- 1Institute for Marine and Antarctic Studies, University of Tasmania, Hobart, TAS, Australia
- 2College of Marine Life Sciences, Institute of Evolution and Marine Biodiversity, Frontiers Science Center for Deep Ocean Multispheres and Earth System, and Key Lab of Polar Oceanography and Global Ocean Change, Ocean University of China, Qingdao, China
- 3Ocean University of China - Universiti Malaysia Terengannu (UMT-OUC) Joint Centre for Marine Studies, Qingdao, China
- 4Haide College, Ocean University of China, Qingdao, China
- 5The Affiliated Hospital of Qingdao University, Qingdao, China
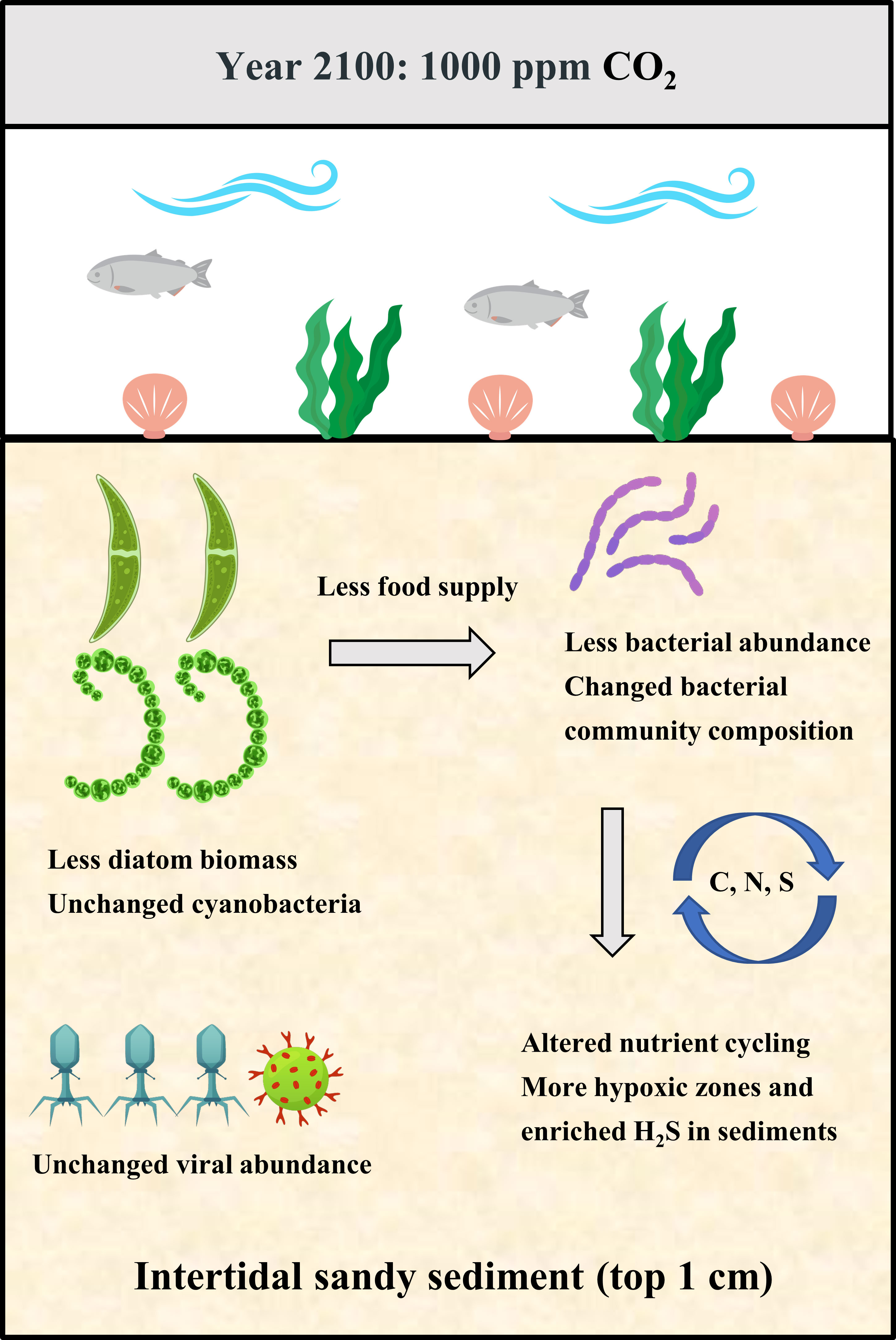
Microphytobenthos (MPB) and bacterial biofilms play crucial roles in primary and secondary production, nutrient cycling and invertebrate settlement in coastal ecosystems, yet little is known of the effects of ocean acidification (OA) on these communities in intertidal soft sediments. To fill in this gap, a 28-day CO2 enhancement experiment was conducted for the benthic biofilms in soft intertidal sediments (muds and sands) from Qingdao, China. This experiment included three CO2 treatments: 400 ppm CO2 (control), 700 ppm CO2 and 1000 ppm CO2 (IPCC predicted value in 2100), which were established in a three-level CO2 incubator that can adjust the CO2 concentration in the overlying air. The effects of OA on benthic biofilms were assessed in the following three aspects: MPB biomass, biofilm community structure and microbial biogeochemical cycling (e.g., C-cycle, N-cycle and S-cycle). This study found that the 700 ppm CO2 treatment did not significantly affect the benthic biofilms in intertidal soft sediments, but the 1000 ppm CO2 treatment significantly altered the biofilm community composition and potentially their role in microbial biogeochemical cyc\ling in sediments (especially in sandy sediments). For the bacterial community in biofilms, the 1000 ppm CO2 enhancement increased the relative abundance of Alteromonadales and Bacillales but decreased the relative abundance of Rhodobacterales and Flavobacteriales. For microbial biogeochemical cycling, the 1000 ppm CO2 treatment enhanced the potential of chemoheterotrophic activity, nitrate reduction and sulfur respiration in sediments, likely resulting in a more stressful environment (hypoxic and enriched H2S) for most benthic organisms. Even though incubations in this study were only 28 days long and thus couldn’t fully accommodate the range of longer-term adaptions, it still suggests that benthic biofilms in intertidal sandy sediments are likely to change significantly near the end of the century if anthropogenic CO2 emissions unmitigated, with profound implications on local ecosystems and biogeochemical cycling.
1 Introduction
As a result of anthropogenic activities such as fossil fuel burning and deforestation, the concentration of atmospheric carbon dioxide (pCO2) has increased from ~280 to ~420 ppm since the industrial revolution (Zhang et al., 2021). This has led to increased concentrations of dissolved CO2 in the oceans which has caused pH levels to drop, a process known as ocean acidification (OA). According to the ‘business as usual scenario’ of IPCC 2019, ocean seawater pH is projected to decrease by 0.3-0.4 units by 2100, which will have extensive biological impacts, ranging from alterations in organism physiology and population dynamics to modified marine communities and ecosystems (Doney et al., 2020; Lutier et al., 2022). Benthic biofilms in coastal regions contain rich communities of photosynthetic microbes (i.e., microphytobenthos, MPB) and heterotrophic bacteria, and these together play crucial roles in coastal ecosystems in terms of primary and secondary production, nutrient cycling and invertebrate settlement (De Carvalho, 2018). MPB is a community of unicellular algae (i.e., diatoms, dinoflagellates) and cyanobacteria, which in coastal regions contribute up to 70% of the diet of farmed oysters, mussels and cockles (Snelgrove et al., 2018). The heterotrophic bacteria in benthic biofilms make an important contribution to nutrient cycling and as a food source for heterotrophic nanoflagellates (Deppeler et al., 2018). In addition, some bacteria in biofilms produce and release chemical cues to attract the settlement of invertebrate larvae and macroalgal propagules (Das and Mangwani, 2015). Given these important ecological services of marine benthic biofilms in coastal regions, it is crucial to better understand how OA is likely to affect these communities.
So far, there have been several studies investigating how OA affects coastal benthic biofilms and these have concluded that OA can alter the biomass and community composition (Kerfahi et al., 2014; Taylor et al., 2014; Kerfahi et al., 2020). However, there are some important gaps in the current understanding. Firstly, most previous studies, solely or independently, investigated the effects of OA on microalgae or bacteria in intertidal benthic biofilms, but failed to consider the impacts of OA at a community-level (Cartaxana et al., 2015; Kerfahi et al., 2020). Thus, to get a better understanding of the implications of OA on benthic biofilms, it is recommended that the effects of OA on microalgae, bacteria and viruses in benthic biofilms are studied simultaneously. Secondly, previous OA research has mainly focused on intertidal epilithic biofilms (i.e., biofilms on rocks), ignoring biofilms in intertidal soft sediments such as sands and muds (Taylor et al., 2014; Kerfahi et al., 2020). Given the distinct environmental gradients in biofilms, it is expected that the effect of OA on biofilms in soft sediment will be different from that of epilithic biofilms (Marques da Silva et al., 2017). Lastly, few studies have assessed how OA affects the functions of microbes in intertidal benthic biofilms (Kerfahi et al., 2020; Sciutteri et al., 2022). Considering the impacts of OA on bacterial community composition in intertidal epilithic biofilms, it is highly likely that OA will also change the ecosystem services (e.g., primary and secondary production, nutrient cycling) provided by those biofilms (Nelson et al., 2020).
To fill in these research gaps, we undertook a 28-day CO2 enhancement experiment to investigate how OA affected the MPB biomass, community structure and functions of benthic biofilms in soft intertidal sediments. The MPB biomass was estimated by chlorophyll a concentration. The biofilm community structure was analysed by 16S rRNA and 18S rRNA sequencing, focusing on α-diversity, community composition and network structure. The FAPROTAX (Functional Annotation of Prokaryotic Taxa) database was used to predict ecologically relevant functions (e.g., biogeochemical cycles) of prokaryotes in biofilms (Louca et al., 2016). It is hypothesized that OA would increase the MPB biomass (microalgal biomass), bacterial abundance and viral abundance in soft intertidal sediments. The increased MPB biomass would probably be attributable to DIC (dissolved inorganic carbon) addition that can enhance the photosynthesis of microalgae (Marques da Silva et al., 2017). The increased bacterial abundance may result from the enhanced MPB biomass because the algal exudates are the primary food sources for bacteria (Haynes et al., 2007). The increased viral abundance will probably result from the elevated bacterial abundance. It is also hypothesized that OA will alter the MPB and bacterial community composition in sediments because of the species-specific responses to OA (Vargas et al., 2017). It is also hypothesized that OA will alter the ecologically relevant functions (e.g., nutrient cycling) of benthic biofilms because of the changed bacterial community composition.
2 Materials and methods
2.1 Field sampling
The conceptual design of the experimental method, including field sampling, incubations, data measurement and statistical analysis is shown in Figure 1. Field sampling was conducted in April 2022 at two sites in the Qingdao area, China (36°04′N, 120°22′E) (Figure S1). Both sites are located in an area characterized by intense urban and industrial development. Muddy sediments were collected at Licun estuary (site 1), a site that features rich molluscan resources (e.g., oysters and clams) and heavy human impacts (e.g., construction and wastewater discharge). Sandy sediments were collected at Zhanqiao Pier (site 2), an area with little molluscan resources and less human disturbance. Both sites are exposed during low tide and submerged during high tide; sampling was conducted at low tide. At each site, a total of 1.5 kg of surface sediments (top 1cm) was collected with a plastic shovel, then filtered through a sieve (mesh size 2.5 mm) to remove the large mollusks. In addition, three separate sediment samples were collected at each site for chl a analysis, with each sample being recovered from a 16 cm2 area to a depth of 1 cm. Sediments were fully mixed and stored in black plastic bags, and promptly returned to the laboratory for the experiments. Lastly, a total of 30 L overlying seawater was collected from each site and the seawater properties (temperature, salinity, pH and dissolved oxygen) were measured by a Pro Plus handheld multiparameter meter (YSI, USA) (Table S1).
