- 1Department of Biological Sciences and Biotechnology, College of Natural Sciences, Chungbuk National University, Cheongju, Republic of Korea
- 2Clinical Research Division, National Institute of Food and Drug Safety Evaluation, Ministry of Food and Drug Safety, Cheongju, Chungcheongbuk-do, Republic of Korea
- 3Department of Anatomy, College of Medicine, Chungbuk National University, Cheongju, Republic of Korea
- 4Department of Life Science, College of Natural Sciences, Chung-Ang University, Seoul, Republic of Korea
- 5Department of Genetic Resources Research, National Marine Biodiversity Institute of Korea, Seocheon, Republic of Korea
The eye of a cephalopod is a well-known example of convergent evolution and resembles the vertebrate eye. Although cephalopods and vertebrates exhibit similar eye form and function, they differ in visual origin and structure. The common long-arm octopus (Octopus minor) is a good model system in evolutionary and developmental studies due to its highly centralized nervous system, shorter life cycle, and specific camera-type eyes that contribute to convergence with vertebrate eye. Lens-containing eyes represent a significant improvement of simple eye and have evolved by convergent mechanisms, a variety of lenses and corneas containing diverse crystallin. The diversity and taxon-specificity of lens crystallin is indicative of convergent evolution of crystallin roles. Previous studies have focused on morphological, ontogenetic and phylogenetic analysis of crystallin to understand the evolution of lens-containing eyes. However, little is known about the functional analysis of taxon-specific crystallin genes at the molecular level in the eye of O. minor. Using an embryonic staging system of Octopus minor as a model system, we investigated fifteen genomes and the structure of eye by immunohistochemistry, phalloidin staining and the three-dimensional structures. We also obtained the crystallin-related genes (i.e., α-, S-, and Ω-crystallin) from the transcriptome data of O. minor. Subsequent molecular phylogenetic analysis based on these genes revealed a distinct divergence pattern among the three gene classes and further suggested the evidence supporting the taxon-specific convergent evolutionary trend. We analyzed the expression pattern of crystallin genes via in situ hybridization during developmental stages. All crystallin genes are commonly expressed in the lentigenic cells of ciliary body. The α-crystallin found in cephalopods was also expressed at the peripheral region of the lens including ciliary body, suggesting a possible role in lens formation in cephalopods. This study will provide information on the eye development of O. minor and support the typical models of convergent evolution by demonstrating independent recruitment of different types of proteins to fulfill their unique visual role.
Introduction
Animal eyes are photoreceptive organs for collecting light from the surrounding environment and elicit response to various stimuli, including visual system. These photoreceptive organs are diverse in structure and range from light-sensitive cells to complex structures for precise images in some group of arthropods, mollusks, and vertebrates (Ogura et al., 2004). Phylum Mollusca has developed different types of eyes such as the concave mirror eye, the pinhole eye, and the camera-type eye with considerable variation (Land, 1965; Land and Fernald, 1992; Serb and Eernisse, 2008; Ogura et al., 2013). Coleoid cephalopods such as octopuses, cuttlefish and squid possess camera-type eye which consists of an iris, a circular lens, vitreous cavity (eye gel), photoreceptor cells, similar to vertebrate eyes (Serb, 2008). Despite the similarities in overall structure, the role of opsin family members in phototransduction within the photoreceptors, the camera-type eyes of vertebrates and cephalopods are thought to have evolved independently and thus represent a classic example of convergent evolution (Yoshida et al., 2015).
There have been reports on the evolution of image-forming eyes in cephalopods of invertebrates with camera-type and in vertebrate (Zinovieva et al., 1999; Gehring, 2005; Yoshida et al., 2015). The Pax6, as common master control gene for eye development, especially in ocular primordia, optic ganglia, and the photoreceptor organ, was expressed in both types of eyes, indicating that the divergence of these two types of eyes from a single prototype eye exists in the common ancestor of cephalopods and vertebrates (Ogura et al., 2004). In addition, a larger number of conserved genes with similar expression, as well as specific genes such as Ets-4, cornichon, and centaurin gamma homologs, may be responsible for the convergent evolution of the camera-type eye (Yoshida et al., 2015). Differences in the composition of lens proteins such as crystallin and their regulators in the camera-type eyes of vertebrates compared with cephalopods are attributed to diverse genes, resulting in convergent morphological evolution of the camera eyes (Jonasova and Kozmik, 2008).
Primitive eyes can only provide information about light intensity and direction. Additional optical elements such as lens and cornea increase light capture and produce an image with dramatically improved optical performance (Jonasova and Kozmik, 2008). The lens evolved initially as the protein-filling structure for concentrating the incident light from the environment onto the receptor cells of the retina in the eye (Chiou, 1988). The transparency and refractive power of lens and cornea are attributed to water-soluble proteins known as crystallin (Piatigorsky, 2003). The diversity and taxon-specificity of lens crystallin throughout the animal kingdom is indicative of convergent evolution of crystallin roles (de Jong et al., 1993). Proteins used as lens crystallin are often related to ubiquitously expressed metabolic enzymes of physiological stress proteins (de Jong et al., 1993). Cephalopod lenses are mainly composed of S-crystallin and Ω-crystallin (minor proteins), which have evolved from glutathione S-transferase (GST) and aldehyde dehydrogenase (ALDH), respectively (Piatigorsky, 1998). The three major gene families of crystallin in vertebrate lenses include α-crystallin, β-crystallin, and γ-crystallin. The β- and the γ-crystallin are evolutionarily related protein families that constitute most of the refractive structure of vertebrate eye lens (Bloemendal et al., 2004). α-crystallins are key members belonging to the superfamily of small heat shock proteins (sHSPs) which have been studied in the ocular lens (Slingsby et al., 2013). The novel functions of α-crystallin has been identified in the retina and in the retinal pigmented epithelium (Kannan et al., 2012) and has a structural function in the lens, contributing to the high protein concentration in the lens fiber cells (Horwitz, 2000).
Among the cephalopods, O. minor is a good model in evolutionary and developmental studies due to its highly centralized nervous system, its relatively small size, shorter life cycle (approximately one year) and specific camera-type eyes that contribute to convergence with vertebrate eyes (Shigeno et al., 2018). In addition, research resources such as whole genome information (Kim et al., 2018) and brain atlas (Jung et al., 2018) make O. minor advantageous for studying convergent evolution. However, in contrast to previous studies investigating the eye development in squid and cuttlefish, studies on the eye development process at the molecular level in octopus until now are insufficient, although many general anatomical and physiological studies of octopus visual system have been reported (Hanke and Kelber, 2020). There have been many comparative genome studies for clarifying genetic mechanism that contributed to the evolution of the cephalopod camera eye within the Mollusca (Yoshida and Ogura, 2011). Recently, the genome of Nautilus pompilius, a non-coleoid cephalopod, was also analyzed to investigate eye evolution within Mollusca (Zhang et al., 2021), suggesting that independent gene losses in nautilus and expansion of crystallins in coleoids may be instrumental in driving eye evolution in cephalopods. However, little is known about the functional analysis of taxon-specific crystallin genes at the molecular level in the eye of O. minor in cephalopods. In this study, we investigate the morphology of eye during the development using histological and molecular assays. We attempted the molecular phylogenetic analysis involving the crystallin-related genes from diverse animal groups to explore an evolutionary trend of divergence or convergence of the genes, and consequently suggested that the cephalopod genome contains S-, Ω-, and α-crystallin. Then, we performed in situ hybridization to determine the spatial and temporal expression of S-, Ω-, and α-crystallin during eye development in O. minor.
Materials and methods
Animal
To obtain O. minor embryos, sexually mature females and males are purchased from commercial catches in the coastal regions of Jeollanam-do, Republic of Korea, in May of 2017-2019. The species and sex were identified by morphology. All experiments were designed to minimize the number of animals and their suffering in accordance with management of cephalopods in the laboratory (Boyle, 1991; Sakaue et al., 2014). After mated with a male octopus for 7 days, each female octopus was isolated in a spawning net and then maintained in the fresh seawater tank at Jeollanam-do Ocean & Fisheries Science Institute (Sinan, Jeollanam-do). Mated females were monitored and fed with Manila clams until they spawned. Spawning dates and seawater conditions were checked daily. At the required developmental stage, embryos were moved and incubated in artificial culture system in dark condition. Temperature was 16°C and Salinity was 33‰ (Practical salinity unit) with artificial sea salt (Reef Crystals, Aquarium systems). Only embryos were separated from yolk and were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (PBS) and stored at 4°C until use.
Histological analysis
Samples (about 5 to 10 samples taken from each stage) were fixed in 4% PFA in PBS for overnight at 4°C. For H&E (hematoxylin and eosin) staining and immunohistochemistry, samples were dehydrated in sucrose series and embedded in O.C.T. compound (VWR) and rapidly frozen in liquified nitrogen (Jung et al., 2018). Cryo-sectioned samples (6 µm) were cut with a CM1520 cryostat (Leica) and stored at –70°C until use.
