Abstract
Seaweed surfaces harbor diverse epibiotic bacterial communities with functions related to morphogenesis, host health, and defense. Among seaweed holobionts, culturable strains can represent innovative sources of bioactive compounds and enzymes. The global industrial demand for microbial enzymes is continually growing in order to improve certain manufacturing processes with new perspectives of industrial exploitation. In this regard, the present study focuses on the enzymatic production and the antimicrobial activities of culturable epibiotic bacteria of Ulva from the Tunisian coast. Culturable associated bacteria were isolated and molecular identification was realized by 16S rRNA gene sequencing. For each strain, eight enzymatic activities were investigated: amylase, hemolysis, DNase, cellulase, lecithinase, lipase, gelatinase, and chitinase. The antimicrobial activity of Ulva-associated bacteria was evaluated against seven pathogenic bacteria, Escherichia coli, Vibrio anguillarum, Vibrio alginoliticus, Pseudomonas aeruginosa, Aeromonas hydrophila, Salmonella typhymurium, and Staphylococcus aureus, and one yeast, Candida albicans. The antibiotic resistance of isolated strains was determined for 15 commonly used antibiotics. The phylogenetic analysis revealed that the isolates belonged to Alphaproteobacteria (3), Gammaproteobacteria (5), Actinobacteria (3), and Firmicutes (4) phyllum. The majority of the isolates (66%) produced simultaneously more than one enzyme. Hemolysis was produced by 46.6% of isolates, while DNase was produced by 33% of strains. On the other hand, 13% of strains produced lecithinase, gelatinase, cellulase, and lipase. No chitinase was produced by the isolated bacteria. In addition, 60% of isolates displayed antimicrobial activity against at least one pathogenic strain. All Ulva ohnoi-associated bacteria were resistant to at least seven commonly used antibiotics. These results highlighted the occurrence of several enzymatic activities within Ulva-associated bacteria that can have potential uses in the industrial sector.
1 Introduction
Bacteria colonize almost all the planet’s ecosystems; within the marine ecosystem, macro-organisms cannot live in total isolation and are exposed to bacteria. Each organism interacts directly and indirectly with its environment, and understanding the nature of interactions between organisms is a key step in the study of ecosystems and biological cycles. Marine bacteria draw the attention of marine specialists in the following aspects: biodiversity, recycling of waste and pollutants, remediation of polluted site, sustainable materials, and even renewable energy. Seaweed-associated bacteria interactions are an important component of the marine ecosystems. Understanding the mechanisms and pathways underlying these interactions is essential to explain the ecological distribution of bacteria, their physiology, and their responses to anthropogenic influences and environmental variations. It has been assumed for a long time that there is an association between algae and bacteria (Falkowski et al., 2004) and that they are the most abundant microorganisms on the algae’s surface (Dobretsov and Qian, 2002). The host organism, also called basibiont, gives a suitable niche for the installation of associated bacteria (Califano et al., 2020) with optimal conditions for growth by providing oxygen, nutrients, and light. The associated bacteria provide hormones, vitamins, minerals, and carbon dioxide to its seaweed host (Martin et al., 2014; Kouzuma and Watanabe, 2015). Prior research confirmed that the composition and the nature of associated bacteria can be different from one seaweed to another and depends on the development stage of the alga (Ghaderiardakani et al., 2020; Qu et al., 2021).
Additionally, Ghaderiardakani et al. (2017) demonstrated that specific bacteria are essential to ensure proper algae growth and differentiation of stem and rhizoid cells. Recently, several studies highlighted the crucial and important role of Ulva-associated bacteria (Wichard et al., 2015; Weiss et al., 2017; Comba-González et al., 2021); in fact, special chemical mediators are used to ensure that the communication between alga and its associated bacteria and those mediators may be an important source of bioactive molecules and enzymes (Alsufyani et al., 2020).
The global industrial enzyme market is expected to grow from $5.5 billion in 2018 to $7.0 billion in 2023, according to Business Communication Company Research (BCC) (Guisan et al., 2020), which demonstrates how much this sector is important and also shows the increasing demand of the industry in enzymes that are recently used for eco-friendly textile processing (Kumar et al., 2021), drugs (Masci and Castagnolo, 2021), and bioremediation (Singh et al., 2021). Marine bacteria associated with algae are a potential source of new biomolecules having several biotechnological applications (Kandasamy et al., 2020; Kumar et al., 2022). They are an untapped source of enzymes; in fact, xylanases, amylases, and glycosidases are enzymes synthesized by bacteria associated with macroalgae (Chandrasekaran and Rajeev, 2010; Bakunina et al., 2012; Hehemann et al., 2014).
Marine bacteria have several uses (Samant et al., 2019); in fact, most enzymes used in different industries are of microbial origin and have specific characteristics, such as stability, high specific activity, and facilitated mass transfer. High-saline marine environments have proven to be a rich source of microorganisms harboring industrially important enzymes. Enzymes produced by marine microorganisms can provide numerous advantages compared to traditional enzymes due to their wide-ranging habitats. The quest for bacterial isolates producing enzymes with high efficacy and of commercial value is still ongoing to exploit enormous marine resources, one of which are marine macroalgae and bacteria associated with their surfaces, seaweeds – epiphytic bacteria are able to produce enzymes that are of biotechnological interest such as cellulases, lipases, amylases, agarases, laccases, and proteases (Comba-González et al., 2016).
Macroalgae, either green, red, or brown, can be the origin of a huge diversity of bacteria living on their surface. Ulva Linnaeus genus (Ulvales, Ulvophyceae) is commonly distributed in oceans and estuaries. It occupies the first 5 m of the marine areas’ surfaces and can be found either in shallow water or in eutrophic zones. It is distinguished by a very thin thallus with a leaf-shaped, ribbon-like, or tubular form and green color that can be transparent under stress (Steinhagen et al., 2019). Currently, 102 species (accepted taxonomically) have been identified worldwide (Guiry and Guiry, 2022). Individuals of this genus are characterized by a broad range of environmental tolerance (Holzinger et al., 2015), high growth rate (Wichard et al., 2015), and photosynthetic activity leading to a relatively abundant natural biomass (Dominguez and Loret, 2019). The phylogenetic analysis of some Ulva sample on the Mediterranean Sea reports the presence of six non-indigenous species: Ulva tepida, Ulva australis, Ulva californica, Ulva lactuca, Ulva chaugulii. and Ulva ohnoi. The latter species U. ohnoi was found in the northern coast of Tunisia and it was previously described by Miladi et al. (2018) in Gabes (Southern cost of Tunisia).
In this study, the screening of Ulva-associated bacteria concerned different aspects from the molecular identification to biological activities. After bacterial sampling, a molecular identification for Ulva-associated bacteria was done, and then the antimicrobial activity was tested against pathogenic strains. In addition, the bioresistance of bacteria was tested and the enzymatic production was performed to test the production of amylase, hemolysis, DNase, cellulase, lecithinase, lipase, gelatinase, and chitinase. The main objectives of the present work are to isolate culturable Ulva-associated bacteria and evaluate their biological activities such as the capacity for enzyme production, antimicrobial activity, and their resistance to antibiotics.
2 Materials and methods
2.1 Isolation of bacterial strains and growth conditions
Ulva samples were collected from Ghar El Melh lagoon (37°10’1.1568” N 10°10’54.9768” E) in May 2018 in shallow waters with an 18°C temperature and with 37‰ salinity. Sterile gloves and bags were used for sampling algae with the surrounding water to preserve the community of associated bacteria, which were then transferred to the laboratory in a cooler. As the morphological identification of Ulva species can be tricky due to their phenotypic plasticity, the samples have been identified through DNA barcoding (Saunders and Kucera, 2010). A voucher specimen was kept in formaldehyde solution (2%).
Once in the laboratory, the samples were washed three times with autoclaved natural seawater to remove the non-attached bacteria (Burgess et al., 2003). Ten grams of seaweed was vortexed in a Stomacher for 6 min to detach surface-associated bacteria. Then, a 1-ml sample was taken for serial dilution up to 10−3 with sterile seawater. Under aseptic conditions, 100 µl of each dilution was spotted on marine agar (MA: Pronadisa Laboratories, CONDA) and then incubated in 20°C for 7 days at least for 1 month (Lemos et al., 1985).
Purification of bacteria was carried out and single colonies were selected based on morphological features and purified until pure cultures were obtained. Storage was done in stock solution made with glycerol 20% at −80°C. Thirty bacteria were isolated and 15 culturable bacteria were selected for the present study to carry out the different tests. Biochemical tests such as oxidase and catalase were performed according to Dalton et al. (1994).
