- 1Department of Biology, Hong Kong Baptist University, Hong Kong, Hong Kong SAR, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 3Laboratory of Marine Organism Taxonomy and Phylogeny, Qingdao Key Laboratory of Marine Biodiversity and Conservation, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 4MLR Key Laboratory of Marine Mineral Resources, Guangzhou Marine Geological Survey, China Geological Survey, Guangzhou, China
Catillopecten is a small genus of deep-sea glass scallops, but its diversity is poorly known in many parts of the world ocean. We described C. margaritatus n. sp. (Pectinoidea: Propeamussiidae), and performed morphological analyses and DNA sequencing, and estimated the divergence time of scallops based on samples collected from Haima cold seep in the South China Sea. Morphologically, the new species can be distinguished from congeneric species by its large shell size, relatively small auricle length, absence of monocrystal aerials, presence of longitudinal radial ridges on the left valve, and the alternated rounded striae and distal and proximal growth lines of prisms on the right valve. Anatomically, this new species can be distinguished from C. vulcani by its anteriorly located auriculate gills, compared to the centrally located lamellar gills of the latter, and the different locations of the pericardium. Sequence comparison and phylogenetic analysis based on the 18S rRNA fragments supported the placement of the new species in Catillopecten. We also report the mitogenome of C. margaritatus n. sp. as the only reported mitogenome of the family Propeamussiidae, which differs from those of other scallops substantially in gene order arrangement. Divergence time estimation revealed that Propeamussiidae and Pectinidae diverged in the early Carboniferous, while Catillopecten and Parvamussium diverged during the late Cretaceous to early Eocene. Finally, we presented a key to the species of Catillopecten.
Introduction
Scallops in the family Propeamussiidae Abbott, 1954, commonly called glass scallops due to their thin and semi-translucent shells, are inhabitants of the deep sea, with species recorded from 200 m to more than 5,000 m (Kamenev, 2018). The genus Catillopecten Iredale, 1939 (Propeamussiidae), including 11 species (MolluscaBase, 2022), is characterized by thin, inequilateral to subequilateral valves, unequal auricles, lack of internal ribs, and a deep byssal notch (Kamenev, 2018). Members of Catillopecten are widely distributed in the global deep oceans, including one species from the Atlantic, four species from the eastern Pacific Ocean, and six species from the western Pacific Ocean (Figure 1, Dall, 1898; Smith, 1885; Dautzenberg and Bavay, 1912; Knudsen, 1970; Bernard, 1978; Schein-Fatton, 1988; Dijkstra and Marshall, 2008; Kamenev, 2018). These species have been reported from a variety of substrates, including rocky bottoms of a hydrothermal vent (Beninger et al., 2003), and muddy bottoms of continental slopes to polymetallic nodule areas in abyssal plains (Table 1).
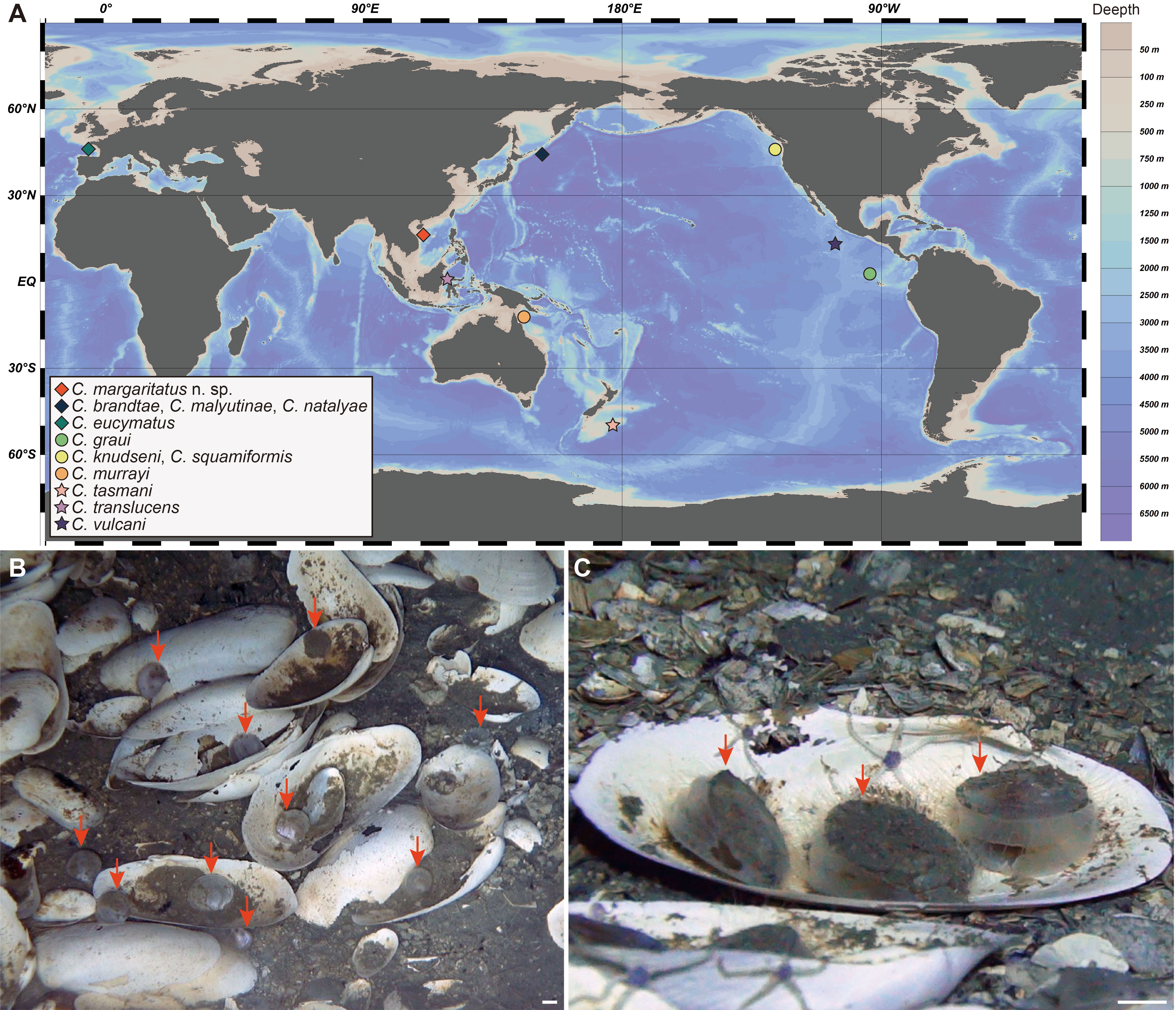
Figure 1 Worldwide geographical distribution of species in the genus Catillopecten and the typical habitat of the new species. (A) Locality of the species of Catillopecten, holotypes only. (B, C) Typical habitat of C. margaritatus n. sp. (indicated by red arrows), which were often found on the empty shells of Archivessica marissinica. Scale bar: (B, C) 10 mm.
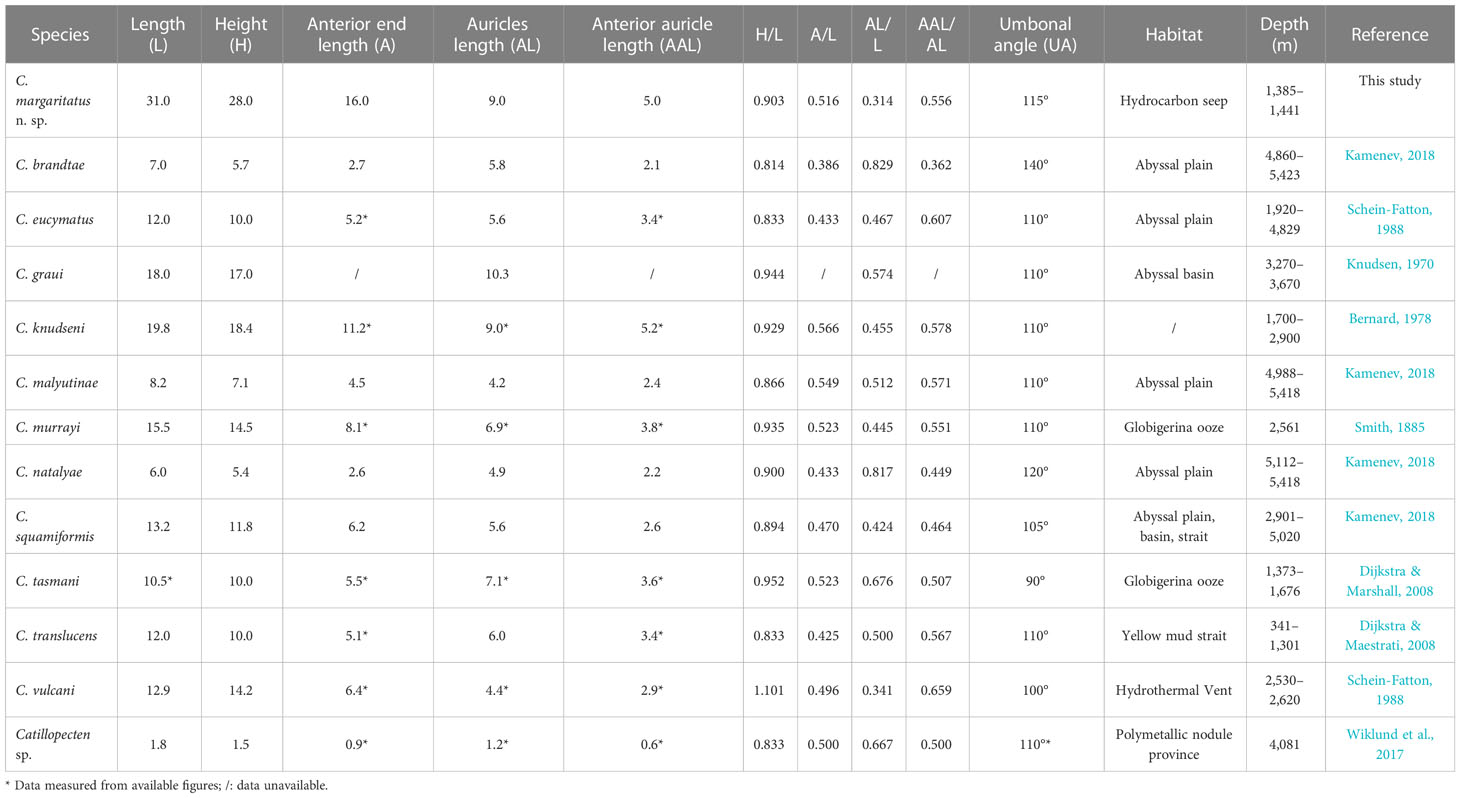
Table 1 Comparison of shell characteristics (the largest specimen of each species) and distribution of Catillopecten species.
