- 1Department of Earth, Environment and Life Science, University of Genova, Genova, Italy
- 2Institute of Physiology of the Czech Academy of Sciences, Praha, Czechia
- 3Institute for Sustainable Plant Protection – SS Turin, CNR, Torino, Italy
The silicification mechanism in sponges is a biologically controlled process where the complex and amazing shape of spicules is the result of the hierarchical assembly of silicon particles to form a composite structure with organic compounds, mainly constituted by proteins. In this work, using an integrated approach of transcriptomic and proteomic analysis, we describe the protein content of sponge spicules in the marine demosponge Petrosia ficiformis (Poiret, 1789). Proteins from spicules were obtained via an ammonium fluoride extraction procedure to remove the inorganic silica followed by SDS-PAGE electrophoresis. The resulting data of LC-MS/MS analysis of the extracted SDS-PAGE bands were then processed with the MASCOT software to search against a database consisting of transcripts and predicted proteins of P. ficiformis. The results revealed a very heterogeneous group of 21 proteins, including silicatein beta, different isoforms of cathepsins, proteins with strong homologies with enzymes like sulphatases, glycosidases, lipid-related proteins, phosphatases, and some others with unknown function. Most of the proteins found here have structures and domains attributable to lysosomes enzymes and for this reason it could be related to these cellular structures the evolutionary origin of the biosilica machinery in sponges.
Introduction
Sponges are very simple and ancient animals, but they have a remarkably complex genetic and molecular organisation (Sebé-Pedrós et al., 2018; Musser et al., 2021). The presence of sponge fossils from more than 600 million years makes palaeontologists consider this phylum as the oldest animal group still living on the earth (Erwin et al., 2011). Sponges are pluricellular organisms with a simple body structure not organised in specialised tissues or organs (Simpson, 1984). Nevertheless, the evolution from unicellular to multi-cellular animals needed a relevant implementation of gene regulation mechanisms (Brunet and King, 2017). Amazingly, there are many molecular analogies between the highly evolved multi-cellular animals and the simple multi-cellular sponges. The presence of RNA based regulatory factors like long noncoding RNA (Gaiti et al., 2018), or signal cascade translation systems based on inflammatory pathway response (Pozzolini et al., 2016; Pozzolini et al., 2017) and growth factors (Pozzolini et al., 2019) are examples showing the great molecular similitudes between sponges and other higher animals. The simplicity of sponges’ body, the direct contact of their cells with the external environment (sponges filter water that permeates in all the body, through a very well organised channels system) and their sessile status induced on the other hand the need to protect from micro-organisms infections and from other animals’ predation. This requirement is managed by the production of a great amount and variety of natural compounds, mainly supplied by symbiotic microorganisms (Brinkmann et al., 2017). All these features make sponges “unique” in the animal kingdom: a fascinating balancing of body simplicity and molecular complexity.
One of the most peculiar aspects of sponges is the structural organisation of their skeleton. The sponge body is supported by organic extracellular matrix (mainly collagen or/and “spongin” in some cases mixed with chitin) (Pozzolini et al., 2012; Ehrlich et al., 2018; Jesionowski et al., 2018) and in many species by inorganic structures called spicules, formed by amorphous silicon dioxide or, in Calcispongiae, calcium carbonate (Simpson, 1984; Ehrlich et al., 2011). Siliceous spicules have different shapes and dimensions with an extremely ordered organisation at a nanometre level (Şen et al., 2016). Biosilica is produced by Hexactinellida, by many Demospongiae and by some Homoscleromorpha (Simpson, 1984). There are many studies on the structural organisation and chemical composition of the biomaterial forming sponge spicules (see for overview Schoeppler et al., 2017; Wysokowski et al., 2018; Görlich et al., 2020; Pisera et al., 2021) and at present time, it seems clear that the features and the properties of biosilica depend on the composition of this material. Sponge biosilica is, in fact, a composite material where organic biomolecules contribute to its synthesis in environmental conditions, to the shape determination of spicules and to their technological properties (Khrunyk et al., 2020), but it is not easy at all to study the dynamics of spicule building. To satisfy this need, a multidisciplinary approach is necessary. Advanced material science technology to analyse in detail the structure and the properties of biosilica and good animal and cellular models to explain the molecular and genetic rules of spicule building are both necessary. At present, Hexactinellida have been mainly studied by material scientists: the production of large and long spicules (up to 2 meters long in Monoraphis chuni) (Müller et al., 2007; Ehrlich et al., 2008; Pisera et al., 2021) and their intriguing technological properties as optic fibre (Cattaneo-Vietti et al., 1996), were the main reasons for the choice of this class of animals. Demospongiae are instead approached by sponge cell biologists and molecular biologists, for the possibility to manage in controlled environment living animals and their sponge cell culture (Valisano et al., 2006; Pozzolini et al., 2014). It is also now clear that Hexactinellida and Demospongiae composite silica-based spicules apparently have common features but also very relevant differences in their composition (Müller et al., 2011; Shimizu et al., 2015). Definitively, many questions are still open to understand exhaustively the biological rules of biosilica production. Specifically, in Demospongiae, siliceous spicules are most commonly formed by monoaxonic or tetraxonic needle-like structures, from some μm to 1-2 mm long (Hooper and Van Soest, 2002). In the Demospongiae Tethya aurantium was for the first time sequenced and cloned silicatein, one key protein involved in the biosilicification process (Shimizu et al., 1998; Cha et al., 1999). In this last two decades, many studies approached from different points of view the role of silicatein in Demospongiae silica biosynthesis as well as its biotechnological applications (for an exhaustive and complete review see Shimizu and Morse, 2018) and animals and 3D cells models have been also developed to study the dynamics of spicule formation (Müller et al., 2006; Valisano et al., 2012; Müller et al., 2013a). In all these works, silicatein was indicated as the main molecular actor of biosilica formation. Silicatein is indeed a very peculiar protein: it has a remarkable sequence homology with the lysosomal protease family of cathepsins, but it is water-insoluble, and it has an enzymatic active site with serine in substitution to cysteine (Shimizu and Morse, 2018). Its insolubility, due to a higher number of hydrophobic regions and different post-translational modifications compared to cathepsins (Armirotti et al., 2009), determines its self-assemblage properties (Murr and Morse, 2005). This evidence clearly supports the assumption of the structural role of silicatein in the formation of the axial filament of spicules as well as its being a key component of biosilica composite structure. The enzymatic active site suggests, in the meantime, the catalytic role of this protein, even if there is not a definitive agreement among scientists on the mechanism. At present time, in fact, two main hypotheses are supported in literature: silicatein as an enzyme able to catalyse the biosilica precipitation from some organosilicon compound (Cha et al., 1999; Povarova et al., 2018a; Povarova et al., 2018b) and silicatein as enzyme able to catalyse the biosilica polymerisation from orthosilicic acid (Müller et al., 2013b). Both models are apparently sustainable and silicatein can be considered one of the major contributors for the biosilica condensation in environmental conditions. It is much more difficult to understand the molecular keys of spicule growth and spicule shape control. Silicatein probably is involved in growth control and shaping, mainly in Demospongiae (Müller et al., 2009), but many other components like collagen, chitin and actin seem to play some fundamental roles (Ehrlich, 2010; Ehrlich et al., 2010; Ehrlich et al., 2016; Ehrlich et al., 2022).
A recent work (Riesgo et al., 2015) further considered the scenario concerning silicatein: a comparative analysis of transcriptomes and genomes datasets puts clearly in evidence that not all biosilica forming sponges have/express silicatein genes. Mainly Hexactinellida and Homoscleromorpha sponges do have not this protein and some silica forming Demospongiae apparently do not have silicatein (Chondrilla nucula). In addition, sponges that do not produce biosilica seem to express silicatein genes. Another interesting and relevant aspect is the evidence of a remarkable expression of cathepsins (specifically cathepsins B and L) in biosilica forming sponges, whose functional role is to be better clarified (Riesgo et al., 2015). In this key work the authors, on the basis of transcriptome analysis results, hypothesized a lysosomal origin of the “silica deposition vesicle” of Demospongiae (Riesgo et al., 2015). It is evident that the biosilicification mechanisms in sponges are far to be understood, and one of the major gaps of knowledge to be filled is to improve the description of the organic content of Demospongiae spicules.
To accomplish this objective, in the present work, an integrated transcriptome and proteome analysis is proposed as a strategy. Here, for the first time, using as experimental model the species Petrosia ficiformis, is described the protein content of Demospongiae spicules by a proteomics analysis followed with protein sequence identification based on the transcriptome dataset matching.
