- 1Department of Life Sciences, Ben-Gurion University of the Negev, Beer-Sheva, Israel
- 2Marine Biotechnology Department, Stazione Zoologica Anton Dohrn, Naples, Italy
- 3The National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer-Sheva, Israel
- 4Department of Life Sciences, Achva Academic College, Shikmim, Israel
Hermaphrodite systems offer unique opportunities to study sexual differentiation, due to their high degree of sexual plasticity and to the fact that, unlike gonochoristic systems, the process is not confined to an early developmental stage. In protandric shrimp species, such as Hippolyte inermis and Pandalus platyceros, male differentiation is followed by transformation to femaleness during adulthood. The mechanisms controlling sexual differentiation have not been fully elucidated in crustaceans, but a key role has been attributed to the insulin-like hormone (IAG) produced by the androgenic gland (AG), a crustacean masculine endocrine organ. To uncover further transcriptomic toolkit elements affecting the sexual differentiation of H. inermis, we constructed eye and whole body RNA libraries of four representative stages during its protandric life cycle (immature, male, young female and mature female). The body libraries contained transcripts related to the reproductive system, among others, while the eye libraries contained transcripts related to the X-organ-sinus gland, a central endocrine complex that regulates crustacean reproduction. Binary pattern analysis, performed to mine for genes expressed differentially between the different life stages, yielded 19,605 and 6,175 transcripts with a specific expression pattern in the eye and body, respectively. Prominent sexually biased transcriptomic patterns were recorded for the IAG and vitellogenin genes, representing, respectively, a key factor within the masculine IAG-switch, and a precursor of the yolk protein, typical of feminine reproductive states. These patterns enabled the discovery of novel putative protein-coding transcripts exhibiting sexually biased expression in the H. inermis body and eye transcriptomes of males and females. Homologs to the above novel genes have been found in other decapod crustaceans, and a comparative study, using previously constructed transcriptomic libraries of another protandric shrimp, P. platyceros, showed similar sexually biased results, supporting the notion that such genes, mined from the H. inermis transcriptome, may be universal factors related to reproduction and sexual differentiation and their control in other crustaceans. This study thus demonstrates the potential of transcriptomic studies in protandric species to uncover unexplored layers of the complex crustacean sex-differentiation puzzle.
Introduction
Naturally occurring sexual shifts during hermaphrodite life cycles offer unique opportunities to study sexual plasticity and sexual differentiation. The latter processes are currently attracting worldwide interest due to accumulating evidence that environmental factors and endocrine-disrupting chemicals affect hormonal regulation, sexual development and fertility in animals (Olmstead and LeBlanc, 2000; Rodriguez et al., 2000, 2007; Ford et al., 2003, 2004; Hayes et al., 2010; Ford, 2012).
Important examples of sexual plasticity may be found among the crustaceans (Pandian, 2016), an ancient highly diverse group of animals in which a variety of reproductive strategies are represented, including gonochorism (separate sexes) (Juchault, 1999), intersexuality (combination of male and female features within a gonochoristic species) (Goldschmidt, 1938; Reinboth, 1975; Sagi et al., 1996; Abdel-Moneim et al., 2015; Levy et al., 2020c), asexual reproduction (e.g., parthenogenesis) (Scholtz et al., 2003; Martin et al., 2007), and different types of hermaphroditism (Levy et al., 2018; Benvenuto and Weeks, 2020). The last of these strategies constitutes a means of reproduction in which a particular individual bears both ovarian and testicular tissues [ovotestis (Hoffman, 1972; Stentiford, 2012)] and produces gametes of both sexes (Benvenuto and Weeks, 2020). Hermaphroditism may be either simultaneous, in which the individual functions both as a male and a female (Bauer and Holt, 1998; Baeza, 2007), or sequential, in which the individual first matures as one sex and then transforms irreversibly into the other (Hoffman, 1968; Subramoniam, 1981; de Almeida and Buckup, 2000). Of relevance to this study, hermaphroditism may be exploited as a valuable model system for studying sexual plasticity, since the sex-differentiation process does not occur as a single event limited to the early developmental stages of the organism.
A fairly well researched case study of hermaphroditism in crustaceans is that of the shrimp Hippolyte inermis, formerly known as H. viridis (Reverberi, 1950), a caridean protandric species inhabiting seagrass (Posidonia oceanica) meadows in shallow waters of the Mediterranean Sea and the Atlantic coasts of Spain (Zupo and Messina, 2007). While protandric species are commonly born as males, followed by a transitional stage before transforming to females (Yaldwyn, 1966), H. inermis diverts from this pattern in that it lacks the ovotestis-containing transitional stage (Cobos et al., 2005; Zupo and Messina, 2007; Mutalipassi et al., 2018) in both of its reproductive cycles, one in the spring and the other in the fall (Figure 1). In the spring, these shrimp exhibit a first reproductive burst, and approximately three months after hatching, both immature males and females are present in the population. In contrast, at the end of the second reproductive burst in the fall, only males are present; these animals transform into females in the following spring (Zupo, 1994). It has been suggested that the spring sexual shift in H. inermis is promoted by a diet based on diatoms (Zupo, 2000), namely, the species of Cocconeis, that are abundant in their seagrass habitat, resulting in an early transformation of males at a younger age and, hence, in the presence of small females in the population by the end of the spring (Zupo, 1994, 2000). However, the lower abundance of these diatoms in the fall leads to a normal process of protandric development, with a predominance of young males in the population following this second reproductive burst. Physiologically, it has been suggested that compounds present in the Cocconeis spp. diatoms cause apoptosis of the crustacean androgenic gland (AG) and, consequently, control the sexual shift from maleness to femaleness (Zupo et al., 2007). This suggestion is in keeping with the universal mechanism of control of crustacean sexual differentiation by the insulin-like androgenic hormone (IAG) – secreted by the AG – in a process involving many upstream and downstream genes, termed the “IAG-switch” (Levy and Sagi, 2020). When the switch is “turned on” (i.e., in normal males or through activation of the AG and expression or induction of IAG), masculinization occurs, whereas when it is “turned off” (i.e., in normal females or through AG ablation or silencing or inactivation of the IAG), the result is feminization (Charniaux-Cotton, 1958, 1962; Nagamine et al., 1980a,b; Ventura et al., 2012; Levy et al., 2016).
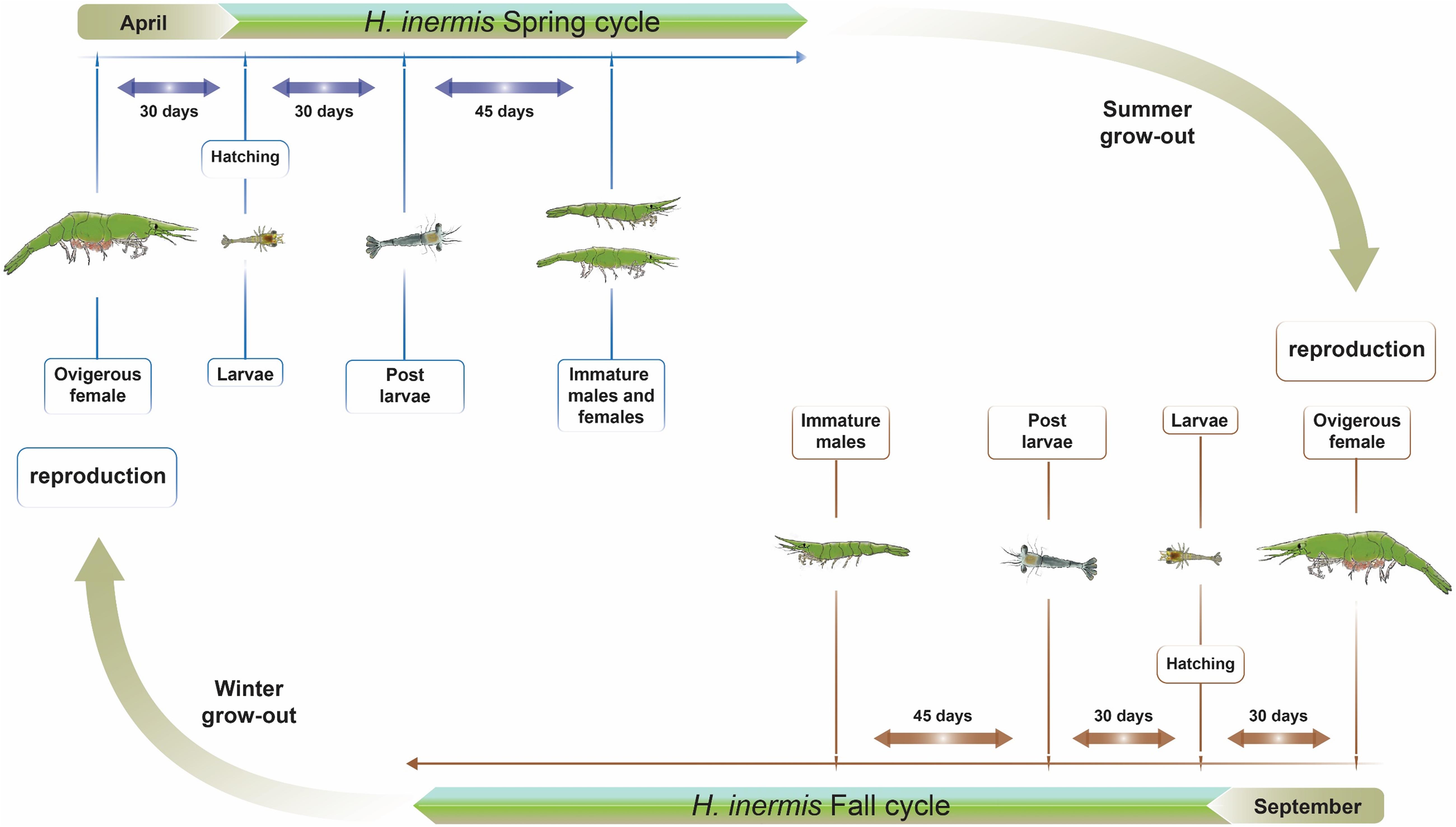
Figure 1. Two reproductive cycles in the life cycle of Hippolyte inermis. Two reproductive seasons are shown. Animals that hatch in April, due to a higher abundance of Cocconeis spp. diatoms, become either immature males or females by July and, following a summer grow-out, reproduce in September. Animals that hatch in September, due to a lower abundance of the diatoms, become males by December and, following a winter grow-out, reproduce in September.
Another case study of crustacean hermaphroditism – one that we also exploit in this study – is that of the protandric Northern spot shrimp, Pandalus platyceros, which is widely distributed in the North Pacific Ocean (Butler, 1964). Unlike H. inermis, in which the transformation from maleness to femaleness takes place up to few months following hatching (Zupo, 1994), the transformation in P. platyceros is slower, occurring at the age of 3 to 5 years (Butler, 1965; King and Moffitt, 1984; Iversen et al., 1993; Kimker et al., 1996). Therefore, unlike gonochoristic species in which the IAG-switch-based sexual differentiation process is limited to early developmental stages, protandric shrimps, such as H. inermis and P. platyceros, may serve as models in the study of sex-controlling toolkits, because in such species the process is not confined to early development but rather occurs later in the life cycle, when adult males transform into females. Protandry may thus be regarded as offering an opportunity to obtain new insights into sexual differentiation that cannot be obtained from studies of gonochoristic species.
In the present study, we collected samples of H. inermis at different stages of the species’ protandric life cycle and constructed RNA libraries that yielded both known and novel, yet unannotated, sex-specific genes that are assumed to be associated with sexual differentiation and reproduction. To obtain insight into the function of these novel genes, we also performed a comparative in silico analysis with P. platyceros, taking advantage of RNA libraries of specific tissues previously obtained over the life cycle of P. platyceros (Levy et al., 2020a). P. platyceros is particularly suitable for such an analysis for two reasons: (i) taxonomically, the two species, H. inermis and P. platyceros, belong to families – Pandalidae and Hippolytidae, respectively (Martin et al., 2009) – that are closely related within the infraorder Caridea (Christoffersen, 1990; Wolfe et al., 2019; Levy et al., 2020c), and (ii) technically, the RNA for the H. inermis libraries was extracted from the entire body of the animal (due to its small size that prevented dissection of specific tissues), while the RNA for constructing P. platyceros libraries was obtained by dissecting out specific tissues (Levy et al., 2020a). Therefore, our working hypothesis for the in silico analysis part of this study was that a search for homologs to the newly discovered H. inermis genes in transcriptomic libraries of P. platyceros (and other decapod crustaceans) might reveal the function of those genes in a more general context that includes a wider range of crustaceans.
Materials and Methods
Animal Collection and Life Stage Definition
Based on the reproductive stages comprising the protandric life history of H. inermis (immature animals, males and females) throughout the course of the year (Zupo, 1994), shrimp were collected during September 2018 and April 2019 in Lacco Ameno d’Ischia (Gulf of Naples, Italy) at depths of 3–15 m by using a plankton net of 400 mm diameter and 100 μm mesh size, as previously described (Mutalipassi et al., 2018; Figure 2). Sorting into the different reproductive stages was performed manually, both visually and under a Leica M16 macroscope, as previously described (Zupo, 1994), based on the total body length and on the presence or absence of the appendix masculina (AM), a prominent external male character (Tombes and Foster, 1979; Nagamine et al., 1980b; Zupo et al., 2008; Mutalipassi et al., 2018; Levy et al., 2020b). The animals were sorted into four stages: I - immature animals (total length of 1–6 mm and the absence of an AM), M – males (total length of 7–11 mm and the presence of an AM), YF – young females (total length of 7–11 mm and the absence of an AM), and MF – mature females (total length of 12–33 mm and the absence of an AM) (Figure 2).
