- 1Institute for Inorganic and Analytical Chemistry, Bioorganic Analytics, Friedrich Schiller University Jena, Jena, Germany
- 2Max Planck Institute for Chemical Ecology, Jena, Germany
Diatoms contribute as a dominant group of microalgae to approximately 20% of the global carbon fixation. In the plankton, these photosynthetic algae are exposed to a plethora of metabolites, especially when competing algae are lysed. It is well established that diatoms can take up specific metabolites, such as vitamins, amino acids as nitrogen source, or dimethylsulfoniopropoionate to compensate for changes in water salinity. It is, however, unclear to which extent diatoms take up other organic resources and if these are incorporated into the cell´s metabolism. Here, we explore the general scope of uptake of metabolites from competitors. Using labeled metabolites released during lysis of algae grown under a 13CO2 atmosphere, we show that the cosmopolitan diatom Chaetoceros didymus takes up even dilute organic substrates from these lysates with little bias for molecular weight or polarity. This is reflected by a high degree of labeling in the metabolome of the exposed cells. The newly developed pulse label/mass spectrometry metabolomics approach reveals that polarity and molecular weight has no detectable influence on uptake. We further show that the taken-up metabolites are partly maintained without metabolic modification within the cells, but also a substantial part is subject to catabolic and anabolic transformation. One of the most dominant phytoplankton groups thus has the potential to compete with heterotrophs, suggesting that the observed osmotrophy may substantially impact organic material fluxes in the oceans. Our findings call for the refinement of our understanding of competition in the plankton.
Introduction
The traditional view on marine plankton distinguishes between phytoplankton as primary producers and zooplankton as consumers (Heesen, 1928). However, many planktonic eukaryotic organisms have been recognized as mixotrophs, which combine autotrophic photosynthesis with organic matter uptake (Selosse et al., 2017). Many microzooplankton grazers are mixotrophic and retain functional algal organelles or even algal endosymbionts. Also photosynthetically active organisms, such as phytoflagellates and dinoflagellates can engulf and consume prey organisms to acquire nutrients (Stoecker et al., 2017). As an additional strategy, the uptake of dissolved organic carbon termed absorbotrophic mixotrophy or osmotrophy can be observed in microalgae. This process seems to be ubiquitous but clearly less understood (Flynn et al., 2013a).
First investigations of osmotrophy in plankton focused on algae growth under extreme darkness in the presence of organic substrates (Neilson and Lewin, 1974). Administration and uptake studies of radiolabeled substrates deepened our mechanistic understanding. However, experiments were always limited to investigating one compound or a compound class, such as specific vitamins or amino acids. It is now clear that mixotrophy is widely distributed among planktonic eukaryotic organisms (Caron, 2016). However, the experiments with single compounds under limiting conditions conducted so far represent an oversimplification and do not reflect the situation in nature, where a cell is exposed to structurally more diverse metabolites. Consequently, the importance of osmotrophy for pelagic food webs and element cycling remains elusive and we are still far from quantitatively deciphering the trophic modes of primary producers forming the phytoplankton (Karlusich et al., 2020).
The availability of organic substrates for uptake will be highly variable. In the plankton mass occurrences of algae, so-called algal blooms can last over days to weeks before the population breaks down and is succeeded by other species that become dominant. Especially during the decay of such algal blooms, the surviving competitors will be exposed to the metabolites of the lysed algae. Also, lysis of specific phytoplankton members by pathogens, such as algicidal bacteria or viruses, results in situations where surviving resistant cells are exposed to the metabolomes released by the lysed species (Meyer et al., 2017). It is entirely unclear if and how all these compounds contribute to the metabolism of the phytoplankton and the potential ecological importance of phytoplankton as consumers of organic material is thus still poorly understood (Burkholder et al., 2008; Flynn et al., 2013b; Borowitzka, 2016; Ferroni et al., 2018). Modeling studies imply a leak-recovery system to be required to initiate a bloom, but the underlying mechanisms are not fully explored (Flynn et al., 2008)
As critical primary producers, diatoms were initially classified as autotrophs, but the uptake capability of specific organic molecules has long been recognized (Raven et al., 2009; Flynn et al., 2013b). These include glucose, small polar organic acids such as acetate, succinate, fumarate, malate and lactate, amino acids, dipeptides and dimethylsulfoniopropionate (DMSP) (Lewin and Lewin, 1960; Hellebust and Guillard, 1967; Hellebust and Lewin, 1972; Vila-Costa et al., 2006; Cochlan et al., 2008; Mulholland and Lee, 2009; Spielmeyer et al., 2011; Petrou and Nielsen, 2018). Most experiments provided mechanistic insight but did not accurately reflect natural conditions, where the water in which the algae live harbors a diverse mixture of organic compounds. Under natural conditions, cells are exposed to these metabolites and also light is available to support photosynthesis. Thus, two competing mechanisms for carbon acquisition, phototrophy and heterotrophy will be active. Here we address osmotrophy in the diatom Chaetoceros didymus under non-limited conditions. The supply of nutrients and light in our study was non-limiting to allow efficient algal growth; organic metabolites were thus offered in addition to available inorganic sources.
We base our experimental setup on a well-investigated multi-partner interaction involving an algicidal bacterium and two cosmopolitan diatom species. Kordia algicida is a marine Flavobacterium that possesses algicidal activity leading to cell lysis of several microalgal species, including the diatom Skeletonema costatum (Paul and Pohnert, 2011; Paul and Pohnert, 2013). C. didymus, in contrast, is a naturally co-occurring diatom that is resistant against K. algicida. The impact of K. algicida was recently shown in field experiments where it induces a population shift in a natural phytoplankton community towards resistant algae (Bigalke et al., 2019). We hypothesize that during bacterial lysis of S. costatum, resistant species can benefit by taking up metabolites of the lysate in an osmotrophic manner. Therefore, we exposed a culture of C. didymus to metabolites from a 50% diluted stationary culture of lysed S. costatum cells. We developed a novel analytical approach to test this hypothesis, including pulse labeling metabolomics and a novel data treatment routine to analyze isotopologue signatures. We show that highly universal uptake and metabolism of metabolites from the environment occur in the resistant alga.
Materials and Methods
Experimental Design
Diatoms were grown under non-limited conditions and exposed to a labeled metabolome released by lysed cells of a competitor. The diatoms were extracted using optimized protocols for metabolomics sampling and extracts were analysed by liquid chromatography mass spectrometry to evaluate the uptake of labeled metabolites.
Algal Culturing
C. didymus was isolated by W. Kooistra from the Gulf of Naples (Stazione Zoologica Anton Dohrn, Naples, Italy) and S. costatum was obtained from the Roscoff Culture Collection (Roscoff, France). Both algae were cultivated in batch culture using nutrient-replete artificial seawater medium (Maier and Calenberg, 1994) in 50 mL Greiner Bio-One cell culture flasks at 11-13°C under a 14:10 h light: dark regime with an illumination of 20-25 µmol photons m-2 s-1. Development of cultures was followed by in-vivo Chl a fluorescence using a Mithras LB 940 plate reader (excitation 430 nm, emission 665 nm).
Global 13C-Labeling
For global 13C-labeling of S. costatum we used autoclaved artificial seawater medium that was prepared without addition of NaHCO3. An aliquot of this medium was utilized to dissolve NaH13CO3 (98 atom %, Sigma-Aldrich, Munich, Germany). This solution, containing sufficient NaH13CO3 to reach a final concentration of 2.38 mM, was sterile filtered (0.2 µm pore size, Sarstedt Filtropur S) and transferred back to the medium bottle. Tissue culture flasks were filled to the neck in order to minimize the area for CO2 exchange with the atmosphere and were inoculated with < 1% (v/v) of a stationary S. costatum culture. After growing to stationary phase, an aliquot was taken and transferred to fresh 13C-enriched medium (< 1% (v/v)). After two of these cycles a plateau in the degree of labeling (verified by mass spectrometry as described below) was reached and the cultures were used for further experiments.
Bacterial Culturing
K. algicida (Sohn et al., 2004) was cultivated on marine broth agar at 30°C for 2 days. The bacterial lawn was removed with a sterile cotton swab and re-suspended in algal culturing medium to an OD550 of 0.5 determined on a Genesys 10S UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Co-Culturing Experiment and Extraction of Released Metabolites
S. costatum cultures reared in medium prepared with (further referred to 12C or 13C medium were co-cultured in triplicates (185 mL each) with the K. algicida added to a final OD550 of 0.01. After 6 days the lysed cultures were gently filtered (0.2 µm pore size) and the filtrate extracted for exometabolome analysis as follows. Solid phase extraction cartridges (Chromabond easy, Macherey-Nagel, Düren, Germany) were equilibrated with 4 mL of methanol (Chromasolv Plus, Sigma-Aldrich, Munich, Germany) and 4 mL of water (Chromasolv Plus, Sigma-Aldrich, Munich, Germany) before the filtrate (170 mL) was applied using vacuum with a flow rate < 1 L h-1. The cartridge was washed with 4 mL water, air-dried and then extracted via gravity flow using 2 mL of methanol followed by 2 mL of methanol/tetrahydrofuran 1:1 (tetrahydrofuran HiPerSolv, VWR, Dresden, Germany). This extract was frozen until further chemical analysis.
