- 1Department of Veterinary Medical Sciences, University of Bologna, Ozzano Emilia, Italy
- 2Prosol S.p.A., Bergamo, Italy
Nowadays functional ingredients have a significant potential for improving current low fish meal (FM) aquafeed formulation in sustaining growth and enhancing animal robustness for Mediterranean aquaculture. Among them, nucleotides (NT) and nucleic acids (NA) drew attention for their application in the last two decades. NT are organic molecules involved in many life-supporting pathways, and are the building blocks of NA, which stand as genetic repositories. NT are naturally present in organic ingredients, and among them FM is known to be one of the highest NT sources. When this NT source is seriously limited, fish might be under the minimum NT requirements, especially in fast growing life stages of carnivorous species. Hence, a trial on European sea bass juveniles was carried out, testing two dietary FM levels (FM10, FM20 as 10% and 20% FM, respectively) supplemented with 500 mg kg-1 yeast-originate NT or NA dose over 80 days. Thereafter, fish were exposed to one week of sub-optimal thermal and dissolved oxygen condition (30°C and 4.0 mg/L O2) to further explore the effect of NT and NA inclusion on immune response and gut microbiome alteration. At the end of the growth period NT increased feed intake at both FM dietary levels. FM20 combined with NA and NT further improved growth performance, enhancing lipid efficiency and increased anti-inflammatory TGF-β. After sub-optimal environmental conditions both NT and NA exerted prebiotic functions on gut microbiome by promoting beneficial lactic acid bacteria such as Weissella and Leuconostoc. At the same time NT in 10% FM diet increased the abundance of Bacillus taxon. In conclusion, the combination of NT/NA included at 500 mg kg-1 was able to promote growth when included in 20% FM level, assuming higher nutritional NT requirement when combined with 10% FM. On the other hand, NT/NA added in 10% FM upregulate proinflammatory IL-1β and favor beneficial gut bacterial taxa.
1 Introduction
To meet the continuously rising demand, Mediterranean marine aquaculture sector urges a continuous production improvement, especially for aquafeed, which covers approximately up to 70% of the total running expenses and is mainly bound to fish meal (FM) as the major but unsustainable protein source, especially for carnivorous fish species (Guillen and Carvalho, 2016; Dawood and Koshio, 2020). So far, the main solution has been the consistent FM replacement with alternative agricultural crop-derived ingredients though with some problems related to anti-nutritional and nutrient requirements issues (Hua and Bureau, 2012; Nasopoulou and Zabetakis, 2012; Bonvini et al., 2018a; Sutili et al., 2018; Parma et al., 2019; Pelusio et al., 2022). As a further step beyond, tailored functional aquafeeds able to improve growth, feed utilization, general health and stress resistance of animals, represent a new emerging paradigm of current fish nutrition to compensate the negative effects of alternative or low FM based diets and improve cost-effectiveness (Hossain and Koshio, 2017a; Dawood et al., 2018). Initially used as feed attractants, nucleotides (NT) and nucleic acids (NA) drew the attention of research and industrial communities as functional ingredients on account of their many features as alternative nitrogen sources and health promoters in the last two decades (Li and Gatlin, 2006; Hossain and Koshio, 2017a). NT are low molecular weight biological compounds involved in most of the biochemical pathways essential for life support. They stand as metabolic energy currency, cell signaling mediators and components of enzyme cofactors. Moreover, they are the constituents of the molecular repositories of genetic information: deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). The structure of every protein, biomolecule, and cellular component, is a product of information programmed into the NT sequence of cellular NA (Bowyer et al., 2019). In feedstuffs, NT are naturally present in all animal and vegetable origin ingredients mostly as nucleoproteins and in low quantities as free NT and among them, FM is one of the richest sources (Hossain et al., 2020). Since NT can be synthesized endogenously by the organism, they are not considered essential nutrients (Low et al., 2003). On the other hand, their synthesis is a metabolically costly process, and the dietary uptake of NT and NA isolated from yeasts may optimize cell proliferation to promote rapid growth, especially during early development stages with high metabolism or fast growth (Ringø et al., 2012; Dawood et al., 2018). Many studies show that NT or NA supplementation can improve growth and nutritional uptake in several aquaculture species such as European sea bass (Oliva-Teles et al., 2006; Rawling et al., 2019), gilthead sea bream (El-Nokrashy et al., 2021), amberjack (Hossain et al., 2017b), Nile tilapia (Barros et al., 2015; Kader et al., 2018), red sea bream (Hossain et al., 2016; Hossain and Koshio, 2017a; Hossain et al., 2017c), Atlantic salmon (Burrells et al., 2001), turbot (Peng et al., 2013) and rainbow trout (Hunt et al., 2014). Moreover, when fish undergo sub-optimal and stressful farming conditions such as handling, elevated temperatures and fluctuating levels of dissolved oxygen, additional NT pools might be required for further signal transduction or immune cell proliferation with a modulatory effect (Carver and Allan Walker, 1995; Cosgrove, 1998; Li and Gatlin, 2006). Many studies reported modulation to stress response (Kenari et al., 2013; Palermo et al., 2013), resistance to disease and pathogens (Burrells et al., 2001; Tahmasebi-Kohyani et al., 2011), lymphocytes activity, macrophage phagocytosis, immunoglobulin and cytokines responses (Gil, 2002; Low et al., 2003; Singhal et al., 2008; Reda et al., 2018). As a further gut health biomarker, gut microbiome composition and its eventual modulation may indicate the status of commensal bacteria related to stressful events or unbalanced diet formulation (Perry et al., 2020). So far, encouraging and consistent NT and NA applications on gut health have been reported in many aquatic species (Burrells et al., 2001; Guo et al., 2019; Rawling et al., 2019; El-Nokrashy et al., 2021). Nowadays, one of the main issues related to their addition in formulated feeds is to establish the right dose for each specific life stage and species. While terrestrial monogastric animals cannot tolerate high levels of dietary NT due to high serum uric acid from purine metabolism and associated toxicity, as well as negative effects on other nutrients pathways, fish such as salmonids and sea bass may tolerate high levels of NT/NA/yeasts thanks to active liver uricase (Hossain et al., 2020). So far, the most promising dose concentration for blend NT has been targeted at 0.5% dietary level, while for single purified nucleotide dosages the range has been between 0.1% and 0.6% of dry weight in different fish species (Hossain et al., 2020). European sea bass (Dicentrarchus labrax) is one of the major Mediterranean aquaculture species in the European Union and it is well known to be a model organism (Vandeputte et al., 2019). In recent years, though its carnivorous regime, sea bass diet formulation has been achieving even low FM contents with commercial potential application (Pelusio et al., 2022). For these reasons, the aim of the present study was to evaluate the effects of dietary NT and NA addition on growth and gut health of European sea bass juveniles fed different low FM level.
2 Materials and methods
2.1 Experimental diets and additive dosages
Ingredients and chemical composition of the experimental diets are presented in Table 1. Six isonitrogenous (45.16% protein) and isolipidic (17.43% lipids) diets were formulated to contain 2 low different FM levels (20 and 10%) as control treatments. Then, nucleotides (NT) and nucleic acid (NA) were added at the same dosage of 500 mg kg feed-1 in accordance with recent recommendations on same and other fish species (De Cruz et al., 2020; El-Nokrashy et al., 2021; Magouz et al., 2021). Experimental diets were produced via extrusion (pellet size = 2.0 mm) by the Danish Technological Institute (Taastrup, Denmark).
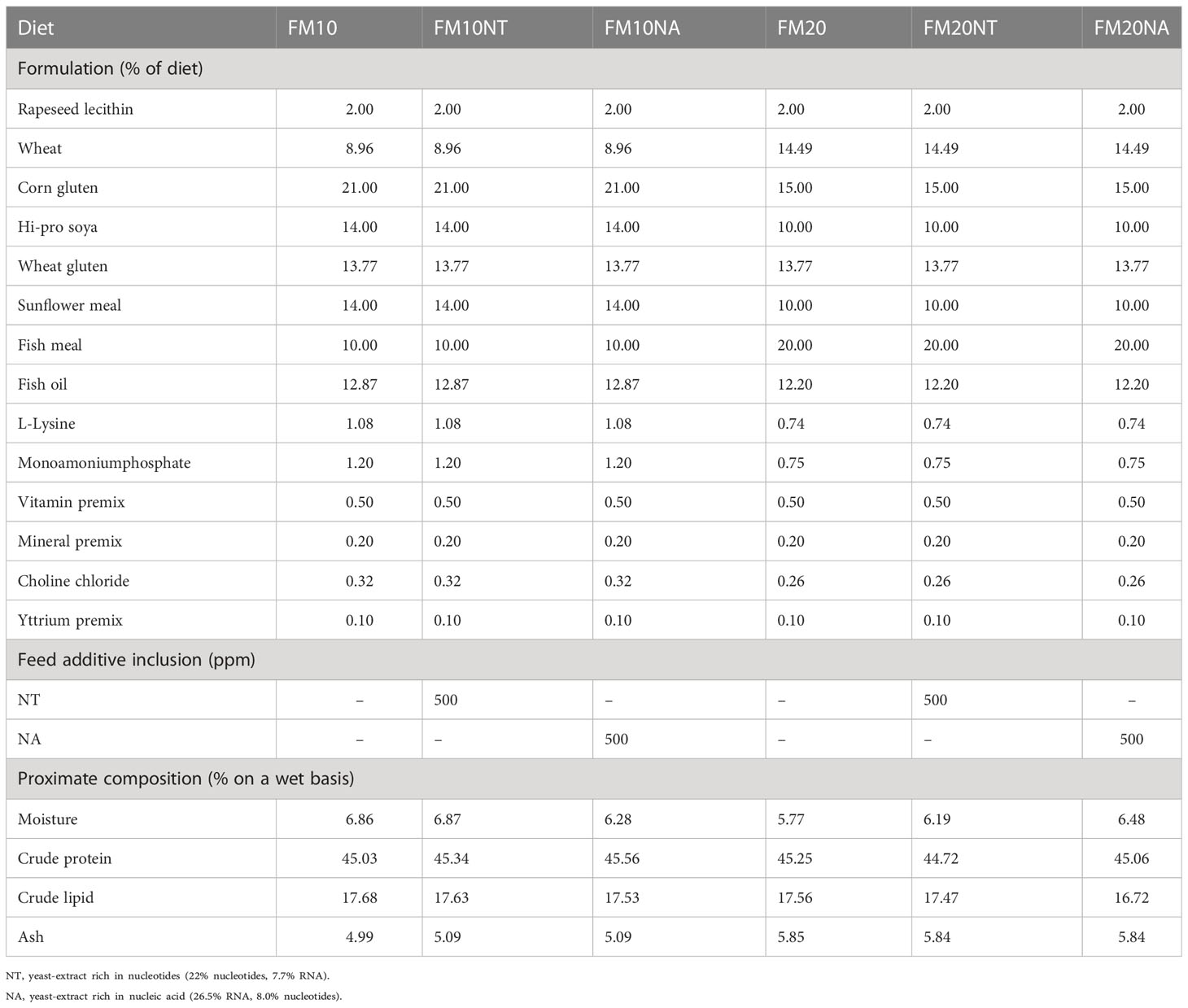
Table 1 Feed formulation and chemical composition of experimental treatments at different fish meal (FM) level and nucleotide (NT) or nucleic acid (NA) inclusion.
2.2 Fish and rearing conditions
The trial was performed at the Laboratory of Aquaculture, Department of Veterinary Medical Sciences of the University of Bologna, Cesenatico, Italy. European sea bass (Dicentrarchus labrax) juveniles were transferred from Ittica Caldoli S.A.R.L (Poggio Imperiale, Foggia, Italy) to the laboratory facilities and adapted for one week before the beginning of the trial. Then, 60 fish tank−1 (initial weight: 14.33 ± 0.18 g, mean ± standard deviation, SD) were randomly allocated in eighteen 800 L tanks. Diets were randomly assigned in triplicate condition and animals were fed for 80 days. Tanks were provided with natural seawater using a closed recirculation system (overall water volume: 20 m3) according to Pelusio et al. (2021). The recirculation system consisted of a mechanical sand filter (PTK 1200, Astralpool, Barcelona, Spain), ultraviolet lights (SH-88, BLUGEO S.r.l., Parma, Italy) and a biofilter (PTK 1200, Astralpool, Barcelona, Spain). The water exchange rate in each tank was 100% every hour, while the overall water renewal in the system was 5% per day. The temperature was kept at 22.0 ± 0.5°C while photoperiod was set at 12 h light and 12 h dark. The oxygen level (8.0 ± 1.0 mg L−1) was provided through a liquid oxygen system regulated by a software programme (B&G Sinergia snc, Chioggia, Italy). Ammonia (total ammonia nitrogen ≤ 0.1 mg L−1) and nitrite (≤ 0.2 mg L−1) were daily measured (Spectroquant Nova 60, Merck, Lab business, Darmstadt, Germany), while salinity (30 g L−1) was analysed using a salt refractometer (106 ATC, Giorgio Bormac S.r.l., Carpi, Italy). Sodium bicarbonate was provided regularly to keep pH at 7.8–8.2 (Pelusio et al., 2021). Fish were fed ad libitum twice a day (8:30 h and 16:30 h) for six days a week by means of automatic feeders using an overfeeding approach with a daily feeding ration 10% higher than the daily ingested ration of the previous days as reported by Bonvini et al. (2018b). Each meal lasted 1 h, after which the uneaten pellets of each tank were collected, dried overnight at 105°C, and weighed for further calculations of feed intake.
