- 1Stazione Zoologica Anton Dohrn, Naples, Italy
- 2Department of Veterinary Medicine, University of Bari Aldo Moro, Bari, Italy
- 3Department of Biology, University of Naples Federico II, Naples, Italy
- 4National Interuniversity Consortium for Marine Sciences (CoNISMa), Rome, Italy
Zooplankton molecular analyses allow for accurate species identification with a proper molecular signature, complementing classic phenotypic-based taxonomy (α taxonomy). For the first time in the Mediterranean Sea, cytochrome oxidase I (COI) gene sequences of calanoid copepods were associated with morphological identification, HD and SEM images, using a fully integrated approach to assess taxonomic diversity. Such method was applied to selected species, generating consensus sequences from the Gulf of Naples (Central Tyrrhenian Sea, Western Mediterranean Sea) also including reference barcodes of three target species (Nannocalanus minor, Pleuromamma gracilis and the non-indigenous species (NIS) Pseudodiaptomus marinus) that are new for the Mediterranean area. The new barcodes were selected including: dominant and rare species; species that were originally described in the study area as type locality, but lacking a molecular description; emergent NIS and potential species complex. The integration between morphological and molecular identification by tree placement, using species-specific highly conserved oligonucleotides, also provided new and high-quality references of the most common and abundant copepod genera and species in the Mediterranean Sea. Our regional reference library was then integrated and analyzed with global data reference available on BOLD database to explore the presence of potential cryptic species and biogeographic patterns and links among geographically distant populations of copepods. Overall, this study provides valuable insight into the actual copepod taxonomic diversity and contributes to building baseline knowledge to monitor coastal biodiversity in neritic areas worldwide, where copepods are of paramount ecological importance, paving the way for future metabarcoding studies.
Introduction
Marine biodiversity is the first of the eleven Descriptors of Good Environmental Status (GES) defined by the EU Marine Strategy Framework Directive (MSFD) (Directive 2008/56/EC). Assessing biodiversity is a challenging task, and molecular taxonomy complements phenotypic approaches, proving reliable for routine identification, once the sequences are assigned to correctly identified phenotypes (Boero and Bernardi, 2014). The taxonomic accuracy of biodiversity datasets, however, varies at both spatial and temporal scales. The zooplankton time-series of IOC-UNESCO International Group for Marine Ecological Time Series (IGMETS) increased our understanding of how marine ecosystems respond to climate change and other environmental variations, and three-quarters of these time series have high taxonomic accuracy (O’Brien et al., 2017; Chiba et al., 2018). Genotypic approaches coupled with traditional (phenotypic-based) taxonomy allow to identify occurring native species and trace non-indigenous ones, estimating the real number of zooplankton species, including the identification of specimens from cryptic species complexes or species groups with high morphological similarity, which are difficult (if not impossible) to discriminate based on morphology only.
Currently, ∼5,700 holoplankton species are described, but it is estimated that ∼1,600 more species are yet to be discovered and described (Bucklin et al., 2010b,2021). Among holoplanktonic organisms, copepods often dominate marine zooplankton assemblages, both in terms of abundance (Humes, 1994) and diversity (Uttieri, 2018; Laakmann et al., 2020; Bucklin et al., 2021; Walter and Boxshall, 2021). The morphological identification and the molecular taxonomy of these microscopic crustaceans are very difficult to address. The morphological taxonomic knowledge of Mediterranean copepods is strongly rooted in the past (Giesbrecht, 1892; Giesbrecht and Schmeil, 1898; Sars, 1924-1925; Rose, 1927a,b), since the end of 19th century when Wilhelm Giesbrecht started the study of planktonic copepods in the Gulf of Naples (Tyrrhenian Sea, Western Mediterranean Sea), by describing and establishing several families, genera and species. Currently, based on morphological characters only, 176 calanoid species referred to 65 genera occur in Italian seas (Mazzocchi and Di Capua, 2010; Di Capua and Mazzocchi, 2021). Of them, 160 species referred to 57 genera are found at the Long Term Ecological Research station MareChiara (LTER-MC) in the Gulf of Naples, routinely sampled since 1984 (Zingone et al., 2019). In this site, new entries of non-indigenous species (NIS) were identified by experienced taxonomists using morphological characters, while processing the regular zooplankton samples (Di Capua and Boxshall, 2008; Kasapidis et al., 2018; Uttieri et al., 2020). In recent years, the introduction of molecular methods provided additional information that robustly supported copepod systematics (Di Capua et al., 2017, 2021). DNA barcoding, based on the sequencing of highly conserved DNA regions with little intraspecific variations of the mitochondrial cytochrome c oxidase I (COI) gene, is a precise and reliable identification tool (Tautz et al., 2002). Despite calanoid copepods are one of the best studied groups of marine zooplankton by using such an approach (Bucklin et al., 2010a,b; Laakmann et al., 2013, 2020; Blanco-Bercial et al., 2014) molecular barcoding is still restricted to few copepod families, mainly from the order Calanoida (42%) (Bucklin et al., 2021), and only 104 calanoid species (26%) presently have COI barcodes.1 Such gap calls for an effort, coupling traditional taxonomy (phenotypic-based) with molecular taxonomy (genotypic-based).
The aim of the present study is to expand present knowledge on the calanoid taxonomic diversity using integrative morphological and molecular approaches and lay the foundation for a reference library for Mediterranean coastal areas, starting from the Gulf of Naples as a case study due to its high representativeness. The taxonomic approach, identifying selected target and key species of the nine most abundant and widespread calanoid families (Acartiidae, Calanidae, Centropagidae, Clausocalanidae, Aetideidae, Paracalanidae, Metridinidae, Pseudodiaptomidae, and Temoridae) using morphological identification, is complemented with high definition (HD) and scanning electron microscopy (SEM) images of the individuals and their DNA barcode using COI genetic marker, with the creation of consensus sequences longer than 500 bp. The selected and analyzed species were chosen according to their taxonomic and ecological features: (1) dominance in neritic areas of the Mediterranean Sea; (2) Mediterranean Sea as type locality, but lacking a molecular description; (3) inclusion of species complex under the same name; (4) rare but cosmopolitan species; (5) emergent NIS, recently spreading in European waters. Molecular dataset was integrated and analyzed with global data reference available on BOLD and NCBI databases for copepod studied to explore the presence of potential cryptic species and biogeographic patterns and links among geographically distant populations of copepods, thus linking individual identification to a wider Ω taxonomy framework.
Materials and Methods
Zooplankton Sampling and Specimens Collection
Since 1984, phyto- and zooplankton are assessed, and environmental variables are measured weekly at the Long-Term Ecological Research MareChiara site (LTER-MC). The station, located two nautical miles off the city of Naples (40°48.5′N, 14°15′E, depth ca. 75 m), is influenced by both the eutrophic inputs from coastal runoffs and the oligotrophic waters of the mid Tyrrhenian Sea (Cianelli et al., 2017; Figure 1). Since 2019, additional samplings are carried out at an offshore mesopelagic station above the Dohrn canyon (40°36.2′N, 14°08′E, depth ca. 700 m) in the framework of the Naples Ecological REsearch and Augmented ocean observation (NEREA) project, an augmented observatory including “omic approaches” (Figure 1).
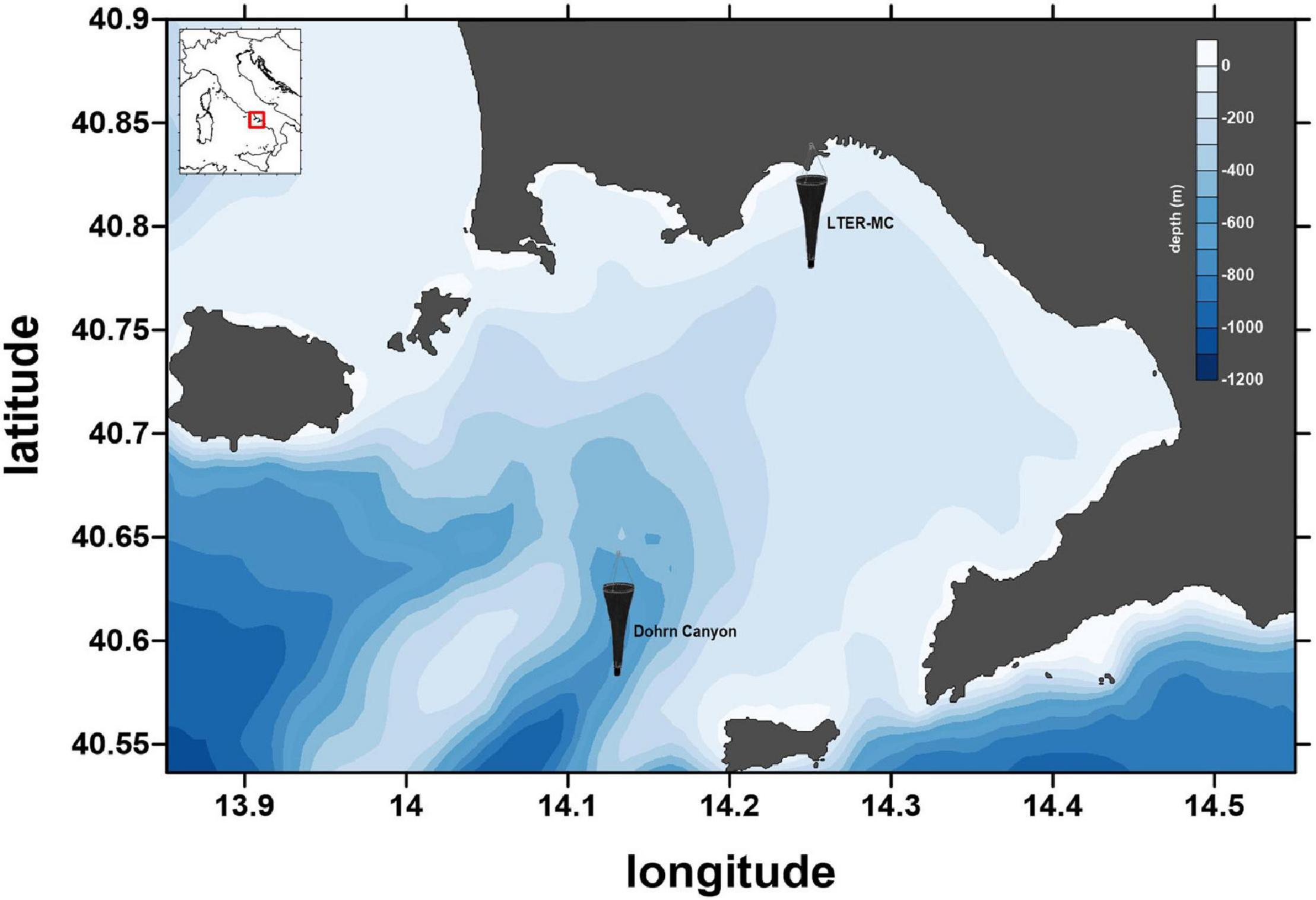
Figure 1. The sampling site, the LTER-MC site and Canyon Dohrn (plankton net icons), in the Gulf of Naples (Central Tyrrhenian Sea, Mediterranean Sea). Coastline data: NOAA National Geophysical Data Center, Coastline extracted: WLC (World Coast Line), Date Retrieved: April 01, 2015, http://www.ngdc.noaa.gov/mgg/shorelines/shorelines.html; bathymetric data from Amante and Eakins (2009).
