- 1Zoological Operations, SeaWorld Parks and Entertainment, Inc., Orlando, FL, United States
- 2Department of Biostatistics, Fielding School of Public Health, University of California, Los Angeles, Los Angeles, CA, United States
- 3Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
- 4Species Preservation Laboratory, SeaWorld San Diego, San Diego, CA, United States
- 5SeaWorld Orlando, Orlando, FL, United States
- 6SeaWorld San Diego, San Diego, CA, United States
- 7SeaWorld San Antonio, San Antonio, TX, United States
- 8Miami Seaquarium, Miami, FL, United States
Accurate identification of individual ages within wild bottlenose dolphins (Tursiops truncatus) is critical for determining population health and the development of population management strategies. As such, we analyzed DNA methylation (DNAm) patterns by applying a custom methylation array (HorvathMammalMethyl40) to both blood (n = 140) and skin samples (n = 87) from known age or approximate age (0–57 years) bottlenose dolphins. We present three bottlenose dolphin specific age estimation clocks using combined blood and skin [48 CpGs, R = 0.93, median absolute error (MAE) = 2.13 years], blood only (64 CpGs, R = 0.97, error = 1.46 years) and skin only (39 CpGs, R = 0.95, error = 2.53). We characterized individual cytosines that correlate with sex and age in dolphins and developed a sex estimator based on 71 CpGs that predicts the sex of any odontocete species with 99.5% accuracy. The presented epigenetic clocks are expected to be useful for conservation efforts and for determining if anthropogenic events affect aging rates in wild populations.
Introduction
Accurate age estimation of wild cetaceans is an important component of any population health assessment and is critical for the development of management plans designed to help wild populations in need (Robeck et al., 2021b). For bottlenose dolphins, standard age estimations of wild animals rely on a combination of techniques including animal length (allometry), long term “capture and recapture” of animals using photo identification, and the counting of tooth growth layer groups (GLGs). Age estimators based on allometry lose accuracy once physical maturity has been reached and surpassed (McFee, 2012; Beal et al., 2019). Although long term photo identification studies have produced the most robust and accurate data concerning life history of a few populations of bottlenose dolphins, these studies face limitations, e.g., the inability to accurately define animals that had already reached physical maturity at the start of the surveys (Würsig and Jefferson, 1990; Wells, 2014). Another challenge faced by long-term photo identification programs is their high economic costs and long-term time commitments. These limitations render them less efficient for rapid and time-sensitive evaluations of critically endangered or threatened populations.
Ages of odontocetes can be estimated based on tooth GLGs (Myrick, 1980). Although GLGs represent the current gold standard for determining the age of bottlenose dolphins, the requirement for a tooth may render this method unacceptably invasive for many research efforts. Finally, some debate exists concerning the accuracy of relying on growth ring deposition for age estimation of older animals that may have experienced significant tooth wear (Hohn and Fernandez, 1999).
As such, other aging methods have been evaluated for use with bottlenose dolphins or other cetaceans including eye lens aspartic acid racemization (Bada et al., 1980; George et al., 1999), fatty acid composition (Koopman et al., 1996; Herman et al., 2009; Marcoux et al., 2015), radiocarbon 14 dating from fallout (Stewart et al., 2006), telomere length (Olsen et al., 2012), radiograph changes in tooth to pulp ratio (Herrman et al., 2020), and pectoral flipper bone ossification (Barratclough et al., 2019). However, all methods vary in inherent accuracy and must be calibrated against some age estimate of the species in question, most often by using GLG counts, and each have limitations for field use and accuracy across different age classes.
Recent efforts at developing less invasive methods with enhanced accuracy for age determination across multiple species, including marine mammals, has led to recent applications of species-specific DNA methylation (DNAm) profiles for the development of epigenetic aging clocks (Polanowski et al., 2014; Cole et al., 2017; Wang et al., 2017; Thompson et al., 2018; Beal et al., 2019; Bors et al., 2020; Prado et al., 2020b; Lu et al., 2021; Robeck et al., 2021b). DNAm is described as an epigenetic modification whereby a transfer of a methyl (CH3) group from S-adenosyl methionine (SAM) to the fifth position of cytosine nucleotides, forming 5-methylcytosine (5 mC) nucleotides (Smith and Meissner, 2013). The degree of methylation, hypo or hyper can be highly correlated with both chronological age and in the case of abnormal physiological conditions, accelerated aging (Rakyan et al., 2010; Teschendorff et al., 2010; Horvath and Raj, 2018; Bell et al., 2019).
An epigenetic aging clock that relied on DNAm changes in skin samples has been published for the bottlenose dolphin [Bottlenose dolphin Epigenetic Aging estimation Tool (BEAT)]. This clock was developed based on methods used for the development of a humpback whale specific aging clock and relied on two CpGs that were associated with chronological age (Polanowski et al., 2014; Beal et al., 2019). In addition to the bottlenose dolphin specific BEAT clock, we recently published a multi-tissue Odontocete Epigenetic Aging Clock (OEAC) which accurately estimates age from blood or skin samples within multiple odontocete species (Robeck et al., 2021b). Although this odontocete clock works well, evidence suggests that both correlation and accuracy could be improved within each individual species by developing species-specific clocks with an increased sample size for the tissues of interest (Field et al., 2018; Zhang et al., 2019). For wild bottlenose dolphins, the majority of tissues sampled are collected via remote biopsy and include both skin and blubber samples. Blubber cells are often used to determine hormone concentrations and to evaluate the bioaccumulation of organochloride toxins, while DNA from skin samples is often used to determine relatedness among conspecifics (Parsons, 2002; Trego et al., 2013; Galligan et al., 2019). Thus, there is an existing, relatively large historical collection of skin samples and precedent for future collection of skin samples that could also be used for age determination.
In addition to clocks for chronological age determination, recent work in humans has indicated that DNAm clocks can be useful for detecting epigenetic accelerated aging (defined as discrepancy between epigenetic and chronologic age), and that the detection of an accelerated age condition can be predictive of multiple disease states and mortality (Marioni et al., 2015; Chen et al., 2016; Perna et al., 2016; Lu et al., 2019). Epigenetic clocks for bottlenose dolphins may be promising tools for quantifying adverse effects that anthropogenic environmental stressors are creating in wild populations (Prado et al., 2020b).
Therefore, the objectives of our research were (1) to apply the mammalian methylation array technology to develop highly accurate bottlenose dolphin clocks based on blood and/or skin samples; (2) to characterize significant aging associated CpGs identified via epigenome-wide association studies (EWAS); (3) to characterize sex linked CpGs, and (4) to develop sex estimators for odontocetes based on DNAm data.
Materials and Methods
Study Animals
For model development, our study population included skin (n = 50) and blood samples (n = 140) from 140 bottlenose dolphins located at three SeaWorld Parks (Orlando, San Antonio, and San Diego) and Discovery Cove (Orlando, FL, United States). The known age animals consisted of 123 zoo born animals (38 male, 85 females) with a median age of 14.5 years (range: 0.57–40.7 years), and 17 wild born animal (6 male, 11 females) with a median age of 36.4 years (range: 3.9–57.0 years). Known (87.9%) or estimated (based on length at capture or rescue for stranded animals) birth dates were used for correlating against methylation predicted age.
Sample Collection
Blood samples were collected either voluntarily from the peripheral periarterial venous rete on the ventral tail fluke using an 18–22 gauge winged blood collection set or attached to a vacutainer collection system during routine physical examination. Blood was collected by either the veterinary technician or staff veterinarian into BD Vacutainers (Becton Dickinson, Franklin Lakes, NJ, United States) containing EDTA. Samples were inverted in the Vacutainer a minimum of 10 times and then frozen at −80°C until further testing.
Skin scrapings were collected either under stimulus control or manual restraint using a sterile disposable dermal curette or 8.0 mm biopsy punch (Miltex, Integra Life Sciences Corp., York, PA, United States) from a location just posterolateral of the dorsal fin overlying the epaxial muscle. Prior to collection, a cold pack was placed on the site for several minutes prior to sampling to numb the sample site. Skin samples were placed into sterile cryovials (Nunc® Cryotubes, Millipore Sigma Corp., St. Louis, MO, United States) and stored at −80°C until shipment on dry ice. Skin samples from non-living animals were obtained from frozen (−80°C) specimens that had been previously collected and stored during standard necropsy procedures.
DNA Extraction
Genomic DNA was extracted from clotted whole blood samples using QIAamp DNA Mini blood kit and following the manufacturer’s instructions (Qiagen, Valencia, CA, United States). Tissue samples were pulverized and broken down manually using a drill and DNA was extracted using DNeasy Tissue kit (Qiagen) and following the manufacturer’s instructions with the exception of extending the proteinase k digestion. DNA was then extracted using the automated nucleic acid extraction platform, Anaprep (Biochain, Newark, CA, United States) that utilizes a magnetic bead extraction process and Tissue DNA Extraction kit (Anaprep).