Tests for Osmotrophy
Stationary cultures (45 mL) of S. costatum reared in 12C or 13C medium were centrifuged (500 x g, 15 min, 10°C) and washed three times by repeated addition of 45 mL of 12C medium to the harvested pellets and centrifugation. After the third washing step the supernatant was collected as control medium (this was processed in parallel to the cells and later served as control for the natural 13CO2) and the cell pellet was re-suspended in 45 mL 12C medium. To disrupt cells, the suspension was frozen at -20°C, thawed and treated in an ice-cold ultrasonic bath for 10 min. The lysate was filtered (1.2 µm pore size, GF/C, Whatman, GE Healthcare, Little Chalfont, United Kingdom), acidified to pH ≤ 1 for sterilization using 30% hydrochloric acid, incubated at 0°C for 10 min and subsequently neutralized under sterile conditions using a saturated sodium hydroxide solution. The solutions were stored at -20°C until use. After thawing, the solutions were diluted 1:1 with 12C medium to yield the final medium for the investigation of osmotrophy (12C organic & 12C inorganic or 13C organic & 12C inorganic). 45 mL aliquots of both media were extracted as described above for exometabolomic analysis to determine the organic metabolites. To determine osmotrophy, 110 mL aliquots of these media were inoculated with 1% (v/v) stationary C. didymus culture in triplicates and cultivated for 3 days. Directly after inoculation and after 3 days of cultivation, samples (45 mL each) for intra- and extracellular metabolomics were processed. Samples were filtered (1.2 µm, GF/C, Whatman, GE Healthcare, Little Chalfont, United Kingdom) and the flow-through was processed for exometabolomic analysis as described above (see co-culturing experiment). The cells were washed off the filter with an ice-cold freshly prepared mixture of methanol/ethanol/chloroform (1:3:1) (ethanol LiChroSolv, Merck, Darmstadt, Germany; chloroform HiPerSolv, VWR, Dresden, Germany). Extracts were treated in an ultrasonic bath for 10 min, centrifuged (30,000 x g, 15 min, 4°C) and the supernatant was stored at -20°C. As a control to prove that no enrichment was present in mixotrophic media, the third wash supernatant (see above) was used as medium to cultivate C. didymus and metabolites were extracted as described. A graphical representation of the experimental setup can be found in Supplementary Figure 1.
Analysis of Exo- and Endometabolomes With LC-MS
Extracts from cells and media (see above) were dried in a nitrogen flow at room temperature and were resuspended in up to 200 µL methanol. Metabolites were separated on an UltiMate 3000 UHPLC (Thermo Fisher Scientific, Waltham, MA, USA) equipped with an Accucore C18 column (100x2.1 mm, 2.6 µm) at 25°C using water with 2% acetonitrile and 0.1% formic acid (A) and pure acetonitrile (B) as mobile phase. The gradient was as follows: 100% A for 0.2 min, linear gradient to 100% B in 7.8 min, 100% B for 3 min, linear gradient to 100% A in 0.1 min, 100% A for 0.9 min. The UHPLC was connected to a QEplus Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with heated electrospray ionization source (capillary temperature 360°C, sheath gas 60 nominal units, aux gas 20 nominal units, sweep gas 5 nominal units, aux gas temperature 400°C, spray voltage 3.3 kV, S-lens RF level 50) operated in positive or negative ion mode. Full scan measurements (m/z 100-1200, resolution 280k, AGC target 3 106, maxIT 900 ms) were performed separately for positive and negative ion mode. All data are available at https://www.ebi.ac.uk/metabolights/MTBLS2333.
Isotopologue Detection
Full scan RAW files were converted to mzXML using ProteoWizard msConvert (Kessner et al., 2008) with the vendor’s algorithm for peak picking. Isotopologue detection was achieved with R-based X13CMS (Huang et al., 2014). The R script can be found in the Supplementary Material. In brief, after peak-picking with centwave (3 ppm, peakwidth 5-20 s) and retention time alignment with orbiwarp, isotopologues with a mass difference of 1.00335 Da were assigned (RTwindow 10 s, 3 ppm). Either all isotopologue groups (α = 1) or only isotopologue groups significantly different from 12C (α = 0.05) were reported. Afterwards, those significantly different isotopologue groups were manually curated with reference to the original spectrum in order to exclude groups that did not contain at least 3 consecutive isotopologues.
Compound Identification
Compounds were identified based on their retention time, high resolution mass to charge ratio and fragmentation pattern. Compound Discoverer (Vers. 2.1, Thermo Fisher Scientific, Waltham, MA, USA) was used to predict sum formula, search an in-house and public databases (ChemSpider and mzCloud) as well as calculate FISh scores of candidates. SIRIUS and CSI : FingerID were used to compute fragmentation trees and search molecular structure databases (Duhrkop et al., 2015). Putatively identified compounds were compared to authentic standards: Arachidonic acid, betaine, carnitine hydrochloride, choline chloride, 11Z-eicosenoic acid, 2-hydroxytetradecanoic acid, isoleucine, leucine and 1-oleoyl-sn-glycero-3-phosphocholine were obtained from Sigma Aldrich (Munich, Germany). Oleic acid was purchased from AppliChem (Darmstadt, Germany). Linoleic acid was from Alfa Aesar (Haverhill, MA, USA). 5Z,8Z,11Z,14Z,17Z-Eicosapentaenoic acid was supplied by Cayman Chemicals (Ann Arbor, MI, USA) and 6Z,9Z,12Z-Hexadecatrienoic acid from Larodan (Solna, Sweden). 6Z,9Z,12Z,15-Hexadecatetraenoic acid has been synthesized according to Pohnert et al. (2004). ß-Alanine betaine has been synthesized according to Chary et al. (2012) Leucine and isoleucine were not baseline separated and are consequently grouped as (iso-)leucine.
Calculation of the Degree of Labeling
To calculate the degree of labeling MS1 data from isotopologues are assumed to have a Bernoulli distribution (see Supplementary Material for formula) (Zachmann, 1994). Measured isotopologue intensities are compared to computed distributions and the squared coefficient of variation between both is minimized in an iterative process.
Results
Generation and Evaluation of a Labeled Metabolome for Uptake Experiments
A complex medium containing the entire labeled metabolome of lysed diatom cells could be generated from 13C-labeled S. costatum. We grew S. costatum in a medium containing as a sole carbon source to obtain a labeled metabolome. With a repeated exchange of the medium, we reached up to 65% labeling in the algal metabolites, sufficient for our analytical protocol. Mechanical lysis of the labeled culture and removing the cell debris gave an axenic medium, rich in organic metabolites (Supplementary Figure 1).
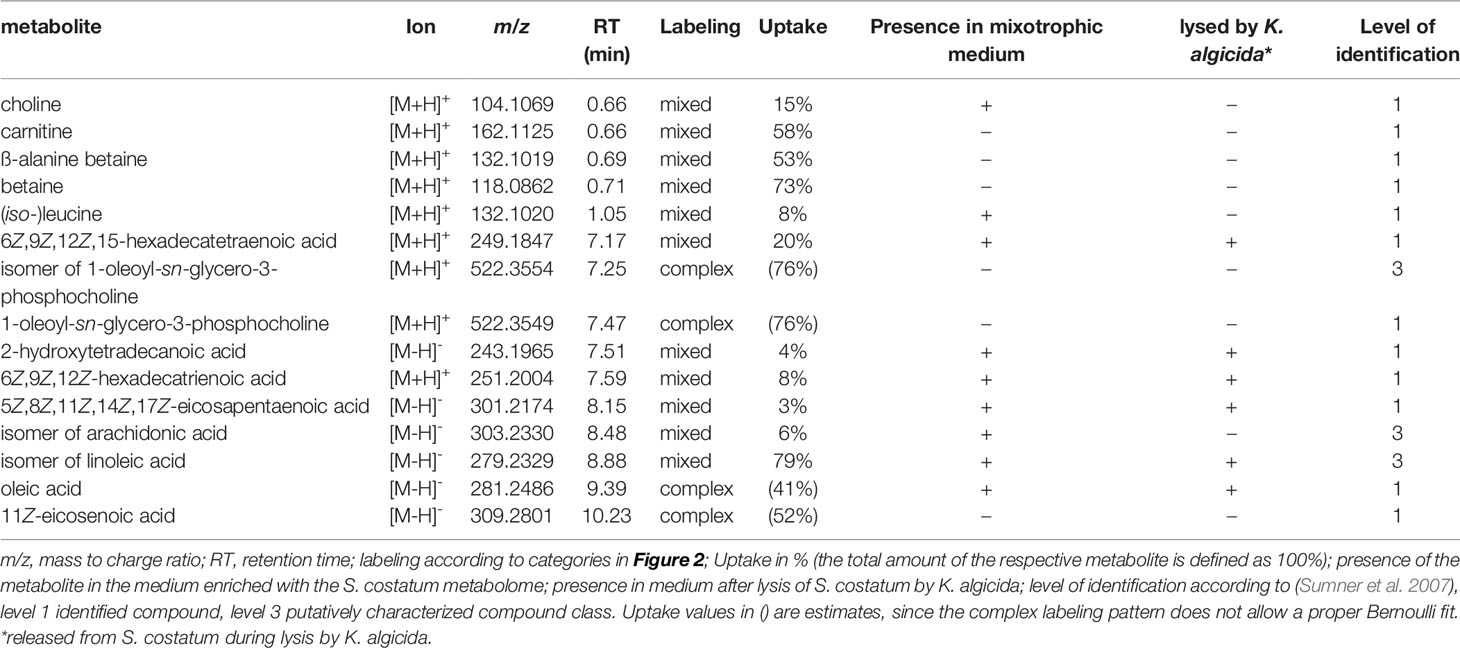
We hypothesized that the resistant C. didymus encounters these metabolites also in the field when algicidal bacteria lyse its competitors. To test this hypothesis, labeled S. costatum cultures were infected with K. algicida, which resulted in lysis of more than half of the diatom cells within six days. Ultrahigh-pressure liquid chromatography-high resolution mass spectrometry (UHPLC-HRMS) of the exometabolome revealed similar metabolic profiles in mechanically lysed S. costatum cells and those lysed by the bacteria which is also reflected in Table 1 where two thirds of the identified metabolites were found in both sample types.
