- Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Guangdong Province Famous Fish Reproduction and Breeding Engineering Technology Research Center, Fisheries College of Guangdong Ocean University, Zhanjiang, China
Spotted scat (Scatophagus argus) is an important mariculture fish that is of great economic significance in East and Southeast Asia. To date, there are no studies on ovary development and regulation in S. argus. Herein, the ovary transcriptome profiles of S. argus at different stages were constructed, and the genes and pathways potentially involved in secondary follicle growth were identified. A total of 25,426 genes were detected by sequencing the mRNAs from the ovary libraries at stage III (n=3) and IV (n=3). Notably, 2950 and 716 genes were up-regulated and down-regulated in the stage IV ovary, respectively, compared to the stage III ovary. The differentially expressed genes (DEGs) were found to be mostly involved in regulating steroidogenesis, vitellogenesis, lipid metabolism, and meiosis. Up-regulation of steroid hormone synthesis pathway genes (fshr, cyp17a1, and foxl2) and insulin-like growth factor pathway genes (igf1r, ifg2r, igfbp1, igfbp3, and igfbp7) in the ovary at stage IV was possibly the reason for the increased serum estrogen. Moreover, ppara, ppard, fabp3, and lpl were up-regulated in the stage IV ovary and were potentially involved in the lipid droplet formation in the oocyte. Many DEGs were involved in the cellular cycle, meiosis, and cAMP or cGMP synthesis and hydrolysis, indicating that meiosis was restarted at stage IV ovary. In addition, numerous TGF-beta signal pathway genes were up-regulated in the stage IV ovary. This ovary transcript dataset forms a baseline for investigating functional genes associated with oogenesis in S. argus.
1 Introduction
In recent years, there has been an increasing demand for newly cultured species, especially fish, because of the reduction of natural resources caused by overfishing. Producing a large number of healthy and viable eggs is pivotal to the success of aquaculture (Lubzens et al., 2010). Mature fertilizable eggs develop from oogonia through oogenesis. A normal oogenesis process is necessary for successful fertilization and embryonic survival. Although oogenesis significantly differs between teleost, the basic stages of oogenesis are: 1) transformation of oogonia into oocytes (onset of meiosis), 2) oocyte growth (under meiotic arrest), 3) maturation (resumption of meiosis), and 4) ovulation. Several articles have detailed the descriptions of these phases (Patiño and Sullivan, 2002; Lubzens et al., 2010; Urbatzka et al., 2011; Breton and Berlinsky, 2014; Sullivan and Yilmaz, 2018; Meng et al., 2022).
Oocyte growth is a critical stage of oogenesis and has been extensively studied in different species. In teleost, oocyte growth may encompass a significant portion of the lifespan and can be divided into the primary growth stage (PG) and secondary growth stage (SG) (Lubzens et al., 2010). SG consists of the cortical alveoli stage (also named previtellogenic stage, PV) and vitellogenic stage, which is further divided into early vitellogenic (EV), mid-vitellogenic (MV), late vitellogenic (LV), and full-grown (FG) stages according to the follicle size and morphological characteristics (Kwok et al., 2005). Primary growth starts with the onset of meiosis when oogonia develop into PG oocytes. The PG oocytes are subsequently arrested in meiotic prophase I and become surrounded by follicular cells (theca and granulosa cells), forming the ovarian follicle structure (Lubzens et al., 2010). The beginning of SG is marked by the appearance and accumulation of cortical alveoli (CA). CA are endogenously synthesized membrane-limited glycoprotein vesicles of variable sizes, which increase in number and size during early SG (Lubzens et al., 2010). CA play an important role in the fertilization response and early embryogenesis. Lipid droplets appear in the ooplasm, and their abundance predominates in the early stages of vitellogenesis over the yolk globules at about the same time as CA appearance or late in the PV stage. The lipids accumulating within the oocyte ooplasm originate from plasma very low-density lipoproteins (VLDL) and vitellogenins (Vtgs) (Lubzens et al., 2010; Sullivan and Yilmaz, 2018; Qu et al., 2022). True vitellogenesis is marked by the formation of yolk granules at the periphery of the cytoplasm as the oocyte grows. During the vitellogenic stage, dramatic follicle growth occurs as the oocyte sequesters Vtgs through receptor-mediated endocytosis (Lubzens et al., 2010; Sullivan and Yilmaz, 2018). The lipid droplets and yolk granules are important nutrient pools for the oocytes to meet the subsequent maturation requirements and early embryonic development (Hiramatsu et al., 2015; Sullivan and Yilmaz, 2018). The FG oocytes undergo meiotic maturation, which includes the resumption of meiosis and the completion of the first meiotic division after growth completion to become fertilizable eggs (Lubzens et al., 2010).
Oocyte growth is a complicated and dynamic process regulated by many circulating and follicle-produced local physiological factors. The follicle-stimulating hormone (Fsh), a pituitary gonadotropin, has been proven to stimulate oocyte growth (vitellogenesis) by activating cyp19a1a transcription and producing estrogen, mainly the estradiol (E2) (Lubzens et al., 2010; Meng et al., 2022), which induces hepatocytes in the liver to synthesize and secrete Vtgs (Sullivan and Yilmaz, 2018). Besides the pituitary gonadotropin hormone (Gth) and gonadal sex steroids hormones, accumulating evidence suggests a cross-talk between the developing oocyte and its surrounding follicle layers in oocyte growth mediated by paracrine factors (Lubzens et al., 2010; Meng et al., 2022). For example, Inha/Inhbb of the TGF-beta signaling pathways and Bmp15 of the Bmp signaling pathway have proven to be required for oocyte growth (Myllymaa et al., 2010; Cook-Andersen et al., 2016). Recently, many genes and pathways that potentially play a role in fish oogenesis have been identified through large-scale transcriptome analyses. These transcriptomic analyses on fish oogenesis have mainly focused on the true vitellogenesis stage (From EV to LV) (Wang et al., 2019; Meng et al., 2022), final maturation and ovulation (Breton and Berlinsky, 2014; Tang et al., 2019; He et al., 2020; Wang et al., 2021a; Meng et al., 2022), and transition from PG to PV (Breton and Berlinsky, 2014; Zhu et al., 2018; Qu et al., 2022). Notably, there is a lack of transcriptome analysis between the CA stage and the true vitellogenic stage, which is critical for understanding the potential mechanisms of secondary follicular growth, especially the production of lipid droplets and yolk granules.
Spotted scat (Scatophagus argus) is an important aquaculture fish with high economic value in East and Southeast Asia. S. argus can eat algae, sick shrimp, parasites on fish bodies, and shellfish attached to the pool wall and cage, making it a good “garbage fish.” It is thus suitable for mixed cultivation with other marine shrimp and fish. Although artificially-induced spawning can be achieved in some laboratories (Cai et al., 2010; Mandal et al., 2021; Washim et al., 2022), the efficiency of artificial propagation is still very low in the actual breeding process, mainly because of the lack of female fish which can produce a large number of healthy and viable eggs. Elucidating the mechanisms underlying oocyte growth may thus help increase the quality and quantity of eggs produced by female S. argus for artificial breeding, thereby promoting its sustainable development in aquaculture. Although numerous studies regarding the reproductive biology of S. argus have been done in recent years (Cui et al., 2017; Chen et al., 2020; Zhai et al., 2021; Jiang et al., 2022), the molecular mechanisms controlling oocyte growth remain poorly understood and are worth further studies. In this study, transcriptomic analyses of the ovary at the PV stage (stage III) and LV stage (stage IV) were performed to identify candidate genes and potential pathways associated with secondary follicular growth in S. argus. The findings of this study provide a solid foundation for further elucidation of the mechanisms of fish oogenesis.
2 Materials and methods
2.1 Experimental fish and sample collection
Twelve one-year-old female S. argus (200-310 g) were collected from Donghai Island, Zhanjiang, Guangdong Province, China. The fish were euthanized with 100 mg·L–1 of tricaine methanesulfonate (MS-222, Sigma, Saint Louis, MO, USA), followed by collecting and storing part of the ovary tissues in Trizol at -80°C for total RNA extraction. The remaining ovary tissues from the same fish were fixed in Bouin’s solution for 12 hours and used for histological identification. Histological characterization of the ovary was performed following the approach of Cui et al. (2017) and Jiang et al. (2022). All experiments were performed according to the requirements of the Animal Research and Ethics Committee of the Institute of Aquatic Economic Animals, Guangdong Ocean University.
2.2 RNA extraction, library construction, and Illumina sequencing
Total RNA was extracted using TRIzol (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. RNA quantity and quality were determined using NanoDrop spectrophotometry (Thermo Scientific, Wilmington, DE, USA) and agarose gel electrophoresis, respectively.
Total RNA extracted from the ovarian tissues of 6 fish (3 at stage III and 3 at stage IV) from the above 12 fish were used for RNA-Seq. The NEBNext UltraTM RNA Library Prep Kit was used for complementary DNA (cDNA) library construction following the manufacturer’s recommendations. Briefly, mRNA was purified from total RNA using Oligo (dT) beads and was then randomly fragmented into short fragments using the fragmentation buffer. The fragments were then used to synthesize the first-strand cDNA by employing random primers. Second strand cDNA was synthesized using DNA polymerase I, dNTP, RNase H, and buffer. The synthesized cDNA was then purified using AMPure XP beads and then end-repaired, poly(A) added, and ligated to Illumina sequencing adapters. The ligation fragments were subsequently size-selected using AMPure XP beads. PCR was then performed to generate cDNA libraries which were sequenced using Illumina HiSeq™ 2000. All clean libraries of sequencing data were submitted to the NCBI Sequence Read Archive (SRA) database (Accession Nos: SRP171076 and PRJNA906196).
2.3 Sequence assembly, annotation, and functional analysis
Clean reads were obtained by removing the reads containing adapters and low-quality reads from the original sequences. The clean reads were then mapped to the S. argus reference genome (Huang et al., 2021) (https://ngdc.cncb.ac.cn/search/?dbId=gwh&q=GWHAOSK00000000.1) using HISAT2.2.4. The Q20, Q30, GC-content, and sequence duplication levels of the clean data were also calculated. The gene expression levels were calculated using the fragments per kilobase of transcript per million mapped reads (FPKM) method. Analysis of the differentially expressed genes (DEGs) between two different groups was performed using the EdgeR package. Genes with a fold change ≥2 (|log2FC|>1) and a false discovery rate (FDR) < 0.05 were highlighted as significant DEGs. The DEGs were annotated by checking their Gene Ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment. The calculated P value of GO and KEGG analyses was determined through FDR correction. GO terms and KEGG pathways with a P-value <0.05 were defined as significantly enriched in the DEGs.
2.4 Validation of DEGs with quantitative real-time PCR (qRT-PCR)
Thirteen DEGs related to ovarian development were randomly selected for qRT-PCR analysis to validate the RNA-seq data. Total RNA was extracted from ovarian tissues of the above 12 fish (6 at stage III and 6 at stage IV), with DNAase I used to digest the genomic DNA. cDNA was synthesized using the TansScript kit (TransGen Biotech, Beijing, China). qRT-PCR was conducted using the qPCRPerfectStart TM Green qPCR SuperMix (TransGen Biotech, Beijing, China) on a Light Cycler 96 (Roche). The PCR reaction (20 μL) contained 1 μL cDNA, 10 μL 2 × PCR mix, 8.2 μL ddH2O, and 0.4 μL of forward and reverse primers (10 μmol·mL–1). The qRT-PCR thermocycling conditions were initial denaturation at 95°C for 1 min and 40 cycles of denaturation, primer annealing, and extension at 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s, respectively. B2m was used as the reference gene. In previous studies, B2M/B2m has been recommended as a stable reference gene for human ovaries (Asiabi et al., 2020), bovine granulosa cells, oocytes and cumulus cells (Baddela et al., 2014; Caetano et al., 2019), and rat ovarian granulosa cells (Cai et al., 2019). The 2–ΔΔCt method was used to calculate the relative expression of the target genes. Each sample was amplified in duplicate. All primers used in the various assays are listed in Supplementary Table 1.