Previous taxonomic studies of Catillopecten heavily relied on shell morphology, such as shell proportion and auricle characters. By contrast, little attention was paid to the anatomic structures. In 1978, anatomic structures of C. knudseni Bernard, 1978 and C. squamiformis Bernard, 1978 were described without specimen images (Bernard, 1978). In C. vulcani Schein-Fatton, 1985, the internal structures were illustrated by a line drawing and briefly described (Beninger et al., 2003). Moreover, Le Pennec et al. (2002) described the spermatogenesis of C. vulcani and proposed that its acrosome features could be applied in studies of scallop evolution. Molecular data of Catillopecten are also limited, with only two 18S rRNA sequences available, one from C. vulcani (Dufour et al., 2006), and the other from Catillopecten sp. (Wiklund et al., 2017), which may be a juvenile of C. malyutinae Kamenev, 2018 (Kamenev, 2019). These two sequences showed a high nucleotide similarity of 99.59% (Wiklund et al., 2017).
The Haima cold seep on the northwestern slope of the South China Sea (SCS) supports a large epibenthic fauna dominated by chemosynthetic bivalves and siboglinid tubeworms (Dong et al., 2021; Ke et al., 2022). In 2020, Ke et al. (2022) collected broken shells of a glass scallop from this area, and identified the specimen as a species of Propeamussium Gregorio, 1884. In 2022, we collected five specimens of propeamussiids from the same area, including three living individuals and two pairs of empty shells. Our preliminary analysis revealed that these glass scallop specimens all belong to a new species of Catillopecten. Therefore, this study aims to (i) describe the new species; (ii) assemble and annotate its mitochondrial genome; and (iii) determine its phylogenetic position. As the first report of propeamussiid species with detailed morphological and molecular information from the South China Sea, our study enhances our knowledge of the biodiversity of this large marginal sea of the western Pacific Ocean, and provides mitogenome and molecular data for a better understanding of the phylogenetic relationships and global biogeography of Propeamussiidae.
Materials and methods
Sample collection
The specimens were captured using a scoop net from the Haima cold seep (16°54.04’N, 110°28.47’E), at 1,433–1,441 m water depth in September 2022, using the ROV Pioneer onboard R/V Xiangyanghong 01 of The First Institute of Oceanography, Ministry of Natural Resources (Qingdao, China). The holotype was frozen with liquid nitrogen and stored at −80°C, while paratypes 1 and 2 were fixed in 4% paraformaldehyde, and paratypes 3 and 4 (empty shells) were kept at room temperature.
Morphological measurement and photography
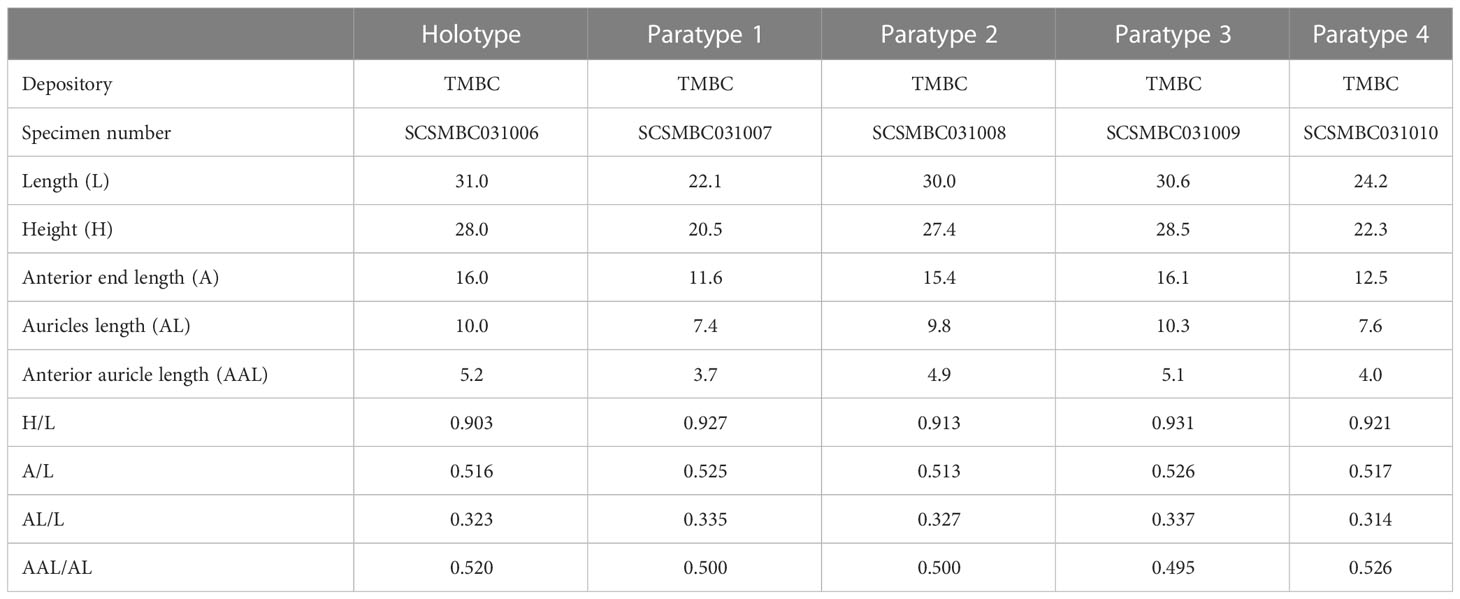
Shell measurements were made according to Kamenev (2018). Shell length (L), height (H), anterior end length (A), auricles length (AL), anterior auricle length (AAL), and umbonal angle (UA) were measured using a vernier caliper. The shell size data of other species of Catillopecten were measured according to the scale or proportion when the specimen images were available. The ratios of these parameters to shell length (i.e., A/L, H/L, AL/L, and AAL/AL) were calculated. An EOS 5D Mark IV camera (Canon, Japan) was used for photography of the whole specimen. A digital Stereomicroscope SMZ1270 Imaging System (Nikon, Japan) was used to document the morphological details. The specimens used in this study were deposited in the Tropical Marine Biodiversity Collections of the South China Sea (TMBC), Chinese Academy of Sciences (Guangzhou, China).
DNA extraction, sequencing, and assembly
Genomic DNA was extracted from the adductor muscle of the holotype using the CTAB method (Stewart and Via, 1993). DNA quality was examined using agarose gel (1.0%) electrophoresis, and the quantity was determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, USA). DNA library was constructed with the NEBNext Ultra DNA Library Prep Kit for Illumina (NEB, United States) with an insert size of 350 bp. Genomic sequencing was conducted on an Illumina NovaSeq 6000 sequencer (Illumina, USA) in Novogene (Tianjin, China) to generate 150-bp paired-end reads. Raw Illumina reads were filtered using Trimmomatic v0.38 (Bolger et al., 2014) to remove adapter and low-quality reads with the following settings: LEADING = 15, TRAILING = 15, SLIDINGWINDOW = 4:20, MINLEN = 40. The clean reads were de novo assembled using SPAdes v3.14.1 (Bankevich et al., 2012) with k-mer sizes of 21, 33, 55, 77, 99, and 127. The nuclear gene 18S rRNA was identified by conducting BLASTn using BLAST v2.11.0+ (Camacho et al., 2009) to search against the available corresponding sequences of Propeamussiidae species from NCBI (https://www.ncbi.nlm.nih.gov/), with an E-value of 1e-10. The mitogenome was assembled using NOVOPlasty v3.2 (Dierckxsens et al., 2016) under default settings with a cox1 sequence of Parvamussium carbaseum Dijkstra, 1991 (AB084106) from NCBI (https://www.ncbi.nlm.nih.gov/) as the seed.
