- 1School of Environment and Science, Griffith University, Nathan, QLD, Australia
- 2Coastal and Marine Research Centre, Griffith University, Gold Coast, QLD, Australia
- 3Australian Rivers Institute, Griffith University, Nathan, QLD, Australia
- 4Griffith Institute for Drug Discovery, Griffith University, Nathan, QLD, Australia
Many pigments, such as melanins, are widely distributed throughout the animal kingdom. Others have arisen as novelties in particular lineages, for example, the Green Fluorescent Protein (GFP) found in cnidarians. While GFPs, widely used as fluorescent tags in biomedical research, are the most famous cnidarian example, other novel proteins have also been identified within this phylum. A blue protein that contains a Kringle (KR) domain inserted within a Frizzled cysteine-rich domain (Fz-CRD) was previously described from the jellyfish Rhizostoma pulmo (named rpulFKz1), however little is known about this pigment’s evolution or distribution among cnidarians. We performed a systematic search for homologs of this protein in published genomes and transcriptomes of 93 cnidarians. Phylogenetic analyses revealed eight predicted proteins that possess both domains in the same arrangement and that fall within the same clade as rpulFKz1. The sequence of one of these proteins contains motifs that match sequenced peptides of Cassio Blue, the blue pigment from Cassiopea xamachana. Another one of these proteins belongs to Stomolophus meleagris, and chemical studies on blue pigments that may occur in this genus have shown similarities to rpulFKz1 and Cassio Blue. Therefore, we hypothesize that the eight rpulFKz1 homologs identified are also pigment precursors. All precursors identified were exclusive to jellyfish in the order Rhizostomeae, so we herein name this new pigment family “rhizostomins.” Not all rhizostomes analyzed are blue, however, so these rhizostomin proteins may also be responsible for other colors, or perform other biochemical and biophysical roles. Previous studies have hypothesized that cnidarian pigments are photoprotective, and this study serves as basis for future investigations not only on the function of rhizostomins, but also on potential biotechnological applications for these proteins.
Introduction
Few features of any organism draw more attention than their color. Color can play an important role in an organism’s survival, providing camouflage and photoprotection, and influencing sexual selection and social interactions (Cuthill et al., 2017). Color can be generated either from physical structures through selective reflection from surfaces or from pigments. Pigments are molecules that selectively absorb light and can either be naturally produced by the organism or acquired by ingestion (Hendry, 1996). Some common types of pigments in animals are carotenoids, melanins, tetrapyrroles and quinones (Hendry, 1996; Bandaranayake, 2006). These pigments can serve a wide range of biophysical and biochemical functions (for an extensive review see Needham, 1974). Research on pigments has led not only to understanding the ecological implications and evolutionary history of color development but also to practical applications. Natural pigments are used as colorants in various textile and cosmetic products, as well as for foods and beverages that benefit from their low toxicity compared to synthetic pigments (Venil et al., 2013). Biotechnology has been revolutionized by Green Fluorescent Proteins (GFP), a family of protein pigments first identified in the cnidarian Aequorea victoria (Murbach and Shaerer, 1902) (Shimomura et al., 1962), and used as fluorescent tags in various in vivo labeling and detection methods (Matz et al., 2002), which led to a Nobel Prize in Chemistry in 2008.
Cnidarian coloration is associated with numerous pigment families, including the GFPs; carotenoids, such as the blue carotenoproteins in Velella velella (Linnaeus, 1758) (Herring, 1971) and the opsin-like pigments in box jellyfish eyes (Martin, 2004); tetrapyrroles, such as the red-brown porphyrins in some deep sea scyphomedusae (Herring, 1972); and melanins, responsible for the magenta and brown color in Pelagia noctiluca (Forsskål, 1775) (Fox and Millott, 1954). More recently, a blue protein was described from the jellyfish Rhizostoma pulmo (Macri, 1778) (Bulina et al., 2004). Blue pigments are relatively uncommon in nature as these compounds require a complex structure to absorb red light (Newsome et al., 2014), and there is a growing need to find more stable blue pigments for industrial purposes (Santos and Bicas, 2021). The blue pigment isolated from the bell margin of R. pulmo was initially hypothesized to be a carotenoid (Christomanos, 1954), but later found to consist of a novel protein, named rpulFKz1, that possesses Frizzled cysteine-rich (Fz-CRD) and Kringle (KR) domains (Bulina et al., 2004). These protein domains had not been previously associated with pigmentation in any organism.
Other studies of blue pigments in jellyfish, such as Cassio Blue in Cassiopea xamachana Bigelow, 1892 (Phelan et al., 2006) and those in Stomolophus spp. such as S2bp (Lugo-Magaña et al., 2018; Martínez-Pérez et al., 2020), have revealed that they have similar spectroscopic and physicochemical properties to rpulFKz1. Phelan et al. (2006) found various transition metals associated with Cassio Blue, indicating that it may serve as a ligand for metal complexation. Cassio Blue may also serve a photoprotective function in these animals, considering its light absorption properties and that it occurs in tissues that have high densities of algal endosymbionts (zooxanthellae, i.e. dinoflagellates of the family Symbiodiniaceae, LaJeunesse et al., 2018), as it could selectively filter harmful ultraviolet radiation while transmitting photosynthetically active wavelengths (Blanquet and Phelan, 1987). This photoprotective function has also been suggested for pigments in other cnidarians such as corals, which express UV-screening GFP-like proteins (Gittins et al., 2015), and for blue carotenoprotein pigments in neustonic animals (Herring, 1965) that may protect them from photooxidative damage (Sommer et al., 2006).
The protein domains that occur in rpulFKz1 (Fz-CRD and KR) are common but usually occur in separate proteins with very different functions. Members of the Frizzled protein family (Fz) are membrane receptors for a variety of signal transduction pathways, such as Wnt, and have a vital role in developmental processes related to cell proliferation, cell fate determination and nervous system development (Huang and Klein, 2004). Secreted Frizzled-related proteins (SFRP) also play a role in Wnt pathways as extracellular modulators (Jones and Jomary, 2002). The Fz-CRD is a ligand binding component within Fz and SFRP proteins, but may also be present in other proteins such as Smoothened (Smo), muscle-skeletal receptor tyrosine kinases (MuSK) and receptor tyrosine kinase-like orphan receptors (ROR) (Yan et al., 2014). KR domains mediate protein binding and commonly occur in proteins involved with blood coagulation and fibrinolysis, such as serine proteases and plasminogens (Patthy et al., 1984), but also in apolipoprotein(a) (Anglés-Cano and Rojas, 2002) and hepatocyte growth factors (Nakamura et al., 1989). Human plasminogen KRs, for example, serve as binding sites for fibrins, other plasma proteins, and a variety of cell types and cell-associated proteins (Chang et al., 1998). Fz-CRD and KR domains have only been reported together within a protein in the extracellular domain of RORs, and these receptors play a critical role in developmental processes linked to Wnt pathways (Green et al., 2008). In cnidarians, Wnt has been associated with patterning of the body axes (Guder et al., 2006; Watanabe et al., 2014; Sanders and Cartwright, 2015), although Wnt signaling pathways may play a role in pigment expression in other animals (Squarzoni et al., 2011; Zhu et al., 2016).
Considering the similarities found between rpulFKz1, Cassio Blue and Stomolophus spp. pigments, we hypothesize that they are part of a novel and unexplored pigment family that commonly appears blue. To explore the presence of potential pigment precursors homologous to rpulFKz1 across cnidarians, we performed a systematic search of Fz-CRD and KR domains and conducted phylogenetic analyses to investigate their relationships. We also analyzed arrangement, amino acid composition and structure of proteins that contained these domains. This gives further insight into the expression and function of these pigments, and provides a basis to understand the evolution of pigments in cnidarians and explore their potential industrial applications.
