- 1Department of Biology, Boston University, Boston, MA, United States
- 2Department of Biology and Marine Biology, Center for Marine Science, University of North Carolina Wilmington, Wilmington, NC, United States
- 3Department of Biology, Marine Biology, and Environmental Science, Roger Williams University, Bristol, RI, United States
Global change is increasing seawater temperatures and decreasing oceanic pH, driving declines of coral reefs globally. Coral ecosystems are also impacted by local stressors, including microplastics, which are ubiquitous on reefs. While the independent effects of these global and local stressors are well-documented, their interactions remain less explored. Here, we examine the independent and combined effects of global change (ocean warming and acidification) and microplastics exposures on gene expression (GE) and microbial community composition in the endangered coral Acropora cervicornis. Nine genotypes were fragmented and maintained in one of four experimental treatments: 1) ambient conditions (ambient seawater, no microplastics; AMB); 2) microplastics treatment (ambient seawater, microplastics; MP); 3) global change conditions (warm and acidic conditions, no microplastics; OAW); and 4) multistressor treatment (warm and acidic conditions with microplastics; OAW+MP) for 22 days, after which corals were sampled for genome-wide GE profiling and ITS2 and 16S metabarcoding. Overall A. cervicornis GE responses to all treatments were subtle; however, corals in the multistressor treatment exhibited the strongest GE responses, and genes associated with innate immunity were overrepresented in this treatment. ITS2 analyses confirmed that all coral were associated with Symbiodinium ‘fitti’ and 16S analyses revealed similar microbiomes dominated by the bacterial associate Aquarickettsia, suggesting that these A. cervicornis fragments exhibited remarkably low variability in algal and bacterial community compositions. Future work should focus on functional differences across microbiomes, especially Aquarickettsia and viruses, in these responses. Overall, results suggest that when local stressors are coupled with global change, these interacting stressors present unique challenges to this endangered coral species.
1 Introduction
Anthropogenic global change represents one of the greatest scientific challenges of our time. As atmospheric concentrations of carbon dioxide (CO2) continue to increase, the world’s ecosystems face unprecedented challenges from the resulting warming, sea level rise, and extreme weather events (Rosenzweig et al., 2008). The effects of global change are being documented across ecosystems, from declines in terrestrial plant diversity (Harrison, 2020) to shifts in species distributions (Cristofari et al., 2018). However, marine environments are faced with additional threats not present in terrestrial ecosystems (e.g., sea-level rise and ocean acidification), and these threats are especially pressing for tropical coral reefs (Hoegh-Guldberg and Bruno, 2010).
Coral reef ecosystems are experiencing major declines under global change stressors, especially ocean warming that causes the breakdown of the symbiosis between the coral host and dinoflagellate algae [family Symbiodiniaceae; (Muller-Parker et al., 2015; LaJeunesse et al., 2018)]. These declines are particularly evident across the Caribbean where reefs have experienced significant coral loss since the late 20th century, largely due to major disease outbreaks, overfishing leading to macroalgal blooms, and thermal stress events (Contreras-Silva et al., 2020; Bove et al., 2022b; Randazzo-Eisemann et al., 2022). In addition to warming, ocean acidification represents a significant challenge to reef-building corals that can lead to reduced growth rates, dissolution of existing reef framework, and reduced holobiont (animal host, algal symbionts, bacteria, viruses, etc.) physiology and metabolic rates (Anthony et al., 2008; Aichelman et al., 2021; Cornwall et al., 2021). While responses of corals under ocean acidification and warming can vary both within and across species (Bove et al., 2022a), it is clear these global stressors represent immediate threats to the future of coral reefs.
Along with changing oceanic conditions, coral reefs are susceptible to pollution via terrestrial input, which includes microplastics (Hall et al., 2015; Soares et al., 2020; Hankins et al., 2021; Oldenburg et al., 2021). Microplastics are small plastics, such as beads or fibers, smaller than 5 mm, that come from a variety of sources, including synthetic clothing and personal care products, that enter the waterways via runoff and wastewater treatment (Ivar do Sul and Costa, 2014). Microplastics are ubiquitous across coastal ecosystems, and pose risk to ecosystems via adsorbed chemical pollutants and novel microbiomes (Browne et al., 2011; Rochman et al., 2019). Microplastic particles have been found within coral gastrovascular cavities, suggesting corals ingest plastics (Hall et al., 2015; Hankins et al., 2018; Rotjan et al., 2019). Active ingestion of microplastic particles can reduce coral growth (Reichert et al., 2019; Hankins et al., 2021; Huang et al., 2021) directly via blockages of coral digestive cavities (Allen et al., 2017). Further, interaction with microplastics may impact the energetic budget of the coral, altering polyp behavior and feeding, impeding gamete fertilization success, and affecting the holobiont immune system and coral disease prevalence and susceptibility (Tang et al., 2018; Berry et al., 2019; Rotjan et al., 2019; Hankins et al., 2021; Huang et al., 2021). Passive surface interactions/surface adhesion with the plastics (Reichert et al., 2018) can cause abrasion and injury, increasing coral susceptibility to infection and disease (Page and Willis, 2008; Lamb et al., 2016). While the impacts of microplastics on corals are an active area of research, little is known about how this emerging stressor interacts with other stressors, especially ocean warming and acidification.
Of the work to date examining global change stressors and microplastics, results are variable and species dependent (Axworthy and Padilla-Gamiño, 2019; Huang et al., 2021). For example, recent work found that warming led to consistent reductions in fitness-related traits in corals, while microplastics resulted in mixed responses (Reichert et al., 2021). Similarly, another study found that microplastics had no effect on Symbiodiniaceae cell density under ambient or elevated temperatures (Plafcan and Stallings, 2022), suggesting that microplastics may represent a minor stressor when compared to warming. Little is known about the combined effects of ocean warming and microplastics on corals and even less work has explored how ocean acidification may interact with microplastic pollution to impact corals. Previous work in other marine invertebrates reports impaired immunity in adult mussels (Huang et al., 2022) and altered larval development in urchins (Bertucci et al., 2022) in response to microplastics and ocean acidification, suggesting that the interaction of these stressors may exacerbate coral stress.
One way to gain a more mechanistic understanding of how an organism is responding to a stressor is to profile their gene expression in response to multiple stressors [reviewed in Rivera et al., (2021)]. Currently, gene expression responses of corals to the combined effects of global change and microplastics remain underexplored. Corals have been shown to shuffle their algal symbionts in response to a variety of stressors (Ros et al., 2021; Rodriguez-Casariego et al., 2022), representing a potential acclimation strategy for corals under stress. In addition to their obligate, intracellular photosymbionts (Symbiodiniaceae), corals maintain diverse but specific microbiomes consisting of bacteria, archaea, fungi, viruses, and protists (Bourne et al., 2016; van Oppen and Blackall, 2019). Research has demonstrated that coral health and survival is mediated by their microbiomes (Glasl et al., 2016; Ziegler et al., 2017; Ricci et al., 2019), and that exposure to stressors is reflected in dysbiosis, or a microbiome-wide disturbance/shift in taxonomic composition (Zaneveld et al., 2016; Apprill, 2017; McDevitt-Irwin et al., 2017). Laboratory experiments have demonstrated the capacity of microplastic particles to transmit bacteria into corals via particle ingestion (Rotjan et al., 2019). Moreover, microplastics are known to harbor microbial biofilms that are taxonomically distinct [the “plastisphere”, (Zettler et al., 2013; Amaral-Zettler et al., 2020)] from those found in naturally occurring particles suspended in seawater, and bacteria associated with coral diseases have been detected on microplastic pieces (Goldstein et al., 2014; Feng et al., 2020). However, the implications of exposure to microplastics and microplastics-associated microbes on the composition and activity of the coral microbiome, including vectoring pathogens to corals, is not yet well understood.
Understanding how all of the members of the coral holobiont respond in highly endangered coral species, such as Acropora cervicornis, remains a top research priority given the current rates of coral decline (Miller et al., 2002; Schutte et al., 2010; Contreras-Silva et al., 2020). Caribbean A. cervicornis populations are experiencing dramatic declines and are the focus of major restoration efforts (Schopmeyer et al., 2017; Ware et al., 2020). Disease susceptibility in A. cervicornis appears to be mediated by host genotype, environment, and the microbiome (Klinges et al., 2020; Williams et al., 2022). The recently described “Candidatus Aquarickettsia rohweri” (hereafter, Aquarickettsia) (Klinges et al., 2019) is extremely widespread across Caribbean acroporids and can represent as much as 99% of the detected sequences in 16S-based studies (Klinges et al., 2020). Aquarickettsia appears to be parasitic to acroporids and is likely an indicator and determinant of disease susceptibility. Conversely, taxa from the genus Endozoicomonas appear to be significantly and directly correlated with disease resistance and resilience (Chu and Vollmer, 2016). After bleaching/thermal stress, Aquarickettsia abundance declines dramatically in the host, suggesting that disease is the indirect result of Aquarickettsia depletion of holobiont nutritional deficiencies, providing a niche for opportunistic pathogens during/following thermal stress (Klinges et al., 2020). Understanding how the microbiomes of these important reef-building corals are involved in holobiont response to stressors is an important consideration for informed conservation, restoration, and management (Ware et al., 2020).
Here, we reared fragments of Acropora cervicornis under projected global change (ocean acidification and warming), microplastic pollution, and their interaction for 22 days. We assessed responses of the holobiont to these stressors via coral gene expression profiling, Symbiodiniaceae community ITS2 metabarcoding, and microbiome community 16S metabarcoding. Overall, we hypothesized that the combined stressor treatment (ocean acidification, warming, and microplastics) would elicit the strongest responses by all holobiont members given that these stressors may interact to cause cellular disruption and limit heterotrophy through microplastic ingestion.
2 Materials and methods
2.1 Coral collection and experimental design
In June 2020, Nova Southeastern University staff collected nine putative Acropora cervicornis genotypes off coastal Fort Lauderdale, FL, USA from the Nova Southeastern University in situ coral nursery (Florida Fish and Wildlife Conservation Commission permit #SAL-19-2200A-SRP). These genotypes were maintained in outdoor tanks for one week in ambient conditions, after which they were transported to the University of North Carolina Wilmington. Fragments were acclimated to recirculating laboratory tank conditions (27.6–28.9 ˚C; 34–36 ppt salinity) for three months. Two weeks before the start of the experiment, four branches (~5 cm) from each genotype were glued to ceramic plugs, allowed to recover for one week, and then acclimated to individual experimental tanks for one week.
Coral fragments were randomly assigned to one of four experimental treatments (mean ± SD): 1) ambient treatment (AMB; temperature: 28.6 ± 0.03°C, pH: 8.07 ± 0.07), 2) microplastics treatment (MP; temperature: 28.6 ± 0.03°C, pH: 8.06 ± 0.06; 1.25 x 10-4 g/ml of microplastics), 3) ocean acidification and warming treatment (OAW; temperature: 30.1 ± 0.57°C, pH: 7.96 ± 0.08), and 4) multistressor treatment (OAW+MP; temperature: 30.1 ± 0.53°C, pH: 7.95 ± 0.11; 1.25 x 10-4 g/ml of microplastics) (Figure 1; see Supplemental Table S1 for details on seawater conditions at tank and treatment levels). Ambient seawater temperature and pH were based on oceanic averages in Fort Lauderdale, FL, while OAW conditions were intermediate predictions for 2075 following IPCC data (Shukla et al., 2019). One branch from each genotype (N=9) was placed in each treatment, resulting in a total of 36 samples. Each fragment was housed in its own non-recirculating 37.85-liter experimental tank that received frequent water changes and thus can be considered a true biological replicate.