DNA Methylation Data
The mammalian DNAm arrays were profiled using a custom Infinium methylation array (HorvathMammalMethylChip40) based on 37,491 CpG sites as previously described (Prado et al., 2020a). Out of these sites, 1951 were selected based on their utility for human biomarker studies; these CpGs, which were previously implemented in human Illumina Infinium arrays (EPIC, 450K), were selected due to their relevance for estimating age, blood cell counts or the proportion of neurons in brain tissue. The remaining 35,540 probes were chosen to assess cytosine DNAm levels in mammalian species (Arneson et al., 2021). The subset of species for each probe is provided in the chip manifest file at the NCBI Gene Expression Omnibus (GEO) platform (GPL28271). Raw data was normalized using the SeSaMe method which assigned a beta value (0–1) for each methylation estimate corresponding to each probe used for every sample (Zhou et al., 2018). Beta values were indicative of methylation rates, with 0 indicating no gene copies were methylated. Unsupervised hierarchical clustering analysis based on the interarray correlation was used to identify technical outliers which were then removed from further analysis.
Penalized Regression
Details on the clocks (CpGs, genome coordinates) and R software code are provided in the Supplementary Material. Penalized regression models were implemented with the R software package “glmnet” (Friedman et al., 2010). The optimal penalty parameters lambda in all cases were determined automatically by using a 10-fold internal cross-validation (cv.glmnet) on the training set. Alpha = 1/2 corresponds to the elastic net penalty that penalizes the coefficients based on their magnitude. We performed a Leave One Out Cross Validation (LOOCV) scheme for arriving at unbiased estimates of the prediction accuracy of the different DNAm aging clocks. The LOOCV, which is based on previously reported methods (Shao, 1993; Zhang, 1993), does the following for each of the N samples: omit one sample from the training set; fit the clock on the training set with (N−1) samples; predict the DNAm age of the omitted sample with the fitted clock. Therefore, the LOOCV allows one to estimate the accuracy of predicting the age for any unknown bottlenose dolphin sample by the clock. The cross-validation study reports unbiased estimates of the age correlation r (defined as Pearson correlation between the DNAm age estimate and chronological age) as well as the median absolute error (MAE). The accuracy of the resulting bottlenose dolphin clocks was assessed via LOOCV estimates of: (1) the correlation r between the predicted epigenetic age and the actual (chronological) age of the animal; and (2) the MAE between DNAm age and chronological age.
Multivariate Sex Predictor
To predict sex (binary variable with values 1 = female, 0 = male), we used a binomial generalized linear model, regularized by elastic net (Zou and Hastie, 2005). The glmnet alpha parameter was set to 0.5, i.e., we used elastic net regression. The lambda parameter of the glmnet function was chosen using 10-fold cross validation. As we observed a reasonably balanced female-male sample sizes in the data set, we set a neutral prediction rule for the binomial generalized linear model. If the predicted probability for a sample being female was >0.5, the sample was predicted to be female. We used all odontocetes samples previously used for DNAm clock development (Robeck et al., 2021b) and the additional samples from bottlenose dolphins, reported herein, in the training data so that the resulting sex estimator would be broadly applicable to odontocetes species.
Results
We obtained DNAm profiles from blood (n = 140) and skin (n = 87) samples from 140 animals (44 males and 96 females) ranging in ages from 0 to 57 years of age (mean: 19.8 years, median: 16.8 years). For each animal, we had either blood or skin samples or both. An unsupervised hierarchical analysis clustered the methylation arrays by tissue type (Supplementary Figure 1).
Epigenetic Aging Models
We developed three bottlenose dolphin clocks for (i) blood + skin, (ii) blood only, and (iii) skin only (Figure 1). To arrive at unbiased estimates of the accuracy, we used LOOCV. We assessed the correlation between predicted age and chronological age, R, and the MAE. LOOCV revealed that all three dolphin clocks were highly accurate: blood and skin clock: R = 0.93, MAE = 2.13 years, blood clock: R = 0.97; MAE = 1.46 years, skin clock: R = 0.95; MAE = 2.53 years (Figures 1B,D,F).
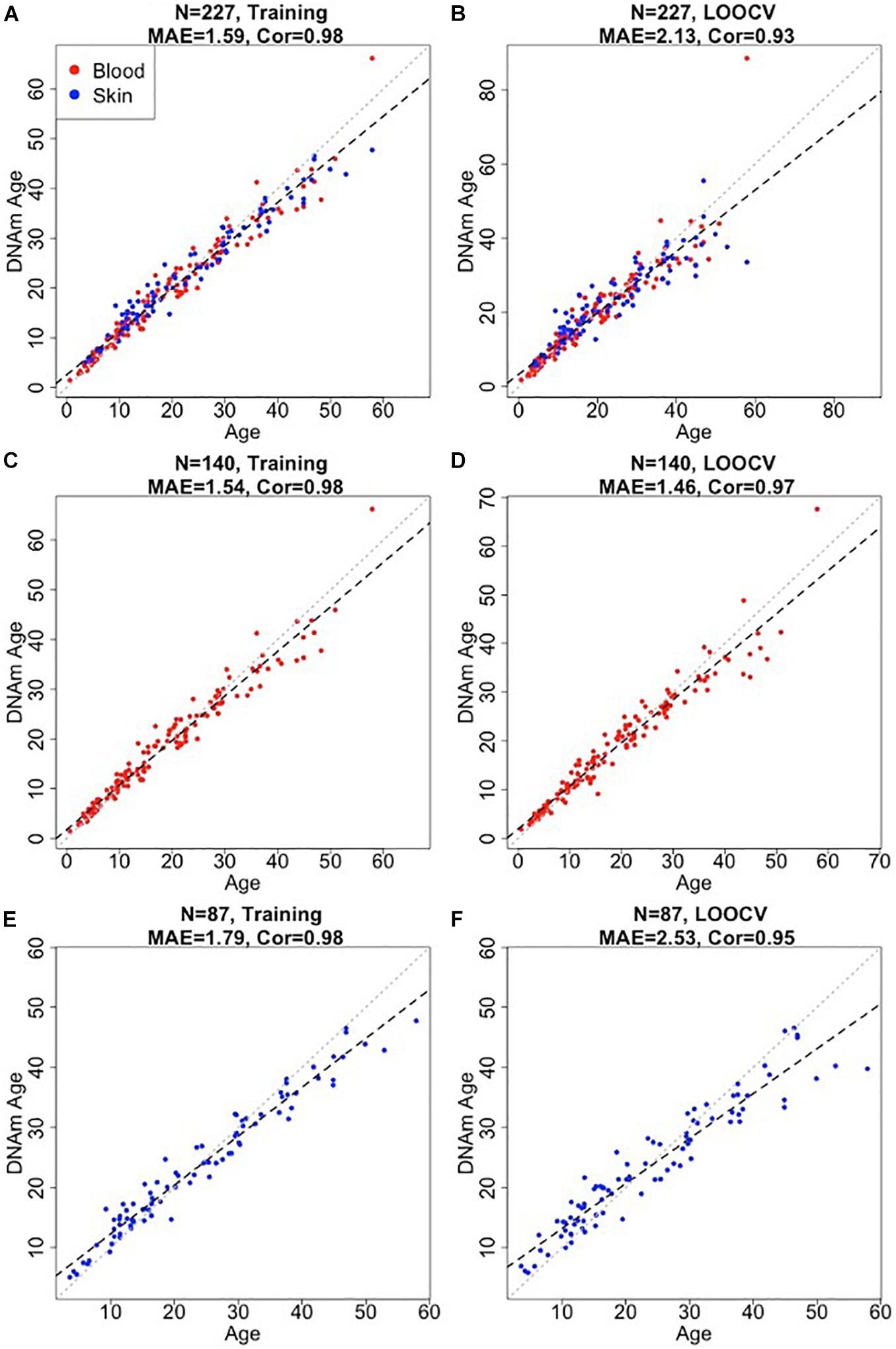
Figure 1. Accuracy of epigenetic clocks for bottlenose dolphin. The panels correspond to three different clocks for bottlenose dolphins: multi tissue clock (A,B); blood clock (C,D); and skin clock (E,F) and the age estimates based on training (A,C,E) and LOOCV (B,D,F). Dots are colored by tissue type (red = blood, blue = skin). The training set estimates (left panels) are highly biased. Each panel depicts a linear regression line (black dashed line), a diagonal line (y = x, dotted line), the sample size (N), Pearson correlation (Cor) across all samples, median absolute error (MAE) across species. Chronological age (x-axis) and DNA methylation age estimates (y-axis) are in units of years.
The final versions of the three clocks (based on all training data) involved 48 CpGs (blood and skin clock), 64 CpGs (blood clock), and 39 CpGs (skin clock) as detailed in Supplementary Table 1. There were 12 common CpGs between the blood and skin clock and the blood clock, and 6 common CpGs between the blood and skin clock and the skin clock (Supplementary Figure 2).
Methylation Estimator of Sex
Methylation based estimators of sex can be useful for identifying human errors in sample labeling or DNA processing. To define a methylation based sex estimator for dolphins and other odontocetes we combined our data with those from eight additional odontocete species (Robeck et al., 2021b). The sex estimator was developed by fitting a binomial elastic net regression model on n = 447 odontocetes samples (n = 276 female and n = 171 male samples). The model only misclassified two male killer whale samples (99.5% accuracy, Table 1). The 71 CpGs of the sex estimator most likely mapped to sex chromosomes in odontocetes because they map to human sex chromosomes (based on human genome hg19).
