- 1Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture, China, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3College of Marine Technology and Environment, Dalian Ocean University, Dalian, China
Portunus trituberculatus is one of the main mariculture crabs of high economic value. To identify genes involved in sex determination, we first performed sex-specific transcriptome sequencing at six larval development stages using a DNA/RNA co-extraction method. A total of 907,952,938 and 828,774,880 reads were obtained from female and male crabs, respectively. 2,379 differentially expressed genes (DEGs) were found between females and males, and were mainly enriched in DNA replication, folate biosynthesis, and retinol metabolism pathways. Furthermore, transcription patterns of genes in the sex-determining region (SD) were analyzed based on the transcriptome data, and one Dmrt gene (PtDMY) was found to be exclusively expressed in males during early developmental stages. Notably, some known sex-related genes, including IAG, Dmrt11E, DmrtB1, and DmrtC2 were significantly down-regulated after knocking down PtDMY. Our results suggested that PtDMY is involved in sex determination and may be one of the key upstream regulators of the sex determination pathway. In addition, the massive volume of transcriptome data obtained in this study provided an important basis for the systematic study of sex determination mechanisms in P. trituberculatus.
Introduction
Crustaceans are the second largest class of arthropods (Alwi et al., 2015). There are multiple sex determination systems in crustaceans that have been identified by karyotype analyses, including XY/ZW systems (Cui et al., 2015; Fang et al., 2020). It is an important material to study the evolution of sex staining and the mechanisms of sex determination (Torrecilla et al., 2017; Toyota et al., 2021). In recent years, with the development of sequencing technologies, sex-related quantitative trait locus (QTL) have been mapped and sex markers have been identified in many crustaceans of economic significance, including Litopenaeus vannamei, Eriocheir sinensis, Scylla paramamosain, and Exopalaemon carinicauda (Cui et al., 2015; Yu et al., 2015; Shi et al., 2018; Fang et al., 2020; Lv et al., 2020a). Multiple studies have contributed to the identification of sex determination genes and have helped clarify the mechanisms of sex determination in crustaceans.
Sex-determining genes, such as sex-determining region of Y-chromosome (Sry) and double-sex and mab-3 related transcription factor (Dmrt), have been identified in mammals, fish, birds, and reptiles (Masaru et al., 2002; Prokop et al., 2020; Han et al., 2021; Naoki and Makoto, 2021). Most sex-determining genes are transcription factors located on the sex chromosomes. For example, Dmrt in Oryzias latipes is located on the Y chromosome (Feng et al., 2021), and Dmrt in Xenopus laevis is located on the W chromosome (Yoshimoto et al., 2008).
Studies have shown that the androgenic gland (AG) is an important organ for sex determination, and the Insulin-like androgenic gland hormone (IAG) secreted by the AG is necessary for male genesis (Sun and Li, 2021). Notably, the knockdown of IAG can lead to sex reversal in Macrobrachium rosenbergii (Lezer et al., 2015). Recently, a Y-linked Dmrt was also identified in Sagmariasus verreauxi (Chandler et al., 2017), which shed new light on the prevalence of Dmrt genes as master sex-determinants in crustaceans. However, studies on sex determination genes in crustaceans are relatively backward, and the regulatory networks of sex determination are not clear, thus, limiting the identification of the mechanisms of sex determination in crustaceans.
P. trituberculatus, known as the swimming crab, is the main mariculture crab in China with important economic value and an annual production of ∼100,000 tons. The growth traits of P. trituberculatus are significantly different between males and females, and therefore, it is of great economic significance to detail the mechanisms of sex determination and establish sex-control technologies. We located the sex QTL and identified sex markers in P. trituberculatus (Lv et al., 2018), and established sex identification technologies based those markers (Zhang et al., 2021). We also confirmed the XY sex determination system in P. trituberculatus according to the differences in sex markers between males and females. However, due to the lack of genomic information, unambiguous sex-determining genes have not been identified.
In this study, we first performed sex-specific transcriptome sequencing of six larval development stages using a DNA/RNA co-extraction method. Through comparative transcriptome analyses, DEGs between males and females were identified and enriched. In addition, we analyzed the transcription patterns of genes in the SD during the early development stages and identified a key candidate gene for sex determination. Our results contribute to the interpretation of sex determination mechanisms in P. trituberculatus.
Materials and Methods
Experimental Materials
P. trituberculatus used in this study was cultured in Changyi Haifeng Aquaculture Co., Ltd., Weifang City, Shandong Province, China. There are 12 developmental stages from embryonic development to adult crab (Figure 1), including the fertilized egg (multicellular stage, Mc; blastula stage, B; gastrulation stage, G; egg-nauplius stage, En; Egg-zoea stage, Ez), zoea (zoea 1, Z1; zoea 2, Z2; zoea 3, Z3; zoea 4, Z4), megalopa (M), and juvenile crab (juvenile crab 1, J1; juvenile crab 2, J2). Samples at each stage were separately picked and placed into RNase-free centrifuge tubes (more than nine individuals per tube and three biological replicates for each period) and immediately placed into liquid nitrogen. Samples were then used to prepare cDNA for qRT-PCR. An additional sample was collected during Z2-J2 (more than 30 individuals per stage), and was immediately soaked in absolute ethanol and placed in liquid nitrogen. Samples were then used for DNA/RNA co-extraction of each individual.
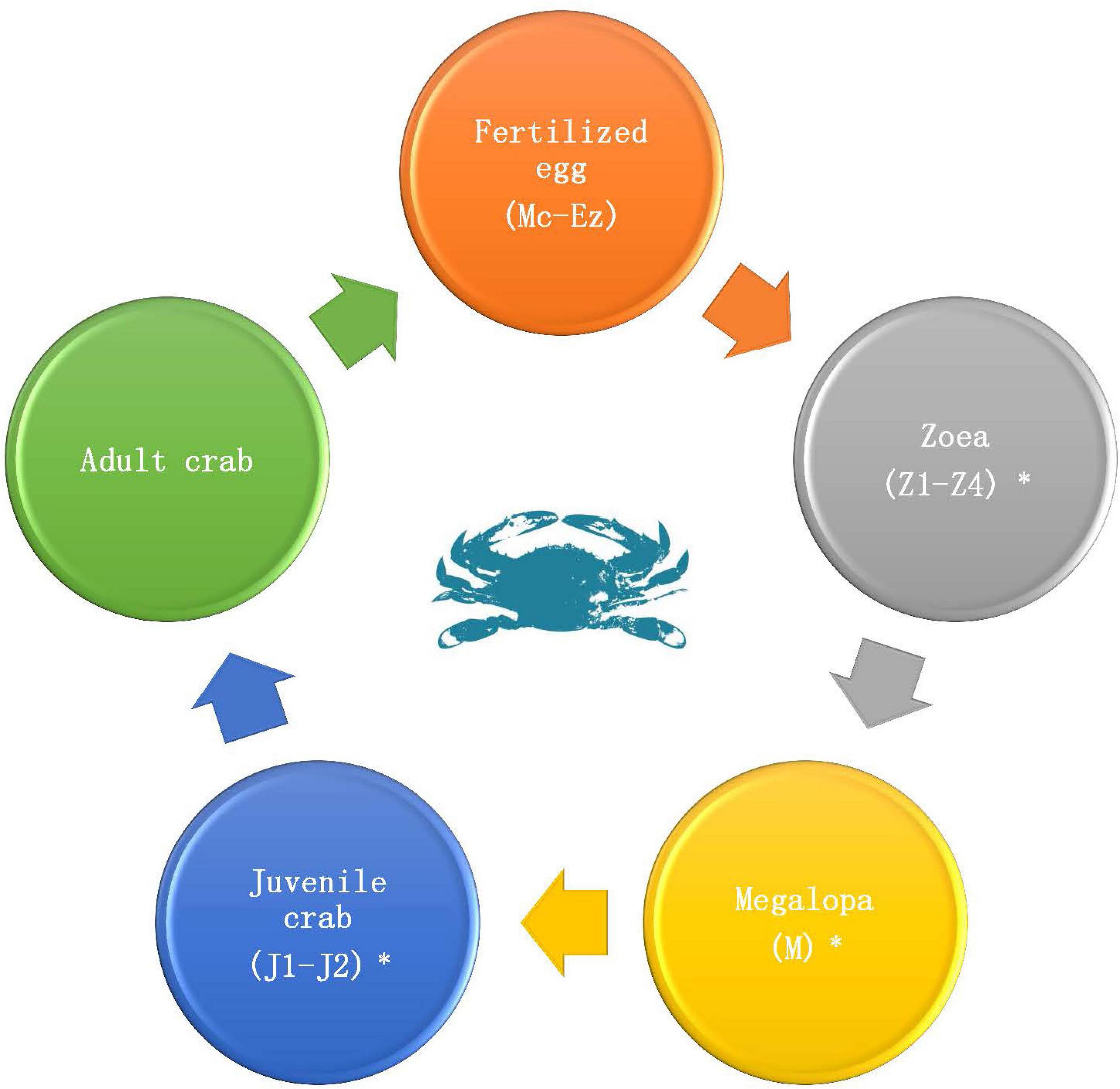
Figure 1. Life cycle of P. trituberculatus. Fertilized egg (Multicellular stage, Mc; Blastula stage, B; Gastrulation stage, G; Egg-nauplius stage, En; Egg-zoea stage, Ez), Zoea (Zoea 1, Z1; Zoea 2, Z2; Zoea 3, Z3; Zoea 4, Z4), Megalopa (M), and Juvenile crab (Juvenile crab 1, J1; Juvenile crab 2, J2). The periods marked with a “*” are the periods at which transcriptome sequencing was performed.
