Abstract
Symbiotic scyphozoan jellyfish are poorly understood in terms of their symbionts and traits, as well as the ecological significance of this association. Dinoflagellate symbionts of the medusae Cotylorhiza tuberculata, Phyllorhiza punctata, and Cassiopea xamachana collected in the Mediterranean Sea and Cabo Frio (Rio de Janeiro, Brazil) were phylogenetically identified based on 28S rDNA and ITS2 haplotypes. The studied medusae harbour only one phylotype of symbionts in a time, but scyphozoan jellyfishes can associate with several types of symbionts. This study confirmed that the main symbionts of investigated scyphozoans belong to the genera Symbiodinium, Philozoon, and Breviolum. The associations between dinoflagellate symbionts and Cotylorhiza tuberculata changed from year to year, hosting Philozoon one year and Breviolum another. Invasive species in the Mediterranean Sea Phyllorhiza punctata harboured dinoflagellate symbionts of genus Symbiodinium as in the native areal. Pigment analysis of two shallow-water symbiont species Breviolum sp. and Philozoon medusarum revealed characteristic profiles for each genus.
Introduction
The mutualistic association between symbiotic dinoflagellates (with trivial name zooxanthellae) and corals, which form the basis of all shallow-water coral reefs on Earth, is one of the most widely studied examples of such a relationship (LaJeunesse, 2020). The biogeography of both corals and their symbionts, as well as the phylogeny of those symbionts, has been extensively studied in recent decades (Baker, 2003; Stat et al., 2006; LaJeunesse et al., 2018). In contrast, less attention has been paid to the association between scyphozoan jellyfishes and their symbionts. This association provide an important key traits as mixotrophic way of nutrition, because zoxanthellate medusae are holobionts and derive their nutrition from predation and photosynthesis. Possession of dinoflagellate symbionts in polyps is rarely necessary for surviving of polyps, but notable trait is a key role of symbionts in the life cycle of the jellyfish by allowing or facilitating strobilation of polyps to secure medusa with suitable symbionts (Djeghri et al., 2019). Of the 79 valid genera in Scyphozoa, only 11 harbour symbionts: Linuche, Nausithoe (Coronamedusae), Bazinga, Cephea, Cassiopea, Cotylorhiza, Netrostoma, Mastigias, Phyllorhiza, Thysanostoma, and Versugia, most belonging to the suborder Kolpophorae and all of which with a metagenic life cycle (Djeghri et al., 2019). The best-studied genus among the zooxanthellate Scyphozoa is Cassiopea, with its symbionts serving as a holobiont model to reveal various aspects of their mutualism (Lampert, 2016; Ohdera et al., 2018). The symbionts of the species Cotylorhiza tuberculata (Macri, 1779) and Phyllorhiza punctata (von Lendenfeld, 1884) have not yet been described in detail (LaJeunesse et al., 2021). The fried egg jellyfish Cotylorhiza tuberculata is one of the rare symbiont-bearing scyphozoans from temperate latitudes, distributed in the Mediterranean Sea and around the Canary Islands (Collins et al., 2021). It is predominantly distributed in oligotrophic environments; however, it can also occur in eutrophic areas, as in the Mar Menor Lagoon in the Balearic Sea (Pérez-Ruzafa et al., 2002). Phyllorhiza punctata is an invasive species introduced into the eastern Mediterranean Sea in the 1990s from oligotrophic tropical seas from the south-central coast of eastern Australia (Galil et al., 1990; Galil et al., 2009). Cassiopea xamachana (Bigelow, 1892) is sedentary in oligotrophic, shallow waters and is distributed in the Gulf of Mexico, the Caribbean Sea, and warmer areas of the western Atlantic Ocean (Verde & McCloskey, 1998). While the symbionts of Cassiopea xamachana have already been studied in terms of their mode of transmission (Thornhill et al., 2006; Mellas et al., 2014; Ohdera et al., 2018) and parasitic potential (Sachs & Wilcox, 2006), the symbionts of Cotylorhiza tuberculata and Phyllorhiza punctata have not yet been characterised in detail (LaJeunesse et al., 2021).
The dinoflagellate family Symbiodiniaceae is characterised by a rich genetic diversity within evolutionary divergent lineages (LaJeunesse et al., 2018; LaJeunesse et al., 2021). Members of this family are symbionts in foraminifera, ciliates, poriforans, cnidarians and molluscs (Venn et al., 2008; Hansen and Daugbjerg, 2009). The taxonomy of the flourishing algae of the family Symbiodiniaceae have recently been resolved by the 28S rDNA and ITS2 regions (LaJeunesse et al., 2018; LaJeunesse et al., 2021) which provide sufficient resolution to species delimitation (Moestrup and Daugbjerg, 2007). In details, the 28S rDNA ribosomal markers and the ITS2 regions of nuclear and chloroplast (cp23S region domain V) DNA have a phylogenetic signal that can resolve clades at the species level (Savage et al., 2002; Casado-Amezúa et al., 2014; LaJeunesse et al., 2018). Both markers (28S rDNA and cp23S region domain V) have similar levels of resolution (Sampayo et al., 2009), although the 28S rDNA region has been widely used to assign clades and phylotypes to zooxanthellae isolates and infer relationships between them (Coffroth and Santos, 2005; Barbrook et al., 2006). ITS markers are more commonly used to obtain phylogenetic resolution at the subclade level, especially at the species level (LaJeunesse, 2001; Rodriguez-Lanetty et al., 2001; Meron et al., 2012; Grajales et al., 2016). The relevance of ITS2 is based on the different secondary structure of ITS2 in the specific clades B, C, F and H (Meron et al., 2012). LaJeunesse et al. (2018) published a phylogenetic analysis of the Symbiodiniaceae (previously assigned Symbiodinium clades from A to H), interpreted the evolution of the group, and provided a redescription of clades A, B, and C into new genera. The former clade A is recognised now as the genus Symbiodinium (Hansen and Daugbjerg, 2009), with Symbiodinium natans as the type species (LaJeunesse et al., 2018). The most studied zooxanthellate jellyfish is Cassiopea xamachana and the majority of symbiont phylotypes associated with Cassiopea xamachana belong to the genera Symbiodinium (previously clade A, especially phylotype A1) (Thornhill et al., 2006; Lampert et al., 2012), Breviolum (previously clade B with common phylotype B1), or Cladocopium (previously clade C with phylotype C3) (LaJeunesse et al., 2003; LaJeunesse et al., 2018). The former phylotype “Temperate A”, which occurs in a variety of hosts from temperate zones, was recently redescribed as Philozoon (Geddes, 1882), containing several species, most closely related to the genus Symbiodinium (LaJeunesse et al., 2021).
Previous studies on mutualistic association between Symbiodiniaceae members pointed out that some of its members are host generalists (Baker, 2003; LaJeunesse et al., 2018), while others are thought to be specialists due to their rarity (LaJeunesse et al., 2004a; LaJeunesse et al., 2004b). Host specialists spread through preferential vertical transmission, while host generalists are transmitted horizontally from the pool of free-living cells into the host (Fabina et al., 2012). Horizontal transmission gives the host the opportunity to acquire locally adapted algal cells (Van Oppen, 2004), thus increasing its fitness to occupy available niches and respond to environmental changes (Bongaerts et al., 2015). This assumption is based on the high genetic diversity of species within the genera of Symbiodiniaceae, which supports their functional diversity as light harvesting and utilization under variable conditions (Stat et al., 2008; see review Suggett et al., 2008). In vertical transmission (Fabina et al., 2012), symbiont diversification is maintained by high genetic plasticity and twinning when the symbiont is isolated from the external population (Sachs & Wilcox, 2006). Meanwhile, horizontally transmitted symbionts are translocated into the host in each generation, limiting the possibility of coevolution.
In the present study, we used a phylogenetic approach using nuclear 28S rDNA and ITS2 markers to identify symbionts in Cotylorhiza tuberculata and Phyllorhiza punctata from the Mediterranean Sea during blooming period and Cassiopea xamachana collected at Cabo Frio (Rio de Janeiro, Brazil). In addition, symbionts were isolated from Cotylorhiza tuberculata and Cassiopea xamachana and cultivated symbiotic cells were used in cloning experiment to reveal the diversity of the ITS2 region of the dinoflagellate cells within individual host medusa. Photosynthetic pigments of two different genera of symbionts from Cotylorhiza tuberculata were characterised.
Material and Methods
Sampling Area
A total of 123 medusae of Cotylorhiza tuberculata were collected in five different locations in the Mediterranean Sea [Gulf of Trieste, Lake Mljet (Adriatic Sea), Vlicho Bay (Ionian Sea), and Mar Menor Lagoon (Balearic Sea)]; seven medusae of Phyllorhiza punctata were also collected in Vlicho Bay. Medusae were collected between November 2009 and September 2013. Three medusae of Cassiopea xamachana were collected from the coast of Cabo Frio (Rio de Janeiro, Brazil) in September 2012 (see map on Figure 1 and Table S1 in Supplementary Material for details on sampling locations). Symbionts were isolated from live medusae by scraping cells from the subumbrella and oral arms immediately after collection. Part of sample was stored for pigment analysis and another part was stored in cryotubes in 96% ethanol and kept at -80°C for DNA extraction.