Immunohistochemistry and phalloidin staining
To detect F-actin, we incubated fixed embryos in Texas-red-conjugated phalloidin (Texas Red™-X phalloidin, Invitrogen T7471: PBS = 1:50) overnight. For immunohistochemistry, fixed samples were incubated in the blocking solution (Sigma, 11096176001, St. Louis, MO, USA) overnight, then incubated with mouse anti-acetylated α-tubulin antibody (1:500 in blocking solution; Sigma, T-7451, St. Louis, MO, USA) at room temperature for 1 day. Samples were washed 3 times for 10 min and overnight in PBT (1× PBS + 0.1% Tween20). After several washes, the secondary antibody (1:1000 in PBT; Alexa Fluor 488 Goat anti-mouse ab150113, Abcam, Cambridge, UK) was applied for 1 day at room temperature. For nuclei staining, samples were incubated in DAPI (4′,6-Diamidino-2-phenylindole dihydrochloride) solution (1:1000 in PBT; Thermo Fisher Scientific, D1306, Waltham, MA, USA) for 30 min. After several times of washing in PBT, samples were mounted in Fluoromount-G (Southern Biotech, Birmingham, AL, USA).
Scanning electron microscope (tissue sampling)
The tissue of eye was cut into 0.5-1mm3 and fixed in 4% PFA for overnight. Thereafter, the tissues were washed with PBS and post-fixed in dark condition with 1% Osmium tetroxide (wPBS). And then the tissues were washed with PBT and dehydrated with EtOH series, followed by Isopentyl acetate (Alfa Aesar): EtOH series, and 100% Isopentyl acetate. And the samples were dried for 3 days under dark condition in a hood. The sample was photographed using a Field Emission Scanning Electron Microscope (ULTRA PLUS) of the Center for Research Facilities of Chungbuk National University.
Crystallin gene search and identification
To obtain the crystallin genes in the transcriptome data of O. minor, the protein sequences of the α-, S- and Ω-crystallin genes from relatively well-studied organisms were priorly acquired through the UniProt database (https://www.uniprot.org): two sequences of α-crystallins from Homo sapiens (human), seven S-crystallins from Enteroctopus dofleini (the giant Pacific octopus) and Octopus vulgaris (the common octopus), and one Ω-crystallin each from Nototodarus sloanii (the New Zealand arrow squid) and E. dofleini (see Supplementary Table 1). Using these sequences as the initial search queries, the putative orthologous genes were sought out from our legacy data of a transcriptome assembly for O. minor using BLAST (standalone tblastn, ver.2.10.0+) software (Altschul et al., 1997; Camacho et al., 2009). The obtained nucleotide sequences were translated into the amino acid sequences on the basis of the standard genetic code, and those amino acid sequences were verified again in another BLAST (blastp, https://blast.ncbi.nlm.nih.gov/Blast.cgi) search at the National Center for Biotechnology Information (NCBI). Additional crystallin gene searches for other 15 animal species in five phyla (Porifera, Cnidaria, Echinodermata, Chordata and Mollusca) were also conducted. For this, the preassembled protein data of these organisms were downloaded from the genome database (https://www.ncbi.nlm.nih.gov/genome) of the GenBank at NCBI. The orthologs of the crystallin genes were sought out from each downloaded dataset based on the initial query sequences and the inspected O. minor sequences using OrthoVenn2 (Xu et al., 2019) (https://orthovenn2.bioinfotoolkits.net). The newly obtained nucleotide sequences of O. minor were submitted to GenBank under following accession numbers: MZ269473-MZ269478.
Phylogenetic analysis
All genes were merged into one set, and the multiple sequence alignment was then generated by MAFFT (ver.7.475) software (Katoh et al., 2002; Katoh and Standley, 2013). The Best-fit model selection for the phylogenetic tree reconstruction was sought out using IQ-TREE2 (ver.2.1.2) software (Kalyaanamoorthy et al., 2017; Minh et al., 2020) based on the corrected Akaike Information Criterion (AICc). The phylogenetic tree was reconstructed with initial 3,000 pseudo-replicates based on “LG+F+G4” model using RAxML-NG (ver. 1.0.1) software (Kozlov et al., 2019). The bootstrap supporting values for the branches were calculated under the transfer bootstrap expectation (Lemoine et al., 2018). The tree visualization was carried out using FigTree (ver. 1.4.4) software.
Developmental and tissue specific semiquantitative RT-PCR
The PCR reactions were performed under the following cycling conditions: pre-denaturation at 98°C for 5 min, followed by 30–40 cycles of denaturation at 98°C for 30 s, elongation at 72°C for each sequence length related times, and a final elongation step at 72°C for 5 min. A 10 μL aliquot of each PCR reaction was removed after 28 cycles, while the remaining material underwent five additional cycles of amplification (for the details of primer information, see Supplementary Table 2).
Gene identification, gene cloning, and probe synthesis
We isolated total RNA from O. minor embryos of different developmental stages using TRIzol (Ambion, Austin, TX, USA). We selected mRNA from RNA using Oligo (dT) primers (Promega, Madison, WI, USA) and then conducted reverse transcription into cDNA (SuperScript II First-Strand Synthesis System for RT-PCR, Invitrogen, Waltham, MA, USA). The primers of the crystallin gene were the same as the primer of Semiquantitative RT-PCR.
The amplified genes fragments were subcloned into pGEM T vectors (Promega, Madison, WI, USA) and sequenced to confirm their identities. Then, these subcloned inserts were amplified using universal SP6 and T7 promoter primers (SP6 primer: 5’- TATTTAGGTGACACTATAG -3’; T7 primer: 5’- TAATACGACTCACTATAGG -3’). The PCR-amplified linear templates were used to synthesize an in vitro-transcribed antisense riboprobe (MEGAscript Transcription kit, Ambion, Austin, TX, USA). The genes are available under accession numbers XM_036502845 (Omi_scry2), XM_029799677.2 (Omi_scry4), XM_036498903.1 (Omi_cryab), and XM_029787702.2 (Omi_crom). Digoxigenin-labeled RNA probes were synthesized from the cloned fragments. The sizes of the probes were Omi_crom 924bp, Omi_scry2 846bp, Omi_scry4 766bp, and Omi_cryab 945bp, respectively.
In situ hybridization in O. minor
Fixation of the embryos was performed with 4% paraformaldehyde (PFA, Sigma-Aldrich, Saint Louis, MO, USA) for overnight. The fixed embryos were washed 5 × 5 min in 0.1% PBT (phosphate-buffered saline and 0.1% Tween 20) and permeabilized by treatment with 0.5 mg/mL proteinase K (Biofact, Daejeon, South Korea) in PBT for 10 min at room temperature. Proteinase K treatment was followed by 3 × 5 min rinses in a solution containing 2 mg/mL glycine in PBT, and 2 × 10 min washes in PBT at room temperature. Following additional 10 min washes in 0.5 mL 0.1 M triethanolamine buffer (TEA, pH 8.0, Sigma-Aldrich, Saint Louis, MO, USA), they were treated for 5 min with 0.5 mL of TEA supplemented with 2.7 µL of acetic anhydride. The embryos and section samples were then rinsed 3 × 5 min in PBT and post-fixed for 1 h in 4% PFA. The embryos and section samples were washed five times for 3 × 5 min each in PBT at room temperature. Further protocol conditions were identical to those described by Cho et al. (Cho et al., 2010).
Results
Embryonic stages of O. minor
The embryogenesis of O. minor shows the typical developmental process of cephalopods such as early embryo is telolecithal, and the movements of gastrulation are epibolic. We investigated and adopted the morphological criteria defined for Octopus bimaculoides (Shigeno et al., 2015) and Octopus vulgaris (Naef, 1928) to stage the embryonic development of O. minor based on morphological characteristics (Figure 1). Embryos attached to the net were cultured in vitro under specific conditions (Figure 1B). Stage 8 embryos developing on day 25 after spawning were found in the lower region of the yolk. The embryos showed peristaltic movements of yolk and the primordia of eye were apparent, but not pigmented. Stage 10 embryos (day 30) showed distinct orange eye pigmentation. At stage 12 (day 33), mantle formation was initiated; however, it did not completely cover the internal organs. In addition, the embryo was located at the far end of the yolk, with suckers developing on the arms, and the eyes gradually darkening from brown to black. The heartbeat was visible in stage 14 embryos (day 35), and arm movement was detected at stage 16 (day 38). The stage 18 (days 40 to 45) embryo showed pigmented internal organs, arm movements, and clearly demarcated brain and optic lobes. Subsequently, the yolk-sac size decreased rapidly from stages 24 (day 50 to 60) to 28 (day 70 to 80), and the pigment in the internal organs and arms increased. Alongside the development of the mantle and tissues, the embryo manifested camouflage and active motility (Figure 1C). The embryos were selected from stage 10, which is suited for cryo-section, to stage 18-20 when the outer segment of the photoreceptor is formed for morphological and molecular studies of O. minor eye.