DNA/RNA Co-extraction and Sex Identification
Individual DNA and RNA were extracted simultaneously using the SteadyPure Universal RNA Extraction kit (Accurate Biology, Hunan). The DNA has been tested by agarose gel electrophoresis to determine the fragment size. And the OD of DNA was measured by uv spectrophotometer, the OD260/OD280 was about 1.8. DNA was used for sex identification using previously established sex identification methods (Zhang et al., 2021). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, United States).
Library Construction and Transcriptome Sequencing
After sex identification, the RNA of three or four individuals of the same sex were mixed into one sample, and three parallel biological samples were prepared for males and females at each developmental stage. To select cDNA fragments that were 370–420 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, United States). Clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit, v3-cBot-HS (Illumina), according to the manufacturer’s instructions. Following cluster generation, the libraries were sequenced on an Illumina Novaseq platform with 150 bp paired-end read datasets generated.
Quality Control
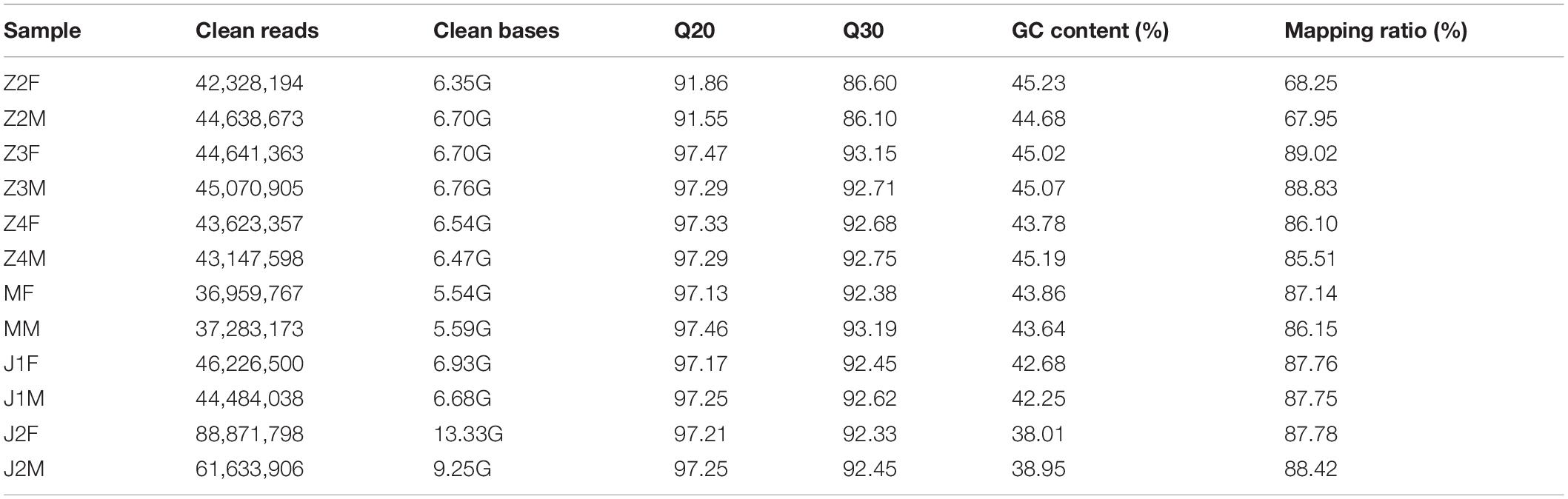
Clean data were obtained by removing low quality reads, and reads containing adapters and ploy-N sequences. The Q20, Q30, and GC contents of the clean data were calculated. The index of the reference genome was built using Hisat2 v2.0.5, and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5.
Quantification of Gene Transcription
The mapped reads of each sample were assembled using StringTie (v1.3.3b) (Pertea et al., 2016) in a reference-based approach. FeatureCounts v1.5.0-p3 was used to count the number of reads mapped to each gene. The fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) of each gene was then calculated based on the length of the gene and read counts mapped to the gene. The FPKM considers the effect of the sequencing depth and gene length for the read counts at the same time.
Differential Transcription Analysis
Differential gene transcription analyses of males and females were performed using the DESeq2 R package (1.20.0) (Love et al., 2014). DESeq2 provides statistical routines to determine the differential transcription of digital gene transcription data using a model based on a negative binomial distribution. Genes with an adjusted p-value ≤ 0.05 and a | log2FoldChange| ≥ 1.0 were considered differentially expressed.
Quantitative Real-Time PCR
Primers were designed using Primer 5.0 software (Li et al., 2020). The volume of the qRT-PCR was 10 μl, containing 5 μL of 2 × SYBR Master Mix (RR420A, Takara Bio, Japan), 0.2 μL of each primer and ROX Reference Dye II, 1 μL of cDNA template, and 3.4 μl of RNase free dH2O. qRT-PCR was carried out in a FAST-7500 system (ABI-7500, Thermo Fisher Scientific, Singapore) with the following parameters: 95°C for 30 s; 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s. The relative quantification of qRT-PCR data was calculated using the 2–Δ Δ Ct method (Livak and Schmittgen, 2001).
RNA Interference
Z1 stage is the first period when eggs hatch into larvae, and the expression of PtDMY was relatively high in the Z1 stage. Therefore, Z1 stage samples were used for RNAi. The PtDMY gene sequence was sent to Shanghai Sangon Biology Co., Ltd. for primer design and preparation of RNAi reagents (G3-Sense, GCGCAUCCAUACUAACAAATT; G3-Antisense, UUUGUUAGUAUGGAUGCGCTT; NC-Sense, UUCUCCGAACGUGUCACGUTT; NC-Antisense, ACGUGACACGUUCGGAGAATT). RNAi was performed by feeding Rotifers soaked in RNAi reagent to crabs. The concentration of RNAi reagent for soaking was 3.3 μg/mL, and feeding was initiated after soaking for 2–3 h. The Rotifers were fed 3 times per day. Crabs at Z1 stage were sampled after feeding for 48 h. RNA extraction and reverse transcription were performed on the collected samples, and the effect of RNAi was detected using qRT-PCR.
Results
Transcriptome Sequencing
Male and female samples from six developmental stages were sequenced on an Illumina platform and 86.84 Gb of clean bases were produced. In total, 907,952,938 and 828,774,880 reads were obtained from females and males, respectively. The mean values of Q20 and Q30 were 96.355 and 91.6175, respectively. The average percentage of genome mapping was 84.21% (Table 1). The raw data were deposited in the Short Read Archive of the National Center for Biotechnology Information (NCBI) with accession number PRJNA631920.