Figure 1
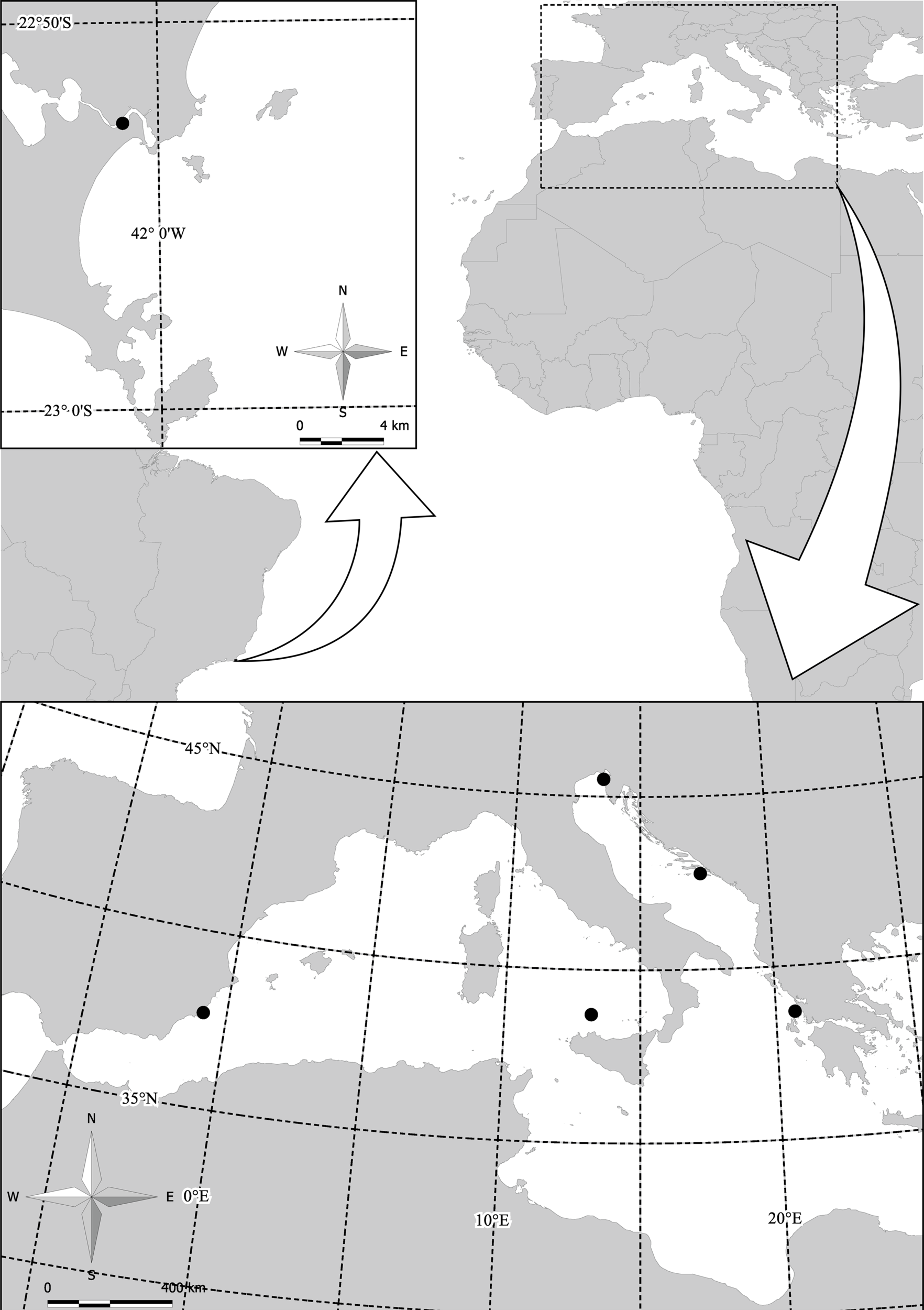
Sampling sites of Cotylorhiza tuberculata and Phylorhiza punctata in the Mediterranean Sea and the sampling site of Cassiopea xamachana from the coast of Cabo Frio (Rio de Janeiro, Brazil).
Isolation and Cultivation of Symbionts From Scyphozoan Hosts
Symbionts were isolated for cultivation from three medusae of Cotylorhiza tuberculata collected in the Gulf of Trieste (Mediterranean Sea) and from one individual of Cassiopea xamachana collected from the coast of Cabo Frio (Rio de Janeiro, Brazil). Tissue samples from Cotylorhiza tuberculata (subumbrella) and Cassiopea xamachana (subumbrella and oral arms) were minced and isolated under a dissecting microscope using a micropipette and then transferred to sterile growth medium B (Agatha et al., 2004). They were washed by re-isolating them twice in sterile growth medium. Following the last passage, an antibiotic mixture (working concentration: 50 mg L-1 kanamycin, 50 mg L-1 streptomycin, and 100 mg L-1 penicillin G) was added and the cultures were transferred to an antibiotic-free medium after four days. A strain obtained from Cassiopea xamachana (strain T4) was isolated without antibiotics to test for a possible selective effect on the cultivation of Symbiodiniaceae strains when adding antibiotics (Santos et al., 2001). In this case, washing of about 200 to 300 cells with fresh sterile medium was continued twice a day for three days. The strains grew best between 20°C and 25°C with a 12 h/12 h light/dark cycle. 28S rDNA and ITS2 markers were amplified from the cultures of the symbionts and used for phylogenetic analysis.
Extraction of DNA, PCR Amplification, and Sequencing of Nuclear Ribosomal Markers From Symbionts
DNA was extracted from scrapings of symbionts living in the oral arms and subumbrella of Cotylorhiza tuberculata, Phyllorhiza punctata and Cassiopea xamachana, as well as from cultured symbionts isolated from Cotylorhiza tuberculata and Cassiopea xamachana. A CTAB-based DNA kit (E.Z.N.A., Omega Bio-Tek, USA) was used for DNA extraction according to the protocol. The symbiont cells removed from the host were stored at -80°C, thawed on ice and ethanol evaporated in a vacuum concentrator before DNA extraction.
The 28S rDNA of the symbionts was amplified with dinoflagellate-specific primers (28Forward: 5’- CCC GCTGAATTTAAGCATATAAGTAAGCGG -3’ and 28Reverse: 5’- GTTAGACTCCTTGGTCCGTGT TTCAAGA -3’) designed by Zardoya and colleagues (Zardoya et al., 1995) at position 26 onward, and reverse primers at position 741 containing the variable domain D1 and D2. The length of the amplified 28S rDNA fragments was approximately 630 base pairs. The major components of the PCR mixture were added at the following concentrations: 0.625 unit TopTaq polymerase (Qiagen), 2 mM MgCl2, 0.05 µg µL-1 bovine serum albumin, and 10 ng DNA µL-1 in 25 µL PCR. The thermal profile included an initial denaturation at 94°C and an annealing temperature of 57°C for a total of 30 amplification cycles. Due to the low amplification efficiency of some samples, re-amplification with a further 25 cycles at an annealing temperature of 60°C was required; 5 µL of the PCR products were used as template, and the final reagent concentrations were the same as for the previous PCR.
Amplification of ITS2 from symbionts was performed using a dinoflagellate-specific primer (ITSintfor2 5’ GAATTGCAGAACTCCGTG-3’), which annealed to a conserved region of the 5.8S rDNA, and the Chlorophyta-specific reverse primer (ITSreverse 5’- GGGATCCATA TGCTTAAGTTCAGCGGGT -3), as described by LaJeunesse (2002). The length of the amplified fragments was between 300 and 330 base pairs. The optimal concentrations for PCR were 0.625 units of GoTaq polymerase (Promega, USA), 2 mM MgCl2, and up to 10 ng of DNA in a volume of 20 µL. The thermal profile of the touch-up PCR began with an initial denaturation step, followed by annealing at a starting temperature of 52°C for 40 seconds, which was then increased to 61°C, with subsequent annealing at this temperature for a further 20 cycles. In some cases, re-amplification was required. For this, 5 µL of the PCR mixture was transferred to a new tube, to which the PCR reagents were added at the same concentration as the first PCR and amplified at an annealing temperature of 54°C for a further 30 cycles. Sanger sequencing was performed at the commercial supplier Macrogen (The Netherlands) using the same primer pairs as the PCR.
Cloning of ITS2 Region From Cotylorhiza tuberculata
Our aim was to reveal the diversity of the ITS2 region within the ribosomal operon of the symbiont harboured by Cotylorhiza tuberculata collected in November 2009 from the Mar Menor Lagoon (Balearic Sea) to verify mixed or homogeneous infection by symbionts in individual medusa. DNA extraction and ITS2 amplification were performed as described above. The PCR product (fragment length 330 base pairs) was cloned into chemically competent One Shot Top 10 cells from the TA Cloning Kit (Invitrogen, USA) according to the manufacturer’s instructions. A total of 273 transformants were screened, of which 184 white transformants were transferred to Luria-Bertani medium with 10% glycerol and allowed to grow overnight before plasmid extraction and Sanger sequencing.
Phylogenetic Analysis
Consensus sequences were generated from both strands using Chromas Pro 1.7.6 (Technelysium, Australia) and manually checked for ambiguous bases. Sequences from our study were compared to those deposited in GenBank by BLAST, and the most similar sequences were added to the dataset for alignment. 28S rDNA and ITS2 sequences were aligned separately using MAFFT v. 7 (Katoh & Standley, 2013); specifically, sequences were aligned according to the clade to which they belonged based on similarity by BLAST, applying a strategy used in a previous study (Correa & Baker, 2009). Subsequently, all groups were aligned together, gaps were removed, and sequences were trimmed to the shortest sequence. Identical haplotypes were checked in both datasets using DAMBE (Xia, 2013), and only unique haplotypes were used for phylogenetic analysis. The substitution models were calculated using jModeltest 2.1.1 (Darriba et al., 2012), which resulted in TIM3+I+G for 28S rDNA and HKY + G for ITS2 datasets. Phylogenetic analyses were performed using MrBayes ver. 3.2.1 (Ronquist and Huelsenbeck, 2003) for 28S rDNA and ITS2 datasets separately. Calculations were performed using 2 000 000 generations with four chains, and a 25% burn-in of the trees and stationarity of the calculations were checked using Tracer (Rambaut et al., 2018) for 28S rDNA and for ITS2 rDNA, respectively. The calculated trees of 28S and ITS2 were visualised in FigTree 1.4.2 (Rambaut, 2014).