Materials and Methods
Protein Extraction
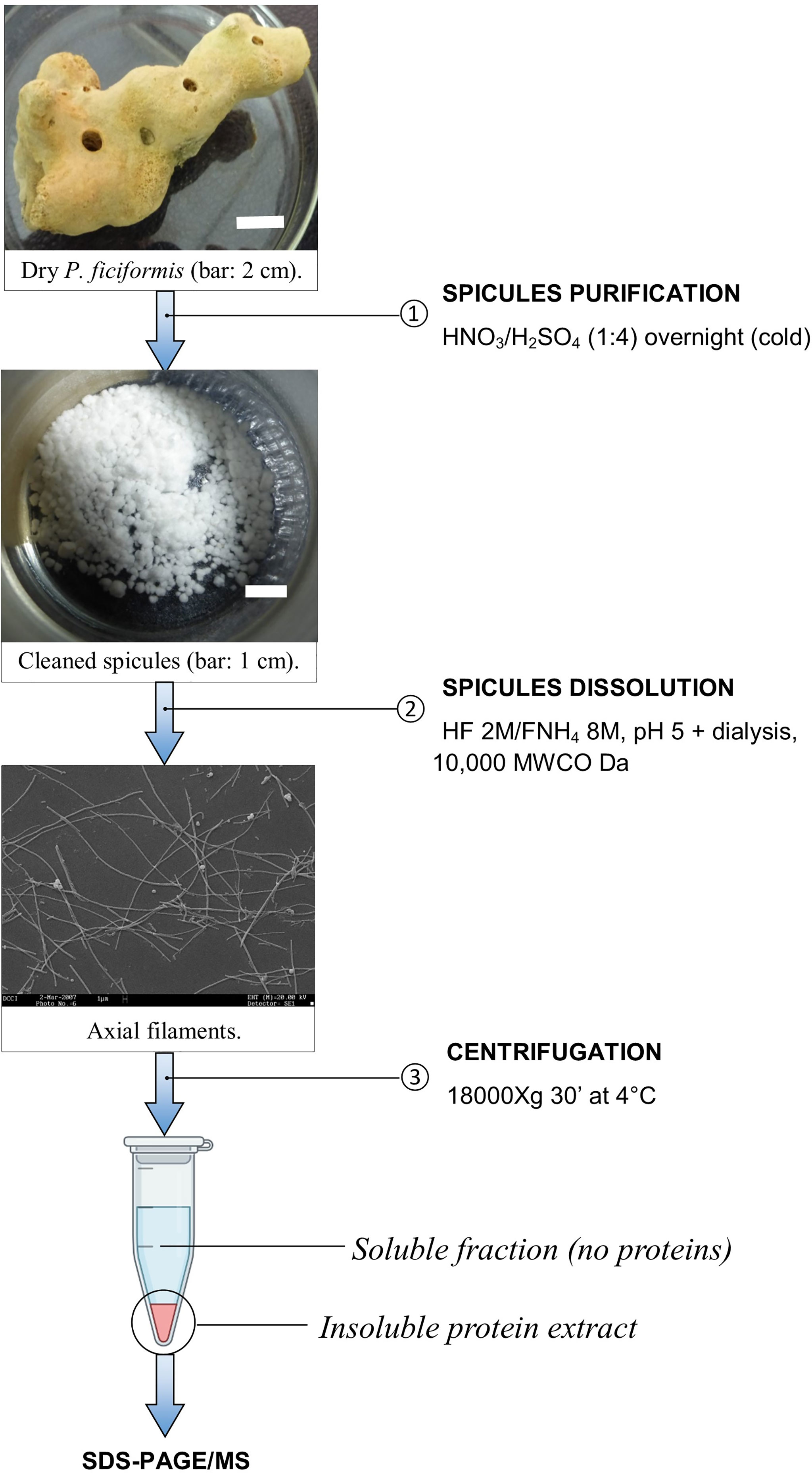
The proteins contained inside the silica spicules were extracted accordingly to the previously described method (Armirotti et al., 2009). Briefly (Scheme 1), 5 g of dry of P. ficiformis collected in the Ligurian sea have been cut into small pieces of 1 cm3 and dissolved in 500 mL of cold HNO3/H2SO4 (1:4) over night. The cleaned spicules have been washed several times in distilled water until the pH was above 6, then once in 100% ethanol and finally air-dried. 1 g of clean spicules were dissolved into 100 ml of 2 M HF/8 M NH4F (pH 5) at room temperature until the complete silica solubilization. The sample was then dialysed (10,000 MWCO Da) against 5 L of Milli-Q water for 4 hours at 4°C (five changes). At the end, the insoluble organic component was recovered by centrifugation for 30 minutes at 18,000 x g at 4°C, resuspended in 3 mL of Milli-Q water and stored at -20°C until use. To check the presence of proteins in the soluble fraction, the supernatant was ten-fold concentrated and analysed using the method of Bradford assay (Bradford, 1976).
To evaluate the total protein concentration in the insoluble fraction, 0.5 mL of protein suspension were centrifuged for 5 minutes at 18,000 x g at room temperature. The supernatant was discarded, and the insoluble pellet was dissolved in 0.5 mL of 8 M urea. The protein content was evaluated by Bradford assay.
Electrophoretic Analysis
20 μg of silica spicule protein extract were separated by SDS-PAGE according to Laemmli (Laemmli, 1970) on 10 × 8 cm vertical 12% polyacrylamide gel at a constant amperage. After electrophoresis, proteins were fixed in 40% ethanol/10% acetic acid for 60 min, washed twice with Milli-Q water, stained over night with colloidal Coomassie blue (Neuhoff et al., 1988) and destained several hours in 5% acetic acid.
Band Cutting and Trypsin Digestion
Protein bands were manually excised from the gel and digested by trypsin at 37°C overnight (Shevchenko et al., 2006). Gel pieces were destained with acetonitrile, reduced in 10 mM DTT/100 mM ammonium bicarbonate and alkylated with 100 mM iodoacetamide/100 mM ammonium bicarbonate. The peptide samples were extracted after digestion by sonicating for 10 minutes, then vacuum dried.
Analysis of Tryptic Digests With LC-MS/MS
The LC-MS/MS method used was similar to the previously published one (Mikšík et al., 2018). The ultrahigh resolution MaXis Q-TOF (quadrupole – time of flight) mass spectrometer (Bruker Daltonics, Bremen, Germany) was coupled by nanoelectrosprayer to nano-HPLC system Proxeon Easy-nLC (Proxeon, Odense, Denmark). All controlled software was from Bruker Daltonics: HyStar (v. 3.2) micrOTOF-control (v. 3.0), ProteinScape (v. 3.0) and DataAnalysis (v. 4.0).
Three microliters of the peptide mixture were injected into trap column NS-MP-10 Biosphere C18 (particle size: 5 µm, pore size: 12 nm, length: 20 mm, inner diameter: 100 µm) and analysed by analytical column NS-AC-11-C18 Biosphere C18 column (particle size: 5 µm, pore size: 12 nm, length: 150 mm, inner diameter: 75 µm), both manufactured by NanoSeparations (Nieuwkoop, Holland).
The separation of peptides was achieved via a linear gradient between mobile phase A (water) and B (acetonitrile), both containing 0.1% (v/v) formic acid. Separation was started by running the system with 5% mobile phase B, followed by a gradient elution to 30% B at 70 min. The next step was a gradient elution to 50% B in 10 min and then a gradient to 100% B in 10 min. Finally, the column was eluted with 100% B for 30 min. Equilibration between the runs was achieved by washing the column with 5% mobile phase B for 10 min. The flow rate was 0.25 µL/min, and the column was held at ambient temperature (25°C).
On-line nano-electrospray ionization (easy nano-ESI) was used in positive mode. The ESI voltage was set to +4.5 kV, scan time: 3 Hz. Operating conditions: drying gas (N2): 4 L/min; drying gas temperature: 180°C; nebulizer pressure: 100 kPa. Experiments were performed by scanning from 50 to 2200 m/z. The reference ion used (internal mass lock) was a monocharged ion of C24H19F36N3O6P3 (m/z 1221.9906). Mass spectra corresponding to each signal from the total ion current chromatogram were averaged, enabling an accurate molecular mass determination. All LC-MS and LC-MS/MS analyses were done in duplicate.
Database Searching
Data were processed using ProteinScape software v. 3.0.0.446 (Bruker Daltonics, Bremen, Germany).
Proteins were identified by correlating tandem mass spectra to the putatively predicted proteins from P. ficiformis transcriptome, using the MASCOT searching engine v. 2.3.0 (http://www.matrixscience.com). Trypsin was chosen as the enzyme parameter. Extract likely coding regions and predicted candidate peptides from transcripts of P. ficiformis (Riesgo et al., 2014) were identified with TRANSDECODER V.5.2.0 (https://github.com/TransDecoder/TransDecoder), as previously described in Pozzolini et al., 2019. Three missed cleavages were allowed, and an initial peptide mass tolerance of ±15.0 ppm was used for MS and ±0.03 Da for MS/MS analysis. Variable modifications were set; proline and lysine were allowed to be hydroxylated, methionine oxidated, whereas asparagine and glutamine deamidated and cysteine carboxymethylated or carbamidomethylated. The monoisotopic peptide charge was set to 1+, 2+, and 3+. The Peptide Decoy option was selected during the data search process to remove false-positive results. Only significant hits were accepted (MASCOT score ≥80 for proteins and MASCOT score ≥20 for peptides), however, all peptides and proteins were additionally manually validated.