Materials and Methods
Publicly available protein sequences predicted from whole genomes of cnidarians were downloaded and this dataset was supplemented by available transcriptomes. In total, 93 cnidarian species were included, encompassing the major orders or suborders from all classes in this phylum (Supplementary Table 1). Sequence reads were downloaded from NCBI SRA, TSA, or Protein databases, and if necessary, assembled de novo using Trinity v. 2.4.0 (Haas et al., 2013) with default parameters and enabling trimmomatic (Bolger et al., 2014), jaccard clip, and without normalization of reads. CD-Hit v. 4.6.8 (Fu et al., 2012) was used to cluster highly similar sequences and reduce redundancy, and sequences were translated where required with TransDecoder (within the Trinity suite), with a minimum peptide length of 50 amino acids, retaining only the single best open-reading frame per transcript.
Predicted proteins were searched for cysteine-rich Frizzled (Fz-CRD), Kringle (KR), and Green Fluorescent Protein (GFP) domains using hmmsearch within HMMER v. 3.1 (Eddy, 1998) based on each domain’s HMM profile from Pfam (Finn et al., 2016) (accessions PF01392, PF00051, and PF01353, respectively). Sequences were further aligned within each domain using hmmalign. Identical sequences were removed and the remaining submitted to BLASTP (NCBI), from which the best hit (determined by E-value) was used to indicate possible protein annotation. Alignments were manually trimmed based on each domain’s HMM logo within Geneious (Kearse et al., 2012). Sequences with less than five amino acids and gaps that were present in more than 80% of sequences were removed. Phylogenetic analyses were performed in IQTREE v. 1.6.12 (Nguyen et al., 2015) under maximum likelihood as the optimality criterion, using ModelFinder (Kalyaanamoorthy et al., 2017) for model selection and measuring ultrafast bootstrap resampling frequencies (Hoang et al., 2018) and SH-aLRT (Guindon et al., 2010) from 1,000 pseudoreplicates. Resulting trees were visualized and edited in FigTree v. 1.4.4.1
Sequences with both Fz-CRD and KR protein domains that fell within the clade of the Rhizostoma pulmo pigment precursor, rpulFKz1 (GenBank accession AAS83462), were investigated further. SignalP v. 5.0 (Almagro Armenteros et al., 2019) was used to predict signal sequences, NetNGlyc v. 1.0 (Gupta et al., 2004) to predict N-glycosylation sites and TMHMM v. 2.0 (Krogh et al., 2001) to predict transmembrane helices. I-TASSER (Yang et al., 2015) was used to predict 3D structure and potential ligand binding sites. To explore previously published peptide data, PeptideCutter (Gasteiger et al., 2005) was used to predict protease cleavage sites and ProteoMapper (Mendoza et al., 2018) was used to map peptide digests to predicted proteins. These sequences were further aligned using MAAFT (Katoh and Standley, 2013). A sequence logo was created for the cnidarian Fz-CRD and KR alignments using WebLogo (Crooks et al., 2004) and matched with the equivalent regions in the sequence alignment that included rpulFKz1, which was manually edited for exact correspondence. Important sites for Wnt-binding and conserved N-glycosilation sites in the Fz-CRD were identified based on Dann et al. (2001) and Yan et al. (2014). Ligand binding sites in the KR domain were identified based on Thewes et al. (1990) and Chang et al. (1998).
All alignments and phylogenetic trees retrieved were deposited in Figshare (Lawley et al., 2021). Bioinformatic analyses were performed on Griffith University’s High Performance Computer Cluster ‘‘Gowonda,’’ and relevant codes used are available in GitHub.2
Results
Within the genetic data of the cnidarians analyzed, 2,524 predicted proteins contained the Fz-CRD protein domain while 1,114 contained the KR domain (Figures 1, 2; see detailed phylogenetic trees in Supplementary Figures 1, 2 for a complete account of sequences analyzed). Of these, 358 predicted proteins possessed both domains (blue terminals in Figures 1, 2). Predicted proteins with both domains derive from representatives of most orders or suborders of cnidarians, except Spirularia, Narcomedusae, Coronatae, Amyostaurida, and both Endocnidozoa (Myxozoa) orders. Nevertheless, only endocnidozoans lacked predicted proteins with the Fz-CRD and only one species in this group had a predicted protein with the KR domain (Thelohanellus kitauei Egusa and Nakajima, 1981). GFP domains were also searched as candidate pigments, but were not found in any of the scyphozoans analyzed and therefore not considered in any further analyses.
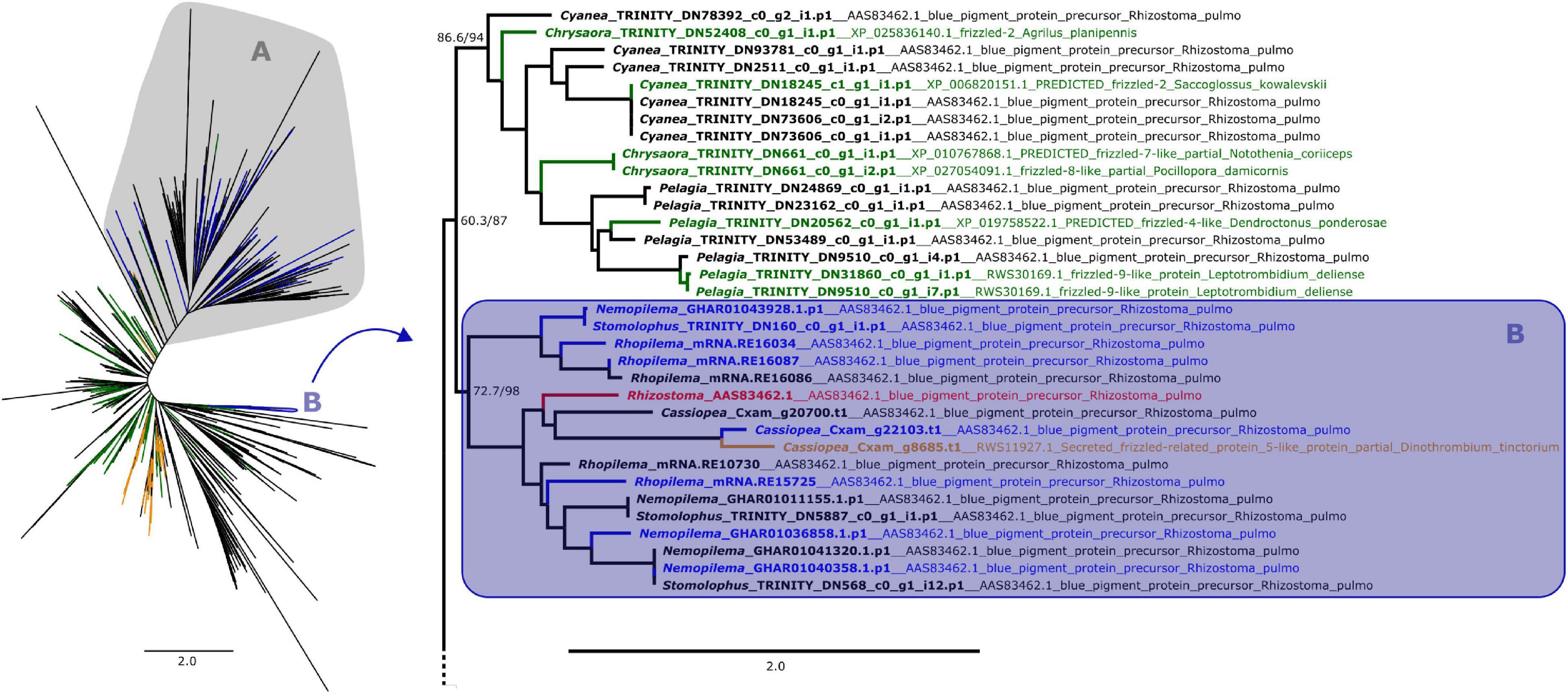
Figure 1. Maximum likelihood phylogenetic relationships between cnidarian Fz-CRD sequences. Green and orange terminals represent sequences that retrieved Fz-like or SFRP-like genes (respectively) as best BLAST hit. Clade A, shown on the left, contains all peptides with ROR receptors as best BLAST hit, which includes most of the predicted proteins that also presented a KR domain (blue terminals). The remaining proteins with both domains are present in clade B (blue terminals), which includes the pigment precursor rpulFKz1 (red terminal). Clade B and its sister clade are shown in detail on the right (as indicated by arrow), with the best BLAST hit for each sequence listed next to the gene identifier. Numbers on nodes represent ultrafast bootstrap resampling frequencies and SH-aLRT. Scale bars represent the number of amino acid substitutions per site. See Supplementary Figure 1 for a detailed Fz-CRD phylogenetic tree.