Before incubation with C. didymus, the metabolites from lysed S. costatum were sterilized and diluted with a medium containing inorganic carbon with normal isotope distribution (1.1% 13C, Supplementary Figure 1). Thereby, we could ensure that 13C labeled organic metabolites taken up from the medium can be distinguished from de novo synthesized compounds. We exposed C. didymus to a three day pulse of labeled metabolites released from lysed cells of S. costatum and then extracted the culture. Pulse labeling for three days (i.e. ca. two cell divisions) instead of prolonged incubation prevents scrambling of isotopes detected in the mass spectra due to excessive cellular metabolism during incubation.
Following the same procedure, a control medium was generated using S. costatum cells grown in a medium with natural isotope distribution that could be used to generate mass spectra for structure elucidation. Using fragmentation trees and database comparison of the unlabeled metabolites and subsequent comparison to authentic standards, we identified several of the labeled metabolites in C. didymus (Table 1).
Our study is limited to the fraction of metabolites that can be extracted and separated in UHPLC (and extracted with SPE in the case of exometabolites) using our standardized protocols. Thus, a broad range of metabolites is investigated, but we cannot claim to cover the entire metabolome (which would be technically not feasible in any metabolomics approach to date). Since we did not use a biasing extraction protocol for the generation of labeled medium, but rather used the cell lysate and since extraction success is identical for isotopologes and the unlabeled compounds we can conclude that our results will not be distorted due to extraction selectivity. We did not aim for a quantification of the respective metabolites in this metabolomics approach but rather surveyed the physiochemical properties of metabolites taken up during exposure to organic metabolites released from a lysed culture.
Evaluation of Uptake
For uptake experiments, C. didymus was cultivated in a medium containing the sterilized labeled S. costatum endometabolome or control medium for three days (n=3). We selected the concentration of added metabolites to be equivalent to 50% of those released by a lysed stationary culture. Cells were then recovered by filtration and washed extensively. After being exposed to this medium under otherwise optimum growth conditions, UHPLC-HRMS analysis of the C. didymus endometabolome revealed that the alga took up substantial amounts of labeled compounds from various metabolic classes. Quantitative analysis of labeling proved to be challenging in terms of chemoinformatic data treatment. Therefore, ions of the same metabolite, only differing in their number of incorporated 13C were summarized in an isotopologue group using the software X13CMS (Huang et al., 2014). Of 5587 isotopologue groups (positive and negative ionization mode) detectable in the endometabolome 2381 were significantly labeled with 13C (Figure 1A). After manual curation, 548 isotopologue groups were categorized according to their labeling pattern and analyzed regarding their retention time and mass to charge ratio. The degree of labeling was estimated using the probability mass function for Bernoulli trials (random experiments with precisely two possible outcomes, “success i.e. incorporation of 13C” and “failure i.e. incorporation of 12C “, in which the probability of success is the same every time the experiment is conducted):
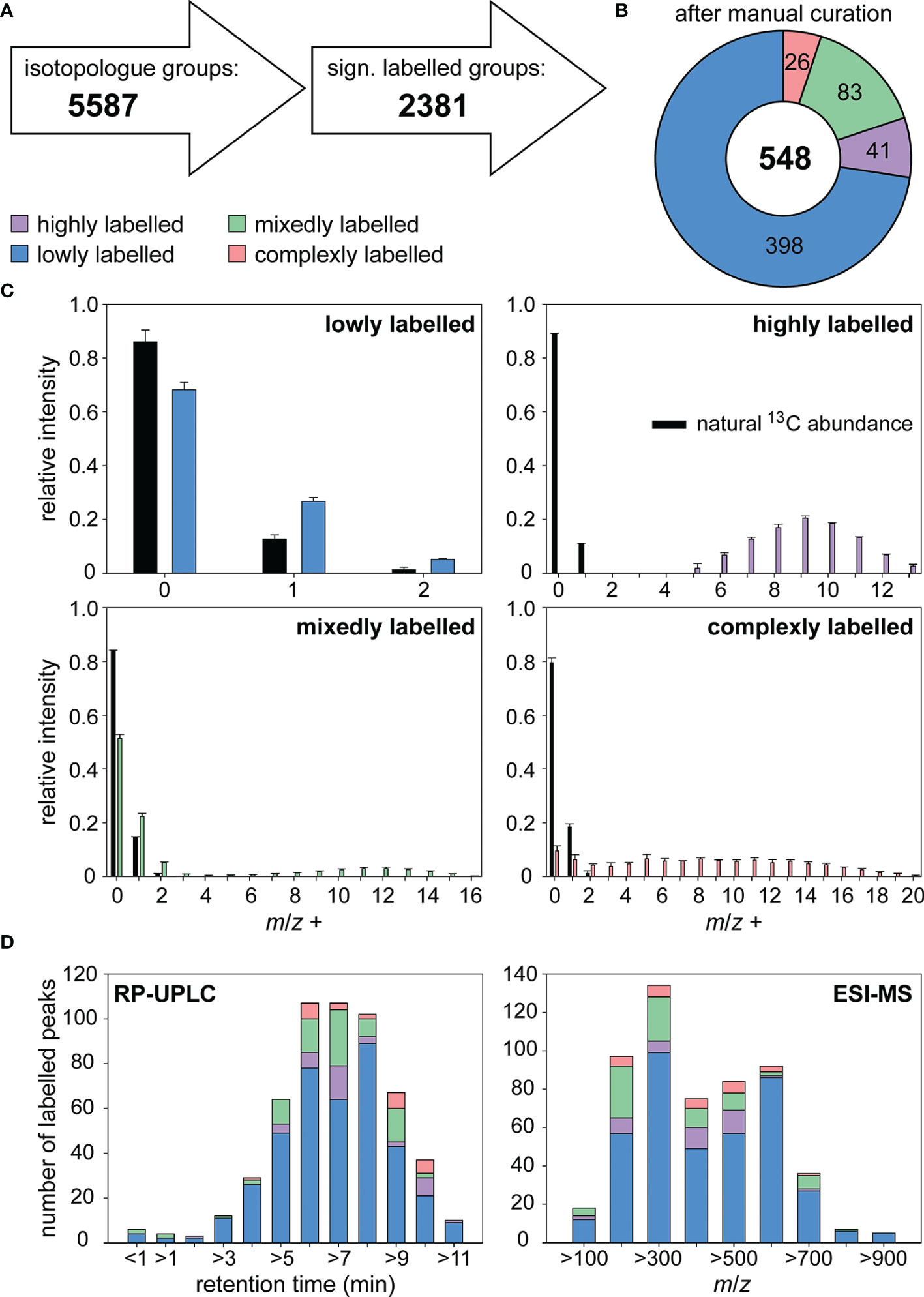
Figure 1 Labeling of metabolites in Chaetoceros didymus endometabolome after exposure to metabolites from lysed labeled S. costatum. (A) Isotopologue groups were detected and tested for significance using X13CMS. (B) After manual curation the remaining 548 isotopologue groups were categorized according to their labeling pattern. (C) Isotopologue distributions in metabolites with natural and enriched 13C abundance are depicted for each category. (D) Isotopologue groups were clustered by retention time and m/z range. Relative intensity refers to the sum of isotopolog ions of the molecular peak. All data are mean ± SD from biological triplicates. A dot plot showing the correlation between retention time and m/z can be found in Supplementary Figure 2.
The degree of labeling p for a metabolite with n carbon atoms is estimated from the intensities of a set of isotopologues I(m) with m 13C atoms (Pohnert and Jung, 2003).
Classification of Taken-Up and Processed Metabolites
We categorized the metabolites according to their degree of labeling in the receiving cells (Figures 1B, C). 73% of the isotopologue groups were lowly labeled with a degree of labeling < 5% indicative of compounds mainly synthesized de novo in C. didymus. The low degree of labeling that still exceeds the natural 13C content of 1.1% can be explained by general utilization of taken-up metabolites in the metabolism: metabolites that are assimilated are catabolized to metabolic building blocks that are used together with the autotrophic metabolic pool and used for anabolism again. Seven percent of the isotopologue groups were highly labeled. They contained a degree of labeling similar to that of metabolites in the medium (ca. 65%). The cellular content of these highly labeled metabolites taken up from lysed S. costatum shows that C. didymus assimilates metabolites that it does not (or only to a minimal extent) produce itself. Certain compounds with a high degree of labeling can be found in C. didymus but not in the DOM label-enriched medium. These could arise from the metabolic transformation of more complex metabolites released by S. costatum. 15% of the isotopologue groups were labeled in a mixed manner. In the mass spectrum both, signals from a lowly labeled and a highly labeled proportion of the respective compound can be detected. This pattern can be explained by metabolites that are biosynthesized by C. didymus and also acquired from the medium. This compound class includes a wide range of natural products, from small charged molecules like choline and carnitine to non-polar lipids and fatty acids. A few signals (5%) had a complex labeling pattern that Bernoulli statistics could not describe. These can be interpreted as compounds that result from metabolites taken up and further metabolized using the pool of de novo synthesized metabolites (Figures 1B, C). For example, intermediate metabolic products like fatty acids can be utilized in the anabolism of more complex ones like lipids.