2.3 Suboptimal rearing conditions
After the end of the feeding trial, fish were exposed to high temperature (28.9 ± 1.8°C) and low oxygen (4.1 ± 0.8 mg L−1, 55.3 ± 4.7% saturation level) for 8 days while keeping the same feeding conditions. The sub-optimal environmental conditions were designed in order to simulate unfavourable Mediterranean summer condition as reported by Busti et al. (2020). Specifically, temperature was increased at a rate of 2-degree day−1 while water oxygen level was decreased (concentration from 8.0 ± 1.0 mg L−1 to 4.6 ± 0.6 mg L−1 and saturation from 105.0 ± 1.0% to 67.6 ± 5.0%, respectively) within 24 h. Temperature and oxygen level were constantly measured through an automatic system regulated by a software programme (B&G Sinergia snc, Chioggia, Italy). The environmental parameters were chosen according to previous studies on oxygen and thermal tolerance of the species (Dülger et al., 2012; Makridis et al., 2018; Busti et al., 2020).
2.4 Sampling
At the beginning and at the end of the feeding trial (day 80) fish were anaesthetised and individually weighed. Specific growth rate (SGR), feed intake (FI) and feed conversion ratio (FCR) were calculated. Proximate carcass composition was determined at the beginning of the trial on a pooled sample of 15 fish and on pooled samples of 5 fish tank−1 at the end of the trial. Protein efficiency ratio (PER), gross protein efficiency (GPE), lipid efficiency ratio (LER), and gross lipid efficiency (GLE) were calculated. At the end of the trial, condition factor (CF), viscerosomatic index (VSI), hepatosomatic index (HSI) and mesenteric fat index (MFI) were also obtained from 3 fish per tank. Concerning immunological parameters, at the beginning (T0, 9 fish in total), at the end of the feeding trial (T1, 6 animals’ diet−1) and at the end of the suboptimal rearing conditions (T2, 6 animals’ diet−1) fish were sampled to assess inflammatory and immune response gene expression of distal intestinal mucosa (1 cm before the rectum) (Busti et al., 2020; Pelusio et al., 2020; Parma et al., 2023). At the end of the feeding and suboptimal rearing conditions periods, samples of feces from distal intestine in 6 fish diet−1 were also individually collected and placed at −80°C for gut bacterial community characterisation (Parma et al., 2019). The experimental design scheme of the feeding trial is reported in Supplementary Figure 1. All experimental procedures were carried in accordance with European directive 2010/63/UE on the protection of animals used for scientific purposes.
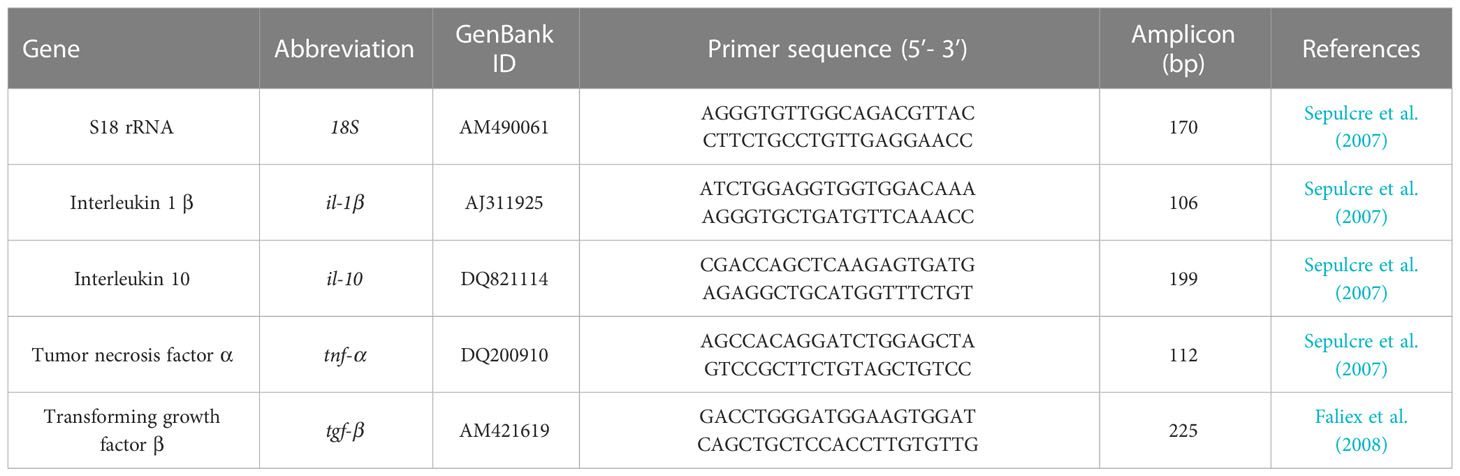
2.5 Cytokines gene expression analyses by real−time polymerase chain reaction
Activation of local (intestine) immune and inflammatory response associated with different diets (FM level and NA/NT supplementation) has been assessed through transcript quantification of target genes in distal intestine. In particular genes of two inflammatory cytokines such as interleukin 1β (IL-1β), tumor necrosis factor α (TNF-α) and two anti-inflammatory cytokines interleukin 10 (IL-10) and transforming growth factor β (TGF-β) were investigated. Total RNA was isolated from 20-30 mg of intestine samples stored in RNA Later (Sigma) using the NucleoSpin RNA extraction kit (Machery-Nagel) following the manufacturer’s instructions. RNA quality was assessed by 260/280 ratio. The RNA extraction protocol includes a treatment with DNAse I in order to remove genomic DNA. The first strand of complementary DNA (cDNA) was synthesised by reverse transcription using the GoScript Reverse Transcriptase (Promega). cDNA concentration was quantified using a Qubit Fluorometer (ThermoFisher). Realtime PCR was performed with an ABI PRISM 7300 instrument (Applied Biosystems) using BRYT Green GoTaq qPCR (Promega). 10 ng of each cDNA sample was added to a reaction mix containing 2 × GoTaq qPCR Master Mix (Promega), 300 nM of CXR and 200 nM of each primer. The primers used for IL-1β, IL-10, TNF-α, TGF-β and the housekeeping S18 rRNA gene, are shown in Table 2. Reaction mixtures were incubated for 2 min at 95°C, followed by 50 cycles of 10 s at 95°C, 30 s at 60°C, and finally 15 s at 95°C, 1 min 60°C and 15 s at 95°C (dissociation stage). Before the experiments, the specificity of each primer pair was studied using positive and negative samples. A melting curve analysis of the amplified products validated the primers for specificity. After these verifications, all cDNA samples were analyzed in triplicate. Negative controls with no template and an internal control were always included in the reactions. For each sample, gene expression was normalized against S18 rRNA and expressed as 2-ΔΔCt, where ΔCt is determined by subtracting the S18 rRNA Ct value from the target Ct. Gene expression of animals fed control and experimental diets collected at the end of the feeding trial (T1) and after suboptimal rearing conditions (T2) were expressed as “fold changes” relative to control animals sampled at T1 and T2, respectively (Pelusio et al., 2020).
2.6 Gut bacterial community DNA extraction and sequencing
The DNA was extracted as described by De Cesare et al. (2017). Distal intestine content aliquots of 0.30 g were initially suspended in 1 mL lysis buffer (500 mM NaCl, 50 mM Tris-Cl, pH 8.0, 50 mM EDTA, 4% SDS) with MagNA Lyser Green Beads (Roche, Milan, Italy) and homogenized on the MagNA Lyser (Roche) at 6500 rpm for 25 s. Afterwards, samples were heated at 70°C for 15 min, followed by centrifugation to split the DNA from the bacterial cellular wreckage. This process was repeated with a second 300 μL aliquot of lysis buffer. The samples were then subjected to 10 M v/v ammonium acetate (Sigma, Milan, Italy) precipitation, followed by isopropanol (Sigma) precipitation and a 70% ethanol (Carlo Erba, Milan, Italy) wash and resuspended in 100 μL 1X Tris-EDTA (Sigma). The samples were then treated with DNase-free RNase (Roche) and incubated overnight at 4°C, before being processed through the QIAmp DNA Stool Mini Kit (Qiagen, Milan, Italy) according to the manufacturer’s directions, with some modifications. Samples were measured on a BioSpectrometer (Eppendorf, Milan, Italy) to assess DNA quantity and quality. Libraries were prepared following the 16S Metagenomic Sequencing Library Preparation protocol (Illumina, San Diego, CA, USA), amplifying the V3 and V4 hypervariable regions of the 16S rRNA gene in order to obtain a single amplicon of approximately 460 bp. Sequencing was performed in paired-end employing MiSeq System (Illumina) with MiSeq Reagent kit v2 500 cycles (Illumina), characterised by a maximum output of 8.5 Gb.
2.7 Analytical methods
Chemical analyses were conducted according to Pelusio et al. (2021). Shortly, diets and fish carcass were analyzed for proximate composition. Moisture was determined by weight loss after drying samples in a stove at 105°C overnight. Crude protein was analysed as total nitrogen (N*6.25) using Kjeldahl’s method according to the AOAC International (AOAC, 2010). Total lipids were measured according to Bligh and Dyer (1959) extraction method. Ash content was determined by incineration in a muffle oven at 450°C overnight.
2.8 Statistical and bioinformatic analysis
For statistical analysis a tank was used as the experimental unit to analyse growth, while pools of five individuals per tank were considered to describe CF, VSI, HSI. Growth, nutritional, and somatometric indices were analyzed by one-way ANOVA, followed by Tukey’s multi-comparison test. Gene expression values have been expressed as mean of fish subjected to each treatment ± standard deviation (SD). Diet effect on gene expression was assessed by two-way ANOVA using NA/NT supplementation (S) and FM level as variables; then Tukey’s post hoc test was performed. The normality and/or homogeneity of variance assumptions were validated for all data preceding ANOVA. Growth, nutritional and somatometric indices and gene expression data were analyzed using GraphPad Prism 9.0 for Windows (Graph Pad Software, San Diego, CA, USA).
Gut microbiome samples before and after suboptimal rearing conditions (T1 and T2) were analysed adopting a bioinformatic pipeline based on QIIME2 (http://qiime.org/). Raw sequences were processed with the dada2 algorithm in order to denoise and to merge forward and reverse sequences per each pair. The taxonomic classification of cleaned data was performed by applying the VSEARCH-based classifier implemented in QIIME and using the Greengenes 13_8 97% OUT dataset as reference. The mean values for the relative frequency of abundance of each taxonomic level in each sample were compared using the the two-way anova followed by the Dunnet post hoc analysis (pakages aov::stats and dunn_test::rstatix respectively). P-values ≤ 0.05 were considered statistically significant. β-diversity was calculated using “vegdist” (vegan “package) and the PCoA analysis using “cmdscale” (“stats” package). Statistical differences were calculated using the “beta_pcoa_stat” (“amplicon”package).
3 Results
3.1 Growth, biometric indices and nutritional uptake
Data on growth performance, somatometric indices, proximate composition and nutritional indices are represented in Table 3. At the end of the growth period the FBW was higher in FM20NT and FM20NA compared to FM20. On WG the lowest level was displayed by FM20 animals, while FM10 was lower than FM20NA. Concerning SGR, FM20 were lower than FM20NT and FM20NA; at the same time FM20NA was higher than FM10. Regarding FCR, FM20 was higher than FM20NA. VFI of FM10 and FM20 fish levels were lower than FM10NT and FM20NT. No significant differences were detected among treatments for survival. Concerning somatometric indices, no significant differences occurred among treatments for CF, VSI, HSI and MFI. Regarding proximate composition, no significant difference was shown for crude protein while the highest crude lipid was measured in fish fed FM20NT. At the same time crude lipid in FM10NT was higher than FM10NA. Moisture was higher in FM10NA than FM10NT, FM10NT, FM20NT, and FM20NA. No significant differences were detected in ash content. Regarding nutritional indices, PER was higher in FM20NA compared to FM20, FM10NA, FM10NT, and FM10. GPE was higher in FM20NA compared to FM10NA and FM10NT. Concerning lipid efficiency, LER was higher in FM20NA compared to the other treatment while GLE was higher in FM20NA than FM20, FM10NA, and FM10.