Zooplankton samples were collected at the LTER-MC and Dohrn canyon sites during 2019 with a WP2 plankton net (mouth diameter: 57 cm; mesh aperture width: 200 μm), towed vertically from −50 m depth to the surface at low speed (0.7–1.0 m s–1). Samples were maintained alive on board at 4°C and transported to the laboratory in a cooler within 2 h. The sample was filtered on a nitex filter (200 μm mesh) to remove the sea water, immediately preserved in 95% ethanol, and stored in the dark at 4°C. The ethanol was replaced after 24 h to remove seawater excess.
Morphological Identification and Occurrence of the Analyzed Species
Mixed zooplankton samples were preserved in ethanol and were morphologically analyzed under a stereomicroscope (Leica M165C). Mature female calanoid specimens were sorted and taxonomically identified at species level based on morphological analysis of diagnostic characters according to Razouls et al. (2005-2021) and WoRMS Editorial Board (2021) and original descriptions and revisions, when relevant.
A total of 81 specimens of calanoid copepods were identified and referred to 16 species: Acartia clausi Giesbrecht, 1889; Calanus helgolandicus Claus, 1863; Calocalanus styliremis Giesbrecht, 1888; Candacia bispinosa Claus, 1863; Candacia simplex Giesbrecht, 1889; Centropages typicus Krøyer, 1849; Clausocalanus parapergens Frost and Fleminger, 1968; Ctenocalanus vanus Giesbrecht, 1888; Euchirella rostrata Claus, 1866; Nannocalanus minor Claus, 1863; Paracalanus indicus Wolfenden, 1903; Paracalanus parvus Claus, 1863; Pleuromamma gracilis Claus, 1863; Pontella mediterranea Claus, 1863; Pseudodiaptomus marinus Sato, 1913; Temora stylifera Dana, 1849. Morphological and morphometric parameters were annotated for each individual, and HD images in lateral and frontal views were taken using a Leica HD camera (DMC 5400). In addition, diagnostic morphological details of Paracalanus parvus and P. indicus were verified using SEM on other specimens selected from the same samples used for morphological and molecular analyses.
A. clausi, C. typicus, T. stylifera, and the P. parvus species complex are the four dominant taxa at LTER-MC, showing a seasonal succession and a recurrent pattern from mid-spring to autumn (Di Capua and Mazzocchi, 2004; Ribera d’Alcalà et al., 2004). C. styliremis, C. parapergens, E. rostrata, N. minor, and P. gracilis are other important contributors to the copepod assemblage at LTER-MC (Mazzocchi et al., 2011). C. helgolandicus is a widespread epipelagic species particularly sensitive to subtle changes in climate (Bonnet et al., 2005); C. vanus was originally described in the Gulf of Naples by Giesbrecht (1888); P. marinus is a global NIS (Sabia et al., 2015; Uttieri et al., 2020), present in the study area since 2011 (Di Capua et al., 2021).
DNA Extraction, Polymerase Chain Reaction Amplification, and DNA Sequencing
DNA was extracted from each specimen using a new cetyltrimethyl ammonium bromide (CTAB) based protocol developed for zooplankton crustacean species. Each specimen was washed with distilled water to remove ethanol, transferred into a 1.5 mL Eppendorf tube, and lysed in 500 μL of AppliChem CTAB extraction buffer (AppliChem, BioChemica, Illinois Tool Works Inc., Chicago, IL, United States, code A4150) and 10 μL of Roche Proteinase K recombinant Polymerase Chain Reaction (PCR) Grade (Roche, code 03115852001) at 60°C overnight. DNA isolation was performed adding 500 μL of SEVAG (chloroformisoamilic-alcohol 24:1 v/v) to the lysate, shaking upside down and incubating for ten minutes at room temperature (RT). Each sample was centrifuged at 14,000 rpm for 30 min at RT. The supernatant was recovered and incubated with a same volume (1:1) of 2-propanol (SIGMA-ALDRICH, code 33539-1L-R) at −20°C for 2 h. The DNA precipitate was centrifuged at 14,000 rpm for 30 min at 4°C. The supernatant was removed without disturbing the pellet, which was washed twice with 100 μL of 75% ice-cold ethanol. The pellet was left to air-dry under the hood and re-suspended in 20 μL of sterile MilliQ water.
Next, as a template for PCR, the extracted DNA was used to amplify the mitochondrial gene cytochrome c oxidase subunit I (COI) 710 bp, in a final volume of 25 μL, using C-1000 TouchTM Thermal Cycler. Primers pairs used for amplification and for the following sequence analysis were the universal COI primers (forward LCO1490: 5’-ggtcaacaaatcataaagatattgg-3’ and reverse HC02198: 5’-taaacttcagggtgaccaaaaaatca-3’) (Folmer et al., 1994), considered a reference standard for marine zooplankton (Bucklin et al., 2010a,2011, 2021). The PCR thermal conditions included an initial denaturation step at 95°C for 5 min, followed by forty cycles at 95°C (denaturation for 5 min), 45°C (annealing for 1 min), and 72°C (extension for 1 min), then a final extension step at 72°C for 7 min. The genomic template not amplified was stored in a genomic collection to preserve DNA vouchers for future additional studies (e.g., amplification of other target genes, such as 18S or ITS).
The PCR products were electrophoresed in a 1% agarose gel in Tris Borate EDTA (TBE) buffer stained with ethidium bromide and visualized under UV light. A DNA ladder 100 bp was used for the estimation of the amplicon length. The obtained PCR products were excised from the gel and isolated for later sequencing using the DNA Isolation Spin-kit Agarose (AppliChem) following the manufacturer’s protocol. Each purified PCR product was sequenced at the Molecular Biology and Sequencing Service of SZN, using Big Dye Terminator Cycle Sequencing Kit (Life Technologies) and analyzed on the Automated Capillary Electrophoresis Sequencer 3730 DNA Analyzer.
Sequences Alignment and Data Analysis
Consensus sequences were obtained from forward and reverse COI sequences using BioEdit Sequence Alignment Editor (Hall, 1999). A molecular identification by similarity approach was performed against Barcode of Life Data Systems database (BOLD)2 (Ratnasingham and Hebert, 2007; Meiklejohn et al., 2019) and National Center for Biotechnology Information (NCBI) GenBank database3 using Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1997). In addition, identification by generation and placement in a tree (phylogenetic approach) was performed. COI reference sequences and metadata associated with the analyzed Calanoida genera were downloaded from BOLD database. Redundancy of molecular data were deleted collapsing the identical sequences using mothur v.1.33.0 (Schloss et al., 2009). Sequences shorter than 500 bp or with low quality (presence of ambiguities) were removed. Moreover, sequences without a complete identification at species level were excluded. A multialignment, including COI reference sequences from BOLD, COI sequences generated in this study and Paracyclopina nana Smirnov, 1935 (NC012455) as outgroup (same as in Blanco-Bercial et al., 2011), was generated using MAFFT (Kuraku et al., 2013; Katoh et al., 2019). The alignment was manually checked using SeaView v.4.0 (Gouy et al., 2010) and a Maximum Likelihood tree (GTR model) was built using Fastree (Price et al., 2010). The phylogenetic tree was visualized in iTOL4 (Letunic and Bork, 2019). The tree was also used as input for species delimitation inference using bPTP (Zhang et al., 2013). This approach uses Poisson Tree Processes (PTP) model to infer putative species boundaries on a given phylogenetic input tree and adds Bayesian Support (BS) values to delimit species on the input tree. A high BS value on a node indicates that all descendants from this node are more likely to be from one species. Finally, phylogenetic networks were generated for the four dominant calanoid species at LTER-MC (A. clausi, C. typicus, P. parvus complex, and T. stylifera), and for the cosmopolitan species C. helgolandicus. Haplotype lists were generated with DnaSp (Rozas et al., 2017), then Median-Joining (MJ) haplotype networks were inferred with Network 105 using default parameters. Copepod sequences from this study are available in GenBank (Table 1).
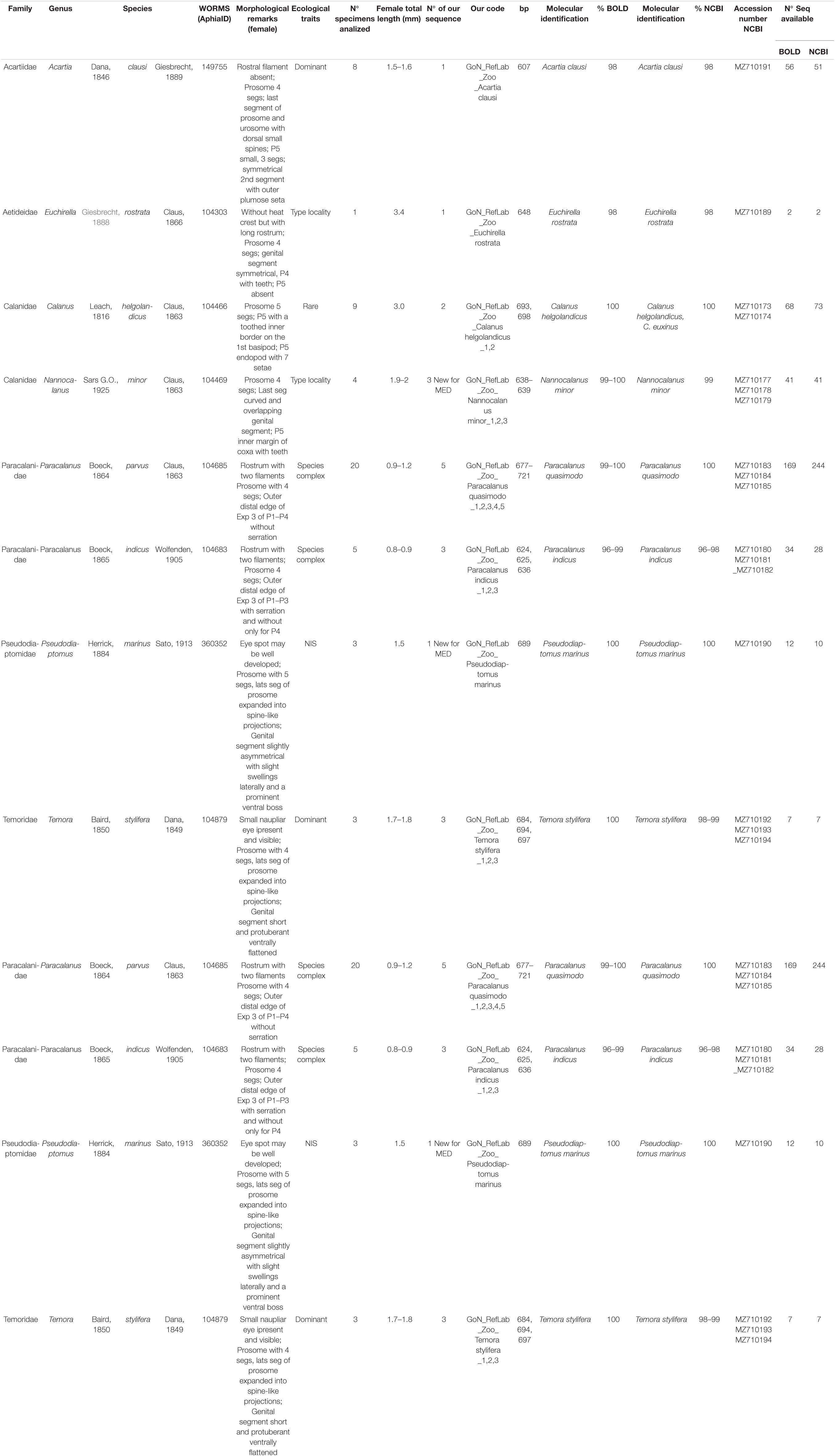
Table 1. Summary of ecological traits, morphological remarks, morphometric analyses and molecular data of the species studied.
