- 1School of Biological Sciences, University of Queensland, Brisbane, QLD, Australia
- 2Flanders Marine Institute, Ostend, Belgium
- 3Faculty of Sciences, Ghent University, Ghent, Belgium
Daily environmental oscillations that follow Earth’s rotation around the Sun set a metronome for life, under which all organisms have evolved. Entrainment to these cues allow organisms to rhythmically set the pace of their own endogenous biological clocks with which the timings of diverse cellular activities are coordinated. In recent years, our knowledge of biological rhythms has extended across all domains of life. This includes both free-living and symbiotic life forms. With the insurgence of metagenomic sequencing tools, the field of holobiont chronobiomics (encompassing chronobiology of host and its associated microbiota) has recently opened and gained significant traction. Here, we review current knowledge regarding free-living prokaryote rhythmic regulation before exploring active areas of research that consider the coordinated rhythmic regulatory activities of hosts and their symbionts as a single entity, i.e., holobiont, and even the extent to which rhythmicity influences virus–host interactions. We describe rhythmicity within non-photosynthetic bacteria, cyanobacteria, and archaea, before investigating the effect of light, and, thus, diel cycle, on viral life cycles and host–virus population dynamics in marine planktonic ecosystems along with their potential to influence host cyanobacterial circadian clocks. We then explore current evidence outlining coordinated rhythmic regulation within marine holobionts and the significance of this for holobiont health and adaptive fitness that, in turn, optimizes their success within their local environments. Finally, we assess the critical role of circadian regulation for holobiont innate immunity and metabolism within well-studied non-marine mammalian systems, and, thus, assess how this can guide us within understudied marine chronobiomics research.
Introduction
The phrase “no man is an island” as put by the 17th-century metaphysical poet John Donne in his prose work ‘Devotions upon Emergent Occasions’ talks about how humans do not do well in isolation and require a community to thrive (Donne, 1987). Similarly, in light of current research, it is becoming increasingly evident that no organism can be considered in isolation. Beyond considering organisms simply within the ecological communities they reside in, it is imperative we also consider the ecological communities they harbor within—their microbiome.
Since the dawn of metagenomic sequencing, research into microbiome community composition has exponentially advanced, shedding light on the complexity of host–symbiont relationships (Huttenhower et al., 2014; Norman et al., 2014; Yoon et al., 2015; Jovel et al., 2016). Yet, much regarding the synchrony of these interactions remains elusive. Rhythmic regulations, including the circadian (Supplementary Box 1) rhythms (∼24-h endogenous biological clocks; Supplementary Box 2) set a metronome for coordinated activity across all domains of life (Edgar et al., 2012), and not only warrant exploration of their critical role in mediating individual cellular activity but also for their role in mediating the rhythmically coordinated activities between host and their symbionts, ultimately facilitating successful adaptive optimization of the holobiont (Supplementary Box 1).
In recent years, the significance of circadian clock regulation has become increasingly apparent. Entrained by external cues, circadian regulation mediates the rhythmic expression of numerous genes under the locally fluctuating environment, including those of innate immunity, metabolism, behavioral responses, cellular division, and global gene expression (Mori et al., 1996; Markson et al., 2013; Thaiss et al., 2016; Parkar et al., 2019). Indeed, the origins of circadian regulation are considered ancient, hypothesized to have first arisen under the strong selecting forces of high ultraviolet (UV-B and UV-C) radiation. Prior to the formation of the Earth’s protective ozone layer, light-vulnerable processes were entrained to the solar night, termed the ‘escape from light’ hypothesis (Pittendrigh, 1993; Nikaido and Johnson, 2000). However, despite these ancient origins, circadian clocks have independently evolved, multiple times across distinct lineages, but with convergent functions (Rosbash, 2009; Takahashi, 2017), and, thus, likely have wide-reaching functional roles in disparate organisms.
Within marine ecosystems, the rhythmic activities and regulation of diverse organisms pertain significantly on a macro-scale (Häfker et al., 2017). The open ocean of the earth’s marine ecosystems comprises vast planktonic communities, important globally for large-scale biogeochemical cycling. These communities comprise a wide variety of organisms from all domains including algae, bacteria, protists, and metazoans. Here, we consider the rhythmically aligned regulation of these communities, entrained to cyclically fluctuating external environmental stimuli. Viruses, though hardly considered living, form an important part of the marine planktonic community and play a huge role in (re)shaping the community structure. Viruses are entirely dependent on their host for proliferation and in the process, often lyse their hosts. The spatiotemporal dynamics of virioplankton communities, like other subsections of the planktonic communities, depend on various direct and indirect biotic and abiotic factors (reviewed in Mojica and Brussaard, 2014). The natural spatiotemporal dynamics of virus–host communities in the marine environment becomes a topic of discussion on the biological rhythms of marine viruses.
Similarly, rhythmic regulation is also significant on a micro-scale, for the nested ecological communities comprising host–symbiont holobionts. Eukaryotic circadian clock mechanisms have been well-studied in diverse multicellular organisms including mammals, arthropods, sponges, and plants (McClung, 2001; Dibner et al., 2010; Jolma et al., 2010; Jindrich et al., 2017; Morioka et al., 2018). Within mammals, a hierarchical system comprises a host of individual circadian clocks that are all centrally aligned by a core circadian clock within the suprachiasmatic nucleus of the hypothalamic region of the brain (Dibner et al., 2010). This allows individual cells of different tissues to independently mediate rhythmic gene expression on diverse periods, appropriate for their cellular function (Dibner et al., 2010). Interestingly, a comparable system has been hypothesized to occur within animal-bacterial holobionts. Here, the central endogenous clock of the host coordinates rhythmic alignment amongst all independent clocks within the microbial symbionts of the microbiome (Lee et al., 2019). Elucidating host and symbiont rhythmic co-regulation could provide further key insights into the establishment of host–symbiont relationships. For instance, whether circadian clock alignment provides an important selection filter in host–symbiont compatibility (temporal mutualism), or whether prokaryotic symbionts first colonize, and later adapt their circadian cycle to match that of their host to stabilize the relationship.
Research into holobiont chronobiology, the dynamic interaction of host–microbe circadian clock regulation, has predominantly focused upon mammalian organisms and provides only tantalizing glimpses at applicable prospects within other holobiont systems. However, considering the global ubiquity and significance of prokaryotes within marine environments, either as symbiotic or free-living forms, and the scarcity of available studies (Sartor et al., 2019), we must extend circadian research to encompass a greater degree of prokaryotic diversity and their crucial functions within holobionts. Here, we outline the currently very limited data from marine holobionts, as well as from more extensively studied terrestrial/mammalian holobionts with the aim that such research can help accelerate that within organisms from marine ecosystems. Indeed, we propose that future chronobiomics (Supplementary Box 1) research should increase its focus upon globally diverse and dominant phyla that comprise basal marine metazoa, such as Porifera, Cnidaria, and Ctenophora. These organisms comprise considerably less complex body plans, making them tractable model systems for the investigation of molecular circadian holobiont regulation. Additionally, since these organisms comprise ancient lineages that were the earliest to diverge from the rest of the animal kingdom (i.e., bilaterians), any shared mechanisms of rhythmic regulation identified may logically be traced back to the last common animal ancestor.
In this review, we first outline the mechanisms of circadian clock regulation within free-living prokaryotes, including non-photosynthetic bacteria, cyanobacteria, and archaea. Secondly, we consider the implication of biological rhythms on marine viruses. We explore the dynamics of host–virus populations and the effect of light in controlling planktonic community dynamics. We also delve into the understudied niche of viral influences upon host biological rhythms by discussing host photosynthesis take-over by the viruses. In the third part, we discuss the significance of circadian rhythm regulation for holobiont health and optimization (using well-studied terrestrial mammalian examples) and, thus, how such chronobiomics research must be considered in the marine environment. We present the currently limited evidence of circadian clocks within marine holobiont systems, demonstrating host–symbiont rhythmically aligned regulatory activity. We aim to provide a comprehensive review of holobiont rhythmic regulation, which promotes the consideration and extension of the universally significant field of chronobiomics research within marine environments.
Biological Rhythms in Prokaryotes
Current knowledge of prokaryotic circadian regulation is severely lacking. To date, prokaryotic circadian clocks have been formally described within the phylum Cyanobacteria only, of which Synechococcus elongatus serves as the current ‘model’ organism (Johnson et al., 2017). That said, evidence of prokaryotic rhythmicity has been around for some time.
Evidence of rhythmic regulation within prokaryotes (outside of Cyanobacteria) dates back as early as the 1930s. Despite early hypotheses that circadian rhythms would not be adaptive for single-celled organisms, for which generation times are shorter than the diel cycle (“circadian-infradian rule”; Ehret and Wille, 1970), studies did report rhythmic periods for bacteria that, remarkably, surpassed the duration of their cell cycles. Within Escherichia coli cultures, Rogers and Greenbank (1930) demonstrated a growth period indicative of long circadian-like rhythms that were later shown to be periods of 20.5 h (Halberg and Conner, 1961). However, later observations demonstrated this was not fully temperature compensated (Supplementary Box 2) and, thus, does not display a fully robust circadian clock (Sturtevant, 1973). Within Alphaproteobacteria Rhodospirillum rubrum, enzymatic activity of hydrogenase (Hup) uptake, which participates in redox metabolism during photosynthesis and nitrogen fixation, demonstrated circadian rhythmic oscillations of 12–15 h (Van Praag et al., 2000). Moreover, non-photosynthetic bacteria Klebsiella pneumoniae and Klebsiella aerogenes displayed rhythmic growth and activity, respectively (Sturtevant, 1973; Paulose et al., 2016); of which K. aerogenes demonstrated a fully temperature-compensated rhythm. Thus, diverse bacteria from multiple phyla, likely, rhythmically regulate their cellular activities on either a circadian-like or ultradian (Supplementary Box 1) period.
Prokaryotic Clock Molecular Mechanisms
Molecular mechanisms of the prokaryotic circadian clock were first described for S. elongatus in 1997, which has since become one of the best-studied circadian systems from all domains (Sartor et al., 2019). Remarkably, the prokaryotic circadian clock is a post-translational system (as opposed to the transcription-translation feedback loop (TTFL), see Supplementary Box 3), which relies solely on the phosphorylation cycles of three proteins (KaiA, KaiB, and KaiC) causing rhythmic oscillations throughout the day (Figure 1; Ishiura et al., 1998). This system has since been more generally classified as a gene expression feedback loop (GEFL).
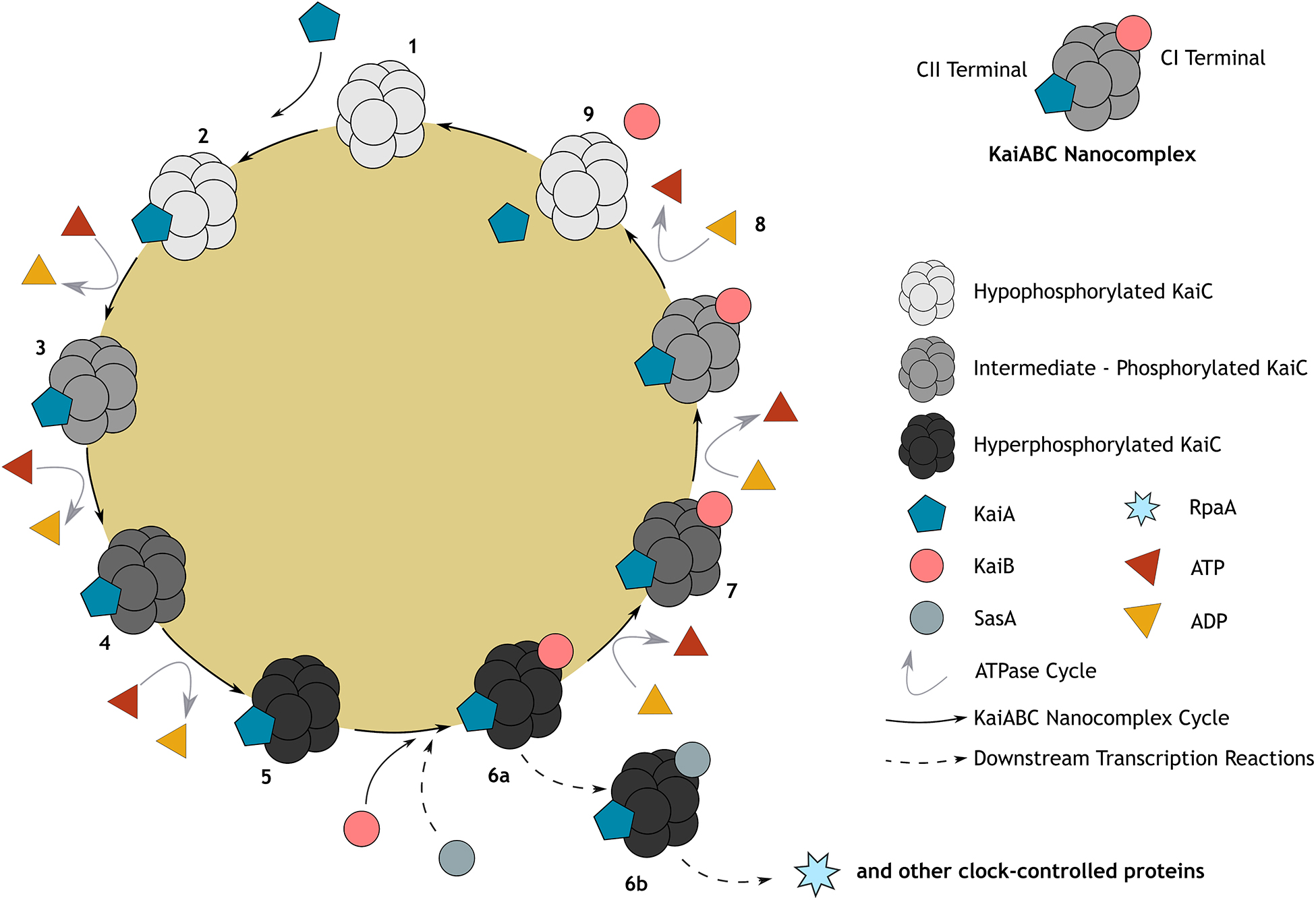
Figure 1. Representation of the canonical cyanobacterial post-translational (KaiABC) circadian clock. Post-translational oscillation occurs through a cycle of association and disassociation from the KaiABC nanocomplex, embedded within a gene expression feedback loop (GEFL). KaiC, a double hexamer (1), serves as the central oscillator and is both an autokinase and autophosphatase, exhibiting ATPase activity (Cohen and Golden, 2015); whilst KaiA and KaiB rhythmically bind to KaiC either leading to its phosphorylation or de-phosphorylation, respectively. Briefly, unphosphorylated KaiC (1) is stimulated through its interaction with KaiA at the CII terminal, leading to KaiC phosphorylation in the CII domain (2–5). At the point at which KaiC reaches hyperphosphorylation (5) of two key residues (T432 followed by S431), KaiB will then bind to the CI terminal of KaiC stably (6a). This sequesters KaiA activity and once again initiates KaiC dephosphorylation (6a–8; Kitayama et al., 2003; Chang et al., 2012). When KaiC is hypophosphorylated, KaiB and KaiA are released, launching a new cycle (9). Because of their similar structure, KaiB and SasA (Kai-C interacting histidine kinase) compete to bind with KaiC (6b), regulating downstream transcriptional factors such as RpaA (transcriptional factor) and global gene expression of clock-controlled genes, including those that drive the essential oscillators, KaiABC (Takai et al., 2006; Taniguchi et al., 2010; Johnson et al., 2011). At the core of this cycle, activity of ATPase (predominantly at the CI domain of KaiC) sustains the robust temperature compensated rhythms through catalyzing a remarkably slow rate of ATP hydrolysis (15–16 ATP molecules hydrolyzed by one KaiC monomer per day). This is hypothesized to be the rate-limiting component of the cycle, mediating KaiABC activity such that they oscillate precisely to produce a period of ∼24 h (Terauchi et al., 2007; Abe et al., 2015).