Results
Analysis of the Proteins on Gel
The strategy of analysis of the proteins extracted from the biosilica was based on HF/NH4F dissolution and dialysis against water (see Scheme 1). This approach allows the recovery of an insoluble fraction of organic material. Typically, 2 g of P. ficiformis clean silica spicules yielded 2 mg of total protein extract. The soluble fraction after dialysis (10,000 MWCO Da) was ten-fold concentrated and the total protein contents was assayed. No evident presence of protein was detected in the soluble fraction. The not-soluble organic residue was resuspended and 20 μg of proteins were run on a 12% polyacrylamide gel, coloured with colloidal Coomassie blue. Figure 1 shows the results, protein separation was not easy, and a substantial swiping of proteins is evident. The scarce solubility of the protein mixture and, probably, the presence of some silica residues could be the cause. We cut anyway the gel into 9 small slices to perform MS analysis of the protein content inside it.
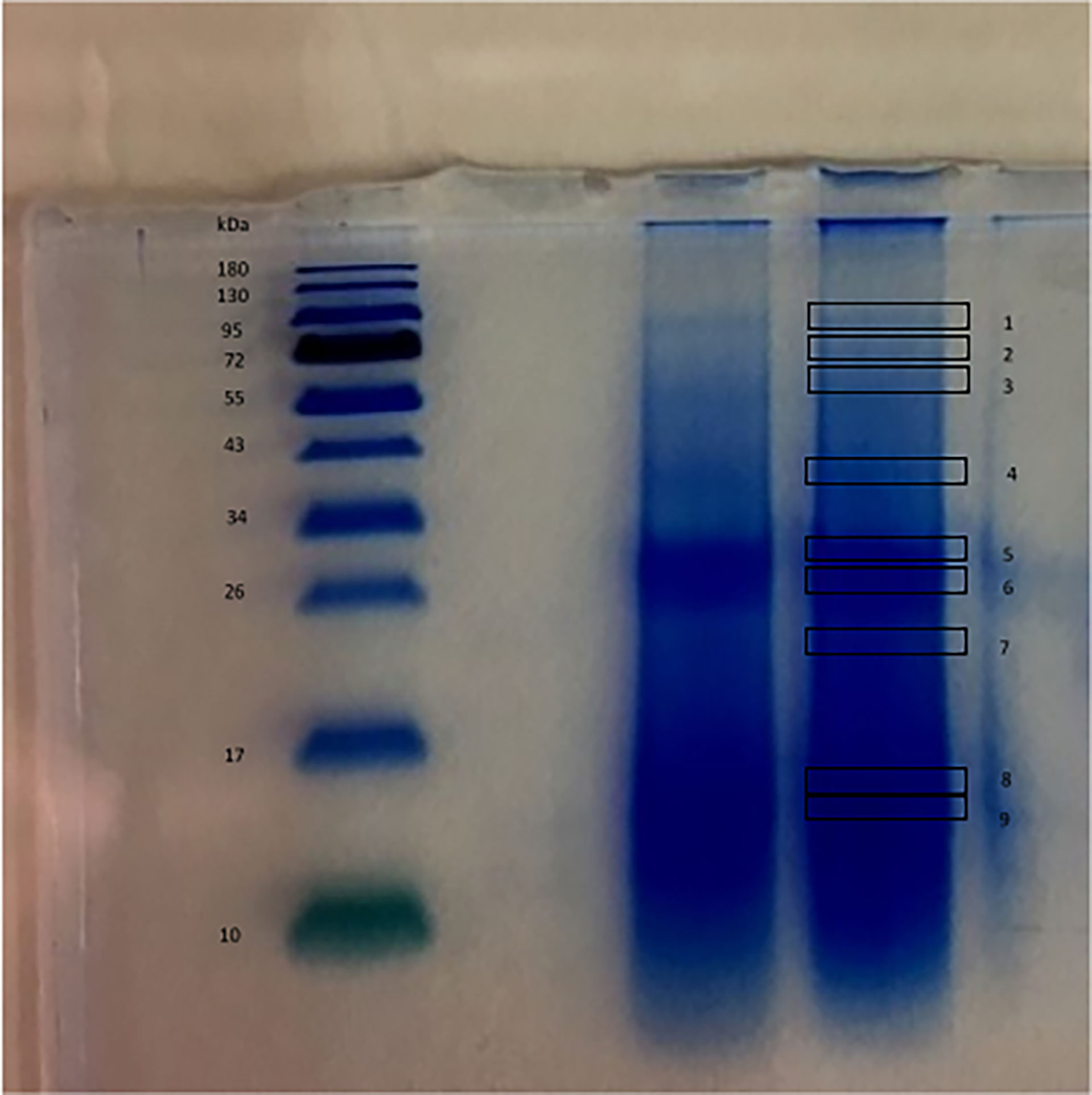
Figure 1 SDS-PAGE of the 20 μg of proteins extracted from the spicules of P. ficiformis. MW standard (left) and two lines containing the proteins extracted (on the right). The numbers indicate the position of the slices cut for proteins extraction and MS analysis.
Identification and Characterisation of Proteins
The LC-MS/MS analysis raw data obtained from the organic components of P. ficiformis spicules were processed with the MASCOT software to search against a database consisting of transcript and predicted proteins of the above-mentioned sponge species here used as a study model.
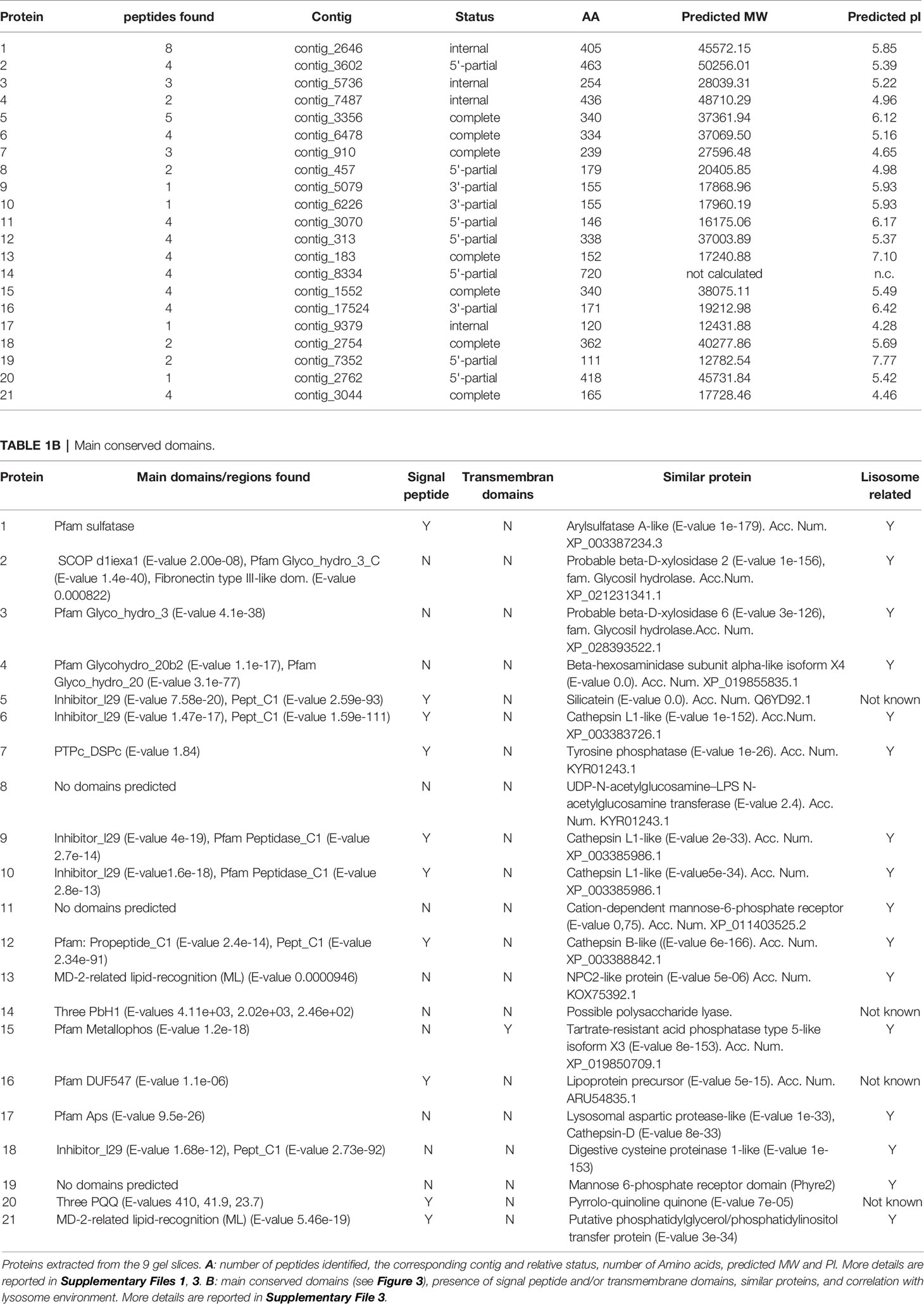
To our knowledge, here we are showing the first case of description of the overall protein content of demosponge biosilica. In our experimental conditions, 21 proteins with a high level of confidence have been identified in their primary structure. The remarkable swiping of proteins in gel electrophoresis reflected its effects in the analytical results, where peptides of the same protein were found in different positions. In Supplementary File 1, the peptides and the related proteins found in the nine slices are listed. The main concentration of different proteins is present on slice 6. Some proteins are present in many or all slices (for example protein 1) while others are better distributed in the lower part of the gel.