Results
Eighty-one specimens referring to 16 species of 14 calanoid genera and 11 families of calanoid copepods were first identified through morphological analysis. For species with unequivocal morphological classification using species-specific morphological keys, the identification was performed through light microscopy and HD images are shown in Figure 2. For the genus Paracalanus a detailed SEM investigation was instead required to resolve finer details undistinguishable through light microscopy. The discrimination between Paracalanus parvus and Paracalanus indicus was based on the absence in the former of serration on the distal edge of the expodite 3 from the second to the fourth leg (Figure 3). Subsequent DNA extraction was successful in 56 specimens referred to 12 species (75% of the total species), but COI sequences were obtained for 27 specimens (47% of extracted specimens) (Table 1). Only high-quality sequences (with barcodes longer than 500 bp) and a high percentage of similarity (98–100%) with reference DNA databases (BOLD and GenBank) were considered robust enough for identification at species level (25 sequences out of 27; 92%). Analysis by similarity confirmed morphological identification for 11 species (91%): Acartia clausi, Calanus helgolandicus, Centropages typicus, Clausocalanus parapergens, Ctenocalanus vanus, Euchirella rostrata, Nannocalanus minor, P. indicus, Pleuromamma gracilis, Pseudodiaptomus marinus, and Temora stylifera (Table 1). COI sequences generated from five specimens morphologically identified as P. parvus were assigned to Paracalanus quasimodo according to molecular identification (Table 1).

Figure 2. HD-images of 10 calanoid copepod species studied in the Gulf of Naples: (A) Temora stylifera; (B) Acartia clausi; (C) Pseudodiaptomus marinus; (D) Centropages typicus; (E) Pleuromamma gracilis; (F) Calanus helgolandicus; (G) Nannocalanus minor; (H) Euchirella rostrata; (I) Clausocalanus parapergens; (J) Ctenocalanus vanus. The total body length of the specimens showed in this Figure are reported in the Table 1.
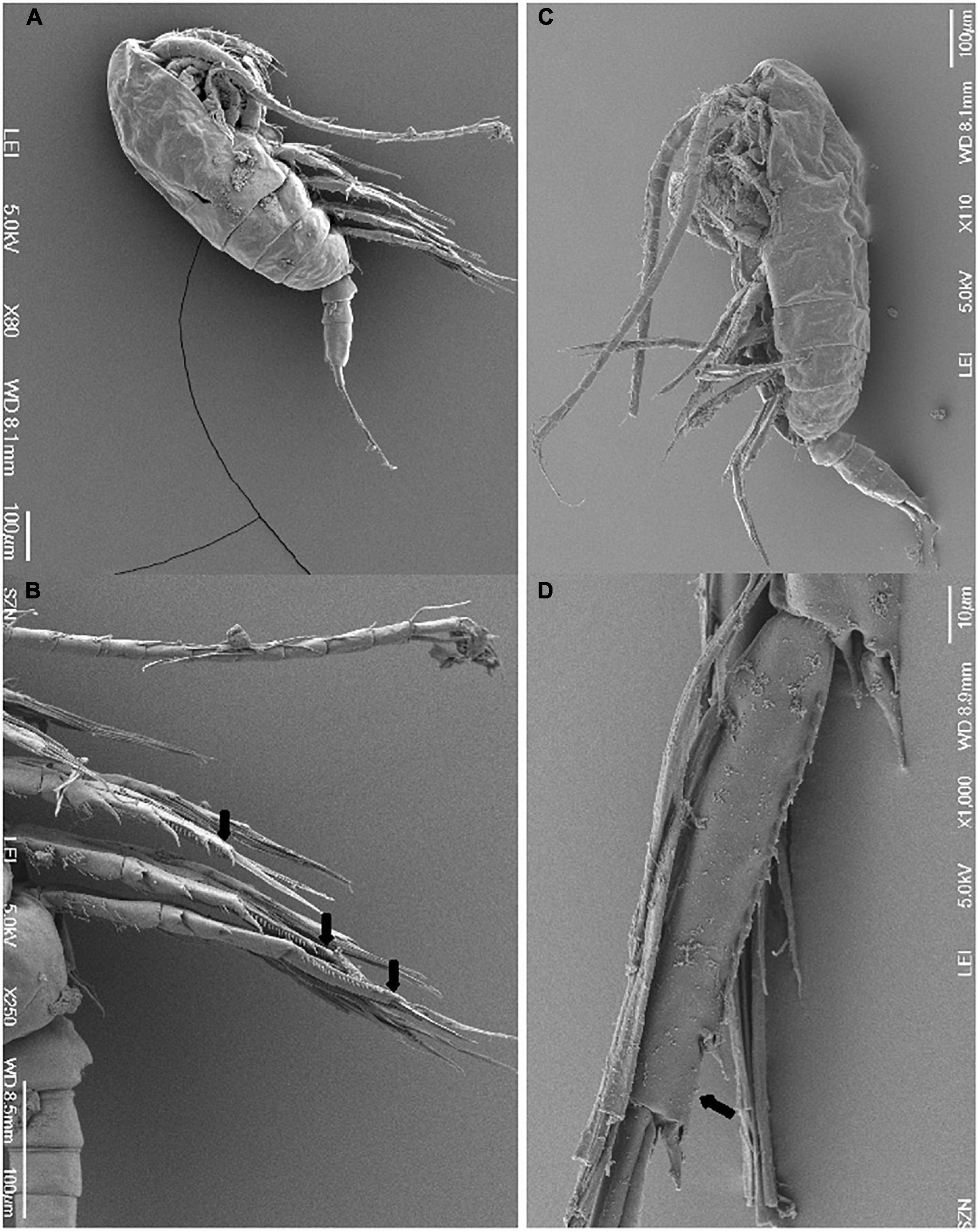
Figure 3. Scanning electron microscopy (SEM) images of (A) entire Paracalanus indicus in later view; (B) details of distal edge of the expodite 3 from the second to the fourth leg (arrowed) of P. indicus; (C) entire Paracalanus parvus in later view; (D) detail of distal edge of the expodite 3 of the fourth leg (arrow) of P. parvus.
A total of 4,400 COI reference sequences belonging to the 11 analyzed calanoid genera were downloaded from BOLD to reconstruct the phylogenetic relationships of the investigated species. Collapse of identical sequences reduced the dataset to 2,490 unique sequences. Filtered sequences generated a final dataset including 717 COI entries. For molecular identification of copepod species by tree placement, a Maximum Likelihood tree was generated using the GTR model (Figure 4), including 743 sequences (717 reference sequences from BOLD, 25 sequences generated in this study, and 1 outgroup). The single marker COI succeeded in identifying the query sequence at the genus and species levels and provided support to discriminate 11 genera with high support (87–98%) (Temora, Acartia, Pseudodiaptomus, Centropages, Pleuromamma, Calanus, Nannocalanus, Euchirella, Clausocalanus, Ctenocalanus, and Paracalanus) (Figure 4 and Supplementary Figure 1).
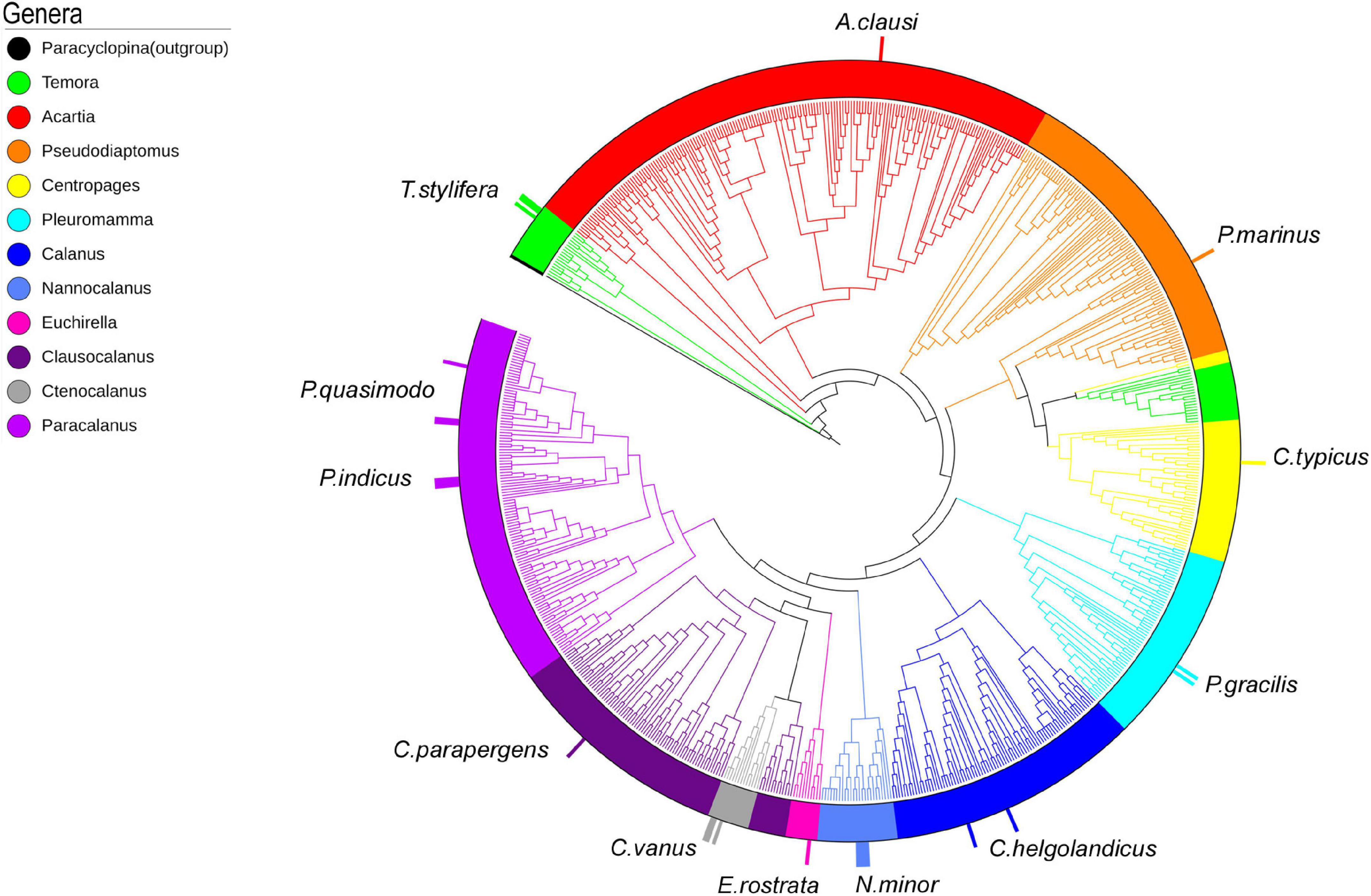
Figure 4. ML tree of 717 COI reference sequences from BOLD and 25 barcodes generated (taller bars). Outgroup: Paracyclopina nana NC012455.
