- 1Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2University of Chinese Academy of Sciences, Huairou District, Beijing, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China
- 4Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese, Academy of Sciences, Guangzhou, China
Identifying the trophic role of primary producers is the basis of assessing seagrass bed functions but remains difficult due to the underdetermined analysis method. Here, we analyzed the multiple isotopes (δ13C, δ15N, and δ34S values) and fatty acid markers of food sources and macrobenthos in a tropical seagrass bed in summer and winter, and tried to combine these indicators to resolve the limitation of δ13C and δ15N values analysis. We found that the δ13C and δ15N values of epiphytes were like that of seagrass and macroalgae, while the δ34S values of epiphytes and macroalgae were significantly different, and the dominant unsaturated Fatty acid markers of seagrass (18:2n6c and 18:3n3) and epiphytes (16:1n7) were obviously different. These results suggest that the combination of multiple isotopes and Fatty acid markers can effectively distinguish the complex food source. In addition, we also found that multiple isotopes were more suitable to identify the food sources of polychaetes and snails with simple diets, fatty acids were more suitable to identify the food sources of crustaceans with complex diets, but their combination is essential in identifying the diets of macrobenthos since the wide range of isotopic values for omnivores crustaceans and the Fatty acid markers transformation during snails and polychaetes assimilation might mislead us when only isotopes or Fatty acid markers were used. Our findings suggest that in tropical seagrass beds, using multiple isotopes and fatty acid markers together can help reduce the uncertainty caused by single markers variation and thus strengthen the separation of food sources and the diets of different consumer species.
1 Introduction
Seagrass beds rank among the most valuable ecosystems on the planet, despite only covering 0.15% of global sea surface area (Saderne et al., 2019; Ivajnšič et al., 2022). They are not only highly productive ecosystems, but also support food webs of coastal habitats (Jiang et al., 2019; Unsworth et al., 2019; Canadell and Jackson, 2021). They provide various food sources including seagrass, epiphytes, macroalgae, suspended particulate organic matter (SPOM), and microphytobenthos in the sediment for various fish and invertebrates, and serve as a shelter, habitat, and nursery ground for adult and juvenile faunas (Liu et al., 2020b; Cui et al., 2021a). However, understanding the function of seagrass bed food webs remains to be a challenge since it requires an effective approach to discriminate the food sources and their potential importance in seagrass beds (Abrantes and Sheaves, 2009; Kohlbach et al., 2021).
Integrating carbon and nitrogen stable isotopes into some kind of modeling, especially the Bayesian mixing models, has been widely employed to evaluate the food source contribution of seagrass ecosystems (Paar et al., 2019; Gagnon et al., 2021). This approach is based on the premise that the carbon and nitrogen stable isotopic values of consumer tissues reflect the diets actually assimilated by consumers over time (Weems et al., 2012). Meanwhile, the isotopic ratio enrichment between each trophic level can be predicted. Previous studies suggested that the δ13C and δ15N values of organisms generally increase by 0.5-1‰ and 2-5‰ relative to their food sources, respectively (Caut et al., 2009; Sun et al., 2020). However, this approach only provides two-dimensional discrimination, which may fail to identify the trophic base of the seagrass bed food webs, especially for tropical seagrass bed food webs, since they usually have more diversified primary producers and the isotopic values of food sources are often similar (Nakamoto et al., 2019). For example, in tropical seagrass ecosystems, the isotopic values of seagrass are usually close to those of epiphytes (Mittermayr et al., 2014; Cui et al., 2021a). In addition, the temporal variances in environmental factors may also result in similar stable isotopic values between seagrass and other food sources (Fritts et al., 2018). Therefore, it is essential to improve this approach.
Recent studies showed that the sulfur stable isotope composition has been successfully used in identifying the consumer diets of saltmarsh food webs (Jinks et al., 2020; Lippold et al., 2020). In saltmarsh ecosystems, the δ34S values of food sources can be clearly separated (seawater column: ~+20‰, anaerobic sediments: ~-24‰, (Valiela et al., 2018b) because different primary producers usually acquire sulfur from different sources. For example, rooted marine vascular plants obtain sulfur most from anaerobic sediments, whereas other food sources, such as algae, obtain sulfur mostly from seawater sulfate. These factors result in a clear dissimilation in their δ34S values (Fritts et al., 2018). Similarly, the δ34S values of rooted seagrass may be also clear dissimilation from that of macroalgae or epiphytes in seagrass beds. Therefore, the sulfur isotopic composition may be proper as a third index (besides δ13C and δ15N) to help us discriminate the food sources of seagrass bed food webs. However, in tropical seagrass beds with various food sources, the δ34S values of primary producers are also largely affected by local sulfur cycling (Guiry et al., 2021). For example, in tropical seagrass beds with various food sources, some food sources (e.g., epiphytes, macroalgae, and phytoplankton) obtain sulfur from seawater sulfate and may have similar isotopes (Valiela et al., 2018b), and the δ34S values of sediment may be more enriched or depleted under microbial metabolisms (e.g., microbial sulfate reduction and microbial sulfide oxidation) (Pellerin et al., 2019). Therefore, the ambiguities associated with similar isotopes in tropical seagrass beds may not be completely overcome by only using carbon, nitrogen, and sulfur stable isotopic composition.
Fatty acid markers may be another helpful tool to overcome the isotopic uncertainties since many primary producers can synthesize high levels of specific fatty acids as their unique identity indicators, and laboratory experiments have proven that the transfer of specific fatty acid markers is conservative in the process of fish diets (Xu et al., 2020). However, we still cannot ignore whether these specific fatty acid markers are similar in primary producers of seagrass beds, and whether they will be affected by the ability of macrobenthos to metabolize and transform fatty acids. Therefore, to overcome the limitation of only using carbon and nitrogen in the modeling, adding the analysis of sulfur isotopes and fatty acids biomarkers may be a promising approach for food web studies in tropical seagrass beds.
Macrobenthos are the key components of the seagrass bed food webs. They not only rely on various primary producers, but also serve as primary food sources for other higher faunas. Therefore, in this study, we made an attempt to identify whether stable isotopes and fatty acids biomarkers can effectively discriminate food sources in complex, tropical seagrass beds, and to explore whether this combination can reliably identify the diets of different macrobenthos.
2 Materials and methods
2.1 Study area
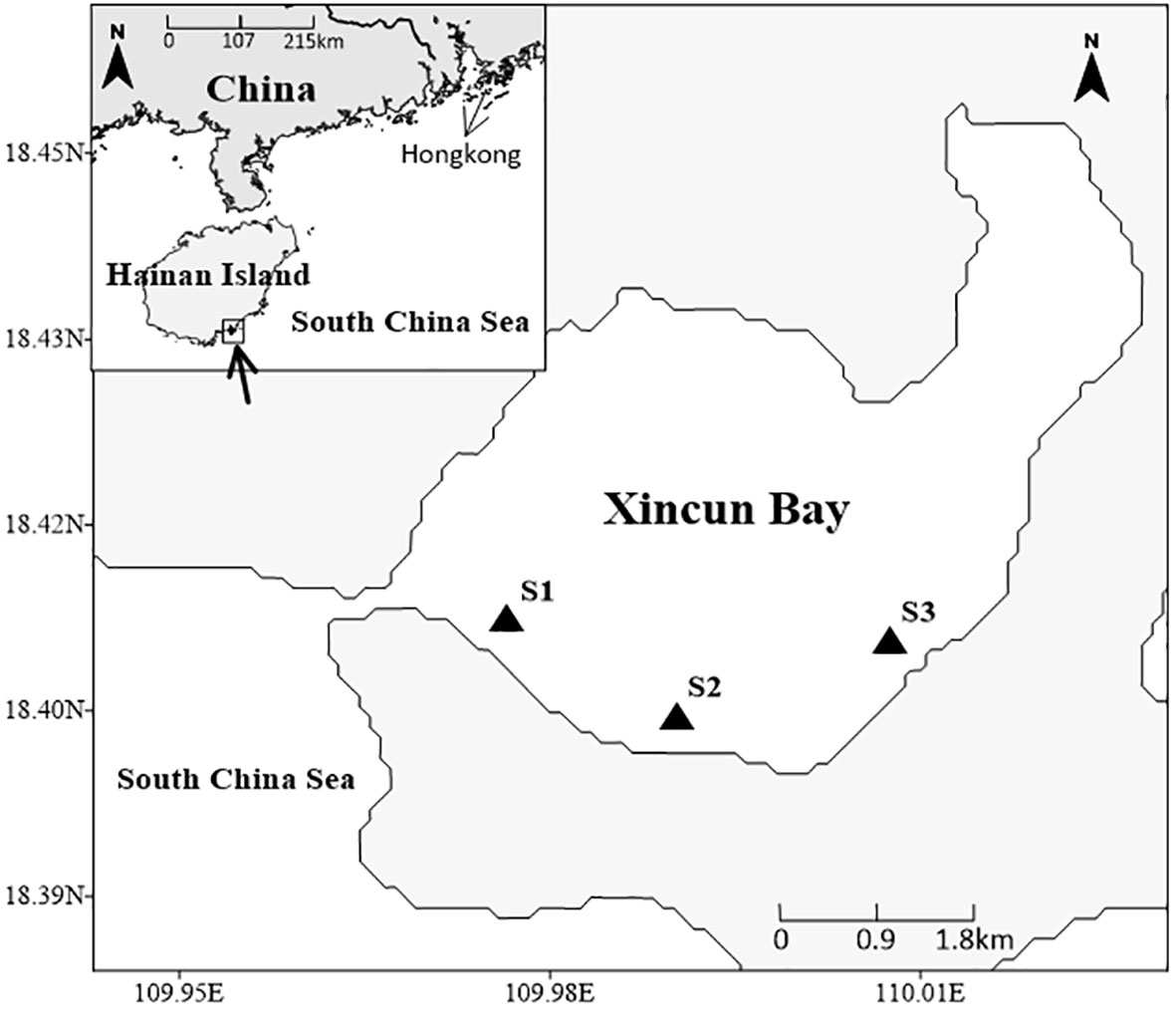
The study region is placed in Xincun Bay, which is located in the southeast coast of Lingshui County, Hainan Island, South China Sea (Figure 1). This bay is an almost entirely closed bay with only one narrow channel connecting to the South China Sea in the southwest. According to previous surveys, the Xincun seagrass bed is a mixed seagrass bed, which occupies an area of approximately 175 ha and is mainly distributed in the shallow waters of southern Xincun Bay (Huang et al., 2006; Huang et al., 2019). Seagrass species in this bay include Enhalus acoroides, Thalassia hemprichii, Cymodocea rotundata, Halodule uninervis, and Halophila ovalis, with E. acoroides and T. hemprichii as the dominant species (Huang et al., 2006; Huang et al., 2019).