2.5 Statistical analyses
Data were analyzed using the independent sample t-test. A probability level of < 0.05 (P < 0.05) was used to indicate statistical significance. The GraphPad Prism 6 software (La Jolla, CA) was used to conduct statistical analyses. All data are presented as means ± standard error of the mean of three replicates.
3 Results
3.1 Characteristics of S. argus ovary development stages
Based on the ovarian histological characteristics (Figure 1), two representative stages of ovary development were identified: stage III (corresponding to the PV stage of zebrafish) and stage IV (corresponding to the LV stage of zebrafish). At stage III, cortical alveoli or lipid droplets appeared within the cytoplasm of some oocytes, and the ovary was mainly composed of primary growth stage oocytes and cortical vesicle stage oocytes. At stage IV, the ovary was mainly filled with preovulatory oocytes, which were significantly larger and contained abundant yolk granules and lipid droplets.
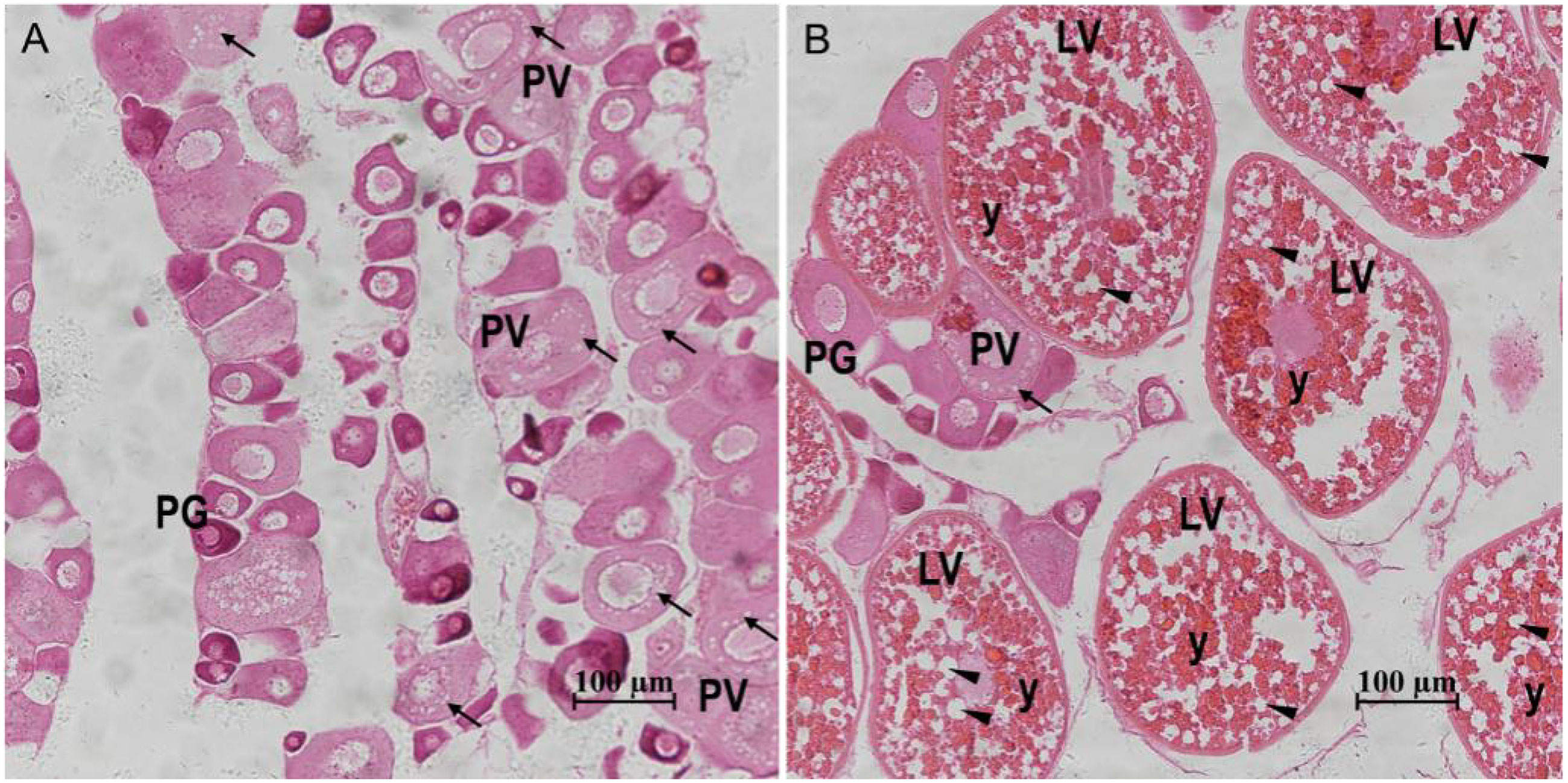
Figure 1 Gonadal histology of the S. argus. (A) Stage III. (B) Stage IV. “↑”: cortical alveoli; “▲”: lipid droplets; “y”: yolk granules; PG: primary growth stage oocyte; PV: previtellogenic stage follicle; LV: late vitellogenic stage follicle; Scale bar: 100 micrometre.
3.2 Transcriptome sequencing and assembly
Sequencing of the mRNAs from the six libraries yielded 312,304,188 raw reads, which were reduced to 310,341,234 clean reads after removing adapter sequences and low-quality reads. The percentage of bases with a Phred value of at least 20 (Q20) and 30 (Q30) were more than 96.71% and 91.71%, respectively (Table 1). The GC contents for all the libraries were around 50% (Table 1). The clean reads were then mapped to the reference genome of S. argus (Huang et al., 2021). Notably, an average of >93.18% of the reads mapped to the S. argus genome (Table 1). A total of 25,426 genes, including 23,690 known genes and 1736 novel genes, accounting for 98.03% of the reference genomes, were detected (Supplementary Table S2 and Table S3).
3.3 Differentially expressed genes of the two libraries
Principal component analysis (PCA) performed before the identification of DEGs revealed that the biological replicates of each group clustered together (Figure 2A), indicating the consistency of the ovary stage grouping. A total of 12,829 genes were detected, with 11,497 genes expressed in both stages, and 448 and 1,332 only detected in stage III and stage IV, respectively (Figure 2B). Comparative expression profiling between stage III and stage IV revealed 3666 DEGs (Figure 2C, Supplementary Table S4). Notably, 2950 and 716 genes were up-regulated and down-regulated, respectively, in the stage IV group compared with the stage III group. A Venn diagram was generated using an online software (https://www.omicshare.com/tools/Home/Soft/venn) to classify all the expressed genes and DEGs. Of note, 210 down-regulated genes and 753 up-regulated genes were detected in stage III and IV ovaries, respectively (Figure 2D).
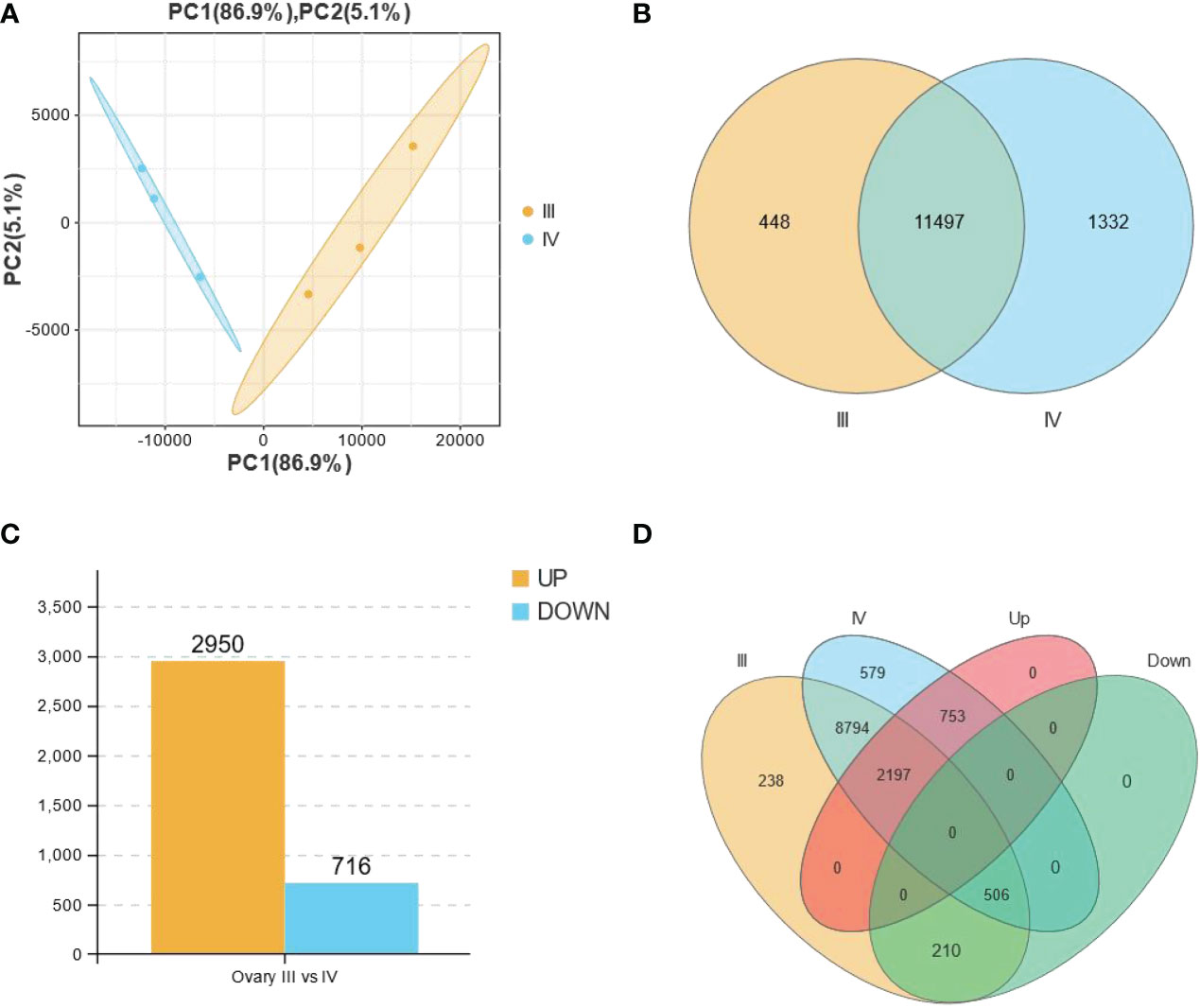
Figure 2 All the expressed and the differentially expressed genes (DEGs) obtained from the RNA-seq data of stage III and IV ovaries of S. argus. (A) Principal component analysis (PCA) showing the differences between biological groups. (B) A Venn diagram depicting the total number of genes expressed in stage III (11945) and stage IV (12829) ovary of S. argus. (C) Differentially expressed genes (DEGs) in stage III and IV ovary of S. argus. (D) A Venn diagram depicting the distribution of DEGs in the different classes, including groups for expressed genes in stage III and stage IV ovaries and up- and down-regulated gene groups.