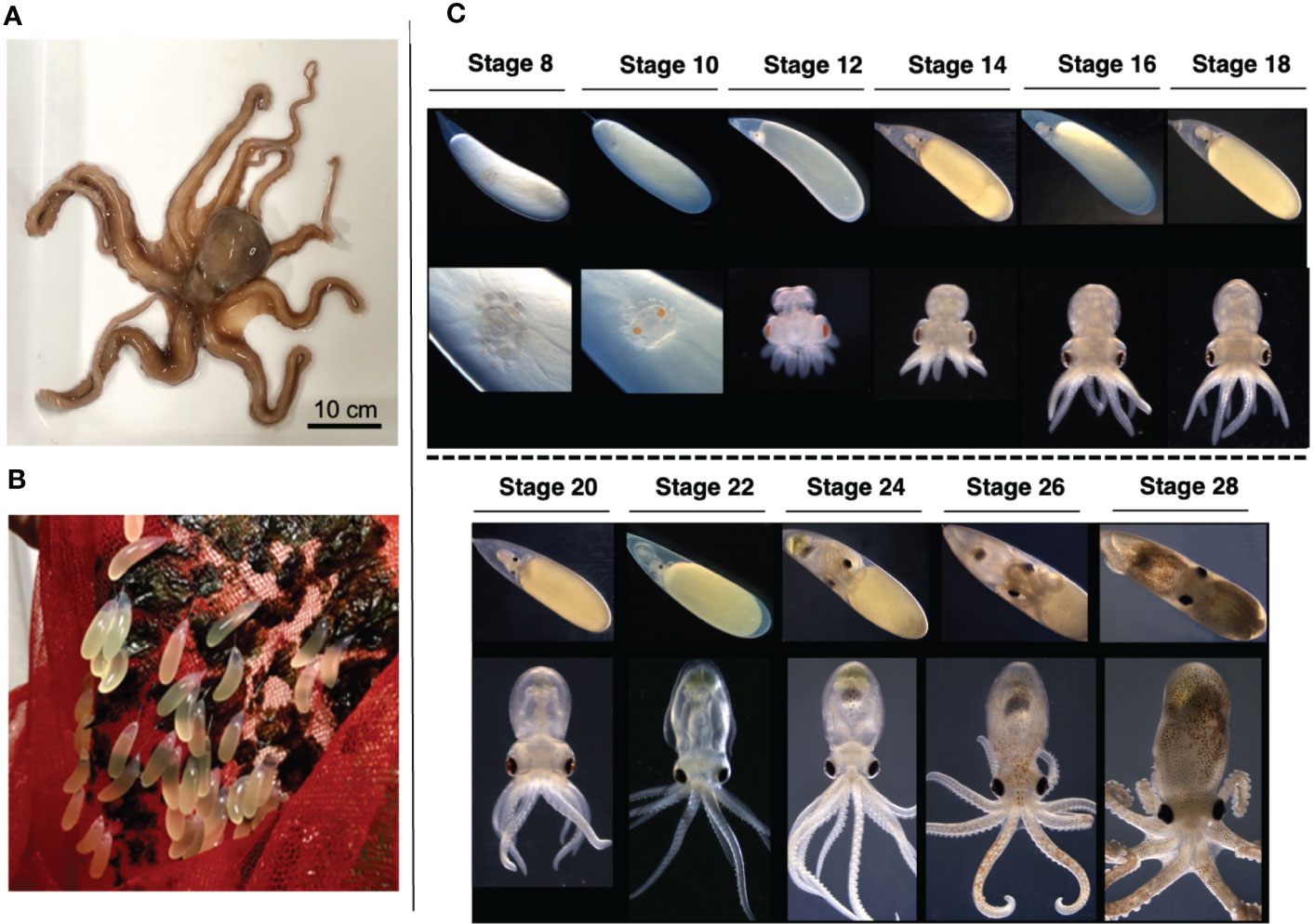
Figure 1 O. minor as an experimental model (A) Adult of an O. minor. (B) Embryo culture system of an O. minor. Cocoon is attached by an attachment substance. Each cocoon has a long diameter of 10–20 mm, a short diameter of 5 mm. (C) Developmental stages of an O. minor. The early embryo of stage 8 was first observed from about 25 days in the lower part of the yolk after spawning. Stage 10 embryos showed distinct orange eye pigmentation. Mantle formation was initiated at stage 12. The heartbeat was visible in stage 14 embryos. Stage 18 embryo showed pigmented internal organs, arm movements, and clearly demarcated brain. Scale bar: 10cm.
Morphological studies of O. minor eye during developmental stages
To investigate the eye structure during the development, we performed H&E staining and immunohistochemical analysis using anti-acetylated α-tubulin and phalloidin staining (Figure 2). The anti-acetylated tubulin and phalloidin staining are used to visualize the axonal processes and general neuroanatomy (Gross and Mayer, 2015; Baldascino et al., 2017), and the distribution of F-actin in muscle cells (Jahnel et al., 2014), respectively.
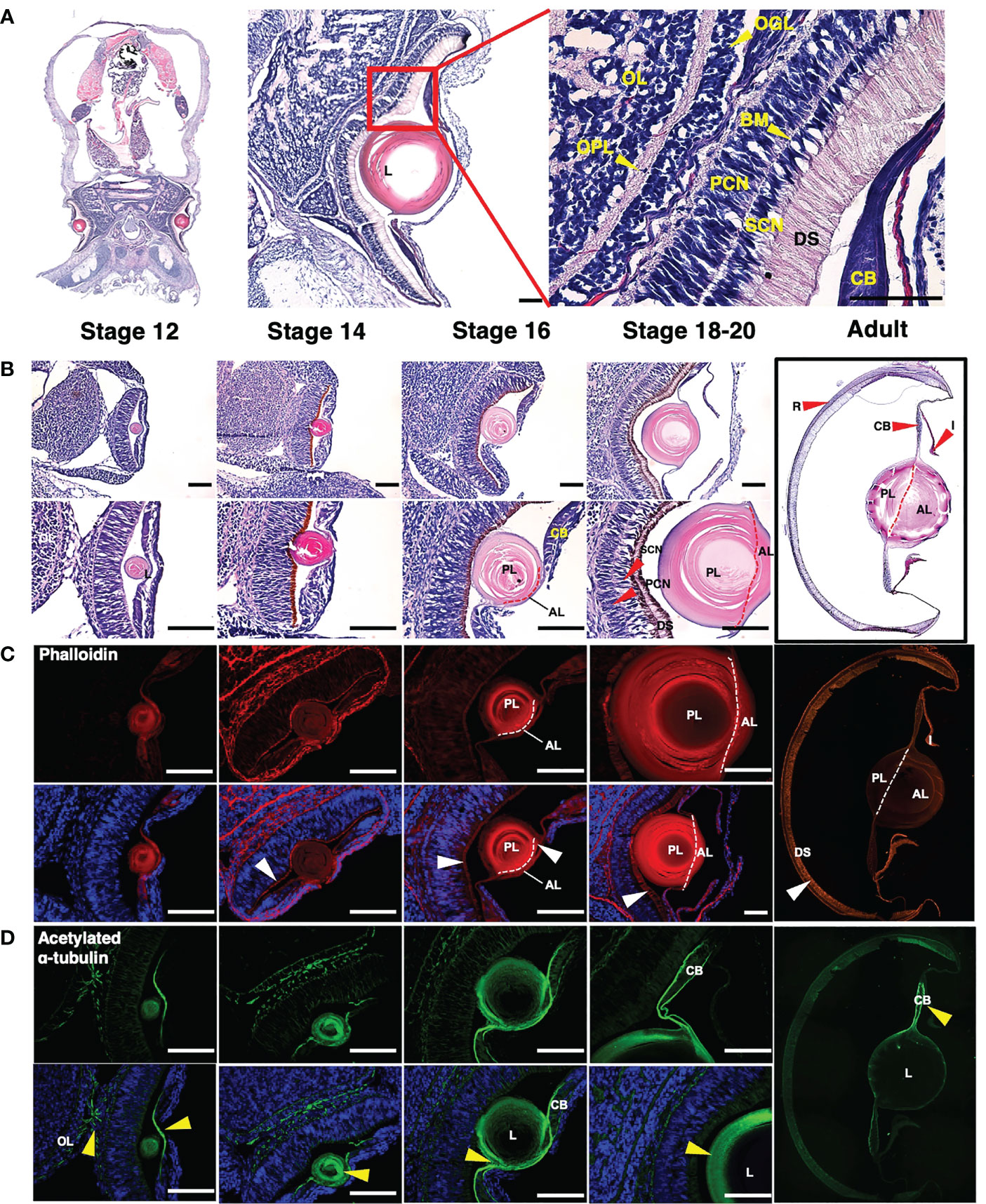
Figure 2 Structural analysis of histological and molecular of an O. minor eye during developmental stages (A, B) Images of H&E staining in O. minor at the longitudinal sectional view. (A) The histological structure of the O. minor visual system is composed of CB, DS, SCN, PCN, BM, OGL, OPL, and OL from the outside to the inside. (B) As the developmental stage progresses, the size of the lens increases, and from stage 16, it is separated into anterior and posterior lenses as indicated by a red dashed line. In addition, in stages 18-20, SCN, PCN, and DS are clearly distinguished. (C) Images of phalloidin staining (red) and DAPI counterstaining (blue) in O. minor eye. As shown in the H&E staining of (B), it was confirmed that it is divided into anterior and posterior lenses at stage 16 as indicated by a white dashed line. (D) Localization of acetylated α-tubulin (green) and DAPI counterstaining (blue) in O. minor eye. Scale bar: 100μm. AL, anterior lens; BM, basal membrane; CB, ciliary body; DS, distal segments of photoreceptors; L, lens; OGL, outer granule cell layer; OPL, outer plexiform layer; OL, outer layer; PCN, photoreceptor cell nuclei; PL, posterior lens; R, retina; SCN, supporting cell nuclei; White arrowhead indicates F-actin enrichment location. Yellow arrowhead indicates acetylated α-tubulin distribution and optic chiasm in eye. Each column in (B–D) represents the same stage. The images were taken with a Leica DM6 B microscope. Mosaic images (2A and adult images) were taken by EVOS FL Auto2 (Invitrogen) microscope.