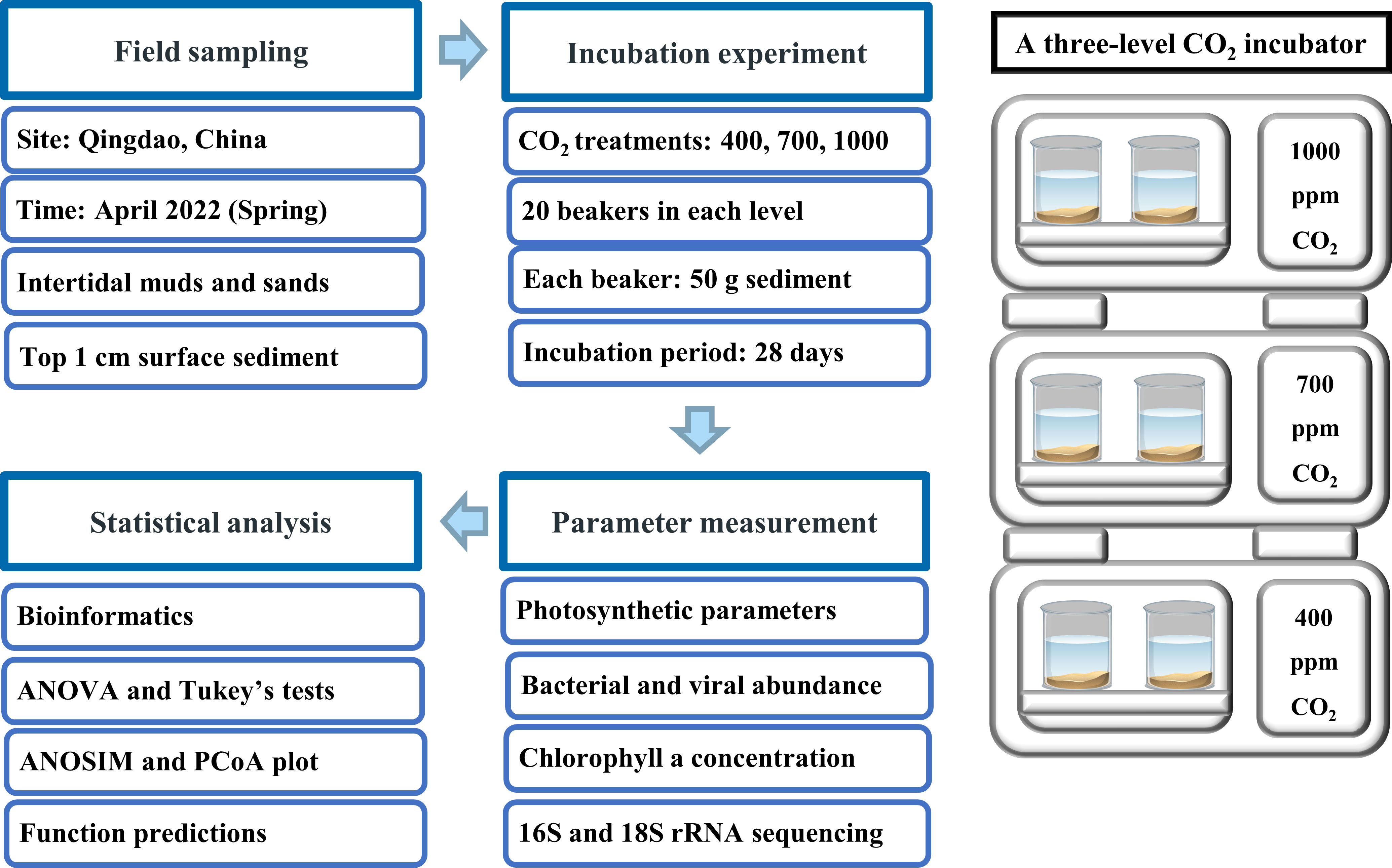
Figure 1 Experimental design of experiment, including field sampling, incubations, parameter measurement and statistical analysis.
2.2 Incubations
To investigate how ocean acidification affects benthic biofilms in intertidal soft sediments, three CO2 treatments were established: 400 ppm (control), 700 ppm (pH reduced by ~ 0.3 units) and 1000 ppm (pH reduced by ~ 0.5 units). These CO2 treatments have been widely used in previous experiments because they reflect the current and future pCO2 levels, according to the IPCC scenario RCP 8.5 (Xu et al., 2018). The three CO2 treatments were established in a three-level CO2 incubator that can automatically adjust the CO2 concentration in the overlying air (HP1000G‐D, Ruihua Instruments, Wuhan, China). For each CO2 treatment, 20 x 500 mL beakers (10 beakers for sands, 10 beakers for muds) were incubated in each level of the CO2 chamber. Each beaker contained ~50 g sediment, 400 mL filtered seawater (filtered by 0.22 um membrane) and 2 mL F/2 medium. F/2 medium was added to ensure adequate nutrient concentration throughout the experiment. Because evaporation caused a drop in water levels, 200 mL of filtered seawater was added to each beaker at day 20 to ensure adequate water was overlying the sediments (seawater had been incubated under relevant CO2 levels for 3 days before adding to beakers). During the incubations there would have been an increase in salinity but this was not measured. The CO2 incubator was set to run on a 12/12 light-dark cycle with an irradiance of 300 μmol photons m−2 s −1 (average sunlight irradiance in April) and 15°C temperature (average coastal seawater temperature in April) throughout the experiment. The incubation experiment lasted 28 days with an extra 3-day acclimation period at the beginning.
2.3 Methodological considerations
Most microbial OA experiments have been based on incubations of less than fourteen days (Hicks et al., 2011; Cartaxana et al., 2015). These have often received criticism for being too short to adequately produce a clear response. However, very long incubations such as that of Torstensson et al. (2015), which ran for 194 days, are likewise inappropriate as they do not capture realistic seasonal environmental change. Here, 28-day incubations were used, a duration selected to try to address both of these concerns.
During these 28-day incubations, the MPB and prokaryote communities were submerged at all times and thus not experiencing natural tidal cycles of immersion and emersion. While this will detract from an ability to predict the future responses of these communities to OA, it still allows for a better understanding of the effects of elevated CO2 concentrations and reduced pH on key metabolic functions.
2.4 Chlorophyll a concentration
Chlorophyll a was measured as a proxy for MPB biomass. Measurements of chlorophyll a concentration were performed on a Turner Designs 10-AU Fluorometer (Sunnyvale, CA, USA) on day 0 and day 28. Before chlorophyll a measurement, 3 x 1 g sediment samples (sediment surface 0 - 1 cm) was collected from three beakers for each treatment. Each sediment sample was then suspended in 10 mL of 90% acetone, followed by a 12 h extraction in the dark at 4°C. The solvent was then decanted off to measure the chlorophyll a concentration and phaeophytin a concentration using acidification methods (Arar and Collins, 1997). The ratio of chlorophyll a to phaeophytin a was calculated as a measure of algal mortality and compromised cell health (Sathish et al., 2020). Lastly, the net growth rates of total MPB in sediments were calculated following the equation: (A28: chl a concentration measured on day 28, A0: chl a concentration measured on day 0).
2.5 Bacterial and viral abundance
Bacterial and viral abundance were counted with a CytoFLEX Platform flow cytometer (Beckman Coulter, USA) on day 0 and day 28. Before counting, 3 x 0.5 g sediment samples (sediment surface 0 - 1 cm) was collected from three beakers for each treatment. Each sediment sample was then treated with tetrasodium pyrophosphate (50 mmol/L), followed by 15 min incubation in the dark at 4°C. The samples then underwent ultrasound treatment (3 x 1 min of ultrasound with 20 s of manual shaking) and centrifugation (800× g, 1 min). The supernatant was collected and diluted 1000× with virus-free seawater. For bacteria enumeration, the diluted supernatant was stained with 1: 1000 dilution SYBR Green 1, with 1h incubation at 37°C in the dark. Supernatants were run for 1 - 2 mins at a high flow rate (∼ 60 µLmin−1) in flow cytometry and bacterial abundance was determined from violet sider scatter vs green fluorescence bivariate scatter plots. For virus enumeration, the diluted supernatant was stained with 1: 1000 dilution SYBR Green 1, with 15 min incubation at 80°C in the dark. Supernatants were run for 1 - 2 mins at a low flow rate (∼ 20 µLmin−1) and virus abundance were determined from violet sider scatter vs green fluorescence bivariate scatter plots. Lastly, the net growth rates of bacteria and viruses in sediments were calculated by the equation: (B28: bacterial or viral abundance measured on day 28, B0: bacterial or viral abundance measured on day 0).
2.6 MPB photosynthetic parameters
Measurements of MPB photosynthetic parameters were performed on a Phyto-PAM II Fluorometer (Walz, Germany) on day 0 and day 28. Before each PAM measurement, 3 x 15 g sediment samples (sediment surface 0 - 1 cm) was collected from three beakers for each treatment. Each sediment sample was mixed with filtered seawater and transferred into a centrifuge tube for a 1h dark-adaptation. The samples were shaken vigorously and then allowed to settle for ~10 s before PAM analysis. The PAM methodology followed McMinn et al. (2005). The Phyto-PAM II uses five measuring light wavelengths (440, 480, 540, 590 and 625 nm) for simultaneous excitation and deconvolution of four algae groups, specifically cyanobacteria, green algae, brown algae (including diatoms) and red algae (Walz, 2022). The different responses were used to measure the phytoplankton community composition, the maximum quantum yield of photosystem II (Fv/Fm) and chl a concentration of each phytoplankton group (Walz, 2019). The chl a concentration of diatoms and cyanobacteria in MPB were measured separately by the PAM fluorometer, providing additional information on how OA affects different components of the MPB community. In addition, rapid light curves (RLC) were constructed in PhytoWin software and described by fitting the model of Platt et al. (1980). α (photosynthetic efficiency) and rETRmax (maximum relative electron transport rate) of each phytoplankton group were calculated from the RLC to assess their photosynthetic activities (Ralph and Gademann, 2005).
2.7 16S and 18S rRNA gene sequencing
16S and 18S rRNA gene sequencing were performed to investigate the effects of OA on the biodiversity, community composition and functions of benthic biofilms. 3 x 10 g sediment samples (sediment surface 0 - 1 cm) was collected on day 28 for each treatment. These samples were then sent to the Majorbio company (Shanghai, China) that conducted polymerase chain reaction (PCR) and sequencing. In brief, for bacteria and the archaea, the V4 region of the 16S rRNA gene was amplified using the PCR primers 515FmodF_806RmodR, and the following PCR conditions: 95°C for 3 min, followed by 27 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 45 s, and a final elongation step at 72°C for 10 min (Walters et al., 2016). For eukaryotes, the V2-V3 regions of the 18rRNA gene were amplified using the PCR primers 82F_516R, and the following PCR conditions: 95°C for 3 min, followed by 32 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 45 s, and a final elongation step at 72°C for 10 min (Amann et al., 1990; López-García et al., 2003). Sequencing of amplified 16S and 18S rRNA genes was performed on an Illumina MiSeq platform.
2.8 Bioinformatics and statistical analysis
Based on 16S rRNA and 18S rRNA sequencing data, the effects of OA on biofilm community structure were analyzed by examining alpha diversity, community composition and network structure. These bioinformatics analyses were performed on the online platform of Majorbio Cloud Platform (www.majorbio.com) which is a one‐stop, comprehensive bioinformatic platform for multiomics analyses (Ren et al., 2022). Alpha diversity (Shannon index) was calculated to determine how OA affected the biodiversity of benthic biofilms. Analysis of similarity (ANOSIM) was performed to determine whether OA altered the bacterial community composition in biofilms. Co-occurrence network analyses were performed to explore how OA affected the co-occurrence patterns of microbial communities in sediments. Using OTU data from gene sequencing, interaction networks were constructed in R software based on Pearson and Spearman correlation values and Bray–Curtis dissimilarity measures. All networks were visualized in Gephi software, and topological features of each network were also calculated in Gephi, including number of network nodes, number of edges, number of positive and negative correlations, average degree, network density, network diameter, network density, modularity, and average clustering coefficient. The details of network analyses referred to Xing et al. (2022). Two functional prediction tools (PICRUST2 and FAPROTAX) were used to analyze the effects of OA on the functional potentials of microbial biofilms. Both tools can predict functional abundances of microbial communities based only on marker gene sequences (e.g., 16S rRNA sequences). PICRUST2 has been widely used to predict functions/biological pathways of both human-related microbes and environmental microbes (Douglas et al., 2020), and FAPROTAX is a specialist in predicting ecological relevant functions of environmental microbes, especially for marine species (Louca et al., 2016). Analysis of variance (ANOVA) tests were performed for the following parameters, including photosynthetic parameters (Fv/Fm, rETRmax, α), chl a/phaeo a, net growth rates (MPB, algae, bacteria, viruses), relative abundance of bacterial taxa and function abundance (Tables S2-S12). Tukey’s test was used post hoc to investigate significant differences between CO2 treatments.
3 Results
3.1 CO2 enhancement and pH reduction
After the 28-day incubation, seawater pH in both muddy and sandy sediments was similar in the 700 ppm CO2 treatment and the 400 ppm CO2 treatment (sandy sediments: pH400 = 8.05 ± 0.02, pH700 = 8.02 ± 0.02; muddy sediments: pH400 = 8.00 ± 0.02, pH700 = 7.97 ± 0.02) (Tukey’s test, sands: p = 0.480, muds: p = 0.502). However, the seawater pH in the 1000 ppm CO2 treatment was lower than the control (sandy sediments: pH1000 = 7.93 ± 0.02; muddy sediments: pH1000 = 7.88 ± 0.02) (Tukey’s test, sands: p < 0.001, muds: p < 0.001). Changes in growth rates, photosynthetic parameters, taxon abundances and functional abundances are summarized in Table 1.
