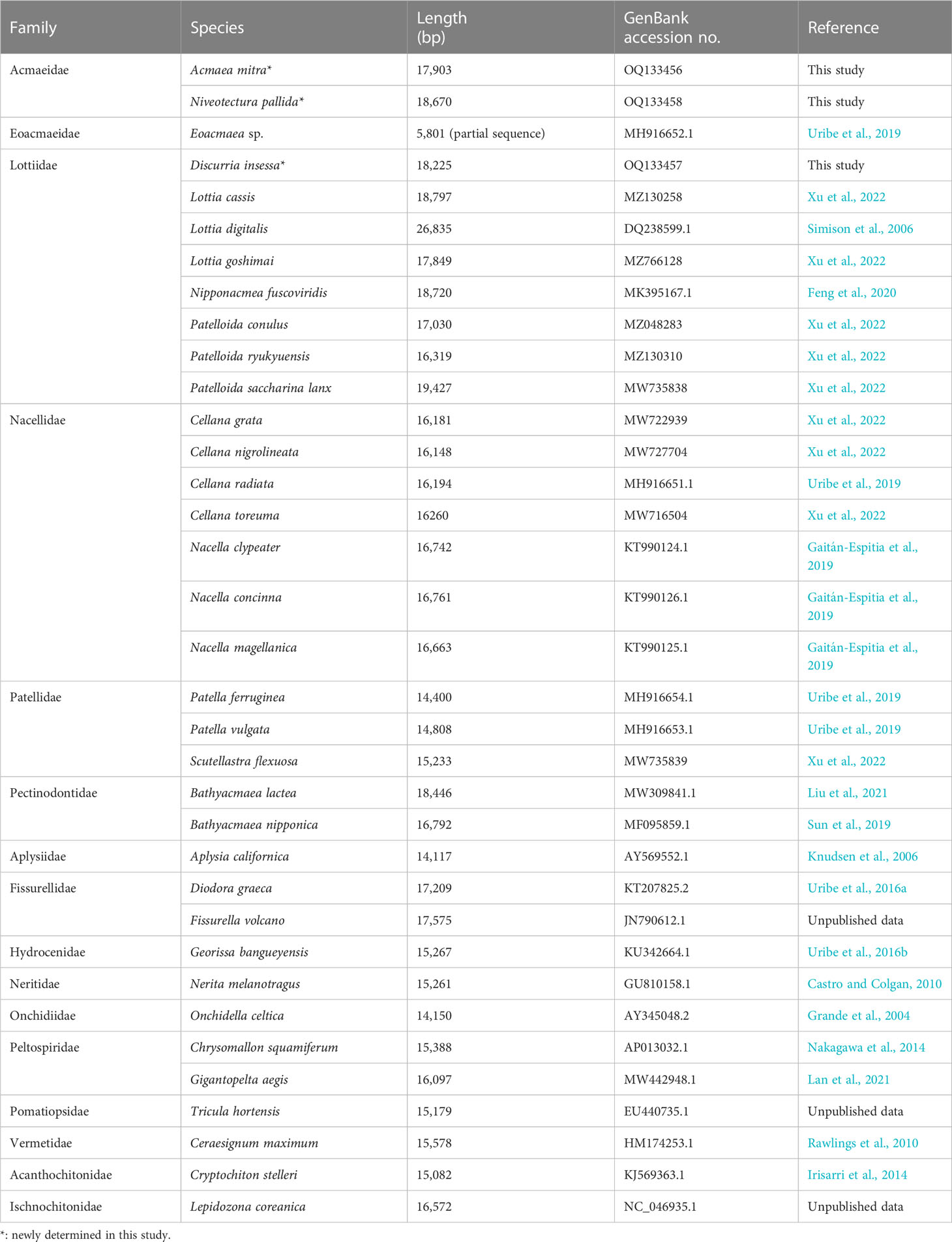
- 1Division of EcoScience, Ewha Womans University, Seoul, Republic of Korea
- 2Seto Marine Biological Laboratory, Kyoto University, Nishimuro, Wakayama, Japan
- 3Natural History Museum, Ewha Womans University, Seoul, Republic of Korea
The subclass Patellogastropoda (called “true limpets”) is one of the most primitive groups of the Gastropoda and contains approximately 350 species worldwide. Within this subclass, internal phylogeny among family members, including relationships of the “Acmaeidae” with other patellogastropod families, remains incompletely clarified. Here, we newly determined two complete mitochondrial genome sequences of “Acmaeidae” (Acmaea mitra and Niveotectura pallida) and one sequence from Lottiidae species (Discurria insessa) and combined them with mitochondrial genome sequences of 20 other published limpet species for phylogenetic analysis of the sequence dataset (nucleotides and amino acids) of 13 protein-coding genes using maximum likelihood and Bayesian inference methods. The resulting phylogenetic trees showed monophyly of Patellogastropoda species that were subsequently subdivided into two clades [clade I (Nacellidae, Pectinodontidae, Acmaeidae, and Patellidae) and clade II (Eoacmaeidae and Lottiidae)]. The sister relationship between the Acmaeidae and Pectinodontidae species revealed by phylogenetic analysis was also supported by sharing their similar gene arrangement patterns, which differ substantially from those of clade II members including the Lottiidae species. The polyphyletic relationship between Acmaeidae (grouped with Pectinodontidae as a sister taxon in clade I) and Lottiidae species (grouped with Eoacmaeidae in clade II) corroborates that they are phylogenetically distinct from each other. This mitochondrial genome phylogeny contradicts previous morphology-based hypotheses, yet highlights that Acmaeidae and Pectinodontidae are the most closely related. Further in-depth analysis of the complete mitochondrial genome sequences based on a broad range of samples including those from relatively unstudied and/or underrepresented taxa is required to fully understand the mitochondrial genome evolution and a more comprehensive phylogeny among the major groups of the Patellogastropoda.