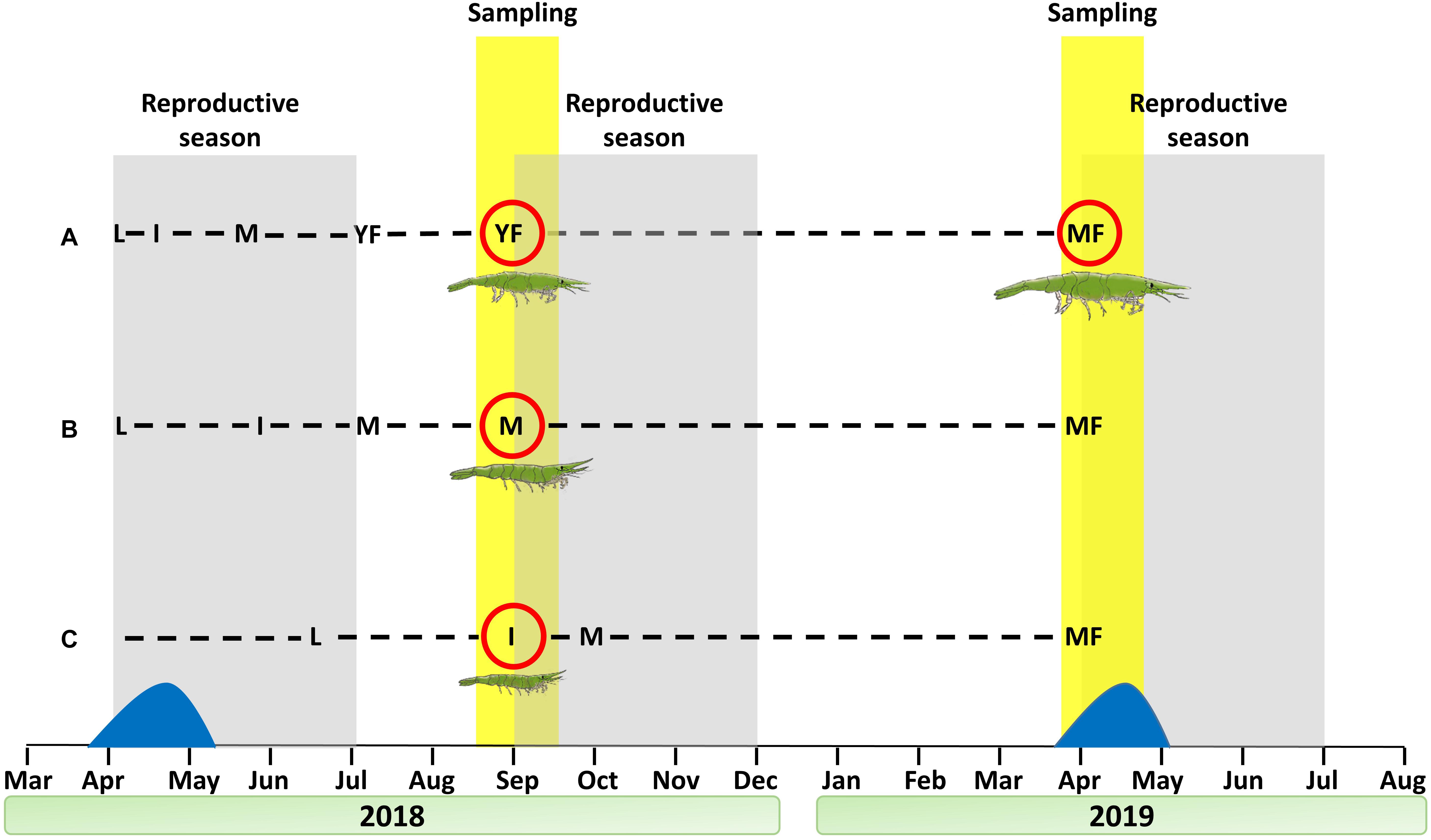
Figure 2. Sampling regimen for the construction of a Hippolyte inermis transcriptomic library. Shrimp were sampled at the beginning of the reproduction seasons of September 2018 and April 2019. Dashed lines represent different cohorts. (A) Larvae hatched in April 2018 and quickly passed through the male stage (due to the diatom peak), becoming young females in September 2018 and mature females in April 2019. (B) Larvae hatched in April 2018 and matured slowly as males, despite the diatom peak, thus remaining males in September 2018 and becoming mature females in April 2019. (C) Larvae hatched in mid-June, thus remaining immature in September 2018 and becoming mature females in April 2019. Sampled stages are ringed with a red circle. Reproductive seasons and sampling periods are indicated with gray and yellow backgrounds, respectively. The diatom peak is represented in blue. L = larvae, I = immature, M = male, YF = young female, MF = mature female.
RNA Library Preparation of H. inermis From the Different Reproductive Stages
To characterize the transcriptional patterns of genes related to sexual development in different stages of the H. inermis life cycle, RNA was extracted using EZ-RNA Isolation kit (BI, Cromwell, CT, United States). Ideally, tissues related to reproduction and its control should be dissected and extracted separately, but due to the small size of H. inermis, it was impossible to separately dissect out tissues such as the AG or gonads. Therefore, we separated the eyestalks, which contain a prominent endocrine controlling organ in crustaceans (Keller, 1992; Wilder et al., 1994), from the rest of the body, which contains the reproductive system, for immature animals (n = 90), males (n = 72), young reproductive females (n = 78), and mature females (n = 15). To compensate for low RNA quantity yielded from a single individual animal, for RNA extraction, three bulks of equal number of different body and eye tissues, for each reproductive stage, were pooled separately. RNA was then extracted and the 24 pooled RNA samples (2 tissues × 3 replicates × 4 stages) were sent for sequencing (Novogene, Hong Kong) on an Illumina NovaSeq 6,000 platform.
Assembly and Annotation of H. inermis RNA Libraries
Bioinformatic analyses were carried out at the Bioinformatics Core Facility of Ben-Gurion University with a NeatSeq-Flow platform (Sklarz et al., 2018) and in-house R scripts. Reads were quality trimmed with TrimGalore1. Ribosomal RNA was filtered out as follows: reads were aligned to a database of crustacean rRNA sequences downloaded from the NCBI with bwa mem (Li, 2013) using default parameters. Reads that aligned to the rRNA database (4.9–18%) were discarded using samtools (Li et al., 2009). A total of 2,458,778,342 clean reads were retained for further analysis. Two reference transcriptomes were de novo assembled, one from the eye samples and the other from the body samples. The transcriptomes were assembled using Trinity version 2.8.4 (Grabherr et al., 2011) and then filtered to exclude transcripts with very low expression. To this end, the clean reads were aligned to the transcriptome, and only transcripts for which at least one of the experimental groups had at least two replicates with counts of three or more transcripts per million were retained. The resulting filtered transcriptomes contained 244,523 and 261,562 transcripts (> 200 bp) from 112,747 and 119,618 putative genes, in the eye and body, respectively. Each transcriptome was quality assessed using Quast (Gurevich et al., 2013) and BUSCO (Simão et al., 2015) vs. the Metazoa_odb9 database. The eye and body transcriptomes included 98.8 and 98.6% of BUSCO proteins, respectively. For transcriptome annotation, the most highly expressed transcript per gene (i.e., the representative transcript) was selected using the filter_low_expr_transcripts.pl script from the Trinity software suite. These representative transcripts were annotated using Trinotate (Trinotate.github.io) by searching Swissprot and PFAM-A, and performing RNAmmer predictions. Best blastp hits of TransDecoder translated transcripts having e-value < 1E-5 were reported. In addition, the representative transcripts were searched against RefSeq Proteins using blastx, and the top 20 best hits of each transcript having e-value < 1E-3 were submitted to Blast2GO “Blast Description Annotator” algorithm.
Differential Expression Analysis of Genes From the RNA Libraries
Clean reads from the eye and body RNA samples were aligned to the respective filtered reference transcriptome (as described above) with bowtie2 (Langmead and Salzberg, 2012), and gene expression was estimated with RSEM (Li and Dewey, 2011). Statistical analyses were carried out for the eye and body libraries separately. For quality assessment, counts were subjected to variance stabilizing transformation [DESeq2; (Love et al., 2014)] and then to sample-wise correlation analysis and principal component analysis (PCA). One eye sample of the immature stage was found to be an outlier and was thus excluded from further analysis. Statistical testing for differential expression was carried out using DESeq2 (Love et al., 2014), a method specifically tailored to count data using negative binomial generalized linear models. All six possible contrasts were assessed (i.e., I vs. M, I vs. YF, I vs. MF, M vs. YF, M vs. MF and YF vs. MF). Genes were considered to be differentially expressed in a certain contrast if they had a false discovery rate (FDR) adjusted p-value of < 0.05 and a linear fold change > 1.3 or < −1.3, where the plus and minus signs denote up- and down-regulation, respectively. A unified list of differentially expressed genes in any of the contrasts was constructed for the eye and body libraries separately. Hierarchical clustering of the differentially expressed genes in each library, after z-scoring of their variance-stabilized expression values, was carried out using the pheatmap function of R. Also, to facilitate comparison between body and eye gene expression, a joint reference transcriptome was constructed from the clean reads of all body and eye samples. Transcriptome assembly and filtering were performed as described above, yielding 296,530 transcripts (> 200 bp) from 161,343 putative genes. Annotation was achieved using Trinotate. Read alignment and quantification of the body and eye samples to the joint transcriptome were done as described above. Counts were normalized using DESeq2 “counts” function with the “normalized” parameter set to TRUE, after excluding the exceptional eye sample. Genes with exclusive expression in either the body or the eye tissues were identified according to the following criteria: (1) All replicates of at least one of the developmental stages in the tissue of interest had more than 300 counts each. (2) All samples (in all stages) of the other tissue had less than 30 counts each.
Functional Enrichment Analyses
GO assignments and gene length data were extracted from trinotate results using trinotate-provided scripts. Gene Ontology (GO) enrichment analysis of the differentially expressed genes was carried out using the GOSeq R package (Young et al., 2010).
Binary Gene Expression Pattern Analysis
The unified list of genes expressed differentially in the eye and that for the body were subjected to binary gene expression pattern analysis, as previously described (Abehsera et al., 2015; Shaked et al., 2020). Briefly, a list of all possible binary patterns was constructed (n = 14), excluding “0000” and “1111” (i.e., low and high expression in all stages, respectively). For each gene, Pearson’s correlation coefficient was computed between the gene’s variance-stabilized counts across all samples and an artificial expression profile of each of the patterns. For example, for the body (having 3 replicate samples per developmental stage), the pattern 0110 was represented by the artificial profile “000111111000.” It is noteworthy that a high correlation of a gene to a pattern does not indicate a “presence” or “absence” status, but rather relatively “high” and “low” expression levels at different stages. Genes were assigned to the pattern with which they had the highest Pearson’s correlation, if the correlation was higher than 0.8. Hierarchical clustering of the genes in each pattern, after z-scoring of their variance-stabilized expression values, was carried out using R’s pheatmap function. To aid in the choice of candidate genes for further study, we subsequently applied an additional cutoff, namely, a “counts cut-off,” requiring that in developmental stages considered as “1” in the binary pattern, the normalized counts in all replicate samples would be larger than 500. Normalized counts were computed using the DESeq2 “counts” function with the “normalized” parameter set to TRUE.
Characterizing the AG and IAG in H. inermis
Even though the AG is prominently situated at the base of the fifth pereiopod in Crustacea (Charniaux-Cotton, 1962), it was nearly impossible to isolate the AG from the very small H. inermis males (total body length of 8–10 mm). Therefore, to enable histological analysis of this organ, the whole body of a male specimen was fixed in 4% buffered formalin for 48 h at room temperature, as previously described (Levy et al., 2020a). Samples were gradually dehydrated through a series of increasing alcohol concentrations (70%, 80%, 90% and 100%), incubated in xylene, and embedded in Paraplast (Kendall, Mansfield, MA, United States). Serial dorsoventral sections of 5 μm were then placed on silane-coated slides (Menzel-Gläser, Braunschweig, Germany) and stained with hematoxylin and eosin, for morphological observations, as follows: The slides were dipped in xylene for 5 min × 2 and then in 100, 90, 80 and 70% EtOH for 1 min each, followed by tap water for 1 min, and hematoxylin for 5 min. The slides were then washed in tap water for 5 min, in acidic 70% EtOH for 10 s (for removing background staining), and again in tap water for 4 min. The final stage comprised dipping the slides in 95% EtOH for 15 s, eosin for 5 min, 95% EtOH for 5 min, 100% EtOH for 5 min × 2, and xylene for 5 min × 2 and then covering the section with a cover slip.