2.2 Molecular identification of Ulva-associated bacteria
The molecular identification was carried out according to Ismail et al. (2018) using a single colony from each bacterium. 16S rRNA gene primers B8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and U1492R (5′-GGTTACCTTGTTACGACTT-3′) were used. PCR reactions were performed using a Gene Biometra T1 DNA Thermal Cycler (Perkin-Elmer, USA) in 25-µl (final volume) reaction mixtures containing 0.1 μl of Hot Start DNA polymerase (Sigma), with each primer at a concentration of 10 pmol μl−1, each deoxynucleoside triphosphate at a concentration of 10 mM, 2.5 μl of PCR buffer (with MgCl2), and 1 μl of DNA template. The PCR protocol consisted of 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 110 s. The cycles were preceded by 15 min of denaturation at 94°C and ended with a final extension for 7 min at 72°C. Negative controls contained all of the components of the PCR mixture except the DNA templates.
2.3 Phylogenetic analysis
The closest phylogenetic relatives of each isolate were identified by comparison of the 16S rRNA gene sequence to the National Center for Biotechnology Information (NCBI) GenBank database using the Basic Local Alignment Search Tool (BLAST) analysis tools (http://www.ncbi.nlm.nih.gov/BLAST). The phylogenetic analysis of isolates and close relative strains present in the NCBI nucleotide database was performed using the neighbor-joining method using MEGA software version 11 (Tamura et al., 2021). Numbers at nodes represent percentages of 1,000 bootstrap replications. The scale bar indicates mutations per nucleotide position.
2.4 Enzymatic production profile of Ulva-associated bacteria
2.4.1 Hemolysis
The hemolysis test was done in triplicate according to Satpute et al. (2008) with some modifications. Agar plates were prepared with horse blood; a streak of bacteria was added and incubated at 37°C. Results are observed after 48 to 72 h and distinguished with a colorless area.
2.4.2 DNase
This test was performed in triplicate on DNA agar medium with a single colony. After incubation for 48 h at 28°C, the surface of the agar was covered with the toluidine blue reagent, which makes it possible to demonstrate the presence or absence of DNA hydrolysis products. The pink halo around the culture means DNase positive; otherwise, it means DNase negative (Schreiber et al., 2016).
2.4.3 Lecithinase
The lecithinase test was carried out according to Garrity et al. (2010) by inoculating bacterial streaks on nutrient agar containing a sterile egg yolk emulsion followed by an incubation of 24 to 72 h at 30°C; this test was done in triplicate. Opaque areas in the transparent halo indicate degradation of the egg yolk lecithin by bacterial production of lecithinase enzyme.
2.4.4 Gelatinase
The gelatinase test was carried out on nutrient agar medium containing 1% gelatin (Smith and Goodner, 1958) in triplicate. Nutrient agar medium containing 1% gelatin was inoculated with the bacterial isolates and plates were incubated at 30 or 37°C for 2 to 5 days. For results, a solution of mercury chloride was used, which makes it possible to highlight the degradation of gelatin with a clear halo around the colonies.
2.4.5 Lipase
In triplicate, agar plates were prepared with tween 80 (Sierra, 1957) and then bacteria were streaked. The results of this test were readable after 24 h at 37°C through the formation of an opaque zone for lipase positive.
2.4.6 Amylase
This enzymatic test was carried out by preparing a specific starch agar according to Krishnan et al. (2011) in triplicate. Bacteria were inoculated in a single streak and then incubated for 24 to 72 h at 30°C. The hydrolysis of amylases was indicated by the presence of a clear zone around the colonies after the addition of lugol. The absence of staining around the culture shows the degradation of starch (amylase-positive bacteria) while areas containing starch stain brown (amylase-negative bacteria).
2.4.7 Cellulase
The cellulase test was carried out on agar prepared according to Sinha and Bedi (2018). The bacterial isolates were streaked on a plate and incubated for 72 h at 30°C. Afterwards, 1% Congo red aqueous solution was added, which makes it possible to demonstrate the degradation of the cellulose after 15 min; this test was done in triplicate.
2.4.8 Chitinase
This test was carried out on nutrient agar supplemented with 1% chitin. After inoculation of bacteria in streaks, plates were incubated for 72 h at 30°C; this test was done in triplicate. The appearance of light areas around the colonies indicates the production of chitinase (Kaur et al., 2012). Diameters for all enzymatic activity were measured in centimeters. All tests were done in triplicate.
2.5 Antimicrobial activity of Ulva-associated bacteria
The drop test assay on Trypto-Casein-Soy agar (TSA) was used to test the antimicrobial activity of Ulva-associated bacteria against Staphylococcus aureus ATCC 25923, Vibrio alginolyticus ATCC 17749T and Vibrio anguillarum ATCC 12964T (clinical strain obtained from Pasteur Institute, Tunis), Escherichia coli ATCC 25922 and Aeromonas hydrophila B3 (fish pathogen obtained from RVAU-Denmark), Salmonella typhimurium C52 and Pseudomonas aeruginosa ATCC 27853 (clinical strain obtained from Laboratoire Hydrobiologie Marine et Continentale, Université de Montpellier II, France), Vibrio alginolyticus ATCC 17749T (clinical strain from Pasteur Institute, Tunis), and Candida albicans (yeast) ATCC 10231.
Fresh bacterial cultures of 24 h for both pathogenic and Ulva-associated strains were prepared for the antagonist test according to Rao et al. (2005) with modifications. The turbidity of the bacterial suspensions was adjusted to 0.5 McFarland. A drop of 0.5 cm of each bacterial suspension was placed in a petri dish containing the pathogenic bacteria grown on TSA. Agar plates were then incubated for 24 h at 20°C. Positive activity was obtained by the presence of a clear zone around the isolate and measured in millimeters. All tests were done in triplicate (n = 3).
2.6 Antibiotic resistance profile of Ulva-associated bacteria
Bacterial suspension in marine broth was prepared and several dilutions were applied to adjust the concentration to 0.5 McFarland. The culture is spread over the entire surface of Mueller-Hinton agar in order to obtain a homogeneous quantity of bacteria and left to dry. Using sterile forceps, antibiotic discs impregnated with standardized and fixed quantities of different antibiotics were placed in a petri dish. The following were the antibiotics used: azithromycin (AZM; 15 μg), spiramycin (SP; 100 µg), ceftazidime (CZD; 10 µg), ceftriaxone (CRO; 30 µg), vancomycin (VA; 30 µg), cefoxitin (FOX; 30 µg), ticarcillin (TIC; 75 µg), sulfonamides (SUL; 200 µg), amikacin (AN; 30 µg), gentamicin (GMN; 10 µg), kanamycin (KMN; 30 µg), chloramphenicol (C; 30 µg), tetracycline (TE; 30 µg), imipenem (IPM; 10 µg), and amoxicillin (AX; 25 µg). The antibiograms were incubated for 24 h at 20°C. Susceptibility measure for inhibitory zones in millimeters was evaluated in accordance with the standard measurement cited in Société Française de Microbiologie (2020) as sensitive (S), intermediary sensitive (I), and resistant (R). The multiple antibiotic resistance (MAR) index of the bacterial isolates was calculated based on the formula (Krumperman, 1983):
where NR = number of antibiotics to which the isolate is resistant and NT = total number of antibiotics tested.
2.7 Statistical analysis
Data were analyzed by one-way analysis of variance (ANOVA), and significant differences at a level of 95% (p< 0.05) were determined by Tukey’s test for the antimicrobial activity. Principal component analysis (PCA) for the enzymatic activity, antimicrobial activity, and antibiotic resistance of Ulva-associated bacteria was carried out to visualize the correlations and identify relationships between the different variables. For each strain, the capacity to give a positive result to a test has been taken into account. For antibiotic resistance, the index was calculated for each isolate by dividing the number of antibiotics to which the isolate is resistant by the total number of antibiotics tested (15), also defined as MAR (multiple antibiotic resistance) (Krumperman, 1983). The same principle has been applied for enzyme production and antibiotic activity. For each strain, “enzymatic capacity” was calculated by dividing the number of produced enzymes by the total number of enzymes tested (8), while “antimicrobial capacity” was calculated by dividing the number of inhibited bacteria by the total pathogenic bacteria used (8).
All statistical analyses were performed using XLSTAT 2021 by Addinsoft.
3 Results
3.1 Phylogenetic characterization of bacteria
Isolation of Ulva-associated bacteria led to the identification of 15 strains based on 16S rRNA gene sequencing. All nucleotide sequences of isolates were assigned to the GenBank database and were accession numbers (Table 1).