1 Introduction
Acmaeidae is a family of Patellogastropoda (called “true limpets”) that represents one of the most primitive groups of the Gastropoda and contains approximately 350 species worldwide. They are found in lower intertidal to subtidal zones and constitute macrobenthic communities on coastal rocky shores, playing an important role in littoral ecosystems (Branch, 1985). This family is characterized by having a foliated shell with a fibrillar prismatic shell microstructure, one feather-shaped ctenidium, and a single pair of tricuspid radular teeth (Lindberg, 1998). Over the last century, external shell morphology has been the feature most widely used for both species-level taxonomy and investigating the internal phylogeny of the Patellogastropoda. However, morphological traits of limpet species are highly variable with different morphs from the ontogenic or ecophenotypic origin, resulting in substantial taxonomic confusion and disagreement on their phylogenetic relationships (Nakano and Ozawa, 2007). Soft body anatomy including radula characters has also been considered phylogenetically informative and widely used to investigate the relationships among the Patellogastropoda families in the past decades (Lindberg, 1998; Sasaki, 1998). Nevertheless, the taxonomic confusion associated with the absence of key characters for defining the family boundaries, and convergent evolution and ecophenotypic plasticity in their morphological features has resulted in different classification systems of the Patellogastropoda over the last decades (Sasaki et al., 2006; Nakano and Sasaki, 2011).
Against this background, phylogenetic relationships including the taxonomic status of the “Acmaeidae” with the Patellogastropoda have remained unresolved from previous studies that employed different sources of taxonomic information, such as shell characters, internal morphology, and molecular sequence data [Dall, 1876; Lindberg, 1998; Sasaki, 1998 (morphology); Harasewych and McArthur, 2000; Nakano and Ozawa, 2007 (molecular data)]. The Acmaeidae was traditionally divided into two subfamilies, Acameinae and Pectinodontinae, based on morphological perspectives (Lindberg, 1998), but these two subfamilies (sensu Lindberg, 1998) were not found as a monophyletic group by subsequent molecular analysis of three mitochondrial genes (mtDNA 12S rDNA, 16S rDNA, and cox1; Nakano and Ozawa, 2007). Moreover, despite this molecular study reporting that the Acmaeidae species (Acmaea mitra, Niveotectura pallida, and Erginus sybaritica) were grouped with Lottiidae species, their relationships were not concordant with other studies using the morphology or using different molecular markers, i.e., nuclear vs. mitochondrial DNA: The Acmaeidae species were included as a subgroup of Lottiidae based on their morphological similarity with lottiid species (e.g., shell microstructure, radula characters, and presence of a ctenidium) (Lindberg, 1998), but they were revived as a separate family (Nakano and Sasaki, 2011) from the Lottiidae. These phylogenetic uncertainties of the “Acmaeidae” necessitate further study using different molecular evidence to elucidate their relationships with other Patellogastropoda families.
During recent decades, comparative analysis of the complete mitochondrial genome information (i.e., nucleotide sequence and gene order data) has been widely used for investigating the phylogenetic relationships in many molluscan groups including Patellogastropoda groups where interrelationships were not fully resolved (Uribe et al., 2019; Feng et al., 2020; Xu et al., 2022). In this study, we aimed to (1) resolve phylogenetic uncertainty of the Acmaeidae and (2) extend our knowledge of mitochondrial genome diversity among major groups of the Patellogastropoda. For this, we determined two complete genome sequences of “Acmaeidae” (Acmaea mitra and Niveotectura pallida), the first representatives from the Acmaeidae, and one complete genome sequence of Lottiidae (Discurria insessa), combined them with the mitochondrial genome sequences of 20 other published limpet species plus 12 non-patellogastropod species (including 2 chiton species), and performed phylogenetic analysis based on nucleotide sequence information and gene order data.
2 Materials and methods
2.1 DNA extraction
Genomic DNA from three limpet species (A. mitra, N. pallida, and D. insessa) was extracted from muscle tissues of an ethanol-fixed single individual using E.Z.N.A. mollusk DNA kit (Omega Biotek, Norcross, GA, USA), in accordance with the manufacturer’s instructions, and subjected to next-generation sequencing (NGS) library construction and sequencing.
2.2 Library preparation and next-generation sequencing
NGS sequencing libraries were prepared using Illumina TruSeq DNA library preparation kit and sequenced with a 151 bp paired-end run of the Illumina HiSeq X sequencing platform (Illumina, San Diego, CA, USA).
2.3 Mitochondrial genome assembly and gene annotation
Low-quality reads and adaptor sequences were trimmed using Trimmomatic v0.39 (Bolger et al., 2014). The clean reads of A. mitra (clean reads: 76,270,083), D. insessa (clean reads: 73,627,056), and N. pallida (clean reads: 14,140,656) were de novo assembled into circular DNAs using NOVOPlasty V.4.2 (Dierckxsens et al., 2017) and their genes were annotated on the MITOS webserver (Bernt et al., 2013). The protein-coding genes (PCGs) and ribosomal RNA (rRNA) genes were also confirmed by comparing them with those of other Patellogastropoda species using Geneious® v.11.0.11. The tRNA genes for each mtDNA sequence were identified using the tRNAscan-SE program (Lowe and Eddy, 1997) or by manually inspecting potential secondary structures and anticodon sequences.