Properties of Taken Up Metabolites
Labeled metabolites span a wide range of polarity from charged small molecules to non-polar fatty acids. Most labeled products are found in the non-polar region of the chromatogram (Figure 1D, Table 1 and Supplementary Figure 2). The identified polar metabolites that are efficiently taken up include the amino acids leucine and/or isoleucine and several small charged molecules, namely glycine betaine, β-alanine betaine, carnitine (58% of total cellular carnitine is labeled), and choline. Also, many fatty acids and lipids are taken up, including oleic acid, 5Z,8Z,11Z,14Z,17Z-eicosapentaenoic acid, 6Z,9Z,12Z-hexadecatrienoic acid, 6Z,9Z,12Z,15-hexadecatetraenoic acid, 2-hydroxytetradecanoic acid and isomers of linoleic and arachidonic acid as well as the lipid 1-oleoyl-sn-glycero-3-phosphocholine.
Metabolites taken up also span a wide m/z range, with a maximum between m/z 200 and m/z 700 (Figure 1D, Table 1 and Supplementary Figure 2). There is thus relatively little size discrimination for the uptake.
Analysis of Selected Metabolites
Detailed analyses of the isotopic pattern enabled us to determine the ratio of heterotrophic uptake to de novo biosynthesis and look for evidence of mixed strategies. For example, the isotopologues of 6Z,9Z,12Z,15-hexadecatetraenoic acid (Figure 2A) originate from two distinct pools (Figure 2D), a lowly labeled pool from de novo biosynthesis and a highly labeled pool from uptake. Modeling with Bernoulli statistics showed that the de novo biosynthesis pool had a degree of labeling of 2.8%, slightly higher than what would be expected from photosynthesis using exclusively natural inorganic carbon (Figure 2B). The highly labeled pool has a very similar isotopologue distribution compared to the fatty acid from the lysed labeled alga (Figure 2C). Modeling a mixture of these two pools showed that ca. 20% of the metabolite in the algae results from uptake, while 80% are synthesized de novo. This complementation of de novo synthesized products with externally available metabolites is observed for several metabolites with variable proportions of the two sources from only a few percent to nearly 80% of the metabolite acquired by uptake (Table 1).
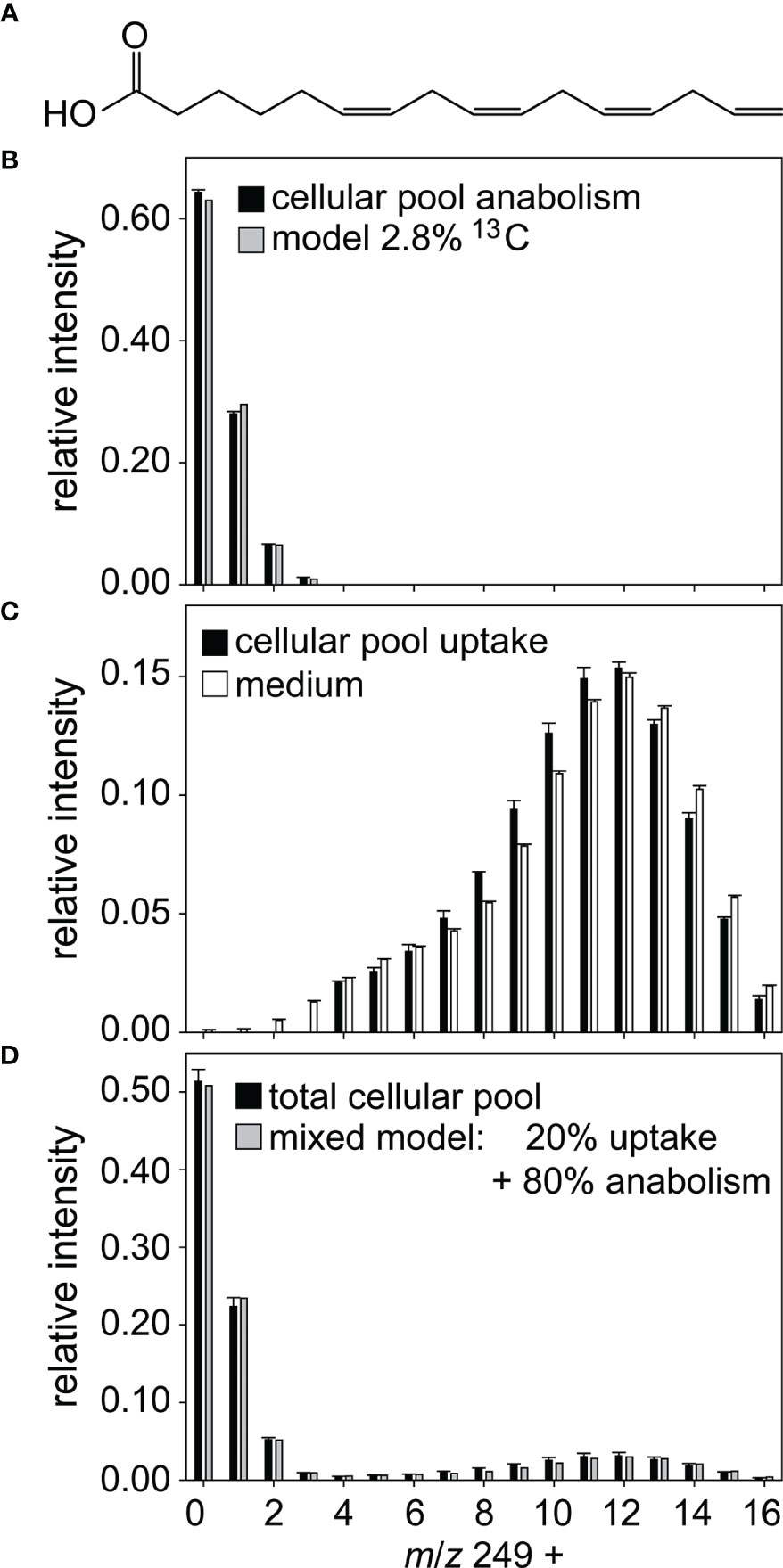
Figure 2 Labeling pattern of hexadecatetraenoic acid. (A) A mixed labeled metabolite was identified as 6Z,9Z,12Z,15-hexadecatetraenoic acid. For data evaluation the measured mass spectrum (black bars in D) was divided into a lowly labeled (B) and a highly labeled pool (C). (B) Modelling (grey bars) shows that the lowly labeled pool contains 2.8% 13C and thus likely derives from anabolism. (C) The highly labeled pool is taken up from the medium, black bars represent measured data of cellular hexadecatetraenoic acid, white bars measured data of hexadecatetraenoic acid in medium. (D) The measured mass spectrum in D can be explained by 20% hexadecatetraenoic acid derived from uptake and 80% from de novo synthesis (grey bars). Relative intensity refers to the sum of isotopolog ions of the molecular peak. All data are mean ± SD from biological triplicates.
The isotopologue patterns of metabolites that can be explained by an uptake of resources from the medium followed by transformations within the cell using the pool of de novo synthesized precursors are more complex. They do not follow the Bernoulli statistics since different resources can be utilized in different relative amounts. An example is the lysophosphatidylcholine shown in Figure 3A. The isotope pattern of the lipid (Figure 3B) cannot be interpreted with the model described above, but tandem MS experiments allow for dissecting the lipid. This reveals a unique labeling pattern for those parts of the molecule that are derived from different biosynthetic pathways. The isotopologue pattern of oleic acid in the lysophosphatidylcholine (Figure 3C) can also not be fitted with a Bernoulli statistic. The fatty acid is thus assembled using resources that were taken up as well as de novo produced. This labeling pattern in the fatty acid moiety is also observed in free oleic acid and 11Z-eicosenoic acid. Lipid assembly thus does not discriminate between acquired and de novo synthesized resources. In contrast, the choline fragment detected in the same substance shows that most choline is highly labeled and therefore taken up (Figure 3D). The glycerol moiety of the molecule is not giving charged fragments, but its labeling could be established indirectly. Therefore, we conducted fragmentation of the M+8 isotopologue of the lysophosphatidylcholine. The MS/MS of this ion gave rise to an oleic acid fragment with isotopologues containing down to zero 13C (Figure 3E). The remaining eight carbon atoms in the uncharged C8-fragment have thus to be labeled in different degrees. The fragments can thus only be derived from a precursor with labeled, thus acquired glycerol.