2.2 Sampling design, field collection, and sample processing
According to the abundance and distribution of seagrass, samples for analysis were collected at stations S1, S2, and S3 (Figure 1) during low tide in summer (August 2018) and winter (January 2019). At each station, one to three samples were collected depending on the abundance and distribution of sampling organisms. Seagrass (E. acoroides and T. hemprichii) and macroalgae Ulva lactuca were collected by hand and washed with distilled water to remove attachments. For seagrass epiphytes, we applied the commonly used scraping method (i.e., removed carefully from the surface of seagrass leaves using a scalpel blade), because of the wideness of seagrass leaves: from 0.8 to 2.1 cm width (Huang et al., 2019), and the relatively high biomass of epiphytes (Cui et al., 2021b). Suspended particulate organic matter (SPOM) samples were obtained by filtering surface seawater onto pretreated (heated 2h at 450°C) Whatman GF/F filters. For sediment organic matter (SOM) samples, only the upper 1cm of the surface sediment layer was collected using a spade. For macroinvertebrates, crustaceans were collected by trammel nets, gastropods were sampled by hand, and polychaetes were sampled by sieving sediments through the 5mm mesh screen. Live invertebrates were put into containers with filtered seawater and kept for 24h to evacuate gut contents. Afterward, the gastropods and crustaceans were sorted, identified, and taken muscle. All the samples were placed in plastic polyethylene bags and stored in a freezer at -20°C (< 1 week) for their transportation to the laboratory.
In the laboratory, samples were freeze-dried at -40°C for 48h, and grounded into a fine homogeneous powder. Before stable isotope analysis, epiphytes, SPOM, and SOM were acidified to remove the effect of inorganic carbon on the δ13C ratio of these samples. SPOM samples were acidified by exposure to hydrochloric acid (HCl) vapor for 24h. Epiphytes and SOM were acidified with a 1 mol/L solution until there were no bubbles. Subsequently, all acidified samples were rinsed with distilled water, freeze-dried again, and stored in the centrifuge tube for subsequent analysis.
Homogenized powder samples and SPOM were transferred to tin capsules, and δ13C, δ15N, and δ34S values of samples were measured using a continuous-flow isotope-ratio mass spectrometer (Delta V Advantage, Thermo Fisher Scientific, Waltham, MA, USA) attached to an elemental analyzer (Flash EA 1112, Thermo Fisher Scientific, Milan, Italy). δ13C, δ15N, and δ34S values were expressed in ‰ relative to the standard reference materials (Vienna Pee Dee Belemnite for δ13C values, atmospheric N2 for δ15N values and Vienna Canyon Diablo Troilite for δ34S values) using the standard δ notation:
where X is δ13C, δ15N or δ34S ratio, and R is 15N/14N, 13C/12C or 34S/32S. The detection limits for δ13C, δ15N and δ34S values were<0.2‰,<0.2‰, and<0.3‰, respectively.
Fatty acids were extracted using a method modified from (Folch et al., 1957). Briefly, a 2:1(v/v) chloroform and methanol mixed solution was used to extract the lipids of each sample (about 0.2-1mg), and a 0.9% sodium chloride solution was used to separate the lipids solution from the mixed solution. The sample was then concentrated using a rotary evaporator to obtain total lipid. The resulting total lipid samples were methylated with 0.5mol/L sodium hydroxide-methanol solution, derivatized with boron trifluoride-methanol solution, extracted with hexane and stratified with saturated sodium chloride to obtain fatty acid methyl ester (FAMEs) samples. To quantification the fatty acid, mixtures of FAMEs: internal standard methyl nonadecanoate (1:1) were run on a Gas Chromatograph (GC-7890B, Agilent Technologies, Inc. USA) attached to a Mass Spectrometer Detector (MSD-5977A, Agilent Technologies, Inc. USA). Helium was the carrier gas, and the thermal gradient was set as from 125°C to 227°C at 2°C min-1. Fatty acid profile identifications were performed by comparison to relative retention times of a known standard (GAQSIQ, 2008).
2.3 Statistical analyses
The contribution proportion of each source to macroinvertebrate diets was estimated based on δ13C, δ15N or δ34S values using the Bayesian stable isotope mixing model SIMMR (Parnell and Inger, 2019), which is a package in R (Team, 2019). Here, the food sources were divided into four groups mainly including seagrass (including E. acoroides and T. hemprichii), epiphytes (including epiphytes attached to E. acoroides and T. hemprichii), macroalgae (U. lactuca), and SPOM. SOM was excluded from SIMMR since that SOM is a mixture of other primary food sources (i.e., seagrass, epiphytes, macroalgae, and POM) (Xu et al., 2018). The δ34S values of SPOM were not detected due to the relatively small amount of collected material and were not used in this model. The trophic enrichment factors (TEFs) in this Bayesian mixing model were δ13C of 0.4 ± 1.3‰ and δ15N of 2.3 ± 1.61‰ for snails, bivalves and polychaetes (Mascart et al., 2018; Débora et al., 2020), and δ13C of 0.5 ± 0.8‰ and δ15N of 3.2 ± 0.26‰ for crustaceans (Mittermayr et al., 2014; Riccialdelli et al., 2017; Beesley et al., 2020). This SIMMR model was based on normal (100,000) interactions, and outputs of the contributional proportion were expressed as mean value, 2.5%, 25%, 50%, 75% and, 99% confidence intervals (CI).
To explore whether the variation of multiple stable isotope ratios (δ13C, δ15N, and δ34S) was greatest amongst food sources (i.e., seagrass, epiphytes, macroalgae, SPOM, and SOM) or between seasons (i.e. summer and winter), all data were firstly checked the normality and/or homogeneity of variance using the Shapiro-Wilk test and Levene’s test, respectively. Subsequently, a two-way ANOVAs was performed. Meanwhile, a post-hoc test (Tukey’s HSD) was also applied in order to identify the specific differences among food source types. Furthermore, we also performed a one-way ANOVA or Mann-Whitney U-test to evaluate the seasonal changes of each food source in detail. Similarly, two-way ANOVAs were performed in order to test the significant differences amongst consumer groups (snails, crustaceans, bivalves, and polychaetes) and between seasons. The variations in the fatty acid markers of food sources and consumers were tested by a distance-based permutational analysis of variance (PERANOVA). This analysis used Euclidean distance resemblance calculated from untransformed data. In addition, to visualize multivariate patterns we generated a generalized discriminant analysis using Canonical Analysis of Principal Coordinates (CAP) (Anderson and Willis, 2003). To explore which fatty acid markers contributed most to the differences among food sources, we correlated the fatty acid data with the canonical axes. ANOVA analyses were performed with SPSS version 22 (IBM Corporation, Chicago, IL, USA), PERMANOVA and CAP were performed with PRIMER v6 software (Clarke and Gorley, 2006).
3 Result
3.1 Multiple stable isotopes of food sources and macrobenthos
The main food sources in Xincun seagrass bed displayed a wide range of δ13C and δ34S values (Figure 2), which correspondingly ranged from -18.6 ± 0.9‰ for winter SPOM to -8.4‰ for summer macroalgae (Appendix S1-Table A1), and from 5.6 ± 2.5‰ for winter SOM to 17.4 ± 0.2‰ for winter macroalgae, respectively. In contrast, the δ15N values showed a narrow range, ranging from 5.5 ± 0.2‰ for winter SPOM to 8.6 ± 0.4‰ for summer macroalgae. Two-way ANOVA analysis showed distinct differences among the five food sources for δ13C, δ15N, and δ34S values, while the clear distinction between seasons could only be found in δ13C and δ34S values (Table 1). Post-hoc test indicated that the difference in δ13C values among the food sources was driven by the OM (organic matter, including SPOM and SOM), which differed from the seagrass, epiphytes, and macroalgae, respectively (Figure 2 and Appendix S1-Table A2). Whereas, significant differences in the δ15N values only existed in seagrass versus macroalgae, epiphytes versus SPOM, and macroalgae versus OM, respectively. By contrast, all other food sources were significantly different for δ34S values.Multiple stable isotope (δ13C, δ15N and δ34S) values of macrobenthos were significantly different among groups (snail, crustaceans, bivalves and polychaetes, Two-way ANOVA, δ13C: F3 = 19.56, p = 0, δ15N: F3 = 20.90, p = 0, δ34S: F3 = 10.22, p = 0), whereas there were no significant differences between seasons (Two-way ANOVA, δ13C: F1 = 0.33, p = 0.565, δ15N: F1 = 0.03, p = 0.861, δ34S: F1 = 8.19, p = 0.01). In contrast to the food sources, macrobenthos had a reduced range in the δ13C and δ34S values, which ranged from -15.8 ± 0.2‰ (crustaceans Calappa xp in winter) to -7.3 ± 0.5‰ (snail Notosinister subaura in winter) for δ13C values and from 8.8 ± 1.9‰ (polychaetes Arenicola cristata in summer) to 18.3 ± 0.5‰ (crustaceans Calappa xp in winter) for δ34S values, redpectively. However, for δ15N values, macrobenthos showed a larger range than that of food sources, ranging from 8‰ (snail Notosinister subaura in summer) to 12.1 ± 0.7‰ (crustaceans Menippe rumphii summer).