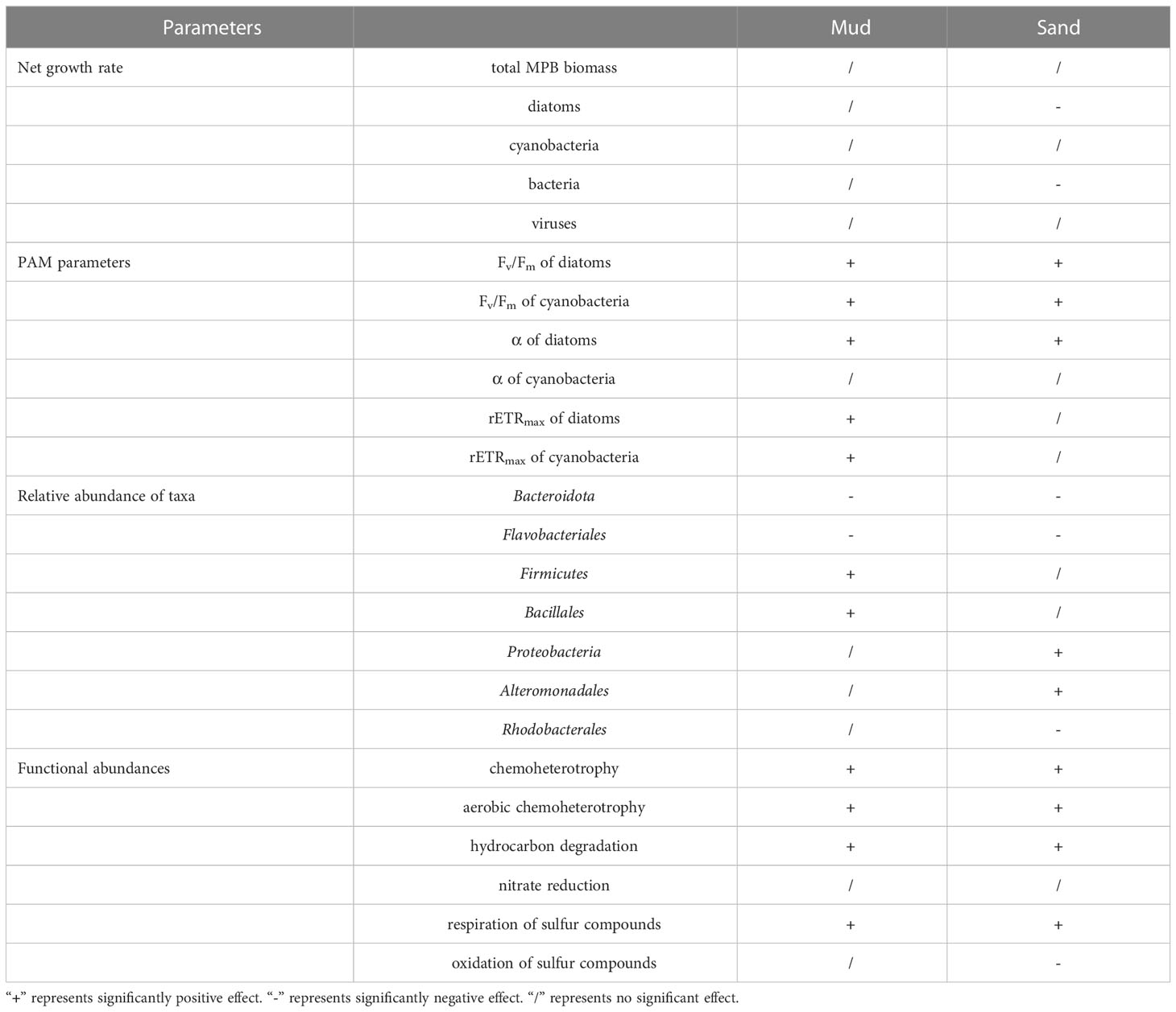
Table 1 Effects of 1000 ppm CO2 treatment on benthic biofilm communities in soft intertidal sediments.
3.2 MPB growth rate
After 28-day incubation, the net growth rate of MPB biomass was negatively affected by OA, but not significantly (ANOVA, sandy sediment: p = 0.197, muddy sediment: p = 0.202; Figure 2). In addition, the net growth rate of cyanobacteria in both muds and sands was not affected by OA, while that of diatoms in sands was negatively affected by the 1000 ppm CO2 treatment (ANOVA, cyanobacteria: Pmud = 0.555, Psand = 0.479; diatoms: Pmud = 0.668, Psand < 0.01; Figure 2). OA did not significantly change the chl a/phaeo a in the sediments, suggesting that the microalgal mortality/cell physiology was not significantly altered by OA (ANOVA, sands: p = 0.860, muds: p = 0.191; Figure S2). Lastly, the 1000 ppm CO2 treatment significantly modified the MPB community composition in sands (few diatoms and more cyanobacteria), but not in muds (ANOVA, sands: p < 0.05, muds: p = 0.705; Figure S3).
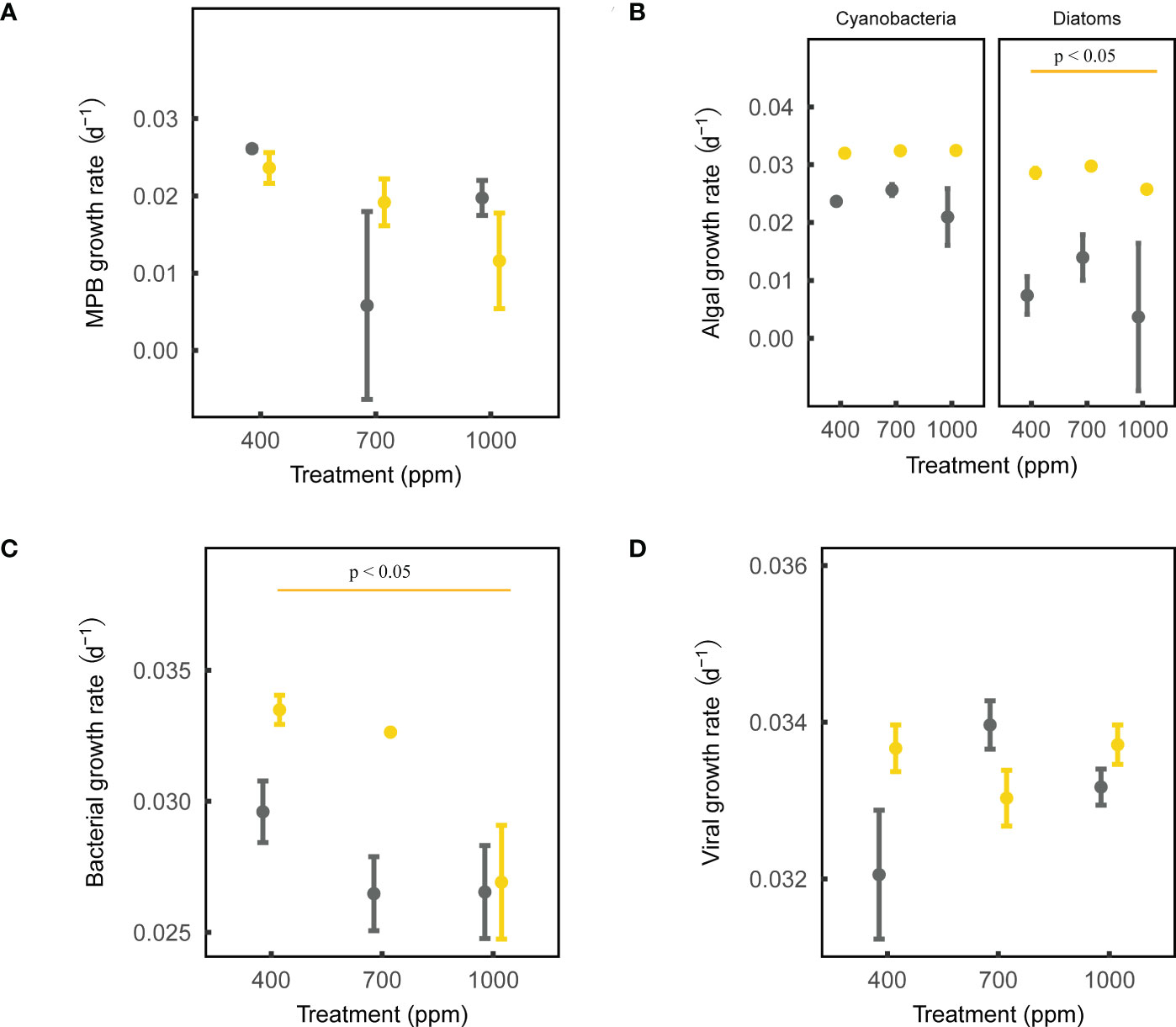
Figure 2 Effects of ocean acidification on the net growth rate parameters for muddy and sandy intertidal sediments. Statistically significant differences were analysed by ANOVA tests and post hoc Tukey’s tests (p < 0.05). Yellow represents sandy sediments, and grey represents muddy sediments. Four parameters are: (A) MPB growth rate, measured by a Phyto-PAM II Fluorometer; (B) algal growth rate, measured by a Turner Designs 10-AU Fluorometer; (C) bacterial growth rate, measured by a CytoFLEX Platform flow cytometer; (D) viral growth rate, measured by a CytoFLEX Platform flow cytometer.
3.3 Bacterial and viral growth rates
After 28-day incubation, the net growth rate of bacteria in sands was negatively affected by the 1000 ppm CO2 treatment, while that in muds was not significantly affected by OA (ANOVA, sands: p < 0.05, muds: p = 0.274; Figure 2). The net growth rate of viruses in both muds and sands was not significantly influenced by OA (ANOVA, sands: p = 0.323, muds: p = 0.239; Figure 2).
3.4 MPB photosynthetic parameters
The Phyto-PAM II uses different emission spectra to differentiate between green (Chlorophyta), blue (Cyanobacteria), brown (diatoms, dinoflagellates, Prymnesiophytes) and red (Rhodophyta, Cryptophyta) microalgae. In this study, the Phyto-PAM measurement showed that the MPB in sediments was dominated by cyanobacteria and brown microalgae. Based on the following 18S rRNA results, the brown microalgae in MPB were dominated by diatoms. Fv/Fm, α and rETRmax of MPB cyanobacteria and diatoms were measured to determine the impacts of OA on MPB photosynthesis.
The maximum quantum yield (Fv/Fm) of both MPB cyanobacteria and diatoms in both muds and sands was positively affected by OA (ANOVA, muds: Pdiatoms < 0.001, Pcyanobacteria < 0.001; sands: Pdiatoms < 0.01, Pcyanobacteria < 0.05; Figure 3). The photosynthetic efficiency (α) of cyanobacteria in both muds and sands was not affected by OA, while that of diatoms was positively affected by OA (ANOVA, cyanobacteria: Pmud = 0.090, Psand = 0.949; diatoms: Pmud < 0.001, Psand < 0.05; Figure 3). The maximum relative electron transport rate (rETRmax) of both MPB cyanobacteria and diatoms in muds was positively influenced by 1000 ppm CO2 enhancement, while the rETRmax values for sands were not affected by OA (ANOVA, muds: Pdiatoms < 0.01, Pcyanobacteria < 0.01; sands: Pdiatoms = 0.346, Pcyanobacteria = 0.109; Figure 3).
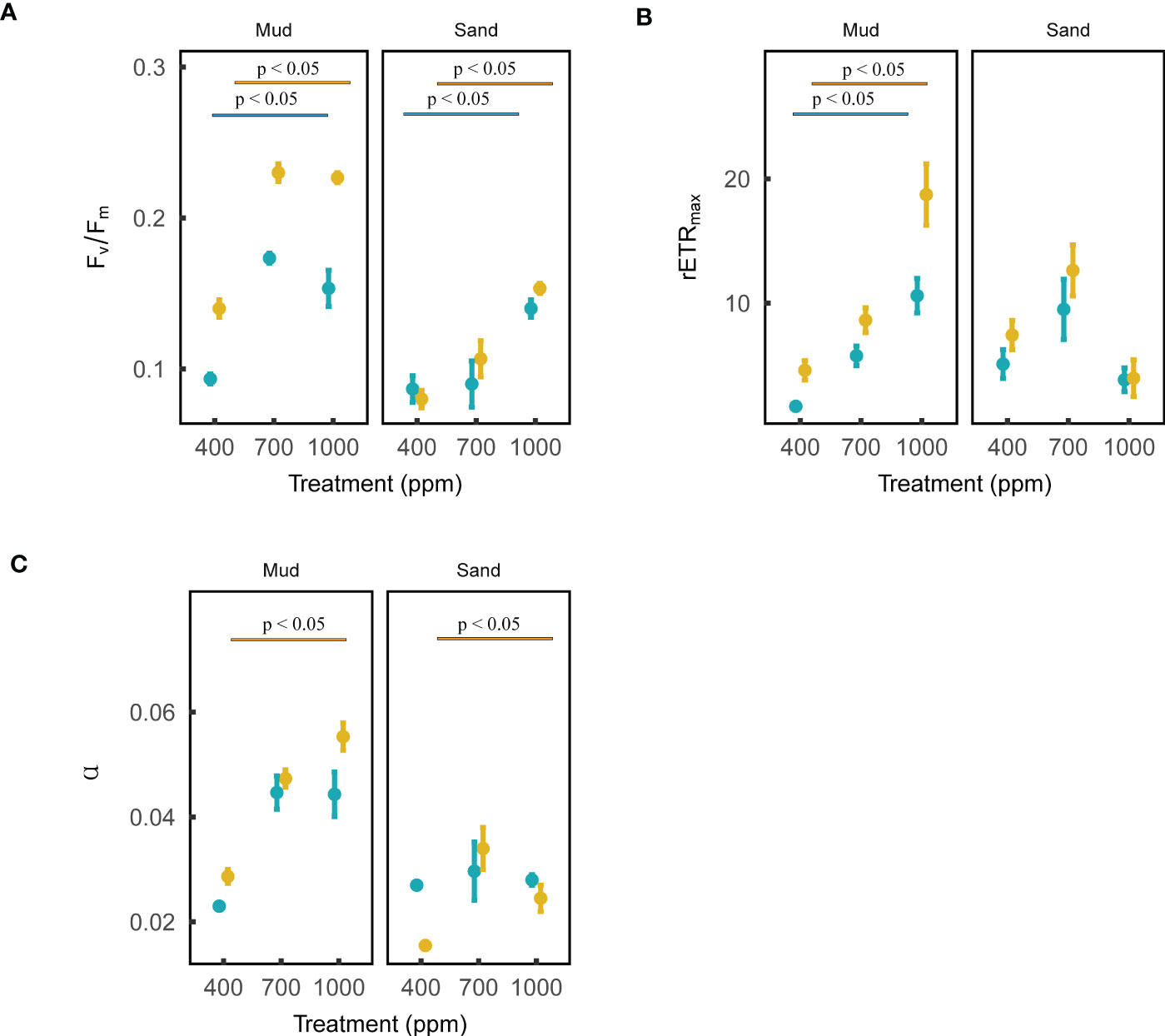
Figure 3 Effects of ocean acidification on the photosynthetic parameters of blue microalgae (i.e., cyanobacteria) and brown microalgae (dominated by diatoms) in muddy and sandy intertidal sediments. Statistically significant differences were analysed by ANOVA tests and post hoc Tukey’s tests. Brown color represents brown microalgae, and blue color represents blue microalgae. Three photosynthetic parameters are: (A) maximum quantum yield (Fv/Fm), (B) relative maximum electron transport rate (rETRmax), (C) photosynthetic efficiency (α). These photosynthetic parameters (mean ± standard error, n = 3) were measured by a Phyto-PAM II Fluorometer.