Differentially Expressed Genes Analysis
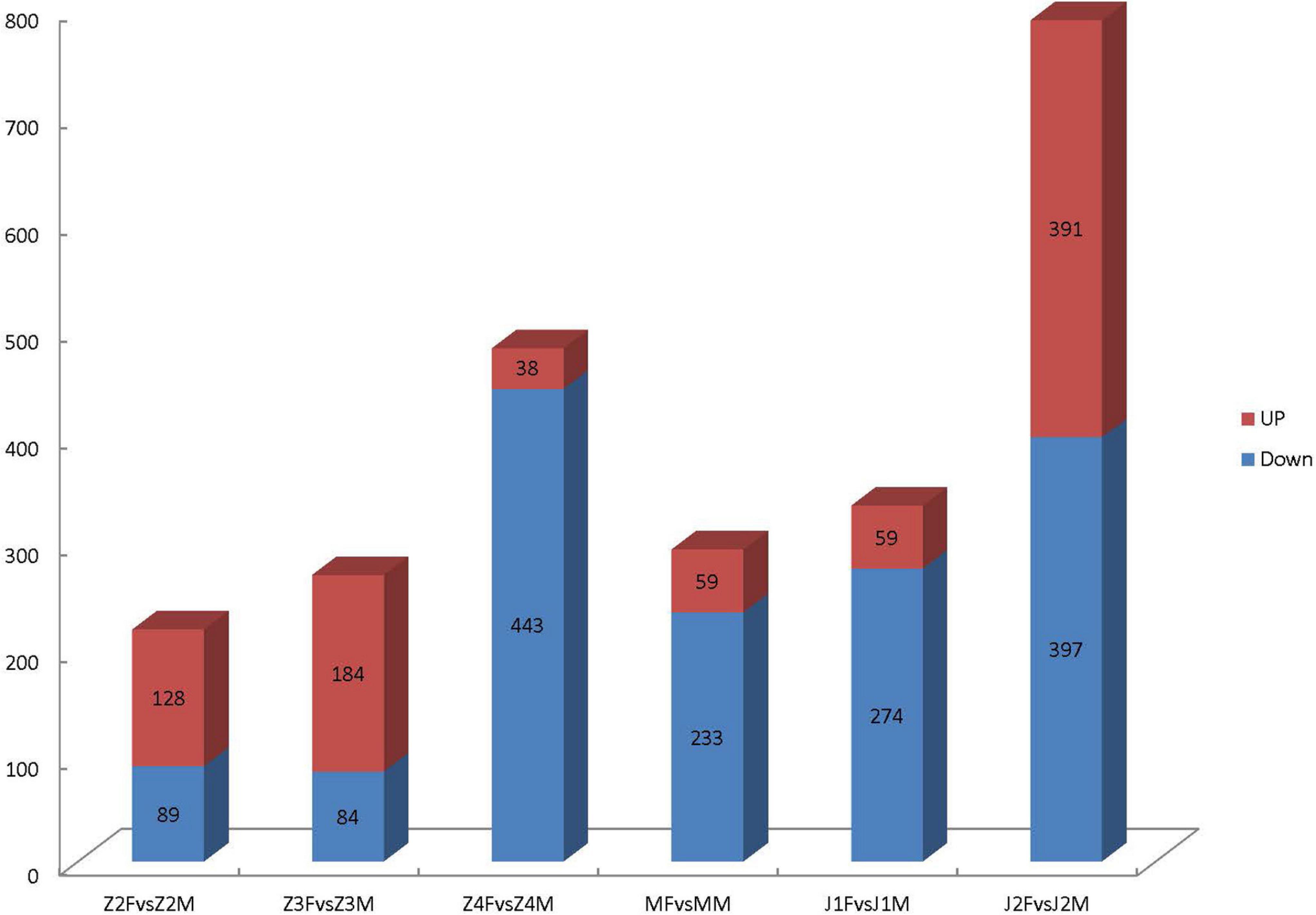
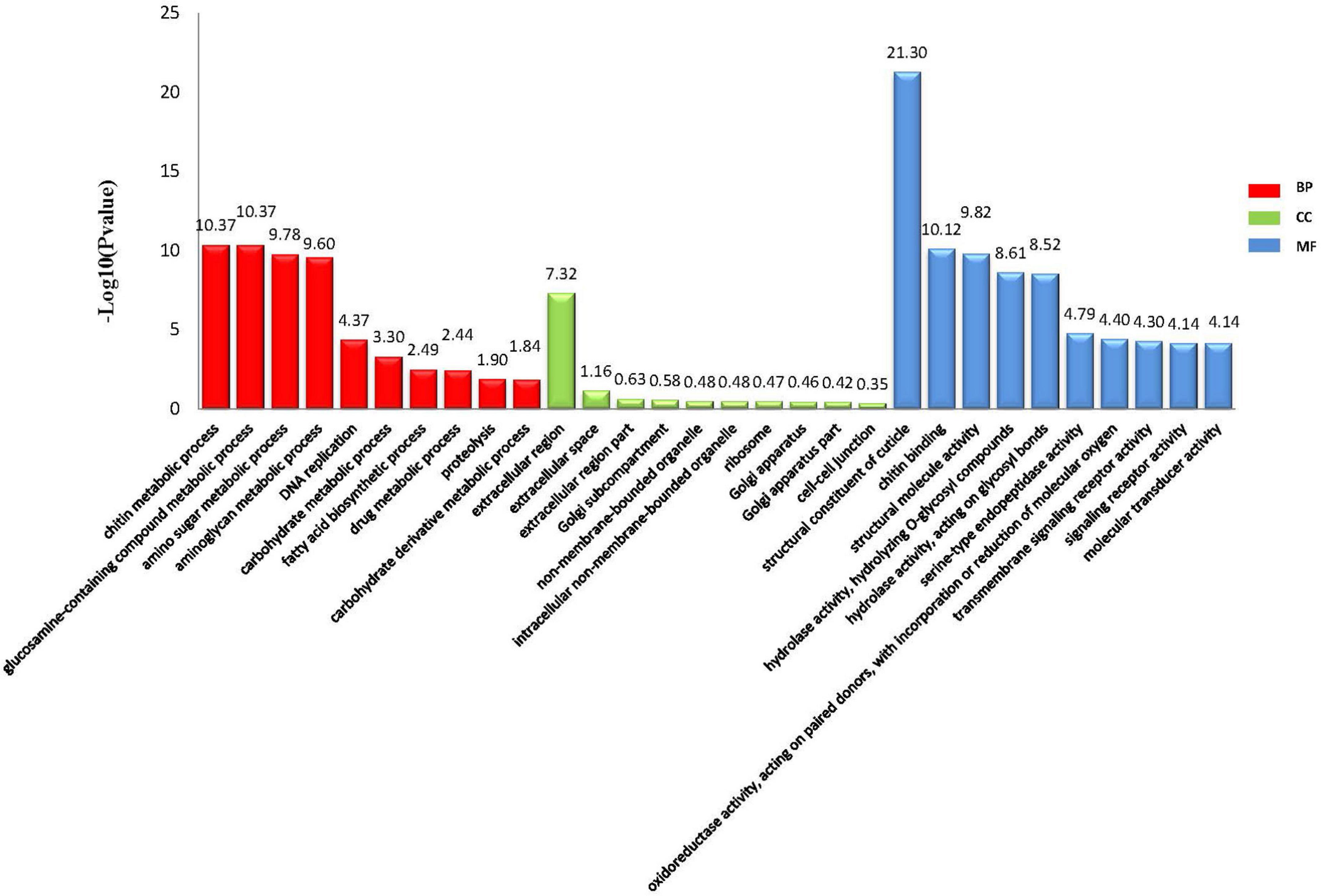
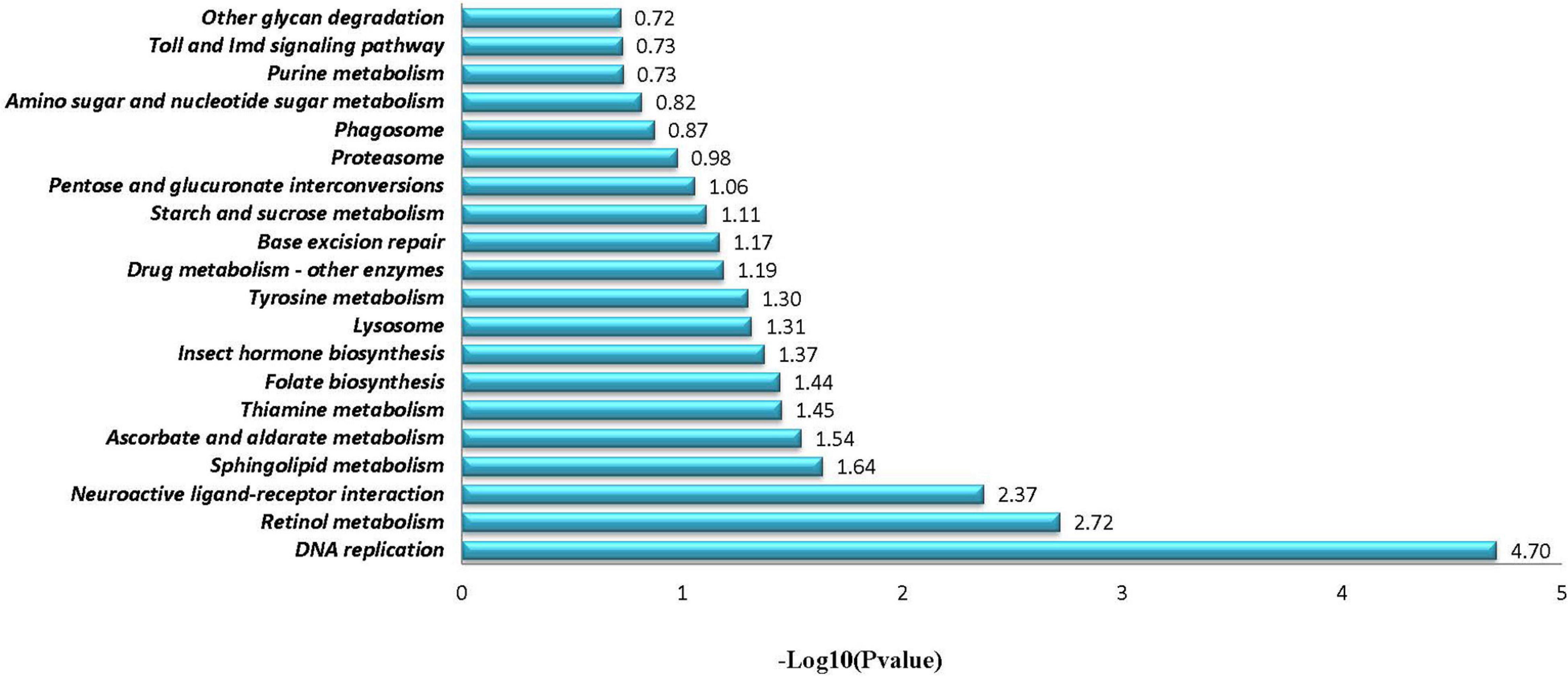
Through comparative analyses, a total of 2,379 significant DEGs were found between females and males (p-value ≤ 0.05 and | log2FoldChange| ≥ 1.0). The number of DEGs were highest in the Z4 and J2 periods, accounting for 20.22 and 33.12%, respectively (Figure 2). Gene Ontology (GO) enrichment analyses showed that chitin metabolic and glucosamine-containing compounds metabolic, among others, were the most significantly enriched (Figure 3). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses revealed that 20 pathways were enriched. The pathways for DNA replication, retinol metabolism, and neuroactive ligand-receptor interactions were the most significantly enriched (Figure 4).