In contrast to vertebrates, the distal segments of the photoreceptors (DS) were arrayed anteriorly in the first region exposed to light, followed by a layer of supporting cell nuclei (SCN), basal membrane (BM) and photoreceptor cell nuclei (PCN) located at the posterior of the retina. O. minor eye also contained an outer plexiform layer (OPL) behind the outer granule layer (OGL) for integration of visual information (Figure 2A).
To elucidate eye development during embryogenesis, we compared the embryos at stages 12 to 20 with adult (Figure 2B). At stage 12, the lens was clearly visible, and the pigment color of the DS turned darker in later stages. The posterior lens formed first, followed by the anterior lens, which was apparent at stages 16-18 (Figures 2B, C). Further, the SCN and PCN layers were clearly distinguished from the basal membrane in the retina with the elongated DS. The lens constituted a high proportion of the adult O. minor eye, and the anterior and posterior lenses were clearly distinguished. Also, compared with the embryonic stage, the DS were predominant in the adult retina (Figure 2B). Next, we investigated the distribution and arrangement of F-actin in muscle during eye development via phalloidin staining (Figure 2C). The DS was stained as described in the previous study of cephalopods (Koenig et al., 2016). The DS carried an increasing number of F-actin fibers according to the developmental stage. The configuration of circular F-actin fibers inside the lens was similar to the F- actin organization in the wild-type lens of zebrafish embryo (Hayes et al., 2012). To identify the distribution and arrangement of nerves during the eye development, we performed immunostaining assays using an acetylated α-tubulin antibody, followed by counterstaining with DAPI (Figure 2D). In stage 12, the optic chiasm, in which nerves intersect with optic lobes, was observed. In addition, nerves were distributed in the area containing lentigenic cells derived from the ectoderm (yellow arrowhead), which is likely a result of the growth of long cytoplasmic projections from the lentigenic cells to make the lens. Acetylated α-tubulin in the lens were gradually concentrated toward the outer side of the lens in stage 14 and 16 and the nerve fibers were distributed on the border of the ciliary body (CB) at stage 18 (Figure 2D).
Morphological analysis of adult O. minor eye
Morphological analysis was performed via SEM (Figures 3B, C) and immunohistochemistry of muscle and nerve in the adult eye (Figure 3A). The nerves of the CB extended from the outer edge to the inside. F-actin extended along the midline and branched between the nerves (Figure 3A). F-actin was enriched in the iris without nerve fibers. F-actin was distributed at a high density in the DS in the retina.
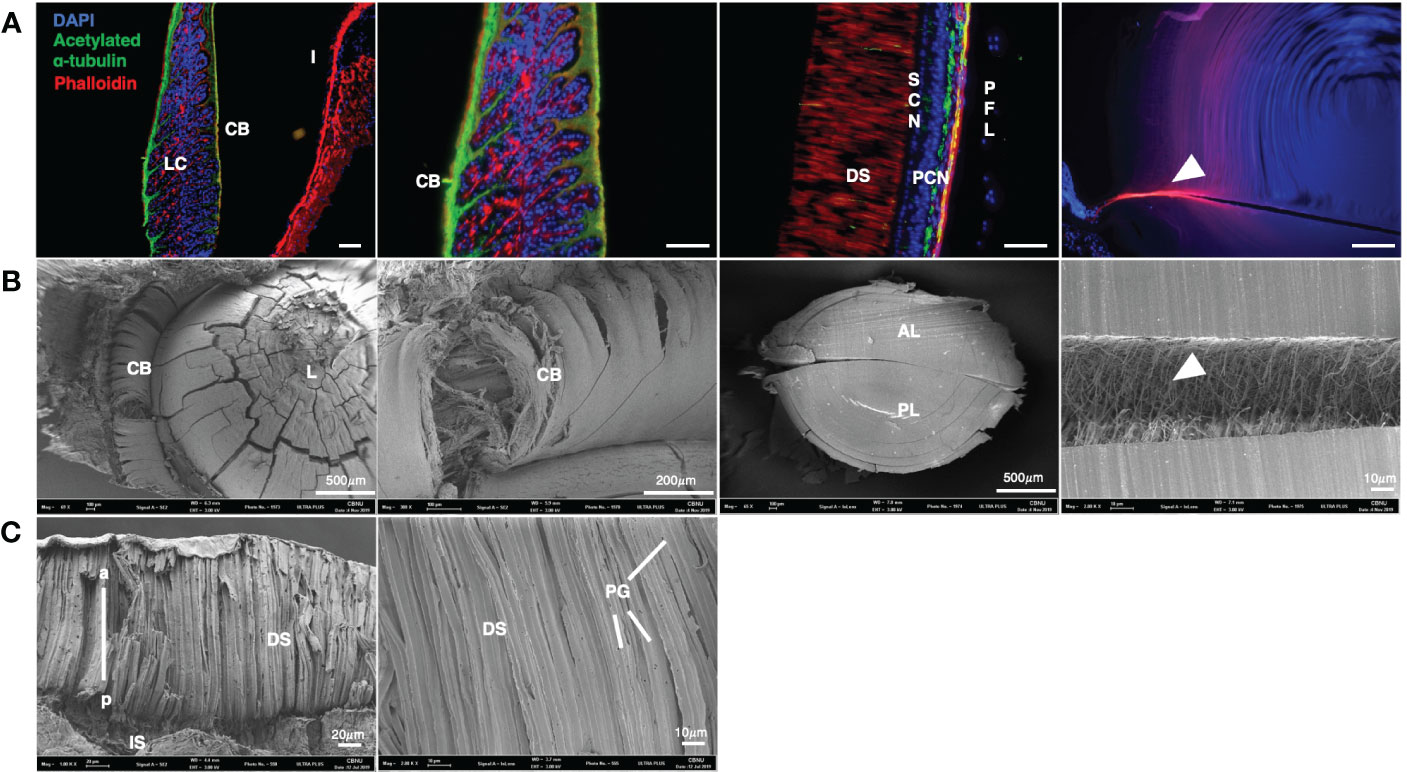
Figure 3 Molecular characterization and SEM images of an O. minor eyes (A) Colocalization of acetylated α- tubulin and F-actin in eye of O. minor at adult stage. The CB is composed of a large amount of F-actin in the structure of the O. minor visual system. (B) Scanning electron microscope images in lens and ciliary body of O. minor at adult stage. It showed that large amount of F-actin is distributed between the anterior and posterior lenses as indicated by white arrowhead in (A, B). (C) Scanning electron microscope images in O. minor retina at adult stage. It is shown that there are PGs in the DS of retina. Scale bar: 100μm. CB, ciliary body; DS, distal segments of photoreceptor; I, iris; LC, lentigenic cells; PCN, photoreceptor cell nuclei; PFL, plexiform layer; PG, pigment granule; SCN, supporting cell nuclei. White arrowhead indicates F-actin distribution. The images were taken with a Leica DM6 B microscope and Field Emission Scanning Electron Microscope (ULTRA PLUS).