2.4 Phylogenetic analyses
We performed phylogenetic analyses for 29 mitochondrial genome sequences from 23 Patellogastropoda species (including three newly sequenced in this study) and 4 non-Patellogastropoda (Heterobranchia and Vetigastropoda) species, as well as 2 chiton species as outgroups (Table 1). Nucleotide sequences of 13 protein-coding genes were individually translated to amino acid sequences and multiple-aligned by ‘reverse translation’ of the aligned protein sequences using RevTrans 1.4 webserver (Wernersson and Pedersen, 2003), and concatenated using MEGA v10.2.6 (Kumar et al., 2018). The best-fit substitution models for nucleotide (NT) and amino acid (AA) sequences for each of the 13 PCGs were identified (see Table 2) using jModelTest (NT; Darriba et al., 2012) and ProtTest 3.4.2 (AA; Darriba et al., 2011) on the CIPRES webserver (Miller et al., 2010). Phylogenetic relationships were reconstructed using Bayesian inference (BI) with MrBayes 3.2.6 (Ronquist et al., 2012) and maximum likelihood (ML) with RAxML V.8.2.9 (Stamatakis, 2014) on the CIPRES portal (Miller et al., 2010). A Markov chain Monte Carlo (MCMC) analysis was performed using two independent runs of 1×106 generations with four chains, sampling every 100 generations for the BI tree. Bayesian posterior probability (BPP) values were estimated after discarding the initial 2×103 trees that reached a stationary stage of the parameters as burn-in. The bootstrap analysis was also performed with 1,000 replicates for the resulting
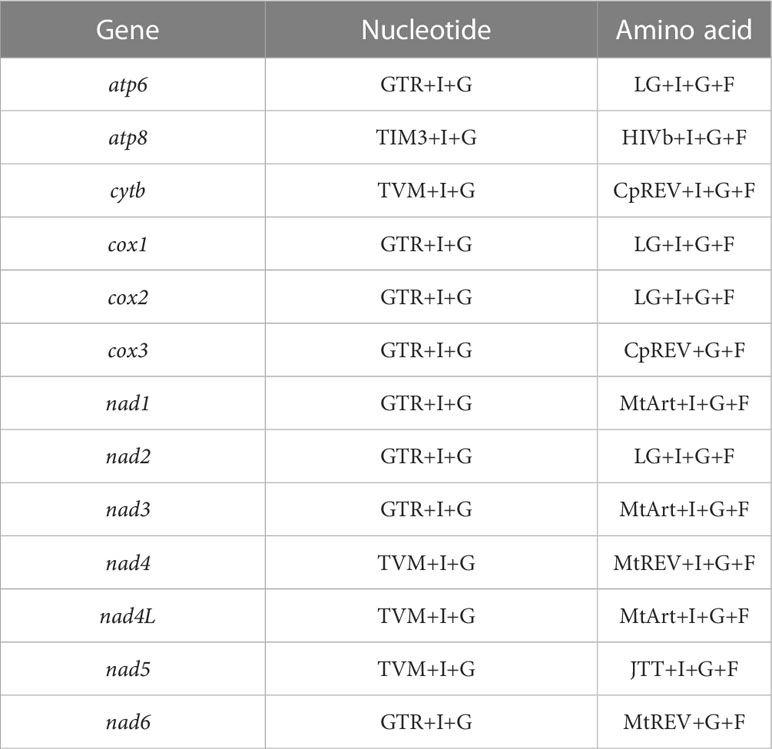
Table 2 The best-fit models estimated from each of the 13 protein-coding genes of 23 patellogastropod and 6 other gastropod species used for phylogenetic analysis.
2.5 Phylogenetic analysis using gene order information
Pairwise comparisons of the gene order among Patellogastropoda species were conducted to infer the parsimonious rearrangement scenarios (e.g., transpositions, reverse transpositions, reversals and tandem-duplication-random-loss events) using CREx (Bernt et al., 2007) based on common intervals. The MLGO web server was used to determine the phylogeny among Patellogastropoda species based on gene order information (PCGs, rRNAs, and tRNAs) (Hu et al., 2014).
2.6 Divergence time estimation
Divergence times among Patellogastropoda species were estimated based on nucleotide dataset of 13 protein-coding genes using an uncorrected lognormal relaxed clock model with a Yule speciation process as a tree prior in BEAST v2.6.7 (Bouckaert et al., 2019). A calibrated Yule speciation model was used as a tree prior, and the best partition schemes and the best evolution models were estimated in Partition Finder 2 based on BIC. We set the time calibration on three points based on Nacellidae (47.8–56 MYA, million years ago) from Nakano & Ozawa (2007), Patellogastropoda (336–339.4 MYA), and Neoloricata (460-466 MYA) data from the Paleobiology Database (https://paleobiodb.org/). We ran Markov chain for three times, each running for 10x107 generations, and sampling every 1,000 generations. The convergence of chains were confirmed with Tracer v1.7.2 (Rambaut et al., 2018) and the first 10% of samples were discarded as burn-in. The results were combined with Log combiner and Tree annotator of the BEAST package. The maximum clade credibility (MCC) tree was visualized in FigTree 1.4.4 (Rambaut, 2018).
3 Results
3.1 Characteristics of mitochondrial genomes of the Acmaeidae
The mitochondrial genomes of the two Acmaeidae species are 17,903 bp (A. mitra), 18,670 bp (N. pallida) and D. insessa mtDNA is 18,225 bp in size, each containing 37 genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes). The D. insessa mtDNA has a duplicated copy of trnM, a universal feature of Lottiidae species that were determined thus far. All tRNA genes from these three newly determined mitochondrial genomes have a typical clover-leaf secondary structure, with the exception of one tRNA in N. pallida (trnS1) and three tRNAs in D. insessa (trnS1, trnS2, and trnE). Both trnS1 and trnS2 lack a DHU stem, whereas the TΨC arm is absent in trnE. This result is consistent with an earlier study that DHU stems are missing in trnS1 and trnS2 in Lottia goshimai and Nipponacmea fuscoviridis (Feng et al., 2020). Information on the genome organization, overall nucleotide composition, start/termination codons, and codon usage of 13 PCGs for the three species is presented in the supplementary data (Supplementary Tables 1–9). Nucleotide compositions of the three newly sequenced species were 26% A, 36.9% T, 24.2% G, and 12.9% C for A. mitra; 28% A, 37.8% T, 22.3% G, and 11.91% C for N. pallida; and 30.1% A, 30.3% T, 17.5% G, and 22.1% C for D. insessa (see Supplementary Tables 2, 5, 8). In their entire genome sequences, there is a strong bias toward A and T, with the A + T content ranging from 60.4% (D. insessa) to 65.8% (N. pallida), which is similar to previously reported findings in other Patellogastropoda species (Feng et al., 2020).