To find the sequence of the IAG mRNA in H. inermis (Hi-IAG), we aligned the IAG sequence from P. platyceros [Pnp-IAG; GenBank accession number: KX619617.1 (Levy et al., 2020a)] to the H. inermis body transcriptome generated in this study. From the IAG sequences that are available for dozens of decapod species, with different reproductive strategies and from various clades within the Crustacea (Levy and Sagi, 2020), Pnp-IAG was chosen for alignment due to the similar protandric nature of P. platyceros and H. inermis and their close evolutionary relationship (Wolfe et al., 2019; Levy et al., 2020c). After finding the Hi-IAG mRNA transcript in the body transcriptome of H. inermis, the predicted structure of the protein was inferred from its deduced amino acid sequence.
Mining for Sex-Related Genes in the Different Stages of the H. inermis Life Cycle and Comparative Transcriptomic Analysis vs. P. platyceros
After successfully sequencing the male-representing IAG gene, as described above, we searched the six IAG-switch related genes previously described in P. platyceros (i.e., Pandalus platyceros IAG-switch-like proteins 1-6; (Levy et al., 2020a)) in the H. inermis body transcriptome. Also, we set out to sequence the female-representing vitellogenin (Vg) gene in H. inermis. Vg is a precursor of vitellin (yolk protein), which is synthesized in the hepatopancreas and, in some species, also in the ovary (Yano and Chinzei, 1987; Quackenbush, 1989; Tsukimura, 2001; Okumura et al., 2007). We note that this protein was found to reflect the physiological reproductive state of females along the life cycle of P. platyceros (Levy et al., 2020b). Guided by the same considerations as those described above for IAG sequencing (Wolfe et al., 2019; Levy et al., 2020c), we aligned the Vg sequence from P. platyceros [Pnp-Vg; GenBank accession number: MK070912.1 (Levy et al., 2020b)] to the H. inermis body transcriptome generated in this study.
In addition, to find novel sex-specific genes in H. inermis (beyond previously characterized sex-related genes), the list of genes (Supplementary Table 1) in the eye and body transcriptomes with binary patterns of 0100 (male-specific) and 0001 (mature female-specific) was investigated, focusing on unannotated transcripts that passed the counts cut-off threshold. Candidate genes with a coding region were investigated for the presence of conserved domains, using SMART2 [(Schultz et al., 2000)], and a signal peptide, using SignalP-5.0 Server (Almagro Armenteros et al., 2019).
As described above, it was not feasible to separate out specific tissues from the bodies of the sampled H. inermis shrimps and to sequence their transcriptomes. Therefore, to determine in which tissue the genes under investigation in this study were expressed, we performed an in silico comparative transcriptomic analysis of the candidate genes between H. inermis and P. platyceros. The coding regions of the IAG and Vg genes in H. inermis and the novel H. inermis male- and female-specific genes found in the present study were aligned to the available P. platyceros transcriptome, by using the tblastn module in BLAST (Altschul et al., 1990; Gertz et al., 2006). The first hit with the highest score from the BLAST alignment of each gene was considered as the homolog sequence in P. platyceros. In addition, to examine whether the novel male- and female-specific genes identified in this study are conserved among decapod crustaceans, blastx of the nucleotide sequences in the NCBI server was performed to Transcriptome Shotgun Assembly proteins (TSA), as was tblastn of the amino acid sequences to TSA (Organism: Decapoda (taxid: 6683)).
In vitro Verification of Sex-Specific Genes in Different Life Cycle Stages
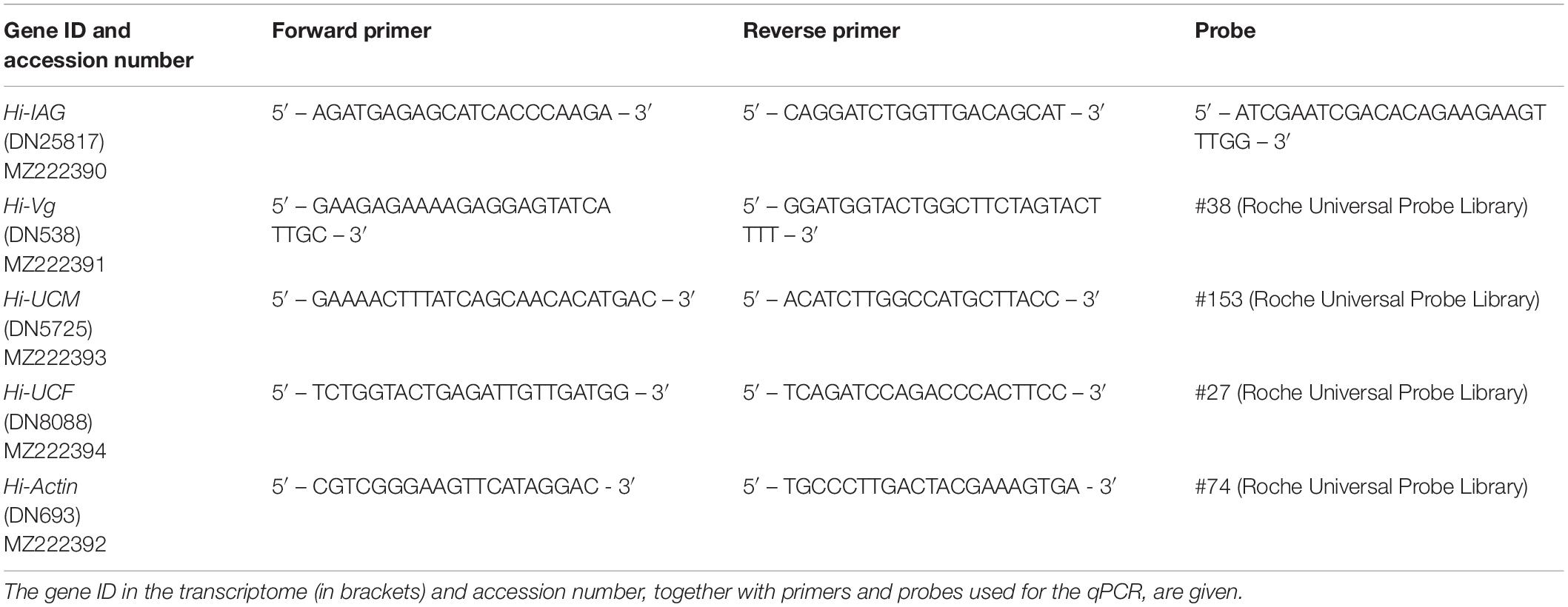
Total RNA was extracted as described above, and cDNA was synthesized using qScript cDNA Synthesis kit (Quanta, Beverly, MA, USA) from the pooled H. inermis body samples at each reproductive stage: I (n = 3), M (n = 3), YF (n = 3) and MF (n = 3). Relative quantification (RQ) of transcript levels was performed using Roche Diagnostics FastStart Universal Probe Master Mix (Basel, Switzerland) and Roche Universal Probe Library probes. The primers and probes that were used for the different qPCR assays are listed in Table 1. The qPCR reactions were performed in the QuantStudio 1 Real-Time PCR System, Applied Biosystems (Foster City, CA, USA). The transcript levels of all samples in each qPCR assay were normalized against the sample with the lowest RQ within the same assay.
Statistical Analyses
For statistical analysis of the qPCR relative transcript levels in all the tested genes, the RQ data was first logarithmically transformed. The transformed data were then analyzed using one-way ANOVA, followed by a post hoc Dunnett test. For Hi-IAG and the tested novel, yet uncharacterized, male gene, the quantification in different stages was compared to the expression in the male stage. For Hi-Vg and the tested novel, yet uncharacterized, female gene, the quantification at different stages was compared to the expression in the mature female stage. Statistical analyses were performed using Statistica v13.3 software (StatSoft Ltd., Tulsa, OK, United States).
Results
H. inermis RNA Library and Representative Sex-Specific Differentially Expressed Genes
De novo assembly of all 2,458,778,342 clean reads yielded filtered transcriptomes with 244,523 and 261,562 contigs in H. inermis eye and body, respectively. Total contig length was 319,146,505 and 349,094,053 bp in the eye and body transcriptomes, respectively. The sequencing depth was 485x and 713x with contig average lengths of 1,305.18 and 1,334.65 bp and N50 values of 2,382 and 2,467 bp in the eye and body transcriptomic libraries, respectively. Following the DESeq2 analysis, totals of 11,821 and 32,317 genes were found to be differentially expressed (FDR adjusted p-value < 0.05 and absolute linear fold change > 1.3) between the reproductive stages (immature, male, young female and mature female) in the body (Figure 3A and Supplementary Table 2) and eye (Figure 3B and Supplementary Table 3), respectively. Alignment of Pnp-IAG to the body transcriptome revealed one transcript (Hippolyte_Body_TRINITY_DN25817_c0_g1; see line 2937 in the “MF vs. M” sheet in Supplementary Table 2) that was found to contain the IAG sequence in H. inermis (Hi-IAG; GenBank accession number: MZ222390) and was annotated as such. Further analysis of the genes differentially expressed between males and mature females in the body transcriptome yielded 11 transcripts that were annotated as vitellogenin. Further analysis revealed that homologs for the five longest transcripts in other decapods could be other copies of vitellogenin as two of them matched to vitellogenin, other two to vitellogenin 2 and one to vitellogenin-like gene. Among them, the transcript with the highest absolute linear fold change (Hippolyte_Body_TRINITY_DN538_c0_g1, Linear FC = 172; see line 7 in the “MF vs. M” sheet in Supplementary Table 2 and Supplementary Figure 1) was defined as Hi-Vg (GenBank accession number: MZ222391).
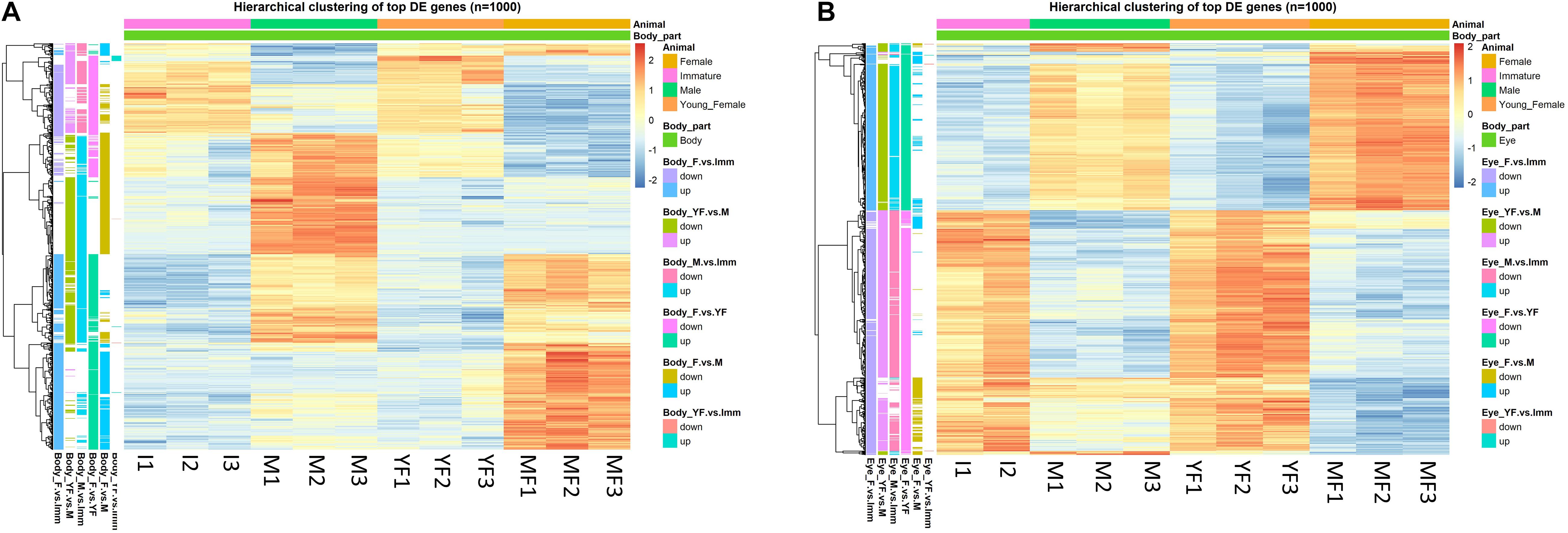
Figure 3. Hierarchical clustering of the top 1000 differentially expressed (DE) genes (lowest p-values) at different reproductive stages of Hippolyte inermis. Heatmaps represent the intensity of variation in gene expression between immature animals (I), males (M), young females (YF) and mature females (MF) in (A) body and (B) eyes. Three pooled samples were used for each reproductive stage in each tissue, except the eyes in immature animals for which two pooled samples were used. A list of all DE genes, including the top 1000 presented in the heatmaps, can be found in Supplementary Tables S2, S3 for the body and eyes, respectively.
Binary Patterning Analysis, GO Enrichment Analysis and Novel Sex-Specific Genes
The binary pattern analysis of the differentially expressed genes in the body (Figure 4 and Table 2) yielded 6,175 significant genes, of which 949 passed the counts cut-off threshold (i.e., the normalized counts in all replicate samples was larger than 500). Similar analysis of the differentially expressed genes in the eye (Figure 5 and Table 2) yielded 19,605 significant genes, of which 2,665 passed the counts cut-off threshold. In the body transcriptome, 819 genes were male specific (0100 pattern); of those, 277 were annotated. Finally, 992 genes were female specific (0001 pattern), and of those, 470 were annotated. In the eye transcriptome, 386 genes were male specific (0100, with 123 being annotated), while 1,368 genes were female specific (0001), with 595 being annotated. In addition, 41 (13 annotated) and 44 (21 annotated) immature-specific genes (1000 pattern) were found in the body and the eye transcriptomes, respectively.