All raw data are available on the following public repository: https://doi.org/10.5281/zenodo.6033766. Table 1 summarizes the structural features of the identified proteins and their theoretical functions based on the predicted primary structure and of the analysis of conserved domains. Spicule protein content in our findings can be divided into functional categories, as shown in Figure 2. Most proteins seem remarkably related to the lysosome environment (see Table 1) and only a few are of unknown origin. In Figure 3 the specific domains of each protein characterized in this work are shown. A detailed overview of the results includes the presence of P. ficiformis silicatein beta, as expected (protein 5, Table 1). Silicatein peptides were detected from bands 4 to 9 with a typical swiping. With similar swiping behaviour, five different proteases of the cathepsin family (proteins 6, 9, 10, 12, 17) have been identified. The larger part of the other proteins in the list has remarkable similarity with lysosome-related polypeptides. Protein 1 shows strong homology with the aryl-sulphatase family enzymes, as in the predicted primary structure are present all the amino acids of the active site included the typical Cys residue normally modified in C-alpha-formyl-glycine by formyl-generating enzymes of rough endoplasmic reticulum. These enzymes are known to catalyse the cleavage of sulphate esters mainly in glycolipids and glycosaminoglycans inside lysosomes (Thompson and Daniel, 1988). Proteins 2, 3, and 4 have their primary structure remarkably homologs to glycosyl hydrolase family 3 (protein 2 and 3), and beta-hexosaminidase group (protein 4). All these proteins are involved in the catabolism of glycosphingolipids inside lysosomes (Ryckman et al., 2020). In mammalians, it is known that glycosyl hydrolases of family 3 are involved in the transxylosylation of cholesterol in lysosomes. Beta-hexosaminidases are involved in the hydrolysis of terminal N-acetyl-D-hexosamine residues in N-acetyl-β-D-hexosaminides and it is typically expressed inside lysosomes for glycosphingolipids catabolism. In the same “group” of proteins related to lipid catabolism it is included protein 13, whose primary sequence is remarkably related to cholesterol transporter from lysosomes as well as protein 21 whose lipid recognition domain is typically present in phosphatidylinositol/phosphatidylglycerol transfer protein, fundamentally found inside lysosomes. The 21 proteins list also includes proteins with possible mannose-6P receptor domain, characteristic of protein trafficking to lysosomes (proteins 11 and 19), in particular interacting with cathepsins and hexosaminidases (Staudt et al., 2017). Tyrosine phosphatases and tartrate-resistant phosphatases (proteins both present in the lysosome environment) are also represented in this very complex pattern of biomolecules found inside biosilica. The list includes one protein with a domain corresponding to pyrrole quinoline quinone dehydrogenase. This protein is not typical of the lysosome environment. Finally, three unknown proteins are also represented.
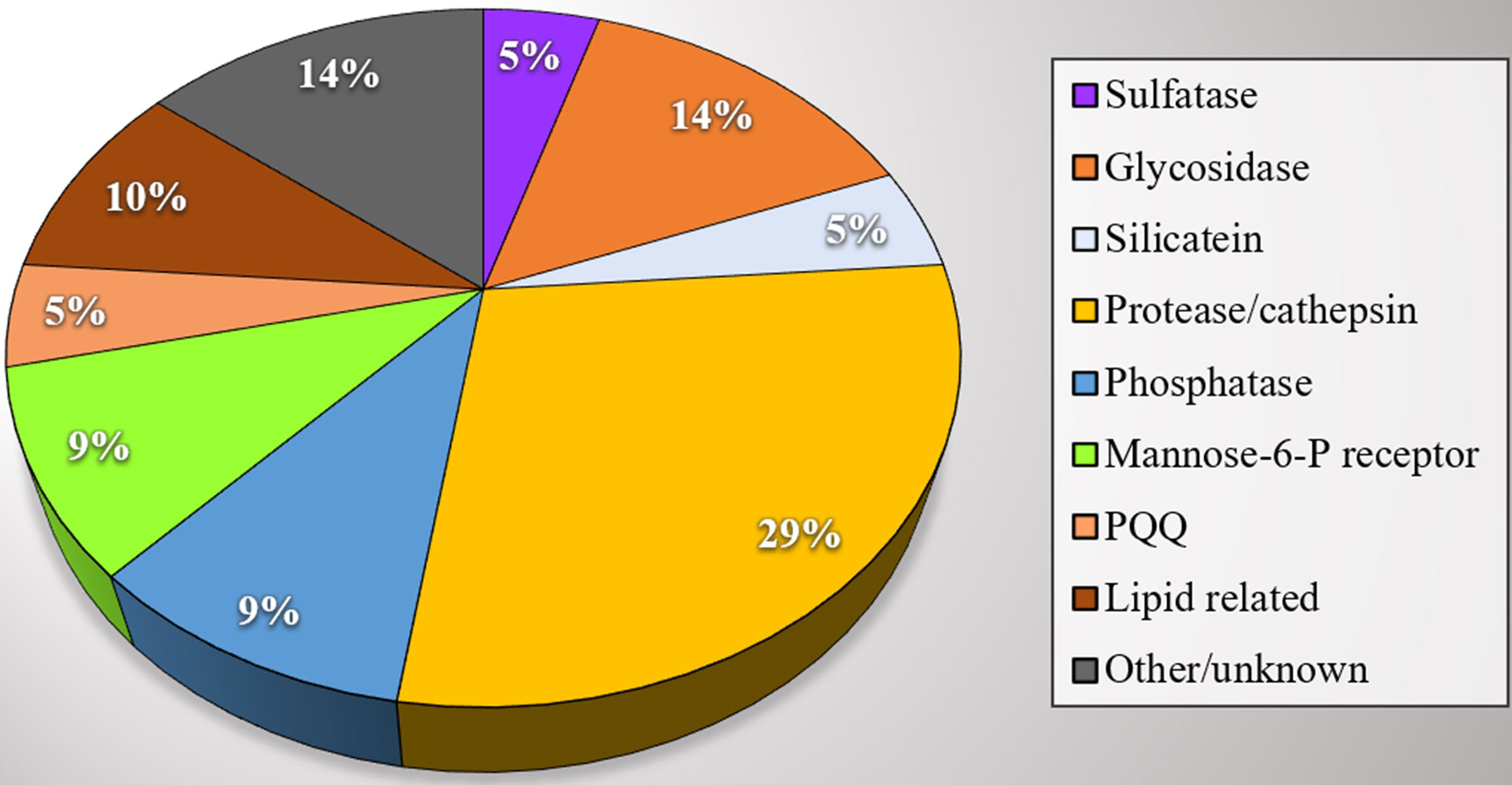
Figure 2 Proteins extracted from P. ficiformis biosilica. Subdivision of proteins in activity categories. Each colour is representative of a category group and the numbers inside the circle areas indicate the numbers of proteins of the same category.