3.5 Biofilm community structure
Based on 16S rRNA and 18S rRNA sequencing data, the effects of OA on biofilm community structure were analyzed by examining α-diversity, network structure and community composition. It is noted that, the eukaryotic community structure was highly variable among sediment replicates (Figure 4). This is probably because the spatial distribution of eukaryotes (e.g., algae and invertebrates) in sediments is notoriously patchy (Pan et al., 2017; Hope et al., 2019), and this will require additional replications in future 18S sequencing tests to adequately address this variability. Thus, in this part, 16S sequencing data were mainly used to investigate the how OA will affect the bacterial community structure in sediments. The α-diversity of the bacterial community in muds was negatively affected by the 1000 ppm CO2 treatment, while that in sands was positively affected by the 700 ppm CO2 treatment (ANOVA, muds: p < 0.05, sands: p < 0.01; Figure 5). Besides, the bacterial community composition in both sands and muds was significantly altered by OA (ANOSIM, muds: p < 0.05 sands: p < 0.01; Figure 5).
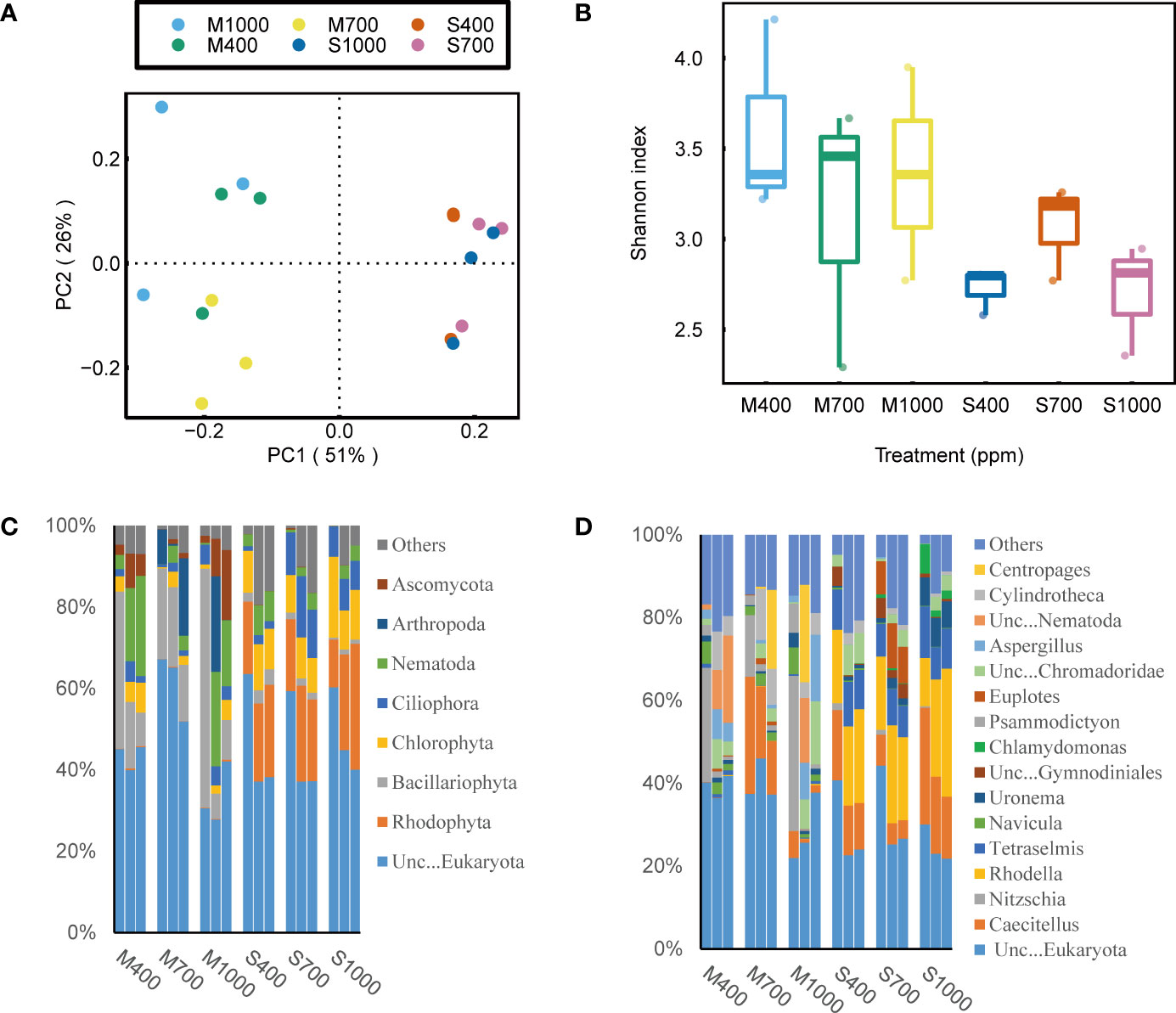
Figure 4 Effects of ocean acidification on the bacterial community in muddy and sandy intertidal sediments. Statistically significant differences were analysed by a ANOSIM test (plot A), ANOVA tests and post hoc Tukey’s tests (plot B, C and D). (A) Principal Coordinate Analysis (PCoA) of bacterial community composition, generated in R software using Bray-Curtis distance on phylum level. (B) Alpha diversity of bacterial community, calculated in Mothur software using OTU data and Shannon index. (C) Relative abundance of bacterial taxa at phylum level, generated in Uparse software based on Silva reference database. (D) Relative abundance of bacterial taxa at order level.
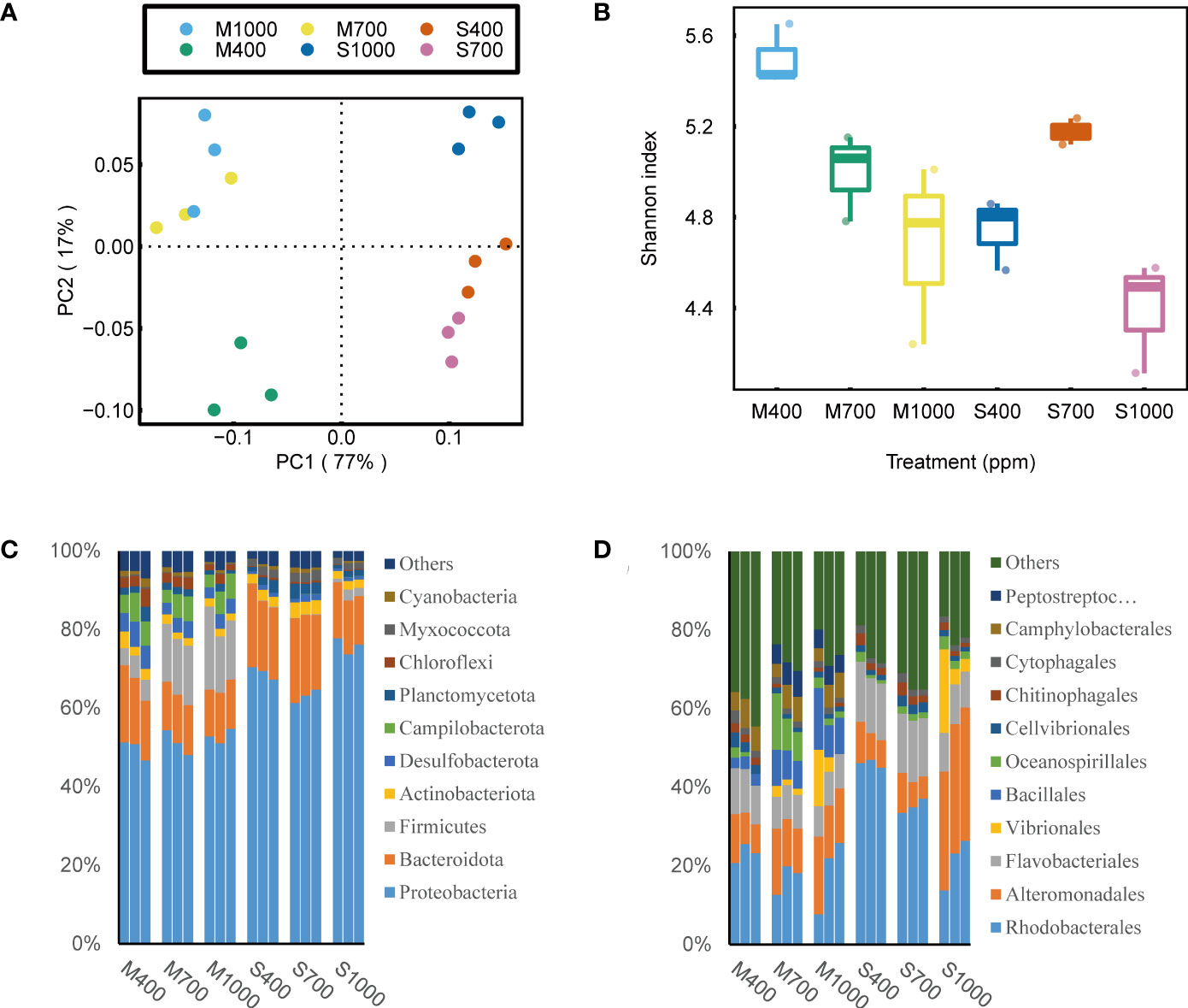
Figure 5 Effects of ocean acidification on the eukaryotic community in muddy and sandy intertidal sediments. Statistically significant differences were analysed by a ANOSIM test (plot A), ANOVA tests and post hoc Tukey’s tests (plot B, C and D). (A) Principal Coordinate Analysis (PCoA) of eukaryotic community composition, generated in R software using Bray–Curtis distance on phylum level. (B) Alpha diversity of eukaryotic community, calculated in Mothur software using OTU data and Shannon index. (C) Relative abundance of eukaryotic taxa at phylum level, generated in Uparse software based on Silva reference database. (D) Relative abundance of eukaryotic taxa at genus level.
Co-occurrence network analyses were performed to explore how OA affected the co-occurrence patterns of microbial communities in sediments. It was found that several topological features of co-occurrence networks were evidently altered by OA, including number of edges (i.e., correlations), average degree, network modularity and density (Table S12). For example, for the biotic networks in sands, OA increased the positive correlations but decreased the network modularity. Network modularity describes the strength of division of a network into modules (Lin et al., 2019). For the biotic networks in muds, OA increased the number of edges, average degree and network density. Average degree is the sum of the degrees (i.e., number of edges) of all its nodes divided by the number of nodes in the network. Network density is defined as the fraction of edges present over all possible edges in the network. Both two parameters describe the connectivity of a network (Xing et al., 2022).
For the bacterial community composition in muds, at the phylum scale, the 1000 ppm CO2 treatment significantly decreased the relative abundance of Bacteroidota but increased the relative abundance of Firmicutes (ANOVA, Bacteroidota: p < 0.01, Firmicutes: p < 0.01; Figure 5). At the order scale, the 1000 ppm CO2 treatment significantly increased the relative abundance of Bacillales and Peptostreptococcales-Tissierellales but decreased the relative abundance of Flavobacteriales (ANOVA, Bacillales: p < 0.05, Peptostreptococcales-Tissierellales: p < 0.001, Flavobacteriales: p < 0.01; Figure 5). For the bacterial community composition in sands, at the phylum scale, the 1000 ppm CO2 treatment significantly decreased the relative abundance of Bacteroidota but increased the relative abundance of Proteobacteria (ANOVA, Bacteroidota: p < 0.01, Proteobacteria: p < 0.001; Figure 5). At the order scale, the 1000 ppm CO2 treatment increased the relative abundance of Alteromonadales but decreased the relative abundance of Flavobacteriales and Rhodobacterales (ANOVA, Alteromonadales: p < 0.001, Flavobacteriales: p < 0.001, Rhodobacterales: p < 0.001; Figure 5).