The cyanobacterial KaiABC system supports a fully robust, self-entrained, temperature-compensated circadian clock (Merrow et al., 2006). Notably, as the only circadian system to have been demonstrated both in vitro and in situ, it has been possible to demonstrate the adaptive benefit of circadian-mediated cellular activity. In vitro growth competition experiments demonstrated that wild-type cyanobacteria outcompeted their arrhythmic counterparts whilst exposed to variable environmental conditions (Ouyang et al., 1998; Woelfle et al., 2004). Indeed, within Cyanobacteria, the circadian clock provides a critical regulatory function for numerous cellular processes. For instance, circadian regulation can mediate cell division (Mori et al., 1996; Mori, 2009), global gene expression (Liu et al., 1995; Markson et al., 2013), chromosome compaction (Smith and Williams, 2006), nitrogen fixation (Grobbelaar et al., 1986; Mitsui et al., 1986), amino acid uptake (Chen et al., 1991), and metabolism (Diamond et al., 2015). Given the importance of such activities to the functions that symbionts perform for their hosts, the circadian regulation of such must also be considered within the context of the holobiont.
However, outside of Cyanobacteria, evidence of fully robust self-entrained endogenous rhythms has yet to be molecularly described within other prokaryotes. Currently, the gammaproteobacterium Enterobacter aerogenes, housed within human gut microbiota, is considered to be the only observation of a fully robust clock outside of Cyanobacteria (Paulose et al., 2016). Specifically, E. aerogenes display robust, temperature-compensated circadian rhythms of luciferase activity that synchronize on daily 24-h cycles with the secretion of the gastrointestinal hormone melatonin (Paulose et al., 2016). Thus, true ∼24-h period rhythms are likely more prevalent amongst prokaryotes, and indeed bacterial symbionts than our current observations suggest. Interestingly, Cyanobacteria are the only bacterial phyla to possess KaiA (Schmelling et al., 2017). Subsequently, it is hypothesized that the evolution of KaiA facilitates fully robust circadian rhythms to sustain regardless of the external environment, and even in the absence of any environmental stimuli (Ouyang et al., 1998; Ma et al., 2016). Indeed, members of the cyanobacterial genus Prochlorococcus, from which KaiA was evolutionarily lost, cannot sustain free-running rhythms under conditions of constant light or dark (Holtzendorff et al., 2008).
Nonetheless, several other key prokaryotic circadian clock components have been identified in diverse bacteria. Several circadian gene homolog searches based on S. elongatus sequences have identified the presence of KaiBC homologs, as well as circadian associated input and output factors amongst diverse prokaryotes (Loza-Correa et al., 2010; Schmelling et al., 2017). Additionally, orthologs of KaiB and KaiC are observed within alphaproteobacteria Rhodobacter sphaeroides and Rhodopseudomonas palustris, both considered to entrain “proto” circadian clocks (Min et al., 2005; Ma et al., 2016), since their rhythmicity is lost under exposure to constant environmental conditions. To encompass the anticipated broad spectrum of rhythmic regulatory mechanisms represented within diverse prokaryotic organisms, a revised, simplified criterion for circadian clock regulation has recently been proposed (Loza-Correa et al., 2010; Johnson et al., 2017; Schmelling et al., 2017). Although we may draw comparisons with the S. elongatus clock model, it is likely that circadian regulation of Proteobacteria (or other bacterial phyla) evolved through distinct environmentally mediated processes (Johnson et al., 2011). As observed in eukaryotes, the core proteins involved in circadian regulatory mechanisms evolved independently several times, despite demonstrating a conserved general mechanism (Rosbash, 2009; Takahashi, 2017).
Redox Clock: An Ancient and Universal Mechanism for Circadian Rhythm Regulation
Recently, it was demonstrated that the cellular redox cycle, which prevents the build-up of reactive oxygen species (ROS), also serves as a circadian oscillator within all domains of life (O’Neill et al., 2011; O’Neill and Reddy, 2011; Edgar et al., 2012). Maintenance of ROS homeostasis is critical to survival since oxidative stress caused from the build-up of toxic ROS (e.g., hydrogen peroxide, superoxide anion radical) can cause a variety of damaging effects, including protein oxidation, lipoprotein modification, lipid peroxidation, and DNA damage (Dalle-Donne et al., 2003; Karlenius and Tonissen, 2010; Hanschmann et al., 2013). Peroxiredoxin (PRX) proteins maintain redox homeostasis by reducing ROS such as hydrogen peroxide to water. In turn, PRXs are reduced themselves, usually consuming a reducing equivalent from nicotinamide adenine dinucleotide phosphate (NADPH). The cyclical activity of peroxiredoxin oxidation-reduction reactions occur rhythmically, overlain by circadian oscillations in the redox state (O’Neill et al., 2011; O’Neill and Reddy, 2011), although the underlying molecular mechanisms for this are not yet fully understood (Xia et al., 2018). ROS is produced as a by-product of metabolic reactions, including respiration and photosynthesis (Turrens, 2003; Pospíšil, 2016), thus, the timing of ROS production, aligned with environmental stimuli may provide an additional circadian zeitgeber (Supplementary Box 1).
The discovery of the redox clock suggests an intricate link between innate immunity (to which ROS contributes) and circadian regulation. Peroxiredoxin proteins protect against stress; for instance, Peroxiredoxin 6 (PRX6) protects supercoiled DNA damage by oxidative stress and negatively regulates toll signaling transduction in red swamp crayfish Procambarus clarkii (Chu et al., 2019). Thus, PRX circadian cycling may maximize efficiency in maintaining ROS cellular homeostasis by aligning PRX activity in anticipation of daily, fluctuating environmental conditions, that may signal innate immune responses. For instance, it has been demonstrated that redox rhythms can be aligned to multiple periods, including both circadian and circatidal (Supplementary Box 1), within the intertidal isopod Eurydice pulchra (O’Neill et al., 2015). Subsequently, an unprecedented environmental stimulus could lead to circadian disturbance and the onset of a ROS-induced stress response/signal.
The PRX active site is highly conserved across all domains of life, representing an ancient, ancestral circadian mechanism that likely predates the canonical circadian clock (Edgar et al., 2012; Hoyle and O’Neill, 2015). Edgar et al. (2012) hypothesize that the redox clock evolved under strong selecting forces during the great oxidation event (∼2.5 billion years ago) when organisms were first exposed to toxic ROS concentrations. They further speculate that, in the absence of such pressure, organisms inhabiting anoxic environments would lack a circadian redox clock (Edgar et al., 2012). Interestingly, the redox clock functions in tandem with, but independently of the GEFL/TTFL canonical circadian clock (Supplementary Box 3). Organisms lacking either endogenously or experimentally (pharmacologically abrogated) canonical circadian clocks can sustain rhythmic cycles of redox regulation, albeit with an altered rhythm (O’Neill et al., 2011; Edgar et al., 2012). This supports the hypothesis that both clock mechanisms evolved independently, but under common selecting pressures, enabling rhythmic synchrony. Since PRXs are present in most organisms (Rosbash, 2009), the redox clock could provide a universal, conserved indicator of non-canonical, post-translational circadian cycling (Edgar et al., 2012).
Circadian Rhythms in Archaea
Archaea are one of the oldest domains of life (Gribaldo and Brochier-Armanet, 2006) and contribute an integral part to the symbiont community of holobionts globally (Bang and Schmitz, 2018). For instance, they may dominate the microbiomes of organisms from environmental extremes, such as highly acidic temperate waters (Korzhenkov et al., 2019). Thus, for a holistic understanding of circadian clock evolution and its role within holobiont systems, rhythmic regulation within archaea must also be explored. To date, archaeon rhythmic activity and circadian clock components have only been identified within the halophilic, aerobic genus Halobacteria (Whitehead et al., 2009; Edgar et al., 2012; Maniscalco et al., 2014; Johnson et al., 2017). Whitehead et al. (2009) first reported diurnal oscillatory transcription (either ultradian or circadian) within Halobacterium salinarum NRC-1, demonstrating rhythmic control for ∼12% of global gene expression under constant conditions after 3 days entrainment in light-dark. Furthermore, Maniscalco et al. (2014) characterized four cyanobacterial circadian clock kaiC homologs (cirA, cirB, cirC, and cirD) within the halophilic, mesophilic archaeon Haloferax volcanii that display rhythmic transcriptional regulation under diel conditions. However, the potential mechanisms underlying archaeal circadian regulatory systems are still far from clear. Although homologs for S. elongatus KaiC are widespread amongst all three archaeal phyla (Maniscalco et al., 2014), they are also much more diversified, with their potential functions mostly unknown. Indeed, Makarova et al. (2017) alternatively demonstrated a proposed central role for archaeal KaiC homologs within signal transduction mechanisms. The largely expanded family of KaiC homologs within archaea lends support to the hypothesis that KaiC first originated within archaea (Makarova et al., 2017) but not necessarily as a circadian oscillator, since to date, this has only been observed within the genus Halobacteria. Considering archaeal and bacterial lineages diverge over distances even greater than that with Eukaryota, it would not be surprising to discover an archaeon circadian mechanism distinct from that observed within bacteria. Thus, considering also the comparatively limited research within this area, it remains feasible that the archaeal canonical circadian oscillators have yet to be identified. That said, evidence of the post-translational, circadian redox clock has recently been observed within archaea and is likely widespread amongst species inhabiting oxygenated environments. This opens new directions to explore rhythmic regulation within this less studied but equally important, prokaryotic domain (Edgar et al., 2012; Hoyle and O’Neill, 2015).
Biological Rhythms and Marine Viruses
Sufficient literature indicates that the cellular regulation of most, if not all, bacterial lineages are rhythmically influenced, albeit with likely diverse periods, but can these rhythms also influence viruses, that are hardly considered living? This is an area of research that has received little attention to date. Thus, we will explore the intriguing subject of biological rhythms concerning viruses in the marine environment. Typically, viruses are considered highly infectious, disease-causing lethal entities that wreak havoc. Indeed, considering the COVID-19 pandemic and its societal impact (Heymann and Shindo, 2020), this view is not entirely wrong. However, knowledge of marine and other aquatic viruses, along with their roles in ecosystem functioning have barely been valorized and are deserving of a more prominent focus.
In 1989, it was discovered that viral abundance within marine environments may be as high as millions within a cubic milliliter (Bergh et al., 1989). Subsequently, this led to the recognition of viruses as the most abundant biological entity within our oceans; totaling up to 1030 virus-like particles (Wommack and Colwell, 2000; Breitbart, 2012). Viruses form the ocean’s second-largest biomass, second only to the prokaryotes (Suttle, 2005). With the capacity to infect all forms of life, viruses lyse considerable volumes of daily prokaryotic production in the marine environment. Thereby, facilitating the flow of nutrients from higher trophic levels back to the lower levels, in what is known as the ‘viral shunt’ (Fuhrman, 1999; Wommack and Colwell, 2000; Koonin et al., 2006; Suttle, 2007; Miranda et al., 2016). Subsequently, one cannot stress enough the ecological importance of viruses for the functioning of marine ecosystems.
To better understand the influence of biological rhythms, we must consider the spatiotemporal community dynamics of viruses within marine planktonic assemblages. In this section, we will discuss the importance of light as a crucial abiotic factor affecting community dynamics through the viral life cycle, focusing specifically on the intensity, duration, and quality of light in determining viral activity and burst size. Lastly, we will discuss the influence of viruses on their cyanobacterial hosts’ biological rhythm through redirecting photosynthesis to enhance viral replication.
Variables Influencing Host–Virus Population Dynamics Within the Marine Environment
Viruses heavily influence the planktonic ecosystems through (re)shaping, and hence mediating, planktonic community dynamics. Two major hypotheses have been proposed to explain the dynamics of marine viral ecology- ‘Kill the Winner’ (KtW) and ‘Piggyback the Winner’ (PtW) (discussed in Supplementary Box 4).
When assessing marine planktonic communities, the significance of cyanobacterial abundance and their role in carbon fixation must not be overlooked (Falkowski, 1994). Marine phytoplankton communities are heavily dominated by photosynthetic cyanobacteria, of which the estimated absolute annual mean cellular abundances of Synechococcus spp. and Prochlorococcus spp. are as high as 7.0 ± 0.3 × 1026 and 2.9 ± 0.1 × 1027 cells across the world’s oceans, respectively (Flombaum et al., 2013). The population dynamics of these cyanobacteria have been studied in a wide range of environments and at different spatial and temporal scales (Flombaum et al., 2013). Brunet et al. (2007) reported a 24 h oscillation in the cell cycle profile for both Synechococcus and Prochlorococcus genera. They observed a peak in the percentage of cells in the G2 phase of the cell cycle at the end of the night for Synechococcus and at the end of the light period for Prochlorococcus at the Strait of Sicily in the Mediterranean Sea (Brunet et al., 2007). Diel dynamics of cyanophage (i.e., phages that infect cyanobacteria) abundance was found to be synchronized with light-dark cycles (Suttle and Chen, 1992). Short-term temporal sampling was conducted at 1 m depth every 3 h for 3 days in the Northwest Mediterranean Sea to explore the activity of various microbial groups including cyanobacteria Synechococcus spp., heterotrophic bacteria, nanoflagellates, and viruses (Bettarel et al., 2002). Synechococcus spp. was observed to reach an abundance peak at midnight and a trough at 9:00 p.m., although the highest number of Synechococcus spp. dividing cells were observed at 9:00 p.m. This diel pattern was consistent throughout the 3-day sampling effort. Viral abundance also showed diel oscillation with a peak at 6:00 p.m. and trough at midnight. Although the authors did not measure Synechococcus-specific viruses, they concluded that the observed decrease in Synechococcus abundance at the time of viral peak was due to viral production (Bettarel et al., 2002).
It is important to note that the abundance and infectivity of marine viruses are also dependent on the loss of viruses via various means of decay, including, but not limited to, removal through adhesion on particulate matters, consumption by heterotrophs such as bacteria and flagellates, and destruction of infectivity by solar radiation (Suttle and Chen, 1992). In the absence of solar radiation, viral decay rates range from 0.009 to 0.028 h–1, but in full sunlight may be as high as 0.4 to 0.8 h–1. Decay rates in full sunlight were demonstrated to be 2 to 10 times higher than those in the absence of sunlight, even when wavelengths of <320 nm were removed, or 80% of the incident irradiance was excluded (Suttle and Chen, 1992). Specifically, UV-B radiation has been shown to have a significant effect in reducing viral infectivity, though a strong link between UV-A radiation and viral infectivity is yet to be described (reviewed by Mojica and Brussaard, 2014). The average decay rate due to flagellate consumption was around 0.15 h–1 (Suttle and Chen, 1992). It is important to note, that viral infectivity is reduced by the solar radiation at a greater rate than the removal of viruses from the water column due to grazing by heteroflagellates, and, thus, a large proportion of viral abundance measured (by electron microscopy, epifluorescence microscopy, and flow cytometry) are at a state of decay and rendered non-infective (Suttle and Chen, 1992). Subsequently, any direct inferences made on viral activity from viral enumeration methods (mentioned above) may not be representative; as they cannot differentiate between infectious viruses, non-infective viruses, and virus-like particles (Supplementary Box 1, Suttle and Chen, 1992). Here, we emphasize the importance of performing viral activity assays in addition to the abundance counts to estimate true proportions of plankton host losses due to viruses (Winget et al., 2005; Baudoux et al., 2006; Evans et al., 2009).
Recently, Kolody et al. (2019) described the expression of viral and host transcripts with a focus on transcripts that exhibited diel oscillatory patterns. Co-expression of host and viral transcripts were observed for diverse reference taxa, including heterotrophic bacteria, cyanobacteria, photosynthetic eukaryotes, and predatory heterotrophic eukaryotes. Interestingly, this opposed Thingstad’s (2000) ‘Kill-the-Winner’ hypothesis. As per the KtW, Kolody et al. (2019) hypothesized an increase in host transcript in the planktonic community should have been preceded by an increase in viral transcripts. An exception to this was the aggregated gene expression of algae Phaeocystis globosa, which peaked during day hours, compared to a synchronized increase in expression of the giant Phaeocystis globosa virus and virophages (Supplementary Box 1). This pattern was not observed for the ssRNA viruses on which viral RNA molecules lagged host transcription (Kolody et al., 2019). Therefore, the KtW hypothesis holds for RNA viruses and viruses of Phaeocystis, whilst the PtW for the other measured groups.