Pigment Analysis
Symbionts were isolated from live medusae by scraping cells from the subumbrella and oral arms from Cotylorhiza tuberculata (sampling sites Mar Menor and Gulf of Trieste), immediately after collection. Symbiont samples were then stored in cryotubes at -80°C until analysis. Phylogenetic analysis revealed that samples of symbionts harboured by Cotylorhiza tuberculata belongs to Breviolum sp. (collected in Mar Menor in 2010) and Philozoon medusarum (collected in the Gulf of Trieste, Adriatic Sea, in 2011). Photosynthetic pigments of the endosymbionts were determined using high-performance liquid chromatography (reversed-phase HPLC) (Mantoura and Llewellyn, 1983; Barlow et al., 1993). Samples of the isolated endosymbionts were extracted by sonication in 90% acetone and centrifuged at 4 000 rpm for 10 min to remove particles. A mixture (1:1) of clarified extract and 1 mol L-1 ammonium acetate was injected into the HPLC system (1260 Infinity, Agilent Technologies) equipped with a 3 µm C18 reversed-phase column (Pecosphere, 35x4.5 mm, Perkin Elmer) to determine the composition of photosynthetic pigments (chlorophylls and carotenoids) in the endosymbionts. Chlorophylls and carotenoids were detected by absorbance at 440 nm using a Diode Array Detector (DAD; Agilent Technologies, model 1290 Infinity).
Results
Cultivation of Symbionts Isolated From Host Medusae
Three cultures (T1, T2, and T3) of symbionts were isolated from three individuals of Cotylorhiza tuberculata, and two cultures (T4 and T5) were isolated from two individuals of Cassiopea xamachana. During cultivation, the organisms were mostly attached to the culture vessel in an immobile form and underwent up to two divisions within the same cyst. Motile dinoflagellate cells were formed almost exclusively during the light phase of the light-dark cycle by one or two binary divisions. The motile forms of the different strains were indistinguishable from one another under light microscopy. Symbionts from all cultures isolated from Cassiopea xamachana and Cotylorhiza tuberculata harboured symbionts of Breviolum sp. as revealed by phylogenetic analysis (see Figure 2 and Figure S1). Strains T1 and T5 are available in the Collection of Sea Microorganisms (CoSMi) at the Istituto Nazionale di Oceanografia e di Geofisica Sperimentale under strain numbers 1062 and 1065, respectively.
Figure 2
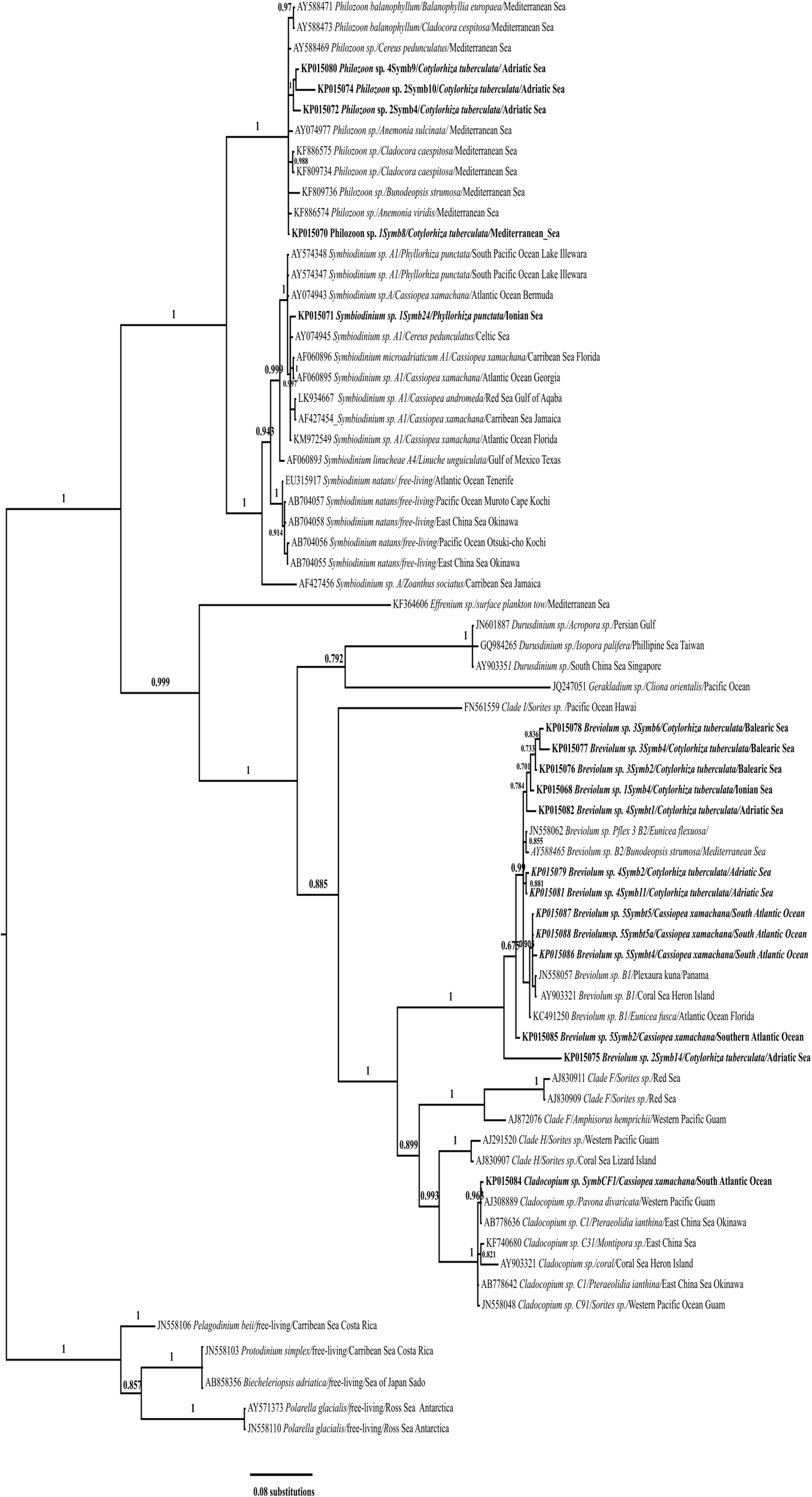
Bayesian inference based on 28S rDNA sequences of symbionts from Cotylorhiza tuberculata, Phylorhiza punctata, and Cassiopea xamachana under the TIM3+I+G model. Numbers at the nodes indicate posterior probabilities. Labels indicate GenBank accession number, genus name, isolate name, host, and sampling site; sequences from this study are in bold.
Sequence Analysis and Phylogenetic Inference of Symbionts 28S rDNA and ITS2
28S rDNA was amplified (D1/D2 hypervariable region) from symbionts of Cotylorhiza tuberculata, Phyllorhiza punctata and Cassiopea xamachana and from cultures of symbionts derived from Cotylorhiza tuberculata (sequences KP015124*, KP015125* and KP015126*) collected in the Gulf of Trieste and from two medusae of Cassiopea xamachana (sequence KP015128* from T4 culture, KP015130* from culture T5 and KP015130* from culture T5A) collected in Cabo Frio (Rio de Janeiro, Brazil). The dataset of 28S rDNA for phylogenetic analysis consisted of 72 sequences (18 unique sequences from this study and 46 sequences from GenBank) of species representing the genera Symbiodinium, Breviolum, Cladocopium, Durusdinium, Effrenium, Fugacium, Gerakladium, and clades H and I as defined by LaJeunesse et al. (2018). The recently redescribed genus Philozoon with the species Philozoon medusarum (Geddes, 1882) was also included in the analysis (see Table S2 in Supplementary Material for details of the sequences used). The outgroup consisted of the following species: Pelagodinium beii (JN558106), Protodinium simplex (JN558103), Biecheleriopsis adriatica (AB858356), and Polarella glacialis (AY571373 and JN558110). The dataset for Symbiodinium contained 29 sequences (five sequences from this study and 24 of the most similar sequences). The dataset for Breviolum contained 17 sequences (12 sequences from this study and five of the most similar sequences), and the dataset for Cladocopium consisted of Cassiopea xamachana sequence KP015084 and six of the most similar sequences. Table S2 shows the full list of sequences and details, and Figure 2 depicts the Bayesian inference tree based on 28S rDNA sequences. Phylogenetic analysis of the 28S rDNA revealed that the symbionts of Cotylorhiza tuberculata, Phyllorhiza punctata, and Cassiopea xamachana belong to the genera Symbiodinium, Philozoon, Breviolum, and Cladocopium. In particular, the sequences of symbionts from Cotylorhiza tuberculata pertain to Philozoon and Breviolum, while those from Phyllorhiza punctata were placed in the genus Symbiodinium; 12 sequences of symbionts from Cotylorhiza tuberculata and Cassiopea xamachana were placed in Breviolum, and one sequence of symbionts from Cassiopea xamachana was placed in Cladocopium (KP015084). Sequence KP015071, belonging to a symbiont of Phyllorhiza punctata, belongs to Symbiodinium sp. type A1, which was recently characterised as Symbidinium microadriaticum (LaJeunesse, 2017). In addition, two sequences (AY574348 and AY574347) of symbionts of Phyllorhiza punctata collected in Australia are in the same group, indicating the wide distribution of this symbiont type (see Figure 2).
Several sequences belong to the genus Philozoon and originate from samples of Cotylorhiza tuberculata collected in the Adriatic Sea: Gulf of Trieste (KP015080), Lake Mljet (KP015074, KP015072), and the Ionian Sea (KP015070), but not from the Balearic Sea. Medusae of Cotylorhiza tuberculata from the Balearic Sea (KP015078, KP015077, and KP015076), Ionian Sea (KP015068), and Adriatic Sea (KP015082, KP015079, and KP015081) harboured symbionts of Breviolum, especially those of type B2. In addition, the symbionts of Cassiopea xamachana collected in Cabo Frio (Rio de Janeiro, Brazil) pertain to this group (KP015087, KP015088, and KP015086). All of the groups mentioned have high bootstrap support of the nodes. Other sequences within the groups are derived from symbionts from a variety of hosts, including corals, anemones, scyphozoans, and free-living species (see Table S2 for more details).