Despite a large variation in the 18S rRNA sequencing data, it is still worth presenting these data because they provided valuable information on how OA altered the eukaryotic community composition in soft intertidal sediments. It was found that in these sediments, the most abundant eukaryote phyla were Unclassified_d_Eukaryota (includes all unknown eukaryotes and some flagellate), Rhodophyta, Bacillariophyta, Chlorophyta, Ciliophora, Nematoda, Arthropoda and Ascomycota. The dominant eukaryotic microalgal phyla were Rhodophyta, Bacillariophyta and Chlorophyta. However, Rhodophyta and Chlorophyta were not detected by the Phyto-PAM-II, which suggests that these microalgae were dead cells or spores. In addition, 18S rRNA results show that brown microalgae in sediments (seen by Phyto-PAM) were dominated by diatoms (Nitzschia, Cylindrotheca and Navicula) with small contributions from dinoflagellates (Unclassified_o_Gymnodiniales). For eukaryotes in muds, at the phylum scale, Unclassified_d_Eukaryota was positively affected by the 700 ppm CO2 treatment (ANOVA, p < 0.01, Figure 4). The increase in Unclassified_d_Eukaryota was mainly attributed to the genus Caecitellus, a small bacterivorous zooflagellate (Figure 4). For eukaryotes in sands, at the phylum scale, OA increased the relative abundance of Ciliophora (dominated by the genus Uronema, Strombidium and Euplotes) but decreased the relative abundance of Bacillariophyta (ANOVA, Ciliophora: p < 0.001, Bacillariophyta: p < 0.01; Figure 4).
3.6 Functional prediction
Two functional prediction tools (FAPROTAX and PICRUST2) were used to investigate the effects of OA on the functional potentials of microbes in sediments. The FAPROTAX was performed to predict ecologically relevant functions (e.g., biogeochemical cycles) of prokaryotes in soft intertidal sediments (Louca et al., 2016). The top 10 ecologically relevant functions predicted were: chemoheterotrophy, aerobic chemoheterotrophy, hydrocarbon degradation, dark oxidation of sulfur compounds, fermentation, dark sulfite oxidation, dark sulfur oxidation, nitrate reduction, respiration of sulfur compounds and thiosulfate respiration (Figure 6). Here, functions related to C-cycle, N-cycle and S-cycle are presented and discussed. For the C-cycle, the 1000 ppm CO2 treatment significantly increased the chemoheterotrophy, aerobic chemoheterotrophy and hydrocarbon degradation in both muds and sands (ANOVA, chemoheterotrophy: Psand < 0.001, Pmud < 0.001; aerobic chemoheterotrophy: Psand < 0.001, Pmud < 0.001; hydrocarbon degradation: Psand < 0.001, Pmud < 0.01; Figure 6). Fermentation was also higher in the 1000 ppm treatment, although this was not significant (ANOVA, muds: p = 0.065; sands: p = 0.115). For the N-cycle, the 1000 ppm CO2 treatment improved nitrate reduction in both muds and sands (ANOVA, not statistically significant, muds: p = 0.087, sands: p = 0.147; Figure 6). Nitrogen fixation, nitrification, ammonification and other denitrification processes were insignificant in the intertidal soft sediments. For the S-cycle, the respiration of sulfur compounds in both muds and sands increased in the 1000 ppm CO2 treatment (ANOVA, muds: p < 0.001, sands: p < 0.001; Figure 6) but dark oxidation of sulfur compounds in sands decreased (not seen in muds) (ANOVA, sands: p < 0.05, muds: p = 0.822; Figure 6).
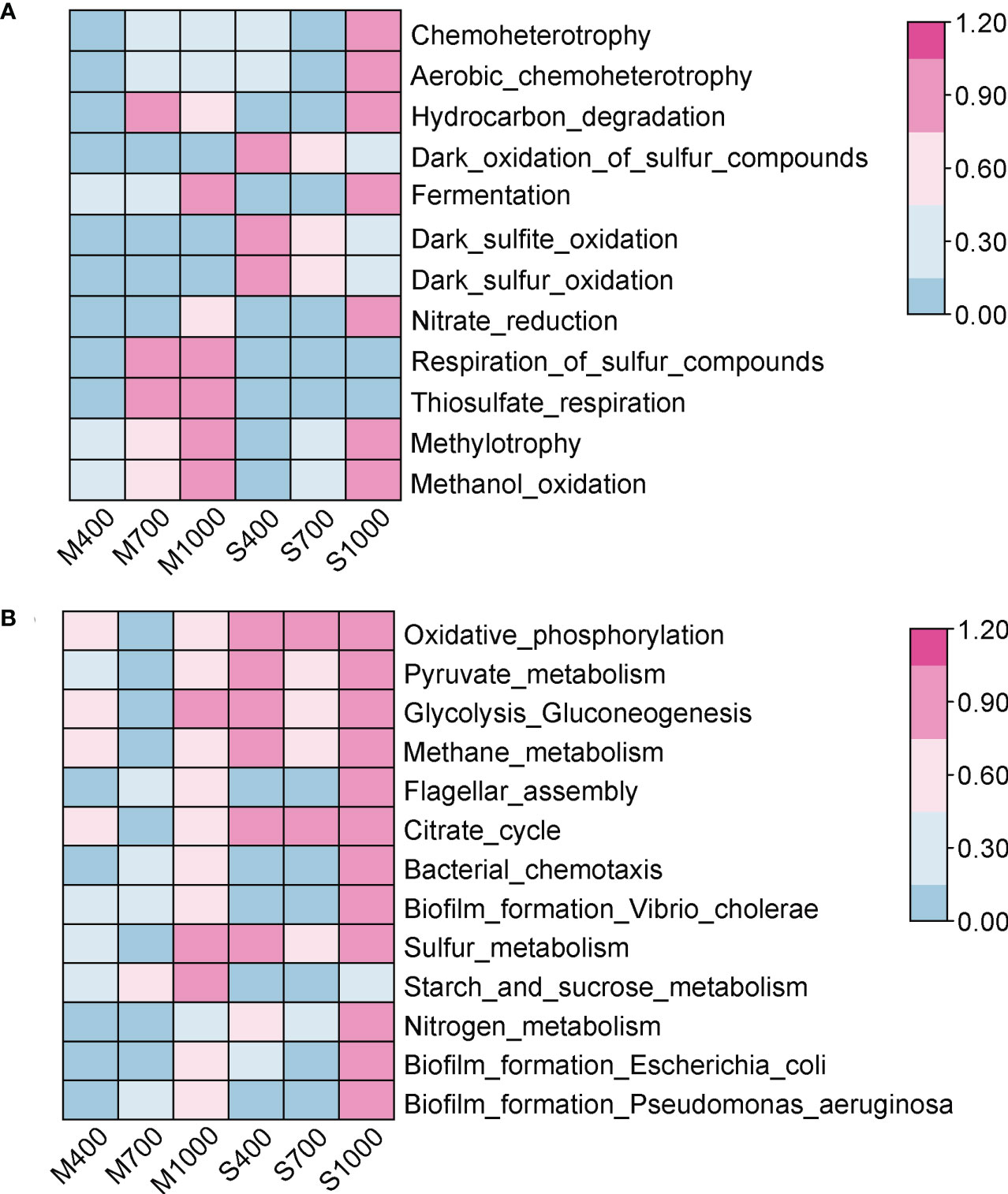
Figure 6 Effects of ocean acidification on the function abundance of prokaryotic community in muddy and sandy intertidal sediments. Statistically significant differences were analysed by ANOVA tests and post hoc Tukey’s tests. Abundance data were standardized to 0 - 1, and heatmaps were generated by TBtool software. (A) The ecologically relevant functions of prokaryotic community, predicted by FAPROTAX. (B) The biological pathways of prokaryotic community, predicted by PICRUSt2 based on KEGG database.
The PICRUSt2 was used to predict the biological pathways of prokaryotes in soft intertidal sediments. Six pathways were selected and analyzed here: cellular respiration, carbohydrate metabolism, nitrogen metabolism, sulfur metabolism, cell motility and biofilm formation (Figure 6). The cellular respiration processes in sediments, including glycolysis, citrate cycle and oxidative phosphorylation, were not significantly affected by OA (ANOVA, glycolysis: Pmud = 0.686, Psand = 0.939; citrate cycle: Pmud = 0.632, Psand = 0.934; oxidative phosphorylation: Pmud = 0.572, Psand = 0.976; Figure 6). The overall carbohydrate metabolism in both sediment types was not significantly altered by CO2 addition (ANOVA, muds: p = 0.479, sands: p = 0.822; Figure 6), while the starch and sucrose metabolism in muds was positively affected by the 1000 ppm CO2 treatment (ANOVA, p < 0.05). Nitrogen metabolism and sulfur metabolism in both sediment types were not significantly affected by OA (ANOVA, nitrogen metabolism: Pmud = 0.509, Psand = 0.133; sulfur metabolism: Pmud = 0.303, Psand = 0.779; Figure 6). Cell motility, including flagellar assembly and bacterial chemotaxis, in both sediment types, was positively influenced in the 1000 ppm CO2 treatment (ANOVA, flagellar assembly: Pmud < 0.05, Psand < 0.01; bacterial chemotaxis: Pmud < 0.01, Psand < 0.001; Figure 6). Biofilm formation by Vibrio cholerae, Escherichia coli and Pseudomonas aeruginosa was also positively affected in the 1000 ppm CO2 treatment (ANOVA, Vibrio: Pmud < 0.05, Psand < 0.001; Escherichia: Pmud < 0.05, Psand < 0.05; Pseudomonas: Pmud < 0.05, Psand < 0.01; Figure 6).
4 Discussion
4.1 Growth rates of MPB, bacteria, viruses
This study carefully investigated the effects of OA on the net growth rates of MPB biomass, bacteria and viruses in benthic biofilms. It was found that the effects of OA were only visible when the pCO2 level reached a concentration of 1000 ppm, which suggests that seawater pH reduction (only happened at 1000 ppm treatment) is a key driver for the effects of OA on benthic biofilm communities. In addition, the impacts of OA were more evident in sandy sediments than muddy sediments. This is probably because sands are more porous and so experience a greater exchange of water with the overlying low pH seawater (Cook and Roy, 2006). The effects of OA on diatoms were also more apparent than on cyanobacteria, confirming a previously noted strong pH-tolerance by cyanobacteria in intertidal sediments (Johnson et al., 2015).
Results produced here show that OA had a minor negative influence on the net growth rate of MPB biomass, which negates our hypothesis that OA would have a positive effect on MPB biomass. It is likely that the negative effect of OA on MPB biomass seen here mostly resulted from the reduced pH of seawater. The physiological effects of OA on microalgae have been well studied (Beardall et al., 2009; Gao et al., 2018). Here, a reduction in seawater pH likely increased the energy expenditure of microalgae because algal cells need to up-regulate their proton pumps to maintain intracellular pH homeostasis (Black et al., 2019). In addition, OA can negatively affect the enzyme activities and gene expressions in microalgae, thereby influencing growth rates (Yu and Chen, 2019). The minimal change in the chl a/phaeo a ratio further suggests that OA has negligible impact on microalgal mortality/cell physiology, caused by grazing (Sathish et al., 2020). Grazing was likely a significant factor in the decline of biomass in all treatments as the sediment was initially filtered through a 2.5 mm mesh, allowing many small invertebrates and protistan grazers to remain. Lastly, the PAM fluorometer results showed that OA improved the photosynthetic performance (α and rETRmax) of diatoms, most likely via DIC addition, potentially boosting the MPB biomass. However, it is suggested that the negative aspects of OA (decline in pH) exceeded the positive aspects (increase in DIC), resulting in an overall negative impact on MPB biomass.
The effect of OA on MPB biomass was also studied by Cartaxana et al. (2015). They reported that OA enhanced MPB biomass of intertidal muddy sediments, a result at variance with those seen here. There are several possible reasons for these inconsistent results, such as different MPB community composition and different experimental conditions. In the present study, the MPB diatom community was dominated by Nitzschia (from 18S results); in the Cartaxana’s study, MPB diatom community was dominated by Navicula. Based on previous findings, compared to Navicula, Nitzschia is more likely to be negatively affected by OA (Johnson et al., 2013; Torstensson et al., 2015). In addition, the experimental conditions (e.g., temperature, nutrient levels, experimental duration) were quite different between studies. In particular, this experiment lasted 28 days, 11 days longer than the Cartaxana et al. (2015) study. Generally, it has been found that experiments with a longer duration provide a more accurate estimate of the impacts of OA. Moreover, long-term CO2 enhancement (months to years) could induce adaptive evolution (genetic changes) of microalgae, thereby altering the MPB growth rate and biomass (Thomas et al., 2022).