To elucidate the 3D structure of O. minor eyes in the adult stage, the retina, lens, and ciliary elements were analyzed by SEM (Figures 3B, C). The lens was ensheathed by the CB consisting of numerous fibers and there were numerous microfibers in the septum. As shown in Figure 3A, the CB is composed of nerves and bundles of F-actin. Phalloidin staining also confirmed the presence of muscle fibers in the microfibers, indicating the connection between anterior and posterior lenses via F-actin fibers (Figures 3A, B; white arrowhead). DS constitutes a large proportion of the adult retina and contains pigment granules (PG) (Figure 3C) (Hanke and Kelber, 2020).
Phylogenetic analysis of crystallin genes
A total 66 crystallin-related genes from diverse animal phyla were obtained and used to reconstruct the phylogenetic tree; Among those genes involved, one α-crystallin B chain (GenBank accession number MZ269473), two S-crystallins 2 and 4 (MZ269474, MZ269475), and one Ω-crystallin (MZ269476) and two closely related retinal dehydrogenases (MZ269477, MZ269478) were newly determined from our transcriptome data of O. minor. The phylogenetic tree resolved three well-diverged gene classes of α-crystallin/small heat shock protein family, glutathione S-transferase protein family and aldehyde dehydrogenase protein with high maximum-likelihood bootstrap (MLB) support values (MLB > 95%). Each gene family carried the newly identified α-, S- and Ω-crystallin genes of O. minor (Figure 4).
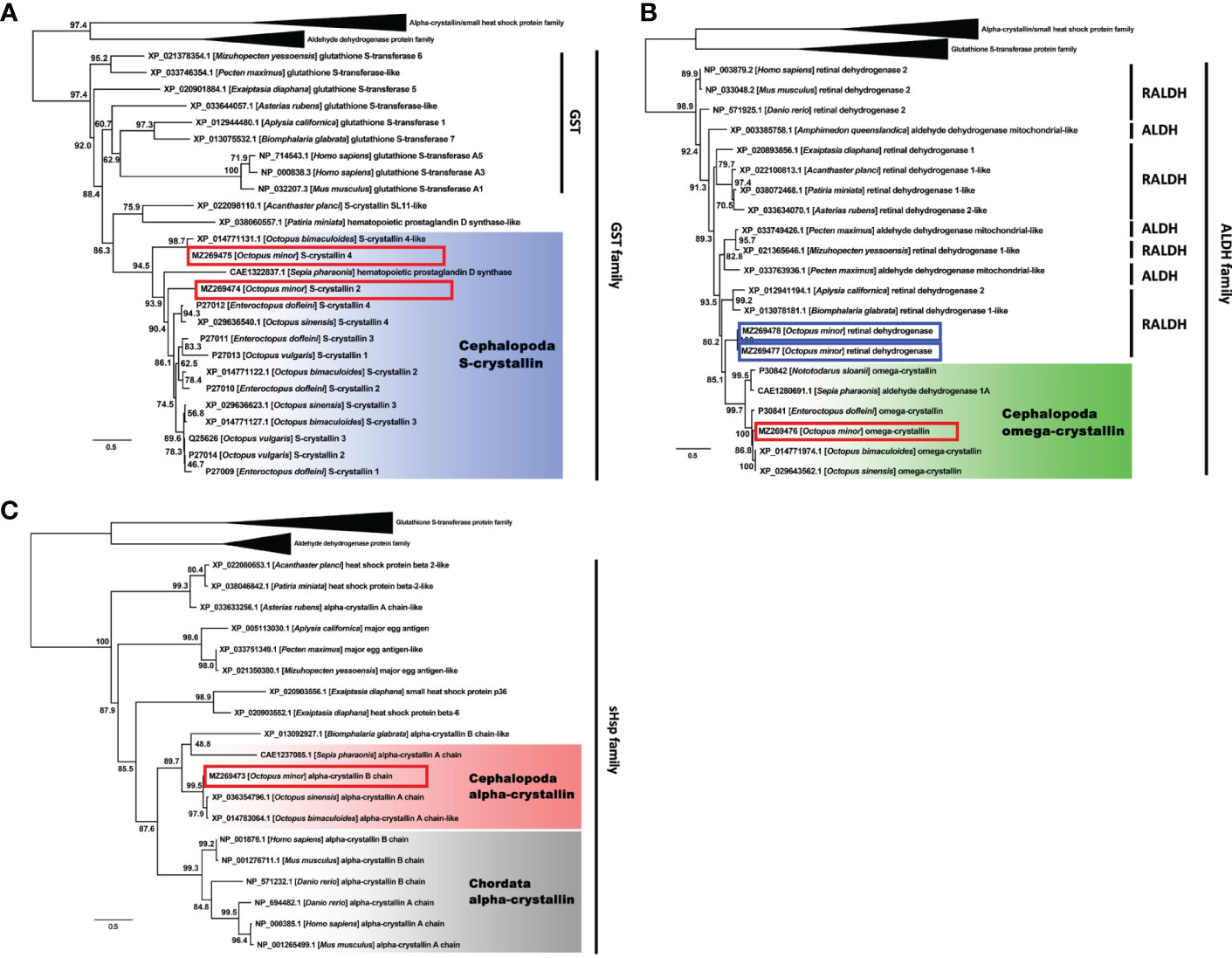
Figure 4 Phylogenetic tree showing a clade containing the S- crystallin (A), Omega- crystallin (B) and α-crystallin (C) of Cephalopoda with the glutathione S-transferase, aldehyde dehydrogenase and small heat shock protein families. Genes from Cephalopoda were highlighted in blue, green and red and those from Chordata were in grey. Red box indicates O. minor gene. Blue box indicates crystallin-related genes, but not crystallin genes. Numbers near branches indicate the bootstrap supporting values.
The sub-phylogenetic tree of α-crystallin contained additional sHSPs (mainly derived from Echinodermata and Cnidaria) and major egg antigens (from Gastropoda and Bivalvia). Excluding those genes, there were two distinct α-crystallin groups belonging to Chordata and Cephalopoda (MLB = 88%; Figure 4C, grey- and red-highlights). Two genes annotated as ‘α-crystallin’ in Asterias rubens (Echinodermata) and Biomphalaris globrata (Gastropoda) were found in unexpected phylogenetic positions. Nonetheless, our α-crystallin B chain gene was still strongly related to the genes derived from the congeneric O. bimaculoides and O. sinensis. The S-crystallin subtree showed similar orthologue clustering for the cephalopod S-crystallins separate from the evolutionarily related glutathione S-transferases (MLB = 86%; Figure 4A, blue-highlight). The subtree for the Ω-crystallins also presented a monophyletic clade of the cephalopod Ω-crystallin genes (MLB = 85%; Figure 4B, green-highlight). Two additional O. minor genes, both identified as the retinal dehydrogenase in this study, were differentiated from the Ω-crystallin gene group. The gene identity of the present O. minor sequence was eventually established as Ω-crystallin.
Temporal expression of crystallin genes during eye development
We suggested the presence of small heat shock protein, glutathione S-transferase and aldehyde dehydrogenase families acting as a crystallin in cephalopods through phylogenetic analysis. Thus, to examine the temporal expression of crystallin genes during eye development, we performed a semi-quantitative RT-PCR using Omi-beta actin as an internal control with RNA samples derived from several stages (10, 14, 18 and 20) (Figures 5A, B) during eye development. Omi-crom was weakly expressed in all stages (10, 14, 18 and 20). Omi-scry2 and Omi-scry4 were strongly expressed in late stages 18, 20 and 20, respectively (Figures 5A, B). Interestingly, unlike S-, Ω-crystallin genes, Omi-cryab was expressed at all stages and gradually increased during eye development, particularly showing strong expression pattern in stage 20 (Figures 5A, B), suggest that α-crystallin, which is known to be the major protein of the mammalian lens in most species (Augusteyn, 2004), is also involved in eye development in cephalopods (Figure 4). All crystallin genes were not expressed in the trunk as a control.