The Temora cluster was separated in two clades, one with a basal position in the tree, and the second including our T. stylifera grouped in the tree at species level (bootstrap value of 98%), confirming their appropriate morphological identifications (Figure 5A). Within the genus Acartia, A. clausi formed two highly supported separate subclades: one comprising specimens coming from the North Sea, and another including specimens from the Mediterranean and the Black Sea (bootstrap value of 100%) (Figure 5B). The first Mediterranean COI sequence for Pseudodiaptomus marinus clustered with other specimens coming from the North Sea, the Pacific Ocean, and the Bilbao Estuary (bootstrap value of 84%) (Figure 5C). Our sequence of Centropages typicus (bootstrap value of 85%) (Figure 5D) was grouped in the tree of the correspondent clade species, confirming the morphological identifications. A similar pattern was found for Pleuromamma gracilis, correctly placed in the tree (bootstrap value of 87%), with specimens from the Atlantic Ocean. However, we observed that some references of Pleuromamma piseki were present in the same clade (Figure 5E). In the Calanus clade (Supplementary Figure 1), Calanus helgolandicus and Calanus euxinus references clustered together with C. helgolandicus from the Gulf of Naples (bootstrap value of 100%) (Figure 5F). The sequences generated for Nannocalanus minor were placed with other references mainly from the Atlantic Ocean; our four references from the Gulf of Naples, new for the Mediterranean Sea, formed a strongly supported subclade (bootstrap value of 100%) with one reference from the Sargasso Sea (Figure 6A). The Euchirella clade comprised the only seven congeneric species available from BOLD, our sequences of E. rostrata and the only two available references for this species grouped in the same subclade (bootstrap value of 100%) (Figure 6B). The Ctenocalanus clade comprised the only two congeneric species C. citer and C. vanus, separately. Our three specimens of C. vanus clustered with specimens coming from Spain, Greece, and the Atlantic Ocean (bootstrap value of 100%), confirming the correct morphological and molecular identification of this species from its type locality (Figure 6C). Clausocalanus parapergens (bootstrap value of 92%) was grouped in the tree of correspondent clade species, confirming the morphological identification (Figure 6D). More complexity was found for the Paracalanus species complex, comprising the nominal species P. indicus, P. quasimodo and P. parvus. P. quasimodo formed a strongly supported clade (bootstrap value of 100%) including specimens from several localities and our samples morphologically identified as P. parvus. Instead, P. indicus and P. parvus BOLD references were mixed in several clades including our three sequences generated from specimens morphologically identified as P. indicus (Figure 6E).
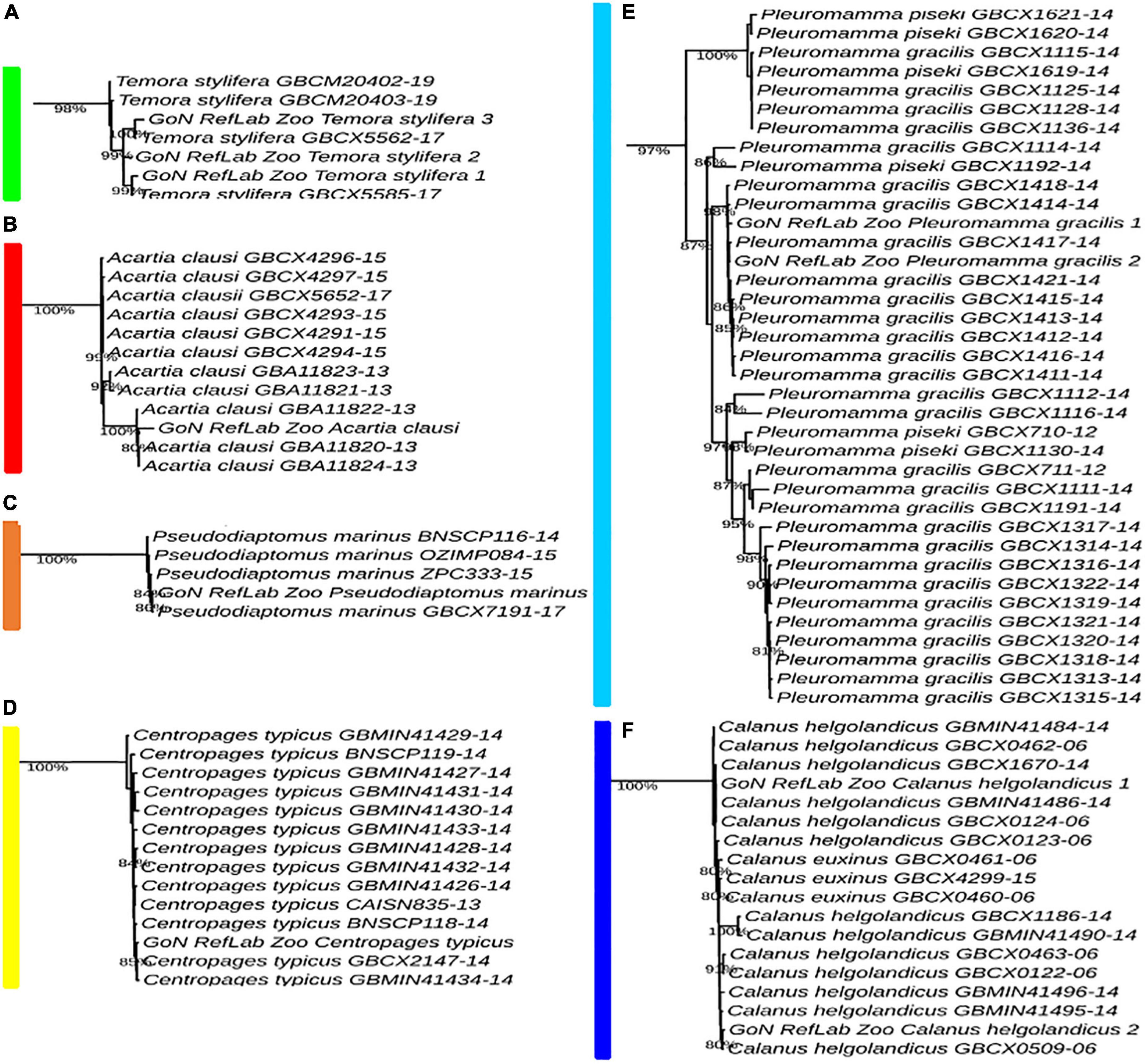
Figure 5. Molecular identification based on ML tree. Portions from the full tree are illustrated: (A) Temora stylifera; (B) Acartia clausi; (C) Pseudodiaptomus marinus; (D) Centropages typicus; (E) Pleuromamma gracilis; (F) Calanus helgolandicus. Full tree available in Supplementary Figure 1.
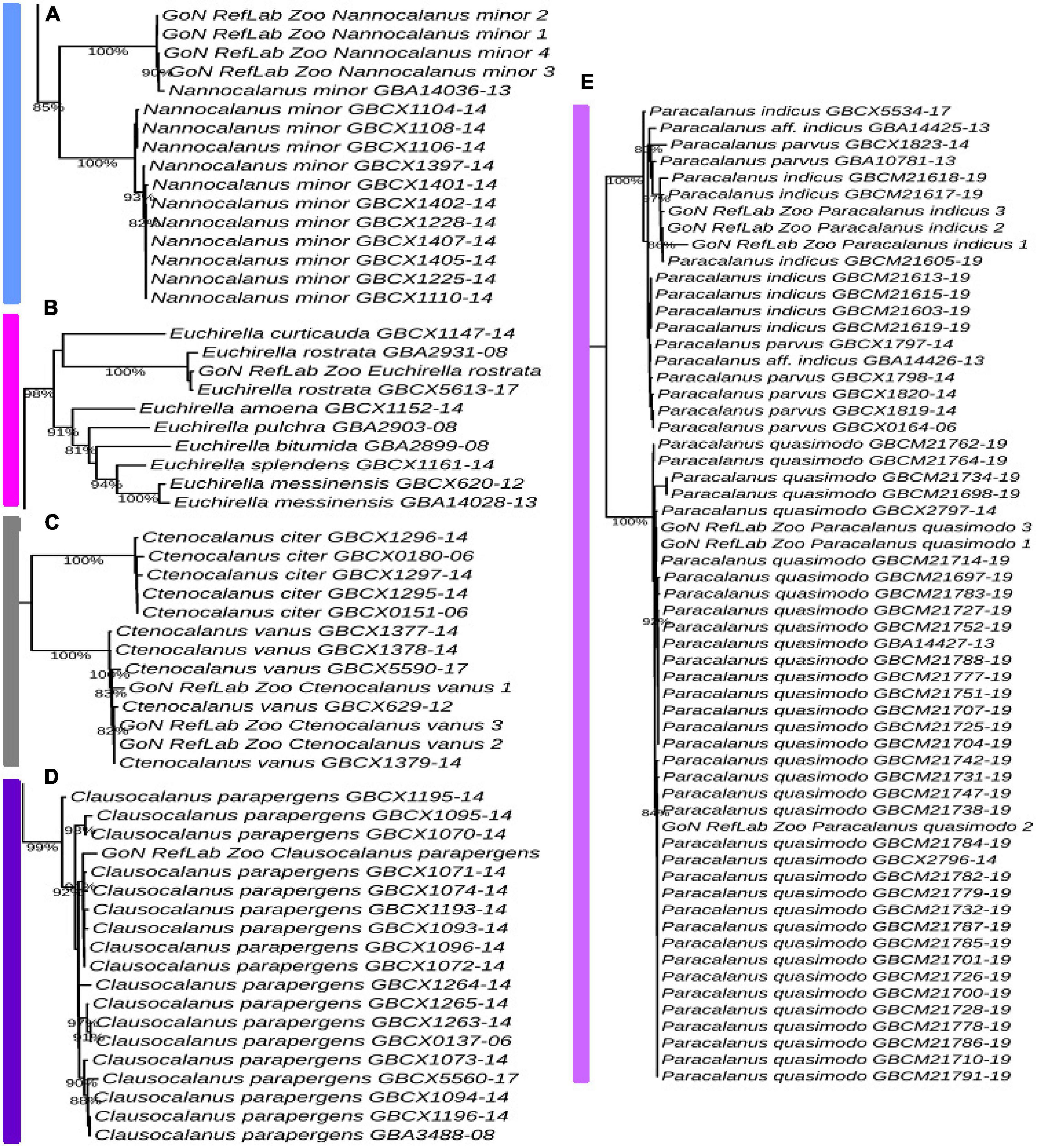
Figure 6. Molecular identification based on ML tree. Portions from the full tree are illustrated: (A) Nannocalanus minor; (B) Euchirella rostrata; (C) Ctenocalanus vanus; (D) Clausocalanus parapergens; (E) Paracalanus parvus. Full tree available in Supplementary Figure 1.