This study also showed that OA caused a decrease in the net growth rate of bacteria in sediments, which again, conflicted with our hypothesis that OA would increase the bacterial abundance. Physiological responses of bacteria to OA have been described by Hartmann et al. (2016). But this study is the first to investigate the impacts of OA on bacterial abundance in intertidal soft sediments. Here, two pathways are suggested to explain how OA caused a decrease in the net growth rate of bacteria: decreasing the food supply for bacteria and increasing the grazing pressure by protozoa. Firstly, OA reduced the net growth rate of diatoms in sands, resulting in a lower organic matter release from diatoms. This organic matter, which includes substances such as extracellular polymeric substances (EPS), are a critical food source for neighboring bacterial communities (Haynes et al., 2007). OA also increased the relative abundance of Caecitellus (a bacterivorous nanoflagellate), and bacterivorous ciliates such as Uronema, Strombidium and Euplotes in the eukaryotic community, suggesting a higher grazing pressure on bacteria under elevated CO2 conditions.
This study also found that OA had a negligible impact on the net growth rate of viruses in sediments, which differed from our hypothesis that OA would increase the viral abundance. Again, this study was the first to investigate the impacts of OA on viral abundance in intertidal soft sediments. However, at this stage, it is unknown how OA influenced the abundance and virulence of algal viruses and bacteriophages respectively, because this study only measured the total viral abundance. It is also unclear why viral abundance was not significantly affected by OA, asking for future research in this area. Previous studies have provided some important insights into the impacts of OA on virus-host interactions. For example, Phaeocystis globosa, a widespread marine phytoplankton, has been shown to suffer a more severe viral infection under elevated CO2 conditions, resulting in a decreased photosynthetic rate and increased respiration rate. In addition, OA might promote lysogenic (i.e., coexist with cells) lifestyles of viruses, thereby influencing the activity and abundance of prokaryotes (Vaqué et al., 2019; Tangherlini et al., 2021).
4.2 MPB photosynthetic parameters
The PAM fluorometer results showed that OA could increase the photosynthetic performance of microalgae in MPB, particularly for diatoms in muds. This study was also the first to assess the impacts of OA on the photosynthetic activity of different microalgae groups within MPB (i.e., cyanobacteria and diatoms), with previous studies only investigating the effects of OA on the whole MPB community (Cartaxana et al., 2015; Vieira et al., 2016). Three photosynthetic parameters derived from rapid light curves (RLC) using the PAM fluorometer were analyzed, including: Fv/Fm, α and rETRmax. Fv/Fm refers to the maximum capacity of the photosystem II to convert light energy into charged states and transfer this into the photochemical pathway. This is often used as a sensitive indicator of photosynthetic stress (McMinn et al., 2005). This study found that Fv/Fm of both cyanobacteria and diatoms in both muds and sands was positively affected by OA, which suggested that OA did not damage the photosystem II of microalgae in MPB. Previous studies have also found that elevated pCO2 conditions did not affect the Fv/Fm of MPB in muddy sediments or sea ice (Cartaxana et al., 2015; McMinn et al., 2017; Black et al., 2019). This is probably because microalgae can up-regulate their proton pumps to maintain intracellular pH homeostasis, thereby maintaining the health of their photosystem II in low pH waters (Cornwall et al., 2014). However, the upregulation of proton pumps causes an extra energetic cost to microalgae, which may impair their growth and other biochemical processes (Yu and Chen, 2019).
In addition to Fv/Fm, this study also investigated the impacts of OA on α and rETRmax. Photosynthetic efficiency (α) is related to the fraction of light energy converted into chemical energy through photosynthesis. rETRmax is often used as a proxy for maximum photosynthetic capacity (Ralph and Gademann, 2005). Results presented here show that OA has a positive effect on α and rETRmax of diatoms, particularly in muds. It is possible that sediment photosynthesis is carbon limited and the addition of CO2 results in an increased photosynthetic rate (Marques da Silva et al., 2017). An alternative explanation for the increased α and rETRmax values is that ocean acidification will first enhance the dark reactions (i.e., Calvin cycle) of diatoms by releasing them from carbon limitation in sediments. The diatoms will then modify their PSII physiology to keep their light reaction and dark reaction in balance, thereby resulting in increased α and rETRmax values. The results presented here show that OA also has a negligible effect on α and rETRmax of cyanobacteria. This is likely to be because cyanobacteria have a very efficient CCM (carbon concentrating mechanism), which is an effective strategy to maximize the efficiency of inorganic carbon uptake and CO2 fixation in low CO2 environments (Marques da Silva et al., 2017).
4.3 Biofilm community structure
This study showed that OA can alter the MPB and bacterial community structures in intertidal soft sediments, consistent with our initial hypothesis. It was found that OA significantly modified the MPB community composition in sands, resulting in less diatoms but more cyanobacteria. Compared to cyanobacteria, diatoms are more nutritious and contain high levels of polyunsaturated fatty acids (Brett and Müller-Navarra, 1997). Thus, this compositional change in MPB will likely impact the nutritional value of this primary food source, and therefore influence the long-term survival and growth of larval invertebrates, finally resulting in significant bottom-up cascading effects in coastal marine ecosystems (Vargas et al., 2013). In addition, an alteration in MPB composition will affect the associated bacterial communities via modified algae-bacteria interactions (Haynes et al., 2007). This compositional shift will decrease diatom-mediated EPS production, negatively influencing the settlement of invertebrate larvae, such as the polychaete Hydroides elegans (Lam et al., 2005).
In addition to MPB community composition, OA also significantly altered the bacterial community structure. The effects of OA on the bacterial community structure were analyzed from three aspects: biodiversity, network structure and community composition. The biodiversity results showed that OA negatively affected the alpha diversity of the bacterial community in muds. This loss of biodiversity may compromise the resilience of ecosystem functions (e.g., nutrient cycling and provision of settlement cues) of coastal marine sediments (Hall-Spencer and Harvey, 2019). Previous studies have also investigated the effect of OA on the intertidal epilithic bacteria diversity, but the results have been inconsistent. OA has been found to have a negative effect (Taylor et al., 2014) or positive effect (Kerfahi et al., 2020) on the intertidal epilithic bacteria diversity. This divergence is possibly due to the different experimental designs (e.g., sampling area, experimental conditions). Thus, it is difficult to make generalizations on the effect of OA on biofilm bacterial diversity.
Apart from biodiversity, OA also changed the biotic network structures in intertidal soft sediments, potentially influencing their linked ecosystem functions (Yuan et al., 2021). For the biotic networks in sands, OA increased the positive correlations (i.e., inter-specific cooperations) but decreased the network modularity (i.e., the strength of division of a network into modules). The increased positive correlations in networks suggests that organisms tended to develop more inter-specific cooperations to mitigate the negative influences of OA (e.g., low pH and less food source). The decreased network modularity suggests that networks had fewer complex correlations within modules, implying a decreased stability of networks and changed ecosystem functioning (Lin et al., 2019). For the biotic networks in muds, OA increased the number of edges (i.e., interactions), average degree and network density, suggesting a more connected biotic network in acidifying sediments. This increased connectivity of network is regarded as compensation for the decreased biodiversity under high CO2 conditions, helping to maintain the essential ecosystem functions (Xing et al., 2022).
The bacterial community composition in intertidal soft sediments was also significantly altered by OA, consistent with previous findings for bacterial biofilms on intertidal rocks (Taylor et al., 2014; Kerfahi et al., 2020). This shift in bacterial community composition may alter the ecological services provided by biofilms (Nelson et al., 2020). For example, changes in bacterial biofilm composition could modify nutrient cycling and sediment oxygenation, thereby affecting the local micro-environment and resident benthic organisms (discussed more in the section “functional prediction”). In addition, a compositional shift may alter its production and release of chemical cues, such as signaling molecules and secondary metabolites, influencing the settlement of invertebrate larvae and macroalgal propagules (Das and Mangwani, 2015). A change in the relative abundance of the genus Pseudoalteromonas for example, might affect the settlement of coral larvae through altering the production of the chemical cue tetrabromopyrrole (TBP) (Sneed et al., 2014).
The effect of OA on the relative abundance of bacterial taxa in intertidal biofilms has been studied previously but the results have been quite variable. This study showed that for bacterial community in muds, OA decreased the relative abundance of Bacteroidota but increased that of Firmicutes; while in sands, OA decreased the relative abundance of Bacteroidota but increased that of Proteobacteria. However, Taylor et al. (2014) observed that within biofilms on intertidal rocks, the relative abundance of phylum Bacteroidota, the class Gammaproteobacteria and Alphaproteobacteria increased while the order Flavobacteriales decreased as CO2 levels increased. Kerfahi et al. (2020) reported that within biofilms on intertidal rocks, the relative abundance of phylum Bacteroidota, the order Alteromonadales decreased while Rhodobacterales remained unchanged as CO2 levels increased. Overall, it is difficult to determine which bacterial taxa in benthic biofilm will benefit from OA and these divergent results can probably be attributed to the use of different substrate (i.e., soft sediments vs. rocks) or different experimental conditions (i.e., aquarium-based experiment vs. natural CO2 vents).
So far, it has been largely unknown how OA altered the bacterial community in intertidal biofilms (Taylor et al., 2014; Kerfahi et al., 2020). Here, this study proposed several mechanisms to explain this compositional shift, including species-specific pH sensitivity, altered diatom-bacteria interactions and modified bacteria-bacteria interactions. Firstly, some bacterial species are more tolerant to a pH reduction than others, giving them a competitive advantage living in acidified seawater. For example, Su et al. (2021a) found that the order Bacillales (in phylum Firmicutes) in estuarine sediments was tolerant to low pH conditions. In this study, the order Bacillales was also positively affected by the 1000 ppm CO2 treatment, which confirmed the finding in Su et al. (2021a) study. Secondly, some bacteria (e.g., Flavobacteriales and Rhodobacterales) have strong positive associations with diatoms (Buchan et al., 2014; Isaac et al., 2021). Thus, in this study, a reduction in diatom biomass in sandy sediments could negatively influence the abundance of Flavobacteriales and Rhodobacterales in benthic biofilms. Thirdly, OA may alter the bacteria-bacteria interactions (e.g., cooperation, competition) in the biofilms, thus modifying their community compositions (Rendueles and Ghigo, 2012). For example, this study showed OA decreased the relative abundance of Flavobacteriales that can degrade many types of complex organic matter and make labile compounds available to other bacterial species (Williams et al., 2013), but increased the relative abundance of Bacillales that can produce antimicrobial compounds to kill competitors within biofilms (Yan et al., 2003).
4.4 Functional prediction
This study performed FAPROTAX and PICRUST2 to predict the functional potentials of prokaryotes in intertidal soft sediments. However, this method only assessed the potential for these microbial processes, without determining rate measurements. To obtain a better understanding of how OA affects these microbial processes in intertidal soft sediments, rate measurements (i.e., quantitative data) are required in future studies. The FAPROTAX results predicted that OA would significantly alter the ecologically relevant functions of prokaryotes in intertidal soft sediments. The effects of OA were only apparent when pCO2 level reached 1000 ppm concentration, suggesting a resistance of ecosystem functions (e.g., nutrient cycling) in intertidal soft sediments responding to elevated CO2 conditions. This study mostly focused on the functions associated with biogeochemical cycles, such as C-cycle, N-cycle and S-cycle. For the C-cycle, the FAPROTAX results predicted that OA would increase chemoheterotrophy, aerobic chemoheterotrophy and hydrocarbon degradation in soft intertidal sediments, suggesting that an increased microbial respiration rate would occur. However, it is still unknown how OA improved microbial respiration in these sediments. There have been several studies investigating the effects of OA on the respiration rate or energy metabolism of microbes in marine sediments or biofilms. Su et al. (2021a) found that OA would decrease the basal microbial respiration rate in submerged estuarine sediments, while Kerfahi et al. (2020) reported that OA could increase the energy metabolism of microbes in intertidal epilithic biofilms. These diverse results can probably be attributed to the use of different substrates (i.e., intertidal sediments vs submerged sediments) or different experimental conditions (i.e., laboratory experiments vs. natural CO2 vents). In the present study, the FAPRTAX results suggest that OA could increase the microbial respiration rate, potentially creating more hypoxic zones in sediments, particularly at night. The enhanced microbial respiration in coastal sediments will possibly weaken the carbon sink capacity of marginal seas (Tao et al., 2016). The increased hypoxic zones in sediments may result in a degradation of the benthic community by reducing the biodiversity of the benthos (Hope et al., 2019).