Additionally, the trophic status of water leads to variation in viral production and decay rates. High nutrient conditions can increase bacterioplankton growth rate and viral burst size (number of viruses released per cell due to viral lysis). Bongiorni et al. (2005) investigated the viral dynamics of production and decay along a trophic gradient in the North Adriatic basin. A decrease in burst size was observed down the trophic gradient, attributable to decreased nutrient availability and smaller cell sizes (Bongiorni et al., 2005). Indeed, a 1.3 to 10.7% viral decay rate of the total viral production was observed in oligotrophic waters, compared with 40% in the eutrophic waters of the Adriatic Sea. But the production rates in Adriatic eutrophic waters were also three to four times higher than that of oligotrophic waters; pointing toward higher viral production and decay rates in eutrophic versus oligotrophic waters (Bongiorni et al., 2005). Grazing by nanoflagellates, dissolved-colloidal substances, and lysogenic infections was responsible for the removal of ca. 66% of viral production in eutrophic waters as compared to 17% in oligotrophic waters (Bongiorni et al., 2005).
Oscillations in marine viral populations are dependent on various direct and indirect factors including, but not limited to, viral inactivation by light, trophic status of the water, abundances of their host, grazing by nanoflagellates, and the latency period (Suttle and Chen, 1992; Bongiorni et al., 2005; Mojica and Brussaard, 2014). These variables pose a significant challenge in the field of marine viral ecology, particularly in the study of spatiotemporal community dynamics of viruses and their host and hence must be meticulously considered.
The Effect of Light on Proliferation of Marine Viruses
Environmental light can significantly influence the viral life cycle, and, thus, is likely the most important factor when considering marine viral chronobiomics. Variation in the duration, intensity, and wavelength of light may all influence various stages of the viral life cycle. In short, a typical life cycle of a lytic virus begins with adsorption on a competent host followed by entry of the viral particle or the genetic material, transcription of the genes required for viral replication, replication of the viral genome, assembly of virions, and finally release through host lysis (Dimmock et al., 2009). Temperate viruses are known to incorporate themselves into the host genome as prophages and proliferate with the host (Dimmock et al., 2009). The effect of light on the adsorption of the virus and replication has been a predominant focus of current research in the field of marine viral ecology.
Viral–Host Adsorption
Host susceptibility to a virus is determined by specific interactions between the receptors present on the host and either the viral-encoded envelope (enveloped viruses), capsid protein (non-enveloped viruses), or proteins present on the tail (bacteriophages) (Rakhuba et al., 2010; Thorley et al., 2010).
Although light is well known to influence downstream viral replication, there is currently no consensus for the dependence of viral adsorption on light (Ni and Zeng, 2016). Assessing the diurnal rhythms of infection by certain cyanophages, Liu et al. (2019) observed three different diel-dependent life-history traits of phages when infecting cultured Prochlorococcus and Synechococcus in the continuous dark. These include the absence of adsorption, presence of adsorption but not viral replication, and presence of both adsorption and viral replication. Specifically, P-HM2 and P-SSM2 cyanomyophages exhibited a diurnal rhythm in adsorption on Prochlorococcus strains MED4 and NATL2A, respectively, with maxima during the day (Liu et al., 2019). On the other hand, cyanomyophage P-SSP7 adsorption on the Prochlorococcus strain MED4 was independent of the light-dark cycle (Liu et al., 2019). Alternatively, Cséke and Farkas (1979), demonstrated that adsorption of AS-1 cyanomyophage on Anacystis nidulans (synonym S. elongatus PCC 6301) does have a light-dependent adsorption pattern that is independent of photosynthesis and host circadian rhythm (discussed later) (Ni and Zeng, 2016). Around 70% of the cyanomyophage AS-1 inoculate titer absorbed to S. elongatus PCC 6301 in the presence of light. This figure was dropped to 40% when the cultures were moved to the dark right after addition of viruses (Cséke and Farkas, 1979; Ni and Zeng, 2016). A similar pattern was also observed for S. elongatus PCC 7942 and AS-1 phage (Kao et al., 2005). It was long believed that the presence of light always facilitated viral adsorption, however, Jia et al. (2010) disproved this in their study comparing the adsorption patterns of nine cyanophages on unsynchronized Synechococcus WH7803 cells. They demonstrated that the cyanomyophages, S-PWM1, S-BM3, S-MM4, S-MM1, and S-MM5 were not affected by light and adsorbed to unsynchronized WH7803 cells to the same extent (∼90%), regardless of light presence; thereby displaying a light-independent adsorption pattern. Conversely, the cyanomyophage S-BNM1, S-PWM3, S-PM2, and cyanopodophage S-BP3 exhibited significantly lower adsorption of ∼10% in the dark as opposed to adsorption of ∼90% in the presence of light (Jia et al., 2010). To test for an influence of host circadian rhythm on phage adsorption, synchronized Synechococcus WH7803 cells (with known circadian rhythm), maintained in 12:12 light-dark (LD), were infected with the light-dependent cyanomyophage S-PM2. Host cultures were sampled at six different timepoints throughout the 24-h LD cycle from two sets (three in the light period, three in the dark period); one incubated with S-PM2 under constant light, and the other under constant dark, for 45 min before measuring the free phage titer. A similar pattern of ∼90% adsorption in the light and ∼10% adsorption in dark was observed for S-PM2 and synchronized WH7803 cells, indicating adsorption of S-PM2 is independent of the host circadian rhythm (Jia et al., 2010; Ni and Zeng, 2016). However, to conclusively demonstrate the independence of viral adsorption from host circadian rhythm, this experiment should also be conducted on light-independent phages (Ni and Zeng, 2016).
Failure of viruses to adsorb in the dark is immediately reversed upon reillumination, indicating a light-dependent temporary change (Jia et al., 2010). This may either be a function of host receptor protein conformational changes or be associated with long-tail fibers (LTFs) of the phages (Jia et al., 2010). In T4 myovirus, protein whiskers (encoded by the wac gene) extending outward from the collar region (Conley and Wood, 1975) control retraction and extension of LTFs and determine the competency of adsorption with respect to environmental abiotic factors, including temperature, ionic strength, pH, and biotic cofactors such as tryptophan (Conley and Wood, 1975). It is hypothesized that the cyanomyovirus S-PM2 possesses an environment sensing mechanism that prevents phage adsorption in the dark by maintaining LTFs in a retracted position (Jia et al., 2010). However, yet no homolog of wac has been found within the genome of S-PM2 (Mann et al., 2005; Clokie et al., 2008). When comparing T4 orthologs of the wac gene, only a short, conserved segment was identified at the N-terminus (Letarov et al., 2005). Subsequently, this leaves the possibility of the discovery of a wac homolog in S-PM2 cyanophage open (Jia et al., 2010). Additionally, Clokie et al. (2010) described the presence of distinct whiskers on a newly isolated cyanomyovirus, strongly suspected to be a potential product from a wac gene homolog. To conclude, it can be noted that though certain wavelengths of the electromagnetic spectrum differentially promote viral decay, light is proven to be essential for adsorption for many viruses. However, to the best of our knowledge, the diel nature of host and viral receptors has not been explored and/or documented.
Viral Replication
Post initial infection, viruses must replicate using their hosts’ replication machinery which, in turn, may be regulated in a rhythmic manner (i.e., circadian or diurnal). A strong dependence of cyanophage replication upon light presence and intensity has been demonstrated in various cyanobacterial hosts (Lindell et al., 2005; Dimmock et al., 2009). For instance, Kao et al. (2005) demonstrated a strong positive correlation in burst size with increasing intensity of light. A two-log fold increase was observed in AS-1 cyanomyophage production in S. elongatus PCC 7982 when light intensity increased from 5 to 25 μMol m–2 s–1. Indeed, the effect of light on viral replication has been shown for various cyanobacteria including, Prochlorococcus spp. (Lindell et al., 2005; Thompson et al., 2011), Nostoc muscorum (Adolph and Haselkorn, 1972), and Plectonema boryanum (Padan et al., 1970). Notably, a minimum exposure time was also required for successful infections and subsequent viral replication; with a 10 times higher viral production rate observed when the host cultures were incubated with viruses for at least 3 h as compared to 1 and 0.5 h, during a 10-h cycle. Notably, a decreased transcript abundance was reported when the T4-like myovirus P-HM2 was incubated under complete darkness with Prochlorococcus MED4 (Thompson et al., 2011). Such observations demonstrate a clear reliance on light for cyanophage replication. However, it must be considered, that this could alternatively be due to the direct influence of light on the photosynthetic capacity of the cyanobacterial host, in turn, influencing host replication machinery, or through the activity of the auxiliary photosynthetic genes brought in by the viruses (discussed below), or indeed a mixture of both.
To confirm the reliance of viral replication on photosynthesis, photosynthetic inhibitors, DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea) and CCCP ([(3-chlorophenyl)hydrazono]malononitrile) were used. A reduction in phage genomic DNA copies was observed by Lindell et al. (2005) when the phage-infected bacterial samples were incubated with DCMU. Moreover, Mackenzie and Haselkorn (1972) found a complete suppression in viral production when cyanosiphophage SM-1 was incubated with Synechococcus strain NRC-1 in the presence of DCMU and CCCP. Subsequently, it can be concluded that cyanophages acquire their energy by exploiting host photosynthetic mechanisms.
Interestingly, phages are also known to carry a myriad of host genes termed as ‘auxiliary metabolic genes’ (AMGs). Mann et al. (2005) reported the presence of psbA and psbD genes encoding proteins D1 and D2 of photosystem II, respectively, within cyanomyophage S-PM2. These two proteins are critical in curbing photoinhibition. Therefore, the presence of these genes in a phage infecting cyanobacteria might prevent the loss of photosynthetic activity due to photodamage. Notably, the D1 protein from S-PM2 is similar to that in Synechococcus sp. WH8102. This likely indicates a direct acquisition from the cyanobacteria through horizontal gene transfer. The psbA and psbD genes were interrupted by an unrelated non-photosynthetic gene in the S-PM2 genome, suggesting independent acquisitions of these two genes (Millard et al., 2004; Mann et al., 2005). In a comparison of genomes from three phages, myovirus P-SSM2, P-SSM4, and podovirus P-SSP7, Lindell et al. (2005) found that all three viral genomes possessed psbA which encodes for D1 as well as a the hli gene encoding high light-inducible protein HLIP. Specifically, the myovirus contained psbD, whereas the podovirus contained photosynthetic electron transport proteins plastocyanin (petE) and ferredoxin (petF). In all cases, the identified genes were uninterrupted and conserved, thus, indicative of functional proteins. Prochlorococcus phage D1, D2, and HLIP cluster together with those from Prochlorococcus. Likewise, Synechococcus phage D1, D2, and HLIP clustered together with those from Synechococcus. This plays in line with the cyanobacterial origin of these cyanophage AMGs. However, their widespread distribution amongst diverse clades also suggests multiple independent acquisition events throughout eons of coevolution (Lindell et al., 2005).
Light-dependent adsorption of P-HM2 and its inability to infect host cells at night is likely a result of the host’s light-sensitive receptors and/or phage tail fibers (Thompson et al., 2011; Liu et al., 2019). Although adsorption itself does not rely on photosynthesis, as demonstrated using photosynthetic inhibitors in the cyanomyovirus S-PM2 and Synechococcus spp. system (Jia et al., 2010; Liu et al., 2019), cyanophage transcription may be regulated by the photosynthetic activity of host cells. Indeed, both photosynthesis inhibitors and the absence of light can reduce cyanophage transcript abundance. P-HM2-like cyanomyophages may only infect during the day, and when host cells are metabolically active (Liu et al., 2019). Conversely, P-SSP7-like cyanopodophages may infect during both day and night, though with a relatively slow replication rate considering the lower availability of cellular resources (Liu et al., 2019). This likely provides a competitive advantage when competing for a limited number of host cells. Interestingly, P-SSM2 cyanomyophages combine both phage strategies, infecting during day and night, but replicating only during the day (Liu et al., 2019). Although to date, no clear pattern in viral adsorption to light has been observed, we can conclude that a minimum threshold of photosynthetic activity is required for successful viral replication. As there are phages that display light-independent and light-dependent patterns of adsorption with adsorption maxima during the day or the night, no generalization can be made on the effect of light on viral adsorption. The certain minimum hours of light required for the viral transcripts to be abundant, the lowered transcript abundance due to the use of photosynthetic inhibitors, and the presence of AMGs in the phage genome point to a strong dependence on light for viral replication.
Viral Interaction With Host Circadian Clock Proteins
The field of marine chronobiomics is relatively new and, thus, research into the influence of host biological rhythms on the viral life cycle and viral community structure is scarce. Viruses completely rely on their host for proliferation. Studies on marine viruses suggest a role of host circadian rhythm in regulating viral infection and progression through the life cycle (Edgar et al., 2012). Research has also been conducted in outlining the role of viruses in influencing the host circadian rhythm (Edgar et al., 2012). In the final part of this section, we discuss the hypothesis on how marine cyanophages influence their host’s circadian rhythm by redirecting host photosynthesis activity toward viral replication.
Previous descriptions that cyanomyophage S-PM2 adsorption patterns are independent of light and rhythmic synchronization are not considered conclusive since light-independent phages were not assessed (Jia et al., 2010). Kao et al. (2005) subjected the cyanomyophage AS-1 to S. elongatus PCC 7942 wild type and mutants (ΔkaiA and ΔkaiB) under constant light to assess the effect of the mutation in the host circadian clock on cyanophage infection. Since no significant difference in burst size was observed, they concluded that an absence of host circadian clock proteins does not influence cyanophage infection (Kao et al., 2005).
Currently, the specific details of the effect a cyanophage infection has on the host metabolism remains unknown. Thompson et al. (2011) explored the hypothesis that phages can influence host metabolism through bringing AMGs and directing the hosts’ metabolic response toward nucleotide synthesis. The light reaction, within uninfected cyanobacteria, yields ATP and NADPH which is consumed by the Calvin cycle to fix CO2 and produce glucose-6-phosphate, stored as glycogen. At night, the Calvin cycle is inhibited by CP12 (host regulatory protein which downregulates the activities of two Calvin cycle-associated enzymes) and the stored glycogen is utilized by the pentose phosphate pathway (PPP) to release further NADPH and ribose skeleton essential for nucleotide synthesis. In an assessment of 20 cyanophage genomes, PPP and light reaction-associated genes were commonly observed whilst those associated with the Calvin cycle were not (Lindell et al., 2005; Thompson et al., 2011). Subsequently, it is hypothesized that phages are responsible for redirecting the carbon flux toward PPP; supported by the discovery of the cp12 gene in both wildtype and cultured cyanophages. An increased NADPH to NADP (electron-deficient NADPH) ratio in cyanobacteria infected with cp12 containing cyanophages leads to the conclusion that cyanophages utilize CP12 to block the Calvin cycle and redirect this energy to the PPP pathway to promote ribose skeleton generation and, thus, nucleotide synthesis, therefore, increasing viral replication rate (Thompson et al., 2011). The glycogen metabolism rhythm (either in the Calvin cycle or PPP) is under host circadian clock regulation and can control the cellular ATP/ADP ratio (Pattanayak et al., 2014). Therefore, circadian clock outputs (glycogen) can feedback through metabolism to control the circadian clock inputs (ATP); as the production of ATP mediates the rhythm of the host circadian clock (as mentioned in section “Prokaryotic Clock Molecular Mechanisms”; Figure 1; Pattanayak et al., 2014; Ni and Zeng, 2016).