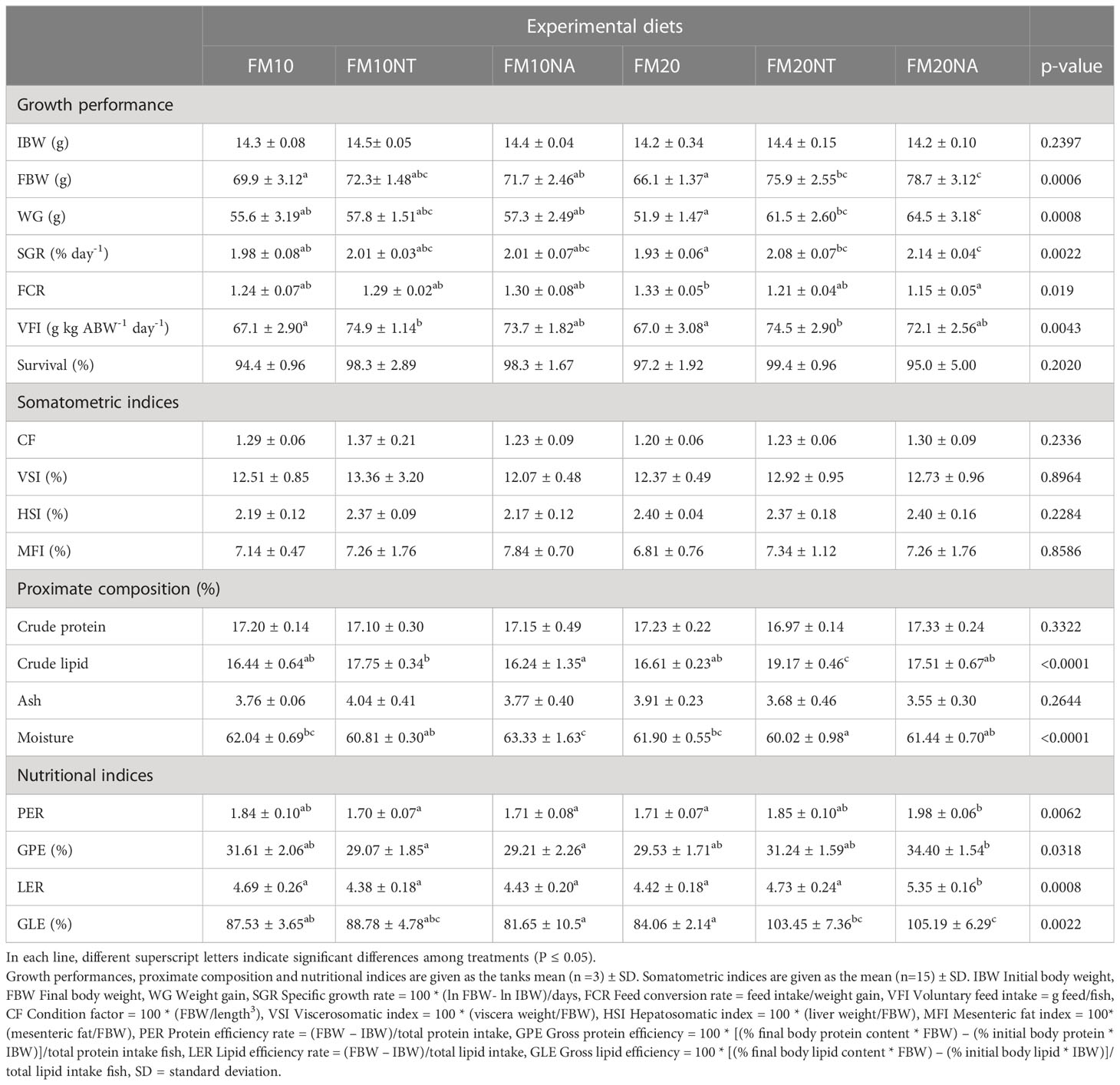
Table 3 Growth performance, somatometric indices, proximate composition and nutritional indices of European sea bass fed the experimental diets over 80 days.
3.2 Cytokines gene expression
The gene expression of four genes involved in the immune and inflammatory response are presented in Figure 1. Significant effect of FM level was found for IL-1 β and TGF-β at the end of the feeding trial (T1). Moreover, a significant interaction between FM level and NA/NT supplementation (FMxS) was observed for TGF-β at the end of the feeding trial (T1). In particular at this time point (T1) the IL-1β was upregulated in animals fed with FM10 diet supplemented with NA or NT compared to FM10 control diet with a significant increase in FM10NA group. Conversely, the TGF-β was downregulated in FM10NA and FM10NT compared to FM10 with a significant decrease in FM10NT group. In FM20 diets supplemented with NA/NT IL-1β was not upregulated and resulted less expressed compared to NA/NT supplemented FM10. Furthermore, a significant difference was observed for TGF-β between FM10NA/NT and FM20NA/NT groups: animals fed FM20NA/NT showed significantly higher expression than those fed FM10NA/NT. After suboptimal rearing condition (T2), no significant differences were observed among experimental groups by two-way ANOVA analysis. However, the post hoc test showed a significant upregulation of IL-1β in FM10 diet fed animals compared to the FM20 group.
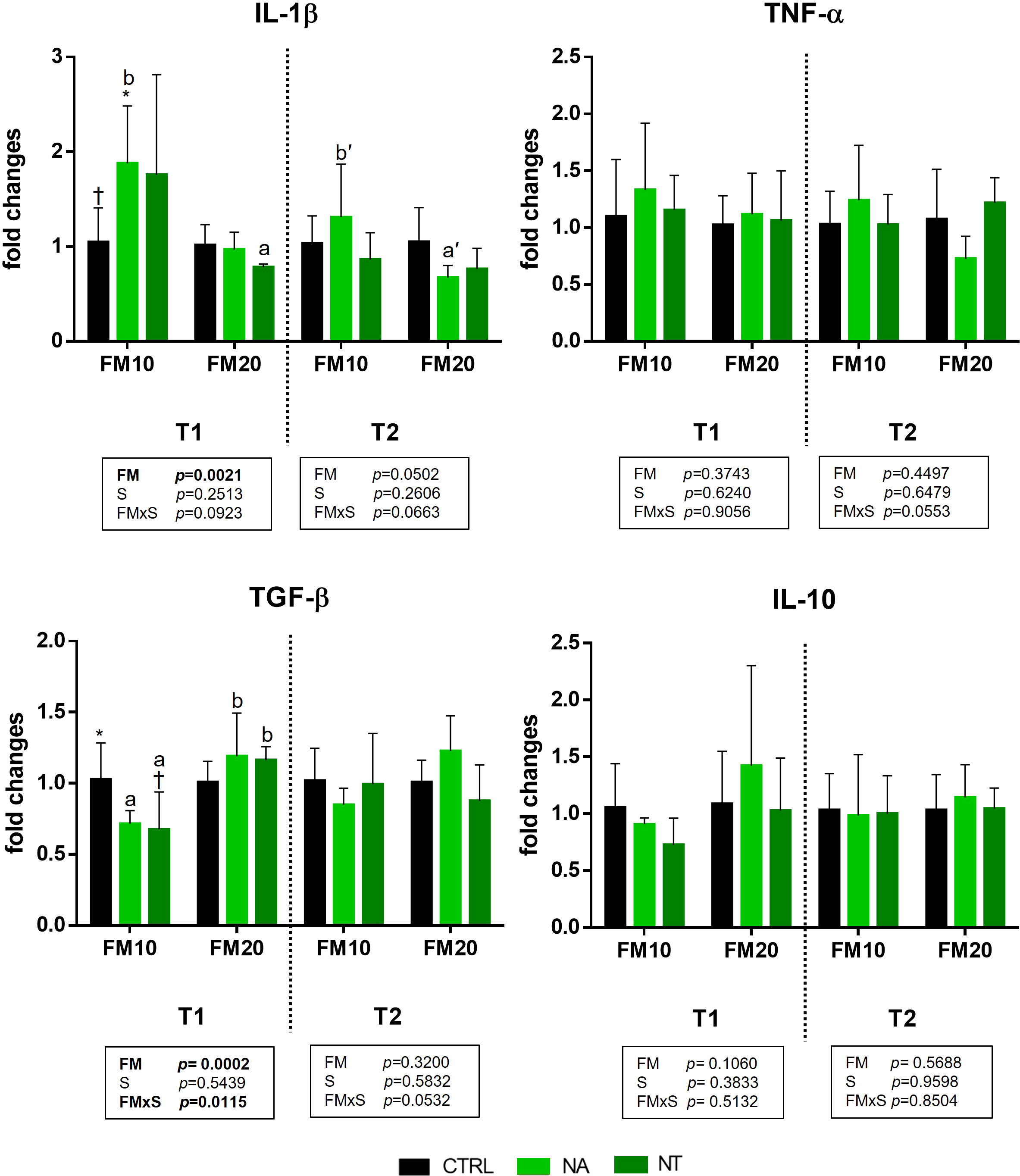
Figure 1 Immune and inflammatory cytokine gene expression in intestinal mucosa of European seabass fed with feed additives (nucleotides and nucleic acids) at two different fish meal levels over 80 days (T1) and after suboptimal rearing condition period (T2). Data are given as 6 individuals per diet. In each graph, significance is attributed to P ≤ 0.05. FM= fish meal level (10% and 20%) with inclusion of nucleotides (NT) or nucleic acids (NA) in diet at 500 ppm. IL-1β = Interleukin 1β; TNFα = Tumor necrosis factor α; TGFβ = Transforming growth factor β; IL-10 = Interleukin 10. Different symbols stand for significant differences among dietary treatments for the same FM level. Different lowercase letters (a/b, T1; a’/b’ T2) stand for significant differences between different FM levels.
3.3 Gut bacterial community
The 16S rRNA gene sequencing was performed on a total of 36 gut contents per each considered timepoint (T1 and T2) to assess how NT or NA dietary inclusion within 10% or 20% FM formulation levels could affect gut bacteria communities and consequently gut health of European sea bass before or after suboptimal rearing conditions. To this aim, samples were evaluated at different phylogenetic levels of phylum (Figure 2), family (Figure 3) and genus (Figure 4), respectively.
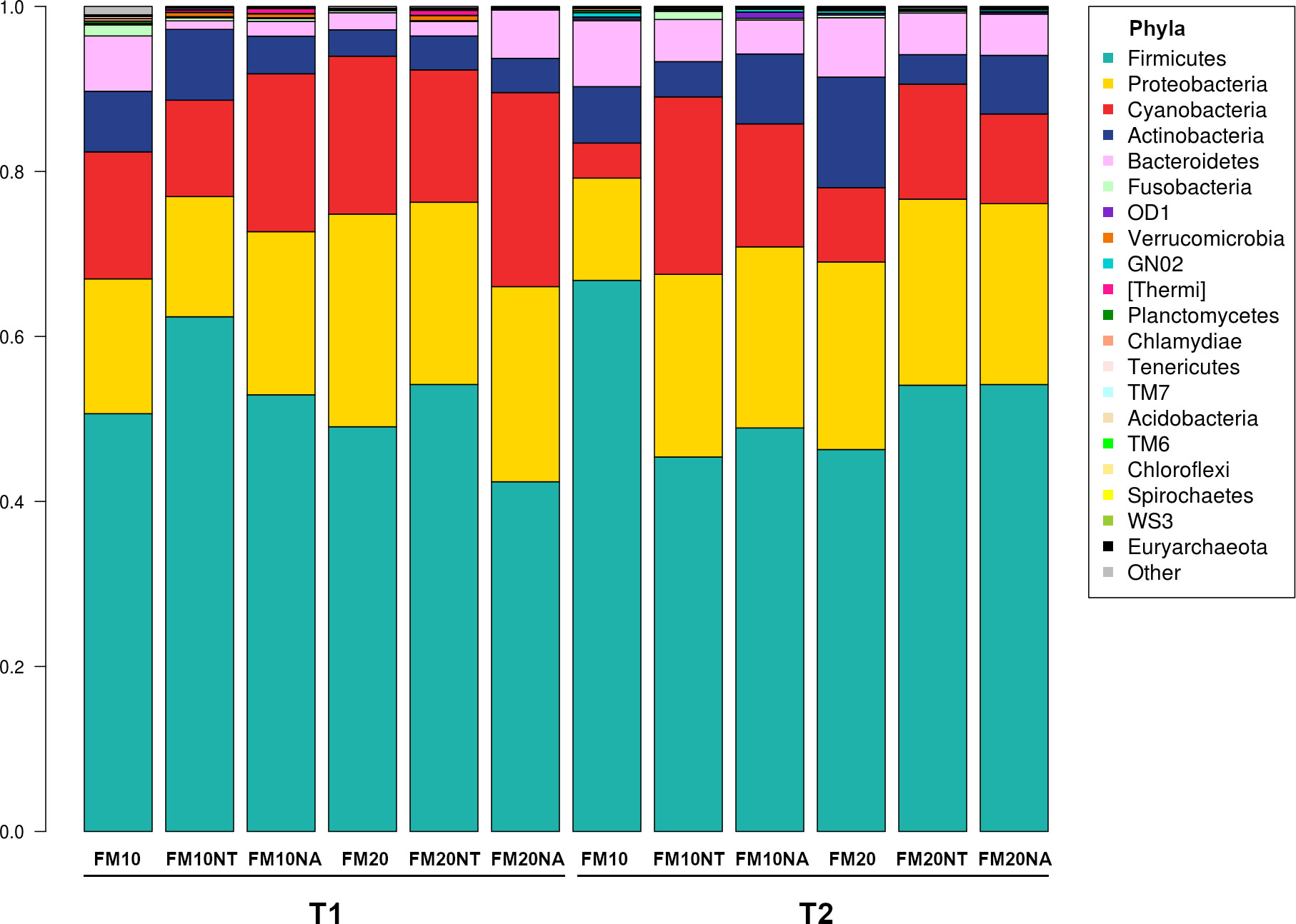
Figure 2 Microbiota composition of distal gut content of European seabass fed feed additives (nucleotides and nucleic acids) at two different fish meal levels over 80 days (T1) and after suboptimal rearing condition period (T2). Bar plot summarizing the microbiota composition at Phylum level of fish intestinal content. FM10 = fish meal 10% diet level; FM20 = fish meal 20% diet level; NT = 500 ppm nucleotide feed inclusion; NA = 500 ppm nucleic acid feed inclusion.