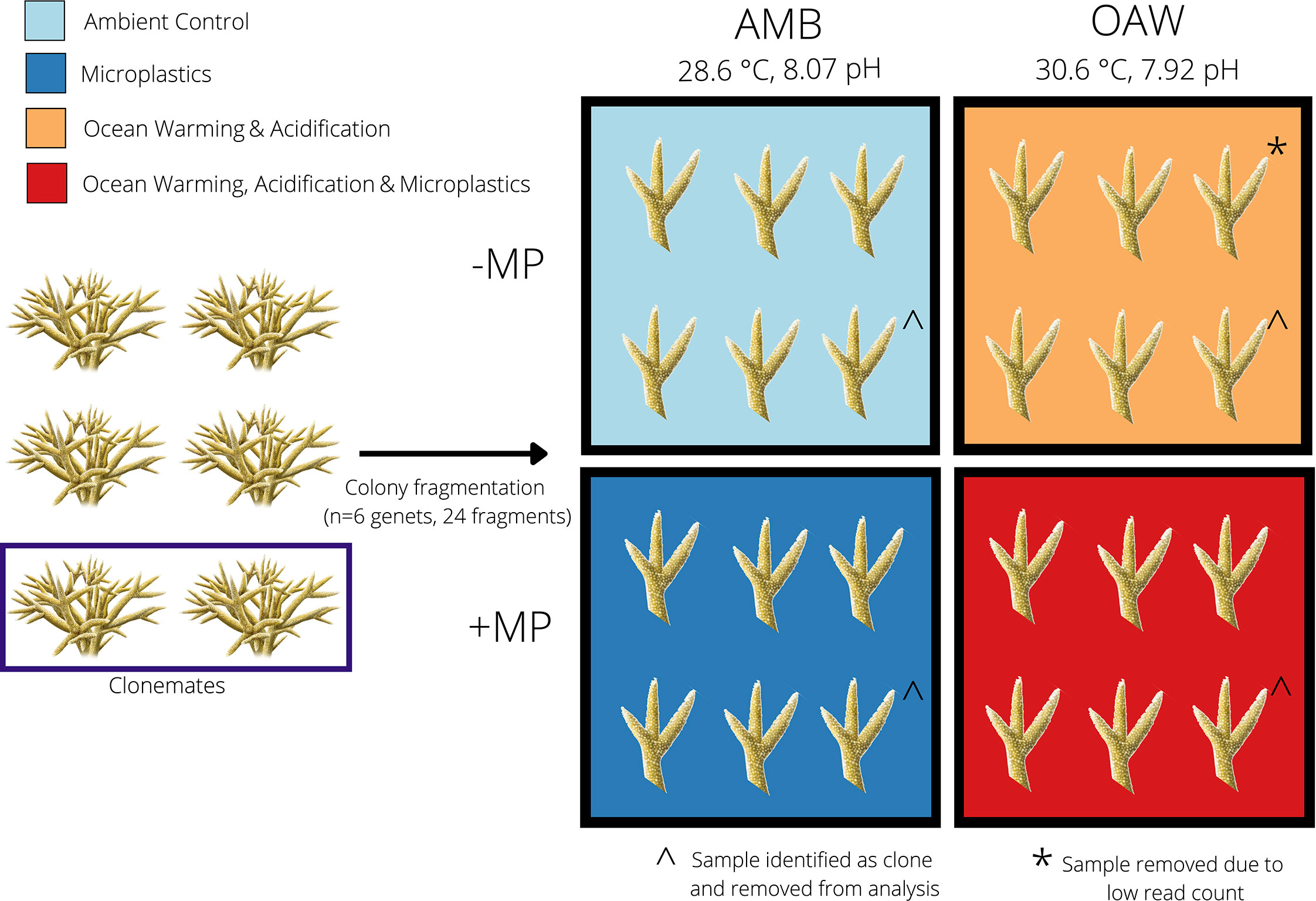
Figure 1 Experimental design to test the effects of ocean warming, acidification, and microplastics on the endangered coral Acropora cervicornis. Four fragments from each of six A. cervicornis genotypes that were used in downstream sequencing were assigned one of four experimental treatments: ambient conditions (AMB; blue), ocean acidification and warming (OAW; orange), microplastics (MP; dark blue), and OAW and MP (OAW+MP; red). Fragments were maintained individually in isolated tanks not depicted in this diagram. The dark blue rectangle (left) showcases two corals that were identified as clones via SNP calling from TagSeq gene expression data and the ^ symbol indicates the clone pair that was removed. The asterisk (*) indicates one gene expression library that was removed from downstream analysis due to low read counts.
The microplastic treatments were achieved by weighing out UV-fluorescent blue polyethylene microspheres in equal amounts of two different size classes (density: 1.13 g/cc, Cospheric, LLC; diameters: 180-212 µm and 355-425 µm) to obtain a concentration of 1.25 x 10-4 g ml-1 per tank. This microplastic concentration was selected based on previous studies to represent moderate microplastic exposure and two size classes were used to ensure the corals could ingest the plastics, which would make the treatment more ecologically relevant (Reichert et al., 2018). The microplastics were added to clean tripour containers within each experimental tank to allow the microplastics to acquire a biofilm over 48 hours before a single coral fragment was placed into each tripour container. The microplastics were then gently resuspended within the individual tripours three times a day using a turkey baster. All tripours were treated the same and this process was repeated throughout the experiment to maintain microplastic densities within each replicate.
Water temperature and pH were controlled using Neptune Systems’ Apex microcontrollers. Each tank contained a pH probe, temperature probe, and aquarium heater to control individual tank conditions. The OAW and OAW+MP tanks also each contained a tube for bubbling in CO2 to control the pH in these systems. The Neptune System allowed for precise control of individual tank parameters by modulating the aquarium heaters and the solenoids connected to the CO2 tubes to achieve desired treatment conditions. Fragments in OAW/OAW+MP treatments were acclimated by starting at 28.6°C and then raising the temperature by 0.5°C each day until 30.5°C was achieved. Similarly, the pH in OAW/OAW+MP treatments started at 8.05 and was reduced by 0.05 pH units each day until 7.94 was reached. Corals were maintained at these experimental conditions for 22 days while monitoring salinity, temperature, pH, and dissolved oxygen daily (Supplemental Figure 1 and Table S1). Ammonia, nitrate, phosphate, and alkalinity were also measured daily in two randomly selected experimental tanks, one ambient and one ocean acidification and warming. Lights were on a 12:12 light:dark schedule with a PAR of 195 μmol m2 S-1 that is similar to those recorded for waters across southeast Florida (Yates et al., 2019). All fragments were fed Reef-Roids coral food following manufacturer’s instructions (2 mL aliquots) three times weekly and 30% water changes were conducted for all tanks the day after feeding. Immediately following the 3-week experiment, tissue samples (1 cm) from each fragment were placed in cryovials, flash frozen in liquid nitrogen, and maintained at –80°C before being transported to Boston University for downstream analyses.
2.2 RNA isolation and TagSeq library preparation
Tissue samples were crushed with a razor blade and RNA was isolated using RNAqueous kits (ThermoFisher Scientific) following manufacturer instructions with the exception of eluting in 30 µl of elution buffer. DNA was then removed using DNA-free DNA Removal kits (ThermoFisher Scientific). Of the nine genotypes, the top six (Figure 1 and Supplemental Table S2) with the highest concentration and quality of RNA were normalized to 15 ng/µl. Normalized samples were sent to UT Austin’s Genome Sequencing and Analysis Facility, where TagSeq libraries were prepared following Meyer et al. (2011) and sequenced (single end 100 bp) on a NovaSeq 6000.
2.3 Identifying Acropora cervicornis clones
Acropora cervicornis is well-known for its ability to reproduce asexually via fragmentation (Tunnicliffe, 1981). To identify potential A. cervicornis clones in our dataset, we called single nucleotide polymorphisms (SNPs) from TagSeq reads. Briefly, 104.64 million raw reads were generated, with individual library counts ranging from 4.17 to 6.58 million reads per sample (mean = 5.23 million reads) (Supplemental Table S2). Fastx_toolkit removed 5’-Illumina leader sequences and poly(A)+ tails. Sequences <20bp in length with <90% of bases having quality cutoff scores <20 were also trimmed. In addition, because degenerate bases were incorporated during cDNA synthesis, PCR duplicates were removed from all libraries. After quality filtering 0.63 to 3.00 million reads remained (mean = 2.07 million) and these resulting quality filtered reads were mapped to the Acropora millepora genome (Fuller et al., 2020) using Bowtie2.2.0 (Langmead and Salzberg, 2012) (Supplemental Table S2). Resulting SAM files were converted to BAM files using samtools (Li et al., 2009). ANGSD (Korneliussen et al., 2014) calculated pairwise identity-by-state (IBS) matrices using a minIndDepth filter of five. IBS matrices were used as input to hclust(), which visualized relatedness of individuals and allowed for the identification and removal of clones. We note here that mapping was also conducted using the Acropora cervicornis transcriptome (Libro et al., 2013) and the A. cervicornis genome (Kitchen et al., 2019), however the transcriptome was found to contain symbiont data and the genome resulted in lower mapping efficiencies relative to the A. millepora genome. Given that results remained the same (e.g., clones were identified and similar overall gene expression patterns were observed), A. millepora has superior annotations, and Cooke et al. (2020) found high synteny among acroporids, we opted to move forward mapping to the A. millepora genome. When clones were detected, only one clonemate was maintained in downstream analyses of gene expression patterns (see Supplemental Table S2).
2.4 Read mapping, differential gene expression and gene ontology enrichment analysis
Reads were trimmed as described above and these reads were mapped to the Acropora millepora genome (Fuller et al., 2020) using Bowtie2 (Langmead and Salzberg, 2012). A custom Perl script (samcount.pl) was used to count the number of reads and the resulting raw counts file was then imported into R v. 3.4.2 (R Core Team (2017) for further analyses. Only samples representing clonemates from SNP analyses above and samples with < 400k counts were removed leaving 19 samples in downstream analyses. Differentially expressed genes (DEGs) were identified using DESeq2 v. 1.32.0 in R, with the model: design = ~ genotype + treatment. A contig was considered significantly differentially expressed if it had an FDR adjusted p-value < 0.1.
Data were rlog-normalized and effects of genotype and treatment on global gene expression profiles were tested using a PERMANOVA [adonis() function; vegan (Oksanen, 2007)] and visualized using principal components analysis (PCA) using Euclidean distances. An additional PCA was computed using only the top 1000 differentially expressed genes for experimental treatment based on raw p-values to further explore sample clustering when only the genes whose profiles were the most divergent were included.
Gene expression plasticity was then calculated using the first two principal component (PC) axes as the distance between a genotype’s gene expression in its treatment condition relative to its expression in ambient conditions using a custom function (Bove, 2022a). The effect of treatment on calculated distances (i.e., gene expression plasticity) was assessed using an analysis of variance (function aov) with treatment as a fixed effect. Genotype was not included in the model because it was accounted for in the PC distance calculations. Differences between treatment levels were tested using Tukey’s Honest Significant Differences (HSD) tests.
To determine whether global gene expression patterns showed enrichment of different gene ontology (GO) classes, the collection of scripts ‘GO_MWU’ from Wright et al. (2015) was used (https://github.com/z0on/GO_MWU). Here, the GO database (go.obo v.1.2) was used to test for enrichment of GO terms based on the ranked −log signed p-values of each gene. Gene ontology terms that were over-represented or under-represented were then visualized in a tree format that groups GO terms with other terms of similar function. All GO enrichment results for all treatment comparisons can be found on the github repository (https://github.com/daviessw/Acer_OAW-Microplastics); however only results of the double stressor treatment (OAW+MP) relative to ambient conditions (AMB) are described in detail here. Within this comparison, a heatmap of genes (raw p-value 0.10) assigned to GO terms associated with immunity was constructed using the R package pheatmap to illustrate gene expression patterns associated with OAW+MP.