3.2 Phylogenetic relationships of the Acmaeidae within the Patellogastropoda
Phylogenetic analyses of nucleotide (NT) and amino acid (AA) sequences using ML and BI methods found an identical tree topology (Figure 1): The resulting trees show that the Patellogastropoda species formed a monophyletic grouping that was strongly supported [76% BP in ML and 1.0 BPP in BI (NT dataset) and 84% BP in ML and 0.99 BPP in BI (AA dataset)], relative to other gastropod groups (e.g., Heterobranchia, Vetigastropoda), and were subsequently subdivided into two groups (clades I and II) with high branch support. Inclusion of other gastropod groups (e.g., Neritimorpha, Caenogastropoda, and Neomphalina) in the analysis did not change the internal relationships among Patellogastropoda species (Supplementary Figure 1). Within the Patellogastropoda, clade I is represented by four monophyletic limpet families (Nacellidae, Pectinodontidae, Acmaeidae, and Patellidae), whereas clade II comprises the two remaining families: Eoacmaeidae and Lottiidae. Compared with clade II, clade I was more strongly supported [92% BP in ML and 1.00 BPP in BI (NT dataset) and 97% BP in ML and 1.00 BPP in BI (AA dataset)]. Within clade I, the Patellidae was positioned basal to the group of families Acmaeidae, Pectinodontidae, and Nacellidae, in which the former two families formed a robust sister relationship. The two Acmaeidae species (A. mitra and N. pallida) were grouped with Pectinodontidae species (Bathyacmaea lactea and B. nipponica) receiving high supporting values from the NT dataset (93% BP in ML and 1.00 BPP in BI; Figure 1), but relatively moderate support in ML analysis for the AA dataset (72% BP in ML; see high branch support of 1.00 BPP in BI). Within clade II, a partial sequence of Eoacmaea sp. representing the family Eoacmaeidae formed a sister relationship with the strongly supported monophyletic Lottiidae including D. insessa (newly sequenced) [70% BP in ML and 1.00 BPP in BI (NT dataset); 80% BP in ML and 0.99 BPP in BI (AA dataset)]. Within Lottiidae, the monophyletic Patelloida species was sister to the group of Lottia, Discurria, and Nipponacmea species with high branch support (100% BP in ML and 1.00 BP in BI). Phylogenetic relationships among the Patellogastropoda species inferred from inclusion of other gastropod groups in phylogenetic analysis (e.g., Neritimorpha, Caenogastropoda, and Neomphalina; Supplementary Figures 1, 2) were identical to those depicted in Figure 1.
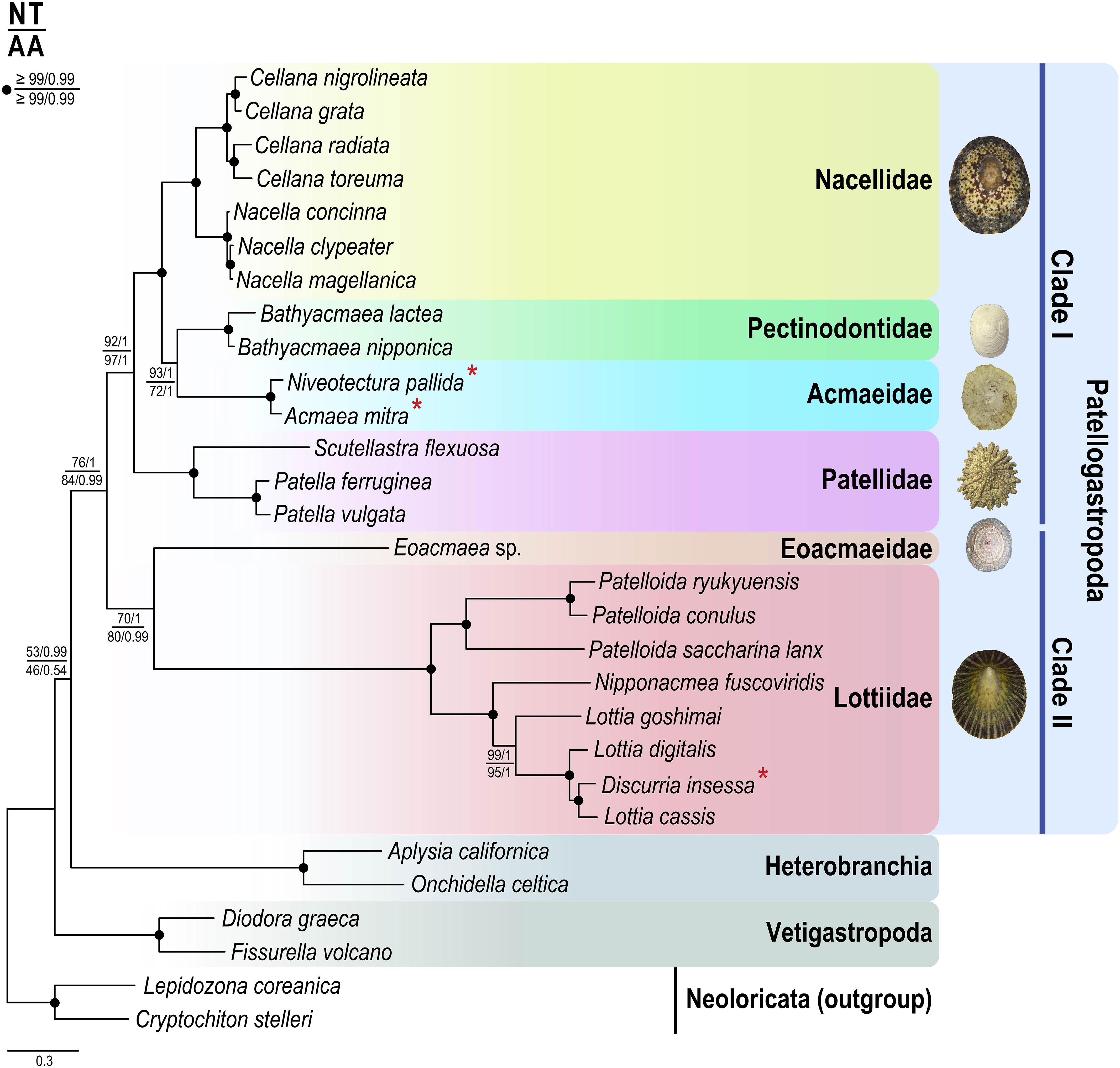
Figure 1 Phylogenetic relationships among Patellogastropoda species inferred from maximum likelihood and Bayesian methods for concatenated nucleotide and amino acid sequences of 13 protein-coding genes of the complete mitochondrial genomes. Node supporting values are indicated as BP (bootstrap percentage)/BP (Bayesian posterior probability) in ML and BI, respectively. An asterisk (*) denotes mtDNA sequences newly determined in this study.