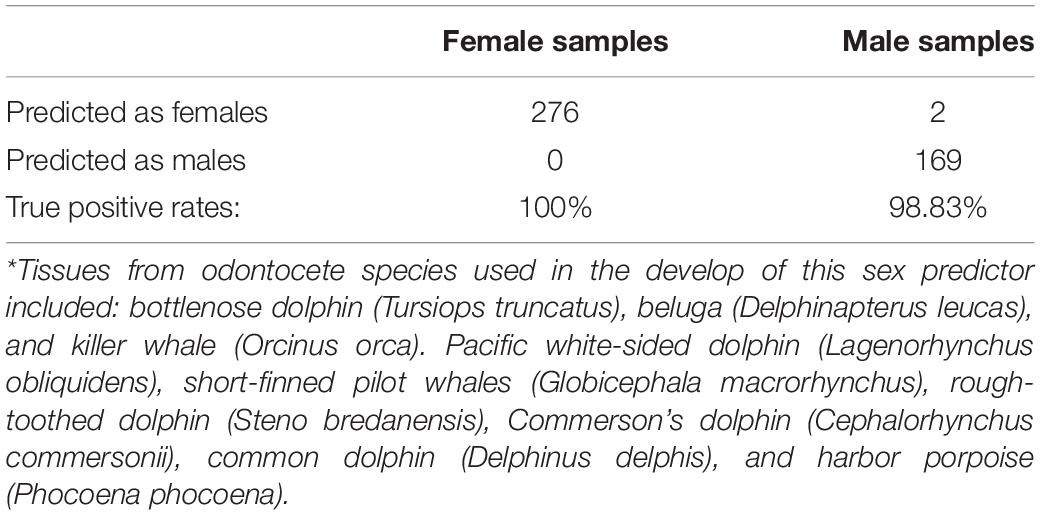
Table 1. Elastic net penalized linear model sex predictions based on odontocetes*, n = 447 (Robeck et al., 2021b).
Epigenome-Wide Association Studies of Age
In total, only 23,005 probes from the mammalian array (HorvathMammalMethylChip40) could be mapped to the bottlenose dolphin genome assembly (turTru1.100). These probes are adjacent to 4,558 unique genes: 65% in gene bodies, 10% in promoters, and 25% in distal regions. The number of aligned probes in the bottlenose dolphin were much lower than other cetacean species (e.g., 34,358 probes in the killer whale; Robeck et al., 2021b), which probably reflects the lower quality of the dolphin genome assembly. A total of 6,073 and 604 CpGs were significantly correlated with age in blood and skin, respectively (p < 10–4, Figure 2A). This difference in CpG counts between sample types reflected differences in statistical power and sample size (blood, n = 140, age range: 1–57 years; skin, n = 87, age range: 4–57 years). The top age-related changes in blood included hypermethylation in LHFPL4 exon and HOXC4 promoter. In skin, the CpGs with the strongest positive correlation with age were located downstream of EVX2 and in an exon of NOL4 exon (Table 2). CpGs in promoters largely gained methylation with age, which matches DNAm aging pattern in other mammals (Figure 2B). CpGs located in CpGs islands had higher age correlations than CpGs located outside of islands (Figure 2C).
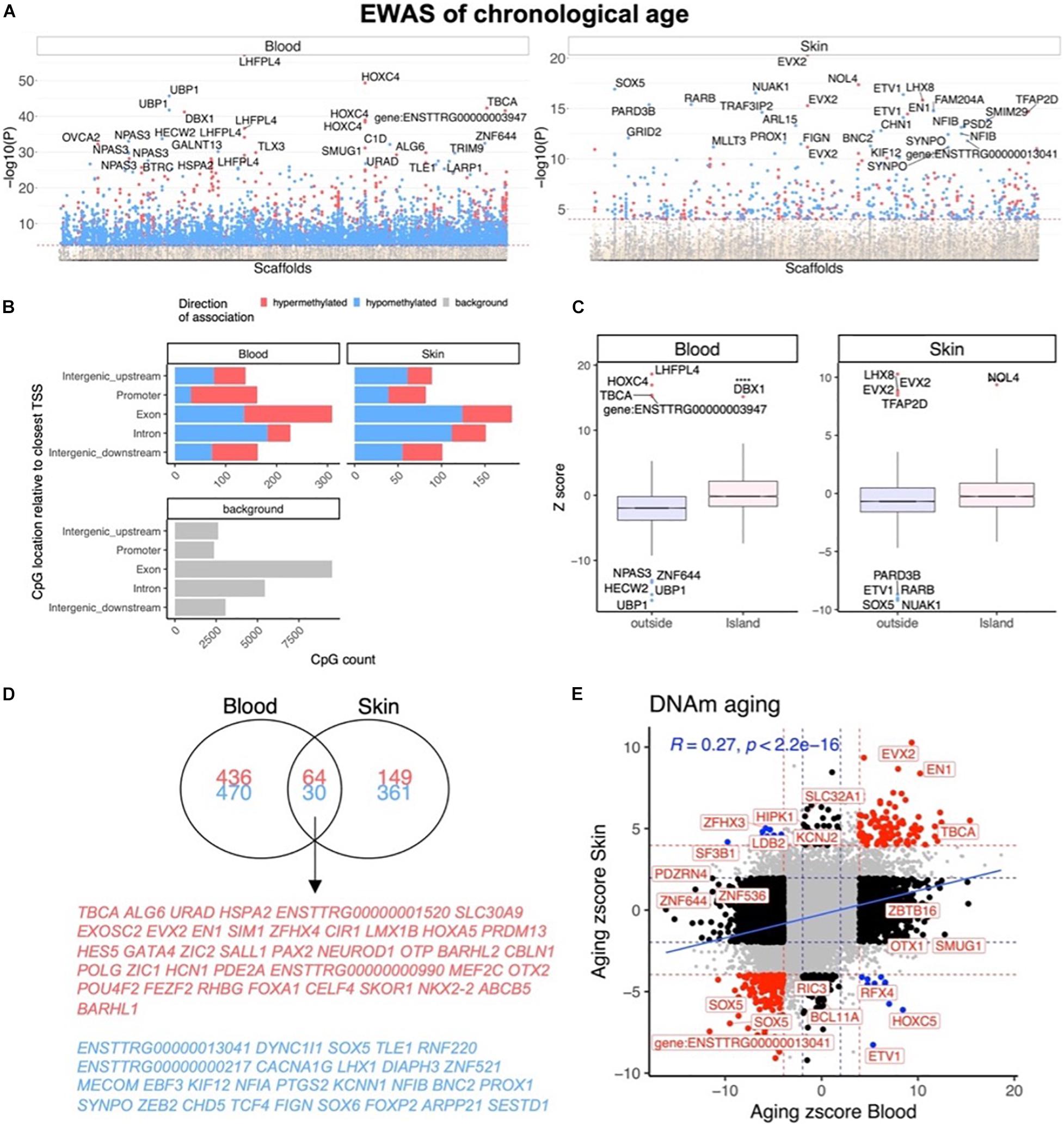
Figure 2. Epigenome-wide association (EWAS) of chronological age in skin and blood of bottlenose dolphin species. (A) Manhattan plots of the EWAS of chronological age. The coordinates are estimated based on the alignment of Mammalian array probes to the bottlenose dolphin (turTru1.100) genome assembly. The direction of associations with P < 0.0001 (red dotted line) is highlighted by red (hypermethylated) and blue (hypomethylated) colors. The 30 most significant CpGs were labeled by their respective neighboring genes. (B) Genomic location analysis of significant (P < 10–4) CpGs. The blood analysis focused on the top 500 positively age related CpGs and the top 500 negatively age related CpGs. The skin analysis focused on 213 positively and 391 negatively age related CpGs. (C) Box plot visualizing the median value (line inside the box) and the 25th and 75th percent quartiles (top and bottom of the box), and the whiskers correspond to the 90th percentile. The x-axis corresponds to CpG island status. (D) Venn diagram of the most significant age related CpGs in blood and skin. (E) Sector plot of DNA methylation aging effects in blood and skin. Each axis reports the Z statistic from a correlation test which follows a standard normal distribution under the null hypothesis. Red dotted line: P < 10–4; blue dotted line: P > 0.05; Red dots: shared CpGs; black dots: tissue specific changes; blue dots: CpGs where age correlation differs between blood and skin tissue. The gray color in the last panel represents the location of 23,005 mammalian array probes mapped to the bottlenose dolphin (turTru1.100) genome.