Table 1
| Isolates | Size (bp) | Closest species in NCBI | Standard strains accession number | Identity (%) | Accession number | Phylum | Oxydase | Catalase |
|---|---|---|---|---|---|---|---|---|
| IH25 | 850 | Dinoroseobacter shibae | JQ660974 | 99 | MZ150448 | α-P | – | – |
| IH1 | 750 | Paracoccus marcusii | MN931275 | 100 | MZ150442 | – | ++ | |
| IH5 | 800 | Yoonia rosea | MT406460 | 99 | MZ150450 | – | – | |
| IH8 | 1,100 | Psychrobacter nivimaris | KP717914 | 96 | MZ150445 | γ-P | – | – |
| IH24 | 850 | Psychrobacter celer | MH256077 | 98 | MZ150449 | – | – | |
| IH19 | 1,000 | Acinetobacter johnsonii | MT226917 | 100 | MZ150447 | – | ++ | |
| IH10 | 800 | Acinetobacter johnsonii | MT226917 | 99 | OL851810 | – | – | |
| IH15 | 1,145 | Acinetobacter johnsonii | MN758792 | 95 | MZ150452 | – | ++ | |
| IH17 | 600 | Exiguobacterium aestuarii | MF426024 | 88 | OL851812 | F | – | ++ |
| IH7 | 800 | Enterococcus faecalis | NR040789 | 100 | MZ150446 | – | – | |
| IH21 | 900 | Alkalibacillus hwajinpoensis | MT510155 | 99 | MZ150451 | – | – | |
| IH3 | 161 | Enterococcus casseliflavus | MT177112 | 92 | OL851823 | – | + | |
| IH2 | 750 | Rathayibacter festucae | MT431571 | 100 | MZ150443 | A | – | + |
| IH9 | 950 | Micrococcus aloeverae | MZ496563 | 99 | MZ150444 | – | – | |
| IH18 | 1,189 | Microbacterium phyllosphaerae | MT533897 | 98 | MZ150453 | – | ++ |
Epibiotic bacteria isolated from the Ulva ohnoi surface.
*α-P: α-proteobacteria, γ-P: γ-proteobacteria, F: firmicutes, A: actinobacteria, bp: base pair
Epibiotic bacteria isolated from the Ulva ohnoi sample were highly diverse. Most of the isolates (eight) belonged to Proteobacteria while four were Firmicutes and three of them belonged to Actinobacteria.
Acinetobacter was the predominant genus of the Gamma-Proteobacteria with three representative species followed by Psychrobacter with two representative species.
Most of Ulva’s isolates shared more than 98% identity with the identified closest matching strain except four strains (IH8, IH15, IH17, and IH3) that showed a lower identity percentage with the closest matching strain, leading to possible novel species or even a novel genus. The topology of the phylogenetic tree in Figure 1 constructed with the neighbor-joining method shows the proximity of the isolated species in this study to the neighboring strains selected from the NCBI database in addition to the eight chosen strains previously isolated by Ismail et al. (2018) from Ulva rigida collected form Ghar El Melh or Cap Zebib. Interestingly, close matching occurred between IH1 and the isolate FN811323 from Ulva Cap Zebib (closely related to Paracoccus sp.). In addition, the position of Acinetobacter strains (IH10, IH15, and IH19) is close to U. rigida isolates from the same geographical area.
Figure 1

Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences and different enzymatic and antimicrobial activities in comparison with isolates of Ulva rigida from Ismail et al. (2018): associated bacteria isolated from Ulva ohnoi (red spots) and Ulva rigida-associated bacteria (blue squares).
3.2 Enzymatic production
The majority of bacteria show enzymatic production. All the results obtained should be considered as preliminary screening. Thus, two-thirds of the isolated bacteria produced at least one enzyme as shown in Figure 2; some of them are able to produce several enzymes with a variety of functions (Supplementary Figure 1). Results of the enzymatic test show the following: 26% of isolates were able to produce amylase while only 33% can produce DNase; 13% of bacteria have lecithinase ability production; hemolysis is produced by 46% of bacteria; gelatinase, lipase, and cellulose were produced by 13% of tested bacteria; and no chitinase production was observed. Strains IH1 and IH2 demonstrated the ability to produce five enzymes among eight tested, followed by IH18, which showed the ability to produce four enzymes (amylase, DNase, hemolysis, and lipase). Seven strains were positive for hemolysis production: IH1, IH7, IH8, IH9, IH15, IH18, and IH19 with diameters of 4.6, 2.3, 2.5, 2.4, 4.5, 2.8, and 2.5 cm, respectively, while five strains were able to degrade DNA (IH1, IH2, IH8, IH18, and IH19) with diameters of 2.5, 2.6, 1.8, 2.7, and 1.9 cm, respectively. IH1 and IH2 are able to produce gelatinase with diameters of 3.9 and 4.4 cm. Cellulase was produced by both IH2 (3 cm) and IH21 (2.7 cm). Lecithinase is produced by IH1 and IH5 with diameters of 4 and 5.4 cm, respectively. IH2 and IH18 are able to produce lipase with diameters of 3.4 and 4 cm. All Ulva isolates were not able to produce chitinase.
Figure 2
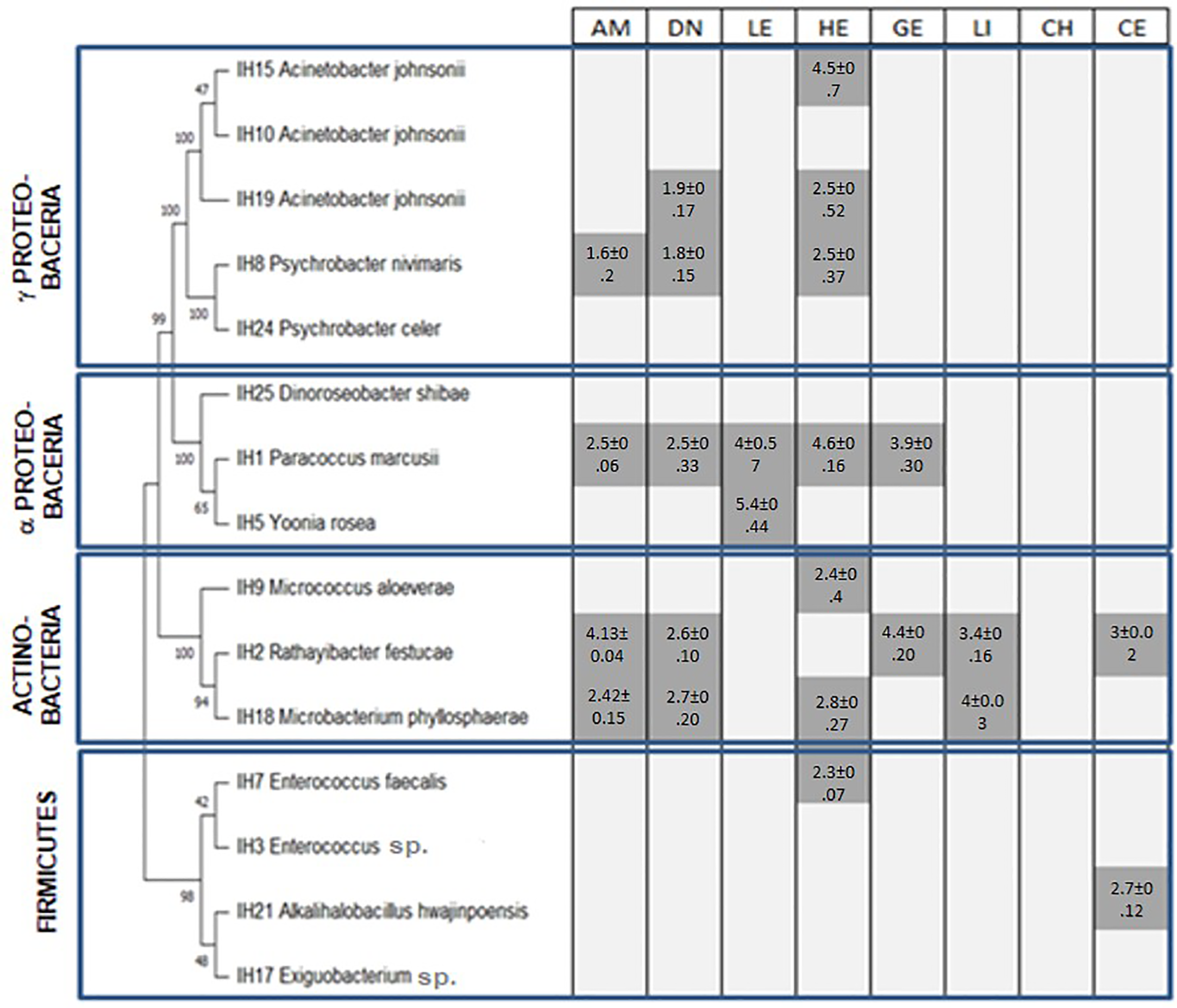
Enzymatic production of Ulva ohnoi-associated bacteria (dark gray is for production of enzyme, light gray is for no enzyme production, numbers are activity diameter expressed in cm) AM, amylase; HE, hemolysis; DN, DNase; CE, cellulase; LE, lecithinase; LI, lipase; GE, gelatinase; and CH, chitinase.
3.3 Antimicrobial activities of Ulva ohnoi-associated bacteria
Antimicrobial activity obtained for isolated strains is presented in Table 2. Out of 15 strains, 9 displayed antimicrobial activity against pathogenic strains: 3 γ-Proteobacteria (IH24, IH10, and IH15), 3 Firmicutes (IH7, IH3, and IH17), 2 α-Proteobacteria (IH5 and IH25), and 1 Actinobacteria (IH9).
