- 1Key Laboratory of Tropical & Subtropical Fishery Resource Application & Cultivation of Ministry of Agriculture and Rural Affairs, Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2National Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai, China
Macrobrachium rosenbergii is an important cultured shrimp worldwide, and its precocious puberty has led to serious economic losses. Although reproductive sterilization can avoid this problem, little is known about the molecular mechanisms underlying gonadal development and gametogenesis in M. rosenbergii. Here, we conducted transcriptome analysis of the ovaries, testes, and male/female brain tissues of M. rosenbergii to discover germ cell development-related genes. A total of 60,607 unigenes were identified, of which 20,963 unigenes could be functionally annotated. Eighteen candidate genes were identified by Venn diagram analysis, keyword, and known marker search, followed by elimination of low-expression and repetitive genes. Subsequent real-time quantitative reverse transcription polymerase chain reaction and in situ hybridization identified five genes (RAD51, vasa, SPDS, MRR, and Fem-1C) associated with germ cell development—RAD51, vasa, and SPDS were expressed in both male and female gonads, whereas Fem-1C was specifically expressed in the ovary and MRR in the testis. In the ovary, vasa, SPDS, and Fem-1C were mainly expressed in stage 1–3 oocytes, while RAD51 was expressed in stage 2–3 oocytes. In testis, vasa were significantly expressed in spermatogonia and primary spermatocytes, whereas RAD51 and SPDS were only enriched in spermatogonia and MRR in vas deferens. Our research indicates that these five genes are important germ cell development-related genes, of which RAD51, SPDS, and Fem-1C were proven to function in germ cells of this species for the first time. The discovery of these genes could help develop molecular breeding techniques to overcome precocious puberty in M. rosenbergii.
1 Introduction
Giant freshwater prawns (Macrobrachium rosenbergii) are native to the Mekong River basin in Southeast Asia. Because of its rapid growth, large size, and delicious flavor, it has gradually become one of the main shrimp species in freshwater aquaculture in Asia. In China, the total annual output reached 171, 263 tons in 2021 (China Fishery Statistical Yearbook, 2022). Recently, the production of M. rosenbergii in China has increased to a certain extent; however, some key problems such as germplasm degradation and various diseases still affect the aquaculture industry of M. rosenbergii in China, and its industrial development has encountered certain bottlenecks.
Initially, our research team conducted a series of market research in China to identify the key bottlenecks in the industrial development of M. rosenbergii. Our research revealed that in addition to the traditional problems such as the lack of seed supply caused by the degradation of germplasm and the low emergence rate, there is a frequent occurrence of gonadal precocity (female shrimp weighing only 6 g already held eggs in their abdomen, and males weighing only 2 g held mature sperm in their testes) was a major factor hindering the healthy development M. rosenbergii. Precocious maturity leads to stagnated growth, compromised health, and flesh quality (O’Brien, 1984; Jin and Xie, 2001), as the resources and energy are diverted to gonadal development, gamete production, mating, and reproduction. Reproductive sterilization strategies such as sterile or single-sex group breeding can preclude the deterioration of growth and stagnated size associated with precocious maturation (Basavaraju et al., 2002). In mollusks and fish such as Crassostrea hongkongensis (Zhang et al., 2017), Crassostrea rivularis (Yang et al., 2008), and Oncorhynchus mykiss (Wang et al., 2005), chemical or physical tools have been used to induce triploids to construct sterile populations. However, the progress in developing a sterile population in M. rosenbergii is limited. M. rosenbergii has a special reproductive mode: egg holding and hatching, and at present, there is no established technology for in vitro culture of fertilized eggs. Although in vitro embryo shock treatment is easy to perform, it may lead to high mortality. In addition, the gonads of artificially induced triploid individuals can still develop normally and produce normal functional aneuploid gametes, which have potential ecological risks. Therefore, researchers attempted to obtain a neo-female M. rosenbergii population by removing the gonads of immature male M. rosenbergii to build a unisexual population (Sagi et al., 1990). However, this method is difficult to operate and has a high threshold, making it difficult to popularize on a large scale.
With the continuous advancements in molecular biotechnology and sequencing technologies, the focus of aquatic animal-related research has shifted to the genome and transcriptome levels. RNA interference (RNAi) has become a new effective tool to induce infertility in aquatic animals and has been widely used in various species. For example, in teleost fish, the agate phenotype of loach (Fujimoto et al., 2010) and sturgeon (Linhartová et al., 2015) has been achieved by inactivating the mRNA necessary for the formation of primordial germ cells using the morpholino method, in which the dsRNA of vasa was injected into the ovaries of the fully mature female Pinctada fucata, and the mother was induced to lay eggs 6 h later. The gonads of the injected offspring were significantly smaller than those of the control group (Iwai et al., 2015). In Eriocheir sinensis, a crustacean, RNAi with the DMRT-like gene inhibited the normal development of the testis (Zhang and Qiu, 2010). In Fenneropenaeus chinensis, closely related to M. rosenbergii, interference with the doublesex (DSX) gene significantly reduced the expression of the androgen-promoting gene (AGH) (Li et al., 2018). In M. rosenbergii, RNAi has been successfully used for silencing of insulin-like AG (Mr-IAG) gene of male M. rosenbergii to reverse its sex into full-functional neo-female individuals, which could mate with normal male individuals to produce an all-male population (Ventura et al., 2012). Levy et al. (2016) successfully constructed an all-female population by injecting hyperplastic gonad (hAG) cells into female juvenile M. rosenbergii to obtain neo-male individuals. Recently, Xu et al. (2022) obtained a fully functional neo-female individual by RNAi-based silencing of MroDmrt11E, which is upstream of Mr-IAG in M. rosenbergii. However, these studies focused on developing parthenogenetic populations of M. rosenbergii, and reports on sterile populations of M. rosenbergii are scarce. Collectively, these results indicate that mRNA inhibition required for reproductive development at various stages of the life cycle of various animals can affect their normal reproductive development and even change their sex phenotypes. However, the limited knowledge of the gonad development and gametogenesis mechanism of M. rosenbergii and the lack of the corresponding breakthrough point hinders the application of potential tools such as RNAi to achieve reproductive sterilization in this species.
Therefore, screening and identifying key genes that play a role in germ cell development, then clarifying the developmental molecular mechanisms of germ cells is an important prerequisite for the future development of sterility induction methods in M. rosenbergii. Recently, related research has made certain progress on other crustacean species: Wei et al. (2021) found that PIWI plays a role in the formation of germ cell nucleus of E. sinensis, and can maintain the apoptosis of abnormal germ cells to remove abnormal germ cells, which is different from its function in mammals; In Penaeus vannamei, a newly discovered maternal gene MnTdrd has been proved to function in the formation and differentiation of reproductive cells in P. vannamei, and can be used as a new germ cell marker to track the origin and migration of germ cells (Dong et al., 2021); Meanwhile, KIFC1 was found to be possibly involved in mitosis of spermatogonia, meiosis of spermatocytes, and acrosomal formation during spermatogenesis in Penaeus japonicus, and its reduced expression may induce abnormal assembly of microtubules and lead to apoptosis of spermatogonia and spermatocytes (Hao and Yang, 2019). Additionally, the research of foxl2 (Wan et al., 2022), IAG2 (Liu et al., 2021) and DDX52 (Li et al., 2018) genes in the germ cells of crustaceans has also made considerable achievements. However, recent research on M. rosenbergii are scarce, and the isolation of germ cell-related key marker genes needs to be further promoted.
Previously, the conventional method for isolating genes in non-model organisms was polymerase chain reaction (PCR) amplification based on the conserved sequences of homologous genes. Because this method is time-consuming, inefficient, and probably not applicable to highly differentiated target genes in different species, screening for target genes has been a major bottleneck in molecular biology research (Bar et al., 2016). In recent years, large-scale parallel transcriptome sequencing platforms, such as ABI solid, Roche 454, and Illumina Solexa platforms, have been gradually popularized, providing new methods and ideas to encounter various research problems (Oghenekaro et al., 2016). In addition to model organisms, large-scale transcriptome sequencing has been applied to different types of research on an increasing number of aquatic species such as Thunnus maccoyii (Bar et al., 2016), Sepiella japonica (Wang et al., 2020), Fenneropenaeus merguiensis (Prasert et al., 2021) and Salmo salar (Trine et al., 2021). In Macrobrachium nipponense, large-scale transcriptome sequencing technology was used to explore the molecular mechanisms of stress resistance (Li et al., 2021), sex differentiation (Abu Abayed et al., 2019), and immunity (Ou et al., 2021). In contrast, transcriptome resources of M. rosenbergii are scarce, which is the main obstacle to the molecular biology research of this species— As of September 27, 2022, there were only 4,576 expressed sequence tags (ESTs) in the National Center for Biotechnology Information (NCBI) database. In this study, we aimed to explore and discover genes in the key regulatory organs of M. rosenbergii, which may be related to germ cell development. To this end, we performed transcriptome sequencing and analysis of the brain and gonad tissues in M. rosenbergii. We employed different strategies, such as various gene screening methods, quantitative real-time PCR (qRT-PCR), and in situ hybridization (ISH), to identify the target genes. The findings will provide key information about germ cell development and contribute to the sustainable development of RNAi research on M. rosenbergii.
2 Materials and methods
2.1 Ethics statement, experimental fish and sample collection
All experiments were conducted at the Pearl River Fisheries Research Institute of the Chinese Academy of Fishery Sciences (Guangzhou, China). The study was approved by the Animal Experiment Ethics Committee of Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences (Guangzhou, China).
To avoid the gonadal atrophy caused by over-ripening, and ensure that the sample contains germ cells at different developmental stages, 30 males and 30 females were obtained from Foshan Sanshui Baijin Seedling Co., Ltd. (Foshan, China) as normal three-month-old M. rosenbergii specimens with a body weight of 10.3 ± 2.1 g and a body length of 7.6 ± 1.5 cm. The brains, gonads, and other tissues (including intestine, gills, muscle, stomach, liver, and heart) of specimens were immediately collected on ice with disinfected scissors and tweezers. Every tissue group had three sets of biological replications; and each replication contained samples from 30 individuals. All tissue samples were soaked in RNAlater (Thermo Scientific, Waltham, MA, USA) and stored at -20°C for transcriptome sequencing and expression analysis. A portion of gonad samples was soaked in Bouin’s solution for 4 h and stored in 70% ethanol at 4°C for ISH.
2.2 RNA extraction, library construction, and illumina sequencing
Total RNA was extracted from tissues using TRIzol reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. Then the sample mRNA was enriched with Oligo (dT) magnetic beads. Fragmentation buffer was added to break mRNA into short segments, mRNA was used as a template to synthesize the first cDNA strand with six-base random primers, and then buffer, dNTPs, RNase H, and DNA polymerase I were added to synthesize the second cDNA strand. After purification with QiaQuick PCR kit (Qiagen, Venlo, Netherlands) and elution by EB buffer, terminal repair, addition of poly(A) and connection of sequencing linker were carried out, then fragment size selection was carried out using agarose gel electrophoresis, and finally PCR amplification was carried out. The built sequencing library was sequenced by Illumina HiSeqTM instrument at SAGENE Biotech Co., Ltd. (Guangzhou, China).