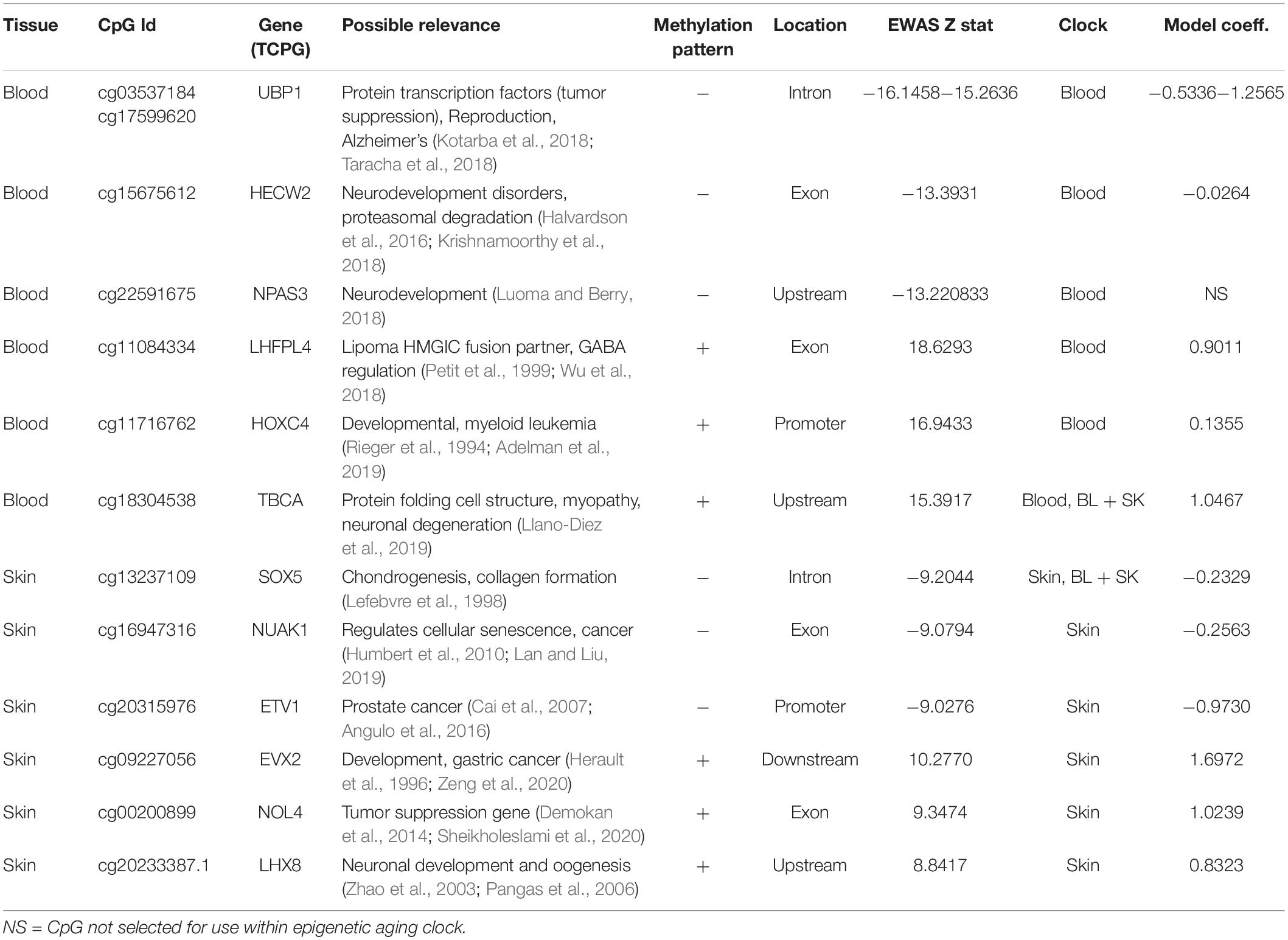
Table 2. Epigenome-wide associated studies (EWAS) identification of top three hypo [−] and hyper [+] methylated age associated CpGs, its proximate gene, the epigenetic clock which included CpG site its coefficients.
We found that 94 CpGs (64 hypermethylated, 30 hypomethylated) had a significant correlation with age in both blood and skin (Figure 2D). Aging effects in blood were moderately correlated with those in skin (R = 0.27, Figure 2E). Functional enrichment studies revealed that significant age-related CpGs are located near genes that play a role in development and with polycomb repressor complex 2 target genes H3K27ME3, EED, PCR2, and SUZ12 (Supplementary Figure 3 and Table 2). Many CpGs correlate with age in a tissue specific manner. As an example, while HOXC4 promoter, suspected to be involved with myeloproliferative disorders and skin tumors (Rieger et al., 1994; Adelman et al., 2019), was hypermethylated with age in blood (z = 4.7), it was hypomethylated in skin (z = −4.5).
Sex Specific DNAm Patterns in Bottlenose Dolphins
Our dataset allowed us to characterize sex differences in baseline methylation levels, and, also, sex effects on aging rates. At a nominal/unadjusted significance threshold of p < 10–4, 1,738 and 742 CpGs were significantly associated with sex in dolphin blood and skin, respectively (Figure 3A). Due to the incomplete genome assembly we were not able to determine whether these CpGs are located on sex chromosomes in bottlenose dolphins. However, we would expect that these CpGs are located on sex chromosomes since more than 70% of these CpGs in blood and 90% in skin were located on the human X chromosome. Some of the top sex-related gene regions were as follows: blood, PTCHD1 exon (hypermethylated in females) and HUWE1 exon (hypomethylated in females); skin, HUWE1 exon (hypomethylated in females) and CNKSR2 exon (hypermethylated in females).
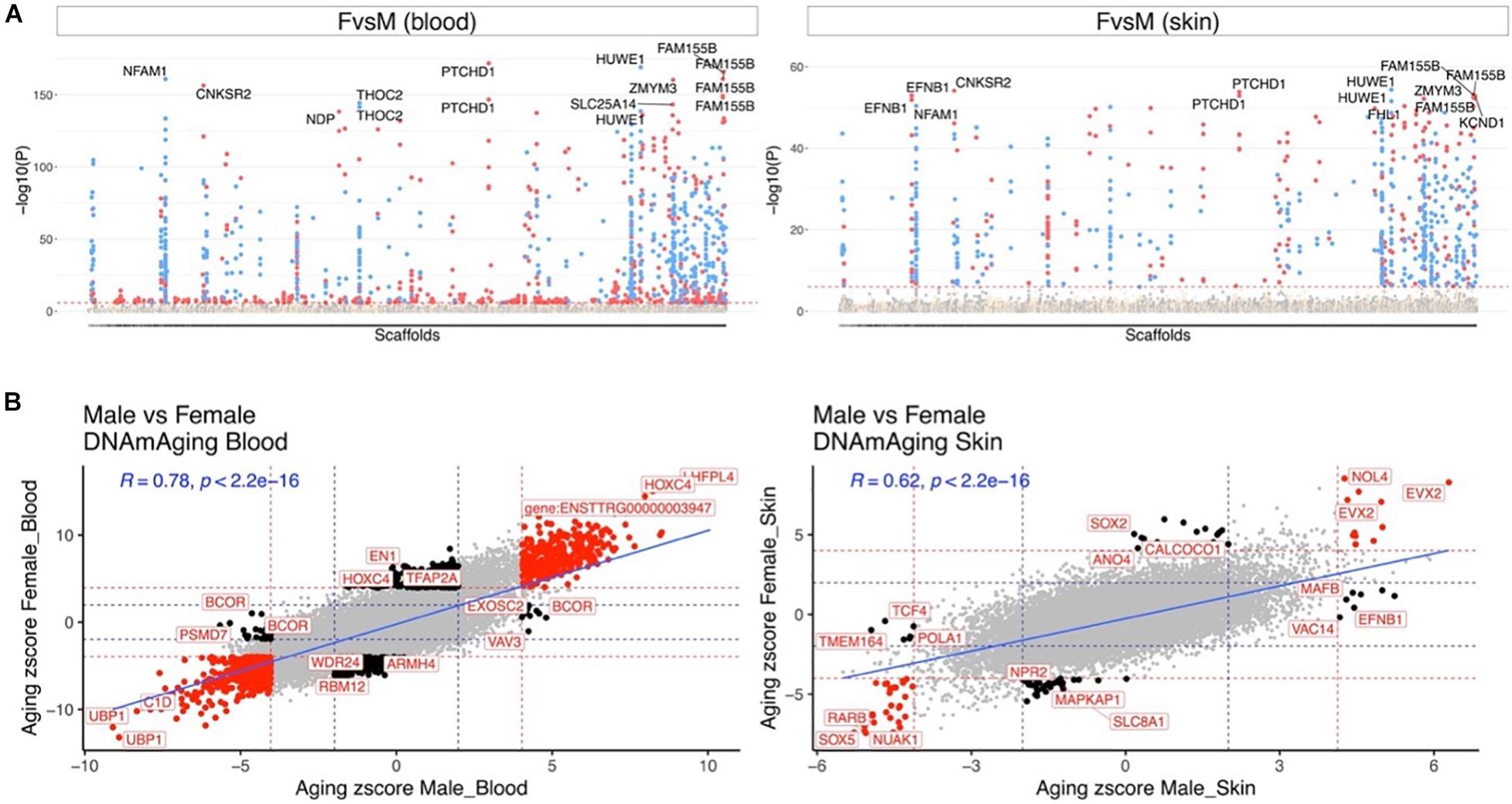
Figure 3. Aging effects in male and female dolphins. (A) Manhattan plot of the epigenome-wide association studies (EWAS) of sex. We adjusted the analysis for chronological age. Sample size: males blood, 45; females blood, 95; males skin, 29; and females skin, 58. The genome coordinates are estimated based on the alignment of Mammalian array probes to the bottle nose dolphin (turTru1.100) genome assembly. The horizontal red dotted line indicate a nominal/uncorrected significance level of P < 10–4. Significant CpGs are colored in red if they are hypermethylated in females, blue hypomethylated in females. The 15 most significant CpGs are labeled by their respective neighboring genes. (B) Sector plot of DNA methylation aging effects by sex in blood (left) and skin (right). Red dotted line: P < 10–4; blue dotted line: P > 0.05; Red dots: shared CpGs; black dots: distinct changes between males and females.