We further evaluated 6 up-regulated genes (cyp17a1, fshr, igfbp7, inha, foxl2, and creb1) and 7 down-regulated genes (zp2, sycp3, dhp3, fabp1, fabp4, epha8 and slc51a) using qRT-PCR to validate the RNA-seq results. Notably, all the genes assayed exhibited a significant difference (P<0.05) in their expression between stage III and stage IV, fully agreeing with the RNA-seq result (Figure 3).
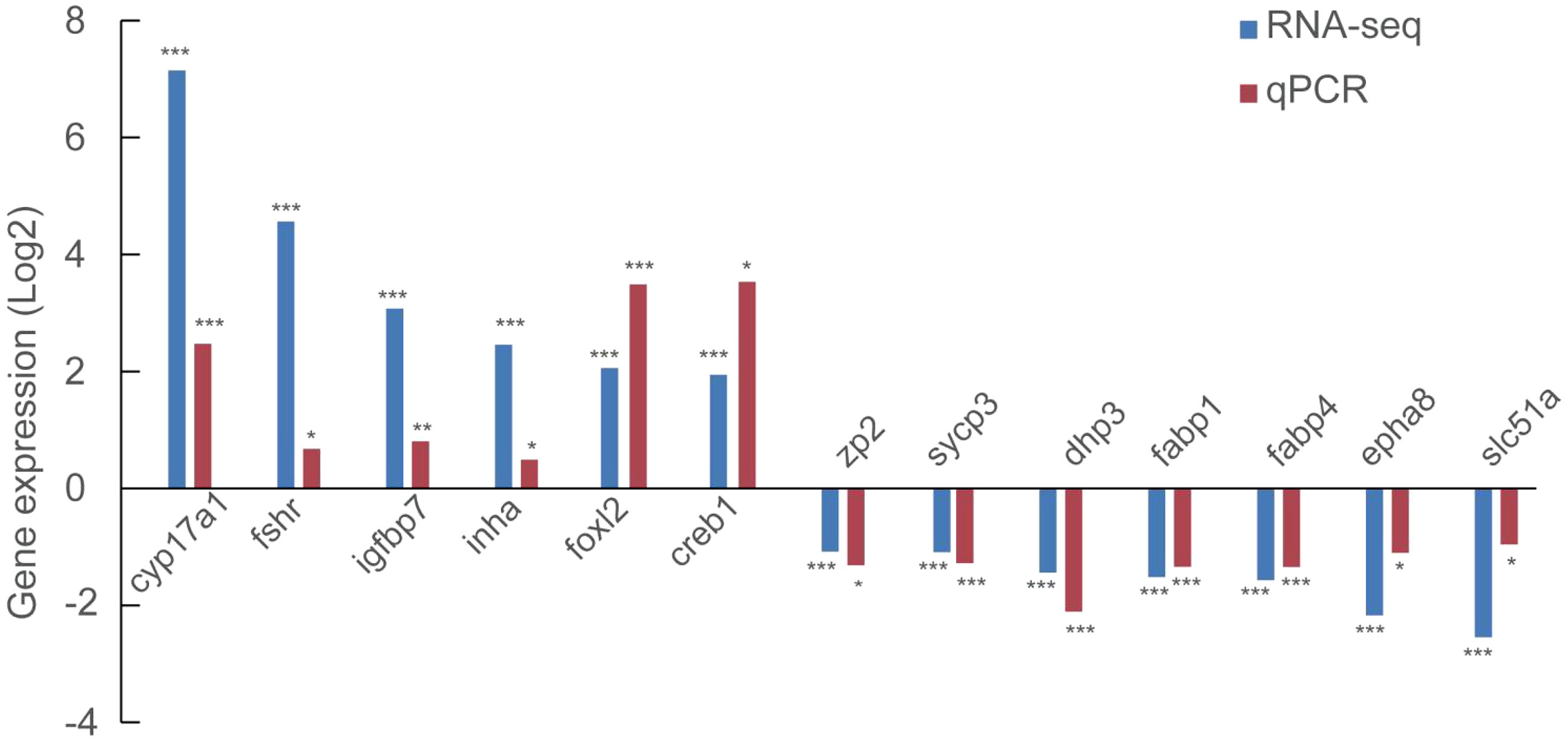
Figure 3 Validation of the expression of 6 up-regulated and 7 down-regulated using qRT-PCR. *, P<0.05; **, P<0.01; ***, P<0.001.
3.4 GO and KEGG classification of DEGs
GO analysis and KEGG enrichment were performed after analysis of the gene expression profiles to better understand the biological functions of the DEGs during secondary ovarian growth in S. argus. GO analysis categorized the DEGs into three basic functional categories: biological processes, cellular components, and molecular functions (Figure 4 and Supplementary Table S5). Cellular process (2163), single-organism process (1992), and metabolic process (1729) were the most enriched GO terms in the biological process category. Binding (1966), catalytic activity (893), and structural molecule activity (187) were the most enriched GO terms in the cellular components category. Cell part (2116), cell (2116), and organelle (1890) were the most enriched GO terms in the molecular functions category.
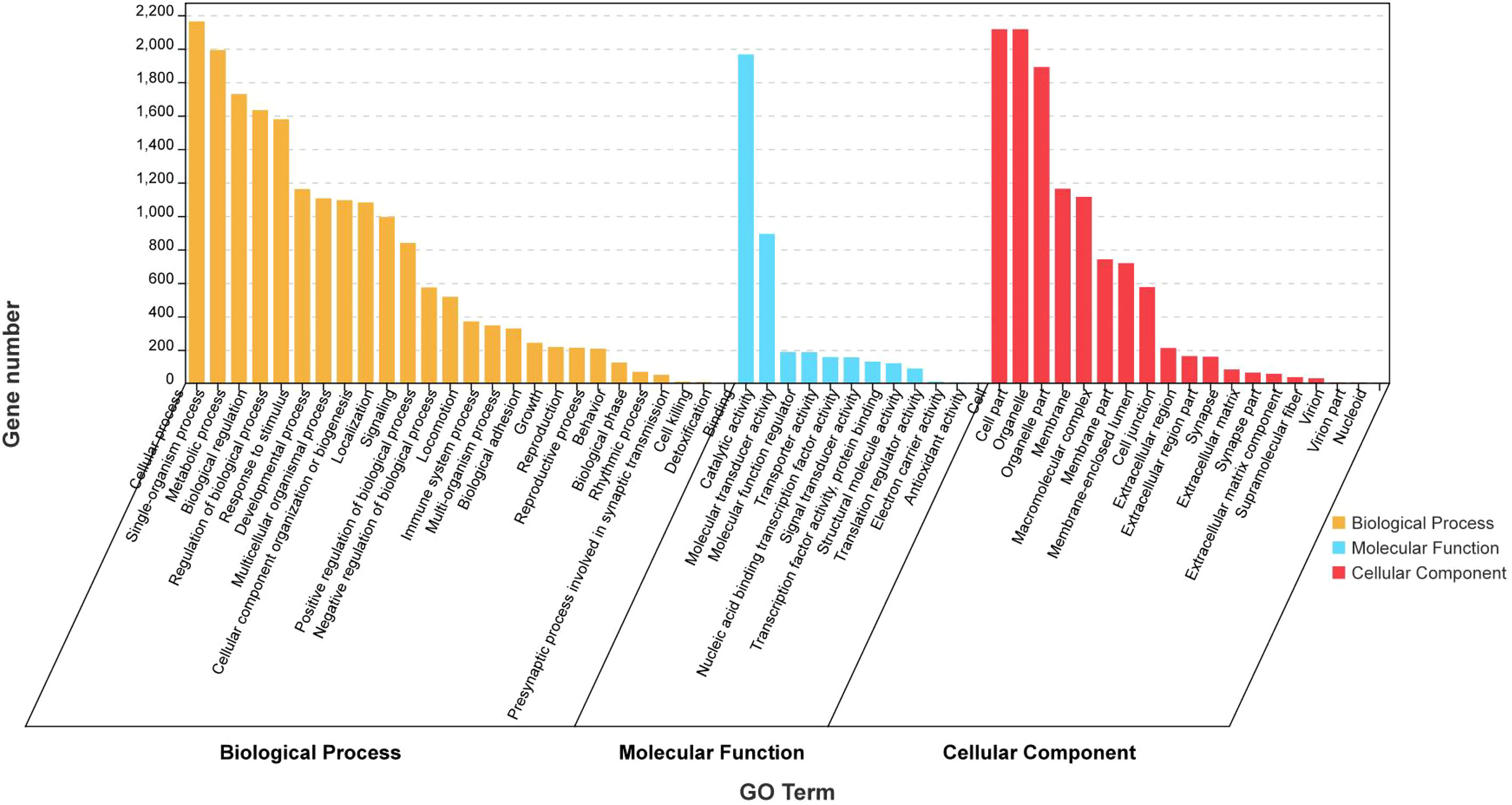
Figure 4 GO analysis of the differentially expressed genes (DEGs) obtained from RNA-seq data of stage III and IV ovary of S. argus.
KEGG pathway enrichment analysis showed that 93 pathways were significantly enriched (P < 0.05) in the up-regulated DEGs (Figure 5A and Supplementary Table S6). The 93 pathways could be divided into six categories: cellular process (eg. Focal adhesion, Adherens junction, Endocytosis, Tight junction, and Cell cycle), environmental information processing (eg. AMPK signaling pathway, Notch signaling pathway, ECM-receptor interaction, MAPK signaling pathway - fly, MAPK signaling pathway, TGF-beta signaling pathway, Hippo signaling pathway, JAK-STAT signaling pathway, Wnt signaling pathway, and FoxO signaling pathway), genetic information processing (eg. Ubiquitin mediated proteolysis, Nucleocytoplasmic transport, and Fanconi anemia pathway), metabolism (eg. Lysine degradation, Glycosaminoglycan biosynthesis - heparan sulfate/heparin, Inositol phosphate metabolism, and Glycerophospholipid metabolism), organismal systems (eg. Axon guidance, Thyroid hormone signaling pathway, Cholesterol metabolism, Insulin signaling pathway, and PPAR signaling pathway), and human diseases (eg. MicroRNAs in cancer, Proteoglycans in cancer, and Pathways in cancer). Similarly, 28 pathways were found to be significantly enriched (P < 0.05) in the down-regulated DEGs and could also be divided into the same six categories (Figure 5B and Supplementary Table S7).