The species delimitation inferred by bPTP predicted 154 species starting from molecular data belonging to 126 nominal species (Supplementary Figure 2 and Supplementary Table 1). Exploration of 12 species morphologically identified in our study revealed the prediction of 26 putative species. Inference from molecular data of eight species (Calanus helgolandicus, Centropages typicus, Ctenocalanus vanus, Euchirella rostrata, Pseudodiaptomus marinus, Temora stylifera, Clausocalanus parapergens, and Paracalanus quasimodo) predicted each of them as unique biological entity, confirming morphological description. Other four species (Acartia clausi, Nannocalanus minor, Paracalanus indicus, and Pleuromamma gracilis) showed instead signals of crypticity since bPTP fragmented the nominal species into different subgroups. Sequences of Paracalanus indicus inferred four potential species (species number 60, 88, 136,137, Supplementary Table 1). Sequences of Acartia clausi were also split and bPTP predicted the presence of five different species (species numbers 26, 91, 92, 93, 94, Supplementary Table 1) with our sequences placed together with other four sequences from a Mediterranean site (France, Sete Channel). Nannocalanus minor sequences were predicted as three species and one of them (species 58) only included our Mediterranean specimens (species numbers 57, 58, 59, Supplementary Table 1). Finally, molecular data from Pleuromamma gracilis have been predicted to include seven species, one of them including P. piseki (species numbers 78, 96, 97, 98, 103, 118, 119, Supplementary Table 1) and the others sequenced from Atlantic and Pacific Oceans.
Phylogenetic networks showed different patterns in the four dominant neritic calanoid copepods at LTER-MC (A. clausi, C. typicus, P. parvus, and T. stylifera), as well as in the offshore species C. helgolandicus (Figures 7A–D). Both in A. clausi (Figure 7A) and T. stylifera (Figure 7B), haplotypes showed a clear separation based on the geographical origin of the specimens, and reticulate structures were absent. In A. clausi, seven haplotypes from the North Sea (H_2-H_6, H_10, and H_13) were separated from four haplotypes from the Mediterranean Sea including our sequences (H_1, H_11, H_12, and H_14) and three from the Black Sea (H_7-H_9). A similar pattern was found in T. stylifera with a clear separation between the three haplotypes from the Atlantic Ocean (H_4, H_5, and H_8) and the five ones from the Mediterranean Sea (H_1-H_3, H_6, and H7). The C. typicus network (Figure 7C) generated a dominant haplotype including different areas connected with several minor haplotypes without any clear geographical signal. The C. helgolandicus phylogeographic network (Figure 7D) showed the presence of a cosmopolitan haplotype (H_1), including individuals from all the geographic areas except the Adriatic Sea: ten from the North Sea, three from the Atlantic Ocean and two from the Mediterranean Sea (one of which isolated from the Gulf of Naples). This haplotype was surrounded by several other unique haplotypes, mainly from the North Sea. Specimens of C. helgolandicus from the Adriatic Sea (H_3, H_8-H_11, H_22, H_25, and H_26) and from the Black Sea (H_37) were not included in the H_1 haplotype, but were found as single haplotypes weakly connected with the other geographical locations. The second most abundant haplotype of the network (H_6) was shared between C. helgolandicus and C. euxinus species, and included two specimens of C. helgolandicus (one from the North Sea and one from the Adriatic Sea) and three specimens of C. euxinus (two from the Black Sea and one from the Adriatic Sea). All other specimens of C. euxinus (H_7, H_13, H_14, H_24, H_27, H_28, and H_38) and C. helgolandicus (H_37) from the Black Sea did not show links with other geographical areas, apart from one haplotype from the Adriatic Sea (H_12). The network for P. parvus revealed an intricate structure owing to the occurrence of a species complex, and consequently to the absence of a commonly agreed morphologically based standard reference (Supplementary Figure 3).
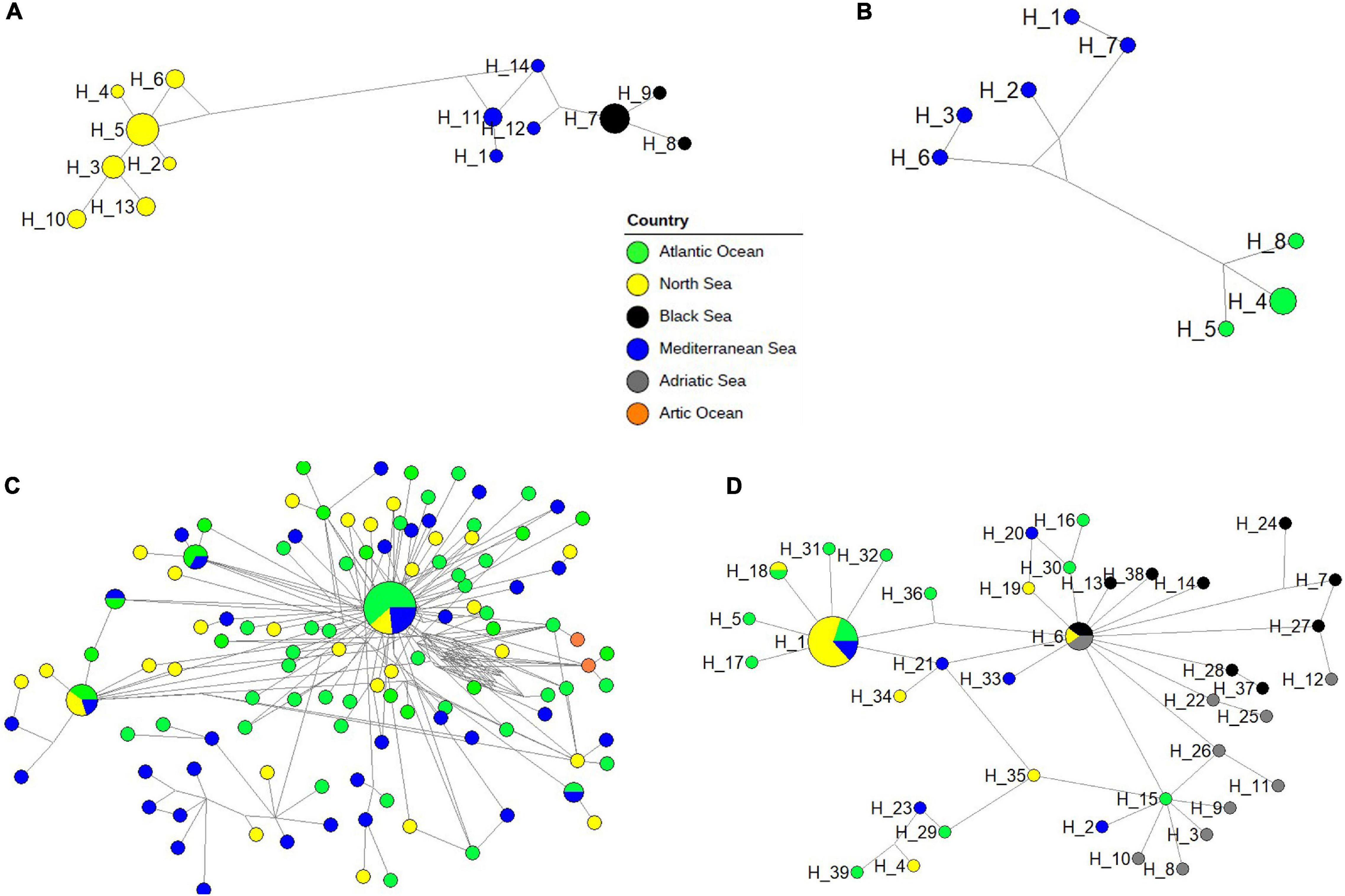
Figure 7. Phylogenetic networks of four species including all intraspecies specimens: (A) Acartia clausi; (B) Temora stylifera; (C) Centropages typicus; (D) Calanus helgolandicus/C. euxinus. Each circle represents a single haplotype and its diameter is proportional to the number of individuals carrying the haplotypes, the smallest circles representing a single individual. The length of connecting lines is proportional to the number of base substitutions between two haplotypes.
Discussion
Our results confirm the mutual dependency of morphological and molecular taxonomy, and support the usefulness of integrative approaches to build a more comprehensive framework not only for species identification (α taxonomy), but also to provide the basic tile to reconstruct the complex mosaic of the ecological, genetic and phylogeographic relationships between different populations (Ω taxonomy).
Studies on the taxonomy of copepods in the Gulf of Naples boast an ancient tradition (Giesbrecht, 1892), and over the years the investigation of planktonic metazoans at LTER-MC has retained a phenotypic approach (Di Capua and Boxshall, 2008; Mazzocchi et al., 2011; Zingone et al., 2019). However regular observations of zooplankton samples and the development of specialized taxonomic expertise have led to a clear demonstration of the high diversity of this assemblage, which also needs investigation at the molecular level (Di Capua et al., 2017, 2021; Kasapidis et al., 2018). Overall, precise identification of calanoid copepods is fully supported by our results using both morphological and molecular approaches, revealing their real taxonomic biodiversity. Such integrated approach is fundamental to deposit only high-quality sequences correctly annotated with proper species identification names in WORMS and BOLD. In addition, the generation of validated sequences is crucial for laying the foundation and keeping updated a functional and reliable barcode reference library for the Mediterranean coastal areas, starting from the Gulf of Naples due to its high representativeness.