The phylogenetic tree for the ITS2 dataset consisted of sequences belonging to species of the genus Symbiodinium (type species Symbiodinium natans (Hansen & Daugbjerg, 2009) and the still unclassified groups A1, A2, A3, A4), the genus Philozoon with the species Philozoon medusarum and Philozoon actiniarum (Geddes, 1882), the genus Breviolum (type species Breviolum minutum (LaJeunesse et al., 2018) and two other unclassified groups, B1 and B2); Polarella glacialis (JN558110) was used as an outgroup. Table 3 (in Supplementary Material) shows the full list of sequences analysed. The ITS2 sequences of symbionts from Cotylorhiza tuberculata samples collected in the Vlicho Bay, Lefkada (Ionian Sea) and the Adriatic Sea (Gulf of Trieste and Lake Mljet) belong to Philozoon medusarum and the Breviolum group, respectively. Furthermore, symbionts collected in samples of Cotylorhiza tuberculata from the Mar Menor pertain only to the Breviolum group. Several symbionts were found in medusae of Cotylorhiza tuberculata from the Gulf of Trieste, the most common being members of genus Breviolum (type B2), though members of genus Philozoon were also found in 2010 and 2011. Symbionts from Phyllorhiza punctata (KP015090 and KP015089) collected in the Ionian Sea were identified as Symbiodinium sp. (type A1). The symbionts of Cassiopea xamachana were identified as Breviolum sp., specifically type B1 (KP015088, KP015087, KP015086). We found that Philozoon medusarum forms a symbiosis with Cotylorhiza tuberculata and anthozoan Cladocora caespitosa (MG991827, KF886573), both collected in the Gulf of Trieste. Members of this large group can form symbioses with various hosts around the Mediterranean Sea. The same group also includes the ITS2 sequences of symbionts from the anthozoan Paranemonia cinerea (Contarini, 1844) from the Mar Menor Lagoon, Balanophyllia europaea (Risso, 1826) from Ischia, and Anemonia viridis (Forsskål, 1775) from the Balearic Sea and the Algerian Basin.
The diversity of the ITS2 operon within the host medusae Cotylorhiza tuberculata was demonstrated using a clone library: among the 20 sequenced clones, we found 16 unique ITS2 haplotypes. Unique ITS2 haplotypes from the clone library were included in the phylogenetic analysis under accession numbers KP015094 – KP015106, KP015109, KP015111, KP015114, and KP015115 (see Table S3). All haplotypes were nearly identical and therefore assigned to the genus Breviolum (see Figure S1).
Pigment Analysis
We estimated the contribution of four different pigments (chlorophyll c2, peridinin, diadinoxanthin, and β,β-carotene) with and without chlorophyll a (Table 1 and Figure 3). Dinoflagellate symbionts collected from two geographically distant populations of Cotylorhiza tuberculata differed in the contribution of pigments (chlorophyll a, chlorophyll c2 and peridinin), while their contributions of diadinoxanthin and β,β-carotene did not differ. In the Mar Menor samples, the contribution of peridinin (38.7% ± 1.5) was greater than in the Gulf of Trieste samples (15.2% ± 2.2), while the latter samples had a greater contribution of chlorophyll a and chlorophyll c2 (Figure 3).
Table 1
| Mar Menor (Balearic Sea) | Gulf of Trieste (North Adriatic Sea) | |||
|---|---|---|---|---|
| with chl a | without chl a | with chl a | without chl a | |
| chlorophyll a | 25-29 % | 36-47 % | ||
| chlorophyll c2 | 23-26 % | 32-36 % | 27-41 % | 50-65 % |
| peridinin | 37-41 % | 52-55 % | 13-18 % | 20-31 % |
| diadinoxanthin | 8-9 % | 11-12 % | 7-9 % | 10-17 % |
| β,β-carotene | 1 % | 1% | 1-2 % | 2-3 % |
Contribution of four different pigments (chlorophyll c2, peridinin, diadinoxanthin, and β,β-carotene) with and without chlorophyll a.
Discussion
We identified symbionts in the scyphomedusae Cotylorhiza tuberculata, Phyllorhiza punctata and Cassiopea xamachana by phylogenetic analyses of 28S rDNA and ITS2 haplotypes as these markers provide sufficient resolution to species delimitation (Moestrup et al., 2007) and have recently been used to resolve the evolution of the flourishing algae of the family Symbiodiniaceae (LaJeunesse et al., 2018; LaJeunesse et al., 2021). In the present study, medusae of Cotylorhiza tuberculata collected in the Mediterranean Sea (Adriatic, Ionian, and Balearic Seas) were confirmed to harbour symbionts of the genus Philozoon or the genus Breviolum. In more details, medusae of Cotylorhiza tuberculata collected at Mar Menor (Balearic Sea) harbour only symbiotic dinoflagellate cells of Breviolum. We also confirmed presence of Breviolum in Cassiopea xamachana (type B1), while Breviolum (type B2) was found in Cotylorhiza tuberculata (Gulf of Trieste). Type B2 is common in temperate latitudes and tolerates a wide range of temperature, light, and other conditions (photosynthetic optimum at 25°C and minimum at 10°C), being able to recover quickly from low temperatures (Thornhill et al., 2006). In this regard, cultures offer the possibility to work on the free-living stage of the species, which are essential for a valid description of dinoflagellate species. Symbiotic dinoflagellates are difficult to isolate from a host in axenic culture (Liu et al., 2017). Therefore, antibiotics are used by default during the cultivation, as it is almost impossible not to transfer the organic matter of the host inhabited with other symbionts and commensals together with symbiotic cells (Agatha et al., 2004). Bacteria that grow on the host’s organic matter usually attack the symbionts as well. We have only managed to isolate one strain without antibiotics. In addition, there are many other conditions that influence the success or failure of isolating dinoflagellates, for example: salinity, light intensity and medium used (Agatha et al., 2004). This means that we could overlooked other types of symbionts. However, we would like to emphasise that the identity of isolated strains and symbionts taken from the host prior the cultivation were confirmed by phylogenetic analysis during study. But no distinctive features at light microscopic level could be found for the isolated strains. For additional morphological information a careful examination at ultrastructural level (SEM) of the different clades and species is indicated as an important completion of the genetic, physiological, and ecological data cited above.
Medusae of the non-native species Phyllorhiza punctata collected in the Mediterranean Sea (Ionian Sea) harbour symbionts of Symbiodinium microadriaticum (type A1), as confirmed in our study. Moreover, our analysis confirmed the presence of the same type of symbionts as in the native range of Phyllorhiza punctata. The sequences of the symbionts from the Mediterranean Sea, together with the sequences AY574347 and AY574348 (from Lake Illawarra, New South Wales, Australia), are combined in a 28S phylogenetic tree (see Figure 2), indicating the widespread occurrence of Symbiodinium microadriaticum. Closely related to the symbionts of Phyllorhiza punctata are those of Cotylorhiza tuberculata (see Figure 2 and Tables S2 and S3 in Supplementary Material), now classified as the genus Philozoon (Geddes, 1882) (LaJeunesse et al., 2021). In our 28S rDNA phylogenetic tree, all sequences of Philozoon are grouped together, though the sequences of Philozoon in the ITS2 tree revealed finer structure into two subgroups (see Figure S1 in Supplementary Material). The first subgroup contained sequences isolated from anemone and coral hosts (Paranemonia and Cladocora), a pattern that has pointed to more specialised group of symbionts (Meron et al., 2012). The other subgroup is formed by Philozoon medusarum from a variety of hosts such as Cotylorhiza tuberculata and hydrozoan species like Anemonia viridis, with the mean distance within this group measured as 0.007 ± 0.003, indicating very little difference between haplotypes of ITS2 (Raspor Dall’Olio, 2016). Philozoon includes earlier groups with the trivial names “A temperate” and “Mediterranean A” (Hunter et al., 2007), both groups are now reclassified into several species (LaJeunesse et al., 2021). They are identified in many hosts (Savage et al., 2002; Lee et al., 2015) with a known range in the Mediterranean Sea and circumglobally in temperate latitudes (Visram et al., 2006; Casado-Amezúa et al., 2014). Well known hosts are the marine snail Pteraeolidia ianthina from South Australia (Loh et al., 2006) and the anemone Anthopleura hermaphroditica along the coast of New Zealand (Howe, 2013).
We noted that the abundant Mediterranean blooming species Cotylorhiza tuberculata can establish a symbiosis with species of Breviolum or Philozoon (see Figures 2, S1). However, the symbiosis in individual medusae only develops with one species at a time, as confirmed for Cotylorhiza tuberculata (Thornhill et al., 2006; Astorga et al., 2012; Newkirk et al., 2018). This is an important indication of the co-existence of free-living species and subsequent selection of symbionts in polyps, with the best-adapted symbionts subsequently transferred by strobilation in ephyra (Astorga et al., 2012; Newkirk et al., 2018). Polyps play a crucial role in infection with symbionts and can be infected with several types of them at the same time, leading to the selection of the most suitable symbiont (Ohdera et al., 2018). To investigate the diversity of the ITS2 haplotypes of dinoflagellate symbionts within the single host medusa Cotylorhiza tuberculata we constructed a clone library from amplified ITS2 region to gain insight into the diversity of ITS2 haplotypes. Cloning of ITS2 haplotypes was performed in a subset of the samples to check for the possibility that more than one type of symbiont is present in the host medusa, which could compromise the analysis and be a source of chimaeras. In addition, cloning all samples would be very tedious and time-consuming (preparation of clone libraries, selection of positive colonies and sequence analysis). The results show that all cloned ITS2 haplotypes form the same phylogenetic group within Breviolum and the differences were negligible and do not reveal differences over the species level. Unique ITS2 haplotypes from the clone library were included into the phylogenetic analysis (see Table S3 in the Supplementary Material).