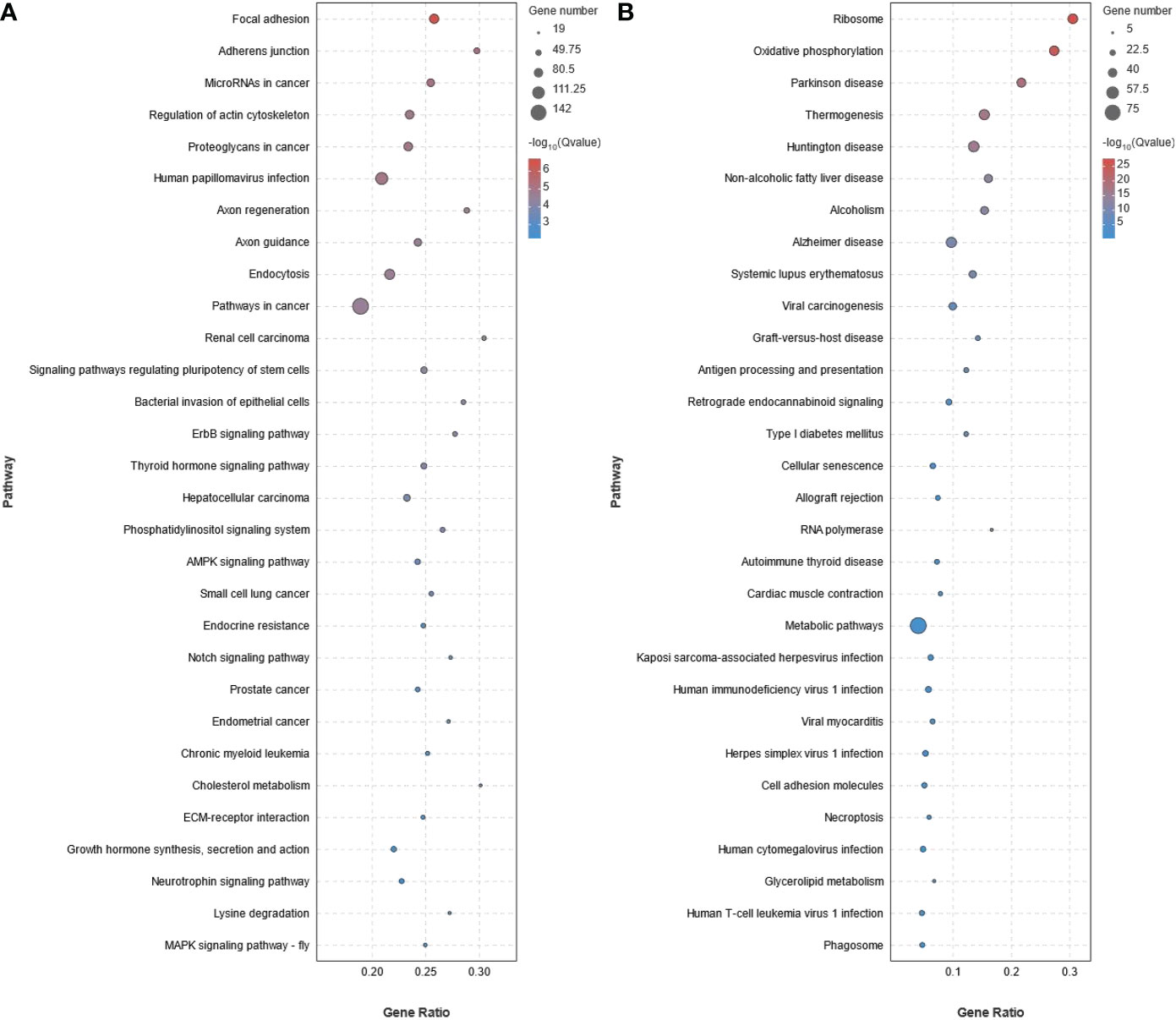
Figure 5 KEGG pathway enrichment analysis of the differentially expressed genes (DEGs) obtained from RNA-seq data of stage III and IV ovary of S. argus. (A) Up-regulated DEGs. (B) Down-regulated DEGs. The diameter of the circles represents the number of genes. The colors bar, from red to blue, represents the significance level of enrichment.
3.5 Expression of representative DEGs in ovarian growth and development
We analyzed some representative DEGs/genes associated with specific aspects based on the transcriptome data to better understand the biological function of DEGs in ovarian growth and development.
3.5.1 Expression of genes associated with steroid hormone biosynthesis
Ten DEGs (fshr, foxl2, cyp17a1, cyp7b1, hsd17b1, hsd3b1, hsd3b7, pgr, esr1, and ar) associated with steroid hormone biosynthesis were identified from the transcriptome data. All the genes were significantly up-regulated in stage IV ovarian growth and development (Figure 6 and Supplementary Table S8).
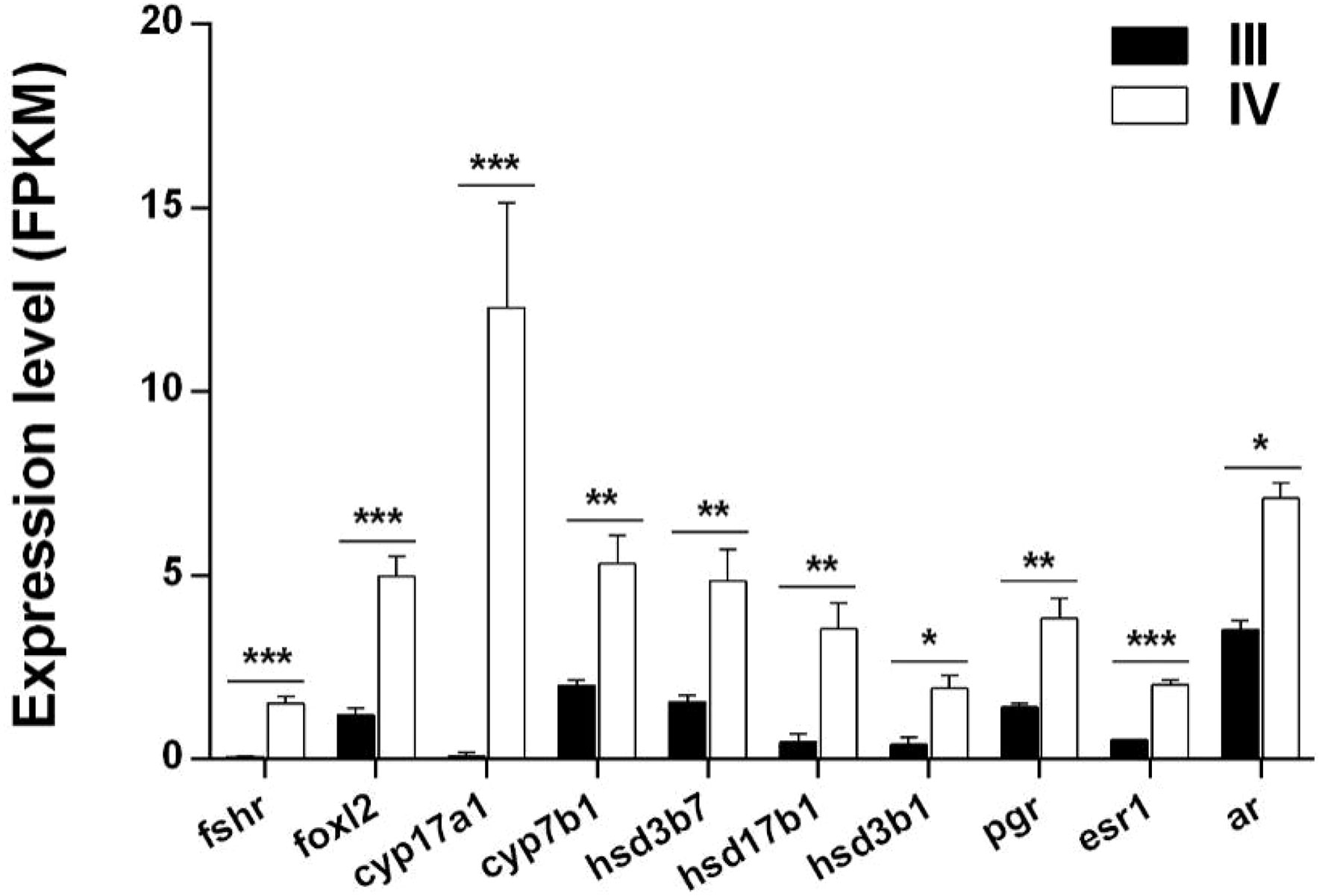
Figure 6 Histogram showing the expression of DEGs (log2 (FC) >1 and FDR <0.05) involved in steroid hormone synthesis in stage III and IV ovarian growth and development of S. argus. The data is based on the FPKM values obtained from the RNA-seq data. *, P<0.05; **, P<0.01; ***, P<0.001.
3.5.2 Expression of genes associated with lipid droplet and yolk production
Thirty-three DEGs associated with lipid droplet and yolk production were identified from the transcriptome data (Supplementary Table S9). Notably, 21 of these genes are shown in Figure 7A. Of the 21 genes, 18 (lpl, lrp1-like, lrp1, lrp5, lrp6, lrp8, lrp10, slc27a4, slc27a2, acsl4, lpcat1, ppara, ppard, plpp5, plpp2, cop1, and copb2) were up-regulated, while three (fabp1, fabp4, and ctss) were down-regulated in stage IV ovaries compared to stage III (Figure 7A).
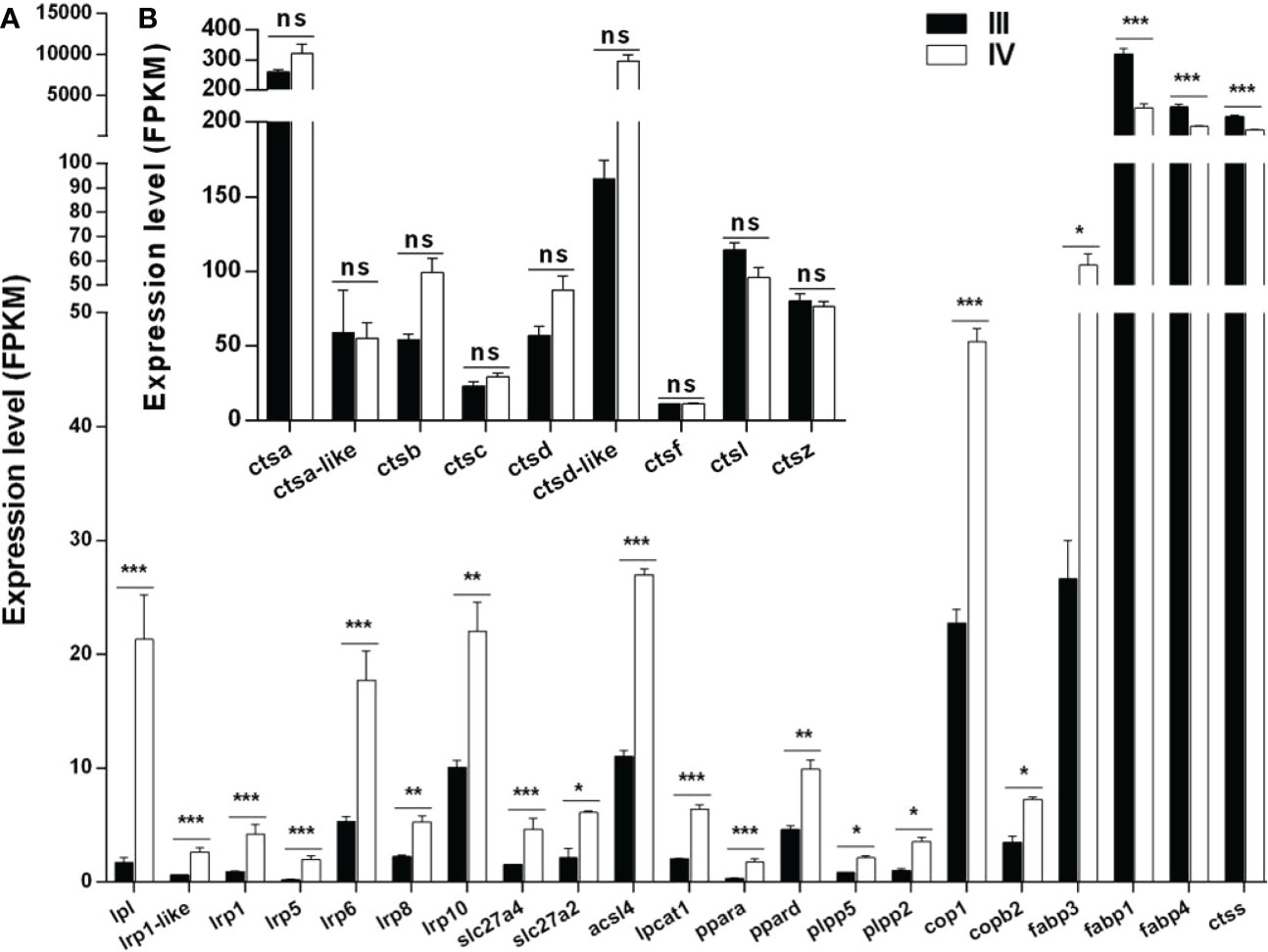
Figure 7 Histogram showing the expression of genes associated with lipid droplet and yolk production. (A) Expression of DEGs (log2 (FC) >1 and FDR <0.05) involved in lipid droplet and yolk production in stage III and IV ovary of S. argus. (B) Expression of genes (log2 (FC) ≤1 or FDR > 0.05) involved in vitellogenin catalysis. The data is based on the FPKM values obtained from the RNA-seq data. *, P<0.05; **, P<0.01; ***, P<0.001; ns, No significant difference.