In addition to a very difficult taxonomic identification, also DNA extraction from calanoid species is challenging (Bucklin and Wiebe, 1998; Bucklin et al., 1999; Lindeque et al., 1999; Cornilis, 2015), due to the small size of the analyzed specimens and to the presence of a chitinous exoskeleton. In the present investigation DNA was successfully extracted and amplified in individual specimens of twelve out of the sixteen species targeted (75%), in line with previous works (Cornils and Held, 2014; Cornilis, 2015; Baek et al., 2016). Our dataset includes females of studied calanoid species due to their high abundance and sex ratio in our samples. However, recent studies (Francis and Nishida, 2018; Francis and Bijoy Nandan, 2019) suggest that morphological and genetic data should be generated and established both for female and male specimens. The mitochondrial gene COI, considered the gold standard for metazoan identification (Hebert et al., 2003a,b), proved itself to be a good marker for species delimitation (Bucklin et al., 2010c,2011, 2021; Blanco-Bercial et al., 2014; Cornils and Held, 2014). To reduce the possible occurrence of false positives, a multimarkers approach has been suggested as a viable solution to permit a more robust identification of organisms at different levels (Zhang et al., 2018; Berry et al., 2019). Most molecular phylogenetic studies in copepods, however, are carried out using single genes, typically COI or 18S (Bernot et al., 2021). Among the two, COI ensures detection of species-level diversity while 18S is more suitable for genus or family level investigations (Stefanni et al., 2018). Based on these premises, the present investigation focused on COI only, without including additional markers, and our sequences confirm that COI barcoding can accurately identify calanoid copepods of widespread different genera, also at species level. We also highlight and detect crypticity, a phenomenon very common in copepod assemblages (Dippenaar et al., 2010).
The phylogenetic tree displays a topology well in line with established cladistics studies based on calanoid morphology (Bradford-Grieve et al., 2010) and in agreement with the evolution of calanoid suborder (with families included) revised by Andronov (1991). The phylogenetic tree generated by the integration of BOLD data represents an overview of the evolutionary relationships among calanoid copepods, and shows a general monophyly and better resolution at the genus level.
DNA taxonomy using different approaches reveals the existence of the complex for Paracalanus spp., Pleuromamma gracilis, Nannocalanus minor, and Acartia clausi. In particular, the molecular identification of the Paracalanus complex confirms the identification of Paracalanus indicus, with our sequences for this species grouped in a big clade, including most of the sequences available from different countries (Australia, Algeria, Spain, Mexico, Tunisia, and Greece), and also supports the occurence of the cryptic species Paracalanus quasimodo in the Gulf of Naples, as reported also by Kasapidis et al. (2018). However, five specimens identified as P. parvus based on detailed morphological characters evident at the SEM were genetically identified as P. quasimodo using BOLD references, confirming the problematic assessment of Paracalanus species. Bowman (1971) already reported on the difficulty to distinguish P. parvus from P. quasimodo based on morphology using light microscopy only. The first description of P. quasimodo was proposed for Paracalanus specimens collected in Florida on few individuals very similar to P. parvus (Bowman, 1971). The name quasimodo derived from the main character of Victor Hugo’s classic novel “The Hunchback of Notre Dame,” alluding to the distinctive hump of the prosome (visible in lateral view). This morphology, however, cannot be considered a diagnostic character, because it could also be due to internal parasites (Ianora et al., 1987). P. parvus, in fact, can be parasitized by Blastodinium and Atelodinium and other endoparasites (Ianora et al., 1987; Ohtsuka et al., 2004), which can induce some prosome modifications (Ianora et al., 1987; Di Capua pers. obs). Detailed molecular COI analyses of P. quasimodo specimens from the type locality (Florida) are needed to disentangle this issue and clarify the taxonomic value of this nominal species, disentangling a species complex comprising 12 putative species of Paracalanus collected around the world (Cornils and Held, 2014).
Other two cases of cryptic species are represented by the two species Pleuromamma gracilis and Nannocalanus minor, described for the first time in the Mediterranean Sea and considered cosmopolitan. Indeed, species delimitation of these two morphological species predicted ten putative species. Even if this species number could be an overestimation, it is a clear signal of a potential cripticity in these genera. Interestengly, putative species delimitated within the same morpho-molecular data correspond to different geographical areas of sampling, suggesting a mechanism of allopatric speciation, across varied pelagic habitats, as observed by Halbert et al. (2013) and Blanco-Bercial et al. (2014).
Our sequences of Pleuromamma gracilis and Nannocalanus minor, the first for the Mediterranean Sea, represent the molecular fingerprint of these species near their type locality (Messina, Southern Tyrrhenian Sea), where Claus made the first descriptions in 1863. Among the seven putative species predicted, our data of P. gracilis clustered with other references, but the same clade included also references of P. piseki, suggesting a morphological misidentification of the latter (Blanco-Bercial et al., 2014). Yet, the prediction of three species for Nannocalanus minor correspond to specimens collected in different areas of Atlantic Ocean. Indeed, as previously described, Nannocalanus minor showed, throughout the Gulf Stream and the Sargasso Sea, two genetically distinct types (distinguished by 10% sequence difference of mtDNA 16S) differing also in size range and in geographic distribution (Bucklin et al., 1996).
Our results, furthermore, may also shed light onto the questionable status of C. helgolandicus and Calanus euxinus. C. helgolandicus is a widespread epipelagic copepod extending from the Atlantic Ocean to the Mediterranean Sea (Fleminger and Hulsemann, 1977), while C. euxinus is a relict species of the Black Sea (Hulsemann, 1991). Fleminger and Hulsemann (1987) were the first to propose the existence of distinct C. helgolandicus populations in the Atlantic Ocean, the Mediterranean Sea and the Black Sea, while Hulsemann (1991) proposed C. euxinus as a distinct species. The two congeneric copepods are distinguished based on subtle morphological differences (Fleminger and Hulsemann, 1987), and show little genotypic difference (Papadopoulos et al., 2005; Unal et al., 2006). Molecular COI analyses supported with 100% similarity confirm the morphological identification of our specimens as C. helgolandicus (based on total body length, habitus, fifth leg and inner margin of basipodite 1 of fifth leg). On the other hand, a close relationship among the genotypes of both species and the topology of the resulting phylogenetic tree suggest a pseudo-cryptic status. In addition, network analyses of C. helgolandicus and C. euxinus show the presence of a shared haplotype (H_6) between specimens of both species, including two individuals of C. helgolandicus collected in the North Sea and the Adriatic Sea, respectively (Lauritano et al., 2013), and three specimens of C. euxinus, two from the Black Sea and one from the Adriatic Sea. The presence of this shared haplotype, reported also by Papadopoulos et al. (2005), suggests that the nominal species C. helgolandicus and C. euxinus might not be distinct, as confirmed by species delimitation analyses.
The phylogenetic networks confirmed the presence, within the species, of biogeographical patterns with different populations, or putative species, according to the different geographic origin of specimens. This confirms the importance of the generation of intraspecies reference barcodes covering their native distribution, not only to evaluate intraspecies diversity, but also to highlight the presence of new emerging lineages. The networks of T. stylifera and A. clausi showed a clear separation between northern and southern populations of both species without signals of gene flow between (reticulation) the two geographic clusters of haplotypes. Such pattern could be due to insufficient references; however, four potential species, corresponding to different geographic areas (North Sea, Black Sea, and Mediterranean Sea) or habitat (Thau lagoon), could be predicted for the complex A. clausi with high support (0.99–1%) through bPTP.
The integration of morphological and molecular taxonomic approaches carried out in the present work provides a further confirmation of the appropriateness of integrated methods for the identification of copepods, as demonstrated also for other organisms (Fontaneto et al., 2015; Avó et al., 2017; Mills et al., 2017; Guimarães-Costa et al., 2020). The success of using barcoding for single-species identification depends on the presence of high-quality reference data available in public sequence libraries (Bucklin et al., 2021). This emphasizes the dual-way benefit of the integrated approach, where a sound phenotypic-based identification can lead to species-specific barcodes, while high-quality sequences can be used for an exact naming of the species (as, for example, in metabarcoding studies). The creation of robust, quality-controlled DNA sequences is therefore fundamental to build reliable libraries to be used as a basis to understand not only the biodiversity of a given region, but also to identify the connections between populations and the evolutionary patterns of species (Djurhuus et al., 2018; Berry et al., 2019). The molecular identification, however, still needs the close verification of the morphological descriptive characters for a perfect pairing of the information (Wiens, 2007; Karanovic et al., 2016). This holds in particular for those species with hardly distinguishable keys, in which case a “turbo taxonomy” approach (i.e., based on few taxonomic details) (Butcher et al., 2012) is not suitable, whilst the support of experienced morphological taxonomists is irreplaceable. The generation of sequences should therefore be promoted not only for the rare, cryptic, and difficult to discriminate species, but also for those easy to identify, common and dominant.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MZ710171-MZ710194.