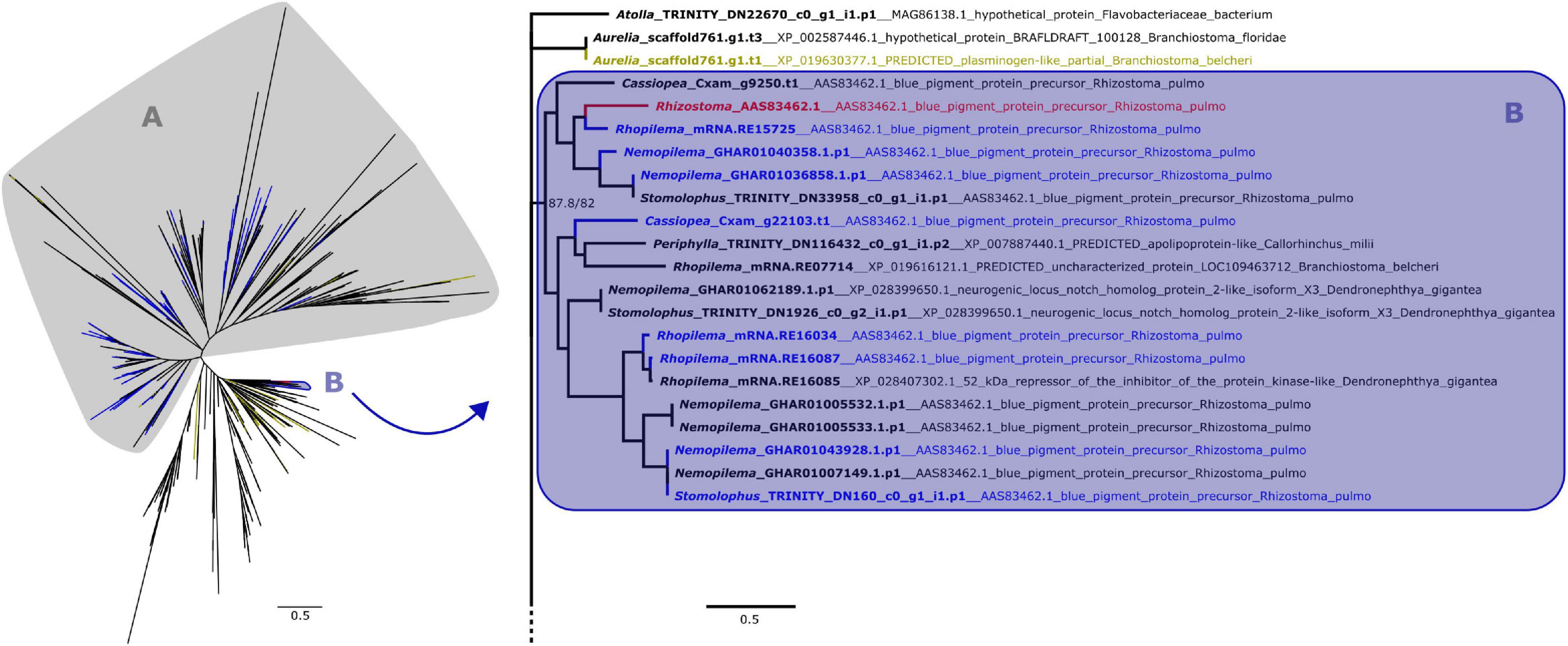
Figure 2. Maximum likelihood phylogenetic relationships between cnidarian KR domain sequences. Yellow terminals represent sequences that retrieved plasminogen-like genes as best BLAST hit. Clade A, shown on the left, contains all peptides with ROR receptors as best BLAST hit, which includes most of the predicted proteins that also presented a Fz-CRD (blue terminals). The remaining proteins with both domains are present in clade B, which includes the pigment precursor rpulFKz1 (red terminal). Clade B and its sister clade are shown in detail on the right (as indicated by arrow), with the best BLAST hit for each sequence listed next to the gene identifier. Numbers on nodes represent ultrafast bootstrap resampling frequencies and SH-aLRT. Scale bars represent the number of amino acid substitutions per site. See Supplementary Figure 2 for a detailed KR phylogenetic tree.
From the 358 predicted proteins with both Fz-CRD and KR domains (blue terminals in Figures 1, 2), 350 are part of a clade that contains all proteins with ROR receptors as best BLAST hit (blue terminals in clade A, Figures 1, 2). The remaining eight are part of a separate clade (blue terminals in clade B, Figures 1, 2) that contains the pigment precursor of Rhizostoma pulmo, rpulFKz1 (red terminal in Figures 1, 2). Each of these eight proteins belong to scyphozoan jellyfishes in the order Rhizostomeae. In both phylogenies there are sequences that did not contain both Fz-CRD and KR domains that fall within this clade (non-blue terminals in clade B, Figures 1, 2), but most still belong to rhizostomes. The only exception to this is a predicted protein that belongs to a coronate scyphozoan, Periphylla periphylla (Péron and Lesueur, 1810), that appears only in the KR phylogeny (clade B, Figure 2). However its sequence contains only 37 of the 76 amino acid positions in the KR alignment, which leads to uncertainty regarding its position in the phylogeny.
In the Fz-CRD phylogeny, all proteins within the clade containing the pigment precursor rpulFKz1 (clade B, Figure 1) return a best BLAST hit to rpulFKz1, except for one of the Cassiopea xamachana sequences that retrieved a SFRP-like gene as best BLAST hit (orange terminal on the right in Figure 1). This clade is most closely related to a clade consisting mainly of predicted proteins with Fz-like genes as their best BLAST hit (green terminals on the right in Figure 1). Outside of the ROR and the pigment precursor’s clades, 78% of the predicted proteins also retrieved Fz-like or SFRP-like genes as best BLAST hit (excluding sequences with uncharacterized and hypothetical proteins as best BLAST hit).