Aging effects were highly correlated between the sexes both in blood (R = 0.78) and skin (R = 0.62, Figure 3B). We highlight a few CpGs with distinct sex specific aging patterns in Figure 4.
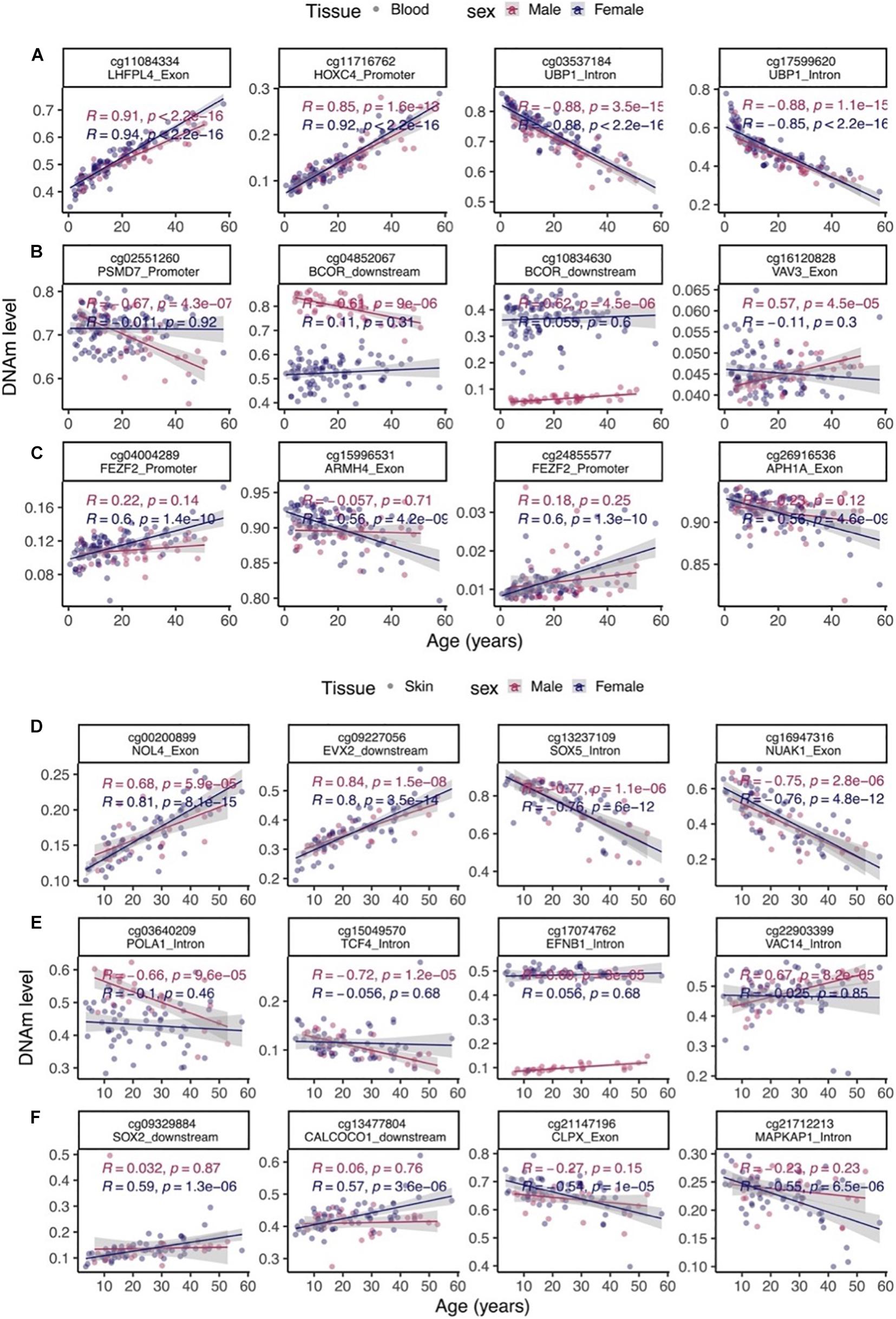
Figure 4. Select age-related CpGs stratified by sex. (A) CpGs that change with age in male and female blood samples from bottlenose dolphins. (B) Male specific changes in blood. (C) Female specific changes in blood. (D) CpGs that change with age in both male and female skin. (E) Male specific changes in skin. (F) Female specific changes in skin. The ordinary least squares regression lines were carried out in males (red) and females (blue) separately.
Discussion
This study describes the development of three highly accurate bottlenose dolphin DNAm epigenetic aging clocks using combined blood and skin, blood only or skin only that were developed with samples collected from known (88%) or approximately known age animals. The mammalian methylation array used in this research profiles cytosines that are conserved across mammalian species (Lu et al., 2021; Robeck et al., 2021b).
Similar to our recently published OEAC (Robeck et al., 2021b), the data were mainly comprised of animals whose ages were known precisely (often the birthday was known) or whose age could be estimated with high accuracy. As such, this dolphin clock is expected to become a useful molecular method for accurate age measurement in dolphins.
Recent efforts to develop rapid, non-invasive methods for age determination include pectoral flipper bone ossification and dental radiographic determination of the pulp to tooth area ratio (Barratclough et al., 2019; Herrman et al., 2020). Although bone ossification and pulp to tooth ratios benefit from portability of radiography units that can be used in the field to estimate age within hours of image collection, they still require animal restraint and suffer from reduced accuracy with animals that have reached physical maturity (Barratclough et al., 2019; Herrman et al., 2020). By contrast, the epigenetic clocks described herein can accurately be applied to the entire life-span of the bottlenose dolphin.
Two previous epigenetic clocks have been applied to Bottlenose dolphins: BEAT and the OEAC (Beal et al., 2019; Robeck et al., 2021b). The OEAC clock is the first multi-tissue clock developed to be used with multiple odontocetes species. For this clock, nine different odontocete species were used across 446 blood and skin samples, and within this data set, there were 181 bottlenose dolphin samples (140 blood and 41 skin samples). Although we obtained very accurate predictive results for the bottlenose dolphin blood (R = 0.95, MAE 1.5 years) and skin samples (R = 0.91, MAE 4.8 years) in our OEAC, predictions based on skin samples were below accuracy levels with a wider variance (MAE) than was found in blood. While still within ranges currently used in forensic sciences for age estimation to within a decade (Maat et al., 2006; Huang et al., 2015; Vidaki et al., 2017), this increase in variance in skin samples as compared to blood was believed to be primarily due to a reduced sample size as compared to blood (40 vs. 140). Additionally, other variables that may have contributed toward an increased predictive variance may have included the types and proportion of cells collected during each skin collections for discussion see (Robeck et al., 2021b) and may possibly reflect a higher sensitivity for skin cells to experience varying epigenetic methylation rates across age (Gronniger et al., 2010). Therefore, we hypothesized that by increasing our skin sample size for bottlenose dolphins, we could improve our predictive ability within skin samples over our previous results presented with the OEAC development (Robeck et al., 2021b). Thus, in our present study, we increased our skin sample size by over 100% and obtained an almost a 50% reduction in MAE (2.5 vs. 4.8) and brought our median predictive ability from skin samples across all age groups to within 5 years (±2.5 years). Although these results cannot take away from the predictive value of the OEAC across multiple species, they do demonstrate the superior accuracy of species-specific clocks.
Direct comparisons of the accuracy between the BDAC and the OEAC are possible because approximately half of the samples were independently analyzed by each clock. However, direct comparison against the only other bottlenose dolphin clock, the BEAT is not possible due to different samples and measurement platforms. In general, our BDAC skin clock had improved accuracy (R = 0.95, MAE = 2.5 years) at predicting ages of bottlenose dolphins when compared to the BEAT results [BEAT, R2 = 0.74, root mean square error = 5.1 years (Beal et al., 2019)]. Important differences between the BDAC and the BEAT clock that have been identified as limitations toward methylation clock development include: (1) the sample number used for the BEAT clock was ∼half (n = 40) the number used for the BDAC; and (2) the number of CpGs used for final clock development were vastly reduced (2 vs. 39). As we have already demonstrated through direct comparisons between the BDAC and OEAC and as was hypothesized by the developers of the BEAT clock, increased sample size has direct effects on clock accuracy (Beal et al., 2019). In addition to sample size, by filtering through a dramatically larger catalog of CpGs we were able to identify and incorporate a wider array of significant age related CpG sites into the predictive model resulting an improved representation of organism age (Field et al., 2018; Zhang et al., 2019; Lu et al., 2021).