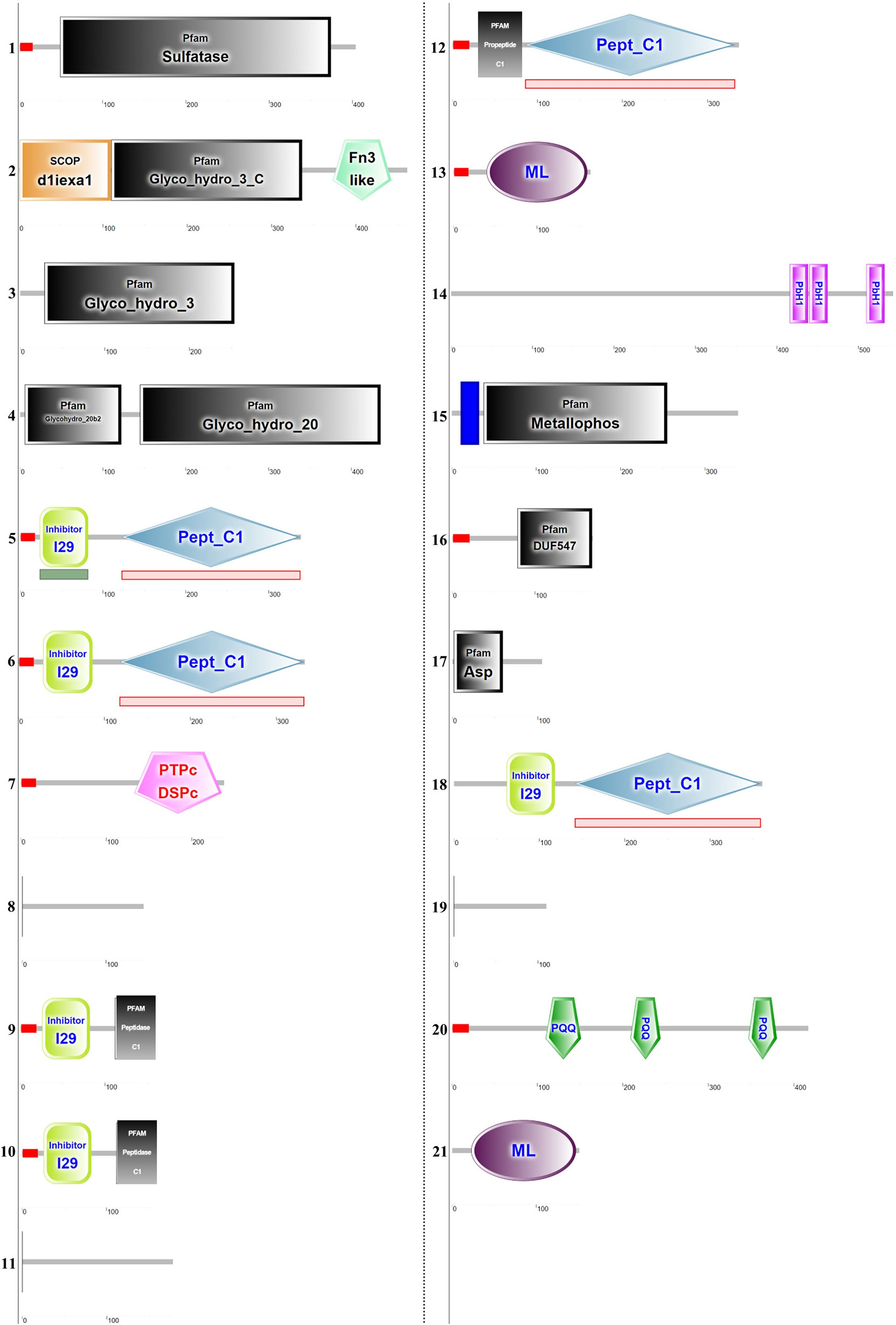
Figure 3 Schematic conserved domain representation of the proteins extracted from the spicules. The analysis was performed using the deduced amino acids sequences obtained after in silico translation of contigs correspondent to MS-identified peptides. The conserved domain has been identified using the Simple Modular Architecture Research Tool (SMART) program (http://smart.embl-heidelberg.de/). The detailed legend is reported in Supplementary File 2. The domains list is reported in Table 1. The contig sequences identified are reported in Supplementary File 3.
Discussion
The biomineralization process in Porifera still has several unsolved questions. Most of the scientists studying biosilicification in demosponges attribute to silicatein a key role in the skeleton forming process, (see the recent review, Shimizu and Morse, 2018). Indeed, silicatein is very abundant inside spicules and its distribution is intimately mixed with the inorganic part to form a composite (Müller et al., 2013; Shimizu and Morse, 2018). Nevertheless, at present, this peculiar protein, with its proposed double role (enzymatic and structural), in our opinion is not enough to fully explain all the aspects of the biosilicification mechanism in these animals. In biological controlled biomineralization, the molecular actors are very numerous, involving proteins and other biomacromolecules (polyamines for example), whose role in the process is often very difficult to identify (Wysokowski et al., 2018). Proteomic analysis in these cases give a relevant contribution to identify the various component of an organic compound and in the past, it has already been used to describe the post-translational modification of silicatein (Armirotti et al., 2009). In the present study we have improved the strategy looking again inside spicules using an integrated approach based on proteome and transcriptome analysis. Recently, similar methodologies were used to describe the organic matrix content of diatoms and corals (Kotzsch et al., 2016; Peled et al., 2020).
Using this technique, we have found together with silicatein a list of proteins until now never described before inside biosilica. The P. ficiformis transcriptome availability (Riesgo et al., 2014) allowed in the present work a fine identification of proteins inside spicules (Figure 2 and Table 1) but, for many of them, it is not easy to attempt a possible explanation of their role in the process. Previous scientific publications demonstrate that the spicule formation in demosponges begins inside cells, but it continues outside, and the process pass through the production of biosilica building units inside cells successively secreted outside (Wang et al., 2011). A last century study (Lévi, 1963) and a more recent description of spicule formation by Schröder and co-workers (Schröder et al., 2007) show the formation of specific intracellular vesicles in which biosilica is precipitated. In accordance with what is described in many other biomineralization processes, the building blocks are organized intracellularly in appropriate compartments where the chemical conditions for mineral precipitation are created and maintained. The ultrastructural analysis reported in the work of Schröder and co-workers gives some explanation of the process: silica is released in extracellular granules outside the sclerocite; these granules are rich in silica, and they are named silicasomes. Morphological analysis showed that silicasomes positively react with antibodies against silicatein, and this reaction allowed to follow the intra- and extracellular evolution of these organelles (Schröder et al., 2007). This evidence suggests a subcellular process involving some systems related to endosome/exosome or microvesicles pathways. Furthermore, it is known that the acidic environment favours biosilica precipitation (Shimizu et al., 2001) and, for this reason, also lysosome or lysosome-related organelles may be involved in the formation of silicasomes. In this conceptual direction it is also placed the co-expression of silicatein and cathepsins recently described in sponge tissues by Riesgo and co-authors, suggesting some specific roles of these proteins in the biomineralisation process as well as clearly remarking the hypothesis of a silica deposition environment of lysosomal origin (Riesgo et al., 2015). In this study Riesgo and co-authors made an accurate analysis of the evolution of cathepsin genes among metazoan and many of them have been described in sponges. Specifically, in P. ficiformis transcriptome they documented the expression of Cathepsin B, L, and D as well as 4 different silicateins (Riesgo et al., 2015). This information derives from comparative transcriptomic analysis of sponges, without any further insights into protein localisation. Our findings demonstrate, for the first time, the presence of cathepsins and silicatein inside the biosilica, fully confirming the lysosome hypothesis. Additionally, in our research results many other proteins are for the first time identified inside P. ficiformis spicules after inorganic silica removal, almost half of them is provided with a signal peptide and shows a strong similarity with proteins normally found inside lysosomes (see Table 1). The scenario is however very complex: from one hand the common lysosomal origin of the majority of proteins listed in our work is something coherent and easy to discuss on the basis of the morphological evidence published (Schröder et al., 2007) and of the evolutionary study cited (Riesgo et al., 2015); on the other hand, it becomes very difficult to attribute a specific role in the process for many of the proteins here identified.
The first aspect emerging from the analysis of biosilica protein content shown in Table 1 of our results is the demonstration of presence of some isoforms of cathepsin family together with silicateins, data strongly congruent with the previously published hypothesis of Riesgo (Riesgo et al., 2015). Riesgo and co-authors already suggested a possible role of cathepsins in the biosilification process (Riesgo et al., 2015). In particular, the fact that one silicatein cited by Riesgo has cysteine inside the active site is suggestive of a more complex scenario where some cathepsins can play some role (Riesgo et al., 2015). In addition, it also known that mutants of L cathepsin produced in recombinant systems confirm the existence of structural issues other than active sites in the biosilica biosynthesis in silicateins and cathepsins (Fairhead et al., 2008). On these bases, the changing of the active site from Cys to Ser is not the only molecular key of the mechanism, but also other regions of the protein can have a role in the catalysis and/or in the biosilica organisation and/or in other proteins assembling (Fairhead et al., 2008). Despite all these considerations, definitively it is not easy to define the exact role of various cathepsins in the biosilica precipitation environment: one additional possibility we suggest is that they can be involved in the specific activation of other proteins relevant in the process (i.e., cleaving signal peptides and\or regulatory peptides) but more studies must be performed to better understand if these roles are related only to catalysis or also to structural organisation. Only future experimental demonstrations based on selective enzymatic inhibition or gene silencing approach in cellular models can give a final answer.
In Figure 3 and in Supplementary File 2 are listed some details of domain and sequence homology also of the other proteins found and, in the results, we have underlined the enzymatic roles of the homologous polypeptides as described in other organisms. But, if it is difficult the possible explanation of cathepsins and silicateins roles, the discussion of other proteins here found in the biosilica matrix only on the basis of their structural similarity with other well-known proteins of lysosomal origin is much more speculative. A further interesting aspect is that many of these proteins have well defined enzymatic domains (see Figure 3), differently from other situations (corals and diatoms for example), where a considerable numbers of proteins with structural functions and apparently lacking catalytic domains have been found (Ramos-Silva et al., 2013; Kotzsch et al., 2016). In our research results we conversely identified aryl sulphatase, glycosyl hydrolase, beta-hexosaminidase, all typical proteins found inside lysosomes. Also, proteins involved in the lysosome cholesterol transport have been found (see Table 1A and Figure 2). Only three proteins are of unknown origin, (with some low similarities to acetylglucosamine transferase, polysaccharide lyase and one lipoprotein).