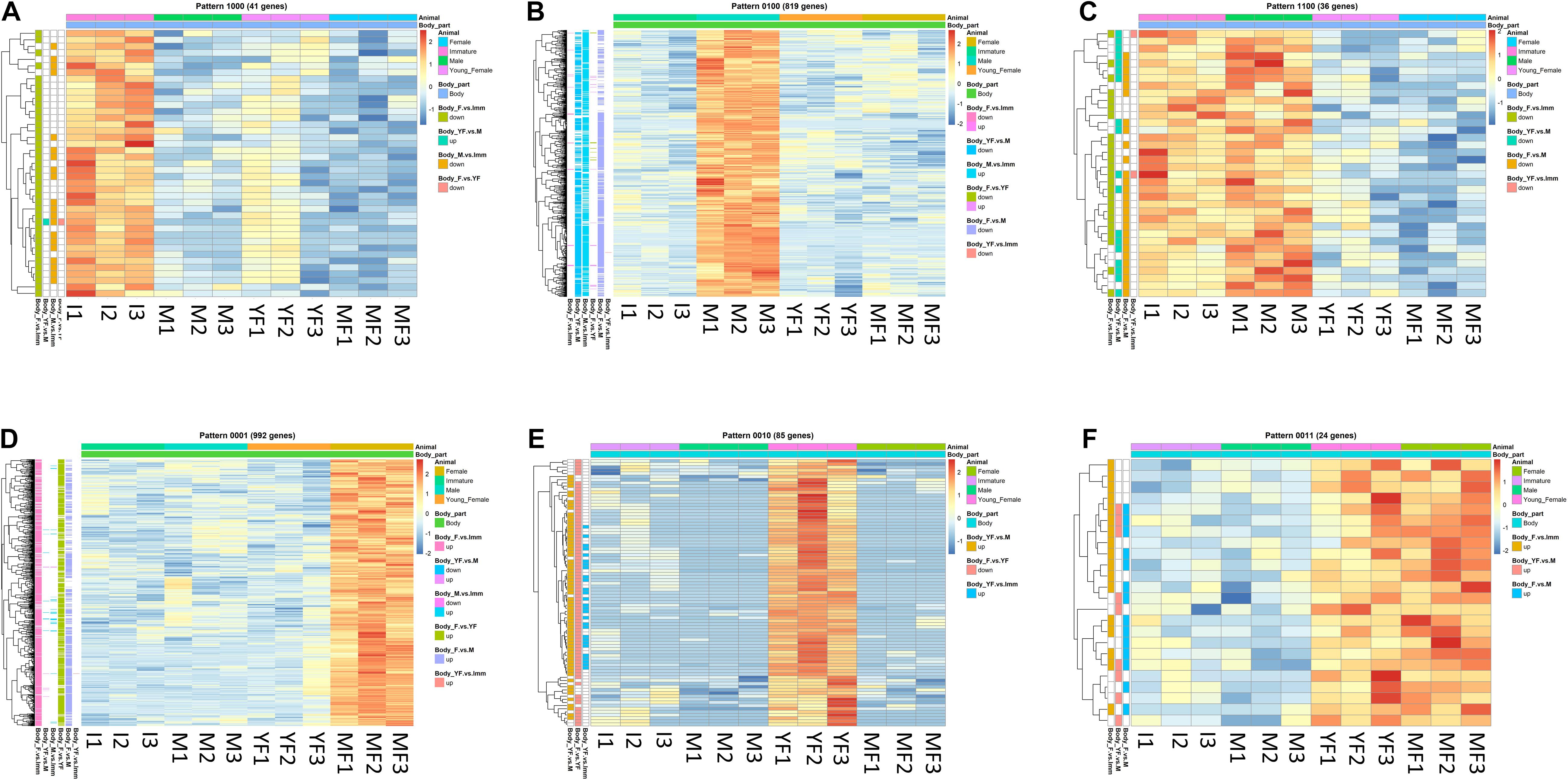
Figure 4. Typical binary patterns of expression in the transcriptomic library of the Hippolyte inermis body. Genes that were differentially expressed (DE; FDR P < 0.05, linear fold change < −1.3 or > 1.3) between any stages were unified and subjected to binary pattern analysis in which 1 = high expression and 0 = low expression. (A) 1000 - immature specific pattern. (B) 0100 - male specific pattern. (C) 1100 - immature and male specific pattern. (D) 0001 - mature female specific pattern. (E) 0010 - young female specific pattern. (F) 0011 - young female and mature female specific pattern. I = immature, M = male, YF = young female, MF = mature female. A list of all DE genes that matched any binary pattern can be found in Supplementary Table 1.
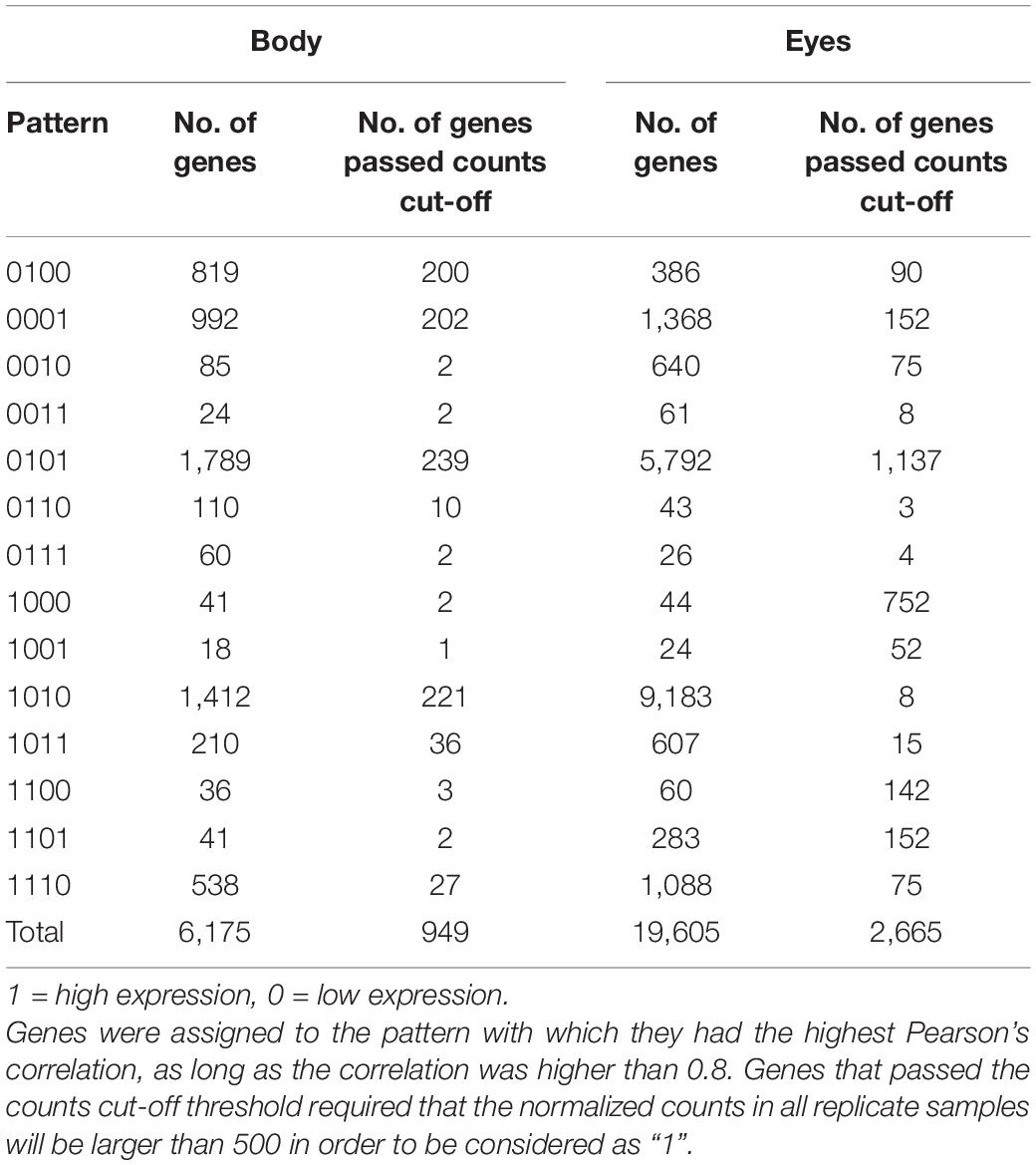
Table 2. Quantity of differentially expressed genes (FDR P < 0.05, linear fold change < −1.3 or > 1.3) between any stages that matched each binary pattern.
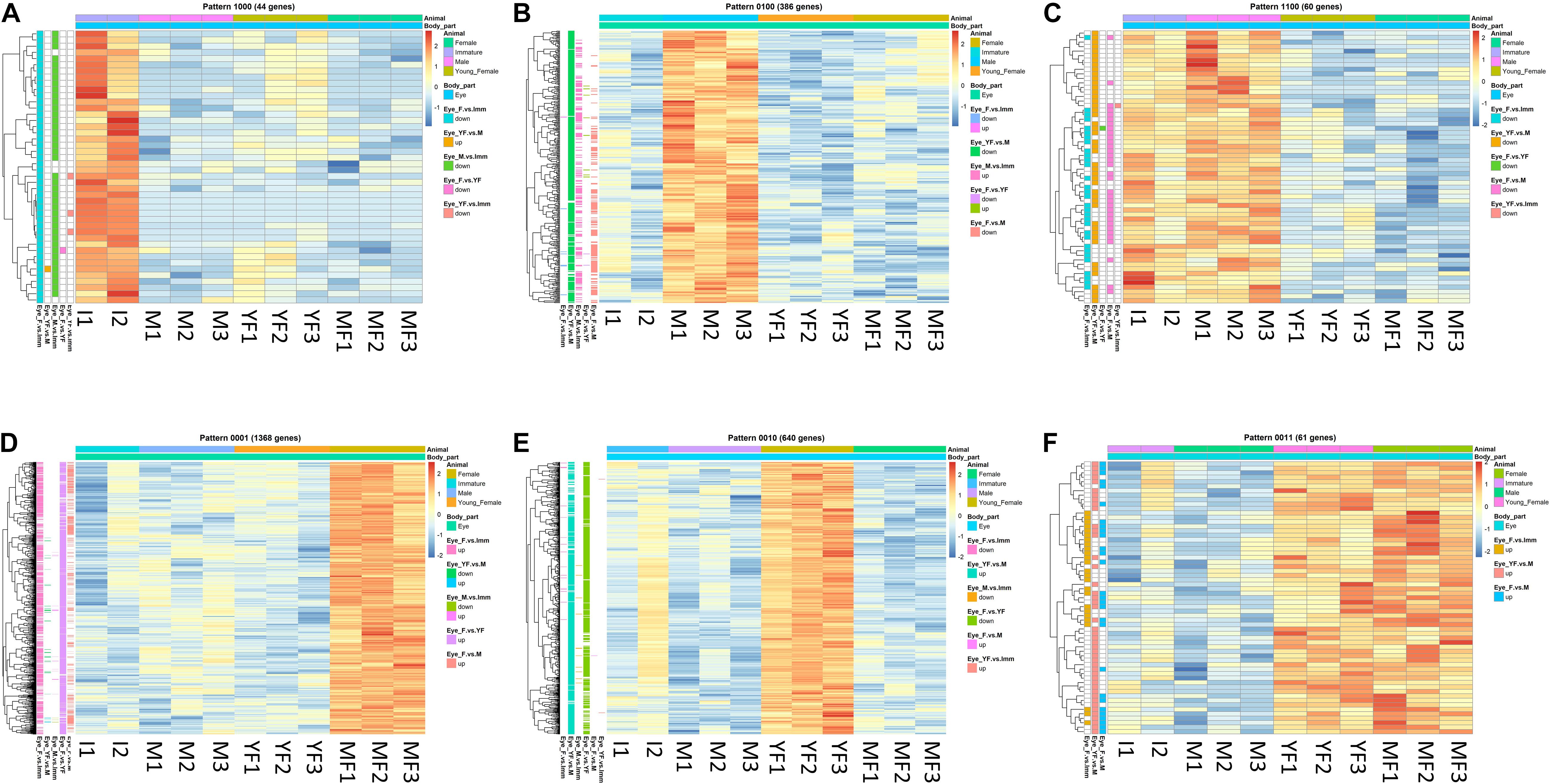
Figure 5. Typical binary patterns of expression in the transcriptomic library of the Hippolyte inermis eye. Genes that were differentially expressed (DE; FDR P < 0.05, linear fold change < –1.3 or > 1.3) between any stages were unified and subjected to binary pattern analysis in which 1 = high expression and 0 = low expression. (A) 1,000 - immature specific pattern. (B) 0100 - male specific pattern. (C) 1100 - immature and male specific pattern. (D) 0001 - mature female specific pattern. (E) 0010 - young female specific pattern. (F) 0011 - young female and mature female specific pattern. I = immature, M = male, YF = young female, MF = mature female. A list of all DE genes that matched any binary pattern can be found in Supplementary Table S1.