Author Contributions
IDC: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, resources, supervision, roles/writing—original draft, and writing—review and editing. RD’A: data curation, formal analysis, investigation, methodology, software, roles/writing—original draft, and writing—review and editing. RP: formal analysis, methodology, software, supervision, and writing—review and editing. CM: methodology, supervision, and writing–review and editing. FB: validation and writing—review and editing. MU and YC: funding acquisition, investigation, resources, supervision, roles/writing–original draft, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Stazione Zoologica Anton Dohrn (SZN) to the research program Long-Term Ecological Research MareChiara (LTER-MC) (LTER, EU-IT061). RD’A was the recipient of a research fellowship financed by ADSP project, Integrative Marine Ecology Dept. and SZN (incentive funds granted to YC and MU).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all LTER-MC and NEREA teams, and the crew of the R/V Vettoria for assistance during the work at sea. We are grateful to Elvira Mauriello and Raimondo Pannone of the Molecular Biology and Sequencing Service for technical assistance and Luca Ambrosino of Bioinformatics of the Marine biology Bioinformatics Services, Andrea Montalbano of Marine Organism Taxonomy Core Facility MOTax. This study benefitted from collaboration with members of the Scientific Committee on Oceanic Research (SCOR) Working Group 157 (MetaZooGene). We thank the WGEUROBUS and WGIMT of the International Council for the Exploration of the Sea (ICES) for facilitating this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.833089/full#supplementary-material
Footnotes
- ^ https://www.st.nmfs.noaa.gov/nauplius/media/metazoogene/atlas/index.html
- ^ https://v3.boldsystems.org/
- ^ http://www.ncbi.nlm.nih.gov
- ^ itol.embl.de
- ^ http://www.fluxus-engineering.com/
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Amante, C., and Eakins, B. W. (2009). ETOPO1 1 Arc-Minute Global Relief Model: Procedures, Data Sources and Analysis. NOAA Technical Memorandum NESDIS NGDC-24. Washington, DC: NOAA. doi: 10.7289/V5C8276M
Avó, A. P., Daniell, T. J., Neilson, R., Oliveira, S., Branco, J., and Adão, H. (2017). DNA barcoding and morphological identification of benthic nematodes assemblages of estuarine intertidal sediments: advances in molecular tools for biodiversity assessment. Front. Mar. Sci 4:66. doi: 10.3389/fmars.2017.00066
Baek, S. Y., Jang, K. H., Choi, E. H., Ryu, S. H., Kim, S. K., Lee, J. H., et al. (2016). DNA Barcoding of metazoan zooplankton copepods from South Korea. PLoS One 11:e0157307. doi: 10.1371/journal.pone.0157307
Bernot, J. P., Boxshall, G. A., and Crandall, K. A. (2021). A synthesis tree of the Copepoda: integrating phylogenetic and taxonomic data reveals multiple origins of parasitism. PeerJ. 9:e12034. doi: 10.7717/peerj.12034
Berry, T. E., Saunders, B. J., Coghlan, M. L., Stat, M., Jarman, S., Richardson, A. J., et al. (2019). Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and Identifies ecosystem responses to anomalous climatic events. PLoS Genet. 15:e1007943. doi: 10.1371/journal.pgen.1007943
Blanco-Bercial, L., Bradford-Grieve, J., and Bucklin, A. (2011). Molecular phylogeny of the Calanoida (Crustacea: Copepoda). Mol. Phylogenet. Evol. 59, 103–113. doi: 10.1016/j.ympev.2011.01.008
Blanco-Bercial, L., Cornils, A., Copley, N., and Bucklin, A. (2014). DNA barcoding of marine copepods: assessment of analytical approaches to species identification. PLoS Curr. 6:ecurrents.tol.cdf8b74881f87e3b01d56b43791626d2. doi: 10.1371/currents.tol.cdf8b74881f87e3b01d56b43791626d2
Boero, F., and Bernardi, G. (2014). Phenotypic vs. genotypic approaches to biodiversity, from conflict to alliance. Mar. Genom. 17, 63–64. doi: 10.1016/j.margen.2014.03.005
Bonnet, D., Richardson, A., Harris, R., Hirst, A., Beaugrand, G., Edwards, M., et al. (2005). An overview of Calanus helgolandicus ecology in European waters. Prog. Oceanogr. 65, 1–53. doi: 10.1016/j.pocean.2005.02.002
Bowman, T. E. (1971). The distribution of calanoid copepods off the Southeastern United States between Cape Hatteras and southern Florida. Smithson. Contrib. Zool. 96, 56–58. doi: 10.5479/si.00810282.96
Bradford-Grieve, J. M., Boxshall, G. A., Ahyong, S. T., and Ohtsuka, S. (2010). Cladistic analysis of the calanoid Copepoda. Invertebr. Syst. 24, 291–321. doi: 10.1071/is10007
Bucklin, A., Guarnieri, M., Hill, R. S., Bentley, A. M., and Kaartvedt, S. (1999). Taxonomic and systematic assessment of planktonic copepods using mitochondrial COI sequence variation and competitive, species-specific PCR. Hydrobiologia 401, 239–254. doi: 10.1023/A:1003790411424
Bucklin, A., Nishida, S., Schnack-Schiel, S., Wiebe, P. H., Lindsay, D., Machida, R. J., et al. (2010b). “A census of zooplankton of the global ocean,” in Marine Life: Diversity, Distribution, and Abundance, ed. A. McIntyre (Oxford: Wiley-Blackwell), 247–265.
Bucklin, A., Hopcroft, R. R., Kosobokova, K. N., Nigro, L. M., Ortman, B. D., Jennings, R. M., et al. (2010a). DNA barcoding of Arctic Ocean holozooplankton for species identifcation and recognition. Deep Sea Res. II 57, 40–48. doi: 10.1016/j.dsr2.2009.08.005
Bucklin, A., LaJeunesse, T. C., Curry, E., Wallinga, J., and Garrison, K. (1996). Molecular diversity of the copepod, Nannocalanus minor: genetic evidence of species and population structure in the North Atlantic Ocean. J. Mar. Res. 54, 285–310. doi: 10.1357/0022240963213385
Bucklin, A., Ortman, B. D., Jennings, R. M., Nigro, L. M., Sweetman, C. J., Copley, N. J., et al. (2010c). A “Rosetta Stone” for metazoan zooplankton: DNA barcode analysis of species diversity of the Sargasso Sea (Northwest Atlantic Ocean). Deep Sea Res. II 57, 2234–2247. doi: 10.1016/j.dsr2.2010.09.025
Bucklin, A., Peijnenburg, K. T. C. A., Kosobokova, K. N., O’Brien, T. D., Blanco−Bercial, L., Cornils, A., et al. (2021). Toward a global reference database of COI barcodes for marine zooplankton. Mar. Biol. 168:78. doi: 10.1007/s00227-021-03887-y
Bucklin, A., Steinke, D., and Blanco-Bercial, L. (2011). DNA barcoding of marine metazoa. Ann. Rev. Mar. Sci. 3, 471–508. doi: 10.1146/annurev-marine-120308-080950
Bucklin, A., and Wiebe, P. H. (1998). Low mitochondrial diversity and small effective population sizes of the copepods Calanus finmarchicus and Nannocalanus minor: possible impact of climatic variation during recent glaciation. J. Hered. 89, 383–392. doi: 10.1093/jhered/89.5.383
Butcher, B. A., Smith, M. A., Sharkey, M. J., and Quicke, D. L. J. (2012). A turbo-taxonomic study of Thai Aleiodes (Aleiodes) and Aleiodes (Arcaleiodes) (Hymenoptera: Braconidae: Rogadinae) based largely on COI barcoded specimens, with rapid descriptions of 179 new species. Zootaxa 3457, 1–232. doi: 10.11646/zootaxa.3457.1.1
Chiba, S., Batten, S., Martin, C. S., Ivory, S., Miloslavich, P., and Weatherdon, L. V. (2018). Zooplankton monitoring to contribute towards addressing global biodiversity conservation challenges. J. Plankton Res. 40, 509–518. doi: 10.1093/plankt/fby030
Cianelli, D., D’Alelio, D., Uttieri, M., Sarno, D., Zingone, A., Zambianchi, E., et al. (2017). Disentangling physical and biological drivers of phytoplankton dynamics in a coastal system. Sci. Rep. 7:15868. doi: 10.1038/s41598-017-15880-x
Cornilis, A. (2015). Non-destructive DNA extraction for small pelagic copepods to perform integrative taxonomy. J. Plankton Res. 35, 6–10. doi: 10.1093/plankt/fbu105
Cornils, A., and Held, C. (2014). Evidence of cryptic and pseudocryptic speciation in the Paracalanus parvus species complex (Crustacea, Copepoda, Calanoida). Front. Zool. 11:19. doi: 10.1186/1742-9994-11-19
Di Capua, I., and Boxshall, G. A. (2008). Records of Oncaeidae (Copepoda) from the Gulf of Naples, with new records of three species of Triconia. J. Mar. Biol. Assoc. U.K. 88, 1407–1415. doi: 10.1017/S002531540800194X
Di Capua, I., Maffucci, F., Pannone, R., Mazzocchi, M. G., Biffali, E., and Amato, A. (2017). Molecular phylogeny of Oncaeidae (Copepoda) using nuclear ribosomal internal transcribed spacer (ITS rDNA). PLoS One 12:e0175662. doi: 10.1371/journal.pone.0175662
Di Capua, I., and Mazzocchi, M. G. (2004). Population structure of the copepods Centropages typicus and Temora stylifera in different environmental conditions. ICES J. Mar. Sci. 61, 632–644.
Di Capua, I., and Mazzocchi, M. G. (2021). “Crustacea Copepoda Calanoida, Cyclopoida, Harpacticoida, Mormonilloida, Siphonostomatoida (marine planktonic),” in Checklist of the Italian Fauna. Version 1.0. Last update: 2021-05-31, eds M. A. Bologna, M. Zapparoli, M. Oliverio, A. Minelli, L. Bonato, and F. Cianferoni, (London, UK: Comitato Scientifico per la Fauna d’Italia).
Di Capua, I., Piredda, R., Mazzocchi, M. G., and Zingone, A. (2021). Metazoan diversity and seasonality through eDNA metabarcoding at a Mediterranean long-term ecological research site. ICES J. Mar. Sci. 78, 3303–3316. doi: 10.1093/icesjms/fsab059
Dippenaar, S. M., Mathibela, R. B., and Bloomer, P. (2010). Cytochrome oxidase I sequences reveal possible cryptic diversity in the cosmopolitan symbiotic copepod Nesippus orientalis Heller, 1868 (Pandaridae: Siphonostomatoida) on elasmobranch hosts from the KwaZulu-Natal coast of South Africa. Exp. Parassitol. 125, 42–50. doi: 10.1016/j.exppara.2009.08.017
Djurhuus, A., Pitz, K., Sawaya, N. A., Rojas-Márquez, J., Michaud, B., Montes, E., et al. (2018). Evaluation of marine zooplankton community structure through environmental DNA metabarcoding. Limnol. Oceanogr. Methods 16, 209–221. doi: 10.1002/lom3.10237
Fleminger, A., and Hulsemann, K. (1977). Geographical range and taxonomic divergence in North Atlantic Calanus (C. helgolandicus. C. finmarchicus and C. glacialis). Mar. Biol. 40, 233–248. doi: 10.1007/BF00390879
Fleminger, A., and Hulsemann, K. (1987). Calanus helgolandicus s.l. (Copepoda. Biol. Oceanogr. 5, 43–81.
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Fontaneto, D., Flot, J. F., and Tang, C. Q. (2015). Guidelines for DNA taxonomy, with a focus on the meiofauna. Mar. Biodivers. 45, 433–451. doi: 10.1007/s12526-015-0319-7
Francis, S. V., and Bijoy Nandan, S. (2019). A new species of Tortanus (Atortus) (Copepoda, Calanoida, Tortanidae) from Minicoy Island, southeastern Arabian Sea. Turk. J. Fish Aquat. Sci 43, 425–436. doi: 10.3906/zoo-1811-29
Francis, S. V., and Nishida, S. (2018). Amendment of the status of Pontella sewelli Heinrich, 1987 as a junior synonym of P. sinica Chen & Zhang, 1965, with confirmation of female-male matching by genetic analysis (Copepoda, Calanoida). Crustaceana 91, 439–449.
Giesbrecht, W. (1888). Elenco dei copepodi pelagici raccolti dal Tenente di vascello Gaetano Chierchia durante il viaggio della R. Corvetta ‘Vettor Pisani’ negli anni 1882-1885 e dal Tenente di vascello Francesco Orsini nel Mar Rosso, nel 1884. Atti Acad. Naz. Lincei. 2, 330–338.
Giesbrecht, W. (1892). Systematik und Faunistik der Pelagischen Copepoden des Golfes von Neapel und der Angrenzenden Meeres-Abschnitte. Berlin: Verlag von R. Friedländer & Sohn.