2.5 ITS2 and 16S metabarcoding
DNA was extracted using an RNAqueous kit (ThermoFisher Scientific) as described above, except samples were not subjected to the DNA removal step. Internal transcribed spacer region 2 (ITS2) PCR amplification was performed using SYM_VAR_5.8S2 and SYM_VAR_REV primers (Hume et al., 2013; Hume et al., 2015) using the following PCR profile: 26 cycles of 95°C for 40 s, 59°C for 2 min, 72°C for 1 min and a final extension of 72°C for 7 min. A negative control was included and failed to amplify so it was not sequenced. PCR products were cleaned using the GeneJET PCR Purification kit (ThermoFisher Scientific) according to the manufacturer’s instructions with the exception of eluting in 30 µl of elution buffer. A second PCR was performed to dual-barcode samples before pooling, which was done based on the visualization of band intensity on a 1% agarose gel. After pooling, the sample was cleaned using the GeneJET PCR Purification kit (ThermoFisher Scientific) according to the manufacturer’s instructions with the exception of using 40 μl of elution buffer. 20 μl of the pool was run on a 2% SYBR Green gel, the target band was excised and placed in 30 μl of Milli-Q water overnight at 4°C before submission for sequencing as detailed below. The V4 region of the 16S rRNA gene was amplified via PCR using Hyb515f (Parada et al., 2016) and Hyb806R (Apprill et al., 2015) primers and the following PCR profile: 30 cycles of 95°C for 40 s, 63°C for 2 min, 72°C for 1 min and a final extension of 7 min. The same procedure as described above for the ITS2 samples was then followed, but with the addition of two negative controls using Milli-Q water which were included in sequencing submission. Concentrations of the ITS2 and 16S pools (via DeNovix DS-11+ Spectrophotometer) were used to combine the two pools in a 1:3 ratio, respectively. Libraries were sequenced on Illumina MiSeq (paired-end 250 bp) at Tufts Genomics Core Facility.
Demultiplexed reads were pre-processed using bbmap (Bushnell, 2014) to split ITS2 and 16S reads based on primers, while tossing reads that included neither primer. Resulting ITS2 reads were then analyzed by submitting paired fastq.gz files directly to SymPortal, which identifies specific sets of defining intragenomic ITS2 sequence variants (DIVs) to define ITS2 type profiles that are indicative of genetically differentiated Symbiodiniaceae taxa (Hume et al., 2019).
16S primers were removed using cutadapt (Martin, 2011), then DADA2 (Callahan et al., 2016) was used to conduct quality filtering and inference of 3,493 amplicon sequence variants (ASVs) (see Supplemental Table S3 to track reads lost through filtering). Taxonomy was assigned with DADA2 using the Silva v. 138.1 database (Quast et al., 2013) and National Center for Biotechnology Information’s nucleotide database using blast+ (Camacho et al., 2009). ASVs matching mitochondria, chloroplasts, or non-bacterial kingdoms were removed (216 total) and 5 ASVs were removed based on negative controls as contaminants [Decontam; (Davis et al., 2018)]. Cleaned counts were rarefied to 9,409 using vegan (Oksanen, 2007) and trimmed using MCMC.OTU (Wright et al., 2015) to remove ASVs present in less than 0.01% of counts, resulting in 260 ASVs across all 36 samples.
Beta diversity of the 16S trimmed counts based on treatment and genotype was assessed using a PCoA on Bray–Curtis dissimilarity [Phyloseq; (McMurdie and Holmes, 2013)] and a PERMANOVA [vegan; (Oksanen, 2007)]. We then calculated alpha diversity (Shannon index, Simpson’s index, ASV richness, and evenness) of the rarefied counts using Phyloseq [function estimate_richness(); (McMurdie and Holmes, 2013)]. Finally, to test for differences in background 16S communities, we removed all ASVs from the dominant genus MD3-55 [Candidatus Aquarickettsia rohweri, referred now as “Aquarickettsia”; (McMurdie and Holmes, 2013; Klinges et al., 2019)] and performed the same beta and alpha diversity assessments. All ITS2 and 16S data, figures, and analyses were completed in R [version 3.6.3; (R Core Team, 2020) and can be found on GitHub (https://github.com/seabove7/acer_microplastics; (Bove, 2022b)].
3 Results
3.1 Identification of coral genotypes
A total of 4,018 SNPs were identified and of the six putative genotypes that were sequenced, one pair of clones was identified (G10a and G12) (Supplemental Figure 2). Thus, genotype G12 samples from each of the four experimental treatments were removed to retain only one representative clone (genotype G10a) in each treatment (Supplemental Table S2).
3.2 All coral fragments are dominated by the algal symbiont Symbiodinium ‘fitti’
Raw ITS2 counts before submission to SymPortal ranged from 4,684 to 613,872 per sample, with a mean of 169,246. After classification by SymPortal, ITS2 counts ranged from 730 to 121,515 per sample across all 36 samples with mean 32,774 counts. All coral fragments hosted Symbiodinium ‘fitti’ (ITS2 type A3) with 44% (N=16) hosting small background amounts (less than 5%) of Breviolum minutum (ITS2 type B2) (Figure 2).
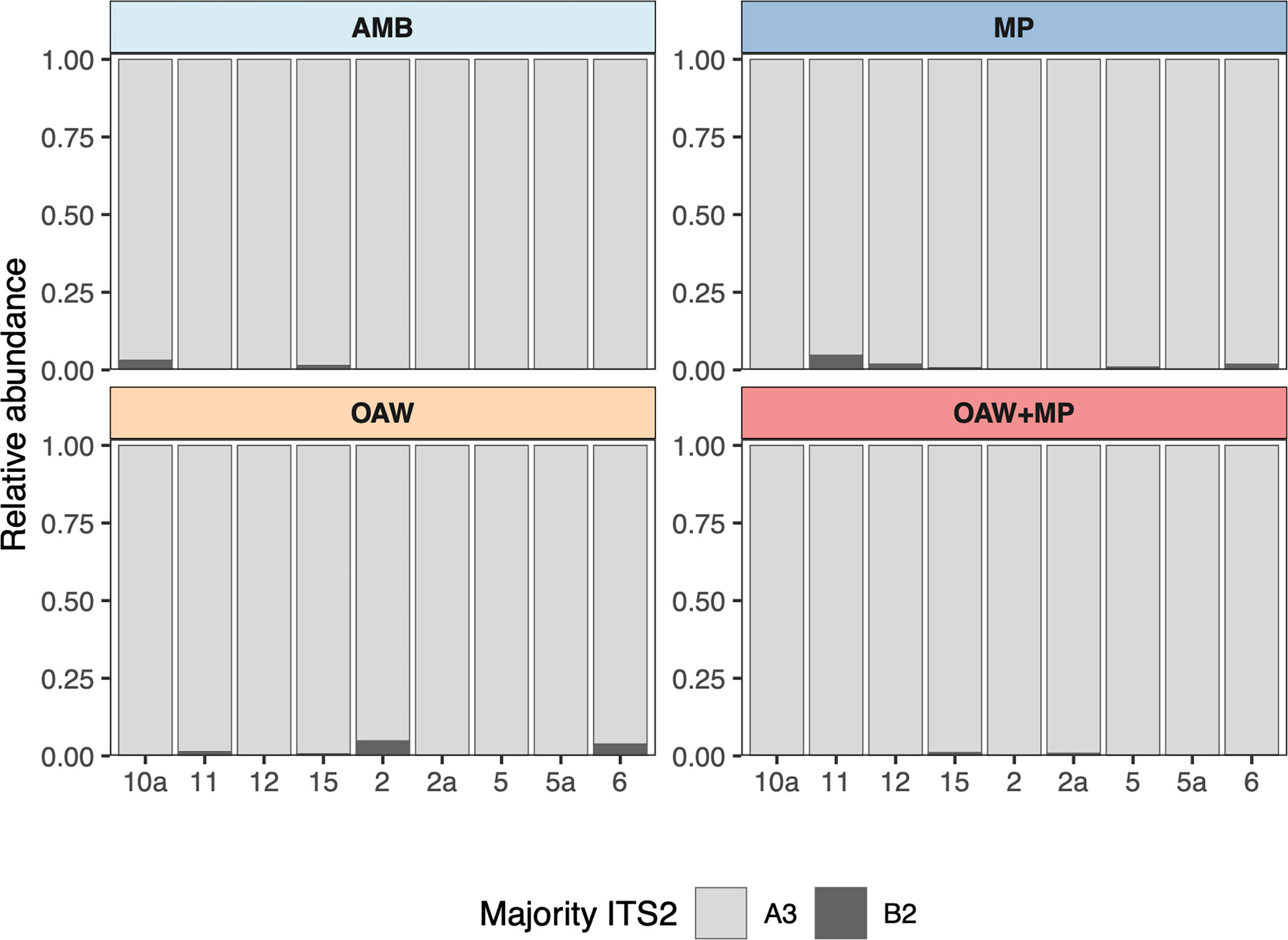
Figure 2 Relative abundance of major ITS2 types across each sample grouped by treatment. Light grey represents Symbiodinium ‘fitti’ (A3) and dark grey represents Breviolum minutum (B2).
3.3 Double stressor treatment elicits strongest gene expression plasticity
A total of 20 samples were sequenced, which resulted in a total of 104.6 million reads, 41.4 million of which remained after trimming, and 13.9 million of which mapped successfully to the A. millepora reference genome. One sample - genotype G2, from the OAW treatment - was removed from the dataset due to low counts (166,828 total; Supplemental Table S2). Mean counts across samples after outlier removal was 661,358.
Pairwise comparisons between each stressor (MP, OAW, OAW+MP) and ambient conditions (AMB) showcased that individuals in the double stressor (OAW + MP) treatment had the highest number of differentially expressed genes (DEGs) (83 up, 60 down), followed by microplastics (48 up, 9 down) and then OAW (18 up, 18 down) with relatively few genes overlapped between treatments (pFDR<0.10, Supplemental Figure 3). A principal component analysis (PCA) on all rlog-transformed counts found that there was no overall effect of treatment on A. cervicornis gene expression; however, there was a significant effect of genotype on overall expression patterns (Figure 3A; pGENOTYPE<0.001). A PCA of the top 1,000 DEGs showcased stronger clustering by experimental treatment and the effect of genotype remained significant (Figure 3B; pGENOTYPE<0.001, pTREATMENT<0.001). This second PCA also showcased that the double stressor (OAW+MP) had the strongest effect on gene expression, which corroborated the numbers of observed DEGs. A gene expression plasticity analysis confirmed this pattern, which showcased a significant effect of treatment on gene expression plasticity (pTREATMENT=0.041), with the OAW+MP treatment exhibiting the highest plasticity (Figure 3C). However, these differences in plasticity between corals in the OAW+MP treatment relative to corals in the OAW and MP treatments were only marginally significant after multiple test correction (OAW+MP~MP, p=0.07; OAW+MP~OAW, p=0.06).
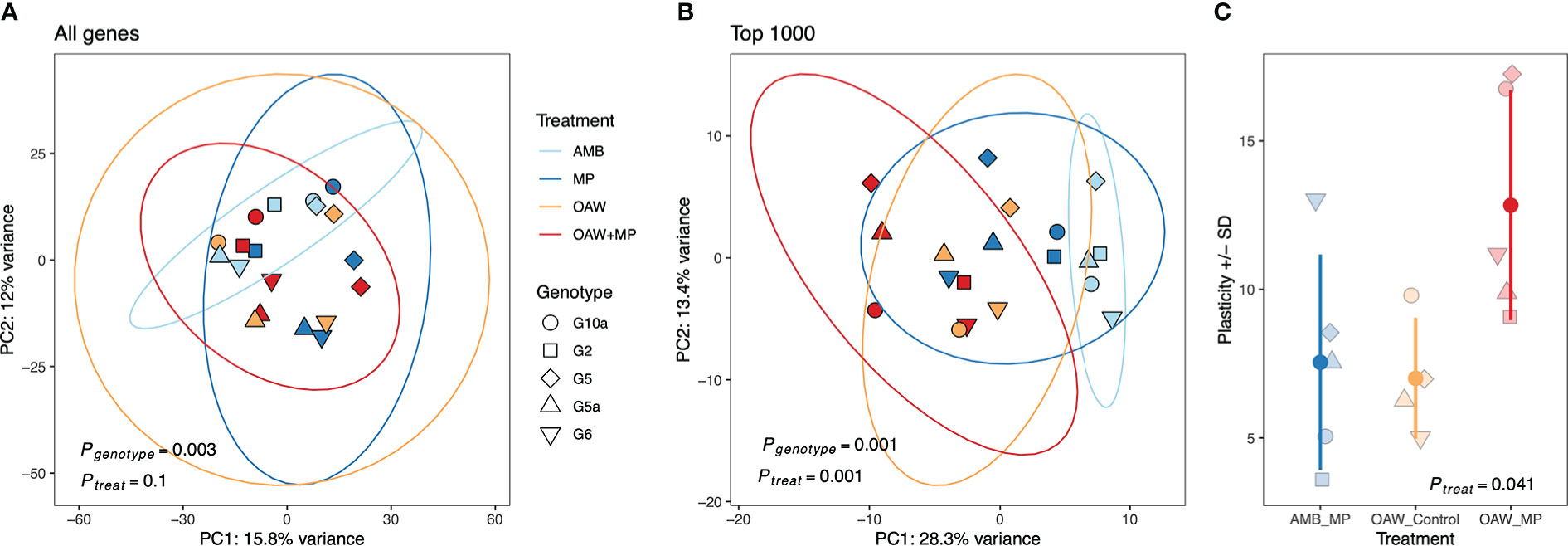
Figure 3 Transcriptomic responses to local and global stressors in Acropora cervicornis. (A) Principal component analysis (PCA) of rlog-transformed counts for all genes and, (B) the top 1000 differentially expressed genes by treatment in DESeq2. PERMANOVA results for the effect of genetic background (genotype) and treatment are included. (C) Gene expression (GE) plasticity +/- standard deviation (SD) estimates calculated from PC distances from (B) where each point is the distance that genotype moved from its ambient fragment (AMB) in PC space.