GO enrichment analysis of the differentially expressed genes in the body transcriptome (Supplementary Table 4) yielded 56 and 70 terms characterized as molecular function (MF) and biological process (BP), respectively, and in the eye transcriptome (Supplementary Table 5), 127 and 191 terms characterized as MF and BP, respectively.
Mining for novel sex-specific genes by focusing on unannotated female-specific genes with a binary pattern of 0001 and male-specific genes with a binary pattern of 0100 that had passed the counts cut-off threshold yielded numerous novel sex-specific genes. Among the novel female-specific genes in the body, a representative unannotated transcript with a clear protein-coding region (Hippolyte_Body_TRINITY_DN8088_c0_g2, Linear FC = 68.9; see line 20 in the “MF vs. M” sheet in Supplementary Table 2) was defined as “Uncharacterized female gene” (Hi-UCF; GenBank accession number: MZ222394). While no predicted domains of the Hi-UCF putative protein were found according to SMART (see Text Footnote 2; (Schultz et al., 2000)), the first 24 amino acids of the predicted protein were found to code a signal peptide according to SignalP-5.0 Server (Almagro Armenteros et al., 2019). Among the novel male-specific genes in the body, a representative unannotated transcript with a clear coding region (Hippolyte_Body_TRINITY_DN5725_c1_g1, Linear FC = −135; see line 2936 in the “MF vs M” sheet in Supplementary Table 2) was defined as “Uncharacterized male gene” (Hi-UCM; GenBank accession number: MZ222393). However, no conserved domains were found within the Hi-UCM putative protein sequence, and the probability of the protein containing a signal peptide was found to be low. In the eye transcriptome, representative unannotated male-specific (Hi-UCMe; GenBank accession number: MZ995266) and female-specific (Hi-UCFe; GenBank accession number: MZ995265) were defined as “Uncharacterized male eye gene” and “Uncharacterized female eye gene”, respectively. Both Hi-UCMe (Hippolyte_Eye_TRINITY_DN22603_c0_g1, Linear FC = −3.09; see line 2066 in the “MF vs M” sheet in Supplementary Table 3) and Hi-UCFe (Hippolyte_Eye_TRINITY_DN39764_c0_g1, Linear FC = 4.19; see line 579 in the “MF vs. M” sheet in Supplementary Table 3) had a clear protein-coding region but only Hi-UCMe contained a signal peptide. The full nucleotide and amino acid sequences of Hi-UCF, Hi-UCM, Hi-UCFe and Hi-UCMe are given in Data S1 and the identified body- and eye-specific genes in the joint reference transcriptome are presented in Supplementary Table 6.
AG and IAG in H. inermis
A dorsoventral histological section of an H. inermis male revealed the AG, as in other crustaceans (Charniaux-Cotton, 1962), at the base of the fifth pereiopod, adjacent to the sperm duct. The AG appeared to consist of dense nucleated cells (Figure 6A). The Hi-IAG mRNA sequence (Figure 6B) included fragments of 50, 426 and 419 bp for the 5′ UTR, ORF and 3′ UTR, respectively. The deduced structure of the Hi-IAG hormone, according to the predicted ORF, was found to contain a signal peptide (19 aa), a B chain (42 aa), an A chain (38 aa), and a C peptide (42 aa).
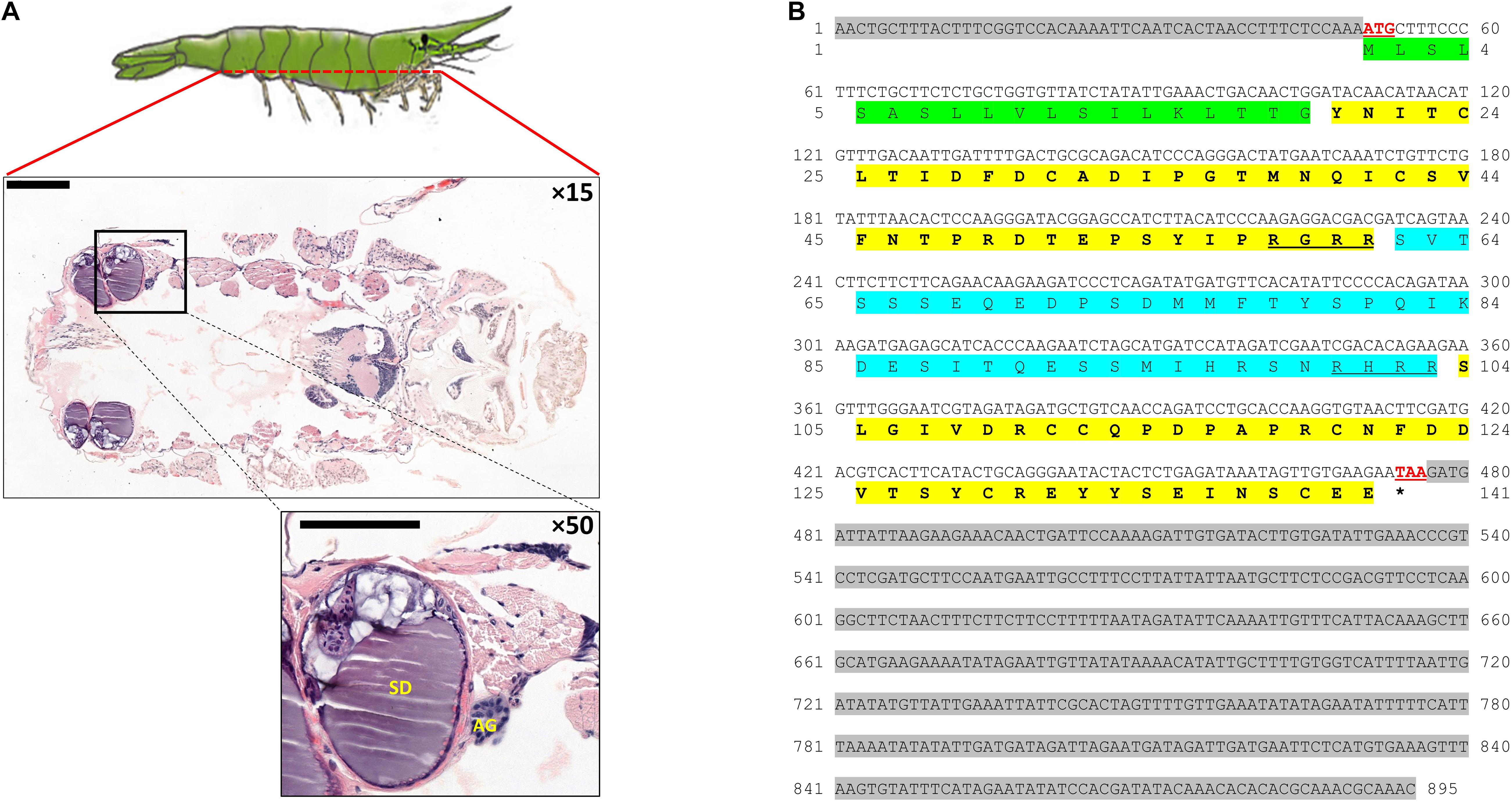
Figure 6. AG and IAG in Hippolyte inermis. (A) Dorsoventral histological section (× 15; top) of a H. inermis male stained with hematoxylin and eosin. The base of the fifth pereiopod is marked out with a black frame in which the androgenic gland (AG) and sperm duct (SD) are visible (× 50; bottom). Bar scales (200 μm) are given. (B) The full sequence of Hi-IAG mRNA and its open reading frame (ORF)-deduced amino acids. The signal peptide is highlighted in green. B chain (first) and A chain (second) are highlighted in bold on a yellow background, with C peptide (blue background), including its cleavage sites (underlined), flanked between them. The start (ATG) and stop (TAA) codons are shown in red and are underlined. The stop codon is also indicated with an asterisk. 5′ (top) and 3′ (bottom) UTRs are highlighted with a gray background.
In silico and in vitro Expression of Sex-Specific Genes Throughout the H. inermis Life Cycle and Their Homologs in P. platyceros
As expected, Hi-IAG and Hi-UCM relative transcript levels (Figure 7 – left panel) were found to be significantly different between the stages sampled in this study (ANOVA, F(3, 8) = 17.775 and F(3, 8) = 18.346, P < 0.05). Hi-Vg and Hi-UCF relative transcript levels (Figure 8 – left panel) were also found to be significantly different between the stages sampled in this study (ANOVA, F(3, 8) = 45.099 and F(3, 8) = 57.704, P < 0.05). More specifically, according to the post hoc Dunnett test, Hi-IAG and Hi-UCM transcript levels were significantly higher in the male stage compared to the immature and female stages, while Hi-Vg and Hi-UCF levels were significantly higher in the mature female stage compared to the immature, male and young female stages (P < 0.05). It is noteworthy that all qPCR results regarding relative transcript levels were consistent with the normalized read counts of the tested genes acquired from the in silico analyses of the body transcriptome (left and middle panels of Figures 7, 8).
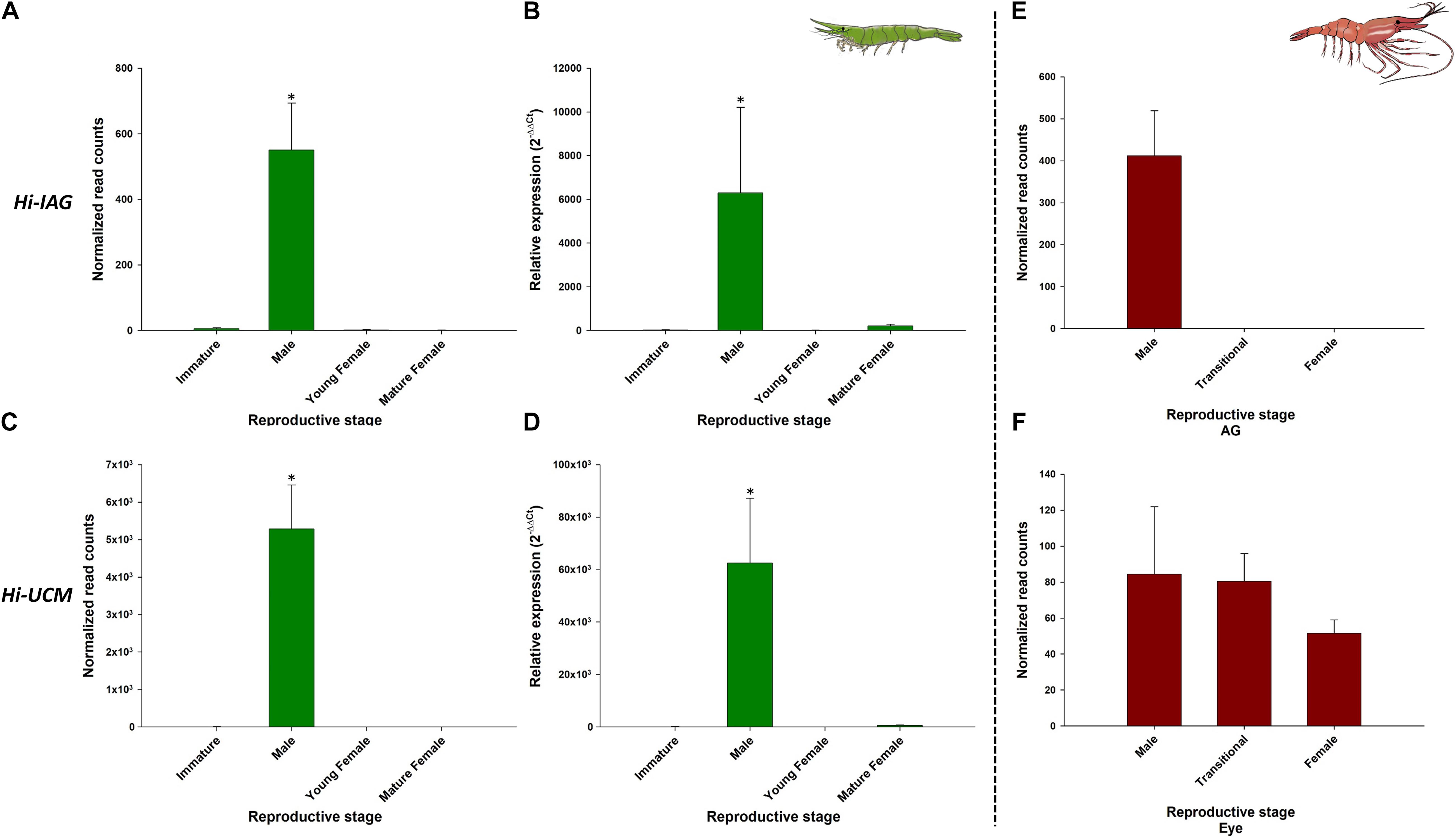
Figure 7. Representative male-related genes in Hippolyte inermis and their homologs in Pandalus platyceros. Left panel - Male related genes in different stages of H. inermis: immature (n = 3), male (n = 3), young female (n = 3) and mature female (n = 3). (A) Normalized read counts of Hi-IAG. (B) Relative expression of Hi-IAG. (C) Normalized read counts of Hi-UCM. (D) Relative expression of Hi-UCM. Right panel – Hi-IAG and Hi-UCM homologs in different stages of P. platyceros: male (n = 2), transitional (n = 2) and female (n = 2). (E) Normalized read counts of Pnp-IAG. (F) Normalized read counts of a Hi-UCM homolog. Read counts of the genes in P. platyceros were obtained from Levy et al. (2020a). Error bars represent standard error of the means. *P ≤ 0.05.