Multiple cathepsins, including cathepsin B, L, and D, play roles in vitellogenin hydrolysis during oocyte growth in oviparous animals (Meng et al., 2022). Interestingly, only the expression of ctss met the condition of log2 (FC) >1 and FDR <0.05 among the 10 cathepsin-encoding genes identified in stage III and IV ovaries (Figure 7A). The other nine cathepsin-encoding genes included ctsd, ctsb, ctsd-like, ctsc, ctsa, ctsf, ctsz, ctsa, and ctsl. The expressions of ctsd, ctsb, ctsd-like, ctsc, and ctsa were up-regulated, while those of ctsf, ctsz, ctsa, and ctsl were down-regulated in stage IV compared to stage III ovarian (Figure 7B). Nonetheless, all the genes generally showed high expression in stage III and stage IV, especially ctsa and ctsd-like.
3.5.3 Expression of local ovarian endocrine factors and cytokines
Numerous local ovarian endocrine factors and cytokines play important roles in ovarian growth and development. In this study, 29 ovarian paracrine factors and their downstream pathway genes were identified from the transcriptome data (Figure 8 and Supplementary Table S10). Notably, all the 29 genes (igf1r, igfbp1, igfbp7, igf2r, igf2bp1, igf2bp3, inha, inhbb, acvr1, acvr1b, acvr2a, acvr2b, tgfa, tgfbr2, smad2, smad4, smad5, egfr, hbegf, megf8, erbb2, bmp1, bmp2k, bmp4, bmpr1a, bmpr2, kita, notch2, and notch3) were significantly up-regulated (Figure 8 and Supplementary Table S10).
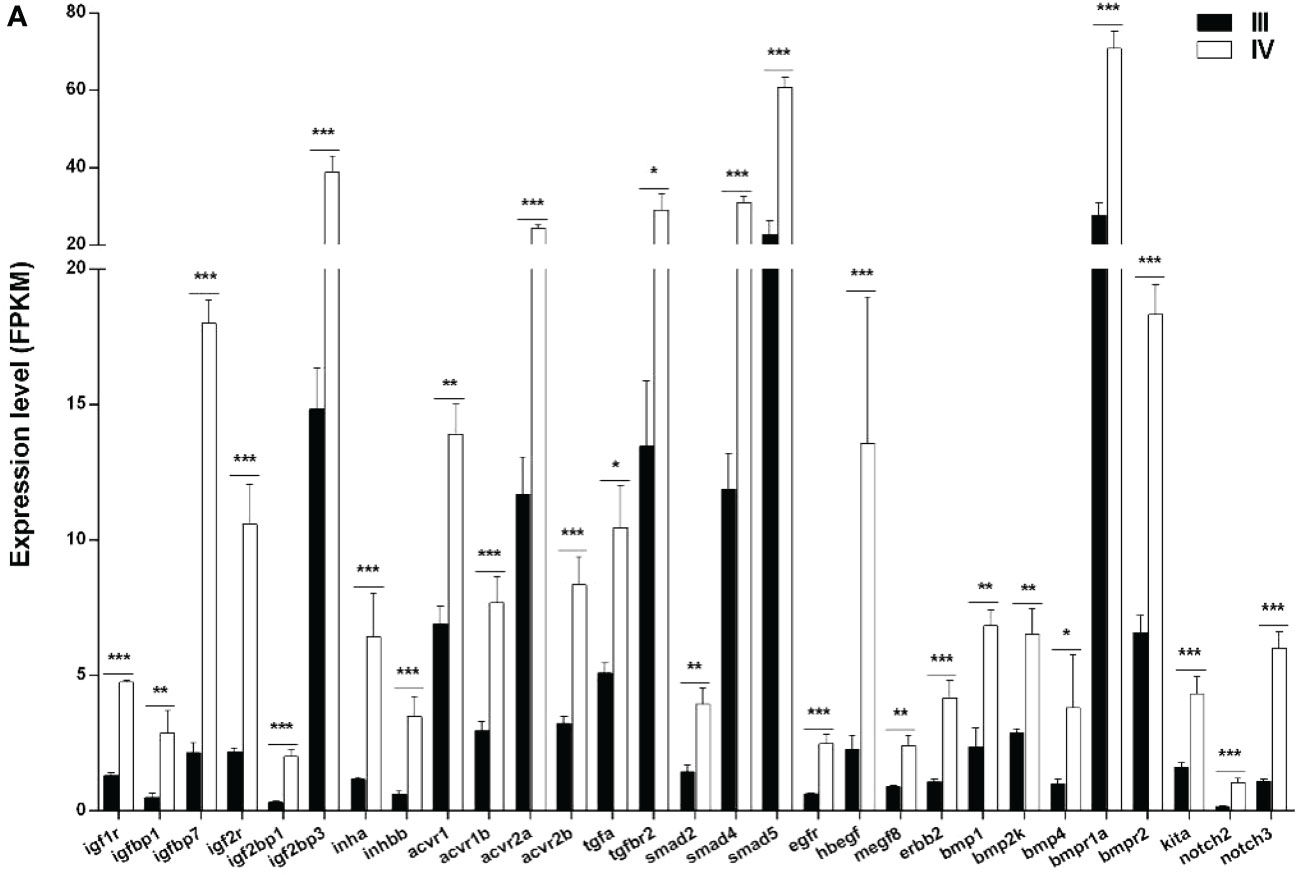
Figure 8 Histogram showing the selected differentially expressed genes (log2 (FC) >1 and FDR <0.05) involved in paracrine signaling in stage III and IV ovary of S. argus. The data is based on the FPKM values obtained from the RNA-seq data. *, P<0.05; **, P<0.01; ***, P<0.001.
3.5.4 Expression of genes potentially involved in meiotic arrest and resumption
The expression of 38 genes potentially involved in the meiotic arrest and resumption was examined using the transcriptomic data (Supplementary Table S11) (Meng et al., 2022). The genes were divided into two categories: genes associated with cell cycle and meiosis (Figure 9A) and those associated with cAMP or cGMP synthesis and hydrolysis (Figure 9B). Of note, 23 (ccnd1, ccne1, ccnf, ccnt2, cdc42, cdc45, cdk12, cdk13, cdk17, cdk17-like, cdkn1c, cenpe, cenpf, cenpi, cenpj, chka, chka-like, g2e3, plk1, plk2, sgk1-like, and sik2) of the 26 genes associated with cell cycle and meiosis were up-regulated, while the other three genes (cdk5r1, cenps, and cenpx) were down-regulated in stage IV compared to stage III. Moreover, all the 12 genes (adcy2, adcy5, adcy9, pde2a, pde4d, pde7a, pde8a, pde8b, atf2, atf6, atf7, and atf7-like) related to cAMP or cGMP synthesis and hydrolysis were up-regulated.
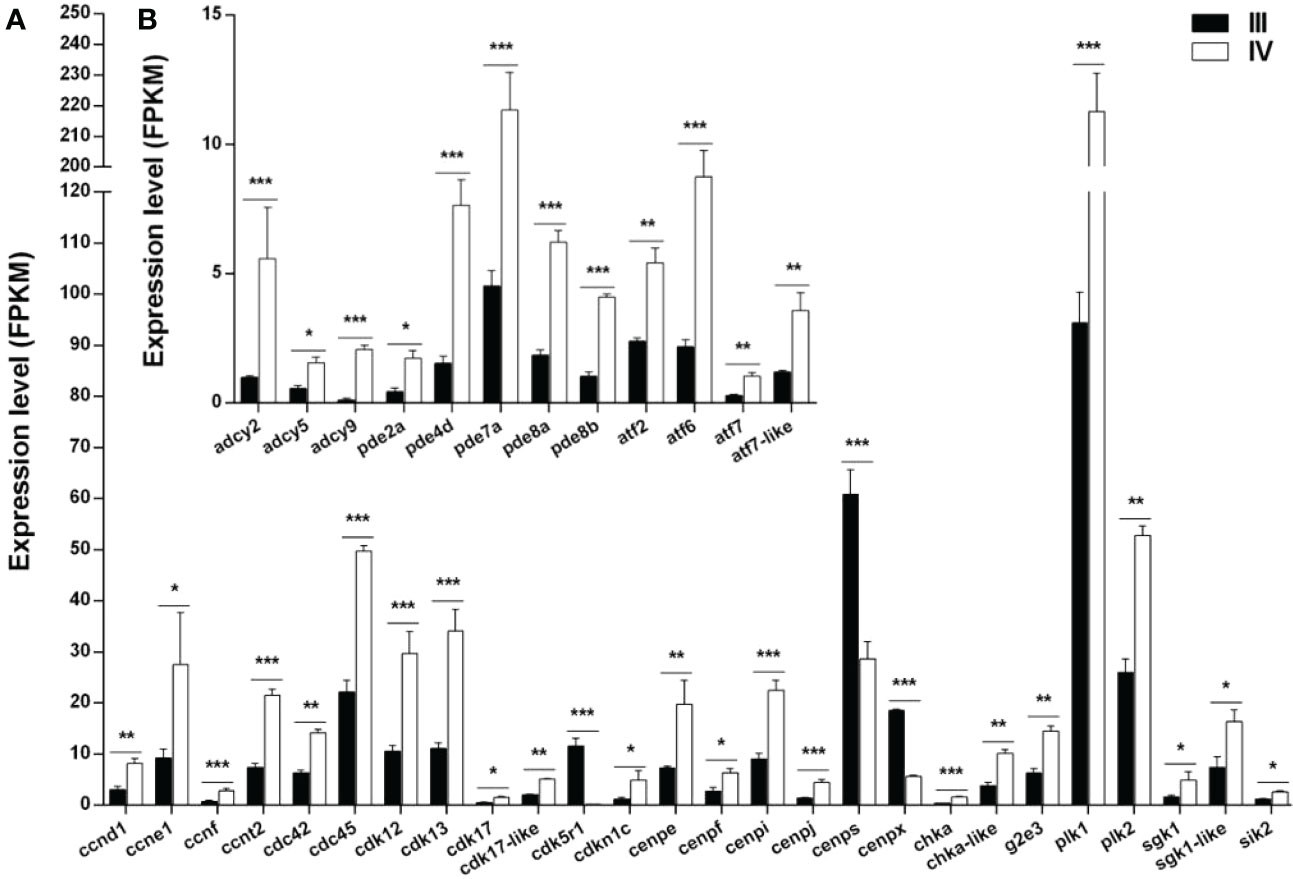
Figure 9 Histogram showing the selected differentially expressed genes (log2 (FC) >1 and FDR <0.05) involved in meiotic regulation in stage III and IV ovaries of S. argus. (A) Expression of genes associated with cell cycle and meiosis. (B) Expression of genes associated with cAMP or cGMP synthesis and hydrolysis. The data is based on the FPKM values obtained from the RNA-seq data. *, P<0.05; **, P<0.01; ***, P<0.001.
