1529-8817.2005.04067.x
Smetacek, V. (1999). Diatoms and the ocean carbon cycle. Protist 150, 25–32. doi: 10.1016/S1434-4610(99)70006-4
Spaulding, S. A., and McKnight, D. M. (1999). “Assessing ecological conditions in rivers and streams with diatoms,” in The Diatoms: Applications to the Environmental and Earth Sciences, eds E. P. Stoermer and J. P. Smol (Cambridge: Cambridge University Press), 245–260.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Trobajoa, R., Mannb, D. G., Claveroa, E., Evans, K. M, Vanormelingenc, P., et al. (2011). The use of partial cox1, rbcL and LSU rDNA sequences for phylogenetics and species identification within the Nitzschia palea species complex (Bacillariophyceae). Eur. J. Phycol. 45, 413–425. doi: 10.1080/09670262.2010.498586
Vanelslander, B., Créach, V., Vanormelingen, P., Ernst, A., Chepurnov, V. A., Sahan, E., et al. (2009). Ecological differentiation between sympatric pseudocryptic species in the estuarine benthic diatom Navicula phyllepta (Bacillariophyceae). J. Phycol. 45, 1278–1289. doi: 10.1111/j.1529-8817.2009.00762.x
Zalack, J. T., and Smucker, N. J., and Vis, M.L. (2010). Development of a diatom index of biotic integrity for acid mine drainage impacted streams. Ecol. Indicat. 10, 287–295. doi: 10.1016/j.ecolind.2009.06.003
Zhang, Y. G., Zhou, X. K., Guo, J. W., et al. (2018). Bacillus tamaricis sp. nov. an alkaliphilic bacterium isolated from a Tamarix cone soil. Int. J. Syst. Evol. MICR 68, 558–563. doi: 10.1099/ijsem.0.002543
Zimmermann, J., GlCkner, G., Jahn, R., Enke, N., and Gemeinholzer, B. (2015). Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. MolEcol. Resourc. 15, 526–542. doi: 10.1111/1755-0998.12336
Zou, S., Fei, C., Song, J. M., Bao, Y., He, M., and Wang, C. (2016a). Combining and comparing coalescent, distance and character-based approaches for barcoding microalgaes: a test with Chlorella-Like Species (Chlorophyta). Plos ONE 11:e0153833. doi: 10.1371/journal.pone.0153833
Zou, S., Fei, C., Wang, C., Gao, Z., Bao, Y., He, M., et al. (2016b). How DNA barcoding can be more effective in microalgae identification: a case of cryptic diversity revelation in Scenedesmus (Chlorophyceae). Sci. Rep. 6:36822. doi: 10.1038/srep36822
Keywords: DNA barcoding, species diversity, diatom, COI, phylogenetic analysis, RBCL, LSU
Citation: Zou S, Bao Y, Wu X and Wang C (2021) DNA Barcoding Diatoms From China With Multiple Genes. Front. Mar. Sci. 8:698331. doi: 10.3389/fmars.2021.698331
Received: 21 April 2021; Accepted: 13 September 2021;
Published: 03 November 2021.
Edited by:
Wen-Jun Li, Sun Yat-sen University, ChinaReviewed by:
Jian-Wei Guo, Kunming Institute of Botany, Chinese Academy of Sciences (CAS), ChinaMo Minghe, Yunnan University, China
Copyright © 2021 Zou, Bao, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanmei Zou, em91c205MTJAbmphdS5lZHUuY24=; Changhai Wang, Y2h3YW5nQG5qYXUuZWR1LmNu
 Shanmei Zou
Shanmei Zou Yachao Bao
Yachao Bao