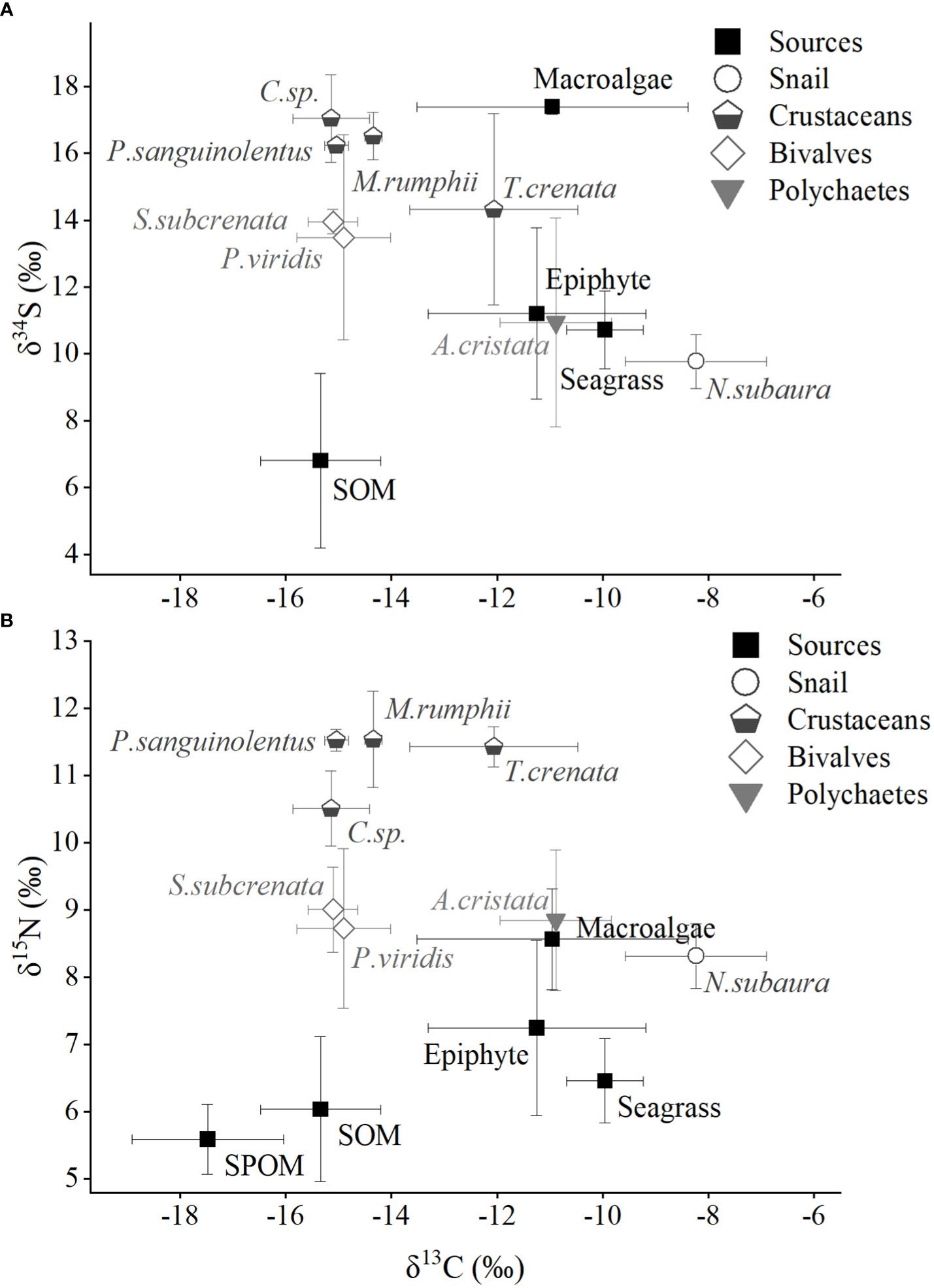
Figure 2 Annual mean values (including summer and winter) of isotopic signatures (A, δ13C and δ15N; B, δ13C and δ34S) for food sources and macroinvertebrates in Xincun seagrass bed. SPOM and SOM represent suspended particulate organic matter and sediment organic matter, respectively. The detailed labels of macroinvertebrates are given in Appendix-Table A1.
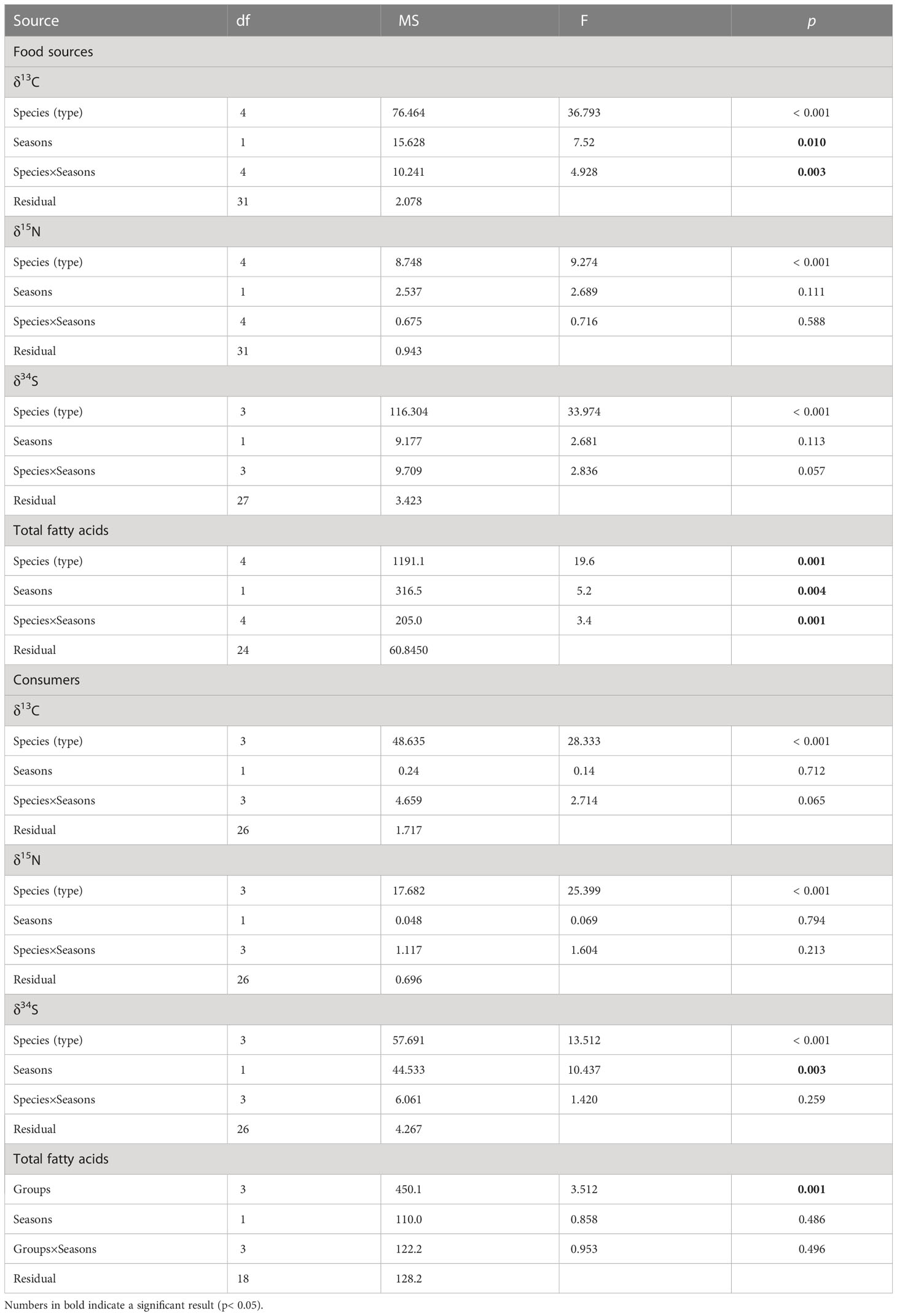
Table 1 Results of two-way ANOVA of multiple stable isotopic compositions (δ13C, δ15N, and δ34S) for food sources and consumers, and PERANOVA of fatty acids for food sources and consumers, (ns, no significant difference p < 0.05).