Successful acquisition of symbionts at the polyp stage also affects the efficiency of strobilation, and in some species (e.g., Cassiopea xamachana), only symbiotic scyphystomae strobilate (Newkirk et al., 2018). Moreover, the efficiency of strobilation depends on the phylotype of the symbiont (cf. Mellas et al., 2014). All of the species investigated in the present study have a metagenic life that includes a scyphistoma, strobilation reproduction, and adult medusae, which reproduce sexually and produce planulae that develop into polyps. According to experimental models, Cassiopea polyps show extreme flexibility in the types of symbionts they acquire from free-living species in their external environment (Mellas et al., 2014; Lampert, 2016; Newkirk et al., 2018). Free-living species of Symbiodiniaceae provide the largest reservoir of symbiotic cells (Coffroth et al., 2006; Pochon & Gates, 2010; Newkirk et al., 2018). Cotylorhiza tuberculata reproduces sexually in the Gulf of Trieste in late August and early September, and polyps are grown from planulae within a few days (A. Ramšak, personal observation). Moreover, environmental conditions like sea temperature during this period are conducive to symbiont infection (hot summer, high light intensity vs colder summer with fewer sunny days). When the polyps acquire symbionts in a temperate climate in a shallow Mediterranean coastal environment is still unknown, though two possibilities emerge: i) immediately after the transformation of planulae into polyps that survive the winter period with colder temperatures, or ii) the polyps acquire the symbionts after the winter. Furthermore, symbiosis is not solely required for strobilation: other factors (e.g., increased temperatures) also trigger strobilation (Prieto et al., 2010; Astorga et al., 2012). Kikinger (1992) observed that the planulae were asymbiotic in a stationary population of Cotylorhiza tuberculata from the Ionian Sea and that the scyphystomae already had symbionts. Besides, the author found that the scyphystomae strobilated only if they had symbionts. In the laboratory, asymbiotic polyps did not strobilate for years and only reproduced by budding (maintaining the population of polyps that can acquire symbionts), and that strobilation occurred after they were fed on tissues of Anemonia sulcata with their symbionts (Kikinger, 1992). During ephyra development, symbionts are clustered in the mesoglea and are particularly numerous along the endodermal lining of the gastrovascular system, indicating their importance for feeding (Kikinger, 1992).
Breviolum psygmophilum has been recorded in Cotylorhiza tuberculata from the Adriatic Sea (Gulf of Trieste), an environment known for its marked temperature variability with minimum values around 8°C and maximum values around 26°C (Boicourt et al., 2021). Most evidence of seasonal and geographical distribution of cnidarian symbionts is available from anthozoan hosts such as Bunodeopsis strumosa (Visram et al., 2006) and Oculina patagonica (Casado-Amezúa et al., 2014). Breviolum psygmophilum is widespread in the temperate latitudes of the western Atlantic Ocean and the Mediterranean Sea (Thornhill et al., 2008; LaJeunesse et al., 2012). We have confirmed that Cassiopea xamachana from the coast of Cabo Frio (Rio de Janeiro, Brazil) harbours symbionts of the genera Breviolum and Cladocopium. Several symbionts harboured by Cassiopea xamachana in different parts of the world (Florida, Hawaii, Bermuda, Japan, and Australia) have been identified as Breviolum minutum (AF333511, JN602457, HQ317740, and AF184940), Breviolum antillogorgium (formerly Symbiodinium antillogorgium KT149341), Breviolum endomadracis (formerly Symbiodinium endomadracis KT149342), and Breviolum pseudominutum (formerly Symbiodinium pseudominutum KT149344). In addition, Symbiodinium type A1 was detected in Cassiopea xamachana and Cassiopea andromeda from the Gulf of Mexico and the Red Sea (see Figure 2). The reported diversity of symbionts of Cassiopea xamachana in Mexican coral reefs was much higher than the present study, and symbiosis with several species of the genus Symbiodinium, Breviolum and Cladocopium has been confirmed (Zardoya et al., 1995; LaJeunesse, 2001; Garcia-Cuetos et al., 2005). Previous studies have found that Symbiodinium type A1 is most likely to be harboured by Cassiopea xamachana in the adult phase, which has been sampled in Puerto Morelos (LaJeunesse, 2002), Florida (LaJeunesse, 2001; Thornhill et al., 2006), the Bahamas (LaJeunesse et al., 2009), Bermuda and the Caribbean (Rowan & Powers, 1991). Furthermore, even clade F symbionts have been detected in Cassiopea xamachana (Santos et al., 2002). Several conditions influence the acquisition of symbionts and their selection in the host. Benthic corals or anemones have different symbionts at the same time e.g., mixed infection with Philozoon sp. and Breviolum psygmophylum detected in Cladocora caespitosa from the island of Ischia (Meron et al., 2012), while scyphozoans have only one type of symbiont. Although exclusive host specificity is very rare (Fabina et al., 2012; Silverstein et al., 2012), some anthozoan hosts are capable of associating with a limited number of species, as in the case of Plesiastrea versipora, which hosts only Cladocopium (Rodriguez-Lanetty et al., 2001) and Echinophyllia aspera (LaJeunesse et al., 2004a). However, other studies have confirmed instances of multiple infections in corals (Coffroth and Santos, 2005; Bongaerts et al., 2011). The fitness of the symbiosis can be optimised in host animals if the symbiont is transferred vertically from the mother to the daughter of the host animal, such that the partners become specialist symbionts (Fabina et al., 2012).
We found only one symbiont species in the host medusae at a time, although many host species in coral reefs are capable of symbiosis with more than one species of Symbiodiniaceae. Mosaicism in coral colonies is a well-known process that depends on local light conditions (Thornhill et al., 2006). Their different environmental preferences have also been noted in cultures and during cultivation (Santos et al., 2001). Such ecological adaptation (mosaicism) is not necessary for individual medusae due to their motile lifestyle. However, adaptation to highly fluctuating light conditions and transition between heterotrophic and mixotrophic states is advantageous. A recent study investigated the adaptability and invasion potential of the holobiont Cassiopea sp. (Mammone et al., 2021). Cassiopea polyps were infected by Cladocopium sp., which showed efficient photosynthetic plasticity at both low and high irradiance as well as during sudden changes in light exposure. Both partners are under strong constraints that lead to the selection of certain morphological, biochemical, and physiological traits (Mammone et al., 2021). The optimisation of photosynthesis is a clear example of these constraints. The dilemma between optimal light exposure and low UV radiation requires balancing and adaptations by both partners.
The advantage of scyphozoan jellyfish with symbiotic medusae is the increase in fitness through physiological changes and their ability to switch between mixotrophic and heterotrophic states. Symbiotic scyphozoans are polytrophic (planktivorous, ingest particulate food, capable of autotrophy) because they grow very fast and have high nutrient requirements to ensure rapid medusa growth (Pitt et al., 2005). Algal endosymbionts accelerate the excretory process of their hosts by recycling ammonium-rich animal wastes (Cates & McLaughlin, 1976) and contributing to the host’s energy requirements through the direct transfer of algal photosynthates (Hofmann & Kremer, 1981). Symbiotic species such as Mastigias sp. probably have little predatory influence on zooplankton and co-occur with the zooplanktivorous Aurelia in the saltwater lakes of Palau (Hamner & Hauri, 1981), while blooms of Cotylorhiza tuberculata also co-exist with zooplanktivorous jellyfish such as Rhizostoma pulmo (Pestorić et al., 2021).
The stability of cnidarian-algal symbiosis depends on symbionts photosystem and on protective role of host mediated by colour pigments, which influence photosynthetic activity in endosymbionts by either providing the photosystem with irradiance of appropriate wavelength or protecting it from excessive and potentially harmful light (Lampert et al., 2012). The main aim of this study was the identification of the symbionts, while the analysis of pigments content and their ratio in the genera Breviolum and Philozoon, which are common symbionts of Cotylorhiza tuberculata from the Adriatic Sea and the Balearic Islands, especially the Mar Menor lagoon, provides valuable complementary data. Details on this part of symbiotic association between symbiotic dinoflagellate cells and Cotylorhiza tuberculata are scarce and limited on analysis of composition and ratio of pigments. Recent advance in taxonomy of Symbiodiniaceae enable to connect knowledge from photobiology with identified symbiont species, in our study case being Cotylorhiza tuberculata and its symbionts Breviolum and Philozoon. The pigment profile of dinoflagellate symbionts Breviolum and Philozoon had a high content of chlorophyll c2, peridinin, and diadinoxanthin, as well as chlorophyll a. The differences observed in the proportions of chlorophyll a, chlorophyll c2, and peridinin were notable between dinoflagellate cells of Breviolum and Philozoon hosted by Cotylorhiza tuberculata collected in two different environments. In details, a higher proportion of peridinin was detected in dinoflagellate cells of Breviolum from Cotylorhiza tuberculata collected at the Mar Menor Lagoon and a higher proportion of chlorophyll c2 was detected in cells of Philozoon from Cotylorhiza tuberculata at the Gulf of Trieste (see Figure 3). Pigment analyses have revealed differences between both genera, and according to the study of Enrique-Navarro et al. (2022), the pigment ratio does not change between free-living symbionts and the cells living “in hospite”, and in this regard we think that the number of samples analysed is sufficient to confirm differences between both symbiotic genera. The recent study provide evidence that the light environment influence on differences in the amount of pigments as chl a, chl c2 and peridinin (Enrique-Navarro et al., 2022) and dinoflagellate cells have adaptations enable them to survive in variable light conditions and intensity as PCPs are light-collecting complexes made of chl a and peridinin, on the other side host provide stable and more protected environment for symbionts. We collected Cotylorhiza tuberculata specimens in two different environments: Mar Menor, a shallow coastal lagoon (4 m average depth), and Gulf of Trieste, the northernmost part of the Mediterranean Sea (15 m average depth). However, the optimal depth for holobiont Cotylorhiza tuberculata can be affected by light penetration, water turbidity, seasonal light intensity, and cloud cover. Therefore, zooxanthellate jellyfish will respond to changes in the vertical water column by selecting a depth that matches their optimal physiological tolerance and maximises photosynthesis (Enrique-Navarro et al., 2022). We pointed out the higher peridinin content in symbionts from Mar Menor. Symbionts therefore occur in both pigmented and non-pigmented host tissues. As reported by Enrique-Navarro et al. (2022) the dinoflagellate symbionts are heterogeneously distributed within the host body being more concentrated in the medusa oral arms as a consequence of the tissue role, their high metabolic activity and their sinuous folding. This study also reveal that the endosymbionts revealed no significant differences in size neither chlorophyll a concentration within the oral arms and the umbrella.