3.4 Corals exhibit mixed functional responses to single stressor treatments
Gene ontology enrichment analysis between microplastics (MP) relative to ambient conditions (AMB) revealed underrepresentation of oxidoreductase (GO: 0016491), suggesting down regulation of stress response in corals in MP treatment. In addition, overrepresentation of GO terms associated with ribosomal processes was observed including structural constituent of ribosome (GO:0003735) and large ribosomal subunit (GO:0015934), suggesting upregulation of terms associated with increased growth under the presence of microplastics.
In response to OAW, we observed an enrichment of regulation of defense response to virus (GO:0050688), terms associated with amino acid catabolism (aromatic amino acid family catabolic process (GO:0009074), cellular amino acid catabolic process (GO:0009063) and alpha-amino acid catabolic process (GO:1901606)), and two terms associated with stress response (chaperone-mediated protein folding (GO:0061077) and unfolded protein binding (GO:0051082)). In addition, we observed downregulation of processes associated with sensory detection including detection of stimulus (GO:0051606), sensory perception (GO:0007600), and cognition (GO:0050890). Lastly, several GO terms associated with synapse were down regulated including synapse (GO:0045202), synaptic membrane (GO:0097060), and cell junction (GO:0030054). All resulting GO trees from these single stressor treatments can be found on the accompanying GitHub repository (https://github.com/daviessw/Acer_OAW-Microplastics).
3.5 Enrichment of immune-related functions under multiple stressors
Gene ontology enrichment analysis between our double stressor (OAW+MP) relative to ambient conditions (AMB) revealed many significantly enriched GO terms within ‘Biological processes’, of which the OAW+MP-enriched categories were dominated by GO terms associated with immunity (green box, Supplemental Figure 4). These enriched GO terms included: regulation of defense response to virus by host (GO:0050691), activation of innate immune response (GO:0002218), regulation of immune effector process (GO:0002697), NIK/NF-kappaB signaling (GO:0038061), regulation of cytokine production (GO:0001819), suggesting that the combined stressor elicited an upregulation of innate immunity in the coral host. When a heatmap was made of all genes belonging to these 30 GO terms that met our alpha cut-off (unadjusted p-value < 0.10), strong differences in gene expression were observed between corals in OAW+MP relative to those in AMB treatments (Figure 4). Of particular interest, many classic stress response genes were upregulated in OAW+MP relative to AMB including: superoxide dismutase, two heat shock proteins, several ubiquitin genes, tumor necrosis factors/receptors, a proto-oncogene, and several genes associated with apoptosis (bcl2-like 1) and response to cytokines (p38 map kinase).
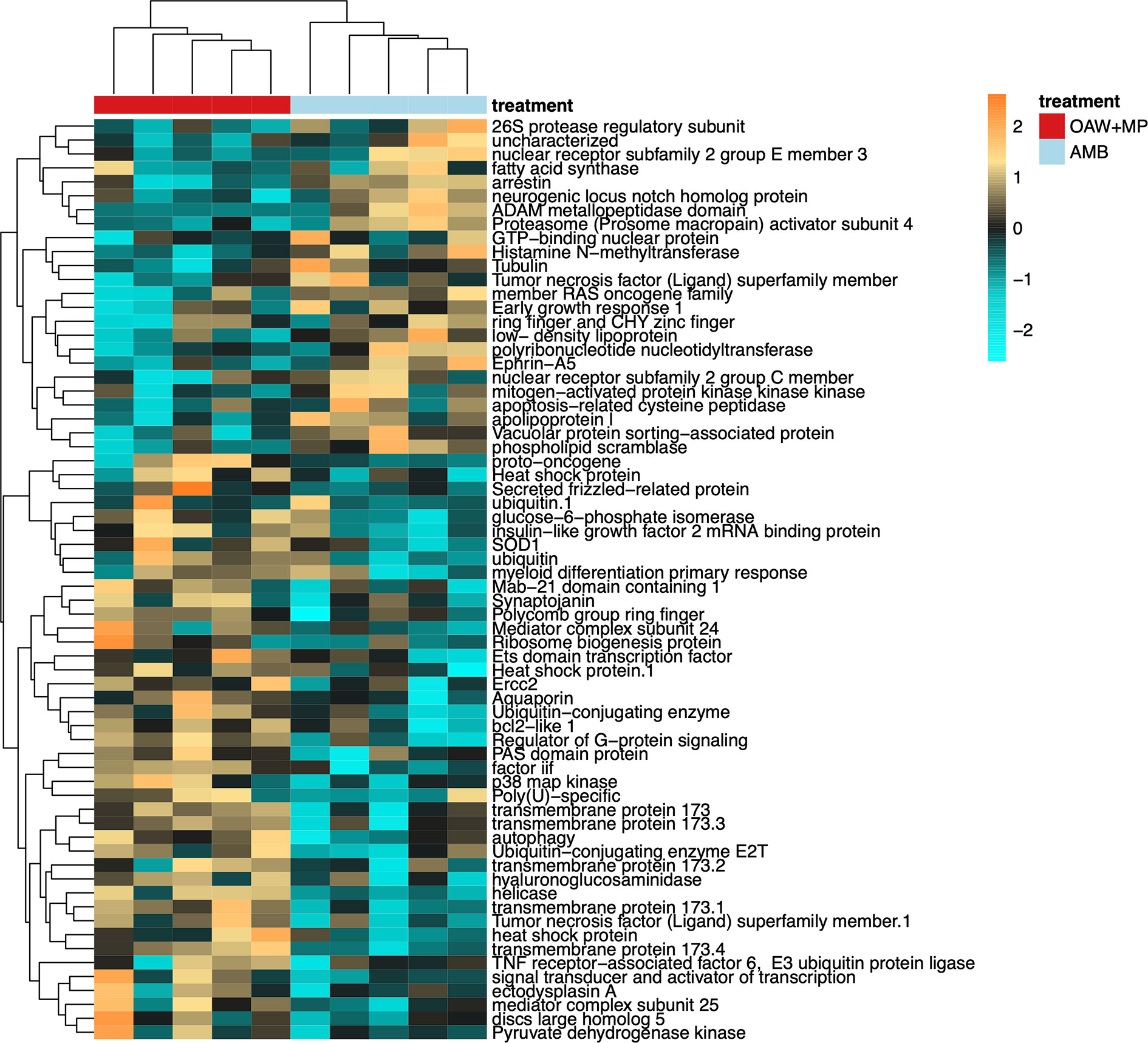
Figure 4 Differentially expressed genes (DEGs, unadjusted p-value < 0.10) with annotations associated with immunity gene ontology (GO) terms from Supplemental Figure 4 (green box). Heatmap showing annotated immunity genes where each row is a gene and each column is a unique gene expression sample. The color scale is in log2 (fold change relative to the gene’s mean) and genes and samples are clustered hierarchically based on Pearson’s correlation of their expression across samples. Colored blocks indicate treatment. Hierarchical clustering of libraries (columns) demonstrates strong differences in gene expression of immunity genes by treatment condition.
3.6 Acropora cervicornis microbiomes dominated by order Rickettsiales
Cleaned 16S reads before trimming and rarefaction averaged 42,577 ± 25,010 ( ± SD) per sample across all A. cervicornis samples with a minimum of 9,409 (Sample 11 A - 1) and a maximum of 107,941 (Sample 10a C - 1). Fragments were dominated by taxa from the phylum Proteobacteria (Figure 5A), which was dominated by a single ASV (Genus MD3-55; “Candidatus Aquarickettsia rohweri”) within the order Rickettsiales (Figure 5B; see Supplemental Figure 5). This dominant ASV was a 100% match to the published 16S rRNA sequence for the putative bacterial parasite “Ca. Aquarickettsia rohweri” (hereafter, Aquarickettsia) (Klinges et al., 2019) that is common in A. cervicornis across the Caribbean (Godoy-Vitorino et al., 2017; Williams et al., 2022).
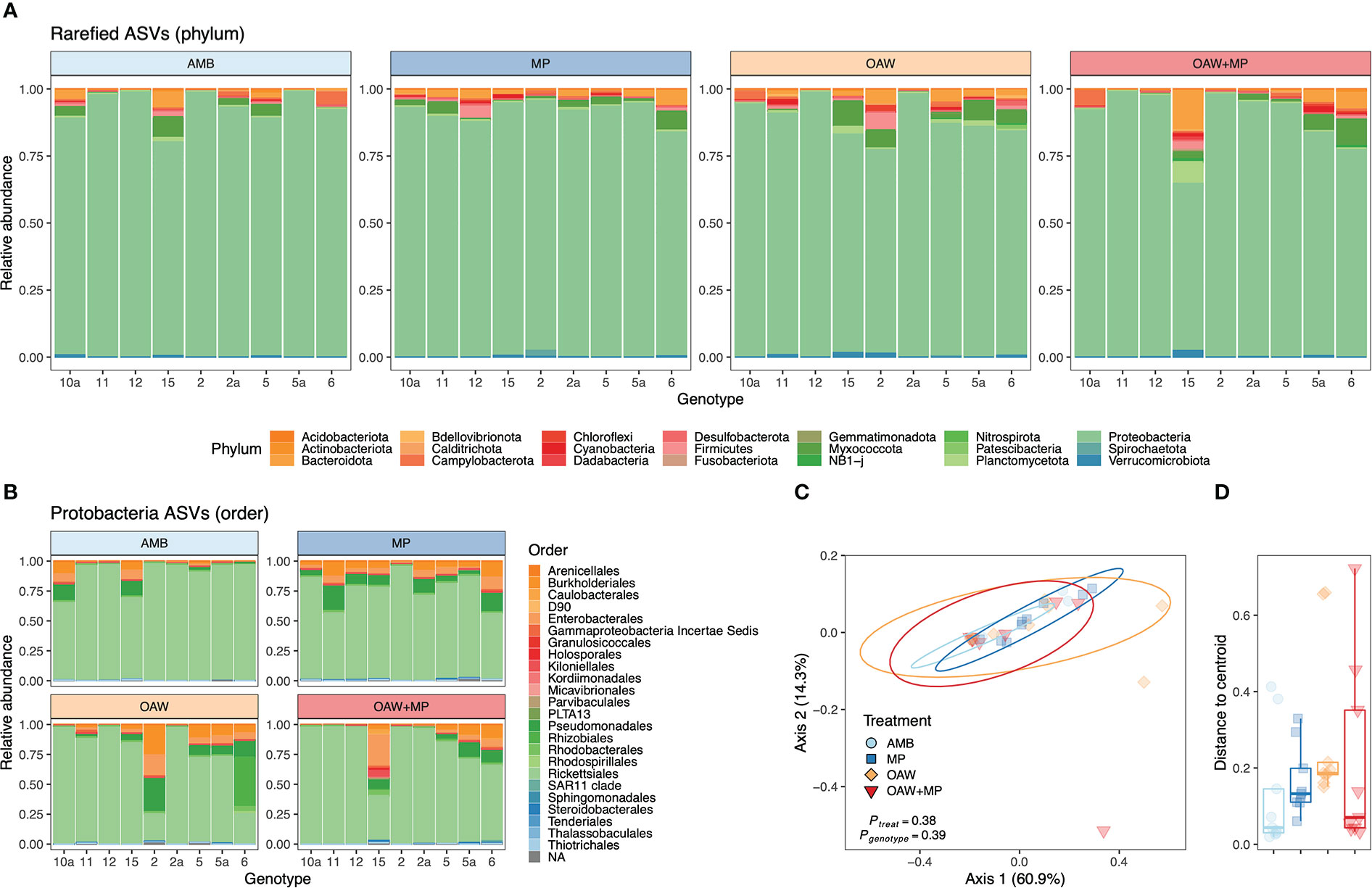
Figure 5 Acropora cervicornis bacterial (16S) relative abundance across fragments and experimental treatments. Diversity at the phylum level (A) depicts dominance of all samples by taxa from Proteobacteria and this (B) phylum was dominated by the order Rickettsiales in most samples. Bacterial diversity color was assigned alphabetically, not based on abundance. Beta diversity was visualized through (C) multivariate ordination plots (PCoA) of between-sample Bray–Curtis dissimilarity of rarefied ASVs of all taxa and (D) distance to treatment centroids. PCoA ellipses depict 95% confidence intervals and p-values indicate significance of treatment and genotype. Treatment is depicte-d by color in all panels: light blue = ambient conditions (AMB), dark blue = ocean acidification and warming (OAW), orange = microplastics (MP), and red = OAW and MP (OAW+MP).