3.2 Fatty acid composition of food sources and macrobenthos
A total of twenty-one fatty acids were used for the profiles of food sources and macrobenthos (Appendix-Table A3). For food sources, percentages of saturated fatty acids accounted for greater than 50% of total fatty acids composition. PERANOVA of the total fatty acid data showed significant differences among food sources and between seasons (Table 1). CAP on total fatty acids showed that seagrass was significantly distinct from epiphytes and OM along the first axis, and the epiphytes were clearly separated from the macroalgae and OM along the second axis (Figure 3A), while SPOM had a greater overlap with SOM. When only poly-unsaturated fatty acids were considered (Figures 3B, 4), Fatty acids 18:2n6c and 18:3n3 for seagrass, 18:1n9c and 16:1n7 for epiphytes, and 18:1n9t, 20:1, 20:5n, 22:6n3, 18:3n6, and 14:1n5 for OM, were significantly higher than those of the other food sources. Macroalgae showed some certain degree of dispersed fatty acid profiles with relatively high levels of 15:1n5 in summer, and 18:1n9c in summer and winter, although these fatty acids also showed similarly high levels in seagrass and epiphytes.
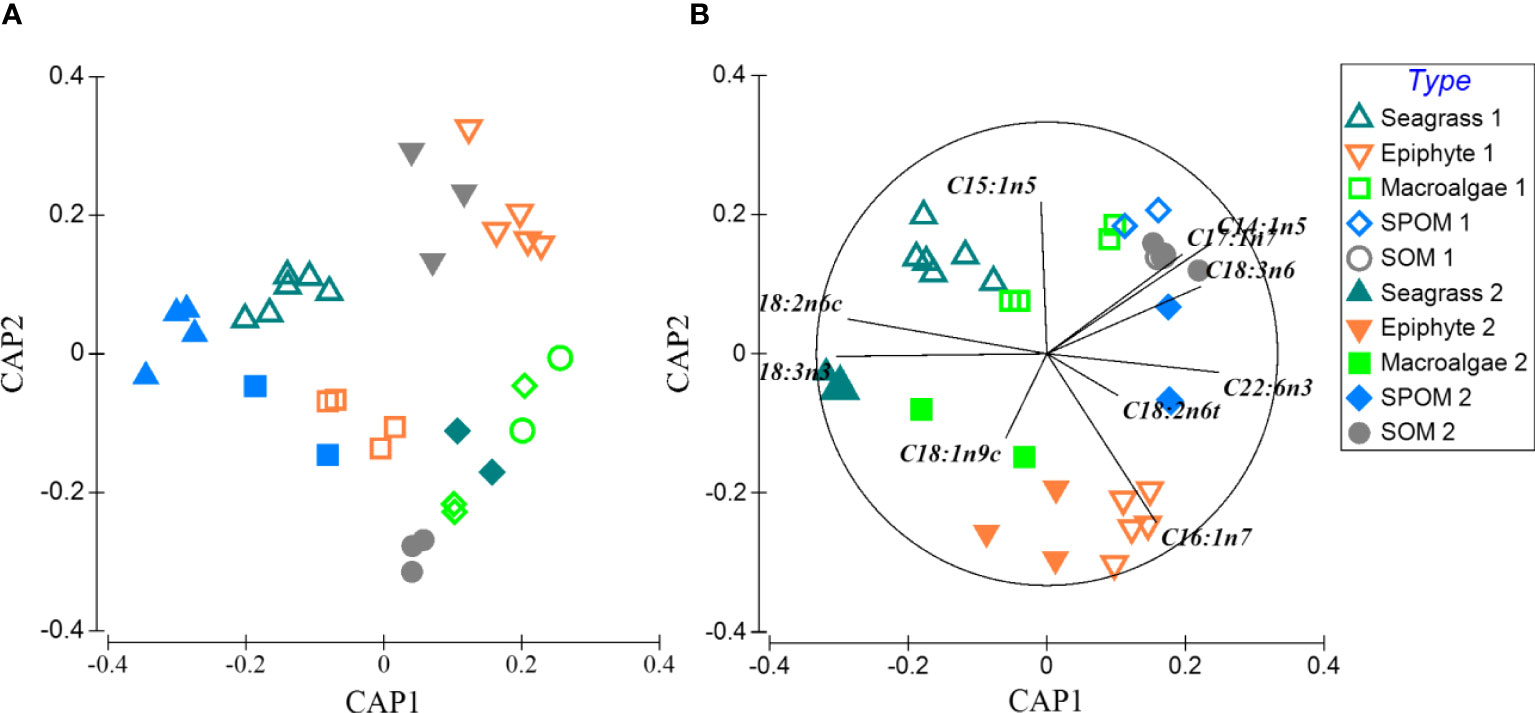
Figure 3 Constrained ordination (canonical analysis of principal coordinates, CAP) of total fatty acid composition from 5 sources in winter and summer of Xincun Bay. Hollow (1) and solid (2) symbols represent sources in summer and winter, respectively. (A, B) display the result of clustering results of total fatty acid composition from 5 sources, and principal component of the 5 sources, respectively.
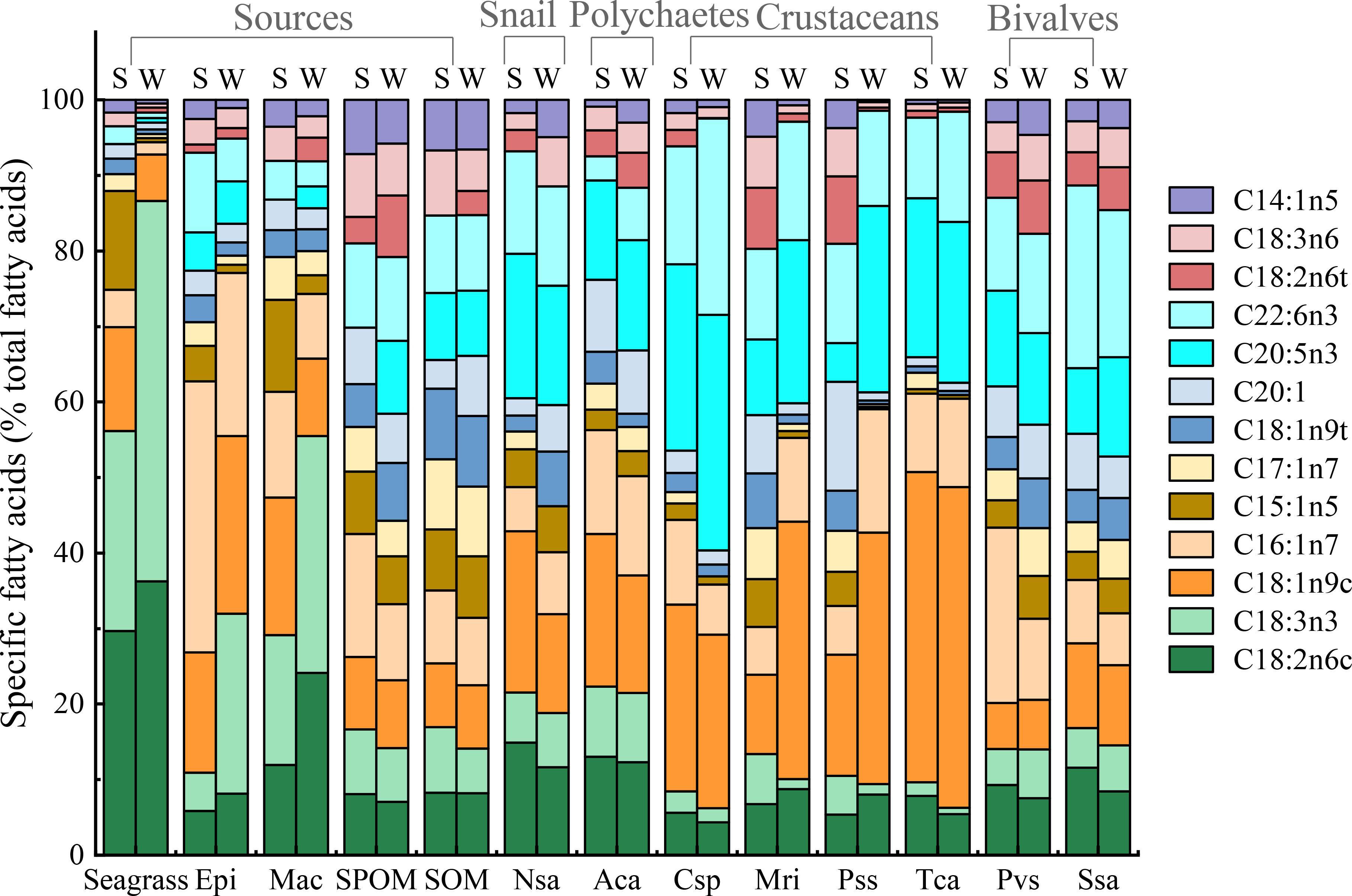
Figure 4 The percentages of specific fatty acids (% of total fatty acids) for the food sources and macroinvertebrates in summer (S) and winter (W) in the Xincun seagrass bed. Epi, Epiphyte; Mac, Macroalgae; Nsa, Notosinister subaura; Aca, Arenicola cristata; Cxp, Calappa xp; Mri, Menippe rumphii; Pss, Portunus sanguinolentus; Tca, Thalamita crenata; Pvs, Perna viridis; Ssa, Scapharca subcrenata.
For consumers, the percentages of saturated fatty acids still accounted for a larger proportion (55-75%) than unsaturated fatty acids (26-47%), but it was lower than that of food sources (Appendix-Table A3). Significant differences in total fatty acids were also evident among groups (Table 1), while differences between seasons were not significant. Especially, for poly-unsaturated fatty acids, gastropods, crustaceans, and polychaetes had a high level of 18:1n9c and 20:5n3, while bivalves contained a high level of 16:1n7 and 22:6n3. In addition, the levels of 18:2n6c and 18:3n3 in snails and polychaetes were higher than those of other macrobenthos.