4 Discussion
Ovary development is pivotal for the successful reproduction of fish and has thus attracted tremendous research interests in aquaculture. Numerous studies have focused on genes regulating ovary development in fish, including S. argus. In recent years, transcriptome analysis has been utilized to identify genes and signal pathways that are critical for special biological processes of oogenesis in several fish species. Herein, we compared the expression of genes in stage III and stage IV ovary in S. argus using RNA-seq. The genes and pathways involved in secondary ovarian growth were then identified. The findings of this study enhance the understanding of the molecular regulation of vitellogenesis, lipid droplet formation, steroidogenesis, and meiosis during ovary growth in S. argus.
The ovary accumulates nutrition factors extrinsically from the blood and produces some structure components or materials intrinsically during the secondary growth stage. The liver is the main source of the nutrition factors, including Vtg, lipid, and zona pellucida (Zp) proteins which are transported into the ovary via the bloodstream. RNA-seq recently revealed that E2 biosynthesis pathway genes were highly activated in ovarian follicles at the late vitellogenic stage in ricefield eel (Monopterus albus) (Meng et al., 2022). Hypothalamic-pituitary-gonadal aix hormones play important roles in ovary development and maturation, including in estrogen-mediated VTG synthesis. Fshr and Lhr are two major receptors that mediate the pituitary Fsh and Lh signals, respectively, in the gonads in mammals. Notably, mutation of Fshr and Lhr results in infertility in female mice (Abel et al., 2000; Jonas et al., 2021). In contrast, fshr mutant female zebrafish exhibit follicle activation failure with follicles arrested at an early stage and became infertile, while the lhr mutant female fish remain fertile (Zhang et al., 2015). Fshr also mediates the signal of Lh in zebrafish (Chu et al., 2014). Loss of fshr also results in blocked ovary development in medaka and a significant decrease in the serum E2 level (Murozumi et al., 2014). The serum E2 and Vtg levels in female individuals at stage IV ovary are significantly higher than those of individuals at stage II and III ovary in S. argus (Cui et al., 2017). The up-regulation of serum E2 is attributed to higher ovary fshr expression in female S. argus at stage IV ovarian growth. Cytochrome P450 (Cyp)17A1 coded by Cyp17a1 has both 17, 20-lyase and 17-hydroxylase activities involved in producing androgens, estrogens, and progestin under Lh regulation (Magoffin, 1989; Zhai et al., 2018). We deduce that cyp17a1 is potentially up-regulated by the fshr pathway in the stage IV ovary, considering that Fsh and Lh possibly share the same receptor, fshr. Up-regulation of cyp17a1 is possibly the direct reason for the increase of estrogen synthesis. In addition, other up-regulated DEGs, including foxl2, hsd17b1, and pgr, involved in steroidogenesis were identified (Figure 5 and Supplementary Table S8). Of note, Foxl2 is an activator of cyp19a1a which codes for armoatase and is critical for estrogen synthesis in fish (Wang et al., 2007; Bertho et al., 2018). Herein, cyp19a1a expression remained unchanged at mRNA level despite foxl2 being up-regulated in stage IV ovary (Supplementary Table S3). Alternatively, the Cyp19a1a protein level possibly increased in stage IV ovary. Future studies should thus determine gene expression at the protein level during ovarian growth and maturation.
Besides regulating Vtgs synthesis via estrogen in the liver, uptake of Vtgs by the oocyte is also an important process during the secondary growth sage (Johnson, 2009). The uptake of Vtgs is fulfilled by its receptor-mediated endocytosis. Lpr8 and Lpr13 are Vtg receptors in fish (Hiramatsu et al., 2015). In orange-spotted grouper (Epinephelus coioides), the lrp13 mRNA level decrease from stage II to stage IV ovary, while western blot (WB) analysis shows that stage IV ovary has the highest Lrp13 protein level (Ye et al., 2022). In this study, numerous lpr genes, including lrp8, were up-regulated at stage IV ovary, while lrp13 (EVM0006844, lrp1b) was highly expressed at both stage III and stage IV (Figure 7 and Supplementary Table S9). Both lrp8 and lrp13 potentially act as Vtgr in S. argus, like in other fish. They could be catalyzed to form yolk granules after Vtg is incorporated into the oocytes. In ricefield eel, RNA-seq analysis revealed that 13 cathepsin genes were highly expressed in the ovarian follicles during some development stages (Meng et al., 2022). Herein, numerous genes involved in Vtg catalyzation (cathepsins genes) were highly expressed at both stage III and IV ovary, implying that they might be critical for vitellogenesis in S. argus (Figure 7 and Supplementary Table S9).
Accumulation of lipid droplets is a major characteristic of stage IV ovary. In Japanese flounder, RNA-seq revealed that numerous genes associated with lipid metabolism in ovaries were up-regulated from the primary growth ovary stage to the late lipid droplet stage (Qu et al., 2022). S. argus is a marine fish, and its oocyte stores lipids forming lipid droplets to ensure egg buoyancy (Zhang et al., 2019). The fatty acid binding protein (Fabp) super-family plays an important role in transporting the lipids to the cellular metabolic target sites (Chmurzyńska, 2006; Lei et al., 2020). The fatty acids and PPAR agonists can up-regulate the regulation of fabp2, fabp3, and fabp6 genes in zebrafish tissues (Venkatachalam et al., 2013). Dietary fish oil supplementation increase fabp2 expression in the liver, indicating that fabp2 potentially participates in the metabolism of long-chain unsaturated fatty acids in S. argus (Wang et al., 2021b). Herein, ppara, ppard, and fabp3 were up-regulated, while fabp1 and fabp4 were down-regulated in the stage IV ovary (Figure 7 and Supplementary Table S9), indicating that different fabps potentially have different functions. However, the critical fabps for lipid droplet acculturation in S. argus remain to be elucidated. In European sea bass (Dicentrarchus labrax L.), lipoprotein lipase (lpl) is highly expressed in the ovary with high gonadal-somatic index in the follicle cells surrounding the oocyte, indicating that it is critical for lipid droplet formation (José Ibáñez et al., 2008). Herein, lpl was significantly up-regulated in stage IV ovary suggesting that it plays a conserved role in lipid absorption in oocytes of S. argus.
Ovaries also produce some egg components, such as Zp, which is critical for fertilizing and protecting eggs (Wu et al., 2018). Twenty Zp family gene members have been identified in teleost and are mostly expressed in fish ovaries (Wu et al., 2018). Notably, six zp gene members are the most significantly expressed genes in the ovary of Japanese flounder (Paralichthys olivaceus). Their expressions gradually decreased from primary growth, early oil droplet to late oil droplet oocyte groups (Qu et al., 2022). In the reference study, the early oocytes needed more Zp protein to meet their oocyte development requirements (Qu et al., 2022). Similarly, both zp2 and zp3 were down-regulated at stage IV compared to stage III ovary in this study (Supplementary Table S12). In black rockfish, immunohistochemistry analysis showed that Zpb2a was detected in the cytoplasm of oocytes at stage III and the region close to the zona pellucida and cell membrane of oocytes at stage IV. The strongest protein signal in the zona pellucida was observed in stage V oocytes (Li et al., 2022). However, Zp expressions at the protein level during ovary development should be studied in the future to elucidate their assembly process at both mRNA and protein levels. The cellular components and preparation of nutrition factors are well organized during ovary development in fish. Of note, the underlying mechanism of this organization would be an important scientific question in the future.
Besides the accumulation of nutrition and structure proteins in the oocyte, a number of meiosis related genes may increase their expression before the end of vitellogenesis and eventually lead to ovarian maturation (resumption of meiosis) (Lubzens et al., 2010; Meng et al., 2022). In ricefield eel ovary, the highest expression of the adenyl cyclase (adcy) genes is at MV and LV. The expression of the cyclic AMP-dependent transcription factor atf-3 gene and the phosphodiesterase (pde) genes observed at full grown stage indicate that cAMP signal pathways potentially play critical roles in oocyte meiotic arrest and resumption (Meng et al., 2022). Herein, many genes involved in the cellular cycle, meiosis, cAMP or cGMP synthesis, and hydrolysis were up-regulated in stage IV ovary compared to stage III ovary, indicating that the oocyte meiosis was potentially restarted, causing them to mature.
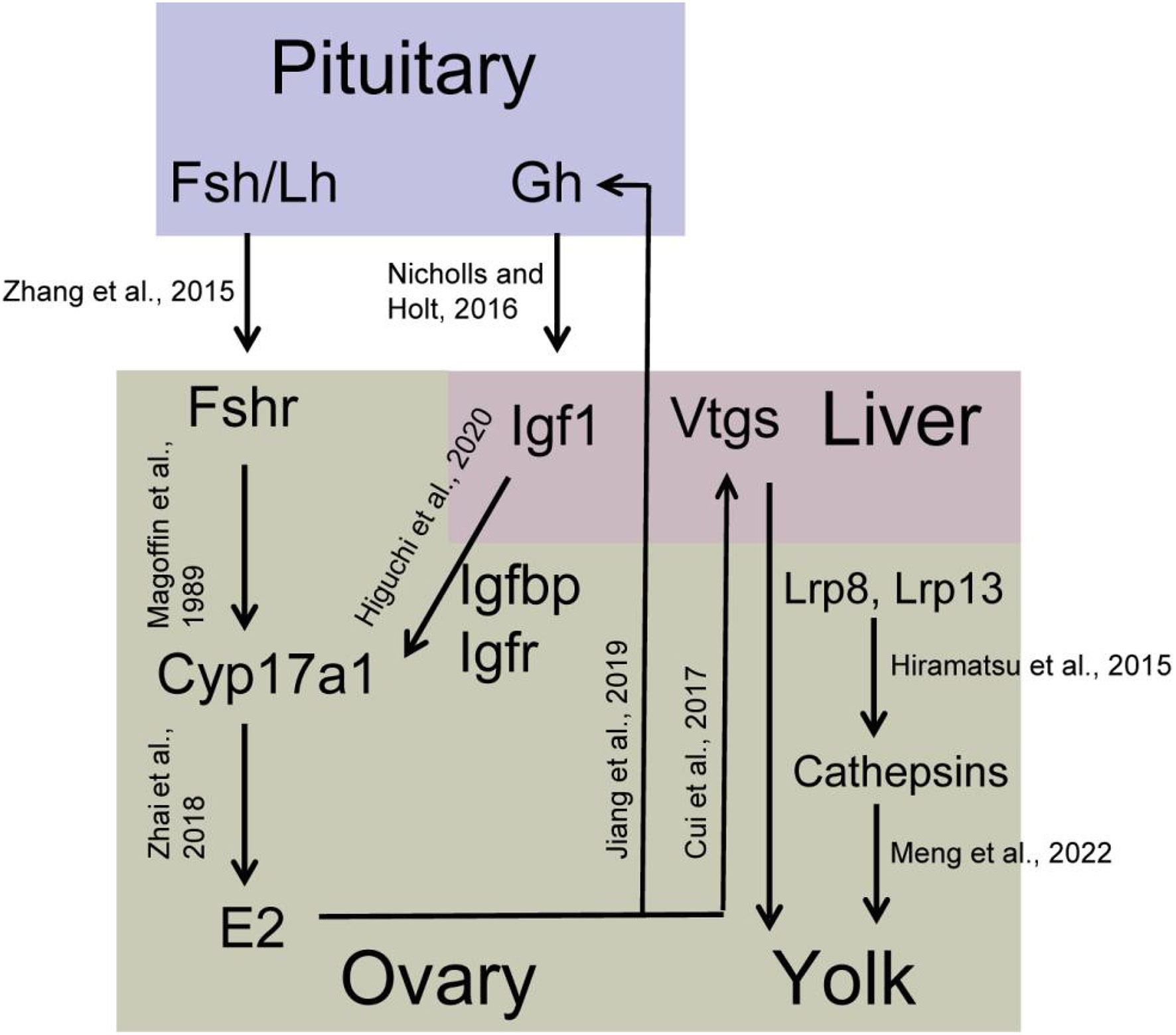
Plenty of endocrine factors and cytokines were involved in regulating multiple cellular processes in the ovary that were identified as DEGs in this study. The insulin like growth factor (Igf) signal pathway plays an important role in ovary development, including in the resumption of meiosis and final maturation in fish (Ndandala et al., 2022). The Igf signal pathway consists of ligands (Igf1, Igf2, and Igf3), Igf binding proteins (Igfbp), and Igf receptors. Herein, igf1r, ifg2r, igfbp1, igfbp3, and igfbp7 were up-regulated in stage IV ovary, indicating that the Igf signal pathway can be potentially enhanced to promote the ovary developmental processes. Igf1 expression and secretion are regulated by the growth hormone (Gh) secreted from the pituitary gland (Nicholls and Holt, 2016). In S. argus, exogenous E2 up-regulates the mRNA expression of pituitary gh (Jiang et al., 2019). Pituitary gh and liver igf1 mRNA expression gradually increase from stage II to IV ovary (Zhang et al., 2019; Ru et al., 2020). We thus deduce that there exists a positive feedback regulation E2 on pituitary gh. Igf1 and Igf2 enhance the expression of cyp17a1 in the vitellogenic ovary via PI3 kinase in yellowtail (Seriola quinqueradiata). However, there is no such up-regulation in pre-vitellogenic ovary (Higuchi et al., 2020). Combined with the steroidogenesis associated genes, we summarize a possible regulation network between Gh-Igf and E2 synthesis pathway during ovarian growth and maturation in S. argus (Figure 10).