Beta diversity analyses did not identify any statistical differences across samples based experimental treatment or genotype (Figures 5C, D and Supplemental Table S4). Similarly, alpha diversity was indistinguishable across treatments (Supplemental Figure 6 and Table S5). After removing Aquarickettsia from all samples to assess background bacterial communities, there were still no differences in alpha or beta diversity across treatments or genotypes (Supplemental Figure 7 and Table S6).
4 Discussion
4.1 Microplastics alone do not drive strong gene expression responses in Acropora cervicornis
Tropical coral reef ecosystems are facing extraordinary challenges from both local and global stressors that are altering their function (Eddy et al., 2021). On the local scale, pollution in the form of microplastics may threaten the health and physiology of reef-building corals (Soares et al., 2020; Nanthini devi et al., 2022). While there is evidence that corals ingest microplastics, which can lead to a variety of physiological responses (Hall et al., 2015; Rotjan et al., 2019), here we detected only a muted gene expression response in A. cervicornis exposed to microplastics. In fact, overall gene expression profiles were indistinguishable between the ambient and microplastics treatments, and an effect of microplastics was only apparent when looking at the expression of top 1,000 genes. This result corroborates previous work reporting negligible effects of microplastics on coral physiology, bleaching susceptibility, and mortality (Reichert et al., 2021; Plafcan and Stallings, 2022) and supports the hypothesis that corals that rely more heavily on heterotrophy may be more impacted by microplastic pollution.
Recent studies have considered the physiological impact of microplastics on tropical corals at the molecular level. These studies, conducted on the tropical coral Pocillopora damicornis (Tang et al., 2018) and the habitat-forming octocoral Corallium rubrum (Corinaldesi et al., 2021), report evidence of elevated stress response after exposure to microplastics in as little as a few hours. In contrast, we identified enrichment of GO terms associated with growth (structural constituent of ribosome, large ribosomal subunit) and an underrepresentation of terms associated with stress response (oxidoreductase). Our gene expression results suggest that A. cervicornis is not exhibiting signs of molecular stress after a 3-week exposure to microplastics, and that these corals may actually be growing more compared to those reared under ambient conditions. It should be noted, however, that the enrichment of growth GO terms in response to microplastics may be due to other compensatory mechanisms, such as increasing photophysiology as a way to counteract declining coral host energy reserves (Reichert et al., 2019; Lanctôt et al., 2020). While we did not quantify photophysiology or gene expression of the algal symbionts in this study, it is possible that algae associated with corals in the microplastic treatment may have exhibited enriched photophysiology, which may have led to the enrichment of molecular signatures of growth. However, it is possible that a longer-term exposure to microplastics may have eventually led to signs of physiological stress (Reichert et al., 2019). While these contrasting results may be due to experimental design differences (microplastic concentration and size, type of microplastics, duration, etc.), it is more likely due to species-specific responses (Hankins et al., 2021; Mendrik et al., 2021). Previous work on A. cervicornis suggests that exposure to microplastics in the presence of warming does not impact bleaching, likely because this species may not ingest the microplastics under otherwise ideal conditions (Plafcan and Stallings, 2022). Instead, A. cervicornis relies on photosynthetically-derived carbon from its algal symbionts as the primary source of nutrition rather than heterotrophy, thus avoiding the potential pitfalls of microplastics ingestion observed in other coral species that are largely heterotrophic (Reichert et al., 2019; Rotjan et al., 2019; Hankins et al., 2021; Mendrik et al., 2021). Overall, we recommend that future work incorporate additional physiological assessments along with coral host and algal symbiont molecular responses to disentangle the role that nutritional source (i.e., dependence on heterotrophy vs. autotrophy) serves in coral physiological response to microplastic pollution.
While these findings suggest that microplastics exposure alone does not elicit a severe stress response in A. cervicornis, it is unclear whether prolonged exposures may eventually impact the host (Hankins et al., 2021) and their algal symbionts (Lanctôt et al., 2020; Ripken et al., 2020). Further, microplastic pollution is occurring alongside global stressors (Soares et al., 2020), including widespread shifts in the microbial landscape of seawater (Cavicchioli et al., 2019) and increased prevalence of pathogens and disease across the Caribbean (van Woesik and Randall, 2017). Because microplastics have been shown to be a vehicle for microbial transmission into a corals (Rotjan et al., 2019), there is emerging concern for microplastics pollution in combination with increased pathogen exposure that has long been a predicted component of global change (Cavicchioli et al., 2019).
4.2 Global change treatment elicits subtle stress signatures
Along with local concerns, global stressors, such as ocean acidification and warming, are contributing to the wide-scale degradation of coral reef ecosystems. Like its response to microplastics, A. cervicornis in the ocean acidification and warming (OAW) treatment exhibited similar overall gene expression profiles to those in the ambient treatment and only gene expression profiles of the top 1,000 genes resulted in a clear response. This subtle response to OAW in A. cervicornis is surprising since this species is known to be sensitive to these global stressors (Enochs et al., 2014; Towle et al., 2015b; Kaufman et al., 2021; Muller et al., 2021). These contrasting results may be due to different experimental design considerations across studies (Bove et al., 2020), especially given that the heat stress temperature employed here (~30.5 ˚C) was lower than most other warming studies. However, there is also significant genetic variation within A. cervicornis (Drury et al., 2017; Million et al., 2022), thus it is also possible that the genotypes used in this study were particularly resistant. Alternatively, these corals are already exposed to warmer seawater conditions where they were collected compared to other Caribbean reefs (Muñiz-Castillo et al., 2019; Bove et al., 2022b), suggesting the potential for acclimatization to elevated temperatures.
While we only observed minor gene expression differences in A. cervicornis reared in the OAW treatment, these shifts were associated with an enrichment of stress-related GO terms. For example, corals in OAW treatment exhibited enrichment of GO terms linked to protein folding and protein binding, which have been reported previously in heat-stressed corals in situ (Ip et al., 2022). This pattern was accompanied by enrichment of terms associated with amino acid catabolism, suggesting A. cervicornis may have mediated physiological stress through protein catabolism (Davies et al., 2016; Aguilar et al., 2019; Rädecker et al., 2021). While we did not assess host or symbiont physiological traits along with these gene expression profiles, it is likely that the growth and/or energy reserves may have been impacted as a result of treatment conditions (Aichelman et al., 2021; Scucchia et al., 2021; Bove et al., 2022a).
Most interestingly, we identified enrichment of GO terms associated with defense against viruses. Previous work has suggested that corals become more susceptible to infection when water temperatures increase (Bruno et al., 2007). Indeed, patterns of Caribbean coral disease have become more prevalent in recent decades (Randazzo-Eisemann et al., 2022) and population declines of A. cervicornis have been specifically linked to increases in disease associated with seawater temperature increases (Muller et al., 2018; Goergen et al., 2019). While the temperature stress applied here was not high enough to induce a classic heat stress response, it suggests that even subtle increases in temperature may make this species of coral more susceptible to infection.
4.3 Global change stressors interact with microplastics to invoke an immune response
Ecological stressors can interact to pose further threats to marine organisms that may not be accounted for when assessing responses to stressors independently (Darling and Côté, 2008; Ellis et al., 2019). Indeed, our multistressor treatment (ocean acidification, warming, and microplastics; OAW+MP) resulted in the strongest gene expression response, suggesting that microplastic pollution may interact with ocean acidification and warming to elicit a more severe molecular response. This strong response to multiple stressors is commonly observed in coral (Coles and Jokiel, 1978; Reynaud et al., 2003; Courtial et al., 2017; Muller et al., 2021) and has been reported previously in studies assessing gene expression as well (Ogawa et al., 2013). The observed response of A. cervicornis to the multistressor treatment in this study may be due to the corals experiencing physiological stress from the acidification and warming exposure (i.e., bleaching), which then led to an increase in reliance on heterotrophy to meet their energetic needs (Ferrier-Pagès et al., 2010; Towle et al., 2015a). This increased heterotrophy in turn may have resulted in the corals consuming more microplastics that do not contribute nutritional value to the coral host, thus furthering the energetic deficit and potentially making them more susceptible to harmful pathogens (Mendrik et al., 2021). While it is unlikely that global change and microplastic pollution are directly interacting, the physiological mechanisms employed by corals in response to these single stressors likely lead to a more severe physiological response.
Here, we identified GO terms enriched in the multistressor treatment related to response to viruses and innate immunity, indicating these stressors may invoke an immune response by the coral. Interestingly, a similar response was observed in OAW alone, however, to a lesser extent, suggesting that the presence of microplastics magnified this stress response. While no studies to date have assessed gene expression responses in corals to the stressors together tested here, previous work in the marine mussel, Mytilus coruscus, exposed to ocean acidification and microplastics identified signals of a repressed immunity that may lead to higher disease susceptibility (Huang et al., 2022). Further, gene expression patterns consistent with an upregulation of immune stress response were reported in the brine shrimp Artemia franciscana exposed to microplastics and warming (Han et al., 2021). This is particularly interesting because Artemia is also frequently used as a nutrition source in coral experiments, including microplastic ingestion experiments (Hall et al., 2015; Axworthy and Padilla-Gamiño, 2019; Lanctôt et al., 2020), potentially altering the nutritional value of the Artemia and impacting coral health. Overall, these results suggest that the combination of global change stressors and microplastics will likely lead to suppressed immunity in corals in tandem with higher disease transmission associated with warming (Randazzo-Eisemann et al., 2022) and microplastic pollution (Rochman et al., 2019; Rotjan et al., 2019).
4.4 Acropora cervicornis microbial communities dominatedby Aquarickettsia
Similar microbiome compositions were observed across the four treatment conditions. A total of 31 of the 36 fragments were high-Aquarickettsia (i.e., greater than 50% dominance), with relative abundances ranging from 17.5% up to 98.8% of the total sequence reads. This sequence dominance in A. cervicornis microbiomes is not uncommon and has been documented in this species in the Florida Keys and throughout the Caribbean (Baker et al., 2022), although previous work has also showcased variation among individual genotypes (Aguirre et al., 2022). Even when Aquarickettsia-affiliated ASVs were removed from analysis, microbiome composition did not significantly differ across treatments. Importantly, the microbiome composition of these corals before collection from the wild is unknown, therefore we cannot determine if the universally high Aquarickettsia levels were a result of captivity muting microbiome signatures (Galand et al., 2018). Future work leveraging experiments on individuals representing a larger number of host genotypes and previously identified as high-Aquarickettsia and low-Aquarickettsia (e.g., (Aguirre et al., 2022)) may further clarify factors that drive differential coral vulnerability to microplastics, acidification, and warming, especially since low-Aquarickettsia individuals exhibit higher resistance to disease, relative to high-Aquarickettsia individuals (Klinges et al., 2022).