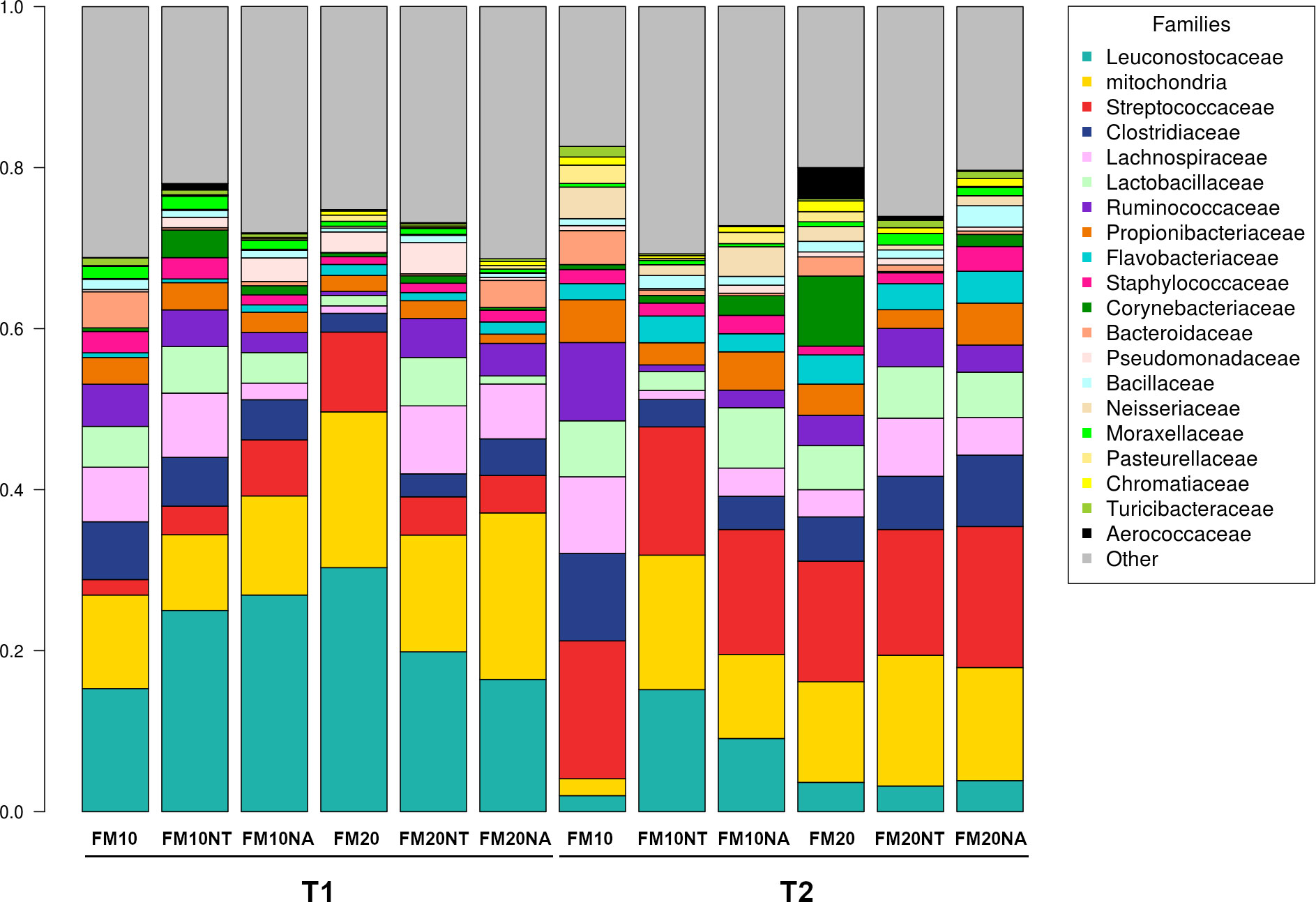
Figure 3 Microbiota composition of distal gut content of European seabass fed feed additives (nucleotides and nucleic acids) at two different fish meal levels over 80 days (T1) and after suboptimal rearing condition period (T2). Bar plot summarizing the microbiota composition at Family level of fish intestinal content. FM10 = fish meal 10% diet level; FM20 = fish meal 20% diet level; NT = 500 ppm nucleotide feed inclusion; NA = 500 ppm nucleic acid feed inclusion.
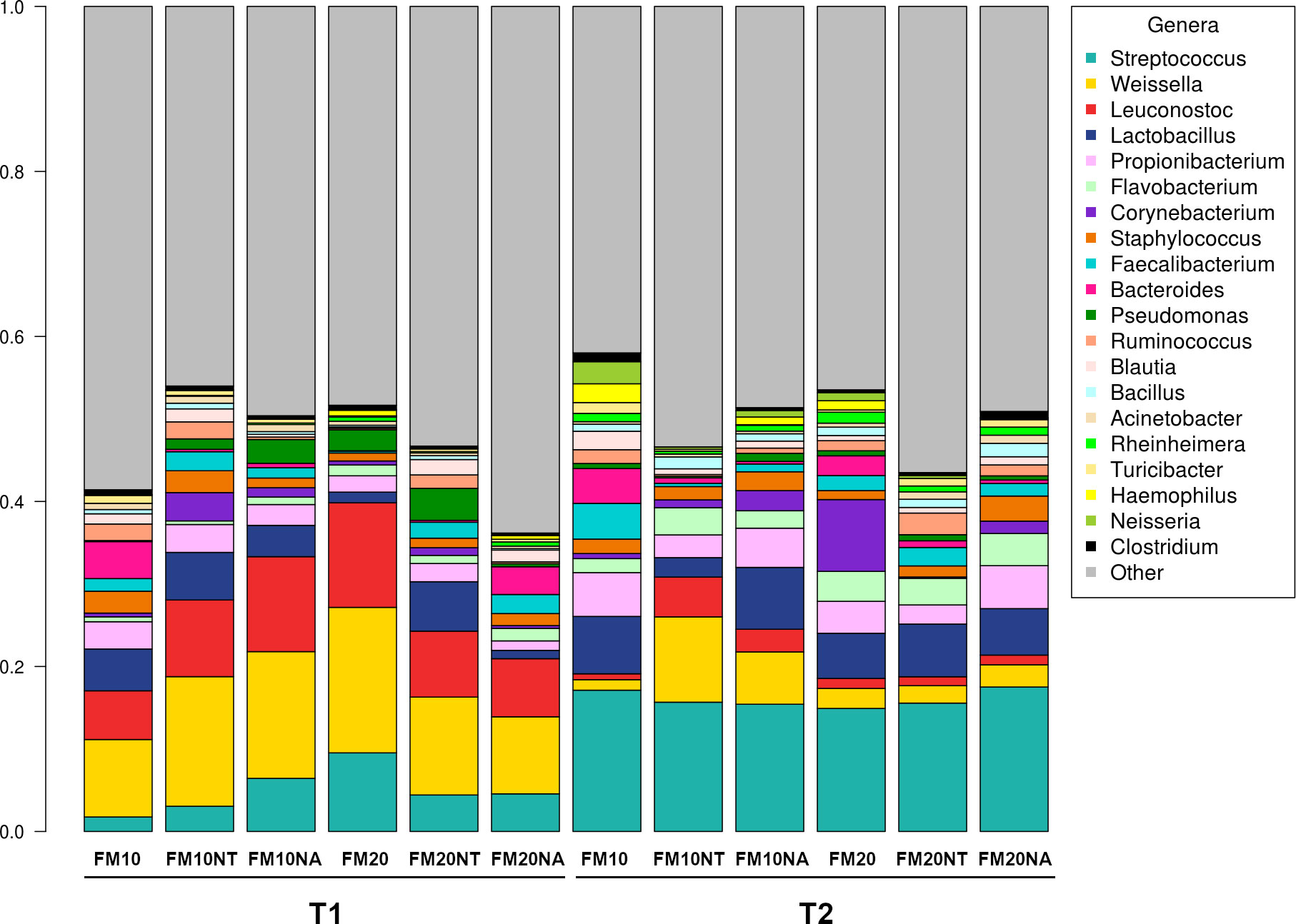
Figure 4 Microbiota composition of distal gut content of European seabass fed feed additives (nucleotides and nucleic acids) at two different fish meal levels over 80 days (T1) and after suboptimal rearing condition period (T2). Bar plot summarizing the microbiota composition at Genus level of fish intestinal content. FM10 = fish meal 10% diet level; FM20 = fish meal 20% diet level; NT = 500 ppm nucleotide feed inclusion; NA = 500 ppm nucleic acid feed inclusion.
At phylum level, the most abundant taxa observed at both considered timepoints were Firmicutes (51.8% at T1, 52.5% at T2), Proteobacteria (20.5% at T1, 20.6% at T2), Cyanobacteria (17.5% at T1, 12.5% at T2), Actinobacteria (5.3% at T1, 7.2% at T2) and Bacteroidetes (3.3% at T1, 5.6% at T2) (Figure 2). The most dominating families were represented by Leuconostocaceae (22.2% at T1, 6.2% at T2), Clostridiaceae (4.6% at T1, 6.5% at T2), Streptococcaceae (5.5% at T1, 16.3% at T2), and Lactobacillaceae (3.8% at T1, 5.5% at T2) (Figure 3). At the end of the feeding trial the genus level abundance was mostly represented by Weissella (13.1% at T1, 4% at T2), followed by Leuconostoc (9.1% at T1), Streptococcus (5% at T1, 16.1% at T2), Lactobacillus (3.8% at T1, 5.5% at T2), and Propionibacterium (2.3% at T1, 4% at T2). (Figure 4).
In Figures 5, 6 are reported the most represented taxa at family and genus level (mean rel. ab. > 1%) showing significant differences among treatments at T1 and T2. Supplementary Figures 2-4 reports differences among treatments of minor represented taxa (mean rel. ab. < 1%).