TGF-beta signal pathway members are also critical during ovary development in fish (Zheng et al., 2018). Herein, numerous TGF-beta signal pathway members involved in ovary development were DEGs. They included inhb, bmp1, bmp2k, bmp4, and TGF-beta pathway receptors and smads (Figure 7 and Supplementary Table S9). However, some TGF-beta members, such as bmp15 and gdf9, were not DEGs between stage III and stage IV ovary in this study (Supplementary Table S12), but we still could not exclude that they play an important role in regulating ovary development because they were highly expressed. Bmp15 is essential for female sex maintenance in adult zebrafish (Dranow et al., 2016). In the same line, Gdf9 regulates the tight junction (TG) related genes in the ovary of zebrafish, thereby influencing cellar permeability (Clelland and Kelly, 2011). Herein, several TG-related genes were differentially expressed and could be regulated by TGF-beta signaling (Supplementary Table S10). These findings collectively suggest that there are plenty of signal pathways and genes that regulate ovary growth and are highly expressed in the ovary in some developmental stages. Future studies should focus more on the detailed function and regulatory networks of these important genes and pathways in ovary development. While the regulation relationships among these genes were mainly dependent on RNA-Seq/RT-qPCR data and the existing research of other species, more experimental evidences are required to certify them.
5 Conclusion
The expression profiles of secondary growth-related genes in the ovaries of S. argus were identified using RNA-seq. There were 3666 DEGs between stage III and IV ovary, regulating steroidogenesis, vitellogenesis, lipid droplet formation, and meiosis. Besides these DEGs, some genes are expressed highly at both ovary stages and are critical for ovary development. The signal pathways and genes associated with ovarian growth are relatively conserved among fish. These results provide baseline data for studying the regulatory mechanisms of oogenesis in S. argus and the artificial propagation of S. argus in aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRP171076, PRJNA906196.
Ethics statement
The animal study was reviewed and approved by Animal Research and Ethics Committee of Guangdong Ocean University, China.
Author contributions
M-YJ: Investigation, Data curation, Formal analysis, Visualization, Writing—original draft and Funding acquisition; Y-FZ, HL, Y-XP, and Y-QH: Investigation, Data curation and Formal analysis; S-PD: Formal analysis and Visualization; YH and GS: Resources; C-HZ and G-LL: Resources, Project administration, Supervision; D-NJ: Conceptualization, Writing—original draft, Writing—review and editing, Funding acquisition, Project administration, Supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Nos. 32002368, 32172971, and 31972775); the Zhan jiang city science and technology projects (2022A01015); Guangdong Basic and Applied Basic Research Foundation (2021A1515010430); the Key Project of “Blue Granary Science and Technology Innovation” of the Ministry of Science and Technology (2018YFD0901203); the program for scientific research start-up funds of Guangdong Ocean University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1114872/full#supplementary-material
References
Abel M. H., Wootton A. N., Wilkins V., Huhtaniemi I., Knight P. G., Charlton H. M. (2000). The effect of a null mutation in the follicle-stimulating hormone receptor gene on mouse reproduction. Endocrinology 141 (5), 1795–1803. doi: 10.1210/endo.141.5.7456
Asiabi P., Ambroise J., Giachini C., Coccia M. E., Bearzatto B., Chiti M. C., et al. (2020). Assessing and validating housekeeping genes in normal, cancerous, and polycystic human ovaries. J. Assist. Reprod. Genet. 37 (10), 2545–2553. doi: 10.1007/s10815-020-01901-8
Baddela V. S., Baufeld A., Yenuganti V. R., Vanselow J., Singh D. (2014). Suitable housekeeping genes for normalization of transcript abundance analysis by real-time RT-PCR in cultured bovine granulosa cells during hypoxia and differential cell plating density. Reprod. Biol. Endocrinol. 12, 118. doi: 10.1186/1477-7827-12-118
Bertho S., Herpin A., Branthonne A., Jouanno E., Yano A., Nicol B., et al. (2018). The unusual rainbow trout sex determination gene hijacked the canonical vertebrate gonadal differentiation pathway. Proc. Natl. Acad. Sci. U. S. A. 115 (50), 12781–12786. doi: 10.1073/pnas.1803826115
Breton T. S., Berlinsky D. L. (2014). Characterizing ovarian gene expression during oocyte growth in Atlantic cod (Gadus morhua). Comp. Biochem. Physiol.,Part D: Genomics Proteomics 9, 1–10. doi: 10.1016/j.cbd.2013.11.001
Caetano L. C., Miranda-Furtado C. L., Batista L. A., Pitangui-Molina C. P., Higa T. T., Padovan C. C., et al. (2019). Validation of reference genes for gene expression studies in bovine oocytes and cumulus cells derived from in vitro maturation. Anim. Reprod. 16 (2), 290–296. doi: 10.21451/1984-3143-AR2018-0064
Cai C., Cai P., Chu G. (2019). Selection of suitable reference genes for core clock gene expression analysis by real-time qPCR in rat ovary granulosa cells. Mol. Biol. Rep. 46 (3), 2941–2946. doi: 10.1007/s11033-019-04755-1
Cai Z. P., Wang Y., Hu J. W., Zhang J. B., Lin Y. G. (2010). Reproductive biology of Scatophagus argus and artificial induction of spawning. J. Trop. Oceanogr. 29 (05), 180–185.
Chen H. P., Cui X. F., Wang Y. R., Li Z. Y., Tian C. X., Jiang D. N., et al. (2020). Identification, functional characterization, and estrogen regulation on gonadotropin-releasing hormone in the spotted scat, Scatophagus argus. Fish Physiol. Biochem. 46 (5), 1743–1757. doi: 10.1007/s10695-020-00825-5
Chmurzyńska A. (2006). The multigene family of fatty acid-binding proteins (FABPs): Function, structure and polymorphism. J. Appl. Genet. 47 (1), 39–48. doi: 10.1007/BF03194597
Chu L., Li J., Liu Y., Hu W., Cheng C. H. K. (2014). Targeted gene disruption in zebrafish reveals noncanonical functions of LH signaling in reproduction. Mol. Endocrinol. 28 (11), 1785–1795. doi: 10.1210/me.2014-1061
Clelland E. S., Kelly S. P. (2011). Exogenous GDF9 but not activin a, BMP15 or TGFβ alters tight junction protein transcript abundance in zebrafish ovarian follicles. Gen. Comp. Endocrinol. 171 (2), 211–217. doi: 10.1016/j.ygcen.2011.01.009
Cook-Andersen H., Curnow K. J., Su H. I., Chang R. J., Shimasaki S. (2016). Growth and differentiation factor 9 promotes oocyte growth at the primary but not the early secondary stage in three-dimensional follicle culture. J. Assist. Reprod. Genet. 33 (8), 1067–1077. doi: 10.1007/s10815-016-0719-z
Cui X. F., Zhao Y., Chen H. P., Deng S. P., Jiang D. N., Wu T. L., et al. (2017). Cloning, expression and functional characterization on vitellogenesis of estrogen receptors in Scatophagus argus. Gen. Comp. Endocrinol. 246, 37–45. doi: 10.1016/j.ygcen.2017.03.002
Dranow D. B., Hu K., Bird A. M., Lawry S. T., Adams M. T., Sanchez A., et al. (2016). Bmp15 is an oocyte-produced signal required for maintenance of the adult female sexual phenotype in zebrafish. PloS Genet. 12 (9), e1006323. doi: 10.1371/journal.pgen.1006323
He S., Du H., Ren P., Leng X.-Q., Tan Q.-S., Li C.-J., et al. (2020). Transcriptome analysis of ovarian maturation in a chondrostei Chinese sturgeon acipenser sinensis. J. Exp. Zool. B Mol. Dev. Evol. 334 (5), 280–293. doi: 10.1002/jez.b.22973
Higuchi K., Kazeto Y., Ozaki Y., Izumida D., Hotta T., Soyano K., et al. (2020). Insulin-like growth factors 1 and 2 regulate gene expression and enzymatic activity of cyp17a1 in ovarian follicles of the yellowtail, Seriola quinqueradiata. Heliyon 6 (6), e04181. doi: 10.1016/j.heliyon.2020.e04181
Hiramatsu N., Todo T., Sullivan C. V., Schilling J., Reading B. J., Matsubara T., et al. (2015). Ovarian yolk formation in fishes: Molecular mechanisms underlying formation of lipid droplets and vitellogenin-derived yolk proteins. Gen. Comp. Endocrinol. 221, 9–15. doi: 10.1016/j.ygcen.2015.01.025
Huang Y., Mustapha U. F., Huang Y., Tian C., Yang W., Chen H., et al. (2021). A chromosome–level genome assembly of the spotted scat (Scatophagus argus). Genome Biol. Evol. 13 (6), evab092. doi: 10.1093/gbe/evab092
Jiang M., Liu J., Jiang D., Pan Q., Shi H., Huang Y., et al. (2022). Characterization and expression analysis of gpr173a and gpr173b revealed their involvement in reproductive regulation in spotted scat (Scatophagus argus). Aquacult. Rep. 25, 101239. doi: 10.1016/j.aqrep.2022.101239
Jiang D., Shi H., Liu Q., Wang T., Huang Y., Huang Y., et al. (2019). Isolation of growth hormone-releasing hormone and its receptor genes from Scatophagus argus and their expression analyses. J. Ocean Univ. China 18 (6), 1486–1496. doi: 10.1007/s11802-019-4110-4
Johnson R. B. (2009). Lipid deposition in oocytes of teleost fish during secondary oocyte growth. Rev. Fisheries Sci. 17 (1), 78–89. doi: 10.1080/10641260802590004
Jonas K. C., Rivero Müller A., Oduwole O., Peltoketo H., Huhtaniemi I. (2021). The luteinizing hormone receptor knockout mouse as a tool to probe the In vivo actions of gonadotropic hormones/receptors in females. Endocrinology 162 (5), 1–13. doi: 10.1210/endocr/bqab035