2.3 Illumina read processing and assembly, functional annotation, and classification
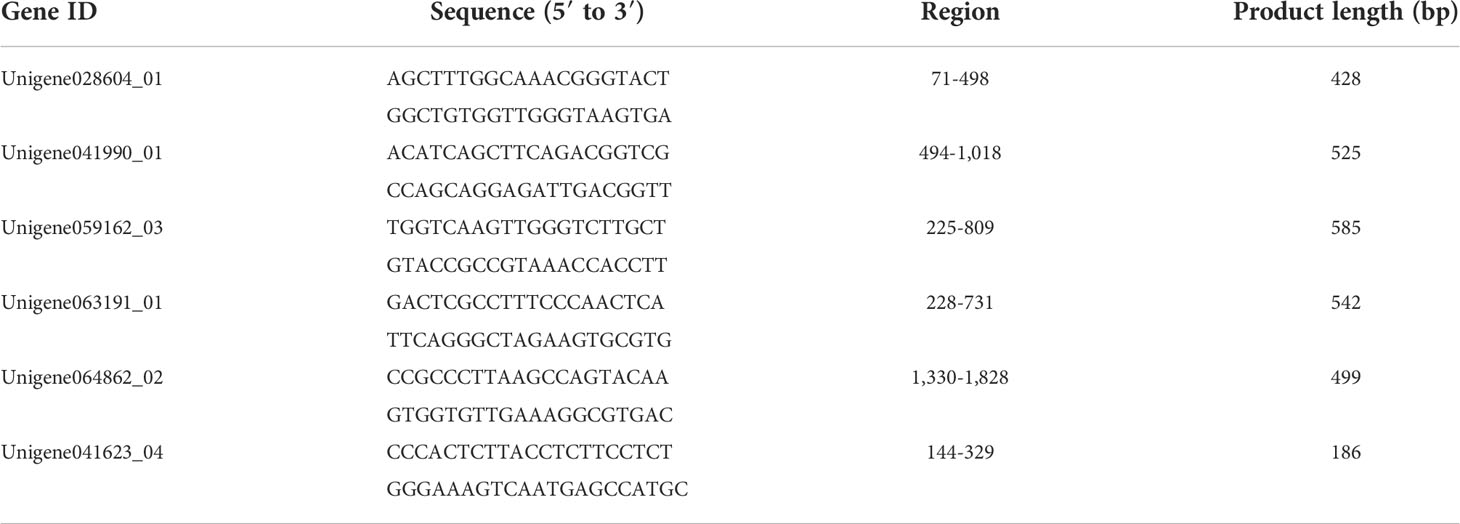
To obtain high-quality reads for further analysis, reads containing adapters or poly(A) sequences and low-quality reads were removed from the raw data using the FastQC v0.11.5 software to generate clean data. The N50 and GC percentages of the cleaned data were calculated. Then, high-quality reads were reconstructed and assembled into a single genome using Trinity v2.2.0 with default parameters (Cabau et al., 2017). First, Trinity was used to connect the reads with a certain length of overlap into longer segments, and these assembled segments without N obtained through the relationship of reads overlap were used as the assembled unigenes. Lastly, the National Center for Biotechnology Information non-redundant (Nr, ftp://ftp.ncbi.nih.gov/blast/db/), Swiss-Protein (SwissProt, https://www.uniprot.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/), and Gene Ontology (GO, http://geneontology.org/) databases were used to obtain the feature and annotation information for all assembled unigenes (Figure 1A).
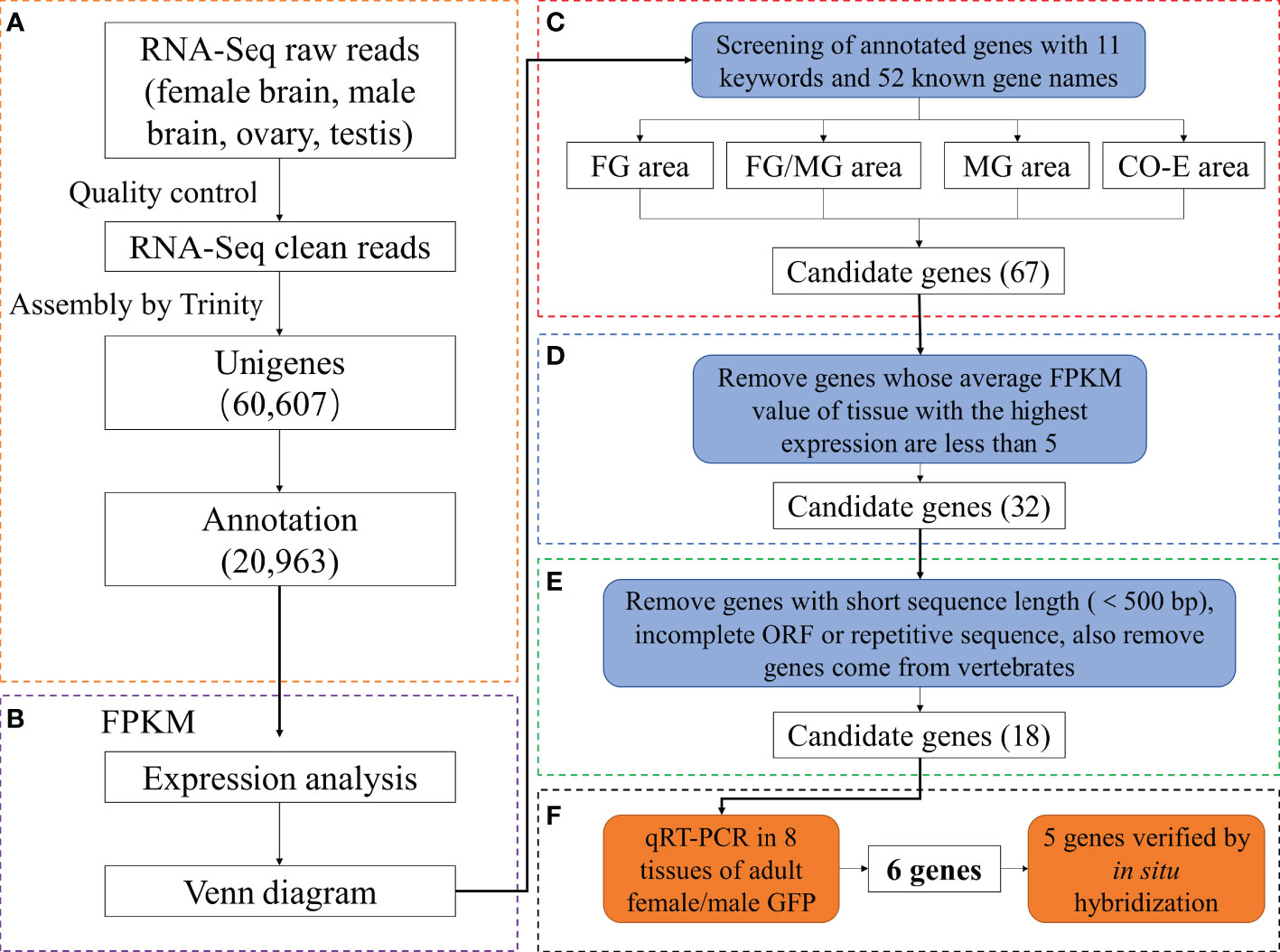
Figure 1 Assembly and analysis pipeline of Giant freshwater prawn Macrobrachium rosenbergii gonadal transcriptome. (A) Workflow of transcriptome data analysis. (B) Expression analysis and construction of Venn diagram. (C) Identification of reproduction-related genes by searching databases with 11 reproduction-related keywords and 52 known gene names (FG/FB and MG/MB areas were not shown because no candidate genes were obtained in them.). (D, E) Filtering the candidate genes by analyzing the FPKM value, sequence length, ORF and source. (F) Obtaining and verifying the final candidate genes using qRT-PCR and in situ hybridization. FG, female gonad (ovary); MG, male gonad (testis); CO-E, Co-expression (ovary, testis, female brain and male brain).
2.4 Gene expression distribution and identification of candidate genes
Reads in RNA-Seq analysis were normalized to the fragments per kilobase of transcript per million mapped reads (FPKM) to estimate gene expression levels (Mortazavi et al., 2008). Results were subjected to multiple-test correction using an error-finding rate (FDR) threshold < 0.05 (Benjamini and Yekutieli, 2001; Lu et al., 2015). A gene is defined as specifically expressed in a single tissue (SEGs) if FDR in that single tissue is ≤ 0.05 and log2 ratio = 0 (the FPKM value of the gene in one sample is at least twice that of the gene in the other sample). Genes with FDR ≤ 0.05 and log2 ratio < 2 in multiple tissues were classified as co-expressed genes (CEGs). The Venn diagrams were constructed from annotated gene expression profiles of single genes in single or multiple tissues. (Figure 1B). Subsequently, candidate genes were obtained by screening keywords (Figure 1C), comparing expression levels (Figure 1D) ,sequences and species origin (Figure 1E).
The FPKM values were also used to estimate gene expression levels using the StringTie software (Pertea et al., 2016). Differentially expressed genes (DEGs) between female brain (FB), female gonad (FG), male brain (MB), and male gonad (MG) groups were analyzed using DESeq 2 (Love et al., 2014) with a threshold FDR < 0.05 and absolute fold change (FC)≥2(log 2 | FC |≥1). Log2(FC) > 0 indicated upregulation, and log2(FC) < 0 indicated downregulation.
2.5 Quantitative real-time PCR (qRT–PCR) analysis
The qRT-PCR was performed to verify the sequencing results of ten randomly selected DEGs and the expression levels of the candidate genes in the male and female gonads, intestine, gills, brain, muscle, stomach, liver, and heart tissues. RNA from each sample (in 3 triplicates) was collected using TRIzol Reagent (Invitrogen, Waltham, MA, USA), as described in section 2.2. First-strand cDNA was synthesized from 1 μg of total RNA using an Evo M-MLV RT Premix for PCR Kit (Accurate Biology, China). The specific qRT–PCR primers shown in Supplementary Tables S1 and S2 were designed using online Primer-BLAST from NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The reaction mix comprised 1 μL (50 ng/μL) cDNA, 10 μL iTaq Universal SYBR® Green Supermix (BIO-RAD, CA, USA), 2 μL of upstream and downstream primer, and 7 μL of nuclease-free water to adjust the final volume to 20 μL. Reactions were performed using the QuantStudio 6 Real-Time PCR System (Applied Biosystems, CA, USA) using the following protocol: 95°C for 5 min; 37 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 20 s; and 72°C for 10 min for data acquisition. Each sample was tested in triplicates. β-Actin gene (AY651918.2) was used as the internal reference to normalize the Ct values of each reaction using the 2−ΔΔCt method (Livak and Schmittgen, 2001) (Figure 1F).