Table 2
| ID | C. a | V. an | S. t | P. a | V. al | E. c | A. h | S. a |
|---|---|---|---|---|---|---|---|---|
| IH3 | 10.6 ± 0.8a | 10.3 ± 2.3a | 13 ± 2.8a | 12.7 ± 1.4a | 12.5 ± 1.7a | 9.9 ± 0.8a | – | – |
| IH5 | – | – | – | – | 11 ± 1.6 | – | – | – |
| IH7 | – | – | – | – | 9 ± 1.1 | – | – | – |
| IH9 | 10 ± 1c | 10.8 ± 1.1bc | 13.2 ± 0.8a | 12.6 ± 0.8ab | 12 ± 0.5ab | 10.9 ± 0.6bc | – | – |
| IH10 | – | – | – | – | – | – | 13.6 ± 1.3 | – |
| IH15 | 9.5 ± 1.2b | – | 10 ± 1.1ab | 11.9 ± 1.3a | 10.3 ± 0.4ab | – | – | – |
| IH17 | 10 ± 0.6b | 11.7 ± 0.7ab | 10.7 ± 2.9ab | 10 ± 1.3b | 11.4 ± 0.4ab | 10.5 ± 1.4ab | 13.8 ± 0.2a | – |
| IH24 | – | – | – | 9.7 ± 0.3 | – | – | – | – |
| IH25 | – | – | – | 9 ± 1 | – | – | – | – |
Antimicrobial activity of Ulva ohnoi-associated bacteria against pathogenic strains (mm).
C. a: Candida albicans; V. an: Vibrio anguillarum; -: no activity; ± standard deviation; and n = 3; Values with the same letter are not significantly different at the level of 5% (p< 0.05).
According to the results presented in Table 2, IH17 (closely related to Exiguobacterium sp.) was the most active stain inhibiting the growth of all the pathogenic tested strains except S. aureus and possesses the strongest antimicrobial activity as shown by the inhibition diameter of 13.8 ± 0.02 against A. hydrophila with high significant variances at 95% confidence level. In addition, isolate IH3 (closely related to Enterococcus casseliflavus) displayed antimicrobial inhibition against five pathogenic bacteria and the yeast C. albicans. Enterococcus faecalis (IH7) inhibited the growth of V. alginoliticus. Fifty-five percent of the total active strains were isolated, and a significant variance exists for Acinetobacter johnsonii (IH10); however, Yoonia rosea Acinetobacter sp. (H15), Psychrobacter celer (IH24), and Dinoroseobacter shibae (IH25) were not significantly different.
Micrococcus aloeverae (IH9) showed a significant antimicrobial inhibition zone with a high significant variance at the level of 5%. C. albicans was inhibited by IH3, IH9, IH15, and IH17; on the other hand, E. coli was sensitive against IH3, IH9, and IH17; S. typhymurium was inhibited by four bacteria: IH3, IH9, IH15, and IH17. The pathogenic strain V. anguillarum was sensitive to three bacteria isolated from Ulva, IH3, IH9, and IH17, while P. aeruginosa and V. alginoliticus were the most sensitive pathogenic strains inhibited by six tested bacteria. A. hydrophila was less sensitive against the tested strains; it was inhibited only by two bacteria, A. johnsonii (IH10) and Exiguobacterium sp. (IH17).
3.4 Antibiotic resistance
Figure 3 presents the antibiotic resistance profile of the U. ohnoi-associated isolates.
Figure 3
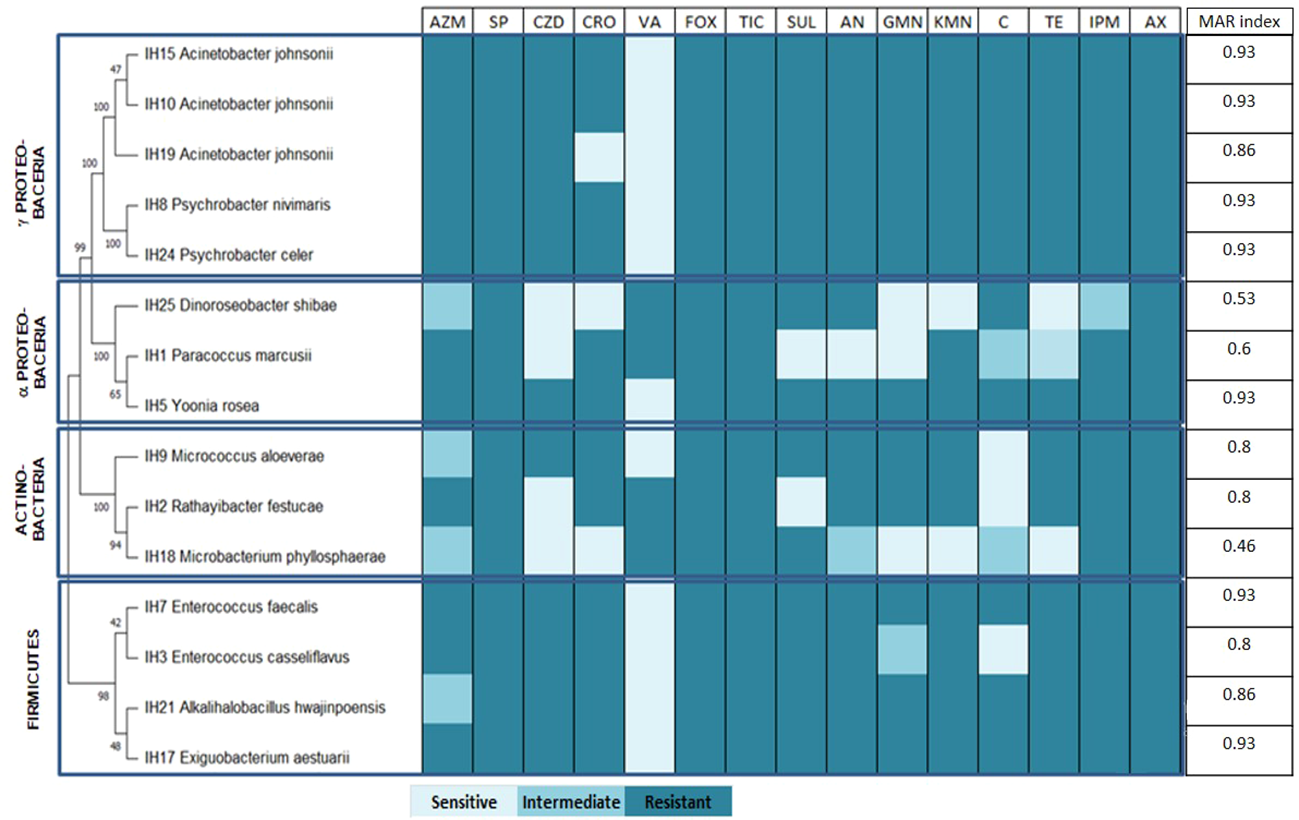
Antibiotic resistance profile of Ulva ohnoi-associated bacteria. AZM, azithromycin; SP, spiramycin; CZD, ceftazidime; CRO, ceftriaxone; VA, vancomycin; FOX, cefoxitin; TIC, ticarcillin; SUL, sulfonamides; AN, amikacin; GMN, gentamicin; KMN, kanamycin; C, chloramphenicol; TE, tetracycline; IPM, imipenem; AX, amoxicillin.
All strains were resistant to spiramycin, cefoxitin, ticarcillin, and amoxicillin, while most of the strains were susceptible to vancomycin except IH25 (D. shibae), IH1 (Paracoccus marcusii), IH2 (Rathayibacter festucae), and IH18 (Microbacterium phyllosphaerae). The calculated MAR index for Ulva-associated bacteria ranged from 0.46 to 0.93, with 50% of the strains showing a very high resistance profile with an MAR index of 0.93 (Figure 3). Strains IH15, IH10, IH8, IH24 IH5, IH7, and IH17 presented the highest MAR index, while strain IH18 had the lowest one.
The PCA performed on enzymatic production (EA), antimicrobial activity (AA), and antibiotic resistance (AR) of bacterial strains led to the opposite loading of EA and AA, depicting an opposite correlation between them (Figure 4). Strains displaying antimicrobial activity presented a lower capacity to produce enzymes while enzyme-producing strains displayed few or no antimicrobial activity against the pathogenic strains tested. The PCA shows that there are three main clusters confirmed by the dissimilarity tree (Figure 4). The first cluster concerns the bacteria that are on the higher side of the second principal component, carried by AR and present high MAR indexes. It is composed of seven strains: IH10 (A. johnsonii), IH5 (Yoonia rosea), IH7 (Enterococcus faecalis), IH24 (Psychrobacter celer), IH21 (Alkalihalobacillus hwajinpoensis), IH19 (A. johnsonii), and IH8 (Psychrobacter nivimaris). The second cluster concerns strains with significant antimicrobial activity and characterized by three strains, IH17 (Exiguobacterium sp.), IH3 (Enterococcus sp.), and IH9 (Micrococcus aloeverae). These strains also have a high MAR index. The last group of isolates is represented by mainly two strains, IH1 (P. marcusii) and IH18 (M. phyllosphaerae), and these strains have a high enzyme production capacity while displaying no antimicrobial activity and the lowest MAR index.