Additionally, genes encoding transaldolase, an enzyme required within the PPP and not shared with the Calvin cycle, have been identified within the cyanophage genome (Thompson et al., 2011). As with other PPP genes, the cyanobacterial host transaldolase gene has a maximal mRNA level in the evening, but transaldolase activity decreases to half of its maximum level at 10:00 a.m. the next day. Meanwhile, the activity of other PPP and Calvin cycle proteins remains relatively constant. Subsequently, a phage transaldolase may be critical in overcoming the host’s low levels of this enzyme during the day to maximally exploit the host’s photosynthetic light reactions (Waldbauer, 2010; Thompson et al., 2011). The cyanophage transaldolase (TalC) is significantly different from the host transaldolase (TalB) due to several deletions in the coding sequence, except for active site residues, essential for catalysis (Thompson et al., 2011). TalC resembles fructose-6-phosphate aldolase (FsaA) of Escherichia coli more closely than the host TalB (Thompson et al., 2011). First, the authors confirmed that the TalC had a transaldolase activity and not F6P aldolase activity, and subsequently hypothesized a higher phage transaldolase activity to support the evolutionary selection of FsaA-like TalC over that of TalB. Surprisingly, they discovered a three times lower kinetic efficiency in TalC than that of TalB. Despite the acquisition of host metabolic genes by viruses contributing to more successful infections, there are fitness trade-offs/compromises involved such as the higher metabolic cost of replicating a larger genome (Bragg and Chisholm, 2008). Reduction in gene size can also be due to the partial limitation exhibited by the capsid size (Russell and Müller, 1984). The trend of shorter phage genes compared to their host orthologs was observed across most host-phage systems (Thompson et al., 2011). Albeit the generalization on kinetic differences cannot be made without performing in vitro studies (Thompson et al., 2011). With the presence of AMGs such as psbA and psbD for increased photosynthesis, cp12 for blocking the Calvin cycle, and transaldolase to maintain the PPP, cyanophages have successfully adapted to exploit host metabolism to increase their fitness. Additionally, through the reversal of the host metabolic rhythm, cyanophages may affect the ATP/ADP ratio, and can, thus, mediate their host’s circadian clock (Ni and Zeng, 2016).
Application of Circadian Rhythms From Mammalian Microbiome Systems
Research from mammalian gut microbial systems provides the backbone to our current understanding of microbiomes. Thus, it is tempting to draw upon mammalian microbiome studies when considering less well-studied species of marine metazoa but this must be done with caution. Mammalian microbiome studies can be extremely useful, considering many microbial symbiont genera are common across diverse host species, and that the mechanisms of both the eukaryote canonical TTFL and peroxiredoxin redox clocks are highly conserved (Jolma et al., 2010; Edgar et al., 2012). However, we must also be careful to consider the disparate influences on microbial communities within divergent host systems. Human gut microbiota is highly diverse and dense, containing up to 1000 horizontally acquired species, the abundances of which go through regular daily fluctuations (Turnbaugh et al., 2009). Thus, the nested ecosystem interactions of mammalian extracellular symbionts, and, hence, influences on their circadian cycling, will be diverse and varied from other host organisms. For instance, host nutrient uptake is known to be one of the most significant influences on human gut community composition and a circadian zeitgeber influencing prokaryotic circadian clocks of the microbiome (Rothschild et al., 2018; Murakami and Tognini, 2019; Perofsky et al., 2019), but whether this is consistent within other organisms remains unknown. Additionally, it was demonstrated that rhythmic cycling of microbial community composition differentiates with host sex (Liang et al., 2015). Ultimately, considering the increased complexity within mammalian study systems, it may prove productive to utilize comparatively less complex, and earlier diverged basal marine metazoa, which can also indicate how conserved the mechanism is.
The Significance of Mammalian Holobiont Rhythms for Innate Immune Response and Metabolic Activity
Circadian regulation has been well established for some time as a significant mediator of host viral, bacterial, and food pathogen responses (Silver et al., 2012; Curtis et al., 2014; Gibbs et al., 2014; Ehlers et al., 2018). However, only recently, it has become clear that holobiont health decline can specifically be induced by rhythmic disturbances to either the host or its symbionts. Mice and human hosts rely on the daily relative abundance fluctuations of around 20% of their total microbiome species (Thaiss et al., 2014). However, under circadian disruption, these delicate gut microbial community compositions are significantly altered, leading to weakened immune defenses and holobiont health decline (Voigt et al., 2016; Deaver et al., 2018; Leach et al., 2019). Specifically, in the absence of rhythmic bacterial abundance fluctuations, hosts exhibit increased susceptibility to disease and decreased metabolomic output (Leone et al., 2015; Montagner et al., 2016; Thaiss et al., 2016).
Rhythmic Regulation of Innate Immunity
A multitude of bilateral mechanisms likely mediates the observed reductions in holobiont health under circadian disruption. For instance, altered community composition may reduce colonization resistance, whereby distinct biological niches, usually occupied by commensal symbionts, are newly opened to potentially harmful pathogens (Mullineaux-Sanders et al., 2018). Reduced symbiont abundances can also lead to a loss of symbiont performed functions, critical to host health. This includes, for example, the onset of inflammation that creates less favorable pathogen colonization conditions (Mullineaux-Sanders et al., 2018), promotion of innate immune cells such as innate lymphoid cells (ILCs) (Mortha et al., 2014), or the production of beneficial metabolites that influence host circadian cycling through histone modification (Thaiss et al., 2016). Conversely, rhythmic disruption to symbionts may also increase symbiont production of harmful compounds, toxic in high concentrations. For example, bacterial production of Lipopolysaccharide (LPS) endotoxin, a key component within the outer membrane of nearly all Gram-negative bacteria (Baltrons and García, 1999; Alexander and Rietschel, 2001), may increase under rhythmic disturbance (Deaver et al., 2018).
Rhythmic disruption to the endogenous rhythms of the holobiont clock can detrimentally moderate holobiont gene expression. Recently, several mammalian studies have demonstrated a connection between core circadian clock components and regulatory feedback of innate immune system pathways. The expression and function of mouse Toll-like receptor (TLR), TLR9, was demonstrated to be controlled by the host molecular circadian clock, in which disease severity varied with infection timing, per daily TLR9 oscillations of expression (Silver et al., 2012). In addition, a core circadian oscillatory protein CLOCK positively upregulates nuclear factor-kappa β (NF-κβ) transcription, whilst the activity of BMAL1 results in down-regulation (Spengler et al., 2012). Moreover, BMAL1 may mediate mRNA expression of NRF2 (basic leucine zipper (bZIP) transcription factor) that reduces inflammation via suppression of ROS and direct inhibition of the proinflammatory cytokines, IL-1β and IL-6. Indeed, under BMAL1 deletion, cell responsiveness to LPS increased (Early et al., 2018).
Though documented to a lesser extent, microbial symbiont circadian rhythmicity feedback to mediate host gene expression has been an actively increasing focus of future research. Within mammalian gut microbial systems, microbe-associated molecular patterns (MAMPs) interact with and induce expression of intestinal epithelial cell wall Pattern Recognition Receptors (PRRs), including TLRs and NOD-like receptors (NLRs) (Abreu, 2010), which, in turn, induce downstream innate immune responses, such as proinflammatory cytokines and NF-κβ (Mukherji et al., 2013). The rhythmic cycling of bacterial abundance and activity, thus, results in oscillatory expression of these innate immune pathways within the host (Mukherji et al., 2013). Additionally, bacterial symbionts produce a range of metabolites to modulate host chromatin and transcriptional oscillations, locally within the large intestine, as well as further afield within the liver (Thaiss et al., 2014, 2016). Specifically, circadian gut microbial disturbance induced gain and loss of histone modifications such as H3K4me3 and H3K27 acetylation at active promoters, and H3K27 acetylation at enhancers, leading to disturbance of circadian regulation within the host (Thaiss et al., 2016). In mammals, the rhythmic oscillation of gut microbiota relative abundance is, at least in part, mediated through transcriptional regulation of the host’s circadian clock (Thaiss et al., 2014); thus, demonstrating the tightly linked reciprocal feedbacks within this tripartite (i.e., host, symbiont, and circadian clock) regulatory system. Subsequently, it seems clear, disruption to any one of the three components is likely to induce detrimental responses within the other.
We also observe evidence of innate immune regulation by the redox circadian clock. Recently, it was shown that PRX6 can induce activation of NF-κβ in HEK293T cells via interaction with the C-terminal TRAF-C domain of TRAF6, which translocates PRX6 into the mitochondria; a significant pathway, in protecting supercoiled DNA from damage by oxidative stress (Chu et al., 2019). Furthermore, single-molecule experiments demonstrated the specific binding of PRDX5 to an innate immune PRR, namely TLR4 (Knoops et al., 2018). That said, specific evidence that demonstrates a link between innate immune responses with redox cycling is sparse, and to date has only been demonstrated within plant holobionts (Karapetyan and Dong, 2018). Zhang et al. (2019) review the independent roles of canonical circadian regulation and redox signaling in pre-empting environmental stressors and appropriate levels of defense, respectively, to facilitate an optimal balance between immune response and growth. They hypothesized the rhythmic activity of NPR1 (nuclear translocation of the non-expresser of PR genes 1), a protein involved with plant systemic acquired resistance (SAR)-mediated defense, to be the core element linking canonical molecular and redox clocks (Zhang et al., 2019). Research regarding the functions of peroxides as signaling molecules is a relatively new field. Only since recognizing that PRX activity is mediated through post-translational modifications, has their involvement with vital cellular functions been hypothesized (Abbas et al., 2019). Indeed, considering the importance of PRX for maintaining cellular ROS homeostasis, rhythmic disturbances to PRX cycling will likely result in damage through oxidative stress. For instance, Khaper et al. (2018) demonstrated the significance of circadian cycling of oxidative stress pathways in contributing to heart disease and decreased brain functionality, especially following myocardial infarction. Considering the widespread significance of oxidative stress across all domains, elucidating the downstream signaling cascades from circadian redox rhythms could provide substantial benefits to understanding innate immune regulation.
Rhythmic Regulation of Metabolism
Host metabolic activity is tightly coupled with circadian regulation and microbiome functioning. Within humans, the connection between circadian disruption (e.g., through jet lag or night-shift workers) and increased levels of obesity, diabetes, or metabolic syndrome has been well documented (Shi et al., 2013; Depner et al., 2014). Similarly, circadian clocks themselves may be food-entrainable. Remarkably, time point restricted feeding was able to restore both transcriptional and behavioral rhythms within mice whose circadian clocks had been genetically impaired (Hughes et al., 2009, 2010; Vollmers et al., 2009). Notably, the timing of nutrient intake dominates over composition since time-restricted feeding restores the adverse metabolic effects from high-fat diets within the host (Chaix et al., 2014). Reciprocally, bacterial symbionts may also modulate host circadian gene expression (reviewed in Nobs et al., 2019). The production of bile salt hydrolase from bacterial symbionts stimulates a significant shift in circadian oscillators and CCGs regulating host lipid and cholesterol metabolism (Joyce et al., 2014). Remarkably, microbial transfer of circadian-disrupted dysbiotic communities from either mice or humans into germ-free mice recapitulates the metabolic derangements exhibited within the donor (Thaiss et al., 2014; Leone et al., 2015). However, the molecular mechanisms of such interactions have only recently been explored. Recently, Kuang et al. (2019) demonstrated that gut microbiota drive genome-wide rhythmic histone acetylation and gene expression within their host. Specifically, through binding with the nuclear receptor co-repressor (NCoR), the gut microbiota is required for diurnal rhythmic recruitment of the histone deacetylase Hdac3 to target gene promoters that regulate processes controlling host nutrient uptake and metabolism (Kuang et al., 2019). Such mechanisms are intricate and complex; thus, it will require extensive research of single pathways to uncover a holistic picture of circadian holobiont mediated activity.
To date, the rhythmic regulatory mechanisms linking host and symbiont activities have largely been demonstrated through correlative studies. Limited research has uncovered the specific molecular pathways by which rhythmic regulation mediates holobiont health (Lee et al., 2019). However, large-scale holobiont studies do not permit the observation of quantifiable cause and effect interactions on the molecular scale, since a multitude of interactions contributes to the intricate web of holobiont rhythmic regulatory pathways (Lee et al., 2019). Thus, the challenge for future chronobiomics research should be to identify localized proteins/genes of interest that can facilitate molecular-level observations. We must demonstrate the environmental stimulus that initiates a primary response through to the resulting cyclical holobiont feedback. It is this reciprocal interaction between host and symbiont circadian rhythms that is most elusive but could prove most useful.
Influence of Viral Infection on Circadian Rhythm of Mice
The timing of infection can play a significant role in viral persistence and success. Recently, Edgar et al. (2016) demonstrated a time-dependent magnitude of response within mice infected with recombinant luciferase-expressing Murid Herpesvirus 4 (MuHV 4). They observed a 10-times higher MuHV 4 replication rate within mice infected intranasally just before their resting period, at subjective night (zeitgeber Time = 0 or ZT0), compared with those infected after the resting period, at the beginning of the subjective day (zeitgeber Time = 10 or ZT10). This indicates a likely dependence of viral infection success on the circadian rhythm of host mice. Within murine organisms, BMAL1, along with NPAS and CLOCK are the activating transcription factors that drive the transcription of repressor proteins CRY1/2 AND PER1/2 (Supplementary Box 3) which together sustain the molecular circadian rhythm. BMAL1 activity reaches a maximum just before the subjective night, and within mutant mice, lacking BMAL1 expression (Bmal1–/–) circadian rhythmicity is disrupted. Notably, mutant mice infected with MuHV 4 virus at the above-mentioned time points (ZT10 and ZT0), demonstrated similar viral replication rates across all time points, thus, indicating an influence upon viral infection success. Interestingly, in an assessment of viral infection persistence, a higher replication rate was observed within mutant mice, showing a peak on day 5, in comparison to day 9 within wild-type mice infected at ZT0. Indeed, wild-type mice showed a significantly lower infection rate at ZT10, 10-times lower than that observed within the mutant mice, and only peaking on day 7. This demonstrates that the effects from variable timing of viral infection are relatively long-term.
However, it is also possible that the observed differences in replication rates at Z10 were a function of an increased host immune response. To test for this, the experiment was repeated using synchronized cell lines with a ∼24-h rhythmic period. For both, MuHV 4 and Herpes Simplex Virus - 1 virus, an increased replication rate was observed in the Bmal1–/– cell lines as compared to the wildtype (WT), indicating a direct influence of host circadian clock on viral replication and persistence. Furthermore, viral infection also affects the host’s circadian rhythm. NIH 3T3 cells with Bmal:luc system, which expresses luciferase under the control of Bmal1 promoter, were infected with MuHV 4 virus at different zeitgeber times. The herpes virus expressed Bmal1 from ∼6 h after infection, irrespective of the time of infection. If indeed, infection occurred at the point of endogenous Bmal1 transcription fall, then a generated Bmal1 peak disturbed the circadian rhythm. However, if infection occurred at another time, then it suggests a higher than usual peak in Bmal1 expression was observed. This suggests that viral infections can modulate host circadian rhythm, with the effect varying depending on the timing of infection.
Inferences From Other Model Organisms
Notably, significant insights may also be gained from other well-studied terrestrial model holobiont systems. Plant-mycorrhizal fungal relationships are one of the most well-known holobiont systems. Here, expression of circadian gene homologs (wc-1, wc-2, and frq) based on Neurospora crassa, the fungal circadian clock model (Grimaldi et al., 2006), were identified within the Arbuscular mycorrhizal fungus (AMF) Rhizoglomus irregularis (Lee et al., 2018). This presents an interesting example, since AMF lives underground, unexposed to sunlight, yet the core circadian oscillator gene frq was still induced by brief light stimulation (Lee et al., 2018). This demonstrates that microbial symbionts may use a diverse range of environmental cues for rhythmic entrainment. Additionally, within the well-established model Drosophila, it was demonstrated that Alphaproteobacteria, Wolbachia endosymbionts support their hosts’ peripheral clock oscillations and diurnal behavioral patterns (Morioka et al., 2018). We must consider these well-studied model organisms as they may provide useful experimental platforms toward understanding molecular mechanisms of circadian clocks within the less well studied, but ecologically significant species from vulnerable marine ecosystems.