2.6 In situ hybridization
Sense and anti-sense RNA probes labeled with digoxigenin were synthesized from the corresponding regions (Table 1). The cDNA fragment of the final candidate gene was subcloned into pGEM T-easy vector. DIG-labeled uridine triphosphate (Roche, Basel, Switzerland) and T7 RNA polymerase (Promega, WI, USA) were used to transcribe sense and antisense probes in vitro. For ISH, tissue samples from gonads were fixed in Bouin’s solution at 4°C for 4 h (GenXion, China). After dehydration under the condition of increasing ethanol concentration, a part of each sample was embedded in paraffin, and cut into 7-μm slices with Leica RM2016 Microtomes (Leica, Weztlar, Germany). Then, paraffin sections were placed on Thermo adhesive glass slides (Thermo Scientific, Waltham, MA, USA), dewaxed, and dehydrated by dipping in xylene-ethanol series. Staining sections with hematoxylin-eosin (HE) solution (Nanjing Jiancheng Bioengineering Institute, China), or ISH treatment with DIG-labeled RNA probes, followed the previously described protocol (Nagasawa et al., 2009). For subcellular localization of the candidate genes, gonad samples were obtained and pretreated as described in 2.1. We divided oogenesis in the obtained samples into four stages and spermatogenesis into three stages, following the method described in previous studies (Luo et al., 1999; Deng and Hu, 2002; Meeratana and Sobhon, 2007). The resulting ISH sections were observed under a Nikon Eclipse ci upright microscope (Nikon, Japan) equipped with a Nikon DS-U3 digital camera (Nikon, Japan).
2.7 Statistical analysis
Microsoft Excel 2021 (Microsoft Corporation, Redmond, WA, USA) and Graphpad Prism 9 (Graphpad Software, San Diego, CA, USA) were used to sort and analyze the experimental data.
3 Results
3.1 Transcriptome sequencing and assembly
For the use of transcriptome databases for M. rosenbergii, four cDNA libraries were produced by applying the stringent quality assessment and data filtering of sequences from the female/male brain, ovary, and testis samples. We obtained 110.7, 104.5, 91.9, and 83.4 million high-quality clean reads, respectively. Using the Trinity assembly program, 60,607 unigenes were generated by hybrid de novo assembly with lengths ranging from 301 to 21,012 bp (average length: 1,420 bp) (Table 2). The raw transcriptome sequences generated in this study were uploaded to the NCBI Sequence Read Archive (SRA) with the accession number PRJNA884099.
3.2 Functional annotation and classification
For functional annotation of the 60,607 unigenes obtained, these genes were used as query sequences and the nr, SwissProt, KEGG, and KOG (evaluate < 1e-5) protein databases were searched using blastx. The results showed that only 34.6% (20,963 in total, NR: 20,792; Swissprot: 14,885; KEGG: 7,387; KOG: 14,063) of the unigenes showed high sequence similarity in these protein databases. The functions of given unigenes were determined by comparing the proteins which had the highest homology with these genes, such as nuclear autoantigenic sperm protein (Unigene045489), round spermatid basic protein 1 (Unigene018151) and male reproductive-related microfibril-associated protein (Unigene026230). Furthermore, using the Blast2GO software, 12.6% of the annotated unigenes (2,642) could be assigned a GO ID (Table 3). According to the GO classification table, the GO annotations of unigenes are analyzed. Among these three ontologies: molecular function, cellular component; biological process, cellular process, cell and catalytic activity are the most gene-rich level 2 GO terms in the three ontologies (Figure S1). Meanwhile, in the KEGG database, the pathways with the most annotated gene entries are Ribosome, RNA transport, and Spliceosome, all of which belong to Genetic Information Processing in KEGG A Class (Figure S2).
3.3 Verification of DEGs using qRT-PCR
DEGs of six comparison groups were acquired by FDR and log2FC (Table S3). Ten DEGs were randomly selected from different groups for the qRT-PCR analysis. All the tested genes showed similar expression patterns (Figure 2) with their FPKM values, indicating the reliability and accuracy of our transcriptome data and subsequent analysis.
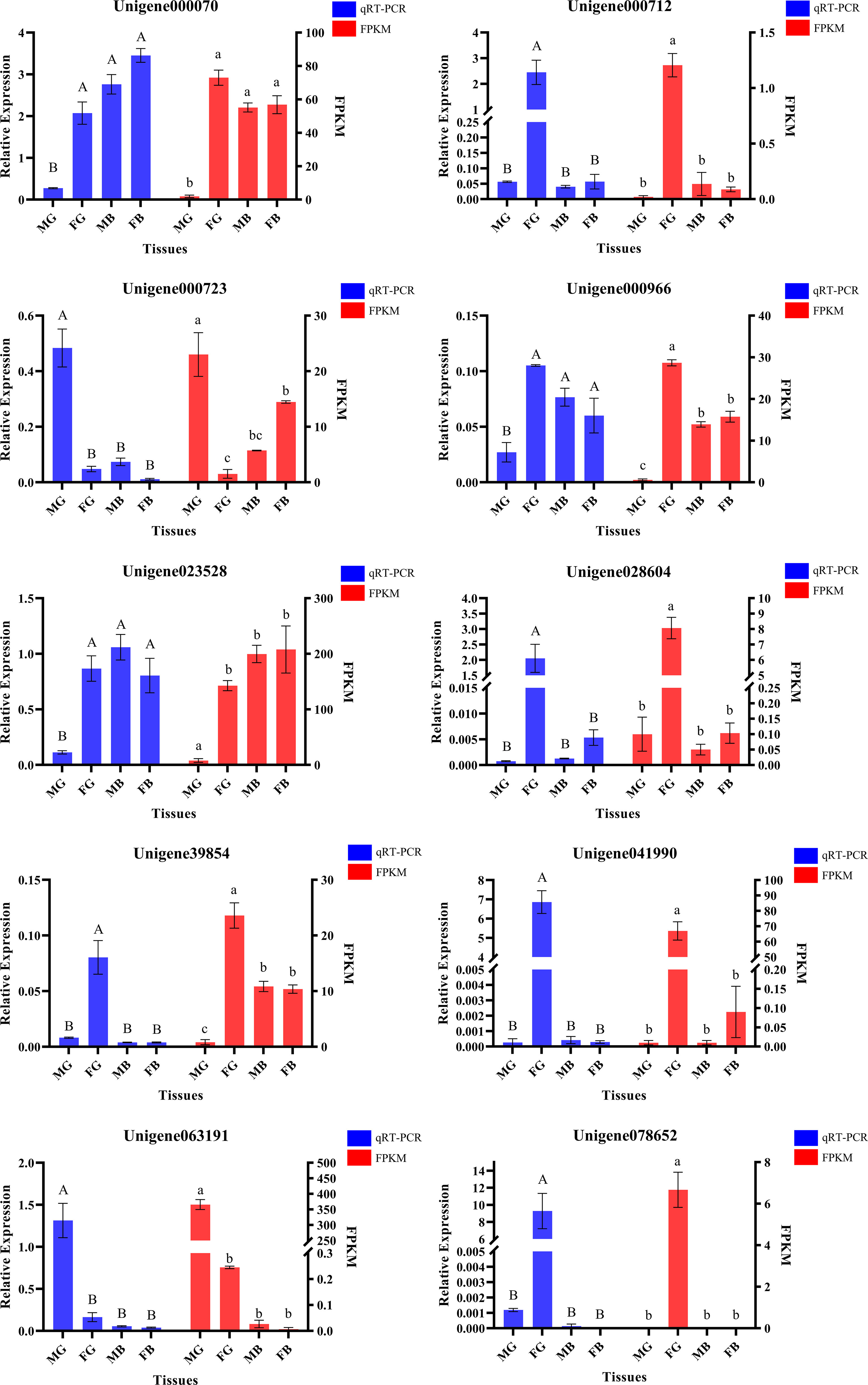
Figure 2 Validation of the 10 differentially expressed genes profiles using quantitative real-time PCR (Columns with the same letter above indicate that there is no significant difference between them, if not, there is significant difference).
3.4 Gene expression distribution and identification and screening of germ cell-related candidate genes
Normalization of the reads in the RNA-Seq analysis to the FPKM identified a total of — SEGs (FG: 755; MG: 197; FB: 32; MB:12) and —– CEGs (15,627) in four tissues: FG, MG, FB, and MB. Venn diagram identified 256 unigenes expressed in FG and MG, 755 specifically expressed in FG, and 197 in MG. In addition, 15,627 unigenes were co-expressed in FG/MG/FB/MB (Figure 3A).
Figure 3 (A) The number of annotated unigenes present in each area of the Venn diagram. (B) Distribution of final candidate genes on Venn diagram (Bold font represents known gene markers).
Next, to identify the germ cell-related candidate genes, genes identified in the four key regions in the Venn diagram (FG/MG, FG, MG, and FG/MG/FB/MB) were screened using 11 gonadal-related keywords, namely, oocyte, sperm, fertilization, sex, meiosis, male, gonad, germ cell, gamete, female, and egg. In addition, 52 known sex-related gene markers and genes related to gonadal development (Matsumoto et al., 2013; Chen et al., 2015; Yue et al., 2015), including vasa, nanos, SPATA, DMC1, Sox, and RAD51, were searched in Venn diagrams and unigene databases to obtain data on homologous genes. The analyses identified 60 candidate genes in FG/MG/FB/MB region, 4 in MG, 2 in FG, and 1 in FG/MG (Figure 3A and Table S4).
Furthermore, to locate candidate genes that participate in germ cell development, genes with low FPKM values were eliminated by querying the statistical data on their expression levels in the transcriptome database, which identified 32 candidate genes (Table S5). Then the sequence assembly of these genes and gene sources were analyzed and compared. Genes with a length of less than 500 bp or without a complete open reading frame were removed. Simultaneously, to further improve the reliability of candidate genes, homologous genes from vertebrates with relatively distant genetic relationships were removed (Table S5). Finally, 18 candidate genes were identified (Figure 3B and Table 4). Among them, 17 genes, including Sox14, sex determination protein fruitless-like isoform X47(SDPF), Fem-1A, RAD51, vasa, spermidine synthase-like (SPDS), and ER degradation-enhancing alpha-mannosidase-like 3(ER3), were found in the FG/MG/FB/MB co-expression region. Only the male reproductive-related (MRR) gene was identified in the MG region. In addition, no qualified candidate genes were found in the FG region after screening.