Mitogenome annotation and analyses
MITOS2 Web Server (Donath et al., 2019) was used to annotate the mitogenome under default settings. The annotated protein coding genes (PCGs) were manually examined and adjusted against the invertebrate genetic codes. The OGDRAW in the Geseq server (Greiner et al., 2019) was used to visualize the mitogenome arrangement. Phylogenetic analyses were conducted to determine the position of the new species in the tree of life of scallops, based on the mitogenome of the new species, as well as available mitogenomes of scallops from the order Pectinida, including Amusium pleuronectes Linnaeus, 1758 (MT419374), Argopecten irradians Lamarck, 1819 (DQ665851), Argopecten purpuratus Lamarck, 1819 (KF601246), Chlamys farreri Jones and Preston, 1904 (EU715252), Crassadoma gigantea Gray, 1825 (MH016739), Mimachlamys crassicostata Sowerby, 1842 (FJ415225), Mimachlamys sanguinea Linnaeus, 1758 (KF214684), Mimachlamys varia Linnaeus, 1758 (MZ520326), Mizuhopecten yessoensis Jay, 1857 (FJ595959), Pecten albicans Schröter, 1802 (KP900974), Pecten maximus Linnaeus, 1758 (KP900975), Placopecten magellanicus Gmelin, 1791 (DQ088274), and Spondylus virgineus Reeve, 1856 (MN019127), with Ostrea edulis Linnaeus, 1758 (NC_016180) as the outgroup. The PCGs and ribosome RNA sequences of these mitogenomes downloaded from NCBI were extracted. The analyses were conducted using PhyloSuite v1.2.2 (Zhang et al., 2020) with the help of several plug-in programs: MAFFT (Katoh and Standley, 2013) under the “auto” option was applied to align each gene fragment, with “Codon alignment mode” for PCGs and “Normal alignment mode” for rRNAs. Gblocks (Talavera and Castresana, 2007) was applied to remove ambiguously aligned fragments in batches, with missing genes or alignment gaps being filled with “-”. Then, the fragments were concatenated and ModelFinder (Kalyaanamoorthy et al., 2017) was used to select the best-fit model based on the BIC criterion. Bayesian inference (BI) and maximum-likelihood (ML) analyses were conducted using MrBayes (Ronquist et al., 2012) and IQ-TREE (Nguyen et al., 2015), under the best-fit GTR+F+I+G4 model for 10 million generations and the GTR+R4+F model for 1,000 ultrafast bootstraps, respectively (Minh et al., 2013). Since the order of the PCGs and tRNAs may contain phylogenetic signal (Shen et al., 2009), we compared the arrangement orders of the mitochondrial genes among the species of Pectinida.
Phylogenetic analyses and divergence time estimation
Since only two 18S rRNA sequences of Catillopecten were available in GenBank (https://www.ncbi.nlm.nih.gov/), we determined the sequence divergence between the new species and these two other Catillopecten sequences, as well as those of other genera of Propeamussiidae based on this gene fragment. In addition, we also obtained 28 fragments of 18S rRNA from the order Pectinida and four from Limidae (outgroup) from GenBank to determine the relationships between Catillopecten and other scallops (Table S1). Fragments with longer than 1,500 bp were kept and the longest sequence was selected when two or more sequences from the same species were available. The number of base substitutions per site among these sequences is calculated using MEGA7 (Kumar et al., 2016) under the Kimura 2-parameter model (Kimura, 1980). The rate variation among sites was modeled with a gamma distribution (shape parameter = 1), and all positions with less than 95% site coverage were eliminated. Phylogenetic analyses were similar to the phylogenetic tree construction of the mitogenome section except the BI and ML analyses were conducted under the best-fit K80+I+G model for 10 million generations and the K80+R2 model for 1,000 ultrafast bootstraps, respectively (Minh et al., 2013).
To estimate the divergence times, fossil ages were incorporated based on available data (Waller, 2006; and the Paleobiology Database, https://paleobiodb.org/). The ages of Pectinidae and Limidae were constrained approximately 251.3–247.2 MYA and 330.9–323.2 MYA, respectively (Smedley et al., 2019). BEAST v.2.6.6 (Bouckaert et al., 2019) was used to estimate the divergence times based on a concatenated dataset of mitochondrial PCGs, under the following settings: (i) HKY+G4 as the partition model; (ii) “strict clock” as the clock model with estimated clock rate; (iii) “calibrated Yule model” as the two prior and fossil calibration dates were set in the Pectinidae clade (“log normal” distribution, M parameter = 5.518, S parameter = 0.0033) and the Limidae clade (“log normal” distribution, M parameter = 5.792, S parameter = 0.0052); and (iv) running chain length as 10,000,000 MCMC generations with 1,000 sample frequencies. The default settings were adopted for other parameters. The convergence of the likelihood parameters in the log file was determined by Tracer v.1.7.2 (Rambaut et al., 2018) with the ESS over 200. The final tree file was generated using TreeAnnotator (Rambaut & Drummond, 2015) with 25% burn-in and plotted with FigTree v1.4.4 (Rambaut & Drummond, 2018).
Results
Systematics
Order: Pectinida, Gray, 1854
Superfamily: Pectinoidea Rafinesque, 1815
Family: Propeamussiidae, Abbott, 1954
Genus: Catillopecten Iredale, 1939
Synonym: Bathypecten Schein-Fatton, 1985
Type species: Catillopecten murrayi Smith, 1885
Diagnosis (adopted from Kamenev, 2018): Propeamussiidae with translucent shells, very thin, fragile and compressed, subequilateral to inequilateral. Valves smooth or undulated, sometimes with weak commarginal ribs and fine radial threads; left valve more convex. Auricles unequal; posterior auricles continuous or weakly separated from shell disc; anterior auricle of right valve distinct and sharply demarcated from shell disc. Internal ribs absent. Byssal notch rather deep; byssal fasciole narrow or absent.
Catillopecten margaritatus n. sp. http://zoobank.org/NomenclaturalActs/396CA196-5051-4A11-975F-D7A74C91568D
Type specimens
Holotype [TMBC (Tropical Marine Biodiversity Collections of the South China Sea), Chinese Academy of Sciences, Guangzhou, SCSMBC031006], paratypes 1–4 (TMBC, SCSMBC031007–SCSMBC031010), Haima cold seep in SCS, 16°54.04’N, 110°28.47’E, 1,433–1,441 m, organic sediment with empty shells of Archivesica marissinica Chen, Okutani, Liang & Qiu, 2018 (Figure 1), collected using the suction sampler of the ROV Pioneer, September 2022, ROV02, Dive 05.
Type locality
Haima cold seep, 1,407–1,441 m water depth, off southern Hainan Island, on the northwestern slope of the South China Sea (Figure 1).
Etymology
The species epithet “margaritatus” comes from “margarita” in Latin, which means pearl and refers to the pearlescent internal surface of the valves of this species.
Diagnosis
Catillopecten with a large shell up to 31 mm in length; internal shell silvery-white and pearlescent. Left valve surface slightly crumpled, with longitudinal radial ridges intersected with conspicuous, rounded, thin ribs. Right valve slightly convex with regular, rippled, and rounded commarginal striae alternated with distal and proximal growth lines of prisms. Auricles small and approximately equal in length. Left valve auricles approximate triangular, with intensive parallel ridges, connected with the rounded ribs of the disc. Right valve anterior auricle with rounded coarse commarginal ribs and ridges; posterior auricle with parallel, dense, indistinct striae and some distinct ridges.
Description
Shell (Figures 2, 3, Table 2) subcircular, external gray, internal silvery-white, iridescent, pearlescent, rayed, without internal ribs (Figures 2C, D). Length up to 31.0 mm, slightly longer than height (H/L = 0.90–0.93) and approximate equilateral (A/L = 0.52–0.53). Shell anterodorsal and ventral margins gracefully curved, posterodorsal margin slightly curved and nearly straight. Auricles small (AL/L = 0.31–0.34), approximately equal in length (AAL/AL = 0.50–0.53). Hinge line straight (Figures 3A, B), subterminal umbones with an angle of 115°.
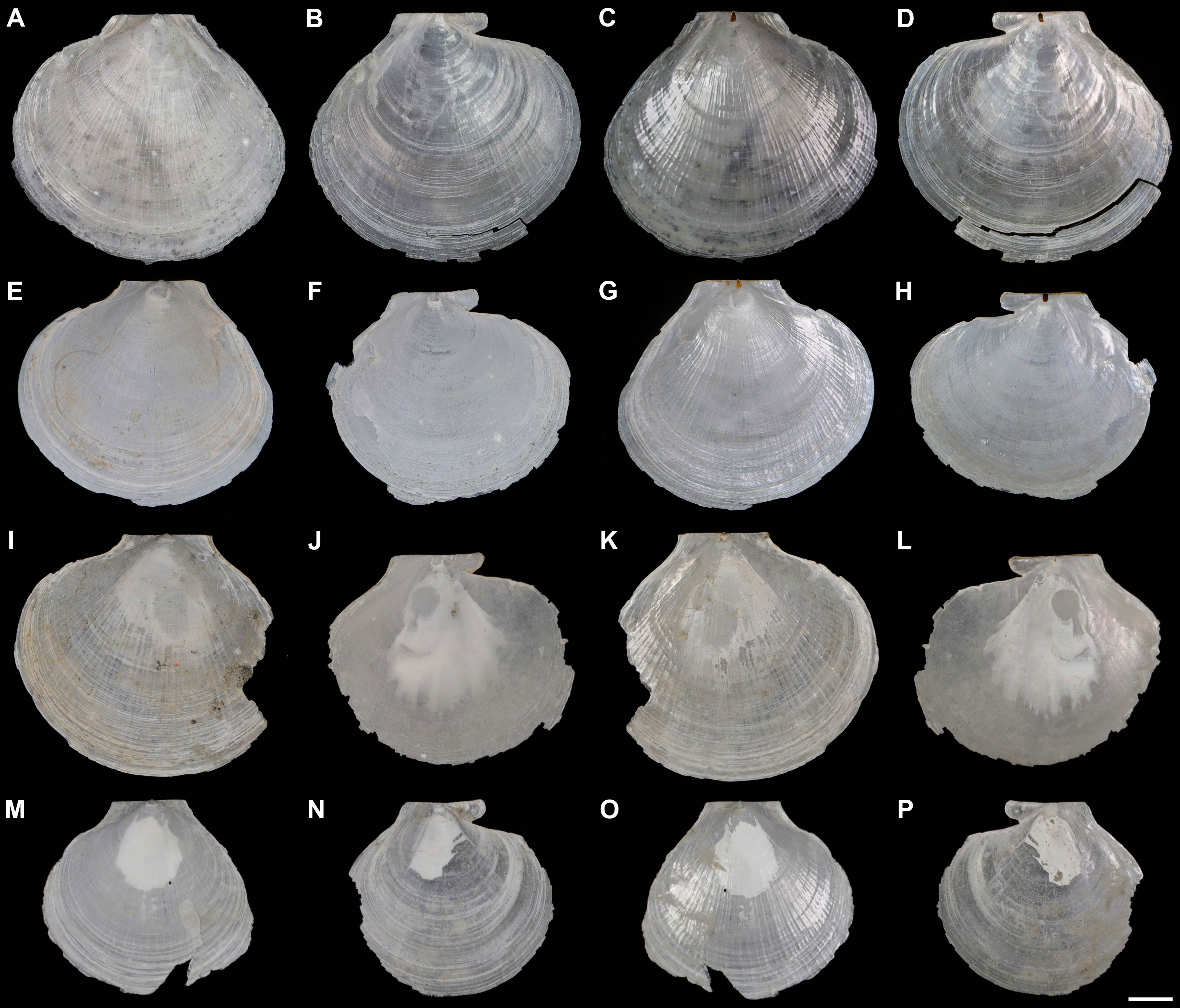
Figure 2 External and internal views of the left and right valves of Catillopecten margaritatus n. sp. specimens. (A–D) Holotype, TMBC, SCSMBC031006; (E–H) paratype 2, TMBC, SCSMBC031008; (I–L) paratype 3, TMBC, SCSMBC031009; (M–P) paratype 4, TMBC, SCSMBC031010. Scale bar: 5 mm.