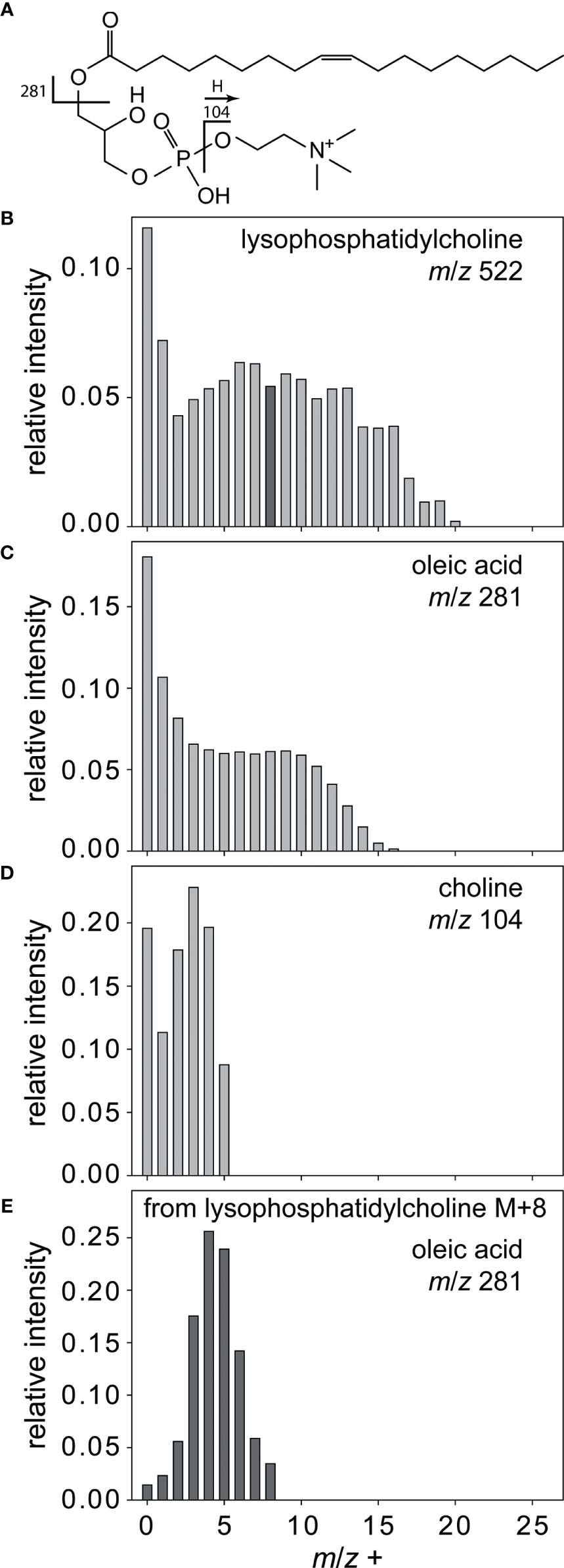
Figure 3 Complex labeling of lysophosphatidylcholine. All isotopologues (B) of lysophosphatidylcholine (A) are fragmented to yield the labeled building blocks oleic acid (C) and choline (D). Fragmentation of lysophosphatidylcholine M+8 isotopologue (black bar in B) yields oleic acid (E) with less than four 13C, thereby indirectly proving the labeling of glycerol. Relative intensity refers to the most sum of isotopolog ions of the molecular peak.
These exemplarily discussed mass spectra stand for several hundreds of labeled peaks in the chromatogram of the endometabolome of exposed C. didymus (see Figure 1 and Supporting information). Using the combined results, we can now draw a picture of the osmotrophic metabolism in C. didymus (Figure 4). The uptake introduced here for C. didymus is not limited to this one species but broader distributed in diatoms. When the diatom Thalassiosira weissflogii was raised in the labeled medium as described for C. didymus, labeling patterns similar to those described above were detected (data not shown).
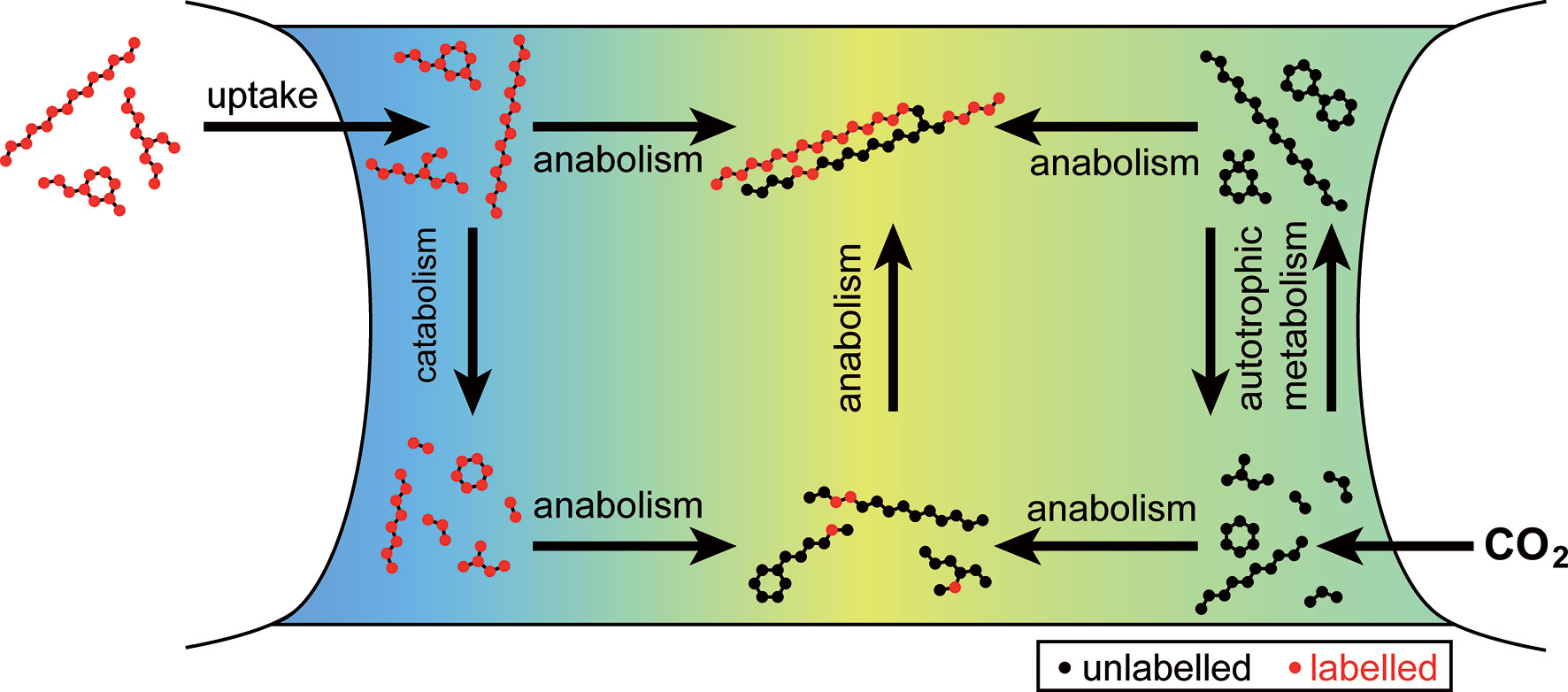
Figure 4 Uptake and metabolism of labeled organic compounds by Chaetoceros didymus. Organic compounds labeled with 13C (red) are taken up and transformed. Labeled compounds and their catabolic products are mixed with unlabeled metabolites (black) from autotrophic metabolism in anabolic reactions.
Discussion
13C Labeling Allows Tracing of Metabolic Shuttling Between Microalgae
In order to study potential osmotrophy in the diatom C. didymus a medium rich in 13C labeled organic metabolites was prepared by mechanical lysis of a globally labeled S. costatum culture and removal of the cell debris by filtration. We diluted this filtrate to reach concentrations for all individual compounds in the incubation that correspond to 50% of those found in a stationary, lysed culture. Given the fact that bacterial lysis of phytoplankton in the lab and field happens within hours and that entire blooms of algae can be lysed, we conclude that the concentrations employed are in the range of those found during lysis of a dense bloom (Bigalke et al., 2019). The employed concentrations will nevertheless be higher than under natural non-bloom conditions. However, we observed uptake of metabolites irrespective of their relative concentration in the extract. Not only abundant metabolites were found to be labeled and therefore acquired compounds. Also, minor and trace signals in the chromatograms indicated the incorporation of labels. Given the sensitivity of the mass spectrometer with detection limits in the femptomolar range, this indicates that the uptake mechanism is not driven by the concentration of the metabolites in the water and does thus not result from a “force-feeding” of C. didymus.
The resulting metabolome enriched medium showed a similar (but labeled) metabolic profile as the medium of an infection experiment where the lytic bacterium K. algicida lysed S. costatum. We thus conclude that K. algicida is a sloppy feeder not utilizing the entire algal metabolome but instead leaving substantial organic resources that it does not require or recover in the water. Surviving algae, such as the resistant competitor C. didymus will be exposed to these metabolites. Earlier, we observed that such exposure to metabolites released from lysed algae supports the growth of C. didymus if administered at low concentrations. However, no information about the underlying mechanism was available (Bigalke and Pohnert, 2019). The ability to take up released organic molecules may counterbalance the metabolic costs to maintain resistance mechanisms and would be highly advantageous, for example, during the collapse of a competing algal bloom as simulated in this study (Sohn et al., 2004; Meyer et al., 2017).