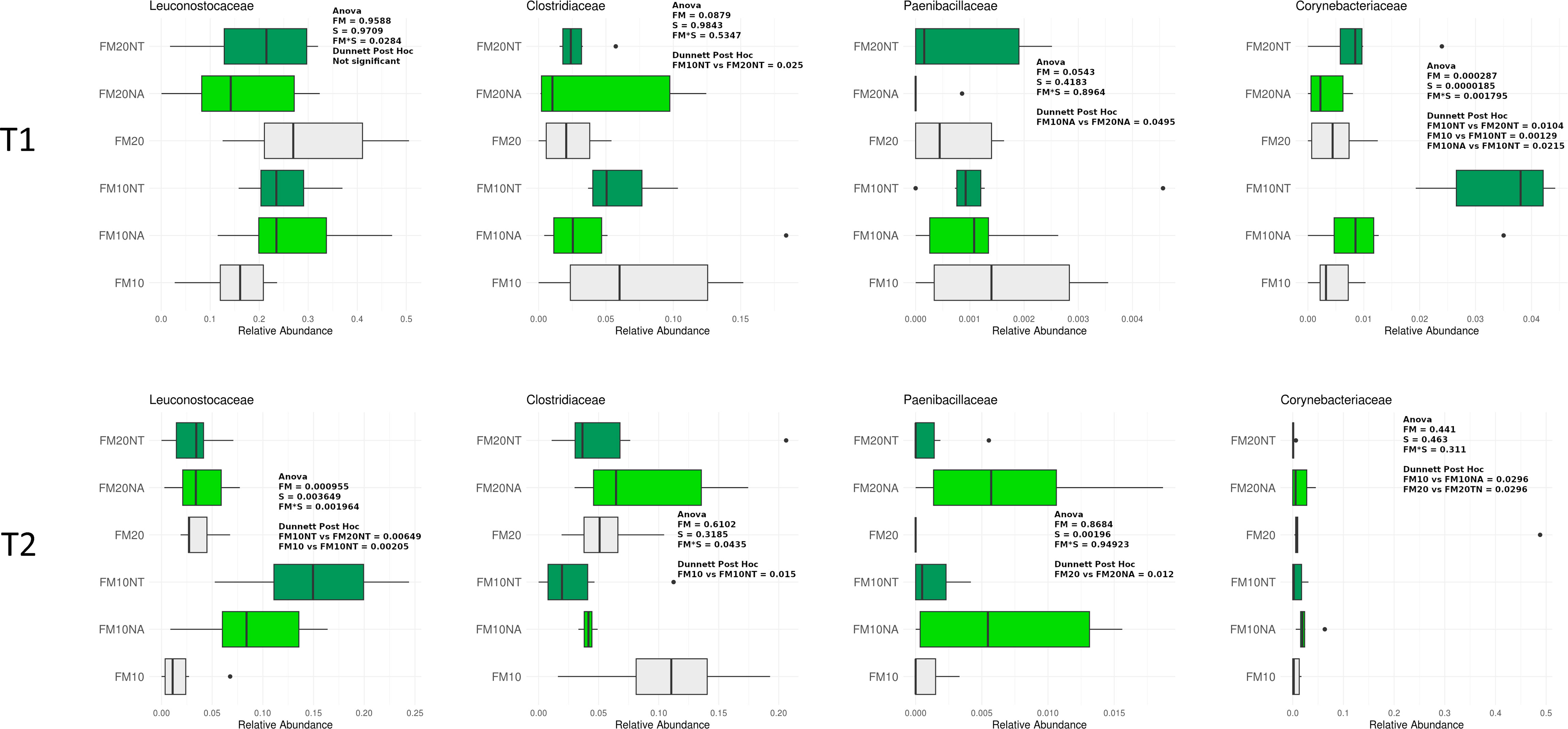
Figure 5 Taxonomic composition of bacterial communities of distal gut content of European seabass fed feed additives (nucleotides and nucleic acids) at two different fish meal levels over 80 days (T1) and after suboptimal rearing condition period (T2). Distributions of relative abundance of family that showed a significant variation between groups fed with different diets (two-way ANOVA, FM= fish meal level; S=feed supplemented with nucleotide, NT or nucleic acid, NA). Only the most abundant families with a mean relative abundance >1% in all groups were represented. FM10 = fish meal 10% diet level; FM20 = fish meal 20% diet level; NT = 500 ppm nucleotide feed inclusion; NA = 500 ppm nucleic acid feed inclusion.
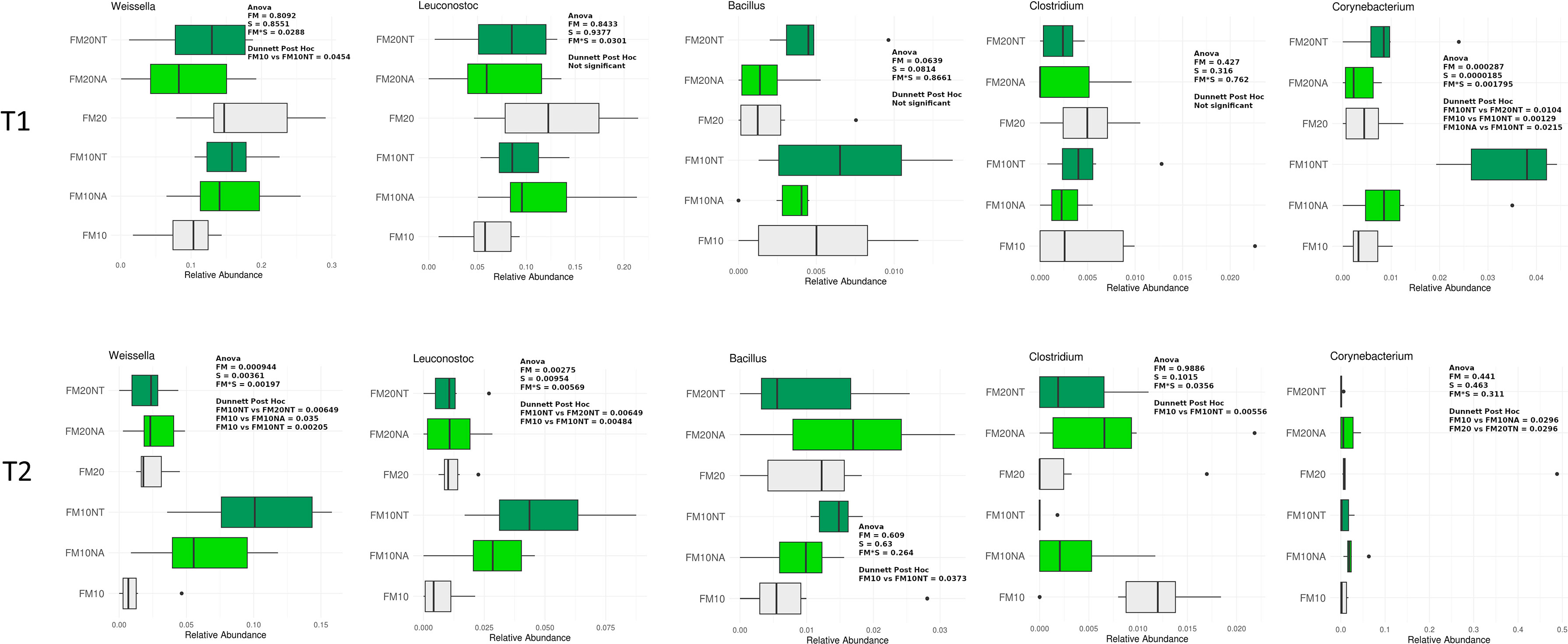
Figure 6 Taxonomic composition of bacterial communities of distal gut content of European seabass fed feed additives (nucleotides and nucleic acids) at two different fish meal levels over 80 days (T1) and after suboptimal rearing condition period (T2). Distributions of relative abundance of genus that showed a significant variation between groups fed with different diets (two-way ANOVA, FM= fish meal level; S=feed supplemented with nucleotide, NT or nucleic acid, NA), only most abundant genus with a mean relative abundance >1% in all groups were represented. FM10 = fish meal 10% diet level; FM20 = fish meal 20% diet level; NT = 500 ppm nucleotide feed inclusion; NA = 500 ppm nucleic acid feed inclusion.
At T1, Corynebacteriaceae was significantly affected by S and FM level, with a significant interaction of SxFM level. In particular, Corynebacteriaceae abundance was higher in FM10NT compared to FM10, FM10NA and FM20NT. At T2, Leuconostocaceae abundance was significantly affected by S and FM level, with a significant interaction of SxFM level. In particular Leuconostocaceae abundance was higher in FM10NT than FM10 and higher in FM10NT than FM20NT. At the same time a significant interaction FMxS affected Clostridiaceae abundance which was lower in FM10NT than FM10. Paenibacillaceae abundance was affected by S, showing higher values in FM20NA compared to FM20 (Figure 5).
At T1 a significant interaction SxFM level affected Weissella abundance which was higher in FM10NT than FM10. At the same time, Corynebacterium was significantly affected by S and FM level, with a significant interaction of SxFM level. Specifically, Corynebacterium abundance was higher in FM10NT compared to FM10, FM10NA and FM20NT. At T2, Weissella and Leuconostoc abundances were significantly affected by S and FM level, with a significant interaction of SxFM level. Specifically, both taxa were higher in FM10NT than FM10 and FM20NT, while Weissella abundance was also higher in FM10NA than FM10. At the same time Bacillus abundance tended to increase in FM10NT compared to FM10, while Clostridium displayed an opposite trend with lower values in FM10NT than FM10 (Figure 6).
At each timepoint (T1 and T2), internal α-diversity gut microbiome was accessed by Shannon, Simpson and Fischer metrics at phylum, family and genus levels (Figure 7) while β-diversity is reported in Supplementary Figure 5. At T1 no significant differences at phylum level were detected while at T2 all the indexes showed a significant FMxS interaction with lower value in FM20NT than FM10NT (Figures 7A–C). At family level, alpha diversity metrics displayed at T1 a significant FM effect and FMxS interaction for Shannon and Simpson indexes, while a significant FM effect was detected in Fisher index. All the alpha diversity indexes at T1 showed at family level higher values in FM10 than FM20 without a significant effect of NT-NA supplementation (Figures 7D-F). At genus level, both Shannon and Simpson indexes displayed at T1 a significant FM and S effects, while a FM effect was observed for Fisher index. Specifically, FM20NT displayed higher values compared to FM20 for all the alpha diversity metrics analysed at T1, while at T1 no differences were detected (Figures 7G-I). Significant separation in the principal coordinate analyses was mostly detected at T2 in FM10 vs FM10NT (Supplementary Figure 5).
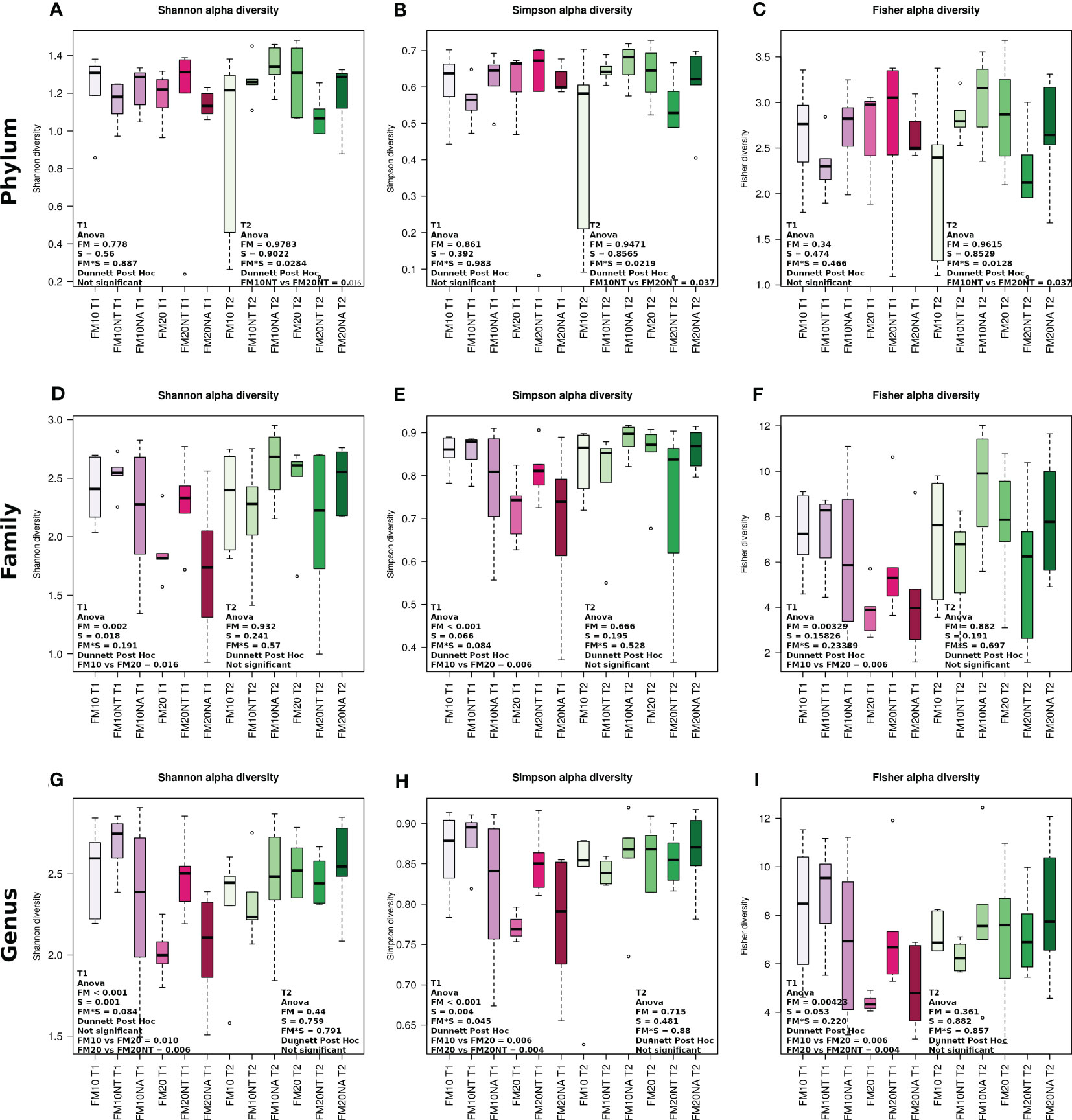
Figure 7 Alpha diversity of bacterial communities of distal gut content of European seabass fed feed additives (nucleotides and nucleic acids) at two different fish meal levels over 80 days (T1) and after suboptimal rearing condition period (T2). Boxplots show alpha diversity values measured by Shannon diversity, Simpson diversity and Fisher diversity at phylum (A–C), family (D–F) and genus (G–I) level Simpson diversity and Fisher diversity. Differences were detected by two-way anova (FM= fish meal level; S=feed supplemented with nucleotide, NT or nucleic acid, NA). FM10 = fish meal 10% diet level; FM20 = fish meal 20% diet level; NT = 500 ppm nucleotide feed inclusion; NA = 500 ppm nucleic acid feed inclusion.