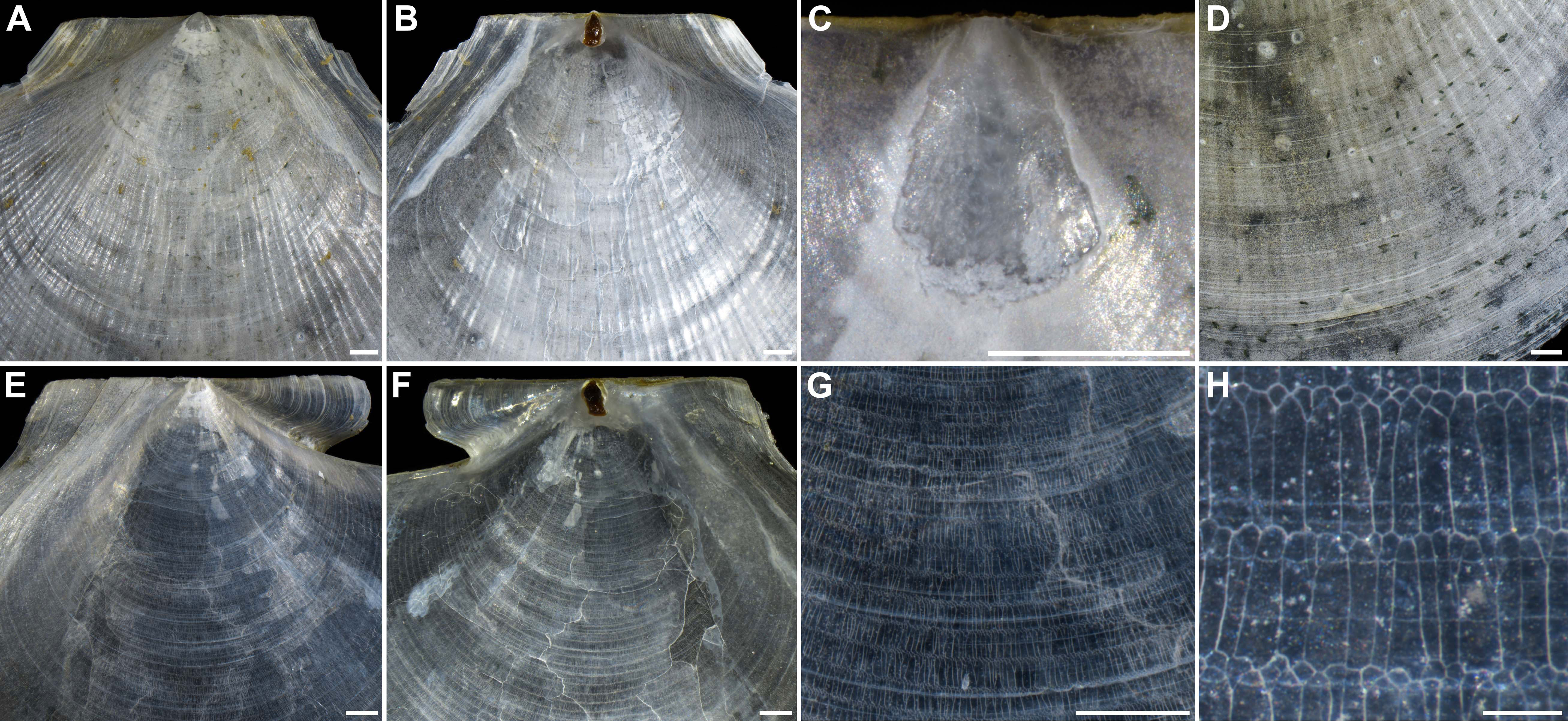
Figure 3 Morphological details of Catillopecten margaritatus n. sp. shells (holotype, TMBC, SCSMBC031006). (A–D) Left valve. (A) External view of auricles. (B) Internal view of auricles. (C) Umbo area of the left valve. (D) External surface commarginal undulations and fine radial threads. (E–H) Right valve. (E) External view of auricles. (F) Internal view of auricles. (G) Sculpture of the center. (H) Prismatic microstructure of the center. Scale bar: (A–G), 1 mm; H, 0.1 mm.
Left valve. Disc convex (Figure 2), surface slightly crumpled, with longitudinal radial ridges intersected with conspicuous, rounded, thin, ribs (Figures 3A, D); distinctly radial ridges and rounded ribs presented on the inner surface. Auricles approximate triangular, with compressed and slightly cambered surface; intensive parallel ridges clear, connected with the rounded ribs of the disc; anterior auricle disconnected with disc and with a shallow notch; posterior auricle continuous with shell edge (Figures 3A, B). Umbo clear, conical shape, convex, distinctively separated from shell disc (Figure 3C). Resilium distinct, teardrop-shaped and brownish black (Figure 3B).
Right valve. Disc slightly convex (Figure 2), thinner, more fragile and transparent than left valve. Regular, rippled, and rounded commarginal striae distinctly presented on the external and internal surfaces, intersected with longitudinal stripes of prisms (Figures 3E–G); prisms distinct, with alternating spherulitic and elongated layers, and clear distal and proximal growth lines, alternated with the rounded striae (Figures 3G, H). Auricles unequal; anterior auricle low-angle sector-shaped and inflated, with clear, rounded, coarse commarginal ribs and ridges, protruding with rounded anterior end, sharply demarcated from shell disc with a deep notch and gorge; posterior auricle triangular, with parallel, dense, indistinct striae and some distinct ridges, continuous connected to shell disc with slight turn (Figures 3E, F). Adductor muscle scar oval and clear (Figures 2D–L, 3F). Umbo very compressed and inconspicuous.
Anatomy (Figure 4). Mantle large, thin and transparent, with a row of long tentacles and velum at the margin. Unilaterally auriculate and large gills at anterior side, consisting of outer and inner demibranches. Rounded digested gland close to resilium, brown to dark color. Rectum extending from digested gland along the dorsal side of adductor muscle. Adductor muscle small, cylindrical, and robust. Foot small, covered by a pair of labial palps, connected to adductor muscle by foot contracted muscle. Gonad long scrotiform, surrounding adductor muscle on anterior side. Pericardium small, located between adductor muscle and gonad.
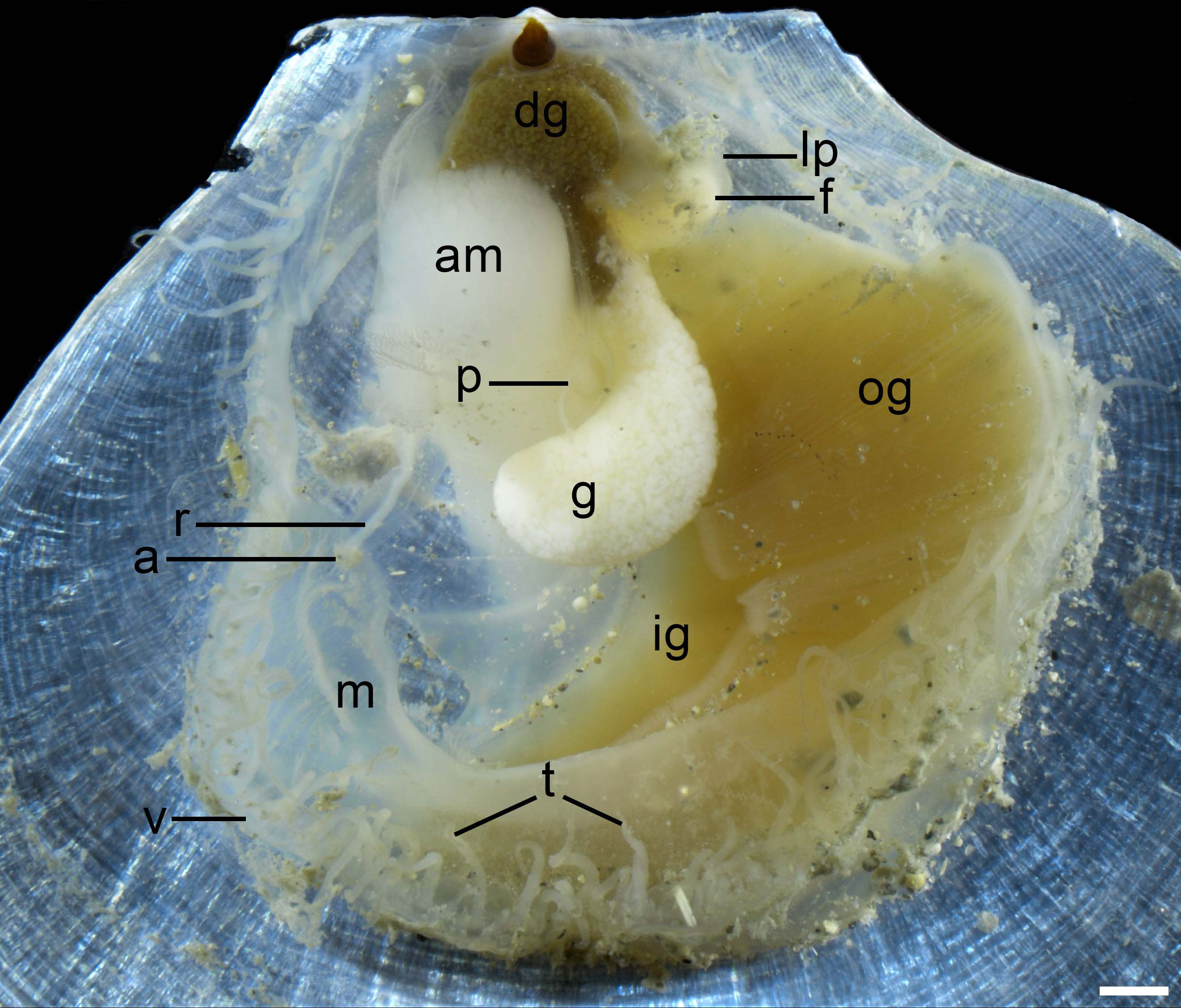
Figure 4 Anatomy features of Catillopecten margaritatus n. sp. of the paratype 1 (TMBC, SCSMBC031007) left valve. Abbreviations: a, anus; am, adductor muscle; dg, digestive gland; f, foot; g, gonad; ig, inner gill; lp, labial palp; m, mantle; og, outer gill; p, pericardium; r, rectum; t, tentacle; v, velum. Scale bar: 1 mm.