The BEAT clock (Beal et al., 2019) identified two CpG sites on two genes, TET2 (CpG site 2) and GRIA2 (CpG site 5) that accounted for 78% of the age-related variation in % DNAm. These same genes were also found to correlate with age in humpback whales (Polanowski et al., 2014). Although we had several probes that mapped to the genomic regions of GRIA2, this gene was not considered to be within the top 10 genes that accounted for age-related methylation changes in neither our present study nor within the OEAC (Robeck et al., 2021b). Our measurement platform (the mammalian array) did not cover the TET2 gene, and we only had probes that mapped to the genomic regions of GRIA2 gene. Interestingly, most of these GRIA2 probes (both exon and intron) were hypomethylated with age in both the blood and skin of bottlenose dolphins (Supplementary Figure 4). This suggests that although promoter of GRIA2 gains methylation with age (Beal et al., 2019), selected CpGs in the gene body lose methylation with age.
Besides epigenetic clock development, the mammalian array is a uniquely reproducible tool for a direct genome-wide comparison of DNAm changes across cetaceans. Our EWAS identified DNAm aging CpGs proximate to genes associated with development (Supplementary Figure 3). The top proximate hypermethylated CpG in blood was associated with LHFPL4, a gene known to be involved with GABA regulation in the central nervous system and peripheral lipoma formation (Petit et al., 1999; Wu et al., 2018). This gene has recently been identified as the most predictive age-related gene across mammalian species and despite its conserved status, its contribution toward age-related functional decline has not be clarified (Lu et al., 2019). The top two hypomethylated CpGs (both within introns) in the blood were proximal to UBP1, a gene that is closely related to TFCP2 and is considered part of the Grainyhead family of transcription factors (Taracha et al., 2018). Although UBPI has been identified as involved in angiogenesis during growth and development and Alzheimer’s disease, its redundancy and homology with TFCP2 make its yet unidentified role in cancer regulation a likely possibility (Kotarba et al., 2018; Taracha et al., 2018). For skin, the top proximate genes were associated with development and energy metabolism (Supplementary Figure 3). In addition to these CpG related genes, multiple CpGs, in both blood and skin, that were associated with genes within the PRC2 complex were hypermethylated (Supplementary Figure 3). The PRC2 is required for embryonic stem cell differentiation but also plays a role in in aging (Pasini et al., 2007; Ito et al., 2018). Overall, we find that age-related methylation changes in bottlenose dolphins mirror those in other odontocete species and mammals in general (Lu et al., 2019; Robeck et al., 2021b).
Our sex related CpGs were largely the same as those found in multiple odontocete species (Robeck et al., 2021b). Therefore, we were able to use these CpGs to develop a highly accurate (99.5%) multivariate predictor of sex across all odontocete species. The ability to determine sex in species that do not have easily identifiable sexual dimorphic patterns across multiple species using the same platform for age determination is an attractive property of cytosine methylation data.
Similar to previous work with other cetaceans, we found no evidence that accounting for sex was necessary for the development of the three BDAC clocks (Bors et al., 2020; Robeck et al., 2021b). Aging effects in males were highly correlated with those in females both in blood (R = 0.78) and skin (R = 0.62). While the majority of sex specific CpGs were found on the X chromosome (78.8%, Supplementary Data File 1), the proximate genes associated with the top non X chromosome CpGs, which were different between the two sexes included in blood, related to cancer (PSMD7, BCOR) and the central nervous system (FEZF2) (Guo et al., 2013; Kao et al., 2016). Noteworthy sex related autosomal genes implicated by the skin methylation data play a role in DNA repair (POLA1), neurodevelopment (TCF), oral cancer (SOX2, higher methylation rates in females) and nutritional or chemical mediated ER autophagy (CALCOCO1) (Dabin et al., 2008; Pisano et al., 2018; de Vicente et al., 2019; Li et al., 2019; Nthiga, 2020). These results suggest a potential difference in aging phenotypes in dolphin sexes, however, this hypothesis should be examined in future studies.
Conclusion
In conclusion, we characterized cytosines that correlate strongly with age and sex in bottlenose dolphins. We present three highly accurate DNAm-based estimators of chronological age for bottlenose dolphins. The high accuracy of these clocks could not have been achieved without knowing the precise ages of a large number of animals which in turn relied on long term efforts of monitoring and caring for these animals.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the SeaWorld Parks and Entertainment Incorporated Research Review Committee and was performed in strict accordance with the United States Animal Welfare Act for the care of marine mammals.
Author Contributions
TR, KS, SD, LS, TS, SO, GM, and MR provided the animal samples and sample curating. ZF, AH, JZ, CL, TR, and SH performed the sample analysis and data modeling, conceived the study, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Paul G. Allen Frontiers Group (SH).
Conflict of Interest
SH is a founder of the non-profit Epigenetic Clock Development Foundation which plans to license several of his patents from his employer UC Regents. TR, KS, SD, LS, TS, SO, and GM are employed by SeaWorld Parks & Entertainment. MR is employed by Miami Seaquarium.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank individuals from all of our zoological parks that contributed to the sample collection and curating for this research. Specifically, from SeaWorld California (SWC), we thank Elsburgh Clarke, Kelsey Herrick, Rachelle Pastorkovich, Sarah McMillen, Melinda Tucker, Kim Regan, and Wendy Ramirez. From SeaWorld of Texas, we thank Jennifer Russell and Tara Klimek. From SeaWorld of Florida, we thank Dana Lindemann, Claire Erlacher-Reid, Jacob Vandenberg, Cynthia O’neil, Megan Fox, Siobhan Diaz, Cynthia Reyes, Stephanie Smith, and Heidy Clifford. Finally, we also thank Species Preservation Laboratory research technicians Amanda McDonnel and Jacqueline Posy. This is a SeaWorld Parks and Entertainment technical manuscript number 2021-7. This submitted manuscript has recently been uploaded to bioRxiv preprint server website at doi: 10.1101/2021.05.03.442523 (Robeck et al., 2021a).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.713373/full#supplementary-material
References
Adelman, E. R., Huang, H.-T., Roisman, A., Olsson, A., Colaprico, A., Qin, T., et al. (2019). Aging human hematopoietic stem cells manifest profound epigenetic reprogramming of enhancers that may predispose to leukemia. Cancer Discov. 9, 1080–1101. doi: 10.1158/2159-8290.cd-18-1474
Angulo, J. C., Andrés, G., Ashour, N., Sánchez-Chapado, M., López, J. I., and Ropero, S. (2016). Development of castration resistant prostate cancer can be predicted by a DNA hypermethylation profile. J. Urol. 195, 619–626. doi: 10.1016/j.juro.2015.10.172
Arneson, A., Haghani, A., Thompson, M. J., Pellegrini, M., Kwon, S. B., Vu, H. T., et al. (2021). A mammalian methylation array for profiling methylation levels at conserved sequences. bioRxiv [Preprint]. doi: 10.1101/2021.01.07.425637
Bada, J., Brown, S., and Masters, P. (1980). Age determination of marine mammals based on aspartic acid racemization in the teeth and lens nucleus. Rep. Intern. Whal. Comm. Spec. 32, 113–118.
Barratclough, A., Sanz-Requena, R., Marti-Bonmati, L., Schmitt, T. L., Jensen, E., and Garcia-Parraga, D. (2019). Radiographic assessment of pectoral flipper bone maturation in bottlenose dolphins (Tursiops truncatus), as a novel technique to accurately estimate chronological age. PLoS One 14:e0222722. doi: 10.1371/journal.pone.0222722
Beal, A. P., Kiszka, J. J., Wells, R. S., and Eirin-Lopez, J. M. (2019). The bottlenose dolphin epigenetic aging tool (BEAT): a molecular age estimation tool for small cetaceans. Front. Mar. Sci. 6:561. doi: 10.3389/fmars.2019.00561
Bell, C. G., Lowe, R., Adams, P. D., Baccarelli, A. A., Beck, S., Bell, J. T., et al. (2019). DNA methylation aging clocks: challenges and recommendations. Genome Biol. 20, 1–24.
Bors, E. K., Baker, C. S., Wade, P. R., O’Neill, K. B., Shelden, K. E., Thompson, M. J., et al. (2020). An epigenetic clock to estimate the age of living beluga whales. Evol. Appl. 14, 1263–1273. doi: 10.1111/eva.13195
Cai, C., Hsieh, C.-L., Omwancha, J., Zheng, Z., Chen, S.-Y., Baert, J.-L., et al. (2007). ETV1 is a novel androgen receptor-regulated gene that mediates prostate cancer cell invasion. Mol. Endocrinol. 21, 1835–1846. doi: 10.1210/me.2006-0480
Chen, B. H., Marioni, R. E., Colicino, E., Peters, M. J., Ward-Caviness, C. K., Tsai, P.-C., et al. (2016). DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 8:1844.
Cole, J. J., Robertson, N. A., Rather, M. I., Thomson, J. P., McBryan, T., Sproul, D., et al. (2017). Diverse interventions that extend mouse lifespan suppress shared age-associated epigenetic changes at critical gene regulatory regions. Genome Biol. 18, 1–16.