In addition to the release of organic material upon cell lysis, intact algae can also be a source of dissolved metabolites. Field studies indicate that natural phytoplankton communities can release 20 to 40% of newly assimilated dissolved inorganic nitrogen in form of dissolved organic nitrogen into the environment (Flynn and Berry, 1999). Also, secretion of dissolved organic carbon e.g., in the form of mucus-forming saccharides is well-documented (Alderkamp et al., 2007). If the metabolic uptake also functions in such situations or phases where dilute phytoplankton abundance results in low concentrations of dissolved metabolites would have to be verified in follow-up studies. Future work focusing on the quantification of selected metabolites could support uptake mechanisms and kinetics. We have, however, no indication from the analysis of labeling that there would be a bias for the uptake of higher concentrated metabolites.
We reached up to 65% labeling in the S. costatum metabolome using repeated exchange of medium containing as an exclusive carbon source. Higher degree of labeling might be reached in gas-tight culturing devices after more generation of culturing, but was not required for the present study. Analysis of the C. didymus endometabolome after being exposed to the labeled metabolome of S. costatum for 3 days under otherwise optimum growth conditions revealed that the alga took up substantial amounts of labeled compounds from various metabolic classes. Given that analysis of every single isotopologue of a labeled metabolite would generate highly complex and partially redundant information, we reverted to a statistical treatment assuming that labeling results in an isotopologue distribution in MS1 data that can be described with a Bernoulli statistic. In the Bernoulli trial a random experiment with exactly two possible outcomes is evaluated. In this case “success” would represent the incorporation of 13C and “failure” that of 12C. We estimate that excess substrate is present, thus the probability of success is the same every time a carbon is incorporated. This data treatment allowed to match labeling patterns to predicted spectra that would result from specific degrees of labeling. Thereby the average 65% labeling of the S. costatum metabolome could be determined.
Labeling Patterns of Metabolites in C. didymus After Exposure to 13C-Labeled S. costatum Extract
Analysis of the endometabolome of the resistant alga C. didymus after exposure to metabolites from 13C-labeled S. costatum revealed unlabeled metabolites, metabolites with unaltered full labeling and those with more complex mass spectra that could be assigned to different metabolic processing (Figure 1). The methodology described here thus not only shows the uptake of one metabolite, but allows simultaneous quantification of the uptake and analysis of the metabolic fate of all taken up metabolites in the receiving alga. This is a valuable expansion of the classical fluxomics approach, where feeding of one single labeled metabolite to heterotrophs can only reveal its uptake and metabolism. Our experimental approach reflects the situation in the plankton with highly complex microbial communities and complex exometabolomes (Barofsky et al., 2010). It allows evaluation of the uptake capacity and incorporation of the taken-up metabolites under ecologically relevant conditions.
General Patterns in the Uptake of Organic Metabolites by the Receiving Diatom C. didymus
Detailed analysis of the mass spectra allows to draw a picture of osmotrophy in C. didymus (Figure 4). More than a quarter of all detected metabolites in C. didymus were labeled to different degrees (Figure 1). Nearly 10% showed the identical labeling pattern as those in the 13C-labeled S. costatum metabolome. These metabolites are not (or only to a minor extent) synthesized by the receiver but taken up and maintained in the cells or the uptake could down-regulate synthesis in the receiver. The major part of taken-up metabolites showed mixed labeling, indicative for the uptake of a metabolite that is also synthesized by the receiver. This includes a wide range of natural products from small charged molecules like choline and carnitine to non-polar lipids and fatty acids. Metabolites acquired from the outside water can thus be metabolized in the same way as the de novo produced compounds, indicating that no compartmentation of assimilated material occurs. Instead, the osmo(hetero)trophic and the photo(auto)trophic pool are used for anabolism and catabolism. For example, the presence of complex labeling patterns can be explained by the use of intermediate fatty acids in the anabolism of more complex lipids. The complex isotopologue pattern of oleic acid and 11Z-eiosenoic acid, for example, might reflect a dynamic system with rounds of beta oxidation releasing labeled acetate and subsequent fatty acid re-assembly from this labeled and the own unlabeled acetate pool. Since the isotopologues of these fatty acids are not Bernoulli-distributed the relative amount of labeled acetate in the total acetate pool used for fatty acid biosynthesis might vary over time.
Utilization of Assimilated Metabolites
The fact that uptake has no apparent preference for nitrogen containing metabolites contradicts the assumption that diatoms use specific heterotrophic mechanisms to acquire reduced nitrogen (Mulholland and Lee, 2009). Since we worked in non-limiting conditions this lack of specificity might also reflect that diatoms do not need additional nitrogen. Flynn and Berry (1999) suggested that these microbes leak metabolites from internal mM levels and that they thus must have some recovery (uptake) potential, else they would ‘bleed to death’. Our findings will likely also hold true for such a recovery of metabolites released by the cells themselves. Thus, such a recovery mechanism would be in place and could be quantified using a modified labeling approach.
Notably, many metabolites with essential physiological functions are thus not exclusively produced de novo but taken up in high proportions. Control of physiological concentrations will thus have to include biosynthesis, catabolism as well as uptake. A universal uptake could be highly advantageous in the event of a lysis of a competing algal bloom (Sohn et al., 2004; Meyer et al., 2017). But even under regular conditions in plankton, algae might encounter metabolites released by other members of the phytoplankton that they can take up and benefit from (Sommer et al., 2012).
The observation of universal uptake of metabolites requires re-thinking of the interpretation of incubation studies with single labeled substrates. It is well documented that specific primary metabolites can be taken up by the cells under limiting conditions. Thus, under darkness, uptake of organic acids like lactate and malate was observed (Hellebust and Lewin, 1972). Amino acid usage is prompted by both N and/or C stress (Flynn and Wright, 1986) and uptake of the essential metabolite dimethylsulfoniopropionate (DMSP) was discussed as a way to compensate for the lack of own DMSP biosynthetic capabilities (Spielmeyer et al., 2011). We now expand this view and introduce that the diatoms C. didymus and T. weissflogii can take up a structural variety of metabolites released from competitors even under non-limiting growth conditions. Thus, a universal complementation of the own metabolome occurs and not a selected uptake of potentially valuable metabolites. This unspecific uptake of metabolites over a wide range of polarities and masses newly defines the investigated diatoms as a general sink of organic carbon in the sea and might be generalized to the entire group of these algae.
Physiological Function of Assimilated Metabolites
Of the multiple labeled metabolites, two groups will be exemplarily highlighted here, polar nitrogen containing metabolites and fatty acids. Polar metabolites that are taken up include glycine betaine, β-alanine betaine, carnitine and choline, with diverse physiological functions. Glycine betaine is a known osmolyte and uptake in diatoms has already been described under N-limited conditions (Keller et al., 1999; Gebser and Pohnert, 2013). Choline is a biosynthetic precursor of glycine betaine (Kageyama et al., 2018) and an important building block of phospholipids (Borowitzka, 2016). In this study, it has been found as a free form and in lysophosphatidylcholine (Table 1). However, the labeling pattern of the choline fragment of lysophosphatidylcholine differs from the free form (Supplementary Figure 3), indicating different origins. β-alanine betaine is a known osmoprotectant in plants and is biosynthesized via the methylation of β-alanine, a building block of coenzyme A (Rathinasabapathi et al., 2001). It has also been found in marine algae (Blunden et al., 1986). Carnitine is a central metabolite in energy metabolism of all eukaryotic cells. It plays an essential role for the transport of fatty acids across the mitochondrial membrane. The utilization of carnitine by diatoms has been reported previously by measuring oxidation rates in a biofilm-forming freshwater diatom (Tuchman et al., 2006). The presence of labeled N-methyl groups (Supplementary Figure 4) excludes lysine degradation (Paik et al., 1977) as a metabolic origin. The high proportion of heterotrophically acquired carnitine and the presence of many fatty acids among the taken-up metabolites is striking. It suggests a highly active transport mechanism for high-energy fatty acids and the N-containing carnitine. It would be interesting to investigate whether an acylcarnitine-type transport system might facilitate the uptake of fatty acids across the cellular membrane. The presence of a protein homologous to a class I carnitine/acylcarnitine translocase in a diatom cell wall proteome supports this hypothesis (Frigeri et al., 2006).
Several labeled fatty acids and derivatives have also been identified (Table 1). Eicosapentaenoic acid is one of the dominant fatty acids in diatoms and precursor of many bioactive oxylipins (Pohnert and Boland, 2002; Borowitzka, 2016; Rettner et al., 2018; Zulu et al., 2018). 6Z,9Z,12Z-hexadecatrienoic acid is the precursor of octadienal, an allelopathic polyunsaturated aldehyde (D’ippolito et al., 2003). Also, oleic acid, 11Z-eicosenoic acid, and 6Z,9Z,12Z,15-hexadecatetraenoic acid are common in diatoms (Ackman et al., 1968; Dunstan et al., 1994; Pohnert et al., 2004; Chen, 2012; Zulu et al., 2018). Two other C18- and C20 polyunsaturated fatty acids were identified according to retention time and mass spectra. Comparison with synthetic standards showed that these fatty acids were isomers of linoleic and arachidonic acid. Fatty acids and their derivatives have multiple physiological functions. The fact that they are present in labeled and unlabeled forms after the pulse labeling experiments indicates an unbiased utilization of de novo and assimilated compounds. This might serve as a mechanism to avoid costly biosynthesis of fatty acids, which is supported by the observation that the fatty acids are not only internalized or adsorbed to lipidic structures due to their physicochemical properties but rather incorporated into the primary metabolism of the diatom.