Rhythms in Marine Holobionts
Current evidence demonstrating circadian regulation within microbial symbionts is largely limited to the mammalian gut microbiota, though a few marine systems have been explored. One of the best-studied examples includes luminous Gammaproteobacterium Aliivibrio fischeri (formerly, Vibrio fischeri) housed within the light organ of Hawaiian bobtail squid Euprymna scolopes. Horizontally acquired A. fischeri accumulate each day in the light organ of E. scolopes whereby nightfall, they reach a high enough density to produce bioluminescence through quorum sensing pathways (Miyashiro and Ruby, 2012). This is suggested to confer an advantage to the host by deterring predators through a mechanism known as counterillumination (Jones and Nishiguchi, 2004). Remarkably, each morning, the host then expels (known as “venting”) 90% of these bacteria and the few remaining regrow throughout the day to again build up sufficient densities. This daily cycle forms the basis of their symbiotic relationship, of which host and symbiont demonstrate rhythmically aligned patterns of gene expression (Wier et al., 2010) underlying day/night activities (Heath-Heckman and McFall-Ngai, 2011). For instance, the transcriptional rhythms of E. scolopes’ circadian associated cryptochrome escry1 are upregulated through the temporal cycling of its symbiont A. fischeri (Heath-Heckman et al., 2013). Likewise, the cyclic provision of chitin by the host regulates symbiont metabolism resulting in the cyclic production of bioluminescence, essential to symbiosis (Schwartzman et al., 2015). Despite being one of the best-studied examples, gammaproteobacterial A. fischeri circadian oscillators have not yet been identified. Thus, indicative of the significant challenges in chronobiomics, limiting our current scope on the potentially widespread prevalence of rhythmic regulatory systems within marine ecosystems.
Circadian-like rhythmic activity has also been described within Cnidarian-Symbiodiniaceae holobionts. Diurnal rhythmic activity within corals has been observed since the 1930s and is perhaps unsurprising given that light, considered the major circadian zeitgeber (McClung, 2001), forms the basis of the coral-Symbiodiniaceae relationship through photosynthetic production of metabolites. Within both free-living Symbiodiniaceae and symbiotic Symbiodiniaceae, the rhythmic activity of photosynthetic processes including, oxygen evolution, PSII yield, and concentrations of pigments, is sustained under constant conditions (Sorek and Levy, 2012a; Sorek et al., 2013). This lends support to the hypothesis that an endogenous circadian clock mediates photosynthesis (Sorek et al., 2014). However, molecular evidence for circadian-driven regulation remains elusive, since uncoupling host–symbiont interactions and reciprocal regulation remains a significant challenge, particularly within vertically inherited intracellular endosymbionts (Sorek and Levy, 2012b). That said, molecular clock mechanisms have been demonstrated within several free-living algae, including Lingulodinium polyedrum, the dinoflagellate clock model (Hastings, 2007; Noordally and Millar, 2015). Additionally, considering the significance of ROS within the coral holobiont from Symbiodiniaceae photosynthesis, we predict an important role for the circadian redox clock in the maintenance of ROS homeostasis. Each day, coral tissue exhibits a significant day–night transition in oxidation state during the switch between daytime photosynthesis and evening respiration (Sorek et al., 2013). This could likely provide a dominant zeitgeber entraining peroxiredoxin circadian rhythms (Edgar et al., 2012). Indeed, evidence links the relationship between circadian regulation and several stress-related proteins. For instance, Heatshock protein 90 (HSP90), coupled to the circadian clock, demonstrates rhythmic expression under both constant and diurnal conditions and modulates the circadian period if inhibited (O’Neill et al., 2011; Hindle et al., 2014). Additionally, toxic stress driven by climate change-induced environmental perturbation results in disruption to host circadian genes, as well as an observed high prevalence of symbiont antioxidant genes (Kaniewska et al., 2015). Coral-Symbiodiniaceae symbioses are one of the best well-studied marine holobionts, yet we understand relatively little of their circadian mediated interactions, demonstrating both the current lack of research within this area and the significant challenge it presents. Given the ecological significance and vulnerability under anthropogenic and climate change-induced pressures, coral holobiont circadian activity ought to be a priority for research.
Closely related sea anemones can also provide tractable cnidarian holobiont study systems since, unlike coral, they exhibit both symbiotic and aposymbiotic morphs. In Aiptasia diaphana, Sorek et al. (2018) demonstrated symbiotic morphs display 24-h circadian rhythms of gene expression, whilst aposymbiotic morphs displayed only 12-h circatidal rhythms. Notably, when aposymbiotic morphs were repopulated with algae, they returned to a 24-h behavioral rhythm. This dramatic rhythmic shift observed between symbiotic and aposymbiotic hosts indicates a strong influence from an endogenous symbiont circadian clock on photosynthesis and host rhythmicity through the transfer of metabolites (Sorek et al., 2018). Moreover, within the sea anemone Nematostella vectensis, significant shifts in the abundance of bacterial community members were observed under constant conditions (light–light LL or dark–dark DD). Under DD, bacterial community diversity increased, as well as upregulation of innate immunity genes, thus, indicative of a symbiont-driven circadian clock (Leach et al., 2019). Outside of Cnidaria, endogenous circadian regulation of Symbiodiniaceae symbionts has also been identified within the giant clam Tridacna crocea, where it was suggested that endogenous symbiont and/or host circadian rhythms, stimulated by light, mediate T. crocea daily trace element incorporation (Warter et al., 2018).
Many marine invertebrate organisms exhibit pelago-benthic life cycles that encompass a free-swimming larval stage, often particularly dependent on environmental cues (reviewed by Hodin et al., 2017). For instance, larvae of the coral reef sponge Amphimedon queenslandica inhabit a shallow-water reef flat where they are highly dependent on regularly oscillating environmental cues for the onset of development and timing of larval settlement to the substrate; occurring 4–6 h post-release, in the late afternoon each day (Degnan and Degnan, 2010; Figure 2). When maintained under conditions of constant environmental light, larvae do not settle, but when re-exposed to darkness, again exhibit settlement (Say and Degnan, 2019). Thus, environmentally entrained clocks can have a profound influence on behavior. As with other larval metazoa, A. queenslandica larvae house a vertically inherited microbiome dominated by proteobacteria (Fieth et al., 2016; Gauthier et al., 2016), which may also be rhythmically regulated. For instance, bacterial production of LPS endotoxin, a key component within the outer membrane of nearly all gram-negative bacteria (Baltrons and García, 1999; Alexander and Rietschel, 2001), may increase under rhythmic disturbance (Deaver et al., 2018). Significant upregulation of the export permease protein (LptF) and an LPS synthesis gene (kdsB) was observed within circadian disrupted gut microbiota (Deaver et al., 2018). As well as the direct toxic effects from LPS, increased production of such compounds may interfere with signaling pathways that sustain crucial holobiont molecular and behavioral activities. Indeed, LPS can interfere with chemosensory G-protein coupled receptors (GPCRs) (Baltrons and García, 1999; Alexander and Rietschel, 2001), which facilitate signaling cascades for an array of diverse host responses. The pelago-benthic life cycle, observed in many marine invertebrates, often relies on GPCR activation to induce settlement of larvae to the substrate (Schneider and Leitz, 1994; Strader et al., 2018; Say and Degnan, 2019), and, thus, may potentially be inhibited through GPCR circadian induced pathway disturbances. The successful development and establishment of free-swimming larvae is a crucial stage in the dispersal of many significant marine holobionts, functional as ecosystem engineers (e.g., coral, sponges, ascidians, bryozoans). Subsequently it is particularly important we understand the rhythmically aligned regulation of these organisms that could likely be disrupted through climate change induced environmental perturbation.
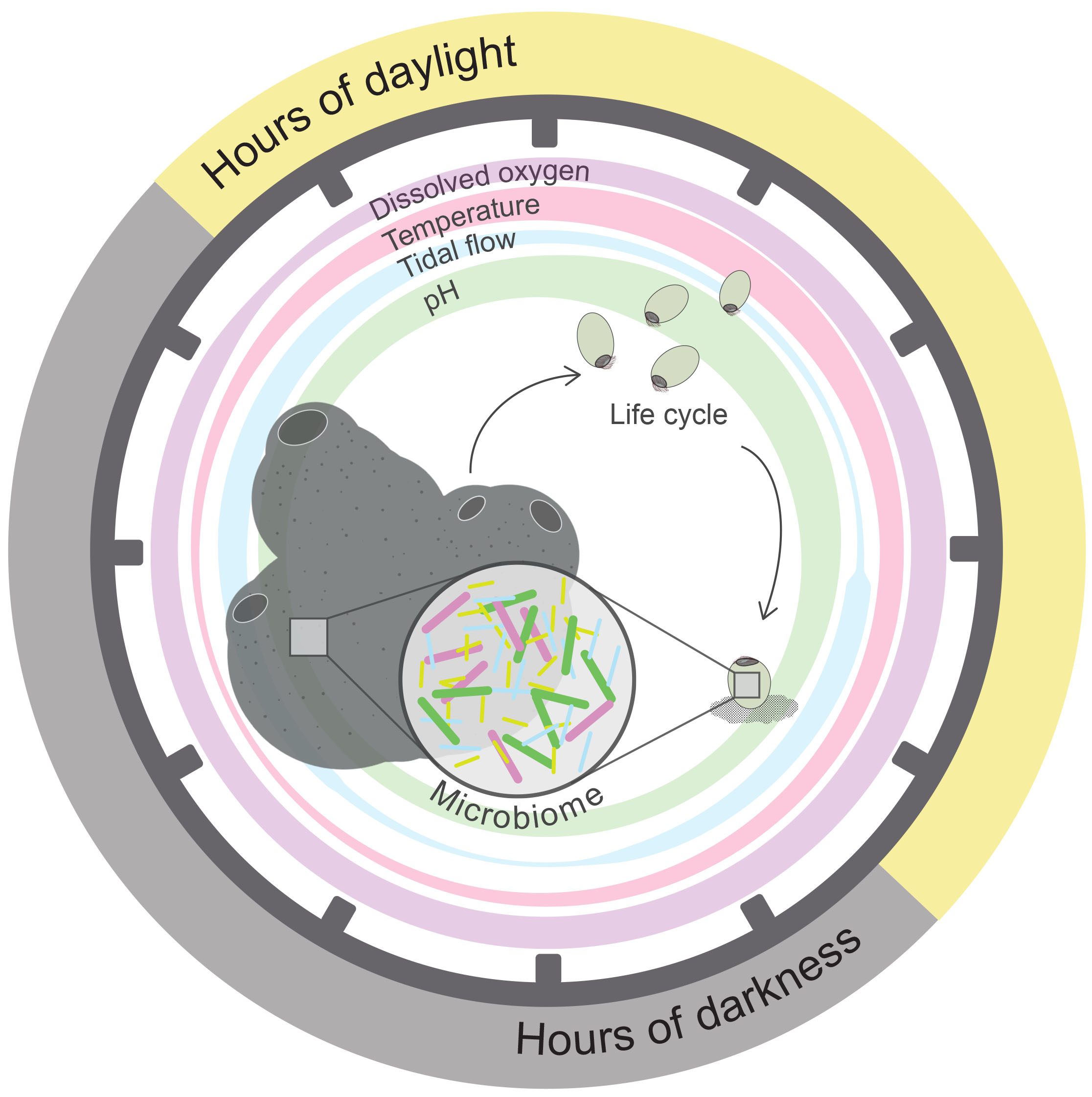
Figure 2. Representation of daily fluctuating environmental factors that may entrain rhythms within a shallow-water marine sponge holobiont. Representation of typical daily fluctuating environmental variables/abiotic parameters within a shallow-water marine habitat (i.e., zeitgebers) that can entrain rhythms for sessile organisms, such as a sponge (shown here). Parameters depicted here by colored circles are hours of daylight, dissolved oxygen, water temperature, tide level, pH, that vary systematically throughout a diel cycle, as indicated here by circle width. The relative influence of independent environmental parameters over an organism’s rhythms will likely vary for the host compared with its microbial symbionts, with symbionts often effected more so. Additionally, each environmental variable may independently entrain diverse rhythms with periods such as circadian, diurnal, ultradian, or circatidal. Successful rhythmic regulation is critical for sponge holobiont development during each stage of its pelago-benthic life cycle. Within the coral reef sponge, A. queenslandica timing of larval release into the water column, development of competency, and larval descent to the sea floor before settlement onto the substrate occurs regularly at the same time each day, in alignment with key environmental cues (Degnan and Degnan, 2010; Jindrich et al., 2017). Importantly, at each life cycle stage this sponge houses vertically and/or horizontally acquired symbionts (Ereskovsky et al., 2005; Maldonado, 2007; Schmitt et al., 2008; Fieth et al., 2016), for which rhythmic regulation must be considered. Dark gray tick marks on circle represent time throughout a 24-h period.
Perspectives on the Holobiont Clock
The bilaterally dependent feedback of host–symbiont rhythmic regulation is an intriguing area of research, presenting many significant challenges. For instance, does each individual within the partnership contribute its own endogenous clock, or do they function cohesively, acting and reacting to rhythmic cues of the other? Regarding the canonical molecular clock within prokaryotic organisms, we observe a loss of features (Schmelling et al., 2017) and less robust rhythms within all non-cyanobacterial species, regardless of whether they are free-living or symbiotic. Thus, microbial symbiont clock features may feasibly have become redundant and led to a shift in reliance upon their hosts for entrainment cues, and even the expression of their clock-controlled genes via host oscillators. Indeed, this may be to a comparatively greater extent within endosymbionts. Evidence indicates free-living ancestral mitochondria and chloroplasts once had their own endogenous clocks before becoming fully obligate within the eukaryotic cell. Likewise, within free-living Symbiodiniaceae, we observe longer-lasting, robust rhythms under constant conditions than within its symbiotic counterpart (Sorek et al., 2014). Robust clocks enable organisms to sustain rhythmic regulation for longer periods, despite variable environmental entrainment cues. Thus, for organisms housed within the relative stability of the microbiome, such a robust clock, that is likely more energetically costly, may not have been selected for. Conversely, considering the redox clock as a conserved and ancient mechanism, that likely predates the canonical clock (Edgar et al., 2012), we hypothesize individuals within the holobiont to each comprise their own endogenous redox clocks, that have optimally aligned through common/shared environmental stimuli. However, it is the interplay between the redox and canonical holobiont clocks that will be an important and challenging aspect of future chronobiomics research to address.
We must also consider the diversity and dominance of environmental cues that a holobiont may use for entrainment. An individual holobiont may each be exposed to a unique complexity of diverse environmental stimuli, specific to its local environment (Figure 1). Thus, as observed within the mantle of the giant clam Tridacna squamous, multiple rhythms set on different periods (e.g., circadian, circatidal) may regulate specific activities (Hiong et al., 2018), such that the efficiency of each is optimized through oscillations under with the relevant environmental stimulus. Furthermore, the relative dominance of specific cues may vary for different members of the holobiont, in accordance with the often-disparate conditions experienced by the microbiome compared with its host. For instance, within human gut microbiota, symbiont rhythms are entrained most dominantly by host nutrient intake, whilst for the host, environmental light provides the dominant zeitgeber (Bass and Takahashi, 2010; Oosterman et al., 2015). In sponges, the ceasing of pumping activity can lead to internal anoxic conditions, which bacterial symbionts of the microbiome are hypothesized to mitigate (Hoffmann et al., 2008; Lavy et al., 2016). Circadian clock timing enables organisms to anticipate daily fluctuating environmental conditions and respond appropriately, thus, maximizing the efficiency of the holobiont; by maintaining an optimum balance in the energy requirements demanded from activities, such as ROS homeostasis, growth, or reproduction, and to help ward off viral infections at a cellular level.
It is widely observed that organisms exhibit thresholds of disturbance, beyond which they are unable to sustain homeostatic cellular functioning and, thus, exhibit, often irreversible, health decline. This can most commonly be observed under climate change-induced disturbances, such as coral holobionts during exposure to degree heating weeks (DHW), whereby different coral holobionts exhibit varying degrees of resistance to the number of DHWs (Sully et al., 2019). Here, we emphasize the importance of understanding circadian clock molecular mechanisms to elucidate the relationship between specific environmental cues and regulatory mechanisms that nudge organisms past their tipping points beyond which they can cope. For instance, in vitro experiments have demonstrated the varied abilities of different organisms to sustain circadian regulation under conditions of prolonged circadian disturbance. We hypothesize that varying degrees of circadian clock robustness equate with longevity until disturbance thresholds are surpassed. That is, in the absence of appropriate environmental zeitgebers, a more robust clock may sustain rhythmic regulation of cellular activities for a longer duration, and, thus, be less likely to exhibit environmental disturbance-induced stress.