In the KR phylogeny, most proteins in the clade containing rpulFKz1 (clade B, Figure 2) return rpulFKz1 as best BLAST hit, yet the clade also includes proteins with a best BLAST hit to notch-like genes, or, in a single case, an apolipoprotein-like gene. This potential apolipoprotein-like gene is the incomplete scyphozoan (Periphylla periphylla) sequence mentioned above. The clade containing rpulFKz1 is most closely related to Atolla and Aurelia predicted proteins, one of which produces a best BLAST hit against a plasminogen-like gene (yellow terminals on the right in Figure 2). Outside of the ROR and the pigment precursor’s clades, 29.3% of the predicted proteins (the highest proportion, excluding sequences with highest BLAST hits to uncharacterized/hypothetical proteins or Symbiodinium microadriaticum LaJeunesse sequences) retrieved plasminogen-like genes as best BLAST hit (LaJeunesse, 2017).
Alignment of rpulFKz1 with the eight rhizostome Fz-CRD and KR domain-containing homologs revealed that, in all cases, the KR domain seems to be inserted within the Fz-CRD domain (Figure 3). Typical features of Frizzled-related domains are present in most cnidarian Fz-CRD domains (indicated by the sequence logo below alignment, Figure 3), and include 10 conserved cysteine residues (highlighted in yellow) and three Wnt interaction sites (red boxes), of which one contains a highly conserved N-glycosylation site that is important for Wnt binding (highlighted in green). Likewise, most cnidarian KR domains contain the six conserved cysteine residues as well as characteristic ligand binding motifs (black boxes in Figure 3). However, the rhizostome Fz-CRD and KR domain-containing sequences lack several conserved features within the Fz-CRD domain, namely two of the conserved cysteines (one of these is within a Wnt interaction region, position 138, Figure 3) and the predicted N-glycosylation site important for Wnt binding (position 89, Figure 3). Each of the sequences, except Nemopilema_GHAR01040358.p1 (sequence 3, Figure 3) and Rhopilema_mRNA.RE16034 (sequence 6, Figure 3), contained a predicted signal peptide.
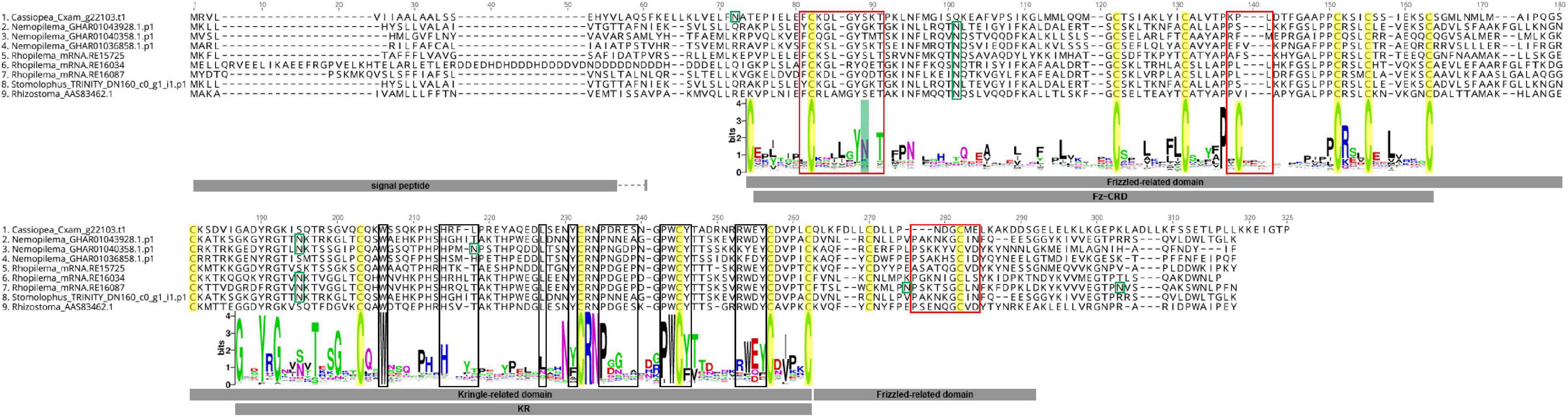
Figure 3. Alignment of rpulFKz1 and homologous rhizostome Fz-CRD and KR domain-containing sequences, showing the insertion of the KR domain within the Fz-CRD (Frizzled-related domains). Red squares indicate regions important for Wnt interaction in Fz-CRD, while black boxes show ligand binding regions in the KR domain. Green boxes show N-glycosylation sites and highlighted in green is a conserved N-glycosylation site that is important for Wnt binding, which is absent in the aligned sequences. Conserved cysteine residues are highlighted in yellow. The sequence logo below the alignment shows the relative frequency of amino acids for each position in the alignment of all cnidarian Fz-CRD and KR domains analyzed. No signal peptides could be predicted for Nemopilema_GHAR01040358.p1 (sequence 3) and Rhopilema_mRNA.RE16034 (sequence 6).
One of the identified rpulFKz1 homologs is from C. xamachana (Figure 3, sequence 1). The alignment of this sequence with C. xamachana Cassio Blue tryptic digests from a previous study (Phelan et al., 2006) revealed that there is an average 78.4% (± 14.4%) match between digest sequences and the protein (Figure 4 and Supplementary Table 2), with digest sequences spanning 3–15 amino acids. Predicted trypsin cleavage sites mostly coincide with ends of tryptic digest sequences (blue squares in Figure 4). When the peptide sequences generated from these Cassio Blue tryptic digests were mapped against the full set of predicted C. xamachana proteins, three mapped to the rpulFKz1 homolog (sequences 8, 10 and 16 in Figure 4); this was the only predicted protein to which more than one peptide mapped.
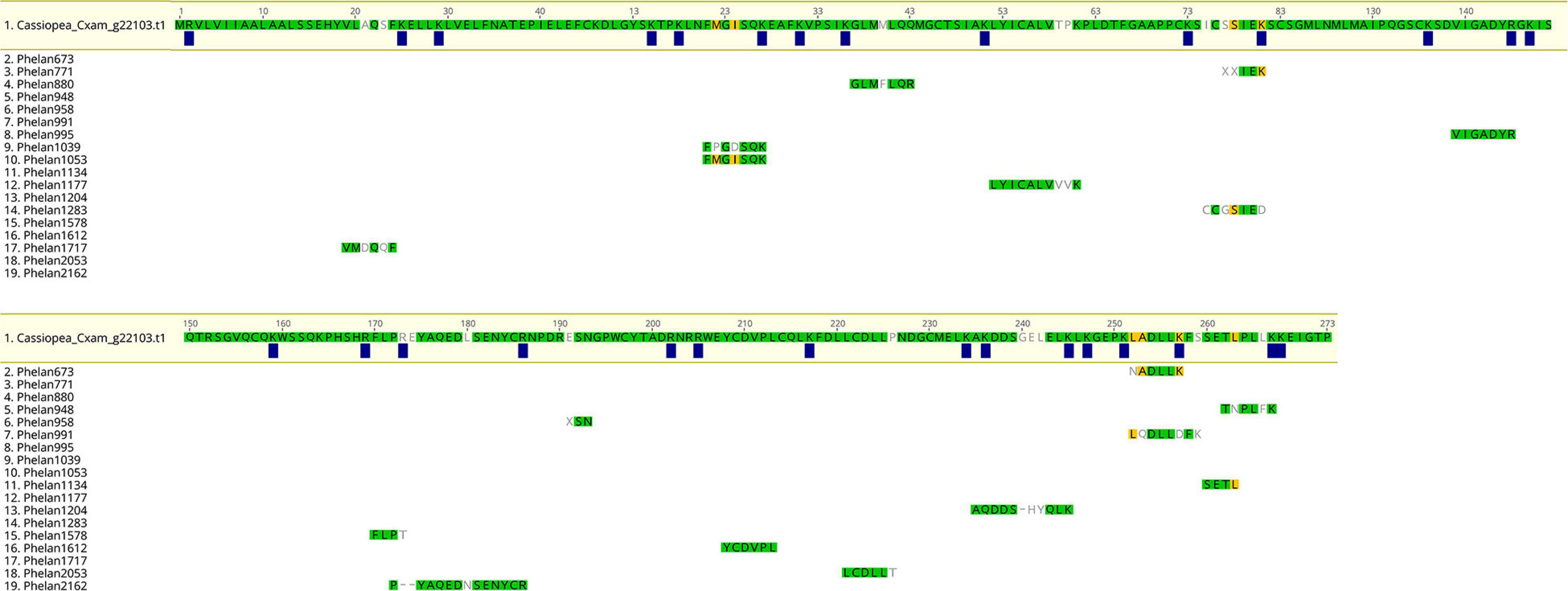
Figure 4. Alignment of predicted protein and tryptic digests of Cassio Blue, the blue pigment from Cassiopea xamachana. Amino acids highlighted in green indicate a perfect match whilst yellow indicates discordance with at least one sequence in alignment. Amino acids that are not highlighted indicate a full discordance. Blue squares represent predicted trypsin cleavage sites.