Transcription Patterns of Genes in the Sex-Determining Region
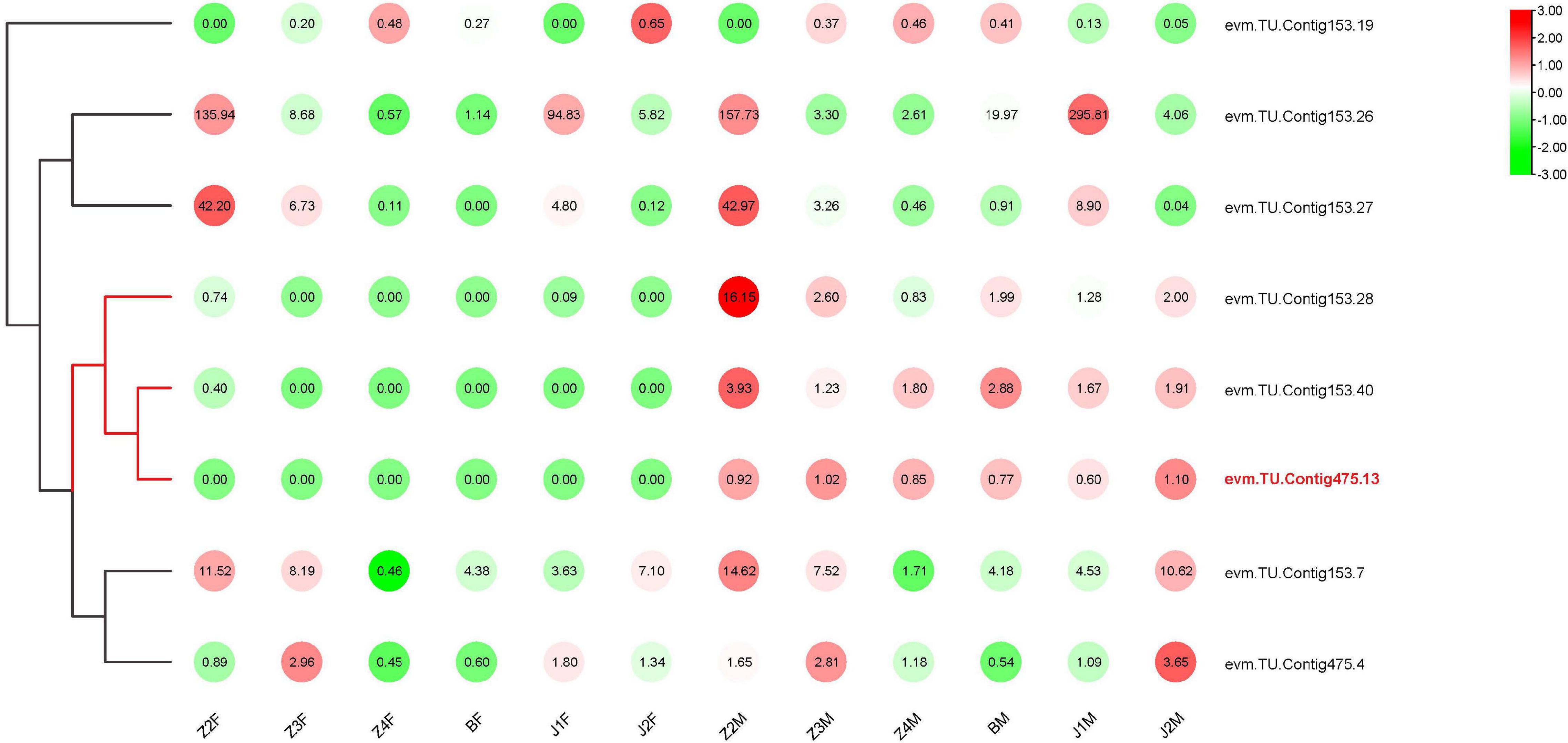
In our previous study, we located the SD region on two contigs in P. trituberculatus (Lv et al., 2020b). Based on the genomic annotations, 41 genes were found in the SD (Supplementary Table 2), among which, eight genes were significantly differentially expressed between females and males. Further analyses showed that the transcription of three genes in males was significantly higher than that in females at all stages of development (Figure 5). In particular, Contig 475.13 was expressed almost exclusively in males, and belonged to the Dmrt family (named PtDMY).