Figure 3
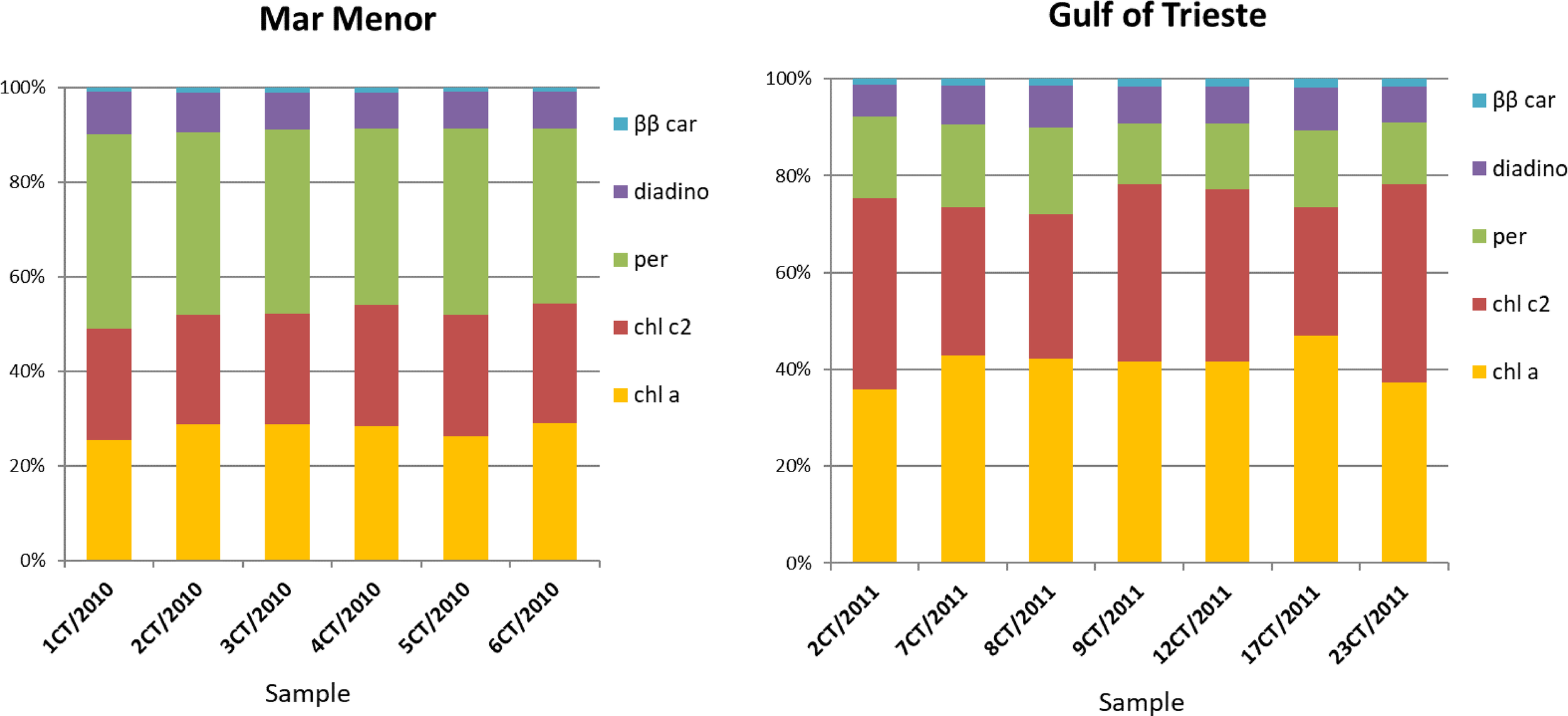
Contribution of five different pigments (chlorophyll a, chl a; chlorophyll c2, chl c2; peridinin, per; diadinoxanthin, diadino; β,β-carotene, ββ car) in the symbionts identified as Breviolum sp. (Mar Menor, Balearic Sea; October 2010) and Philozoon medusarum (Gulf of Trieste, Adriatic Sea, August 2011) isolated from Cotylorhiza tuberculata.
Conclusions
Symbionts in medusae of Cotylorhiza tuberculata, Phyllorhiza punctata, and Cassiopea xamachana collected in the Mediterranean Sea and from the coast of Cabo Frio (Rio de Janeiro, Brazil) were identified by haplotypes of 28S rDNA and ITS2. We confirmed that the predominant symbionts in the investigated scyphozoan jellyfishes Cotylorhiza tuberculata belong to genera Breviolum and Philozoon, while non-indigenous species Phyllorhiza punctata harbour Symbiodinium. Cassiopea xamachana hosted Breviolum and Cladocopium symbiotic dinoflagellates. The individual medusae studied harbour only one phylotype of symbionts at a time as confirmed with detailed analysis of the ITS2 region within the ribosomal operon of the symbiont, although symbionts in zooxanthellate jellyfish can be different between years as we confirmed in Cotylorhiza tuberculata from the eastern Mediterranean Sea. The population of medusae from Mar Menor hosted only symbionts of Breviolum, while Breviolum and Philozoon were present in medusae from the Adriatic Sea. Furthermore, the possession of zooxanthellae is a unique feature of scyphozoans jellyfish that allows them to occupy several niches compared to non-zooxanthellate species due to mixotrophic way of nutrition.
Funding
The work of A. Ramšak, A. Malej, and V. Flander-Putrle was funded by the ARRS Research Program P1-0237 (Marine Coastal Research) and by the bilateral cooperation between Slovenia and Brazil (agreements BI-BR /10-12-005) and project RI-SI-2 LifeWatch financed by Ministry of Education, Science and Sport of Slovenia and the European Regional Development Fund. The work of L. Raspor Dall’Olio was funded by the ARRS grant for young researchers (MR -33223).
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
Haplotypes from this study are deposited in GeneBank under accession numbers listed in Tables S2, S3 in the Supplementary Material.
Author contributions
Conceptualisation, AR and AM. Data analysis, AR, LD’O, and VF-P. Writing, AR, AB, VF-P, LD’O and AM. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Andre C. Morandini, Sergio Stampar, Laura Prieto, Reinhard and Lili Kikinger, G. Aglieri, Valentina Turk, and Massimo Avian, who kindly provided samples for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.867554/full#supplementary-material
References
1
Agatha S. Strüder-Kypke M. C. Beran A. (2004). Morphologic and Genetic Variability in the Marine Planktonic Ciliate Laboea Strobila Lohmann 1908 (Ciliophora, Oligotrichia), With Notes on its Ontogenesis. J. Eukaryot. Microbiol.51, 267–281. doi: 10.1111/j.1550-7408.2004.tb00567.x
2
Astorga D. Ruiz J. Prieto L. (2012). Ecological Aspects of Early Life Stages of Cotylorhiza Tuberculata (Scyphozoa: Rhizostomae) Affecting its Pelagic Population Success. Hydrobiologia690, 141–155. doi: 10.1007/s10750-012-1036-x
3
Baker A. C. (2003). Flexibility and Specificity in Coral-Algal Symbiosis: Diversity, Ecology, and Biogeography of Symbiodinium. Annu. Rev. Ecol. Syst.34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
4
Barbrook A. C. Visram S. Douglas A. E. Howe C. J. (2006). Molecular Diversity of Dinoflagellate Symbionts of Cnidaria: The psbA Minicircle of Symbiodinium. Protist157, 159–171. doi: 10.1016/j.protis.2005.12.002
5
Barlow R. G. Mantoura R. F. C. Gough M. A. Fileman T. W. (1993). Pigment Signatures of the Phytoplankton Composition in the Northeastern Atlantic During the 1990 Spring Bloom. Deep-Sea Res.40 (1/2), 459–477. doi: 10.1016/0967-0645(93)90027-K
6
Boicourt W. C. Ličer M. Li M. Vodopivec M. Malačič V. (2021). “Sea State: Recent Progress in the Context of Climate Change. In Coastal Ecosystems in Transition,” in A Comparative Analysis of the Northern Adriatic and Chesapeake Bay, vol. 256 . Eds. MaloneT. C.MalejA.FaganeliJ. (Hoboken, NY, USA: Geophysical Monograph). doi: 10.1002/9781119543626.ch3
7
Bongaerts P. Carmichael M. Hay K. B. Tonk L. Frade P. R. Hoegh-Guldberg O. (2015). Prevalent Endosymbiont Zonation Shapes the Depth Distributions of Scleractinian Coral Species. R. Soc Open. Sci.2, 140297. doi: 10.1098/rsos.140297
8
Bongaerts P. Sampayo E. M. Bridge T. C. L. Ridgway T. Vermeulen F. Englebert N. et al . (2011). Symbiodinium Diversity in Mesophotic Coral Communities on the Great Barrier Reef: A First Assessment. Mar. Ecol. Prog. Ser.439, 117–126. doi: 10.3354/meps09315
9
Casado-Amezúa P. Machordom A. Bernardo J. González-Wangüemert M. (2014). New Insights Into the Genetic Diversity of Zooxanthellae in Mediterranean Anthozoans. Symbiosis63, 41–46. doi: 10.1007/s13199-014-0286-y
10
“Cassiopea and its zooxanthellae.” In The Cnidaria, Past, Present and Future: The World of Medusa and Her Sisters (eBook), EdsS. GoffredoDubinskyZ. (Switzerland: Springer International Publishing AG), 415–423.
11
Cates N. McLaughlin J. J. (1976). Differences of Ammonia Metabolism in Symbiotic and Aposymbiotic Condylactus and Cassiopea Spp. J. Exp. Mar. Biol. Ecol.21, 1–5. doi: 10.1016/0022-0981(76)90065-4
12
Coffroth M. A. Lewis C. F. Santos S. R. Weaver J. L. (2006). Environmental Populations of Symbiotic Dinoflagellates in the Genus Symbiodinium can Initiate Symbioses With Reef Cnidarians. Curr. Biol.16, R985–R987. doi: 10.1016/j.cub.2006.10.049
13
Coffroth M. A. Santos S. R. (2005). Genetic Diversity of Symbiotic Dinoflagellates in the Genus Symbiodinium. Protist156, 19–34. doi: 10.1016/j.protis.2005.02.004
14
Collins A. G. Jarms G. Morandini A. C. (2021) World List of Scyphozoa. Cotylorhiza Tuberculata (Macri 1778). Available at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=135297on2021-12-26.