Structural predictions of rpulFKz1 and its homologs were attempted, but these could not be predicted confidently, with C-scores < −1.03 (most < −1.5, see Supplementary Table 3). In most cases, the closest structural analog in the Protein Data Bank (PDB) to these proteins was the type II human plasminogen, chain A (PDB ID: 4DURA, see Supplementary Table 3), which had a higher sequence identity to the KR domain. Transmembrane helices could also not be predicted for most sequences except Rhopilema_mRNA.RE16087, for which one transmembrane helix is predicted (from positions 25 to 61 of the alignment in Figure 3), although it overlaps with the signal peptide.
Discussion
Our search demonstrated that Fz-CRD and KR domains are ubiquitous among cnidarians, yet only eight protein sequences were discovered that fall within the same clade and have the same domain structure as rpulFKz1 (Bulina et al., 2004). Among these proteins, the sequence that belongs to Cassiopea xamachana has motifs that mostly match Cassio Blue tryptic digests (Phelan et al., 2006). Mismatches between digests and the rpulFKz1 homolog potentially come from amino acid modifications in the chromophore or even due to sequencing error, and we propose that the digest sequences derive from the rpulFKz1 homolog. Additionally, we also identified a rpulFKz1 homolog from Stomolophus meleagris Agassiz, 1860, and previous studies on the blue pigments of Stomolophus spp. have found similar spectroscopic and physicochemical properties to rpulFKz1 and Cassio Blue (Lugo-Magaña et al., 2018; Martínez-Pérez et al., 2020). Therefore, we hypothesize that the eight proteins identified as rpulFKz1 homologs are also pigment precursors. These precursors are exclusive to jellyfish that belong to the order Rhizostomeae. Of the rhizostomes included in this study, only C. xamachana possesses zooxanthellae (Djeghri et al., 2019), and Cassio Blue was not present in isolated and purified algae homogenates (Blanquet and Phelan, 1987), so it is unlikely that the pigment precursors identified derive from zooxanthellae or other contaminants. All of these findings support the establishment of a novel pigment family that we herein name as rhizostomins, which includes rpulFKz1 and Cassio Blue. Recent studies show rhizostomes as a monophyletic group (Gómez Daglio and Dawson, 2017; Kayal et al., 2018; although see Bayha et al., 2010), therefore we hypothesize a single origin of rhizostomins among cnidarians (Figure 5). These homologous proteins do not appear in a monophyletic group in the phylogenetic trees herein due to independent analysis of protein domains.
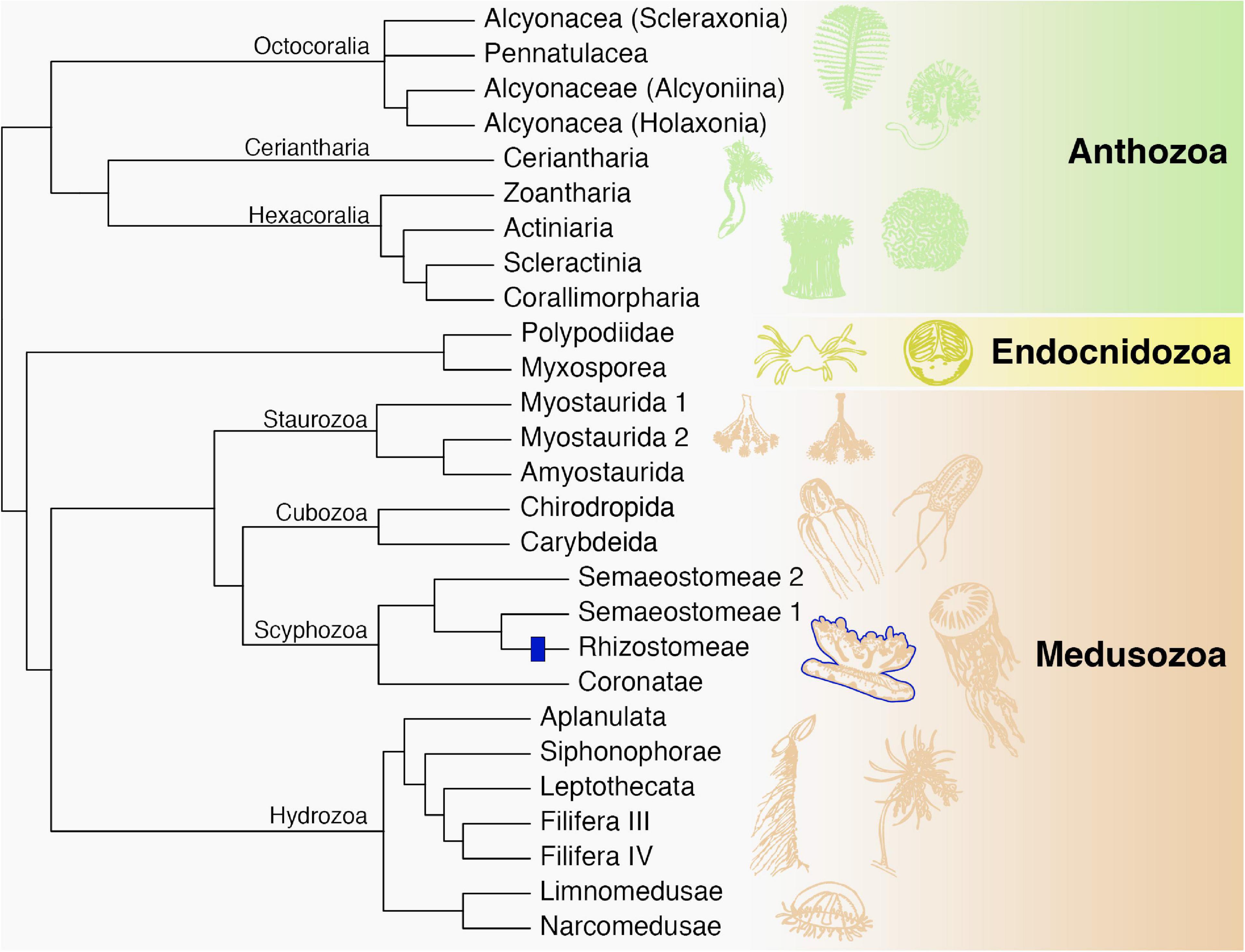
Figure 5. Phylogenetic relationships among major cnidarian lineages that are included in this study (except Polypodiidae). We propose a single origin of rhizostomins within the Rhizostomeae lineage (blue). Modified from Kayal et al. (2018).