For the N-cycle, the FAPROTAX results predicted that OA would increase nitrate reduction in soft intertidal sediments, although not significantly so. Previous studies also found that OA can enhance the bacterial denitrification in estuarine sediments (Su et al., 2021b), but not change the nitrogen metabolism in microbial biofilms on intertidal or submerged rocks (Kerfahi et al., 2020; Sciutteri et al., 2022). These divergent findings mainly resulted from different sampling areas (i.e., sediments vs. rocks). In the present study, OA is likely to enhance nitrate reduction in sediments by stimulating denitrification and/or dissimilatory nitrate reduction to ammonium (DNRA). Firstly, OA might boost the growth of N2O-producing bacterial denitrifies, thereby promoting the denitrification in sediments (i.e., NO3- to N2O) (Su et al., 2021b). Besides, as discussed above, under elevated pCO2 conditions, our incubated sediments possibly had more hypoxic zones and contained more sulfide, which would have benefited the DNRA, a critical nitrate reduction pathway in sediments (Papaspyrou et al., 2014). This increased nitrate reduction activity in sediments might help remove excess nitrogen and control eutrophication in coastal ecosystems (Herbert, 1999), but it could also reduce the amount of nitrogen available to primary producers in low nutrient systems (Sundbäck and Miles, 2002). Furthermore, the enhanced nitrate reduction in sediments may stimulate the emission of N2O (a potent greenhouse gas, 298 times more powerful than CO2), potentially accelerating global climate change (Su et al., 2021b).
For the S-cycle, the FAPROTAX results predicted that OA could increase the respiration of sulfur compounds but decrease the dark oxidation of sulfur compounds in soft intertidal sediments. Previous studies also found that OA would increase sulfur metabolism in microbial biofilms on intertidal rocks (Kerfahi et al., 2020), and alter the taxonomic composition of S cycle-related microbes in coastal sediments (Hassenrück et al., 2016). However, those studies solely used 16S analysis to predict the sulfur-related activates of microbes, without any quantitative data (i.e., rate measurements) to back up these predictions. Thus, more rate measurements (e.g., sulfate reduction rates) of microbes are recommended in future OA studies. In the present study, the increased sulfur respiration (mostly anaerobic process) and decreased sulfur oxidation (mostly aerobic process) were mainly due to the modification of physicochemical properties within the sediments, i.e., the presence of more hypoxic zones (Fuseler et al., 1996; Hedderich et al., 1998). An increased respiration of sulfur compounds (SO4- or S2O32- → H2S) with decreased oxidation of sulfur compounds (HS- or H2S → S0 or SO4-) under higher CO2 conditions will result in an accumulation of H2S in intertidal soft sediments. H2S has many cytotoxic effects, for example, impairing the ATP production in mitochondrial respiratory chain, and thus the accumulated H2S in sediments will heavily influence the survival and reproduction of resident benthic organisms (Cooper and Brown, 2008).
Interestingly, unlike the FAPROTAX results, the PICRUST2 results suggest that OA would not affect these biological pathways (e.g., cellular respiration, nitrogen metabolism, sulfur metabolism) of prokaryotes in intertidal soft sediments. These conflicting findings can mostly be attributed to the different characteristics of the two prediction tools, i.e., FAPROTAX annotates functions based on published metabolic and ecological functions, while PICRUST2 predicts metagenomic functions based on an arbitrary OTU/ASV table (Liu et al., 2020; Yang et al., 2022). Based on previous research, FAPROTAX works especially well in predicting ecologically relevant functions (e.g., biogeochemical cycles) of marine microbes (Louca et al., 2016; Sansupa et al., 2021). Thus, it is concluded that OA can alter the microbial functional potentials in biogeochemical cycles in intertidal soft sediments. More studies are needed to determine why these two prediction tools produced divergent results.
The PICRUST2 results further showed that OA increased biofilm formation by Vibrio cholerae, Escherichia coli and Pseudomonas aeruginosa. This is because OA favored the growth of biofilm-forming species, thereby boosting biofilm formation. These biofilms provide bacteria with more protection from environmental stresses, such as pH reduction, protozoa predation and viral infection (Teschler et al., 2015). In addition, increased bacterial biofilm formation is likely to promote the settlement of some invertebrate larvae such as Bugula stolonifera (bryozoans) and Hydroides elegans (polychaete), but block the settlement of other invertebrates, such as Balanus amphitrite (barnacle) (Maki et al., 1988; Lau et al., 2005). The PICRUST2 results also showed that OA increased cell motility in sediments, including flagellar assembly and bacterial chemotaxis. It is thus suggested that OA is likely to select motile bacteria for the following three reasons. Firstly, motile bacteria are able to migrate to neutral locations in sediments, escaping the negative impacts of low pH (Tohidifar et al., 2020). Secondly, under elevated CO2 conditions, microalgae in sediments (e.g., diatoms) might migrate more frequently to avoid exposure to low pH porewater and motile bacteria would follow these microalgae to acquire their organic-rich exudates. Finally, OA favored some aggressive bacteria, such as Bacillus, that are motile and can produce antimicrobial compounds (Yan et al., 2003). These bacteria can easily kill their neighboring non-motile competitors, while the motile counterparts would have a higher survival rate through movement or chemotaxis.
5 Conclusion
This study provides important insights into the effects of OA on benthic biofilms (MPB and heterotrophic bacteria) in soft intertidal sediments. It was found that the 700 ppm CO2 treatment did not significantly affect the benthic biofilms in intertidal soft sediments, but the 1000 ppm CO2 treatment significantly altered the community composition and potential functions of biofilms in sediments (especially in sandy sediments). For the MPB community in sandy sediments, cyanobacteria became more abundant at 1000 ppm CO2 treatment, which potentially alters the nutritional value of MPB and thus influences the growth and development of larval invertebrates. For the bacterial community in biofilms, OA increased the relative abundance of Alteromonadales and Bacillales but decreased the relative abundance of Rhodobacterales and Flavobacteriales, potentially causing an alteration in biogeochemical cycling in coastal sediments. The FAPROTAX analysis predicted that OA could enhance the potential of chemoheterotrophic activity, nitrate reduction and sulfur respiration in sediments, likely resulting in a more stressful environment (hypoxic and enriched H2S) for most benthic organisms. Overall, this paper suggests that benthic biofilms in intertidal sandy sediments are likely to change significantly near the end of the century if anthropogenic CO2 emissions unmitigated, with profound implications on local ecosystems and biogeochemical cycling. Nevertheless, this study represented a short-term period of rapid change in OA and it is possible that over a longer time period the benthic biofilms could acclimate or evolve to cope with mild OA (i.e., a more realistic condition).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
AM, YL and MW contributed to conception and design of the study. CZ and HY conducted the field sampling. CZ, HY, XC and YJ measured the relevant parameters in the experiment. CZ performed the statistical analysis and wrote the draft of the manuscript. AM, YL, MW and CZ contributed to manuscript revision, read, and approved the submitted version. All authors contributed to the article and approved the submitted version
Funding
This work was supported by the Laoshan Laboratory (No. LSKJ202203201), National Key Research and Development Program of China (2022YFC2807500), Natural Science Foundation of China (No. 41976117, 42120104006, 42176111 and 42188102), and the Fundamental Research Funds for the Central Universities (202172002, 201812002, 202072001 and AM).
Acknowledgments
We thank Prof. Naihao Ye and Dr. Dong Xu from Yellow Sea Fisheries Research Institute for their assistance on the CO2 incubator operation. We thank our colleges from Ocean University of China for their great help: Yue Dong for seawater filtration, Tong Jiang for flow cytometric analyses, Yan Zhang for mathematics help, Ziyue Wang and Hongbing Shao for technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1117826/full#supplementary-material
References
Amann R. J., Binder B. L., Chisholm S. W., Devereux R., Stahl D. A. (1990). Combination of 16S rRNA targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990
Arar E. J., Collins G. B. (1997). Method 445.0: In vitro determination of chlorophyll a and pheophytin a in marine and freshwater algae by fluorescence (Washington DC: US Environmental Protection Agency).
Beardall J., Stojkovic S., Larsen S. (2009). Living in a high CO2 world: impacts of global climate change on marine phytoplankton. Plant Ecol. Diversity 2, 191–205. doi: 10.1080/17550870903271363
Black J., Stark J., Johnstone G., McMinn A., Boyd P., McKinlay J., et al. (2019). In-situ behavioral and physiological responses of Antarctic microphytobenthos to ocean acidification. Sci. Rep. 9, 1890. doi: 10.1038/s41598-018-36233-2
Brett M. T., Müller-Navarra D. C. (1997). The role of highly unsaturated fatty acids in aquatic foodweb processes. Freshw. Biol. 38, 483–499. doi: 10.1046/j.1365-2427.1997.00220.x
Buchan A., LeCleir G., Gulvik C., González J. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Cartaxana P., Vieira S., Ribeiro L., Rocha R. J., Cruz S., Calado R., et al. (2015). Effects of elevated temperature and CO2 on intertidal microphytobenthos. BMC Ecol. 15, 10. doi: 10.1186/s12898-015-0043-y
Cook P. L. M., Roy H. (2006). Advective relief of CO2 limitation in microphytobenthos in highly productive sandy sediments. Limnol. Oceanogr. 51, 1594–1601. doi: 10.4319/lo.2006.51.4.1594
Cooper C. E., Brown G. C. (2008). The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 40, 533–539. doi: 10.1007/s10863-008-9166-6
Cornwall C. E., Boyd P. W., McGraw C. M., Hepburn C. D., Pilditch C. A. (2014). Diffusion boundary layers ameliorate the negative effects of ocean acidification on the temperate coralline macroalga Arthrocardia corymbosa. PLoS. One 9, e97235. doi: 10.1371/journal.pone.0097235
Das S., Mangwani N. (2015). Ocean acidification and marine microorganisms: responses and consequences. Oceanologia 57, 349–361. doi: 10.1016/j.oceano.2015.07.003
De Carvalho C. C. (2018). Marine biofilms: a successful microbial strategy with economic implications. Front. Mar. Sci. 5, 126. doi: 10.3389/fmars.2018.00126
Deppeler S., Petrou K., Schulz K. G., Westwood K., Pearce I., McKinley J., et al. (2018). Ocean acidification of a coastal Antarctic marine microbial community reveals a critical threshold for CO2 tolerance in phytoplankton productivity. Biogeosciences 15, 209–231. doi: 10.5194/bg-15-209-2018
Doney S., Busch D., Cooley S., Kroeker K. (2020). The impacts of ocean acidification on marine ecosystems and reliant human communities. Annu. Rev. Environ. Resour. 45, 83–112. doi: 10.1146/annurev-environ-012320-083019
Douglas G., Maffei V., Zaneveld J., Yurgel S., Brown J., Taylor C., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Fuseler K., Krekeler D., Sydow U., Cypionka H. (1996). A common pathway of sulfide oxidation by sulfate-reducing bacteria. FEMS Microbiol. Lett. 144 (2-3), 129–134. doi: 10.1111/j.1574-6968.1996.tb08518.x
Gao K. S., Beardall J., Hader D. P., Hall-Spencero J. M., Jason M., Gao G., et al. (2018). Ffects of ocean acidification on marine photosynthetic organisms under the concurrent influences of warming, UV Radiation, And deoxygenation. Front. Mar. Sci. 6, 322. doi: 10.3389/fmars.2019.00322
Hall-Spencer J. M., Harvey B. P. (2019). Ocean acidification impacts on coastal ecosystem services due to habitat degradation. Emerging. Top. Life. Sci. 3, 197–206. doi: 10.1042/ETLS20180117
Hartmann M., Hill P. G., Tynan E., Achterberg E. P., Eric P., Leakey R. J. G., et al. (2016). Resilience of SAR11 bacteria to rapid acidification in the high-latitude open ocean. FEMS Microbial Ecol. 92, 161. doi: 10.1093/femsec/fiv161
Hassenrück C., Fink A., Lichtschlag A., Tegetmeyer H. E., de Beer D., Ramette A. (2016). Quantification of the effects of ocean acidification on sediment microbial communities in the environment: The importance of ecosystem approaches. FEMS Microbiol. Ecol. 92, fiw027. doi: 10.1093/femsec/fiw027