Figure 4
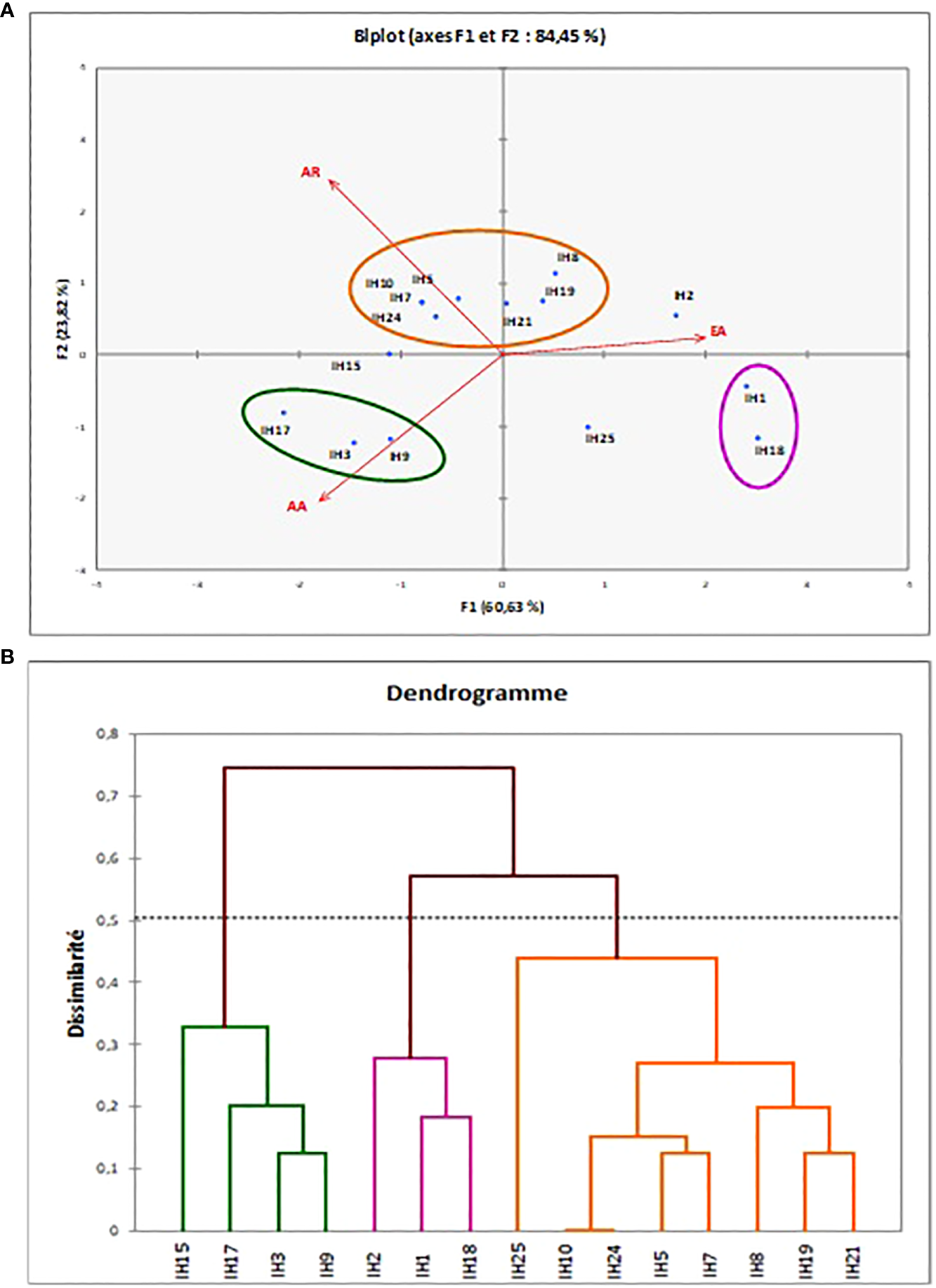
Different clusters obtained for the enzymatic, antimicrobial, and antibiotic resistance tests of Ulva ohnoi-associated bacteria strains obtained by PCA (A) and dissimilarity tree (B).
4 Discussion
The Ulva surface represents a large and precious ecological niche for the installation of biofilm. Algae thallus can be colonized by Proteobacteria, Firmicutes, and Actinobacteria.
In the present study, Proteobacteria was the most represented taxonomic group (53%) of the total identified strains. This result is in accordance with literature reporting Proteobacteria generally being the most abundant taxonomic group on seaweed surfaces (Ismail et al., 2018). More specifically, Burke et al. (2011) showed that the clade of Proteobacteria are more likely associated with green macroalgae.
Two subclasses were found to be associated with U. ohnoi, the α-Proteobacteria represented by strains closely related to D. shibae, P. marcusii, and Yoonia rosea, and the γ-Proteobacteria represented by isolates closely related to A. johnsonii. Psychrobacter celer and Psychrobacter nivimaris Actinobacteria representatives occupied a minor proportion of isolated bacteria with 3 strains out of 15. These findings are in agreement with those of Choi et al. (2016) who found a few representatives of Actinobacteria (4%) among Ulva pertusa-associated strains.
In a previous study conducted by Ismail et al. (2018), epiphytic bacteria isolated from U. rigida collected from the same sampling site, Ghar El Melh, were identified. Among the isolated strains, these authors identified one strain closely related to Paracoccus sp. Interestingly, Paracoccus sp. have been described to have growth-promoting and morphogenesis-inducing function in Ulva (Ghaderiardakani et al., 2017). In this study, isolate IH1 has been found to belong to Paracoccus genus, supporting that species of this genus belong to the resident flora of epiphytic community of Ulva sp. in Ghar El Melh Lagoon.
Similarly, strains closely related to D. shibae were isolated from both Ghar El Melh Lagoon samples U. rigida (Ismail et al., 2018) and U. ohnoi (present study). This species belongs to the Roseobacter group, which have been described as playing an important role in host health by producing bioactive secondary metabolites (Tujula et al., 2010; Wienhausen et al., 2017).
Except for these two species described above (Paracoccus sp. and D. shibae), all other isolates were different from Ismail et al. (2018)U. rigida isolates. This finding is in accordance with Lachnit et al. (2011) who demonstrated that at different sampling intervals, epibacterial community profiles differ significantly at a bacterial phylum level among algal species.
As most enzymes used in different industries are of microbial origin, associated bacteria with sedentary marine organisms including seaweeds represent an important reservoir as a novel source of industrial enzymes with possible commercial applications (Mohapatra et al., 2003). In this study, majority of U. ohnoi culturable associated isolates had the capacity to produce at least one extracellular enzyme. Enzymes isolated from marine bacteria attract the attention of several researchers, which is due to their availability and cheap production from marine bacteria.
Comba-González et al. (2016) highlighted several enzymes of biotechnological interest potentially produced by epiphytic bacteria associated with Ulva including lipase and amylase. In the present study, R. festucae and M. phyllosphaerae displayed lipolytic ability and can be a potential source for lipase production in several fields. For instance, lipases are used in nanomedicine (Khan et al., 2017), the cheese industry (Kumar and Ray, 2014), animal feed (Ferreira-Dias et al., 2013), and the biodiesel industry (Najjar et al., 2021). Joseph and Ramteke (2013) reported the production of lipase by M. phyllosphaerae and its possible use as a detergent additive. To the best of our knowledge, R. festucae is described for the first time as capable of synthesizing lipase.
Four isolates were found to be capable of producing amylase. Microbial amylases have a wide range of applications in several industries such as food, textile, or biofuel (Raveendran et al., 2018). Amylases are enzymes used in starch conversion (Van der Maarel et al., 2002), the detergent industry (Gupta et al., 2003), and the textile industry (Feitkenhauer, 2003). Matsumoto et al. (2003) highlighted the potential use amylase produced by the marine bacteria Pseudoalterimonas undina isolated from green microalgae for ethanol production. On the other hand, Kalpana et al. (2012) confirm that α-amylase from Bacillus subtilis is used against biofilm-forming human bacterial pathogens.
Another enzyme of interest is extracellular cellulase, which has several biotechnological applications in the food industry and textile (Raveendran et al., 2018). Two isolates, IH2 and IH21, closely related to R. festucae and Alkalibacillus hwajinpoensis presented cellulase production ability. All these findings confirm the potential use of U. ohnoi epiphytic bacteria for several industrial applications.
Isolates IH1 and IH5 identified as P. marcusii and Yoonia rosea, respectively, produce lecithinase and the two isolates IH1 and IH2 were found to be positive for gelatinase. Lecithinase is a type of phospholipase that acts upon lecithin; in the present study, two strains produce this enzyme, which are P. marcusii and Yoonia rosea. This enzyme is used as antibodies in serum according to the study of Kyle et al. (1971). For gelatinase, it has several applications in the culinary industry and pharmaceutical products, and it is also an essential ingredient in the food industry. To the best of our knowledge, this is the first mention of gelatinase production from Ulva-associated bacteria.