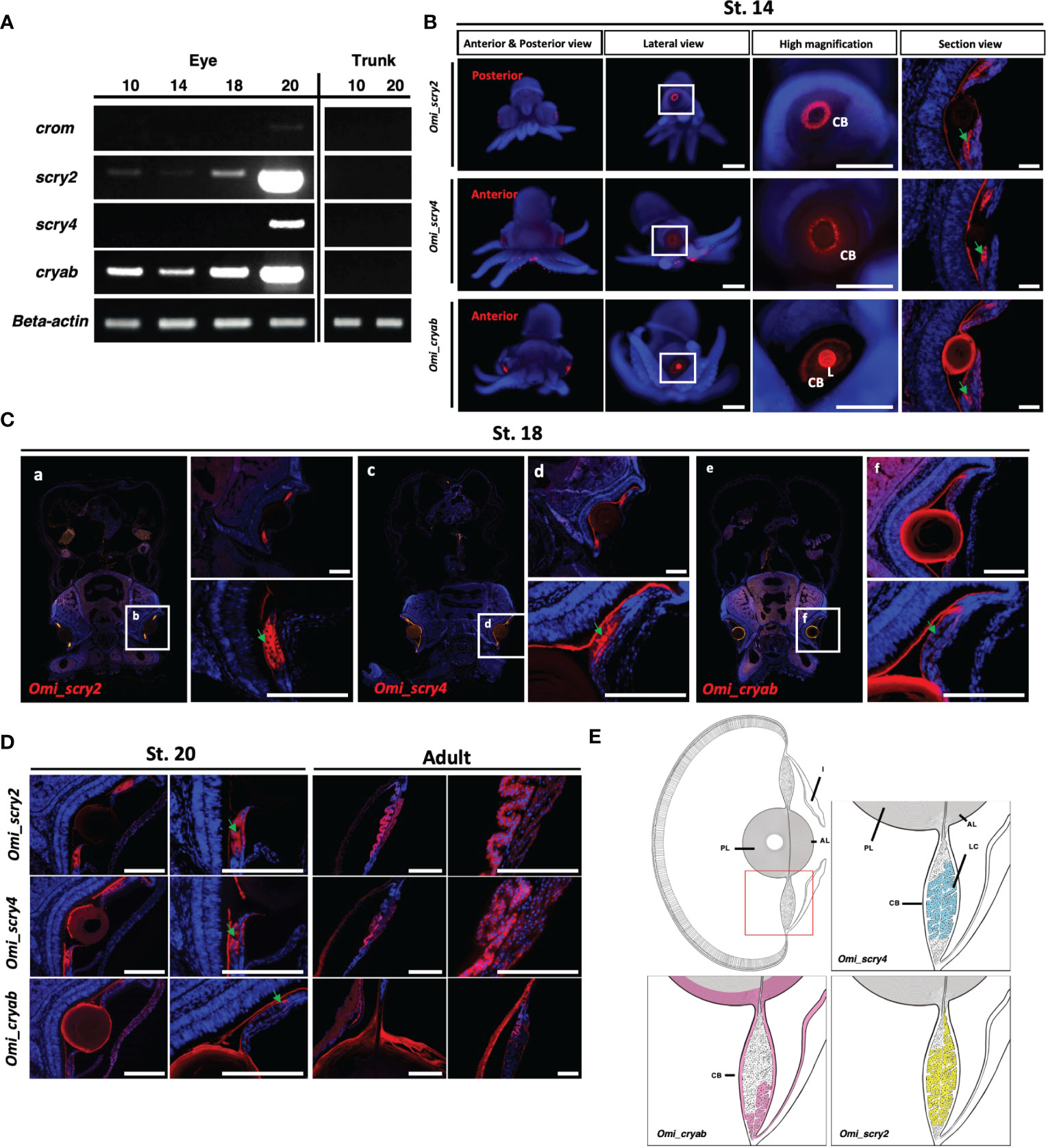
Figure 5 Expression analysis of crystallin genes in O. minor(A) Expression level confirmation from semi-quantitative RT-PCR. Standard deviations were calculated from three independent sq RT-PCR experiments. (B) Highly expressed genes in situ hybridization at St. 14 crystallin. (C, D) Section in situ hybridization during developmental stages (St. 18, St. 20, and adult) in O. minor. (E) Schematic drawings of crystallin gene expression in O. minor. AL, anterior lens; CB, ciliary body; I, iris; L, lens; LC, lentigenic cells; PL, posterior lens. Green arrow indicates the lentigenic cell. The section images were taken with a Leica DM6 B microscope. Scale bar: 50μm. Images of whole-mount in situ hybridization were taken by Zeiss microscope. Scale bar: 500μm.
Spatial expression of crystallin genes during eye development
Cephalopod lenses are mainly composed of S-crystallin and Ω-crystallin (a water soluble protein), which are evolved from glutathione S-transferase (GST) and aldehyde dehydrogenase (ALDH), respectively [12]. Although there have been many studies on the structure and composition of S-crystallin (Tomarev and Zinovieva, 1988; Piatigorsky and Wistow, 1991; West et al., 1994; Tomarev et al., 1995; West et al., 1995; Tomarev and Piatigorsky, 1996), the functional study of crystallin genes at the molecular level has not been studied yet. To determine their spatial expression pattern of cephalopod-specific crystallin genes, such as S-crystallin (Omi_scry2, Omi_scry4), showing strong expression pattern in the late stage of eye development, we performed whole mount in situ hybridization during the developmental stage and observed the expression of Omi_scry2 and Omi_scry4 in the eye of stage 14 embryos. As shown in the cross-sectional view (Figure 5C), both Omi_scry2 and Omi_scry4 were found to be broadly expressed in LC in the ciliary body at stage 14, 18, 20 and adult.
Mishra et al. (2018) reported that α-crystallin B chain play a critical role in the development of the lens and stress resistance of the heart through α-crystallin B mutants in zebrafish (Mishra et al., 2018). The expression pattern of α-crystallin was analyzed through whole-mount in situ hybridization to see if this gene, which plays a very important role in lens development in vertebrates, is also conserved in the cephalopod octopus. The α-crystallin was strongly expressed in the lens, showing weak expression in the ciliary body at stage 14 embryos. As the cross-sectional view (Figures 5B–D), α-crystallin gene (Omi_cryab) was strongly expressed along the peripheral regions of the lens and at the tip of LC from stage 14 to adult.
The schematic diagram summarizes the expression patterns of crystallin genes (Figure 5E). Since crystallin is the major protein constituting lens, all crystallin genes were expressed in lentigenic cells, and particularly α-crystallin gene in the peripheral region of the lens.
Discussion
The camera-type eye of octopus is similar to that of vertebrates including humans. However, phylogenetic analyses and developmental studies suggested independent acquisition of camera eyes, which is a typical example of convergent evolution. Several studies have reported the visual system of octopus (Ogura et al., 2004; Yoshida et al., 2015); however, little is known about the molecular mechanisms or the diversity of cell types and structure such as muscles and nerves during eye development. First, we characterized the developmental processes in O. minor as a model system based on the morphogenesis of the embryo. Eye development during embryogenesis was investigated by selecting stage 12-20 embryos, in which the pigmentation of eye was apparent (Figure 1).
Cephalopods carry everted retina with rhabdomeric photoreceptors towards the light in contrast to the inverted retina carrying ciliary photoreceptors in vertebrates. We also showed that the DS faces the front of the eye, followed by SCN, BM and PCN, which is consistent with previous studies involving cephalopods (Figure 2A).
In contrast to fish, the lens of octopus consists of anterior and posterior elements divided by a septum, as reported by Budelmann, 1995 (Figures 2B, C). The circular F-actin arrangement inside the lens showed a pattern similar to the wild-type lens of zebrafish embryo (Hayes et al., 2012) (Figure 2C). The acetylated α-tubulin in the lens were gradually concentrated toward the exterior of the lens and distributed on the periphery of the lens and CB during the developmental process (Figure 2D). Our results showed that the DS and iris were rich in F-actin in the adult stage, and the lens showed a high density of circular F-actin, which is essential for strong refraction in an aquatic environment (Cheng et al., 2017). Based on immunostaining and SEM findings, the CB was composed of nerves and muscle fibers, and both the anterior and posterior lenses were connected by F-actin fibers.
The diversity and taxon specificity of lens crystallin throughout the animal kingdom is indicative of their convergent evolution. Lens crystallin is often related to ubiquitously expressed metabolic enzymes or physiological stress proteins. All vertebrate lenses contain α-crystallin belonging to the family of small heat shock proteins (Ingolia and Craig, 1982; de Jong et al., 1993) and β/γ-crystallins are related to microbial stress (Wistow, 1990; D'Alessio, 2002). The present molecular phylogenetic analyses were performed to establish the taxon specificity of the three groups of crystallin genes and provide further evidence supporting their convergent evolution. The newly identified genes from O. minor were orthologous to α-, S- and Ω-crystallin. The α-crystallin was found in both vertebrate and cephalopod group, but were still divided into two subgroups by taxon. The α-crystallin is the main lens protein in mammalians contributing to lens transparency and refractive index. As reported previously, α-crystallin also exhibits a protective function similar to other small heat shock proteins (sHSPs) in terms of thermostability and inhibition of denatured protein precipitation (Bakthisaran et al., 2015). Sharing the functional properties and the highly conserved domain with the other sHSPs molecules suggests a probable divergence from a common ancestor, despite acquisition or loss of specialized roles via evolutionary processes. However, the distinct gene clusters of S- and Ω-crystallin establish their taxon specificity only to the cephalopods. This finding supports the typical models of convergent evolution by demonstrating independent recruitment of different types of proteins to fulfill their unique visual role.