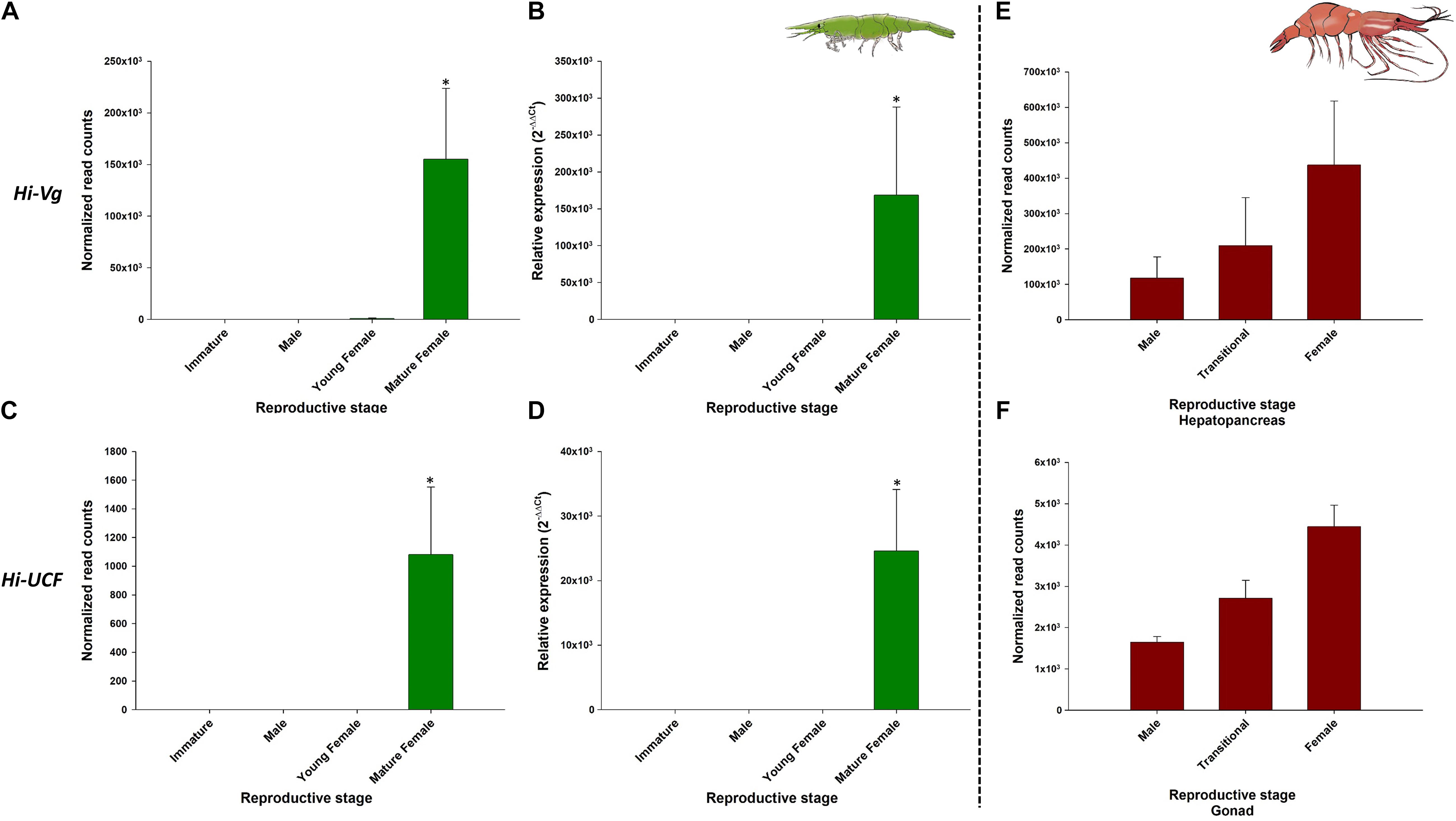
Figure 8. Representative female-related genes in Hippolyte inermis and their homologs in Pandalus platyceros. Left panel - Female-related genes in different stages of H. inermis: immature (n = 3), male (n = 3), young female (n = 3) and mature female (n = 3). (A) Normalized read counts of Hi-Vg. (B) Relative expression of Hi-Vg. (C) Normalized read counts of Hi-UCF. (D) Relative expression of Hi-UCF. Right panel – Hi-Vg and Hi-UCF homologs in different stages of P. platyceros: male (n = 2), transitional (n = 2) and female (n = 2). (E) Normalized read counts of Pnp-Vg. (F) Normalized read counts of a Hi-UCF homolog. Read counts of the genes in P. platyceros were obtained from Levy et al. (2020a). Error bars represent standard error of the means. *P ≤ 0.05.
The comparative analysis of the above H. inermis genes with the P. platyceros transcriptome revealed that the Hi-IAG homolog (Pnp-IAG) is exclusively expressed in the AG of P. platyceros males, while the Hi-UCM homolog is expressed in the P. platyceros eye with the highest transcript level in males, with the level decreasing as the animal transforms toward the female stage (Figure 7 – right panel). In contrast, the Hi-Vg homolog (Pnp-Vg) is expressed in the hepatopancreas of P. platyceros with a relatively low transcription level in males, with the level increasing as the animal transforms toward femaleness. The Hi-UCF homolog was found to be expressed in the gonad of P. platyceros with the highest transcript levels in females compared to male and transitional stages (Figure 8 – right panel). Furthermore, according to the blastx and tblasn results against the TSA database, Hi-UCF protein homolog sequences were found in 21 other decapod species, including prawns and shrimps belonging to the families Palaemonidae, Lysmatidae, Alpheidae, Alvinocarididae, Atyidae, and Penaeidae, while the homolog sequence for Hi-UCM was found in 8 other decapod species, including prawns, shrimps and a crab from the families Palaemonidae, Lysmatidae, Alpheidae, and Portunidae (Table 3). All corresponding sequences to Hi-IAG, Hi-Vg, Hi-UCF and Hi-UCM in P. playceros are given in Data S2. Moreover, homolog sequence for Hi-UCFe was found in 26 other decapods including prawns, shrimps, crayfish, lobsters and crabs from the families Portunidae, Penaeidae, Cancridae, Nephropidae, Lithodidae, Varunidae, Palaemonidae, Atyidae, Grapsidae, Astacidae, Gecarcinidae and Cambaridae while homolog sequence for Hi-UCMe was found in seven other shrimps and prawns from the families Palaemonidae and Lysmatidae (Supplementary Figure 2).

Table 3. Homolog sequences of Hi-UCM and Hi-UCF in other decapod species found by blastx and tblastn in the NCBI server.
As per the IAG-switch-like protein from P. platyceros, only IAG-switch-like protein 1, 4, 5 and 6 corresponded to homologs in the H. inermis body transcriptome. Among them, only the homolog for IAG-switch-like 1 was male-specific (Hippolyte_Body_TRINITY_DN12510_c0_g1, Linear FC = −2.4; see line 1376 in the “MF vs. M” sheet in Supplementary Table 2) while the rest were not sexually biased (Supplementary Table 7).
Discussion
In gonochoristic species, sexual differentiation processes are confined to early developmental stages – from embryogenesis to early post-larvae – while in hermaphrodite species they also occur during adult stages (Levy et al., 2018; Benvenuto and Weeks, 2020; Levy and Sagi, 2020). Indeed, in the present study, thousands of genes were recorded as differentially expressed and sexually biased during the life cycle of the protandric shrimp H. inermis, and homologs to representative genes were also found in P. platyceros and other decapod crustaceans. While the gene list generated in this study includes previously known key genes, such as the male-specific IAG and the female-specific Vg genes, which were used as reference genes, Hi-UCM, Hi-UCF, Hi-UCMe and Hi-UCFe were just four, among numerous novel (not yet annotated) candidate genes from the body and eye transcriptomes of H. inermis, that appear to be promising for further studies.
Hi-Vg, which was used as a female reference gene had, as expected, a transcriptional pattern that was negligible in H. inermis immature individuals and males but increased when the younger shrimp began to mature as females, similar to other protandric shrimps, such as P. platyceros (Levy et al., 2020b) and P. hypsinotus (Tsutsui et al., 2004; Okumura et al., 2005). In these two Pandalus species there is a clear transitional stage, in which vitellogenin gene and protein levels are intermediate between those during maleness and those during femaleness. In contrast, in H. inermis the transitional stage is absent (Cobos et al., 2005; Zupo and Messina, 2007; Mutalipassi et al., 2018), and consequently the Hi-Vg level begins to rise at the young female stage and a sharp increase is observed in the mature female stage, as shown in this study. Also, the fact that 11 transcripts were annotated as vitellogenin in the H. inermis body transcriptome and some of them corresponded to vitellogenin 2 or vitellogenin-like genes supports the previous evidence that multiple copies of vitellogenin may be found in decapods (Zhao et al., 2021). In the male stage, the transcriptional pattern of Hi-IAG, which controls male differentiation and served as a male reference gene in H. inermis, differed from the pattern in P. platyceros: In the latter species, the IAG transcript level was sixfold higher in juveniles than in males (Levy et al., 2020a), but in H. inermis the IAG transcript level was negligible in immature animals and high in males.
The above findings could be explained by the differences in the lengths of the maturation period from juveniles to males: in H. inermis, this period lasts only about a month and is contained within a single molt cycle (Reverberi, 1950; Zupo et al., 2008), whereas in P. platyceros the maturation period lasts at least three years and thus extends over several molt cycles (Butler, 1965; King and Moffitt, 1984; Iversen et al., 1993; Kimker et al., 1996; Levy et al., 2020b). The very short maturation period in H. inermis makes it easy to miss the immature stage during which the IAG transcript level begins to rise. In addition, for this reason, AMs may be absent in immature stages of H. inermis, because sex maturation is a process expressed and contained in a very short period—a character unique to H. inermis among the crustaceans. Indeed, taken together, the absence of the AM in the immature H. inermis samples collected in the present study and the presence of a small AM in the juvenile P. platyceros samples collected in a previous study (Levy et al., 2020a) may imply that the immature H. inermis individuals in this study were collected at a very early stage before the development of the AG, compared to the juvenile P. platyceros in which the development of the AG already begun. Also, the fact that H. inermis homologs to four out of the six conserved IAG-switch like proteins that were previously described in P. platyceros, one of them with the same male-specific pattern, supports the claim that IAG-switch related genes are conserved among the Crustacea (Levy et al., 2020a).
While the IAG and Vg genes in H. inermis are reported here for the first time, the vast potential of the transcriptomic approach presented in this study is exemplified by the discovery of four representative novel transcripts, two male-specific and two female-specific, out of numerous potential candidates from the H. inermis body and eye transcriptomes that were found to be highly conserved among decapods. This is supported by the fact that the transcriptional pattern of Hi-UCF in the H. inermis body is positively correlated with the Hi-Vg transcript. Moreover, the in silico transcript level of the Hi-UCF homolog in the P. platyceros gonad (low in males and then increasing along the transition to the female stage) had similar transcription pattern to the Vg gene in the hepatopancreas of P. platyceros. However, although Vg is a prominent glycolipoprotein that is expressed in some decapods in both the hepatopancreas and the ovary of females (Okumura et al., 2007; Bai et al., 2015) and the Hi-UCF homolog in P. platyceros is expressed in the ovary and correlated with Vg transcription in the hepatopancreas, Hi-UCF is clearly different from Vg. Hi-UCF thus could be part of the vitellogenin toolkit or a controlling element in the vitellogenesis process.
A homolog to the representative body male-specific gene in H. inermis, Hi-UCM, was also expressed in the transcriptome of P. platyceros (Levy et al., 2020a), exclusively in the eye, with the expression being the highest in males and transitionals and decreasing as the animals transformed towards femaleness. Generally, a novel sex-specific gene found to be exclusively expressed in crustacean eyes could potentially be a sex-controlling-related candidate gene, since the X-organ-sinus gland complex of the eyestalk in crustaceans produces numerous neurohormones that regulate key physiological processes, including molting, reproduction, and even AG development (Webster and Keller, 1986; Keller, 1992; Aguilar et al., 1996; Khalaila et al., 2002; Chang and Mykles, 2011; Katayama et al., 2013; Li et al., 2015; Pitts et al., 2017). However, unlike the behavior of its homolog in P. platyceros, Hi-UCM transcript levels were negligible in all stages sampled for the H. inermis eye transcriptome. Thus, Hi-UCM might be produced in the thoracic ganglia (TG), a component of the central nervous system (CNS), found in the crustacean body (Suwansa-Ard et al., 2015; Nguyen et al., 2018) rather than in the eyestalk. But, since Hi-UCM was not annotated as one of the well-known neurohormones that modulate reproduction and development [e.g., gonad-inhibiting hormone (GIH)/vitellogenesis-inhibiting hormone (VIH), the molt-inhibiting hormone (MIH), the crustacean hyperglycemic hormone (CHH) and the mandibular organ inhibiting hormone (MOIH) (Webster and Keller, 1986; Keller, 1992; Aguilar et al., 1996; Khalaila et al., 2002; Chang and Mykles, 2011; Katayama et al., 2013; Li et al., 2015; Pitts et al., 2017)], it might be a part of the genomic male-differentiation toolkit, upstream or downstream to the IAG-switch (which governs the activity of such hormones) rather than being a neurohormone per se.