3.3 Gene arrangement patterns among major Patellogastropoda groups
Comparison of gene order information among Patellogastropoda species indicates that the gene arrangement patterns among clade I members are relatively conserved, compared to the clade II members in which gene rearrangements are very substantial (Figure 2). Excluding tRNA genes that are highly variable in their position even between closely related species, the gene arrangement pattern of 13 PCGs and two rRNA genes among the group of three families Nacellidae, Pectinidontidae, and Acmaeidae is identical, but radically differs from that of their sister group Patellidae. By contrast, an extensive gene order rearrangement was observed among clade II members, even between congeneric species belonging to the genera Patelloida and Lottia. Phylogenetic tree from parsimony analysis based on gene order data (Supplementary Figure 3) is generally consistent with relationships among the family members inferred from phylogenetic analysis of the protein-coding gene sequence data (see Discussion section for the details of their relationships).

Figure 2 Linearized mitochondrial gene arrangement of Patellogastropoda and other gastropod (Heterobranachia and Vetigastropoda) species lay alongside the phylogenetic tree (see Figure 1 for their detailed relationships). The tRNA genes are labeled by single-letter abbreviations of the amino acid code. An asterisk (*) denotes mtDNA sequences newly determined in this study.
3.4 Estimated divergence times among major Patellogastropoda groups
Divergence time estimation using mitochondrial 13 PCGs inferred the origin of Patellogastropoda was dated 337.20 MYA (95% HPD interval 332.60–341.81 MYA) in the middle Carboniferous period (Figure 3). Subsequent divergence among major family groups belonging to clade I and clade II was estimated to begin in the Permian-Triassic boundary (clade I; 257.67 MYA; 185.20–325.34) and in the early Permian period (clade II; 295.84 MYA; 95% HPD interval 255.72–329.43 MYA), respectively. Within clade I, the family Patellidae started branching off first from the common ancestor of the remaining families (Nacellidae, Pectinodontidae, and Acmaeidae) in the Permian-Triassic boundary, followed by lineage splitting of the Acmaeidae from the Pectinodontidae in the Jurassic-Cretaceus boundary (132.62 MYA; 95% HPD interval 65.84–206.46 MYA).
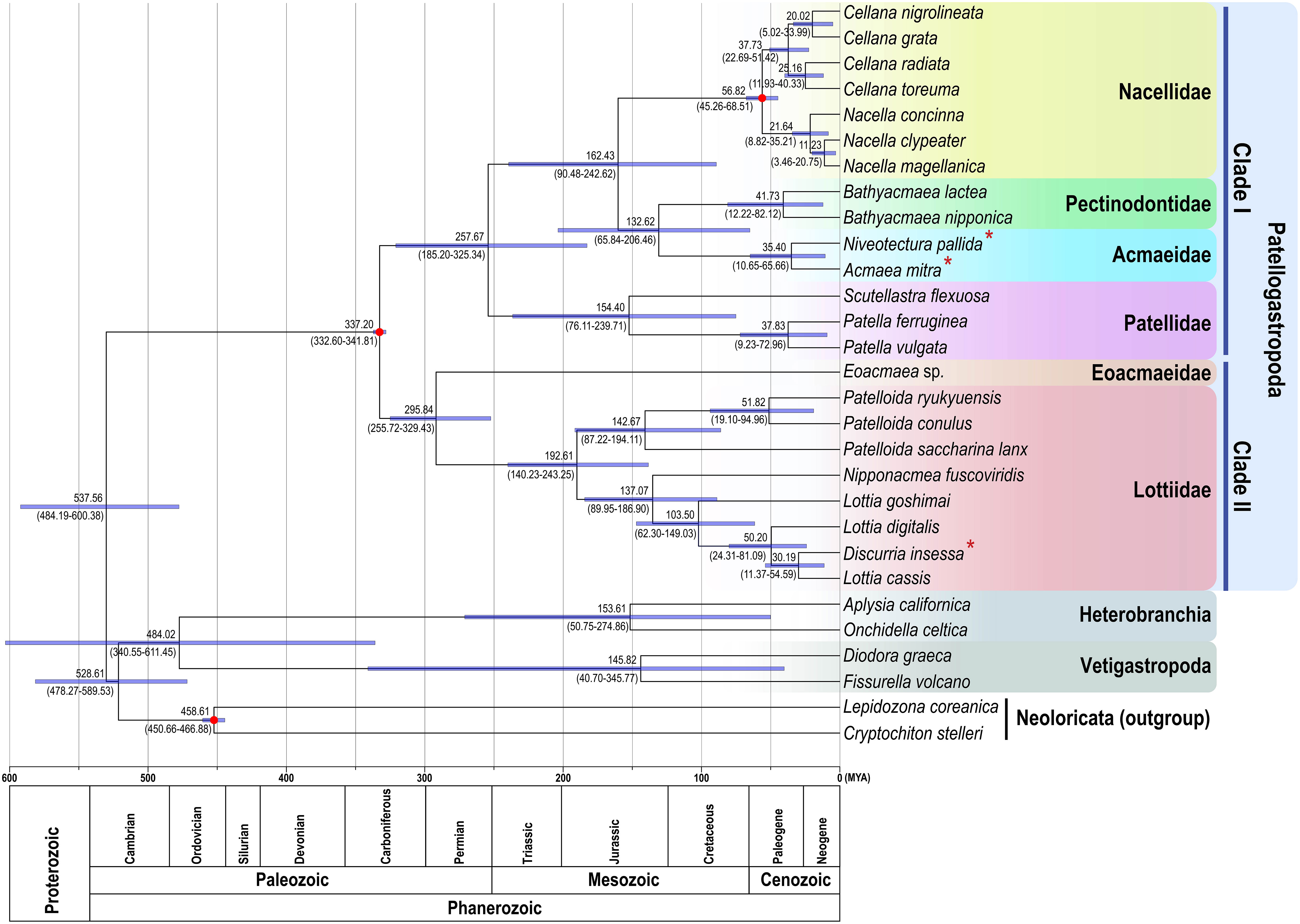
Figure 3 Time-calibrated phylogenetic chronogram showing the mean age estimates (with 95% HPD intervals) based on 13 protein coding genes of patellogastropod mitochondrial genomes. Newly determined mtDNA sequences in this study are marked with an asterisk (*), and calibration constraints are represented by red dots on the corresponding nodes.