To determine the temporal expression of crystallin genes during eye development, we performed semi-quantitative RT-PCR using Omi-beta actin as an internal control with RNA samples at several stages (10, 14, 18 and 20) (Figures 5A, B). The levels of all crystallin genes were increased at later stages, demonstrating differential expression patterns due to the accumulation of lens proteins (Arnold, 1967; West et al., 1994). We then visualized the expression of cephalopod crystallin genes, such as the S-crystallin genes (Omi_scry2, Omi_scry4), and α-crystallin gene (Omi_cryab) via in situ hybridization during the developmental stages. All crystallin genes are commonly expressed in lentigenic cells of ciliary body. The α-crystallin was also expressed at the peripheral region of the lens including LC (Figure 5C). The expression of all crystallin genes in the lentigenic cells suggests that crystallin continues to be synthesized during eye development and is important in lens formation in cephalopods (Figure 5E) (West et al., 1994).
Conclusions
In this study, we constructed an embryonic staging system of Octopus minor as a model system to investigate the morphology of eye during the development using histological and molecular assays.
The present molecular phylogenetic results confirmed our previous knowledge that the S- and Ω-crystallins appear cephalopod specific. In addition, the α-crystallin was also found in the genomes of the cephalopods such as Sepia pharaonis, Octopus sinensis, O. bimaculoides and O. minor. Interestingly, the α-crystallin of the cephalopods formed a distinct phylogenetic clade separate from those of the vertebrates and thus this further implies that the gene evolution in each animal groups has independently been proceeded through the convergence process. Our in situ hybridization show that crystallin genes such as S- and α-crystallin are commonly expressed in the lentigenic cells of ciliary body, which means that crystallin continues to be synthesized during eye development. In addition, the α-crystallin of O. minor was spatiotemporally expressed in the eye, suggesting a possible role in lens formation during eye development in cephalopods.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical review and approval was not required for the animal study because Based on ‘The regulation of Institutional Animal Care and Use Committee (IACUC) in CBNU (Chungbuk national University)’ and ‘The related laws (animal welfare act, laboratory animal act)’, the IACUC in CBNU review only the protocols of the experiments using vertebrates animals including mammals, reptiles, amphibians, and etc. and it doesn’t review any protocols of the experiments doing invertebrates animals including mollusks like octopus, squids, etc.
Author contributions
K-BR, G-HJ, Y-CG, S-iE, H-YL, and S-JC conceived the project and designed the experiments. K-BR, S-HJ, SJ, HSA, and G-HJ documented the embryonic development of O. minor embryos by microscopy. K-BR, N-RC, and G-HJ performed cloning and sequencing of crystallin, elaborated the embryogenesis timetable for this species. K-BR, Y-CG, S-iE, G-HJ, H-YL, and S-JC was a major contributor in writing the manuscript. DJ and S-iE performed phylogenetic tree analysis. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grant from the National Marine Biodiversity Institute of Korea funded by the Ministry of Oceans and Fisheries (2023M00400). This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A06046235). This work was supported by Korea Institute of Marine Science & Technology Promotion funded by the Ministry of Oceans and Fisheries (20220596).
Acknowledgments
We thank members of the Cho Laboratory for valuable comments. We also thank the Resources Management Section of Jeollanam-do Ocean & Fisheries Science Institute (Sinan, Jeollanam-do) for providing tanks and daily check of octopuses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1136602/full#supplementary-material
ALDH, aldehyde dehydrogenase; BM, basal membrane; CB, ciliary body; DS, distal segments of the photoreceptors; GST, glutathione S-transferase; H&E, hematoxylin and eosin; MLB, maximum-likelihood bootstrap; OGL, outer granule layer; OPL, outer plexiform layer; PCN, photoreceptor cell nuclei; PG, pigment granules; SCN, supporting cell nuclei; SHSPs, small heat shock proteins.
References
Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 (17), 3389–3402. doi: 10.1093/nar/25.17.3389
Arnold J. M. (1967). Fine structure of the development of the cephalopod lens. J. Ultrastruct Res. 17 (5), 527–543. doi: 10.1016/s0022-5320(67)80139-4
Augusteyn R. C. (2004). α-crystallin: a review of its structure and function. Clin. Exp. Optom. 87 (6), 356–366. doi: 10.1111/j.1444-0938.2004.tb03095.x
Bakthisaran R., Tangirala R., Rao C. M. (2015). Small heat shock proteins: role in cellular functions and pathology. Biochim. Biophys. Acta (BBA)-Proteins Proteomics 1854 (4), 291–319. doi: 10.1016/j.bbapap.2014.12.019
Baldascino E., Di Cristina G., Tedesco P., Hobbs C., Shaw T. J., Ponte G., et al. (2017). The gastric ganglion of octopus vulgaris: preliminary characterization of gene-and putative neurochemical-complexity, and the effect of aggregata octopiana digestive tract infection on gene expression. Front. Physiol. 8, 1001. doi: 10.3389/fphys.2017.01001
Bloemendal H., de Jong W., Jaenicke R., Lubsen N. H., Slingsby C., Tardieu A. (2004). Ageing and vision: structure, stability and function of lens crystallins. Prog. biophysics Mol. Biol. 86 (3), 407–485. doi: 10.1016/j.pbiomolbio.2003.11.012
Boyle P. R. (1991). The UFAW handbook on the care and management of cephalopods in the laboratory (Potters Bar) (Universities Federation for Animal Welfare).
Budelmann B. U. (1995). Cephalopod sense organs, nerves and the brain: adaptations for high performance and life style. Mar. Freshw. Behav. Physiol. 25 (1-3), 13–33. doi: 10.1080/10236249409378905
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Cheng C., Nowak R. B., Fowler V. M. (2017). The lens actin filament cytoskeleton: diverse structures for complex functions. Exp. Eye Res. 156, 58–71. doi: 10.1016/j.exer.2016.03.005
Chiou S.-H. (1988). A novel crystallin from octopus lens. FEBS Lett. 241 (1-2), 261–264. doi: 10.1016/0014-5793(88)81073-1
Cho S. J., Valles Y., Giani V. C. Jr., Seaver E. C., Weisblat D. A. (2010). Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective. Mol. Biol. Evol. 27 (7), 1645–1658. doi: 10.1093/molbev/msq052
D'Alessio G. (2002). The evolution of monomeric and oligomeric betagamma-type crystallins. facts and hypotheses. Eur. J. Biochem. 269 (13), 3122–3130. doi: 10.1046/j.1432-1033.2002.03004.x
de Jong W. W., Leunissen J. A., Voorter C. E. (1993). Evolution of the alpha-crystallin/small heat-shock protein family. Mol. Biol. Evol. 10 (1), 103–126. doi: 10.1093/oxfordjournals.molbev.a039992
Gehring W. J. (2005). New perspectives on eye development and the evolution of eyes and photoreceptors. J. Hered. 96 (3), 171–184. doi: 10.1093/jhered/esi027
Gross V., Mayer G. (2015). Neural development in the tardigrade hypsibius dujardini based on anti-acetylated α-tubulin immunolabeling. EvoDevo 6 (1), 1–15. doi: 10.1186/s13227-015-0008-4
Hanke F. D., Kelber A. (2020). The eye of the common octopus (Octopus vulgaris). Front. Physiol. 10, 1637. doi: 10.3389/fphys.2019.01637
Hayes J. M., Hartsock A., Clark B. S., Napier H. R., Link B. A., Gross J. M. (2012). Integrin alpha5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish. Mol. Biol. Cell 23 (24), 4725–4738. doi: 10.1091/mbc.E12-09-0672
Horwitz J. (2000). The function of alpha-crystallin in vision. Semin. Cell Dev. Biol. 11, 53–60. doi: 10.1006/scdb.1999.0351
Ingolia T. D., Craig E. A. (1982). Four small drosophila heat shock proteins are related to each other and to mammalian alpha-crystallin. Proc. Natl. Acad. Sci. 79 (7), 2360–2364. doi: 10.1073/pnas.79.7.2360