3.3 Isotopic mixing model
The SIMMR mixing models showed that the contribution of the food sources to most macrobenthos was relatively similar between seasons in this seagrass bed (Figure 5). For the snail Notosinister subaura and the polychaete Arenicola cristata, seagrass contributed the highest to their diet, with the corresponding mean value of 30-35% and 29-31%, respectively, while epiphytes contributed subordinately, with the corresponding mean value of 25-26% and 24-29%, respectively. For the crustaceans, SPOM contributed significantly to Calappa sp and Menippe rumphii in summer, while macroalgae and SPOM were the main contributors to their diets in winter. The diet of Portunus sanguinolentus and Thalamita crenata had significantly seasonal variation, with a high contribution of epiphytes to P.sanguinolentus (67%) and macroalgae (42%) to T.crenata in summer, as well as the high contribution of macroalgae to their diets in winter (Figure 5). While for the bivalves Perna viridis and Scapharca subcrenata, SPOM showed a consistently high contribution to their diets in both seasons (mean value, 34-44% and 33-41%, respectively).
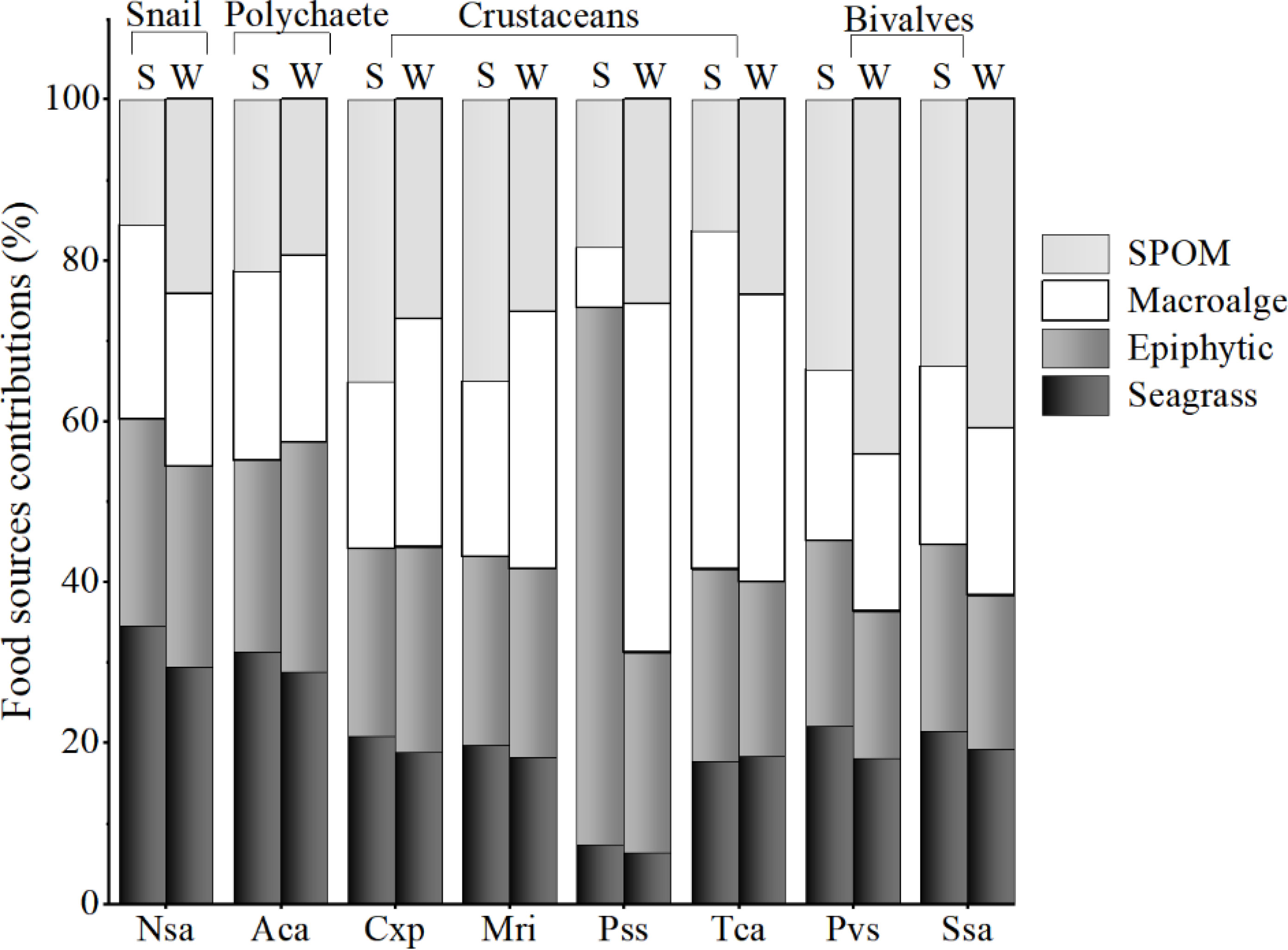
Figure 5 Mean relative contributions of food sources to the diets of macroinvertebrates in summer (S) and winter (W) in the Xincun seagrass bed. Nsa, Notosinister subaura; Aca, Arenicola cristata; Csp, Calappa sp.; Mri, Menippe rumphii; Pss, Portunus sp.; Tca, Thalamita crenata; Pvs, Perna viridis; Ssa, Scapharca subcrenata. The detailed outputs of contribution proportion calculated by SIMMR are given in Supporting information Table A5.
4 Discussion
4.1 Differences in the multiple stable isotopes and fatty acids among food sources
In the present study of tropical primary produces, the main variance of food sources for stable isotopes and fatty acids was amongst source types rather than seasons, except for the obvious seasonal variation in δ13C values of macroalgae. In particular, the significant difference in δ13C values between seagrass and OM allows this indicator to distinguish between vascular and organic matter. However, the wide range of δ13C (from -14‰ to -7.9‰) and δ15N (from 4.1‰ to 8.8‰) values for epiphytes overlapped with that of seagrass, which suggested the difficulty in using these biomarkers to identify the diets of consumers. The epiphytes attached to seagrass are mainly composed of bryozoans, diatoms Cocconeis scutellum, red algae, brown algae, small crustose coralline alga Pneophyllum lejolisii, bacteria, and fungi (Kharlamenko et al., 2001). The variation of environmental factors caused by stations (e.g. salinity and nutrient availability) and seasons (e.g. temperature and light) may lead to differences in the composition of epiphytes, and thus result in a large isotopic range (Nichols et al., 1985; Gacia et al., 2009; Liu et al., 2017). Similarly, the wide range of δ13C values for macroalgae, overlapped with that of seagrass, might also be caused by these variations in environmental factors. This is mainly because the macroalgae Ulva sp. have been proven to be capable of both C3 and C4 photosynthesis (Valiela et al., 2018a), which may be affected by environmental factors (Xu et al., 2012). However, the δ 15N values of seagrass and macroalgae were significantly different, which indicated that we can use this indicator to distinguish macroalgae from seagrass. This may be reasonable because macroalgae are generally considered to assimilate significantly sufficient amounts of dissolved inorganic nitrogen from highly nutritious seawater columns in a shorter time than seagrass and OM (Gartner et al., 2002; Thornber et al., 2008), and previous study have shown that this area was subject to considerable anthropogenic nitrogen source input (Liu et al., 2020a).
Sulfur isotope was considered to be the most likely biomarker to effectively distinguish food sources since different food sources have distinct sulfur sources (Moncreiff and Sullivan, 2001). In the present study, significant differences in the δ34S values of food sources among food sources helped distinguish food sources. In particular, the obvious difference in δ34S values between macroalgae and epiphytes can make up for the limitation in the use of δ13C and δ15N values. Algae usually obtain more sulfur from seawater sulfate (δ34S =20‰) than seagrass, which results in enriched δ34S values (Moncreiff and Sullivan, 2001). In our study, the δ34S values of macroalgae were the most enriched, which was similar to those of macroalgae in other seagrass beds (Holmer and Nielsen, 2007; Oduro et al., 2012). However, similar to δ13C and δ15N values, the δ34S values of epiphytes and seagrass also exhibited significant overlap. This obvious overlap might be attributed to the difference in the composition of epiphytes among stations or the resuspension of benthic microalgae in some stations (Jorge and Van Beusekom, 1995; Kasim and Mukai, 2006), which led to great changes in the sulfur isotopes of epiphytes, ranging from 5.8‰ (close to the δ34S values of 6.8‰ SOM in this area) to 15.5‰. In addition, significant seasonal differences in δ34S values of seagrass and epiphytes may also increase the overlapping probability of their δ34S values. Therefore, the combination of multiple stable isotopes cannot completely distinguish the food sources in this area.