4 Discussion
The Acmaeidae is one of the most taxonomically debated Patellogastropoda families, particularly in terms of their taxonomic status and phylogenetic relationships with other groups. Nonetheless, to date there has been only minimal study of this group using molecular data (Nakano and Sasaki, 2011). The relationships inferred in many previous studies did not agree with each other, according to the data sources used in the analysis, such as morphology vs. molecular data, or even between molecular markers (i.e., nuclear vs. mitochondrial DNA). Members of the family Acmaeidae were originally assigned to many lottiid genera and species (e.g., Pilsbry, 1891). However, this scheme was substantially revised based on the shell structure (Lindberg, 1986). After this revision, the genus Acmaea was restricted to the type species Acmaea mitra, and the rest of the species formerly belonging to Acmaeidae were transferred to the Lottiidae (Lindberg, 1986). The mitochondrial genome phylogeny reconstructed in this study provides new and invaluable insights into the internal phylogeny of the Patellogastropoda. The monophyletic grouping of Acmaeidae + Pectinodontidae species contradicts the previous analysis of partial gene fragments of mtDNA (Nakano and Ozawa, 2007), in which the “Acmaeinae” was nested within the Lottiidae, whereas the “Pectinodontinae” was clustered with Lepetidae species. These inferred relationships provided a basis of taxonomic treatment in which the “Pectinodontinae” was elevated to the familial rank (Nakano and Ozawa, 2007). Nakano and Sasaki (2011) later corrected their earlier erroneous interpretation derived from sequence contamination of Acmaea mitra by placing it as a junior synonym of Lottiidae. The Pectinodontidae still retained its familial status thereafter, awaiting taxonomic clarification from further analysis. In this regard, our mitochondrial genome tree highlights that Acmaeidae and Pectinodontidae are the most closely related, and their close affinity lends strong support to the previous morphology-based classification in which the pectinodontid species were treated as a subfamily of the family Acmaeidae (Lindberg, 1998). Nevertheless, further extensive studies based on re-evaluation of their morphological characters including shell and soft body anatomy will be necessary to clarify the taxonomic status of the pectinodontid species as a subgroup or confirm them as a taxonomically valid, separate family from the Acmaeidae.
Historically, phylogenetic relationships of the Acmaeidae with other groups have not yet clearly defined (Nakano and Sasaki, 2011). Based on morphological similarity [e.g., shell microstructure, radula characters (Lindberg, 1998), and presence of a left ctenidium (Sasaki, 1998)] and molecular analysis [12S and 16S mtDNA sequence data (Nakano and Ozawa, 2004)], Acmaeidae was once considered more closely related to Lottiidae than to Patellidae, Nacellidae, and Lepetidae, and was included as a subgroup of Lottiidae. In contrast, later analysis of mtDNA cox1 and histone H3 sequence data revealed that they were distinct from Lottiidae and were subsequently revived as a separate family from Lottiidae (Nakano and Sasaki, 2011). Polyphyletic relationships between Acmaeidae and Lottiidae species discovered from this mitochondrial genome phylogeny provide strong taxonomic justification of their independent family status. Moreover, the clade I monophyly (Patellidae, Nacellidae, Pectinodontidae + Acmaeidae) that is separated from clade II (Eoacmaeidae + Lottiidae) indicates that Acmaeidae is a member of the superfamily Patelloidea along with other clade I family members, not belongs to the superfamily Lottioidea. Note that the time-calibrated phylogenetic tree (Figure 3) indicates that the divergence of the Acmaeidae from the Pectinodontidae is estimated to begin in the Jurassic-Cretaceus boundary (132.62 MYA), being much more recent event than the clade I/clade II separation (in the middle Carboniferous; 337.20 MYA) and the Lottiidae/Eoacmaeidae splitting (in the Carboniferous-Permian boundary; 295.84 MYA).