Dabin, W., Cossais, F., Pierce, G. J., and Ridoux, V. (2008). Do ovarian scars persist with age in all Cetaceans: new insight from the short-beaked common dolphin (Delphinus delphis Linnaeus, 1758). Mar. Biol. 156, 127–139. doi: 10.1007/s00227-008-1070-4
de Vicente, J. C., Donate-Pérez del Molino, P., Rodrigo, J. P., Allonca, E., Hermida-Prado, F., Granda-Díaz, R., et al. (2019). SOX2 expression is an independent predictor of oral cancer progression. J. Clin. Med. 8:1744. doi: 10.3390/jcm8101744
Demokan, S., Chuang, A. Y., Pattani, K. M., Sidransky, D., Koch, W., and Califano, J. A. (2014). Validation of nucleolar protein 4 as a novel methylated tumor suppressor gene in head and neck cancer. Oncol. Rep. 31, 1014–1020. doi: 10.3892/or.2013.2927
Field, A. E., Robertson, N. A., Wang, T., Havas, A., Ideker, T., and Adams, P. D. (2018). DNA methylation clocks in aging: categories, causes, and consequences. Mol. Cell 71, 882–895. doi: 10.1016/j.molcel.2018.08.008
Friedman, J., Hastie, T., and Tibshirani, R. (2010). Regularization paths for generalized linear models via coordinate descent. J. Statist. Softw. 33, 1–22.
Galligan, T. M., Balmer, B. C., Schwacke, L. H., Bolton, J. L., Quigley, B. M., Rosel, P. E., et al. (2019). Examining the relationships between blubber steroid hormones and persistent organic pollutants in common bottlenose dolphins. Environ. Pollut. 249, 982–991. doi: 10.1016/j.envpol.2019.03.083
George, J. C., Bada, J., Zeh, J., Scott, L., Brown, S. E., O’Hara, T., et al. (1999). Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can. J. Zool. 77, 571–580. doi: 10.1139/cjz-77-4-571
Gronniger, E., Weber, B., Heil, O., Peters, N., Stab, F., Wenck, H., et al. (2010). Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 6:e1000971. doi: 10.1371/journal.pgen.1000971
Guo, C., Eckler, M. J., McKenna, W. L., McKinsey, G. L., Rubenstein, J. L., and Chen, B. (2013). Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron 80, 1167–1174. doi: 10.1016/j.neuron.2013.09.037
Halvardson, J., Zhao, J. J., Zaghlool, A., Wentzel, C., Georgii-Hemming, P., Månsson, E., et al. (2016). Mutations in HECW2 are associated with intellectual disability and epilepsy. J. Med. Genet. 53, 697–704.
Herault, Y., Hraba-Renevey, S., Van der Hoeven, F., and Duboule, D. (1996). Function of the Evx-2 gene in the morphogenesis of vertebrate limbs. EMBO J. 15, 6727–6738. doi: 10.1002/j.1460-2075.1996.tb01062.x
Herman, D. P., Ylitalo, G. M., Robbins, J., Straley, J. M., Gabriele, C. M., Clapham, P. J., et al. (2009). Age determination of humpback whales Megaptera novaeangliae through blubber fatty acid compositions of biopsy samples. Mar. Ecol. Prog. Ser. 392, 277–293. doi: 10.3354/meps08249
Herrman, J. M., Morey, J. S., Takeshita, R., De Guise, S., Wells, R. S., McFee, W., et al. (2020). Age determination of common bottlenose dolphins (Tursiops truncatus) using dental radiography pulp:tooth area ratio measurements. PLoS One 15:e0242273. doi: 10.1371/journal.pone.0242273
Hohn, A. A., and Fernandez, S. (1999). Biases in dolphin age structure due to age estimation technique. Mar. Mamm. Sci. 15, 1124–1132. doi: 10.1111/j.1748-7692.1999.tb00881.x
Horvath, S., and Raj, K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384. doi: 10.1038/s41576-018-0004-3
Huang, Y., Yan, J., Hou, J., Fu, X., Li, L., and Hou, Y. (2015). Developing a DNA methylation assay for human age prediction in blood and bloodstain. Forens. Sci. Intern. Genet. 17, 129–136. doi: 10.1016/j.fsigen.2015.05.007
Humbert, N., Navaratnam, N., Augert, A., Da Costa, M., Martien, S., Wang, J., et al. (2010). Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 29, 376–386. doi: 10.1038/emboj.2009.342
Ito, T., Teo, Y. V., Evans, S. A., Neretti, N., and Sedivy, J. M. (2018). Regulation of cellular senescence by polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent pathways. Cell Rep. 22, 3480–3492. doi: 10.1016/j.celrep.2018.03.002
Kao, Y.-C., Sung, Y.-S., Zhang, L., Jungbluth, A. A., Huang, S.-C., Argani, P., et al. (2016). BCOR overexpression is a highly sensitive marker in round cell sarcomas with BCOR genetic abnormalities. Am. J. Surg. Pathol. 40:1670.
Koopman, H. N., Iverson, S. J., and Gaskin, D. E. (1996). Stratification and age-related differences in blubber fatty acids of the male harbour porpoise (Phocoena phocoena). J. Comp. Physiol. B 165, 628–639. doi: 10.1007/BF00301131
Kotarba, G., Krzywinska, E., Grabowska, A. I., Taracha, A., and Wilanowski, T. (2018). TFCP2/TFCP2L1/UBP1 transcription factors in cancer. Cancer Lett. 420, 72–79. doi: 10.1016/j.canlet.2018.01.078
Krishnamoorthy, V., Khanna, R., and Parnaik, V. K. (2018). E3 ubiquitin ligase HECW2 targets PCNA and lamin B1. Biochim. Biophys. Acta Mol. Cell Res. 1865, 1088–1104. doi: 10.1016/j.bbamcr.2018.05.008
Lan, X., and Liu, X. (2019). LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J. Cell. Mol. Med. 23, 2351–2361. doi: 10.1111/jcmm.13497
Lefebvre, V., Li, P., and de Crombrugghe, B. (1998). A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 17, 5718–5733. doi: 10.1093/emboj/17.19.5718
Li, H., Zhu, Y., Morozov, Y. M., Chen, X., Page, S. C., Rannals, M. D., et al. (2019). Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol. Psychiatry 24, 1235–1246. doi: 10.1038/s41380-019-0353-0
Llano-Diez, M., Fury, W., Okamoto, H., Bai, Y., Gromada, J., and Larsson, L. (2019). RNA-sequencing reveals altered skeletal muscle contraction, E3 ligases, autophagy, apoptosis, and chaperone expression in patients with critical illness myopathy. Skelet. Mus. 9:9. doi: 10.1186/s13395-019-0194-1
Lu, A. T., Fei, Z., Haghani, A., Robeck, T. R., Zoller, J. A., Li, C. Z., et al. (2021). Universal DNA methylation age across mammalian tissues. bioRxiv [Preprint]. doi: 10.1101/2021.01.18.426733v1
Lu, A. T., Quach, A., Wilson, J. G., Reiner, A. P., Aviv, A., Raj, K., et al. (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11:303. doi: 10.18632/aging.101684
Luoma, L. M., and Berry, F. B. (2018). Molecular analysis of NPAS3 functional domains and variants. BMC Mol. Biol. 19:14. doi: 10.1186/s12867-018-0117-4
Maat, G. J., Maes, A., Aarents, M. J., and Nagelkerke, N. J. (2006). Histological age prediction from the femur in a contemporary dutch sample. the decrease of nonremodeled bone in the anterior cortex. J. Forens. Sci. 51, 230–237. doi: 10.1111/j.1556-4029.2006.00062.x
Marcoux, M., Lesage, V., Thiemann, G. W., Iverson, S. J., and Ferguson, S. H. (2015). Age estimation of belugas, Delphinapterus leucas, using fatty acid composition: a promising method. Mar. Mamm. Sci. 31, 944–962. doi: 10.1111/mms.12212
Marioni, R. E., Shah, S., McRae, A. F., Ritchie, S. J., Muniz-Terrera, G., Harris, S. E., et al. (2015). The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Intern. J. Epidemiol. 44, 1388–1396. doi: 10.1093/ije/dyu277
McFee, W. (2012). Age distribution and growth of two bottlenose dolphin (Tursiops truncatus) populations from capture-release studies in the Southeastern United States. Aquat. Mamm. 38, 17–30. doi: 10.1578/am.38.1.2012.17
Myrick, A. C. (1980). “Examination of layered tissues of Odontocetes for age determination using polarized light microscopy,” in Age Determination of Toothed Whales and Sirenians, eds W. F. Perrin and A. C. Myrick Jr. (Cambridge: International Whaling Commission), 105–112.
Nthiga, T. M. (2020). Role of CALCOCO1 in Scaling Down Endoplasmic Reticulum and Golgi by Autophagy. Tromsø, NO: UiT The Arctic University of Norway.