Remarks
Catillopecten is similar to Propeamussium and other genera of Propeamussiidae in having translucent shells. However, Propeamussium can be distinguished from Catillopecten by the almost equivalve and laterally compressed shell, weak byssal notch, and distinct internal ribs (Dijkstra, 2013). Based on the shell characteristics, our glass scallop specimens, as well as those empty shells described as Propeamussium sp. by Ke et al. (2022) all belong to Catillopecten.
Within Catillopecten, shell shape, shell and auricle proportion, monocrystal aerials and sculpture on shell surface, and byssal notch shape have been used to distinguish the species. Among the described species, C. margaritatus n. sp. most closely resembles C. tasmani Dijkstra and Marshall, 2008—a species reported from Bounty Trough, southern New Zealand at depths of 1,373 to 1,676 m—in shell shape, proportions, absence of monocrystal aerials, auricle features, deep and sharp byssal notch, and the prism structure of the right valve (Dijkstra and Marshall, 2008). Nevertheless, C. margaritatus n. sp. can be easily distinguished from C. tasmani with its larger shell size (L: 22.1−31.0 mm vs. 10.5 mm), smaller auricle length (AL/L: 0.31-0.34 vs. 0.68), and the wide-angled umbones (UA: 115° vs. 90°). Catillopecten margaritatus n. sp. has relatively small auricles, which are about one-third of the total shell length, similar to those of C. vulcani (Schein-Fatton, 1985). Nevertheless, C. vulcani differs from the former with its small size and unique slim shell profile (H/L: 0.90−0.93 vs. 1.10). In addition, the shell surface of C. margaritatus n. sp. lacks monocrystal aerials, which differentiates it clearly from C. brandtae Kamenev, 2018, C. malyutinae Kamenev, 2018, C. natalyae Kamenev, 2018, and Catillopecten sp. (may be a synonym of C. malyutinae) (Wiklund et al., 2017; Kamenev, 2018; Kamenev, 2019; Checa et al., 2022). Significantly, in C. margaritatus n. sp., the presence of longitudinal radial ridges on the left valve and the alternated rounded striae and distal and proximal growth lines of prisms on the right valve, which are absent in all of its congeneric species, allow this new species to be distinguished from its congeneric species.
Although anatomical features have been used in the taxonomy of deep-sea bivalves (Xu et al., 2019), available data are too limited to judge whether they are useful for species delimitation in Catillopecten (Le Pennec et al., 2002; Beninger et al., 2003). Our comparison of the anatomical features with the drawing of Beninger et al. (2003) shows that C. vulcani has centrally located lamellar gills with a straight axis. By contrast, C. margaritatus n. sp. possesses a pair of large and unilaterally auriculate gills with a straight axis, located at the anterior side of the shell. Furthermore, the pericardium of C. margaritatus n. sp. is located between the adductor muscle and gonad, whereas the pericardium and heart of C. vulcani were speculated to be located at the dorsal side of the rectum (Beninger et al., 2003). Therefore, C. margaritatus n. sp. and C. vulcani can be distinguished by anatomical features. When anatomical data from other species become available, studies should be conducted to further examine their potential use in the taxonomy of these glass scallops.
Mitogenomic features, phylogenetic position, and divergence
Illumina sequencing generated 29.21 million paired-end reads. Our assembly produced a complete, circular mitogenome of C. margaritatus with a size of 17,979 bp, and an overall base composition of 40.24% for T, 10.01% for C, 27.78% for G, and 21.97% for A (Table S2). This is the first assembled mitogenome of the family Propeamussiidae. It contains 13 PCGs, namely, cox1–3, nad1–6, nad4l, atp6, atp8, and cytb; two rRNA genes; and 22 tRNA genes (Figure 5A, Table S3). There are three overlap regions between adjacent genes, including 1 bp between trnN and trnS1, 2 bp between nad2 and trnL, and 23 bp between rrnL and trnV. All these genes are encoded in the same strand (Figure 5A), similar to other species of Pectinidae.
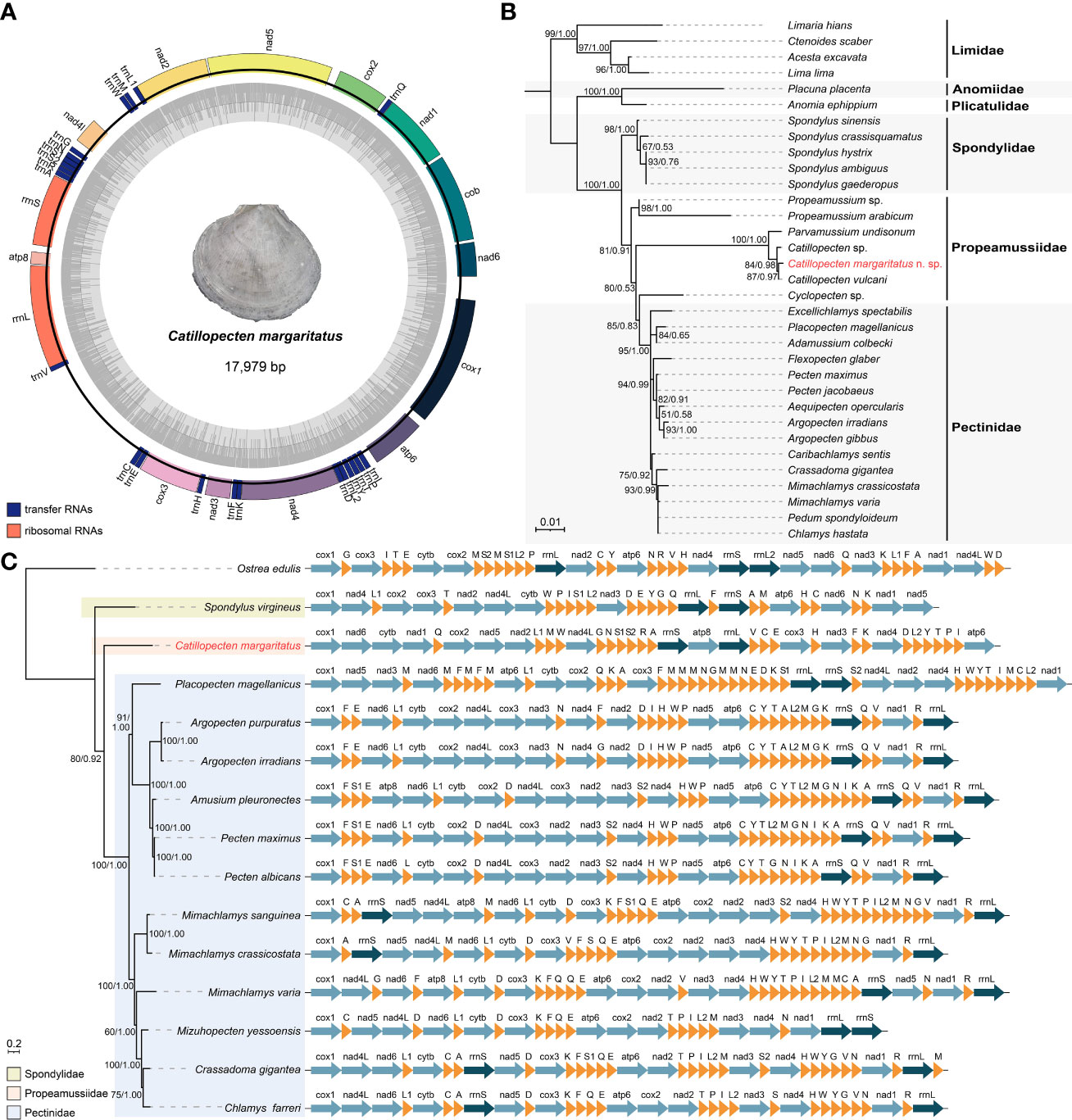
Figure 5 Mitogenome features of Catillopecten margaritatus n. sp. and its phylogenetic position. (A) Mitogenomic gene orders and relative lengths. (B) Phylogenetic tree of the order Pectinida based on 18S rRNA fragments. The topology is based on BI analysis, with Limidae as the outgroup. The accession numbers are presented in Table S1. (C) Mitogenomic phylogenetic tree based on protein coding genes and rRNAs and the mitochondrial gene arrangements of the superfamily Pectinidea species. The topology is based on BI analysis, with Ostrea edulis as the outgroup. Posterior probabilities values (>0.50) from BI analysis and bootstraps values (>50) from the ML analysis are given at the nodes.