DNase is also recommended for the degradation of biofilm associated with the most common upper respiratory tract diseases, which is chronic rhinosinusitis (Shields et al., 2013), and interestingly, in the present study, five bacteria produce DNase and can be valuable source for the extraction of this enzyme.
Seaweed-associated bacteria have the ability to produce active enzyme carbohydrates; in fact, seaweeds present approximately half of the global primary production and contribute to the production of large amounts of polysaccharides that will be consumed by microbes that utilize carbohydrate-active enzymes (CAZymes) able to degrade agars, carrageenan, alginate, and ulvan-polysaccharides from red, brown, and green algae as described by Hehemann et al. (2014).
Hemolytic ability screening allowed the identification of seven isolates with the ability to destroy red blood cells. Among these strains, P. marcusii (IH1) is in agreement with the hemolysis ability being commonly observed within Paracoccus spp. (Leinberger et al., 2021). The screening of the hemolysis ability degradation of 15 bacterial isolates collected from seawater, sediment, and shell samples from the Indian coasts shows that 12 isolates have α-hemolytic activity according to Satpute et al. (2008), which in accordance with our finding where majority of the strains (8 out of 15) demonstrated hemolysis capacity.
U. ohnoi-associated strains in the present study were not able to produce chitinase. Naik et al. (2019) obtained chitinase production from Ulva-associated bacteria and suggested the possible use of bacterial genes to enhance the degradation of crab shell.
Concerning the antimicrobial activity of U. ohnoi-associated bacteria, nine strains displayed positive activity in the present study. This result is in agreement with that found by Ismail et al. (2018) and Wu et al. (2014) who showed that several bacterial strains isolated from Ulva surface have an important antimicrobial activity. Accordingly, these authors described significant antibacterial activity for the isolate D. shibae, which has been found to be closely related to IH25. Contrary to our findings, P. marcusii isolated from a crustacean displayed antibacterial activity (Leinberger et al., 2021). This difference may be due to the biotic or abiotic conditions as bacterial metabolism is dynamic and flexible, depending on the host and environmental conditions, and will lead to different metabolic capacities (Passalacqua et al., 2016).
Dhanya et al. (2016) and Trivedi et al. (2011) demonstrated not only that Ulva-associated bacteria have antimicrobial activity but also that its endophytes can display significant antibacterial properties against human pathogens like Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Salmonella typhymurium, or Bacillus subtilis and maybe a new source for drug research against multidrug-resistant pathogens.
The intense use of antibiotics allows the bacteria to acquire a certain resistance due to gene modifications (Munita and Arias, 2016). Antibiotic resistance is a global phenomenon; resistance to anti-infectives could be responsible for more than 10 million deaths per year and thus might become the major cause of death by 2050 according to O’Neill (2016). Currently, humans are consuming 37 antibiotics used in aquaculture farms via seafood composed of shrimp, crab, mollusks, and fish (Hui et al., 2015), which increase bioresistance to these antibiotics.
On the other hand, not only did bioresistance increase but also some pathogenic bacteria became resistant due to the wide use of antibiotics (Miranda et al., 2018). Until now, the antibiotic resistance of bacteria from marine environment is very little studied. In particular, this is the first report of antibiotic resistance of bacterial strains associated with the green alga U. ohnoi. A better knowledge of environmental resistance could be of great importance to predict the emergence of new resistance of clinical concern (Lupo et al., 2012). Results obtained demonstrated the multiple drug resistance profile of Ulva-associated bacteria, suggesting their adaptation and ability to encode antibiotic resistance genes. In the present study, three Acinetobacter of marine origin were mentioned, and they are all resistant to all the antibiotics except vancomycin. The same genus is clinically important in terms of causing several infections; in fact, Gupta et al. (2015) studied this genus isolated from human origin, and they show that this group is highly resistant to several antibiotics especially ceftriaxone and ceftazidime.
The Acinetobacter genus either of marine origin or of clinical origin is always resistant to antibiotics. Tetracyclines are antibiotics used in aquaculture especially for fish farming of salmon (Higuera-Llanten et al., 2018); in the present study, all γ-Proteobacteria and Firmicutes were resistant to this antibiotic.
Compared with Ktari et al. (2022), the MAR index of A. taxiformis-associated bacteria ranged from 0.11 to 0.78, with more than 60% of strains showing resistance to antibiotics. However, in the present study, the MAR index was between 0.46 and 0.93, which shows that bacteria isolated from Ulva thallus were more resistant to antibiotics; this resistance may be due to a high level of expose to antibiotics through anthropic pollution characterizing the sampling site Ghar El Melh lagoon. In fact, wastewater from the urban area of the Ghar El Melh region is poured directly into the lagoon without treatment, and the water quality and the vegetation of the lagoon have changed considerably over the years according to Moussa et al. (2005).
The functional role of associated bacteria in maintaining their host health has been described in several studies leading to the holobiont concept (Egan et al., 2013; Ghaderiardakani et al., 2020). Ghaderiardakani et al. (2017) demonstrated in Ulva holobiont that there are always specific strains with functions related to growth and morphological development. These authors identified Microbacterium sp. and Paracoccus sp. among the strains that have an algal morphogenesis-inducing function. Interestingly, our finding showed that IH1 and IH18 closely related to P. marcusii and M. phyllosphaerae, respectively, were grouped in the same cluster. These two strains displayed high enzymatic production capacity and no antimicrobial activity and had a lower multidrug-resistant index. In fact, enzymes produced by bacteria do not necessarily have antibiotic activity, and each strain has a particular interest that benefits the host.
5 Conclusion
U. ohnoi-associated bacteria were screened for their enzymatic production, antimicrobial potential, and antibiotic resistance. Fifteen strains were phylogenetically identified and analyzed. Isolates belonged to three major phyla: Proteobacteria, Firmicutes, and Actinobacteria. The enzymatic production of Ulva epiphytes was studied and showed that 67% of the tested bacteria produce enzymes that can be useful in several industrial fields like food, pharmacology, textile, and biofuel. Nevertheless, the development and application of the enzymes need further quantitative evaluation. On the other hand, nine bacteria strains displayed antibacterial activity. This study highlighted that Ulva-associated bacteria can potentially contribute to the development of different industrial sectors.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
IH – carried out the experiment and drafted the first version of the manuscript. AI – supervised molecular analysis and provided critical feedback. LK and MEB supervised, reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1042527/full#supplementary-material
References
1
Alsufyani T. Califano G. Deicke M. Grueneberg J. Weiss A. Engelen A. H. et al . (2020). Macroalgal–bacterial interactions: identification and role of thallusin in morphogenesis of the seaweed ulva (Chlorophyta). J. Exp. Bot.71, 3340–3349. doi: 10.1093/jxb/eraa066
2
Bakunina I. Nedashkovskaya O. Kim S. Zvyagintseva T. Mikhailov V. (2012). Distribution of α-n-acetylgalactosaminidases among marine bacteria of the phylum bacterioidetes, epiphytes of marine algae of the seas of Okhotsk and Japan. Microbiology81, 373–378. doi: 10.1134/S0026261712030022
3
Burgess J. G. Boyd K. G. Armstrong E. Jiang Z. Yan L. Berggren M. et al . (2003). The development of a marine natural product-based antifouling paint. Biofouling19, 197–205. doi: 10.1080/0892701031000061778
4
Burke C. Steinber P. Rusch D. Kjelleberg S. Thomas T. (2011). Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. U.S.A.108, 14288–14293. doi: 10.1073/pnas.1101591108
5
Califano G. Kwantes M. Abreu M. H. Costa R. Wichard T. (2020). Cultivating the macroalgal holobiont: effects of integrated multi-trophic aquaculture on the microbiome of Ulva rigida (chlorophyta). Front. Mar. Sci.7. doi: 10.3389/fmars.2020.00052
6
Chandrasekaran M. Rajeev K. (2010). Marine microbial enzymes. Biotechnology.9, 1–15.