Haynes K., Hofmann T. A., Smith C. J., Ball A. S., Underwood G. J., Osborn A. M. (2007). Diatom-derived carbohydrates as factors affecting bacterial community composition in estuarine sediments. Appl. Environ. Microbiol. 73, 6112–6124. doi: 10.1128/AEM.00551-07
Hedderich R., Klimmek O., Kröger A., Dirmeier R., Keller M., Stetter K. (1998). Anaerobic respiration with elemental sulfur and with disulfides. FEMS. Microbiol. Rev. 22, 353–381. doi: 10.1111/j.1574-6976.1998.tb00376.x
Herbert R. (1999). Nitrogen cycling in coastal marine ecosystems. FEMS. Microbiol. Rev. 23, 563–590. doi: 10.1111/j.1574-6976.1999.tb00414.x
Hicks N., Bulling M. T., Solan M., Raffaelli D., White P. C. L., Paterson D. M. (2011). Impact of biodiversity-climate futures on primary production and metabolism in a model benthic estuarine system. BMC Ecol. 11, 7. doi: 10.1186/1472-6785-11-7
Hope J., Paterson D., Thrush S. (2019). The role of microphytobenthos in soft-sediment ecological networks and their contribution to the delivery of multiple ecosystem services. J. Ecol. 108, 815–830. doi: 10.1111/1365-2745.13322
Isaac A., Francis B., Amann R., Amin S. (2021). Tight adherence (Tad) pilus genes indicate putative niche differentiation in phytoplankton bloom associated Rhodobacterales. front. Microbiol. 12, 718297. doi: 10.3389/fmicb.2021.718297
Johnson V. R., Brownlee C., Milazzo M., Hall-Spencer J. M. (2015). Marine microphytobenthic assemblage shift along a natural shallow-water CO2 gradient subjected to multiple environmental stressors. J. Mar. Sci. Eng. 3, 1425–1447. doi: 10.3390/jmse3041425
Johnson V. R., Brownlee C., Rickaby R. E. M., Graziano M., Milazzo M., Hall-Spencer J. M. (2013). Responses of marine benthic microalgae to elevated CO2. Mar. Biol. 160, 1813–1824. doi: 10.1007/s00227-011-1840-2
Kerfahi D., Hall-Spenser J. M., Tripathy B. M., Milazzo M., Lee J., Adams J. M. (2014). Shallow Water Marine Sediment Bacterial Community Shifts Along a Natural CO2 Gradient in the Mediterranean Sea Off Vulcano, Italy. Microb. Ecol. 67, 819–828. doi: 10.1007/s00248-014-0368-7
Kerfahi D., Harvey B., Agostini S., Kon K., Huang R., Adams J., et al. (2020). Responses of intertidal bacterial biofilm communities to increasing pCO2. Mar. Biotechnol. 22, 727–738. doi: 10.1007/s10126-020-09958-3
Lam C., Harder T., Qian P. (2005). Induction of larval settlement in the polychaete Hydroides elegans by extracellular polymers of benthic diatoms. Mar. Ecol. Prog. Ser. 286, 145–154. doi: 10.3354/meps286145
Lau S. C. K., Thiyagarajan V., Cheung S. C. K., Qian P. Y. (2005). Roles of bacterial community composition in biofilms as a mediator for larval settlement of three marine invertebrates. Aquat. Microb. Ecol. 38, 41–51. doi: 10.3354/ame038041
Lin Y., Zhao D., Zeng J., Cao X., Jiao C. (2019). Network analysis reveals seasonal patterns of bacterial community networks in lake taihu under aquaculture conditions. Water. 11, 1868. doi: 10.3390/w11091868
Liu Y., Qin Y., Chen T., Lu M., Qian X., Guo X., et al. (2020). A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell. 12, 315–330. doi: 10.1007/s13238-020-00724-8
López-García P., Philippe H., Gail F., Moreira D. (2003). Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the mid-Atlantic ridge. PNAS. U.S.A. 100, 697–702. doi: 10.1073/pnas.0235779100
Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science. 353, 1272–1277. doi: 10.1126/science.aaf4507
Lutier M., Poi C. D., Gazeau F., Appolis A., Le Luyer J., Pernet F. (2022). Revisiting tolerance to ocean acidification: Insights from a new framework combining physiological and molecular tipping points of pacific oyster. Glob. Change. Biol. 28, 3333–3348. doi: 10.1111/gcb.16101
Maki J. S., Rittschof D., Costlow J. D., Mitchell R. (1988). Inhibition of attachment of larval barnacles, Balanus amphitrite, by bacterial surface films. Mar. Biol. 97, 199–206. doi: 10.1007/BF00391303
Marques da Silva J., Cruz S., Cartaxana P. (2017). Inorganic carbon availability in benthic diatom communities: photosynthesis and migration. Phil. Trans. R. Soc B. 372, 20160398. doi: 10.1098/rstb.2016.0398
McMinn A., Muller M., Martin A., Ryan K. G. (2017). Effects of changing pH and CO2 concentration on a late summer surface sea ice community. Mar. Biol. 164, 86–96. doi: 10.1007/s00227-017-3102-4
McMinn A., Sellah S., Llah W., Mohammad M. (2005). Quantum yield of the marine benthic microflora of near-shore coastal penang, Malaysia. Mar. Freshw. 56, 1047–1053. doi: 10.1071/MF05007
Nelson K., Baltar F., Lamare M., Morales S. (2020). Ocean acidification affects microbial community and invertebrate settlement on biofilms. Sci. Rep. 10, 3274. doi: 10.1038/s41598-020-60023-4
Pan J., Cuadrado D., Bournod C. (2017). Diatom-driven recolonization of microbial mat-dominated siliciclastic tidal flat sediments. FEMS. Microbiol. Ecol. 93, fix111. doi: 10.1093/femsec/fix111
Papaspyrou S., Smith C. J., Dong L. F., Whitby C., Dumbrell A. J., Nedwell D. B. (2014). Nitrate reduction functional genes and nitrate reduction potentials persist in deeper estuarine sediments. Why? PLoS. One 9, e94111. doi: 10.1371/journal.pone.0094111
Platt T., Gallegos C. L., Harrison W. G. (1980). Photoinhibition of photosynthesis in natural assemblages of marine phytoplankton. J. Mar. Res. 38, 687–701.
Ralph P., Gademann R. (2005). Rapid light curves: a powerful tool to assess photosynthetic activity. Aquat. Bot. 82, 222–237. doi: 10.1016/j.aquabot.2005.02.006
Ren Y., Yu G., Shi C., Liu L., Guo Q., Han C., et al. (2022). Majorbio cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta. 1, e12. doi: 10.1002/imt2.12
Rendueles O., Ghigo J. (2012). Multi-species biofilms: how to avoid unfriendly neighbors. FEMS. Microbiol. Rev. 36, 972–989. doi: 10.1111/j.1574-6976.2012.00328.x
Sansupa C., Wahdan S., Hossen S., Disayathanoowat T., Wubet T., Purahong W. (2021). Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria? Appl. Sci. 11, 688. doi: 10.3390/app11020688
Sathish K., Patil J., Anil A. (2020). Phytoplankton chlorophyll-breakdown pathway: Implication in ecosystem assessment. J. Environ. Manage. 258, 109989. doi: 10.1016/j.jenvman.2019.109989
Sciutteri V., Smedile F., Vizzini S., Mazzola A., Vetriani C. (2022). Microbial biofilms along a geochemical gradient at the shallow-water hydrothermal system of vulcano island, Mediterranean Sea. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.840205
Sneed J. M., Sharp K. H., Ritchie K. B., Paul V. J. (2014). The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc. Biol. Sci. 281, 20133086. doi: 10.1098/rspb.2013.3086
Snelgrove P. V. R., Soetaert K., Solan M., Thrush S., Wei C., Danovaro R., et al. (2018). Global carbon cycling on a heterogeneous seafloor. Trends. Ecol. Evol. 33, 96–105. doi: 10.1016/j.tree.2017.11.004
Su X., Wen T., Wang Y., Xu J., Cui L., Zhang J., et al. (2021b). Stimulation of N2O emission via bacterial denitrification driven by acidification in estuarine sediments. Glob. Change Biol. 27, 5564–5579. doi: 10.1111/gcb.15863
Su X., Yang X., Li H., Wang H., Wang Y., Xu J., et al. (2021a). Bacterial communities are more sensitive to ocean acidification than fungal communities in estuarine sediments. FEMS. Microbiol. Ecol. 97, fiab058. doi: 10.1093/femsec/fiab058
Sundbäck K., Miles A. (2002). Role of microphytobenthos and denitrification for nutrient turnover in embayments with floating macroalgal mats: A spring situation. Aquat. Microb. Ecol. 30, 91–101. doi: 10.3354/ame030091
Tangherlini M., Corinaldesi C., Ape F., Greco S., Romeo T., Andaloro F., et al. (2021). Ocean acidification induces changes in virus–host relationships in Mediterranean benthic ecosystems. Microorganisms. 9, 769. doi: 10.3390/microorganisms9040769
Tao S. Q., Eglinton T. I., Montluçon D. B., McIntyre C., Zhao M. X. (2016). Diverse origins and pre-depositional histories of organic matter in contemporary Chinese marginal sea sediments. Geochim. Cosmochim. Acta 191, 70–88. doi: 10.1016/j.gca.2016.07.019
Taylor J., Ellis R., Milazzo M., Hall-Spencer J., Cunliffe M. (2014). Intertidal epilithic bacteria diversity changes along a naturally occurring carbon dioxide and pH gradient. FEMS. Microbiol. Ecol. 89, 670–678. doi: 10.1111/1574-6941.12368
Teschler J., Zamorano-Sánchez D., Utada A., Warner C., Wong G., Linington R., et al. (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268. doi: 10.1038/nrmicro3433
Thomas A., Ramkumar A., Shanmugam A. (2022). CO2 acidification and its differential responses on aquatic biota - a review. Environ. Adv. 8, 100219. doi: 10.1016/j.envadv.2022.100219
Tohidifar P., Plutz M., Ordal G., Rao C. (2020). The mechanism of bidirectional pH taxis in Bacillus subtilis. J. Bacteriol. Res. 202, e00491-19. doi: 10.1128/JB.00491-19
Torstensson A., Hedblom M., Mattsdotter Bjo¨rk M., Chierici M., Wulff A. (2015). Long-term acclimation to elevated pCO2 alters carbon metabolism and reduces growth in the Antarctic diatom Nitzschia lecointei. Proc. R. Soc B. 282, 20151513. doi: 10.1098/rspb.2015.1513
Vaqué D., Lara E., Arrieta J., Holding J., Sà E., Hendriks I., et al. (2019). Warming and CO2 enhance Arctic heterotrophic microbial activity. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00494
Vargas C., de la Hoz M., Aguilera V., Martín V., Manríquez P., Navarro J., et al. (2013). CO2-driven ocean acidification reduces larval feeding efficiency and changes food selectivity in the mollusk Concholepas concholepas. J. Plankton. Res. 35, 1059–1068. doi: 10.1093/plankt/fbt045
Vargas C., Lagos N., Lardies M., Duarte C., Manríquez P. H., Aguilera V. M., et al. (2017). Species-specific responses to ocean acidification should account for local adaptation and adaptive plasticity. Nat. Ecol. Evol. 1, 0084. doi: 10.1038/s41559-017-0084
Vieira S., Cartaxana P., Ma´guas C., Marques da Silva J. (2016). Photosynthesis in estuarine intertidal microphytobenthos is limited by inorganic carbon availability. Photosynth. Res. 128, 85–92. doi: 10.1007/s11120-015-0203-0
Walters W., Hyde E. R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. Msystems. 1, e00009–e00015. doi: 10.1128/mSystems.00009-15
Walz (2019) Principles of operation: Phytoplankton analyzer PHYTO-PAM-II and PhytoWin_3 software V 3. Available at: https://www.walz.com/files/downloads/manuals/phyto-pam/PhytoPamII_3.pdf (Accessed December 30, 2022).
Walz (2022) Heinz Walz gamb h, phyto-PAM-II topics website. Available at: https://www.walz.com/products/chl_p700/phyto-pam/introduction.html (Accessed March 15, 2022).
Williams T., Wilkins D., Long E., Evans F., DeMaere M., Raftery M., et al. (2013). The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 15, 1302–1317. doi: 10.1111/1462-2920.12017
Xing X., Xu H., Wang D., Yang X., Qin H., Zhu B. (2022). Nitrogen use aggravates bacterial diversity and network complexity responses to temperature. Sci. Rep. 12, 13989. doi: 10.1038/s41598-022-15536-5
Xu D., Brennan G., Xu L., Zhang X., Fan X., Han W., et al. (2018). Ocean acidification increases iodine accumulation in kelp-based coastal food webs. Glob. Change Biol. 25, 629–639. doi: 10.1111/gcb.14467
Yan L., Boyd K. G., Adams D. R., Burgess J. G. (2003). Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl. Environ. Microb. 69, 3719–3727. doi: 10.1128/AEM.69.7.3719-3727.2003
Yang Z., Peng C., Cao H., Song J., Gong B., Li L., et al. (2022). Microbial functional assemblages predicted by the FAPROTAX analysis are impacted by physicochemical properties, but c, n and s cycling genes are not in mangrove soil in the beibu gulf, China. Ecol. Indic. 139, 108887. doi: 10.1016/j.ecolind.2022.108887
Yu T., Chen Y. (2019). Effects of elevated carbon dioxide on environmental microbes and its mechanisms: A review. Sci. Total. Environ. 655, 865–879. doi: 10.1016/j.scitotenv.2018.11.301
Yuan M. M., Guo X., Wu L., Zhang Y., Xiao N., Ning D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Chan 11, 343–348. doi: 10.1038/s41558-021-00989-9
Keywords: ocean acidification, biofilms, microphytobenthos, bacteria, intertidal, sediments
Citation: Zhai C, Liang Y, Yu H, Ji Y, Chen X, Wang M and McMinn A (2023) Ocean acidification alters the benthic biofilm communities in intertidal soft sediments. Front. Mar. Sci. 10:1117826. doi: 10.3389/fmars.2023.1117826
Received: 06 December 2022; Accepted: 20 March 2023;
Published: 04 April 2023.
Edited by:
Jin Zhou, Tsinghua University, ChinaCopyright © 2023 Zhai, Liang, Yu, Ji, Chen, Wang and McMinn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yantao Liang, bGlhbmd5YW50YW9Ab3VjLmVkdS5jbg==; Min Wang, bWluZ3dhbmdAb3VjLmVkdS5jbg==; Andrew McMinn, YW5kcmV3Lm1jbWlubkB1dGFzLmVkdS5hdQ==
 Chuan Zhai1
Chuan Zhai1 Yantao Liang
Yantao Liang Yan Ji
Yan Ji Xuechao Chen
Xuechao Chen Min Wang
Min Wang Andrew McMinn
Andrew McMinn