Expression Analysis of PtDMY
To further clarify the expression pattern of PtDMY in early development, qRT-PCR was performed using samples from 12 developmental periods from fertilized egg to juvenile crab, including Z1-J2 which were sex-specific. The results showed that PtDMY was almost undetectable in early embryonic development (Mc-G) and was up-regulated from the En phase to a peak at the Ez-Z1, before being down-regulated. It is worth noting that the expression of PtDMY in Z1-J2 was only detected in males, with almost no expression in females. Thus, those results were consistent with the transcriptome results (Figure 6).
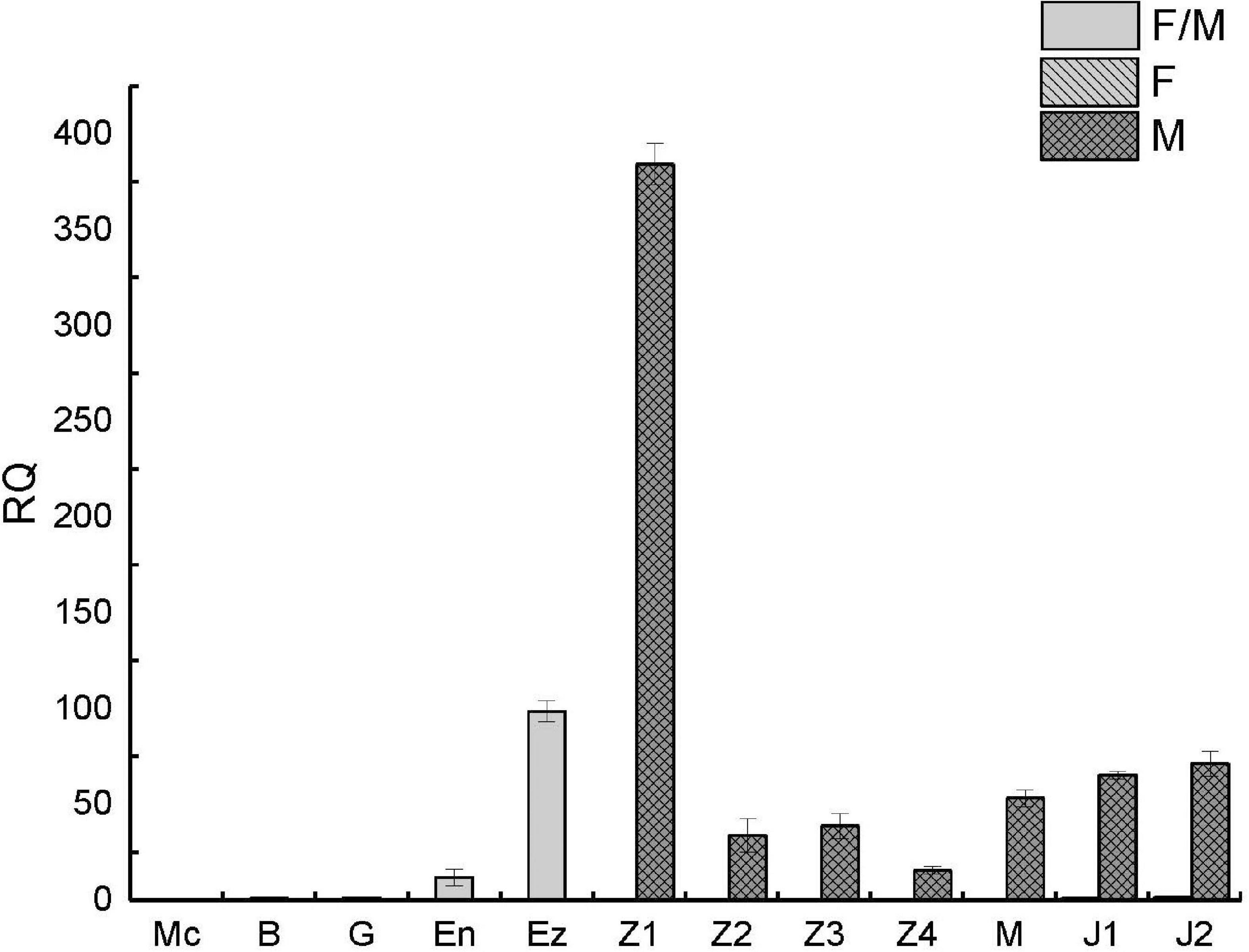
Figure 6. qRT-PCR of PtDMY at different developmental stages. Fertilized egg (Multicellular stage, Mc; Blastula stage, B; Gastrulation stage, G; Egg-nauplius stage, En; Egg-zoea stage, Ez), Zoea (Zoea 1, Z1; Zoea 2, Z2; Zoea 3, Z3; Zoea 4, Z4), Megalopa (M), and Juvenile crab (Juvenile crab 1, J1; Juvenile crab 2, J2).
RNAi for PtDMY
In this study, we successfully knocked down the expression of PtDMY in zoea larva after 48 h of RNAi (Figure 7). Furthermore, we analyzed the expression of known sex-related genes after PtDMY knockdown, including IAG, Dmrt, and SPATA. The results showed that most genes were significantly down-regulated, among which, IAG decreased by 78.48% and Dmrt decreased by an average of 66.49% (Figure 8).
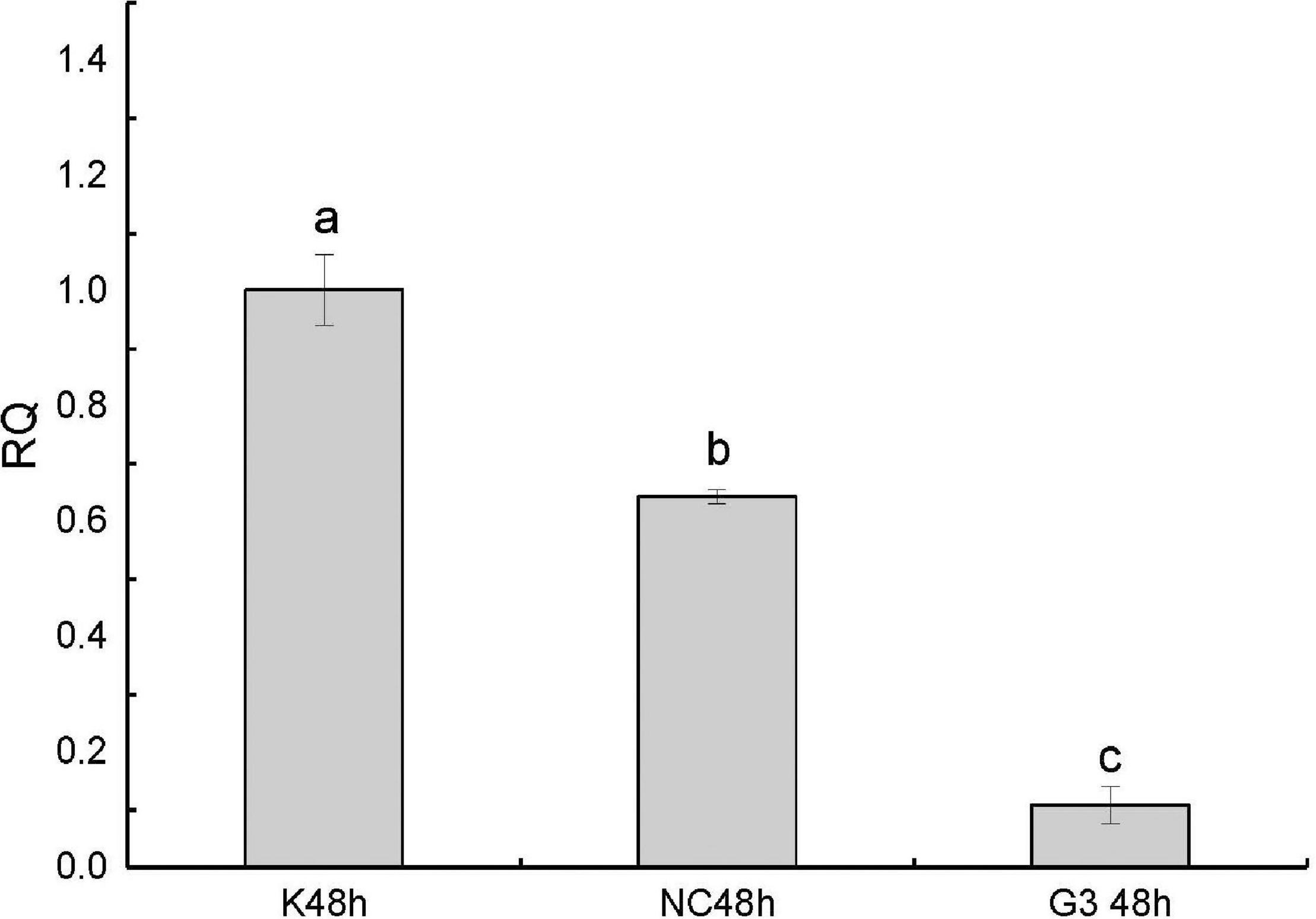
Figure 7. qRT-PCR of PtDMY following 48 h of RNAi. K48, Blank control without any treatment; NC48, Inject a meaningless chain; G3, Inject interference to knock down the PtDMY. Means with different letters are significantly different (P < 0.05).