Phylogenetic analyses based on the 18S rRNA fragment showed that the three Catillopecten sequences, including the one from C. margaritatus, formed a clade, which is sister to Parvamussium undisonum Dijkstra, 1995 (Figure 5B). Among the three Catillopecten species, C. margaritatus is more closely related to C. vulcani. Among the 1,835 bp aligned 18S rRNA fragment, C. margaritatus only showed 7 bp (0.38%) difference from C. vulcani, with a K2P pairwise distance of 0.18% between the two species (Table S4), while C. margaritatus showed only 0.59% divergence from Catillopecten sp. and 1.25% from P. undisonum in 18S rRNA, with genetic distances of 0.36% and 1.03%, respectively. Furthermore, the family Propeamussiidae appears polyphyletic, with Cyclopecten Verrill, 1897, currently accepted as a genus of Propeamussiidae, being placed in Pectinidae, consistent with previous studies (Combosch et al., 2017; Smedley et al., 2019). Phylogenetic analysis of 14 species of Pectinoidea based on mitogenomic PCGs showed that Spondylus virgineus is sister to a large clade of all other pectinoids, and within this clade, C. margaritatus is sister to a clade of all other 12 species (Figure 5C). Moreover, the C. margaritatus mitogenome differs substantially from those of Spondylidae and Pectinidae in the order of PCGs, rRNAs, and tRNAs (Figure 5C). Furthermore, even within Pectinidae, the gene order arrangements were very different among the species available for comparison.
The divergence times among Pectinoidea were estimated based on mitochondrial PCGs, using two fossil calibration points with horizontal bars representing the 95% highest posterior density (HPD) interval for each node (Figure 6). The results revealed the origin of the superfamily Pectinoidea in the Devonian (384.11–360.24 MYA, median value 371.42 MYA). Propeamussiidae is sister to Pectinidae and they diverged in the early Carboniferous (351.20–331.77 MYA, median value 341.03 MYA). Catillopecten and Parvamussium Sacco, 1897 diverged during the late Cretaceous to early Eocene (70.51–46.63 MYA, median value 57.68 MYA).
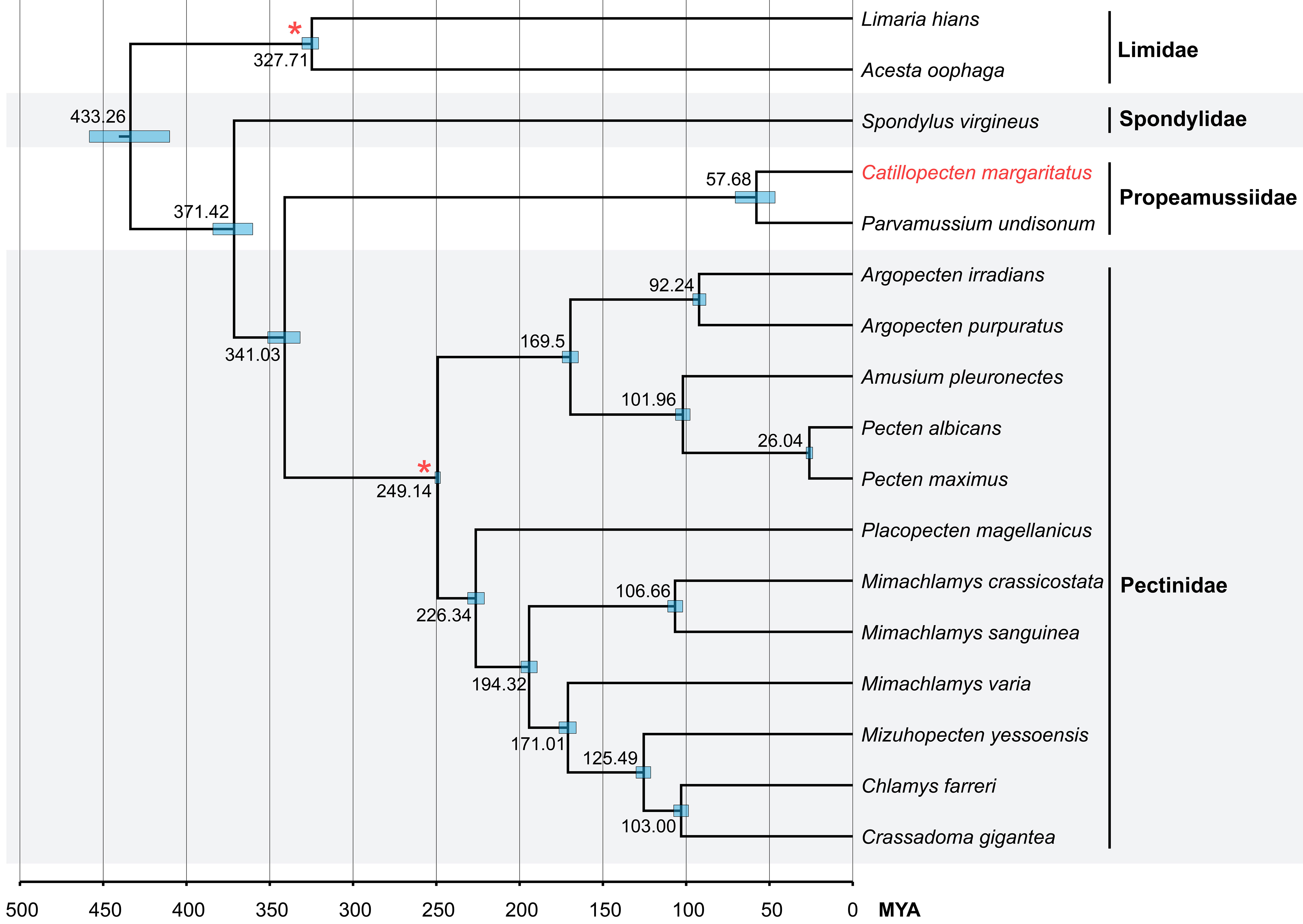
Figure 6 Divergence time estimation analysis of Pectinoidea based on concatenated mitochondrial PCG gene sequences. Bottom axis represents millions of years and the internal nodes calibrated by fossil records are represented by red asterisks. The 95% highest posterior density (HPD) is indicated as node blue bars and the Bayesian posterior probabilities of all nodes are 1.00.
Discussion
Since most of the Catillopecten species lack gene sequences, and the only gene sequence available in two Catillopecten species was the 18S rRNA, a conserved gene marker that is more appropriate for deep phylogenetic study than species delimitation (Zhan et al., 2019), it was not surprising that we only found 0.38%−0.59% divergences in 18S rRNA among the three species of Catillopecten in this study. Nevertheless, the result of C. margaritatus and the other two species of Catillopecten forming a clade that is sister to P. undisonum supports that the new species belongs to Catillopecten. When sequences of more sensitive genetic markers, such as the mitochondrial cytochrome subunit I (COI) or 16S ribosomal RNA sequences, from other Catillopecten species become available in the future, molecular delimitation should be conducted in future studies to determine the genetic distances between C. margaritatus and its congeneric species.
In this study, we presented the first mitogenome of Propeamussidae. Its length (17,979 bp) is similar to that of Crassadoma gigantea (18,495 bp), Mimachlamys sanguinea (17,383 bp), and Pecten maximus (17,252 bp), but much smaller than that of Placopecten magellanicus (32,115 bp) in Pectinidae and Spondylus virgineus (30,160 bp) in Spondylidae. Among the sequenced mitogenomes of the three families of Pectinoidea, C. margaritatus presents the highest A+T bias with a proportion of 62.21%, while that of the others is from 55.39% to 58.77% (Table S2). Arrangements of PCGs may imply common ancestry (Shen et al., 2009), and such information may be useful to understand the transition from shallow-water to deep-sea habitats (Zhang et al., 2018). However, the substantially different orders of PCGs between the deep-sea C. margaritatus and other pectinoids and the bias in sequencing efforts (i.e., only one mitogenome each in Spondylidae and Propeamussiidae) make it difficult to associate these changes with the invasion from shallow water to the deep sea.
Catillopecten margaritatus is the first reported extant seep-dwelling species of this genus. These scallops usually stay on the empty shells of the vesicomyid clam Archivesica marissinica (Figures 1B, C), one of the dominant species in the Haima cold seep (Chen et al., 2018). In this seep area, bathymodioline mussel Gigantidas haimaensis Xu et al., 2019, and siboglinid tubeworms Paraescarpia echinospica Southward et al., 2002 (Sun et al., 2021) and Sclerolinum annulatum Xu et al., 2022 form huge colonies on the sea bed. Other low-density populations of bivalves were also reported in this area, including Gigantidas platifrons Hashimoto & Okutani, 1994, “Bathymodiolus” aduloides Hashimoto and Okutani, 1994, Nypamodiolus samadiae Lin et al., 2022, and other undetermined species of Malletia and Acharax (Xu et al., 2019; Guan et al., 2022; Ke et al., 2022). These siboglinid and bivalve species are endosymbiotic with chemosynthetic bacteria in their trophosome and gill epithelial cells, respectively, to provide energy and materials for the holobiont (Ip et al., 2021; Sun et al., 2021; Lin et al., 2023). These chemosymbioses allow vent and seep annelids and bivalves to flourish in the deep sea (Sen et al., 2018; Osman and Weinnig, 2021). Nevertheless, no deep-sea scallops have been reported to be symbiotic with chemosynthetic bacteria, including C. vulcani, which inhabits hydrothermal vent sites in the eastern Pacific Ocean (Schein-Fatton, 1985). This is reflected in the lack of morphological specialization of the flat and homorhabdic gill of C. vulcani, as well as the abundant particles on the gill indicating its suspension feeding trophic pattern (Beninger et al., 2003), although Le Pennec et al. (1988) observed a few bacteria in the gill filaments of this species. The gill of C. margaritatus is similar to that of C. vulcani, and therefore, we speculate that C. margaritatus also relies on filter-feeding for nutrition.