4 Discussion
Since exogenous NT and NA supply have attracted attention in the last two decades, the present study is an attempt to fulfil some of the gaps in aquaculture research. Our results showed that 500 ppm of NT enhanced feed intake in 10 and 20% FM diet. Whether used as NA or NT, composition of nucleotides and their molecular form is crucial, since some compounds can stand as feed enhancer or feed intake inhibitors like free-adenine (Rumsey et al., 1992). The palatability mechanism is still not well known; however free 5’-NT are known as powerful gustatory stimulants on their own (Morais, 2017). Fish gustatory receptors (taste buds) have been found to have different positions on body surface among species and also attractiveness to different NT molecules is species-specific (Morais, 2017). For example, jack mackerel displayed appetite for IMP, GMP, UMP, UDP, UTP, while it was not attracted by nucleosides such as inosine, adenosine, guanosine and uridine, or other NT (AMP, ADP, ATP, IDP, GDP, xanthosine 5V-monophosphate, 3VIMP, 3V-UMP, 2-deoxy-IMP, allyltio-IMP), (Ikeda et al., 1991). In largemouth bass dietary IMP supply (2800 mg kg-1) displayed an FI increase of 46% to soybean meal-based diet without additive, but FI of animals fed both IMP dose diets (2800 or 5600 mg kg-1) was lower than fish fed 10% FM diet (Kubitza et al., 1997). The authors explained that probably FM is a relatively high source of IMP, therefore, the beneficial influence of IMP supplementation generally is not noticeable when fish meal is added to aquafeed formulations (Li and Gatlin, 2006). Concerning NA palatability effect, similar observations were reported by Peres and Oliva-Teles (2003), where low and high protein diets (39% and 50%, with 62.1% FM and 46.6% FM, respectively) supplied with yeast-RNA at 6.2% and 12.4% increased feed intake in European sea bass. Accordingly, the inclusion of 5.8% RNA extract combined with 44.4% FM led to a significantly higher FI and growth in gilthead sea bream (Oliva-Teles et al., 2006). On the contrary, RNA extract included at dietary levels of 3-6% in rainbow trout or 4-8% in turbot had no significant effect on FI (Fournier et al., 2002). In the present study, FCR was consistently reduced by 25.6% in FM20NA compared to FM20 control diet. Consequently, SGR of FM20 met a major enhancement by 11.0% in FM20NA diet. These findings are consistent with Magouz et al. (2021), where sea bass specimens fed increasing doses of NT blend (500, 1000, 1500 mg kg-1) had improved SGR, especially at 500 mg kg-1. On the same species another recent study testing diets containing 0%, 0.15% and 0.3% inclusion of a NT mixture found a positive effect on growth (Bowyer et al., 2019). However, rainbow trout fed 0, 8, 16 and 24 mg kg diet-1 of same NT blend supplied in diets with low FM levels of 0% and 5% were found to have a lesser growth and feed efficiency compared to a control group fed 25% FM without NT inclusion (Liu, 2016). Similar growth and feed efficiency improvements were also found in red drum Sciaenops ocellatus (Cheng et al., 2011), greater amberjack Seriola dumerili (Hossain and Koshio, 2017a), red sea bream Pagrus major (Hossain et al., 2016; Hossain and Koshio, 2017a), zebrafish Danio rerio (Guo et al., 2017), turbot Scophtalmus maximus (Peng et al., 2013), and gilthead sea bream Sparus aurata (Peres and Oliva-Teles, 2003; El-Nokrashy et al., 2021). In our study, the synergic growth and feed intake improvements of animals fed FM20NT may reveal a satisfied NT exogenous requirement for the considered life stage of this species. This hypothesis might confirm that exogenous supply of NT may promote growth of juvenile fish meeting their high rate of cell replication, especially when dietary FM is heavily limited (Li and Gatlin, 2006). In addition, the differences in feed and nutrient efficiency observed at the different FM levels may indicate that the requirements are met when 20% of FM is included, thus indicating that a possible higher NT/NA inclusion level should be considered for dietary FM level of 10%. Although both NT and NA at 20% FM promoted the overall growth, it seemed that NT had a greater role in enhancing feed palatability, while NA promoted feed efficiency. Though no significant differences occurred, HSI values were found to be in alignment with previous studies on same species and size (Peres and Oliva-Teles, 2003; Busti et al., 2020; Magouz et al., 2021). The liver size is an indicator of the nutritional status of fish (Hossain and Koshio, 2017a). A study on rainbow trout reported that dietary NT tended to change liver status of fish fed low FM diet by decreasing HSI and plasma alanine aminotransferase (ALT) activity (Ridwanudin et al., 2019). The increase of HSI was attributed as a biomarker of proper nutrient storage and health when red sea bream was fed diets with added UMP (Hossain and Koshio, 2017a). NA and NT diet showed the highest lipid efficiency when added to 20% FM. These outcomes are in contrast with findings described in European sea bass (Magouz et al., 2021 and Bowyer et al., 2019) and gilthead sea bream (Oliva-Teles et al., 2006). However, according to the present results, increased body lipid content was described in red drum juveniles fed a blend of NT supplemented diets (Li et al., 2004). The increase in carcass lipid content may indicate the involvement of NT in lipid metabolism or nutrient uptake at enterocytes level as stated by Li et al. (2015).
The fish intestinal surface represents the first line of defense against potentially harmful exogenous factors such as pathogens, antinutrients and environmental stressors; and eu- or dis-byosis assessment of gut microbiome (GM), and expression of innate immune genes changes are key actors of gut-health status (Busti et al., 2020). As immune biomarkers, cytokines are low molecular weight glycoproteins involved in the regulation of immune response, and can be elicited by immunostimulants (Zou and Secombes, 2016). They are mainly secreted by cells of both the innate and acquired immune system in response to microbial invasion and tissue injury, and act as signal cell messengers via specific-receptors on target cell surfaces, thus inducing physiological response (Sakai and Kono, 2021). When inflammatory response occurs, tumor necrosis factor alpha (TNF-α) secretion sparks the cytokine cascade with IL-1β followed by the release of a myriad of other molecules and chemoattractant to promote neutrophil and macrophage migration in loco, acting as pro-inflammatory markers (Pelusio et al., 2020). Transforming growth factor-β (TGF-β) is one of the anti-inflammatory cytokines that depicts a down-regulated effect on the expression of numerous cytokines and is associated with the deactivation of macrophages. Interleukin-10 (IL-10) is another anti-inflammatory cytokine that inhibits bacterial lipopolysaccharide (LPS) affecting pro-inflammatory cytokines (Reda et al., 2018). In the present study the gene expression analysis showed a significant effect of diets on the activation of local immune response on some intestinal cytokines. In fish fed FM10 groups an upregulation of the proinflammatory cytokine IL-1β and a concurrent downregulation of the anti-inflammatory TGF-β gene was observed prominently for fish fed the NA/NT supplemented diets signaling an activation of the immune response. Previous studies showed the upregulation of cytokines as an effect associated to nucleotide supplementation in turbot (Scophthalmus maximus) and Nile tilapia (Oreochromis niloticus) (Reda et al., 2018). NT enhanced the expression of immune genes from different organs involved in nonspecific defenses of fish like gills, kidney and intestine (Hossain et al., 2020). In particular, mRNA levels of intestinal cytokines including IL-1β, IL-10, TNF-α and TGF-β were significantly higher in Nile tilapia fed dietary NT (Reda et al., 2018). A later study on guanidinoacetic acid supplementation in the same species showed a different effect on liver proinflammatory and anti-inflammatory cytokines (Aziza and El-Wahab, 2019). In particular, IL-1β and TGF-β1 mRNA level were respectively downregulated and upregulated in Nile tilapia fed guanidinoacetic acid supplemented diet for 60 days. Similarly, anti-inflammatory effect was observed in zebrafish (Danio rerio) fed a diet supplemented with RNA yeast extract; in this case proinflammatory cytokines such as IL-1, IL-8 and TNF-α were downregulated, whereas the anti-inflammatory cytokine IL-10 was upregulated or not modulated depending on the growth stage of the investigated larvae (Falcinelli et al., 2018). In our study, we observed opposite expression trends of IL-1β and TGF-β in accordance to their pro- and anti-inflammatory roles respectively. As a matter of fact, in NA/NT supplemented FM10 diets an upregulation of the pro-inflammatory cytokine and a downregulation of the anti-inflammatory cytokine was observed. Moreover, European sea bass TGF- β modulation seems affected mainly by the FM and supplementation interaction. In effect, this anti-inflammatory cytokine was significantly more expressed in animals fed FM20NT and FM20NA than those fed with FM10NA and FM10NT at the end of the feeding trial. Actually, the combination of high FM diet and NA/NT supplementation showed the highest expression of TGF-β gene, depicting an increased ability to regulate inflammatory process. The upregulation of anti-inflammatory cytokines such as TGF-β could also contribute to protect the epithelial integrity decreasing intestinal epithelial permeability (Busti et al., 2020). On the other hand, IL-1β expression was affected only by FM level. Moreover, the NA/NT supplementation seems not to affect the gene expression after the suboptimal rearing conditions. Indeed, the two-way ANOVA analysis showed no significant influence of diets. However, a FM-associated effect was observed for IL-1β expression levels limited to NA supplemented animals at this time point with the FM20NA group showing the lowest IL-1β expression.
Though many studies have turned their attention on NT and NA, to the best of our knowledge the present study is the first to investigate their effects on gut microbiota on this species. A different response in the microbial internal diversity was observed. At the end of the growing period, NT increased internal diversity at genus level when included in 20% FM diet. Increase in GM diversity may have positive implications for gut health due to increased competition against opportunistic pathogens (Parma et al., 2020; Moroni et al., 2021). In addition, a wider range of bacteria supported by a higher diversity may promote a more diverse number of host functions (Solé-Jiménez et al., 2021). In accordance, an increase in gut microbiome alpha diversity was also detected in gilthead sea bream fed 20% inclusion of bacterial single cell protein (Marchi et al., 2023).
At phylum level the gut bacterial community of sea bass was mainly represented by Firmicutes followed by Proteobacteria, Cyanobacteria and Actinobacteria, findings which are consistent with previous works on the same species (Parma et al., 2019; Busti et al., 2020). After the suboptimal environmental condition NT and NA added to 10% FM diet were able to promote beneficial taxa such as Weissela and Leuconostoc, when both taxa displayed a general decreasing trend. At this time a significant separation in the PCoA analyses also indicated an overall impact of NT and NA supplemented diet on the gut microbiome layout. These lactic acid bacteria genera are well known to have positive influence on host gut intestinal health in fish species. For instance, Leuconostoc has been described to be able to produce bacteriocins tolerant to high thermal stress and wide range of pH, which form pores in the target membrane of eventual bacteria pathogens (Ringo et al., 2018). Beneficial probiotic effects reported on Weissella in aquaculture have been associated with intestinal histology improvement by increasing height, width, number and mucosal productivity of villi and has been proposed for its application as a probiotic in aquaculture (Ringo et al., 2018; Pelusio et al., 2021). A similar overall decrease of LAB species after suboptimal rearing conditions (high temperature and low oxygen) was also described in previous studies (Busti et al., 2020) on European sea bass. In aquaculture species, LAB feed supplementation or their proliferation in GM has been attributed to positive outputs such as improved performance (LeBlanc et al., 2017; Xia et al., 2020), immune response and disease resistance, though their action mechanisms are still poorly understood (Ringo et al., 2018; Sun et al., 2022). After suboptimal condition, dietary NT was also able to increase Bacillus abundance in low fish meal diet. Bacillus is a well-known probiotics taxon in fish species and recently its abundance was found to significantly increase in gilthead sea bream fed bacterial single cell protein, raw material which are rich in nucleic acid (Marchi et al., 2023). In addition, after the suboptimal environmental condition, dietary nucleotides inclusion at 10% FM was also able to counteract Clostridiaceae abundance compared to its control group. The growth of this anaerobic bacteria family was observed under similarly high temperature and low oxygen; conditions that were able to negatively affect the overall GM structure of the same species (Busti et al., 2020).