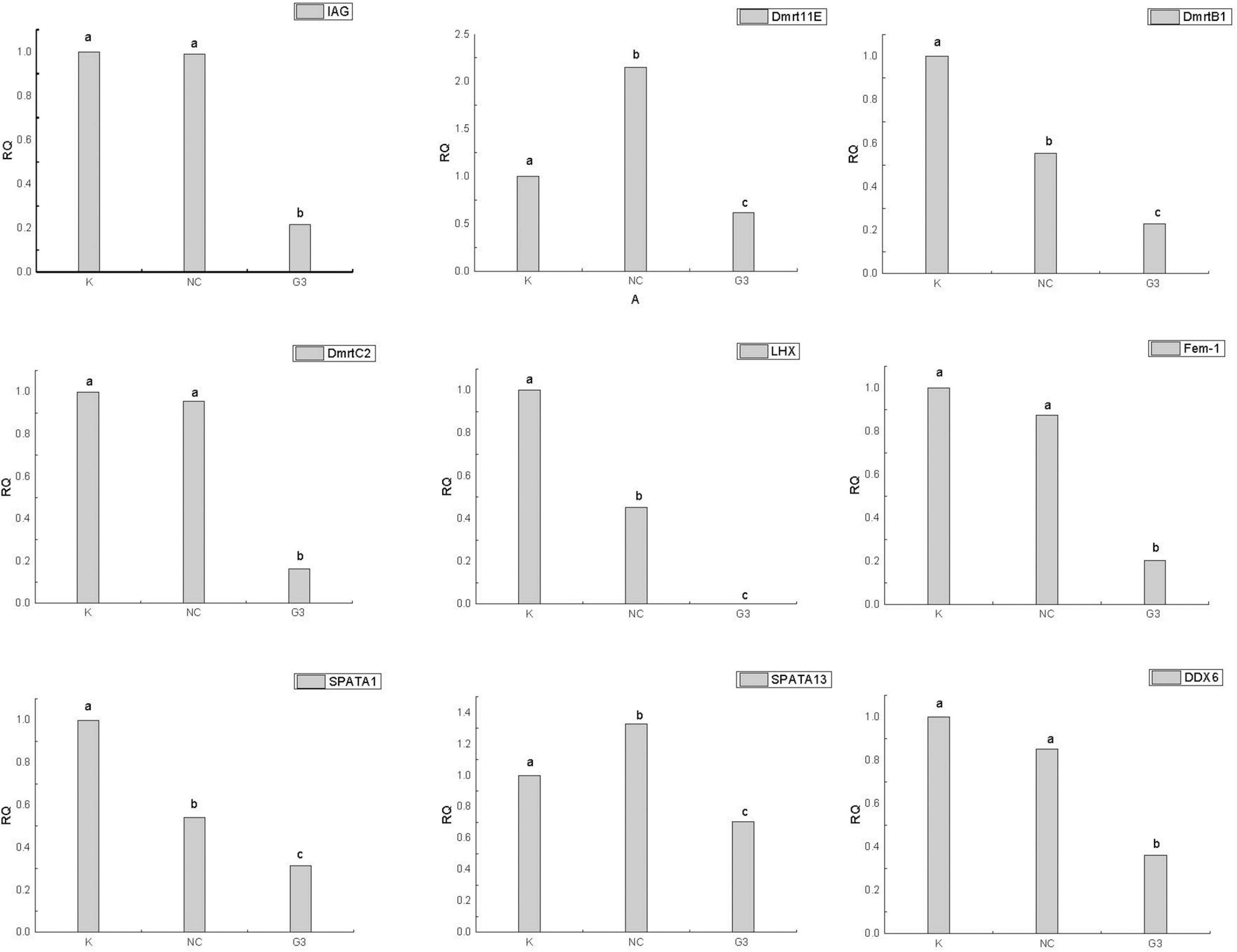
Figure 8. Expression of other sex-related genes after PtDMY knockdown. Means with different letters are significantly different (P < 0.05).
Discussion
Sex determination and differentiation are among the most interesting topics in biology, however, such processes remain unclear for most crustaceans (Wang et al., 2021). P. trituberculatus is one of the main mariculture crabs worldwide, with high economic value and differences in values between males and females. Thus, the identification of sex determination mechanisms in P. trituberculatus will facilitate sex control technologies for monosex breeding during production.
Sex determination in crustaceans generally occurs during early developmental stages (Aflalo et al., 2006). Therefore, sex-specific transcriptome sequencing at early developmental stage will provide the necessary information for the identification of genes related to sex determination. In fact, the sex determining genes in the silkworm moth, Bombyx mori, have been successfully identified using this approach (Takashi et al., 2014). Beside, DEGs between female and male L. vannamei at the Z1 stage have also been obtained using transcriptome sequencing (Wang et al., 2021). However, there have been no reports on sex-specific transcriptome sequencing at that early developmental stage in crabs, and thus, research on genetic sex markers is currently lacking. For the first time, this study sequenced and analyzed the sex-specific transcriptomes of crabs at early developmental stages. Compared to previous work in shrimp (Wang et al., 2021), our study included six developmental stages and obtained more sequencing data (272 G), which allowed us to identify sex-related genes more robustly.
A total of 2,379 DEGs were identified between female and male transcriptomes from Z2 to J2, and suggested that many genes may be involved in sex determination or differentiation. GO enrichment results showed that genes involved in chitin metabolic processes (GO:0006030), glucosamine-containing compound metabolic processes (GO:1901071), and amino sugar metabolic processes (GO:0006040) were the most significantly enriched in biological process (BP), which were related to molting, development, and growth (Kimura et al., 2008; Rocha et al., 2012). Crustacean growth requires molting, and the growth rate of male and female crabs differs between young crabs to adult crabs (Zhang et al., 2021). DEGs were enriched in processes related to molting and development, which indicated that the differentiation of growth and development of male and female crabs may have begun in the early stage of larval development.
The Dmrt, which is characterized by one or more highly conserved DNA-binding (DM) domains, was involved in sex determination (Meng et al., 1999). In Medaka (XX/XY sex-determination system), the Y-linked DMY gene was characterized as sex-determining genes, leading to male sexual development (Masaru et al., 2002). In X. laevis (ZZ/ZW sex-determination system), one W-linked DM-W gene was identified as a likely sex-determining gene (Yoshimoto et al., 2008). In Chicken and Flatfish (ZZ/ZW sex-determination system), Dmrt was located on the Z chromosome, and the level of expression may induce male development (Nanda et al., 2000; Chen et al., 2014). Recently, an invertebrate sex-linked Dmrt was found in Sagmariasus verreauxi (Chandler et al., 2017), suggesting that it also plays an important role in sex determination in crustaceans. In this work, one Dmrt gene in the SD showed obvious male-specific transcription, which was consistent with the characteristics of closely linked with males (Lv et al., 2020b). We named that gene PtDMY and hypothesized that it played a key role in sex determination.