Genes implicated in the coral immune response were upregulated in response to the OAW and multistressor treatment, suggesting that some sort of external stimulus triggered this response. Microbiome taxonomic profiling did not reveal significant differences in microbiomes across treatments, so we suggest that further research should investigate the transcriptomic and metabolomic responses of the bacterial and viral fractions across these stressors. For example, it is possible that while Aquarickettsia remained dominant, it may have differentially regulated genes in response to OAW conditions and/or OAW+MP. Alternatively, the immune response may reflect host response to physical injury/insult by contact with the plastics (van de Water et al., 2015), but not necessarily microbial infection.
Another potential explanation for the observed immune response is a host response to viruses. Indeed, GO categories consistent with a viral response were enriched in OAW+MP treatments (i.e., cellular response to virus). Coral viromes have been implicated in directing holobiont response to environmental stress (Thurber and Correa, 2011; Leruste et al., 2012; Nguyen-Kim et al., 2015; Thurber et al., 2017). It is thought that viruses may play a role in coral bleaching under thermal stress through host lysis of algal symbiont cells that precedes bleaching signals (Lawrence et al., 2014; Grupstra et al., 2022). Additionally, previous work has demonstrated increased expression of anti-viral transcripts in hosted algal symbionts (Cladocopium, formerly Clade C) in response to thermal stress (Levin et al., 2017). Taken together, it is possible that the A. cervicornis immune response may suggest a virally driven response that precedes the classic bleaching response (Barshis et al., 2013; Dixon et al., 2020).There is also evidence that microplastics and the microbial plastisphere can facilitate survival of certain viruses (Moresco et al., 2021). It is plausible that increased levels of virus-coral interactions could elicit a host immune response, if not also facilitate viral infection of coral cells (Bowley et al., 2021; Loiseau and Sorci, 2022).
We recommend that future work investigating the effects of microplastic pollution on reef-building corals, especially in combination with global change stressors, specifically target the viral dynamics and diversity in the seawater relative to those associated with the coral holobiont. Additionally, assessing the microbiome and virome at several time points throughout stress challenge, coupled with holobiont gene expression and metabolome profiles, will be crucial for understanding the timeline and underlying causes of potential assemblage shifts that inform holobiont phenotypes. Overall, our results highlight the importance of applying multi-omic approaches to better understand the consequences of local and global stressors on reef-building corals.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI PRJNA888134 and GitHub https://github.com/seabove7/acer_microplastics & https://github.com/daviessw/Acer_OAW-Microplastics.
Author contributions
SS, NF, and SD conceived the research. SS and NF conducted the experiment. KG, NK, AKH, AMH, and CB performed molecular lab work. KG, NK, and SD completed the gene expression analyses. CB, AKH, and AMH completed the 16S and ITS2 analyses. CB and KS interpreted the 16S analyses. CB, KG, and SD wrote the manuscript with help from all co-authors. Funding acquisition, project administration, and resources was led by SD, NF, and SS. All authors contributed to the article and approved the submitted version.
Funding
Funding for this project was facilitated by Boston University (BU) Biology Department start-up funds to SD and by keystone project funds to KG from BU’s Kilachand Honors College. CB was funded through a BU Microbiome Initiative Accelerator grant. Funding for the experimental portion was supplied to NF by the UNCW College of Arts and Sciences Research Initiative Award and SS by the Explorers Club. KS was supported by the Institutional Development Award (IDeA) Network for Biomedical Research Excellence from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103430.
Acknowledgments
All computational work was conducted on Boston University’s Shared Computing Cluster (SCC). We thank the numerous UNCW undergraduate students and graduate students, Chad Campbell, Reanna Jeanes, and Bryce Corbett for assisting with the experimental portion. Thank you to Nova Southeastern University staff, particularly Dave Gilliam and his lab for collecting the coral and Joana Figueiredo and her lab for quarantining the corals used in this experiment. Thank you to Florida Fish and Wildlife Conservation Commission for providing permit #SAL-19-2200A-SRP for this collection. We thank two reviewers for their helpful feedback during the review process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1037130/full#supplementary-material
References
Aguilar C., Raina J.-B., Fôret S., Hayward D. C., Lapeyre B., Bourne D. G., et al. (2019). Transcriptomic analysis reveals protein homeostasis breakdown in the coral acropora millepora during hypo-saline stress. BMC Genomics 20, 148. doi: 10.1186/s12864-019-5527-2
Aguirre E. G., Million W. C., Bartels E., Krediet C. J., Kenkel C. D. (2022). Host-specific epibiomes of distinct acropora cervicornis genotypes persist after field transplantation. Coral Reefs 41, 265–276. doi: 10.1007/s00338-022-02218-x
Aichelman H. E., Bove C. B., Castillo K. D., Boulton J. M., Knowlton A. C., Nieves O. C., et al. (2021). Exposure duration modulates the response of Caribbean corals to global change stressors. Limnol. Oceanogr. 66, 3100–3115. doi: 10.1002/lno.11863
Allen A. S., Seymour A. C., Rittschof D. (2017). Chemoreception drives plastic consumption in a hard coral. Mar. pollut. Bull. 124, 198–205. doi: 10.1016/j.marpolbul.2017.07.030
Amaral-Zettler L. A., Zettler E. R., Mincer T. J. (2020). Ecology of the plastisphere. Nat. Rev. Microbiol. 18, 139–151. doi: 10.1038/s41579-019-0308-0
Anthony K. R. N., Kline D. I., Diaz-Pulido G., Dove S., Hoegh-Guldberg O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. U. S. A. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Apprill A. (2017). Marine animal microbiomes: Toward understanding host–microbiome interactions in a changing ocean. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00222
Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Axworthy J. B., Padilla-Gamiño J. L. (2019). Microplastics ingestion and heterotrophy in thermally stressed corals. Sci. Rep. 9, 18193. doi: 10.1038/s41598-019-54698-7
Baker L. J., Reich H. G., Kitchen S. A., Grace Klinges J., Koch H. R., Baums I. B., et al. (2022). The coral symbiont candidatus aquarickettsia is variably abundant in threatened Caribbean acroporids and transmitted horizontally. ISME J. 16, 400–411. doi: 10.1038/s41396-021-01077-8
Barshis D. J., Ladner J. T., Oliver T. A., Seneca F. O., Traylor-Knowles N., Palumbi S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U. S. A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Berry K. L. E., Epstein H. E., Lewis P. J., Hall N. M., Negri A. P. (2019). Microplastic contamination has limited effects on coral fertilisation and larvae. Diversity 11, 228. doi: 10.3390/d11120228
Bertucci J. I., Juez A., Bellas J. (2022). Impact of microplastics and ocean acidification on critical stages of sea urchin (Paracentrotus lividus) early development. Chemosphere 301, 134783. doi: 10.1016/j.chemosphere.2022.134783
Bourne D. G., Morrow K. M., Webster N. S. (2016). Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bove C. (2022b). Data and code: Exposure to global change and microplastics elicits an immune response in an endangered coral (Version 1.0.0). doi: 10.5281/zenodo.7396265
Bove C. B., Davies S. W., Ries J. B., Umbanhowar J., Thomasson B. C., Farquhar E. B., et al. (2022a). Global change differentially modulates Caribbean coral physiology. PloS One 17, e0273897. doi: 10.1371/journal.pone.0273897
Bove C. B., Mudge L., Bruno J. F. (2022b). A century of warming on Caribbean reefs. PloS Climate 1, e0000002. doi: 10.1371/journal.pclm.0000002
Bove C. B., Umbanhowar J., Castillo K. D. (2020). Meta-analysis reveals reduced coral calcification under projected ocean warming but not under acidification across the Caribbean sea. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00127
Bowley J., Baker-Austin C., Porter A., Hartnell R., Lewis C. (2021). Oceanic hitchhikers - assessing pathogen risks from marine microplastic. Trends Microbiol. 29, 107–116. doi: 10.1016/j.tim.2020.06.011
Browne M. A., Crump P., Niven S. J., Teuten E., Tonkin A., Galloway T., et al. (2011). Accumulation of microplastic on shorelines woldwide: Sources and sinks. Environ. Sci. Technol. 45, 9175–9179. doi: 10.1021/es201811s
Bruno J. F., Selig E. R., Casey K. S., Page C. A., Willis B. L., Harvell C. D., et al. (2007). Thermal stress and coral cover as drivers of coral disease outbreaks. PloS Biol. 5, e124. doi: 10.1371/journal.pbio.0050124
Bushnell B. (2014) BBMap: A fast, accurate, splice-aware aligner (Berkeley, CA (United States: Lawrence Berkeley National Lab. (LBNL). Available at: https://www.osti.gov/biblio/1241166 (Accessed August 2, 2022).