The presence of enzymes related to glycosaminoglycans (GAGs) and polysaccharides catabolism inside biosilica is indeed intriguing. The sugars, the polyol and the GAGs interaction with silicon recently suggested by Nielsen (Nielsen, 2014) and the regulation of connective tissue formation in mammals influenced by silicon is suggestive of some biochemical mechanism involving silicon and GAGs and other polysaccharides in mammalians (Götz et al., 2019). The presence of these enzymes inside of silica sponge spicules suggest a possible sugars-silicon interaction originally involved in biomineralization. A future explanation of their role can give important answers not only in biosilicification process in sponges but also in cartilage and bone formation in mammalians and humans. The results here shown undoubtedly confirm the lysosome hypothesis since the list of proteins here found are in their majority clearly related to lysosomes, but at the same time opens the door to many unanswered questions, specifically on the metabolic management of silicon inside cells. The silicon uptake in sponges is under the control of specific transporters (Schröder et al., 2004; Maldonado et al., 2020), but the intracellular destiny of silicon is not clear. The hypothesis of one organosilicon compound already emerged from the beginning of studies of silicatein (see Shimizu et al., 1998) but, at the same time no specific ideas have already been confirmed. Bio-organic chemistry works suggested the general possibility to have silica derivatives with organic molecules of biological relevance (Kinrade et al., 1999; Vis et al., 2020). The presence of many enzymes related to sugar metabolism could be suggestive of silicon-organic compounds specifically involved in this process, while cathepsins and silicateins can play a role both in the enzymatic control of biosilica precipitation and in the structural organisation of spicules.
Much more difficult is to give some functional hypothesis of other proteins. Another finding is a protein with pyrrolo-quinoline quinone (PQQ) dehydrogenase domain. This family of proteins are normally present in prokaryotes but recently they have been described also in Eukaryotic, involved in sugars oxidoreductases process (Matsumura et al., 2014).
In this scenario, the apparent absence of silintaphins (Wiens et al., 2009) and galectins (Schröder et al., 2006) is surprising. In the first case, also inside the template transcriptome (Riesgo et al., 2014) we have not found silintaphins. We cannot currently confirm if this is due to incompleteness in transcriptome database used in this study or to some specific genetic differences that occurred in this sponge species. Different considerations should be made regarding galectins. In this second case, in the transcriptome database used, the presence of galectin transcript is confirmed, but any traces of these proteins in the proteomic analysis was not found, despite the presence of galectins in the biosilica matrix was proposed using specific antibodies in a previous work on S. domuncula (Schröder et al., 2006). In our experimental conditions, we performed a strong acid spicules cleaning before silica dissolution. Differently, in the study of Schröder and co-workers the extraction procedure was completely different and milder (Schröder et al., 2006) and maybe some proteins involved in the biosilica building not intimately mixed inside biosilica in our approach can be missed. The methodological approach in the extraction procedure without doubt can affect the results. Here we have used this very strong cleaning process (cold HNO3/H2SO4) of spicules in order to avoid contaminations. MS analysis, in fact, is highly sensitive and it is not easy to evaluate proteins at the quantitative level. This sensitivity on the other hand allows to identify many proteins not found before with different analytical approaches. Here we decide to focus our attention to those proteins very intimately mixed with the inorganic silica lattice.
Conclusions
Biomineralization in sponges is a process not completely explained. In this work, using a strong spicules cleaning procedure, we have extracted the organic proteinaceous material embedded inside biosilica spicules from the marine sponge P. ficiformis and analysed by MS analysis. The results obtained confirmed the hypothesis of lysosome origin of silicasomes, due to the identification of various cathepsins together with silicateins and surprisingly we found many proteins similar to lysosome enzymes, several of them related to polysaccharide and sugar metabolism. Taken together, these results open a new perspective in the study of biosilica forming in demosponges, suggesting a more complex scenario where sugars and maybe lipids can have key roles.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.5281/zenodo.6033766.
Author Contributions
Conceptualization: MP and MG. Methodology: MP, IM, SG, CO, ET, and MG. Investigation: MP, IM, SG, CO, and ET. Writing: MP and MG. Review and editing: MG and ET. Supervision: MP and MG. Funding acquisition: MP, IM, and MG. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding
This work was partially supported by University of Genova funds to MP and MG. IM acknowledges funding from the Czech Science Foundation (grant number 20-03899S).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Hermann Ehrlich for the critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.850653/full#supplementary-material
References
Armirotti A., Damonte G., Pozzolini M., Mussino F., Cerrano C., Salis A., et al. (2009). Primary Structure and Post-Translational Modifications of Silicatein Beta From the Marine Sponge Petrosia Ficiformis (Poiret 1789). J. Proteome Res. 8, 3995–4004. doi: 10.1021/pr900342y
Bradford M. M. (1976). A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Analytical Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brinkmann C. M., Marker A., Kurtböke Dİ. (2017). An Overview on Marine Sponge-Symbiotic Bacteria as Unexhausted Sources for Natural Product Discovery. Diversity 9, 40. doi: 10.3390/d9040040
Brunet T., King N. (2017). The Origin of Animal Multicellularity and Cell Differentiation. Dev. Cell 43, 124–140. doi: 10.1016/j.devcel.2017.09.016
Cattaneo-Vietti R., Bavestrello G., Cerrano C., Sarà M., Benatti U., Giovine M., et al. (1996). Optical Fibres in an Antarctic Sponge. Nature 383, 397–398. doi: 10.1038/383397b0
Cha J. N., Shimizu K., Zhou Y., Christiansen S. C., Chmelka B. F., Stucky G. D., et al. (1999). Silicatein Filaments and Subunits From a Marine Sponge Direct the Polymerization of Silica and Silicones In Vitro. PNAS 96, 361–365. doi: 10.1073/pnas.96.2.361
Ehrlich H. (2010). Chitin and Collagen as Universal and Alternative Templates in Biomineralization. Int. Geology Rev. 52, 661–699. doi: 10.1080/00206811003679521
Ehrlich H., Brunner E., Simon P., Bazhenov V. V., Botting J. P., Tabachnick K. R., et al. (2011). Calcite Reinforced Silica–Silica Joints in the Biocomposite Skeleton of Deep-Sea Glass Sponges. Advanced Funct. Mater. 21, 3473–3481. doi: 10.1002/adfm.201100749
Ehrlich H., Deutzmann R., Brunner E., Cappellini E., Koon H., Solazzo C., et al. (2010). Mineralization of the Metre-Long Biosilica Structures of Glass Sponges Is Templated on Hydroxylated Collagen. Nat. Chem. 2, 1084–1088. doi: 10.1038/nchem.899
Ehrlich H., Janussen D., Simon P., Bazhenov V. V., Shapkin N. P., Erler C., et al. (2008). Nanostructural Organization of Naturally Occurring Composites–Part II: Silica-Chitin-Based Biocomposites. J. Nanomater. 2008, 670235. doi: 10.1155/2008/670235
Ehrlich H., Luczak M., Ziganshin R., Mikšík I., Wysokowski M., Simon P., et al. (2022). Arrested in Glass: Actin Within Sophisticated Architectures of Biosilica in Sponges. Adv. Sci. 2105059. doi: 10.1002/advs.202105059
Ehrlich H., Maldonado M., Parker A. R., Kulchin Y. N., Schilling J., Köhler B., et al. (2016). Supercontinuum Generation in Naturally Occurring Glass Sponges Spicules. Advanced Optical Mater. 4, 1608–1613. doi: 10.1002/adom.201600454
Ehrlich H., Wysokowski M., Żółtowska-Aksamitowska S., Petrenko I., Jesionowski T. (2018). Collagens of Poriferan Origin. Marine Drugs 16, 79. doi: 10.3390/md16030079
Erwin D. H., Laflamme M., Tweedt S. M., Sperling E. A., Pisani D., Peterson K. J. (2011). The Cambrian Conundrum: Early Divergence and Later Ecological Success in the Early History of Animals. Science 334, 1091–1097. doi: 10.1126/science.1206375
Fairhead M., Johnson K. A., Kowatz T., McMahon S. A., Carter L. G., Oke M., et al. (2008). Crystal Structure and Silica Condensing Activities of Silicatein α–Cathepsin L Chimeras. Chem. Commun. 15, 1765–1767. doi: 10.1039/B718264C
Gaiti F., Degnan B. M., Tanurdžić M. (2018). Long Non-Coding Regulatory RNAs in Sponges and Insights into the Origin of Animal Multicellularity. RNA Biol. 15, 696–702. doi: 10.1080/15476286.2018.1460166
Görlich S., Samuel A. J., Best R. J., Seidel R., Vacelet J., Leonarski F. K., et al. (2020). Natural Hybrid Silica/Protein Superstructure at Atomic Resolution. PNAS 117, 31088–31093. doi: 10.1073/pnas.2019140117
Götz W., Tobiasch E., Witzleben S., Schulze M. (2019). Effects of Silicon Compounds on Biomineralization, Osteogenesis, and Hard Tissue Formation. Pharmaceutics 11, 117. doi: 10.3390/pharmaceutics11030117
Hooper J. N. A., Van Soest R. W. M. (2002). “Class Demospongiae Solla,” in In Systema Porifera: A Guide to the Classification of Sponges;. Eds. Hooper J. N. A., Van Soest R. W. M., Willenz P. (Boston, MA: Springer US), pp 15–pp 51. doi: 10.1007/978-1-4615-0747-5_3
Jesionowski T., Norman M., Żółtowska-Aksamitowska S., Petrenko I., Joseph Y., Ehrlich H. (2018). Marine Spongin: Naturally Prefabricated 3d Scaffold-Based Biomaterial. Marine Drugs 16, 88. doi: 10.3390/md16030088
Khrunyk Y., Lach S., Petrenko I., Ehrlich H. (2020). Progress in Modern Marine Biomaterials Research. Marine Drugs 18, 589. doi: 10.3390/md18120589
Kinrade S. D., Del Nin J. W., Schach A. S., Sloan T. A., Wilson K. L., Knight C. T. G. (1999). Stable Five- and Six-Coordinated Silicate Anions in Aqueous Solution. Science 285, 1542–1545. doi: 10.1126/science.285.5433.1542
Kotzsch A., Pawolski D., Milentyev A., Shevchenko A., Scheffel A., Poulsen N., et al. (2016). Biochemical Composition and Assembly of Biosilica-Associated Insoluble Organic Matrices From the Diatom Thalassiosira Pseudonana. J. Biol. Chem. 291, 4982–4997. doi: 10.1074/jbc.M115.706440
Laemmli U. K. (1970). Cleavage of Structural Proteins During the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lévi C. (1963). Cytologie-Scléroblastes Et Spiculogenèse Chez Une Éponge Siliceuse. C. R. Hebd. Séances Acad. sci. 256, 497–498.