To verify the function of PtDMY, we successfully knocked down its expression during larval development using RNAi technology. We found multiple known sex-related genes were significantly down-regulated after PtDMY knockdown, including IAG and Dmrt (Figure 8). Previous studies showed that IAG secreted by the AG is necessary for male genesis (Sun and Li, 2021), and is regulated by Dmrt in P. trituberculatus (Wang et al., 2020). It should be noted that three Dmrt genes were significantly down regulated after PtDMY knockdown, which strongly suggested that PtDMY may be a key upstream factor in the sex determination pathway. Furthermore, we systematically analyzed the expression pattern of PtDMY from the multicellular stage of crab to the juvenile stage. The results showed that its expression peaked in the Ez to Z1 stages (Figure 6), indicating that this period may be key for sex determination.
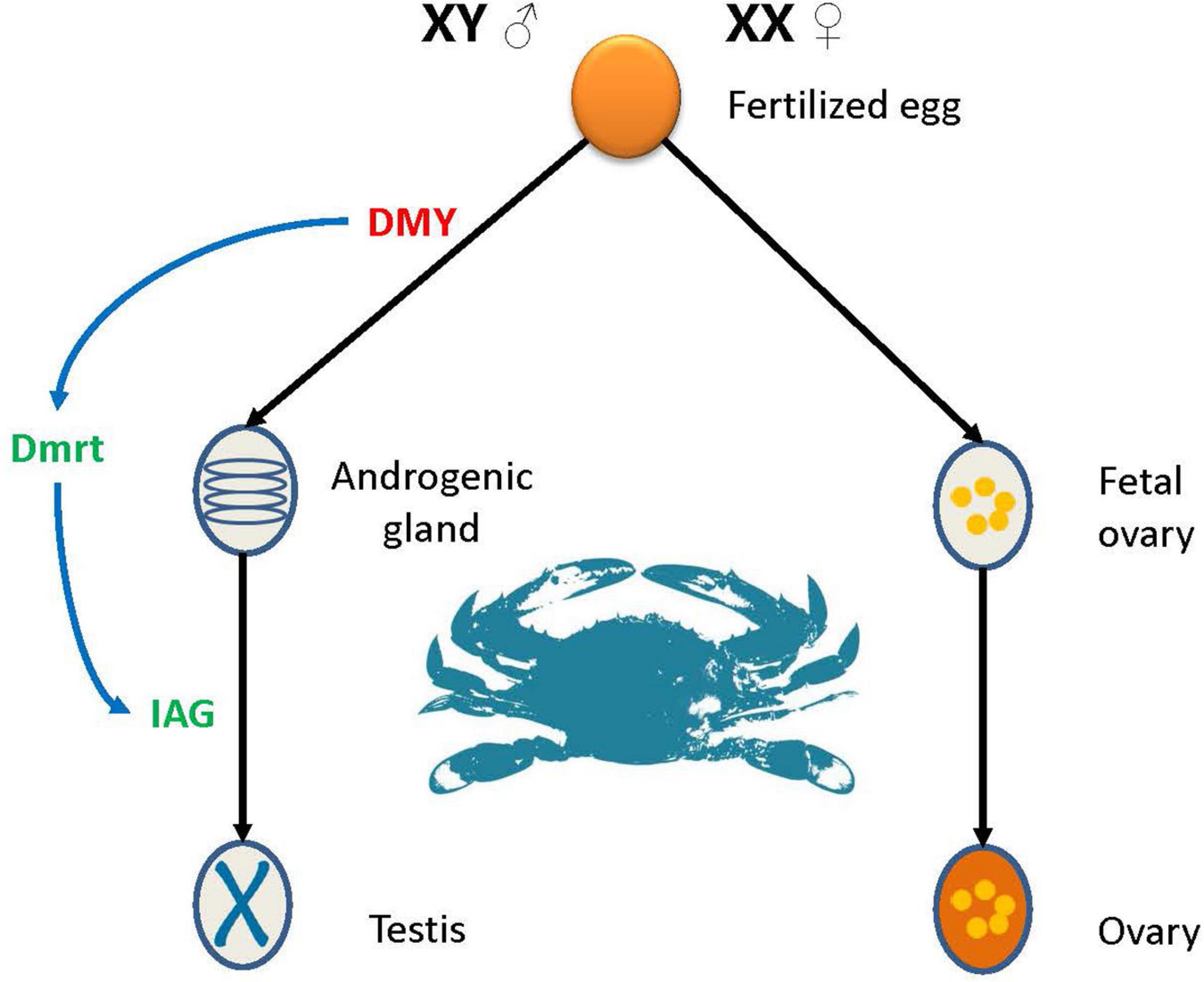
Interestingly, no genes were continuously highly expressed during female development. There are several possibilities for this observation: (1) The low expression leads to the female related genes have not been found; (2) the effect of genes related to female genesis is short, and there is no need for long-term high expression; or (3) there are no genes in P. trituberculatus that determine female sex. Combining the results of this and previous studies (Takashi et al., 2014; Wang et al., 2020), a model for sex determination and differentiation in P. trituberculatus was preliminarily outlined (Figure 9). The species belongs to the male heterogametic XY system (Lv et al., 2019), where PtDMY is the key gene for sex determination, and is expressed almost exclusively in males during larval development (Z1-J2). Dmrt and IAG are important sex differentiation-related genes, and are regulated by PtDMY. Notably, multiple aspects of the sex determination mechanism in P. trituberculatus remain unknown, and our research is just begin. In follow-up studies, further research is required to more fully detail the sex-determination pathway in crab.
Conclusion
In this work, sex-specific transcriptome sequencing at six larval development stages was performed and 2,379 DEGs were found between females and males. one Dmrt gene located in SD was exclusively expressed in males during early developmental stages, which was considered to be a key upstream regulators of the sex determination pathway. Our results here provided an important basis for clarifying the sex determination mechanisms of P. trituberculatus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA631920).
Ethics Statement
The animal study was reviewed and approved by the Yellow Sea Fisheries Research Institute. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
WZ conducted experiments, analyzed data and results, and manuscript writing. JL was experiment design, analyzed data and results, and manuscript revising. WL and BG conducted experiments. PL was experiment design and review manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key R&D Program of China (2018YFD0900303), the National Natural Science Foundation of China (42076116), and the Ministry of Agriculture of China and the China Agriculture Research System (CARS-48).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.811052/full#supplementary-material
Supplementary Table 1 | Primers used in this study.
Supplementary Table 2 | Gene information of the 41 genes in the SD region.
Supplementary Table 3 | Annotation information for the detected sex-related genes.
References
Aflalo, E. D., Hoang, T. T. T., Nguyen, V. H., Lam, Q., Nguyen, D. M., Trinh, Q. S., et al. (2006). A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 256, 468–478. doi: 10.1016/j.aquaculture.2006.01.035
Alwi, W., Firdaus, M. L., Syakura, H., Arief, S., Darajat, S., and Ghufira. (2015). “Utilization of shrimp shells as biosorbent to remove heavy metal Cu and Zn in aquatic samples,” in Proceedings of the 3rd International Conference on Biological, Chemical & Environmental Sciences (BCES-2015), Kuala Lumpur, 5–8.