3.5 Validation of target genes related to germ cell development
Next, to validate the 18 candidate genes, their specific expression in the tissues of adult male and female M. rosenbergii were verified using qRT-PCR. As shown in Figure 4, 12 genes, including ER (Unigene038520_03), ER3 (Unigene045914_01), NAS (Unigene045489_07), SN1A (Unigene037486_01), SPATA7 (Unigene052170_01) and SPDSCRF (Unigene052832_05) showed significantly lower expression in ovary and testis tissues than those in some other hermaphroditic tissues. Only RAD51 (Unigene041623_04), vasa (Unigene059162_03), and SPDS (Unigene064862_02) showed specific expression in both FG and MG, and their expression in other body tissues was extremely low or undetectable. Fem-1C (Unigene041990_01) and SDPF (Unigene028604_01) specifically expressed in FG were only slightly expressed in the liver, heart, and other body tissues of male M. rosenbergii. In addition, the expression of MRR (Unigene063191_01) was more significant in the testis than in other hermaphroditic body tissues.
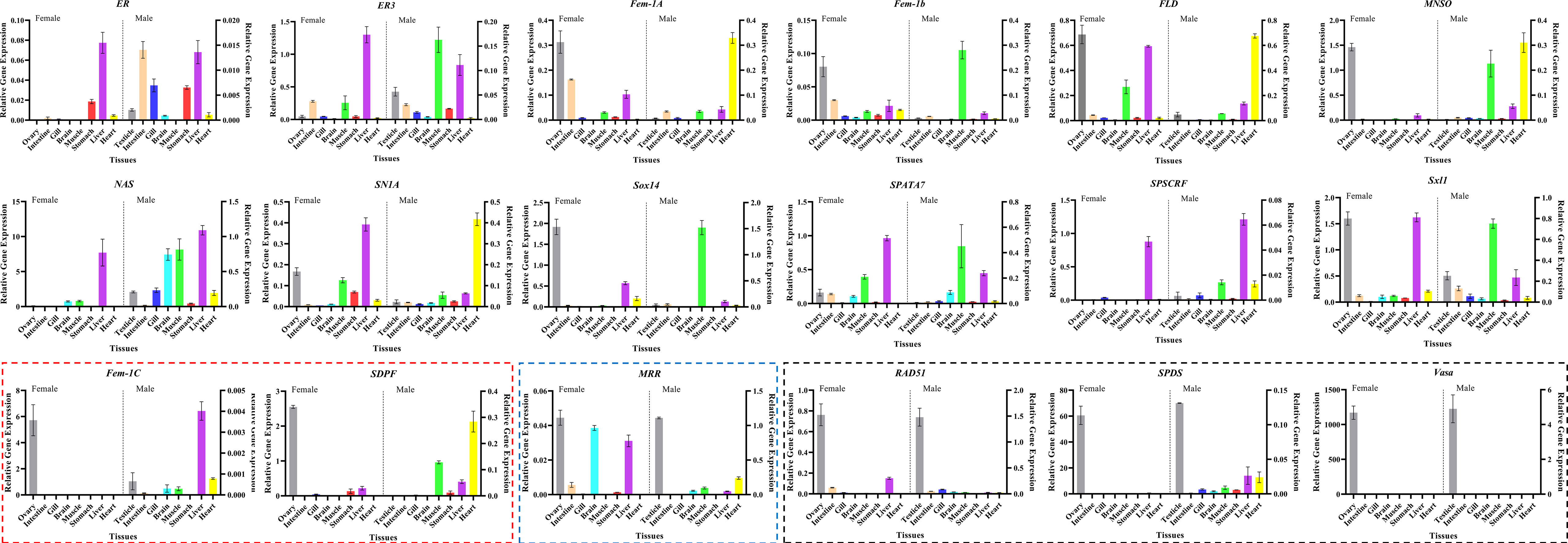
Figure 4 Tissue expression pattern of the final candidate genes in adult female/male M. rosenbergii using qRT-PCR (Boxes indicate that these genes are specifically expressed in female/male gonads. Red box represents the genes with high expression only in ovary, blue box represents the genes with high expression only in testis, black box represents the genes with high expression in both ovary and testis).
According to the qRT-PCR results, five candidate genes (Fem-1C, SDPF, RAD51, vasa, and SPDS) were selected for further analysis to determine the subcellular localization of these genes in the gonads. A previous study in M. rosenbergii has shown that MRR is specifically expressed in secretory cells in the inner wall of vas deferens, which may be related to sperm maturation and sperm pod formation (Cao, 2006). Therefore, we did not analyze the localization of MRR. Compared with gonad sections after HE staining (Figures 5A, D, G, J; Figures 6A, D, G, J; Figures 7A, D, G, J; Figures 8A, D, G, J;), ISH results showed hybridization signals with the anti-sense probes of RAD51, vasa, SPDS, and Fem-1C in the germ cells of M. rosenbergii. However, no signals were observed in any germ cells using the anti-sense probe of SDPF (data not shown). Vasa mRNA was detected in stage 1–3 oocytes of the ovaries (Figures 8B, E) and spermatogonia and primary spermatocytes of the testes (Figures 8H, K). RAD51 mRNA was detected in stage 2–3 oocytes (Figures 5B, E) and spermatogonia (Figures 5H, K). SPDS mRNA was mainly enriched in the cytoplasm of stage 1–3 oocytes (Figures 6B, E) and spermatogonia (Figures 6H, K). Fem-1C mRNA was only detected in stage 1–3 oocytes (Figures 7B, E), whereas no signal was detected in any germ cells in the testes (Figures 7H, K). In addition, hybridization signals were not observed in any germ cells using the vasa, RAD51, Fem-1C, and SPDS sense probes (Figures 5C, F, I, L; Figure 8C, F, I, L; Figures 6C, F, I, L; Figures 7C, F, I, L).
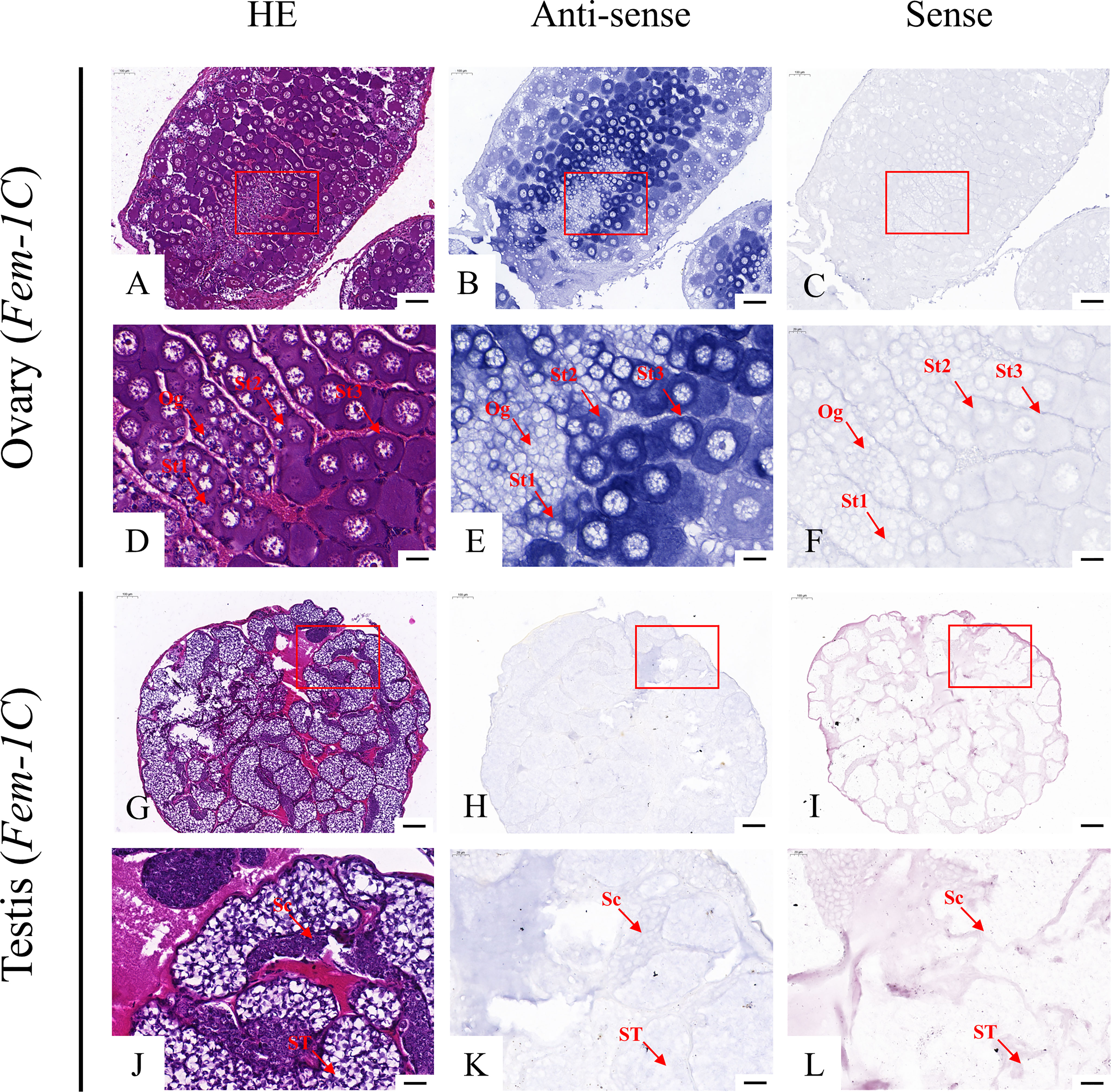
Figure 5 Localization of vasa mRNA in the ovary (A–F) and testis (G–L) of M. rosenbergii. HE staining of ovary (A, D) and testis (G, J) used for in situ hybridization. Anti-DIG signals obtained by vasa-antisense probes (B, E, H, K) and vasa-sense probes (C, F, I, L). (D–F, J–L) High-magnification images of areas enclosed in red boxes in the images to the top (A–C, G–I). Og, oogonia; St1, stage-1 oocytes; St2, stage-2 oocytes; St3, stage-3 oocytes. Sc: spermatogonia; Sg: primary spermatocyte; ST: spermatids. Scale bars 100 μm (A–C, G–I); 20 μm (D–F, J–L).