Conclusion
We have only just begun to touch the surface of understanding rhythmic regulations and the extent to which they can mediate cellular, organismal, or ecosystem-level functioning. Yet, circadian-driven regulatory systems, through both canonical molecular and redox clocks, have evolutionary ancient origins, of which the environmental zeitgebers entraining such rhythms are ubiquitous, although nuanced, for all organisms. Thus, we cannot stress enough the importance of elucidating the mechanisms by which organisms perceive entrainment cues and utilize them to optimize rhythmically coordinated regulation within their local environment.
The trophic interactions within large-scale planktonic systems, including the viral shunt, are extremely important for global biogeochemical cycling on a humungous scale, with numerous downstream effects for energy transfer within locally significant ecosystems, such as tropical coral reefs or deep-sea habitats. Dominant cues such as environmental light entrain the rhythms for free-living prokaryotes and viruses within large-scale planktonic communities. However, the molecular mechanisms by which these organisms coordinate such rhythms are less clear. To date, molecular evidence describing prokaryote circadian regulation is limited to Cyanobacteria and a few proteobacterial species. Though within the last decade, research has demonstrated that a circadian redox clock is ubiquitous across all domains, as well as less robust, simplified forms of the canonical molecular circadian clocks are also plentiful. Future research should expand our understanding of such mechanisms outside of Cyanobacteria to other important bacterial and archaeal phyla, which make up a significant part of these macro-scale ecosystems.
In parallel, rhythmically coordinated regulation of cellular activity is equally significant within micro-scale nested ecosystems of marine holobionts. Holobionts, which comprise a host and complex cohort of prokaryotic communities, interacting either competitively or beneficially with one another, are considered within an ecological context. Each member within this nested ecosystem will regulate activities on an independent endogenous clock. However, remarkably, the interdependent relationship between a host and its symbionts is also rhythmically coordinated, through both redox and canonical circadian clocks. This association is complex, particularly as there are likely numerous entrainment cues, of which the relative importance of each will differ for host and symbiont. Untangling the links between host and symbiont-driven circadian regulation will require extensive research. Currently, the best models for this are from mammals and a few terrestrial organisms. Whilst studies from these models are insightful, they are not necessarily representative and we, therefore, encourage further research on marine metazoan, particularly from earlier diverged lineages such as Cnidaria, Porifera, and Ctenophora. This will facilitate greater evolutionary insight into the origins of such mechanisms since they provide the earliest sister group to all other bilaterians. Additionally, these organisms are very important parts of reef ecosystems in marine habitats currently facing climate change-induced stressors.
It is important to recognize the rhythmically interconnected nature of both micro-and macro-scale ecosystems. Indeed, many of the above-mentioned reef organisms (e.g., coral, sponges) help couple benthic and pelagic zones themselves through their filter-feeding, meroplanktonic life cycle, provision of larval settlement cues, and providing breeding grounds for a remarkably high number of species. Within each ecosystem, every organism is regulated by its endogenous rhythms, but crucially, those rhythms must also be aligned with both the abiotic cues and other clock-controlled biotic cues from within their local environment.
With this review, we have introduced the relatively new field of marine chronobiomics within both macro-scale planktonic ecosystems and micro-scale holobiont nested ecosystems. We highlighted the interplay of biological rhythms across the living domains, as well as the not-so-living viruses, both within and between micro-and macro-scale ecosystems. Our review provides a baseline in which we emphasize the currently large gaps in knowledge and active research, to help guide future exploration and recognition of chronobiomics within the marine domain.
Author Contributions
OH introduced the subject for this review. HS and OH drew Figures 1, 2, respectively, contributed equally toward the conception of this review, and provided essential and critical feedback throughout the writing process. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge all coordinators of the International Conference for Young Marine Researchers (ICYMARE) for inviting us with the opportunity to write this review. We would like to thank Douglas Campbell for providing us with essential feedback, that lead to the final form of this review. We also acknowledge and thank the State of Bremen for providing us with the funds necessary to publish this review in an open access format.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.744169/full#supplementary-material
References
Abbas, M. N., Kausar, S., and Cui, H. (2019). The biological role of peroxiredoxins in innate immune responses of aquatic invertebrates. Fish Shellfish Immunol. 89, 91–97. doi: 10.1016/j.fsi.2019.03.062
Abe, J., Hiyama, T. B., Mukaiyama, A., Son, S., Mori, T., Saito, S., et al. (2015). Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349, 312–316. doi: 10.1126/science.1261040
Abreu, M. T. (2010). Toll-like receptor signaling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 10, 131–144. doi: 10.1038/nri2707
Adolph, K. W., and Haselkorn, R. (1972). Photosynthesis and the development of blue-green algal virus N-1. Virology 47, 370–374. doi: 10.1016/0042-6822(72)90503-X
Alexander, C., and Rietschel, E. T. (2001). Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 7, 167–202. doi: 10.1177/09680519010070030101
Baltrons, M., and García, A. (1999). Nitric oxide-independent down-regulation of soluble guanylyl cyclase by bacterial endotoxin in astroglial cells. J. Neurochem. 73, 2149–2157.
Bang, C., and Schmitz, R. A. (2018). Archaea: forgotten players in the microbiome. Emerg. Top. Life Sci. 2, 459–468. doi: 10.1042/etls20180035
Bass, J., and Takahashi, J. S. (2010). Circadian integration of metabolism and energetics. Science 330, 1349–1354. doi: 10.1126/science.1195027
Baudoux, A. C., Noordeloos, A. A., Veldhuis, M. J., and Brussaard, C. P. (2006). Virally induced mortality of Phaeocystis globosa during two spring blooms in temperate coastal waters. Aquat. Microb. Ecol. 44, 207–217. doi: 10.3354/ame044207
Bergh, Ø, Børsheim, K. Y., Bratbak, G., and Heldal, M. (1989). High abundance of viruses found in aquatic environments. Nature 340, 467–468. doi: 10.1038/340467a0
Bettarel, Y., Dolan, J. R., Hornak, K., Lemee, R., Masin, M., Pedrotti, M. L., et al. (2002). Strong, weak, and missing links in a microbial community of the N.W. Mediterranean Sea. FEMS Microbiol. Ecol. 42, 451–462. doi: 10.1016/S0168-6496(02)00383-5
Bongiorni, L., Magagnini, M., Armeni, M., Noble, R., and Danovaro, R. (2005). Viral production, decay rates, and life strategies along a trophic gradient in the North Adriatic Sea. Appl. Environ. Microbiol. 71, 6644–6650. doi: 10.1128/AEM.71.11.6644-6650.2005
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226. doi: 10.1371/journal.pbio.1002226
Bosch, T. C., and McFall-Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology 114, 185–190. doi: 10.1016/j.zool.2011.04.001
Bragg, J. G., and Chisholm, S. W. (2008). Modeling the fitness consequences of a cyanophage-encoded photosynthesis gene. PLoS One 3:e3550. doi: 10.1371/journal.pone.0003550
Breitbart, M. (2012). Marine viruses: truth or dare. Ann. Rev. Mar. Sci. 4, 425–448. doi: 10.1146/annurev-marine-120709-142805
Breitbart, M., and Rohwer, F. (2005). Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284. doi: 10.1016/j.tim.2005.04.003
Brunet, C., Casotti, R., Vantrepotte, V., and Conversano, F. (2007). Vertical variability and diel dynamics of picophytoplankton in the Strait of Sicily, Mediterranean Sea, in summer. Mar. Ecol. Prog. Ser. 346, 15–26. doi: 10.3354/meps07017
Chaix, A., Zarrinpar, A., Miu, P., and Panda, S. (2014). Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell. Metab. 20, 991–1005. doi: 10.1016/j.cmet.2014.11.001
Chang, Y. G., Tseng, R., and Kuo, N. W. (2012). Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl. Acad. Sci. U.S.A. 109, 16847–16851. doi: 10.1073/pnas.1211508109
Chen, T. H., Chen, T. L., Hung, L. M., and Huang, T. C. (1991). Circadian rhythm in amino acid uptake by Synechococcus RF-11. Plant Physiol. 97, 55–59. doi: 10.1104/pp.97.1.55
Chu, S. H., Liu, L., Abbas, M. N., Li, Y. Y., Kausar, S., Qian, X. Y., et al. (2019). Peroxiredoxin 6 modulates Toll signaling pathway and protects DNA damage against oxidative stress in red swamp crayfish (Procambarus clarkii). Fish Shellfish Immunol. 89, 170–178. doi: 10.1016/j.fsi.2019.03.055
Clokie, M. R. J., Millard, A. D., and Mann, N. H. (2010). T4 genes in the marine ecosystem: studies of the T4-like cyanophages and their role in marine ecology. Virol. J. 7:291. doi: 10.1186/1743-422X-7-291
Clokie, M. R., Thalassinos, K., Boulanger, P., Slade, S. E., Stoilova-McPhie, S., Cane, M., et al. (2008). A proteomic approach to the identification of the major virion structural proteins of the marine cyanomyovirus S-PM2. Microbiology 154, 1775–1782. doi: 10.1099/mic.0.2007/016261-0
Cohen, S. E., and Golden, S. S. (2015). Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 79, 373–385. doi: 10.1128/MMBR.00036-15
Conley, M. P., and Wood, W. B. (1975). Bacteriophage T4 whiskers: a rudimentary environment-sensing device. Proc. Natl. Acad. Sci. U.S.A. 72, 3701–3705. doi: 10.1073/pnas.72.9.3701
Coutinho, F. H., Silveira, C. B., Gregoracci, G. B., Thompson, C. C., Edwards, R. A., Brussaard, C. P., et al. (2017). Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat. Commun. 8:15955. doi: 10.1038/ncomms15955
Cséke, C., and Farkas, G. L. (1979). Effect of light on the attachment of cyanophage AS-1 to Anacystis nidulans. J. Bacteriol. 137, 667–669. doi: 10.1128/jb.137.1.667-669.1979
Curtis, A. M., Bellet, M. M., Sassone-Corsi, P., and O’Neill, L. A. (2014). Circadian clock proteins and immunity. Immunity 40, 178–186. doi: 10.1016/j.immuni.2014.02.002
Dalle-Donne, I., Giustarini, D., Colombo, R., Rossi, R., and Milzani, A. (2003). Protein carbonylation in human diseases. Trends Mol. Med. 9, 169–176. doi: 10.1016/s1471-4914(03)00031-5
Deaver, J. A., Eum, S. Y., and Toborek, M. (2018). Circadian disruption changes gut microbiome taxa and functional gene composition. Front. Microbiol. 9:737. doi: 10.3389/fmicb.2018.00737
Degnan, S. M., and Degnan, B. M. (2010). The initiation of metamorphosis as an ancient polyphenic trait and its role in metazoan life-cycle evolution. Philos. Trans. R. Soc. B Biol. Sci. 365, 641–651.
Depner, C. M., Stothard, E. R., and Wright, K. P. (2014). Metabolic consequences of sleep and circadian disorders. Curr. Diab. Rep. 14:507. doi: 10.1007/s11892-014-0507-z
Diamond, S., Jun, D., Rubin, B. E., and Golden, S. S. (2015). The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc. Natl. Acad. Sci. U.S.A. 112, E1916–E1925. doi: 10.1073/pnas.1504576112
Dibner, C., Schibler, U., and Albrecht, U. (2010). The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549. doi: 10.1146/annurev-physiol-021909-135821
Dimmock, N. J., Easton, E., and Leppard, K. N. (2009). Introduction to Modern Virology. Chichester: WILEY Blackwell. doi: 10.1007/s13398-014-0173-7.2
Donne, J. (1987). No Man is an Island, Meditation 17, Devotions Upon Emergent Occasions. Oxford: Oxford University Press.
Early, J. O., Menon, D., Wyse, C. A., Cervantes-Silva, M. P., Zaslona, Z., Carroll, R. G., et al. (2018). Circadian clock protein BMAL1 regulates IL-1beta in macrophages via NRF2. Proc. Natl. Acad. Sci. U.S.A. 115, E8460–E8468. doi: 10.1073/pnas.1800431115
Edgar, R. S., Green, E. W., Zhao, Y., Van Ooijen, G., Olmedo, M., Qin, X., et al. (2012). Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464. doi: 10.1038/nature11088
Edgar, R. S., Stangherlin, A., Nagy, A. D., Nicoll, M. P., Efstathiou, S., O’Neill, J. S., et al. (2016). Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 113, 10085–10090. doi: 10.1073/pnas.1601895113
Ehlers, A., Xie, W., Agapov, E., Brown, S., Steinberg, D., Tidwell, R., et al. (2018). BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal Immunol. 11, 97–111. doi: 10.1038/mi.2017.24
Ehret, C. F., and Wille, J. J. (1970). “The photobiology of circadian rhythms in protozoa and other eukaryotic microorganisms,” in Photobiology of Microorganisms, ed. P. Halldal (New York, NY: Wiley-Interscience), 369–416.
Ereskovsky, A., Gonobobleva, E., and Vishnyakov, A. (2005). Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarcida). Mar. Biol. 146, 869–875. doi: 10.1007/s00227-004-1489-1
Evans, C., Pearce, I., and Brussard, C. (2009). Viral-mediated lysis of microbes and carbon release in the sub-Antarctic and polar frontal zones of the Australian Southern Ocean. Environ. Microbiol. 11, 2924–2934. doi: 10.1111/j.1462-2920.2009.02050
Falkowski, P. G. (1994). The role of phytoplankton photosynthesis in global biogeochemical cycles. Photosynth. Res. 39, 235–258. doi: 10.1007/BF00014586
Fieth, R. A., Gauthier, M. E. A., Bayes, J., Green, K. M., and Degnan, S. M. (2016). Ontogenetic changes in the bacterial symbiont community of the tropical demosponge Amphimedon queenslandica: metamorphosis is a new beginning. Front. Mar. Sci. 3:228. doi: 10.3389/fmars.2016.00228
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincón, J., Zabala, L. L., Jiao, N., et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. doi: 10.1073/pnas.1307701110
Fuhrman, J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
Gauthier, M. E. A., Watson, J. R., and Degnan, S. M. (2016). Draft genomes shed light on the dual bacterial symbiosis that dominates the microbiome of coral reef sponge Amphimedon queenslandica. Front. Mar. Sci. 3:196. doi: 10.3389/fmars.2016.00196
Gibbs, J., Ince, L., Matthews, L., Mei, J., Bell, T., Yang, N., et al. (2014). An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 20, 919–926. doi: 10.1038/nm.3599
Gribaldo, S., and Brochier-Armanet, C. (2006). The origin and evolution of Archaea: a state of the art. Philos. Trans. R. Soc. B Biol. Sci. 361, 1007–1022. doi: 10.1098/rstb.2006.1841
Grimaldi, B., Coiro, P., Filetici, P., Berge, E., Dobosy, J. R., Freitag, M., et al. (2006). The Neurospora crassa White Collar-1 dependent blue light response requires acetylation of histone H3 lysine 14 by NGF-1. Mol. Biol. Cell 17, 4576–4583. doi: 10.1091/mbc.e06-03-0232
Grobbelaar, N., Huang, T. C., Lin, H. Y., and Chow, T. J. (1986). Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol. Lett. 37, 173–177.
Häfker, N. S., Meyer, B., Last, K. S., Pond, D. W., Hüppe, L., and Teschke, M. (2017). Circadian clock involvement in zooplankton diel vertical migration. Curr. Biol. 27, 2194–2201. doi: 10.1016/j.cub.2017.06.025
Halberg, F., and Conner, R. L. (1961). Circadian organization and microbiology: variance spectra and a periodogram on behavior of Escherichia coli growing in fluid culture. Proc. Minn. Acad. Sci. U.S.A. 29, 227–239.