Rhizostomins that have been studied to date are all blue pigments. Nevertheless, we found pigment precursors in species that do not have blue coloration, such as Nemopilema nomurai Kishinouye, 1922 and Rhopilema esculentum Kishinouye, 1891 (Prof. Shin-ichi Uye and Prof. Hiroshi Miyake, personal communication; Figures 6E,G), as well as S. meleagris (Figure 6C). Similar to S. meleagris and Stomolophus sp. 2 sensu (Gómez Daglio and Dawson, 2017) (Figure 6D), R. esculentum has a congener that possesses blue coloration, Rhopilema asamushi Uchida, 1927 (Figure 6F). It is possible that modifications in the protein structure of rhizostomins, especially in the chromophore, could result in different colors. This is the case, for example, for GFP-like proteins, which despite chromophore diversity in the four color classes (green, yellow, orange-red and non-fluorescent purple-blue), share the same “beta-can” fold of the polypeptide chain (Matz et al., 2002). The structure of rhizostomins is yet to be elucidated, even though there are indications of where the covalent bonds that form the chromophore might be (Bulina et al., 2004). Our attempts at predicting protein structure were unsuccessful. The closest structural analog in most cases was a human plasminogen, likely because it possesses a KR domain, which was the most conserved domain of the pigment precursors.

Figure 6. Rhizostome jellyfish from genera that possess the rhizostomin pigment precursors identified in this study. (A) Rhizostoma pulmo, (B) Cassiopea xamachana, (C) Stomolophus meleagris, (D) Stomolophus sp. 2, (E) Rhopilema esculentum, (F) Rhopilema asamushi and (G) Nemopilema nomurai. Photos by Prof. André Morandini (A,B), Prof. Ronald Larson (C), Dr. Liza Gómez Daglio (D), Mr. Takashi Murai (E,G) and Tsuruoka City Kamo Aquarium (F).
Phylogenetic analyses of the cnidarian Fz-CRD and KR protein domains indicate that pigment precursors likely evolved from an insertion of a KR domain from plasminogen-like genes into a Fz-CRD domain from Fz-like or SFRP-like genes. Fz proteins, however, are transmembrane receptors (Huang and Klein, 2004), whereas most rhizostomins did not possess transmembrane helices and are likely secreted (Blanquet and Phelan, 1987; Bulina et al., 2004). Rhizostomins are therefore more similar to SFRPs, which are also secreted (Jones and Jomary, 2002). Fz-CRD and KR domains also appear in ROR receptors, however, in this case they occur intact and alongside other protein domains. Previously, only domain fusion was reported in the evolution of proteins with Fz-CRDs (including ROR) (Yan et al., 2014), and fusion is one of the most common domain rearrangements in protein evolution (Björklund et al., 2005). Domain rearrangements often occur in the evolution of multi-domain proteins (Björklund et al., 2005), although an insertion of a domain into another is unusual (Aroul-Selvam et al., 2004). When it occurs, it is more often a single domain insertion and the inserted domain is usually smaller than the parent domain (Aroul-Selvam et al., 2004), which is what we observed for the rhizostome pigment precursors. Rhizostomins are the only reported case of a domain insertion within a parent Fz-CRD.
Both Fz-CRD and KR domains are involved in ligand binding, and genes with a Fz-CRD, such as ROR, are associated with Wnt signaling pathways (Dann et al., 2001). The KR domain is well conserved across cnidarians, especially the ligand binding regions. Yet, the Fz-CRD of the rhizostomins lacks two of the ten typical C residues, which usually form one of the disulfide bonds (Dann et al., 2001; Yan et al., 2014). The N-glycosylation site that may be important for Wnt binding (Yan et al., 2014), situated within a Wnt interaction region, is also missing from rhizostomins. These modifications may indicate that these molecules are no longer involved, or have a distinctive role, in Wnt pathways. However, tissue-specific expression patterns of Wnt genes in jellyfish (Khalturin et al., 2019) resemble some of the color patterns observed in the rhizostomes used in this study (Figure 6). For example, the expression of Wnt-11a around the bell margin of Aurelia ephyrae (see Figure 4j–n in Khalturin et al., 2019) is similar to that of the blue pigment rpulFKz1, which appears around the bell margin of Rhizostoma pulmo (Bulina et al., 2004; Figure 6A). Some Wnt genes in Aurelia are highly upregulated in the endodermal gastrovascular system, while others are predominantly expressed in both the bell margin and oral arms (Khalturin et al., 2019), the latter of which matches the locality of the red-brown coloration in N. nomurai (Figure 6G). Considering the apparent similarity in tissue-specific expression of some Wnt genes and rhizostomins, it is possible that their expression is co-regulated.
There are over 70 species in the order Rhizostomeae that range in color not only from red-brown to blue, but also white, green, pink and purple (Jarms and Morandini, 2019). Some of these colors might even vary within a single species (Lampert et al., 2012). Some rhizostomes harbor zooxanthellae that usually give them a yellow-brown color (e.g., Phyllorhiza punctata von Lendenfeld, 1884), yet others may appear brown but not harbor zooxanthellae [e.g., Catostylus mosaicus (Quoy and Gaimard, 1824)]. This demonstrates that color variation in this group of jellyfish is not directly linked to the presence of endosymbionts (Lampert et al., 2012). Cassio Blue, the rhizostomin of C. xamachana, has been hypothesized to function as a metal reservoir (Phelan et al., 2006) and to confer photoprotection (Blanquet and Phelan, 1987; Phelan et al., 2006). The latter case is not unprecedented for cnidarians, as in corals such as Acropora millepora (Ehrenberg, 1834), differential expression of a red-fluorescent GFP-like pigment (amilFP597) is correlated with light exposure and reduced photodamage of zooxanthellae (Gittins et al., 2015).
Conclusion
Rhizostome jellyfish harbor unique pigments with Fz-CRD and KR domains, which we herein term rhizostomins. We hypothesize these pigments have a single origin and likely evolved from an insertion of a KR domain, derived from a plasminogen-like gene, into a Fz-CRD, derived from a Fz-like or SFRP-like gene. There are modifications in important Wnt binding and interactions sites in the Fz-CRD of rhizostomins, indicating that they may not play a role in Wnt pathways. We also show that pigment precursors occur in species that are not blue, and we hypothesize that this pigment family may include other colors. Rhizostomins may confer photoprotection in a similar manner to other cnidarian pigments. Future studies may not only demonstrate their function and color variation, but also their potential biomedical and industrial applications.
Data Availability Statement
Publicly available datasets were analyzed in this study. Alignments and phylogenetic trees can be accessed through Figshare (Lawley et al., 2021) and relevant codes are available in GitHub (github.com/lawleyjw/Rhizostomins).
Author Contributions
All authors contributed to the conception and design of this study. JL undertook data collection and analyses with assistance from all other authors. JL led the writing of the manuscript. All authors contributed with revisions and approved the submitted version.