In comparison to the multiple stable isotope data, the fatty acid compositions of food sources are generally considered to be significantly different (Crawley et al., 2009; Madgett et al., 2019). In our study, all food sources were rich in saturated fatty acids (61%-77%), especially 16:0, which was similar to the research results in other seagrass beds (Crawley et al., 2009; Park et al., 2013). In contrast, the unsaturated fatty acid compositions among food sources were clearly distinct. The much higher level of 18:2n6c and 18:3n3 in seagrass, 16:1n7 in epiphytes and 18:1n9t, 20:1, 20:5n3, 22:6n3 18:2n6t,18:2n6 and 14:1n5 in OM can effectively separate seagrass, epiphytes, and OM from each other. It is worth noting that 20:5n3 and 22:6n3 were also abundant in epiphytes. The unsaturated fatty acids 16:1n7, 20:5n3, and 22:6n3 are generally biosynthesized by diatoms (Coelho et al., 2011) and have been used as benthic diatom biomarkers (Kharlamenko et al., 2001; Belicka et al., 2012; Jankowska et al., 2018). Therefore, diatoms mainly originated from the resuspension of SOM might have an important effect on the component of epiphytes, which can also explain the wide range of δ13C values for epiphytes and the overlap in δ15N and δ34S values for epiphytes and SOM in this area. In addition, the fatty acid compositions of macroalgae were also similar to that of seagrass and epiphytes, with a high level of 18:2n6c and 18:3n3. Such fatty acid composition characteristics of seagrass and macroalgae were also evidenced in other temperate bays (Meziane and Tsuchiya, 2000; Lebreton et al., 2011). The most likely explanation was that macroalgae and seagrass are in the same lineage with the same suite of photosynthetic pigments and similar biochemical pathways (Galloway et al., 2012). These characteristics limit us to distinguish seagrass and macroalgae based on their specific fatty acid compositions.
4.2 Application of multiple stable isotopes and fatty acids markers in different macrobenthos
Despite the combination of multiple stable isotopes and fatty acid biomarkers can make a clear distinction for food sources, caution is still required with interpretations of field-based biomarker studies, as some factors such as growth rates, metabolism, and mixed diets will affect the isotope values and fatty acid compositions of consumers (Hanson et al., 2010; Pecquerie et al., 2010). In this study, the accuracy of using multiple stable isotopes and fatty acid biomarkers to identify the diets of different trophic groups was significantly different. The filter-feeder bivalves are the only trophic group, of which diets can be accurately identified by the stable isotopes and fatty acid biomarkers. This has been proved by the similar δ13C values for these bivalves (P. sanguinolentus and S. subcrenata) and OM, and the high level of OM FA markers 18:2n6c, 22:6n3, and 20:5n3 for these bivalves, which all indicated the significant OM contribution to their diets. However, for the grazer snail N. subaura, its δ13C, δ15N, and δ34S values significantly overlapped with that of seagrass and epiphytes. This indicated that their main food sources were more likely to be seagrass, which was also confirmed by the SIMMR results. Accordingly, the seagrass FA marker 18:2n6c in the snail was indeed higher than that of other consumers, whereas, the most abundant fatty acid composition in the snail was OM indicators 20:5n3 and 22:6n3, which may mislead us to identify the diet of this snail. The most likely reason is that there are some transformations in the process of assimilation of fatty acids by the snails. Evidence to date indicates that some primary consumers, such as herbivores, have the ability to convert C18 fatty acids to 20:5n3 and 22:6n3 (Caramujo et al., 2008; McLeod et al., 2013), therefore a high level of 20:5n3 and 22:6n3 in these consumers may be transformed by the seagrass biomarker 18:2n6c. Similarly, although the δ13C, δ15N, and δ34S values of deposit feeder polychaete A. cristata were more similar to that of seagrass and epiphytes, the level of 18:2n6c in the polychaete was not clearly higher than that of other unsaturated fatty acids (i.g., 20:5n3 and 20:6n3), which indicated that the polychaete may also have the ability to convert these fatty acids.
Most crustaceans are known to be omnivores or generalized predators and have mixed diets, therefore there may be great obscure when using δ13C and δ15N values to identify their diets (Reaka-Kudla, 2001). In this study, the δ13C values of predators P. sanguinolentus and M. rumphii overlapped more with those of OM in both seasons, which is often mistakenly believed that their carbon sources were mainly OM in both seasons (Fan et al., 2011; Du et al., 2019). The mixing model also showed a high POM contribution to their diets. Whereas, their δ13C values were also located in the middle of OM and epiphytes, which also indicated they might feed on OM and epiphytes together. This possibility has also been confirmed by their fatty acid composition. The high level of OM FA marker 18:2n6c in summer, and OM FA markers (20:5n3, 22:6n3, and 18:2n6t) and epiphytes FA markers (18:1n9c and 16:1n7) in winter indicated that their food sources were OM in summer and OM and epiphytes in winter, respectively. Some researchers also suggested that OM and epiphytes had a high contribution to the predator crustaceans (Ning et al., 2019; Paar et al., 2019).
Similarly, it is also difficult to accurately distinguish the diets of omnivores crab C. sp. and T. crenata because of their wide range of δ13C and δ15N values. The SIMMR results showed the relative average coverage contribution of each food source to C. sp. and the high contribution of macroalgae to T. crenata. Relatively higher levels of epiphytes FA marker 16:1n7 and OM FA markers 20:5n3 and 22:6n3 in these omnivores indicated a mixed diets contribution of epiphytes and OM. Nevertheless, the significantly high level of FA marker 18:1n9 in the omnivores during summer suggested that their chief food source was probably macroalgae. Therefore, it is necessary to further distinguish epiphytes from macroalgae by the δ34S values. Considering the trophic fraction of δ34S values was from 0 to 1.9 (McCutchan et al., 2003; Mittermayr et al., 2014), and the higher δ34S values of macroalgae were higher than that of these omnivores. Therefore, the food sources of these omnivores were more likely to be epiphytes and OM.
Overall, for diverse primary producers in our tropical seagrass bed food webs study, multiple stable isotopes usually change obviously among primary producer species and seasons within a region. This characteristic would affect the application of multiple isotopes to identify the food sources of dispersed consumers. Whereas, for snails, polychaetes and bivalves, multiple isotopes usually could discriminate their food sources, because limited mobile ability induced their diets to be often relatively single and fixed within a period and region (Fukumori et al., 2008; Tue et al., 2012; Cui et al., 2021b). By contrast, fatty acids generally provide clearer separation of primary producer species than multiple stable isotopes by providing specific FA biomarkers for each food source. This characteristic would allow FAs to identify the food sources of crustaceans with complex diets. However, the specific FA marker of each species might be the same due to they might have the same suite of photosynthetic pigments and similar biochemical pathways (i.g., seagrass and macroalgae in this study) (Galloway et al., 2012), and its dynamic is always linked with the metabolic conditions of the consumers (i.g., snails in this study). Therefore, using both types of biomarkers together may reduce the uncertainty caused by natural variation and thus strengthen the separation of food sources and the diets of different consumer species. Further study is needed to apply multiple isotopes and fatty acids into the mixing model to quantify the diet contribution of consumers. In addition, in our study, most consumers have a high level of diatom biomarkers, which indicated microphytobenthos might also be their important food source. Therefore, further study in seagrass ecosystems is also needed to consider microphytobenthos as an independent carbon source.
5 Conclusions
Our study has evaluated various food sources and macrobenthos in a tropical seagrass bed, and has shown that the combination of multiple stable isotopes and fatty acids is essential in identifying the diets of consumers. We showed that the use of a combination of multiple stable isotopes and fatty acids can effectively distinguish different food sources, among which multiple stable isotopic compositions (particularly δ13C and δ34S) can distinguish seagrass and epiphytes from other food sources, while the application of fatty acid markers can distinguish seagrass from epiphytes. In addition, we evaluated for the first time the feasibility of applying this combination to identify the food sources of macrobenthos in tropical seagrass beds. Our results indicated that multiple stable isotopic compositions are more useful than fatty acid markers in identifying the diets of grazers snails and deposit feeder polychaetes due to the transformation in the process of fatty acids assimilation. In contrast, fatty acid markers are more useful in identifying the diets of predatory crustaceans due to their mixed diets. Whereas the identification of omnivorous crustaceans food sources requires the combination of multiple stable isotopes and fatty acids biomarkers due to their wide range of isotope values. The bivalves are the only consumer that can be effectively evaluated by both stable isotopes and fatty acids biomarkers.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the South China Sea Institute of Oceanology, Chinese Academy of Sciences.
Author contributions
LC: Conceptualization, Methodology, Investigation, Formal analysis, Visualization, Writing - original draft. ZJ: Conceptualization, Investigation, Writing - review & editing. XH: Supervision, Conceptualization Writing - review & editing, Funding acquisition. SL: Investigation, Writing – review. YW: Investigation. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Nansha District Science and Technology Plan Project Incentive Fund, National Natural Science Foundation of China (nos. 41730529, U1901221, 42176158), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0405), the Natural Science Fund of Guangdong (2019A1515010552, 2020A1515010907, 2018A030310043), and the Science and Technology Planning Project of Guangdong Province, China (2020B1212060058).