The broad range of polarity and molecular weight of assimilated compounds raises the question of the uptake mechanism. Candidate systems for fatty acid shuttling include specific transporters and genome data shows homolog candidate sequences (Frigeri et al., 2006; Indiveri et al., 2011). But indeed, also unspecific osmotrophy and incorporation mechanisms that do not require transporters will most likely be involved. Nevertheless, the question of how diatoms acquire exogenous metabolites is still open.
Concluding Remarks
The diatom C. didymus takes up and incorporates metabolites from lysed competitors with surprising little bias. Preliminary results suggest that the same is true for the diatom T. weissflogii (data not shown), which indicates a rather universal mechanism. The multitude of cellular functions in which these metabolites are involved suggests that uptake complements the internal metabolic pool from many different biosynthetic pathways.
The universal osmotrophic lifestyle of members of one of the most abundant algal classes in the oceans substantially changes our view of the metabolic shuttling in phytoplankton communities. These algae take up more metabolites than just a few highly polar compounds for e.g. nitrogen supply. They rather complement their metabolism quite universally with resources from the surrounding seawater. This process occurs even under illumination and is not related to the complementation of lacking photosynthate in the dark. It will have to be verified if this is a universal property of diatoms and if also other members of the phytoplankton utilize similar strategies, as it is already indicated by work on Emiliania huxleyi (Godrijan et al., 2021). In addition to bacteria, diatoms and other phytoplankton compete for the dissolved organic carbon in the plankton (Berge et al., 2012; Georges et al., 2014). Our study thus has consequences for element cycling in the oceans and ecosystem dynamics that will have to be addressed in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: MetaboLights [accession: MTBLS2333].
Author Contributions
NM developed the pipeline and analyzed the data. AR acquired and analyzed data. NM and GP conceived the study, directed all experiments, and wrote the manuscript with contributions from the co-author. All authors approved the manuscript.
Funding
Deutsche Forschungsgemeinschaft (DFG) - SFB 1127/2 ChemBioSys – 239748522. Deutsche Forschungsgemeinschaft (DFG) EXC 2051 – Project-ID 390713860.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank W. Kooistra for providing diatom cultures. David Russo is acknowledged for helpful comments on an initial draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.821167/full#supplementary-material
References
Ackman R. G., Tocher C. S., Mclachla J. (1968). Marine Phytoplankter Fatty Acids. J. Fisher. Res. Board Canada 25, 1603–1608. doi: 10.1139/f68-145
Alderkamp A. C., Buma A. G. J., van Rijssel M. (2007). The Carbohydrates of Phaeocystis and Their Degradation in the Microbial Food Web. Biogeochemistry 83, 99–118. doi: 10.1007/s10533-007-9078-2
Barofsky A., Simonelli P., Vidoudez C., Troedsson C., Nejstgaard J. C., Jakobsen H. H., et al. (2010). Growth Phase of the Diatom Skeletonema Marinoi Influences the Metabolic Profile of the Cells and the Selective Feeding of the Copepod Calanus Spp. J. Plankton. Res. 32, 263–272. doi: 10.1093/plankt/fbp121
Berge T., Poulsen L. K., Moldrup M., Daugbjerg N., Hansen P. J. (2012). Marine Microalgae Attack and Feed on Metazoans. ISME J. 6, 1926–1936. doi: 10.1038/ismej.2012.29
Bigalke A., Meyer N., Papanikolopoulou L. A., Wiltshire K. H., Pohnert G. (2019). The Algicidal Bacterium Kordia Algicida Shapes a Natural Plankton Community. Appl. Env. Microbiol. 85 (7), e02779–18. doi: 10.1128/AEM.02779-18
Bigalke A., Pohnert G. (2019). Algicidal Bacteria Trigger Contrasting Responses in Model Diatom Communities of Different Composition. MicrobiologyOpen 8 (8), e00818. doi: 10.1002/mbo3.818
Blunden G., Gordon S. M., Crabb T. A., Roch O. G., Rowan M. G., Wood B. (1986). Nmr-Spectra of Betaines From Marine-Algae. Magn. Reson. Chem. 24, 965–971. doi: 10.1002/mrc.1260241108
Burkholder J. M., Glibert P. M., Skelton H. M. (2008). Mixotrophy, a Major Mode of Nutrition for Harmful Algal Species in Eutrophic Waters. Harm. Algae. 8, 77–93. doi: 10.1016/j.hal.2008.08.010
Caron D. A. (2016). Mixotrophy Stirs Up Our Understanding of Marine Food Webs. Proc. Nat. Acad. Sci. U.S.A 113, 2806–2808. doi: 10.1073/pnas.1600718113
Chary V. N., Kumar C. D., Vairamani M., Prabhakar S. (2012). Characterization of Amino Acid-Derived Betaines by Electrospray Ionization Tandem Mass Spectrometry. J. Mass. Spectrom. 47, 79–88. doi: 10.1002/jms.2029
Chen Y. C. (2012). The Biomass and Total Lipid Content and Composition of Twelve Species of Marine Diatoms Cultured Under Various Environments. Food Chem. 131, 211–219. doi: 10.1016/j.foodchem.2011.08.062
Cochlan W. P., Herndon J., Kudela R. M. (2008). Inorganic and Organic Nitrogen Uptake by the Toxigenic Diatom Pseudo-Nitzschia Australis (Bacillariophyceae). Harm. Algae. 8, 111–118. doi: 10.1016/j.hal.2008.08.008
D’ippolito G., Romano G., Caruso T., Spinella A., Cimino G., Fontana A. (2003). Production of Octadienal in the Marine Diatom Skeletonema Costatum. Org. Lett. 5, 885–887. doi: 10.1021/ol034057c
Duhrkop K., Shen H., Meusel M., Rousu J., Bocker S. (2015). Searching Molecular Structure Databases With Tandem Mass Spectra Using CSI:FingerID. Proc. Natl. Acad. Sci. U.S.A 112 (41), 12580–12585. doi: 10.1073/pnas.1509788112
Dunstan G. A., Volkman J. K., Barrett S. M., Leroi J. M., Jeffrey S. W. (1994). Essential Polyunsaturated Fatty-Acids From 14 Species of Diatom (Bacillariophyceae). Phytochemistry 35, 155–161. doi: 10.1016/S0031-9422(00)90525-9
Ferroni L., Giovanardi M., Poggioli M., Baldisserotto C., Pancaldi S. (2018). Enhanced Photosynthetic Linear Electron Flow in Mixotrophic Green Microalga Ettlia Oleoabundans UTEX 1185. Plant Phys. Biochem. 130, 215–223. doi: 10.1016/j.plaphy.2018.07.005
Flynn K. J., Berry L. S. (1999). The Loss of Organic Nitrogen During Marine Primary Production may be Significantly Overestimated When Using 15n Substrates. Proc. R. Soc London. Ser. B 266, 641–647. doi: 10.1098/rspb.1999.0684
Flynn K. J., Clark D. R., Xue Y. (2008). Modelling the Release of Dissolved Organic Matter by Phytoplankton. J. Phycol. 44, 1171–1187. doi: 10.1111/j.1529-8817.2008.00562.x
Flynn K. J., Stoecker D. K., Mitra A., Raven J. A., Glibert P. M., Hansen P. J., et al. (2013a). Misuse of the Phytoplanktonzooplankton Dichotomy: The Need to Assign Organisms as Mixotrophs Within Plankton Functional Types. J. Plankton. Res. 35, 3–11. doi: 10.1093/plankt/fbs062
Flynn K. J., Stoecker D. K., Mitra A., Raven J. A., Glibert P. M., Hansen P. J., et al. (2013b). Misuse of the Phytoplankton–Zooplankton Dichotomy: The Need to Assign Organisms as Mixotrophs Within Plankton Functional Types. J. Plankton. Res. 35, 3–11. doi: 10.1093/plankt/fbs062
Flynn K. J., Wright C. R. N. (1986). The Simultaneous Assimilation of Ammonium and L-Arginine by the Marine Diatom Phaeodactylum Tricornutum Bohlin. J. Exp. Mar. Biol. Ecol. 95, 257–269. doi: 10.1016/0022-0981(86)90258-3
Frigeri L. G., Radabaugh T. R., Haynes P. A., Hildebrand M. (2006). Identification of Proteins From a Cell Wall Fraction of the Diatom Thalassiosira Pseudonana - Insights Into Silica Structure Formation. Mo. Cell. Proteom. 5, 182–193. doi: 10.1074/mcp.M500174-MCP200
Gebser B., Pohnert G. (2013). Synchronized Regulation of Different Zwitterionic Metabolites in the Osmoadaption of Phytoplankton. Mar. Drugs 11, 2168–2182. doi: 10.3390/md11062168
Georges A. A., El-Swais H., Craig S. E., Li W. K. W., Walsh D. A. (2014). Metaproteomic Analysis of a Winter to Spring Succession in Coastal Northwest Atlantic Ocean Microbial Plankton. ISME J. 8, 1301–1313. doi: 10.1038/ismej.2013.234
Godrijan J., Drapeau D. T., Balch W. M. (2021). Osmotrophy of Dissolved Organic Carbon by Coccolithophores in the Darkness. New Phytol. 233, 781–794. doi: 10.1111/nph.17819
Heesen W. (1928). Planktonkunde - Eine Einführung in Die Ökologie Der Im Wasser Schwebenden Kleinwelt. (Berlin: Verlag Otto Salle).