Jahnel S. M., Walzl M., Technau U. (2014). Development and epithelial organisation of muscle cells in the sea anemone nematostella vectensis. Front. Zool. 11 (1), 1–15. doi: 10.1186/1742-9994-11-44
Jonasova K., Kozmik Z. (2008). Eye evolution: lens and cornea as an upgrade of animal visual system. Semin Cell Dev Biol. (Elsevier), 71–81. doi: 10.1016/j.semcdb.2007.10.005
Jung S.-H., Song H. Y., Hyun Y. S., Kim Y.-C., Whang I., Choi T.-Y., et al. (2018). A brain atlas of the long arm octopus, octopus minor. Exp. Neurobiol. 27 (4), 257. doi: 10.5607/en.2018.27.4.257
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi: 10.1038/nmeth.4285
Kannan R., Sreekumar P. G., Hinton D. R. (2012). Novel roles for α-crystallins in retinal function and disease. Prog. Retinal Eye Res. 31 (6), 576–604. doi: 10.1016/j.preteyeres.2012.06.001
Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 (14), 3059–3066. doi: 10.1093/nar/gkf436
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kim B.-M., Kang S., Ahn D.-H., Jung S.-H., Rhee H., Yoo J. S., et al. (2018). The genome of common long-arm octopus octopus minor. Gigascience 7 (11), giy119. doi: 10.1093/gigascience/giy119
Koenig K. M., Sun P., Meyer E., Gross J. M. (2016). Eye development and photoreceptor differentiation in the cephalopod doryteuthis pealeii. Development 143 (17), 3168–3181. doi: 10.1242/dev.134254
Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35 (21), 4453–4455. doi: 10.1093/bioinformatics/btz305
Land M. F. (1965). Image formation by a concave reflector in the eye of the scallop, pecten maximus. J. Physiol. 179 (1), 138–153. doi: 10.1113/jphysiol.1965.sp007653
Land M. F., Fernald R. D. (1992). The evolution of eyes. Annu. Rev. Neurosci. 15, 1–29. doi: 10.1146/annurev.ne.15.030192.000245
Lemoine F., Domelevo Entfellner J. B., Wilkinson E., Correia D., Davila Felipe M., De Oliveira T., et al. (2018). Renewing felsenstein's phylogenetic bootstrap in the era of big data. Nature 556 (7702), 452–456. doi: 10.1038/s41586-018-0043-0
Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi: 10.1093/molbev/msaa015
Mishra S., Wu S.-Y., Fuller A. W., Wang Z., Rose K. L., Schey K. L., et al. (2018). Loss of αB-crystallin function in zebrafish reveals critical roles in the development of the lens and stress resistance of the heart. J. Biol. Chem. 293 (2), 740–753. doi: 10.1074/jbc.M117.808634
Naef A. (1928). Die cephalopoden. embryologie. Die Fauna Flora Golf Neapel 35 (2), 1–357. doi: 10.5962/bhl.title.11215
Ogura A., Ikeo K., Gojobori T. (2004). Comparative analysis of gene expression for convergent evolution of camera eye between octopus and human. Genome Res. 14 (8), 1555–1561. doi: 10.1101/gr.2268104
Ogura A., Yoshida M. A., Moritaki T., Okuda Y., Sese J., Shimizu K. K., et al. (2013). Loss of the six3/6 controlling pathways might have resulted in pinhole-eye evolution in nautilus. Sci. Rep. 3, 1432. doi: 10.1038/srep01432
Piatigorsky J. (1998). Gene sharing in lens and cornea: facts and implications. Prog. Retinal Eye Res. 17 (2), 145–174. doi: 10.1016/S1350-9462(97)00004-9
Piatigorsky J. (2003). Gene sharing, lens crystallins and speculations on an eye/ear evolutionary relationship. Integr. Comp. Biol. 43 (4), 492–499. doi: 10.1093/icb/43.4.492
Piatigorsky J., Wistow G. (1991). The recruitment of crystallins: new functions precede gene duplication. Science 252 (5009), 1078–1079. doi: 10.1126/science.252.5009.1078
Sakaue Y., Bellier J.-P., Kimura S., D’Este L., Takeuchi Y., Kimura H. (2014). Immunohistochemical localization of two types of choline acetyltransferase in neurons and sensory cells of the octopus arm. Brain Structure Funct. 219, 323–341. doi: 10.1007/s00429-012-0502-6
Serb J. M. (2008). Toward developing models to study the disease, ecology, and evolution of the eye in Mollusca. Am. Malacological Bull. 26 (1/2), 3–18. doi: 10.4003/006.026.0202
Serb J. M., Eernisse D. J. (2008). Charting evolution’s trajectory: using molluscan eye diversity to understand parallel and convergent evolution. Evol. Educ. Outreach 1 (4), 439–447. doi: 10.1007/s12052-008-0084-1
Shigeno S., Andrews P. L. R., Ponte G., Fiorito G. (2018). Cephalopod brains: an overview of current knowledge to facilitate comparison with vertebrates. Front. Physiol. 9. doi: 10.3389/fphys.2018.00952
Shigeno S., Parnaik R., Albertin C. B., Ragsdale C. W. (2015). Evidence for a cordal, not ganglionic, pattern of cephalopod brain neurogenesis. Zoological Lett. 1 (1), 1–13. doi: 10.1186/s40851-015-0026-z
Slingsby C., Wistow G. J., Clark A. R. (2013). Evolution of crystallins for a role in the vertebrate eye lens. Protein Sci. 22 (4), 367–380. doi: 10.1002/pro.2229
Tomarev S. I., Chung S., Piatigorsky J. (1995). Glutathione s-transferase and s-crystallins of cephalopods: evolution from active enzyme to lens-refractive proteins. J. Mol. Evol. 41 (6), 1048–1056. doi: 10.1007/BF00173186
Tomarev S. I., Piatigorsky J. (1996). Lens crystallins of invertebrates: diversity and recruitment from detoxification enzymes and novel proteins. Eur. J. Biochem. 235 (3), 449–465. doi: 10.1111/j.1432-1033.1996.00449.x
Tomarev S. I., Zinovieva R. D. (1988). Squid major lens polypeptides are homologous to glutathione s-transferases subunits. Nature 336 (6194), 86–88. doi: 10.1038/336086a0
West J. A., Sivak J. G., Doughty M. J. (1995). Microscopical evaluation of the crystalline lens of the squid (Loligo opalescens) during embryonic development. Exp. Eye Res. 60 (1), 19–35. doi: 10.1016/S0014-4835(05)80080-6
West J. A., Sivak J. G., Pasternak J., Piatigorsky J. (1994). Immunolocalization of s-crystallins in the developing squid (Loligo opalescens) lens. Dev. Dyn. 199 (2), 85–92. doi: 10.1002/aja.1001990202
Wistow G. (1990). Evolution of a protein superfamily: relationships between vertebrate lens crystallins and microorganism dormancy proteins. J. Mol. Evol. 30 (2), 140–145. doi: 10.1007/BF02099940
Xu L., Dong Z., Fang L., Luo Y., Wei Z., Guo H., et al. (2019). OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47 (W1), W52–W58. doi: 10.1093/nar/gkz333
Yoshida M.-a., Ogura A. (2011). Genetic mechanisms involved in the evolution of the cephalopod camera eye revealed by transcriptomic and developmental studies. BMC Evol. Biol. 11 (1), 1–11. doi: 10.1186/1471-2148-11-180
Yoshida M. A., Ogura A., Ikeo K., Shigeno S., Moritaki T., Winters G. C., et al. (2015). Molecular evidence for convergence and parallelism in evolution of complex brains of cephalopod molluscs: insights from visual systems. Integr. Comp. Biol. 55 (6), 1070–1083. doi: 10.1093/icb/icv049
Zhang Y., Mao F., Mu H., Huang M., Bao Y., Wang L., et al. (2021). The genome of nautilus pompilius illuminates eye evolution and biomineralization. Nat. Ecol. Evol. 5 (7), 927–938. doi: 10.1038/s41559-021-01448-6
Keywords: cephalopod, octopus, eye development, crystallin genes, Octopus minor
Citation: Ryu K-B, Jo G-H, Gil Y-C, Jeon D, Choi N-R, Jung S-H, Jo S, An HS, Lee H-Y, Eyun S-i and Cho S-J (2023) Eye development and developmental expression of crystallin genes in the long arm octopus, Octopus minor. Front. Mar. Sci. 10:1136602. doi: 10.3389/fmars.2023.1136602
Received: 03 January 2023; Accepted: 05 May 2023;
Published: 17 May 2023.
Edited by:
Tomer Ventura, University of the Sunshine Coast, AustraliaReviewed by:
Kyudong Han, Dankook University, Republic of KoreaHuarong Guo, Ocean University of China, China
Gianluca Polese, University of Naples Federico II, Italy
Copyright © 2023 Ryu, Jo, Gil, Jeon, Choi, Jung, Jo, An, Lee, Eyun and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung-Jin Cho, c2pjaG9iaW9AY2h1bmdidWsuYWMua3I=; Hae-Youn Lee, aHlsZWU3MUBjaHVuZ2J1ay5hYy5rcg==; Seong-il Eyun, ZXl1bkBjYXUuYWMua3I=
†These authors have contributed equally to this work
 Kyoung-Bin Ryu1,2†
Kyoung-Bin Ryu1,2† Donggu Jeon
Donggu Jeon Seong-il Eyun
Seong-il Eyun Sung-Jin Cho
Sung-Jin Cho