15
Correa A. M. S. Baker A. C. (2009). Understanding Diversity in Coral-Algal Symbiosis: A Cluster-Based Approach to Interpreting Fine-Scale Genetic Variation in the Genus Symbiodinium. Coral Reefs28, 81–93. doi: 10.1007/s00338-008-0456-6
16
Darriba D. Taboada G. L. Doallo R. R. Posada D. (2012). Jmodeltest 2: More Models, New Heuristics and Parallel Computing. Nat. Method9, 772. doi: 10.1038/nmeth.2109
17
Djeghri N. Pondaven P. Stibor H. Dawson M. N. (2019). Review of the Diversity, Traits, and Ecology of Zooxanthellate Jellyfishes. Mar. Biol.166, 1–19. doi: 10.1007/s00227-019-3581-6
18
Enrique-Navarro A. Huertas E. Flander-Putrle V. Bartual A. Navarro G. Ruiz J. et al . (2022). Living Inside a Jellyfish: The Symbiosis Case Study of Host-Specialized Dinoflagellates, “Zooxanthellae”, and the Scyphozoan Cotylorhiza Tuberculata. Front. Mar. Sci.9. doi: 10.3389/fmars.2022.817312
19
Fabina N. S. Putnam H. M. Franklin E. C. Stat M. Gates R. D. (2012). Transmission Mode Predicts Specificity and Interaction Patterns in Coral-Symbiodinium Networks. PloS One7, e44970. doi: 10.1371/journal.pone.0044970
20
Galil B. S. Shoval L. Goren M. (2009). Phyllorhiza Punctata Von Lendenfeld 1884 (Scyphozoa: Rhizostomeae: Mastigiidae) Reappeared Off the Mediterranean Coast of Israel. Aquat Invasions4, 381–389. doi: 10.3391/ai.2009.4.3.6
21
Galil B. S. Spanier E. Ferguson W. W. (1990). The Scyphomedusae of the Mediterranean Coast of Israel, Including Two Lessepsian Migrants New to the Mediterranean. Zool. Med.64, 95–105.
22
Garcia-Cuetos L. Pochon X. Pawlowski J. (2005). Molecular Evidence for Host–Symbiont Specificity in Soritid Foraminifera. Protist156, 399–412. doi: 10.1016/J.PROTIS.2005.08.003
23
Grajales A. Rodríguez E. Thornhill D. J. (2016). Patterns of Symbiodinium Spp. Associations Within the Family Aiptasiidae, a Monophyletic Lineage of Symbiotic of Sea Anemones (Cnidaria, Actiniaria). Coral Reefs35, 345–355. doi: 10.1007/s00338-015-1352-5
24
Hamner W. M. Hauri I. R. (1981). Effects of Island Mass: Water Flow and Plankton Pattern Around a Reef in the Great Barrier Reef Lagoon, Australia. Limnol. Oceanogr.26, 1084–1102. doi: 10.4319/lo.1981.26.6.1084
25
Hansen G. Daugbjerg N. (2009). Symbiodinium Natans Sp. Nov.: A “Free-Living” Dinoflagellate From Tenerife (Northeast-Atlantic Ocean) 1. J. Phycol45, 251–263. doi: 10.1111/j.1529-8817.2008.00621.x
26
Hofmann D. K. Kremer B. P. (1981). Carbon Metabolism and Strobilation in Cassiopea Andromeda (Cnidaria: Scyphozoa): Significance of Endosymbiotic Dinoflagellates. Mar. Biol.65, 25–33. doi: 10.1007/BF00397064
27
Howe J. N. (2013). The Genetic and Physiological Characteristics of the Symbiodinium Spp. In the Endemic Anemone Anthopleura Aureoradiata. [Master’s Thesis] (Wellington, Australia: Victoria University of Wellington).
28
Hunter R. L. LaJeunesse T. C. Santos S. R. (2007). Structure and Evolution of the rDNA Internal Transcribed Spacer (ITS) Region 2 in the Symbiotic Dinoflagellates (Symbiodinium, Dinophyta). J. Phycol43, 120–128. doi: 10.1111/j.1529-8817.2006.00309.x
29
Katoh K. Standley D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol.30, 772–780. doi: 10.1093/molbev/mst010
30
Kikinger R. (1992). Cotylorhiza Tuberculata (Cnidaria: Scyphozoa) - Life History of a Stationary Population.Mar. Ecol13, 333–362. doi: 10.1111/j.1439-0485.1992.tb00359.x
31
LaJeunesse T. C. (2001). Investigating the Biodiversity, Ecology, and Phylogeny of Endosymbiotic Dinoflagellates in the Genus Symbiodinium Using the ITS Region: In Search of a “Species” Level Marker. J. Phycol37, 866–880. doi: 10.1046/j.1529-8817.2001.01031.x
32
LaJeunesse T. C. (2002). Diversity and Community Structure of Symbiotic Dinoflagellates From Caribbean Coral Reefs. Mar. Biol.141, 387–400. doi: 10.1007/s00227-002-0829-2
33
LaJeunesse T. C. (2020). Zooxanthellae. Curr. Biol.30, R1110–R1113. doi: 10.1016/j.cub.2020.03.058
34
LaJeunesse T. C. Bhagooli R. Hidaka M. Done T. Schmidt G. W. Fitt W. K. (2004b). Closely Related Symbiodinium Spp. Differ in Relative Dominance in Coral Reef Host Communities Across Environmental, Latitudinal and Biogeographic Gradients. Mar. Ecol. Prog. Ser.284, 147–161. doi: 10.3354/meps284147
35
LaJeunesse T. C. Loh W. Trench R. K. (2009). Do Introduced Endosymbiotic Dinoflagellates “Take” to New Hosts? Biol. Invasions11, 995–1003. doi: 10.1007/s10530-008-9311-5
36
LaJeunesse T. C. Loh W. K. Van Woesik R. Hoegh-Guldberg O. Schmidt G. W. Fitt W. K. (2003). Low Symbiont Diversity in Southern Great Barrier Reef Corals, Relative to Those of the Caribbean. Limnol. Oceanogr.48, 2046–2054. doi: 10.4319/lo.2003.48.5.2046
37
LaJeunesse T. C. Parkinson J. E. Gabrielson P. W. Jeong H. J. Reimer J. D. Voolstra C. R. et al . (2018). Systematic Revision of Symbiodiniaceae Highlights the Antiquity and Diversity of Coral Endosymbionts. Curr. Biol.28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
38
LaJeunesse T. C. Parkinson J. E. Reimer J. D. (2012). A Genetics–Based Description ofSymbiodinium Minutum Sp. Nov. And S. Psygmophilum Sp. Nov. (Dinophyceae), Two Dinoflagellates Symbiotic With Cnidaria. J. Phycol.48, 1380–1391. doi: 10.1111/j.1529-8817.2012.01217.x
39
LaJeunesse T. C. Thornhill D. J. Cox E. F. Stanton F. G. Fitt W. K. Schmidt G. W. (2004a). High Diversity and Host Specificity Observed Among Symbiotic Dinoflagellates in Reef Coral Communities From Hawaii. Coral Reefs23, 596–603. doi: 10.1007/s00338-004-0428-4
40
LaJeunesse T. C. Wiedenmann J. Casado-Amezúa P. D’Ambra I. Turnham K. E. Nitschke M. R. et al . (2021). Revival of Philozoon Geddes for Host-Specialized Dinoflagellates, ‘Zooxanthellae’, in Animals From Coastal Temperate Zones of Northern and Southern Hemispheres. Eur. J. Phycol.00, 1–15. doi: 10.1080/09670262.2021.1914863
41
Lampert K. P. (2016). ““Cassiopea and its Zooxanthellae.”,” in The Cnidaria, Past, Present and Future: The World of Medusa and Her Sisters. Eds. GoffredoS.DubinskyZ. (Springer International Publishing), 415–423.