In many metazoan groups including molluscan species, comparison of the gene arrangement of the complete mitochondrial genome has been widely used and sharing similar synteny pattern observed among certain taxonomic groups can often be interpreted as additional evidence of their phylogenetic affinity (Lee et al., 2019). Likewise, gene order comparison across the limpet species provides additional insights into the phylogenetic relationships and mitochondrial genome evolution among the family members of the Patellogastropoda. The gene arrangement patterns of the two Acmaeidae species (A. mitra and N. pallida) showed the same gene order. Excluding the tRNAs that are often highly mobile even between closely related species, the arrangement patterns of PCGs and rRNA genes of the clade I members are highly conserved showing very similar synteny blocks within each family member (Figure 2). Notably, the three families Acmaeidae, Pectinodontidae, and Nacellidae display the identical gene arrangement in PCGs and rRNA genes, lending additional support to their phylogenetic relatedness. By contrast, the gene order of the Lottiidae species belonging to clade II is highly variable according to the genus and even between congeneric species (see Lottia species), as previously reported (Xu et al., 2022). Moreover, the gene order of Lottiidae species shows substantially different patterns from those of clade I members (see Figure 2 for more details). Inferred relationships among Patellogastropoda species from gene order analysis (Supplementary Figure 3) agreed with tree topology from the sequence analysis (Figure 1 and Supplementary Figures 1, 2) in that Patellogastropoda species were subdivided into two clades (clade I and clade II) and inferred relationships among clade I family members. One exception is that the two Pectinodontidae species (B. lactea and B. nipponica) were not grouped as monophyletic in their gene order tree: the former was clustered with Nacellidae species, whereas the latter was grouped with Acmaeidae species (Supplementary Figure 3). This discrepancy results from B. lactea species shows an intermediate pattern between the Nacellidae and Acmaeidae in some tRNA synteny blocks. Within clade II, most of branches leading to the Lottiidae species were unresolved and showed extremely long branches that are ascribed to their extensive gene rearrangement events between the Lottiidae species. In addition, it is also interesting to note that the Acmaeidae and one Pectinodontidae species (B. nipponica) retain the same consecutive tRNA block of “tRNA KARNI” (trnK, trnA, trnR, trnN, and trnI), which has long been considered an ancestral state across the major clades of the Gastropoda including other subclasses (Uribe et al., 2016b; Gaitán-Espitia et al., 2019) and outgroup (chiton) species (Irisarri et al., 2014). At present, we are unable to precisely address the evolutionary history of gene arrangement patterns across the patellogastropods. To this end, in-depth analysis of complete mitochondrial genome sequences based on a full breadth of taxon coverage from relatively unstudied and/or underrepresented taxa is required to fully understand the mitochondrial genome evolution and a more comprehensive phylogeny among the major groups of the Patellogastropoda.
Data availability statement
The data presented in the study are deposited on GenBank (accession numbers: OQ133456, OQ133457 and OQ133458).
Author contributions
J-KP designed and led the study. EP, DL, HK, YK collected samples, conducted the laboratory work, and performed phylogenetic analysis of the sequence data. EP, TN, and J-KP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the management of Marine Fishery Bio-resources Center (2023) funded by the National Marine Biodiversity Institute of Korea (MABIK) and the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. 2020R1A2C2005393).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1134991/full#supplementary-material
References
Bernt M., Merkle D., Ramsch K., Fritzsch G., Perseke M., Bernhard D., et al. (2007). CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 23 (21), 2957–2958. doi: 10.1093/bioinformatics/btm468
Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., et al. (2013). MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69 (2), 313–319. doi: 10.1016/j.ympev.2012.08.023
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouckaert R., Vaughan T. G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., et al. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PloS Comput. Biol. 15 (4), 1–28. doi: 10.1371/journal.pcbi.1006650
Branch G. M. (1985). “‘Limpets: their role in littoral and sublittoral community dynamics.’,” in The ecology of rocky coasts (London: Hodder and Stoughton Educational), 97–116.
Castro L. R., Colgan D. J. (2010). The phylogenetic position of neritimorpha based on the mitochondrial genome of nerita melanotragus (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 57 (2), 918–923. doi: 10.1016/j.ympev.2010.08.030
Dall W. H. (1876). On the extrusion of the seminal products in limpets, with some remarks on the phyllogeny of the docoglossa. Acad. Nat. Sci. 28, 34–35.
Darriba D., Taboada G. L., Doallo R., Posada D. (2011). ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27 (8), 1164–1165. doi: 10.1093/bioinformatics/btr088
Darriba D., Taboada G. L., Doallo R., Posada D. (2012). JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9 (8), 772. doi: 10.1038/nmeth.2109
Dierckxsens N., Mardulyn P., Smits G. (2017). NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45 (4), 1–9. doi: 10.1093/nar/gkw955
Feng J. T., Guo Y. H., Yan C. R., Ye Y. Y., Li J. J., Guo B. Y., et al. (2020). Comparative analysis of the complete mitochondrial genomes in two limpets from lottiidae (Gastropoda: Patellogastropoda): rare irregular gene rearrangement within Gastropoda. Sci. Rep. 10 (1), 19277. doi: 10.1038/s41598-020-76410-w
Gaitán-Espitia J. D., González-Wevar C. A., Poulin E., Cardenas L. (2019). Antarctic And sub-Antarctic nacella limpets reveal novel evolutionary characteristics of mitochondrial genomes in patellogastropoda. Mol. Phylogenet. Evol. 131, 1–7. doi: 10.1016/j.ympev.2018.10.036
Grande C., Templado J., Cervera J. L., Zardoya R. (2004). Molecular phylogeny of euthyneura (Mollasca: Gastropoda). Mol. Biol. Evol. 21 (2), 303–313. doi: 10.1093/molbev/msh016
Harasewych M. G., McArthur A. G. (2000). A molecular phylogeny of the patellogastropoda (Mollusca: Gastropoda). Mar. Biol. 137 (2), 183–194. doi: 10.1007/s002270000332
Hu F., Lin Y., Tang J. (2014). MLGO: Phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinf. 15 (1), 1–6. doi: 10.1186/s12859-014-0354-6
Irisarri I., Eernisse D. J., Zardoya R. (2014). Molecular phylogeny of acanthochitonina (Mollusca: Polyplacophora: Chitonida): three new mitochondrial genomes, rearranged gene orders and systematics. J. Nat. Hist. 48 (45–48), 2825–2853. doi: 10.1080/00222933.2014.963721
Knudsen B., Kohn A. B., Nahir B., McFadden C. S., Moroz L. L. (2006). Complete DNA sequence of the mitochondrial genome of the sea-slug, aplysia californica: Conservation of the gene order in euthyneura. Mol. Phylogenet. Evol. 38 (2), 459–469. doi: 10.1016/j.ympev.2005.08.017
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Lan Y., Sun J., Chen C., Sun Y., Zhou Y., Yang Y., et al. (2021). Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail gigantopelta aegis. Nat. Commun. 12 (1), 1–15. doi: 10.1038/s41467-021-21450-7
Lee Y., Kwak H., Shin J., Kim S. C., Kim T., Park J. K. (2019). A mitochondrial genome phylogeny of mytilidae (Bivalvia: Mytilida). Mol. Phylogenet. Evol. 139, 106533. doi: 10.1016/j.ympev.2019.106533
Lindberg D. R. (1998). “Subclass eogastropoda and order patellogastropoda,” in Mollusca: The southern synthesis, fauna of Australia (Melbourne: CSIRO).