Callahan B. J., Sankaran K., Fukuyama J. A., McMurdie P. J., Holmes S. P. (2016). Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Res 5, 1492. doi: 10.12688/f1000research.8986.2
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST: architecture and applications. BMC Bioinf. 10. doi: 10.1186/1471-2105-10-421
Cavicchioli R., Ripple W. J., Timmis K. N., Azam F., Bakken L. R., Baylis M., et al. (2019). Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586. doi: 10.1038/s41579-019-0222-5
Chu N. D., Vollmer S. V. (2016). Caribbean Corals house shared and host-specific microbial symbionts over time and space. Environ. Microbiol. Rep. 8, 493–500. doi: 10.1111/1758-2229.12412
Coles S. L., Jokiel P. L. (1978). Synergistic effects of temperature, salinity and light on the hermatypic coral montipora verrucosa. Mar. Biol. 49, 187–195. doi: 10.1007/BF00391130
Contreras-Silva A. I., Tilstra A., Migani V., Thiel A., Pérez-Cervantes E., Estrada-Saldívar N., et al. (2020). A meta-analysis to assess long-term spatiotemporal changes of benthic coral and macroalgae cover in the Mexican Caribbean. Sci. Rep. 10, 8897. doi: 10.1038/s41598-020-65801-8
Cooke I., Ying H., Forêt S., Bongaerts P., Strugnell J. M., Simakov O., et al. (2020). Genomic signatures in the coral holobiont reveal host adaptations driven by Holocene climate change and reef specific symbionts. Sci. Adv. 6. doi: 10.1126/sciadv.abc6318
Corinaldesi C., Canensi S., Dell’Anno A., Tangherlini M., Di Capua I., Varrella S., et al. (2021). Multiple impacts of microplastics can threaten marine habitat-forming species. Commun. Biol. 4, 431. doi: 10.1038/s42003-021-01961-1
Cornwall C. E., Comeau S., Kornder N. A., Perry C. T., van Hooidonk R., DeCarlo T. M., et al. (2021). Global declines in coral reef calcium carbonate production under ocean acidification and warming. Proc. Natl. Acad. Sci. U. S. A. 118. doi: 10.1073/pnas.2015265118
Courtial L., Roberty S., Shick J. M., Houlbrèque F., Ferrier-Pagès C. (2017). Interactive effects of ultraviolet radiation and thermal stress on two reef-building corals. Limnol. Oceanogr. 62, 1000–1013. doi: 10.1002/lno.10481
Cristofari R., Liu X., Bonadonna F., Cherel Y., Pistorius P., Le Maho Y., et al. (2018). Climate-driven range shifts of the king penguin in a fragmented ecosystem. Nat. Clim. Change 8, 245–251. doi: 10.1038/s41558-018-0084-2
Darling E. S., Côté I. M. (2008). Quantifying the evidence for ecological synergies. Ecol. Lett. 11, 1278–1286. doi: 10.1111/j.1461-0248.2008.01243.x
Davies S. W., Marchetti A., Ries J. B., Castillo K. D. (2016). Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 3. doi: 10.3389/fmars.2016.00112
Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226. doi: 10.1186/s40168-018-0605-2
Dixon G., Abbott E., Matz M. (2020). Meta-analysis of the coral environmental stress response: Acropora corals show opposing responses depending on stress intensity. Mol. Ecol. 29, 2855–2870. doi: 10.1111/mec.15535
Drury C., Manzello D., Lirman D. (2017). Genotype and local environment dynamically influence growth, disturbance response and survivorship in the threatened coral, acropora cervicornis. PloS One 12, e0174000. doi: 10.1371/journal.pone.0174000
Eddy T. D., Lam V. W. Y., Reygondeau G., Cisneros-Montemayor A. M., Greer K., Palomares M. L. D., et al. (2021). Global decline in capacity of coral reefs to provide ecosystem services. One Earth 4, 1278–1285. doi: 10.1016/j.oneear.2021.08.016
Ellis J. I., Jamil T., Anlauf H., Coker D. J., Curdia J., Hewitt J., et al. (2019). Multiple stressor effects on coral reef ecosystems. Glob. Change Biol. 25, 4131–4146. doi: 10.1111/gcb.14819
Enochs I. C., Manzello D. P., Carlton R., Schopmeyer S., van Hooidonk R., Lirman D. (2014) Effects of light and elevated pCO2 on the growth and photochemical efficiency of acropora cervicornis. Coral Reefs 33, 477–485. doi: 10.1007/s00338-014-1132-7
Feng L., He L., Jiang S., Chen J., Zhou C., Qian Z.-J., et al. (2020). Investigating the composition and distribution of microplastics surface biofilms in coral areas. Chemosphere 252, 126565. doi: 10.1016/j.chemosphere.2020.126565
Ferrier-Pagès C., Rottier C., Beraud E., Levy O. (2010). Experimental assessment of the feeding effort of three scleractinian coral species during a thermal stress: Effect on the rates of photosynthesis. J. Exp. Mar. Biol. Ecol. 390, 118–124. doi: 10.1016/j.jembe.2010.05.007
Fuller Z. L., Mocellin V. J. L., Morris L. A., Cantin N., Shepherd J., Sarre L., et al. (2020). Population genetics of the coral Acropora millepora: Toward genomic prediction of bleaching. Science 369. doi: 10.1126/science.aba4674
Galand P. E., Chapron L., Meistertzheim A.-L., Peru E., Lartaud F. (2018). The effect of captivity on the dynamics of active bacterial communities differs between two deep-Sea coral species. Front. Microbiol. 9, 2565. doi: 10.3389/fmicb.2018.02565
Glasl B., Herndl G. J., Frade P. R. (2016). The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10, 2280–2292. doi: 10.1038/ismej.2016.9
Godoy-Vitorino F., Ruiz-Diaz C. P., Rivera-Seda A., Ramírez-Lugo J. S., Toledo-Hernández C. (2017). The microbial biosphere of the coral in northeastern Puerto Rico. PeerJ 5, e3717. doi: 10.7717/peerj.3717
Goergen E. A., Moulding A. L., Walker B. K., Gilliam D. S. (2019). Identifying causes of temporal changes in acropora cervicornis populations and the potential for recovery. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00036
Goldstein M. C., Carson H. S., Eriksen M. (2014). Relationship of diversity and habitat area in north pacific plastic-associated rafting communities. Mar. Biol. 161, 1441–1453. doi: 10.1007/s00227-014-2432-8
Grupstra C. G. B., Howe-Kerr L. I., Veglia A. J., Bryant R. L., Coy S. R., Blackwelder P. L., et al. (2022). Thermal stress triggers productive viral infection of a key coral reef symbiont. ISME J. 16, 1430–1441. doi: 10.1038/s41396-022-01194-y
Hall N. M., Berry K. L. E., Rintoul L., Hoogenboom M. O. (2015). Microplastic ingestion by scleractinian corals. Mar. Biol. 162, 725–732. doi: 10.1007/s00227-015-2619-7
Hankins C., Duffy A., Drisco K. (2018). Scleractinian coral microplastic ingestion: Potential calcification effects, size limits, and retention. Mar. pollut. Bull. 135, 587–593. doi: 10.1016/j.marpolbul.2018.07.067
Hankins C., Moso E., Lasseigne D. (2021). Microplastics impair growth in two atlantic scleractinian coral species, pseudodiploria clivosa and acropora cervicornis. Environ. pollut. 275, 116649. doi: 10.1016/j.envpol.2021.116649
Han X., Zheng Y., Dai C., Duan H., Gao M., Ali M. R., et al. (2021). Effect of polystyrene microplastics and temperature on growth, intestinal histology and immune responses of brine shrimp artemia franciscana. J. Oceanol. Limnol. 39, 979–988. doi: 10.1007/s00343-020-0118-2
Harrison S. (2020). Plant community diversity will decline more than increase under climatic warming. Philos. Trans. R. Soc Lond. B Biol. Sci. 375, 20190106. doi: 10.1098/rstb.2019.0106
Hoegh-Guldberg O., Bruno J. F. (2010). The impact of climate change on the world’s marine ecosystems. Science 328, 1523–1528. doi: 10.1126/science.1189930
Huang W., Chen M., Song B., Deng J., Shen M., Chen Q., et al. (2021). Microplastics in the coral reefs and their potential impacts on corals: A mini-review. Sci. Total Environ. 762, 143112. doi: 10.1016/j.scitotenv.2020.143112
Huang X., Leung J. Y. S., Hu M., Xu E. G., Wang Y. (2022). Microplastics can aggravate the impact of ocean acidification on the health of mussels: Insights from physiological performance, immunity and byssus properties. Environ. pollut. 308, 119701. doi: 10.1016/j.envpol.2022.119701
Hume B., D’Angelo C., Burt J., Baker A. C., Riegl B., Wiedenmann J. (2013). Corals from the Persian/Arabian gulf as models for thermotolerant reef-builders: Prevalence of clade C3 symbiodinium, host fluorescence and ex situ temperature tolerance. Mar. pollut. Bull. 72, 313–322. doi: 10.1016/j.marpolbul.2012.11.032
Hume B. C. C., D’Angelo C., Smith E. G., Stevens J. R., Burt J., Wiedenmann J. (2015). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian gulf. Sci. Rep. 5, 8562. doi: 10.1038/srep08562
Hume B. C. C., Smith E. G., Ziegler M., Warrington H. J. M., Burt J. A., LaJeunesse T. C., et al. (2019). SymPortal: A novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063–1080. doi: 10.1111/1755-0998.13004
Ip J. C.-H., Zhang Y., Xie J. Y., Yeung Y. H., Qiu J.-W. (2022). Comparative transcriptomics of two coral holobionts collected during the 2017 El niño heat wave reveal differential stress response mechanisms. Mar. pollut. Bull. 182, 114017. doi: 10.1016/j.marpolbul.2022.114017
Ivar do Sul J. A., Costa M. F. (2014). The present and future of microplastic pollution in the marine environment. Environ. pollut. 185, 352–364. doi: 10.1016/j.envpol.2013.10.036
Kaufman M. L., Watkins E., van Hooidonk R., Baker A. C., Lirman D. (2021). Thermal history influences lesion recovery of the threatened Caribbean staghorn coral acropora cervicornis under heat stress. Coral Reefs 40, 289–293. doi: 10.1007/s00338-020-02025-2
Kitchen S. A., Ratan A., Bedoya-Reina O. C., Burhans R., Fogarty N. D., Miller W., et al. (2019). Genomic variants among threatened acropora corals. G3 9, 1633–1646. doi: 10.1534/g3.119.400125
Klinges G., Maher R. L., Vega Thurber R. L., Muller E. M. (2020). Parasitic “Candidatus aquarickettsia rohweri” is a marker of disease susceptibility in acropora cervicornis but is lost during thermal stress. Environ. Microbiol. 22, 5341–5355. doi: 10.1111/1462-2920.15245
Klinges J. G., Patel S. H., Duke W. C., Muller E. M., Vega Thurber R. L. (2022). Phosphate enrichment induces increased dominance of the parasite aquarickettsia in the coral acropora cervicornis. FEMS Microbiol. Ecol. 98. doi: 10.1093/femsec/fiac013
Klinges J. G., Rosales S. M., McMinds R., Shaver E. C., Shantz A. A., Peters E. C., et al. (2019). Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated rickettsiales parasite, candidatus aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 13, 2938–2953. doi: 10.1038/s41396-019-0482-0
Korneliussen T. S., Albrechtsen A., Nielsen R. (2014). ANGSD: Analysis of next generation sequencing data. BMC Bioinf. 15, 356. doi: 10.1186/s12859-014-0356-4
LaJeunesse T. C., Parkinson J. E., Gabrielson P. W., Jeong H. J., Reimer J. D., Voolstra C. R., et al. (2018). Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.e6. doi: 10.1016/j.cub.2018.07.008
Lamb J. B., Wenger A. S., Devlin M. J., Ceccarelli D. M., Williamson D. H., Willis B. L. (2016). Reserves as tools for alleviating impacts of marine disease. Philos. Trans. R. Soc. B: Biol. Sci. 371, 20150210. doi: 10.1098/rstb.2015.0210
Lanctôt C. M., Bednarz V. N., Melvin S., Jacob H., Oberhaensli F., Swarzenski P. W., et al. (2020). Physiological stress response of the scleractinian coral stylophora pistillata exposed to polyethylene microplastics. Environ. pollut. 263, 114559. doi: 10.1016/j.envpol.2020.114559
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lawrence S. A., Wilson W. H., Davy J. E., Davy S. K. (2014). Latent virus-like infections are present in a diverse range of symbiodinium spp. (Dinophyta). J. Phycol. 50, 984–997. doi: 10.1111/jpy.12242
Leruste A., Bouvier T., Bettarel Y. (2012). Enumerating viruses in coral mucus. Appl. Environ. Microbiol. 78, 6377–6379. doi: 10.1128/AEM.01141-12
Levin R. A., Voolstra C. R., Weynberg K. D., van Oppen M. J. H. (2017). Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J. 11, 808–812. doi: 10.1038/ismej.2016.154
Libro S., Kaluziak S. T., Vollmer S. V. (2013). RNA-Seq profiles of immune related genes in the staghorn coral acropora cervicornis infected with white band disease. PloS One 8, e81821. doi: 10.1371/journal.pone.0081821
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Loiseau C., Sorci G. (2022). Can microplastics facilitate the emergence of infectious diseases? Sci. Total Environ. 823, 153694. doi: 10.1016/j.scitotenv.2022.153694
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
McDevitt-Irwin J. M., Baum J. K., Garren M., Vega Thurber R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00262
McMurdie P. J., Holmes S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Mendrik F. M., Henry T. B., Burdett H., Hackney C. R., Waller C., Parsons D. R., et al. (2021). Species-specific impact of microplastics on coral physiology. Environ. pollut. 269, 116238. doi: 10.1016/j.envpol.2020.116238
Meyer E., Aglyamova G. V., Matz M. V. (2011). Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol. Ecol. 20, 3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x
Miller M., Bourque A., Bohnsack J. (2002). An analysis of the loss of acroporid corals at looe key, Florida, USA: 1983–2000. Coral Reefs 21, 179–182. doi: 10.1007/s00338-002-0228-7
Million W. C., Ruggeri M., O’Donnell S., Bartels E., Krediet C. J., Kenkel C. D. (2022). Evidence for adaptive morphological plasticity in the Caribbean coral, Acropora cervicornis. Proc Natl Acad Sci U S A. 119(49), e2203925119. doi: 10.1073/pnas.2203925119
Moresco V., Oliver D. M., Weidmann M., Matallana-Surget S., Quilliam R. S. (2021). Survival of human enteric and respiratory viruses on plastics in soil, freshwater, and marine environments. Environ. Res. 199, 111367. doi: 10.1016/j.envres.2021.111367
Muller E. M., Bartels E., Baums I. B. (2018). Bleaching causes loss of disease resistance within the threatened coral species. Elife 7. doi: 10.7554/eLife.35066
Muller E. M., Dungan A. M., Million W. C., Eaton K. R., Petrik C., Bartels E., et al. (2021). Heritable variation and lack of tradeoffs suggest adaptive capacity in despite negative synergism under climate change scenarios. Proc. Biol. Sci. 288, 20210923. doi: 10.1098/rspb.2021.0923
Muller-Parker G., D’Elia C. F., Cook C. B. (2015). ““Interactions between corals and their symbiotic algae,”,” in Coral reefs in the anthropocene. Ed. Birkeland C. (Dordrecht: Springer Netherlands), 99–116.