José Ibáñez A., Peinado-Onsurbe J., Sánchez E., Cerdá-Reverter J. M., Prat F. (2008). Lipoprotein lipase (LPL) is highly expressed and active in the ovary of European sea bass (Dicentrarchus labrax l.), during gonadal development. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 150 (3), 347–354. doi: 10.1016/j.cbpa.2008.04.598
Kwok H.-F., So W.-K., Wang Y., Ge W. (2005). Zebrafish gonadotropins and their receptors: I. cloning and characterization of zebrafish follicle-stimulating hormone and luteinizing hormone receptors— evidence for their distinct functions in follicle development1. Biol. Reprod. 72 (6), 1370–1381. doi: 10.1095/biolreprod.104.038190
Lei C.-x., Li M.-m., Tian J.-j., Wen J.-k., Li Y.-y. (2020). Transcriptome analysis of golden pompano (Trachinotus ovatus) liver indicates a potential regulatory target involved in HUFA uptake and deposition. Comp. Biochem. Physiol.,Part D: Genomics Proteomics 33, 100633. doi: 10.1016/j.cbd.2019.100633
Li R., Qu J., Huang D., He Y., Niu J., Qi J. (2022). Expression analysis of ZPB2a and its regulatory role in sperm-binding in viviparous teleost black rockfish. Int. J. Mol. Sci. 23 (16), 9498. doi: 10.3390/ijms23169498
Lubzens E., Young G., Bobe J., Cerdà J. (2010). Oogenesis in teleosts: How fish eggs are formed. Gen. Comp. Endocrinol. 165 (3), 367–389. doi: 10.1016/j.ygcen.2009.05.022
Magoffin D. A. (1989). Evidence that luteinizing hormone-stimulated differentiation of purified ovarian thecal-interstitial cells is mediated by both type I and type II adenosine 3′,5′-monophosphate-dependent protein kinases. Endocrinology 125 (3), 1464–1472. doi: 10.1210/endo-125-3-1464
Mandal B., Kailasam M., Bera A., Sukumaran K., Hussain T., Biswas G., et al. (2021). Standardization of oocyte size during artificial fertilization and optimization of stocking density during indoor larval and outdoor nursery rearing of captive spotted scat (Scatophagus argus) for a viable juvenile production system. Aquaculture 534, 736262. doi: 10.1016/j.aquaculture.2020.736262
Meng F., Sun S., Xu X., Yu W., Gan R., Zhang L., et al. (2022). Transcriptomic analysis provides insights into the growth and maturation of ovarian follicles in the ricefield eel (Monopterus albus). Aquaculture 555, 738251. doi: 10.1016/j.aquaculture.2022.738251
Murozumi N., Nakashima R., Hirai T., Kamei Y., Ishikawa-Fujiwara T., Todo T., et al. (2014). Loss of follicle-stimulating hormone receptor function causes masculinization and suppression of ovarian development in genetically female medaka. Endocrinology 155 (8), 3136–3145. doi: 10.1210/en.2013-2060
Myllymaa S., Pasternack A., Mottershead D. G., Poutanen M., Pulkki M. M., Pelliniemi L. J., et al. (2010). Inhibition of oocyte growth factors in vivo modulates ovarian folliculogenesis in neonatal and immature mice. Reproduction 139 (3), 587–598. doi: 10.1530/rep-09-0391
Ndandala C. B., Dai M., Mustapha U. F., Li X., Liu J., Huang H., et al. (2022). Current research and future perspectives of GH and IGFs family genes in somatic growth and reproduction of teleost fish. Aquacult.Rep. 26, 101289. doi: 10.1016/j.aqrep.2022.101289
Nicholls A. R., Holt R. I. (2016). Growth hormone and insulin-like growth factor-1. Front. Horm. Res. 47, 101–114. doi: 10.1159/000445173
Patiño R., Sullivan C. V. (2002). Ovarian follicle growth, maturation, and ovulation in teleost fish. Fish Physiol. Biochem. 26 (1), 57–70. doi: 10.1023/A:1023311613987
Qu J., Li R., Xie Y., Liu Y., Liu J., Zhang Q. (2022). Differential transcriptomic profiling provides new insights into oocyte development and lipid droplet formation in Japanese flounder (Paralichthys olivaceus). Aquaculture 550, 737843. doi: 10.1016/j.aquaculture.2021.737843
Ru X.-Y., Shi H.-J., Wang T., Liu Q.-Q., Jiang D.-N., Peng Y.-H., et al. (2020). Effects of 17β-estradiol on growth-related genes expression in female and male spotted scat (Scatophagus argus). Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 250, 110492. doi: 10.1016/j.cbpb.2020.110492
Sullivan C. V., Yilmaz O. (2018). Vitellogenesis and yolk proteins, fish” in Encyclopedia of reproduction, 2nd ed. Ed. Skinner M. K. (Oxford: Academic Press), 266–277.
Tang L., Chen J., Ye Z., Zhao M., Meng Z., Lin H., et al. (2019). Transcriptomic analysis revealed the regulatory mechanisms of oocyte maturation and hydration in orange-spotted grouper (Epinephelus coioides). Mar. Biotechnol. 21 (4), 537–549. doi: 10.1007/s10126-019-09902-0
Urbatzka R., Rocha M. J., Rocha E. (2011). “Chapter 4 - regulation of ovarian development and function in teleosts,” in Hormones and reproduction of vertebrates. Eds. Norris D. O., Lopez K. H. (London: Academic Press), 65–82.
Venkatachalam A. B., Sawler D. L., Wright J. M. (2013). Tissue-specific transcriptional modulation of fatty acid-binding protein genes, fabp2, fabp3 and fabp6, by fatty acids and the peroxisome proliferator, clofibrate, in zebrafish (Danio rerio). Gene 520 (1), 14–21. doi: 10.1016/j.gene.2013.02.034
Wang T., Jiang D., Shi H., Mustapha U. F., Deng S., Liu Z., et al. (2021b). Liver transcriptomic analysis of the effects of dietary fish oil revealed a regulated expression pattern of genes in adult female spotted scat (Scatophagus argus). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.784845
Wang D.-S., Kobayashi T., Zhou L.-Y., Paul-Prasanth B., Ijiri S., Sakai F., et al. (2007). Foxl2 up-regulates aromatase gene rranscription in a female-specific manner by binding to the promoter as well as interacting with Ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 21 (3), 712–725. doi: 10.1210/me.2006-0248
Wang C., Qin H., Zhao C., Yang L., Yu T., Zhang Y., et al. (2021a). Whole-genome re-sequencing and transcriptome reveal oogenesis-related genes in autotetraploid Carassius auratus. Mar. Biotechnol. 23 (2), 233–241. doi: 10.1007/s10126-021-10018-7
Wang Y., Zhang M., Qin Q., Peng Y., Huang X., Wang C., et al. (2019). Transcriptome profile analysis on ovarian tissues of autotetraploid fish and diploid red crucian carp. Front. Genet. 10. doi: 10.3389/fgene.2019.00208
Washim M. R., Ahmmed S., Rubel A. S. A., Mondal D., Begum N., Islam M. L. (2022). The effects of synthetic gonadotropin releasing hormone analogue (S-GnRHa) on artificial propagation of spotted scat, Scatophagus argus (Linnaeus 1766). South Asian J. Exp. Biol. 12 (2), 203–212. doi: 10.38150/sajeb.12(2).p203-212
Wu T., Cheng Y., Liu Z., Tao W., Zheng S., Wang D. (2018). Bioinformatic analyses of zona pellucida genes in vertebrates and their expression in Nile tilapia. Fish Physiol. Biochem. 44 (2), 435–449. doi: 10.1007/s10695-017-0434-4
Ye Z., Zhao T., Zhang J., Huang H., Huang C., Chen M., et al. (2022). Ovarian expression and localization of a low-density lipoprotein receptor related protein 13 (Lrp13) in orange-spotted grouper (Epinephelus coioides). Aquacult. Rep. 27, 101416. doi: 10.1016/j.aqrep.2022.101416
Zhai Y., Deng S. P., Liu J. Y., Jiang D. N., Huang Y., Zhu C. H., et al. (2021). The reproductive regulation of LPXRFa and its receptor in the hypothalamo-pituitary-gonadal axis of the spotted scat (Scatophagus argus). Fish Physiol. Biochem. 47 (1), 93–108. doi: 10.1007/s10695-020-00898-2
Zhai G., Shu T., Xia Y., Lu Y., Shang G., Jin X., et al. (2018). Characterization of sexual trait development in cyp17a1-deficient zebrafish. Endocrinology 159 (10), 3549–3562. doi: 10.1210/en.2018-00551
Zhang Z., Lau S.-W., Zhang L., Ge W. (2015). Disruption of zebrafish follicle-stimulating hormone receptor (fshr) but not luteinizing hormone receptor (lhcgr) gene by TALEN leads to failed follicle activation in females followed by sexual reversal to males. Endocrinology 156 (10), 3747–3762. doi: 10.1210/en.2015-1039
Zhang K.-W., Wu T.-L., Chen H.-P., Jiang D.-N., Zhu C.-H., Deng S.-P., et al. (2019). Estradiol-17β regulates the expression of insulin-like growth factors 1 and 2 via estradiol receptors in spotted scat (Scatophagus argus). Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 237, 110328. doi: 10.1016/j.cbpb.2019.110328
Zheng S., Long J., Liu Z., Tao W., Wang D. (2018). Identification and evolution of TGF-β signaling pathway members in twenty-four animal species and expression in tilapia. Int. J. Mol. Sci. 19 (4), 1154. doi: 10.3390/ijms19041154
Keywords: Scatophagus argus, ovarian follicle, transcriptomic analysis, secondary follicle growth, steroid hormone
Citation: Jiang M-Y, Zhou Y-F, Liu H, Peng Y-X, Huang Y-Q, Deng S-P, Huang Y, Shi G, Zhu C-H, Li G-L and Jiang D-N (2023) Transcriptomic analysis provides new insights into the secondary follicle growth in spotted scat (Scatophagus argus). Front. Mar. Sci. 10:1114872. doi: 10.3389/fmars.2023.1114872
Received: 03 December 2022; Accepted: 10 February 2023;
Published: 21 February 2023.
Edited by:
Eduardo Almansa, Spanish Institute of Oceanography (IEO), SpainCopyright © 2023 Jiang, Zhou, Liu, Peng, Huang, Deng, Huang, Shi, Zhu, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Neng Jiang, ZG5qaWFuZ0BnZG91LmVkdS5jbg==
 Mou-Yan Jiang
Mou-Yan Jiang Yi-Fan Zhou
Yi-Fan Zhou You-Xing Peng
You-Xing Peng Yuan-Qing Huang
Yuan-Qing Huang Chun-Hua Zhu
Chun-Hua Zhu Guang-Li Li
Guang-Li Li Dong-Neng Jiang
Dong-Neng Jiang