Liu Y., Li Y., Wang M., Bao Z., Wang S. (2021). The complete mitochondrial genome and phylogenetic analysis of the deep-sea limpet bathyacmaea lactea. Mitochondrial DNA Part B: Resour. 6 (7), 2090–2091. doi: 10.1080/23802359.2021.1923422
Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 (5), 955–964. doi: 10.1093/nar/25.5.955
Miller M. A., Pfeiffer W., Schwartz T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees (Accessed 2010 Gateway Computing Environments Workshop, GCE 2010).
Nakagawa S., Shimamura S., Takaki Y., Suzuki Y., Murakami S. I., Watanabe T., et al. (2014). Allying with armored snails: The complete genome of gammaproteobacterial endosymbiont. ISME J. 8 (1), 40–51. doi: 10.1038/ismej.2013.131
Nakano T., Ozawa T. (2004). Phylogeny and historical biogeography of limpets of the order patellogastropoda based on mitochondrial DNA sequences. J. Molluscan Stud. 70 (1), 31–41. doi: 10.1093/mollus/70.1.31
Nakano T., Ozawa T. (2007). Worldwide phylogeography of limpets of the order patellogastropoda: Molecular, morphological and palaeontological evidence. J. Molluscan Stud. 73 (1), 79–99. doi: 10.1093/mollus/eym001
Nakano T., Sasaki T. (2011). Recent advances in molecular phylogeny, systematics and evolution of patellogastropod limpets. J. Molluscan Stud. 77 (3), 203–217. doi: 10.1093/mollus/eyr016
Pilsbry H. (1891). “Acmaeidae, lepetidae, patellidae, titiscaniidae,” in Manual of conchology; structural and systematic Vol. 13 . Ed. Pilsbry H. A. (Philadelphia: Academy of Natural Sciences).
Rambaut A. (2018) ‘FigTree v. 1.4.4’. Available at: http://tree.bio.ed.ac.uk/software/figtree/.
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67 (5), 901–904. doi: 10.1093/sysbio/syy032
Rawlings T. A., MacInnis M. J., Bieler R., Boore J. L., Collins T. M. (2010). Sessile snails, dynamic genomes: Gene rearrangements within the mitochondrial genome of a family of caenogastropod molluscs. BMC Genomics 11 (1), 440. doi: 10.1186/1471-2164-11-440
Ronquist F., Teslenko M., Van Der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 (3), 539–542. doi: 10.1093/sysbio/sys029
Sasaki T. (1998). Comparative anatomy and phylogeny of the recent archaeogastropoda (Mollusca: Gastropoda). Bulletin 38, 1–223.
Sasaki T., Okutani T., Fujikura K. (2006). Anatomy of bathyacmaea secunda okutani, fujikura & sasaki 1993 (Patellogastropoda: Acmaeidae). J. Molluscan Stud. 72 (3), 295–309. doi: 10.1093/mollus/eyl007
Simison W. B., Lindberg D. R., Boore J. L. (2006). Rolling circle amplification of metazoan mitochondrial genomes. Mol. Phylogenet. Evol. 39 (2), 562–567. doi: 10.1016/j.ympev.2005.11.006
Stamatakis A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. doi: 10.1093/bioinformatics/btu033
Sun J., Liu Y., Xu T., Zhang Y., Chen C., Qiu J. W., et al. (2019). The mitochondrial genome of the deep-sea limpet bathyacmaea nipponica (Patellogastropoda: Pectinodontidae). Mitochondrial DNA Part B: Resour. 4 (2), 3175–3176. doi: 10.1080/23802359.2019.1668732
Uribe J. E., Colgan D., Castro L. R., Kano Y., Zardoya R. (2016b). Phylogenetic relationships among superfamilies of neritimorpha (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 104, 21–31. doi: 10.1016/j.ympev.2016.07.021
Uribe J. E., Irisarri I., Templado J., Zardoya R. (2019). New patellogastropod mitogenomes help counteracting long-branch attraction in the deep phylogeny of gastropod mollusks. Mol. Phylogenet. Evol. 133, 12–23. doi: 10.1016/j.ympev.2018.12.019
Uribe J. E., Kano Y., Templado J., Zardoya R. (2016a). Mitogenomics of vetigastropoda: Insights into the evolution of pallial symmetry. Zool. Scr. 45 (2), 145–159. doi: 10.1111/zsc.12146
Wernersson R., Pedersen A. G. (2003). RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 31 (13), 3537–3539. doi: 10.1093/nar/gkg609
Keywords: mitochondrial genome, Acmaeidae, Pectinodontidae, Lottiidae, Patellogastropoda, phylogeny
Citation: Putri ET, Lee D, Kwak H, Kim Y, Nakano T and Park J-K (2023) Complete mitochondrial genomes of the “Acmaeidae” limpets provide new insights into the internal phylogeny of the Patellogastropoda (Mollusca: Gastropoda). Front. Mar. Sci. 10:1134991. doi: 10.3389/fmars.2023.1134991
Received: 31 December 2022; Accepted: 20 March 2023;
Published: 26 April 2023.
Edited by:
Khor Waiho, University of Malaysia Terengganu, MalaysiaReviewed by:
Yingying Ye, Zhejiang Ocean University, ChinaLeila Belén Guzmán, Instituto de Biología Subtropical, Argentina
Jun Liu, Institute of Deep-Sea Science and Engineering, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Putri, Lee, Kwak, Kim, Nakano and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joong-Ki Park, amtwYXJrQGV3aGEuYWMua3I=
†These authors have contributed equally to this work
 Eggy Triana Putri
Eggy Triana Putri Damin Lee
Damin Lee Haena Kwak
Haena Kwak Yukyung Kim
Yukyung Kim Tomoyuki Nakano
Tomoyuki Nakano Joong-Ki Park
Joong-Ki Park