Muñiz-Castillo A. I., Rivera-Sosa A., Chollett I., Eakin C. M., Andrade-Gómez L., McField M., et al. (2019). Three decades of heat stress exposure in Caribbean coral reefs: A new regional delineation to enhance conservation. Sci. Rep. 9, 11013. doi: 10.1038/s41598-019-47307-0
Nanthini devi K., Raju P., Santhanam P., Perumal P. (2022). Impacts of microplastics on marine organisms: Present perspectives and the way forward. Egypt. J. Aquat. Res. 48, 205–209. doi: 10.1016/j.ejar.2022.03.001
Nguyen-Kim H., Bouvier T., Bouvier C., Bui V., Le-Lan H., Bettarel Y. (2015). Viral and bacterial epibionts in thermally-stressed corals. J. Mar. Sci. Eng. 3, 1272–1286. doi: 10.3390/jmse3041272
Ogawa D., Bobeszko T., Ainsworth T., Leggat W. (2013). The combined effects of temperature and CO2 lead to altered gene expression in acropora aspera. Coral Reefs 32, 895–907. doi: 10.1007/s00338-013-1046-9
Oksanen J. (2007) Vegan: Community ecology package. Available at: http://www.cran.r-project.orghttps://ci.nii.ac.jp/naid/10020010631/ (Accessed August 2, 2022).
Oldenburg K. S., Urban-Rich J., Castillo K. D., Baumann J. H. (2021). Microfiber abundance associated with coral tissue varies geographically on the Belize mesoamerican barrier reef system. Mar. pollut. Bull. 163, 111938. doi: 10.1016/j.marpolbul.2020.111938
Page C. A., Willis B. L. (2008). Epidemiology of skeletal eroding band on the great barrier reef and the role of injury in the initiation of this widespread coral disease. Coral Reefs 27, 257–272. doi: 10.1007/s00338-007-0317-8
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Plafcan M. M., Stallings C. D. (2022). Microplastics do not affect bleaching of acropora cervicornis at ambient or elevated temperatures. PeerJ 10, e13578. doi: 10.7717/peerj.13578
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rädecker N., Pogoreutz C., Gegner H. M., Cárdenas A., Roth F., Bougoure J., et al. (2021). Heat stress destabilizes symbiotic nutrient cycling in corals. Proc. Natl. Acad. Sci. U. S. A. 118. doi: 10.1073/pnas.2022653118
Randazzo-Eisemann Á., Garza-Pérez J. R., Figueroa-Zavala B. (2022). The role of coral diseases in the flattening of a Caribbean coral reef over 23 years. Mar. pollut. Bull. 181, 113855. doi: 10.1016/j.marpolbul.2022.113855
R Core Team (2017) R a language and environment for statistical computing. Available at: https://www.scirp.org/.
R Core Team (2020). R: A language and environment for statistical computing (Vienna, Austria:R Foundation for Statistical Computing).
Reichert J., Arnold A. L., Hoogenboom M. O., Schubert P., Wilke T. (2019). Impacts of microplastics on growth and health of hermatypic corals are species-specific. Environ. pollut. 254, 113074. doi: 10.1016/j.envpol.2019.113074
Reichert J., Schellenberg J., Schubert P., Wilke T. (2018). Responses of reef building corals to microplastic exposure. Environ. pollut. 237, 955–960. doi: 10.1016/j.envpol.2017.11.006
Reichert J., Tirpitz V., Anand R., Bach K., Knopp J., Schubert P., et al. (2021). Interactive effects of microplastic pollution and heat stress on reef-building corals. Environ. pollut. 290, 118010. doi: 10.1016/j.envpol.2021.118010
Reynaud S., Leclercq N., Romaine-Lioud S., Ferrier-Pagés C., Jaubert J., Gattuso J.-P. (2003). Interacting effects of CO2 partial pressure and temperature on photosynthesis and calcification in a scleractinian coral. Glob. Change Biol. 9, 1660–1668. doi: 10.1046/j.1365-2486.2003.00678.x
Ricci F., Rossetto Marcelino V., Blackall L. L., Kühl M., Medina M., Verbruggen H. (2019). Beneath the surface: community assembly and functions of the coral skeleton microbiome. Microbiome 7, 159. doi: 10.1186/s40168-019-0762-y
Ripken C., Khalturin K., Shoguchi E. (2020). Response of coral reef dinoflagellates to nanoplastics under experimental conditions suggests downregulation of cellular metabolism. Microorganisms 8. doi: 10.3390/microorganisms8111759
Rivera H. E., Aichelman H. E., Fifer J. E., Kriefall N. G., Wuitchik D. M., Wuitchik S. J. S., et al. (2021). A framework for understanding gene expression plasticity and its influence on stress tolerance. Mol. Ecol. 30, 1381–1397. doi: 10.1111/mec.15820
Rochman C. M., Brookson C., Bikker J., Djuric N., Earn A., Bucci K., et al. (2019). Rethinking microplastics as a diverse contaminant suite. Environ. Toxicol. Chem. 38, 703–711. doi: 10.1002/etc.4371
Rodriguez-Casariego J. A., Cunning R., Baker A. C., Eirin-Lopez J. M. (2022). Symbiont shuffling induces differential DNA methylation responses to thermal stress in the coral montastraea cavernosa. Mol. Ecol. 31, 588–602. doi: 10.1111/mec.16246
Rosenzweig C., Karoly D., Vicarelli M., Neofotis P., Wu Q., Casassa G., et al. (2008). Attributing physical and biological impacts to anthropogenic climate change. Nature 453, 353–357. doi: 10.1038/nature06937
Ros M., Suggett D. J., Edmondson J., Haydon T., Hughes D. J., Kim M., et al. (2021). Symbiont shuffling across environmental gradients aligns with changes in carbon uptake and translocation in the reef-building coral pocillopora acuta. Coral Reefs 40, 595–607. doi: 10.1007/s00338-021-02066-1
Rotjan R. D., Sharp K. H., Gauthier A. E., Yelton R., Lopez E. M. B., Carilli J., et al. (2019). Patterns, dynamics and consequences of microplastic ingestion by the temperate coral, astrangia poculata. Proc. Biol. Sci. 286, 20190726. doi: 10.1098/rspb.2019.0726
Schopmeyer S. A., Lirman D., Bartels E., Gilliam D. S., Goergen E. A., Griffin S. P., et al. (2017). Regional restoration benchmarks for acropora cervicornis. Coral Reefs 36, 1047–1057. doi: 10.1007/s00338-017-1596-3
Schutte V. G. W., Selig E. R., Bruno J. F. (2010). Regional spatio-temporal trends in Caribbean coral reef benthic communities. Mar. Ecol. Prog. Ser. 402, 115–122. doi: 10.3354/meps08438
Scucchia F., Malik A., Putnam H. M., Mass T. (2021). Genetic and physiological traits conferring tolerance to ocean acidification in mesophotic corals. Glob. Change Biol. 27, 5276–5294. doi: 10.1111/gcb.15812
Shukla P. R., Skeg J., Buendia E. C., Masson-Delmotte V., Pörtner H.-O., Roberts D. C., et al. (2019). Climate change and land: An IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems.
Soares M., de O., Matos E., Lucas C., Rizzo L., Allcock L., et al. (2020). Microplastics in corals: An emergent threat. Mar. pollut. Bull. 161, 111810. doi: 10.1016/j.marpolbul.2020.111810
Tang J., Ni X., Zhou Z., Wang L., Lin S. (2018). Acute microplastic exposure raises stress response and suppresses detoxification and immune capacities in the scleractinian coral pocillopora damicornis. Environ. pollut. 243, 66–74. doi: 10.1016/j.envpol.2018.08.045
Thurber R. L. V., Correa A. M. S. (2011). Viruses of reef-building scleractinian corals. J. Exp. Mar. Bio. Ecol. 408, 102–113. doi: 10.1016/j.jembe.2011.07.030
Thurber R. V., Payet J. P., Thurber A. R., Correa A. M. S. (2017). Virus-host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 15, 205–216. doi: 10.1038/nrmicro.2016.176
Towle E. K., Enochs I. C., Langdon C. (2015a). Correction: Threatened Caribbean coral is able to mitigate the adverse effects of ocean acidification on calcification by increasing feeding rate. PloS One 10, e0139398. doi: 10.1371/journal.pone.0139398
Towle E. K., Enochs I. C., Langdon C. (2015b). Threatened Caribbean coral is able to mitigate the adverse effects of ocean acidification on calcification by increasing feeding rate. PloS One 10, e0123394. doi: 10.1371/journal.pone.0123394
Tunnicliffe V. (1981). Breakage and propagation of the stony coral Acropora cervicornis. Proc. Natl. Acad. Sci. U. S. A. 78, 2427–2431. doi: 10.1073/pnas.78.4.2427
van de Water J. A. J. M., Leggat W., Bourne D. G., van Oppen M. J. H., Willis B. L., Ainsworth T. D. (2015). Elevated seawater temperatures have a limited impact on the coral immune response following physical damage. Hydrobiologia 759, 201–214. doi: 10.1007/s10750-015-2243-z
van Oppen M. J. H., Blackall L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
van Woesik R., Randall C. J. (2017). Coral disease hotspots in the Caribbean. Ecosphere 8, e01814. doi: 10.1002/ecs2.1814
Ware M., Garfield E. N., Nedimyer K., Levy J., Kaufman L., Precht W., et al. (2020). Survivorship and growth in staghorn coral (Acropora cervicornis) outplanting projects in the Florida keys national marine sanctuary. PloS One 15, e0231817. doi: 10.1371/journal.pone.0231817
Williams S. D., Klinges J. G., Zinman S., Clark A. S., Bartels E., Villoch Diaz Maurino M., et al. (2022). Geographically driven differences in microbiomes of originating from different regions of florida’s coral reef. PeerJ 10, e13574. doi: 10.7717/peerj.13574
Wright R. M., Aglyamova G. V., Meyer E., Matz M. V. (2015). Gene expression associated with white syndromes in a reef building coral, acropora hyacinthus. BMC Genomics 16, 371. doi: 10.1186/s12864-015-1540-2
Yates K. K., Moore C. S., Smiley N. A. (2019). Time series of autonomous carbonate system parameter measurements from crocker reef (Florida, USA:U.S. Geological Survey). doi: 10.5066/P90NCI8T
Zaneveld J. R., Burkepile D. E., Shantz A. A., Pritchard C. E., McMinds R., Payet J. P., et al. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 7, 11833. doi: 10.1038/ncomms11833
Zettler E. R., Mincer T. J., Amaral-Zettler L. A. (2013). Life in the “Plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Keywords: global change, microplastics, gene expression, Acropora cervicornis, coral microbiome, ocean acidification, ocean warming
Citation: Bove CB, Greene K, Sugierski S, Kriefall NG, Huzar AK, Hughes AM, Sharp K, Fogarty ND and Davies SW (2023) Exposure to global change and microplastics elicits an immune response in an endangered coral. Front. Mar. Sci. 9:1037130. doi: 10.3389/fmars.2022.1037130
Received: 05 September 2022; Accepted: 06 December 2022;
Published: 13 January 2023.
Edited by:
Michael E. Hellberg, Louisiana State University, United StatesReviewed by:
Jessica Reichert, University of Giessen, GermanyCheryl Hankins, United States Environmental Protection Agency (EPA), United States
Copyright © 2023 Bove, Greene, Sugierski, Kriefall, Huzar, Hughes, Sharp, Fogarty and Davies. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Colleen B. Bove, Y29sbGVlbmJvdmVAZ21haWwuY29t; Sarah W. Davies, ZGF2aWVzc3dAYnUuZWR1
 Colleen B. Bove
Colleen B. Bove Katharine Greene
Katharine Greene Sharla Sugierski2
Sharla Sugierski2 Nicola G. Kriefall
Nicola G. Kriefall Koty Sharp
Koty Sharp Nicole D. Fogarty
Nicole D. Fogarty Sarah W. Davies
Sarah W. Davies