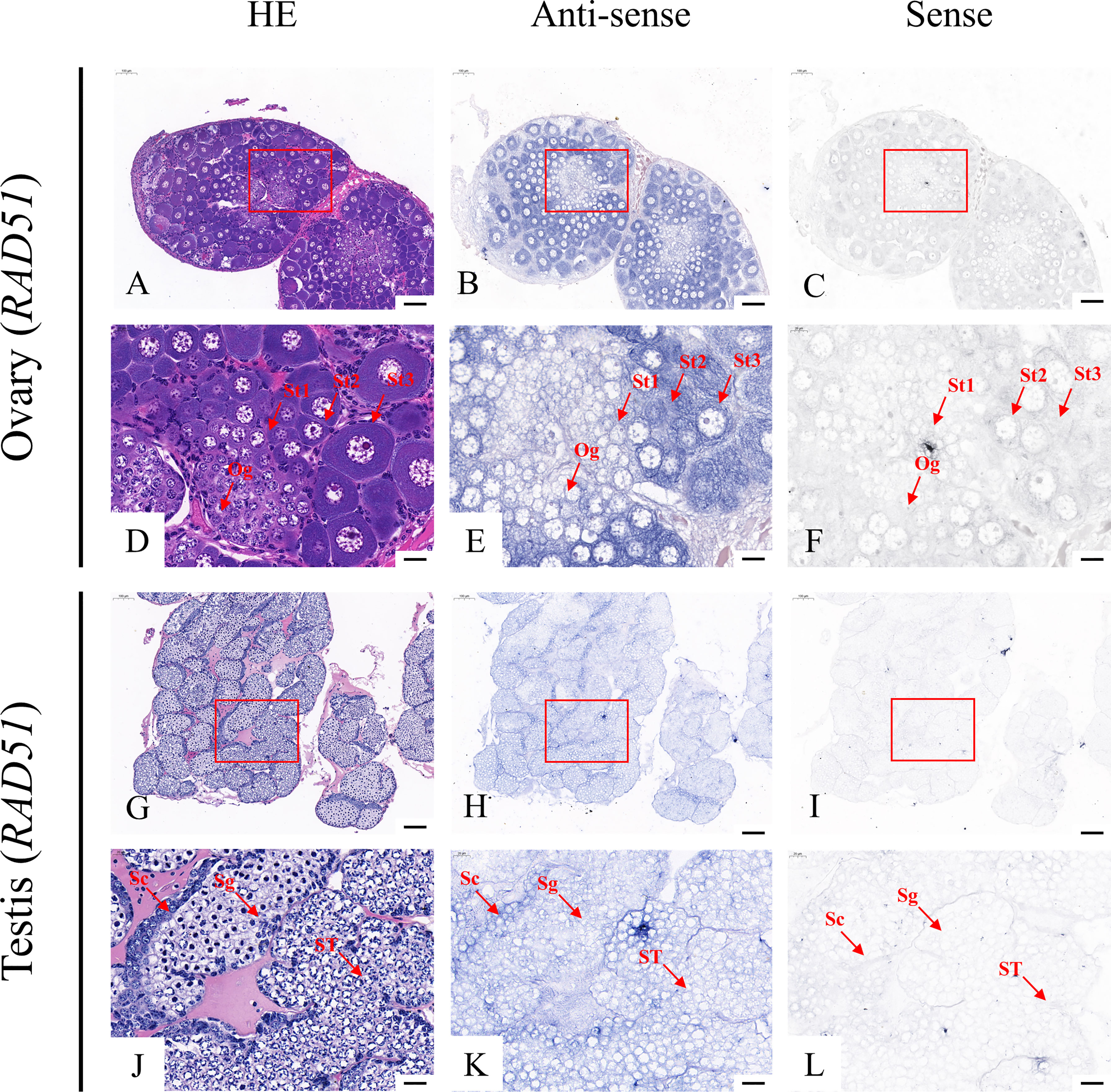
Figure 6 Localization of Rad51 mRNA in the ovary (A–F) and testis (G–L) of M. rosenbergii. HE staining of the ovary (A, D) and testis (G, J) used for in situ hybridization. Anti-DIG signals obtained by Rad51-antisense probes (B, E, H, K) and Rad51-sense probes (C, F, I, L). (D–F, J–L) High-magnification images of areas enclosed in red boxes in the images at the top (A–C, G–I). Og, oogonia; St1, stage-1 oocytes; St2, stage-2 oocytes; St3, stage-3 oocytes. Sc, spermatogonia; Sg, primary spermatocyte; ST, spermatids. Scale bars 100 μm (A–C, G–I); 20 μm (D–F, J–L).
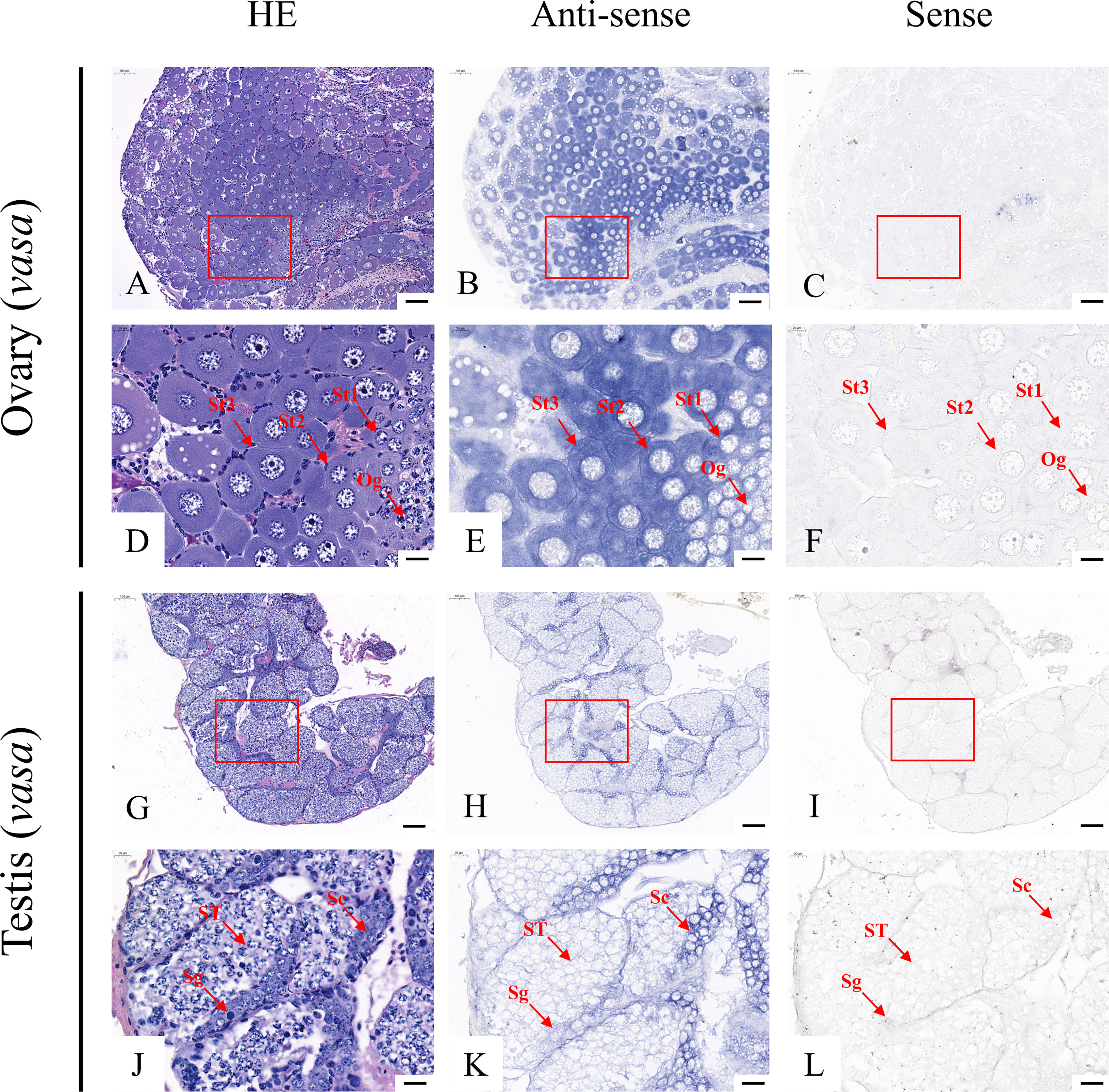
Figure 7 Localization of SPDS mRNA in the ovary (A–F) and testis (G–L) of M. rosenbergii. HE staining of the ovary (A, D) and testis (G, J) used for in situ hybridization. Anti-DIG signals obtained by SPDS-antisense probes (B, E, H, K) and SPDS-sense probes (C, F, I, L). (D–F, J–L) High-magnification images of areas enclosed in red boxes in the images to the top (A–C, G–I). Og, oogonia; St1, stage-1 oocytes; St2, stage-2 oocytes; St3, stage-3 oocytes. Sc: spermatogonia; ST: spermatids. Scale bars 100 μm (A–C, G–I); 20 μm (D–F, J–L).
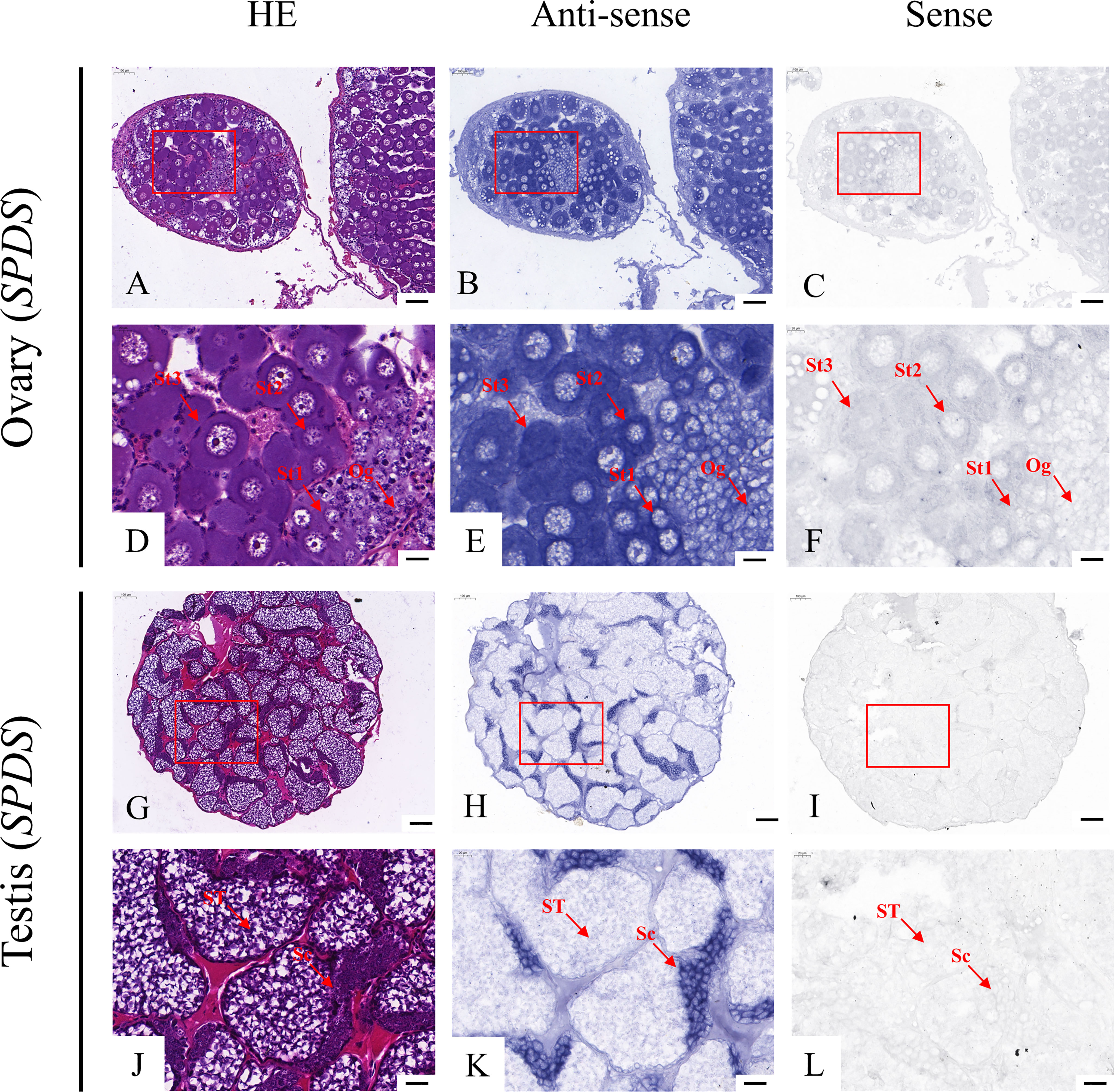
Figure 8 Localization of Fem-1C mRNA in the ovary (A–F) and testis (G–L) of M. rosenbergii. HE staining of ovary (A, D) and testis (G, J) used for in situ hybridization. Anti-DIG signals obtained by Fem-1C-antisense probes (B, E, H, K) and Fem-1C-sense probes (C, F, I, L). (D–F, J–L) High-magnification images of areas enclosed in red boxes in the images to the top (A–C, G–I). Og, oogonia; St1, stage-1 oocytes; St2, stage-2 oocytes; St3, stage-3 oocytes. Sc: spermatogonia; ST: spermatids. Scale bars 100 μm (A–C, G–I); 20 μm (D–F, J–L).