Hanschmann, E. M., Godoy, J. R., Berndt, C., Hudemann, C., and Lillig, C. H. (2013). Thioredoxins, glutaredoxins, and peroxiredoxins—molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid. Redox Signal. 19, 1539–1605. doi: 10.1089/ars.2012.4599
Hastings, J. W. (2007). The gonyaulax clock at 50: translational control of circadian expression. Cold Spring Harb. Symp. Quant. Biol. 72, 141–144. doi: 10.1101/sqb.2007.72.026
Heath-Heckman, E. A., and McFall-Ngai, M. J. (2011). The occurrence of chitin in the hemocytes of invertebrates. Zoology 114, 191–198. doi: 10.1016/j.zool.2011.02.002
Heath-Heckman, E. A., Peyer, S. M., Whistler, C. A., Apicella, M. A., Goldman, W. E., and McFall-Ngai, M. J. (2013). Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio 4:e00167-13. doi: 10.1128/mBio.00167-13
Heymann, D. L., and Shindo, N. (2020). COVID-19: what is next for public health? Lancet 395, 542–545. doi: 10.1016/S0140-6736(20)30374-3
Hindle, M. M., Martin, S. F., Noordally, Z. B., Van Ooijen, G., Barrios-Llerena, M. E., Simpson, T. I., et al. (2014). The reduced kinome of Ostreococcus tauri: core eukaryotic signalling components in a tractable model species. BMC Genomics 15:640. doi: 10.1186/1471-2164-15-640
Hiong, K. C., Koh, C. Z., Boo, M. V., Choo, C. Y., Wong, W. P., Chew, S. F., et al. (2018). The colorful mantle of the giant clam, Tridacna squamosa, expresses a light-dependent manganese superoxide dismutase to ameliorate oxidative stresses due to its symbiotic association with zooxanthellae. Coral Reefs 37, 1039–1051. doi: 10.1007/s00338-018-01738-9
Hodin, J., Ferner, M. C., Heyland, A., Gaylord, B., Carrier, T. J., and Reitzel, A. M. (2017). “I feel that! Fluid dynamics and sensory aspects of larval settlement across scales,” in Evolutionary Ecology of Marine Invertebrate Larvae, eds T. Carrier, A. Reitzel, and A. Heyland (Oxford: Oxford University Press), 190–207.
Hoffmann, F., Røy, H., Bayer, K., Hentschel, U., Pfannkuchen, M., Brümmer, F., et al. (2008). Oxygen dynamics and transport in the Mediterranean sponge Aplysina aerophoba. Mar. Biol. 153, 1257–1264. doi: 10.1007/s00227-008-0905-3
Holtzendorff, J., Partensky, F., Mella, D., Lennon, J. F., Hess, W. R., and Garczarek, L. (2008). Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythm 23, 187–199. doi: 10.1177/0748730408316040
Hoyle, N. P., and O’Neill, J. S. (2015). Oxidation-reduction cycles of peroxiredoxin proteins and nontranscriptional aspects of timekeeping. Biochemistry 54, 184–193. doi: 10.1021/bi5008386
Hughes, M. E., DiTacchio, L., Hayes, K. R., Vollmers, C., Pulivarthy, S., Baggs, J. E., et al. (2009). Harmonics of circadian gene transcription in mammals. PLoS Genet. 5:e1000442. doi: 10.1371/journal.pgen.1000442
Hughes, M. E., Hogenesch, J. B., and Kornacker, K. (2010). JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythm 25, 372–380. doi: 10.1177/0748730410379711
Huttenhower, C., Knight, R., Brown, C. T., Caporaso, J. G., Clemente, J. C., Gevers, D., et al. (2014). Advancing the microbiome research community. Cell 159, 227–230. doi: 10.1016/j.cell.2014.09.022
Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Ersson, C. R., Tanabe, A., et al. (1998). Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523. doi: 10.1126/science.281.5382.1519
Jia, Y., Shan, J., Millard, A., Clokie, M. R., and Mann, N. H. (2010). Light-dependent adsorption of photosynthetic cyanophages to Synechococcus sp. WH7803. FEMS Microbiol. Lett. 310, 120–126. doi: 10.1111/j.1574-6968.2010.02054.x
Jindrich, K., Roper, K. E., Lemon, S., Degnan, B. M., Reitzel, A. M., and Degnan, S. M. (2017). Origin of the animal circadian clock: diurnal and light-entrained gene expression in the sponge Amphimedon queenslandica. Front. Mar. Sci. 4:327. doi: 10.3389/fmars.2017.00327
Johnson, C. H., Stewart, P. L., and Egli, M. (2011). The cyanobacterial circadian system: from biophysics to bioevolution. Annu. Rev. Biophys. 40, 143–167. doi: 10.1146/annurev-biophys-042910-155317
Johnson, C. H., Zhao, C., Xu, Y., and Mori, T. (2017). Timing the day: what makes bacterial clocks tick? Nat. Rev. Microbiol. 15, 232–242. doi: 10.1038/nrmicro.2016.196
Jolma, I. W., Laerum, O. D., Lillo, C., and Ruoff, P. (2010). Circadian oscillators in eukaryotes. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 533–549. doi: 10.1002/wsbm.81
Jones, B. W., and Nishiguchi, M. K. (2004). Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol. 144, 1151–1155. doi: 10.1007/s00227-003-1285-3
Jovel, J., Patterson, J., Wang, W., Hotte, N., O’Keefe, S., Mitchel, T., et al. (2016). Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 7:459. doi: 10.3389/fmicb.2016.00459
Joyce, S. A., MacSharry, J., Casey, P. G., Kinsella, M., Murphy, E. F., Shanahan, F., et al. (2014). Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. U.S.A. 111, 7421–7426. doi: 10.1073/pnas.1323599111
Kaniewska, P., Chan, C. K. K., Kline, D., Ling, E. Y. S., Rosic, N., Edwards, D., et al. (2015). Transcriptomic changes in coral holobionts provide insights into physiological challenges of future climate and ocean change. PLoS One 10:e0139223. doi: 10.1371/journal.pone.0139223
Kao, C. C., Green, S., Stein, B., and Golden, S. S. (2005). Diel infection of a cyanobacterium by a contractile bacteriophage. Appl. Environ. Microbiol. 71, 4276–4279. doi: 10.1128/AEM.71.8.4276-4279.2005
Karapetyan, S., and Dong, X. (2018). Redox and the circadian clock in plant immunity: a balancing act. Free Radic. Biol. Med. 119, 56–61. doi: 10.1016/j.freeradbiomed.2017.12.024
Karlenius, T. C., and Tonissen, K. F. (2010). Thioredoxin and cancer: a role for thioredoxin in all states of tumor oxygenation. Cancers 2, 209–232. doi: 10.3390/cancers2020209
Khaper, N., Bailey, C. D., Ghugre, N. R., Reitz, C., Awosanmi, Z., Waines, R., et al. (2018). Implications of disturbances in circadian rhythms for cardiovascular health: a new frontier in free radical biology. Free Radic. Biol. Med. 119, 85–92. doi: 10.1016/j.freeradbiomed.2017.11.006
Kitayama, Y., Iwasaki, H., Nishiwaki, T., and Kondo, T. (2003). KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22, 2127–2134. doi: 10.1093/emboj/cdg212
Knoops, B., Becker, S., Poncin, M. A., Glibert, J., Derclaye, S., Clippe, A., et al. (2018). Specific interactions measured by afm on living cells between peroxiredoxin-5 and tlr4: relevance for mechanisms of innate immunity. Cell Chem. Biol. 25, 550–559.e3. doi: 10.1016/j.chembiol.2018.02.006
Knowles, B., Bailey, B., Boling, L., Breitbart, M., Cobián-Güemes, A., Del Campo, J., et al. (2017). Variability and host density independence in inductions-based estimates of environmental lysogeny. Nat. Microbiol. 2:17064. doi: 10.1038/nmicrobiol.2017.64
Knowles, B., Silveira, C. B., Bailey, B. A., Barott, K., Cantu, V. A., Cobián-Güemes, A. G., et al. (2016). Lytic to temperate switching of viral communities. Nature 531, 466–470. doi: 10.1038/nature17193
Ko, C. H., and Takahashi, J. S. (2006). Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277. doi: 10.1093/hmg/ddl207
Kolody, B. C., McCrow, J. P., Allen, L. Z., Aylward, F. O., Fontanez, K. M., Moustafa, A., et al. (2019). Diel transcriptional response of a California Current plankton microbiome to light, low iron, and enduring viral infection. ISME J. 13, 2817–2833. doi: 10.1038/s41396-019-0472-2
Koonin, E. V., Senkevich, T. G., and Dolja, V. V. (2006). The ancient virus world and evolution of cells. Biol. Direct 1:29. doi: 10.1186/1745-6150-1-29
Korzhenkov, A. A., Toshchakov, S. V., Bargiela, R., Gibbard, H., Ferrer, M., Teplyuk, A. V., et al. (2019). Archaea dominate the microbial community in an ecosystem with low-to-moderate temperature and extreme acidity. Microbiome 7:11. doi: 10.1186/s40168-019-0623-8
Kuang, Z., Wang, Y., Li, Y., Ye, C., Ruhn, K. A., Behrendt, C. L., et al. (2019). The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science 365, 1428–1434. doi: 10.1126/science.aaw3134
Lavy, A., Keren, R., Yahel, G., and Ilan, M. (2016). Intermittent hypoxia and prolonged suboxia measured in situ in a marine sponge. Front. Mar. Sci. 3:263. doi: 10.3389/fmars.2016.00263
Leach, W. B., Carrier, T. J., and Reitzel, A. M. (2019). Diel patterning in the bacterial community associated with the sea anemone Nematostella vectensis. Ecol. Evol. 9, 9935–9947. doi: 10.1002/ece3.5534
Lee, S. J., Kong, M., Morse, D., and Hijri, M. (2018). Expression of putative circadian clock components in the arbuscular mycorrhizal fungus Rhizoglomus irregulare. Mycorrhiza 28, 523–534. doi: 10.1007/s00572-018-0843-y
Lee, S. J., Morse, D., and Hijri, M. (2019). Holobiont chronobiology: mycorrhiza may be a key to linking aboveground and underground rhythms. Mycorrhiza 29, 403–412. doi: 10.1007/s00572-019-00903-4
Leone, V., Gibbons, S. M., Martinez, K., Hutchison, A. L., Huang, E. Y., Cham, C. M., et al. (2015). Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17, 681–689. doi: 10.1016/j.chom.2015.03.006
Letarov, A., Manival, X., Desplats, C., and Krisch, H. M. (2005). gpwac of the T4-type bacteriophages: structure, function, and evolution of a segmented coiled-coil protein that controls viral infectivity. J. Bacteriol. 187, 1055–1066. doi: 10.1128/JB.187.3.1055-1066.2005
Liang, X., Bushman, F. D., and FitzGerald, G. A. (2015). Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. U.S.A. 112, 10479–10484. doi: 10.1073/pnas.1501305112
Lindell, D., Jaffe, J. D., Johnson, Z. I., Church, G. M., and Chisholm, S. W. (2005). Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438, 86–89. doi: 10.1038/nature04111
Liu, R., Liu, Y., Chen, Y., Zhan, Y., and Zeng, Q. (2019). Cyanobacterial viruses exhibit diurnal rhythms during infection. Proc. Natl. Acad. Sci. U.S.A. 116, 14077–14082. doi: 10.1073/pnas.1819689116
Liu, Y., Tsinoremas, N. F., Johnson, C. H., Lebedeva, N. V., Golden, S. S., Ishiura, M., et al. (1995). Circadian orchestration of gene expression in cyanobacteria. Gene Dev. 9, 1469–1478. doi: 10.1101/gad.9.12.1469
Loza-Correa, M., Gomez-Valero, L., and Buchrieser, C. (2010). Circadian clock proteins in prokaryotes: hidden rhythms? Front. Microbiol. 1:130. doi: 10.3389/fmicb.2010.00130
Ma, P., Mori, T., Zhao, C., Thiel, T., and Johnson, C. H. (2016). Evolution of KaiC-dependent timekeepers: a proto-circadian timing mechanism confers adaptive fitness in the purple bacterium Rhodopseudomonas palustris. PLoS Genet. 12:e1005922. doi: 10.1371/journal.pgen.1005922
Mackenzie, J. J., and Haselkorn, R. (1972). An electron microscope study of infection by the blue-green algal virus SM-1. Virology 49, 505–516. doi: 10.1016/0042-6822(72)90502-8
Makarova, K. S., Galperin, M. Y., and Koonin, E. V. (2017). Proposed role for KaiC-Like ATPases as major signal transduction hubs in archaea. MBio 8:e01959-17. doi: 10.1128/mBio.01959-17
Maldonado, M. (2007). Intergenerational transmission of symbiotic bacteria in oviparous and viviparous demosponges, with emphasis on intracytoplasmically-compartmented bacterial types. J. Mar. Biol. Assoc. U. K. 87, 1701–1713. doi: 10.1017/S0025315407058080
Maniscalco, M., Nannen, J., Sodi, V., Silver, G., Lowrey, P., and Bidle, K. (2014). Light-dependent expression of four cryptic archaeal circadian gene homologs. Front. Microbiol. 5:79. doi: 10.3389/fmicb.2014.00079
Mann, N. H., Clokie, M. R., Millard, A., Cook, A., Wilson, W. H., Wheatley, P. J., et al. (2005). The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187, 3188–3200. doi: 10.1128/JB.187.9.3188-3200.2005
Markson, J. S., Piechura, J. R., Puszynska, A. M., and O’Shea, E. K. (2013). Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell 155, 1396–1408. doi: 10.1016/j.cell.2013.11.005
McClung, C. (2001). Circadian rhythms in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 139–162. doi: 10.1146/annurev.arplant.52.1.139
Merrow, M., Mazzotta, G., Chen, Z., and Roenneberg, T. (2006). The right place at the right time: regulation of daily timing by phosphorylation. Gene Dev. 20, 2629–2633. doi: 10.1101/gad.1479706
Millard, A., Clokie, M. R., Shub, D. A., and Mann, N. H. (2004). Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. U.S.A. 101, 11007–11012. doi: 10.1073/pnas.0401478101
Min, H., Guo, H., and Xiong, J. (2005). Rhythmic gene expression in a purple photosynthetic bacterium, Rhodobacter sphaeroides. FEBS Lett. 579, 808–812. doi: 10.1016/j.febslet.2005.01.003
Miranda, J. A., Culley, A. I., Schvarcz, C. R., and Steward, G. F. (2016). RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 18, 3714–3727. doi: 10.1111/1462-2920.13291
Mitsui, A., Kumazawa, S., Takahashi, A., Ikemoto, H., Cao, S., and Arai, T. (1986). Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature 323, 720–722. doi: 10.1038/323720a0
Miyashiro, T., and Ruby, E. G. (2012). Shedding light on bioluminescence regulation in Vibrio fischeri. Mol. Microbiol. 84, 795–806. doi: 10.1111/j.1365-2958.2012.08065.x
Mojica, K. D. A., and Brussaard, C. P. D. (2014). Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 89, 495–515. doi: 10.1111/1574-6941.12343
Montagner, A., Korecka, A., Polizzi, A., Lippi, Y., Blum, Y., Canlet, C., et al. (2016). Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci. Rep. 6:20127. doi: 10.1038/srep20127
Mori, T. (2009). “Cell division and circadian clock,” in Bacterial Circadian Programs, eds J. L. Ditty, S. Mackey, and C. H. Johnson (Berlin: Springer-Verlag), 241–258. doi: 10.1007/978-3-540-88431-6_11
Mori, T., Binder, B., and Johnson, C. H. (1996). Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. U.S.A. 93, 10183–10188.