Chandler, J. C., Fitzgibbon, Q. P., Smith, G. S., Elizur, A., and Ventura, T. (2017). Y-linked iDmrt1 paralogue (iDMY) in the Eastern spiny lobster, Sagmariasus verreauxi: the first invertebrate sex-linked Dmrt. Dev. Biol. 430, 337–345. doi: 10.1016/j.ydbio.2017.08.031
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole-genome sequence of a Flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253–260. doi: 10.1038/ng.2890
Cui, Z., Hui, M., Liu, Y., Song, C., Li, X., Li, Y., et al. (2015). High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 115, 206–215. doi: 10.1038/hdy.2015.26
Fang, S., Zhang, Y., Shi, X., Zheng, H., Li, S., Zhang, Y., et al. (2020). Identification of male-specific SNP markers and development of PCR-based genetic sex identification technique in crucifix crab (Charybdis feriatus) with implication of an XX/XY sex determination system. Genomics 112, 404–411. doi: 10.1016/j.ygeno.2019.03.003
Feng, B., Li, S., Wang, Q., Tang, L., Huang, F., Zhang, Z., et al. (2021). lncRNA DMRT2-AS acts as a transcriptional regulator of Dmrt2 involving in sex differentiation in the Chinese tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 253:110542. doi: 10.1016/J.CBPB.2020.110542
Han, C., Wang, C., Ouyang, H., Zhu, Q., Huang, J., Han, L., et al. (2021). Characterization of Dmrts and their potential role in gonadal development of mandarin fish (Siniperca chuatsi). Aquac. Rep. 21:100802. doi: 10.1016/j.aqrep.2021.100802
Kimura, K., Iwata, H., and Thompson, J. G. (2008). The effect of glucosamine concentration on the development and sex ratio of bovine embros. Anim. Reprod. Sci. 103, 228–238. doi: 10.1016/j.anireprosci.2006.12.014
Lezer, Y., Aflalo, E. D., Manor, R., Sharabi, O., Abilevich, L. K., and Sagi, A. (2015). On the safety of RNAi usage in aquaculture: the case of all-male prawn stocks generated through manipulation of the insulin-like androgenic gland hormone. Aquaculture 435, 157–166. doi: 10.1016/j.aquaculture.2014.09.040
Li, C., Li, G., Wang, Y., and Peng, Y. (2020). Development of SSR markers based on transcriptome sequences of the wolf spider Pardosa pseudoannulata (Araneae: lycosidae). Entomol. News 129, 6–15. doi: 10.3157/021.129.0102
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I, Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lv, J., Lu, X., Ti, X., Liu, P., Li, J., and Li, J. (2020a). QTL mapping and marker identification for sex determination in the ridgetail white prawn, Exopalaemon carinicauda. Genomics 112, 5240–5247. doi: 10.1016/j.ygeno.2020.09.037
Lv, J., Su, Z., Gao, B., Ti, X., Yan, D., Liu, G., et al. (2020b). A chromosome-level genome of Portunus trituberculatus provides insights into its evolution, salinity adaptation, and sex determination. Authorea 1–39. doi: 10.22541/au.159646761.15797764
Lv, J., Sun, D., Huan, P., Song, L., Liu, P., and Li, J. (2018). QTL mapping and marker identification for sex-determining: indicating XY sex determination system in the Swimming Crab (Portunus trituberculatus). Front. Genet. 9:337. doi: 10.3389/fgene.2018.00337
Lv, J., Sun, D., Yan, D., Ti, X., Liu, P., and Li, J. (2019). Quantitative trait loci mapping and marker identification for low salinity tolerance trait in the Swimming Crab (Portunus trituberculatus). Front. Genet. 10:1193. doi: 10.3389/fgene.2019.01193
Masaru, M., Yoshitaka, N., Ai, S., Tadashi, S., Chika, M., Tohru, K., et al. (2002). DMY is a Y-specific DM-domain gene required for male development in the Medaka fish. Nature 417, 559–563. doi: 10.1038/nature751
Meng, A., Moore, B., Tang, H., Yuan, B., and Lin, S. (1999). A Drosophila doublesex-related gene, terra, is involved in somitogenesis in vertebrates. Development 126, 1259–1268.
Nanda, I., Zend-Ajusch, E., Shan, Z., Grutzner, F., Schartl, M., Burt, D. W., et al. (2000). Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: a comparative (re)view on avian sex determination. Cytogenet. Cell Genet. 89, 67–78. doi: 10.1159/000015567
Naoki, O., and Makoto, T. (2021). Transcriptional regulation of the Y-linked mammalian testis-determining gene SRY. Sex Dev. 15, 351–359. doi: 10.1159/000519217
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with hisat, stringtie and ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Prokop, J. W., Chhetri, S. B., van Veen, J. E., Chen, X., Underwood, A. C., Uhl, K., et al. (2020). Transcriptional analysis of the multiple Sry genes and developmental program at the onset of testis differentiation in the Rat. Biol. Sex Differ. 11:28. doi: 10.1186/s13293-020-00305-8
Rocha, J., Garcia-Carreno, F. L., Muhlia-Almazan, A., Peregrino-Uriarte, A. B., Yepiz-Plascencia, G., and Cordova-Murueta, J. H. (2012). Cuticular chitin synthase and chitinase mRNA of whiteleg shrimp Litopenaeus vannamei during the molting cycle. Aquaculture 330-333, 111–115. doi: 10.1016/j.aquaculture.2011.12.024
Shi, X., Waiho, K., Li, X., Ikhwanuddin, M., Miao, G., Lin, F., et al. (2018). Female-specific SNP markers provide insights into a WZ/ZZ sex determination system for mud crabs Scylla paramamosain, S. tranquebarica and S. serrata with a rapid method for genetic sex identification. BMC Genomics 19:981. doi: 10.1186/s12864-018-5380-8
Sun, R., and Li, Y. (2021). A sex-reversing factor: insulin-like androgenic gland hormone in decapods. Rev. Aquac. 13, 1352–1366. doi: 10.1111/raq.12525
Takashi, K., Hikaru, K., Munetaka, K., Keisuke, S., Hiroki, S., Yuji, A., et al. (2014). A single female-specific piRNA is the primary determiner of sex in the Silkworm. Nature 509, 633–636. doi: 10.1038/nature13315
Torrecilla, Z., Martinez-Lage, A., Perina, A., Gonzalez-Ortegon, E., and Gonzalez-Tizon, A. M. (2017). Comparative cytogenetic analysis of marine palaemon species reveals a X1X1X2X2/X1X2Y sex chromosome system in Palaemon elegans. Front. Zool. 14:47. doi: 10.1186/s12983-017-0233-x
Toyota, K., Miyakawa, H., Hiruta, C., Sato, T., Katayama, H., Ohira, T., et al. (2021). Sex determination and differentiation in decapod and Cladoceran crustaceans: an overview of endocrine regulation. Genes 12:305. doi: 10.3390/GENES12020305
Wang, Y., Xie, X., Wang, M., Zheng, H., Zheng, L., and Zhu, D. (2020). Molecular characterization and expression analysis of the inverbrate Dmrt1 homologs in the Swimming Crab, Portunus Trituberculatus (MIERS, 1876) (Decapoda, Portunidae). Crustaceana 93, 851–866. doi: 10.1163/15685403-bja10025
Wang, Y., Yu, Y., Li, S. H., Zhang, X. J., Xiang, J. H., and Li, F. H. (2021). Sex-specific transcriptome sequencing of Zoea I larvae and identification of sex-linked genes using bulked segregant analysis in pacific white shrimp Litopenaeus vannamei. Mar. Biotechnol. 22, 423–432. doi: 10.1007/S10126-021-10038-3
Yoshimoto, S., Okada, E., Umemoto, H., Tamura, K., Uno, Y., Nishida-Umehara, C., et al. (2008). A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 105, 2469–2474. doi: 10.1073/pnas.0712244105
Yu, Y., Zhang, X., Yuan, J., Li, F., Chen, X., Zhao, Y., et al. (2015). Genome survey and high-density genetic map construction provide genomic and genetic resources for the pacific white shrimp Litopenaeus vannamei. Sci. Rep. 5:15612. doi: 10.1038/srep15612
Keywords: Portunus trituberculatus, transcriptome, sex-determining, Dmrt, PtDMY
Citation: Zhang W, Lv J, Lan W, Gao B and Liu P (2022) Discovery of Sex-Determining Genes in Portunus trituberculatus: A Comparison of Male and Female Transcriptomes During Early Developmental Stages. Front. Mar. Sci. 8:811052. doi: 10.3389/fmars.2021.811052
Received: 08 November 2021; Accepted: 14 December 2021;
Published: 05 January 2022.
Edited by:
Yuan Liu, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Changkao Mu, Ningbo University, ChinaQiong Shi, Beijing Genomics Institute (BGI), China
Copyright © 2022 Zhang, Lv, Lan, Gao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Liu, bGl1cGluZ0B5c2ZyaS5hYy5jbg==; Jianjian Lv, bHZqakB5c2ZyaS5hYy5jbg==
†These authors have contributed equally to this work
 Wen Zhang
Wen Zhang Jianjian Lv
Jianjian Lv Weikang Lan1,3
Weikang Lan1,3 Ping Liu
Ping Liu