4 Discussion
The development of hermaphroditic germ cells is an important physiological process in the life history of sexually reproducing animals. In the recent decade, large-scale transcriptome sequencing has gradually become a powerful, highly repeatable, and cost-effective tool for biomolecular research to understand various molecular mechanisms involved in the development of gonadal germ cells. This study obtained more than 300 million high-quality reads and assembled them into more than 60,000 unigenes. Of these, approximately 20,000 were functionally annotated in the protein database. Compared to other recent studies on the M. rosenbergii transcriptome (Jiang et al., 2019; Yang et al., 2022), the average length of unigenes obtained in this study was longer (Our study: 1,420 bps; Jiang et al.’s: 1,196 bps; Yang et al.’s: 1,133 bps). However, the total number of unigenes obtained was slightly lower than those obtained in previous studies (Our study: 60,607; Jiang et al.’s: 115,338; Yang et al.’s: 75,887). This difference could be due to the different versions of the protein database used for functional annotation or different criteria to delete short and partially overlapping sequences to obtain non-redundant sequences.
The construction of the Venn diagram provides a convenient way to query the tissue distribution of target genes and identify candidate genes in key regions. In this study, we used two methods to search the Venn map to screen sex-related candidate genes involved in gonadal development in M. rosenbergii. As our purpose was to identify genes involved in gonadal development and sex, we focused on genes that were mainly or specifically expressed in the FG and MG regions. First, based on their functional classification, genes were searched using gonadal, reproductive, or gender-based keywords. Second, based on genes related to germ cell development found in other species, potential cognate genes for these genes were obtained in our species. Some genes are obtained by both methods, such as sex-lethal 1 (Sxl1), gametocyte-specific factor 1(GTSF1), MRR, and Sperm-associated antigen 7-like (SPATA7). Nevertheless, the first method complemented the second; it identified additional genes, such as SPDS, which could be important for screening new target genes. In addition, it was noted that most of the final candidate genes (17/18) were distributed in the FG/MG/FB/MB region, which might be because the brain of M. rosenbergii had certain regulatory effects on gonadal development. The role of brains in gonadal development has previously been reported in several species, such as mammals (Plant, 2015), birds (Tsutsui et al., 2003), fish (Zohar et al., 2010), and insects (Van Wielendaele et al., 2013), including sex differentiation and regulatory maturation; however, related topics have not been well studied in aquatic crustaceans. The transcriptome data of this study (PRJNA884099) can provide helpful information for future relevant studies on the neuroendocrine regulation of reproduction of M. rosenbergii.
Through the series of studies described above, we finally obtained five genes that are closely related to germ cells. RAD51 is one of the genes that has been shown for the first time to play a role in the germ cells of M. rosenbergii. RAD51 originates from the RecA of cell biological ancestors (Lin et al., 2006). It participates in DNA damage repair during meiosis and plays a key role in guiding recombination. When double-strand breaks (DSBs) occur, 5’-3’ nucleic acid exonuclease hydrolyzes the single strand end of the broken DNA, and replication protein A (RPA) binds to DSBs (Lewandoski, 2001). RAD51 replaces RPA and combines with the 3’ end of DNA to form ssDNA-RAD51 nucleoprotein filament, recognizing and mediating the recombinant exchange between the two chains (Takzare et al., 2016). Previous studies on the effect of RAD51 on meiosis have mostly been conducted in plants and a few model organisms, and deletion of RAD51 has been confirmed to cause chromosome pairing and synapse defects in Arabidopsis thaliana (Li et al., 2004) and Zea Mays (Franklin et al., 2003). It also affects the meiotic recombination of Saccharomyces cerevisiae and causes cell cycle arrest (Rockmill et al., 1995). The directional knockout of RAD51 in male germ cells of Mus musculus inhibited meiosis and led to the development of reproduction sterility. These studies have revealed multiple effects of RAD51 on meiotic recombination in various organisms. In this study, the existence of RAD51 in M. rosenbergii was first discovered, and its tissue expression and subcellular localization were obtained, preliminarily revealing the expression pattern of RAD51 in M. rosenbergii. RAD51 was widely expressed in male and female germ cells of M. rosenbergii, especially in spermatogonia and stage 2–3 oocytes, suggesting that this gene plays a functional role in gamete production of M. rosenbergii, which would be of great value for further research.
Another gene that specifically develops color in germ cells through ISH experiments is vasa. It was first discovered in Drosophila melanogaster by Schüpbach in 1986 and is involved in forming abdominal segments and developing germ cells in Drosophila (Schüpbach and Wieschaus, 1986). Since then, due to its conservation, vasa has been successively identified in several other species, including Caenorhabditis elegans (CEU62772), Fenneropenaeus chinensis (EF206693), Danio rerio (AF461759), Parhyale hawaiensis (EU289291), and Scylla paramamosain (GU187045) and is expressed specifically in germ cells in the vast majority of species. Previously, Nakkrasae and Damrongphol (2007) discovered a vasa-like gene (ABC87271) in M. rosenbergii, and the results of the neighbor-joining tree showed that the vasa-like gene in M. rosenbergii was adjacent to D. rerio and Mus musculus. Molecular phylogenetic analysis of the amino acid sequences of vasa family genes, including vasa, obtained using the neighbor-joining method, showed that vasa in our study was clustered with those of Macrobrachium nipponense and P. vannamei (Figure S3). This result indicates that vasa identified in this study was the actual vasa of M. rosenbergii. Moreover, vasa has been used as a reliable molecular marker for germ cell lineages (Raz, 2000). In this study, we demonstrated the subcellular localization of vasa in the mature gonads of M. rosenbergii, and its expression pattern was similar to that of homologous genes of D. rerio (Sun et al., 2013), F. chinensis (Zhou, 2007), Chlamys farreri (Shao, 2007), and S. paramamosain (Zhang et al., 2010). Taken together, these results indicated that vasa played a vital role in the development of the germ cell lineage of M. rosenbergii and could be used as a marker for germ cells in this species.
Fem-1 (Feminization-1) gene family, first discovered in Caenorhabditis elegans, plays a crucial role in regulating spermatogenesis in male/hermaphroditic reproductive lines (Doniach and Hodgkin, 1984). The Fem-1C gene obtained in this study is a member of the Fem-1 family. A few studies have shown that Fem-1C is involved in the sex regulation of some species and is highly conserved, but its role and mechanism in different organisms are quite different. For example, Xiong et al. (2014) found that HsFem-1C was highly expressed in the testes of Hyriopsis schlegelii, but it was also expressed in oocytes of Hyriopsis cumingii (Wang, 2018). Here, we confirmed that Fem-1C was significantly enriched in the ovaries of M. rosenbergii using qRT-PCR and ISH. This result is similar to those of previous studies on Fem-1C in Procambarus clarkii (Zheng et al., 2022) and Fem-1 in M. rosenbergii (Zhou et al., 2020). These findings suggest that Fem-1C probably plays a key role in the ovary of M. rosenbergii by regulating the development of female germ cells in the middle and late stages.
Spermidine, a natural polyamine, plays an important regulatory role in the animal breeding process, together with its upstream product putrescine and its downstream product spermine. Polyamine levels in human and rat seminal plasma are much higher than those in any other body fluid or tissue (Mann, 1974). Polyamines are widely present in germ cells, Sertoli cells, and Leydig cells of the rat testis, and their concentrations increase during puberty (Shubhada et al., 1989). Furthermore, the activity of ornithine decarboxylase (ODC), the rate-limiting enzyme for polyamine synthesis in female mammals, can be regulated by changes in related estrogen hormones (Moulton and Leonard, 1969; Ramos et al., 2014). Injection of exogenous polyamines into aging female mice significantly reduced their embryonic chromosomal aneuploidy, indicating the importance of polyamines in maintaining female reproductive capacity (Liu et al., 2017). These studies suggested that spermidine and SPDS play an important role in the formation of male and female gametes in mammals. The qRT-PCR results of this study showed that SPDS was specifically expressed in the gonads of M. rosenbergii, but its expression in the ovary was significantly higher than that in the testes. However, ISH showed that SPDS mRNA could also be detected in the testis of M. rosenbergii. These results indicate that SPDS may be conservative and participate in the generation of male and female gametes in this species.