42
Lee S. Y. Jeong H. J. Kang N. S. Jang T. Y. Jang S. H. Lajeunesse T. C. (2015). Symbiodinium Tridacnidorum Sp. Nov., a Dinoflagellate Common to Indo-Pacific Giant Clams, and a Revised Morphological Description of Symbiodinium Microadriaticum Freudenthal, Emended Trench & Blank. Eur. J. Phycol.50, 155–172. doi: 10.1080/09670262.2015.1018336
43
Liu C.-H. Place A. R. Jagus R. (2017). Use of Antibiotics for Maintenance of Axenic Cultures of Amphidinium Carterae for the Analysis of Translation. Mar. Drugs15, 242. doi: 10.3390/md15080242
44
Loh W. K. W. Cowlishaw M. Wilson N. G. (2006). Diversity of Symbiodinium Dinoflagellate Symbionts From the Indo-Pacific Sea Slug Pteraeolidia Ianthina (Gastropoda: Mollusca). Mar. Ecol. Prog. Ser.320, 177–184. doi: 10.3354/meps320177
45
Mammone M. Ferrier-Pagés C. Lavorano S. Rizzo L. Piraino S. Rossi S. (2021). High Photosynthetic Plasticity may Reinforce Invasiveness of Upside-Down Zooxanthellate Jellyfish in Mediterranean Coastal Waters. PloS One16, e0248814. doi: 10.1371/journal.pone.0248814
46
Mantoura R. F. C. Llewellyn C. A. (1983). "The Rapid Determination of Algal Chlorophyll and Carotenoid Pigments and Their Breakdown Products in Natural Waters by Reverse-Phase High-Performance Liquid Chromatography.". Anal. Chim. Acta151, 297–314. doi: 10.1016/S0003-2670(00)80092-6
47
Mellas R. E. McIlroy S. E. Fitt W. K. Coffroth M. A. (2014). Variation in Symbiont Uptake in the Early Ontogeny of the Upside-Down Jellyfish, Cassiopea Spp. J. Exp. Mar. Biol. Ecol.459, 38–44. doi: 10.1016/j.jembe.2014.04.026
48
Meron D. Rodolfo-Metalpa R. Cunning R. Baker A. C. Fine M. Banin E. (2012). Changes in Coral Microbial Communities in Response to a Natural pH Gradient. ISME J.6, 1775–1785. doi: 10.1038/ismej.2012.19
49
Moestrup O. Daugbjerg N. Taylor and Francis Group (2007). “On Dinoflagellate Phylogeny and Classification,” in Unravelling the Algae: The Past, Present, and Future of Algal Systematics. Eds. BrodieJ.LewisJ.. (Boca Raton: CRC Press). 215–230. doi: 10.1201/9780849379901
50
Newkirk C. R. Frazer T. K. Martindale M. Q. (2018). Acquisition and Proliferation of Algal Symbionts in Bleached Polyps of the Upside-Down Jellyfish, Cassiopea Xamachana. J. Exp. Mar. Biol. Ecol.508, 44–51. doi: 10.1016/j.jembe.2018.08.010
51
Ohdera A. H. Abrams M. J. Ames C. L. Baker D. M. Suescún-Bolívar L. P. Collins A. G. et al . (2018). Upside-Down But Headed in the Right Direction: Review of the Highly VersatileCassiopea Xamachana System. Front. Ecol. Evol.6. doi: 10.3389/fevo.2018.00035
52
Pérez-Ruzafa A. Gilabert J. Gutiérrez J. M. Fernández A. I. Marcos C. Sabah S. (2002). Evidence of a Planktonic Food Web Response to Changes in Nutrient Input Dynamics in the Mar Menor Coastal Lagoon, Spain. Hydrobiologia475–476, 359–369. doi: 10.1023/A:1020343510060
53
Pestorić B. Lučić D. Bojanić N. Vodopivec M. Kogovšek T. Violić I. et al . (2021). Scyphomedusae and Ctenophora of the Eastern Adriatic: Historical Overview and New Data. Diversity13, 186. doi: 10.3390/d13050186
54
Pitt K. A. Koop K. Rissik D. (2005). Contrasting Contributions to Inorganic Nutrient Recycling by the Co-Occuring Jellyfishes, Catostylus Mosaicus and Phyllorhiza Punctata (Scyphozoa, Rhizostomeae). J. Exp. Mar. Biol. Ecol.315, 71–86. doi: 10.1016/j.jembe.2004.09.007
55
Pochon X. Gates R. D. (2010). A New Symbiodinium Clade (Dinophyceae) From Soritid Foraminifera in Hawai’i. Mol. Phylogenet. Evol.56, 492–497. doi: 10.1016/j.ympev.2010.03.040
56
Prieto L. Astorga D. Navarro G. Ruiz J. (2010). Environmental Control of Phase Transition and Polyp Survival of a Massive-Outbreaker Jellyfish. PloS One5, e13793. doi: 10.1371/journal.pone.0013793
57
Rambaut A. (2014) FigTree V1.4.2, A Graphical Viewer of Phylogenetic Trees. Available at: https://github.com/rambaut/figtree/releases.
58
Rambaut A. Drummond A. J. Xie D. Baele G. Suchard M. A. (2018). Posterior Summarisation in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol.67, 901–904. doi: 10.1093/sysbio/syy032
59
Raspor Dall’Olio L. (2016). Symbiosis Ecology of Selected Scyphozoa (Nova Gorica: Doctoral dissertation, University of Nova Gorica). Available at: https://repozitorij.ung.si/IzpisGradiva.php?id=2622&lang=eng&prip=rup:9058647:d4.
60
Rodriguez-Lanetty M. Loh W. Carter D. Hoegh-Guldberg O. (2001). Latitudinal Variability in Symbiont Specificity Within the Widespread Scleractinian Coral Plesiastrea Versipora. Mar. Biol.138, 1175–1181. doi: 10.1007/s002270100536
61
Ronquist F. Huelsenbeck J. P. (2003). MRBAYES 3: Bayesian Phylogenetic Inference Under Mixed Models. Bioinformatics19, 1572–1574. doi: 10.1093/bioinformatics/btg180
62
Rowan R. Powers D. A. (1991). A Molecular Genetic Classification of Zooxanthellae and the Evolution of Animal-Algal Symbioses. Science251, 1348–1351. doi: 10.1126/science.251.4999.1348
63
Sachs J. L. Wilcox T. P. (2006). A Shift to Parasitism in the Jellyfish Symbiont Symbiodinium Microadriaticum. Proc. Biol. Sci.273, 425–429. doi: 10.1146/annurev.mi.31.100177.000543
64
Sampayo E. Dove S. LaJeunesse T. (2009). Cohesive Molecular Genetic Data Delineate Species Diversity in the Dinoflagellate Genus Symbiodinium. Mol. Ecol.18, 500–519. doi: 10.1111/j.1365-294X.2008.04037.x
65
Santos S. R. Taylor D. J. Coffroth M. A. (2001). Genetic Comparisons of Freshly Isolated Versus Cultured Symbiotic Dinoflagellates: Implications for Extrapolating to the Intact Symbiosis. J. Phycol.37, 900–912. doi: 10.1046/j.1529-8817.2001.00194.x
66
Santos S. R. Taylor D. J. Kinzie R. A. III Hidaka M. Sakai K. Coffroth M. A. (2002). Molecular Phylogeny of Symbiotic Dinoflagellates Inferred From Partial Chloroplast Large Subunit (23S)-rDNA Sequences. Mol. Phylogenet. Evol.23, 97–111. doi: 10.1016/s1055-7903(02)00010-6
67
Savage A. Goodson M. Visram S. Trapido-Rosenthal H. Wiedenmann J. Douglas A. (2002). Molecular Diversity of Symbiotic Algae at the Latitudinal Margins of Their Distribution: Dinoflagellates of the GenusSymbiodinium in Corals and Sea Anemones. Mar. Ecol. Prog. Ser.244, 17–26. doi: 10.3354/meps244017
68
Silverstein R. N. Correa A. M. S. Baker A. C. (2012). Specificity is Rarely Absolute in Coral-Algal Symbiosis: Implications for Coral Response to Climate Change. Proc. R. Soc. Lond. Ser. B Biol. Sci.279, 2609–2618. doi: 10.1098/rspb.2012.0055
69
Stat M. Carter D. Hoegh-Guldberg O. (2006). The Evolutionary History of Symbiodinium and Scleractinian Hosts—Symbiosis, Diversity, and the Effect of Climate Change. Perspect. Plant Ecol. Evol. Syst.8, 23–43. doi: 10.1016/j.ppees.2006.04.001
70
Stat M. Morris E. Gates R. D. (2008). Functional Diversity in Coral-Dinoflagellate Symbiosis. Proc. Natl. Acad. Sci.105, 9256–9261. doi: 10.1073/pnas.0801328105
71
Suggett D. J. Warner M. E. Smith D. J. Davey P. Hennige S. Baker N. R. (2008). Photosynthesis and Production of Hydrogen Peroxide by Symbiodinium (Pyrrhophyta) Phylotypes With Different Thermal Tolerances. J. Phycol44, 948–956. doi: 10.1111/j.1529-8817.2008.00537.x
72
Thornhill D. J. Daniel M. W. LaJeunesse T. C. Schmidt G. W. Fitt W. K. (2006). Natural Infections of Aposymbiotic Cassiopea Xamachana Scyphistomae From Environmental Pools of Symbiodinium. J. Exp. Mar. Biol. Ecol.338, 50–56. doi: 10.1016/j.jembe.2006.06.032
73
Thornhill D. J. Kemp D. W. Bruns B. U. Fitt W. K. Schmidt G. W. (2008). Correspondence Between Cold Tolerance and Temperate Biogeography in a Western Atlantic Symbiodinium (Dinophyta) Lineage. J. Phycol44, 1126–1135. doi: 10.1111/j.1529-8817.2008.00567.x
74
Van Oppen M. J. (2004). Mode of Zooxanthella Transmission Does Not Affect Zooxanthella Diversity in Acroporid Corals. Mar. Biol.144, 1–7. doi: 10.1007/s00227-003-1187-4
75
Venn A. A. Loram J. E. Douglas A. E. (2008). Photosynthetic Symbioses in Animals. J. Exp. Biol.59, 1069–1080. doi: 10.1093/jxb/erm328
76
Verde E. A. McCloskey L. R. (1998). Production, Respiration, and Photophysiology of the Mangrove Jellyfish Cassiopea Xamachana Symbiotic With Zooxanthellae: Effect of Jellyfish Size and Season. Mar. Ecol. Prog. Ser.168, 147–162. doi: 10.3354/meps168147
77
Visram S. Wiedenmann J. Douglas A. (2006). Molecular Diversity of Symbiotic Algae of the Genus Symbiodinium (Zooxanthellae) in Cnidarians of the Mediterranean Sea. J. Mar. Biol. Assoc. U K86, 1281–1283. doi: 10.1017/S0025315406014299
78
Xia X. (2013). DAMBE5: A Comprehensive Software Package for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol.30, 1720–1728. doi: 10.1093/molbev/mst064
79
Zardoya R. Costas E. López-Rodas V. Garrido-Pertierra A. Bautista J. M. (1995). Revised Dinoflagellate Phylogeny Inferred From Molecular Analysis of Large-Subunit Ribosomal RNA Gene Sequences. J. Mol. Evol.41, 637–645. doi: 10.1007/BF00175822
Summary
Keywords
Symbiodiniaceae, Scyphozoa, 28S rDNA, ITS2, cultivation, pigments
Citation
Dall’Olio LR, Beran A, Flander-Putrle V, Malej A and Ramšak A (2022) Diversity of Dinoflagellate Symbionts in Scyphozoan Hosts From Shallow Environments: The Mediterranean Sea and Cabo Frio (Rio de Janeiro, Brazil). Front. Mar. Sci. 9:867554. doi: 10.3389/fmars.2022.867554
Received
01 February 2022
Accepted
12 April 2022
Published
13 May 2022
Volume
9 - 2022
Edited by
Gleyci A. O. Moser, Rio de Janeiro State University, Brazil
Reviewed by
Christine Ferrier-Pagès, Centre Scientifique de Monaco, Monaco; Catharina Alves-de-Souza, University of North Carolina Wilmington, United States
Updates
Copyright
© 2022 Dall’Olio, Beran, Flander-Putrle, Malej and Ramšak.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreja Ramšak, andreja.ramsak@nib.si
This article was submitted to Marine Ecosystem Ecology, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.