Key to species of Catillopecten Iredale, 1939:
1a. External shell surface with monocrystal aerials……2
1b. External shell surface without monocrystal aerials……4
2a. Shell rounded-triangular ……C. malyutinae Kamenev, 2018
2b. Shell D-shaped……3
3a. Shell with fine and closely located commarginal riblets……C. natalyae Kamenev, 2018
3b. Shell with irregular commarginal and radial undulations……C. brandtae Kamenev, 2018
4a. Shell height larger than length……C. vulcani Schein-Fatton, 1985
4b. Shell height slightly smaller than length……5
5a. Byssal notch shallow and rounded ……C. knudseni Bernard, 1978
5b. Byssal notch moderately deep or deep ……6
6a. Byssal notch sharp……7
6b. Byssal notch rounded……9
7a. Small auricles, length about one-third of shell length……C. margaritatus
7b. Large auricles, length more than half of shell length……8
8a. Right valve with obscure radial lines……C. tasmani Dijkstra and Marshall, 2008
8b. Right valve with sharp ribs and fine radial lines……C. graui Knudsen, 1970
9a. Auricles subequal in length……10
9b. Auricles unequal in length……11
10a. Dorsal shell margin straight……C. squamiformis Bernard, 1978
10b. Dorsal shell margin round……C. eucymatus Dall, 1898
11a. Anterior auricles longer than posterior auricles……C. murrayi Smith, 1885
11b. Posterior auricles slightly longer than anterior auricles……C. translucens Dautzenberg and Bavay, 1912
Conclusions and perspectives
We reported Catillopecten margaritatus as the first species of seep-inhabiting Catillopecten and described its morphological features of both shell and anatomy, mitogenome, and phylogenetic position. Morphologically, C. margaritatus can be easily distinguished from congeneric species with its largest shell size, smaller auricle length, and wide-angled umbones. Our anatomic description of the new species provides useful characteristics for the further taxonomic study of the genus Catillopecten. Our phylogenetic analyses confirm the placement of this new species in Catillopecten. As the only species of Catillopecten distributed within 10°N and 30°N in the western Pacific Ocean, our discovery of C. margaritatus enhances our understanding of seep fauna and fills a gap in the geographic distribution of Catillopecten in this region.
Data availability statement
The datasets presented in this study can be found in online repositories. Raw Illumina sequencing data were deposited in the National Centre for Biotechnology Information (NCBI) under the BioProject PRJNA934135. Sequence of the mitogenome and the 18S rRNA sequence were deposited in the GenBank with the accession number of OQ434059 and OQ427653, respectively.
Ethics statement
Ethical review and approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
Author contributions
J-WQ conceived and designed this project. Y-TL, Y-XL, and JT collected and photographed the samples. Y-TL performed morphological examination and description. Y-TL, Y-XL and YS conducted DNA extraction and molecular analyses. Y-TL drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (SMSEGL20SC02) and Research Grants Council of Hong Kong (12101021).
Acknowledgments
We thank the First Institute of Oceanography, MNR (Qingdao, China) for organizing the cruise, as well as the captains and crews of the Xiangyanghong 01 for their supports.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1168991/full#supplementary-material
References
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), 455–477. doi: 10.1089/cmb.2012.0021
Beninger P. G., Dufour S. C., Decottignies P., Le Pennec M. (2003). Particle processing mechanisms in the archaic, peri-hydrothermal vent bivalve Bathypecten vulcani, inferred from cilia and mucocyte distributions on the gill. Mar. Ecol. Prog. Ser. 246, 183–195. doi: 10.3354/meps246183
Bernard F. R. (1978). New bivalve molluscs, subclass pteriomorphia, from the northeastern pacific. Venus 37, 61–75. doi: 10.18941/venusjjm.37.2_61
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouckaert R., Vaughan T. G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., et al. (2019). BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PloS Comput. Biol. 15 (4), e1006650. doi: 10.1371/journal.pcbi.1006650
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10 (421), 1–9. doi: 10.1186/1471-2105-10-421
Checa A. G., Salas C., Varela-Feria F. M., Rodríguez-Navarro A. B., Grenier C., Kamenev G. M., et al. (2022). Crystallographic control of the fabrication of an extremely sophisticated shell surface microornament in the glass scallop Catillopecten. Sci. Rep. 12 (1), 11510. doi: 10.1038/s41598-022-15796-1
Chen C., Okutani T., Liang Q., Qiu J. W. (2018). A noteworthy new species of the family vesicomyidae from the south China Sea (Bivalvia: glossoidea). Venus 76 (1-4), 29–37. doi: 10.18941/venus.76.1-4_29
Combosch D. J., Collins T. M., Glover E. A., Graf D. L., Harper E. M., Healy J. M., et al. (2017). A family-level tree of life for bivalves based on a Sanger-sequencing approach. Mol. Phylogenet. Evol. 107, 191–208. doi: 10.1016/j.ympev.2016.11.003
Dall W. H. (1898). Contributions to the tertiary fauna of Florida: with especial reference to the Miocene silex-beds of Tampa and the pliocene beds of the caloosahatchee river. Wagner Free Inst. Sci. 3 (4), 571–947. doi: 10.5962/bhl.title.29760
Dautzenberg P., Bavay A. (1912). Les Lamellibranches de l’expédition du « siboga ». Systématique I Pectinidés SibogaExpeditie 53b, 1–41.
Dierckxsens N., Mardulyn P., Smits G. (2016). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Dijkstra H. H. (1991). A contribution to the knowledge of the pectinacean Mollusca (Bivalvia: propeamussiidae, entoliidae, pectinidae) from the Indonesian archipelago. Zool. Verh. 271 (1), 1–54.
Dijkstra H. H. (1995). Bathyal pectinoidea (Bivalvia: propeamussiidae, entoliidae, pectinidae) from new Caledonia and adjacent areas. Mém. Mus. Natl. hist. nat. Sér. C Géol. 167, 9–73.
Dijkstra H. H. (2013). Pectinoidea (Bivalvia: propeamussiidae and pectinidae) from the panglao region, Philippine islands. Vita. Malacologica 10, 1–108.
Dijkstra H. H., Maestrati P. (2008). New species and new records of deep-water pectinoidea (Bivalvia: propeamussiidae, entoliidae and pectinidae) from the south pacific, in tropical deep-Sea benthos Vol. 25. Eds. Héros V., Cowie R., Bouchet P. (Paris: Publications Scientifique du Muséum), 77–113.
Dijkstra H. H., Marshall B. A. (2008). The recent pectinoidea of the new Zealand region (Mollusca: bivalvia: propeamussiidae, pectinidae and spondylidae. Molluscan Res. 28 (1), 1–88.
Donath A., Jühling F., Al-Arab M., Bernhart S. H., Reinhardt F., Stadler P. F., et al. (2019). Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47 (20), 10543–10552. doi: 10.1093/nar/gkz833
Dong D., Li X., Yang M., Gong L., Li Y., Sui J., et al. (2021). Report of epibenthic macrofauna found from haima cold seeps and adjacent deep-sea habitats, south China Sea. Mar. Life Sci. Technol. 3, 1–12. doi: 10.1007/s42995-020-00073-9
Dufour S. C., Steiner G., Beninger P. G. (2006). Phylogenetic analysis of the peri-hydrothermal vent bivalve Bathypecten vulcani based on 18S rRNA. Malacologia 48 (1-2), 35–42.
Gmelin J. F. (1791). “Vermes,” in Caroli a linnaei, systema naturae per regna tria naturae, vol. 1. (Lipsiae (Leipzig: Editio decima tertia, Aucta, Reformata), 3021–3910.
Gray J. E. (1825). A list and description of some species of shells not taken notice of by lamarck. Ann. Philos. 9 (2), 134–140.
Gray J. E. (1854). A revision of the arrangement of the families of bivalve shells (Conchifera). Ann. Mag. Nat. 13 (77), 408–418. doi: 10.1080/03745485709496364
Gregorio A. (1884). Nota intorno ad alcune nuove conchiglie mioceniche di sicilia. Nat. Sici. 3, 119–120.
Greiner S., Lehwark P., Bock R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, 59–64. doi: 10.1093/nar/gkz238
Guan H., Feng D., Birgel D., Kiel S., Peckmann J., Li S., et al. (2022). Lipid biomarker patterns reflect nutritional strategies of seep-dwelling bivalves from the south China Sea. Front. Mar. Sci. 9, 831286. doi: 10.3389/fmars.2022.831286
Hashimoto J., Okutani T. (1994). Four new mytilid mussels associated with deepsea chemosynthetic communities around Japan. Venus 53 (2), 61–83. doi: 10.18941/venusjjm.53.2_61
Ip J. C. H., Xu T., Sun J., Li R., Chen C., Lan Y., et al. (2021). Host-endosymbiont genome integration in a deep-sea chemosymbiotic clam. Mol. Biol. Evol. 38 (2), 502–518. doi: 10.1093/molbev/msaa241
Iredale T. (1939). Mollusca. part I, in British museum (Natural history) great barrier reef expedition 1928–29. Sci. Rep. 5, 209–425.
Jay J. C. (1857). Report on the shells collected by the Japan expedition together with a list of Japan shells Vol. 2. Eds. Perry M. C., Hawks F. L. (Washington: Tucker), 289–297.