7
Choi H. R. Park S. H. Kim D. H. Kim J. Y. Heo M. S. (2016). Analysis of the phylogenetic diversity and community structure of marine bacteria inhabiting Ulva pertusa. J. Life. Sci.26, 819–825. doi: 10.5352/JLS.2016.26.7.819
8
Comba-González N. B. Niño Corredor A. N. López Kleine L. Castaño D. M. (2021). Temporal changes of the epiphytic bacteria community from the marine macroalga Ulva lactuca (Santa Marta, Colombian-Caribbean). Curr. Microbiol.78, 534–543. doi: 10.1007/s00284-020-02302-x
9
Comba-González N. B. Ruiz-Toquica J. S. López-Kleine L. Montoya-Castaño D. (2016). Epiphytic bacteria of macroalgae of the genus ulva and their potential in producing enzymes having biotechnological interest. J. Mar. Biol. Oceanogr.5, 2. doi: 10.4172/2324-8661.1000153
10
Dalton H. M. Poulsen L. K. Halasz P. Angles M. L. Goodman A. E. Marshall K. C. (1994). Substratum induced morphological changes in a marine bacterium and their relevance to bioflm structure. J. Bacteriol176, 6900–6906. doi: 10.1128/jb.176.22.6900-6906.1994
11
Dhanya K. I. Swati V. I. Vanka K. Osborne J. (2016). Antimicrobial activity of Ulva reticulata and its endophytes. J. Ocean. Univ. China.15, 363–369. doi: 10.1007/s11802-016-2803-7
12
Dobretsov S. V. Qian P. Y. (2002). Effect of bacteria associated with the green alga Ulva reticulata on marine micro-and macrofouling. Biofouling.18, 217–228. doi: 10.1080/08927010290013026
13
Dominguez H. Loret E. P. (2019). Ulva lactuca, a source of troubles and potential riches. Mar. Drugs17, 357. doi: 10.3390/md17060357
14
Egan S. Harder T. Burke C. Steinberg P. Kjelleberg S. Thomas T. (2013). The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS. Microbiol. Rev.37, 462–476. doi: 10.1111/1574-6976.12011
15
Falkowski P. G. Katz M. E. Knoll A. H. Quigg A. Raven J. A. Schofield O. et al . (2004). The evolution of modern eukaryotic phytoplankton. Science305, 354–360. doi: 10.1126/science.1095964
16
Feitkenhauer H. (2003). Anaerobic digestion of desizing wastewater: influence of pretreatment and anionic surfactant on degradation and intermediate accumulation. Enzyme. Microb. Technol.33, 250–258. doi: 10.1016/S0141-0229(03)00125-X
17
Ferreira-Dias S. Sandoval G. Plou F. Valero F. (2013). The potential use of lipases in the production of fatty acid derivatives for the food and nutraceutical industries. Electron J. Biotechnol.16 (3), 12. doi: 10.2225/vol16-issue3-fulltext-5
18
Garrity G. De Vos P. Jones D. Kreig N. Ludwig W. Rainey F. et al . (2010). Bergey’s manual of systematic bacteriology: The firmicutes (USA: University of Georgia).
19
Ghaderiardakani F. Coates J. Wichard T. (2017). Bacteria-induced morphogenesis of Ulva intestinalis and Ulva mutabilis (Chlorophyta): a contribution to the lottery theory. FEMS Microbiol. Ecol.93, 8. doi: 10.1093/femsec/fix094
20
Ghaderiardakani F. Quartino M. Wichard T. (2020). Microbiome-dependent adaptation of seaweeds under environmental stresses: A perspective. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.575228
21
Guiry M. D. Guiry G. M. (2022) AlgaeBase. Available at: https://www.algaebase.org (Accessed December 22, 2021).
22
Guisan J. M. Bolivar J. M. López-Gallego F. Rocha-Martín J. (2020). Methods in molecular biology immobilization of enzymes and cells (New Yorks: Humana New York).
23
Gupta N. Gandham N. Jadhav S. Mishra R. N. (2015). Isolation and identification of acinetobacter species with special reference to antibiotic resistance. J. Nat. Sc. Biol. Med.6, 159–162. doi: 10.4103/0976-9668.149116
24
Gupta R. Gigras P. Mohapatra H. Goswami V. K. Chauhan B. (2003). Microbial -amylases: a biotechnological perspective. Process. Biochem.38, 1599–1616. doi: 10.1016/S0032-9592(03)00053-0
25
Hehemann J. Boraston A. Czjzek M. (2014). A sweet new wave: structures and mechanism of enzymes that digest polysaccharides from marine algae. Curr. Opin. Struc. Biol.28, 77–86. doi: 10.1016/j.sbi.2014.07.009
26
Higuera-Llanten S. Vasquez-Ponce F. Barrientos-Espinoza B. Mardones F. O. Marshall S. H. Olivares-Pacheco J. (2018). Extended antibiotic treatment in salmon farms select multiresistant gut bacteria with a high prevalence of antibiotic resistance genes. PloS One13, e0203641. doi: 10.1371/journal.pone.0203641
27
Holzinger A. Herburger K. Kaplan F. Louise A. L. (2015). Desiccation tolerance in the chlorophyte green alga Ulva compressa: does cell wall architecture contribute to ecological success? Planta.242, 477–492. doi: 10.1007/s00425-015-2292-6
28
Hui C. Shan L. Xiang-Rong X. Shuang-Shuang L. Guang-Jie Z. Kai-Feng S. et al . (2015). Antibiotics in typical marine aquaculture farms surrounding hailing island, south China: Occurrence, bioaccumulation and human dietary exposure. Mar. pollut. Bull.90, 181–187. doi: 10.1016/j.marpolbul.2014.10.053
29
Ismail A. Ktari L. Ahmed M. Bolhuis H. Bouhaouala-Zahar B. Stal L. et al . (2018). Heterotrophic bacteria associated with the green alga Ulva rigida: identification and antimicrobial potential. J. Appl. Phycol.30, 2883–2899. doi: 10.1007/s10811-018-1454-x
30
Joseph B. Ramteke P. W. (2013). Extracellular solvent stable cold active lipase from psychrotrophic bacillus sphaericus MTCC 7526: partial purification and characterization. Ann. Microbiol.63, 363–370. doi: 10.1007/s13213-012-0483-y
31
Kalpana B. J. Aarthy S. Pandian S. K. (2012). Antibiofilm activity of α-amylase from bacillus subtilis S8-18 against biofilm forming human bacterial pathogens. Appl. Biochem. Biotechnol.167, 1778–1794. doi: 10.1007/s12010-011-9526-2
32
Kandasamy P. Radhesh K. S. Radhakrishnan S. Sengali R. Balaji G. Gracy M. et al . (2020). Shewanella algae and microbulbifer elongatus from marine macro-algae – isolation and characterization of agarhydrolysing bacteria. Access Microbiol.10, 2. doi: 10.1099/acmi.0.000170
33
Kaur K. Dattajirao V. Shrivastava V. Bhardwaj U. (2012). Isolation and characterization of chitosan-producing bacteria from beaches of chennai, India. Enzyme. Res.2012, 421683. doi: 10.1155/2012/421683
34
Khan I. Dutta J. R. Ganesan R. (2017). Lactobacillus sps. lipase mediated poly (ϵ-caprolactone) degradation. Int. J. Biol. Macromol.95, 126–131. doi: 10.1016/j.ijbiomac.2016.11.040
35
Kouzuma A. Watanabe K. (2015). Exploring the potential of algae/bacteria interactions. Curr. Opin. Biotechnol.33, 125–129. doi: 10.1016/j.copbio.2015.02.007
36
Krishnan A. Kumar G. Loganathan K. Rao B. (2011). Optimization, production and partial purification of extracellular αamylase from Bacillus sp. Marini. Arch. Appl. Sci. Res.3, 33–42.
37
Krumperman P. H. (1983). Multiple antibiotic resistance indexing of escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol.46, 165–170. doi: 10.1128/aem.46.1.165-170.1983
38
Ktari L. Ismail A. Selmi H. Hmani I. Elbour M. (2022). Biological potential of the alien red alga asparagopsis taxiformis and characterization of its culturable associated bacteria. J. Appl. Phycol34, 2769–2782. doi: 10.1007/s10811-022-02818-8
39
Kumar D. Bhardwaj R. Jassal S. Goyal A. Khullar A. Gupta N. (2021). Application of enzymes for an eco-friendly approach to textile processing. Environ. Sci. pollut. Res. doi: 10.1007/s11356-021-16764-4
40
Kumar D. S. Ray S. (2014). Fungal lipase production by solid state fermentation–an overview. J. Anal. Bioanal. Tech6, 1–10. doi: 10.4172/2155-9872.1000230
41
Kumar P. Verma A. Sundharam S. S. Ojha A. K. Krishnamurthi S. (2022). Exploring diversity and polymer degrading potential of epiphytic bacteria isolated from marine macroalgae. Microorganisms10 (12), 2513. doi: 10.3390/microorganisms10122513
42
Kyle H. S. Freddie A. B. Karen D. P. Robert L. V. (1971). Lecithin-agar assay for lecithinase antibodies serum. Appl. Microbiol.21, 98–103. doi: 10.1128/am.21.1.98-103.1971
43
Lachnit T. Meske D. Wahl M. Harder T. Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol.13 (3), 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
44
Leinberger J. Holste J. Bunk B. Freese H. M. Spröer C. Dlugosch L. et al . (2021). High potential for secondary metabolite production of paracoccus marcusii CP157, isolated from the crustacean cancer pagurus. Front. Microbiol.12. doi: 10.3389/fmicb.2021.688754
45
Lemos M. L. Toranzo A. E. Barja J. L. (1985). Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microb. Ecol.11, 149–163. doi: 10.1007/BF02010487
46
Lupo A. Coyne S. Berendonk T. U. (2012). Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front. Microbiol.3. doi: 10.3389/fmicb.2012.00018
47
Martin M. Portetelle D. Michel G. Vandenbol M. (2014). Microorganisms living on macroalgae: diversity, interactions, and biotechnological applications. Appl. Microbiol. Biotech.98, 2917–2935. doi: 10.1007/s00253-014-5557-2
48
Masci D. Castagnolo D. (2021). “An introduction. exploiting enzymes as green catalysts in the synthesis of chemicals and drugs,” in Book the royal society of chemistry, vol. 68. , 118.