In conclusion, the present study reports the transcriptome of the gonad and brain tissues of M. rosenbergii. It provides an in-depth insight into the process of germ cell development in M. rosenbergii at the molecular level. It identified 18 germ cell development-related genes, of which five genes were validated. These validated genes include one testis-related gene (MRR), one ovary-related gene (Fem-1C), and three genes (RAD51, vasa, and SPDS) related to both male and female germ cells in M. rosenbergii. Among these five genes, RAD51, SPDS, and Fem-1C were proven to function in germ cells of this species for the first time. The findings of this study will assist in developing suitable methods for producing sterile populations, which can be applied in practical production to preclude precocious puberty. In the future, we plan to silence these genes using RNAi and assess their effect on the development of germ cells and upstream and downstream genes. These potential studies will contribute to exploring a theoretical foundation for future related research in M. rosenbergii. However, the research progress is still a long way from causing sexual reversal or infertility in M. rosenbergii at present. In addition, the way to obtain sterile population on a large scale continuously and stably has not been clarified, and a lot of exploration is still needed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA884099.
Author contributions
LY and XZ conceived and designed the experiments. JW, KH, and QZ performed experiments. XL, XH, and WL analyzed the data. JW and LY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Central Public-interest Scientific Institution Basal Research Fund, CAFS (Nos. 2021SJXK3, 2022SJ-XT1, 2020TD35 and 2020ZJTD01), China-ASEAN Maritime Cooperation Fund (No. CAMC-2018F), National Freshwater Genetic Resource Center (No. NFGR-2020) and the National key R&D Program of China (SQ2022YFD2400003).
Acknowledgments
We would like to thank Editage [webshop.elsevier.com/language-editingservices/] for English language editing. I also want to thank Mikoto Misaka for her emotional support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1060594/full#supplementary-material
Supplementary Figure 1 | Level2 GO terms of four tissues in Macrobrachium rosenbergii.
Supplementary Figure 2 | Primary KEGG Pathway of four tissues in Macrobrachium rosenbergii.
Supplementary Figure 3 | Molecular phylogenetic analysis of the amino acid sequences of vasa family genes using the neighbor joining method. Sources (accession number) for vasa gene sequences: Macrobrachium rosenbergii (DQ339110), Macrobrachium nipponense (JF917240), Penaeus monodon (HQ385221), Litopenaeus vannamei (DQ095772), Carassius auratus (AY773078), Danio rerio (NM_131057), Mus musculus (NM_001145885). Sources (accession number) for PL10 gene sequences: Carassius auratus (AY842133), Macrobrachium nipponense (JF917241), Danio rerio (Y12819) and Mus musculus (J04847).
References
Abu Abayed F. A., Manor R., Aflalo E. D., Sagi A. (2019). Screening for dmrt genes from embryo to mature Macrobrachium rosenbergii prawns. Gen. Comp. Endocrinol. 282, 113205. doi: 10.1016/j.ygcen.2019.06.009
Bar I., Cummins S., Elizur A. (2016). Transcriptome analysis reveals differentially expressed genes associated with germ cell and gonad development in the southern bluefin tuna (Thunnus maccoyii). BMC Genomics 17, 217. doi: 10.1186/s12864-016-2397-8
Basavaraju Y., Mair G. C., Kumar H. M. M., Kumar S. P., Keshavappa G. Y., Penman D. J. (2002). An evaluation of triploidy as a potential solution to the problem of precocious sexual maturation in common carp, Cyprinus carpio, in karnataka, India. Aquaculture 204, 407–418. doi: 10.1016/S0044-8486(01)00827-4
Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. doi: 10.1214/aos/1013699998
Cabau C., Escudié F., Djari A., Guiguen Y., Bobe J., Klopp C. (2017). Compacting and correcting trinity and oases RNA – seq de novo assemblies. PeerJ 5, e2988. doi: 10.7717/peerj.2988
Cao J. X. (2006). Molecular mechanism of Male reproductive system of macrobrachium rosenbergii. [dissertation/doctor’s thesis] ([Hangzhou]: Zhejiang University (in Chinese).
Chen X., Mei J., Wu J. J., Jing J., Ma W. G., Zhang J., et al. (2015). A comprehensive transcriptome provides candidate genes for sex determination/differentiation and SSR/SNP markers in yellow catfish. Mar. Biotechnol. (NY). 17, 190–198. doi: 10.1007/s10126-014-9607-7
Deng D. G., Hu Y. L. (2002). Histology of the testis development of Macrobrachium crassipes. J. Zool. 03, 62–64. doi: 10.13859/j.cjz.2002.03.018
Dong Y. T., Feng H. Y., Tian X. Q., Wang Q. L., Zhang S. F., Ma K. Y., et al. (2021). Identification of a novel germ cell marker MnTdrd from the oriental river prawn Macrobrachium nipponense. Dev. Genes Evol. 231 (1-2), 11–19. doi: 10.1007/s00427-020-00671-8
Doniach T., Hodgkin J. (1984). A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev. Biol. 106, 223–235. doi: 10.1016/0012-1606(84)90077-0
Franklin A. E., Golubovskaya I. N., Bass H. W., Cande W. Z. (2003). Improper chromosome synapsis is associated with elongated RAD51 structures in the maize desynaptic2 mutant. Chromosoma 112, 17–25. doi: 10.1007/s00412-003-0242-8
Fujimoto T., Nishimura T., Goto-Kazeto R., Kawakami Y., Yamaha E., Arai K. (2010). Sexual dimorphism of gonadal structure and gene expression in germ cell-deficient loach, a teleost fish. Proc. Natl. Acad. Sci. U.S.A. 107, 17211–17216. doi: 10.1073/pnas.1007032107
Hao S. L., Yang W. X. (2019). KIFC1 is essential for normal spermatogenesis and its depletion results in early germ cell apoptosis in the kuruma shrimp, Penaeus (Marsupenaeus) japonicus. Aging-US 11 (24), 12773–12792. doi: 10.18632/aging.102601
Iwai T., Takahashi M., Ido A., Miura C., Miura T. (2015). Effect of gender on Akoya pearl quality. Aquaculture 437, 333–338. doi: 10.1016/j.aquaculture.2014.12.028
Jiang J. P., Yuan X., Qiu Q. Q., Huang G. H., Jiang Q. Y., Fu P. H., et al. (2019). Comparative transcriptome analysis of gonads for the identification of sex-related genes in giant freshwater prawns (Macrobrachium rosenbergii) using RNA sequencing. Genes 10, 1035. doi: 10.3390/genes10121035
Jin G., Xie P. (2001). The growth patterns of juvenile and precocious Chinese mitten crabs, Eriocheir sinensis (Decapoda, grapsidae), stocked in freshwater lakes of China. Crustaceana 13, 261–273. doi: 10.1163/156854001505505
Levy T., Rosen O., Eilam B., Azulay D., Aflalo E. D., Manor R., et al. (2016). A single injection of hypertrophied androgenic gland cells produces all-female aquaculture. Mar. Biotechnol. (NY). 18, 554–563. doi: 10.1007/s10126-016-9717-5
Lewandoski M. (2001). Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2, 743–755. doi: 10.1038/35093537
Li W. X., Chen C. B., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., et al. (2004). The arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. U.S.A. 101, 10596–10601. doi: 10.1073/pnas.0404110101
Li F., Fu C., Li Z., Wang A. (2021). MicroRNA transcriptome analysis of oriental river prawn Macrobrachium nipponense in responding to starvation stress. Comp. Biochem. Physiol. Part D Genomics Proteomics 38, 100820. doi: 10.1016/j.cbd.2021.100820
Li S. H., Li F. H., Yu K. J., Xiang J. H. (2018). Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 649, 1–7. doi: 10.1016/j.gene.2018.01.043
Linhartová Z., Saito T., Kašpar V., Rodina M., Prášková E., Hagihara S., et al. (2015). Sterilization of sterlet Acipenser ruthenus by using knockdown agent, antisense morpholino oligonucleotide, against dead end gene. Theriogenology 84, 1246–1255.e1. doi: 10.1016/j.theriogenology.2015.07.003
Lin Z. G., Kong H. Z., Nei M., Ma H. (2006). Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl. Acad. Sci. U.S.A. 103, 10328–10333. doi: 10.1073/pnas.0604232103
Liu D. D., Mo G. L., Tao Y., Wang H. M., Liu X. J. (2017). Putrescine supplementation during in vitro maturation of aged mouse oocytes improves the quality of blastocysts. Reprod. Fertil. Dev. 29, 1392–1400. doi: 10.1071/RD16061
Liu F., Shi W. Y., Ye H. H., Liu A., Zhu Z. H. (2021). RNAi reveals role of insulin-like androgenic gland hormone 2 (IAG2) in sexual differentiation and growth in hermaphrodite shrimp. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.666763
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-delta delta c) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li Q., Yang H. D., He L., Wang Q. (2018). Characterization of the Es-DDX52 involved in the spermatogonial mitosis and spermatid differentiation in Chinese mitten crab (Eriocheir sinensis). Gene 646, 106–119. doi: 10.1016/j.gene.2017.12.044
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo Y. L., Wu Z. X., Chen X. X., Shen X. H. (1999). Histological study on testis development of red crayfish. J. Huazhong Agric. Univ. 01, 83–84.