Olsen, M. T., Berube, M., Robbins, J., and Palsboll, P. J. (2012). Empirical evaluation of humpback whale telomere length estimates; quality control and factors causing variability in the singleplex and multiplex qPCR methods. BMC Genet. 13:77. doi: 10.1186/1471-2156-13-77
Pangas, S. A., Choi, Y., Ballow, D. J., Zhao, Y., Westphal, H., Matzuk, M. M., et al. (2006). Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc. Natl. Acad. Sci. U.S.A. 103, 8090–8095. doi: 10.1073/pnas.0601083103
Parsons, K. M. (2002). The Use of Molecular and Observational Data to Infer the Structuring of Bottlenose Dolphin Populations. Aberdeen: University of Aberdeen.
Pasini, D., Bracken, A. P., Hansen, J. B., Capillo, M., and Helin, K. (2007). The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27, 3769–3779. doi: 10.1128/mcb.01432-06
Perna, L., Zhang, Y., Mons, U., Holleczek, B., Saum, K.-U., and Brenner, H. (2016). Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin. Epigenet. 8, 1–7.
Petit, M. M., Schoenmakers, E. F., Huysmans, C., Geurts, J. M., Mandahl, N., and Van de Ven, W. J. (1999). LHFP, a novel translocation partner gene ofHMGICin a lipoma, is a member of a new family of LHFP-like genes. Genomics 57, 438–441. doi: 10.1006/geno.1999.5778
Pisano, C., Merlini, L., Penco, S., Cincinelli, R., Darwiche, N., Guglielmi, M. B., et al. (2018). Preclinical antitumor activity of novel DNA polymerase 1 (POLA1) inhibitors. Cancer Res. 8(13 Suppl.), 4848–4848.
Polanowski, A. M., Robbins, J., Chandler, D., and Jarman, S. N. (2014). Epigenetic estimation of age in humpback whales. Mol. Ecol. Resourc. 14, 976–987. doi: 10.1111/1755-0998.12247
Prado, N. A., Brown, J. L., Zoller, J. A., Haghani, A., Yao, M., Bagryanova, L. R., et al. (2020a). Epigenetic clock and methylation studies in elephants. bioRxiv [Preprint]. doi: 10.1101/2020.09.22.308882
Prado, N. A., Brown, J. L., Zoller, J. Z., Haghani, A., Yao, M., Bagryanova, L., et al. (2020b). Epigenetic clock and methylation studies in elephants. Aging Cell 20:e13414.
Rakyan, V. K., Down, T. A., Maslau, S., Andrew, T., Yang, T.-P., Beyan, H., et al. (2010). Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 20, 434–439. doi: 10.1101/gr.103101.109
Rieger, E., Bijl, J. J., van Oostveen, J. W., Soyer, H. P., Oudejans, C. B., Jiwa, N. M., et al. (1994). Expression of the homeobox gene HOXC4 in keratinocytes of normal skin and epithelial skin tumors is correlated with differentiation. J. Invest. Dermatol. 103, 341–346. doi: 10.1111/1523-1747.ep12394888
Robeck, T. R., Fei, Z., Haghani, A., Zoller, J. A., Li, C. Z., Steinman, K. J., et al. (2021a). Multi-tissue methylation clocks for age estimation in the common bottlenose dolphin. bioRxiv [Preprint]. doi: 10.1101/2021.05.03.442523
Robeck, T. R., Fei, Z., Lu, A. T., Haghani, A., Jourdain, E., Zoller, J. A., et al. (2021b). Multi-species and multi-tissue methylation clocks for age estimation in toothed whales and dolphins. Commun. Biol. 4:642.
Sheikholeslami, S., Azizi, F., Ghasemi, A., Alibakhshi, A., Parsa, H., Tavangar, S. M., et al. (2020). NOL4 is downregulated and hyper-methylated in papillary thyroid carcinoma suggesting its role as a tumor suppressor gene. Intern. J. Endocrinol. Metabol. 18:e108510.
Smith, Z. D., and Meissner, A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220. doi: 10.1038/nrg3354
Stewart, R., Campana, S., Jones, C., and Stewart, B. (2006). Bomb radiocarbon dating calibrates beluga (Delphinapterus leucas) age estimates. Can. J. Zool. 84, 1840–1852. doi: 10.1139/z06-182
Taracha, A., Kotarba, G., and Wilanowski, T. (2018). Neglected functions of TFCP2/TFCP2L1/UBP1 transcription factors may offer valuable insights into their mechanisms of action. Intern. J. Mol. Sci. 19:2852. doi: 10.3390/ijms19102852
Teschendorff, A. E., Menon, U., Gentry-Maharaj, A., Ramus, S. J., Weisenberger, D. J., Shen, H., et al. (2010). Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 20, 440–446. doi: 10.1101/gr.103606.109
Thompson, M. J., Chwiałkowska, K., Rubbi, L., Lusis, A. J., Davis, R. C., Srivastava, A., et al. (2018). A multi-tissue full lifespan epigenetic clock for mice. Aging 10:2832. doi: 10.18632/aging.101590
Trego, M. L., Kellar, N. M., and Danil, K. (2013). Validation of blubber progesterone concentrations for pregnancy determination in three dolphin species and a porpoise. PLoS One 8:e69709. doi: 10.1371/journal.pone.0069709
Vidaki, A., Ballard, D., Aliferi, A., Miller, T. H., Barron, L. P., and Syndercombe Court, D. (2017). DNA methylation-based forensic age prediction using artificial neural networks and next generation sequencing. Forens. Sci. Int. Genet. 28, 225–236. doi: 10.1016/j.fsigen.2017.02.009
Wang, T., Tsui, B., Kreisberg, J. F., Robertson, N. A., Gross, A. M., Yu, M. K., et al. (2017). Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 18, 1–11.
Wells, R. S. (2014). “Social structure and life history of common bottlenose dolphins near Sarasota Bay, Florida: Insights from four decades and five generations,” in Primates and Cetaceans: Field Research and Conservation of Complex Mammalian Societies, Primatology Monographs, eds J. Yamagiwa and L. Karczmarski (Berlin: Springer), 149–172. doi: 10.1007/978-4-431-54523-1_8
Wu, M., Tian, H.-L., Liu, X., Lai, J. H. C., Du, S., and Xia, J. (2018). Impairment of inhibitory synapse formation and motor behavior in mice lacking the NL2 binding partner LHFPL4/GARLH4. Cell Rep. 23, 1691–1705. doi: 10.1016/j.celrep.2018.04.015
Würsig, B. G., and Jefferson, T. A. (1990). Methods of photo-identification for small cetaceans. Rep. Intern. Whal. Comm. 12, 42–43.
Zeng, Z., Xie, D., and Gong, J. (2020). Genome-wide identification of CpG island methylator phenotype related gene signature as a novel prognostic biomarker of gastric cancer. PeerJ 8:e9624. doi: 10.7717/peerj.9624
Zhang, Q., Vallerga, C. L., Walker, R. M., Lin, T., Henders, A. K., Montgomery, G. W., et al. (2019). Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 11:54. doi: 10.1186/s13073-019-0667-1
Zhao, Y., Marín, O., Hermesz, E., Powell, A., Flames, N., Palkovits, M., et al. (2003). The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc. Natl. Acad. Sci. U.S.A. 100, 9005–9010. doi: 10.1073/pnas.1537759100
Zhou, W., Triche, T. J. Jr., Laird, P. W., and Shen, H. (2018). SeSAMe: reducing artifactual detection of DNA methylation by infinium BeadChips in genomic deletions. Nucleic Acids Res. 46:e123. doi: 10.1093/nar/gky691
Keywords: Tursiops truncatus, bottlenose dolphin, epigenetic clock, DNA methylation, sex determination, accelerated aging
Citation: Robeck TR, Fei Z, Haghani A, Zoller JA, Li CZ, Steinman KJ, Dirocco S, Staggs L, Schmitt T, Osborn S, Montano G, Rodriguez M and Horvath S (2021) Multi-Tissue Methylation Clocks for Age and Sex Estimation in the Common Bottlenose Dolphin. Front. Mar. Sci. 8:713373. doi: 10.3389/fmars.2021.713373
Received: 24 May 2021; Accepted: 24 August 2021;
Published: 14 September 2021.
Edited by:
Lars Bejder, The University of Hawai‘i at Mānoa, United StatesReviewed by:
Vera Gorbunova, University of Rochester, United StatesCharles Rice, Clemson University, United States
Copyright © 2021 Robeck, Fei, Haghani, Zoller, Li, Steinman, Dirocco, Staggs, Schmitt, Osborn, Montano, Rodriguez and Horvath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Todd R. Robeck, dG9kZC5yb2JlY2tAc2Vhd29ybGQuY29t; Steve Horvath, c2hvcnZhdGhAbWVkbmV0LnVjbGEuZWR1
†These authors have contributed equally to this work
 Todd R. Robeck
Todd R. Robeck Zhe Fei
Zhe Fei Amin Haghani
Amin Haghani Joseph A. Zoller
Joseph A. Zoller Caesar Z. Li2
Caesar Z. Li2 Karen J. Steinman
Karen J. Steinman Stacy Dirocco
Stacy Dirocco Todd Schmitt
Todd Schmitt Magdalena Rodriguez
Magdalena Rodriguez Steve Horvath
Steve Horvath