49
Matsumoto M. Yokouchi H. Suzuki N. Ohata H. Matsunaga T. (2003). Saccharification of marine microalgae using marine bacteria for ethanol production. Appl. Biochem. Biotechnol.5, 247–254. doi: 10.1385/ABAB:105:1-3:247
50
Miladi R. Manghisi A. A. Minicante S. Genovese G. Abdelkafi S. Morabito M. (2018). A DNA barcoding survey of Ulva (Chlorophyta) in Tunisia and Italy reveals the presence of the overlooked alien u. ohnoi. Cryptogamie Algologie.39, 85–107. doi: 10.7872/crya/v39.iss1.2018.85
51
Miranda C. D. Godoy F. A. Lee M. R. (2018). Current status of the use of antibiotics and the antimicrobial resistance in the Chilean salmon farms. Front. Microbiol.9. doi: 10.3389/fmicb.2018.01284
52
Mohapatra B. R. Bapuji M. Sree A. N. (2003). Production of industrial enzymes (Amylase, carboxymethylcellulase and protease) by bacteria isolated from marine sedentary organisms. Acta Biotechnol.23, p75–pp84. doi: 10.1002/abio.200390011
53
Moussa M. Baccar L. Ben Khemis R. (2005). La lagune de ghar El melh : Diagnostic écologique et perspectives d'aménagement hydraulique. Rev. Des. Sci. l'Eau / J. Water Sci.18, 13–26.
54
Munita J. M. Arias C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr.4, 10. doi: 10.1128/microbiolspec.VMBF-0016-2015
55
Naik S. Sanika Naik M. Vaingankar D. Mujawar S. Parab P. Meena S. (2019). Biodegradation of seafood waste by seaweed-associated bacteria and application of seafood waste for ethanol production. Adv. Biol. Sci. Res., 149–159.
56
Najjar A. Hassan E. A. Zabermawi N. Saber S. H. Bajrai L. H. Almuhayawi M. S. et al . (2021). Optimizing the catalytic activities of methanol and thermotolerant Kocuria flava lipases for biodiesel production from cooking oil wastes. Sci. Rep.11, 13659. doi: 10.1038/s41598-021-93023-z
57
O’Neill J. (2016). Tackling drug-resistant infections globally: Final report and recommendations. Microbiol. Spectr.4, 10. doi: 10.1128/microbiolspec.VMBF-0016-2015
58
Passalacqua K. D. Charbonneau M. E. O'Riordan M. X. D. (2016). Bacterial metabolism shapes the host-pathogen interface. Microbiol. Spectr.4, 10. doi: 10.1128/microbiolspec.VMBF-0027-2015
59
Qu T. Chengzong H. Xinyu Z. Yi Z. Chen G. Zhihao L. et al . (2021). "Bacteria associated with Ulva prolifera: A vital role in green tide formation and migration.". Harmful Algae108 (2021), 102104. doi: 10.1016/j.hal.2021.102104
60
Rao D. Webb J. S. Kjelleberg S. (2005). Competitive interactions in mixedspecies biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol.71, 1729–1736. doi: 10.1128/AEM.71.4.1729-1736.2005
61
Raveendran S. Parameswaran B. Ummalyma S. B. (2018). Applications of microbial enzymes in food industry. Food Technol. Biotechnol.56 (1), 16–30. doi: 10.17113/ftb.56.01.18.5491
62
Samant S. Naik M. M. Vaingankar D. C. Mujawar S. Y. Parab P. Meena S. N. (2019). “Biodegradation of seafood waste by seaweed-associated bacteria and application of seafood waste for ethanol production,” in Advances in biological science research (Academic Press), 149–159.
63
Satpute S. Bhawsar B. D. Dhakephalkar P. Chopade B. (2008). Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Indian. J. Mar. Sci.37, 243–250.
64
Saunders G. W. Kucera H. (2010). An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie Algologie31 (4), 487.
65
Schreiber L. Kjeldsen K. U. Funch P. Jensen J. Obst M. López-Legentil S. et al . (2016). Endozoicomonas are specific, facultative symbionts of Sea squirts. Front. Microbiol.7. doi: 10.3389/fmicb.2016.01042
66
Shields R. C. Mokhtar N. Ford M. Hall M. J. Burgess J. G. ElBadawey M. R. et al . (2013). Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS. One8, 55339. doi: 10.1371/journal.pone.0055339
67
Sierra G. (1957). A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie. Van. Leeuwenhoek.23, 15–22. doi: 10.1007/BF02545855
68
Singh A. K. Bilal M. Hafiz M. N. I. Anne S. Meyer Abhay R. (2021). Bioremediation of lignin derivatives and phenolics in wastewater with lignin modifying enzymes: Status, opportunities and challenges. Sci. Total. Environ.777, 1459883. doi: 10.1016/j.scitotenv.2021.145988
69
Sinha P. Bedi S. (2018). Production of sugars from cellulosic wastes by enzymatic hydrolysis. Int. J. Curr. Res.10, 72141–72144. doi: 10.24941/ijcr.31694.08.2018
70
Smith H. L. Goodner K. (1958). Detection of bacterial gelatinases by gelatin-agar plate methods. J. Bacteriol.76, 662–665. doi: 10.1128/jb.76.6.662-665.1958
71
Société Française de Microbiologie (2020). “Comité de l’antibiogramme de la société française de microbiologie,” in CASFM / EUCAST : Société française de microbiologie, 6–181.
72
Steinhagen S. Weinberger F. Karez R. (2019). Molecular analysis of Ulva compressa (Chlorophyta, Ulvales) reveals its morphological plasticity, distribution and potential invasiveness on German north Sea and Baltic Sea coasts. Eur. J. Phycol.54, 102–114. doi: 10.1080/09670262.2018.1513167
73
Tamura K. Stecher G. Kumar S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol.38, 3022–3027. doi: 10.1093/molbev/msab120
74
Trivedi N. Gupta V. Kumar M. Kumari P. Reddy B. (2011). An alkali-halotolerant cellulase from bacillus flexus isolated from green seaweed Ulva lactuca. carbohyd. Polym.83, 891–897. doi: 10.1016/j.carbpol.2010.08.069
75
Tujula N. A. Crocetti G. R. Burke C. Thomas T. Holmström C. Kjelleberg S. (2010). Variability and abundance of the epiphytic bacterial community associated with a green marine ulvacean alga. ISME J.4 (2), 301–311. doi: 10.1038/ismej.2009.107
76
Van der Maarel M. J. van der Veen B. Uitdehaag J. C. Leemhuis H. Dijkhuizen L. (2002). Properties and applications of starch-converting enzymes of the alpha-amylase family. J. Biotechnol.94, 137–155. doi: 10.1016/s0168-1656(01)00407-2
77
Weiss A. Costa R. Wichard T. (2017). Morphogenesis of Ulva mutabilis (Chlorophyta) induced by maribacter species (Bacteroidetes, flavobacteriaceae). Botanica. Marina.60, 197–206. doi: 10.1515/bot-2016-0083
78
Wichard T. Charrier B. Mineur F. Bothwell J. H. De Clerck O. Coates J. C. (2015). The green seaweed Ulva: a model system to study morphogenesis. Front. Plant Sci.6. doi: 10.3389/fpls.2015.00072
79
Wienhausen G. Noriega-Ortega B. E. Niggemann J. Dittmar T. Simon M. (2017). The exometabolome of two model strains of the roseobacter group: a marketplace of microbial metabolites. Front. Microbiol.8, 1985. doi: 10.3389/fmicb.2017.01985
80
Wu G. Qin Y. Cheng Q. Liu Z. (2014). Characterization of a novel alkali-stable and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda. J. Mol. Catal. B-Enzym110, 8–15. doi: 10.1016/j.molcatb.2014.08.023
Summary
Keywords
seaweed-epibiont, enzyme production, antimicrobial activity, bacterial phylogeny, antibiogram, Ulva ohnoi , antibioresistance
Citation
Hmani I, Ktari L, Ismail A and EL Bour M (2023) Biotechnological potential of Ulva ohnoi epiphytic bacteria: enzyme production and antimicrobial activities. Front. Mar. Sci. 10:1042527. doi: 10.3389/fmars.2023.1042527
Received
12 September 2022
Accepted
20 March 2023
Published
19 April 2023
Volume
10 - 2023
Edited by
Ming Ma, School of Pharmaceutical Sciences, Peking University, China
Reviewed by
Satya P. Singh, Saurashtra University, India; Peng Jiang, Institute of Oceanology (CAS), China
Updates
Copyright
© 2023 Hmani, Ktari, Ismail and EL Bour.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. EL Bour, monia.elbour@instm.rnrt.tn
This article was submitted to Aquatic Microbiology, a section of the journal Frontiers in Marine Science
†ORCID: L. Ktari, orcid.org/0000-0002-0515-4135
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.