Funding
The publication of this manuscript was kindly funded by the state of Bremen. JL acknowledges financial support through Postgraduate Research Scholarships awarded by Griffith University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are very grateful to Kylie Pitt for providing advice on this research, especially with insights on rhizostome ecology, on which she has been working for over 20 years, as well as comments on a first version of this manuscript. We sincerely thank Tessa Page for her help with transcriptomic analyses and Indy Silva for his assistance on running these analyses in Griffith University’s HPC cluster “Gowonda.” We also thank Profs. André Morandini, Shin-ichi Uye, Hiroshi Miyake, Ronald Larson, Mr. Takashi Murai and Dr. Liza Gómez Daglio for providing beautiful jellyfish photographs as well as information on species considered in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.752949/full#supplementary-material
Footnotes
References
Agassiz, L. (1860). Contributions to the Natural History of the United States of America. Beston: Brown and Company.
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Anglés-Cano, E., and Rojas, G. (2002). Apolipoprotein(a): structure-Function Relationship at the Lysine-Binding Site and Plasminogen Activator Cleavage Site. Biol. Chem. 383, 93–9. doi: 10.1515/BC.2002.009
Aroul-Selvam, R., Hubbard, T., and Sasidharan, R. (2004). Domain Insertions in Protein Structures. J. Mol. Biol. 338, 633–641. doi: 10.1016/j.jmb.2004.03.039
Bandaranayake, W. M. (2006). The nature and role of pigments of marine invertebrates. Nat. Prod. Rep. 23, 223–255. doi: 10.1039/B307612C
Bayha, K. M., Dawson, M. N., Collins, A. G., Barbeitos, M. S., and Haddock, S. H. D. (2010). Evolutionary Relationships Among Scyphozoan Jellyfish Families Based on Complete Taxon Sampling and Phylogenetic Analyses of 18S and 28S Ribosomal DNA. Integr. Compar. Biol. 50, 436–455. doi: 10.1093/icb/icq074
Björklund, ÅK., Ekman, D., Light, S., Frey-Skött, J., and Elofsson, A. (2005). Domain Rearrangements in Protein Evolution. J. Mol. Biol. 353, 911–923. doi: 10.1016/j.jmb.2005.08.067
Blanquet, R. S., and Phelan, M. A. (1987). An unusual blue mesogleal protein from the mangrove jellyfish Cassiopea xamachana. Mar. Biol. 94, 423–430.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bulina, M. E., Lukyanov, K. A., Yampolsky, I. V., Chudakov, D. M., Staroverov, D. B., Shcheglov, A. S., et al. (2004). New Class of Blue Animal Pigments Based on Frizzled and Kringle Protein Domains. J. Biol. Chem. 279, 43367–43370. doi: 10.1074/jbc.C400337200
Chang, Y., Mochalkin, I., McCance, S. G., Cheng, B., Tulinsky, A., and Castellino, F. J. (1998). Structure and Ligand Binding Determinants of the Recombinant Kringle 5 Domain of Human Plasminogen †, ‡. Biochemistry 37, 3258–3271. doi: 10.1021/bi972284e
Christomanos, A. (1954). A Violet Pigment from the Mediterranean Medusa Rhizostoma pulmo. Nature 173:875. doi: 10.1038/173875a0
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a Sequence Logo Generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Cuthill, I. C., Allen, W. L., Arbuckle, K., Caspers, B., Chaplin, G., Hauber, M. E., et al. (2017). The biology of color. Science 357:6350. doi: 10.1126/science.aan0221
Dann, C. E., Hsieh, J.-C., Rattner, A., Sharma, D., Nathans, J., and Leahy, D. J. (2001). Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412, 86–90. doi: 10.1038/35083601
Djeghri, N., Pondaven, P., Stibor, H., and Dawson, M. N. (2019). Review of the diversity, traits, and ecology of zooxanthellate jellyfishes. Mar. Biol. 166:147. doi: 10.1007/s00227-019-3581-6
Eddy, S. R. (1998). Profile hidden Markov models. Bioinformatics 14, 755–763. doi: 10.1093/bioinformatics/14.9.755
Egusa, S., and Nakajima, K. (1981). A New Myxozoa Thelohanellus kitauei, the Cause of Intestinal Giant Cystic Disease of Carp. Fish. Pathol. 15, 213–218. doi: 10.3147/jsfp.15.213
Ehrenberg, C. G. (1834). Beiträge zur physiologischen Kenntniss der Corallenthiere im allgemeinen, und besonders des rothen Meeres, nebst einem Versuche zur physiologischen Systematik derselben. Abhandlungen der Königlichen Akademie der Wissenschaften, Berlin 1, 225–380.
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Forsskål, P. (1775). Pelagia noctiluca. Available online at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=135305
Fox, D. L., and Millott, N. (1954). The pigmentation of the jellyfish, Pelagia noctiluca (Forskål) var. panopyra Péron & Lesueur. Proceedings of the Royal Society of London. Ser. B Biol. Sci. 142, 392–408.
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). Protein Identification and Analysis Tools on the ExPASy ServerJ. M.Walker. Germany: Springer. 571–607. doi: 10.1385/1-59259-890-0:571
Gittins, J. R., D’Angelo, C., Oswald, F., Edwards, R. J., and Wiedenmann, J. (2015). Fluorescent protein-mediated colour polymorphism in reef corals: multicopy genes extend the adaptation/acclimatization potential to variable light environments. Mol. Ecol. 24, 453–465. doi: 10.1111/mec.13041
Gómez Daglio, L., and Dawson, M. N. (2017). Species richness of jellyfishes (Scyphozoa: discomedusae) in the Tropical Eastern Pacific: missed taxa, molecules, and morphology match in a biodiversity hotspot. Invert. Syst. 31:635. doi: 10.1071/IS16055
Green, J. L., Kuntz, S. G., and Sternberg, P. W. (2008). Ror receptor tyrosine kinases: orphans no more. Trends Cell Biol. 18, 536–544. doi: 10.1016/j.tcb.2008.08.006
Guder, C., Philipp, I., Lengfeld, T., Watanabe, H., Hobmayer, B., and Holstein, T. W. (2006). The Wnt code: cnidarians signal the way. Oncogene 25, 7450–7460. doi: 10.1038/sj.onc.1210052
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: assessing the Performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gupta, R., and Brunak, S. (2002). Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 310–322.
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hendry, G. A. F. (1996). “Natural pigments in biology” in Natural Food Colorants. eds G. A. F. Hendry and J. D. Houghton (Boston: Springer). 1–39. doi: 10.1007/978-1-4615-2155-6_1
Herring, P. J. (1965). Blue Pigment of a Surface-living Oceanic Copepod. Nature 205, 103–104. doi: 10.1038/205103a0
Herring, P. J. (1971). Stability of the blue pigment of Velella and Porpita (Coelenterata: Siphonophora). Compar. Biochem. Physiol. Part B Compar. Biochem. 39, 1039–1043. doi: 10.1016/0305-0491(71)90127-1
Herring, P. J. (1972). Porphyrin Pigmentation in Deep-sea Medusae. Nature 238:276. doi: 10.1038/238276a0
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Huang, H.-C., and Klein, P. S. (2004). The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 5:234.
Jarms, G., and Morandini, A. C. (2019). World Atlas of Jellyfish: Scyphozoa Except Stauromedusae. Germany: Dölling und Galitz Verlag.
Jones, S. E., and Jomary, C. (2002). Secreted Frizzled-related proteins: searching for relationships and patterns. BioEssays 24, 811–820. doi: 10.1002/bies.10136
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kayal, E., Bentlage, B., Sabrina Pankey, M., Ohdera, A. H., Medina, M., Plachetzki, D. C., et al. (2018). Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 18:68. doi: 10.1186/s12862-018-1142-0
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Khalturin, K., Shinzato, C., Khalturina, M., Hamada, M., Fujie, M., Koyanagi, R., et al. (2019). Medusozoan genomes inform the evolution of the jellyfish body plan. Nat. Ecol. Evol. 3, 811–822. doi: 10.1038/s41559-019-0853-y
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
LaJeunesse, T. C. (2017). Validation and description of Symbiodinium microadriaticum, the type species of Symbiodinium (Dinophyta). J. Phycol. 53, 1109–1114. doi: 10.1111/jpy.12570
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.