Hellebust J. A., Guillard R. R. (1967). Uptake Specificity for Organic Substrates by the Marine Diatom Melosira Nummuloides(1). J. Phycol. 3, 132–136. doi: 10.1111/j.1529-8817.1967.tb04646.x
Hellebust J. A., Lewin J. (1972). Transport Systems for Organic Acids Induced in the Marine Pennate Diatom, Cylindrotheca Fusiformis. Can. J. Microbiol. 18, 225–233. doi: 10.1139/m72-035
Huang X. J., Chen Y. J., Cho K., Nikolskiy I., Crawford P. A., Patti G. J. (2014). (XCMS)-C-13: Global Tracking of Isotopic Labels in Untargeted Metabolomics. Anal. Chem. 86, 1632–1639. doi: 10.1021/ac403384n
Indiveri C., Iacobazzi V., Tonazzi A., Giangregorio N., Infantino V., Convertini P., et al. (2011). The Mitochondrial Carnitine/Acylcarnitine Carrier: Function, Structure and Physiopathology. Mol. Aspect. Med. 32, 223–233. doi: 10.1016/j.mam.2011.10.008
Kageyama H., Tanaka Y., Takabe T. (2018). Biosynthetic Pathways of Glycinebetaine in Thalassiosira Pseudonana; Functional Characterization of Enzyme Catalyzing Three-Step Methylation of Glycine. Plant Phys. Biochem. 127, 248–255. doi: 10.1016/j.plaphy.2018.03.032
Karlusich J. J. P., Ibarbalz F. M., Bowler C. (2020). “Phytoplankton in the Tara Ocean,” in Ann. Rev. Mar. Sci, vol. Vol 12 . Eds. Carlson C. A., Giovannoni S. J., 233–265.
Keller M. D., Kiene R. P., Matrai P. A., Bellows W. K. (1999). Production of Glycine Betaine and Dimethylsulfoniopropionate in Marine Phytoplankton. II. N-Limited Chemostat Cultures. Mar. Biol. 135, 249–257. doi: 10.1007/s002270050622
Kessner D., Chambers M., Burke R., Agusand D., Mallick P. (2008). ProteoWizard: Open Source Software for Rapid Proteomics Tools Development. Bioinformatics 24, 2534–2536. doi: 10.1093/bioinformatics/btn323
Lewin J. C., Lewin R. A. (1960). Auxotrophy and Heterotrophy in Marine Littoral Diatoms. Can. J. Microbiol. 6, 127–134. doi: 10.1139/m60-015
Maier I., Calenberg M. (1994). Effect of Extracellular Ca2+ and Ca2+-Antagonists on the Movement and Chemoorientation of Male Gametes of Ectocarpus-Siliculosus (Phaeophyceae). Bot. Acta 107, 451–460. doi: 10.1111/j.1438-8677.1994.tb00820.x
Meyer N., Bigalke A., Kaulfuss A., Pohnert G. (2017). Strategies and Ecological Roles of Algicidal Bacteria. FEMS Microbiol. Rev. 41, 880–899. doi: 10.1093/femsre/fux029
Mulholland M. R., Lee C. (2009). Peptide Hydrolysis and the Uptake of Dipeptides by Phytoplankton. Limnol. Oceanogr. 54, 856–868. doi: 10.4319/lo.2009.54.3.0856
Neilson A. H., Lewin R. A. (1974). The Uptake and Utilization of Organic Carbon by Algae: An Essay in Comparative Biochemistry. Phycologia 13, 227–264. doi: 10.2216/i0031-8884-13-3-227.1
Paik W. K., Nochumson S., Kim S. (1977). Carnitine Biosynthesis Via Protein Methylation. Trends Biochem. Sci. 2, 159–161. doi: 10.1016/0968-0004(77)90365-6
Paul C., Pohnert G. (2011). Interactions of the Algicidal Bacterium Kordia Algicida With Diatoms: Regulated Protease Excretion for Specific Algal Lysis. PloS One 6, e21032. doi: 10.1371/journal.pone.0021032
Paul C., Pohnert G. (2013). Induction of Protease Release of the Resistant Diatom Chaetoceros Didymus in Response to Lytic Enzymes From an Algicidal Bacterium. PloS One 8, e57577. doi: 10.1371/journal.pone.0057577
Petrou K., Nielsen D. A. (2018). Uptake of Dimethylsulphoniopropionate (DMSP) by the Diatom Thalassiosira Weissflogii: A Model to Investigate the Cellular Function of DMSP. Biogeochemistry 141, 265–271. doi: 10.1007/s10533-018-0507-1
Pohnert G., Adolph S., Wichard T. (2004). Short Synthesis of Labeled and Unlabeled 6z,9z,12z,15-Hexadecatetraenoic Acid as Metabolic Probes for Biosynthetic Studies on Diatoms. Chem. Phys. Lipids 131, 159–166. doi: 10.1016/j.chemphyslip.2004.04.011
Pohnert G., Boland W. (2002). The Oxylipin Chemistry of Attraction and Defense in Brown Algae and Diatoms. Nat. Prod. Rep. 19, 108–122. doi: 10.1039/a806888g
Pohnert G., Jung V. (2003). Intracellular Compartmentation in the Biosynthesis of Caulerpenyne: Study on Intact Macroalgae Using Stable-Isotope-Labeled Precursors. Org. Lett. 5, 5091–5093. doi: 10.1021/ol036163k
Rathinasabapathi B., Fouad W. M., Sigua C. A. (2001). Beta-Alanine Betaine Synthesis in the Plumbaginaceae. Purification and Characterization of a Trifunctional, S-Adenosyl-L-Methionine-Dependent N-Methyltransferase From Limonium Latifolium Leaves. Plant Phys. 126, 1241–1249. doi: 10.1104/pp.126.3.1241
Raven J. A., Beardall J., Flynn K. J., Maberly S. C. (2009). Phagotrophy in the Origins of Photosynthesis in Eukaryotes and as a Complementary Mode of Nutrition in Phototrophs: Relation to Darwin's Insectivorous Plants. J. Exp. Bot. 60, 3975–3987. doi: 10.1093/jxb/erp282
Rettner J., Werner M., Meyer N., Werz O., Pohnert G. (2018). Survey of the C20 and C22 Oxylipin Family in Marine Diatoms. Tetrahed. Lett. 59, 828–831. doi: 10.1016/j.tetlet.2018.01.057
Selosse M. A., Charpin M., Not F. (2017). Mixotrophy Everywhere on Land and in Water: The Grand Ecart Hypothesis. Ecol. Lett. 20, 246–263. doi: 10.1111/ele.12714
Sohn J. H., Lee J. H., Yi H., Chun J., Bae K. S., Ahn T. Y., et al. (2004). Kordia Algicida Gen. Nov., Sp Nov., an Algicidal Bacterium Isolated From Red Tide. I. J. Systemat. Evol. Microbiol. 54, 675–680. doi: 10.1099/ijs.0.02689-0
Sommer U., Adrian R., Domis L. D., Elser J. J., Gaedke U., Ibelings B., et al. (2012). Beyond the Plankton Ecology Group (PEG) Model: Mechanisms Driving Plankton Succession. Ann. Rev. Ecol. Evol. Systemat. 43, 429–448. doi: 10.1146/annurev-ecolsys-110411-160251
Spielmeyer A., Gebser B., Pohnert G. (2011). Investigations of the Uptake of Dimethylsulfoniopropionate by Phytoplankton. ChemBioChem 12, 2276–2279. doi: 10.1002/cbic.201100416
Stoecker D. K., Hansen P. J., Caron D. A., Mitra A., Annual R. (2017). “Mixotrophy in the Marine Plankton,” in Ann. Rev. Mar. Sci, vol. Vol 9. , 311–335.
Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., Daykin C. A., et al. (2007). Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221. doi: 10.1007/s11306-007-0082-2
Tuchman N. C., Schollett M. A., Rier S. T., Geddes P. (2006). Differential Heterotrophic Utilization of Organic Compounds by Diatoms and Bacteria Under Light and Dark Conditions. Hydrobiologia 561, 167–177. doi: 10.1007/s10750-005-1612-4
Vila-Costa M., Simo R., Harada H., Gasol J. M., Slezak D., Kiene R. P. (2006). Dimethylsulfoniopropionate Uptake by Marine Phytoplankton. Science 314, 652–654. doi: 10.1126/science.1131043
Keywords: phytoplankton interactions, diatoms, absorbotrophy, mixotrophy, mass spectrometry, Osmotrophy, metabolomics, isotopic lableing
Citation: Meyer N, Rydzyk A and Pohnert G (2022) Pronounced Uptake and Metabolism of Organic Substrates by Diatoms Revealed by Pulse-Labeling Metabolomics. Front. Mar. Sci. 9:821167. doi: 10.3389/fmars.2022.821167
Received: 23 November 2021; Accepted: 24 March 2022;
Published: 25 April 2022.
Edited by:
Susana Agusti, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Kevin John Flynn, Plymouth Marine Laboratory, United KingdomErin McParland, Woods Hole Oceanographic Institution, United States
Copyright © 2022 Meyer, Rydzyk and Pohnert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georg Pohnert, Z2VvcmcucG9obmVydEB1bmktamVuYS5kZQ==
 Nils Meyer
Nils Meyer Aljoscha Rydzyk1
Aljoscha Rydzyk1 Georg Pohnert
Georg Pohnert