Acknowledgments
The authors thank Shouhui Dai and Qiming Chen for performing the stable isotope analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1093181/full#supplementary-material
References
Abrantes K., Sheaves M. (2009). Food web structure in a near-pristine mangrove area of the Australian wet tropics. Estuarine Coast. Shelf Sci. 82, 597–607. doi: 10.1016/j.ecss.2009.02.021
Anderson M. J., Willis T. J. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84, 511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2
Beesley L. S., Pusey B. J., Douglas M. M., Gwinn D. C., Canham C. A., Keogh C. S., et al. (2020). New insights into the food web of an Australian tropical river to inform water resource management. Sci. Rep. 10, 14294. doi: 10.1038/s41598-020-71331-0
Belicka L. L., Burkholder D., Fourqurean J. W., Heithaus M. R., MacKo S. A., Jaff́ R. (2012). Stable isotope and fatty acid biomarkers of seagrass, epiphytic, and algal organic matter to consumers in a pristine seagrass ecosystem. Mar. Freshw. Res. 63, 1085–1097. doi: 10.1071/MF12027
Canadell J. G., Jackson R. B. (2021). Ecosystem collapse and climate change: an introduction (Cham: Springer International Publishing). doi: 10.1007/978-3-030-71330-0_1
Caramujo M. J., Boschker H. T. S., Admiraal W. (2008). Fatty acid profiles of algae mark the development and composition of harpacticoid copepods. Freshw. Biol. 53, 77–90. doi: 10.1111/j.1365-2427.2007.01868.x
Caut S., Angulo E., Courchamp F. (2009). Variation in discrimination factors (Δ15N and Δ13C): the effect of diet isotopic values and applications for diet reconstruction. J. Appl. Ecol. 46, 443–453. doi: 10.1111/j.1365-2664.2009.01620.x
Coelho H., Lopes da Silva T., Reis A., Queiroga H., Serôdio J., Calado R. (2011). Fatty acid profiles indicate the habitat of mud snails hydrobia ulvae within the same estuary: mudflats vs. seagrass meadows. Estuarine Coast. Shelf Sci. 92, 181–187. doi: 10.1016/j.ecss.2011.01.005
Crawley K. R., Hyndes G. A., Vanderklift M. A., Revill A. T., Nichols P. D. (2009). Allochthonous brown algae are the primary food source for consumers in a temperate, coastal environment. Mar. Ecol. Prog. Ser. 376, 33–44. doi: 10.3354/meps07810
Cui L., Jiang Z., Huang X., Chen Q., Wu Y., Liu S., et al. (2021a). Eutrophication reduces seagrass contribution to coastal food webs. Ecosphere 12:e03626. doi: 10.1002/ecs2.3626
Cui L., Jiang Z., Huang X., Wu Y., Liu S., Chen Q., et al. (2021b). Carbon transfer processes of food web and trophic pathways in a tropical eutrophic seagrass meadow. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.725282
Débora R., de C., Carlos B. M. A., Marcelo Z. M., Paulo S. P. (2020). Trophic diversity and carbon sources supporting fish communities along a pollution gradient in a tropical river. Sci. Total Environ. 738, 139878. doi: 10.1016/j.scitotenv.2020.139878
Du J., Chen Z., Xie M., Chen M., Zheng X., Liao J., et al. (2019). Analysis of organic carbon sources in tropical seagrass fish: a case study of the east coast of Hainan Province. Mar. Biol. Res. 15, 513–522. doi: 10.1080/17451000.2019.1673896
Fan M., Huang X., Zhang D., Zhang J., Jiang Z., Zeng Y. (2011). Food sources of fish and macro-invertebrates in a tropical seagrass bed at Xincun Bay, Southern China. Shengtai Xuebao/ Acta Ecologica Sin. 31, 31–38 (in Chinese).
Folch J., Lees M., Sloane stanley G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509. doi: 10.1016/s0021-9258(18)64849-5
Fritts A. K., Knights B. C., Lafrancois T. D., Bartsch L. A., Vallazza J. M., Bartsch M. R., et al. (2018). Spatial and temporal variance in fatty acid and stable isotope signatures across trophic levels in large river systems. River Res. Appl. 34, 834–843. doi: 10.1002/rra.3295
Fukumori K., Oi M., Doi H., Takahashi D., Okuda N., Miller T. W., et al. (2008). Bivalve tissue as a carbon and nitrogen isotope baseline indicator in coastal ecosystems. Estuarine Coast. Shelf Sci. 79, 45–50. doi: 10.1016/j.ecss.2008.03.004
Gacia E., Costalago D., Prado P., Piorno D., Tomas F. (2009). Mesograzers in posidonia oceanica meadows: an update of data on gastropod-epiphyte-seagrass interactions. Botanica Marina 52, 439–447. doi: 10.1515/BOT.2009.054
Gagnon K., Gustafsson C., Salo T., Rossi F., Gunell S., Richardson J. P., et al. (2021). Role of food web interactions in promoting resilience to nutrient enrichment in a brackish water eelgrass (Zostera marina) ecosystem. Limnol. Oceanogr. 66, 2810–2826. doi: 10.1002/lno.11792
Galloway A. W. E., Britton-Simmons K. H., Duggins D. O., Gabrielson P. W., Brett M. T. (2012). Fatty acid signatures differentiate marine macrophytes at ordinal and family ranks. J. Phycol. 48, 956–965. doi: 10.1111/j.1529-8817.2012.01173.x
GAQSIQ (2008). Determination of total fat, saturated fat, and unsaturated fat in foods: hydrolytic extraction-gas chromatography (Beijing: Standards Press of China).
Gartner A., Lavery P., Smit A. J. (2002). Use of δ15N signatures of different functional forms of macroalgae and filter-feeders to reveal temporal and spatial patterns in sewage dispersal. Mar. Ecol. Prog. Ser. 235, 63–73. doi: 10.3354/meps235063
Guiry E., Noël S., Fowler J. (2021). Archaeological herbivore δ13C and δ34S provide a marker for saltmarsh use and new insights into the process of 15N-enrichment in coastal plants. J. Archaeol. Sci. 125:14750–14760. doi: 10.1016/j.jas.2020.105295
Hanson C. E., Hyndes G. A., Wang S. F. (2010). Differentiation of benthic marine primary producers using stable isotopes and fatty acids: implications to food web studies. Aquat. Bot. 93, 114–122. doi: 10.1016/j.aquabot.2010.04.004
Holmer M., Nielsen R. M. (2007). Effects of filamentous algal mats on sulfide invasion in eelgrass (Zostera marina). J. Exp. Mar. Biol. Ecol. 353, 245–252. doi: 10.1016/j.jembe.2007.09.010
Huang X., Huang L., Li Y., Xu Z., Fong C. W., Huang D., et al. (2006). Main seagrass beds and threats to their habitats in the coastal sea of South China. Chin. Sci. Bull. 51, 136–142. doi: 10.1007/s11434-006-9136-5
Huang X., Jiang Z., Liu S., Yu S., Wu Y., Zhang J., et al. (2019). Study on ecology of tropical seagrass in China (Beijing: Science China Press (in Chinese).
Ivajnšič D., Orlando-Bonaca M., Donša D., Grujić V. J., Trkov D., Mavrič B., et al. (2022). Evaluating seagrass meadow dynamics by integrating field-based and remote sensing techniques. Plants 11, 1196. doi: 10.3390/plants11091196
Jankowska E., De Troch M., Michel L. N., Lepoint G., Włodarska-Kowalczuk M. (2018). Modification of benthic food web structure by recovering seagrass meadows, as revealed by trophic markers and mixing models. Ecol. Indic. 90, 28–37. doi: 10.1016/j.ecolind.2018.02.054
Jiang Z., Zhao C., Yu S., Liu S. (2019). Contrasting root length, nutrient content and carbon sequestration of seagrass growing in offshore carbonate and onshore terrigenous sediments in the South China Sea. Sci. Total Environ. 662, 151–159. doi: 10.1016/j.scitotenv.2019.01.175
Jinks K. I., Rasheed M. A., Brown C. J., Olds A. D., Schlacher T. A., Sheaves M., et al. (2020). Saltmarsh grass supports fishery food webs in subtropical Australian estuaries. Estuarine Coast. Shelf Sci. 238, 106719. doi: 10.1016/j.ecss.2020.106719
Jorge V. N., Van Beusekom J. E. E. (1995). Wind- and tide-induced resuspension of sediment and microphytobenthos from tidal flats in the Ems estuary. Limnol. Oceanogr. 40, 776–778. doi: 10.4319/lo.1995.40.4.0776
Kasim M., Mukai H. (2006). Contribution of benthic and epiphytic diatoms to clam and oyster production in the Akkeshi-ko estuary. J. Oceanogr. 62, 267–281. doi: 10.1007/s10872-006-0051-9
Kharlamenko V. I., Kiyashko S. I., Imbs A. B., Vyshkvartzev D. I. (2001). Identification of food sources of invertebrates from the seagrass Zostera marina community using carbon and sulfur stable isotope ratio and fatty acid analyses. Mar. Ecol. Prog. Ser. 220, 103–117. doi: 10.3354/meps220103
Kohlbach D., Hop H., Wold A., Schmidt K., Smik L., Belt S. T., et al. (2021). Multiple trophic markers trace dietary carbon sources in barents sea zooplankton during late summer. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.610248
Lebreton B., Richard P., Galois R., Radenac G., Pfléger C., Guillou G., et al. (2011). Trophic importance of diatoms in an intertidal Zostera noltii seagrass bed: evidence from stable isotope and fatty acid analyses. Estuarine Coast. Shelf Sci. 92, 140–153. doi: 10.1016/j.ecss.2010.12.027