Lu J. G., Zheng M., Zheng J. J., Liu J., Liu Y. Z., Peng L. N., et al. (2015). Transcriptomic analyses reveal novel genes with sexually dimorphic expression in yellow catfish (Pelteobagrus fulvidraco) brain. Mar. Biotechnol. (NY). 17, 613–623. doi: 10.1007/s10126-015-9650-z
Mann T. (1974). Secretory function of the prostate, seminal vesicle and other male accessory organs of reproduction. J. Reprod. Fertil. 37, 179–188. doi: 10.1530/jrf.0.0370179
Matsumoto T., Masaoka T., Fujiwara A., Nakamura Y., Satoh N., Awaji M. (2013). Reproduction-related genes in the pearl oyster genome. Zool. Sci. 30, 826–850. doi: 10.2108/zsj.30.826
Meeratana P., Sobhon P. (2007). Classification of differentiating oocytes during ovarian cycle in the giant freshwater prawn, Macrobrachium rosenbergii de man. Aquaculture 270, 249–258. doi: 10.1016/j.aquaculture.2007.03.009
Ministry of Agriculture and rural affairs of the People’s Republic of China (2022). China Fishery statistical yearbook. (China: China Agriculture Press)
Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Moulton B. C., Leonard S. L. (1969). Hormonal effects on spermidine levels in male and female reproductive organs of the rat. Endocrinology 84, 1461–1465. doi: 10.1210/endo-84-6-1461
Nagasawa K., Takeuchi Y., Miwa M., Higuchi K., Morita T., Mitsuboshi T., et al. (2009). cDNA cloning and expression analysis of a vasa-like gene in pacific bluefin tuna Thunnus orientalis. Fish. Sci. 75, 71–79. doi: 10.1007/s12562-008-0021-9
Nakkrasae L. I., Damrongphol P. (2007). A vasa-like gene in the giant freshwater prawn, Macrobrachium rosenbergii. Mol. Reprod. Dev. 74, 835–842. doi: 10.1002/mrd.20680
O’Brien J. (1984). Precocious maturity of the majid crab, Pugettia producta, parasitized by the rhizocephalan barnacle, Heterosaccus californicus. Biol. Bull. 166, 384–395. doi: 10.2307/1541224
Oghenekaro A. O., Raffaello T., Kovalchuk A., Asiegbu F. O. (2016). De novo transcriptomic assembly and profiling of Rigidoporus microporus during saprotrophic growth on rubber wood. BMC Genomics 17, 234. doi: 10.1186/s12864-016-2574-9
Ou J. T., Luan X. Q., Chen H., Zhou K. Y., Wang Z. S., Wang H., et al. (2021). Transcriptome in combination with experimental validation unveils hub immune-related genes in oriental river prawn Macrobrachium nipponense against Spiroplasma eriocheiris challenge. Aquaculture 539 (9), 736625. doi: 10.1016/j.aquaculture.2021.736625
Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L. (2016). Transcriptlevel expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 11 (9), 1650–1667. doi: 10.1038/nprot.2016.095
Plant T. M. (2015). 60 YEARS OF NEUROENDOCRINOLOGY: The hypothalamo-pituitary-gonadal axis. J. Endocrinol. 226, T41–T54. doi: 10.1530/JOE-15-0113
Prasert Y., Jiratchaya N., Ponsit S., Unitsa S. (2021). RNA-Seq dataset of thoracic ganglia transcriptome across four ovarian development stages in Fenneropenaeus merguiensis. Data Brief. 36, 107053. doi: 10.1016/j.dib.2021.107053
Ramos R. S., Mesquita F. S., D’Alexandri F. L., Gonella-Diaza A. M., Papa P. C., Binelli M. (2014). Regulation of the polyamine metabolic pathway in the endometrium of cows during early diestrus. Mol. Reprod. Dev. 81, 584–594. doi: 10.1002/mrd.22323
Raz E. (2000). The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 1, REVIEWS1017. doi: 10.1186/gb-2000-1-3-reviews1017
Rockmill B., Sym M., Scherthan H., Roeder G. S. (1995). Roles for 2 RECA homologs in promoting meiotic chromosome synapsis. Genes Dev. 9, 2684–2695. doi: 10.1101/gad.9.21.2684
Sagi A., Cohen D., Milner Y. (1990). Effect of androgenic gland ablation on morphotypic differentiation and sexual characteristics of male freshwater prawns, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 77, 15–22. doi: 10.1016/0016-6480(90)90201-v
Schüpbach T., Wieschaus E. (1986). Maternal-effect mutations altering the anterior-posterior pattern of the drosophila embryo. Rouxs Arch. Dev. Biol. 195, 302–317. doi: 10.1007/BF00376063
Shao M. Y. (2007). cDNA cloning and developmental expression pattern of the DEAD-box family and boule genes related to reproduction in chlamys farreri [dissertation/doctor’s thesis] ([Qingdao]: Ocean University of China (in Chinese).
Shubhada S., Lin S. N., Qian Z. Y., Steinberger A., Tsai Y. H. (1989). Polyamine profiles in rat testis, germ cells and sertoli cells during testicular maturation. J. Androl. 10, 145–151. doi: 10.1002/j.1939-4640.1989.tb00076.x
Sun D. J., Wang C., Yan J. Z. (2013). Vasa expression analysis of VASA gene in zebrafish at different development stages. Guangdong Agric. Sci. 40, 132–134. doi: 10.16768/j.issn.1004-874x.2013.13.066
Takzare N., Samizadeh E., Shoar S., Majidi Zolbin M., Naderan M., Lashkari A., et al. (2016). Impacts of morphine addiction on spermatogenesis in rats. Int. J. Reprod. Biomed. 14, 303–308. doi: 10.29252/ijrm.14.5.303
Trine Y., Sergey A., Bente R., Bjarne H., Kari Ø.T., Aleksei K. (2021). Transcriptome and functional responses to absence of astaxanthin in Atlantic salmon fed low marine diets. Comp. Biochem. Physiol. 39, 100841. doi: 10.1016/j.cbd.2021.100841
Tsutsui K., Matsunaga M., Ukena K. (2003). Biosynthesis and biological actions of neurosteroids in the avian brain. Avian Poult. Biol. Rev. 14, 63–78. doi: 10.3184/147020603783641297
Van Wielendaele P., Badisco L., Vanden Broeck J. V. (2013). Neuropeptidergic regulation of reproduction in insects. Gen. Comp. Endocrinol. 188, 23–34. doi: 10.1016/j.ygcen.2013.02.005
Ventura T., Manor R., Aflalo E. D., Weil S., Rosen O., Sagi A. (2012). Timing sexual differentiation: Full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 86, 90. doi: 10.1095/biolreprod.111.097261
Wang Y. Y. (2018). Preliminary studies on identification, expression and function of sex differentiation genes fem-1b, fem-1c and ank of hyriopsis cumingii. [dissertation/master’s thesis] ([Shanghai]: Shanghai Ocean University (in Chinese).
Wang L. Y., Li S. G., Xu L. L., Li Y. Q., Chen H. X., Chen D. H. (2020). De novo transcriptome sequencing and analysis of the cuttlefish (Sepiella japonica) with different embryonic developmental stages. Anim. Biotechnol. 8, 8. doi: 10.1080/10495398.2020.1735406
Wang B. Q., Xu L. W., Jia Z. H., Mou Z. B., Fan Z. T. (2005). Heat shock induction of triploid female rainbow trout. J. Fish. 02, 22–27. doi: 10.3969/j.issn.1005-3832.2005.02.005
Wan H. F., Zhong J. Y., Zhang Z. P., Zou P. F., Wang Y. L. (2022). Comparative transcriptome reveals the potential modulation mechanisms of spfoxl-2 affecting ovarian development of Scylla paramamosain. Mar. Biotechnol. 24 (1), 125–135. doi: 10.1016/j.gene.2017.12.044
Wei B. H., Ni J. H., Yang T., Hao S. L., Yang W. X. (2021). PIWIs maintain testis apoptosis to remove abnormal germ cells in Eriocheir sinensis. Reproduction 162 (3), 193–207. doi: 10.1530/rep-21-0157
Xiong W. F., Shi J. W., Jiang M. Y., Chen Y. L., Peng K., Sheng J. Q., et al. (2014). Structural characteristics analysis of fem-1c and its protein molecules, a male-causing gene of Hyriopsis cumingii. Aquat. Biol. 38, 1002–1007. doi: 10.7541/2014.148
Xu H. J., Chen Y. L., Wang Y. M., Luo J. Y., Li J. W., Shen S. Q., et al. (2022). Full functional sex reversal achieved through silencing of MroDmrt11E gene in Macrobrachium rosenbergii: Production of all-male monosex freshwater prawn. Front. Endocrinol. 12 (15). doi: 10.3389/fendo.2021.772498
Yang C. L., Chen X. H., Li Y. M., Peng M., Jiang W. M., Wei P. Y., et al. (2008). A preliminary study on artificial induction of triploid Crassostrea rivularis by 6-DMAP. Southwest Agric. J. 03, 825–828. doi: 10.16213/j.cnki.scjas.2008.03.072
Yang G., Qin Z. D., Lu Z. J., Liang R. S., Zhao L. J., Pan G., et al. (2022). Comparative transcriptomics of gonads reveals the molecular mechanisms underlying gonadal development in giant freshwater prawns (Macrobrachium rosenbergii). J. Mar. Sci. Eng. 10 (13), 737. doi: 10.3390/jmse10060737
Yue H. M., Li C. J., Du H., Zhang S. H., Wei Q. W. (2015). Sequencing and de novo assembly of the gonadal transcriptome of the endangered Chinese sturgeon (Acipenser sinensis). PLoS One 10, e0127332. doi: 10.1371/journal.pone.0127332
Zhang Y. H., Li J., Qin Y. P., Zhou Y. G., Zhang Y., Yu Z. N. (2017). A comparative study of the survival, growth and gonad development of the diploid and triploid Hong Kong oyster, Crassostrea hongkongensis (Lam & Morton 2003). Aquacult. Res. 48, 2453–2462. doi: 10.1111/are.13081
Zhang F. Y., Ma L. B., Jiang K. J., Zhang D., Chen L. Q. (2010). Characteristic analysis and gonadal specific expression of vasa gene in Scylla paramamosain. mar. Fish 32, 361–367. doi: 10.13233/j.cnki.mar.fish.2010.04.014
Zhang E. F., Qiu G. F. (2010). A novel dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis. Dev. Genes Evol. 220, 151–159. doi: 10.1007/s00427-010-0336-2
Zheng J. B., Chen L. R., Jia Y. Y., Chi M. L., Li F., Cheng S., et al. (2022). Genomic structure, expression, and functional characterization of the fem-1 gene family in the redclaw crayfish, Cherax quadricarinatus. Gen. Comp. Endocrinol. 316, 113961. doi: 10.1016/j.ygcen.2021.113961
Zhou Q. R. (2007). Cloning and expression analysis of the two DEAD-box genes fc-vasa and fc-PL10α in fenneropenaeus chinensis. [dissertation/master’s thesis] ([Qingdao]: Ocean University of China (in Chinese).
Zhou L. X., Liu X., Ye B. Q., Liu Y., Tan S. P., Ma K. Y., et al. (2020). Molecular characterization of ovary-specific gene Mrfem-1 and siRNA-mediated regulation on targeting mrfem-1 in the giant freshwater prawn, Macrobrachium rosenbergii. Gene 754, 144891. doi: 10.1016/j.gene.2020.144891
Keywords: macrobrachium rosenbergii, transcriptome, germ cell, RAD51, Fem-1C, SPDS, vasa
Citation: Wei J, Hong K, Zhou Q, Wang Y, Li W, Liu X, Hong X, Chen C, Yu L and Zhu X (2022) Transcriptome analysis of gonads and brain of giant freshwater prawn (Macrobrachium rosenbergii): screening and validation of genes related to germ cell development. Front. Mar. Sci. 9:1060594. doi: 10.3389/fmars.2022.1060594
Received: 03 October 2022; Accepted: 25 October 2022;
Published: 10 November 2022.
Edited by:
Xianyun Ren, Yellow Sea Fisheries Research Institute (CAFS), ChinaReviewed by:
Shubo Jin, Freshwater Fisheries Research Center (CAFS), ChinaCuimin Mu, Shandong Agricultural University, China
Copyright © 2022 Wei, Hong, Zhou, Wang, Li, Liu, Hong, Chen, Yu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingyun Yu, bHlzbnBAMTYzLmNvbQ==; Xinping Zhu, emh1eGlucGluZ18xOTY0QDE2My5jb20=
 Jie Wei
Jie Wei Kunhao Hong1
Kunhao Hong1 Chen Chen
Chen Chen Lingyun Yu
Lingyun Yu Xinping Zhu
Xinping Zhu