Lampert, K. P., Bürger, P., Striewski, S., and Tollrian, R. (2012). Lack of association between color morphs of the jellyfish Cassiopea andromeda and zooxanthella clade. Mar. Ecol. 33, 364–369.
Lawley, J., Carroll, A. R., and McDougall, C. (2021). Sequence Data for Rhizostomin Pigment Analyses. figshare Dataset. doi: 10.6084/m9.figshare.16563876.v1
Linnaeus, C. (1758). Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis., Edition Decima, Reformata. Vienna: Typis Ioannis Thomae.
Lugo-Magaña, O., García, X. G., Barrientos, R. G., and Legarreta, I. G. (2018). Preliminary Characterization of a Blue Pigment Extracted from “Cannon Ball” Jellyfish Stomolophus meleagris. Russ. J. Mar. Biol. 44, 477–483.
Macri, S. (1778). Nuove osservazioni intorno la storia naturale del Polmone marino degli antichi. Naples.
Martínez-Pérez, R. B., Díaz-Tenorio, L. M., Leyva Soto, L. A., Gortáres-Moroyoqui, P., García-Rico, L., and Rodríguez, J. A. (2020). Characterization of cannonball jellyfish (Stomolophus sp. 2) blue protein: a pH-stable pigment. Environ. Sci. Pollut. Res. 27, 28597–28606. doi: 10.1007/s11356-020-08689-1
Matz, M. V., Lukyanov, K. A., and Lukyanov, S. A. (2002). Family of the green fluorescent protein: journey to the end of the rainbow. BioEssays 24, 953–959. doi: 10.1002/bies.10154
Mendoza, L., Deutsch, E. W., Sun, Z., Campbell, D. S., Shteynberg, D. D., and Moritz, R. L. (2018). Flexible and fast mapping of peptides to a proteome with ProteoMapper. J. Proteome Res. 17, 4337–4344. doi: 10.1021/acs.jproteome.8b00544
Murbach, L., and Shaerer, C. (1902). Preliminary report on a collection of medusae from the coast of British Columbia and Alaska. Ann. Magaz. Nat. Hist. 9, 71–73.
Nakamura, T., Nishizawa, T., Hagiya, M., Seki, T., Shimonishi, M., Sugimura, A., et al. (1989). Molecular cloning and expression of human hepatocyte growth factor. Nature 342, 440–443. doi: 10.1038/342440a0
Newsome, A. G., Culver, C. A., and van Breemen, R. B. (2014). Nature’s Palette: the Search for Natural Blue Colorants. J. Agric. Food Chem. 62, 6498–6511. doi: 10.1021/jf501419q
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Patthy, L., Trexler, M., Váli, Z., Bányai, L., and Váradi, A. (1984). Kringles: modules specialized for protein binding: homology of the gelatin-binding region of fibronectin with the kringle structures of proteases. FEBS Lett. 171, 131–136. doi: 10.1016/0014-5793(84)80473-1
Péron, F., and Lesueur, C. A. (1810). Tableau des caractères génériques et spécifiques de toutes les espèces de méduses connues jusqu’à ce jour. Ann. Mus. Natl. Nat. Paris 14, 325–366.
Phelan, M. A., Matta, J. L., Reyes, Y. M., Fernando, R., Boykins, R. A., and Blanquet, R. S. (2006). Associations between metals and the blue mesogleal protein of Cassiopea xamachana. Mar. Biol. 149, 307–312.
Quoy, J. R. C., and Gaimard, J. P. (1824). Voyage au Tour du Monde Fait par Ordre du Roi, Sur Les Corvettes De S. M: l’Uranie et la Physicienne Pendant les Années 1817, 1818, 1819 et 1820. Paris: Chez Pillet aîné. 1–328.
Sanders, S. M., and Cartwright, P. (2015). Patterns of Wnt signaling in the life cycle of Podocoryna carnea and its implications for medusae evolution in Hydrozoa (Cnidaria). Evol. Dev. 17, 325–336. doi: 10.1111/ede.12165
Santos, M. C. D., and Bicas, J. L. (2021). Natural blue pigments and bikaverin. Microbiol. Res. 244:126653. doi: 10.1016/j.micres.2020.126653
Shimomura, O., Johnson, F. H., and Saiga, Y. (1962). Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Compar. Physiol. 59, 223–239. doi: 10.1002/jcp.1030590302
Sommer, F., Agurto, C., Henriksen, P., and Kiørboe, T. (2006). Astaxanthin in the calanoid copepod Calanus helgolandicus: dynamics of esterification and vertical distribution in the German Bight, North Sea. Mar. Ecol. Prog. Ser. 319, 167–173. doi: 10.3354/meps319167
Squarzoni, P., Parveen, F., Zanetti, L., Ristoratore, F., and Spagnuolo, A. (2011). FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development 138, 1421–1432. doi: 10.1242/dev.057323
Thewes, T., Constantine, K., Byeon, I.-J. L., and Llinás, M. (1990). Ligand interactions with the Kringle 5 domain of Plasminogen. J. Biochem. 265, 3906–3915.
Uchida, T. (1927). Report of the biological survey of Mutsu Bay. 2. Medusae of Mutsu Bay. Sci. Rep. Tohoku Imp. Univ. 2, 215–238.
Venil, C. K., Zakaria, Z. A., and Ahmad, W. A. (2013). Bacterial pigments and their applications. Process Biochem. 48, 1065–1079. doi: 10.1016/j.procbio.2013.06.006
von Lendenfeld, R. (1884). The scyphomedusae of the southern hemisphere. Part III. Proc. Linnean Soc. N. South Wales 9, 259–306.
Watanabe, H., Schmidt, H. A., Kuhn, A., Höger, S. K., Kocagöz, Y., Laumann-Lipp, N., et al. (2014). Nodal signalling determines biradial asymmetry in Hydra. Nature 515, 112–115. doi: 10.1038/nature13666
Yan, J., Jia, H., Ma, Z., Ye, H., Zhou, M., Su, L., et al. (2014). The evolutionary analysis reveals domain fusion of proteins with Frizzled-like CRD domain. Gene 533, 229–239. doi: 10.1016/j.gene.2013.09.083
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Keywords: blue pigment, coloration, protein domain, Frizzled (Fz), Kringle, genomics, transcriptomics
Citation: Lawley JW, Carroll AR and McDougall C (2021) Rhizostomins: A Novel Pigment Family From Rhizostome Jellyfish (Cnidaria, Scyphozoa). Front. Mar. Sci. 8:752949. doi: 10.3389/fmars.2021.752949
Received: 03 August 2021; Accepted: 03 September 2021;
Published: 24 September 2021.
Edited by:
Simon Jungblut, University of Bremen, GermanyReviewed by:
Stefania De Domenico, University of Salento, ItalyShannon Grace Klein, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2021 Lawley, Carroll and McDougall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan W. Lawley, am9uYXRoYW4ud2FuZGVybGV5bGF3bGV5QGdyaWZmaXRodW5pLmVkdS5hdQ==
 Jonathan W. Lawley
Jonathan W. Lawley Anthony R. Carroll
Anthony R. Carroll Carmel McDougall
Carmel McDougall