5 Conclusions
NT and NA feed inclusion at 500 ppm enhanced voluntary feed intake when included in vegetable-based diet containing 10% and 20% dietary FM. A further improvement of feed efficiency was displayed in fish fed FM20 which overall led to the highest growth performance parameters. NT and NA combined with FM20 improved lipid efficiency. Furthermore, NT and NA combined with FM20 had also positive impact on intestinal mucosa by increasing anti-inflammatory TGF-β and decreasing pro-inflammatory IL-1 β, possibly translated in an improved gut-health functionality with a consequent improved nutritional uptake and growth performance. On gut microbiome, NT and NA when included in 10% FM diet were able to exert prebiotic properties stimulating the development of beneficial bacteria taxa such as Weissella and Leuconostoc when fish underwent suboptimal elevated temperature and low oxygen levels.
In conclusion, the combination of NT/NA included at 500 ppm was able to promote growth and lipid efficiency when included in 20% FM level, assuming higher nutritional NT requirement when combined with 10% FM. On the other hand, NT/NA added in 10% FM upregulate proinflammatory IL-1β and downregulate anti-inflammatory TGF- β intestinal level and favour beneficial gut bacterial taxa after the exposure to high temperature and low oxygen.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA933537.
Author contributions
AB, LP, PC, OM, PPG, conceived and designed the experiment. NFP, LP, AB, EV, SC, FE wrote the first draft of the manuscript. NFP, SN, conducted the fish rearing and sampling. SC, EV, FE performed cytokine gene expression analyses, ADC, VI carried out microbiota and bioinformatic analysis. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors would like to thank Gillian Forlivesi Heywood for English language editing
Conflict of interest
Author PC is employed by Prosol S.p.A.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1145660/full#supplementary-material
References
AOAC. (2010). Officials methods of analysis. 17th Edition (Washington DC, USA: Association of Official Analytical Chemists).
Aziza A., El-Wahab A. A. (2019). Impact of partial replacing of dietary fish meal by different protein sources on the growth performance of Nile tilapia (Oreochromis niloticus) and whole body composition. J. Appl. Sci. 19 (5), 384–391. doi: 10.3923/jas.2019.384.391
Barros M. M., Guimarães I. G., Pezzato L. E., de Oliveira Orsi R., Fernandes Junior A. C., Teixeira C. P., et al. (2015). The effects of dietary nucleotide mixture on growth performance, haematological and immunological parameters of Nile tilapia. Aquac. Res. 46, 987–993. doi: 10.1111/ARE.12229
Bligh E. G., Dyer W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bonvini E., Bonaldo A., Parma L., Mandrioli L., Sirri R., Grandi M., et al. (2018b). Feeding European sea bass with increasing dietary fibre levels: impact on growth, blood biochemistry, gut histology, gut evacuation. Aquaculture 494, 1–9. doi: 10.1016/j.aquaculture.2018.05.017
Bonvini E., Parma L., Badiani A., Fontanillas R., Gatta P. P., Sirri F., et al. (2018a). Integrated study on production performance and quality traits of European sea bass (Dicentrarchus labrax) fed high plant protein diets. Aquaculture 484, 126–132. doi: 10.1016/j.aquaculture.2017.10.041
Bowyer P. H., El-Haroun E. R., Hassaan M., Salim H., Davies S. J. (2019). Dietary nucleotides enhance growth performance, feed efficiency and intestinal functional topography in European seabass (Dicentrarchus labrax). Aquac. Res. 50, 1921–1930. doi: 10.1111/ARE.14078
Burrells C., Williams P. D., Southgate P. J., Wadsworth S. L. (2001). Dietary nucleotides: a novel supplement in fish feeds: effects on vaccination, salt water transfer, growth rates and physiology of Atlantic salmon (Salmo salar l.). Aquaculture. 199, 171–184. doi: 10.1016/S0044-8486(01)00576-2
Busti S., Rossi B., Volpe E., Ciulli S., Piva A., D’Amico F., et al. (2020). Effects of dietary organic acids and nature identical compounds on growth, immune parameters and gut microbiota of European sea bass. Sci. Rep 10 (1), 21321. doi: 10.1038/s41598-020-78441-9
Carver J. D., Allan Walker W. (1995). The role of nucleotides in human nutrition. J. Nutr. Biochem. 6, 58–72. doi: 10.1016/0955-2863(94)00019-I
Cheng Z., Buentello A., Gatlin D. M. (2011). Dietary nucleotides influence immune responses and intestinal morphology of red drum sciaenops ocellatus. Fish Shellfish Immunol. 30, 143–147. doi: 10.1016/j.fsi.2010.09.019
Cosgrove M. (1998). Perinatal and infant nutrition. Nucleotides. Nutr. 14, 748–751. doi: 10.1016/S0899-9007(98)00075-6
Dawood M. A. O., Koshio S. (2020). Application of fermentation strategy in aquafeed for sustainable aquaculture. Rev. Aquac 12 (2020), 987–1002. doi: 10.1111/RAQ.12368
Dawood M. A. O., Koshio S., Esteban M.Á. (2018). Beneficial roles of feed additives as immunostimulants in aquaculture: a review. Rev. Aquac. 10, 950–974. doi: 10.1111/RAQ.12209
De Cesare A., Sirri F., Manfreda G., Moniaci P., Giardini A., Zampiga M., et al. (2017). Effect of dietary supplementation with lactobacillus acidophilus D2/CSL (CECT 4529) on caecum microbioma and productive performance in broiler chickens. PLoS One 12, 1–21. doi: 10.1371/journal.pone.0176309
De Cruz C. R., Castillo S., Yamamoto F. Y., Gatlin D. M. (2020). Exploring potential synergistic effects between dietary adenosine and inosine monophosphate on growth performance and acute stress-induced immune responses of hybrid striped bass morone chrysops × morone saxatilis. Aquac. Nutr. 26, 2195–2210. doi: 10.1111/anu.13157
Dülger N., Kumlu M., Türkmen S., Ölçülü A., Tufan Eroldoĝan O., Asuman Yilmaz H., et al. (2012). Thermal tolerance of European Sea bass (Dicentrarchus labrax) juveniles acclimated to three temperature levels. J. Therm. Biol. 37, 79–82. doi: 10.1016/j.jtherbio.2011.11.003
El-Nokrashy A. M., El-Banna R. A., Edrise B. M., Abdel-Rahim M. M., Jover-Cerdá M., Tomás-Vidal A., et al. (2021). Impact of nucleotide enriched diets on the production of gilthead seabream, sparus aurata fingerlings by modulation of liver mitochondrial enzyme activitity, antioxidant status, immune gene expression, and gut microbial ecology. Aquaculture. 535, 736398. doi: 10.1016/J.AQUACULTURE.2021.736398
Falcinelli S., Randazzo B., Vargas Abúndez J. A., Cangiotti G., Olivotto I., Carnevali O. (2018). Kluyveromyces fragilis RNA extract supplementation promotes growth, modulates stress and inflammatory response in zebrafish. Aquaculture Res. 49 (4), 1521–1534. doi: 10.1111/are.13607
Faliex E., Da Silva C., Simon G., Sasal P. (2008). Dynamic expression of immune response genes in the sea bass, dicentrarchus labrax, experimentally infected with the monogenean diplectanum aequans. Fish Shellfish Immunol. 24 (2008), 759–767. doi: 10.1016/j.fsi.2008.02.011
Fournier V., Gouillou-Coustans M. F., Métailler R., Vachot C., Moriceau J., Le Delliou C., et al. (2002). Nitrogen utilisation and ureogenesis as affected by dietary nucleic acid in rainbow trout (Oncorhynchus mykiss) and turbot (Psetta maxima). Fish Physiol. Biochem. 26 (2002), 177–188. doi: 10.1023/A:1025465603604
Gil A. (2002). Modulation of the immune response mediated by dietary nucleotides. Eur. J. Clin. Nutr. 56, 1–4. doi: 10.1038/sj.ejcn.1601475
Guillen J., Carvalho N. (2016). Scientific, technical and economic committee for fisheries (STECF) economic report of EU aquaculture secto. doi: 10.2788/677322
Guo X., Ran C., Zhang Z., He S., Jin M., Zhou Z. (2017). The growth-promoting effect of dietary nucleotides in fish is associated with an intestinal microbiota-mediated reduction in energy expenditure. J. Nutr. 174, 781–788.
Guo X., Li J., Ran C., Wang A., Xie M., Xie Y., et al. (2019). Dietary nucleotides can directly stimulate the immunity of zebrafish independent of the intestinal microbiota. Fish Shellfish Immunol. 86, 1064–1071. doi: 10.1016/J.FSI.2018.12.058
Hossain M. S., Koshio S. (2017a). Dietary substitution of fishmeal by alternative protein with guanosine monophosphate supplementation influences growth, digestibility, blood chemistry profile, immunity, and stress resistance of red sea bream, pagrus major. Fish Physiol. Biochem. 43, 1629–1644. doi: 10.1007/s10695-017-0398-4
Hossain M. S., Koshio S., Ishikawa M., Yokoyama S., Sony N. M. (2016). Dietary nucleotide administration influences growth, immune responses and oxidative stress resistance of juvenile red sea bream (Pagrus major). Aquaculture 455, 41–49. doi: 10.1016/j.aquaculture.2016.01.008
Hossain M. S., Koshio S., Ishikawa M., Yokoyama S., Sony N. M. (2017c). Dietary supplementation of uridine monophosphate enhances growth, hematological profile, immune functions and stress tolerance of juvenile red sea bream, pagrus major. Aquaculture 475, 29–39. doi: 10.1016/j.aquaculture.2017.04.008
Hossain M. S., Koshio S., Ishikawa M., Yokoyama S., Sony N. M., Kader M. A., et al. (2017b). Effects of dietary administration of inosine on growth, immune response, oxidative stress and gut morphology of juvenile amberjack, seriola dumerili. Aquaculture 468, 534–544. doi: 10.1016/j.aquaculture.2016.11.020
Hossain M. S., Koshio S., Kestemont P. (2020). Recent advances of nucleotide nutrition research in aquaculture: a review. Rev. Aquac. 12, 1028–1053. doi: 10.1111/RAQ.12370
Hua K., Bureau D. P. (2012). Exploring the possibility of quantifying the effects of plant protein ingredients in fish feeds using meta-analysis and nutritional model simulation-based approaches. Aquaculture 356–357, 284–301. doi: 10.1016/J.AQUACULTURE.2012.05.003
Hunt A.Ö., Özkan Yilmaz F., Engin K., Berköz M., Gündüz S. G., Yalin S., et al. (2014). The effects of fish meal replacement by yeast based nucleotides on growth, body composition and digestive enzyme activity in rainbow trout juveniles (Onchorchyncus mykiss). Isr. J. Aquac. - Bamidgeh. 66, 1-10. doi: 10.46989/001c.20745
Ikeda I., Hosokawa H., Shimeno S., Takeda M. (1991). Feeding stimulant activity of nucleotides, tryptophan, and their related compounds for jack mackerel, bull. Japanese Soc Fish. Ocean. 57, 1539–1542.