Maldonado M., López-Acosta M., Beazley L., Kenchington E., Koutsouveli V., Riesgo A. (2020). Cooperation Between Passive and Active Silicon Transporters Clarifies the Ecophysiology and Evolution of Biosilicification in Sponges. Sci. Adv. 6, 9322. doi: 10.1126/sciadv.aba9322
Matsumura H., Umezawa K., Takeda K., Sugimoto N., Ishida T., Samejima M., et al. (2014). Discovery of a Eukaryotic Pyrroloquinoline Quinone-Dependent Oxidoreductase Belonging to a New Auxiliary Activity Family in the Database of Carbohydrate-Active Enzymes. PloS One 9, e104851. doi: 10.1371/journal.pone.0104851.g002
Mikšík I., Paradis S., Eckhardt A., Sedmera D. (2018). Analysis of Siamese Crocodile (Crocodylus Siamensis) Eggshell Proteome. Protein J. 37, 21–37. doi: 10.1007/s10930-017-9750-x
Müller W. E. G., Belikov S. I., Tremel W., Perry C. C., Gieskes W. W. C., Boreiko A., et al. (2006). Siliceous Spicules in Marine Demosponges (Example Suberites Domuncula). Micron 37, 107–120. doi: 10.1016/j.micron.2005.09.003
Müller W. E. G., Eckert C., Kropf K., Wang X., Schloßmacher U., Seckert C., et al. (2007). Formation of Giant Spicules in the Deep-Sea Hexactinellid Monorhaphis Chuni (Schulze 1904): Electron-Microscopic and Biochemical Studies. Cell Tissue Res. 329, 363–378. doi: 10.1007/s00441-007-0402-x
Müller W. E. G., Mugnaioli E., Schröder H. C., Schloßmacher U., Giovine M., Kolb U., et al. (2013a). Hierarchical Composition of the Axial Filament From Spicules of the Siliceous Sponge Suberites Domuncula: From Biosilica-Synthesizing Nanofibrils to Structure- and Morphology-Guiding Triangular Stems. Cell Tissue Res. 351, 49–58. doi: 10.1007/s00441-012-1519-0
Müller W. E. G., Schröder H. C., Burghard Z., Pisignano D., Wang X. (2013b). Silicateins-A Novel Paradigm in Bioinorganic Chemistry: Enzymatic Synthesis of Inorganic Polymeric Silica. Chem. Eur. J. 19, 5790–5804. doi: 10.1002/chem.201204412
Müller W. E. G., Wang X., Chen A., Hu S., Gan L., Schröder H. C., et al. (2011). “The Unique Invention of the Siliceous Sponges: Their Enzymatically Made Bio-Silica Skeleton,” in Molecular Biomineralization: Aquatic Organisms Forming Extraordinary Materials;. Ed. Müller W. E. G. (Berlin, Heidelberg: Springer), pp 251–pp 281. doi: 10.1007/978-3-642-21230-7_9
Müller W. E. G., Wang X., Cui F. Z., Jochum K. P., Tremel W., Bill J., et al. (2009). Sponge Spicules as Blueprints for the Biofabrication of Inorganic–Organic Composites and Biomaterials. Appl. Microbiol. Biotechnol. 83, 397–413. doi: 10.1007/s00253-009-2014-8
Murr M. M., Morse D. E. (2005). Fractal Intermediates in the Self-Assembly of Silicatein Filaments. PNAS 102, 11657–11662. doi: 10.1073/pnas.0503968102
Musser J. M., Schippers K. J., Nickel M., Mizzon G., Kohn A. B., Pape C., et al. (2021). Profiling Cellular Diversity in Sponges Informs Animal Cell Type and Nervous System Evolution. Science 374, 717–723. doi: 10.1126/science.abj2949
Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988). Improved Staining of Proteins in Polyacrylamide Gels Including Isoelectric Focusing Gels With Clear Background at Nanogram Sensitivity Using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262. doi: 10.1002/elps.1150090603
Nielsen F. H. (2014). Update on the Possible Nutritional Importance of Silicon. J. Trace. Elem. Med. Biol. 28, 379–382.