To summarize, crustacean sexual development and differentiation and their control have largely been studied in gonochoristic species and, within these species, mostly in species of global importance to the growing aquaculture industry. However, to study genes that control sex-differentiation in such gonochoristic species, one must investigate the animals at very early developmental stages, which is not always easy and is sometimes impossible. According to the data generated from this study (i.e., a long list of novel sex-specific unannotated genes), the main finding of this study is that the solution to this problem may lie in investigating protandric species, because in these species the expression of genes related to the sexual differentiation mechanism are not limited to early developmental stages. Also, as exemplified here, it is likely that candidate genes found in protandric species (with or without intermediate stages) are conserved among crustaceans. Hence, a gene could be discovered in a hermaphrodite model species and then studied further in gonochoristic species. Eventually, the strategy adopted successfully in the current study, expands the debate over protandric life histories, previously based solely on histological evidence, by involving molecular tools utilizing protandric species to uncover parts of the crustacean sex-differentiation puzzle.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found within the article or below: NCBI (accession: PRJNA736313).
Author Contributions
This study was conceived and designed by TL, VZ, MC, AS, and EDA. EDA, VZ, MM, and ES collected the animals for the study. VZ, EDA, and MM dissected the animals, collected biometric data, and fixed individuals for further extractions. NR and MC extracted the RNA for the libraries. AS, EDA, MC, and VZ reviewed and analyzed the data. TL performed the in vitro analysis. TL, SA, and RM performed the in silico analyses. VC-C performed all bioinformatics analyses. All authors analyzed and interpreted the data. The manuscript was written by TL and reviewed and approved by all co-authors.
Funding
This study was generously supported by the Israel Ministry of Science and Technology, grant no. 3-15151 under the Italy-Israel collaboration program, in cooperation with the Italian Ministry for Foreign Affairs (Research Project Excites) and by grant no. 2015073 from the United States-Israel Binational Science Foundation (BSF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work is dedicated to the memory of our dear student and friend SA. We would like to thank Olabiyi Obayomi for his bioinformatics work related to this project. Samples were collected by Cpt. V. Rando on board the vessel Phoenicia of the Stazione Zoologica Anton Dohrn. Rearing of live samples was possible thanks to the technical assistance of A. Macina and D. Caramiello, personnel of the MARE unit of Stazione Zoologica.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.745540/full#supplementary-material
Supplementary Figure 1 | Vitellogenin in Hippolyte inermis. (A) The five longest transcripts that were annotated as vitellogenin/vitellogenin-like genes in decapods. The gene ID in the H. inermis body transcriptome is given along with the length of the transcript, best blastx hit and accession number. (B) The full sequence of Hi-Vg mRNA and its open reading frame (ORF)-deduced amino acids. The signal peptide is highlighted in green. Lipoprotein N-terminal Domain is highlighted in bold on a yellow background, DUF1943 is highlighted in bold on a blue background, DUF1081 is highlighted in bold on a pink background and von Willebrand factor (vWF) type D domain is highlighted in bold on a red background. The start (ATG) and stop (TAA) codons are shown in red and are underlined. The stop codon is also indicated with an asterisk. 5′ (top) and 3′ (bottom) UTRs are highlighted with a gray background. The predicted domains of the putative protein were inferred from its deduced amino acids sequence using SMART (http://SMART.embl-heidelberg.de).
Supplementary Figure 2 | Uncharacterized sex-specific genes in the eye Hippolyte inermis. (A) Homolog sequences of Hi-UCMe and Hi-UCFe in other decapod species found by blastx and tblastn in the NCBI server. Accession numbers of the homolog sequences from each species are given for each gene ID. (B) Normalized read counts of (Left) Male-related gene (Hi-UCMe) and (Right) Female-related gene (Hi-UCFe) in different stages of H. inermis: immature (n = 2), male (n = 3), young female (n = 3) and mature female (n = 3). Error bars represent standard error of the means. ∗P ≤ 0.05.
Supplementary Table 1 | Differentially expressed transcripts that were assigned a binary pattern in H. inermis body and eye transcriptomes. Trinotate and Blast2GO annotations are given.
Supplementary Table 2 | Differentially expressed transcripts between every possible contrast (I vs. M, I vs. YF, I vs. MF, M vs. YF, M vs. MF and YF vs. MF) in the H. inermis body transcriptome. Read counts in each sample in each stage, linear fold change, pattern (upregulation/downregulation), Trinotate and Blast2GO annotations are given.
Supplementary Table 3 | Differentially expressed transcripts between every possible contrast (I vs. M, I vs. YF, I vs. MF, M vs. YF, M vs. MF and YF vs. MF) in the H. inermis eye transcriptome. Read counts in each sample in each stage, linear fold change, pattern (upregulation/downregulation), Trinotate and Blast2GO annotations are given.
Supplementary Table 4 | Gene Ontology (GO) enrichment analysis of the differentially expressed genes between every possible contrast (I vs. M, I vs. YF, I vs. MF, YF vs. M, MF vs. M and YF vs. MF) in the H. inermis body transcriptome. The genes that were assigned to each term within the molecular function (MF; yellow) and biological process (BP; blue) aspects are indicated along with the fold enrichment and FDR adjusted p-values for each GO term.
Supplementary Table 5 | Gene Ontology (GO) enrichment analysis of the differentially expressed genes between every possible contrast (I vs. M, I vs. YF, I vs. MF, YF vs. M, MF vs. M and YF vs. MF) in the H. inermis eye transcriptome. The genes that were assigned to each term within the molecular function (MF; yellow) and biological process (BP; blue) aspects are indicated along with the fold enrichment and FDR adjusted p-values for each GO term.
Supplementary Table 6 | Body and eye-specific genes found in the joint reference transcriptome. Read counts in each stage (I, M, YF and MF) as well as Trinotate annotations are given.
Supplementary Table 7 | H. inermis homologs to IAG-switch-like proteins in P. platyceros. GenBank accession numbers of the genes in P. platyceros as well as the corresponding transcripts in the H. inermis body transcriptome are given.
Supplementary Data 1 | The full nucleotide (red) and amino acid (black) sequences of Hi-UCF, Hi-UCM, Hi-UCFe and Hi-UCMe.
Supplementary Data 2 | The full nucleotide corresponding sequences of Hi-IAG, Hi-Vg, Hi-UCF and Hi-UCM in P. playceros.
Footnotes
References
Abdel-Moneim, A., Coulter, D. P., Mahapatra, C. T., and Sepulveda, M. S. (2015). Intersex in fishes and amphibians: population implications, prevalence, mechanisms and molecular biomarkers. J. Appl. Toxicol. 35, 1228–1240. doi: 10.1002/jat.3204
Abehsera, S., Glazer, L., Tynyakov, J., Plaschkes, I., Chalifa-Caspi, V., Khalaila, I., et al. (2015). Binary gene expression patterning of the molt cycle: the case of chitin metabolism. PLoS One 10:e0122602. doi: 10.1371/journal.pone.0122602
Aguilar, M. B., Falchetto, R., Shabanowitz, J., Hunt, D. F., and Huberman, A. (1996). Complete primary structure of the molt-inhibiting hormone (MIH) of the Mexican crayfish Procambarus bouvieri (Ortmann). Peptides 17, 367–374. doi: 10.1016/0196-9781(96)00010-1
Almagro Armenteros, J. J., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Baeza, J. A. (2007). Sex allocation in a simultaneously hermaphroditic marine shrimp. Evolution 61, 2360–2373. doi: 10.1111/j.1558-5646.2007.00199.x
Bai, H. K., Qiao, H., Li, F. J., Fu, H. T., Sun, S. M., Zhang, W. Y., et al. (2015). Molecular characterization and developmental expression of vitellogenin in the oriental river prawn Macrobrachium nipponense and the effects of RNA interference and eyestalk ablation on ovarian maturation. Gene 562, 22–31. doi: 10.1016/j.gene.2014.12.008
Bauer, R. T., and Holt, G. J. (1998). Simultaneous hermaphroditism in the marine shrimp Lysmata wurdemanni (Caridea : Hippolytidae): an undescribed sexual system in the decapod Crustacea. Mar. Biol. 132, 223–235. doi: 10.1007/s002270050388
Benvenuto, C., and Weeks, S. C. (2020). Hermaphroditism and gonochorism. Nat. Hist. Crustacea Reproduct. Biol. 6:197.
Butler, T. H. (1964). Growth, reproduction, and distribution of pandalid shrimps in British Columbia. J. Fish. Board Canada 21, 1403–1452.
Butler, T. H. (1965). Synopsis of Biological Data on The Prawn Pandalus Platyceros Brandt, 1851. Rome: Food and Agriculture Organization of the United Nations.
Chang, E. S., and Mykles, D. L. (2011). Regulation of crustacean molting: a review and our perspectives. Gen. Comp. Endocrinol. 172, 323–330. doi: 10.1016/j.ygcen.2011.04.003
Charniaux-Cotton, H. (1958). Contrôle hormonal de la différenciation du sexe et de la reproduction chez les Crustacés supérieurs. Bull. Soc. Zool. 82, 314–336.
Charniaux-Cotton, H. (1962). Androgenic gland of crustaceans. Gen. Comp. Endocrinol. Suppl. 1, 241–247.
Christoffersen, M. L. (1990). A new superfamily classification of the Caridea (Crustacea: Pleocyemata) based on phylogenetic pattern. Z. Fur Zool. Syst. Evol. 28, 94–106.
Cobos, V., Diaz, V., Raso, G., Enrique, J., and Manjon-Cabeza, M. E. (2005). Insights on the female reproductive system in Hippolyte inermis (Decapoda, Caridea): is this species really hermaphroditic? Invertebr. Biol. 124, 310–320. doi: 10.1111/j.1744-7410.2005.00029.x
de Almeida, A. O., and Buckup, L. (2000). Occurrence of protandric hermaphroditism in a population of the neotropical freshwater crayfish Parastacus brasiliensis (Parastacidae). J. Crustacean Biol. 20, 224–230.
Ford, A. T. (2012). Intersexuality in Crustacea: an environmental issue? Aquat. Toxicol. 108, 125–129. doi: 10.1016/j.aquatox.2011.08.016
Ford, A. T., Fernandes, T. F., Rider, S. A., Read, P. A., Robinson, C. D., and Davies, I. M. (2003). Measuring sublethal impacts of pollution on reproductive output of marine Crustacea. Mar. Ecol. Prog. Ser. 265, 303–309. doi: 10.3354/meps265303
Ford, A. T., Fernandes, T. F., Rider, S. A., Read, P. A., Robinson, C. D., and Davies, I. M. (2004). Endocrine disruption in a marine amphipod? Field observations of intersexuality and de-masculinisation. Mar. Environ. Res. 58, 169–173. doi: 10.1016/j.marenvres.2004.03.013
Gertz, E. M., Yu, Y. K., Agarwala, R., Schaffer, A. A., and Altschul, S. F. (2006). Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4:41. doi: 10.1186/1741-7007-4-41
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hayes, T. B., Khoury, V., Narayan, A., Nazir, M., Park, A., Brown, T., et al. (2010). Atrazine induces complete feminization and chemical castration in male African clawed frogs (Xenopus laevis). Proc. Natl. Acad. Sci. U.S.A. 107, 4612–4617. doi: 10.1073/pnas.0909519107
Hoffman, D. L. (1968). Seasonal eyestalk inhibition on androgenic glands of a protandric shrimp. Nature 218, 170–187. doi: 10.1038/218170a0
Hoffman, D. L. (1972). Development of ovotestis and copulatory organs in a population of protandric shrimp, Pandalus platyceros Brandt from Lopez Sound, Washington. Biol. Bull. 142, 251–269. doi: 10.2307/1540229
Iversen, E. S., Allen, D. M., and Higman, J. B. (1993). Shrimp Capture and Culture Fisheries of the United States. New York, NY: Halsted Press.
Juchault, P. (1999). Hermaphroditism and gonochorism. A new hypothesis on the evolution of sexuality in Crustacea. Comptes Rendus l’Academie Sci. Ser. III Sci. de la Vie 322, 423–427. doi: 10.1016/S0764-4469(99)80078-X
Katayama, H., Ohira, T., and Nagasawa, H. (2013). Crustacean peptide hormones: structure, gene expression and function. Aqua Biosci. Monogr. 6, 49–90.
Keller, R. (1992). Crustacean neuropeptides - structures, functions and comparative aspects. Experientia 48, 439–448. doi: 10.1007/Bf01928162
Khalaila, I., Manor, R., Weil, S., Granot, Y., Keller, R., and Sagi, A. (2002). The eyestalk-androgenic gland-testis endocrine axis in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 127, 147–156.
Kimker, A., Donaldson, W., and Bechtol, W. R. (1996). Spot shrimp growth in Unakwik Inlet, Prince William Sound, Alaska. Alaska Fish. Res. Bull. 3, 1–8.
King, M. G., and Moffitt, R. B. (1984). The sexuality of tropical deep-water shrimps (Decapoda: Pandalidae). J. Crustacean Biol. 4, 567–571. doi: 10.2307/1548071
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Levy, T., Aflalo, E. D., and Sagi, A. (2018). “Sex control in cultured decapod crustaceans,” in Sex Control in Aquaculture, eds F. Piferrer, Z.-G. Shen, H. Wang, and S. Chen (Hoboken, NJ: John Wiley & Sons), 689–704.