Jones K. H., Preston H. B. (1904). List of Mollusca collected during the commission of H.M.S. "Waterwitch" in the China seas 1900-1903, with descriptions of new species. Proc. Malacol. Soc Lond. 6, 138–154. doi: 10.1093/oxfordjournals.mollus.a066053
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kamenev G. M. (2018). Four new species of the family propeamussiidae (Mollusca: bivalvia) from the abyssal zone of the northwestern pacific, with notes on Catillopecten squamiformis (Bernard 1978). Mar. Biodivers. 48 (1), 647–676. doi: 10.1007/s12526-017-0821-1
Kamenev G. M. (2019). Bivalve mollusks of the kuril-kamchatka trench, Northwest pacific ocean: species composition, distribution and taxonomic remarks. Prog. Oceanogr. 176, 102127. doi: 10.1016/j.pocean.2019.102127
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Ke Z., Li R., Chen Y., Chen D., Chen Z., Tan Y. (2022). A preliminary study of macrofaunal communities and their carbon and nitrogen stable isotopes in the haima cold seeps, south China Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 184, 103774. doi: 10.1016/j.dsr.2022.103774
Kimura M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Knudsen J. (1970). The systematics and biology of abyssal and hadal bivalvia. Galathea. Rep. 11, 1–241.
Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Le Pennec M., Herry A., Lutz R., Fiala-Médioni A. (1988). Premières observations ultrastructurales de la branchie d'un bivalve pectinidae hydrothermal profond. Comptes. rendus. l'Académie. Des. Sci. Sér 3 Sci. Vie. 307 (11), 627–633.
Le Pennec G., Le Pennec M., Beninger P., Dufour S. (2002). Spermatogenesis in the archaic hydrothermal vent bivalve, Bathypecten vulcani, and comparison of spermatozoon ultrastructure with littoral pectinids. Invertebr. Reprod. Dev. 41 (1-3), 13–19. doi: 10.1080/07924259.2002.9652730
Lin Y. T., Kiel S., Xu T., Qiu J. W. (2022). Phylogenetic placement, morphology and gill-associated bacteria of a new genus and species of deep-sea mussel (Mytilidae: bathymodiolinae) from the south China Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 190, 103894. doi: 10.1016/j.dsr.2022.103894
Lin Y. T., Xu T., Ip J. C. H., Sun Y., Fang L., Luan T., et al. (2023). Interactions among deep-sea mussels and their epibiotic and endosymbiotic chemoautotrophic bacteria: insights from multi-omics analysis. Zool. Res. 44 (1), 104–123. doi: 10.24272/j.issn.2095-8137.2022.279
Linnaeus C. (1758). Systema naturæ per regna tria naturæ, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Editio decima reformata. Salvius Holmiæ. 1, 824. doi: 10.5962/bhl.title.542
Minh B. Q., Nguyen M. A. T., von Haeseler A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
MolluscaBase (2022). Available at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=156983 (Accessed November 28, 2022).
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Osman E. O., Weinnig A. M. (2021). Microbiomes and obligate symbiosis of deep-sea animals. Annu. Rev. Anim. Biosci. 10, 151–176. doi: 10.1146/annurev-animal-081621-112021
Rafinesque C. S. (1815). Analyse de la nature, ou tableau de l'univers et des corps organisés (Palerme: Aux dépens de l'auteur).
Rambaut A., Drummond A. J. (2015) TreeAnnotator v1.8.2. MCMC output analysis. Available at: http://beast.bio.ed.ac.uk.
Rambaut A., Drummond A. J. (2018). Available at: https://github.com/rambaut/figtree.
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67 (5), 901–904. doi: 10.1093/sysbio/syy032
Reeve L. A. (1856). “Monograph of the genus spondylus,” in Conchologia iconica, or, illustrations of the shells of molluscous animals (London: L. Reeve & Co.).
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sacco F. (1897). I Molluschi dei terreni terziarii del piemonte e della liguria. part 24 (Pectinidae) Vol. 116 (Torino: Stamperia Reale).
Schein-Fatton E. (1985). Découverte sur la ride du pacifique oriental à 13° n d'un pectinidae (Bivalvia, pteromorphia) d'affinités paléozoïques. Comptes. rendus. l'Académie. Des. Sci. Sér 3 Sci. Vie. 301 (10), 491–496.
Schein-Fatton E. (1988). Un pectinacea (Bivalvia) tres primitif: Bathypecten vulcani du site hydrothermal de 13 degree n (Pacifique oriental). Oceanol. Acta Special issue 8, 83–98.
Schröter J. S. (1802). Neue conchylienarten und abänderungen, anmerkungen, und berichtigungen nach dem linnéischen system der XII ausgabe. Archiv für Zoologie und Zootomie. 3 (1), 125–166.
Sen A., Åström E. K., Hong W. L., Portnov A., Waage M., Serov P., et al. (2018). Geophysical and geochemical controls on the megafaunal community of a high Arctic cold seep. Biogeosciences 15 (14), 4533–4559. doi: 10.5194/bg-15-4533-2018
Shen X., Tian M., Liu Z., Cheng H., Tan J., Meng X., et al. (2009). Complete mitochondrial genome of the sea cucumber Apostichopus japonicus (Echinodermata: holothuroidea): the first representative from the subclass aspidochirotacea with the echinoderm ground pattern. Gene 439 (1-2), 79–86. doi: 10.1016/j.gene.2009.03.008
Smedley G. D., Audino J. A., Grula C., Porath-Krause A., Pairett A. N., Alejandrino A., et al. (2019). Molecular phylogeny of the pectinoidea (Bivalvia) indicates propeamussiidae to be a non-monophyletic family with one clade sister to the scallops (Pectinidae). Mol. Phylogenet. Evol. 137, 293–299. doi: 10.1016/j.ympev.2019.05.006
Smith E. A. (1885). Report on the lamellibranchiata collected by H.M.S. challenger during the years 1873-76, in report on the scientific results of the voyage of H.M.S. challenger during the years 1873-1876 Vol. 13. Eds. Thomson C. W., Murray J. (Edinburgh, Neill: Zoology), 1–341.
Southward E. C., Schulze A., Tunnicliffe V. (2002). Vestimentiferans (Pogonophora) in the pacific and Indian oceans: a new genus from lihir island (Papua new Guinea) and the Java trench, with the first report of Arcovestia ivanovi from the north Fiji basin. J. Nat. 36 (10), 1179–1197. doi: 10.1080/00222930110040402
Stewart C., Via L. E. (1993). A rapid CTAB DNA isolation technique useful forRAPD fingerprinting and other PCR applications. Biotechniques 14, 748–751
Sun Y., Sun J., Yang Y., Lan Y., Ip J. C. H., Wong W. C., et al. (2021). Genomic signatures supporting the symbiosis and formation of chitinous tube in the deep-sea tubeworm Paraescarpia echinospica. Mol. Biol. Evol. 38, 4116–4134. doi: 10.1093/molbev/msab203
Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Verrill A. E. (1897). A study of the family pectinidae, with a revision of their genera and subgenera. Trans. Conn. Acad. 10, 41–95.
Waller T. R. (2006). Phylogeny of families in the pectinoidea (Mollusca: bivalvia): importance of the fossil record. Zool. J. Linn. Soc 148 (3), 313–342. doi: 10.1111/j.1096-3642.2006.00258.x
Wiklund H., Taylor J. D., Dahlgren T. G., Todt C., Ikebe C., Rabone M., et al. (2017). Abyssal fauna of the UK-1 polymetallic nodule exploration area, clarion-clipperton zone, central pacific ocean: Mollusca. ZooKeys 707), 1–46. doi: 10.3897/zookeys.707.13042
Xu T., Feng D., Tao J., Qiu J. W. (2019). A new species of deep-sea mussel (Bivalvia: mytilidae: Gigantidas) from the south China Sea: morphology, phylogenetic position, and gill-associated microbes. Deep Sea Res. Part I Oceanogr. Res. Pap. 146, 79–90. doi: 10.1016/j.dsr.2019.03.001
Xu T., Sun Y., Wang Z., Sen A., Qian P. Y., Qiu J. W. (2022). The morphology, mitogenome, phylogenetic position, and symbiotic bacteria of a new species of Sclerolinum (Annelida: siboglinidae) in the south China Sea. Front. Mar. Sci. 8, 793645. doi: 10.3389/fmars.2021.793645
Zhan Z., Li J., Xu K. (2019). Ciliate environmental diversity can be underestimated by the V4 region of SSU rDNA: insights from species delimitation and multilocus phylogeny of Pseudokeronopsis (Protist, ciliophora). Microorganisms 7 (11), 493. doi: 10.3390/microorganisms7110493
Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W. X., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20 (1), 348–355. doi: 10.1111/1755-0998.13096
Keywords: Bivalvia, Catillopecten, cold seep, deep sea, scallop, Pectinoidea
Citation: Lin Y-T, Li Y-X, Sun Y, Tao J and Qiu J-W (2023) A new species of the genus Catillopecten (Bivalvia: Pectinoidea: Propeamussiidae): morphology, mitochondrial genome, and phylogenetic relationship. Front. Mar. Sci. 10:1168991. doi: 10.3389/fmars.2023.1168991
Received: 18 February 2023; Accepted: 05 April 2023;
Published: 27 April 2023.
Edited by:
Andrew R. Thurber, Oregon State University, United StatesReviewed by:
Gennady Kamenev, Far Eastern Branch (RAS) (NSCMB FEB RAS), RussiaHelena Wiklund, University of Gothenburg, Sweden
Copyright © 2023 Lin, Li, Sun, Tao and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Wen Qiu, cWl1andAaGtidS5lZHUuaGs=
 Yi-Tao Lin
Yi-Tao Lin Yi-Xuan Li
Yi-Xuan Li Yanan Sun3
Yanan Sun3 Jian-Wen Qiu
Jian-Wen Qiu