Giesbrecht, W., and Schmeil, O. (1898). Copepoda I. Gymnoplea. Tierreich 6, 1–169. doi: 10.1038/s41598-017-06656-4
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Guimarães-Costa, A., Machado, F. S., Reis-Filho, J. A., Andrade, M., Araújo, R. G., Corrêa, E. M. R., et al. (2020). DNA barcoding for the assessment of the taxonomy and conservation status of the fish bycatch of the northern Brazilian shrimp trawl fishery. Front. Mar. Sci. 7:566021. doi: 10.3389/fmars.2020.566021
Halbert, K. M. K., Goetze, E., and Carlon, D. B. (2013). High cryptic diversity across the global range of the migratory planktonic copepods Pleuromamma piseki and P. gracilis. PLoS One 8, 1–18. doi: 10.1371/journal.pone.0077011
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003a). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert, P. D. N., Ratnasingham, S., and deWaard, J. R. (2003b). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B 270, S96–S99. doi: 10.1098/rsbl.2003.0025
Hulsemann, K. (1991). Calanus euxinus, new name, a replacement name for Calanus ponticus Karavaev, 1894 (Copepoda : Calanoida). Proc. Biol. Soc. Wash. 104, 620–621.
Ianora, A., Mazzocchi, M. G., and Scotto di Carlo, B. (1987). Impact of parasitism and intersexuality on Mediterranean populations of Paracalanus parvus (Copepoda: Calanoida). Dis. Aquat. Organ. 3, 29–36.
Karanovic, T., Djurakic, M., and Eberhard, S. M. (2016). Cryptic species or inadequate taxonomy? Implementation of 2D geometric morphometrics based on integumental organs as landmarks for delimitation and description of copepod taxa. Syst. Biol. 65, 304–327. doi: 10.1093/sysbio/syv088
Kasapidis, P., Siokou, I., Khelifi-Touhami, M., Mazzocchi, M. G., Matthaiaki, M., Chiristou, E., et al. (2018). Revising the taxonomic status and distribution of the Paracalanus parvus species complex (Copepoda, Calanoida) in the Mediterranean and Black Seas through an integrated analysis of morphology and molecular taxonomy. J. Plankton Res. 40, 595–605. doi: 10.1093/plankt/fby036
Katoh, K., Rozewicki, J., and Yamad, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kuraku, S., Zmasek, C. M., Nishimura, O., and Katoh, K. (2013). aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41, W22–W28. doi: 10.1093/nar/gkt389
Laakmann, S., Blanco-Bercial, L., and Cornils, A. (2020). The crossover from microscopy to genes in marine diversity: from species to assemblages in marine pelagic copepods. Phil. Trans. R. Soc, B 375:20190446. doi: 10.1098/rstb.2019.0446
Laakmann, S., Gerdts, G., Erler, R., Knebelsberger, T., Martínez Arbizu, P., and Raupach, M. J. (2013). Comparison of molecular species identifcation for North Sea calanoid copepods (Crustacea) using proteome fngerprints and DNA sequences. Mol. Ecol. Res. 13, 862–876. doi: 10.1111/1755-0998.12139
Lauritano, C., Carotenuto, Y., Procaccini, G., Turner, J. T., and Ianora, A. (2013). Changes in expression of stress genes in copepods feeding upon a non-brevetoxin-producing strain of the dinoflagellate Karenia brevis. Harmful Algae 28, 23–30. doi: 10.1016/j.hal.2013.05.004
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Lindeque, P. K., Harris, R. P., Jones, M. B., and Smerdon, G. R. (1999). Simple molecular method to distinguish the identity of Calanus species (Copepoda: Calanoida) at any developmental stage. Mar. Biol. 133, 91–96. doi: 10.1007/s002270050446
Mazzocchi, M. G.Di Capua, I. (2010). Copepodi planctonici/planktonic copepods: Calanoida, Cyclopoida, Harpacticoida, Mormonilloida, Siphonostomatoida. Biol. Mar. Mediterr. 17, 420–431.
Mazzocchi, M. G., Licandro, P., Dubroca, L., Di Capua, I., and Saggiomo, V. (2011). Zooplankton associations in a Mediterranean long-term time-series. J. Plankton Res. 33, 1163–1181. doi: 10.1093/plankt/fbr017
Meiklejohn, K. A., Damaso, N., and Robertson, J. M. (2019). Assessment of BOLD and GenBank – Their accuracy and reliability for the identification of biological materials. PLoS One 14:e0217084. doi: 10.1371/journal.pone.0217084
Mills, S., Alcántara-Rodríguez, J., Ciros-Pérez, J., Gómez, A., Hagiwara, A. K., Galindo, K. H., et al. (2017). Fifteen species in one: deciphering the Brachionus plicatilis species complex (Rotifera, Monogononta) through DNA taxonomy. Hydrobiologia 796, 39–58. doi: 10.1007/s10750-016-2725-7
O’Brien, T. D., Lorenzoni, L., Isensee, K., and Valdes, L. (eds) (2017). What are Marine Ecological Time Series telling us about the ocean? A status report. Paris: IOC-UNESCO, IOC Technical Series.
Ohtsuka, S., Hora, M., Suzaki, T., Arikawa, M., Omura, G., and Yamada, K. (2004). Morphology and host-specificity of the apostome ciliate Vampyrophrya pelagica infecting pelagic copepods in the Seto Inland Sea. Japan. Mar. Ecol. Prog. Ser. 282, 129–142. doi: 10.3354/meps282129
Papadopoulos, L. N., Peijnenburg, K. T. C. A., and Luttikhuizen, P. C. (2005). Phylogeography of the calanoid copepods Calanus helgolandicus and C. euxinus suggests Pleistocene divergences between Atlantic, Mediterranean, and Black Sea populations. Mar. Biol. 147, 1353–1365. doi: 10.1007/s00227-005-0038-x
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Ratnasingham, S., and Hebert, P. D. N. (2007). BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Razouls, C., de Bovée, F., Kouwenberg, J., and Desreumaux, N. (2005-2021). Biodiversity of Marine Planktonic Copepods (morphology, geographical distribution and biological data). [Online]. Available online at: http://copepodes.obs-banyuls.fr/en [accessed on Aug, 12, 2021]
Ribera d’Alcalà, M., Conversano, F., Corato, F., Licandro, P., Mangoni, O., Marino, D., et al. (2004). Seasonal patterns in plankton communities in a pluriannual time series at a coastal Mediterranean site (Gulf of Naples): an attempt to discern recurrences and trends. Sci. Mar. 68, 65–83. doi: 10.3989/scimar.2004.68s165
Rose, M. (1927a). Comparaison entre le plankton d’Alger et celui des côtes Méditerranéennes françaises. C. r. Ass. fr. Avanc. Sci. 51, 542–543.
Rose, M. (1927b). Considérations générales sur le plankton de la Méditerranée occidentale. C. r. Ass. fr. Avanc. Sci. 51:544.
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Sabia, L., Zagami, G., Mazzocchi, M. G., Zambianchi, E., and Uttieri, M. (2015). Spreading factors of a globally invading coastal copepod. Medit. Mar. Sci. 16, 460–471. doi: 10.12681/mms.1154
Sars, G. O. (1924-1925). Copépodes particulièrement bathypélagiques provenant des campagnes scientifiques du Prince Albert Ier de Monaco (Résultats des Campagnes Scientifiques accomplies sur son yacht par Albert Ier, Prince Souverain de Monaco, Fascicule LXIX). Paris: PSL University.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported soft ware for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541.
Stefanni, S., Stanković, D., Borme, D., de Olazabal, A., Juretić, T., Pallavicini, A., et al. (2018). Multi-marker metabarcoding approach to study mesozooplankton at basin scale. Sci. Rep 8:12085. doi: 10.1038/s41598-018-30157-7
Tautz, D., Arctander, P., Minelli, A., Thomas, R. H., and Vogler, P. A. (2002). DNA points the way ahead in taxonomy. Nature 418, 479. doi: 10.1038/418479a
Unal, E., Frost, B. W., Armbrust, V., and Kideysa, A. E. (2006). Phylogeography of Calanus helgolandicus and the Black Sea copepod Calanus euxinus, with notes on Pseudocalanus elongatus (Copepoda, Calanoida). Deep Sea Res. II 53, 1961–1975. doi: 10.1016/j.dsr2.2006.03.017
Uttieri, M. (2018). “Trends in Copepod Studies,” in Trends in Copepod Studies - Distribution, Biology and Ecology, ed. M. Uttieri (New York, NY: Nova Science Publishers, Inc), 1–11.
Uttieri, M., Aguzzi, L., Aiese Cigliano, R., Amato, A., Bojanić, N., Brunetta, M., et al. (2020). WGEUROBUS – Working Group “Towards a EURopean OBservatory of the non-indigenous calanoid copepod Pseudodiaptomus marinUS”. Biol. Invasions 22, 885–906. doi: 10.1007/s10530-019-02174-8
Walter, T. C., and Boxshall, G. (2021). World of Copepods Database [Online]. Available online at: http://www.marinespecies.org/copepoda [accessed July-12;2021]
Wiens, J. J. (2007). Species delimitation: new approaches for discovering diversity. Syst. Biol. 56, 875–878. doi: 10.1080/10635150701748506
WoRMS Editorial Board. (2021). World Register of Marine Species (WoRMS) [Online]. WoRMS Editorial Board. Available online at: http://www.marinespecies.org at VLIZ [accessed Aug-12;2021]
Zhang, G. K., Chain, F. J. J., Abbott, C. L., and Cristescu, M. E. (2018). Metabarcoding using multiplexed markers increases species detection in complex zooplankton communities. Evol. Appl. 11, 1901–1914. doi: 10.1111/eva.12694
Zhang, J., Kapli, P., Pavlidis, P., and Stamatakis, A. (2013). A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876. doi: 10.1093/bioinformatics/btt499
Keywords: integrated taxonomy, DNA barcoding, biodiversity, calanoid copepods, Mediterranean Sea
Citation: Di Capua I, D’Angiolo R, Piredda R, Minucci C, Boero F, Uttieri M and Carotenuto Y (2022) From Phenotypes to Genotypes and Back: Toward an Integrated Evaluation of Biodiversity in Calanoid Copepods. Front. Mar. Sci. 9:833089. doi: 10.3389/fmars.2022.833089
Received: 10 December 2021; Accepted: 17 January 2022;
Published: 25 February 2022.
Edited by:
Yong Wang, Institute of Deep-Sea Science and Engineering (CAS), ChinaReviewed by:
S. Bijoy Nandan, Cochin University of Science and Technology, IndiaFrancesca Leasi, University of Tennessee at Chattanooga, United States
Xinzheng Li, Institute of Oceanology (CAS), China
Copyright © 2022 Di Capua, D’Angiolo, Piredda, Minucci, Boero, Uttieri and Carotenuto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iole Di Capua, aW9sZS5kaWNhcHVhQHN6bi5pdA==
 Iole Di Capua
Iole Di Capua Rosa D’Angiolo
Rosa D’Angiolo Roberta Piredda
Roberta Piredda Carmen Minucci1
Carmen Minucci1 Ferdinando Boero
Ferdinando Boero Marco Uttieri
Marco Uttieri Ylenia Carotenuto
Ylenia Carotenuto