Levy, T., Rosen, O., Eilam, B., Azulay, D., Aflalo, E. D., Manor, R., et al. (2016). A single injection of hypertrophied androgenic gland cells produces all-female aquaculture. Mar. Biotechnol. 18, 554–563. doi: 10.1007/s10126-016-9717-5
Levy, T., and Sagi, A. (2020). The ‘IAG-switch’ - a key controlling element in decapod crustacean sex differentiation. Front. Endocrinol. 11:651. doi: 10.3389/fendo.2020.00651
Levy, T., Ventura, T., De Leo, G., Grinshpan, N., Amterat Abu Abayed, F., Manor, R., et al. (2020c). Two homogametic genotypes - one crayfish: on the consequences of intersexuality. iScience 23:101652. doi: 10.1016/j.isci.2020.101652
Levy, T., Tamone, S. L., Manor, R., Aflalo, E. D., Sklarz, M. Y., Chalifa-Caspi, V., et al. (2020a). The IAG-switch and further transcriptomic insights into sexual differentiation of a protandric shrimp. Front. Mar. Sci. 7:587454. doi: 10.3389/fmars.2020.587454
Levy, T., Tamone, S. L., Manor, R., Bower, E. D., and Sagi, A. (2020b). The protandric life history of the Northern spot shrimp Pandalus platyceros: molecular insights and implications for fishery management. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-58262-6
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, F., Bai, H., Zhang, W., Fu, H., Jiang, F., Liang, G., et al. (2015). Cloning of genomic sequences of three crustacean hyperglycemic hormone superfamily genes and elucidation of their roles of regulating insulin-like androgenic gland hormone gene. Gene 561, 68–75. doi: 10.1016/j.gene.2015.02.012
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint]. doi: 10.6084/M9.FIGSHARE.963153.V1
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Martin, J. W., Crandall, K. A., and Felder, D. L. (2009). Decapod Crustacean Phylogenetics. Boca Raton: CRC Press.
Martin, P., Kohlmann, K., and Scholtz, G. (2007). The parthenogenetic Marmorkrebs (marbled crayfish) produces genetically uniform offspring. Naturwissenschaften 94, 843–846. doi: 10.1007/s00114-007-0260-0
Mutalipassi, M., Maibam, C., and Zupo, V. (2018). The sex change of the caridean shrimp Hippolyte inermis Leach: temporal development of the gonopore morphology. Zoomorphology 137, 377–388. doi: 10.1007/s00435-018-0405-z
Nagamine, C., Knight, A. W., Maggenti, A., and Paxman, G. (1980a). Effects of androgenic gland ablation on male primary and secondary sexual characteristics in the Malaysian prawn, Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae), with first evidence of induced feminization in a nonhermaphroditic decapod. Gen. Comp. Endocrinol. 41, 423–441. doi: 10.1016/0016-6480(80)90048-9
Nagamine, C., Knight, A. W., Maggenti, A., and Paxman, G. (1980b). Masculinization of female Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae) by androgenic gland implantation. Gen. Comp. Endocrinol. 41, 442–457. doi: 10.1016/0016-6480(80)90049-0
Nguyen, T. V., Rotllant, G. E., Cummins, S. F., Elizur, A., and Ventura, T. (2018). Insights into sexual maturation and reproduction in the Norway lobster (Nephrops norvegicus) via in silico prediction and characterization of neuropeptides and G protein-coupled receptors. Front. Endocrinol. 9:430. doi: 10.3389/fendo.2018.00430
Okumura, T., Nikaido, H., Yoshida, K., Kotaniguchi, M., Tsuno, Y., Seto, Y., et al. (2005). Changes in gonadal development, androgenic gland cell structure, and hemolymph vitellogenin levels during male phase and sex change in laboratory-maintained protandric shrimp, Pandalus hypsinotus (Crustacea : Caridea : Pandalidae). Mar. Biol. 148, 347–361. doi: 10.1007/s00227-005-0073-7
Okumura, T., Yamano, K., and Sakiyama, K. (2007). Vitellogenin gene expression and hemolymph vitellogenin during vitellogenesis, final maturation, and oviposition in female kuruma prawn, Marsupenaeus japonicus. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 147, 1028–1037. doi: 10.1016/j.cbpa.2007.03.011
Olmstead, A. W., and LeBlanc, G. A. (2000). Effects of endocrine-active chemicals on the development of sex characteristics of Daphnia magna. Environ. Toxicol. Chem. 19, 2107–2113. doi: 10.1002/etc.5620190821
Pitts, N. L., Schulz, H. M., Oatman, S. R., and Mykles, D. L. (2017). Elevated expression of neuropeptide signaling genes in the eyestalk ganglia and Y-organ of Gecarcinus lateralis individuals that are refractory to molt induction. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 214, 66–78. doi: 10.1016/j.cbpa.2017.09.011
Quackenbush, L. S. (1989). Vitellogenesis in the shrimp, Penaeus vannamei: in vitro studies of the isolated hepatopancreas and ovary. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 94, 253–261. doi: 10.1016/0305-0491(89)90342-8
Reverberi, G. (1950). La situazione sessuale di Hyppolyte viridis e le condizioni che la reggono. Boll. Zool. 17, 91–94.
Rodriguez, E. M., Lopez Greco, L. S., and Fingerman, M. (2000). Inhibition of ovarian growth by cadmium in the fiddler crab, Uca pugilator (Decapoda, ocypodidae). Ecotoxicol. Environ. Saf. 46, 202–206. doi: 10.1006/eesa.1999.1896
Rodriguez, E. M., Medesani, D. A., and Fingerman, M. (2007). Endocrine disruption in crustaceans due to pollutants: a review. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 146, 661–671. doi: 10.1016/j.cbpa.2006.04.030
Sagi, A., Khalaila, I., Barki, A., Hulata, G., and Karplus, I. (1996). Intersex red claw crayfish, Cherax quadricarinatus (von Martens): functional males with pre-vitellogenic ovaries. Biol. Bull. 190, 16–23. doi: 10.2307/1542672
Scholtz, G., Braband, A., Tolley, L., Reimann, A., Mittmann, B., Lukhaup, C., et al. (2003). Ecology: parthenogenesis in an outsider crayfish. Nature 421, 806–808. doi: 10.1038/421806a
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P., and Bork, P. (2000). SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. doi: 10.1093/nar/28.1.231
Shaked, S. A., Abehsera, S., Levy, T., Chalifa-Caspi, V., and Sagi, A. (2020). From sporadic single genes to a broader transcriptomic approach: insights into the formation of the biomineralized exoskeleton in decapod crustaceans. J. Struct. Biol. 212:107612. doi: 10.1016/j.jsb.2020.107612
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212.
Sklarz, M. Y., Levin, L., Gordon, M., and Chalifa-Caspi, V. (2018). Neatseq-flow# lightweight high throughput sequencing workflow platform for non-programmers and programmers alike. bioRxiv [Preprint]. doi: 10.1101/173005v3
Stentiford, G. D. (2012). Histological intersex (ovotestis) in the European lobster Homarus gammarus and a commentary on its potential mechanistic basis. Dis. Aquat. Organ. 100, 185–190. doi: 10.3354/dao02455
Subramoniam, T. (1981). Protandric hermaphroditism in a mole crab, Emerita asiatica (Decapoda: Anomura). Biol. Bull. 160, 161–174. doi: 10.2307/1540910
Suwansa-Ard, S., Thongbuakaew, T., Wang, T., Zhao, M., Elizur, A., Hanna, P. J., et al. (2015). In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of eyestalk, central nervous system and ovary. PLoS One 10:e0123848. doi: 10.1371/journal.pone.0123848
Tombes, A. S., and Foster, M. W. (1979). Growth of appendix masculina and appendix interna in juvenile Macrobrachium rosenbergii (De Man)(Decapoda, Caridea). Crustaceana 5, 179–184.
Tsukimura, B. (2001). Crustacean vitellogenesis: its role in oocyte development. Am. Zool. 41, 465–476.
Tsutsui, N., Saido-Sakanaka, H., Yang, W. J., Jayasankar, V., Jasmani, S., Okuno, A., et al. (2004). Molecular characterization of a cDNA encoding vitellogenin in the coonstriped shrimp, Pandalus hypsinotus and site of vitellogenin mRNA expression. J. Exp. Zoolog. Part A Comp. Exp. Biol. 301, 802–814. doi: 10.1002/jez.a.53
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Rosen, O., and Sagi, A. (2012). Timing sexual differentiation: full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 86, 1–6. doi: 10.1095/biolreprod.111.097261
Webster, S. G., and Keller, R. (1986). Purification, characterization and amino-acid-composition of the putative molt-inhibiting hormone (MIH) of Carcinus maenas (Crustacea, Decapoda). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 156, 617–624. doi: 10.1007/Bf00692738
Wilder, M. N., Okumura, T., Suzuki, Y., Fusetani, N., and Aida, K. (1994). Vitellogenin production induced by eyestalk ablation in juvenile giant freshwater prawn Macrobrachium rosenbergii and trial methyl farnesoate administration. Zool. Sci. 11, 45–53.
Wolfe, J. M., Breinholt, J. W., Crandall, K. A., Lemmon, A. R., Lemmon, E. M., Timm, L. E., et al. (2019). A phylogenomic framework, evolutionary timeline and genomic resources for comparative studies of decapod crustaceans. Proc. R. Soc. B Biol. Sci. 286:20190079. doi: 10.1098/rspb.2019.0079
Yaldwyn, J. C. (1966). Protandrous hermaphroditism in decapod prawns of the families Hippolytidae and Campylonotidae. Nature 209:1366.
Yano, I., and Chinzei, Y. (1987). Ovary is the site of vitellogenin synthesis in kuruma prawn, Penaeus japonicus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 86, 213–218. doi: 10.1016/0305-0491(87)90280-X
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhao, J., Wang, W., Wang, C., Shi, L., Wang, G., Sun, C., et al. (2021). The presence of multiple copies of the vitellogenin gene in Fenneropenaeus merguiensis (De Man, 1888)(Decapoda: Dendrobranchiata: Penaeidae): evidence for gene expansion and functional diversification in shrimps. J. Crustacean Biol. 41:ruaa100. doi: 10.1093/jcbiol/ruaa100
Zupo, V. (1994). Strategies of sexual inversion in Hippolyte inermis Leach (Crustacea, Decapoda) from a Mediterranean seagrass meadow. J. Exp. Mar. Biol. Ecol. 178, 131–145. doi: 10.1016/0022-0981(94)90229-1
Zupo, V. (2000). Effect of microalgal food on the sex reversal of Hippolyte inermis (Crustacea : Decapoda). Mar. Ecol. Prog. Ser. 201, 251–259. doi: 10.3354/meps201251
Zupo, V., and Messina, P. (2007). How do dietary diatoms cause the sex reversal of the shrimp Hippolyte inermis Leach (Crustacea, Decapoda). Mar. Biol. 151, 907–917.
Zupo, V., Messina, P., Buttino, I., Sagi, A., Avila, C., Nappo, M., et al. (2007). Do benthic and planktonic diatoms produce equivalent effects in crustaceans? Mar. Freshw. Behav. Physiol. 40, 169–181. doi: 10.1080/10236240701592930
Keywords: androgenic gland, hermaphrodite, Hippolyte inermis, IAG-switch, Pandalus platyceros, protandry, reproductive physiology, sex-differentiation
Citation: Levy T, Zupo V, Mutalipassi M, Somma E, Ruocco N, Costantini M, Abehsera S, Manor R, Chalifa-Caspi V, Sagi A and Aflalo ED (2021) Protandric Transcriptomes to Uncover Parts of the Crustacean Sex-Differentiation Puzzle. Front. Mar. Sci. 8:745540. doi: 10.3389/fmars.2021.745540
Received: 22 July 2021; Accepted: 21 September 2021;
Published: 08 October 2021.
Edited by:
Valerio Matozzo, University of Padua, ItalyReviewed by:
Tereza Manousaki, Hellenic Centre for Marine Research (HCMR), GreeceShihao Li, Institute of Oceanology, Chinese Academy of Sciences (CAS), China
Naoaki Tsutsui, Mie University, Japan
Copyright © 2021 Levy, Zupo, Mutalipassi, Somma, Ruocco, Costantini, Abehsera, Manor, Chalifa-Caspi, Sagi and Aflalo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valerio Zupo, dnp1cG9Ac3puLml0; Amir Sagi, c2FnaWFAYmd1LmFjLmls; Eliahu D. Aflalo, YWZsYWxvZUBiZ3UuYWMuaWw=
†ORCID: Tom Levy, orcid.org/0000-0003-1484-0310; Amir Sagi, orcid.org/0000-0002-4229-1059; Eliahu D. Aflalo, orcid.org/0000-0003-1385-2387
 Tom Levy
Tom Levy Valerio Zupo
Valerio Zupo Mirko Mutalipassi2
Mirko Mutalipassi2 Emanuele Somma
Emanuele Somma Vered Chalifa-Caspi
Vered Chalifa-Caspi Amir Sagi
Amir Sagi Eliahu D. Aflalo
Eliahu D. Aflalo