Lippold A., Aars J., Andersen M., Aubail A., Derocher A. E., Dietz R., et al. (2020). Two decades of mercury concentrations in barents sea polar bears (Ursus maritimus) in relation to dietary carbon, sulfur, and nitrogen. Environ. Sci. Technol. 54, 7388–7397. doi: 10.1021/acs.est.0c01848
Liu S., Deng Y., Jiang Z., Wu Y., Huang X., Macreadie P. I. (2020a). Nutrient loading diminishes the dissolved organic carbon drawdown capacity of seagrass ecosystems. Sci. Total Environ. 740, 140185. doi: 10.1016/j.scitotenv.2020.140185
Liu S., Jiang Z., Zhang J., Wu Y., Huang X., Macreadie P. I. (2017). Sediment microbes mediate the impact of nutrient loading on blue carbon sequestration by mixed seagrass meadows. Sci. Total Environ., 599–600 1479–1484. doi: 10.1016/j.scitotenv.2017.05.129
Liu S., Trevathan-Tackett S. M., Ewers Lewis C. J., Huang X., Macreadie P. I. (2020b). Macroalgal blooms trigger the breakdown of seagrass blue carbon. Environ. Sci. Technol. 54, 14750–14760. doi: 10.1021/acs.est.0c03720
Madgett A. S., Yates K., Webster L., McKenzie C., Moffat C. F. (2019). Understanding marine food web dynamics using fatty acid signatures and stable isotope ratios: improving contaminant impacts assessments across trophic levels. Estuarine Coast. Shelf Sci. 227, 106327. doi: 10.1016/j.ecss.2019.106327
Mascart T., De Troch M., Remy F., Michel L. N., Lepoint G. (2018). Seasonal dependence on seagrass detritus and trophic niche partitioning in four copepod eco-morphotypes. Food Webs 16, e00086. doi: 10.1016/j.fooweb.2018.e00086
McCutchan J. H., Lewis W. M., Kendall C., McGrath C. C. (2003). Variation in trophic shift for stable isotope ratios of carbon, nitrogen, and sulfur. Oikos 102, 378–390. doi: 10.1034/j.1600-0706.2003.12098.x
McLeod R. J., Hyndes G. A., Hurd C. L., Frew R. D. (2013). Unexpected shifts in fatty acid composition in response to diet in a common littoral amphipod. Mar. Ecol. Prog. Ser. 479, 1–12. doi: 10.3354/meps10327
Meziane T., Tsuchiya M. (2000). Fatty acids as tracers of organic matter in the sediment and food web of a mangrove/intertidal flat ecosystem, Okinawa, Japan. Mar. Ecol. Prog. Ser. 200, 49–57. doi: 10.3354/meps200049
Mittermayr A., Fox S. E., Sommer U. (2014). Temporal variation in stable isotope composition (δ13C, δ15N and δ34S) of a temperate Zostera marina food web. Mar. Ecol. Prog. Ser. 505, 95–105. doi: 10.3354/meps10797
Moncreiff C. A., Sullivan M. J. (2001). Trophic importance of epiphytic algae in subtropical seagrass beds: evidence from multiple stable isotope analyses. Mar. Ecol. Prog. Ser. 215, 93–106. doi: 10.3354/meps215093
Nakamoto K., Hayakawa J., Kawamura T., Ohtsuchi N., Yamada H., Kitagawa T., et al. (2019). Seasonal fluctuation in food sources of herbivorous gastropods in a subtropical seagrass bed estimated by stable isotope analysis. J. Mar. Biol. Assoc. United Kingdom 99, 1119–1125. doi: 10.1017/S0025315418001108
Nichols P. D., Klumpp D. W., Johns R. B. (1985). A study of food chains in seagrass communities iii. stable carbon isotope ratios. Mar. Freshw. Res. 36, 683–690. doi: 10.1071/MF9850683
Ning J., Du F., Wang X., Wang L., Li Y. (2019). Trophic connectivity between intertidal and offshore food webs in Mirs Bay, China. Oceanologia 61, 208–217. doi: 10.1016/j.oceano.2018.10.001
Oduro H., Van Alstyne K. L., Farquhar J. (2012). Sulfur isotope variability of oceanic DMSP generation and its contributions to marine biogenic sulfur emissions. Proc. Natl. Acad. Sci. United States America 109, 9012–9016. doi: 10.1073/pnas.1117691109
Paar M., Lebreton B., Graeve M., Greenacre M., Asmus R., Asmus H. (2019). Food sources of macrozoobenthos in an Arctic kelp belt: trophic relationships revealed by stable isotope and fatty acid analyses. Mar. Ecol. Prog. Ser. 615, 31–49. doi: 10.3354/meps12923
Park H. J., Choy E. J., Lee K., Kang C. (2013). Trophic transfer between coastal habitats in a seagrass-dominated macrotidal embayment system as determined by stable isotope and fatty acid signatures. Mar. Freshw. Res. 64, 1169–1183. doi: 10.1071/MF12327
Parnell A., Inger R. (2019) Stable isotope mixing models in R with simmr. Available at: https://cran.r-project.org/web/packages/simmr/vignettes/simmr.html.
Pecquerie L., Nisbet R. M., Fablet R., Lorrain A., Kooijman S. A. L. M. (2010). The impact of metabolism on stable isotope dynamics: a theoretical framework. Philos. Trans. R. Soc. B: Biol. Sci. 365, 3455–3468. doi: 10.1098/rstb.2010.0097
Pellerin A., Antler G., Holm S. A., Findlay A. J., Crockford P. W., Turchyn A. V., et al. (2019). Large Sulfur isotope fractionation by bacterial sulfide oxidation. Sci. Adv. 5, 1–7. doi: 10.1126/sciadv.aaw1480
Reaka-Kudla M. L. (2001). Crustaceans. Encyclopedia Biodivers.: Second Edition 1, 396–418. doi: 10.1016/B978-0-12-384719-5.00165-9
Riccialdelli L., Newsome S. D., Fogel M. L., Fernández D. A. (2017). Trophic interactions and food web structure of a subantarctic marine food web in the Beagle Channel: Bahía Lapataia, Argentina. Polar Biol. 40, 807–821. doi: 10.1007/s00300-016-2007-x
Saderne V., Geraldi N. R., Macreadie P. I., Maher D. T., Middelburg J. J., Serrano O., et al. (2019). Role of carbonate burial in Blue Carbon budgets. Nat. Commun. 10, 1–9. doi: 10.1038/s41467-019-08842-6
Sun H., Li Y., Hao Y., Zhu Y., Yang R., Wang P., et al. (2020). Bioaccumulation and trophic transfer of polybrominated diphenyl ethers and their hydroxylated and methoxylated analogues in polar marine food webs. Environ. Sci. Technol. 54, 15086–15096. doi: 10.1021/acs.est.0c05427
Team R. C. (2019). R: a language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria).
Thornber C. S., DiMilla P., Nixon S. W., McKinney R. A. (2008). Natural and anthropogenic nitrogen uptake by bloom-forming macroalgae. Mar. pollut. Bull. 56, 261–269. doi: 10.1016/j.marpolbul.2007.10.031
Tue N. T., Hamaoka H., Sogabe A., Quy T. D., Nhuan M. T., Omori K. (2012). Food sources of macro-invertebrates in an important mangrove ecosystem of Vietnam determined by dual stable isotope signatures. J. Sea Res. 72, 14–21. doi: 10.1016/j.seares.2012.05.006
Unsworth R. K. F., Nordlund L. M., Cullen-Unsworth L. C. (2019). Seagrass meadows support global fisheries production. Conserv. Lett. 12, 1–8. doi: 10.1111/conl.12566
Valiela I., Liu D., Lloret J., Chenoweth K., Hanacek D. (2018a). Stable isotopic evidence of nitrogen sources and C4 metabolism driving the world’s largest macroalgal green tides in the Yellow Sea. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-35309-3
Valiela I., Pascual J., Giblin A., Barth-Jensen C., Martinetto P., Otter M., et al. (2018b). External and local controls on land-sea coupling assessed by stable isotopic signatures of mangrove producers in estuaries of Pacific Panama. Mar. Environ. Res. 137, 133–144. doi: 10.1016/j.marenvres.2018.03.003
Weems J., Iken K., Gradinger R., Wooller M. J. (2012). Carbon and nitrogen assimilation in the Bering Sea clams Nuculana radiata and Macoma moesta. J. Exp. Mar. Biol. Ecol., 430–431, 32–42. doi: 10.1016/j.jembe.2012.06.015
Xu W.-Z., Cheung S. G., Zhang Z.-N., Shin P. K. S. (2018). Dual isotope assessment of trophic dynamics of an intertidal infaunal community with seasonal shifts in food sources. Mar. Biol. 165, 21. doi: 10.1007/s00227-017-3278-7
Xu J., Fan X., Zhang X., Xu D., Mou S., Cao S., et al. (2012). Evidence of coexistence of C3 and C4 photosynthetic pathways in a green-tide-forming alga, ulva prolifera. PloS One 7, 1–10. doi: 10.1371/journal.pone.0037438
Keywords: trophic role, macrobenthos, sulfur stable isotope, fatty acids, tropical seagrass beds
Citation: Cui L, Jiang Z, Huang X, Liu S and Wu Y (2023) Identification of food sources in tropical seagrass bed food web using triple stable isotopes and fatty acid signatures. Front. Mar. Sci. 10:1093181. doi: 10.3389/fmars.2023.1093181
Received: 08 November 2022; Accepted: 23 June 2023;
Published: 21 July 2023.
Edited by:
Monica Montefalcone, University of Genoa, ItalyReviewed by:
Yanyi Zeng, Chinese Academy of Fishery Sciences, ChinaYibo Liao, Ministry of Natural Resources, China
Copyright © 2023 Cui, Jiang, Huang, Liu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijian Jiang, amlhbmd6ajE5ODJAc2NzaW8uYWMuY24=; Xiaoping Huang, eHBodWFuZ0BzY3Npby5hYy5jbg==
 Lijun Cui
Lijun Cui Zhijian Jiang
Zhijian Jiang Xiaoping Huang
Xiaoping Huang Songlin Liu
Songlin Liu Yunchao Wu
Yunchao Wu