Peled Y., Drake J. L., Malik A., Almuly R., Lalzar M., Morgenstern D., et al. (2020). Optimization of Skeletal Protein Preparation for LC–MS/MS Sequencing Yields Additional Coral Skeletal Proteins in Stylophora Pistillata. BMC Mater. 2, 8. doi: 10.1186/s42833-020-00014-x
Pisera A., Łukowiak M., Masse S., Tabachnick K., Fromont J., Ehrlich H., et al. (2021). Insights Into the Structure and Morphogenesis of the Giant Basal Spicule of the Glass Sponge Monorhaphis Chuni. Front. Zool. 18, 58. doi: 10.1186/s12983-021-00440-x
Povarova N. V., Barinov N. A., Baranov M. S., Markina N. M., Varizhuk A. M., Pozmogova G. E., et al. (2018b). Efficient Silica Synthesis From Tetra(Glycerol)Orthosilicate With Cathepsin- and Silicatein-Like Proteins. Sci. Rep. 8, 16759. doi: 10.1038/s41598-018-34965-9
Povarova N. V., Markina N. M., Baranov M. S., Barinov N. A., Klinov D. V., Kozhemyako V. B., et al. (2018a). A Water-Soluble Precursor for Efficient Silica Polymerization by Silicateins. Biochem. Biophys. Res. Commun. 495, 2066–2070. doi: 10.1016/j.bbrc.2017.12.075
Pozzolini M., Bruzzone F., Berilli V., Mussino F., Cerrano C., Benatti U., et al. (2012). Molecular Characterization of a Nonfibrillar Collagen From the Marine Sponge Chondrosia Reniformis Nardo 1847 and Positive Effects of Soluble Silicates on Its Expression. Mar Biotechnol. 14, 281–293. doi: 10.1007/s10126-011-9415-2
Pozzolini M., Gallus L., Ghignone S., Ferrando S., Candiani S., Bozzo M., et al. (2019). Insights Into the Evolution of Metazoan Regenerative Mechanisms: Roles of TGF Superfamily Members in Tissue Regeneration of the Marine Sponge Chondrosia Reniformis. J. Exp. Biol. 222, 207894. doi: 10.1242/jeb.207894
Pozzolini M., Mussino F., Cerrano C., Scarfì S., Giovine M. (2014). Sponge Cell Cultivation: Optimization of the Model Petrosia Ficiformis (Poiret 1789). J. Exp. Marine Biol. Ecol. 454, 70–77. doi: 10.1016/j.jembe.2014.02.004
Pozzolini M., Scarfì S., Gallus L., Ferrando S., Cerrano C., Giovine M. (2017). Silica-Induced Fibrosis: An Ancient Response From the Early Metazoans. J. Exp. Biol. 220, 4007–4015. doi: 10.1242/jeb.166405
Pozzolini M., Scarfì S., Ghignone S., Mussino F., Vezzulli L., Cerrano C., et al. (2016). Molecular Characterization and Expression Analysis of the First Porifera Tumor Necrosis Factor Superfamily Member and of Its Putative Receptor in the Marine Sponge Chondrosia Reniformis. Dev. Comp. Immunol. 57, 88–98. doi: 10.1016/j.dci.2015.12.011
Ramos-Silva P., Kaandorp J., Huisman L., Marie B., Zanella-Cléon I., Guichard N., et al. (2013). The Skeletal Proteome of the Coral Acropora Millepora: The Evolution of Calcification by Co-Option and Domain Shuffling. Mol. Biol. Evol. 30, 2099–2112. doi: 10.1093/molbev/mst109
Riesgo A., Farrar N., Windsor P. J., Giribet G., Leys S. P. (2014). The Analysis of Eight Transcriptomes From All Poriferan Classes Reveals Surprising Genetic Complexity in Sponges. Mol. Biol. Evol. 31, 1102–1120. doi: 10.1093/molbev/msu057
Riesgo A., Maldonado M., López-Legentil S., Giribet G. (2015). A Proposal for the Evolution of Cathepsin and Silicatein in Sponges. J. Mol. Evol. 80, 278–291. doi: 10.1007/s00239-015-9682-z
Ryckman A. E., Brockhausen I., Walia J. S. (2020). Metabolism of Glycosphingolipids and Their Role in the Pathophysiology of Lysosomal Storage Disorders. Int. J. Mol. Sci. 21, 6881. doi: 10.3390/ijms21186881
Schoeppler V., Reich E., Vacelet J., Rosenthal M., Pacureanu A., Rack A., et al. (2017). Shaping Highly Regular Glass Architectures: A Lesson From Nature. Sci. Adv. 3, 2047. doi: 10.1126/sciadv.aao2047
Schröder H. C., Boreiko A., Korzhev M., Tahir M. N., Tremel W., Eckert C., et al. (2006). Co-Expression and Functional Interaction of Silicatein With Galectin: Matrix-Guided Formation of Siliceous Spicules in the Marine Demosponge Suberites Domuncula. J. Biol. Chem. 281, 12001–12009. doi: 10.1074/jbc.M512677200
Schröder H. C., Natalio F., Shukoor I., Tremel W., Schloßmacher U., Wang X., et al. (2007). Apposition of Silica Lamellae During Growth of Spicules in the Demosponge Suberites Domuncula: Biological/Biochemical Studies and Chemical/Biomimetical Confirmation. J. Struct. Biol. 159, 325–334. doi: 10.1016/j.jsb.2007.01.007
Schröder H. C., Perović-Ottstadt S., Rothenberger M., Wiens M., Schwertner H., Batel R., et al. (2004). Silica Transport in the Demosponge Suberites Domuncula: Fluorescence Emission Analysis Using the PDMPO Probe and Cloning of a Potential Transporter. Biochem. J. 381, 665–673. doi: 10.1042/BJ20040463
Sebé-Pedrós A., Chomsky E., Pang K., Lara-Astiaso D., Gaiti F., Mukamel Z., et al. (2018). Early Metazoan Cell Type Diversity and the Evolution of Multicellular Gene Regulation. Nat. Ecol. Evol. 2, 1176–1188. doi: 10.1038/s41559-018-0575-6
Şen E. H., Ide S., Bayari S. H., Hill M. (2016). Micro- and Nano-Structural Characterization of Six Marine Sponges of the Class Demospongiae. Eur. Biophys. J. 45, 831–842. doi: 10.1007/s00249-016-1127-0
Shevchenko A., Tomas H., Havli J., Olsen J. V., Mann M. (2006). In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 1, 2856–2860. doi: 10.1038/nprot.2006.468
Shimizu K., Amano T., Bari M. R., Weaver J. C., Arima J., Mori N. (2015). Glassin, a Histidine-Rich Protein From the Siliceous Skeletal System of the Marine Sponge Euplectella, Directs Silica Polycondensation. PNAS 112, 11449–11454. doi: 10.1073/pnas.1506968112
Shimizu K., Cha J., Stucky G. D., Morse D. E. (1998). Silicatein α: Cathepsin L-Like Protein in Sponge Biosilica. PNAS 95, 6234–6238. doi: 10.1073/pnas.95.11.6234
Shimizu K., Del Amo Y., Brzezinski M. A., Stucky G. D., Morse D. E. (2001). A Novel Fluorescent Silica Tracer for Biological Silicification Studies. Chem. Biol. 8, 1051–1060. doi: 10.1016/s1074-5521(01)00072-2
Shimizu K., Morse D. E. (2018). “Chapter Fourteen - Silicatein: A Unique Silica-Synthesizing Catalytic Triad Hydrolase From Marine Sponge Skeletons and Its Multiple Applications,” in Methods in Enzymology, vol. Vol. 605 . Ed. Moore B. S.. Cambridge. Massachussets USA: (Academic Press), pp 429–pp 455. Available at: http://doi.org/10.1016/bs.mie.2018.02.025.
Simpson T. L. (1984). The Cell Biology of Sponges. Ed. Simpson T. L. (New York, NY: Springer), 216–254. Available at: https://doi.org/10.1007/978-1-4612-5214-6_5.
Staudt C., Puissant E., Boonen M. (2017). Subcellular Trafficking of Mammalian Lysosomal Proteins: An Extended View. Int. J. Mol. Sci. 18, 47. doi: 10.3390/ijms18010047
Thompson D. B., Daniel W. L. (1988). Comparative Biochemistry of Mammalian Arylsulfatases A and B. Comp. Biochem. Physiol. B 90, 823–831. doi: 10.1016/0305-0491(88)90340-9
Valisano L., Bavestrello G., Giovine M., Arillo A., Cerrano C. (2006). Seasonal Production of Primmorphs From the Marine Sponge Petrosia Ficiformis (Poiret 1789) and New Culturing Approaches. J. Exp. Marine Biol. Ecol. 337, 171–177. doi: 10.1016/j.jembe.2006.06.030
Valisano L., Pozzolini M., Giovine M., Cerrano C. (2012). “Biosilica Deposition in the Marine Sponge Petrosia Ficiformis (Poiret 1789): The Model of Primmorphs Reveals Time Dependence of Spiculogenesis,” in Ancient Animals, New Challenges: Developments in Sponge Research. Eds. Maldonado M., Turon X., Becerro M., Uriz M. J. (Dordrecht: Springer Netherlands), 259–273. doi: 10.1007/978-94-007-4688-6_22
Vis B. M., Wen J., Mellerup S. K., Merchant R. D., Mawhinney R. C., Kinrade S. D. (2020). Silicon Forms a Rich Diversity of Aliphatic Polyol Complexes in Aqueous Solution. J. Am. Chem. Soc. 142, 9188–9202. doi: 10.1021/jacs.9b10701
Wang X., Wiens M., Schröder H. C., Schloßmacher U., Pisignano D., Jochum K. P., et al. (2011). Evagination of Cells Controls Bio-Silica Formation and Maturation During Spicule Formation in Sponges. PloS One 6 (6), e20523. doi: 10.1371/journal.pone.0020523
Wiens M., Bausen M., Natalio F., Link T., Schlossmacher U., Müller W. E. G. (2009). The Role of the Silicatein-α Interactor Silintaphin-1 in Biomimetic Biomineralization. Biomaterials 30, 1648–1656. doi: 10.1016/j.biomaterials.2008.12.021
Keywords: marine sponges, Petrosia ficiformis, biosilica, spicules, lysosomes, silicatein, cathepsins
Citation: Pozzolini M, Mikšík I, Ghignone S, Oliveri C, Tassara E and Giovine M (2022) The Lysosome Origin of Biosilica Machinery in the Demospongiae Model Petrosia ficiformis (Poiret, 1789). Front. Mar. Sci. 9:850653. doi: 10.3389/fmars.2022.850653
Received: 08 January 2022; Accepted: 14 March 2022;
Published: 13 April 2022.
Edited by:
Rebecca Metzler, Colgate University, United StatesReviewed by:
Manuel Maldonado, Center for Advanced Studies of Blanes (CSIC), SpainMaria Costantini, Anton Dohrn Zoological Station, Italy
Katsuhiko Shimizu, Tottori University, Japan
Copyright © 2022 Pozzolini, Mikšík, Ghignone, Oliveri, Tassara and Giovine. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Giovine, bWdpb3ZpbmVAdW5pZ2UuaXQ=
 Marina Pozzolini
Marina Pozzolini Ivan Mikšík
Ivan Mikšík Stefano Ghignone
Stefano Ghignone Caterina Oliveri
Caterina Oliveri Eleonora Tassara
Eleonora Tassara Marco Giovine
Marco Giovine