Morioka, E., Oida, M., Tsuchida, T., and Ikeda, M. (2018). Nighttime activities and peripheral clock oscillations depend on Wolbachia endosymbionts in flies. Sci. Rep. 8:15432. doi: 10.1038/s41598-018-33522-8
Mortha, A., Chudnovskiy, A., Hashimoto, D., Bogunovic, M., Spencer, S. P., Belkaid, Y., et al. (2014). Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343:1249288. doi: 10.1126/science.1249288
Mukherji, A., Kobiita, A., Ye, T., and Chambon, P. (2013). Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153, 812–827. doi: 10.1016/j.cell.2013.04.020
Mullineaux-Sanders, C., Suez, J., Elinav, E., and Frankel, G. (2018). Sieving through gut models of colonization resistance. Nat. Microbiol. 3, 132–140. doi: 10.1038/s41564-017-0095-1
Murakami, M., and Tognini, P. (2019). The circadian clock as an essential molecular link between host physiology and microorganisms. Front. Cell. Infect. Microbiol. 9:469. doi: 10.3389/fcimb.2019.00469
Ni, T., and Zeng, Q. (2016). Diel infection of cyanobacteria by cyanophages. Front. Mar. Sci. 2:123. doi: 10.3389/fmars.2015.00123
Nijus, D., McMurry, L., and Hastings, J. W. (1977). Conditionality of circadian rhythmicity: synergistic action of light and temperature. J. Comp. Physiol. 117, 335–344. doi: 10.1007/BF00691559
Nikaido, S., and Johnson, C. (2000). Daily and circadian variation in survival from ultraviolet radiation in Chlamydomonas reinhardtii. Photochem. Photobiol. 71, 758–765. doi: 10.1562/0031-865520000710758DACVIS2.0.CO2
Nobs, S. P., Tuganbaev, T., and Elinav, E. (2019). Microbiome diurnal rhythmicity and its impact on host physiology and disease risk. EMBO Rep. 20, e47129. doi: 10.15252/embr.201847129
Noordally, Z. B., and Millar, A. J. (2015). Clocks in algae. Biochemistry 54, 171–183. doi: 10.1021/bi501089x
Norman, J. M., Handley, S. A., and Virgin, H. W. (2014). Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology 146, 1459–1469. doi: 10.1053/j.gastro.2014.02.001
O’Neill, J. S., Lee, K. D., Zhang, L. Z., Feeney, K., Webster, S. G., Blades, M. J., et al. (2015). Metabolic molecular markers of the tidal clock in the marine crustacean Eurydice pulchra. Curr. Biol. 25, R301–R327. doi: 10.1016/j.cub.2015.02.052
O’Neill, J. S., Van Ooijen, G., Dixon, L. E., Troein, C., Corellou, F., Bouget, F. Y., et al. (2011). Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558. doi: 10.1038/nature09654
O’Neill, J. S., and Reddy, A. B. (2011). Circadian clocks in human red blood cells. Nature 469, 498–503. doi: 10.1038/nature09702
Oosterman, J. E., Kalsbeek, A., La Fleur, S. E., and Belsham, D. D. (2015). Impact of nutrients on circadian rhythmicity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 308, R337–R350. doi: 10.1152/ajpregu.00322.2014
Ouyang, Y., Ersson, C. R., Kondo, T., Golden, S. S., and Johnson, C. H. (1998). Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 95, 8660–8664. doi: 10.1073/pnas.95.15.8660
Padan, E., Ginzburg, D., and Shilo, M. (1970). The reproductive cycle of cyanophage LPP1-G in Plectonema boryanum and its dependence on photosynthetic and respiratory systems. Virology 40, 514–521. doi: 10.1016/0042-6822(70)90194-7
Parkar, S. G., Kalsbeek, A., and Cheeseman, J. F. (2019). Potential role for the gut microbiota in modulating host circadian rhythms and metabolic health. Microorganisms 7:41. doi: 10.3390/microorganisms7020041
Pattanayak, G. K., Phong, C., and Rust, M. J. (2014). Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr. Biol. 24, 1934–1938. doi: 10.1016/j.cub.2014.07.022
Paulose, J. K., Wright, J. M., Patel, A. G., and Cassone, V. M. (2016). Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS One 11:e0146643. doi: 10.1371/journal.pone.0146643
Perofsky, A. C., Lewis, R. J., and Meyers, L. A. (2019). Terrestriality and bacterial transfer: a comparative study of gut microbiomes in sympatric Malagasy mammals. ISME J. 13, 50–63. doi: 10.1038/s41396-018-0251-5
Pittendrigh, C. (1993). Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 17–54. doi: 10.1146/annurev.ph.55.030193.000313
Pospíšil, P. (2016). Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 7:1950. doi: 10.3389/fpls.2016.01950
Rakhuba, D. V., Kolomiets, E. I., Dey, E. S., and Novik, G. I. (2010). Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Polish J. Microbiol. 59, 145–155. doi: 10.33073/pjm-2010-023
Reppert, S. M., and Weaver, D. R. (2002). Coordination of circadian timing in mammals. Nature 418, 935–941. doi: 10.1038/nature00965
Rogers, L. A., and Greenbank, G. R. (1930). The intermittent growth of bacterial cultures. J. Bacteriol. 19, 181–189. doi: 10.1128/jb.19.3.181-190.1930
Rosbash, M. (2009). The implications of multiple circadian clock origins. PLoS Biol. 7:e62. doi: 10.1371/journal.pbio.1000062
Rothschild, D., Weissbrod, O., Barkan, E., Kurilshikov, A., Korem, T., Zeevi, D., et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. doi: 10.1038/nature25973
Russell, P. W., and Müller, U. R. (1984). Construction of bacteriophage luminal diameterX174 mutants with maximum genome sizes. J. Virol. 52, 822–827. doi: 10.1128/jvi.52.3.822-827.1984
Sartor, F., Eelderink-Chen, Z., Aronson, B., Bosman, J., Hibbert, L. E., Dodd, A. N., et al. (2019). Are there circadian clocks in non-photosynthetic bacteria? Biology 8:41. doi: 10.3390/biology8020041
Sato, T. K., Panda, S., Miraglia, L. J., Reyes, T. M., Rudic, R. D., McNamara, P., et al. (2004). A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537. doi: 10.1016/j.neuron.2004.07.018
Say, T. E., and Degnan, S. M. (2019). Molecular and behavioural evidence that interdependent photo – and chemosensory systems regulate larval settlement in a marine sponge. Mol. Ecol. 29, 247–261. doi: 10.1111/mec.15318
Schmelling, N. M., Lehmann, R., Chaudhury, P., Beck, C., Albers, S. V., Axmann, I. M., et al. (2017). Minimal tool set for a prokaryotic circadian clock. BMC Evol. Biol. 17:169. doi: 10.1186/s12862-017-0999-7
Schmitt, S., Angermeier, H., Schiller, R., Lindquist, N., and Hentschel, U. (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74, 7694–7708. doi: 10.1128/AEM.00878-08
Schneider, T., and Leitz, T. (1994). Protein kinase C in hydrozoans: involvement in metamorphosis of Hydractinia and in pattern formation of Hydra. Roux’s Arch. Dev. Biol. 203, 422–428. doi: 10.1007/BF00188691
Schwartzman, J. A., Koch, E., Heath-Heckman, E. A., Zhou, L., Kremer, N., McFall-Ngai, M. J., et al. (2015). The chemistry of negotiation: rhythmic, glycan-driven acidification in a symbiotic conversation. Proc. Natl. Acad. Sci. U.S.A. 112, 566–571. doi: 10.1073/pnas.1418580112
Shi, S. Q., Ansari, T. S., McGuinness, O. P., Wasserman, D. H., and Johnson, C. H. (2013). Circadian disruption leads to insulin resistance and obesity. Curr. Biol. 23, 372–381. doi: 10.1016/j.cub.2013.01.048
Silver, A. C., Arjona, A., Walker, W. E., and Fikrig, E. (2012). The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36, 251–261. doi: 10.1016/j.immuni.2011.12.017
Smith, R. M., and Williams, S. B. (2006). Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 103, 8564–8569. doi: 10.1073/pnas.0508696103
Sorek, M., and Levy, O. (2012a). Influence of the quantity and quality of light on photosynthetic periodicity in coral endosymbiotic algae. PLoS One 7:e43264. doi: 10.1371/journal.pone.0043264
Sorek, M., and Levy, O. (2012b). The effect of temperature compensation on the circadian rhythmicity of photosynthesis in Symbiodinium, coral-symbiotic alga. Sci. Rep. 2:536. doi: 10.1038/srep00536
Sorek, M., Díaz-Almeyda, E. M., Medina, M., and Levy, O. (2014). Circadian clocks in symbiotic corals: the duet between Symbiodinium algae and their coral host. Mar. Genomics 14, 47–57. doi: 10.1016/j.margen.2014.01.003
Sorek, M., Schnytzer, Y., Waldman Ben-Asher, H., Caspi, V. C., Chen, C. S., Miller, D. J., et al. (2018). Setting the pace: host rhythmic behaviour and gene expression patterns in the facultatively symbiotic cnidarian Aiptasia are determined largely by Symbiodinium. Microbiome 6:83. doi: 10.1186/s40168-018-0465-9
Sorek, M., Yacobi, Y. Z., Roopin, M., Berman-Frank, I., and Levy, O. (2013). Photosynthetic circadian rhythmicity patterns of Symbiodinium, the coral endosymbiotic algae. Proc. R. Soc. B Biol. Sci. 280:20122942. doi: 10.1098/rspb.2012.2942
Spengler, M. L., Kuropatwinski, K. K., Comas, M., Gasparian, A. V., Fedtsova, N., Gleiberman, A. S., et al. (2012). Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc. Natl. Acad. Sci. U.S.A. 109, E2457–E2465. doi: 10.1073/pnas.1206274109
Strader, M. E., Aglyamova, G. V., and Matz, M. V. (2018). Molecular characterization of larval development from fertilization to metamorphosis in a reef-building coral. BMC Genomics 19:17. doi: 10.1186/s12864-017-4392-0
Sully, S., Burkepile, D. E., Donovan, M. K., Hodgson, G., and Van Woesik, R. (2019). A global analysis of coral bleaching over the past two decades. Nat. Commun. 10:1264. doi: 10.1038/s41467-019-09238-2
Suttle, C. A. (2007). Marine viruses – major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Suttle, C. A., and Chen, F. (1992). Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58, 3721–3729. doi: 10.1128/aem.58.11.3721-3729.1992
Takahashi, J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. doi: 10.1038/nrg.2016.150
Takai, N., Nakajima, M., Oyama, T., Kito, R., Sugita, C., Sugita, M., et al. (2006). A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 12109–12114. doi: 10.1073/pnas.0602955103
Taniguchi, Y., Takai, N., Katayama, M., Kondo, T., and Oyama, T. (2010). Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 3263–3268. doi: 10.1073/pnas.0909924107
Terauchi, K., Kitayama, Y., Nishiwaki, T., Miwa, K., Murayama, Y., Oyama, T., et al. (2007). ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 16377–16381.
Thaiss, C. A., Levy, M., and Elinav, E. (2015). Chronobiomics: the biological clock as a new principle in host-microbial interactions. PLoS Pathog. 11:e1005113. doi: 10.1371/journal.ppat.1005113
Thaiss, C. A., Levy, M., Korem, T., Dohnalová, L., Shapiro, H., Jaitin, D. A., et al. (2016). Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510.e12. doi: 10.1016/j.cell.2016.11.003
Thaiss, C. A., Zeevi, D., Levy, M., Zilberman-Schapira, G., Suez, J., Tengeler, A. C., et al. (2014). Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529. doi: 10.1016/j.cell.2014.09.048
Thingstad, T. F. (2000). Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328. doi: 10.4319/lo.2000.45.6.132
Thingstad, T. F., Vague, S., Storesund, J. E., Sandaa, R. A., and Giske, J. (2014). A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc. Natl. Acad. Sci. U.S.A. 111, 7813–7818. doi: 10.1073/pnas.1400909111
Thompson, L. R., Zeng, Q., Kelly, L., Huang, K. H., Singer, A. U., Stubbe, J., et al. (2011). Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. U.S.A. 108, E757–E764. doi: 10.1073/pnas.1102164108
Thorley, J. A., McKeating, J. A., and Rappoport, J. Z. (2010). Mechanisms of viral entry: sneaking in the front door. Protoplasma 244, 15–24. doi: 10.1007/s00709-010-0152-6
Turnbaugh, P. J., Ridaura, V. K., Faith, J. J., Rey, F. E., Knight, R., and Gordon, J. I. (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14. doi: 10.1126/scitranslmed.3000322
Turrens, J. F. (2003). Mitochondrial formation of reactive oxygen species. J. Physiol. 552(Pt 2), 335–344. doi: 10.1113/jphysiol.2003.049478
Van Praag, E., Agosti, R. D., and Bachofen, R. (2000). Rhythmic activity of uptake hydrogenase in the prokaryote Rhodospirillum rubrum. J. Biol. Rhythm 15, 218–224. doi: 10.1177/074873040001500303
Voigt, R. M., Summa, K. C., Forsyth, C. B., Green, S. J., Engen, P., Naqib, A., et al. (2016). The circadian clock mutation promotes intestinal dysbiosis. Alcohol. Clin. Exp. Res. 40, 335–347. doi: 10.1111/acer.12943
Vollmers, C., Gill, S., Di Tacchio, L., Pulivarthy, S. R., Le, H. D., and Panda, S. (2009). Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Biochem. Natl. Acad. Sci. U.S.A. 106, 21453–21458. doi: 10.1073/pnas.0909591106
Waldbauer, J. R. (2010). Molecular Biogeochemistry of Modern and Ancient Marine Microbes. [dissertation]. Falmouth, MA: Massachusetts Institute of Technology and Woods Hole Oceanographic Institution. doi: 10.1575/1912/3238
Warter, V., Erez, J., and Müller, W. (2018). Environmental and physiological controls on daily trace element incorporation in Tridacna crocea from combined laboratory culturing and ultra-high resolution LA-ICP-MS analysis. Palaeogeogr. Palaeoclimatol. Palaeoecol. 496, 32–47. doi: 10.1016/j.palaeo.2017.12.038
Whitehead, K., Pan, M., and Masumura, K. (2009). Diurnally entrained anticipatory behaviour in archaea. PLoS One 4:e5485. doi: 10.1371/journal.pone.0005485
Wier, A. M., Nyholm, S. V., Mandel, M. J., Massengo-Tiassé, R. P., Schaefer, A. L., Koroleva, I., et al. (2010). Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107, 2259–2264. doi: 10.1073/pnas.0909712107
Winget, D. M., Williamson, K. E., Helton, R. R., and Wommack, K. E. (2005). Tangential flow diafiltration: an improved technique for estimation of virioplankton production. Aquat. Microb. Ecol. 41, 221–232. doi: 10.3354/ame041221
Woelfle, M. A., Ouyang, Y., Phanvijhitsiri, K., and Johnson, C. H. (2004). The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486. doi: 10.1016/j.cub.2004.08.023
Wommack, K. E., and Colwell, R. R. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114. doi: 10.1128/mmbr.64.1.69-114.2000
Xia, Y., Cheng, Z., and Wang, S. (2018). Modelling the crosstalk between the circadian clock and ROS in Neurospora crassa. J. Theor. Biol. 458, 125–132. doi: 10.1016/j.jtbi.2018.09.013
Yoon, S. S., Kim, E. K., and Lee, W. J. (2015). Functional genomic and metagenomic approaches to understanding gut microbiota-animal mutualism. Curr. Opin. Microbiol. 24, 38–46. doi: 10.1016/j.mib.2015.01.007
Keywords: circadian clock, chronobiomics, microbiome, rhythmic regulation, marine plankton, viruses, symbioses, nested ecosystems
Citation: Hewitt OH and Shaikh HM (2021) The Rhythm of Many: Biological Rhythms in the Marine Environment, From Macro-Scale Planktonic Ecosystems to Micro-Scale Holobionts. Front. Mar. Sci. 8:744169. doi: 10.3389/fmars.2021.744169
Received: 19 July 2021; Accepted: 13 October 2021;
Published: 18 November 2021.
Edited by:
Simon Jungblut, University of Bremen, GermanyReviewed by:
Shengwei Hou, Southern University of Science and Technology, ChinaSoon-Jae Lee, University of Lausanne, Switzerland
Copyright © 2021 Hewitt and Shaikh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hisham M. Shaikh, aGlzaGFtLnNoYWlraEB2bGl6LmJl
 Olivia H. Hewitt
Olivia H. Hewitt Hisham M. Shaikh
Hisham M. Shaikh