Kader M. A., Bulbul M., Abol-Munafi A. B., Asaduzzaman M., Mian S., Noordin N. B. M., et al. (2018). Modulation of growth performance, immunological responses and disease resistance of juvenile Nile tilapia (Oreochromis niloticus) (Linnaeus 1758) by supplementing dietary inosine monophosphate. Aquac. Rep. 10, 23–31. doi: 10.1016/j.aqrep.2018.03.003
Kenari A. A., Mahmoudi N., Soltani M., Abediankenari S. (2013). Dietary nucleotide supplements influence the growth, haemato-immunological parameters and stress responses in endangered Caspian brown trout (Salmo trutta caspius kessler 1877). Aquac. Nutr. 19, 54–63. doi: 10.1111/J.1365-2095.2012.00938.X
Kubitza F., Lovshin L. L., Lovell R. T. (1997). Identification of feed enhancers for juvenile largemouth bass micropterus salmoides. Aquaculture 148, 191–200. doi: 10.1016/S0044-8486(96)01417-2
LeBlanc J. G., Chain F., Martín R., Bermúdez-Humarán L. G., Courau S., Langella P. (2017). Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 16, 1–10. doi: 10.1186/s12934-017-0691-z
Li P., Gatlin D. M. (2006). Nucleotide nutrition in fish: current knowledge and future applications. Aquaculture 251, 141–152. doi: 10.1016/J.AQUACULTURE.2005.01.009
Li P., Lewis D. H., Gatlin D. M. (2004). Dietary oligonucleotides from yeast RNA influence immune responses and resistance of hybrid striped bass (Morone chrysops X morone saxatilis) to streptococcus iniae infection. Fish Shellfish Immunol. 16, 561–569. doi: 10.1016/j.fsi.2003.09.005
Li P., Zhao J., Gatlin D. M. (2015). Chapter 12, diet. nutr. addit. Fish Heal, 249–269. doi: 10.1002/9781119005568.CH12
Liu B. (2016). he effect of dietary nucleotide supplementation on growth and feed efficiency of rainbow trout (Oncorhynchus mykiss) fed fish meal-free and animal protein-free diets. M.S thesis. Guelph, ON: University of Guelph, 74.
Low C., Wadsworth S., Burrells C., Secombes C. J. (2003). Expression of immune genes in turbot (Scophthalmus maximus) fed a nucleotide-supplemented diet. Aquaculture 221, 23–40. doi: 10.1016/S0044-8486(03)00022-X
Magouz F. I., Abdel-Rahim M. M., Lotfy A. M., Mosbah A., Alkafafy M., Sewilam H., et al. (2021). Dietary nucleotides enhanced growth performance, carcass composition, blood biochemical, and histology features of European sea bass, dicentrarchus labrax l. Aquac. Rep. 20, 100738. doi: 10.1016/J.AQREP.2021.100738
Makridis P., Mente E., Grundvig H., Gausen M., Koutsikopoulos C., Bergheim A. (2018). Monitoring of oxygen fluctuations in seabass cages (Dicentrarchus labrax l.) in a commercial fish farm in Greece. Aquac. Res. 49 (2018), 684–691. doi: 10.1111/are.13498
Marchi A., Bonaldo A., Scicchitano D., Candela M., De Marco A., Falciglia S., et al. (2023). Feeding gilthead sea bream with increasing dietary bacterial single cell protein level: implication on growth, plasma biochemistry, gut histology, and gut microbiota. Aquaculture 565, 739132. doi: 10.1016/j.aquaculture.2022.739132
Morais S. (2017). The physiology of taste in fish: potential implications for feeding stimulation and gut chemical sensing. Rev. Fish. Sci. Aquac. 25, 133–149. doi: 10.1080/23308249.2016.1249279
Moroni F., Naya-Català F., Piazzon M. C., Rimoldi S., Calduch-Giner J., Giardini A., et al. (2021). The effects of nisin-producing lactococcus lactis strain used as probiotic on gilthead Sea bream (Sparus aurata) growth, gut microbiota, and transcriptional response. Front. Mar. Sci. 8, 659519. doi: 10.3389/fmars.2021.659519
Nasopoulou C., Zabetakis I. (2012). Benefits of fish oil replacement by plant originated oils in compounded fish feeds. A review LWT. 47, 217–224. doi: 10.1016/J.LWT.2012.01.018
Oliva-Teles A., Guedes M. J., Vachot C., Kaushik S. J. (2006). The effect of nucleic acids on growth, ureagenesis and nitrogen excretion of gilthead sea bream sparus aurata juveniles. Aquaculture 253, 608–617. doi: 10.1016/j.aquaculture.2005.09.010
Palermo F. A., Cardinaletti G., Cocci P., Tibaldi E., Polzonetti-Magni A., Mosconi G. (2013). Effects of dietary nucleotides on acute stress response and cannabinoid receptor 1 mRNAs in sole, solea solea, comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 164, 477–482. doi: 10.1016/J.CBPA.2012.12.005
Parma L., Busti S., Ciulli S., Volpe E., Errani F., Oterhals Å., et al. (2023). Growth, plasma biochemistry and immune-related gene expression of European sea bass (Dicentrarchus labrax) fed bioactive peptides from farmed salmon by-products. Aquaculture 563, 738982. doi: 10.1016/j.aquaculture.2022.738982
Parma L., Pelusio N. F., Gisbert E., Esteban M. A., D’Amico F., Soverini M., et al. (2020). Effects of rearing density on growth, digestive conditions, welfare indicators and gut bacterial community of gilthead sea bream (Sparus aurata, l. 1758) fed different fishmeal and fish oil dietary levels. Aquaculture 518, 734854. doi: 10.1016/j.aquaculture.2019.734854
Parma L., Yúfera M., Navarro-Guillén C., Moyano F. J., Soverini M., D’Amico F., et al. (2019). Effects of calcium carbonate inclusion in low fishmeal diets on growth, gastrointestinal pH, digestive enzyme activity and gut bacterial community of European sea bass (Dicentrarchus labrax l.) juveniles. Aquaculture 510, 283–292. doi: 10.1016/J.AQUACULTURE.2019.05.064
Pelusio N. F., Bonaldo A., Gisbert E., Andree K. B., Esteban M. A., Dondi F., et al. (2022). Different fish meal and fish oil dietary levels in European Sea bass: welfare implications after acute confinement stress. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.779053
Pelusio N. F., Rossi B., Parma L., Volpe E., Ciulli S., Piva A., et al. (2020). Effects of increasing dietary level of organic acids and nature-identical compounds on growth, intestinal cytokine gene expression and gut microbiota of rainbow trout (Oncorhynchus mykiss) reared at normal and high temperature. Fish Shellfish Immunol. 107 (2020), 324–335. doi: 10.1016/j.fsi.2020.10.021
Pelusio N. F., Scicchitano D., Parma L., Dondi F., Brini E., D’Amico F., et al. (2021). Interaction between dietary lipid level and seasonal temperature changes in gilthead Sea bream sparus aurata: effects on growth, fat deposition, plasma biochemistry, digestive enzyme activity, and gut bacterial community. Front. Mar. Sci. 8, 664701. doi: 10.3389/fmars.2021.664701
Peng M., Xu W., Ai Q., Mai K., Liufu Z., Zhang K. (2013). Effects of nucleotide supplementation on growth, immune responses and intestinal morphology in juvenile turbot fed diets with graded levels of soybean meal (Scophthalmus maximus l.). Aquaculture 392–395, 51–58. doi: 10.1016/j.aquaculture.2013.02.002
Peres H., Oliva-Teles A. (2003). The effect of dietary ribonucleic acid incorporation in performance of European sea bass (Dicentrarchus labrax) juveniles. Aquaculture 215 (2003), 245–253. doi: 10.1016/S0044-8486(02)00130-8
Perry W. B., Lindsay E., Payne C. J., Brodie C., Kazlauskaite R. (2020). The role of the gut microbiome in sustainable teleost aquaculture. Proc. R. Soc B Biol. Sci. 287, 1–10. doi: 10.1098/rspb.2020.0184
Rawling M. D., Pontefract N., Rodiles A., Anagnostara I., Leclercq E., Schiavone M., et al. (2019). The effect of feeding a novel multistrain yeast fraction on European seabass (Dicentrachus labrax) intestinal health and growth performance. J. World Aquacult Soc. 50, 1108–1122. doi: 10.1111/jwas.12591
Reda R. M., Selim K. M., Mahmoud R., El-Araby I. E. (2018). Effect of dietary yeast nucleotide on antioxidant activity, non-specific immunity, intestinal cytokines, and disease resistance in Nile tilapia. Fish Shellfish Immunol. 80, 281–290. doi: 10.1016/J.FSI.2018.06.016
Ridwanudin A., Haga Y., Katagiri T., Satoh S. (2019). Effect of nucleotides supplementation to low-fish meal feed on long-chain polyunsaturated fatty acid composition of juvenile rainbow trout oncorhynchus mykiss. Aquac. Res. 50, 2218–2230. doi: 10.1111/ARE.14103
Ringo E., Hossein S., Ghosh K., Doan H. V., Beck B. R., Song S. (2018). Lactic acid bacteria in finfish–an update. Front. Microbiol. 9, 1818. doi: 10.3389/fmicb.2018.01818
Ringø E., Olsen R. E., Vecino J. L. G., Wadsworth S., Song S. K. (2012). Use of immunostimulants and nucleotides in aquaculture: a review. J. Mar. Sci. Res. Dev. 1, 104. doi: 10.4172/2155-9910.1000104
Rumsey G. L., Winfree R. A., Hughes S. G. (1992). Nutritional value of dietary nucleic acids and purine bases to rainbow trout (Oncorhynchus mykiss). Aquaculture. 108, 97–110. doi: 10.1016/0044-8486(92)90321-B
Sakai M., Kono T. (2021). Fish cytokines: current research and applications. Fish. Sci. 87, 1–9. doi: 10.1007/s12562-020-01476-4
Sepulcre M. P., Sarropoulou E., Kotoulas G., Meseguer J., Mulero V. (2007). Vibrio anguillarum evades the immune response of the bony fish sea bass (Dicentrarchus labrax l.) through the inhibition of leukocyte respiratory burst and down-regulation of apoptotic caspases. Mol. Immunol. 44, 3751–3757. doi: 10.1016/j.molimm.2007.03.021
Singhal A., Macfarlane G., Macfarlane S., Lanigan J., Kennedy K., Elias-Jones A., et al. (2008). Dietary nucleotides and fecal microbiota in formula-fed infants: a randomized controlled trial. Am. J. Clin. Nutr. 87, 1785–1792. doi: 10.1093/AJCN/87.6.1785
Solé-Jiménez P., Naya-Català F., Piazzon M. C., Estensoro I., Calduch-Giner J.À., Sitjà-Bobadilla A., et al. (2021). Reshaping of gut microbiota in gilthead Sea bream fed microbial and processed animal proteins as the main dietary protein source. Front. Mar. Sci. 8, 705041. doi: 10.3389/fmars.2021.705041
Sun Y.-Z., Zhang Y., Ringø E. (2022). Editorial: composition, functions and modulation of gut microbiota in maricultural animals. Front. Mar. Sci. 9, 985012. doi: 10.3389/fmars.2022.985012
Sutili F. J., Gatlin D. M., Heinzmann B. M., Baldisserotto B. (2018). Plant essential oils as fish diet additives: benefits on fish health and stability in feed. Rev. Aquac. 10, 716–726. doi: 10.1111/raq.12197
Tahmasebi-Kohyani A., Keyvanshokooh S., Nematollahi A., Mahmoudi N., Pasha-Zanoosi H. (2011). Dietary administration of nucleotides to enhance growth, humoral immune responses, and disease resistance of the rainbow trout (Oncorhynchus mykiss) fingerlings. Fish Shellfish Immunol. 30, 189–193. doi: 10.1016/J.FSI.2010.10.005
Vandeputte M., Gagnaire P.-A., Allal F. (2019). The European sea bass: a key marine fish model in the wild and in aquaculture. Anim. Genet. 50 (3), 195–206. doi: 10.1111/age.12779
Xia Y., Yu E. M., Lu M., Xie J. (2020). Effects of probiotic supplementation on gut microbiota as well as metabolite profiles within Nile tilapia, oreochromis niloticus. Aquaculture 527, 735428. doi: 10.1016/j.aquaculture.2020.735428
Keywords: aquaculture, feed additives, nucleotides, fishmeal, growth, cytokines, gut microbiome
Citation: Pelusio NF, Parma L, Volpe E, Ciulli S, Errani F, Natale S, De Cesare A, Indio V, Carcano P, Mordenti O, Gatta PP and Bonaldo A (2023) Yeast-extracted nucleotides and nucleic acids as promising feed additives for European sea bass (Dicentrarchus labrax) juveniles. Front. Mar. Sci. 10:1145660. doi: 10.3389/fmars.2023.1145660
Received: 16 January 2023; Accepted: 09 May 2023;
Published: 18 May 2023.
Edited by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaReviewed by:
Carmen G Feijoo, Andres Bello University, ChileH-Michael Habte-Tsion, University of Maine, United States
Adnan Gora, Central Marine Fisheries Research Institute (ICAR), India
Copyright © 2023 Pelusio, Parma, Volpe, Ciulli, Errani, Natale, De Cesare, Indio, Carcano, Mordenti, Gatta and Bonaldo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luca Parma, bHVjYS5wYXJtYUB1bmliby5pdA==
 Nicole Francesca Pelusio1
Nicole Francesca Pelusio1 Luca Parma
Luca Parma Sara Ciulli
Sara Ciulli Silvia Natale
Silvia Natale Alessandra De Cesare
Alessandra De Cesare Valentina Indio
Valentina Indio Pier Paolo Gatta
Pier Paolo Gatta Alessio Bonaldo
Alessio Bonaldo